Abstract
Human herpesvirus 8 (HHV-8; also designated Kaposi’s sarcoma-associated herpesvirus) is the likely etiological agent of Kaposi’s sarcoma (KS). HHV-8 encodes a latent nuclear antigen (LNA) which is the product of the viral gene orf 73. LNA is recognized by most infected patient sera and is the basis of current immunofluorescence assays used in epidemiological studies of HHV-8 infection. Here we describe the characterization of four monoclonal antibodies raised to the C-terminal third of LNA–glutathione S-transferase fusion proteins. These monoclonal antibodies recognized discrete linear epitopes within the C terminus and repetitive region of LNA, detected antigen in primary effusion lymphoma (PEL) cells, and precipitated a 220- to 230-kDa protein doublet corresponding to LNA from HHV-8-infected PEL cell lines. In situ immunocytochemistry of KS lesions with these antibodies show that LNA is extensively expressed in KS spindle cells.
Human herpesvirus 8 (HHV-8; also designated Kaposi’s sarcoma-associated herpesvirus) is the first known human member of the genus Rhadinovirus within the Herpesviridae (9). HHV-8 is related to the New World primate rhadinovirus herpesvirus saimiri (1), a rhesus macaque rhadinovirus (10), and macaque retroperitoneal fibromatosis virus isolates Mn and Mm (25). More recently, equine herpesvirus 2, bovine herpesvirus 4, alcelaphine herpesvirus 1, and murine herpesvirus 68 have been tentatively classified as rhadinoviruses. HHV-8 is associated with all epidemiological forms of Kaposi’s sarcoma (KS) (4, 6, 28). HHV-8 is also associated with primary effusion lymphoma (PEL) (7) and a subset of multicentric Castleman’s disease (12, 30). Epidemiological studies using PCR detection of specific HHV-8 genomic sequences, and immunological assays for antibodies to the major latent nuclear antigen (LNA) and other antigens, have shown that HHV-8 largely fulfills epidemiological criteria for causation in KS (5, 13, 14, 16, 19, 20, 22, 29, 32).
Sera from HHV-8-infected individuals react with a specific nuclear antigen in latently infected PEL cell lines which is characterized by a punctate nuclear immunofluorescence pattern (19). Screening of cDNA libraries with an HHV-8-positive patient serum identified this nuclear antigen as the product of the viral gene open reading frame 73 (orf73), and the encoded protein was designated LNA or LANA (17, 18, 24). LNA is expressed from a latently controlled 5.32-kb transcript that also encodes the viral cyclin (v-Cyc) and v-FLIP (11, 18). The 5.32-kb latent transcript is spliced to form a 1.7-kb transcript that encodes only v-Cyc and v-FLIP (11, 18, 27). The predicted LNA protein is 1,162 amino acids long and has a theoretical molecular mass of 135 kDa. In contrast to LNA’s predicted molecular size, the protein has an apparent molecular size of 220 to 230 kDa when analyzed by Western blotting (13, 18, 24). LNA can be divided into three domains: an N-terminal 337-amino-acid domain; an extremely hydrophilic central domain of 585 amino acids consisting of multiple repeat elements predominantly containing the charge polar amino acids glutamine, glutamic acid, and asparagine; and a C-terminal 240-amino-acid domain. In contrast to the central domain, the N- and C-terminal domains are rich in basic amino acids. The differently charged domains of the protein may in part explain the aberrant running of LNA on sodium dodecyl sulfate (SDS)-polyacrylamide gels.
LNA is thus far the only latent nuclear antigen described for HHV-8 and has no obvious sequence identity to known proteins. However, the immunogenic nature, nuclear localization, and latent expression of LNA suggest that this protein is analogous to Epstein-Barr virus (EBV) nuclear antigens (EBNAs). EBNAs have a pivotal role in maintenance and replication of the virus episome and transforming cells. Insights into the function of LNA could be essential in the understanding of HHV-8 latency and tumor formation. Here we describe the characterization of four monoclonal antibodies (MAbs) to the C terminus of orf73. These antibodies recognize distinct linear epitopes of LNA, confirm that orf73 encodes the major latent nuclear antigen of HHV-8, and detect LNA expression in PEL cell lines and in KS nodules.
Generation of MAbs against LNA.
