Abstract
How populations adapt to their environment is a fundamental question in biology. Yet we know surprisingly little about this process, especially for endangered species such as non-human great apes. Chimpanzees, our closest living relatives, are particularly interesting because they inhabit diverse habitats, from rainforest to woodland-savannah. Whether genetic adaptation facilitates such habitat diversity remains unknown, despite having wide implications for evolutionary biology and conservation. Using 828 newly generated exomes from wild chimpanzees, we find evidence of fine-scale genetic adaptation to habitat. Notably, adaptation to malaria in forest chimpanzees is mediated by the same genes underlying adaptation to malaria in humans. This work demonstrates the power of non-invasive samples to reveal genetic adaptations in endangered populations and highlights the importance of adaptive genetic diversity for chimpanzees.
One-Sentence Summary:
Chimpanzees show evidence of local genetic adaptation to habitat, particularly to pathogens, such as malaria, in forests.
Understanding how primates are adapted to their environments provides insights into our own evolution and vital information for conservation efforts. This is particularly relevant and urgent for our closest living relatives, non-human great apes, all of which are either endangered or critically endangered. Chimpanzees (Pan troglodytes) have the largest geographic and ecological range of any non-human ape (2.6 million km² (1)) spanning a variety of environments across Equatorial Africa, from dense tropical rainforest to open woodland-savannah mosaics (hereafter ‘savannah’ for simplicity (2)). Aside from humans, they are the only great apes that inhabit savannah habitats (2). Yet, each of the four subspecies of chimpanzee (central (P. t. troglodytes), eastern (P. t. schweinfurthii), Nigeria-Cameroon (P. t. ellioti) and western (P. t. verus) (3–5)) are endangered (westerns critically so) with numbers continuing to decline due to hunting, habitat destruction and infectious diseases (1, 6–8). This decline has widespread negative impacts, as chimpanzees are important conservation flagship species for biodiversity protection and crucial ecosystem engineers (9–11).
Between the forest and savannah extremes, chimpanzees occupy a gradient of habitats known as forest-savannah mosaics (12). Forests, which are likely closest to chimpanzee ancestral habitats (3, 13), have closed canopies with high availability of food and water throughout the year, and therefore tend to support high population densities (2). Forests also harbour a great diversity of pathogens and disease vectors (14). Conversely, savannahs are on the edge of chimpanzee distribution in East and West Africa and are characterised by open canopies, higher temperatures, lower annual rainfall and higher rainfall seasonality (2, 15).
The occupation of such a range of habitats is facilitated by chimpanzee behavioural diversity (16). Savannah chimpanzees exhibit unique thermoregulatory behaviours (17, 18) and on average trend towards greater behavioural diversity than forest chimpanzees (16)–a potential adaptation to higher environmental variability. Behavioural adaptations also include tool use in a range of contexts such as foraging (19–21), water extraction (22–24) and communication (25). Nevertheless, behaviour does not fully compensate for stressors, as shown by physiological stress in response to pathogens (26–29) and environmental pressures (15, 30). Another mechanism that can facilitate the occupation of diverse habitats is genetic adaptation, just as local adaptation has contributed to genetic population differentiation in humans (31) despite great behavioural flexibility (32, 33). In fact, humans have evolved local genetic adaptations to environmental pressures that differ between forest and savannah habitats, including pathogens (34–36) such as malaria (37, 38), and climatic variables such as temperature and water availability (39, 40), diet (41–43), and solar exposure (44). Culture can also promote genetic adaptations, such as in human adaptations to diet and zoonotic diseases associated with animal domestication (43, 45).
Establishing if genetic differences underlie local adaptation in chimpanzees is important to understanding primate evolution and critical for chimpanzee conservation. If adaptive genetic differences exist among populations, this genetic diversity must be conserved to maintain existing adaptations and adaptive potential (46, 47). Additionally, recent genetic adaptations highlight key selective pressures that likely shape fitness in the wild today, and can help establish which populations may be more vulnerable to environmental change (46). This is particularly relevant in the face of anthropogenic climate change, which is increasing temperatures and precipitation seasonality within the chimpanzees’ range (2). Furthermore, chimpanzees are excellent models for understanding our own evolution, particularly in savannah regions, which resemble early hominin habitats (2, 48–52). Lastly, the close genetic similarity between humans and other great apes (53) has resulted in zoonotic disease transmissions (54, 55) such as HIV-1/AIDS (56) and malaria (57). Understanding how chimpanzees have evolved to reduce the pathogenicity of microorganisms can thus reveal potential targets for treatments and vaccines (58–61).
We have a growing understanding of chimpanzee demographic history thanks to population genomics studies (3, 5, 62), which have identified genetic differentiation among populations within each subspecies. However, our knowledge of genetic adaptation lags behind, largely due to sample limitations. Because existing genomic datasets include only dozens of captive chimpanzees of unknown geographic origin (5, 62–64), previous studies investigated adaptation only at the subspecies level (63, 65–73), revealing interesting subspecies-level adaptations e.g., to pathogens such as SIV (65, 66). However, habitats vary greatly within chimpanzee subspecies ranges (12); therefore, subspecies comparisons are uninformative on adaptations to many potential selective pressures. Investigation of fine-scale local adaptation is essential for understanding adaptation in chimpanzees but requires large numbers of DNA samples from wild individuals of known geographic origin, coupled with detailed environmental data.
Non-invasive sampling is the only ethical and feasible option for studying wild populations of many protected species (74), including non-human apes; however, recent technical and analytical advancements are beginning to enable population genomic analyses on such samples (3, 75–80). Using faecal samples from wild individuals collected as part of the Pan African Programme: The Cultured Chimpanzee (PanAf) (3, 81), we have generated full exome (protein-coding regions of the genome) sequences from hundreds of chimpanzees across their geographic and environmental range. We demonstrate that genomic data from non-invasive samples can be used to reveal the fine-scale adaptive history of endangered primates. Specifically, when integrated with environmental data, the exomes reveal evidence of local genetic adaptation to habitat conditions in chimpanzees. In forests, we find evidence of pathogen-mediated adaptation, including to malaria via the same loci that mediate malaria adaptation in humans.
Samples and sequences
Faecal samples of 828 unique individuals were collected from 52 sampling sites across the geographic range of all four chimpanzee subspecies as part of PanAf (3, 81). This represents a ten-fold increase in sample size and a massive increase in geographic coverage over existing genome-wide datasets (5, 62) of any non-human ape. The scale and resolution of the dataset are only comparable to the chromosome 21 (chr21) sequences of the same individuals (3).
Non-invasive samples typically contain low levels of endogenous DNA. Thus, we target-captured and sequenced full exomes (akin to chr21 (3)) because they are informative for the vast majority of functional sites in the genome, including both sequenced protein-coding and linked regulatory regions (e.g., promoters). Samples were strictly filtered to omit those with first-order relatives, contamination or low read depth (Supplemental Note 3). To mitigate the potential effects of the moderate read depth and take advantage of the large sample size, we used genotype likelihoods and allele frequency-based methods, which minimised the effects of individual sequencing errors.
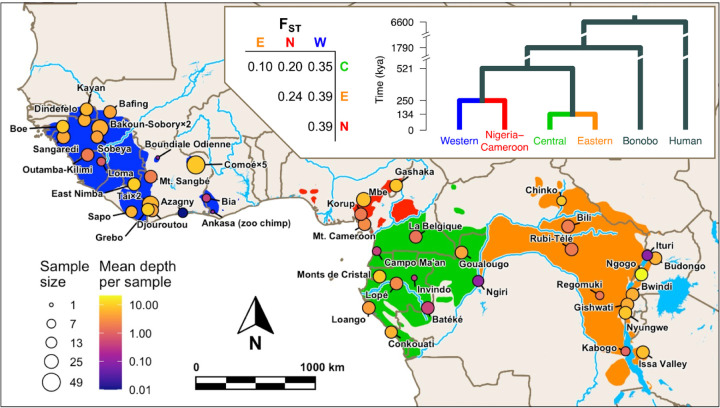
As expected, using either exomes or chr21 (3), population structure analyses separate samples into four subspecies (Figs. S12, S14 and S15), and within-subspecies population structure inferred with the exomes closely matches results from chr21 (3) (Figs. S12–16). Each sample site was considered a genetic unit, which we refer to as a ‘population’, except for four populations formed by combining very closely related sample sites (details in Materials and Methods and Supplemental Note 4). After removing populations with fewer than eight samples, the final dataset contains 388 exomes (385 chr21) from 30 populations: 5 central, 9 eastern, 2 Nigeria-Cameroon and 14 western. The resulting exomes have a median read depth per sample of 5.30-fold (0.51- to 52.27-fold) in the exome target space (60Mbp). The signatures of local adaptation within and across subspecies were investigated in four ‘subspecies-datasets’ containing populations from all subspecies (All), central and eastern together (Central-Eastern), Nigeria-Cameroon (Nigeria-Cameroon) and western (Western). Central and eastern were combined because of their low genetic differentiation (FST=0.10, Fig. 1) (65). The unfolded site frequency spectra (SFS) conform to expectations given the inferred demographic history of these subspecies (Fig. S18).
Fig. 1. Chimpanzee exome dataset distribution, sample size and coverage.
Top panel: Hominini phylogenetic tree highlighting chimpanzee subspecies with estimated evolutionary split times (62, 82) (thousands of years ago (kya)) and pairwise FST between subspecies (65). Main panel: map of West, Central and East Africa indicating the location of sample sites, sample sizes and mean exome sequencing read depth per sample. Each point represents a sample site except for five geographically close sites sampled at Comoé, two at Taï and two at Bakoun-Sobory. The geographic distribution of each subspecies is shown (green=central, orange=eastern, red=Nigeria-Cameroon, blue=western) (1) with major rivers and lakes indicated in light blue. The sample sizes and populations in the final filtered dataset used for selection analyses are shown in Fig. 3.
Allele frequency population differentiation
Local adaptation increases the frequency of alleles only where they are beneficial, generating large allele frequency differences among populations. We first investigated local positive selection by analysing population differentiation with a genetics-only hypothesis-free analysis using the BayPass (83) core model. BayPass estimates the genome-wide population allele frequency covariance matrix, which is used to standardise allele frequencies for each single-nucleotide polymorphism (SNP) with respect to population structure (BayPass effectively accounts for population structure here, see Supplemental Note 4); the variance across populations of these standardised frequencies is summarised in the test statistic XtX* (84). SNPs under local adaptation are expected to have exceptionally large population allele frequency differentiation, and therefore the highest XtX* values in the genome. Null expectations under neutrality were generated using the non-genic regions of chr21 (non-genic-chr21) in these samples (3). Generating an empirical null frees the analysis from demographic assumptions and accounts for many potential confounding factors because non-genic-chr21 has an almost identical demographic history, sample size and read depth to the exome and has been processed in the exact same way (Supplemental Note 6.2). Candidate targets of positive selection (hereafter ‘candidate SNPs’) were defined as the exome SNPs with higher XtX* than the values corresponding to estimated false positive rates (FPR) of 0.5%, 0.1% and 0.05% using the non-genic-chr21 XtX* distribution while accounting for read depth (details in Materials and Methods and Supplementary Note 6.3.2).
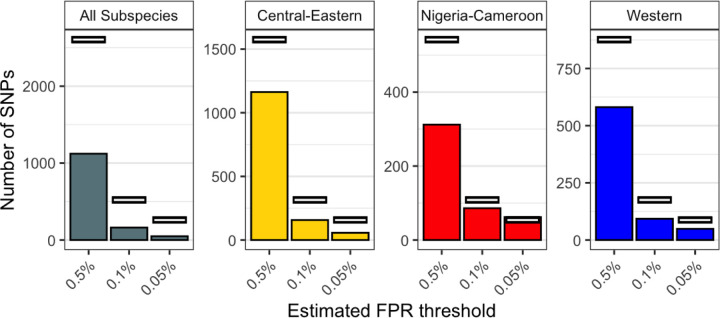
If local adaptation drives population differentiation, we expect exomes to show an excess of highly differentiated SNPs compared to neutral expectations. Contrary to this expectation, there are fewer SNPs with very large XtX* values in the exome compared to null expectations (Fig. 2, Fig. S27), potentially reflecting the effects of purifying selection in the exome. It remains possible that specific selection pressures have driven local genetic adaptation in chimpanzees but we do not observe evidence of this at the genome level using exomes alone.
Fig. 2. Number of genetics-only candidate SNPs.
The number of candidate SNPs from the genetics-only test (bars) compared to the null expectation (white lines) at XtX* thresholds corresponding to estimated FPRs of 0.5%, 0.1% and 0.05%, for each subspecies-dataset. Note that y-axis scales are not consistent across panels.
Genetic adaptation to habitat
Integrating genetic and environmental data increases the power to detect signatures of local adaptation (85, 86) and allows us to directly test the hypothesis that chimpanzees have adapted to selection pressures that vary between habitats. We thus performed a genotype-environment association (GEA) test by integrating an environmental covariable into the analysis using the BayPass AUX model (83). BayPass calculates a Bayes factor (BF) for each SNP that indicates the strength of evidence for the linear correlation between population allele frequencies and the environmental covariable while accounting for population structure (Supplemental Note 6.4.5). SNPs evolving under local adaptation are expected to be highly correlated with the relevant environmental covariable and therefore have the highest BF in the genome.
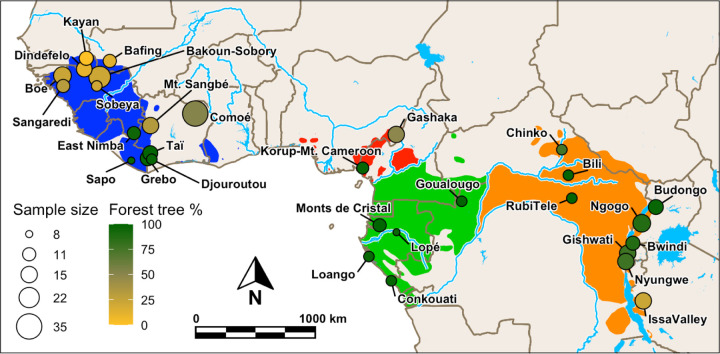
Environmental metrics based on temperature, precipitation or land cover do not correspond well with researcher-defined forest and savannah regions (12). Therefore, we used a floristic measure informed by a large-scale biogeographic analysis which identified very different tree species compositions between forest and savannah regions and has been shown to produce more accurate maps of habitat distributions across Africa (87). Specifically, we used the percentage of trees identified as ‘forest specialists’ (87) among all the classified trees recorded at each sample site (hereafter ‘forest-tree-percentage’) (Fig. 3) (see Materials and Methods and Supplemental Note 2 for details). This variable is ideal because the data was collected within the known ranges of the sampled populations and the same field protocol can be applied to new sample sites, making our data and results comparable to future studies incorporating additional sample sites. This variable is not used here to test for adaptation to tree species compositions per se; rather, it is used to describe the chimpanzee habitat gradient, which summarises many potential selective pressures. The GEA analysis was run with this covariable in each subspecies-dataset except Nigeria-Cameroon, as it has only two populations. Candidate SNPs were selected as in the genetics-only test (details in Supplementary Note 6.4.1).
