Abstract
The overuse and misuse of antibiotics have accelerated the selection of antibiotic-resistant bacteria, significantly impacting human, animal, and environmental health. As aquatic environments are vulnerable to antibiotic resistance, suitable management practices should be adopted to tackle this phenomenon. Here we show an effective, nature-based solution for reducing antibiotic resistance from actual wastewater. We utilize a bioreactor that relies on benthic (biofilms) and planktonic microbial communities to treat secondary effluent from a small urban wastewater treatment plant (<10,000 population equivalent). This treated effluent is eventually released into the local aquatic ecosystem. We observe high removal efficiency for genes that provide resistance to commonly used antibiotic families, as well as for mobile genetic elements that could potentially aid in their spread. Importantly, we notice a buildup of sulfonamide (sul1 and sul2) and tetracycline (tet(C), tet(G), and tetR) resistance genes specifically in biofilms. This advancement marks the initial step in considering this bioreactor as a nature-based, cost-effective tertiary treatment option for small UWWTPs facing antibiotic resistance challenges.
Keywords: ARGs, MGEs, UWWTPs, NBS, Biofilms
Graphical abstract
Highlights
-
•
A nature-based solution reduces antimicrobial resistance at a real scale.
-
•
ARGs and MGEs are removed by biofilms and plankton reactors.
-
•
Metagenomics offers a comprehensive insight into antimicrobial resistance.
-
•
Scientific advances support the implementation of small urban wastewater treatment plants.
1. Introduction
The growth of the urban population has led to a significant increase in wastewater production, generating a serious problem in its management [1]. The current regulation in the European Union (EU) (e.g., Council Directive 91/271/EEC [2] concerning urban wastewater treatment) establishes a mandatory control of total nitrogen and phosphorus loads in urban wastewater treatment plants (UWWTPs) over 100,000 population equivalents (P.E).
The latest data reveal that 96% of the wastewater generated by the population of the European Union is treated in UWWTPs, of which 7.75% is processed only up to secondary treatment, while the large majority (86.14%) undergoes much more stringent treatment (understood as at least tertiary treatment) [3]. Contrastingly, the small sewage facilities in the EU (<10,000 P.E) have not incorporated a tertiary treatment due to high energetic and economic costs [1,4].
The proposal for a new EU Directive concerning UWWTPs is to apply this strict control between 10,000 and 100,000 P.E [5]. Nevertheless, UWWTPs have presented an important issue in coping with bioavailable nutrients [6]. Despite the modern advanced technologies applied in UWWTPs, different global scenarios predict an increase in nutrient loads from their effluents into aquatic ecosystems [7], with the consequent impact associated with their ecosystem services and biodiversity, thereby affecting the environmental, economic, and social pathways [8]. Water quality regulators have been increasingly concerned about contaminants of emerging concern (CECs) detected in aquatic ecosystems [9]. However, the limits of CEC release are not yet embedded under the current EU directive.
The CECs monitoring list in the EU (Directive 2008/105/EC [10] amended in Decision 2015/495/EU [11]) refers to monitoring different micropollutants, including some antibiotics such as macrolides (i.e., azithromycin, clarithromycin, and erythromycin) and sulfonamides (i.e., sulfamethoxazole), because of their occurrence, resistance and resilience, and ecotoxicological and health-related issues in the environment.
The major issue associated with human health is the development of anti-microbial resistance (AMR), which is primarily caused by antibiotic-resistant bacteria (ARB) characterised by the presence of resistome describing antibiotic resistance genes (ARGs) commonly associated with mobilome capable of transmitting them. The mobilome associated with the mobile genetic elements (MGEs) facilitated the horizontal gene transfer (HGT), enhancing direct contact between bacterial cells with the ability to transfer ARGs by different processes (i.e., conjugation, transformation, and transduction) [12]. Recent studies have demonstrated that one of the major hotspots for ARGs and ARB emergence and spread to the environment is represented by wastewater treatment plants and their effluents [13,14].
The magnitude of the problem is undeniable. World antimicrobial resistance is estimated to produce 64.0 (95% uncertainty interval [UI] = 46.8–84.9) deaths per 100,000 inhabitants, whereas in central and eastern Europe is estimated at 57.9 (95% UI = 41.6–77.6) deaths per 100,000 inhabitants [15]. Furthermore, resistance to β-lactams and fluoroquinolones has been considered remarkable due to the efficiency of these antibiotic families in the initial treatments of severe infections [16], considerably reducing more than 70% of the deaths of AMR infections [15].
ARGs are considered to bond with MGEs due to their high plasticity in interacting with bacterial communities, favouring their acquisition and spread [12]. MGEs are the precursors of horizontal genetic transfer, an important role in bacterial communities considered as potential hotspots of transferring ARGs [17].
This public health concern has increased in recent years, and therefore, further research is needed to improve the treatment, such as using advanced technologies to reduce ARB, ARGs, and MGEs in UWWTP effluents [[18], [19], [20]]. Different technologies have been evaluated in laboratory conditions and under more realistic scenarios in pilot plant approaches [21,22]. However, the economic and energetic costs of these advanced technologies are generally high (i.e., ozonation, ultraviolet (UV)/H2O2, nanofiltration, and reverse osmosis [23]), limiting their potential application, particularly in small facilities with infrastructural and economic constraints.
Recent studies explored the possibility of implementing nature-based solutions (NBSs), such as constructed wetlands [[24], [25], [26]], to improve the overall quality of UWWTP effluent released to the environment and consequently protect the receptor ecosystems at lower costs. NBSs are considered a key tool for improving waste and sewage management at lower economic and energy costs, leveraging ecosystem services [27].
Benthic and planktonic freshwater microbial communities have shown the capacity to attenuate nutrient concentrations by direct uptake for growth [28,29], metabolizing some CECs [30,31] and, less studied, reducing ARB and ARGs [32], with biofilms being particularly considered as potential ARB reservoirs [33,34]. Reactors mimicking aquatic ecosystems in which these microbial communities actively develop can be a potential NBS to reduce contamination in UWWTP effluents before their release into aquatic environments, specifically acting as a trap for ARB, ARGs, and MGEs. In this study, the efficiency of an NBS bioreactor, in which microbial benthonic and planktonic communities’ composition and activity were monitored, was evaluated to reduce AMR loads in a UWWTP effluent. Furthermore, advanced analytical methods, grounded on next-generation sequencing (NGS) approaches [35], have been applied to analyse microbial communities, resistome, and mobilome variation in treated wastewater after its pass through the tested NBS bioreactor. Targeted ARGs and MGEs have been previously analysed using PCR-based methods [36]. However, metagenomic approaches have been applied to obtain a wider range of ARGs and MGEs [[37], [38], [39]], providing a full picture of the resistome and mobilome in a given ecosystem.
Therefore, this study aims to assess the efficiency of this benthic and planktonic nature-based bioreactor, called biofilm and plankton reactor (BPR), to remove ARGs and MGEs as an additional treatment step in small sewage facilities. This experiment evaluates the efficiency of the biological system in reducing the AMR load of urban wastewater in the operational environment of a small UWWTP facility, directly treating its secondary effluent. We expected the BPR to effectively reduce the abundance of most ARGs and MGEs, optimizing the efficiency and performance of the bioreactor. We also expected an accumulation of ARGs and MGEs within the benthic compartment of the reactor as a consequence of the removal of ARGs and MGEs from the system.
2. Methodology
2.1. Experimental design
The BPR was operated in the UWWTP of Quart, Girona (41°57′59.3″ N, 2°50′41.3″ E). The UWWTP treats 2750 P.E with an average input flow of 600 m3 day−1 and a maximal removal potential of 70 mg nitrogen L−1, 20 mg phosphorus L−1, and a maximal chemical oxygen demand (COD) of 495 mg L−1 (data from the Catalan Water Agency). The BPR mimicked lotic and lentic freshwater ecosystems composed of a stream-pond sequence in which the most relevant active biological compartments are benthic and planktonic microbial communities. The upper part contained the stream-based component of the BPR, in which aquatic biofilms (benthic community) were grown over artificial substrata (sand-rugous glasses) of different sizes (4, 9, 36, and 90 cm2), covering an overall surface of 120 × 380 cm (Fig. 1). On the lower part, the pond-based component of the BPR consisted of a water tank in which the planktonic communities were allowed to grow. The total volume of the tank was 1.96 m3 (length × depth × width: 4.05 × 0.40 × 1.21 m). The input wastewater effluent treated from the secondary treatment entered the system into a recirculation tank of 0.027 m3 (Fig. 1). The system was set in recirculation mode during the first 15 days (colonization phase) to favour biofilm settlement in glass substrata and planktonic community development in a pond-based system before switching to the operational phase. In the first stages of the operational phase, the secondary effluent inflow to the reactor was set at 0.76 m3 day−1 for 15 days (acclimatation stage) and then increased at 1.20 m3 day−1 for a resultant hydraulic time residence (HTR) of 1.6 days. The system was operational from early May until the end of June 2022.
Fig. 1.
Real image and scheme of the bioreactor. Dark blue: input effluent; red: output effluent; green: benthic compartment; Light blue: output/planktonic compartment; yellow: recirculating compartment; orange: water pumps.
2.2. Sampling performance and DNA extraction
Samples from input and output (100 mL) were collected considering the HRT of the system (to calculate the removal efficiency of the BPR) at days 19–21, 33–35, and 47–49 during the operational phase (T0, T14, and T28, respectively) and assembled by filtration through a 0.20 μm hydrophilic polycarbonate membrane filters (Merck; Darmstadt, Germany). Biofilm samples were scrapped directly from glass tiles (9 cm2) and centrifugated at 800 g for 2 min to remove the supernatant. Triplicates were collected and stored at −20 °C until DNA extraction in both cases. Firstly, 900 μL of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) buffer and 40 μL of lysozyme (40 mg mL−1) were added to the filters (cut in small pieces), and biofilm samples briefly agitated and incubated for 1 h at 37 °C and 300 rpm in a heat incubator. We added 40 μL of proteinase K (20 mg mL−1) and 100 μL of SDS 10% for incubation at 300 rpm and 56 °C for 1.5 h. Afterwards, 3 μL of RNase A was added to digest RNA and incubated briefly at 56 °C for 5 min. We used Maxtract High Density (MHD) tubes (Qiagen, Hilden, Germany), previously centrifugated at 800 g for 2 min at room temperature, into which the lysate was transferred, and DNA was extracted by adding 1 mL of phenol-chloroform-isoamyl alcohol (25:24:1) (Sigma Aldrich, St.Louis, MO, USA). The MHD tubes were centrifugated at 200 g for 5 min at room temperature, followed by three cycles of mixing and centrifugation. Phases were separated by decantation, followed by adding 10% volume of sodium acetate (3 M, pH 5.3) and two volumes of ethanol 100% stored at −20 °C overnight. The next day, we centrifugated the extracts at 15,000 g at 4 °C for 30 min, and we removed the supernatant to add 1 mL of ethanol 70%. The previous centrifugation was also set for 10 min with removing the supernatant. Finally, the extracts were dried using a speed vacuum concentrator at 30 °C for 15 min to further mix the pellet into 60 μL of TE buffer and stored at −20 °C for sequencing analysis. DNA concentration of the extracts was quantified using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA), and their quality was examined using agarose gel electrophoresis.
2.3. Metagenomics sequencing
Metagenomic analysis was conducted using the TruSeq Nano DNA High Throughput Library Prep Kit (Illumina, San Diego, CA, USA) at Macrogen, South Korea. The sequencing library was prepared by randomly fragmenting the DNA sample, followed by 5′ and 3′ adapter ligation. Specifically, 100 ng of genomic DNA was sheared using adaptive focused acoustic technology (Covaris, Woburn, MA, USA), and the fragmented DNA was end-repaired to create 5′-phosphorylated, blunt-ended dsDNA molecules. Following end-repair, DNA was size-selected using the bead-based method. These DNA fragments go through adding a single ‘A’ base and ligating the TruSeq DNA UD Indexing adapters. The products were then purified and enriched with PCR to create the final DNA library. The library was quantified using qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and qualified using the Agilent Technologies 4200 TapeStation D1000 screentape (Agilent Technologies, Santa Clara, CA, USA). Afterwards, the library was loaded onto a flow cell, where fragments were captured on a surface coated with oligonucleotides complementary to the library adapters. Each fragment was then amplified into distinct clonal clusters through bridge amplification. After cluster generation was completed, the templates were prepared for sequencing. Then, the paired-end (2 × 150 bp) sequencing was performed by NovaSeq 6000 (Illumina, San Diego, CA, USA) synthesis technology using a proprietary reversible terminator-based method. This method detected single bases as they were incorporated into DNA template strands. As all 4-reversible, terminator-bound dNTPs were presented during each sequencing cycle, natural competition minimized incorporation bias and greatly reduced raw error rates compared to other technologies. Accurate base-by-base sequencing was obtained, eliminating sequence-context-specific errors, even within repetitive sequence regions and homopolymers. Raw data was generated using sequencing control software for system control and base calling, facilitated by integrated primary Real Time Analysis software. The BCL/cBCL (base call) binary files were converted into FASTQ files using bcl2fastq (Illumina-provided package). Total read sequences obtained for different samples ranged between 13,658,860 and 54,003,130 reads. Specifically, there were 26,692.013 ± 13,638,656 reads in the input samples compared to 33,275,531 ± 18,148,755 in the output samples and 23,466,640 ± 15,056,764 in the benthic compartment.
