Abstract
Objective
An enlarged cup-to-disc ratio (CDR) is a hallmark of glaucomatous optic neuropathy. Manual assessment of the CDR may be less accurate and more time-consuming than automated methods. Here, we sought to develop and validate a deep learning–based algorithm to automatically determine the CDR from fundus images.
Design
Algorithm development for estimating CDR using fundus data from a population-based observational study.
Participants
A total of 181 768 fundus images from the United Kingdom Biobank (UKBB), Drishti_GS, and EyePACS.
Methods
FastAI and PyTorch libraries were used to train a convolutional neural network–based model on fundus images from the UKBB. Models were constructed to determine image gradability (classification analysis) as well as to estimate CDR (regression analysis). The best-performing model was then validated for use in glaucoma screening using a multiethnic dataset from EyePACS and Drishti_GS.
Main Outcome Measures
The area under the receiver operating characteristic curve and coefficient of determination.
Results
Our gradability model vgg19_batch normalization (bn) achieved an accuracy of 97.13% on a validation set of 16 045 images, with 99.26% precision and area under the receiver operating characteristic curve of 96.56%. Using regression analysis, our best-performing model (trained on the vgg19_bn architecture) attained a coefficient of determination of 0.8514 (95% confidence interval [CI]: 0.8459–0.8568), while the mean squared error was 0.0050 (95% CI: 0.0048–0.0051) and mean absolute error was 0.0551 (95% CI: 0.0543–0.0559) on a validation set of 12 183 images for determining CDR. The regression point was converted into classification metrics using a tolerance of 0.2 for 20 classes; the classification metrics achieved an accuracy of 99.20%. The EyePACS dataset (98 172 healthy, 3270 glaucoma) was then used to externally validate the model for glaucoma classification, with an accuracy, sensitivity, and specificity of 82.49%, 72.02%, and 82.83%, respectively.
Conclusions
Our models were precise in determining image gradability and estimating CDR. Although our artificial intelligence–derived CDR estimates achieve high accuracy, the CDR threshold for glaucoma screening will vary depending on other clinical parameters.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: Computer Vision, Deep Learning, Glaucoma, Fundus Image, UK Biobank
Primary open-angle glaucoma is one of the most common glaucoma subtypes, with a global prevalence of 2.4%.1 Accurate detection of glaucoma is essential to prevent irreversible damage to the optic nerve head (ONH), and this often involves clinically assessing the cup-to-disc ratio (CDR). The CDR is a morphological characteristic of the ONH that can estimate the risk of developing glaucoma.2 A larger CDR or interocular asymmetry >0.2 is one of the key risk factors for the development and progression of glaucoma.3, 4, 5 In patients with advanced glaucoma, even small alterations in CDR may lead to considerable loss of retinal ganglion cells.6 Furthermore, many genetic variants are associated with CDR, with a subset of these variants also associated with primary open-angle glaucoma risk.7, 8, 9, 10 Therefore, accurate assessment of CDR is important for glaucoma screening and tracking progression in a clinical setting.
Fundus photography is a noninvasive imaging technique frequently used in glaucoma practice for documenting the condition of the optic nerve and retina, including CDR, neuroretinal rim, disc hemorrhages, and longitudinal monitoring of the optic nerve and surrounding retinal structures. Manual assessment of the CDR is also possible but may be challenging and time-consuming for clinicians. Even among glaucoma specialists, CDR measurement is subject to interobserver and intraobserver variability.11, 12, 13 Advanced imaging technologies, such as OCT and confocal scanning laser ophthalmoscopy, have been observed to provide different CDR values for an individual due to distinct imaging technologies and analysis methodologies.14,15 To overcome these difficulties, CDR estimation can be automated through leveraging advances in artificial intelligence (AI) and computer vision.16
Several studies have precisely segmented the optic disc and cup with deep learning–based techniques for calculating CDR using shape-based methods (circular or elliptic ONH) or appearance-based approaches (texture, color, and intensity of the optic disc).17, 18, 19 Accurately and precisely segmenting the optic cup and disc is challenging because of the variability in ONH morphology, which can be influenced by the patient’s ethnicity, age, disease conditions, and image quality.20, 21, 22 Additionally, segmentation techniques that require masking for both optic disc and cup can be more time-consuming and costly.23 In contrast, nonsegmentation techniques automatically learn the features (disc and cup) from the images. A limited number of studies have used convolutional neural network (CNN)–based regression analysis to estimate CDR using fundus image data.24, 25, 26 Therefore, we developed and validated a CNN-based regression model on a large cohort of retinal images to compute CDR without applying the segmentation techniques for diagnosing glaucoma in clinical and community settings. This selection was primarily driven by the novel application of CNNs for regression analysis in CDR determination, diverging from their common use in image classification due to the considerable amount of resources needed to accurately grade a large dataset in terms of time and costs.