Fig. 3. BayPass analysis dataset.
Map of West, Central and East Africa showing the location, sample size (after filtering) and forest-tree-percentage for each population in the BayPass analyses. The ranges of the four subspecies are shown (green=central, orange=eastern, red=Nigeria-Cameroon, blue=western) (1) with major rivers and lakes indicated in light blue.
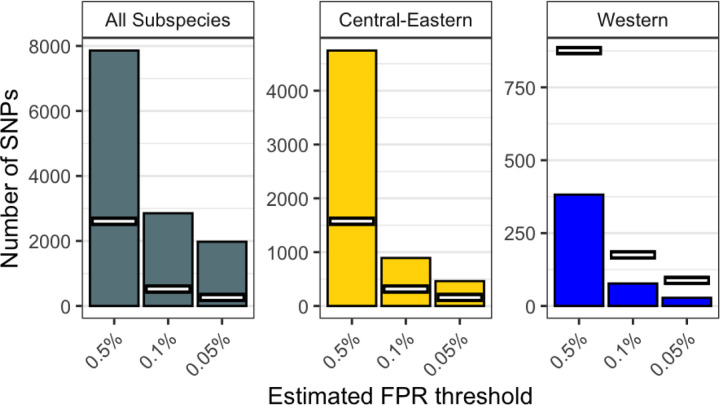
In contrast to the genetics-only results, the GEA shows a substantial excess of SNPs strongly associated with forest-tree-percentage in the exome when compared with neutral expectations in All and Central-Eastern (Fig. 4). This excess is what we expect under local adaptation associated with habitat. This excess is not present in Western, as expected if the strong population bottleneck experienced by this subspecies (3, 5, 62) increased drift and reduced the efficacy of natural selection, or if recent gene flow (3) inhibited the evolution of local adaptation (Fig. 4). Nevertheless, this does not exclude the possibility that strong selective forces may have driven local adaptation in the western subspecies, and the SNPs with the highest BFs in the exome are the best candidate targets of positive selection. Read depth and population substructure do not drive our candidates, and independent tests confirm that candidate allele frequencies correlate strongly with forest-tree-percentage (Figs. S39 and S44–46). For all thresholds and subspecies-datasets, the minimum BF is very high: over 14.7 for FPR<0.5%, over 18.3 for FPR<0.1% and over 19.5 for FPR<0.05% (Fig. S38). Jeffrey’s rule (88) defines 15<BF<20 as ‘very strong evidence’ and BF>20 as ‘decisive evidence’, demonstrating that a vast majority of candidate SNPs have very strong evidence of being associated with habitat, with almost all SNPs in the 0.1% tail having decisive evidence (Fig. S37).
Fig. 4. Number of GEA candidate SNPs.
The number of candidate SNPs from the GEA (bars) compared to the null expectation (white lines) at BF thresholds corresponding to estimated FPRs of 0.5%, 0.1% and 0.05%, for each subspecies-dataset tested. Note that y-axis scales are not consistent across panels.
These results provide strong evidence for local genetic adaptation to habitat in chimpanzees, revealing the presence of important genetic differences among wild populations, even within subspecies, that likely shape fitness in an environment-dependent way.
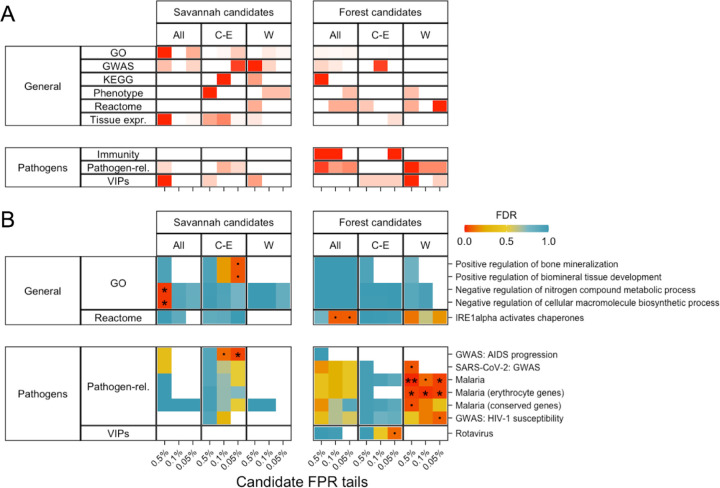
Under the reasonable assumption that novel adaptations are more commonly mediated by the novel, derived allele than the ancestral one, we assigned SNPs as associated with forest or savannah adaptations according to the sign of their correlation coefficient. There is an excess of SNPs with high BFs in the exome for both savannah and forest candidates (Fig. S42) in All and Central-Eastern, suggesting that adaptation in either direction contributes to the overall excess. To interpret these loci biologically, we investigated the genes the candidate SNPs fall within (hereafter ‘candidate genes’) by testing for an overrepresentation of functional categories in hypothesis-free gene set enrichment analyses. Given the relevance of pathogens as selective pressures (65, 66), we also performed a hypothesis-driven enrichment analysis of pathogen-related genes (details in Supplemental Note 7). Interestingly, these analyses point to potential differential adaptations in savannah and forest chimpanzees.
Adaptations to savannah
Savannah candidate genes belong to many categories associated with physiological traits when compared to forest candidate genes (Fig. 5A). There are over six times more “General” gene categories with an FDR<0.5 in savannah than forest candidates (260 vs 42, Figs. 5A and S50), although only two of these categories are significantly enriched (FDR < 0.05) (Fig. 5B) (negative regulation of nitrogen compound metabolic processes and negative regulation of cellular macromolecule biosynthetic processes). This would be compatible with a large degree of polygenicity in the genetic adaptation of chimpanzees to the environmental extremes of savannahs. The diversity of categories and their overlap in genes makes it difficult to infer selection pressures that may drive this signal.
Fig. 5. GEA candidate gene set enrichment results.
Results for 0.5%, 0.1% and 0.05% FPR tails for savannah and forest candidate SNPs are shown. Vertical panels indicate results from each subspecies-dataset. Horizontal panels show the broad categories that the gene sets belong to. Multiple testing correction was done within each gene set enrichment analysis run (i.e., each tail and gene set database such as ‘Pathogen-related’, ‘GWAS’, ‘Phenotype’ etc.). (A) the number of gene sets with FDR<0.5, cells are coloured in a gradient from white (0) to red (the largest value per row) (Fig. S50 shows the numbers in each cell). (B) shows the FDR values for the most enriched gene sets with FDR<0.1 for at least one candidate tail in at least one subspecies-dataset (‘.’ FDR<0.1, ‘*’ FDR<0.05, ‘**’ FDR<0.01).
Restricted availability of water in savannahs during the dry season is a potential selection pressure (17, 30) that could partially explain the enrichment of physiological categories in our candidates. Nevertheless, there is no significant enrichment in the two dehydration response gene categories we analysed (89, 90) (Supplemental Note 7), neither in the GEA nor genetics-only candidates (Fig. S51). Chimpanzees may thus have adapted to dehydration stress through genes not included in these categories or through behavioural adaptations (e.g., well digging (22)). Alternatively, dehydration stress may be present but independent of habitat (15).
There is limited evidence of adaptation to pathogens in savannah populations (Fig. 5A, left bottom corner). Savannah candidates are significantly enriched (FDR < 0.05) for only one pathogen-related gene set: genes associated with AIDS progression in GWAS at the Central-Eastern 0.05% tail (Fig. 5B), which contains only two candidate genes (Table S2). Viruses similar to SIV/HIV are not known to be associated with savannahs, instead, this result may be explained by adaptation in Issa Valley, which has a high prevalence of SIV (91, 92) and a particularly low forest-tree-percentage in Central-Eastern (Fig. 3). Being extreme in forest-tree-percentage means that Issa Valley weighs heavily on the Central-Eastern savannah candidates, although without fully driving them (Supplemental Note: 6.4.6). Analysis of additional populations will help establish to what extent the evidence of adaptation is general across central and eastern savannah populations, and identify the specific adaptive mechanisms and selective factors. In any case, the excess of exonic savannah candidate SNPs in All and Central-Eastern suggests that chimpanzees do harbour genetic adaptations to savannah habitats.
Adaptations to forest
While forest candidates show weak enrichment in general physiological categories, they show a pattern of stronger enrichment in pathogen-related genes as shown in stronger enrichment for general “immunity genes” (93) and “innate immunity genes” (94) in the forest than savannah candidates in All and Central-Eastern (Fig. 5A). This pattern is also evident for individual pathogen categories in All and especially in Western, although not in Central-Eastern (Fig. 5). This is consistent with the higher population densities (2) and increased pathogen exposure (14) in forests resulting in a greater infectious disease burden. In humans, local adaptation has likely also been driven by high pathogen diversity (95), particularly in tropical forests (34–36). Central-Eastern does not show this pattern, likely due to the presence of eastern populations from montane forests, which are considerably cooler than lowland forests and therefore have lower levels of vector-borne diseases such as malaria (14) (Supplemental Note 7.2.1). Enrichment of pathogen-related categories in the Western forest candidates suggests that, although we do not see evidence of positive selection on the genome-scale, strong selection at a limited number of pathogen-related genes is likely driving local adaptation in this subspecies.
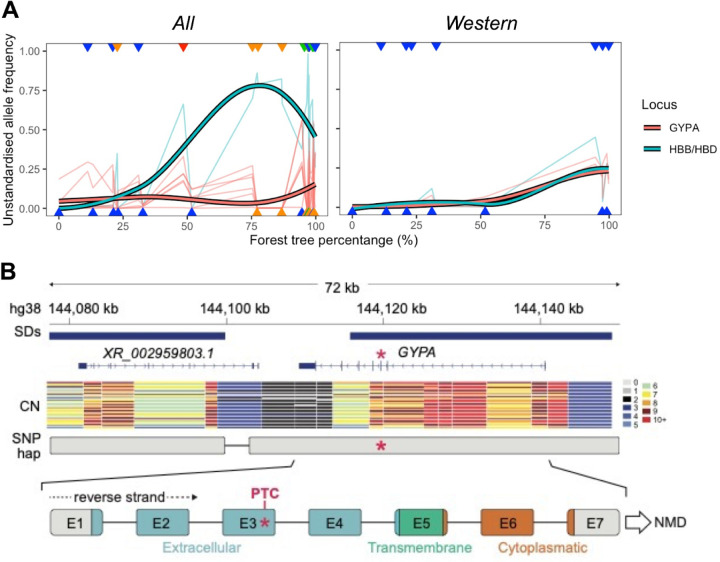
Focusing on individual pathogens, the strongest and clearest signal is enrichment for malaria-related categories in Western forest candidates (Fig. 5B, Table S2). They are significantly enriched (FDR<0.05) in “Malaria related genes” (96) at the 0.5% and 0.05% tails and “Erythrocyte genes related to malaria” (97) at all three tails; enrichment in Plasmodium-interacting proteins that are conserved across mammals (98) (thus excluding haemoglobin and glycophorin genes, see below) very narrowly exceeds the significance threshold (FDR=0.050) in the 0.5% tail. Interestingly, malaria infection probability in chimpanzees is closely correlated with canopy cover (14), which is itself highly correlated with forest-tree-percentage in our dataset (Pearson r = 0.92, p = 9.044×10−13) (99). Malaria is a major selection pressure and has driven some of the clearest examples of local adaptation in humans (37, 38). Only five genetic variants have been significantly associated with severe malaria in human GWAS (100, 101). Strikingly, two of these loci, which encode for haemoglobin (HB) and glycophorin (GYP) genes (Fig. 6A), contain chimpanzee forest candidate SNPs; both of which also underlie adaptations to malaria in humans (102–105).
Fig. 6. Key malaria-related forest candidate genes.
(A) Derived allele frequencies of candidate SNPs at the HBB/HBD (green) and GYPA (red) loci plotted against forest-tree-percentage, with population values indicated with triangles coloured according to subspecies (green=central, orange=eastern, red=Nigeria-Cameroon, blue=western) arbitrarily assigned to the top or bottom of the graph to reduce overlap. Left for candidate SNPs and populations from All. Right for candidate SNPs and populations from Western. Thin lines represent the estimated population allele frequencies for each candidate SNP, thick lines show the smoothed pattern of all candidate SNPs per locus using LOESS. (B) Diagram of the GYPA locus in hg38 coordinates, including segmental duplications (SDs), copy numbers (CN) across captive chimpanzees, representative long-read sequencing haplotype containing candidate SNP C>A at chr4:145040845 in hg19 coordinates (red asterisk), and schematic representation of the candidate SNP location within GYPA exons (E1-7). PTC: premature termination codon. NMD: Nonsense-mediated mRNA decay. The PTC SNP is 210 exonic base pairs upstream of the last exon-exon junction (between exons 6 and 7), and therefore, likely to cause NMD according to the 50-55nt rule (116).
For HB, candidate SNP chr11:5254366 (Western 0.5% and All 0.05% tails) lies within an intron of Haemoglobin Subunit Delta (HBD), less than 5kb upstream of the adjacent paralogue Haemoglobin Subunit Beta (HBB). While mutations in HBD have little effect on malaria resistance due to low expression in adults (106), the HbS mutation in HBB is a classic example of balancing selection in humans, as heterozygotes are protected against severe malaria (102, 103). Therefore, the signatures that we observe may reflect selection on a linked variant within HBB, the regulation of HBB or HBD itself. In any case, it is striking that this locus shows evidence of local adaptation in both chimpanzees and humans.
For GYP, two candidate SNPs, chr4:145040845 and chr4:145039806 (Western 0.5% and 0.05% tails respectively) lie within Glycophorin A (GYPA) (Ensemble hg19 also places them within an intron of Glycophorin B (GYPB) likely due to an annotation error, see Supplemental Note 8.2) (Fig. S52). The evidence for selection at this locus is strong with chr4:145039806 having the 23rd highest BF in Western accounting for read depth (FPR≤2.91×10−4). In All, there are six forest candidate SNPs in GYPA including chr4:145040845 (0.05% tail) and chr4:145039806 (0.5% tail), and another GYPA SNP is a candidate in the genetics-only 0.5% tail (Fig. S52).
