Summary
Maternal obesity has long-term effects on offspring metabolic health. Among the potential mechanisms, prior research has indicated potential disruptions in circadian rhythms and gut microbiota in the offspring. To challenge this hypothesis, we implemented a maternal high fat diet regimen before and during pregnancy, followed by a standard diet after birth. Our findings confirm that maternal obesity impacts offspring birth weight and glucose and lipid metabolisms. However, we found minimal impact on circadian rhythms and microbiota that are predominantly driven by the feeding/fasting cycle. Notably, maternal obesity altered rhythmic liver gene expression, affecting mitochondrial function and inflammatory response without disrupting the hepatic circadian clock. These changes could be explained by a masculinization of liver gene expression similar to the changes observed in polycystic ovarian syndrome. Intriguingly, such alterations seem to provide the first-generation offspring with a degree of protection against obesity when exposed to a high fat diet.
Subject areas: neuroscience, behavioral neuroscience, molecular neuroscience, omics, transcriptomics
Graphical abstract
Highlights
-
•
Maternal obesity has little impact on the rhythmic physiology of the offspring
-
•
The impact on the microbiome composition appears mainly driven by feeding
-
•
Liver inflammation and mitochondrial activity are impacted by maternal obesity
-
•
This likely results from the masculinization of the offspring similar to PCOS models
Circadian rhythms; Behavioral neuroscience; Molecular neuroscience; Omics; Transcriptomics; Endocrinology.
Introduction
The global obesity epidemic has resulted in a rise in metabolic disorders among women of reproductive age. Currently, about 30–50% of women of reproductive age falling into the overweight or obese range.1,2 This rise has also profound implications for offspring metabolism spanning from the fetal stage to adulthood.3,4,5 Both epidemiological and animal studies have shown that maternal high fat diet (HFD) and obesity increase the offspring’s risk of developing diabetes, obesity, and cardiometabolic diseases.6,7,8,9 This phenomenon of metabolic imprinting has given rise to the concept of Developmental Origins of Health and Disease (DOHaD).10,11
To uncover the mechanisms behind this transgenerational effect, recent hypotheses propose an intergenerational alteration of the interplay between gut microbiota and the circadian clock. The circadian clock is an endogenous oscillator that orchestrates most aspects of mammalian physiology and behavior in the anticipation of environmental day-night changes. This includes the sleep/wake cycle, feeding/fasting cycles, and rhythmic hormone secretion. Organized in a hierarchical manner, the central circadian clock localized in the suprachiasmatic nuclei (SCN) of the hypothalamus synchronizes peripheral clocks present in virtually every cell of the organism.12,13 While the central clock is synchronized by light through direct connections between the retina and the SCN, peripheral clocks can be also synchronized by feeding and other systemic cues.14 The importance of the circadian clock for physiology and metabolism is underscored by its disruption in scenarios such as shift work, that lead to metabolic diseases.15,16 Conversely, HFD and obesity have been observed to disrupt the circadian clock.17,18 Accordingly, a few studies have reported a transgenerational impact of maternal HFD and obesity on the circadian rhythm of the offspring in rodents and primates.19,20,21,22
The feeding/fasting cycle not only synchronizes peripheral clocks but also leads to changes in daily fluctuations in the composition of the gut microbiota, further influencing host physiology.23,24 Given that obesity is known to impact the gut microbiome and is associated with dysbiosis,25 alterations in gut microbiota composition are suggested to mediate the metabolic consequences of maternal HFD.26,27,28 A disrupted rhythmic gut microbiome could also potentially impact the diurnal physiology of the offspring. For these reasons, we explored whether the metabolic effects of maternal HFD on offspring could be attributed to disturbances in rhythmic gut microbiota and liver physiology. While the liver circadian clock and feeding rhythms appear not impacted by maternal HFD, we observed alterations in the rhythmic expression of genes involved in pathways related to mitochondrial activity or ribosome biogenesis. Further analysis revealed an increased expression of genes involved in pathways related to inflammation, potentially associated with perturbations in endocrine regulations. These findings could help to elucidate the transgenerational effects of maternal HFD on offspring physiology.
Results
Maternal high fat diet induces glucose and cholesterol metabolism alterations in the offspring
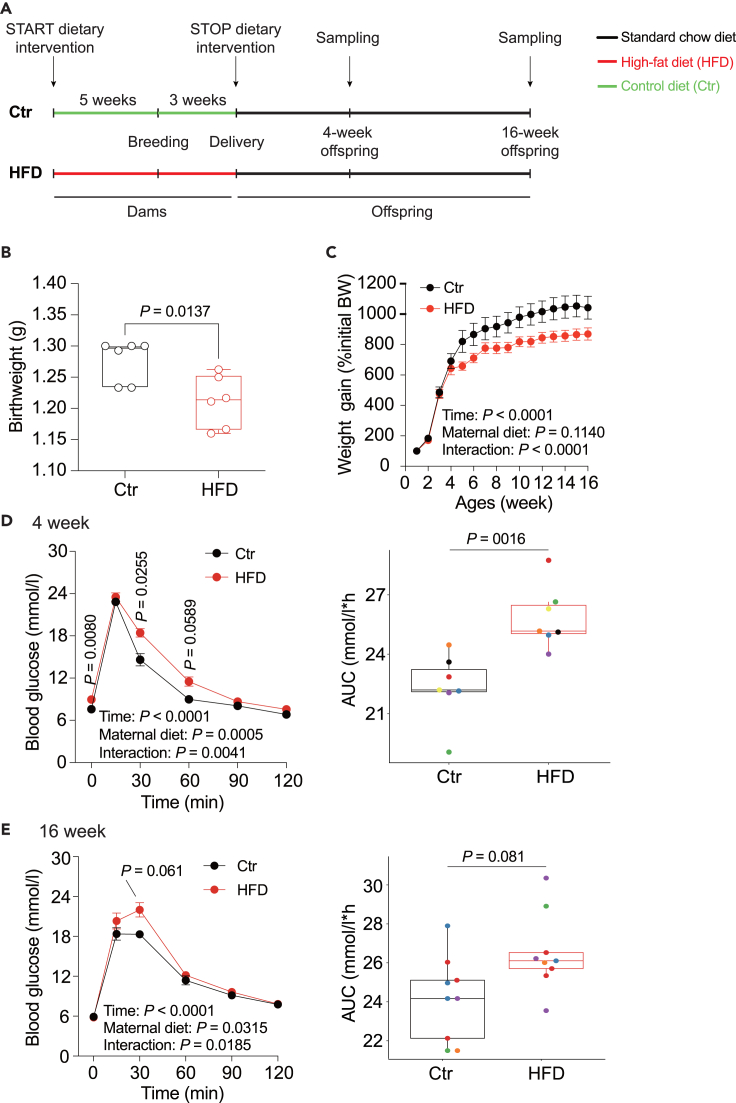
To determine the impact of maternal HFD on offspring metabolism, we established an intergenerational mouse model by feeding female mice a control (Ctr) diet or HFD starting 5 weeks before mating and throughout pregnancy. The induced metabolic phenotype caused by this treatment on the dams was described elsewhere.29 All dams and litters were transferred to a normal chow diet after delivery. To avoid nutritional bias between litters, the litter sizes were homogenized to 6 pups for each dam within the first 3 post-natal days. Male offspring were weaned at postnatal day 21 and fed a standard chow diet until 16 weeks of age (Figure 1A). The results showed that maternal HFD results in reduced body weight of the offspring at birth (Figure 1B). This phenotype is commonly seen in studies using similar mouse models but also exhibits high experimental variability.8,30 While body weight gain was not statistically different between animals from Ctr or HFD fed mothers, there was a significant interaction between time and maternal diet and animals from HFD mothers were lighter throughout the experiment (Figure 1C). While neither liver weight of the offspring at 4 and 16 weeks (Figure S1A) nor liver triglyceride concentration were statistically different, we observed an increase in liver cholesterol concentration in 16-week-old offspring (Figures S1B and S1C). We also did not detect significant alterations in serum total cholesterol (TC), triglycerides (TG), free fatty acids (FFAs), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), alanine transaminase (ALT), and aspartate aminotransferase (AST) in the offspring, but observed a significant increase in glucose levels at 4 weeks, suggesting impaired glucose tolerance (Figures S2).
Figure 1.
Maternal HFD impacts glucose and cholesterol metabolism in offspring
(A) Experimental setup assessing the effects of maternal HFD before and throughout pregnancy on metabolic and circadian changes in offspring.
(B) Offspring birthweight.
(C) Percentage change in offspring body weight throughout the study.
(D and E) Glucose tolerance test (left) and the corresponding area under the curve (right, each color represents different litter) for 4-week-old offspring (D) and 16-week-old offspring (E). N = 5–6 mice (from different litters) per experimental group. All boxplots are Tukey boxplots and data is assessed with a Student’s t-test; line chart data are presented as mean ± S.E.M. and are analyzed via a repeated measure two-way ANOVA or mixed linear model followed by Šídák post hoc tests. The details of the statistical analysis results are available in Table S6. Ctr: maternal control diet (black); HFD: maternal high-fat diet (red).
Contrary to previous studies suggesting that glucose tolerance is often unaltered when a Ctr diet is provided after birth,31,32,33 our data revealed a slightly delayed and impaired glucose tolerance in the offspring. This is evidenced by elevated blood glucose levels 30 min post glucose load and a larger AUC at 4 weeks and 16 weeks compared to the Ctr group (Figure 1D and 1E). These findings are similar to studies that maintained HFD during lactation.34,35 However, there were no significant changes in insulin tolerance (Figures S1D and S1E). Taken together, the data suggest that prenatal exposure to maternal HFD, followed by a control chow diet postnatally, slightly impairs glucose and cholesterol metabolism, but overall physiology appears largely unaffected, aligning with prior findings.31,32,33
Diurnal rhythms in food intake and energy expenditure are subtly altered by maternal high fat diet
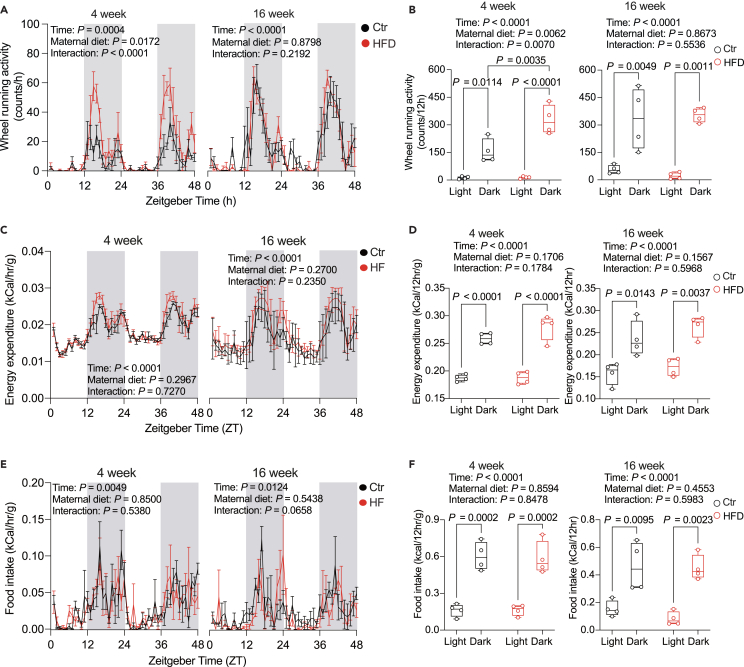
Mice fed an HFD exhibit attenuated rhythms of the circadian clock and feeding rhythms.18,36 A previous study linked gestational obesity with altered daily rhythms of activity and food intake in 15 weeks old offspring.20 We thus measured the diurnal rhythms of running wheel locomotor activity, energy expenditure, and feeding in the offspring of Ctr and HFD dams at 4 and 16 weeks of age. At 4 weeks, we observed an increase in total locomotor activity and a mild but not significant increase in energy expenditure. However, these differences were not present at 16 weeks (Figure 2A–2D), diverging from a previous report.20 In addition, there were no observed differences in feeding rhythms (Figures 2D and 2E), suggesting that metabolic alterations were not associated with changes in diurnal feeding behavior.
