Abstract
To elucidate the relationship between early viral infection events and immunodeficiency virus disease progression, quantitative-competitive and branched-DNA methods of simian immunodeficiency virus (SIV) RNA quantitation were cross-validated and used to measure viremia following infection of rhesus macaques with the pathogenic SIVmac251 virus isolate. Excellent correlation between the methods suggests that both accurately approximate SIV copy number. Plasma viremia was evident 4 days postinfection, and rapid viral expansion led to peak viremia levels of 107 to 109 SIV RNA copies/ml by days 8 to 17. Limited resolution of primary viremia was accompanied by relatively short, though variable, times to the development of AIDS (81 to 630 days). The persistent high-level viremia observed following intravenous inoculation of SIVmac251 explains the aggressive disease course in this model. Survival analyses demonstrated that the disease course is established 8 to 17 days postinfection, when peak viremia is observed. The most significant predictor of disease progression was the extent of viral decline following peak viremia; larger decrements in viremia were associated with both lower steady-state viremia (P = 0.0005) and a reduced hazard of AIDS (P = 0.004). The data also unexpectedly suggested that following SIVmac251 infection, animals with the highest peak viremia were better able to control virus replication rather than more rapidly developing disease. Analysis of early viral replication dynamics should help define host responses that protect from disease progression and should provide quantitative measures to assess the extent to which protective responses may be induced by prophylactic vaccination.
Human immunodeficiency virus (HIV) infection can result in a spectrum of outcomes varying from rapid development of AIDS to long-term nonprogressive infection. Studies suggest that both viral and host factors influence the variable course of lentiviral disease. Viral pathogenicity, inoculum size, viral complexity, and route of inoculation may have roles in disease progression (14, 36, 46, 74, 79). Divergent patterns of disease progression following infection from a common source virus have been cited as evidence of differential host responses to HIV (45, 52, 57). Host factors that could affect the disease course include the susceptibility of host target cells to HIV infection, innate immunity, and acquired immune resistance. Inherited polymorphisms in genes encoding chemokine receptors and chemokines have also been linked to variable HIV disease courses (4, 15, 21, 27, 37, 48, 70, 73, 87). HIV-specific aspects of host resistance could include variable effectiveness of host CD4+-T-helper cells, cytolytic or noncytolytic CD8+ T lymphocytes, and humoral immune responses that limit HIV infection of target cells and delay disease progression (24, 28, 66, 89). Restricted expansions of CD8+ T cells, as determined by the usage of variable domains of the beta chain of the T-cell receptor, have been linked with a less effective immune containment of HIV replication and faster disease progression (60–62). Other factors associated with rates of disease progression include age at time of infection, HLA haplotype, natural killer cell activity, and levels of host immune system activation (1, 3, 5, 13, 33, 58).
Primary HIV infection is marked by a burst of viral replication, followed by variable declines in virus load within the following weeks to months. The emergence of HIV-specific cytotoxic T lymphocytes (CTLs) is temporally correlated with control of HIV replication, although mathematical models have also been proposed that suggest that the initial decline in levels of HIV replication during primary infection could also be explained by depletion of susceptible target cells (38, 39, 54, 63, 67). Whereas measures of early HIV replication (usually obtained within the first months of infection) do not appear to be associated with disease progression, the plateau concentration of plasma viral RNA achieved 6 to 12 months after primary HIV infection is highly predictive of long-term clinical outcome (49, 51, 60). Although host factors have been correlated with this viral “set point,” their mechanism of action or effects on the primary virus-host interaction remain to be fully elucidated (66, 90).
Critical host responses to HIV infection are likely to occur within days of virus entry, and careful analyses of the initial lentivirus-host interaction should help elucidate the relationship between early viral infection events and subsequent outcome. However, analyses of early correlates of HIV disease progression are hampered by the inability to control for the virus isolate and the timing, amount, and route of virus inoculation. Therefore, investigations of early viral host dynamics must be performed in animal models, where standardization of virus inocula and the route of infection, combined with an ability to perform early and frequent monitoring of infection, allow the opportunity to test hypotheses and the impact of interventions. Infection of macaques with simian immunodeficiency viruses (SIV) has been established as a model in which to study the natural history and pathogenesis of immunodeficiency virus infection. Transfer of SIV from natural African mangabey hosts to various Asian macaque species results in an AIDS-like syndrome in the new host (reviewed in reference 19). The resultant SIV strains isolated from immunodeficient macaques vary in pathogenicity. It appears that increased passage of SIV in vivo results in increased virulence as evidenced by significant shortening of the asymptomatic period (83). The isolates of SIV commonly used have been selected for their ability to rapidly and reproducibly result in disease. The SIVmac251 strain that has been used in many studies, including this one, typically causes fatal disease within 1 to 2 years rather than the decade of infection that represents a median time to progression to AIDS in humans (6, 25, 43, 55).
Intensive analysis of primary SIV replication is made possible by the recent development of sensitive quantitative methods for the measurement of plasma SIV RNA levels (11, 26, 75, 77, 85). Frequent, quantitative sampling during primary SIV infection not only can indicate when the disease course is established, it may also provide important clues to critical features of the initial virus-host interaction that determine disease outcome (44). Although studies suggest roles for both cellular and humoral immune responses in protection from immunodeficiency virus-induced disease, rigorous demonstration of the precise host protective determinants remains a challenge of AIDS pathogenesis and vaccine research (31). Intensive virologic analyses could help to identify protective virologic correlates of primary SIV infection; such virologic correlates could serve as surrogate markers of host susceptibility and antiviral responses and help to identify candidate protective host-response mechanisms worthy of further investigation.
In this study, SIV quantitation assays based on different principles were cross-validated and applied to the sensitive virologic monitoring of primary SIVmac251 infection. The excellent correspondence between results obtained with the quantitative-competitive RNA PCR (QC-PCR) and branched-DNA (bDNA) assays suggests that both accurately measure absolute copy number. The rapid and persistent high-level viremia resulting from intravenous inoculation of the pathogenic SIVmac251 isolate exceeds that observed in most HIV infections of humans and explains the more rapid disease course observed in macaques. Survival analysis demonstrated early virologic predictors of disease progression, pointing to potential host response markers and suggesting avenues for future virologic and immunologic studies. However, these data suggest that definition of more subtle aspects of the host-virus relationship, including those more relevant to HIV infection of humans, will likely require the use of less pathogenic viruses than those commonly in use.
MATERIALS AND METHODS
Virus stocks.
SIVmac251, generously provided by Ronald Desrosiers (New England Regional Primate Research Center) was propagated on activated human peripheral blood lymphocytes as previously described (20). Titration of the virus stock in a limited number of animals demonstrated that the 1-ml inoculum used contained approximately 10 50% animal infectious doses. Titration on cells indicated the inoculating stock contained ∼50 50% tissue culture infectious doses (TCID50)/ml on human peripheral blood mononuclear cells and 1 to 10 TCID50/ml on established T-cell lines (CEMx174, SupT1, and Hut78).
Animals and infections.
Rhesus macaques (Macaca mulatta) were housed in accordance with American Association for Accreditation of Laboratory Animal Care standards and cared for according to standards set forth in the Guide for the Care and Use of Laboratory Animals of the Institute of Laboratory Resources (National Research Council, Washington, D.C.). Two groups of six rhesus macaques (groups 1 and 2) were analyzed as part of a study of the effects of immunization with commonly used vaccines on the T-cell repertoire of antigen recognition in SIV-infected animals. Group 1 animals were 2 to 12 years of age and weighed 3.2 to 14.7 kg. Group 2 animals were 3 to 14 years of age and weighed 2.7 to 8.8 kg. Group 1 animals were immunized three times at biweekly intervals with diphtheria-pertussis-tetanus (DPT) vaccine, with the last immunization being administered two weeks prior to SIV inoculation. The animals in both groups were intravenously inoculated with ten 50% monkey infectious doses of the same stock of SIVmac251. Following SIV infection, group 1 animals were immunized with tetanus vaccine at weeks 10, 14, and 18. Group 2 animals were immunized with tetanus vaccine at weeks 13 and 24 following SIV infection. Due to the rapid development of fulminant SIV disease in several of the animals and the overall rapid disease progression, we found it difficult to obtain meaningful assessments of vaccine antigen-specific immune function; therefore, this report focuses solely on early viral replication parameters.
Specimen collection.
