Abstract
Herpesvirus ateles is a gamma-2-herpesvirus which naturally infects spider monkeys (Ateles spp.) and causes malignant lymphoproliferative disorders in various other New World primates. The genomic sequence of herpesvirus ateles strain 73 revealed a close relationship to herpesvirus saimiri, with a high degree of variability within the left terminus of the coding region. A spliced mRNA transcribed from this region was detected in New World monkey T-cell lines transformed by herpesvirus ateles in vitro or derived from T cells of infected Saguinus oedipus. The encoded viral protein, termed Tio, shows restricted homology to the oncoprotein StpC and to the tyrosine kinase-interacting protein Tip, two gene products responsible for the T-cell-transforming and oncogenic phenotype of herpesvirus saimiri group C strains. Tio was detectable in lysates of the transformed T lymphocytes. Dimer formation was observed after expression of recombinant Tio. After cotransfection, Tio was phosphorylated in vivo by the protein tyrosine kinases Lck and Src and less efficiently by Fyn. Stable complexes of these Src family kinases with the viral protein were detected in lysates of the transfected cells. Binding analyses indicated a direct interaction of Tio with the SH3 domains of Lyn, Hck, Lck, Src, Fyn, and Yes. In addition, tyrosine-phosphorylated Tio bound to the SH2 domains of Lck, Src, or Fyn. Thus, herpesvirus ateles-encoded Tio may contribute to viral T-cell transformation by influencing the function of Src family kinases.
Two simian viruses, herpesvirus saimiri (HVS) and herpesvirus ateles (HVA), have been isolated from squirrel (Saimiri sciureus) and spider (Ateles spp.) monkeys, respectively. They have proven unique in the ability to induce T-cell lymphomas and leukemias in several New World primate species cognate to the natural hosts. HVS and HVA are related viruses of the genus Rhadinovirus (gamma-2-herpesviruses) which differ in their biological properties (reviewed in reference 28). While genomic sequences in general are well conserved among these rhadinoviruses, the far left ends of the coding sequences display a pronounced variability (1, 2, 23, 58, 66, 67, 73). In the case of HVS, this led to the classification of the viral isolates into subgroups A, B, and C (53). The divergence correlated with differences in the ability of HVS strains to immortalize T lymphocytes in vitro (4, 72).
Sequence analyses of this variable region revealed open reading frames for all of the HVS isolates examined. Subgroup A strains code for StpA (saimiri transformation-associated protein of group A) (48, 57), the corresponding reading frame of subgroup B isolate SMHI was designated StpB (3), and two open reading frames in subgroup C genomes give rise to StpC and Tip (5, 31). The viral proteins have only limited sequence similarities. A hydrophobic carboxy terminus is common to all of these proteins and probably serves as a membrane anchor (3, 38, 42, 51). The amino-terminal parts of StpA, StpC, and Tip are rich in acidic amino acids (42). The central region of StpC consists of 18 consecutive collagen-like triplets (Gly-X-Y, where X and/or Y represent Pro), and individual triplets are spread over the amino-terminal halves of StpA and StpB (3, 42). Spontaneous and targeted viral deletion mutants of subgroup A and C strains indicated that these proteins are not required for virus replication but are responsible for the oncogenic phenotype of these viruses in vitro and in vivo (13, 14, 18, 43, 44, 52). StpA and StpC were also shown to be oncogenic in the absence of other viral factors. Both proteins were found to be sufficient to transform rodent fibroblast cells in vitro (42). In addition, mice carrying an StpC-encoding transgene developed malignant epithelial tumors (56), while animals with a StpA-encoding transgene developed peripheral T-cell lymphomas (46).
The transforming effects are supposed to be mediated by cellular factors interacting with the viral proteins. StpA was found to interact with cellular Src and, after phosphorylation by Src, bound to Lck and Fyn kinase Src homology 2 (SH2) domains in vitro (48). SH2 domains generally consist of about 150 amino acids and directly bind to phosphotyrosine residues. Specificity of distinct SH2 domains is determined by flanking sequences of the tyrosine residues (69, 70). The tyrosine-containing motif of StpA (V/IYAEV/I) represents the consensus for optimal binding to the SH2 domain. The tyrosine residue within this domain is required for interaction with Src (48). In contrast, StpC associates with cellular Ras and activates mitogen-activated protein kinase (39). In the viral context, active Ras may substitute for StpC in viral transformation (34), indicating that Ras is indeed a major effector of the transforming functions of StpC. However, StpC alone did not induce detectable alterations of the T-cell compartment in transgenic mice (56). Additional viral factors were suspected to determine the cell tropism of viral transformation. This function was attributed to the tyrosine kinase-interacting protein Tip, which has been shown to associate with the lymphocyte-specific kinase Lck (6, 51).
This interaction was found to be mediated by a region of Tip containing a proline-rich Src homology 3 (SH3)-binding (SH3B) motif, a motif referred to as the C-terminal Src kinase homology (CSKH) motif, and a spacer sequence between SH3B and CSKH (40). On the other hand, the SH3 domain of Lck has been shown to be sufficient to form a stable complex with Tip in vitro (41). SH3 domains are protein modules mediating protein-protein interactions with short, specific, proline-rich sequences that possess a left-handed polyproline type II helix structure (25, 26, 65). The consequences of the Tip-Lck interaction have been controversial, ranging from inhibition of Lck-mediated signal transduction by Tip to dramatic stimulation of Lck signaling and increased Lck phosphotransferase activity in cell-free systems (41, 50, 59, 75). HVS expressing an SH3B mutant form of Tip (Pro to Ala) effectively transformed T lymphocytes in vivo and in vitro, leading to the assumption that Lck interaction is not necessary for the Tip effector function (19). A novel Tip-associated protein, Tap, might instead be the mediator of Tip functions essential for transformation. Stable expression of Tap together with Tip in Jurkat cells induced the surface expression of adhesion molecules, leading to lymphocyte aggregation and NF-κB activation (77).
While HVS has been studied extensively during the last 2 decades, very little is known about HVA. In this report, we describe the HVA strain 73-encoded protein Tio, which shares homologies with HVS oncoprotein StpC and with Lck-interacting protein Tip. This protein appears to be functionally related to HVS proteins StpA and StpB, as well as to Tip. Thus, Tio is considered to be the prime candidate for a novel oncogene encoded by HVA strain 73.
MATERIALS AND METHODS
Cells, virus, and cell culture.
Owl monkey kidney (OMK) cells (12) (OMK 637 [ATCC CRL 1556]) and 293T cells (20, 63) were kept in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, l-glutamine, streptomycin, and penicillin. Virus was propagated on permissive OMK cells by following standard protocols (27). HVA strain 73 was originally isolated from lymphocytes by cocultivation with permissive cell cultures from a healthy spider monkey (Ateles paniscus) imported from Colombia. Marmoset cell line A661 was isolated after in vitro transformation of cotton-topped marmoset (Saguinus oedipus) lymphocytes (28), cell line A1022 was obtained after isolation of lymphocytes from an S. oedipus marmoset infected with HVA strain 73 (24). Lymphoid cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, l-glutamine, streptomycin, and penicillin but without exogenous interleukin-2. Cell line A17 was established from the spleen of an S. oedipus marmoset infected with HVS strain A11, cell line B1 was isolated from an S. oedipus marmoset after infection with HVS group B strain SMHI, and cell line C37 was established from a lymph node of an S. oedipus marmoset infected with HVS strain C-488.
Molecular cloning of Tio, expression cloning, and recombinant proteins.
