Abstract
Ubiquitination, a pivotal posttranslational modification of proteins, plays a fundamental role in regulating protein stability. The dysregulation of ubiquitinating and deubiquitinating enzymes is a common feature in various cancers, underscoring the imperative to investigate ubiquitin ligases and deubiquitinases (DUBs) for insights into oncogenic processes and the development of therapeutic interventions. In this review, we discuss the contributions of the ubiquitin–proteasome system (UPS) in all hallmarks of cancer and progress in drug discovery. We delve into the multiple functions of the UPS in oncology, including its regulation of multiple cancer-associated pathways, its role in metabolic reprogramming, its engagement with tumor immune responses, its function in phenotypic plasticity and polymorphic microbiomes, and other essential cellular functions. Furthermore, we provide a comprehensive overview of novel anticancer strategies that leverage the UPS, including the development and application of proteolysis targeting chimeras (PROTACs) and molecular glues.
Keywords: Ubiquitination, Cancer hallmarks, Molecular mechanisms, Targeted therapies, Immunotherapies
Introduction
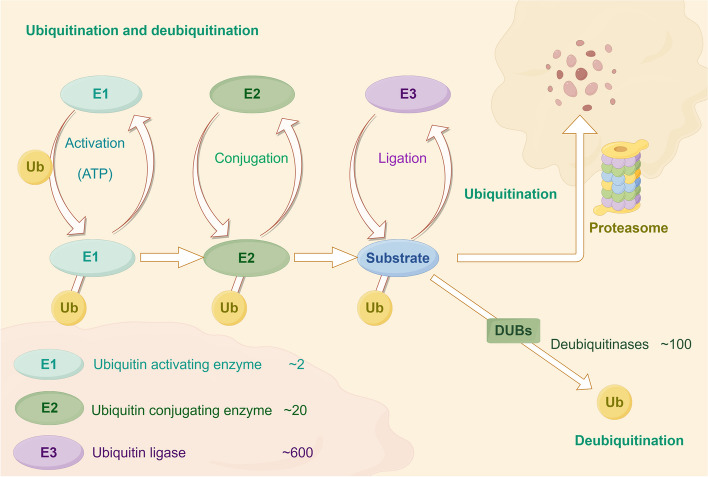
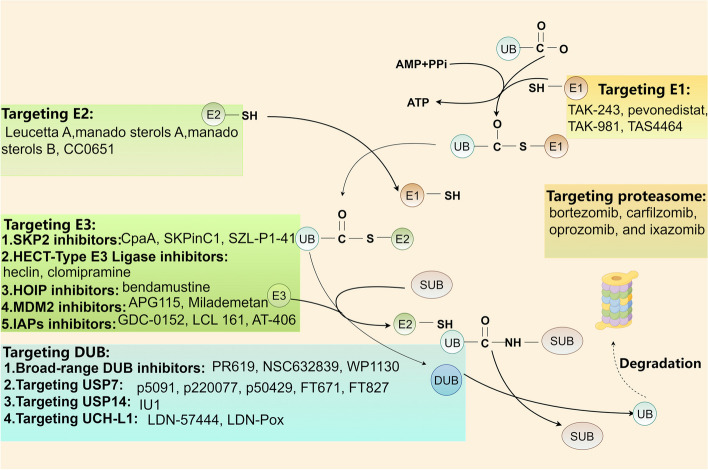
Ubiquitin (Ub) is composed of 76 amino acids and is found in all eukaryotic tissues [1]. Ubiquitination is the second most common posttranslational modification of proteins following phosphorylation [2]. Ubiquitination is a highly specific process of ATP-dependent cascade labeling substrate proteins with ubiquitin [3]. Moreover, ubiquitin and its degradation by the proteasome constitute the ubiquitin–proteasome system (UPS), which is responsible for 80–90% of cellular proteolysis and 10–20% of autophagy [4]. The ubiquitination modification involves a series of reactions mediated by a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3) (Fig. 1) [5]. Ubiquitination plays a crucial regulatory role in the modulation of tumors, impacting cellular survival, proliferation, and differentiation. At the same time, ubiquitination is reversible, and ubiquitin or ubiquitin chains linked to substrate proteins can be removed by deubiquitinases (DUBs).
Fig. 1.
The processes of ubiquitination and deubiquitination occur within the ubiquitin–proteasome system (UPS). Ubiquitination involves the sequential action of three enzyme classes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin-protein ligases (E3). Initially, E1 enzymes activate ubiquitin (Ub) in an ATP-dependent process. Subsequently, the activated Ub is transferred to E2 enzymes via a thioester bond. The final step is catalyzed by E3 ligases, which facilitate the transfer of Ub from E2 to the target substrate protein, marking it for degradation
The emerging functions of ubiquitination and deubiquitination in regulating cancer hallmarks, including “evading growth suppressors,” “reprogramming energy metabolism,” “unlocking phenotypic plasticity,” “polymorphic microbiomes,” and “senescent cells,” have been revealed [6, 7]. The UPS can regulate the protein levels of programmed cell death 1/programmed cell death ligand 1 (PD-1/PD-L1) in the tumor microenvironment (TME) and enhance the effectiveness of immunotherapy [8]. For example, ubiquitin-specific protease 2 (USP2), a DUB, can stabilize PD-1 and promote tumor immune escape through deubiquitination [9]. Moreover, the UPS also regulates tumor metabolic reprogramming. Recent investigations have revealed that the E3 ligase Parkin can facilitate the ubiquitination of pyruvate kinase M2 (PKM2) [10]. In addition, the DUB OTU domain-containing ubiquitin aldehyde-binding protein 2 (OTUB2) interacts with PKM2 to inhibit PKM2 ubiquitination by the E3 ligase Parkin, enhancing glycolysis and accelerating colorectal cancer progression [11]. Many new reports have recently revealed novel ubiquitination methods for cancer treatment, such as PROTACs and molecular glues. PROTAC technology is a valuable platform for driving the degradation of target proteins. ARV-110 (alias bavdegalutamide) and ARV-471 (alias vepdegestrant) represent the forefront of PROTAC drug development in clinical trials and have progressed to phase II trials [12]. Compared to PROTACs, molecular glues have smaller molecular dimensions, simplifying the optimization of their chemical characteristics. A few molecular glue degradants have been identified. Notably, CC-90009 facilitates the ubiquitination-mediated degradation of G1-to-S phase transition 1 (GSPT1) by recruiting the E3 ligase complex CUL4-DDB1-CRBN-RBX1 (CRL4CRBN). It is in phase II clinical trials for leukemia therapy [13]. ARV-110 is designed to selectively target and bind to the androgen receptor (AR), facilitating its degradation by recruiting an E3 ubiquitin ligase. Early data from the first-in-human phase I study revealed the safety and tolerability of ARV-110 in patients diagnosed with metastatic castration-resistant prostate cancer [14]. Our research team recently identified a drug that promotes protein ubiquitination and degradation. Indomethacin, for instance, diminishes the growth and recurrence of esophageal squamous cell carcinoma (ESCC) by enhancing the E3 ligase synovial apoptosis inhibitor 1 (SYVN1)-mediated ubiquitination of integrin αv (ITGAV) [15]. We also discovered that honokiol directly interacts with keratin 18 (KRT18), inhibiting melanoma growth by inducing KRT18 ubiquitination and degradation [16].
In this review, we integrate the ubiquitination and deubiquitination processes with the 14 hallmarks of cancer. We clarify the fundamental mechanisms and functions of ubiquitination and deubiquitination in tumor suppression, metabolic reprogramming, immune evasion, phenotypic plasticity, polymorphic microbiomes, and other essential cellular functions, focusing on recent developments. Finally, we explore the therapeutic potential of targeting the UPS in cancer therapy.
Functions of various types of ubiquitination in cancer
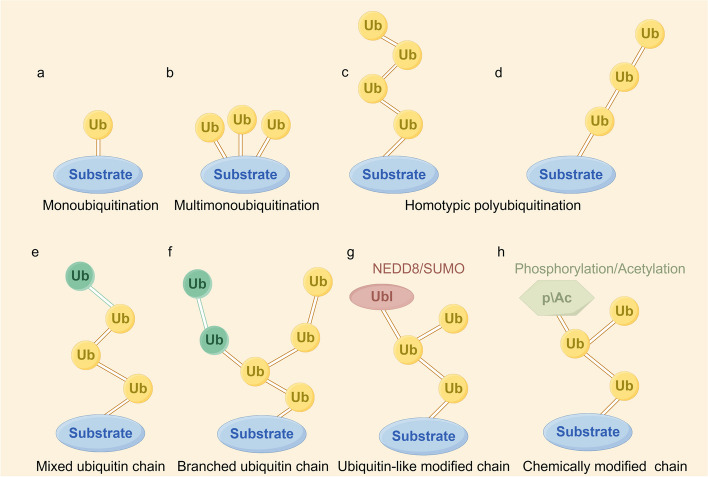
Ubiquitination can be categorized into monoubiquitination, multimonoubiquitination, homotypic polyubiquitination, and heterotypic polyubiquitination (Fig. 2) [4]. Monoubiquitination refers to the attachment of a single ubiquitin protein to a substrate protein. When multiple ubiquitin proteins are attached to different lysine residues on the same substrate protein, it is termed multimonoubiquitination. Ubiquitin itself contains eight potential linkage sites, which include seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) and one N-terminal methionine residue (M1) [17]. These sites allow for the formation of polyubiquitin chains through further ubiquitin attachment. Homotypic polyubiquitination occurs when ubiquitin proteins are linked through the same residue type, creating a uniform chain [4]. In contrast, heterotypic polyubiquitination involves ubiquitin proteins linked through different residue types, resulting in mixed or branched chains [18]. When more than one ubiquitin molecule is simultaneously linked to a single ubiquitin molecule, the resulting ubiquitin chain is referred to as a branched ubiquitin chain [18]. A family of proteins, known as ubiquitin-like proteins (UBLs), shares structural and functional characteristics with ubiquitin, playing similar roles in modulating protein activity and cellular processes. This family encompasses proteins such as neural precursor cell-expressed developmentally downregulated 8 (NEDD8), small ubiquitin-related modifier (SUMO), and interferon-stimulated gene 15 (ISG15) [17]. A ubiquitin-like modified chain means that the substrate or ubiquitin is modified by a ubiquitin-like protein. In addition, ubiquitin can also be posttranslationally modified through phosphorylation and acetylation, which is called a chemically modified ubiquitin chain. The formation of mixed ubiquitin chains, branched ubiquitin chains, ubiquitin-like modified chains, and chemically modified ubiquitin chains are collectively referred to as heterotypic polyubiquitination [4].
Fig. 2.
The various types of ubiquitin (Ub) linkages are as follows. a Mono-ubiquitination: A single ubiquitin protein is attached to a substrate protein. b Multi-monoubiquitination: Multiple ubiquitin proteins are each linked to different sites on the same substrate protein. c Homotypic polyubiquitination: Ubiquitin can bind to another ubiquitin through one of its seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) or the N-terminal methionine residue (M1). Multiple identical ubiquitin proteins form a chain, which is then attached to a substrate protein. d Linear ubiquitination: A specific form of homotypic polyubiquitination where ubiquitin molecules are connected via Met1 linkages. e Mixed ubiquitin chain: A ubiquitin can be linked by two or more different connection methods within the same polymerization reaction, resulting in mixed ubiquitin chains. f Branched ubiquitin chain: Ubiquitin proteins in a chain are modified by adding more ubiquitin proteins at different binding sites. g Ubiquitin-like modified chain: A ubiquitin protein in the chain is linked to a ubiquitin-like protein. h Chemically modified chain: Ubiquitin proteins in the chain are modified by other protein modifications, such as phosphorylation or acetylation. The formation of mixed ubiquitin chains, branched ubiquitin chains, ubiquitin-like modified chains, and chemically modified ubiquitin chains are collectively referred to as heterotypic polyubiquitination
Monoubiquitination
Previous studies have shown that monoubiquitination of proteins acts as a signal for DNA repair, signal transduction, and phagocytosis in vivo [19, 20]. Recently, histone monoubiquitination has been widely studied. Histone monoubiquitination often occurs on H2A and H2B. Ring finger protein 2 (RNF2) is an E3 ligase with a RING domain [21]. RNF2 facilitates the monoubiquitination of histone H2A at lysine 119, leading to the recruitment of E-cadherin to the promoter region and subsequent transcriptional repression of E-cadherin. This mechanism contributes to enhancing the metastatic potential of hepatocellular carcinoma [22]. In addition, ubiquitin-conjugating enzyme E2T (UBE2T) regulates the monoubiquitination of the histone variant H2AX (γH2AX). This process induces the phosphorylation of cell cycle checkpoint kinase 1 (CHK1), thereby enhancing the radioresistance of hepatocellular carcinoma cells [23]. Monoubiquitination also plays an essential role in immune escape. Metastasis suppressor protein 1 (MTSS1) promotes the monoubiquitination of the immune checkpoint PD-L1 at K263 mediated by the E3 ligase atrophin-interacting protein 4 (AIP4), which leads to the internalization of PD-L1, endosomal transport, and lysosomal degradation, thus inhibiting the immune escape of lung adenocarcinoma [24]. Additionally, the ubiquitin-binding enzyme E2B (UBE2B) can facilitate the monoubiquitination of the transcription regulator zinc finger MYM-type protein 2 (ZMYM2) mediated by the ubiquitin E3 ligase ring finger protein 73 (RNF73), thereby promoting the growth of ovarian cancer [25]. These observations suggest that the monoubiquitination of proteins primarily affects the growth, metastasis, radiation resistance, and immune escape of cancer cells by affecting DNA repair and gene transcription.
