Abstract
Metabolic-associated fatty liver disease (MAFLD) is predominantly associated with metabolic disturbances representing aberrant liver function and increased uric acid (UA) levels. Growing evidences have suggested a close relationship between metabolic disturbances and the gut microbiota. A placebo-controlled, double-blinded, randomized clinical trial was therefore conducted to explore the impacts of daily supplements with various combinations of the probiotics, Lactobacillus fermentum TSF331, Lactobacillus reuteri TSR332, and Lactobacillus plantarum TSP05 with a focus on liver function and serum UA levels. Test subjects with abnormal levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), and UA were recruited and randomly allocated into six groups. Eighty-two participants successfully completed the 60-day intervention without any dropouts or occurrence of adverse events. The serum AST, ALT, and UA levels were significantly reduced in all treatment groups (P < 0.05). The fecal microbiota analysis revealed the intervention led to an increase in the population of commensal bacteria and a decrease in pathobiont bacteria, especially Bilophila wadsworthia. The in vitro study indicated the probiotic treatments reduced lipid accumulation and inflammatory factor expressions in HepG2 cells, and also promoted UA excretion in Caco-2 cells. The supplementation of multi-strain probiotics (TSF331, TSR332, and TSP05) together can improve liver function and UA management and may have good potential in treating asymptomatic MAFLD.
Trial registration. The trial was registered in the US Library of Medicine (clinicaltrials.gov) with the number NCT06183801 on December 28, 2023.
Introduction
Urbanization leads to higher rates of metabolic disturbances due to changes in diet, reduced physical activity, and increased stress. Metabolic disturbances commonly result in obesity, insulin resistance, and dyslipidemia. Fatty liver disease (FLD) is a condition where excess fat builds up in the liver, and there are two types of FLDs: non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD) [1]. FLD is a spectrum, with the presence of fat accumulation being the mildest form and steatohepatitis and cirrhosis being the worst form [2]. While ALD and NAFLD share similar pathological spectra, their etiological factors diverge significantly. ALD is primarily driven by excessive alcohol intake, whereas NAFLD is associated with overconsumption of food [3]. There is growing evidence that NAFLD is a multisystem disease, affecting extra-hepatic organs and regulatory pathways [4]. Recently, the new term ’metabolic-associated fatty liver disease’ (MAFLD) was proposed to better reflect the metabolism-related etiology [5]. NAFLD, or MAFLD, are asymptomatic, so the combination of serum ALT and AST levels, age, body mass index (BMI), and sex is an important biomarker in the diagnosis of hepatic steatosis [6].
In addition to the three high-risk factors, abnormal UA levels were found to be involved in implications for various medical conditions and became the fourth factor for metabolic syndrome [7]. The relationship between high UA levels and gouty arthritis was well-known for centuries. Recently, high UA levels have been observed to affect inflammation, oxidative stress, as well as enzymes related to lipid and glucose metabolism [8]. Both hyperuricemia and NAFLD are associated with lipid metabolism, and their correlation has been observed in both animal and human studies [9, 10]. Moreover, the association was identified in individuals both with and without obesity. Thus, serum UA levels can be an early indicator for metabolic dysregulation [11, 12]. In other words, effectively managing UA levels is as crucial to human health as managing blood pressure, blood sugar, and blood lipids [13].
Although conflicting results existed in different human clinical studies due to the difficulty to identify all intestinal microbes, the evidence still implicated a link between the gut microbiome and metabolic events [14, 15]. The animal studies demonstrated that FLD development was determined by gut bacteria, and dysbiosis of the gut microbiota was observed in NAFLD patients [16]. Studies have observed different changes in specific taxa correlated with obesity, hyperglycemia, dyslipidemia, hypertension, hyperuricemia, and NAFLD [17]. The development of high-throughput sequencing technologies further illustrates microbial composition differences in diseases [18]. Recent research reveals that dietary lipids favor the growth of Bilophila, especially the pathobiont B. wadsworthia, which can synergize with high fat diet and lead to higher glucose dysmetabolism and hepatic steatosis [19, 20]. The supplementation of prebiotic and probiotic functional foods has drawn much attention as an alternative microbial medicine to maintain healthy metabolism [21]. The health conditions of patients can be improved by probiotic supplementation via reducing serum UA level, fatty liver index, and body weight in clinical trials [22–24]. However, no significant impact of probiotics was reported on the modulation of Bilophila or B. wadsworthia. The positive effects of symbiotics appear to be strain-specific, meaning that a particular symbiotic strain may enhance intestinal health by reshaping the gut microbiota composition without necessarily exhibiting any beneficial effects on liver fat [25, 26]. Therefore, clinical studies are imperative to validate the influence of probiotic functional foods on maintaining human health and microbiota balance, specifically by reducing potential pathobionts.
Prevention is better than cure. A probiotic was defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [27]. In recent years, probiotic supplements have been widely used to prevent diseases. Lactobacillus fermentum TSF331 and Lactobacillus reuteri TSR332 were isolated from healthy human gut. Lactobacillus plantarum TSP05 was isolated from Taiwanese pickled cabbage. A study demonstrated L. fermentum TSF331, L. reuteri TSR332, and L. plantarum TSP05 had high antioxidant capacity in vitro, and reduced oxidative stress and inflammatory responses in ethanol-induced liver damaged mice [28]. Another study presented L. fermentum TSF331 and L. reuteri TSR332 assimilated purine nucleoside without UA generation in vitro, and stabilized serum UA levels in oxonate-induced hyperuricemia rats [29]. Therefore, these three Lactobacillus strains displayed good potential for liver health and UA management. In addition to probiotics, growing studies demonstrated the postbiotics (probiotic metabolites) played an important role in modulating intestinal microbiota [30, 31].
The aim of this study was to understand whether the supplementation of mono-strain probiotic, multi-strain probiotics, and multi-strain probiotics plus postbiotics displayed different improvements on liver function and UA levels in subjects with a potential risk of MAFLD. Blood samples were collected to monitor the biochemistry profiles, such as AST, ALT and UA levels. Fecal samples were collected to investigate the gut microbiota modulation using NGS analysis. Lipid accumulation and gene expressions of inflammatory factors were examined to elucidate the liver-protective mechanisms in HepG2 cells. UA transporter activity was analyzed to assess UA excretion in Caco-2 cells.
Materials and methods
Clinical study
Study population
This was a placebo-controlled, double-blinded, randomized clinical study. One hundred and twenty participants were recruited according to their latest physical examination report. The inclusion criteria were ≥ 18 years old, AST > 38 U/L, ALT > 44 U/L, UA > 7 mg/dL for male, and > 6 mg/dL for female. Besides, participants should not have any history of severe liver, cardiovascular, respiratory, kidney disorders or malignancies. Participants were excluded under the following conditions: Firstly, if their serum AST, ALT, or UA levels did not meet the inclusion criteria on day 0. Secondly, if their informed consents were not correctly signed. Thirdly, if their blood test results on day 0 indicated underlying health conditions, such as infections or anemia.
Ethical approval
Informed consent was obtained in written form from every subject randomized in the study. The study protocol was approved by the Ethics Committee of Aging and Disease Prevention Research Center at Fooyin University Hospital (FYH-IRB-110-01-02). The recruitment period for this study was from June 1 to December 15, 2021.
Study design and sample collection
The placebo capsule contained 500 mg maltodextrin. The mono-strain probiotic capsule contained 6.7 × 109 cfu of either L. fermentum TSF331, L. reuteri TSR332, or L. plantarum TSP05. The 3 Mix capsule contained a total 6.7 × 109 cfu, combining L. fermentum TSF331, L. reuteri TSR332, and L. plantarum TSP05. The 3 Mix+PE0401 capsule was composed of a total 6.7 × 109 cfu, including L. fermentum TSF331, L. reuteri TSR332, L. plantarum TSP05, and 200 mg Totipro® PE0401 postbiotic powder. L. fermentum TSF331 (BCRC 910815 = CGMCC 15527) and L. reuteri TSR332 (BCRC 910816 = CGMCC 15528) were isolated from the gut of healthy humans, whereas L. plantarum TSP05 (BCRC 910855 = CGMCC 16710) was isolated from kimchi. Totipro® PE0401 was a fermentation product derived from probiotics [32]. Capsules were prepared with the same appearance by Glac Biotech Co., Ltd., Taiwan. Every subject was not aware of his/her treatment group, and took 3 capsules per day for 60 days. The blood and fecal samples were collected on day 0 and 60.
Blood biochemistry
The blood samples were analyzed by Everest Inspection International Co., Ltd. Seventeen tests were performed to examine the effect of the intervention on physical indexes. The panel of tests measuring liver function included AST, ALT, albumin (ALB), total bilirubin (TBIL), gamma-glutamyl transferase (GGT), and alkaline phosphatase (ALKP). The panel of tests measuring kidney function included UA, creatinine (CREA), and blood urea nitrogen (BUN). The panel of tests measuring energy metabolism included blood glucose (GLU), triglycerides (TG), cholesterol (CHOL), high-density lipoprotein (HDL), low-density lipoprotein (LDL). The panel of tests measuring body damages or inflammation included lactate dehydrogenase (LDH), creatine kinase (CK), high-sensitive C-reactive protein (hs-CRP).
