Abstract
We show the presence of numerous short tandem repeats in the human cytomegalovirus (HCMV) genome and assess their usefulness as molecular markers. The genome is shown to contain at least 24 microsatellite regions that exhibit length polymorphisms. Insertion-deletion polymorphisms at these short tandem repeats are common (80% of repeats examined are polymorphic among two laboratory strains and 10 clinical isolates). This is the first report of widespread microsatellite length polymorphism in a viral genome. Some regions are highly polymorphic: one was revealed by DNA sequencing to contain length variants at five closely linked sites, which combined resulted in 10 variants for this region among the 12 strains and isolates examined. This study not only provides a new molecular marker system for this virus but also extends our understanding of microsatellite polymorphism in two important ways. First, variable-length repeats in HCMV can be considerably shorter than polymorphic repeats previously found in other organisms. Second, highly variable microsatellite repeats are not confined to prokaryotes and eukaryotes, as previously assumed. This variation provides a useful marker system for distinguishing viral isolates, and similar markers are also likely to be found in other large-genome DNA viruses.
Short tandem repeats, or microsatellites, defined as iterations of short motifs made up of one to six bases, are known to be hot spots of length mutation in higher (42) and lower (14) eukaryotes. Even prokaryotic species have short iterations of short tandem repeats (14, 15) that can vary in length (16, 33), as well as longer hypermutable repeats that regulate virulence factors (28). Insertions and deletions at microsatellite loci are generally considered to be due to replication slippage errors (36, 42). Polymorphisms that result from these slippage errors are valuable in pathogenic species because of their use as epidemiological markers (13, 26, 27).
Microsatellites are unusual for the following reasons. First, because of replication slippage, they undergo a much higher rate of both insertion and deletion mutations than nonrepetitive regions. Second, unlike point mutations and insertions-deletions in nonrepetitive DNA, microsatellite mutations are highly reversible, with back mutation rates similar to forward mutation rates (8, 25, 46, 48). In some cases, microsatellites can provide pathogenic microorganisms with functional variation adaptive in rapidly changing environments (28, 29).
It has been suspected that viral genomes may also be prone to the accumulation of replication slippage errors (23). Variation has been found at the minisatellite level (defined as repeats of a motif of 11 to 60 bp [30]) in herpes simplex virus, although no clinical correlates of this variation have been discovered (44). Such large, minisatellite repeats have also been found within Epstein-Barr virus (4, 20, 21, 24, 46) and have proven useful in epidemiological studies of this virus (12, 37). These mutations affect very long repetitive regions, but other mutations that affect repeats of very short motifs have been found.
Several examples of functional microsatellite tracts have been found among different classes of viruses. Functional mononucleotide repeats have been found in mengovirus (10, 22), vesicular stomatitis virus (VSV) (2), hepatitis C virus (50), and human respiratory syncytial virus (18). Changes in the length of trinucleotide and hexanucleotide repeats at the hemagglutinin cleavage site in avian influenza virus have been associated with increased virulence (31, 49). While these microsatellite tracts function in different ways within each virus, their presence in a variety of lengths in several types of viral populations demonstrates that mutational processes leading to expansion and contraction of nucleotide repeats do occur in viruses.
Although some effects of repetitive DNA sequences within viruses have been demonstrated, the full extent of naturally occurring variation at a variety of repeat loci, particularly at short repeats, has not been investigated for any virus. Studies in humans have shown that repeats less than five units long are typically monomorphic, and it has been generally assumed that extremely short iterations do not undergo replication slippage (47). Long repeats have been shown experimentally to be more mutable than short ones in yeast (43). Extremely short repeats, however, have not been well investigated in any organism. In this study, we have investigated the prevalence of length polymorphisms at short tandem repeats in human cytomegalovirus (HCMV). HCMV is a large double-stranded DNA virus of clinical importance. HCMV infection is considered endemic throughout the world, with seroprevalence in adults ranging from 50 to 90%, and infections are associated with a diverse range of clinical syndromes (39). We have examined the potential of insertion-deletion events at short tandem repeats to differentiate clinical viral isolates. It might seem unlikely that the short repeats in this genome would be sites of length polymorphisms. However, abundant length polymorphisms were discovered at many of these repeats.