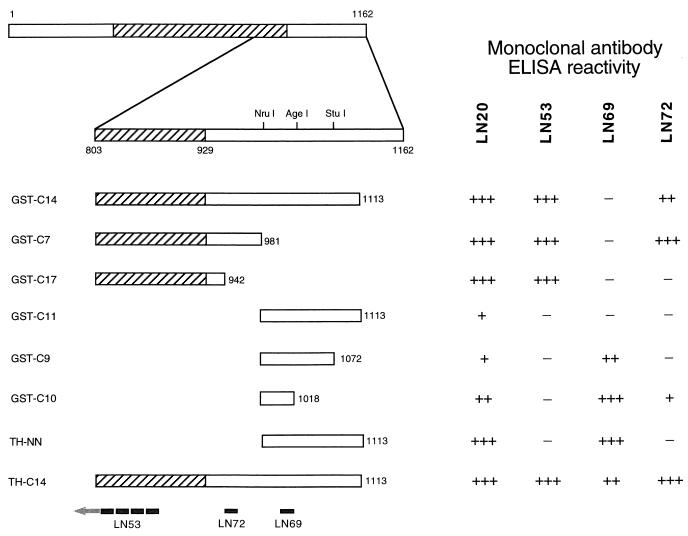
Glutathione S-transferase (GST) proteins of the C terminus of orf73 (GST-C14 and GST-C17) were described previously (18). Briefly, GST-C14 contains amino acids 803 to 1113 of orf73 encompassing 126 amino acids of the central repeat domain as well as 184 amino acids of the C-terminal basic domain (Fig. 1). GST-C17 contains amino acids 803 to 942 comprised almost entirely of repetitive coding sequence. GST-C14 served as the backbone for making further defined deletions of the C terminus of orf73 by using three unique restriction enzyme sites. GST-C7 was constructed by digesting GST-C14 with NruI (in orf73) and NotI (in pGEX-4T3 [Pharmacia]), followed by end repair and ligation. GST-C11 was constructed by digesting GST-C14 with NruI and NotI and cloning the restriction fragment into the SmaI and NotI sites of the vector pGEX-4T1 (Pharmacia). GST-C11 served as the backbone for two further deletion clones. GST-C9 and GST-C10 were constructed by restriction enzyme digestion with StuI plus NotI and AgeI plus NotI, respectively, followed by end repair and ligation. Recombinant protein-expressing clones were confirmed by DNA sequence analysis. In addition, two polyhistidine fusion proteins were constructed. The entire BamHI-to-NotI C-terminal restriction fragment from GST-C14 was cloned into the polyhistidine fusion protein vector pTrcHis2C (Invitrogen) via BamHI and HindIII following conversion of the NotI and HindIII sites to blunt ends to produce clone TH-C14. The NruI-to-NotI fragment corresponding to GST-C11 was PCR amplified and cloned to produce TH-NN. Large-scale production and single-step affinity purification of fusion proteins (GST [Pharmacia] and polyhistidine [Invitrogen]) were performed according to the manufacturer’s instructions. All fusion proteins were recognized by antibodies to GST (B14; Santa Cruz) or to the Myc epitope present in the polyhistidine fusion proteins (data not shown). Pools of affinity-purified GST fusion proteins were used for intraperitoneal immunization of rats at 3-week intervals. Three days following the final boost, rats were sacrificed to obtain splenocytes for hybridoma production by standard procedures. Hybridomas were screened by enzyme-linked immunosorbent assay (ELISA) and Western blotted against the polyhistidine fusion protein TH-C14, and positive hybridomas were subcloned. Following a second ELISA screening, reactive MAbs were bulk cultured and antibodies were purified from culture supernatants by antibody affinity chromatography. This procedure produced MAbs ranging in concentration from 1 to 2 mg/ml. The isotype of each MAb was determined by standard methods. Four MAbs, LN20, LN53, LN69, and LN72, with isotypes immunoglobulin M (IgM), IgG2c, IgG1 and IgG2b, respectively, were raised to these GST fusion proteins.
FIG. 1.
Fusion protein mapping of LNA MAbs. The C-terminal locations of six GST-LNA fusion proteins (GST-C14, -C7, -C17, -C11, -C9, and -C10) and two histidine-tagged fusion proteins (TH-C14 and TH-NN) are shown relative to that of full-length LNA. Numbers represent amino acids relative to the putative start methionine at codon 127296 of the published HHV-8 genome (26). The hatched box represents the extent of the repetitive coding region of LNA. Restriction enzyme sites NruI, AgeI, and StuI used to construct the fusion protein clones are indicated. The reactivity of each MAb to each fusion protein is shown as A410 above background of greater than 0.5 absorbance units (+++), between 0.1 and 0.5 absorbance units (++), or between 0.02 and 0.1 absorbance units (+). The epitope-mapped position of each MAb is indicated on TH-C14. The epitopes are EQEQE for LN53, EVDYPV for LN72, and THPKKPHPRYQQ for LN69.