2.4. Bioinformatic analyses
After sequencing libraries on a NovaSeq 6000 system (Illumina, San Diego, CA, USA) using paired-end 2 × 150 bp reads, low-quality reads were filtered out using the FASTX-Toolkit [40], applying a quality cut-off value of 20 for reads covering more than 90% of their length. In each metagenome, the total number of 16S rRNA gene reads was identified using METAXA2 [41], which was used for taxonomic classification and data normalization. Identification of ARGs and MGEs in each compartment (biofilm, plankton, and input) was conducted using the BLASTX tool [42] implemented in DIAMOND [43], aligning high-quality reads against the ARGminer database [44] and an in-house database [45], respectively. The ARGminer database contains sequences from CARD [46], DeepARG-DB [47], ARDB [48], MEGARes [49], UniProt [50], the National Database of Antibiotic Resistant Organisms (NDARO), SARG [51], ResFinder [52], and ARG-ANNOT [53], whereas our in-house database contains non-redundant sequences from INTEGRALL [54], InterPro [55], Isfinder [56], MobileElementFinder [57], MOBscan [58], and the Transposon Registry [59]. In both cases, a read was annotated as a resistance gene or mobile genetic element if the best BLAST hit (BLASTX) demonstrated a minimum identity of 90% amino acid over 90% of the length of the query sequence (see pipeline details in Supplementary material B). This conservative threshold was selected to minimize false positives [45]. Detected ARGs were also assessed against the list of “critically important antimicrobials” [60] and classified based on their respective gene family, resistance antibiotic class, and membership in the same protein complex, using DIAMOND as described above. The abundance of reads annotated as ARGs or MGEs was normalised to the total number of reads annotated as 16S rRNA genes from each metagenome, using METAXA2 as previously described. The normalised abundance of ARGs and MGEs was compared between different compartments and sampling uptake times to determine removal efficiency rates. The removal efficiencies of ARGs and MGEs were then assessed between input and output (equation (1)) [61].
| (1) |
2.5. Accession number
Sequence data obtained in this study were submitted to the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) under the Bioproject accession number PRJNA1049147.
2.6. Statistical analyses
Previously, selection and reorder of the most abundant ARGs and MGEs were set at 0.1% and 0.01% of relative abundances normalised to the total 16S rRNA genes of different compartments (input, benthic, and output) and sampling uptake times. ARGs and MGEs below this limit were classified as minority groups.
To determine differences between compartments of the bioreactor and sampling uptake times in ARGs and MGEs, permutational multivariate tests and analysis of similarity by Bray-Curtis dissimilarity distance were performed by vegan [62] and PERMANOVA [63] R packages. Additionally, ARGs and MGEs removal efficiency in the bioreactor and the accumulation in the benthic compartment were determined to interpret the efficiency of the bioreactor. We also performed a non-metric multidimensional scaling (NMDS) with the vegan [62] R package to visualise the differences between treatments.
Each most representative family in antibiotic families, ARGs and MGEs groups were tested by non-parametric Kruskal-Wallis by rstatix [64] R package with pairwise Dunn post-hoc tests by dunn.test [65] R package, to evaluate differences between different compartments and time due to the lack of normality and homoscedasticity of data, the signification level was set at 0.05. The alpha diversity of ARGs and MGEs was determined using vegan [62] R packages. This parameter was also analysed following the same statistical approach.
A heatmap was achieved to interpret the relative abundance of ARGs and MGEs and to facilitate the interpretation of the differences between compartments and sampling uptake times using the pheatmap [66] R package.
Similarly, the most abundant bacterial families were set at 5% of relative abundances normalised to the total 16S rRNA genes of the input and output samples and sampling uptake times. This classification was essential for the interpretation graph and the NMDS analysis. Spearman correlations were calculated between the most relevant bacterial families and ARGs and MGEs using the Hmisc package [67] in R. All statistical analyses were performed using Rstudio in an R version 2022.12.0 + 353.
3. Results
3.1. BPR removal efficiency: resistome
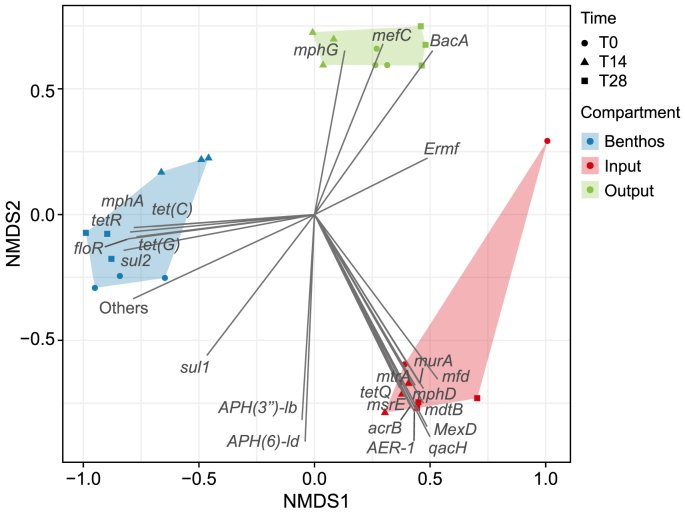
The resistome, containing the most abundant and frequently observed ARGs in the metagenomes analysed in the BPR, showed clear differences among the different compartments considered (Input: secondary effluent from UWWTP; benthos; and Output: effluent treated from BPR), as observed in the permutational multivariate analysis of variance and similarity distance results (Fig. 2; Table 1). A total of 419 ARGs were detected in this study. The number of ARGs detected in each compartment provides initial evidence of the high efficiency of the BPR in achieving an overall reduction of AMR in urban treated wastewater. Specifically, 400 ARGs were detected in the input, whereas only 119 were observed in the output, indicating a 70% reduction. The benthic compartment (biofilms) hosted only 56 ARGs, corresponding to 13% of the overall resistome (419 ARGs).
Fig. 2.
NMDS of the most abundant and frequently occurring ARGs reads normalised by 16S rRNA reads in different compartments and sampling times.
Table 1.
PERMANOVA and ANOSIM multivariant similarity analyses of ARGs and MGEs reads normalised to the 16S rRNA reads between compartments and sampling times. Df Num: numerator degree of freedom; df Denom: denominator degree of freedom; R2: determination coefficient. n.s: not significant.
| Variables | Factors | PERMANOVA |
ANOSIM |
|||||
|---|---|---|---|---|---|---|---|---|
| Explained | Residual | df Num | df Denom | F (p-value) | R2 (%) | Significance | ||
| ARGs | Compartment:Time | 516.95 | 133.05 | 8 | 18 | 8.74 (0.0001) | 84.13 | 0.0001 |
| Compartment | 327.88 | 322.12 | 2 | 24 | 12.21 (0.0001) | 87.46 | 0.0001 | |
| Time | 52.72 | 597.28 | 2 | 24 | 1.06 (n.s) | −0.03 | 0.6119 | |
| MGEs | Compartment:Time | 499.14 | 46.86 | 8 | 18 | 23.97 (0.0001) | 95.75 | 0.0001 |
| Compartment | 372.64 | 173.36 | 2 | 24 | 25.79 (0.0001) | 99.82 | 0.0001 | |
| Time | 43.75 | 502.25 | 2 | 24 | 1.05 (n.s) | 0.87 | 0.3569 | |
Our results also revealed a significant decrease (X2 = 16.53, p-value = 0.0003) in the relative abundance of ARGs (normalised to the total 16S rRNA) in the output (4.8 ± 1.8% of total 16S rRNA) compared to the input (16.9 ± 3.9% of total 16S rRNA) (Dunn tests p-value < 0.05). Furthermore, the benthic compartment exhibited a significantly higher relative abundance of ARGs (13.4 ± 6.6% of total 16S rRNA) compared to the output and contrastingly to a slight reduction in the input (Dunn tests p-value < 0.05), indicating an accumulation of ARGs within the system. Additionally, the α-diversity of ARGs was significantly higher in the input (65.5 ± 2.7) compared to the output (12.0 ± 2.2) and the benthic compartment (4.8 ± 1.5) (X2 = 23.14, p-value < 0.0001; Dunn tests p-value < 0.05; Supplementary Material Tables S4 and S5), indicating a reduction in the diversity of ARGs after the treatment.
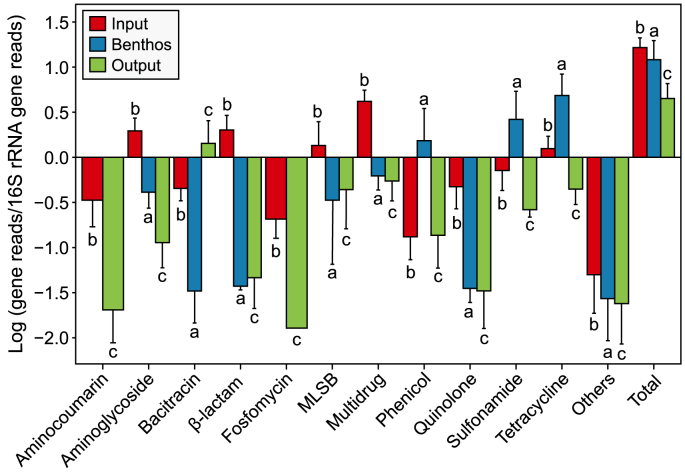
The most relevant ARGs detected in the BPR (n = 24), selected for relative abundances higher than 0.1%, encompassed a diverse array of resistances to different antibiotic families, including aminocoumarin, aminoglycoside, bacitracin, β-lactam, fosfomycin, macrolides, lincosamides, and streptogramines B (MLSB), multidrug, phenicol, quinolone, sulfonamide, and tetracycline (Fig. 3; Supplementary Material Table S1). Specifically, aminocoumarin, beta-lactam, fosfomycin, and quinolone ARG families were predominantly detected in the input, significantly lower in the output (Dunn tests p-value < 0.05; Fig. 3). In contrast, the bacitracin ARG family showed a significant increase in its relative abundance in the output compared to its levels in the input (Dunn tests p-value < 0.05; Fig. 3). The benthic compartment served as a reservoir for specific ARG families, such as sulfonamide and tetracycline resistances, whose relative abundances were significantly higher than the other compartments analysed (Dunn tests p-value < 0.05; Fig. 3). The most relevant ARG relative abundances revealed clear differences among the analysed compartments (Fig. 2). The input resistome was characterised by multidrug (acrB, MexD, and mtrA), quinolone (mfd and qacH), MLSB (mphD and msrE), fosfomycin (murA), aminocoumarin (mdtB), β-lactam (AER-1), and tetracycline (tetQ), whereas the output was dominated by MLSB (mphG and mefC) and bacitracin (BacA) ARGs (Fig. 2). The mentioned ARGs were detected in significantly higher abundance in the input than the output (Fig. 4; Dunn tests p-value < 0.05, Supplementary Material Table S2) and represented nearly all detected antibiotic families within the BPR system.
Fig. 3.
The most abundant and frequently occurring logarithmic mean relative abundances of antibiotic families with minority groups and total relative abundances between different compartments. Letters represented the significance between compartments in each group.
Fig. 4.
Pheatmap of the most abundant and frequently occurring antibiotic resistance genes (ARGs) from logarithmic relative abundance ARG reads normalised to the 16S rRNA gene reads at different compartments (input, benthos and output) and sampling times (T0, T14, and T28). Antibiotic families are represented with different colours on the right axis of the pheatmap.
