A revolution is occurring in the detection, identification, and characterisation of pathogens by the combining of the seemingly disparate fields of nucleic acid analysis, bioinformatics, data storage and retrieval, nanotechnology, physics, microelectronics, and polymer, solid state, and combinatorial chemistry. The scenario of taking a drop of blood, urine, or saliva and within an hour knowing whether a pathogen is present and its antimicrobial resistance potential is no longer science fiction but will soon be reality. These developments, particularly with regard to near patient testing, have important implications for the delivery of health care. They will affect primary care, prescribing practice, organisation of pathology laboratories, counselling services, surveillance and epidemiology, and medicolegal practice.
Near patient testing could be implemented in various settings—at hospital bedside, in an outpatient clinic, in a dental or general practice surgery, or in a patient’s home. Testing kits might be complete diagnostic units, needing no processing other than application of test material and yielding instant results, or they may need manipulation of test material or use of other equipment for the test to be read and interpreted.
Developments enabling near patient testing
The main driving forces behind the development of such testing kits have been the search for life in space exploration and the military’s need to detect agents of biological warfare. In both cases miniaturisation and robustness of detection systems have been necessary. Systems for detecting biological weapons must be able to rapidly detect and identify a variety of pathogens or their virulence factors, particularly toxins.
Antibody based systems
Many new test kit technologies coming on to the market for patient diagnosis are still based on antigen-antibody interactions, an old diagnostic technology. The developments that have permitted near patient testing are in new detection systems for antigen-antibody complexes, allowing results to be read by eye, use of a control that is built into the kit, and incorporation of all reagents and diluents into the kit. Such kits include those for detecting the flu virus, respiratory syncytial virus, and group A streptococci.
In many currently available immunoassays—such as the Clearview C.DIFF A kit (Oxoid) for detecting Clostridium difficile toxin A in faeces—the antigen bound to a specific capture antibody is detected by a second specific antibody that is tagged so that its accumulation yields a visible colour. Sensitivity is improved by labelling the second antibody with a fluorescent dye. The antigen-antibody complex can be detected with a bidirectional optic fibre that carries laser light to excite the fluorescent label and recovers the fluorescent signal.1
Predicted developments
Increasing use of specific antigen detectors other than antibodies
Microminiaturisation will allow integration of diagnostic procedures in order to produce a “laboratory on a chip”
Incorporation of diagnostic tests into healthcare products such as wound dressings
Coupling of over the counter diagnostic kits and treatments
Test kits that can be swallowed or added to body fluids are coupled to data transmitters so that results can be sent to remote site for analysis
Techniques that permit detection of the antigen-antibody complex without use of a second antibody or any other reagent can simplify such kits (see figure). In this situation, binding of antigen to the capture antibody will alter the properties of the matrix holding the capture antibody. For example, when a layer of antibodies is immobilised on to a gold surface subsequent capture of antigen causes detectable changes in the refractive index at the surface layer, giving it a different appearance. This approach has been used for the development of an optical immunoassay for group A streptococci.2
Other natural receptor molecules
Antibodies are not the only biological materials with characteristics of specificity. Use is now being made of other receptors for pathogens and toxins in experimental biosensing kits. One of the most exciting developments is incorporation of receptors into artificial membranes with new optical and interfacial properties so that the membranes’ colour changes after the target molecules bind to the capture probes. This method has been used for detecting cholera toxin, Escherichia coli heat labile enterotoxin, and botulinum neurotoxin.3 Such approaches raise the possibility of diagnostic plasters, swabs, or dipsticks, with the sampling procedure also being the diagnostic step. An exciting possibility is to combine different capture probes in the same membrane so that different pathogens cause different colour changes.
A possible problem with using natural receptors for detecting infectious agents is their lack of specificity. Pathogens often “hijack” host receptors that have other functions—such as viral use of cytokine receptors. Another potential problem is that any natural receptor molecules in clinical material might compete with the kit receptors for binding of the target.
RNA fragments
RNA fragments are another possible alternative to monoclonal antibodies that may have the same specificity without the problems of maintaining hybridoma cells for producing antibody and the ethics of animal experimentation. Sequences of RNA can adopt tertiary configurations with specific receptor properties,4 and it is possible to generate millions of fragments of RNA of random sequence that could be screened for specific binding to a target. These binding stretches of RNA (aptamers) can be sequenced, thereby facilitating their specific production. Aptamers can therefore be considered as synthesisable equivalents to monoclonal antibody.
