Abstract
Background
Rodents are recognized as major reservoirs of numerous zoonotic pathogens and are involved in the transmission and maintenance of infectious diseases. Furthermore, despite their importance, diseases transmitted by rodents have been neglected. To date, there have been limited epidemiological studies on rodents, and information regarding their involvement in infectious diseases in the Republic of Korea (ROK) is still scarce.
Methodology/Principal findings
We investigated rodent-borne pathogens using nested PCR/RT-PCR from 156 rodents including 151 Apodemus agrarius and 5 Rattus norvegicus from 27 regions in eight provinces across the ROK between March 2019 and November 2020. Spleen, kidney, and blood samples were used to detect Anaplasma phagocytophilum, Bartonella spp., Borrelia burgdorferi sensu lato group, Coxiella burnetii, Leptospira interrogans, and severe fever with thrombocytopenia syndrome virus (SFTSV). Of the 156 rodents, 73 (46.8%) were infected with Bartonella spp., 25 (16.0%) with C. burnetii, 24 (15.4%) with L. interrogans, 21 (13.5%) with A. phagocytophilum, 9 (5.8%) with SFTSV, and 5 (3.2%) with Borrelia afzelii. Co-infections with two and three pathogens were detected in 33 (21.1%) and 11 rodents (7.1%), respectively. A. phagocytophilum was detected in all regions, showing a widespread occurrence in the ROK. The infection rates of Bartonella spp. were 83.3% for B. grahamii and 16.7% for B. taylorii.
Conclusions/Significance
To the best of our knowledge, this is the first report of C. burnetii and SFTSV infections in rodents in the ROK. This study also provides the first description of various rodent-borne pathogens through an extensive epidemiological survey in the ROK. These results suggest that rodents harbor various pathogens that pose a potential threat to public health in the ROK. Our findings provide useful information on the occurrence and distribution of zoonotic pathogens disseminated among rodents and emphasize the urgent need for rapid diagnosis, prevention, and control strategies for these zoonotic diseases.
Author summary
Rodents live almost everywhere in the world, adapt to extremely diverse habitats, and transmit various infectious diseases to humans and other animals. All six pathogens were detected in rodents. Our findings demonstrated that 66.7% (104/156) of rodents were infected with at least one pathogen. We also observed differences in the pathogens detected in rodents by province. These results provide evidence that rodents play an important role in the transmission of SFTSV. Although we did not screen for all rodent-borne diseases, these data provide information about emerging rodent-borne diseases disseminated in the ROK and emphasize the risk of occurrence of rodent-borne diseases.
Introduction
Rodents are globally abundant and well-known reservoirs and vectors of infectious diseases that affect both livestock and humans [1, 2]. The current global change context (e.g., land-use change, urbanization, and temperature increase) is particularly suitable for the expansion of several rodent species beyond their natural distribution areas [3,4]. Rodents are widespread in rural and urban areas and cause numerous human infections in regions where humans are in close contact with them. Rodents are reservoir hosts for at least 60 zoonotic diseases and play a vital role in disease transmission by spreading disease directly through contact or bites or indirectly through arthropods or food contamination [5–7]. Despite their potential threat to public health, there has not been much focus on diseases transmitted by rodents [8,9]. Moreover, the control of rodents is difficult due to their behavioral plasticity, life history traits, and high breeding potential [3].
Anaplasma phagocytophilum is a tick-transmitted, obligatory intracellular zoonotic bacterium that infects the neutrophils of various hosts, including humans, dogs, cats, horses, domestic, and wild animals [10–13]. The clinical signs of A. phagocytophilum infection range from asymptomatic to serious symptoms of veterinary and public health importance. The occurrence of A. phagocytophilum infection is increasing along with climate change worldwide. A broad variety of animal species are known to harbor A. phagocytophilum, and humans are incidental dead-end hosts [14]. Vertebrate hosts are crucial for the maintenance and circulation of this pathogen in enzootic foci. In particular, small rodents and wild ruminants have been suggested as primary reservoirs [15–19]. In the United States, the white-footed mouse (Peromyscus leucopus) is considered a well-established reservoir species [20,21]. In the Republic of Korea (ROK), A. phagocytophilum has also been detected in small mammals such as rodents and shrews (Crocidura lasiura) [22,23].
Bartonella spp. are facultative intracellular bacteria that cause persistent infections in the erythrocytes and endothelial cells of mammalian hosts [24]. The clinical manifestations caused by these species are characterized by fever, endocarditis, myocarditis, neuroretinitis, lymphadenopathy, and a range of vascular pathologies [24–28]. More than 30 Bartonella spp. and three subspecies are currently recognized [29], and at least 20 species are associated with rodents, indicating that rodents serve as potential reservoirs for zoonotic Bartonella spp. [30–32]. Among the rodent-adapted Bartonella spp., B. elizabethae, B. grahamii, B. rochalimae, B. tribocorum, B. vinsonii, and B. washoensis have been found to cause human infections [32,33]. In general, Bartonella spp. have been considered to be transmitted by arthropods [24,31]. Although Bartonella infections are widely distributed in rodents in different geographic regions [34–41], there is very little information on the distribution and prevalence of these species in rodents in the ROK [22,42,43].
Lyme borreliosis (LB) is one of the most common vector-borne diseases in North America and Eurasia and is caused by a spirochete belonging to the Borrelia burgdorferi sensu lato (s.l.) group [44]. Among this group, B. burgdorferi sensu stricto (s.s.), B. afzelii, and B. garinii are the major causative agents of LB in humans and exhibit different geographical distributions [45,46]. These species are transmitted between vertebrate hosts and tick vectors [47]. B. burgdorferi s.s. occurs in North America and Europe and has various reservoir hosts (e.g., rodents and birds), whereas B. afzelii and B. garinii occur in Eurasia and can only use specific vertebrate hosts, namely, rodents and birds, respectively [44,45]. Different Borrelia species cause different symptoms in humans. For instance, B. burgdorferi s.s. infection is associated with Lyme arthritis, whereas B. garinii is mostly linked to neuroborreliosis, and B. afzelii infection is related to a chronic skin condition known as acrodermatitis [44,48–50]. In the ROK, B. burgdorferi s.l. was first detected in 1993 and has been sporadically identified in ticks, dogs, horses, wild rodents, and humans [51–56].
Coxiella burnetii is an obligate intracellular bacterium with a worldwide distribution and is the causative agent of Q fever in humans and a wide range of animals [57]. It is highly infectious and has the ability to form spore-like particles that can withstand harsh environmental conditions and can be easily dispersed by airflow [58]. Humans acquire C. burnetii infection through inhalation of contaminated aerosols or dust particles [59]. Q fever is a public health concern as it ranks as one of the 13 leading global priority zoonoses. Moreover, it has been considered a potential biological weapon due to its widespread availability, aerosolized spread, and environmental stability [60]. The clinical manifestations of C. burnetii infection include fever and flu-like symptoms. The major sources of these infections are infected ruminants, which experience issues of abortion and infertility. Ticks and rodents are also known as natural reservoirs of C. burnetii [61]. Other studies have recently performed molecular characterization of this pathogen in domestic animals in the ROK [57,62]; however, these studies had a limited spatial distribution and were species-specific.
Leptospirosis is a zoonotic infectious disease with a global distribution and is caused by a spirochete of the genus Leptospira [63,64]. It infects more than one million people annually, with 60,000 deaths recorded [65]. Leptospira is maintained in several wild and domestic animal hosts through renal carriage and is excreted in the urine for several months [66,67]. Infection in humans and animals primarily occurs through direct contact with the urine of infected hosts or indirect exposure to contaminated water, soil, or food [68]. Its clinical manifestations in humans range from a mild febrile illness to life-threatening renal failure, pulmonary hemorrhage, and/or cardiac complications [69]. Recent studies have suggested that an increase in the incidence of leptospirosis in humans is often associated with heavy rainfall and flooding [70,71]. Rodents are considered the most important reservoir of pathogenic Leptospira spp. and contribute to disease transmission because of their close contact with humans and domestic animals [72]. L. interrogans, L. borgpetersenii, and L. kirschneri are the most abundant species circulating in humans and animals worldwide [73], with L. interrogans being the most commonly described in rodents [72].
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging tick-borne viral disease that has been primarily reported in China, the ROK, Japan, Vietnam, and Taiwan [74–78]. SFTS is formally caused by Bandavirus dabieense [also commonly known as SFTS virus (SFTSV)], which belongs to the genus Bandavirus in the family Phenuiviridae. SFTSV infections are characterized by high fever, fatigue, myalgia, gastrointestinal symptoms, thrombocytopenia, and multiorgan failure [74,79]. SFTSV can also spread from person to person through exposure to the blood of an infected individual [80]. Due to its potential as a public health threat, SFTS was chosen as one of nine emerging diseases given a priority for research by the World Health Organization in 2017 [81]. As humans are often in close contact with domestic animals and may encounter rodents when they work outdoors, transmission between animals and humans is another possible major transmission route [82]. The overall mortality rate of this disease has been reported to be 3–30% in different countries [74,83,84]. Although SFTSV has been identified in various animals, its natural reservoir hosts have not been determined.
As demonstrated in the abovementioned studies, rodents may be involved in the transmission cycles of various diseases. Recently, the incidences of several infectious diseases have been rapidly increasing worldwide due to climate warming. Rodent populations are also growing exponentially due to climate change and urbanization. To date, most studies on rodent-borne diseases in the ROK have been primarily focused on identifying hantavirus infections. Although rodents are considered important reservoirs of zoonotic infectious pathogens, epidemiological information regarding their involvement in infectious diseases is limited in the ROK. Therefore, the aims of this study were to investigate the occurrence of some rodent-borne diseases, characterize their genetic relationships, and determine the roles of rodents as reservoir hosts for these diseases.
Methods
Ethical statement
Rodent collection was approved by the Seoul National University Institutional Animal Care and Use Committee (No. SNU-190524-2-1) and performed according to the Seoul National University Guidelines on the care and use of laboratory animals.
Sample collection
Rodents were captured using Sherman traps (3 × 3.5 × 9-inch folding traps; H.B. Sherman Traps, Tallahassee, FL, USA) from 27 regions in eight provinces across the country between March 2019 and November 2020. Rodents were captured from at least one region in each province (S1 Table). The traps were set in locations where human infections with SFTSV had been reported based on statistical data from the Korea Disease Control and Prevention Agency. To capture rodents, rural (agricultural land such as regions with rivers, valleys, mountains, and lakes) and peri-urban (human residential and farm) areas were selected. Rural areas were defined as regions with natural landscapes and minimal urban impact, whereas peri-urban areas were defined as transitional zones between urban and rural areas. A total of 60 traps were installed in each capture region in lines in 3-m intervals between 5 p.m. and 6 p.m. and retrieved the next day between 9 a.m. and 10 a.m. after setup: 60 traps per night were set in each region. The capture duration for every site was 14–15 h, and each capture site was sampled only once. The captured rodents [rural (n = 127) and peri-urban (n = 29)] were transported to the laboratory in an icebox with the traps, morphologically identified, and euthanized using CO2. Thereafter, blood, spleen, and kidney samples were collected from each animal. A whole blood sample was also collected in a serum separation tube, and the serum was separated and used for RNA extraction.
DNA/RNA extraction and PCR analysis
DNA was extracted from spleen (10 mg) and kidney (25 mg) samples using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and stored at −20°C until analysis. Splenic DNA was subjected to PCR amplification to detect A. phagocytophilum, Bartonella spp., Borrelia spp., and C. burnetii, and kidney DNA was subjected to PCR to detect L. interrogans. These pathogens were screened using each specific primer with the nested PCR method under the following conditions: 93–95°C for 5 min, followed by 30–40 cycles of 93–95°C for 1 min, the annealing temperature of each pathogen, 72°C for 1 min, and a final extension step at 72°C for 10 min (Table 1). Distilled water was used as a negative control in all PCR analyses. Secondary PCR products were visualized on 1.5% agarose gels stained with ethidium bromide.
