Abstract
Previous biochemical and genetic evidence indicated that the functional form of retroviral integrase protein (IN) is a multimer. A direct demonstration of IN oligomerization during the infectious cycle was, however, missing, due to the absence of a sensitive detection method. We describe here the generation of infectious human immunodeficiency virus type 1 (HIV-1) viral clones carrying IN protein tagged with highly antigenic epitopes. In this setting, we could readily visualize IN both in producer cells and in viral particles. More interestingly, we detected IN oligomers, the formation of which was dependent on disulfide bridges and took place inside virions. Additionally, expression of a tagged HIV-1 IN in the absence of other viral components resulted in almost exclusive nuclear accumulation of the protein. Mutation of a conserved cysteine in the proposed dimer interface determined the loss of viral infectivity, associated with a reduction of IN oligomer formation and the redistribution of the mutated protein in the nucleus and cytoplasm. Epitope tagging of HIV-1 IN expressed alone or in the context of a replication-competent viral clone provides powerful tools to validate debated issues on the implication of this enzyme in different steps of the viral cycle.
Retroviral integrase (IN) is the enzyme responsible for the insertion of a DNA copy of the viral genome in the host DNA, a crucial step of the retroviral cycle (see references 2, 3, and 6 for reviews). IN catalyzes the two steps of the retroviral integration process. The first step consists in the elimination of 2 nucleotides from each 3′ end of the proviral DNA. In the second step, the resulting 3′-OH ends of the viral DNA are covalently joined to newly created 5′ ends in the target DNA (12, 18, 22, 45, 47, 57, 61). Viral IN is expressed and incorporated into viral particles as part of a large Gag-Pol polyprotein precursor, which is processed by the viral protease in the late stages of assembly (39, 58). After viral entry in the target cell, IN remains associated with a large nucleoprotein preintegration complex (PIC), which delivers the viral genome to the nucleus (5, 11, 13, 23, 44, 48, 52). Besides IN, other viral proteins that were found to cofractionate with human immunodeficiency virus type 1 (HIV-1) PICs are the matrix, the reverse transcriptase, the viral protein R (Vpr), and, to a lesser extent, the nucleocapsid (NC) and capsid (CA) proteins (11, 23, 25, 38, 43, 44, 52). In addition, host proteins such as HMG I(Y) and Ini1, the human homologue of yeast transcription factor SNF5, although dispensable for IN activity in vitro, can stimulate IN activity under certain conditions (43) or restore function to salt-treated PICs (24). Access of PICs to the nuclear compartment differentiates oncoretroviruses, which need mitotic nuclear envelope breakdown (37, 54), from lentiviruses, which are capable of infecting nonmitotic cells (10, 49, 63). Three viral proteins, matrix (MA), IN, and Vpr, have been suggested to be implicated in the nuclear transport of HIV-1 PICs; however, their roles remain disputed (8–10, 15, 25, 28–35, 38, 62). Nuclear localization signals (NLSs) were described within the sequence of HIV-1 IN, but alteration of these putative signals was reported to interfere with a phase of the viral cycle different from the nuclear import of PICs (16, 32). Expression of HIV-1 IN fused to β-Gal failed to reveal the intrinsic nuclear tropism of IN probably because of the large size of the resulting fusion protein (46). Indeed, microinjected IN–glutathione S-transferase fusion proteins readily migrated to the nuclear compartment (32).
Another intriguing issue of retroviral IN proteins consists in the oligomerization status of IN. Detailed biochemical and genetic analysis indicates that IN proteins form multimers; however, the number of monomers involved and the conditions under which different-order oligomerization takes place are debated (4). Retroviral IN proteins comprise three functionally distinct domains: the N-terminal domain, which contains a zinc finger-like motif; the catalytic core domain; and the C-terminal DNA-binding domain (6). Sequences throughout IN have been pointed out as important for multimerization (1, 19, 21, 65). Dimers and/or tetramers of recombinant IN proteins were observed in solution (40, 41, 57, 60). The use of the yeast two-hybrid system, besides revealing homomeric interactions for HIV-1 IN, helped in defining some of the sequences involved (42). The most convincing argument that the functional form of IN is a multimer is provided by the complementation of pairs of inactive proteins carrying mutations or deletions in separate functional domains (20, 21, 27, 59). The crystal structure of the core domain of HIV-1 IN (19) shows that it consists of a five-strand β-sheet and six α-helices. In the crystal, the contact between two monomers formed a large solvent-excluded interface involving four helices and one β-strand. The crystal structure was found to be compatible with a model in which the functional unit for integration is composed of two dimers of IN (19). The more recently described structure of the N-terminal domain indicates the presence of additional multimerization determinants (14). Furthermore, zinc binding to the HHCC sequence located in this domain favors the formation of tetramers and increases the efficiency of integration in vitro (20, 65). Despite such detailed biochemical characterization of retroviral IN, oligomers of the enzyme were not directly visualized during virus replication because of the lack of a sensitive detection method.
