ABSTRACT
Biological valorization of lignin, the second most abundant biopolymer on Earth, is an indispensable sector to build a circular economy and net-zero future. However, lignin is recalcitrant to bioupcycling, demanding innovative solutions. We report here the biological valorization of lignin-derived aromatic carbon to value-added chemicals without requesting extra organic carbon and freshwater via reprogramming the marine Roseobacter clade bacterium Roseovarius nubinhibens. We discovered the unusual advantages of this strain for the oxidation of lignin monomers and implemented a CRISPR interference (CRISPRi) system with the lacI-Ptrc inducible module, nuclease-deactivated Cas9, and programmable gRNAs. This is the first CRISPR-based regulatory system in R. nubinhibens, enabling precise and efficient repression of genes of interest. By deploying the customized CRISPRi, we reprogrammed the carbon flux from a lignin monomer, 4-hydroxybenzoate, to achieve the maximum production of protocatechuate, a pharmaceutical compound with antibacterial, antioxidant, and anticancer properties, with minimal carbon to maintain cell growth and drive biocatalysis. As a result, we achieved a 4.89-fold increase in protocatechuate yield with a dual-targeting CRISPRi system, and the system was demonstrated with real seawater. Our work underscores the power of CRISPRi in exploiting novel microbial chassis and will accelerate the development of marine synthetic biology. Meanwhile, the introduction of a new-to-the-field lineage of marine bacteria unveils the potential of blue biotechnology leveraging resources from the ocean.
IMPORTANCE
One often overlooked sector in carbon-conservative biotechnology is the water resource that sustains these enabling technologies. Similar to the “food-versus-fuel” debate, the competition of freshwater between human demands and bioproduction is another controversial issue, especially under global water scarcity. Here, we bring a new-to-the-field lineage of marine bacteria with unusual advantages to the stage of engineering biology for simultaneous carbon and water conservation. We report the valorization of lignin monomers to pharmaceutical compounds without requesting extra organic substrate (e.g., glucose) or freshwater by reprogramming the marine bacterium Roseovarius nubinhibens with a multiplex CRISPR interference system. Beyond the blue lignin valorization, we present a proof-of-principle of leveraging marine bacteria and engineering biology for a sustainable future.
KEYWORDS: CRISPRi, marine bacteria, Roseovarius nubinhibens, valorization, lignin
INTRODUCTION
Reverting carbon emissions back into resources for carbon-conservative bioproduction is an important route toward a net-zero future. Lignin, a prevalent aromatic polymer, represents a substantial fraction, typically ranging from 15% to 40%, of the dry mass of terrestrial plants on Earth (1–3). However, lignin is largely underutilized due to its inherent recalcitrance to biodegradation and heterogeneity in composition (4–7). Consequently, lignin-containing biomass (e.g., plant cell walls and agriculture wastes) often demands disposal via, for instance, incineration, causing CO2 emission and environmental pollution. Innovations for the valorization of lignin are imperative and urgent (8). Given the revolutionary advances in engineering biology, bioconversion of lignin-derived aromatic compounds to value-added chemicals, e.g., cis,cis-muconic acids, pyridine-dicarboxylic acids, and β-ketoadipic acids, has been achieved in engineered Pseudomonas putida (9–11), Corynebacterium glutamicum (12), and Rhodococcus jostii (13), illustrating a promising avenue for the sustainable valorization of lignin-derived aromatics.
One often overlooked sector, however, is the source of water to sustain these enabling biotechnologies for, but not limited to, lignin valorization. A large amount of freshwater is commonly necessary to cultivate engineered microbes (14, 15), competing with human needs for freshwater. This will be a controversial issue, especially with the fact of global water scarcity (16–18). To tackle this conflict, we propose blue valorization of lignin-derived monomers, which leverages resources from the ocean including seawater and marine bacteria, to alleviate the dependence on freshwater during the biological processes (19). Marine bacteria usually survive or even thrive in environments with limited nutrients, extreme temperatures, and high salinity, which endow them with versatile metabolic capabilities and tolerances to various stressors (20–22). Besides, marine bacteria inherently grow in seawater avoiding direct competition for freshwater resources with humans. Therefore, marine bacteria can be engineered as promising microbial chassis for lignin biovalorization with seawater, resolving the challenges in carbon and water conservation simultaneously.
Roseovarius nubinhibens is a member of the marine Roseobacter clade with diverse metabolic capacities, including the degradation of aromatics through the β-ketoadipate pathway (23). The natural properties enable R. nubinhibens a potential whole-cell catalyst for the conversion of lignin-derived aromatic compounds into value-added chemicals. Though bacteria in this clade have been intensively interrogated for the ecological and evolutionary dynamics (24–26), the potential of these bacteria for biocatalysis has rarely been discovered. Notably, genetic systems are available for strain engineering, including the delivery of foreign DNAs, replicable shuttle vectors, and “knock-in” gene disruption (27–29). In a previous work, we established a CRISPR-Cas-based genome editing tool at a single-nucleotide resolution for R. nubinhibens, and efficient and precise gene inactivation can be realized, paving the way for the design and construction of strains capable of lignin upcycling (30).
Here, we report a customized CRISPR interference (CRISPRi) system to reprogram the marine bacterium R. nubinhibens for the biological valorization of lignin-derived monomers. We revealed the unusual potentials of this strain for the oxidation of the lignin-derived monomers. We modularly designed the CRISPRi system with nuclease-deactivated Cas9 (dCas9) from Streptococcus pyogenes and programmable gRNAs driven by an inducible system. Then, we selected 4-hydroxybenzoate (4HB) as a proof-of-principle lignin monomer and protocatechuate (PCA), a pharmaceutical compound, as the product to evaluate the bioproduction by R. nubinhibens with CRISPRi in both defined synthetic medium and seawater. We present here a new-to-the-field lineage of marine bacteria as whole-cell biocatalyst for lignin valorization and a paradigm of leveraging marine bacteria with cutting-edge synthetic biology tools for sustainable biorefinery. Beyond carbon, we are appealing for more attention to the water requirement of bioinnovations to ensure a sustainable future.