Table 1. Primer information used for PCR analysis.
| Pathogen | Target genes | Sequences (5′–3′) | Sizes (bp) |
Annealing temp./Time |
|---|---|---|---|---|
| Anaplasma phagocytophilum | 16S rRNA | TCCTGGCTCAGAACGAACGCTGGCGGC | 1,433 | 50°C/30 s |
| AGTCACTGACCCAACCTTAAATGGCTG | ||||
| GTCGAACGGATTATTTTTATAGCTTGC | 926 | 56°C/30 s | ||
| CCCTTCCGTTAAGAAGGATCTAATCTCC | ||||
| Bartonella spp. | ITS | TTCAGATGATGATCCCAAGC | 639 | 55°C/30 s |
| AACATGTCTGAATATATCTTC | ||||
| CCGGAGGGCTTGTAGCTCAG | 499 | 55°C/30 s | ||
| CACAATTTCAATAGAAC | ||||
| Borrelia spp. | ospA | GGGAATAGGTCTAATATTAGCC | 665 | 42°C/60 s |
| CACTAATTGTTAAAGTGGAAGT | ||||
| GCAAAATGTTAGCAGCCTTGAT | 392 | 56°C/60 s | ||
| CTGTGTATTCAAGTCTGGC | ||||
| Coxiella burnetii | IS1111 | TATGTATCCACCGTAGCCAGTC | 687 | 54°C/30 s |
| CCCAACAACAACCTCCTTATTC | ||||
| GAGCGAACCATTGGTATCG | 203 | 54°C/30 s | ||
| CTTTAACAGCGCTTGAACGT | ||||
| Leptospira interrogans | rpoB | GTTCCAACATGCAACGYCAR | 1,649 | 52°C/60 s |
| GTTGAAGGATTCRGGRATAC | ||||
| TYATGCCKTGGGAAGGWTAC | 1,023 | 56°C/30 s | ||
| GCATRTCRTCKGACTTGATG | ||||
| SFTSV | S | CATCATTGTCTTTGCCCTGA | 461 | 52°C/40 s |
| AGAAGACAGAGTTCACAGCA | ||||
| AAYAAGATCGTCAAGGCATCA | 346 | 55°C/40 s | ||
| TAGTCTTGGTGAAGGCATCTT |
*SFTSV: severe fever with thrombocytopenia syndrome virus
RNA was extracted from 200-μL aliquots of serum using the Gene-spin Viral DNA/RNA Extraction Kit (iNtRON Biotechnology, Seongnam, ROK) according to the manufacturer’s instructions. The viral RNA was stored at − 80°C until use. Each RNA sample was tested using nested reverse transcription-polymerase chain reaction (RT-PCR) assays to detect the small (S) segment of SFTSV. Primary PCR was performed using one-step RT-PCR premix (Solgent, Daejeon, ROK) under the following conditions: initial step of 30 min at 50°C and 15 min at 95°C for denaturation, followed by 40 cycles of 20 s at 95°C, 40 s at 52°C, and 30 s at 72°C, with a final extension step of 5 min at 72°C. Nested PCR was conducted using 1 μL of the primary PCR product as a template (BIOFACT, Daejeon, ROK). The reaction for the nested PCR consisted of 25 cycles of 20 s at 94°C, 40 s at 55°C, and 30 s at 72°C. The primer information used to detect SFTSV is listed in Table 1. Secondary PCR products were visualized on 1.5% agarose gels stained with ethidium bromide.
Phylogenetic analysis
The secondary PCR products were purified using the AccuPrep PCR Purification Kit (Bioneer, Daejeon, ROK) according to the manufacturer’s instructions and directly sequenced (Macrogen Inc., Seoul, ROK). Less than five PCR-positive samples from each pathogen were utilized for all sequencing analyses. In cases in which there were more than five positive samples, we selected only a few samples and used them for the sequencing analysis. All the nucleotide sequences obtained for each pathogen were aligned using the BioEdit software and then compared with reference sequences from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) to determine their similarity using Geneious Prime 2022.2 software (http://www.geneious.com). Phylogenetic analysis of each pathogen was performed using the maximum-likelihood method implemented in MEGA11 using the best substitution model. Bootstrap values were calculated by analyzing 1,000 replicates to evaluate the reliability of clusters. The models used in this study were K2 + G for A. phagocytophilum, Tamura 3-parameter + G + I for Bartonella spp., Tamura-Nei for Borrelia spp., and the Kimura 2-parameter model for C. burnetii, L. interrogans, and SFTSV. The nucleotide sequences obtained in this study were assigned the following accession numbers: OR287077-OR287091 for A. phagocytophilum, OR288176-OR288190 for B. grahamii, OR288191-OR288193 for B. taylorii, OR284310-OR284311 for B. afzelii, OR284312-OR284321 for C. burnetii, OR284322-OR284324 for L. interrogans, and OR257718-OR257726 for SFTSV.
Statistical analysis
The infection rates were calculated with 95% confidence intervals (CIs). The PCR results for each rodent sample were recorded as negative or positive and were categorized as a single infection or a co-infection with two or three pathogens. Statistical analysis was performed using the SPSS 29.0 software package for Windows (SPSS Inc., Chicago, IL, USA). The association between sex and the infection rate for each pathogen was determined using the chi-square test. A P-value ≤ 0.05 was considered statistically significant.
Results
Sample collection
A total of 175 rodents were captured and morphologically classified as follows: 151 Apodemus agrarius (striped field mouse) (70 males and 81 females), 5 Rattus norvegicus (Norway rat) (2 males and 3 females), and 19 unknown. A. agrarius was found in most of the capture sites in the ROK, whereas R. norvegicus was captured only in two provinces (Table 2). Unknown samples were excluded from this study, and the remaining 156 rodents were used for data analysis.
Table 2. Number of rodents captured by province.
| Province | Apodemus agrarius | Rattus norvegicus | Total |
|---|---|---|---|
| Gyeonggi | 12 | - | 12 |
| Gangwon | 18 | - | 18 |
| Chungbuk | 19 | - | 19 |
| Chungnam/Daejeon | 4 | - | 4 |
| Jeonbuk | 13 | - | 13 |
| Jeonnam | 4 | - | 4 |
| Gyeongbuk | 76 | 4 | 80 |
| Gyeongnam | 5 | 1 | 6 |
| Total | 151 | 5 | 156 |
“-”: no rodents captured
Prevalence of pathogens detected in the captured rodents
The presence of six pathogens was investigated by PCR analysis from the two species, A. agrarius and R. norvegicus. Of the 156 rodents, 104 (66.7%; 95% CI: 59.3–74.1) were infected with at least one pathogen. The infection rate was 64.3% in females (54/84) and 66.6% in males (48/72). There was no significant difference between single infection or co-infections and sex (P = 0.886). The infection rates of each pathogen by sex are shown in Table 3. In all the six pathogens, no significant difference was found in the infection rate between the sexes (Table 3).
Table 3. Association between each pathogen and the sex of the captured rodents.
| Pathogens | Male (n = 72) | Female (n = 84) | χ2 (P-value) |
|---|---|---|---|
| A. phagocytophilum | 9 | 12 | 0.008 (0.928) |
| Bartonella spp. | 40 | 33 | 3.494 (0.062) |
| Borrelia spp. | 2 | 3 | 0.000 (1.000) |
| C. burnetii | 9 | 16 | 0.796 (0.372) |
| L. interrogans | 12 | 12 | 0.035 (0.851) |
| SFTSV | 4 | 5 | 0.000 (1.000) |
| Total | 48 | 54 | 0.020 (0.886) |
*P < 0.05
**P < 0.005
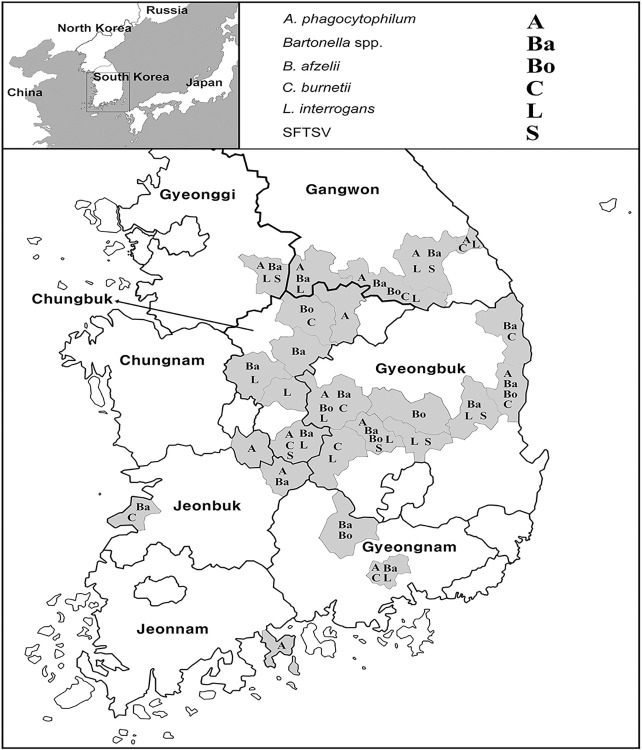
In terms of pathogens, Bartonella spp. were frequently detected (46.8%; 95% CI: 39.0–54.6), followed by C. burnetii (16.0%; 95% CI: 10.2–21.8), L. interrogans (15.4%; 95% CI: 9.7–21.1), A. phagocytophilum (13.5%; 95% CI: 8.1–18.9), SFTSV (5.8%; 95% CI: 2.1–9.5), and Borrelia spp. (3.2%; 95% CI: 0.4–6.0) (Table 4). The details of the pathogens identified by province are shown in Table 4. All six pathogens were detected in Gangwon, Chungbuk, and Gyeongbuk provinces. Five of the six pathogens, excluding SFTSV, were found in Gyeongnam province, whereas only one pathogen was detected in Chungnam and Jeonnam provinces (Table 4). Co-infections with two and three pathogens from the captured rodents were also detected in 33 (21.2%; 95% CI: 14.8–27.6) and 11 (7.1%; 95% CI: 3.1–11.1) animals, respectively (Table 5), and co-infections with Bartonella spp. and L. interrogans were the most frequently detected (Table 5). Information regarding the pathogens identified by region is shown in the map in (Fig 1).
Table 4. Number of positive samples in which pathogens were identified in captured rodents according to each province.
| Province | A. phagocytophilum | Bartonella spp. | Borrelia spp. | C. burnetii | L. interrogans | SFTSV |
|---|---|---|---|---|---|---|
| Gyeonggi (n = 12) | 1 | 5 | - | - | 2 | 2 |
| Gangwon (n = 18 | 4 | 12 | 1 | 5 | 6 | 1 |
| Chungbuk (n = 19) | 3 | 10 | 1 | 2 | 6 | 2 |
| Chungnam/Daejeon (n = 4) | 1 | - | - | - | - | - |
| Jeonbuk (n = 13) | 3 | 8 | - | 3 | - | - |
| Jeonnam (n = 4) | 1 | - | - | - | - | - |
| Gyeongbuk (n = 76) | 7 | 32 | 2 | 14 | 6 | 4 |
| Gyeongnam (n = 5) | 1 | 6 | 1 | 1 | 4 | - |
| Total (n = 151) | 21 (13.5%) | 73 (46.8%) | 5 (3.2%) | 25 (16.0%) | 24 (15.4%) | 9 (5.8%) |
“-”: not detected.