To follow IN through the viral cycle and directly visualize possible multimeric forms of the enzyme, we constructed a replication-competent HIV-1 clone carrying a tagged IN. We show here that HIV-1 IN forms dimers and higher-order multimers in viral particles. Interestingly, oligomerization of IN was found to be dependent on disulfide bridge formation. We also confirmed the intrinsic karyophilic properties of HIV-1 IN, which displayed an almost exclusively nuclear localization when expressed in the absence of other viral products. Additionally, we report that mutation of a conserved cysteine (C130) in the proposed dimer interface results in loss of viral infectivity, reduced dimer formation, and altered subcellular localization of the enzyme. Tagged viruses provide a valuable tool for additional characterization of IN, for studying the intracellular trafficking of the PICs, and for PIC isolation and in vitro analysis.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
The HIV-1 BRU infectious molecular clone was used to express the viral IN fused to the hemagglutinin (HA) or the Flag epitope. Both epitopes were fused either at the N or at the C terminus of IN. The procedure consisted of amplification by PCR with two sets of primers. For fusion at the C terminus of IN, we used a primer encoding part of the sequence of the fusion epitope in its 5′ portion and complementary to the C-terminal sequence of IN in the 3′ portion (primer HA1 for the HA epitope or Flag1 for the Flag epitope) in combination with a primer overlapping the PflMI restriction site located upstream of the IN sequence (primer Pflm1). In a separate reaction, a primer encoding the remaining portion of the tagging epitope including a short overlap with the tagging primer and complementary to the sequence downstream of IN (HA2 or Flag2) was used with the Pflm2 primer, which overlaps the PflMI site downstream of IN. Equimolar PCR products obtained in the two initial reactions were mixed and amplified with the Pflm1 and Pflm2 primers, producing a fragment which upon digestion with PflMI could be used to replace the original BRU sequence. A similar approach was used to insert the tagging sequences near the N terminus of the IN coding region. In this setting, the Pflm1 primer was used in combination with the HA.N1 or the Flag.N1 primer, while the Pflm2 primer was combined with the HA.N2 or the Flag.N2 primer. Addition of the HA tag creates an NheI site, while the Flag sequence forms a ClaI site if inserted at the N terminus and a SpeI site if inserted at the C terminus of IN. All constructions were confirmed by DNA sequencing of the entire PCR-amplified fragment. The sequences of the primers used were as follows: Pflm1, 5′ TTCTAAAAGAACCAGTACATGGAGTGTATTAT 3′; Pflm2, 5′ CTCTTTTTCCTCCATTCTATGGAGACTCCCTG 3′; HA1, 5′ ACC TAGGCTAGCGTAATCTGGAACATCGTATGGGTAATCCTCATCCTGT CTACTTGCC 3′; HA2, 5′ GTTCCAGATTACGCTAGCCTAGGTTAGAACATGGAA 3′; HA.N1, 5′ ATCCAAGCTAGCGTAATCTGGAACATCGTATGGGTATATTCCATCTAAAAATAGTAC 3′; HA.N2, 5′ GTTCCAGATTACGCTAGCTTGGATAAGGCCCAA 3′; Flag1, 5′ CCACTAGTTACTTGTCATCGTCGTCCTTGTAATCCTCATCCTGTC 3′; Flag2, 5′ ACGACGATGACAAGTAACTAGTGGAAAAGTTTAGTAAAAC 3′; Flag.N1, 5′ ATCGTCGTCCTTGTAATCGATTCCATCTAAAAAT 3′; and Flag.N2, 5′ GAATCGATTACAAGGACGACGATGATAAGGCCCAAGATG 3′. To express the IN in the absence of other viral products, we used the pFlag expression vector (55), a kind gift from Serge Benichou (Institut Cochin, Paris, France), in which we inserted the IN sequence under the control of the simian virus 40 promoter. The IN sequence was amplified by PCR with the following primers, which created a BamHI and an XhoI restriction site at the 5′ and 3′ ends, respectively, of the IN sequence: Flagint 1, 5′ CCAGGATCCTTTTTAGATGGAATAGATAAG 3′; and Flagint 2, 5′ CTGGCTCGAGCTAATCCTCATCCTGTCTAC 3′. The Flag-INT expression vector was used for the C130-G mutagenesis, with the Quick Change mutagenesis kit (Stratagene) and the primers Cys130+ and Cys130−; Cys130+, 5′ GTACTACGGTTAAGGCAGCTGGTTGGTGGGCGGGAATC 3′; and Cys130−, 5′ GATTCCCGCCCACCAACCAGCTGCCTTAACCGTAGTAC 3′. After the mutagenesis, the BspMI fragment from the mutated Flag-INT vector was substituted for the corresponding sequence of the BRU clone, to obtain the mutated BRU-HA.Cys− and BRU-Flag.Cys− full-length clones.
Cells, virus infection, and reagents.
Human epithelial HeLa and P4 (HeLa-CD4, long terminal repeat [LTR]-LacZ) cells were grown in Dulbecco’s modified Eagle medium (Gibco), supplemented with glutamine, antibiotics, and 10% fetal calf serum. P4 cells were grown in the presence of G418 (1 mg/ml). MT4 lymphoid cells were grown in RPMI medium supplemented with glutamine, antibiotics, and 10% fetal calf serum. Viruses were produced by transfection of the plasmids as described elsewhere (56). Supernatants were analyzed for HIV-1 p24 antigen content by enzyme-linked immunosorbent assay (Dupont). P4 cells, plated in 96-well plates, were infected with different viral doses. Infectivity was measured as previously described (51). MT4 cells (106) were infected with viral supernatant corresponding to 30 ng of p24 antigen. Accumulation of p24 in the culture supernatant was measured every 2 or 3 days. The anti-HA monoclonal antibody (MAb) 12CA5 was a kind gift from Jacob Seeler (Institut Pasteur, Paris, France). The anti-Gag MAb 25A (anti-CA) and MAb 18A (anti-MA) were a kind gift from François Traincart (Institut Pasteur). The rabbit polyclonal anti-Flag antibodies were from Zymed Laboratories (San Francisco, Calif.), and the mouse anti-Flag MAb M2 was from Kodak. Cy3-conjugated anti-mouse immunoglobulin G (IgG) (heavy plus light chain) and peroxidase-conjugated anti-mouse or anti-rabbit IgG were from Amersham Life Science. N-Ethylmaleimide (NEM) was from Pierce.
Western blot analysis.
Viral supernatants were collected from transfected HeLa cells. Cells were lysed in lysis buffer (20 mM HEPES, 150 mM NaCl, 0.5% Triton X-100) containing standard protease inhibitors. Proteins corresponding to 30 μg of lysate were analyzed. Viruses were pelleted by ultracentrifugation (15 min at 60,000 rpm in a Beckman TL100 centrifuge), and pellets were resuspended in lysis buffer. For virion lysates, proteins corresponding to the indicated amounts of p24 were analyzed. When stated, NEM (freshly dissolved in dimethyl sulfoxide as a 2 M stock solution) was added in lysis buffer at a 20 mM final concentration. Samples were diluted in loading buffer, in the absence or in the presence of dithiothreitol (DTT) (60 mM final concentration), and boiled for 5 min. Samples were then run on a 10 or 12% polyacrylamide gel electrophoresis and transferred to a nitrocellulose filter (Hybond-C Super; Amersham Life Science) with a semidry electrotransfer apparatus (Bio-Rad). Filters were blocked with 5% nonfat dry milk in phosphate-buffered saline (PBS)–0.5% Tween 20 and incubated 1 h at room temperature or overnight at 4°C in PBS-Tween-milk with the indicated antibodies (final concentrations for each antibody are as follows: anti-Gag 25A plus 18A, 0.5 μg/ml; anti-HA 12CA5, 2 μg/ml; anti-Flag, 2 μg/ml). After three washes with PBS–0.1% Tween, the filters were incubated with a 1:2,500 solution of horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG in PBS-Tween-milk. After three washes, bound peroxidase activity was revealed with the ECL chemiluminescence kit (Amersham Life Science) and exposure on Hyperfilm (Amersham Life Science).
Immunoprecipitation.
Viral lysates were prepared as described above. Rabbit anti-Flag polyclonal antibodies or mouse anti-HA MAbs were covalently cross-linked to protein A-Sepharose with dimethyl pimelimidate. Ten microliters of a packed volume of protein A beads bound to antibody was incubated with viral lysates containing 200 ng of p24 at room temperature. The beads were then washed three times with lysis buffer, resuspended in denaturing loading buffer, and boiled for 10 min. Samples were then analyzed by Western blotting as described above.
Pulse-chase immunoprecipitation.
