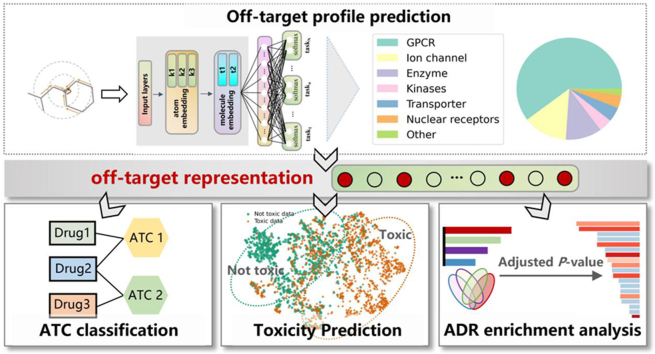
Abstract
Ensuring drug safety in the early stages of drug development is crucial to avoid costly failures in subsequent phases. However, the economic burden associated with detecting drug off-targets and potential side effects through in vitro safety screening and animal testing is substantial. Drug off-target interactions, along with the adverse drug reactions they induce, are significant factors affecting drug safety. To assess the liability of candidate drugs, we developed an artificial intelligence model for the precise prediction of compound off-target interactions, leveraging multi-task graph neural networks. The outcomes of off-target predictions can serve as representations for compounds, enabling the differentiation of drugs under various ATC codes and the classification of compound toxicity. Furthermore, the predicted off-target profiles are employed in adverse drug reaction (ADR) enrichment analysis, facilitating the inference of potential ADRs for a drug. Using the withdrawn drug Pergolide as an example, we elucidate the mechanisms underlying ADRs at the target level, contributing to the exploration of the potential clinical relevance of newly predicted off-target interactions. Overall, our work facilitates the early assessment of compound safety/toxicity based on off-target identification, deduces potential ADRs of drugs, and ultimately promotes the secure development of drugs.
Key words: Drug safety, Off-target prediction, Adverse drug reactions, Toxicity, Molecular representation, Artificial intelligence
Graphical abstract
An advanced framework for drug safety assessment, encompassing precise drug off-target profile prediction and its utilization in ATC classification, toxicity prediction, and ADR enrichment analysis.
1. Introduction
Ensuring drug safety is of utmost importance for individuals using medications, encompassing measures to minimize the risks of adverse drug reactions (ADRs) and potential harms associated with drug utilization. This commitment contributes significantly to the overall success and viability of pharmaceutical endeavors1, 2, 3, 4. Traditional approaches to safety evaluation and toxicity prediction for compounds have relied on costly in vitro methods (e.g., organ-on-a-chip) and in vivo methods (e.g., animal models) that may not accurately reflect human responses5. ADRs are influenced by factors such as drug dose, drug metabolism and patient individual differences6,7. Categorically, about 75% of ADRs fall into Type A, which is dose-dependent and predictable based on the pharmacological characteristics of the drug. Notably, off-target interactions primarily constitute secondary pharmacological features leading to Type A ADRs1, making off-target toxicity a substantial contributor to drug attrition8, 9, 10. This underscores the importance of identifying undesired drug targets1,11, presenting a relatively cost-effective approach for evaluating drug safety.
Pharmaceutical companies commonly employ in vitro pharmacological assays to profile compounds against a comprehensive panel of unsafe off-targets to reduce the number of molecules tested in subsequent assays1. Based on the internal off-target panels of four pharmaceutical companies―AstraZeneca, GlaxoSmithKline, Novartis, and Pfizer, Bowes et al. proposed 44 early drug safety targets that include the toxicity of the central nervous system, immune system, gastrointestinal tract, and heart1. AbbVie obtained 70 safety-related targets via a literature search, most of which are included in Eurofins' safety panel12. Roche utilized experimental data based on the Bioprint® database and employed a statistical ranking method, resulting in a panel of 50 safety targets13. However, compared to screening compounds on known therapeutic targets, off-target screening of compounds is challenging due to the lack of boundaries.
Conducting extensive experimental screening on numerous targets can be cost-prohibitive. Therefore, employing in-silico predictions to assess compound–target interactions provides a cost–effective approach to investigating off-target compound safety13,14. The traditional target prediction methods rely on chemical similarity search, where many studies use multi-target structure–activity relationship (SAR) models to retrieve putative targets of compounds. With the advent of the big data era, integrating artificial intelligence (AI) methods into this process offers further cost reduction opportunities. Mayr et al.15 employed data collected from ChEMBL to construct a series of supervised binary classification models, such as Random Forest (RF), K-Nearest Neighbor (KNN), and Deep Neural Network. In a recent study by Roche, researchers proposed a suite of off-target prediction models, including Neural Networks, RF, Auto-Sklearn, AutoGluon, and H2O. These models were assessed using a dataset of 4000 compounds from the company, enabling a thorough exploration and comparison of neural networks and machine learning methods in constructing off-target prediction models for 50 distinct targets, each with varying dataset sizes and imbalances16. Lunghini et al.17 introduced ProfhEX, a platform that utilizes tree-based Gradient Boosting (GB) and RF algorithms to establish prediction models for 46 off-targets. The platform also provides a comprehensive mechanistically-driven liability profile of small molecules.
Previous research related to compound safety has predominantly concentrated on singular aspects. Apart from off-target prediction, ADR prediction and toxicity prediction are also commonly employed for evaluating compound safety18. Various machine learning algorithms have been applied to ADR prediction, leveraging features such as drug phenotype, chemical and biological information, and target proteins to model the complex drug‒ADR relationships19, 20, 21. Liu et al. integrated phenotype, chemical and biological information of drugs and tested various classifiers such as Logistic Regression, Naive Bayes, KNN, RF and SVM for each ADR22. Zhang et al. modeled drugs and their side effects as a multi-label task, using various information such as chemical substructures, target proteins, and indications to represent drugs23. Zhang et al. constructed a KG containing four types of nodes (drug, indication, target, and side effect), and proposed a novel knowledge graph embedding method combined with a logistic regression classification model to predict whether a given drug has a certain ADR24. For compound toxicity prediction, computational toxicology, an emerging field, offers numerous models for large-scale virtual screening to identify candidates for subsequent experimental testing25, 26, 27. These models can be expert-designed, involving techniques like structural alerts28,29 or read-cross30, or they can be created automatically using machine learning techniques. While expert-designed rules provide some guidance for toxicity prediction, they often exhibit excessive sensitivity, leading to numerous false-positive outcomes. Machine learning methods primarily rely on quantitative structure–activity relationship (QSAR)31, which characterizes a drug's chemical structure and combines it with relevant supervised learning algorithms such as RF, XGBoost, and SVM, among others, for toxicity modeling8,25,32. Hao et al.33 introduced the drug toxicity prediction model DTox which is based on Visible Neural Network (VNN), established a meaningful network structure through a hierarchical approach. Sharma et al.34 employed pre-trained SMILES embeddings as molecular representations in deep networks. This approach allows simultaneous learning of in vitro, in vivo, and clinical toxicity tasks, thereby enhancing toxicity prediction.
However, ADR and toxicity data are primarily derived from limited clinical sources, posing challenges to traditional prediction methods, especially those relying on marketed and clinical compound structures, which could exhibit poor generalizability. Moreover, considering the inherent link between drug off-target effects and ADRs, as well as toxicity3,35, characterizing the compound's off-targets becomes a critical determinant of its safety. In light of this, we propose predicting a drug's off-target profile and utilizing it as a compound representation for subsequent tasks, including ATC classification, toxicity prediction, and ADR enrichment analysis. Initially, using a comprehensive compound–protein interaction database, we construct a multi-task graph neural network model to predict compounds' off-target profiles based on their chemical structures and compare it with several previous off-target prediction models to demonstrate its performance. The off-target prediction results for any given molecule can then serve as a molecular representation, capturing the molecule's off-target and subsequent ADR or toxicity effects. We explored the use of these representations in drug ATC classification and toxicity prediction, comparing their performance with that of ECFP-based models to showcase their effectiveness. Furthermore, ADR enrichment analysis is employed to leverage the off-target profile, identifying crucial ADRs at the target level, particularly severe ADRs leading to drug withdrawal. Using Pergolide as a case study, we predicted its off-target profile and subsequently utilized the off-target representation to elucidate its ADR mechanisms, attempting to provide the potential explanations for drug‒target‒ADR correlations of Pergolide. Hence, initiating from the molecular structures, we can obtain the molecules' off-target representations, which provide valuable information for safety-related prediction tasks. This early safety assessment protocol can steer a rational drug development process, facilitating the discovery of safe compounds.
2. Method
2.1. Collection and processing of compound–target interaction datasets
Initially, our project created an off-target panel consisting of 90 targets. More information about these targets can be found in Supporting Information Text S1 and Table S1. According to the gene names of the targets, we collected compounds associated with the corresponding targets from ChEMBL36 and PubChem37, following the steps outlined in Supporting Information Text S2. All databases used in our study are presented in Supporting Information Table S2. We eliminated experiment indicators with insufficient data and mainly retained the following six indicators: Ki, Kd, IC50, EC50, %activity, and %inhibition. To classify compounds under a specific target as active or inactive, we applied the threshold settings introduced in the Illuminating the Druggable Genome (IDG) project (Supporting Information Table S3)38,39. To facilitate subsequent training needs, we merged the data obtained from ChEMBL and PubChem and only selected targets containing at least 10 positive compounds. In total, 242 multi-species targets were retained, including 90 human targets, screening against approximately 320,000 unique compounds. The statistic of the processed data is shown in Table 1.
Table 1.