MATERIALS AND METHODS
Computer survey of the genome.
A computer survey of the HCMV AD169 genome (7) (GenBank entry HEHCMVCG) was carried out by using a program written in True Basic for the Macintosh and available from D.F. on request (12a). The program searches for all possible mono- through hexanucleotide repeats and has been previously described (14). Expected repeat distributions are derived from the mean distribution of repeat sequences in randomly generated genomes of identical length and base content (the HCMV genome is 57% G+C). We targeted our attention on sites that had at least four trinucleotide repeat units or five dinucleotide repeats or where the mononucleotide tract was nine or more bases long. This resulted in 85 potentially polymorphic repeat sites.
Source of strains.
A total of 12 isolates were used to test for polymorphisms at short repeats. HCMV laboratory strains AD169 (34) and Towne (32) were obtained from the American Type Culture Collection (Rockville, Md.). Of the 10 clinical isolates examined, 8 were cultured from five urine specimens and three blood specimens of seven individuals diagnosed with AIDS-related CMV retinitis. Two isolates were cultured from bronchial alveolar lavage of bone marrow transplant recipients who were receiving treatment for CMV pneumonitis. CMV cultures were expanded on human foreskin fibroblasts maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, 0.01% streptomycin, and 20 μg of amphotericin B per ml. DNA was purified from fully cytopathic CMV-infected human foreskin fibroblasts by phenol extraction and ethanol precipitation as described previously (38).
PCR and analysis.
Primer pairs were designed to amplify 42 of the 85 longest mono-, di-, and trinucleotide repeats in the HCMV genome. A total of 30 primer pairs were manually designed, each pair amplifying a product containing from one to four target repeats (Table 1) and purchased from Research Genetics (Huntsville, Ala.). PCR was performed as previously described (13), with the addition of 0.1 μl of fluorescence-labeled dCTP from Perkin-Elmer (Foster City, Calif.). A Perkin-Elmer 2400 thermocycler was used for the reactions, with a PCR program of 40 cycles of 1-min denaturation at 94°C, 1-min annealing at various temperatures depending on primer (Table 1), and 1-min extension at 72°C. All products were analyzed on an AB1 373 automated DNA sequencer (Applied Biosystems, Inc.) using labeled dCTP incorporation. Genescan software was used to search for polymorphisms by length. All polymorphisms were confirmed by analysis of independently amplified PCR products. Sequencing to confirm length changes was carried out on an ABI 373 sequencer, and all reported sequences were determined twice in each direction. Before sequencing, all reaction products were cleaned by using a QiaQuick PCR Clean-up kit from Qiagen (Santa Clarita, Calif.).
TABLE 1.
Primer pairs found to be polymorphic in this survey
| Name | Sequencea | Expected size (bp)b | Target repeat(s)c | Position in genomed | Temp (°C)e | No. of variantsf |
|---|---|---|---|---|---|---|
| HCMVTRL8C | F, cccccgagacgagagccaccc | 252 | (GGC)4 | 7287 | 70 | 2 |
| R, ccatcctcgccttcggacgcccc | ||||||