Gut microbiota analysis
DNA extraction and next generation sequencing (NGS)
Bacteria DNA extraction was performed on the fecal samples using the QIAamp® DNA Mini Kit (QIAGEN Canada, Mississauga, ON, Canada), following the manufacturer’s protocol. After extraction and purification, the DNA was used as the polymerase chain reaction (PCR) template for amplification. The bacterial V3-V4 region of 16S rRNA was amplified using the specific primer pair 314F (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3’) and 805R (5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’) [33, 34]. The amplification was performed with KAPA HiFi HotStart ReadyMix [Roche Sequencing Solutions, Pleasanton, CA, USA (KK2601)] through the following steps: 95°C for 5 min; followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and final extension at 72°C for 5 min. The PCR products were stored at 4°C and then used as templates for Index PCR, which was run under the following conditions: 95°C for 30 s; 8 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and final extension at 72°C for 5 min. DNA samples were paired-end sequenced (2 × 300 bp) on an Illumina MiSeq platform (Illumina, San Diego, USA) by Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
Flora diversity and statistical analysis
Sequence data were performed using 16S Metagenomics apps on Basespace (Illumina, San Diego, CA, USA) and the reads were clustered to operational taxonomic units (OTUs) with Illumina-curated version of May 2013 Greengenes taxonomic database for downstream analysis. The Simpson index was used to indicate the alpha diversity of bacteria, in which a larger index represents higher community diversity. Beta diversity was estimated with Jaccard index using MicrobiomeAnalyst 2.0 [35], and principal coordinate analysis (PCoA) was calculated to analyze changes in species composition on time and space scales.
Heatmap of microbiota modulation and statistical analysis DNA extraction and next generation sequencing (NGS)
The percentage of each bacterium was calculated by dividing its individual hit number by the total hit number. The difference between the end and the beginning of the intervention was assessed using Student’s t-test. Statistical significance is denoted as the P-value.
The THP-1-HepG2 cell system analysis
The co-culture supernatant of probiotic bacteria and THP-1-differentiated macrophages (SPT) preparation.
THP-1 monocytic leukemia cells (ATCC TIB-202) were inoculated in a 75T flask at a density of 2 × 105 cells/ml and grown to a density of 107 cells/ml. The THP-1 cells were differentiated into macrophages using a 24-hours phorbol-12-myristate-13-acetate (PMA, Sigma, United States) treatment. After resting in fresh medium for 24 hours, the THP-1 macrophages were detached from the culture plate by adding 5 ml 0.25% trypsin. Then, the THP-1-differentiated macrophages were seeded into a 12-well plate at a density of 8 × 105 cells/ml/well, and co-cultured for 24 hours with probiotic bacteria at a cell to bacteria ratio of 1:50 after cell attachment. The supernatant was collected by centrifugation at 4000 rpm for 10 min and 0.22 μm filtration. The SPT was store at −20°C for further use. The supernatant of THP-1-differentiated macrophages served as a blank SPT.
Oil Red O staining in HepG2 cells
The free fatty acid (FFA) medium was prepared by adding 186 μL 354 mM oleic acid and 846 μL 39 mM palmitic acid in Dulbecco’s Modified Eagle Medium (DMEM, Cytiva, United States) medium containing 10% fetal bovine sera (FBS, Gibco, United States), 1% penicillin-streptomycin (PS, Cytiva, United States), 2% bovine serum albumin (BSA, EMD Millipore, United States). HepG2 human hepatoma cells (BCRC 60177) were seeded into a 6-well plate and incubated for 3 days before further treatments. Then, the HepG2 cells were treated with 1 mM FFA medium for 24 hours to induce hepatic steatosis [36]. After washing with phosphate-buffered saline (PBS) twice, the HepG2 cells were treated for 48 hours with either 1mM FFA medium, 1 μg/mL lipopolysaccharide (LPS, Sigma, United States) or 50 μL/mL SPT mentioned above. After washing with PBS twice, the HepG2 cells were fixed with 10% formalin, washed with 60% isopropanol, and stained with 0.5% oil red O (Sigma-Aldrich, United States). After the stained HepG2 cells were observed at 100x magnification under a microscope (Nikon, Japan), the oil red O dye was eluted by 100% isopropanol and measured at OD520 by a CLARIOstar® Plus (BMGlabtech, Germany).
The uric acid excretion in Caco-2 cells
Caco-2 human colon carcinoma cells (BCRC 67001) were seeded and grown as a monolayer on a 0.4 μm Falcon® permeable support (CORNING, United States). Before treatments, the trans-epithelial electric resistance (TEER) across the Caco-2 monolayer was measured with fresh DMEM medium by a EVOM2 Epithelial Voltohmmeter (World Precision Instruments, United States). Then, the Caco-2 cells were treated with DMEM medium containing probiotic bacteria in the upper compartment, and DMEM medium containing 10 μM UA in the lower compartment. The UA concentration in the upper compartment was measured at 30 minutes as the starting point and at 24 hours as the ending point with a UA assay kit (Cayman Chemical, United States) by a CLARIOstar® Plus microplate reader (BMG LABTECH, Germany). After washing with PBS twice, the TEER across the Caco-2 monolayer was measure with fresh DMEM medium again at the end of the treatment.
Gene expression analysis
For cytokine gene expression (IL-6, IL-8 and CCL2), the HepG2 cells grown in a 6-well culture plate for 3 days, and then treated with 1 mM FFA medium for 24 hours. After washing with PBS twice, the HepG2 cells were treated for 48 hours with either 1 mM FFA medium or 50 μL/mL SPT mentioned above. Except for the FFA group, all groups were treated with 1 μg/mL LPS for further 3 hours, and cells were harvested for qPCR analysis.
For UA transporter gene expression (ABCG2 and SLC2A9), the Caco-2 cell was grown as a monolayer for 6 days on a 0.4 μm Falcon® permeable support (CORNING, United States). The Caco-2 monolayer was treated for 6 hours with DMEM medium containing probiotic bacteria in the upper compartment, and DMEM medium containing 10 μM UA in the lower compartment. After removing medium in the upper compartment, Caco-2 cells were harvested for qPCR analysis.
RNA was extracted with Trizol (Invitrogen, United States). The cDNA was generated with a GoScript™ Reverse Transcriptase (Promega, United States), and qPCR was performed with a PB20.12–05 qPCRBio SyGreen Mix Hi-ROX (PCR Biosystems, United States) by an ABI StepOnePlus™ qPCR machine (Applied Biosystems, United States). The primer pair 5’-CATCCTCGACGGCATCTCAG-3’ and 5’-TGCCTCTTTGCTGCTTTCAC-3’ was used to detect IL-6 expression [37]. The primer pair 5’-CTGGCCGTGGCTCTCTTG-3’ and 5’-CCTTGGCAAAACTGCACCTT-3’ was used to detect IL-8 expression [36]. The primer pair 5’-CTCAGCCAGATGCAATCAATG-3’ and 5’-AGATCACAGCTTCTTTGGGACAC-3’ was used to detect CCL2 expression [36]. The primer pair 5’-AATACATCAGCGGATACTA-3’ and 5’-AATAAGCCACCATCATAAG-3’ was used to detect ABCG2 expression [38]. The primer pair 5’-CAATAGACCCAGACACTCTGACT-3’ and 5’-TCTTCACAATTAACGTCCCCAC-3’ was used to detect SLC2A9 expression [38]. The primer pair 5’-GAAGATGGTGATGGGATTTC-3’ and 5’-GAAGGTGAAGGTCGGAGT-3’ was used to detect GAPDH expression as the internal control [39].
Statistical analysis
One-way analysis of variance (ANOVA) was used to examine the differences among all groups, and Student’s t-test was used to compare the differences between two groups. The paired t-test was used to compare the difference before and after the intervention within each group. The difference of P < 0.05 was considered statistically significant. The change rate of the test item was presented as the standardized mean percentage (ValueEnd/ValueStart). Figs were generated by using Graphpad prism 8 (Graphpad Software, San Diego, CA, United States). The statistical analyses were performed utilizing SPSS software (IBM, Armonk, NY, USA).
Results
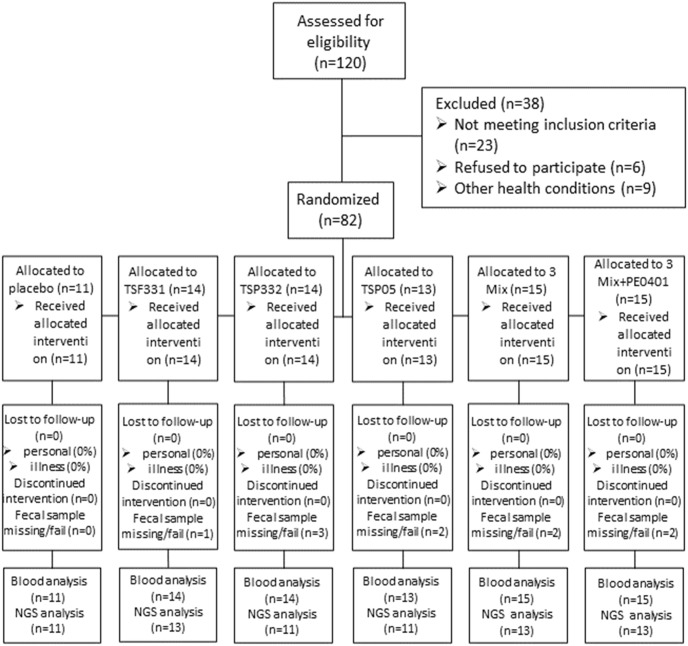
The clinical study was carried out as the flowchart in Fig 1.
Fig 1. The subject enrollment, randomization, and disposition were presented in the diagram.
One hundred and twenty participants were recruited according to their latest physical examination report. Twenty-three participants were excluded due to unqualified blood biochemistry profiles on day 0. Additionally, six participants were excluded because of improperly signed informed consents, while nine others were excluded due to other underlying health conditions. Eighty-two participants were randomized into 6 groups, and all subjects completed the intervention (11 in the placebo group, 41 in the mono-strain group, 15 in the 3 Mix groups, and 15 in the 3 Mix+PE0401 group). Ten fecal samples were excluded due to poor storage or bad sample quality. In the end, the blood chemistry was analyzed in 82 subjects, and gut microbiota was analyzed in 72 subjects.