Removal rates were calculated to better describe the system's efficiency in reducing ARG levels. The results confirmed that most of these ARGs were drastically reduced by the BPR (Fig. 5), averaging LRV = 2.3 ± 1.2 (93.8 ± 8.6%; Supplementary Material Table S6). However, specific ARGs exhibited lower removal rates, notably observed in aminoglycoside [APH(3″)-Ib and APH(6)-Id], sulfonamide (sul1), and tetracycline [tet(C)] ARGs, averaging LRV = 0.9 ± 1.2 (78.3 ± 21.1; Supplementary Material Table S6). In contrast, the output displayed an increase of MLSB (mphG and mefC) and bacitracin (BacA) ARGs compared to the input (Fig. 4), reflecting substantial negative LRVs (Fig. 5; Supplementary Material Table S6).
Fig. 5.
The logarithmic removal rate of the most abundant and frequently occurring ARG reads normalised to the 16S rRNA reads in the bioreactor.
Nevertheless, certain ARGs presented high variability, showing a reduced removal, specifically for MLSB (ErmF, mphA, and mphG), phenicol (floR), sulfonamide (sul2), and tetracycline [tet(G), tet(C), and tetR] ARGs (Fig. 5; Supplementary Material Table S6). Furthermore, these genes exhibited higher relative abundances within the benthic compartment (Fig. 2), indicating a clear biofilm accumulation (Fig. 4). Notably, MLSB (mphA, mefC, and ErmF), sulfonamide (sul2) and tetracycline [tet(C)] were not differentiated between the input and output (Dunn tests p > 0.05; Supplementary Material Table S2). Additionally, aminoglycoside [APH(3″)-Ib and APH(6)-Id], MLSB (mefC), sulfonamide (sul1), and other ARGs (n = 395) were also notably accumulated within the benthic compartment (Fig. 4; Dunn tests p-value <0.05, Supplementary Material Table S2). However, the ARGs representing bacitracin (BacA) and MLSB (ErmF) demonstrated a tendency towards either removal in the BPR or accumulation in the benthic compartment, consequently increasing their relative abundances in the output (Fig. 4). Specifically, BacA presented a notable increment in relative abundances in the output (Dunn tests p-value <0.05, Supplementary Material Table S2) concerning the input.
The BPR system performance did not show significant changes across different sampling times (0, 14, and 28 days) (Supplementary Material Tables S1 and S2). However, specific ARGs displayed an accumulation trend within the benthos compartment between day 0 and day 14, notably tetracycline [tet(C)] and MLSB (mefC) (Dunn tests p-value <0.05). Furthermore, MLSB (mphG), sulfonamide (sul1) and tetracycline [tet(C)] ARGs demonstrated a slight increase in the benthic compartment on the final day (day 28) of the BPR operation (Dunn tests p-value < 0.05).
3.2. BPR removal efficiency: mobilome
The mobilome, encompassing the most abundant and frequently occurring MGEs, exhibited clear differences across different compartments, as evidenced by permutational multivariate analysis of variance and similarity distance results (Fig. 6; Table 1) in contrast to the sampling times. The metagenomic analysis revealed the occurrence of 43 distinct MGEs. Most of these MGEs were detected in the input (n = 41, 94% of total MGEs), with a noticeable decrease observed in the output (n = 31, −25%). In the benthic compartment (biofilm), 61% of the MGEs identified were detected (n = 27).
Fig. 6.
NMDS of the most abundant and frequently occurring MGEs read normalised to the 16S rRNA reads in different compartments and sampling times.
Our results also revealed a significant decrease (X2 = 21.80, p-value <0.0001; Dunn tests p-value <0.05) of the relative MGEs abundances (normalised to the total 16S rRNA) in the output (4.2 ± 0.8% of the total 16S rRNA) compared with the input of the BPR in which MGEs represented the 71.2 ± 20.4% of the total 16S rRNA. Conversely, the benthos compartment exhibited a significantly lower relative abundance of MGEs (32.8 ± 16.0% of the total 16S rRNA) compared to BPR the input and output (Dunn tests p-value <0.05).
Furthermore, the α-diversity of MGEs was slightly, but significantly, higher in the input (5.28 ± 0.62) concerning the output (4.26 ± 0.88) (X2 = 19.70, p-value <0.0001; Dunn tests p-value >0.05; Supplementary Material Tables S4 and S5). Additionally, the α-diversities of MGEs in the input and output effluents were significantly higher compared to the benthic compartment (1.83 ± 0.54) (Dunn tests p-value <0.05; Supplementary Material Tables S4 and S5), indicating a relevant decrease in the diversity of MGEs within the biofilms.
The most prevalent MGEs (relative abundances higher than 0.01%) in the BPR (n = 20) were similarly represented by the total number of MGEs detected in the system (n = 43). These selected MGEs corresponded to various groups related to integron integrases, insertion sequences (IS), insertion sequence common region (ISCR), mobilization (MOB) relaxases, and transposons (Tn).
The relative abundance of these MGEs exhibited significant differences among the analysed compartments within the BPR (Fig. 6). The input mobilome primarily featured a multitude of MGEs: integron integrases (intl1 and intl3), IS and Tn transposases (IS3, IS4, IS5, IS6, IS30, Tn3, and Tn7), ISCR elements (ISCR8, ISCR9, ISCR15, ISCR21, and ISCR22), MOB relaxases (MOBP and MOBF), and others. In contrast, the output of the BPR was dominated by the MOB relaxase MOBQ and, to a lesser extent, the IS transposase IS256 (Fig. 6). These MGEs were notably more abundant in the BPR input than the output (Fig. 7, Dunn tests p-value <0.05, Supplementary Material Table S3), representing most detected MGE groups in the BPR.
Fig. 7.
Pheatmap of the most abundant and frequently occurring mobile genetic elements (MGEs) from logarithmic relative abundance MGEs reads normalised to the 16S rRNA gene reads at different compartments (input, benthos and output) and sampling times (T0, T14, and T28). MGE groups are represented with different colours on the right axis of the pheatmap.
The BPR showed a very good efficiency, achieving a substantial reduction of these MGEs (Fig. 8), resulting in an average LRV of about 2.3 ± 0.9, which corresponded to the 96.4 ± 4.9% removal of the total MGEs load detected in the BPR input (Supplementary Material Table S7). Distinct MGE groups were reduced over 90%, in particular different transposase families [Tn (96.0 ± 1.6%) and IS (94.8 ± 9.2%)], MOB relaxases (98.4 ± 3.1%), ISCR elements (97.1 ± 3.4%), and integron integrases (97.4 ± 3.1%) (Fig. 8). In contrast, some MGEs exhibited lower removal rates as observed for the IS transposases (IS1380 and IS256), the ISCR element ISCR2, the MOB relaxase MOBQ, and other MGEs averaging LRV = 0.7 ± 0.4 (67.0 ± 17.7%; Table S7). However, all MGEs analysed exhibited positive LRV (Fig. 8; Supplementary Material Tables S3 and S7, Dunn tests p-value <0.05), confirming the very good efficiency of the BPR in reducing the mobile load in treated urban wastewater.
Fig. 8.
The logarithmic removal rate of the most abundant and frequently occurring MGEs reads normalised to the 16S rRNA reads in the bioreactor.
A noticeable accumulation in biofilms was evident for the transposase IS1380, ISCR2, and MOBQ (Fig. 7), corresponding to a lower reduction of their abundance between the BPR input and output (Fig. 8; Supplementary Material Tables S3 and S7, Dunn tests p-value <0.05). Specifically, IS1380 and ISCR2 showed a reduced removal achieved by the BPR, resulting on average LRV = 0.5 ± 0.2 (64.5 ± 12.5%), in comparison with MOBQ which reduction was substantial, resulting in a LRV = 1.4 ± 0.9 corresponding to 90.8 ± 7.6% reduction.
The BPR performance did not show any significant variation across different sampling times (0, 14, and 28 days) concerning the MGEs removal efficiency (Supplementary Material Table S3) nor showed accumulation into the benthic compartment over time (Dunn tests p-value >0.05, Supplementary Material Table S3).
3.3. The interplay of microbial communities with ARGs and MGEs
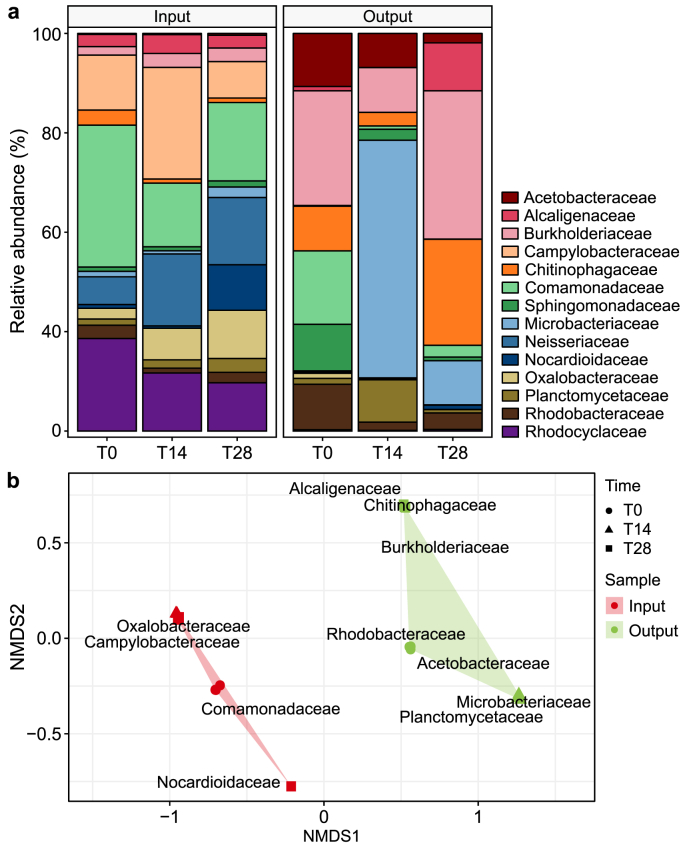
The bacterial communities exhibited clear differences between the input and output samples (Fig. 9b). The most abundant and frequently occurring bacterial families in the input samples were Comamonadaceae (6.8 ± 2.3%), Campylobacteraceae (5.2 ± 3.3%), Rhodocyclaceae (4.8 ± 1.6), and Neisseriaceae (4.1 ± 2.5%) in the Proteobacteria phylum, as well as Nocardioidaceae (1.2 ± 1.9%) in the Actinobacteria phylum (Fig. 9a). Conversely, Burkholderiaceae (13.6 ± 5.4%), Acetobacteraceae (4.5 ± 2.8%), and Rhodobacteraceae (3.2 ± 2.5%) in the Proteobacteria phylum, Microbacteriaceae (13.5 ± 16.2) in the Actinobacteria phylum, and Chitinophagaceae (7.1 ± 5.0%) in the Bacteroidetes phylum, were predominantly detected in the output samples respect to the input samples (Fig. 9a).
Fig. 9.
Relative abundance of the most abundant and frequently occurring bacterial families normalised to the 16S rRNA gene reads in the input and output samples and sampling times (T0, T14, and T28) (a) and non-metric multi-dimensional scaling (NMDS) (b).
The most abundant and frequently occurring bacterial families in the input samples revealed robust positive correlations with various ARG families (r > 0.7, p-value <0.001; Supplementary Material Fig. S1). Notably, Campylobacteraceae (Proteobacteria) demonstrated strong positive correlations with several ARGs, including aminocoumarin mdtB, aminoglycoside APH(6)-Id, MLSB mphD and msrE, multidrug MexD and acrB, quinolone qacH and mfd, and tetracycline tetQ (Supplementary Material Fig. S1). Similarly, other families from the Proteobacteria phylum, such as Comamonadaceae, Neisseriaceae, Oxalobacteraceae, and Rhodocyclaceae exhibited similar positive correlations across these ARGs (Supplementary Material Fig. S1). The most abundant and frequently occurring bacterial families in the output samples correlated differently with ARGs. Rhodobacteraceae displayed negative correlations with these ARG families (Supplementary Material Fig. S1). Conversely, this family showed strong positive correlations with sulfonamide and tetracycline ARGs, except for tetQ (Supplementary Material Fig. S1). Burkholderiaceae and Chitinophagaceae were strongly associated with the bacitracin BacA and the MLSB mfD. In contrast, Acetobacteraceae exhibited negative correlations with a broad range of ARGs, including aminocoumarin mdtB, β-lactam AER-1, MLSB mphD and msrE, multidrugs MexD and acrB, quinolone qacH and mfd, and tetracycline tetQ (Supplementary Material Fig. S1).
