Abstract
Progressive changes in host mRNA expression can illuminate crucial pathogenetic pathways in infectious disease. We examined general and specific approaches to mRNA expression in three rodent models of Creutzfeldt-Jakob disease (CJD). Each of these models displays distinctive neuropathology. Although mRNAs for the chemokine receptor CCR5, the lysosomal protease cathepsin S, and the pleiotropic cytokine transforming growth factor β1 (TGF-β1) were progressively upregulated in rodent CJD, the temporal patterns and peak magnitudes of each of these transcripts varied substantially among models. Cathepsin S and TGF-β1 were elevated more than 15-fold in mice and rats infected with two different CJD strains, but not in CJD-infected hamsters. In rats, an early activation of microglial transcripts preceded obvious deposits of prion protein (PrP) amyloid. However, in each of the three CJD models, the upregulation of CCR5, cathepsin S, and TGF-β1 was variable with respect to the onset of PrP pathology. These results show glial cell involvement varies as a consequence of the agent strain and species infected. Although neurons are generally assumed to be the primary sites for agent replication and abnormal PrP formation, microglia may be targeted by some agent strains. In such instances, microglia can both process PrP to become amyloid and can enhance neuronal destruction. Because microglia can participate in agent clearance, they may also act as chronic reservoirs of infectivity. Finally, the results here strongly suggest that TGF-β1 can be an essential signal for amyloid deposition.
There are two fundamentally different concepts of the infectious agents that cause Creutzfeldt-Jakob disease (CJD), bovine spongiform encephalopathy (BSE), and scrapie. The prion hypothesis stipulates that some conformational form of the host prion protein (PrP) is infectious (1, 53). However, no detectable form of this protein reproducibly correlates with infectious titers (reviewed in reference 33), and purified recombinant or transgenic PrP has failed to yield significant infectivity (19). An alternate view is that the agent is likely to contain a nucleic acid genome. This is based on biological characteristics of agent replication and strain variation (5, 34). Attenuated strains of CJD can dramatically suppress the replication of more virulent strains without detectable changes in PrP (34). Additionally, infectivity displays virus-like physical characteristics that are distinct from those of PrP (2, 54, 56, 57), and disruption of virus-like structures leads to a loss of infectivity (39). Nonetheless, a viral sequence has not been defined, even though nucleic acids exceeding 1 kb in size can be retrieved from more purified infectious preparations (3, 11).
PrP is clearly important in disease pathogenesis and susceptibility to infection (6). Abnormal PrP accumulation is often postulated to be the earliest and most central event in the infection, ultimately leading to secondary changes in glial cells. Perhaps this is not universally true. Using different agent strain-host combinations, recent experiments with animal models of CJD have suggested that pathogenetic events may be more complex. For example, microglial and astrocytic changes can be observed before major PrP pathology in a rat CJD model (SY-Rat), whereas the same strain in hamsters (SY-Ha) shows major astrocytic changes only after large accumulations of pathologic PrP (35, 36). In these instances, the abnormal PrP level was determined by a sensitive Western blot assay based on abnormal PrP’s relative proteinase K resistance (PrP-res).
To begin to define the cellular basis of developing pathology in different CJD models, we used an arbitrary screen for alterations in mRNA expression in late-stage SY-Ha CJD. Using differential display (DD), we identified only a few true differentially expressed transcripts as assessed by Northern blot studies. Because these were not highly informative, we ultimately targeted genes that might be indicative of microglial and astrocytic changes, including those that could differ in three distinct CJD models. The current study evaluated the SY-Rat and SY-Ha models, as well as the FU strain of CJD passaged in mice (FU-Mo). These models encompass a broad spectrum of pathologic sequelae, including wide variations in amyloid plaque formation and incubation times. Incubation times generally indicate differences in agent virulence (34). Our current mRNA expression data confirm remarkable differences in the pattern of glial activation in these models. The microglial activation in particular emphasizes the importance of bone marrow-derived cells in host responses to different agent strains. Indeed, different strains may target different cell types in the immune system. Historically the role of the immune system in CJD has largely been ignored, although the spleen, lymph nodes, and leukocytes have long been known to harbor these infectious agents (13, 31).
MATERIALS AND METHODS
Disease models and agent strains.
The SY-Ha model was from the 32nd serial passage of the SY CJD strain in hamsters and has been characterized for temporal increases in infectivity as well as PrP changes (35). Although first established in guinea pigs in 1976, SY is representative of other common CJD strains in the United States directly passaged from humans to hamsters and mice (38). SY-Rat was from the fourth serial passage as described previously (36), allowing direct comparison with the same material previously analyzed for other molecular and histological changes. FU-Mo was derived from an Asiatic strain of CJD and has considerably greater virulence in mice than standard U.S. CJD strains (34). Mice were inoculated intracerebrally with 30 μl of a 1% brain homogenate in isotonic saline, whereas rats and hamsters were inoculated with 50 μl of a 10% brain homogenate.
RNA isolation and DNase treatment.
Fresh tissue was homogenized in Trizol (Gibco Life Technologies, Gaithersburg, Md.) at 1 ml/100 mg of tissue (wet weight), and RNA was extracted according to the manufacturer’s instructions or was purified by guanidinium isothiocyanate-cesium chloride gradient centrifugation (40). RNA was resuspended in diethyl pyrocarbonate-treated water at concentrations of >5 μg/μl and then digested with 40 U of DNase I (Boehringer Mannheim, Indianapolis, Ind.) for 1 h at 37°C in a 50-μl reaction mixture containing 50 mM Tris (pH 7.5), 10 mM MgCl2, and 20 U of RNase inhibitor (RNasin; Boehringer Mannheim). Samples were then reextracted with Trizol, and RNA was stored at −70°C.
Arbitrarily-primed DD RT-PCR.
DD with arbitrary primers was conducted by using reagents provided in the DisplaySystems kit (Display Systems Biotech, Los Angeles, Calif.). For each reaction, 200 ng of total RNA was reverse transcribed in a 30-μl reaction mixture containing 50 mM Tris (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 20 mM (each) deoxynucleoside triphosphates (dNTPs), 2.5 μM anchored primer (T11GG, T11CC, T11GC, or T11CG), 28 U of RNasin, and 450 U of Superscript II reverse transcriptase (Gibco Life Technologies). RNA was denatured for 2 min at 70°C in the presence of the anchored primer and then quickly chilled on ice. After the addition of reaction buffers, dNTPs, and RNasin, samples were incubated at 25°C for 10 min. Reverse transcriptase was then added and the reactions were incubated for 5 min at 25°C, 1 h at 37°C, and 5 min at 95°C. Separate reactions were used for each anchored primer, and control reactions were performed without reverse transcriptase to assess genomic DNA contamination.
One-microliter aliquots of reverse transcription (RT) reaction mixtures were used as templates for PCR containing buffer A [67 mM Tris (pH 8.8), 4 mM MgCl2, 16 mM (NH4)2SO4, 33.2 μg of purified bovine serum albumin per ml] (28) with 2 μM each dNTP, 0.5 μM arbitrary decamer, 2.5 μM anchored primer, 1 μCi of [α-33P]dATP (∼3,000 Ci/mmol; Amersham, Arlington Heights, Ill.), and 2 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.) in a final volume of 20 μl. The hot start procedure included denaturation for 1 min at 95°C and equilibration to 80°C for 1 min prior to the addition of polymerase. Samples were then cycled for 20 to 30 times (see Results section) of 30 s at 95°C, 1 min at 40°C, and 1 min at 72°C with a final extension for 5 min at 72°C. The resulting PCR products were separated by electrophoresis on 5% polyacrylamide–7.3 M urea gels with 1× TBE running buffer and exposed to BioMax MR film (Kodak, Rochester, N.Y.) at room temperature.
