Abstract
Accurate quantification of transcript isoforms is crucial for understanding gene regulation, functional diversity, and cellular behavior. Existing RNA sequencing methods have significant limitations: short-read (SR) sequencing provides high depth but struggles with isoform deconvolution, whereas long-read (LR) sequencing offers isoform resolution at the cost of lower depth, higher noise, and technical biases. Addressing this gap, we introduce Multi-Platform Aggregation and Quantification of Transcripts (MPAQT), a generative model that combines the complementary strengths of different sequencing platforms to achieve state-of-the-art isoform-resolved transcript quantification, as demonstrated by extensive simulations and experimental benchmarks. By applying MPAQT to an in vitro model of human embryonic stem cell differentiation into cortical neurons, followed by machine learning-based modeling of transcript abundances, we show that untranslated regions (UTRs) are major determinants of isoform proportion and exon usage; this effect is mediated through isoform-specific sequence features embedded in UTRs, which likely interact with RNA-binding proteins that modulate mRNA stability. These findings highlight MPAQT’s potential to enhance our understanding of transcriptomic complexity and underline the role of splicing-independent post-transcriptional mechanisms in shaping the isoform and exon usage landscape of the cell.
Introduction
Nearly all protein-coding genes encode multiple transcript isoforms, resulting from a wide array of alternative transcription start sites (TSSs), transcription termination sites (TTSs), and/or alternative splicing of exons1. This isoform variation is a significant source of molecular and functional diversity, as proteins produced from different isoforms of the same gene can have distinct (and even opposite2) functions. Even transcript isoforms that encode the same protein can differentially affect cellular functions due to variations in mRNA localization, stability, and/or translation3–5. Thus, isoform abundances can reflect the biological state better than the gene-level aggregation of expression profiles.
Given the sequence similarity of transcript isoforms, accurate quantification of isoform abundances by RNA sequencing remains a major challenge. Methods based on short-read (SR) sequencing can generate a large number of reads at increasingly low costs, providing high sequencing depth for reproducible quantification. However, the vast majority of short reads cannot be unambiguously assigned to a single isoform6. On the other hand, by generating reads that almost capture full-length transcripts, long-read (LR) RNA sequencing offers the ability to unambiguously resolve the isoform of origin of most reads. However, at comparable costs, current LR sequencing platforms produce 1–2 orders of magnitude fewer reads compared to SR sequencing, increasing the noise and sacrificing the accuracy of quantification. Their complementary abilities suggest the potential for highly accurate, isoform-resolved, transcript quantification by combining short- and long-read sequencing strategies. However, while several existing tools leverage SR and LR data for a range of upstream tasks—such as splice site identification and splice junction refinement7, alternative polyadenylation site identification8, and de novo transcriptome assembly9, 10—none provide a principled statistical framework for transcript quantification from joint analysis of SR and LR data.
Here, we introduce Multi-Platform Aggregation and Quantification of Transcripts (MPAQT), a probabilistic framework for the inference of isoform-resolved transcript abundances. By integrating the quantification information across multiple platforms with different data-generating processes, such as SR and LR sequencing, MPAQT leverages their complementary advantages to obtain highly accurate isoform abundance profiles, as shown by extensive simulations and benchmarking experiments. By applying MPAQT to matching SR and LR data from an in vitro model of neuronal differentiation, we provide a high-resolution picture of isoform abundance changes that accompany the differentiation of human embryonic stem cells (hESCs) into cortical neurons. Machine learning-based models of the determinants of isoform abundance, trained using MPAQT measurements, revealed the role of alternative mRNA untranslated regions (UTRs) in determining the abundances of isoforms with different cassette exons, highlighting a previously overlooked relationship between cassette exon inclusion rate and distal sequence elements located in the mRNA UTRs.
Results
The MPAQT framework
At the core of MPAQT is a generative model that connects the latent abundances of the transcripts to the observed counts of the “observation units” (OUs) (Figure 1a). Here, an observation unit is any entity that we can directly quantify from RNA-seq reads, defined in a technology-dependent manner. For example, in long-read (LR) sequencing data, in which most reads can each be unambiguously assigned to one transcript, we can simply define each transcript as one OU, resulting in a one-to-one relationship between the transcripts and the OUs. The expected count of each OU, thus, scales linearly with the abundance of its corresponding transcript, with factors such as transcript length or GC content affecting the slope of this relationship. The observed count is then modeled as a sample from a Poisson distribution whose mean is the expected count determined by the transcript abundance and transcript-level covariates.
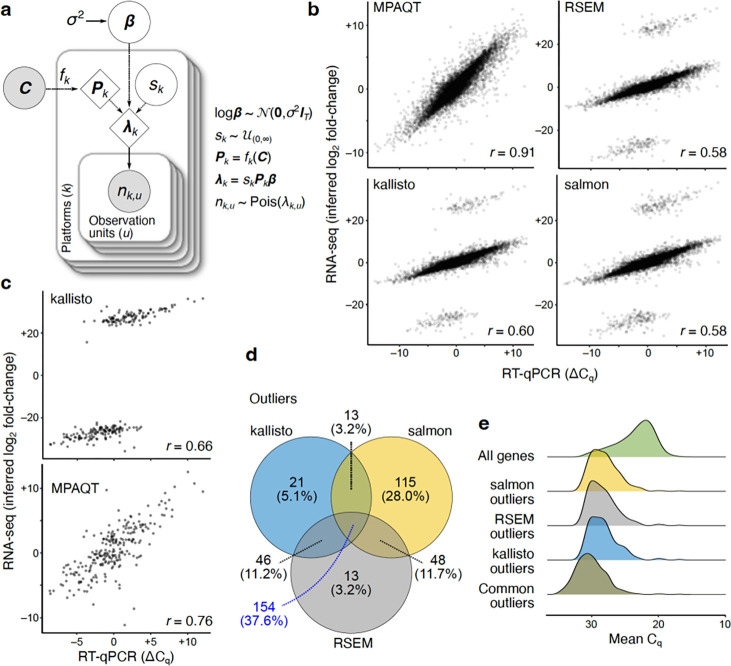
Figure 1.
Overview of MPAQT and its performance for inference of gene-level abundances. (a) The generative model of MPAQT. Open circles, closed circles, and the diamonds represent latent variables, observed variables, and deterministic computations, respectively. : vector of transcript abundances; : library size for platform : set of transcript sequences; : number of reads mapping to the observation unit in platform . See Methods for description of other variables. (b) Inferred log fold-change between MAQCA and MAQCB, calculated from TPM predictions by MPAQT, RSEM, kallisto, and salmon, plotted against the ground truth qPCR difference. Each point is one gene . (c) Top: kallisto’s outliers, isolated from other genes. Bottom: MPAQT’s inferences for kallisto’s outlier genes . (d) Venn diagram showing overlap of outliers among kallisto, salmon, and RSEM. (e) Density curves of mean of MAQCA and MAQCB Cq-values for each tool’s outliers and for all genes. For benchmarking results with the expanded filtering range, see Supplementary Figure 2. Data underlying this figure can be found in Supplementary Data Table 1.
In short-read (SR) RNA-seq data, in which most reads can be mapped to multiple transcripts (or even multiple genes), the relationship between transcripts and OUs can be more complex. Here, we use the equivalence classes (ECs)6 as the OUs. In this case, each transcript may be connected to multiple OUs, and each OU may be connected to multiple transcripts (it is possible to extend the concept of EC’s to long-read data as well; see ref11). MPAQT models the expected count of each OU as a linear function of the abundances of all transcripts that may contribute to that OU (Figure 1a), with the parameters of this function (i.e., the transcript-OU weights) obtained through analysis of simulated short reads (see Methods for details).