Microminiaturisation
Microminiaturisation will revolutionise diagnostics tests by making existing technology (such as the polymerase chain reaction) more compatible with near patient testing. Much of the technology and language has been adopted from the microelectronics industry, yielding such terms as “DNA chips,” “addresses” for capture probes, and “microfluidic integrated circuits.” As procedures become reduced to the microscale, it becomes possible to sequentially integrate them in order to reproduce a complete process—that is, to develop a “laboratory on a chip.”
The DNA chip has become synonymous with future diagnostic technology. In essence, this is a high density array of DNA capture probes that allows a range of target DNA to be bound and recognised. It is already possible to manufacture single grids with over 400 000 different hybridisation probes. It is also now possible, taking advantage of DNA’s strong electronegative charge, to use electronics to accelerate and enhance hybridisation of capture probes to target DNA. DNA chips can be used for sequencing5 and have already been used to determine mutations in a 382 base pair region of the HIV-1 protease gene,6 which will help in understanding resistance to therapeutic protease inhibitors, and for monitoring gene expression.7
The first application of DNA chips in near patient testing will probably be for identifying pathogens and their antimicrobial resistance potential, which would require amplification of the pathogen nucleic acid for it to be detectable. Such chips therefore require the development of “on chip” cell lysis and nucleic acid amplification.8,9 Cheng et al separated Escherichia coli from whole blood by dielectrophoresis and subjected the E coli to high voltage shock, to cause lysis, and subsequent proteinase K digestion, all on a single chip.10 The lysate was then examined on a separate DNA chip. The prospect of separating pathogens from biological material, releasing and amplifying the nucleic acid, and using the amplified nucleic acid to identify and characterise the pathogen on a single chip is now a possibility. This process may be enhanced by replacing the polymerase chain reaction, which needs alternate heating and cooling, with transcription mediated amplification, which can be undertaken at a fixed temperature.
Indirect methods
The above techniques all relate to direct detection of a pathogen. There have also been developments in technology enabling detection of metabolic end-products for indirect detection and identification of bacteria. One promising development for near patient testing is that of artificial olfaction (the “artificial nose”). Artificial olfaction is used extensively in the food and beverage industry and is now being applied to wound infections.11 The normal, “bench top” size devices have shrunk to the size of a silicon chip, heralding the prospect of near patient breath tests for respiratory or gastric infection, or even urine analysis. For example, it may be possible to detect breath ammonia produced by Helicobacter pylori infection of the stomach.
Implications of near patient testing
Near patient testing will undoubtedly change clinical practice. Although such tests will bring many benefits, there are also some associated risks (table).
Speed of diagnosis
A major advantage of near patient testing is the potential for rapid accurate diagnosis and associated appropriate treatment. At its simplest level, detection of the flu virus, respiratory syncytial virus, or group A streptococci (kits already available) in general practice or at home should improve antibiotic prescribing,2 enable targeted use of antiviral drugs, and improve patient compliance with the proposed treatment because of the “kit evidence.” Further advantages are that near patient testing kits would improve the quality of service offered by clinicians who are remote from major diagnostic facilities and be useful to emergency, prison, and immigration services, for which rapid determination of HIV infection, hepatitis, tuberculosis, or infection with methicillin resistant Staphylococcus aureus (MRSA) may be beneficial.
General practitioners, outpatient clinics, and those involved in controlling communicable disease in the community would also benefit from having rapid diagnostic kits for diseases such as diphtheria, tuberculosis, or salmonella infection and for acute infections such as meningococcal meningitis or septicaemia, for which rapid treatment is essential. The ability to determine quickly whether to instigate community contact tracing and screening could be one of the most important public health consequences of accurate and rapid near patient testing. Such kits could also be used to target vaccination to high risk groups, particularly those who are hard to reach and often do not present for a delayed test result. An example would be near patient testing of drug misusers for hepatitis antibody status and targeted vaccination.
However, the development of kits will be market driven. Thus, development will probably be favoured for infections where diagnosis can be coupled to specific treatment—such as an anticandidal drug for thrush—and alliances can be formed between kit manufacturers and the pharmaceutical companies. There may be little incentive to develop kits for which the perceived market is small even though the need is great—such as diagnosis of meningococcal infection by general practitioners in patients’ homes or rapid determination of HIV status in pregnant women in developing countries. A challenge will be to identify how development of such kits can be encouraged or commissioned.
