Summary
Structural analyses of large, complex noncoding RNAs continue to lag behind their rapid discovery and functional descriptions. Site-specifically incorporated, minimally invasive fluorescent probes such as 2-aminopurine (2AP) and pyrrolo-cytosine (PyC) have provided essential complementary information about local RNA structure, conformational dynamics and interactions. Here I describe a protocol that benchmarks and correlates local RNA conformations with their respective fluorescence lifetimes, as a general technique that confers key advantages over fluorescence intensity-based methods. The observation that fluorescence lifetimes are more sensitive to local structures than sequence contexts suggest broad utility across diverse RNA and ribonucleoprotein systems.
Keywords: RNA structure, fluorescence, RNA-RNA interactions, riboswitches, tRNA, T-box
1. Introduction
Emerging enabling technologies such as RNA-seq and Nanopore have transformed the landscape of RNA biology in the past two decades, and greatly accelerated the discovery and functional description of the noncoding genome. However, high-resolution structural analyses of biologically important noncoding RNAs and their protein complexes remain technically challenging. Effective analyses are hampered by RNA’s intrinsic conformational heterogeneity and flexibility, highly charged, non-specific and repetitive surfaces, and non-native, solid-phase imaging environments that either densely pack RNAs in crystals or embed them in thin ice on Cryo-EM grids [1, 2]. Therefore, complementary solution-based methods are essential to corroborate and extend structural findings, and to provide insights into the dynamic property of the RNA structures that frequently underlies their biological functions.
Fluorescent nucleoside analogues such as 2-aminopurine (2AP) and pyrrolo-cytosine (PyC) have been widely used to probe nucleic acid structure and interactions, as they are capable of forming Watson-Crick base pairs, minimally perturb the geometry of local structures, and exhibit quantum yields that are sensitive to local environments [3-9] (Fig. 1). Most such applications are based on measurements of fluorescence intensity, whose precision and reproducibility are subject to a number of external variables including sample concentrations, incident light intensities, detector gain and sensitivities, etc. By contrast, fluorescence lifetime is an intrinsic property of the fluorophore in a given chemical environment. It is independent of sample concentration, sample absorption, excitation light intensity, detector sensitivity, fluorescence emission intensity, and is unaffected by photo-bleaching. Nevertheless, it is still sensitive to temperature, polarity, and quenchers in solution. Fluorescence lifetime is reduced by Förster resonance energy transfer (FRET) and can quantitatively report FRET efficiency using a single variable that can be accurately measured and compared. Further, it can even be used where the fluorescence acceptor is not a fluorophore but a quencher, such as with molecular beacons.
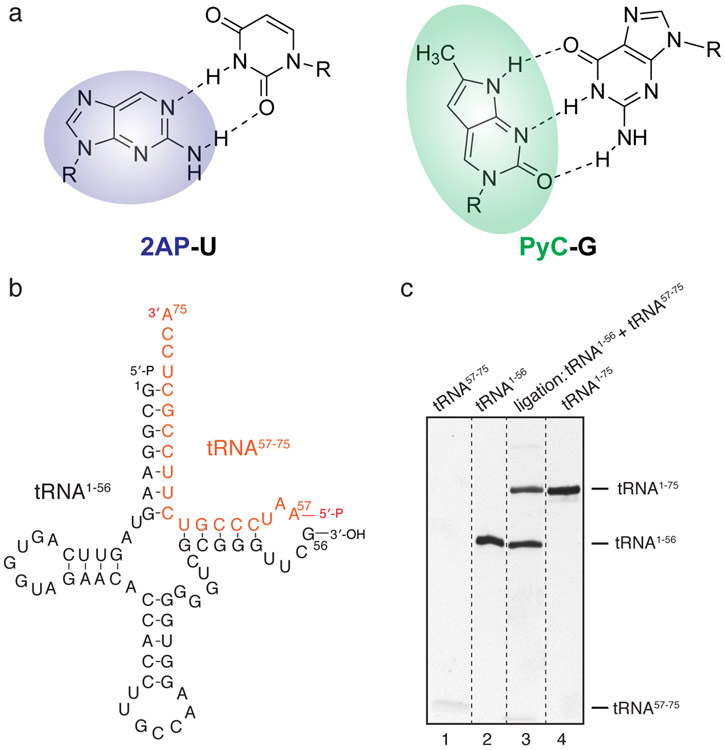
Fig. 1. Semi-enzymatic synthesis of 2AP-labelled tRNA.