The data volume corresponding to each target type before and after negative sampling.
| GPCR | Ion channel | Enzyme | Kinase | Transporter | Nuclear receptor | Others | ||
|---|---|---|---|---|---|---|---|---|
| Number of targets | 146 (49 human) | 33 (16 human) | 27 (9 human) | 10 (6 human) | 11 (4 human) | 10 (4 human) | 5 (2 human) | |
| Before negative sampling | Positive | 114 k | 23 k | 19 k | 9 k | 20 k | 10 k | 6 k |
| Negative | 177 k | 21 k | 47 k | 15 k | 11 k | 17 k | 2 k | |
| Pos: Neg | 0.64 | 1.08 | 0.40 | 0.62 | 1.69 | 0.58 | 2.68 | |
| After negative sampling | Positive | 114 k | 23 k | 19 k | 9 k | 20 k | 10 k | 6 k |
| Negative | 500 k | 130 k | 103 k | 42 k | 100 k | 46 k | 34 k | |
| Pos: Neg | 0.23 | 0.18 | 0.18 | 0.22 | 0.20 | 0.22 | 0.20 | |
To expose the model to a larger number of negative samples and facilitate a more comprehensive understanding of the negative sample space40, mitigate false positives and enhance the model's usability, we employed a negative sampling approach to augment the negative samples. Under each target class, we selected compounds as negative sampled decoys based on their similarity to positive samples in terms of physical and chemical properties and molecular fingerprint similarity of less than 0.6. The sampling ratio differed among target classes to maintain a positive-to-negative compound ratio of approximately 1:5 in the data after negative sampling. This ensured that the model had enough exposure to negative samples. The data amounts after negative sampling are detailed in Table 1. Supporting Information Fig. S1 depicts the chemical spatial distribution of the sampled decoys and positive molecules under different target classes. The similarity in chemical spatial distribution indicates the reasonableness of the sampling approach. Employing these decoys, we aimed to demonstrate the model's ability to distinguish between the two molecule types based on their structural characteristics, rather than solely considering their physical and chemical properties41.
The compound–target interaction data division used in this project adopts the drug-blind mode, which does not consider the target and groups all small molecules together for classification. We performed random stratified partitioning with a ratio of 0.1 to obtain the test set first and then performed five-fold cross-validation on the remaining data to obtain train and validation datasets for each fold. We then trained and validated the model five times and evaluated the model by computing the average of the results obtained from the five experiments.
2.2. Construction of multi-task GNN models
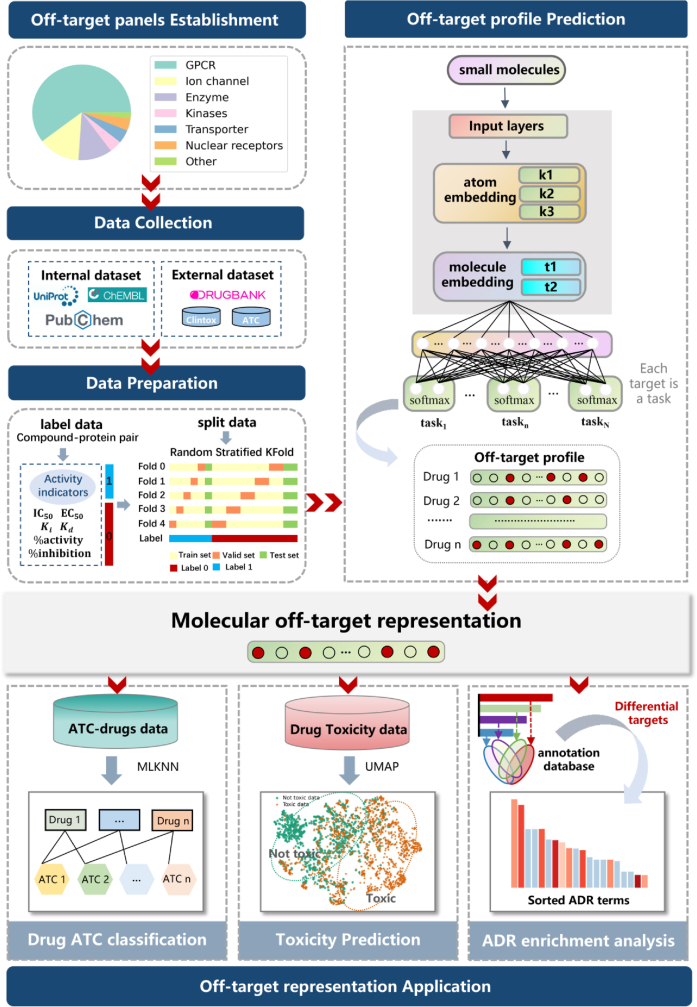
Multitask learning is a machine learning approach that utilizes a shared representation, training multiple related tasks simultaneously42,43. We employ a hard parameter sharing GNN model for multitask learning. Throughout the training process, the tasks share common hidden layers and hyperparameters, with a distinct separation occurring into individual tasks at the output of the network's fully connected layer, as depicted in Fig. 144. The model incorporates implicit data augmentation by exploiting similar structural features of molecules45. This approach enhances the model's focus on influential features, as evidence from other tasks is effectively utilized.
Figure 1.
Illustration of off-target prediction model for drugs and the utilization of the off-target representation.
A graph neural network, Attentive FP, is used for molecular representation, which leverages the graph attention mechanism to represent molecules and learn related tasks46. Specifically, we adopted the Attentive FP network architecture, based on DeepChem, for predicting drugs off-targets (Fig. 1). The network consists of three Attentive FP convolutional layers for extracting atomic features and two readout layers for extracting molecular features, culminating in a fully connected layer to produce prediction values. To reduce the bias caused by the imbalance label (negative samples are much more than positive samples), we used a weighted cross-entropy loss function, where the class weights were set as the inverse of class frequencies47. Early stopping and hyperparameter search strategy were used for model optimization. The hyperparameter search range of the multi-task GNN can be found in Supporting Information Table S4.
2.3. Execution of ADR enrichment analysis
The ADR enrichment analysis approach is essentially a bioinformatics-inspired strategy, similar to gene set enrichment analysis. Initially, we established ADR-targets mappings, creating an annotation database akin to gene ontology. The foundation for the pairs of ADR-targets was built on studies by Novartis48, Jeffrey et al.3 and other drug safety researchers1,49. These comprehensive connections between ADRs and off-targets were established through rigorous testing and analysis of commercially available drugs. Leveraging their data, we created a mapping linking each ADR with its related off-targets. To ensure precision in subsequent ADR enrichment analysis, ADRs corresponding to off-targets of fewer than 3 were excluded, and those with severity scores less than 0.1 were filtered to retain more hazardous ADRs. ADR severity scores ranging from 0 to 1 were obtained from Stanford's study, which ranked 2929 ADRs through crowdsourcing18. Consequently, we obtained 358 ADR terms associated with 193 off-targets (all included in our off-target panel). The process is detailed in Supporting Information Fig. S2. These ADR-targets mappings served as an annotation database like gene ontology (GO) used in gene enrichment analysis, providing prior information for ADR enrichment analysis.
The enrichment analysis utilized the hypergeometric distribution. For a given ADR, the probability that predicted off-targets for a drug are included in the ADR-related off-target set is calculated by Eq. (1):
| (1) |
where N represents the total number of targets in the annotation database (193 targets); n denotes the number of predicted positive targets for a drug, and K denotes the number of targets belonging to the specific ADR term, of which, k is predicted as positive targets for the drug. In essence, this equation signifies the probability that the predicted off-target profile for a drug is enriched in a specific ADR. For enrichment analysis, P-values are calculated based on the binomial approximation of the hypergeometric distribution, with Bonferroni correction and multiple hypothesis testing performed by FDR adjustment50,51 (See Supporting Information Text S3 for detailed instructions). ADR enrichment analysis was conducted using the enrichr function in the GSEApy Python package (version 1.0.6).
2.4. Evaluation metrics
The classification model's performance is assessed using AUROC, Balanced Accuracy (BACC), Matthews Correlation Coefficient (MCC), and F1 score—critical indicators for evaluating classifiers in imbalanced data scenarios. These metrics are computed from the confusion matrix: True Positives (TP), False Positives (FP), False Negatives (FN), and True Negatives (TN). Specifically, BACC, MCC, and F1 are calculated using Eqs. (2), (3), (4) respectively. The Area Under the Receiver Operating Characteristic Curve (AUROC) is the area beneath the ROC curve, which consists of the True Positive Rate (TPR) (Eq. (5)) against the False Positive Rate (FPR) (Eq. (6)) at different thresholds, the AUROC can be calculated using Eq. (7).
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
For multi-label learning, two prominent ranking-based metrics hold significance: Mean Average Precision (mAP) and Rank Loss. mAP, particularly, functions as an indicator of the ranking quality reflected in prediction outcomes. In the context of C classes of labels, mAP is calculated as the average of the Average Precision (AP) across all classes (Eq. (8)), where AP is determined by calculating the area under the Precision-Recall curve for each class i (Eq. (9)).
| (8) |
| (9) |
where Precision and Recall are computed at each threshold k, i.e., a specific point along the Precision-Recall curve; n is the number of thresholds, i.e., the total number of positive instances.
Rank Loss is a metric employed in multi-label classification to quantify the quality of the ranking assigned to positive labels for each instance. The calculation involves sorting the predicted probabilities of positive labels in descending order for each instance, forming a ranked list. Subsequently, the algorithm counts the number of inversions, representing instances where the predicted ranking contradicts the true ranking of positive labels. This count is then divided by the total number of possible pairs of positive labels, calculated as Eq. (10):
| (10) |
where the total number of possible pairs is calculated as , and n is the number of positive labels for the instance. The process is repeated for all instances, and the average Rank Loss is determined by averaging the obtained values. A lower Rank Loss value signifies superior performance, with an optimal score of 0, indicating perfect agreement between predicted and true rankings.