Methods
Study Design
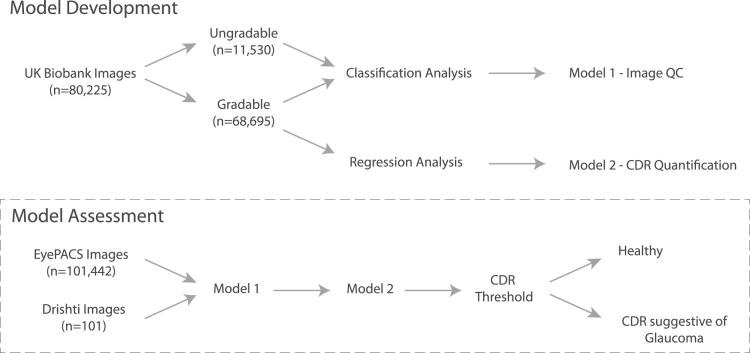
A comprehensive overview of this study is shown in Figure 1. An extensive dataset from the United Kingdom Biobank (UKBB) was used, with each retinal image in the study independently assessed and graded by 2 ophthalmologists (A.W.H. and J.E.C.). We utilized CNN-based models for classification, and regression analyses to determine the gradability and estimate the CDR from 80 225 and 68 695 colored fundus images, respectively.
Figure 1.
Overview of our study design used for model development. A model for gradability assessment was developed (model 1), and then a model for cup-to-disc ratio quantification was generated based on gradable images (model 2). These 2 models were then combined to assess the utility of diagnosing glaucoma in 2 separate cohorts. CDR = cup-to-disc ratio; QC = quality control; UK = United Kingdom.
The UKBB study received ethical approval from the local research ethics boards, and all participants gave informed written consent. This study's procedures were performed in accordance with the ethical guidelines for medical research as described by the World Medical Association Declaration of Helsinki.
Participants and Imaging Modality
Training Dataset
All the participants' data were from the UKBB dataset, which is a large population-based observational study in the United Kingdom that began in 2006 and recruited >500 000 participants aged between 40 to 69 years at the time of recruitment; 67,040 participants had ≥1 gradable retinal image.27,28 The images were available from both left and right eyes from baseline and repeat assessment visits. The retinal images were obtained with a Topcon 3D OCT 1000 Mark II (Topcon Corp).29 This study included both eyes of the participants during 2 visits, irrespective of ancestry.
As described previously, a total of 80 225 fundus images from the UKBB fundus dataset were evaluated and had CDR assessed.28 Of these images, 68 695 (82.73%) were gradable, and 2812 (3.38%) duplicate images were removed. Two independent graders exhibited a strong positive correlation (Pearson correlation coefficient [R] = 0.70) and high consistency (intraclass correlation coefficient, 3k = 0.82) on a shared set of 2812 images (Fig S1).
External Validation
The models were externally validated on the EyePACS dataset—an excellent and reliable resource for creating AI-powered tools to diagnose glaucoma.30 This collection of 101 442 fundus images represents a diverse population of 60 357 individuals who visited various centers across the EyePACS network in the United States.31 This dataset is of exceptional quality and accuracy, given a team of 20 expert graders thoroughly graded the complete dataset, with ≥2 graders reviewing each image. These graders have demonstrated excellent proficiency in detecting glaucoma from fundus photographs, with a minimum sensitivity of 80% and a specificity of 95%. Our models underwent additional validation using the Drishti dataset from India,32 allowing us to assess the consistency and effectiveness of our models across different geographical populations. It includes 70 images from people diagnosed with glaucoma and 31 normal images, all with a resolution of 2047 × 1760 pixels in portable network graphics format.
Image Grading and Preprocessing
In our previous study, 2 fellowship-trained ophthalmologists independently viewed and graded the retinal images.28 All the fundus images were cropped to a pixel ratio of 1080 × 800 before the training or validation of the gradability assessment. However, for regression analysis, we further cropped and removed the noninformative areas, background, or margin around the main content of the images based on their pixel intensity using the OpenCV library.33 The final downsized images were 512 × 512 pixels. As undertaken previously, retinal photographs with low-quality images (ungradable and artifacts) were removed.34,35
Model Selection
Twelve pretrained models from the FastAI (TorchVision)36,37 were utilized and tested on grader “A” data (larger dataset) for CDR estimation using regression analysis. The potential model (vgg19) with batch normalization (bn) layers was selected based on the model’s performance (Fig S2) for both classification and regression analyses. The vgg19_bn pretrained model was initially trained on ImageNet.38 This pretrained model automatically learns high-level features from a large variety of images; thus, it offers a robust feature extractor for classification and regression tasks. We trained and fine-tuned the CNN regression model on a combined dataset of 68 695 images to estimate CDR. The models for detecting glaucoma, using the overall cut-off CDR (≥0.60),39 were extensively validated on 2 publicly available datasets including different ethnicities.