(a) Chemical structure of 2-aminopurine and pyrrolo-cytosine with their respective Watson-Crick base pairs. (b) General schematic of assembly and enzymatic ligation from two RNA fragments to produce full-length tRNAs that contain specific labels near its 3′ region. (c) Urea-PAGE analysis of the ligation reaction in (b). Note the lower band of tRNA57-75 oligo stains weakly.
Site-specific labeling of long RNA with fluorophores, especially at internal positions, remains a technical issue. While custom short RNA oligonucleotides under study can be chemically synthesized by several commercial sources, longer RNAs (> 60 nucleotides, or nts) generally require hybridization-based assembly of two or more RNA fragments with optional enzymatic ligation to seal nicks. Notably, recent technological developments, such as the position-selective labelling of RNA (PLOR) method, enables site-specific labeling of long RNAs by “walking” the RNA polymerase along the DNA template discontinuously [10]. PLOR operates by immobilizing the transcription elongation complex (TEC) on solid support, withholding one or more nucleotide triphosphates (NTP) in each cycle of transcript extension to allow partial extension, and iterative rounds of washing to supply only the appropriate subsets of NTPs [11, 12]. Multiple strategies for specific tRNA labeling have been developed [9, 13-15]. Here, we label the 3′-end of tRNAs with 2AP employing a flexible two-piece ligation scheme, where an unlabeled, in vitro transcribed 5′ fragment (nts 1-56) is enzymatically ligated with a partially complementary, chemically synthesized 3′ fragment (nts 57-75) bearing a terminal 2AP, forming the full-length tRNA [16, 17]. Similarly, we label a Bacillus subtilis glyQS full-length T-box riboswitch by annealing an in vitro transcribed 5′ fragment (nts 1-170) with a chemically synthesized 3′ fragment (nts 171-182) bearing an internal PyC.
In the method below, I describe the use of 2AP and PyC lifetime analyses to probe local RNA structural contexts, as well as RNA-RNA interactions that mediate the gene regulation by the T-box riboswitches. Using 2AP or PyC-containing benchmarking oligonucleotides derived from the tRNA and T-box riboswitch sequences, we demonstrate the general applicability of the method and consistency of lifetime values that correspond to structural contexts as opposed to sequence contexts. Critical insights from such lifetime measurements were instrumental in deriving correct structural models that were recently verified by crystallographic and Cryo-EM analyses [2, 17-19].
2. Materials
1. Custom oligonucleotides labelled with 2-aminopurine (2AP) or pyrrolocytosine (PyC) (IDT, Horizon, Trilink, etc.)
2. Taq DNA polymerase, 5,000 U/mL (New England Biolabs).
3. T7 RNA polymerase, 50,000 U/mL (New England Biolabs).
4. Diethylpyrocarbonate (DEPC)-treated water.
5. RNA Binding Buffer: 50 mM HEPES-KOH, pH 7.0, 100 mM KCl, 20 mM MgCl2, 5 mM tris (2-carboxyethyl) phosphine (TCEP).
6. EasyLife-LS system (Photon Technology International) or FluoroMax Plus Spectrofluorometer equipped with DeltaTime TCSPC (Time Correlated Single Photon Counting) module (Horiba Scientific).
7. LUDOX colloidal silica solution (Sigma-Aldrich).
8. T4 RNA ligase 1 (ssRNA ligase).
9. 2AP-triphosphate (2AP-TP) (Jena Bioscience).
10. Elutrap® Electroelution System (Cytiva).
3. Methods
3. 1. Semi-enzymatic synthesis of 2AP-labelled tRNA
1. Perform in vitro transcription of tRNA 5′ fragment (1-56 nts) using T7 RNA Polymerase (RNAP). 5′ monophosphates are installed on tRNAs by “GMP priming”, where a mixture of GMP (20 mM) and GTP (2 mM) is used instead of GTP, producing ~91% 5′ monophosphorylated tRNAs [20, 21]. Purify the desired RNA fragment (tRNA1-56) by preparative denaturing Urea-PAGE, electroelution on an Elutrap device (Cytiva), washing with 1 M KCl and extensive exchange into DEPC-treated water, and store RNA samples at −20° C or −80° C before use [22-24].
