Abstract
We developed a bio-cheminformatics method, exploring disease inhibition mechanisms using machine learning-enhanced quantitative structure-activity relationship (ML-QSAR) models and knowledge-driven neural networks. ML-QSAR models were developed using molecular fingerprint descriptors and the Random Forest algorithm to explore the chemical spaces of Chalcones inhibitors against diverse disease properties, including antifungal, anti-inflammatory, anticancer, antimicrobial, and antiviral effects. We generated and validated robust machine learning-based bioactivity prediction models (https://github.com/RatulChemoinformatics/QSAR) for the top genes. These models underwent ROC and applicability domain analysis, followed by molecular docking studies to elucidate the molecular mechanisms of the molecules. Through comprehensive neural network analysis, crucial genes such as AKT1, HSP90AA1, SRC, and STAT3 were identified. The PubChem fingerprint-based model revealed key descriptors: PubchemFP521 for AKT1, PubchemFP180 for SRC, PubchemFP633 for HSP90AA1, and PubchemFP145 and PubchemFP338 for STAT3, consistently contributing to bioactivity across targets. Notably, chalcone derivatives demonstrated significant bioactivity against target genes, with compound RA1 displaying a predictive pIC50 value of 5.76 against HSP90AA1 and strong binding affinities across other targets. Compounds RA5 to RA7 also exhibited high binding affinity scores comparable to or exceeding existing drugs. These findings emphasize the importance of knowledge-based neural network-based research for developing effective drugs against diverse disease properties. These interactions warrant further in vitro and in vivo investigations to elucidate their potential in rational drug design. The presented models provide valuable insights for inhibitor design and hold promise for drug development. Future research will prioritize investigating these molecules for mycobacterium tuberculosis, enhancing the comprehension of effectiveness in addressing infectious diseases.
Keywords: Chalcone; Gene (AKT, SRC, HSP90AA1, and STAT3); Systems biology; Ml-QSAR; Molecular docking; Dynamics
Graphical Abstract
1. Introduction
Chalcones, an important subclass within the flavonoid family of organic compounds, exhibit a unique structural arrangement, characterized by three aromatic rings (A, B, and C) linked through an α, β-unsaturated carbonyl system[1], [2]. This distinctive arrangement not only defines their chemical identity but also plays a pivotal role in their diverse biological activities. Naturally occurring in various plants, chalcones have attracted considerable attention due to their wide-ranging biological effects[3]. These compounds exhibit a broad spectrum of biological activities, including cytoprotective and regulatory functions, which are essential for their diverse therapeutic applications. Their roles in reducing inflammation and combating cancer, malaria, tuberculosis, and microbial infections highlight their therapeutic potential, particularly in developing new treatments for infectious diseases [4], [5], [6], [7].
Chalcones with specific substitutions on their aromatic rings have shown significant biological effects. Chalcones with a trifluoromethyl group in ring B and a 3,4,5-trimethoxy substitution in ring A have shown strong antiproliferative effects against cancer cells. Similarly, derivatives with certain substitutions have exhibited notable antifungal and antibacterial activities [8], [9]. The presence of electron-withdrawing groups can enhance anti-inflammatory activity, while specific substitutions on the phenyl ring can improve anti-hyperglycemic effects. Recently, chalcones have also shown promise against mycobacterium tuberculosis, indicating their potential in developing new tuberculosis treatments. The growing interest in their anti-mycobacterial capabilities further indicates their potential to develop new treatments for infectious diseases [10], [11].
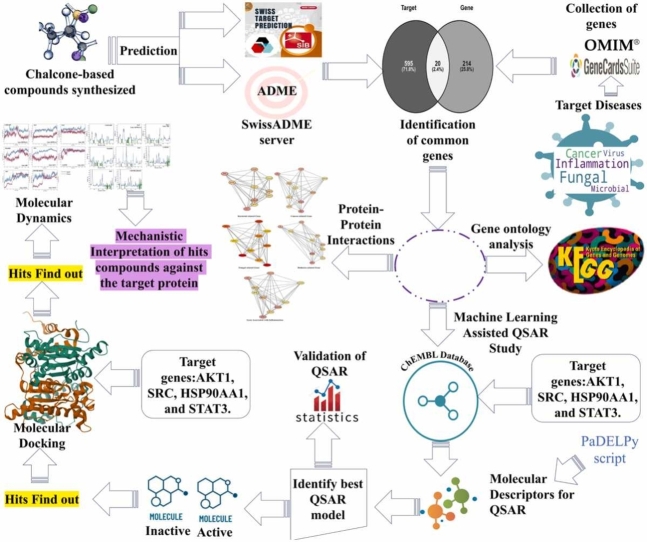
Previous studies have highlighted the diverse biological activities of chalcones, yet the mechanisms underlying their interactions with biological systems remain to be fully elucidated. The limitations of prior approaches include a lack of comprehensive analysis integrating both biological and chemical perspectives. To address these gaps, we used the knowledge-based system biology network and molecular modeling methods to identify chemical and biological co-interactions and to create mechanistic modeling of the study. In this study, a three-step approach was used to investigate chalcone-based small molecules and their interactions with biological systems. Primarily, we employed the systems biology network approach for identifying crucial cellular components, biological processes, and pathways interacted by genes in a polypharmacology analysis. Then, we used a random forest machine learning algorithm to build “Quantitative Structure-Activity Relationship” (QSAR) models that helped unravel chemical mechanisms associated with genes identified in the previous step. Besides, the bioactivity of chalcone derivatives was predicted and compared with FDA-approved drugs based on previously observed inhibitory activity. Finally, molecular docking studies were carried out to uncover structure-function relationships and biological mechanisms of chalcones derivatives. In addition, to gain insight into their behavior we subjected the most promising molecules to molecular dynamics simulations examining their interactions with target proteins over time. Thus, the objectives of this work provided valuable insights into how these molecules interact with specific genes and biological processes, revealing potential mechanisms of action.
2. Methods and materials
In this manuscript, we utilized a bio-cheminformatics method to analyze the spectrum of genes, expressed pathways, and cellular components. Our methodology involved identifying common genes involved in fungal, inflammatory, cancer, microbial, and viral diseases. Key biological pathways, processes, cellular components, and KEGG pathways showed significant biological interactions with these common genes. We performed molecular modeling studies on our target genes (AKT1, SRC, HSP90AA1, and STAT3) to establish mechanistic interpretations of compounds using an integrated QSAR approach. Furthermore, molecular dynamics simulations were conducted to understand the molecular behavior of these target proteins, providing mechanistic insights. Table 1 shows a representation of the computational tools and algorithms used for mechanistic bio-chemoinformatic insights.
Table 1.
Computational tools and algorithms for mechanistic bio-cheminformatics insights.
| Tools / Methods used | Background of Methodology | Key Function and Motive |
|---|---|---|
| Swiss ADME http://www.swissadme.ch/ |
Online tool for predicting ADME properties of small molecules | To assess the absorption, distribution, metabolism, and excretion properties of compounds |
| Swiss Target Prediction http://www.swisstargetprediction.ch/ |
Online tool for predicting the targets of bioactive small molecules | To identify potential molecular targets for chalcone derivatives |
| Venn Plot https://bioinformatics.psb.ugent.be/webtools/Venn/ |
Online tool for creating Venn diagrams | To visualize overlapping genes involved in multiple diseases |
| ShinyGO/ DAVID / FunRich 3.1.3 | Functional annotation tools for bioinformatics and microarray analysis | To identify and analyze biological pathways, processes, and cellular components |
| Machine Learning Assisted QSAR Study (Random Forest) | Weka software for machine learning | To develop QSAR models predicting the bioactivity of chalcone derivatives |
| PaDEL package (PaDELpy-0.1.13) | PaDELpy-0.1.13 for descriptor calculation and QSAR modeling | To calculate molecular descriptors and perform QSAR modeling |
| StreamLit web App | Web application for bioactivity prediction | To provide a user-friendly interface for predicting the bioactivity of compounds |
| Cytoscape 3.10.2 | Software for network analysis and visualization | To visualize and analyze biological networks and interactions |
| Auto Dock | Automated docking tools suite using a genetic algorithm for simulations | To predict binding affinities of compounds to protein targets and screen large libraries |
| Desmond (Schrödinger Software) | Software for molecular dynamics simulations | To simulate the molecular behavior and interactions of compounds with target proteins |
2.1. Data set collection
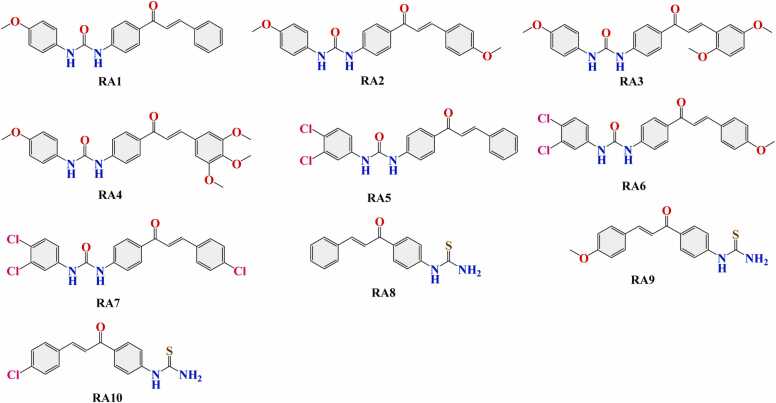
We collected a comprehensive dataset of chalcone-based compounds synthesized in our laboratory. These compounds were selected based on their structural diversity and potential biological activities (Fig. 1) [12], [13].
Fig. 1.
Structural diagrams of chalcone-based derivatives (RA1-RA10).
2.2. ADME prediction and drug likeliness
To understand the pharmacokinetics study of the synthesized compounds, it is essential to evaluate their physicochemical properties profile, which includes Absorption, Distribution, Metabolism, and Excretion (ADME). We utilized the SMILES data of the compounds and curated ADME physicochemical properties using the SwissADME server (https://www.swissadme.ch). These properties demonstrated various parameters such as lipophilicity, water solubility, drug-likeness rules, gastrointestinal (GI) absorption, blood-brain barrier (BBB) permeation, P-gp substrate status, cytochrome-P enzymes inhibition, and PAINS (Pan-Assay Interference Compounds) of the selected compounds[14] [15].
2.3. Identification of gene targets
Firstly, the molecular targets of the synthesized compounds were determined using the Swiss Target Prediction server (https://www.swisstargetprediction.ch). [15]. After that, we filter out genes based on a probability score that is greater than or equal to 0.40. Secondly, we curated disease-associated genes from the “Human Gene Database” (GeneCards, http://www.genecards.org)[16] and the “Online Mendelian Inheritance in Man” (OMIM, https://www.omim.org/)database [17]. Finally, we plotted the Venn diagram to identify common genes between the compounds and disease-specific genes. These combined approaches provided valuable insights into potential molecular targets of the compounds and their relevance to different target genes.
2.4. Constructing and visualizing protein-protein interaction networks using STRING and cytoscape
To explore how proteins interact with each other, We began by integrating the common genes identified from both compound targets and disease-specific targets into the STRING database to map out protein-protein interactions (PPI) (https://string-db.org/)[18]. After mapping, we took this data for further analysis using the Cytoscape plug-in, along with an extension called CytoHubba. These tools were instrumental in helping us visualize the complex network of interactions among the compounds and their target proteins. In this network, nodes represent the compounds and targets, while the edges depict the interactions between them [19].
2.5. Gene ontology (GO) analysis
To examine cellular components, biological pathways, and processes, we utilized FunRich 3.1.3 for gene ontology analysis[20]. Additionally, the Kyoto Encyclopedia of Genes and Genomes (KEGG)[21] was employed to identify common pathways associated with the selected targets. We considered a p-value lower than 0.05 to be statistically significant, indicating the relevance of our findings. We also used ShinyGO 0.80 for KEGG pathway network analysis[22].
2.6. Machine learning assisted QSAR study
All modeling processes are done using the Python programming language in Google Colab, facilitated by the Scikitlearn package (version 1.0.2).
2.6.1. Data collection and pre-processing
We used the panda’s library for effective manipulation of structured data including data frames. The chembl_webresource_client library, which is specifically made to query bioactive molecules and biological activities, helped us access the ChEMBL database. This technique generated a complete dataset of inhibitors against AKT1, SRC, HSP90AA1, and STAT3. For each gene, there were 4170 AKT1 inhibitors in the dataset, 5172 SRC inhibitors, 1369 HSP90AA1 inhibitors, and 1437 STAT3 inhibitors along with their corresponding IC50 values. These molecules were divided into three categories based on their IC50 values: active (IC50 <1000 nM), intermediate (1000 nM< IC50 <10 μM), and inactive (IC50 >10 μM). The next step involved further exploratory data analysis regarding the inhibitory characteristics of these compounds that included the molecular weight, and ALogP among others as considered by Lipinski’s rule of five descriptors. We could understand more about the distribution as well as properties of molecules in our research through this analysis.
2.6.2. Molecular fingerprints calculation
For generating the molecular features of compounds, we employed the PaDELPy script. This script utilizes the PubChem fingerprints provided by the PaDEL package (version PaDELpy-0.1.13) to model the compounds[23]. The fingerprint set comprises 881 binary representations that capture various chemical structural fragments recognized by PubChem. To optimize the accuracy and efficiency of feature generation, we configured several parameters within the PaDEL package. These settings included enabling the detection of aromaticity, standardization of nitrogen and tautomers, and removal of salts. Additionally, we set the number of processing threads to 2 to balance computational load and speed[24], [25], [26].
2.6.3. Feature selection
During the feature selection phase, we aimed to refine our dataset by eliminating less informative features. Specifically, we removed features with a variance lower than 0.1 and those demonstrating high correlation (greater than 0.95) to reduce redundancy and enhance model performance. We also processed descriptors with high inter-correlation, using a specified cut-off value for the inter-correlation coefficient to ensure that our models used the most informative and independent features. This careful selection of descriptors is critical for the reliable prediction of inhibitor efficacy. The impact of this process varied across different target genes: for AKT1, out of the original 881 features, only 213 remained after removing those with low variance and high correlation. Similarly, for SRC, 241 features were retained; for HSP90AA1, the process left 253 features; and for STAT3, 262 features remained. This selective approach ensured that only the most relevant and distinct features were carried forward for further analysis, significantly enhancing the robustness and interpretability of our results.
2.6.4. QSAR model construction
We constructed QSAR models for four different targets: AKT1, SRC, HSP90AA1, and STAT3, using the ChEMBL dataset. Each dataset was divided into training and test sets using the Kennington Stone algorithm, with an 80:20 split ratio (DTClab.Https://Dtclab.Webs.Com/Software-Tools; Github.Https://Github.Com/Dataprofessor/Code/Tree/Master/Python;Padel.Http://Www.Yapcwsoft.Com/Dd/Padel descriptor/) [27].
2.6.5. Development and validation of random forest-based QSAR models
We developed Random Forest (RF)-based QSAR models for datasets from ChEMBL, specifically targeting AKT1, SRC, HSP90AA1, and STAT3. These models were constructed using the Weka software suite ((Weka.Https://Www.Cs.Waikato.Ac.Nz/Ml/Weka/) [28]. RF, a supervised machine learning technique, utilizes an ensemble of decision trees to enhance prediction accuracy and robustness, addressing the common issue of overfitting in standalone decision trees. The training of the RF models was performed using the Bagging or Bootstrap aggregation technique. This method not only helps in reducing variance but also improves the generalization of the model over unseen data. To validate the performance of these QSAR models, we compared the correlation coefficient (R2) between the training and test datasets. The most valuable features were identified through the application of the RF Regressor algorithms for the RF models, which were then depicted in Variance Importance Plots (VIP). A graphical comparison of experimental versus predicted values for each QSAR model was conducted using the matplotlib Python package(Github. Https://Github.Com/Vappiah/Machine-Learning-Tutorials).
In addition to these methods, the performance of the QSAR models was also evaluated using receiver operating characteristic (ROC) graphs, generated by a pre-existing Python script designed for multi-class model classification. The ROC plots are crucial for visualizing the trade-off between true positive (TP) and false positive (FP) rates, with the area under the curve (AUC) serving as a quantitative measure of model discriminative ability. An AUC closer to 1 indicates a highly effective model, while an AUC near 0 suggests total misclassification [29]. Lastly, the applicability domain (AD) of the QSAR models was assessed using the bounding box technique via principal component analysis (PCA). This technique involved a PCA examination of the scores plot to compare the chemical space of molecules from the training and test sets. The AD was determined using the PCA function from the sklearn decomposition module of the scikit-learn machine learning toolkit [30] (Scikit-Learn. Https://Github.Com/Scikit-Learn/Scikit-Learn.Git.) [31], [32], [33].