Figure 2.
Maternal HFD marginally affects diurnal food consumption and energy expenditure
(A, C, and E) Diurnal rhythms of running wheel activity (A), energy expenditure (C), and food intake (E) in 4-week-old (left) and 16-week-old offspring (right).
(B, D and F) Daily alterations in running wheel activity (B), energy expenditure (D), and food intake (F) in 4-week-old (left) and 16-week-old offspring (right). Zeitgeber time (ZT) indicates light entrainment periods (ZT0-12: lights on; ZT12-24: lights off). N = 4 mice (from different litters) per group. All boxplots are Tukey boxplots and line chart data are presented as mean ± S.E.M. Data are analyzed via a repeated measure two-way ANOVA followed by Šídák post hoc tests. The details of the results of the statistical analyses are available in Table S6. Ctr: maternal control diet (black); HFD: maternal high-fat diet (red).
Maternal high fat diet modifies the rhythmic gut microbiome of the offspring
Given that maternal HFD before and during pregnancy leads to changes in the rhythmic cecal microbiome composition of the dams29 and microbiome alterations are transmitted to the offspring,27,37 we aimed to investigate potential alterations in the rhythmic gut microbiome of the offspring through the analysis of 16S rRNA gene amplicon profiles. At the global level, α-diversity analysis revealed that the expected altered Simpson index diversity observed in dams under HFD was still present in 4-weeks old offspring, albeit less pronounced. However, this difference disappears in 16-weeks old offspring, suggesting that the Ctr diet after birth normalized the altered diversity caused by maternal HFD (Figure S3A). Accordingly, while the Shannon diversity index was also different in dams, this difference disappears in the offspring (Figure S3A). Overall, the 16S rRNA analyses revealed that microbiota communities were dominated by Firmicutes in all mice and time points (Figure S3B and Table S1) and that the difference in gut microbiota abundance observed in HFD fed dams is normalized under Ctr diet after birth.
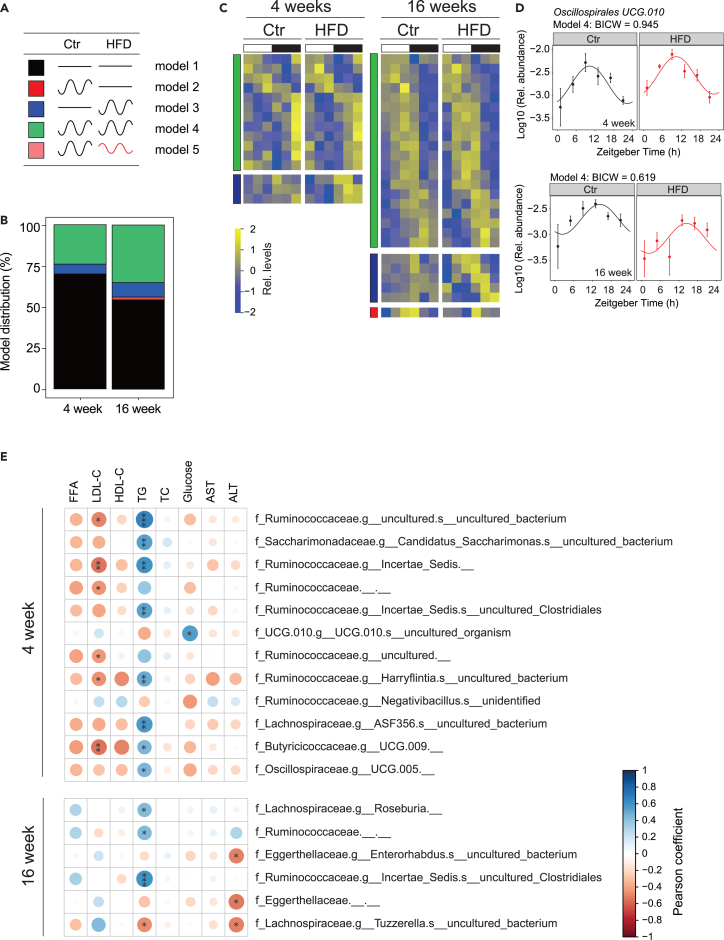
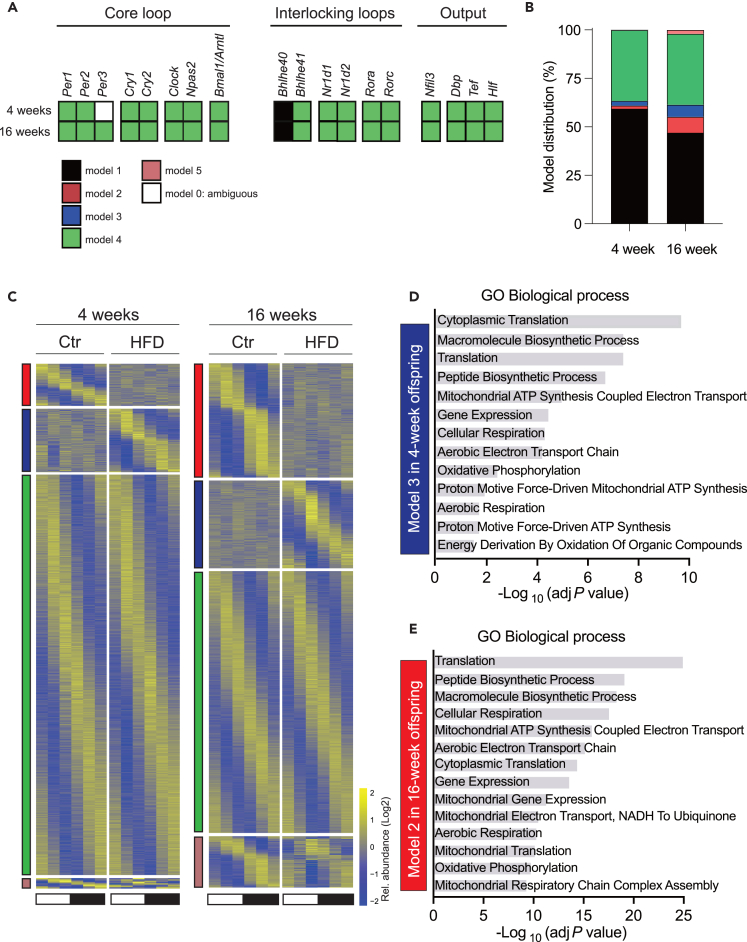
We employed dryR38 to analyze the rhythmic amplicon sequence variants (ASV) identified in the 16S profiles that were among the ones present in at least 50% of samples in Ctr and HFD offspring. This resulted in the characterization of five models defined by the differential rhythmicity between the groups (Figure 3A and Table S1). At both 4 and 16 weeks, most ASVs were found in model 1 (not rhythmic) and model 4 (similar rhythmicity in Ctr and HFD offspring) (Figures 3B and 3C). This observation aligns with previous studies indicating that feeding rhythm, which shows no difference between Ctr and HFD offspring (Figures 2E and 2F), is the primary driver of microbiome rhythmicity, with only a limited influence from other factors.39,40,41 Among the rhythmic microbiome, we found that Oscillospirales exhibited robust rhythmicity in 4- and 16-week offspring across all feeding conditions (Figure 3D). We identified only a few bacterial groups with different rhythmicity between Ctr and HFD offsprings that globally showed a gain in rhythmicity in the HFD offspring (Figure 3B, Table S1). To determine the association between the microbiome composition and metabolism, we performed a correlation analysis between the relative abundance of each bacterial group with the measured metabolic parameters (Figure 3E). Circulating triglyceride levels were the only parameter that showed a broad correlation with ASVs, likely reflecting that feeding rhythm is the main drivers of the microbiome and triglyceride rhythms.42
Figure 3.
Maternal exposure to HFD alters gut microbiome composition in offspring
(A) Model selection for rhythmic ASVs in male offspring from maternal HFD and maternal Ctr groups: black line, non-rhythmic ASVs; black sinus wave: rhythmic ASVs; red sinus wave, rhythmic ASVs with different phase and/or amplitude.
(B) Model distribution percentage of ASVs across models 1–5, with different colors indicating the respective model as illustrated in (A).
(C) Heatmap showing rhythmic ASVs in 4-week-old (left) and 16-week-old offspring (right). Standardized relative ASV abundance is indicated in blue (low) and yellow (high). The white and black bars denote light conditions. Different color indicates the corresponding model as shown in (A).
(D) Exemple of rhythmic ASV in 4-week-old (top) and 16-week-old offspring (bottom). Each dot represents the mean ASV abundance for each zeitgeber time (ZT) with the line illustrating the cosinor regression fit. The ZT defines the timing of entrainment by light (ZT0: lights on; ZT12: lights off).
(E) Correlation plots based on Pearson coefficient between serum metabolic profiles and ASV abundance. Only correlations with a Pearson coefficient that had an associated Benjamini-Hochberg adjusted p-value of less than 0.05 (determined through Fisher’s Z transform) were deemed statistically significant. The details of the results of the statistical analyses are available in Table S6. Colors represent positive (blue) and negative (red) correlation. Size of the circles indicates the corresponding p-value. N = 4 mice (from different litters) per group. Ctr: maternal control diet (black); HFD: maternal high-fat diet (red).
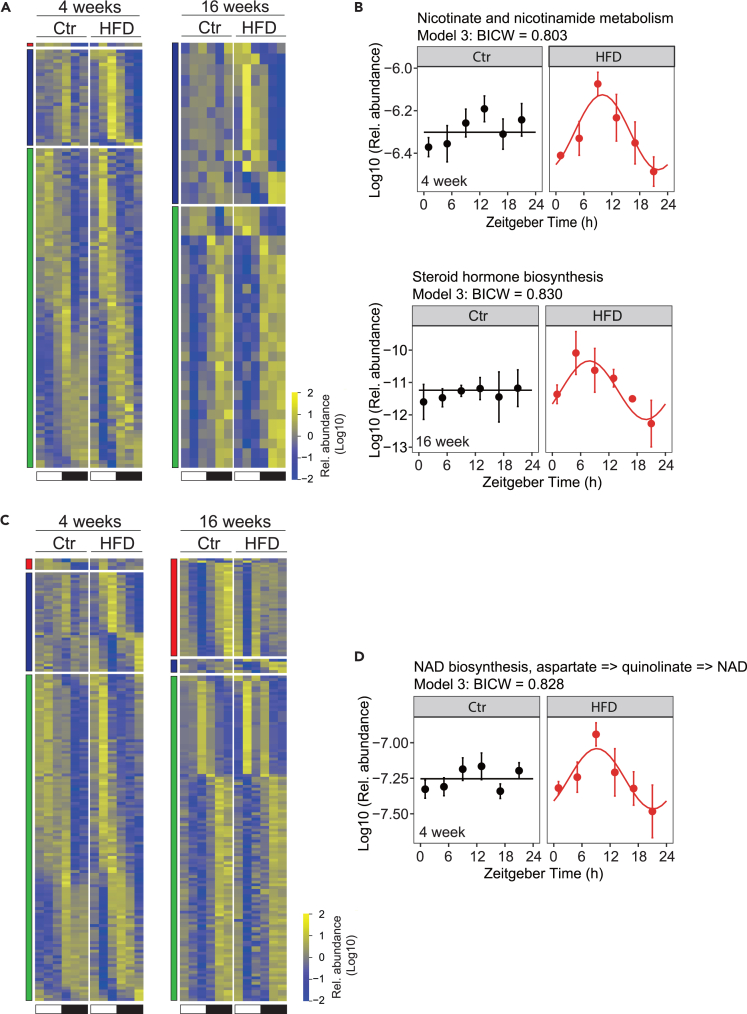
Interestingly, a similar rhythmic analysis of the metagenomic data and the predicted metabolic functions of the cecal microbiome showed a slightly different result. While most of the KEGG pathways were still in models 1 and 4, we detected a gain of rhythmicity (model 3) in HFD offspring at both 4 and 16 weeks for some pathways, in agreement with the analysis of 16S rRNA (Figures 4A, S4A, and Table S2). Among the KEGG pathways that showed conserved rhythmicity at both 4 and 16 weeks (model 4) were pathways involved in cell motility or environmental adaptation (Figure S4B), confirming that such general functions are mainly driven by feeding rhythms.40 Many of the pathways that gained rhythmicity under a maternal HFD were involved in metabolism, including metabolisms of nicotinamide at 4 weeks and steroid hormones at 16 weeks, potentially influencing in this way the rhythmic host metabolism (Figure 4B). Our observation was different for KEGG modules that showed a gain of rhythmicity at 4 weeks (model 3) but a loss of rhythmicity at 16 weeks (model 2) (Figures 4C, S4C, and Table S2). Among those modules with conserved rhythmicity at 4 and 16 weeks were those involved with hydrogen sulfide (H2S) and methane metabolism (Figure S4D). Both these modules can support important hydrogen “sinks” during fermentation: sulfide in particular can be produced and released by some gut bacteria during their metabolism of sulfur-containing dietary proteins and/or other endogenous sulfated compounds (e.g., sulfomucins and/or some secondary bile acids) and has a broad physiological role, including influencing local microbiome.43,44 Interestingly, we also noticed in the HFD offspring at 4 weeks a gain of rhythmicity for nicotinamide adenine dinucleotide (NAD+) synthesis which can play a central role in hydrogen (proton) transactions (Figure 4D).