2. Ligate the tRNA1-56 with an tRNA57-75 oligonucleotide (IDT) bearing a 5′ monophosphate and 3′ 2AP modification in place of A75, using T4 RNA ligase 1 (ssRNA ligase), which seals the nick in the TΨC-loop with ~50% efficiency (Fig. 1). Gel-purify the ligated full-length tRNA1-75(2AP75) as described in Step 1.
3. 2. Instrument setup and validation
1. On a fluorescence lifetime instrument, such as an EasyLife-LS system (Photon Technology International) used in this chapter, or a FluoroMax Plus Spectrofluorometer equipped with DeltaTime TCSPC (Time Correlated Single Photon Counting) module (Horiba Scientific), install a pulse excitation diode or laser light source at 280-310 nm (for 2AP) or 340 nm (for PyC) and set emission monochromators or bandpass emission filters at 376/20 nm (for 2AP) or 448/20 nm (for PyC).
2. Measure the Instrument Response Function (IRF, also known as “prompt”) using a 0.1-1% LUDOX colloidal silica solution (Sigma). To measure short lifetimes (< 100 ps) accurately, laser sources that have shorter pulse widths and faster detectors (such as Hybrid Picosecond Photon Detectors, or HPPDs) is generally needed.
3. Verify the instrument setup by measuring the lifetime of a sample of free 2AP-triphosphate (2AP-TP). The expected average lifetime of 2AP-TP is ~ 8-10 ns in aqueous solution (Fig. 2a).
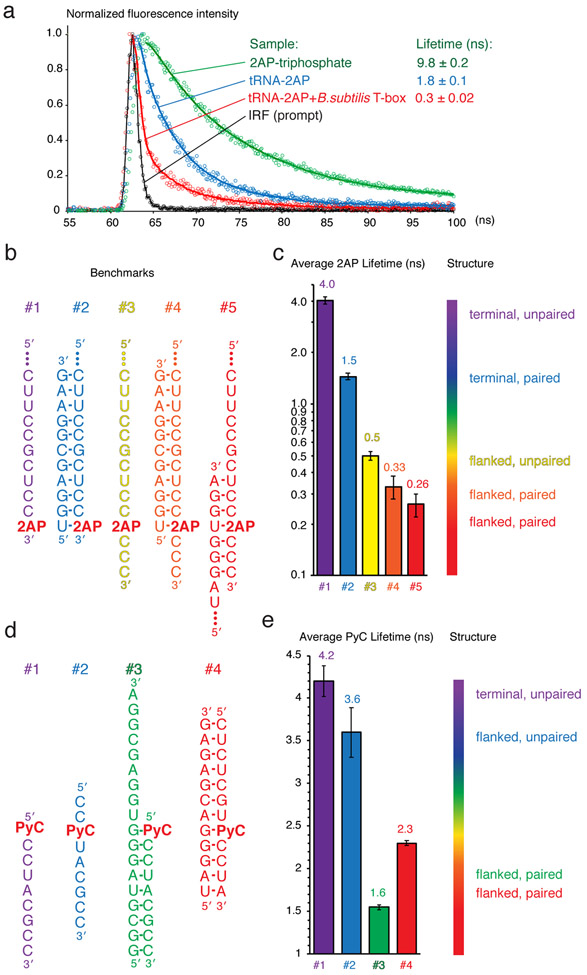
Fig. 2. 2AP and PyC lifetime analyses and benchmarks.
(a) Representative curves of the Instrument Response Function (IRF, or Prompt, black), and 2AP fluorescence decay curves of 2AP-triphosphate (green), 2AP-labeled tRNA alone (blue) and in complex with a full-length T-box riboswitch (red). (b) 2AP-containing benchmark oligos derived from the Bacillus subtilis glyQS T-box riboswitch and tRNAGly sequences. (c) Average 2AP lifetimes of the samples in (b) and a spectrum bar correlating local structure of 2AP with its lifetimes. (d) PyC-containing benchmark oligos derived from the B. subtilis glyQS T-box riboswitch. (e) PyC lifetimes of the samples in (d) and a spectrum bar correlating local structure of PyC with its lifetimes. All values reported throughout the chapter were measured on a PTI EasyLife-LS system.