Products of interest were excised from the polyacrylamide gels and eluted into 100 μl of Tris-EDTA by incubating for 3 h at 65°C. One microliter of the eluate was reamplified by using buffer A, 50 μM each dNTP, 0.2 μM arbitrary primer, 0.2 μM anchored primer, and 2 U of Taq for 30 cycles as described above. Reamplification products were analyzed by agarose gel electrophoresis to confirm the size and purity of the DD product.
For DD targeted to the hamster glial fibrillary acidic protin (GFAP) gene, forward (F) 5′-AGCCTCAAGG-3′ and reverse complement (RC) 5′-TGACACGGAC-3′ arbitrary decamers were designed based on nucleotides 10 to 19 and 192 to 201 of the hamster GFAP cDNA sequence (GenBank accession no. J03847). These primers are equivalent to the arbitrary primers used in standard DD (10-mers; 50 to 60% GC content). RT was performed as described above with 2.5 μM of the GFAP reverse arbitrary decamer. PCR amplification was also done as previously described, except that the GFAP forward and reverse arbitrary decamers were each used at a final concentration of 0.5 μM.
Targeted DD.
One microgram of total RNA was reverse transcribed in a 20-μl reaction mixture containing 50 mM Tris (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 20 μM each dNTP, 250 ng of random hexamers, 20 U of RNasin, and 300 U of Superscript II reverse transcriptase. RNA was denatured for 2 min at 70°C in the presence of random hexamers and then quickly chilled on ice. After the addition of reaction buffers, dNTPs, and RNasin, the reaction was incubated at 25°C for 10 min. Reactions were then equilibrated at 42°C for 2 min prior to the addition of reverse transcriptase. First-strand cDNA synthesis proceeded for 50 min at 42°C, and enzymes were inactivated by heating to 95°C for 5 min. Genomic DNA contamination was monitored as described in the previous section.
Design of degenerate primers for PCR was based on the conserved amino acid motifs present in particular gene families. The F primer 5′-YMGHTACCTGGCYATTGTSC-3′ and RC primer 5′-AYNGGRTTNACGCAGCARTG-3′ were designed to prime from the DRYLAIV and HCCVNPL motifs present in various chemokine receptors. For other G protein-coupled receptors, the transmembrane domain sequences IYIFNLA and NPVLYAF were targeted with F primer 5′-ATHTAYATHTTYAAYCTNGC-3′ and RC primer 5′-AANGCRTANAGNACGGGRTT-3′. The zinc binding motif VGHNFG and the disintegrin domain sequence EECDPG of certain metalloproteinases were targeted with F primer 5′-GGTNGGNCAYAAYTTYGG-3′ and RC primer 5′-GCCNGGRTCRCAYTCYTC-3′. For zinc finger proteins, the motifs CPECGK and HTGEKP were targeted with F primer 5′-TGYCCNGARTGYGGNAA-3′ and RC primer 5′-GGYTTYTCNCCNGTRTG-3′. RT reaction mixtures were diluted 10-fold, and 1-μl aliquots were used as templates for hot start PCR containing buffer A, 2 μM each dNTP, 1 μM F primer, 1 μM RC primer, 1 μCi of [α-33P]dATP (∼3,000 Ci/mmol), and 2 U of Taq in a final volume of 20 μl. The cycling protocol was two cycles of 30 s at 95°C, 1 min at 40°C, and 1 min at 72° followed by 25 cycles of 30 s at 95°C, 1 min at 65°C, and 1 min at 72°C, with a final extension for 5 min at 72°C. Products were separated and purified by gel electrophoresis as described above. Reamplification was performed in buffer A, 50 μM dNTP, 1 μM F primer, 1 μM RC primer, and 2 U of Taq.
Selection of differentially expressed sequences for cloning.
All DD reactions were performed in duplicate to demonstrate the reproducibility of the PCR product representation. To purify products of interest from contaminating PCR products of the same size that are not differentially expressed, we used a modified single-stranded conformation polymorphism (SSCP) approach. This SSCP technique provides an additional method for eliminating potential false positives from further analysis (41). Ten microliters of reamplified PCR product was combined with 10 μl of loading buffer (95% formamide, 20 mM EDTA [pH 8.0], 0.05% bromophenol blue, 0.05% xylene cyanol), heated for 2 min at 94°C, and quickly chilled on ice. Products were then applied to a 0.5× Mutation Detection Enhancer gel (J. T. Baker, Phillipsburg, N.J.) in 0.6× TBE. After electrophoresis for approximately 2 h at 6 W constant power, the gel was stained with SYBR Gold (Molecular Probes, Eugene, Oreg.), and the products of interest were purified from the gel by using the QIAEX II gel purification kit (Qiagen, Valencia, Calif.). The purified material was amplified again by 25 cycles of PCR for cloning with the Perfectly Blunt PCR cloning kit (Novagen, Madison, Wis.).
In some cases, products of interest were purified by an analogous technique using agarose gels (61). Ten microliters of reamplified product was loaded onto a 3% agarose gel containing 1 U of Resolver Gold (Novagen) per ml. Gels were run for 60 to 90 min at 5 V/cm in 0.5× TBE (pH 7.5) and stained after electrophoresis with ethidium bromide. Products were extracted from the gels by using the Qiaquick gel purification kit (Qiagen) and also reamplified by 25 PCR cycles for cloning as described above.
RNA expression analysis.
Portions of the contactin, CCR5, CCR3, cathepsin S, and transforming growth factor β1 (TGF-β1) sequences were cloned by RT-PCR to generate probes for Northern blots of total RNA. One microgram of rat brain total RNA was reverse transcribed with random hexamers as described in the “Targeted DD” section above. One-microliter aliquots of the RT reaction mixtures were used as templates for PCR containing 50 mM KCl, 10 mM Tris (pH 8.3), 1.5 mM MgCl2, 20 μM dNTP, 1 μM F primer, 1 μM RC primer, and 2 U of Taq. The F and RC primers used were as follows: contactin, F, 5′-TGCCATTGCTGGTCAGCCATCTCC-3′, and RC, 5′-CCGGCAGTTGAGTGACACTTTTCC-3′; CCR5, F, 5′-AAGAGAAGGTGAGACATCCGTTCCC-3′, and RC, 5′-AAACTTCCTGTTCTCCTGTGGACCG-3′; CCR3, F, 5′-CCTTTGAGACCACACCCTATG-3′, and RC, 5′-CTGTGGAAAAAGAGCCGAAGG-3′; cathepsin S, F, 5′-CAACTGCAGAGAGACCTACCCTGG-3′, and RC, 5′-AGAGGAAGAAGGAGGAATGGCTGG-3′; and TGF-β1, F, 5′-CAGTGCCAGAACCCCCATTG-3′, and RC, 5′-GAAGGGTCGGTTCATGTCATG-3′. The PCR products were also cloned with the Perfectly Blunt PCR cloning kit. In all cases, the fidelity of the probe was confirmed by automated sequencing prior to labeling.
RNA glyoxylation, electrophoresis, blotting, and hybridization were performed as previously described (35, 40). Each time point on any Northern blot was a pooled RNA sample from two animal brains. Hybridization probes were generated by random priming of double-stranded DNA templates with [α-32P]dCTP by using the Decaprime II labeling kit (Ambion, Austin, Tex.). Blots were exposed to BioMax MS film at −70°C with TransScreen HE intensifying screens (Kodak). For quantitation, densitometry of linear-range film exposures with optical density standards was performed by using NIH Image version 1.61 (available at ftp://codon.nih.gov/pub/nih-image) on direct scans that had not been subjected to any image processing. Levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH; clone kindly provided by Adrian Hayday) were used to normalize the expression of all other genes studied; GAPDH expression has previously been shown to remain constant through the course of the CJD infection, regardless of the animal model examined (35, 36).
