Abstract
Background
The changes in the tumor microenvironment of high-grade serous ovarian carcinomas following neoadjuvant chemotherapy are a complex area of study. Previous research underscores the importance of investigating the immune and collagen components within the tumor microenvironment for prognostic implications.
Methods
In this study, we utilized computational pathology techniques with Hematoxylin and Eosin-stained images to quantitatively characterize the immune and collagen architecture within the tumor microenvironment of patients with high-grade serous ovarian carcinoma.
Results
Our analysis of 12 pre- and post-neoadjuvant chemotherapy images revealed an increase in immune infiltrate, primarily within the epithelial region. Additionally, post-neoadjuvant chemotherapy images exhibited chaotic collagen architecture compared to pre-neoadjuvant chemotherapy images. Importantly, features extracted from post-neoadjuvant chemotherapy images showed associations with overall survival, potentially aiding in the selection of patients for immunotherapy trials.
Conclusions
These findings offer critical insights into the changes in the tumor microenvironment of high-grade serous ovarian carcinomas following neoadjuvant chemotherapy and their potential implications for clinical outcomes.
Abbreviations
- TME
Tumor microenvironment
- HGSOC
High-grade serous ovarian carcinoma
- NACT
Neoadjuvant chemotherapy
- H&E
Hematoxylin and eosin
- WSI
Whole-slide image
- TIL:
Tumor-infiltrating lymphocyte
- ECM
Extracellular matrix
- HR
Hazard ratio
- CI
Confidence interval
- FIGO
Federation of Gynecology and Obstetrics
- UPMC
University of Pittsburgh Medical Center Magee Women's Hospital
1. Introduction
The standard treatment approach for high-grade serous ovarian carcinomas (HGSOC) typically involves either cancer-directed debulking surgery followed by adjuvant platinum-taxane chemotherapy or neoadjuvant chemotherapy (NACT) followed by interval cytoreductive surgery [1]. NACT, also known as preoperative chemotherapy, is widely used in managing HGSOC [2]. Furthermore, immunotherapy employing immune checkpoint inhibition has gained increasing popularity as a treatment option due to its durable effects and lower toxicity [[3], [4], [5]].
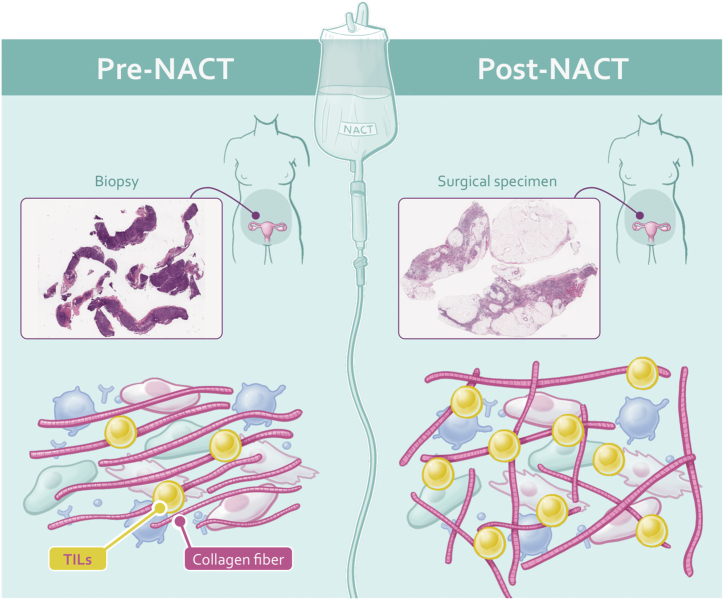
Recent studies have shed light on changes in the tumor microenvironment (TME) composition in HGSOC patients after NACT. This insight arises from analyzing paired samples collected before and after NACT administration in the same patient [6,7], raising crucial questions about the dynamic changes within the TME of HGSOC patients post-NACT that could potentially influence clinical outcomes (Fig. 1).
Fig. 1.
Illustrates the comparison of tumor microenvironment (TME) characteristics in patients with high-grade serous ovarian carcinoma before and after neoadjuvant chemotherapy (NACT). Pre-NACT H&E images show lower TIL density and less disorder in collagen fiber orientations, whereas post-NACT H&E images demonstrate higher TIL density and higher disorder in collagen fiber orientations. TME, Tumor Microenvironment; TIL, Tumor-Infiltrating Lymphocytes.