3. 3. Benchmarking fluorescence lifetimes
1. Measure 2AP fluorescence lifetimes on a set of benchmark oligonucleotides that hybridize into known ssRNA and dsRNA structures at 5-10 μM (Fig. 2b). Less material would be required using TCSPC-capable instruments with enhanced sensitivity. A typical decay curve of 2AP fluorescence has four distinct components including a very short-lived species (< 100 ps) corresponding to highly stacked conformation, and a long-lived species (~10 ns) corresponding to a mobile, unstacked conformation [7, 8]. Depending on the specific environment of 2AP, 1-3 decaying species are often sufficient to satisfactorily fit the decay curve, producing reasonable χ2 values (< 1.0-1.5). Collected traces are corrected by accounting for the IRF, normalized, and fit to minimal number of exponential decay components that produced acceptable χ2 and random residuals, using Felix32 spectroscopy software (Photon Technology International), or DAS6 decay analysis software (Horiba). Then a linear average lifetime is calculated using the fractional amplitudes and lifetimes of each component. As distinct decay curves and lifetime distributions can by chance produce very similar amplitude-weighted average lifetimes, it is important to examine the full decaying curves, in addition to the reduced average lifetime numeric values.
2. Correlate 2AP benchmark lifetimes with their respective structural contexts. Using different sets of 2AP benchmarks using distinct sequences, we generally obtained comparable lifetime values, which are more sensitive to local structural contexts than to sequence contexts (Fig. 2b-c). The extent of quenching is more dependent on stacking than base pairing, consistent with stacking with the nearest neighbor being the principal quenching mechanism [4, 7, 8]. Terminally located 2AP exhibit average lifetimes of ~1.5 and 4.0 ns, when paired with a uridine or unpaired respectively (Fig. 2b-c, samples #1 and #2). This is likely due to the complete absence of stacking on one face of 2AP, as well as transient fraying of the duplex termini, which exposes both faces of 2AP to the solvent. Notably, an unpaired 2AP, when flanked by nucleobases on both sides, exhibited a much shorter lifetime of 0.5 ns (Fig. 2b-c, sample #3), even as both neighbors are pyrimidines that stack more weakly than purines. Importantly, the 2AP lifetime analysis is sensitive enough to distinguish between stacking on one face of 2AP versus stacking on both flanks, which can yield crucial information regarding local RNA structure, organization or interaction [1]. When 2AP is located in the middle of a stable duplex, it exhibits a short lifetime of ~0.25 ns (Fig. 2b-c, sample #5). Interestingly, when a paired 2AP is located at the boundary between double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA), it yields an intermediate lifetime of ~0.33 ns (Fig. 2b-c, sample #5). Together, these benchmarks demonstrate remarkable sensitivity and consistency of 2AP lifetimes as reporters of local RNA structure. It is important to note that sequence context of 2AP, especially adjacent guanosines in the same or opposing strand, can significantly impact 2AP lifetime through charge transfer [25, 26].
3. Correlate PyC benchmark lifetimes with their respective structural contexts. When the desired probe location is normally occupied by a pyrimidine rather than a purine, PyC can serve as a more appropriate probe than 2AP. Compared to 2AP, PyC has the advantage of longer excitation and emission wavelengths and longer lifetimes, reducing the technical requirements for instrumentation [13, 27]. The benchmarks of PyC lifetimes showed a pattern of sensitivity that is distinct from and more complex than that of 2AP (Fig. 2d, e). The data suggest that PyC lifetime appears more sensitive to base pairing than stacking with nearest nucleobases, which can either increase or decrease the lifetime of PyC depending on the structural and sequence context. This is consistent with theoretic calculations and previous reports that solvent shielding by neighboring nucleobases can enhance PyC fluorescence [5, 28, 29]. Nonetheless, the benchmarks suggest that PyC lifetime analyses effectively distinguish between ssRNA and dsRNA structural contexts, but are unable to reliably distinguish between one-sided stacking and double-sided stacking, as 2AP can [17].
4. (Optional) Examine the dependency of lifetimes on buffer solute conditions, especially divalent cation concentrations such as Mg2+. The conformations and dynamics of RNA structures can be modulated by the presence and concentrations of monovalent and divalent cations, in particular of Mg2+ [30-32].
