Abstract
The properties of infectious prions and the pathology of the diseases they cause are dependent upon the unique conformation of each prion strain. How the pathology of prion disease correlates with different strains and genetic backgrounds has been investigated via in vivo assays, but how interactions between specific prion strains and cell types contribute to the pathology of prion disease has been dissected more effectively using in vitro cell lines. Observations made through in vivo and in vitro assays have informed each other with regards to not only how genetic variation influences prion properties, but also to how infectious prions are taken up by cells, modified by cellular processes and propagated, and the cellular components they rely on for persistent infection. These studies suggest that persistent cellular infection results from a balance between prion propagation and degradation. This balance may be shifted depending upon how different cell lines process infectious prions, potentially altering prion stability, and how fast they can be transported to the lysosome. Thus, in vitro studies have given us a deeper understanding of the interactions between different prions and cell types and how they may influence prion disease phenotypes in vivo.
Introduction
Transmissible spongiform encephalopathies (TSEs or prion diseases) are a family of diseases with wide ranging strain dependent pathologies (Aguzzi et al., 2007) caused by the deposition of infectious prions in tissue (Beekes et al., 2007) and subsequent interference with vital cellular processes, primarily in the brain (Harris et al., 2006). Infectious prions, known as PrPSc, exist almost exclusively as insoluble and partially protease resistant aggregates of non-native prion protein that propagate via a process of seeded polymerization whereby the native, soluble, and protease-sensitive conformation of cellular prion protein, known as PrPC, interacts with PrPSc and is converted into PrPSc with the same non-native conformation (Collinge et al., 1996). PrPSc is found in several cellular compartments along the endocytic pathway where conversion occurs, including early endosomes and endolysosomes, and accumulates in lysosomes (Priola, 2018). In prion disease, PrPSc is the main, and possibly sole, component of the infectious prion (Collinge et al., 1996). While this conversion happens without guidance from coding nucleic acid (Prusiner, 1982), genetic polymorphism in the prion protein gene PRNP can still play a critical role in prion disease as point mutations in PrPC can alter prion infectivity and prion disease pathogenesis (Parchi et al., 1996).
A wide range of non-native conformations in the prion protein are infectious and able to promote the conversion of PrPC into their own distinctive PrPSc conformation. This variation in PrPSc conformation leads to infectious prions with different properties that may explain the existence of prion strains, which are defined in vivo by variations in prion disease presentation and pathology. The unique conformational structure adopted by PrPSc leads to its ordered assembly into large aggregates, the properties of which likely contribute to the prion disease phenotypes (Morales et al., 2007). For example, the structure of PrPSc aggregates (Kraus et al., 2021) are believed to contribute to the specific and reproducible pathologies associated with different prion strains (Bessen et al., 1992; Caughey et al., 1998; Parchi et al., 1996; Spassov et al., 2006), the cellular uptake routes that best facilitate prion propagation (Fehlinger et al., 2017), the rates of prion degradation by cells (Choi et al., 2013; Shoup et al., 2021), and the susceptibility of different cells to prion infection (Nishida et al., 2000). While the phenotypic differences between strains of prions is thought to primarily be the result of conformational differences in PrPSc, post-translational modifications to PrPC may alter the phenotypic properties of prion strains, including their ability to form PrPSc (Chesebro et al., 2005; Dear et al., 2007).
PrPC is expressed ubiquitously expressed throughout the body but is present at higher levels in cells of the central nervous system, particularly neurons, as well as cells in the spleen and heart (Castle et al., 2017). Mature PrPC has two N-linked glycosylation sites and is found in di-glycosylated, mono-glycosylated, and un-glycosylated forms (Haraguchi et al., 1989). In addition, PrPC contains a glycophosphatidylinositol (GPI) membrane anchor that tethers it to the cell surface (Puig et al., 2019). PrPC can also be cleaved by cellular proteases at three different sites, although the purpose of these cleavages is unknown (Linsenmeier et al., 2017). Since PrPSc is derived from host PrPC, its post-translational modifications may also be present in PrPSc. The amount of each glycoform (Tuzi et al., 2008), the sialylation pattern (Katorcha et al., 2015), and truncation state of PrPSc (Frankenfield et al., 2005; Notari et al., 2008) can vary between prion strains and may directly influence the properties of PrPSc (Collinge et al., 1996) and the pathology of prion disease (Nicot et al., 2010). Experimental modification of the properties of PrPC through truncation (Flechsig et al., 2000), removal of the GPI anchor (Chesebro et al., 2005), or removal of the glycosylation sites (Tuzi et al., 2008) can change how PrPSc deposits in vivo, as well as other aspects of prion pathogenesis, without necessarily leading to new prion strains. Thus, if the prevalence of different post-translational modifications varies between cells, the host PrPC molecule may help to determine the phenotypes of different prion strains, suggesting that prion disease may be influenced not only by PrPSc conformation but also by the host cell type.
In contrast to PrPC, PrPSc is found in a limited number of cell types, primarily neurons in the brain and follicular dendritic cells in the spleen (Beekes et al., 2007). The intracellular concentration of PrPSc throughout the body of an infected host varies with cell type and prion strain (Beekes et al., 2007; Parchi et al., 1996; Parchi et al., 1999) and the properties of PrPSc, such as sialyation pattern, vary between originating cell types (Srivastava et al., 2015), suggesting that the properties of different cell types may play a role in prion pathology. For example, PrPSc produced at high titer in muscle tissue or with high amounts of sialyation in the spleen may migrate through blood or lymphatic fluids to other organs including the brain, potentially contributing to the rapid degeneration of the brain during late-stage prion disease (Beekes et al., 2007; Bosque et al., 2002; Srivastava et al., 2015). Although it has been historically difficult to identify cell lines that are susceptible to prions (Piccardo et al., 2011), the eventual development of multiple in vitro cell culture models of prion infection has allowed the underlying mechanisms that enable infection of a cell, as well as other aspects of prion infection, to be studied in more detail. This review will explore how cell line models have furthered our understanding of prion biology and clarified in vivo observations regarding the role of sequence variation in the prion protein gene (PRNP) on both prion disease pathology and species barriers to prion disease. In addition, this review will focus on how cell culture models have provided new insights into the establishment and persistence of prion infection, in particular the ways in which different prion strains interact with and potentially infect cells. While many mysteries still persist, such as why some prion strains cannot persistently infect cells in vitro or how the structure of a prion strain dictates how it interacts with cellular machinery, it’s possible that the development of new in vitro cell systems will eventually allow us to map the molecular interactions between PrPSc and the cell, potentially providing novel targets for drug development.
From in vivo to in vitro: dissecting how genetic variation influences prion disease
Certain mutations in human PRNP lead to the spontaneous formation of PrPSc and the development of human prion diseases such as Creutzfeldt-Jakob disease (CJD) and Gerstmann-Sträussler-Schienker (GSS) disease (Gambetti et al., 2003). However, only 10 % of all CJD cases arise from heritable mutations that reproducibly lead to PrPSc formation, which are referred to as familial CJD (fCJD). About 90 % of all CJD cases arise spontaneously and are referred to as sporadic CJD (sCJD). Less than 1 % of human CJD cases are acquired through contact with prion contaminated materials such as prion-contaminated medical equipment or tissues, which results in iatrogenic CJD (iCJD). Exposure to BSE contaminated materials results in variant CJD (vCJD) (Weinstein et al., 2001), a so far unique instance where a prion disease has crossed species barriers to cause disease in humans. While the most common type of prion disease in humans is sCJD, iCJD and vCJD can provide clearer information about human prion pathogenesis, especially with regard to disease incubation times and prion spread. This is in part because the source of infection and exposure can often be determined, especially in the case of iCJD, and in part because the route of infection is known. Polymorphisms in human PRNP can both alter an individual’s susceptibility to sCJD (Collinge, 2005; Sangeetham et al., 2021) and fCJD (Monari et al., 1994) (Parchi et al., 1996). For example, the D178N mutation leads to a heritable form of prion disease but the type of disease depends upon a polymorphism at codon 129, with the presence of methionine at codon 129 leading to the development of fatal familial insomnia (FFI) and the presence of valine leading to development of fCJD (Monari et al., 1994). D178N PrPSc can also have different sizes after proteinase K (PK) treatment depending on the combination of polymorphisms at codon 129, indicating that genetic polymorphism in PRNP can also influence the structure and properties of PrPSc (Monari et al., 1994; Parchi et al., 1996). In this area of prion research, in vivo work has been instrumental in identifying the combination of genetic backgrounds and strains that allow for prion disease development and the characterization of their unique pathologies. However, the properties of different PrPSc aggregates, and how they may contribute to the underlying biochemical and molecular interactions that drive disease pathology, can be examined under more controlled conditions using in vitro cell line models for prion infection.
Properties of genetic variants of PrPC that lead to spontaneous formation prion disease
Several transgenic mouse lines have been developed in recent years that appear to be promising models for the study of spontaneous prion disease formation. For example, mouse models have been generated by replacement of mouse PRNP with either bovine PRNP bearing a leucine mutation at codon 113, which models the human GSS mutation P102L (Torres et al., 2013), or bank vole PRNP bearing a lysine mutation at codon 200, which models the E200K mutation associated familial CJD (Watts et al., 2016). To date, however, no in vitro cell culture model has been successfully developed for the study of spontaneous PrPSc formation and prion infectivity. Mutations associated with fCJD were shown to lead to increased aggregation and protease resistance in PrPC, though none led to the spontaneous formation of fully-protease resistant and infectious PrPSc (Lehmann et al., 1996; Priola et al., 1998). For instance, cerebral organoid cultures derived from patients carrying the E200K mutation do not spontaneously form PrPSc (Foliaki et al., 2020) despite the E200K mutation in PrP driving PrPSc formation in vivo. Interestingly prion strains that are unable to infect cells in vitro are still able to convert PrPC into PrPSc upon initial cellular interaction (Vorberg, Raines, & Priola, 2004), possibly indicating that the intercellular spaces found in vivo may promote the persistence of certain prion strains by allowing for the accumulation of PrPSc outside of the cell. While it’s unclear why aggregation prone mutant PrPC does not spontaneously form PrPSc in cells in vitro, it may be that other cellular deficits, perhaps in the proteostasis machinery that can occur with natural cell aging, are required for the formation of PrPSc.
