Abstract
Human brain organoids are self-organizing three-dimensional structures that emerge from human pluripotent stem cells and mimic aspects of the cellular composition and functionality of the developing human brain. Despite their impressive self-organizing capacity, organoids lack the stereotypic structural anatomy of their in vivo counterpart, making conventional analysis techniques underpowered to assess cellular composition and gene network regulation in organoids. Advances in single cell transcriptomics have recently allowed characterization and improvement of organoid protocols, as they continue to evolve, by enabling identification of cell types and states along with their developmental origins. In this review, we summarize recent approaches, progress and challenges in resolving brain organoids complexity through single-cell transcriptomics. We then discuss emerging technologies that may complement single-cell RNA sequencing by providing additional readouts of cellular states to generate organ-level view of developmental processes. Altogether, these integrative technologies will allow monitoring of global gene regulation in thousands of individual cells and will offer unprecedented opportunity to investigate features of human brain development and disease, across multiple cellular modalities and with cell-type resolution.
Keywords: human brain organoids, single-cell transcriptomics, single-cell multi-omics, integrative technologies, cell-states, cell-type
1. Introduction
Human brain development is an astonishing process. Starting in the third gestational week and within a relatively short timeframe, a homogenous population of neural progenitor cells differentiates into the billions of interconnected cells that make up the human brain. Diverse combinations of thousands of genes identify the many cell types of the human brain, arguably the most diverse and complex of all biological systems. How does this remarkable diversity of human brain cell types arise? What is the function of each type? How do different types interact, and how does the transcriptome of individual cell changes in disease state? Although much has been learned using model organisms, many of these fundamental questions are still unanswered given the limited availability of developing human tissue. Human pluripotent- stem cell-derived brain organoids have opened, for the first time a window into aspects of human brain development and disease that were not accessible before. However, this young field is challenged by difficulties in data analysis and interpretation that result from a lack of reproducible anatomy across individual organoids [1]. Classic analysis techniques, including immunohistochemistry and bulk sequencing, are low resolution and not effective for elucidation of cellular composition [2] and for localization and targeting of selected cell types in organoids. Recent advances in single cell-transcriptomics have revolutionize our ability to resolve cellular heterogeneity and to access cell-type specific gene expression in organoids. Here we summarize recent findings in the brain organoid field that have leveraged advances in single cell-genomics, and we discuss how emerging technologies that complement single-cell transcriptomics will provide additional information across cellular modalities to substantially advance our understanding of human brain development and disease.
2. Overview on single cell transcriptomics
Up until the last few years, bulk sequencing has been the primary approach to obtain a low-resolution, average molecular RNA profile of a given sample. However, the advent of single cell sequencing has revolutionized the study of the transcriptome by allowing researchers to analyze the composition of a sample at single cell resolution. This single cell approach has given researchers insight to the diverse cellular heterogeneity that exist with an unprecedented level of detail.
The process of obtaining a single cell transcriptomic profile starts by (1) isolating single-cell or single-nuclei from the sample into discrete compartments. The sequential steps following the isolation remain well conserved between methodologies: (2) RNA extraction, (3) reverse transcription, (4) amplification, (5) and finally, next-generation sequencing of library. Each cDNA molecule in the expansive library is barcoded with short sequences to give it a unique identity and to further identify the single cell it came from. Unfortunately, there is no gold standard method when it comes to the analysis of resulting single cell RNA sequencing (scRNA-seq) expression data. However, the success of these various analysis techniques is determined by their ability to classify the cell types present, to determine the developmental trajectory of each cell, and to infer the putative receptor-ligand regulatory networks involved. In this review, we provide an overview on how the use of this technology has shaped the study of brain organoids and cortical developmental.
3. ScRNA-seq pipeline to deconvolute brain organoids
In the past five years, there has been an immense increase in wet lab techniques used to capture and reverse transcribe the RNA of single cells, which has been thoroughly annotated in previous reviews [3–5]. Each of these techniques comes with their own advantages and disadvantages. These methodologies vary by transcript length coverage (full length, 3’-only, or 5’-only), use of unique molecular identifier (UMI), and strand specificity. Droplet-based methods (Drop-seq and Chromium 10X Genomics) and multi-well sorting methods (Smart-seq, Smart-seq2, BD-Resolve/Rhapsody) are platforms that have been used to characterize brain organoids (Figure 1). Various papers have been published testing an array of scRNA-seq techniques to determine the pros and cons of each platform [6–10]. This thorough analysis gives insight into the potential applications of each of the various scRNA-seq techniques and what questions can be answered from them. With all of these different platforms, one must consider a few options: intensity of throughput, depth of sequencing (how many transcripts per cell), and the range of the captured transcriptome.
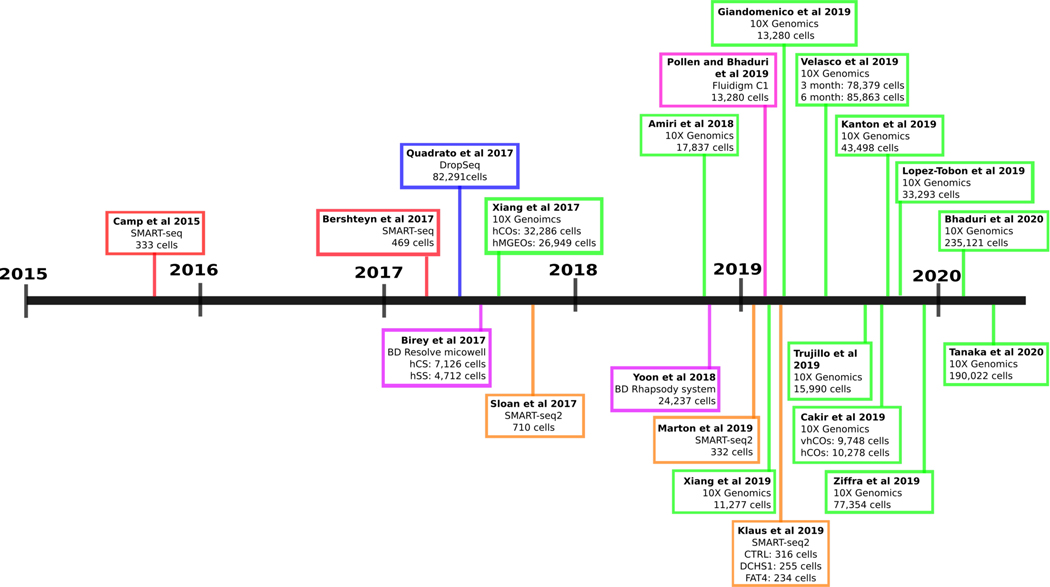
Fig. 1. Timeline of all papers that have used scRNA-seq to characterize hPSC derived brain organoids.
Each colored bubble contains author, year of publication, single cell platform used, and number of cells characterized after passing quality control. SMART-seq (red), DropSeq (blue), BD Resolve/Rhapsody microwell (purple), SMART-seq2 (orange), Chromium by 10X Genomics (green), Fluidigm C1 (pink). Human Cortical Spheres (hCS); human Striatal Spheres (hSS); human Cortical Organoids (hCOs); human Medial Ganglionic Emience Organoids (hMGEOs); vascularized human Cortical Organoids (vhCOs).