3. 4. Measure fluorescence lifetimes of the experimental systems under study.
1. With an empirical correlation between fluorescence lifetimes and local RNA structures established for a given sequence context on a particular instrument, actual experimental samples are ready to be measured. We illustrate this using samples of a free tRNA2AP75, a free T-box riboswitch labelled by PyC, and their complexes.
2. Measure the lifetime of 2AP in free tRNA2AP75. We observed a lifetime of ~1.8 ns, substantially shorter than the ~4.0 ns observed for a ssRNA oligo of the same sequence as the tRNA. This finding is consistent with a partially stacked conformation of the 3′-NCCA terminus [33, 34].
3. Assemble a native tRNA-Bacillus subtilis glyQS full-length T-box riboswitch complex sample by transcribing excess T-box RNA in the presence of purified tRNA2AP75. Remove free labelled tRNA from the complexes using size-exclusion chromatography on a Superdex 200 Increase column. It is important to remove any labelled tRNA that is not in the complex, as they would obscure the lifetime analysis.
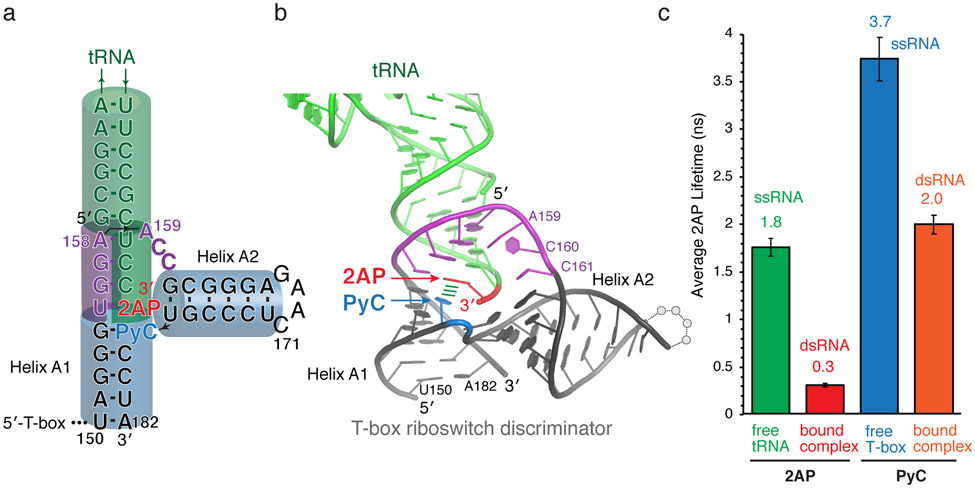
4. Measure the 2AP lifetime of the tRNA2AP75 in the context of the T-box riboswitch-tRNA complex. We observed a highly quenched lifetime of 0.3 ns (Fig. 2a, 3a-c), which matches the lifetimes of 2AP in the middle of an RNA duplex from the benchmarks (0.26-0.33 ns; Fig. 2b-c). This finding provided a key clue to the structural organization of the T-box riboswitch-tRNA complex before structures were available (Fig. 3a-c). Namely, the tRNA 3′-adenosine, tA75, resides in a dsRNA duplex environment and is stacked on both sides [17]. As the tA75 is the terminal tRNA residue and stacks with tC74, then the other stacking must stem from the T-box. This critical insight suggests that the intermolecular helix formed between the tRNA 3′-NCCA and the T-box bulge stacks directly with Helix A1 of the T-box antiterminator, which allows the tRNA to stabilize the “ON” conformation of the T-box riboswitch (Fig. 3a, b). This exact structural arrangement has recently been validated by two independently solved co-crystal structures as well as a cryo-EM structure [2, 19, 35].
Fig. 3. 2AP and PyC lifetime analyses of T-box riboswitch-tRNA complexes suggest structural model.