After starvation in methionine- and cysteine-free medium, transfected HeLa cells were metabolically labeled for 1 h with 100 μCi of 35S Promix (Amersham) per ml. One sample was lysed immediately (time zero), while others were chased by an excess of unlabeled amino acids for various lengths of time (1, 2, and 6 h). Cells were lysed in RIPA buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate [SDS]) containing standard protease inhibitors. Cell extracts (corresponding to 106 cells for each time point) were clarified by centrifugation and incubated overnight with 1 μg of the rabbit polyclonal anti-Flag antibodies (Zymed), and immune complexes were precipitated with Dynabeads (DYNAL) coated with anti-rabbit IgG and washed in RIPA buffer. Samples were subjected to SDS–12% polyacrylamide gel electrophoresis. After film exposure, the gel was analyzed with a PhosphorImager (Molecular Dynamics).
Indirect immunofluorescence staining.
HeLa cells (2 × 105) were spread on glass coverslips in 24-well plates, transfected with the indicated plasmids, and stained for immunofluorescence 24 to 40 h later. Cells were fixed in 3.7% formaldehyde–PBS for 20 min, washed three times in PBS, and incubated for 10 min in 50 mM NH4Cl to quench free aldehydes. After one wash in PBS and a 15-min incubation in permeabilization buffer (0.05% saponin, 0.01% Triton X-100, 2% bovine serum albumin, PBS), cells were incubated for 1 h with the first MAb (M2 anti-Flag MAb at 7.5 μg/ml) in permeabilization buffer. Cells were then washed three times in permeabilization buffer and incubated with Cy3-conjugated anti-mouse MAbs (Amersham) at a final dilution of 1:200. Cells were washed three times in permeabilization buffer and once in PBS and mounted in 133 mg of Mowiol (Hoechst) per ml–33% glycerol–133 mM Tris HCl (pH 8.5). Confocal microscopy was performed with a Leica TCS4D microscope. Series of optical sections at 0.7-μm intervals were recorded. One representative medial section was mounted by using Adobe Photoshop software.
RESULTS
Construction of integrase-tagged viral clones.
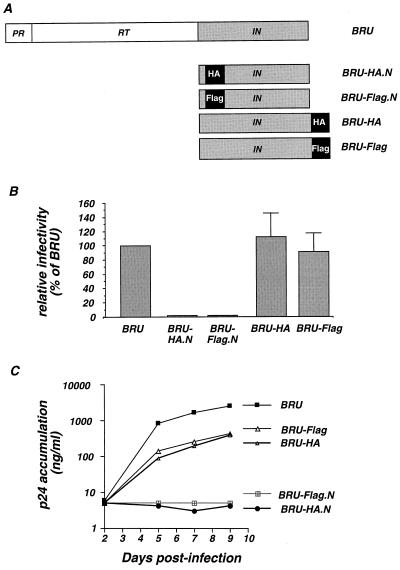
One of the major obstacles to the visualization and characterization of HIV-1 integrase protein is the absence of strongly reactive antibodies. To circumvent this problem, we constructed four proviral clones of HIV-1 in which an antigenic epitope was fused either near the N or at the C terminus of the IN protein. We chose the influenza virus HA epitope and the Flag peptide. To reduce perturbation of the proteolytic cleavage by the protease, the N-terminal fusion inserted the sequence of the epitopes after the fifth amino acid of IN. Epitope fusion at the C terminus of IN disrupted the vif open reading frame after amino acid 17; however, virus production and infectivity are not sensitive to Vif in the cell lines used in our study. We obtained the clones BRU-HA.N and BRU-Flag.N, carrying tagging epitopes fused near the N terminus of IN, and the clones BRU-HA and BRU-Flag, in which the epitopes are fused to the C terminus of IN (Fig. 1A).
FIG. 1.
Construction and infectivity of proviral clones carrying a tagged IN. (A) Sequences encoding the epitope tags were inserted by PCR either near the N or at the C terminus of IN with the HIV-1 BRU infectious molecular clone, yielding the indicated proviral clones. RT, reverse transcriptase. (B) Infectivity of the wild-type BRU and of the tagged clones was measured in a single-cycle assay on P4 (HeLa-CD4+, LTR-LacZ) cells. Values are expressed as percentages of the BRU virus and represent the means and the standard deviations of four independent experiments. (C) Viral infectivity measured as the accumulation of the HIV-1 antigen p24 in the supernatant of MT4 cells.
Effects of IN-epitope fusion on viral infectivity.
To analyze the impact of the presence of the tags on viral replication, we transfected HeLa cells with the original BRU and the four tagged clones. Viral particle production, measured as HIV-1 p24 antigen release in the culture supernatant, was not affected in the four tagged viral clones (data not shown). Culture supernatants were normalized for HIV-1 p24 viral antigen and used to infect P4 (HeLa-CD4+, LTR-LacZ) and MT4 (T-lymphoblastoid) target cells. The expression of β-galactosidase in P4 cells is strictly inducible by the HIV transactivator protein Tat, thereby allowing precise quantification of HIV-1 infectivity based on a single cycle of replication. As shown in Fig. 1B, the presence of either tag fused at the C terminus of IN did not perturb single-cycle viral infectivity. In contrast, exposure of P4 cells to the supernatant from HeLa cells transfected with the N-terminally tagged IN clones did not produce detectable infectious events. Accordingly, when the transfected cell supernatant was used to infect MT4 cells (Fig. 1C), HIV p24 antigen readily accumulated in the MT4 culture supernatant infected with the original BRU clone, as well as in that infected with the C-terminally tagged clones, indicating ongoing viral replication. The growth kinetics of these three viral clones were similar, although the amounts of p24 that accumulated in the culture supernatants of the two tagged viruses were decreased six- to ninefold with respect to the wild-type virus (Fig. 1C). In agreement with the single-cycle data, exposure of MT4 cells to the supernatant from HeLa cells transfected with BRU-HA.N and BRU-Flag.N did not result in detectable accumulation of p24 antigen in the culture supernatant, even after 9 days (Fig. 1C). To summarize, while C-terminal tagging of IN had only a minor effect on viral infectivity and growth kinetics, the same epitopes fused at the N terminus of IN resulted in noninfectious viral progeny.
Viral protein synthesis and maturation of IN-tagged viral clones.
To study viral protein synthesis for the HA-tagged proviral clones, transfected HeLa cells were lysed and analyzed by Western blotting with MAbs for the HIV-1 matrix (MA; p17) and capsid (CA; p24) proteins. Mature p17 and p24 proteins as well as partially cleaved precursor proteins were readily detected after transfection with the wild-type BRU as well as with BRU-HA and BRU-HA.N plasmid (Fig. 2A). Quantitatively and qualitatively similar Gag protein profiles were observed for the wild type and the BRU-HA clone both in cell lysates and in released viral particles (Fig. 2A), indicating that Gag protein synthesis and maturation were not affected by addition of the HA epitope at the C terminus of IN. Interestingly, despite the lack of infectivity of the N-tagged clone, we noticed only an accumulation of incompletely cleaved CA protein (still bound to the p2 spacer peptide) in the cell lysate from the BRU-HA.N clone, while no difference was noticed in the viral-particle-associated Gag protein content (Fig. 2A). Fusion of the HA epitope at the N or C terminus had no major effect on Gag protein production, particle release, and maturation (Fig. 2A), although N-terminal fusion was associated with loss of viral infectivity. Transfection of the two viral clones encoding the Flag fusion epitope at the N or C terminus of IN (BRU-Flag.N and BRU-Flag) produced Western blot protein profiles similar to those observed with the HA-tagged viruses (data not shown). This indicates that the minor proteolytic defect associated with the N-terminally tagged viruses is independent of the nature of the tag.