Despite the inability of in vitro cell culture models to spontaneously form PrPSc, they have still proven useful in dissecting the molecular biology of PrPSc bearing the mutations that drive spontaneous PrPSc formation in vivo. For example, brain homogenates from mice infected with the M1000 mouse adapted GSS strain were used to infect rabbit kidney (RK-13) cells expressing mouse PrPC with the GSS causing mutations G114V and A117V (positions 113 and 116, respectively, in mouse PrPC). The cells produced aggregates of PrPSc that were both smaller and less PK resistant than PrPSc produced by RK-13 cells expressing wild-type mouse PrPC (Coleman et al., 2014). Despite the smaller aggregate size and lower resistance to PK, PrPSc from the RK-13 cells expressing mutant PrPC was more infectious when inoculated into mice than PrPSc from RK-13 cells expressing wild-type PrPC (Coleman et al., 2014). The increased infectivity of G113V and A116V PrPSc aggregates may be the result of their smaller size as smaller prion aggregates have been found to be more infectious and possess a higher capacity for both prion conversion and neuroinvasion (Bett et al., 2017; Kim et al., 2012; Silveira et al., 2005). These studies illustrate how in vitro and in vivo data combined can contribute to a better understanding of sporadic prion disease, even when de novo infectious prion formation is difficult to observe.
The spontaneous transformation of either wild-type or mutant PrPC into PrPSc is poorly understood, but it is believed that a critical first step may be the failure of cellular systems to clear misfolded and aggregated prion protein (Thody et al., 2018). In addition, mutations in PRNP that are associated with the formation of infectious prions in vivo are known to alter the properties of PrPC in ways that may lead to the spontaneous formation of infectious prions. For example, in M17 human neuroblastoma cells, un-glycosylated PrPC bearing the Q217R, E200K, and D178N mutations displays aberrant transport to the cell surface. This appears to be the result of conformational instability in the PrPC molecules bearing these mutations driving misfolding and aggregation of prion protein in the absence of glycosylation (Capellari et al., 2000; Petersen et al., 1996; Singh et al., 1997). However, the misfolded and un-glycosylated prion protein that results from these mutations may only contribute to the formation of infectious prions if it is trafficked to the cell surface, as intracellular un-glycosylated PrPC, formed via mutations that remove glycosylation, was not found to support prion infection in vitro (Salamat et al., 2011). Fully glycosylated forms of PrPC bearing the Q217R, E200K, and D178N mutations that are properly trafficked to the surface of cells may also be conformationally unstable and able to misfold and spontaneously form PrPSc. Residues 217, 200, and 178 reside in the C-terminal region of PrPC which makes up the alpha-helical core of PrPC, and the amino acids at these residues either participate in potentially critical ionic interactions across PrPC or reside next to amino acids that do (Pastore et al., 2007). This may explain why Q217R, E200K, D178N, and other mutations in this region (Mead, 2006) potentially destabilize the structure of PrPC, promoting aggregation and the eventual conversion into infectious prions.
Mutations in PrPC that result in C-terminal truncation, such as those that terminate PrPC at amino acid residues 226 (Race et al., 2018), 145 (Ghetti et al., 1996; Kitamoto et al., 1993), and 163 (Revesz et al., 2009), also result in spontaneous formation of PrPSc in humans but with pathological features, such as perivascular amyloid formation, that are distinct from those associated with PrPSc formed from full length PrPC. Similarly, prion infection of transgenic mice expressing mouse PrPC with a mutation to prevent the attachment of the GPI anchor leads to the development of large, extracellular, perivascular deposits of amyloid and a prolonged disease course when compared to prion infected mice expressing GPI anchored PrPC, where PrPSc was found in non-amyloid deposits and disease incubation times were much shorter (Chesebro et al., 2005). PrPSc without an anchor also appears to be more infectious and less affected by the species barrier between mice and humans (Race et al., 2015). Studies in vitro have shown that neural cells expressing anchorless PrPC cannot be persistently infected with prions unless they also express wild-type, anchored PrPC (McNally et al., 2009; Priola et al., 2009), suggesting that PrPSc infection in mice expressing PrPC without the GPI anchor is mostly extracellular. Overall, the combined in vitro and in vivo data suggest that the GPI anchor in PrPC is needed to infect a cell and can determine the localization and type of PrPSc deposited, as well as influence disease incubation times and the transmission of prions across species.
Modeling prion species barriers in vitro
In a manner analogous to how mutations in PrP can alter the susceptibility of an organism to prion infection, genetic variation between species can form a ‘species barrier’ that hinders prion transmission between species. Thus, effective transmission of prions between species is often dependent upon the sequence of PrPC in the species infected and the species from which PrPSc originates (Priola et al., 1995; Scott et al., 1993). White-tail deer and humans share 90% PRNP sequence identity and yet that small sequence difference appears to prevent the transmission of chronic wasting disease (CWD) from deer to humans (Waddell et al., 2018). However, the amount of sequence similarity between PRNP from different species is not the critical determining factor for transmission between species. Bovines and humans share only 86% PRNP sequence identity yet the prion strain that causes BSE has been transmitted to humans, likely through the consumption of BSE contaminated, bovine derived material (Will et al., 1999). While the species barrier between humans and certain animals can protect human populations from prion diseases in specific animal populations, species barriers also pose a problem for the study of human prions as commonly used animal models can be resistant to human prion strains. This barrier was largely overcome with the discovery that transgenic mice expressing PrPC with the same amino acid sequence as PrPSc were susceptible to infection with prions from another species (Prusiner et al., 1990). With regard to other species of mammals, there is a strong species barrier between mice and hamsters (Prusiner et al., 1990) while rabbits and dogs appear to be resistant to prion infection. However, this resistance is not absolute as transgenic mice expressing rabbit PrPC, but not mouse PrPC, were susceptible to BSE and mouse prions (Vidal et al., 2015), indicating that non-prion factors specific to rabbits may contribute to their resistance to prion disease. By contrast, bank voles are infectable with prions from a wide range of species, earning them the title ‘universal prion acceptor’ (Watts et al., 2014).
In vitro, many of these in vivo species barriers have been studied using cell lines expressing species-specific and/or chimeric PrPC and determining which PrPC molecules are able to convert to PrPSc when exposed to prions from different species. This has allowed the mapping of key amino acid residues involved in the species-specific formation of PrPSc and expanded our understanding of how infectious prions transmit between species. (Priola, 2013). While non-prion factors may contribute to species barriers to prion disease, the majority of work using in vitro systems has focused on the influence of the PrPC amino acid sequence on cross-species transmission of prions. For example, in vitro prion infection of canine kidney cells as well as in vitro prion amplification assays using canine PrP showed no signs of PrPSc formation when exposed to mouse, human, cervid, and sheep prions (Fernández-Borges et al., 2017; Polymenidou et al., 2008), suggesting that the dog prion protein was highly resistant to conversion by PrPSc. A mutation replacing asparagine at codon 163 with an aspartic acid was determined to be responsible for the resistance of dog PrPC to conversion to PrPSc and the correlating mutation in mouse PrPC also inhibited prion infection (Fernández-Borges et al., 2017; Vidal et al., 2020). While the N163D mutation in dogs creates a barrier to infection by all prion strains that have been tested, single codon mutations in PRNP can create a barrier to infection by specific strains from specific species (Priola et al., 1995). For example, replacing an isoleucine in mouse PrPC at residue 138 with a methionine found at the corresponding residue of the hamster PrPC sequence was sufficient to prevent PrPSc formation in mouse prion infected mouse neuroblastoma (N2A) cells (Priola et al., 1995). Similar studies between different species have enabled the mapping of specific regions of PrPC, in many cases down to a single amino acid residue, that are responsible for the species-specific formation of PrPSc (Priola, 2013). For example, a proline to leucine mutation in mouse PRNP at codon 101 can significantly slow the progression of vCJD in mice and valine at codon 129 in human PRNP protected against the transmission of bovine spongiform encephalopathy (BSE) while the methionine polymorphism at codon 129 did not (Barron et al., 2001; Collinge, 2005; Parchi et al., 1996; Wadsworth et al., 2004). These studies have shown that the most important regions and amino acids for interspecies prion transmission differ depending upon the species involved in transmission and whether the polymorphism is present in PrPSc or the PrPC molecule being converted to PrPSc. This suggests that conformational differences associated with these amino acid variations are the driving force behind the species-specific formation of PrPSc.
Factors that mediate cellular association and uptake of PrPSc and their role in disease
The interaction of PrPSc with cellular membrane and its subsequent internalization by cells are important steps in prion biology that are mediated by various biological and biochemical factors like the GPI anchor (McNally et al., 2009) and membrane bound biomolecules (Gauczynski et al., 2006; Hijazi et al., 2005; Horonchik et al., 2005; Morel et al., 2005). PrPC primarily localizes to membranes via the GPI anchor which has been found to both facilitate persistent infection of in vitro cell lines and to alter the pathology of prion disease in vivo (Chesebro et al., 2005; McNally et al., 2009; Priola et al., 2009). However, other membrane biomolecules, such as glycosaminoglycans (GAGs), also participate in membrane association, guide the cellular uptake, and influence the properties of PrPSc in a strain and cell type dependent manner (Hijazi et al., 2005; Horonchik et al., 2005; Imamura et al., 2016; Mayer-Sonnenfeld et al., 2008; Vieira et al., 2014; Wong et al., 2001). These factors were found to be so critical for the propagation of PrPSc that early drug treatments for prion disease were based around mimetics for these biomolecules (Adjou et al., 2003). Continued study of these factors may help us to better understand how PrPSc relies on different cellular components for propagation, fostering the advancement of mimetic drugs to fight prion disease.
The influence of GPI anchoring on the persistence, propagation, and spread of PrPSc
The C-terminal GPI anchor that tethers PrPC to cell membranes is also present in aggregates of PrPSc (Stahl et al., 1990) and may facilitate attachment of PrPSc to membranes. Enzymes like phospholipase C normally cleave the GPI anchor from PrPC but, in the prion strains tested thus far, the GPI anchor of PrPSc is protected from cleavage by enzymes like phospholipase C (Stahl et al., 1990) which may allow PrPSc to stay attached to membrane for long periods of time. However, GPI mediated cell surface association is not absolutely required for prion infection in vivo as mice bearing mutations in PRNP that remove the GPI anchor can still sustain prion infection (Chesebro et al., 2010; Chesebro et al., 2005). Enzymatically removing the GPI anchor from PrPSc does not significantly alter its conversion capability or infectivity (Lewis et al., 2006), and anchorless PrPSc derived from prion infected mice expressing anchorless PrPC is able to infect cell lines expressing GPI-anchored mouse PrPC (McNally et al., 2009). However, prion strains that are normally able to persistently infect cell lines in vitro fail to do so when the cells express anchorless PrPC alone (McNally et al., 2009; Priola et al., 2009). The failure of prions to infect cells expressing only anchorless PrPC can be corrected by co-expression of anchored PrPC, which results in persistent infection and formation of both anchored and anchorless PrPSc (McNally et al., 2009; Priola et al., 2009).