Linear ubiquitination
The ubiquitin chains assembled by M1 are called linear ubiquitin chains. These chains are assembled exclusively by the E3 ligase linear ubiquitin chain assembly complex (LUBAC) and are disassembled by OTU deubiquitinase with linear linkage specificity (OTULIN) and cylindromatosis (CYLD) [26, 27]. LUBAC consists of HOIL-1 interacting protein (HOIP), heme-oxidized IRP2 ubiquitin ligase 1 (HOIL-1L), and SHANK-associated RH domain-interacting protein (SHARPIN) [28]. The impact of linear ubiquitin chains on cancer has been extensively investigated. Met1-Ub signaling plays a vital role in many aspects of cancer through NF-κB regulation. HOIP promotes lymphoma by activating NF-κB signal transduction, indicating that LUBAC is a viable therapeutic target for B-cell lymphoma [29]. Epsin, a member of the ubiquitin-binding endocytosis adaptor protein family, engages with the linear ubiquitin chain assembly complex (LUBAC) to facilitate the linear ubiquitination of the NF-κB essential modulator (NEMO). This interaction is implicated in the progression of breast cancer [30]. Protein kinase transforming growth factor β-activated kinase 1 (TAK1) is the main mediator of NF-κB activation in the LUBAC-dependent mechanism. Targeting LUBAC or TAK1 may be an attractive therapeutic strategy for A20-mutant Hodgkin’s lymphoma [31]. RANBP2-type and C3HC4-type zinc finger containing 1 (RBCK1) was first identified as an essential component of LUBAC and promoted NF-κB signal transduction during the immune response [32]. Furthermore, the phosphorylation of OTULIN facilitates the activation of the genotoxic Wnt/β-catenin pathway, thereby augmenting drug resistance in breast cancer [28]. Consequently, the Met1-linked linear ubiquitin chain acts as an essential positive modulator of NF-κB signaling pathways, playing pivotal roles in oncogenesis, inflammation, and immune regulation.
K48-linked polyubiquitination
K48-linked polyubiquitination is the most widely studied type and the main connection type in cells. It mainly marks proteins that are recognized and degraded by the 26S proteasome and targets proteins for proteasomal degradation [18]. The E3 ligase tripartite motif protein 7 (TRIM7) can directly interact with the tyrosine kinase Src, induce the ubiquitination of Lys48-linked Src, reduce the abundance of the Src protein in hepatocellular carcinoma cells, and inhibit the progression of hepatocellular carcinoma [33]. Recently, a new circRNA involved in hypoxia reactions named circular insulin-induced gene 1 (circINSIG1) was identified. CircINSIG1 encodes the protein circINSIG1-121, which has 121 amino acids. By recruiting the E3 ligase CUL5-ASB6 complex, circINSIG1-121 promotes the ubiquitination of the critical cholesterol metabolism regulator INSIG1 at the K48 linkage of lysine 156 and lysine 158, thus inducing cholesterol biosynthesis and promoting colorectal cancer proliferation and metastasis [34]. In addition, the E3 ligase MG53 catalyzes the K48-linked ubiquitination and subsequent degradation of cyclin D1, thus inhibiting the growth of colorectal cancer [35]. However, in the ubiquitinating enzyme family, studies have shown that methyltransferase 5, N6-adenosine (METTL5) regulates the translation of USP5 and suppresses K48-linked ubiquitination of c-Myc, thus reprogramming glucose metabolism and promoting the progression of hepatocellular carcinoma [36]. Therefore, as the most widely studied ubiquitination form, K48-linked polyubiquitination plays a key role in various aspects of cancer by promoting the degradation of corresponding proteins.
K63-linked polyubiquitination
K63-linked polyubiquitination participates in signal assembly and promotes the autophagic degradation of protein substrates. It can also regulate nondegradative processes, such as protein transport, DNA repair, and protein kinase activation [37]. The AB22A-NeoF1 fusion gene encodes the Rab22a-NeoF1 fusion protein, which coordinates various mechanisms to facilitate lung metastasis in osteosarcoma [38]. The E3 ligase STIP1 homology and U-box-containing protein 1 (STUB1) catalyzes the K63-linked ubiquitination of K112 of the Rab22a-NeoF1 fusion protein, which promotes the lung metastasis of osteosarcoma [39]. K63-linked polyubiquitination also plays an important role in immune escape. For instance, the E3 ligase TRIM28 promotes the K63-linked ubiquitination of TANK-binding kinase 1 (TBK1). It activates the TBK1-IRF1 and TBK1-mTOR pathways, thus enhancing the transcription of PD-L1 and promoting the escape of gastric cancer cells from immune surveillance [40]. In addition, mind bomb homolog 2 (MIB2) catalyzes the ubiquitination of PD-L1 at the K63 linkage, but not its degradation, and promotes tumor immune escape [41]. In addition, anillin (ANLN) is a mitotic protein that can promote the formation of contractile rings and cell division. The results showed that USP10 removes the K11- and K63-linked ubiquitin chains of ANLN through its ubiquitinating enzyme activity and prevents the ubiquitin-mediated degradation of ANLN, effectively inhibiting the cell cycle procession of ESCC [42]. Taken together, K63-linked polyubiquitination plays an important role in cancer metastasis, immune escape, and the cell cycle.
Other types of polyubiquitination
Relatively few modified substrates and functions of “atypical” ubiquitin chains (K6, K11, K27, K29, K33, and M1 chains) are known [43, 44]. The ubiquitination of K11 is mainly related to UBE2S. Previous research revealed that UBE2S stabilizes β-catenin via K11-linked ubiquitination, contributing to the development of colorectal cancer [45]. In addition, UBE2S interacts with TRIM21, which degrades lipoma preferred partner (LPP) through ubiquitination linked with K11 and promotes the lymphatic metastasis of bladder cancer [46]. The K29-linked ubiquitin chain plays a significant role in driving cancer invasion and metastasis and in the positive regulation of immunity [47]. Recent studies have demonstrated that ring finger protein 167 (RNF167) activates mTORC1 and promotes the occurrence of breast cancer by targeting and degrading K29-linked ubiquitinated cytosolic arginine sensor for mTORC1 subunit 1 (CASTOR1). In addition, this observation confirmed that RNF167 is a therapeutic target of breast cancer [48].
In addition to eight homotypic polyubiquitination modifications, heterotypic polyubiquitination modifications also occur widely in cells [18]. These modifications predominantly involve mixed and branched polyubiquitination, characterized by the formation of polyubiquitin chains on substrates that feature two distinct types of lysine linkages, resulting in complex ubiquitin chain configurations [49]. Poly(A)-binding protein, cytoplasmic 1 (PABPC1), is an extensively studied protein, and recent research has revealed its involvement in the tumorigenesis of numerous cancers. CDC2-like kinase 2 (CLK2) is a bispecific kinase, that facilitates the phosphorylation of diverse proteins, and an increasing amount of data indicate that CLK2 functions as an oncogenic kinase [50]. USP10 can reverse K27/29-linked ubiquitination of PABPC1 and upregulate the translation of CLK2, thus promoting tumor development of pancreatic ductal adenocarcinoma (PDAC) [51].
Ubiquitination and deubiquitination regulate the hallmarks of cancer
Sustained proliferative signaling
The ability to maintain cancer cell proliferation is a fundamental characteristic of cancer cells. Normal cells can control the production and release of growth-promoting signals. However, cancer cells can escape the control of these signals and obtain sustained proliferative stimulation (Fig. 3) [6].
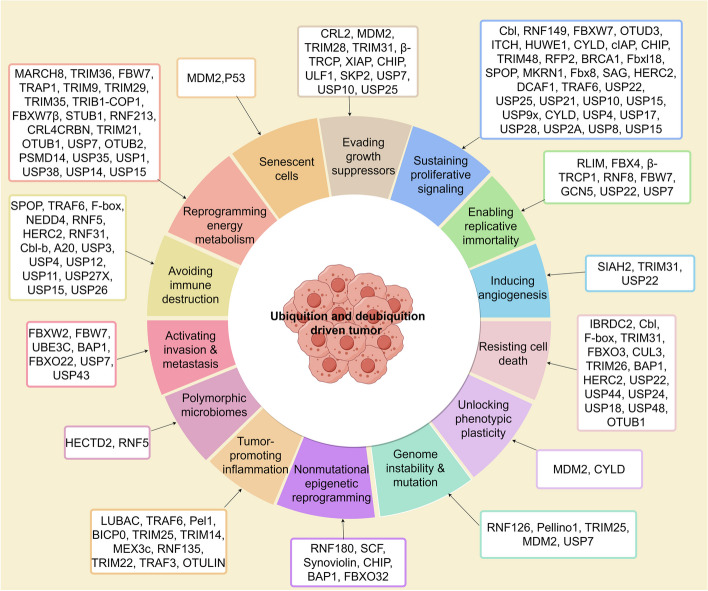
Fig. 3.
Ubiquitination and deubiquitination regulation of the hallmarks of cancer. E3 ubiquitin ligases and deubiquitinating enzymes, by regulating the degradation and stability of proteins, significantly influence the hallmarks of malignant tumors, which include sustained proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, genome instability and mutation, tumor-promoting inflammation, reprogramming energy metabolism, evading immune destruction, unlocking phenotypic plasticity, nonmutational epigenetic reprogramming, polymorphic microbiomes, and senescent cells. Each cancer hallmark-associated E3 ligase and deubiquitinating enzyme (DUB) is listed in the corresponding box
Epidermal growth factor receptor (EGFR) signaling pathway
The EGFR-dependent signaling pathway maintains cell proliferation, and its dysregulation increases cancer cell proliferation [52, 53]. EGFRvIII is the most common mutation in gliomas [54]. Tumors with EGFRvIII mutations exhibit an increase in phosphorylation at Y371 of casitas B-lineage lymphoma (Cbl), a critical residue regulating E3 activity. The conformational changes in Cbl are essential for EGFR ubiquitination in vitro. EGFRvIII mutations can reduce EGFR protein degradation in tumors by inhibiting Cbl activation (Table 1) [55, 56]. It was indicated that β-Element, a traditional Chinese herb, played an anti-tumor and anti-metastatic role in multidrug-resistant (MDR) gastric cancer by suppressing EGFR levels through Cbl-b upregulation [57]. Additionally, a high expression level of Cbl-b is significantly associated with improved prognosis in patients with lung adenocarcinoma, suggesting its potential as a prognostic biomarker for better clinical outcomes [58]. Recent studies have reported that the overexpression of DUBs such as Cezanne-1, USP22, and USP25 in cancer cells prevents EGFR degradation [59–61]. In clinical studies, Cezanne-1 is often amplified in tumor samples from breast cancer patients, with elevated levels of its mRNA associated with a poor prognosis [62]. Similarly, there is a significant association between USP22 levels and poor prognosis in neuroblastoma [63]. Notably, an in vivo study suggested that USP25/28 inhibitor showed a potent anti-tumor effect on pancreatic cell-derived xenograft (CDX) mouse model, implying combining DUB inhibitors with EGFR inhibitors or chemotherapeutic agents may enhance therapeutic efficacy [64]. In conclusion, these observations suggest that E3 ligases and DUBs regulate EGFR signaling, opening new avenues for targeted therapeutic strategies against cancer cell proliferation.
Table 1.