Once the single cell RNA transcript counts has been assembled into an expression matrix, the dataset is subject to normalization [11] and possibly batch correction [12] prior to decoding the cell types present based on the count matrix [13]. A commonly used method based on the variance is principal component analysis (PCA), which determines patterns of gene expression that vary between single cells. This information can also be subject to other forms of dimensionality reduction as a t-distributed stochastic neighbor embedding (t-SNE) or uniform manifold approximation and projection (UMAP) [14]. Although the way to identify the biologically proper number of different cell subtypes from these graphs is somewhat arbitrary, the use of other algorithms such as K-means [15] to determine the “nearest neighbor” allows for an approximation for the number of cell types available in the sample of interest. Further details about clustering techniques are found in a recent review by Petegrosso et al. [16]
Cells are captured within a snapshot of their developmental trajectory when they are dissociated for scRNA-seq. Within an organoid, the various cell states, from a progenitor to a fully differentiated cell are found within this single time frame. Pseudotime is a way of inferring the actual temporal differentiation trajectory by ordering the cells in a “pseudo”time that tries to capture the progression of transcriptional changes for one cell to the next. The various lineage trajectory platforms can be categorized into seven trajectory types: cycle, linear, bifurcation, multifurcation, tree, connected graph, and disconnected graph [16]. A thorough analysis on the various platforms and their categorization has been examined in Saelens et al. [17]. One of the “tree type” platforms, Monocle2 [18] has been most commonly used to study brain organoids. Specifically, it was used to study the developmental trajectory of all cell types in dorsal forebrain [19], oligodendrocytic [20], and astrocytic [21] organoids. The previous version, Monocle1 [22] was incorporated into Camp et al to study the lineage trajectory from apical progenitors all the way to the final resting place of the cortical plate [23]. Another lineage trajectory techniques named RNA velocity, looks into the directionality of the lineage progression by observing the levels of certain spliced isoforms of a transcript. [24]. In addition to these platforms, probabilistic models including Diffusion Pseudotime (DPT) [25] and Waddington-OT [26] could also potentially be used to study organoid cell type trajectories based on the probability of a certain cell’s fate. In addition to the cell type identification, uncovering the lineage trajectory gives further insight into human brain development through the use of these techniques on brain organoids. The use of single cell transcriptomics to characterize brain organoids has only been conducted in a handful of papers. In the following sections, we highlight these papers, discuss emerging technologies and determine how these techniques can move the brain organoid field forward.
4. Progress in understanding brain organoids through scRNA-seq
The progress made in characterizing human brain organoids through the use of scRNA-seq has been exponential over the last five years. Camp et al. pioneered the use of scRNA-seq technology to characterize brain organoids [23]. This work used the single cell RNA-sequencing platform called SMARTer [27] to compare individual cells generated in cortical organoids ranging from 33–65 days old with cortical cells derived from 12–13-week-old human fetal tissue. The organoids were developed using the whole brain organoid protocol established by Madeline Lancaster in the Knoblich laboratory [28] while a reference transcriptomic atlas was built using cell composition and lineage trajectory of human fetal cortical cells at time points comparable to the organoid developmental stage. Due to the low throughput method, only 333 cells were captured by this technique, but nonetheless the authors were able to provide the first evidence that brain organoids recapitulate gene expression program of human fetal cortex development.
Quadrato et al. carried on the first systematic study of the cellular composition of whole brain organoids after extended time in culture. Using high-throughput droplet based microfluidics technique known as Drop-seq [29], they analyzed more than 80,000 individual cells, from 31 individual organoids, providing the first molecular map of the diversity and reproducibility of cell types generated in brain organoids. They discovered that organoids can make a large diversity of cell classes from distinct regions of the brain and from the retina, and that that the cellular composition of organoids diversifies over time in culture and displays progressive levels of maturity. This was reflected in the acquisition of structural traits characteristic of mature neurons, including dendritic spine-like structures, which have been notoriously difficult to generate by directed differentiation in culture. In agreement with an advanced state of maturation, they showed that whole brain organoids progressively generate spontaneously-active neuronal networks and that during the same period of time, photoreceptor-like cells mature substantially and become responsive to non-invasive, light-based sensory stimulation that appears capable of affecting neuronal activity.
By shifting focus from whole brain organoids, Birey et al [30] and Xiang et al 2017 [31] generated assembloids by fusing patterned dorsal and ventral forebrain organoids derived from human pluripotent stem cells (hPSCs). The aim was to better recapitulate early development and migration of interneurons from ventral forebrain to dorsal forebrain as seen in vivo. Birey et al used the BD-Resolve platform [32], a non-droplet based microwell system, while Xiang et al 2017 used the 10X Genomics system [33] to obtain cells from these dorsal and ventral organoids. These cells, through downstream analysis techniques, were identified as having proper transcriptional profiles of early developing dorsal and ventral forebrain. In addition to dorsal and ventral forebrain organoids, telencephalic organoids, which contain both excitatory and inhibitory neurons, have been characterized in recent studies using the 10X Genomics system as well [34,35].
One key question still remained unanswered in the brain organoid field. Can these organoids be reproducibly made? This point was addressed by Yoon et al. that showed a reliable and consistent generation of cortical spheroids collected at 4 different stages of differentiation across 6 hiPSC lines over 100 days in cultures. Velasco et al. brought this characterization to the next level by analyzing 166,242 cells with the 10X Genomics system from 21 patterned dorsal forebrain organoids generated accordingly to a modified version of the Kadoshima protocol published by the Sasai lab in 2013 [36]. They found that organoids generated across different batches and cell lines, reproducibly generate a large variety of progenitor and postmitotic cell types of the human cerebral cortex. Through pseudotime analysis, cells in organoids follow a precise developmental trajectory that is consistent between organoids and comparable to the endogenous cerebral cortex.
In addition to cortical neurons, glial cells, a late born cell type of the developing brain, grown in 3D neural spheroids were thoroughly analyzed in Sloan et al. and Marton et al. using large scale single cell transcriptomics. Specifically, Sloan et al. analyzed the development and maturation of astrocytes cultured for up to 590 day and demonstrated functional changes and overtime through synaptosome phagocytic assays and calcium imaging on co-cultured neurons purified from human cortical spheroids [21]. Following comparison with human primary samples, single cell analysis of cortical spheroid-derived astrocytes with SMART-seq2 [37] revealed a shift in their transcriptional profile from fetal to postnatal-like. The publication by Marton et al. focused on the differentiation and maturation of another glial subpopulation, the oligodendrocytes, cultured in 3D neural cultures [20]. Using SMART-seq2, the authors captured 295 cells derived from neural spheroid containing oligodendrocytes and showed similarity in both molecular identity and timing of developmental progression to human primary oligodendrocytes. Over time, oligodendrocytes in culture successfully myelinated neuronal axons [20]
Despite the vast amount of information that one can obtain on the identity of individual cells from a comprehensive RNA profile, epigenetic heterogeneity plays a fundamental role in establishing the diversity of cell states found in transcriptionally identical cells. Amiri et al. conducted the first epigenetic characterization of organoids by comparing the transcriptome and the epigenome of cortical organoids at 0, 11, and 30 days in vitro to previously published data as well as to human primary fetal tissue ranging from 15 and 17 PCW [38]. Although this pioneering work on investigating brain organoids with assay for transposase-accessible chromatin sequencing (ATAC-seq) was not at a single cell resolution, it identified autism gene modules by investigating patient derived organoids. Recently, Kanton et al. conducted the first foundational tour de force to characterize putative regulatory mechanisms over the human and chimpanzee organoid differentiation trajectory using bulk and single cell ATAC-seq (sc-ATACseq). In addition to discovering that human development occurs at a slower pace relative to chimpanzee and macaque, the authors revealed human specific chromatin accessibility dynamics during cortex development and evolution [39]
This seminal work builds on an elegant study by Pollen and Bhaduri et al., that for the first time employed scRNAseq to establish brain organoids as models of human-specific brain evolution. Specifically, the authors identified 261 differentially expressed genes in human cortical development compared to non-human primates using brain organoids [40]. Of those differentially expressed genes, key players of the PI3K-AKT-mTOR signaling were upregulated in human outer radial glial cells compared to other non-human primates.