RESULTS
Effectiveness of DD.
We first tested the ability of the DD procedure to identify tissue-specific patterns of gene expression. DD using standard arbitrary and anchored primers generated reproducible band patterns from hamster brain and liver RNA; several examples of putative differential expression were apparent in each tissue (Fig. 1A). These results indicated that the procedure was adequate for identifying qualitative examples of differential expression.
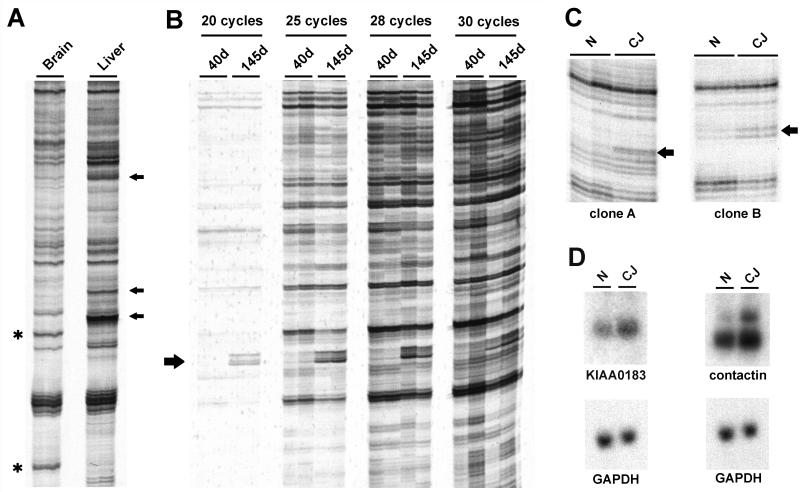
FIG. 1.
DD of control and CJD samples. (A) cDNA samples in duplicate from hamster brain and liver showing typical analysis with a standard arbitrary primer pair. Several brain-enriched and liver-enriched DD products are indicated by asterisks and arrows, respectively. (B) DD using primers targeted to the hamster GFAP cDNA sequence. RNA samples in duplicate from CJD-infected hamsters 40 and 145 days after inoculation were amplified by RT-PCR for 20, 25, 28, or 30 PCR cycles. Each PCR was performed in duplicate to verify reproducibility of the PCR products. The 192-bp GFAP product is indicated by an arrow. (C) DD of normal (N) and terminal CJD (CJ) of the SY-Ha model verified in duplicate. Differentially expressed products are indicated by arrows. (D) Northern blots of total RNA from normal and terminal SY-Ha CJD confirm differential expression compared to that of the steady-state GAPDH control.
To assess the accuracy and sensitivity of DD we compared GFAP expression in early and terminal SY-Ha infection (40 and 145 days postinoculation, respectively). GFAP mRNA levels differ by about 10-fold between these two time points, as assessed by Northern blotting (35). To find the best differential sensitivity, 20 to 30 cycles of PCR were used with the designed GFAP primers. The expected 192-bp product was increased in terminal disease (Fig. 1B). However, with increasing numbers of cycles, the differences in GFAP expression diminished. Because an intermediate number of cycles offered greater sensitivity than fewer cycles, but still retained the stoichiometry of differential GFAP expression, we used 25 PCR cycles in subsequent experiments. These results concur with those of other investigators indicating a trade-off between sensitivity and quantitative accuracy in DD (42).
Arbitrarily primed DD.
End-stage CJD hamsters were compared with control uninoculated hamsters of comparable age by standard DD procedures. Unlike the liver-versus-brain patterns, the vast majority of bands were identical in control and infected hamster brains. Figure 1C shows sample reactions with apparent differentially expressed products. To more rigorously evaluate altered mRNA levels in the SY-Ha model, Northern blots of total RNA from another group of hamsters were probed with cloned DD products. The majority of the products cloned (17 of 19) did not demonstrate any differential expression in SY-Ha total RNA as compared to uninfected controls. The remaining clones showed a modest increase in end-stage SY-Ha RNA (Fig. 1D). Sequence analysis revealed that clone A was 99% identical to the sequence of the human KIAA0183 gene, a cDNA identified by sequencing of a human myeloblast cDNA library (47).
Clone B was 95% identical to the mouse sequence for contactin (also known as the F3 antigen), a neuronal cell-surface molecule of the immunoglobulin superfamily. On Northern blots, contactin hybridized to two different RNA species of about 7 and 10 kb. Although the intensity of the 7-kb band was unchanged in terminal SY-Ha brain, densitometric analysis of Northern blots demonstrated an approximately threefold increase in expression of the 10-kb mRNA (Fig. 1D). The detection of the 7-kb transcript was in agreement with the results of other investigators, but whether the 10-kb band was an alternative splice variant or was derived from another gene product with considerable sequence homology to the contactin probe could not be determined. To test these possibilities, another region of the hamster contactin gene was amplified, cloned, and used to probe a Northern blot. The results were indistinguishable from those obtained with the original contactin probe, suggesting that the 10-kb band seen on these blots was indeed the result of alternative processing of a contactin transcript (data not shown). Consistent with this interpretation, a human mRNA of similar length has been detected (4). Northern blot analysis across the time course of SY-Ha infection demonstrated that contactin mRNA upregulation was confined to the terminal stage of disease (data not shown). Contactin is thought to mediate interactions between neurons and glia (48, 50) and may be evoked in the final stages of repair and scarring in CJD-infected hamsters. In summary, whereas standard DD with 96 arbitrary primer combinations was reasonably robust for comparing different tissues, it was not particularly useful for revealing abnormalities in CJD-infected brain.
Targeted DD reveals differences in chemokine receptor expression.
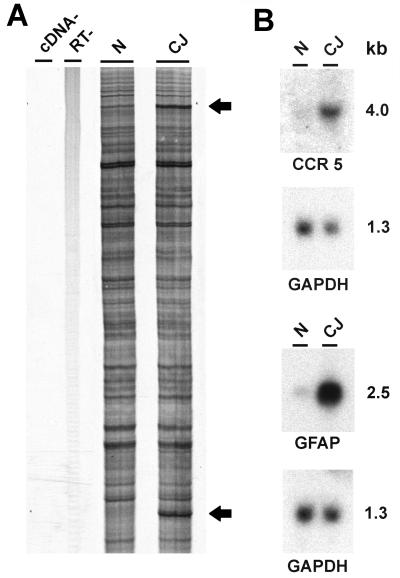
DD was combined with degenerate primer PCR to examine a range of specific gene families for altered expression in SY-Ha. Metalloproteinases are involved in the processing of molecules such as tumor necrosis factor alpha (TNFα) and TGF-α (49), cytokines that might contribute to alterations in neurons and astroglia. Degenerate primers were targeted to the zinc binding domain and disintegrin domain present in these proteases. We also exploited well-known conserved regions in zinc finger proteins and G protein-coupled receptors. None of these primer sets identified differentially expressed transcripts (data not shown). However, primers targeted to conserved motifs in both the CCR and CXCR subfamilies of chemokine receptors, known to be highly concentrated in cells of the immune system, revealed two DD bands that were reproducibly upregulated in terminal SY-Ha infection (Fig. 2A). These products were reamplified, purified by SSCP electrophoresis, and used to confirm a greater than threefold increased expression in the SY-Ha model by Northern blotting (Fig. 2B). The 550-bp product was within the size range expected for these degenerate primers, and sequencing revealed that this product was 92% identical to the mouse β-chemokine receptor CCR5.
FIG. 2.