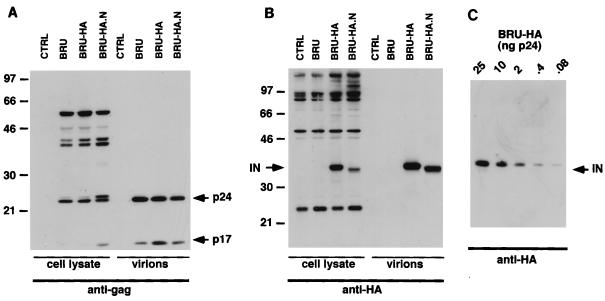
FIG. 2.
Detection of the tagged HIV-1 IN. (A and B) Western blot analysis of transfected cells and viral particles with antibody specific for the HIV-1 gag-encoded MA and CA proteins (A) and for the tagging epitope HA (B). Molecular masses (in kilodaltons) are indicated on the left side of panels A and B. Cell lysates containing 30 μg of total proteins and virion preparations containing 50 ng of p24 were analyzed. CTRL, control. (C) High sensitivity of Western blot detection of the HA-tagged IN present in viral particles. The p24 antigen content of viral particles (in nanograms) is indicated.
To detect IN in producer cell lines and in virions, we performed Western blot analysis with a MAb specific for the HA epitope. The IN-HA fusion protein was easily detected in lysates from BRU-HA.N- and BRU-HA-transfected cells (Fig. 2B). Besides the mature form of the fusion proteins, we could distinguish higher-molecular-weight bands specifically reacting with the anti-HA MAb, likely corresponding to the immature and partially cleaved Gag-Pol precursor proteins (Fig. 2B). The intensity of the signal corresponding to the mature IN was lower for the BRU-HA.N virus preparations, while accumulation of higher-molecular-weight protein bands suggested that the N-terminal fusion of the HA epitope altered processing of IN from the reverse transcriptase boundary (Fig. 2B). Of note, the apparent molecular weight of the N-terminally tagged IN protein was lower than that of the C-terminally tagged one. Such finding may reflect either a different conformation of the two proteins or the elimination of few residues from the N-terminally tagged protein. More interestingly, the mature tagged IN proteins could readily be detected in viral particle preparations from both HA-tagged viral clones (Fig. 2B). Qualitatively similar results were obtained with the BRU-Flag.N and BRU-Flag viral clones, with antibodies specific for the Flag epitope (data not shown), although the sensitivity was clearly lower than that attained with the HA epitope. Indeed, visualization of the HA fusion protein by Western blotting was remarkably sensitive, since viral preparations of the BRU-HA clone corresponding to as little as 80 pg of HIV-1 p24 antigen tested positive with the anti-HA antibody (Fig. 2C). By comparison, the IN-Flag fusion protein was detected in viral preparations corresponding to 50 ng of HIV-1 p24 antigen (data not shown).
Given the above results, we limited further analysis to the infectious C-terminally tagged viral clones BRU-HA and BRU-Flag. In particular, for the Western blot analysis, the BRU-HA clone was preferentially used.
Integrase forms protein complexes by disulfide bridges in viral particles.
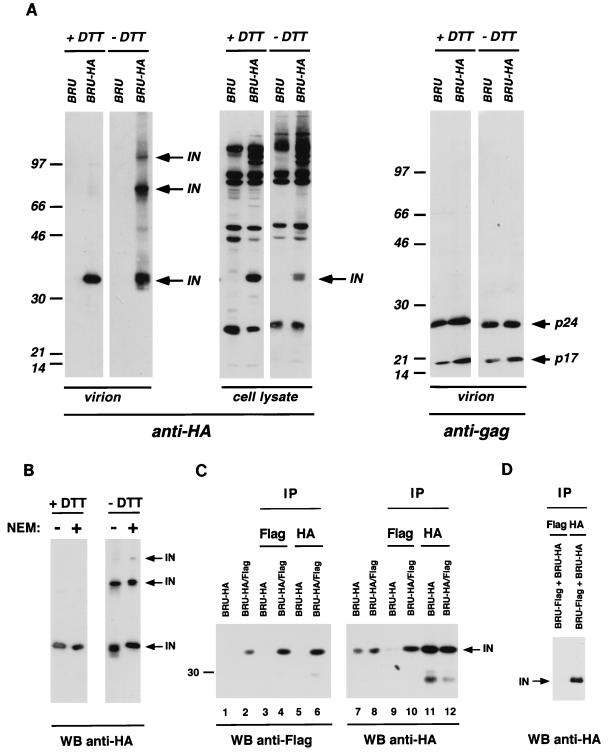
The high sensitivity of Western blot detection shown for the BRU-HA viral clone provides a tool for the biochemical characterization of IN. We investigated the possibility that IN molecules multimerize or form stable protein complexes with viral or cellular proteins. After transfection of HeLa cells with the IN-tagged BRU-HA molecular clone, we analyzed cell lysates and viral particle protein content by Western blotting, in the presence or in the absence of the reducing agent DTT (Fig. 3A). As described above, under reducing conditions the IN-HA fusion protein was readily detected in the viral particles produced by the BRU-HA clone (Fig. 3A, left panel). When the same viral preparations were analyzed under nonreducing conditions, two additional IN-specific, strongly reactive protein bands were observed (Fig. 3A, left panel), indicating that disulfide bridges are required to form two different protein complexes that contain the IN-HA fusion protein. The apparent molecular mass of the prevalent protein complex (70 kDa) would precisely match the size of an IN dimer, while the electrophoretic mobility of the larger product is compatible with a multimeric form of the IN protein or with heterologous protein complexes. A similar protein profile was observed for particles produced by the BRU-Flag clone (data not shown), demonstrating that IN complexes reflect an intrinsic property of IN and are independent of the nature of the fusion epitope.
FIG. 3.