Immune cells, particularly tumor-infiltrating lymphocytes (TILs), are essential components in the battle against cancer [8]. While previous research indicates increased TIL infiltration after NACT, there remains a need for a comprehensive quantitative assessment of the immune milieu in these samples [6,7]. Machine learning methods have become effective tools for quantifying the immune milieu from Hematoxylin and Eosin (H&E) Whole Slide Images (WSIs), demonstrating potential in prognostic evaluation [9]. Moreover, collagen, the predominant protein found in the extracellular matrix (ECM), significantly influences cancer progression and metastasis [10]. Recent developments in computational pathology have facilitated the quantitative assessment of collagen structure within the TME using H&E WSIs [11].
In this study, we adopt a unique computational pathology approach using H&E WSIs to quantitatively characterize the immune and collagen architecture within the TME of HGSOC patients. Our main objectives are threefold: a) to quantitatively analyze the immune architecture; b) to quantitatively analyze the disorder in collagen architecture within the TME in pre- and post-NACT H&E WSIs of HGSOC patients; and c) to investigate the association of the extracted features from post-NACT H&E WSIs with patient survival outcomes in HGSOC patients.
2. Methods
2.1. Dataset
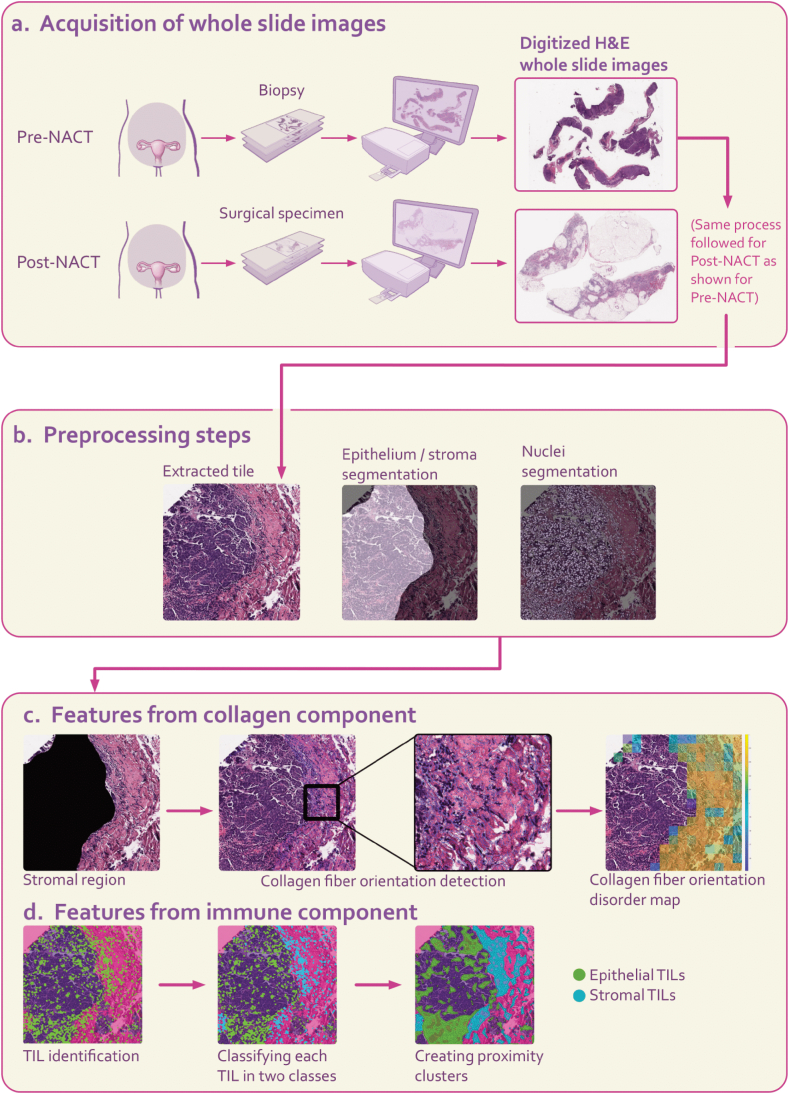
This study focused on HGSOC samples obtained from the peritoneum region and adhered to specific inclusion criteria, including the following: 1) cases with a preoperative pathological diagnosis of HGSOC confirmed through diagnostic laparoscopic biopsy; 2) cases that underwent at least one cycle of NACT followed by interval debulking surgery; 3) stage III/IV disease as classified by the International Federation of Gynecology and Obstetrics (FIGO) 2014 criteria; 4) cases for which overall survival information was available. This dataset underwent meticulous curation with active involvement from an expert pathologist with extensive experience in ovarian cancer. Consequently, we collected 12 pairs of pre- and post-NACT H&E WSIs from patients diagnosed and treated at the University of Pittsburgh Medical Center Magee Women's Hospital (UPMC) between 2004 and 2016 (Fig. 2a).