2.7. Molecular docking
A molecular docking approach was employed to evaluate the inhibitory potential of chalcone derivatives across various activities, including antibacterial, anticancer, antidiabetes, anti-inflammation, and antifungal effects. The protein structures pertinent to the investigation, namely AKT1 (PDB ID: 4EJN)[34], SRC (PDB ID: 2OIQ)[35], HSP90AA1 (PDB ID: 3O0I)[36], and STAT3 (PDB ID: 6NJS)[37], were sourced from the Protein Data Bank in PDB format. Active sites within each protein structure were pre-delineated to facilitate docking by constructing grid boxes around the co-crystallized ligand. The AutoDock Tools software[38] was then employed to prepare the protein molecules. This process involved rectifying missing residues, eliminating water molecules, adding polar hydrogens, and applying Kollman charges. The resulting protein structures were saved in pqbqt format. Ligand molecules' 2D structures underwent conversion to 3D structures using the MMFF94 force field within the AutoDock Vina software. These transformed ligand structures were saved and converted to pdbqt format utilizing the Open Babel GUI. In the final step, a Perl script, in conjunction with Perl software, facilitated the docking of all ligand molecules against the protein structures. The resulting binding affinities or docking scores for each ligand molecule and respective target receptor were quantified in kcal/mol units. To glean insights into the molecular interactions, Pymol and Discovery Studio Visualizer were employed, enabling an in-depth exploration of ligand binding interactions with the most favorably binding proteins.
2.8. Molecular dynamic
To explore the stability of the most promising molecule in biological conditions, we carried out molecular dynamics (MD) simulations. These simulations are essential for understanding how the molecule behaves in a solvent environment. We set up the simulation in an orthorhombic box with dimensions of 12 Å on each side, using the buffer size method to optimize the volume of the box. The simulations were conducted using the TIP3P water model and the OPLS3e force field by Schrodinger Inc., which are standards for simulating proteins and ions. Sodium chloride was added to the system at a concentration of 0.15 M to mimic physiological conditions, with sodium (Na+) and chloride (Cl-) ions. The simulations ran for 200 nanoseconds using the Desmond Molecular Dynamics module, producing around 1000 snapshots of the system's behavior[39]. These were performed under the NPT ensemble, maintaining a constant temperature of 300 K and a pressure of 1 bar, ensuring the system was equilibrated before the simulations began.
3. Results
3.1. ADME prediction and drug likeliness
ADME parameters are crucial for predicting the pharmacokinetic behavior of compounds in drug discovery. Using the SwissADME database, we evaluated the structural and physicochemical properties of our selected compounds to determine their drug-like potential and pharmacokinetic profiles. The physicochemical properties and pharmacokinetic profile of the selected compounds are depicted in Table s1. (See Supplementary file s1 for more information on Table s1). All combinations shared an identical bioavailability score of 0.55, indicating moderate bioavailability. None of the compounds exhibited any PAINS alerts, suggesting a lack of common structural motifs associated with assay interference. Most of the compounds exhibited inhibitory activity against different Cytochrome P450 (CYPs) enzymes, which could have an impact on drug metabolism and potential drug-drug interactions. Nonetheless, these compounds were predicted not to be substrates for P-glycoprotein (Pgp), thus reducing the risk of interference by efflux transporter that is mediated through this protein. All the compounds presented high gastrointestinal (GI) absorption implying good absorption in the gastrointestinal tract while none of them was expected to cross the blood-brain barrier (BBB). All tested compounds were Lipinski's Rule of Five compliant which suggests that they possess favorable drug-like characteristics concerning their oral absorption and distribution. The Silicos-IT classification categorized most compounds as poorly soluble solvents except Compound RA8 which was moderately soluble. RA4 compound had the highest Molar refractivity of 130.94 indicating a likelihood for considerable intermolecular interaction as well as polarizability whereas compound RA8 had the least with 86.05 molar refractivity. Furthermore, there was a variation in the topological polar surface area where Compound RA4 showed the highest value (95.12 Ų) increasing its solubility and permeability while Compound RA5 had the lowest value (58.2 Ų) affecting its pharmacokinetic properties.
3.2. Compound and disease targets
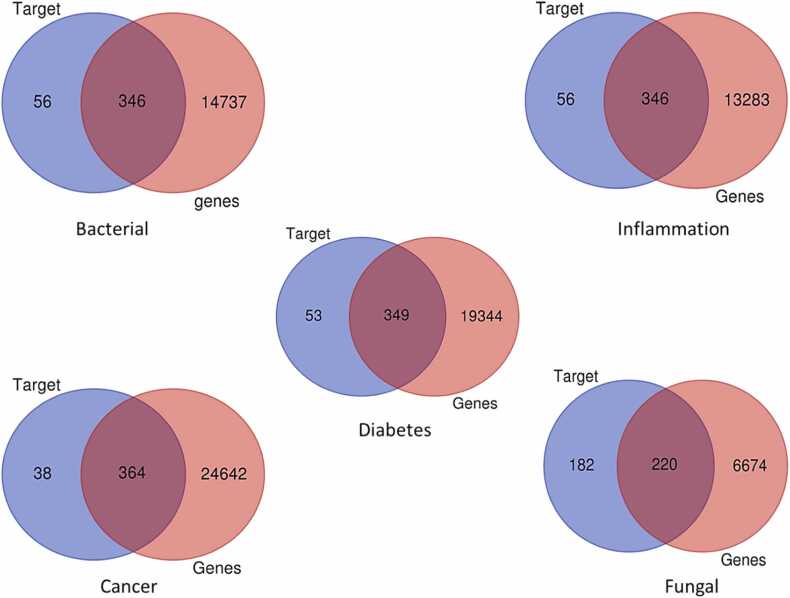
We curated molecular targets of compounds from the Swiss target prediction server and disease-associated genes of four diseases from the Human gene and OMIM databases (See Supplementary file, s1). The Venn diagram demonstrates the intersection of 346 targets related to bacterial diseases, 346 targets associated with inflammation, 364 targets linked to cancer, 349 targets relevant to diabetes, and 220 targets about fungal diseases. These intersections represent the common gene targets shared between compounds and the specified diseases’ s genes (Fig. 2).
Fig. 2.
Overlapping targets between the potential compound’s targets and disease-related genes using Ven Plot Diagram.
3.3. PPI network analysis
The PPI network was constructed using the STRING database, explicitly focusing on target organisms from Homo sapiens. This analysis compares five biological networks: bacterial, cancer, diabetes, fungal, and inflammation. The networks were constructed using data from the STRING database and visualized in Cytoscape after importing the data in a.tsv format[40]. For each network, the number of nodes and edges was recorded: bacterial network (345 nodes, 4342 edges), cancer network (363 nodes, 4555 edges), diabetes network (348 nodes, 4412 edges), fungal network (220 nodes, 2814 edges), and inflammation network (345 nodes, 4457 edges) (See Supplementary file, s2 for Hub gene_string data).
The analysis revealed a substantial number of network properties. The average local clustering coefficient, which is an indication of the level to which nodes tend to show mutual clustering, was found to be 0.458 for the bacterial network, 0.459 for the cancer network, 0.462 for the diabetes network, 0.52 for the fungal network, and 0.457 for the inflammation network. These high clustering coefficients suggest strong local connectivity within each network, indicating robust interactions among the proteins involved. Additionally, the PPI enrichment p-value for all networks was less than 1.0 * 10−16, highlighting the statistically significant enrichment of protein-protein interactions compared to a random network. The average node degree, which represents the average number of connections per node, was 25.2 for the bacterial network, 25.1 for the cancer network, 25.4 for the diabetes network, 25.6 for the fungal network, and 25.8 for the inflammation network. These values suggest a high level of interaction and potential functional importance of the proteins within these networks. The average shortest path length, indicating the average number of steps required to connect any two nodes, was 2.46 for the bacterial network, 2.48 for the cancer network, 2.46 for the diabetes network, 2.26 for the fungal network, and 2.44 for the inflammation network. This indicates efficient communication and the potential for rapid signal transduction within these networks. Furthermore, we analyzed the network diameter and radius which indicate the maximum and minimum path in a network respectively. The bacterial, cancer, and inflammation networks had a diameter of 6 units and a radius of 3 units while diabetes and fungal networks had a diameter of 6 units and a radius of 4 units. These structural characteristics provide insights into the overall connectivity and resilience of the networks. Detailed analysis of these network properties provides valuable insights into complex interactions and the functional significance of the proteins involved. The high clustering coefficients as well as node degrees suggest that these networks are highly interactive; this is important when it comes to understanding underlying biological processes as well as possible therapeutic targets. Besides, the average shortest path lengths that indicated efficient communication pathways further emphasize the relevance of these networks in maintaining cellular functions and responses.
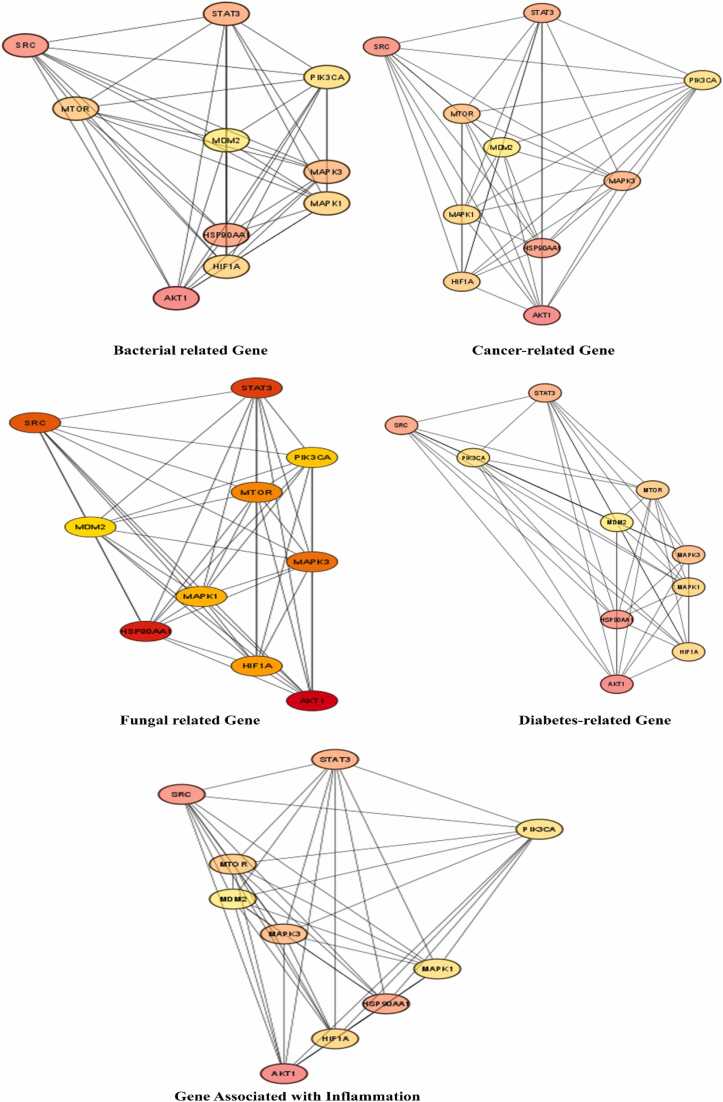
Fig. s1A and s1B display the common genes of compound and target interactions that were constructed using String. The top 10 targets were analyzed for network analysis, and the degree of freedom for each target was reported in Table s2 and Fig. 3 and visualized from Cytoscope. The top ten genes shared across the five diseases, AKT1, SRC1, HSP90AA1, and STAT3 exhibited strong associations in all five diseases. These four genes were selected based on their degree scores, ranging between 91 and 148. Genes such as AKT1, SRC, HSP90AA1, and STAT3 consistently displayed the highest degrees in each disease network, exceeding 110, except in the fungal network where their degrees were above 90. Notably, AKT1 demonstrated significant prominence, ranking first in bacterial, inflammation, and cancer networks with scores of 148, 149, and 149, respectively. In the fungal and diabetes networks, AKT1 also maintained high significance, ranking first with scores of 119 and 148, respectively. AKT1, a serine/threonine kinase, plays a crucial role in various cellular processes including cell survival, proliferation, and metabolism [41]. In bacterial infections, for example, the host immune response and pathogen invasion mechanisms are modulated by AKT1 signaling pathway activation. In cancer, AKT1 dysregulation frequently leads to tumor growth and reduced responsiveness to conventional treatments. Additionally, its involvement in glucose metabolism and insulin signaling makes it an important target for the treatment of diabetes. Furthermore, antifungal immune responses are modulated by AKT1 signaling but also could be used as a potential host enhancer through its inhibition within this system [42].
Fig. 3.
Top 10 Gene Target Interactions in Four Diseases Visualized through Cytoscape and Analyzed Using Network Analysis.
SRC, a non-receptor tyrosine kinase, plays a pivotal role in signal transduction pathways that govern cell growth, motility, and invasion. In bacterial infections, SRC has been linked to host cell invasion and the intracellular survival of pathogens [43]. In the realm of cancer, SRC is frequently overexpressed, promoting tumor progression and metastasis. Furthermore, SRC participates in insulin signaling and glucose metabolism, making it relevant to diabetes research. Additionally, SRC activation is involved in inflammation-related processes, contributing to the pathogenesis of various diseases[44]. SRC, a key cellular motility and adhesion regulator, contributes to fungal invasion and dissemination. Targeting SRC using specific inhibitors may impede fungal spread and improve treatment outcomes. HSP90AA1, a heat shock protein, is a molecular chaperone that plays a pivotal role in protein folding and stability. In bacterial infections, HSP90AA1 facilitates bacterial virulence by promoting the stability of bacterial effectors.
Within the cancer domain, HSP90AA1 acts as a critical chaperone for oncoproteins and proteins associated with drug resistance[45]. Its implication in insulin resistance and β-cell dysfunction highlights its relevance in diabetes disease. Moreover, HSP90AA1 is involved in inflammatory responses across various diseases. HSP90AA1 functions as a chaperone for fungal proteins, essential for fungal survival and virulence. Disrupting HSP90AA1's function has been explored as a strategy to weaken fungal pathogens. STAT3, a transcription factor, is essential in cell survival, proliferation, and immune responses[46]. The inflammation and immune responses of the host to bacterial invasion are modulated by the STAT3 signaling. Tumor growth and immune evasion in cancer are also promoted by STAT3 activation. In diabetes, pancreatic β-cell function and insulin signaling are impacted by STAT3. It also mediates inflammatory processes which contribute to disease pathogenesis. During fungal infections, STAT3 plays a vital role in orchestrating immune responses, and inhibiting its activity could enhance the host's antifungal defense mechanisms [47], [48]. The analysis of protein ranking across diverse biological networks offers valuable insights into the relative significance of AKT1, SRC, HSP90AA1, and STAT3 in various cellular processes and disease contexts. The consistently high rankings of these proteins suggest their crucial roles in cellular regulation, signal transduction, and disease development. Based on the Cytohubba analysis, all synthesized compounds exhibited the highest score, indicating their interactions with the maximum number of identified targets in all five diseases, achieving a score of 100. Previous research has highlighted the potential of chalcone-based novel phenyl ureas as effective antihyperglycemic agents with a likely PPAR gamma agonistic action.