The genomic sequence of the Tio-encoding gene was obtained in the course of the sequencing of the complete genome of HVA strain 73 as previously described (1). mRNA was isolated from cell lines A661 and A1022, reverse transcribed, and amplified by PCR using primers corresponding to the 5′ end of orf2 and oligo(dT) or a primer derived from the 3′ end of orf1. To facilitate cloning, restriction enzyme recognition sites and sequences corresponding to the Flag or AU1 epitope tag were attached to the primer sequences, resulting in N-terminally tagged proteins. Identity to the genomic sequence was confirmed for all derived clones by automated DNA sequencing on an ABI377 sequencer. Tio cDNA was cloned into pGEX-2TK with or without a Flag epitope tag to give glutathione S-transferase (GST) fusion proteins. These proteins were expressed in standard Escherichia coli XL2-Blue bacteria or in bacteria expressing a constitutively active Elk-1 tyrosine kinase (47) (TKX1; Stratagene, La Jolla, Calif.). After affinity chromatography using glutathione-Sepharose CL4B, Flag epitope-tagged Tio (Flag-Tio) and untagged Tio were purified by thrombin cleavage of GST-Flag-Tio in accordance with the manufacturer’s (Amersham Pharmacia Biotech, Freiburg, Germany) instructions. Eukaryotic expression clones were constructed by cloning of Tio, Flag-Tio, or AU1 epitope-tagged Tio (AU-Tio) into pcDNA3 (Invitrogen, Carlsbad, Calif.). Human Lck was cloned into the pFJ expression vector (41), pFJ-Src was obtained from S. M. Lang, Erlangen, Germany (17), and human Fyn (21) was cloned into the pcDNA-3.1(−)/Myc-HisA vector (Invitrogen). GST-SH3 fusion proteins used for fluorescence spectrometry were expressed and purified as previously described (60). The functionality of these fusion proteins has been previously described.
Antisera, antibodies, and synthetic peptides.
Anti-Tio serum was raised in rabbits immunized with purified GST-Tio or with Tio protein whose GST tag had been removed through site-specific proteolysis with thrombin. The antiserum was used at a dilution of 1:5,000. Antibodies against Src family kinases were purchased from Santa Cruz Biotechnologies (Santa Cruz, Calif.), Pharmingen (San Diego, Calif.), Upstate Biotechnology Inc. (Lake Placid, N.Y.), or Transduction Laboratories (Lexington, Ky.). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Dako (Hamburg, Germany), Jackson Immunoresearch Laboratories (Dianova, Hamburg, Germany), or Medac (Hamburg, Germany). Anti-myc hybridoma cell line 9E10 was obtained from the American Type Culture Collection (ATCC CRL-1729), and tissue culture supernatants were used at a dilution of 1:100. Antiphosphotyrosine monoclonal antibody PY99 (unconjugated or HRP coupled) was purchased from Santa Cruz Biotechnologies and used at a dilution of 1:3,000. Synthetic peptides were synthesized and purified at the Institute for Biochemistry, University of Erlangen. The authentic sequences of the peptides were determined by mass spectrometry.
Transient transfection, immunoprecipitation, and immunoblotting.
293T cells were transfected with plasmid DNA by using a CaCl2 transfection method. Briefly, cells were distributed among the wells of six-well plates. The next day, each well was fed with 3.6 ml of complete medium. DNA (1 to 5 μg) was diluted in 180 μl of H2O, 20 μl of 2.5 M CaCl2 was added, and the combination was mixed with 200 μl of BES buffer [50 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid, 280 mM NaCl, 1.5 mM Na2HPO4, pH 6.96]. This mixture was applied to the cells, which were then incubated at 37°C overnight. The cells were washed twice with phosphate-buffered saline (PBS), pH 7.4, fed with 2 ml of complete medium, and incubated for 16 to 24 h. Cells were harvested and frozen at −80°C or lysed for further analyses. For immunoprecipitation assays, cells were lysed in TNE buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40) supplemented with 1 mM Na3VO4, 5 mM NaF, and 10 μg each of aprotinin and leupeptin (Sigma, St. Louis, Mo.) per ml for 20 min on ice. Lysates were cleared by centrifugation at 14,000 × g for 10 min, and the protein concentration of the supernatants was determined. For each experiment, the same amount of total protein was used. A 5-μl volume of antiserum/mg of protein or 1 to 2 μg of monoclonal antibody was added, and the mixture was incubated for at least 1 h at 4°C to allow complex formation. Flag epitope-tagged proteins were precipitated by using monoclonal antibody M2 covalently bound to agarose (Kodak, New Haven, Conn.). Immunoprecipitation with uncoupled antibodies was completed by incubation with protein A-Sepharose or with rabbit anti-mouse antibodies coupled to protein A-Sepharose. The immunoprecipitates were washed at least five times in TNE buffer. For immunoblotting, cell lysates or immunoprecipitates were separated by sodium dodecyl sulfate (SDS)–8, 9, 10, or 12% polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (PVDF) membrane filters (Amersham Pharmacia Biotech, Freiburg, Germany). The PVDF membrane filters were incubated for 1 h at room temperature in blocking buffer (PBS [pH 7.4], 0.1% Tween 20, 5% [wt/vol] nonfat dried milk powder) and then incubated with antiserum or antibody diluted in blocking buffer. After thorough washing in PBS containing 0.1% Tween 20, immunoblots were incubated with secondary antibodies coupled to HRP. Bands were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech) in accordance with the manufacturer’s instructions. Antiphosphotyrosine immunoblotting was performed in accordance with the same protocol but without milk powder.
Fluorescence spectrometry and calculation of the binding constant.
Fluorescence measurements were based on the interaction of peptides with aromatic residues in the SH3 domains, predominantly tryptophan. Measurements were performed essentially as previously described (64) in a Perkin-Elmer 760-40 fluorescence spectrophotometer at an excitation wavelength of 290 nm (slit width, 2 nm) and an emission wavelength of 345 nm (slit width, 17 nm). A mini magnetic stirrer was used to mix the solution in a 1-cm2 quartz fluorescence cell. A circulating water bath was used to maintain the sample temperature at 18°C. To obtain the titration curves for calculation of the binding constants, peptides from a stock solution of 5 mg/ml in PBS–1 mM dithiothreitol were added in small increments to 1 ml of PBS–1 mM dithiothreitol containing 50 μg of GST-SH3 domain fusion proteins. Upon addition of the peptide solution, changes in fluorescence were measured. Since the concentration of the SH3 domain-containing protein was low, the experimental data were fitted to the following equation: F = Fmax × [peptide]/(Kd + [peptide]), where [peptide] is the final peptide concentration at each measurement point, F is the measured protein fluorescence intensity at the particular peptide concentration, and Fmax is the observed maximal fluorescence intensity of the protein when saturated with the peptide. Nonlinear regression curve fitting was carried out to fit the experimental data to the equation, with Fmax and Kd as fitted parameters. The change in protein concentration that occurred as a result of peptide addition was properly corrected.
In vitro binding assays.
One microgram of GST-SH2 domain fusion proteins was incubated with 500 ng of purified Flag-Tio protein in 1 ml of TNE for 1 h at 4°C. Complexes were precipitated with glutathione-Sepharose (Amersham Pharmacia Biotech) for 1 or 2 h at 4°C, washed five times with RIPA buffer (1% [wt/wt] Nonidet P-40, 1% [wt/vol] sodium deoxycholate, 0.1% [wt/vol] SDS, 0.15 M NaCl, 0.01 M sodium phosphate [pH 7.2], 2 mM EDTA), resolved on by SDS–9% PAGE, and transferred onto a PVDF membrane filter. Immunoblot detection was performed as described above.
Nucleotide sequence accession number.
The nucleotide sequence of the gene for Tio is available under GenBank accession no. AF083423.
RESULTS
Cloning and prediction of the tio gene product of herpesvirus ateles.