Similarly, significant positive correlations were observed between the most relevant bacterial families and MGEs detected. Specifically, Campylobacteraceae, Comamonadaceae, Neisseriaceae, Oxalobacteraceae, and Rhodocyclaceae exhibited strong positive correlations with most MGEs (Supplementary Material Fig. S1). In contrast, Acetobacteraceae, Planctomycetaceae, and Rhodobacteraceae exhibited negative correlations with these MGEs (Supplementary Material Fig. S1). Additionally, Alcaligenaceae and Microbacteriaceae showed weaker correlations with the ARGs and MGEs (Supplementary Material Fig. S1).
4. Discussion
4.1. BPR potentiality for future urban wastewater treatment plants
The growing global impact of ARB and ARGs on human and animal health demands immediate attention from policymakers. In particular, there is an urgent need to maximise the removal of antibiotic resistance determinants from UWWTP effluents, recognized as one of the main dissemination sources of AMR to aquatic environments worldwide [68,69]. Consequently, there is an increasing interest in applied research focusing on potential cost-effective solutions, as the NBS tested in this study. These solutions aim to develop and demonstrate new, advanced, and more sustainable systems utilising natural processes occurring in simulated ecosystems to efficiently reduce ARGs (i.e., constructed wetlands (CWs) [70,71]).
In our study of the sequential microbial BPR, notable removal efficiencies were obtained for most of the relevant ARGs and MGEs families identified. These findings highlight the BPR's potential as a nature-based tertiary treatment for UWWTPs, especially beneficial for facilities managing wastewater from smaller agglomerations (<10,000 inhabitants).
Research on streambed biofilms’ effectiveness in reducing ARGs has gained traction in the last decade [72,73]. However, the collective exploration of benthic and planktonic communities operating as a consortium at the macrocosm scale remains unexplored. These microbial communities are considered potential reservoirs of ARGs in freshwater ecosystems [34,74,75]. Interactions within the biofilm and planktonic communities, composed of hundreds of bacterial and algal species, lead to complex co-aggregation, thereby enhancing the potential for HGT events and amplifying AMR spread [76]. Moreover, anthropogenic antimicrobial compounds accumulate in wastewater through faecal pollution, forming aggregates and assemblages with bacterial and algal consortia, creating new adhesion sites. This situation introduces a surveillance bias and may increase AMR spread from UWWTPs, also depending on the socioeconomic conditions of the country [69]. Despite achieving high removal efficiencies of ARB, especially after secondary and tertiary treatments [77], the high loads of antibiotic resistance determinants discharged into freshwater ecosystems from UWWTP effluents raise concerns about maximising the resistome and mobilome removal performance [78].
For instance, the prevalent UWWTP secondary treatment strategy uses Fe3Cl as a coagulant for nutrient removal, serving as a trap for ARGs [79]. Furthermore, reliance on chlorination alone (<4 mg L−1) has proven insufficient for reducing ARGs in UWWTPs [80,81]. However, high dosages of chlorine (>80 mg Cl2 min L−1) may conduce to better ARG removal efficiencies but can produce excessive chlorine residues, leading to corrosion and human toxicity [80]. Fortunately, this practice has not been implemented in sewage facilities. Some advanced treatments (i.e., UV and ozonation [82]) are directly proportional to the reduction of ARGs in treated wastewater. However, the release of free DNA into water could enhance other environmental risks, contributing to the persistence of ARGs [82]. Other advanced treatments (i.e., nanofiltration [83], reverse osmosis [84], and advanced oxidation processes [85]) are considered highly efficient for reducing ARGs and ARB in UWWTPs, despite their high economic and energetic demands [86], representing new challenges in smaller UWWTPs. Additionally, some researchers suggest that post-treatment, lower bacteria densities and diversities could increase the availability of ecological niches occupied by highly resistant ARB [87,88]. The BPR developed in this study offers a cost-effective NBS to reduce resistome and mobilome threats, specifically for these small communities.
4.2. Antimicrobial resistance reduction: resistome removal efficiency
The BPR achieved a broad spectrum of removal rates for ARGs conferring resistance to various antibiotic families. Removal performance was particularly evident for multidrug (acrB, MexD, and mtrA), MLSB (mphD and msrE), quinolone (mfd and qacH), β-lactam (AER-1), fosfomycin (murA), and tetracycline (tetQ) genes. Notably, the BPR achieved, on average, a 93.8% reduction for these ARGs, surpassing the overall efficiency of other advanced treatments such as A2/O + ozonation, which exhibited an average 95.0% reduction of the total ARGs abundance [89], considering MLSB (i.e., msrE, mphE), tetracyclines [i.e., tet(C)] and cephalosporin (i.e., acrB, AAC(6′)-Ie-APH(2″)-Ia) [89]. The latter data was determined by metagenomic analysis, highlighting the enhanced analytical depth of our study (n = 420) compared to quantitative PCR (qPCR) assays (n = 4 [[90], [91]]) and high-throughput qPCR (HT-qPCR) (n = 283 [92]) techniques used in previous studies.
However, while the majority of ARGs displayed substantial reduction, the efficacy of the BPR to reduce aminocoumarin (mdtB), quinolone (mfd), and MLSB (msrE) genes did not align with prior reviewed studies. Conversely, low removal efficiencies were observed for aminoglycosides [APH(3″)-Ib and APH(6)-Id], sulfonamide (sul1), and tetracycline [tet(C)] ARGs, in contrast to the high variability in the removal rates for MLSB (mphA, mphG, mefC, and ErmF), sulfonamide (sul2), and tetracyclines [tet(G) and tetR] ARGs with less reduction in the BPR. Moreover, ARGs conferring resistance to the aminoglycosides [APH(3″)-Ib and APH(6)-Id], MLSB (mphA and mefC), sulfonamide (sul2), and tetracyclines [tet(C), tet(G), and tetR] exhibited relevant accumulation within the benthic compartment. This accumulation was also described in activated sludge and conventional treatments, indicating an overall increase in tetracycline (tet genes) and sulfonamide (sul1 and sul2) [13] ARGs in wastewater treatment processes. Otherwise, other aminoglycosides (i.e., AacAad, StrA, and AadA) showed high resistance in UWWTPs [93], potentially explaining the accumulation in biological treatments also observed in our study. Furthermore, MLSB is one of the most abundant groups of ARGs in UWWTPs (i.e., Erm, mphE, and msrE), and the biofilm enrichment observed in our study for these ARGs can be explained by their generally high occurrence in wastewaters [89].
Interestingly, sulfonamide sul2 exhibited a relevant reduction in advanced treatments [e.g., UV disinfection and biological activated carbon (BAC) [13] compared to low removal and substantial accumulation in biofilms detected in our study. UV disinfection process achieves an intracellular photochemical degradation of DNA into ARB. It can eventually lead to the release of free DNA. In contrast, the BAC mode of action relies on the adsorption of the ARB containing ARGs (and other bacteria) as the main efficient removal method [13,82]. In our system, the UV effect of solar radiation has not been considered but may have played a role in removing ARGs, considering that the BPR has been operational during late spring-early summer. On the other hand, biofilms could have acted as active and/or passive traps for ARB (as demonstrated by the enrichment sul2 in biofilms), performing somehow similarly to BAC technology. This element suggests that biofilm could incorporate planktonic ARB released from WWTP effluents. It has been demonstrated that AR determinants may migrate from the water column into biofilms [94], and field studies confirmed that biofilms could act as a sink for ARGs [95]. Similarly, sulfonamides were not reduced from CWs and accumulated in the sediments [96]. Comparable observations were reported for sulfonamides (sul1 and sul2) in drinking water treatments [97], emphasising their persistence post-disinfection. These sulfonamide ARGs were extensively analysed in the literature and are considered one of the most relevant ARGs released from UWWTPs. Contrastingly, tetracycline tet(C) exhibited a lower reduction in our system compared to these advanced treatments.
Likewise, the minimal removal rates observed in the BPR for MLSB (ErmF, mefC, and mphG), bacitracin (BacA), phenicol (floR), and aminocoumarin (mdtB) align with findings from other studies. For instance, BacA [98] and ErmF [13] showed similar patterns in drinking water and conventional and advanced processes in wastewater treatments, respectively, in which a multiresistance combination with multidrug (mexT) and β-lactam (blaOXA-12) ARGs was observed. Moreover, phenicol floR exhibited enrichment in biofilms and lesser removal in the BPR compared to its removal in advanced ultrafiltration and UV radiation treatments [99].
The observed variations may be attributed to the BPR's enhanced performance compared to conventional treatments and its slightly lower efficiency than some advanced treatments. Additionally, differences in accumulation and removal patterns in biofilms could be related to unique system-specific traits as a trap for ARGs.
Moreover, a global study of freshwater biofilms detected a wide range of ARGs [73], reminiscent of the diversity observed in our investigation. In that research, the mdtC, kdpE, and emrB ARGs were used as indicators in freshwater biofilms to further evaluate the other co-occurring resistance genes. However, in our experiment, only mdtC and emrB were detected and classified as other minority ARG groups. Specifically, bacitracin (BacA) represented the most resistant antibiotic class in our study and on a cross-regional scale [73]. These discrepancies in the occurrence and resistance levels of specific ARGs among studies could be associated with (i) the sample collection (grab or composite sampling, filtration or centrifugation), (ii) the DNA extraction (low or high-molecular weight kit procedures or phenol-chloroform method), (iii) the DNA characterization and quantification (gel electrophoresis, and Nanodrop or QUBIT DNA quantification), (iv) metagenomics sequencing (library preparation and short- or long-read sequencing), (v) integrity of ARGs (different referenced databases, alignment software) and (vi) corresponding analysis [35,100,101] that demonstrate a lack of global standardisation on next-generation sequencing (NGS) approaches for AMR detection and research. Another reason could be related to the regional specificity of our study compared to the global research reviewed in the cited literature.
The identification of prevalent clinical ARGs in our system, predominantly beta-lactam (i.e., AER-1), quinolone (i.e., qacH), and sulfonamides (i.e., sul1 and sul2), diverges from the broader diversity observed globally [102]. This divergence could be linked to treated population density, antibiotic consumption [87], and socioeconomic characteristics specific to our study location [73].
4.3. Horizontal gene transfer reduction connected to the resistome
The BPR achieved high removal efficiencies for a wide spectrum of MGEs, especially the integron-associated integrase genes (i.e., intI1 and intI3), considered one of the most important MGEs with high capacity to spread AMR into the environment [13,103,104]. The prevalence of high removal rates for whole MGEs (i.e., transposases families, MOB relaxases, ISCR elements, and integron integrases) achieved by the BPR demonstrated its high capacity to reduce the risk of transferring ARGs into the wastewater treatment systems and, more importantly, to the environment. The MGEs may represent the HGT in ARB, which, in our study, was highly reduced compared to an analogous study on activated sludge [13]. Additionally, HRT has been associated with HGT [105], suggesting a potential relationship between low HRT and limited adaptation time for ARB communities, possibly reducing their proliferation in the BPR and leading to the observed results. However, a lower HRT might compromise the removal efficiency for certain ARGs [106], as observed in some specific groups in our system, highlighting that an increase in HRT could also indirectly reduce HGT by lowering the persistence of faecal bacteria associated with ARB and ARGs into the system. It is known that these enteric bacteria are not well adapted to aquatic life, and it has been demonstrated that 90% of culturable Escherichia coli would not survive more than three days in freshwater environments at temperatures lower than 25 °C [107].
The BPR achieved remarkable removal of various MGEs such as transposons (i.e., Tn3), IS (i.e., IS3, IS4, IS5, IS6, IS30, IS256, IS1380), ISCR (i.e., ISCR2, ISCR8, ISCR9, ISCR15, ISCR21, ISCR22), and MOB relaxases (i.e., MOBF, MOBP, MOBQ) from secondary effluent in the UWWTP. The notable removal of Tn3 might be linked to the reduced occurrence of β-lactam genes, suggesting a possible association between Tn3 and β-lactam genes (i.e., blaKPC) [108] as observed in other studies. Contrastingly, transposon (Tn3) and IS (IS3 and IS5) were considered dominant in UWWTPs and strictly associated with HGT of intI1 [37], emphasising that our system is highly efficient for the removal of these elements, achieving an overall reduction of the risk associated with the AMR spread from treated urban wastewaters. Other detected MGEs related to IS (IS4 and IS256) were associated with tetracycline resistance genes [38], along with IS6 and IS1380, which were also linked to MLSB and phenicol resistance genes, respectively [38]. Other MGEs related to IS30, ISCR8, and MOBP and MOBF were highly removed from our system and are described as highly prevalent in other UWWTPs [45,109]. These patterns underscore the variability in the removal efficiency of ARGs, facilitating their reduction in our system by curbing their HGT.