GYPA and GYPB encode glycophorins used by Plasmodium falciparum to enter erythrocytes (107). In humans, structural variants associated with this locus appear to mediate adaptation to malaria (104, 105, 108–112). It is therefore interesting to investigate structural variation in chimpanzees. Read depth in the PanAf exomes is not unusual at this locus, but low-coverage target capture data is not ideal for investigating structural variation. Copy-number (CN) estimates from high-coverage short-read (n=60) (5, 62) and long-read data (n=2) (113, 114) from captive chimpanzees confirm that, in addition to the full-length and likely ancestral GYPA, chimpanzees also carry 2–9 copies of truncated paralogues lacking the last two exons of GYPA, which encode for the cytoplasmic domain (115) (Supplemental Note 8.1, Fig. 6B, Fig. S53D). Thus, like in humans, structural variants contribute to the complexity of the locus in chimpanzees. We note that the GYPA candidate SNPs are present in both the long-read (113, 114) and high-coverage short-read (5, 62) data (Fig. S55), confirming them to be true polymorphisms; further, there is no evidence of an association between forest-tree-percentage and read-depth in the PanAf exomes at these SNPs (Fig. S54, details in Supplemental Note 8.1).
The long-read data (113, 114) show the GYPA candidate SNPs residing in a single haplotype spanning the full-length gene (Fig. 6B, Fig. S53C). The candidate allele at chr4:145040845 introduces a premature stop gain in exon 3 of GYPA (E76X) predicted to result in degradation of the mRNA by nonsense-mediated decay (116); even if the truncated protein was translated, it would encode only a partial extracellular domain and be missing the remaining extracellular and entire transmembrane and cytoplasmic domains (115), resulting in non-functional GYPA. Thus, as suggested for GYPA deletions in humans (109), this SNP may be the target of natural selection, by preventing the expression of a key receptor protein used by the malaria parasite to enter erythrocytes (107).
Conclusions
We present the largest population genomic study of natural selection in a non-human ape to date, capturing and sequencing the exome from non-invasive samples of hundreds of wild chimpanzees and integrating these exomes with previously published full chr21 sequences from the same samples (3). Even in the face of limitations from non-invasive sampling (Supplemental Note 1), this work demonstrates that population genomics can reveal the presence of local genetic adaptation in an endangered species.
The genotype-environment association analysis provides strong, genome-scale evidence of local adaptation to habitat in All and Central-Eastern–although not in Western, likely because of their small long-term Ne (5, 62). This demonstrates the power of genotype-environment associations (GEA) to identify positive selection by revealing signatures of local adaptation in the form of subtle allele frequency changes correlating with a relevant covariable (Supplemental Note 6.4.4). Indeed, the GEA candidate SNPs differ consistently in allele frequency with respect to habitat but generally do not have large frequency differences between populations (Fig. S45A). This is consistent with local adaptation in chimpanzees being mostly polygenic and driven by soft sweeps as observed in humans (32, 117, 118), and suggests the presence of complex genetic adaptations even in the absence of fixed differences among populations.
Our findings suggest that while behaviours such as tool use (16, 119) and thermoregulatory behaviours (17) are important in mitigating environmental stressors, selective pressures associated with habitat still appear to drive genetic adaptation in chimpanzees. Thus, both behavioural flexibility and genetic adaptation may explain how chimpanzees inhabit such a range of habitats. Far from replacing genetic adaptation, behavioural adaptations may drive genetic changes via gene-culture coevolution (120), as seen with human diets (43), whereby behavioural flexibility facilitates exposure to novel selection pressures that later drive genetic adaptations.
The evidence of genetic adaptation in forests demonstrates the importance of novel adaptations even in habitats with high availability of resources that support high population densities. This is perhaps because the struggle against the high pathogen load of lowland forests shapes the evolution of these populations. This is not surprising, as pathogens have been important selective pressures for chimpanzees over longer time scales (65, 66). Today, infectious diseases are major causes of chimpanzee population decline(1) and recent increased exposure to humans has led to an increase in deadly outbreaks caused by cross-species transmission (121). Our findings highlight the importance of genetic adaptation in shaping infectious disease mortality in chimpanzees and suggest that individuals are adapted to the pathogens present in their local habitat, emphasising the dangers of displacement and environmental change.
Evidence of adaptation to malaria in forests is particularly interesting. A range of malaria parasites infect wild chimpanzees, including three Laverania species closely related to P. falciparum, which originated in gorillas (57, 122) and is now responsible for 90% of global malaria mortality in humans (123). However, the fitness effects of malaria in wild chimpanzees are poorly understood (124). Its high prevalence in wild populations (122) and the few studies of captive chimpanzees suggest that severe effects are rare (125–128). However, fitness effects in the wild may be more severe than in captivity, as demonstrated by the largely asymptomatic SIV infections in captivity (58, 129, 130) but fitness effects in the wild (29, 131). Young chimpanzees and pregnant mothers are particularly susceptible to malaria infection (132, 133), which may lead to higher morbidity/mortality as observed in humans (134). Our findings indicate that malaria has been an important selection pressure in the recent past and may have important fitness effects in present-day wild populations. That adaptation appears to be mediated by the same few genes in chimpanzees and humans is striking from an evolutionary point of view; further, it demonstrates how understanding chimpanzee evolution can inform human medicine.
Chimpanzees also appear to have adapted genetically to savannah habitats, although identifying key selective pressures and adaptive traits is harder. Studying additional savannah populations would help address this question. This would provide insights into how our ancestors may have adapted to similar habitats, and have important implications for the conservation of wild chimpanzees as their habitats become hotter and more seasonal under climate change (2).
Just as previous studies highlighted the importance of conserving behavioural diversity (16, 135, 136), we emphasise the importance of conserving adaptive genetic diversity across chimpanzees’ ecological range to maintain their adaptive potential and ensure long-term survival in the wild (46, 74). This is important because chimpanzee habitats are changing rapidly due to direct anthropogenic destruction (1, 137, 138), climate change (139) and disease transmission (121, 140). We emphasise the need to protect all chimpanzee habitats and to consider local genetic adaptations when planning conservation efforts to ensure that individuals are adapted to the local environment. Finally, our study demonstrates the value and promise of non-invasive sampling to investigate genetic adaptation in endangered species.
Supplementary Material
Acknowledgements:
We would like to thank Laia Llovera for assistance in the laboratory. We thank Thierry Aebischer, Floris Aubert, Emmanuel Ayuk Ayimisin, Arcel Bamba, Matthieu Bonnet, Chloe Cipoletta, Katherine Corogenes, Bryan Curran, Lucy D’Auvergne, Jean Claude Dengui, Theophile Desarmeaux, Karsten Dierks, Emmanuel Dilambaka, Dervla Dowd, Andrew Dunn, Manasseh Eno-Nku, Theo Freeman, Annemarie Goedmakers, John Hart, Martijn Ter Heegde, Inaoyom Imong, Mohamed Kambi, Laura Kehoe, Vincent Lapeyre, Vera Leinert, Joshua M Linder, Sergio Marrocoli, Michael Masozera, Richard McElreath, Vianet Mihindou, Geoffrey Muhanguzi, Felix Mulindahabi, Mizuki Murai, Protais Niyigaba, Nadege Wangue Njomen, Nicolas Ntare, Bruno Perodeau, Alhaji Malikie Siaka, Alexander Tickle, Els Ton, Richard Tshombe, Hilde Vanleeuwe, Paula Álvarez Varona, Virginie Vergnes, Magloire Kambale Vyalengerera and Klaus Zuberbuehler for assistance in field site coordination and sample collection. We thank the field assistants and volunteers from the Jane Goodall Institute Spain in Senegal for their help with sample collection in Dindefelo. We also thank the Greater Mahale Ecosystem Research and Conservation (GMERC) field assistants who provide crucial support and data collection at the Issa Valley field site in Tanzania.
We thank the following government agencies for their support in conducting field research in their countries: Ministère de la Recherche Scientifique et de l’Innovation, Cameroon; Ministère des Forêts et de la Faune, Cameroon; Ministère des Eaux et Forêts, Cote d’Ivoire; Ministère de l’Enseignement Supérieur et de la Recherche Scientifique in Côte d’Ivoire; Institut Congolais pour la Conservation de la Nature, DR-Congo; Ministère de la Recherche Scientifique, DR-Congo; Agence Nationale des Parcs Nationaux, Gabon; Center National de la Recherche Scientifique (CENAREST), Gabon; Société Equatoriale d’Exploitation Forestière (SEEF), Gabon; Department of Wildlife and Range Management, Ghana; Forestry Commission, Ghana; Ministère de l’Agriculture de l’Elevage et des Eaux et Forets, Guinea; Instituto da Biodiversidade e das Áreas Protegidas (IBAP), Guinea-Bissau; Ministro da Agricultura e Desenvolvimento Rural, Guinea-Bissau; Forestry Development Authority, Liberia; Eaux et Forets, Mali; Ministre de l’Environnement et de l’Assainissement et du Developpement Durable du Mali; Conservation Society of Mbe Mountains (CAMM), Nigeria; National Park Service, Nigeria; Ministère de l’Economie Forestière, R-Congo; Ministère de le Recherche Scientifique et Technologique, R-Congo; Ministry of Education, Rwanda; Rwanda Development Board, Rwanda; Direction des Eaux, Forêts, et Chasses, Senegal; Reserve Naturelle Communautaire de Dindefelo, Senegal; Ministry of Agriculture, Forestry, and Food Security, Sierra Leone; National Protected Area Authority, Sierra Leone; Tanzania Commission for Science and Technology, Tanzania; Tanzania Wildlife Research Institute, Tanzania; Makerere University Biological Field Station (MUBFS), Uganda; Uganda National Council for Science and Technology (UNCST), Uganda; Uganda Wildlife Authority, Uganda; National Forestry Authority, Uganda; Agence Congolaise de la Faune et des Aires Protégées; and The Wild Chimpanzee Foundation (WCF).
We thank Dean Lavelle for assistance in re-base calling ONT data using guppy 5.0.11, as well as Agilent for their collaboration in the project. We thank Hernán Burbano, Max Reuter and all the members of the Andrés and Burbano groups for useful comments and suggestions. Finally, we thank Jinliang Wang for his support and supervision.
Funding:
HJO is supported by the Natural Environment Research Council through the London NERC Doctoral Training Programme scholarship, grant no. NE/L002485/1. AMA is supported by UCL’s Wellcome Trust ISSF3 award no. 204841/Z/16/Z, the BBSRC grant number BB/W007703/1, and the Leverhulme trust grant number RPG-2021-414. The Pan African Program: The Cultured Chimpanzee (PanAf) is generously funded by the Max Planck Society, the Max Planck Society Innovation Fund, and the Heinz L. Krekeler Foundation. TM-B is supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 864203), PID2021-126004NB-100 (MICIIN/FEDER, UE) and Secretaria d’Universitats i Recerca and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2021 SGR 00177). We also acknowledge funding from the ‘‘la Caixa’’ Foundation doctoral fellowship program LCF/BQ/DE15/ 10360006 (to CF), U.S. National Institutes of Health (NIH) grants from the Office of the Director and National Institute of Mental Health (DP2MH119424 and R01MH132818 to MYD), FPI (Formación de Personal Investigador) PRE2018- 083966 from Ministerio de Ciencia, Universidades e Investigacion (to MA-E) and PID2020-116908GB-I00 (MCIN) (to EL). FAS and AKP are grateful to the UCSD/Salk Center for Academic Research and Training in Anthropogeny (CARTA) and Department of Human Origins, Max Plank Institute for Evolutionary Anthropology for support of GMERC, the Issa Valley field site, and sample collection in Tanzania
Funding Statement
HJO is supported by the Natural Environment Research Council through the London NERC Doctoral Training Programme scholarship, grant no. NE/L002485/1. AMA is supported by UCL’s Wellcome Trust ISSF3 award no. 204841/Z/16/Z, the BBSRC grant number BB/W007703/1, and the Leverhulme trust grant number RPG-2021-414. The Pan African Program: The Cultured Chimpanzee (PanAf) is generously funded by the Max Planck Society, the Max Planck Society Innovation Fund, and the Heinz L. Krekeler Foundation. TM-B is supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 864203), PID2021-126004NB-100 (MICIIN/FEDER, UE) and Secretaria d’Universitats i Recerca and CERCA Programme del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2021 SGR 00177). We also acknowledge funding from the ‘‘la Caixa’’ Foundation doctoral fellowship program LCF/BQ/DE15/ 10360006 (to CF), U.S. National Institutes of Health (NIH) grants from the Office of the Director and National Institute of Mental Health (DP2MH119424 and R01MH132818 to MYD), FPI (Formación de Personal Investigador) PRE2018- 083966 from Ministerio de Ciencia, Universidades e Investigacion (to MA-E) and PID2020-116908GB-I00 (MCIN) (to EL). FAS and AKP are grateful to the UCSD/Salk Center for Academic Research and Training in Anthropogeny (CARTA) and Department of Human Origins, Max Plank Institute for Evolutionary Anthropology for support of GMERC, the Issa Valley field site, and sample collection in Tanzania
Footnotes
Competing interests:
The authors declare no competing interests.
Data and materials availability:
All data and code will be made available on publication.