All animals were frequently phlebotomized during the first three weeks following infection; group 1 animals were phlebotomized twice weekly. The weight of each animal in group 2 was used to determine the maximum allowable blood volume that could be collected. Five of six group 2 animals were phlebotomized at least three times weekly (animal 27141 was only bled twice a week). The largest animals were phlebotomized almost daily (see Fig. 2 and Results). Blood was collected in tubes containing either acid citrate dextrose or EDTA as an anticoagulant for the QC-PCR and bDNA assays, respectively. Plasma was processed within 3 h of blood draw as previously described and frozen at −80°C (75). Serum for Western blot analysis was collected and processed as previously described (9). Blood for flow cytometry was collected in heparin tubes and processed immediately.
FIG. 2.
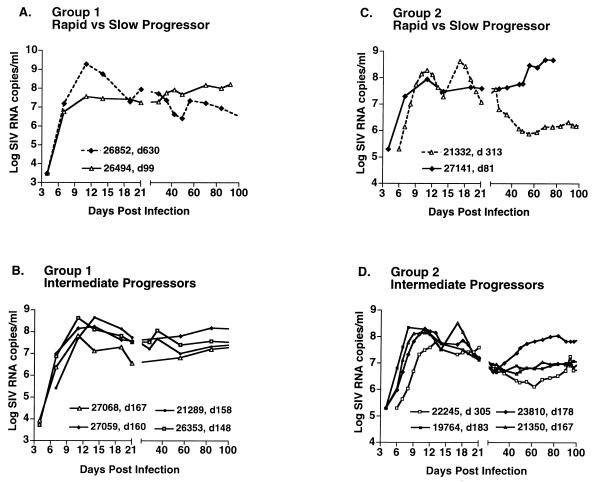
Plasma SIV RNA copy numbers during the first 100 days following primary infection of rhesus macaques inoculated with SIVmac251. For clarity, animals manifesting the slowest courses of disease (26852 and 21332; slow progressors) are compared to the most rapid progressors within each group (26494 and 27141). Animals manifesting intermediate survival times are plotted separately as intermediate progressors. Only SIV copy numbers above 3 to 5 log units of SIV RNA/ml are shown in order to adequately represent different patterns of peak viremia and viral decline observed during early infection. The survival time for each animal is indicated as the day (d) post-SIV infection when euthanasia was performed. (A and B) Rhesus macaques from group 1. Animal 26494 did not seroconvert to SIV antigens and was terminated with fulminant SIV infection 99 days postinoculation. The remaining five animals seroconverted and were euthanized between 148 and 630 days postinfection. For animal 26852, the plasma virus load after day 100 decreased to approximately 5 × 105 SIV RNA copies/ml and remained fairly steady until termination at day 630. (C and D) Rhesus macaques from group 2. Animals 27141, 21350, and 23810 did not seroconvert and were euthanized with progressive disease at 81, 167, and 178 days, respectively, postinfection. The remaining three animals seroconverted and were euthanized between 183 and 312 days postinfection. Animal 21332 was terminated with a plasma virus load of 8.7 × 105 SIV RNA copies/ml.
SIV QC-PCR.
Plasma SIV RNA levels were measured in specimens corresponding to early time points (through day 70) from group 1 animals by the SIV QC-PCR assay (described in reference 75). In this method, a synthetic competitive RNA template, which is derived from viral sequences but is distinguishable from the real viral RNA by virtue of an internal 40-bp insertion, is included in the reverse transcription and amplification steps to provide a stringent internal copy number control. A 162-bp region in the SIVmac239 gag gene, flanked by sequences conserved among almost all HIV type 2 (HIV-2)-related SIV isolates, serves as the target sequence for amplification. Virions from 0.5 to 1 ml of plasma are concentrated by centrifugation, SIV RNA is extracted, and the RNA is resuspended in 1/10 volume of H2O. The linear range of the assay is ≤50 to 50,000 copies, and the interassay variation in this range is <25%. Typically, RNA from 0.05 ml of plasma is loaded into each amplification reaction, giving a 1,000 copy/ml quantitation limit. As few as 10 copies of SIV RNA per amplification reaction can be detected, giving an assay detection limit of 200 copies/ml of plasma, comparable with high-sensitivity HIV RNA assays. The assay variance is greater near the limits of assay sensitivity. In order to quantitate SIV RNA from specimens containing low copy numbers during the first days after infection, the extracted RNA was resuspended in a smaller volume to achieve greater RNA input per amplification reaction.
bDNA quantitation of plasma SIV RNA.
Plasma SIV RNA determinations corresponding to late time points for the group 1 animals and to all time points for the group 2 animals were performed with a bDNA signal amplification assay (11). This assay is similar to the Quantiplex HIV RNA assay except that target probes were designed to hybridize with the pol region of the SIVmac group of strains, including SIVmac239, SIVmac251, SIVmac142, and SIVmne (59). SIV RNA associated with viral particles was quantified after centrifugation and concentration of SIV from 1-ml plasma aliquots (23,000 × g; 1 h) or by direct lysis of 50-μl plasma aliquots. Quantitative SIV determinations were made by comparison with a standard curve produced by using serial dilutions of cell-free SIV-infected tissue culture supernatant. (The cell-free SIV standard curve was previously validated by direct comparison with purified, quantified, in vitro-transcribed SIVmac239 pol RNA.) The lower quantitation limits of the 1-ml and 50-μl assay formats were 10,000 and 200,000 SIV RNA copies per sample, respectively.
Comparison of the SIV QC-PCR and bDNA assays.
A panel of 41 paired plasma specimens from SIV-infected animals (group 2) was measured by both the QC-PCR and bDNA assays. The correspondence of results was analyzed by Pearson’s product-moment correlation test and the t test for dependent (paired) samples. All statistical analyses were performed after log10 transformation of data.
Humoral SIVmac responses.
Sequential serum samples were assessed for antibodies to SIVmac251 proteins by Western blotting as previously described (9).
Calculation of the rates of virus growth and decay during primary infection.
Initial viral replication rates were calculated by using the exponential growth rate equation: r = (lnY1 − lnY2)/(t1 − t2), where Y1 and Y2 are the virus load values at times t1 and t2, respectively. The plasma virus load doubling time, T2, in days was calculated by using the equation T2 = (ln2)/r. Initial growth rates and T2 were calculated with data from days 4 and 7 from group 1 animals and days 6 and 7 for group 2 animals. Rates of viral decay were calculated by using the decay rate equation: α = (lnY1 − lnY2)/(t1 − t2). The maximum plasma viral decay half-life for free virus and productively infected cells, T1/2, in days, was calculated by using the equation T1/2 = (ln2)/α. Viral decay rates and T1/2 were calculated with the steepest interval of the decay curve following the peak datum point.
Statistical analyses.
Statistical analyses were performed on log10-transformed SIV RNA values, or ln values in the case of viral growth and decay rates.
(i) Survival analyses.
Proportional hazards modeling was used for analyses of survival; this approach was chosen in lieu of linear regression analysis due to the nonnormal distribution of survival times. The endpoint for survival analyses was the development of simian AIDS, which prompted euthanasia. Proportional hazards modeling was used to test associations of predictor variables with survival times (which were nonnormally distributed) (10). The method used assumes that the effect of a 1-unit increase in the value of a predictor is to multiply the hazard rate at each time by a constant, called the hazard ratio (HR). Hazard ratios of greater than 1.0 indicate that higher values of the predictor are associated with greater hazard and therefore shorter survival; hazard ratios of less than 1.0 indicate a protective effect of higher values of the predictor; hazard ratios near 1.0 indicate no association of the predictor with survival. Because the two groups of animals used in these studies received vaccinations with DPT or tetanus vaccine on different schedules and had certain measurements performed by different methods, multivariate models were used to evaluate predictors while controlling for any group effect (23). Tests of the assumption that the hazard ratios are the same at all possible failure times found no statistically significant evidence against the assumption for the analyses presented here.
(ii) Regression methods.
Regression analysis was used to look for associations among the various viremia variables under study. For this analysis, datum points that were common to the two groups of animals were used to avoid spurious associations due to the more frequent sampling of group 2 animals. In order to determine whether two viremia variables were associated, it was necessary to adjust the association for the potential group effect. Therefore, multivariate linear regression models were used, with group as one of the predictors, and fitted by the method of least squares (17). This fit is obtained by minimizing the sum of squared distances of the line from the actual response. To perform tests on the slope, it is necessary to assume that these distances are normally distributed and independent of one another, with a mean of 0 and unknown constant variance. An informal test of this assumption was performed by checking the normality of the responses by the Shapiro-Wilk test (71). There was no statistically significant evidence against the assumption of normality for these analyses.