Mapping of the epitopes recognized by the MAbs.
The antigenic epitopes recognized by these MAbs were mapped by using the panel of GST and polyhistidine fusion proteins (Fig. 1) in an ELISA format. Briefly, fusion proteins were immobilized onto 96-well plates (Immulon 2; Dynex Technologies) in 10 mM sodium phosphate buffer (pH 7.0) by overnight incubation at 4°C. A standard ELISA protocol was followed, using appropriate dilutions of anti-LNA MAbs and 1:2,000 dilution of goat anti-rat alkaline phosphatase-conjugated secondary antibody (Sera-Lab) as described elsewhere (15). Reactions were visualized by using p-nitrophenyl phosphate (Sigma Fast) according to the manufacturer’s instructions. Antibody LN20 recognized all recombinant proteins to various degrees, as shown by reactivity to clones of the repetitive and nonrepetitive C-terminal regions (Fig. 1). The recombinant proteins represent a nonoverlapping set and contain no apparent common epitopes, which suggests that LN20 is a mixed antibody population, requiring further rounds of monocloning. Antibody LN53 recognized recombinant proteins GST-C14, GST-C17, GST-C7, and TH-C14 (Fig. 1), demonstrating that the minimum epitope recognized by this antibody is the repetitive region of orf73 encompassing GCT-C17 or the 13 amino acids C terminal to the repetitive region. Antibody LN69 recognized only GST-C9, GST-C10, TH-C14, and TH-NN. This mapped the LN69 minimal epitope to the 37 amino acids (amino acids 981 to 1018 of LNA) present in GST-C10 (Fig. 1). It is unclear why LN69 did not recognize GST-C14 or GST-C11, as both contained the minimal epitope. Antibody LN72 recognized GST-C14 and -C7 but not GST-C17 and thus recognizes a 39-amino-acid region (from amino acids 942 to 981) of LNA. To confirm and further refine the minimal epitopes for each MAb, multiple overlapping peptides corresponding to the regions of LNA identified by ELISA were synthesized by using the SPOTS system (Genosys). Peptide mapping revealed that LN69 recognized the epitope THPKKPHPRYQQ, LN72 recognized the epitope EVDYPV, and LN53 recognized the epitope EQEQE. These epitopes are contained within the minimal LNA recombinant proteins identified by ELISA, thus confirming the binding sites of the three MAbs. The LN53 epitope is contained within the repetitive region of LNA, resulting in 23 copies of the LN53 epitope in full-length LNA. Database searching revealed that this epitope is present in human proteins, including the alpha-type calcitonin gene-related peptide precursor (P06881), drebrin (Q16643), translation initiation factor IF-2 (P46199), and aldehyde dehydrogenase (P30838), but only in single copy. The LN69 and LN72 epitopes have only partial homologies to known cellular proteins.
Immunoprecipitation and immunofluorescence assay (IFA) of HHV-8 LNA.