Furthermore, IS1380, ISCR2, and MOBQ were accumulated in the benthos compartment with lower removal rates than the other most occurring and detected MGEs in the BPR. Notably, MOBQ contributed to a high potential for conjugation [110]. Although intI1 exhibited high removal efficiency in the BPR, IS1380 was associated with intI1 and tet genes in aerobic biofilm reactors [111], suggesting a potential correlation between low HRT and reduced reduction of these ARGs in our system. Additionally, ISCR2 was associated with multiple types of ARGs, including MLSB resistance genes (i.e., ErmF) [112] that were also accumulated in our system and the benthic compartment.
Studies focused on removing MGEs from treated wastewater have primarily targeted intI1 [99]. Various advanced treatments, such as UV disinfection, ozonation, and different biological reactors (i.e., membrane bioreactors and constructed wetlands), have demonstrated similar removal efficiencies to those observed in the BPR. Furthermore, chlorination systems in UWWTPs have also exhibited efficient reductions in insertion intI1, IS (i.e., IS1216 and IS613), and different transposons [81].
4.4. The BPR influencing the bacterial communities, ARGs, and MGEs
The BPR induced significant changes in the bacterial communities between the input and output samples, indicating distinct microbial compositions associated with different types of effluent [113]. Notably, Comamonadaceae, Campylobacteraceae, Rhodocyclaceae, and Neisseriaceae (Proteobacteria), along with the Nocardioidaceae (Actinobacteria), were more abundant in the input samples compared to the output, a trend consistent with observations in wastewater [114] and streams impacted by UWWTP effluents [115]. These findings imply that these bacterial communities of wastewater origin are still dominant in the current wastewater effluent released to the environment (the input water to the BPR system). Conversely, in the output of the BPR, the dominance of Burkholderiaceae and Rhodobacteraceae (both belonging to the Proteobacteria phylum), along with Chitinophagaceae (Bacteroidetes), indicates their higher similarity with the microbiota detected in the streams [116]. Additionally, members of the Microbacteriaceae family (Actinobacteria) may be associated with planktonic microalgae communities [117]. This finding evidences that the BPR system also greatly reduces the bacterial families associated with wastewater, releasing an effluent with a microbiome much more similar to those described in freshwater ecosystems. This implies an overall reduction of the impacts of WWTP effluent release on the microbial biodiversity of the receiving aquatic ecosystems.
Moreover, the positive correlations observed between the most relevant bacterial families and several ARGs underscore the potential role of specific bacterial communities in harbouring and disseminating antibiotic resistance [92]. Nevertheless, the reduction in relative abundances of some of these families (Comamonadaceae, Campylobacteraceae, Rhodocyclaceae, and Neisseriaceae) in the output of the BPR suggests that the bacterial and resistome dynamics may be altered, potentially resulting in a decrease in potential pathogens [118,119]. Otherwise, the bacterial families negatively correlated with several ARGs, particularly Acetobacteraceae could be associated with reducing ARGs within the BPR. A similar pattern was observed with MGEs, in which negatively and non-correlated bacterial families with the most relevant MGEs detected in the output of the BPR were also associated with their removal from the system. These evidences demonstrate the reduced AMR potential spread from treated wastewater to the environment achieved by the BPR system. These results underscore the complexity of the interplay between microbial communities and antibiotic resistance elements, highlighting the need for further research to understand the mechanisms underlying antibiotic resistance dissemination within wastewater treatment systems and to the aquatic environment [120].
5. Conclusions
Given the limited studies utilising next-generation sequencing (NGS) approaches to investigate AMR spread reduction from UWWTPs, our comprehensive detection of a wide range of MGEs and ARGs, along with the overall resistome and mobilome reduction achieved by the BPR, signifies pioneering work with implications for future studies in biological wastewater treatments, particularly nature-based solutions (NBS) for AMR spread mitigation. The BPR system substantially reduced the diversity of ARGs and MGEs, highlighting its potential to mitigate AMR release from UWWTPs into freshwater ecosystems [88]. Particularly, this system holds promise for socio-economically challenged countries grappling with limited wastewater treatment capacities [121], where an increase in AMR would hinder the achievement of a One Health approach. For facilities treating wastewater from small village populations (<10,000 inhabitants) in the EU, we strongly advocate additional research and monitoring of the overall quality of treated wastewater effluents from conventional treatments, in which the new Directive proposal should establish the monitoring of antimicrobial resistance and consequent removal capacity. This study has demonstrated the high potential of the BPR as a viable secondary/tertiary treatment option with expected low energetic demand and economic costs for UWWTPs lacking biological and/or advanced treatments and exhibiting low and uncontrolled AMR removal efficiencies. Nonetheless, to validate this nature-based technology and boost its implementation on a wider scale, a techno-economic life cycle assessment would be recommended to confirm and quantify its overall sustainability.
CRediT authorship contribution statement
Lluís Bertrans-Tubau: Writing - Original Draft, Visualization, Methodology, Investigation, Formal Analysis, Data Curation, Conceptualization. Sergio Martínez-Campos: Writing - Review & Editing, Methodology, Data Curation, Conceptualization. Julio Lopez-Doval: Writing - Review & Editing, Methodology, Investigation, Conceptualization. Meritxell Abril: Writing - Review & Editing, Methodology, Investigation, Conceptualization. Sergio Ponsá: Supervision. Victoria Salvadó: Writing - Review & Editing, Investigation. Manuela Hidalgo: Writing - Review & Editing, Investigation. Anna Pico-Tomàs: Investigation. Jose Luis Balcazar: Writing - Review & Editing, Supervision, Methodology, Funding Acquisition, Data Curation, Conceptualization. Lorenzo Proia: Writing - Review & Editing, Supervision, Methodology, Funding Acquisition, Data Curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Firstly, L.B.T would like to express gratitude to the ICRA staff and colleagues (J.L.B, A.P and others) for their support and help in the collaboration work of this research article. To BETA colleagues (J.L.D, M.A, and L.P) for their support and contribution during the experiment in the pilot plant, and in the review of the manuscript. S.M.C to his help with metagenomic analysis and the review of the manuscript. UdG colleagues (A.M.R, A.D, V.A, M.H., A.C.S) and other BETA colleagues (M.R and B.P) for other chemical and biologic analyses, and other issues not documented in this research article but who participated during the operational period of the experiment. This research was funded by the Spanish Ministry of Science and Innovation (Project PID2021-127326OB-100) and by the Spanish Ministry of Science and Innovation, the State Investigation Agency and the Regional Development of European Funding (FRENAWASTE Project: PID2022-1385630A-100). L.P has received funding from the Spanish Ministry of Science and Innovation through a Ramón y Cajal contract ‘[RYC 2020-029829-I]’. L.B.T has received funding from the European Social Funding Plus (FSE+) and the Generalitat de Catalunya (grant number 2022_FI_B2 00 027). S.M.C has an Investigo Programme contract funded by the European Union, Next Generation EU.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ese.2024.100445.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Zagklis D.P., Bampos G. Tertiary wastewater treatment technologies: a review of technical, economic, and life cycle aspects. Processes. 2022;10(11):2304. doi: 10.3390/pr10112304. [DOI] [Google Scholar]
- 2.Directive Council . Vol. 34. 1991. (Urban Wastewater Treatment Directive 91/271/EEC of the European Parliament and of the Council Concerning Urban Waste-Water Treatment). [Google Scholar]
- 3.European Environment Agency . 2023. Waterbase - UWWTD: Urban Waste Water Treatment Directive – Reported Data.https://www.eea.europa.eu/en/datahub/datahubitem-view/6244937d-1c2c-47f5-bdf1-33ca01ff1715 [Google Scholar]
- 4.Ferreiro C., Villota N., de Luis A., Lombraña J.I., Etxebarria N., Lomas J.M. Water reuse study from urban wwtps via C-ultrafiltration and ozonation technologies: basis for resilient cities and agriculture. Agronomy. 2021;11(2) doi: 10.3390/agronomy11020322. [DOI] [Google Scholar]
- 5.European Commission . Vol. 345. 2022. (Proposal for a Directive of the European Parliament and of the Council Concerning Urban Wastewater Treatment; Brussels). [Google Scholar]
- 6.Preisner M., Neverova-Dziopak E., Kowalewski Z. Analysis of eutrophication potential of municipal wastewater. Water Sci. Technol. 2020;81(9):1994–2003. doi: 10.2166/wst.2020.254. [DOI] [PubMed] [Google Scholar]
- 7.van Puijenbroek P.J.T.M., Beusen A.H.W., Bouwman A.F. Global nitrogen and phosphorus in urban waste water based on the shared socio-economic pathways. J. Environ. Manag. 2019;231(October 2018):446–456. doi: 10.1016/j.jenvman.2018.10.048. [DOI] [PubMed] [Google Scholar]
- 8.Shortle J.S., Mihelcic J.R., Zhang Q., Arabi M. Nutrient control in water bodies: a systems approach. J. Environ. Qual. 2020;49(3):517–533. doi: 10.1002/jeq2.20022. [DOI] [PubMed] [Google Scholar]
- 9.Noguera-Oviedo K., Aga D.S. Lessons learned from more than two decades of research on emerging contaminants in the environment. J. Hazard Mater. 2016;316:242–251. doi: 10.1016/j.jhazmat.2016.04.058. [DOI] [PubMed] [Google Scholar]
- 10.European Parliament and the Council . Vol. 16. EEC, 84/491/EEC, 86/280/EEC and Amending Directive 2000/60/EC of the European Parl; 2008. p. 84/156. (DIRECTIVE 2008/105/EC on Environmental Quality Standards in the Field of Water Policy, Amending and Subsequently Repealing Council Directives 82/176/EEC, 83/513/EEC). [Google Scholar]
- 11.European Commission . Vol. 2015. 2015. (Commission Implementing Decision (EU) 2015/495 of 20 March 2015 Establishing a Watch List of Substances for Union-Wide Monitoring in the Field of Water Policy Pursuant to Directive 2008/105/EC of the European Parliament and of the Council). [Google Scholar]
- 12.Lekunberri I., Balcázar J.L., Borrego C.M. Metagenomic exploration reveals a marked change in the river resistome and mobilome after treated wastewater discharges. Environ. Pollut. 2018;234:538–542. doi: 10.1016/j.envpol.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Han Y., Li L., Liu J., Yan X. Distribution, sources, and potential risks of antibiotic resistance genes in wastewater treatment plant: a review. Environ. Pollut. 2022;310(July) doi: 10.1016/j.envpol.2022.119870. [DOI] [PubMed] [Google Scholar]