3.4. Gene ontology
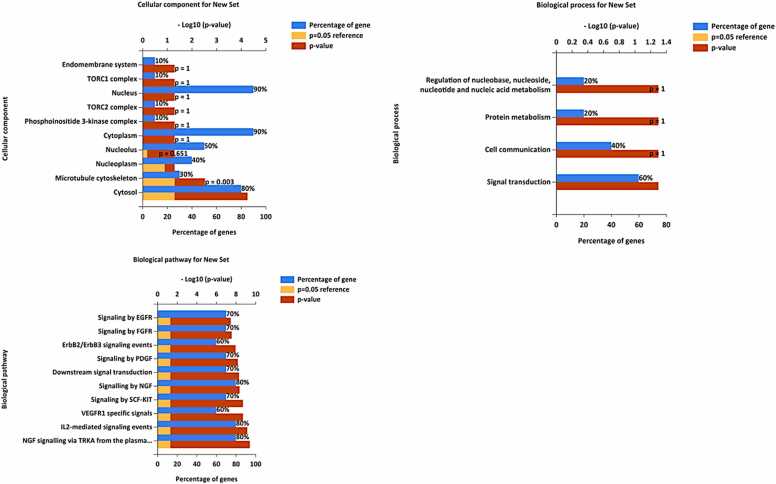
We performed a functional enrichment analysis using the FunRich software on the top 10 targets selected based on their degree. However, the degree of gene targets has different ranks; all targets, diabetes, inflammation, fungal, bacterial, and cancer, have almost the same 10 ten degrees of genes identified. Based on the data analysis of target genes, all diseases have the same cellular component, biological pathway, and process. Fig. 4 illustrates the top 10 Biological Pathway Annotations, Cellular Component Annotations, and Biological Process Annotations. Among the top 10 biological pathways identified, the following pathways were found: NGF signaling via TRKA from the plasma membrane 80 %, Signaling by EGFR 70 %, Signaling by FGFR 70 %, ErbB2/ErbB3 signaling events 60 %, Signaling by PDGF 70 %, Downstream signal transduction 70 %, Signalling by NGF 80 %, Signaling by SCF-KIT 70 %, VEGFR1 specific signals 60 %, IL2-mediated signaling events 80 %.
Fig. 4.
Gene ontology analysis: cellular components, biological processes, and biological pathway.
From previous studies, we can conclude that these pathways are involved in the development of diabetes, inflammation, fungal infections, bacterial infections, and cancer. Nerve Growth Factor (NGF) is a neurotrophic factor involved in neurons' development, survival, and function. Enriching genes in this pathway suggests their potential roles in mediating NGF signaling through its receptor TRKA (NTRK1). SRC, STAT3, and MAPK1, in particular, are known to be involved in neuronal signaling and synaptic plasticity, and they may play important roles in the downstream events of NGF-TRKA signaling[49]. Interleukin-2 (IL-2) is a cytokine central to regulating immune responses. Enriching genes in this pathway suggests their potential roles in mediating IL-2 signaling events. SRC and STAT3 are known to be involved in immune cell signaling and activation. In cancer, IL-2 has been used as an immunotherapy to stimulate the immune system's anti-tumor response, and SRC and STAT3 may be involved in the downstream effects of IL-2-mediated immune activation[50]. EGFR signaling is closely linked to various types of cancers, including lung cancer, breast cancer, colorectal cancer, and head and neck cancer. Dysregulation of EGFR, such as overexpression or activating mutations, can lead to uncontrolled cell proliferation, invasion, and metastasis in these malignancies[51]. EGFR signaling, while not a central factor in the development of diabetes, may impact certain cellular responses associated with complications of the disease, such as diabetic retinopathy. Similarly, while EGFR signaling doesn't directly correlate with bacterial or fungal infections, it might indirectly affect the immune responses to these infections. This is possible due to the expression of EGFR in diverse immune cells and tissues, suggesting its involvement in modulating host responses to various health challenges. FGFR (Fibroblast Growth Factor Receptor) is another family of receptor tyrosine kinases involved in cell proliferation, migration, and differentiation. Fibroblast growth factors (FGFs) binding to FGFR leads to receptor dimerization and activation of downstream signaling pathways.
FGFR signaling is critical in development, tissue repair, and angiogenesis. Aberrant FGFR signaling has been implicated in various cancers and developmental disorders [52]. FGFR signaling, while not directly involved in bacterial or fungal infections nor a primary contributor to diabetes development, may have a role in inflammation. However, its explicit involvement in inflammation-associated diseases warrants further study. VEGFR1 signaling plays a crucial role in angiogenesis, forming new blood vessels. It is expressed in tumor cells and various immune cells, making it relevant to several diseases, including cancer, inflammation, diabetes, and vascular diseases. In cancer, particularly colorectal and breast cancer, VEGFR1 signaling contributes to tumor angiogenesis and growth. High levels of VEGFR1 expression are associated with poor prognosis in these cancers. As a result, targeting VEGFR1-specific signals is being investigated as a potential strategy for cancer treatment. In inflammatory diseases like rheumatoid arthritis and inflammatory bowel disease, VEGFR1 gene-mediated signals play a role in recruiting immune cells and promoting angiogenesis to facilitate tissue repair. Consequently, interventions focused on regulating VEGFR1 are under investigation to manage the progression of diabetic retinopathy. VEGFR1 signaling also affects the progression of various vascular diseases, such as atherosclerosis and vascular malformations. It can modulate angiogenesis within atherosclerotic plaques and contribute to abnormal vessel development in vascular malformations[53], [54].
PDGF (Platelet-Derived Growth Factor) is a growth factor in cell proliferation and wound healing. It signals through two receptor tyrosine kinases, PDGFRα and PDGFRβ. Upon ligand binding, PDGF receptors undergo autophosphorylation and activate downstream signaling pathways, including the PI3K-AKT and MAPK pathways. PDGF signaling is important in tissue repair, angiogenesis, and development. Aberrant PDGF signaling has been implicated in cancer and fibrotic diseases[55]. While not directly associated with fungal infections, PDGF signaling may be involved in regulating inflammation and tissue repair. Additionally, it may hold relevance for diabetic complications, including nephropathy and retinopathy. The downstream Signal Transduction pathway involves the transmission of signals from activated cell surface receptors (such as EGFR, FGFR, and PDGFR) to intracellular effectors. Downstream signal transduction pathways include MAPK/ERK, PI3K-AKT, and JAK-STAT. These pathways regulate gene expression and modulate cellular responses, such as proliferation, survival, and differentiation. Dysregulation of downstream signal transduction can lead to various diseases, including cancer and inflammatory disorders[56]. ErbB2 (HER2) and ErbB3 (HER3) are members of the EGFR family of receptor tyrosine kinases. They form heterodimers and activate downstream signaling pathways upon ligand binding or through other mechanisms. ErbB2 does not bind a specific ligand but can enhance signaling by forming heterodimers with other ErbB family members. ErbB2/ErbB3 signaling plays crucial roles in the cell proliferation, survival, and metastasis of various cancers. Abnormal ErbB2 (HER2) expression is closely linked with aggressive forms of breast cancer, and targeted treatments focusing on ErbB2 have demonstrated clinical effectiveness [57]. SCF (Stem Cell Factor) and KIT (KIT proto-oncogene) are involved in hematopoiesis, melanogenesis, and cell survival. The binding of SCF to its receptor KIT activates downstream signaling events. KIT signaling is crucial for stem cell development and hematopoiesis aberrant KIT signaling[58].
These proteins SRC, HSP90AA1, STAT3, MAPK3, MTOR, HIF1A, MAPK1, PIK3CA, and MDM2 are involved in several important biological pathways that have relevance to various diseases, including cancer, diabetes, inflammation, and other disorders. These proteins are crucial in signal transduction, growth regulation, immune responses, and cellular metabolism. When these pathways and proteins become dysregulated, they can contribute to the development and progression of diseases. In cancer, these proteins often promote cell growth, survival, and metastasis. Dysregulation of these pathways can lead to uncontrolled cell proliferation and tumor formation. For example, the MAPK pathway (involving proteins like MAPK1 and MAPK3) is frequently altered in cancer, leading to excessive cell division and tumor growth. In diabetes, proteins like MTOR and PIK3CA are involved in insulin signaling and glucose metabolism. Dysfunctional signaling in these pathways can affect insulin sensitivity and glucose regulation, contributing to diabetes and its complications. In inflammation, proteins like STAT3 and HIF1A are key players in immune responses and inflammation regulation. Aberrant activation of these proteins can lead to chronic inflammation associated with various inflammatory diseases.
The gene ontology analysis uncovers critical biological processes associated with the top five targets for diseases, including diabetes, inflammation, fungal, bacterial, and cancer. Notable processes include signal transduction (60 %), protein metabolism (20 %), cell communication (40 %), energy pathways (10 %), and regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (20 %) (Fig. 4). These insights are invaluable, shedding light on the molecular mechanisms driving disease development and progression. Additionally, identified cellular components associated with the top ten disease targets reveal where these elements predominantly exist within cells. These locations include the phosphoinositide 3-kinase complex (10 %), TORC1 and TORC2 (10 %), the nucleus (90 %), nucleoplasm (40 %), endomembrane system (10 %), the nucleolus (50 %), the TORC2 complex (10 %), the cytoplasm (90 %), microtubules, and the cytosol (80 %) (Fig. 4). Understanding the cellular location of these targets provides crucial insights into their functional roles in specific diseases, allowing for more targeted and precise intervention strategies.
3.5. KEGG pathway
In this study, the examined compounds demonstrated distinctive impacts on target disease. These molecules showed promising effects on neoplastic cells through their interaction with the MAPK signaling pathways, particularly focusing on the ERK element and MAPK receptors [59]. This observation aligns with the well-established role of MAPK receptors in fostering tumor proliferation and survival. Additionally, the compounds impacted the mTOR signaling pathway, targeting elements such as PI3K, AKT1, and mTOR receptors, which are crucial for cell proliferation and survival[60]. Similarly, these compounds affected the JAK-STAT signaling pathway, explicitly targeting components like STAT3 receptors[61]. The compounds influenced the MAPK signaling pathway by activating components via the EGFR receptors. RTKs activate HIF1 alpha, so by targeting them, the compounds could potentially deregulate their activity.
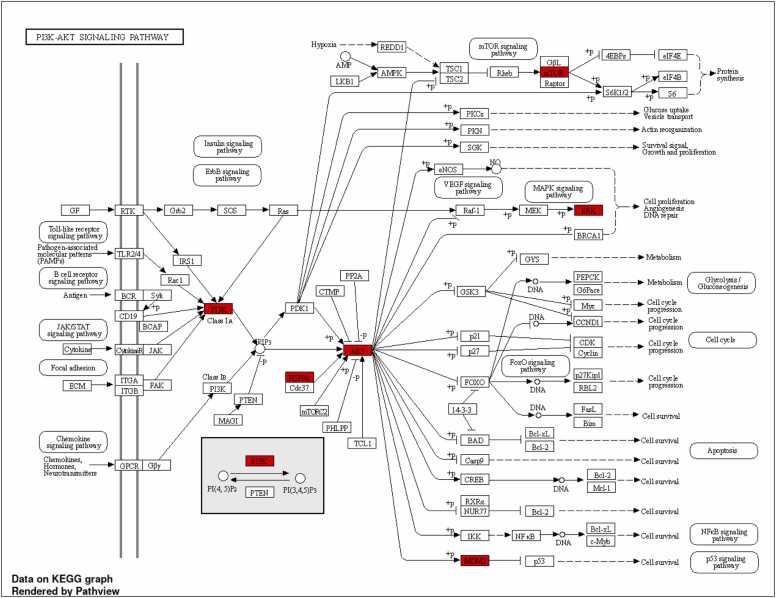
The PI3K-AKT signaling pathway is a crucial intracellular signaling pathway implicated in multiple cellular functions, such as cell growth, proliferation, angiogenesis, and survival. It is activated by various types of cellular stimuli or toxic insults[62], [63]. The PI3K-AKT pathway is central to insulin signaling. When insulin binds to its receptor, it triggers the activation of PI3K, leading to the activation of AKT. AKT subsequently stimulates glucose uptake by promoting the translocation of the glucose transporter GLUT4 to the cell membrane. This pathway's alteration can lead to insulin resistance, a key factor in developing type 2 diabetes. Dysregulation in PI3K/AKT signaling has been associated with diabetic complications, including nephropathy and retinopathy[64], [65] (Fig. 5A). In inflammation disease, the activation of the NF-κB pathway, including the resultant upregulation of BCL-XL and c-Myb, can contribute to inflammation[4], [66]. This pathway plays a critical role in cell cycle regulation and is heavily involved in cancer pathogenesis due to its influence on cell proliferation and apoptosis. In the MAPK signaling pathway context, the PI3K-AKT pathway can influence cell proliferation and angiogenesis, mainly through the ERK component. The PI3K-AKT pathway's interaction with the mTOR, JAK/STAT3, chemokine, and Toll-like receptor signaling pathways allows for a complex network of regulation, further expanding its role in various cellular processes. Pathogen-associated molecular patterns (PAMPs) can directly influence TLR2/4 and activate the small GTPase Rac1. This activation triggers the PI3K, producing PIP3, a crucial second messenger in the PI3K-AKT pathway. PIP3 then stimulates the kinase AKT1, which is critical for cell survival, primarily through its influence on the MDM2 gene. Furthermore, the chaperone protein HSP90AA1 also activates AKT1, adding another level of regulation to this pathway. This complexity contributes to the range of cellular processes the PI3K-AKT pathway influences, reinforcing its importance in understanding disease pathogenesis, particularly in cancer and inflammatory conditions[63], [64], [65].
Fig. 5A.
KEGG pathway analysis of the top 10 targets, with special emphasis on PI3K-AKT signaling pathway.
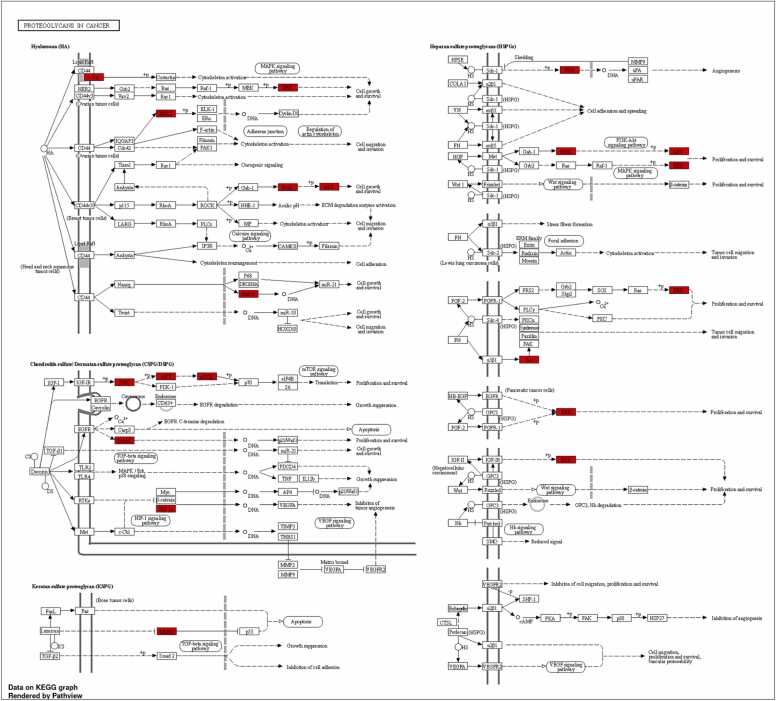
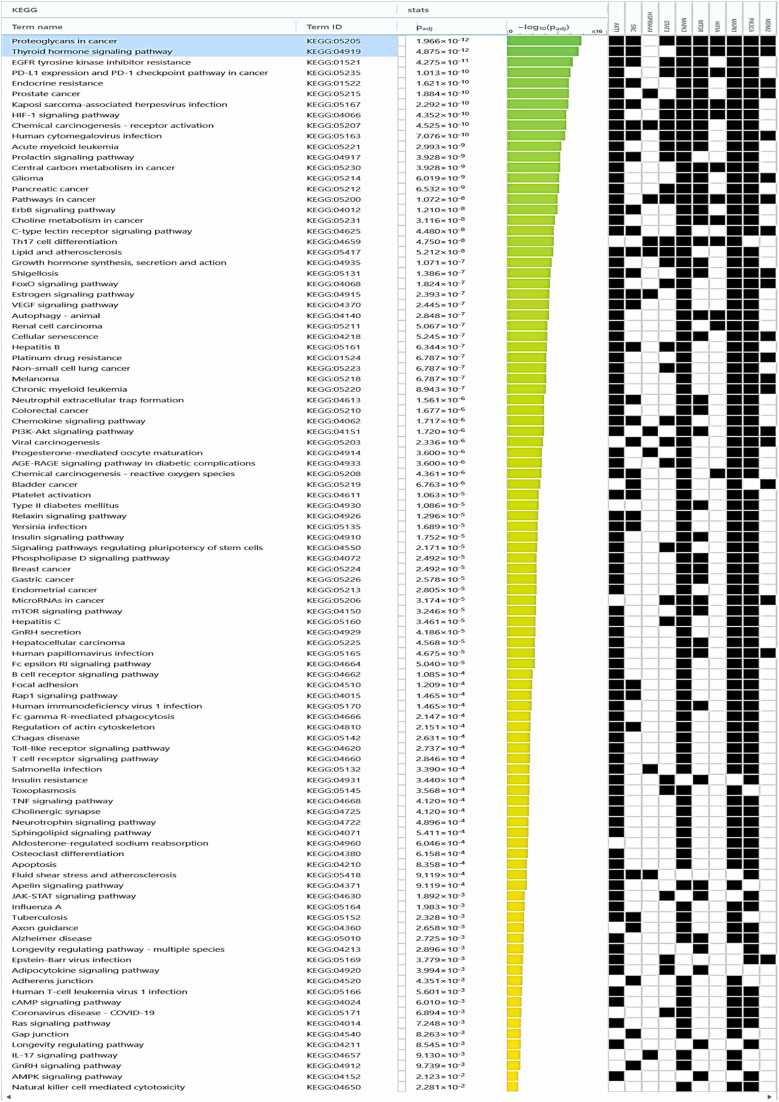
The pathway "Proteoglycans in cancer" (KEGG:05205) has a higher negative p-value of 11.70, making it the most significant path in the dataset. Proteoglycans are a group of glycosylated proteins mainly present in the extracellular matrix. They play crucial roles in many biological processes, including cell proliferation, migration, and angiogenesis, all of which are integral to cancer development and progression (Fig. 5B and Fig. 6)[67]. Several genes from data, such as AKT1, SRC, STAT3, MAPK3, and PIK3CA, are implicated in this pathway, indicating a potential role in cancer-related processes. The "Thyroid hormone signaling pathway" (KEGG:04919) is the second most significant pathway, with a negative p-value of 11.31. The thyroid hormone signaling pathway regulates metabolism, growth, and development. It involves several critical genes from the data set, including SRC, AKT1, and PIK3CA. Dysregulation in this pathway may lead to various disorders, ranging from developmental issues to metabolic diseases and certain cancers[48]. The pathway "EGFR tyrosine kinase inhibitor resistance" (KEGG:01521) also shows high significance with a negative p-value of 10.36. EGFR, a key receptor tyrosine kinase, regulates cellular activities, including proliferation and survival (See Fig. 6). EGFR mutations often result in over-activated EGFR pathways, causing uncontrolled cell growth, which is common in various cancers like NSCLC. EGFR tyrosine kinase inhibitors (TKIs) can hinder tumor growth by inhibiting EGFR's tyrosine kinase activity. However, resistance to these drugs often develops through mechanisms like secondary EGFR mutations or changes in other growth factor receptors. Key genes in data, such as EGFR, AKT1, PIK3CA, ERBB2, MET, and FGFR1, can contribute to EGFR TKI resistance, either by direct alterations in EGFR or by influencing related signaling pathways[68]. Specific genes, like AKT1, MAPK3, MAPK1, PIK3CA, etc., appear frequently across many routes. These genes could be essential nodes in biological networks and serve as potential targets for broad-spectrum treatments.