E2 enzymes, E3 ligases, and DUBs in the regulation process of cancer hallmarks
| Cancer hallmarks | E2s/E3s/DUBs | Name | Substrate | Cancer type | Outcomes | References |
|---|---|---|---|---|---|---|
| 1.Sustaining proliferative signaling | E3s | RNF7 | PI3K/AKT signaling pathway | Pancreatic cancer | Activates the PI3K/AKT signaling pathway and promotes tumor formation | [65] |
| RNF41 | HER3 | Breast cancer | Degrades HER3 and promotes tumor proliferation | [66] | ||
| MARCH6 | DHX9 | Primary papillary thyroid cancers | Activates AKT/mTOR signaling pathway and promotes tumor proliferation and metastasis | [67] | ||
| ANKRD9 | IMPDH | Gastric cancer | Promotes ubiquitination and proteasomal degradation of IMPDH to suppress tumor growth | [68] | ||
| Cbl | EGFR | Lung cancer | Promotes tumor growth | [55, 56] | ||
| MKRN1 | PTEN | Cervical cancer | Ubiquitinates PTEN protein and promotes cancer proliferation | [69] | ||
| BCR-ABL | SHIP | Hematopoietic tumors | Promotes SHIP proteasomal degradation | [70] | ||
| FBX8 | mTOR | Colorectal cancer | Targets mTOR for degradation | [71] | ||
| SAG | PHLPP1/DEPTOR | Prostate cancer | Inactivates the PI3K/AKT/mTOR axis | [72] | ||
| MULAN | AKT | Head and neck cancer | Promotes the ubiquitination and degradation of AKT1 and AKT2 | [73] | ||
| RFP2 | AKT/MDM2 | Multiple myeloma | Degrades AKT and MDM2 | [74] | ||
| BRCA1 | AKT | Breast cancer | Ubiquitinates and directly degrades AKT1 | [75] | ||
| CHIP | AKT | Cervical Cancer | Ubiquitinates and directly degrades ASK1 | [76] | ||
| SKP2 | AKT | Breast cancer | Promotes tumor occurrence and metastasis | [77] | ||
| TRAF4 | AKT | Lung cancer | Promotes tumorigenesis | [78] | ||
| FBXL18 | AKT | Glioma | Promotes tumor proliferation and development | [79] | ||
| DUBs | USP17/USP4 | PDFGRβ | Osteosarcoma | Promotes aberrant STAT3 transcription | [80] | |
| USP15 | ERα | Breast cancer | Blocks the ubiquitination and degradation of ERα | [81] | ||
| USP4 | TAK1 | Esophageal squamous cell carcinoma | Stabilizes the TAK1 protein level | [82] | ||
| USP7 | ERα | Breast cancer | Deubiquitinates ERα and promotes tumor proliferation | [83] | ||
| Ataxin-3 | PTEN | Lung cancer | Restricts PTEN transcription | [84] | ||
| USP13 | PTEN | Breast cancer | Deubiquitinates PTEN | [85] | ||
| USP10 | PTEN | Lung cancer/ hepatocellular carcinoma | Upregulates PTEN | [86] | ||
| USP10 | PTEN | Lung cancer | Inhibits lung cancer cell growth and invasion by upregulating PTEN | [87] | ||
| OTUD3 | PTEN | Breast cancer | Upregulates PTEN and suppresses tumorigenesis | [88] | ||
| USP46 | PHLPP1 | Colon cancer | Functions as a tumor suppressor by controlling PHLPP-dependent attenuation of AKT signaling | [89] | ||
| USP1 | PHLPP1 | Lung cancer | Regulates AKT phosphorylation by modulating the stability of PHLPP1 | [90] | ||
| USP21 | MEK2 | Hepatocellular carcinoma | Maintains MEK2 stability and activates ERK signaling | [91] | ||
| USP12/WRD48 | PHLPP1 | Colon cancer | Suppresses AKT-dependent cell survival signaling by stabilizing PHLPP1 | [92] | ||
| USP12/UAF-1/WDR20 | PHLPP1 | Prostate cancer | Regulates the interaction between the androgen receptor and the AKT pathway | [93] | ||
| 2. Evading growth suppressors | E3s | MDM2 | p53 | Multiple cancers | Promotes tumor development | [94] |
| TRIM28 | p53 | Lung cancer | Promotes tumor development | [95] | ||
| TRIM31 | p53 | Pancreatic cancer/Lung cancer | Promotes tumor development | [96] | ||
| TRIM28 | RB | Multiple cancers | Promotes tumor development | [95] | ||
| CRL2 | ARF | Multiple cancers | Promotes tumor development | [97] | ||
| ULF1 | ARF | Hepatocarcinoma | Promotes tumor development | [98] | ||
| Β-TRCP | β-catenin | Multiple cancers | Promotes tumor development | [99] | ||
| F-box | Β-catenin | Colorectal cancer | Promotes tumor development | [100] | ||
| SKP2 | NBS1 | Prostate cancer | Promotes tumor development | [101] | ||
| CUL1 | CHK2 | Breast cancer/Ovarian cancer | Promotes DDR | [102] | ||
| DUBs | USP10 |
p53 /ARF |
Multiple cancers | Inhibits tumor development | [98, 103] | |
| USP7 | p53 | Multiple cancers | Inhibits tumor development | [104] | ||
| 3. Resisting cell death | E3s | IBRDC2 | BAX | Colorectal cancer | Promotes apoptosis | [105, 106] |
| Cbl | BimEL | Multiple cancers | Inhibits apoptosis | [105] | ||
| Peli1 | RIPK1 | Breast cancer/Lung cancer/Lymphoma | Promotes apoptosis | [107] | ||
| TRIM31 | NLRP3 | Colitis-associated cancer | Promotes NLRP3 inflammasome | [96, 108] | ||
| CUL3 | BECN1 | Breast cancer/Ovarian cancer |
Promotes autophagy Promotes tumor development |
[109] | ||
| TRIM26 | SLC7A11 | Liver cancer | Promotes tumor development | [110] | ||
| DUBs | A20 | RIPK3 | Colorectal cancer | Promotes necroptosis | [105, 111] | |
| USP22 | RIPK3 | Colorectal cancer | Promotes necroptosis | [105, 111] | ||
| USP24 | GSDMB | Bladder cancer | Inhibits pyroptosis | [112] | ||
| USP48 | GSDME | Pancreatic cancer | Promotes pyroptosis | [113] | ||
| OTUB1 | GPX4 | Gastric cancer | Inhibits ferroptosis | [114] | ||
| 4. Enabling replicative immortality | E3s | Rlim | TRF1 | Renal cell carcinoma | Binds to the region between TRFl dimerization and Myb domains to promote tumor growth | [115] |
| FBX4 | TRF1 | Lung cancer | Binds to the N-terminal region of the TRFH dimerization domain of free TRF1 to promote tumor growth | [116] | ||
| β-TrCP1 | TRF1 | Leukemia | Ubiquitinates TRF1 and promotes tumor growth | [117] | ||
| FBW7 | TPP1 | Lung cancer | Ubiquitinates TPP1 and promotes tumor growth | [118] | ||
| SIAH1 | TRF2 | Colorectal cancer | Targets TRF2 degradation promotes tumor proliferation | [119] | ||
| DUBs | USP7 | TPP1 | Lung cancer | Promotes tumor growth | [120] | |
| 5. Inducing angiogenesis | E3 | SIAH2 | NRF-1 | Breast cancer | Decreased activity of SIAH2 and promotes cancer development | [121] |
| 6. Activating invasion and metastasis | E3s | FBXW2 | SKP2/β-catenin |
Prostate cancer/Lung cancer/Hepatocellular carcinoma Non-small cell lung cancer/Gastric cancer/Ovarian cancer |
Promotes ubiquitination and degradation of oncogenic proteins and inhibits tumor migration, invasion, and metastasis | [122–125] |
| FBW7 | Brg1 | Ubiquitinates a variety of oncogenic proteins | [126–130] | |||
| UBE3C | AHNAK | Breast cancer/Hepatocellular carcinoma/Renal cell carcinoma | Promotes tumor invasion and metastasis | [131–133] | ||
| FBXO22 | PTEN | Colorectal cancer/Hepatoma | Promotes tumor invasion and metastasis | [134, 135] | ||
| DUBs | BAP1 | PTEN | Prostate cancer/Intrahepatic cholangiocarcinoma | Promotes tumor proliferation | [136, 137] | |
| USP7 | EZH2 | Prostate cancer/Breast cancer | Enhances the stability of FOXA 1 protein and promotes tumor proliferation | [138–140] | ||
| USP43 | NUDR | Breast cancer | Promotes tumor invasion and metastasis | [141] | ||
| 7. Genome instability and mutation | E3s | RNF126 | MRE11 | Triple-negative breast cancer | Confers resistance of triple-negative breast cancer to radiotherapy | [142] |
| TRIM25 | Ku80 | Esophageal cancer/Pancreatic cancer | Intensifies DNA damage | [143] | ||
| MDM2 | DICER | Breast cancer | Impairs DDR and promotes cancer progression | [144] | ||
| DUBs | USP44 | TRIM25 | Nasopharyngeal carcinoma | Intensifies DNA damage | [143] | |
| USP7 | SAMHD1 | Colonic adenocarcinoma/Lung adenocarcinoma/Glioblastoma/Glioma | Repairs DNA damage induced by ROS or genotoxic insults | [145] | ||
| 8. Tumor-promoting inflammation | E3s | TRAF6/PEL1 | TAK1 | Lung cancer | Activates TAK1 complex and IKK complex and promotes tumor proliferation | [146] |
| BICP0 | TRAF6 | Cervical Cancer | Promotes K48-ubiquitination of TRAF6 and promotes tumor proliferation | [147] | ||
| TRIM25/TRIM14/MEX3c/RNF135 | RIG-I | Breast cancer | Promotes polyubiquitination of RIG-I and promotes tumor proliferation | [148] | ||
| TRIM22 | NOD2/NF-κB pathway | Endometrial cancer | Inhibits tumor progression through the NOD2/ NFκB pathway | [149] | ||
| TRAF3 | NF-κB | Gastric cancer | Inhibits tumor proliferation | [150] | ||
| LUBAC | NF-κB | Lymphoma | Enhances NF-κB activation | [29] | ||
| LUBAC | NEMO | Breast cancer | Promotes breast cancer development | [30] | ||
| 26S proteasome | MDA-7/IL-24 | Breast cancer/Lung cancer | Ubiquitinates MDA-7/IL-24 and reduces its antitumor activity | [151] | ||
| OTULIN | M1-linked polyubiquitin signaling | Hepatocellular carcinoma | Promotes tumor growth | [152] | ||
| USP 7 | MDM2 | Breast cancer | Impairs DDR and promotes cancer progression | [144] | ||
| 9. Reprogramming energy metabolism | E3s | TRIM36 | HK2 | Prostate cancer | Inhibits the neuroendocrine differentiation of prostate cancer | [153] |
| MARCH8 | HK2 | Colorectal cancer | Inhibits glycolysis | [154] | ||
| FBW7 | C-Myc | Oral squamous cell carcinoma | Inhibits oral squamous cell carcinoma | [155] | ||
| STUB1 | PKM2 | Colorectal cancer | Inhibits the progress of colorectal cancer | [156] | ||
| TRIM9 | PKM2 | Triple-negative breast cancer | Promotes glycolysis | [157] | ||
| TRIM29 | PKM2 | Colorectal cancer | Promotes colorectal cancer carcinogenesis | [158] | ||
| TRIM35 | PKM2 | Breast cancer | Inhibits the malignant behaviour of breast cancer | [159] | ||
| DLG4 | G-6-PD | Colorectal cancer | Inhibits the progress of colorectal cancer | [160] | ||
| STUB1 | GDH1 | Lung adenocarcinoma | Inhibits the proliferation of cancer cells and cancer growth | [65] | ||
| RNF213 | GDH1 | Kidney renal clear cell carcinoma | Maintains the survival of cancer cells after amino acid deprivation | [161] | ||
| Trib1-COP1 | ACC1 | Myeloid leukemia | Promotes the occurrence of myeloid leukemia | [162] | ||
| FBXW7β | FASN | Colorectal cancer | Promotes the growth of colorectal cancer | [163] | ||
| TRIM21 | GAC | Non-small cell lung cancer | Promotes the occurrence of non-small cell lung cancer | [164] | ||
| DUBs | USP7 | HK2 | Gastric cancer | Promotes aerobic glycolysis | [165] | |
| OTUB1 | C-Myc | Breast cancer | Inhibits oral squamous cell carcinoma | [166] | ||
| USP7 | C-Abl | Non-small-cell lung cancer | Promotes glycolysis and survival of non-small cell lung cancer cells | [167] | ||
| TRAP1 | PFK1 | Colorectal cancer | Enhances Warburg metabolism | [168] | ||