Directed DD and confirmation of CCR5 upregulation in SY-Ha CJD. (A) Degenerate primers directed to chemokine receptor motifs were used to amplify normal (N) and terminal CJD (CJ) hamster cDNA. The upper and lower bands (arrows) correspond to CCR5 and GFAP, respectively. The GFAP band had been amplified from the 3′-untranslated region of the transcript by the degenerate primers. No bands are seen in the two negative control experiments performed in parallel, one without cDNA (cDNA− lane) and the second without reverse transcriptase (RT− lane). (B) Northern blots of total hamster brain RNA confirm upregulation of sequences identified by DD. Approximate molecular weights (kilobases) are indicated.
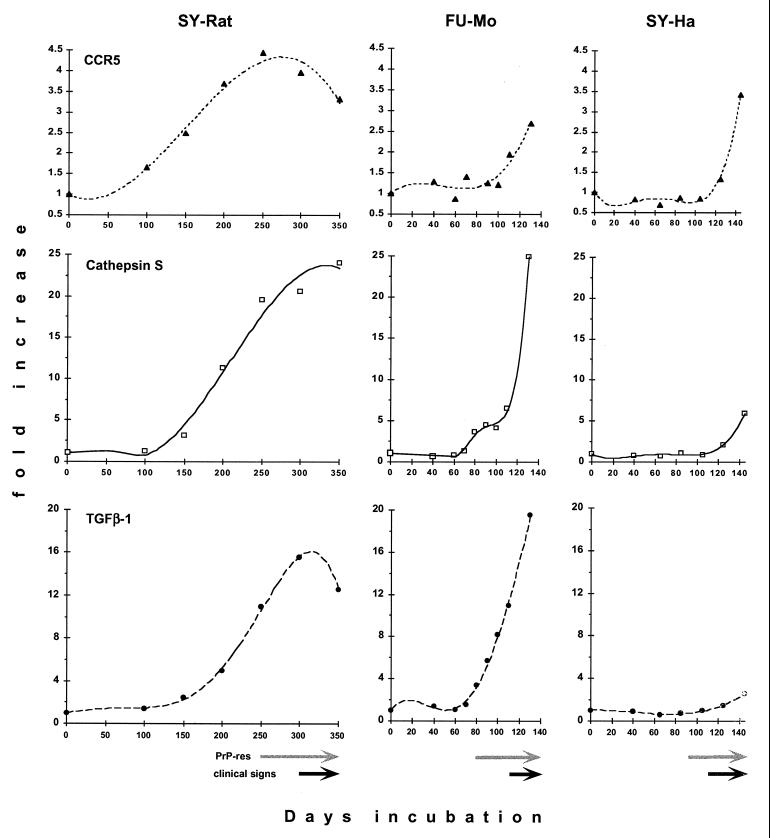
The relevance of this transcript was further documented by examining a complete time course in three distinct CJD models. In the SY-Rat model, CCR5 levels were increased 1.6-fold as early as 100 days after inoculation and continued to rise thereafter, peaking at a level of about 4.5-fold after 250 days. CCR5 expression remained elevated throughout disease, but did decline somewhat during the final stages (Fig. 3 and 4). The initial increase in CCR5 mRNA correlated with the activation of astrocytes, as assessed by previous GFAP mRNA studies, and microglial activation, as indicated by keratan sulfate immunohistochemistry (36). In contrast, FU-Mo CCR5 was upregulated only at the terminal stage of disease, with twofold elevations at 110 days and only a threefold elevation at 120 days (Fig. 4). Similarly, SY-Ha upregulation of CCR5 was only detectable at 125 days and reached maximal levels of ∼3.6-fold when hamsters were moribund at 145 days (Fig. 4).
FIG. 3.
Time course of gene expression in the SY-Rat CJD model. Northern blots of total rat brain RNA isolated at the indicated times (days [d]) after inoculation were sequentially hybridized with the CCR5, cathepsin S, TGF-β1, and GAPDH probes. Hamster and mouse Northern blots were analyzed in the same manner (data not shown). GAPDH expression was used as a control to normalize for sample loading.
FIG. 4.
Quantitative analysis of Northern blot transcripts showing marked differences between models. Each time point represents a pooled RNA sample from two animal brains. CCR5 (upper row, solid triangles), cathepsin S (middle row, open squares), and TGF-β1 (lower row, solid circles) are shown in the SY-Rat, FU-Mo, and SY-Ha models of CJD. Values are normalized to levels of GAPDH mRNA and expressed in terms of the fold increase over the corresponding normal brain sample (indicated here as day 0). The curve fits shown for mRNA increases all have r2 values greater than 0.977, except for CCR5 expression in the FU-Mo model (r2 = 0.896). Note the different scales for the expression levels of the three transcripts. PrP-res levels and clinical signs were assessed with the same animals used for these RNA studies. The onset of major PrP-res accumulation is indicated by gray arrows, and the durations of clinical symptoms are indicated by black arrows as previously documented in the SY-Rat and SY-Ha models (35, 36), with PrP-res increasing ∼20-fold after 200 days in SY-Rat infection and >30-fold after 87 days in SY-Ha infection. Figure 5 documents the major deposition of PrP amyloid in the FU-Mo model after 80 days.
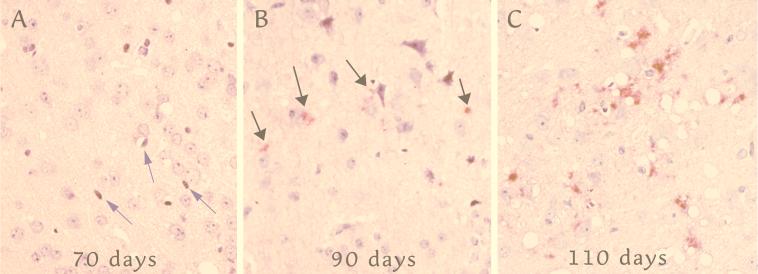
We wanted to examine the relationship between CCR5 upregulation and levels of PrP-res, the more protease-resistant form of host PrP. The detection of PrP-res by Western blotting or immunohistochemistry yields comparable results as previously demonstrated (36). In SY-Rat infection, PrP-res levels are very low until 250 days postinoculation (36). After 200 days, PrP-res levels increase ∼20-fold, reaching final plateau levels by 300 days. In hamsters, PrP-res levels are very low before 87 days and increase more than 30-fold thereafter (35). Maximal PrP-res levels are reached rapidly. Figure 5 shows representative pertinent sections during FU-Mo disease progression demonstrating abnormal PrP deposits. For this sequence, sections through the cortex and caudate were assessed at each time point, and foci with the maximal PrP-res accumulation and vacuolization were photographed. Even at 90 days, there was only a rare solitary focus of PrP-res (Fig. 5B). More foci with abundant PrP deposits and vacuoles were seen only after 90 days (e.g., Fig. 5C). Because a small focus of PrP-res was seen in the subiculum in the FU-Mo model at 80 days, we scored the onset of PrP-res accumulation at this time point. Arrows at the bottom of Fig. 4 summarize these major accumulations of PrP-res in each model.
FIG. 5.
Maximal levels of PrP-res in representative cortical sections taken every 10 days in the FU-Mo model (see text). (A) At 70 days, a few elongated nuclei typical of microglia (arrows) are seen with very rare vacuoles and no detectable PrP-res. (B) A rare focus at 90 days showing few small, granular, abnormal PrP deposits (red; arrows). (C) By 110 days, vacuoles and abundant larger deposits of abnormal PrP are seen. The sections were stained in parallel as described in reference 36 and were counterstained with hematoxylin.
It is clear that CCR5 mRNA preceded significant PrP-res accumulation in SY-Rat infection. In contrast, CCR5 upregulation followed PrP-res increases in the FU-Mo and SY-Ha models. These differences in CCR5 expression among the three CJD models demonstrated that particular agent strain-host combinations lead to specific patterns of pathology and gene expression. The CCR5 changes in SY-Rat infection independently confirmed the relatively early and central activation of glial cells that has been previously noted in this model (36). Because microglial activation is less pronounced in the SY-Ha model than in SY-Rat, we considered the possibility that the SY agent was targeting rat microglia for infection or that the rat was recognizing the presence of the agent through a microglial response.