These data suggest that the GPI anchor may influence the localization of PrPSc and that the presence of an anchor on PrPC may facilitate the spread of infectious prions from cell to cell. This latter prediction was supported through subsequent studies in vivo which showed that anchored PrPSc propagated along neurons (Klingeborn et al., 2011) while anchorless PrPSc propagated along interstitial fluid drainage systems (Rangel et al., 2014). While PrPC is initially produced with a GPI anchor, PrPC in vivo exists as both anchored and anchorless PrPC as enzymes like phospholipase C and the ADAM 10 protease both remove its GPI anchor (Altmeppen et al., 2015; Stahl et al., 1990). Interestingly, when ADAM10 activity was inhibited in vivo reducing the amount of PrPC with cleaved GPI anchors, the spread of PrPSc was hindered (Altmeppen et al., 2015). These data suggest that the spread of PrPSc in the brain may also be facilitated by the presence of PrPC with enzymatically removed GPI anchors. In this case, anchorless PrPC would not be restricted to the cell surface and would be able to move more freely, likely via the interstitial fluid (Rangel et al., 2014), thus facilitating the spread of PrPSc throughout the brain (Chesebro et al., 2005).
The role of GAGs in prion biology
In animal tissue, GAGs are ubiquitously present linear polysaccharides that associate with membranes, attach to proteins to form proteoglycans, and regulate interactions between proteins. (Horonchik et al., 2005; Li et al., 2016; Shyng et al., 1995; Simon Davis et al., 2013; Warner et al., 2002). GAGs also appear to play an important role in prion biology as binding of the GAG heparan sulfate to PrPC stimulates the internalization of PrPC by cultured cells (Gabizon et al., 1993; Shyng et al., 1995) and heparan sulfate is associated with amyloid plaques of PrPSc from prion infected mice (McBride et al., 1998). In addition, under-sulfation of heparan sulfate, which resulted from Papss2 gene silencing, was found to increase the deposition of PrPC in the extracellular matrix and facilitate the propagation of PrPSc (Marbiah et al., 2014). Incorporation of heparan sulfate into PrPSc aggregates likely happens during the aggregation process as it was found to promote prion replication and infectivity (Imamura et al., 2016; Shaked et al., 2001; Wong et al., 2001), and GAG mimetic compounds can slow the progression of prion disease in vivo (Ehlers et al., 1984; Farquhar et al., 1986). While heparan sulfate facilitates endocytosis for a wide range of biomolecules (Christianson et al., 2014) and has been observed facilitating internalization of PrPSc (Hijazi et al., 2005; Horonchik et al., 2005), it is not always required for PrPSc uptake by cells (Paquet et al., 2007; Wolf et al., 2015). One possible explanation for this discrepancy is that in studies where GAGs promoted PrPSc uptake, purification of the PrPSc used may have removed potential cofactors that promote an association between PrPSc, GAGS, and the cellular membrane. Alternatively, in studies that did not observe GAG mediated cellular uptake of PrPSc, GAG depleted cells were inoculated with PrPSc infected brain homogenate which may have contained enough brain-derived GAGs, or other cofactors, to facilitate cellular uptake.
Regardless of whether GAGs are critical for prion uptake or whether GAG independent uptake mechanisms exist, GAGs such as heparan sulfate clearly impact prion disease progression in vivo and can influence prion infection of cells in a strain dependent manner (Das et al., 2020; Das et al., 2017; Wolf et al., 2015). The prion protein has three binding sites for heparan sulfate that can be found at amino acids 23 to 52, 53 to 93, and 110 to 128 in human PrPC (Warner et al., 2002). These sites may play different roles in the propagation of different prion strains as conformational variations between prion strains may influence how well heparan sulfate associates with these sites. This idea is supported by studies in C57BL/6 mice expressing PrPC with mutated heparan sulfate binding sites that were infected with either the 22L or RML prion strains, both of which are mouse adapted sheep scrapie strains that have similar incubations times in C57BL/6 mice (Das et al., 2020; Das et al., 2017). Mutations in PrPC that disrupted heparan sulfate binding to the N-terminal amino acids from 25 to 90 had different impacts on disease progression in the two strains, with RML infected mice having longer incubation times than 22L infected mice (Das et al., 2020; Das et al., 2017). Mutations in the heparan sulfate binding sites of PrPC may lengthen the incubation times of the RML strain by reducing the amount of RML PrPSc maintained by cells. This possible explanation is supported by in vitro studies in L929 mouse fibroblast cells expressing wild-type mouse PrPC, which can be infected by both the RML and 22L prion strains (Vorberg, Raines, Story, et al., 2004). A reduction in sulfated GAG levels, including heparan sulfate, through GAG mimetic dextran sulfate 500 (DS-500) and sodium chlorate treatment resulted in a greater decrease in cellular RML PrPSc concentrations when compared to 22L (Wolf et al., 2015). Heparan sulfate has also been found to promote the uptake of PrPSc and it may be that the RML strain is more dependent upon it for internalization when compared to the 22L strain. Alternatively, it is possible that heparan sulfate is more important for the structural stability of the RML strain than the 22L strain. PrPSc from these strains varies in its structural stability, with PrPSc from the 22L strain being thermally more stable than PrPSc from the RML strain (Marín-Moreno et al., 2019). Heparan sulfate has been found to stabilize the structure of RML prions which may explain why, when heparan sulfate is forcibly accumulated in cells, the ability of the lysosome to clear RML PrPSc is delayed (Mayer-Sonnenfeld et al., 2008; Vieira et al., 2014). The strain specific interaction of GAGs with PrPSc may also help to explain the pattern of PrPSc deposition in the host. The concentration and characteristics of heparan sulfate and other GAGS varies between tissues (Clark et al., 2011; Warda et al., 2006) and PrPSc from prion strains like RML may be more stable and may accumulate to higher concentrations in cells and areas of the tissue with higher heparan sulfate concentrations. While variation in GAG distribution and characteristics between cell lines may promote strain-specific differences in PrPSc accumulation in different cells, variability in the way different cell types treat different prion strains, such as differences in the endocytic uptake route of PrPSc, may also contribute (Fehlinger et al., 2017).
Potential protein cofactors for cell association and uptake of PrPSc
Another way in which GAGs may influence PrP uptake is by altering interactions between clathrin coated pits and prions (Figure 1A). Clathrin-mediated uptake of PrPC requires interactions with both heparan sulfate and the laminin receptor protein (LRP) (Gauczynski et al., 2001; Hundt et al., 2001), and there is evidence that LRP may facilitate internalization of PrPSc with some cell lines depending upon LRP for prion uptake more than others (Gauczynski et al., 2006; Morel et al., 2005). Two cell lines with seemingly different dependencies on LRP for uptake of PrPSc are Baby Hamster Kidney (BHK) cells and the human colon adenocarcinoma cell line Caco-2TC7. Inhibition of GAGs in BHK cells inhibited PrPSc uptake independent of LRP concentration, while antibody mediated inhibition of LRP in Caco-2TC7 cells inhibited PrPSc uptake (Gauczynski et al., 2006; Morel et al., 2005). This may mean that in some cell types LRP is more critical for PrPSc uptake while in other cell lines GAGs are more critical. Alternatively, the low-density lipoprotein receptor related protein (LRP1) may also facilitate uptake of PrPSc as inhibition of LRP1 in both primary neuron and hippocampal cell cultures prevented uptake of PrPSc from the prion strain ME7, which is a mouse adapted sheep scrapie strain (Jen et al., 2010). While only the single study shows that LRP1 contributes to PrPSc uptake, numerous studies have shown that LRP1 may be involved in Alzheimer’s disease as LRP1 regulates uptake of Aβ and tau aggregates (Lane-Donovan et al., 2014; Rauch et al., 2020; Zhang et al., 2020), demonstrating that it can facilitate the uptake of a multiple non-native aggregates. Interestingly, PrPC is also involved in tau aggregate uptake, which can inhibit the accumulation of PrPSc (De Cecco et al., 2020). This may indicate that membrane proteins, other than LRP1, that are critical for membrane interaction and cellular uptake of Aβ and tau aggregates in Alzheimer’s disease may also be important for membrane interaction and cellular uptake of PrPSc.
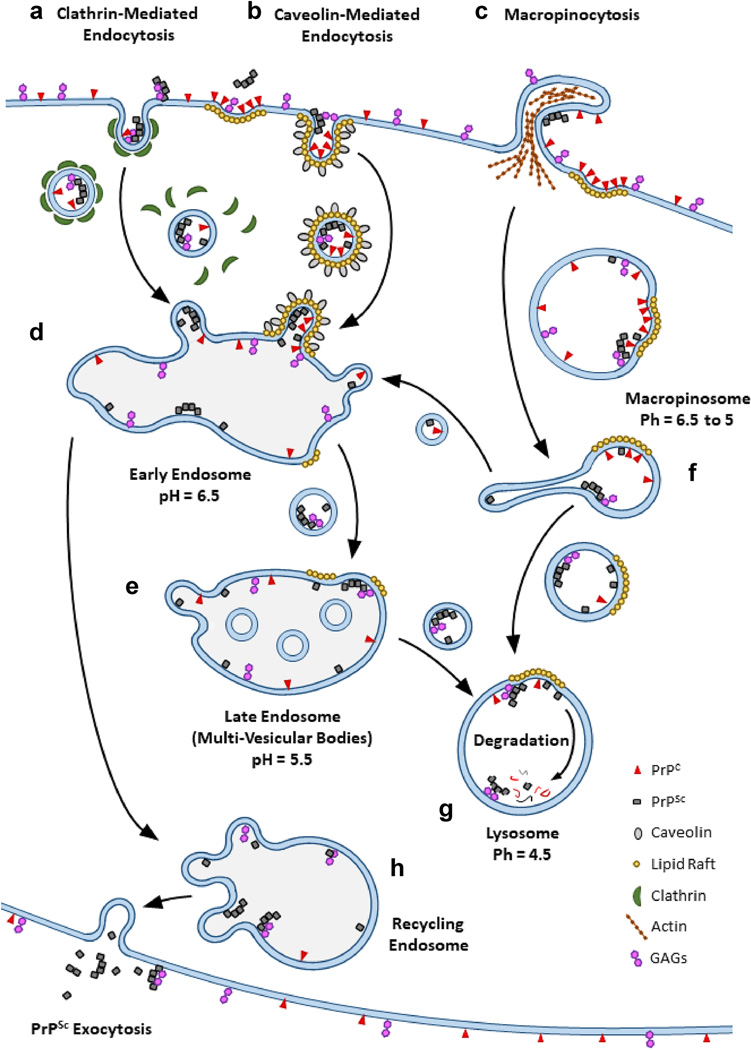
Figure 1 -. Uptake, trafficking, conversion, and degradation of PrPSc -.