3. Results and discussion
3.1. The overall workflow of drug safety analysis based on off-target prediction
The workflow of drug safety analysis based on off-target prediction is shown in Fig. 1. Initially, we constructed an off-target panel and curated corresponding target–compound interaction data. Based on these data, we built 7 ligand-based off-target prediction models through multi-task GNNs, corresponding to 7 target families, i.e., GPCR, ion channel, enzyme, kinase, transporter, nuclear receptor and others. Consequently, the binding probabilities against the off-target panel can be obtained for each compound. The predicted off-target profiles can then be employed as molecular representations for the subsequent classification of a drug's ATC, toxicity, as well as ADR enrichment analysis.
3.2. Classification performance of off-target prediction models
We constructed multi-task GNNs (MTGNN) for the ligand-based prediction of off-target profiles. The panel for off-target analysis comprises 90 protein targets of Homo sapiens collected from the previous research1,12,13, comprising 50 GPCR targets, 16 ion channel targets, 9 enzyme targets, 6 kinase targets, 4 nuclear receptor targets, 4 transporter targets and 2 other targets. Subsequently, seven distinct multi-task GNN models were created, one for each of the seven target families. The multi-task strategy was employed to leverage shared information among tasks within each target family44 resulting in an enhancement of model quality and robustness, while simultaneously preventing cross-family negative transfer. Additionally, corresponding protein targets from other species were included in the training data to improve the multitask model, as these targets could serve as valuable sources of inductive bias. The hyperparameter values for these seven multi-task GNN models were individually optimized through grid searching (see Supporting Information Table S5). As a result, for each compound, the interaction probabilities with the 242 targets can be inferred from these seven multi-task GNNs (see Method; the information for the off-targets can be found in Table S1).
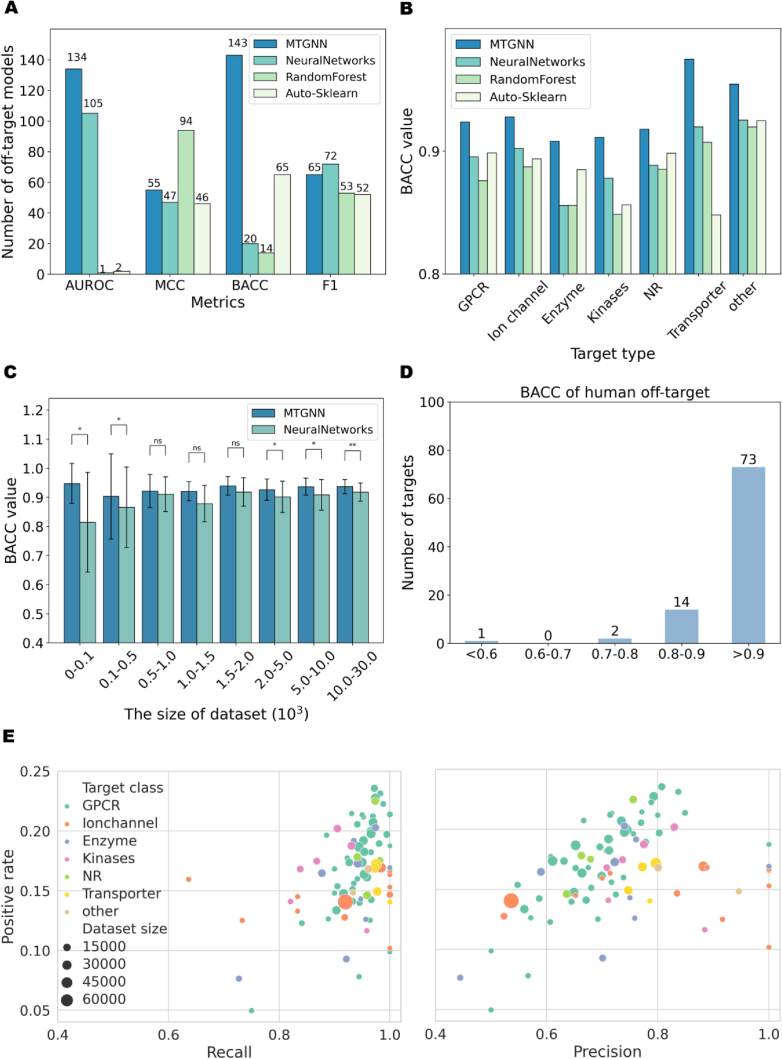
We compare the performance of our off-target prediction MTGNN model with Roche's off-target prediction methods, including NeuralNetworks, RandomForest, and Auto-Sklearn. Detailed information on the models and their parameter settings can be found in Roche's work16 and Supporting Information Table S6. Fig. 2A provides an overview of the highest-scoring models based on AUROC, BACC, MCC, and F1 scores. In terms of AUROC and BACC, MTGNN scored higher than the other models for 134 and 143 targets, respectively. In terms of MCC, MTGNN outperformed NeuralNetworks and Auto-Sklearn. Regarding F1 score, RandomForest had the best MCC for up to 94 targets, but its overall performance was suboptimal due to poor AUROC and BACC. Regarding F1 score, MTGNN outperformed RandomForest and Auto-Sklearn but was inferior to NeuralNetworks. Given the importance of distinguishing true positives and true negatives in off-target modeling, we considered that BACC serves as the primary evaluation indicator16. As can be observed from the BACC results in Fig. 2B, MTGNN demonstrates the highest average BACC than the other three models for each target family. Furthermore, when considering the average performance for all targets together while disregarding their target type, MTGNN outperformed all Roche's off-target models in all indicators (Supporting Information Fig. S3). Further details and evaluation metric values for these methods under each target type are available in Supporting Information Table S7.
Figure 2.
Performance comparison among MTGNN, NeuralNetworks, RandomForest, and Auto-Sklearn in constructing off-target models. (A) A bar chart compares the number of tasks (y-axis) corresponding to the maximum scores achieved by each method on AUROC, MCC, BACC, and F1 metrics. The number above each bar indicates the tasks with the highest score for that method. (B) Average performance, measured by BACC, is depicted for the seven types of target models under four off-target prediction models. (C) The performance of MTGNN and Neural Networks in tasks with different data volumes. The bar chart shows the average BACC (y-axis) for tasks with corresponding data volumes (x-axis). ManneWhitney U test is used to test for significant differences, where: ns indicates no significant difference; 0.01<∗P < 0.05; 0.001<∗∗P < 0.01. (D) The histogram of the number of human target tasks (y-axis) corresponding to different interval ranges (x-axis) of BACC. (E) Scatter plots depict Recall and Precision values for human target tasks. Each color represents a different target type, and dot size corresponds to the amount of available data for that target, with larger dots indicating larger datasets. The y-axis represents the positive rate of the overall data volume, while the x-axis represents the respective indicator value.
In the specific context of this study, it is evident that MTGNN exhibits superior overall performance, thus positioning it as a top-ranked method. The robust predictive capacity of MTGNN is attributed to its integration of domain information from related tasks, thereby enhancing its performance on tasks characterized by limited data availability. As illustrated in Fig. 2C, MTGNN consistently outperforms Neural Networks across a range of dataset sizes, which was identified as the best-performing model by Roche. This is particularly pronounced when dealing with data insufficiency, as seen in the dataset size intervals of [1, 100] and [100, 500]. This observation is of paramount significance for the extension of target range research aimed at broad and comprehensive predictions of off-target effects.
It's noteworthy that experts advocate the utilization of human targets, rather than animal homologs, in constructing off-target panel for predicting ADRs in humans1. Thus, we focus on the predicted outcomes for 90 human targets. As depicted in Fig. 2D, these human targets consistently exhibit robust classification performance, with a significant majority achieving BACC scores exceeding 0.7. In Fig. 2E–a detailed exploration of precision and recall metrics for each of the 90 human targets reveals high recall values (>0.8) for most tasks, coupled with moderate precision levels ranging from 0.4 to 0.6. Despite the low positive rate (<25%) for these tasks due to augmented negative samples, the models maintain their sensitivity to potentially unsafe compound–target interactions. Given the paramount importance of avoiding false negatives in early-stage drug development to prevent the oversight of unsafe molecules during safety assessments, our model holds considerable value in mitigating research and development failures arising from unsafe off-target interactions.
As the off-target prediction task essentially involves protein‒ligand interaction, we conducted a comparison between our off-target prediction models and benchmark models for drug–target interaction prediction, namely GraphDTA52, TransformerCPI53 and TransformerCPI2.054. To ensure an unbiased assessment, we retrained these benchmark models using our dataset, maintaining consistency in the training data. Each benchmark model utilized the original hyperparameters as provided in their respective publications, implemented an early stopping strategy, and underwent training until convergence. Our findings indicate that our multitask off-target prediction model outperforms or performs comparably to the baseline models across AUROC, MCC, F1, and BACC metrics (Supporting Information Table S8). Despite not incorporating protein information, our multitask off-target prediction model effectively learns patterns from off-target data containing sufficient compound structure information, enabling it to describe drug–target interactions under specific targets. Moreover, compared to drug–target interaction models, the multitask algorithm reduces the model's complexity, facilitating easier training and faster convergence.
3.3. Application of off-target prediction panel
The off-target prediction model offers the capability to infer interaction probabilities against 242 targets for any given molecule. This can be regarded as a 242-dimensional representation that characterizes the off-target-related molecular features. We explore the utilization of these representations in drug ATC classification, toxicity prediction and ADR enrichment analysis.
3.3.1. Drug ATC classification
The Anatomical Therapeutic Chemical (ATC) classification system, was proposed by the World Health Organization (WHO) in 1981 (https://www.whocc.no/atc/structure_and_principles/).