HIV-1 IN oligomer formation in viral particles. (A) The left and middle panels show that oligomeric forms of HIV-1 IN protein were observed only in Western blot analyses performed under nonreducing conditions (− DTT). The right panel shows that migration of gag-encoded proteins is independent of the reducing conditions. Numbers at left of panels show molecular masses in kilodaltons. (B) Treatment of viral particles with the thiol-blocking agent NEM has no effect on the oligomerization status of HIV-1 IN. (C) Coimmunoprecipitation and Western blot analysis of IN oligomers formed by IN molecules carrying different tagging epitopes. BRU-HA/Flag chimeric virus was produced by the cotransfection of BRU-HA and BRU-Flag proviral clones. Antibodies used for immunoprecipitation are indicated above the panel, while antibodies used to reveal the Western blots are indicated below the panel. (D) Independently expressed BRU-HA and BRU-Flag viral particles were lysed, mixed before immunoprecipitation with the indicated antibodies, and analyzed by Western blotting with anti-HA antibody. For the direct analysis of cellular lysate and viral particle preparation, 30 μg of total protein and 30 ng of p24 were used, respectively. Viral lysates corresponding to 200 ng of p24 were analyzed in the coimmunoprecipitation experiments. WB, Western blot; IP, immunoprecipitation.
Detection of protein complexes was limited to the viral-particle-associated material, while the only HA-specific signals in the cell lysate from BRU-HA-transfected cells corresponded to the monomeric form of the IN-HA fusion protein and the above-described Gag-Pol precursor or cleavage intermediates (Fig. 3A, middle panel). This finding suggests that the formation of protein complexes involving the HIV-1 IN protein is a late event taking place inside the viral particles.
By comparison, the gag-encoded MA and CA proteins are observed in viral particle preparations only in their monomeric forms, both in the presence and in the absence of DTT (Fig. 3A, right panel), showing that disulfide bridges are not involved in intermolecular Gag product interactions.
To verify that the disulfide bridges responsible for the additional protein bands observed in the absence of DTT were actually present inside viral particles, we lysed BRU-HA virions in the presence of NEM, to block free thiol groups (17, 53). Occupation of free thiols by NEM eliminates the possibility that new disulfide bridges are formed during the lysis procedure. As shown in Fig. 3B, high-molecular-weight protein complexes were observed in the absence of DTT also when NEM was present in the lysis buffer, consistent with the interpretation that IN can be found in viral particles in at least two major protein complexes which involve disulfide bridges.
Protein complexes involving IN observed in viral particles are IN multimers.
Several reports based on trans-complementation of IN mutants or on structural considerations suggested that the functional form of retroviral integrase is a multimer (20, 21, 27, 59). We used the two replication-competent viral clones BRU-HA and BRU-Flag to investigate the nature of the disulfide-bound protein complexes described above; more specifically, we wanted to determine whether they represent IN homomers. With the aim of producing viral particles carrying different IN molecules that could be distinguished, we cotransfected HeLa cells with equal amounts of BRU-HA and BRU-Flag clones. The resulting virus, named BRU-HA/Flag, was as infectious as the parental tagged viruses in a single-cycle assay (data not shown). We then immunoprecipitated viral proteins with antibodies specific for one tagging epitope and analyzed by Western blotting the immunoprecipitated material with antibodies specific for the other tagging epitope. Protein complexes, immunoprecipitated by one antitag antibody, should be recognized by the other antitag antibody used in the Western blotting only if they contain the two differently tagged IN molecules. As controls, Western blots of nonimmunoprecipitated viral particles produced after transfection of one or both tagged viral clones were run alongside, as well as Western blots of proteins immunoprecipitated with the same antibody used for the detection (Fig. 3C).
Western blot analysis with anti-Flag serum is shown in Fig. 3C, left panel. The anti-Flag serum did not cross-react with HA-tagged IN (lane 1) and could readily detect IN-Flag protein in BRU-HA/Flag viral particles, produced by cotransfection of BRU-HA and BRU-Flag plasmids (lane 2). As expected, immunoprecipitation of BRU-HA viral proteins with anti-Flag serum or anti-HA antibody tested negative on Western blots performed with anti-Flag serum (lanes 3 and 5). Instead, BRU-HA/Flag viral particles were efficiently immunoprecipitated and subsequently revealed by the anti-Flag serum (lane 4). Most importantly, these BRU-HA/Flag viral particles tested strongly positive after immunoprecipitation with anti-HA antibody and Western blot detection with anti-Flag serum (lane 6). Such finding demonstrates that after cotransfection of BRU-HA and BRU-Flag viral clones, particles carrying both IN-HA and IN-Flag fusion proteins are formed and these proteins form multimers.
The above results were confirmed by anti-HA Western blot analysis of immunoprecipitated viral proteins (Fig. 3C, right panel). IN-HA proteins produced after transfection of BRU-HA or after BRU-HA/Flag cotransfection were easily detected in viral lysates by Western blots with anti-HA antibody (lanes 7 and 8, respectively). Predictably, immunoprecipitation of BRU-HA viral proteins with anti-Flag serum tested negative in Western blots (lane 9), while the same BRU-HA particle preparation immunoprecipitated and revealed with anti-HA antibody was strongly positive (lane 11). Proteins from BRU-HA/Flag virus could be efficiently immunoprecipitated with both anti-HA antibody (lane 10) and anti-Flag serum (lane 11), validating our finding that IN multimers are abundant in viral particle preparations.
To verify that the observed IN multimers are formed in virions that carry the two differently tagged IN proteins and not after particle lysis, culture supernatants produced after independent transfection of BRU-HA and BRU-Flag clones were lysed, mixed, and immunoprecipitated with anti-Flag serum or with anti-HA antibody. As expected, Western blot analysis performed with anti-HA antibody demonstrated that mixed IN multimers were not produced under these conditions (Fig. 3D).
Mutation of a conserved cysteine perturbs IN structure and multimerization.