Although there is evidence for CCR5 expression in certain populations of neurons (15, 44, 62), previous in vivo and in vitro studies have demonstrated CCR5 predominantly in microglia (18, 20, 58, 69). We also attempted to examine the expression of CCR3, another β-chemokine receptor reported to be coexpressed with CCR5 on human microglia (18). CCR3 expression was undetectable by Northern blotting in any of our models (data not shown). This result is consistent with the failure of others to detect CCR3 mRNA by Northern blots of rat spinal cord extracts or of purified rat microglia or astrocytes in culture (20).
Targeted studies of glial mRNA expression.
Because previous studies as well as the data presented above strongly implicated glial cells in rat CJD pathogenesis, we began to focus on other microglial and astrocytic markers to further clarify their contribution to progressive disease. These studies again underscored the differences among the CJD models. The lysosomal protease cathepsin S is an important component of antigen presentation in bone marrow-derived cells (52, 59), and we had previously proposed that lysosomes could be an important site for agent sequestration (35). Cathepsin S mRNA was elevated in all animal models tested, reaching peak levels of about 24-fold in the SY-Rat model, 25-fold in FU-Mo, and 5.6-fold in SY-Ha (Fig. 4). Cathepsin S elevations began relatively early in SY-Rat infection (at 100 to 150 days) and progressively increased thereafter (Fig. 3 and 4). Unlike CCR5, rat cathepsin S levels did not plateau, but continued to increase and reached magnitudes of 24-fold by 350 days. Again, this pattern of expression closely paralleled the activation of microglia and astrocytes in this model. Cathepsin S mRNA in the FU-Mo model was upregulated more than threefold at 80 days and progressively increased to end stage disease (Fig. 4). Unlike that in the SY-Rat model, the onset of cathepsin S changes in FU-Mo infection more closely coincided with the extracellular PrP deposition occurring after 80 days. In contrast, cathepsin S mRNA was elevated well after maximal PrP-res levels were achieved in the SY-Ha model at 125 and 145 days.
Given the results presented above, we chose to investigate other candidate molecules that could be linked to a microglial response to infection. In the central nervous system (CNS), the cytokine TGF-β1 typically indicates a microglial response to injury (reviewed in references 23 and 43). Thus, TGF-β1 levels might be expected to increase with the development of obvious spongiform change seen in the SY-Rat model by 150 days (36). Consistent with this hypothesis, TGF-β1 mRNA in SY-Rat infection was elevated more than 2-fold at 150 days, and peak expression levels exceeded 15-fold at 300 days (Fig. 3 and 4). The upregulation of TGF-β1 was independently confirmed by semiquantitative RT-PCR using primers specific for the TGF-β1 sequence (data not shown).
Because TGF-β1 had been previously implicated in β-amyloid formation (43, 66), we examined the relationship between TGF-β1 expression and ubiquitin, a protein showing pronounced changes earlier than abnormal PrP accumulation in SY-Rat infection (36). The TGF-β1 curve in the SY-Rat model corresponded to the early microglial and ubiquitin changes rather than to PrP amyloid deposition. Thus, TGF-β1 is likely to have a formative role in the development of PrP amyloid, a role consistent with that postulated for β-amyloid in Alzheimer’s disease.
Similarly, in FU-Mo infection, initial threefold increases in TGF-β1 mRNA were detected at 80 days, and upregulation correlated with or slightly preceded the development of extracellular punctate PrP deposits (Fig. 4 and 5). Final ∼20-fold increases in TGF-β1 coincided with robust PrP deposition. In contrast, PrP deposits are not seen in SY-Ha infection except at the inoculation site (35), and TGF-β1 upregulation was remarkably low (Fig. 4). These data further substantiate a crucial role for TGF-β1 and microglia in plaque deposition in CJD.
DISCUSSION
The preceding results clearly demonstrate distinct patterns of gene expression in each of three animal models of CJD. Although we detected mRNA increases for cathepsin S and CCR5 in all models, the onset and peak magnitude of upregulation varied substantially. The initial increases in cathepsin S and CCR5 mRNA in SY-Rat infection occurred before major accumulation of PrP-res. In contrast, cathepsin S and CCR5 upregulation in SY-Ha infection occurred well after major changes in PrP-res were detectable (35, 36). Thus, the modulation of cathepsin S and CCR5 expression was not universally linked to the amount of PrP-res. The degree of mRNA upregulation was also not determined by the length of the incubation period. The model with the longest incubation (SY-Rat) had peak mRNA levels comparable to those of a short-incubation model (FU-Mo), whereas the two models with similar incubation periods (FU-Mo and SY-Ha) had dramatically different mRNA expression patterns. Indeed, generalizations from any one model of CJD or other transmissible spongiform encephalopathy may be inappropriate and may lead to unjustified conclusions.
While the present research was in progress, another DD study also found only three transcripts of 72 potential DD bands that were upregulated early (at 120 days of an ∼165-day disease course) in a murine scrapie model (10). One of the early upregulated mRNAs was cathepsin S, which was found to be elevated approximately fivefold. Although we detected a similar cathepsin S increase in our SY-Ha model, a far more substantial 25-fold increase was observed in both rats and mice infected with two very different CJD strains. Because the previous study evaluated only 40-day intervals in a relatively short incubation model and had minimal reference to corresponding histopathology or PrP-res levels at each time point, it is difficult to evaluate their conclusion that cathepsin S upregulation is a consequence of PrP change. The current demonstration of variable cathepsin S profiles with respect to PrP-res indicates that cathepsin S and microglial activation are not necessarily linked in a direct and predictable way to pathologic PrP.
In the CNS, the vast majority of cathepsin S and CCR5 expression is restricted to microglial cells (18, 20, 51, 58, 69). Activation of microglia in vitro with gamma interferon results in upregulation of CCR5 mRNA (20), and other soluble or intracellular factors probably also influence CCR5 expression. Additionally, cathepsin S mRNA in SY-Rat infection had the same temporal curve previously shown for microglial activation in this model. Together, these results indicate that the altered gene expression seen here reflects activation of microglia and suggest that different magnitudes and temporal patterns of microglial involvement occur in each CJD model. The early activation of microglia with concurrent astrocytic responses also suggests that both cell types can contribute to early vacuolar changes seen in SY-Rat infection, in a cascade analogous to that seen with human immunodeficiency virus (16).
Because neurons express high levels of PrP, they are often assumed to be the principal targets of the CJD and scrapie agents as well as the major source of PrP-res. However, the actual cell types that are infected in vivo are unknown and may vary with the strain of agent. Our previous immunocytochemical and current gene expression data suggest that the SY agent targets microglia in the rat. Alternatively, SY may induce a host response that leads to early microglial activation. The absence of such responses in the SY-Ha model may indicate a lack of sequestration or replication of the SY agent in hamster microglia. The variable recruitment of microglia in different CJD models is reminiscent of murine leukemia virus studies, in which the extent of spongiform neurodegeneration is positively correlated with viral tropism for microglia (9). Furthermore, spongiform neurodegeneration induced by the CasBrE leukemia virus occurs only after viral replication in microglia, whereas neuronal infection is insufficient to induce neuropathology (22, 30). The development of pathological change in CJD is also likely to be mediated by the types of cells that are recruited during the infection. Studies of rat but not hamster CJD showed migration or accumulation of microglia within brain regions not normally rich in these cells (36). Because microglia can derive from blood-borne macrophages, the idea that macrophages act as “Trojan horses” for delivering infectivity to the CNS must be considered, especially since this is a classic route for many types of infectious agents. Additionally, macrophages, dendritic cells, and other leukocytes that express CCR5 will migrate in response to the chemokines MIP-1α, MIP-1β, and RANTES (reviewed in reference 29). The production of these ligands by reactive glia, as implied by our studies of SY-Rat infection and documented in Alzheimer’s disease and AIDS-associated dementia, suggests that CCR5-mediated chemotaxis may be a primary means to regulate the trafficking of microglia within the brain parenchyma (55, 68). Microglia and macrophages may also transport infectivity if they are involved in agent clearance. This clearance may be abortive or incomplete, or it may be part of an attempt at antigen presentation. Clearance is not a hypothetical concept, because infectivity studies have demonstrated that these agents can be cleared from both spleen (13) and brain (35).