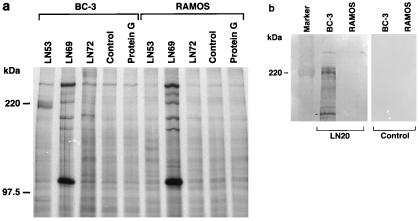
Previous studies have shown that the authentic LNA protein encoded in PEL cell lines is a doublet with an apparent molecular mass of 220 to 230 kDa (13, 18, 24), in contrast to the predicted molecular mass of 135 kDa. The reasons for the size difference are unclear. MAbs LN53, LN69, and LN72 produced in this study were used to immunoprecipitate LNA from the PEL cell line BC-3 (2). Briefly, cells were starved for 1 h in methionine- and cysteine-free RPMI (Sigma) supplemented with 10% dialyzed fetal calf serum (FCS; Gibco), washed, and then labeled for 4 h in fresh medium supplemented with a mixture of [35S]methionine and [35S]cysteine (70 μCi/ml in medium; Amersham Pro-mix) plus 10% dialyzed FCS. After being washed, the cell pellets were lysed for 30 min on ice in 1 ml of lysis buffer (150 mM NaCl, 1.0% Nonidet P-40, 50 mM Tris-HCl [pH 8]) supplemented with the protease inhibitor phenylmethylsulfonyl fluoride (100 μg/ml), pepstatin A (0.7 μg/ml), and leupeptin (5 μg/ml), and lysates were cleared by centrifugation at 14,000 × g for 10 min at 4°C. Extracts from the equivalent of 107 cells were precleared for 1 h with protein G-Sepharose beads (Sigma) equilibrated in PBST (phosphate-buffered saline [PBS] plus 1% Triton X-100) and then immunoprecipitated overnight at 4°C with protein G-Sepharose beads precoated with saturating amounts of the indicated MAbs. Immunoprecipitates were washed and loaded onto an SDS–8% polyacrylamide gel. Only MAb LN53 was able to precipitate a doublet protein of 220 to 230 kDa. LN69 and LN72 precipitated proteins present in both BC-3 and control Ramos cells but did not precipitate LNA (Fig. 2a). The identity of the major 105- to 110-kDa protein precipitated by LN69 from both cell types is unclear, and a database search using the LN69 epitope failed to identify any candidate proteins. Antibodies LN20, LN53, and LN72 reacted with a protein doublet of 220 to 230 kDa by Western blotting (Fig. 2b). In addition, a protein of approximately 180 kDa was recognized by the antibodies by Western blotting (Fig. 2b). Reactivity to proteins of this size had previously been seen in Western blots with HHV-8-positive patient sera but had not been assigned as LNA specific (13, 18, 24).
FIG. 2.
Immunoprecipitation and Western blot analyses using LNA MAbs. (a) Immunoprecipitates from 107 BC-3 cells or Ramos cells per lane were resolved on SDS–8% polyacrylamide gels. LN53 was able to precipitate a 220- to 230-kDa doublet corresponding to LNA. Pooled antibody isotype controls to HIV gp120 for each LNA MAb as well as Protein G-Sepharose-only immunoprecipitation controls (Control and Protein G lanes, respectively) were used in identical immunoprecipitations. (b) Western blot of 2 × 105 cell equivalents of total protein extract from BC-3 (2) and Ramos cells detected with LNA MAb LN20. A doublet of 220 to 230 kDa, and a smaller reactive band of approximately 180 kDa (marked on blot) are seen. Identical lanes were detected with an anti-rat alkaline phosphatase-conjugated secondary antibody as a control. The same results were obtained for LN53 and LN72 (data not shown).
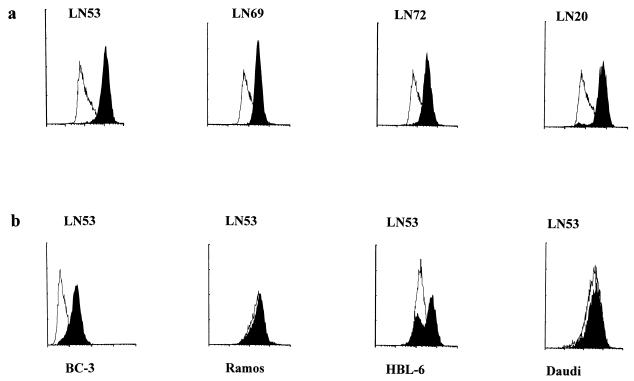
IFAs are used extensively in the seroepidemiology of HHV-8 (6). Under standard IFA conditions, MAbs LN20, LN53, and LN72 were able to produce the punctate nuclear fluorescence pattern associated with HHV-8 LNA in PEL cell lines (Fig. 3a to c). The antibodies were specific for HHV-8 LNA, with no apparent cross-reaction to other proteins present in control B-cell lines Daudi and Ramos (Fig. 3d and e). The best antibody for IFA was LN53, which could be diluted to 1:10,000 without significantly affecting the characteristic LNA IFA pattern in BCP-1 cells. All antibodies were also able to recognize LNA expressed in BCP-1 cells by fluorescence-activated cell sorting analysis (FACS) analysis (Fig. 4a). No cross-reactivity was observed for each antibody with the control B-cell lines Ramos (EBV negative) and Daudi (EBV positive) (representative controls [Fig. 4b]). FACS analysis was also performed with LN53 on one other PEL, BC-3, and on the HHV-8/EBV-coinfected B-cell lymphoma line HBL-6 (Fig. 4b). The BC-3 cell line exhibited the characteristic FACS pattern showing that the majority of cells express LNA. Analysis of HBL-6 showed that most cells expressed LNA, but a subpopulation appeared to be nonexpressing. Whether this is a consequence of HHV-8/EBV coinfection is currently being studied.
