ABSTRACT
Treatment options for Epstein-Barr virus (EBV)-cancers are limited, underscoring the need for new therapeutic approaches. We have previously shown that EBV-transformed cells and cancers lack homologous recombination (HR) repair, a prominent error-free pathway that repairs double-stranded DNA breaks; instead, EBV-transformed cells demonstrate genome-wide scars of the error-prone microhomology-mediated end joining (MMEJ) repair pathway. This suggests that EBV-cancers are vulnerable to synthetic lethal therapeutic approaches that target MMEJ repair. Indeed, we have previously found that targeting PARP, an enzyme that contributes to MMEJ, results in the death of EBV-lymphoma cells. With the emergence of clinical resistance to PARP inhibitors and the recent discovery of inhibitors of Polymerase theta (POLθ), the polymerase essential for MMEJ, we investigated the role of POLθ in EBV-lymphoma cells. We report that EBV-transformed cell lines, EBV-lymphoma cell lines, and EBV-lymphomas in AIDS patients demonstrate greater abundance of POLθ, driven by the EBV protein EBNA1, compared to EBV-uninfected primary lymphocytes and EBV-negative lymphomas from AIDS patients (a group that also abundantly expresses POLθ). We also find POLθ enriched at cellular DNA replication forks and exposure to the POLθ inhibitor Novobiocin impedes replication fork progress, impairs MMEJ-mediated repair of DNA double-stranded breaks, and kills EBV-lymphoma cells. Notably, cell killing is not due to Novobiocin-induced activation of the lytic/replicative phase of EBV. These findings support a role for POLθ not just in DNA repair but also DNA replication and as a therapeutic target in EBV-lymphomas and potentially other EBV-cancers as EBNA1 is expressed in all EBV-cancers.
IMPORTANCE
Epstein-Barr virus (EBV) contributes to ~2% of the global cancer burden. With a recent estimate of >200,000 deaths a year, identifying molecular vulnerabilities will be key to the management of these frequently aggressive and treatment-resistant cancers. Building on our earlier work demonstrating reliance of EBV-cancers on microhomology-mediated end-joining repair, we now report that EBV lymphomas and transformed B cell lines abundantly express the MMEJ enzyme POLθ that likely protects cellular replication forks and repairs replication-related cellular DNA breaks. Importantly also, we show that a newly identified POLθ inhibitor kills EBV-cancer cells, revealing a novel strategy to block DNA replication and repair of these aggressive cancers.
KEYWORDS: Epstein-Barr virus, lymphoma, polymerase theta, microhomology-mediated end joining, synthetic lethal, PARP inhibitor
INTRODUCTION
Human herpesvirus 4 or Epstein-Barr virus (EBV) is an orally acquired pathogen that can cause infectious mononucleosis. The first tumor virus to be identified, EBV is also causal to several cancers including endemic Burkitt lymphoma (BL), lymphomas in HIV/AIDS patients, lymphomas in other immunosuppressed hosts, and nasopharyngeal cell carcinoma (1–3). While EBV infects B and epithelial cells, it persists in a latent state in B lymphocytes. Upon infection of B cells, EBV oncoproteins drive host DNA replication and cell proliferation, essential for the establishment of latency. In immunocompetent hosts, viral gene expression is then systematically turned down to establish silent persistence in newly-infected cells. In the absence of T cells, e.g., in culture, infected B cells continue to express all EBV latency genes and grow into proliferating lymphoblastoid cell lines (LCL). LCL are an important model for DLBCL in HIV/AIDS patients and lymphoproliferative diseases (LPD)/lymphomas in transplant recipients (1, 4)—both groups lack functional EBV-specific T cells. Similarly, cell lines derived from explants of EBV+ Burkitt lymphomas provide excellent model systems to understand EBV-B lymphocyte relationships and EBV-associated cancer-related processes.
Treatment of EBV lymphomas, especially those that occur during immunosuppression, is fraught with challenges. While reduction of immunosuppression remains the mainstay, it can lead to graft versus host disease or rejection/loss of the transplanted organ. With many of these tumors expressing CD20, ablation of CD20+ B cells is a frequently used option. However, administration of the anti-CD20 monoclonal antibody Rituximab often causes long-term B cell immunodeficiency. Furthermore, although adoptive T cell therapy targeting EBV antigens is an attractive therapeutic option, these are not readily available while chemotherapy, radiation, and surgery are effective in selected cases (4–8). There is, therefore, a need for identifying additional strategies to treat EBV-cancers especially since vaccines and EBV-specific antiviral agents do not exist.
DNA repair continues to be a major area of interest for treating cancer. This is because cancer cells rely on DNA repair mechanisms to mitigate damage to their genomes from intrinsic (e.g., DNA replication stress and reactive oxygen species) and extrinsic (e.g., radiation) sources. EBV-cancers are no different. That said, loss of DNA repair pathways is a common feature of cancers and likely provides a growth advantage (9). However, this loss also makes cancer cells dependent on the remaining DNA repair pathways, rendering them vulnerable to inhibition of these remaining repair pathways. This concept of synthetic lethality has been used successfully to treat breast and ovarian cancers with inactivating mutations in genes such as BRCA1 and BRCA2 that result in loss of homologous recombination (HR) repair, the most error-free form of repair during the S and G2 phases of the cell cycle. Indeed, synthetic lethally-acting PARP inhibitors, directed against the enzyme PARP, target the repair of single-stranded DNA breaks, double-stranded DNA breaks, and the microhomology-mediated end-joining (MMEJ) pathway, an S-, G2-, and M-phase associated error-prone mechanism of repair (10–14). This reliance on DNA repair mechanisms is a vulnerability that has been minimally explored in EBV-cancers. Like other cancers, EBV-transformed and EBV-cancer cells sustain damage to the cellular genome by a variety of mechanisms (15–22)—and our investigations have revealed that as expected, EBV oncogene-induced replication stress activates the cellular kinase ATR (22). However, we have also found that EBV, by activating the cellular protooncogene STAT3, ensures that ATR is unable to phosphorylate Chk1, an important downstream target. As a result, the intra-S phase cell cycle checkpoint is relaxed, enabling the proliferation of EBV+ cells despite DNA damage (22–24). An important consequence of STAT3 activation and defective Chk1 phosphorylation is loss of HR repair; this is because p(phospho)Chk1 facilitates recruitment of the RAD51 recombinase to DNA double-strand breaks (25, 26), a key step in HR repair. Our studies have further revealed that upon losing HR, EBV-cancer cells and LCLs become dependent on PARP-mediated activities, carry genome-wide scars of MMEJ repair, and succumb to PARP inhibition (27).
Given that EBV-cancer cells rely on MMEJ repair and are susceptible to PARP inhibitors, we investigated the role of Polymerase theta (POLθ) in EBV-lymphoma cells. POLθ is the polymerase responsible for MMEJ repair; this error-prone enzyme is often upregulated in cancer, particularly in HR-deficient tumors (28). We report that POLθ is upregulated by EBNA1 in EBV-transformed and -cancer cell lines as well as in EBV+ AIDS lymphomas (more so than in EBV− AIDS lymphomas)—indicating addiction to POLθ in these HR-deficient cancers. Our experiments also reveal that POLθ supports cellular DNA replication and contributes to MMEJ-mediated repair of DNA lesions and that blocking POLθ with a newly discovered inhibitor prevents proliferation and outgrowth of EBV-transformed and EBV+ lymphoma cells.