The contribution of nonneuronal cells to the spread of these agents should not be underestimated, since both spleen tissue and peripheral blood are known to harbor infectivity (13, 31, 32). In addition to the transport or presentation of these agents by various cells of the immune system (24, 46), the present study continues to define important nonneuronal host cell responses that can have major effects on disease progression. Microglia and astrocytes may produce soluble factors that modulate pathology, and previous studies have shown that upregulation of major histocompatibility complex class II molecules occurs in hamster scrapie (12), human CJD (60), and the BSE-linked human variant CJD (36). The increased expression of cathepsin S shown in the current study is relevant because cathepsin S is an important regulator of major histocompatibility complex class II trafficking within antigen-presenting cells (52, 59). A local CNS inflammatory response, most likely mediated by astrocytes and microglia, is evident from increased expression of interleukin-1α (IL-1α), IL-1β, IL-6, TNFα, and inducible nitric oxide synthase during the final stages of murine scrapie (7, 21, 25, 64, 65). Pathologic effects of microglia have also been linked to neuronal apoptosis in scrapie (17, 63). However, because CCR5 and cathepsin S upregulation occurred ∼200 days before the onset of clinical disease in the SY-Rat model, early glial activation could have beneficial consequences for the host. In addition to producing trophic or neuroprotective factors, activated microglia may also participate in processing and clearance of the infectious agent. Such clearance mechanisms are also implicated by the accumulation of PrP-res in rat microglia in vivo (36). In this context, microglia may also have a role in preserving the immunological privilege of the brain. Interestingly, the anti-inflammatory role of TGF-β1 in immune-privileged tissues (8) makes it a candidate molecule for such immunosuppressive functions of microglia.
One of the major characteristics that distinguishes the SY-Rat model from other strains of CJD passaged in laboratory animals is the presence of large PrP amyloid plaques that are distinct from the smaller PrP deposits seen in the FU-Mo model (34, 36). Such deposits are completely absent in SY-Ha infection (35), and large PrP amyloid plaques are also minimal or absent in most human cases of CJD. The kinetics of microglial activation in SY-Rat infection suggest that these cells may be required for plaque development. Because CCR5 upregulation occurred 50 to 100 days before PrP amyloid deposition, CCR5 may be an important factor in the recruitment of microglia to sites destined for plaque formation. Furthermore, both cathepsin S and TGF-β1 have been implicated in the development of amyloid plaques (27, 43, 45, 66). The current analyses strongly indicate a crucial role for microglia and specific products elaborated by these cells (e.g., TGF-β1 and cathepsin S) in the formation of amyloid deposits. A model of CJD without these mRNA changes (SY-Ha) showed a lack of PrP amyloid.
Our studies underscore the importance of detailed temporal analysis for understanding the progression of CJD in its various pathologic costumes. Correlations between immunocytochemical changes, mRNA expression, and infectivity data are necessary for the correct interpretation of pathogenetic events. These events include the formation of PrP amyloid plaques and the coincident accumulation of other molecules commonly associated with a variety of plaques, including those found in Alzheimer’s disease. It is also crucial not to limit studies to PrP, since several investigators have observed neuropathology and infectivity that cannot be quantitatively correlated with pathologic PrP (14, 26, 36, 67). Such broad-based studies have the power to identify potential therapeutic targets. The chronic administration of dapsone, an anti-inflammatory drug thought to affect macrophage (and possibly microglial) function, significantly prolongs the incubation time to clinical signs in SY-Rat infection without affecting the rate of PrP plaque formation (37). Our findings further underscore microglia as potential targets for therapeutic intervention in at least some forms of CJD.
ACKNOWLEDGMENTS
We thank William Fritch for excellent assistance with animal inoculation and tissue collection.
This research was supported by NIH grants NS12674 and NS34569 and by NIH training grant GM07527.
REFERENCES
- 1.Aguzzi A, Weissmann C. Prion research: the next frontiers. Nature. 1997;389:795–798. doi: 10.1038/39758. [DOI] [PubMed] [Google Scholar]
- 2.Akowitz A, Sklaviadis T, Manuelidis E E, Manuelidis L. Nuclease-resistant polyadenylated RNAs of significant size are detected by PCR in highly purified Creutzfeldt-Jakob disease preparations. Microb Pathog. 1990;9:33–45. doi: 10.1016/0882-4010(90)90038-r. [DOI] [PubMed] [Google Scholar]
- 3.Akowitz A, Sklaviadis T, Manuelidis L. Endogenous viral complexes with long RNA cosediment with the agent of Creutzfeldt-Jakob disease. Nucleic Acids Res. 1994;22:1101–1107. doi: 10.1093/nar/22.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berglund E O, Ranscht B. Molecular cloning and in situ localization of the human contactin gene (CNTN1) on chromosome 12q11-q12. Genomics. 1994;21:571–582. doi: 10.1006/geno.1994.1316. [DOI] [PubMed] [Google Scholar]
- 5.Bruce M E, McConnell I, Fraser H, Dickinson A G. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 6.Büeler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Auget M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 7.Campbell I L, Eddleston M, Kemper P, Oldstone M B A, Hobbs M V. Activation of cerebral cytokine gene expression and its correlation with onset of reactive astrocyte and acute-phase response gene expression in scrapie. J Virol. 1994;68:2383–2387. doi: 10.1128/jvi.68.4.2383-2387.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J J, Sun Y N, Nabel G J. Regulation of the proinflammatory effects of Fas ligand (CD95L) Science. 1998;282:1714–1717. doi: 10.1126/science.282.5394.1714. [DOI] [PubMed] [Google Scholar]
- 9.Czub M, Czub S, Rappold M, Mazgareanu S, Schwender S, Demuth M, Hein A, Dörries R. Murine leukemia virus-induced neurodegeneration of rats: enhancement of neuropathogenicity correlates with enhanced viral tropism for macrophages, microglia, and brain vascular cells. Virology. 1995;214:239–244. doi: 10.1006/viro.1995.0027. [DOI] [PubMed] [Google Scholar]
- 10.Dandoy-Dron F, Guillo F, Benboudjema L, Deslys J-P, Lasmezas C, Dormont D, Tovey M G, Dron M. Gene expression in scrapie: cloning of a new scrapie-responsive gene and the identification of increased levels of seven other mRNA transcripts. J Biol Chem. 1998;273:7691–7697. doi: 10.1074/jbc.273.13.7691. [DOI] [PubMed] [Google Scholar]
- 11.Diringer H, Beekes M, Ozel M, Simon D, Queck I, Cardone F, Pocchiari M, Ironside J W. Highly infectious purified preparations of disease-specific amyloid of transmissible spongiform encephalopathies are not devoid of nucleic acids of viral size. Intervirology. 1997;40:238–246. doi: 10.1159/000150553. [DOI] [PubMed] [Google Scholar]