Although the main focus of the brain organoid field has been on thoroughly characterizing forebrain organoids, Xiang et al. [41] recently took on the challenge to generate and characterize a new protocol to generate organoids with a thalamic regional identity. With the 10X Genomics scRNA-seq platform, they showed generation of distinct thalamic lineages, however, these and organoids of other brain regions including, hippocampus [42], midbrain [43,44], hypothalamus [43] and cerebellum [45] still necessitate a thorough analysis of their cell type composition and lineage trajectory.
5. Disease modeling using brain organoids coupled with single cell transcriptomics
To date, a number of studies have modeled neurodevelopmental disorders including microcephaly, autism, schizophrenia and zika virus infection, as thoroughly addressed in recent reviews [46–48]. However, large scale single-cell multiomics approach combined with brain organoid models could revolutionize our understanding of neurodevelopmental disease by revealing cell-state specific changes along progenitor to neuron lineages. To our knowledge, only two papers have applied scRNA-seq to characterize disease phenotypes in brain organoids to date.
Impairments of molecular mechanisms involved in human corticogenesis leads to a broad range of gross morphological defects ranging from microcephaly or lissencephaly to subcortical or periventricular heterotopia (PE) [49–53]. Recently, Klaus et al. [54] showed that mutations in the protocadherins DCSH1 and FAT4 result in cortical heterotopia phenotype associated with PH. Through scRNA-seq and pseudotime analysis they identified a subset of neurons with defective neuronal migration pattern that was not generated in the control organoids, likely due to impaired morphology of progenitor cells. This subset of neurons had a unique transcriptional profile defined for its upregulation of axon guidance and migration genes and downregulation of synapse formation genes, suggesting a causal link between mutations in the DCHS1 and FAT4 genes and dysregulation of the proteins essential for cell guidance and cell-cell communication.
Lopez-Tobon et al. [55] studied the role of GSK3 pathway in human cortical organoid development. Disruption of this signaling pathway is of great interest as it has been associated to multiple neurodevelopmental disorders including fragile-X, autism, bipolar syndrome and schizophrenia. Despite this, few advances have been made in untangling the impact of GSK3 activity during human brain development. In this study, Lopez-Tobon et al. found that chronic inhibition of the GSK3 pathway leads to morphological alteration of cortical organoids. Through scRNA-seq they identified a cell-type specific role of GSK3 signaling in human corticogenesis. Specifically, followed GSK3 inhibition they found impairment of NEUROD1/2 lineages and overall decrease in outer radial glia (oRG) production. To our knowledge this is the first paper that attempts mechanistic dissection human corticogenesis in brain organoids, which demonstrates the potential of this technology to uncover cell type specific disease phenotype in brain organoids.
6. Limitations of single cell RNA transcriptomics
Despite the ability to probe cellular heterogeneity at a single cell resolution, some considerations need to be made into the limitations of this method prior to investing time, energy, and finances into its utilization. Some technical limitations of single cell RNA sequencing are low capture efficiency, high dropout, low depth, and dissociation bias. In addition to these technical issues, a lack of spatial context, proteomic, and splicing isoform variation information inherently limit the types of questions that single cell RNA transcriptomics can answer.
One of the main technical limitation is the high dropout rate, a feature of scRNA-seq experiments whereby only a small portion of an entire transcriptome from a single cell is captured [4]. Some reasons for a high dropout rate includes the inability to properly capture the mRNA from the cell (only 10–20% of transcripts are reverse transcribed) [58], the low levels of the transcript, as well as the highly stochastic nature of mRNA expression within each cell [59]. A high number of dropouts can skew the data set by artificially inflating the number of zero expression genes. This could be a major issue when opting for low depth sequencing to decrease cost and increase practicality of the experiment. The low depth, in combination with the high dropout rate could prevent the identification of rare cell types and detection of subtle changes in mRNA levels.
Another limitation that may skew the data is the dissociation bias. This comes about from samples undergoing an enzymatic isolation that results in a selection against certain cell types due to the higher stress experienced by these cells due to the digestion conditions. The selection of more resilient population of cells could give a skewed perspective on the actual organoid composition. Finally, it is important to recognized that single cell disassociation leads to the loss of spatial context within the organoid, and it is this important piece of information showing direct cell-to-cell interactions within the organoid that could reveal how communication between key partners regulate various processes including maturation, pruning, and migration. Currently, putative cell-cell, and receptor-ligand interactions based on around scRNA data is inferred information through computational modeling.
In addition to the inherent limitation to directly probe cell-cell and receptor-ligand interactions, scRNA seq does not offer any information about the proteome within these single cells. Due to the lack of high throughput proteomics conducted on human brain organoids, it is difficult to determine whether the levels of a certain receptor or ligand are at comparable levels based only in the RNA presence. Previous work has shown that the levels of mRNA do not necessarily linearly correlate to protein levels [60]. The variable relationship between mRNA and protein levels forces only inferences to be made about the complex interactions at the protein level.
Finally, current techniques are mostly aimed toward polyT priming, which initiates the coverage of a transcript from the 3’ end in order to avoid capturing ribosomal RNA. This method excludes certain isoforms and thus results in a biased view of the transcriptome. However, efforts have been made to address this particular issue by modifying platforms to capture various isoforms of RNA, which indicates that the field is heading in the right direction [61](cite Hagemann-Jensen et al). As the field develops, it is clear that these limitations must be addressed in order to take advantage of the full potential of scRNA-seq.
7. Multi-omics approach to complement brain organoid characterization
Although attempts are being made to amend some of the previously mentioned limitations, alternative methods are also being devised that can be used in conjunction with scRNA-seq to get a clearer picture of what a human brain organoid is made up of and how it functions. The next step forward in characterizing brain organoids would be to understand the epigenetic modifications that regulate key transcriptional programs. Another step would be to probe alternative RNA sequencing techniques that incorporate other aspects such as spatial, functional, or lineage information. Finally, incorporating single cell proteomics, and ligand-receptor interaction via the use of synthetically engineering receptors/ligands will be essential to answering our question concerning cellular functionality and connectivity within organoids (Figure 2).