Fig. 5B.
KEGG pathway analysis of the top 10 targets, with special emphasis on proteoglycans in cancer.
Fig. 6.
The analysis of KEGG pathways, along with their corresponding Predictive p-values and the genes they interact with.
3.6. Machine learning QSAR analysis
3.6.1. Exploratory data analysis
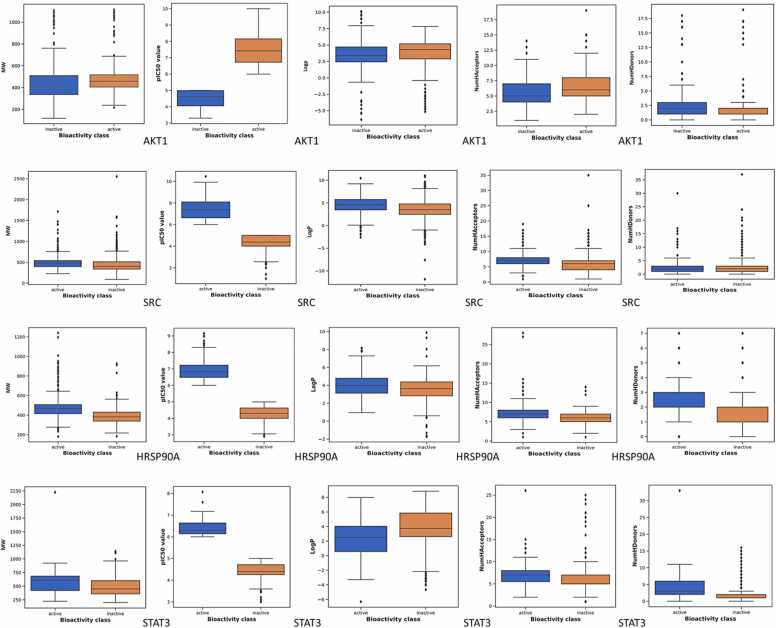
Further analysis for the QSAR model focused on four target genes: AKT1, SRC, HSP90AA1, and STAT3. For the AKT1 gene data, after data pre-processing and the elimination of missing values, 2876 compounds were selected for study. Exploratory data analysis revealed a greater proportion of active compounds compared to inactive ones. The pIC50 values of active compounds ranged from 6 to 10, whereas those of inactive compounds ranged from 3.30 to 5. The Mann-Whitney U test showed statistical significance between active and inactive groups. Active molecules generally had larger pIC50, MW, and NumHAcceptors values, while inactive molecules had slightly smaller MW and fewer NumHDonors. LogP values remained identical between active and inactive molecules (Fig. 7). For the SRC gene data, after data pre-processing and the elimination of missing values, 3177 compounds were selected for study. Exploratory data analysis also revealed a greater proportion of active compounds compared to inactive ones. The pIC50 values of active compounds ranged from 6 to 10.45, whereas those of inactive compounds ranged from 1 to 5. The Mann-Whitney U test confirmed statistical significance between active and inactive groups. Active molecules exhibited higher pIC50 values, while inactive molecules had slightly elevated LogP values. MW, NumHDonors, and NumHAcceptors values were nearly indistinguishable between active and inactive molecules (Fig. 7).
Fig. 7.
Exploratory data analysis for four genes, AKT1, SRC, HSP90AA1, and STAT3 inhibitors’ dataset from the ChEMBL database.
For the HSP90AA1 gene data, after data pre-processing, curation, and the elimination of missing values, 1009 compounds were selected for study. Exploratory data analysis revealed a greater proportion of active compounds compared to inactive ones. The pIC50 values of active compounds ranged from 6 to 9.15, whereas those of inactive compounds ranged from 2.90 to 5. The Mann-Whitney U test showed statistical significance between active and inactive groups. Active molecules typically had slightly elevated LogP and NumHAcceptors values compared to inactive molecules, and higher values for MW, pIC50, and NumHDonors (Fig. 7). For the STAT3 gene data, after data pre-processing and the elimination of missing values, 640 compounds were selected for study. Exploratory data analysis also revealed a greater proportion of active compounds compared to inactive ones. The pIC50 values of active compounds ranged from 6 to 8.07, whereas those of inactive compounds ranged from 3 to 5. The Mann-Whitney U test confirmed statistical significance between active and inactive groups. Active molecules exhibited significantly higher pIC50 values and a greater number of NumHAcceptors compared to inactive molecules. The values for MW, LogP, and NumHDonors remained nearly unchanged between active and inactive compounds (Fig. 7).
3.6.2. Machine learning QSAR model predication
To predict the activity of the compounds, we first calculated their descriptors using PaDELPy in the PaDEL software. We generated PubChem fingerprint descriptors, which resulted in 881 attributes. Using Data Pre-treatment GUI 1.2, we filtered out constant descriptors based on their correlation coefficient and variance scores. This resulted in 213, 241, 253, and 262 attributes for AKT1, SRC, HSP90AA1, and STAT3, respectively. Next, we used the Kennard Stone algorithm to split the dataset into training and evaluation sets in an 80:20 ratio. For the training phase, we developed QSAR models with the following attributes: 2255 for AKT1, 2481 for SRC, 791 for HSP90AA1, and 504 for STAT3. The testing phase included 531 attributes for AKT1, 576 for SRC, 186 for HSP90AA1, and 120 for STAT3. We then loaded the training and test datasets into WEKA to build the QSAR models (Supplementary File S3).
The WeKa model analysis for the AKT1 gene demonstrated strong predictive performance during both the training and testing phases. During the training set, the model achieved a high correlation coefficient of 0.9802, indicating a close fit to the actual data. The mean absolute error (MAE) was 0.2225, and the root mean squared error (RMSE) was 0.2937, showing that the model's predictions were relatively accurate. The relative absolute error (RAE) of 20.1691 % suggests that the model's predictions were, on average, about 20 % off from the actual values, while the root relative squared error (RRSE) of 21.3556 % reflects the consistency of these errors. In test set data, the model maintained a decent performance with an MAE of 0.5868 and an RMSE of 0.7371. The RAE and RRSE for the test set were around 58–59 %, indicating a moderate increase in prediction error compared to the training phase. Cross-validation, which helps evaluate how well the model generalizes, resulted in correlation coefficients of 0.8634, an MAE of 0.5299, and an RMSE of 0.702. The cross-validation RAE and RRSE were approximately 48 % and 51 %, respectively indicating that it does moderately well with data. For the SRC gene, the QSAR model showed similarly strong performance with a high training correlation coefficient of 0.9867, indicating its reliability in predicting this gene's activity.
The HSP90AA1 model also showed comparable results, further validating the model's robustness. The mean absolute error and root mean squared error are 0.2227 and 0.3, respectively. The relative absolute error and root relative squared errors are 14.8335 % and 17.3816 %, respectively. The model maintains strong performance during testing, with a correlation coefficient of 0.9147. The mean absolute error and root mean squared error are 0.5065 and 0.6198, respectively. The relative absolute and root relative squared errors are around 36 %, suggesting good generalization. Cross-validation results also show high predictive ability, with a correlation coefficient of 0.8983. The mean absolute error and root mean squared error are 0.5677 and 0.7648, respectively. The relative absolute error and root relative squared error are approximately 37 %, indicating consistent and reliable performance. The model analysis for the HSP90AA1 gene demonstrates high predictive accuracy during training, with a correlation coefficient of 0.9867. The mean absolute error and root mean squared error are 0.1403 and 0.1993, respectively. The relative absolute and root relative squared errors are 16.1236 % and 17.1802 %, respectively. The model maintains strong performance during testing, with a correlation coefficient of 0.9295. The mean absolute error and root mean squared error are 0.3371 and 0.4213, respectively. The relative absolute and root relative squared errors are around 37 %, indicating good generalization. Cross-validation results also show high predictive ability, with a correlation coefficient of 0.9011. The mean absolute error and root mean squared error are 0.3553 and 0.5073, respectively. The relative absolute error and root relative squared error are approximately 40 %, suggesting consistent performance across different folds.
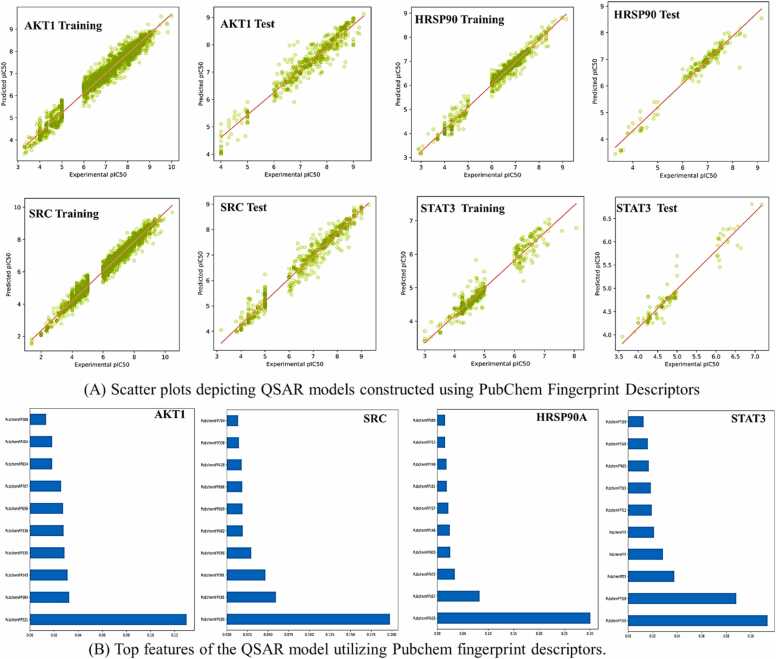
For the STAT3, the QSAR model achieves a high correlation coefficient of 0.9713 during training, indicating predictive solid ability. The mean absolute and root mean squared errors are 0.1719 and 0.2541, respectively, implying accurate predictions. The relative absolute and root relative squared errors are 23.079 % and 27.1873 %, respectively. However, on the test set, the model's performance slightly decreases, with a correlation coefficient of 0.783. The mean absolute error and root mean squared error increase to 0.3219 and 0.4605, respectively. The relative absolute and root relative squared errors are around 59 % and 70 %, respectively. Cross-validation results show a correlation coefficient of 0.7102, suggesting good performance compared with training. The mean absolute error and root mean squared error are 0.4488 and 0.6589, respectively. The relative absolute and root relative squared errors are approximately 60 % and 70 %, respectively. The robustness of the QSAR models was inferred from the high correlation coefficients observed in both the training and test sets, suggesting a high degree of reliability. Additionally, the outcomes of tenfold cross-validation for each model demonstrated a notable level of satisfaction, further affirming the models' performance (Fig. 8A).
Fig. 8.
Scatter plots of QSAR models utilizing pubchem fingerprint descriptors for training and test sets, and VIP plot illustrating the key features of the QSAR model incorporating pubchem fingerprint descriptors against four genes.
To discern the pivotal molecular fingerprints and their respective contributions to bioactivity within QSAR models, a comprehensive feature importance analysis was conducted. This investigation involved the utilization of the Random Forest regressor algorithm to pinpoint the top ten molecular fingerprints for each QSAR model. The Variance Importance Plots (VIP) were generated using the matplotlib package in Python, providing a visual representation of the significance of these fingerprints (Fig. 8B). The most significant descriptors in the Pubchem fingerprint-based model were identified as follows: PubchemFP521 (C:N-C-[#1]) in AKT1, PubchemFP180 (containing at least one saturated or aromatic nitrogen-containing ring of size 6) in SRC, PubchemFP633 (N-C-C:C-C) in HSP90AA1, and PubchemFP145 (including at least one saturated or aromatic nitrogen-containing ring of size 5) and PubchemFP338 (C(∼C)(∼C)(∼H)(∼N)) in STAT3. For the PubChem fingerprints-based model targeting the AKT1 gene, the VIP analysis highlighted PubchemFPs 143, 184, 186, 335, 338, 404, 521, 614, 696, and 707 as the most influential molecular fingerprints. Similarly, in the context of the SRC gene, the VIP plot identified PubchemFPs 180, 181, 338, 391, 439, 590, 609, 682, 696, and 704 as the key contributors to bioactivity. Moving to the HSP90AA1 gene, the VIP analysis underscored the significance of PubchemFPs 146, 181, 357, 380, 633, 672, 712, 737, 749, and 800. Lastly, within the context of the STAT3 gene, PubchemFPs 1, 2, 21, 145, 146, 180, 181, 338, 685, and 712 were identified as critical molecular fingerprints. Based on feature selection, structural insights for the best descriptor-containing compounds were investigated for both models individually (Fig. 8B).