- 12.Duguid J, Trzepacz C. Major histocompatibility complex genes have an increased brain expression after scrapie infection. Proc Natl Acad Sci USA. 1993;90:114–117. doi: 10.1073/pnas.90.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eklund C M, Kennedy R C, Hadlow W J. Pathogenesis of scrapie virus infection in the mouse. J Infect Dis. 1967;117:15–22. doi: 10.1093/infdis/117.1.15. [DOI] [PubMed] [Google Scholar]
- 14.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissman C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 15.Galasso J M, Harrison J K, Silverstein F S. Excitotoxic brain injury stimulates expression of the chemokine receptor CCR5 in neonatal rats. Am J Pathol. 1998;153:1631–1640. doi: 10.1016/S0002-9440(10)65752-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gendelman H E, Lipton S A, Tardieu M, Bukrinsky M I, Nottet H S. The neuropathogenesis of HIV-1 infection. J Leukocyte Biol. 1994;56:389–398. doi: 10.1002/jlb.56.3.389. [DOI] [PubMed] [Google Scholar]
- 17.Giese A, Brown D R, Groschup M H, Feldmann C, Haist I, Kretzschmar H A. Role of microglia in neuronal cell death and prion disease. Brain Pathol. 1998;8:449–457. doi: 10.1111/j.1750-3639.1998.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 19.Hsiao K K, Groth D, Scott M, Yang S-L, Serban H, Rapp D, Foster D, Torchia M, DeArmond S J, Prusiner S B. Serial transmission in rodents of neurodegeneration from transgenic mice expressing mutant prion protein. Proc Natl Acad Sci USA. 1994;91:9126–9130. doi: 10.1073/pnas.91.19.9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, Salafranca M N, Adhikari S, Xia Y, Feng L, Sonntag M K, deFiebre C M, Pennell N A, Streit W J, Harrison J K. Chemokine receptor expression in cultured glia and rat experimental allergic encephalomyelitis. J Neuroimmunol. 1998;86:1–12. doi: 10.1016/s0165-5728(98)00005-8. [DOI] [PubMed] [Google Scholar]
- 21.Ju W K, Park K J, Choi E K, Kim J, Carp R I, Wisniewski H M, Kim Y S. Expression of inducible nitric oxide synthase in the brains of scrapie-infected mice. J Neurovirol. 1998;4:445–450. doi: 10.3109/13550289809114544. [DOI] [PubMed] [Google Scholar]
- 22.Kay D G, Gravel C, Robitaille Y, Jolicoeur P. Retrovirus-induced spongiform myeloencephalopathy in mice: regional distribution of infected target cells and neuronal loss occurring in the absence of viral expression in neurons. Proc Natl Acad Sci USA. 1991;88:1281–1285. doi: 10.1073/pnas.88.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiefer R, Streit W J, Toyka K V, Kreutzberg G W, Hartung H-P. Transforming growth factor-β1: a lesion-associated cytokine of the nervous system. Int J Dev Neurosci. 1995;13:331–339. doi: 10.1016/0736-5748(94)00074-d. [DOI] [PubMed] [Google Scholar]
- 24.Klein M A, Frigg R, Flechsig E, Raeber A J, Kalinke U, Bluethmann H, Bootz F, Suter M, Zinkernagel R M, Aguzzi A. A crucial role for B cells in neuroinvasive scrapie. Nature. 1997;390:687–690. doi: 10.1038/37789. [DOI] [PubMed] [Google Scholar]
- 25.Kordek R, Nerurkar V R, Liberski P P, Isaacson S, Yanagihara R, Gajdusek D C. Heightened expression of tumor necrosis factor-α, interleukin-1α, and glial fibrillary acidic protein in experimental Creutzfeldt-Jakob disease in mice. Proc Natl Acad Sci USA. 1996;93:9754–9758. doi: 10.1073/pnas.93.18.9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasmezas C I, Deslys J P, Robain O, Jaegly A, Beringue V, Peyrin J M, Fournier J G, Hauw J J, Rossier J, Dormont D. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science. 1997;275:402–405. doi: 10.1126/science.275.5298.402. [DOI] [PubMed] [Google Scholar]
- 27.Lemere C A, Munger J S, Shi G-P, Natkin L, Haass C, Chapman H A, Selkoe D J. The lysosomal cysteine protease, cathepsin S, is increased in Alzheimer’s disease and Down syndrome brain. Am J Pathol. 1995;146:848–860. [PMC free article] [PubMed] [Google Scholar]
- 28.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 29.Luster A D. Chemokines—chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 30.Lynch W P, Snyder E Y, Qualtiere L, Portis J L, Sharpe A H. Late virus replication events in microglia are required for neurovirulent retrovirus-induced spongiform neurodegeneration: evidence from neural progenitor-derived chimeric mouse brains. J Virol. 1996;70:8896–8907. doi: 10.1128/jvi.70.12.8896-8907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manuelidis E E, Gorgacs E J, Manuelidis L. Viremia in experimental Creutzfeldt-Jakob disease. Science. 1978;200:1069–1071. doi: 10.1126/science.349691. [DOI] [PubMed] [Google Scholar]
- 32.Manuelidis E E, Kim J H, Mericangas J R, Manuelidis L. Transmission to animals of Creutzfeldt-Jakob disease from human blood. Lancet. 1985;2:896–897. doi: 10.1016/s0140-6736(85)90165-5. [DOI] [PubMed] [Google Scholar]
- 33.Manuelidis L. The dimensions of Creutzfeldt-Jakob disease. Transfusion. 1994;34:915–928. doi: 10.1046/j.1537-2995.1994.341095026981.x. [DOI] [PubMed] [Google Scholar]
- 34.Manuelidis L. Vaccination with an attenuated Creutzfeldt-Jakob disease strain prevents expression of a virulent agent. Proc Natl Acad Sci USA. 1998;95:2520–2525. doi: 10.1073/pnas.95.5.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manuelidis L, Fritch W. Infectivity and host responses in Creutzfeldt-Jakob disease. Virology. 1996;215:46–59. doi: 10.1006/viro.1996.0033. [DOI] [PubMed] [Google Scholar]
- 36.Manuelidis L, Fritch W, Xi Y-G. Evolution of a strain of CJD that induces BSE-like plaques. Science. 1997;277:94–98. doi: 10.1126/science.277.5322.94. [DOI] [PubMed] [Google Scholar]
- 37.Manuelidis L, Fritch W, Zaitsev I. Dapsone to delay symptoms in Creutzfeldt-Jakob disease. Lancet. 1998;352:456. doi: 10.1016/S0140-6736(05)79191-1. [DOI] [PubMed] [Google Scholar]
- 38.Manuelidis L, Murdoch G, Manuelidis E. Possible involvement of retroviral elements in human dementias. CIBA Found Symp. 1988;135:117–134. doi: 10.1002/9780470513613.ch8. [DOI] [PubMed] [Google Scholar]
- 39.Manuelidis L, Sklaviadis T, Akowitz A, Fritch W. Viral particles are required for infection in neurodegenerative Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 1995;92:5124–5128. doi: 10.1073/pnas.92.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manuelidis L, Tesin D M, Sklaviadis T, Manuelidis E E. Astrocyte gene expression in Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA. 1987;84:5937–5941. doi: 10.1073/pnas.84.16.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathieu-Daude F, Cheng R, Welsh J, McClelland M. Screening of differentially amplified cDNA products from RNA arbitrarily primed PCR fingerprints using single strand conformation polymorphism (SSCP) gels. Nucleic Acids Res. 1996;24:1504–1507. doi: 10.1093/nar/24.8.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathieu-Daude F, Welsh J, Vogt T, McClelland M. DNA rehybridization during PCR: the ’CoT effect’ and its consequences. Nucleic Acids Res. 1996;24:2080–2086. doi: 10.1093/nar/24.11.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattson M P, Barger S W, Furukawa K, Bruce A J, Wyss-Coray T, Mark R J, Mucke L. Cellular signaling roles of TGFβ, TNFα and β-APP in brain injury responses and in Alzheimer’s disease. Brain Res Rev. 1997;23:47–61. doi: 10.1016/s0165-0173(96)00014-8. [DOI] [PubMed] [Google Scholar]
