Abstract
We have developed a reconstituted system which models the events associated with human immunodeficiency virus type 1 (HIV-1) plus-strand transfer. These events include synthesis of plus-strand strong-stop DNA [(+) SSDNA] from a minus-strand DNA donor template covalently attached to human tRNA3Lys, tRNA primer removal, and annealing of (+) SSDNA to the minus-strand DNA acceptor template. Termination of (+) SSDNA synthesis at the methyl A (nucleotide 58) near the 3′ end of tRNA3Lys reconstitutes the 18-nucleotide primer binding site (PBS). Analysis of (+) SSDNA synthesis in vitro and in HIV-1 endogenous reactions indicated another major termination site: the pseudouridine at nucleotide 55. In certain HIV-1 strains, complementarity between nucleotides 56 to 58 and the first three bases downstream of the PBS could allow all of the (+) SSDNA products to be productively transferred. Undermodification of the tRNA may be responsible for termination beyond the methyl A. In studies of tRNA removal, we find that initial cleavage of the 3′ rA by RNase H is not sufficient to achieve successful strand transfer. The RNA-DNA hybrid formed by the penultimate 17 bases of tRNA still annealed to (+) SSDNA must also be destabilized. This can occur by removal of additional 3′-terminal bases by RNase H (added either in cis or trans). Alternatively, the nucleic acid chaperone activity of nucleocapsid protein (NC) can catalyze this destabilization. NC stimulates annealing of the complementary PBS sequences in (+) SSDNA and the acceptor DNA template. Reverse transcriptase also promotes annealing but to a lesser extent than NC.
During retrovirus replication, a linear, double-stranded DNA copy of the viral RNA genome is synthesized in a complex series of steps catalyzed by the virus-encoded enzyme reverse transcriptase (RT) (25; for reviews, see references 3 and 12). Two strand transfer reactions, minus- and plus-strand DNA transfer, are required for elongation of each strand and for synthesis of the long terminal repeat present at the ends of proviral DNA (reviewed in reference 68).
Reverse transcription is initiated by a cellular tRNA primer which is bound to the primer binding site (PBS) located near the 5′ end of viral RNA (12). The first product formed, a short DNA termed minus-strand strong-stop DNA [(−) SSDNA], is translocated to the 3′ end of the genome (either by intra- or intermolecular strand transfer) in a reaction facilitated by base pairing of the repeat (R) region at the 3′ terminus of viral RNA and its complement in (−) SSDNA (reviewed in reference 68). RNase H degradation of the viral genome (10), including removal of the 5′-terminal 14 to 18 bases by the polymerase-independent mode of RNase H cleavage (22, 26, 55) (what we have previously termed 3′-OH-independent RNase H activity [55]), frees (−) SSDNA for the annealing reaction (5, 11, 18, 24, 54).
In addition to work demonstrating the important role of RNase H, many studies have shown that human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein (NC) dramatically stimulates HIV-1 minus-strand transfer (2, 15, 27, 41, 53, 60, 75). NC is able to exert this effect because it exhibits nucleic acid chaperone activity (32, 59), i.e., it is able to catalyze conformational rearrangements of nucleic acids which result in more thermodynamically stable structures having the maximal number of base pairs (6, 16, 17, 27, 33, 37, 40, 46, 51, 69, 73, 75; also see reference 13 and references therein). Thus, during HIV-1 minus-strand transfer, NC transiently destabilizes the complementary TAR structures at the 3′ ends of (−) SSDNA and viral RNA, thereby preventing self-priming reactions (27, 45, 47) and promoting annealing (28, 44, 46, 75).
Interestingly, the strategy RT uses to make plus-strand DNA has many similarities to the strategy employed for synthesis of minus-strand DNA. For example, RT also initiates plus-strand DNA synthesis with an RNA primer, in this case, a purine-rich sequence known as the polypurine tract (PPT) (for references before 1993, see reference 10; also see references 21, 56, and 58). Plus-strand priming occurs while minus-strand DNA is being elongated; the template is minus-strand DNA which has the tRNA primer (for HIV-1, tRNA3Lys) still attached to its 5′ end. The initial product is a short DNA termed plus-strand SSDNA [(+) SSDNA]. Synthesis of (+) SSDNA continues until the 18 nucleotides (nt) at the 3′ end of the tRNA are copied, which reconstitutes the PBS.
Since the 19th base from the 3′ end of all retroviral tRNA primers is a methyl A (reviewed in reference 49) and RT-catalyzed elongation is blocked at modified bases (see the review by Marquet [49] and references therein), current models for reverse transcription specify that synthesis of (+) SSDNA terminates when RT encounters the first modified base in the tRNA (25). For example, the murine leukemia virus (MuLV) (+) SSDNA intermediate generated in vivo stops precisely after RT has copied the first 18 nt at the 3′ end of tRNAPro (61). It was also reported that the methyl A (nt 58) of tRNA3Lys is the major termination site for synthesis of HIV-1 (+) SSDNA synthesis in vitro (8). However, another study with an HIV-1 reconstituted system showed that (+) SSDNA synthesis is not terminated exclusively at the methyl A position (4). If tRNA3Lys is completely unmodified, the entire molecule is reverse transcribed and plus-strand transfer will not take place (4, 8, 76).
Before the actual plus-strand transfer event can occur, RNase H cleavage is again required. Here, the tRNA must be removed to allow (+) SSDNA to anneal to the 3′ end of minus-strand DNA, a step mediated by base pairing of the 18-nt complementary PBS sequences. Analysis of the sequences at HIV-1 circle junctions (34, 43, 65, 70) and characterization of in vitro HIV-1 RNase H reactions (23, 57, 66) indicate that the initial cleavage is not at the RNA-DNA junction but instead occurs between the 3′-terminal rA and penultimate rC in tRNA3Lys, leaving the rA at the 5′ end of (−) SSDNA. Recently, similar observations have been reported for the initial cleavage of the MuLV primer, tRNAPro, during MuLV plus-strand synthesis; in this case, the rA is ultimately removed from the 5′ end of minus-strand DNA (63, 64).
In the studies of HIV-1 RNase H activity, in vitro secondary cleavage of an 18-nt RNA oligonucleotide representing the 3′-terminal sequences of tRNA3Lys was observed (11, 66). However, the question of whether this event is obligatory for complete removal of the primer was not addressed. In addition, a direct role for NC in HIV-1 plus-strand transfer has not been demonstrated; in one study, it was reported that mutations in the N-terminal zinc finger of HIV-1 NC did not affect plus-strand transfer in vivo (67).
In this work, we have investigated three aspects of plus-strand transfer: (i) termination of (+) SSDNA synthesis, (ii) tRNA primer removal, and (iii) a possible role for NC. To clarify these issues, we have developed a reconstituted system which models the events associated with plus-strand transfer and includes the use of a donor DNA template that contains human tRNA3Lys. Analysis of (+) SSDNA synthesis in this system and in endogenous RT assays with detergent-treated HIV-1 virions clearly demonstrates that position 58 in the tRNA3Lys primer is not an exclusive termination site: there are additional (+) SSDNA products which terminate at or near two other modified bases in the tRNA. Moreover, we find that in certain HIV-1 strains, products terminated before the second modified base (a pseudouridine [Ψ]) at the 3′ end of the tRNA (48) may be productively transferred. We also show that initial cleavage of rA from the tRNA is not sufficient for successful strand transfer. The RNA-DNA hybrid formed by the penultimate 17 tRNA bases still annealed to (+) SSDNA has a high melting temperature (Tm) and must therefore be destabilized to allow complete removal of the tRNA primer. This can be accomplished by RNase H-catalyzed secondary cleavage to remove additional bases from the 3′ end of the tRNA or by the nucleic acid chaperone activity of NC. Finally, as is the case for minus-strand transfer (14, 28, 44, 46, 75), we demonstrate that HIV-1 NC stimulates the annealing reaction in plus-strand transfer.
MATERIALS AND METHODS
Materials.
Purified tRNA3Lys from human placenta was obtained from Bio S&T (Lachine, Quebec, Canada). DNA oligonucleotides were purchased from Lofstrand (Gaithersburg, Md.) or from Oligo’s Etc., Inc. (Wilsonville, Oreg.). RNA and chimeric DNA-RNA oligonucleotides were obtained from Oligo’s Etc., Inc. An RNase H-minus HIV-1 RT having a point mutation changing residue E478 to Q (62) was a gift from Stuart Le Grice (HIV Drug Resistance Program, NCI-Frederick Cancer Research and Development Center, Frederick, Md.). Escherichia coli RNase H was kindly supplied by Robert Crouch (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Md.). The sources of all other materials are given in Guo et al. (27).
Preparation of the minus-strand DNA donor template containing tRNA3Lys.
A threefold excess of a 50-nt plus-strand DNA oligonucleotide template (JL278), representing (+) SSDNA and having the sequence 5′-CCC TTT TAG TCA GTG TGG AAA ATC TCT AGC AGT GGC GCC CGA ACA GGG AC-3′, nt 604 to 653 in NL4.3 (1), was annealed to tRNA3Lys. The annealing was carried out at 65°C for 5 min in buffer consisting of 50 mM Tris-HCl (pH 8.0) and 75 mM KCl; the mixture was then gradually cooled to 37°C for about 30 min. A 32-nt minus-strand donor DNA [corresponding to the 5′-terminal sequences of (−) SSDNA] was synthesized with T4 DNA polymerase (Boehringer Mannheim), with tRNA3Lys as the primer and the 32-nt unannealed portion of the 50-nt (+) SSDNA as the template. Following termination of the reaction, unextended tRNA3Lys (76 nt), unreacted 50-nt (+) SSDNA, and tRNA3Lys covalently attached to the DNA donor (108 nt) were separated by electrophoresis in a 6% sequencing gel. The 108-nt product was eluted from the gel, further purified with a GENECLEAN II kit (BIO 101), and stored at −80°C. For RNase H cleavage assays, the tRNA3Lys donor template was dephosphorylated with calf intestinal phosphatase (1 U/μg of tRNA3Lys; Boehringer Mannheim) at 50°C for 2 h, extracted several times with phenol-chloroform, precipitated with alcohol, and then labeled with 32P at its 5′ end, as described previously (29). Due to steric hindrance, the 5′ phosphorylation of tRNA3Lys was less efficient than the corresponding reaction with oligonucleotides.