Figure 4.
Maternal HFD alters the functional attributes of the offspring’s gut microbiome
(A and C) Heatmap for rhythmic KEGG pathway (A) and KEGG module (C) in 4-week-old offspring (left) and 16-week-old offspring (right). Standardized relative pathway/module abundance is indicated in blue (low) and yellow (high). White and black bars indicate light conditions. Different color indicates the corresponding model as shown in Figure 3A.
(B) Nicotinate and nicotinamide metabolism pathway in 4-week-old (top) and steroid hormone biosynthesis pathway in 16-week-old offspring (bottom).
(D) NAD+ biosynthesis module in 4-week-old offspring. The dots mark values of inferred functional activity for each zeitgeber time (ZT) with the line illustrating the cosinor regression fit. The ZT defines the timing of entrainment by light (ZT0: lights on; ZT12: lights off). N = 3 mice (from different litters) per group. Ctr: maternal control diet (black); HFD: maternal high-fat diet (red).
Impact of maternal high fat diet on liver gene expression
To gain more insight into the impact of maternal HFD on rhythmic liver gene expression, we quantified mRNA by RNA sequencing and analyzed differential rhythmic gene expression with dryR. A focused analysis of the rhythmic expression of the core circadian clock genes shows that the temporal expression profiles of liver circadian clock genes are globally unaffected by maternal obesity (Figures 5A, S5A, and Table S3), in contrast to previous reports.19,22 A more global analysis of rhythmicity shows that most of the rhythmic genes exhibited a similar rhythmicity in Ctr and HFD mice at both 4 and 16 weeks (Figures 5B and 5C). Nevertheless, more differentially expressed genes were found in 16-week-old mice compared to 4-week-old. Among all the genes that showed rhythmicity, 15.6 and 11.7% of genes showed a loss (model 2) or gain (model 3) of rhythmicity, respectively, at 16 weeks compared to 4.1 and 6.0% at 4 weeks. 0.2 and 3.7% of genes also showed differential rhythmicity (model 5, altered acrophase and/or amplitude) at 4 and 16 weeks, respectively. Interestingly, at 4 weeks, we found enrichment for biological processes only among the genes that gain rhythmicity (Figure 5D, Table S4). Most of these enriched genes are involved in ribosome biogenesis and mitochondrial function (Figure S5B), processes regulated by the circadian clock and feeding rhythms.45,46,47 Surprisingly, the exact same pathways were enriched among the genes that lost rhythmicity at 16 weeks (Figures 5E, S5C, and Table S4). Interestingly, both ribosome biogenesis48,49 and mitochondrial activity50,51,52 are regulated by NAD+-dependent SIRT7 and SIRT3. This observation may align with the rhythmic NAD+ metabolism observed through metagenomics at 4 weeks, highlighting the significant role of the microbiome in regulating host NAD+ levels.53,54
Figure 5.
Impact of maternal HFD on rhythmic liver gene expression
(A) Hepatic circadian clock genes show an unaltered temporal expression profile under maternal HFD, with most genes assigned to model 4.
(B) Model distribution percentage of genes in model 1–5. Different color indicates the corresponding model as shown in (A).
(C) Heatmap for rhythmic genes in 4-week-old offspring (left) and 16-week-old offspring (right). Standardized relative gene expression is indicated in blue (low) and yellow (high). White and black bars indicate light conditions. Different color indicates the corresponding model as shown in (A).
(D and E) Enrichment of GO biological process for genes in model 3 in 4-week-old offspring (D) and in model 2 in 16-week-old offspring (E). N = 2–3 mice (from different litters) per group. Ctr: maternal control diet (black); HFD: maternal high-fat diet (red).
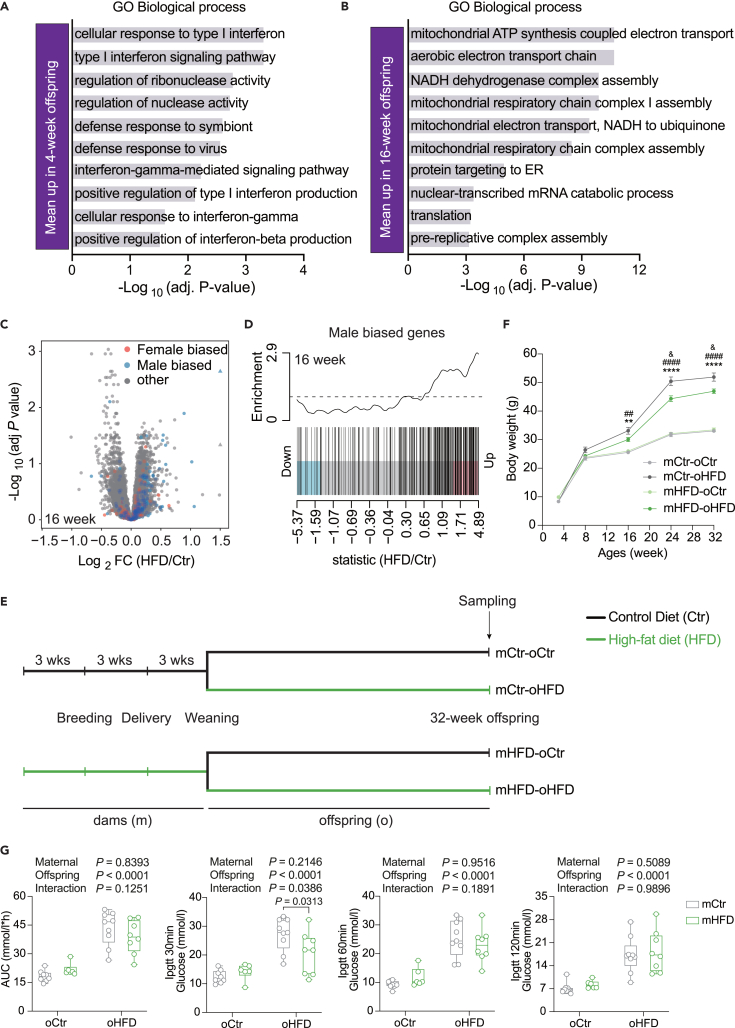
Focusing on differential gene expression at the mean level, upregulated genes at 4 weeks showed a strong enrichment for the type I interferon (IFN) pathway and genes upregulated at 16 weeks exhibited a strong enrichment in mitochondria-associated processes (Figures 6A, 6B, S6A–S6C, Tables S3, and S4). This correlates with a transcriptional signature associated with the IFN-activated IRF9 and STAT1/2 transcription factors at 4 weeks (Figure S6D, Table S5). Previous studies reported that the activation of IFN signaling decreases mitochondrial activity and gene expression and induces mitochondrial stress,55,56 a process that protects against HFD induced obesity57,58,59 and alters mitochondrial gene expression.60,61 This could therefore constitute an adaptation process to protect against HFD induced obesity.
Figure 6.
Differential gene expression in offspring exposed to maternal HFD
(A and B) GO enrichment analysis of hepatic genes showing a mean increase in expression in 4-week-old (A) and 16-week-old offspring upon maternal HFD (B).
(C) Volcano plots illustrating sex-biased differentially expressed genes in the livers of 16-week-old offspring upon maternal HFD.
(D) Barcode plots for male-biased genes in the liver of 16-week-old offspring with genes ordered from most down to most upregulated.
(E) Experimental design of Zheng et al.33
(F) Changes in offspring bodyweight from 4 to 32 weeks of age.
(G) Area under the curve for the glucose tolerance test and blood glucose levels at 30-, 60-, and 120-min post-glucose load (2 g/kg) in 32-week-old offspring. N = 2–3 mice (from different litters) per group. All boxplots are Tukey boxplots and line chart data are presented as mean ± S.E.M. Data are analyzed via two-way ANOVA followed by Holm-Šídák post hoc tests. The details of the results of the statistical analyses are available in Table S6. ∗∗p < 0.05 and ∗∗∗∗p < 0.0001, mHF-oHF vs. mHF-oCtr; ##p < 0.01 and ####p < 0.0001, mCtr-oHF vs. mCtr-oCtr; & p < 0.05, mCtr-oHF vs. mHF-oHF. mCtr, maternal control diet; mHFD, maternal high-fat diet; oCtr, offspring control diet; oHFD, offspring high-fat diet.
At 16 weeks, we noticed a transcriptional signature corresponding to the activation of the SUZ12 transcription factor. SUZ12 is one of the three components of the polycomb repressive complex-2 (PRC2)62 that plays a role in the liver sex-biased gene expression.63 Sex-biased liver gene expression develops after puberty and is regulated by the interaction between growth hormone (GH) and sex hormones.64,65,66 Therefore, we investigated the sex-biased gene expression and identified a striking increase in male-biased gene at 16 weeks (Figures 6C and 6D). Interestingly, a similar analysis of differential gene expression from a related study using female mice67 revealed a similar increase in GH-male induced genes associated with a similar increase of IFN-regulated genes (Figure S6F and Table S5). Therefore, it is plausible that maternal obesity impacts these pathways, influencing in this way the sexual development and fertility of the offspring.68,69 Interestingly, similar sexual development phenotypes are observed in the offspring of female with polycystic ovary syndrome (PCOS),70,71,72 a condition defined by a combination of androgen excess and ovarian dysfunction from complex etiology.73 We therefore analyzed the liver gene expression of a mouse model of PCOS74 and found a similar masculinization phenotype of liver gene expression, with an increase of GH male-induced genes and a decrease of GH male repressed genes (Figure S6G). This suggests that these two phenomena, despite having different etiology, may represent the manifestation of a similar physiological condition.
Sex difference influences mitochondrial activity, that is higher in female,75,76 as well as inflammation, with prepubertal male displaying an increase inflammatory response.77 It is therefore conceivable that this “masculinization” plays a role in the observed increase in inflammation and disrupted mitochondrial activity, contributing to the observed glucose intolerance. Nevertheless, because these processes have also been shown to protect against HFD-induced obesity,57,58,59 we hypothesized that it can somehow protect against metabolic syndrome. To test this assumption, we reanalyzed our previously published study with the goal of elucidating the effects of maternal obesity on the metabolism of the offspring33 (Figure 6E). While still obese and showing impaired metabolism when exposed to HFD throughout lactation and weaning, male offspring from obese dams unexpectedly exhibited a lower body weight gain (Figures 6F and S6H), improved glucose tolerance (Figure 6G), and insulin sensitivity33 compared to animals from Ctr dams. These results suggest that maternal HFD can confer short-term moderate transgenerational protection against metabolic syndrome through endocrine adaptation.