Fig. 2.
Overall framework for extracting features from the collagen and tumor-infiltrating lymphocyte (TIL) components within the tumor microenvironment (TME) of H&E images. a H&E images from both pre- and post-NACT stages of the same patient were acquired. The pre-NACT H&E image was derived from a biopsy, whereas the post-NACT H&E image originated from a surgical specimen. b Tiles of size 3000x3000-pixel were extracted from each pre- and post-NACT H&E image. These tiles underwent preprocessing steps for feature extraction, including epithelium/stroma segmentation and nuclei segmentation. c The first set of features focused on quantitative aspects of Collagen Fiber Orientation Disorder (CFOD) in stromal regions. Collagen fiber orientations were determined using a derivative-of-Gaussian model. An orientation co-occurrence matrix was generated, where brighter on-diagonal cells indicated higher co-occurrence of collagen fibers with similar orientations. A feature quantifying the disorder of collagen fiber orientations was computed from this matrix. Warmer colors in the feature map represented greater disorder in collagen fiber orientations. d Another set of features characterized TIL architecture quantitatively in epithelial and stromal regions. HGSOC, high grade serous ovarian carcinoma; TIL, tumor-infiltrating lymphocytes; CFOD, collagen fiber orientation disorder.
2.2. Clinicopathologic variables of the cohort
A total of 12 patients with HGSOC underwent NACT followed by interval debulking surgery, with pre- and post-NACT H&E WSIs available (Supplementary Table 1). The median age of this cohort was 67 years, with an interquartile range of 60.5–73.5 years. Most of the patients (10 out of 12, or 83.3 %) had FIGO stage III disease.
2.3. Feature extraction process
-
-
Preprocessing steps
We extracted non-overlapping 3000x3000-pixel tiles from H&E WSIs. To segment epithelial and stromal regions within each tile, we employed a pretrained U-Net architecture model. This model demonstrated pixel accuracy ranging from 95.8 % to 96.6 % and a dice coefficient ranging from 82.3 % to 89 % [12]. For nuclei segmentation within each tile, we used the Hovernet model [13,14] (Fig. 2b).
-
-
Collagen component within the TME
Fig. 2c illustrates how collagen fiber orientation disorder is quantified in stromal regions of H&E WSIs. To achieve this, each tile was divided into 200x200-pixel neighborhoods, and a derivative-of-Gaussian model was applied to capture fiber orientations by identifying linear structures within the stromal regions. An orientation co-occurrence matrix was then created for each neighborhood, which captures the distribution of these orientations. The disorder in collagen fiber orientations within the stromal regions was quantitatively measured using entropy theory, where entropy signifies the uncertainty in fiber orientation within each tumor neighborhood. This approach is further detailed in the work by Li et al. [11]. From these feature maps, first-order statistics (mean, minimum, and maximum) were computed, resulting in three features per patient.
-
-
Immune component within the TME
Fig. 2d depicts the process of feature extraction for the immune component, focusing on the geospatial architecture of TIL nuclei on the H&E WSI. After segmenting nuclei regions on each tile, the subsequent step involved classifying each nucleus as TIL or non-TIL. This classification was accomplished using a pre-trained support vector machine with a linear kernel, achieving a precision of 89.12 %, recall of 83.57 %, and an F1 score of 86.31 % [15]. The spatial arrangement of TILs was characterized by forming clusters of nuclei classes (epithelial TILs and stromal TILs) based on proximity. The methodology is described in more detail in the study by Corredor et al. [16]. For each patient, we derived three features from both the pre- and post-NACT H&E WSIs, representing TIL density, the mean density of epithelial TIL clusters, and the mean density of stromal TIL clusters.