| DDX39B | PKM2 | Colorectal cancer | Promotes the progress of colorectal cancer | [156] | ||
| OTUB2 | PKM2 | Colorectal cancer | Exacerbates the progression of colorectal cancer by promoting PKM2 activity and glycolysis | [11] | ||
| PSMD14 | PKM2 | Ovarian cancer | Promotes ovarian cancer progression by decreasing enzymatic activity of PKM2 | [169] | ||
| USP35 | PKM2 | Hepatocellular carcinoma | Promotes hepatocellular carcinoma progression | [170] | ||
| USP15 | GS | Multiple myeloma | Promotes amino acid metabolism | [171] | ||
| USP22 | PPARγ | Hepatocellular carcinoma | Promotes tumorigenesis | [172] | ||
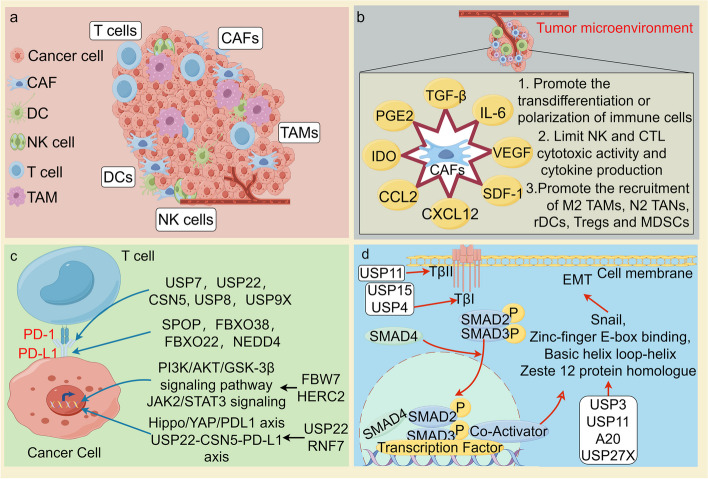
| 10. Avoiding immune destruction | E3s | SPOP protein | PD-L1 | Primary human prostate cancer | Promotes degradation of PD-L1 | [173] |
| SPOP protein | PD-L1 | Colorectal cancer | Mediates degradation of PD-L1 | [174] | ||
| SPOP protein | PD-L1 | Breast cancer | Ubiquitinates PD-L1 | [175] | ||
| FBW7 | NFAT1 | Metastatic renal cell carcinoma | Reduces the expression of PD-L1 by down-regulating NFAT1 | [176] | ||
| FBXO38 | PD-1 | Melanoma | Regulates PD-1 on the cell surface through the polyubiquitination linked with K48 | [177] | ||
| FBXO22 | PD-L1 | Non-small cell lung cancer | Promotes ubiquitination and degradation of PD-L1 | [178] | ||
| NEDD4 | PD-L1 | Urothelial carcinoma | Catalyzes the polyubiquitination of K48 linkage in PD-L1 | [179] | ||
| Cbl-b | TCR | Lymphoma | Downregulates TCR expression | [180] | ||
| HERC2 | JAK2/STAT3 pathway | Hepatocellular carcinoma | Mediates immune escape through the JAK2/STAT3 pathway | [181] | ||
| RNF31 | PD-L1 | Triple-negative breast cancer | Inhibits the expression of PD-L1 by inhibiting the Hippo/YAP/PD-L1 axis | [182] | ||
| RNF5 | PTEN | Pancreatic ductal adenocarcinoma | Promotes tumor advancement | [183] | ||
| A20 | Snail1 | Breast cancer | Promotes tumor metastasis | [184] | ||
| TRAF6 | CTLA-4 | Melanoma | Promotes the ubiquitination and degradation of CTLA-4 | [185] | ||
| DUBs | USP7 | PD-L1 | Gastric cancer | Interacts with PD-L1 to stabilize PD-L1 | [186] | |
| USP22 | PD-L1 | Liver cancer | Interacts with the C terminus of PD-L1, inducing its deubiquitination and stabilization | [187] | ||
| USP22 | PD-L1 | Non-small cell lung cancer | Regulates the level of PD-L1 protein through USP22-CSN5 -PD-L1 axis | [188] | ||
| CSN5 | PD-L1 | Breast cancer | Interacts with PD-L1 and deubiquitinates PD-L1 protein | [189] | ||
| USP8 | PD-L1 | Pancreatic cancer | Deubiquitinates PD-L1 | [190] | ||
| USP14 | IDO1 | Colonic adenocarcinoma | Stabilizes IDO1 and reduces anti-PD-1 responsiveness | [191] | ||
| USP12 | PD-1 | Lung cancer | Deubiquitinates PD-1 | [192] | ||
| USP9X | PD-L1 | Oral squamous cell carcinoma | Combines with PD-L1 to induce its ubiquitination and stabilize its protein expression in oral squamous cell carcinoma | [193] | ||
| USP3 | SUZ12 | Gastric carcinoma | Stabilizes SUZ12 through deubiquitination to promote EMT of tumor cells | [194] | ||
| USP11 | Snail | Breast cancer | Deubiquitinates Snail and enhances tumor EMT and metastatic capacity | [195] | ||
| USP11 | TGF-βRII | Breast cancer | Promotes breast cancer metastasis by stabilizing TGF-βRII | [196] | ||
| USP26 | SMAD7 | Glioblastoma | Negatively regulates TGF-β signaling by deubiquitinating and stabilizing SMAD7 | [197] | ||
| USP15 | TGF-β I | Glioblastoma | Enhances the tumorigenic effect of TGF-β in glioblastoma | [198] | ||
| USP4 | TGF-β I | Breast cancer | Maintains the stability of TGF-βRI | [199] | ||
| USP27X | Snail1 | Breast cancer/Pancreatic cancer | Maintains the stability of Snail1 | [200] | ||
| DUB3 | Snail1 | Breast cancer | Inhibits breast cancer invasion and metastasis by promoting Snail1 degradation | [201] | ||
| 11. Unlock surface plasticity | E3s | MDM2 | p53 | Dedifferentiated liposarcoma | Targets tumor suppressor p53 | [202] |
| CYLD | NOX4 | Abdominal aortic aneurysm | Ubiquitinates NOX4 | [203] | ||
| USP38 | FASN | Gastric cancer | Increases the production of triglycerides | [204] | ||
| 12. Nonmutational epigenetic reprogramming | E2 | RAD6a | H2b | Esophageal squamous cell carcinoma | Promotes the proliferation of cancer cells | [205] |
| E3s | RNF180 | DNMT1 | Gastric cancer | Inhibits the proliferation of cancer cells | [206] | |
| RNF180 | DNMT3A | Gastric cancer | Inhibits the vitality and motility of cancer cells | [207] | ||
| DDB1-Cul4A | H2A | Osteosarcoma | Inhibits osteosarcoma progression | [208] | ||
| SCFFBW7 | Brg1 | Gastric cancer | Promotes cancer metastasis | [127] | ||
| Β-TRCP | ARID1A | Gastric cancer | Promotes the destruction of cancer cells | [209] | ||
| SCF | ARID1A | Hepatocellular carcinoma | Enhances the growth of cancer cells in vitro and tumor growth in vivo | [186] | ||
| CHIP | INO80 | Colorectal cancer | Achieves effective DNA replication | [210] | ||
| DUBs | USP7 | FBP1 | Pancreatic cancer | Increases the sensitivity of pancreatic cancer to PARP inhibitors | [211] | |
| USP22 | H2A | Osteosarcoma | Promotes the progress of osteosarcoma | [208] | ||
| OTUD6A | Brg1 | Prostate cancer | Promotes tumorigenesis | [212] | ||
| 13. Polymorphic microbiomes | E3s | PSMB4 | NIK/NF-κb pathway | Colorectal cancer | Activates inflammatory response through NIK/NF- κB pathway | [213] |
| HECTD2 | EHMT2 | Colorectal cancer | Promotes proteasomal degradation of EHMT2 | [214] | ||
| 14. Senescent cells | E3 | MDM2 | p53 | Multiple cancers |
Under low-stress conditions, p53 will initiate repair and cell cycle arrest mechanisms that will promote cell survival Under acute stress conditions, p53 eliminates the damaged cells from the proliferative pool through apoptosis and senescence |
[214] |
MAPK signaling pathway
The MAPK pathway includes the RAS-RAF-MEK-ERK pathway and the JNK and p38 pathways [215]. In the ERK1/2 signaling pathway, E3 ligases RNF149 and FBXW7 regulate the stability of B-Raf in colon adenocarcinoma (COAD), leading to its degradation. This degradation inhibits ERK1/2 signaling and tumor cell growth [216, 217]. Additionally, the deubiquitinating enzyme USP10 protects C-Raf from degradation in ectopic endometrial stromal cells [218]. The UPS also regulates the ERK signaling pathway by affecting MEK1/2 expression, with USP21 involved in maintaining MEK2 stability and activating ERK signaling in hepatocellular carcinoma. The high expression of USP21 in hepatocellular carcinoma is associated with a lower survival rate among hepatocellular carcinoma patients. The research identified new clinical treatment strategies targeting the USP21-MEK2 interaction and its functions [91]. Activation of the MAPK pathway is known to promote the progression of hepatocellular carcinoma [219]. Targeted therapies against the MAPK pathway have become a focal point, with inhibitors targeting this pathway currently undergoing clinical trials [220, 221]. Deubiquitinating enzymes like USP2A, USP8, and USP15 have been identified as crucial modulators that promote MAPK pathway molecules deubiquitination [222]. Interestingly, USP8 knockdown can overcome gefitinib and erlotinib resistance [223, 224]. However, only a few USP8 inhibitors have been identified. Tian et al. discovered that DC-U4106 effectively binds to USP8 with a KD value of 4.7 μM and significantly suppresses breast cancer tumor growth while exhibiting minimal toxicity in a xenograft model [225].
The JNK1/2/3 and p38 signaling cascades involve multiple MAPKKKs and MAPKKs, including MEKK1/2/3/4, TAK1, and ASK1, which can be activated by various stimuli [226]. USP4 stabilizes the TAK1 protein level in ESCC cells through deubiquitination [82]. USP4/TAK1 plays a critical role in the progression of esophageal squamous cell carcinoma (ESCC) by regulating proliferation, migration, and invasion. Silencing USP4 has been shown to inhibit tumor proliferation in ESCC nude mouse models. Moreover, the USP4 inhibitor, Neutral Red, can suppress ESCC progression both in vitro and in vivo [82]. Another UDB molecule, USP15, can also target TAK1 and inhibit the proteasomal degradation of TAK1-binding protein 2 (TA B2) [227]. Apoptosis signal-regulating kinase 1 (ASK1) is a MAPKKK that initiates cell death and inflammatory responses by activating the p38 and JNK signaling pathways [228, 229]. The E3 ligase inhibitor of apoptosis protein (IAP) can directly bind to ASK1 and induce its degradation via the E2 enzyme UbcH5. IAP depletion increases TNF receptor 2 (TNFR2)-mediated activation of p38 and JNK, increasing tumor cell proliferation [230]. These research findings reveal the role of the UPS in regulating important signaling pathways in various cancers, providing new insights for the future development of cancer therapeutics.
PI3K/AKT/mTOR signaling pathway
AKT, also known as phosphokinase B (PKB), plays a central role in the PI3K/AKT/mTOR signaling pathway [231]. Ubiquitin-conjugating enzyme E2S (UBE2S), mitochondrial ubiquitin ligase activator NF-κB (MULAN), ret finger protein 2 (RFP2), breast cancer susceptibility gene 1 (BRCA1), speckle-type POZ protein (SPOP), TNF receptor-associated factor 4 (TRAF4), and F-box and leucine-rich repeat protein 18 (FBXL18), regulate AKT through ubiquitination, affecting its degradation or activation in various cancers (Table 1) [74–76, 78, 79, 232]. UBE2S has been shown to be associated with AKT phosphorylation [233]. One study found that UBE2S is highly expressed in epithelial ovarian cancer and induces cisplatin resistance by activating the PI3K/AKT/mTOR signaling pathway and inhibiting autophagy. Knocking down of UBE2S can inhibit the proliferation and migration of cisplatin-resistant ovarian cancer cells, providing new insights for the evaluation and treatment of high-risk ovarian cancer patients with cisplatin resistance [234]. SPOP, an E3 ligase, inhibits the activity of AKT kinase and its oncogenic function by mediating the ubiquitination and degradation of phosphatidylinositol-dependent protein kinase 1 (PDK1) (upstream protein of AKT). Cancer patients with PDK1 mutations exhibit oncogenic effects by evading SPOP recognition. This could be an attractive therapeutic direction [235].