Deep Learning Algorithm and Training
All the models were trained via the FastAI Framework—a deep learning library built on PyTorch that provides a high-level application programming interface and allows deep learning architectures to be trained quickly to achieve state-of-the-art results.40 We used 2 separate CNN-based models from the FastAI library for the gradability assessment of CDR and regression analysis for estimating CDR using fundus images. The binary cross entropy was utilized for the loss function for the gradability task, and the mean squared error loss function was used for regression analysis.
Image gradability was defined as binary classification (gradable or ungradable) based on the CDR that clinicians can obtain. Two experienced fellowship-trained ophthalmologists (A.W.H. and J.E.C.) independently assessed the gradability of the images. Images were randomly (80:20) separated into 2 groups, to train and validate the models for classification and regression analyses. The classification model was trained on a dataset of 64 180 fundus images, while the regression model was trained on a dataset of 48 734. The validation sets for the classification and regression model comprised 16 045 and 12 183 images, respectively.
The class weights were balanced for the gradability task, and in-built data augmentation techniques were implemented for both classification (softmax probability >0.5 was designated as gradable) and regression analysis to improve the accuracy for generalizing the models' utility.41 The augmentation parameters are illustrated in Table S1. Model training was completed in 2 steps: in the first step, we used fine-tuning with frozen layers of the pretrained weights (transfer learning) for 5 to 10 epochs using the Fastai library “valley” function to find the optimal learning rate, validation loss was monitored, and the task was executed until it reached the point where validation loss stopped decreasing. After this, the model unfroze all their layers and used a 1-cycle policy for 10 epochs with the “slice” function to implement a strategy known as discriminative learning rates during the model training.40,42 The callback function was employed to monitor the validation loss during the training and stop training if the loss failed to improve by ≥0.1 (minimum delta) at patience 2 for a maximum of 10 epochs. The regularization technique (weight decay = 1e-3) was also operated to prevent overfitting. This was repeated 10 times with data shuffling, whereby the images were randomly separated into training and validation datasets, and prior to each iteration the top 1% of images found to be in the “top losses” category were removed.
The experiment was conducted on virtual Ubuntu (22.04) desktop with NVIDIA A100 with 40GB of graphics processing unit RAM at Nectar Research Cloud43 using Python (3.10.6) programming language with PyTorch (2.0.0+cu117), FastAI (2.7.12), TorchVision (0.15.1+cu117), Matplotlib (3.5.1), and Scikit-learn (1.2.2) libraries.44, 45, 46
Model Evaluation
Models were evaluated on their area under the receiver operating characteristic curve (AUROC), sensitivity, specificity, precision, recall, and F1-score for gradability assessment. Mean absolute error, mean squared error, R, and coefficient of determination (R2) were used for regression analysis with a 95% confidence interval (CI) for each outcome.47 These metrics assessed model performance by quantifying the difference between actual (graded by clinicians) and predicted CDR by the models. Furthermore, the classification metrics were measured by setting a tolerance bin around each regression point and seeing if the truth value fell within tolerance.
Results
Gradability Assessment
Our classification model obtained a high accuracy of 97.13% on the internal validation set of 20% (16 045) of the total data (80 225) for gradability assessment: gradable and ungradable. The CI with 95% for each metric was estimated through Bootstrap resampling48 with 4000 iterations to ensure the reliability of the results (Table 1). The model underwent testing on random images from the validation set, and the outcome from the classification model is visualized in Fig S3.
Table 1.
Performance of Our Classification Model (Model 1) for Both Gradable and Ungradable Fundus Images in the Validation Dataset
| Classification Metrics |
Gradable (n = 13 717) Mean [95% CI] |
Ungradable (n = 2328) Mean [95% CI] |
|---|---|---|
| AUROC | 0.9656 [0.9614, 0.9697] | 0.9656 [0.9613, 0.9699] |
| Accuracy | 0.9713 [0.9688, 0.9738] | 0.9712 [0.9687, 0.9738] |
| Sensitivity (recall) | 0.9736 [0.9710, 0.9763] | 0.9575 [0.9499, 0.9652] |
| Specificity | 0.9574 [0.9491, 0.9657] | 0.9736 [0.9709, 0.9763] |
| Precision | 0.9926 [0.9912, 0.9941] | 0.8605 [0.8471, 0.8738] |
| F1-score | 0.9830 [0.9815, 0.9846] | 0.9063 [0.8981, 0.9145] |
AUROC = area under the receiver operating characteristic curve; CI = confidence interval; n = number of fundus images.