2.4. Statistical analysis
We employed the Wilcoxon matched-pairs signed-rank test for paired comparisons [17]. Each feature extracted from the post-NACT H&E WSIs was treated as a continuous risk score for subsequent survival analysis. This score was then split into high and low values using the median threshold. To compare the time-to-event data between these groups, we utilized Kaplan-Meier survival analysis in conjunction with the log-rank test [17]. Model performance was assessed using Hazard Ratio (HR) and their corresponding 95 % Confidence Interval (CI). Statistical significance was determined using a two-sided threshold of p < 0.05.
Ethical statement
This research was conducted following Emory University Institutional Review Board (IRB) protocol STUDY00005888, which was sanctioned as a non-human study and ensuring adherence to all applicable ethical guidelines. De-identified human samples were sourced from UPMC, gathered under the same IRB-approved protocol STUDY00005888. UPMC obtained the specimens with informed consent from the participants.
3. Results
3.1. Changes in the immune component following NACT
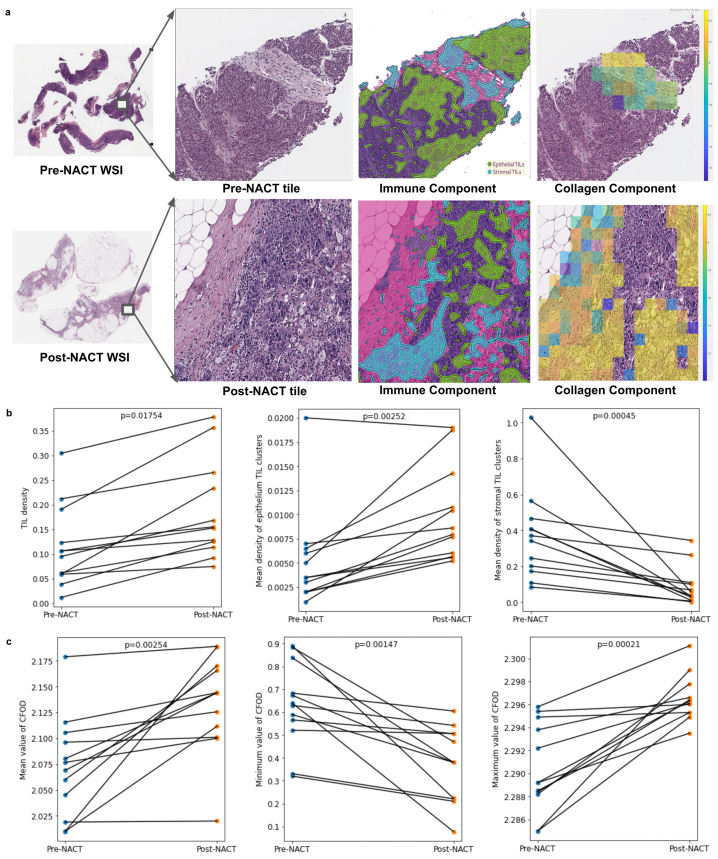
We conducted an experiment to explore the impact of NACT on immune architecture using pre- and post-NACT H&E WSIs. Our results showed a significant overall increase in TIL density (p-value = 0.01754), especially in the epithelial region which had a higher mean density of TIL clusters (p-value = 0.00252). Additionally, we observed a decrease in the mean density of stromal TIL clusters (p-value = 0.00045), indicating a shift in the immune response within stromal regions due to NACT (Methods, Fig. 3a–b).
Fig. 3.
Assessment of changes in collagen and tumor-infiltrating lymphocyte (TIL) components within the tumor microenvironment (TME) of patients with high-grade serous ovarian carcinoma following neoadjuvant chemotherapy (NACT). a Qualitative visualization of changes in the collagen and TIL components within the TME of pre- and post-NACT H&E images. b Quantitative changes in the TIL component (TIL density, mean density of epithelial TIL clusters, mean density of stromal TIL clusters) within the TME of pre- and post-NACT H&E images. c Quantitative changes in the collagen component (mean, minimum, and maximum entropy values of collagen fiber orientation disorder) within the TME of pre- and post-NACT H&E images. TIL, tumor-infiltrating lymphocytes; TME, tumor microenvironment; NACT, neoadjuvant chemotherapy; CFOD, collagen fiber orientation disorder. P-value, Wilcoxon matched-paired signed-ranked test for paired samples.