- 14.Gwenzi W., Musiyiwa K., Mangori L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: a hotspot reservoir. J. Environ. Chem. Eng. 2020;8(1) doi: 10.1016/j.jece.2018.02.028. [DOI] [Google Scholar]
- 15.Murray C.J., Ikuta K.S., Sharara F., Swetschinski L., Robles Aguilar G., Gray A., Han C., Bisignano C., Rao P., Wool E., Johnson S.C., Browne A.J., Chipeta M.G., Fell F., Hackett S., Haines-Woodhouse G., Kashef Hamadani B.H., Kumaran E.A.P., McManigal B., Agarwal R., Akech S., Albertson S., Amuasi J., Andrews J., Aravkin A., Ashley E., Bailey F., Baker S., Basnyat B., Bekker A., Bender R., Bethou A., Bielicki J., Boonkasidecha S., Bukosia J., Carvalheiro C., Castañeda-Orjuela C., Chansamouth V., Chaurasia S., Chiurchiù S., Chowdhury F., Cook A.J., Cooper B., Cressey T.R., Criollo-Mora E., Cunningham M., Darboe S., Day N.P.J., De Luca M., Dokova K., Dramowski A., Dunachie S.J., Eckmanns T., Eibach D., Emami A., Feasey N., Fisher-Pearson N., Forrest K., Garrett D., Gastmeier P., Giref A.Z., Greer R.C., Gupta V., Haller S., Haselbeck A., Hay S.I., Holm M., Hopkins S., Iregbu K.C., Jacobs J., Jarovsky D., Javanmardi F., Khorana M., Kissoon N., Kobeissi E., Kostyanev T., Krapp F., Krumkamp R., Kumar A., Kyu H.H., Lim C., Limmathurotsakul D., Loftus M.J., Lunn M., Ma J., Mturi N., Munera-Huertas T., Musicha P., Mussi-Pinhata M.M., Nakamura T., Nanavati R., Nangia S., Newton P., Ngoun C., Novotney A., Nwakanma D., Obiero C.W., Olivas-Martinez A., Olliaro P., Ooko E., Ortiz-Brizuela E., Peleg A.Y., Perrone C., Plakkal N., Ponce-de-Leon A., Raad M., Ramdin T., Riddell A., Roberts T., Robotham J.V., Roca A., Rudd K.E., Russell N., Schnall J., Scott J.A.G., Shivamallappa M., Sifuentes-Osornio J., Steenkeste N., Stewardson A.J., Stoeva T., Tasak N., Thaiprakong A., Thwaites G., Turner C., Turner P., van Doorn H.R., Velaphi S., Vongpradith A., Vu H., Walsh T., Waner S., Wangrangsimakul T., Wozniak T., Zheng P., Sartorius B., Lopez A.D., Stergachis A., Moore C., Dolecek C., Naghavi M. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization . World Health Organization; 2021. The Selection And Use Of Essential Medicines: Report Of the WHO Expert Committee On Selection And Use Of Essential Medicines, 2021 (Including The 22nd WHO Model List Of Essential Medicines And the 8th WHO Model List Of Essential Medicines For Children); WHO Technical Report Series;1035. [Google Scholar]
- 17.Felis E., Kalka J., Sochacki A., Kowalska K., Bajkacz S., Harnisz M., Korzeniewska E. Antimicrobial pharmaceuticals in the aquatic environment - occurrence and environmental implications. Eur. J. Pharmacol. 2020;866(November 2019) doi: 10.1016/j.ejphar.2019.172813. [DOI] [PubMed] [Google Scholar]
- 18.Pan D., Shao S., Zhong J., Wang M., Wu X. Performance and mechanism of simultaneous nitrification–denitrification and denitrifying phosphorus removal in long-term moving bed biofilm reactor (MBBR) Bioresour. Technol. 2022;348(January) doi: 10.1016/j.biortech.2022.126726. [DOI] [PubMed] [Google Scholar]
- 19.Li P., Li K., Xu P., Liu X., Pu Y. Treatment of wastewater with high carbon-to-nitrogen ratio using a waterfall aeration biofilm reactor combined with sequencing batch reactor: microbial community structure and metabolism analysis. Bioresour. Technol. 2021;337(June) doi: 10.1016/j.biortech.2021.125450. [DOI] [PubMed] [Google Scholar]
- 20.Ng C., Tan B., Jiang X.T., Gu X., Chen H., Schmitz B.W., Haller L., Charles F.R., Zhang T., Gin K. Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Front. Microbiol. 2019;10(FEB):1–13. doi: 10.3389/fmicb.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulas A., Belhadi D., Descamps A., Andremont A., Benoit P., Courtois S., Dagot C., Grall N., Makowski D., Nazaret S., Nélieu S., Patureau D., Petit F., Roose-Amsaleg C., Vittecoq M., Livoreil B., Laouénan C. How effective are strategies to control the dissemination of antibiotic resistance in the environment ? A systematic review. Environ. Evid. 2021:1–32. doi: 10.1186/s13750-020-0187-x. [DOI] [Google Scholar]
- 22.Pinheiro E., Vieira M.C., Martins V., Amorim C.C. Simultaneous removal of emerging contaminants and disinfection for municipal wastewater treatment plant effluent quality improvement : a systemic analysis of the literature. Environ. Sci. Pollut. Res. 2021:24092–24111. doi: 10.1007/s11356-021-12363-5. [DOI] [PubMed] [Google Scholar]
- 23.Rizzo L., Malato S., Antakyali D., Beretsou V.G., Đolić M.B., Gernjak W., Heath E., Ivancev-Tumbas I., Karaolia P., Lado Ribeiro A.R., Mascolo G., McArdell C.S., Schaar H., Silva A.M.T., Fatta-Kassinos D. Consolidated vs new advanced treatment methods for the removal of contaminants of emerging concern from urban wastewater. Sci. Total Environ. 2019;655(August 2018):986–1008. doi: 10.1016/j.scitotenv.2018.11.265. [DOI] [PubMed] [Google Scholar]
- 24.Dell'Osbel N., Colares G.S., Oliveira G.A., Rodrigues L.R., da Silva F.P., Rodriguez A.L., López D.A.R., Lutterbeck C.A., Silveira E.O., Kist L.T., Machado Ê.L. Hybrid constructed wetlands for the treatment of urban wastewaters: increased nutrient removal and landscape potential. Ecol. Eng. 2020;158(July) doi: 10.1016/j.ecoleng.2020.106072. [DOI] [Google Scholar]
- 25.Hassan I., Chowdhury S.R., Prihartato P.K., Razzak S.A. Wastewater treatment using constructed wetland: current trends and future potential. Processes. 2021;9(11):1–27. doi: 10.3390/pr9111917. [DOI] [Google Scholar]
- 26.Vymazal J. Removal of nutrients in constructed wetlands for wastewater treatment through plant harvesting – biomass and load matter the most. Ecol. Eng. 2020;155(April) doi: 10.1016/j.ecoleng.2020.105962. [DOI] [Google Scholar]
- 27.McPhearson T., Cook E.M., Berbés-Blázquez M., Cheng C., Grimm N.B., Andersson E., Barbosa O., Chandler D.G., Chang H., Chester M.V., Childers D.L., Elser S.R., Frantzeskaki N., Grabowski Z., Groffman P., Hale R.L., Iwaniec D.M., Kabisch N., Kennedy C., Markolf S.A., Matsler A.M., Mcphillips L.E., Miller T.R., Muñoz-Erickson T.A., Rosi E., Troxler T.G. A social-ecological-technological systems framework for urban ecosystem services. One Earth. 2022:505–518. doi: 10.1016/j.oneear.2022.04.007. [DOI] [Google Scholar]
- 28.Acuña V., Casellas M., Font C., Romero F., Sabater S. Nutrient attenuation dynamics in effluent dominated watercourses. Water Res. 2019;160:330–338. doi: 10.1016/j.watres.2019.05.093. [DOI] [PubMed] [Google Scholar]
- 29.Bergbusch N.T., Hayes N.M., Simpson G.L., Leavitt P.R. Unexpected shift from phytoplankton to periphyton in eutrophic streams due to wastewater influx. Limnol. Oceanogr. 2021;66(7):2745–2761. doi: 10.1002/lno.11786. [DOI] [Google Scholar]
- 30.Edwards S.J., Kjellerup B.V. Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl. Microbiol. Biotechnol. 2013;97(23):9909–9921. doi: 10.1007/s00253-013-5216-z. [DOI] [PubMed] [Google Scholar]
- 31.Sousa H., Sousa C.A., Simões L.C., Simões M. Microalgal-based removal of contaminants of emerging concern. J. Hazard Mater. 2022;423(September 2021) doi: 10.1016/j.jhazmat.2021.127153. 0–2. [DOI] [PubMed] [Google Scholar]
- 32.Flores-Vargas G., Bergsveinson J., Lawrence J.R., Korber D.R. Environmental biofilms as reservoirs for antimicrobial resistance. Front. Microbiol. 2021;12(December) doi: 10.3389/fmicb.2021.766242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matviichuk O., Mondamert L., Geffroy C., Gaschet M., Dagot C., Labanowski J. River biofilms microbiome and resistome responses to wastewater treatment plant effluents containing antibiotics. Front. Microbiol. 2022;13(February):1–14. doi: 10.3389/fmicb.2022.795206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balcázar J.L., Subirats J., Borrego C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015;6(OCT):1–9. doi: 10.3389/fmicb.2015.01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garner E., Davis B.C., Milligan E., Forrest M., Keenum I., Maile-Moskowitz A., Pan J., Gnegy M., Liguori K., Gupta S., Prussin A.J., Marr L.C., Heath L.S., Vikesland P.J., Zhang L., Pruden A. Next generation sequencing approaches to evaluate water and wastewater quality. Water Res. 2021;194 doi: 10.1016/j.watres.2021.116907. [DOI] [PubMed] [Google Scholar]
- 36.Abramova A., Berendonk T.U., Bengtsson-Palme J. A global baseline for QPCR-determined antimicrobial resistance gene prevalence across environments. Environ. Int. 2023;178(March) doi: 10.1016/j.envint.2023.108084. [DOI] [PubMed] [Google Scholar]
- 37.Ping Q., Zhang Z., Ma L., Yan T., Wang L., Li Y. The prevalence and removal of antibiotic resistance genes in full-scale wastewater treatment plants: bacterial host, influencing factors and correlation with nitrogen metabolic pathway. Sci. Total Environ. 2022;827 doi: 10.1016/j.scitotenv.2022.154154. [DOI] [PubMed] [Google Scholar]
- 38.Zhang R.M., Liao M.N., Wu J. en, Lu X. qing, Tan H. zhen, Sun J., Liao X.P., Liu Y.H. Metagenomic insights into the influence of mobile genetic elements on ARGs along typical wastewater treatment system on pig farms in China. Sci. Total Environ. 2022;839(May) doi: 10.1016/j.scitotenv.2022.156313. [DOI] [PubMed] [Google Scholar]
- 39.Liu L., Zhang Y., Chen H., Teng Y. Fate of resistome components and characteristics of microbial communities in constructed wetlands and their receiving river. Sci. Total Environ. 2022;844(May) doi: 10.1016/j.scitotenv.2022.157226. [DOI] [PubMed] [Google Scholar]
- 40.Gordon A., Hannon G. 2010. Fastx-Toolkit, FASTQ/A Short-Reads Preprocessing Tools.http://hannonlab.cshl.edu/fastx_toolkit [Google Scholar]
- 41.Bengtsson J., Eriksson K.M., Hartmann M., Wang Z., Shenoy B.D., Grelet G.A., Abarenkov K., Petri A., Alm Rosenblad M., Nilsson R.H. Metaxa: a software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2011;100(3):471–475. doi: 10.1007/s10482-011-9598-6. [DOI] [PubMed] [Google Scholar]
- 42.Ye J., McGinnis S., Madden T.L. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:6–9. doi: 10.1093/nar/gkl164. (WEB. SERV. ISS.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2014;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 44.Arango-Argoty G.A., Guron G.K.P., Guron G.K.P., Garner E., Riquelme M.V., Heath L.S., Pruden A., Vikesland P.J., Zhang L. ARGminer: a web platform for the crowdsourcing-based curation of antibiotic resistance genes. Bioinformatics. 2020;36(9):2966–2973. doi: 10.1093/bioinformatics/btaa095. [DOI] [PubMed] [Google Scholar]
- 45.Gionchetta G., Fillol M., López N., Kassotaki E., Sànchez-Melsió A., Gutiérrez C., Gutiérrez O., Luis Balcázar J., Borrego C.M. Impact of nitrate addition on the resistome and mobilome from a full-scale sewer. Chem. Eng. J. 2022;439(February) doi: 10.1016/j.cej.2022.135653. [DOI] [Google Scholar]