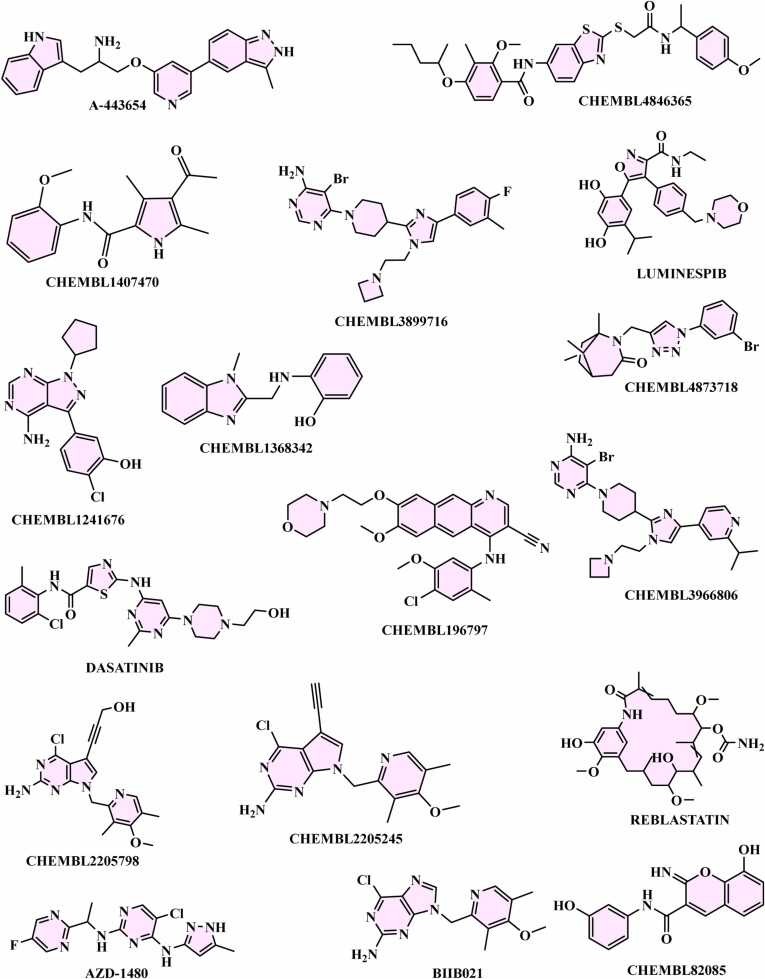
In the context of AKT1, specific analysis has revealed that clinical drugs 443654 (CHEMBL379300), CHEMBL3899716, and CHEMBL3966806 exhibit consistent fingerprints associated with distinct molecular features. These fingerprints include PUbchemFP143 (greater than or equal to 1, any ring size 5) and PUbchemFP521 (C:N-C-[#1]). Experimentally determined pIC50 values for these compounds were 9.796, 10, and 9.824, respectively. For SRC, quantitative structure-activity relationship (QSAR) data analysis was conducted on the VIP plot. The FDA-approved drug DASATINIB (CHEMBL1421) and Chembl IDs CHEMBL1241676 and CHEMBL196797 were observed to possess common PubChem fingerprints. These fingerprints, specifically PubchemFPs 180 (greater than or equal to 1 saturated or aromatic nitrogen-containing ring size 6), 181 (greater than or equal to 1 saturated or aromatic heteroatom-containing ring size 6), and PubChem Fp696 (C-C-C-C-C-C-C-C), were reflected in experimental pIC50 values of 9.301, 9.921, and 9.824. These findings suggest particular structural attributes contributing to the compound's bioactivity. In the case of HSP90AA1, QSAR data analysis of the VIP plot revealed shared Pubchem fingerprint attributes in FDA approved drugs REBLASTATIN (CHEMBL267792), BIIB021 (CHEMBL467399), LUMINESPIB (CHEMBL252164), and Chembl IDs CHEMBL2205798, CHEMBL4873718, and CHEMBL2205245 (Fig. 9). The common characteristics include PubChem146 (greater than or equal to 1 saturated or aromatic heteroatom-containing ring size 5), PubChem181 (greater than or equal to 1 saturated or aromatic heteroatom-containing ring size 6), PubChem357 (C(∼C)(:C)(:N)), and PubChem633 (N-C-C:C-C). The corresponding experimental pIC50 values are 8.30, 8.29, 8.10, 9.15, 9.14, and 9, reinforcing the structural attributes responsible for their bioactivity (Fig. 9).
Fig. 9.
Identification of structural insights for PubChem Fingerprint Descriptors through analysis of top-performing molecules.
Lastly, in STAT3, a QSAR analysis of the VIP plot was performed for FDA-approved drug AZD-1480 (CHEMBL1231124) and Chembl IDs CHEMBL1368342, CHEMBL1407470 and CHEMBL4846365. Shared PubChem fingerprints were identified, such as PubChem146 (greater than or equal to 1 saturated or aromatic nitrogen-containing ring size 5), PubChem146 (greater than or equal to 1 saturated or aromatic heteroatom-containing ring size 5), PubChem181 (greater than or equal to 1 saturated or aromatic heteroatom-containing ring size 6), PubChem357 (C(∼C)(:C)(:N)), and PubChem633 (N-C-C:C-C). The experimental pIC50 values were measured at 7.097, 8.071, 7.593, and 7.17, further elucidating the structural attributes that contribute to the bioactivity of these compounds (Fig. 9 and Table 2). Information regarding the training set and testing of PubChem fingerprint and CHEMBL molecules for each target gene can be found in Supplementary file s3.
Table 2.
Interpretation for the most significant PubChem and substructure fingerprints.
| Best Features | Interpretation |
|---|---|
| PubchemFP1 | > = 8 H |
| PubchemFP2 | > = 16 H |
| PubchemFP21 | > = 8 O |
| PubchemFP143 | > = 1 any ring size 5 |
| PubchemFP145 | > = 1 saturated or aromatic nitrogen-containing ring size 5 |
| PubchemFP146 | > = 1 saturated or aromatic heteroatom-containing ring size 5 |
| PubchemFP180 | > = 1 saturated or aromatic nitrogen-containing ring size 6 |
| PubchemFP181 | > = 1 saturated or aromatic heteroatom-containing ring size 6 |
| PubchemFP184 | > = 1 unsaturated non-aromatic heteroatom-containing ring size 6 |
| PubchemFP186 | > = 2 saturated or aromatic carbon-only ring size 6 |
| PubchemFP335 | C(∼C)(∼C)(∼C)(∼H) |
| PubchemFP338 | C(∼C)(∼C)(∼H)(∼N) |
| PubchemFP357 | C(∼C)(:C)(:N) |
| PubchemFP380 | C(∼O)(∼O) |
| PubchemFP391 | N(∼C)(∼C)(∼C) |
| PubchemFP404 | N(:C)(:C)(:C) |
| PubchemFP439 | C(-C)(-N)(=O) |
| PubchemFP521 | C:N-C-[#1] |
| PubchemFP590 | C-C:C-O-[#1] |
| PubchemFP609 | Cl-C-C-N-C |
| PubchemFP614 | C-C-O-C-C |
| PubchemFP633 | N-C-C:C-C |
| PubchemFP672 | O C-C C-[#1] |
| PubchemFP682 | O-C-C-C-C-N |
| PubchemFP685 | O C-C-C-C-N |
| PubchemFP696 | C-C-C-C-C-C-C-C |
| PubchemFP704 | O C-C-C-C-C-C-C |
| PubchemFP707 | O C-C-C-C-C(N)-C |
| PubchemFP712 | C-C(C)-C(C)-C |
| PubchemFP737 | Cc1cc(N)ccc1 |
| PubchemFP749 | Nc1cc(N)ccc1 |
| PubchemFP800 | CC1CC(N)CCC1 |
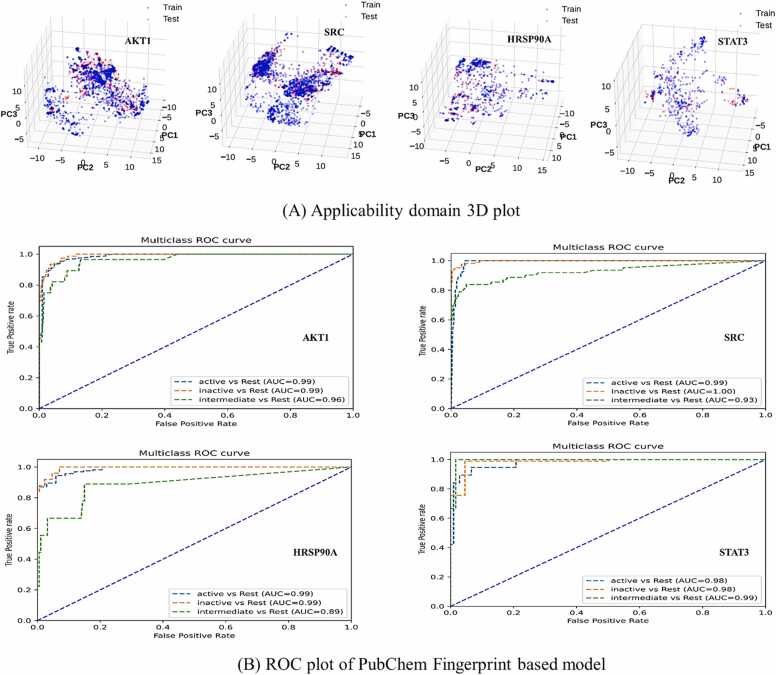
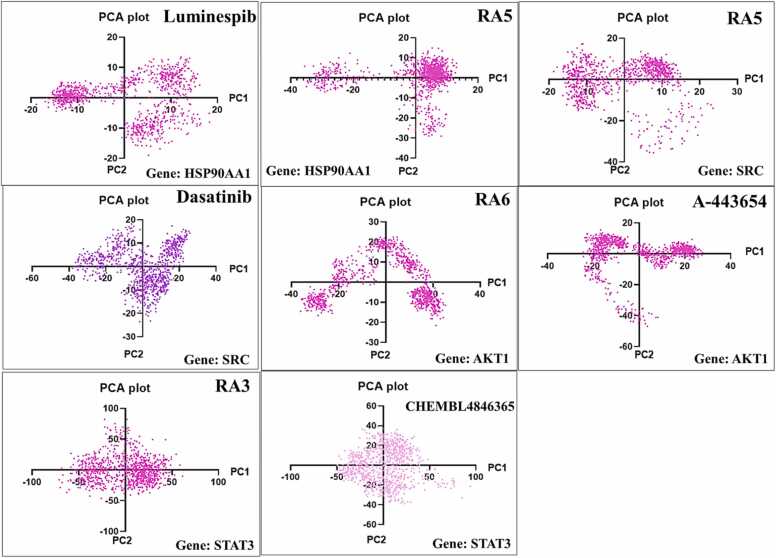
In the context of validation parameters, a comparative analysis was conducted to assess the chemical space encompassed by the training and test sets. This evaluation involved the application of the PCA bounding box method, aiming to determine the applicability domain of the molecular fingerprint datasets developed within this study. The method's efficacy in detecting outliers within both the fingerprint models was examined. The PCA analysis was executed during the training phase, encompassing the descriptors/attributes for AKT1 (2255), SRC (2481), HSP90AA1 (791), and STAT3 (504). Subsequently, in the testing phase, distinct attributes were employed for model evaluation, namely 531 for AKT1, 576 for SRC, 186 for HSP90AA1, and 120 for STAT3, utilizing the PubChem fingerprint dataset. The outcomes of this analysis revealed that the chemical space spanned by the test set remained within the boundaries of the chemical space occupied by the training set. Consequently, it was determined that the developed fingerprint datasets exhibited applicability domains encompassing the test set. Furthermore, an examination of the PCA scores plot indicated a significant similarity in the relative chemical space occupied by compounds within both the training and test sets, as depicted in Fig. 10.
Fig. 10.
Applicability domain assessed through PCA application and ROC plot generated for PubChem fingerprint descriptor-implemented QSAR models, respectively.
The QSAR models were validated by applying Receiver Operating Characteristic (ROC) analysis, yielding pertinent insights into the predictive performance of the four target genes. Specifically, for the AKT1 gene, the computed Area Under the Curve (AUC) values were 0.99, 0.99, and 0.96 for active, inactive, and intermediate molecules, respectively. Similarly, for the SRC gene, the ROC analysis yielded AUC values of 0.99, 1.00, and 0.93 for the respective molecular classes. The HSP90AA1 gene demonstrated AUC values of 0.99, 0.99, and 0.89 for active, inactive, and intermediate molecules. In contrast, the validation of the QSAR model for the STAT3 gene revealed AUC values of 0.98, 0.98, and 0.99 for the corresponding molecular categories. These AUC values collectively underscore the commendable and dependable performance of the QSAR models in accurately predicting molecular interactions (Fig. 10).
3.7. Prediction of bioactivity of phytochemicals using generated machine learning models
We developed a Python-based web application called ASHS-Pred using the Streamlit library. This application leverages established molecular fingerprint-based models for the AKT1, HSP90AA1, STAT3, and SRC genes. To build the web application, we utilized various Python libraries including sci-kit-learn, pandas, subprocess, os, base64, and pickle. ASHS-Pred operates by processing the SMILES representations of multiple molecules and their corresponding names or IDs provided by the user in a text file. Upon uploading this text file, the application predicts the inhibition activity (pIC50) of the loaded molecules against the specified genes. The application calculates the pertinent molecular fingerprints for the molecules using established fingerprint-based random forest models and presents the predicted activity as pIC50 values alongside their respective molecule names. Users can download the activity values and molecule names in CSV format directly from the application. The complete source code for ASHS-Pred is openly accessible on GitHub at the following URL: https://github.com/RatulChemoinformatics/QSAR. To use the application, users need to have the Anaconda Navigator interface installed on their systems, along with Streamlit and other necessary package dependencies. The installation process is detailed in the readme file available on the GitHub repository. Following these instructions, users can accurately predict molecular activity against the four target genes using the ASHS-Pred application.
For further analysis, chalcone derivatives identified through intensive network pharmacology screening were assessed for bioactivity prediction using the fingerprint-based machine learning models developed in ASHS-Pred. Notably, RA1 displayed strong interactions with HSP90AA1, indicating potential as a potent inhibitor for this gene. Multi-target potential was evident in several derivatives, including RA1, RA2, and RA10, highlighting their adaptability across various pathways. Compound RA1, with its notable pIC50 value of 5.76 against HSP90AA1, displays promising inhibitory effects, indicating its potential for diverse applications. Additionally, RA1 exhibited substantial activity against AKT1, SRC, and STAT3 (pIC50: 4.89, 4.36, and 5.09), showcasing multi-target capability. Compound RA2 exhibited significant interactions with HSP90AA1 (pIC50 = 5.62) and STAT3 (pIC50 = 5.09), indicating modulation potential (See Table 5). While interactions with AKT1 and SRC (pIC50 = 4.85 and 4.43) were slightly lower, RA2's multi-target potential was evident. Compound RA3 showed meaningful interactions with HSP90AA1 (pIC50 = 5.48) and STAT3 (pIC50 = 4.82), suggesting inhibitory effects. Interactions with AKT1 and SRC (pIC50 = 4.81 and 4.5) contribute to its diverse bioactivity (Table 3). Compound RA1 and RA2 consistently exhibited higher pIC50 values, indicating relatively stronger inhibition against most target genes. In contrast, Compound RA10 displayed lower activity across all genes. Subsequently, the chalcone derivatives analyzed molecular docking and dynamic studies.
Table 3.