(a) Secondary structure model of a Bacillus subtilis glyQS T-box riboswitch-tRNA complex proposed based on lifetime analyses. The locations of 2AP (red) and PyC (blue) are indicated. (b) Co-crystal structure of a Geobacillus Kaustophilus T-box riboswitch discriminator domain-tRNA complex (PDB: 6PMO [2]). Stem III region is omitted for clarity. The locations of 2AP (red) and PyC (blue) are indicated. Numbering is the same as in (a). (c) 2AP lifetimes of tRNA2AP75 either alone (green), or in complex with a full-length T-box riboswitch (red), and PyC lifetimes of T-box171-182PyC178 RNA oligo either alone (blue), or in the context of a full-length T-box-tRNA complex (orange).
5. Similarly to 2AP, measure the lifetimes of PyC in a free RNA oligo corresponding to T-box171-182, as well as when the oligo is annealed with the rest of the T-box1-170 and in complex with unlabeled tRNA. Upon tRNA binding, the PyC lifetime was reduced from 3.7 ns to 2.0 ns (Fig. 3c), suggesting that the local environment of PyC changed from ssRNA to dsRNA, consistent with the 2AP findings and corroborated the structural model from the T-box side [17].
4. Notes
1. When measuring 2AP or PyC lifetimes in a binary or ternary complex, 1:1 stoichiometry is not required. The non-labeled partner protein or RNA can be in excess, and should be, if the complex is unstable or has relatively low affinities. The excess binding partner will ensure that the labelled RNA remains in the complex, and will not interfere with lifetime measurements.
2. One key consideration for choosing between 2AP or PyC, when either nucleoside analogue is compatible with the biological system under study, is that 2AP fluorescence is more sensitive to nearest neighbor stacking than PyC. Using PyC as a stacking probe would generally require careful controls that take into account the specific sequence and structural contexts of PyC, with a particular emphasis on solvent shielding effects.
3. Since 2AP and PyC are most useful as a probe of local structure and direct interactions, it is advisable to be mindful that the global RNA conformations may change in unexpected ways when titrating divalent cations, ligands or proteins, which may in turn cause drastic changes of the probe’s local environment and fluorescence properties.
4. While 2AP and PyC substitutions rarely impact the biological function of the RNAs to significant extents, it is advisable to verify experimentally that these chemical modifications are indeed tolerated.
5. Absolute lifetime measurements depend significantly on the specific instrumentation used and its technical parameters, in particular, on the spectral width and pulse profile of the excitation light source. All values reported here were measured on a PTI EasyLife-LS system. For comparison, measurements of the same RNA samples on a Horiba FluoroMax Plus Spectrofluorometer equipped with DeltaTime TCSPC module generally produce longer lifetimes. Thus, extensive benchmarking on the specific instrument used for the experiments is required.
ACKNOWLEDGEMENTS
I thank G. Piszczek and D. Wu for support with biophysical analyses, and A. R. Ferré-D’Amaré, C. Bou-Nader, N. Baird, M. Lau, S. Li, K. Suddala, and K. Sapkota for discussions. This work was supported by the Intramural Research Program of the NIH, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (ZIADK075136 to J.Z.), and a NIH Deputy Director for Intramural Research (DDIR) Challenge Award to J.Z. The author declares no conflicts of interests.
References
- 1.Zhang J and Ferré-D'Amaré AR, (2014) New molecular engineering approaches for crystallographic studies of large RNAs. Curr Opin Struct Biol. 26:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S., et al. , (2019) Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions. Nat Struct Mol Biol. 26:1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun A., et al. , (2019) SAM-VI riboswitch structure and signature for ligand discrimination. Nat Commun. 10:5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jean JM and Hall KB, (2001) 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc Natl Acad Sci U S A. 98:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardman SJ and Thompson KC, (2006) Influence of base stacking and hydrogen bonding on the fluorescence of 2-aminopurine and pyrrolocytosine in nucleic acids. Biochemistry. 45:9145–55. [DOI] [PubMed] [Google Scholar]
- 6.Lenz T., et al. , (2007) 2-Aminopurine flipped into the active site of the adenine-specific DNA methyltransferase M.TaqI: crystal structures and time-resolved fluorescence. J Am Chem Soc. 129:6240–8. [DOI] [PubMed] [Google Scholar]
- 7.Jones AC and Neely RK, (2015) 2-Aminopurine as a fluorescent probe of DNA conformation and the DNA-enzyme interface. Q Rev Biophys. 48:244–79. [DOI] [PubMed] [Google Scholar]
- 8.Xia T., (2008) Taking femtosecond snapshots of RNA conformational dynamics and complexity. Curr Opin Chem Biol. 12:604–11. [DOI] [PubMed] [Google Scholar]