RESULTS
PCA accumulation during cell growth on 4-hydroxybenzoate
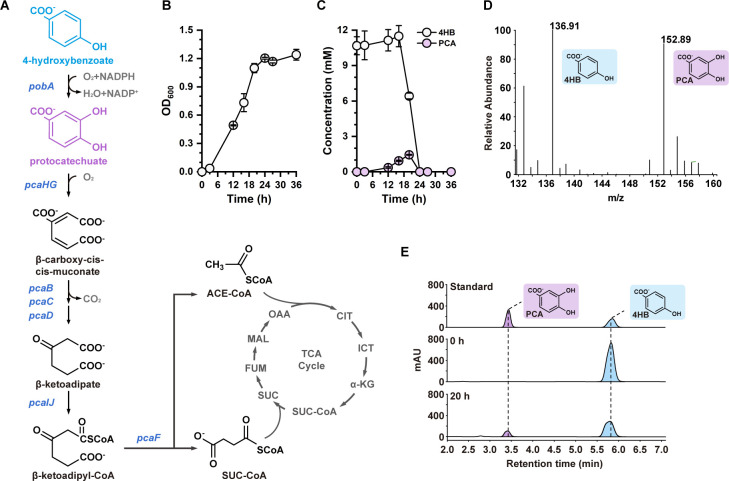
The protocatechuate branch of the β-ketoadipate pathway is the main pathway for the metabolism of lignin-derived aromatic molecules (31, 32). Going through this pathway, the aromatic carbon goes to the tricarboxylic acid (TCA) cycle with two key metabolites, succinyl-CoA and acetyl-CoA, as the nodes, providing carbon and energy for the anabolism and catabolism in the cell (Fig. 1A). To evaluate the potentials of R. nubinhibens for the degradation of lignin-derived monomers, we selected 4HB, a representative and readily available lignin-derived aromatic compound monomer, as the substrate and grew the strain in a marine basal medium (MBM). Notably, R. nubinhibens demonstrated robust growth with 4HB as the sole carbon source without extra organic substrates, such as glucose, to power the metabolism (Fig. 1B and C). Interestingly, during the cultivation of the wild-type R. nubinhibens in the basal medium with 4HB, a gradual accumulation of a violet compound was observed with the growth of cells in the first 20 h (Fig. S1). Subsequent characterization via mass spectrometry (MS) and high performance liquid chromatography (HPLC) confirmed the identity of this compound as PCA (Fig. 1D and E; Fig. S2), a central metabolite in the β-ketoadipate pathway and a pharmaceutical compound with antibacterial, antioxidant, anticancer, and anti-inflammatory potentials, which has been demonstrated both in vitro and in vivo (33, 34). PCA can also be a platform compound for the production of diverse value-added compounds with significance in chemical, food, and pharmaceutical industries, such as cis,cis-muconic acid, adipic acid, and levulinic acid (35–37). However, conventional PCA production relies on extraction from plants with low yield and high cost (38). This observation implied an unusually high catalytic capability of the native 4-hydroxybenzoate 3-monooxygenase, PobA, for 4HB hydroxylation to PCA, which is usually a limiting step for aromatic utilization (39–41). Therefore, a rational allocation of carbon flux from 4HB would be possible to maximize the biosynthesis of PCA and sustain cell growth simultaneously. This can be realized using a CRISPRi system as a carbon-flux-limiting stopcock to restrain the flux toward the TCA cycle by downregulating relevant genes downstream of pobA (Fig. 1A).
Fig 1.
PCA accumulation during R. nubinhibens growth on 4HB. (A) The protocatechuate branch of β-ketoadipate metabolic pathway in R. nubinhibens. CIT, citrate; ICT, isocitrate; α-KG, alpha-ketoglutarate; SUC-CoA, succinyl-CoA; SUC, succinate; FUM, fumarate; MAL, malate; OAA, oxaloacetate; ACE-CoA, acetyl-CoA. (B) Growth profile. (C) Utilization of 4HB and accumulation of PCA in the wild-type R. nubinhibens with 4HB as the sole carbon source. Experiments were carried out in triplicate, and the error bars represented the standard deviations of the means of three biological replicates. (D) MS analysis of samples. (E) HPLC profile of standard and samples.
Characterization of a lacI-Ptrc inducible system in R. nubinhibens
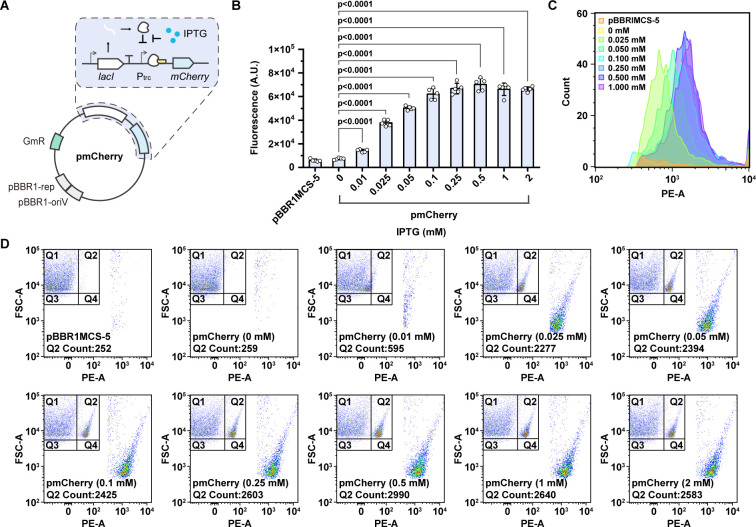
One of the key advantages of CRISPRi is that the repression of target genes is tunable, which usually relies on an inducible system based on transcriptional regulators. Currently, only countable inducible systems have been reported for the Roseobacter clade bacteria (42), and no inducible system has been established for the marine R. nubinhibens. As such, we designed and constructed a lacI-Ptrc inducible system consisting of the lacI gene coding for the repressor LacI and the corresponding promoter Ptrc driving the expression of red fluorescent protein, mCherry, as a reporter. Without inducers, LacI would bind to the operator region of the Ptrc promoter blocking the expression of mCherry, while, when isopropyl-β-D-thiogalactopyranoside (IPTG) is supplemented, the repressor would ligand to IPTG instead and release the promoter for the transcription of mCherry (Fig. 2A).
Fig 2.
The lacI-Ptrc inducible system for R. nubinhibens. (A) Schematic representation of the pmCherry. The lacI gene is driven by a constitutive promoter, and the mCherry fluorescence gene is driven by the Ptrc inducible promoter. (B) Fluorescence of strains with pBBR1MCS-5, as a control, and pmCherry under different IPTG concentrations. The value of fluorescence intensity (A.U., arbitrary unit) was normalized by optical density at 600 nm with a final OD600 of 0.01. Experiments were carried out in quintuplicate, and the error bars represented the standard deviations of the means of five biological replicates. The differences were statistically evaluated by the t-test. (C) Count of mCherry fluorescent cells. (D) Distribution of mCherry fluorescent cells in the population. For each sample, 10,000 events were analyzed.
To assess the functionality of the lacI-Ptrc inducible system, we monitored the fluorescence of R. nubinhibens with pmCherry (Table S1) and the control plasmid (pBBR1MCS-5; Table S1) via fluorimetry and flow cytometry. Without IPTG, both strains showed a similarly low level of background fluorescence. We tested eight different concentrations of IPTG from 0.01 to 2 mM. The strain displayed a gradually increased signal of fluorescence along with the rising concentration of IPTG from 0.01 to 0.5 mM, and the signals indicated a saturation of IPTG induction when the concentration reached 0.5 mM (Fig. 2B). The proportion of fluorescent cells reached saturation at 29.9% with 0.5 mM of IPTG (Fig. 2C and D). The partially induced cell population might result from the heterologously adapted transcriptional regulator or the distinct uptake behavior of IPTG in this marine chassis. This is the first demonstration of an inducible system in R. nubinhibens, making dynamic regulation of designed genetic modules possible.