Table 5. Co-infections of two or three pathogens detected in captured rodents.
| Pathogens | No. of positive samples |
|---|---|
| A. phagocytophilum + Bartonella spp. | 7 |
| A. phagocytophilum + Borrelia spp. | 1 |
| A. phagocytophilum + C. burnetii | 1 |
| Bartonella spp. + Borrelia spp. | 2 |
| Bartonella spp. + C. burnetii | 7 |
| Bartonella spp. + L. interrogans | 10 |
| Bartonella spp. + SFTSV | 3 |
| C. burnetii + L. interrogans | 2 |
| A. phagocytophilum + Bartonella spp.+ C. burnetii | 1 |
| A. phagocytophilum + Bartonella spp.+ L. interrogans | 3 |
| A. phagocytophilum + Bartonella spp.+ SFTSV | 2 |
| A. phagocytophilum + C. burnetii + L. interrogans | 1 |
| Bartonella spp. + Borrelia spp. + C. burnetii | 1 |
| Bartonella spp. + C. burnetii + L. interrogans | 1 |
| Bartonella spp. + L. interrogans + SFTSV | 2 |
Fig 1. Maps showing the regions where rodent-borne pathogens were detected in the Republic of Korea.
The word symbol is indicated differently according to each pathogen. Maps were created using NGII [https://www.data.go.kr/data/15062309/fileData.do] and are Korea Open Government License Type 1, which can be freely used without any permission.
Phylogenetic trees of rodent-associated pathogens
Anaplasma phagocytophilum
A. phagocytophilum was detected in rodents from all the examined provinces, indicating that this pathogen is spread throughout the ROK. Of 21 positive samples, 15 were sequenced and confirmed to be A. phagocytophilum by a phylogenetic tree analysis based on the 16S rRNA gene (Fig 2). Our sequences exhibited 97.6–99.9% identity with each other and 95.6–100% identity with those reported in the ROK. These 15 sequences were similar to those previously reported from several different hosts such as cattle, dogs, horses, humans, ticks, and rodents in other countries, sharing 96.9–100% nucleotide identities with these. Furthermore, several variants co-existed in the same geographical area. According to the phylogenetic tree, A. phagocytophilum was divided into clades 1 and 2, and all our sequences from A. agrarius belonged to clade 1 (Fig 2). The difference in sequences between clades 1 and 2 revealed 94.7–98.5% nucleotide identities. Clade 2 had 10 nucleotide differences compared with clade 1. Genetic variants were detected in A. phagocytophilum circulating in the ROK.
Fig 2. Phylogenetic tree inferred by maximum-likelihood analysis using the K2 + G model of the 16S rRNA gene sequence of Anaplasma phagocytophilum.
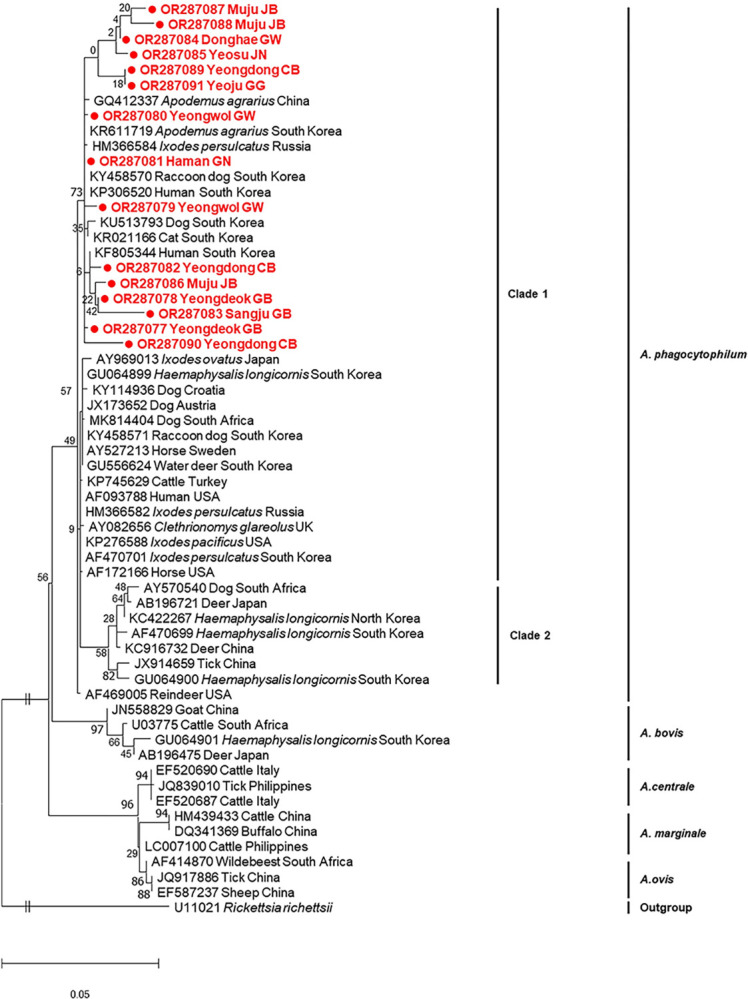
The numbers at the nodes are bootstrap values expressed as a percentage of 1,000 replicates. The scale bar indicates nucleotide substitution per site. Samples sequenced from Apodemus agrarius are shown in filled circles.
Bartonella spp.
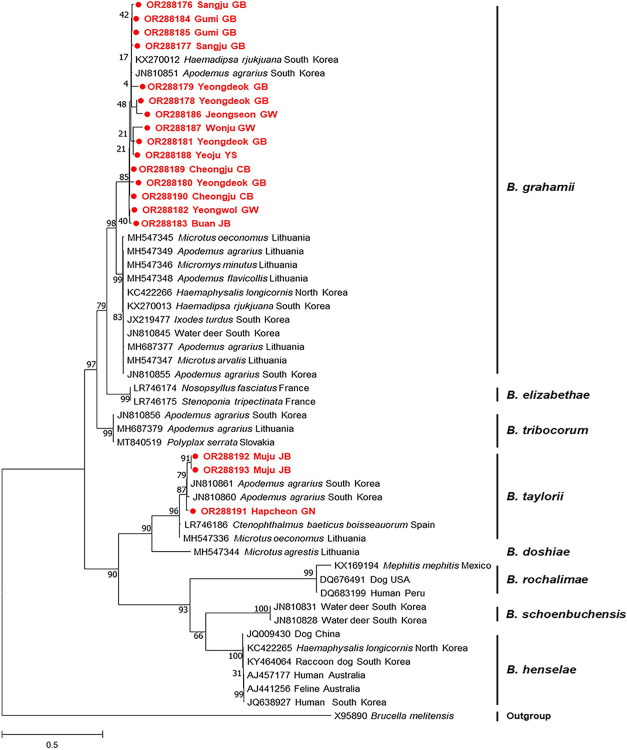
Bartonella spp. were frequently detected in A. agrarius in the ROK, but they were not found in all provinces (Table 4). Bartonella spp. were detected in both A. agrarius and R. norvegicus. Of 73 internal transcribed spacer (ITS) PCR-positive samples, 18 sequences were included in a phylogenetic analysis. Two species of Bartonella spp. were identified circulating in the examined rodents using the phylogenetic tree based on the ITS gene: B. grahamii and B. taylorii (Fig 3). The prevalence of B. grahamii and B. taylorii was 83.3% (15/18) and 16.7% (3/18), respectively. The 15 sequences belonging to B. grahamii showed 94.9–100% identity with each other and formed the same group as B. grahamii found in leeches (KX270012) and A. agrarius (JN810851) reported in the ROK, exhibiting 95.9–99.8% identity with those sequences. Furthermore, another sequence (JN810855) reported from A. agrarius in the ROK demonstrated 87.1–90.8% similarity to the sequences reported in our study. The three sequences classified as B. taylorii exhibited 100% identity with each other and shared 92.5–100% identity with those belonging to this species.
Fig 3. Phylogenetic analysis based on the ITS region of Bartonella spp. (maximum-likelihood analysis using the Tamura 3-parameter + G + I model with 1,000 replicates).
The scale bar indicates nucleotide substitution per site. Sequences determined from A. agrarius are indicated in filled circles.
Borrelia spp.
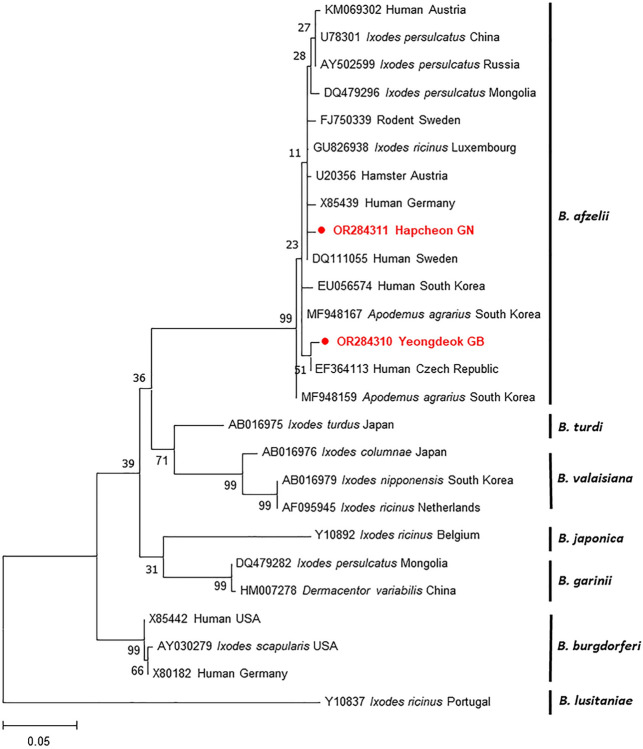
Borrelia spp. were found in four provinces (Table 4), and the infection rate of Borrelia spp. was the lowest (3.2%) compared with those of the other pathogens identified. Among the five PCR-positive samples, only two sequences were obtained and demonstrated 98.6% identity with each other. A phylogenetic analysis based on the outer surface protein A (ospA) gene revealed that our sequences were assigned to B. afzelii (Fig 4). The two sequences exhibited 98.9–100% homology with those identified previously in A. agrarius in the ROK. Our sequences showed 97.8–100% identity with those belonging to this group. Furthermore, these sequences displayed 98.2–99.6% similarity to those reported in humans from Austria, Germany, the Czech Republic, the ROK, and Sweden.
Fig 4. Maximum-likelihood phylogenetic tree using the Tamura-Nei model based on the ospA gene of Borrelia spp. Bootstrap values were calculated with 1,000 replicates of the alignment.
The scale bar indicates nucleotide substitution per site. Sequences obtained from A. agrarius are symbolized in filled circles.
Coxiella burnetii
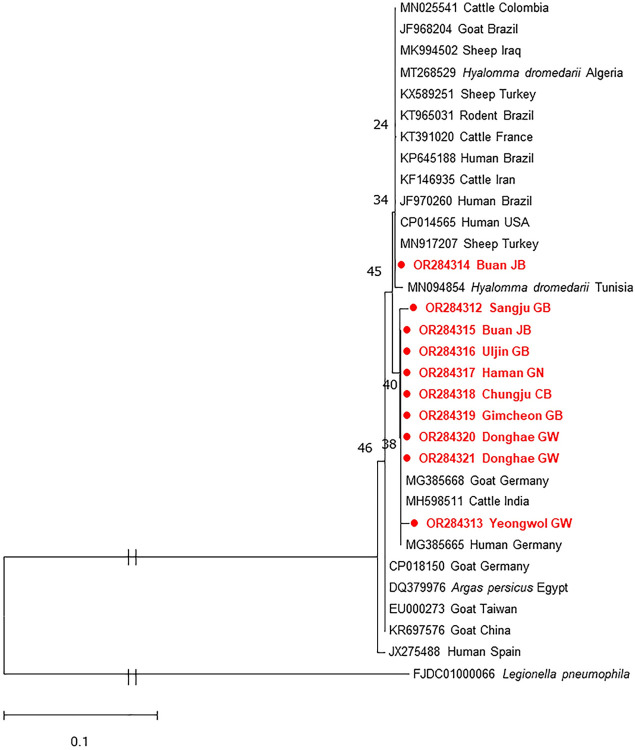
C. burnetii was the second most frequently detected pathogen and was identified in both A. agrarius and R. norvegicus. However, it was found in five different provinces. Of 25 positive samples, 10 sequences were included in a phylogenetic tree based on the IS1111 gene. These sequences showed 97.5–100% identity with each other. Only one sequence (OR284314) had the closest genetic relationship with strains identified in febrile and pneumonic patients (KP645188 and JF970260), which are known as virulent strains, exhibiting 100% homology with those (Fig 5). The others formed a separate branch, exhibiting 99.0–99.5% identity with two human isolates (KP645188 and JF970260). The phylogenetic tree revealed the presence of genetic variations within the C. burnetii sequences.