Regression Analysis
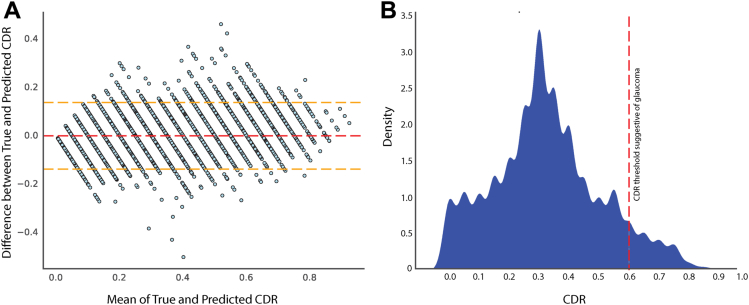
Our regressing model achieved an R2 of 0.71 on the validation set. To further enhance its performance, we excluded 7780 images from the “top losses” category as predicted by the model using the ImageClassifierCleaner from the FastAI library.49 We then retrained the regression model on the final dataset of 60 917 images, with 80% used for training and 20% for validation, resulting in a 19.92% improvement in the R2. The Bland-Altman plot displays the accuracy of the CDR estimation on a validation dataset. The plot includes the corresponding CDR ground truth, which ranges from 0 to 0.95 with 0.05 intervals. Figure 2A exhibits the mean offsets, agreement limits with a 95% CI.
Figure 2.
Comparative analysis and final dataset distribution. The Bland-Altman plot displays the cup-to-disc ratio (CDR) estimation on the validation dataset with the corresponding CDR ground truth shown in (A). The mean difference between true and predicted CDR was 0.022 (95% confidence interval: 0.009–0.034). The red dashed line represents the mean difference, while the yellow dashed lines represent the 95% confidence interval for the mean difference for upper and lower limits. The distribution of the CDR in the final analysis data is graphically presented in (B).
The mean CDR values from graders A and B were 0.36 and 0.31, respectively, but the final mean and standard deviation for the analyzed data were 0.33 and 0.18. Figure 2B visualizes the distribution of CDR from the final analyzed dataset, while Fig S4 shows the random images taken from the validation set to estimate CDR by the regression model.
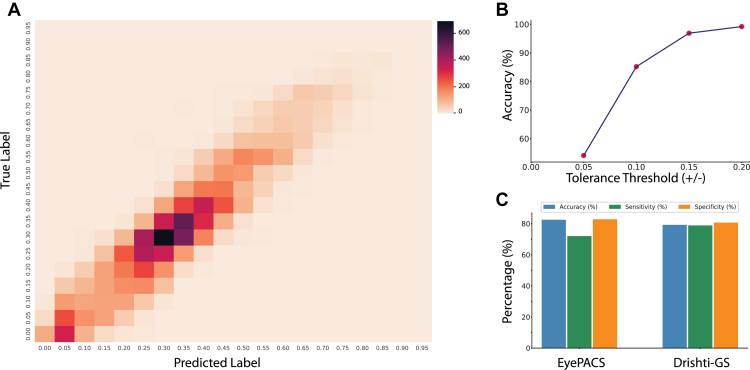
The classification metrics were also assessed using a tolerance-based approach, wherein a predefined tolerance bin was established around each regression point. Subsequently, the true value was evaluated to determine if it fell within the established tolerance bin. Thus, we examined the model’s classification metrics using 4 tolerance thresholds (0.05, 0.10, 0.15, and 0.20) around the ground truth values (Table S2). The regression point was converted into classification metrics with a tolerance of 0.2 for 20 classes, and the classification metrics achieved an accuracy of 99.20%. This approach facilitated the rigorous evaluation of the model's accuracy in correctly assigning class labels; these 20 classes are displayed in the confusion matrix in Figure 3A.
Figure 3.
The confusion matrix for cup-to-disc ratio classification is illustrated in (A). In contrast, (B) presents the conversion of regression points into classification metrics utilizing a particular threshold. Publicly available datasets were used for external validation of the glaucoma screening at the globally accepted cut-off threshold, as shown in (C).
Glaucoma Screening
Our models were externally validated on EyePACS and Drishti datasets with promising results for screening glaucoma. On EyePACS, our models achieved 82.49% accuracy, 72.02% sensitivity, and 82.83% specificity. On Drishti, the models achieved 79.21% accuracy, 78.87% sensitivity, and 80.65% specificity. These models predicted <1% and 0% ungradable images on the EyePACS and Drishti datasets, respectively, as shown in Table S3. Our AI system completed gradability, CDR estimation, and classification within 0.3 seconds, which is approximately 10 times faster with more accurate and consistent results than experienced human graders. We also found our regression model could accurately estimate the CDR even from poor-quality retinal images (Fig S5).
After analyzing the EyePACS and Drishti datasets, the optimal CDR threshold for detecting glaucoma was 0.56 (Figs S6 and S7) as based on receiver operating characteristic curve analysis.50 At this threshold, AUROC was 0.87, suggesting this model can appropriately distinguish fundus images with and without glaucomatous optic discs.