Modular design of a CRISPR interference system for R. nubinhibens
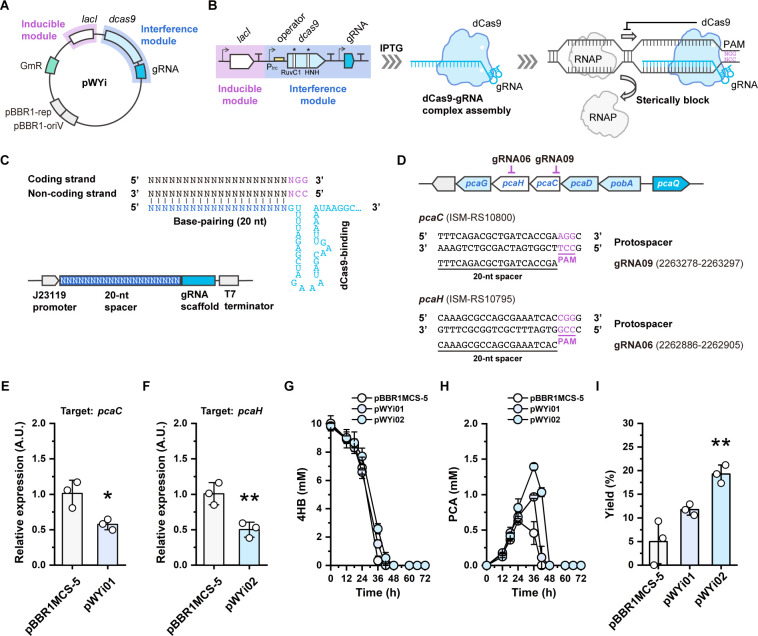
To implement a CRISPRi system in R. nubinhibens, we modularly assembled the inducible module based on the lacI-Ptrc system and an interference module containing dcas9 and the gRNA cassette, carried by the pBBR1MCS-5 vector plasmid (Fig. 3A). In the design, dCas9 driven by the lacI-Ptrc system reserves the ability to bind the target DNA (43–45), and navigated by a programmable gRNA, it binds to the target and sterically blocks RNA polymerase (RNAP), thereby interfering the transcription process and eventually repressing the expression of the target gene (Fig. 3B) (46–48). The gRNA cassette can be programmed for any genes of interest by substitution of the 20-nt spacer via a one-step PCR protocol (Fig. 3C).
Fig 3.
CRISPRi system for R. nubinhibens. (A) Design of the working plasmid pWYi. The pWYi plasmid was comprised of the inducible module (lacI-Ptrc inducible system) and the CRISPR interference module (dcas9 and the gRNA cassette). (B) Schematic representation of the working principle of CRISPRi. The gRNA and dCas9 (D10A and H840A) complex bind to the target DNA and sterically block the RNAP. The site of protospacer adjacent motif was highlighted in violet. (C) Structure of the gRNA cassette targeting the coding strand. The sequences of the 20-nt base-pairing region of the gRNA are identical to the coding strand. The base-pairing region was indicated in navy blue, and the dCas9 binding region was indicated in sky blue. (D) Positions of the gRNAs targeting the pca gene cluster. (E) Relative expression of target genes in the strain with pWYi01 and (F) pWYi02. Samples were taken at the exponential phase (OD600 of 0.5) with 0.5 mM IPTG, and the strain with pBBR1MCS-5 was included as the control. (G) Utilization of 4HB. (H) Titer of PCA. (I) Molar yield (%) of PCA. Experiments were carried out in triplicate, and the error bars represented the standard deviations of the means of three biological replicates. The differences were statistically evaluated by the t-test (*P < 0.05; **P < 0.01).
As a proof of concept, we attempted to target two genes, pcaC and pcaH, in the genome, respectively. Both genes are involved in the β-ketoadipate pathway, which has been identified as the primary route to degrade aromatic compounds in R. nubinhibens (23). Specifically, the pcaC gene encodes the γ-carboxymuconolactone decarboxylase and pcaH encodes the protocatechuate 3,4-dioxygenase β-subunit catalyzing the ring cleavage together with PcaG (Fig. 1A). We constructed two working plasmids, pWYi01 (with gRNA09; Table S1) and pWYi02 (with gRNA06; Table S1) targeting pcaC and pcaH, respectively (Fig. 3D). IPTG (0.25 and 0.5 mM) were added to induce the expression of dCas9, and the expression of target genes was determined by quantitative real-time PCR (RT-qPCR). As expected, the expression of pcaC and pcaH in strains with pWYi01 or pWYi02 significantly decreased by 43.13% and 50.26%, respectively, compared to strains with the control plasmid under 0.5 mM IPTG induction (Fig. 3E and F). The repression degree with 0.5 mM IPTG was, though marginal, higher than that with 0.25 mM IPTG, implying a higher expression level of dCas9, which is in agreement with our characterization above (Fig. S3). These results demonstrated that the CRISPRi system was successfully established.
The pcaH and pcaC genes, which we targeted with CRISPRi, are located downstream of pobA in the pca gene cluster. The repression of these downstream genes is expected to promote the accumulation of upstream metabolites (Fig. 1A and 3D). So, we tested the strains with the CRISPRi system targeting pcaH and pcaC, respectively, to evaluate the conversion of 4HB to PCA. Both strains were cultivated in shake flasks with 4HB as the sole substrate and induced with 0.5 mM IPTG. Compared with the control, both strains with CRISPRi showed slower growth rates and 4HB consumption (Fig. 3G; Fig. S4). The strain with the control plasmid pBBR1MCS-5 produced PCA at a titer of 0.46 ± 0.43 mM and a molar yield of 5.00 ± 4.69% at 36 h (Fig. 3H and I; Table S2). As expected, the strains with CRISPRi displayed higher performance with improved titer and yield of PCA. The strain expressing pWYi02 (targeting pcaH) exhibited a 3.85-fold increase in PCA yield, while that of the strain expressing pWYi01 (targeting pcaC) increased by 2.35-fold than the control strain (Fig. 3I). Specifically, the strain with CRISPRi repressing pcaH and pcaC produced 1.39 ± 0.06 and 0.97 ± 0.03 mM of PCA with a molar yield of 19.27 ± 1.93% and 11.74 ± 1.16% at 36 h, respectively (Fig. 3H and I; Table S2). Under this condition, strains with 0.25 and 0.5 mM IPTG showed similar performance in the growth rate (Fig. S5) and 4HB consumption and accumulation of PCA (Fig. S6 and S7), indicating that the difference between these two inducible strengths is marginal. These results demonstrated that the CRISPRi system efficiently allocated the carbon flux and could be modified as a powerful tool for the dynamical regulation of bioproduction in R. nubinhibens. However, the CRISPRi system with only one target was not sufficient, and further improvement is necessary.
Multiplex CRISPRi enables enhanced biosynthesis of PCA
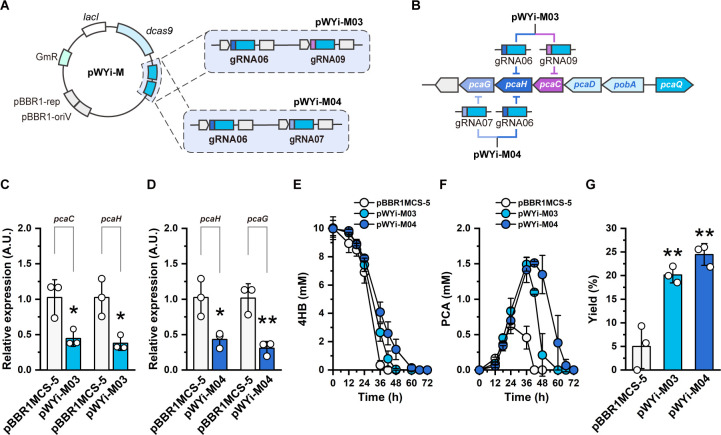
To further enhance the accumulation of PCA, we developed a multiplex CRISPRi system with tandem gRNA cassettes targeting different genes in the operon simultaneously. We selected pcaG, encoding the protocatechuate 3,4-dioxygenase α-subunit, which directly catalyzes the PCA ring-opening process together with PcaH, as another target. The tandem gRNA modules were generated by assembling two gRNA cassettes (Fig. 4A). The resulting working plasmids pWYi-M03 (with gRNA09 and gRNA06; Table S1) and pWYi-M04 (with gRNA06 and gRNA07; Table S1) target pcaC and pcaH or pcaH and pcaG, respectively (Fig. 4B). We first evaluated the expression of the target genes at the transcriptional level. As intended, all strains with CRISPRi exhibited a downregulated expression of the target genes. The expression of target genes in strains harboring the multiplex CRISPRi system was lower than that of strains with a single target (Fig. 4C and D). The strain with pWYi-M03 descended the expression of pcaC and pcaH by 22.38% and 24.47%, respectively, compared to the strain with one gRNA (Fig. 4C). The multiplex CRISPRi system regulating the pcaHG genes displayed the strongest interference and declined the expression of pcaH and pcaG by 57.80% and 69.69% than the control, respectively (Fig. 4D). These data indicated the stronger interference of genes at the RNA level with multiple locus targeting than single sites.