RESULTS AND DISCUSSION
Polymerase theta is abundantly expressed in EBV-transformed cells, EBV+ BL cells, and EBV+ DLBCL tumors
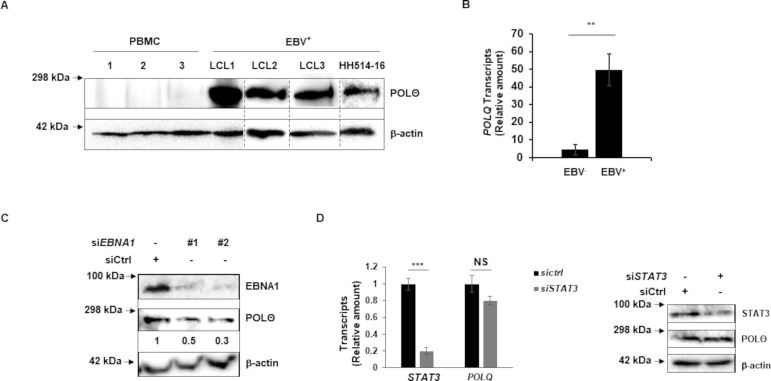
We have shown previously that EBV, by activating STAT3, is able to circumvent replication stress-induced apoptosis but in doing so, renders HR inactive as STAT3-mediated activities block the key step of Chk1 phosphorylation (22–24, 27). The loss of HR, which repairs DNA double-strand breaks in the S and G2 phases, leads to a dependence on other DNA repair pathways including those that use PARP (for single-strand and double-strand break repair) and the S/G2/M phase-associated error-prone MMEJ pathway that repairs DNA double-strand breaks. A key enzyme in the MMEJ pathway is POLθ whose abundance has been observed to be significantly higher in cancers that are dependent on MMEJ (13, 28, 29). Given our prior observation of genome-wide scars of MMEJ in EBV-transformed cells, we investigated if LCL and BL lines also demonstrated POLθ overexpression. We tested PBMC from three healthy subjects (as control), three LCL, and one EBV+ BL line using immunoblotting (Fig. 1A). We observed strong POLθ expression in these EBV+ cell lines compared to essentially no expression in healthy PBMC. The overexpression of POLθ in these lines suggests a dependence on MMEJ repair and points to POLθ as a potential synthetic lethal target. We also analyzed tumor biopsies from three EBV+ and three EBV− AIDS-DLBCL for the abundance of POLQ transcripts; transcripts of POLθ are referred to as POLQ. We found that EBV+ tumors exhibited higher levels of POLQ transcripts compared to EBV− tumors (Fig. 1B). Together, these data suggest that compared to EBV− tumors that abundantly express POLQ transcripts, EBV-transformed/cancer cell lines and EBV+ cancers demonstrate an even greater abundance of POLQ gene products. Additionally, these data suggest that examining expression of POLθ may be useful in recognizing EBV+ cancers that are dependent on MMEJ.
Fig 1.
Polymerase theta is abundantly expressed in EBV-transformed/cancer cell lines and EBV-cancers. (A) Healthy subject-derived PBMC, LCL, and HH514-16 BL cells were immunoblotted with indicated antibodies. (B) Tumor biopsies from three EBV− and three EBV+ DLBCL in AIDS patients were analyzed for relative POLQ transcript levels using reverse transcriptase-qPCR (RT-qPCR). (C) HH514-16 BL cells were transfected with scrambled/control siRNA versus two siRNAs targeting BKRF1 (EBNA1). Cells were collected and immunoblotted with indicated antibodies; numbers indicate the abundance of POLθ relative to β-actin. (D) LCL were transfected with scrambled siRNA or siRNA targeting STAT3 and cells were collected 20 h later for RT-qPCR to assay STAT3 and POLQ transcripts and for immunoblotting with indicated antibodies. Error bars, SEM of technical replicates; **P < 0.01; ***P < 0.001. Experiments were performed twice.
Greater abundance of POLθ in EBV+ cells and EBV+ lymphomas suggested viral protein-mediated regulation of POLθ expression. Since EBNA1 is expressed in all types of EBV cancers and latency, we depleted EBNA1 and examined POLθ. As shown in Fig. 1C, siRNA-mediated knockdown of EBNA1 reduced the abundance of POLθ. Because STAT3-mediated loss of HR results in dependence on MMEJ and since STAT3 is a prominent transcription factor that is upregulated following EBV infection (23), we also depleted STAT3 but were unable to detect a change in abundance of POLQ transcripts or POLθ in LCL (Fig. 1D). Thus, while EBV− tumors express POLθ, EBNA1 upregulates POLθ further in EBV+ tumors. EBNA3 proteins and the leader protein might also contribute to POLθ expression but EBNA2 and LMP1 likely do not since HH514-16 BL cells that abundantly express POLθ lack the BYRF1 gene (that encodes EBNA2) (30, 31) and do not express LMP1 during latency.
Polymerase theta enriches at replication forks and contributes to DNA replication and EBV+ cancer cell proliferation
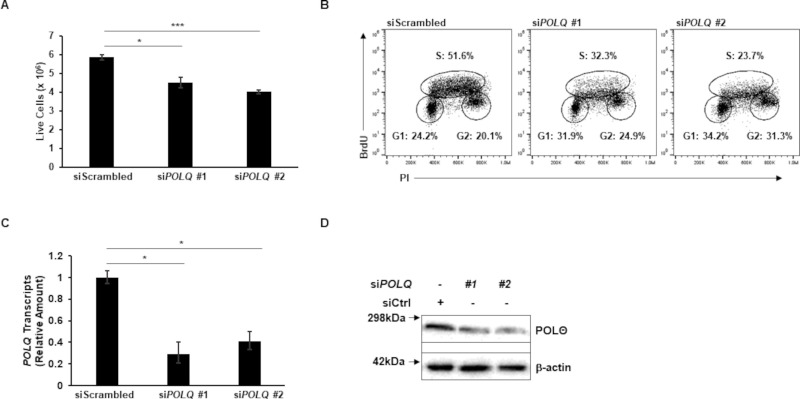
We next investigated the effect of POLθ knockdown in EBV+ cells. HH514-16 BL cells were nucleofected with two siRNAs targeting POLQ or a scrambled siRNA as control. To determine the effect on cell growth in culture, we counted live cells 48 h after nucleofection and found significantly fewer live cells in the POLQ knockdown conditions compared to scrambled control (Fig. 2A). To assess cell proliferation, 24 h after transfection, cells were treated with BrdU for 1 h, and BrdU uptake and DNA content, measured by propidium iodide staining of fixed cells, were plotted (Fig. 2B). Cells were gated into G1, S, and G2 phases based on DNA content and BrdU uptake. The cells transfected with siRNAs targeting POLQ displayed substantially fewer cells in the S phase, indicating that POLθ may contribute to DNA replication. POLQ knockdown was confirmed in Fig. 2C.
Fig 2.
Polymerase theta contributes to cellular DNA replication. HH514-16 BL cells were transfected with two siRNAs targeting POLQ versus scrambled siRNA. (A) At 48 h post-transfection, cells were counted. (B) At 24 h post-transfection, cells were exposed to BrdU for 1 h, and BrdU uptake was measured using flow cytometry. These cells were also stained with propidium iodide to measure DNA content. (C and D) Cells were harvested 24 h after transfection for RT-qPCR to analyze relative POLQ transcript levels (C) and immunoblotting with indicated antibodies (D) Error bars, SEM of technical replicates; *P < 0.05; ***P < 0.001. Experiments were performed twice.