Discussion
In this study, we conducted a regression analysis on fundus images utilizing deep learning architectures to estimate the CDR without employing segmentation techniques from image gradability to glaucoma screening in a single study. Our classification model was first trained for the gradability task and successfully predicted gradable fundus images based on CDR with remarkable accuracy, precision, and recall.
Deep learning-based models to directly estimate CDR are a more efficient and reliable method than clinical assessment. Our results are comparable to Yuen et al's fundus image quality assessment study, which reported an accuracy of 92.5%, a sensitivity of 92.1%, and a specificity of 98.3% on internal validation with 11.6 times fewer validation data than ours.51 However, both studies demonstrate that the deep learning–based model has the potential to grade fundus images accurately.
Accurate estimation of the CDR is useful in diagnosing and monitoring glaucoma. Conventionally, AI-based CDR estimation relies on segmentation techniques.22,52, 53, 54, 55 This technique can be computationally expensive, particularly when dealing with a large volume of medical images. While advancements in graphics processing units have improved computational efficiency, training deep learning models on a significant number of images can still be resource-intensive and costly. Aljazaeri et al reported that a CNN-based regression model performs better than the standard segmentation technique for calculating CDR.25 Our study utilized a deep learning-based model to directly estimate CDR from the fundus images using feature extraction by pretrained weights (vgg19_bn) without applying segmentation techniques. This approach provides a more efficient and reliable method for estimating CDR.
Interestingly, Hemelings et al also used CNN regression analysis to estimate CDR from fundus images, and their findings were notable. However, our results surpassed theirs with an R2 value of 86% compared with their 77% [95% CI 0.77–0.79] on the test set of 4765 for CDR estimation. We observed that their mean of CDR was 0.67, approximately twice our dataset's CDR mean.24 Their different regression model exhibited a strong Pearson's R of 0.88 when predicting the CDR against expert grader.26 Another study reported that the model achieved an R of 0.89 on a dataset of 2115 fundus images from the UKBB cohort.56 In contrast, our model achieved a higher R of 0.93, indicating a strong correlation between the predicted and expert-graded CDR.
It is important to note that our present model is not trained to detect glaucoma but to estimate the CDR, which is just one of several features that can be used to predict the presence of glaucoma. Further to this, screening of glaucoma on a global scale poses challenges due to the variability of optic disc sizes among different ethnic groups.57,58 This creates obstacles in standardizing screening techniques using computer vision based on CDR. Hemelings et al reported an impressive AUROC for external validation but altered in both sensitivity and specificity values across the different datasets when using a fixed threshold (0.7) for glaucoma.26 The highest specificity was observed in the Retinal Fundus Glaucoma Challenge 1 dataset at 0.99 [0.98–0.99], whereas the PAPILA dataset exhibited the lowest specificity at 0.70 [0.63–0.76]. Regarding sensitivity, the Gutenberg Health Study, PAPILA, and Artificial Intelligence for Robust Glaucoma Screening datasets achieved the highest value of 0.94, with the Online Retinal Fundus Image Database for Glaucoma Analysis and Research dataset presenting the lowest sensitivity at 0.68 [0.61–0.75]. They also validated the model externally on the EyePACS dataset; sensitivity was achieved 0.68 and 0.89, with thresholds set at 0.82 and 0.72, respectively. Bhuiyan et al noted 80.11% of sensitivity and 84.96% of specificity on the Online Retinal Fundus Image Database for Glaucoma Analysis and Research dataset for screening glaucoma suspects using retinal images, with a cut-off point of CDR >0.5.59 Consequently, depending exclusively on CDR for glaucoma screening may overlook the complexity of glaucoma (Table S3). A robust and comprehensive screening model for glaucoma at a population level requires integrating glaucomatous phenotypic and genotypic information with advanced AI algorithms.
This research has some important limitations. Firstly, the dataset used in the training of the models was mainly from the healthy White population—the mean of CDR was 0.37—potentially biased for glaucomatous discs; the distribution of CDR can be seen in Figure 2B. The sample we used to train our models may not represent other age groups or populations from different ethnic or geographical backgrounds. Second, we downsized the input images (224 × 224 × 3) to train the regression model, which could lose some meaningful information for accurately estimating the CDR. Finally, dropping poor-performing images (i.e., the top 1% categorized as “top losses” during each of 10 rounds) during model construction could have led to imbalance classes. Nevertheless, these exclusions were made when our initial model's predictions of CDR values exhibited high errors, indicating substantial deviations from the ground truth values.
In summary, we developed a fully automated end-to-end computer vision model for estimating CDR from the fundus images using CNN regression analysis for glaucoma care. The models were externally validated on 2 publicly available databases for glaucoma screening based on CDR. Deep learning–based models to directly estimate CDR are a more efficient and reliable method than clinical assessment.60 However, a generalized model for screening glaucoma based on CDR is challenging because of uncertainty in what the optimal CDR cut-off should be for diagnosing glaucoma.