FIG. 3.
LNA MAbs react with nuclear bodies in PEL cells. MAbs LN20 (a), LN53 (b), and LN72 (c) all react by IFA to nuclear antigenic structures in BCP-1 cells (3, 14) identical to those detected with HHV-8-positive KS patient serum (f). Antibody LN53 did not react with nuclear antigens present in control Daudi (d) or Ramos (e) cells. LN20 and LN72 were also negative on control cells (not shown). The bar in panel a applies to all panels.
FIG. 4.
FACS profile of LNA MAbs on HHV-8-infected cells. The MAbs were tested by FACS on cell lines BCP-1, BC-3, HBL-6, Daudi, and Ramos; 5 × 105 cells were permeabilized, and LNA antibodies were used at the dilution of 1:100. The cells were incubated for 1 h with individual antibodies in PBS–0.05% saponin–1% FCS), washed three times in PBS–0.1% (vol/vol) Tween, and incubated for 1 h with rabbit anti-rat fluorescein isothiocyanate-conjugated (DAKO) secondary antibody. Analysis was performed on a FACS 440 (Becton Dickinson). Fluorescent intensity is expressed on an arbitrary logarithmic scale. Black histograms represent antibody-stained cells, and white histograms represent secondary antibody conjugate controls. (a) All antibodies detected the LNA-expressing cell population in BCP-1 cells. (b) Antibody LN53 does not detect LNA in the negative control cells Ramos (EBV negative) and Daudi (EBV positive) but does detect the LNA-positive population in the HHV-8-infected BC-3 and HBL-6 cell lines. All other antibodies are negative for the control cell lines (data not shown).
HHV-8 LNA is abundantly expressed in KS lesions.
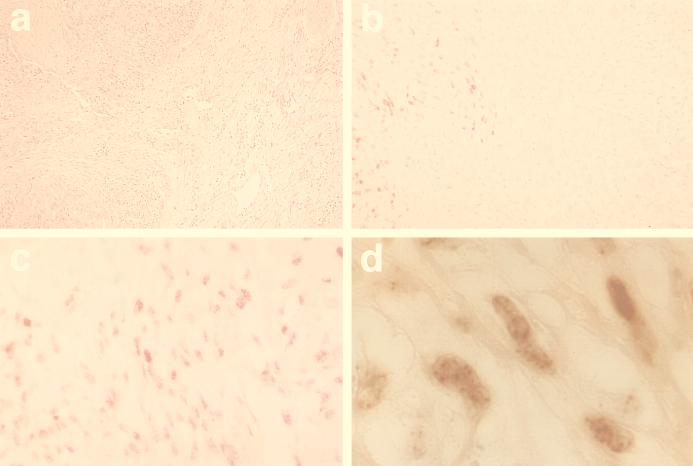
To date few protein-specific antisera or MAbs have been raised to HHV-8 proteins. Antibodies to virus-encoded interleukin-6 (vIL6) (21) and to orf26 (23) and orf59 (8) protein products have been used to study protein expression in a variety of HHV-8-infected cell types. However, no antibodies to latent proteins have been used in such studies. LN53 and LN72 but not LN20 and LN69 reacted with antigen in paraffin-embedded KS tissue. As MAb LN53 was assessed to be the best antibody raised, it was used further in the examination of the expression of HHV-8 LNA in KS lesions. LNA was extensively expressed in late-stage nodular KS, predominantly in cells of spindle-shaped morphology (Fig. 5b to d). LNA-positive cells account for more than 90% of all cells present in nodular KS lesions. The staining was exclusively nuclear, and in most cases a punctate nuclear staining, reminiscent of the nuclear IFA staining seen in PEL cell lines (Fig. 3), was seen (Fig. 5d). No expression of LNA was detected in surrounding normal dermis (Fig. 5b). No staining was observed in the control tumors angiosarcoma and hemangioma (not shown).
FIG. 5.