RESULTS
This study aimed to elucidate variable host responses to a constant virus inoculum. Within the group of 12 rhesus macaques studied, diverse clinical outcomes were observed, with survival times of infected animals ranging from 81 to 630 days following SIVmac251 infection. In order to test the hypothesis that events occurring shortly after infection dictated subsequent disease outcome, we performed early and frequent virologic monitoring following SIVmac251 infection.
Ten of the 12 animals studied were euthanized according to clinical and laboratory criteria established for terminally ill immunodeficient macaques by the California Regional Primate Research Center. These criteria include a number of conditions defining late-stage simian AIDS, such as progressive weight loss (>25% of baseline), severe opportunistic infections unresponsive to specific therapy, persistent anemia (hemoglobin, <10; hematocrit, <30) and other hematologic abnormalities, persistent anorexia, intractable diarrhea, and the presence of neurologic signs. The two longest-surviving animals in group 2 (22245 and 21332 [see below]) were euthanized with early-to-intermediate symptoms of gross SIV infection, including splenomegaly and generalized lymphadenopathy with histopathological evidence of widespread lymphofollicular hyperplasia. For statistical analyses, these animals were considered right censored at the time of euthanasia. In the statistical analyses discussed below, the P values depend only on the fact that these two animals lived longer than the other animals in the group. There were no statistically significant differences in survival or viremia levels between the two groups of animals studied. Fluorescence-activated cell sorter analyses of CD4 cell counts in a subset of animals demonstrated median 50 and 43% declines in absolute CD4 counts and percentage, respectively, by 6 months postinfection (data not shown). Among these animals, there was no relationship of CD4 decline to survival.
The animals described in this study participated in a study of the effects of commonly used vaccines (DPT and tetanus) on the T-cell repertoire of specific-antigen recognition in SIV-infected animals. Due to the rapid development of fulminant SIV disease in several of the animals, and the overall rapid disease progression, we found it difficult to obtain meaningful assessments of vaccine antigen-specific immune responses. The observed high-level viremia, accompanied by substantial generalized immune activation, rendered the surviving animals unresponsive to subsequent immunization with specific antigens (data not shown). These observations are consistent with our previous work, in which HIV-infected humans with high baseline viremia were the least responsive to influenza vaccine (76). Using historical controls infected with the same virus inoculum, we could not detect a statistically significant effect of vaccination on disease progression; however, this study was not designed to test the effects of vaccination on SIV disease progression. The use of a less pathogenic SIV infection model and larger numbers of animals will likely be needed to ascertain whether more subtle aspects of the host-virus interaction, such as exposure to environmental antigens or immunizations, accelerate development of immunodeficiency disease (see below). Therefore, this report focuses solely on the relationship between early viral replication parameters and survival.
Application of the SIV bDNA and QC-PCR assays to the measurement of plasma SIV load demonstrates excellent correlation.
Because these studies relied extensively on the use of recently developed quantitative SIV RNA assays, we first validated the research utility of these assays on a panel of relevant research specimens. The “target amplification” SIV QC-PCR method was compared with the “signal amplification” SIV bDNA method (11, 75). The SIV bDNA assay has a sensitivity of 10,000 copies/ml with a 1-ml input and has a mean coefficient of variation (CV) of 25%. The SIV QC-PCR assay can detect as few as 200 copies of SIV RNA per ml of plasma and can reliably quantitate 1,000 copies/ml with <25% interassay variation. Assignment of absolute copy number for the bDNA assay is based on in vitro RNA transcripts of the pol gene, whereas the QC-PCR assay utilizes in vitro transcripts from the gag gene. The comparison of two assays based on different principles and internal standards offered the opportunity to further validate the reliability of the absolute copy number determinations obtained by these different approaches.
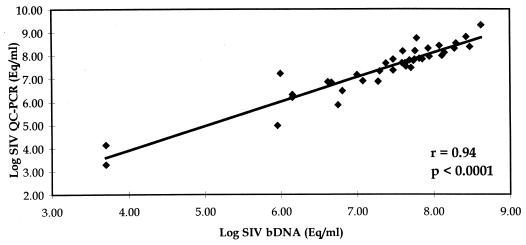
Cross-validation of the assays was performed through the analysis of 41 paired plasma specimens. The specimens represented a range of plasma viral RNA concentrations (range, >103 to 109 SIV RNA copies/ml of plasma [Fig. 1]). Statistical analyses of the assay comparison gave the following correlations: Pearson (parametric) correlation, r = 0.94 and P < 0.0001; Spearman (nonparametric) correlation, R = 0.93 and P < 0.0001. A t test for dependent samples indicated that bDNA and QC-PCR values were not significantly different (P = 0.21). QC-PCR results were, on average, 0.08 log units, or 1.2-fold, higher. (The correction factor, 1/1.2, is used to adjust QC-PCR values to match bDNA values.) This cross-validation of the bDNA and QC-PCR assays increases the confidence that the absolute copy number values obtained are accurate and supports the observation that typical plasma SIVmac251 RNA levels observed in experimentally inoculated macaques are substantially higher than the HIV-1 levels typically observed in infected humans.
FIG. 1.
Comparison of QC-PCR and bDNA assays for SIV (n = 41). Eq, equivalents.
Intravenous inoculation of SIVmac251 results in rapid, high peak viremia, followed by limited restriction of virus growth.
In order to characterize the early course of viremia following intravenous inoculation of SIVmac251, frequent measurements of plasma virus concentration were performed. Two groups of six rhesus macaques were infected. The animals in the first group (group 1) had plasma viral RNA levels monitored twice weekly during the first 3 weeks following inoculation, followed by weekly measurements thereafter (Fig. 2A and B). Animals in the second group (group 2) were phlebotomized more frequently; the four largest animals were phlebotomized at daily or near-daily intervals for 2 to 3 weeks postinfection, followed by weekly bleeds thereafter (Fig. 2C and D).
All group 1 animals manifested levels of SIV RNA that could be readily detected or quantified by the SIV QC-PCR assay by day 4, the first day of sampling, with copy numbers ranging from ∼2 × 102 to 8.6 × 103 SIV RNA copies/ml of plasma (Table 1, group 1). This suggested early and rapid growth of SIVmac251 in the newly infected hosts. At day 7, the measured copy numbers ranged from 2.6 × 105 to 1.6 × 107. Measurements at days 4, 7, 11, 14, and 19 demonstrated single peaks of viremia at days 11 or 14 in all of the animals (Fig. 2A and B). The maximum SIV RNA copy numbers attained were high, varying from 3.6 × 107 to 1.9 × 109 SIV RNA copies/ml. Thereafter, SIV RNA levels decreased by various extents, ranging from 0.3 to 2.0 log10 in the 1 to 2 weeks after peak viremia, followed by a continued slower decline in some animals. The animals appeared to establish quasi-steady-state levels of plasma viremia by 4 to 6 weeks postinfection, with levels ranging from 105 to >108 SIV RNA copies/ml of plasma (Fig. 2A and B). These “postacute” viremia levels are substantially higher than those usually seen in humans. Western blot analysis of anti-SIV antibodies at 6 weeks postinfection demonstrated that five of the six animals had seroconverted (data not shown). Animal 26494, which did not seroconvert, developed fulminant SIV disease and was the first animal to be euthanized, with signs of simian AIDS at 99 days postinfection.
TABLE 1.
Viral replication dynamics parametersa before and after peak viremia
| Group | Mean growth rateb (SD) | Rangec | Doubling time (days) | Range | Mean decay rated (SD) | Range | Half-life (days) | Range |
|---|---|---|---|---|---|---|---|---|
| 1 | 2.55 (0.46) | 1.86–3.22 (n = 6) | 0.27 | 0.21–0.37 | 0.48 (0.25) | 0.21–0.85 (n = 6) | 1.44 | 0.82–3.3 |
| 2 | 2.00 (0.53) | 1.47–2.53 (n = 5) | 0.35 | 0.27–0.47 | 0.51 (0.32) | 0.05–0.99 (n = 6) | 1.36 | 0.70–13.9 |
In ln units.
Based on earliest available time points (days 4 and 7 for group 1 and days 6 and 7 for group 2).
Ranges of observed values.
Based on the steepest interval of the decay curve following peak viremia.