Fig 5. Maximum-likelihood phylogenetic tree from the IS1111 gene of Coxiella burnetii.
The evolutionary analysis was inferred using the Kimura 2-parameter model. Bootstrap values (1,000 replicates) are indicated in each node. The scale bar indicates nucleotide substitution per site. Sequences determined from A. agrarius are highlighted in filled circles.
Leptospira interrogans
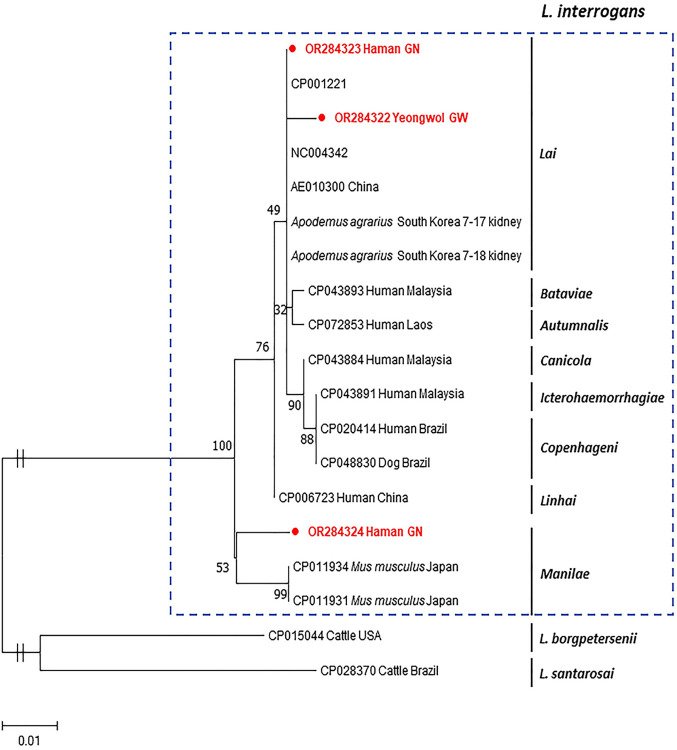
L. interrogans was the third most commonly detected pathogen and was also found in both A. agrarius and R. norvegicus. Of 24 positive samples, only three sequences were obtained, and they had 97.7–99.5% identity with each other. A phylogenetic tree based on the RNA polymerase subunit beta (rpoB) gene revealed that these sequences belonged to L. interrogans (Fig 6). Two sequences (OR284322 and OR284323) were classified into L. interrogans serovar Lai and showed 99.2–100% identity with those reported in China and 99.4–100% identity with those reported in A. agrarius in the ROK. The other sequence (OR284324) belonged to L. interrogans serovar Manilae, which has detected in Mus musculus in Japan, exhibiting 98.2% similarity (Fig 6). At least two serovars of L. interrogans were found to be circulating in A. agrarius in the ROK.
Fig 6. Phylogenetic analysis based on the rpoB gene of Leptospira interrogans.
The tree was inferred in MEGA X using maximum-likelihood and Kimura 2-parameter with 1,000 replicates. The scale bar implies nucleotide substitution per site. The box of dash lines indicates L. interrogans. Sequences obtained from A. agrarius are shown in filled circles.
Severe fever with thrombocytopenia syndrome virus
SFTSV was detected in nine A. agrarius samples (5.8%) and found in four different provinces (Table 4). Of the nine SFTSV infections, a single infection with SFTSV was detected only in two A. agrarius samples, and the remaining samples were primarily co-infected with other pathogens such as Bartonella spp. and L. interrogans (Table 5). Nine sequences were obtained and included in the phylogenetic tree. These sequences demonstrated 95.95–100.0% identity with each other. The phylogenetic analysis based on the S segments revealed that five and four sequences were classified into subgenotype B-2 and genotype D, respectively (Fig 7). The sequences belonging to genotype B-2 exhibited 94.5–97.4% homology with those identified in human and other animal samples in the ROK, whereas four sequences showed 99.7–100.0% identity with those identified in human samples. These results revealed that genotype B-2 is prevalent in the ROK and that genetic variants exist within genotype B-2.
Fig 7. Phylogenetic tree of the severe fever with thrombocytopenia syndrome virus based on the analysis of partial sequences of small segments.
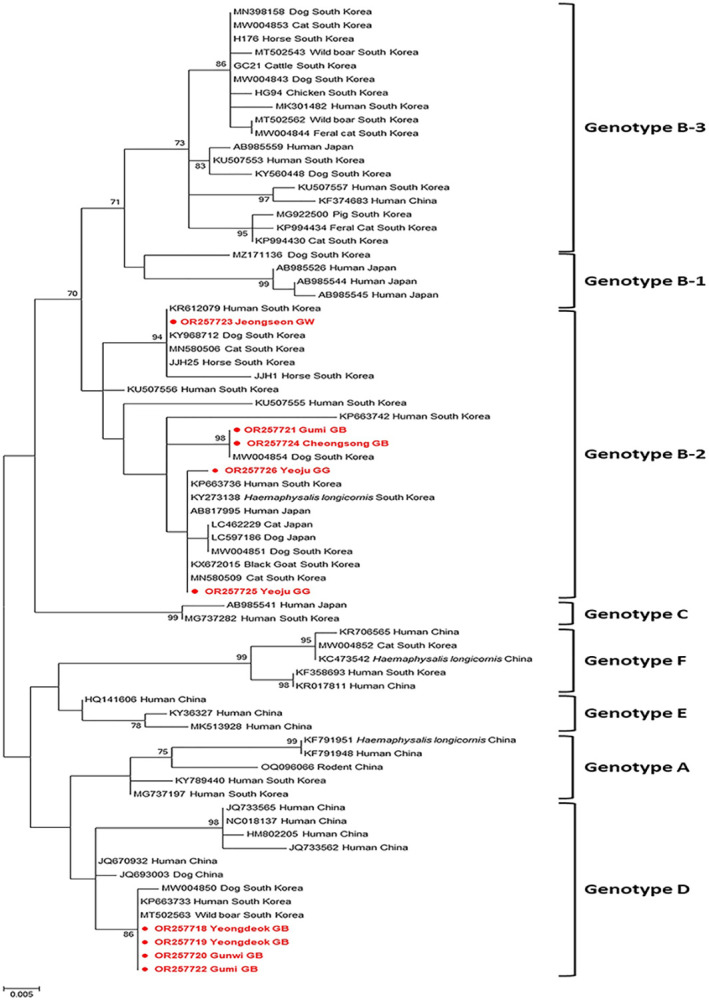
Maximum-likelihood analysis was used to construct the Kimura 2-parameter model (1,000 bootstrap replicates). The scale bar implies nucleotide substitution per site. Sequences identified from A. agrarius are indicated in filled circles.
Discussion
This study demonstrated the infection rate and genetic characterization of zoonotic pathogens by molecular analysis in rodents captured from throughout the ROK. Rodents were trapped from rural and peri-urban areas with frequent movement of people, which may be associated with a high probability of disease transmission because humans and rodents share the same space. In the present study, all six pathogens were detected in rodents. The results demonstrated that 66.7% (104/151) of rodents were infected with at least one pathogen. According to our findings, Bartonella spp. were most frequently detected, and Borrelia spp. were the least commonly detected in rodents. Moreover, by sex, there was no difference in the infection rate and no statistically significant differences between single infection and co-infections. A limitation of this study is that the number of captured rodents in each province varied. Nevertheless, five pathogens were identified in Gyeongnam province, which had the second lowest number of rodents captured. In contrast, although more rodents were captured in Gyeonggi and Jeonbuk provinces than in Gyeongnam province, fewer pathogens were detected. These results imply that increased sample numbers do not necessarily correlate with the probability of pathogen detection. Moreover, because most rodents were captured in rural areas, it is impossible to compare the infection rates between rural and peri-urban areas. At this time, we cannot conclude differences in pathogen detection, but this finding may be influenced by the sampling site where the rodents were captured. Although the infection rate was not very high, A. phagocytophilum was found in all provinces. Considering that the number of rodents captured varied by province and that few rodents were captured in some provinces, A. phagocytophilum may be the most widespread pathogen in the ROK. Furthermore, to the best of our knowledge, this is the first study to report C. burnetii and SFTSV infections in rodents in the ROK and to conduct an extensive study to investigate infections with several pathogens. These data provide valuable information for evaluating the potential risk of rodents in public health.
In this study, all six of the evaluated pathogens were detected using the nested PCR method. In general, nested PCR is more sensitive than qPCR but is also prone to contamination and is more cumbersome [85]. There were the discrepancies between PCRs and sequencing results. We cannot address the reason why the sequencing results were not good at this time. This may be the need for more amounts of genetic material to enable proper sequencing. Nonetheless, we considered PCR-positives to be truly positive, regardless of whether the amplicons could be sequenced. It seems difficult to consider them negative only because some amplicons could not be sequenced.
Anaplasma phagocytophilum is known as the third most common tick-borne pathogen in the USA and Europe [86] and has been detected in 20 different rodent species [87]. A. phagocytophilum infection rates range considerably in rodent species [87]; this may be explained by differences in small mammals that maintain tick species. In this study, the prevalence of A. phagocytophilum infections in A. agrarius was 13.5%, which was rather lower than reported in a previous study conducted in the ROK (19.1%) [88]. To date, there has been no report of A. phagocytophilum infection in Rattus spp. in the ROK; however, a high infection rate (31.5%) of A. phagocytophilum has been reported in Rattus spp. from China [89]. A. phagocytophilum has been detected in a variety of animals, including ticks in the ROK, but its pathogenicity still remains unclear. The sequences obtained from rodents showed 97.9–100% identity to A. agrarius (KR611719) reported in the ROK. According to the phylogenetic analysis, there are several genetic variants among A. phagocytophilum circulating in the ROK. Haemaphysalis longicornis, which is found primarily in the ROK, tends to use A. agrarius as the major host to maintain A. phagocytophilum [23,89], indicating that A. agrarius is an enzootic reservoir. Thus, further studies are required to determine the pathogenicity of A. phagocytophilum variants circulating in the ROK.
The overall prevalence of Bartonella spp. in rodents was 46.8%, which was the highest of the prevalence rates of all the other pathogens examined in this study. However, the detection rate in the present study was lower than that described in a previous report (62.0%) based on the ITS gene [43]; this difference may be because of the location where the rodents were captured. Moreover, the prevalence of Bartonella spp. in rodents varies across countries, e.g., 5.5% in Turkey [37], 23.7% in Lithuania [90], 36.3% in Chile [32], 40.4% in Slovenia [34], and 65.8% in Eastern Germany [41]. In addition, the Bartonella spp. that are prevalent in each country are different [27,32,34,38,90–93]. According to the phylogenetic tree, B. grahamii has two distinct groups, and the sequences obtained in this study formed the same group and diverged from some sequences previously identified in the ROK. This indicates that genetic variations of B. grahamii exist in the ROK. R. norvegicus and R. rattus are known as major reservoirs for Bartonella spp. in several countries [27,94–96], but Bartonella spp. have not been detected in other rodent species or in R. norvegicus in the ROK [22]. The analysis of R. norvegicus in this study was unfortunately limited because only five animals were captured, and their data were excluded. Further studies are necessary to investigate Bartonella spp. infection in R. norvegicus. The present results demonstrated that B. grahamii was the most predominant species, and B. taylorii was found in three rodents, which is consistent with the findings of a previous study [43]. B. grahamii is a zoonotic pathogen that is associated with neuroretinitis and retinal artery occlusion in humans [25]. B. taylorii can cause infection in animals [90], but its pathogenicity remains unclear. In Europe, B. taylorii is the predominant species found in rodents [24,37]. In this study, the Bartonella spp. detected were different by province, and in Jeonbuk province, both B. grahamii and B. taylorii were identified, indicating that the pathogens present in each province are different. Although B. taylorii has been detected in some A. agrarius in the ROK, its transmission route remains unknown. Considering its high infection rate in A. agrarius, potential vectors of this pathogen should be identified to prevent infection.