3.2. Changes in the collagen component following NACT
To assess the impact on collagen architecture, we compared entropy values representing the disorder in collagen fiber orientations within stromal regions among patients with both pre- and post-NACT H&E WSIs. Our analysis revealed significant differences in the mean (p-value = 0.00254), maximum (p-value = 0.00021), and minimum (p-value = 0.00147) entropy values. These findings indicate that collagen fiber orientations within stromal regions are more disordered in post-NACT H&E WSIs compared to pre- NACT H&E WSIs (Methods, Fig. 3c).
3.3. Association of post-NACT features with prognosis
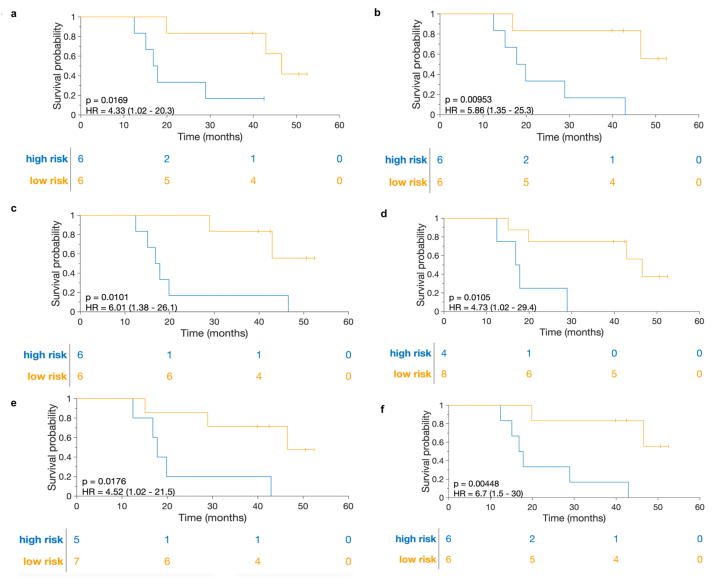
Each extracted feature from post-NACT H&E WSIs demonstrated significant associations with overall survival. These features include TIL density (p-value = 0.017, HR = 4.33, 95 % CI = 1.02–20.3), mean density of epithelial TIL clusters (p-value = 0.0095, HR = 5.86, 95 % CI = 1.35–25.3), mean density of stromal TIL clusters (p-value = 0.01, HR = 6.01, 95 % CI = 1.38–26.1), mean entropy value of collagen fiber orientation disorder (p-value = 0.01, HR = 4.73, 95 % CI = 1.02–29.4), minimum entropy value of collagen fiber orientation disorder (p-value = 0.018, HR = 4.52, 95 % CI = 1.02–21.5), and maximum entropy value of collagen fiber orientation disorder (p-value = 0.0045, HR = 6.7, 95 % CI = 1.5–30) (Methods, Fig. 4a–f).
Fig. 4.
Kaplan-Meier curves illustrating the prediction of overall survival based on features extracted from post-neoadjuvant chemotherapy (NACT) H&E images of patients with high-grade serous ovarian carcinoma. a Tumor-infiltrating lymphocyte (TIL) density. b mean density of epithelium TIL clusters. c mean density of stromal TIL clusters. d mean entropy value of the disorder in collagen fiber orientations. e minimum entropy value of the disorder in collagen fiber orientations. f maximum entropy value of the disorder in collagen fiber orientations. NACT, neoadjuvant chemotherapy; TIL, tumor-infiltrating lymphocytes.
4. Discussion
Recent studies have provided valuable insights into changes in the composition of the TME in HGSOC patients following NACT [6,7]. Mesnage et al. observed an increase in TIL levels following NACT in individuals with epithelial ovarian cancer, suggesting a potential enhancement of the immune response within the TME [7]. Lee et al. reported dynamic shifts in the TME following NACT, characterized by elevated TIL levels [6]. Moreover, it is crucial to investigate whether this heightened presence of TILs disrupts the organization of collagen fibers, as noted in previous studies [18,19]. Thus, understanding these mechanisms can provide insights into the complex interplay between NACT, TME, and tumor progression among HGSOC patients.
In this study, we employed a computational pathology approach using H&E WSIs to examine the changes in the immune and collagen components within the TME following NACT. It is important to note that our analysis is centered around exploring correlations, and establishing a causative association is beyond the scope of our current study's objectives. We observed a significant increase in TIL density (p-value = 0.01754), particularly in the epithelial region with higher mean density of TIL clusters (p-value = 0.00252). Conversely, we observed a decrease in the mean density of stromal TIL clusters (p-value = 0.00045), indicating a shift in the immune response within stromal regions following NACT. Furthermore, our study highlighted a higher disorder in collagen fiber orientations in the stromal regions in post-NACT H&E WSIs compared to pre-NACT H&E WSIs. Importantly, we also found that each extracted feature from post-NACT H&E WSIs of HGSOC patients was associated with overall survival, which may help select patients for immunotherapy trials.