Group 2 animals were monitored with the SIV bDNA assay throughout the entire course of SIV infection. The bDNA assay was chosen because it readily allows simultaneous quantitation of a large number of specimens from the same animals. Because limited plasma volumes were available, the 50-μl format of this assay was used, limiting SIV RNA detection to 200,000 copies per ml and precluding viral RNA quantitation at the earliest time point (day 4). At day 6, SIV was quantifiable in three of five animals tested (range, 9.7 × 105 to 6.4 × 106), and it was quantifiable in all six animals by day 7 (range, 4.4 × 105 to 4.0 × 107) (Fig. 2C and D). The group 2 animals manifested early viral growth comparable to that in the animals in group 1, with the exception of animal 22245, which demonstrated a lag in early replication (Fig. 2D). With the more frequent monitoring of viremia in group 2, initial peaks of viremia were observed between days 8 and 13 in all of the animals, with copy numbers ranging from 6.1 × 107 to 2.2 × 108 SIV RNA copies/ml. Two animals manifested two peaks of viremia at days 11 and 17 postinfection. The second peaks, at 3.3 × 108 and 4.2 × 108 copies/ml, were approximately twofold higher than the first peaks observed in the same animals (confirmed by application of both the QC-PCR and bDNA assays [Fig. 2C and D]). It is important to note that such double peaks were not observed in group 1 animals, monitored less frequently. Viral titers decreased by 0.4 to 1.8 log10 units in the 1 to 2 weeks following peak viremia, followed by a more gradual decline. The quasi-steady-state viremia levels established in these animals ranged from 105 to >108 SIV RNA copies/ml of plasma (Fig. 2C and D). Overall, the measured levels of early viremia in group 2 animals were not statistically different from the values obtained for the group 1 animals. Three of six animals in group 2 seroconverted to anti-SIV antibodies by week 12 postinfection. The three animals that did not seroconvert (26141, 21350, and 23810) were the first animals to be euthanized, at days 81, 167, and 178 postinfection, respectively.
Prior to clinical decline and euthanasia, levels of SIV replication increased by approximately 1 to 2 log units above the nadir attained in most animals (not shown). None of the animals was able to suppress viral replication to the levels typically seen in HIV-infected humans. Together, these observations show that intravenous inoculation of SIVmac251 can result in rapid, high peak viremia, followed by a variable but rather limited restriction of virus growth.
Similar initial virus growth rates are followed by more variable virus decay rates.
In order to more precisely characterize initial virus growth and subsequent decay, growth and decay slopes were calculated. Early restrictions in viral replication would manifest as variable virus growth in the newly infected host. Early virus replication in individual hosts was assessed by estimation of mean rate (r) and standard deviation (SD) of virus growth and the corresponding doubling time, T2 (described in Materials and Methods). Growth slopes, in ln units, were calculated by using the initial two quantifiable datum points for each animal. For group 1 animals, these datum points corresponded to days 4 and 7 postinfection and gave a mean growth rate (r) of 2.55 (SD = 0.46; range, 1.86 to 3.22) (Table 1). This growth slope corresponds to a T2 of 0.27 days, or 6.5 h. Since the first two quantifiable datum points for group 2 animals were on days 6 and 7, these time points were used to estimate growth slopes, giving a mean r value of 2.00 (SD = 0.53; range, 1.47 to 2.53) and a T2 value of 0.35 days, or 8.4 h (Table 1). The lower growth rate calculated with later time points for group 2 may reflect deviation from exponential growth as the growth curves approach peak viremia (Fig. 2). Variabilities in growth slopes, calculated as the CV (CV = SD/mean), were 18 and 26% for group 1 and 2 animals, respectively, suggesting modest variation in initial growth slopes (Table 1). Although this analysis does not address the determinants of the initial rates of SIV replication, it suggests that SIVmac251 infection of rhesus macaques results in similar rates of early virus growth in different hosts.
The determinants of the magnitude of reduction of viremia following peak viremia are poorly understood. The two prevailing models of viral suppression invoke either target cell availability or antivirus immune responses as the primary determinants of a decline in peak viremia (38, 39, 63). This phase of SIVmac251 infection was analyzed by calculation of slopes of viral decay and plasma virus load half-life, T1/2 (see Materials and Methods). Decay slopes, in ln units, were calculated from the steepest interval of the decay curve following peak viremia for each animal. (The steepest interval was chosen as an approximation of the fastest rate of viral decay in each animal. This approach does not account for potentially changing rates of viral decay during resolution of primary infection.) For group 1 animals, the mean decay rate was 0.48 (SD = 0.25; range, 0.21 to 0.85) (Table 1), corresponding to a T1/2 of 1.44 days. Group 2 animals manifested a mean of 0.51 (SD = 0.32; range, 0.05 to 0.99) and a corresponding T1/2 of 1.36 days. In contrast to more similar initial virus growth slopes prior to peak viremia, the CVs associated with the slopes of viral decline were larger (52 and 64% for groups 1 and 2, respectively), and a broad range of viral T1/2s were observed (1 to 14 days). These data suggest differential host responses to virus during this phase of the early virus-host interaction (Fig. 2), although the mechanisms of virus reduction remain to be elucidated.
Patterns of early viremia are associated with differences in levels of postpeak viremia and survival.
The animals in this study manifested variable courses of disease and were terminated with signs and symptoms of AIDS at widely varying times, ranging from 81 to 644 days postinfection. In this small group, neither age nor baseline CD4 count was predictive of disease course. In order to ascertain whether the character of early primary viremia predicted disease course, statistical analyses were performed. The primary viremia variables chosen for survival analyses included early viral growth, peak viremia, the decline in viremia, and postacute viremia. Associations between these viremia parameters were also assessed. Proportional hazards modeling was used to test associations of putative predictor variables with survival (10). Because the two groups of animals used in these studies were handled differently and had a subset of virus load measurements performed by different methods, multivariate models were used to evaluate predictors while controlling for any group effect. Linear regression analyses were performed to look for associations between viremia variables.
(i) Early viremia (days 4, 6, and 7) and virus growth rates.
Because the extent of early virus growth might influence the resulting host-virus equilibrium, or set point, we looked for an association between viremia levels at days 4, 6, or 7 and postacute viremia or survival. There was no correlation among these parameters. There also did not appear to be any correlation between early virus growth rates and subsequent virologic events or survival. These data are consistent with the lack of large variation in early SIVmac251 growth rates (Table 1), and they differ from the variable early virus growth observed for less virulent SIVsmE660 infection (44).
(ii) Peak viremia.
Peak viremia might reflect the extent of activated cell targets available for SIV replication, as well as ensuing factors which abrogate SIV replication. Interestingly, the highest peak viremia was observed in animal 26852, which thereafter exhibited relative control of virus replication and the longest survival (630 days [Fig. 2A]). These data suggest the possibility of a more optimally activated CD4+-T-lymphocyte-based antiviral immune response in this animal (see Discussion). Hazards modeling, used to test the putative association between peak viremia and survival, suggested that each 1-log10-unit increase in peak viremia was associated with a 0.19 relative hazard ratio (HR) of progression to AIDS, or an approximately fivefold-decreased risk (P = 0.026) (Table 2). When the two animals with the most extreme values were removed from this analysis, (animals 26852 and 26494), the association between peak viremia and survival was no longer statistically significant; thus, studies with larger numbers of animals will be needed to confirm these provocative results. Higher peak viremia was also associated with larger subsequent declines in viremia, calculated as the difference between peak viremia and average postacute viremia over days 35 to 60 (P = 0.009). Conversely, animals with the lowest peak viremia exhibited an inability to control virus replication thereafter (Fig. 2A and C).
TABLE 2.
Early plasma viremia correlates of SIV disease progression (n = 12)
| Predictora | HRb (95% CI) | P value |
|---|---|---|
| Peak viremia (days 8–17) | 0.19 (0.05, 0.82) | 0.026 |
| Postacute viremia (days 35–60)c | 12.6 (2.0, 80) | 0.007 |
| Absolute decline in viremiad | 0.11 (0.02, 0.49) | 0.004 |
| Day 35 viremia | 3.5 (1.2, 10) | 0.021 |
| Day 42 viremia | 30.2 (1.3, 684) | 0.032 |
All viremia measurements were analyzed as the log10 (SIV RNA).
HR for progression to AIDS, determined by proportional hazards modeling controlled for group in a bivariate (two-predictor) proportional hazards model. HR corresponds to each 1-log10-unit increase in SIV RNA for all viremia measures.
Calculated as the average of the log10 SIV RNA levels on days 35 to 60 postinfection.
Log10 peak viremia − log10 postacute viremia.
(iii) Decline in viremia.