Changing workload
Although the development of near patient testing kits is expected to reduce the number of specimens sent from the community (and possibly hospital departments) to microbiology laboratories for testing, it is possible that laboratories’ workload would increase if kit users considered results to be indicative as opposed to definitive and send material for confirmation. Whatever the outcome, redistribution of funds will need to be considered to ensure that money follows the activity. There are other financial consequences, such as the reduction in specimen flow to a diagnostic centre reducing the opportunity for large scale automation and associated efficiency gains.
Proper use
For use of kits to become used widespread, there must be confidence in the operators as well as in the kits: people must be confident that the correct specimen was obtained, the kit was used appropriately, the result was interpreted correctly, and that any machine used for part of the process is appropriately maintained and used. These concerns indicate that quality control and assurance, and possibly accreditation, need to be considered12 and raise questions about whether testing kits should be available as over the counter diagnostics, although similar concerns were initially raised about home pregnancy testing kits. As kits will become more accessible and easier to use, their potential for misuse will increase, especially in the absence of expert explanation or counselling. Potential problems include testing of others without their knowledge, misinterpretation of the meaning of positive results, deliberate infection of others after infection has been confirmed, and self administration of remedies.
Safe disposal of used kits will be an additional responsibility for general practitioners and poses a possible problem for home use.
Loss of information
An often overlooked problem with near patient testing is the potential loss of epidemiological data and associated surveillance information. The use of kits will shift the traditional base of diagnostic testing and reporting from diagnostic laboratories to the community, with the risk that delivery of individual patient information will be at the expense of population information. The challenge is to capture the data. Such data collection will place an extra burden on general practice. Over the counter diagnostics pose a further complication, especially in countries where kits could be linked to over the counter sale of treatments. If patients have no contact with health services, either for diagnosis or treatment, there will be no record of infection. Alternatively, if such data are captured, results from patients referred for confirmatory tests may be counted as separate events, again leading to inaccurate data.
Near patient tests should also be considered as near “target” tests: their use in veterinary medicine will have a similar effect on that discipline, with corresponding potential loss of epidemiological data and impact on public health for zoonotic infections.
Diagnosing infections without isolating the pathogens will result in pathogens not being characterised for attributes such as type, virulence, and mechanisms of antimicrobial resistance. Such information is invaluable in understanding pathogenesis, detecting outbreaks, and informing vaccine development and implementation. This would be particularly true for advising on the annual composition of flu vaccine.
A glimpse into the future
As the technology develops, some of the suggested advantages and problems of near patient testing will not materialise, while unexpected problems and benefits will become apparent. In the medium term future the introduction and use of near patient testing kits will be gradual, and factors such a market size, unit cost, and healthcare infrastructure will have as much influence as the convenience, accuracy, rapidity, and desirability of the tests. The most obvious market is for non-invasive kits available in general practice and over the counter for diagnosing respiratory, enteric, and urinary tract infections.
In the long term it seems inevitable that near patient testing kits and machines will be developed that will make diagnoses and identify and characterise the causative pathogen. Further, it will become possible to transmit this information automatically to a central database to help inform regional, national, and international policy as well as commercial decisions. These systems for analysis and data transmission will probably be developed by the commercial sector. It is also likely that near patient testing will increasingly mean remote patient testing. For example, it may be possible to develop a small capsule that can be swallowed and that has the ability to detect different enteric pathogens and transmit the data to a physician or advice centre, which will then transmit advice back to the patient. Such diagnostic capsules or chips could also be added to sputum or urine in the home. It may also be possible to connect breath analysers directly to telephones or computers to transmit diagnostic information or to perform in-house diagnosis.
Overall, we should be optimistic that technological developments will bring great improvements in public health through advances in detecting and controlling infections.
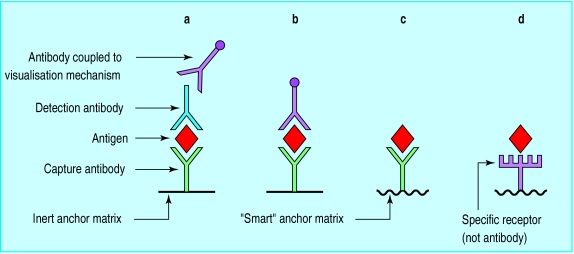
Figure.