Looking ahead, we anticipate rapid evolution in the field of computational pathology-based prognostic biomarkers for HGSOC over the next five years. Furthermore, integrating multi-omics data and immune profiling approaches will facilitate a more comprehensive characterization of the TME, laying the groundwork for personalized treatment strategies in HGSOC [20]. Overall, our study contributes to this evolving landscape and sets the stage for further exploration of the intricacies of the TME in HGSOC.
We acknowledge several limitations in our study. Firstly, our small sample size is a notable constraint that could potentially impact the generalizability of our findings. Future work will focus on expanding the dataset to overcome the limitation of the small sample size. Additionally, we did not distinguish tertiary lymphoid structures from stromal TIL clusters within the TME of HGSOC patients [21]. Further investigation into this aspect could be crucial, as tertiary lymphoid structures could be indicative of an active immune response, while stromal TIL clusters could signify a more chronic inflammatory process. Another limitation was the lack of specialized staining techniques like Mason's trichrome or Mallory's trichrome, which would have enabled a quantitative analysis of collagen fiber segmentation. This limitation arose because Mason's trichrome or Mallory's trichrome-stained WSIs were not available for the cohort used in our study. Despite the absence of these staining techniques, we validated the results of our method through input from pathologists. This validation helped ensure the accuracy and reliability of our findings.
In conclusion, while our study contributes to the growing body of knowledge on TME dynamics post-NACT in HGSOC, there is immense potential for further exploration and translation into clinical practice. By addressing knowledge gaps, leveraging advanced technologies, and fostering interdisciplinary collaborations, we envision a future where TME-focused therapies revolutionize the management of ovarian cancer and other solid tumors, leading to improved patient outcomes and survival rates.
Data availability statement
All data required to reproduce the results presented here are available at https://github.com/arp95/pre_post_nact_hgsoc. The H&E images associated with the study have not been deposited into a publicly available repository. The authors do not have permission to share data. Requests to access the dataset from UPMC (used with permission for this study) should be made directly to the institutions via their data access request forms.
CRediT authorship contribution statement
Arpit Aggarwal: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Formal analysis, Conceptualization. Germán Corredor: Writing – review & editing, Supervision, Methodology, Conceptualization. Pingfu Fu: Writing – review & editing, Investigation. Tilak Pathak: Writing – review & editing, Resources, Project administration, Investigation. Tuomas Mirtti: Writing – review & editing, Resources, Project administration, Investigation. Susan Modesitt: Writing – review & editing, Project administration, Investigation. T. Rinda Soong: Writing – review & editing, Resources, Project administration, Investigation, Data curation. Anant Madabhushi: Writing – review & editing, Supervision, Project administration, Investigation, Funding acquisition.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Anant Madabhushi reports a relationship with Picture Health, Elucid Bioimaging, and Inspirata Inc that includes: equity or stocks. Anant Madabhushi reports a relationship with Picture Health, Aiforia Inc, and SimBioSys that includes: board membership. Anant Madabhushi reports a relationship with AstraZeneca, Boehringer-Ingelheim, Eli-Lilly and Bristol Myers-Squibb that includes: consulting or advisory. Anant Madabhushi has patent #R01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01, 1U54CA254566-01 issued to National Cancer Institute. Anant Madabhushi has patent #1R01HL15127701A1, R01HL15807101A1 issued to National Heart, Lung and Blood Institute. Anant Madabhushi has patent #1R43EB028736-01 issued to National Institute of Biomedical Imaging and Bioengineering. Anant Madabhushi has patent #1 C06 RR12463-01 issued to National Center for Research Resources. Declaration of interests Dr. Madabhushi is an equity holder in Picture Health, Elucid Bioimaging, and Inspirata Inc. Currently he serves on the advisory board of Picture Health, Aiforia Inc, and SimBioSys. He also currently consults for SimBioSys. He also has sponsored research agreements with AstraZeneca, Boehringer-Ingelheim, Eli-Lilly and Bristol Myers-Squibb. His technology has been licensed to Picture Health and Elucid Bioimaging. He is also involved in 3 different R01 grants with Inspirata Inc. He also serves as a member for the Frederick National Laboratory Advisory Committee.