The multi-strain probiotics plus postbiotics enhanced the protective effect on liver health subsection
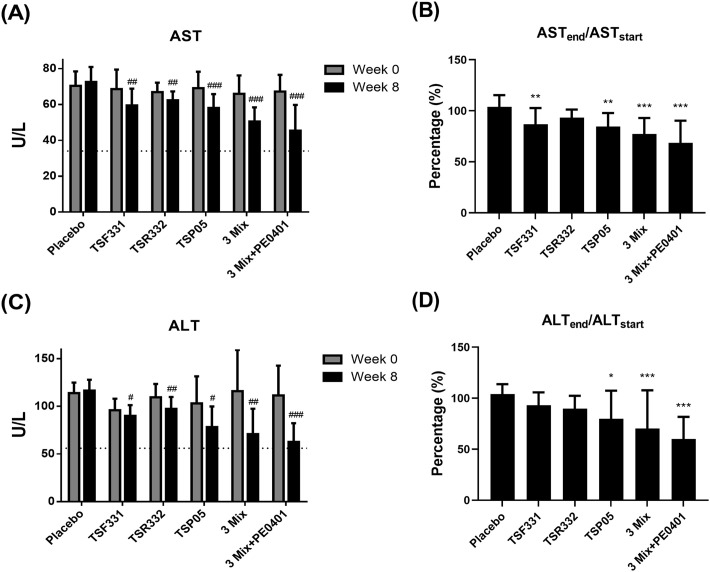
To assess the impact of the intervention on liver health, a comprehensive panel of tests measuring liver function was conducted using blood samples obtained from the subjects. Before the initiation of the intervention on day 0, there was no significant difference in the serum AST levels among the groups (P = 0.735, Fig 2A). After the intervention on day 60, a notable disparity in the serum AST levels emerged among the groups (P < 0.001, Fig 2A). All treatment groups demonstrated a marked reduction in serum AST levels compared to the control group (P < 0.05). Remarkably, within each treatment group, serum AST levels exhibited a significant reduction compared to their respective baseline levels (P < 0.01). The placebo group showed a marginal increase of 3.84 ± 11.53%. In contrast, with the exception of the TSR332 group, the TSF331, TSP05, 3 Mix, and 3 Mix+PE0401 groups demonstrated substantial reductions of 13.09 ± 15.82%, 15.46 ± 13.29%, 22.61 ± 15.57%, and 31.43 ± 21.69%, respectively (Fig 2B). Particularly noteworthy is the multi-probiotics plus postbiotics group, which exhibited the most significant reduction in AST levels among all the groups.
Fig 2. The effect of probiotic supplementation on liver health was evaluated in liver enzymes.
(A) The AST levels were recorded on day 0 and 60. (B) The difference of serum AST levels between before and after the intervention was converted to the change rate in each group. (C) The serum ALT levels were recorded on day 0 and 60. (D) The difference of serum ALT levels between before and after the intervention was converted to the change rate in each group. The dotted lines denote the upper limit of normal serum AST and ALT levels. Data are presented as mean ± standard deviation (SD) of the results from each subject. #The paired t-test was used to compare the difference before and after the intervention within each group, #P < 0.05, ##P < 0.01, ###P < 0.001. The one-way analysis of variance (ANOVA) was utilized to compare the change rate with the control group, followed by the least significant difference (LSD) post hoc test for subsequent analysis. Statistical significance was denoted as *P < 0.05, **P < 0.01, and ***P < 0.001.
Similarly, there was no discernible difference in the serum ALT levels among the groups prior to the initiation of the intervention on day 0 (P = 0.683, Fig 2C). A notable divergence in the serum ALT levels emerged among the groups, showing a significant contrast post-intervention on day 60 (P < 0.001, Fig 2C). Notably, within each treatment group, serum ALT levels showed a substantial reduction compared to their respective baseline levels (P < 0.05). The placebo group experienced a marginal increase of 4.01 ± 9.68%. In contrast, with the exception of the TSF331 and TSR332 groups, the TSP05, 3 Mix, and 3 Mix+PE0401 groups demonstrated noteworthy reductions of 20.26 ± 27.59%, 29.74 ± 37.43%, 40.00 ± 21.72%, respectively (Fig 2D). Particularly remarkable is the multi-probiotics plus postbiotics group, showcasing the most significant reduction in ALT levels among all the groups.
The multi-strain probiotics plus postbiotics enhanced the UA lowing effect
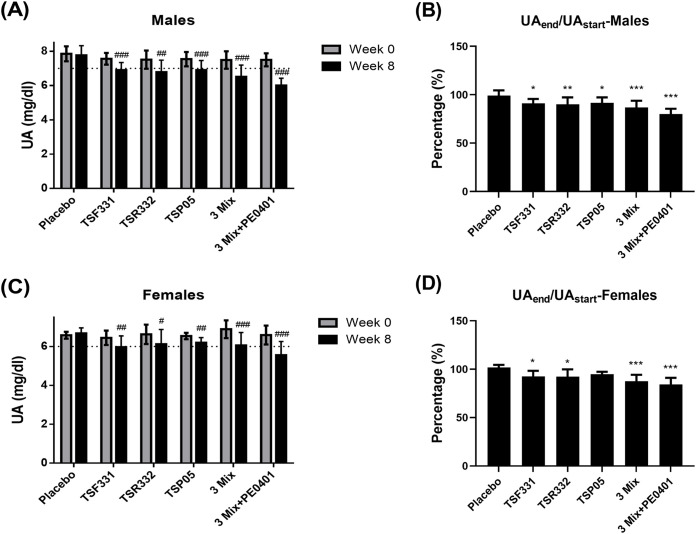
UA levels can vary between males and females, partly due to the sex hormone. To assess the impact of the intervention on UA management, the serum UA levels were separately analyzed in the male and female subjects. In males, there was no significant difference in the serum UA levels among the groups before the intervention on day 0 (P = 0.535, Fig 3A). After the intervention on day 60, a significant difference in the serum UA levels appeared among the groups (P < 0.001, Fig 3A). All treatment groups demonstrated a marked reduction in serum UA levels compared to the control group (P < 0.01). Notably, within each treatment group, serum UA levels exhibited a significant reduction compared to their respective baseline levels (P < 0.01). The placebo group showed a marginal reduction of 0.97 ± 5.59%, while the TSF331, TSR332, TSP05, 3 Mix, and 3 Mix+PE0401 groups demonstrated substantial reductions of 8.77 ± 4.44%, 9.76 ± 7.04%, 8.29 ± 5.61%, 13.01 ± 6.81%, 19.93 ± 5.43%, respectively (Fig 3B). The serum UA levels for males were significantly reduced in all probiotic groups and, remarkably, the greatest reduction was observed in the multi-probiotics plus postbiotics group.
Fig 3. The effect of probiotic supplementation on UA management was investigated in mono- and multi-strain groups.
(A) The serum UA levels were recorded in male on day 0 and 60. (B) The difference of serum UA levels between before and after the intervention was converted to the change rate in male subjects in each group. (C) The serum UA levels were recorded in female on day 0 and 60. (D) The difference of serum UA levels between before and after the intervention was converted to the change rate in female subjects in each group. The dotted lines denote the upper limit of normal serum UA levels. Data are presented as mean ± SD of the results from each subject. # The paired t-test was used to compare the difference before and after the intervention within each group, #P < 0.05, ##P < 0.01, ###P < 0.001. The one-way ANOVA was utilized to compare the change rate with the control group, followed by the LSD post hoc test for subsequent analysis. Statistical significance was denoted as *P < 0.05, **P < 0.01, and ***P < 0.001.
In females, no discernible difference was observed in the serum UA levels among the groups prior to the initiation of the intervention on day 0 (P = 0.793, Fig 3C). There was no statistical significance in the serum UA levels was detected among the groups after the intervention on day 60 (P = 0.188, Fig 3C). Remarkably, within each treatment group, serum UA levels underwent a notable reduction compared to their respective baseline levels (P < 0.05). The placebo group displayed a slight increase of 1.69 ± 2.79%, while the TSF331, TSR332, 3 Mix, and 3 Mix+PE0401 groups exhibited significant reduction of 7.47 ± 5.77%, 7.77 ± 7.58%, 12.46 ± 6.62%, 15.76 ± 6.81%, respectively (Fig 3D). The serum UA levels for females were significantly reduced in all probiotic groups, with the exception of the TSP05 group. Notably, the greatest reduction was observed in the multi-probiotics plus postbiotics group.
The multi-strain probiotics plus postbiotics enhanced the improvement on energy metabolism
Despite the wide age range among the subjects, the allocation was evenly distributed (P = 0.161, S1 Table). The blood chemistry profiles were initially comparable, with measurements, e.g., TBIL, GGT, and ALKP, in the liver function panel falling within the normal range across all groups on day 0 (S1 Table). Remarkably, by day 60, the blood chemistry profiles continued to remain within the normal range (Table 1).