By explicitly modeling the OU counts as probabilistic functions of transcript abundances, MPAQT provides a natural framework for joint analysis of data across multiple platforms that assay the same RNA sample, such as short- and long-read sequencing data (Figure 1a)—MPAQT infers the transcript abundances by maximum a posteriori (MAP) estimation given the observed OU counts across all platforms, along with optimization of platform- and experiment-specific model parameters such as the length- or GC-biases and the library size.
Improved gene-level quantification with MPAQT
To examine whether MPAQT might be broadly applicable to gene expression quantification, we began by benchmarking its performance against that of three leading SR analysis tools, salmon12, kallisto6, and RSEM13, for gene-level quantification using SR data alone. For this purpose, we used data from the MicroArray Quality Control (MAQC) project14. This dataset consists of single-end RNA-seq data for two MAQC samples15: MAQCA (Universal Human Reference RNA, pool of 10 cell lines) and MAQCB (Human Brain Reference RNA). Each MAQC dataset is accompanied by RT-qPCR expression measurements for 18,080 protein-coding genes in the form of Cq-values (representing the number of PCR cycles before a signal is seen for a given gene; higher Cq-values correspond to lower abundances). Of the genes that could be readily matched to quantifications from the RNA-seq-based methods based on their IDs, 14,956 genes had Cq-values between 11 and 32, a range deemed reliable in the original report15. For this subset, the ground truth differential expression (DE) was calculated as the difference of RT-qPCR Cq-values between MAQCA and MAQCB (representing log2 fold-change of expression), which was then compared to gene-level log fold-change of TPM (transcripts per million) calculated from SR RNA-seq data by different tools (gene-level TPM values in each sample were calculated by summing up transcript-level TPMs for each gene).
We observed excellent agreement between log fold-changes inferred by MPAQT and the ground truth DE values (Pearson r=0.91, Figure 1b). In contrast, for all other existing tools, we saw distinct outliers that were visibly separated from the remainder of the data points (Figure 1b). Figure 1c shows kallisto’s outliers, isolated from other, well-behaving genes. Interestingly, MPAQT preserves differential expression information for these outlier genes (Figure 1c). A similar trend is observed for outliers from salmon and RSEM (Supplementary Figure 1). These outliers represent ~2–3% of genes in our data (Figure 1d), and correspond to genes with higher mean Cq-values (Figure 1e), suggesting they are genes with low expression; this trend toward higher mean Cq-values is even stronger for the 154 outliers shared among kallisto, salmon, and RSEM (Figure 1e), suggesting that low-abundance genes are commonly mis-quantified by existing tools. We also examined an extended filtering range (Cq-value between 8–35) to assess MPAQT’s performance on noisier qPCR measurements, allowing for an additional 1148 genes to be included in the analysis. Surprisingly, MPAQT’s performance remained comparable to the more conservative filtering range (Cq-value between 11–32), whereas the number of outliers for the other SR tools approximately doubled (Supplementary Figure 2), highlighting the volatility of existing methods in the presence of very high- or very low-abundance genes.
MPAQT improves isoform-level quantification with short- and long-read data
Since no transcriptome-wide benchmarking datasets exist for isoform-level quantification, we used simulated data to benchmark the ability of MPAQT and other existing tools for isoform-level quantification, starting with SR data alone. We used six different simulated datasets: three with ground truth TPM values sampled randomly from an exponential distribution, and another three with ground truth TPM values sampled from a distribution that was modeled after measurements from real RNA-seq data (see Methods for details). In both simulations, we observed that MPAQT substantially outperforms the other tools in terms of Pearson correlation and Root Mean Square Deviation (RMSD) (Figure 2a–b). The amount of variance in the ground truth log-TPMs that is captured by MPAQT (i.e., R2 between MPAQT inferences and ground truth) is ~13%–32% higher than the next best method (13% for the data simulated for TPMs that are modeled after real RNA-seq measurements, and 32% for the data simulated for TPMs that are exponentially distributed). Together, these simulation experiments suggest that, even without LR data, MPAQT outperforms the state-of-the-art in transcript quantification from SR data alone.
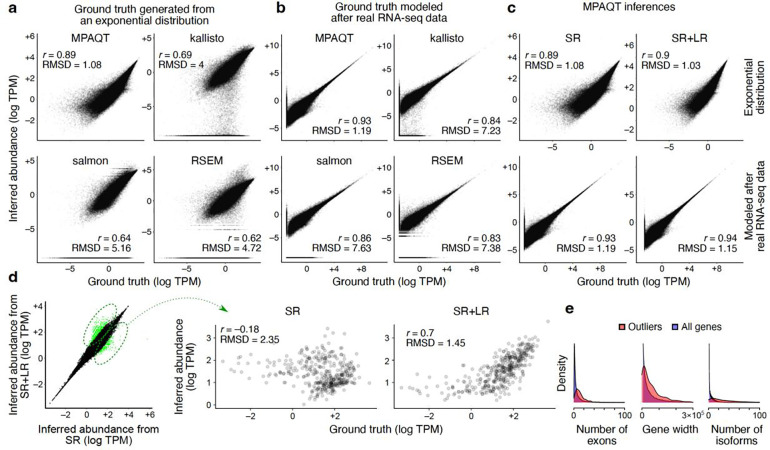
Figure 2.
Accurate inference of transcript abundances using MPAQT. (a) Benchmarking of MPAQT and three other tools using simulated SR data with ground-truth TPMs generated from an exponential distribution. (b) Similar to (a), but for a simulated sample with ground-truth TPM values modeled after real data (see Methods for details). (c) Performance of MPAQT using SR data alone (left) or SR+LR data (right), on simulated data generated from ground truth TPMs with an exponential distribution (top) or modeled after real RNA-seq data (bottom). SR plots are identical to the top-left plots in panels (a) and (b) and are repeated here for easier comparison to SR+LR. (d) Left: Comparison of SR vs. SR+LR inferences for one dataset simulated from an exponential distribution (see Supplementary Figure 3 for more simulation repeats). Transcripts with substantially different inferences are highlighted (outlier analysis based on Mahalanobis distance >6.36, equivalent to upper-tail P<10−10 for normally distributed data). Right: Comparison of inferred vs. ground truth TPMs for transcripts that are significantly differentially quantified (i.e., transcripts highlighted in the left panel). (e) Distributions of number of exons, gene width, and number of isoforms for genes encoding the transcripts that are differentially quantified between SR and SR+LR analyses. The distributions for all genes are also shown, for comparison.
Next, we set out to examine whether LR data can further improve MPAQT’s estimates of transcript abundances. We used the same ground truth TPM sets that we created for SR data simulation and generated a simulated LR dataset with moderate coverage for each replicate. Specifically, we simulated the LR full-length transcript counts by sampling from independent Poisson distributions, with the ground truth TPM of each transcript as the mean of its Poisson distribution (adjusted to obtain ~200K transcript counts per sample). Then, the combination of simulated SR data and LR counts was used as input to MPAQT, and the results were compared between SR-alone and SR+LR quantifications (Figure 2c). When MPAQT’s inferences from SR data are directly compared to those from SR+LR data, we can identify a subset of transcripts whose quantified abundances differ substantially between the two measurements (Figure 2d), despite the moderate depth of the simulated LR datasets. For this subset, we see substantial improvement in SR+LR data in terms of agreement with ground truth (Pearson correlation 0.67–0.71 for SR+LR, compared to −0.26 to −0.18 for SR data alone, Figure 2d and Supplementary Figure 3a), suggesting that LR data can substantially improve transcript quantification when combined with SR data.
Interestingly, although the simulations were based on independent (uncorrelated) ground truths, among the 490 transcripts for which inclusion of LR data had a significant effect in at least one simulation, ~30% were identified in more than one simulation (Supplementary Figure 3b), suggesting that these transcripts may have intrinsic features that make them sensitive to the presence/lack of LR data. The genes encoding these transcripts have significantly more exons, are longer, and have more isoforms compared to other genes (Figure 2e). Furthermore, pathway enrichment analysis revealed a significant and recurrent enrichment of “nervous system development” among the LR-sensitive genes in all replicates (P<7×10−5 for the three replicates, based on g:Profiler16). This finding is consistent with previous reports showing that genes preferentially expressed in the nervous system tend to be longer, have more exons, and exhibit more complex splicing patterns compared to other tissues 4, 17, and suggests that joint analysis of SR and LR data using MPAQT is particularly beneficial to the quantification of isoforms involved in the nervous system development.