Discussion
The present study corroborates that maternal HFD before and during pregnancy alters physiology in male offspring, reinforcing the DOHaD concept. This includes a lower body weight at birth, associated with later metabolic defect.78 However, while this phenomenon is often observed in mice,8,30 it is less common in humans where maternal obesity is rather associated with increased body weight at birth.79 In contrast to previous publications,19,20,21,22 maternal obesity was not associated with a significant alteration of the diurnal circadian clock genes expression and behavior. Nevertheless, our findings are consistent with previous reports showing that changes in diurnal behavior and gene expression are absent when offspring from obese rat dams were nursed by non-obese foster dams during lactation and later switched to standard chow diet post-weaning.19 Alterations in the expression patterns of circadian clock genes observed in these studies were consistently associated with obesity in the offspring, while a functional circadian clock correlated with normal body weight.19,80 Therefore, the disruption of the circadian clock in these cases is likely a direct consequence of the observed obesity in the offspring rather than a result of maternal obesity. Indeed, obesity induced by an HFD is associated with disruptions of circadian rhythms and circadian clock genes expression.18,36 However, these alterations are absent in mice protected from HFD induced obesity.17,81 This underscores the notion that maternal HFD during lactation and after weaning is an independent risk factor for offspring health outcomes.82 Hence, extending maternal dietary intervention to the lactation period likely obscures the effects caused by in utero exposure to maternal HFD. In this study, we restricted maternal HFD to the prenatal period to avoid potential confounding effects of lactational exposure, which is generally associated with greater body weight gain in the offspring.82 Our findings suggest that while maternal obesity does not directly impact offspring diurnal feeding behavior, it can predispose offspring to alteration induced by HFD through changes in synaptic connections in the hypothalamus.83,84
Analysis of the 24h-rhythmicity of the microbiome confirms the predominant influence of feeding rhythms on microbiota composition and abundance.39,40,41 Although dysbiosis is associated with obesity, increasing evidence suggests that these changes are primarily driven by caloric load and food composition.85,86 In our study, all animals were fed an identical standard chow diet and exhibited similar feeding rhythms (Figure 2E and 2F). Under these conditions, we observed that the dysbiosis in 4-weeks old offspring from HFD dams gradually resolves, and by 16 weeks, the microbiome composition is similar across all animals. Similarly, another study found little difference in microbiota diversity in adult offspring87 and an analysis of various factors influencing microbiota composition highlighted a significant impact of sex which correlates with the obese phenotype observed only in males.88,89 Interestingly, a study using a cross-fostering paradigm demonstrated that the maternal feeding regimen is the main driver of the offspring’s microbiota composition and diversity,90 reinforcing the notion that microbiome diversity is largely dictated by feeding. Nevertheless, we found a gain in rhythmicity for a few bacterial groups and associated functions, such as NAD+ metabolism, in HFD offspring. This correlates with altered rhythmicity of NAD+-regulated ribosome biogenesis and mitochondrial function in the liver of the offspring. Further research will be required to establish a causal relationship between these two phenomena.
One important observation of the present study includes the activation of the type I interferon pathway in 4-week-old HF offspring and mitochondrial pathway activation at 16 weeks of age. The role of the IFN pathway in obesity is complex. While the inactivation of the type I IFN pathway exacerbates metabolic dysfunction associated with obesity,91,92 the activation of the type II IFN pathway protect against HFD induced obesity by altering mitochondrial activity.93,94 Interestingly, recent studies suggest that the simultaneous activation of both IFN pathway and mitochondrial stress can confer protection against HFD-induced obesity.57,58,59 This might contribute to the partial resistance observed in male offspring from HFD-fed mothers to HFD induced weight gain and glucose intolerance, potentially due to a predictive adaptive response (PAR).95 The PAR hypothesis describes a type of developmental plasticity where early life signals shape the development of a phenotype that helps the offspring to adapt better in their future environment. In light of the PAR hypothesis, male offspring in the present study would benefit from maternal HFD due to a match between the in utero environment (maternal HFD) and the likely future environment of postnatal HFD exposure. However, published data demonstrate that mitochondrial dysfunction is a risk factor for liver disease and metabolic dysfunction-associated steatohepatitis96 and increases the transgenerational susceptibility to metabolic dysfunction-associated steatohepatitis.97 It is therefore likely that the short-term benefits of maternal HFD have long-term detrimental effects. In addition, this transgenerational mitochondrial dysfunction might be a contributing factor to the recently described global decrease in basal energy expenditure over the past thirty years, as documented in the study of a human cohort from the United States and Europe.98
Another striking observation of the present study is the “masculinization” of liver gene expression of 16 weeks offspring that displays some similarities with a PCOS mouse model. Similar to what is detected in animal models of maternal obesity8,30 or in utero exposure to testosterone,99,100 offspring from PCOS women show a lower gestational body weight101 that correlates with maternal testosterone levels.102 Offspring from PCOS females also show similar metabolic dysfunction70,72 and perturbed sexual development and fertility,71,72 raising the question about what could explain these similarities between maternal obesity and PCOS. Among the different animal models of PCOS,103 prenatal exposure to testosterone in primates,104 sheep,105 mice,106 and rats107 induces phenotypes similar to PCOS, associated with an increased circulating testosterone level in the offspring. Interestingly, although testosterone levels rise during pregnancy in humans,108 this surge in testosterone is more pronounced during the last trimester of pregnancy in obese women109 and obese rats.110 Therefore, it appears that high testosterone during the prenatal period of the pregnancy of obese female could play a role in the DOHaD associated with maternal obesity during pregnancy. Additionally, testosterone impacts glucose metabolism107 and locomotor activity,111,112 which could explain the observed increases in glucose intolerance (Figure 1D), glucose levels (Figure S2A), and locomotor activity at 4 weeks (Figure 2A). In addition, testosterone exposure in utero could play a role in the reported alteration of DNA methylation observed in the offspring of obese mothers,113 testosterone inducing DNA demethylation after puberty.114 Altogether, these results demonstrate similarities between PCOS and the intergenerational effect of maternal obesity, suggesting that both conditions are part of the same spectrum of endocrine perturbations.
Limitations of the study
Given the differences between the analysis of 4 and 16-week offspring data, particularly in gene expression, including intermediate time points and additional data on protein-level changes could have provided deeper insights into the underlying phenomena and the dynamics of the transition. Furthermore, while an increasing body of literature points to an impact of paternal obesity on the metabolic trajectory of the offspring,115,116,117,118 this aspect was not explored in our study. Although other metabolic tissues, such as adipose tissue and muscle, are significant in metabolic physiology, our study focused solely on rhythmic liver physiology and microbiota. While 16S rRNA gene amplicon sequencing is a common method for characterizing microbiome composition, the resolution is typically at the genus level, and thereby precludes a precise assessment of the functional attributes of the communities, hence the value and utility of the metagenomic data generation and analyses included here.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| E.Z.N.A.® Total RNA Kit II | Omega Bio-Tek | R6934 |
| Qiagen Gel Extraction Kit | Qiagen | 28706 |
| Illumina DNA Prep Kit | Illumina | 20060059 |
| RNA Nano 6000 Assay Kit | Agilent | 5067–1511 |
| NEBNext® UltraTM RNA Library Prep Kit | Illumina | E7770 |
| Triglycerides content kit | Michybio | M1009A |
| Total cholesterol content kit | Michybio | M1010A |
| Deposited data | ||
| Raw data of 16S rRNA sequencing and metagenomics | This paper | SRA: PRJNA881293 |
| Raw and processed data of RNA-seq | This paper | GEO: GSE240147 |
| GH induced genes in male GF mice | Weger et al.66 | GEO: GSE77221 |
| Liver gene expression in female offspring from obese mother | Savva et al.67 | SRA: PRJNA723771 |
| Liver gene expression in female mice treated with dihydrotestosterone, a PCOS model | Roy et al.74 | GEO: GSE197765 |
| Transcription factors ChIP-Atlas data | Oki et al.119 | https://chip-atlas.org/ |
| ChIP-seq IRF9 in male mouse macrophages | Platanitis et al.120 | SRA: SRX4178758 |
| ChIP-seq STAT2 in male mouse macrophages | Platanitis et al.120 | SRA: SRX4178757 |
| ChIP-seq SUZ12 in male mouse ES cells | Hu et al.121 | SRA: SRX111886 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Huafukang Biological Technology | SCXK-2020-0026 |
| Oligonucleotides | ||
| Forward_16S_rRNA: 5′-CCTAYGGGRBGCASCAG-3′ | Xing et al.122 | N/A |
| Reverse_16S_rRNA: 5′-GGACTACNNGGGTATCTAAT-3′ | Xing et al.122 | N/A |
| Software and algorithms | ||
| R | R Foundation123 | https://www.r-project.org |
| DESeq2 | Love et al.124 | https://doi.org/10.18129/B9.bioc.DESeq2 |
| Enrichr | Kuleshov et al.125 | http://amp.pharm.mssm.edu/Enrichr/ |
| dryR | Weger et al.38 | https://github.com/naef-lab/dryR |
| QIIME 2 | Bolyen et al.126 | https://qiime2.org/ |
| Silva | Quast et al.127 | http://www.arb-silva.de/ |
| Microeco | Liu et al.128 | https://github.com/ChiLiubio/microeco |
| Readfq | Chen et al.129 | https://github.com/cjfields/readfq |
| MetaGeneMark | Zhu et al.130 | http://topaz.gatech.edu/GeneMark/ |
| MEGAHIT | Karlsson et al.131 | N/A |
| CD-HIT | Li et al.132 | http://www.bioinformatics.org/cd-hit/ |
| Bowtie2 | Langmead et al.133 | https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.4.4/ |
| DIAMOND | Buchfink et al.134 | https://github.com/bbuchfink/diamond/ |
| Hisat2 | Kim et al.135 | https://daehwankimlab.github.io/hisat2/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Frédéric Gachon (f.gachon@uq.edu.au).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The RNA-Seq data have been stored in the NCBI Gene Expression Omnibus136 and are publicly accessible through GEO Series accession number GSE240147 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE240147). The raw data for the 16S rRNA and metagenomics data have been deposited in the NCBI Sequence Read Archive137 and are publicly accessible under the accession number PRJNA881293 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881293).
This paper does not report any original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contacts upon request.
Experimental model and study participant details
Mice
All animal studies were approved by the Animal Care and Ethics Committee at Peking Union Medical College Hospital (Beijing, China, XHDW-2020-041) and compliant with the National Institutes of Health guide for the care and use of laboratory animals.
As previously described,29,138 4-week-old female C57BL/6 mice were purchased from Huafukang Biological Technology Co. Ltd (Beijing, China, SCXK-2020-0026). Mice were housed in a specific pathogen-free (SPF) environment. The temperature was maintained at 22 ± 2°C, and the lighting followed a 12:12 light/dark cycle (lights on at 06:00 h = zeitgeber time (ZT) 0). After a 7-day acclimation period, 5-week-old mice were randomized according to body weight into two groups: the control group (Ctr, n = 40) fed a standard chow diet (AIN-93G, Research Diets, 15.8 kcal% fat), and the HFD group (HFD, n = 40) fed a fat-rich diet (D12492, Research Diets, 60 kcal% fat). After 5 weeks on these diets, female mice were bred with 9-week-old C57BL/6 male mice, fed a control diet, in trio configuration (one male and two females) for 5 days, maintaining the allocated dietary regimen [i.e., HFD or Ctr diet]. Post-delivery, both dams and offspring were transitioned to the same control chow diet. Body weights were recorded weekly.