| HCMVUL23C | F, ggtcgcgacgtcttaggagg | 229 | (A)10 | 28762 | 70 | 4 |
| R, cttcccgtttgactcgcgtgc | ||||||
| HCMVUL30C | F, cctccacacgctcagccgc | 282 | (TG)5 | 37254 | 70 | 2 |
| R, gcgccgtcaacagcgtgcc | ||||||
| HCMVUL37C | F, ggcgtcgatgggttcacctcgggcg | 157 | (TTC)4 | 52405 | 60 | 2 |
| R, ggagaccgagcggtcccgtggacttgg | ||||||
| HCMVUL38N | F, gcagcatgtggggctaatagg | 400 | (A)10 | 51083 | 70 | 3 |
| R, gctgcgagccaattcgttgg | ||||||
| HCMVUL46C | F, cctcgcggtttggctttgagc | 138 | (CG)5 | 60374 | 68 | 2 |
| R, cccgcagctgctctatcaac | ||||||
| HCMVUL50C | F, ggcggatgacggtgatgggtcg | 348 | (CTC)4...(GGC)4...(GAG)4 | 72429 | 70 | 3 |
| R, ggcgggaaagcggtcctctcgg | ||||||
| HCMVUL67AN | F, gcctttggtgctcgttgagcc | 280 | (T)10 | 96977 | 63 | 2 |
| R, gtggggacgggaatcgatgtc | ||||||
| HCMVUL67BC | F, gacatcgattcccgtccccac | 303 | (GCG)4 | 97476 | 65 | 2 |
| R, gaggacgaggagacgacgacc | ||||||
| HCMVUL68C | F, gggagggagaggacgtggggctcg | 414 | (T)11 | 97928 | 60 | 7 |
| R, cgctgccagacgcccgattacgagg | ||||||
| HCMVUL69C | F, gcttaacttgatgacgccgtcgcc | 209 | (GGT)5(GGC)4(TGC)3 | 98290 | 65 | 5/9 |
| R, cctcgtaatcgggcgtctggcagcg | ||||||
| HCMVUL95C | F, ggctgagctgtacgtctttgtttgg | 388 | (GGT)5 | 139531 | 60 | 4/4 |
| R, ggcgcagtcttggataacgatgggg | ||||||
| HCMVUL96N | F, cggtttcggcgacgtgcgatttgg | 297 | (A)10 | 140406 | 70 | 3 |
| R, gcgcagcttcgcgcatccactgg | ||||||
| HCMVUL102C | F, cctccaccgcttccaccacc | 167 | (TTC)4 | 148933 | 60 | 2 |
| R, ccgccagcgaacgcaccc | ||||||
| HCMVTRAN1N | F, cccgctaaggacttacc | 397 | (A)11 | 156864 | 55 | 3 |
| R, gctgtaagtctcacctagc | ||||||
| HCMVTRAN2N | F, ccatccgtctagtttttcg | 307 | (TGTA)4 | 157332 | 55 | 2 |
| R, ccaccgtgatagaagacacc | ||||||
| HCMVTRAN3C | F, cctctcctccagtggtagtcgtg | 342 | (TGC)4...(T)10...(TAT)4 | 157835 | 60 | 4/10 |
| R, cctcctctccccatcatcttctcc | ||||||
| HCMVUL111C | F, ccatgatcaattaagcccaccacc | 431 | (GGT)4...(A)10...(A)10...(CGA)4 | 159451 | 62 | 4/5 |
| R, cgtaatcctctggacgacactgcg | ||||||
| HCMVUL112AC | F, ggaggaacagcaacggcagagg | 233 | (GGT)3 | 161192 | 63 | 2 |
| R, ggactcaccgtcgttctcggagg | ||||||
| HCMVUL122N | F, ggctgagaacagtgatcagg | 330 | (AT)5 | 170988 | 60 | 3/5 |
| R, gcgtgacacgtttattgagtagg | ||||||
| HCMVUL123I2N | F, gcacatgggtcacatacaggag | 295 | (A)9 | 172660 | 60 | —/6 |
| R, ggtgctgttaacggtggaggg | ||||||
| HCMVUL123I1N | F, ccctccaccgttaacagcacc | 265 | (TGG)4 | 173066 | 70 | 3 |
| R, gactcatggtcgctcggcag | ||||||
| HCMVUL132C | F, ggaatctgccagctggtgctgttc | 307 | (CCT)4 | 177227 | 65 | 2/2 |
| R, gccggattttgttcctcgagccc | ||||||
| HCMVUS35C | F, gacgaagatgccgatgtgtgac | 325 | (G)12 | 225448 | 65 | —/6 |
| R, cgtagcgtacgcccaacggc |
F, forward; R, reverse.
Expected length of the PCR fragment in the AD169 genome (GenBank entry HEHCMVCG).
Ellipses indicate repeats separated by a stretch of nonrepetitive sequence; contiguous repeats are not separated by ellipses. Repeats in bold are those found within open reading frames according to the GenBank annotation.
Nucleotide number of the first repeat within the primer from the same reference sequence.
Annealing temperature used for PCR.
The number of variant alleles as determined by length analysis and sequencing (number of length variants/number of sequence variants).