The detection rate of Borrelia spp. in A. agrarius was 3.2% and was the lowest infection rate compared with those of the other pathogens examined in this study. Our result was different from those of previous studies that evaluated heart samples from A. agrarius (29.6%) [56] and ticks (33.6%) collected from wild rodents in the ROK [97]. This finding can be explained by the differences in the genes and samples used. For instance, Kim et al [56] reported that B. burgdorferi s.s. and B. garinii infected the spleen and that B. afzelii exhibited a high detection rate in the heart; however, B. burgdorferi s.s. and B. garinii were not detected in the spleen. It is speculated that the number of positive samples was small and could not be detected. Among the Borrelia burgdorferi s.l. group, only B. afzelii was identified in A. agrarius, which supports previous findings that B. afzelii is the predominant species in the ROK [54,97]. Furthermore, our results were significantly lower than those reported in rodents in other countries: 24% in Austria [98], 16% in the Czech Republic [99], and 6.3% in Spain [100]. These differences in prevalence may be due to the tick vectors; the common tick vectors of Borrelia spp. in the ROK are Ixodes persulcatus, I. nipponensis, and I. granulatus [101]. B. afzelii is transmitted by I. ricinus and hosted by small mammals, and it is the most common causative agent of human LB [45,102]. The sequences from rodents showed 98.2–99.6% identity with those identified in ticks and humans in several countries. In the ROK, B. afzelii has been primarily reported in ticks [54,97,103] and rarely in humans [104]. Nevertheless, information on B. afzelii is still lacking. Although the infection rate of B. afzelii in rodents was the lowest, our findings suggest that B. afzelii may act as a causative agent of LB in the ROK.
This is the first report of C. burnetii in A. agrarius in the ROK. In this study, C. burnetii exhibited the second highest infection rate (16.0%). Nevertheless, our results were lower than those reported in China (18%) [105], Senegal (22.4%) [106], and Zambia (45%) [107] but higher than those reported in Brazil (4.6%) [93], Egypt (6.7%) [58], and Italy (1.4%) [61]. These differences may be explained by the rodent species and samples used for detection. In prior studies, C. burnetii detection was performed using various sample types such as blood, spleen, liver, and fecal samples. Consequently, the liver and spleen are considered suitable for the identification of C. burnetii. According to a previous study, the infection rate of C. burnetii in domestic livestock ranged from 6% to 22.7% depending on the species [57]. Although C. burnetii is a tick-borne pathogen, there are only a few reports of C. burnetii in ticks in the ROK [108,109]. Recent studies have reported co-infections of C. burnetii and SFTSV in ticks and humans [110, 111]; however, there was no co-infection with these two pathogens identified in rodents in this study. Despite the small number of R. norvegicus captured, C. burnetii infection was mostly detected in R. norvegicus; this could be because R. norvegicus may also serve as a reservoir in the ROK. A phylogenetic analysis based on the IS1111 gene revealed the presence of genetic variations within the sequences identified in A. agrarius. One sequence formed the same clade with the virulent strains reported in Brazil, whereas the other exhibited high similarity to strains reported in different countries. The disadvantage of the IS1111 gene is that it does not provide exact information, such as pathogenicity and species specificity (Fig 5); thus, we cannot draw any conclusions about what the separate groupings within these C. burnetii sequences might represent. Further research is necessary to determine the pathogenicity of C. burnetii circulating in the ROK. The results obtained in the present study suggest that A. agrarius plays a role in the transmission of C. burnetii in humans and animals.
Leptospira interrogans is a rodent-borne pathogen, and accordingly, it was the third most frequently detected (15.4%) in this study. Our results demonstrated a relatively high prevalence compared with that reported in previous studies [88,112]. This is the first time that Leptospira spp. have been investigated in rodents through sampling of extensive regions in the ROK. Compared with those reported in other countries, the infection rates ranged from 1.3% to 35.2% [113–117]. R. norvegicus is also an important reservoir of this pathogen [72]; however, L. interrogans was detected in only one R. norvegicus and mostly detected in A. agrarius. Considering that R. norvegicus is commonly found around barns and farmhouses, this species also plays a critical role in the transmission of leptospirosis in domestic animals and humans. To date, L. interrogans has been divided into 23 serogroups based on serological methods and subdivided into more than 300 serovars [72]. The serovars circulating in each country are different, but the most frequently reported serovar worldwide is Icterohaemorrhagiae [72]. In the ROK, only a few studies have been conducted on serovar Lai [88, 118]. The phylogenetic analysis revealed that of the three sequences from rodents, two were classified as serovar Lai and one as serovar Manila, consistent with the findings from a previous study [88]. Consequently, Lai and Manilae are considered epidemic serovars in the ROK. The most important limitations of this study are that a serological analysis such as a microscopic agglutination test was not performed and that only a small number of samples were sequenced; therefore, serovar comparisons by province were not conducted. Although the number of the sequenced samples was low, the rpoB gene used in this study can be applicable for detection and serovar identification of L. interrogans. Furthermore, for accurate identification of L. interrogans serovars, serological testing along with the PCR method is absolutely necessary. These results suggest that the existence of various serovars in each province cannot be ruled out.
Since its first identification in China, SFTSV has been primarily detected in Asia [74–78]. Due to its associated high mortality rate, there is significant interest in SFTSV [74,83,84]. In the present study, the infection rate of SFTSV in A. agrarius was 5.8%, and this is the first report to describe SFTSV infection in A. agrarius in the ROK. Our results were significantly lower than those reported in China (32.3%) [119]. Compared with the infection rates reported in other animals in the ROK, the prevalence of SFTSV in rodents was similar to that in wild boars (5.2%) [120] and ticks (6.0%) [121], but higher than that in cats (4.0%) [122], dogs (2.9%) [123], pigs (1.7%) [124], black goats (2.4%) [125], and wild animals (3.3%) [126]. However, the prevalence of SFTSV was the highest in feral cats (17.5%) in the ROK [127]. There has been a recent increase in the populations of feral cats, and they share habitats with wildlife, domestic animals, and humans. Several studies have demonstrated that SFTSV is transmitted to humans through direct contact with cats [128,129], suggesting that feral cats are infected by rodents. It is believed that SFTSV circulates in a zoonotic cycle between ticks and vertebrates [130]. Rodents are considered the representative reservoirs in maintaining tick-borne pathogens and may play a vital role in the transmission of SFTSV. The sequences obtained from A. agrarius belonged to subgenotype B-2 and D genotype; these results revealed a similar distribution in both genotypes. Sequences belonging to subgenotype B-2 are the most prevalent and associated with the highest mortality rate (43.8%) in the ROK [131], whereas genotype D is primarily found in China. Four sequences belonging to genotype D were identical to those found in a human patient in the ROK, suggesting that this genotype is pathogenic. Different genotypes of SFTSV have been shown to trigger different clinical manifestations in a ferret model [130]; however, although the clinical manifestations have not been confirmed in rodents, they may be pathogenic to humans. To date, SFTSV has been detected in diverse animals, but no conclusions can be drawn on how the virus is transmitted to these animals. The results of the present study provide a clue for elucidating the transmission route of SFTSV, suggesting the need to establish a continuous monitoring and surveillance systems to minimize a serious risk of SFTSV infection.
Conclusions
Urbanization and climate change affect not only on humans but also wildlife. The most significant concern caused by these changes is that the probability of disease transmission through ecosystem destruction has been significantly increasing compared with that observed in the past. This study investigated the prevalence of zoonotic pathogens in rodent populations through a systematic epidemiological investigation. Although we did not screen for all rodent-borne pathogens, the results indicated that, at least, rodents may act as critical reservoirs for A. phagocytophilum, Bartonella spp., B. afzelii, C. burnetii, L. interrogans, and SFTSV in the ROK. Our findings also demonstrated that rodents harbor several pathogens, implying the possibility of simultaneous transmission to humans. Most importantly, except for SFTSV, the pathogens investigated in this study are commonly misdiagnosed or underdiagnosed in the ROK; thus, their importance is neglected. Our findings indicate that rodents pose a potential risk to public health. Overall, our study provides useful information on rodent-borne pathogens and underscores the urgent need for rapid diagnosis, prevention, and control strategies for zoonotic diseases.
Supporting information
(DOCX)
Acknowledgments
We are very grateful to Dong-Hun Jang, Eun-Mi Kim, and Seung-Uk Shin for their valuable help in sample collection. All authors read and approved the final version.
Data Availability
All data generated during this study are included in the article and supplementary material. Raw sequencing files were deposited and are available in the repository, WEB LINK for A. phagocytophilum (https://www.ncbi.nlm.nih.gov/nuccore/OR287077.1, OR287091.1), for B. grahamii (https://www.ncbi.nlm.nih.gov/nuccore/OR288176.1, OR288190.1), for B. taylorii (https://www.ncbi.nlm.nih.gov/nuccore/OR288191.1, OR288193.1), for B. afzelii (https://www.ncbi.nlm.nih.gov/nuccore/OR284310.1, OR284311.1), for C. burnetii (https://www.ncbi.nlm.nih.gov/nuccore/OR284312.1, OR284321.1), for L. interrogans (https://www.ncbi.nlm.nih.gov/nuccore/OR284322.1, OR284324.1), and for SFTSV (https://www.ncbi.nlm.nih.gov/nuccore/OR257718.1, OR257726.1).