The postpeak decline in viremia might represent the best measure of host ability to suppress virus replication. Viremia decline, measured as the difference between peak and average postacute viremia (days 35 to 56), was significantly correlated with both lower postacute viremia (P = 0.0005) and increased survival (P = 0.004) (Table 2). Each 1-log10-unit increment in the difference between peak and postacute viremia was associated with an HR of 0.11, or an approximately ninefold-reduced hazard of AIDS. Thus, the magnitude of early viral decline is positively linked to longer survival.
(iv) Postacute viremia.
Higher postacute viremia, defined as the average viremia over days 35 to 60 postinfection, was significantly correlated with an increased risk of progression to AIDS (HR = 12.6; P = 0.007) (Table 2). In order to determine how soon after the initial decline in viremia postacute viremia measures predicted survival, the predictive value of viremia at days 28, 35, and 42 was analyzed. Viremia levels 28 days postinfection did not correlate with survival, consistent with some animals not yet having reached a nadir in viremia. A significant correlation between postacute viremia and survival was first detected at day 35, with each 1-log10-unit increase in viremia associated with a 3.5-fold-increased hazard of progression to AIDS (95% confidence interval [CI], 1.2, 10.3; P = 0.021) (Fig. 2). By day 42 postinfection, the HR associated with each 1-log10-unit increase in viremia had risen to 30.2 (95% CI, 1.3, 684; P = 0.032).
Although either peak viremia or postacute viremia alone predicted survival (Table 2), there did not appear to be a significant correlation between these two parameters. After adjustment for postacute viremia, the P value of peak viremia as a survival predictor was 0.07, with an HR of 0.17, suggesting the possibility that peak viremia might be an independent predictor of survival.
DISCUSSION
This study analyzed different patterns of virus replication following inoculation of SIVmac251 into 12 rhesus macaques. A constant viral inoculum was used in order to elucidate variation in host response to primary SIV infection. The ability to control the virus inoculum and timing of infection allowed us to make observations not feasible in studies of primary HIV-1 infection of humans.
We applied two sensitive methods for the analysis of SIV replication following primary infection of rhesus macaques. The comparison of two methods for plasma SIV RNA quantitation, target amplification (QC-PCR) and direct detection (bDNA) (11, 75), demonstrated excellent concordance between the assays (Fig. 1), suggesting that the measured high levels of SIV plasma viremia are a close approximation of the absolute copy number. Frequent virus load monitoring elucidated aspects of the dynamic primary viremia period that were not observed in other longitudinal disease progression studies with less intensive monitoring (26, 85) and allowed the correlation of early virologic events to subsequent disease course and survival time.
Differential host responses to viral infection could be due to genetic factors affecting the differing ability of host cells to support SIV replication, as recognized previously for HIV and as reported recently for SIVsm infection (44, 88). Host cells could also vary in their activation or proliferation state, depending on baseline immune activation in the animal (18) as well as the extent of T-cell activation induced by SIV antigens. Individual hosts likely vary in their ability to mount innate or adaptive immune responses. Antiviral immune responses, such as CTLs, could be influenced by macaque major histocompatibility complex haplotype or other genetic factors (42).
SIV was detected in the plasma as early as 4 days postinfection (the first time point at which blood was collected) in all animals monitored by QC-PCR, suggesting that SIVmac251 rapidly infected all animals with similar efficiency. This initial appearance of virus is faster than that reported in a study of SIVsmE660 infection of Macaca nemestrina (56). Estimation of rates of initial virus replication demonstrated that circulating virus levels double approximately every 7 to 8 h during this phase (Table 1). These growth rates are similar to those recently reported for SIVsmE660 infection of M. nemestrina, despite the reduced magnitude of early SIVsmE660 viremia relative to that observed here for SIVmac251 (56). Modest host-to-host variation in initial SIVmac251 growth rates suggests limited differences in the initial virus-host interaction, perhaps due to the overwhelming nature of the virus inoculum.
The timing of initial peak viremia, occurring between 8 and 14 days postinfection, is similar to, or slightly earlier than, that reported following SIVsmE660 infection of M. mulatta or M. nemestrina (56, 85). Peak viremia levels ranged from 3.6 × 107 to 1.9 × 109 SIV RNA copies/ml. These peak values are higher than those observed for SIVmac239 and SIVsm and are consistent with the known increased virulence of SIVmac251 relative to SIVmac239 (26, 34, 56, 85). This peak SIVmac251 viremia is also higher than that reported in most primary HIV-1 infection studies, although human studies can only rarely precisely pinpoint peak viremia (35, 49, 68, 69). Viral diversity analysis in a subset of animals (based on the SIV env V1-V2 region) indicated that the SIV quasispecies detected early after intravenous inoculation resembled the virus inoculum, suggesting that little selection or “filtering” of viral variants had occurred (22). This is in contrast to mucosal transmission of HIV or SIV, in which a subset of variants in the donor may become the major quasispecies in the newly infected recipient (74, 91). Taken together, these data suggest that SIVmac251 is an exceptionally virulent virus which, upon intravenous inoculation, grows explosively in macaques. The seroconversion rates and survival times of the animals described in this study are similar to those observed by other investigators with the SIVmac251 isolate.
Following peak viremia, the rate and extent of viral decay observed in the different animals was quite variable, with the magnitude of early virus load decreases ranging from 0.3 to 2.0 log10 units. Following initial rapid declines in viremia, most animals continued to manifest more-gradual decreases in viremia over the course of several weeks, with individual animals establishing different quasi-steady-state levels of viremia 4 to 6 weeks postinfection. However, a prolonged steady-state viremia, similar to that observed in many HIV-infected humans, was not observed. Most animals established postacute viremia levels ranging from 106 to 108 SIV RNA copies/ml, in contrast to the levels typically seen in HIV infection (≤105 copies/ml). The inability to gain substantial control of initial viral replication and the continued high postacute viremia explain the accelerated disease course observed in rhesus macaques, nonnatural hosts of SIV, intravenously inoculated with SIVmac251.
More frequent sampling of group 2 animals indicated the presence of two peaks of viremia in the first 3 weeks of infection in some animals. Double peaks were not observed in animals monitored less frequently, emphasizing the importance of near-daily monitoring for the accurate characterization of early primary viremia events. The observed double peaks may reflect temporal variations in target cell availability and in the course of virus dissemination within the host, as well as the kinetics of host response to infection. However, the appearance of a second, higher peak in some animals is difficult to reconcile with acquired immunity being the sole or perhaps even predominant driving force in the resolution of the first peak.
The outcome of SIVmac251 infection was related to viral replication dynamics occurring during the first 3 weeks after infection. Following peak viremia, there was considerable variation in viral decay or suppression (Table 1), and this variation was related to the disease course. The mean T1/2 of decline of plasma SIVmac251 in this study (1.4 days) is comparable to that observed following resolution of primary SIVsm infection of M. nemestrina (56). The most significant virologic correlate of slower disease progression was the magnitude of postpeak decline in viremia; larger differences between the peak and postacute viremia levels predicted both lower postacute viremia and better survival. Other studies have correlated the appearance of SIV-specific CTLs or NK cells with the decline in virus load, suggesting a role for these cellular immune responses in early virus suppression (31, 65, 72). If “protective” patterns of early viral replication can be reproducibly linked to identifiable immune responses, then measures of early viral dynamics might serve as surrogates for monitoring the effects of interventions. Vaccine study design could include well-characterized virologic markers to assess how well ex vivo measures of immune function correlate with the in vivo effectiveness of vaccine-elicited host immune responses in controlling the extent of virus replication.
Consistent with other studies of HIV and SIV, postacute viremia was found to significantly predict the risk of disease progression (26, 78, 85). However, in contrast with previous studies, the correlation between higher postacute viremia and greater risk of SIV disease progression could be detected even earlier, as soon as 35 days postinfection (Table 2). Each 1-log-unit increase in postacute SIV viremia was associated with a 3.5- to 30-fold-increased hazard of progression to AIDS, a risk comparable to that associated with increased HIV levels in people (Table 2) (50).