Researchers leverage the hierarchical structure of ATC codes as features to improve the performance of ADR prediction models. Drugs categorized under different ATC codes often manifest specific ADRs, influenced by their off-target profiles55.
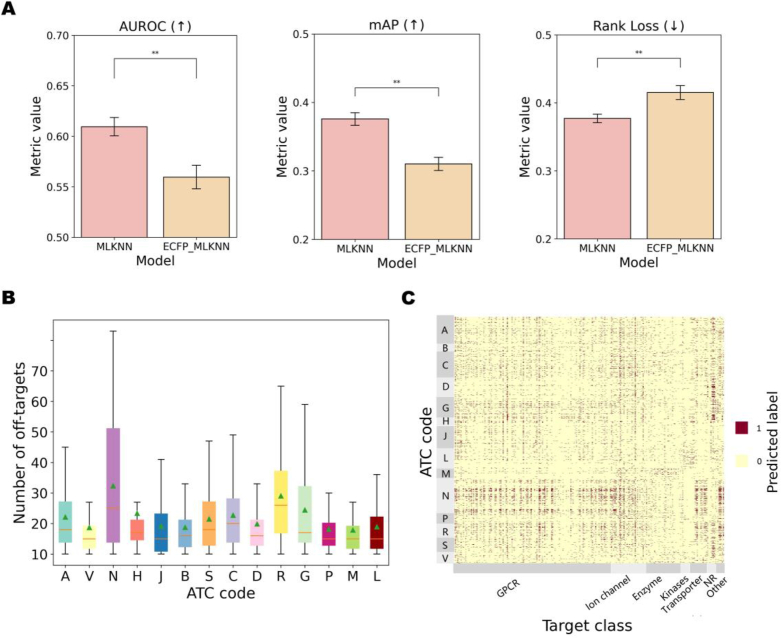
To assess how the off-target representation characterizes the ATC code, as described in Supporting Information Text S4, we modeled the ATC classification as a multi-label problem, where each compound corresponds to 14 ATC labels. From the ATC-SMILES dataset, a benchmark collection designed for the ATC classification task56, we curated a total of 3491 compounds spanning 14 categories. For precise counts of compounds under each ATC code, refer to Supporting Information Table S9. The impact of off-target representation on ATC classification was demonstrated through a comparative experiment involving two models, MLKNN and ECFP_MLKNN, using the compound's off-target representation and molecular fingerprint features (1024-dimensional ECFP4 fingerprint) as characteristics, respectively. Conducting five-fold cross-training and evaluation on the same test set revealed that MLKNN outperformed ECFP_MLKNN, as indicated by superior AUROC, mAP, and Rank Loss metrics (Fig. 3A, and Supporting Information Table S10). This underscores the efficacy of the off-target representation-based multi-label model in accurately ordering ATC codes for compounds, surpassing the performance of conventional molecular fingerprint features.
Figure 3.
The performance comparison of ATC classification models and the off-target prediction results analysis of different ATC codes compounds/drugs. (A) The bars depict performances of MLKNN and ECFP_MLKNN models, where higher AUROC and mAP indicate better model performance, and lower rank loss indicates superior performance. Different colored bars represent different models, and y-axis represents the mean metric values of the five-fold cross-training. ManneWhitney U test is used to test for significant differences, where: ns indicates no significant difference; 0.001< ∗∗P < 0.01. (B) A bar chart displays the number of binding off-targets (y-axis) for the 14 categories of compounds (x-axis). (C) The heat map showcases the off-target panel prediction results for all study compounds. ATC codes (A–V) are represented on the y-axis, while target points are on the x-axis. Dark colors (value of 1.0) indicate binding, and light colors (value of 0.0) indicate no binding.
It's noteworthy that compounds classified under “Nervous system (N)” manifest a higher frequency of off-target bindings (Fig. 3B), as depicted in Fig. 3C, in contrast to other drug categories, evident in denser and darker points on the heatmap. Prior investigations have highlighted that in drugs exhibiting fatal toxicity, 78.6% acted on the Nervous system (N)57. This occurrence can be attributed to the fact that many drugs in the N category often display pharmacological promiscuity, targeting GPCR receptors58,59, including adenosine receptors, acetylcholine receptors, serotonin receptors, along with potassium ion channels, voltage-gated sodium ion channels, and specific transporter targets60, 61, 62, 63. These characteristics are reflected in the off-target representation of these drugs.
3.3.2. Toxicity prediction
The off-target representation is considered a crucial feature for toxicity prediction, forming the basis for an off-target-based toxicity prediction approach applicable to any given drug. To conduct the experiment, we curated a drug toxicity dataset from different datasets: (1) The Clintox dataset, obtained from MoleculeNet64, comprises 108 toxic and 1365 non-toxic compounds after data cleaning. (2) From DrugBank and online resources, we collected 107 drugs withdrawn due to toxic side effects. (3) Onakpoya et al.65 compiled 462 drugs withdrawn globally for toxic side effects, resulting in 390 small molecules. (4) ChEMBL provided 865 compounds flagged with ‘Black Box Warnings’, identifying 505 toxic compounds. After merging and deduplication, the dataset includes 877 toxic (labeled as 1) and 1229 non-toxic compounds (labeled as 0).
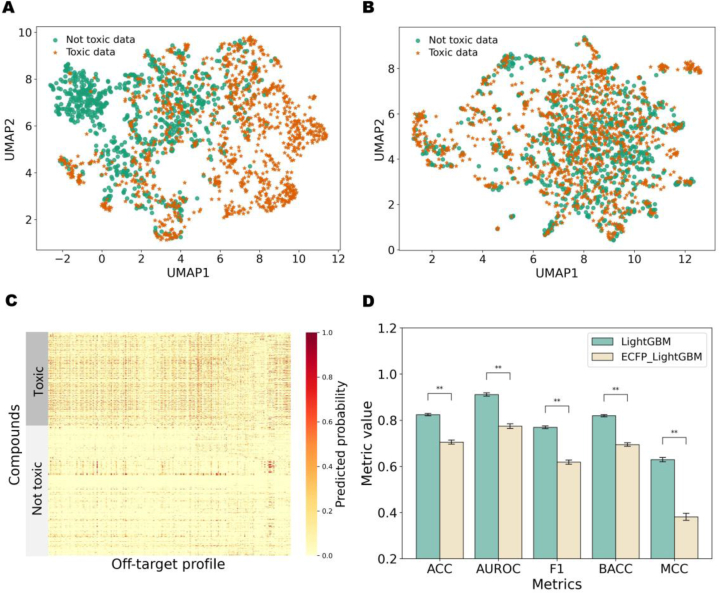
UMAP visualization was employed to illustrate the relationship between off-target representation and compound toxicity. As shown in Fig. 4A, the off-target representation exhibited clearer discrimination between safe and unsafe compounds, outperforming molecular fingerprint features (Fig. 4B). This observation may be attributed to the evidently fewer bound safety-related off-targets for most safe drugs, a phenomenon illustrated by the denser and darker points in the heatmap for toxic drugs in contrast to non-toxic drugs (Fig. 4C). Moreover, to gauge the impact of off-target and structural representation on the compound toxicity prediction model, we employed a toxicity classifier using LightGBM based on the off-target representation (Supporting Information Text S5). LightGBM demonstrated superior performance compared to various machine learning models (RF, SVM, and XGBoost). We also implemented ECFP_LightGBM, where the off-target representation was substituted with a molecular fingerprint feature (1024-dimensional ECFP4 fingerprint). The best hyperparameter settings for LightGBM and ECFP_LightGBM are shown in Supporting Information Table S11. Compared to LightGBM, the performance of ECFP_LightGBM exhibited a noticeable decline on the same test set (Fig. 4D, and Supporting Information Table S12). Moreover, we conducted a comparison with two recent toxicity prediction models, DTox33 and STDNN-SE34, which characterized compounds using MACCS fingerprints and pre-trained SMILES embeddings, respectively. Our findings indicate that LightGBM, based on off-target representations, outperformed on the same test dataset (Table S12). This further affirms the rationality and effectiveness of using our off-target panel predictions as representations for predicting compound toxicity-related properties.
Figure 4.
The visualization of toxic compounds and not toxic compounds and the performance comparison of toxicity prediction models. (A) UMAP plot representing off-target panel prediction results for Toxic and Non-toxic data. (B) UMAP plot of ECFP fingerprint characterization for Toxic data and Not toxic data. (C) Heat map displaying off-target panel prediction results fotextr Toxic and Non-toxic data. (D) The performance of LightGBM and ECFP_LightGBM for toxicity prediction. The bar chart shows the mean value of the five-fold cross-training (y-axis) under different metrics (x-axis). ManneWhitney U test is used to test for significant differences, where: ns indicates no significant difference; 0.001< ∗∗P < 0.01.
3.3.3. ADR enrichment analysis
The relationship between an Adverse Drug Reaction (ADR) and its corresponding off-target can be analogized to that of a biological pathway and its associated gene set. Upon obtaining the predicted off-target profile of a queried drug, potential ADRs can be inferred through enrichment analysis, employing an annotation database that correlates each ADR with its corresponding off-target set. Our study established mappings between 358 ADR terms and 193 off-targets, serving as an annotation database for prior information in ADR enrichment analysis, conducted using the hypergeometric distribution (refer to Method).
We conducted ADR enrichment analysis on four withdrawn drugs due to safety concerns—Pergolide, Phenylpropanolamine, Sibutramine, and Sertindole, evaluating the efficacy of the enriched ADRs compared to the known relevant ADRs of these drugs (Supporting Information Table S13). Positive off-target predictions for these drugs were obtained, analogous to “differential genes”, with prediction values exceeding 0.3 to broaden the scope of enriched ADRs beyond the conventional threshold of 0.5. Additional analyses employing various positive off-target thresholds of 0.2, 0.5 and 0.8 demonstrate that a threshold of 0.3 facilitates a more significant enrichment of ADRs (Supporting Information Fig. S4).