References and Notes:
- 1.IUCN, Pan troglodytes: Humle T., maisels F., Oates J.f., plumptre A. & Williamson E.a. IUCN Red List of Threatened Species (2016), , doi: 10.2305/iucn.uk.2016-2.rlts.t15933a17964454.en. [DOI] [Google Scholar]
- 2.Lindshield S., Hernandez-Aguilar R. A., Korstjens A. H., Marchant L. F., Narat V., Ndiaye P. I., Ogawa H., Piel A. K., Pruetz J. D., Stewart F. A., van Leeuwen K. L., Wessling E. G., Yoshikawa M., Chimpanzees (Pan troglodytes) in savanna landscapes. Evol. Anthropol. 30, 399–420 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Fontsere C., Kuhlwilm M., Morcillo-Suarez C., Alvarez-Estape M., Lester J. D., Gratton P., Schmidt J. M., Dieguez P., Aebischer T., Álvarez-Varona P., Agbor A., Angedakin S., Assumang A. K., Ayimisin E. A., Bailey E., Barubiyo D., Bessone M., Carretero-Alonso A., Chancellor R., Cohen H., Danquah E., Deschner T., Dunn A., Dupain J., Egbe V. E., Feliu O., Goedmakers A., Granjon A.-C., Head J., Hedwig D., Hermans V., Hernandez-Aguilar R. A., Imong I., Jones S., Junker J., Kadam P., Kaiser M., Kambere M., Kambale M. V., Kalan A. K., Kienast I., Kujirakwinja D., Langergraber K., Lapuente J., Larson B., Laudisoit A., Lee K., Llana M., Llorente M., Marrocoli S., Morgan D., Mulindahabi F., Murai M., Neil E., Nicholl S., Nixon S., Normand E., Orbell C., Ormsby L. J., Pacheco L., Piel A., Riera L., Robbins M. M., Rundus A., Sanz C., Sciaky L., Sommer V., Stewart F. A., Tagg N., Tédonzong L. R., Ton E., van Schijndel J., Vergnes V., Wessling E. G., Willie J., Wittig R. M., Yuh Y. G., Yurkiw K., Zuberbuehler K., Hecht J., Vigilant L., Boesch C., Andrés A. M., Hughes D. A., Kühl H. S., Lizano E., Arandjelovic M., Marques-Bonet T., Cell Genom, in press, doi: 10.1016/j.xgen.2022.100133. [DOI] [Google Scholar]
- 4.Gonder M. K., Oates J. F., Disotell T. R., Forstner M. R., Morales J. C., Melnick D. J., A new west African chimpanzee subspecies? Nature. 388, 337 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Prado-Martinez J., Sudmant P. H., Kidd J. M., Li H., Kelley J. L., Lorente-Galdos B., Veeramah K. R., Woerner A. E., O’Connor T. D., Santpere G., Cagan A., Theunert C., Casals F., Laayouni H., Munch K., Hobolth A., Halager A. E., Malig M., Hernandez-Rodriguez J., Hernando-Herraez I., Prüfer K., Pybus M., Johnstone L., Lachmann M., Alkan C., Twigg D., Petit N., Baker C., Hormozdiari F., Fernandez-Callejo M., Dabad M., Wilson M. L., Stevison L., Camprubí C., Carvalho T., Ruiz-Herrera A., Vives L., Mele M., Abello T., Kondova I., Bontrop R. E., Pusey A., Lankester F., Kiyang J. A., Bergl R. A., Lonsdorf E., Myers S., Ventura M., Gagneux P., Comas D., Siegismund H., Blanc J., Agueda-Calpena L., Gut M., Fulton L., Tishkoff S. A., Mullikin J. C., Wilson R. K., Gut I. G., Gonder M. K., Ryder O. A., Hahn B. H., Navarro A., Akey J. M., Bertranpetit J., Reich D., Mailund T., Schierup M. H., Hvilsom C., Andrés A. M., Wall J. D., Bustamante C. D., Hammer M. F., Eichler E. E., Marques-Bonet T., Great ape genetic diversity and population history. Nature. 499, 471–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plumptre A., Hart J. A., Hicks T. C., Nixon S., Piel A. K., Pan troglodytes ssp. Schweinfurthii. The IUCN Red List of Threatened Species. International Union for. [Google Scholar]
- 7.Iucn, IUCN, Pan troglodytes ssp. verus: Humle T., Boesch C., Campbell G., Junker J., Koops K., Kuehl H. & Sop T. IUCN Red List of Threatened Species (2016), , doi: 10.2305/iucn.uk.2016-2.rlts.t15935a17989872.en. [DOI] [Google Scholar]
- 8.Iucn, IUCN, Pan troglodytes ssp. ellioti: Oates J.F., Doumbe O., Dunn A., Gonder M.K., Ikemeh R., Imong I., Morgan B.J., Ogunjemite B. & Sommer V. IUCN Red List of Threatened Species (2015), , doi: 10.2305/iucn.uk.2016-2.rlts.t40014a17990330.en. [DOI] [Google Scholar]
- 9.Gross-Camp N. D., Masozera M., Kaplin B. A., Chimpanzee seed dispersal quantity in a tropical montane forest of Rwanda. Am. J. Primatol. 71, 901–911 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Wrangham R. W., Chapman C. A., Chapman L. J., Seed dispersal by forest chimpanzees in Uganda. J. Trop. Ecol. 10, 355–368 (1994). [Google Scholar]
- 11.Aguado W. D., Rogers H. S., Pruetz J. D., Chimpanzees as ecosystem service providers: Seed dispersal of an economically important plant resource. Biotropica. 54, 656–669 (2022). [Google Scholar]
- 12.van Leeuwen K. L., Hill R. A., Korstjens A. H., Classifying Chimpanzee (Pan troglodytes) Landscapes Across Large-Scale Environmental Gradients in Africa. Int. J. Primatol. 41, 800–821 (2020). [Google Scholar]
- 13.Barratt C. D., Lester J. D., Gratton P., Onstein R. E., Kalan A. K., McCarthy M. S., Bocksberger G., White L. C., Vigilant L., Dieguez P., Abdulai B., Aebischer T., Agbor A., Assumang A. K., Bailey E., Bessone M., Buys B., Carvalho J. S., Chancellor R., Cohen H., Danquah E., Deschner T., Dongmo Z. N., Doumbé O. A., Dupain J., Duvall C. S., Eno-Nku M., Etoga G., Galat-Luong A., Garriga R., Gatti S., Ghiurghi A., Goedmakers A., Granjon A.-C., Hakizimana D., Head J., Hedwig D., Herbinger I., Hermans V., Jones S., Junker J., Kadam P., Kambi M., Kienast I., Kouakou C. Y., N Goran K. P., Langergraber K. E., Lapuente J., Laudisoit A., Lee K. C., Maisels F., Mirghani N., Moore D., Morgan B., Morgan D., Neil E., Nicholl S., Nkembi L., Ntongho A., Orbell C., Ormsby L. J., Pacheco L., Piel A. K., Pintea L., Plumptre A. J., Rundus A., Sanz C., Sommer V., Sop T., Stewart F. A., Sunderland-Groves J., Tagg N., Todd A., Ton E., van Schijndel J., VanLeeuwe H., Vendras E., Welsh A., Wenceslau J. F. C., Wessling E. G., Willie J., Wittig R. M., Yoshihiro N., Yuh Y. G., Yurkiw K., Boesch C., Arandjelovic M., Kühl H., Quantitative estimates of glacial refugia for chimpanzees (Pan troglodytes) since the Last Interglacial (120,000 BP). Am. J. Primatol. 83, e23320 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Scully E. J., Liu W., Li Y., Ndjango J.-B. N., Peeters M., Kamenya S., Pusey A. E., Lonsdorf E. V., Sanz C. M., Morgan D. B., Piel A. K., Stewart F. A., Gonder M. K., Simmons N., Asiimwe C., Zuberbühler K., Koops K., Chapman C. A., Chancellor R., Rundus A., Huffman M. A., Wolfe N. D., Duraisingh M. T., Hahn B. H., Wrangham R. W., The ecology and epidemiology of malaria parasitism in wild chimpanzee reservoirs. Commun Biol. 5, 1020 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wessling E. G., Deschner T., Mundry R., Pruetz J. D., Wittig R. M., Kühl H. S., Seasonal Variation in Physiology Challenges the Notion of Chimpanzees (Pan troglodytes verus) as a Forest-Adapted Species. Frontiers in Ecology and Evolution. 6 (2018), doi: 10.3389/fevo.2018.00060. [DOI] [Google Scholar]
- 16.Kalan A. K., Kulik L., Arandjelovic M., Boesch C., Haas F., Dieguez P., Barratt C. D., Abwe E. E., Agbor A., Angedakin S., Aubert F., Ayimisin E. A., Bailey E., Bessone M., Brazzola G., Buh V. E., Chancellor R., Cohen H., Coupland C., Curran B., Danquah E., Deschner T., Dowd D., Eno-Nku M., Michael Fay J., Goedmakers A., Granjon A.-C., Head J., Hedwig D., Hermans V., Jeffery K. J., Jones S., Junker J., Kadam P., Kambi M., Kienast I., Kujirakwinja D., Langergraber K. E., Lapuente J., Larson B., Lee K. C., Leinert V., Llana M., Marrocoli S., Meier A. C., Morgan B., Morgan D., Neil E., Nicholl S., Normand E., Ormsby L. J., Pacheco L., Piel A., Preece J., Robbins M. M., Rundus A., Sanz C., Sommer V., Stewart F., Tagg N., Tennie C., Vergnes V., Welsh A., Wessling E. G., Willie J., Wittig R. M., Yuh Y. G., Zuberbühler K., Kühl H. S., Environmental variability supports chimpanzee behavioural diversity. Nat. Commun. 11, 4451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruetz J., Bertolani P., Chimpanzee (Pan troglodytes verus) Behavioral Responses to Stresses Associated with Living in a Savannah-Mosaic Environment: Implications for Hominin Adaptations to Open Habitats. PaleoAnthropology. 2009 (2009), pp. 252–262. [Google Scholar]
- 18.Tagg N., McCarthy M., Dieguez P., Bocksberger G., Willie J., Mundry R., Stewart F., Arandjelovic M., Widness J., Landsmann A., Agbor A., Angedakin S., Ayimisin A. E., Bessone M., Brazzola G., Corogenes K., Deschner T., Dilambaka E., Eno-Nku M., Eshuis H., Goedmakers A., Granjon A.-C., Head J., Hermans V., Jones S., Kadam P., Kambi M., Langergraber K. E., Lapeyre V., Lapuente J., Lee K., Leinert V., Maretti G., Marrocoli S., Meier A., Nicholl S., Normand E., Ormsby L. J., Piel A., Robinson O., Sommer V., Ter Heegde M., Tickle A., Ton E., van Schijndel J., Vanleeuwe H., Vergnes V., Wessling E., Wittig R. M., Zuberbuehler K., Kuehl H., Boesch C., Nocturnal activity in wild chimpanzees (Pan troglodytes): Evidence for flexible sleeping patterns and insights into human evolution. Am. J. Phys. Anthropol. 166, 510–529 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Pruetz J. D., Bertolani P., Ontl K. B., Lindshield S., Shelley M., Wessling E. G., New evidence on the tool-assisted hunting exhibited by chimpanzees (Pan troglodytes verus) in a savannah habitat at Fongoli, Sénégal. R Soc Open Sci. 2, 140507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boesch C., Boesch C., Boesch-Achermann H., The Chimpanzees of the Taï Forest: Behavioural Ecology and Evolution (Oxford University Press, 2000; https://play.google.com/store/books/details?id=6drtXIjlPZgC). [Google Scholar]
- 21.Luncz L. V., Mundry R., Boesch C., Evidence for cultural differences between neighboring chimpanzee communities. Curr. Biol. 22, 922–926 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Péter H., Zuberbühler K., Hobaiter C., Well-digging in a community of forest-living wild East African chimpanzees (Pan troglodytes schweinfurthii). Primates. 63, 355–364 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hockings K. J., Bryson-Morrison N., Carvalho S., Fujisawa M., Humle T., McGrew W. C., Nakamura M., Ohashi G., Yamanashi Y., Yamakoshi G., Matsuzawa T., Tools to tipple: ethanol ingestion by wild chimpanzees using leaf-sponges. R Soc Open Sci. 2, 150150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tutin C. E. G., Boesch, McGrew W. C., Wrangham R. W., Sugiyama, Goodall, Reynolds, Whiten, Nishida, CHARTING CULTURAL VARIATION IN CHIMPANZEES. Behaviour. 138, 1481–1516 (2001). [Google Scholar]
- 25.Kühl H. S., Kalan A. K., Arandjelovic M., Aubert F., D’Auvergne L., Goedmakers A., Jones S., Kehoe L., Regnaut S., Tickle A., Ton E., van Schijndel J., Abwe E. E., Angedakin S., Agbor A., Ayimisin E. A., Bailey E., Bessone M., Bonnet M., Brazolla G., Buh V. E., Chancellor R., Cipoletta C., Cohen H., Corogenes K., Coupland C., Curran B., Deschner T., Dierks K., Dieguez P., Dilambaka E., Diotoh O., Dowd D., Dunn A., Eshuis H., Fernandez R., Ginath Y., Hart J., Hedwig D., Ter Heegde M., Hicks T. C., Imong I., Jeffery K. J., Junker J., Kadam P., Kambi M., Kienast I., Kujirakwinja D., Langergraber K., Lapeyre V., Lapuente J., Lee K., Leinert V., Meier A., Maretti G., Marrocoli S., Mbi T. J., Mihindou V., Moebius Y., Morgan D., Morgan B., Mulindahabi F., Murai M., Niyigabae P., Normand E., Ntare N., Ormsby L. J., Piel A., Pruetz J., Rundus A., Sanz C., Sommer V., Stewart F., Tagg N., Vanleeuwe H., Vergnes V., Willie J., Wittig R. M., Zuberbuehler K., Boesch C., Chimpanzee accumulative stone throwing. Sci. Rep. 6, 22219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur T., Singh J., Tong S., Humphrey C., Clevenger D., Tan W., Szekely B., Wang Y., Li Y., Alex Muse E., Kiyono M., Hanamura S., Inoue E., Nakamura M., Huffman M. A., Jiang B., Nishida T., Descriptive epidemiology of fatal respiratory outbreaks and detection of a human-related metapneumovirus in wild chimpanzees (Pan troglodytes) at Mahale Mountains National Park, Western Tanzania. Am. J. Primatol. 70, 755–765 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuzawa T., Jokro: The death of a wild infant chimpanzee from respiratory disease. Primates. 61, 339–346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbian H. J., Li Y., Ramirez M., Klase Z., Lipende I., Mjungu D., Moeller A. H., Wilson M. L., Pusey A. E., Lonsdorf E. V., Bushman F. D., Hahn B. H., Destabilization of the gut microbiome marks the end-stage of simian immunodeficiency virus infection in wild chimpanzees. Am. J. Primatol. 80 (2018), doi: 10.1002/ajp.22515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keele B. F., Jones J. H., Terio K. A., Estes J. D., Rudicell R. S., Wilson M. L., Li Y., Learn G. H., Beasley T. M., Schumacher-Stankey J., Wroblewski E., Mosser A., Raphael J., Kamenya S., Lonsdorf E. V., Travis D. A., Mlengeya T., Kinsel M. J., Else J. G., Silvestri G., Goodall J., Sharp P. M., Shaw G. M., Pusey A. E., Hahn B. H., Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 460, 515–519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wessling E. G., Kühl H. S., Mundry R., Deschner T., Pruetz J. D., The costs of living at the edge: Seasonal stress in wild savanna-dwelling chimpanzees. J. Hum. Evol. 121, 1–11 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Key F. M., Fu Q., Romagné F., Lachmann M., Andrés A. M., Human adaptation and population differentiation in the light of ancient genomes. Nat. Commun. 7, 10775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rees J. S., Castellano S., Andrés A. M., The Genomics of Human Local Adaptation. Trends Genet. 36, 415–428 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Fan S., Hansen M. E. B., Lo Y., Tishkoff S. A., Going global by adapting local: A review of recent human adaptation. Science. 