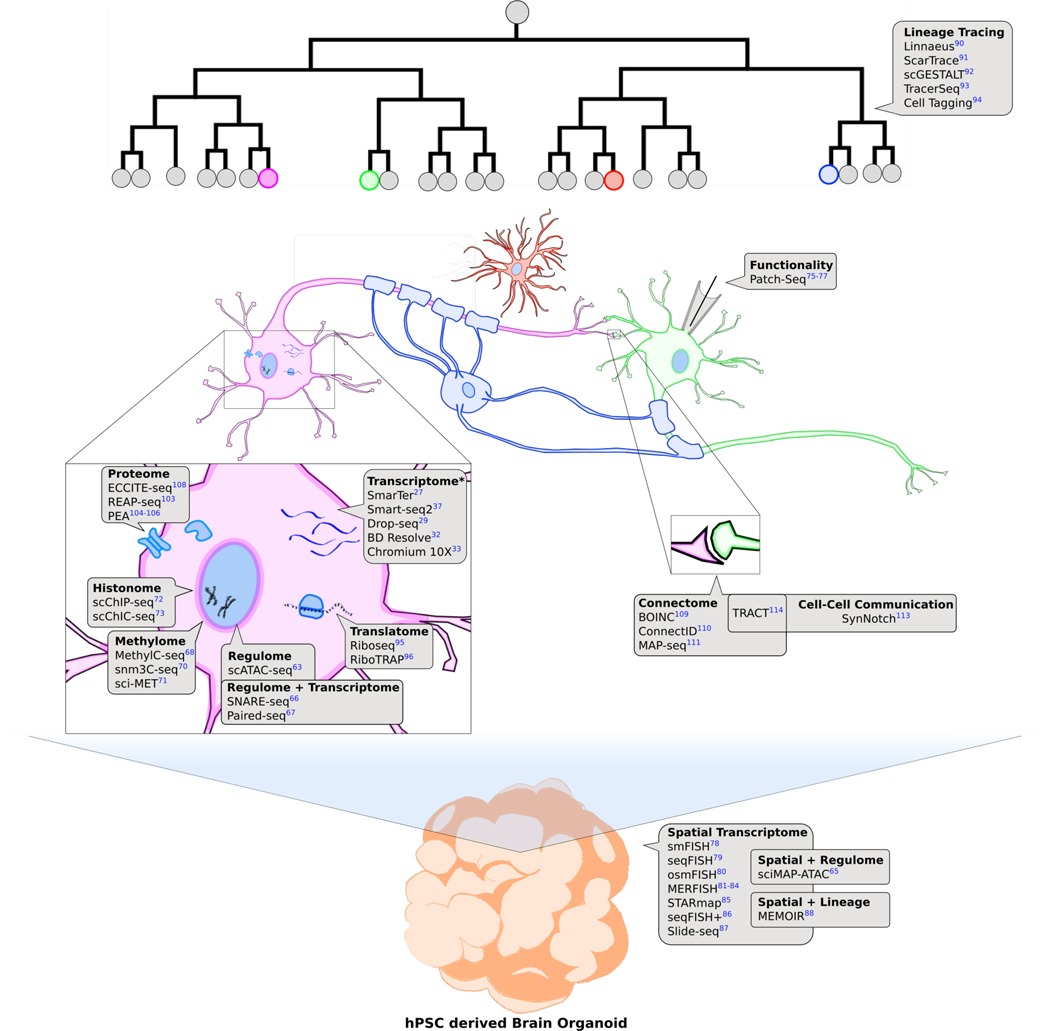
Fig. 2. Multimodal approach to characterize hPSC derived brain organoids.
An overview on the current methods that are available for characterizing brain organoids at a single cell resolution. Cell types include astrocyte (red), oligodendrocyte (blue), neuron A (purple), neuron B (green). *the platforms listed under “Transcriptome” only include ones used to study hPSC derived brain organoids
7.1. Probing the Epigenetic Landscape
DNA methylation and histone modifications are two mechanisms that contribute to a complex epigenetic landscape. The combinatorial activity between these modifications and transcription factors control DNA accessibility and ultimately govern the plethora of states that a near homogenous ball of pluripotent stem cells can achieve. It is thus essential to characterize how epigenetic regulation contributes to generating the highly diverse cell types found in human brain organoids. Amiri et al used transposase-accessible chromatin sequencing (ATAC-seq) to look into the epigenetic landscape governing chromatin accessibility of their brain organoids. However, more recently a method known as single-cell transposase-accessible chromatin sequencing (scATAC-seq) aims to address this broad stroke approach offered by ATAC-seq by peering into the “regulome” at a finer resolution [62,63]. Kanton et al. used scATAC-seq to identified human specific differences in chromatin accessibility by comparing human vs chimpanzee derived brain organoids [39]. Through measuring the genome-wide chromatin accessibility, the variability between cell-type specific epigenomes could be determined and thus aid in identification of cell types. The field is also moving toward integrating a spatial component to single cell ATAC seq (sciMAP-ATAC) [63,64], as well as combining the ATAC-seq platform with single cell RNA-seq to obtain the transcriptome and the DNA chromatin accessibility by sequencing (SNARE-seq) [65].
Other methods for studying DNA methylation signatures have also come forward. MethylC-seq [66], a methylome characterization technique, has been used to identify the high degree of similarity between the epigenetic signature of human fetal brain compared to cerebral organoids developed using the Lancaster protocol [67]. As interest in this field grows, newer, emerging techniques, such as snmC-seq2 [68] and sci-MET [63] will be used to study the methylome of single cells within brain organoids.
In addition to the methylome, histone modifications can be interrogated through single cell chromatin immunoprecipitation sequencing (scChIP-seq) [69] or single cell chromatin immunocleavage sequencing (scChIC-seq) [70], which offer the potential to study cellular subpopulations at a single cell resolution. In the study conducted by Rotem et al, the use of droplet based technology combined with CHIP allowed for the identification of cell types in a mixed population of mouse embryonic stem cells (ESC), embryonic fibroblasts (MEF), and hematopoetic progenitor cells (EML) based on of known epigenetic landscapes [71]. By bringing these technologies to the organoid field, cell type identification and characterization at the single cell level could be observed from an epigenetic standpoint.
7.2. Combining RNAseq with Functional, Spatial, and Lineage Information
Classical functional analysis techniques have involved measuring the electrical activity of living neurons by recording the flux of intra- and extracellular ions. Patch Clamp recordings are a type of intracellular functional assays done to record the firing patterns of neurons. Given the vast amount of information that can be obtained from this functional analysis in term of characterizing different cells based on their firing patterns, integrating this technique with current methods of characterization cell types based on scRNA-seq would be a great step forward for the field. Patch-seq addresses this point by combining the elegant recording techniques of Patch Clamp with scRNA-seqto capture the transcriptomic information from within a patched cell [72–74].
One major drawback to scRNAseq, which include the techniques listed above, is the loss of spatial information. The ability of combining the data between the spatial orientation or morphology of a specific cell with that same cell and its transcriptomic profile is not a trivial task. Modified versions of fluorescence in situ hybridization (FISH) such as smFISH [59], seq-FISH [75], and osmFISH [76] have the potential to overcome this issue by capturing multiple transcripts within a cell without losing its spatial context. As technology developed, the ability to capture more RNA with a lower error rate and lower off-targets emerged as was demonstrated with MERFISH [77–80]. Next, STARmap [48] came along, and afterward to improve the ability of MERFISH, which only captures long RNA species. Further development has led to improvements to previously published platforms such as seqFISH+ [81], which addresses the limitation of the previous method to capture a lower number of genes within a cell. One of the most recent studies, Slide-seq [82], aimed to provide single cell spatial transcriptomics information by placing a sample on a slide covered in DNA barcoded beads. As newer platforms of higher resolution, lower cost, and increased specificity come about, the ability to clarify the spatial orientation of cell types within brain organoids will become more commonplace.