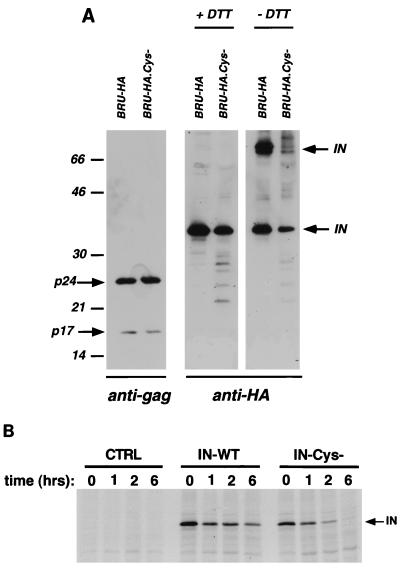
The above-described results demonstrate that HIV-1 IN forms stable multimers involving disulfide bridges in viral particles. With the aim of perturbing the structure and the multimeric status of the IN enzyme, we mutated a conserved cysteine into a glycine (C130-G) in the third α-helix of IN, in the proposed dimer interface (19), and analyzed the consequences for viral infectivity and IN multimerization. Such a mutation was inserted in both replication-competent C-terminally tagged viral clones, producing BRU-HA.Cys− and BRU-Flag.Cys−. Strikingly, mutation of the central cysteine completely abolished viral infectivity in both P4 and MT4 cells (data not shown), demonstrating that conservation of this residue is essential for viral replication. The C130-G mutation was inserted also in a nontagged virus clone, with consequent total loss of infectivity. To elucidate the impact of C130-G mutation on viral particle protein content and IN multimerization, p24 antigen-normalized amounts of particles produced by transfection of BRU-HA and BRU-HA.Cys− were analyzed by Western blotting (Fig. 4A). When the Western blot was revealed with anti-Gag antibodies, similar protein profiles were observed (Fig. 4A, left panel), demonstrating that these particles contain properly processed Gag proteins. When viral particles were lysed in the presence of DTT (Fig. 4A, middle panel), slightly less IN protein was detected for the BRU-HA.Cys− viral clone than for BRU-HA. In addition, lower-molecular-weight protein degradation products were observed for BRU-HA.Cys− virus, suggesting that the reduced IN-HA detection could be due at least in part to degradation of the mutant IN protein. Since lysis of viral particles was performed in the presence of cellular protease inhibitors, it is likely that the viral protease was responsible for the observed degradation. When viral pellets were lysed in the absence of DTT, signals of similar intensity were observed for the monomeric and dimeric forms of IN for the BRU-HA virus (Fig. 4A, right panel). Under the same conditions, the BRU-HA.Cys− virus produced a lower-intensity signal corresponding to monomers of IN and a markedly decreased signal corresponding to dimeric IN protein. Densitometric analysis of three independent experiments revealed that while for the BRU-HA virus the band corresponding to IN dimers represented approximately 60% of the intensity of the total IN signal (monomer plus dimer), for the BRU-HA.Cys− virus the dimer band corresponded to 20% of the total value. The significant difference in the ratio of the dimer to monomer signals between BRU-HA and BRU-HA.Cys− viruses indicates that cysteine 130 is important for IN dimer formation. Similar observations were made with viruses carrying Flag-IN (data not shown), indicating that the defect is independent of the tagging epitope. Cysteine 130 could be one of the residues directly involved in disulfide bridge formation between two monomers, or it could act indirectly, by determining the structure of each monomer. A possible partner for cysteine 130 could be the highly conserved cysteine 65 or cysteine 56, while it is unlikely that cysteine 130 on each monomer would interact directly, considering the relative positions of the monomers in the published structure (19).
FIG. 4.
(A) The IN mutation C130-G reduces the amount of IN oligomers in viral particles. Western blot analysis of viral particle content was performed as described in the legend to Fig. 2, with antibodies specific for the Gag products MA and CA (left panel) or with anti-HA antibody (middle and right panels) under reducing and nonreducing conditions. IN monomers and oligomers are indicated. Numbers at left show molecular masses in kilodaltons. (B) Pulse-chase labeling of IN expressed in the absence of other viral proteins. Wild-type (IN-WT) and C130-G mutant (IN-Cys−) Flag-tagged IN proteins were transiently expressed in HeLa cells. Transfected cells were metabolically labeled for 1 h and chased by an excess of unlabeled amino acids for the indicated time intervals. Cell lysates were immunoprecipitated with anti-Flag antibody and run on an SDS-polyacrylamide gel electrophoresis gel. CTRL, mock-transfected cells.
In contrast with the results illustrated in Fig. 3C for the wild-type IN, we could not detect oligomers of C130-G IN by coimmunoprecipitation followed by Western blotting (data not shown). Although this result would indeed support the hypothesis of reduced or even abolished dimer formation for the C130-G mutant, it should not be considered definitive, since mutated IN protein was poorly immunoprecipitated (data not shown).
To evaluate the stability of the mutated IN, we expressed the Flag-tagged IN proteins (wild type and C130-G mutant) in the absence of other viral proteins in HeLa cells and analyzed the cell lysates by a pulse-chase immunoprecipitation experiment. The kinetics of disappearance of the C130-G IN protein (Fig. 4B, IN-Cys- lanes) was similar to that of the wild-type IN (Fig. 4B, IN-WT lanes). Quantification by phosphorimager of two independent experiments showed that on average after a 1-h chase, 40 and 36% of wild-type and mutated IN proteins, respectively, were present. At the end of our kinetic analysis, after 6 h of chase, the percentages for wild-type and mutated IN proteins were 13 and 5%, respectively. This slight decrease in the stability of the cysteine-mutated IN may not be the sole element responsible for the marked reduction of the Western blot dimer signal observed in Fig. 4A for the BRU-HA.Cys− virus.
The data described above demonstrate that the C130-G substitution abolished viral infectivity and resulted in partial degradation of IN in viral particles, suggesting that the viral protease participates in the degradation process. Our data also indicate that the C130-G mutation reduces IN multimer formation.
Nuclear accumulation of HIV-1 IN protein and altered subcellular localization of the C130-G mutant.
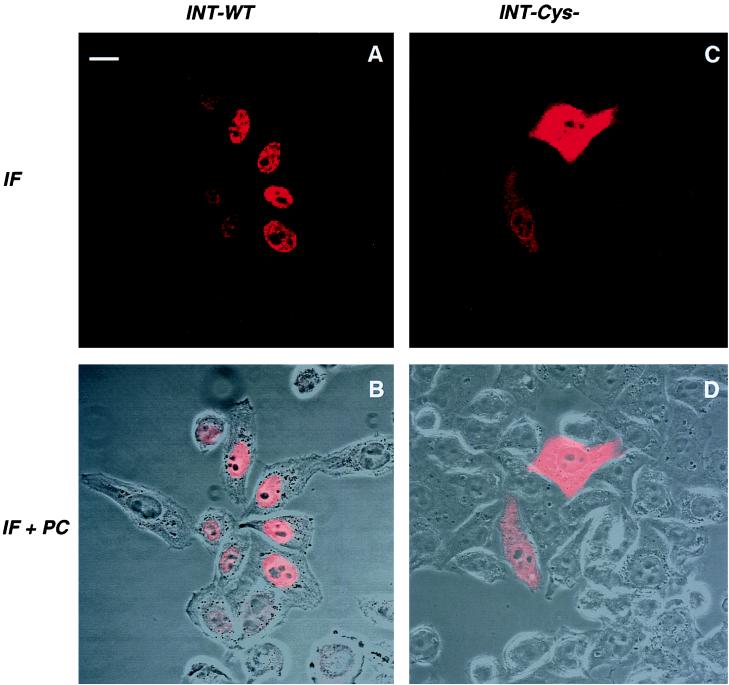
The integration process requires that PICs migrate to the nucleus of the newly infected cell. IN contains at least three sequences that resemble typical NLSs (32), suggesting a selective pressure for the accumulation of this enzyme in the nucleus or even the participation of IN in the transport of the large PIC from the cytoplasm to the nucleus. To analyze the intrinsic karyophilic properties of HIV IN, we expressed HIV-1 IN-Flag fusion protein by transfection in HeLa cells, in the absence of other viral proteins. Cells were stained with an anti-Flag antibody and analyzed by confocal microscopy. An intense nuclear staining was observed (Fig. 5A and B), demonstrating that HIV-1 IN expressed in the absence of other viral products specifically accumulates in the nuclear compartment. Since the size of the HIV-1 IN monomer is compatible with its passive diffusion through the nuclear pores (26), this observation implies an active retention of IN in the nucleus.