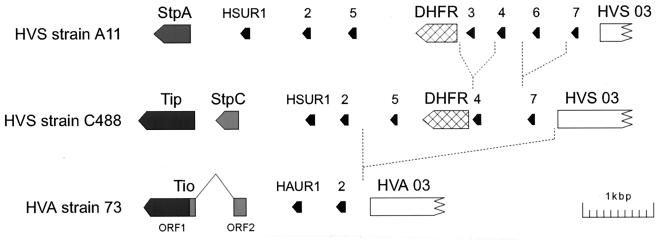
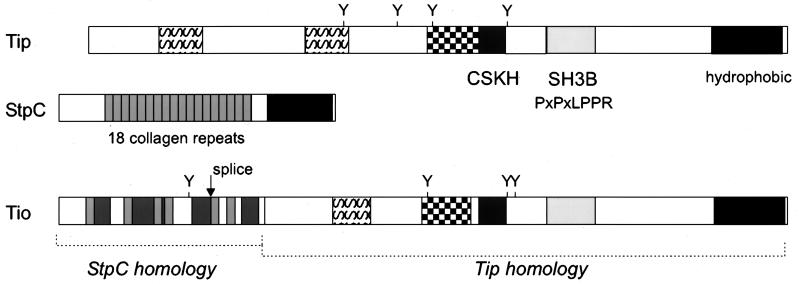
The complete nucleotide sequence of the coding unique DNA of herpesvirus ateles has been determined recently (1). The transformation-relevant region at the left terminus of HVS unique DNA displays a strong variability among HVS subgroups and HVA, while the rest of the genome of HVA shows a high degree of conservation with respect to the complete sequence of HVS strain A11 (Fig. 1) (1, 2). Initially, an open reading frame (ORF1) encoding 193 amino acids was identified at the genomic position of the transformation-associated proteins of HVS. The amino acid sequence predicted from ORF1 showed significant similarities to the product of the tip gene (Fig. 1 to 3). The homologies found were restricted to definite domains of Tip, namely, a serine-rich stretch of amino acids duplicated in Tip of strain C488, a domain surrounding a conserved tyrosine residue (Y-127), the CSKH motif which is homologous to the αI helix of Src (6, 76), the proline-rich SH3B motif, and a C-terminal transmembrane domain (Fig. 2).
FIG. 1.
Left-terminal genome variation among strains of HVS and HVA. The variable regions of HVS subgroups A and C encode the proteins StpA, StpC, and Tip, respectively, which are necessary for the transforming phenotype of the virus. Two open reading frames (ORF1 and ORF2) were identified in the corresponding genomic region of HVA strain 73. This region was transcribed into an mRNA of about 1 kb. The transcript was found to be spliced and to code for a single protein, Tio, that has homology to Tip and StpC. The splice occurs within the StpC-homologous portion of the Tio mRNA. Two of the small U RNAs of HVS (HSUR and HAUR) and reading frame 3 (HVS and HVA 03) are conserved. The other HSUR copies and the reading frame for dihydrofolate reductase (DHFR) found in HVS are not present in the genome of HVA.
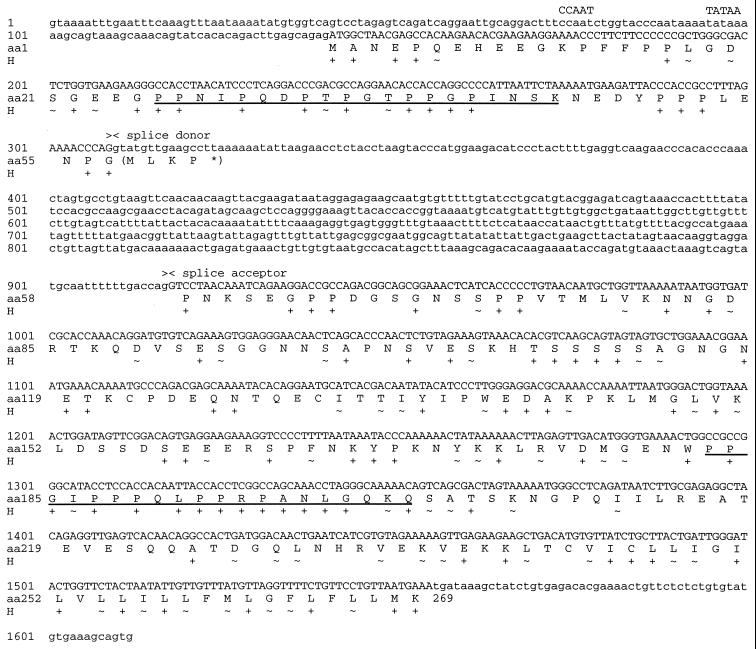
FIG. 3.
Primary structure of the gene for Tio. The presentation is inverted relative to the standard orientation of the HVA strain 73 genome. The nucleotide sequence starts with a region homologous to the StpC promoter and ends with the first nucleotide of the nonrepetitive DNA. The splice donor and acceptor sites of the mRNA are indicated above the nucleotide sequence. The amino acid translation of Tio (amino acids [aa] 1 to 269) is given in single-letter code. The line below the amino acid sequence (H) shows the homology of Tio to StpC and Tip of HVS strain C488. Identical amino acids are marked by plus signs, and similar amino acids are marked by tilde symbols. Similar amino acids were hydrophobic (L, I, V, M, F, Y, and W), basic (R and K), acidic (D and E), polar (N and Q), or small and neutral (G, A, S, and T). The underlined amino acid sequences are those of the synthetic peptides used for fluorescence spectrometry.
FIG. 2.
Modular structure of Tio. The Tio molecule can be divided into two sections. The N-terminal third displays 36% amino acid identity to HVS StpC. This homology relies on richness in glycine and proline residues which are also common to collagen repeats. The perfect repetitive collagen-like structure of StpC (small boxes, 30% grey) was not found in Tio. Instead, individual collagen triplets are dispersed between positions 11 and 77 and connected by proline-rich sequences (60% grey boxes). The C-terminal two-thirds of Tio shows 33% identity to Tip. The most prominent feature here is the conservation of the SH3B domain (10% grey) and its functionally associated domain, CSKH (80% grey). The distance between these domains is also conserved. Other Tip-related regions of Tio include a serine-rich motif (S pattern box) and the region around a conserved tyrosine residue (checkerboard pattern box). The hydrophobic C terminus (black box) probably serves as a membrane anchor.
Further analysis of the left terminal nucleotide sequence of HVA suggested that a second reading frame (ORF2) encoding 61 amino acids may have homology to StpC. While StpC consists primarily of 18 consecutive collagen-like motifs (Gly-X-Y, where X or Y is Pro) (5), the deduced amino acid sequence of ORF2 displays a dispersed pattern of four collagen-like motifs that are flanked by multiple proline-rich sequences (Fig. 2 and 3). Within the nucleotide sequence of ORF2, we recognized a possible splice donor consensus sequence, and consequently, we found a possible splice acceptor site which lies within the 5′ untranslated region of ORF1. Computational translation of this region again revealed a high proline content and two additional collagen-like motifs.
Finally, we examined mRNA from HVA-transformed monkey T cells by reverse transcription and PCR. Analysis of the resulting cDNA clones uncovered a single, spliced mRNA where a 608-bp intron has been spliced out (Fig. 3). This mRNA encodes a 269-amino-acid protein that displays homology to StpC in the amino-terminal third of its amino acid sequence and to Tip in the C-terminal half of its amino acid sequence (Fig. 2 and 3); thus, we refer to it as the two-in-one (Tio) protein of herpesvirus ateles strain 73.
Identification of Tio in transformed monkey T-cell lines.