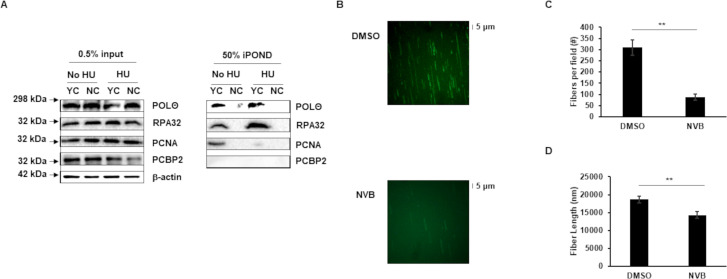
Given our observation of POLθ’s contribution to the S phase of the cell cycle, we next asked if POLθ was enriched at DNA replication forks. To address this question, we enriched cellular DNA replication forks using isolation of proteins on nascent DNA (iPOND) followed by immunoblotting (32–34). We identified POLθ at actively replicating cellular DNA forks (Fig. 3A). Notably, we also detected POLθ at forks that had been stalled using hydroxyurea (HU). As expected, control proteins RPA32 and PCNA were both at replication forks and while RPA32, as expected, continued to be enriched at stalled forks, PCNA, consistent with its predominant function in active DNA replication, was not. In contrast, the RNA-binding protein PCBP2 (35), used as a negative control, was not enriched at replication forks.
Fig 3.
Polymerase theta is enriched at and contributes to the function of cellular DNA replication forks. (A) HH514-16 BL cells were exposed to EdU-containing medium for 15 min prior to performing iPOND for replication forks. To stall replication forks, EdU exposure was followed by exposure to hydroxyurea (HU) for 2 h followed by iPOND. Input and iPOND samples were subjected to immunoblotting with the indicated antibodies. YC, yes click group, was processed with biotin-azide; NC, no click control group, was performed without biotin-azide; 0.5% of input samples and 50% of iPOND samples were loaded. (B–D) HH514-16 cells were harvested after 144 h of treatment with 50 μM Novobiocin (NVB) for DNA fiber analysis. Representative images (B), fiber counts (C), and fiber lengths (D) are shown. Three fields of fibers were counted in each condition. Partial fields are shown in B. Error bars, SEM; **P < 0.01. Experiments were performed twice.
To obtain definitive evidence of POLθ’s role in DNA replication, we visually examined replication forks using the single molecule fluorescent DNA fiber assay. Compared to cells in which POLθ was functionally intact, cells exposed to Novobiocin, a newly discovered selective POLθ inhibitor, demonstrated significantly fewer and shorter DNA fibers (Fig. 3B through D). These results suggest that POLθ enriches and functions at cellular DNA replication forks, contributing to proliferation of EBV+ cells.
Novobiocin, a newly discovered selective inhibitor of polymerase theta, kills MMEJ-reliant EBV-transformed cells and EBV+ BL cells
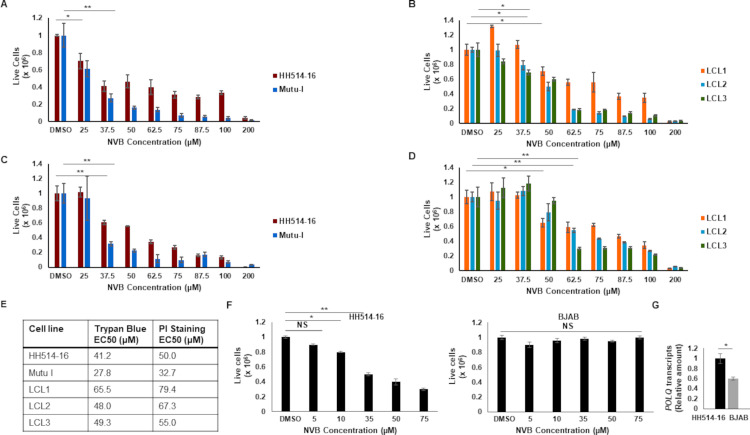
Recent clinical observations of the emergence of resistance of HR-deficient cancers to PARP inhibitors have led to pharmacologic targeting of POLθ. A small-molecule screen identified the antibiotic Novobiocin as a selective inhibitor of POLθ able to kill HR-deficient tumor cells in vitro and in patient-derived xenograft models. Novobiocin binds to the ATPase domain of POLθ, inhibits its ATPase activity, and phenocopies POLθ depletion (36). With our results pointing toward a role for POLθ in the survival and growth of EBV-transformed and EBV+ cancer cells, and with an increased abundance of POLθ observed in EBV+ cell lines and cancers, we examined the susceptibility of EBV+ BL cell lines and EBV-transformed cells (LCL) to Novobiocin. Cells were exposed to various concentrations (comparable to those used in the original study) of Novobiocin, and live cells were counted manually using Trypan blue exclusion as well as via flow cytometric enumeration of propidium iodide-negative cells (Fig. 4A through D). We found that Novobiocin is indeed toxic to EBV+ BL cell lines and EBV-transformed cells, with EC50 values in the double-digit micromolar ranges regardless of the assay used (Fig. 4E). Notably, these EC50 values are comparable to those observed by Zhou et al. in HR-deficient breast and ovarian cancer cells (36). To address if the abundance of POLθ correlated with susceptibility to Novobiocin, we exposed high POLQ-expressing EBV+ BL (HH514-16) cells versus low POLQ-expressing EBV− B lymphoma (BJAB) cells to Novobiocin and assayed cell survival. In contrast to the EBV+ BL cells, the EBV− B lymphoma cells were impervious to Novobiocin, indicating a correlation between the abundance of POLθ and susceptibility to its inhibitor (Fig. 4F and G).
Fig 4.
An inhibitor of polymerase theta impairs the growth of LCL and BL cell lines. Two BL lines (A, C, F left panel), three LCL (B, D), and an EBV-negative B lymphoma cell line (F right panel; BJAB) were treated with increasing concentrations of Novobiocin (NVB) for 144 h. Live cells were enumerated via Trypan blue counts (A, B) or propidium iodide staining and flow cytometry (C, D, F); data were plotted after normalizing cell counts for DMSO-treated cells to 1 million to enable direct comparison across cell lines. EC50 values are shown in E. POLQ transcript levels in EBV+ HH514-16 BL cells and EBV− BJAB cells analyzed by RT-qPCR are shown in G. Error bars, SEM of biological triplicates in A, B, C, D, and F, and SEM of technical triplicates within biological duplicates in G; *P < 0.05; **P < 0.01.