Fig 4.
Multiplex CRISPRi for R. nubinhibens. (A) Design of pWYi-M. The multiple gRNAs module consisted of two tandem gRNA cassettes. (B) Positions of gRNAs targeting in the pca gene cluster. (C) Relative expression of target genes in the strain with pWYi-M03 and (D) pWYi-M04. Samples were taken at the exponential phase with 0.5 mM IPTG induction, and the strain with pBBR1MCS-5 was included as the control. (E) Utilization of 4HB. (F) Titer of PCA. (G) Molar yield (%) of PCA. Experiments were carried out in triplicate, and the error bars represented the standard deviations of the means of three biological replicates. The differences were statistically evaluated by the t-test (*P < 0.05; **P < 0.01).
With the multiplex CRISPRi system, we found that the growth of strains with all designed CRISPRi systems was repressed, while the multiplex CRISPRi targeting pcaHG resulted in the lowest growth rate (Fig. S8). Consistent with the growth rate, multiplex CRISPRi showed stronger interference in the utilization of 4HB than single-targeting CRISPRi (Fig. 4E; Table S3). PCA production was raised in the strain with pWYi-M03 (targeting pcaC and pcaH) at a titer of 1.49 ± 0.12 mM at 36 h, an increase of 1.54-fold and 1.07-fold over the strain with pWYi01 and pWYi02, respectively (Fig. 4F; Table S3). The maximum titer of PCA appeared in the strain harboring pWYi-M04 (targeting pcaH and pcaG) with 1.51 ± 0.03 mM at 42 h and raised 13.45-fold and 1.46-fold than the strain individually targeting pcaC and pcaH, respectively (Fig. 4F; Table S3). We calculated the molar yields of each strain at 36 h, and the strain with pWYi-M04 exhibited the highest yield at 24.47 ± 2.30%, a 2.08-fold and 1.27-fold increase over the strain with pWYi01 and pWYi02, respectively (Fig. 4G; Table S3). Yet, little improvement of the yield was observed in the strain with pWYi-M03 repressing pcaC and pcaH when compared to the strain only repressing pcaH alone (Tables S2 and S3). This can be explained by analyzing the β-ketoadipate pathway. PCA was catalyzed into β-carboxy-cis-cis-muconate by PcaG and PcaH. As a result, interference of these enzymes, compared to PcaC, would directly impede the aromatic ring cleavage, therefore promoting the accumulation of PCA. While the strain with the control plasmid only produced PCA with the maximum yield at 5.00 ± 4.69% at 36 h, the strain with pWYi-M04 showed an increase of 4.89-fold than the control (Fig. 4G; Table S3). To be noticed, we also observed that all PCA produced by the control strain was consumed between 36 and 42 h. In contrast, PCA synthesized by the reprogrammed strain with pWYi-M04 was retained in the culture for 72 h. We prolonged nearly double the accumulation time of the target product compared to the control strain, providing biosynthesis with an extended time to harvest the bioproducts or terminate the process at the maximal titer of bioproducts.
Blue valorization of 4HB in real seawater
Real seawater is more complex than defined marine synthetic medium, and it is essential to evaluate our system in real seawater for blue valorization. To do so, we collected seawater in the coastal area in Qingdao city, China, and we cultivated the strain harboring pWYi-M04 with tandem gRNAs targeting pcaH and pcaG in seawater medium. Similar to the phenomenon in the defined synthetic medium, the strain with multiplex CRISPRi showed higher PCA production compared to the strain with control plasmid pBBR1MCS-5 in seawater (Fig. 5A and B; Fig. S9). As expected, the strain harboring pWYi-M04 exhibited increased PCA titer with the maximum titer at 0.92 ± 0.11 mM appearing at 60 h and raised 2.09-fold than the maximum titer of the control strain (0.44 ± 0.07 mM PCA) (Fig. 5C). In addition, our results showed that the strain with repressed pcaH and pcaG displayed higher molar yield of PCA (16.07 ± 5.29%) with a 2.10-fold increase (P = 0.09) over the control of which the yield is 7.64 ± 1.47% (Fig. 5D). Notably, the rate of 4HB consumption and PCA production in seawater medium was slower than that in marine basal medium, and we observed a slight drop of the production of PCA compared to that in the marine basal media by the same strain with pWYi-M04, which may be a result of the complexity of seawater. These results demonstrated the potential of the oceans as a water resource for bioprocesses, indicating the feasibility of blue valorization by reprogramming marine bacteria.
Fig 5.
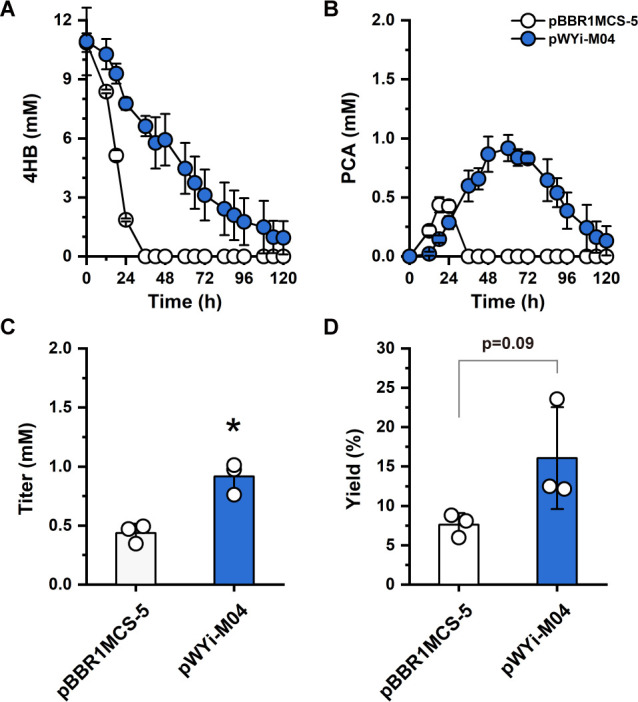
PCA production of the strain with pWYi-M04 in real seawater. (A) Utilization of 4HB. (B) Titer of PCA. (C) The maximum titer of PCA. (D) Molar yield (%) of PCA. Experiments were carried out in triplicate, and the error bars represented the standard deviations of the means of three biological replicates. The differences were statistically evaluated by the t-test (*P < 0.05).
DISCUSSION
In this study, we report a customized CRISPRi system to reprogram the marine bacterium R. nubinhibens for the biological valorization of lignin-derived aromatic carbon into value-added chemicals without requesting extra organic carbon and freshwater. The strategy was demonstrated in both defined synthetic medium, such as MBM, and real seawater. Our findings highlight the unique advantages offered by Roseobacter clade bacteria, particularly in the context of lignin valorization. An obvious benefit is the high catalytical activity of PobA in R. nubinhibens, which is no longer a bottleneck but leads to the accumulation of PCA in the β-ketoadipate pathway (Fig. 1C). By using CRISPRi, we successfully targeted the key genes (pcaC, pcaH, and pcaG) in the β-ketoadipate pathway individually or in tandem, generating a carbon-flux-limiting stopcock for rational carbon reallocation. By doing so, we maximized the carbon accumulation before the ring-cleavage step as PCA and allowed minimal flux going to the TCA cycle to sustain cell growth and provide energy for biocatalysis. When targeting pcaH and pcaG simultaneously, the conversion of 4HB to PCA exhibited a 4.89-fold increase in molar yield (Fig. 4G), showing a great potential of the blue biovalorization of lignin monomers via the dynamic regulation of marine bacteria with multiplex CRISPRi.