Table 1. The blood biochemical profile on day 60.
| Placebo (N = 11) | TSF331 (N = 14) | TSR332 (N = 14) | TSP05 (N = 13) | 3 Mix (N = 15) | 3 Mix +PE0401 (N = 15) | P-value | |
|---|---|---|---|---|---|---|---|
| TBIL | 0.82 | 0.58 | 0.82 | 0.80 | 0.59# | 0.81# | 0.136 |
| (mg/dL) | ± 0.35 | ± 0.26 | ± 0.32 | ± 0.36 | ± 0.25 | ± 0.40 | |
| GGT | 23.18 | 29.43 | 27.07 | 27.85 | 23.13### | 20.40*### | 0.394 |
| (U/L) | ± 13.28 | ± 16.35 | ± 10.99 | ± 5.87 | ± 6.51 | ± 17.72 | |
| ALKP | 54.36 | 62.00 | 58.21 | 63.23 | 51.33 | 52.00## | 0.358 |
| (U/L) | ± 7.57 | ± 13.52 | ± 24.18 | ± 20.82 | ± 15.57 | ± 19.54 | |
| ALB | 4.46# | 4.83 | 4.51 | 4.57 | 4.41 | 4.41 | 0.076 |
| (g/dL) | ± 0.24 | ± 0.48 | ± 0.39 | ±0.42 | ± 0.22 | ± 0.22 | |
| CREA | 0.85 | 0.85 | 0.75 | 0.77 | 0.74 | 0.74 | 0.096 |
| (mg/dL) | ± 0.10 | ± 0.14 | ± 0.14 | ± 0.11 | ± 0.14 | ± 0.14 | |
| BUN | 13.55 | 14.61 | 13.61 | 15.62 | 11.92 | 11.92 | 0.466 |
| (mg/dL) | ± 5.40 | ± 6.26 | ± 4.24 | ± 6.83 | ± 2.60 | ± 2.60 | |
| GLU | 88.91 | 80.36*# | 77.79*# | 77.92*# | 78.27*# | 78.73*# | 0.206 |
| (mg/dL) | ± 7.08 | ± 12.11 | ± 10.42 | ± 12.44 | ± 15.33 | ± 11.98 | |
| TG | 124.55 | 110.64 | 116.79# | 111.92## | 104.33*## | 102.20*### | 0.256 |
| (mg/dL) | ± 39.06 | ± 15.00 | ± 22.11 | ± 20.72 | ± 31.97 | ± 18.92 | |
| CHOL | 183.82 | 163.29 | 176.29 | 160.08# | 160.27# | 160.80*## | 0.341 |
| (mg/dL) | ± 25.98 | ± 38.42 | ± 37.99 | ± 33.53 | ± 31.13 | ± 29.99 | |
| LDL | 115.25 | 100.64 | 84.16 | 99.13 | 90.59 | 95.31# | 0.295 |
| (mg/dL) | ± 30.80 | ± 29.72 | ± 34.88 | ± 36.48 | ± 30.33 | ± 35.89 | |
| HDL | 51.55 | 54.61 | 53.23 | 51.91 | 49.72 | 51.36 | 0.909 |
| (mg/dL) | ± 12.09 | ± 12.17 | ± 10.53 | ± 13.72 | ± 10.00 | ± 10.90 | |
| Hs-CRP | 0.19a | 0.26a | 0.32b | 0.27a | 0.15a | 0.19a | 0.042 |
| (mg/dL) | ± 0.08 | ± 0.18 | ± 0.16 | ± 0.15 | ± 0.14 | ± 0.17 | |
| LDH | 109.27 | 123.43 | 97.93 | 110.62 | 99.07 | 89.00**## | 0.058 |
| (U/L) | ± 28.99 | ± 32.52 | ± 30.62 | ± 36.50 | ± 32.12 | ± 19.78 | |
| CK | 112.55 | 111.29 | 102.71 | 80.69 | 81.67 | 98.27 | 0.362 |
| (U/L) | ± 65.94 | ± 52.05 | ± 44.59 | ± 40.27 | ± 36.22 | ± 50.05 |
Data are presented as mean ± SD of the results from each subject. The P-value presented the difference among groups using One-way ANOVA. a,b Groups with different letters are considered significantly different from each other, P < 0.05. Change rate was calculated as valueend/valuestart × 100% in the same subject. * The change rate was compared between placebo and treatment groups using Student’s t-test, *P < 0.05, and **P < 0.01. # The difference before and after the intervention was compared within each group using a paired t-test, #P < 0.05, ##P < 0.01, ###P < 0.001.
Notably, the probiotic intervention exerted discernible effects on various energy metabolic panels. Specifically, the serum glucose (GLU) levels exhibited a reduction in all probiotic groups. Additionally, the blood lipid profile, including TG, CHOL, and LDL, showed a decrease in the multi-probiotics plus postbiotics group.
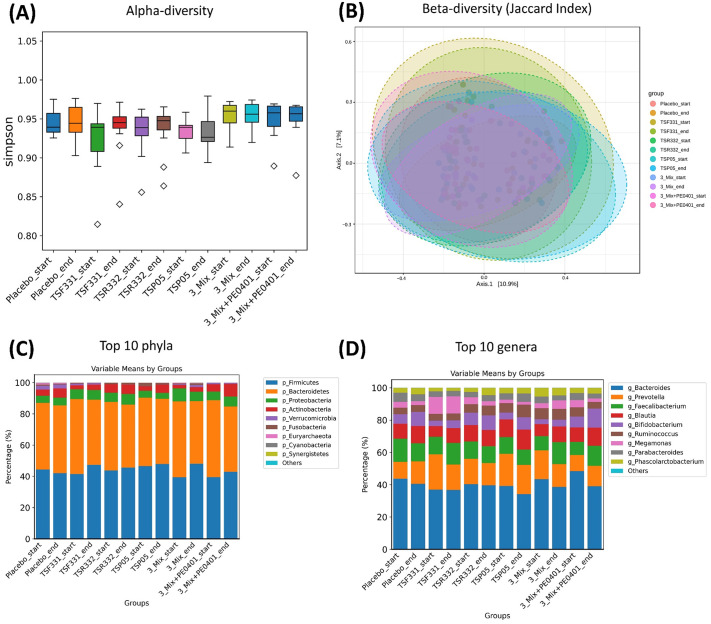
Probiotic supplementation modulated gut microbiota without affecting diversity
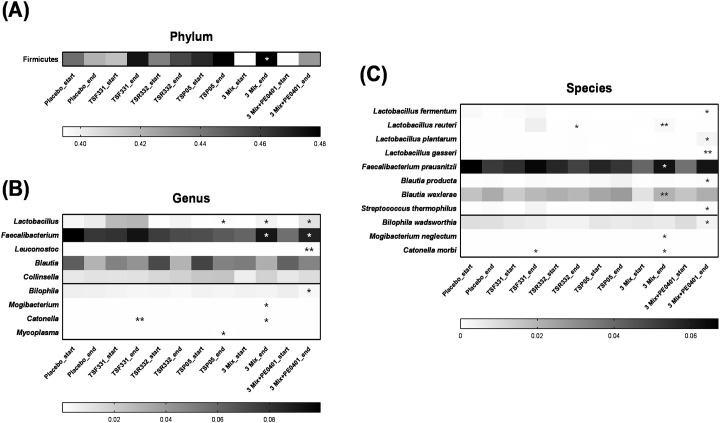
To assess the modulation of gut microbiota through probiotic supplementation, the microbial composition in fecal samples was compared between day 0 and day 60. No significant alteration was detected in alpha and beta diversity (Fig 4A and 4B). Two phyla Firmicutes and Bacteroidetes composed more than 80% of the flora (Fig 4C). Five genera Bacteroides, Prevotella, Faecalibacterium, Blautia, and Bifidobacterium represented more than 80% of the population (Fig 4D). Prior to the intervention, the patterns of the 10 most abundant phyla and genera were similar but not identical among the groups, and the probiotic supplementation did not dramatically alter these patterns in any of the groups.
Fig 4. The effect of probiotic supplementation on gut microbiota was analyzed by NGS.
No significant alteration was detected in (A) alpha diversity and (B) beta diversity. The composition of the 10 most abundant (C) phyla and (D) genera was not affected by the intervention in all groups.
While preserving diversity, several microbial changes occurred. The abundance of the Firmicutes phylum significantly increased in the 3 Mix group (Fig 5A). Notably, the 3 Mix and 3 Mix+PE0401 groups displayed the best efficiency on the modulation. Two probiotic genera (Lactobacillus and Faecalibacterium) showed significant increases in the 3 Mix group, while two pathobiont genera (Mogibacterium and Catonella) decreased (Fig 5B). Three probiotic genera (Lactobacillus, Faecalibacterium, and Leuconostoc) showed significant increases in the 3 Mix+PE0401 group, while one pathobiont genus, Bilophila, decreased (Fig 5B). Additionally, three probiotic species (L. reuteri, F. prausnitzii, and B. wexlerae) significantly increased, and two pathobiont species (M. neglectum and C. morbi) decreased in the 3 Mix group (Fig 5C). Five probiotic species (L. fermentum, L. plantarum, L. gasseri, B. producta, and S. thermophilus) significantly increased, and one pathobiont species, B. wadsworthia, decreased in the 3 Mix+PE0401 group (Fig 5C).
Fig 5. The probiotic supplementation promoted the growth of commensal bacteria and inhibited pathobiont bacteria in gut microbiota.
The intervention resulted in significant alterations to the abundance of (A) one phylum, (B) seven genera, and (C) eleven species. * The paired t-test was employed to assess the differences in abundance within the group before and after the intervention, *P < 0.05, and **P < 0.01.