Acknowledgments
This research has been conducted using the UK Biobank Resource under Application Number 25331. S.M., D.A.M., and A.W.H. acknowledge program grants from the National Health and Medical Research Council (GNT1150144) and Centre of Research Excellence (1116360) and funding from the Australian National Health and Medical Research Council (NHMRC). S.M. and D.A.M. are supported by research fellowships from the Australian National Health and Medical Research Council (NHMRC). P.G. (#1173390) and A.W.H. are supported by NHMRC Investigator Grants and Research Training Program Scholarship from the University of Tasmania (A.K.C.).
Manuscript no. XOPS-D-23-00210.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s):
D. A. M.: Support – Australian National Health and Medical Research Council (NHMRC) Fellowship, NHMRC Partnership Grant, NHMRC Program Grant.
A. W. H.: Support – Australian NHMRC Fellowship, NHMRC Partnership Grant, NHMRC Program Grant.
J.E.C.: Support – Australian NHMRC Fellowship, NHMRC Partnership Grant.
S. M.: Support – Australian NHMRC Fellowship, NHMRC Partnership Grant.
P.G. (#1173390) and A.W.H. are supported by NHMRC Investigator Grants and Research Training Program Scholarship from the University of Tasmania (A.K.C.).
HUMAN SUBJECTS: Human subjects data were included in this study. The UKBB study received ethical approval from the local research ethics boards. All research adhered to the tenets of the Declaration of Helsinki. All participants provided informed consent.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Chaurasia, Hewitt
Data collection: Chaurasia, Gharahkhani, MacGregor, Craig, Hewitt
Analysis and interpretation: Chaurasia, Greatbatch, Han, Gharahkhani, Mackey, MacGregor, Hewitt
Obtained funding: Mackey, MacGregor, Chaurasia
Overall responsibility: Chaurasia, Greatbatch, Gharahkhani, MacGregor, Hewitt
Supplementary Data
References
- 1.Zhang N., Wang J., Li Y., Jiang B. Prevalence of primary open angle glaucoma in the last 20 years: a meta-analysis and systematic review. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-021-92971-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damms T., Dannheim F. Sensitivity and specificity of optic disc parameters in chronic glaucoma. Invest Ophthalmol Vis Sci. 1993;34:2246–2250. [PubMed] [Google Scholar]
- 3.Gordon M.O., Beiser J.A., Brandt J.D., et al. The ocular hypertension treatment study: baseline factors that predict the onset of primary open-angle glaucoma. Arch Ophthalmol. 2002;120:714–720. doi: 10.1001/archopht.120.6.714. [DOI] [PubMed] [Google Scholar]
- 4.Mwanza J.C., Tulenko S.E., Barton K., et al. Eight-Year incidence of open-angle glaucoma in the tema eye survey. Ophthalmology. 2019;126:372–380. doi: 10.1016/j.ophtha.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 5.BMJ Publishing Group Ltd. BMA House. Square T. London & 9jr, W. European glaucoma society terminology and guidelines for glaucoma, 4th edition - Part 1supported by the EGS foundation. Br J Ophthalmol. 2017;101:1–72. [Google Scholar]
- 6.Tatham A.J., Weinreb R.N., Zangwill L.M., et al. The relationship between cup-to-disc ratio and estimated number of retinal ganglion cells. Invest Ophthalmol Vis Sci. 2013;54:3205–3214. doi: 10.1167/iovs.12-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Springelkamp H., Iglesias A.I., Mishra A., et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum Mol Genet. 2017;26:438. doi: 10.1093/hmg/ddw399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han X., Steven K., Qassim A., et al. Automated AI labeling of optic nerve head enables insights into cross-ancestry glaucoma risk and genetic discovery in >280,000 images from UKB and CLSA. Am J Hum Genet. 2021;108:1204–1216. doi: 10.1016/j.ajhg.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlesworth J., Kramer P.L., Dyer T., et al. The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol Vis Sci. 2010;51:3509. doi: 10.1167/iovs.09-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nannini D.R., Kim H., Fan F., Gao X. Genetic risk score is associated with vertical cup-to-disc ratio and improves prediction of primary open-angle glaucoma in latinos. Ophthalmology. 2018;125:815–821. doi: 10.1016/j.ophtha.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper R, Reeves B, Smith G. Observer variability in optic disc assessment: implications for glaucoma shared care. Ophthalmic and Physiological Optics. 2000;20:265–273. [PubMed] [Google Scholar]