Cellular uptake of PrPSc through three different routes and subsequent trafficking pathways to the lysosome are shown. PrPSc (grey squares) may be taken up via (A) clathrin-mediated endocytosis, (B) caveolin-mediated endocytosis, and (C) macropinocytosis alongside PrPC. Endosomal vesicles formed via clathrin-mediated and caveolin-mediated endocytosis are typically directed to the (D) early endosomal vesicles before PrPSc, and other vesicle cargo, is trafficked into (E) late endosomal vesicles, also known as multi-vesicular bodies. Vesicles formed via macropinocytosis become a part of the (F) macropinosome where the cargo of large vesicles can be separated into different smaller vesicles by tubulation and shuttled to early endosomal vesicles. Over time, the remains of tubulated macropinosomal vesicles and the contents of late endosomal vesicles are shuttled to the (G) lysosome for degradation. Contents of the early endosome may also be trafficked to the (H) recycling endosome and then onto the cell surface for expulsion. Molecules such as PrPC (red triangles) and GAGs (purple hexagons) can co-migrate with PrPSc through different routes of uptake and endosomal trafficking. GAGs promote the uptake, conversion, and the stability of PrPSc while PrPC can associate with PrPSc at numerous cellular locations from the cell surface to the pre-lysosomal vesicles. While a significant amount of prion conversion can happen on the surface of cells, conversion of PrPC to PrPSc has been observed in several pre-lysosomal vesicles (shaded vesicles). GPI anchored PrPC can also accumulate on lipid rafts (gold circles), making lipid rafts potentially important sites of prion conversion at numerous cellular locations.
Endocytosis pathways and endosomal trafficking of PrPSc
Determining which cell types can be infected by prions and why is critical for understanding how prion infection is established as well as how prions replicate and spread. Upon initial interaction with the cell, PrPC is rapidly converted into PrPSc regardless of the prion strain or its ability to persistently infect the cell with which it is interacting (Goold et al., 2011; Vorberg, Raines, & Priola, 2004). Prion conversion can happen across the cell surface (Goold et al., 2011) but PrPC can also accumulate within cell surface features, such as lipid rafts, which may be critical sites of prion conversion (Hooper, 2011; Lewis et al., 2011; Vey et al., 1996). In vitro, the amount of PrPSc taken up by cells is also independent of strain and whether a cell line can be infected by a specific prion strain (Greil et al., 2008). However, the rate at which PrPSc is degraded by the cell is strain dependent (Choi et al., 2013; Shoup et al., 2021) and may be determined by the stability of the PrPSc aggregate (Shoup et al., 2021). This suggests that the ability of a strain to infect a cell may be in part the result of cellular factors that control how rapidly PrPSc is trafficked to the lysosome for degradation versus how well newly formed PrPSc is trafficked away from the lysosome. How aggregates of PrPSc from different strains interact with cells may influence the rate at which they are trafficked, as fluorescent PrPSc aggregates that were found to form strings on the cell surface were taken up very slowly (Rouvinski et al., 2014). Thus, the route by which PrPSc is endocytosed as well as how PrPSc is trafficked through the endocytic pathway are potentially important factors that are likely to vary with cell type, as they do with the uptake of other molecules (Douglas et al., 2008), and may help determine whether a particular cell can be infected by a specific strain of prion (Fehlinger et al., 2017).
Routes of endocytosis for PrPSc and PrPC
Endocytosis is critical for maintaining homeostasis in multicellular organisms and thus can be difficult to manipulate and study in detail using in vivo models. As such, in vitro cell line models have been particularly useful in furthering our understanding of how cells internalize and traffic PrPC and PrPSc. How cells internalize PrPC is an important consideration in prion disease as endocytic vesicles that contain PrPC may be central sites of prion conversion if cells take up PrPSc via the same route that PrPC is taken up. Cell surface associated PrPC is predominately internalized via clathrin-coated pits and caveolae before trafficking to the early endosome (Figure 1A,B, and D) with uptake of PrPC by these mechanisms being modulated by association of PrPC with lipid rafts and metal ions (Hooper et al., 2008; Peters et al., 2003; Sarnataro et al., 2009). The exact uptake pathway for PrPC can also be cell type dependent. L929 cells, a murine fibroblast cell line, predominately internalize PrPC via clathrin-coated pits as opposed to utilizing caveolae (Fehlinger et al., 2017). While PrPC uptake in L929 cells was independent of caveolin-mediated endocytosis, knocking down caveolin led to cell surface accumulation of PrPC, indicating that caveolin may play a role in PrPC uptake even if it is not required for internalization (Fehlinger et al., 2017). Since GPI anchored proteins like PrPC can accumulate on lipid rafts, inhibition of caveolin-mediated uptake may cause PrPC to remain on the cell surface with no way to leave lipid rafts or be internalized, which would explain why PrPC accumulates on the surface of cells when caveolin-mediated uptake is inhibited. Thus, even though PrPC is predominately taken up by clathrin-mediated endocytosis in L929 cells, PrPC that becomes trapped in lipid rafts is likely only taken up by caveolae mediated endocytosis. As previous studies have shown that PrPC and PrPSc co-localize to lipid rafts (Hooper, 2011; Lewis et al., 2011; Vey et al., 1996) it may be that caveolae-mediated uptake plays a role in persistent infection of L929 cells. Lipid rafts may also facilitate the spread of PrPSc between cells as anchorless PrPC has also been found to associate with membranes via lipid rafts and is capable of being converted by PrPSc while attached to lipid rafts in vitro (Baron et al., 2003). In addition, lipid rafts may only function as sites of prion conversion on the surface of cells, or in early endosomal compartments, as in vitro prion conversion assays on lipid rafts showed that conversion of GPI anchored PrPC was optimal between a pH of 6 to 7 (Baron et al., 2002).
Cellular uptake of PrPSc via macropinocytosis was observed in N2a cells (Wadia et al., 2008), and clathrin-mediated endocytosis may participate in uptake of PrPSc as laminin receptors were found to be involved in PrPSc uptake in Caco-2/TC7 cells (Morel et al., 2005). Caveolae may also either directly internalize PrPSc or assist PrPSc uptake through other endocytic pathways. PrPSc has been found associated with caveolae on the surface of N2a cells, and both inhibition of caveolin in N2a cells and knock down of caveolin in L929 cells reduced the internal accumulation of PrPSc in chronically infected cells (Fehlinger et al., 2017; Marella et al., 2002; Vey et al., 1996). One possible explanation for how knocking down or inhibiting caveolin can reduce intracellular accumulation of PrPSc without interfering with cellular internalization may be that inhibition of one endocytic route often causes upregulation of other endocytic routes to compensate for the lost endocytic pathway (Fehlinger et al., 2017). Some endocytic routes may provide more favorable cellular microenvironments for establishing persistent cellular infection. Evidence supporting this was derived from studies in L929 cells which showed that inhibition of clathrin-mediated endocytosis increased uptake of FITC-dextran via macropinocytosis as well as PrPSc (Figure 1C) (Fehlinger et al., 2017). While these data, in conjunction with data from primary neuronal culture (Bett et al., 2017), suggests that uptake via macropinocytosis may facilitate persistent infection of cells by 22L prions, the level of RML PrPSc in chronically infected L929 cells decreases with a reduction in clathrin mediated endocytosis activity (Fehlinger et al., 2017). Since both RML and 22L prions can infect L929 cells, these data suggest that they may do so most efficiently via different endocytic routes.
Variations in vesicle microenvironment and their influence in prion biology
The microenvironment of endocytic vesicles and the treatment of the cargo they carry varies between different endocytic routes early in the endocytosis process. Variable treatment of prion protein cargo between different endocytic routes may therefore promote or disrupt prion replication, stability, and aggregation. One example of an endocytic vesicle component that influences prion biology is the lipid raft. Lipid rafts are sites of PrPC localization and potentially prion replication as PrPSc can co-localize with PrPC in the lipid raft (Hooper, 2011; Lewis et al., 2011; Vey et al., 1996). Lipid rafts are not necessary for clathrin-mediated endocytosis or macropinocytosis, though lipids rafts can be taken up through macropinocytosis (El-Sayed et al., 2013). However, caveolae formation does require lipid rafts, which are drawn into the vesicles during endocytosis and may thus facilitate prion replication if both PrPC and PrPSc are present (Kiss et al., 2009; Lajoie et al., 2010). While lipid rafts may facilitate prion conversion by bringing PrPC and PrPSc together (Baron et al., 2003; Baron et al., 2002), other characteristics of endocytic vesicles that may vary during early endocytosis, such as intravesicular pH and ion concentration, (Maxfield, 2014; Sonawane et al., 2002; Tsang et al., 2000) may influence prion conversion by altering prion structure (DeMarco et al., 2007; Nandi et al., 2002; Zanusso et al., 2001). Endocytic vesicles that result from caveolae- and clathrin-mediated endocytosis transition through early, late, and lysosomal vesicles where the general pH of each intracellular compartment is generally 6.5, 5.5, and 4.5 respectively (Hu et al., 2015) (Figure 1D,E, and G). However, the pH of vesicles formed from macropinocytosis can drop to a pH of 5.1 within 10 minutes of vesicle formation and stay at that low pH until the vesicle merges with the lysosome (Tsang et al., 2000) (Figure 1G). The decreasing pH experienced by vesicle cargo at different steps of endocytosis and endosomal trafficking may promote prion conversion as PrPC has been found to misfold at lower pH values which may facilitate conversion into non-native PrPSc conformations (DeMarco et al., 2007; Thompson et al., 2018). In addition, changes in pH that have been found to alter the stability of PrPSc in a strain specific manner may also facilitate PrPSc formation (Zanusso et al., 2001). The potential influence of pH on prion conversion and stability is further compounded by changes in vesicle pH being accompanied by intravesicular increases in anionic salts (Sonawane et al., 2002), which can alter the structural stability of PrPSc (Concha-Marambio et al., 2014; Nandi et al., 2002). Thus, differences in the chemical environment of vesicles as a direct result of cellular internalization via different endocytic pathways, including how fast the environments change and how long prion cargo is incubated in each microenvironment, may either facilitate or hinder the ability of a particular prion strain to establish a persistent infection by affecting the conversion of PrPC to PrPSc.