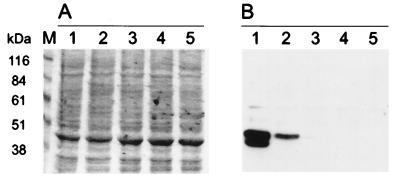
In order to identify the predicted protein in HVA-transformed monkey cell lines, an antiserum against a GST-Tio-fusion protein was raised in rabbits. Upon Western blot analysis, the antiserum specifically recognized a number of proteins in the 44- to 46-kDa range in lysates of cell line A661, which was generated by in vitro transformation of S. oedipus peripheral lymphocytes, and of cell line A1022, which was isolated from an S. oedipus marmoset infected with HVA strain 73. The main difference between these cell lines is in the numbers of genome equivalents they carry (28). While A661 cells harbor approximately 103 genome copies per diploid cell, A1022 cells have only 4 genome copies per cell, which might reflect differences in expression levels as detected by Western blotting (Fig. 4B, lanes 1 and 2). The appearance of Tio as at least a doublet of 43 and 46 kDa is thought to be related to posttranslational modifications. The specificity of the antiserum was confirmed by incubating anti-Tio serum with lysates of S. oedipus T cells which were transformed by an HVS subgroup A, B, or C strain (Fig. 4, lanes 3 to 5). Expression cloning of Tio and transfection into 293T cells gave rise to a protein pattern with the same electrophoretic mobility as the proteins identified in the transformed monkey T cells, confirming the identity of Tio (data not shown).
FIG. 4.
Expression of Tio in transformed monkey T cells. (A) Coomassie-stained SDS-PAGE gel loaded with 15 μg of total cell lysate of transformed S. oedipus T cells in each lane. Lanes: 1, cell line A661; 2, cell line A1022; 3 to 5, cell lines established with HVS subgroups A, B, and C, respectively. (B) Parallel gel transferred to a PVDF membrane filter and incubated with rabbit anti-Tio serum. Molecular size standards are given on the left.
Multimerization of Tio.
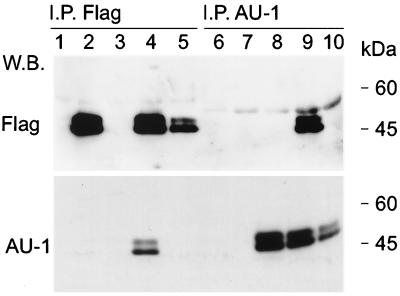
The viral protein was also expressed in 293T cells as a fusion protein with an N-terminal Flag epitope tag (Flag-Tio) which caused a mobility shift of Tio to a 45- and 47-kDa doublet. Flag-specific Western blot analyses of immunoprecipitated Flag-Tio often revealed an additional band twice the size of the Tio band which was sensitive to high concentrations of β-mercaptoethanol and extended boiling (data not shown). This band was not observed in control reactions and raised the question of whether Tio dimerizes or multimerizes when expressed in 293T cells. To assess the possibility of Tio homodimer formation, an additional expression plasmid was constructed with an AU1 epitope tag fused to the N terminus of Tio (AU-Tio). When Flag-Tio and AU-Tio were expressed in the same cells, the Flag and AU1 epitope-tagged proteins coprecipitated (Fig. 5, lanes 4 and 9). However, mixture of separately expressed Flag-Tio and AU-Tio proteins did not lead to coprecipitation of the respective proteins (Fig. 5, lanes 5 and 10). We concluded that Tio forms homodimers or even multimers in transfected 293T cells. The fact that Flag-Tio and AU-Tio did not aggregate in vitro argues against an artifact due to overexpression or gel overloading. Furthermore, this observation suggests that Tio homodimers or multimers are very stable and have a low exchange rate.
FIG. 5.
Tio forms homodimers. 293T cells were transfected with expression vectors (lanes 1 and 6), with an expression construct for Flag-Tio (lanes 2 and 7) or AU-Tio (lanes 3 and 8), or with a mixture of both Tio expression plasmids (lanes 4 and 9). In addition, lysates containing either Flag-Tio or AU-Tio were mixed prior to immunoprecipitation (I.P.) (lanes 5 and 10). Precipitations were performed with anti-Flag agarose (lanes 1 to 5) or with anti-AU1 antibody bound to protein A-Sepharose (lanes 6 to 10). Coprecipitated proteins and their controls were detected by Western blotting (W.B.) with anti-Flag (upper panel) or anti-AU1 (lower panel) antibodies. Molecular mass standards are given on the right.
Tio is phosphorylated on tyrosine when coexpressed with Lck, Src, or Fyn.
HVS-C488 Tip is known to be associated with Lck, and StpA of HVS strain A11 interacts with cellular Src. To compare the functional relatedness of Tio to Tip and StpA, we performed cotransfection experiments with 293T cells by employing Flag-Tio and Src family kinase Lck, Src, or Fyn. Crude lysates of transfected cells were analyzed by Western blotting using phosphotyrosine-specific, kinase-specific, or anti-Flag antibodies (Fig. 6). The kinases were expressed at comparable levels in the appropriate samples (Fig. 6B) and were phosphorylated on tyrosine (Fig. 6A, lanes 3 and 4). After coexpression of Tio, an additional phosphoprotein was detected in the presence of Lck and Src. A corresponding protein was not observed in the absence of Tio or when Tio was expressed alone or together with Fyn (Fig. 6A). The total amount of Flag-Tio detected by the epitope-specific antibody was approximately constant in all Tio-transfected samples, but in the presence of Lck or Src, a shift to the slower-migrating forms was observed (Fig. 6C, compare lanes 2 to lanes 4) which was most likely due to tyrosine phosphorylation (Fig. 6A, lanes 4).
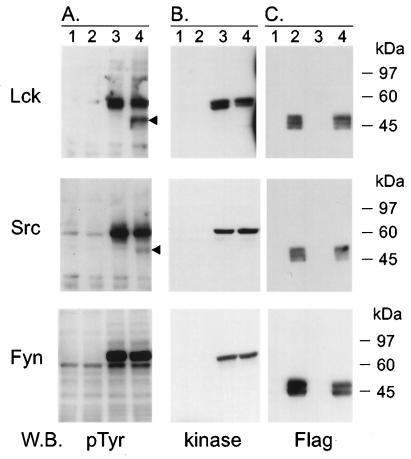
FIG. 6.
Tio is tyrosine phosphorylated by Src family kinases in vivo. 293T cells were transfected with expression vectors (lane 1) or an expression plasmid for Flag-Tio (lane 2), Lck, Src, or myc-tagged Fyn kinase (lane 3) or for Flag-Tio plus the respective kinase (lane 4). Twenty micrograms of total cell lysates was separated by SDS–9% PAGE and analyzed for tyrosine-phosphorylated proteins (A) by Western blotting (W.B.). Expression of the transfected plasmids was controlled by detection of each individual kinase with specific antibodies (B) or, for Flag-Tio, with epitope-specific antibodies (C). Arrowheads to the right of the phosphotyrosine blots indicate a protein detected only after cotransfection of Flag-Tio with Lck or Src. Molecular mass standards are given on the right.
While phosphorylation of Flag-Tio by Fyn was not detected in standard cell lysates (Fig. 6A), treatment of the transfected cells with orthovanadate prior to lysis revealed tyrosine phosphorylation of Tio by endogenous kinases with an increase after coexpression of Fyn. Antiphosphotyrosine Western blotting after Flag immunoprecipitation of untreated cell lysates confirmed tyrosine phosphorylation of Flag-Tio in the presence of Fyn. However, compared to that by Lck and Src, phosphorylation of Tio by Fyn was significantly lower (data not shown).
Thus, Tio was found to be tyrosine phosphorylated in vivo in the presence of Lck, Src, or Fyn. Two of these Src family kinases (Lck and Fyn) play an important role in T-cell signaling and might link Tio to lymphocyte growth regulation.
Association of Tio with Lck, Src, and Fyn in transfected 293T cells.