LN53 reactivity to LNA in nodular KS. Immunocytochemistry was performed on a paraffin-embedded tumor from classical KS. Permeabilization was performed by microwave. Negative control biopsies were from angiosarcomas and hemangiomas (not shown). After incubation with normal rabbit serum (DAKO), MAbs were applied for 1 h at 22°C followed by two washes in PBS–0.1% (vol/vol) Tween. The secondary biotin-conjugated antibody (rabbit anti-rat; DAKO) was applied for 30 min followed by washing. Antibody reactions were visualized with streptavidine-alkaline phosphatase (Vector Laboratories) and a substrate red chromogen (Vector). Adjacent sections were stained by standard methods. (a) Hematoxylin and eosin staining showing KS nodule (left) and surrounding dermis (right). (b) Adjacent sections stained with LN53 showing clear demarcation between LNA expression in the KS lesion and not in the surrounding dermis (magnification, ×40). Almost all spindle-shaped cells are positive for LNA (×60) (c), with a stippling pattern reminiscent of the staining of PEL cells (×160) (d).
Conclusions.
One of the predominant proteins encoded by HHV-8 during the latent phase of the virus life cycle is LNA, the product of orf73 (17, 18, 24). Here we describe the characterization of four MAbs to the C terminus of LNA. The properties of the MAbs are summarized in Table 1. These antibodies recognize LNA in immunofluorescence, immunoprecipitation, in situ immunocytochemistry, and FACS analyses. Three of the MAbs recognized different discrete epitopes in the C terminus of LNA. The most reactive MAb, LN53, recognized the minimal epitope EQEQE present as 23 copies in LNA. This provides an explanation for this antibody’s high reactivity in all assays used, as the repeat units of LNA should provide multiple epitopes for the antibody. The majority of the EQEQE epitopes are located in the extensive leucine zipper motif of LNA. Structure predictions of this region and knowledge of coiled-coil leucine zipper structures suggest that the EQEQE motif is located in a regular pattern on the leucine zipper alpha helices, allowing accessibility of the antibody under native protein folding conditions. This correlates with the ability of LN53 to immunoprecipitate LNA from cell extracts.
TABLE 1.
Properties of MAbs to LNA
| Antibody | Isotype | Reaction
|
||||
|---|---|---|---|---|---|---|
| ELISA | Western blottinga | Immunoprecipitationa | IFAb | Immunocytochemistryc | ||
| LN20 | IgM | + | + | NDd | + | − |
| LN53 | IgG2c | + | + | + | + | + |
| LN69 | IgG1 | + | − | − | − | − |
| LN72 | IgG2b | + | + | − | + | + |
BC-3 cells.
BCP-1 cells.
Paraffin-embedded KS lesion.
ND, not determined.
Immunoprecipitation studies confirmed that LNA exists as a protein doublet of 220 to 230 kDa in BC-3 cells (Fig. 2a). A protein of similar size was recognized by Western blotting (Fig. 2b). As with HHV-8-positive patient sera, the LNA antibodies produce a punctate immunofluorescence pattern in PEL cell lines (Fig. 3). Using LN53, we demonstrated extensive LNA expression in nodular KS. There is a clear demarcation between the KS nodule and surrounding dermis (Fig. 5b). The majority of cells showed punctate nuclear staining reminiscent of the nuclear staining in HHV-8-positive PEL cells (Fig. 5d), which suggests that the nuclear structures containing LNA present in PEL cells are also present in HHV-8-infected KS spindle cells. LNA expression has recently been demonstrated in all tissue and cell types associated with HHV-8 infection and disease (12). At present it is unclear what LNA associates with in the nuclei of latently infected cells. Studies have shown that LNA remains associated with chromosomes during mitosis (31). This feature is reminiscent of EBNA1, which is chromatin associated in both interphase and metaphase nuclei (31). However, in cells coinfected with EBV and HHV-8, EBNA1 and LNA do not colocalize, indicating that they occupy different nuclear domains (31). A careful study of colocalization and protein association with LNA by using MAb LN53 should provide insights into the function of LNA in HHV-8 latency.
The expression pattern of all known HHV-8 genes in the PEL cell line BC-1 has shown that relatively few transcripts are abundantly produced during HHV-8 latency (27). Using antibody LN53, we show expression of one such latent protein, LNA, in both PEL cell lines and KS tumors. Antibodies to other, predominantly lytic HHV-8 proteins, namely, the products of orf59 (8), a putative DNA polymerase accessory protein, and orf26, a 32-kDa inducible protein (23), have been described. Antisera to vIL6 showed expression in approximately 50% of uninduced BCP-1 cells and primary ascitic PELs, suggesting constitutive vIL6 expression in latently infected PEL cells (21). However, vIL6 protein was rarely detected in KS lesions, and when detected, it accounted for less than 2% of HHV-8-infected cells, indicating that vIL6 could be differentially expressed in hematopoietic versus mesenchymal cells. Whether LNA is involved, through interaction with different tissue-specific cellular factors, in the differential expression of HHV-8 genes and whether this contributes to HHV-8 pathogenesis remain to be determined. LNA is the first latent protein described for HHV-8, and use of our MAbs to understand its function is likely to be crucial to the understanding of HHV-8 cellular transformation and virus pathogenesis.