To investigate the effects of maternal HFD consumption before and during pregnancy on the diurnal physiology of juvenile and adult offspring, male offspring aged 4 and 16 weeks were sacrificed every 4 h (n = 4 per time point) over a 24-h period at six time points: 07:00 (ZT1), 11:00 (ZT5), 15:00 (ZT9), 19:00 (ZT13), 23:00 (ZT17), and 03:00 (ZT21). To minimize the influence of litter on experimental outcomes, we randomized across the six time points. Mice were sacrificed, and livers, blood serum, and cecal content were collected and snap-frozen in liquid nitrogen and stored at −80°C until further processing.
Method details
Indirect calorimetry and behavior
Male offspring at 4 weeks old and 16 weeks old were randomly picked from each group. Indirect calorimetry, feeding rhythm, and locomotor activity were measured using Promethion 8-channel respirometry cages (Sable Systems International). After acclimation for 24 h to the metabolic cage, they were monitored for 2 consecutive days (24 h per day). Measurements were taken every 5 min at a 12:12 light/dark cycle. Data were averaged and graphed at 1-h intervals. Indirect calorimetry and feeding rhythm data were corrected for body weight.
Glucose and insulin tolerance tests
For glucose tolerance test, mice were fasted for 6 h and injected intraperitoneally with a glucose load (2 g/kg of body weight) at ZT8. For insulin tolerance test, mice were fasted for 4 h and injected intraperitoneally with insulin (Humulin R; 1.0 U/kg of body weight) at ZT6. Blood glucose levels were measured from tail bleeding before intervention and 15, 30, 60, 90, and 120 min after intervention using a glucometer (Bayer).
Serum biochemical parameters measurement
Blood samples were collected from the intraorbital retrobulbar plexus of mice. All blood samples were separated by centrifugation at 3,000g for 10 min at 4°C, and the serum was stored in aliquots at −80°C. Serum total cholesterol (TC), total triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), free fatty acids (FFAs), alanine transaminase (ALT), and aspartate aminotransferase (AST) were measured using chemistry analyzer (Beckman Coulter, AU5800) as previously described.33
Liver total cholesterol and triglyceride
Levels of liver TC and TG were determined using the TC (M1010A) and TG (M1009A) assay kits from Michybio. Briefly, lipids were extracted from approximately 100 mg of frozen liver tissue using 1 mL of isopropanol. Subsequently, 3–4 mm steel beads were added, and the mixture was homogenized for two rounds of 30 s each, with a 10-s pause in between, using a homogenizer (KZ-III-F, Servicebio) at 4°C. After homogenization, the mixture was centrifuged at 8,000g for 10 min at 4°C. The supernatant was then collected and used for measurements, following the manufacturer’s instructions.
Cecal 16S ribosomal RNA and metagenomics sequencing and analysis
16S rRNA gene amplicon sequencing and metagenomic analysis were conducted as previously described122 with minor modifications. Genomic DNA was extracted from the cecal contents using the cetyltrimethyl ammonium bromide (CTAB) method.139 The V3-V4 regions of the 16S rRNA genes were amplified using barcoded region-specific primers (341F, 5′-CCTAYGGGRBGCASCAG-3’; 806R, 5′-GGACTACNNGGGTATCTAAT-3′). The PCR products were purified using a Qiagen Gel Extraction Kit. Sequencing libraries were prepared using the TruSeq DNA PCR-Free Sample Preparation Kit (Illumina) according to the manufacturer’s instructions. Sequencing was performed on an Illumina NovaSeq platform (250 bp paired-end reads). Raw sequence reads were processed and analyzed using quantitative insights into microbial ecology (QIIME) 2 version 2022.8126 according to the developer’s recommendations. Sequence quality control was carried out using DADA2 algorithm and taxonomies were assigned based on the SILVA_138 library.127 The comparison of alpha diversity indexes between subject categories were performed using R studio with microeco package128 according to the developer’s operating manual, with those reads annotated as “unassigned” excluded from these specific analyses.
We utilized 1 μg of DNA as starting material from each sample to prepare the metagenomic sequencing libraries. To ensure a sufficient amount of genomic DNA for metagenomic analysis, certain cecal samples had to be reextracted. Following the manufacturer’s instructions, we employed the Next Ultra DNA Library Prep Kit for Illumina (NEB) to generate sequencing libraries. This involved sonication to fragment the DNA to approximately 350 bp, followed by end-polishing, A-tailing, and adapter ligation for Illumina sequencing. After PCR amplification, the PCR products were purified using the AMPure XP beads (Beckman-Coulter). We then assessed the size distribution of the libraries using the Agilent 2100 Bioanalyzer and quantified them via real-time PCR. Quantified libraries were equimolarly pooled and clustered on a cBot Cluster Generation System, according to the manufacturer’s protocol. Subsequent sequencing was performed on the Illumina NovaSeq platform (250 bp paired end reads).
Raw sequencing data was cleaned using Readfq (V8) with subsequent host contamination checks using Bowtie2 (version 2.2.4).133 Assembly of metagenome was performed using MEGAHIT (v1.0.4-beta),131 followed by gene prediction and abundance analysis with MetaGeneMark (V3.05)130 and CD-HIT (V4.5.8).132 Unigenes were annotated for species using DIAMOND (v0.9.9,25402007)134 against NCBI’s Nucleotide (non-redundant) database and for functional characteristics using Kyoto Encyclopedia of Genes and Genomes (KEGG).140 Differential rhythmicity analysis was performed using the drylm function of dryR.38 For the metagenomics data, we introduced a batch effect to account for effects caused by the re-extraction of some samples.
RNA extraction, sequencing, and analysis
Total RNA was extracted from liver tissues using the E.Z.N.A. Total RNA Kit II (OMEGA, R6934). Briefly, 500 μL of RNA-Solv Reagent was added to approximately 50 mg of frozen liver tissue. We then added 3–4 mm zirconia beads. This mixture was subjected to homogenization in two 30s cycles, interspersed with a 10s interval, at 4°C using a KZ-III-F homogenizer (Servicebio). Following this, 200 μL of chloroform was added, and the solution was vigorously shaken for 20s. The mixture was then allowed to settle for 3 min at room temperature before being centrifuged at 12,000g for 15 min at 4°C. The resulting RNA supernatant was collected and subsequently purified using the columns provided with the kit, in accordance with the manufacturer’s protocol. RNA integrity of all samples was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies). Only RNA with a RIN above 8.0 were further processed. mRNA was purified from total RNA (3 μg of total RNA) using poly-T oligo-attached magnetic beads. Sequencing libraries were obtained using the NEBNext UltraTM RNA Library Prep Kit for Illumina following the manufacturer’s instructions.141 Libraries were then sequenced by the Illumina NovaSeq 6000. Raw data from published RNA-Seq studies were retrieved from NCBI GEO (accession number: GSE197765)74 and NCBI SRA (accession number: PRJNA723771).67
Reads were trimmed from adapters and mapped with Hisat2 (v2.0.5)135 to align the reads to the mouse reference genome (GRCm38/mm10). Uniquely mapped reads per genes were counted using FeatureCounts (v1.5.0-p3)142 based on the annotation from Ensembl release 94. We tested for functional enrichment using annotated gene sets from Gene Ontology (GO)143 using Enrichr.125 To statistically assess mean differences between two treatment groups we employed DESeq2,124 incorporating time as a covariate in the statistical model when available. We conducted differential rhythmicity analysis using dryR.38
Transcription factor activity analysis
Target genes of transcription factors were sourced from ChIP-Atlas119 for the mouse genome (mm10). Peak-calls overlapping a 5,000 bp window around a gene’s TSS were attributed to that gene. Hepatic sex-biased genes and genes that are up (GH_induced_male) or downregulated (GH_induced_male) upon injection of GH in germ-free male mice were taken from.66 To conduct an enrichment analysis of the genesets containing sex-specific biased genes or predicted target genes of transcription factors, we used the geneSetTest function from the limma package. This analysis was applied to gene lists that were pre-ranked defined based on results from DESeq2.124
Quantification and statistical analysis
For statistical analysis, two groups with a normal distribution were assessed using the Student’s t test, while the Mann–Whitney U test was used for non-normally distributed data. When examining a 2 × 2 factorial design, we utilized a two-way ANOVA. If values matched, a repeated measures two-way ANOVA or a mixed linear model was used, complemented by the indicated post hoc test. The details of the results of the statistical analyses are available in Table S6. Differential rhythmicity analyses were performed with dryR38 using the drylm function for normally distributed data and the dryseq function for count data. Only features (e.g., gene expression profiles, ASV abundance) with a signal in ≥50% of samples per condition were included in the analysis.
The correlation matrix between serum metabolic profile measures and ASV abundance was determined using the corrplot R package. Only correlations with a Pearson coefficient that had an associated Benjamini-Hochberg adjusted P-value of less than 0.05 (determined through Fisher’s Z transform) were deemed statistically significant. We used R v.1.4.1717 with ggplot2 v 3.4.4 and GraphPad Prism version 8.0 to perform the described statistical analysis and data visualization. If not otherwise stated, data is represented as mean and error bars indicate the standard error of the mean (S.E.M).
Acknowledgments
We would like to acknowledge Zhiyue Gu, Ling Zhong, Xiaorui Lyu, Baoluhe Zhang, and Lixin Gong for excellent technical assistance and scientific discussions.
This work was supported by the grants from National Natural Science Foundation of China (No. 81870579, 82170854, 82200903, 81900723), National High Level Hospital Clinical Research Funding (2022-PUMCH-C-019, 2022-PUMCH-C-023), Fundamental Research Funds for the Central Universities (3332021094), Beijing Municipal Science & Technology Commission (Z201100005520011), China Endocrinology & Metabolism Young Scientific Talent Research Project (2022-N-02-05), National Key Research and Development Program of China (2018YFC2001100), CAMS Innovation Fund for Medical Sciences (CIFMS2017-I2M-1–008), Australian National Health and Medical Research Council (Synergy grant #2019260 to FG, Investigator grant #2016334 to BDW, and Centres of Research Excellence grant #1170893 to MM) and Alzheimer’s Association Research Fellowship (#AARF-22-917584 to BDW), and The University of Queensland. Research by YL and MM was carried out at the Translational Research Institute, Woolloongabba, QLD 4102, Australia. The Translational Research Institute is supported by a grant from the Australian Government.
Author contributions
Conceptualization: LD, JL, LZ, XX, and FG; methodology: LD, BDW, DW, ZX, JL, JR, JZ, QZ, XX, and FG; software: BDW and YL.; validation: LD and BDW; formal analysis: LD, BDW, and YL; investigation: LD, BDW, DW, ZX, JL, JR, JZ, QZ, and MW; resources: DW, ZX, JL, JR, JZ, and QZ, and MY; data curation: LD and BDW; writing - original draft: LD, BDW, JL, XX, and FG; writing - review and editing: LD, BDW, MW, MM, XX, and FG; visualization: LD, BDW, and YL; supervision: BDW, JL, LZ, MM, XX, and FG; project administration: JL, LZ, XX, and FG; funding acquisition: BDW, JL, LZ, MY, MM, XX, and FG.
Declaration of interests
Mark Morisson has received consultancy fees from Bayer Steigerwald Arzneimittelwerk (Bayer Consumer Health), Sanofi Australia, and Danone-Nutricia Australia, and serves on the science advisory board (non-remunerated) for GenieBiome, Hong Kong SAR. All other authors report no potential conflicts of interest.
Published: June 21, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110343.
Contributor Information
Xinhua Xiao, Email: xiaoxh@pumch.cn.
Frédéric Gachon, Email: f.gachon@uq.edu.au.