354, 54–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez M., Choin J., Sikora M., Siddle K., Harmant C., Costa H. A., Silvert M., Mouguiama-Daouda P., Hombert J.-M., Froment A., Le Bomin S., Perry G. H., Barreiro L. B., Bustamante C. D., Verdu P., Patin E., Quintana-Murci L., Genomic Evidence for Local Adaptation of Hunter-Gatherers to the African Rainforest. Curr. Biol. 29, 2926–2935.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Couto-Silva C. M., Nunes K., Venturini G., Araújo M. Silva Castro E, Pereira L. V., Comas D., Pereira A., Hünemeier T., Indigenous people from Amazon show genetic signatures of pathogen-driven selection. Sci Adv. 9, eabo0234 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison G. F., Sanz J., Boulais J., Mina M. J., Grenier J.-C., Leng Y., Dumaine A., Yotova V., Bergey C. M., Nsobya S. L., Elledge S. J., Schurr E., Quintana-Murci L., Perry G. H., Barreiro L. B., Natural selection contributed to immunological differences between hunter-gatherers and agriculturalists. Nat Ecol Evol. 3, 1253–1264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamid I., Korunes K. L., Beleza S., Goldberg A., Rapid adaptation to malaria facilitated by admixture in the human population of Cabo Verde. Elife. 10 (2021), doi: 10.7554/eLife.63177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kariuki S. N., Williams T. N., Human genetics and malaria resistance. Hum. Genet. 139, 801–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malaspinas A.-S., Westaway M. C., Muller C., Sousa V. C., Lao O., Alves I., Bergström A., Athanasiadis G., Cheng J. Y., Crawford J. E., Heupink T. H., Macholdt E., Peischl S., Rasmussen S., Schiffels S., Subramanian S., Wright J. L., Albrechtsen A., Barbieri C., Dupanloup I., Eriksson A., Margaryan A., Moltke I., Pugach I., Korneliussen T. S., Levkivskyi I. P., Moreno-Mayar J. V., Ni S., Racimo F., Sikora M., Xue Y., Aghakhanian F. A., Brucato N., Brunak S., Campos P. F., Clark W., Ellingvåg S., Fourmile G., Gerbault P., Injie D., Koki G., Leavesley M., Logan B., Lynch A., Matisoo-Smith E. A., McAllister P. J., Mentzer A. J., Metspalu M., Migliano A. B., Murgha L., Phipps M. E., Pomat W., Reynolds D., Ricaut F.-X., Siba P., Thomas M. G., Wales T., Wall C. M., Oppenheimer S. J., Tyler-Smith C., Durbin R., Dortch J., Manica A., Schierup M. H., Foley R. A., Lahr M. M., Bowern C., Wall J. D., Mailund T., Stoneking M., Nielsen R., Sandhu M. S., Excoffier L., Lambert D. M., Willerslev E., A genomic history of Aboriginal Australia. Nature. 538, 207–214 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Qi X., Chan W. L., Read R. J., Zhou A., Carrell R. W., Temperature-responsive release of thyroxine and its environmental adaptation in Australians. Proc. Biol. Sci. 281, 20132747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry G. H., Dominy N. J., Claw K. G., Lee A. S., Fiegler H., Redon R., Werner J., Villanea F. A., Mountain J. L., Misra R., Carter N. P., Lee C., Stone A. C., Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P., Jørgensen M. E., Korneliussen T. S., Gerbault P., Skotte L., Linneberg A., Christensen C., Brandslund I., Jørgensen T., Huerta-Sánchez E., Schmidt E. B., Pedersen O., Hansen T., Albrechtsen A., Nielsen R., Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 349, 1343–1347 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Evershed R. P., Davey Smith G., Roffet-Salque M., Timpson A., Diekmann Y., Lyon M. S., Cramp L. J. E., Casanova E., Smyth J., Whelton H. L., Dunne J., Brychova V., Šoberl L., Gerbault P., Gillis R. E., Heyd V., Johnson E., Kendall I., Manning K., Marciniak A., Outram A. K., Vigne J.-D., Shennan S., Bevan A., Colledge S., Allason-Jones L., Amkreutz L., Anders A., Arbogast R.-M., Bălăşescu A., Bánffy E., Barclay A., Behrens A., Bogucki P., Carrancho Alonso Á., Carretero J. M., Cavanagh N., Claßen E., Collado Giraldo H., Conrad M., Csengeri P., Czerniak L., Dębiec M., Denaire A., Domboróczki L., Donald C., Ebert J., Evans C., Francés-Negro M., Gronenborn D., Haack F., Halle M., Hamon C., Hülshoff R., Ilett M., Iriarte E., Jakucs J., Jeunesse C., Johnson M., Jones A. M., Karul N., Kiosak D., Kotova N., Krause R., Kretschmer S., Krüger M., Lefranc P., Lelong O., Lenneis E., Logvin A., Lüth F., Marton T., Marley J., Mortimer R., Oosterbeek L., Oross K., Pavúk J., Pechtl J., Pétrequin P., Pollard J., Pollard R., Powlesland D., Pyzel J., Raczky P., Richardson A., Rowe P., Rowland S., Rowlandson I., Saile T., Sebők K., Schier W., Schmalfuß G., Sharapova S., Sharp H., Sheridan A., Shevnina I., Sobkowiak-Tabaka I., Stadler P., Stäuble H., Stobbe A., Stojanovski D., Tasić N., van Wijk I., Vostrovská I., Vuković J., Wolfram S., Zeeb-Lanz A., Thomas M. G., Dairying, diseases and the evolution of lactase persistence in Europe. Nature. 608, 336–345 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jablonski N. G., Chaplin G., Human skin pigmentation as an adaptation to UV radiation. Proceedings of the National Academy of Sciences. 107, 8962–8968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfe N. D., Dunavan C. P., Diamond J., Origins of major human infectious diseases. Nature. 447, 279–283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira J. C., Huber C. D., The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. U. S. A. 118 (2021), doi: 10.1073/pnas.2015096118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flanagan S. P., Forester B. R., Latch E. K., Aitken S. N., Hoban S., Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evol. Appl. 11, 1035–1052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore J., 20• Savanna chimpanzees, referential models. Great ape societies, 275 (1996). [Google Scholar]
- 49.Marchant L. F., Wessling E. G., Lindshield S. M., Introduction to the Special Issue on Savanna Chimpanzees. Int. J. Primatol. 41, 767–774 (2020). [Google Scholar]
- 50.Potts R., Environmental hypotheses of hominin evolution. Am. J. Phys. Anthropol. Suppl 27, 93–136 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Domínguez-Rodrigo M., Is the “savanna hypothesis” a dead concept for explaining the emergence of the earliest hominins? Curr. Anthropol. (2014) (available at 10.1086/674530?casa_token=uKJG4VJc_EEAAAAA:3BIaHORr6254E2omxEQb0Sg-GLHocNI5Nfpag0j-jSUJ9NyHobwD_byUWCNheDsEyK_oFrKDIms). [DOI] [Google Scholar]
- 52.Drummond-Clarke R. C., Kivell T. L., Sarringhaus L., Stewart F. A., Humle T., Piel A. K., Wild chimpanzee behavior suggests that a savanna-mosaic habitat did not support the emergence of hominin terrestrial bipedalism. Sci Adv. 8, eadd9752 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chimpanzee Sequencing and Analysis Consortium, Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 437, 69–87 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Calvignac-Spencer S., Leendertz S. A. J., Gillespie T. R., Leendertz F. H., Wild great apes as sentinels and sources of infectious disease. Clin. Microbiol. Infect. 18, 521–527 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Azevedo D. S., Duarte J. L. C., Freitas C. F. G., Soares K. L., Sousa M. S., Sousa E. S. S., Lucena R. B., One Health Perspectives on New Emerging Viral Diseases in African Wild Great Apes. Pathogens. 10 (2021), doi: 10.3390/pathogens10101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keele B. F., Van Heuverswyn F., Li Y., Bailes E., Takehisa J., Santiago M. L., Bibollet-Ruche F., Chen Y., Wain L. V., Liegeois F., Loul S., Ngole E. M., Bienvenue Y., Delaporte E., Brookfield J. F. Y., Sharp P. M., Shaw G. M., Peeters M., Hahn B. H., Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 313, 523–526 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharp P. M., Plenderleith L. J., Hahn B. H., Ape Origins of Human Malaria. Annu. Rev. Microbiol. 74, 39–63 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharp P. M., Shaw G. M., Hahn B. H., Simian immunodeficiency virus infection of chimpanzees. J. Virol. 79, 3891–3902 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silvestri G., Paiardini M., Pandrea I., Lederman M. M., Sodora D. L., Understanding the benign nature of SIV infection in natural hosts. J. Clin. Invest. 117, 3148–3154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sodora D. L., Allan J. S., Apetrei C., Brenchley J. M., Douek D. C., Else J. G., Estes J. D., Hahn B. H., Hirsch V. M., Kaur A., Kirchhoff F., Muller-Trutwin M., Pandrea I., Schmitz J. E., Silvestri G., Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat. Med. 15, 861–865 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paiardini M., Pandrea I., Apetrei C., Silvestri G., Lessons learned from the natural hosts of HIV-related viruses. Annu. Rev. Med. 60, 485–495 (2009). [DOI] [PubMed] [Google Scholar]
- 62.de Manuel M., Kuhlwilm M., Frandsen P., Sousa V. C., Desai T., Prado-Martinez J., Hernandez-Rodriguez J., Dupanloup I., Lao O., Hallast P., Schmidt J. M., Heredia-Genestar J. M., Benazzo A., Barbujani G., Peter B. M., Kuderna L. F. K., Casals F., Angedakin S., Arandjelovic M., Boesch C., Kühl H., Vigilant L., Langergraber K., Novembre J., Gut M., Gut I., Navarro A., Carlsen F., Andrés A. M., Siegismund H. R., Scally A., Excoffier L., Tyler-Smith C., Castellano S., Xue Y., Hvilsom C., Marques-Bonet T., Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science. 354, 477–481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bataillon T., Duan J., Hvilsom C., Jin X., Li Y., Skov L., Glemin S., Munch K., Jiang T., Qian Y., Hobolth A., Wang J., Mailund T., Siegismund H. R., Schierup M. H., Inference of purifying and positive selection in three subspecies of chimpanzees (Pan troglodytes) from exome sequencing. Genome Biol. Evol. 7, 1122–1132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hvilsom C., Qian Y., Bataillon T., Li Y., Mailund T., Sallé B., Carlsen F., Li R., Zheng H., Jiang T., Jiang H., Jin X., Munch K., Hobolth A., Siegismund H. R., Wang J., Schierup M. H., Extensive X-linked adaptive evolution in central chimpanzees. Proc. Natl. Acad. Sci. U. S. A. 109, 2054–2059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt J. M., de Manuel M., Marques-Bonet T., Castellano S., Andrés A. M., The impact of genetic adaptation on chimpanzee subspecies differentiation. PLoS Genet. 15, e1008485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pawar H., Ostridge H. J., Schmidt J. M., Andrés A. M., Genetic adaptations to SIV across chimpanzee populations. PLoS Genet. 18, e1010337 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cagan A., Theunert C., Laayouni H., Santpere G., Pybus M., Casals F., Prüfer K., Navarro A., Marques-Bonet T., Bertranpetit J., Andrés A. M., Natural Selection in the Great Apes. Mol. Biol. Evol. 33, 3268–3283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nye J., Laayouni H., Kuhlwilm M., Mondal M., Marques-Bonet T., Bertranpetit J., Selection in the Introgressed Regions of the Chimpanzee Genome. Genome Biol. Evol. 10, 1132–1138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nye J., Mondal M., Bertranpetit J., Laayouni H., A fully integrated machine learning scan of selection in the chimpanzee genome. NAR Genom Bioinform. 2, lqaa061 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kovalaskas S., Rilling J. K., Lindo J., Comparative analyses of the Pan lineage reveal selection on gene pathways associated with diet and sociality in bonobos. Genes Brain Behav. 20, e12715 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Han S., Andrés A. M., Marques-Bonet T., Kuhlwilm M., Genetic Variation in Pan Species Is Shaped by Demographic History and Harbors Lineage-Specific Functions. Genome Biol. Evol. 11, 1178–1191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nam K., Munch K., Mailund T., Nater A., Greminger M. P., Krützen M., Marquès-Bonet T., Schierup M. H., Evidence that the rate of strong selective sweeps increases with population size in the great apes. Proc. Natl. Acad. Sci. U. S. A. 114, 1613–1618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nam K., Munch K., Hobolth A., Dutheil J. Y., Veeramah K. R., Woerner A. E., Hammer M. F., Great Ape Genome Diversity Project, Mailund T., Schierup M. H., Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc. Natl. Acad. Sci. U. S. A. 112, 6413–6418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hohenlohe P. A., Funk W. C., Rajora O. P., Population genomics for wildlife conservation and management. Mol. Ecol. (2021) (available at 10.1111/mec.15720). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White L. C., Fontsere C., Lizano E., Hughes D. A., Angedakin S., Arandjelovic M., Granjon A.-C., Hans J. B., Lester J. D., Rabanus-Wallace M. T., Rowney C., Städele V., Marques-Bonet T., Langergraber K. E., Vigilant L., A roadmap for high-throughput sequencing studies of wild animal populations using noninvasive samples and hybridization capture. Mol. Ecol. Resour. 19, 609–622 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Frandsen P., Fontsere C., Nielsen S. V., Hanghøj K., Castejon-Fernandez N., Lizano E., Hughes D., Hernandez-Rodriguez J., Korneliussen T. S., Carlsen F., Siegismund H. R., Mailund T., Marques-Bonet T., Hvilsom C., Targeted conservation genetics of the endangered chimpanzee. Heredity . 125, 15–27 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fontsere C., Alvarez-Estape M., Lester J., Arandjelovic M., Kuhlwilm M., Dieguez P., Agbor A., Angedakin S., Ayuk Ayimisin E., Bessone M., Brazzola G., Deschner T., Eno-Nku M., Granjon A.