Acknowledgements Research reported in this publication was supported by the National Cancer Institute under award numbers R01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01, 1U54CA254566-01, National Heart, Lung and Blood Institute 1R01HL15127701A1, R01HL15807101A1, National Institute of Biomedical Imaging and Bioengineering 1R43EB028736-01, National Center for Research Resources under award number 1 C06 RR12463-01, VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404, W81XWH-21-1-0345, W81XWH-21-1-0160), the Mayo Clinic Breast Cancer SPORE grant P50 CA116201 from the NIH, the Kidney Precision Medicine Project (KPMP) Glue Grant, and sponsored research agreements from Bristol Myers-Squibb, Boehringer-Ingelheim, Eli-Lilly and AstraZeneca. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is made possible by the National Cancer Institute under award numbers R01CA249992-01A1, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, R01CA257612-01A1, U01CA239055–01, U01CA248226–01, U54CA254566-01, National Heart, Lung and Blood Institute R01HL151277-01A1, R01HL158071-01A1, National Institute of Biomedical Imaging and Bioengineering R43EB028736-01, National Center for Research Resources under award number C06 RR12463-01, VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-18-1-0404, W81XWH-21-1-0345), the Kidney Precision Medicine Project (KPMP) Glue Grant, DoD Prostate Cancer Research Program Idea Development Award W81XWH-18-1-0524, Clinical and Translational Science Collaborative (CTSC) Cleveland Annual Pilot Award 2020 UL1TR002548 and sponsored research agreements from Bristol Myers-Squibb, Boehringer-Ingelheim, Eli-Lilly and Astrazeneca, DoD Peer Reviewed Cancer Research Program (W81XWH-22-1-0236), Winship Invest$ Pilot Grant, American Cancer Society Institutional Research Grant from the Winship Cancer Institute.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e33618.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Penn C.A., Alvarez R.D. Current issues in the management of patients with newly diagnosed advanced-stage high-grade serous carcinoma of the ovary. JCO Oncol Pract. 2023 Jan 5;19(3):116–122. doi: 10.1200/OP.22.00461. Available from: [DOI] [PubMed] [Google Scholar]
- 2.Patel A., Iyer P., Matsuzaki S., Matsuo K., Sood A.K., Fleming N.D. Emerging trends in neoadjuvant chemotherapy for ovarian cancer. Cancers. MDPI AG. 2021;13:1–19. doi: 10.3390/cancers13040626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mollica V., Rizzo A., Marchetti A., Tateo V., Tassinari E., Rosellini M., et al. The impact of ECOG performance status on efficacy of immunotherapy and immune-based combinations in cancer patients: the MOUSEION-06 study. Clin. Exp. Med. 2023 Dec 1;23(8):5039–5049. doi: 10.1007/s10238-023-01159-1. [DOI] [PubMed] [Google Scholar]
- 4.Ricci A.D., Rizzo A., Novelli M., Tavolari S., Palloni A., Tober N., et al. Vol. 40. 2020. Specific toxicity of maintenance olaparib versus placebo in advanced malignancies: a systematic review and meta-analysis; pp. 597–608. (Anticancer Research. International Institute of Anticancer Research). [DOI] [PubMed] [Google Scholar]
- 5.Santoni M., Rizzo A., Mollica V., Matrana M.R., Rosellini M., Faloppi L., et al. Vol. 170. Elsevier Ireland Ltd; 2022. The impact of gender on the efficacy of immune checkpoint inhibitors in cancer patients: the MOUSEION-01 study. (Critical Reviews in Oncology/Hematology). [DOI] [PubMed] [Google Scholar]
- 6.Lee Y.J., Woo H.Y., Kim Y.N., Park J., Nam E.J., Kim S.W., et al. Dynamics of the tumor immune microenvironment during neoadjuvant chemotherapy of high-grade serous ovarian cancer. Cancers. 2022 May 1;14(9) doi: 10.3390/cancers14092308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesnage S.J.L., Auguste A., Genestie C., Dunant A., Pain E., Drusch F., et al. Neoadjuvant chemotherapy (NACT) increases immune infiltration and programmed death-ligand 1 (PD-L1) expression in epithelial ovarian cancer (EOC) Ann. Oncol. 2017 Mar 1;28(3):651–657. doi: 10.1093/annonc/mdw625. [DOI] [PubMed] [Google Scholar]