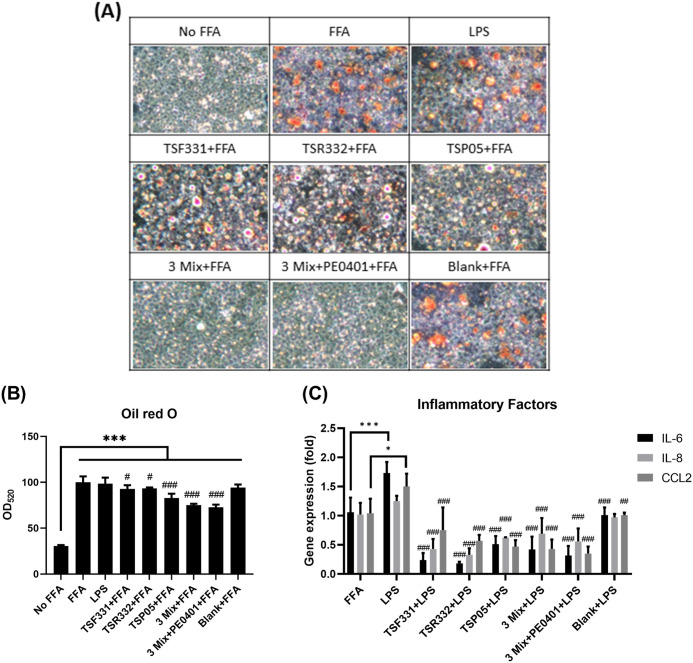
Lactobacillus strains reduced the lipid accumulation and inflammatory factor expressions in a gut-liver axis system
To ascertain whether the oral probiotic supplementation reduced serum AST and ALT levels by mitigating lipid accumulation and inflammation, the oil red O staining and the expression of inflammatory factor genes were conducted in a THP-1-HepG2 cell system. The oil red O staining reveled a red color for the lipid accumulation in both FFA and LPS treated HepG2 cells (Fig 6A). The lipid accumulation was quantified by eluting the cellular oil red O dye, and the dye concentration was significantly reduced in all treatment groups (Fig 6B). The LPS treatment significantly elevated the expression of inflammatory factors IL-6 and chemokine (C-C motif) ligand 2 (CCL2) in FFA-treated HepG2 cells. Conversely, the expression of IL-6, IL-8, and CCL2 was significantly reduced in all SPT treatment groups (Fig 6C).
Fig 6. The SPT treatments reduced lipid droplet accumulation and inflammatory factor expression in HepG2 cells.
(A) The lipid droplet accumulation in HepG2 cells was observed at 100x magnification under microscope. (B) The oil red O dye was eluted and measured at OD520. Data were analyzed using one-way ANOVA, followed by the LSD post hoc test. Significance levels were indicated as follows: ***P < 0.001 compared to No FFA; #P < 0.05, and ###P < 0.001 compared to FFA. (C) The gene expression of IL-6, IL-8, and CCL2 was analyzed by RT-qPCR. Blank: The supernatant of THP-1-differentiated macrophages without co-culturing with probiotic bacteria. Data were analyzed using one-way ANOVA, followed by the LSD post hoc test. Significance levels were denoted as follows: for the difference between FFA and LPS groups, *P < 0.05 and ***P < 0.001; for the difference between LPS and SPT treatment groups, ##P < 0.01 and ###P < 0.001. All experiments were performed in triplicate and data are presented as mean ± SD.
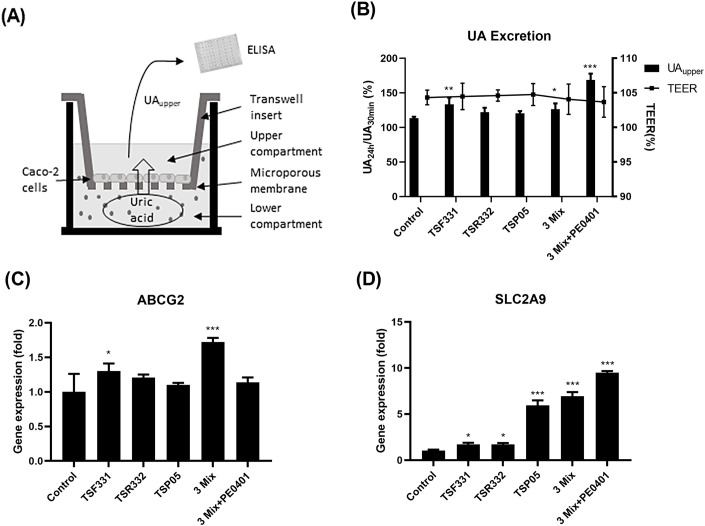
Lactobacillus strains enhanced UA transporter expressions and heightened the UA excretion in Caco-2 cells
To confirm whether oral probiotic supplementation reduces serum UA levels by enhancing UA excretion in the gastrointestinal tract, the directional UA transport experiments were conducted using the Transwell system as indicated in Fig 7A. In the presence of a well-maintained Caco-2 monolayer with good integrity, efficient transport of UA from the lower compartment to the upper compartment was observed in the TSF331, 3 Mix, and 3 Mix+PE0401 groups (Fig 7B). ABCG2 is a membrane transporter involving in the efflux of UA from intestinal cells, and its expression was enhanced in the TSF331 and 3 Mix groups (Fig 7C). SLC2A9 is another transporter playing a role in the regulation of UA levels in the body, and its expression significantly increased in all treatment groups (Fig 7D).
Fig 7. The probiotic bacteria promoted UA excretion and the gene expression of UA transporters in Caco-2 cells.
(A) UA was introduced into the lower compartment, and the concentration of UA in the upper compartment was analyzed using enzyme-linked immunosorbent assay (ELISA). (B) The UA concentration in the upper compartment was recorder at 30 min and 24 hours. At the end of 24 hours incubation, the TEER was recorded to exam the integrity of Caco-2 monolayer. The gene expression of UA transporters, (C) ABCG2 and (D) SLC2A9, was analyzed by RT-qPCR. All experiments were performed in triplicate and data are presented as mean ± SD. Data were analyzed using one-way ANOVA, followed by the LSD post hoc test. Significance levels were indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001 compared to control.
Discussion
This pilot study sought to assess the varying efficacy of mono-strain, multi-strain, and multi-strain plus postbiotics in the individuals at high risk of MAFLD. The findings should be interpreted cautiously owing to the limited sample size within each group. Nonetheless, our results indicate that further investigation into the efficacy of multi-strain and multi-strain plus postbiotics is warranted. In the context of treating irritable bowel syndrome (IBS), a systematic review revealed more pronounced beneficial effects in trials utilizing multi-strain supplements as opposed to mono-strain alternatives [40]. The gut microbiota is a microbial ecosystem containing a complex and diverse community of microorganisms. Gut microbiota dysbiosis is disruptions in the balance, which may lead to neurodegenerative diseases, cardiovascular diseases, metabolic diseases and gastrointestinal diseases [41]. Hence, the synergistic effect of combining probiotics and postbiotics may be more effective than mono-strain probiotics in creating a harmonious environment within the gut, resulting in enhanced health benefits.
As of now, the information establishing a clear relationship between a specific genus or species and metabolic disturbances remains limited. Our study sheds the light on a potential target for reshaping the gut microbiota. Particularly in the 3 Mix+PE0401 group, we observed a significant inhibition of the growth of Bilophila, especially the pathobiont B. wadsworthia. In addition, several animal investigations have indicated an increase in the abundance of Bilophila within the gut microbiota of high-fat diet (HFD)-fed rats, implying an association between this bacterial genus and high-fat dietary intake [42, 43]. Enrichment of B. wadsworthia in the host’s intestinal tract may precipitate disruptions in bile acid metabolism, inflammation, and compromise intestinal barrier integrity. Consequently, these disturbances may exacerbate glucose metabolism disorders and hepatic steatosis [43–45]. Totipro® PE0401 is postbiotics consisted of metabolites from Lactobacillus salivarius AP-32, L. acidophilus TYCA06, L. plantarum LPL28, Bifidobacterium longum subsp. infantis BLI-02. Totipro® PE0401 displayed the unique synergistic effect on the expression of tight junction proteins (TJPs) and the growth promoting effect on various probiotic bacteria strains [32]. Therefore, Totipro® PE0401 may be able to enhance the health effect of probiotic bacteria via promoting intestinal barrier and beneficial bacteria growth. While some weak inhibition was observed in other treatment groups, the effect was not profound. Instead, there was a reduction in the abundance of genera Mogibacterium, Catonella, and Mycoplasma in certain groups. Mogibacterium exhibited a positive correlation with the pro-inflammatory factors interferon-γ and tumor necrosis factor-α in individuals diagnosed with medication-related osteonecrosis of the jaw (MRONJ) [46]. Catonella bacteria typically inhabit the gastrointestinal tract, but the presence of this genus, especially C. morbi species, in the oral cavity is associated with oral microbial dysbiosis [47, 48]. Mycoplasmas have a unique cell membrane and lack a rigid cell wall [49]. Although not all Mycoplasma species are naturally pathobionts, some commensals can become opportunistic pathobionts under certain conditions, especially when the host’s immune system is compromised [50]. In this study, varying degrees of growth inhibition were observed among pathobiont populations in different combinations of probiotics and postbiotics. The findings indicated that the combination of multi-strain probiotics with postbiotics exhibited the most effective inhibition of B. wadsworthia, a bacterium associated with glucose dysmetabolism and hepatic steatosis.