Novobiocin increases DNA damage and reduces MMEJ activity
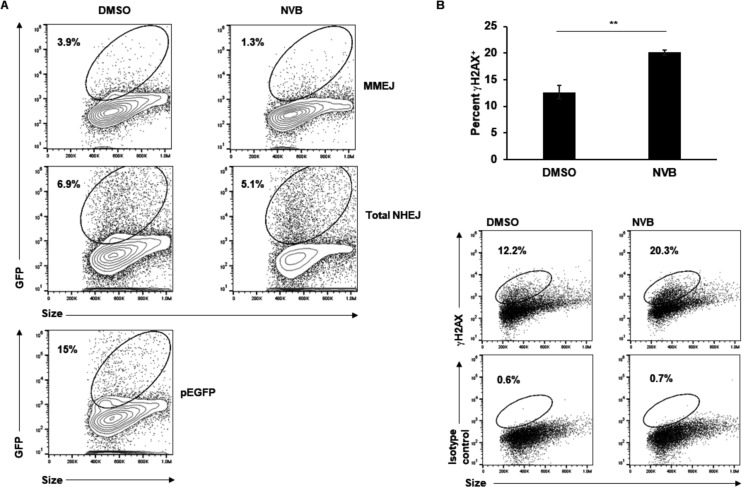
The literature supports a role for POLθ in DNA replication though the precise nature of this function remains ill defined (29). The presence of POLθ at DNA replication forks and its impairment resulting in fewer and shorter replication forks (shown in Fig. 3) support a function for POLθ in DNA replication in EBV+ cells. That said, POLθ also functions as a gap-filling polymerase that repairs DNA double-strand breaks via MMEJ. Indeed, using plasmid-based single molecule reporter assays, we had previously demonstrated that of 20%–25% of EBV+ cells that had taken up the assay plasmids by nucleofection, ~4% demonstrated repair of a double-strand break in the MMEJ reporter plasmid compared to ≤1% in the HR reporter plasmid (27). Using the same reporter assay, we now asked if exposure to the POLθ inhibitor Novobiocin dampened MMEJ-mediated repair of double-strand breaks. We performed two parallel assays using an EJ2 or EJ5 reporter plasmid. These plasmids carry a defective GFP gene containing an I-Sce1 cut site that can only be repaired to regain function through MMEJ (EJ2) or NHEJ (non-homologous end-joining; EJ5); thus, while the EJ2 assay measures MMEJ-mediated repair, EJ5 measures all types of NHEJ-repair with MMEJ being a subtype (37). Cells were pre-treated with Novobiocin for 2 days, transfected with these plasmids along with an I-Sce1 plasmid, and harvested at 3 days post-transfection (Fig. 5A). Compared to solvent control, the cells treated with Novobiocin demonstrated a near-70% reduction in MMEJ activity and a smaller decrease in total NHEJ activity. This supports Novobiocin’s inhibition of MMEJ; the smaller decrease in repair of the EJ5 plasmid can be attributed to the decrease in MMEJ, as EJ5 assesses all NHEJ.
Fig 5.
Polymerase theta inhibition reduces MMEJ activity and increases DNA damage. (A) HH514-16 BL cells were treated with Novobiocin (NVB) or DMSO (control) for 48 h and then nucleofected with EJ2 + ISce1 (top panels) to assay MMEJ reporter activity, EJ5 + ISce1 plasmids (middle panels) to assay total NHEJ reporter activity, or pEGFP to monitor transfection efficiency (bottom panel). After another 72 h, cells were harvested for GFP expression using flow cytometry. GFP+ gates were placed after comparing with identically-treated cells lacking ISce1 expression (for the top and middle panels) or untransfected cells (for the bottom panel); experiment was performed twice. (B) HH514-16 BL cells were exposed to Novobiocin (NVB) for 72 h and harvested for γH2AX staining and flow cytometry; dotplots show representative results. Error bars, SEM of biological triplicates; **, P < 0.01.
To assess if Novobiocin-exposed cells indeed experienced DNA damage, whether through unrepaired DNA breaks (Fig. 5A) or enhanced replication stress (Fig. 3B through D), we treated cells with Novobiocin for 3 days and quantified γH2AX+ cells using flow cytometry. We found significantly more cells positive for γH2AX in the Novobiocin-treated condition compared to the solvent control (Fig. 5B). This indicates that Novobiocin causes an accumulation of DNA lesions likely from unrepaired DNA intermediates such as overly resected ssDNA as has been shown by Zhou et al. (36)—this would result in an increase in the DNA damage sensor γH2AX.
Novobiocin does not activate the EBV lytic phase
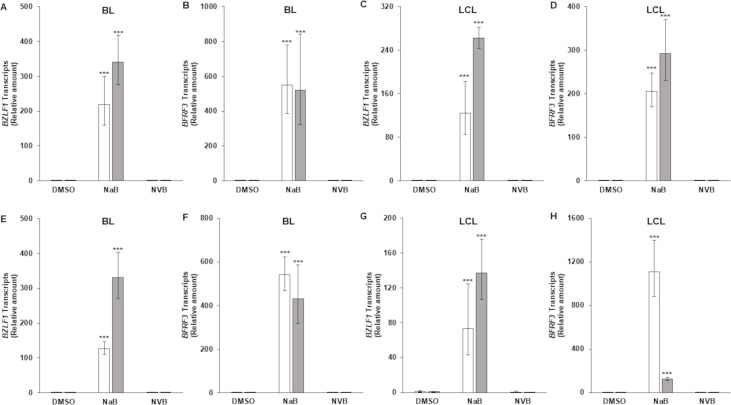
Others have previously demonstrated that DNA damaging agents such as Gemcitabine and Doxorubicin activate the EBV lytic cycle (38). We, therefore, addressed if Novobiocin contributes to death of EBV+ cells by turning the lytic phase on, potentially through damaged DNA activating the inflammasome; activation of the inflammasome causes EBV reactivation (39). As shown in Fig. 6, while exposure to sodium butyrate, a known lytic cycle activator, turned the expression of the latent-to-lytic switch gene BZLF1 and the late gene BFRF3 on, Novobiocin failed to do so in BL cells and LCL. We looked for BZLF1 and BFRF3 transcripts at both early and late times following Novobiocin treatment, i.e., 3 and 6 days, to ensure that we did not miss reactivation that may have occurred very early or very late after drug exposure. Thus, when used at EC50 concentrations, Novobiocin does not activate the lytic cycle and further, lytic activation does not contribute to cell killing by Novobiocin.
Fig 6.
An inhibitor of polymerase theta does not induce the lytic cycle of EBV. HH514-16 BL cells (A, B, E, F) and an LCL (C, D, G, H) were exposed to DMSO control, sodium butyrate (NaB; 3 mM; positive control), or Novobiocin (NVB; 50μM for BL cells and 70μM for LCL) for 3 days (A–D) or 6 days (E–H). Samples were analyzed for BZLF1 (NaB samples harvested at 24 h) or BFRF3 (NaB samples harvested at 48 h) transcripts by RT-qPCR. Error bars, 95% CI of ΔΔCT extended to relative transcripts; ***P < 0.001, compared to DMSO. Open and gray bars represent biological replicates.
Inhibiting the synthetic lethal target PARP1 alongside Novobiocin is additive
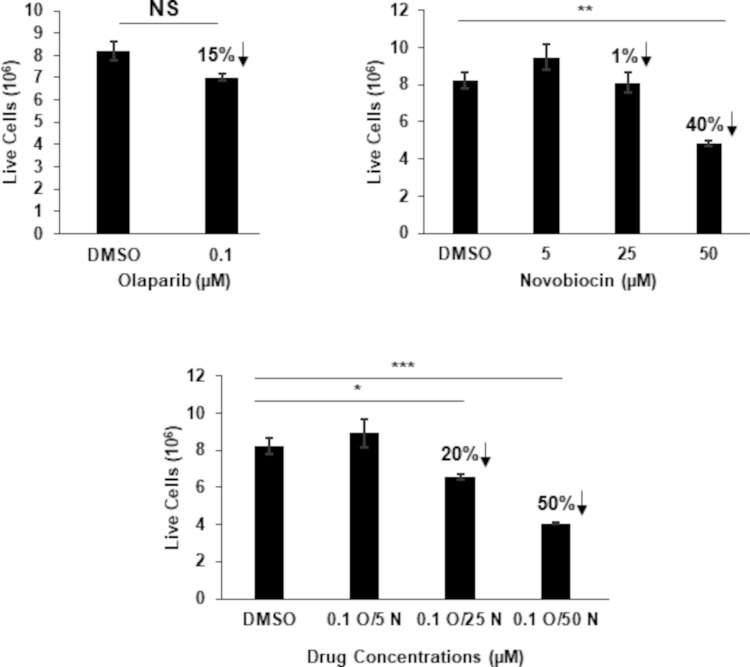
We have previously shown that EBV-cancer cells are susceptible to PARP inhibitors, agents that are synthetic lethal in the context of impaired HR or NHEJ. Given that POLθ contributes to MMEJ repair and DNA replication, and PARP1 contributes to the repair of single- and double-stranded DNA breaks through several mechanisms (13, 14), we asked if the effective concentration of Novobiocin could be reduced in the presence of the PARP inhibitor Olaparib. We treated BL cells with three concentrations (up to the EC50 values) of Novobiocin alongside 0.1 μM, a suboptimal concentration, of Olaparib; we had previously established 0.5 μM Olaparib to be the EC50 for these cells (27). As shown in Fig. 7, combining the two drugs showed additive inhibitory effects on growth of the BL cell line particularly when both drugs were used at suboptimal concentrations.