FIG. 5.
Confocal microscopy analysis of wild-type (WT) HIV-1 IN (A and B) and of mutant C130-G IN (C and D). HeLa cells were transfected with plasmids expressing the wild-type or mutant IN, fused to the Flag epitope. After 24 h, cells were fixed, permeabilized, and stained with anti-Flag antibodies. Series of optical sections at 0.7-μm intervals were recorded. One representative medial section is shown. Panels B and D represent a superposition of immunofluorescence (IF) staining and a phase-contrast (PC) picture of the same field. Bar = 15 μm.
We described the detrimental effects of the IN mutation C130-G on viral infectivity and on the multimerization properties of IN. We suspected that the structural alteration associated with this mutation could perturb the subcellular localization of IN, even if C130 is not part of a known NLS. Expression of mutated IN-Flag protein indeed generated a strong and diffuse staining in both the nucleus and the cytoplasm (Fig. 5C and D), which strikingly contrasts with the preferential nuclear accumulation of the wild-type IN protein (Fig. 5A and B). Thus, the C130-G mutation, which abrogates viral infectivity and decreases the efficiency of IN multimerization, also redirects IN subcellular localization.
DISCUSSION
The main goal of our study was to investigate the multimeric status of HIV-1 IN in infectious viral particles. Previous biochemical analysis evidenced homomeric interactions for several retroviral IN proteins, expressed in the absence of other viral components (40–42, 57, 60). trans-complementation of IN proteins affected in different functional domains provided genetic evidence that the functional form of retroviral IN is multimeric (20, 21, 27, 59). Determination of the crystal structure of the core and the N-terminal domains added a new dimension to the evaluation of functional models of IN (14, 19). Direct visualization of multimers in a replication-competent viral setting was hampered by the lack of a sensitive detection method.
The epitope tagging strategy adopted here allowed us to overcome the sensitivity problem and provides a powerful tool for further investigation of IN properties. Two different epitopes were fused to HIV-1 IN in order to rule out the possibility that a specific tagging sequence could be responsible for the observed phenotypes. We showed that while N-terminal tagging abolished infectivity, C-terminal tagging of HIV-1 IN had only a minor effect on viral infectivity. We could readily visualize HIV-1 IN by Western blotting performed on infectious viral particles and producer cell lysates. By coimmunoprecipitation experiments, we demonstrated that HIV-1 IN forms homomers in viral particles. Virions carrying differently tagged IN molecules were produced by cotransfection of the two C-terminally tagged viral clones. Maturation of these precursors led to the formation of chimeric IN multimers, which we could immunoprecipitate with antibody specific for one epitope and subsequently reveal on a Western blot with antibody that recognized the other epitope. Dimers were shown to be the prevalent multimeric form, by Western blot analysis. However, high-molecular-weight products, which could represent higher-order multimers or IN molecules strongly interacting with other proteins, were readily detectable. The amount of multimers present in intact viral particles could even be higher than that observed in our study, since the particle disruption procedure may have perturbed certain interactions.
Interestingly, we found that multimerization takes place inside viral particles and is dependent on disulfide bridges. However, we could not determine whether monomers are held together by intermolecular disulfide bridges or whether disulfide interaction is required to stabilize the structure of each monomer, allowing exposure of interacting surfaces. Of note, these two alternatives are not mutually exclusive. It is also likely that various molecular interactions participate in oligomerization of IN.
Several pieces of evidence provided here argue for the formation of IN oligomers inside viral particles, after proteolytic maturation of the Gag-Pol precursor. First of all, the coimmunoprecipitation experiments clearly demonstrated that chimeric IN oligomers were present in viral particles, while separately produced viral lysates mixed after particle lysis were devoid of chimeric IN oligomers. Oligomers of IN were unaffected by the addition of the thiol-blocking agent NEM to the lysis buffer, confirming that oligomer formation preceded lysis. Mature IN molecules in the cytoplasm of cells transfected with full-length viral clones were found only as monomers, possibly reflecting incompatible redox conditions of this environment. The confined space of the viral particle could also provide favorable conditions for homomeric interaction given the high local concentration of mature monomers. Our observation that IN multimerization takes place within viral particles is compatible with previous models of viral protein interaction. The contribution of Pol sequences to viral precursor assembly is considered to be negligible, and there is no evidence of homomeric interaction at the level of IN sequences in the Gag-Pol precursor (27, 39, 50, 58, 64). The finding that premature termination of protein synthesis in the IN domain resulted in improper PR activation actually suggested a role for the IN domain in delaying protease activation by reducing intermolecular interaction (7).
We also investigated the role of a highly conserved cysteine in IN multimerization. In an attempt to affect dimer formation, we mutated the cysteine 130 in the third α-helix of HIV-1 IN, which is part of the proposed dimer interacting surface (19). Such substitution abolished viral replication, although it was compatible with viral particle production and maturation. Lysed viral particles carrying mutated IN sequence were characterized by a decrease of both multimeric forms of IN. The ratio of monomers to oligomers clearly differentiated mutant and wild-type viruses. Western blot analysis of viral particles carrying the C130-G mutant IN revealed the presence of IN degradation products. Decreased stability of mutant IN protein was probably also due to structural perturbation.
The use of a tagging epitope to study the karyophilic properties of HIV-1 IN was the third aim of our study. It was previously demonstrated that avian sarcoma virus IN efficiently penetrated the nuclear envelope, under the form of a β-Gal fusion protein (46). In the same study, HIV-1 IN fused to β-Gal localized in the cytoplasm of transfected cells (46). As already suggested by those authors, the large size of the fusion protein may be responsible for the nuclear exclusion, since monomers of IN would be compatible with passive diffusion through the nuclear pores (26). We describe here an almost exclusively nuclear accumulation of a tagged IN in the absence of other viral components. Tagging the isolated IN provides a convenient tool to determine the residues responsible for nuclear import and retention. NLSs have been described within the HIV-1 IN sequence, which may be validated by using the IN-Flag fusion protein. Although the C130-G mutation does not directly affect any of the potential NLSs, the mutated proteins were evenly distributed in the nucleus and the cytoplasm of transfected cells. In the absence of specific NLS, passive diffusion of IN through the nuclear pores would generate a staining pattern similar to the one observed for the C130-G mutant. We propose that the loss of nuclear retention reflects the lesser ability of the C130-G IN protein to form dimers. With respect to size, IN dimers (or oligomers) must enter the nucleus through active transport (36). It is also conceivable that the C130-G mutation indirectly perturbed motifs that are implicated in nuclear targeting.