The dynamic chemical microenvironment of vesicles traversing the endosomal network from different cellular internalization pathways undoubtedly influences prion biology but morphological variations in vesicles, such as those observed in the macropinosome, may also play a role (Buckley et al., 2017) (Figure 1F). Work in human embryonic kidney (HEK) cells revealed macropinosome vesicles are divided into smaller vesicles through constriction of portions of the membrane into long tubes (Kerr et al., 2006). These tubulated macropinosome vesicles migrate to early endosomal (Figure 1D) vesicles while the remainder of the un-tubulated vesicle migrates toward the lysosome (Buckley et al., 2017; Kerr et al., 2006). Prion proteins caught in macropinosome tubules may be forced into close proximity, promoting both conversion into and aggregation of PrPSc. Membrane tubulation can also occur in multi-vesicular bodies, also known as the late endosome, (Woodman et al., 2008) and these vesicle compartments have been identified as key sites for prion conversion (Yim et al., 2015). Other intracellular compartments, such as the trans-golgi network or recycling endosome (Figure 1H), also possess unique morphologies or undergo morphological transformations such as tubulation (Rambourg et al., 1979; Van Ijzendoorn, 2006) that may alter aggregated PrPSc or promote the propagation of PrPSc trafficked to these compartments (Goold et al., 2013; Yamasaki et al., 2018). Trafficking of PrPSc between endosomal compartments may influence prion propagation by exposing PrPSc to new endosomal microenvironments but may also provide a way for newly formed PrPSc to escape trafficking to the lysosomal as intracellular compartments, such as the recycling endosome, can traffic PrPSc back to the cells surface (Figure 1H) (Grant et al., 2009). In vivo, data consistent with localization of PrPSc to the lysosome has been reported in the brains of human CJD and scrapie infected mice (Ironside et al., 1993; Kovács et al., 2007; Laszlo et al., 1992). PrPSc has also been found trafficked back to the surface of cells via exosomes (Hartmann et al., 2017) and may have experienced different vesicle microenvironments compared to PrPSc released from cells via endosomal recycling vesicles. In either case, passage thru cells may alter the properties of PrPSc which may contribute to the progression of prion disease in other cells in unique ways after release from one cell and re-uptake by another.
PrPSc interferes with trafficking of vesicles to the lysosome
Regardless of the route of endocytosis, in vitro most PrPSc is ultimately trafficked to the lysosome (Caughey et al., 1991) (Figure 1G), a finding consistent with in vivo observations suggesting that PrPSc can be found in lysosomes in the brains of human CJD and scrapie infected mice (Ironside et al., 1993; Kovács et al., 2007; Laszlo et al., 1992). However, PrPSc may be able to delay trafficking to the lysosome by interfering with the proteins that regulate normal endosomal trafficking, potentially allowing it more time to convert PrPC before it degrades. In N2a cells infected with 22L and RML prions, the amount of membrane bound Rab7, a protein that regulates trafficking to lysosomal vesicles through association with the vesicle membrane, was decreased. This indicated that lysosomal activity was impaired in N2a cells chronically infected with prions (Shim et al., 2016). However, a comparison of global protein degradation rates in N2a cells that were or were not infected with RML prions found that proteasomal degradation rates actually increased during infection and that RML PrPSc had a half-life 1.7 times greater than PrPC in chronically infected cells (Hutti et al., 2020). This suggests that Rab7 may only fail to associate with vesicles containing PrPSc, thus preventing trafficking of PrPSc to the lysosome. Rab proteins have been found to co-purify with PrPSc (Moore et al., 2010), suggesting that interference with the association of Rab7 with membrane vesicles may result from a direct interaction between PrPSc and Rab proteins. Blocking of the Rab7 association with lysosomal vesicles may also result from interference with other endosomal proteins, such as sortilin, which is a sorting receptor for endocytosis. In N2a cells, sortilin overexpression reduces the amount of intracellular 22L and RML PrPSc and under expression increases the internal concentration of both proteins (Uchiyama et al., 2017). Sortilin also appears to direct PrPSc to lysosomal vesicles (Uchiyama et al., 2017). However, the expression of sortilin is decreased during prion infection, suggesting that while sortilin may play a role in directing PrPSc to the lysosome, PrPSc may interfere with sortilin function during prion infection (Uchiyama et al., 2017). Again, disrupting the normal transition of PrPSc to the lysosome may give it more time to convert PrPC into PrPSc leading to the accumulation of more PrPSc and potentially giving newly formed PrPSc more time to escape vesicles bound for the lysosome.
Conclusions
In vitro studies have proven critical in uncovering the molecular mechanisms by which different prion strains produce distinct disease pathologies in vivo. The mechanistic insights gained from in vitro studies have furthered our understanding of how different means of cellular interaction, uptake, and intracellular trafficking contribute to prion spread, uptake, degradation, and the ability to persistently infect cells. In addition, in vitro studies have clarified how genetic variation leads to transmission barriers between species and effects infectious prion formation. Further development of more complex, three-dimensional cell systems, such as cerebral organoids (Foliaki et al., 2020) may help emulate the extracellular spaces found in vivo, providing the proper environment to maintain and study extracellular PrPSc formation. Prion strains that cannot infect cell lines can still be taken up and degraded by cells, making the processes of cellular uptake, endosomal trafficking, and lysosomal degradation a common denominator in the biology of all prion strains. How prions interfere with or bypass normal trafficking to the lysosome may be exploited for the development of drugs that prevent prions from escaping lysosomal degradation. Taken together, the mechanistic insights into prion biology provided by in vitro studies have deepened our understanding of prion biology, providing multiple new avenues for the development of anti-prion therapeutics.
Acknowledgements
This research was supported by a grant from the National Institutes of Health, National Institute of Allergy and Infectious Diseases Division of Intramural Research.
References
- Adjou KT, Simoneau S, Sales N, Lamoury F, Dormont D, Papy-Garcia D, Barritault D, Deslys J-P, & Lasmezas CI (2003). A novel generation of heparan sulfate mimetics for the treatment of prion diseases. Journal of General Virology, 84, 2595–2603. [DOI] [PubMed] [Google Scholar]
- Aguzzi A, Heikenwalder M, & Polymenidou M. (2007). Insights into prion strains and neurotoxicity. Nature reviews Molecular cell biology, 8, 552–561. [DOI] [PubMed] [Google Scholar]
- Altmeppen HC, Prox J, Krasemann S, Puig B, Kruszewski K, Dohler F, Bernreuther C, Hoxha A, Linsenmeier L, & Sikorska B. (2015). The sheddase ADAM10 is a potent modulator of prion disease. Elife, 4, e04260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron GS, & Caughey B. (2003). Effect of glycosylphosphatidylinositol anchor-dependent and-independent prion protein association with model raft membranes on conversion to the protease-resistant isoform. Journal of Biological Chemistry, 278, 14883–14892. [DOI] [PubMed] [Google Scholar]
- Baron GS, Wehrly K, Dorward DW, Chesebro B, & Caughey B. (2002). Conversion of raft associated prion protein to the protease-resistant state requires insertion of PrP-res (PrPSc) into contiguous membranes. The EMBO journal, 21, 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron RM, Thomson V, Jamieson E, Melton DW, Ironside J, Will R, & Manson JC (2001). Changing a single amino acid in the N-terminus of murine PrP alters TSE incubation time across three species barriers. The EMBO journal, 20, 5070–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekes M, & McBride PA (2007). The spread of prions through the body in naturally acquired transmissible spongiform encephalopathies. The FEBS journal, 274, 588–605. [DOI] [PubMed] [Google Scholar]
- Bessen RA, & Marsh RF (1992). Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. Journal of General Virology, 73, 329–334. [DOI] [PubMed] [Google Scholar]
- Bett C, Lawrence J, Kurt TD, Orru C, Aguilar-Calvo P, Kincaid AE, Surewicz WK, Caughey B, Wu C, & Sigurdson CJ (2017). Enhanced neuroinvasion by smaller, soluble prions. Acta neuropathologica communications, 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosque PJ, Ryou C, Telling G, Peretz D, Legname G, DeArmond SJ, & Prusiner SB (2002). Prions in skeletal muscle. Proceedings of the National Academy of Sciences, 99, 3812–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CM, & King JS (2017). Drinking problems: mechanisms of macropinosome formation and maturation. The FEBS journal, 284, 3778–3790. [DOI] [PubMed] [Google Scholar]
- Capellari S, Parchi P, Russo CM, Sanford J, Sy M-S, Gambetti P, & Petersen RB (2000). Effect of the E200K mutation on prion protein metabolism: comparative study of a cell model and human brain. The American Journal of Pathology, 157, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle AR, & Gill AC (2017). Physiological functions of the cellular prion protein. Frontiers in molecular biosciences, 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Raymond GJ, & Bessen RA (1998). Strain-dependent differences in β-sheet conformations of abnormal prion protein. Journal of Biological Chemistry, 273, 32230–32235. [DOI] [PubMed] [Google Scholar]
- Caughey B, Raymond GJ, Ernst D, & Race RE (1991). N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease (s): implications regarding the site of conversion of PrP to the protease-resistant state. Journal of virology, 65, 6597–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Race B, Meade-White K, LaCasse R, Race R, Klingeborn M, Striebel J, Dorward D, McGovern G, & Jeffrey M. (2010). Fatal transmissible amyloid encephalopathy: a new type of prion disease associated with lack of prion protein membrane anchoring. PLoS pathogens, 6, e1000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, & Priola S. (2005). Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science, 308, 1435–1439. [DOI] [PubMed] [Google Scholar]