Fig 7.
Inhibiting PARP1 alongside polymerase theta is additive. HH514-16 BL cells were treated with indicated concentrations of the PARP inhibitor Olaparib (O), Novobiocin (N), or both. After 144 h, live cells were enumerated using Trypan Blue staining. Error bars, SEM of biological triplicates; *P < 0.05; **P < 0.01; ***P < 0.001. Arrows indicate percent reduction in live cells compared to solvent-treated cells.
Polymerase theta function and as a predictive biomarker for synthetic lethality in EBV-transformed/cancer cells
Studies in the context of tumors unrelated to EBV have shown that cancers with abundant expression of POLθ are susceptible to inhibitors of POLθ. Our results show that EBV-transformed and cancer cells abundantly express POLθ and rely on POLθ’s contribution to DNA replication and repair for survival and proliferation. The prominent expression of POLθ in EBV+ cell lines and cancers parallels observations by others of POLθ overexpression in HR-deficient or p53-deficient cancers (29, 40–42); in contrast, POLθ expression is highly limited in normal tissues (29). Indeed, cancers overexpressing POLθ are thought to be dependent on POLθ activity—and this overexpression, assessed by immunoassays or reverse transcriptase-PCR (RT-qPCR), can serve as a predictive biomarker of susceptibility to POLθ/MMEJ inhibition. MMEJ-specific mutational signatures such as the insertion-deletion signature 6 (43) or identification of MMEJ-scars in EBV-transformed cells (27) may also be useful as biomarkers but require whole genome sequencing. Indicators of HR-deficiency (or deficiencies in other types of DNA repair) may serve as a third type of biomarker: in earlier work, we identified a STAT3-dependent gene expression signature that predicts HR deficiency in EBV+ cell lines and EBV-unrelated cancers with overactive STAT3 (27).
Mechanistically, in EBV+ cells, POLθ contributes to MMEJ-mediated DNA repair and DNA replication. While POLθ is also known to inhibit HR (29), we do not believe this to be a prominent function in EBV+ cells as we have shown previously that these cells already lack HR (27). As for DNA replication, POLθ plays an as yet ill-defined role in mitigating replication stress. Studies have found that POLθ mediates end-joining of replication-associated DNA breaks in C. elegans (44) and helps reinitiate replication following fork collapse in mammalian cells (45–47). That said, these and other studies place POLθ at replication forks after their collapse into double-strand breaks (48, 49)—in agreement with these studies, we find POLθ at stalled/collapsed replication forks (Fig. 3A, +HU condition) but we also find it at actively replicating forks (Fig. 3A, no HU condition), suggesting that POLθ travels with the replication machinery and is readily available to re-establish fork function, presumably before forks collapse. This idea is further supported by the observation that under baseline conditions, exposure to a selective inhibitor of POLθ results in fewer and shorter DNA fibers indicative of fork collapse (Fig. 3B through D). Parsing out POLθ’s exact function at forks is an active area of investigation. What signals and other proteins recruit POLθ to replication forks before versus after dissolution of the replisome complex is one of several questions of interest. Whether POLθ uses its polymerase function at active forks is also an important question. What contributes to cell death when POLθ is inhibited is another such question. Experiments with Novobiocin point to toxicity from unrepaired DNA double-strand breaks (related or unrelated to DNA replication) due to increased end-resection leading to increased single-stranded DNA intermediates (36). The increase in γH2AX that we observe following exposure to Novobiocin can be attributed to the overly-resected single-stranded DNA. POLθ’s role in replication stress tolerance suggests that loss of replication fork stability further contributes to toxic DNA lesions. While loss of POLθ also results in toxic accumulation of RAD51 on single-stranded DNA, cell death may not be attributable to this mechanism in EBV+ cells as these cells are defective in recruiting RAD51 to DNA breaks (27).
In therapeutic settings, combining POLθ inhibitors with other agents including PARP inhibitors may be additive. This prediction is supported by our findings and work from other groups in the context of EBV-unrelated cancers (29, 36). Whether POLθ inhibitors may be toxic to normal cells remains unclear; although experiments in mammalian cells with intact HR and NHEJ indicate that MMEJ usage is at 10%–20% of HR, these experiments were done in cancer cells and not in normal cells (50).
While Novobiocin was reported in 1978 to inhibit DNA gyrase in bacteria (51), it was subsequently shown to be minimally effective against topoisomerase II in eukaryotic cells (52); the reported IC50 value against topoisomerase II was 300 microM which was far higher than the EC/IC50 values in our study and that of Zhou et al. (36). Furthermore, Zhou et al. demonstrated that the effects of Novobiocin and a topoisomerase II inhibitor were additive, suggesting that topoisomerase II was not a shared target between the two inhibitors and that the effects of Novobiocin are unrelated to topoisomerase II inhibition.
We have previously shown that LCLs and EBV+ BL cells are susceptible to PARP inhibition. While important in sensing and mediating repair of single- and double-strand DNA breaks, until recently, PARP was also thought to significantly contribute to MMEJ-mediated repair (10, 13). However, it was recently demonstrated that PARP only plays a limited role in MMEJ (14). Therefore, targeting PARP, classical NHEJ, and the ATR-dependent replication stress response pathway are likely to reveal further synthetic lethality when combined with MMEJ inhibition. Inhibiting multiple DNA repair pathways at the same time could produce an effect that is stronger than each drug alone while staving off the emergence of drug resistance.
MATERIALS AND METHODS
Study subjects and ethics statement
Peripheral blood mononuclear cells (PBMC) were isolated from three young healthy subjects at the University of Florida. Blood was drawn after obtaining written informed consent. The study of human subjects was approved by the Institutional Review Board at the University of Florida.
DLBCL tumors
RNA isolated from frozen diffuse large B cell lymphomas (DLBCL), three EBV-positive and three EBV-negative, were subjected to RT-qPCR as described below. These specimens were provided by the AIDS and Cancer Specimen Resource (ACSR), funded by the National Cancer Institute. To determine if tumors were EBV-positive or EBV-negative, multiplexed PCRs were used to amplify EBNA1, LMP1, and LMP2 genes using an established assay at the Mayo Clinic based on the work of Ryan et al. (53).