The lack of a requirement for other viral components to determine nuclear accumulation of IN shown here may suggest an active role of IN in the nuclear targeting of the PICs. Mutational analysis performed on the tagged IN expressed alone and in a replication-competent viral clone could help determine the role of IN at the level of PICs. Our current goal is to follow the intracellular trafficking of the PICs by using the tagged IN as a marker.
Given the implication of IN in different steps of the viral cycle, it is often difficult to associate a single functional role with a protein domain. The combined use of the two tagged-IN expression vectors could facilitate the evaluation of the impact of specific mutations on the dimerization and the nuclear accumulation properties. We consider that the significantly increased immunogenicity of the tagged IN provides a valuable tool for the study of several currently debated aspects of the retroviral cycle in which IN is implicated.
ACKNOWLEDGMENTS
We thank Jean Michel Heard and Francois Clavel for their support and critical suggestions. We thank Emmanuelle Perret for confocal microscopy analysis and Jacob Seeler, François Traincart, and Serge Benichou for the kind gifts of reagents.
F.M. is a fellow of the Agence Nationale de Recherche sur le SIDA (ANRS). C.P. is a fellow of SIDACTION. This work was supported by grants from the ANRS and the Pasteur Institute.
REFERENCES
- 1.Andrake M D, Skalka A M. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 2.Andrake M D, Skalka A M. Retroviral integrase, putting the pieces together. J Biol Chem. 1996;271:19633–19636. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 3.Asante-Appiah E, Skalka A M. Mini-review: molecular mechanisms in retrovirus DNA integration. Antiviral Res. 1997;36:139–156. doi: 10.1016/s0166-3542(97)00046-6. [DOI] [PubMed] [Google Scholar]
- 4.Barsov E V, Huber W E, Marcotrigiano J, Clark P K, Clark A D, Arnold E, Hughes S H. Inhibition of human immunodeficiency virus type 1 integrase by the Fab fragment of a specific monoclonal antibody suggests that different multimerization states are required for different enzymatic functions. J Virol. 1996;70:4484–4494. doi: 10.1128/jvi.70.7.4484-4494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 6.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–204. [PubMed] [Google Scholar]
- 7.Bukovsky A, Gottlinger H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol. 1996;70:6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Haffar O K. HIV-1 nuclear import: matrix protein is back on center stage, this time together with Vpr. Mol Med. 1998;4:138–143. [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukrinsky M I, Sharova N, Dempsey M P, Stanwick T L, Bukrinskaya A G, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 13.Bushman F D, Miller M D. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai M L, Zheng R L, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 15.Camaur D, Gallay P, Swingler S, Trono D. Human immunodeficiency virus matrix tyrosine phosphorylation: characterization of the kinase and its substrate requirements. J Virol. 1997;71:6834–6841. doi: 10.1128/jvi.71.9.6834-6841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon P M, Byles E D, Kingsman S M, Kingsman A J. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J Virol. 1996;70:651–657. doi: 10.1128/jvi.70.1.651-657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carleton M, Brown D T. The formation of intromolecular disulfide bridges is required for induction of the Sindbis virus mutant ts23 phenotype. J Virol. 1997;71:7696–7703. doi: 10.1128/jvi.71.10.7696-7703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 19.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 20.Ellison V, Gerton J, Vincent K A, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 21.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 23.Farnet C, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farnet C M, Bushman F D. HIV-1 cDNA integration—requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 25.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finlay D R, Forbes D J. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- 27.Fletcher T M, Soares M A, McPhearson S, Hui H X, Wiskerchen M A, Muesing M A, Shaw G M, Leavitt A D, Boeke J D, Hahn B H. Complementation of integrase function in HIV-1 virions. EMBO J. 1997;16:5123–5138. doi: 10.1093/emboj/16.16.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouchier R A, Meyer B E, Simon J H, Fischer U, Albright A V, Gonzalez-Scarano F, Malim M H. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fouchier R A, Meyer B E, Simon J H, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed E, Englund G, Maldarelli F, Martin M A. Phosphorylation of residue 131 of HIV-1 matrix is not required for macrophage infection. Cell. 1997;88:171–174. doi: 10.1016/s0092-8674(00)81836-x. [DOI] [PubMed] [Google Scholar]
- 31.Freed E O, Martin M. HIV-1 infection of non-dividing cells. Nature. 1994;369:107–108. doi: 10.1038/369107b0. [DOI] [PubMed] [Google Scholar]
- 32.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- 35.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 36.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hajihosseini M, Iavachev L, Price J. Evidence that retroviruses integrate into post-replication host DNA. EMBO J. 1993;12:4969–4974. doi: 10.1002/j.1460-2075.1993.tb06190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 40.Jenkins T M, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 41.Jones K S, Coleman J, Merkel G W, Laue T M, Skalka A M. Retroviral integrase functions as a multimer and can turn over catalytically. J Biol Chem. 1992;267:16037–16040. [PubMed] [Google Scholar]
- 42.Kalpana G V, Goff S P. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 44.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 45.Katzman M, Mack J P, Skalka A M, Leis J. A covalent complex between retroviral integrase and nicked substrate DNA. Proc Natl Acad Sci USA. 1991;88:4695–4699. doi: 10.1073/pnas.88.11.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kukolj G, Jones K S, Skalka A M. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J Virol. 1997;71:843–847. doi: 10.1128/jvi.71.1.843-847.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leavitt A D, Rose R B, Varmus H E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee M S, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis P, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H M, Wu X Y, Xiao H L, Conway J A, Kappes J C. Incorporation of functional human immunodeficiency virus type 1 integrase into virions independent of the Gag-Pol precursor protein. J Virol. 1997;71:7704–7710. doi: 10.1128/jvi.71.10.7704-7710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mammano F, Petit C, Clavel F. Resistance-associated loss of viral fitness in human immunodeficiency virus: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J Virol. 1998;72:7632–7637. doi: 10.1128/jvi.72.9.7632-7637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Opstelten D J E, Wallin M, Garoff H. Moloney murine leukemia virus envelope protein subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J Virol. 1998;72:6537–6545. doi: 10.1128/jvi.72.8.6537-6545.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rousset R, Desbois C, Bantignies F, Jalinot P. Effects of NF-kappa B1/p105 processing of the interaction between the HTLV-1 transactivator Tax and the proteasome. Nature. 1996;381:328–331. doi: 10.1038/381328a0. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz O, Maréchal V, Danos O, Heard J M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–204. [PubMed] [Google Scholar]
- 59.van Gent D C, Vink C, Groeneger A A, Plasterk R H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vink C, van Gent D C, Elgersma Y, Plasterk R H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Schwedler U, Kornblut R S, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinberg J B, Matthews T J, Cullen B R, Malim M H. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferative human monocytes. J Exp Med. 1991;88:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu X Y, Liu H M, Xiao H L, Conway J A, Hunter E, Kappes J C. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 1997;16:5113–5122. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng R L, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]