To investigate the direct association of Tio with the tyrosine kinases, cDNA expression constructs of Flag-Tio and of the Src kinases were cotransfected into 293T cells and coimmunoprecipitation assays were performed. Immune complexes from mock-transfected cells did not react with any of the antibodies used in these assays (Fig. 7, lanes 1 and 5). An anti-Flag monoclonal antibody directly precipitated Flag-Tio (Fig. 7, lanes 2) and was not cross-reactive with the tyrosine kinases (Fig. 7, lanes 3). On the other hand, antibodies directed against the recombinant kinases were specific (Fig. 7, lanes 7), as no binding to Flag-Tio was observed (Fig. 7, lanes 6).
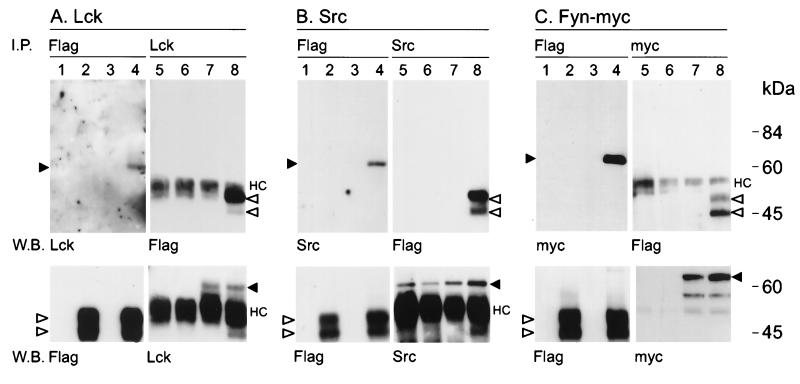
FIG. 7.
Tio coprecipitates with Src family kinases. 293T cells were transfected with expression vectors (lanes 1 and 5) or an expression plasmid for Flag-Tio (lanes 2 and 6), Lck, Src, or myc-tagged Fyn kinase (lanes 3 and 7), or Flag-Tio plus the respective kinase (lanes 4 and 8). Flag-Tio was immunoprecipitated (I.P.) by anti-Flag (lanes 1 to 4) antibody, and Western blotting (W.B.) was performed by using specific antibodies to detect coprecipitated Lck (A), Src (B), or myc-tagged Fyn (C). The reverse experiments were performed by using kinase-specific or anti-myc antibodies for immunoprecipitation (lanes 5 to 8) and anti-Flag antibody for Western blotting. The lower panel shows the results of control Western blotting after reprobing of the membranes with the antibodies used for immunoprecipitation. Open arrowheads, Flag-Tio; black arrowheads, Lck, Src, or Fyn-myc; HC, immunoglobulin heavy chains. Molecular mass standards are given on the right.
After coexpression of Flag-Tio with Lck, anti-Flag, as well as anti-Lck, immune complexes contained both Lck and Flag-Tio (Fig. 7A, lanes 4 and 8), indicating direct binding of Tio to Lck. An analogous experiment was performed with expression constructs for Flag-Tio and c-Src (Fig. 7B). Exogenous Src associated with Tio and vice versa (Fig. 7B, lanes 4 and 8). In contrast, endogenous Src was detectable in Src immunoprecipitates (Fig. 7B, lanes 5 and 6), but Flag-Tio was not coprecipitated (Fig. 7B, lane 6). Furthermore, Flag-Tio did not coprecipitate endogenous Src (Fig. 7B, lane 2). This is consistent with the finding that no tyrosine phosphorylation of Tio by endogenous Src was detected (Fig. 6). The third tyrosine kinase tested was Fyn. To facilitate the experimental procedure, myc epitope-tagged Fyn was used. Coprecipitation analysis revealed that Tio associates with Fyn and vice versa (Fig. 7C). Remarkably, Fyn appeared to favor association with a 45-kDa Flag-Tio phosphoprotein while Lck and Src associated preferentially with a 47-kDa Flag-Tio phosphoprotein. This observation may be due to differential phosphorylation by Lck, Src, or Fyn. Alternatively, the individual kinases may display different affinities for the Tio variants.
Tio interacts with the SH3 domains of Src family tyrosine kinases.
Sequence analysis and comparison with Tip suggested that Tio interacts with Src family kinases through their SH3 domains and that this interaction is mediated by its class II SH3B motif. To test this hypothesis, we performed GST affinity chromatography experiments. A GST-Lck-SH3 fusion protein and Flag-Tio were purified from E. coli. GST-Lck-SH3 specifically bound Flag-Tio as monitored by Flag Western blotting. In a reverse assay, Flag-Tio coimmunoprecipitated GST-Lck-SH3 (data not shown). These experiments confirmed that Tio is comparable to HVS Tip with respect to Lck-SH3 interaction.
To investigate the direct interaction of Tio with other Src family tyrosine kinases, a synthetic peptide corresponding to the most obvious candidate SH3B motif of Tio was synthesized and its affinity to SH3 domains was measured by fluorescence spectrometry. This assay revealed that the predicted SH3B domain of Tio is sufficient to bind directly to the SH3 domains of all of the Src family members tested (Table 1). Surprisingly, the affinity of the peptide for the Lyn and Hck SH3 domains was high, while it was moderate for the Lck SH3 domain and relatively low for the Fyn, Src, and Yes SH3 domains. The SH3 domains of Grb2, Abl, and phosphatidylinositol 3-kinase (PI3K) subunit p85-alpha did not bind to the Tio SH3B peptide. No binding to any of the SH3 domains tested was observed with a synthetic peptide from the proline-rich N terminus of Tio. This indicates that the interaction of the class II SH3B motif of Tio is specific for Src family kinases.
TABLE 1.
Measurement of the affinity of proline-rich Tio peptides for GST-SH3 domains
| GST-SH3 domain |
Kd (μM) of:
|
||
|---|---|---|---|
| Tio-C20a
|
Tio-N20b | ||
| Expt 1 | Expt 2 | ||
| Lyn | 5.88 ± 0.17 | 5.75 ± 0.34 | NBc |
| Hck | 9.30 ± 0.36 | 8.75 ± 0.16 | NB |
| Lck | 25.67 ± 0.73 | 26.24 ± 0.43 | NB |
| Fyn | 41.36 ± 1.33 | 39.41 ± 2.15 | NB |
| Src | 45.03 ± 2.73 | 44.45 ± 0.75 | NB |
| Yes | 76.46 ± 1.39 | 72.05 ± 2.12 | NB |
| Grb2 | NB | NB | NB |
| Abl | NB | NB | NB |
| PI3K | NB | NB | NB |
Corresponding to amino acids 183 to 202 of Tio. Sequence: PPGIPPPQLPPRPANLGQKQ (molecular weight, 2,101).
Corresponding to amino acids 26 to 45 of Tio. Sequence: PPNIPQDPTPGTPPGPINSK (molecular weight, 2,023).
NB, no binding.
Tyrosine-phosphorylated Tio binds directly to the SH2 domains of Lck, Src, and Fyn.
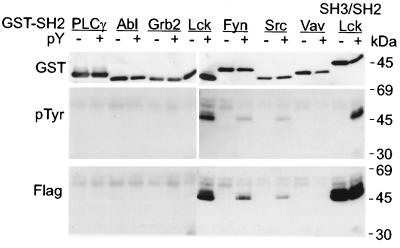
The direct interaction of Tio with the SH3 domains of Src kinases and the tyrosine phosphorylation of Tio in vivo raised the possibility that binding to the SH2 domains of these kinases might be allowed. To determine the capability of Tio to bind to SH2 domains, tyrosine-phosphorylated or nonphosphorylated Flag-Tio was incubated with various GST-SH2 fusion proteins. The resulting complexes were purified by GST affinity chromatography, resolved by SDS-PAGE, and analyzed for the presence of Tio bound to GST-SH2 domains (Fig. 8). No binding of nonphosphorylated Tio to SH2 domains was observed, but complex formation of phosphorylated Tio with the SH2 domains of Lck, Src, and Fyn was detected. A GST fusion protein containing the SH2 and SH3 domains of Lck served as a positive control, demonstrating phosphorylation-independent binding of Tio to the Lck SH3 domain. Similar to the SH3 interaction, SH2 binding seemed to be specific for Src family kinases, as no association of phosphorylated Tio was observed with the SH2 domain of phospholipase C-γ, Abl, Grb2, or Vav.