Isoform quantification during neuronal differentiation with MPAQT
The analyses presented above suggest that profiling the transcriptome using a combination of SR and LR sequencing can substantially improve isoform quantification for genes related to neuronal differentiation. To further investigate the landscape of isoform usage during neuronal cell differentiation, we analyzed human embryonic stem cells (hESCs) undergoing in vitro differentiation toward cortical neurons (Supplementary Figure 4), by joint SR and LR RNA-seq from cells collected at days 0, 41, and 61 since the start of growth in neural induction medium (see Methods for details).
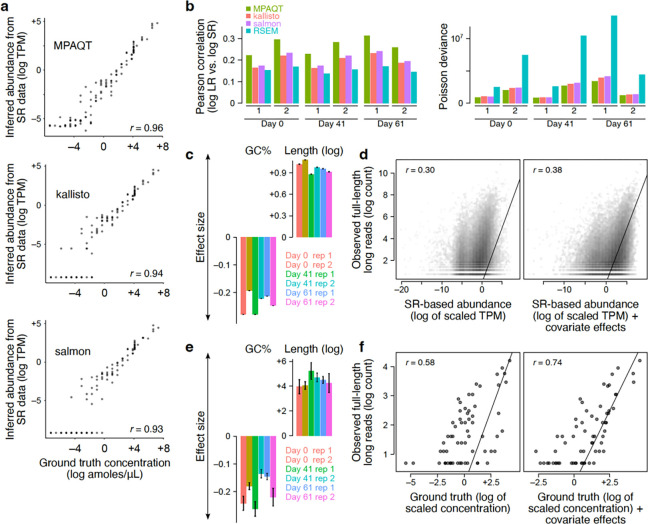
We first used MPAQT to analyze the SR data of each sample, which provided further evidence demonstrating superior performance of MPAQT over state-of-the-art SR-based quantification tools: first, MPAQT’s SR-based quantifications show a minor but consistent improvement in accuracy compared to other tools for synthetic mRNAs that were spiked in the samples at known concentrations (Figure 3a); secondly, MPAQT’s SR-based quantifications of gene isoforms are more consistent than other tools with full-length LR counts obtained from the same sample (Figure 3b). However, we generally see only a moderate correlation between SR quantifications and LR counts, suggesting the presence of potential biases in LR RNA-seq data. We found that the deviation between LR and SR data can be at least partially explained by transcript length and GC content: longer and GC-poor transcripts are more likely to be captured by LR sequencing (Figure 3c). Once we account for the biases introduced by these factors, we see a far larger agreement between SR and LR data (Poisson likelihood ratio test P<10−16, Figure 3d). Importantly, the same biases can be replicated when we compare the LR counts of spike-in RNAs to their ground truth concentrations (Figure 3e–f). We note, however, that these LR biases might be due to experiment-, instrument-, and/or protocol-specific factors. MPAQT’s statistical model enables the inference of sample-specific sources of bias and incorporates them in its framework for integration of LR and SR data (Figure 1a; see Methods for details).
Figure 3.
Application of MPAQT to SR and LR data collected from cells undergoing differentiation toward neurons. (a) Performance of MPAQT, kallisto, and salmon based on 107 spike-in transcripts with known abundances. Each point represents one spike-in RNA in one sample/replicate. (b) Comparison of SR-based abundances and full-length LR counts for genomic transcripts. Left: Pearson correlation between LR log-counts and SR-based inferences, for each tool and each time point/replicate separately. Right: Poisson deviance is shown as an alternative measure of goodness of fit. (c) The effect of GC content and length on LR counts, estimated by including them as covariates in a Poisson regression with log-scale SR-based inferences as the regressor. (d) The scatterplots show the relationship between full-length LR counts and SR-based abundances (left) or SR-based abundances after adding the effect of covariates (transcript GC content and length). The SR-based abundances are scaled separately for each sample and each model to maximize the likelihood of LR counts. Each point in each scatterplot shows one transcript in one sample (six samples combined in each plot). Data points with LR count of zero were removed to allow log-scale plotting of the y-axis. (e and f) Similar to (c-d), showing the coefficients of GC content and length for models that are fitted to spiked-in transcript LR counts with log-scale ground-truth RNA concentration as the regressor (e), and the scatterplots of LR counts vs. ground truth concentration without (left) and with (right) the covariate effects (f). Data underlying this figure can be found in Supplementary Data Table 2.
Next, we used MPAQT to jointly analyze the LR and SR (SR+LR) data obtained from neuronal differentiation samples at days 0, 41, and 61, while accounting for the biases described above. We found 6309 transcripts whose inferred abundances based on SR+LR analysis deviated substantially from abundances inferred from SR-only analysis in at least one of the three time points (Mahalanobis distance >2.32, equivalent to upper-tail P<0.01 for normally distributed data). Even at a substantially stricter cutoff (Mahalanobis distance >6.36, equivalent to upper-tail P<10−10), there were still 2459 transcripts whose SR+LR and SR-only quantifications differed significantly in at least one time point (Figure 4a). Interestingly, these transcripts corresponded to larger genes with more exons and more isoforms (Supplementary Figure 5), which is in line with the findings from our simulations (Figure 2e).
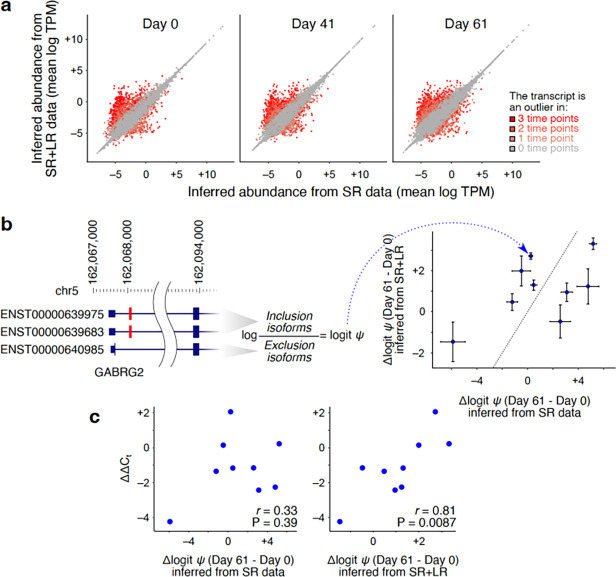
Figure 4.
Inclusion of LR data significantly improves transcript abundance quantification in neuronal differentiation models. (a) Comparison of inferred TPMs based on SR data alone (x-axis) vs. SR+LR data (y-axis) in each of the three time points during neuronal differentiation. For each measurement, the mean of two replicates is used. Each data point is one transcript, with the dot color representing the number of time points in which the inferred abundance of the transcript differs significantly between SR and SR+LR measurements (Mahalanobis distance >6.36). (b) Quantification of cassette exon percent-spliced-in (PSI) from transcript isoform abundances. Left: An example cassette exon for gene GABRG2, shown in red. PSI (shown with ) is calculated as the sum of abundances of the isoforms that include the exon divided by that of all isoforms. We use the logit of PSI for regression analysis and compatibility with qPCR , which is equal to the logarithm of the sum of abundances of inclusion isoforms divided by that of exclusion isoforms. Right: Top cassette exons for which the inferred change in PSI between days 0 and 61 differ significantly depending on whether SR or SR+LR quantifications are used. (c) Scatterplot of qPCR-based differential PSI quantification (y-axis) vs. differential PSI values inferred from SR data alone (x-axis of the left plot) or SR+LR data (x-axis of the right plot). Data underlying this figure can be found in Supplementary Data Table 3.