- 46.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., Pascale G. De, Ejim L., Kalan L., King A.M., Koteva K., Morar M., Mulvey M.R., O'Brien J.S., Pawlowski A.C., Piddock L.J.V., Spanogiannopoulos P., Sutherland A.D., Tang I., Taylor P.L., Thaker M., Wang W., Yan M., Yu T., Wright G.D. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57(7):3348–3357. doi: 10.1128/aac.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arango-Argoty G., Garner E., Pruden A., Heath L.S., Vikesland P., Zhang L. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 2018;6(1):1–15. doi: 10.1186/s40168-018-0401-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B., Pop M. ARDB--Antibiotic resistance genes database. Nucleic Acids Res. 2009;37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lakin S.M., Dean C., Noyes N.R., Dettenwanger A., Ross A.S., Doster E., Rovira P., Abdo Z., Jones K.L., Ruiz J., Belk K.E., Morley P.S., Boucher C. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017;45(D1):D574–D580. doi: 10.1093/nar/gkw1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bateman A., Martin M.J., O'Donovan C., Magrane M., Apweiler R., Alpi E., Antunes R., Arganiska J., Bely B., Bingley M., Bonilla C., Britto R., Bursteinas B., Chavali G., Cibrian-Uhalte E., Da Silva A., De Giorgi M., Dogan T., Fazzini F., Gane P., Castro L.G., Garmiri P., Hatton-Ellis E., Hieta R., Huntley R., Legge D., Liu W., Luo J., Macdougall A., Mutowo P., Nightingale A., Orchard S., Pichler K., Poggioli D., Pundir S., Pureza L., Qi G., Rosanoff S., Saidi R., Sawford T., Shypitsyna A., Turner E., Volynkin V., Wardell T., Watkins X., Zellner H., Cowley A., Figueira L., Li W., McWilliam H., Lopez R., Xenarios I., Bougueleret L., Bridge A., Poux S., Redaschi N., Aimo L., Argoud-Puy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.C., Boeckmann B., Bolleman J., Boutet E., Breuza L., Casal-Casas C., De Castro E., Coudert E., Cuche B., Doche M., Dornevil D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Jungo F., Keller G., Lara V., Lemercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T., Nouspikel N., Paesano S., Pedruzzi I., Pilbout S., Pozzato M., Pruess M., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A.L., Wu C.H., Arighi C.N., Arminski L., Chen C., Chen Y., Garavelli J.S., Huang H., Laiho K., McGarvey P., Natale D.A., Suzek B.E., Vinayaka C.R., Wang Q., Wang Y., Yeh L.S., Yerramalla M.S., Zhang J. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(D1):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y., Jiang X., Chai B., Ma L., Li B., Zhang A., Cole J.R., Tiedje J.M., Zhang T. ARGs-OAP: online analysis pipeline for antibiotic resistance genes detection from metagenomic data using an integrated structured ARG-database. Bioinformatics. 2016;32(15):2346–2351. doi: 10.1093/bioinformatics/btw136. [DOI] [PubMed] [Google Scholar]
- 52.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L., Rolain J.-M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014;58(1):212–220. doi: 10.1128/aac.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moura A., Soares M., Pereira C., Leitão N., Henriques I., Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25(8):1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.Y., El-Gebali S., Fraser M.I., Gough J., Haft D.R., Huang H., Letunic I., Lopez R., Luciani A., Madeira F., Marchler-Bauer A., Mi H., Natale D.A., Necci M., Nuka G., Orengo C., Pandurangan A.P., Paysan-Lafosse T., Pesseat S., Potter S.C., Qureshi M.A., Rawlings N.D., Redaschi N., Richardson L.J., Rivoire C., Salazar G.A., Sangrador-Vegas A., Sigrist C.J.A., Sillitoe I., Sutton G.G., Thanki N., Thomas P.D., Tosatto S.C.E., Yong S.Y., Finn R.D. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47(D1):D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siguier P., Varani A., Perochon J., Chandler M. Exploring bacterial insertion sequences with ISfinder: objectives, uses, and future developments. Methods Mol. Biol. 2012;859:91–103. doi: 10.1007/978-1-61779-603-6_5. [DOI] [PubMed] [Google Scholar]
- 57.Johansson M.H.K., Bortolaia V., Tansirichaiya S., Aarestrup F.M., Roberts A.P., Petersen T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021;76(1):101–109. doi: 10.1093/JAC/DKAA390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcillán-Barcia M.P., Redondo-Salvo S., Vielva L., de la Cruz F. MOBscan: automated annotation of MOB relaxases. Methods Mol. Biol. 2020;2075:295–308. doi: 10.1007/978-1-4939-9877-7_21. [DOI] [PubMed] [Google Scholar]
- 59.Tansirichaiya S., Rahman M.A., Roberts A.P. The transposon Registry. Mobile DNA. 2019;10(1):1–6. doi: 10.1186/s13100-019-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.World Health Organization . World Health Organization; 2018. Critically Important Antimicrobials For Human Medicine, 6th Revisi. [Google Scholar]
- 61.Thai-Hang L., Charmaine N., Ngoc Han T., Hongjie C., Karina Yew-Hoong G. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018 doi: 10.1016/j.watres.2018.08.060. [DOI] [PubMed] [Google Scholar]
- 62.Oksanen J., Blanchet G.F., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., Wagner H. Package “vegan.”. R Proj. Stat. Comput. 2019 [Google Scholar]
- 63.Vicente-Gonzalez L., Vicente-Villardon J.L. PERMANOVA: multivariate analysis of variance based on distances and permutations. 2021. https://cran.r-project.org/package=PERMANOVA
- 64.Kassambara A. Rstatix: pipe-friendly framework for basic statistical tests. 2022. https://cran.r-project.org/package=rstatix
- 65.Dinno A. Dunn. Test: dunn's test of multiple comparisons using rank sums. 2017. https://cran.r-project.org/package=dunn.test
- 66.Kolde R. R Package “Pheatmap.”. 2022 https://github.com/raivokolde/pheatmap [Google Scholar]
- 67.Harrell Jr, Hmisc F.E. 2023. Harrell Miscellaneous.https://cran.r-project.org/package=Hmisc [Google Scholar]
- 68.European Centre for Disease Prevention and Control and World Health Organisation . 2023. Antimicrobial Resistance Surveillance In Europe 2023: 2021 Data; Stockholm. [Google Scholar]
- 69.Collignon P., Beggs J.J., Walsh T.R., Gandra S., Laxminarayan R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet. Health. 2018;2(9):e398–e405. doi: 10.1016/S2542-5196(18)30186-4. [DOI] [PubMed] [Google Scholar]
- 70.Ma J., Cui Y., Li A., Zou X., Ma C., Chen Z. Antibiotics and antibiotic resistance genes from wastewater treated in constructed wetlands. Ecol. Eng. 2022;177(18) doi: 10.1016/j.ecoleng.2022.106548. [DOI] [Google Scholar]
- 71.Felis E., Sochacki A., Bajkacz S., Łuczkiewicz A., Jóźwiakowski K., García J., Vymazal J. Removal of selected sulfonamides and sulfonamide resistance genes from wastewater in full-scale constructed wetlands. Sci. Total Environ. 2024;912(October 2023):169–195. doi: 10.1016/j.scitotenv.2023.169195. [DOI] [PubMed] [Google Scholar]
- 72.Subirats J., Triadó-Margarit X., Mandaric L., Acuña V., Balcázar J.L., Sabater S., Borrego C.M. Wastewater pollution differently affects the antibiotic resistance gene pool and biofilm bacterial communities across streambed compartments. Mol. Ecol. 2017;26(20):5567–5581. doi: 10.1111/mec.14288. [DOI] [PubMed] [Google Scholar]
- 73.Yao Y., Liu Z., Yip K.K., Pu Y., Cheng W., Li M., Habimana O. Cross-regional scale pollution of freshwater biofilms unveiled by antibiotic resistance genes. Sci. Total Environ. 2022;818 doi: 10.1016/j.scitotenv.2021.151835. [DOI] [PubMed] [Google Scholar]
- 74.Kneis D., Berendonk T.U., Forslund S.K., Hess S. Antibiotic resistance genes in river biofilms: a metagenomic approach toward the identification of sources and candidate hosts. Environ. Sci. Technol. 2022;56(21):14913–14922. doi: 10.1021/acs.est.2c00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xue X., Wang L., Xing H., Zhao Y., Li X., Wang G., Wang Z. Characteristics of phytoplankton-zooplankton communities and the roles in the transmission of antibiotic resistance genes under the pressure of river contamination. Sci. Total Environ. 2021;780 doi: 10.1016/j.scitotenv.2021.146452. [DOI] [PubMed] [Google Scholar]
- 76.Baquero F., Coque T.M., Guerra-Pinto N., Galán J.C., Jiménez-Lalana D., Tamames J., Pedrós-Alió C. The influence of coalescent microbiotic particles from water and soil on the evolution and spread of antimicrobial resistance. Front. Environ. Sci. 2022;10(April):1–13. doi: 10.3389/fenvs.2022.824963. [DOI] [Google Scholar]
- 77.Wang J., Chu L., Wojnárovits L., Takács E. Occurrence and fate of antibiotics , antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant : an overview. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140997. [DOI] [PubMed] [Google Scholar]
- 78.Karkman A., Do T.T., Walsh F., Virta M.P.J. Antibiotic-resistance genes in waste water. Trends Microbiol. 2018;26(3):220–228. doi: 10.1016/j.tim.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 79.Li N., Sheng G.P., Lu Y.Z., Zeng R.J., Yu H.Q. Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res. 2017;111:204–212. doi: 10.1016/j.watres.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 80.Pei M., Zhang B., He Y., Su J., Gin K., Lev O., Shen G., Hu S. State of the art of tertiary treatment technologies for controlling antibiotic resistance in wastewater treatment plants. Environ. Int. 2019;131(July) doi: 10.1016/j.envint.2019.105026. [DOI] [PubMed] [Google Scholar]
- 81.Lin W., Zhang M., Zhang S., Yu X. Can chlorination Co-select antibiotic-resistance genes? Chemosphere. 2016;156:412–419. doi: 10.1016/j.chemosphere.2016.04.139. [DOI] [PubMed] [Google Scholar]
- 82.Zheng J., Su C., Zhou J., Xu L., Qian Y., Chen H. Effects and mechanisms of ultraviolet, chlorination, and ozone disinfection on antibiotic resistance genes in secondary effluents of municipal wastewater treatment plants. Chem. Eng. J. 2017;317:309–316. doi: 10.1016/j.cej.2017.02.076. [DOI] [Google Scholar]
- 83.Oliveira M., Leonardo I.C., Silva A.F., Crespo J.G., Nunes M., Crespo M.T.B. Nanofiltration as an efficient tertiary wastewater treatment: elimination of total bacteria and antibiotic resistance genes from the discharged effluent of a full-scale wastewater treatment plant. Antibiotics. 2022;11(5) doi: 10.3390/antibiotics11050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Slipko K., Reif D., Wögerbauer M., Hufnagl P., Krampe J., Kreuzinger N. Removal of extracellular free DNA and antibiotic resistance genes from water and wastewater by membranes ranging from microfiltration to reverse osmosis. Water Res. 2019;164 doi: 10.1016/j.watres.2019.114916. [DOI] [PubMed] [Google Scholar]
- 85.Wang J., Zhuan R., Chu L. The occurrence, distribution and degradation of antibiotics by ionizing radiation: an overview. Sci. Total Environ. 2019;646:1385–1397. doi: 10.1016/j.scitotenv.2018.07.415. [DOI] [PubMed] [Google Scholar]
- 86.Pesqueira J.F.J.R., Pereira M.F.R., Silva A.M.T. Environmental impact assessment of advanced urban wastewater treatment technologies for the removal of priority substances and contaminants of emerging concern : a review. J. Clean. Prod. 2020;261 doi: 10.1016/j.jclepro.2020.121078. [DOI] [Google Scholar]
- 87.Ding D., Wang B., Zhang X., Zhang J., Zhang H., Liu X., Gao Z., Yu Z. The spread of antibiotic resistance to humans and potential protection strategies. Ecotoxicol. Environ. Saf. 2023;254(March) doi: 10.1016/j.ecoenv.2023.114734. [DOI] [PubMed] [Google Scholar]
- 88.Klümper U., Gionchetta G., Catao E.C.P., Bellanger X., Dielacher I., Fang P., Galazka S., Goryluk-Salmonowicz A., Kneis D., Okoroafor U., Radu E., Szadziul M., Szekeres E., Teban-Man A., Coman C., Kreuzinger N., Popowska M., Vierheilig J., Walsh F., Woegerbauer M., Bürgmann H., Merlin C., Berendonk T.U. Microbiome diversity : a barrier to the environmental spread of antimicrobial resistance. bioRxiv. 2023;1–36 doi: 10.1101/2023.03.30.534382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen M., Hu X., Li M., Lyu C., Hu Y., Bu X., Chen T., Cai H., Li C., Liu J., Fan K. Distribution of antibiotic resistance genes and their association with microbes in wastewater treatment plants: a metagenomics analysis. Water. 2023;15(8) doi: 10.3390/w15081587. [DOI] [Google Scholar]