RT assays. (i) In vitro strand transfer assay.
The assay was performed with either of two chimeric DNA-RNA donor templates: (i) a 32-nt minus-strand DNA oligonucleotide representing (−) SSDNA, covalently attached to tRNA3Lys (see above) and having the sequence 5′-CTG CTA GAG ATT TTC CAC ACT GAC TAA AAG GG-3′, complementary to nt 604 to 635 (1), and (ii) a synthetic 50-nt chimeric DNA-RNA oligonucleotide (JL237) containing the same 32-nt DNA sequence covalently attached to RNA consisting of the first 18 bases at the 3′ end of tRNA3Lys (5′-GUC CCU GUU CGG GCG CCA-3′, complementary to the PBS, nt 636 to 653 in NL4.3 [1]). DNA synthesis was initiated with a 20-nt DNA primer (JL239), 5′-CCC TTT TAG TCA GTG TGG AA-3′, nt 604 to 623 in NL4.3 (1), labeled at its 5′ end with 32P, as described previously (29). The donor DNA (0.2 pmol) was annealed to the 5′-end-labeled DNA primer (0.4 pmol, ∼2 × 106 cpm) at 65°C for 5 min, followed by slow cooling to 37°C for ∼30 min, as described by Guo et al. (27). A detailed description of the procedures used in the assay is given by Guo et al. (27), except that in the present work, the acceptor template (0.2 pmol) is a 48-nt minus-strand DNA (JL238): 5′-GAT CTC CTC TGG CTT TAC TTT CGC TTT CAA GTC CCT GTT CGG GCG CCA-3′, complementary to nt 636 to 683 in NL4.3 (1). The final concentration of the 20-nt DNA primer was 20 nM; the donor and acceptor templates as well as RT were present at final concentrations of 10 nM. Reactions (final volume, 20 μl) were initiated by addition of MgCl2 and the four deoxynucleoside triphosphates (dNTPs) and, unless specified otherwise, were incubated for 60 min at 37°C. Termination of reactions and electrophoresis of the DNA products in 6% sequencing gels were performed as previously described (27). Radioactive DNA products were quantified by using a PhosphorImager (Molecular Dynamics) and ImageQuant software; calculations were performed as indicated in reference 27.
(ii) RNase H cleavage assay.
Reactions were performed under the conditions of the in vitro strand transfer assay (see above), except that the donor templates containing either tRNA3Lys (see above) or the 18-nt RNA sequence (3′ end of tRNA3Lys) were labeled with 32P at their 5′ ends (29); the specific activities for the two donors were ∼4.8 × 103 cpm/0.2 pmol and ∼6.5 × 105 cpm/0.2 pmol, respectively. The 20-nt DNA primer was unlabeled. RNA cleavage products produced from the tRNA3Lys-containing donor or from the 18-nt RNA-containing donor were resolved by electrophoresis in a 6 or 12.5% sequencing gel, respectively, and were quantified as described above.
(iii) Assay for secondary RNase H cleavage during plus-strand transfer.
For this assay, we used oligonucleotides representing the intermediates which are present after the initial RNase H cleavage at the 3′ end of tRNA3Lys: (i) the 32-nt minus-strand DNA donor template with an rA attached at its 5′ end (JL354; 5′-rA CTG CTA GAG ATT TTC CAC ACT GAC TAA AAG GG, complementary to nt 604 to 636 in NL4.3 [1], (ii) a 17-nt RNA oligonucleotide representing the 17 remaining bases at the 3′ end of tRNA3Lys (JL353; 5′-GUC CCU GUU CGG GCG CC-3′, complementary to nt 637 to 653 in NL4.3 [1]), and (iii) a 50-nt oligonucleotide (JL278) representing (+) SSDNA (see above). For the experiments in which RNase H cleavage was measured directly, the 17-nt RNA was labeled at its 5′ end with 32P (29). In experiments in which synthesis of the 80-nt strand transfer product was measured, a fivefold excess of the chimeric 33-nt DNA-RNA oligonucleotide and the 17-nt RNA oligonucleotide (each at 1 pmol) were annealed to (+) SSDNA (0.2 pmol) labeled at its 5′ end with 32P (5.8 × 106 cpm/0.2 pmol), as described previously (27). In all cases, reactions were continued following addition of the acceptor DNA template, MgCl2, dNTPs, and RT, as indicated above (section i), except that the enzymes added were either wild-type HIV-1 RT, RNase H-minus HIV-1 RT (62), or RNase H-minus HIV-1 RT plus E. coli RNase H (each at 0.2 pmol). Incubation was for 1 h at 37°C. Where specified, NC was added following annealing of the chimeric DNA-RNA oligonucleotide and 17-nt RNA to 32P-labeled (+) SSDNA; in these experiments, reaction mixtures contained either wild-type RT or RNase H-minus RT (62). Note that the DNA primer was not present in this assay.
Southern blot hybridization.
The procedures used for Southern blot analysis were performed essentially as described by Miller et al. (50). Briefly, a HindIII digest of DNA synthesized in a 6-h endogenous RT reaction with four unlabeled dNTPs (27) was subjected to electrophoresis in a 6% sequencing gel. The gel was transferred by electroblotting to a Hybond-N+ filter (Amersham Life Sciences, Inc.). The filter was rinsed one time with 10× SSC (1× SSC is 0.15 M NaCl and 0.015 sodium citrate), cross-linked as described by Miller et al. (50), and then air dried. Prehybridization was carried out at 65°C for 4 h in 25 to 30 ml of buffer (Quality Biological, Inc., Gaithersburg, Md.) containing 6× SSC, 5× Denhardt’s solution, and 0.5% sodium dodecyl sulfate (SDS); herring sperm DNA (Sigma), at a final concentration of 100 μg/ml, was also present. Hybridization to a 32P-labeled DNA oligonucleotide probe (total of 2 × 108 cpm) which detects sequences at the 3′ end of (+) SSDNA was performed by further incubation at 65°C overnight. The filter was washed twice for 20 min at 60°C with 2× SSC and 0.1% SDS and for 20 min at 60°C with 0.5× SSC and 0.1% SDS; it was then air dried and exposed to Kodak XAR-5 film. A sequencing ladder was generated with the M13 template supplied in a Sequenase kit (U.S. Biochemical Corp.).
Recombinant HIV-1 NC.
Methods for cloning, expression, and purification of recombinant HIV-1 NC having 55 amino acid residues are described by Wu et al. (73).
RESULTS
Reconstituted plus-strand transfer system.
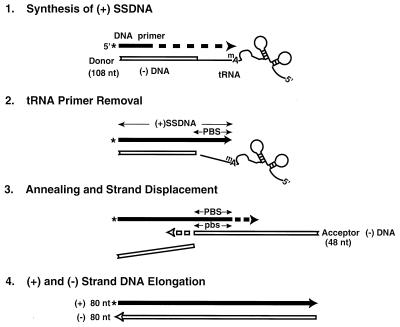
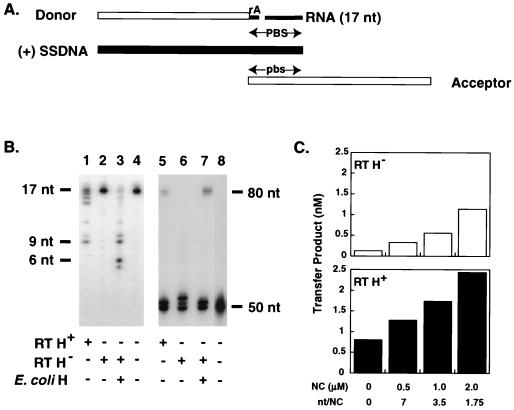
To investigate the mechanism of plus-strand transfer, we designed a model system which mimics events which occur at this stage in reverse transcription. As shown in Fig. 1, reaction 1, synthesis of (+) SSDNA, is carried out with a donor template containing a 32-nt minus-strand DNA oligonucleotide [representing the 5′-terminal sequences of (−) SSDNA] covalently attached to the 76-nt natural tRNA3Lys primer. A 32P-labeled 20-nt DNA oligonucleotide (rather than the PPT in genomic RNA) serves as the primer for (+) SSDNA synthesis in these experiments. After copying 12 nt of DNA, RT copies the 18-nt sequence at the 3′ end of the tRNA and thereby reconstitutes the PBS. In the next step (reaction 2), the tRNA primer is removed by RNase H cleavage. The actual transfer event (reaction 3) occurs when the 18-nt PBS sequence in (+) SSDNA and its complement in the 48-nt minus-strand DNA acceptor template are annealed. As the acceptor strand is elongated, RT also catalyzes strand displacement of the donor DNA template. (Strand displacement has been extensively studied by others [7, 20, 35, 36, 71, 72] and is not directly examined here.) In this system, elongation (reaction 4) of the plus- and minus-DNA strands yields an 80-nt, linear, double-stranded DNA product.
FIG. 1.
Schematic diagram illustrating the HIV-1 model system for plus-strand transfer. (Reaction 1) Synthesis of (+) SSDNA. Plus-strand DNA synthesis is initiated with a 32P-labeled 20-nt DNA primer; the label is indicated by an asterisk. Experiments were carried out with a donor template containing a 32-nt minus-strand DNA [representing the 32 nt at the 5′ end of (−) SSDNA] attached to tRNA3Lys (template size, 108 nt). During DNA synthesis, RT first copies 12 nt of DNA and then copies the 18-nt sequence from the 3′ end of tRNA3Lys, thereby reconstituting the PBS. (Reaction 2) tRNA primer removal. In this step, RNase H cleavage occurs at the 3′ end of tRNA3Lys. (Reaction 3) Annealing and strand displacement. (+) SSDNA containing the reconstituted PBS is annealed to the complementary PBS sequence (pbs) in the minus-strand acceptor DNA (48 nt). As the acceptor strand is elongated, there is also displacement of the DNA moiety of the donor template. (Reaction 4) Elongation of plus- and minus-strand DNA chains. The final product is an 80-nt linear, double-stranded DNA. Filled arrow, plus-strand DNA; open arrow, minus-strand DNA. The tRNA structure is depicted schematically and the first modified base (a methyl A) is shown. The figure is not drawn to scale. mA, methyl A.