Lipid accumulation in the liver, also known as hepatic steatosis, occurs when there is an excessive build-up of fats (lipids), primarily TG, within liver cells (hepatocytes) [51]. In some cases, hepatic steatosis can progress to non-alcoholic steatohepatitis, which is characterized by not only fat accumulation but also liver inflammation and damage. Elevated levels of ALT and AST in the blood can be indicative of liver damage. Recently, accumulating data indicated that FFAs played a damaging role on liver cells and involved in the cross-talk between the gut and the liver [52]. A meta-analysis of randomized controlled trials concluded microbial therapies could improve liver steatosis and ALT levels [53]. Furthermore, an additional systematic review, encompassing 10 studies, assessed the influence of probiotics on liver function tests, demonstrating a beneficial effect of probiotics on ALT, AST, and GGT levels among patients with NAFLD [54]. In an animal study, feeding mice a HFD along with L. paracasei CCFM1224-derived postbiotics (800 mg/kg/day) was found to mitigate weight gain [55]. Simultaneously, it inhibited the accumulation of epididymal white adipose tissue and insulin resistance, improved serum biochemical indicators related to lipid metabolism, and alleviated hepatic steatosis and inflammation. The postbiotics were capable of modulating the gut microbiota of HFD-fed mice, thereby increasing the relative abundance of Akkermansia. Remarkably, our study demonstrated the combination of probiotic microbials (L. fermentum TSF331, L. reuteri TSR332, and L. plantarum TSP05) and postbiotics (Totipro® PE0401) exhibited more effective ALT and AST reduction than the probiotics alone. This combination also displayed the lowest lipid accumulation and the enhanced anti-inflammatory effect in vitro. Symbiotic metabolites can modulate the composition and activity of the gut microbiota, promoting a balanced and diverse microbial community [56]. Moreover, symbiotic metabolites have been shown to possess anti-inflammatory properties, helping to mitigate excessive immune responses and reduce inflammation in the body [57]. Our study uncovered that the beneficial effects of combining probiotics and postbiotics are additive.
Presently, there are safety and tolerability concerns associated with urate-lowering drugs [58]. Drugs aimed at promoting UA excretion primarily target the kidneys, posing a heightened risk for kidney burden, particularly in chronic kidney disease patients. Hence, there is a potential shift towards the intestine as a safer and more effective target organ for urate-lowering drugs due to its substantial excretory capacity [59]. Remodeling the gut microbiota has been suggested as a promising therapeutic strategy to manage hyperuricemia and gout [60]. An animal study revealed that oral administration of Lacticaseibacillus paracasei MJM60396 (3 × 109 cfu/mice/day) for three weeks significantly reduced serum uric acid levels to within normal range [61]. This effect was achieved through the inhibition of xanthine oxidase (XO) to decrease UA synthesis, as well as the modulation of uric acid transporters to enhance UA excretion. Another animal study showed that mice with hyperuricemia experienced a notable 35.5% decrease in serum uric acid concentration after consuming Lactiplantibacillus plantarum X7022 for four weeks [62]. Concurrently, levels of propionate and butyrate in their feces were elevated. These physiological changes could be attributed to the suppression of XO activity and the regulation of UA transport protein expression to normal levels. Furthermore, probiotics also ameliorated dysbiosis in the gut microbiota of hyperuricemia mice and promoted the production of SCFA-related microbiota. The supplementation of probiotic microbials contributed to the nucleoside degeneration in the gastrointestinal tract and the UA excretion in the feces and urine [63–66]. Interestingly, the combination of probiotic microbials (L. fermentum TSF331, L. reuteri TSR332, and L. plantarum TSP05) and postbiotics (Totipro® PE0401) displayed more effective UA reduction compared to probiotics alone in our study. This combination exhibited the best UA excretion and UA transporter SLC2A9 upregulation in Caco-2 cells. However, the regulation may not be synchronized in different UA transporter genes, such as ABCG2. Further investigations are needed to illustrate the UA lowering mechanism by the combined action of probiotics and postbiotics in details.
Three Lactobacillus strains were introduced in this study, and several microbial changes in respond to the intervention. The rise in abundance within the phylum Firmicutes, as well as in the genera Lactobacillus and the specific species L. fermentum, L. reuteri, and L. plantarum, signifies the successful implementation of the intervention. Notably, the genera Faecalibacterium and Leuconostoc increased in the 3 Mix and 3 Mix+PE0401 groups. Low levels of Faecalibacterium spp. are reported to correlate with inflammatory conditions, and numerous studies have demonstrated the important role of Faecalibacterium prausnitzii in human health [67–70]. The Leuconostoc spp. are commonly used as starter bacteria in some dairy fermentations, and members in this genus were found to improve UA and liver metabolism in animals [71, 72]. While the overall increase in the Blautia genus did not reach statistical significance, noteworthy elevations were observed in the specific species B. producta within the 3 Mix+PE0401 group and B. wexlerae within the 3 Mix group. Recently, a substantial body of research has concentrated on exploring the probiotic effects of the Blautia genus, particularly its potential in alleviating metabolic syndrome [73]. B. wexlerae demonstrated efficacy in improving both obesity and type 2 diabetes [74], while B. producta emerged as a promising candidate for probiotic use in preventing acute liver injury [75]. Overall, our results indicated the multi-strain probiotics together with postbiotics was able to elevate commensal populations in the gut microbiota.
Conclusions
The supplementation of multi-strain probiotics (L. fermentum TSF331, L. reuteri TSR332, and L. plantarum TSP05) together was able to prevent fatty liver by reducing lipid accumulation and inflammation. This combination was also more efficient on UA management by promoting UA excretion in the gut. The synergy between probiotics and postbiotics has the potential to establish a harmonious environment, specifically reshaping dysbiosis in metabolic disturbances.
Supporting information
(DOC)
(PDF)
(PDF)
(DOCX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This study was supported by grant from Glac Biotech Co., Ltd., Tainan, Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Singh S, Osna NA, Kharbanda KK. Treatment options for alcoholic and non-alcoholic fatty liver disease: A review. World J Gastroenterol. 2017;23(36):6549–70. Epub 2017/11/01. doi: 10.3748/wjg.v23.i36.6549 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antunes C, Azadfard M, Hoilat GJ, Gupta M. Fatty Liver. StatPearls. Treasure Island (FL)2022. [PubMed]
- 3.Toshikuni N, Tsutsumi M, Arisawa T. Clinical differences between alcoholic liver disease and nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20(26):8393–406. Epub 2014/07/16. doi: 10.3748/wjg.v20.i26.8393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62(1 Suppl):S47–64. Epub 2015/04/29. doi: 10.1016/j.jhep.2014.12.012 . [DOI] [PubMed] [Google Scholar]
- 5.Heeren J, Scheja L. Metabolic-associated fatty liver disease and lipoprotein metabolism. Mol Metab. 2021;50:101238. Epub 2021/04/24. doi: 10.1016/j.molmet.2021.101238 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sattar N, Forrest E, Preiss D. Non-alcoholic fatty liver disease. BMJ. 2014;349:g4596. Epub 2014/09/23. doi: 10.1136/bmj.g4596 declaration of interests and declare the following interests: none. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King C, Lanaspa MA, Jensen T, Tolan DR, Sanchez-Lozada LG, Johnson RJ. Uric Acid as a Cause of the Metabolic Syndrome. Contrib Nephrol. 2018;192:88–102. Epub 2018/02/03. doi: 10.1159/000484283 . [DOI] [PubMed] [Google Scholar]
- 8.Lima WG, Martins-Santos ME, Chaves VE. Uric acid as a modulator of glucose and lipid metabolism. Biochimie. 2015;116:17–23. Epub 2015/07/03. doi: 10.1016/j.biochi.2015.06.025 . [DOI] [PubMed] [Google Scholar]
- 9.Xie D, Zhao H, Lu J, He F, Liu W, Yu W, et al. High uric acid induces liver fat accumulation via ROS/JNK/AP-1 signaling. Am J Physiol Endocrinol Metab. 2021;320(6):E1032–E43. Epub 2021/04/27. doi: 10.1152/ajpendo.00518.2020 . [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Ma X, Xing J, Shi H, Yang R, Jiao Y, et al. Serum Uric Acid Is a Mediator of the Association Between Obesity and Incident Nonalcoholic Fatty Liver Disease: A Prospective Cohort Study. Front Endocrinol (Lausanne). 2021;12:657856. Epub 2021/06/01. doi: 10.3389/fendo.2021.657856 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Bonito P, Valerio G, Licenziati MR, Miraglia Del Giudice E, Baroni MG, Morandi A, et al. High uric acid, reduced glomerular filtration rate and non-alcoholic fatty liver in young people with obesity. J Endocrinol Invest. 2020;43(4):461–8. Epub 2019/10/23. doi: 10.1007/s40618-019-01130-6 . [DOI] [PubMed] [Google Scholar]