-C., Head J., Kadam P., Kalan A. K., Kambi M., Langergraber K., Lapuente J., Maretti G., Jayne Ormsby L., Piel A., Robbins M. M., Stewart F., Vergnes V., Wittig R. M., Kühl H. S., Marques-Bonet T., Hughes D. A., Lizano E., Maximizing the acquisition of unique reads in noninvasive capture sequencing experiments. Mol. Ecol. Resour. 21, 745–761 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Ozga A. T., Webster T. H., Gilby I. C., Wilson M. A., Nockerts R. S., Wilson M. L., Pusey A. E., Li Y., Hahn B. H., Stone A. C., Urine as a high-quality source of host genomic DNA from wild populations. Mol. Ecol. Resour. 21, 170–182 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perry G. H., Marioni J. C., Melsted P., Gilad Y., Genomic-scale capture and sequencing of endogenous DNA from feces. Mol. Ecol. (2010) (available at 10.1111/j.1365-294X.2010.04888.x?casa_token=iUVLfym_ND8AAAAA:K-T_LBMjybL0aGTbLxO0bqKFgCJn-MMIXcFfMQKwOt0Ozf74EmBuCrElhFHBM_JdPtX5j9A67AlYg_0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hernandez-Rodriguez J., Arandjelovic M., Lester J., de Filippo C., Weihmann A., Meyer M., Angedakin S., Casals F., Navarro A., Vigilant L., Others, The impact of endogenous content, replicates and pooling on genome capture from faecal samples. Mol. Ecol. Resour. 18, 319–333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lester J. D., Vigilant L., Gratton P., McCarthy M. S., Barratt C. D., Dieguez P., Agbor A., Álvarez-Varona P., Angedakin S., Ayimisin E. A., Bailey E., Bessone M., Brazzola G., Chancellor R., Cohen H., Danquah E., Deschner T., Egbe V. E., Eno-Nku M., Goedmakers A., Granjon A.-C., Head J., Hedwig D., Hernandez-Aguilar R. A., Jeffery K. J., Jones S., Junker J., Kadam P., Kaiser M., Kalan A. K., Kehoe L., Kienast I., Langergraber K. E., Lapuente J., Laudisoit A., Lee K., Marrocoli S., Mihindou V., Morgan D., Muhanguzi G., Neil E., Nicholl S., Orbell C., Ormsby L. J., Pacheco L., Piel A., Robbins M. M., Rundus A., Sanz C., Sciaky L., Siaka A. M., Städele V., Stewart F., Tagg N., Ton E., van Schijndel J., Vyalengerera M. K., Wessling E. G., Willie J., Wittig R. M., Yuh Y. G., Yurkiw K., Zuberbuehler K., Boesch C., Kühl H. S., Arandjelovic M., Recent genetic connectivity and clinal variation in chimpanzees. Commun Biol. 4, 283 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Besenbacher S., Hvilsom C., Marques-Bonet T., Mailund T., Schierup M. H., Direct estimation of mutations in great apes reconciles phylogenetic dating. Nat Ecol Evol. 3, 286–292 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Gautier M., Genome-Wide Scan for Adaptive Divergence and Association with Population-Specific Covariates. Genetics. 201, 1555–1579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olazcuaga L., Loiseau A., Parrinello H., Paris M., Fraimout A., Guedot C., Diepenbrock L. M., Kenis M., Zhang J., Chen X., Borowiec N., Facon B., Vogt H., Price D. K., Vogel H., Prud’homme B., Estoup A., Gautier M., A Whole-Genome Scan for Association with Invasion Success in the Fruit Fly Drosophila suzukii Using Contrasts of Allele Frequencies Corrected for Population Structure. Mol. Biol. Evol. 37, 2369–2385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahrens C. W., Rymer P. D., Stow A., Bragg J., Dillon S., Umbers K. D. L., Dudaniec R. Y., The search for loci under selection: trends, biases and progress. Mol. Ecol. 27, 1342–1356 (2018). [DOI] [PubMed] [Google Scholar]
- 86.De Mita S., Thuillet A.-C., Gay L., Ahmadi N., Manel S., Ronfort J., Vigouroux Y., Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol. Ecol. 22, 1383–1399 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Aleman J. C., Fayolle A., Favier C., Staver A. C., Dexter K. G., Ryan C. M., Azihou A. F., Bauman D., Te Beest M., Chidumayo E. N., Comiskey J. A., Cromsigt J. P. G. M., Dessard H., Doucet J.-L., Finckh M., Gillet J.-F., Gourlet-Fleury S., Hempson G. P., Holdo R. M., Kirunda B., Kouame F. N., Mahy G., Gonçalves F. M. P., McNicol I., Quintano P. N., Plumptre A. J., Pritchard R. C., Revermann R., Schmitt C. B., Swemmer A. M., Talila H., Woollen E., Swaine M. D., Floristic evidence for alternative biome states in tropical Africa. Proc. Natl. Acad. Sci. U. S. A. 117, 28183–28190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeffreys H., Theory of Probability, Ed. 3 Oxford University Press; (1961). [Google Scholar]
- 89.Kordonowy L., MacManes M., Characterizing the reproductive transcriptomic correlates of acute dehydration in males in the desert-adapted rodent, Peromyscus eremicus. BMC Genomics. 18, 473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Giorello F. M., Feijoo M., D’Elía G., Naya D. E., Valdez L., Opazo J. C., Lessa E. P., An association between differential expression and genetic divergence in the Patagonian olive mouse (Abrothrix olivacea). Mol. Ecol. (2018), doi: 10.1111/mec.14778. [DOI] [PubMed] [Google Scholar]
- 91.Rudicell R. S., Piel A. K., Stewart F., Moore D. L., Learn G. H., Li Y., Takehisa J., Pintea L., Shaw G. M., Moore J., Sharp P. M., Hahn B. H., High prevalence of simian immunodeficiency virus infection in a community of savanna chimpanzees. J. Virol. 85, 9918–9928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Y., Ndjango J.-B., Learn G. H., Ramirez M. A., Keele B. F., Bibollet-Ruche F., Liu W., Easlick J. L., Decker J. M., Rudicell R. S., Inogwabini B.-I., Ahuka-Mundeke S., Leendertz F. H., Reynolds V., Muller M. N., Chancellor R. L., Rundus A. S., Simmons N., Worobey M., Shaw G. M., Peeters M., Sharp P. M., Hahn B. H., Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J. Virol. 86, 10776–10791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klunk J., Vilgalys T. P., Demeure C. E., Cheng X., Shiratori M., Madej J., Beau R., Elli D., Patino M. I., Redfern R., DeWitte S. N., Gamble J. A., Boldsen J. L., Carmichael A., Varlik N., Eaton K., Grenier J.-C., Golding G. B., Devault A., Rouillard J.-M., Yotova V., Sindeaux R., Ye C. J., Bikaran M., Dumaine A., Brinkworth J. F., Missiakas D., Rouleau G. A., Steinrücken M., Pizarro-Cerdá J., Poinar H. N., Barreiro L. B., Evolution of immune genes is associated with the Black Death. Nature. 611, 312–319 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Deschamps M., Laval G., Fagny M., Itan Y., Abel L., Casanova J.-L., Patin E., Quintana-Murci L., Genomic Signatures of Selective Pressures and Introgression from Archaic Hominins at Human Innate Immunity Genes. Am. J. Hum. Genet. 98, 5–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fumagalli M., Sironi M., Pozzoli U., Ferrer-Admetlla A., Pattini L., Nielsen R., Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 7, e1002355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daub J. T., Hofer T., Cutivet E., Dupanloup I., Quintana-Murci L., Robinson-Rechavi M., Excoffier L., Evidence for polygenic adaptation to pathogens in the human genome. Mol. Biol. Evol. 30, 1544–1558 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Ebel E. R., Kuypers F. A., Lin C., Petrov D. A., Egan E. S., Common host variation drives malaria parasite fitness in healthy human red cells. eLife. 10 (2021), , doi: 10.7554/elife.69808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ebel E. R., Telis N., Venkataram S., Petrov D. A., Enard D., High rate of adaptation of mammalian proteins that interact with Plasmodium and related parasites. PLoS Genet. 13, e1007023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hansen M. C., Potapov P. V., Moore R., Hancher M., Turubanova S. A., Tyukavina A., Thau D., Stehman S. V., Goetz S. J., Loveland T. R., Kommareddy A., Egorov A., Chini L., Justice C. O., Townshend J. R. G., High-resolution global maps of 21st-century forest cover change. Science. 342, 850–853 (2013). [DOI] [PubMed] [Google Scholar]
- 100.M. G. E. Network, Malaria Genomic Epidemiology Network, Band G., Le Q. S., Clarke G. M., Kivinen K., Hubbart C., Jeffreys A. E., Rowlands K., Leffler E. M., Jallow M., Conway D. J., Sisay-Joof F., Sirugo G., d’Alessandro U., Toure O. B., Thera M. A., Konate S., Sissoko S., Mangano V. D., Bougouma E. C., Sirima S. B., Amenga-Etego L. N., Ghansah A. K., Hodgson A. V. O., Wilson M. D., Enimil A., Ansong D., Evans J., Ademola S. A., Apinjoh T. O., Ndila C. M., Manjurano A., Drakeley C., Reyburn H., Phu N. H., Quyen N. T. N., Thai C. Q., Hien T. T., Teo Y. Y., Manning L., Laman M., Michon P., Karunajeewa H., Siba P., Allen S., Allen A., Bahlo M., Davis T. M. E., Simpson V., Shelton J., Spencer C. C. A., Busby G. B. J., Kerasidou A., Drury E., Stalker J., Dilthey A., Mentzer A. J., McVean G., Bojang K. A., Doumbo O., Modiano D., Koram K. A., Agbenyega T., Amodu O. K., Achidi E., Williams T. N., Marsh K., Riley E. M., Molyneux M., Taylor T., Dunstan S. J., Farrar J., Mueller I., Rockett K. A., Kwiatkowski D. P., Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa, Asia and Oceania. Nature Communications. 10 (2019), , doi: 10.1038/s41467-019-13480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ebel E. R., Uricchio L. H., Petrov D. A., Egan E. S., Revisiting the malaria hypothesis: accounting for polygenicity and pleiotropy. Trends Parasitol. 38, 290–301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allison A. C., Protection afforded by sickle-cell trait against subtertian malareal infection. Br. Med. J. 1, 290–294 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pauling L., Itano H. A., Sickle cell anemia a molecular disease. Science. 110, 543–548 (1949). [DOI] [PubMed] [Google Scholar]
- 104.Leffler E. M., Band G., Busby G. B. J., Kivinen K., Le Q. S., Clarke G. M., Bojang K. A., Conway D. J., Jallow M., Sisay-Joof F., Bougouma E. C., Mangano V. D., Modiano D., Sirima S. B., Achidi E., Apinjoh T. O., Marsh K., Ndila C. M., Peshu N., Williams T. N., Drakeley C., Manjurano A., Reyburn H., Riley E., Kachala D., Molyneux M., Nyirongo V., Taylor T., Thornton N., Tilley L., Grimsley S., Drury E., Stalker J., Cornelius V., Hubbart C., Jeffreys A. E., Rowlands K., Rockett K. A., Spencer C. C. A., Kwiatkowski D. P., Malaria Genomic Epidemiology Network, Resistance to malaria through structural variation of red blood cell invasion receptors. Science. 356 (2017), doi: 10.1126/science.aam6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Algady W., Louzada S., Carpenter D., Brajer P., Färnert A., Rooth I., Ngasala B., Yang F., Shaw M.-A., Hollox E. J., The Malaria-Protective Human Glycophorin Structural Variant DUP4 Shows Somatic Mosaicism and Association with Hemoglobin Levels. Am. J. Hum. Genet. 103, 769–776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Steinberg M. H., Adams J. G. 3rd, Hemoglobin A2: origin, evolution, and aftermath. Blood. 78, 2165–2177 (1991). [PubMed] [Google Scholar]
- 107.Jaskiewicz E., Jodłowska M., Kaczmarek R., Zerka A., Erythrocyte glycophorins as receptors for Plasmodium merozoites. Parasit. Vectors. 12, 317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kariuki S. N., Marin-Menendez A., Introini V., Ravenhill B. J., Lin Y.-C., Macharia A., Makale J., Tendwa M., Nyamu W., Kotar J., Carrasquilla M., Rowe J. A., Rockett K., Kwiatkowski D., Weekes M. P., Cicuta P., Williams T. N., Rayner J. C., Red blood cell tension protects against severe malaria in the Dantu blood group. Nature. 585, 579–583 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gassner C., Denomme G. A., Portmann C., Bensing K. M., Mattle-Greminger M. P., Meyer S., Trost N., Song Y.-L., Engström C., Jungbauer C., Just B., Storry J. R., Forster M., Franke A., Frey B. M., Two prevalent ∼100-kb GYPB deletions causative of the GPB-deficient blood group MNS phenotype S-s-U- in black Africans. Transfus. Med. Hemother. 47, 326–336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tokunaga E., Sasakawa S., Tamaka K., Kawamata H., Giles C. M., Ikin E. W., Poole J., Anstee D. J., Mawby W., Tanner M. J., Two apparently healthy Japanese individuals of type MkMk have erythrocytes which lack both the blood group MN and Ss-active sialoglycoproteins. J. Immunogenet. 6, 383–390 (1979). [DOI] [PubMed] [Google Scholar]
- 111.Hadley T. J., Klotz F. W., Pasvol G., Haynes J. D., McGinniss M. H., Okubo Y., Miller L. H., Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk). Strain differences indicate receptor heterogeneity and two pathways for invasion. J. Clin. Invest. 80, 1190–1193 (October 1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hollox E. J., Louzada S., Genetic variation of glycophorins and infectious disease. Immunogenetics. 75, 201–206 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Soto D. C., Shew C., Mastoras M., Schmidt J. M., Sahasrabudhe R., Kaya G., Andrés A. M., Dennis M. Y., Identification of structural variation in chimpanzees using optical mapping and nanopore sequencing. Genes . 11, 276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kronenberg Z. N., Fiddes I. T., Gordon D., Murali S., Cantsilieris S., Meyerson O. S., Underwood J. G., Nelson B. J., Chaisson M. J. P., Dougherty M. L., Munson K. M., Hastie A. R., Diekhans M., Hormozdiari F., Lorusso N., Hoekzema K., Qiu R., Clark K., Raja A., Welch A. E., Sorensen M., Baker C., Fulton R. S., Armstrong J., Graves-Lindsay T. A., Denli A. M., Hoppe E. R., Hsieh P., Hill C. M., Pang A. W. C., Lee J., Lam E. T., Dutcher S. K., Gage F. H., Warren W. C., Shendure J., Haussler D., Schneider V. A., Cao H., Ventura M., Wilson R. K., Paten B., Pollen A., Eichler E. E., High-resolution comparative analysis of great ape genomes. Science. 360 (2018), doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bigham A. W., Magnaye K., Dunn D. M., Weiss R. B., Bamshad M., Complex signatures of natural selection at GYPA. Hum. Genet. 137, 151–160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nagy E., Maquat L. E., A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–199 (1998). [DOI] [PubMed] [Google Scholar]