This is the first report of a CRISPR-based regulatory system for the marine Roseobacter clade bacteria, underscoring the power of CRISPRi in exploiting novel microbial chassis (49–51). Our work also provides a paradigm of expanding cutting-edge synthetic biology tools to marine bacteria and other non-traditional microbial hosts, which are exhibiting increasingly important roles in next-generation industrial biotechnology (52–54). Nevertheless, CRISPRi cannot achieve the insertion and deletion of DNA fragments. As a result, the accumulated PCA was eventually metabolized for cell growth if it was not harvested in a certain time frame, even though our CRISPRi system expanded the operational window to around 36–48 h without sacrificing the maximum yield and titer. A combination of gene regulation and editing would be beneficial for further advances via the introduction of heterologous metabolic pathways or the termination of unwanted endogenous pathways, and R. nubinhibens may be further equipped with designed new features (e.g., unnatural substrates or products) for enhanced upcycling of lignin-derived monomers. Besides, we would like to acknowledge that the maximum titer of PCA of R. nubinhibens was lower than that of traditional microbial chassis, such as C. glutamicum and P. putida. However, members of the Roseobacter clade can use lignin-derived aromatic compounds as the sole carbon source without demanding any extra carbon, and most of the strains are naturally tolerant to high salinity, making it possible and preferable to use seawater for cultivation and fermentation. Finally, the development of this novel microbial chassis is only in a very early stage, and the performance would be improved significantly with the advances in synthetic biology and the understanding of its metabolisms.
Upcycling carbon lost to the environment is vital for a net-zero future. Lignin-rich biomass commonly needs conventional disposal via incineration, leading to considerable carbon loss and air pollution, for instance, the emission of greenhouse gases CO2, NOx, and inhalable particles (e.g., PM10 and PM2.5) (55–57). In contrast, biological valorization has been considered a carbon-efficient, economically viable, and environmentally friendly alternative to upcycle lignin to high-performance bioproducts. Two long-standing but often overlooked issues in, but not limited to, lignin biovalorization remain. One is the requirement of extra organic carbon sources, e.g., glucose, during fermentation (12, 39, 58), aggravating the “food-versus-fuel” debate. The other is the demand for water to cultivate engineered microbes, potentially rousing the competition of freshwater between humans and biosynthesis, especially under global water scarcity (59–61). Inspiringly, the development of marine microbial chassis for the valorization of lignin-derived carbon, to some extent, provides a solution to tackle these issues at once (62). We believe that blue biovalorization by engineered marine microbes will be a new wave for building a circular economy and contributing to multiple sustainable development goals (SDGs) related to the climate, carbon, and water, such as SDG6 (clean water and sanitation), SDG12 (responsible consumption and production), and SDG13 (climate action). Beyond these, blue biovalorization depending on seawater rather than the unequally distributed freshwater may also contribute to reducing inequality (SDG10) in the wave of biotechnological revolution.
MATERIALS AND METHODS
Strains and media
Escherichia coli DH5α (Takara Bio) was used for molecular cloning and cultivated in Luria–Bertani (LB) liquid media containing 10 g NaCl, 10 g tryptone, and 5 g yeast extract per liter or on LB solid agar (1.5%) plates at 37°C. When screening E. coli transformants and maintaining plasmids, appropriate antibiotics were added (100 µg mL−1 ampicillin and 20 µg mL−1 gentamicin). R. nubinhibens ISM (DSM 15170) was grown at 30°C and 180 rpm in Difco Marine Broth 2216 (MB2216) liquid media or on solid plates for general cultivation and in the MBM containing 200 mM NaCl, 50 mM MgSO4, 10 mM CaCl2, 10 mM KCl, 10 mM NH4Cl, 1 mM K2HPO4, 0.1 mM FeEDTA, 0.05% yeast extract, and 0.1% vitamin with 10 mM 4HB as the carbon source for biosynthesis. Wolfe’s Vitamin Solution (0.1%) was used in the MBM to provide vitamins for microbial growth. It contains 2.0 mg/L biotin, 2.0 mg/L folic acid, 10.0 mg/L pyridoxine hydrochloride, 5.0 mg/L thiamine⋅HCl, 5.0 mg/L riboflavin, 5.0 mg/L nicotinic acid, 5.0 mg/L calcium D-(+)-pantothenate, 0.1 mg/L vitamin B12, 5.0 mg/L p-aminobenzoic acid, and 5.0 mg/L thioctic acid. Seawater was collected in the coastal area at E120°69′96″ and N36°37′72″ near our campus in Qingdao city, China, and after filtration (0.22 µm), 0.05% yeast extract, 0.1% vitamin, and 10 mM 4HB were supplemented for cell growth and PCA biosynthesis. Gentamicin (20 µg mL−1) was used for isolating clones of R. nubinhibens.
Plasmid construction and electroporation
All plasmids used in this study are summarized in Table S1, and all primers used in this study are summarized in Table S4. All the gRNA cassettes were summarized in Table S5. The synthesized sequence of mCherry is listed in Table S6. The mCherry fragment carried by pQLL plasmid with AmpR was synthesized by Beijing Liuhe BGI and then amplified using PrimeSTAR Max DNA Polymerase (Takara Bio). The plasmid pmCherry was generated by fusing the lacI-Ptrc inducible system and mCherry into the vector pBBR1MCS-5 using In-Fusion Snap Assembly Master Mix (Takara Bio). All plasmids were extracted using the QIAprep Spin Miniprep Kit (Qiagen). The gRNA09 cassette targeting pcaC was built via inverse PCR using back-to-back primers containing the 20 bp spacer. Based on the plasmid pWY06 (30), we constructed pWYi02 by eliminating the deaminase, and then, pWYi01 was constructed by substituting the gRNA cassette. To generate multiplex CRISPRi working plasmids, the gRNA09 and gRNA07 cassettes were assembled in the pWYi02, forming pWYi-M03 and pWYi-M03, respectively.
For the transformation of working plasmids, the electrocompetent cells were prepared by washing twice using the buffered sucrose solution and stored at −80°C before use (30). The working plasmids were added to 80 µL competent cells in a pre-cooled 0.1 cm MicroPulser Electroporation Cuvette (Bio-Rad), and the mixture was treated in MicroPulser Electroporator (Bio-Rad) at the pulse intensity of 1.8 kV and resulting pulse length of 1.3–1.5 ms. Cells after electroporation were transferred to fresh media for recovery and then plated on a solid medium supplemented with 20 µg mL−1 gentamicin.
Fluorimetry and flow cytometry
The strains transformed with pBBRIMCS-5 and pmCherry were cultivated overnight in MB2216 medium with 20 µg mL−1 gentamicin. Next, aliquots of 2 mL suspensions were induced for 4 h with IPTG at different concentrations from 0 to 2 mM. After induction, the cells were harvested and washed twice using phosphate-buffered saline. For the fluorimetry assay, the resuspended cells with an OD600 of 1.0 were diluted 102 times to reach an OD600 of 0.01, and 200 µL of each bacterial suspension was transferred to 96-well microplates (Corning Costar). The OD600 and fluorescence intensity at an excitation wavelength of 590 nm and an emission wavelength of 645 nm were determined using Spark Multimode Microplate Reader (Tecan). The fluorescence intensity (arbitrary unit) was normalized for fair comparison. For flow cytometry, the resuspended cells were diluted by 103 times and then stored at 4°C until analysis. The mCherry fluorescence distribution at the population level was analyzed using BD FACS Aria Fusion (BD Biosciences) equipped with a 561 nm laser. Ten thousand single-cell events were collected for each sample. Data were processed using FlowJo (TreeStar Inc.).