Cell lines and chemical treatment
The EBV-positive endemic BL cell line HH514-16 was a kind gift from Dr. George Miller (Yale University). Mutu I was a kind gift from Dr. Erik Flemington (Tulane University). Three different LCLs (LCL1, LCL2, and LCL3) were generated as described before (54). All cell lines were maintained in RPMI1640 containing 10% fetal bovine serum and 1% penicillin/streptomycin in the presence of 5% CO2. For chemical treatment, cells were sub-cultured at 5 × 105 cells/mL in the presence of indicated concentrations of Novobiocin (HY-B0425A, MedChemExpress) alone or with 0.1 μM Olaparib (HY-10162, MedChemExpress) with supplementation at the initial concentration every 72 h; in DNA repair assays, 50 µM (EC50 for HH514-16 cells) Novobiocin was used. To enumerate live/dead cells, cells were stained with Trypan blue for manual counting under a light microscope or with propidium iodide at 2 µg/mL in PBS with 2% FBS and analyzed via flow cytometry.
Immunoblotting and antibodies
Immunoblotting was performed as described previously (55). In short, cells were lysed with RIPA buffer on ice for 15 min and spun at 4°C for 5 min at high speed to remove debris. Cell extracts were then electrophoresed using SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were probed using the following antibodies: rabbit anti-human POLθ antibody (PA5-69577, ThermoFisher Scientific), mouse anti-β-actin antibody (AC-15) (A1978, Sigma-Aldrich), rabbit anti-PCNA antibody (A300-277A, Bethyl Laboratories), rabbit anti-RPA32 antibody (A300-244A, Bethyl Laboratories), rabbit anti-STAT3 antibody (4904S, Cell Signaling Technology), mouse anti-EBNA1 antibody (sc-81581, Santa Cruz Biotechnology), mouse anti-PCBP2 antibody (sc-101136; Santa Cruz Biotechnology), HRP-conjugated goat anti-mouse IgG (626520, ThermoFisher Scientific), and HRP-conjugated goat anti-rabbit IgG (31460, ThermoFisher Scientific).
Flow cytometry
Flow cytometry was performed as described previously (55). In brief, cells were fixed with Cytofix/Cytoperm solution (554722, BD Bioscience) for 15 min at room temperature, washed twice with Perm/Wash buffer (554723, BD Bioscience), and incubated with indicated primary and secondary antibodies prior to performing flow cytometry. For cell cycle analysis, cells were incubated with 100 µM BrdU for 1 h, fixed with ice cold 70% Ethanol in PBS at −20°C for 1 h, and denatured with 2 N HCl/0.5% Triton X- for 30 min followed by two washes with 1% BSA-containing PBS. Cells were then incubated with indicated primary and secondary antibodies. After two washes with 1% BSA/PBS, cells were suspended in PBS containing 10 µg/mL RNase A (EN0531, Thermo Scientific) and 20 µg/mL propidium iodide (PI, P4864, Sigma-Aldrich) for 30 min at RT before analysis using flow cytometry. Antibodies used for flow cytometry include mouse anti-BrdU antibody (555627, BD Bioscience), rabbit anti-phospho histone H2AX (Ser139) antibody (9718S, Cell Signaling), mouse IgG1 (557273, BD Bioscience), and FITC-conjugated goat anti-mouse IgG (F0257, Sigma-Aldrich).
Transfection, plasmids, and siRNAs
Nucleofection was performed as described previously (55). For each nucleofection, one million cells were washed twice with PBS and resuspended in 100 µL Ingenio solution (MIR50117, Mirus) containing 20 µg plasmids or 200 pmol siRNA; 3 million cells were transfected with each siRNA. Cells were transferred to cuvette and transfected by using Amaxa Nucleofector II (program A-024).
Plasmids DR-GFP, pCBASce (encoding I-Sce1 enzyme), and pCAGGS (empty vector control) were kind gifts from Dr.Maria Jasin (56). EJ2-GFP and EJ5-GFP were gifts from Dr. Jeremy Stark (via Addgene) (42). The siRNAs used in this study include control siRNA (D001810-01-20, Dharmacon), si-STAT3 (J003544-07, Dharmacon), si-POLQ #1 (122556, ThermoFisher Scientific), and si-POLQ #2 (122557, ThermoFisher Scientific). Previously published siRNAs targeting EBNA1 (si-EBNA1 #1, GGAGGUUCCAACCCGAAAUTT; si-EBNA1 #2, GGACTACCGACGAAGGAACTT) (57) were synthesized by Thermo Fisher Scientific.
Isolation of proteins on nascent DNA
iPOND was performed as described previously (34). Briefly, 100 million cells were pulsed with 10 µM EdU for 15 min. For stalling DNA replication, 3 mM hydroxyurea (HU) was added to cells after the 15 min EdU labeling step. Cells were fixed with 1% formaldehyde for 20 min, quenched with 0.125 M glycine for 5 min, and permeabilized in 0.25% Triton X-100/PBS for 30 min. After two washes, cells were incubated with click reaction buffer (10 µM biotin-azide, 10 mM sodium ascorbate, 2 mM CuSO4) for 2 h and washed twice with PBS. Nuclei were extracted and re-suspended in cold lysis buffer (1% SDS, 50 mM Tris, pH 8.0). After sonication and debris removal, lysates were incubated with 100 µL of streptavidin agarose beads (69203, EMD Millipore) overnight at 4°C. After three washes with lysis buffer and one wash with 1M NaCl, 2× Laemmli buffer was added. Lysates were heated for 25 min at 95°C prior to immunoblotting with indicated antibodies.
Quantitative reverse transcriptase-PCR
RT-qPCR was performed as described previously (58). In short, 1 µg RNA was used as template to synthesize cDNA using MuLV Reverse Transcriptase (M0253L, New England Biolabs). Quantitative PCR was used to assay relative transcript levels of each gene using the following primers: 5′GTAACCCGTTGAACCCCATT3′ (forward) and 5′CCATCCAATCGGTAGTAGCG3′ (reverse) for 18S rRNA; 5′ GAGGACTGAGCATCGAGCAGC3′ (forward) and 5′ TTAGCCCATGTGATCTGACACCC3′ (reverse) for STAT3; 5′ CAGCATCTTGTCAGGCAGATCT3′ (forward) and 5′ CGGTACAGGGCGAAAGTCGG3′ (reverse) for POLQ. Data were analyzed using the ΔΔCT method after normalization to 18S rRNA.
DNA fiber assay
DNA fiber assay was performed as described previously (59). Briefly, cells were treated with or without 50 µM Novobiocin for 96 h. Cells were then incubated with 250 µM IdU (I7125, Sigma-Aldrich) for 30 min. Two hundred thousand cells were resuspended in low-melting point agarose and 0.5% trypsin and transferred to plug mold. After proteinase K digestion (50°C overnight) followed by four washes with 0.01 M Tris-EDTA, DNA fibers were combed to coverslips (COV-002-RUO, Genomic Vision) and baked at 60°C for 2 h. DNA fibers were then denatured with 0.5 M NaOH/1 M NaCl for 8 min, washed twice with PBS, and incubated with indicated antibodies (mouse anti-BrdU antibody: 347580, BD Biosciences; goat anti-mouse Alexa Fluor 488 antibody: A11001, Thermo fisher scientific). Images of DNA fibers were acquired using a fluorescence microscope (OLYMPUS). DNA fibers were analyzed using Image J software (60).
Statistical analysis
P values were calculated using unpaired Student t test. For DNA fiber analysis, P values were calculated using two tailed Mann-Whitney U tests.