Synthesis of plus-strand DNA with the tRNA3Lys donor template.
The time course of plus-strand DNA synthesis (reaction 1 in Fig. 1) was investigated with a donor DNA template having the 5′-terminal sequences of (−) SSDNA covalently attached to the tRNA3Lys primer and the DNA primer labeled with 32P (Fig. 2). Under the conditions of the assay, (+) SSDNA was synthesized to a significant extent within the first 5 min of incubation (lane 1) whereas the plus-strand transfer product [(+) 80 nt] was not detected before 10 min (lane 2). This lag in synthesis of the (+) 80 nt DNA is reminiscent of the 5- to 10-min delay between the detection of (−) SSDNA and the appearance of the minus-strand DNA transfer product, which we observed previously in an HIV-1 reconstituted minus-strand transfer system (27). Note that there was no synthesis of the (+) 80-nt DNA when the acceptor DNA template was omitted from the reaction (Fig. 2, lane C).
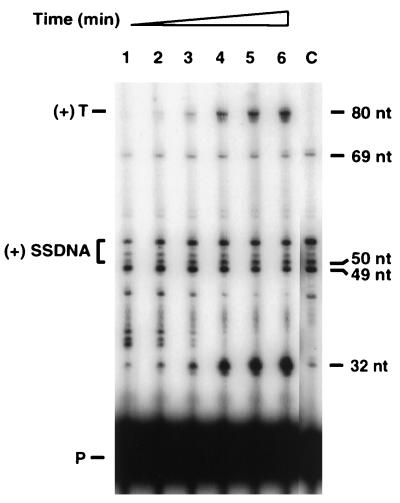
FIG. 2.
Plus-strand transfer with the tRNA3Lys-containing donor template. Plus-strand transfer was assayed as described in Materials and Methods. At the specified times, 20-μl portions were removed and analyzed according to the procedures given in Materials and Methods. Lanes: 1, 5 min; 2, 10 min; 3, 15 min; 4, 30 min; 5, 45 min; 6, 60 min; C (control reaction without acceptor), 60 min. The designations to the left of the gel are as follows: (+) T, plus-strand transfer product; (+) SSDNA, (+) SSDNAs of 50 to 53 nt (bracketed); P, primer. The positions of 80-, 69-, 50-, 49-, and 32-nt bands indicated on the right were determined by running an M13 sequencing ladder together with the samples.
The data in Fig. 2 show that in addition to the expected HIV-1 DNA products, there were prominent bands at 32 and 49 nt. The 32-nt product is the plus-strand copy of the DNA portion of the donor template. Once RT begins to copy bases at the 3′ end of the tRNA moiety, the enzyme apparently has more difficulty in traversing the template, as evidenced by significant accumulation of the 32-nt DNA (representing termination at the U5-PBS junction) and by many DNA products greater than 32 nt. The latter were presumably pause products since they diminished with increasing time of incubation. The 49-nt DNA also resulted from RT pausing, which occurred 1 base upstream of the termination site at nt 58 in the tRNA. Pausing within the PBS sequence during HIV-1 plus-strand DNA synthesis was also observed in the system described by Ben-Artzi et al. (4). It is perhaps even more striking that products larger than the 50-nt (+) SSDNA were also made. These included DNAs of 51 to 53 nt (the 53-nt DNA being the most prominent) and a product of 69 nt.
To account for these findings, we considered the possibility that (+) SSDNA synthesis does not terminate solely at the methyl A at position 58 in tRNA3Lys (Fig. 3A) but instead continues past this base, with a major termination site at the Ψ at position 55 and another termination site at a second Ψ at position 39. This possibility would result in synthesis of 53- and 69-nt products, respectively. Figure 3B illustrates the 3′-terminal bases expected to be present in each of the 51- to 53-nt products. For many of the HIV-1 strains in a class designated “consensus B” (42), including NL4.3 (1) and MN (30), both of which we have used in this study, each of the three additional bases (5′-TTG) in the 53-nt (+) SSDNA is also immediately downstream of the PBS in the viral genome. Thus, in our system, when RT copies the 3′-AAC sequence in tRNA3Lys (positions 58 to 56), all of the 50- to 53-nt (+) SSDNAs may undergo productive strand transfer. In contrast, the 69-nt DNA is a dead-end product which increased only during the first 5 to 10 min of incubation, before the 80-nt transfer product was detectable (Fig. 2); the explanation for this result is not clear.
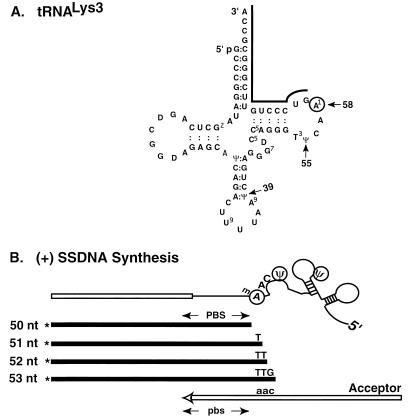
FIG. 3.
Schematic diagram illustrating multiple termination sites for (+) SSDNA synthesis. (A) Nucleotide sequence of tRNA3Lys. The sequence is taken from a review by Litvak et al. (48). A black line is shown above the 18 bases at the 3′ end of the tRNA. The methyl A at position 58 is circled. Arrows point to the modified bases which represent termination sites: methyl A at position 58 and two Ψ bases at positions 55 and 39. (B) (+) SSDNA products synthesized in vitro with tRNA3Lys attached to the 32-nt minus-strand donor template. The donor template (open rectangle) is shown attached to a schematic representation of tRNA3Lys; the 18 3′ bases of the tRNA are shown as a thin black line. Four plus-strand DNA products (filled rectangles), each labeled at the 5′ end with 32P (indicated by an asterisk), are shown. The 50- and 53-nt products terminate before modified bases (methyl [m] A and Ψ, respectively). The DNA product which terminates at the Ψ at position 39 is not shown. The 3′-terminal bases of the 51-, 52-, and 53-nt products are complementary to positions 58 to 56 in tRNA3Lys and to bases downstream of the PBS in the minus-strand acceptor template (open arrow) of several HIV-1 strains, including MN (30) and NL4.3 (1). Lowercase letters indicate minus-strand sequences.
To determine whether the phenomenon of multiple termination sites is reproduced during synthesis of plus-strand DNA in virions, we investigated the size of (+) SSDNA made in an endogenous RT reaction with HIV-1 MN (30) virions (Fig. 4). [It is known that (+) SSDNA is present among the DNA products made in the endogenous reaction (12; also see references 27 and 28)]. Our experimental approach was to digest the endogenous DNA preparation with HindIII and then to analyze the digest by gel electrophoresis and Southern hybridization, using a minus-strand probe to detect the product(s) originating from the 3′ end of the (+) SSDNA. Two different amounts of the digest were added to the gel (Fig. 4B, lanes 1 and 2).
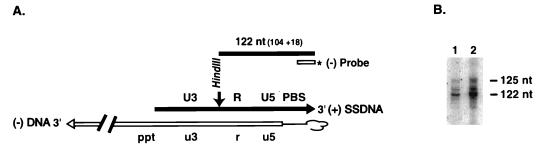
FIG. 4.
Termination of (+) SSDNA synthesis in the HIV-1 endogenous RT reaction. HIV-1 MN virions (from infected H9 cells) were incubated for 6 h in an endogenous RT reaction mixture, as described by Guo et al. (27), except that all four dNTPs were unlabeled. The DNA products were isolated according to the procedures given by Guo et al. (27) and then digested with HindIII. Portions of the digest were subjected to electrophoresis in a 6% sequencing gel. The gel was transferred to a Hybond-N+ filter by electroblotting and analyzed by Southern hybridization, as described in Materials and Methods. (A) Strategy for Southern blot analysis. (+) SSDNA contains the U3, R, U5, and reconstituted PBS sequences. Regions of minus-strand DNA with tRNA3Lys attached at its 5′ end are indicated with lowercase letters; synthesis is presumed to have continued beyond the minus-strand PPT. The HindIII restriction site (nt 9635 in HIV-1 MN [30]) is denoted by the vertical arrow. A minus-strand probe was used to analyze the DNA digestion products downstream of the HindIII site (see Materials and Methods). If termination of (+) SSDNA synthesis occurs as expected at a methyl A, position 58 in tRNA3Lys, the fragment detected will be 122 nt. Plus- and minus-strand DNAs are represented by filled and open arrows, respectively. tRNA is represented by a thin black line. The figure is not drawn to scale. (B) Analysis of endogenous (+) SSDNA products. A 32P-labeled minus-strand probe (JL303; 5′-GTC CCT GTT CGG GCG CCA CTG CTA GAG-3′, complementary to nt 626 to 652 in HIV-1 MN [30]) was used to detect (+) SSDNA. The samples loaded on the gel contained 3.2 μg (lane 1) and 6.4 μg (lane 2) of DNA. The positions of the 122- and 125-nt bands are shown to the right of the gel. An M13 sequencing ladder was used to determine the sizes of the DNA products.
The results of hybridization with a minus-strand DNA probe (see Materials and Methods) showed that several bands were detected: a major band of 122 nt and significant amounts of larger bands of 123 to 125 nt (Fig. 4B). The 122-nt band would be produced if (+) SSDNA synthesis terminated at the methyl A at position 58 in tRNA3Lys, whereas the 125-nt product would result from termination at the Ψ at position 55. Hybridization with a plus-strand probe for sequences in minus-strand U5 revealed a single fragment (data not shown), indicating that the multiple bands detected with the minus-strand probe were not the result of incomplete digestion with HindIII.