Cultivation experiment
For biosynthesis, the strains from glycerol stocks were cultivated in MB2216 media overnight with appropriate antibiotics. Next, the culture was inoculated into 250 mL shake flasks containing 50 mL fresh marine basal medium with 4HB (10 mM) as the carbon source by 1:50 dilution and cultivated at 30°C and 180 rpm. Gentamicin (20 µg mL−1) was added to maintain the working plasmids, and IPTG (0.25 or 0.5 mM) was supplemented for induction. Samples were taken at different intervals and stored at −20°C until analysis. OD600 was measured to determine the cell growth profiles of each sample. The concentration of 4HB and PCA was quantified using HPLC as described below. At least two parallel biological experiments for each strain were performed, and the mean value with standard deviation was calculated and reported.
Quantification of mRNA expression levels
After induction for 24 h, cells at the exponential phase (OD600 of 0.5) with the CRISPRi systems were harvested by centrifugation at 10,000 rpm and frozen with liquid nitrogen and then stored at −80°C until extraction. Total RNA was extracted using the RNAprep Pure Cell/Bacteria Kit (Tiangen) and reverse-transcribed into cDNA using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio). The resulting cDNA was utilized for analysis using TB Green Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio). Quantitative PCR was performed using Applied Biosystems QuantStudio 5 (Thermo Fisher Scientific). To normalize the gene expression between different samples, the gene encoding for the 16S ribosomal RNA was employed as the housekeeping gene. The primers used for RT-qPCR are listed in Table S4.
Identification and quantification of 4HB and PCA
Samples were centrifuged at 10,000 rpm for 1 min, and the supernatants were sterilely filtered by 0.22 µm filter and diluted 10 times. For the identification of products, samples were demineralized by solid phase extraction using HyperSep C18 (Thermo Scientific) and analyzed using HPLC-MS (LCQ Fleet, Thermo Fisher Scientific). For quantification of 4HB and PCA, samples were detected by Agilent 1260 HPLC equipped with a UV detector (Agilent Technologies) at a wavelength of 210 nm with an injection volume of 10 µL. Chromatographic separation was realized by an EC-C18 column 4 µm, 4.6 × 100 µm (Agilent Technologies) at 25°C with the mobile phase comprised of 90% formic acid (0.1%) and 10% acetonitrile at a constant flow rate of 0.8 mL per min. The molar yield (%) of PCA was calculated by dividing the titers of PCA by 4HB.
ACKNOWLEDGMENTS
The authors thank Dr. Haiyan Yu for the assistance with flow cytometry analysis and Dr. Fanping Zhu for the help with HPLC-MS analysis. This work was supported by the National Natural Science Foundation of China (22278246, U20A20146, and 22378233), the Department of Science and Technology of Shandong Province (2022HWYQ-017), the Natural Science Foundation of Shandong Province (ZR2021ME066), the Qilu Young Scholar Program of Shandong University (to P.-F.X.), and the Taishan Scholars Project of Shandong Province (No. tstp20230604).
Contributor Information
Peng-Fei Xia, Email: pfxia@sdu.edu.cn.
Pablo Ivan Nikel, Danmarks Tekniske Universitet The Novo Nordisk Foundation Center for Biosustainability, Kgs. Lyngby, Denmark.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00890-24.
Tables S1 to S6; Figures S1 to S9.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Bugg TDH, Ahmad M, Hardiman EM, Rahmanpour R. 2011. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896. doi: 10.1039/c1np00042j [DOI] [PubMed] [Google Scholar]
- 2. Dong L, Wang Y, Dong Y, Zhang Y, Pan M, Liu X, Gu X, Antonietti M, Chen Z. 2023. Sustainable production of dopamine hydrochloride from softwood lignin. Nat Commun 14:4996. doi: 10.1038/s41467-023-40702-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lan HN, Liu RY, Liu ZH, Li X, Li BZ, Yuan YJ. 2023. Biological valorization of lignin to flavonoids. Biotechnol Adv 64:108107. doi: 10.1016/j.biotechadv.2023.108107 [DOI] [PubMed] [Google Scholar]
- 4. Beckham GT, Johnson CW, Karp EM, Salvachúa D, Vardon DR. 2016. Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53. doi: 10.1016/j.copbio.2016.02.030 [DOI] [PubMed] [Google Scholar]
- 5. De Wild PJ, Huijgen WJJ, Gosselink RJA. 2014. Lignin pyrolysis for profitable lignocellulosic biorefineries. Biofuels Bioprod Bioref 8:645–657. doi: 10.1002/bbb.1474 [DOI] [Google Scholar]
- 6. Wu X, Fan X, Xie S, Lin J, Cheng J, Zhang Q, Chen L, Wang Y. 2018. Solar energy-driven lignin-first approach to full utilization of lignocellulosic biomass under mild conditions. Nat Catal 1:772–780. doi: 10.1038/s41929-018-0148-8 [DOI] [Google Scholar]
- 7. Liu ZH, Hao N, Wang YY, Dou C, Lin F, Shen R, Bura R, Hodge DB, Dale BE, Ragauskas AJ, Yang B, Yuan JS. 2021. Transforming biorefinery designs with 'plug-in processes of lignin' to enable economic waste valorization. Nat Commun 12:3912. doi: 10.1038/s41467-021-23920-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu ZH, Li BZ, Yuan JS, Yuan YJ. 2022. Creative biological Lignin conversion routes toward lignin valorization. Trends Biotechnol 40:1550–1566. doi: 10.1016/j.tibtech.2022.09.014 [DOI] [PubMed] [Google Scholar]
- 9. Johnson CW, Salvachúa D, Rorrer NA, Black BA, Vardon DR, St. John PC, Cleveland NS, Dominick G, Elmore JR, Grundl N, Khanna P, Martinez CR, Michener WE, Peterson DJ, Ramirez KJ, Singh P, VanderWall TA, Wilson AN, Yi X, Biddy MJ, Bomble YJ, Guss AM, Beckham GT. 2019. Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule 3:1523–1537. doi: 10.1016/j.joule.2019.05.011 [DOI] [Google Scholar]
- 10. Ling C, Peabody GL, Salvachúa D, Kim Y-M, Kneucker CM, Calvey CH, Monninger MA, Munoz NM, Poirier BC, Ramirez KJ, St John PC, Woodworth SP, Magnuson JK, Burnum-Johnson KE, Guss AM, Johnson CW, Beckham GT. 2022. Muconic acid production from glucose and xylose in Pseudomonas putida via evolution and metabolic engineering. Nat Commun 13:4925. doi: 10.1038/s41467-022-32296-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Werner AZ, Cordell WT, Lahive CW, Klein BC, Singer CA, Tan ECD, Ingraham MA, Ramirez KJ, Kim DH, Pedersen JN, Johnson CW, Pfleger BF, Beckham GT, Salvachúa D. 2023. Lignin conversion to β-Ketoadipic acid by Pseudomonas putida via metabolic engineering and bioprocess development. Sci Adv 9:eadj0053. doi: 10.1126/sciadv.adj0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weiland F, Barton N, Kohlstedt M, Becker J, Wittmann C. 2023. Systems metabolic engineering upgrades Corynebacterium glutamicum to high-efficiency cis,cis-muconic acid production from lignin-based aromatics. Metab Eng 75:153–169. doi: 10.1016/j.ymben.2022.12.005 [DOI] [PubMed] [Google Scholar]