Supplemental information
References
- 1.Driscoll A.K., Gregory E.C.W. NCHS Data Brief; 2020. Increases in Prepregnancy Obesity: United States, 2016-2019; pp. 1–8. [PubMed] [Google Scholar]
- 2.Hales C.M., Carroll M.D., Fryar C.D., Ogden C.L. NCHS Data Brief; 2020. Prevalence of Obesity and Severe Obesity Among Adults: United States, 2017-2018; pp. 1–8. [PubMed] [Google Scholar]
- 3.Fernandez-Twinn D.S., Hjort L., Novakovic B., Ozanne S.E., Saffery R. Intrauterine programming of obesity and type 2 diabetes. Diabetologia. 2019;62:1789–1801. doi: 10.1007/s00125-019-4951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godfrey K.M., Reynolds R.M., Prescott S.L., Nyirenda M., Jaddoe V.W.V., Eriksson J.G., Broekman B.F.P. Influence of maternal obesity on the long-term health of offspring. Lancet Diabetes Endocrinol. 2017;5:53–64. doi: 10.1016/s2213-8587(16)30107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poston L., Harthoorn L.F., van der Beek E.M., Contributors to the ILSI Europe Workshop Obesity in Pregnancy: Implications for the Mother and Lifelong Health of the Child. A Consensus Statement. Pediatr. Res. 2011;69:175–180. doi: 10.1203/PDR.0b013e3182055ede. [DOI] [PubMed] [Google Scholar]
- 6.Perng W., Oken E., Dabelea D. Developmental overnutrition and obesity and type 2 diabetes in offspring. Diabetologia. 2019;62:1779–1788. doi: 10.1007/s00125-019-4914-1. [DOI] [PubMed] [Google Scholar]
- 7.Warrington N.M., Beaumont R.N., Horikoshi M., Day F.R., Helgeland Ø., Laurin C., Bacelis J., Peng S., Hao K., Feenstra B., et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menting M.D., Mintjens S., van de Beek C., Frick C.J., Ozanne S.E., Limpens J., Roseboom T.J., Hooijmans C.R., van Deutekom A.W., Painter R.C. Maternal obesity in pregnancy impacts offspring cardiometabolic health: Systematic review and meta-analysis of animal studies. Obes. Rev. 2019;20:675–685. doi: 10.1111/obr.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman J.E. Developmental Programming of Obesity and Diabetes in Mouse, Monkey, and Man in 2018: Where Are We Headed? Diabetes. 2018;67:2137–2151. doi: 10.2337/dbi17-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barker D.J.P. The origins of the developmental origins theory. J. Intern. Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman D.J., Powell T.L., Barrett E.S., Hardy D.B. Developmental origins of metabolic diseases. Physiol. Rev. 2021;101:739–795. doi: 10.1152/physrev.00002.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaput J.-P., McHill A.W., Cox R.C., Broussard J.L., Dutil C., da Costa B.G.G., Sampasa-Kanyinga H., Wright K.P. The role of insufficient sleep and circadian misalignment in obesity. Nat. Rev. Endocrinol. 2023;19:82–97. doi: 10.1038/s41574-022-00747-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lane J.M., Qian J., Mignot E., Redline S., Scheer F.A.J.L., Saxena R. Genetics of circadian rhythms and sleep in human health and disease. Nat. Rev. Genet. 2023;24:4–20. doi: 10.1038/s41576-022-00519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koronowski K.B., Sassone-Corsi P. Communicating clocks shape circadian homeostasis. Science. 2021;371 doi: 10.1126/science.abd0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khosravipour M., Khanlari P., Khazaie S., Khosravipour H., Khazaie H. A systematic review and meta-analysis of the association between shift work and metabolic syndrome: The roles of sleep, gender, and type of shift work. Sleep Med. Rev. 2021;57 doi: 10.1016/j.smrv.2021.101427. [DOI] [PubMed] [Google Scholar]
- 16.Vetter C., Dashti H.S., Lane J.M., Anderson S.G., Schernhammer E.S., Rutter M.K., Saxena R., Scheer F.A.J.L. Night Shift Work, Genetic Risk, and Type 2 Diabetes in the UK Biobank. Diabetes Care. 2018;41:762–769. doi: 10.2337/dc17-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckley T.N., Omotola O., Archer L.A., Rostron C.R., Kamineni E.P., Llanora J.D., Chalfant J.M., Lei F., Slade E., Pendergast J.S. High-fat feeding disrupts daily eating behavior rhythms in obesity-prone but not in obesity-resistant male inbred mouse strains. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2021;320:R619–R629. doi: 10.1152/ajpregu.00150.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohsaka A., Laposky A.D., Ramsey K.M., Estrada C., Joshu C., Kobayashi Y., Turek F.W., Bass J. High-Fat Diet Disrupts Behavioral and Molecular Circadian Rhythms in Mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Borengasser S.J., Kang P., Faske J., Gomez-Acevedo H., Blackburn M.L., Badger T.M., Shankar K. High Fat Diet and In Utero Exposure to Maternal Obesity Disrupts Circadian Rhythm and Leads to Metabolic Programming of Liver in Rat Offspring. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cleal J.K., Bruce K.D., Shearer J.L., Thomas H., Plume J., Gregory L., Shepard J.N., Spiers-Fitzgerald K.L., Mani R., Lewis R.M., et al. Maternal Obesity during Pregnancy Alters Daily Activity and Feeding Cycles, and Hypothalamic Clock Gene Expression in Adult Male Mouse Offspring. Int. J. Mol. Sci. 2019;20:5408. doi: 10.3390/ijms20215408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suter M., Bocock P., Showalter L., Hu M., Shope C., McKnight R., Grove K., Lane R., Aagaard-Tillery K. Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. FASEB J. 2011;25:714–726. doi: 10.1096/fj.10-172080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D., Chen S., Liu M., Liu C. Maternal obesity disrupts circadian rhythms of clock and metabolic genes in the offspring heart and liver. Chronobiol. Int. 2015;32:615–626. doi: 10.3109/07420528.2015.1025958. [DOI] [PubMed] [Google Scholar]
- 23.Choi H., Rao M.C., Chang E.B. Gut microbiota as a transducer of dietary cues to regulate host circadian rhythms and metabolism. Nat. Rev. Gastroenterol. Hepatol. 2021;18:679–689. doi: 10.1038/s41575-021-00452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weger B.D., Rawashdeh O., Gachon F. At the Intersection of Microbiota and Circadian Clock: Are Sexual Dimorphism and Growth Hormones the Missing Link to Pathology? Bioessays. 2019;41 doi: 10.1002/bies.201900059. [DOI] [PubMed] [Google Scholar]
- 25.Van Hul M., Cani P.D. The gut microbiota in obesity and weight management: microbes as friends or foe? Nat. Rev. Endocrinol. 2023;19:258–271. doi: 10.1038/s41574-022-00794-0. [DOI] [PubMed] [Google Scholar]
- 26.Soderborg T.K., Clark S.E., Mulligan C.E., Janssen R.C., Babcock L., Ir D., Young B., Krebs N., Lemas D.J., Johnson L.K., et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat. Commun. 2018;9:4462. doi: 10.1038/s41467-018-06929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., Sonnenburg J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura I., Miyamoto J., Ohue-Kitano R., Watanabe K., Yamada T., Onuki M., Aoki R., Isobe Y., Kashihara D., Inoue D., et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. 2020;367 doi: 10.1126/science.aaw8429. [DOI] [PubMed] [Google Scholar]
- 29.Ding L., Liu J., Zhou L., Jia X., Li S., Zhang Q., Yu M., Xiao X. A high-fat diet disrupts the hepatic and adipose circadian rhythms and modulates the diurnal rhythm of gut microbiota-derived short-chain fatty acids in gestational mice. Front. Nutr. 2022;9 doi: 10.3389/fnut.2022.925390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christians J.K., Lennie K.I., Wild L.K., Garcha R. Effects of high-fat diets on fetal growth in rodents: a systematic review. Reprod. Biol. Endocrinol. 2019;17:39. doi: 10.1186/s12958-019-0482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhaphong B., Gregg B., Kumusoglu D., Jo S., Singer K., Scheys J., DelProposto J., Lumeng C., Bernal-Mizrachi E., Alejandro E.U. Maternal High-Fat Diet During Pre-Conception and Gestation Predisposes Adult Female Offspring to Metabolic Dysfunction in Mice. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.780300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang E., Hafner H., Varghese M., Griffin C., Clemente J., Islam M., Carlson Z., Zhu A., Hak L., Abrishami S., et al. Programming effects of maternal and gestational obesity on offspring metabolism and metabolic inflammation. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-52583-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J., Xiao X., Zhang Q., Yu M., Xu J., Qi C., Wang T. The effects of maternal and post-weaning diet interaction on glucose metabolism and gut microbiota in male mice offspring. Biosci. Rep. 2016;36 doi: 10.1042/BSR20160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuelsson A.-M., Matthews P.A., Argenton M., Christie M.R., McConnell J.M., Jansen E.H.J.M., Piersma A.H., Ozanne S.E., Twinn D.F., Remacle C., et al. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: a novel murine model of developmental programming. Hypertension. 2008;51:383–392. doi: 10.1161/HYPERTENSIONAHA.107.101477. [DOI] [PubMed] [Google Scholar]
- 35.Zheng J., Zhang L., Wang Z., Zhang J. Maternal high-fat diet regulates glucose metabolism and pancreatic β cell phenotype in mouse offspring at weaning. PeerJ. 2020;8 doi: 10.7717/peerj.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pendergast J.S., Branecky K.L., Yang W., Ellacott K.L.J., Niswender K.D., Yamazaki S. High-fat diet acutely affects circadian organisation and eating behavior. Eur. J. Neurosci. 2013;37:1350–1356. doi: 10.1111/ejn.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myles I.A., Fontecilla N.M., Janelsins B.M., Vithayathil P.J., Segre J.A., Datta S.K. Parental Dietary Fat Intake Alters Offspring Microbiome and Immunity. J. Immunol. 2013;191:3200–3209. doi: 10.4049/jimmunol.1301057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weger B.D., Gobet C., David F.P.A., Atger F., Martin E., Phillips N.E., Charpagne A., Weger M., Naef F., Gachon F. Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2015803118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks J.F., Behrendt C.L., Ruhn K.A., Lee S., Raj P., Takahashi J.S., Hooper L.V. The microbiota coordinates diurnal rhythms in innate immunity with the circadian clock. Cell. 2021;184:4154–4167.e12. doi: 10.1016/j.cell.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thaiss C.A., Zeevi D., Levy M., Zilberman-Schapira G., Suez J., Tengeler A.C., Abramson L., Katz M.N., Korem T., Zmora N., et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 41.Zarrinpar A., Chaix A., Yooseph S., Panda S. Diet and Feeding Pattern Affect the Diurnal Dynamics of the Gut Microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adamovich Y., Rousso-Noori L., Zwighaft Z., Neufeld-Cohen A., Golik M., Kraut-Cohen J., Wang M., Han X., Asher G. Circadian Clocks and Feeding Time Regulate the Oscillations and Levels of Hepatic Triglycerides. Cell Metab. 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magee E.A., Richardson C.J., Hughes R., Cummings J.H. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 2000;72:1488–1494. doi: 10.1093/ajcn/72.6.1488. [DOI] [PubMed] [Google Scholar]