We conclude from these results that termination of (+) SSDNA synthesis occurs at multiple sites in both the reconstituted and endogenous assay systems.
RNase H-catalyzed removal of tRNA3Lys from donor DNA.
As illustrated in Fig. 1 (reaction 2), once the PBS has been reconstituted in plus-strand DNA, tRNA3Lys should be removed by the RNase H activity of RT. To verify the requirement for the RNase H step, we performed control experiments with RNase H-minus RT (62) (data not shown). We found that strand transfer did not occur with the mutant RT; however, the RNase H function could be supplied in trans by E. coli RNase H (data not shown; also see Fig. 7B).
FIG. 7.
Requirement for secondary RNase H cleavage and/or NC activity to completely remove the tRNA3Lys primer. The assay was performed as described in Materials and Methods. In panel B, lanes 5 to 8, and panel C, the chimeric DNA-RNA oligonucleotide and the 17-nt RNA were added in fivefold excess of the amount of (+) SSDNA (see the text). [However, note that essentially the same results were obtained with a 1:1 ratio of these oligonucleotides to (+) SSDNA (data not shown).] (A) Nucleic acid strand transfer intermediates present in the reaction mixture. The donor DNA template with a single rA at its 5′ end, a 17-nt RNA representing the 17 bases remaining at the 3′ end of tRNA3Lys after the initial RNase H cleavage, (+) SSDNA, and the acceptor DNA template are shown. (+) SSDNA and the minus-strand donor and acceptor DNAs are represented by filled and open rectangles, respectively; the rA attached to the donor DNA and the 17-nt RNA are indicated by narrow filled rectangles. (B) Gel analysis of RNase H-mediated cleavage of 17-nt RNA and synthesis of the 80-nt strand transfer product. For analysis of RNase H cleavage (lanes 1 to 4) and plus-strand transfer (lanes 5 to 8), the 17-nt RNA and the 50-nt (+) SSDNA were 5′ end labeled with 32P (29), respectively. The positions of 17-, 9-, and 6-nt RNAs shown to the left of lanes 1 to 4 were determined as described in the legend to Fig. 6C. The positions of the (+) 80-nt transfer product and the 50-nt (+) SSDNA are shown to the right of lanes 5 to 8. The enzymes added to the reaction mixtures are shown underneath each lane. Reactions were carried out with wild-type HIV-1 RT (RT H+) (lanes 1 and 5), RNase H-minus HIV-1 RT (RT H−) (62) (lanes 2 and 6), and RNase H-minus HIV-1 RT plus E. coli RNase H (E. coli H) (lanes 3 and 7). Lanes 4 and 8 contain reaction mixtures without enzyme and indicate the positions of the 32P-labeled 17-nt RNA oligonucleotide and the 32P-labeled 50-nt (+) SSDNA oligonucleotide, respectively. (C) Effect of NC on the ability of RNase H-minus (RT H−) and wild-type (RT H+) RT to catalyze plus-strand transfer. The reaction conditions were the same as those used for the gel in panel B, lanes 5 and 6. The amounts of 80-nt transfer product are shown for reaction mixtures without NC and with 0.5, 1.0, and 2.0 μM NC. The open and filled bars represent results with RNase H-minus RT (62) and wild-type RT, respectively.
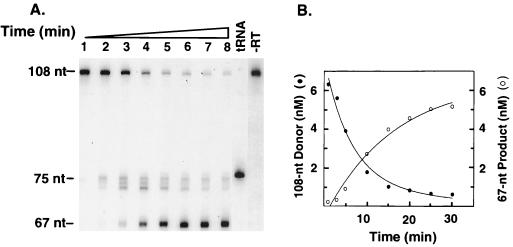
To measure the time course of primer removal in our system (Fig. 5), the experimental conditions were the same as those used in Fig. 2, except that the 5′ end of the tRNA moiety in the donor template was labeled with 32P and the 20-nt DNA primer was unlabeled. The RNA cleavage products were analyzed in a 6% sequencing gel, as described in Materials and Methods. The migrations of labeled, 76-nt tRNA3Lys and the labeled, 108-nt tRNA donor template are shown in Fig. 5, lanes tRNA and −RT, respectively. Two classes of RNA cleavage products were observed: RNAs of 73 to 75 nt and an RNA of 67 nt (Fig. 5A). The 75-nt RNA was detectable on long exposures at 1 min (lane 1) and easily detectable by 3 min (lane 2), but the 73- and 74-nt RNAs were more visible at 5 min (lanes 3); all of these bands decreased substantially by 15 to 20 min (lanes 5 and 6). The 75-nt product results from the initial RNase H cleavage at the 3′ end of the tRNA, which leaves one base (an rA) attached to the minus-strand donor template (23, 57, 66, 70). The appearance of the smaller 74- and 73-nt products may be due to polymerase-independent cleavages of the 75-nt RNA. Further cleavage of the initial RNA products would account for the production of a 67-nt RNA; this product was detectable by 3 min (lane 2) but became more prominent during further incubation from 10 to 30 min (lanes 4 to 8). Quantification of the data by PhosphorImager analysis (Fig. 5B) demonstrated that virtually all of the 108-nt tRNA donor template gave rise to a 67-nt RNA. The significant reduction in the amount of the 75-nt product and almost complete disappearance of the 74- and 73-nt RNAs at later times during incubation indicate that the 73- to 75-nt RNAs are transient intermediates which are ultimately converted to the final 67-nt RNA product.
FIG. 5.
RNase H-mediated removal of tRNA3Lys. In vitro plus-strand transfer with the 32P-labeled tRNA3Lys donor template was carried out as described in Materials and Methods. (A) Time course for RNase H cleavage. Lanes: 1, 1 min; 2, 3 min; 3, 5 min; 4, 10 min; 5, 15 min; 6, 20 min; 7, 25 min; 8, 30 min. The lanes labeled “tRNA” and “−RT” contain controls and refer to tRNA3Lys (indicating the position of the 76-nt full-length tRNA) and to a reaction mixture incubated without RT (indicating the position of the 108-nt labeled tRNA3Lys donor), respectively. The positions of the 75- and 67-nt cleavage products are shown to the left of the gel. (B) Quantitative PhosphorImager analysis. The gel data shown in panel A were analyzed with a PhosphorImager, as described in Materials and Methods. The concentrations of the 108-nt donor template (filled circles) and the 67-nt product (open circles) were plotted against time of incubation.
The early appearance of RNase H cleavage products (Fig. 5), in contrast to the lag in the appearance of the transfer product (Fig. 2), suggests that RNase H-catalyzed removal of the primer must occur prior to the actual strand transfer reaction (Fig. 1, reaction 3). The data in Fig. 5 also suggest that secondary cleavage at the 3′ end of the tRNA may be important for plus-strand transfer.
Plus-strand transfer with an 18-nt RNA-DNA donor template.
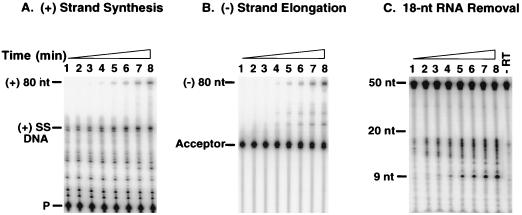
To investigate the question of whether secondary cleavage is required for plus-strand transfer, we realized that it would be necessary to use a simpler system, e.g., one in which the donor template has the 32-nt minus-strand DNA linked to an RNA oligonucleotide with the 18-nt sequence present at the 3′ end of tRNA3Lys (Fig. 6), rather than to the entire tRNA, as shown in Fig. 1; all other reaction constituents were unchanged. To determine whether such a system would also mimic the events which occur during plus-strand transfer, we analyzed three reactions in parallel: plus-strand DNA synthesis (Fig. 6A), minus-strand elongation (Fig. 6B), and removal of the 18-nt RNA (Fig. 6C).
FIG. 6.
Plus-strand transfer with an 18-nt RNA-containing donor template. In vitro plus-strand transfer was assayed as described in Materials and Methods. Each panel illustrates the time course for three reactions which occur during plus-strand transfer. The time points were the same for all panels. Lanes: 1, 1 min; 2, 3 min; 3, 5 min; 4, 10 min; 5, 20 min; 6, 30 min; 7, 60 min; 8, 120 min. (A) Plus-strand synthesis. The positions of the 32P-labeled DNA primer (P), (+) SSDNA, and the (+) 80-nt transfer product are shown to the left of the gel. (B) Minus-strand elongation. The acceptor DNA template was 5′ end labeled with 32P (29) and the 20-nt DNA primer was unlabeled. The positions of the acceptor and the (−) 80-nt elongation product are shown to the left of the gel. The nature of the two intermediate products which accumulated during the incubation is not known. (C) Removal of the 18-nt RNA. The RNase H assay was performed as described in Materials and Methods. The lane labeled “−RT” refers to a reaction mixture incubated without RT and shows the size of the 50-nt labeled 18-nt RNA donor template. An alkaline hydrolysate of a 20-nt RNA oligonucleotide labeled at its 5′ end with 32P was used to determine the sizes of the nucleotide bands. The positions of the 20- and 9-nt RNAs are indicated to the left of the gel.
The time course of plus-strand DNA synthesis with the 18-nt RNA-DNA donor template (Fig. 6A) was analyzed with the DNA primer labeled with 32P at its 5′ end; the results were similar to those observed with the tRNA system (Fig. 2). Thus, (+) SSDNA synthesis could be detected at early times during the incubation (Fig. 6A, lanes 1 to 3; compare with Fig. 2, lane 1), whereas the plus-strand 80-nt transfer product was detected after a lag of 10 min (Fig. 6A, lane 4; compare with Fig. 2, lane 2). The 49-nt pause product running slightly ahead of the 50-nt (+) SSDNA product appeared early (Fig. 6A, lanes 1 to 3) but was less prominent at later times of incubation (Fig. 6A, lanes 7 to 8). Smaller pause products occurred during copying of both the DNA and RNA portions of the template; these products were present to a greater extent than seen above with the tRNA system (Fig. 2). In contrast with these results, when the minus-strand donor template is covalently attached to a DNA version of the 18-nt RNA sequence, no pause products are observed (see Fig. 1A in reference 28). This suggests that RT catalyzes continuous synthesis with a DNA template more efficiently than with a chimeric DNA-RNA template.