- 117.Pritchard J. K., Pickrell J. K., Coop G., The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20, R208–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schrider D. R., Kern A. D., Soft Sweeps Are the Dominant Mode of Adaptation in the Human Genome. Mol. Biol. Evol. 34, 1863–1877 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Whiten A., Goodall J., McGrew W. C., Nishida T., Reynolds V., Sugiyama Y., Tutin C. E., Wrangham R. W., Boesch C., Cultures in chimpanzees. Nature. 399, 682–685 (1999). [DOI] [PubMed] [Google Scholar]
- 120.Whitehead H., Laland K. N., Rendell L., Thorogood R., Whiten A., The reach of gene-culture coevolution in animals. Nat. Commun. 10, 2405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gilardi K., Gillespie T., Leendertz F., Macfie E., Travis D., Whittier C., Williamson E., Best practice guidelines for health monitoring and disease control in great ape populations (IUCN/SSC Primate Specialist Group; ) (2015). [Google Scholar]
- 122.Rayner J. C., Liu W., Peeters M., Sharp P. M., Hahn B. H., A plethora of Plasmodium species in wild apes: a source of human infection? Trends Parasitol. 27, 222–229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Snow R. W., Global malaria eradication and the importance of Plasmodium falciparum epidemiology in Africa. BMC Med. 13, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.De Nys H. M., Löhrich T., Wu D., Calvignac-Spencer S., Leendertz F. H., Wild African great apes as natural hosts of malaria parasites: current knowledge and research perspectives. Primate Biol. 4, 47–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rodhain J., Van Den Berghe L., Contribution à l’étude des plasmodiums des singes africains. Ann. Soc. Belg. Med. Trop. [Google Scholar]
- 126.Rodhain J., Others, “Plasmadia of Central African Apes and their Relationship to Human Plasmodia” in Annales de la Societe Belge de Medecine Tropicale (Societe Belge de Medecine Tropicale, 1940; https://www.cabdirect.org/cabdirect/abstract/19432900605), vol. 20. [Google Scholar]
- 127.Hayakawa T., Arisue N., Udono T., Hirai H., Sattabongkot J., Toyama T., Tsuboi T., Horii T., Tanabe K., Identification of Plasmodium malariae, a human malaria parasite, in imported chimpanzees. PLoS One. 4, e7412 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Herbert A., Boundenga L., Meyer A., Moukodoum D. N., Okouga A. P., Arnathau C., Durand P., Magnus J., Ngoubangoye B., Willaume E., Ba C. T., Rougeron V., Renaud F., Ollomo B., Prugnolle F., Malaria-like symptoms associated with a natural Plasmodium reichenowi infection in a chimpanzee. Malar. J. 14, 220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.de Groot N. G., Heijmans C. M. C., Bontrop R. E., AIDS in chimpanzees: the role of MHC genes. Immunogenetics. 69, 499–509 (2017). [DOI] [PubMed] [Google Scholar]
- 130.Gilden R. V., Arthur L. O., Robey W. G., Kelliher J. C., Graham C. E., Fischinger P. J., HTLV-III antibody in a breeding chimpanzee not experimentally exposed to the virus. Lancet. 1, 678–679 (1986). [DOI] [PubMed] [Google Scholar]
- 131.Rudicell R. S., Holland Jones J., Wroblewski E. E., Learn G. H., Li Y., Robertson J. D., Greengrass E., Grossmann F., Kamenya S., Pintea L., Mjungu D. C., Lonsdorf E. V., Mosser A., Lehman C., Collins D. A., Keele B. F., Goodall J., Hahn B. H., Pusey A. E., Wilson M. L., Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 6, e1001116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.De Nys H. M., Calvignac-Spencer S., Thiesen U., Boesch C., Wittig R. M., Mundry R., Leendertz F. H., Age-related effects on malaria parasite infection in wild chimpanzees. Biol. Lett. 9, 20121160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Nys H. M., Calvignac-Spencer S., Boesch C., Dorny P., Wittig R. M., Mundry R., Leendertz F. H., Malaria parasite detection increases during pregnancy in wild chimpanzees. Malar. J. 13, 413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.World Health Organization, World malaria report 2022 (World Health Organization, 2022; https://play.google.com/store/books/details?id=ST-hEAAAQBAJ). [Google Scholar]
- 135.Kühl H. S., Boesch C., Kulik L., Haas F., Arandjelovic M., Dieguez P., Bocksberger G., McElreath M. B., Agbor A., Angedakin S., Ayimisin E. A., Bailey E., Barubiyo D., Bessone M., Brazzola G., Chancellor R., Cohen H., Coupland C., Danquah E., Deschner T., Dowd D., Dunn A., Egbe V. E., Eshuis H., Goedmakers A., Granjon A.-C., Head J., Hedwig D., Hermans V., Imong I., Jeffery K. J., Jones S., Junker J., Kadam P., Kambere M., Kambi M., Kienast I., Kujirakwinja D., Langergraber K. E., Lapuente J., Larson B., Lee K., Leinert V., Llana M., Maretti G., Marrocoli S., Martin R., Mbi T. J., Meier A. C., Morgan B., Morgan D., Mulindahabi F., Murai M., Neil E., Niyigaba P., Ormsby L. J., Orume R., Pacheco L., Piel A., Preece J., Regnaut S., Rundus A., Sanz C., van Schijndel J., Sommer V., Stewart F., Tagg N., Vendras E., Vergnes V., Welsh A., Wessling E. G., Willie J., Wittig R. M., Yuh Y. G., Yurkiw K., Zuberbühler K., Kalan A. K., Human impact erodes chimpanzee behavioral diversity. Science. 363, 1453–1455 (2019). [DOI] [PubMed] [Google Scholar]
- 136.Carvalho S., Wessling E. G., Abwe E. E., Almeida-Warren K., Arandjelovic M., Boesch C., Danquah E., Diallo M. S., Hobaiter C., Hockings K., Humle T., Ikemeh R. A., Kalan A. K., Luncz L., Ohashi G., Pascual-Garrido A., Piel A., Samuni L., Soiret S., Sanz C., Koops K., Using nonhuman culture in conservation requires careful and concerted action. Conserv. Lett. 15 (2022), doi: 10.1111/conl.12860. [DOI] [Google Scholar]
- 137.Laporte N. T., Stabach J. A., Grosch R., Lin T. S., Goetz S. J., Expansion of industrial logging in Central Africa. Science. 316, 1451 (2007). [DOI] [PubMed] [Google Scholar]
- 138.Junker J., Blake S., Boesch C., Campbell G., du Toit L., Duvall C., Ekobo A., Etoga G., Galat-Luong A., Gamys J., Ganas-Swaray J., Gatti S., Ghiurghi A., Granier N., Hart J., Head J., Herbinger I., Hicks T. C., Huijbregts B., Imong I. S., Kuempel N., Lahm S., Lindsell J., Maisels F., McLennan M., Martinez L., Morgan B., Morgan D., Mulindahabi F., Mundry R., N’Goran K. P., Normand E., Ntongho A., Okon D. T., Petre C.-A., Plumptre A., Rainey H., Regnaut S., Sanz C., Stokes E., Tondossama A., Tranquilli S., Sunderland-Groves J., Walsh P., Warren Y., Williamson E. A., Kuehl H. S., Recent decline in suitable environmental conditions for African great apes. Divers. Distrib. 18, 1077–1091 (2012). [Google Scholar]
- 139.Graham T. L., Matthews H. D., Turner S. E., A global-scale evaluation of primate exposure and vulnerability to climate change. Int. J. Primatol. 37, 158–174 (2016). [Google Scholar]
- 140.Köndgen S., Kühl H., N’Goran P. K., Walsh P. D., Schenk S., Ernst N., Biek R., Formenty P., Mätz-Rensing K., Schweiger B., Junglen S., Ellerbrok H., Nitsche A., Briese T., Lipkin W. I., Pauli G., Boesch C., Leendertz F. H., Pandemic human viruses cause decline of endangered great apes. Curr. Biol. 18, 260–264 (2008). [DOI] [PubMed] [Google Scholar]
- 141.Carøe C., Gopalakrishnan S., Vinner L., Mak S. S. T., Sinding M. H. S., Samaniego J. A., Wales N., Sicheritz-Pontén T., Gilbert M. T. P., Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 9, 410–419 (2018). [Google Scholar]
- 142.Meisner J., Albrechtsen A., Inferring Population Structure and Admixture Proportions in Low-Depth NGS Data. Genetics. 210, 719–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kuhlwilm M., Fontsere C., Han S., Alvarez-Estape M., Marques-Bonet T., HuConTest: Testing Human Contamination in Great Ape Samples. Genome Biol. Evol. 13 (2021), doi: 10.1093/gbe/evab117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hanghøj K., Moltke I., Andersen P. A., Manica A., Korneliussen T. S., Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. Gigascience. 8 (2019), doi: 10.1093/gigascience/giz034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Skotte L., Korneliussen T. S., Albrechtsen A., Estimating individual admixture proportions from next generation sequencing data. Genetics. 195, 693–702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Quinlan A. R., Hall I. M., BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stekhoven D. J., Bühlmann P., MissForest--non-parametric missing value imputation for mixed-type data. Bioinformatics. 28, 112–118 (2012). [DOI] [PubMed] [Google Scholar]
- 149.Kofler R., Schlötterer C., Gowinda: unbiased analysis of gene set enrichment for genome-wide association studies. Bioinformatics. 28, 2084–2085 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G., Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fabregat A., Korninger F., Viteri G., Sidiropoulos K., Marin-Garcia P., Ping P., Wu G., Stein L., D’Eustachio P., Hermjakob H., Reactome graph database: Efficient access to complex pathway data. PLOS Computational Biology. 14 (2018), p. e1005968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Buniello A., MacArthur J. A. L., Cerezo M., Harris L. W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., Suveges D., Vrousgou O., Whetzel P. L., Amode R., Guillen J. A., Riat H. S., Trevanion S. J., Hall P., Junkins H., Flicek P., Burdett T., Hindorff L. A., Cunningham F., Parkinson H., The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ramos E. M., Hoffman D., Junkins H. A., Maglott D., Phan L., Sherry S. T., Feolo M., Hindorff L. A., Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Hum. Genet. 22, 144–147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Karlsson M., Zhang C., Méar L., Zhong W., Digre A., Katona B., Sjöstedt E., Butler L., Odeberg J., Dusart P., Edfors F., Oksvold P., von Feilitzen K., Zwahlen M., Arif M., Altay O., Li X., Ozcan M., Mardinoglu A., Fagerberg L., Mulder J., Luo Y., Ponten F., Uhlén M., Lindskog C., A single-cell type transcriptomics map of human tissues. Sci Adv. 7 (2021), doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Enard D., Cai L., Gwennap C., Petrov D. A., Viruses are a dominant driver of protein adaptation in mammals. eLife. 5 (2016), , doi: 10.7554/elife.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Enard D., Petrov D. A., Evidence that RNA Viruses Drove Adaptive Introgression between Neanderthals and Modern Humans. Cell. 175, 360–371.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Svardal H., Jasinska A. J., Apetrei C., Coppola G., Huang Y., Schmitt C. A., Jacquelin B., Ramensky V., Müller-Trutwin M., Antonio M., Weinstock G., Grobler J. P., Dewar K., Wilson R. K., Turner T. R., Warren W. C., Freimer N. B., Nordborg M., Ancient hybridization and strong adaptation to viruses across African vervet monkey populations. Nat. Genet. 49, 1705–1713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jacquelin B., Petitjean G., Kunkel D., Liovat A.-S., Jochems S. P., Rogers K. A., Ploquin M. J., Madec Y., Barré-Sinoussi F., Dereuddre-Bosquet N., Lebon P., Le Grand R., Villinger F., Müller-Trutwin M., Innate immune responses and rapid control of inflammation in African green monkeys treated or not with interferon-alpha during primary SIVagm infection. PLoS Pathog. 10, e1004241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jacquelin B., Mayau V., Targat B., Liovat A.-S., Kunkel D., Petitjean G., Dillies M.-A., Roques P., Butor C., Silvestri G., Giavedoni L. D., Lebon P., Barré-Sinoussi F., Benecke A., Müller-Trutwin M. C., Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. Journal of Clinical Investigation (2009), , doi: 10.1172/jci40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rogers L. R. K., de Los Campos G., Mias G. I., Microarray Gene Expression Dataset Re-analysis Reveals Variability in Influenza Infection and Vaccination. Front. Immunol. 10, 2616 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C., SARS-CoV-2 infected host cell proteomics reveal potential therapy targets (2020) (available at https://www.researchsquare.com/article/rs-17218/latest.pdf). [DOI] [PubMed] [Google Scholar]
- 163.Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., O’Meara M. J., Guo J. Z., Swaney D. L., Tummino T. A., Hüttenhain R., Kaake R. M., Richards A. L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B. J., Braberg H., Fabius J. M., Eckhardt M., Soucheray M., Bennett M. J., Cakir M., McGregor M. J., Li Q., Naing Z. Z. C., Zhou Y., Peng S., Kirby I. T., Melnyk J. E., Chorba J. S., Lou K., Dai S. A., Shen W., Shi Y., Zhang Z., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Mathy C. J. P., Perica T., Pilla K. B., Ganesan S. J., Saltzberg D. J., Ramachandran R., Liu X., Rosenthal S. B., Calviello L., Venkataramanan S., Lin Y., Wankowicz S. A., Bohn M., Trenker R., Young J. M., Cavero D., Hiatt J., Roth T., Rathore U., Subramanian A., Noack J., Hubert M., Roesch F., Vallet T., Meyer B., White K. M., Miorin L., Agard D., Emerman M., Ruggero D., García-Sastre A., Jura N., von Zastrow M., Taunton J., Schwartz O., Vignuzzi M., d’Enfert C., Mukherjee S., Jacobson M., Malik H. S., Fujimori D. G., Ideker T., Craik C. S., Floor S., Fraser J. S., Gross J., Sali A., Kortemme T., Beltrao P., Shokat K., Shoichet B. K., Krogan N. J., A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. bioRxiv (2020), doi: 10.1101/2020.03.22.002386. [DOI] [Google Scholar]
- 164.Blanco-Melo D., Nilsson-Payant B. E., Liu W.-C., Møller R., Panis M., Sachs D., Albrecht R. A., tenOever B. R., SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv (2020), p. 2020.03.24.004655. [Google Scholar]