The mTOR signaling pathway plays a vital role in regulating essential cellular functions such as cell growth, autophagy, metabolism, and DNA damage [236]. The lipid phosphatase PTEN can antagonize PI3K [237]. In cervical cancer cells, the E3 ligase makorin ring finger protein 1 (MKRN1) ubiquitinates and degrades the PTEN protein. In cervical cancer patients exhibiting high expression levels of MKRN1, the protein level of PTEN is found to be lower, which is associated with a decreased 5-year survival rate [69]. Additionally, another study identified that the deubiquitinase OTUD3 interacts with the substrate KPTN to regulate the mTORC1 signaling pathway, significantly inhibiting tumor cell proliferation and growth. By uncovering OTUD3’s essential role in cancer, this research provides crucial insights for developing novel cancer treatment strategies targeting OTUD3 or its regulatory pathways [238]. Additionally, the E3 ligase FBX8 partially achieves its tumor suppressor function by degrading mTOR in colorectal cancer. Low expression levels of FBX8 are correlated with poor prognosis in colorectal cancer patients [71]. In prostate adenocarcinoma (PRAD), the E3 ligase sensitive to apoptosis gene (SAG) targets DEPTOR for degradation, activating the mTORC2/AKT signaling pathway and promoting tumorigenesis. The SAG conditional KO mouse model was employed with PTEN deletion in the prostate to assess the in vivo function of SAG in prostate cancer development, indicating that targeting the SAG E3 ligase could be beneficial in prostate cancer therapy [72]. These experiments demonstrate that targeting the E3 ligases that regulate key proteins in the PI3K/AKT/mTOR signaling pathway offers promising therapeutic avenues for various cancers, providing a new direction for developing more effective cancer treatment strategies.
Evading growth suppressors
Inactivation of tumor suppressors eliminates the negative regulation of cell growth and proliferation to promote cancer development [239]. In addition to inducing and maintaining positive growth-stimulating factors, cancer cells must evade growth suppressors. Typical tumor suppressors encode retinoblastoma (RB) and p53 proteins, which regulate cell proliferation and apoptosis (Table 1) [6, 103].
p53
p53 has the highest frequency of mutations in human cancers and is usually expressed at low levels in cancer cells [172]. p53 regulates the cell cycle, induces apoptosis in response to DNA damage, and contributes to genomic stability by promoting DNA repair [172]. MDM2 functions as a p53 monoubiquitinating E3 ligase, facilitating the ubiquitination and subsequent degradation of p53 [104]. Currently, activating p53 by antagonizing MDM2 involves several approaches: (a) reducing MDM2 expression; (b) inhibiting its ubiquitin ligase function; and (c) blocking interactions between MDM2 and p53 [104]. The strategy of disrupting MDM2-p53 interactions using small molecules has been extensively pursued. For instance, AMG232 triggered apoptosis and inhibited cell proliferation in glioblastoma and multiple myeloma. The observations also indicated a great specificity for p53 wild-type cells compared to p53 mutant stem cells in glioblastoma [240]. AMG232 has also been studied in clinical trials [241]. E3 ligases TRIM28 and TRIM31 are also reported to promote p53 degradation. In osteosarcoma cells, TRIM28 cooperates with MDM2 to regulate the ubiquitination and degradation of p53, promoting tumor proliferation [95]. A high level of TRIM31 correlates with shorter overall survival (OS) in lung cancer patients [242]. Elevated levels of TRIM31 are associated with more aggressive characteristics and unfavorable outcomes in pancreatic cancer patients. Inhibition of TRIM31 increases the sensitivity of gemcitabine in pancreatic cancer cells, indicating suppressing TRIM31 could be an effective approach to improve the efficacy of gemcitabine in overcoming chemotherapy resistance in pancreatic cancer [243]. Furthermore, USP7 is identified to directly deubiquitinate p53, inhibiting tumor proliferation. High levels of USP7 and MDM2 are implicated in the onset and development of various cancers, playing a critical role by suppressing p53 activities. Inhibiting these proteins can reactivate p53 pathways, leading to the halting of the cell cycle and programmed cell death. Studies emphasize the pharmacological properties, potential therapeutic uses, and the action mechanisms of small molecule inhibitors targeting USP7 and MDM2 [104]. Moreover, USP25 has been shown to be an important upstream regulator of the MDM2-p53 signaling pathway and has the potential to be a novel target gene for developing new therapeutic applications [244].
RB
The dysregulation of the RB pathway is frequently observed in cancer. The impairment of RB function, frequently due to mutations or mechanisms that induce hyperphosphorylation, allows uncontrolled cell cycle progression [245]. This process can result in excessive cell proliferation and contribute to tumor development. In addition to being phosphorylated, RB can be ubiquitinated, sumoylated, acetylated, or methylated [246]. TRIM28 binds to the phosphorylated RB protein (p-RB), promoting its ubiquitination and degradation [95]. On the other hand, SETDB1, a binding partner of TRIM28, protects p-RB from degradation, which is particularly notable in prostate cancer [247]. Inhibiting SETDB1 expression reduces tumor growth but accelerates the degradation of RB protein. Notably, combined use with the CDK4/6 inhibitor palbociclib can block SETDB1 inhibition-induced RB degradation and demonstrate stronger anticancer effects. These research findings reveal the potential value of using a combination strategy of CDK4/6 and SETDB1 inhibition to reduce RB degradation and suppress cancer growth [248].
ADP-ribosylation Factor (ARF)
ARF is a tumor suppressor encoded by the cyclin-dependent kinase inhibitor 2A (CDKN2A) locus and primarily exerts its tumor suppressive effects through the MDM2-P53 axis [98]. Under normal conditions, oncogenic signals induced by MYC, RAS, and E2Fs lead to the upregulation of ARF. ARF subsequently inhibits MDM2, thereby activating the tumor suppressor function of p53 [98, 249]. ARF function, stability, and cellular localization are tightly regulated by posttranslational modifications such as phosphorylation and ubiquitination [98]. Elongin B (ELOB), as a core component of the Cullin2-RBX1-ELOB E3 ligase (CRL2) complex, regulates ubiquitination and degradation of the oncoprotein p14/ARF [250]. Research indicates that a peptide strongly adheres to the ELOB/C dimer, disrupting the binding of ELOB/C to its binding molecules. Treatment of cancer cells with this peptide inhibitor led to reduced cell survival, heightened apoptosis, and altered gene activity. Consequently, these findings suggest that targeting the BC-box-binding pocket of ELOB/C is a viable method for disrupting its activity and inhibiting the proliferation of cancer cells [251]. Prame is overexpressed in tumor tissues compared to paired adjacent tissues and is associated with poor prognosis in cancer patients. As a substrate recognition receptor protein of Cullin RING E3 ligases (CRLs), Prame regulates the ubiquitination and subsequent degradation of ARF through the Cullin2-RBX1-ELOB E3 ligase complex, making it a potential novel therapeutic target [252].
Resist cell death
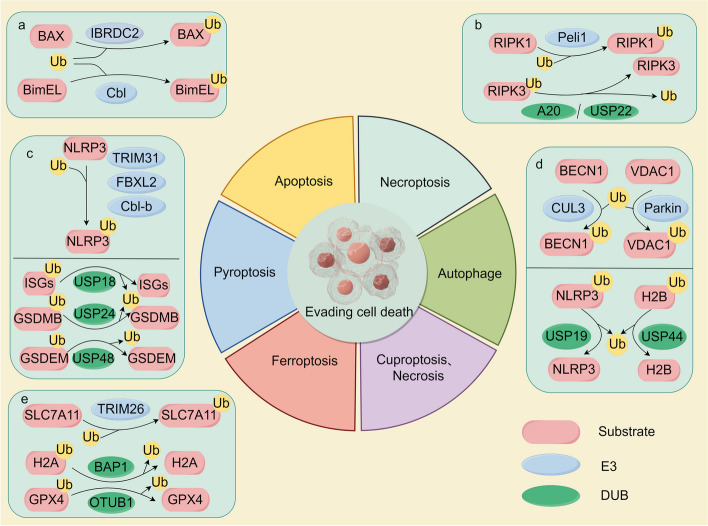
Cell death is a normal physiological process in all living organisms and plays essential roles in embryonic development, homeostatic maintenance, aging, and immune coordination [253]. Cell death includes apoptosis, necrotizing apoptosis, pyroptosis, autophagy, ferroptosis, cuproptosis [182, 253, 254]. Ubiquitination also plays an essential role in resisting cell death. In the following paragraphs, we will briefly introduce the types of cell death and describe the functions of ubiquitination and deubiquitination in these types of cell death (Fig. 4).
Fig. 4.
The ubiquitin–proteasome system (UPS) plays a vital role in resisting cell death through various mechanisms. a IBR domain containing 2 (IBRDC2) can target Bcl-2-associated X protein (BAX) for ubiquitination and degradation, which can prevent mitochondrial outer membrane permeabilization (MOMP) induced by active BAX and reduce cell apoptosis. Additionally, Cbl can target BimEL for ubiquitination and degradation, thereby inhibiting apoptosis. b A20 and USP22 can deubiquitinate receptor-interacting protein kinase 3 (RIPK3) to stabilize it, thus suppressing TNF-α-induced necroptosis. Pellino1 (Peli1) can mediate K63 ubiquitination on K115 of RIPK1 in a kinase-dependent manner, promoting the formation of necrosomes and facilitating necroptosis. c Tripartite motif 31 (TRIM31), F-box and leucine-rich repeat protein 2 (FBXL2), and casitas b-lineage lymphoma-b (Cbl-b) promote NLRP3 inflammasome protein 3 (NLRP3) polyubiquitination at different sites, thereby inhibiting the process of pyroptosis. USP18 inhibits pyroptosis in cancer cells via enhancing interferon-stimulated genes (ISGs), while USP48 promotes pyroptosis by stabilizing gasdermin E (GSDEM), and USP24 promotes pyroptosis by stabilizing gasdermin B (GSDEB). (d) Cullin3 (CUL3) and Parkin are responsible for ubiquitinating beclin 1 (BECN1) and voltage-dependent anion channel 1 (VDAC1), respectively, whereas USP19 and USP44 can deubiquitinate NLRP3 and H2B, respectively. e TRIM26 targets solute carrier family 7 member 11 (SLC7A11) for ubiquitination and degradation, promoting cellular ferroptosis. BRCA1-associated protein 1 (BAP1) removes H2A ubiquitination from the SLC7A11 promoter, resulting in decreased cystine uptake and increased ferroptosis. OTU deubiquitinase ubiquitin aldehyde-binding 1 (OTUB1) promotes glutathione peroxidase 4 (GPX4) deubiquitination, inhibiting ferroptosis in gastric cancer cells
Apoptosis
Apoptosis is the most common noninflammatory form of programmed cell death [182, 255]. It produces remnants of apoptotic cells, such as cytoplasm, organelles, and the contents of some nuclei, which are randomly sorted into each apoptotic body [105]. Two main pathways of apoptosis have been identified: the intrinsic apoptosis pathway and the extrinsic pathway initiated by death receptors [105, 182]. The intrinsic and extrinsic apoptosis pathways depend on the activation of the caspase family [256]. BAX is the main pro-apoptotic executioner protein. The E3 ligase IBR domain containing 2 (IBRDC2) can target BAX for ubiquitination-mediated degradation, thereby preventing mitochondrial outer membrane permeabilization (MOMP) induced by active BAX [105]. When inducing apoptosis, IBRDC2 accumulates in BAX-rich mitochondrial structures, allowing the accumulation of BAX to occur simultaneously with its activation [106]. BimEL, belonging to the Bcl-2 protein family, crucially promotes apoptosis by inducing mitochondrial outer membrane permeabilization (MOMP) and activating the caspase cascade. The E3 ligase Cbl can degrade the extralong splice variant of Bim (BimEL) and is cell type-specific [105]. Icotinib is a specific tyrosine kinase inhibitor (TKI) targeting the epidermal growth factor receptor (EGFR). Treatment with Icotinib significantly reduces the levels of p-EGFR (phosphorylated EGFR), p-ERK (phosphorylated extracellular signal-regulated kinase), and c-Cbl in HCC827 lung cancer cells, leading to inhibited proliferation and induced apoptosis of HCC827 lung cancer cells [257]. Aurora A phosphorylates BimEL, enhancing binding to the F-box protein β-transducin, which contains the E3 ligase. This interaction facilitates the ubiquitination and subsequent degradation of BimEL [258]. The E3 ligases mentioned above all decrease the levels of pro-apoptotic proteins; therefore, finding drugs that can inhibit the binding affinity of these ligases with their target proteins can maintain a certain level of apoptosis in cancer cells, effectively inhibiting the development of cancer.