- 13. Spence EM, Calvo-Bado L, Mines P, Bugg TDH. 2021. Metabolic engineering of Rhodococcus jostii RHA1 for production of pyridine-dicarboxylic acids from lignin. Microb Cell Fact 20:15. doi: 10.1186/s12934-020-01504-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye JW, Lin YN, Yi XQ, Yu ZX, Liu X, Chen GQ. 2023. Synthetic biology of extremophiles: a new wave of biomanufacturing. Trends Biotechnol 41:342–357. doi: 10.1016/j.tibtech.2022.11.010 [DOI] [PubMed] [Google Scholar]
- 15. Chen GQ, Jiang XR. 2018. Next generation industrial biotechnology based on extremophilic bacteria. Curr Opin Biotechnol 50:94–100. doi: 10.1016/j.copbio.2017.11.016 [DOI] [PubMed] [Google Scholar]
- 16. Scanlon BR, Fakhreddine S, Rateb A, de Graaf I, Famiglietti J, Gleeson T, Grafton RQ, Jobbagy E, Kebede S, Kolusu SR, Konikow LF, Long D, Mekonnen M, Schmied HM, Mukherjee A, MacDonald A, Reedy RC, Shamsudduha M, Simmons CT, Sun A, Taylor RG, Villholth KG, Vörösmarty CJ, Zheng C. 2023. Global water resources and the role of groundwater in a resilient water future. Nat Rev Earth Environ 4:87–101. doi: 10.1038/s43017-022-00378-6 [DOI] [Google Scholar]
- 17. Schyns JF, Hoekstra AY, Booij MJ, Hogeboom RJ, Mekonnen MM. 2019. Limits to the world’s green water resources for food, feed, fiber, timber, and bioenergy. Proc Natl Acad Sci U S A 116:4893–4898. doi: 10.1073/pnas.1817380116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu J, Yang H, Gosling SN, Kummu M, Flörke M, Pfister S, Hanasaki N, Wada Y, Zhang X, Zheng C, Alcamo J, Oki T. 2017. Water scarcity assessments in the past, present and future. Earths Future 5:545–559. doi: 10.1002/2016EF000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paoli L, Ruscheweyh H-J, Forneris CC, Hubrich F, Kautsar S, Bhushan A, Lotti A, Clayssen Q, Salazar G, Milanese A, et al. 2022. Biosynthetic potential of the global ocean microbiome. Nature 607:111–118. doi: 10.1038/s41586-022-04862-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Q, Hotta K, Deng Y, Yuan R, Quan S, Chen X. 2021. Advances in biosynthesis of natural products from marine microorganisms. Microorganisms 9:2551. doi: 10.3390/microorganisms9122551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lyu C, Chen T, Qiang B, Liu N, Wang H, Zhang L, Liu Z. 2021. CMNPD: a comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res 49:D509–D515. doi: 10.1093/nar/gkaa763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imhoff JF, Labes A, Wiese J. 2011. Bio-mining the microbial treasures of the ocean: new natural products. Biotechnol Adv 29:468–482. doi: 10.1016/j.biotechadv.2011.03.001 [DOI] [PubMed] [Google Scholar]
- 23. Buchan A, Neidle EL, Moran MA. 2004. Diverse organization of genes of the β-ketoadipate pathway in members of the marine Roseobacter lineage. Appl Environ Microbiol 70:1658–1668. doi: 10.1128/AEM.70.3.1658-1668.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A. 2007. Ecological genomics of marine Roseobacters. Appl Environ Microbiol 73:4559–4569. doi: 10.1128/AEM.02580-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo H, Moran MA. 2014. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78:573–587. doi: 10.1128/MMBR.00020-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chu X, Li S, Wang S, Luo D, Luo H. 2021. Gene loss through pseudogenization contributes to the ecological diversification of a generalist Roseobacter lineage. ISME J 15:489–502. doi: 10.1038/s41396-020-00790-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piekarski T, Buchholz I, Drepper T, Schobert M, Wagner-Doebler I, Tielen P, Jahn D. 2009. Genetic tools for the investigation of Roseobacter clade bacteria. BMC Microbiol 9:265. doi: 10.1186/1471-2180-9-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith AF, Rihtman B, Stirrup R, Silvano E, Mausz MA, Scanlan DJ, Chen Y. 2019. Elucidation of glutamine lipid biosynthesis in marine bacteria reveals its importance under phosphorus deplete growth in Rhodobacteraceae. ISME J 13:39–49. doi: 10.1038/s41396-018-0249-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li C-Y, Mausz MA, Murphy A, Zhang N, Chen X-L, Wang S-Y, Gao C, Aguilo-Ferretjans MM, Silvano E, Lidbury IDEA, Fu H-H, Todd JD, Chen Y, Zhang Y-Z. 2023. Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria. ISME J 17:579–587. doi: 10.1038/s41396-023-01375-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei Y, Feng LJ, Yuan XZ, Wang SG, Xia PF. 2023. Developing a base editing system for marine Roseobacter clade bacteria. ACS Synth Biol 12:2178–2186. doi: 10.1021/acssynbio.3c00259 [DOI] [PubMed] [Google Scholar]
- 31. Alejandro-Marín CM, Bosch R, Nogales B. 2014. Comparative genomics of the protocatechuate branch of the β-ketoadipate pathway in the Roseobacter lineage. Mar Genomics 17:25–33. doi: 10.1016/j.margen.2014.05.008 [DOI] [PubMed] [Google Scholar]
- 32. Yamanashi T, Kim SY, Hara H, Funa N. 2015. In vitro reconstitution of the catabolic reactions catalyzed by PcaHG, PcaB, and PcaL: the protocatechuate branch of the β-ketoadipate pathway in Rhodococcus jostii RHA1. Biosci Biotechnol Biochem 79:830–835. doi: 10.1080/09168451.2014.993915 [DOI] [PubMed] [Google Scholar]
- 33. Krzysztoforska K, Mirowska-Guzel D, Widy-Tyszkiewicz E. 2019. Pharmacological effects of protocatechuic acid and its therapeutic potential in neurodegenerative diseases: review on the basis of in vitro and in vivo studies in rodents and humans. Nutr Neurosci 22:72–82. doi: 10.1080/1028415X.2017.1354543 [DOI] [PubMed] [Google Scholar]
- 34. Song J, He Y, Luo C, Feng B, Ran F, Xu H, Ci Z, Xu R, Han L, Zhang D. 2020. New progress in the pharmacology of protocatechuic acid: a compound ingested in daily foods and herbs frequently and heavily. Pharmacol Res 161:105109. doi: 10.1016/j.phrs.2020.105109 [DOI] [PubMed] [Google Scholar]
- 35. Shanks BH, Keeling PL. 2017. Bioprivileged molecules: creating value from biomass. Green Chem 19:3177–3185. doi: 10.1039/C7GC00296C [DOI] [Google Scholar]
- 36. Lo TM, Hwang IY, Cho HS, Fedora RE, Chng SH, Choi WJ, Chang MW. 2021. Biosynthesis of commodity chemicals from oil palm empty fruit bunch lignin. Front Microbiol 12:663642. doi: 10.3389/fmicb.2021.663642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kohlstedt M, Starck S, Barton N, Stolzenberger J, Selzer M, Mehlmann K, Schneider R, Pleissner D, Rinkel J, Dickschat JS, Venus J, B J H van Duuren J, Wittmann C. 2018. From lignin to nylon: cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab Eng 47:279–293. doi: 10.1016/j.ymben.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 38. Kogure T, Suda M, Hiraga K, Inui M. 2021. Protocatechuate overproduction by Corynebacterium glutamicum via simultaneous engineering of native and heterologous biosynthetic pathways. Metab Eng 65:232–242. doi: 10.1016/j.ymben.2020.11.007 [DOI] [PubMed] [Google Scholar]