- 12.Reus NJ, Lemij HG, Garway-Heath DF, et al. Clinical assessment of stereoscopic optic disc photographs for glaucoma: the European Optic Disc Assessment Trial. Ophthalmology. 2010;117:717–723. doi: 10.1016/j.ophtha.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 13.Hertzog LH, Albrecht KG, LaBree L, Lee PP. Glaucoma care and conformance with preferred practice patterns: examination of the private, community-based ophthalmologist. Ophthalmology. 1996;103:1009–1013. doi: 10.1016/s0161-6420(96)30573-3. [DOI] [PubMed] [Google Scholar]
- 14.Perera SA, Foo LL, Cheung CY, et al. Cup-to-disc ratio from Heidelberg retina tomograph 3 and high-definition optical coherence tomography agrees poorly with clinical assessment. J Glaucoma. 2016;25:198–202. doi: 10.1097/IJG.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 15.Chan PP, Chiu V, Wong MO. Variability of vertical cup to disc ratio measurement and the effects of glaucoma 5-year risk estimation in untreated ocular hypertensive eyes. Br J Ophthalmol. 2019;103:361–368. doi: 10.1136/bjophthalmol-2017-311841. [DOI] [PubMed] [Google Scholar]
- 16.Chaurasia A.K., Greatbatch C.J., Hewitt A.W. Diagnostic accuracy of artificial intelligence in glaucoma screening and clinical practice. J Glaucoma. 2022;31:285–299. doi: 10.1097/IJG.0000000000002015. [DOI] [PubMed] [Google Scholar]
- 17.Fu H, Cheng J, Xu Y, Wong DW, Liu J, Cao X. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans Med Imaging. 2018;37:1597–1605. doi: 10.1109/TMI.2018.2791488. [DOI] [PubMed] [Google Scholar]
- 18.Veena H., Muruganandham A., Senthil Kumaran T. A novel optic disc and optic cup segmentation technique to diagnose glaucoma using deep learning convolutional neural network over retinal fundus images. Int J Comput Assist Radiol Surg. 2022;34:6187–6198. [Google Scholar]
- 19.Gao Y, Yu X, Wu C, Zhou W, Wang X, Chu H. Accurate and efficient segmentation of optic disc and optic cup in retinal images integrating multi-view information. IEEE. 2019;7 doi: 10.1109/ACCESS.2019.2946374. 148183–97. [DOI] [Google Scholar]
- 20.Veena H.N., Muruganandham A., Kumaran T.S. A Review on the optic disc and optic cup segmentation and classification approaches over retinal fundus images for detection of glaucoma. SN Appl Sci. 2020;2:1–15. [Google Scholar]
- 21.Almazroa A, Burman R, Raahemifar K, Lakshminarayanan V. Optic disc and optic cup segmentation methodologies for glaucoma image detection: a survey. J Ophthalmol. 2015;2015 doi: 10.1155/2015/180972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y., Zhang X., Xu X., et al. Deep level set method for optic disc and cup segmentation on fundus images. Biomed Opt Express. 2021;12:6969. doi: 10.1364/BOE.439713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong DWK, Liu J, Tan NM, Yin F, Lee BH, Wong TY. 2010. Learning-based approach for the automatic detection of the optic disc in digital retinal fundus photographs; pp. 5355–5358. 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina. [DOI] [PubMed] [Google Scholar]
- 24.Hemelings R., Elen B., Barbosa-Breda J., et al. Deep learning on fundus images detects glaucoma beyond the optic disc. Sci Rep. 2021;11:1–12. doi: 10.1038/s41598-021-99605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aljazaeri M, Bazi Y, Almubarak H, Alajlan N. 2020. Faster R-CNN and DenseNet regression for glaucoma detection in retinal fundus images; pp. 1–4. 2020 2nd International Conference on Computer and Information Sciences (ICCIS) [Google Scholar]
- 26.Hemelings R, Elen B, Schuster AK, et al. A generalizable deep learning regression model for automated glaucoma screening from fundus images. NPJ Digit Med. 2023;6:112. doi: 10.1038/s41746-023-00857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warwick AN, Curran K, Hamill B, et al. UK Biobank retinal imaging grading: methodology, baseline characteristics and findings for common ocular diseases. Eye. 2023;37:2109–2116. doi: 10.1038/s41433-022-02298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craig J.E., Han X., Qassim A., et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet. 2020;52:160. doi: 10.1038/s41588-019-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.AIROGS - Grand Challenge. grand-challenge.org. https://airogs.grand-challenge.org/data-and-challenge/ Available at:
- 31.Lemij HG, de Vente C, Sánchez CI, Vermeer KA. Characteristics of a large, labeled data set for the training of artificial intelligence for glaucoma screening with fundus photographs. Ophthalmol Sci. 2023;3:100300. doi: 10.1016/j.xops.2023.100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drishti-GS Dataset Webpage. http://cvit.iiit.ac.in/projects/mip/drishti-gs/mip-dataset2/Home.php Available at:
- 33.OpenCV Library OpenCV - open computer vision library. 2021. https://opencv.org/ OpenCV. Available at:
- 34.Dodge S, Karam L. Vol. 6. IEEE; 2016. Understanding how image quality affects deep neural networks; pp. 1–6. 2016 eighth international conference on quality of multimedia experience (QoMEX) [Google Scholar]