- 44.Meucci O, Fatatis A, Simen A A, Bushell T J, Gray P W, Miller R J. Chemokines regulate hippocampal neuronal signaling and gp120 neurotoxicity. Proc Natl Acad Sci USA. 1998;95:14500–14505. doi: 10.1073/pnas.95.24.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munger J S, Haass C, Lemere C A, Shi G-P, Wong W S F, Teplow D B, Selkoe D J, Chapman H A. Lysosomal processing of amyloid precursor protein to Aβ peptides: a distinct role for cathepsin S. Biochem J. 1995;311:299–305. doi: 10.1042/bj3110299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muramoto T, Kitamoto T, Hoque M Z, Tateishi J, Goto I. Species barrier prevents an abnormal isoform of prion protein from accumulating in follicular dendritic cells of mice with Creutzfeldt-Jakob disease. J Virol. 1993;67:6808–6810. doi: 10.1128/jvi.67.11.6808-6810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- 48.Peles E, Nativ M, Lustig M, Grumet M, Schilling J, Martinez R, Plowman G D, Schlessinger J. Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 1997;16:978–988. doi: 10.1093/emboj/16.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peschon J J, Slack J L, Reddy P, Stocking K L, Sunnarborg S W, Lee D C, Russell W E, Castner B J, Johnson R S, Fitzner J N, Boyce R W, Nelson N, Kozlosky C J, Wolfson M F, Rauch C T, Ceretti D P, Paxton R J, March C J, Black R A. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 50.Pesheva P, Gennarini G, Goridis C, Schachner M. The F3/11 cell adhesion molecule mediates the repulsion of neurons by the extracellular matrix glycoprotein J1-160/180. Neuron. 1993;10:69–82. doi: 10.1016/0896-6273(93)90243-k. [DOI] [PubMed] [Google Scholar]
- 51.Petanceska S, Canoll P, Devi L A. Expression of rat cathepsin S in phagocytic cells. J Biol Chem. 1996;271:4403–4409. doi: 10.1074/jbc.271.8.4403. [DOI] [PubMed] [Google Scholar]
- 52.Pierre P, Mellman I. Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Cell. 1998;93:1135–1145. doi: 10.1016/s0092-8674(00)81458-0. [DOI] [PubMed] [Google Scholar]
- 53.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 54.Riesner D, Kellings K, Post K, Wille H, Serban H, Groth D, Baldwin M A, Prusiner S B. Disruption of prion rods generates 10-nm spherical particles having high α-helical content and lacking scrapie infectivity. J Virol. 1996;70:1714–1722. doi: 10.1128/jvi.70.3.1714-1722.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidtmayerova H, Nottet H S, Nuovo G, Raabe T, Flanagan C R, Dubrovsky L, Gendelman H E, Cerami A, Bukrinsky M, Sherry B. Human immunodeficiency virus type 1 infection alters chemokine β peptide expression in human monocytes: implications for recruitment of leukocytes into brain and lymph nodes. Proc Natl Acad Sci USA. 1996;93:700–704. doi: 10.1073/pnas.93.2.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sklaviadis T, Akowitz A, Manuelidis E E, Manuelidis L. Nuclease treatment results in high specific purification of Creutzfeldt-Jakob disease infectivity with a density characteristic of nucleic acid-protein complexes. Arch Virol. 1990;112:215–228. doi: 10.1007/BF01323166. [DOI] [PubMed] [Google Scholar]
- 57.Sklaviadis T, Dreyer R, Manuelidis L. Analysis of Creutzfeldt-Jakob disease infectious fractions by gel permeation chromatography and sedimentation field flow fractionation. Virus Res. 1992;26:241–254. doi: 10.1016/0168-1702(92)90016-3. [DOI] [PubMed] [Google Scholar]
- 58.Vallat A-V, De Girolami U, He J, Mhashilkar A, Marasco W, Shi B, Gray F, Bell J, Keohane C, Smith T W, Gabuzda D. Localization of HIV-1 co-receptors CCR5 and CXCR4 in the brain of children with AIDS. Am J Pathol. 1998;152:167–178. [PMC free article] [PubMed] [Google Scholar]
- 59.Villadangos J A, Riese R J, Peters C, Chapman H A, Ploegh H L. Degradation of mouse invariant chain: roles of cathepsins S and D and the influence of major histocompatibility complex polymorphism. J Exp Med. 1997;186:549–560. doi: 10.1084/jem.186.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Eitzen U, Egensperger R, Kosel S, Grasbon-Frodl E M, Imai Y, Bise K, Kohsaka S, Mehraein P, Graeber M B. Microglia and the development of spongiform change in Creutzfeldt-Jakob disease. J Neuropathol Exp Neurol. 1998;57:246–256. doi: 10.1097/00005072-199803000-00006. [DOI] [PubMed] [Google Scholar]
- 61.Wawer C, Rüggeberg H, Meyer G, Muyzer G. A simple and rapid electrophoresis method to detect sequence variation in PCR-amplified DNA fragments. Nucleic Acids Res. 1995;23:4928–4929. doi: 10.1093/nar/23.23.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westmoreland S V, Rottman J B, Williams K C, Lackner A A, Sasseville V G. Chemokine receptor expression on resident and inflammatory cells in the brain of macaques with simian immunodeficiency virus encephalitis. Am J Pathol. 1998;152:659–665. [PMC free article] [PubMed] [Google Scholar]
- 63.Williams A, Lucassen P J, Ritchie D, Bruce M. PrP deposition, microglial activation, and neuronal apoptosis in murine scrapie. Exp Neurol. 1997;144:433–438. doi: 10.1006/exnr.1997.6424. [DOI] [PubMed] [Google Scholar]
- 64.Williams A, Van Dam A-M, Ritchie D, Eikelenboom P, Fraser H. Immunocytochemical appearance of cytokines, prostaglandin E2 and lipocortin-1 in the CNS during the incubation period of murine scrapie correlates with progressive PrP accumulations. Brain Res. 1997;754:171–180. doi: 10.1016/s0006-8993(97)00067-x. [DOI] [PubMed] [Google Scholar]
- 65.Williams A E, Van Dam A-M, Man-A-Hing W K H, Berkenbosch F, Eikelenboom P, Fraser H. Cytokines, prostaglandins, and lipocortin-1 are present in the brains of scrapie-infected mice. Brain Res. 1994;654:200–206. doi: 10.1016/0006-8993(94)90480-4. [DOI] [PubMed] [Google Scholar]
- 66.Wyss-Coray T, Masliah E, Mallory M, McConlogue L, Johnson-Wood K, Lin C, Mucke L. Amyloidogenic role of cytokine TGF-β1 in transgenic mice and in Alzheimer’s disease. Nature. 1997;389:603–606. doi: 10.1038/39321. [DOI] [PubMed] [Google Scholar]
- 67.Xi Y-G, Ingrosso L, Ladogana A, Masullo C, Pocchiari M. Amphotericin B treatment dissociates in vivo replication of the scrapie agent from PrP accumulation. Nature. 1992;356:598–601. doi: 10.1038/356598a0. [DOI] [PubMed] [Google Scholar]
- 68.Xia M, Qin S, Wu L, Mackay C R, Hyman B T. Immunohistochemical study of the β-chemokine receptors CCR3 and CCR5 and their ligands in normal and Alzheimer’s disease brains. Am J Pathol. 1998;153:31–37. doi: 10.1016/s0002-9440(10)65542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, He T, Talal A, Wang G, Frankel S S, Ho D D. In vivo distribution of the human immunodeficiency virus/simian immunodeficiency virus coreceptors: CXCR4, CCR3, and CCR5. J Virol. 1998;72:5035–5045. doi: 10.1128/jvi.72.6.5035-5045.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]