- 9.Bou-Nader C., et al. , (2021) HIV-1 matrix-tRNA complex structure reveals basis for host control of Gag localization. Cell Host Microbe. 29:1421–1436 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stagno JR, et al. , (2017) Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature. 541:242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., et al. , (2018) Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA. Nat Protoc. 13:987–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Li M, and Liu Y, (2020) Optimization and characterization of position-selective labelling of RNA (PLOR) for diverse RNA and DNA sequences. RNA Biol. 17:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CM, et al. , (2008) Pyrrolo-C as a molecular probe for monitoring conformations of the tRNA 3' end. RNA. 14:2245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betteridge T., et al. , (2007) Fluorescent labeling of tRNAs for dynamics experiments. RNA. 13:1594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda I., et al. , (2017) A genetically encoded fluorescent tRNA is active in live-cell protein synthesis. Nucleic Acids Res. 45:4081–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beuning PJ and Musier-Forsyth K, (1999) Transfer RNA recognition by aminoacyl-tRNA synthetases. Biopolymers. 52:1–28. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J and Ferré-D’Amaré AR, (2014) Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol Cell. 55:148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suddala KC and Zhang J, (2019) High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove. Nat Struct Mol Biol. 26:1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battaglia RA, Grigg JC, and Ke A, (2019) Structural basis for tRNA decoding and aminoacylation sensing by T-box riboregulators. Nat Struct Mol Biol. 26:1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J and Ferré-D'Amaré AR, (2013) Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 500:363–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin CT and Coleman JE, (1989) T7 RNA polymerase does not interact with the 5'-phosphate of the initiating nucleotide. Biochemistry. 28:2760–2. [DOI] [PubMed] [Google Scholar]
- 22.Hood IV, et al. , (2019) Crystal structure of an adenovirus virus-associated RNA. Nat Commun. 10:2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein DJ and Ferré-D'Amaré AR, (2006) Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 313:1752–1756. [DOI] [PubMed] [Google Scholar]
- 24.Milligan JF and Uhlenbeck OC, (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180:51–62. [DOI] [PubMed] [Google Scholar]
- 25.Kelley SO and Barton JK, (1999) Electron transfer between bases in double helical DNA. Science. 283:375–81. [DOI] [PubMed] [Google Scholar]
- 26.O'Neill MA and Barton JK, (2002) Effects of strand and directional asymmetry on base-base coupling and charge transfer in double-helical DNA. Proc Natl Acad Sci U S A. 99:16543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tinsley RA and Walter NG, (2006) Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 12:522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen QL, Spata VA, and Matsika S, (2016) Photophysical properties of pyrrolocytosine, a cytosine fluorescent base analogue. Phys Chem Chem Phys. 18:20189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardman SJ, Botchway SW, and Thompson KC, (2008) Evidence for a nonbase stacking effect for the environment-sensitive fluorescent base pyrrolocytosine--comparison with 2-aminopurine. Photochem Photobiol. 84:1473–9. [DOI] [PubMed] [Google Scholar]
- 30.Misra VK and Draper DE, (2001) A thermodynamic framework for Mg2+ binding to RNA. Proc Natl Acad Sci U S A. 98:12456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grilley D, Soto AM, and Draper DE, (2006) Mg2+-RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc Natl Acad Sci U S A. 103:14003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merriman DK, et al. , (2018) Increasing the length of poly-pyrimidine bulges broadens RNA conformational ensembles with minimal impact on stacking energetics. RNA. 24:1363–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limmer S., et al. , (1993) The 3'-terminal end (NCCA) of tRNA determines the structure and stability of the aminoacyl acceptor stem. Proc Natl Acad Sci U S A. 90:6199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagan MC, et al. , (2000) Importance of discriminator base stacking interactions: molecular dynamics analysis of A73 microhelix(Ala) variants. Nucleic Acids Res. 28:2527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., (2020) Unboxing the T-box riboswitches-A glimpse into multivalent and multimodal RNA-RNA interactions. Wiley Interdiscip Rev RNA. 11:e1600. [DOI] [PMC free article] [PubMed] [Google Scholar]