Acknowledgments
We thank, Y. Chang, S. J. Gao, and P. S. Moore for providing the BCP-1 cell line. BC-3 cells were kindly provided by E. Cesarman. The LNA MAbs were provided by Advanced Biotechnologies Inc., Columbia, Md.
This work was supported by the Cancer Research Campaign and GlaxoWellcome.
REFERENCES
- 1.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Boshoff C, Gao S J, Healy L E, Matthews S, Thomas A J, Coignet L, Warnke R A, Strauchen J A, Matutes E, Kamel O W, Moore P S, Weiss R A, Chang Y. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 4.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O’Leary J J. Kaposi’s sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff C, Talbot S, Kennedy M, O’Leary J, Schulz T, Chang Y. HHV8 and skin cancers in immunosuppressed patients. Lancet. 1996;347:338–339. doi: 10.1016/s0140-6736(96)90524-3. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff C, Weiss R A. Kaposi’s sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Chan S R, Bloomer C, Chandran B. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology. 1998;240:118–126. doi: 10.1006/viro.1997.8911. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, Van Marck E, Salmon D, Gorin I, Escande J-P, Weiss R A, Alitalo K, Boshoff C. Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease and in primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 14.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians, and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 15.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 16.Kedes D H, Ganem D, Ameli N, Bacchetti P, Greenblatt R. The prevalence of serum antibody to human herpesvirus 8 (Kaposi sarcoma-associated herpesvirus) among HIV-seropositive and high-risk HIV-seronegative women. JAMA. 1997;277:478–481. [PubMed] [Google Scholar]
- 17.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 19.Lennette E T, Blackbourn D J, Levy J A. Antibodies to human herpesvirus type 8 in the general population and in Kaposi’s sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- 20.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 21.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 22.Moore P S, Chang Y. Kaposi’s sarcoma (KS), KS-associated herpesvirus, and the criteria for causality in the age of molecular biology. Am J Epidemiol. 1998;147:217–221. doi: 10.1093/oxfordjournals.aje.a009440. [DOI] [PubMed] [Google Scholar]
- 23.O’Neill E, Douglas J L, Chien M L, Garcia J V. Open reading frame 26 of human herpesvirus 8 encodes a tetradecanoyl phorbol acetate- and butyrate-inducible 32-kilodalton protein expressed in a body cavity-based lymphoma cell line. J Virol. 1997;71:4791–4797. doi: 10.1128/jvi.71.6.4791-4797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Jr, Thouless M E, Tsai C C, Bosch M L. Identification of two homologs of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schalling M, Ekman M, Kaaya E E, Linde A, Biberfeld P. A role for a new herpes virus (KSHV) in different forms of Kaposi’s sarcoma. Nat Med. 1995;1:707–708. doi: 10.1038/nm0795-707. [DOI] [PubMed] [Google Scholar]
- 29.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V, Weiss R A, Moore P S. Prevalence of Kaposi’s sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 30.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 31.Szekely L, Chen F, Teramoto N, Ehlin Henriksson B, Pokrovskaja K, Szeles A, Manneborg Sandlund A, Lowbeer M, Lennette E T, Klein G. Restricted expression of Epstein-Barr virus (EBV)-encoded, growth transformation-associated antigens in an EBV- and human herpesvirus type 8-carrying body cavity lymphoma line. J Gen Virol. 1998;79:1445–1452. doi: 10.1099/0022-1317-79-6-1445. [DOI] [PubMed] [Google Scholar]
- 32.Whitby D, Luppi M, Barozzi P, Boshoff C, Weiss R A, Torelli G. Human herpesvirus 8 seroprevalence in blood donors and lymphoma patients from different regions of Italy. J Natl Cancer Inst. 1998;90:395–397. doi: 10.1093/jnci/90.5.395. [DOI] [PubMed] [Google Scholar]