RESULTS
The computer survey revealed that the HCMV genome contains only repeats with very short iterations (two to five units), and there is a slight excess of the longer mono-, di-, and trinucleotide repeats compared to randomized genomes of equivalent nucleotide composition and size (14). Observed and expected repeat distributions are compared in Table 2. For all repeat classes, longer repeats are more common than expected; however, in the HCMV genome the maximum number of repeat units of a di- or trinucleotide motif is only five, and the longest mononucleotide tract is 13 bases.
TABLE 2.
Observed and expected distributions of mono-, di-, and trinucleotide repeats in the HCMV AD169 genomea
| No. of repeat units | Mononucleotides
|
Dinucleotides
|
Trinucleotides
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. observed | No. expected | Deviation | No. observed | No. expected | Deviation | No. observed | No. expected | Deviation | |
| 2 | 31,203 | 32,112 ± 30.8 | − | 8,209 | 7,394 ± 13.2 | + | 4,307 | 2,518 ± 8.8 | + |
| 3 | 7,152 | 8,241 ± 18.9 | − | 644 | 486 ± 5.8 | + | 275 | 43.7 ± 1.3 | + |
| 4 | 2021 | 2,153 ± 7.3 | − | 70 | 33 ± 1.1 | + | 27 | 1.15 ± 0.3 | + |
| 5 | 659 | 572 ± 5.3 | + | 5 | 1.9 ± 0.4 | + | 2 | ||
| 6 | 199 | 154 ± 2.3 | + | ||||||
| 7 | 69 | 42 ± 1.6 | + | ||||||
| 8 | 24 | 11 ± 0.6 | + | ||||||
| 9 | 27 | 3 ± 0.4 | + | ||||||
| 10 | 17 | ||||||||
| 11 | 2 | ||||||||
| 12 | 3 | ||||||||
| 13 | 2 | ||||||||
Target repeats for study are in boldface. The expected number of repeats is the mean of 20 genomes with standard error. All observed values were significantly different from expectation (in all cases, P ≤ 0.001). “Deviation” indicates whether the observed number was significantly more (+) or less (−) than expectation. Tetra-, penta-, and hexanucleotide repeats are extremely rare and match random expectations and are therefore not shown.
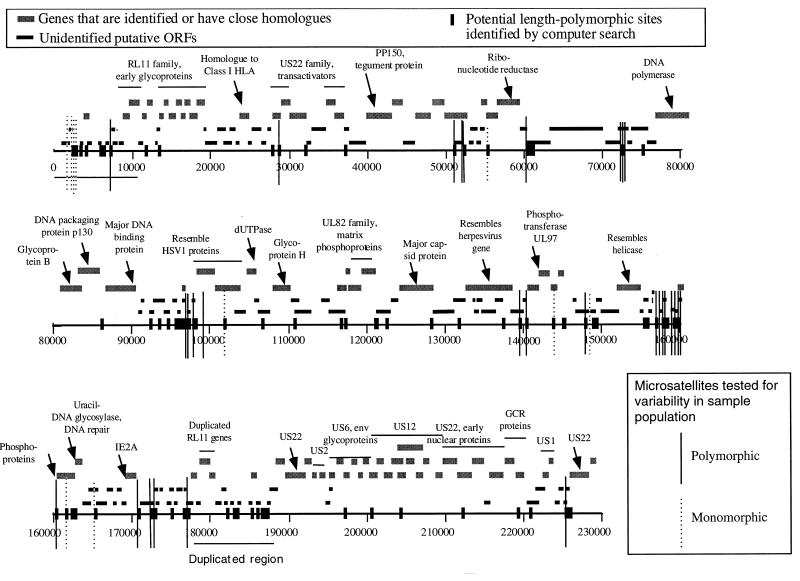
We surveyed 42 of the longest tandem repeats in 12 HCMV isolates, using 30 PCR primer pairs. Despite the short lengths of the repeats analyzed, 24 of 30 PCR primers (80%) amplified multiple length forms (Table 1), demonstrating repeat polymorphism, in this small group. All 12 isolates could be uniquely identified by using the 24 polymorphic regions. Figure 1 is a diagram of the HCMV AD169 genome showing positions of the polymorphic and monomorphic microsatellites that have been identified in this study.