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Ecke F, Han BA, Hornfeldt B, Khalil H, Magnusson M, Singh NJ, et al. Population fluctuations and synanthropy explain transmission risk in rodent-borne zoonoses. Nat Commun. 2022;13(1):7532. doi: 10.1038/s41467-022-35273-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sumangali K, Rajapakse R, Rajakaruna R. Urban rodents as potential reservoirs of zoonoses: a parasitic survey in two selected areas in Kandy district. Ceylon J Sci. (Bio Sci.) 2012;41(1):71–7. 10.4038/cjsbs.v41i1.4539. [DOI] [Google Scholar]
- 3.Capizzi D, Bertolino S, Mortelliti A. Rating the rat: global patterns and research priorities in impacts and management of rodent pests. Mamm Rev. 2014;44(2):148–62. 10.1111/mam.12019. [DOI] [Google Scholar]
- 4.Dalecky A, Bâ K, Piry S, Lippens C, Diagne CA, Kane M, et al. Range expansion of the invasive house mouse Mus musculus domesticus in Senegal, West Africa: a synthesis of trapping data over three decades, 1983–2014. Mamm Rev. 2015;45(3):176–90. 10.1111/mam.12043. [DOI] [Google Scholar]
- 5.Banda A, Gandiwa E, Muposhi VK, Muboko N. Ecological interactions, local people awareness and practices on rodent-borne diseases in Africa: a review. Acta Trop. 2023;238:106743. doi: 10.1016/j.actatropica.2022.106743 [DOI] [PubMed] [Google Scholar]
- 6.Lu M, Tang G, Ren Z, Zhang J, Wang W, Qin X, et al. Ehrlichia, Coxiella and Bartonella infections in rodents from Guizhou Province, Southwest China. Ticks Tick Borne Dis. 2022;13(5):101974. doi: 10.1016/j.ttbdis.2022.101974 . [DOI] [PubMed] [Google Scholar]
- 7.Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol. 2009;35(3):221–70. doi: 10.1080/10408410902989837 . [DOI] [PubMed] [Google Scholar]
- 8.Morand S, Jittapalapong S, Kosoy M. Rodents as hosts of infectious diseases: biological and ecological characteristics. Vector Borne Zoonotic Dis. 2015;15(1):1–2. doi: 10.1089/vbz.2015.15.1.intro . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selmi R, Belkahia H, Dhibi M, Abdelaali H, Lahmar S, Ben Said M, et al. Zoonotic vector-borne bacteria in wild rodents and associated ectoparasites from Tunisia. Infect Genet Evol. 2021;95:105039. doi: 10.1016/j.meegid.2021.105039 . [DOI] [PubMed] [Google Scholar]
- 10.Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32(3):589–95. doi: 10.1128/jcm.32.3.589-595.1994 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ’HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51(Pt 6):2145–65. doi: 10.1099/00207713-51-6-2145 . [DOI] [PubMed] [Google Scholar]
- 12.Lappin MR, Breitschwerdt EB, Jensen WA, Dunnigan B, Rha JY, Williams CR, et al. Molecular and serologic evidence of Anaplasma phagocytophilum infection in cats in North America. J Am Vet Med Assoc. 2004;225(6):893–6. doi: 10.2460/javma.2004.225.893 . [DOI] [PubMed] [Google Scholar]
- 13.Saleem S, Ijaz M, Farooqi SH, Ghaffar A, Ali A, Iqbal K, et al. Equine granulocytic anaplasmosis 28 years later. Microb Pathog. 2018;119:1–8. doi: 10.1016/j.micpath.2018.04.001 . [DOI] [PubMed] [Google Scholar]
- 14.Dumler JS, Choi KS, Garcia-Garcia JC, Barat NS, Scorpio DG, Garyu JW, et al. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg Infect Dis. 2005;11(12):1828–34. doi: 10.3201/eid1112.050898 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson WL, Muir S, Sumner JW, Childs JE. Serologic evidence of infection with Ehrlichia spp. in wild rodents (Muridae: Sigmodontinae) in the United States. J Clin Microbiol. 1998;36(3):695–700. doi: 10.1128/jcm.36.3.695-700.1998 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alberdi MP, Walker AR, Urquhart KA. Field evidence that roe deer (Capreolus capreolus) are a natural host for Ehrlichia phagocytophila. Epidemiol Infect. 2000;124(2):315–23. doi: 10.1017/s0950268899003684 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bown KJ, Lambin X, Telford GR, Ogden NH, Telfer S, Woldehiwet Z, et al. Relative importance of Ixodes ricinus and Ixodes trianguliceps as vectors for Anaplasma phagocytophilum and Babesia microti in field vole (Microtus agrestis) populations. Appl Environ Microbiol. 2008;74(23):7118–25. doi: 10.1128/AEM.00625-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrovec M, Sixl W, Schweiger R, Mikulasek S, Elke L, Wust G, et al. Infections of wild animals with Anaplasma phagocytophila in Austria and the Czech Republic. Ann N Y Acad Sci. 2003;990:103–6. doi: 10.1111/j.1749-6632.2003.tb07345.x . [DOI] [PubMed] [Google Scholar]
- 19.Scharf W, Schauer S, Freyburger F, Petrovec M, Schaarschmidt-Kiener D, Liebisch G, et al. Distinct host species correlate with Anaplasma phagocytophilum ankA gene clusters. J Clin Microbiol. 2011;49(3):790–6. doi: 10.1128/JCM.02051-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravyn MD, Kodner CB, Carter SE, Jarnefeld JL, Johnson RC. Isolation of the etiologic agent of human granulocytic ehrlichiosis from the white-footed mouse (Peromyscus leucopus). J Clin Microbiol. 2001;39(1):335–8. doi: 10.1128/JCM.39.1.335-338.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telford SR, 3rd, Dawson JE. Persistent infection of C3H/HeJ mice by Ehrlichia chaffeensis. Vet Microbiol. 1996;52(1–2):103–12. doi: 10.1016/0378-1135(96)00064-8 . [DOI] [PubMed] [Google Scholar]
- 22.Chae JS, Yu DH, Shringi S, Klein TA, Kim HC, Chong ST, et al. Microbial pathogens in ticks, rodents and a shrew in northern Gyeonggi-do near the DMZ, Korea. J Vet Sci. 2008;9(3):285–93. doi: 10.4142/jvs.2008.9.3.285 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim CM, Yi YH, Yu DH, Lee MJ, Cho MR, Desai AR, et al. Tick-borne rickettsial pathogens in ticks and small mammals in Korea. Appl Environ Microbiol. 2006;72(9):5766–76. doi: 10.1128/AEM.00431-06 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolkacz K, Alsarraf M, Kowalec M, Dwuznik D, Grzybek M, Behnke JM, et al. Bartonella infections in three species of Microtus: prevalence and genetic diversity, vertical transmission and the effect of concurrent Babesia microti infection on its success. Parasit Vectors. 2018;11(1):491. doi: 10.1186/s13071-018-3047-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37(12):4034–8. doi: 10.1128/jcm.37.12.4034-4038.1999 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosoy M, Murray M, Gilmore RD, Jr., Bai Y, Gage KL. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol. 2003;41(2):645–50. doi: 10.1128/JCM.41.2.645-650.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krugel M, Pfeffer M, Krol N, Imholt C, Baert K, Ulrich RG, et al. Rats as potential reservoirs for neglected zoonotic Bartonella species in flanders, Belgium. Parasit Vectors. 2020;13(1):235. doi: 10.1186/s13071-020-04098-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oksi J, Rantala S, Kilpinen S, Silvennoinen R, Vornanen M, Veikkolainen V, et al. Cat scratch disease caused by Bartonella grahamii in an immunocompromised patient. J Clin Microbiol. 2013;51(8):2781–4. doi: 10.1128/JCM.00910-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okaro U, Addisu A, Casanas B, Anderson B. Bartonella species, an emerging cause of blood-culture-negative endocarditis. Clin Microbiol Rev. 2017;30(3):709–46. doi: 10.1128/CMR.00013-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goncalves LR, Favacho AR, Roque AL, Mendes NS, Fidelis Junior OL, Benevenute JL, et al. Association of Bartonella species with wild and synanthropic rodents in different Brazilian Biomes. Appl Environ Microbiol. 2016;82(24):7154–64. doi: 10.1128/AEM.02447-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutierrez R, Krasnov B, Morick D, Gottlieb Y, Khokhlova IS, Harrus S. Bartonella infection in rodents and their flea ectoparasites: an overview. Vector Borne Zoonotic Dis. 2015;15(1):27–39. doi: 10.1089/vbz.2014.1606 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller A, Gutierrez R, Seguel M, Monti G, Otth C, Bittencourt P, et al. Molecular survey of Bartonella spp. in rodents and fleas from Chile. Acta Trop. 2020;212:105672. doi: 10.1016/j.actatropica.2020.105672 . [DOI] [PubMed] [Google Scholar]
- 33.Buffet JP, Kosoy M, Vayssier-Taussat M. Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Future Microbiol. 2013;8(9):1117–28. 10.2217/fmb.13.77. . [DOI] [PubMed] [Google Scholar]
- 34.Knap N, Duh D, Birtles R, Trilar T, Petrovec M, Avsic-Zupanc T. Molecular detection of Bartonella species infecting rodents in Slovenia. FEMS Immunol Med Microbiol. 2007;50(1):45–50. doi: 10.1111/j.1574-695X.2007.00226.x . [DOI] [PubMed] [Google Scholar]
- 35.Kosoy MY, Regnery RL, Tzianabos T, Marston EL, Jones DC, Green D, et al. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am J Trop Med Hyg. 1997;57(5):578–88. doi: 10.4269/ajtmh.1997.57.578 . [DOI] [PubMed] [Google Scholar]
- 36.Pretorius AM, Beati L, Birtles RJ. Diversity of bartonellae associated with small mammals inhabiting free state province, South Africa. Int J Syst Evol Microbiol. 2004;54(Pt 6):1959–67. doi: 10.1099/ijs.0.03033-0 . [DOI] [PubMed] [Google Scholar]
- 37.Polat C, Celebi B, Irmak S, Karatas A, Colak F, Matur F, et al. Characterization of Bartonella taylorii strains in small mammals of the Turkish Thrace. Ecohealth. 2020;17(4):477–86. doi: 10.1007/s10393-021-01518-y . [DOI] [PubMed] [Google Scholar]
- 38.Qin XR, Liu JW, Yu H, Yu XJ. Bartonella species detected in rodents from eastern China. Vector Borne Zoonotic Dis. 2019;19(11):810–4. doi: 10.1089/vbz.2018.2410 . [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Pastor R, Escudero R, Lambin X, Vidal MD, Gil H, Jado I, et al. Zoonotic pathogens in fluctuating common vole (Microtus arvalis) populations: occurrence and dynamics. Parasitology. 2019;146(3):389–98. doi: 10.1017/S0031182018001543 . [DOI] [PubMed] [Google Scholar]
- 40.Rothenburger JL, Himsworth CG, Nemeth NM, Pearl DL, Jardine CM. Beyond abundance: how microenvironmental features and weather influence Bartonella tribocorum infection in wild Norway rats (Rattus norvegicus). Zoonoses Public Health. 2018;65(3):339–51. doi: 10.1111/zph.12440 . [DOI] [PubMed] [Google Scholar]
- 41.Silaghi C, Pfeffer M, Kiefer D, Kiefer M, Obiegala A. Bartonella, rodents, fleas and ticks: a molecular field study on host-vector-pathogen associations in Saxony, Eastern Germany. Microb Ecol. 2016;72(4):965–74. doi: 10.1007/s00248-016-0787-8 . [DOI] [PubMed] [Google Scholar]
- 42.Kim CM, Kim JY, Yi YH, Lee MJ, Cho MR, Shah DH, et al. Detection of Bartonella species from ticks, mites and small mammals in Korea. J Vet Sci. 2005;6(4):327–34. . [PubMed] [Google Scholar]
- 43.Ko S, Kang JG, Kim HC, Klein TA, Choi KS, Song JW, et al. Prevalence, isolation and molecular characterization of Bartonella species in Republic of Korea. Transbound Emerg Dis. 2016;63(1):56–67. doi: 10.1111/tbed.12217 . [DOI] [PubMed] [Google Scholar]
- 44.Zhong X, Nouri M, Raberg L. Colonization and pathology of Borrelia afzelii in its natural hosts. Ticks Tick Borne Dis. 2019;10(4):822–7. doi: 10.1016/j.ttbdis.2019.03.017 . [DOI] [PubMed] [Google Scholar]