FIG. 8.
Tyrosine-phosphorylated Tio binds to GST-SH2 domains of Src family kinases. Flag-tagged Tio fusion proteins were expressed in bacteria with or without an active tyrosine kinase. Both phosphorylated (pY+) and unphosphorylated (pY−) Tio proteins were purified and tested for the ability to bind to GST-SH2 domains derived from the proteins given at the top. Western blotting was performed to control the amount of input GST fusion proteins (GST) and to detect tyrosine-phosphorylated (pTyr), as well as total (Flag), Flag-Tio bound by the GST-SH2 proteins. A GST-SH3/SH2 fusion protein of Lck was used to analyze the binding capacity of the unphosphorylated Tio preparation. Molecular mass standards are given on the right.
In conclusion, our experiments demonstrate that Tio can bind to Src family kinases via their SH3 domains and, upon tyrosine phosphorylation, via their SH2 domains. Thus, Tio combines properties of Tip, which binds directly to the Lck SH3 domain, and StpA, which interacts with Src kinases in a phosphotyrosine dependent manner.
DISCUSSION
We have identified Tio as a protein encoded by the putative transformation-relevant region of HVA strain 73. Expression of Tio in virus-transformed monkey T cells hints at a functional role for this viral protein in lymphocyte growth transformation. Its potential to form dimers or multimers and its association with nonreceptor tyrosine kinases of the Src family are reminiscent of oncoproteins of other DNA tumor viruses (16).
Homodimerization of viral effector proteins was shown to be essential for the oncogenic properties of several tumor viruses. A prominent example is the bovine papillomavirus (BPV). BPV oncoprotein E5 acts as a disulfide-linked homodimer which induces dimerization of platelet-derived growth factor receptor β in the absence of ligand (16). Autophosphorylation of dimerized platelet-derived growth factor receptor β augments intrinsic kinase activity (35). Further tyrosine phosphorylation generates specific binding sites for cellular signaling molecules containing SH2 or phosphotyrosine-binding (PTB) domains and initiates a sustained mitogenic signal through activation of the ras (32), mitogen-activated protein kinase (33), and PI3K pathways (9, 32). Finally, BPV E5 expression results in transformation of cultured fibroblasts (16). Dimeric protein gp55 of spleen focus-forming virus appears to use a similar mechanism of cellular transformation. This protein activates the erythropoietin receptor and is responsible for the induction of erythroleukemia by spleen focus-forming virus (16).
Polyomavirus middle T antigen (mT) is also able to form dimers or multimers, but this property is not necessary for the transforming phenotype (68). Like Tio, mT associates with Src kinase family members (reviewed in reference 54), namely, with Src (11), Yes (45), and Fyn (10, 36). The interaction has been mapped to the kinase domain of Src (22) and results in increased Src (7) and Yes (45) kinase activities. Tyrosine phosphorylation of mT generates binding sites for SH2 and PTB domains and results in the recruitment of PI3K (74), phospholipase C-γ1 (71), and the adapter molecule Shc (15). Furthermore, mT associates with 14-3-3 proteins (61) and with protein phosphatase 2A (62). In addition, proline-rich sequences of mT might serve as interaction sites for SH3 domains. These properties of mT suggest that the viral oncoprotein leads to transformation of rodent cells by inducing the constitutive dimerization-independent formation of signaling complexes.
Association with tyrosine kinases has also been described for transformation-related proteins of other gammaherpesviruses and was the basis of our experiments. The Tio-related protein Tip of HVS strain C488 binds to the T-cell-specific tyrosine kinase Lck (6). This interaction has been mapped to a proline-rich region of the viral protein and to the SH3 domain of the kinase (40, 41). However, mutation of the SH3B site of Tip enhanced the transforming phenotype of HVS strain C488 (18), indicating that SH3-mediated Lck interaction of Tip is not essential for T-cell growth transformation. The absolute requirement for Tip in this system (19) suggests that Tip employs other mechanisms to promote T-cell transformation. Association of Tio with Src also hints at a functional relationship with StpA of HVS strain A11. This protein is required for the transforming phenotype of the virus (57) and is oncogenic by itself when expressed in rodent fibroblasts or in transgenic mice (42, 46). While association of StpA with Src was found to be mediated by an SH2-binding site (48), the significance of this interaction for transformation has not been analyzed so far. Finally, Epstein-Barr virus latent membrane protein 2A (LMP2A) associates with several tyrosine kinases expressed in transformed B cells. LMP2A interacts with the B-cell-specific kinase Lyn via the SH2 domain, and its binding to Syk kinase depends on phosphorylation of an immuno tyrosine-based activation motif (29, 30). LMP2A appears not to be required for B-cell transformation but is thought to maintain latency by downregulation of Lyn and prevent Epstein-Barr virus from reactivation by blocking B-cell receptor signaling (49, 55). However, expression in transgenic mice recently revealed novel B-cell-stimulatory effects of LMP2A (8).
In this context, our findings of Tio dimerization and association with Src family kinases support the hypothesis that Tio is the oncoprotein of HVA, which was initially based on sequence homologies. Through dimerization and/or simultaneous binding of SH3 and SH2 domains, Tio might assemble signaling complexes which finally initiate sustained mitogenic signals leading to T-cell transformation. Most critical in this context appears to be the involvement of the protein tyrosine kinases Lck and Fyn, which are key enzymes in T-cell activation. In contrast, signals mediated by Lyn, Hck, Src, or Yes in T cells are not described. Initial binding of Tio to SH3 domains of tyrosine kinases enables Tio phosphorylation and subsequent binding to SH2 or PTB domains. Thereby, Tio might serve as an adapter either among Src family kinases or between these kinases and other signaling molecules. Besides SH3- and phosphotyrosine-dependent interactions, sequence homologies suggest that Tio also might recruit cellular factors known to associate with the HVS oncoprotein StpC. These proteins, the GTPase Ras and the NFκB-inducing tumor necrosis factor receptor-associated factors (reviewed in reference 37), could link Tio to additional growth-promoting cellular pathways.
Our experiments demonstrate that the HVA protein Tio is expressed in virus-transformed T lymphocytes and has the potential to interfere with cellular growth regulation. Further analyses are necessary to determine the transforming properties of Tio and the composition of the Tio complexes and to identify the signaling pathways leading to T-cell transformation by HVA.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 466 (B2), and a grant of the Wilhelm-Sander Stiftung to S.M.F.
We thank F. Friedrich for technical assistance and S. M. Lang for providing 9E10 hybridoma supernatants and for critical reading of the manuscript.
REFERENCES
- 1.Albrecht, J. C., and B. Fleckenstein. Unpublished data. GenBank accession no. AF083424.
- 2.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biesinger, B. Unpublished data.