To validate the higher accuracy of SR+LR transcript quantifications, we first used the inferred transcript abundances to estimate the usage of alternatively spliced exons, and then selected nine cassette exons for which the change in percent-spliced-in (Ψ) between day 0 and day 61 was significantly different between SR and SR+LR measurements (Figure 4b)—more accurate transcript quantifications are expected to result in more accurate Ψ quantifications, of which the latter can be validated by RT-qPCR using junction-specific primer pairs. The selected cassette exons were mostly from genes that are neuron-specific and often associated with nervous system and/or mental disorders (Supplementary Figure 6). For these exons, we quantified the true change in Ψ using RT-qPCR and compared to differential Ψ measurements from SR or SR+LR analyses. As shown in Figure 4c, we found that SR+LR analysis provides substantially more accurate estimates of differential Ψ (r=0.81, P=0.0087) compared to SR-only analysis (r=0.33, P=0.39). These results further support the notion that joint analysis of SR+LR data is crucial for accurate estimation of the abundances of many transcripts, especially those involved in neuronal function and related diseases.
Sequence determinants of mRNA abundances during neuronal differentiation
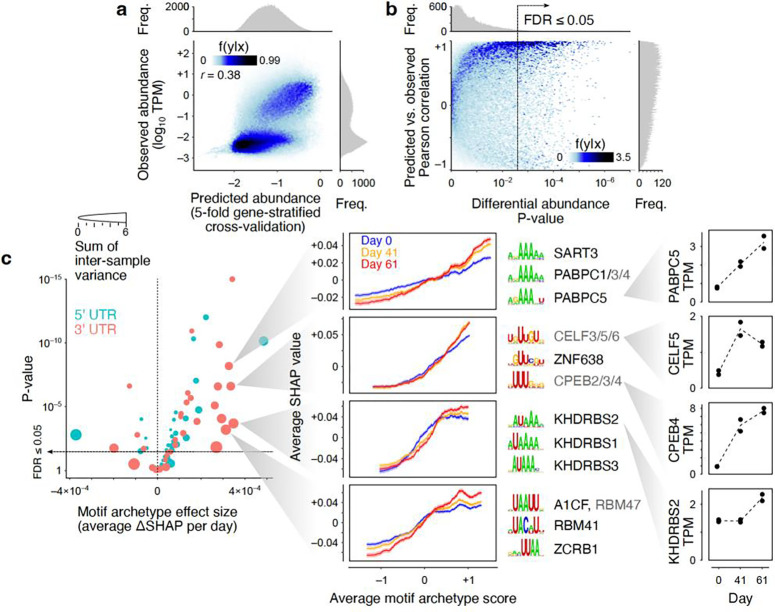
Accurate measurement of isoform abundances in differentiating neurons provides the opportunity to study the sequence determinants of mRNA abundance in these cells. We sought to examine the extent to which the observed mRNA abundances in progenitor and differentiated neuronal cells could be explained by the binding sites of sequence-specific RNA-binding proteins (RBPs). To this end, we predicted the affinities18 of 128 RBPs, based on their known motifs19, toward the 5’ and 3’ untranslated regions (UTRs) of each mRNA isoform, removed redundancies by grouping the motifs with similar affinity profiles into 35 motif archetypes, and, for each differentiation time point and replicate, developed a random forest machine learning (ML) model that could predict the abundance of each mRNA from its motif archetype scores (see Methods for details). Based on gene-stratified five-fold cross-validation experiments, we found that our ML models achieved an overall Pearson correlation of 0.38 (Figure 5a, minimum and maximum r of 0.36 and 0.40 across individual timepoints/replicates).
Figure 5.
Sequence-based prediction of transcript abundances in neuronal differentiation samples. (a) Observed vs. predicted transcript abundances (5-fold gene-stratified cross-validation). Each point represents one transcript in one time point/replicate. Histograms represent marginal distribution of predicted (x-axis) and observed (y-axis) log-TPM values. (b) Correlation of predicted vs. observed log-fold changes across differentiation. Each point represents one transcript. The x-axis represents the statistical significance for differential log-TPM across time points (one-way ANOVA test). The y-axis represents the Pearson correlation between predicted and observed abundances across time points/replicates for each transcript. (c) Left: Volcano plot of the differentiation-associated change in SHAP value per motif archetype. The x-axis shows the effect size obtained by modeling the SHAP value as a function of differentiation time point and motif archetype score (see Methods for details). The y-axis shows the p-value associated with the regression coefficient. The size of each circle represents the sum of transcript-wise variances of the SHAP values across time points/replicates. Middle: Example motif archetypes with the largest effect sizes and sample-to-sample variances. For each motif archetype, the moving average chart of SHAP vs. motif archetype score is shown (transcripts were sorted by their motif archetype scores, following by mean calculation over sliding windows of 500 transcripts). Each curve represents one time point. The shaded areas correspond to the standard error of mean of SHAP values per sliding window. The top three motifs associated with each motif archetype are shown next to each chart, along with the RBPs that recognize each motif (RBPs shown in grey are inferred to recognize the motif based on homology19). Right: Gene-level TPM profiles for example RBPs across differentiation time points. Each replicate is shown with a separate point. Data underlying this figure can be found in Supplementary Data Table 4.
Interestingly, for mRNAs with significant differential abundance across time points (one-way ANOVA), the sequence-based predictions correlated strongly with the observed differential abundances (mean r of 0.35 and 0.64 for isoforms with significant differential expression at FDR ≤0.05 and ≤0.01, respectively; Figure 5b). Analysis of the Shapley additive explanations (SHAP) suggests that several 3’ UTR motif archetypes dominate the top features that are differentially used by ML models across time points (Figure 5c), nominating these motifs as the main drivers of differential mRNA abundance. As shown in Figure 5c, presence of these motifs in the 3’ UTRs is generally associated with higher mRNA abundance—on average, larger motif archetype scores correspond to larger positive SHAP values. For the mRNAs with the highest scores for these motif archetype, the SHAP values increase even further at later time points, consistent with the up-regulation of these mRNAs during differentiation. Figure 5c shows the top individual motifs associated with these motif archetypes and the RBPs that recognize them. For each motif archetype, at least one RBP can be identified whose expression pattern and known function in mRNA regulation is consistent with the increased expression of the mRNAs associated with that motif archetype. For example, one motif archetype represents several A-rich motifs recognized by various poly-A binding proteins, including PABPC5, which is known to stabilize the mRNAs it binds to20, and whose expression increases during neuronal differentiation in our dataset. Similar observations nominate the CELF21, CPEB22, 23, and KHDRBS24 families of proteins as major regulators of differential mRNA abundance during neuronal differentiation (Figure 5c).