- 45.Kurtenbach K, Hanincova K, Tsao JI, Margos G, Fish D, Ogden NH. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat Rev Microbiol. 2006;4(9):660–9. doi: 10.1038/nrmicro1475 . [DOI] [PubMed] [Google Scholar]
- 46.Stanek G, Reiter M. The expanding Lyme Borrelia complex-clinical significance of genomic species? Clin Microbiol Infect. 2011;17(4):487–93. doi: 10.1111/j.1469-0691.2011.03492.x . [DOI] [PubMed] [Google Scholar]
- 47.Rollins RE, Yeyin Z, Wyczanska M, Alig N, Hepner S, Fingerle V, et al. Spatial variability in prevalence and genospecies distributions of Borrelia burgdorferi sensu lato from ixodid ticks collected in southern Germany. Ticks Tick Borne Dis. 2021;12(1):101589. doi: 10.1016/j.ttbdis.2020.101589 . [DOI] [PubMed] [Google Scholar]
- 48.Balmelli T, Piffaretti JC. Association between different clinical manifestations of Lyme disease and different species of Borrelia burgdorferi sensu lato. Res Microbiol. 1995;146(4):329–40. doi: 10.1016/0923-2508(96)81056-4 . [DOI] [PubMed] [Google Scholar]
- 49.Wang G, van Dam AP, Schwartz I, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin Microbiol Rev. 1999;12(4):633–53. doi: 10.1128/cmr.12.4.633 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheffold N, Herkommer B, Kandolf R, May AE. Lyme carditis-diagnosis, treatment and prognosis. Dtsch Arztebl Int. 2015;112(12):202–8. doi: 10.3238/arztebl.2015.0202 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kee S, Hwang KJ, Oh HB, Park KS. Identification of Borrelia burgdorferi isolated in Korea using outer surface protein A (OspA) serotyping system. Microbiol Immunol. 1994;38(12):989–93. doi: 10.1111/j.1348-0421.1994.tb02157.x . [DOI] [PubMed] [Google Scholar]
- 52.Bell DR, Berghaus RD, Patel S, Beavers S, Fernandez I, Sanchez S. Seroprevalence of tick-borne infections in military working dogs in the Republic of Korea. Vector Borne Zoonotic Dis. 2012;12(12):1023–30. doi: 10.1089/vbz.2011.0864 . [DOI] [PubMed] [Google Scholar]
- 53.Lee SH, Yun SH, Choi E, Park YS, Lee SE, Cho GJ, et al. Serological detection of Borrelia burgdorferi among horses in Korea. Korean J Parasitol. 2016;54(1):97–101. doi: 10.3347/kjp.2016.54.1.97 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.VanBik D, Lee SH, Seo MG, Jeon BR, Goo YK, Park SJ, et al. Borrelia species detected in ticks feeding on wild Korean Water Deer (Hydropotes inermis) using molecular and genotypic analyses. J Med Entomol. 2017;54(5):1397–402. doi: 10.1093/jme/tjx106 . [DOI] [PubMed] [Google Scholar]
- 55.Moon S, Hong Y, Hwang KJ, Kim S, Eom J, Kwon D, et al. Epidemiological features and clinical manifestations of Lyme borreliosis in Korea during the period 2005–2012. Jpn J Infect Dis. 2015;68(1):1–4. doi: 10.7883/yoken.JJID.2013.462 . [DOI] [PubMed] [Google Scholar]
- 56.Kim CM, Park SY, Kim DM, Park JW, Chung JK. First report of Borrelia burgdorferi sensu stricto detection in a commune genospecies in Apodemus agrarius in Gwangju, South Korea. Sci Rep. 2021;11(1):18199. doi: 10.1038/s41598-021-97411-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho HC, Hwang S, Kim EM, Park YJ, Shin SU, Jang DH, et al. Prevalence and molecular characterization of Coxiella burnetii in cattle, goats, and horses in the Republic of Korea. Vector Borne Zoonotic Dis. 2021;21(7):502–8. doi: 10.1089/vbz.2020.2764 . [DOI] [PubMed] [Google Scholar]
- 58.Abdel-Moein KA, Hamza DA. Rat as an overlooked reservoir for Coxiella burnetii: a public health implication. Comp Immunol Microbiol Infect Dis. 2018;61:30–3. doi: 10.1016/j.cimid.2018.11.002 . [DOI] [PubMed] [Google Scholar]
- 59.Chochlakis D, Santos AS, Giadinis ND, Papadopoulos D, Boubaris L, Kalaitzakis E, et al. Genotyping of Coxiella burnetii in sheep and goat abortion samples. BMC Microbiol. 2018;18(1):204. doi: 10.1186/s12866-018-1353-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. Q fever: a biological weapon in your backyard. Lancet Infect Dis. 2003;3(11):709–21. doi: 10.1016/s1473-3099(03)00804-1 . [DOI] [PubMed] [Google Scholar]
- 61.Pascucci I, Di Domenico M, Dall’Acqua F, Sozio G, Camma C. Detection of lyme disease and Q fever agents in wild rodents in central Italy. Vector Borne Zoonotic Dis. 2015;15(7):404–11. doi: 10.1089/vbz.2015.1807 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang S, Cho HC, Shin SU, Kim HY, Park YJ, Jang DH, et al. Seroprevalence and molecular characterization of Coxiella burnetii in cattle in the Republic of Korea. Pathogens. 2020;9(11):890. doi: 10.3390/pathogens9110890 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esteves LM, Bulhoes SM, Branco CC, Carreira T, Vieira ML, Gomes-Solecki M, et al. Diagnosis of human leptospirosis in a clinical setting: real-time PCR high resolution melting analysis for detection of Leptospira at the onset of disease. Sci Rep. 2018;8(1):9213. doi: 10.1038/s41598-018-27555-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karpagam KB, Ganesh B. Leptospirosis: a neglected tropical zoonotic infection of public health importance-an updated review. Eur J Clin Microbiol Infect Dis. 2020;39(5):835–46. doi: 10.1007/s10096-019-03797-4 . [DOI] [PubMed] [Google Scholar]
- 65.Lopez-Osorio S, Molano DA, Lopez-Arias A, Rodriguez-Osorio N, Zambrano C, Chaparro-Gutierrez JJ. Seroprevalence and molecular characterization of Leptospira spp. in rats captured near pig farms in Colombia. Int J Environ Res Public Health. 2022;19(18):11539. doi: 10.3390/ijerph191811539 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3(12):757–71. doi: 10.1016/s1473-3099(03)00830-2 . [DOI] [PubMed] [Google Scholar]
- 67.Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009;7(10):736–47. doi: 10.1038/nrmicro2208 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ellis WA. Animal leptospirosis. Curr Top Microbiol Immunol. 2015;387:99–137. doi: 10.1007/978-3-662-45059-8_6 [DOI] [PubMed] [Google Scholar]
- 69.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14(2):296–326. doi: 10.1128/CMR.14.2.296-326.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lau CL, Smythe LD, Craig SB, Weinstein P. Climate change, flooding, urbanisation and leptospirosis: fuelling the fire? Trans R Soc Trop Med Hyg. 2010;104(10):631–8. doi: 10.1016/j.trstmh.2010.07.002 . [DOI] [PubMed] [Google Scholar]
- 71.Schonning MH, Phelps MD, Warnasekara J, Agampodi SB, Furu P. A case-control study of environmental and occupational risks of leptospirosis in Sri Lanka. Ecohealth. 2019;16(3):534–43. doi: 10.1007/s10393-019-01448-w . [DOI] [PubMed] [Google Scholar]
- 72.Boey K, Shiokawa K, Rajeev S. Leptospira infection in rats: a literature review of global prevalence and distribution. PLoS Negl Trop Dis. 2019;13(8):e0007499. doi: 10.1371/journal.pntd.0007499 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1(1):e56. doi: 10.1371/journal.pntd.0000056 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364(16):1523–32. doi: 10.1056/NEJMoa1010095 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, et al. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg Infect Dis. 2013;19(11):1892–4. doi: 10.3201/eid1911.130792 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, et al. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis. 2014;209(6):816–27. doi: 10.1093/infdis/jit603 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tran XC, Yun Y, Van An L, Kim SH, Thao NTP, Man PKC, et al. Endemic severe fever with thrombocytopenia syndrome, Vietnam. Emerg Infect Dis. 2019;25(5):1029–31. doi: 10.3201/eid2505.181463 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin TL, Ou SC, Maeda K, Shimoda H, Chan JP, Tu WC, et al. The first discovery of severe fever with thrombocytopenia syndrome virus in Taiwan. Emerg Microbes Infect. 2020;9(1):148–51. doi: 10.1080/22221751.2019.1710436 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu JW, Zhao L, Luo LM, Liu MM, Sun Y, Su X, et al. Molecular evolution and spatial transmission of severe fever with thrombocytopenia syndrome virus based on complete genome sequences. PLoS One. 2016;11(3):e0151677. doi: 10.1371/journal.pone.0151677 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bao CJ, Guo XL, Qi X, Hu JL, Zhou MH, Varma JK, et al. A family cluster of infections by a newly recognized bunyavirus in eastern China, 2007: further evidence of person-to-person transmission. Clin Infect Dis. 2011;53(12):1208–14. doi: 10.1093/cid/cir732 . [DOI] [PubMed] [Google Scholar]
- 81.Mehand MS, Millett P, Al-Shorbaji F, Roth C, Kieny MP, Murgue B. World health organization methodology to prioritize emerging infectious diseases in need of research and development. Emerg Infect Dis. 2018;24(9):e171427. doi: 10.3201/eid2409.171427 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng J, Zhang L, Hu B, Wang Q, Wu R, Zhan F, et al. Prevalence and molecular phylogenetic analysis of severe fever with thrombocytopenia syndrome virus in domestic animals and rodents in Hubei Province, China. Virol Sin. 2019;34(5):596–600. doi: 10.1007/s12250-019-00119-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi SJ, Park SW, Bae IG, Kim SH, Ryu SY, Kim HA, et al. Severe fever with thrombocytopenia syndrome in South Korea, 2013–2015. PLoS Negl Trop Dis. 2016;10(12):e0005264. doi: 10.1371/journal.pntd.0005264 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kobayashi Y, Kato H, Yamagishi T, Shimada T, Matsui T, Yoshikawa T, et al. Severe fever with thrombocytopenia syndrome, Japan, 2013–2017. Emerg Infect Dis. 2020;26(4):692–9. doi: 10.3201/eid2604.191011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Homan WL, Vercammen M, De Braekeleer J, Verschueren H. Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int J Parasitol. 2000;30(1):69–75. doi: 10.1016/s0020-7519(99)00170-8 . [DOI] [PubMed] [Google Scholar]
- 86.Dumler JS. The biological basis of severe outcomes in Anaplasma phagocytophilum infection. FEMS Immunol Med Microbiol. 2012;64(1):13–20. doi: 10.1111/j.1574-695X.2011.00909.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum—a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol. 2013;3:31. doi: 10.3389/fcimb.2013.00031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bang MS, Kim CM, Park JW, Chung JK, Kim DM, Yun NR. Prevalence of Orientia tsutsugamushi, Anaplasma phagocytophilum and Leptospira interrogans in striped field mice in Gwangju, Republic of Korea. PLoS One. 2019;14(8):e0215526. 10.1371/journal.pone.0215526. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zheng W, Liu Y, Tao H, Li Z, Xuan X, Liu X, et al. First Molecular evidence of Anaplasma phagocytophilum in rodent populations of Nanchang, China. Jpn J Infect Dis. 2018;71(2):129–33. doi: 10.7883/yoken.JJID.2017.301 . [DOI] [PubMed] [Google Scholar]
- 90.Kim HC, Han SH, Chong ST, Klein TA, Choi CY, Nam HY, et al. Ticks collected from selected mammalian hosts surveyed in the Republic of Korea during 2008–2009. Korean J Parasitol. 2011;49(3):331–5. doi: 10.3347/kjp.2011.49.3.331 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lipatova I, Paulauskas A, Puraite I, Radzijevskaja J, Balciauskas L, Gedminas V. Bartonella infection in small mammals and their ectoparasites in Lithuania. Microbes Infect. 2015;17(11–12):884–8. doi: 10.1016/j.micinf.2015.08.013 . [DOI] [PubMed] [Google Scholar]