- 35.Chalakkal R.J., Abdulla W.H., Thulaseedharan S.S. Quality and content analysis of fundus images using deep learning. Comput Biol Med. 2019;108:317–331. doi: 10.1016/j.compbiomed.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 36.vision.models. https://fastai1.fast.ai/vision.models.html Available at:
- 37.vision.models. https://fastai1.fast.ai/vision.models.html#:∼:text=The%20fastai%20library%20includes%20several,resnet34%2C%20resnet50%2C%20resnet101%2C%20resnet152 Available at:
- 38.ImageNet. https://www.image-net.org/ Available at:
- 39.Soh ZD, Chee ML, Thakur S, et al. Asian-specific vertical cup-to-disc ratio cut-off for glaucoma screening: An evidence-based recommendation from a multi-ethnic Asian population. Clin Exp Ophthalmol. 2020;48:1210–1218. doi: 10.1111/ceo.13836. [DOI] [PubMed] [Google Scholar]
- 40.Howard J., Gugger S. Fastai: a layered API for deep learning. Information. 2020;11:108. [Google Scholar]
- 41.Vision augmentation. https://docs.fast.ai/vision.augment.html Available at:
- 42.Smith LN. A disciplined approach to neural network hyper-parameters: Part 1--learning rate, batch size, momentum, and weight decay. arXiv preprint. 2018 arXiv:1803.09820. [Google Scholar]
- 43.Login - Nectar Dashboard. https://dashboard.rc.nectar.org.au/dashboard_home/ Available at:
- 44.PyTorch 2.0. https://pytorch.org/get-started/pytorch-2.0/ Available at:
- 45.torchvision PyPI. https://pypi.org/project/torchvision/ Available at:
- 46.Installing. scikit-learn. https://scikit-learn.org/stable/install.html Available at:
- 47.Chicco D, Warrens MJ, Jurman G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput Sci. 2021;7:e623. doi: 10.7717/peerj-cs.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulesa A., Krzywinski M., Blainey P., Altman N. Sampling distributions and the bootstrap. Nat Methods. 2015;12:477–478. doi: 10.1038/nmeth.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vision widgets. https://docs.fast.ai/vision.widgets.html Available at:
- 50.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4:627. [PMC free article] [PubMed] [Google Scholar]
- 51.Yuen V, Ran A, Shi J, et al. Deep-learning–based pre-diagnosis assessment module for retinal photographs: A multicenter study. Transl Vis Sci Tech. 2021;10:16. doi: 10.1167/tvst.10.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian Z., Zheng Y., Li X., et al. Graph convolutional network based optic disc and cup segmentation on fundus images. Biomed Opt Express. 2020;11:3043. doi: 10.1364/BOE.390056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Tran L, Peto T, Chew EY. Identifying those at risk of glaucoma: a deep learning approach for optic disc and cup segmentation and their boundary analysis. Diagnostics. 2022;12:1063. doi: 10.3390/diagnostics12051063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacCormick IJ, Williams BM, Zheng Y, et al. Accurate, fast, data efficient and interpretable glaucoma diagnosis with automated spatial analysis of the whole cup to disc profile. PloS One. 2019;14 doi: 10.1371/journal.pone.0209409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alawad M., Aljouie A., Alamri S., et al. Machine learning and deep learning techniques for optic disc and cup segmentation – a review. Clin Ophthalmol. 2022;16:747. doi: 10.2147/OPTH.S348479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alipanahi B., Hormozdiari F., Behsaz B., et al. Large-scale machine-learning-based phenotyping significantly improves genomic discovery for optic nerve head morphology. Am J Hum Genet. 2021;108:1217. doi: 10.1016/j.ajhg.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee RY, Kao AA, Kasuga T, et al. Ethnic variation in optic disc size by fundus photography. Curr Eye Res. 2013;38:1142–1147. doi: 10.3109/02713683.2013.809123. [DOI] [PubMed] [Google Scholar]
- 58.Marsh BC, Cantor LB, WuDunn D, et al. Optic nerve head (ONH) topographic analysis by stratus OCT in normal subjects: correlation to disc size, age, and ethnicity. J Glaucoma. 2010;19:310–318. doi: 10.1097/IJG.0b013e3181b6e5cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhuiyan A, Govindaiah A, Smith RT. An artificial-intelligence-and telemedicine-based screening tool to identify glaucoma suspects from color fundus imaging. J Ophthalmol. 2021 doi: 10.1155/2021/6694784. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mvoulana A, Kachouri R, Akil M. Fully automated method for glaucoma screening using robust optic nerve head detection and unsupervised segmentation based cup-to-disc ratio computation in retinal fundus images. Comput Med Imaging Graph. 2019;77:101643. doi: 10.1016/j.compmedimag.2019.101643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.