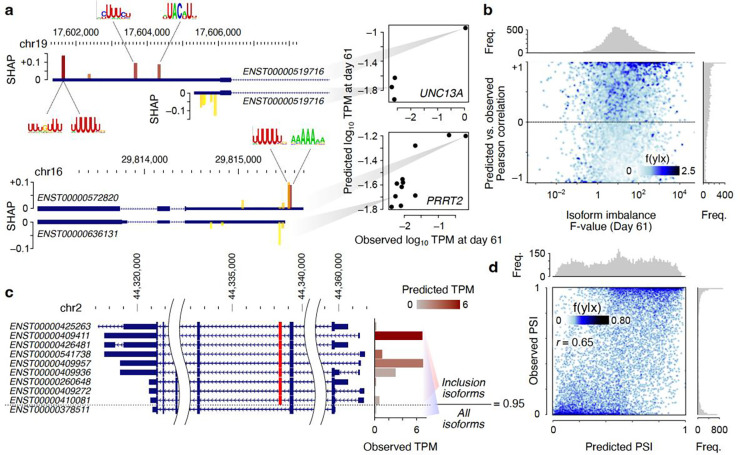
Surprisingly, even though our models do not include features that are directly attributable to alternative splicing, we found that they can explain the isoform usage patterns of a large number of genes within each time point. Figure 6a shows a few examples, wherein differences in the 3’ UTR sequences of the isoforms of the same gene can predict the differences in the relative abundances of those isoforms in terminally differentiated neurons. In these examples, the region that is unique to the longer 3’ UTR contains instances of motifs with large positive SHAP values, suggesting that the higher relative abundance of the dominant isoform is due to higher stability conferred by the binding of stabilizing RBPs to its 3’ UTR, as opposed to preferential splicing. Overall, for genes with more than two isoforms, differences in UTR sequences can predict the isoform usage in differentiated neurons with mean r=0.23. For 25% of such genes (2508 out of 10,010), the correlation between UTR-based predictions and isoform usage in differentiated neurons exceeds 0.7. This fraction increases to 41% if we focus on the subset of genes with the most reproducible isoform usage profiles (166 out of 405 genes with isoform imbalance F-value >500; Figure 6b). For genes with two isoforms and highly reproducible isoform imbalances (F-value >500), in 73% of cases (377 out of 517) the UTR-based predictions can correctly identify the dominant isoform. Overall, these results suggest that alternative UTR usage is a major determinant of isoform ratios. This raises the possibility that alternative UTR usage may also be responsible for variations in other, locally measured, metrics of alternative splicing, such as percent-spliced-in (PSI) of cassette exons. Figure 6c shows an example cassette exon that is excluded in an isoform that also harbors short 5’ and 3’ UTRs. In contrast, the isoforms in which this cassette exon is included have longer UTRs—this association between cassette exon inclusion and the choice of UTR enables accurate exon inclusion prediction based on UTR sequences alone (predicted PSI of 0.963 vs. observed PSI of 0.956 for the example cassette exon shown in Figure 6c), without any knowledge of the local sequence features of the cassette exon itself. When we expanded this analysis to all expressed cassette exons (inclusion + exclusion TPM ≥1), we observed that our UTR-based ML models can predict PSI with a Pearson correlation of 0.65 (Figure 6d), and can separate “included” exons (defined as those with PSI≥0.8) from “excluded” exons (PSI≤0.2) at AUROC (area under the receiver operating characteristic curve) of 0.92 (Supplementary Figure 7). These observations suggest that the sequence determinants of exon inclusion rate are not limited to the local context of the exon, and distal elements in the UTRs contribute significantly to the exon usage landscape, potentially through non-splicing mechanisms such as regulation of mRNA stability.
Figure 6.
UTR features predict isoform-level and exon-level splicing. (a) Example isoforms of UNC13A (top) and PRRT2 (bottom). For each gene, one dominant isoform and one low-abundance isoform is shown, along with the top five motifs whose presence in the 3’ UTR explains the higher abundance of the dominant isoform. For each motif, the position of the best-matching sequence in each isoform is shown, along with the SHAP value of the associated motif archetype (shown with the bar height) and the motif hit score (yellow: low-scoring hit; red: high-scoring hit). The predicted and observed abundances of the highlighted isoforms (along with other isoforms of each gene) are shown in the scatterplot on the right. (b) Pearson correlation of predicted vs. observed isoform abundances (log10TPM) for each gene. Each point represents one gene with at least three isoforms. The x-axis shows the F-value from one-way ANOVA test for unequal abundances of isoforms. (c) The isoforms associated with inclusion or exclusion of an example cassette exon (shown in red) for gene PREPL. The observed TPM of each isoform is shown using the bar graph on the right (the color gradient specifies the predicted TPM). (d) The scatter plot of predicted vs. observed PSI. Each point represents one cassette exon in one time point/replicate (cassette exons for which the sum of TPMs of inclusion and exclusion isoforms was <1 were excluded). Data underlying this figure can be found in Supplementary Data Table 5.
Discussion
MPAQT’s generative model can effectively combine sequencing data from multiple platforms to enhance gene- and isoform-level mRNA quantification. This superior performance stems from MPAQT’s ability to leverage the complementary strengths of each platform, while overcoming the limitations posed by the inherent properties of each sequencing technology. Compared to SR data alone, we show that combining SR and LR data with MPAQT leads to more accurate isoform-level quantifications, owing to the unambiguity of LR-transcript assignments. Compared to LR data alone, combining LR data with SR data provides the coverage needed to obtain low-uncertainty measurements across the spectrum of mRNA abundances. In this work, we simulated LR data with a library size of ~200K reads per sample to emphasize the information gained even by inclusion of low-to-moderate amounts of LR data (relative to SR-only). Nonetheless, even in our neuronal differentiation dataset, with a mean library size of ~1.1M mappable full-length long reads per time point, only the most highly abundant transcripts are quantified accurately with LR data alone (e.g., see spike-in measurements in Figure 3f). Other recent studies also report similar sequencing depths for LR-RNA-seq (e.g., ~1.4M full-length long reads per sample in ENCODE4 human LR data25), falling considerably short of the sequencing depths routinely obtained from SR-RNA-seq.
In addition, combining LR and SR data overcomes sequence-dependent biases that may be introduced by reliance on LR data alone: analysis of spike-in mRNAs with known concentrations suggests that LR sequencing data may be biased toward AT-rich and/or longer transcripts, while SR data appear to be unaffected by these factors (Figure 3a,f). The LR biases may be study- and/or protocol-specific (e.g., see ref26 for nucleotide composition biases that are different from our observations), underlining the need to learn such study- and/or experiment-specific biases from the data. When the ground-truth abundances of the mRNAs are not known, however, it may not be feasible to distinguish technical biases from biologically relevant phenomena; for example, if we see higher long read counts for low-GC transcripts, is it because these transcripts are truly expressed at higher levels, or is it a bias introduced by the sequencing procedure? This unidentifiability issue, however, is alleviated when unbiased SR data are combined with LR data, since SR data provide an implicit reference for MPAQT to learn the source and magnitude of biases in LR quantifications. Thus, even without considering the higher cost (and, thus, lower depth) of LR sequencing, combining LR data with SR data is still advantageous.
We show that the increased accuracy that we gain from combining SR and LR data is especially important when quantifying mRNAs from longer genes with complex alternative splicing landscapes, a common feature of genes expressed in neuronal systems. By applying this approach to an in vitro model of neuronal differentiation, followed by ML-based modeling of the measured transcript quantities, we uncovered the sequence determinants of isoform abundance within and across differentiation time points. The most important sequence features correspond to RBP recognition sequences located in 3’ UTRs (Figure 5c), suggesting a critical role for post-transcriptional mechanisms in shaping the mRNA landscape of differentiating neurons. Most surprisingly, we observed that these UTR-based features are also strong predictors of within-gene isoform usages, and can even predict, to a large extent, the usage of cassette exons without any knowledge of the local features surrounding such exons (Figure 6b,d). A substantial body of work has been dedicated to identifying the determinants of cassette exon usage, including ML-based modelling of the sequences that surround these exons (e.g., see refs27–30). However, recent work has highlighted the challenges of predicting cell type-specific exon inclusion using only the local sequence features, especially in neurons31. Our results underline the importance of considering global mRNA sequence features in models of splicing regulation and exon usage, given that non-local sequence features can also affect exon inclusion levels through splicing-independent mechanisms, such as isoform-specific regulation of mRNA stability. Identification of such splicing-independent determinants of exon inclusion can also have implications in the design of therapeutics aimed at modulating disease-associated exons32.