- 8.Galli F., Aguilera J.V., Palermo B., Markovic S.N., Nisticò P., Signore A. Vol. 39. Journal of Experimental and Clinical Cancer Research. BioMed Central Ltd.; 2020. (Relevance of Immune Cell and Tumor Microenvironment Imaging in the New Era of Immunotherapy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azarianpour S., Corredor G., Bera K., Leo P., Fu P., Toro P., et al. Computational image features of immune architecture is associated with clinical benefit and survival in gynecological cancers across treatment modalities. J Immunother Cancer. 2022 Feb 3;10(2) doi: 10.1136/jitc-2021-003833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu K.S., Dunleavey J.M., Szot C., Yang L., Hilton M.B., Morris K., et al. Cancer cell survival depends on collagen uptake into tumor-associated stroma. Nat. Commun. 2022 Dec 1;13(1) doi: 10.1038/s41467-022-34643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Bera K., Toro P., Fu P.F., Zhang Z., Lu C., et al. Collagen fiber orientation disorder from H&E images is prognostic for early stage breast cancer: clinical trial validation. NPJ Breast Cancer. 2021 Dec 1;7(1) doi: 10.1038/s41523-021-00310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y., Koyuncu C.F., Toro P., Corredor G., Feng Q., Buzzy C., et al. A machine learning model for separating epithelial and stromal regions in oral cavity squamous cell carcinomas using H&E-stained histology images: a multi-center, retrospective study. Oral Oncol. 2022 Aug 1:131. doi: 10.1016/j.oraloncology.2022.105942. [DOI] [PubMed] [Google Scholar]
- 13.Graham S., Vu Q.D., Raza S.E.A., Azam A., Tsang Y.W., Kwak J.T., et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 2019 Dec 1;58 doi: 10.1016/j.media.2019.101563. [DOI] [PubMed] [Google Scholar]
- 14.Gamper J., Koohbanani N.A., Benes K., Graham S., Jahanifar M., Khurram S.A., et al. PanNuke dataset extension, insights and baselines. 2020 Mar 24. http://arxiv.org/abs/2003.10778 Available from:
- 15.Romero Castro E., Corredor G., Lu C., Madabhushi A., Wang X., Velcheti V. In SPIE-Intl Soc Optical Eng; 2018. A Watershed and Feature-Based Approach for Automated Detection of Lymphocytes on Lung Cancer Images; p. 26. [Google Scholar]
- 16.Corredor G., Wang X., Zhou Y., Lu C., Fu P., Syrigos K., et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non–small cell lung cancer. Clin. Cancer Res. 2019 Mar 1;25(5):1526–1534. doi: 10.1158/1078-0432.CCR-18-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macfarland TW, Yates JM. Introduction to Nonparametric Statistics for the Biological Sciences Using R.
- 18.Edsparr K., Basse P.H., Goldfarb R.H., Albertsson P. vol. 4. Cancer Microenvironment; 2011. pp. 351–360. (Matrix Metalloproteinases in Cytotoxic Lymphocytes Impact on Tumour Infiltration and Immunomodulation). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pires A., Greenshields-Watson A., Jones E., Smart K., Lauder S.N., Somerville M., et al. Immune remodeling of the extracellular matrix drives loss of cancer stem cells and tumor rejection. Cancer Immunol. Res. 2020 Dec 1;8(12):1520–1531. doi: 10.1158/2326-6066.CIR-20-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao G., Hua D., Li J., Zhang X., Zhang Z., Zhang B., et al. Tumor immune microenvironment changes are associated with response to neoadjuvant chemotherapy and long-term survival benefits in advanced epithelial ovarian cancer: a pilot study. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1022942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou Y., Qiao S., Li M., Han X., Wei X., Pang Y., et al. The gene signature of tertiary lymphoid structures within ovarian cancer predicts the prognosis and immunotherapy benefit. Front. Genet. 2023 Jan;10:13. doi: 10.3389/fgene.2022.1090640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data required to reproduce the results presented here are available at https://github.com/arp95/pre_post_nact_hgsoc. The H&E images associated with the study have not been deposited into a publicly available repository. The authors do not have permission to share data. Requests to access the dataset from UPMC (used with permission for this study) should be made directly to the institutions via their data access request forms.