Unexpectedly, among these animals with overall high-level viremia, the animals with the lowest peak viremia were unable to control virus replication and experienced a rapid disease course whereas animals with higher peak viremia (occurring between days 8 and 17 postinfection) manifested larger postpeak declines in viremia and better survival (Fig. 2). For a given virus strain, a positive correlation between peak viremia and increased survival has not been reported, although most published studies have used less frequent monitoring and therefore would not precisely identify the timing and magnitude of peak viremia (26, 85). A recent SIV study which did include frequent virologic determinations used the apparently less pathogenic SIVsmE660 and a molecularly cloned variant, SIVsmE543-3; the variable early growth of these isolates appeared to be the primary predictor of lower postacute viremia (44). When early viral growth is restricted or variable, as is seen in comparisons of attenuated and pathogenic SIVs, then this early variable growth may be a predominant determinant of the subsequent disease course (16). The level of peak viremia will be influenced by significant variation in early virus growth. For example, in this study, animal 22245 manifested a lag in the first appearance of viremia, followed by lower peak and postacute viremia levels (Fig. 2D). Thus, less vigorous early virus replication could give the immune response an advantage in the dynamic host-virus interaction that occurs very early after infection. In this study, very similar and robust initial SIVmac251 growth rates were followed by varying levels of peak viremia. It remains to be determined which features, namely variable early virus growth or variable peak viremia, are predominant determinants of subsequent outcome in HIV-infected humans.
Although either peak viremia or postacute viremia alone predicted survival (Table 2), a correlation between these parameters was not detected, suggesting that peak viremia may either be an independent predictor of survival or represent one of two or more factors that influence postacute viremia. This latter possibility is supported by the results in the SIVsm model, which identify early virus growth (day 7 viremia) as a predictor of steady-state viremia (44). Better understanding of the various determinants of infection outcome will require further analysis of the various SIV infection models and larger sample sizes.
The SIVmac251 isolate used in these studies was originally selected for its dramatic disease-causing properties in rhesus macaques (12, 43) (although the passage history and culture conditions of SIVmac251 stocks used by different investigators may vary in ways that affect the relative virulence of a particular stock). For as yet unknown reasons, sequential intravenous passage of SIV or simian-human immunodeficiency virus (SHIV) recombinants in macaques or HIV in chimpanzees can select for extremely pathogenic viruses that cause rapid CD4 decline, even when the initial virus strain replicated poorly in vivo and was of limited pathogenicity (32, 83). This type of rapid, extreme selection does not appear to occur in natural (as opposed to experimental) HIV transmission. Furthermore, the size of the inoculum typically used to achieve 100% infection of animals is likely much larger than that involved in natural host-to-host transmission events. Finally, the intravenous inoculation route used to introduce virus bypasses mucosal barriers, which nascent HIV infections must traverse, including tissue dendritic and antigen-presenting cells. The consequently rapid delivery of virus to the bloodstream and to central and peripheral lymphatic tissues, where numerous susceptible target cells reside and where activation of the immune system in response to virus infection takes place, may facilitate the magnitude of virus growth and the associated pathogenic consequences. Thus, the rapid disease course following SIVmac251 infection may bear limited resemblance to the viruses selected during the course of natural HIV transmission or to the disease course following HIV infection of humans. Because highly pathogenic viruses may obscure more subtle aspects of the host virus relationship, they should be used cautiously in studies designed to test concepts relevant to HIV-induced disease. For example, in this study, the rapid disease progression and high-level immune activation resulting from SIVmac251 infection compromised our ability to analyze the impact of immunization with specific antigen on the CD4+-T-cell repertoire of antigen recognition and response. Furthermore, the use of SIVmac251 and similar or derivative viruses (e.g., SHIV89.6P) as vaccine challenge stocks may present an overwhelming infection against which it is difficult to mount immunologic protection (29, 30, 32, 64).
The development of optimal antiviral immunity is dependent upon the magnitude and quality of the primary T-cell immune response to viral infection (reviewed in reference 2), with a majority of responding cells being virus specific as opposed to non-antigen-specific bystanders (7, 8, 41, 53). Because HIV and SIV replication are driven by T-cell activation (86), it will be important to determine how primary SIV-specific CD4+-T-cell proliferative responses contribute to the pool of susceptible target cells available to support SIV production (66) and the relationship of primary CD4 responses to CD8+-CTL responses. HIV-specific CD4+-T-cell proliferative responses have been reported to be found only in rare individuals with slowly progressing or nonprogressing HIV infection, where they are seen in conjunction with HIV-specific CTL responses and low viremia (40, 66). Administration of antiretroviral therapy or anti-SIV antibodies near the time of primary immunodeficiency virus infection may partially recapitulate this beneficial host-virus equilibrium, perhaps by inhibiting the earliest rounds of virus replication, giving the immune response an opportunity to establish more effective antiviral immunity before irreversible immune system damage occurs (47, 80–82, 84). In this study, the observation that the animal with the lowest peak viremia was unable to mount humoral immune responses and rapidly succumbed to disease suggests that future studies should investigate the fate of virus-specific immune effector cells during the course of SIV infection.
This study highlights the role of very early events in the establishment of the SIV disease course. If the HIV disease course is established at a similarly early time, then alteration of the natural host-virus equilibrium may only be possible through prior vaccine intervention or antiretroviral therapy applied within days of HIV exposure. It is important to fully define the window of opportunity within which potentially protective interventions can alter the host-virus equilibrium. The simian model will be essential for the further elucidation of early determinants of disease progression, their time course of expression, and how these determinants are influenced by vaccines and other interventions. Future study design should include the use of more natural virus stock challenges, which may present more realistic tests of immunodeficiency virus replication dynamics in vivo and the impact that vaccine-induced immune responses can exert to prevent immunodeficiency virus infection or disease.
ACKNOWLEDGMENTS
We thank Ronald Desrosiers for providing the initial SIVmac251 stock, Peter Bachetti and the UCSF Biostatistics Core for assistance with statistical analyses, Theodore Voronov for assistance on the analysis of viral dynamics, and Angela McLean, Marc Lipschitz, and Rustom Antia for thoughtful comments.
This work was supported by American Foundation for AIDS Research grant 02364-17-RG (to M.B.F.); a Pediatric AIDS Foundation Scholar Award (to S.I.S.), NIH-NCRR RR00169; a grant from the National Institutes of Health, University of California, San Francisco Center for AIDS Research, P30 AI27763; and UCSF AIDS Clinical Research Center grant no. CC93-SF-130. M.B.F. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
REFERENCES
- 1.Ahmad A, Menezes J. Defective killing activity against gp120/41-expressing human erythroleukaemic K562 cell line by monocytes and natural killer cells from HIV-infected individuals. AIDS. 1996;10:143–149. doi: 10.1097/00002030-199602000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 3.Bentwich Z, Kalinkovich A, Weisman Z. Immune activation is a dominant factor in the pathogenesis of African AIDS. Immunol Today. 1995;16:187–191. doi: 10.1016/0167-5699(95)80119-7. [DOI] [PubMed] [Google Scholar]
- 4.Blaak H, van’t Wout A B, Brouwer M, Cornelissen M, Kootstra N A, Albrecht-van Lent N, Keet R P M, Goudsmit J, Coutinho R A, Schuitemaker H. Infectious cellular load in human immunodeficiency virus type 1 (HIV-1)-infected individuals and susceptibility of peripheral blood mononuclear cells from their exposed partners to non-syncytium-inducing HIV-1 as major determinants for HIV-1 transmission in homosexual couples. J Virol. 1998;72:218–224. doi: 10.1128/jvi.72.1.218-224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaxhult A, Granath F, Lidman K, Giesecke J. The influence of age on the latency period to AIDS in people infected by HIV through blood transfusion. AIDS. 1990;4:125–129. doi: 10.1097/00002030-199002000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Buchbinder, S. P., T. Elbiek, E. Vittinghoff, S. Staprans, and M. B. Feinberg. Unpublished data.