Although MPAQT offers state-of-the-art inference of isoform abundances, it also comes with current limitations that motivate further work. For example, at the core of MPAQT’s generative model are platform-specific matrices whose elements represent the probabilities of transcript-OU associations—for short-read data, the current implementation of MPAQT relies on generation and analysis of a large number of simulated reads to obtain this matrix, which is computationally expensive. While this matrix only needs to be generated once per reference transcriptome, more efficient approaches for its derivation could expedite the analysis of new reference transcriptomes. On the other hand, MPAQT’s reliance on simulated data to construct the OU-transcript association matrix provides advantages, such as awareness of the probabilities that reads get assigned to the wrong OU by the read-OU mapping tool. This ability to implicitly account for the errors introduced by the read-OU mapping algorithms (such as kallisto) may underlie MPAQT’s superior performance even when applied to short-read data alone, although this speculation needs further examination.
Another limitation of our current implementation lies in our assumption that read-isoform assignments are unambiguous for long reads. While this assumption may be true for a large fraction of long reads, factors such as transcript degradation, read truncation, and other sequencing or alignment artifacts can introduce ambiguities in at least a fraction of read-transcript assignments11. The statistical framework of MPAQT in principle allows for these ambiguities to be taken into consideration, for example by using “read classes”11 as OUs, each of which may be compatible with multiple isoforms. With tools that are capable of simulating long read data33, a simulation-based strategy can be used to construct the transcript-OU association matrix for long-read data, which may further increase the accuracy of MPAQT’s inferences.
Methods
MPAQT generative model
As described in the Results section, MPAQT’s generative model connects the latent transcript abundances to the expected counts of a set of “observation units” (OUs), which are defined based on the technology/platform. Consider an RNA-seq dataset, generated by different platforms from sequencing the same mixture of transcripts from the set , with each transcript having the relative abundance so that . We also define to be the vector of relative abundances; . For each platform and each transcript , let’s define the “effective length6” to be a normalization factor such that , where is the probability of observing a read (or fragment) from transcript in platform . In other words, is the expected proportion of reads that originated from transcript , given the transcript abundance profile and the platform.
Each read from each platform is assigned to one OU from the set . For a read that originated from a given transcript , the probability of being assigned to a given observation unit is represented by . In other words, is the probability of a read mapping to conditional on that read having been selected from . We also define . Note that does not depend on the abundance of transcript , and is rather a function of transcript properties/sequence and the platform . It follows that where is the probability that a read in platform is assigned to the observation unit (i.e., is the expected proportion of reads in the dataset that map to . Subsequently, the expected number of reads from platform mapping to is given by , where is the total number of reads obtained from platform . In turn, the observed number of reads from platform mapping to is drawn from a Poisson distribution (which is commonly used to model count data, e.g., see ref34), with expectation . This generative model can be summarized as follows:
Here, is a platform-specific matrix whose elements, , are described above, is a column vector of scaled abundances for transcripts , and is a platform-specific scaling factor. Note that and are differently scaled representations of and , respectively, so that . Since and together form an underdetermined system, we impose a log-normal prior for with a mean of zero to enforce a unique solution:
Given , which is the set of observed OU counts across platforms, and , which is the set of matrices that connect transcript identities to OU probabilities in each platform as described above, MPAQT finds the maximum a posteriori (MAP) estimate of and using an expectation maximization (EM) algorithm, as described in Supplementary Methods. The MAP estimate of is then used to obtain the vector of relative abundances (and TPM) by rescaling.
Obtaining the platform-specific matrices
For any platform k that represents short-read (SR) data, we obtain the matrix by simulation from a reference transcriptome in which all transcripts have exactly equal abundances, using the Rsubread “simReads” function35, followed by read-EC assignment using kallisto6. To make sure that accurately approximates the EC probabilities, we simulate 24 replicates of 100 million reads with the same length as those of the query platform, for a total of 2.4 billion reads. Since each simulated read is tagged with its transcript of origin and is mapped to a unique EC , we can calculate the proportion of reads that originate from transcript and map to EC , i.e., In practice, however, we do not calculate the proportions , but instead directly use the read count . Since is proportional to , it only affects the scale . The scripts for these steps are available at https://github.com/csglab/MPAQT.
For long-read data, in the simplest scenario, we can assume that each long read is unambiguously assigned to one transcript; thus, , and when and zero otherwise. Furthermore, we can assume that read counts are proportional to the transcript abundances, i.e., no biases exist and all transcripts have the same effective length . The matrix for long-read sequencing data is then simply the identity matrix . However, both these assumptions can be violated in real-life applications. Particularly, as discussed in the Results section, we have found that substantial length and GC-biases exist in PacBio Sequel II data. Therefore, MPAQT provides the option to explicitly learn these biases from data and incorporate them in the matrix . We model , the effective length of transcript in long-read platform , as a function of transcript-level variables (, where is the set of transcript-level covariates whose effects we want to model). Specifically, , where · is the dot product, and is the vector of coefficients representing the effect of the covariates on the propensity of the transcripts to be captured by long-read platform . The log-link ensures that is restricted to the domain . The MAP estimate of is obtained during model fitting as described in Supplementary Methods.
Cortical neuron differentiation and RNA-seq data generation
Cortical neuron differentiation
The hESC SOX10::GFP bacterial artificial chromosome reporter line (in the H9 background) was used for neural differentiation according to the protocol adapted from a study of brain organoids36. In brief, the hESC line was maintained in feeder-free conditions with the E8 medium. Neural differentiation was initiated when the cells reached 90–100% confluency. From days 0–11, the cells were maintained in neural induction medium (10 μM SB431542 and 100 nM LDN193189 in E6 medium) with medium change every two days. From day 12, the cells were fed the cortical neuron medium (10 ng/mL GDNF, 100 μM ascorbic acid, 1x Glutagro, 1x N2 supplement, 1x B27 without vitamin A in neurobasal medium) with medium change every other day until rosette structures became visible. Then, neurons were detached using Accutase and replated on poly-L-ornithin/fibronection/laminin-coated plates. Neurons were maintained in cortical neuron medium with medium change every other day. On days 22–24, neurons were checked for the presence of axonal projections and 10 μM DAPT was included in the cortical neuron medium until the projections appeared. From day 30, neurons were considered mature with the medium feeding frequency reduced to 1–2 times per week.
RNA extraction, short-/long-read RNA-seq library prep, and sequencing
Cells were harvested at days 0, 41, and 61, followed by RNA extraction using Zymo Quick-RNA Microprep kit according to the manufacturer’s protocol. SIRV set 4 (Lexogen) was spiked at 1% in the hESC and differentiated neuron-derived RNA samples. Short-read RNA-seq libraries were prepared using the SMARTer Stranded Total RNA-Seq Kit v3. Libraries were sequenced on a NextSeq 550 sequencer (2×75 bp paired-end). PacBio Iso-seq libraries from the same RNA samples were generated using the NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module, PacBio Iso-Seq Express Oligo Kit and SMRTbell express template prep kit 2.0 according to the manufacturer’s protocol. The libraries were sequenced on a PacBio Sequel IIe.
Processing of short-read RNA-seq data
All SR data, including those generated from the neuronal differentiation model (above), those obtained from publicly available data, and simulated data (below) were processed using RSEM 13 (version 1.3.3, following alignment with bowtie2 version 2.4.2), salmon 12 (version 1.3.0, with the --validateMappings and --gcBias flags ), kallisto 6 (version 0.48.0) and MPAQT.
The MAQC data was taken from GEO accession GSE83402, and single-end samples MAQCA_1 (four technical replicates: SRR3670977, SRR3670978, SRR3670979, SRR3670980) and MAQCB_1 (four technical replicates: SRR3670985, SRR3670986, SRR3670987, SRR3670988) were processed with the above SR quantification tools (RSEM, salmon, kallisto and MPAQT). Technical replicates for each sample were combined (at the level of FASTQ files) during quantification. Differential expression was calculated as the logarithm of fold-change (logFC) between MAQCB_1 and MAQCA_1, separately for each quantification tool.