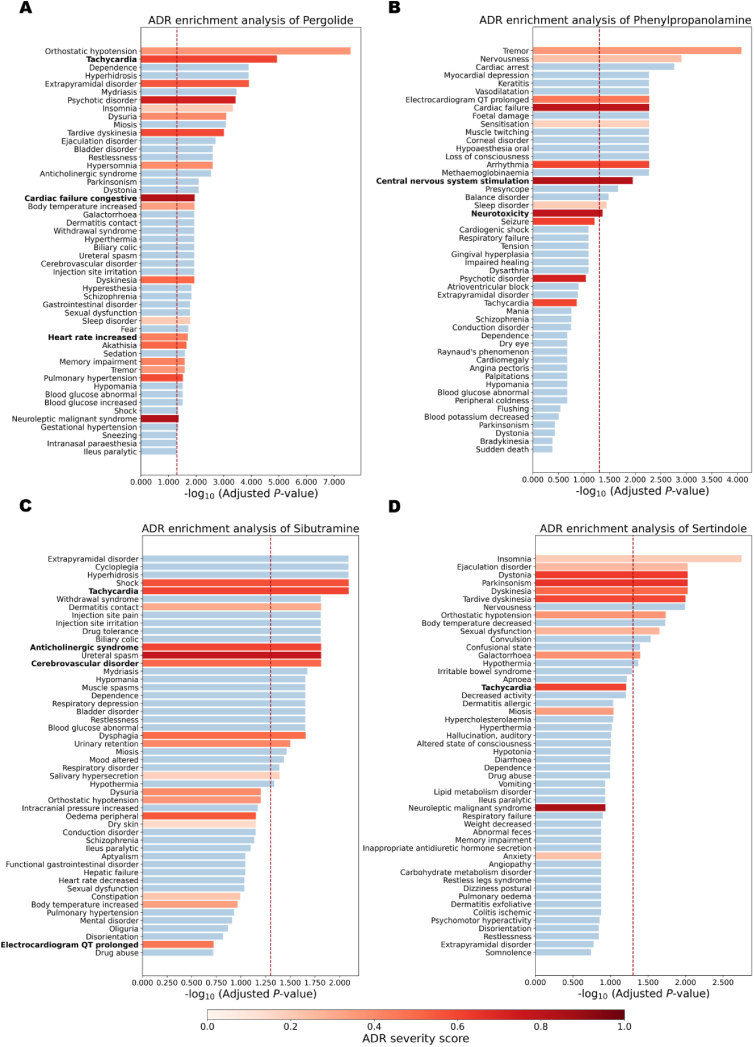
Fig. 5 illustrates the ranking of ADR enrichment analysis results for each drug based on Adjusted P-value. Detailed enrichment analysis ranking results for each drug can be found in Supporting Information Tables S14‒S17.
-
(1)
Pergolide is a dopamine receptor agonist commonly utilized in the treatment of Parkinson's disease and other conditions66, has been associated with an increased risk of cardiac valvulopathy, leading to its withdrawal from the US and Canadian markets in 2007. Among the 42 pertinent ADRs for Pergolide, 18 are significantly enriched (P < 0.05) out of a total of 358 ADRs (Fig. 5A). These include high-frequency ADR terms associated with pergolide such as Orthostatic hypotension (frequent, 9%), Extrapyramidal disorder (frequent, 1.6%), Insomnia (frequent, 7.9%), and Dyskinesia (frequent, 62.4%). Notably, consistent with Pergolide's withdrawal due to cardiotoxicity, several cardiotoxic-related ADR terms were significantly enriched, including Tachycardia, Cardiac failure congestive, and Heart rate increased.
-
(2)
Similarly, phenylpropanolamine has 25 relevant ADRs, and 9 of them are significantly enriched (Fig. 5B). Neurotoxicity and Central nervous system stimulation, two significantly enriched ADR terms, are associated with hemorrhagic stroke67, 68, 69, the primary reason for the withdrawal of Phenylpropanolamine from the market. Apart from its impacts on the nervous system, Phenylpropanolamine also induced a series of cardiac side effects70, including significantly enriched Arrhythmia, Cardiac failure, and Prolonged electrocardiogram QT. The recognition of critical and potentially fatal adverse drug reactions, such as prolonged Electrocardiogram QT, in the early stages of drug discovery is imperative for prioritizing human safety and identifying potential risks.
-
(3)
Sibutramine was withdrawn from the Canadian and U.S. markets due to the increased risk of heart attacks and strokes in patients with a history of heart disease. It is associated with 35 known ADRs, and the enrichment results reveal 9 significant ADRs among them (Fig. 5C). Notably, Tachycardia, Anticholinergic syndrome, and Cerebrovascular disorder71,72, which are related to its withdrawal, are significantly enriched.
-
(4)
Sertindole, withdrawn from the market due to cardiotoxicity, did not show significant enrichment in cardiotoxic-related Tachycardia. However, 9 out of 25 ADRs associated with this drug were significantly enriched, with all of the top six enrichment results being known ADRs of this drug (Fig. 5D). These examples underscore the effectiveness of off-target prediction results in enriching relevant ADRs, particularly severe ADRs leading to drug withdrawal, thereby highlighting the validity of off-target representation in characterizing ADRs.
Figure 5.
The ADR enrichment analysis results of four drugs. The graph illustrates the top 50 significant ADR terms, with the red dotted line representing the position where the P-value is 0.05. Known literature-reported drug-ADR associations are highlighted in red, and the intensity of the color reflects the severity score of the corresponding ADR, with darker shades indicating higher severity scores. The bars present the top 50 ADR terms (y-axis) and their corresponding enrichment results (x-axis) for each drug—Pergolide (A), Phenylpropanolamine (B), Sibutramine (C), and Sertindole (D).
Additionally, ML-based models for ADR prediction were established and compared with ADR enrichment analysis. As described in Supporting Information Text S6, the off-target profiles or ECFP are used as drug features to train MLKNN models and RF models for ADR prediction. Among these models, the off-target-based MLKNN model showcased an optimal AUROC of 0.905 (Supporting Information Table S18), and thus, the ADRs of the aforementioned four drugs were predicted based on it. The classification results indicate that among the 42 ADRs related to Pergolide, 11 were identified; for Phenylpropanolamine, 5 out of 25 ADRs were identified; for Sibutramine, 8 out of 35 ADRs were identified; and for Sertindole, 6 out of 25 ADRs were identified. The fewer ADRs identified by the off-target-based MLKNN model compared with ADR enrichment analysis indicates that the latter is more effective, possibly due to sparse training data causing difficulties in ML model training and potential overfitting. Therefore, ADR enrichment analysis is theoretically more reasonable and provides more accurate results than ADR prediction ML modeling.
3.4. Drug–target–ADR networks
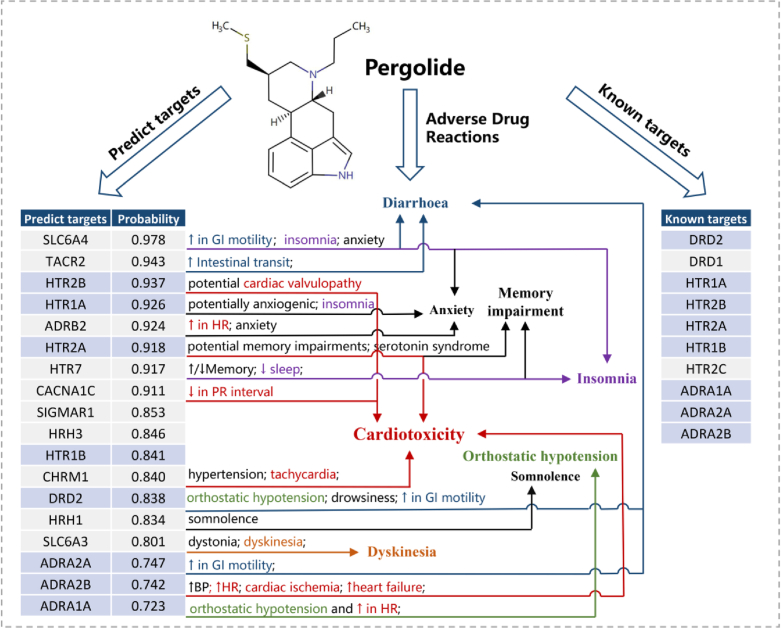
Through off-target prediction and subsequent ADR enrichment analysis, we can establish correlations between a drug's off-targets and the corresponding ADRs, providing an off-target-based explanation for ADRs. Using Pergolide as a case study, we predicted its off-target profile and correlated the known ADRs of the drugs with the predicted off-targets to create a drug‒target‒ADR correlation network.
Firstly, the accurate prediction of Pergolide's off-target profile was achieved (known off-targets are obtained from databases such as ChEMBL, PubChem and DrugBank, overlapping with our off-target panel). Fig. 6 demonstrates that out of the 10 known targets of Pergolide predictable by the off-target model, 8 were appropriately included in the predicted off-target profile. Subsequently, using the off-target representation, the ADR enrichment analysis correctly identified its crucial ADRs related to Cardiotoxicity, Orthostatic hypotension, Insomnia, etc. (see ADR enrichment analysis). Furthermore, the off-target model indicated the presence of additional potential off-targets related to Pergolide's ADRs, providing the potential explanation for its drug‒target‒ADR correlation. For instance, cardiac-related toxicities associated with Pergolide were linked to both its known targets (HTR2A, ADRA2B, and HTR2B73) and predicted targets (CHRM1 and CACNA1C). Diarrhoea, a prevalent side effect of Pergolide, can be ascribed not only to Pergolide's known target ADRA2A but also possibly to the predicted targets SLC6A474 and TACR275. For Insomnia, besides the known target HTR1A of Pergolide, two predicted new targets, SLC6A476 and HTR777, were correlated with this ADR. Similarly, the drug–target–network diagram for Sertindole can be found in Supporting Information Fig. S5, and the relationship between side effects and targets can be analyzed in a similar manner as for Pergolide (Supporting Information Text S7).