Predicted bioactivity of chalcone derivatives using generated machine learning models.
| Genes / Chalcone derivatives |
AKT1 |
SRC |
HSP90AA1 |
STAT3 |
|---|---|---|---|---|
| pIC50 | ||||
| RA1 | 4.89 | 4.36 | 5.76 | 5.09 |
| RA2 | 4.85 | 4.43 | 5.62 | 5.09 |
| RA3 | 4.81 | 4.5 | 5.48 | 4.82 |
| RA4 | 4.77 | 4.57 | 5.34 | 4.73 |
| RA5 | 4.73 | 4.64 | 5.2 | 4.59 |
| RA6 | 4.69 | 4.71 | 5.06 | 4.46 |
| RA7 | 4.65 | 4.78 | 4.92 | 4.32 |
| RA8 | 4.61 | 4.85 | 4.78 | 4.19 |
| RA9 | 4.57 | 4.92 | 4.64 | 4.05 |
| RA10 | 4.53 | 4.99 | 4.5 | 3.92 |
3.8. Molecular docking
A molecular docking approach was employed to investigate the mechanisms underlying chalcone-based derivatives' anti-inflammatory, antibacterial, anticancer, antidiabetic, and antifungal activities. The docking was performed against four target proteins, namely AKT1, SRC, HSP90AA1, and STAT3. Additionally, a set of ten chalcone derivatives and compounds with Chembl IDs were included in the study. The results of the docking analysis revealed that compounds RA1 to RA7 exhibited superior binding affinities compared to other compounds across the four target genes (Table 4). Notably, chalcone derivatives RA1 to RA7 demonstrated comparable binding affinities to the clinical drug A-443654 (dock score = −10.9 Kcal/mol) against the AKT1 gene. Among these derivatives, Compound RA6 displayed exceptionally high binding affinity (dock score = −10.7 Kcal/mol) towards the AKT1 target gene. “Dasatinib”, a known FDA drug, exhibited significant binding affinity against the SRC target gene with a docking score of − 10.5 Kcal/mol (See Table 6). Interestingly, Compound R5 showed an even better dock score of − 10.7 Kcal/mol, surpassing the previously mentioned drug. Furthermore, among the studied compounds, Compound RA5 demonstrated the strongest affinity against HSP90AA1 with a docking score of − 10.9 Kcal/mol, outperforming the FDA-approved drug “Luminespib”, which achieved a docking score of − 9.6 Kcal/mol. Compound R5 showed the highest docking scores for SRC and HSP90AA1, suggesting its potential to interact with these target genes. Compound R6 demonstrated the highest docking score for AKT1, making it a potential candidate for targeting this gene. The docking scores indicated that “Luminespib” has a notable affinity for HSP90AA1. The docking scores for “Dasatinib” indicated a strong interaction with the SRC target gene. The docking scores point to a potential interaction between CHEMBL4846365 and STAT3. The docking scores for A-443654 indicated a strong interaction with the AKT1 target gene.
Table 4.
Binding affinity scores of all the chalcone derivatives against four distinct targets.
| Compound Name | Target Genes |
|||
|---|---|---|---|---|
|
AKT1 |
SRC |
HSP90AA1 |
STAT3 |
|
| Dock score (Kcal/mol) | ||||
| RA1 | -10.4 | -9.8 | -10.2 | -7.4 |
| RA2 | -10.4 | -10.2 | -10.2 | -7.0 |
| RA3 | -10.5 | -10.2 | -9.7 | -7.5 |
| RA4 | -10.1 | -10.1 | -9.8 | -7.3 |
| RA5 | -10.6 | -10.7 | -10.9 | -7.1 |
| RA6 | -10.7 | -10.6 | -10.5 | -7.4 |
| RA7 | -10.5 | -10.6 | -10.5 | -7.2 |
| RA8 | -8.7 | -8.3 | -8.6 | -6.1 |
| RA9 | -8.9 | -8.2 | -8.6 | -6.2 |
| RA10 | -8.7 | -8.5 | -8.6 | -6.2 |
| A-443654 | -10.9 | - | - | - |
| CHEMBL3899716 | -10.9 | - | - | - |
| CHEMBL3966806 | -10.8 | - | - | - |
| CHEMBL1241676 | - | -8.7 | - | - |
| CHEMBL196797 | - | -10.2 | - | - |
| CHEMBL82085 | - | -9.5 | - | - |
| DASATINIB | - | -10.5 | - | - |
| BIIB021 | - | - | -8.8 | - |
| CHEMBL2205245 | - | - | -9.3 | - |
| CHEMBL2205798 | - | - | -9.1 | - |
| CHEMBL4873718 | - | - | -9.7 | - |
| LUMINESPIB | - | - | -9.6 | - |
| REBLASTATIN | - | - | -7.6 | - |
| AZD-1480 | - | - | - | -6.5 |
| CHEMBL1368342 | - | - | - | -6.1 |
| CHEMBL1407470 | - | - | - | -6.1 |
| CHEMBL4846365 | - | - | - | -7.0 |
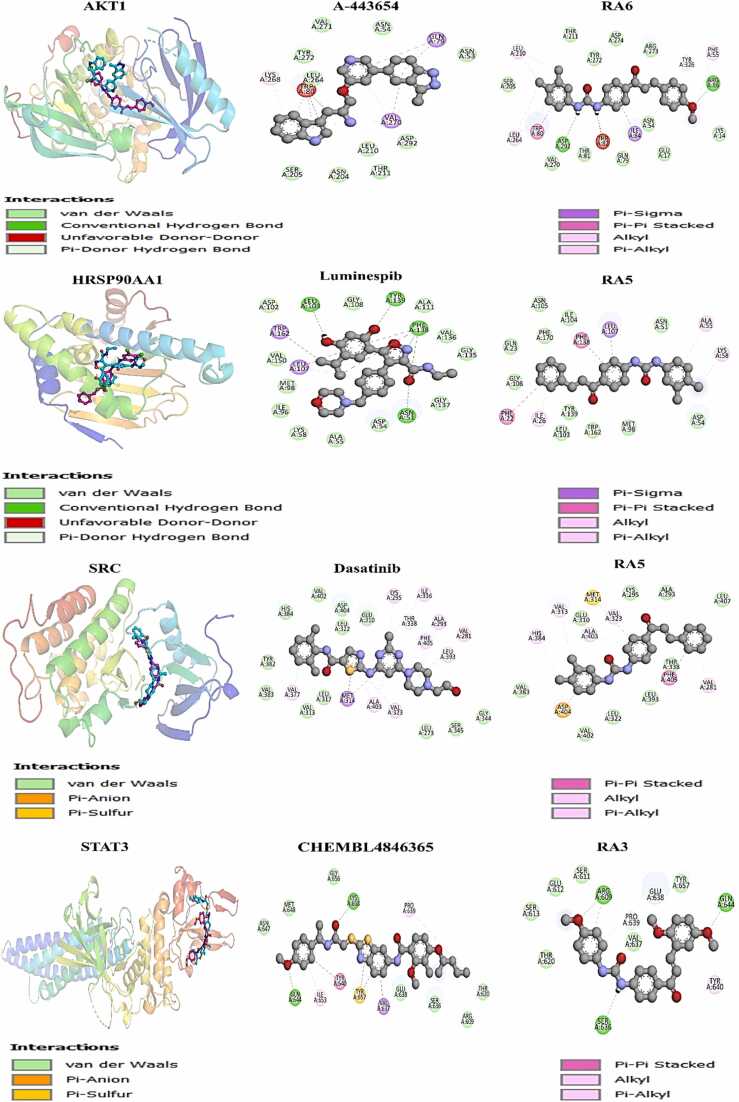
In the context of AKT1, the clinical drug A-443654 did not engage in hydrogen bonding interactions. However, it establishes notable molecular interactions through pi-sigma interactions at Gln79 and Val270, alongside pi-pi stacked formations at Gln79. Additionally, alkyl and pi-alkyl interaction are formatted with Lys268, Val270, and Trp80. In contrast, Compound R6 formed two hydrogen bonding interactions: one with Asp292 involving the urea moiety's NH group, and another involving an oxygen atom and the methoxy group with Arg86. A further interaction is observed with Tyr326 through van der Waals interactions. Notably, Compound R6 exhibited alkyl and pi-alkyl interaction formations at Leu264, Leu210, and Phe55. In contrast, a pi-stacked interaction occurs at Trp80 (Fig. 11). For SRC, the drug “Dasatinib” did not interact as hydrogen bonding interactions. Nevertheless, it demonstrated molecular interactions, such as pi-sigma interactions at Met314, along with alkyl and pi-alkyl interaction formations at Val377, Val323, Ala403, Leu393, Phe405, Ala293, Val281, Ile336, and Lys295. Conversely, Compound R5 did not display hydrogen bonding interactions but presented alkyl and pi-alkyl interactions at Val323, Ala403, Val313, His384, Val281, and pi-pi stacked formation at Phe405 (Fig. 11).
Fig. 11.
3D visualization of compound-protein interactions and 2D analysis for selected compounds (RA3, RA5, RA6, CHEMBL4846365, Dasatinib, Luminespib, A-443654) with the protein.
In the case of HSP90AA1, the drug “Luminespib” formed four hydrogen bonding interactions: the isoxazole ring's nitrogen atom interacted with Phe138, while the carboxamide oxygen atom interacted with Asn51 and Phe138. Another interaction was shown between the oxygen atom of the 4-isopropylbenzene-1,3-diol moiety and Tyr139, as well as Leu103. Further interactions included pi-sigma interactions at Trp162 and Phe138, pi-pi stacked formations at Phe138, and alkyl and pi-alkyl interaction formations at Leu107. In contrast, Compound R5 did not engage in hydrogen bonding interactions. Still, it presented alkyl and pi-alkyl interactions at Ile26, Ala55, and Lys58, along with pi-stacked interactions at Phe22 and Phe138, and a pi-sigma interaction at Leu107 (Fig. 11). Regarding STAT3 gene, Compound RA3 exhibited remarkable binding affinity with a docking score of − 7.5 Kcal/mol compared to CHEMBL4846365 (−7.0 Kcal/mol) and the clinical drug AZD-1480 (−6.5 Kcal/mol). Compound RA3 displayed the highest docking score for STAT3, indicating its strong potential as a candidate for targeting this gene. Compound CHEMBL4846365 formed two hydrogen bonding interactions: one with the methoxy-substituted benzene ring's oxygen atom and Gln644 and another between the urea group's oxygen atom and Lys658. Furthermore, pi-sigma interactions occur at Val637, while pi-pi stacked formations manifest at Tyr640 and Tyr657. Alkyl and pi-alkyl interaction formations are evident at Ile653 and Pro639. In contrast, Compound R3 showed three hydrogen bonding interactions: the oxygen atom of the methoxy-substituted benzene ring interacts with Arg609, while the second di-methoxy benzene-substituted ring interacts with Gln644, and the urea's NH group interacts with Ser636. Moreover, alkyl and pi-alkyl interactions occur with Tyr640 and Pro639 (Fig. 11).
3.9. Molecular dynamics analysis
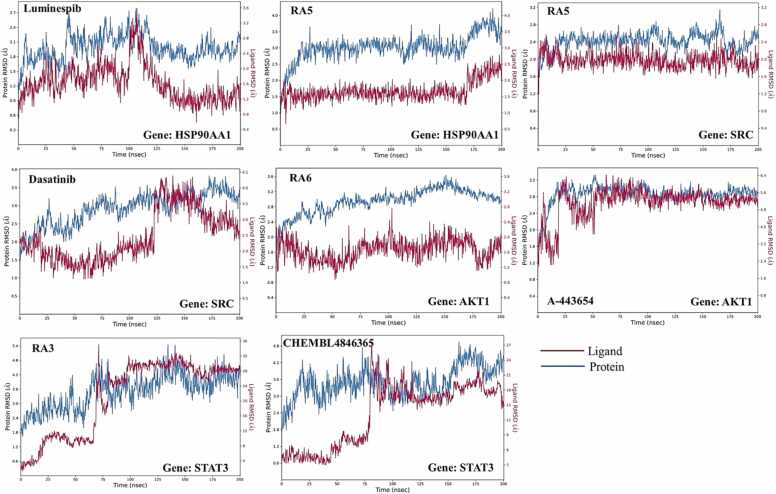
A comprehensive molecular dynamics simulation running 200 nanoseconds was conducted using Desmond software to meticulously evaluate the formation of an optimal complex involving compounds RA3, RA5, RA6, CHEMBL4846365, Dasatinib, Luminespib, A-443654, and the target protein. The analysis focused on critical parameters such as root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and essential interactions between the protein and ligands.
The simulation results indicated that compound RA5 achieved a state of stability in terms of the RMSD values of the C-alpha atoms within the protein complex after the 10-nanosecond threshold, maintaining steady values around 2.0 Angstroms in the SRC protein and 1.5 Angstroms in HSP90AA1 protein throughout the simulation. For the SRC target protein, ligand RA5 exhibited an initial equilibration phase lasting approximately 20 nanoseconds, subsequently maintaining stability within the binding pocket up to the 200-nanosecond mark (Fig. 12). The RMSD of the protein also fluctuated with the ligand and, after 175 ns, slightly decreased, mirroring the initial running time from 25 ns to 172 ns. For the HSP90AA1 target, ligand RA5 displayed the same stable profile, with an upward trend towards stability between 12 and 172 nanoseconds, showing steady RMSD values around 1.5 Angstroms post the initial equilibration phase of 10 nanoseconds. After 175 ns, the ligand showed smaller fluctuations until 200 ns with RMSD values around 1.7 Angstroms. Meanwhile, the protein also showed less fluctuation throughout, remaining within 3.5 Angstroms from 25 to 172 ns, but after that, it showed a conformational shift and slightly increased until 200 ns (Fig. 12). Moreover, the RMSD of the known drug Luminespib initially fluctuated until 120 ns after which it stabilized and remained stable from 130 to 200 ns against the HSP90AA1 gene. Dasatinib showed that the RMSD of the protein backbone of all the complexes stabilized at approximately 1.5 Angstrom before 100 ns of simulation and then from 125 to 150 ns it increased and became more fluctuated; however, after 150 ns, Dasatinib became stable throughout the period against the SRC gene.
Fig. 12.
Analysis of the Root Mean Square Deviation (RMSD) of the hit compounds obtained from molecular docking studies against the target gene through Molecular Dynamics (MD) simulation.
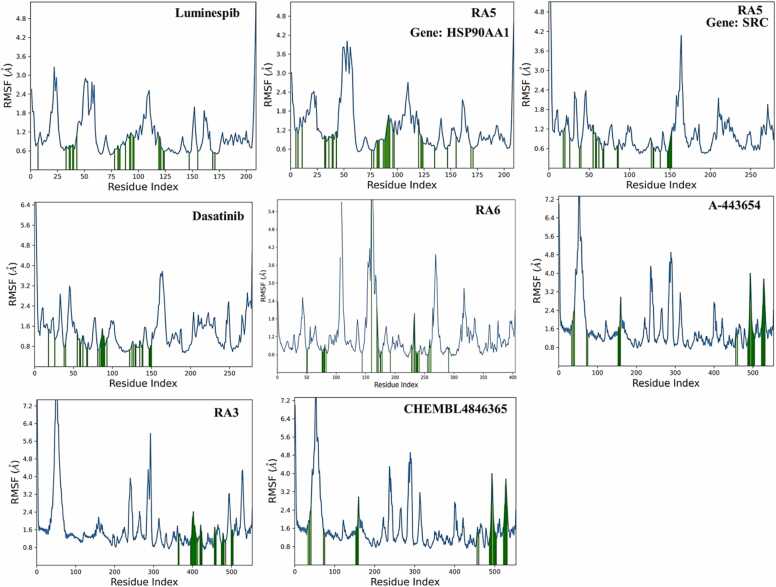
RMSF values for compound RA5 for both targets, SRC and HSP90AA1, highlighted significant fluctuations primarily in the protein's loop and terminal regions, while lower RMSF values at the binding site indicated stable interactions between the protein and ligands. Additionally, the secondary structural composition of the protein was analyzed. For compound RA5 against the HSP90AA1 target, the structural elements, including alpha-helices and beta-strands, constituted 46.35 % of the protein's structure, thereby contributing to its structural stability and functional efficacy. Specifically, helices and strands accounted for 25.51 % and 20.84 % of the total structure, respectively. In the case of SRC, these elements comprised 39.76 % of the protein's structure, with helices and strands representing 26.34 % and 13.42 %, respectively (Fig. 13).
Fig. 13.
Analysis of the root mean square fluctuation (RMSF) of the hit compounds obtained from molecular docking studies against the target gene through molecular dynamics (MD) simulations.