- Choi YP, & Priola SA (2013). A specific population of abnormal prion protein aggregates is preferentially taken up by cells and disaggregated in a strain-dependent manner. Journal of virology, 87, 11552–11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson HC, & Belting M. (2014). Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biology, 35, 51–55. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Keenan TD, Fielder HL, Collinson LJ, Holley RJ, Merry CL, van Kuppevelt TH, Day AJ, & Bishop PN (2011). Mapping the differential distribution of glycosaminoglycans in the adult human retina, choroid, and sclera. Investigative ophthalmology & visual science, 52, 6511–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman BM, Harrison CF, Guo B, Masters CL, Barnham KJ, Lawson VA, & Hill AF (2014). Pathogenic mutations within the hydrophobic domain of the prion protein lead to the formation of protease-sensitive prion species with increased lethality. Journal of virology, 88, 2690–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J. (2005). Molecular neurology of prion disease. Journal of Neurology, Neurosurgery & Psychiatry, 76, 906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Sidle KC, Meads J, Ironside J, & Hill AF (1996). Molecular analysis of prion strain variation and the aetiology of’new variant’CJD. Nature, 383, 685–690. [DOI] [PubMed] [Google Scholar]
- Concha-Marambio L, Diaz-Espinoza R, & Soto C. (2014). The extent of protease resistance of misfolded prion protein is highly dependent on the salt concentration. Journal of Biological Chemistry, 289, 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das NR, Miyata H, Hara H, Chida J, Uchiyama K, Masujin K, Watanabe H, Kondoh G, & Sakaguchi S. (2020). The N-Terminal Polybasic Region of Prion Protein Is Crucial in Prion Pathogenesis Independently of the Octapeptide Repeat Region. Molecular neurobiology, 57, 1203–1216. [DOI] [PubMed] [Google Scholar]
- Das NR, Miyata H, Hara H, Uchiyama K, Chida J, Yano M, Watanabe H, Kondoh G, & Sakaguchi S. (2017). Effects of prion protein devoid of the N-terminal residues 25–50 on prion pathogenesis in mice. Archives of virology, 162, 1867–1876. [DOI] [PubMed] [Google Scholar]
- De Cecco E, Celauro L, Vanni S, Grandolfo M, Bistaffa E, Moda F, Aguzzi A, & Legname G. (2020). The uptake of tau amyloid fibrils is facilitated by the cellular prion protein and hampers prion propagation in cultured cells. Journal of neurochemistry, 155, 577–591. [DOI] [PubMed] [Google Scholar]
- Dear DV, Young DS, Kazlauskaite J, Meersman F, Oxley D, Webster J, Pinheiro TJ, Gill AC, Bronstein I, & Lowe CR (2007). Effects of post-translational modifications on prion protein aggregation and the propagation of scrapie-like characteristics in vitro. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1774, 792–802. [DOI] [PubMed] [Google Scholar]
- DeMarco ML, & Daggett V. (2007). Molecular mechanism for low pH triggered misfolding of the human prion protein. Biochemistry, 46, 3045–3054. [DOI] [PubMed] [Google Scholar]
- Douglas KL, Piccirillo CA, & Tabrizian M. (2008). Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. European Journal of Pharmaceutics and Biopharmaceutics, 68, 676–687. [DOI] [PubMed] [Google Scholar]
- Ehlers B, & Diringer H. (1984). Dextran sulphate 500 delays and prevents mouse scrapie by impairment of agent replication in spleen. Journal of General Virology, 65, 1325–1330. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, & Harashima H. (2013). Endocytosis of gene delivery vectors: from clathrin-dependent to lipid raft-mediated endocytosis. Molecular Therapy, 21, 1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar C, & Dickinson A. (1986). Prolongation of scrapie incubation period by an injection of dextran sulphate 500 within the month before or after infection. Journal of General Virology, 67, 463–473. [DOI] [PubMed] [Google Scholar]
- Fehlinger A, Wolf H, Hossinger A, Duernberger Y, Pleschka C, Riemschoss K, Liu S, Bester R, Paulsen L, & Priola SA (2017). Prion strains depend on different endocytic routes for productive infection. Scientific reports, 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Borges N, Parra B, Vidal E, Eraña H, Sánchez-Martín MA, de Castro J, Elezgarai SR, Pumarola M, Mayoral T, & Castilla J. (2017). Unraveling the key to the resistance of canids to prion diseases. PLoS pathogens, 13, e1006716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechsig E, Shmerling D, Hegyi I, Raeber AJ, Fischer M, Cozzio A, von Mering C, Aguzzi A, & Weissmann C. (2000). Prion protein devoid of the octapeptide repeat region restores susceptibility to scrapie in PrP knockout mice. Neuron, 27, 399–408. [DOI] [PubMed] [Google Scholar]
- Foliaki ST, Groveman BR, Yuan J, Walters R, Zhang S, Tesar P, Zou W, & Haigh CL (2020). Pathogenic prion protein isoforms are not present in cerebral organoids generated from asymptomatic donors carrying the E200K mutation associated with familial prion disease. Pathogens, 9, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenfield KN, Powers ET, & Kelly JW (2005). Influence of the N-terminal domain on the aggregation properties of the prion protein. Protein Science, 14, 2154–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabizon R, Meiner Z, Halimi M, & Ben-Sasson SA (1993). Heparin-like molecules bind differentially to prion-proteins and change their intracellular metabolic fate. Journal of cellular physiology, 157, 319–325. [DOI] [PubMed] [Google Scholar]
- Gambetti P, Kong Q, Zou W, Parchi P, & Chen SG (2003). Sporadic and familial CJD: classification and characterisation. British medical bulletin, 66, 213–239. [DOI] [PubMed] [Google Scholar]
- Gauczynski S, Nikles D, El-Gogo S, Papy-Garcia D, Rey C, Alban S, Barritault D, Lasmézas CI, & Weiss S. (2006). The 37-kDa/67-kDa laminin receptor acts as a receptor for infectious prions and is inhibited by polysulfated glycanes. The Journal of infectious diseases, 194, 702–709. [DOI] [PubMed] [Google Scholar]
- Gauczynski S, Peyrin JM, Haïk S, Leucht C, Hundt C, Rieger R, Krasemann S, Deslys JP, Dormont D, & Lasmézas CI (2001). The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. The EMBO journal, 20, 5863–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti B, Piccardo P, Spillantini MG, Ichimiya Y, Porro M, Perini F, Kitamoto T, Tateishi J, Seiler C, & Frangione B. (1996). Vascular variant of prion protein cerebral amyloidosis with tau-positive neurofibrillary tangles: the phenotype of the stop codon 145 mutation in PRNP. Proceedings of the National Academy of Sciences, 93, 744–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold R, McKinnon C, Rabbanian S, Collinge J, Schiavo G, & Tabrizi SJ (2013). Alternative fates of newly formed PrPSc upon prion conversion on the plasma membrane. Journal of cell science, 126, 3552–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold R, Rabbanian S, Sutton L, Andre R, Arora P, Moonga J, Clarke A, Schiavo G, Jat P, & Collinge J. (2011). Rapid cell-surface prion protein conversion revealed using a novel cell system. Nature communications, 2, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, & Donaldson JG (2009). Pathways and mechanisms of endocytic recycling. Nature reviews Molecular cell biology, 10, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil CS, Vorberg IM, Ward AE, Meade-White KD, Harris DA, & Priola SA (2008). Acute cellular uptake of abnormal prion protein is cell type and scrapie-strain independent. Virology, 379, 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Fisher S, Olofsson S, Endo T, Groth D, Tarentino A, Borchelt DR, Teplow D, Hood L, & Burlingame A. (1989). Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Archives of biochemistry and biophysics, 274, 1–13. [DOI] [PubMed] [Google Scholar]
- Harris DA, & True HL (2006). New insights into prion structure and toxicity. Neuron, 50, 353–357. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Muth C, Dabrowski O, Krasemann S, & Glatzel M. (2017). Exosomes and the prion protein: more than one truth. Frontiers in neuroscience, 11, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijazi N, Kariv-Inbal Z, Gasset M, & Gabizon R. (2005). PrPSc incorporation to cells requires endogenous glycosaminoglycan expression. Journal of Biological Chemistry, 280, 17057–17061. [DOI] [PubMed] [Google Scholar]
- Hooper NM (2011). Glypican-1 facilitates prion conversion in lipid rafts. Journal of neurochemistry, 116, 721–725. [DOI] [PubMed] [Google Scholar]
- Hooper NM, Taylor DR, & Watt NT (2008). Mechanism of the metal-mediated endocytosis of the prion protein. Biochemical Society Transactions, 36, 1272–1276. [DOI] [PubMed] [Google Scholar]
- Horonchik L, Tzaban S, Ben-Zaken O, Yedidia Y, Rouvinski A, Papy-Garcia D, Barritault D, Vlodavsky I, & Taraboulos A. (2005). Heparan sulfate is a cellular receptor for purified infectious prions. Journal of Biological Chemistry, 280, 17062–17067. [DOI] [PubMed] [Google Scholar]
- Hu Y-B, Dammer EB, Ren R-J, & Wang G. (2015). The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Translational neurodegeneration, 4, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundt C, Peyrin JM, Haïk S, Gauczynski S, Leucht C, Rieger R, Riley ML, Deslys JP, Dormont D, & Lasmézas CI (2001). Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. The EMBO journal, 20, 5876–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutti CR, Welle KA, Hryhorenko JR, & Ghaemmaghami S. (2020). Global analysis of protein degradation in prion infected cells. Scientific reports, 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M, Tabeta N, Kato N, Matsuura Y, Iwamaru Y, Yokoyama T, & Murayama Y. (2016). Heparan sulfate and heparin promote faithful prion replication in vitro by binding to normal and abnormal prion proteins in protein misfolding cyclic amplification. Journal of Biological Chemistry, 291, 26478–26486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ironside J, McCardle L, Hayward P, & Bell J. (1993). Ubiquitin immunocytochemistry in human spongiform encephalopathies. Neuropathology and applied neurobiology, 19, 134–140. [DOI] [PubMed] [Google Scholar]
- Jen A, Parkyn CJ, Mootoosamy RC, Ford MJ, Warley A, Liu Q, Bu G, Baskakov IV, Moestrup S, & McGuinness L. (2010). Neuronal low-density lipoprotein receptor-related protein 1 binds and endocytoses prion fibrils via receptor cluster 4. Journal of cell science, 123, 246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katorcha E, Makarava N, Savtchenko R, & Baskakov IV (2015). Sialylation of the prion protein glycans controls prion replication rate and glycoform ratio. Scientific reports, 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MC, Lindsay MR, Luetterforst R, Hamilton N, Simpson F, Parton RG, Gleeson PA, & Teasdale RD (2006). Visualisation of macropinosome maturation by the recruitment of sorting nexins. Journal of cell science, 119, 3967–3980. [DOI] [PubMed] [Google Scholar]
- Kim C, Haldiman T, Surewicz K, Cohen Y, Chen W, Blevins J, Sy M-S, Cohen M, Kong Q, & Telling GC (2012). Small protease sensitive oligomers of PrPSc in distinct human prions determine conversion rate of PrPC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AL, & Botos E. (2009). Endocytosis via caveolae: alternative pathway with distinct cellular compartments to avoid lysosomal degradation? Journal of cellular and molecular medicine, 13, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T, Iizuka R, & Tateishi J. (1993). An amber mutation of prion protein in Gerstmann-Sträussler syndrome with mutant PrP plaques. Biochemical and biophysical research communications, 192, 525–531. [DOI] [PubMed] [Google Scholar]