- 92.Pangjai D, Maruyama S, Boonmar S, Kabeya H, Sato S, Nimsuphan B, et al. Prevalence of zoonotic Bartonella species among rodents and shrews in Thailand. Comp Immunol Microbiol Infect Dis. 2014;37(2):109–14. doi: 10.1016/j.cimid.2013.12.001 . [DOI] [PubMed] [Google Scholar]
- 93.Malania L, Bai Y, Osikowicz LM, Tsertsvadze N, Katsitadze G, Imnadze P, et al. Prevalence and diversity of Bartonella species in rodents from Georgia (Caucasus). Am J Trop Med Hyg. 2016;95(2):466–71. doi: 10.4269/ajtmh.16-0041 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rozental T, Ferreira MS, Guterres A, Mares-Guia MA, Teixeira BR, Goncalves J, et al. Zoonotic pathogens in Atlantic Forest wild rodents in Brazil: Bartonella and Coxiella infections. Acta Trop. 2017;168:64–73. doi: 10.1016/j.actatropica.2017.01.003 . [DOI] [PubMed] [Google Scholar]
- 95.Bai Y, Kosoy MY, Lerdthusnee K, Peruski LF, Richardson JH. Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am J Trop Med Hyg. 2009;81(5):811–6. doi: 10.4269/ajtmh.2009.09-0294 . [DOI] [PubMed] [Google Scholar]
- 96.Gundi VA, Billeter SA, Rood MP, Kosoy MY. Bartonella spp. in rats and zoonoses, Los Angeles, California, USA. Emerg Infect Dis. 2012;18(4):631–3. doi: 10.3201/eid1804.110816 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Su Q, Chen Y, Wang B, Huang C, Han S, Yuan G, et al. Epidemiology and genetic diversity of zoonotic pathogens in urban rats (Rattus spp.) from a subtropical city, Guangzhou, southern China. Zoonoses Public Health. 2020;67(5):534–45. doi: 10.1111/zph.12717 . [DOI] [PubMed] [Google Scholar]
- 98.Kim SY, Kim TK, Kim TY, Lee HI. Geographical distribution of Borrelia burgdorferi sensu lato in ticks collected from wild rodents in the Republic of Korea. Pathogens. 2020;9(11):866. doi: 10.3390/pathogens9110866 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khanakah G, Kocianova E, Vyrostekova V, Rehacek J, Kundi M, Stanek G. Seasonal variations in detecting Borrelia burgdorferi sensu lato in rodents from north eastern Austria. Wien Klin Wochenschr. 2006;118(23–24):754–8. doi: 10.1007/s00508-006-0730-y . [DOI] [PubMed] [Google Scholar]
- 100.Kybicova K, Kurzova Z, Hulinska D. Molecular and serological evidence of Borrelia burgdorferi sensu lato in wild rodents in the Czech Republic. Vector Borne Zoonotic Dis. 2008;8(5):645–52. doi: 10.1089/vbz.2007.0249 . [DOI] [PubMed] [Google Scholar]
- 101.Barandika JF, Hurtado A, Garcia-Esteban C, Gil H, Escudero R, Barral M, et al. Tick-borne zoonotic bacteria in wild and domestic small mammals in northern Spain. Appl Environ Microbiol. 2007;73(19):6166–171. doi: 10.1128/AEM.00590-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moon S, Gwack J, Hwang KJ, Kwon D, Kim S, Noh Y, et al. Autochthonous lyme borreliosis in humans and ticks in Korea. Osong Public Health and Research Perspectives. 2013;4(1):52–6. doi: 10.1016/j.phrp.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hanincova K, Schafer SM, Etti S, Sewell HS, Taragelova V, Ziak D, et al. Association of Borrelia afzelii with rodents in Europe. Parasitology. 2003;126(Pt 1):11–20. doi: 10.1017/s0031182002002548 . [DOI] [PubMed] [Google Scholar]
- 104.Seo MG, Kwon OD, Kwak D. Molecular Identification of Borrelia afzelii from ticks parasitizing domestic and wild animals in South Korea. Microorganisms. 2020;8(5):649. doi: 10.3390/microorganisms8050649 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi YJ, Han SH, Park JM, Lee KM, Lee EM, Lee SH, et al. First molecular detection of Borrelia afzelii in clinical samples in Korea. Microbiol Immunol. 2007;51(12):1201–7. doi: 10.1111/j.1348-0421.2007.tb04015.x . [DOI] [PubMed] [Google Scholar]
- 106.Liu L, Baoliang X, Yingqun F, Ming L, Yu Y, Yong H, et al. Coxiella burnetii in rodents on Heixiazi Island at the Sino-Russian border. Am J Trop Med Hyg. 2013;88(4):770–3. doi: 10.4269/ajtmh.12-0580 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mangombi-Pambou J, Granjon L, Labarrere C, Kane M, Niang Y, Fournier PE, et al. New genotype of Coxiella burnetii causing epizootic Q fever outbreak in rodents, northern Senegal. Emerg Infect Dis. 2023;29(5):1078–81. doi: 10.3201/eid2905.221034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chitanga S, Simulundu E, Simuunza MC, Changula K, Qiu Y, Kajihara M, et al. First molecular detection and genetic characterization of Coxiella burnetii in Zambian dogs and rodents. Parasit Vectors. 2018;11(1):40. doi: 10.1186/s13071-018-2629-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoo JR, Kim MS, Heo ST, Oh HJ, Oh JH, Ko SY, et al. Seroreactivity to Coxiella burnetii in an agricultural population and prevalence of Coxiella burnetii infection in ticks of a non-endemic region for Q fever in South Korea. Pathogens. 2021;10(10):1337. doi: 10.3390/pathogens10101337 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Truong AT, Yun BR, Lim J, Min S, Yoo MS, Yoon SS, et al. Real-time PCR biochip for on-site detection of Coxiella burnetii in ticks. Parasit Vectors. 2021;14(1):239. doi: 10.1186/s13071-021-04744-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoo JR, Heo ST, Kim M, Kim M, Kang MJ, Kim ET, et al. Coinfection of severe fever with thrombocytopenia syndrome virus and Coxiella burnetii in developmental stage of hard ticks in subtropical region of Korea. J Korean Med Sci. 2023;38(20):e156. doi: 10.3346/jkms.2023.38.e156 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim JH, Choi YJ, Lee KS, Kim JE, Oh JW, Moon JH. Severe fever with thrombocytopenia syndrome with Q fever coinfection in an 8-year-old girl. Pediatr Infect Dis J. 2021;40(1):e31–4. doi: 10.1097/INF.0000000000002948 . [DOI] [PubMed] [Google Scholar]
- 113.Dobigny G, Garba M, Tatard C, Loiseau A, Galan M, Kadaoure I, et al. Urban market gardening and rodent-borne pathogenic Leptospira in arid zones: a case study in niamey, Niger. PLoS Negl Trop Dis. 2015;9(10):e0004097. doi: 10.1371/journal.pntd.0004097 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Samir A, Soliman R, El-Hariri M, Abdel-Moein K, Hatem ME. Leptospirosis in animals and human contacts in Egypt: broad range surveillance. Rev Soc Bras Med Trop. 2015;48(3):272–7. doi: 10.1590/0037-8682-0102-2015 . [DOI] [PubMed] [Google Scholar]
- 115.Halliday JEB, Knobel DL, Allan KJ, de CBBM, Handel I, Agwanda B, et al. Urban leptospirosis in Africa: a cross-sectional survey of Leptospira infection in rodents in the Kibera urban settlement, Nairobi, Kenya. Am J Trop Med Hyg. 2013;89(6):1095–102. doi: 10.4269/ajtmh.13-0415 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Izquierdo-Rodriguez E, Fernandez-Alvarez A, Martin-Carrillo N, Marchand B, Feliu C, Miquel J, et al. Pathogenic Leptospira species in rodents from Corsica (France). PLoS One. 2020;15(6):e0233776. doi: 10.1371/journal.pone.0233776 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vitale M, Agnello S, Chetta M, Amato B, Vitale G, Bella CD, et al. Human leptospirosis cases in Palermo Italy. The role of rodents and climate. J Infect Public Health. 2018;11(2):209–14. doi: 10.1016/j.jiph.2017.07.024 . [DOI] [PubMed] [Google Scholar]
- 118.Cho MK, Kee SH, Song HJ, Kim KH, Song KJ, Baek LJ, et al. Infection rate of Leptospira interrogans in the field rodent, Apodemus agrarius, in Korea. Epidemiol Infect. 1998;121(3):685–90. doi: 10.1017/s0950268898001691 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gu XL, Su WQ, Zhou CM, Fang LZ, Zhu K, Ma DQ, et al. SFTSV infection in rodents and their ectoparasitic chiggers. PLoS Negl Trop Dis. 2022;16(8):e0010698. doi: 10.1371/journal.pntd.0010698 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rim JM, Han SW, Cho YK, Kang JG, Choi KS, Jeong H, et al. Survey of severe fever with thrombocytopenia syndrome virus in wild boar in the Republic of Korea. Ticks Tick Borne Dis. 2021;12(6):101813. doi: 10.1016/j.ttbdis.2021.101813 . [DOI] [PubMed] [Google Scholar]
- 121.Kang JG, Cho YK, Jo YS, Han SW, Chae JB, Park JE, et al. Severe fever with thrombocytopenia syndrome virus in ticks in the Republic of Korea. Korean J Parasitol. 2022;60(1):65–71. doi: 10.3347/kjp.2022.60.1.65 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kang JG, Cho YK, Han SW, Jeon K, Choi H, Kim JH, et al. Molecular and serological investigation of severe fever with thrombocytopenia syndrome virus in cats. Vector Borne Zoonotic Dis. 2020;20(12):916–20. doi: 10.1089/vbz.2020.2649 . [DOI] [PubMed] [Google Scholar]
- 123.Kang JG, Cho YK, Jo YS, Chae JB, Joo YH, Park KW, et al. Severe fever with thrombocytopenia syndrome virus in dogs, South Korea. Emerg Infect Dis. 2019;25(2):376–8. doi: 10.3201/eid2502.180859 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kang JG, Oh SS, Jo YS, Chae JB, Cho YK, Chae JS. Molecular detection of severe fever with thrombocytopenia syndrome virus in Korean domesticated pigs. Vector Borne Zoonotic Dis. 2018;18(8):450–2. doi: 10.1089/vbz.2018.2310 . [DOI] [PubMed] [Google Scholar]
- 125.Kang JG, Cho YK, Jo YS, Chae JB, Oh SS, Kim KH, et al. Prevalence of severe fever with thrombocytopenia syndrome virus in black goats (Capra hircus coreanae) in the Republic of Korea. Ticks Tick Borne Dis. 2018;9(5):1153–7. doi: 10.1016/j.ttbdis.2018.04.018 . [DOI] [PubMed] [Google Scholar]
- 126.Oh SS, Chae JB, Kang JG, Kim HC, Chong ST, Shin JH, et al. Detection of severe fever with thrombocytopenia syndrome virus from wild animals and Ixodidae ticks in the Republic of Korea. Vector Borne Zoonotic Dis. 2016;16(6):408–14. doi: 10.1089/vbz.2015.1848 . [DOI] [PubMed] [Google Scholar]
- 127.Hwang J, Kang JG, Oh SS, Chae JB, Cho YK, Cho YS, et al. Molecular detection of severe fever with thrombocytopenia syndrome virus (SFTSV) in feral cats from Seoul, Korea. Ticks Tick Borne Dis. 2017;8(1):9–12. doi: 10.1016/j.ttbdis.2016.08.005 . [DOI] [PubMed] [Google Scholar]
- 128.Tsuru M, Suzuki T, Murakami T, Matsui K, Maeda Y, Yoshikawa T, et al. Pathological characteristics of a patient with severe fever with thrombocytopenia syndrome (SFTS) infected with SFTS Virus through a sick cat’s bite. Viruses. 2021;13(2):204. doi: 10.3390/v13020204 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kida K, Matsuoka Y, Shimoda T, Matsuoka H, Yamada H, Saito T, et al. A case of cat-to-human transmission of severe fever with thrombocytopenia syndrome virus. Jpn J Infect Dis. 2019;72(5):356–8. doi: 10.7883/yoken.JJID.2018.526 . [DOI] [PubMed] [Google Scholar]
- 130.Seo JW, Kim D, Yun N, Kim DM. Clinical update of severe fever with thrombocytopenia syndrome. Viruses. 2021;13(7):1213. doi: 10.3390/v13071213 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yun SM, Park SJ, Kim YI, Park SW, Yu MA, Kwon HI, et al. Genetic and pathogenic diversity of severe fever with thrombocytopenia syndrome virus (SFTSV) in South Korea. JCI Insight. 2020;5(2):e129531. doi: 10.1172/jci.insight.129531 . [DOI] [PMC free article] [PubMed] [Google Scholar]