ACKNOWLEDGMENTS
S.B.-M. was supported by the Children’s Miracle Network and NIH (U01 CA275310 and R01 DE032623); M.T.M. was supported by the Children’s Miracle Network, NIH (U01 CA275310 and R01 DE032623), DHS (70 RSAT 19 CB 0000027), and USDA-APHIS (6000027242).
Contributor Information
Sumita Bhaduri-McIntosh, Email: sbhadurimcintosh@ufl.edu.
Felicia Goodrum, The University of Arizona, Tucson, Arizona, USA.
DATA AVAILABILITY
All data supporting the conclusions are presented in the paper. There are no large data sets associated with this work.
REFERENCES
- 1. Thorley-Lawson DA, Gross A. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med 350:1328–1337. doi: 10.1056/NEJMra032015 [DOI] [PubMed] [Google Scholar]
- 2. Farrell PJ. 2019. Epstein-Barr virus and cancer. Annu Rev Pathol 14:29–53. doi: 10.1146/annurev-pathmechdis-012418-013023 [DOI] [PubMed] [Google Scholar]
- 3. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. doi: 10.1038/nrc1452 [DOI] [PubMed] [Google Scholar]
- 4. Gottschalk S, Rooney CM, Heslop HE. 2005. Post-transplant lymphoproliferative disorders. Annu Rev Med 56:29–44. doi: 10.1146/annurev.med.56.082103.104727 [DOI] [PubMed] [Google Scholar]
- 5. Allen UD, Preiksaitis JK, AST Infectious Diseases Community of Practice . 2019. Post-transplant lymphoproliferative disorders, Epstein-Barr virus infection, and disease in solid organ transplantation: guidelines from the American society of transplantation infectious diseases community of practice. Clin Transplant 33:e13652. doi: 10.1111/ctr.13652 [DOI] [PubMed] [Google Scholar]
- 6. Al-Mansour Z, Nelson BP, Evens AM. 2013. Post-transplant lymphoproliferative disease (PTLD): risk factors, diagnosis, and current treatment strategies. Curr Hematol Malig Rep 8:173–183. doi: 10.1007/s11899-013-0162-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crombie JL, LaCasce AS. 2019. Epstein Barr virus associated B-cell lymphomas and iatrogenic lymphoproliferative disorders. Front Oncol 9:109. doi: 10.3389/fonc.2019.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Styczynski J, Einsele H, Gil L, Ljungman P. 2009. Outcome of treatment of Epstein-Barr virus-related post-transplant lymphoproliferative disorder in hematopoietic stem cell recipients: a comprehensive review of reported cases. Transpl Infect Dis 11:383–392. doi: 10.1111/j.1399-3062.2009.00411.x [DOI] [PubMed] [Google Scholar]
- 9. Kass EM, Moynahan ME, Jasin M. 2016. When genome maintenance goes badly awry. Mol Cell 62:777–787. doi: 10.1016/j.molcel.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sfeir A, Symington LS. 2015. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci 40:701–714. doi: 10.1016/j.tibs.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brambati A, Sacco O, Porcella S, Heyza J, Kareh M, Schmidt JC, Sfeir A. 2023. RHINO directs MMEJ to repair DNA breaks in mitosis. Science 381:653–660. doi: 10.1126/science.adh3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gelot C, Kovacs MT, Miron S, Mylne E, Haan A, Boeffard-Dosierre L, Ghouil R, Popova T, Dingli F, Loew D, Guirouilh-Barbat J, Del Nery E, Zinn-Justin S, Ceccaldi R. 2023. Polθ is phosphorylated by PLK1 to repair double-strand breaks in mitosis. Nature 621:415–422. doi: 10.1038/s41586-023-06506-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wicks AJ, Krastev DB, Pettitt SJ, Tutt ANJ, Lord CJ. 2022. Opinion: PARP inhibitors in cancer-what do we still need to know? Open Biol 12:220118. doi: 10.1098/rsob.220118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luedeman ME, Stroik S, Feng W, Luthman AJ, Gupta GP, Ramsden DA. 2022. Poly(ADP) ribose polymerase promotes DNA polymerase theta-mediated end joining by activation of end resection. Nat Commun 13:4547. doi: 10.1038/s41467-022-32166-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frappier L. 2019. Epstein–Barr virus: overcoming the DNA damage response. Future Virol 14:349–360. doi: 10.2217/fvl-2019-0015 [DOI] [Google Scholar]
- 16. McFadden K, Luftig MA. 2013. Interplay between DNA tumor viruses and the host DNA damage response. Curr Top Microbiol Immunol 371:229–257. doi: 10.1007/978-3-642-37765-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pancholi NJ, Price AM, Weitzman MD. 2017. Take your PIKK: tumour viruses and DNA damage response pathways. Philos Trans R Soc Lond B Biol Sci 372:20160269. doi: 10.1098/rstb.2016.0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, Hu K, Guo J, Tainter D, Rusyn E, Luftig MA. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522. doi: 10.1016/j.chom.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jha HC, A J MP, Saha A, Banerjee S, Lu J, Robertson ES. 2014. Epstein-Barr virus essential antigen EBNA3C attenuates H2AX expression. J Virol 88:3776–3788. doi: 10.1128/JVI.03568-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. 2009. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc Natl Acad Sci U S A 106:2313–2318. doi: 10.1073/pnas.0810619106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kamranvar SA, Masucci MG. 2011. The Epstein-Barr virus nuclear antigen-1 promotes telomere dysfunction via induction of oxidative stress. Leukemia 25:1017–1025. doi: 10.1038/leu.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koganti S, Hui-Yuen J, McAllister S, Gardner B, Grasser F, Palendira U, Tangye SG, Freeman AF, Bhaduri-McIntosh S. 2014. STAT3 interrupts ATR-Chk1 signaling to allow oncovirus-mediated cell proliferation. Proc Natl Acad Sci U S A 111:4946–4951. doi: 10.1073/pnas.1400683111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koganti S, de la Paz A, Freeman AF, Bhaduri-McIntosh S. 2014. B lymphocytes from patients with a hypomorphic mutation in STAT3 resist Epstein-Barr virus-driven cell proliferation. J Virol 88:516–524. doi: 10.1128/JVI.02601-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koganti S, Burgula S, Bhaduri-McIntosh S. 2020. STAT3 activates the anti-apoptotic form of caspase 9 in oncovirus-infected B lymphocytes. Virology 540:160–164. doi: 10.1016/j.virol.2019.11.017 [DOI] [PubMed] [Google Scholar]
- 25. Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ. 2008. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene 27:3977–3985. doi: 10.1038/onc.2008.17 [DOI] [PubMed] [Google Scholar]
- 26. Sørensen CS, Hansen LT, Dziegielewski J, Syljuåsen RG, Lundin C, Bartek J, Helleday T. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7:195–201. doi: 10.1038/ncb1212 [DOI] [PubMed] [Google Scholar]
- 27. McIntosh MT, Koganti S, Boatwright JL, Li X, Spadaro SV, Brantly AC, Ayers JB, Perez RD, Burton EM, Burgula S, MacCarthy T, Bhaduri-McIntosh S. 2020. STAT3 imparts BRCAness by impairing homologous recombination repair in Epstein-Barr virus-transformed B lymphocytes. PLoS Pathog 16:e1008849. doi: 10.1371/journal.ppat.1008849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schrempf A, Slyskova J, Loizou JI. 2021. Targeting the DNA repair enzyme polymerase θ in cancer therapy. Trends Cancer 7:98–111. doi: 10.1016/j.trecan.2020.09.007 [DOI] [PubMed] [Google Scholar]