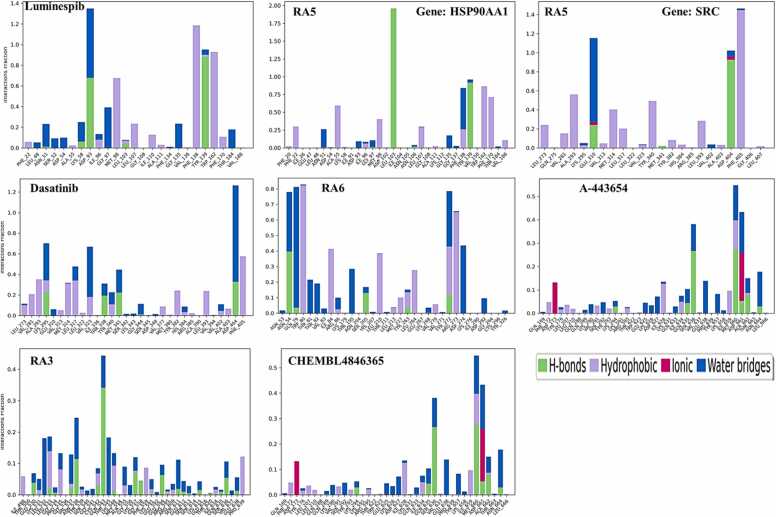
The detailed analysis further explored the interactions between the ligands and the protein's amino acid residues, illustrated through a histogram plot in Fig. 14. This plot clearly showed the different types of interactions hydrogen bonding (marked in green), water bridges (in blue), and hydrophobic interactions (in purple), highlighting their importance in the binding process. Compound RA5 exhibited four hydrogen bonds against HSP90AA1, particularly with amino acids Tyr139 (oxygen atom of the urea group with 91 %), Leu103 (two hydrogen bonds, NH atom of the urea group with 96 % and 99 %), and Phe138 (with a water molecule, and those water molecules interact with the oxygen atom). A di-substituted chlorobenzene ring interacted with Phe170 residue as a hydrophobic interaction with 37 %. On the other hand, Compound RA5 exhibited three hydrogen bonds against SRC, particularly with amino acids Asp404 (oxygen atom of the urea group with 92 %), Glu310 (two hydrogen bonds with two water molecules, and those water molecules interact with the NH atom of the urea group with 37 % and 41 %). A di-substituted methoxy-containing benzene ring interacted with Phe405 residue as a hydrophobic interaction with 53 % (Fig. s2).
Fig. 14.
Analysis of the 2D histogram of protein-ligand contact for the hit compounds derived from molecular docking studies against the target gene via molecular dynamics (MD) simulations.
The clinical drug A-443654 demonstrated greater stability in the AKT1 gene, with RMSD values reaching 2.8 Angstroms, higher than those of compound R6. Compound R6 maintained stability over time within 1.6 Angstroms, while A-443654 showed more consistent stability after 70 ns up to 200 ns, exhibiting steady RMSD values (Fig. 12). RMSF values for both compounds highlighted significant fluctuations primarily in the protein's loop and terminal regions, while lower RMSF values at the binding site indicated stable interactions between the protein and ligands. Overall, both compound R6 and drug A-443654 displayed similar secondary structural composition in the protein, with structural elements, including alpha-helices and beta-strands, constituting 40.52 % of the protein's structure. This composition contributes to its structural stability and functional efficacy, with helices and strands accounting for 18.50 % and 22.01 % of the total structure, respectively. The detailed analysis further explored the interactions between the ligands and the protein's amino acid residues, as illustrated through a histogram plot in Fig. 14. Compound R6 exhibited all hydrogen bonding with water molecules, with 54 % of interactions with Gln79 facilitated by the oxygen atom attached to the benzene ring, and 36 % and 30 % of interactions with Asp274 and Tyr272, respectively, facilitated by the NH atom of the urea group. Asn54 directly interacted with the oxygen atom of the urea group by 39 % and connected with water molecules by 37 %, which in turn are connected with the oxygen atom. Arg273 and Trp84 were linked with di-substituted chloro and methoxy-containing benzene rings through pi-cation and hydrophobic interactions, at 64 % and 43 %, respectively. Conversely, drug A-443654 displayed only two hydrogen bonding interactions: one from the NH group of the pyridine ring with Ser205 at 36 % and the other with Asn53 from the NH group of the benzimidazole ring at 45 % (Fig. s2).
Fig. 12 illustrates the RMSD plot for compound CHEMBL4846365, showing the RMSD of the STAT3 protein and the ligand over time. Initially, both protein and ligand RMSD values raised, typical as the system equilibrates. After this period, the ligand RMSD stabilized, indicating that the compound has found a relatively stable conformation within the binding site. However, the protein RMSD continues to exhibit some fluctuations, suggesting that while the ligand may be stable, the protein is still undergoing conformational changes, possibly adjusting to the ligand's presence or due to its dynamic nature. In the case of compound R3, the RMSD plot showed a change over time, maintaining stability in the binding pocket from 100 ns to 150 ns, with some conformational shifts observed around the 10 to 100 nanosecond range, followed by stability from 125 to 150 ns (Fig. 12). The protein's RMSD, while fluctuating, did not show a pronounced rise, implying a more rigid structure or less conformational change in response to ligand binding compared to the complex with compound CHEMBL4846365. RMSF values, demonstrated in Fig. 13, exhibit significant fluctuations mainly in the protein's loop and terminal regions, with lower RMSF values at the binding site suggesting stable interactions. The structural elements of compound R3, including alpha-helices and beta-strands, constituted 57.15 % of the protein's structure, contributing to its structural stability and functional efficacy, with helices and strands accounting for 40.63 % and 16.51 %, respectively. In contrast, for compound CHEMBL4846365, these elements comprised 57.43 % of the protein's structure, with helices and strands representing 40.42 % and 17.02 %, respectively. Compound R3 exhibited one hydrogen bonding interaction with the oxygen atom of the urea group by Gln543 at 33 %. Conversely, compound CHEMBL4846365 did not show any significant contribution to interaction with the STAT3 protein target. This comprehensive interaction analysis underscores the specificity and diversity of ligand-protein interactions and emphasizes the role of molecular dynamics simulations in uncovering intricate details of binding mechanisms, invaluable in the rational design of therapeutics for optimizing ligand efficacy and specificity.
3.9.1. Principal component analysis (PCA)
Principal Component Analysis (PCA) provided a detailed view of the interaction dynamics between diverse compounds and their target proteins throughout Molecular Dynamics (MD) simulations (Fig. 15). This technique captures key aspects of the compounds' stability and the range of their motion when bound to protein targets. In the case of the Luminespib drug in the HSP90AA1 protein, the PCA plot shows data points tightly grouped near the origin for both principal components. This clustering signifies a consistent interaction dynamic, with the compound maintaining a stable conformation throughout the simulation process. Conversely, Dasatinib displayed a distinct pattern when bound to the SRC protein, with data points scattered more widely along the principal component one (PC1) axis. This spread indicated a broader range of conformational states that Dasatinib may adopt during its interaction, implying a higher degree of flexibility and dynamic behavior in its binding conformations. RA5 presents an interesting case; when tested against both HSP90AA1 and SRC targets, there is a noticeable dispersal along the PC1 axis for each. This observation suggests that RA5 can induce diverse conformational states within these protein complexes. Notably, when bound to SRC, the spread along the principal component two (PC2) axis is relatively constrained, hinting that while RA5 may exhibit a variety of shapes, these conformations likely change within a limited range within the multidimensional conformational landscape.
Fig. 15.
Principal component analysis (PCA) of hit compounds in protein-ligand complexes.
Examining the interactions of compound RA6 and A-443654 with AKT1 further expanded our understanding. Both compounds share a pattern of greater distribution along PC1 than PC2, which may suggest a significant conformational diversity that unfolds along a particular dimension of the interaction. Compound A-443654 showed a pronounced distribution along PC2 as well, suggesting that it can move through an even more varied range of conformations, possibly affecting different domains of the AKT1 protein. The interaction of RA3 with STAT3 is characterized by the widest distribution, especially along PC1, indicating that RA3 might access a considerable array of conformational states (Fig. 15). This wide range might represent various modes of binding or a high degree of structural flexibility within the ligand when it is associated with the protein. CHEMBL4846365 engagement with STAT3 is also depicted with a substantial spread along PC1, which is indicative of notable conformational dynamics. However, its moderate dispersal along PC2, particularly when contrasted with RA3, suggests that the diversity of its conformational changes might be less extreme across the entire structure of the complex.
3.9.2. MMGBSA
The Molecular Mechanics Generalized Born Surface Area (MM-GBSA) methods have been applied to calculate the free energy of binding for a series of compounds against various protein targets, providing insights into the potential efficacy of these compounds as inhibitors. The analysis presents the binding free energies along with contributions from Coulombic, covalent, hydrogen bonding, lipophilic, packing, self-contact, solvation, and van der Waals interaction. For compound A443654 targeting AKT1, the MM-GBSA binding energy is notably high at − 64.31 kcal/mol, with significant contributions from lipophilic interactions at − 22.74 kcal/mol and van der Waals forces at − 59.68 kcal/mol. This suggests a substantial nonpolar interaction component, complemented by Coulombic interactions at − 11.06 kcal/mol. Similarly, CHEMBL4846365 against STAT3 shows a binding energy of − 31.80 kcal/mol, with a relatively lower van der Waals contribution of − 30.78 kcal/mol, indicating a slightly less hydrophobic interaction compared to A443654 with AKT1. The lipophilic interactions for CHEMBL4846365 are also lower at − 8.58 kcal/mol. Dasatinib's binding to SRC is characterized by a binding energy of − 83.46 kcal/mol, with a large negative contribution from lipophilic interactions at − 29.06 kcal/mol and a significant van der Waals term at − 73.20 kcal/mol, reflecting strong hydrophobic and van der Waals interactions within the binding site. Luminespib shows a strong affinity for HSP90AA1 with a binding energy of − 85.86 kcal/mol. The notable lipophilic and van der Waals contributions of − 26.50 and − 62.68 kcal/mol, respectively, highlight the compound's strong hydrophobic binding character.
For RA3 against STAT3, the binding energy is − 45.08 kcal/mol. This is paired with a hydrogen bond contribution of − 0.57 kcal/mol and a notable van der Waals term of − 36.51 kcal/mol, indicating a good balance of polar and nonpolar interactions. Compound RA5 targeting HSP90AA1 exhibits a particularly strong binding energy of − 96.26 kcal/mol, with the highest lipophilic contribution among the compounds at − 34.71 kcal/mol and a substantial van der Waals component at − 66.31 kcal/mol, suggesting a potent interaction with the protein. RA6 interacting with AKT1 has a binding energy of − 66.30 kcal/mol. Its lipophilic and van der Waals contributions are significant at − 28.07 and − 66.11 kcal/mol, respectively, indicative of favorable hydrophobic interactions. Lastly, RA5 against SRC shows the most potent binding energy of − 100.01 kcal/mol within this dataset. The lipophilic term is extremely high at − 38.98 kcal/mol, coupled with a large van der Waals contribution of − 75.37 kcal/mol, which could be reflective of a tight and efficient binding to the active site.
4. Discussion
The focus of our recent study was to identify key chalcone compounds and Chembl libraries aimed at AKT1, SRC, HSP90AA1, and STAT3 genes. These targets, by potentially inhibiting their metabolic pathways, were chosen for their capacity to act against cancer, diabetes, inflammation, and fungal and bacterial infections. This versatility makes chalcones a valuable candidate for drug development because they can target multiple disease pathways. The approach to achieving this objective involved the integration of machine learning QSAR, molecular mechanisms, and systems biology techniques. This approach involved ligand and structure-based screening of small molecule databases targeting these four genes, followed by pharmacokinetic screening and docking. We also identified potential small molecule inhibitors that could inhibit the binding sites of these target gene pathways using machine learning-assisted Quantitative Structure-Activity Relationship (QSAR) modeling and web-based bioactivity prediction. A multifaceted approach such as this demonstrates the potential of integrated computational methodologies for advancing the discovery and development of drugs. Thorough exploratory data analysis (EDA) in the initial phase of this study played a pivotal role in shaping the subsequent phases. We meticulously curated and preprocessed the dataset, revealing significant distinctions between active and inactive compounds across various molecular properties. The Mann-Whitney U test highlighted the statistical significance of these differences, underscoring the potential of chalcone derivatives as bioactive compounds.
Central to our study was the creation of Random Forest-based QSAR models for each target gene. These models exhibited commendable predictive performance, characterized by high correlation coefficients and acceptable error rates during both training and testing. Notably, the feature importance analysis identified specific molecular descriptors crucial for predicting bioactivity, providing vital insights into the structural determinants of chalcone derivatives' effectiveness. Extensive analysis uncovered a promising group of compounds, particularly RA1 to RA7, which demonstrated exceptional bioactivity against the target genes. For instance, compound RA1 displayed a remarkable pIC50 value of 5.76 against HSP90AA1, making it a candidate for further exploration. This correlation between compound structure and bioactivity emphasizes the potential utility of chalcone derivatives in drug discovery. Molecular docking studies further elucidated the binding interactions between chalcone derivatives and the target genes. Compounds RA5, RA6, and RA7 exhibited significant binding affinities, equalling or exceeding those of existing drugs, indicating their promise as inhibitory compounds. The detailed analysis of these binding interactions reveals the specific structural features responsible for bioactivity, aiding in a rational approach to drug design. The fingerprint-based predictive models for the top genes AKT1, SRC, HSP90AA1, and STAT3 were further deployed as the ASHS-Pred web-based application. The source codes (https://github.com/RatulChemoinformatics/QSAR) and data sets were made available on GitHub to facilitate further extension or modification of the web server. It is important to observe that as new experimental data on individual gene inhibitors become available, the predictive model proposed here could be continuously updated to increase its coverage and accuracy. In molecular dynamic study, particularly highlighting compound RA5's stability with SRC and HSP90AA1 targets. This stability, evidenced by consistent RMSD values, suggests potential therapeutic efficacy. Comparatively, the differences in RMSD and RMSF values between compounds, including A-443654 and R6 against the AKT1 gene, underscore the unique interaction dynamics each compound exhibits with its target. Furthermore, detailed analyses of secondary structures and ligand-protein interactions, such as hydrogen bonding and hydrophobic contacts, offer a view of binding affinities and specificities.
Discussing the results of MMGBSA, the particularly high van der Waals and lipophilic interaction energies observed for most compounds suggest that these compounds may have substantial hydrophobic contacts within the binding sites of their respective targets, which is often a mark of drug-like molecules. For example, the compound RA5 against the SRC protein exhibited the most potent binding energy at − 100.01 kcal/mol, marked by the highest lipophilic contribution at − 38.98 kcal/mol among the dataset, underscoring its strong affinity and specificity towards the target. This suggests that RA5 could robustly occupy the hydrophobic pockets within SRC, maximizing van der Waals contacts and potentially leading to high inhibitory activity. Moreover, the compounds targeting HSP90AA1, specifically Luminespib and RA5, demonstrated high binding energies and substantial lipophilic contributions, indicating effective hydrophobic interactions that could stabilize the inhibitor within the binding domain. These interactions, coupled with the observed hydrogen bonds, are essential for a stable drug-protein complex, enhancing the efficacy of the drug. In contrast, the lower binding energies seen with compounds like CHEMBL4846365 against STAT3 suggest weaker interactions, which could be due to less optimal alignment within the binding pocket or insufficient hydrophobic contact, potentially leading to reduced inhibitory activity. Comparing the PCA plots collectively, it is evident that the conformational stability and flexibility of these compounds when interacting with their respective targets vary. Compounds such as Luminespib exhibit a more constrained range of motion, indicative of a stable interaction, while others like RA3 and CHEMBL4846365 demonstrate significant conformational diversity, which might correlate with multiple binding modes or interactions with the protein targets. These observations are critical for understanding the dynamic nature of protein-ligand interactions and can have implications for the optimization of these hit compounds for potential therapeutic applications.
This study concisely demonstrates the significant potential of chalcone derivatives in targeting key genes, with a focus on high-efficacy compounds RA1 to RA7. It underscores the relevance of structural factors in drug design and advocates for further experimental validation. Integrating machine learning and knowledge-based neural network insights with molecular docking and dynamics simulations, this work offers a promising direction for developing treatments in anti-inflammatory, antibacterial, anticancer, antidiabetic, antifungal, and antituberculosis areas. This approach has the potential to address critical medical needs and advance drug discovery. In future efforts, the aim will be to expand the research focus on studying the efficacy of these molecules specifically for tuberculosis treatment. This will involve further exploration of their interactions with Mycobacterium tuberculosis and understanding their potential mechanisms of action in combating tuberculosis infections. Additionally, we plan to conduct safety studies of chalcones using the zebrafish larval model and perform in vitro and in vivo studies using M. marinum and zebrafish to validate the safety and effectiveness of chalcone derivatives as potential anti-tuberculosis agents [69], [70], [71], [72].