To analyze the time course of minus-strand DNA elongation (Fig. 6B), the acceptor template was labeled at its 5′ end with 32P and an unlabeled DNA primer was used. Interestingly, as shown above for synthesis of the plus-strand 80-nt transfer product, the full-length 80-nt minus-strand DNA was not detected until after 10 min of incubation (Fig. 6B, lane 4).
For analysis of RNase H-mediated removal of the 18-nt RNA moiety in the donor template (Fig. 6C), the RNA was labeled at its 5′ end with 32P and the donor, acceptor, and DNA primer were unlabeled. Multiple cleavages yielding RNAs of sizes between 12 and 17 nt were detected at the earliest times during the incubation (Fig. 6C, lanes 1 to 3). As the time of incubation was increased, a 9-nt cleavage product accumulated. These results indicate that initial RNase H cleavages occur to a greater extent in this system than in the original system with a tRNA-containing donor template (Fig. 5A). However, it is striking that in both systems, a small product (9 nt in Fig. 6C and 67 nt in Fig. 5A), presumably representing the final product generated by polymerase-independent RNase H activity (22, 26, 55), is the major RNA present at late times during the incubation period. In addition, the overall similarity of the cleavage patterns (compare Fig. 6C with 5A) indicates that interaction of RT with the structural elements of tRNA3Lys is not required for the primer removal step in plus-strand transfer.
Requirement for secondary RNase H cleavage or NC activity for complete removal of tRNA3Lys.
The results showing that secondary RNase H cleavage takes place in both the standard (Fig. 5) and modified (Fig. 6) systems suggested that the 17-nt RNA remaining after the initial cleavage, which is still hybridized to (+) SSDNA, must be shortened by 8 nt to completely remove the tRNA primer from minus-strand DNA and allow (+) SSDNA to anneal to the acceptor template. To investigate this possibility, we calculated the predicted Tm values for hybrids of different sizes using the MacVector program. Thus, the Tm values for 14- to 18-nt RNA-DNA hybrids with sequences from the 3′ end of the tRNA are significantly greater than 37°C (ranging from 58 to 71°C); however, the values for 11- and 8-nt hybrids are 38 and 20°C, respectively. These data indicate that hybrids smaller than 11 nt will dissociate spontaneously during incubation at 37°C but that hybrids larger than 11 nt will not.
For a more direct test of a possible requirement for secondary RNase H cleavage in plus-strand transfer, we performed the experiment illustrated in Fig. 7. In this experiment, we modeled the step in which RNase H has already removed one base from the 18 nt at the 3′ end of tRNA3Lys (Fig. 5) (see also references 23, 57, 66, and 70), leaving the minus-strand DNA donor with an rA at its 5′ end and a 17-nt RNA (representing the 17 penultimate bases still attached to the rest of tRNA3Lys). Both the chimeric DNA-RNA oligonucleotide and the 17-nt RNA were hybridized to the 50-nt (+) SSDNA (see the schematic diagram in Fig. 7A). RNase H cleavage of the 17-nt RNA, labeled at its 5′ end with 32P (Fig. 7B, lanes 1 to 4), and transfer of labeled (+) SSDNA (Fig. 7B, lanes 5 to 8) were examined in reactions with wild-type RT or RNase H-minus RT (62) with or without E. coli RNase H. The ability of NC to substitute for secondary RNase H cleavage was also determined (Fig. 7C).
In the presence of wild-type RT, the labeled 17-nt RNA was cleaved to a number of small products ranging primarily from 14 to 16 nt in size. There was also a significant amount of a 9-nt RNA (Fig. 7B, lane 1), the same size as that of the product which accumulates in the strand transfer system having an 18-nt RNA-DNA donor template (Fig. 6C). With RNase H-minus RT, no cleavage occurred (lane 2) and only the 17-nt RNA could be detected (compare with the control in lane 4). However, when the RNase H-minus RT was added together with E. coli RNase H, the major cleavage products were 6 and 9 nt in size (lane 3); in addition, under these conditions, cleavage was also more pronounced than with wild-type RT (compare lane 3 with lane 1). These results reflect the fact that E. coli RNase H has ∼300-fold higher specific activity than RT-associated RNase H (55). Moreover, the specificity of E. coli RNase H cleavage differs from that of retroviral RNase H; with the bacterial RNase H, cleavage occurs closer to the 5′ end of the RNA (11, 29, 55, 58, 77).
To determine whether cleavage of the 17-nt RNA is in fact correlated with transfer of (+) SSDNA to the acceptor DNA template, we assayed plus-strand transfer with the intermediates shown in Fig. 7A. In this case, (+) SSDNA was labeled. It is critical for the interpretation of the results that in the initial step, all of the (+) SSDNA is annealed to the chimeric DNA-RNA oligonucleotide and the 17-nt RNA. Only if this condition is fulfilled will the availability of free (+) SSDNA depend upon cleavage; for this reason, a fivefold excess of the chimeric and RNA oligonucleotides was used. With wild-type RT, there was an 80-nt plus-strand DNA product (Fig. 7B, lane 5), as expected. In the presence of the RNase H-minus RT alone, no transfer product could be detected (lane 6); if E. coli RNase H was also added to the reaction mixture, however, the 80-nt DNA was synthesized (lane 7). Note that the amount of the 80-nt product was greater in the presence of E. coli RNase H than in the presence of wild-type RT (compare lane 7 with lane 5), consistent with the greater amount of RNase H cleavage activity seen with E. coli RNase H (Fig. 7B, compare lane 3 with lane 1). The bands observed in addition to the 50-nt (+) SSDNA (lane 8) in lanes 5 to 7 may be the result of blunt-end addition (54).
We also investigated the possibility that destabilization of the 17-nt RNA-DNA hybrid by HIV-1 NC replaces the secondary cleavage step catalyzed by RNase H and allows synthesis of the 80-nt transfer product (Fig. 7C). NC was added after the hybridization step in which the two donor fragments and (+) SSDNA were annealed (Fig. 7A). In the absence of NC, only background radioactivity was detected in the RNase H-minus RT reaction. However, when NC was added, the 80-nt transfer product was detected and the extent of strand transfer increased with increasing concentrations of NC (Fig. 7C, upper graph). In reactions with wild-type RT, NC exerted a modest stimulatory effect (Fig. 7C, lower graph). In this case, plus-strand transfer was stimulated by ∼1.6-, 2.1-, and 3-fold with NC concentrations of 0.5, 1.0, and 2.0 μM, respectively. (These concentrations of NC correspond to 7, 3.5, and 1.75 nt per NC molecule, respectively.)
The results of the experiment shown in Fig. 7 taken together with the data in Fig. 5 and 6 demonstrate that removal of the 3′-terminal base from the 18 nt at the 3′ end of tRNA3Lys is required, but not sufficient, for successful plus-strand transfer. There must be additional RNase H cleavage and/or NC activity, both of which result in destabilization of the remaining RNA-DNA hybrid and allow the complete dissociation of the tRNA primer. These findings indicate that secondary RNase H cleavage is not obligatory for plus-strand transfer.
Stimulation of the annealing step by HIV-1 NC.
In view of the fact that NC can play a role in tRNA primer removal (see above and Fig. 7C) and is known to be a critical accessory protein for efficient HIV-1 minus-strand transfer (2, 15, 27, 41, 53, 60, 75), it was of interest to determine whether NC has other effects on plus-strand transfer. Initial experiments with the standard tRNA3Lys donor system (Fig. 1) indicated that NC stimulated synthesis of the plus-strand 80-nt transfer product by only 1.5- to 2-fold (data not shown). We therefore decided to investigate the influence of HIV-1 NC on individual steps which occur in the plus-strand transfer system (Fig. 1). When the experiments shown in Fig. 5A and 6C were repeated in the presence of NC, there was no significant effect on RNase H-catalyzed tRNA primer removal (data not shown). The data are also in accord with our earlier observation that NC has little effect on the rate and extent of degradation of the donor RNA template by RNase H (unpublished observation cited in reference 28) or on the nature of the RNase H cleavage pattern (28, 41) produced during minus-strand transfer.
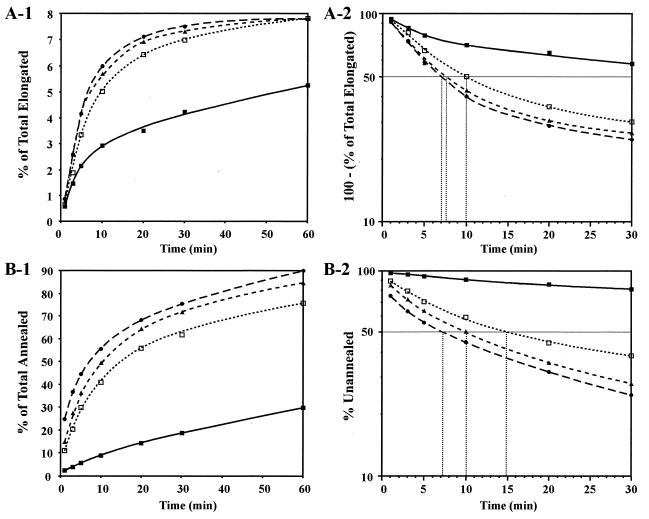
To determine whether NC affects steps following primer removal, we used a partial system containing the 50-nt (+) SSDNA labeled with 32P and the acceptor DNA template but no donor DNA template or DNA oligonucleotide primer (Fig. 8). Reaction mixtures were incubated for various times up to 1 h, without NC or with three different NC concentrations: 0.14, 0.28, and 0.56 μM. [Since a 1:1 ratio of (+) SSDNA to acceptor was used, the numbers of nucleotides per NC molecule were 7, 3.5, and 1.75, respectively.]