FIG. 1.
Diagram of the HCMV AD169 genome showing positions of polymorphic and monomorphic satellites identified in this study. ORFs, open reading frames; HSV1, herpes simplex virus type 1.
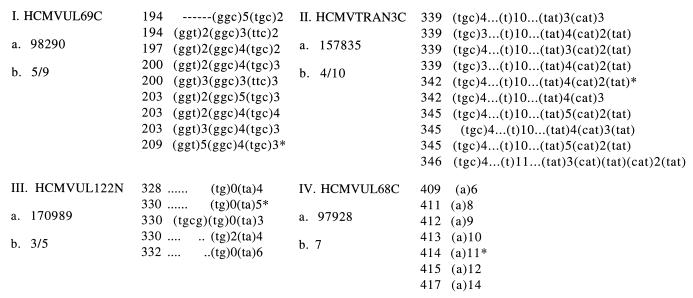
Variability in the number of repeat units at some of the targeted short tandem repeats was confirmed by sequencing, as shown in Fig. 2. In most cases, for the di- and trinucleotide repeats, more variants were revealed by sequencing than by size separation. For example, separation by length at the TRAN3 locus showed 4 length variants, while sequencing revealed a total of 10 different variants among the 12 strains. The microsatellites in UL69 and TRAN3 are both strikingly complex, with polymorphisms in more than one contiguous or neighboring repeat. Further, some of the polymorphic repeats in these clustered microsatellites (13) have as few as two repeat units, similar to the pattern seen in short clustered microsatellites in Candida albicans (27).
FIG. 2.
Length variants from all 12 strains were sequenced at four loci (I to IV), and the short tandem repeats contributing polymorphisms to each unique length variant found are shown. The number at the beginning of each sequence refers to PCR product length. a, location in genome (HEHCMVCG); b, number of variants by length/by sequence divergence. Variants from reference sequence AD169 are marked by asterisks.
DISCUSSION
The 12 HCMV isolates examined were distinguished from one another by variability at 24 repeat regions. Therefore, this study provides a substantial panel of molecular markers with which to investigate evolutionary and epidemiological patterns in this important pathogen. Because the genome of HCMV is smaller than those of the eukaryotes and prokaryotes in which microsatellite variation has been previously utilized, it will be possible to survey virtually all microsatellite variation in HCMV populations. Our results suggest that this will also be a useful approach to the development of molecular markers in similar viruses.
The microsatellites screened here do not yet constitute an exhaustive survey of the HCMV genome; we surveyed only 49% of the total repeats longer than the chosen cutoff (tracts of four trinucleotide, five dinucleotide, or nine mononucleotide repeats), and these excluded the regions of the Toledo genome that are not found in Towne or AD169 (6). Therefore, a variety of additional length polymorphisms may be present throughout the rest of the genome. Examination of these additional repetitive regions will increase the likelihood of finding specific polymorphic markers that are linked to biological or pathological phenotypes.
A single set of primers often flanks multiple short polymorphic iteractions, and sequencing reveals that length variants from different isolates with the same overall length are often made up of more than one sequence (Fig. 2). This phenomenon, in which apparently equivalent length variants can be shown to have different mutational histories, is an example of homoplasy (40), and many cases of homoplasy in microsatellite length variants can be resolved by sequencing (11, 19, 27). Thus, variation in length offers only a first order of discrimination. Sequencing of alleles or viral length variants adds a second level and is necessary to obtain maximum resolution between viral isolates.
Sequencing to confirm length variation at tandem repeats also revealed a variety of point mutations. It has been shown that there are more point mutations in regions around microsatellites than in nonrepetitive regions (9). Targeting of microsatellites therefore permits the detection of more point mutations than would be found if random segments of the genome of the same length were sequenced.
Many of the HCMV repeats are found in noncoding regions and are associated with a high local level of base pair polymorphism. This suggests that these polymorphisms are evolutionarily neutral or nearly so. They are therefore likely to provide appropriate markers for epidemiology and strain identification. Other repeats are found in known or putative coding regions, promoters, operators, and other functional DNA. These may not be neutral and may therefore be limited in utility as markers. These microsatellites, however, may provide adaptive variation important to viral evolution and genetic variability, perhaps similar to the functionally important mononucleotide runs found in VSV (2) and respiratory syncytial virus (18). If changes in these microsatellites are found to directly influence the expression of a gene or function of a gene product, studies of these polymorphisms may provide novel insight into gene regulation in the virus.