In conjunction with spatial transcriptomics, lineage tracing can be done with a technique called memory by engineered mutagenesis with optical in situ readout (MEMOIR). This is a system that stably integrates 28 barcoded scratchpads into a target genome that is susceptible to editing by the expression of an inducible sgRNA and a degradable Cas9 protein. This unique system is coupled with the spatial transcriptomic readout obtained with seqFISH, which is based on sequential in situ hybridization using a scratchpad oligo probe set, a barcode-specific oligo probe set and the corresponding set of transcript-specific probes [81,83,84]. Other recent lineage tracing techniques that make use of Cas9 “scars” include Lineage tracing by nuclease-activated editing of ubiquitous sequences (Linnaeus) [85], ScarTrace [86], and Single-cell genome editing of synthetic target arrays for lineage tracing (scGESTALT) [87]. These techniques offer the ability to directly track the fate of a specific cells and overcomes the post hoc strategy offered by pseudotemporal analysis, which limits the identification of the causal factors that drive a cell down to a specific trajectory [22]. In Linnaeus, a red fluorescent protein (RFP) transgene is integrated into 16–32 different loci to ensure that Cas9 “scar” created on this transgene by injecting the Cas9 protein and targeting sgRNA is not deleted or overwritten. This “scar” is detected once the sample undergoes two rounds of amplification during sc-RNAseq with primers specific to the RFP transgene. The ScarTrace method uses 8 copies of the green fluorescent protein (GFP) repeated in tandem within the construct are scarred upon the injection of the Cas9 protein and sgRNA targeting the GFP. This method tries to overcome the ability of a scar to be overwritten or deleted by sorting by robot-assisted transcriptome sequencing (SORT-seq) into a 384 well-plate and has the ability to detect both RNA and DNA scars. ScGESTALT, like Linnaeus, has the ability to make use of scRNA-seq and can edit at two time points. While the previous methods allow for scaring with one initial injection, this technique allows for tracking heritable changes and progression at later stages with an inducible second scarring.
Non-Cas9 barcoding techniques such as transposon-based barcoding, (TracerSeq) [88], or a viral delivery approach, Cell Tagging [89], offer an alternative to Cas9 “scars”. TracerSeq integrates GFP fused to a scratchpad of 20 nucleotides into the genome and generates higher combinatorial diversity of barcodes compared to Cas9 methods. Cell Tagging transduces viral vector containing a specific library of an 8 nucleotide random sequence in the 3’ UTR of GFP and delivered at targeted developmental time points. The amalgamation of scRNA-seq into these two techniques results in the emergence of a map of a cell’s divergent or convergent paths toward a committed cellular fate.
7.3. Investigating Cell-Type Specific Translatome and Proteome
The critical step of translation from mRNA to protein offers the potential to explore cellular diversity by capturing ribosomes that are in the process of generating the proteome of a cell through a technique called Riboseq or active mRNA translation sequencing (ARTseq). [90,91]. There are papers that have been published that make use of this technology in order to better understand the translational changes found in fragile X syndrome model mice [92,93]. Taking this one step further, Furlanis et al. was able to combine this platform of Riboseq with ribosomes conditionally tagged in known cell types (RiboTRAP), which gives a clearer picture of the translatome of specific cell populations [94]. Unfortunately, this technology does not exist at the single cell level yet. One l future avenue is to combine it with a labeling technique that would allow for the identification and FACS sorting of cell types [95,96]. This would then lead to the ability to uncover the cell type specific “translatome”.
Given the discrepancy found between mRNA and protein levels, proteomics takes Riboseq one-step further by characterizing cells by their protein composition. CITE-seq is a step in the right direction by integrating transcriptomic and cell surface protein measurements into a single single cell output [97]. Other antibody linked oligonucleotide techniques such as REAP-seq [98] and proximity extension assay (PEA) [99–101] also make use of this multiplexing technique by combining the proteomic and transcriptomic profiles to get a better picture. FACS sorting for cell surface proteins is also proving to be a useful tool with a recent study sorting 7000 proteins across 12 immune cell populations and elucidating more cell type specific proteins in the process [102].
8. Conclusions and future outlook
Despite the excitement and increased popularity of brain organoid as a model system, their complex structure, heterogenous cell types, and lack of reproducible anatomical organization, poses an incredible challenge to scientists wishing to interrogate developmental process and disease phenotypes. In recent years, advances in single cell-genomics have given us the means to partially tame this complexity, and we anticipate that in the coming years integration of single-cell multi-omics techniques will became commonplace. For example, a much-needed characterization of the wiring diagram of monosynaptic connections in the organoids could be achieved by combining pseudotyped rabies virus tracing with large-scale spatial transcriptomics.
Recently, approaches to characterize cell-cell communication mediated by ligand-receptor interactions across individual cell-types, using single cell RNA sequencing, have been developed [103]. However, these methods, due to cell dissociation, do not preserve the spatial and morphological information present in the sample. We envision that human brain organoids, in combination with spatial transcriptomics and other rapidly expanding technologies, including tools to engineering human pluripotent stem cells with synthetic receptors and ligands in which communication between cells can be easily monitored, [104] and 4D live-cell imaging techniques [105,106] will, in the near future enable the study of dynamic interactions between individual cell types as well as uncover dysfunctional communication signals in disease states with single-cell resolution.
Acknowledgments
We thank members of the Quadrato lab for insightful discussions. Tuan Nguyen for editing the manuscript. This work was supported by a grant from the Baxter foundation. We apologize to colleagues whose work we could not cite because of space limitations.