- 4.Biesinger B, Muller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus samiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biesinger B, Trimble J J, Desrosiers R C, Fleckenstein B. The divergence between two oncogenic Herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology. 1990;176:505–514. doi: 10.1016/0042-6822(90)90020-r. [DOI] [PubMed] [Google Scholar]
- 6.Biesinger B, Tsygankov A Y, Fickenscher H, Emmrich F, Fleckenstein B, Bolen J B, Broker B M. The product of the Herpesvirus saimiri open reading frame 1 (tip) interacts with T cell-specific kinase p56lck in transformed cells. J Biol Chem. 1995;270:4729–4734. doi: 10.1074/jbc.270.9.4729. [DOI] [PubMed] [Google Scholar]
- 7.Bolen J B, Thiele C J, Israel M A, Yonemoto W, Lipsich L A, Brugge J S. Enhancement of cellular src gene product associated tyrosyl kinase activity following polyoma virus infection and transformation. Cell. 1984;38:767–777. doi: 10.1016/0092-8674(84)90272-1. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell R G, Wilson J B, Anderson S J, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 9.Cantley L C, Auger K R, Carpenter C, Duckworth B, Graziani A, Kapeller R, Soltoff S. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. . (Erratum, 65:914–915, 1991.) [DOI] [PubMed] [Google Scholar]
- 10.Cheng S H, Harvey R, Espino P C, Semba K, Yamamoto T, Toyoshima K, Smith A E. Peptide antibodies to the human c-fyn gene product demonstrate pp59c-fyn is capable of complex formation with the middle-T antigen of polyomavirus. EMBO J. 1988;7:3845–3855. doi: 10.1002/j.1460-2075.1988.tb03270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtneidge S A, Smith A E. Polyoma virus transforming protein associates with the product of the c-src cellular gene. Nature. 1983;303:435–439. doi: 10.1038/303435a0. [DOI] [PubMed] [Google Scholar]
- 12.Daniel M D, Melendez L V, Hunt R D, King N W, Anver M, Fraser C E, Barahona H, Baggs R B. Herpesvirus saimiri. VII. Induction of malignant lymphoma in New Zealand White rabbits. J Natl Cancer Inst. 1974;53:1803–1807. [PubMed] [Google Scholar]
- 13.Desrosiers R C, Bakker A, Kamine J, Falk L A, Hunt R D, King N W. A region of the Herpesvirus saimiri genome required for oncogenicity. Science. 1985;228:184–187. doi: 10.1126/science.2983431. [DOI] [PubMed] [Google Scholar]
- 14.Desrosiers R C, Silva D P, Waldron L M, Letvin N L. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986;57:701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dilworth S M, Brewster C E, Jones M D, Lanfrancone L, Pelicci G, Pelicci P G. Transformation by polyoma virus middle T-antigen involves the binding and tyrosine phosphorylation of Shc. Nature. 1994;367:87–90. doi: 10.1038/367087a0. [DOI] [PubMed] [Google Scholar]
- 16.DiMaio D, Lai C C, Klein O. Virocrine transformation: the intersection between viral transforming proteins and cellular signal transduction pathways. Annu Rev Microbiol. 1998;52:397–421. doi: 10.1146/annurev.micro.52.1.397. [DOI] [PubMed] [Google Scholar]
- 17.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilyinskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 18.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duboise S M, Lee H, Guo J, Choi J K, Czajak S, Simon M, Desrosiers R C, Jung J U. Mutation of the Lck-binding motif of Tip enhances lymphoid cell activation by herpesvirus saimiri. J Virol. 1998;72:2607–2614. doi: 10.1128/jvi.72.4.2607-2614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunant N M, Messerschmitt A S, Ballmer-Hofer K. Functional interaction between the SH2 domain of Fyn and tyrosine 324 of hamster polyomavirus middle-T antigen. J Virol. 1997;71:199–206. doi: 10.1128/jvi.71.1.199-206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunant N M, Senften M, Ballmer-Hofer K. Polyomavirus middle-T antigen associates with the kinase domain of Src-related tyrosine kinases. J Virol. 1996;70:1323–1330. doi: 10.1128/jvi.70.3.1323-1330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ensser A, Pflanz R, Fleckenstein B. Primary structure of the alcelaphine herpesvirus 1 genome. J Virol. 1997;71:6517–6525. doi: 10.1128/jvi.71.9.6517-6525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falk L A, Nigida S M, Deinhardt F, Wolfe L G, Cooper R W, Hernandez-Camacho J I. Herpesvirus ateles: properties of an oncogenic herpesvirus isolated from circulating lymphocytes of spider monkeys (Ateles sp.) Int J Cancer. 1974;14:473–482. doi: 10.1002/ijc.2910140407. [DOI] [PubMed] [Google Scholar]
- 25.Feng S, Kapoor T M, Shirai F, Combs A P, Schreiber S L. Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis. Chem Biol. 1996;3:661–670. doi: 10.1016/s1074-5521(96)90134-9. [DOI] [PubMed] [Google Scholar]
- 26.Feng S, Kasahara C, Rickles R J, Schreiber S L. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc Natl Acad Sci USA. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fickenscher H, Fleckenstein B. Generation of human T-cell lines using lymphotropic herpesviruses. Methods Mol Genet. 1994;4:345–362. [Google Scholar]
- 28.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and herpesvirus ateles. In: Roizman B, editor. The herpesviruses. Vol. 1. New York, N.Y: Plenum Press; 1982. pp. 253–332. [Google Scholar]
- 29.Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology. 1997;235:241–251. doi: 10.1006/viro.1997.8690. [DOI] [PubMed] [Google Scholar]
- 30.Fruehling S, Swart R, Dolwick K M, Kremmer E, Longnecker R. Tyrosine 112 of latent membrane protein 2A is essential for protein tyrosine kinase loading and regulation of Epstein-Barr virus latency. J Virol. 1998;72:7796–7806. doi: 10.1128/jvi.72.10.7796-7806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geck P, Whitaker S A, Medveczky M M, Medveczky P G. Expression of collagenlike sequences by a tumor virus, herpesvirus saimiri. J Virol. 1990;64:3509–3515. doi: 10.1128/jvi.64.7.3509-3515.1990. . (Erratum, 65:7084, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghai J, Ostrow R S, Tolar J, McGlennen R C, Lemke T D, Tobolt D, Liu Z, Faras A J. The E5 gene product of rhesus papillomavirus is an activator of endogenous Ras and phosphatidylinositol-3′-kinase in NIH 3T3 cells. Proc Natl Acad Sci USA. 1996;93:12879–12884. doi: 10.1073/pnas.93.23.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu Z, Matlashewski G. Effect of human papillomavirus type 16 oncogenes on MAP kinase activity. J Virol. 1995;69:8051–8056. doi: 10.1128/jvi.69.12.8051-8056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J, Williams K, Duboise S M, Alexander L, Veazey R, Jung J U. Substitution of ras for the herpesvirus saimiri STP oncogene in lymphocyte transformation. J Virol. 1998;72:3698–3704. doi: 10.1128/jvi.72.5.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heldin C H. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 36.Horak I D, Kawakami T, Gregory F, Robbins K C, Bolen J B. Association of p60fyn with middle tumor antigen in murine polyomavirus-transformed rat cells. J Virol. 1989;63:2343–2347. doi: 10.1128/jvi.63.5.2343-2347.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung, J. U., J. K. Choi, A. Ensser, and B. Biesinger. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin. Cancer Biol., in press. [DOI] [PubMed]