Necroptosis
Necroptosis, a type of programmed cell death, involves cell and organelle swelling, membrane rupture, and the release of cellular contents [259]. Necrotizing apoptosis is a receptor-interacting protein kinase 1 (RIPK1)-RIPK3-mixed lineage kinase domain-like protein (MLKL) pathway triggered by death and Toll-like receptor 3/4 [260]. Pellino1 (Peli1) can mediate K63 ubiquitination on K115 of RIPK1 in a kinase-dependent manner, promoting the formation of necrosomes and facilitating necroptosis [23]. OTULIN can remove the M1 chain from the necroptosis pathway, enhancing TNF-α-induced necrotizing apoptosis [25]. Ubiquitin-editing enzyme A20 removes K63-linked ubiquitin chains from RIPK3 [22], inhibiting RIPK3 ubiquitination and reducing RIPK1:RIPK3 interactions. This inhibition effectively restrains TNF-α-induced necrotizing apoptosis, which can be reversed by USP22 [18]. Interestingly, several cases of solid tumors with high A20 expression are associated with lower survival rates [261]. Knocking down of A20 reduces cell growth and enhances sensitivity to agents that induce apoptosis [262]. Moreover, researchers found that A20 plays a vital role in drug resistance, and they established a direct link between elevated A20 levels and increased in vitro resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) [263].
Pyroptosis
Pyroptosis is the programmed death of cells caused by the activation of inflammasome sensors. It results in plasma membrane lysis, cell swelling, chromatin fragmentation, and the release of intracellular proinflammatory contents [253, 264]. Pyroptosis mainly relies on the activation of caspase family proteins by inflammasomes to cause various physiological responses, and NOD-like receptor protein 3 (NLRP3) is a typical inflammasome [265]. The E3 ligase TRIM31 can act as a feedback inhibitor for the NLRP3 inflammasome, directly binding to NLRP3, promoting K48-linked polyubiquitination, and facilitating its degradation via the proteasome [96]. E3 ligase F-box and leucine rich repeat protein 2 (FBXL2) interact with Trp73 within the NLRP3 protein specifically by targeting the ubiquitination and subsequent degradation of Lys689 [266]. The E3 ligase Cbl-b binds to K63 ubiquitin chains on the leucine-rich repeat domain (LRR) of NLRP3 and then targets the K496 site to link with K48 ubiquitin chains and mediate proteasomal degradation [267, 268]. Notably, by comparing the effects of the NLRP3 inducer Nigericin across various tumor types and normal fibroblast controls, it was discovered that Nigericin may represent a novel therapeutic approach for controlling the growth of tumors that produce low levels of IL-1β and IL-18 [269]. According to recent literature, several DUBs were reported to regulate pyroptosis in cancer, including USP18 [270], USP24 [112], and USP48 [113]. Mechanistically, USP18 inhibits pyroptosis in cancer cells via enhancing ISGs, while USP48 promotes pyroptosis by stabilizing gasdermin E (GSDEM). An in vivo study indicates that upregulating USP48 can enhance the antitumor activity of PD-1 inhibitor [113], suggesting that USP48 activation pharmacologically could be a promising approach to enhance cancer cell sensitivity to pyroptosis and improve immunotherapy outcomes.
Autophagy
Autophagy is initiated in response to various signals, including nutrient deprivation, the absence of growth factors, hypoxia (low oxygen levels), and exposure to pathogens [271]. Autophagy has been shown to play a critical role in tumor maintenance, even with elevated basal autophagy levels in many tumors under nutrient-adequate conditions. Autophagy inhibition or systemic autophagy inhibition in tumor cells disrupts tumor metabolism, resulting in antitumor effects [272]. Autophagy can promote tumor cell survival by providing nutrients during periods of stress, such as nutrient deprivation or hypoxia. USP19 plays a significant role in autophagy regulation. It cleaves the ubiquitin chain of NLRP3, inhibiting proteasomal degradation and transforming NLRP3 from a proinflammatory to an anti-inflammatory state [273]. Beclin 1 (BECN1) is an important member of the autophagy-related protein family, primarily involved in regulating the formation of autophagic vesicles [109]. In various cancers such as breast cancer, ovarian cancer, and colorectal cancer, the expression level of BECN1 is significantly reduced, which is positively correlated with poor prognosis for patients [109]. In breast cancer and ovarian cancer, the E3 ligase cullin-3 (CUL3) interacts with BECN1, promoting its K48 ubiquitination and downregulating BECN1, ultimately enhancing tumor cell proliferation and resulting in a poor prognosis [274]. Therefore, searching for inhibitors of CUL3 may be an important approach for treating breast cancer and ovarian cancer. A research team has also targeted the functions of Kelch-like (KLHL) family proteins, which are substrate adaptor proteins of Cullin3-RING ligase (CRL3), in order to disrupt the function of CRL3 [97].
Ferroptosis
Ferroptosis is a recently discovered form of cellular death triggered by the excessive accumulation of iron-dependent lipid peroxidation products [275, 276]. It primarily occurs through exogenous and endogenous pathways involving transporter-dependent and enzyme-regulated mechanisms [276]. The regulation of ferroptosis involves a balance between tumor promotion and inhibition. Genetic ablation of solute carrier family seven member 11 (SLC7A11) or glutathione peroxidase 4 (GPX4) induces ferroptosis in cancer cells, leading to significant tumor suppression [277]. The E3 ligase TRIM26 mediates the ubiquitination of SLC7A11, targeting it for proteasomal degradation and inducing ferroptosis. This process inhibits hepatic stellate cell activation and reduces liver fibrosis [110]. Searching for agonists of TRIM26 might be a strategy for treating early-stage liver cancer. The deubiquitinase BRCA1-associated protein 1 (BAP1) plays an essential role in regulating ferroptosis by removing H2A ubiquitination (H2Aub). BAP1 forms a polycomb repressive deubiquitinase (PR-DUB) complex that deubiquitinates H2Aub on the SLC7A11 promoter, leading to decreased cystine uptake and increased ferroptosis [278]. DUB enzyme OTU deubiquitinase ubiquitin aldehyde-binding 1 (OTUB1) promotes GPX4 deubiquitination, thereby inhibiting ferroptosis in gastric cancer cells [279]. OTUB1 is a distinctive target because of its conventional and unconventional functions. The compound PR-619 is anticipated to decrease OTUB1 activity, as evidenced by its capacity to prevent OTUB1 from binding to an active site probe [114].
Enabling replicative immortality
Telomeres, conserved nucleoprotein structures found at the termini of linear eukaryotic chromosomes, consist of repetitive sequences (TTAGGG)n in humans [280]. They interact with six protein species that form a “shelterin complex” [281]. As a result of repeated rounds of replication in eukaryotic cells, telomeres are shortened continuously. Therefore, during tumor development, cells must utilize a telomere DNA maintenance mechanism (TMM) to counteract telomere shortening, protect telomeres from the influence of the DNA damage repair system, and avoid telomere-mediated aging and apoptosis [282]. Changes in telomere structure are intricately linked to the onset and progression of tumors. Telomere repeats, and the involvement of TRF1 and TRF2 serve as guardians of telomeres, whose expression levels are disrupted across diverse cancer forms [283]. In renal cell carcinoma (RCC), the telomere proteins TRF1 and TRF2 are overexpressed, and their inhibition by siRNAs can induce apoptosis, reducing cell proliferation and migration [284].
Recent studies have shown that the ubiquitin mechanism can regulate elements of telomeres. Furthermore, the UPS influences cancer progression by impacting telomeres. Ubiquitin-mediated degradation of telomere associated protein TRF1 levels is facilitated by E3 ligases such as repeatability limit (RLIM), β-TRCP1, and FBX4 (Table 1) [115–117]. Clinical studies have shown that TRF1 upregulation in glioblastoma multiforme (GBM) contributes to tumor initiation and progression. This was demonstrated by the inhibition of tumor growth and extended survival in GBM mouse models following brain-specific TRF1 genetic deletion. Additionally, chemical inhibitors of TRF1 in human GBM cells blocked tumor sphere formation and slowed growth in patient-derived GSC xenografts [285]. These studies suggest the direction of future clinical research on ubiquitination. Conversely, members of the chromatin-modifying complex family, such as general control nonderepressible-5 (GCN5) and USP22, have been reported to facilitate the deubiquitination of TRF1 [286]. GCN5 is necessary for the binding of USP22 to Spt-Ada-Gcn5 acetyltransferase (SAGA) complexes, enabling the deubiquitination of TRF1 and preventing its turnover [286]. Research has demonstrated that eliminating USP22 from pancreatic tumor cells enhances the immune response by decreasing suppressive myeloid cells and increasing cytotoxic T cells and natural killer cells. Additionally, USP22 influences the cancer cell transcriptome, thereby modifying the immune tumor microenvironment. Targeting USP22 in pancreatic cancer can enhance the effectiveness of immunotherapy and improve treatment outcomes [287].
TPP1, another shelterin protein subunit, also undergoes ubiquitin-mediated proteolysis, which has been evidenced by the stability of TPP1 protein levels after proteasome inhibition. In mice, the stabilization of TPP1 at telomeres requires its ubiquitination by the E3 ligase RNF8 [120]. Recent research has indicated that F-box and WD repeat domain-containing 7 (FBW7) can promote cell senescence and tissue fibrosis by facilitating telomere decapitation [118]. The deubiquitinase USP7 interacts with human TPP1 and removes ubiquitin chains. Although the degradation of USP7 does not impact the level of TPP1 regulated by the proteasome, USP7 might interact with other deubiquitinases redundantly to stabilize TPP1 [288]. By now, P22077 has been extensively studied and has become a prevalent tool compound for inhibiting USP7 in biological research. For example, it has been shown that P22077 can effectively trigger p53-dependent apoptosis in neuroblastoma (NB) cells and markedly reduce tumor growth in xenograft models of three NB cell types [289]. Additionally, certain natural compounds have also been identified as USP7 inhibitors. Notably, Spongiacidin C, a pyrrole alkaloid from the marine sponge Stylissa massa, was identified as a USP7 inhibitor with an IC50 of 3.8 μM, though its precise in vivo effects need further investigation [290].
Inducing angiogenesis
Tumor angiogenesis refers to the process of forming new blood vessels within and surrounding tumors. This phenomenon is critical for tumor growth and progression, as it provides the necessary nutrients and oxygen supply to sustain rapidly dividing cancer cells [291]. The rapid growth of tumors leads to areas with low oxygen concentrations, known as hypoxia. In response to hypoxia, cells release hypoxia-inducible factors (HIFs) that stimulate the expression of proangiogenic factors, including vascular endothelial growth factor (VEGF), to form new blood vessels [292]. The specific prolyl-4-hydroxylase enzyme continuously hydroxylates HIF1α. Once hydroxylated, HIF1α is recognized by the E3 ligase complex, leading to its polyubiquitination and subsequent degradation by the proteasome [291, 293]. VEGF is highly expressed in most human tumors [294]. Under normoxic conditions, the E3 ligase von hippel lindau (VHL) ubiquitinates HIF-1, thereby preventing the dimerization and binding of HIF-1 to the promoter of the VEGF gene and inhibiting its transcription and translation. Under hypoxic conditions, HIF-1 dimerizes and stimulates VEGF production and angiogenesis [295]. By regulating the levels of HIF1α, this process directly influences the cellular response to low oxygen conditions. A study has revealed that USP22 promotes the stemness of hepatocellular carcinoma induced by hypoxia-inducible factors through a HIFα/USP22 positive feedback loop after TP53 inactivation [296]. This process contributes to promoting angiogenesis, tumor invasion and metastasis, and tumor drug resistance [296]. The research team achieved high tumor suppression and increased sensitivity to sorafenib in mice with hepatocellular carcinoma by targeting USP22 with a lipid-polymeric complex. This further indicates that USP22 is a highly promising therapeutic target for hepatocellular carcinoma [296]. Additionally, seven in absentia homology 2 (SIAH2) can target HIF for ubiquitination and degradation, thereby modulating the cellular response to hypoxic conditions. HIF-mediated inhibition of nuclear respiratory factor 1 (NRF-1) reduces the transcription of mitochondrial genes and inhibits the activity of the E3 ligase SIAH2 [121].