- 12.Oral A, Sahin T, Turker F, Kocak E. Relationship Between Serum Uric Acid Levels and Nonalcoholic Fatty Liver Disease in Non-Obese Patients. Medicina (Kaunas). 2019;55(9). Epub 2019/09/20. doi: 10.3390/medicina55090600 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanai H, Adachi H, Hakoshima M, Katsuyama H. Molecular Biological and Clinical Understanding of the Pathophysiology and Treatments of Hyperuricemia and Its Association with Metabolic Syndrome, Cardiovascular Diseases and Chronic Kidney Disease. Int J Mol Sci. 2021;22(17). Epub 2021/09/11. doi: 10.3390/ijms22179221 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Festi D, Schiumerini R, Eusebi LH, Marasco G, Taddia M, Colecchia A. Gut microbiota and metabolic syndrome. World J Gastroenterol. 2014;20(43):16079–94. Epub 2014/12/05. doi: 10.3748/wjg.v20.i43.16079 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabke K, Hendrick G, Devkota S. The gut microbiome and metabolic syndrome. J Clin Invest. 2019;129(10):4050–7. Epub 2019/10/02. doi: 10.1172/JCI129194 Institute and a named inventor on a patent application related to intestinal microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Safari Z, Gerard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD). Cell Mol Life Sci. 2019;76(8):1541–58. Epub 2019/01/27. doi: 10.1007/s00018-019-03011-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang PX, Deng XR, Zhang CH, Yuan HJ. Gut microbiota and metabolic syndrome. Chin Med J (Engl). 2020;133(7):808–16. Epub 2020/02/28. doi: 10.1097/CM9.0000000000000696 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssens Y, Nielandt J, Bronselaer A, Debunne N, Verbeke F, Wynendaele E, et al. Disbiome database: linking the microbiome to disease. BMC Microbiol. 2018;18(1):50. Epub 2018/06/06. doi: 10.1186/s12866-018-1197-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahl WJ, Rivero Mendoza D, Lambert JM. Diet, nutrients and the microbiome. Prog Mol Biol Transl Sci. 2020;171:237–63. Epub 2020/06/02. doi: 10.1016/bs.pmbts.2020.04.006 . [DOI] [PubMed] [Google Scholar]
- 20.Natividad JM, Lamas B, Pham HP, Michel ML, Rainteau D, Bridonneau C, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun. 2018;9(1):2802. Epub 2018/07/20. doi: 10.1038/s41467-018-05249-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green M, Arora K, Prakash S. Microbial Medicine: Prebiotic and Probiotic Functional Foods to Target Obesity and Metabolic Syndrome. Int J Mol Sci. 2020;21(8). Epub 2020/04/25. doi: 10.3390/ijms21082890 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamanaka H, Taniguchi A, Tsuboi H, Kano H, Asami Y. Hypouricaemic effects of yoghurt containing Lactobacillus gasseri PA-3 in patients with hyperuricaemia and/or gout: A randomised, double-blind, placebo-controlled study. Mod Rheumatol. 2019;29(1):146–50. doi: 10.1080/14397595.2018.1442183 . [DOI] [PubMed] [Google Scholar]
- 23.Kobyliak N, Abenavoli L, Mykhalchyshyn G, Kononenko L, Boccuto L, Kyriienko D, et al. A Multi-strain Probiotic Reduces the Fatty Liver Index, Cytokines and Aminotransferase levels in NAFLD Patients: Evidence from a Randomized Clinical Trial. J Gastrointestin Liver Dis. 2018;27(1):41–9. Epub 2018/03/21. doi: 10.15403/jgld.2014.1121.271.kby . [DOI] [PubMed] [Google Scholar]
- 24.Mohammadi-Sartang M, Bellissimo N, Totosy de Zepetnek JO, Brett NR, Mazloomi SM, Fararouie M, et al. The effect of daily fortified yogurt consumption on weight loss in adults with metabolic syndrome: A 10-week randomized controlled trial. Nutr Metab Cardiovasc Dis. 2018;28(6):565–74. Epub 2018/05/05. doi: 10.1016/j.numecd.2018.03.001 . [DOI] [PubMed] [Google Scholar]
- 25.Kechagia M, Basoulis D, Konstantopoulou S, Dimitriadi D, Gyftopoulou K, Skarmoutsou N, et al. Health benefits of probiotics: a review. ISRN Nutr. 2013;2013:481651. Epub 2013/01/01. doi: 10.5402/2013/481651 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scorletti E, Afolabi PR, Miles EA, Smith DE, Almehmadi A, Alshathry A, et al. Synbiotics Alter Fecal Microbiomes, But Not Liver Fat or Fibrosis, in a Randomized Trial of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2020;158(6):1597–610 e7. Epub 2020/01/29. doi: 10.1053/j.gastro.2020.01.031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–14. doi: 10.1038/nrgastro.2014.66 . [DOI] [PubMed] [Google Scholar]
- 28.Hsieh PS, Chen CW, Kuo YW, Ho HH. Lactobacillus spp. reduces ethanol-induced liver oxidative stress and inflammation in a mouse model of alcoholic steatohepatitis. Exp Ther Med. 2021;21(3):188. Epub 2021/01/26. doi: 10.3892/etm.2021.9619 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo YW, Hsieh SH, Chen JF, Liu CR, Chen CW, Huang YF, et al. Lactobacillus reuteri TSR332 and Lactobacillus fermentum TSF331 stabilize serum uric acid levels and prevent hyperuricemia in rats. PeerJ. 2021;9:e11209. Epub 2021/05/15. doi: 10.7717/peerj.11209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. 2021;18(9):649–67. Epub 2021/05/06. doi: 10.1038/s41575-021-00440-6 Initiative and has been a speaker in meetings funded by industry, Nestle Nutrition Institute and Institute Danone. M.C.C. has participated as a speaker for HIPP, Danone, Nutricia, Nestle Nutrition Institute and Mead Johnson. A.E. has led industry-sponsored research projects with support from B Food Science and Takanashi Milk Products, and has been a speaker for the companies. C.H. serves on the board of ISAPP, is a consultant to Artugen Therapeutics developing a live biotherapeutic, and has received research grants from several industry partners, including ADARE Pharmaceuticals, manufacturers of Lacteol. S.L. serves on the academic board of ISAPP and has received research grants from several industry partners, such as Yun. She has been compensated for speaking by Yakult. E.M.M.Q. serves on the board of ISAPP, as a consultant to 4D Pharma, Alimentary Health, Allergan, Biocodex, Ironwood, Salix, Takeda and Vibrant, and has research support from 4D Pharma, Biomerica and Vibrant. M.E.S. has been compensated for speaking engagements or for consulting from Associated British Foods, California Dairy Research Foundation, Cargill, Danone Research, Danone USA, Fairlife, General Mills, GlaxoSmithKline, JJ Heimbach, Kellogg, Kerry, Mead Johnson, Medscape, PepsiCo, Pfizer, Probi, Procter & Gamble, Sanofi, Trouw Nutrition, Visalia Dairy Company, Winclove Probiotics and Yakult. R.S. has participated as a clinical investigator, and/or advisory board member, and/or consultant, and/or speaker for Abbott, Danone and Nestle. J.R.S. has led industry-sponsored research projects with support from AstraZeneca, Danone, Servier and Vitacress. H.S. serves on the board of ISAPP and has participated as a clinical investigator, and/or advisory board member, and/or consultant, and/or speaker for Arla, Biogaia, Biocodex, Ch. Hansen, Danone, Nestle, Nestle Nutrition Institute, Nutricia and Merck. G.V. has led industry-sponsored research projects on dairy products and probiotics. These projects were independently carried out and had no influence on the content of this manuscript. He is member of the Argentinian board of the Yoghurt in Nutrition Initiative (YINI Danone Argentina) and serves on the board of ISAPP. He was not a member of ISAPP Board at the time of the meeting, but has been elected as a board member as of June 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott E, De Paepe K, Van de Wiele T. Postbiotics and Their Health Modulatory Biomolecules. Biomolecules. 2022;12(11). Epub 2022/11/12. doi: 10.3390/biom12111640 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin WY, Kuo Y.W., Chen C.W., Hsu Y.C., Huang Y.F., Hsu C.H., et al. The Function of Mixed Postbiotic PE0401 in Improving Intestinal Health via Elevating Anti-inflammation, Anti-oxidation, Epithelial Tight Junction Gene Expression and Promoting Beneficial Bacteria Growth. J Pure Appl Microbiol. 2022;16(3):1771–82. doi: 10.22207/JPAM.16.3.19 [DOI] [Google Scholar]
- 33.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic acids research. 2013;41(1):e1–e. doi: 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amplicon P, Clean-Up P, Index P. 16S Metagenomic sequencing library preparation. Illumina: San Diego, CA, USA. 2013. [Google Scholar]
- 35.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45(W1):W180–W8. Epub 2017/04/28. doi: 10.1093/nar/gkx295 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanmani P, Kim H. Protective Effects of Lactic Acid Bacteria Against TLR4 Induced Inflammatory Response in Hepatoma HepG2 Cells Through Modulation of Toll-Like Receptor Negative Regulators of Mitogen-Activated Protein Kinase and NF-kappaB Signaling. Front Immunol. 2018;9:1537. Epub 2018/07/20. doi: 10.3389/fimmu.2018.01537 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grondona P, Bucher P, Schmitt A, Schönfeld C, Streibl B, Müller A, et al. Threonine phosphorylation of IκBζ mediates inhibition of selective proinflammatory target genes. Journal of Investigative Dermatology. 2020;140(9):1805–14. e6. [DOI] [PubMed] [Google Scholar]
- 38.Chen M, Ye C, Zhu J, Zhang P, Jiang Y, Lu X, et al. Bergenin as a novel urate-lowering therapeutic strategy for hyperuricemia. Frontiers in Cell and Developmental Biology. 2020;8:703. doi: 10.3389/fcell.2020.00703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tőkés A-M, Kulka J, Paku S, Szik Á, Páska C, Novák PK, et al. Claudin-1,-3 and-4 proteins and mRNA expression in benign and malignant breast lesions: a research study. Breast Cancer Research. 2005;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dale HF, Rasmussen SH, Asiller OO, Lied GA. Probiotics in Irritable Bowel Syndrome: An Up-to-Date Systematic Review. Nutrients. 2019;11(9). Epub 2019/09/05. doi: 10.3390/nu11092048 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Zhou J, Wang L. Role and Mechanism of Gut Microbiota in Human Disease. Front Cell Infect Microbiol. 2021;11:625913. Epub 2021/04/06. doi: 10.3389/fcimb.2021.625913 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun W-L, Hua S, Li X-Y, Shen L, Wu H, Ji H-F. Microbially produced vitamin B12 contributes to the lipid-lowering effect of silymarin. Nature Communications. 2023;14(1):477. doi: 10.1038/s41467-023-36079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan W, Tang K, Deng Y, Zheng C, Pan M, Pi D, et al. Bifidobacterium lactis Probio-M8 prevents nonalcoholic fatty liver disease in high-fat diet-fed rats: The potential role in modulating gut microbiota. Food Bioengineering. 2024. [Google Scholar]