- 38.Jung J U, Desrosiers R C. Identification and characterization of the herpesvirus saimiri oncoprotein STP-C488. J Virol. 1991;65:6953–6960. doi: 10.1128/jvi.65.12.6953-6960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung J U, Desrosiers R C. Association of the viral oncoprotein STP-C488 with cellular ras. Mol Cell Biol. 1995;15:6506–6512. doi: 10.1128/mcb.15.12.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung J U, Lang S M, Friedrich U, Jun T, Roberts T M, Desrosiers R C, Biesinger B. Identification of Lck-binding elements in tip of herpesvirus saimiri. J Biol Chem. 1995;270:20660–20667. doi: 10.1074/jbc.270.35.20660. [DOI] [PubMed] [Google Scholar]
- 41.Jung J U, Lang S M, Jun T, Roberts T M, Veillette A, Desrosiers R C. Downregulation of Lck-mediated signal transduction by tip of herpesvirus saimiri. J Virol. 1995;69:7814–7822. doi: 10.1128/jvi.69.12.7814-7822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of Herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamine J, Bakker A, Desrosiers R C. Mapping of RNA transcribed from a region of the herpesvirus saimiri genome required for oncogenicity. J Virol. 1984;52:532–540. doi: 10.1128/jvi.52.2.532-540.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koomey J M, Mulder C, Burghoff R L, Fleckenstein B, Desrosiers R C. Deletion of DNA sequence in a nononcogenic variant of herpesvirus saimiri. J Virol. 1984;50:662–665. doi: 10.1128/jvi.50.2.662-665.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornbluth S, Sudol M, Hanafusa H. Association of the polyomavirus middle-T antigen with c-yes protein. Nature. 1987;325:171–173. doi: 10.1038/325171a0. [DOI] [PubMed] [Google Scholar]
- 46.Kretschmer C, Murphy C, Biesinger B, Beckers J, Fickenscher H, Kirchner T, Fleckenstein B, Ruther U. A Herpes saimiri oncogene causing peripheral T-cell lymphoma in transgenic mice. Oncogene. 1996;12:1609–1616. [PubMed] [Google Scholar]
- 47.Larose L, Gish G, Shoelson S, Pawson T. Identification of residues in the beta platelet-derived growth factor receptor that confer specificity for binding to phospholipase C-gamma 1. Oncogene. 1993;8:2493–2499. [PubMed] [Google Scholar]
- 48.Lee H, Trimble J J, Yoon D-W, Regier D, Desrosiers R C, Jung J U. Genetic variation of herpesvirus saimiri subgroup A transforming protein and its association with cellular src. J Virol. 1997;71:3817–3825. doi: 10.1128/jvi.71.5.3817-3825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longnecker R, Miller C L. Regulation of Epstein-Barr virus latency by latent membrane protein 2. Trends Microbiol. 1996;4:38–42. doi: 10.1016/0966-842x(96)81504-6. [DOI] [PubMed] [Google Scholar]
- 50.Lund T, Medveczky M M, Medveczky P G. Herpesvirus saimiri Tip-484 membrane protein markedly increases p56lck activity in T cells. J Virol. 1997;71:378–382. doi: 10.1128/jvi.71.1.378-382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lund T, Medveczky M M, Neame P J, Medveczky P G. A herpesvirus saimiri membrane protein required for interleukin-2 independence forms a stable complex with p56lck. J Virol. 1996;70:600–606. doi: 10.1128/jvi.70.1.600-606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medveczky M M, Geck P, Sullivan J L, Serbousek D, Djeu J Y, Medveczky P G. IL-2 independent growth and cytotoxicity of herpesvirus saimiri-infected human CD8 cells and involvement of two open reading frame sequences of the virus. Virology. 1993;196:402–412. doi: 10.1006/viro.1993.1495. [DOI] [PubMed] [Google Scholar]
- 53.Medveczky P, Szomolanyi E, Desrosiers R C, Mulder C. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J Virol. 1984;52:938–944. doi: 10.1128/jvi.52.3.938-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Messerschmitt A S, Dunant N, Ballmer-Hofer K. DNA tumor viruses and Src family tyrosine kinases, an intimate relationship. Virology. 1997;227:271–280. doi: 10.1006/viro.1996.8316. [DOI] [PubMed] [Google Scholar]
- 55.Miller C L, Burkhardt A L, Lee J H, Stealey B, Longnecker R, Bolen J B, Kieff E. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity. 1995;2:155–166. doi: 10.1016/s1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 56.Murphy C, Kretschmer C, Biesinger B, Beckers J, Jung J, Desrosiers R C, Muller-Hermelink H K, Fleckenstein B W, Ruther U. Epithelial tumours induced by a herpesvirus oncogene in transgenic mice. Oncogene. 1994;9:221–226. [PubMed] [Google Scholar]
- 57.Murthy S C, Trimble J J, Desrosiers R C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noraz N, Saha K, Ottones F, Smith S, Taylor N. Constitutive activation of TCR signaling molecules in IL-2-independent Herpesvirus saimiri-transformed T cells. J Immunol. 1998;160:2042–2045. [PubMed] [Google Scholar]
- 60.Oehrl W, Kardinal C, Ruf S, Adermann K, Groffen J, Feng G S, Blenis J, Tan T H, Feller S M. The germinal center kinase (GCK)-related protein kinases HPK1 and KHS are candidates for highly selective signal transducers of Crk family adapter proteins. Oncogene. 1998;17:1893–1901. doi: 10.1038/sj.onc.1202108. [DOI] [PubMed] [Google Scholar]
- 61.Pallas D C, Fu H, Haehnel L C, Weller W, Collier R J, Roberts T M. Association of polyomavirus middle tumor antigen with 14-3-3 proteins. Science. 1994;265:535–537. doi: 10.1126/science.8036498. [DOI] [PubMed] [Google Scholar]
- 62.Pallas D C, Shahrik L K, Martin B L, Jaspers S, Miller T B, Brautigan D L, Roberts T M. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 63.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Posern G, Zheng J, Knudsen B S, Kardinal C, Muller K B, Voss J, Shishido T, Cowburn D, Cheng G, Wang B, Kruh G D, Burrell S K, Jacobson C A, Lenz D M, Zamborelli T J, Adermann K, Hanafusa H, Feller S M. Development of highly selective SH3 binding peptides for Crk and CRKL which disrupt Crk-complexes with DOCK180, SoS and C3G. Oncogene. 1998;16:1903–1912. doi: 10.1038/sj.onc.1201714. [DOI] [PubMed] [Google Scholar]
- 65.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 66.Richter J, Puchtler I, Fleckenstein B. Thymidylate synthase gene of herpesvirus ateles. J Virol. 1988;62:3530–3535. doi: 10.1128/jvi.62.9.3530-3535.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Senften M, Dilworth S, Ballmer-Hofer K. Multimerization of polyomavirus middle-T antigen. J Virol. 1997;71:6990–6995. doi: 10.1128/jvi.71.9.6990-6995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 70.Songyang Z, Shoelson S E, McGlade J, Olivier P, Pawson T, Bustelo X R, Barbacid M, Sabe H, Hanafusa H, Yi T. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su W, Liu W, Schaffhausen B S, Roberts T M. Association of polyomavirus middle tumor antigen with phospholipase C-gamma 1. J Biol Chem. 1995;270:12331–12334. doi: 10.1074/jbc.270.21.12331. [DOI] [PubMed] [Google Scholar]
- 72.Szomolanyi E, Medveczky P, Mulder C. In vitro immortalization of marmoset cells with three subgroups of herpesvirus saimiri. J Virol. 1987;61:3485–3490. doi: 10.1128/jvi.61.11.3485-3490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whitman M, Kaplan D R, Schaffhausen B, Cantley L, Roberts T M. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 75.Wiese N, Tsygankov A Y, Klauenberg U, Bolen J B, Fleischer B, Broker B M. Selective activation of T cell kinase p561ck by Herpesvirus saimiri protein tip. J Biol Chem. 1996;271:847–852. doi: 10.1074/jbc.271.2.847. [DOI] [PubMed] [Google Scholar]
- 76.Xu W, Harrison S C, Eck M J. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 77.Yoon D W, Lee H, Seol W, DeMaria M, Rosenzweig M, Jung J U. Tap: a novel cellular protein that interacts with tip of herpesvirus saimiri and induces lymphocyte aggregation. Immunity. 1997;6:571–582. doi: 10.1016/s1074-7613(00)80345-3. [DOI] [PubMed] [Google Scholar]