FIG. 8.
Effect of HIV-1 NC on annealing and elongation and on annealing alone. Reaction mixtures were incubated at 37°C from 1 to 60 min without NC or with three different concentrations of NC; RT was added where indicated. (A) Annealing plus elongation. The 50-nt (+) SSDNA oligonucleotide, labeled at its 5′ end with 32P (29), was incubated with the acceptor DNA template and HIV-1 RT as described in Materials and Methods (under “RT assays. (i) In vitro strand transfer assay”), except that the donor DNA template and DNA oligonucleotide primer were not present. At each time point, a 20-μl aliquot was removed and was then processed and analyzed as described in Materials and Methods. (A-1) The fraction of total (+) SSDNA elongated (percentage of total elongated) was plotted against the time of incubation. (A-2) The data in panel A-1 were replotted as a semilogarithmic plot of 100 minus the percentage of total (+) SSDNA elongated against the time of incubation. (B) Annealing only. Labeled (+) SSDNA was incubated with the acceptor DNA template as described for panel A except that HIV-1 RT was also omitted from the reaction mixture. A 20-μl aliquot was removed, and an equal volume of STOP solution (27) was added. Four microliters was then analyzed in a 6% native polyacrylamide gel (acrylamide-bis, 19:1). (B-1) The percentage of (+) SSDNA annealed was plotted against the time of incubation. (B-2) The data in panel B-1 were replotted as a semilogarithmic plot of the percentage of unannealed 50-nt (+) SSDNA versus the time of incubation. In panels A-2 and B-2, the t1/2 values for the reactions with NC were calculated by determining where the 50% point on each curve intersects the x axis (dotted lines); in reactions without NC, the t1/2 values were estimated by extrapolating the data to 50% and determining where a vertical line from the 50% point intersects the x axis (not shown). Symbols: filled squares and solid line, no NC; open squares and line with short dashes, 0.14 μM NC; filled triangles and line with medium dashes, 0.28 μM NC; filled circles and line with long dashes, 0.56 μM NC.
In the presence of RT, both the annealing and elongation steps can occur in the partial system and the 80-nt product should be synthesized. PhosphorImager analysis showed that under these conditions, i.e., annealing plus elongation, HIV-1 NC stimulated the rate and extent of formation of the 80-nt DNA (Fig. 8A-1). In other experiments in which (+) SSDNA was annealed to the acceptor template by prior incubation at 65°C for 5 min and then incubated with RT in the presence or absence of NC, i.e., elongation only, no effect of NC could be observed (data not shown). This result suggests that in this partial system, the effect of NC is on the annealing reaction and not on elongation.
Since NC is known to catalyze annealing of complementary sequences (see reviews by Darlix et al. [13] and Rein et al. [59]), we expected that it would have a similar effect on the annealing step in plus-strand transfer. We tested the effect of NC on annealing only by incubating reaction mixtures as described for Fig. 8A-1, except that RT was omitted (Fig. 8B-1). Annealing was very inefficient in the absence of NC, and only 20% of the initially added (+) SSDNA formed a DNA duplex at the end of the 1-h incubation. Addition of NC at the concentrations given above significantly increased both the rate and the extent of annealing. At 1 h, annealing reached 90% with the highest concentration of NC tested (0.56 μM).
The kinetic data in Fig. 8A-1 and B-1 were replotted as semilogarithmic plots (Fig. 8A-2 and B-2, respectively). These plots all show curvature, as previously observed with typical annealing reactions (reference 16 and references therein). Half-life (t1/2) values for the NC-containing reaction mixtures were calculated by determining where a stippled line from the 50% point on the curves intersects the x axis (time); in reaction mixtures without NC, the values were estimated by extrapolation of the data to 50% (not shown).
In the absence of NC, the reaction with annealing plus elongation had a t1/2 of ∼56 min (Fig. 8A-2) whereas the reaction with annealing alone had a t1/2 of ∼127 min (Fig. 8B-2). The higher rate achieved in the presence of RT leads us to speculate that RT is able to facilitate the annealing of single-stranded nucleotides. The t1/2 values for reactions with RT and NC were also lower than the values for the corresponding annealing reactions with NC alone (compare Fig. 8A-2 with B-2). Thus, the rate of the annealing reaction increased with increasing concentrations of NC and had a t1/2 value of ∼7.2 min at the highest NC concentration (0.56 μM) (Fig. 8B-2). In contrast, in the presence of 0.28 μM NC, the rate of the annealing-plus-elongation reaction had already increased to a maximum t1/2 value of ∼7.2 min (Fig. 8A-2). These observations are consistent with the annealing step being rate limiting for RT-catalyzed elongation between the concentrations of 0 and 0.28 μM NC under our experimental conditions.
These results demonstrate that NC-catalyzed annealing of (+) SSDNA to the acceptor DNA template represents a major effect of NC on events associated with plus-strand transfer. This conclusion is in accord with the large stimulatory effect of NC-catalyzed annealing during minus-strand transfer (14, 28, 44, 46, 75).
DISCUSSION
The present study describes a reconstituted system which is designed to accurately reproduce the events associated with HIV-1 plus-strand transfer. Using this system, we were able to analyze in detail individual steps which contribute to the overall plus-strand transfer process. These include generation of (+) SSDNA, tRNA primer removal, annealing of (+) SSDNA and minus-strand acceptor DNA, and elongation of plus- and minus-strand DNA products. The goal of this work was to address several important questions. (i) Does termination of (+) SSDNA synthesis occur exclusively at the first modified base (a methyl A) at the 3′ end of the tRNA primer? (ii) Does complete removal of the tRNA primer require secondary cleavage of bases at the 3′ end of the tRNA? (iii) Does NC have a role in plus-strand transfer?
In the standard system, we used a 1:1 ratio of the donor and acceptor templates to mimic the situation which exists during virus replication. In addition, the donor DNA template consisted of the 5′-terminal sequences of (−) SSDNA covalently attached to human tRNA3Lys (Fig. 1). This template allowed us to determine which bases in tRNA are termination sites for (+) SSDNA synthesis. Our results demonstrate that these sites include the methyl A at position 58 as well as two Ψ bases at positions 55 and 39. Increasing the ratio of acceptor to donor or changing the RT-to-template ratio did not lead to detectable changes in the pattern of (+) SSDNA products or to any significant quantitative effect (data not shown). The existence of multiple termination sites was also observed in earlier studies with other reconstituted systems (4, 8). However, we have now shown that termination at the methyl A and at (or near) the Ψ at position 55 also occurs in reactions with detergent-treated HIV-1 virions (Fig. 4). This result indicates that termination at multiple sites is not simply an in vitro artifact.
The use of multiple termination sites raises the question of whether this phenomenon adversely affects plus-strand transfer (reviewed in reference 68; see also reference 8). To examine this issue, we searched the database for the genomic sequences of HIV-1 strains in the PBS region. We found that for certain strains of HIV-1 in the consensus B class, including NL4.3 (1) and MN (30) used in this study, and for the ANT70 strain in the consensus O class, the three bases immediately downstream of the PBS (5′-TTG) are complementary to the 3′-AAC bases at positions 58 to 56, respectively, in the tRNA (42). In the HIV-1 consensus U MAL strain there is a match between two of three base pairs: the first two 5′ bases downstream of the PBS are TT, but the 3′ base is a T instead of a G (42). This means that for these viruses, (+) SSDNA products which terminate after the methyl A, but before the Ψ at position 55, may be productively transferred (Fig. 3). Since RT is unable to copy modified bases (reference 49 and references therein), it appears likely that termination of (+) SSDNA synthesis at sites beyond the methyl A is due to incomplete modification of the tRNA in the host cell. Since the degree of tRNA modification may vary depending on the source of the tRNA, the extent to which multiple (+) SSDNA products are formed will also vary (e.g., compare results of Fig. 2 and 4, where the tRNA3Lys primers are from human placenta and HIV-1-infected H9 cells, respectively). It is of interest that in an earlier study of hypomodification of tRNAs required for HIV ribosomal frameshifting, it was reported that the chromatographic profiles of tRNA3Lys in HIV-1-infected and uninfected H9 cells differ (30a). It was suggested that this difference may reflect hypomodification of bases in the anticodon loop of tRNA3Lys from infected cells (30a), but conceivably, other bases such as the methyl A at position 58 may be undermodified as well. Another group has also observed that there are multiple forms of tRNA3Lys in HIV-1 virions (47a).
The annealing of (+) SSDNA to the minus-strand DNA acceptor is expected to take place only if the tRNA primer is completely removed from the 5′ end of (−) SSDNA (reviewed in references 10 and 68). This idea is supported experimentally by the finding that RNase H cleavage begins early in the incubation, i.e., between 1 and 3 min (Fig. 5A and 6C), while the 80-nt transfer product appears after a lag of 10 min (Fig. 2 and 6A and B). Furthermore, if the tRNA removal step is bypassed, no lag phase is detected (Fig. 8A-1).
Since initial cleavage of only 1 base from the tRNA leaves a 17-nt RNA-DNA hybrid which has a high Tm, under these conditions, the tRNA is unable to completely dissociate from the complementary DNA bases in (+) SSDNA. To solve this problem, the virus must have a way to destabilize the remaining RNA-DNA hybrid. The appearance of a secondary cleavage product (67 nt [Fig. 5A] or 9 nt [Fig. 6C]) prior to detection of the 80-nt transfer product suggested that additional cleavage may be required for successful strand transfer. In the experiment illustrated in Fig. 7, we found that plus-strand transfer could occur when the polymerase-independent RNase H activity of RT (or RNase H activity provided in trans, e.g., by E. coli RNase H) could remove additional bases from the 3′ end of the tRNA; with an RNase H-minus RT (62), plus-strand transfer was not detected (Fig. 7B). Apparently, in the absence of RNase H activity, RNA strand displacement by RT (19, 39) is not effective in this system. Interestingly, with avian myeloblastosis virus, the tRNATrp primer is removed intact by a single cleavage at the RNA-DNA junction (52), suggesting that, in this case, RNA strand displacement and/or NC (see below) may play a role in primer removal.