References
- [1].Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Yang SM, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P, Cell diversity and network dynamics in photosensitive human brain organoids, Nature. 545 (2017) 48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Camp J. Gray, Treutlein B, Human organomics: A fresh approach to understanding human development using single-cell transcriptomics, Dev. 144 (2017) 1584–1587. doi: 10.1242/dev.150458. [DOI] [PubMed] [Google Scholar]
- [3].Hwang B, Lee JH, Bang D, Single-cell RNA sequencing technologies and bioinformatics pipelines, Exp. Mol. Med. 50 (2018). doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haque A, Engel J, Teichmann SA, Lönnberg T, A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications, Genome Med. (2017). doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hedlund E, Deng Q, Single-cell RNA sequencing: Technical advancements and biological applications, Mol. Aspects Med. (2018). doi: 10.1016/j.mam.2017.07.003. [DOI] [PubMed] [Google Scholar]
- [6].Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W, Comparative Analysis of Single-Cell RNA Sequencing Methods, Mol. Cell. (2017). doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- [7].Tian L, Dong X, Freytag S, Lê Cao KA, Su S, JalalAbadi A, Amann-Zalcenstein D, Weber TS, Seidi A, Jabbari JS, Naik SH, Ritchie ME, Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments, Nat. Methods. (2019). doi: 10.1038/s41592-019-0425-8. [DOI] [PubMed] [Google Scholar]
- [8].Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z, Huang Y, Wang J, Comparative Analysis of Droplet-Based Ultra-High-Throughput Single-Cell RNA-Seq Systems, Mol. Cell. (2019). doi: 10.1016/j.molcel.2018.10.020. [DOI] [PubMed] [Google Scholar]
- [9].Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND, Hughes TK, Wadsworth MH, Burks T, Nguyen LT, Kwon JYH, Barak B, Ge W, Kedaigle AJ, Carroll S, Li S, Hacohen N, Rozenblatt-Rosen O, Shalek AK, Villani A-C, Regev A, Levin JZ, Systematic comparative analysis of single cell RNA-sequencing methods, BioRxiv. (2019). doi: 10.1101/632216. [DOI] [Google Scholar]
- [10].Vieth B, Parekh S, Ziegenhain C, Enard W, Hellmann I, A Systematic Evaluation of Single Cell RNA-Seq Analysis Pipelines, Nat. Commun. (2019). doi: 10.1038/s41467-019-12266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cole MB, Risso D, Wagner A, DeTomaso D, Ngai J, Purdom E, Dudoit S, Yosef N, Performance Assessment and Selection of Normalization Procedures for Single-Cell RNA-Seq, Cell Syst. (2019). doi: 10.1016/j.cels.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Haghverdi L, Lun ATL, Morgan MD, Marioni JC, Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors, Nat. Biotechnol. (2018). doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Duò A, Robinson MD, Soneson C, A systematic performance evaluation of clustering methods for single-cell RNA-seq data., F1000Research. 7 (2018) 1141. doi: 10.12688/f1000research.15666.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW, Dimensionality reduction for visualizing single-cell data using UMAP, Nat. Biotechnol. (2019). doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- [15].Selim SZ, Ismail MA, K-Means-Type Algorithms: A Generalized Convergence Theorem and Characterization of Local Optimality, IEEE Trans. Pattern Anal. Mach. Intell. (1984). doi: 10.1109/TPAMI.1984.4767478. [DOI] [PubMed] [Google Scholar]
- [16].Petegrosso R, Li Z, Kuang R, Machine learning and statistical methods for clustering single-cell RNA-sequencing data, Brief. Bioinform. (2019). doi: 10.1093/bib/bbz063. [DOI] [PubMed] [Google Scholar]
- [17].Saelens W, Cannoodt R, Todorov H, Saeys Y, A comparison of single-cell trajectory inference methods, Nat. Biotechnol. 37 (2019) 547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- [18].Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C, Reversed graph embedding resolves complex single-cell trajectories, Nat. Methods. 14 (2017) 979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, Levin JZ, Arlotta P, Individual brain organoids reproducibly form cell diversity of the human cerebral cortex, Nature. 570 (2019) 523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, Huguenard JR, Pașca SP, Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures, Nat. Neurosci. 22 (2019) 484–491. doi: 10.1038/s41593-018-0316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, Paşca SP, Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells, Neuron. 95 (2017) 779–790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells, Nat. Biotechnol. (2014). doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Pääbo S, Huttner WB, Treutlein B, Human cerebral organoids recapitulate gene expression programs of fetal neocortex development, Proc. Natl. Acad. Sci. 112 (2015) 201520760. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundström E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, Kharchenko PV, RNA velocity of single cells, Nature. (2018). doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ, Diffusion pseudotime robustly reconstructs lineage branching, Nat. Methods. (2016). doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- [26].Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, Lee L, Chen J, Brumbaugh J, Rigollet P, Hochedlinger K, Jaenisch R, Regev A, Lander ES, Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming, Cell. (2019). doi: 10.1016/j.cell.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R, Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells, Nat. Biotechnol. (2012). doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA, Cerebral organoids model human brain development and microcephaly, Nature. 501 (2013) 373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets, Cell. (2015). doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O’Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Pasca SP, Assembly of functionally integrated human forebrain spheroids, Nature. 545 (2017) 54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, Zhong M, Stanley EG, Elefanty AG, Naegele JR, Lee SH, Weissman SM, Park IH, Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration, Cell Stem Cell. 21 (2017) 383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fan HC, Fu GK, Fodor SPA, Combinatorial labeling of single cells for gene expression cytometry, Science (80-. ). (2015). doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- [33].Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, Massively parallel digital transcriptional profiling of single cells, Nat. Commun. (2017). doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, Paulsen O, Lakatos A, Lancaster MA, Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output, Nat. Neurosci. (2019). doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, Domissy A, Vandenberghe M, Devor A, Yeo GW, Voytek B, Muotri AR, Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development, Cell Stem Cell. (2019). doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y, Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex, Proc. Natl. Acad. Sci. 110 (2013) 20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R, Smart-seq2 for sensitive full-length transcriptome profiling in single cells, Nat. Methods. 10 (2013) 1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- [38].Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, Pochareddy S, Shin Y, Safi A, Song L, Zhu Y, Sousa AMM, Gerstein M, Crawford GE, Sestan N, Abyzov A, Vaccarino FM, Transcriptome and epigenome landscape of human cortical development modeled in organoids, Science (80-. ). 362 (2018). doi: 10.1126/science.aat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Calleja FS, Sidow L, Fleck J, Guijarro P, Han D, Qian Z, Heide M, Huttner W, Khaitovich P, Pääbo S, Treutlein B, Camp JG, Single-cell genomic atlas of great ape cerebral organoids uncovers human-specific features of brain development, BioRxiv. (2019). doi: 10.1101/685057. [DOI] [PubMed] [Google Scholar]