- Klingeborn M, Race B, Meade-White KD, Rosenke R, Striebel JF, & Chesebro B. (2011). Crucial role for prion protein membrane anchoring in the neuroinvasion and neural spread of prion infection. Journal of virology, 85, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács GG, Gelpi E, Ströbel T, Ricken G, Nyengaard JR, Bernheimer H, & Budka H. (2007). Involvement of the endosomal-lysosomal system correlates with regional pathology in Creutzfeldt-Jakob disease. Journal of Neuropathology & Experimental Neurology, 66, 628–636. [DOI] [PubMed] [Google Scholar]
- Kraus A, Hoyt F, Schwartz CL, Hansen B, Artikis E, Hughson AG, Raymond GJ, Race B, Baron GS, & Caughey B. (2021). High-resolution structure and strain comparison of infectious mammalian prions. Molecular Cell. [DOI] [PubMed] [Google Scholar]
- Lajoie P, & Nabi IR (2010). Lipid rafts, caveolae, and their endocytosis. International review of cell and molecular biology, 282, 135–163. [DOI] [PubMed] [Google Scholar]
- Lane-Donovan C, Philips GT, & Herz J. (2014). More than cholesterol transporters: lipoprotein receptors in CNS function and neurodegeneration. Neuron, 83, 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo L, Lowe J, Self T, Kenward N, Landon M, McBride T, Farquhar C, McConnell I, Brown J, & Hope J. (1992). Lysosomes as key organelles in the pathogenesis of prion encephalopathies. The Journal of pathology, 166, 333–341. [DOI] [PubMed] [Google Scholar]
- Lehmann S, & Harris DA (1996). Two mutant prion proteins expressed in cultured cells acquire biochemical properties reminiscent of the scrapie isoform. Proceedings of the National Academy of Sciences, 93, 5610–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PA, Properzi F, Prodromidou K, Clarke AR, Collinge J, & Jackson GS (2006). Removal of the glycosylphosphatidylinositol anchor from PrPSc by cathepsin D does not reduce prion infectivity. Biochemical Journal, 395, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis V, & Hooper NM (2011). The role of lipid rafts in prion protein biology. Front Biosci, 16, 151–168. [DOI] [PubMed] [Google Scholar]
- Li J-P, & Kusche-Gullberg M. (2016). Heparan sulfate: biosynthesis, structure, and function. International review of cell and molecular biology, 325, 215–273. [DOI] [PubMed] [Google Scholar]
- Linsenmeier L, Altmeppen HC, Wetzel S, Mohammadi B, Saftig P, & Glatzel M. (2017). Diverse functions of the prion protein–Does proteolytic processing hold the key? Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1864, 2128–2137. [DOI] [PubMed] [Google Scholar]
- Marbiah MM, Harvey A, West BT, Louzolo A, Banerjee P, Alden J, Grigoriadis A, Hummerich H, Kan HM, & Cai Y. (2014). Identification of a gene regulatory network associated with prion replication. The EMBO journal, 33, 1527–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella M, Lehmann S, Grassi J, & Chabry J. (2002). Filipin prevents pathological prion protein accumulation by reducing endocytosis and inducing cellular PrP release. Journal of Biological Chemistry, 277, 25457–25464. [DOI] [PubMed] [Google Scholar]
- Marín-Moreno A, Aguilar-Calvo P, Moudjou M, Espinosa JC, Béringue V, & Torres JM (2019). Thermostability as a highly dependent prion strain feature. Scientific reports, 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR (2014). Role of endosomes and lysosomes in human disease. Cold Spring Harbor perspectives in biology, 6, a016931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Sonnenfeld T, Avrahami D, Friedman-Levi Y, & Gabizon R. (2008). Chemically induced accumulation of GAGs delays PrP Sc clearance but prolongs prion disease incubation time. Cellular and molecular neurobiology, 28, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride P, Wilson M, Eikelenboom P, Tunstall A, & Bruce M. (1998). Heparan sulfate proteoglycan is associated with amyloid plaques and neuroanatomically targeted PrP pathology throughout the incubation period of scrapie-infected mice. Experimental neurology, 149, 447–454. [DOI] [PubMed] [Google Scholar]
- McNally KL, Ward AE, & Priola SA (2009). Cells expressing anchorless prion protein are resistant to scrapie infection. Journal of virology, 83, 4469–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S. (2006). Prion disease genetics. European Journal of Human Genetics, 14, 273–281. [DOI] [PubMed] [Google Scholar]
- Monari L, Chen SG, Brown P, Parchi P, Petersen RB, Mikol J, Gray F, Cortelli P, Montagna P, & Ghetti B. (1994). Fatal familial insomnia and familial Creutzfeldt-Jakob disease: different prion proteins determined by a DNA polymorphism. Proceedings of the National Academy of Sciences, 91, 2839–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Timmes A, Wilmarth PA, & Priola SA (2010). Comparative profiling of highly enriched 22L and Chandler mouse scrapie prion protein preparations. Proteomics, 10, 2858–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales R, Abid K, & Soto C. (2007). The prion strain phenomenon: molecular basis and unprecedented features. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease, 1772, 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel E, Andrieu T, Casagrande F, Gauczynski S, Weiss S, Grassi J, Rousset M, Dormont D, & Chambaz J. (2005). Bovine prion is endocytosed by human enterocytes via the 37 kDa/67 kDa laminin receptor. The American Journal of Pathology, 167, 1033–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi P, Leclerc E, & Marc D. (2002). Unusual property of prion protein unfolding in neutral salt solution. Biochemistry, 41, 11017–11024. [DOI] [PubMed] [Google Scholar]
- Nicot S, & Baron TG (2010). Strain-specific proteolytic processing of the prion protein in prion diseases of ruminants transmitted in ovine transgenic mice. Journal of General Virology, 91, 570–574. [DOI] [PubMed] [Google Scholar]
- Nishida N, Harris DA, Vilette D, Laude H, Frobert Y, Grassi J, Casanova D, Milhavet O, & Lehmann S. (2000). Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. Journal of virology, 74, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notari S, Strammiello R, Capellari S, Giese A, Cescatti M, Grassi J, Ghetti B, Langeveld JP, Zou W-Q, & Gambetti P. (2008). Characterization of truncated forms of abnormal prion protein in Creutzfeldt-Jakob disease. Journal of Biological Chemistry, 283, 30557–30565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet S, Daude N, Courageot M-P, Chapuis J, Laude H, & Vilette D. (2007). PrPc does not mediate internalization of PrPSc but is required at an early stage for de novo prion infection of Rov cells. Journal of virology, 81, 10786–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen SG, Farlow M, Dickson DW, Sima AA, & Trojanowski JQ (1996). Molecular basis of phenotypic variability in sporadc creudeldt-jakob disease. Annals of neurology, 39, 767–778. [DOI] [PubMed] [Google Scholar]
- Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, Zerr I, Budka H, Kopp N, & Piccardo P. (1999). Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Annals of neurology, 46, 224–233. [PubMed] [Google Scholar]
- Pastore A, & Zagari A. (2007). A structural overview of the vertebrate prion proteins. Prion, 1, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Mironov A Jr, Peretz D, van Donselaar E, Leclerc E, Erpel S, DeArmond SJ, Burton DR, Williamson RA, & Vey M. (2003). Trafficking of prion proteins through a caveolae-mediated endosomal pathway. The Journal of cell biology, 162, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RB, Parchi P, Richardson SL, Urig CB, & Gambetti P. (1996). Effect of the D178N Mutation and the Codon 129 Polymorphism on the Metabolism of the Prion Protein (∗). Journal of Biological Chemistry, 271, 12661–12668. [DOI] [PubMed] [Google Scholar]
- Piccardo P, Cervenakova L, Vasilyeva I, Yakovleva O, Bacik I, Cervenak J, McKenzie C, Kurillova L, Gregori L, & Pomeroy K. (2011). Candidate cell substrates, vaccine production, and transmissible spongiform encephalopathies. Emerging infectious diseases, 17, 2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Trusheim H, Stallmach L, Moos R, Julius C, Miele G, Lenz-Bauer C, & Aguzzi A. (2008). Canine MDCK cell lines are refractory to infection with human and mouse prions. Vaccine, 26, 2601–2614. [DOI] [PubMed] [Google Scholar]
- Priola SA (2013). Species barriers in prion disease. In Prions and Diseases (pp. 139–154): Springer. [Google Scholar]
- Priola SA (2018). Cell biology of prion infection. In Handbook of clinical neurology (Vol. 153, pp. 45–68): Elsevier. [DOI] [PubMed] [Google Scholar]
- Priola SA, & Chesebro B. (1995). A single hamster PrP amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. Journal of virology, 69, 7754–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priola SA, & Chesebro B. (1998). Abnormal properties of prion protein with insertional mutations in different cell types. Journal of Biological Chemistry, 273, 11980–11985. [DOI] [PubMed] [Google Scholar]
- Priola SA, & McNally KL (2009). The role of the prion protein membrane anchor in prion infection. Prion, 3, 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB (1982). Novel proteinaceous infectious particles cause scrapie. Science, 216, 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, & Carlson GA (1990). Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell, 63, 673–686. [DOI] [PubMed] [Google Scholar]
- Puig B, Altmeppen HC, Linsenmeier L, Chakroun K, Wegwitz F, Piontek UK, Tatzelt J, Bate C, Magnus T, & Glatzel M. (2019). GPI-anchor signal sequence influences PrPC sorting, shedding and signalling, and impacts on different pathomechanistic aspects of prion disease in mice. PLoS pathogens, 15, e1007520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Phillips K, Meade-White K, Striebel J, & Chesebro B. (2015). Increased infectivity of anchorless mouse scrapie prions in transgenic mice overexpressing human prion protein. Journal of virology, 89, 6022–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Williams K, Hughson AG, Jansen C, Parchi P, Rozemuller AJ, & Chesebro B. (2018). Familial human prion diseases associated with prion protein mutations Y226X and G131V are transmissible to transgenic mice expressing human prion protein. Acta neuropathologica communications, 6, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, & Hermo L. (1979). Three-dimensional architecture of the Golgi apparatus in Sertoli cells of the rat. American Journal of Anatomy, 154, 455–475. [DOI] [PubMed] [Google Scholar]