Basic developments in antigen detection technology. In earlier systems (a and b) visualisation of the antibody-antigen complex depends on at least one additional antibody: the visualisation mechanism might be an enzyme or coloured microparticle and might be coupled to the detection antibody itself (b) or to an antibody to the detection antibody (a). With a “smart” anchor matrix (c and d), recognition of the antibody-antigen complex is based on changes to the matrix−such as change in colour or refractive index change. The capture antibody may be replaced with a specific receptor such as a toxin receptor (d)
Table.
Pros and cons of near patient testing
| Advantages | Disadvantages |
|---|---|
| More appropriate prescribing and targeted vaccination | Misuse or misinterpretation of test result (especially if used in the home)—Such as accusations of infidelity, adoption of inappropriate self administered remedies, deliberate infection of others |
| Rapid instigation of infection control measures and appropriate treatment | Potential loss of epidemiological data |
| Decreased dependency of remote areas on distant diagnostic facilities | Less opportunity for large scale automation |
| Rapid diagnosis, alleviating unnecessary anxiety | Inadequate discussion or counselling |
| Reduced burden on microbiology laboratories | Increased burden on microbiology laboratories (from demands for confirmatory tests) |
| Decreased overall cost of health care (more appropriate prescribing, fewer laboratory tests) | Increased overall cost of health care (more tests performed) |
| Collection of specimen in privacy of one’s own home; no need for transport of specimen | Reduced opportunity for internal and external quality assurance, with associated risk of misdiagnosis |
| Increased risk of inappropriate disposal of diagnostic kits (especially if used in the home) | |
| Medicolegal implications |
Acknowledgments
I thank Dr Painter, Dr Zambon and Professor Boseley for helpful discussion and Lynne Foster for preparing the manuscript.
Footnotes
Competing interest: None declared.
References
- 1.Cao LK, Anderson GP, Ligler FS, Ezzell J. Detection of Yersinia pestis fraction 1 antigen with a fiber optic biosensor. J Clin Microbiol. 1995;2:336–341. doi: 10.1128/jcm.33.2.336-341.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Needham CA, McPherson KA, Webb KH. Streptococcal pharyngitis: impact of a high-sensitivity antigen test on physician outcome. J Clin Microbiol. 1998;36:3468–3473. doi: 10.1128/jcm.36.12.3468-3473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charych D, Cheng Q, Reichert A, Kuziemko G, Stroh M, Nagy JO, et al. A “litmus test” for molecular recognition using artificial membranes. Chem Biol. 1996;3:113–120. doi: 10.1016/s1074-5521(96)90287-2. [DOI] [PubMed] [Google Scholar]
- 4.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 5.Southern EM. DNA chips: analysing sequence by hybridization to oligonucleotides on a large scale. Trends Genetics. 1996;12:110–115. doi: 10.1016/0168-9525(96)81422-3. [DOI] [PubMed] [Google Scholar]
- 6.Kozal MJ, Shah N, Shen N, Yang R, Fucini R, Merigan TC, et al. Extensive polymorphisms observed in HIV-1 clade B protease gene using high density oligonucleotide arrays. Nature Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 7.Shena M, Shalon D, David RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 8.Wilding P, Kricka LJ, Cheng J, Krichia G, Hvichia G, Fortina P. Integrated cell isolation and PCR analysis using silicon microfilter-chambers. Anal Biochem. 1998;257:95–100. doi: 10.1006/abio.1997.2530. [DOI] [PubMed] [Google Scholar]
- 9.Cheng J, Waters LC, Fortina P, Hvichia G, Jacobson SC, Ramsey JM, et al. Degenerate oligonucleotide primed-polymerase chain reaction and capillary electrophoretic analysis of human DNA on microchip-based devices. Anal Biochem. 1998;257:101–106. doi: 10.1006/abio.1997.2531. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Sheldon EL, Wu L, Uribe A, Gerrue LO, Carrino J, et al. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nature Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 11.Greenwood JE, Crawley BA, Clark SL, Chadwick PR, Ellison DA, Oppenheim BA. Monitoring wound healing by odour. J Wound Care. 1997;6:219–221. doi: 10.12968/jowc.1997.6.5.219. [DOI] [PubMed] [Google Scholar]
- 12.Bevan VM, Bullock DG, Hueney M, Dhell J. The application of near patient testing to microbiology. Commun Dis Public Health. 1999;2:14–21. [PubMed] [Google Scholar]