- [40].Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, Fiddes IT, Kronenberg ZN, Shuga J, Leyrat AA, West JA, Bershteyn M, Lowe CB, Pavlovic BJ, Salama SR, Haussler D, Eichler EE, Kriegstein AR, Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution, Cell. 176 (2019) 743–756.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, Lee SH, Weissman SM, Park IH, hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids, Cell Stem Cell. 24 (2019) 487–497.e7. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y, Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue, Nat. Commun. 6 (2015) 1–11. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL, Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure, Cell. 165 (2016) 1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Göke J, Tan ZY, Saw TY, Tan CP, Lokman H, Lee Y, Kim D, Ko HS, Kim SO, Park JH, Cho NJ, Hyde TM, Kleinman JE, Shin JH, Weinberger DR, Tan EK, Je HS, Ng HH, Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons, Cell Stem Cell. 19 (2016) 248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y, Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells, Cell Rep. (2015). doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- [46].Amin ND, Paşca SP, Building Models of Brain Disorders with Three-Dimensional Organoids, Neuron. 100 (2018) 389–405. doi: 10.1016/j.neuron.2018.10.007. [DOI] [PubMed] [Google Scholar]
- [47].Quadrato G, Brown J, Arlotta P, The promises and challenges of human brain organoids as models of neuropsychiatric disease, Nat. Med. 22 (2016) 1220–1228. doi: 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- [48].Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava FA, Deisseroth K, Three-dimensional intact-tissue sequencing of single-cell transcriptional states, Science (80-. ). (2018). doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Broix L, Jagline H, Ivanova EL, Schmucker S, Drouot N, Clayton-Smith J, Pagnamenta AT, Metcalfe KA, Isidor B, Louvier UW, Poduri A, Taylor JC, Tilly P, Poirier K, Saillour Y, Lebrun N, Stemmelen T, Rudolf G, Muraca G, Saintpierre B, Elmorjani A, Moïse M, Weirauch NB, Guerrini R, Boland A, Olaso R, Masson C, Tripathy R, Keays D, Beldjord C, Nguyen L, Godin J, Kini U, Nischké P, Deleuze JF, Bahi-Buisson N, Sumara I, Hinckelmann MV, Chelly J, Mutations in the HECT domain of NEDD4L lead to AKT-mTOR pathway deregulation and cause periventricular nodular heterotopia, Nat. Genet. (2016). doi: 10.1038/ng.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cappello S, Gray MJ, Badouel C, Lange S, Einsiedler M, Srour M, Chitayat D, Hamdan FF, Jenkins ZA, Morgan T, Preitner N, Uster T, Thomas J, Shannon P, Morrison V, Di Donato N, Van Maldergem L, Neuhann T, Newbury-Ecob R, Swinkells M, Terhal P, Wilson LC, Zwijnenburg PJG, Sutherland-Smith AJ, Black MA, Markie D, Michaud JL, Simpson MA, Mansour S, McNeill H, Götz M, Robertson SP, Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development, Nat. Genet. 45 (2013) 1300–1310. doi: 10.1038/ng.2765. [DOI] [PubMed] [Google Scholar]
- [51].Kielar M, Tuy FPD, Bizzotto S, Lebrand C, De Juan Romero C, Poirier K, Oegema R, Mancini GM, Bahi-Buisson N, Olaso R, Le Moing AG, Boutourlinsky K, Boucher D, Carpentier W, Berquin P, Deleuze JF, Belvindrah R, Borrell V, Welker E, Chelly J, Croquelois A, Francis F, Mutations in Eml1 lead to ectopic progenitors and neuronal heterotopia in mouse and human, Nat. Neurosci. 17 (2014) 923–933. doi: 10.1038/nn.3729. [DOI] [PubMed] [Google Scholar]
- [52].Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, Parrini E, Valence S, Saint Pierre B, Oger M, Lacombe D, Geneviève D, Fontana E, Darra F, Cances C, Barth M, Bonneau D, Bernadina BD, N’Guyen S, Gitiaux C, Parent P, Des Portes V, Pedespan JM, Legrez V, Castelnau-Ptakine L, Nitschke P, Hieu T, Masson C, Zelenika D, Andrieux A, Francis F, Guerrini R, Cowan NJ, Bahi-Buisson N, Chelly J, Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly, Nat. Genet. 45 (2013) 639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Reiner O, Sapir T, LIS1 functions in normal development and disease, Curr. Opin. Neurobiol. 23 (2013) 951–956. doi: 10.1016/j.conb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- [54].Klaus J, Kanton S, Kyrousi C, Ayo-Martin AC, Di Giaimo R, Riesenberg S, O’Neill AC, Camp JG, Tocco C, Santel M, Rusha E, Drukker M, Schroeder M, Götz M, Robertson SP, Treutlein B, Cappello S, Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia, Nat. Med. 25 (2019) 561–568. doi: 10.1038/s41591-019-0371-0. [DOI] [PubMed] [Google Scholar]
- [55].López-tobón A, Villa CE, Cheroni C, Trattaro S, Human cortical organoids expose a differential function of GSK3 on direct and indirect neurogenesis, (2018). doi: 10.1101/48474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Iefremova V, Manikakis G, Krefft O, Jabali A, Weynans K, Wilkens R, Marsoner F, Brändl B, Müller FJ, Koch P, Ladewig J, An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome, Cell Rep. 19 (2017) 50–59. doi: 10.1016/j.celrep.2017.03.047. [DOI] [PubMed] [Google Scholar]
- [57].Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, Parrini E, Valence S, Saint Pierre B, Oger M, Lacombe D, Geneviève D, Fontana E, Darra F, Cances C, Barth M, Bonneau D, Bernadina BD, N’Guyen S, Gitiaux C, Parent P, Des Portes V, Pedespan JM, Legrez V, Castelnau-Ptakine L, Nitschke P, Hieu T, Masson C, Zelenika D, Andrieux A, Francis F, Guerrini R, Cowan NJ, Bahi-Buisson N, Chelly J, Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly, Nat. Genet. 45 (2013) 639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S, Quantitative single-cell RNA-seq with unique molecular identifiers, Nat. Methods. (2014). doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- [59].Raj A, van Oudenaarden A, Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences, Cell. (2008). doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vogel C, Marcotte EM, Insights into the regulation of protein abundance from proteomic and transcriptomic analyses, Nat. Rev. Genet. (2012). doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hagemann-Jensen R.S. Michael, Ziegenhain Christoph, Chen Ping, Ramsköld Daniel, Hendriks Gert-Jan, Larsson Anton J.M., Faridani Omid R., Single-cell RNA counting at allele- and isoform-resolution using Smart-seq3, BioRxiv. (2019). doi: 10.1101/817924. [DOI] [PubMed] [Google Scholar]
- [62].Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ, Single-cell chromatin accessibility reveals principles of regulatory variation, Nature. (2015). doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mulqueen RM, DeRosa BA, Thornton CA, Sayar Z, Torkenczy KA, Fields AJ, Wright KM, Nan X, Ramji R, Steemers FJ, O’Roak BJ, Adey AC, Improved single-cell ATAC-seq reveals chromatin dynamics of in vitro corticogenesis, BioRxiv. (2019). doi: 10.1101/637256. [DOI] [Google Scholar]
- [64].Thornton A.C.A. Casey A., Mulqueen Ryan M., Torkenczy Kristof A., Lowenstein Eve G., Fields Andrew J., Steemers Frank J., Wright Kevin M., Spatially-mapped single-cell chromatin accessibility, BioRxiv. (2019). doi: 10.1101/815720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chen S, Lake BB, Zhang K, High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell, Nat. Biotechnol. (2019). doi: 10.1038/s41587-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Urich MA, Nery JR, Lister R, Schmitz RJ, Ecker JR, MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing, Nat. Protoc. (2015). doi: 10.1038/nprot.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR, Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain, Cell Rep. 17 (2016) 3369–3384. doi: 10.1016/j.celrep.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Luo C, Rivkin A, Zhou J, Sandoval JP, Kurihara L, Lucero J, Castanon R, Nery JR, Pinto-Duarte A, Bui B, Fitzpatrick C, O’Connor C, Ruga S, Van Eden ME, Davis DA, Mash DC, Behrens MM, Ecker JR, Robust single-cell DNA methylome profiling with snmC-seq2, Nat. Commun. (2018). doi: 10.1038/s41467-018-06355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gomez D, Shankman LS, Nguyen AT, Owens GK, Detection of histone modifications at specific gene loci in single cells in histological sections, Nat. Methods. (2013). doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ku WL, Nakamura K, Gao W, Cui K, Hu G, Tang Q, Ni B, Zhao K, Single-cell chromatin immunocleavage sequencing (scChIC-seq) to profile histone modification, Nat. Methods. (2019). doi: 10.1038/s41592-019-0361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE, Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state, Nat. Biotechnol. (2015). doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fuzik J, Zeisel A, Mate Z, Calvigioni D, Yanagawa Y, Szabo G, Linnarsson S, Harkany T, Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes, Nat. Biotechnol. (2016). doi: 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bardy C, Van Den Hurk M, Kakaradov B, Erwin JA, Jaeger BN, Hernandez RV, Eames T, Paucar AA, Gorris M, Marchand C, Jappelli R, Barron J, Bryant AK, Kellogg M, Lasken RS, Rutten BPF, Steinbusch HWM, Yeo GW, Gage FH, Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology, Mol. Psychiatry. (2016). doi: 10.1038/mp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, Sandberg R, Tolias AS, Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq, Nat. Biotechnol. (2016). doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shah S, Lubeck E, Zhou W, Cai L, In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus, Neuron. (2016). doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Codeluppi S, Borm LE, Zeisel A, La Manno G, van Lunteren JA, Svensson CI, Linnarsson S, Spatial organization of the somatosensory cortex revealed by osmFISH, Nat. Methods. (2018). doi: 10.1038/s41592-018-0175-z. [DOI] [PubMed] [Google Scholar]