- Rangel A, Race B, Phillips K, Striebel J, Kurtz N, & Chesebro B. (2014). Distinct patterns of spread of prion infection in brains of mice expressing anchorless or anchored forms of prion protein. Acta neuropathologica communications, 2, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch JN, Luna G, Guzman E, Audouard M, Challis C, Sibih YE, Leshuk C, Hernandez I, Wegmann S, & Hyman BT (2020). LRP1 is a master regulator of tau uptake and spread. Nature, 580, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revesz T, Holton JL, Lashley T, Plant G, Frangione B, Rostagno A, & Ghiso J. (2009). Genetics and molecular pathogenesis of sporadic and hereditary cerebral amyloid angiopathies. Acta neuropathologica, 118, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinski A, Karniely S, Kounin M, Moussa S, Goldberg MD, Warburg G, Lyakhovetsky R, Papy-Garcia D, Kutzsche J, & Korth C. (2014). Live imaging of prions reveals nascent PrPSc in cell-surface, raft-associated amyloid strings and webs. Journal of Cell Biology, 204, 423–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamat MK, Dron M, Chapuis J, Langevin C, & Laude H. (2011). Prion propagation in cells expressing PrP glycosylation mutants. Journal of virology, 85, 3077–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangeetham SB, Engelke AD, Fodor E, Krausz SL, Tatzelt J, & Welker E. (2021). The G127V variant of the prion protein interferes with dimer formation in vitro but not in cellulo. Scientific reports, 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnataro D, Caputo A, Casanova P, Puri C, Paladino S, Tivodar SS, Campana V, Tacchetti C, & Zurzolo C. (2009). Lipid rafts and clathrin cooperate in the internalization of PrPC in epithelial FRT cells. PLoS One, 4, e5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Groth D, Foster D, Torchia M, Yang S-L, DeArmond SJ, & Prusiner SB (1993). Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell, 73, 979–988. [DOI] [PubMed] [Google Scholar]
- Shaked GM, Meiner Z, Avraham I, Taraboulos A, & Gabizon R. (2001). Reconstitution of prion infectivity from solubilized protease-resistant PrP and nonprotein components of prion rods. Journal of Biological Chemistry, 276, 14324–14328. [DOI] [PubMed] [Google Scholar]
- Shim SY, Karri S, Law S, Schatzl HM, & Gilch S. (2016). Prion infection impairs lysosomal degradation capacity by interfering with rab7 membrane attachment in neuronal cells. Scientific reports, 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoup D, & Priola SA (2021). The Size and Stability of Infectious Prion Aggregates Fluctuate Dynamically during Cellular Uptake and Disaggregation. Biochemistry, 60, 398–411. [DOI] [PubMed] [Google Scholar]
- Shyng S-L, Lehmann S, Moulder KL, & Harris DA (1995). Sulfated Glycans Stimulate Endocytosis of the Cellular Isoform of the Prion Protein, PrPC, in Cultured Cells (∗). Journal of Biological Chemistry, 270, 30221–30229. [DOI] [PubMed] [Google Scholar]
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, & Caughey B. (2005). The most infectious prion protein particles. Nature, 437, 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon Davis DA, & Parish CR (2013). Heparan sulfate: a ubiquitous glycosaminoglycan with multiple roles in immunity. Frontiers in immunology, 4, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Zanusso G, Chen SG, Fujioka H, Richardson S, Gambetti P, & Petersen RB (1997). Prion protein aggregation reverted by low temperature in transfected cells carrying a prion protein gene mutation. Journal of Biological Chemistry, 272, 28461–28470. [DOI] [PubMed] [Google Scholar]
- Sonawane N, Thiagarajah JR, & Verkman A. (2002). Chloride concentration in endosomes measured using a ratioable fluorescent Cl− indicator: evidence for chloride accumulation during acidification. Journal of Biological Chemistry, 277, 5506–5513. [DOI] [PubMed] [Google Scholar]
- Spassov S, Beekes M, & Naumann D. (2006). Structural differences between TSEs strains investigated by FT-IR spectroscopy. Biochimica et Biophysica Acta (BBA)-General Subjects, 1760, 1138–1149. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Makarava N, Katorcha E, Savtchenko R, Brossmer R, & Baskakov IV (2015). Post-conversion sialylation of prions in lymphoid tissues. Proceedings of the National Academy of Sciences, 112, E6654–E6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N, Borchelt DR, & Prusiner SB (1990). Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry, 29, 5405–5412. [DOI] [PubMed] [Google Scholar]
- Thody SA, Mathew M, & Udgaonkar JB (2018). Mechanism of aggregation and membrane interactions of mammalian prion protein. Biochimica et Biophysica Acta (BBA)-Biomembranes, 1860, 1927–1935. [DOI] [PubMed] [Google Scholar]
- Thompson HN, Thompson CE, Andrade Caceres R, Dardenne LE, Netz PA, & Stassen H. (2018). Prion protein conversion triggered by acidic condition: a molecular dynamics study through different force fields. Journal of computational chemistry, 39, 2000–2011. [DOI] [PubMed] [Google Scholar]
- Torres J-M, Castilla J, Pintado B, Gutiérrez-Adan A, Andréoletti O, Aguilar-Calvo P, Arroba A-I, Parra-Arrondo B, Ferrer I, & Manzanares J. (2013). Spontaneous generation of infectious prion disease in transgenic mice. Emerging infectious diseases, 19, 1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AW, Oestergaard K, Myers JT, & Swanson JA (2000). Altered membrane trafficking in activated bone marrow-derived macrophages. Journal of leukocyte biology, 68, 487–494. [PubMed] [Google Scholar]
- Tuzi NL, Cancellotti E, Baybutt H, Blackford L, Bradford B, Plinston C, Coghill A, Hart P, Piccardo P, & Barron RM (2008). Host PrP glycosylation: a major factor determining the outcome of prion infection. PLoS biology, 6, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Tomita M, Yano M, Chida J, Hara H, Das NR, Nykjaer A, & Sakaguchi S. (2017). Prions amplify through degradation of the VPS10P sorting receptor sortilin. PLoS pathogens, 13, e1006470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ijzendoorn SC (2006). Recycling endosomes. Journal of cell science, 119, 1679–1681. [DOI] [PubMed] [Google Scholar]
- Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, & Prusiner SB (1996). Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proceedings of the National Academy of Sciences, 93, 14945–14949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E, Fernández-Borges N, Pintado B, Eraña H, Ordóñez M, Márquez M, Chianini F, Fondevila D, Sánchez-Martín MA, & Andreoletti O. (2015). Transgenic mouse bioassay: evidence that rabbits are susceptible to a variety of prion isolates. PLoS pathogens, 11, e1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E, Fernández-Borges N, Eraña H, Parra B, Pintado B, Sánchez-Martín MA, Charco JM, Ordóñez M, Pérez-Castro MA, & Pumarola M. (2020). Dogs are resistant to prion infection, due to the presence of aspartic or glutamic acid at position 163 of their prion protein. The FASEB Journal, 34, 3969–3982. [DOI] [PubMed] [Google Scholar]
- Vieira TC, Cordeiro Y, Caughey B, & Silva JL (2014). Heparin binding confers prion stability and impairs its aggregation. The FASEB Journal, 28, 2667–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorberg I, Raines A, & Priola SA (2004). Acute formation of protease-resistant prion protein does not always lead to persistent scrapie infection in vitro. Journal of Biological Chemistry, 279, 29218–29225. [DOI] [PubMed] [Google Scholar]
- Vorberg I, Raines A, Story B, & Priola SA (2004). Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. The Journal of infectious diseases, 189, 431–439. [DOI] [PubMed] [Google Scholar]
- Waddell L, Greig J, Mascarenhas M, Otten A, Corrin T, & Hierlihy K. (2018). Current evidence on the transmissibility of chronic wasting disease prions to humans—a systematic review. Transboundary and emerging diseases, 65, 37–49. [DOI] [PubMed] [Google Scholar]
- Wadia JS, Schaller M, Williamson RA, & Dowdy SF (2008). Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS One, 3, e3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth JD, Asante EA, Desbruslais M, Linehan JM, Joiner S, Gowland I, Welch J, Stone L, Lloyd SE, & Hill AF (2004). Human prion protein with valine 129 prevents expression of variant CJD phenotype. Science, 306, 1793–1796. [DOI] [PubMed] [Google Scholar]
- Warda M, Toida T, Zhang F, Sun P, Munoz E, Xie J, & Linhardt RJ (2006). Isolation and characterization of heparan sulfate from various murine tissues. Glycoconjugate journal, 23, 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner RG, Hundt C, Weiss S, & Turnbull JE (2002). Identification of the heparan sulfate binding sites in the cellular prion protein. Journal of Biological Chemistry, 277, 18421–18430. [DOI] [PubMed] [Google Scholar]
- Watts JC, Giles K, Bourkas ME, Patel S, Oehler A, Gavidia M, Bhardwaj S, Lee J, & Prusiner SB (2016). Towards authentic transgenic mouse models of heritable PrP prion diseases. Acta neuropathologica, 132, 593–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JC, Giles K, Patel S, Oehler A, DeArmond SJ, & Prusiner SB (2014). Evidence that bank vole PrP is a universal acceptor for prions. PLoS pathogens, 10, e1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein RA, Rutala WA, & Weber DJ (2001). Creutzfeldt-Jakob disease: recommendations for disinfection and sterilization. Clinical Infectious Diseases, 32, 1348–1356. [DOI] [PubMed] [Google Scholar]
- Will R, Cousens S, Farrington C, Smith P, Knight R, & Ironside J. (1999). Deaths from variant Creutzfeldt-Jakob disease. The Lancet, 353, 979. [DOI] [PubMed] [Google Scholar]
- Wolf H, Graßmann A, Bester R, Hossinger A, Möhl C, Paulsen L, Groschup MH, Schätzl H, & Vorberg I. (2015). Modulation of glycosaminoglycans affects PrPSc metabolism but does not block PrPSc uptake. Journal of virology, 89, 9853–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. n., Xiong LW, Horiuchi M, Raymond L, Wehrly K, Chesebro B, & Caughey B. (2001). Sulfated glycans and elevated temperature stimulate PrPSc-dependent cell-free formation of protease-resistant prion protein. The EMBO journal, 20, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman PG, & Futter CE (2008). Multivesicular bodies: co-ordinated progression to maturity. Current opinion in cell biology, 20, 408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Suzuki A, Hasebe R, & Horiuchi M. (2018). Retrograde Transport by Clathrin-Coated Vesicles is Involved in Intracellular Transport of PrP Sc in Persistently Prion-Infected Cells. Scientific reports, 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim Y-I, Park B-C, Yadavalli R, Zhao X, Eisenberg E, & Greene LE (2015). The multivesicular body is the major internal site of prion conversion. Journal of cell science, 128, 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanusso G, Farinazzo A, Fiorini M, Gelati M, Castagna A, Righetti PG, Rizzuto N, & Monaco S. (2001). pH-dependent prion protein conformation in classical Creutzfeldt-Jakob disease. Journal of Biological Chemistry, 276, 40377–40380. [DOI] [PubMed] [Google Scholar]
- Zhang H, Chen W, Tan Z, Zhang L, Dong Z, Cui W, Zhao K, Wang H, Jing H, & Cao R. (2020). A role of low-density lipoprotein receptor-related protein 4 (LRP4) in astrocytic Aβ clearance. Journal of Neuroscience, 40, 5347–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]