- 165.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D., Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses. 12 (2020), doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A. A., Zimmerman M. G., Bedoya S., Aoued H., Tharp G. M., Pellegrini K. L., Manfredi C., Sorscher E., Mainou B., Lobby J. L., Kohlmeier J. E., Lowen A. C., Shi P.-Y., Menachery V. D., Anderson L. J., Grakoui A., Bosinger S. E., Suthar M. S., Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 94 (2020), doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Drayman N., Patel P., Vistain L., Tay S., HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations. Elife. 8 (2019), doi: 10.7554/eLife.46339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Booth J. L., Duggan E. S., Patel V. I., Wu W., Burian D. M., Hutchings D. C., White V. L., Coggeshall K. M., Dozmorov M. G., Metcalf J. P., Gene expression profiling of primary human type I alveolar epithelial cells exposed to Bacillus anthracis spores reveals induction of neutrophil and monocyte chemokines. Microb. Pathog. 121, 9–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fontsere C., Frandsen P., Hernandez-Rodriguez J., Niemann J., Scharff-Olsen C. H., Vallet D., Le Gouar P., Ménard N., Navarro A., Siegismund H. R., Hvilsom C., Gilbert M. T. P., Kuhlwilm M., Hughes D., Marques-Bonet T., The genetic impact of an Ebola outbreak on a wild gorilla population. BMC Genomics. 22, 735 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shafin K., Pesout T., Lorig-Roach R., Haukness M., Olsen H. E., Bosworth C., Armstrong J., Tigyi K., Maurer N., Koren S., Sedlazeck F. J., Marschall T., Mayes S., Costa V., Zook J. M., Liu K. J., Kilburn D., Sorensen M., Munson K. M., Vollger M. R., Monlong J., Garrison E., Eichler E. E., Salama S., Haussler D., Green R. E., Akeson M., Phillippy A., Miga K. H., Carnevali P., Jain M., Paten B., Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. (2020), doi: 10.1038/s41587-020-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shumate A., Salzberg S. L., Liftoff: accurate mapping of gene annotations. Bioinformatics. 37, 1639–1643 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pendleton A. L., Shen F., Taravella A. M., Emery S., Veeramah K. R., Boyko A. R., Kidd J. M., Comparison of village dog and wolf genomes highlights the role of the neural crest in dog domestication. BMC Biol. 16, 64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hach F., Sarrafi I., Hormozdiari F., Alkan C., Eichler E. E., Sahinalp S. C., mrsFAST-Ultra: a compact, SNP-aware mapper for high performance sequencing applications. Nucleic Acids Res. 42, W494–500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Poplin R., Ruano-Rubio V., DePristo M. A., Fennell T. J., Carneiro M. O., Van der Auwera G. A., Kling D. E., Gauthier L. D., Levy-Moonshine A., Roazen D., Shakir K., Thibault J., Chandran S., Whelan C., Lek M., Gabriel S., Daly M. J., Neale B., MacArthur D. G., Banks E., Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv (2017), , doi: 10.1101/201178. [DOI] [Google Scholar]
- 175.Wang K., Li M., Hakonarson H., ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chang X., Wang K., wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 49, 433–436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bolger A. M., Lohse M., Usadel B., Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.1000 Genomes Project Consortium, Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., Marchini J. L., McCarthy S., McVean G. A., Abecasis G. R., A global reference for human genetic variation. Nature. 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jacquard A., The Genetic Structure of Populations (Springer Science & Business Media, 2012; https://play.google.com/store/books/details?id=jsrsCAAAQBAJ). [Google Scholar]
- 181.Ahrens C. W., Jordan R., Bragg J., Regarding the F-word: The effects of data filtering on inferred genotype-environment associations. Mol. Ecol. (2021) (available at 10.1111/1755-0998.13351?casa_token=ucpfMkTimdgAAAAA:iWFoMmXxnynGIbdl2gQvf9BjZLm-nNdulqUpv1a6GQPbBBfJUcZFL5NUfy0Ds3WH87KWmdOHh2NPVhY). [DOI] [PubMed] [Google Scholar]
- 182.Mank J. E., Vicoso B., Berlin S., Charlesworth B., Effective population size and the Faster-X effect: empirical results and their interpretation. Evolution. 64, 663–674 (2010). [DOI] [PubMed] [Google Scholar]
- 183.Tataru P., Mollion M., Glémin S., Bataillon T., Inference of Distribution of Fitness Effects and Proportion of Adaptive Substitutions from Polymorphism Data. Genetics. 207, 1103–1119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Förstner W., Moonen B., “A Metric for Covariance Matrices” in Geodesy-The Challenge of the 3rd Millennium, Grafarend E. W., Krumm F. W., Schwarze V. S., Eds. (Springer Berlin Heidelberg, Berlin, Heidelberg, 2003; 10.1007/978-3-662-05296-9_31), pp. 299–309. [DOI] [Google Scholar]
- 185.Frachon L., Bartoli C., Carrère S., Bouchez O., Chaubet A., Gautier M., Roby D., Roux F., A Genomic Map of Climate Adaptation in Arabidopsis thaliana at a Micro-Geographic Scale. Front. Plant Sci. 9, 967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gao Y., Gautier M., Ding X., Zhang H., Wang Y., Wang X., Faruque M. O., Li J., Ye S., Gou X., Han J., Lenstra J. A., Zhang Y., Species composition and environmental adaptation of indigenous Chinese cattle. Sci. Rep. 7, 16196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Eoche-Bosy D., Gautier M., Esquibet M., Legeai F., Bretaudeau A., Bouchez O., Fournet S., Grenier E., Montarry J., Genome scans on experimentally evolved populations reveal candidate regions for adaptation to plant resistance in the potato cyst nematode Globodera pallida. Mol. Ecol. 26, 4700–4711 (2017). [DOI] [PubMed] [Google Scholar]
- 188.Rohfritsch A., Galan M., Gautier M., Gharbi K., Olsson G., Gschloessl B., Zeimes C., VanWambeke S., Vitalis R., Charbonnel N., Preliminary insights into the genetics of bank vole tolerance to Puumala hantavirus in Sweden. Ecol. Evol. 8, 11273–11292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Luikart G., England P. R., Tallmon D., Jordan S., Taberlet P., The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4, 981–994 (2003). [DOI] [PubMed] [Google Scholar]
- 190.Günther T., Coop G., Robust identification of local adaptation from allele frequencies. Genetics. 195, 205–220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Weir B. S., Cockerham C. C., ESTIMATING F-STATISTICS FOR THE ANALYSIS OF POPULATION STRUCTURE. Evolution. 38, 1358–1370 (1984). [DOI] [PubMed] [Google Scholar]
- 192.Hennig C., fpc: flexible procedures for clustering (Version 2.2–9). Bologna BO: UNIBO. [Google Scholar]
- 193.R. Core Team, R: A language and environment for statistical computing. Version 3.6. 0. Vienna, Austria. /ra-language-and-environment-forstatistical-computing. [Google Scholar]
- 194.Todesco M., Owens G. L., Bercovich N., Légaré J.-S., Soudi S., Burge D. O., Huang K., Ostevik K. L., Drummond E. B. M., Imerovski I., Lande K., Pascual-Robles M. A., Nanavati M., Jahani M., Cheung W., Staton S. E., Muños S., Nielsen R., Donovan L. A., Burke J. M., Yeaman S., Rieseberg L. H., Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. 584, 602–607 (2020). [DOI] [PubMed] [Google Scholar]
- 195.Gautier M., Yamaguchi J., Foucaud J., Loiseau A., Ausset A., Facon B., Gschloessl B., Lagnel J., Loire E., Parrinello H., Severac D., Lopez-Roques C., Donnadieu C., Manno M., Berges H., Gharbi K., Lawson-Handley L., Zang L.-S., Vogel H., Estoup A., Prud’homme B., The Genomic Basis of Color Pattern Polymorphism in the Harlequin Ladybird. Curr. Biol. 28, 3296–3302.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Stern D. B., Lee C. E., Evolutionary origins of genomic adaptations in an invasive copepod. Nat Ecol Evol. 4, 1084–1094 (2020). [DOI] [PubMed] [Google Scholar]
- 197.Dauphin B., Wüest R. O., Brodbeck S., Zoller S., Fischer M. C., Holderegger R., Gugerli F., Rellstab C., Disentangling the effects of geographic peripherality and habitat suitability on neutral and adaptive genetic variation in Swiss stone pine. Mol. Ecol. 29, 1972–1989 (2020). [DOI] [PubMed] [Google Scholar]
- 198.Flori L., Moazami-Goudarzi K., Alary V., Araba A., Boujenane I., Boushaba N., Casabianca F., Casu S., Ciampolini R., Coeur D’Acier A., Coquelle C., Delgado J.-V., El-Beltagi A., Hadjipavlou G., Jousselin E., Landi V., Lauvie A., Lecomte P., Ligda C., Marinthe C., Martinez A., Mastrangelo S., Menni D., Moulin C.-H., Osman M.-A., Pineau O., Portolano B., Rodellar C., Saïdi-Mehtar N., Sechi T., Sempéré G., Thévenon S., Tsiokos D., Laloë D., Gautier M., A genomic map of climate adaptation in Mediterranean cattle breeds. Mol. Ecol. 28, 1009–1029 (2019). [DOI] [PubMed] [Google Scholar]
- 199.Formenty P., Boesch C., Wyers M., Steiner C., Donati F., Dind F., Walker F., Le Guenno B., Ebola virus outbreak among wild chimpanzees living in a rain forest of Cote d’Ivoire. J. Infect. Dis. 179, S120–S126 (1999). [DOI] [PubMed] [Google Scholar]
- 200.Leendertz F. H., Ellerbrok H., Boesch C., Couacy-Hymann E., Mätz-Rensing K., Hakenbeck R., Bergmann C., Abaza P., Junglen S., Moebius Y., Vigilant L., Formenty P., Pauli G., Anthrax kills wild chimpanzees in a tropical rainforest. Nature. 430, 451–452 (2004). [DOI] [PubMed] [Google Scholar]
- 201.Locatelli S., Harrigan R. J., Sesink Clee P. R., Mitchell M. W., McKean K. A., Smith T. B., Gonder M. K., Why Are Nigeria-Cameroon Chimpanzees (Pan troglodytes ellioti) Free of SIVcpz Infection? PLoS One. 11, e0160788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.COVID-19 Host Genetics Initiative, The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur. J. Hum. Genet. 28, 715–718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Vanderheiden A., Ralfs P., Chirkova T., Upadhyay A. A., Zimmerman M. G., Bedoya S., Aoued H., Tharp G. M., Pellegrini K. L., Lowen A. C., Menachery V. D., Anderson L. J., Grakoui A., Bosinger S. E., Suthar M. S., Type I and Type III IFN Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures, , doi: 10.1101/2020.05.19.105437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rappaport N., Twik M., Plaschkes I., Nudel R., Iny Stein T., Levitt J., Gershoni M., Morrey C. P., Safran M., Lancet D., MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 45, D877–D887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Patrono L. V., Samuni L., Corman V. M., Nourifar L., Röthemeier C., Wittig R. M., Drosten C., Calvignac-Spencer S., Leendertz F. H., Human coronavirus OC43 outbreak in wild chimpanzees, Côte dÍvoire, 2016. Emerg. Microbes Infect. 7, 1–4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Scully E. J., Basnet S., Wrangham R. W., Muller M. N., Otali E., Hyeroba D., Grindle K. A., Pappas T. E., Thompson M. E., Machanda Z., Watters K. E., Palmenberg A. C., Gern J. E., Goldberg T. L., Lethal Respiratory Disease Associated with Human Rhinovirus C in Wild Chimpanzees, Uganda, 2013. Emerg. Infect. Dis. 24, 267–274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Köndgen S., Schenk S., Pauli G., Boesch C., Leendertz F. H., Noninvasive monitoring of respiratory viruses in wild chimpanzees. Ecohealth. 7, 332–341 (2010). [DOI] [PubMed] [Google Scholar]
- 208.Negrey J. D., Reddy R. B., Scully E. J., Phillips-Garcia S., Owens L. A., Langergraber K. E., Mitani J. C., Emery Thompson M., Wrangham R. W., Muller M. N., Otali E., Machanda Z., Hyeroba D., Grindle K. A., Pappas T. E., Palmenberg A. C., Gern J. E., Goldberg T. L., Simultaneous outbreaks of respiratory disease in wild chimpanzees caused by distinct viruses of human origin. Emerg. Microbes Infect. 8, 139–149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hanamura S., Kiyono M., Lukasik-Braum M., Mlengeya T., Fujimoto M., Nakamura M., Nishida T., Chimpanzee deaths at Mahale caused by a flu-like disease. Primates. 49, 77–80 (2008). [DOI] [PubMed] [Google Scholar]
- 210.Souilmi Y., Lauterbur M. E., Tobler R., Huber C. D., Johar A. S., Moradi S. V., Johnston W. A., Krogan N. J., Alexandrov K., Enard D., An ancient viral epidemic involving host coronavirus interacting genes more than 20,000 years ago in East Asia. Curr. Biol. 31, 3704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Enard D., Petrov D. A., Ancient RNA virus epidemics through the lens of recent adaptation in human genomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Uricchio L. H., Petrov D. A., Enard D., Exploiting selection at linked sites to infer the rate and strength of adaptation. Nat Ecol Evol. 3, 977–984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Louzada S., Algady W., Weyell E., Zuccherato L. W., Brajer P., Almalki F., Scliar M. O., Naslavsky M. S., Yamamoto G. L., Duarte Y. A. O., Passos-Bueno M. R., Zatz M., Yang F., Hollox E. J., Structural variation of the malaria-associated human glycophorin A-B-E region. BMC Genomics. 21, 446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mao Y., Harvey W. T., Porubsky D., Munson K. M., Hoekzema K., Lewis A. P., Audano P. A., Rozanski A., Yang X., Zhang S., Gordon D. S., Wei X., Logsdon G. A., Haukness M., Dishuck P. C., Jeong H., del Rosario R., Bauer V. L., Fattor W. T., Wilkerson G. K., Lu Q., Paten B., Feng G., Sawyer S. L., Warren W. C., Carbone L., Eichler E. E., Structurally divergent and recurrently mutated regions of primate genomes. bioRxiv (2023), p. 2023.03.07.531415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and code will be made available on publication.