- 44.Wallace J.L., Motta J.-P., Buret A.G. Hydrogen sulfide: an agent of stability at the microbiome-mucosa interface. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;314:G143–G149. doi: 10.1152/ajpgi.00249.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F., Gachon F. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. USA. 2015;112:E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bignon Y., Wigger L., Ansermet C., Weger B.D., Lagarrigue S., Centeno G., Durussel F., Götz L., Ibberson M., Pradervand S., et al. Multiomics reveals multilevel control of renal and systemic metabolism by the renal tubular circadian clock. J. Clin. Invest. 2023;133 doi: 10.1172/JCI167133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jouffe C., Cretenet G., Symul L., Martin E., Atger F., Naef F., Gachon F. The Circadian Clock Coordinates Ribosome Biogenesis. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S., Blank M.F., Iyer A., Huang B., Wang L., Grummt I., Voit R. SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat. Commun. 2016;7 doi: 10.1038/ncomms10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai Y.-C., Greco T.M., Cristea I.M. Sirtuin 7 Plays a Role in Ribosome Biogenesis and Protein Synthesis. Mol. Cell. Proteomics. 2014;13:73–83. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dittenhafer-Reed K.E., Richards A.L., Fan J., Smallegan M.J., Fotuhi Siahpirani A., Kemmerer Z.A., Prolla T.A., Roy S., Coon J.J., Denu J.M. SIRT3 Mediates Multi-Tissue Coupling for Metabolic Fuel Switching. Cell Metab. 2015;21:637–646. doi: 10.1016/j.cmet.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B., Grueter C.A., Harris C., Biddinger S., Ilkayeva O.R., et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. http://www.nature.com/nature/journal/v464/n7285/suppinfo/nature08778_S1.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirschey M.D., Shimazu T., Jing E., Grueter C.A., Collins A.M., Aouizerat B., Stančáková A., Goetzman E., Lam M.M., Schwer B., et al. SIRT3 Deficiency and Mitochondrial Protein Hyperacetylation Accelerate the Development of the Metabolic Syndrome. Mol. Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chellappa K., McReynolds M.R., Lu W., Zeng X., Makarov M., Hayat F., Mukherjee S., Bhat Y.R., Lingala S.R., Shima R.T., et al. NAD precursors cycle between host tissues and the gut microbiome. Cell Metab. 2022;34:1947–1959.e5. doi: 10.1016/j.cmet.2022.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shats I., Williams J.G., Liu J., Makarov M.V., Wu X., Lih F.B., Deterding L.J., Lim C., Xu X., Randall T.A., et al. Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway. Cell Metab. 2020;31:564–579.e7. doi: 10.1016/j.cmet.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewis J.A., Huq A., Najarro P. Inhibition of Mitochondrial Function by Interferon. J. Biol. Chem. 1996;271:13184–13190. doi: 10.1074/jbc.271.22.13184. [DOI] [PubMed] [Google Scholar]
- 56.Olson G.S., Murray T.A., Jahn A.N., Mai D., Diercks A.H., Gold E.S., Aderem A. Type I interferon decreases macrophage energy metabolism during mycobacterial infection. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCabe K.M., Hsieh J., Thomas D.G., Molusky M.M., Tascau L., Feranil J.B., Qiang L., Ferrante A.W., Jr., Tall A.R. Antisense oligonucleotide treatment produces a type I interferon response that protects against diet-induced obesity. Mol. Metab. 2020;34:146–156. doi: 10.1016/j.molmet.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodríguez-Nuevo A., Díaz-Ramos A., Noguera E., Díaz-Sáez F., Duran X., Muñoz J.P., Romero M., Plana N., Sebastián D., Tezze C., et al. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. EMBO J. 2018;37 doi: 10.15252/embj.201796553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pereira R.O., Tadinada S.M., Zasadny F.M., Oliveira K.J., Pires K.M.P., Olvera A., Jeffers J., Souvenir R., McGlauflin R., Seei A., et al. OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. EMBO J. 2017;36:2126–2145. doi: 10.15252/embj.201696179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forsström S., Jackson C.B., Carroll C.J., Kuronen M., Pirinen E., Pradhan S., Marmyleva A., Auranen M., Kleine I.-M., Khan N.A., et al. Fibroblast Growth Factor 21 Drives Dynamics of Local and Systemic Stress Responses in Mitochondrial Myopathy with mtDNA Deletions. Cell Metab. 2019;30:1040–1054.e7. doi: 10.1016/j.cmet.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 61.Weger M., Alpern D., Cherix A., Ghosal S., Grosse J., Russeil J., Gruetter R., de Kloet E.R., Deplancke B., Sandi C. Mitochondrial gene signature in the prefrontal cortex for differential susceptibility to chronic stress. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-75326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laugesen A., Højfeldt J.W., Helin K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell. 2019;74:8–18. doi: 10.1016/j.molcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lau-Corona D., Bae W.K., Hennighausen L., Waxman D.J. Sex-biased genetic programs in liver metabolism and liver fibrosis are controlled by EZH1 and EZH2. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lichanska A.M., Waters M.J. How growth hormone controls growth, obesity and sexual dimorphism. Trends Genet. 2008;24:41–47. doi: 10.1016/j.tig.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 65.Waxman D.J., O’Connor C. Growth Hormone Regulation of Sex-Dependent Liver Gene Expression. Mol. Endocrinol. 2006;20:2613–2629. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- 66.Weger B.D., Gobet C., Yeung J., Martin E., Jimenez S., Betrisey B., Foata F., Berger B., Balvay A., Foussier A., et al. The Mouse Microbiome Is Required for Sex-Specific Diurnal Rhythms of Gene Expression and Metabolism. Cell Metab. 2019;29:362–382.e8. doi: 10.1016/j.cmet.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Savva C., Helguero L.A., González-Granillo M., Melo T., Couto D., Angelin B., Domingues M.R., Li X., Kutter C., Korach-André M. Molecular programming modulates hepatic lipid metabolism and adult metabolic risk in the offspring of obese mothers in a sex-specific manner. Commun. Biol. 2022;5:1057. doi: 10.1038/s42003-022-04022-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobs S., Teixeira D.S., Guilherme C., da Rocha C.F.K., Aranda B.C.C., Reis A.R., de Souza M.A., Franci C.R., Sanvitto G.L. The impact of maternal consumption of cafeteria diet on reproductive function in the offspring. Physiol. Behav. 2014;129:280–286. doi: 10.1016/j.physbeh.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 69.Rodríguez-González G.L., Vega C.C., Boeck L., Vázquez M., Bautista C.J., Reyes-Castro L.A., Saldaña O., Lovera D., Nathanielsz P.W., Zambrano E. Maternal obesity and overnutrition increase oxidative stress in male rat offspring reproductive system and decrease fertility. Int. J. Obes. 2015;39:549–556. doi: 10.1038/ijo.2014.209. [DOI] [PubMed] [Google Scholar]
- 70.Huang Y., Gao J.-M., Zhang C.-M., Zhao H.-C., Zhao Y., Li R., Yu Y., Qiao J. Assessment of growth and metabolism characteristics in offspring of dehydroepiandrosterone-induced polycystic ovary syndrome adults. Reproduction. 2016;152:705–714. doi: 10.1530/REP-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kauffman A.S., Thackray V.G., Ryan G.E., Tolson K.P., Glidewell-Kenney C.A., Semaan S.J., Poling M.C., Iwata N., Breen K.M., Duleba A.J., et al. A Novel Letrozole Model Recapitulates Both the Reproductive and Metabolic Phenotypes of Polycystic Ovary Syndrome in Female Mice. Biol. Reprod. 2015;93:69. doi: 10.1095/biolreprod.115.131631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Risal S., Li C., Luo Q., Fornes R., Lu H., Eriksson G., Manti M., Ohlsson C., Lindgren E., Crisosto N., et al. Transgenerational transmission of reproductive and metabolic dysfunction in the male progeny of polycystic ovary syndrome. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2023.101035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Escobar-Morreale H.F. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat. Rev. Endocrinol. 2018;14:270–284. doi: 10.1038/nrendo.2018.24. [DOI] [PubMed] [Google Scholar]
- 74.Roy S., Abudu A., Salinas I., Sinha N., Cline-Fedewa H., Yaw A.M., Qi W., Lydic T.A., Takahashi D.L., Hennebold J.D., et al. Androgen-mediated Perturbation of the Hepatic Circadian System Through Epigenetic Modulation Promotes NAFLD in PCOS Mice. Endocrinology. 2022;163 doi: 10.1210/endocr/bqac127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Norheim F., Hasin-Brumshtein Y., Vergnes L., Chella Krishnan K., Pan C., Seldin M.M., Hui S.T., Mehrabian M., Zhou Z., Gupta S., et al. Gene-by-Sex Interactions in Mitochondrial Functions and Cardio-Metabolic Traits. Cell Metab. 2019;29:932–949.e4. doi: 10.1016/j.cmet.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Justo R., Boada J., Frontera M., Oliver J., Bermúdez J., Gianotti M. Gender dimorphism in rat liver mitochondrial oxidative metabolism and biogenesis. Am. J. Physiol. Cell Physiol. 2005;289:C372–C378. doi: 10.1152/ajpcell.00035.2005. [DOI] [PubMed] [Google Scholar]
- 77.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 78.Mericq V., Martinez-Aguayo A., Uauy R., Iñiguez G., Van der Steen M., Hokken-Koelega A. Long-term metabolic risk among children born premature or small for gestational age. Nat. Rev. Endocrinol. 2017;13:50–62. doi: 10.1038/nrendo.2016.127. [DOI] [PubMed] [Google Scholar]
- 79.Heslehurst N., Vieira R., Akhter Z., Bailey H., Slack E., Ngongalah L., Pemu A., Rankin J. The association between maternal body mass index and child obesity: A systematic review and meta-analysis. PLoS Med. 2019;16 doi: 10.1371/journal.pmed.1002817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borengasser S.J., Faske J., Kang P., Blackburn M.L., Badger T.M., Shankar K. In utero exposure to prepregnancy maternal obesity and postweaning high-fat diet impair regulators of mitochondrial dynamics in rat placenta and offspring. Physiol. Genomics. 2014;46:841–850. doi: 10.1152/physiolgenomics.00059.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Omotola O., Legan S., Slade E., Adekunle A., Pendergast J.S. Estradiol regulates daily rhythms underlying diet-induced obesity in female mice. Am. J. Physiol. Endocrinol. Metab. 2019;317:E1172–E1181. doi: 10.1152/ajpendo.00365.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ribaroff G.A., Wastnedge E., Drake A.J., Sharpe R.M., Chambers T.J.G. Animal models of maternal high fat diet exposure and effects on metabolism in offspring: a meta-regression analysis. Obes. Rev. 2017;18:673–686. doi: 10.1111/obr.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shrivastava K., Swaminathan T., Barlotta A., Athreya V., Choudhry H., Rossi M.A. Maternal overnutrition is associated with altered synaptic input to lateral hypothalamic area. Mol. Metab. 2023;71 doi: 10.1016/j.molmet.2023.101702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Y., Yang D., Wang L., Król E., Mazidi M., Li L., Huang Y., Niu C., Liu X., Lam S.M., et al. Maternal High Fat Diet in Lactation Impacts Hypothalamic Neurogenesis and Neurotrophic Development, Leading to Later Life Susceptibility to Obesity in Male but Not Female Mice. Adv. Sci. 2023;10 doi: 10.1002/advs.202305472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jumpertz R., Le D.S., Turnbaugh P.J., Trinidad C., Bogardus C., Gordon J.I., Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singh R.K., Chang H.-W., Yan D., Lee K.M., Ucmak D., Wong K., Abrouk M., Farahnik B., Nakamura M., Zhu T.H., et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Newman T.M., Clear K.Y.J., Wilson A.S., Soto-Pantoja D.R., Ochs-Balcom H.M., Cook K.L. Early-life dietary exposures mediate persistent shifts in the gut microbiome and visceral fat metabolism. Am. J. Physiol. Cell Physiol. 2023;324:C644–C657. doi: 10.1152/ajpcell.00380.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wankhade U.D., Zhong Y., Kang P., Alfaro M., Chintapalli S.V., Piccolo B.D., Mercer K.E., Andres A., Thakali K.M., Shankar K. Maternal High-Fat Diet Programs Offspring Liver Steatosis in a Sexually Dimorphic Manner in Association with Changes in Gut Microbial Ecology in Mice. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wankhade U.D., Zhong Y., Kang P., Alfaro M., Chintapalli S.V., Thakali K.M., Shankar K. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Daoust L., Choi B.S.Y., Lacroix S., Rodrigues Vilela V., Varin T.V., Dudonné S., Pilon G., Roy D., Levy E., Desjardins Y., et al. The postnatal window is critical for the development of sex-specific metabolic and gut microbiota outcomes in offspring. Gut Microb. 2021;13 doi: 10.1080/19490976.2021.2004070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wieser V., Adolph T.E., Grander C., Grabherr F., Enrich B., Moser P., Moschen A.R., Kaser S., Tilg H. Adipose type I interferon signalling protects against metabolic dysfunction. Gut. 2018;67:157–165. doi: 10.1136/gutjnl-2016-313155. [DOI] [PubMed] [Google Scholar]