- 90.Martínez-Campos S., González-Pleiter M., Rico A., Schell T., Vighi M., Fernández-Piñas F., Rosal R., Leganés F. Time-course biofilm formation and presence of antibiotic resistance genes on everyday plastic items deployed in river waters. J. Hazard Mater. October 2022;2023:443. doi: 10.1016/j.jhazmat.2022.130271. [DOI] [PubMed] [Google Scholar]
- 91.Proia L., von Schiller D., Sànchez-Melsió A., Sabater S., Borrego C.M., Rodríguez-Mozaz S., Balcázar J.L. Occurrence and persistence of antibiotic resistance genes in river biofilms after wastewater inputs in small rivers. Environ. Pollut. 2016;210:121–128. doi: 10.1016/j.envpol.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 92.Yu K., Li P., Chen Y., Zhang B., Huang Y., Huang F.Y., He Y. Antibiotic resistome associated with microbial communities in an integrated wastewater reclamation system. Water Res. 2020;173 doi: 10.1016/j.watres.2020.115541. [DOI] [PubMed] [Google Scholar]
- 93.Conco T., Kumari S., Awolusi O.O., Allam M., Ismail A., Stenström T.A., Bux F. Profiling of emerging pathogens, antibiotic resistance genes and mobile genetic elements in different biological wastewater treatment plants. J. Environ. Chem. Eng. 2022;10(3) doi: 10.1016/j.jece.2022.107596. [DOI] [Google Scholar]
- 94.Engemann C.A., Keen P.L., Knapp C.W., Hall K.J., Graham D.W. Fate of tetracycline resistance genes in aquatic systems: migration from the water column to peripheral biofilms. Environ. Sci. Technol. 2008;42(14):5131–5136. doi: 10.1021/es800238e. [DOI] [PubMed] [Google Scholar]
- 95.Guo X. pan, Yang Y., Lu D. pei, Niu Z. shun, Feng J. nan, Chen Y. ru, Tou F. yun, Garner E., Xu J., Liu M., Hochella M.F. Biofilms as a sink for antibiotic resistance genes (ARGs) in the yangtze estuary. Water Res. 2018;129:277–286. doi: 10.1016/j.watres.2017.11.029. [DOI] [PubMed] [Google Scholar]
- 96.Sabri N.A., Schmitt H., van der Zaan B.M., Gerritsen H.W., Rijnaarts H.H.M., Langenhoff A.A.M. Performance of full scale constructed wetlands in removing antibiotics and antibiotic resistance genes. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147368. [DOI] [PubMed] [Google Scholar]
- 97.Su H.C., Liu Y.S., Pan C.G., Chen J., He L.Y., Ying G.G. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: from drinking water source to tap water. Sci. Total Environ. 2018;616–617:453–461. doi: 10.1016/j.scitotenv.2017.10.318. [DOI] [PubMed] [Google Scholar]
- 98.Jia S., Bian K., Shi P., Ye L., Liu C.H. Metagenomic profiling of antibiotic resistance genes and their associations with bacterial community during multiple disinfection regimes in a full-scale drinking water treatment plant. Water Res. 2020;176 doi: 10.1016/j.watres.2020.115721. [DOI] [PubMed] [Google Scholar]
- 99.Pei M., Zhang B., He Y., Su J., Gin K., Lev O., Shen G., Hu S. State of the art of tertiary treatment technologies for controlling antibiotic resistance in wastewater treatment plants. Environ. Int. 2019;131(March) doi: 10.1016/j.envint.2019.105026. [DOI] [PubMed] [Google Scholar]
- 100.Davis B.C., Brown C., Gupta S., Calarco J., Liguori K., Milligan E., Harwood V.J., Pruden A., Keenum I. Recommendations for the use of metagenomics for routine monitoring of antibiotic resistance in wastewater and impacted aquatic environments. Crit. Rev. Environ. Sci. Technol. 2023;53(19):1731–1756. doi: 10.1080/10643389.2023.2181620. [DOI] [Google Scholar]
- 101.Nguyen A.Q., Vu H.P., Nguyen L.N., Wang Q., Djordjevic S.P., Donner E., Yin H., Nghiem L.D. Monitoring antibiotic resistance genes in wastewater treatment: current strategies and future challenges. Sci. Total Environ. 2021;783 doi: 10.1016/j.scitotenv.2021.146964. [DOI] [PubMed] [Google Scholar]
- 102.Li B., Yang Y., Ma L., Ju F., Guo F., Tiedje J.M., Zhang T. Metagenomic and network analysis reveal wide distribution and Co-occurrence of environmental antibiotic resistance genes. ISME J. 2015;9(11):2490–2502. doi: 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao Y.X., Li X., Fan X.Y., Zhao J.R., Zhang Z.X. Wastewater treatment plants as reservoirs and sources for antibiotic resistance genes: a review on occurrence, transmission and removal. J. Water Process Eng. 2022;46(100) doi: 10.1016/j.jwpe.2021.102539. [DOI] [Google Scholar]
- 104.Uyaguari-Díaz M.I., Croxen M.A., Luo Z., Cronin K.I., Chan M., Baticados W.N., Nesbitt M.J., Li S., Miller K.M., Dooley D., Hsiao W., Isaac-Renton J.L., Tang P., Prystajecky N. Human activity determines the presence of integron-associated and antibiotic resistance genes in southwestern British columbia. Front. Microbiol. 2018;9(MAY):1–20. doi: 10.3389/fmicb.2018.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Korzeniewska E., Harnisz M. Relationship between modification of activated sludge wastewater treatment and changes in antibiotic resistance of bacteria. Sci. Total Environ. 2018;639:304–315. doi: 10.1016/j.scitotenv.2018.05.165. [DOI] [PubMed] [Google Scholar]
- 106.Pallares-Vega R., Blaak H., van der Plaats R., de Roda Husman A.M., Hernandez Leal L., van Loosdrecht M.C.M., Weissbrodt D.G., Schmitt H. Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: a cross-sectional study. Water Res. 2019;161:319–328. doi: 10.1016/j.watres.2019.05.100. [DOI] [PubMed] [Google Scholar]
- 107.Servais P., Garcia-Armisen T., George I., Billen G. Fecal bacteria in the rivers of the seine drainage network (France): sources, fate and modelling. Sci. Total Environ. 2007;375(1–3):152–167. doi: 10.1016/j.scitotenv.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 108.Huang J., Hu X., Zhao Y., Shi Y., Ding H., Wu R., Zhao Z., Ji J. Comparative analysis of BlaKPC expression in Tn4401 transposons and the tn3-tn4401 chimera. Antimicrob. Agents Chemother. 2019;63(5) doi: 10.1128/AAC.02434-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ajayi A.O., Perry B.J., Yost C.K., Jamieson R.C., Hansen L.T., Rahube T.O. Comparative genomic analyses of β-lactamase (BlaCMY-42)-Encoding plasmids isolated from wastewater treatment plants in Canada. Can. J. Microbiol. 2021;67(10):737–748. doi: 10.1139/cjm-2021-0012. [DOI] [PubMed] [Google Scholar]
- 110.Garcillán-Barcia M.P., Cuartas-Lanza R., Cuevas A., de la Cruz F. Cis-acting relaxases guarantee independent mobilization of MOBQ4 plasmids. Front. Microbiol. 2019;10(November):1–11. doi: 10.3389/fmicb.2019.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huyan J., Tian Z., Zhang Y., Zhang H., Shi Y., Gillings M.R., Yang M. Dynamics of class 1 integrons in aerobic biofilm reactors spiked with antibiotics. Environ. Int. 2020;140(January) doi: 10.1016/j.envint.2020.105816. [DOI] [PubMed] [Google Scholar]
- 112.Lund D., Kieffer N., Parras-Moltó M., Ebmeyer S., Berglund F., Johnning A., Joakim Larsson D.G., Kristiansson E. Large-scale characterization of the macrolide resistome reveals high diversity and several new pathogen-associated genes. Microb. Genom. 2022;8(1) doi: 10.1099/mgen.0.000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huo Y., Bai Y., Qu J. Unravelling riverine microbial communities under wastewater treatment plant effluent discharge in large urban areas. Appl. Microbiol. Biotechnol. 2017;101(17):6755–6764. doi: 10.1007/s00253-017-8384-4. [DOI] [PubMed] [Google Scholar]
- 114.Numberger D., Ganzert L., Zoccarato L., Mühldorfer K., Sauer S., Grossart H.P., Greenwood A.D. Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S RRNA sequencing. Sci. Rep. 2019;9(1):1–14. doi: 10.1038/s41598-019-46015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mansfeldt C., Deiner K., Mächler E., Fenner K., Eggen R.I.L., Stamm C., Schönenberger U., Walser J.C., Altermatt F. Microbial community shifts in streams receiving treated wastewater effluent. Sci. Total Environ. 2020;709 doi: 10.1016/j.scitotenv.2019.135727. [DOI] [PubMed] [Google Scholar]
- 116.Chung T., Weller D.L., Kovac J. The composition of microbial communities in six streams, and its association with environmental conditions, and foodborne pathogen isolation. Front. Microbiol. 2020;11(July):1–16. doi: 10.3389/fmicb.2020.01757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paddock M.B., Fernández-Bayo J.D., VanderGheynst J.S. The effect of the microalgae-bacteria microbiome on wastewater treatment and biomass production. Appl. Microbiol. Biotechnol. 2020;104(2):893–905. doi: 10.1007/s00253-019-10246-x. [DOI] [PubMed] [Google Scholar]
- 118.Tang J., Bu Y., Zhang X.X., Huang K., He X., Ye L., Shan Z., Ren H. Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water. Ecotoxicol. Environ. Saf. 2016;132:260–269. doi: 10.1016/j.ecoenv.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 119.Tiwari A., Kurittu P., Al-Mustapha A.I., Heljanko V., Johansson V., Thakali O., Mishra S.K., Lehto K.M., Lipponen A., Oikarinen S., Pitkänen T., Heikinheimo A. Wastewater surveillance of antibiotic-resistant bacterial pathogens: a systematic review. Front. Microbiol. 2022;13(December):1–19. doi: 10.3389/fmicb.2022.977106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yin X., Yang Y., Deng Y., Huang Y., Li L., Chan L.Y.L., Zhang T. An assessment of resistome and mobilome in wastewater treatment plants through temporal and spatial metagenomic analysis. Water Res. 2022;209(November 2021) doi: 10.1016/j.watres.2021.117885. [DOI] [PubMed] [Google Scholar]
- 121.Hendriksen R.S., Munk P., Njage P., van Bunnik B., McNally L., Lukjancenko O., Röder T., Nieuwenhuijse D., Pedersen S.K., Kjeldgaard J., Kaas R.S., Clausen P.T.L.C., Vogt J.K., Leekitcharoenphon P., van de Schans M.G.M., Zuidema T., de Roda Husman A.M., Rasmussen S., Petersen B., Bego A., Rees C., Cassar S., Coventry K., Collignon P., Allerberger F., Rahube T.O., Oliveira G., Ivanov I., Vuthy Y., Sopheak T., Yost C.K., Ke C., Zheng H., Baisheng L., Jiao X., Donado-Godoy P., Coulibaly K.J., Jergović M., Hrenovic J., Karpíšková R., Villacis J.E., Legesse M., Eguale T., Heikinheimo A., Malania L., Nitsche A., Brinkmann A., Saba C.K.S., Kocsis B., Solymosi N., Thorsteinsdottir T.R., Hatha A.M., Alebouyeh M., Morris D., Cormican M., O'Connor L., Moran-Gilad J., Alba P., Battisti A., Shakenova Z., Kiiyukia C., Ng’eno E., Raka L., Avsejenko J., Bērziņš A., Bartkevics V., Penny C., Rajandas H., Parimannan S., Haber M.V., Pal P., Jeunen G.J., Gemmell N., Fashae K., Holmstad R., Hasan R., Shakoor S., Rojas M.L.Z., Wasyl D., Bosevska G., Kochubovski M., Radu C., Gassama A., Radosavljevic V., Wuertz S., Zuniga-Montanez R., Tay M.Y.F., Gavačová D., Pastuchova K., Truska P., Trkov M., Esterhuyse K., Keddy K., Cerdà-Cuéllar M., Pathirage S., Norrgren L., Örn S., Larsson D.G.J., Heijden T. Van der, Kumburu H.H., Sanneh B., Bidjada P., Njanpop-Lafourcade B.M., Nikiema-Pessinaba S.C., Levent B., Meschke J.S., Beck N.K., Van C.D., Phuc N. Do, Tran D.M.N., Kwenda G., Tabo D., Wester A.L., Cuadros-Orellana S., Amid C., Cochrane G., Sicheritz-Ponten T., Schmitt H., Alvarez J.R.M., Aidara-Kane A., Pamp S.J., Lund O., Hald T., Woolhouse M., Koopmans M.P., Vigre H., Petersen T.N., Aarestrup F.M. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019;10(1) doi: 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.