Figure 6.
Drug‒target‒ADR association diagram for Pergolide. The left table enumerates the known targets of the drug, while the right table lists the predicted off-targets along with their respective probability values. Targets with a blue background represent overlapping targets between predicted and known targets. Side effect descriptions associated with each target are provided adjacent to them, with arrows indicating the corresponding ADRs of Pergolide. Colored ADRs are linked to predicted new targets. (e.g., Cardiotoxicity is marked in red, corresponding to “↑ in HR”, “tachycardia”, “↑heart failure” and so on in the target's side effect description; Insomnia is marked in purple, corresponding to “insomnia” and “↓sleep” in the target's side effect description). BP: blood pressure; HR: heart rate; GI: glycemic index; PR interval: the time from the beginning of the P wave (atrial depolarization) to the beginning of the QRS complex (ventricular depolarization).
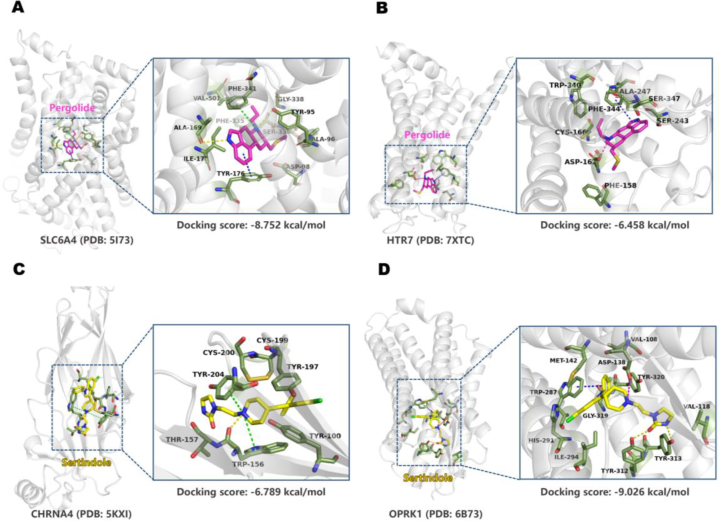
Based on the predicted off-target profiles for Pergolide and Sertindole, we conducted molecular docking for each drug's newly predicted targets, following the outlined procedure in Supporting Information Text S8. The binding modes of the two drugs with their corresponding targets are illustrated in Fig. 7. Molecular docking of Pergolide with SLC6A4 and HTR7, and Sertindole with CHRNA4 and OPRK1, revealed substantial binding energies. The strongest binding occurred within the Sertindole‒OPRK1 complex, with a binding energy of −9.026 kcal/mol, indicating a robust interaction. Specifically, key interactions were observed in these complexes, suggesting a close and potentially highly active interaction between the drugs and the targets.
Figure 7.
Molecular docking-predicted binding mode. (A) Molecule docking of Pergolide binding to SLC6A4 (PDB: 5I73). (B) Molecule docking of Pergolide binding to HTR7 (PDB: 7XTC). (C) Molecule docking of Sertindole binding to CHRNA4 (PDB: 5KXI). (D) Molecule docking of Sertindole binding to OPRK1 (PDB: 6B73). Left panel: overall view; right panel: partial view; magenta stick: Pergolide; yellow stick: Sertindole; light gray cartoon: protein; yellow dotted line: hydrogen bond; blue dotted line: pi–pi stacking; green dotted line: pi‒cation; magenta dotted line: salt bridge.
In the SLC6A4 protein, crucial hydrogen bonds and hydrophobic interactions played a significant role in securing the attachment of small molecules to proteins. Pergolide formed key hydrogen bonds with TYR9578 and ALA169 on the helix. The indole moiety exhibited a pi–pi stacking interaction with TYR176, and the quinoline ring's nitrogen cation interacts with PHE341 through pi–cation interactions78. Additionally, valuable hydrophobic interactions occurred79 (Fig. 7A). In the HTR7 protein Pergolide forms a pi–pi stacking interaction with PHE344 and establishes a salt bridge with ASP16280. Moreover, valuable hydrophobic interactions further strengthen the binding of the molecule to the HTR7 protein (Fig. 7B).
In the CHRNA4‒Sertindole complex, nitrogen cations on the pyridine ring formed pi–cation interactions with TPR156 and TYR20481, and it also formed a hydrogen bond interaction with TPR156 (Fig. 7C). In the OPRK1 protein, Sertindole bound within the helical structure, forming hydrogen bonds with TYR312 through the carbonyl group on the imidazole ring and with TYR313 through the nitrogen on the imidazole ring82. It also exhibited pi–pi stacking interaction with TRP287 and salt-bridge interaction with ASP138 (Fig. 7D).
In summary, the molecular docking analysis for predicted off-targets of Pergolide and Sertindole unveiled robust binding energies and essential molecular interactions within the respective protein complexes, confirming the high prediction accuracy of our model in identifying potential off-targets. This approach, combining off-target prediction and subsequent ADR enrichment analysis, facilitates the establishment of an off-target-based explanation for ADRs.
3.5. Code availability
All data and scripts to build the models are available in the GitHub repository: https://github.com/myzhengSIMM/Offtarget_drugsafety.
4. Conclusions
Off-target interactions frequently occur with drug usage and are a major cause of drug side effects and candidate failure during drug discovery. We employed a multi-task GNN to accurately predict these off-target interactions derived from molecular graphs. These predictions were then utilized to comprehensively assess drug safety from multiple aspects, including ATC catalogs, toxicity, and ADRs, providing a valuable supplement to traditional, time-consuming, and labor-intensive safety pharmacology experiments. Notably, in ADR enrichment analysis, based on differential targets for each drug derived from off-target prediction results, severe ADRs leading to drug withdrawal were significantly enriched in our cases, further illustrating the effectiveness of the off-target panel for drug safety assessment.
One limitation of our study lies in the reliance on ligand similarity for off-target prediction, excluding protein-related information. The variability in predictive performance across different protein targets and families indicated potential disparities driven by data or biological factors. Therefore, incorporating protein-related insights into off-target predictions holds promise for enhancing the accuracy of our predictions. Additionally, it's important to note that our model is not universally applicable for off-target prediction, and re-modeling may be necessary when the number of targets changes. Inspired by the application of pre-training and transfer learning in the field of medicine83, 84, 85, we consider building a general paradigm for off-target panel prediction by simultaneously incorporating information from both proteins and compounds in future work. In terms of safety assessment, the free plasma concentration of drugs significantly impacts adverse reactions and drug safety3,86,87. Compounds predicted to have multiple off-target bindings pose lower risks when they have a low free plasma concentration. Conversely, compounds with fewer target bindings but high free plasma concentrations can be more hazardous13. Hence, evaluating drug safety and explaining ADRs should consider the free plasma concentration during therapeutic use. It's noteworthy that drugs used to treat severe, refractory diseases might result in more frequent and severe side effects, which are deemed acceptable32.
Overall, our work aims to predict drug off-target interactions to assess drug safety. Utilize the compound's off-target representation to deduce ATC catalogs, toxicity, and ADR offers a valuable framework and methodology for the preclinical identification of compound toxicity. Future steps include expanding the off-target panel, optimizing off-target and ADR prediction models, and refining the safety prediction model to contribute to the development of safer pharmaceuticals.
Author contributions
Xutong Li and Mingyue Zheng conceived the project. Jin Liu implemented the off-target prediction model and conducted the computational analysis. Yike Gui, Jingxin Rao, Jingjing Sun, Gang Wang, Qun Ren, Ning Qu, Buying Niu, Zhiyi Chen, Xia Sheng and Yitian Wang collected and analyzed the data. Jin Liu, Yike Gui and Jingxin Rao wrote the paper. All authors discussed the results and commented on the manuscript.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
This work was supported by National Key Research and Development Program of China (2022YFC3400504 to Mingyue Zheng), National Natural Science Foundation of China (T2225002 and 82273855 to Mingyue Zheng, 82204278 to Xutong Li), Lingang Laboratory (LG202102-01-02 to Mingyue Zheng), SIMM-SHUTCM Traditional Chinese Medicine Innovation Joint Research Program (E2G805H to Mingyue Zheng), and Shanghai Municipal Science and Technology Major Project.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2024.03.002.
Contributor Information
Mingyue Zheng, Email: myzheng@simm.ac.cn.
Xutong Li, Email: lixutong@simm.ac.cn.