- 92.Chan C.C., Damen M.S.M.A., Moreno-Fernandez M.E., Stankiewicz T.E., Cappelletti M., Alarcon P.C., Oates J.R., Doll J.R., Mukherjee R., Chen X., et al. Type I interferon sensing unlocks dormant adipocyte inflammatory potential. Nat. Commun. 2020;11:2745. doi: 10.1038/s41467-020-16571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alsaggar M., Mills M., Liu D. Interferon beta overexpression attenuates adipose tissue inflammation and high-fat diet-induced obesity and maintains glucose homeostasis. Gene Ther. 2017;24:60–66. doi: 10.1038/gt.2016.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bradley D., Smith A.J., Blaszczak A., Shantaram D., Bergin S.M., Jalilvand A., Wright V., Wyne K.L., Dewal R.S., Baer L.A., et al. Interferon gamma mediates the reduction of adipose tissue regulatory T cells in human obesity. Nat. Commun. 2022;13:5606. doi: 10.1038/s41467-022-33067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bateson P., Gluckman P., Hanson M. The biology of developmental plasticity and the Predictive Adaptive Response hypothesis. J. Physiol. 2014;592:2357–2368. doi: 10.1113/jphysiol.2014.271460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benegiamo G., von Alvensleben G.V.G., Rodríguez-López S., Goeminne L.J.E., Bachmann A.M., Morel J.-D., Broeckx E., Ma J.Y., Carreira V., Youssef S.A., et al. The genetic background shapes the susceptibility to mitochondrial dysfunction and NASH progression. J. Exp. Med. 2023;220 doi: 10.1084/jem.20221738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bruce K.D., Cagampang F.R., Argenton M., Zhang J., Ethirajan P.L., Burdge G.C., Bateman A.C., Clough G.F., Poston L., Hanson M.A., et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology. 2009;50:1796–1808. doi: 10.1002/hep.23205. [DOI] [PubMed] [Google Scholar]
- 98.Speakman J.R., de Jong J.M.A., Sinha S., Westerterp K.R., Yamada Y., Sagayama H., Ainslie P.N., Anderson L.J., Arab L., Bedu-Addo K., et al. Total daily energy expenditure has declined over the past three decades due to declining basal expenditure, not reduced activity expenditure. Nat. Metab. 2023;5:579–588. doi: 10.1038/s42255-023-00782-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sathishkumar K., Elkins R., Chinnathambi V., Gao H., Hankins G.D.V., Yallampalli C. Prenatal testosterone-induced fetal growth restriction is associated with down-regulation of rat placental amino acid transport. Reprod. Biol. Endocrinol. 2011;9:110. doi: 10.1186/1477-7827-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wolf C.J., Hotchkiss A., Ostby J.S., LeBlanc G.A., Gray L.E., Jr. Effects of Prenatal Testosterone Propionate on the Sexual Development of Male and Female Rats: A Dose-Response Study. Toxicol. Sci. 2002;65:71–86. doi: 10.1093/toxsci/65.1.71. [DOI] [PubMed] [Google Scholar]
- 101.Sir-Petermann T., Hitchsfeld C., Maliqueo M., Codner E., Echiburú B., Gazitúa R., Recabarren S., Cassorla F. Birth weight in offspring of mothers with polycystic ovarian syndrome. Hum. Reprod. 2005;20:2122–2126. doi: 10.1093/humrep/dei009. [DOI] [PubMed] [Google Scholar]
- 102.Carlsen S.M., Jacobsen G., Romundstad P. Maternal testosterone levels during pregnancy are associated with offspring size at birth. Eur. J. Endocrinol. 2006;155:365–370. doi: 10.1530/eje.1.02200. [DOI] [PubMed] [Google Scholar]
- 103.van Houten E.L.A.F., Visser J.A. Mouse models to study polycystic ovary syndrome: A possible link between metabolism and ovarian function? Reprod. Biol. 2014;14:32–43. doi: 10.1016/j.repbio.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 104.Eisner J.R., Dumesic D.A., Kemnitz J.W., Abbott D.H. Timing of Prenatal Androgen Excess Determines Differential Impairment in Insulin Secretion and Action in Adult Female Rhesus Monkeys. J. Clin. Endocrinol. Metab. 2000;85:1206–1210. doi: 10.1210/jcem.85.3.6453. [DOI] [PubMed] [Google Scholar]
- 105.Sullivan S.D., Moenter S.M. Prenatal androgens alter GABAergic drive to gonadotropin-releasing hormone neurons: Implications for a common fertility disorder. Proc. Natl. Acad. Sci. USA. 2004;101:7129–7134. doi: 10.1073/pnas.0308058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roland A.V., Nunemaker C.S., Keller S.R., Moenter S.M. Prenatal androgen exposure programs metabolic dysfunction in female mice. J. Endocrinol. 2010;207:213–223. doi: 10.1677/JOE-10-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.More A.S., Mishra J.S., Gopalakrishnan K., Blesson C.S., Hankins G.D., Sathishkumar K. Prenatal Testosterone Exposure Leads to Gonadal Hormone-Dependent Hyperinsulinemia and Gonadal Hormone-Independent Glucose Intolerance in Adult Male Rat Offspring1. Biol. Reprod. 2016;94:1–11. doi: 10.1095/biolreprod.115.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.O'Leary P., Boyne P., Flett P., Beilby J., James I. Longitudinal assessment of changes in reproductive hormones during normal pregnancy. Clin. Chem. 1991;37:667–672. doi: 10.1093/clinchem/37.5.667. [DOI] [PubMed] [Google Scholar]
- 109.Maliqueo M., Cruz G., Espina C., Contreras I., García M., Echiburú B., Crisosto N. Obesity during pregnancy affects sex steroid concentrations depending on fetal gender. Int. J. Obes. 2017;41:1636–1645. doi: 10.1038/ijo.2017.159. [DOI] [PubMed] [Google Scholar]
- 110.Crew R.C., Mark P.J., Clarke M.W., Waddell B.J. Obesity Disrupts the Rhythmic Profiles of Maternal and Fetal Progesterone in Rat Pregnancy. Biol. Reprod. 2016;95:55. doi: 10.1095/biolreprod.116.139451. [DOI] [PubMed] [Google Scholar]
- 111.Butler M.P., Karatsoreos I.N., LeSauter J., Silver R. Dose-Dependent Effects of Androgens on the Circadian Timing System and Its Response to Light. Endocrinology. 2012;153:2344–2352. doi: 10.1210/en.2011-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karatsoreos I.N., Wang A., Sasanian J., Silver R. A Role for Androgens in Regulating Circadian Behavior and the Suprachiasmatic Nucleus. Endocrinology. 2007;148:5487–5495. doi: 10.1210/en.2007-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seki Y., Suzuki M., Guo X., Glenn A.S., Vuguin P.M., Fiallo A., Du Q., Ko Y.-A., Yu Y., Susztak K., et al. In Utero Exposure to a High-Fat Diet Programs Hepatic Hypermethylation and Gene Dysregulation and Development of Metabolic Syndrome in Male Mice. Endocrinology. 2017;158:2860–2872. doi: 10.1210/en.2017-00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reizel Y., Spiro A., Sabag O., Skversky Y., Hecht M., Keshet I., Berman B.P., Cedar H. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes Dev. 2015;29:923–933. doi: 10.1101/gad.259309.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cropley J.E., Eaton S.A., Aiken A., Young P.E., Giannoulatou E., Ho J.W.K., Buckland M.E., Keam S.P., Hutvagner G., Humphreys D.T., et al. Male-lineage transmission of an acquired metabolic phenotype induced by grand-paternal obesity. Mol. Metab. 2016;5:699–708. doi: 10.1016/j.molmet.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fullston T., Ohlsson Teague E.M.C., Palmer N.O., DeBlasio M.J., Mitchell M., Corbett M., Print C.G., Owens J.A., Lane M. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013;27:4226–4243. doi: 10.1096/fj.12-224048. [DOI] [PubMed] [Google Scholar]
- 117.Ng S.-F., Lin R.C.Y., Laybutt D.R., Barres R., Owens J.A., Morris M.J. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 118.Raad G., Serra F., Martin L., Derieppe M.-A., Gilleron J., Costa V.L., Pisani D.F., Amri E.-Z., Trabucchi M., Grandjean V. Paternal multigenerational exposure to an obesogenic diet drives epigenetic predisposition to metabolic diseases in mice. Elife. 2021;10 doi: 10.7554/eLife.61736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Oki S., Ohta T., Shioi G., Hatanaka H., Ogasawara O., Okuda Y., Kawaji H., Nakaki R., Sese J., Meno C. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018;19 doi: 10.15252/embr.201846255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Platanitis E., Demiroz D., Schneller A., Fischer K., Capelle C., Hartl M., Gossenreiter T., Müller M., Novatchkova M., Decker T. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat. Commun. 2019;10:2921. doi: 10.1038/s41467-019-10970-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu G., Cui K., Northrup D., Liu C., Wang C., Tang Q., Ge K., Levens D., Crane-Robinson C., Zhao K. H2A.Z Facilitates Access of Active and Repressive Complexes to Chromatin in Embryonic Stem Cell Self-Renewal and Differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xing J., Ying Y., Mao C., Liu Y., Wang T., Zhao Q., Zhang X., Yan F., Zhang H. Hypoxia induces senescence of bone marrow mesenchymal stem cells via altered gut microbiota. Nat. Commun. 2018;9:2020. doi: 10.1038/s41467-018-04453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Team R.C. R Foundation for Statistical Computing; 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 124.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu C., Cui Y., Li X., Yao M. microeco: an R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021;97 doi: 10.1093/femsec/fiaa255. [DOI] [PubMed] [Google Scholar]
- 129.Chen K., Pachter L. Bioinformatics for Whole-Genome Shotgun Sequencing of Microbial Communities. PLoS Comput. Biol. 2005;1:106–112. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhu W., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Karlsson F.H., Tremaroli V., Nookaew I., Bergström G., Behre C.J., Fagerberg B., Nielsen J., Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 132.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 133.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 135.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Katz K., Shutov O., Lapoint R., Kimelman M., Brister J.R., O’Sullivan C. The Sequence Read Archive: a decade more of explosive growth. Nucleic Acids Res. 2022;50:D387–D390. doi: 10.1093/nar/gkab1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ding L., Liu J., Zhou L., Zhang Q., Yu M., Xiao X. Maternal High-Fat Diet Results in Long-Term Sex-Specific Alterations to Metabolic and Gut Microbial Diurnal Oscillations in Adult Offspring. Mol. Nutr. Food Res. 2023;67 doi: 10.1002/mnfr.202200753. [DOI] [PubMed] [Google Scholar]
- 139.Waterhouse R.N., Glover L.A. Differences in the hybridization pattern of Bacillus subtilis genes coding for rDNA depend on the method of DNA preparation. Appl. Environ. Microbiol. 1993;59:919–921. doi: 10.1128/aem.59.3.919-921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 143.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data have been stored in the NCBI Gene Expression Omnibus136 and are publicly accessible through GEO Series accession number GSE240147 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE240147). The raw data for the 16S rRNA and metagenomics data have been deposited in the NCBI Sequence Read Archive137 and are publicly accessible under the accession number PRJNA881293 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881293).
This paper does not report any original code.
Any additional information required to reanalyse the data reported in this paper is available from the lead contacts upon request.