- 44.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57(2):601–9. doi: 10.1002/hep.26093 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q, Xing W, Wang Q, Tang Z, Wang Y, Gao W. Gut microbiota–mitochondrial inter-talk in non-alcoholic fatty liver disease. Frontiers in Nutrition. 2022;9:934113. doi: 10.3389/fnut.2022.934113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Q, Pu Y, Lu H, Zhao N, Wang Y, Guo Y, et al. Porphyromonas, Treponema, and Mogibacterium promote IL8/IFNgamma/TNFalpha-based pro-inflammation in patients with medication-related osteonecrosis of the jaw. J Oral Microbiol. 2020;13(1):1851112. Epub 2021/01/05. doi: 10.1080/20002297.2020.1851112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu M, Shi Y, Wu K, Xie W, Ser HL, Jiang Q, et al. From Mouth to Brain: Distinct Supragingival Plaque Microbiota Composition in Cerebral Palsy Children With Caries. Front Cell Infect Microbiol. 2022;12:814473. Epub 2022/04/29. doi: 10.3389/fcimb.2022.814473 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao H, Chu M, Huang Z, Yang X, Ran S, Hu B, et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017;7(1):11773. Epub 2017/09/20. doi: 10.1038/s41598-017-11779-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razin S, Hayflick L. Highlights of mycoplasma research—an historical perspective. Biologicals. 2010;38(2):183–90. Epub 2010/02/13. doi: 10.1016/j.biologicals.2009.11.008 . [DOI] [PubMed] [Google Scholar]
- 50.Zuo LL, Wu YM, You XX. Mycoplasma lipoproteins and Toll-like receptors. J Zhejiang Univ Sci B. 2009;10(1):67–76. Epub 2009/02/07. doi: 10.1631/jzus.B0820256 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ipsen DH, Lykkesfeldt J, Tveden-Nyborg P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol Life Sci. 2018;75(18):3313–27. Epub 2018/06/25. doi: 10.1007/s00018-018-2860-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marra F, Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J Hepatol. 2018;68(2):280–95. Epub 2017/11/21. doi: 10.1016/j.jhep.2017.11.014 . [DOI] [PubMed] [Google Scholar]
- 53.Xing W, Gao W, Lv X, Zhao Z, Mao G, Dong X, et al. The effects of supplementation of probiotics, prebiotics, or synbiotics on patients with non-alcoholic fatty liver disease: A meta-analysis of randomized controlled trials. Front Nutr. 2022;9:1024678. Epub 2022/11/18. doi: 10.3389/fnut.2022.1024678 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Musazadeh V, Roshanravan N, Dehghan P, Ahrabi SS. Effect of probiotics on liver enzymes in patients with non-alcoholic fatty liver disease: an umbrella of systematic review and meta-analysis. Frontiers in Nutrition. 2022;9:844242. doi: 10.3389/fnut.2022.844242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan Z, Mao B, Zhang Q, Tang X, Yang B, Zhao J, et al. Postbiotics prepared using Lactobacillus paracasei CCFM1224 prevent nonalcoholic fatty liver disease by modulating the gut microbiota and liver metabolism. International Journal of Molecular Sciences. 2022;23(21):13522. doi: 10.3390/ijms232113522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li HY, Zhou DD, Gan RY, Huang SY, Zhao CN, Shang A, et al. Effects and Mechanisms of Probiotics, Prebiotics, Synbiotics, and Postbiotics on Metabolic Diseases Targeting Gut Microbiota: A Narrative Review. Nutrients. 2021;13(9). Epub 2021/09/29. doi: 10.3390/nu13093211 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ho HH, Chen CW, Yi TH, Huang YF, Kuo YW, Lin JH, et al. Novel application of a Co-Fermented postbiotics of TYCA06/AP-32/CP-9/collagen in the improvement of acne vulgaris-A randomized clinical study of efficacy evaluation. J Cosmet Dermatol. 2022;21(11):6249–60. Epub 2022/07/13. doi: 10.1111/jocd.15228 . [DOI] [PubMed] [Google Scholar]
- 58.Strilchuk L, Fogacci F, Cicero AF. Safety and tolerability of available urate-lowering drugs: a critical review. Expert Opin Drug Saf. 2019;18(4):261–71. Epub 2019/03/28. doi: 10.1080/14740338.2019.1594771 . [DOI] [PubMed] [Google Scholar]
- 59.Yin H, Liu N, Chen J. The Role of the Intestine in the Development of Hyperuricemia. Front Immunol. 2022;13:845684. Epub 2022/03/15. doi: 10.3389/fimmu.2022.845684 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Li Y, Liao W, Huang J, Liu Y, Li Z, et al. Gut microbiota remodeling: A promising therapeutic strategy to confront hyperuricemia and gout. Front Cell Infect Microbiol. 2022;12:935723. Epub 2022/08/30. doi: 10.3389/fcimb.2022.935723 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee Y, Werlinger P, Suh J-W, Cheng J. Potential probiotic lacticaseibacillus paracasei MJM60396 prevents hyperuricemia in a multiple way by absorbing purine, suppressing xanthine oxidase and regulating urate excretion in mice. Microorganisms. 2022;10(5):851. doi: 10.3390/microorganisms10050851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou Y, Ro K-S, Jiang C, Yin D, Zhao L, Zhang D, et al. The anti-hyperuricemic and gut microbiota regulatory effects of a novel purine assimilatory strain, Lactiplantibacillus plantarum X7022. European Journal of Nutrition. 2023:1–15. doi: 10.1007/s00394-023-03291-w [DOI] [PubMed] [Google Scholar]
- 63.Zhao S, Feng P, Hu X, Cao W, Liu P, Han H, et al. Probiotic Limosilactobacillus fermentum GR-3 ameliorates human hyperuricemia via degrading and promoting excretion of uric acid. iScience. 2022;25(10):105198. Epub 2022/10/18. doi: 10.1016/j.isci.2022.105198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H, Mei L, Deng Y, Liu Y, Wei X, Liu M, et al. Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis. Nutrition. 2019;62:63–73. Epub 2019/03/11. doi: 10.1016/j.nut.2018.11.018 . [DOI] [PubMed] [Google Scholar]
- 65.Li M, Wu X, Guo Z, Gao R, Ni Z, Cui H, et al. Lactiplantibacillus plantarum enables blood urate control in mice through degradation of nucleosides in gastrointestinal tract. Microbiome. 2023;11(1):153. Epub 2023/07/20. doi: 10.1186/s40168-023-01605-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rodriguez JM, Garranzo M, Segura J, Orgaz B, Arroyo R, Alba C, et al. A randomized pilot trial assessing the reduction of gout episodes in hyperuricemic patients by oral administration of Ligilactobacillus salivarius CECT 30632, a strain with the ability to degrade purines. Front Microbiol. 2023;14:1111652. Epub 2023/03/04. doi: 10.3389/fmicb.2023.1111652 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin R, Rios-Covian D, Huillet E, Auger S, Khazaal S, Bermudez-Humaran LG, et al. Faecalibacterium: a bacterial genus with promising human health applications. FEMS Microbiol Rev. 2023;47(4). Epub 2023/07/15. doi: 10.1093/femsre/fuad039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16(3):255–61. Epub 2013/07/09. doi: 10.1016/j.mib.2013.06.003 . [DOI] [PubMed] [Google Scholar]
- 69.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11(4):841–52. Epub 2017/01/04. doi: 10.1038/ismej.2016.176 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Effendi R, Anshory M, Kalim H, Dwiyana RF, Suwarsa O, Pardo LM, et al. Akkermansia muciniphila and Faecalibacterium prausnitzii in Immune-Related Diseases. Microorganisms. 2022;10(12). Epub 2022/12/24. doi: 10.3390/microorganisms10122382 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun M, Wang Q, Zhang M, Zhang G, Wu T, Liu R, et al. Leuconostoc pseudomesenteroides improves microbiota dysbiosis and liver metabolism imbalance and ameliorates the correlation between dihydroceramide and strains of Firmicutes and Proteobacteria in high fat diet obese mice. Food Funct. 2020;11(8):6855–65. Epub 2020/07/16. doi: 10.1039/d0fo01009j . [DOI] [PubMed] [Google Scholar]
- 72.Liang L, Meng Z, Zhang F, Jianguo Z, Fang S, Hu Q, et al. Lactobacillus gasseri LG08 and Leuconostoc mesenteroides LM58 exert preventive effect on the development of hyperuricemia by repairing antioxidant system and intestinal flora balance. Front Microbiol. 2023;14:1211831. Epub 2023/06/28. doi: 10.3389/fmicb.2023.1211831 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, et al. Blautia-a new functional genus with potential probiotic properties? Gut Microbes. 2021;13(1):1–21. Epub 2021/02/03. doi: 10.1080/19490976.2021.1875796 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hosomi K, Saito M, Park J, Murakami H, Shibata N, Ando M, et al. Oral administration of Blautia wexlerae ameliorates obesity and type 2 diabetes via metabolic remodeling of the gut microbiota. Nat Commun. 2022;13(1):4477. Epub 2022/08/19. doi: 10.1038/s41467-022-32015-7 interest: M.S., Y.O., H.S., and Y.Y. are employees of Noster, Inc. (Kyoto, Japan). Other authors declare no competing interests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao B, Guo W, Liu X, Cui S, Zhang Q, Zhao J, et al. Potential Probiotic Properties of Blautia producta Against Lipopolysaccharide-Induced Acute Liver Injury. Probiotics Antimicrob Proteins. 2023;15(3):785–96. Epub 2023/02/16. doi: 10.1007/s12602-023-10044-y . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(PDF)
(PDF)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.