Appendix A. Supporting information
The following is the Supporting information to this article:
References
- 1.Bowes J., Brown A.J., Hamon J., Jarolimek W., Sridhar A., Waldron G., et al. Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat Rev Drug Discov. 2012;11:909–922. doi: 10.1038/nrd3845. [DOI] [PubMed] [Google Scholar]
- 2.Whitebread S., Hamon J., Bojanic D., Urban L. Keynote review: in vitro safety pharmacology profiling: an essential tool for successful drug development. Drug Discov Today. 2005;10:1421–1433. doi: 10.1016/S1359-6446(05)03632-9. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland J.J., Yonchev D., Fekete A., Urban L. A preclinical secondary pharmacology resource illuminates target-adverse drug reaction associations of marketed drugs. Nat Commun. 2023;14:4323. doi: 10.1038/s41467-023-40064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun D., Gao W., Hu H., Zhou S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm Sin B. 2022;12:3049–3062. doi: 10.1016/j.apsb.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hao Y., Moore J.H. TargetTox: a feature selection pipeline for identifying predictive targets associated with drug toxicity. J Chem Inf Model. 2021;61:5386–5394. doi: 10.1021/acs.jcim.1c00733. [DOI] [PubMed] [Google Scholar]
- 6.Edwards I.R., Aronson J.K. Adverse drug reactions: definitions, diagnosis, and management. Lancet. 2000;356:1255–1259. doi: 10.1016/S0140-6736(00)02799-9. [DOI] [PubMed] [Google Scholar]
- 7.Aronson J.K., Ferner R.E. Joining the DoTS: new approach to classifying adverse drug reactions. BMJ. 2003;327:1222. doi: 10.1136/bmj.327.7425.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.John L., Mahanta H.J., Soujanya Y., Sastry G.N. Assessing machine learning approaches for predicting failures of investigational drug candidates during clinical trials. Comput Biol Med. 2023;153 doi: 10.1016/j.compbiomed.2022.106494. [DOI] [PubMed] [Google Scholar]
- 9.Siramshetty V.B., Nickel J., Omieczynski C., Gohlke B.O., Drwal M.N., Preissner R. WITHDRAWN‒a resource for withdrawn and discontinued drugs. Nucleic Acids Res. 2016;44:D1080–D1086. doi: 10.1093/nar/gkv1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vo A.H., Van Vleet T.R., Gupta R.R., Liguori M.J., Rao M.S. An overview of machine learning and big data for drug toxicity evaluation. Chem Res Toxicol. 2020;33:20–37. doi: 10.1021/acs.chemrestox.9b00227. [DOI] [PubMed] [Google Scholar]
- 11.Cook D., Brown D., Alexander R., March R., Morgan P., Satterthwaite G., et al. Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat Rev Drug Discov. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 12.Lynch J.J., 3rd, Van Vleet T.R., Mittelstadt S.W., Blomme E.A.G. Potential functional and pathological side effects related to off-target pharmacological activity. J Pharmacol Toxicol Methods. 2017;87:108–126. doi: 10.1016/j.vascn.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Bendels S., Bissantz C., Fasching B., Gerebtzoff G., Guba W., Kansy M., et al. Safety screening in early drug discovery: an optimized assay panel. J Pharmacol Toxicol Methods. 2019;99 doi: 10.1016/j.vascn.2019.106609. [DOI] [PubMed] [Google Scholar]
- 14.Li Y., Fan Z., Rao J., Chen Z., Chu Q., Zheng M., et al. An overview of recent advances and challenges in predicting compound–protein interaction (CPI) Mediev Rev (2021) 2023;(3):465–486. doi: 10.1515/mr-2023-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayr A., Klambauer G., Unterthiner T., Steijaert M., Wegner J.K., Ceulemans H., et al. Large-scale comparison of machine learning methods for drug target prediction on ChEMBL. Chem Sci. 2018;9:5441–5451. doi: 10.1039/c8sc00148k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naga D., Muster W., Musvasva E., Ecker G.F. Off-targetP ML: an open source machine learning framework for off-target panel safety assessment of small molecules. J Cheminf. 2022;14:27. doi: 10.1186/s13321-022-00603-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lunghini F., Fava A., Pisapia V., Sacco F., Iaconis D., Beccari A.R. ProfhEX: AI-based platform for small molecules liability profiling. J Cheminf. 2023;15:60. doi: 10.1186/s13321-023-00728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb A., Hoehndorf R., Dumontier M., Altman R.B. Ranking adverse drug reactions with crowdsourcing. J Med Internet Res. 2015;17:e80. doi: 10.2196/jmir.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ietswaart R., Arat S., Chen A.X., Farahmand S., Kim B., DuMouchel W., et al. Machine learning guided association of adverse drug reactions with in vitro target-based pharmacology. EBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Nueno V.I., Souchet M., Karaboga A.S., Ritchie D.W. GESSE: predicting drug side effects from drug‒target relationships. J Chem Inf Model. 2015;55:1804–1823. doi: 10.1021/acs.jcim.5b00120. [DOI] [PubMed] [Google Scholar]
- 21.Dey S., Luo H., Fokoue A., Hu J., Zhang P. Predicting adverse drug reactions through interpretable deep learning framework. BMC Bioinf. 2018;19:476. doi: 10.1186/s12859-018-2544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M., Wu Y., Chen Y., Sun J., Zhao Z., Chen X.W., et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inf Assoc. 2012;19:e28–e35. doi: 10.1136/amiajnl-2011-000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W., Liu F., Luo L., Zhang J. Predicting drug side effects by multi-label learning and ensemble learning. BMC Bioinf. 2015;16:365. doi: 10.1186/s12859-015-0774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F., Sun B., Diao X., Zhao W., Shu T. Prediction of adverse drug reactions based on knowledge graph embedding. BMC Med Inf Decis Making. 2021;21:38. doi: 10.1186/s12911-021-01402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemmerich J., Ecker G.F. In silico toxicology: from structure‒activity relationships towards deep learning and adverse outcome pathways. Wiley Interdiscip Rev Comput Mol Sci. 2020;10:e1475. doi: 10.1002/wcms.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raies A.B., Bajic V.B. In silico toxicology: computational methods for the prediction of chemical toxicity. Wiley Interdiscip Rev Comput Mol Sci. 2016;6:147–172. doi: 10.1002/wcms.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo Y.C., Rensi S.E., Torng W., Altman R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov Today. 2018;23:1538–1546. doi: 10.1016/j.drudis.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen T.E.H., Goodman J.M., Gutsell S., Russell P.J. Using 2D structural alerts to define chemical categories for molecular initiating events. Toxicol Sci. 2018;165:213–223. doi: 10.1093/toxsci/kfy144. [DOI] [PubMed] [Google Scholar]
- 29.Metivier J.P., Lepailleur A., Buzmakov A., Poezevara G., Cremilleux B., Kuznetsov S.O., et al. Discovering structural alerts for mutagenicity using stable emerging molecular patterns. J Chem Inf Model. 2015;55:925–940. doi: 10.1021/ci500611v. [DOI] [PubMed] [Google Scholar]
- 30.Mellor C.L., Marchese Robinson R.L., Benigni R., Ebbrell D., Enoch S.J., Firman J.W., et al. Molecular fingerprint-derived similarity measures for toxicological read-across: recommendations for optimal use. Regul Toxicol Pharmacol. 2019;101:121–134. doi: 10.1016/j.yrtph.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Cherkasov A., Muratov E.N., Fourches D., Varnek A., Baskin, Cronin M., et al. QSAR modeling: where have you been? Where are you going to? J Med Chem. 2014;57:4977–5010. doi: 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gayvert K.M., Madhukar N.S., Elemento O. A data-driven approach to predicting successes and failures of clinical trials. Cell Chem Biol. 2016;23:1294–1301. doi: 10.1016/j.chembiol.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao Y., Romano J.D., Moore J.H. Knowledge-guided deep learning models of drug toxicity improve interpretation. Patterns (N Y) 2022;3 doi: 10.1016/j.patter.2022.100565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma B., Chenthamarakshan V., Dhurandhar A., Pereira S., Hendler J.A., Dordick J.S., et al. Accurate clinical toxicity prediction using multi-task deep neural nets and contrastive molecular explanations. Sci Rep. 2023;13:4908. doi: 10.1038/s41598-023-31169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen D.A., Nguyen C.H., Mamitsuka H. A survey on adverse drug reaction studies: data, tasks and machine learning methods. Briefings Bioinf. 2021;22:164–177. doi: 10.1093/bib/bbz140. [DOI] [PubMed] [Google Scholar]
- 36.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Felix E., et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47:D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodgers G., Austin C., Anderson J., Pawlyk A., Colvis C., Margolis R., et al. Glimmers in illuminating the druggable genome. Nat Rev Drug Discov. 2018;17:301–302. doi: 10.1038/nrd.2017.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oprea T.I., Bologa C.G., Brunak S., Campbell A., Gan G.N., Gaulton A., et al. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov. 2018;17:317–332. doi: 10.1038/nrd.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nath A., Leier A. Improved cytokine-receptor interaction prediction by exploiting the negative sample space. BMC Bioinf. 2020;21:493. doi: 10.1186/s12859-020-03835-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imrie F., Bradley A.R., Deane C.M. Generating property-matched decoy molecules using deep learning. Bioinformatics. 2021;37:2134–2141. doi: 10.1093/bioinformatics/btab080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen D., Gao K., Nguyen D.D., Chen X., Jiang Y., Wei G.W., et al. Algebraic graph-assisted bidirectional transformers for molecular property prediction. Nat Commun. 2021;12:3521. doi: 10.1038/s41467-021-23720-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crawshaw M. 2020. Multi-task learning with deep neural networks: a survey. Available from: https://arxiv.org/pdf/2009.09796. [Google Scholar]