Activating invasion and metastasis
The invasion and metastasis of tumors include the movement of tumor cells, infiltration into neighboring tissues, circulation, and extravasation to distant organs, which are the leading causes of cancer-mediated damage to the body [297]. Ubiquitination and deubiquitination are pivotal in numerous protein modification and regulatory processes and often influence tumor invasion and metastasis. This section aims to explore the role of the ubiquitin mechanism in tumor invasion and metastasis and analyze recent findings related to E3 ligases and DUBs and their potential mechanisms (Fig. 3).
The role of E3 ligases in cancer metastasis
E3 ligases F-Box and WD repeat domain containing 2 (FBXW2) [122], FBW7 [127], Ub-protein ligase E3C (UBE3C) [131], and F-Box protein 22 (FBXO22) [298] play vital roles in cancer metastasis. For instance, FBXW2 functions as a tumor suppressor by facilitating the ubiquitination and degradation of oncogenic proteins such as SKP2 [123] and β-catenin [122], thus impeding cancer migration, invasion, and metastasis. FBXW2 can be ubiquitinated and degraded as a substrate of β-TrCP1 [123]. Additionally, overexpression of FBXW2 decreases β-catenin-driven transactivation and suppresses invasion, while depletion enhances β-catenin stability and promotes lung cancer metastasis [122]. FBW7 acts as a tumor suppressor by promoting the degradation of cancer-related proteins like Snail [126], Brahma-related gene 1 (Brg1) [127], and YTH N6-methyladenosine RNA Binding Protein F2 (YTHDF2) [128], thereby inhibiting metastasis in various cancers including non-small cell lung cancer, gastric cancer, and ovarian cancer. It also modulates the HIF-1α/CEACAM5 axis in colorectal cancer and potentially predicts immunotherapy response in thymic cancer [129]. In clinical studies, it has been shown that low expression of FBW7 in breast cancer cells leads to resistance to the BET inhibitor JQ1, but combining JQ1 with a Mcl-1 inhibitor can overcome this resistance. This finding suggests that enhancing the effectiveness of BET inhibitors in patients with low FBW7 expression is a promising clinical strategy [299].
UBE3C is a tumor promoter that ubiquitinates substrates such as neuroblast differentiation-associated protein (AHNAK), disrupting the p53-AHNAK complex and enhancing stem cell-like properties in non-small cell lung cancer [300, 301]. It also promotes RCC growth and metastasis by upregulating β-catenin and activating the Wnt/β-catenin pathway [132]. In non-small cell lung cancer, FBXO22 promotes Lys63-linked polyubiquitination of liver kinase B1 (LKB1), reducing its activity and impeding the LKB1-AMPK-mTOR pathway, thereby enhancing cell proliferation. Clinically, elevated FBXO22 levels in lung adenocarcinoma patients indicate a poor prognosis [302]. FBXO22 promotes angiogenesis and tumor cell migration by increasing the levels of vascular endothelial growth factor A and HIF-1α expression [303]. Recent research suggests that FBXO22 may facilitate the ubiquitin-mediated degradation of cyclin G-associated kinase (GAK), thereby inhibiting the proliferation and metastasis of cervical cancer cells [304]. Additionally, clinical studies have shown that FBXO22 negativity significantly affects survival in breast cancer patients, especially those with invasive lobular carcinoma (ILC), and leads to poorer outcomes in patients treated with selective estrogen receptor modulators (SERMs) [305]. These findings suggest the need for tailored therapeutic strategies based on histopathological types when considering adjuvant endocrine therapy.
The role of DUBs in cancer metastasis
The deubiquitinating enzymes BRCA1-associated Protein 1 (BAP1), USP7, and USP43 are the primary focus of the discussion below. BAP1, characterized by its UCH domain, is a crucial tumor suppressor across various malignancies. In breast cancer, BAP1 promotes tumorigenesis by stabilizing Kruppel-like factor 5 (KLF5) through deubiquitination, facilitating cell cycle progression, while its depletion inhibits tumorigenesis and lung metastasis [306]. BAP1 holds significant potential in clinical research. A study on Pembrolizumab efficacy in thymic cancer found that PD-L1 expression, along with alterations in genes or pathways like BAP1, may predict patient response or resistance to immunotherapy [307]. USP43 mediates Cav2.2 function by regulating cortical actin stability, extracellular matrix degradation, and migration, with Cav2.2 enhancing USP43 expression through NFAT2 activation, thus promoting breast cancer metastasis [308]. USP43 is markedly expressed in epithelial ovarian cancer, fostering cell proliferation, migration, invasion, and cisplatin resistance by stabilizing HDAC2 and activating the Wnt/β-catenin pathway. These discoveries underscore the clinical importance of USP43 in epithelial ovarian cancer, accentuating its potential as a therapeutic target to manage cancer progression, increase sensitivity to cisplatin chemotherapy, and ultimately enhance patient outcomes [309].
Genome instability and mutation
Genome instability is the core of carcinogenesis in multicellular organisms and is characterized by a high frequency of mutations in cell lineage genomes. High-frequency DNA damage and epigenetic or mutation-induced reductions of DNA repair gene expression may contribute to genome instability [310, 311].
DDR pathways are complex and intricate. Thousands of endogenous and exogenous DNA damage events occur daily [312, 313]. The MRE11-RAD50-NBS1 (MRN) complex first recognizes the repair factors recruited at DNA fragmentation sites [314]. The E3 ligase RNF126 ubiquitinates meiotic recombination 11 (MRE11) at K339 and K480, activating the DDR and conferring resistance to radiotherapy in triple-negative breast cancer (Table 1) [142]. A member of the PI3/PI4-kinase family, ataxia-telangiectasia mutated (ATM) is a protein kinase that is essential for the cellular response to DNA damage, specifically double-strand breaks (DSBs), and is mainly involved in preserving genomic integrity. [315]. Research has found that the E3 ubiquitin ligase Peli1 is activated by ATM-mediated phosphorylation, promoting the ubiquitination of NBS1 and enhancing the accumulation of ATM and the MRN complex at DSB sites [316, 317]. SAM and HD domain containing protein 1 (SAMHD1) combines with the DSB repair initiator CtBP-interacting protein (CtIP) to promote DNA repair [318]. It is worth noting that USP7 interacts with SAMHD1 and deubiquitinates the K421 site, thus reducing its degradation by the proteasome to stabilize SAMHD1. Consequently, it repairs DNA damage induced by ROS or genotoxic insults, overcoming carcinogenic stress and influencing chemotherapy sensitivity [145].
Tumor-promoting inflammation
Chronic inflammation is an essential factor in cancer development and is associated with approximately 20% of human cancers [319]. Cancer often occurs in inflamed tissues, suggesting that local inflammation plays an essential role in cancer initiation and progression. Moreover, ubiquitination can contribute to tumors initiated by chronic inflammation through the regulation of transcription factors and cytokines, thereby inducing cancer development, maintenance, and metastasis (Fig. 3) [320].
Chronic inflammation and NF-κB activation are closely associated with cancer progression and spread. Linear ubiquitination of key NF-κB regulators by LUBAC plays an essential role. Abnormally regulated linear ubiquitin signaling is associated with cancer initiation and progression [321]. For example, elevated LUBAC expression enhances NF-κB activation, accelerating the development of somatic mutations and lymphoma pathogenesis [29]. The natural compound thiolutin, which specifically inhibits LUBAC, has been shown to inhibit tumor growth in mouse xenograft models, indicating that LUBAC could be a viable therapeutic target for B-cell lymphoma [29]. However, the deubiquitinase OTULIN negatively regulates linear ubiquitin signaling. In hepatocytes, OTULIN deficiency contributes to hepatocellular carcinoma development [152].
In addition, the upstream signaling activator pattern recognition receptor (PRR) is also regulated by E3 ligases [322, 323]. Toll-like receptors (TLRs) are essential components of the immune system, that can activate NF-κB and induce interferon (IFN) production [323, 324]. Alternatively, the c-Cbl ubiquitin ligase is involved in TRAF6 ubiquitination and negatively regulates NF-κB activity [325]. In the context of nucleotide-binding oligomerization domain (NOD)-like receptor (NLR), overexpression of TRIM22 reduces the occurrence and development of endometrial cancer, and its inhibition is mediated by the NOD-NF-κB pathway, which may be one of the mechanisms of NLR [149], indicating TRIM22 may emerge as a valuable prognostic indicator in endometrial cancer patients. TRIM22 can interact with IKKγ, an upstream molecule in the NF-κB pathway, increasing the K63-linked polyubiquitination of IKKγ, thereby activating the NF-κB pathway in GBM. This study indicates that inhibiting the E3 ligase activity of TRIM22 or blocking its interaction with the IκBα or IKKγ proteins could have significant implications for the development of potential therapeutic drugs for GBM [326]. In addition, NOD1 protects intestinal cells from precancerous lesions by inhibiting the NF-κB signaling pathway through the induction of TRAF3 [150]. Cytokines play a crucial role in the tumor microenvironment, promoting communication between malignant cells and surrounding cells. The UPS can influence cancer progression by regulating cytokines [320]. For example, SAG plays an important role in chronic inflammation-induced cancers by ubiquitylating key apoptotic factors such as SARM and Noxa, regulating the ratio of pro- and antiapoptotic factors. Therefore, SAG-UPS may serve as an early diagnostic marker for liver cancer and a potential target for therapeutic development [327, 328]. In conclusion, chronic inflammation, closely linked to aberrant ubiquitination pathways, is a significant driving force in cancer initiation and progression, highlighting potential targets for future cancer treatments and prognostic indicators.
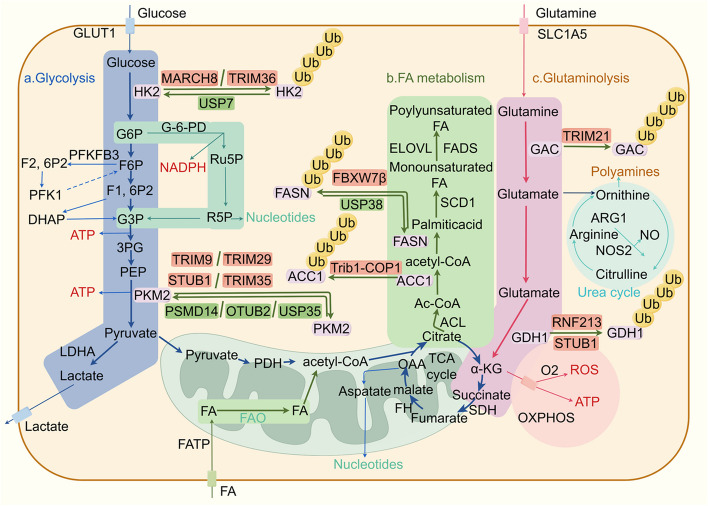
Reprogramming energy metabolism
Glucose metabolism
Cancer tissues need to be reprogrammed in terms of both matter and energy to maintain or further enhance the progress of cancer. Otto Warburg was the first to discover the unique metabolism of cancer cells. He observed that cancer cells exhibit a greater tendency toward glycolysis under aerobic conditions, which is called the “Warburg effect” or “aerobic glycolysis” [329].
Hexokinase 2 (HK2) phosphorylates glucose to produce glucose-6-phosphate (G-6-P), regulating glucose metabolism [330]. The E3 ligase TRIM36 ubiquitinates HK2 to inhibit the neuroendocrine differentiation (NED) of prostatic cancer (Fig. 5) [153]. Additionally, studies have shown that TRIM36 can enhance the efficacy of anti-androgen drugs in treating prostate cancer. Therefore, adding TRIM36 during androgen deprivation therapy (ADT) could be a novel therapeutic approach to better suppress castration-resistant prostatic cancer [331]. Recent research has discovered that the E3 ligase membrane-associated RING-CH protein (MARCH8) is a novel glycolysis repressor that inhibits glycolysis in colorectal cancer through the ubiquitination and degradation of HK2 (Fig. 5) [154]. However, clinical studies on MARCH8’s role in cancer metabolism are lacking, with most research focusing on its involvement in cancer cell apoptosis [332]. Future studies could explore therapeutic strategies targeting MARCH8 and its regulatory mechanisms in cancer metabolism, such as combination therapy, to enhance treatment efficacy. In turn, the circular RNA derived from ribosomal protein S19 (circRPS19) upregulates USP7 expression, leading to an increase in HK2 protein levels and the promotion of aerobic glycolysis in gastric cancer cells [165]. Therefore, targeting the circRPS19-USP7-HK2 pathway presents a promising therapeutic strategy for treating gastric cancer. PKM2 converts phosphoenolpyruvate (PEP) to pyruvate in the last step of glycolysis. On the one hand, several E3 ligases within the TRIM family, including TRIM9, TRIM29, and TRIM35, have been found to ubiquitinate PKM2 in tumor cells (Fig. 5) [157–159]. On the other hand, deubiquitinases such as OTUB2, proteasome non-ATPase regulatory subunit 14 (PSMD14) and USP35 enhance the activity and stability of PKM2, thereby promoting glycolysis in tumor cells (Fig. 5) [11, 169, 170]. Additionally, some lncRNAs including the lncRNA LINC01554 and lncRNA UCA1 have been proven to facilitate the ubiquitination of PKM2, thereby suppressing the Warburg effect [333, 334]. Therefore, developing targeted inhibitors for these enzymes and combining them with traditional chemotherapy could enhance treatment efficacy by addressing multiple aspects of tumor metabolism and growth. In conclusion, ubiquitination plays a crucial role in glucose metabolism by regulating key enzymes. This posttranslational modification influences the activity and stability of enzymes involved in glucose metabolism, impacting the overall cellular energy balance in cancer.