In a report by Cameron et al. (9), it was shown that an HIV-1 RT mutant having a deletion of the 13 C-terminal residues from the p51 subunit and lacking the ability to catalyze polymerase-independent RNase H cleavage could not support efficient minus-strand transfer in vitro. However, when the 71-amino-acid form (NC71) (but not the 55-amino-acid form) of HIV-1 NC was added, the defect in strand transfer was suppressed (9). This observation was interpreted as evidence for the formation of a specific complex between RT and the C-terminal residues in NC71 (9).
In this work on HIV-1 plus-strand transfer, we find that addition of HIV-1 NC (having 55 amino acids [31]) and the E478Q RNase H-minus RT (62) to a reaction mixture in which the initial cleavage has already occurred (Fig. 7A) results in production of the 80-nt strand transfer product; this effect is dependent on the concentration of NC (Fig. 7C). While our data do not address the question of whether an NC-RT complex is involved in this phenomenon, it seems plausible that the nucleic acid chaperone activity of NC (32, 59) is responsible for destabilization of the RNA-DNA hybrid remaining after the initial cleavage event.
How might this occur? One can view the annealing of (+) SSDNA to the 18-nt sequence at the 3′ end of the tRNA and to the minus-strand PBS sequence in the acceptor DNA template as a strand exchange reaction. NC establishes an equilibrium between these two hybrids. However, the presence of (+) SSDNA in the DNA duplex (rather than the RNA-DNA hybrid) is ultimately favored: both strands of the duplex can be elongated by the polymerase activity of RT, and this elongation leads to formation of a structure with a greater number of base pairs and greater thermodynamic stability than in the RNA-DNA hybrid. Thus, as more of the (+) SSDNA is annealed to the acceptor, less (+) SSDNA is available to hybridize to the tRNA and the tRNA will eventually be released. (Note that, according to these considerations, NC does not need to have a direct effect on RNase H cleavage and that in fact, as mentioned above, it does not have such an effect [data not shown].) This situation resembles the NC-catalyzed strand exchange reaction described in the experiments of Tsuchihashi and Brown (69), which showed that annealing of DNA strands with a large number of complementary base pairs was favored over annealing to DNA strands with a smaller number of base-paired regions. It is also possible that in our experiment, there may be a small contribution to primer removal by NC-stimulated RNA strand displacement activity (4, 39).
The finding that NC can substitute for secondary cleavage at the 3′ end of the tRNA raises another issue: does NC facilitate primer removal when wild-type RT is available? In the experiment illustrated in Fig. 7, we found that when wild-type RT was used instead of the mutant version, addition of NC stimulated synthesis of the 80-nt DNA by two- to threefold (Fig. 7C). These observations suggest that during normal HIV-1 replication, both polymerase-independent RNase H cleavage and NC activity can contribute to removal of the primer and formation of the DNA duplex between (+) SSDNA and the minus-strand acceptor DNA template.
Although NC has been shown to promote DNA extension with a long DNA template (37), in our system, elongation of a relatively small number of bases (30 nt) after synthesis of (+) SSDNA is not affected by NC (data not shown). This is presumably due to the fact that the sequence being extended does not have secondary structures which could interfere with RT movement along the DNA template.
What we do observe is a large stimulatory effect of NC on the annealing of the complementary PBS sequences in (+) SSDNA and the minus-strand DNA acceptor template (Fig. 8). Several studies have shown that NC stimulates DNA duplex formation with non-PBS-containing reactants (14, 16, 44, 46, 69), but to our knowledge, this effect of NC on DNA annealing has not been previously demonstrated with DNA molecules representing the actual intermediates in plus-strand transfer. The data of Fig. 8 show that RT also facilitates annealing; however, in the presence of a high concentration of NC, the contribution of RT is less significant.
Semilogarithmic plots of the kinetics of annealing plus elongation and of annealing only show curvature (Fig. 8A-2 and B-2) and are consistent with the possibility that the annealing reaction follows second-order kinetics, as is typical for hybridization reactions (reference 16 and references therein). In minus-strand transfer, however, annealing of the R regions in (−) SSDNA and the 3′ acceptor RNA template follows pseudo first-order kinetics and is independent of nucleic acid concentration (75). Since approximately two-thirds of R consists of the stable, 59-nt TAR stem-loop, it has been proposed that an unfolding step is rate limiting (75). The apparent difference between the annealing reactions in plus- and minus-strand transfer may reflect the fact that the 18-nt PBS sequence is smaller and much less structured than the TAR sequence in R, thereby precluding the requirement for a rate-limiting, unfolding step (75). Rather, with PBS annealing, it is more likely that formation of the DNA duplex is a bimolecular reaction, with rate-limiting nucleation followed by fast zippering (16).
Our results (Fig. 8B-1) demonstrate that the stimulatory effect of NC on annealing is concentration dependent. Under the conditions of our experiment, there is already a significant effect when NC is added at a ratio of 7 nt to 1 NC molecule (0.14 μM); this amount of NC corresponds to the estimated ratio of RNA nucleotides to NC molecules in the HIV-1 particle (31) and also to the size of the binding site calculated from in vitro experiments (16, 38, 74). An increase in NC concentration to as high a ratio as 1.75 nt to 1 NC molecule (0.56 μM) leads to an even greater increase in the rate of annealing, i.e., ∼20-fold greater than the rate achieved in the absence of NC (Fig. 8B-2).
In summary, we have modeled the events associated with HIV-1 plus-strand transfer and have defined the individual steps in molecular terms. We find that in two respects, the strategy employed by the virus for plus-strand transfer strikingly parallels that used for minus-strand transfer. (i) Removal of the tRNA primer or degradation of the RNA donor template, respectively, is required, and in both cases, there must be destabilization of a terminal RNA-DNA hybrid by the polymerase-independent RNase H activity of RT and/or by the action of NC. (ii) Annealing of complementary bases (PBS or R sequences) in the initial copy of the donor template [i.e., (+) or (−) SSDNA, respectively] with the acceptor molecule is required for the actual strand transfer event. Moreover, NC functioning as a nucleic acid chaperone has a profound stimulatory effect on each of these annealing reactions.
ACKNOWLEDGMENTS
We thank Stuart Le Grice for his generous gift of an RNase H-minus mutant RT, Robert Crouch for kindly supplying E. coli RNase H, and Bradley Kane for graciously providing HIV-1 NC protein. We are also indebted to Robert Crouch, Stephen Hughes, and Michael Powell for valuable suggestions concerning the experiment described in Fig. 7 and to Alan Rein for a thoughtful reading of the manuscript.
This work was supported in part by the National Institutes of Health Intramural AIDS Targeted Antiviral Program and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-56000.
ADDENDUM IN PROOF
After this work was accepted for publication, we became aware of a report (S. Auxilien, G. Keith, S. F. J. Le Grice, and J.-L. Darlix, J. Biol. Chem. 274:4412–4420, 1999) describing the relationship between multiple termination sites for (+) SSDNA synthesis and the fidelity of HIV-1 plus-strand DNA transfer.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arts E J, Wainberg M A. Human immunodeficiency virus type 1 reverse transcriptase and early events in reverse transcription. Adv Virus Res. 1996;46:97–163. doi: 10.1016/s0065-3527(08)60071-8. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Artzi H, Shemesh J, Zeelon E, Amit B, Kleiman L, Gorecki M, Panet A. Molecular analysis of the second template switch during reverse transcription of the HIV RNA template. Biochemistry. 1996;35:10549–10557. doi: 10.1021/bi960439x. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Artzi H, Zeelon E, Amit B, Wortzel A, Gorecki M, Panet A. RNase H activity of reverse transcriptases on substrates derived from the 5′ end of retroviral genome. J Biol Chem. 1993;268:16465–16471. [PubMed] [Google Scholar]
- 6.Bertrand E L, Rossi J J. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boone L R, Skalka A M. Viral DNA synthesized in vitro by avian retrovirus particles permeabilized with melittin. II. Evidence for a strand displacement mechanism in plus-strand synthesis. J Virol. 1981;37:117–126. doi: 10.1128/jvi.37.1.117-126.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnett B P, McHenry C S. Posttranscriptional modification of retroviral primers is required for late stages of DNA replication. Proc Natl Acad Sci USA. 1997;94:7210–7215. doi: 10.1073/pnas.94.14.7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameron C E, Ghosh M, Le Grice S F J, Benkovic S J. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc Natl Acad Sci USA. 1997;94:6700–6705. doi: 10.1073/pnas.94.13.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champoux J J. Roles of ribonuclease H in reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 103–117. [Google Scholar]
- 11.Cirino N M, Cameron C E, Smith J S, Rausch J W, Roth M J, Benkovic S J, Le Grice S F J. Divalent cation modulation of the ribonuclease functions of human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:9936–9943. doi: 10.1021/bi00031a016. [DOI] [PubMed] [Google Scholar]
- 12.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 13.Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 14.Davis W R, Gabbara S, Hupe D, Peliska J A. Actinomycin D inhibition of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase and nucleocapsid protein. Biochemistry. 1998;37:14213–14221. doi: 10.1021/bi9814890. [DOI] [PubMed] [Google Scholar]
- 15.DeStefano J J. Human immunodeficiency virus nucleocapsid protein stimulates strand transfer from internal regions of heteropolymeric RNA templates. Arch Virol. 1995;140:1775–1789. doi: 10.1007/BF01384341. [DOI] [PubMed] [Google Scholar]