- 44.Ruder S. An overview of multi-task learning in deep neural networks. arXiv preprint arXiv:170605098. 2017 [Google Scholar]
- 45.Xu Y., Ma J., Liaw A., Sheridan R.P., Svetnik V. Demystifying multitask deep neural networks for quantitative structure–activity relationships. J Chem Inf Model. 2017;57:2490–2504. doi: 10.1021/acs.jcim.7b00087. [DOI] [PubMed] [Google Scholar]
- 46.Xiong Z., Wang D., Liu X., Zhong F., Wan X., Li X., et al. Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J Med Chem. 2020;63:8749–8760. doi: 10.1021/acs.jmedchem.9b00959. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z., Clark N.R., Ma'ayan A. Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics. 2016;32:2338–2345. doi: 10.1093/bioinformatics/btw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lounkine E., Keiser M.J., Whitebread S., Mikhailov D., Hamon J., Jenkins J.L., et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang L.H., He Q.S., Liu K., Cheng J., Zhong M.D., Chen L.S., et al. ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018;46:D911–D917. doi: 10.1093/nar/gkx899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rappoport N., Fromer M., Schweiger R., Linial M. PANDORA: analysis of protein and peptide sets through the hierarchical integration of annotations. Nucleic Acids Res. 2010;38:W84–W89. doi: 10.1093/nar/gkq320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korthauer K., Kimes P.K., Duvallet C., Reyes A., Subramanian A., Teng M., et al. A practical guide to methods controlling false discoveries in computational biology. Genome Biol. 2019;20:118. doi: 10.1186/s13059-019-1716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen T., Le H., Quinn T.P., Nguyen T., Le T.D., Venkatesh S. GraphDTA: predicting drug-target binding affinity with graph neural networks. Bioinformatics. 2021;37:1140–1147. doi: 10.1093/bioinformatics/btaa921. [DOI] [PubMed] [Google Scholar]
- 53.Chen L., Tan X., Wang D., Zhong F., Liu X., Yang T., et al. TransformerCPI: improving compound-protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics. 2020;36:4406–4414. doi: 10.1093/bioinformatics/btaa524. [DOI] [PubMed] [Google Scholar]
- 54.Chen L., Fan Z., Chang J., Yang R., Hou H., Guo H., et al. Sequence-based drug design as a concept in computational drug design. Nat Commun. 2023;14:4217. doi: 10.1038/s41467-023-39856-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henriksson A., Zhao J., Dalianis H., Bostrom H. Ensembles of randomized trees using diverse distributed representations of clinical events. BMC Med Inf Decis Making. 2016;16(Suppl 2):69. doi: 10.1186/s12911-016-0309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao Y., Yang Z.Q., Zhang X.L., Fan W., Wang Y., Shen J., et al. Identifying the kind behind SMILES-anatomical therapeutic chemical classification using structure-only representations. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbac346. [DOI] [PubMed] [Google Scholar]
- 57.Ojanpera I., Kriikku P., Vuori E. Fatal toxicity index of medicinal drugs based on a comprehensive toxicology database. Int J Leg Med. 2016;130:1209–1216. doi: 10.1007/s00414-016-1358-8. [DOI] [PubMed] [Google Scholar]
- 58.Hauser A.S., Attwood M.M., Rask-Andersen M., Schioth H.B., Gloriam D.E. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wendell S.G., Fan H., Zhang C. G protein-coupled receptors in asthma therapy: pharmacology and drug action. Pharmacol Rev. 2020;72:1–49. doi: 10.1124/pr.118.016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dolphin A.C., Insel P.A., Blaschke T.F., Meyer U.A. Introduction to the theme "ion channels and neuropharmacology: from the past to the future". Annu Rev Pharmacol Toxicol. 2020;60:1–6. doi: 10.1146/annurev-pharmtox-082719-110050. [DOI] [PubMed] [Google Scholar]
- 61.Belvisi M.G., Birrell M.A. The emerging role of transient receptor potential channels in chronic lung disease. Eur Respir J. 2017;50 doi: 10.1183/13993003.01357-2016. [DOI] [PubMed] [Google Scholar]
- 62.Ayala-Lopez N., Watts S.W. Physiology and pharmacology of neurotransmitter transporters. Compr Physiol. 2021;11:2279–2295. doi: 10.1002/cphy.c200035. [DOI] [PubMed] [Google Scholar]
- 63.Pera T., Penn R.B. Bronchoprotection and bronchorelaxation in asthma: new targets, and new ways to target the old ones. Pharmacol Ther. 2016;164:82–96. doi: 10.1016/j.pharmthera.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu Z., Ramsundar B., Feinberg E.N., Gomes J., Geniesse C., Pappu A.S., et al. MoleculeNet: a benchmark for molecular machine learning. Chem Sci. 2018;9:513–530. doi: 10.1039/c7sc02664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Onakpoya I.J., Heneghan C.J., Aronson J.K. Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: a systematic review of the world literature. BMC Med. 2016;14:10. doi: 10.1186/s12916-016-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clarke C.E., Speller J.M. Pergolide for levodopa-induced complications in Parkinson's disease. Cochrane Database Syst Rev. 2000;(2) doi: 10.1002/14651858.CD000235. [DOI] [PubMed] [Google Scholar]
- 67.Puy L., Parry-Jones A.R., Sandset E.C., Dowlatshahi D., Ziai W., Cordonnier C. Intracerebral haemorrhage. Nat Rev Dis Prim. 2023;9:14. doi: 10.1038/s41572-023-00424-7. [DOI] [PubMed] [Google Scholar]
- 68.Walker J.S. Phenylpropanolamine potentiates caffeine neurotoxicity in rats. J Pharmaceut Sci. 1989;78:986–989. doi: 10.1002/jps.2600781203. [DOI] [PubMed] [Google Scholar]
- 69.Dietz A.J., Jr. Amphetamine-like reactions to phenylpropanolamine. JAMA. 1981;245:601–602. [PubMed] [Google Scholar]
- 70.Lake C.R., Gallant S., Masson E., Miller P. Adverse drug effects attributed to phenylpropanolamine: a review of 142 case reports. Am J Med. 1990;89:195–208. doi: 10.1016/0002-9343(90)90299-s. [DOI] [PubMed] [Google Scholar]
- 71.Caprio F.Z., Sorond F.A. Cerebrovascular disease: primary and secondary stroke prevention. Med Clin. 2019;103:295–308. doi: 10.1016/j.mcna.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 72.Liu L., Chen W., Zhou H., Duan W., Li S., Huo X., et al. Chinese Stroke Association guidelines for clinical management of cerebrovascular disorders: executive summary and 2019 update of clinical management of ischaemic cerebrovascular diseases. Stroke Vasc Neurol. 2020;5:159–176. doi: 10.1136/svn-2020-000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hutcheson J.D., Setola V., Roth B.L., Merryman W.D. Serotonin receptors and heart valve disease‒it was meant 2B. Pharmacol Ther. 2011;132:146–157. doi: 10.1016/j.pharmthera.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camilleri M. Is there a SERT-ain association with IBS? Gut. 2004;53:1396–1399. doi: 10.1136/gut.2004.039826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tack J., Schumacher K., Tonini G., Scartoni S., Capriati A., Maggi C. The neurokinin-2 receptor antagonist ibodutant improves overall symptoms, abdominal pain and stool pattern in female patients in a phase II study of diarrhoea-predominant IBS. Gut. 2017;66:1403–1413. doi: 10.1136/gutjnl-2015-310683. [DOI] [PubMed] [Google Scholar]
- 76.Perlis M.L., Corbitt C.B., Kloss J.D. Insomnia research: 3Ps and beyond. Sleep Med Rev. 2014;18:191–193. doi: 10.1016/j.smrv.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 77.Hedlund P.B., Huitron-Resendiz S., Henriksen S.J., Sutcliffe J.G. 5-HT7 receptor inhibition and inactivation induce antidepressantlike behavior and sleep pattern. Biol Psychiatr. 2005;58:831–837. doi: 10.1016/j.biopsych.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 78.Coleman J.A., Green E.M., Gouaux E. X-ray structures and mechanism of the human serotonin transporter. Nature. 2016;532:334–339. doi: 10.1038/nature17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ravna A.W., Jaronczyk M., Sylte I. A homology model of SERT based on the LeuT(Aa) template. Bioorg Med Chem Lett. 2006;16:5594–5597. doi: 10.1016/j.bmcl.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 80.Thirumaran S.L., Lepailleur A., Rochais C. Structure‒activity relationships of serotonin 5-HT(7) receptors ligands: a review. Eur J Med Chem. 2019;183 doi: 10.1016/j.ejmech.2019.111705. [DOI] [PubMed] [Google Scholar]
- 81.Morales-Perez C.L., Noviello C.M., Hibbs R.E. X-ray structure of the human alpha4beta2 nicotinic receptor. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Che T., Majumdar S., Zaidi S.A., Ondachi P., McCorvy J.D., Wang S., et al. Structure of the nanobody-stabilized active state of the Kappa opioid receptor. Cell. 2018;172:55–67.e15. doi: 10.1016/j.cell.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Theodoris C.V., Xiao L., Chopra A., Chaffin M.D., Al Sayed Z.R., Hill M.C., et al. Transfer learning enables predictions in network biology. Nature. 2023;618:616–624. doi: 10.1038/s41586-023-06139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kroll A., Ranjan S., Engqvist M.K.M., Lercher M.J. A general model to predict small molecule substrates of enzymes based on machine and deep learning. Nat Commun. 2023;14:2787. doi: 10.1038/s41467-023-38347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dalkıran A., Atakan A., Rifaioğlu A.S., Martin M.J., Atalay R.Ç., Acar A.C., et al. Transfer learning for drug–target interaction prediction. Bioinformatics. 2023;39:i103–i110. doi: 10.1093/bioinformatics/btad234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dudley J.T., Deshpande T., Butte A.J. Exploiting drug-disease relationships for computational drug repositioning. Briefings Bioinf. 2011;12:303–311. doi: 10.1093/bib/bbr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao W., Hu H., Dai L., He M., Yuan H., Zhang H., et al. Structure‒tissue exposure/selectivity relationship (STR) correlates with clinical efficacy/safety. Acta Pharm Sin B. 2022;12:2462–2478. doi: 10.1016/j.apsb.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.