While microsatellites are frequently used as markers in mapping, gene function, and population studies of higher eukaryotes (1, 5, 17), the full extent of such repeats has not been determined in any virus. Other classes of repeats have been examined. Many studies have used polymorphic restriction enzyme sites as epidemiological markers. Herpesvirus restriction enzyme polymorphisms have been examined in several studies, some of which have found relatively little variation (3, 45). Some restriction enzyme variants have been of use in distinguishing among different strains of VZV (41) and even in detecting different patterns of herpesvirus evolution in different parts of the world (35). Microsatellites are likely to evolve more quickly than restriction sites. This will make them useful for following changes in pathogen populations over short time scales.
None of the repeats examined in this study are long enough to have been included in a similar survey of a eukaryotic genome for microsatellite length variants (48). Despite previous assumptions that such polymorphisms should be extremely rare, this study demonstrates the potential of very short repeats to provide polymorphic molecular markers. It also reveals an important role for insertion-deletion mutations as a mechanism for producing genetic variation in viral genomes, as previously demonstrated for eukaryotic and prokaryotic genomes. This study provides an important new type of molecular marker with which to investigate questions of epidemiology and evolution within this virus. While these short tandem repeats are shorter than those frequently examined for studies of microsatellite variation, the insertion-deletion events at these repeats clearly classifies them as microsatellites.
ACKNOWLEDGMENTS
This work was supported by grant 1-RO3 AI41370 from the National Institute of Allergy and Infectious Diseases (to C.W.). D.M. is an NSCORT graduate fellow, and D.F. is a Markey graduate fellow.
We are most grateful to Chris Lambros for making this study possible. We thank Frank Huang and Tamarah Westmoreland for assistance.
REFERENCES
- 1.Ashley M, Dow B. The use of microsatellite analysis in population biology: background, methods and potential applications. EXS. 1994;69:185–201. doi: 10.1007/978-3-0348-7527-1_10. [DOI] [PubMed] [Google Scholar]
- 2.Barr J N, Whelan S P J, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black D H, Eberle R. Detection and differentiation of primate alpha-herpesviruses by PCR. J Vet Diagn Investig. 1997;9:225–231. doi: 10.1177/104063879700900301. [DOI] [PubMed] [Google Scholar]
- 4.Bornkamm G, Delius H, Zimber U, Hudewentz J, Epstein M. Comparison of Epstein-Barr virus strains of different origin by analysis of the viral DNAs. J Virol. 1980;35:603–618. doi: 10.1128/jvi.35.3.603-618.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caskey C, Pizzuti A, Fu Y H, Fenwick R G J, Nelson D L. Triplet repeat mutations in human disease. Science. 1992;256:784–789. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- 6.Cha T, Tom E, Kemble G, Duke G, Mocarski E, Spaette R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee M, Bankier A, Beck S, Bohni R, Brown C, Cerny R, Horsnell T, Hutchison C, Kouzarides T, Martignetti J, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 8.Di Rienzo A, Peterson A C, Garza J C, Valdes A M, Slatkin M, Freimer N B. Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA. 1994;91:3166–3170. doi: 10.1073/pnas.91.8.3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djian P, Hancock J, Chana H. Codon repeats in genes associated with human diseases: fewer repeats in the genes of nonhuman primates and nucleotide substitutions concentrated at the sites of reiteration. Proc Natl Acad Sci USA. 1996;93:417–421. doi: 10.1073/pnas.93.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duke G M, Osorio J E, Palmenberg A C. Attenuation of Mengo virus through genetic engineering of the 5′ noncoding poly(C) tract. Nature. 1990;343:474–476. doi: 10.1038/343474a0. [DOI] [PubMed] [Google Scholar]
- 11.Estoup A, Tailliez C, Cornuet J M, Solignac M. Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae) Mol Biol Evol. 1995;12:1074–1084. doi: 10.1093/oxfordjournals.molbev.a040282. [DOI] [PubMed] [Google Scholar]
- 12.Falk K, Gratama J, Rowe M, Zou J, Khanim F, Young L, Oosterveer M. The role of repetitive DNA sequences in the size variation of Epstein-Barr virus (EBV) nuclear antigens, and the identification of different EBV isolates using RFLP and PCR analysis. J Gen Virol. 1995;76:779–790. doi: 10.1099/0022-1317-76-4-779. [DOI] [PubMed] [Google Scholar]
- 12a.Field, D.dfield@ucsd.edu.