- 7.Butz E A, Bevan M J. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callan M F, Tan L, Annels N, Ogg G S, Wilson J D, O’Callaghan C A, Steven N, McMichael A J, Rickinson A B. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus in vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson J R, McGraw T P, Keddie E, Yee J L, Rosenthal A, Langlois A J, Dickover R, Donovan R, Luciw P A, Jennings M B. Vaccine protection of rhesus macaques against simian immunodeficiency virus infection. AIDS Res Hum Retroviruses. 1990;6:1239–1246. doi: 10.1089/aid.1990.6.1239. [DOI] [PubMed] [Google Scholar]
- 10.Cox D. Regression models and life-tables. J R Stat Soc Ser B. 1972;34:187–220. [Google Scholar]
- 11.Dailey P J, Zamroud M, Kelso R, Kolberg J, Urdea M. Quantitation of simian immunodeficiency virus (SIV) RNA in plasma of acute and chronically infected rhesus macaques using a branched DNA (bDNA) signal amplification assay. J Med Primatol. 1995;24:209. [Google Scholar]
- 12.Daniel M D, Desrosiers R C, Letvin N L, King N W, Schmidt D K, Sehgal P, Hunt R D. Simian models for AIDS. Cancer Detect Prev Suppl. 1987;1:501–507. [PubMed] [Google Scholar]
- 13.Darby S C, Ewart D W, Giangrande P L, Spooner R J, Rizza C R. Importance of age at infection with HIV-1 for survival and development of AIDS in UK haemophilia population. UK Haemophilia Centre Directors’ Organisation. Lancet. 1996;347:1573–1579. doi: 10.1016/s0140-6736(96)91073-9. [DOI] [PubMed] [Google Scholar]
- 14.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasispecies of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 15.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CCR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. . (Erratum, 274:1069.) [DOI] [PubMed] [Google Scholar]
- 16.Desrosiers R C, Lifson J D, Gibbs J S, Czajak S C, Howe A Y M, Arthur L O, Johnson R P. Identification of highly attenuated mutants of simian immunodeficiency virus. J Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draper N, Smith H. Applied regression analysis. 2nd ed. New York, N.Y: John Wiley and Sons; 1981. pp. 1–55. [Google Scholar]
- 18.Folks T, Rowe T, Villinger F, Parekh B, Mayne A, Anderson D, McClure H, Ansari A A. Immune stimulation may contribute to enhanced progression of SIV induced disease in rhesus macaques. J Med Primatol. 1997;26:181–189. doi: 10.1111/j.1600-0684.1997.tb00050.x. [DOI] [PubMed] [Google Scholar]
- 19.Gardner M, Endres M, Barry P. The simian retroviruses: SIV and SRV. In: Levy J, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 133–277. [Google Scholar]
- 20.Gardner M, Rosenthal A, Jennings M, Yee J, Antipa L, Robinson E., Jr Passive immunization of rhesus macaques against SIV infection and disease. AIDS Res Hum Retroviruses. 1995;11:843–854. doi: 10.1089/aid.1995.11.843. [DOI] [PubMed] [Google Scholar]
- 21.Garred P, Eugen-Olsen J, Iversen A K, Benfield T L, Svejgaard A, Hofmann B. Dual effect of CCR5 delta 32 gene deletion in HIV-1-infected patients. Copenhagen AIDS Study Group. Lancet. 1997;349:1884. doi: 10.1016/s0140-6736(05)63874-3. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 22.Grant, R. M., and M. B. Feinberg. Unpublished data.
- 23.Harrell F, Lee K L. Proceedings of the 11th Annual SAS User’s Group International Conference. Cary, N.C: SAS Institute; 1986. Verifying assumptions of the Cox proportional hazards model. [Google Scholar]
- 24.Harrer T, Harrer E, Kalams S A, Elbeik T, Staprans S I, Feinberg M B, Cao Y, Ho D D, Yilma T, Caliendo A M, Johnson R P, Buchbinder S P, Walker B D. Strong cytotoxic T cell and weak neutralizing antibody responses in a subset of persons with stable nonprogressing HIV type 1 infection. AIDS Res Hum Retroviruses. 1996;12:585–592. doi: 10.1089/aid.1996.12.585. [DOI] [PubMed] [Google Scholar]
- 25.Hessol N A, Koblin B A, van Griensven G J, Bacchetti P, Liu J Y, Stevens C E, Coutinho R A, Buchbinder S P, Katz M H. Progression of human immunodeficiency virus type 1 (HIV-1) infection among homosexual men in hepatitis B vaccine trial cohorts in Amsterdam, New York City, and San Francisco, 1978–1991. Am J Epidemiol. 1994;139:1077–1087. doi: 10.1093/oxfordjournals.aje.a116951. [DOI] [PubMed] [Google Scholar]
- 26.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Yang L C, Goldstein S, Piatak M, Jr, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, Ho T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 28.Jassoy C, Walker B D. HIV-1-specific cytotoxic T lymphocytes and the control of HIV-1 replication. Springer Semin Immunopathol. 1997;18:341–354. doi: 10.1007/BF00813502. [DOI] [PubMed] [Google Scholar]
- 29.Joag S V, Li Z, Foresman L, Pinson D M, Raghavan R, Zhuge W, Adany I, Wang C, Jia F, Sheffer D, Ranchalis J, Watson A, Narayan O. Characterization of the pathogenic KU-SHIV model of acquired immunodeficiency syndrome in macaques. AIDS Res Hum Retroviruses. 1997;13:635–645. doi: 10.1089/aid.1997.13.635. [DOI] [PubMed] [Google Scholar]
- 30.Joag S V, Li Z, Foresman L, Stephens E B, Zhao L-J, Adany I, Pinson D M, McClure H M, Narayan O. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson R P, Glickman R L, Yang J Q, Kaur A, Dion J T, Mulligan M J, Desrosiers R C. Induction of vigorous cytotoxic T-lymphocyte responses by live attenuated simian immunodeficiency virus. J Virol. 1997;71:7711–7718. doi: 10.1128/jvi.71.10.7711-7718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlsson G B, Halloran M, Li J, Park I-W, Gomila R, Reimann K A, Axthelm M K, Iliff S A, Letvin N L, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaslow R A, Carrington M, Apple R, Park L, Munoz A, Saah A J, Goedert J J, Winkler C, O’Brien S J, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann D L. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 34.Kaur A, Grant R M, Means R E, McClure H, Feinberg M, Johnson R P. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J Virol. 1998;72:9597–9611. doi: 10.1128/jvi.72.12.9597-9611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinloch-De Loes S, Hirschel B J, Hoen B, Cooper D A, Tindall B, Carr A, Saurat J H, Clumeck N, Lazzarin A, Mathiesen L. A controlled trial of zidovudine in primary human immunodeficiency virus infection. N Engl J Med. 1995;333:408–413. doi: 10.1056/NEJM199508173330702. . (Erratum, 333:1367.) [DOI] [PubMed] [Google Scholar]
- 36.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 37.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 38.Koup R A, Ho D D. Shutting down HIV. Nature. 1994;370:416. doi: 10.1038/370416a0. [DOI] [PubMed] [Google Scholar]
- 39.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krowka J F, Stites D P, Jain S, Steimer K S, George-Nascimento C, Gyenes A, Barr P J, Hollander H, Moss A R, Homsy J M. Lymphocyte proliferative responses to human immunodeficiency virus antigens in vitro. J Clin Investig. 1989;83:1198–1203. doi: 10.1172/JCI114001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuroda M J, Schmitz J E, Barouch D H, Craiu A, Allen T M, Sette A, Watkins D L, Forman M A, Letvin N L. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lekutis C, Letvin N L. HIV-1 envelope-specific CD4+ T helper cells from simian/human immunodeficiency virus-infected rhesus monkeys recognize epitopes restricted by MHC class II DRB1*0406 and DRB*W201 molecules. J Immunol. 1997;159:2049–2057. [PubMed] [Google Scholar]
- 43.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 44.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S-L, Schacker T, Musey L, Shriner D, McElrath M J, Corey L, Mullins J I. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marthas M L, Van Rompay K K A, Otsyula M, Miller C J, Canfield D R, Pedersen N C, McChesney M B. Viral factors determine progression to AIDS in simian immunodeficiency virus-infected newborn rhesus macaques. J Virol. 1995;69:4198–4205. doi: 10.1128/jvi.69.7.4198-4205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin L N, Murphey-Corb M, Soike K F, Davison-Fairburn B, Baskin G B. Effects of initiation of 3′-azido,3′-deoxythymidine (zidovudine) treatment at different times after infection of rhesus monkeys with simian immunodeficiency virus. J Infect Dis. 1993;168:825–835. doi: 10.1093/infdis/168.4.825. [DOI] [PubMed] [Google Scholar]
- 48.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N L, Lee B, Doms R W, Margolick J, Buchbinder S, Goedert J J, O’Brien T R, Hilgartner M W, Vlahov D, O’Brien S J, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 49.Mellors J W, Kingsley L A, Rinaldo C R, Jr, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 50.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R., Jr Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 51.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. . (Erratum, 275:14, 1997.) [DOI] [PubMed] [Google Scholar]