5. Conclusion
We identified significant chalcone derivatives and ChEMBL libraries targeted at AKT1, SRC, HSP90AA1, and STAT3. The ability of chalcones to target multiple disease pathways underscores their potential for drug development. An integrated approach, combining machine learning QSAR, molecular mechanisms, and knowledge-based neural network techniques, has advanced drug discovery. Notably, chalcone derivatives RA1 to RA7 exhibited substantial bioactivity against key target genes, with RA1 showing the most promising pIC50 value, particularly against HSP90AA1. Docking scores corroborated these findings, with RA1 displaying robust binding affinities across all genes. Remarkably, compounds RA5, RA6, and RA7 exhibited docking scores comparable to RA1, indicating similar potential. However, a decline in activity was observed from RA8 to RA10, consistent with pIC50 trends. Further supporting these findings, comprehensive molecular dynamics simulations provided deeper insights into the dynamic interactions and stability of these compounds, particularly RA5, with target proteins SRC and HSP90AA1. The simulations of 200 nanoseconds highlighted the compounds' stability and interaction dynamics, crucial for understanding their therapeutic potential highlighted the compounds' stability and interaction dynamics, crucial for understanding their therapeutic potential. The consistent RMSD values of compound RA5 after the initial equilibration phase illustrated a stable interaction with the proteins, potentially contributing to its efficacy. This dynamic analysis enhanced the insights provided by static docking scores and bioactivity findings, giving a crucial understanding of how the compounds interact with their target proteins. Compared to established drugs and ChEMBL compounds, chalcone derivatives demonstrated promising results, with some outperforming known drugs in binding affinity. Specifically, compound RA5 exhibited exceptional binding affinity against HSP90AA1, surpassing Luminespib, an FDA-approved drug. Compound RA3 exhibited significant binding to STAT3, highlighting the potential of chalcone derivatives, as evidenced by their encouraging binding scores with crucial genes. Additional research into these derivatives, encompassing both in vitro and in vivo studies, is necessary to confirm their effectiveness in treating diseases related to AKT1, HSP90AA1, SRC, and STAT3. Insights from machine learning models provide a robust foundation for future research in chalcone-based small molecule binding and drug discovery.
CRediT authorship contribution statement
Mohd. Imran: Visualization, Validation. Bijo Mathew: Visualization, Validation, Resources. Seppo Parkkila: Writing – review & editing, Visualization, Validation. Ashok Aspatwar: Writing – review & editing, Visualization, Validation, Supervision, Project administration, Investigation, Funding acquisition. Sunil Kumar: Visualization, Software, Resources. Satarupa Acharjee: Writing – review & editing, Visualization, Validation, Supervision, Investigation, Conceptualization, Project administration, Resources. Sameer Sharma: Visualization, Formal analysis. Ajay Manaithiya: Writing – review & editing, Writing – original draft, Visualization, Software, Resources, Methodology, Formal analysis, Data curation, Conceptualization, Investigation, Supervision, Validation. Ratul Bhowmik: Writing – review & editing, Writing – original draft, Software, Resources, Methodology, Formal analysis, Data curation, Conceptualization, Investigation, Supervision, Validation, Visualization.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgment
The authors extend their sincere appreciation to the following organizations for their invaluable support and funding: 1) Finnish Cultural Foundation, 2) Tampere Tuberculosis Foundation, (Ashok Aspatwar); 3) Academy of Finland, and 4) Jane and Aatos Erkko Foundation (Seppo Parkkila).
Footnotes
Elucidating molecular mechanism of chalcones through biological networks and machine learning approaches
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.07.006.
Contributor Information
Satarupa Acharjee, Email: satarupa.acharjee@nshm.com.
Ashok Aspatwar, Email: ashok.aspatwar@tuni.fi.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Ferrer J.L., Austin M.B., Stewart C., Noel J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem. 2008;46:356–370. doi: 10.1016/J.PLAPHY.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.K. Sahu N., S. Balbhadra S., Choudhary J., V. Kohli D. Exploring pharmacological significance of chalcone scaffold: a review. Curr Med Chem. 2012;19:209–225. doi: 10.2174/092986712803414132. [DOI] [PubMed] [Google Scholar]
- 3.Mahapatra D.K., Bharti S.K., Asati V. Anti-cancer chalcones: structural and molecular target perspectives. Eur J Med Chem. 2015;98:69–114. doi: 10.1016/J.EJMECH.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/CSHPERSPECT.A001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivakumar P.M., Geetha Babu S.K., Mukesh D. QSAR studies on chalcones and flavonoids as anti-tuberculosis agents using genetic function approximation (GFA) method. Chem Pharm Bull. 2007;55:44–49. doi: 10.1248/CPB.55.44. [DOI] [PubMed] [Google Scholar]
- 6.Dhaliwal J.S., Moshawih S., Goh K.W., Loy M.J., Hossain M.S., Hermansyah A., et al. Pharmacotherapeutics applications and chemistry of chalcone derivatives. Mol. 2022;Vol 27:7062. doi: 10.3390/MOLECULES27207062. Page 7062 2022;27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura T.L.B., Calixto S.D., De Azevedo, Abrahim-Vieira B., De Souza A.M.T., Mello M.V.P., et al. Antimycobacterial and anti-inflammatory activities of substituted chalcones focusing on an anti-tuberculosis dual treatment approach. Mol. 2015;Vol 20 doi: 10.3390/MOLECULES20058072. Pages 8072-8093 2015;20:8072–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thapa P., Upadhyay S.P., Suo W.Z., Singh V., Gurung P., Lee E.S., et al. Chalcone and its analogs: therapeutic and diagnostic applications in Alzheimer’s disease. Bioorg Chem. 2021;108 doi: 10.1016/J.BIOORG.2021.104681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karthikeyan C., Narayana Moorthy N.S.H., Ramasamy S., Vanam U., Manivannan E., Karunagaran D., et al. Advances in chalcones with anticancer activities. Recent Pat Anticancer Drug Discov. 2014;10:97–115. doi: 10.2174/1574892809666140819153902. [DOI] [PubMed] [Google Scholar]
- 10.Chiaradia L.D., Martins P.G.A., Cordeiro M.N.S., Guido R.V.C., Ecco G., Andricopulo A.D., et al. Synthesis, biological evaluation, and molecular modeling of chalcone derivatives as potent inhibitors of Mycobacterium tuberculosis protein tyrosine phosphatases (PtpA and PtpB) J Med Chem. 2012;55:390–402. doi: 10.1021/jm2012062. [DOI] [PubMed] [Google Scholar]
- 11.Mishra S., Jana P. Chalcones as anti-infective agents for effective management of tuberculosis. Polycycl Aroma Compd. 2023 doi: 10.1080/10406638.2023.2261593. [DOI] [Google Scholar]
- 12.Sengupta S.A., Maity T.K., Samanta S. Synthesis, biological screening and in silico studies of chalcone based novel phenyl urea derivatives as potential antihyperglycemics. J Pharm Res. 2017;16:237. doi: 10.18579/JPCRKC/2017/16/3/118765. [DOI] [Google Scholar]
- 13.Acharjee S., Maity T.K., Samanta S., Mana S., Chakraborty T., Singha T., et al. Antihyperglycemic activity of chalcone based novel 1-{3-[3-(substituted phenyl) prop-2-enoyl] phenyl} thioureas. Synth Commun. 2018;48:3015–3024. doi: 10.1080/00397911.2018.1539178. [DOI] [Google Scholar]
- 14.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7 doi: 10.1038/SREP42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathak M., Ojha H., Tiwari A.K., Sharma D., Saini M., Kakkar R. Design, synthesis and biological evaluation of antimalarial activity of new derivatives of 2,4,6-s-triazine. Chem Cent J. 2017;11 doi: 10.1186/S13065-017-0362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinforma. 2016;54(1) doi: 10.1002/CPBI.5. 30.1-1.30.33. [DOI] [PubMed] [Google Scholar]
- 17.Amberger J.S., Hamosh A. Searching online mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinforma. 2017;58(1) doi: 10.1002/CPBI.27. 2.1-1.2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/NAR/GKAA1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/GR.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonseka P., Pathan M., Chitti S.V., Kang T., Mathivanan S. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol. 2021;433 doi: 10.1016/J.JMB.2020.166747. [DOI] [PubMed] [Google Scholar]
- 21.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:1–11. 10.1186/GB-2003-4-9-R60/TABLES/3. [DOI] [PubMed]
- 22.Ge S.X., Jung D., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/BIOINFORMATICS/BTZ931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap C.W. PaDEL-descriptor: An open source software to calculate molecular descriptors and fingerprints. J Comput Chem. 2011;32:1466–1474. doi: 10.1002/JCC.21707. [DOI] [PubMed] [Google Scholar]
- 24.DTClab-〈https://dtclab.webs.com/software-tools〉.
- 25.Padel. 〈http://www.yapcwsoft.com/dd/padeldescriptor/〉.
- 26.Github. 〈https://github.com/dataprofessor/code/tree/master/python〉.
- 27.De P., Kar S., Ambure P., Roy K. Prediction reliability of QSAR models: an overview of various validation tools. Arch Toxicol. 2022;96:1279–1295. doi: 10.1007/S00204-022-03252-Y/METRICS. [DOI] [PubMed] [Google Scholar]
- 28.Hall M., Frank E., Holmes G., Pfahringer B., Reutemann P., Witten I.H. The WEKA data mining software. ACM SIGKDD Explor Newsl. 2009;11:10–18. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- 29.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2012;12:2825–2830. [Google Scholar]
- 30.Sahigara F., Mansouri K., Ballabio D., Mauri A., Consonni V., Todeschini R. Comparison of different approaches to define the applicability domain of QSAR models. Molecules. 2012;17:4791–4810. doi: 10.3390/MOLECULES17054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weka.〈https://www.cs.waikato.ac.nz/ML/weka/〉.
- 32.Github. 〈https://github.com/vappiah/Machine-Learning-Tutorials〉.
- 33.scikit-learn. 〈https://github.com/scikit-learn/scikit-learn.git〉.
- 34.Ashwell M.A., Lapierre J.M., Brassard C., Bresciano K., Bull C., Cornell-Kennon S., et al. Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b] pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors. J Med Chem. 2012;55:5291–5310. doi: 10.1021/jm300276x. [DOI] [PubMed] [Google Scholar]
- 35.Seeliger M.A., Nagar B., Frank F., Cao X., Henderson M.N., Kuriyan J. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure. 2007;15:299–311. doi: 10.1016/J.STR.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Patel P.D., Yan P., Seidler P.M., Patel H.J., Sun W., Yang C., et al. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat Chem Biol. 2013;9:677–684. doi: 10.1038/NCHEMBIO.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai L., Zhou H., Xu R., Zhao Y., Chinnaswamy K., McEachern D., et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell. 2019;36:498–511.e17. doi: 10.1016/J.CCELL.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/JCC.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowers K.J., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proc 2006 ACM/IEEE Conf Supercomput SC’06 2006. 10.1145/1188455.1188544. [DOI]
- 40.Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D., et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/BIOINFORMATICS/BTQ430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiliro C., Firestein B.L. Mechanisms of metabolic reprogramming in cancer cells supporting enhanced growth and proliferation. Cells. 2021;10 doi: 10.3390/CELLS10051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fayard E., Xue G., Parcellier A., Bozulic L., Hemmings B.A. Protein kinase B (PKB/Akt), a key mediator of the PI3K signaling pathway. Curr Top Microbiol Immunol. 2010;346:31–56. doi: 10.1007/82_2010_58. [DOI] [PubMed] [Google Scholar]
- 43.Siddiqui R., Iqbal J., Maugueret M.J., Khan N.A. The role of Src kinase in the biology and pathogenesis of Acanthamoeba castellanii. Parasit Vectors. 2012;5 doi: 10.1186/1756-3305-5-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu S.T., Pham H., Pandol S.J., Ptasznik A. Src as the link between inflammation and cancer. Front Physiol. 2014;4 doi: 10.3389/FPHYS.2013.00416/BIBTEX. JAN:78701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Backe S.J., Sager R.A., Woodford M.R., Makedon A.M., Mollapour M. Post-translational modifications of Hsp90 and translating the chaperone code. J Biol Chem. 2020;295:11099–11117. doi: 10.1074/JBC.REV120.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/NRC2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vella V., Lappano R., Bonavita E., Maggiolini M., Clarke R.B., Belfiore A., et al. Insulin/IGF axis and the receptor for advanced glycation end products: role in meta-inflammation and potential in cancer therapy. Endocr Rev. 2023;44:693–723. doi: 10.1210/ENDREV/BNAD005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y.C., Yeh C.T., Lin K.H. Molecular functions of thyroid hormone signaling in regulation of cancer progression and anti-apoptosis. Int J Mol Sci. 2019;20 doi: 10.3390/IJMS20204986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chao M.V. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/NRN1078. [DOI] [PubMed] [Google Scholar]
- 50.Rosenberg S.A., Restifo N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/SCIENCE.AAA4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/J.CELL.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner N., Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/NRC2780. [DOI] [PubMed] [Google Scholar]
- 53.Bollenbecker S., Barnes J.W., Krick S. Fibroblast growth factor signaling in development and disease. Int J Mol Sci. 2023;24 doi: 10.3390/IJMS24119734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shibuya M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: a crucial target for anti- and pro-angiogenic therapies. Genes Cancer. 2011;2:1097–1105. doi: 10.1177/1947601911423031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heldin C.H., Lennartsson J. Structural and functional properties of platelet-derived growth factor and stem cell factor receptors. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/CSHPERSPECT.A009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen S., Wang F., Fernandez A., Hu W. Role of platelet-derived growth factor in type II diabetes mellitus and its complications. Diabetes Vasc Dis Res. 2020;17 doi: 10.1177/1479164120942119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hynes N.E., Lane H.A. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/NRC1609. [DOI] [PubMed] [Google Scholar]
- 58.Lennartsson J., Rönnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/PHYSREV.00046.2011. [DOI] [PubMed] [Google Scholar]
- 59.Dhillon A.S., Hagan S., Rath O., Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/SJ.ONC.1210421. [DOI] [PubMed] [Google Scholar]
- 60.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/J.CELL.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Shea J.J., Schwartz D.M., Villarino A.V., Gadina M., McInnes I.B., Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/ANNUREV-MED-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/J.CELL.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Porta C., Paglino C., Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014;4 doi: 10.3389/FONC.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadikot R.T., Blackwell T.S., Christman J.W., Prince A.S. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171:1209–1223. doi: 10.1164/RCCM.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oeckinghaus A., Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/CSHPERSPECT.A000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reece M.D., Song C., Hancock S.C., Pereira Ribeiro S., Kulpa D.A., Gavegnano C. Repurposing BCL-2 and Jak 1/2 inhibitors: Cure and treatment of HIV-1 and other viral infections. Front Immunol. 2022;13 doi: 10.3389/FIMMU.2022.1033672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahrens T.D., Bang-Christensen S.R., Jørgensen A.M., Løppke C., Spliid C.B., Sand N.T., et al. The role of proteoglycans in cancer metastasis and circulating tumor cell analysis. Front Cell Dev Biol. 2020;8 doi: 10.3389/FCELL.2020.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji W., Choi C.M., Rho J.K., Jang S.J., Park Y.S., Chun S.M., et al. Mechanisms of acquired resistance to EGFR-tyrosine kinase inhibitor in Korean patients with lung cancer. BMC Cancer. 2013;13 doi: 10.1186/1471-2407-13-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aspatwar A., Hammaren M.M., Parikka M., Parkkila S. Rapid evaluation of toxicity of chemical compounds using zebrafish embryos. J Vis Exp. 2019;2019 doi: 10.3791/59315. [DOI] [PubMed] [Google Scholar]
- 70.Aspatwar A., Hammarén M., Koskinen S., Luukinen B., Barker H., Carta F., et al. β-CA-specific inhibitor dithiocarbamate Fc14-584B: a novel antimycobacterial agent with potential to treat drug-resistant tuberculosis. J Enzym Inhib Med Chem. 2017;32:832–840. doi: 10.1080/14756366.2017.1332056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aspatwar A., Kairys V., Rala S., Parikka M., Bozdag M., Carta F., et al. Mycobacterium tuberculosis β-Carbonic Anhydrases: Novel Targets for Developing Antituberculosis Drugs. Int J Mol Sci. 2019;20 doi: 10.3390/IJMS20205153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aspatwar A., Winum J.Y., Carta F., Supuran C.T., Hammaren M., Parikka M., et al. Carbonic anhydrase inhibitors as novel drugs against mycobacterial β-carbonic anhydrases: an update on in vitro and in vivo studies. Molecules. 2018;23 doi: 10.3390/MOLECULES23112911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material