- 13.Field D, Metzgar D, Eggert L, Rose R, Wills C. The use of short, clustered polymorphic microsatellite loci to distinguish strains of Candida albicans. FEMS Lett. 1996;15:73–79. doi: 10.1111/j.1574-695X.1996.tb00056.x. [DOI] [PubMed] [Google Scholar]
- 14.Field D, Wills C. Abundant microsatellite polymorphisms in S. cerevisiae and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae results from strong mutation pressures and a variety of selection forces. Proc Natl Acad Sci USA. 1998;95:1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field D, Wills C. Long, polymorphic microsatellites in simple organisms. Proc R Soc Lond. 1996;263:209–215. doi: 10.1098/rspb.1996.0033. [DOI] [PubMed] [Google Scholar]
- 16.Foster P L, Trimarchi J M. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freimer N B, Slatkin M. Microsatellites: evolution and mutational processes. Variation in the human genome. Ciba Found Symp. 1996;197:51–72. doi: 10.1002/9780470514887.ch4. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Barreno B, Delgado T, Melero J A. Oligo(A) sequences of human respiratory syncytial virus G protein gene: assessment of their genetic stability in frameshift mutants. J Virol. 1994;68:5460–5468. doi: 10.1128/jvi.68.9.5460-5468.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garza J C, Freimer N B. Homoplasy for size at microsatellite loci in humans and chimpanzees. Genome Res. 1996;6:211–217. doi: 10.1101/gr.6.3.211. [DOI] [PubMed] [Google Scholar]
- 20.Gergely L, Sternas L, Dillner J, Klein G. Molecular size variation of EBNA is determined by the EB viral genome. Intervirology. 1984;22:85–96. doi: 10.1159/000149538. [DOI] [PubMed] [Google Scholar]
- 21.Given D, Kieff E. DNA of Epstein-Barr virus. VI. Mapping of the internal tandem reiteration. J Virol. 1979;31:315–324. doi: 10.1128/jvi.31.2.315-324.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn H, Palemberg A C. Encephalomyocarditis viruses with short poly(C) tracts are more virulent than their mengovirus counterparts. J Virol. 1995;69:2697–2699. doi: 10.1128/jvi.69.4.2697-2699.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock J M, Chaleeprom W, Chaleeprom W, Dale J, Gibbs A. Replication slippage in the evolution of potyviruses. J Gen Virol. 1995;76:3229–3232. doi: 10.1099/0022-1317-76-12-3229. [DOI] [PubMed] [Google Scholar]
- 24.Hayward S, Nogee L, Hayward G. Organization of repeated regions within the Epstein-Barr virus DNA molecule. J Virol. 1980;33:507–521. doi: 10.1128/jvi.33.1.507-521.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruglyak S, Durrett R, Schug M, Aquadro C. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc Natl Acad Sci USA. 1998;95:10774–10778. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzgar D, van Belkum A, Field D, Haubrich R, Wills C. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J Clin Microbiol. 1998;36:2308–2313. doi: 10.1128/jcm.36.8.2308-2313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzgar D, Field D, Haubrich R, Wills C. Sequence analysis of a compound coding-region microsatellite in Candida albicans resolves homoplasies and provides a high-resolution tool for genotyping. FEMS Immunol Med Microbiol. 1998;20:103–109. doi: 10.1111/j.1574-695X.1998.tb01116.x. [DOI] [PubMed] [Google Scholar]
- 28.Moxon E, Rainey P, Nowak M, Lenski R. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 29.Moxon R, Wills C. DNA microsatellites: agents of evolution? Sci Am. 1999;280:94–99. doi: 10.1038/scientificamerican0199-94. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura Y, Leppert M, O’Connell P, Wolff R, Holm T, Culver M, Martin C, Fujimoto E, Hoff M, Kumlin E, White R. Variable number tandem repeat (VNTR) markers for human gene mapping. Science. 1987;235:1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- 31.Perdue M L, Garcia M, Senne D, Fraire M. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 1997;49:173–186. doi: 10.1016/s0168-1702(97)01468-8. [DOI] [PubMed] [Google Scholar]