Simulated datasets for benchmarking, in the form of paired-end FASTQ files, were generated from ground truth TPM values using the simReads function from the Rsubread R package 35. Rsubread takes as input the number of reads to simulate and a list of transcripts with their desired TPMs. We generated two simulated datasets using two different sets of ground truth TPMs. For the first dataset, ground truth TPMs were sampled from an exponential distribution (using rexp R function with default rate=1). For the second dataset, we first used kallisto to quantify transcript abundances from RNA-seq data of the MDA-MB-231 cancer cell line (GEO entries GSM4886854, GSM4886855) 37 and then used the resulting TPMs as the ground truth for read simulation. For the rexp.sim dataset, three simulated “replicates” were generated, and one sample was generated for the MDA-MB-231-based dataset, each with 30 million paired-end reads of 75 bp. These samples were processed with RSEM, salmon, kallisto and MPAQT, as described above.
SR sequencing data for the neuronal differentiation samples were processed using the paired-end options for above tools. For this dataset, we added the spike-ins to the reference transcriptome of the above tools to enable their quantification. Each spike-in was added in as its own separate chromosome, and 1000 “N” spacer nucleotides were added on either side of each spike-in sequence. As described above, we used the SIRV-Set 4 from Lexogen, which contains 114 spike-in transcripts. We used 107 in this analysis, since SIRV-403 to SIRV-410 were not included in the reference FASTA provided by Lexogen.
Processing of long-read RNA-seq data
The IsoSeq pipeline (Pacific Biosciences) was used to process the neuronal differentiation LR data and generate circular consensus sequence (CCS) reads, which were stored in uBAM (unaligned BAM) format. Next, lima (PacBio) was used to remove primer sequences. IsoSeq3 ‘refine’ command was used to remove poly-A tails and concatemers (reads which are attached end-to-end), followed by the ‘cluster’ command to cluster reads that represent the same transcript (i.e. make them adjacent). The ‘align’ command of pbmm2 (PacBio) was then used to align reads to the reference genome, followed by the Isoseq3 ‘collapse’ command to condense the data into a transcriptome (fasta + GFF) and provide an abundance file containing full length counts (FL counts).
The quality control script from SQANTI338 (sqanti3_qc.py) was used together with supporting data types (CAGE peak, polyA motif list, polyA peaks file, and Intropolis splice junctions), removing low-quality transcripts according to SQANTI’s quality criteria. Next, the rules filter script (sqanti3_RulesFilter.py) was run to further filter transcripts based on the following criteria: if a transcript is a full splice match (FSM), then it is kept unless the 3’ end is unreliable (intrapriming); if a transcript is not a FSM, then it is kept only if all of below are true: (a) 3’ end is reliable; (b) the transcript does not have a junction that is labeled as RT Switching; and (c) all junctions are canonical. Finally, the full-length (FL) LR counts from SQANTI3 output that had the same “associated_transcript” were combined, providing transcript counts for input to MPAQT and for use in benchmarking.
For all analyses, reference transcriptome and genome annotations from GENCODE39 v38 was used, corresponding to human genome assembly GRCh38.p13.
RT-qPCR measurement of differential cassette exon inclusion
Selection of cassette exons
To aggregate isoform-level abundances into cassette exon-level measurements, we obtained the annotation of cassette exons using the “generateEvents” command from SUPPA2 v.2.340 for the reference transcript annotations. The abundances of all transcripts supporting each of the two possible outcomes of every event were then aggregated, providing, for each cassette exon in each sample, the sum-TPM of isoforms supporting the inclusion of the cassette exon and the sum TPM of isoforms supporting exon exclusion. This process was repeated separately for TPM inferences obtained by MPAQT using SR data and MPAQT using SR+LR data. We then fitted a limma41 model (using “limma” package v3.56.2) with the following formula for each cassette exon across all samples/measurement types: . Here, is the log-sum-TPM, is a multi-level sample indicator, is a binary indicator of whether the measurement belongs to the inclusion or exclusion set of transcripts, is a binary indicator of whether the measurement is based on SR+LR data or SR-only data , and is a multi-level timepoint indicator, with as the reference level. The coefficient of the interaction term indicates the degree to which Δlogit-PSI between days 0 and 61 changes if we switch from SR-only measurements to SR+LR measurements (note that logit-PSI of each exon is equal to log-sum-TPM of inclusion minus log-sum-TPM of exclusion transcripts). We used the empirical Bayes functionality of limma to extract the coefficient and associated P-value of this coefficient, followed by selection of the top nine cassette exons with the smallest P-values (P<4×10−5).
RT-qPCR measurements
Transcript levels were measured using RT–qPCR by reverse transcribing total RNA to complementary DNA (Maxima H Minus RT, Thermo), then using PerfeCTa SYBR Green SuperMix (QuantaBio) per the manufacturer’s instructions (for primer sequences, see Supplementary Data Table 3).
Sequence-based prediction of mRNA abundances
RNA-binding protein (RBP) motifs
Human RBP motifs were downloaded from CISBP-RNA19 (v0.6) and filtered to include only motifs obtained by RNA compete assays, encompassing 99 direct and 116 indirect (homology-based) motif-RBP associations, with 128 unique motifs and 128 unique RBPs (Supplementary Data Table 4). We used AffiMx18 to scan the 5’ and 3’ UTRs of all isoforms in GENCODE v38 with the 128 RNAcompete motifs, limiting to transcript isoforms with both annotated 5’ and 3’ UTRs. For each of the 5’ and 3’ UTR sets, the affinities were log-transformed and scaled for each motif separately (mean=0, variance=1), followed by application of convex nonnegative matrix factorization42 to obtain a non-redundant set of 35 “motif archetypes” and the score of each UTR for each archetype—each motif archetype is a convex combination of several motifs (usually with similar affinity profiles across the UTRs); in turn, the affinity profile of each motif can be reconstructed by a convex combination of motif archetypes. The number of motif archetypes was selected so as to result in a minimum Pearson correlation of 0.9 between the original and reconstructed affinity profiles per motif.
Training/validation of machine learning models
The 5’ and 3’ UTR motif archetypes were collated to obtain 70 motif archetype scores per transcript, which were used as predictive features for construction of machine learning models of isoform abundances. Specifically, for each neuronal differentiation time point (days 0, 41, and 61) and each of the two replicates, random forest models were constructed to predict log10 TPM of each isoform from its 5’ and 3’ UTR motif archetype scores, using a 5-fold gene-stratified cross-validation approach. In other words, genes were randomly assigned to five different folds, each time all isoforms of the genes in one of the folds were held out, a random forest model was trained on the remaining isoforms (using R “ranger” package v0.16.0 with default parameters), and the model was used to predict the abundances of held-out transcripts. SHAP values were also calculated on held-out transcripts, using the “explain” function from “fastshap” package v0.1.0.
Identification of differentiation-associated motif archetypes
For each motif archetype, we tested its contribution to differential mRNA abundances by examining how the relationship between the motif archetype scores and SHAP values change as a function of differentiation time point. For example, for transcripts with high motif archetype scores, if the SHAP value of that motif archetype increases through differentiation, it signifies increased stability of those transcripts due to the contribution of that motif archetype. In order to identify such associations, for each motif archetype, we concatenated the SHAP values across all transcripts and time points, and fitted a linear regression model of the form , where y is the SHAP value, is the differentiation time point (0, 41, or 61), and is the binary variable indicating whether a transcript is among the top 500 transcripts with the largest score for the motif archetype of interest or not . The coefficient of the interaction term represents the change in the SHAP value of top-scoring transcripts as a function of time, which we used to identify differentiation-associated motif archetypes.
Supplementary Material
Supplementary Methods.
Supplementary Figure 1. MPAQT’s performance on salmon and RSEM’s outliers.
Supplementary Figure 2. Comparison of quantification tools after widening the range of acceptable Cq values.
Supplementary Figure 3. Transcripts differentially quantified by MPAQT upon addition of LR data.
Supplementary Figure 4. Differentiation of hESCs to neurons.
Supplementary Figure 5. Differentially quantified transcripts between SR-only and LR+SR inferences at day 61.