- [77].Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X, High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing, Proc. Natl. Acad. Sci. 113 (2016) 14456–14461. doi: 10.1073/pnas.1617699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X, High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization, Proc. Natl. Acad. Sci. U. S. A. (2016). doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Moffitt JR, Zhuang X, RNA Imaging with Multiplexed Error-Robust Fluorescence in Situ Hybridization (MERFISH), in: Methods Enzymol, 2016. doi: 10.1016/bs.mie.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Xia C, Fan J, Emanuel G, Hao J, Zhuang X, Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression., Proc. Natl. Acad. Sci. U. S. A. (2019). doi: 10.1073/pnas.1912459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Eng CHL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, Cai L, Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+, Nature. (2019). doi: 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ, Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution, Science (80-. ). 363 (2019) 1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Frieda KL, Linton JM, Hormoz S, Choi J, Chow KHK, Singer ZS, Budde MW, Elowitz MB, Cai L, Synthetic recording and in situ readout of lineage information in single cells, Nature. (2017). doi: 10.1038/nature20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Shah S, Takei Y, Zhou W, Lubeck E, Yun J, Eng CHL, Koulena N, Cronin C, Karp C, Liaw EJ, Amin M, Cai L, Dynamics and Spatial Genomics of the Nascent Transcriptome by Intron seqFISH, Cell. 174 (2018) 363–376.e16. doi: 10.1016/j.cell.2018.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP, Simultaneous lineage tracing and cell-type identification using CrIsPr-Cas9-induced genetic scars, Nat. Biotechnol. (2018). doi: 10.1038/nbt.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Alemany A, Florescu M, Baron CS, Peterson-Maduro J, Van Oudenaarden A, Whole-organism clone tracing using single-cell sequencing, Nature. (2018). doi: 10.1038/nature25969. [DOI] [PubMed] [Google Scholar]
- [87].Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF, Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain, Nat. Biotechnol. (2018). doi: 10.1038/nbt.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM, Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo, Science (80-. ). (2018). doi: 10.1126/science.aar4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Biddy BA, Kong W, Kamimoto K, Guo C, Waye SE, Sun T, Morris SA, Single-cell mapping of lineage and identity in direct reprogramming, Nature. (2018). doi: 10.1038/s41586-018-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Eastman G, Smircich P, Sotelo-Silveira JR, Following Ribosome Footprints to Understand Translation at a Genome Wide Level, Comput. Struct. Biotechnol. J. (2018). doi: 10.1016/j.csbj.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Calviello L, Ohler U, Beyond Read-Counts: Ribo-seq Data Analysis to Understand the Functions of the Transcriptome, Trends Genet. (2017). doi: 10.1016/j.tig.2017.08.003. [DOI] [PubMed] [Google Scholar]
- [92].Das Sharma S, Metz JB, Li H, Hobson BD, Hornstein N, Sulzer D, Tang G, Sims PA, Widespread Alterations in Translation Elongation in the Brain of Juvenile Fmr1 Knockout Mice, Cell Rep. (2019). doi: 10.1016/j.celrep.2019.02.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Liu B, Molinaro G, Shu H, Stackpole EE, Huber KM, Richter JD, Optimization of ribosome profiling using low-input brain tissue from fragile X syndrome model mice, Nucleic Acids Res. (2019). doi: 10.1093/nar/gky1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Furlanis E, Traunmüller L, Fucile G, Scheiffele P, Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs, Nat. Neurosci. (2019). doi: 10.1038/s41593-019-0465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD, Novel Subtype-Specific Genes Identify Distinct Subpopulations of Callosal Projection Neurons, J. Neurosci. 29 (2009) 12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Molyneaux BJ, Goff LA, Brettler AC, Chen H-H, Brown JR, Hrvatin S, Rinn JL, Arlotta P, DeCoN: Genome-wide Analysis of In Vivo Transcriptional Dynamics during Pyramidal Neuron Fate Selection in Neocortex, Neuron. 85 (2015) 275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P, Simultaneous epitope and transcriptome measurement in single cells, Nat. Methods. (2017). doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, Mcclanahan TK, Sadekova S, Klappenbach JA, Multiplexed quantification of proteins and transcripts in single cells, Nat. Biotechnol. (2017). doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- [99].Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, Eriksson A, Dickens ER, Ohlsson S, Edfeldt G, Andersson AC, Lindstedt P, Stenvang J, Gullberg M, Fredriksson S, Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability, PLoS One. (2014). doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, Fredriksson S, Assarsson E, Lundberg M, Nelander S, Westermark B, Landegren U, Simultaneous Multiplexed Measurement of RNA and Proteins in Single Cells, Cell Rep. (2016). doi: 10.1016/j.celrep.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Genshaft AS, Li S, Gallant CJ, Darmanis S, Prakadan SM, Ziegler CGK, Lundberg M, Fredriksson S, Hong J, Regev A, Livak KJ, Landegren U, Shalek AK, Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction, Genome Biol. (2016). doi: 10.1186/s13059-016-1045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Myers SA, Rhoads A, Cocco AR, Peckner R, Haber AL, Schweitzer LD, Krug K, Mani DR, Clauser KR, Rozenblatt-Rosen O, Hacohen N, Regev A, Carr SA, Streamlined protocol for deep proteomic profiling of FAC-sorted cells and its application to freshly isolated murine immune cells, Mol. Cell. Proteomics. (2019). doi: 10.1074/mcp.RA118.001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, Kageyama J, Damm G, Seehofer D, Belicova L, Bickle M, Barsacchi R, Okuda R, Yoshizawa E, Kimura M, Ayabe H, Taniguchi H, Takebe T, Treutlein B, Multilineage communication regulates human liver bud development from pluripotency, Nature. (2017). doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- [104].Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, Lim WA, Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors, Cell. (2016). doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen BC, Chen B-C, Legant WR, Legant WR, Wang K, Wang K, Shao L, Shao L, Milkie DE, Milkie DE, Davidson MW, Davidson MW, Janetopoulos C, Janetopoulos C, Wu XS, Wu XS, Hammer JA, Hammer JA, Liu Z, Liu Z, English BP, English BP, Mimori-Kiyosue Y, Mimori-Kiyosue Y, Romero DP, Romero DP, Ritter AT, Ritter AT, Lippincott-Schwartz J, Lippincott-Schwartz J, Fritz-Laylin L, Fritz-Laylin L, Mullins RD, Mullins RD, Mitchell DM, Mitchell DM, Bembenek JN, Bembenek JN, Reymann A-C, Reymann AC, Bohme R, Böhme R, Grill SW, Grill SW, Wang JT, Wang JT, Seydoux G, Seydoux G, Tulu US, Tulu US, Kiehart DP, Kiehart DP, Betzig E, Betzig E, Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution., Science (80-. ). (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Schöneberg J, Dambournet D, Liu TL, Forster R, Hockemeyer D, Betzig E, Drubin DG, 4D cell biology: big data image analytics and lattice light-sheet imaging reveal dynamics of clathrin-mediated endocytosis in stem cell–derived intestinal organoids, Mol. Biol. Cell. (2018). doi: 10.1091/mbc.E18-06-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]