- 32.Plotkin S A, Farukama T, Zygraich N, Huggelen C. Candidate cytomegalovirus strain for human vaccination. Infect Immun. 1975;12:521–527. doi: 10.1128/iai.12.3.521-527.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg S M, Longerich S, Gee P, Harris R S. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 34.Rowe E A, Hartley J W, Waterman S, Turner H C, Huebner R J. Cytopathic agent resembling common salivary gland virus recovered from tissue cultures of human adenoids. Proc Soc Exp Biol Med. 1956;92:418. [PubMed] [Google Scholar]
- 35.Sakaoka H, Kurita K, Iida Y, Takada S, Umene K, Kim Y T, Ren C S, Nahmias A J. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: studies of molecular evolution and molecular epidemiology of the virus. J Gen Virol. 1994;75:513–527. doi: 10.1099/0022-1317-75-3-513. [DOI] [PubMed] [Google Scholar]
- 36.Schlotterer C, Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992;20:211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sidagis J, Ueno K, Tokunaga M, Ohyama M, Eizuru Y. Molecular epidemiology of Epstein-Barr virus (EBV) in EBV-related malignancies. Int J Cancer. 1997;72:72–76. doi: 10.1002/(sici)1097-0215(19970703)72:1<72::aid-ijc11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 38.Smith I L, Cherrington J M, Jiles R E, Fuller M D, Freeman W R, Spector S A. High-level resistance of cytomegalovirus to glanciclovir is associated with alterations in both UL97 and DNA polymerase genes. J Infect Dis. 1997;176:69–77. doi: 10.1086/514041. [DOI] [PubMed] [Google Scholar]
- 39.Spector S. Spectrum and treatment of cytomegalovirus disease in persons with AIDS. J Int Soc Phys AIDS Care. 1996;2:9–22. [PubMed] [Google Scholar]
- 40.Stewart C-B. The powers and pitfalls of parsimony. Nature. 1993;361:603–617. doi: 10.1038/361603a0. [DOI] [PubMed] [Google Scholar]
- 41.Takayama M, Takayama N, Inoue N, Kameoka Y. Application of long PCR method of identification of variations in nucleotide sequences among varicella-zoster virus isolates. J Clin Microbiol. 1996;34:2869–2874. doi: 10.1128/jcm.34.12.2869-2874.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tautz D, Schlotterer C. Simple sequences. Curr Opin Genet Dev. 1994;4:832–837. doi: 10.1016/0959-437x(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 43.Tran H, Keen J, Kricker J, Resnick M, Gordenin D. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umene K, Watson R, Enquist L. Tandem repeated DNA in an intergenic region of Herpes simplex virus type 1 (Patton) Gene. 1984;30:33–39. doi: 10.1016/0378-1119(84)90102-1. [DOI] [PubMed] [Google Scholar]
- 45.Umene K, Yoshida M. Genomic characterization of two predominant genotypes of herpes simplex virus type 1. Arch Virol. 1993;131:29–46. doi: 10.1007/BF01379078. [DOI] [PubMed] [Google Scholar]
- 46.Vaisanen M, Haataja R, Leisti J. Decrease in the CGGn trinucleotide repeat mutation of the fragile X syndrome to normal size range during parental transmission. Am J Hum Genet. 1996;59:540–546. [PMC free article] [PubMed] [Google Scholar]
- 47.Valdes A, Slatkin M, Freimer N. Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber J L. Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics. 1990;524:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- 49.Webster R G, Bean W, Gorman O T, Kawaoka Y. Ecology of avian influenza viruses. Microbiol Rev. 1992;56:151–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsitsumi M, Shimomura H, Tsuji T, Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]