- 52.Michael N L, Brown A E, Voigt R F, Frankel S S, Mascola J R, Brothers K S, Louder M, Birx D L, Cassol S A. Rapid disease progression without seroconversion following primary human immunodeficiency virus type 1 infection—evidence for highly susceptible human hosts. J Infect Dis. 1997;175:1352–1359. doi: 10.1086/516467. [DOI] [PubMed] [Google Scholar]
- 53.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 54.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath M J. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997;337:1267–1274. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 55.Naidu Y M, Kestler III H W, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, Desrosiers R C. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nowak M A, Lloyd A L, Vasquez G M, Wiltrout T A, Wahl L M, Bischofberger N, Williams J, Kinter A, Fauci A S, Hirsch V M, Lifson J D. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J Virol. 1997;71:7518–7525. doi: 10.1128/jvi.71.10.7518-7525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Operskalski E A, Busch M P, Mosley J W, Stram D O. Comparative rates of disease progression among persons infected with the same or different HIV-1 strains. The Transfusion Safety Study Group. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;15:145–150. doi: 10.1097/00042560-199706010-00008. [DOI] [PubMed] [Google Scholar]
- 58.Operskalski E A, Mosley J W, Busch M P, Stram D O. Influences of age, viral load, and CD4+ count on the rate of progression of HIV-1 infection to AIDS. Transfusion Safety Study Group. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;15:243–244. doi: 10.1097/00042560-199707010-00009. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 59.Pachl C, Todd J A, Kern D G, Sheridan P J, Fong S J, Stempien M, Hoo B, Besemer D, Yeghiazarian T, Irvine B. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA signal amplification assay. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;8:446–454. doi: 10.1097/00042560-199504120-00003. [DOI] [PubMed] [Google Scholar]
- 60.Pantaleo G, Demarest J F, Schacker T, Vaccarezza M, Cohen O J, Daucher M, Graziosi C, Schnittman S S, Quinn T C, Shaw G M, Perrin L, Tambussi G, Lazzarin A, Sekaly R P, Soudeyns H, Corey L, Fauci A S. The qualitative nature of the primary immune response to HIV infection is a prognosticator of disease progression independent of the initial level of plasma viremia. Proc Natl Acad Sci USA. 1997;94:254–258. doi: 10.1073/pnas.94.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sekaly R P. Major expansion of CD8+ T cells with a predominant V beta usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 62.Pantaleo G, Fauci A S. New concepts in the immunopathogenesis of HIV infection. Annu Rev Immunol. 1995;13:487–512. doi: 10.1146/annurev.iy.13.040195.002415. [DOI] [PubMed] [Google Scholar]
- 63.Phillips A N. Reduction of HIV concentration during acute infection: independence from a specific immune response. Science. 1996;271:497–499. doi: 10.1126/science.271.5248.497. [DOI] [PubMed] [Google Scholar]
- 64.Reimann K A, Li J T, Veazey R, Halloran M, Park L-W, Karlsson G B, Sodroski J, Letvin N L. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reimann K A, Tenner-Racz K, Racz P, Montefiori D C, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 67.Safrit J T, Koup R A. The immunology of primary HIV infection: which immune responses control HIV replication? Curr Opin Immunol. 1995;7:456–461. doi: 10.1016/0952-7915(95)80088-3. [DOI] [PubMed] [Google Scholar]
- 68.Schacker T, Collier A C, Hughes J, Shea T, Corey L. Clinical and epidemiologic features of primary HIV infection. Ann Intern Med. 1996;125:257–264. doi: 10.7326/0003-4819-125-4-199608150-00001. . (Erratum, 126:174, 1997.) [DOI] [PubMed] [Google Scholar]
- 69.Schacker T W, Hughes J P, Shea T, Coombs R W, Corey L. Biological and virologic characteristics of primary HIV infection. Ann Intern Med. 1998;128:613–620. doi: 10.7326/0003-4819-128-8-199804150-00001. [DOI] [PubMed] [Google Scholar]
- 70.Schwartz D H, Castillo R C, Arango-Jaramillo S, Sharma U K, Song H F, Sridharan G. Chemokine-independent in vitro resistance to human immunodeficiency virus (HIV-1) correlating with low viremia in long-term and recently infected HIV-1-positive persons. J Infect Dis. 1997;176:1168–1174. doi: 10.1086/514109. [DOI] [PubMed] [Google Scholar]
- 71.Shapiro S, Wilk M. An analysis of variance test for normality. Biometrika. 1965;52:591–611. [Google Scholar]
- 72.Shieh T, Carter D, Chadwick K, Margolick J, Zink M, Clements J. Keystone Symposia on Molecular and Cellular Biology: HIV Pathogenesis and Treatment. Drawer 1630, Silverthorne, Colo.: Keystone Symposia; 1998. Role of NK cells in control of acute SIV infection, abstr. 4085; p. 102. [Google Scholar]
- 73.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 74.Sodora D L, Lee F, Dailey P J, Marx P A. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res Hum Retroviruses. 1998;14:171–181. doi: 10.1089/aid.1998.14.171. [DOI] [PubMed] [Google Scholar]
- 75.Staprans S, Corliss B, Guthrie J, Feinberg M B. Quantitative methods to monitor viral load in simian immunodeficiency virus infections. In: Adolph K, editor. Viral genome methods. Boca Raton, Fla: CRC Press; 1996. pp. 167–184. [Google Scholar]
- 76.Staprans S I, Hamilton B L, Follansbee S E, Elbeik T, Barbosa P, Grant R M, Feinberg M B. Activation of virus replication after vaccination of HIV-1-infected individuals. J Exp Med. 1995;182:1727–1737. doi: 10.1084/jem.182.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suryanarayana K, Wiltrout T A, Vasquez G M, Hirsch V M, Lifson J D. Plasma SIV RNA viral load determination by real-time quantification of product generation in reverse transcriptase-polymerase chain reaction. AIDS Res Hum Retroviruses. 1998;14:183–189. doi: 10.1089/aid.1998.14.183. [DOI] [PubMed] [Google Scholar]
- 78.Ten Haaft P, Verstrepen B, Überla K, Rosenwirth B, Heeney J. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J Virol. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trivedi P, Horejsh D, Hinds S B, Hinds II P W, Wu M S, Salvato M S, Pauza C D. Intrarectal transmission of simian immunodeficiency virus in rhesus macaques: selective amplification and host responses to transient or persistent viremia. J Virol. 1996;70:6876–6883. doi: 10.1128/jvi.70.10.6876-6883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsai C-C, Emau P, Follis K E, Beck T W, Benveniste R E, Bischofberger N, Lifson J D, Morton W R. Effectiveness of postinoculation (R)-9-(2-phosphonylmethoxypropyl)adenine treatment for prevention of persistent simian immunodeficiency virus SIVmne infection depends critically on timing of initiation and duration of treatment. J Virol. 1998;72:4265–4273. doi: 10.1128/jvi.72.5.4265-4273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsai C C, Follis K E, Grant R, Sabo A, Nolte R, Bartz C, Bischofberger N, Benveniste R. Comparison of the efficacy of AZT and PMEA treatment against acute SIVmne infection in macaques. J Med Primatol. 1994;23:175–183. doi: 10.1111/j.1600-0684.1994.tb00119.x. [DOI] [PubMed] [Google Scholar]
- 82.Tsai C C, Follis K E, Sabo A, Beck T W, Grant R F, Bischofberger N, Benveniste R E, Black R. Prevention of SIV infection in macaques by (R)-9-(2-phosphonylmethoxypropyl)adenine. Science. 1995;270:1197–1199. doi: 10.1126/science.270.5239.1197. [DOI] [PubMed] [Google Scholar]
- 83.Valli P J S, Lukashov V V, Heeney J L, Goudsmit J. Shortening of the symptom-free period in rhesus macaques is associated with decreasing nonsynonymous variation in the env variable regions of simian immunodeficiency virus SIVsm during passage. J Virol. 1998;72:7494–7500. doi: 10.1128/jvi.72.9.7494-7500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Rompay K K A, Otsyula M G, Marthas M L, Miller C J, McChesney M B, Pedersen N C. Immediate zidovudine treatment protects simian immunodeficiency virus-infected newborn macaques against rapid onset of AIDS. Antimicrob Agents Chemother. 1995;39:125–131. doi: 10.1128/aac.39.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S-L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weissman D, Barker T D, Fauci A S. The efficiency of acute infection of CD4+ T cells is markedly enhanced in the setting of antigen-specific immune activation. J Exp Med. 1996;183:687–692. doi: 10.1084/jem.183.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O’Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien S J. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 88.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang O O, Walker B D. CD8+ cells in human immunodeficiency virus type 1 pathogenesis: cytolytic and noncytolytic inhibition of viral replication. Adv Immunol. 1997;66:273–311. doi: 10.1016/s0065-2776(08)60600-8. [DOI] [PubMed] [Google Scholar]
- 90.Zaitseva M, Blauvelt A, Lee S, Lapham C K, Klaus-Kovtun V, Mostowski H, Manischewitz J, Golding H. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat Med. 1997;3:1369–1375. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 91.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]