- 16.Dib-Hajj F, Khan R, Giedroc D P. Retroviral nucleocapsid proteins possess potent nucleic acid strand renaturation activity. Protein Sci. 1993;2:231–243. doi: 10.1002/pro.5560020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y-X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu T-B, Taylor J. When retroviral reverse transcriptases reach the end of their RNA templates. J Virol. 1992;66:4271–4278. doi: 10.1128/jvi.66.7.4271-4278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuentes G M, Fay P J, Bambara R A. Relationship between plus strand DNA synthesis and removal of downstream segments of RNA by human immunodeficiency virus, murine leukemia virus and avian myeloblastoma virus reverse transcriptases. Nucleic Acids Res. 1996;24:1719–1726. doi: 10.1093/nar/24.9.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuentes G M, Palaniappan C, Fay P J, Bambara R A. Strand displacement synthesis of the central polypurine tract region of HIV-1 promotes DNA to DNA strand transfer recombination. J Biol Chem. 1996;271:29605–29611. doi: 10.1074/jbc.271.47.29605. [DOI] [PubMed] [Google Scholar]
- 21.Fuentes G M, Rodríguez-Rodríguez L, Fay P J, Bambara R A. Use of an oligoribonucleotide containing the polypurine tract as a primer by HIV reverse transcriptase. J Biol Chem. 1995;270:28169–28176. doi: 10.1074/jbc.270.47.28169. [DOI] [PubMed] [Google Scholar]
- 22.Furfine E S, Reardon J E. Reverse transcriptase · RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem. 1991;266:406–412. [PubMed] [Google Scholar]
- 23.Furfine E S, Reardon J E. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNALys3-primer excision. Biochemistry. 1991;30:7041–7046. doi: 10.1021/bi00243a001. [DOI] [PubMed] [Google Scholar]
- 24.Gao H-Q, Boyer P L, Arnold E, Hughes S H. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 25.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 26.Gopalakrishnan V, Peliska J A, Benkovic S J. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci USA. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Wu T, Bess J, Henderson L E, Levin J G. Actinomycin D inhibits human immunodeficiency virus type 1 minus-strand transfer in in vitro and endogenous reverse transcriptase assays. J Virol. 1998;72:6716–6724. doi: 10.1128/jvi.72.8.6716-6724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Wu W, Yuan Z Y, Post K, Crouch R J, Levin J G. Defects in primer-template binding, processive DNA synthesis, and RNase H activity associated with chimeric reverse transcriptases having the murine leukemia virus polymerase domain joined to Escherichia coli RNase H. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 30.Gurgo C, Guo H-G, Franchini G, Aldovini A, Collalti E, Farrell K, Wong-Staal F, Gallo R C, Reitz M S., Jr Envelope sequences of two new United States HIV-1 isolates. Virology. 1988;164:531–536. doi: 10.1016/0042-6822(88)90568-5. [DOI] [PubMed] [Google Scholar]
- 30a.Hatfield D, Feng Y-X, Lee B J, Rein A, Levin J G, Oroszlan S. Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology. 1989;173:736–742. doi: 10.1016/0042-6822(89)90589-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 33.Herschlag D, Khosla M, Tsuchihashi Z, Karpel R L. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong T, Drlica K, Pinter A, Murphy E. Circular DNA of human immunodeficiency virus: analysis of circle junction nucleotide sequences. J Virol. 1991;65:551–555. doi: 10.1128/jvi.65.1.551-555.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hottiger M, Podust V N, Thimmig R L, McHenry C, Hübscher U. Strand displacement activity of the human immunodeficiency virus type 1 reverse transcriptase heterodimer and its individual subunits. J Biol Chem. 1994;269:986–991. [PubMed] [Google Scholar]
- 36.Huber H E, McCoy J M, Seehra J S, Richardson C C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989;264:4669–4678. [PubMed] [Google Scholar]
- 37.Ji X, Klarmann G J, Preston B D. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry. 1996;35:132–143. doi: 10.1021/bi951707e. [DOI] [PubMed] [Google Scholar]
- 38.Karpel R L, Henderson L E, Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 39.Kelleher C D, Champoux J J. Characterization of RNA strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase. J Biol Chem. 1998;273:9976–9986. doi: 10.1074/jbc.273.16.9976. [DOI] [PubMed] [Google Scholar]
- 40.Khan R, Giedroc D P. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J Biol Chem. 1992;267:6689–6695. [PubMed] [Google Scholar]
- 41.Kim J K, Palaniappan C, Wu W, Fay P J, Bambara R A. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J Biol Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 42.Korber B, Foley B, Leitner T, McCutchan F, Hahn B, Mellors J W, Myers G, Kuiken C, editors. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Los Alamos National Laboratory; 1997. [Google Scholar]
- 43.Kulkosky J, Katz R A, Skalka A M. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J Acquired Immune Defic Syndr. 1990;3:852–858. [PubMed] [Google Scholar]
- 44.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J-L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lapadat-Tapolsky M, Gabus C, Rau M, Darlix J-L. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol. 1997;268:250–260. doi: 10.1006/jmbi.1997.0978. [DOI] [PubMed] [Google Scholar]
- 46.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix J-L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Quan Y, Arts E J, Li Z, Preston B D, De Rocquigny H, Roques B P, Darlix J-L, Kleiman L, Parniak M A, Wainberg M A. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Li Z, Shalom A, Huang Y, Mak J, Arts E, Wainberg M A, Kleiman L. Multiple forms of tRNALys3 in HIV-1. Biochem Biophys Res Commun. 1996;227:530–540. doi: 10.1006/bbrc.1996.1541. [DOI] [PubMed] [Google Scholar]
- 48.Litvak S, Sarih-Cottin L, Fournier M, Andreola M, Tarrago-Litvak L. Priming of HIV replication by tRNALys3: role of reverse transcriptase. Trends Biochem Sci. 1994;19:114–118. doi: 10.1016/0968-0004(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 49.Marquet R. Importance of modified nucleotides in replication of retroviruses, plant pararetroviruses, and retrotransposons. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 517–533. [Google Scholar]
- 50.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller G, Strack B, Dannull J, Sproat B S, Surovoy A, Jung G, Moelling K. Amino acid requirements of the nucleocapsid protein of HIV-1 for increasing catalytic activity of a Ki-ras ribozyme in vitro. J Mol Biol. 1994;242:422–429. doi: 10.1006/jmbi.1994.1592. [DOI] [PubMed] [Google Scholar]
- 52.Omer A C, Faras A J. Mechanism of release of the avian retrovirus tRNATrp primer molecule from viral DNA by ribonuclease H during reverse transcription. Cell. 1982;30:797–805. doi: 10.1016/0092-8674(82)90284-7. [DOI] [PubMed] [Google Scholar]
- 53.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 54.Peliska J A, Benkovic S J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 55.Post K, Guo J, Kalman E, Uchida T, Crouch R J, Levin J G. A large deletion in the connection subdomain of murine leukemia virus reverse transcriptase or replacement of the RNase H domain with Escherichia coli RNase H results in altered polymerase and RNase H activities. Biochemistry. 1993;32:5508–5517. doi: 10.1021/bi00072a004. [DOI] [PubMed] [Google Scholar]
- 56.Powell M D, Levin J G. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J Virol. 1996;70:5288–5296. doi: 10.1128/jvi.70.8.5288-5296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pullen K A, Ishimoto L K, Champoux J J. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:367–373. doi: 10.1128/jvi.66.1.367-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Randolph C A, Champoux J J. The use of DNA and RNA oligonucleotides in hybrid structures with longer polynucleotide chains to probe the structural requirements for Moloney murine leukemia virus plus-strand priming. J Biol Chem. 1994;269:19207–19215. [PubMed] [Google Scholar]
- 59.Rein A, Henderson L E, Levin J G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 60.Rodríguez-Rodríguez L, Tsuchihashi Z, Fuentes G M, Bambara R A, Fay P J. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 61.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 62.Schatz O, Cromme F V, Grüninger-Leitch F, Le Grice S F J. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 63.Schultz S J, Whiting S H, Champoux J J. Cleavage specificities of Moloney murine leukemia virus RNase H implicated in the second strand transfer during reverse transcription. J Biol Chem. 1995;270:24135–24145. doi: 10.1074/jbc.270.41.24135. [DOI] [PubMed] [Google Scholar]
- 64.Smith C M, Potts III W B, Smith J S, Roth M J. RNase H cleavage of tRNAPro mediated by M-MuLV and HIV-1 reverse transcriptases. Virology. 1997;229:437–446. doi: 10.1006/viro.1997.8454. [DOI] [PubMed] [Google Scholar]
- 65.Smith J S, Kim S, Roth M J. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J Virol. 1990;64:6286–6290. doi: 10.1128/jvi.64.12.6286-6290.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith J S, Roth M J. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNALys3. J Biol Chem. 1992;267:15071–15079. [PubMed] [Google Scholar]
- 67.Tanchou V, Decimo D, Péchoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J-L. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Telesnitsky A, Goff S P. Strong-stop strand transfer during reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 49–83. [Google Scholar]
- 69.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitcomb J M, Kumar R, Hughes S H. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J Virol. 1990;64:4903–4906. doi: 10.1128/jvi.64.10.4903-4906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whiting S H, Champoux J J. Strand displacement synthesis capability of Moloney murine leukemia virus reverse transcriptase. J Virol. 1994;68:4747–4758. doi: 10.1128/jvi.68.8.4747-4758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whiting S H, Champoux J J. Properties of strand displacement synthesis by Moloney murine leukemia virus reverse transcriptase: mechanistic implications. J Mol Biol. 1998;278:559–577. doi: 10.1006/jmbi.1998.1720. [DOI] [PubMed] [Google Scholar]
- 73.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.You J C, McHenry C S. HIV nucleocapsid protein. Expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 75.You J C, McHenry C S. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J Biol Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 76.Yusupova G, Lanchy J-M, Yusupov M, Keith G, Le Grice S F J, Ehresmann C, Ehresmann B, Marquet R. Primer selection by HIV-1 reverse transcriptase on RNA-tRNA3Lys and DNA-tRNA3Lys hybrids. J Mol Biol. 1996;261:315–321. doi: 10.1006/jmbi.1996.0463. [DOI] [PubMed] [Google Scholar]
- 77.Zhan X, Crouch R J. The isolated RNase H domain of murine leukemia virus reverse transcriptase. Retention of activity with concomitant loss of specificity. J Biol Chem. 1997;272:22023–22029. doi: 10.1074/jbc.272.35.22023. [DOI] [PubMed] [Google Scholar]