- 29. Patterson-Fortin J, D’Andrea AD. 2020. Exploiting the microhomology-mediated end-joining pathway in cancer therapy. Cancer Res 80:4593–4600. doi: 10.1158/0008-5472.CAN-20-1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gradoville L, Kwa D, El-Guindy A, Miller G. 2002. Protein kinase C-independent activation of the Epstein-Barr virus lytic cycle. J Virol 76:5612–5626. doi: 10.1128/jvi.76.11.5612-5626.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rooney C, Howe JG, Speck SH, Miller G. 1989. Influence of Burkitt's lymphoma and primary B cells on latent gene expression by the nonimmortalizing P3J-HR-1 strain of Epstein-Barr virus. J Virol 63:1531–1539. doi: 10.1128/JVI.63.4.1531-1539.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25:1320–1327. doi: 10.1101/gad.2053211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sirbu BM, McDonald WH, Dungrawala H, Badu-Nkansah A, Kavanaugh GM, Chen Y, Tabb DL, Cortez D. 2013. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J Biol Chem 288:31458–31467. doi: 10.1074/jbc.M113.511337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu H, Perez RD, Frey TR, Burton EM, Mannemuddhu S, Haley JD, McIntosh MT, Bhaduri-McIntosh S. 2019. Novel replisome-associated proteins at cellular replication forks in EBV-transformed B lymphocytes. PLoS Pathog 15:e1008228. doi: 10.1371/journal.ppat.1008228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao H, Wei Z, Shen G, Chen Y, Hao X, Li S, Wang R. 2022. Poly(rC)-binding proteins as pleiotropic regulators in hematopoiesis and hematological malignancy. Front Oncol 12:1045797. doi: 10.3389/fonc.2022.1045797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou J, Gelot C, Pantelidou C, Li A, Yücel H, Davis RE, Färkkilä A, Kochupurakkal B, Syed A, Shapiro GI, Tainer JA, Blagg BSJ, Ceccaldi R, D’Andrea AD. 2021. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat Cancer 2:598–610. doi: 10.1038/s43018-021-00203-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bennardo N, Cheng A, Huang N, Stark JM. 2008. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4:e1000110. doi: 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feng W, Hong G, Delecluse H-J, Kenney SC. 2004. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J Virol 78:1893–1902. doi: 10.1128/jvi.78.4.1893-1902.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burton EM, Goldbach-Mansky R, Bhaduri-McIntosh S. 2020. A promiscuous inflammasome sparks replication of a common tumor virus. Proc Natl Acad Sci U S A 117:1722–1730. doi: 10.1073/pnas.1919133117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carvajal-Garcia J, Cho JE, Carvajal-Garcia P, Feng W, Wood RD, Sekelsky J, Gupta GP, Roberts SA, Ramsden DA. 2020. Mechanistic basis for microhomology identification and genome scarring by polymerase theta. Proc Natl Acad Sci U S A 117:8476–8485. doi: 10.1073/pnas.1921791117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MIR, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD. 2015. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518:258–262. doi: 10.1038/nature14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. 2015. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518:254–257. doi: 10.1038/nature14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, Islam SMA, Lopez-Bigas N, Klimczak LJ, McPherson JR, Morganella S, Sabarinathan R, Wheeler DA, Mustonen V, Getz G, Rozen SG, Stratton MR, PCAWG Mutational Signatures Working Group, PCAWG Consortium . 2020. The repertoire of mutational signatures in human cancer. Nature 578:94–101. doi: 10.1038/s41586-020-1943-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roerink SF, van Schendel R, Tijsterman M. 2014. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res 24:954–962. doi: 10.1101/gr.170431.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feng W, Simpson DA, Carvajal-Garcia J, Price BA, Kumar RJ, Mose LE, Wood RD, Rashid N, Purvis JE, Parker JS, Ramsden DA, Gupta GP. 2019. Genetic determinants of cellular addiction to DNA polymerase theta. Nat Commun 10:4286. doi: 10.1038/s41467-019-12234-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Z, Song Y, Li S, Kurian S, Xiang R, Chiba T, Wu X. 2019. DNA polymerase θ (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J Biol Chem 294:3909–3919. doi: 10.1074/jbc.RA118.005188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yousefzadeh MJ, Wyatt DW, Takata K-I, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublié S, Johansson E, Ramsden DA, McBride KM, Wood RD. 2014. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet 10:e1004654. doi: 10.1371/journal.pgen.1004654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deng L, Wu RA, Sonneville R, Kochenova OV, Labib K, Pellman D, Walter JC. 2019. Mitotic CDK promotes replisome disassembly, fork breakage, and complex DNA rearrangements. Mol Cell 73:915–929. doi: 10.1016/j.molcel.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kais Z, Rondinelli B, Holmes A, O’Leary C, Kozono D, D’Andrea AD, Ceccaldi R. 2016. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep 15:2488–2499. doi: 10.1016/j.celrep.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Truong LN, Li Y, Shi LZ, Hwang P-H, He J, Wang H, Razavian N, Berns MW, Wu X. 2013. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A 110:7720–7725. doi: 10.1073/pnas.1213431110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sugino A, Higgins NP, Brown PO, Peebles CL, Cozzarelli NR. 1978. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A 75:4838–4842. doi: 10.1073/pnas.75.10.4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pocklington MJ, Jenkins JR, Orr E. 1990. The effect of novobiocin on yeast topoisomerase type II. Mol Gen Genet 220:256–260. doi: 10.1007/BF00260491 [DOI] [PubMed] [Google Scholar]
- 53. Ryan JL, Fan H, Glaser SL, Schichman SA, Raab-Traub N, Gulley ML. 2004. Epstein-Barr virus quantitation by real-time PCR targeting multiple gene segments: a novel approach to screen for the virus in paraffin-embedded tissue and plasma. J Mol Diagn 6:378–385. doi: 10.1016/S1525-1578(10)60535-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hui-Yuen J, McAllister S, Koganti S, Hill E, Bhaduri-McIntosh S. 2011. Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. J Vis Exp 57:3321. doi: 10.3791/3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li X, Burton EM, Koganti S, Zhi J, Doyle F, Tenenbaum SA, Horn B, Bhaduri-McIntosh S. 2018. KRAB-ZFP repressors enforce quiescence of oncogenic human herpesviruses. J Virol 92:e00298-18. doi: 10.1128/JVI.00298-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, Brunet E, Jasin M. 2011. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol 18:500–503. doi: 10.1038/nsmb.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sivachandran N, Wang X, Frappier L. 2012. Functions of the Epstein-Barr virus EBNA1 protein in viral reactivation and lytic infection. J Virol 86:6146–6158. doi: 10.1128/JVI.00013-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Burton EM, Akinyemi IA, Frey TR, Xu H, Li X, Su LJ, Zhi J, McIntosh MT, Bhaduri-McIntosh S. 2021. A heterochromatin inducing protein differentially recognizes self versus foreign genomes. PLoS Pathog 17:e1009447. doi: 10.1371/journal.ppat.1009447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sheng Y, Wei J, Yu F, Xu H, Yu C, Wu Q, Liu Y, Li L, Cui X-L, Gu X, Shen B, Li W, Huang Y, Bhaduri-McIntosh S, He C, Qian Z. 2021. A critical role of nuclear M6A reader YTHDC1 in leukemogenesis by regulating MCM complex-mediated DNA replication. Blood 138:2838–2852. doi: 10.1182/blood.2021011707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the conclusions are presented in the paper. There are no large data sets associated with this work.