- 39. Kuatsjah E, Johnson CW, Salvachúa D, Werner AZ, Zahn M, Szostkiewicz CJ, Singer CA, Dominick G, Okekeogbu I, Haugen SJ, Woodworth SP, Ramirez KJ, Giannone RJ, Hettich RL, McGeehan JE, Beckham GT. 2022. Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate. Metab Eng 70:31–42. doi: 10.1016/j.ymben.2021.12.010 [DOI] [PubMed] [Google Scholar]
- 40. Salvachúa D, Johnson CW, Singer CA, Rohrer H, Peterson DJ, Black BA, Knapp A, Beckham GT. 2018. Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem 20:5007–5019. doi: 10.1039/C8GC02519C [DOI] [Google Scholar]
- 41. Sonoki T, Morooka M, Sakamoto K, Otsuka Y, Nakamura M, Jellison J, Goodell B. 2014. Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J Biotechnol 192:71–77. doi: 10.1016/j.jbiotec.2014.10.027 [DOI] [PubMed] [Google Scholar]
- 42. Schuster LA, Reisch CR. 2021. A plasmid toolbox for controlled gene expression across the proteobacteria. Nucleic Acids Res 49:7189–7202. doi: 10.1093/nar/gkab496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196. doi: 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu J, Cheng ZH, Min D, Cheng L, He RL, Liu DF, Li WW. 2020. CRISPRi system as an efficient, simple platform for rapid identification of genes involved in pollutant transformation by Aeromonas hydrophila. Environ Sci Technol 54:3306–3315. doi: 10.1021/acs.est.9b07191 [DOI] [PubMed] [Google Scholar]
- 45. Huang H-H, Bellato M, Qian Y, Cárdenas P, Pasotti L, Magni P, Del Vecchio D. 2021. dCas9 regulator to neutralize competition in CRISPRi circuits. Nat Commun 12:1692. doi: 10.1038/s41467-021-21772-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Todor H, Silvis MR, Osadnik H, Gross CA. 2021. Bacterial CRISPR screens for gene function. Curr Opin Microbiol 59:102–109. doi: 10.1016/j.mib.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peters JM, Koo BM, Patino R, Heussler GE, Hearne CC, Qu J, Inclan YF, Hawkins JS, Lu CHS, Silvis MR, Harden MM, Osadnik H, Peters JE, Engel JN, Dutton RJ, Grossman AD, Gross CA, Rosenberg OS. 2019. Enabling genetic analysis of diverse bacteria with mobile-CRISPRi. Nat Microbiol 4:244–250. doi: 10.1038/s41564-018-0327-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu Y, Pinto F, Wan X, Yang Z, Peng S, Li M, Cooper JM, Xie Z, French CE, Wang B. 2022. Reprogrammed tracrRNAs enable repurposing of RNAs as crRNAs and sequence-specific RNA biosensors. Nat Commun 13:1937. doi: 10.1038/s41467-022-29604-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kozaeva E, Nielsen ZS, Nieto-Domínguez M, Nikel PI. 2024. The pAblo.pCasso self-curing vector toolset for unconstrained cytidine and adenine base-editing in Gram-negative bacteria. Nucleic Acids Res 52:e19. doi: 10.1093/nar/gkad1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martínez-García E, Fraile S, Algar E, Aparicio T, Velázquez E, Calles B, Tas H, Blázquez B, Martín B, Prieto C, Sánchez-Sampedro L, Nørholm MHH, Volke DC, Wirth NT, Dvořák P, Alejaldre L, Grozinger L, Crowther M, Goñi-Moreno A, Nikel PI, Nogales J, deLorenzo V. 2023. SEVA 4.0: an update of the standard European vector architecture database for advanced analysis and programming of bacterial phenotypes. Nucleic Acids Research 51:D1558–D1567. doi: 10.1093/nar/gkac1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Volke DC, Orsi E, Nikel PI. 2023. Emergent CRISPR-Cas-based technologies for engineering non-model bacteria. Curr Opin Microbiol 75:102353. doi: 10.1016/j.mib.2023.102353 [DOI] [PubMed] [Google Scholar]
- 52. Chen Y, Chen XY, Du HT, Zhang X, Ma YM, Chen JC, Ye JW, Jiang XR, Chen GQ. 2019. Chromosome engineering of the TCA cycle in Halomonas bluephagenesis for production of copolymers of 3-hydroxybutyrate and 3-hydroxyvalerate (PHBV). Metab Eng 54:69–82. doi: 10.1016/j.ymben.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 53. Jiang XR, Yan X, Yu LP, Liu XY, Chen GQ. 2021. Hyperproduction of 3-hydroxypropionate by Halomonas bluephagenesis. Nat Commun 12:1513. doi: 10.1038/s41467-021-21632-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Z, Qin Q, Zheng Y, Li F, Zhao Y, Chen GQ. 2021. Engineering the permeability of Halomonas bluephagenesis enhanced its chassis properties. Metab Eng 67:53–66. doi: 10.1016/j.ymben.2021.05.010 [DOI] [PubMed] [Google Scholar]
- 55. Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE. 2014. Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. doi: 10.1126/science.1246843 [DOI] [PubMed] [Google Scholar]
- 56. Liu Q, Kawai T, Inukai Y, Aoki D, Feng Z, Xiao Y, Fukushima K, Lin X, Shi W, Busch W, Matsushita Y, Li B. 2023. A lignin-derived material improves plant nutrient bioavailability and growth through its metal chelating capacity. Nat Commun 14:4866. doi: 10.1038/s41467-023-40497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi SY, Lee Y, Yu HE, Cho IJ, Kang M, Lee SY. 2023. Sustainable production and degradation of plastics using microbes. Nat Microbiol 8:2253–2276. doi: 10.1038/s41564-023-01529-1 [DOI] [PubMed] [Google Scholar]
- 58. Johnson CW, Salvachúa D, Khanna P, Smith H, Peterson DJ, Beckham GT. 2016. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab Eng Commun 3:111–119. doi: 10.1016/j.meteno.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Greve P, Kahil T, Mochizuki J, Schinko T, Satoh Y, Burek P, Fischer G, Tramberend S, Burtscher R, Langan S, Wada Y. 2018. Global assessment of water challenges under uncertainty in water scarcity projections. Nat Sustain 1:486–494. doi: 10.1038/s41893-018-0134-9 [DOI] [Google Scholar]
- 60. He C, Liu Z, Wu J, Pan X, Fang Z, Li J, Bryan BA. 2021. Future global urban water scarcity and potential solutions. Nat Commun 12:4667. doi: 10.1038/s41467-021-25026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoon J, Klassert C, Selby P, Lachaut T, Knox S, Avisse N, Harou J, Tilmant A, Klauer B, Mustafa D, Sigel K, Talozi S, Gawel E, Medellín-Azuara J, Bataineh B, Zhang H, Gorelick SM. 2021. A coupled human-natural system analysis of freshwater security under climate and population change. Proc Natl Acad Sci U S A 118:e2020431118. doi: 10.1073/pnas.2020431118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schada von Borzyskowski L. 2023. Taking synthetic biology to the seas: from blue chassis organisms to marine aquaforming. ChemBioChem 24:e202200786. doi: 10.1002/cbic.202200786 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S6; Figures S1 to S9.