Supplementary Figure 6. Cell type and disease associations of top genes whose cassette exons are differentially quantified between SR-only and LR+SR analyses.
Supplementary Figure 7. Predicting cassette exon inclusion.
Supplementary Data Table 1. Inference of gene-level expression using SR data from MAQC.
Supplementary Data Table 2. SR- and LR-based quantification of spike-in and endogenous mRNAs during hESC differentiation into cortical neurons.
Supplementary Data Table 3. RT-qPCR analysis of exon inclusion in hESCs and differentiated cortical neurons.
Supplementary Data Table 4. ML-based modeling of isoform abundances during hESC-to-neuron differentiation.
Supplementary Data Table 5. ML-based prediction of cassette exon inclusion during hESC-to-neuron differentiation.
Acknowledgements
We thank Aldo H. Corchado for technical support. This work was supported by the Canadian Institutes of Health Research (CIHR) grant PJT-173317 and resource allocations from Digital Research Alliance of Canada to HSN. MA was supported by a Canada Graduate Scholarships-Master’s award from CIHR. HG is an Era of Hope Scholar (W81XWH-2210121) and supported by grants from the National Cancer Institute (R01CA240984 and R01CA244634). HSN holds a CIHR Canada Research Chair.
Footnotes
Competing interests
The authors declare no competing interests.
Code availability
MPAQT is available at https://github.com/csglab/MPAQT.
Data availability
All processed data generated as part of this study are provided as Supplementary Datasets. ML models are available via Zenodo (DOI: 10.5281/zenodo.12637434). Raw RNA-sequencing data generated in this study are available via GEO (accession numbers GSE271530).
References
- 1.Arzalluz-Luque A. & Conesa A. Single-cell RNAseq for the study of isoforms-how is that possible? Genome Biol 19, 110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belluti S., Rigillo G. & Imbriano C. Transcription Factors in Cancer: When Alternative Splicing Determines Opposite Cell Fates. Cells 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tushev G. et al. Alternative 3’ UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments. Neuron 98, 495–511 e496 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Clark M.B. et al. Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain. Mol Psychiatry 25, 37–47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su C.H., D D. & Tarn W.Y. Alternative Splicing in Neurogenesis and Brain Development. Front Mol Biosci 5, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray N.L., Pimentel H., Melsted P. & Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Tang A.D. et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat Commun 11, 1438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdel-Ghany S.E. et al. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun 7, 11706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu S. et al. IDP-denovo: de novo transcriptome assembly and isoform annotation by hybrid sequencing. Bioinformatics 34, 2168–2176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shumate A., Wong B., Pertea G. & Pertea M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput Biol 18, e1009730 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y. et al. Context-aware transcript quantification from long-read RNA-seq data with Bambu. Nat Methods 20, 1187–1195 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patro R., Duggal G., Love M.I., Irizarry R.A. & Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B. & Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium S.M.-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol 32, 903–914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everaert C. et al. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep 7, 1559 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolberg L., Raudvere U., Kuzmin I., Vilo J. & Peterson H. gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zylka M.J., Simon J.M. & Philpot B.D. Gene length matters in neurons. Neuron 86, 353–355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert S.A., Albu M., Hughes T.R. & Najafabadi H.S. Motif comparison based on similarity of binding affinity profiles. Bioinformatics 32, 3504–3506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray D. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jing F. et al. The PABPC5/HCG15/ZNF331 Feedback Loop Regulates Vasculogenic Mimicry of Glioma via STAU1-Mediated mRNA Decay. Mol Ther Oncolytics 17, 216–231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlasova-St Louis I., Dickson A.M., Bohjanen P.R. & Wilusz C.J. CELFish ways to modulate mRNA decay. Biochim Biophys Acta 1829, 695–707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Guijarro E. et al. Lineage-specific roles of the cytoplasmic polyadenylation factor CPEB4 in the regulation of melanoma drivers. Nat Commun 7, 13418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suner C. et al. Macrophage inflammation resolution requires CPEB4-directed offsetting of mRNA degradation. Elife 11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q. et al. Pseudogene ACTBP2 increases blood-brain barrier permeability by promoting KHDRBS2 transcription through recruitment of KMT2D/WDR5 in Abeta(1-)(42) microenvironment. Cell Death Discov 7, 142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reese F. et al. The ENCODE4 long-read RNA-seq collection reveals distinct classes of transcript structure diversity. bioRxiv, 2023.2005.2015.540865 (2023). [Google Scholar]
- 26.Browne P.D. et al. GC bias affects genomic and metagenomic reconstructions, underrepresenting GC-poor organisms. Gigascience 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong H.Y., Barash Y. & Frey B.J. Bayesian prediction of tissue-regulated splicing using RNA sequence and cellular context. Bioinformatics 27, 2554–2562 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Barash Y. et al. Deciphering the splicing code. Nature 465, 53–59 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Xiong H.Y. et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 347, 1254806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jha A., Gazzara M.R. & Barash Y. Integrative deep models for alternative splicing. Bioinformatics 33, i274–i282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michielsen L. et al. Predicting cell-type-specific exon inclusion in the human brain reveals more complex splicing mechanisms in neurons than glia. bioRxiv, 2024.2003.2018.585465 (2024). [Google Scholar]
- 32.Schneider-Poetsch T., Chhipi-Shrestha J.K. & Yoshida M. Splicing modulators: on the way from nature to clinic. J Antibiot (Tokyo) 74, 603–616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardo-Palacios F.J. et al. Systematic assessment of long-read RNA-seq methods for transcript identification and quantification. Nat Methods (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Townes F.W., Hicks S.C., Aryee M.J. & Irizarry R.A. Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model. Genome Biol 20, 295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y., Smyth G.K. & Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47, e47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian A., Muffat J. & Li Y. Studying Human Neurodevelopment and Diseases Using 3D Brain Organoids. J Neurosci 40, 1186–1193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fish L. et al. A prometastatic splicing program regulated by SNRPA1 interactions with structured RNA elements. Science 372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardo-Palacios F.J. et al. SQANTI3: curation of long-read transcriptomes for accurate identification of known and novel isoforms. Nat Methods 21, 793–797 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frankish A. et al. Gencode 2021. Nucleic Acids Res 49, D916–D923 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trincado J.L. et al. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol 19, 40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie M.E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding C.H.Q., Li T. & Jordan M.I. Convex and Semi-Nonnegative Matrix Factorizations. IEEE Transactions on Pattern Analysis and Machine Intelligence 32, 45–55 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods.
Supplementary Figure 1. MPAQT’s performance on salmon and RSEM’s outliers.
Supplementary Figure 2. Comparison of quantification tools after widening the range of acceptable Cq values.
Supplementary Figure 3. Transcripts differentially quantified by MPAQT upon addition of LR data.
Supplementary Figure 4. Differentiation of hESCs to neurons.
Supplementary Figure 5. Differentially quantified transcripts between SR-only and LR+SR inferences at day 61.
Supplementary Figure 6. Cell type and disease associations of top genes whose cassette exons are differentially quantified between SR-only and LR+SR analyses.
Supplementary Figure 7. Predicting cassette exon inclusion.
Supplementary Data Table 1. Inference of gene-level expression using SR data from MAQC.
Supplementary Data Table 2. SR- and LR-based quantification of spike-in and endogenous mRNAs during hESC differentiation into cortical neurons.
Supplementary Data Table 3. RT-qPCR analysis of exon inclusion in hESCs and differentiated cortical neurons.
Supplementary Data Table 4. ML-based modeling of isoform abundances during hESC-to-neuron differentiation.
Supplementary Data Table 5. ML-based prediction of cassette exon inclusion during hESC-to-neuron differentiation.
Data Availability Statement
All processed data generated as part of this study are provided as Supplementary Datasets. ML models are available via Zenodo (DOI: 10.5281/zenodo.12637434). Raw RNA-sequencing data generated in this study are available via GEO (accession numbers GSE271530).