Fig. 5.
The ubiquitin–proteasome system regulates tumor metabolism in several pathways. a Glycolysis: Hexokinase 2 (HK2) can be ubiquitinated by E3 ligases membrane-associated RING-CH protein 8 (MARCH8) and tripartite motif protein 36 (TRIM36), a process that can be reversed by the deubiquitinase ubiquitin-specific protease 7 (USP7). Pyruvate kinase M2 (PKM2) can be ubiquitinated by E3 ligases TRIM9, TRIM29, STIP1 homology and U-box-containing protein 1 (STUB1), and TRIM35. Conversely, PKM2 can be deubiquitinated by proteasome non-ATPase regulatory subunit 14 (PSMD14), OTU domain-containing ubiquitin aldehyde-binding protein 2 (OTUB2), and USP35. b Fatty acid (FA) metabolism: Acetyl-CoA carboxylase 1 (ACC1) and fatty acid synthase (FASN), both involved in fatty acid metabolism, can be ubiquitinated by E3 ligases tribbles pseudokinase 1 (Trib1)-constitutive photomorphogenic 1 (COP1), and F-box and WD repeat domain containing 7β (FBXW7β), respectively. In contrast, FASN can be deubiquitinated by USP38. c Glutaminolysis: Glutaminase C (GAC), which catalyzes the initial step of glutamine decomposition into glutamic acid and ammonia, can be ubiquitinated by the E3 ligase TRIM21. This leads to K63-linked ubiquitination that inhibits GAC activity. Glutamate dehydrogenase (GDH), another key enzyme in glutamine catabolism, can be ubiquitinated by E3 ligases ring finger protein 213 (RNF213) and STUB1
Fatty acid metabolism
Fatty acid metabolism is regulated by three rate-limiting enzymes: ATP-citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), and fatty acid synthase (FASN) [335, 336]. Peroxisome proliferator-activated receptor γ (PPARγ) is a pivotal regulatory protein that is highly expressed in adipocytes and participates in lipid uptake, synthesis, and storage [337]. USP22 upregulates the expression of ACC and ACLY by promoting the K48-linked ubiquitination of PPARγ, ultimately fostering lipid accumulation and tumorigenesis in hepatocellular carcinoma cells [172]. Furthermore, its overexpression has been linked to a poor prognosis for a number of cancers [338–340]. This suggests that USP22 could serve as a novel tumor marker for cancer prognosis. The E3 ligase Trib1-COP1 complex targets ACC1 for ubiquitination and degradation, inhibiting leukemia-initiating cells and promoting myeloid differentiation in AML, protecting against leukemia-related mortality and impeding the progression of acute myeloid leukemia (Fig. 5) [162]. Hence, identifying a ubiquitination enzyme that counteracts the ubiquitination of Trib1-COP1 is crucial for targeting the fatty acid metabolism pathway for cancer treatment. Another study found that CSN6 can antagonize the activity of the E3 ligase FBXW7β, preventing FBXW7β-mediated ubiquitination and degradation of FASN, thereby positively regulating lipogenesis in colorectal cancer (Fig. 5) [163]. Importantly, the study demonstrated that combining cetuximab with orlistat can inhibit the growth of CSN6-high patient-derived xenograft (PDX) tumors. These findings hold key prognostic and therapeutic significance for colorectal cancer patients [163]. On the other hand, USP38 can deubiquitinate and stabilize FASN in gastric cancer, increasing triglyceride production and promoting growth and migration in gastric cancer cells [204]. In addition, orilistat, an inhibitor of USP38, can reverse the phenotype of USP38 overexpressed gastric cancer cells [204]. Therefore, targeting FASN with USP38 inhibitors can be used as a potential treatment for gastric cancer patients with high expression of USP38. Moreover, previous reports have indicated that FASN serves as a substrate for USP14 in hepatocytes, but other reports suggest that FASN levels in cancer cells are not considerably impacted by the USP14 inhibitor IU1, suggesting that FASN may not be a direct substrate of USP14 in cancer cells [341, 342]. Given the above, further investigations are needed to clarify the functions of the ubiquitination and deubiquitination of pivotal enzymes in fatty acid metabolism.
Amino acid metabolism
Amino acid metabolism in cancer is influenced by ubiquitination and deubiquitination. Glutamate dehydrogenase (GDH) catalyzes the deamination of glutamic acid and has two subtypes, GDH1 and GDH2, of which GDH1 is mainly degraded by the ubiquitin–proteasome pathway [343]. The E3 ligase RNF213 mediates GDH1 degradation in kidney renal clear cell carcinoma (KIRC) (Fig. 5) [161]. Researchers found that the loss of GDH1 promotes tumor formation after amino acid deprivation by reducing α-ketoglutarate (αKG) levels and αKG-dependent lysine demethylase (KDM) activity [161]. Additionally, another study in hepatocellular carcinoma identified two GDH1 inhibitors: Quercetin and Permethylated Anigopreissin A [344]. We can hypothesize that applying these drugs to KIRC could maintain αKG levels and KDM activity, potentially preventing the progression of KIRC. The only enzyme in mammals capable of eliminating ammonia and glutamic acid and synthesizing glutamine de novo is glutamine synthetase (GS) [345]. In multiple myeloma (MM), USP15 controls the ubiquitination of GS, which is mediated by the E3 ligase complex Cul4-DDB1-CRBN-RBX1 (CRL4CRBN) [171]. Interestingly, immunomodulatory drug (IMiD)-resistant cells have high expression of USP15, and lenalidomide, an immunomodulatory medication, can sensitize these cells when USP15 is depleted [171]. Thus, focusing on USP15 offers a significant therapeutic potential to improve the efficacy of CRBN-based PROTAC treatments for the treatment of cancer. A preclinical study mentioned a small molecule inhibitor of USP15 (USP15-Inh) provided by Forma Therapeutics [346]. Glutaminase (GAC) catalyzes the initial step of glutamine decomposition, converting it into glutamic acid and ammonia [347]. The E3 ligase TRIM21 promotes K63-linked ubiquitination of GAC, inhibiting its activity in non-small cell lung cancer (Fig. 5) [164]. Furthermore, acetylation of Lys311 on GAC further enhances this inhibitory process, thereby suppressing non-small cell lung cancer progression and offering new insights for targeting TRIM21 in lung cancer therapy [164].
Evading immune destruction
The crosstalk between the ubiquitin protein system and the TME
Tumors are closely related to the surrounding microenvironment and constantly interact with each other [348]. Ubiquitination is a common posttranslational modification that plays a vital role in regulating cellular signal transduction pathways in the TME [320]. This modification effectively stimulates antitumor immunity and modulates the balance between tumor suppressors and oncoproteins by modulating the immune response [349]. The function of the UPS is to influence the TME by directly or indirectly regulating the degradation of immune checkpoint molecules and the release of oncogenic cytokines [8]. In conclusion, TME is an important component affecting tumor growth and development. The UPS affects tumor progression by regulating the interaction between tumors and the TME (Fig. 3).
The role of ubiquitination in crosstalk between CAFs and tumor cells
Cancer-associated fibroblasts (CAFs) are the most common cells in the TME. CAFs regulate the activities of tumor cells and other stromal cells through direct contact and by secreting regulatory factors, especially TGF-β, IL-6, and CC-chemokine ligand 2 (CCL2). Therefore, CAFs play an essential role in tumor progression [350].
TGF-β plays a vital role in the epithelial mesenchymal transition (EMT), promoting the transition of epithelial cells to motile mesenchymal cells and thereby promoting the migration and invasion of tumor cells (Fig. 6) [351]. The USP family is involved in regulating the TGF-β-induced EMT. According to recent literature, USP3, USP4, USP11, USP15, and USP26 positively regulated TGF-β signaling in various cancer types [194, 197–199]. For example, USP11 regulates TGF-β-induced plasticity and promotes breast cancer metastasis by stabilizing TGF-βRII [196]. High expression of USP11 was found in gastric cancer patients’ tumor samples, and its upregulation promoted gastric cancer tumor growth and metastasis. Interestingly, suppression of USP11 enhanced the sensitivity of GC cells to chemotherapy [352]. Additionally, in human basal-like basal cells, overexpression of the ubiquitin-editing enzyme A20 amplifies the TGF-β1-induced epithelial-mesenchymal transition by enhancing the polyubiquitination of Snail1. Knockdown of A20 reduces cancer metastasis in mouse xenograft tumors and an orthotopic breast cancer model, suggesting that the polymononubiquitination of A20 and Snail1 plays a key role in the metastasis process [184]. In addition, the DUB USP27X is regulated by TGF-β during the EMT and maintains the stability of Snail1 in breast cancer and prostate cancer. Inhibition of USP27X leads to the destabilization of Snail1, inhibits the EMT process, and enhances the sensitivity of tumor cells to chemotherapy [200].
Fig. 6.
The ubiquitin–proteasome system regulates tumor immunity. a Components of the tumor microenvironment include cancer-associated fibroblasts (CAFs), dendritic cells (DCs), natural killer (NK) cells, tumor-associated macrophages (TAMs), and T lymphocytes. b CAFs secrete a variety of chemokines, cytokines, and other effector molecules, such as transforming growth factor-β (TGF-β), interleukin-6 (IL-6), C-X-C chemokine ligand 12 (CXCL12), C–C chemokine ligand 2 (CCL2), stromal cell-derived factor 1 (SDF-1), vascular endothelial growth factor (VEGF), indoleamine 2,3-dioxygenase (IDO), and prostaglandin E2 (PGE2). These molecules regulate the function of immune cell populations in the TME, mediated by immune cells to inhibit immune responses. c E3 ligases and deubiquitinases that directly target PD-L1. E3 ligases USP7, USP22, CSN5, USP8, and USP9X stabilize PD-L1. Conversely, deubiquiting enzymes SPOP, FBXO38, FBXO22, and NEDD4 degrade PD-L1 through ubiquitination. The following list comprises the E3 ligases and deubiquitinating enzymes involved in processes that affect PD-L1 transcription. FBW7 and RNF31 inhibit PD-L1 transcription through the PI3K/AKT/GSK-3β signaling pathway and the Hippo/YAP/PD-L1 axis. USP22 and HERC2 promote PD-L1 transcription through the USP22-CSN5-PD-L1 axis and the JAK2/STAT3 signaling pathway, respectively. d The involvement of the ubiquitin–proteasome system in the TGF-β signaling pathway includes USP11 acting on the TGF-β type II receptor, USP15, and USP4 acting on the TGF-β type I receptor, and USP3, USP11, A20, and USP27X acting on EMT transcription factors
The role of the ubiquitin protein system in immune checkpoints
Increasing evidence has shown that the UPS plays an important role in immune checkpoints [353]. The primary focus is on the role of the UPS in the PD-1/PD-L1 pathway [8]. These findings suggest that the UPS is a novel approach for enhancing antitumor immunity. E3 ligases and deubiquitinases are pivotal in modulating the stability of PD-1 and PD-L1 [176]. SPOP proteins regulate various cancer-related substrates and play a crucial role in mediating PD-L1 degradation [354]. Research has found that CDK4/6 increases PD-L1 protein levels by inhibiting the phosphorylation of SPOP mediated by cyclin D-CDK4. The combination of CDK4/6 inhibitors with anti-PD-1 immunotherapy has been shown to reduce tumors and significantly improve overall survival rates in mouse tumor models [173]. Additionally, as a subunit of the SCF E3 ligase complex, the F-box protein plays various roles in human tumors, including mediating the ubiquitination of PD-1 [176, 177]. One experiment showed that FBXO22 could activate the ubiquitination of PD-L1 to increase the sensitivity of non-small cell lung cancer cells to DNA damage, with CDK5 acting as an upstream regulator of FBXO22. Research suggests that combining CDK5 inhibitors with immune checkpoint inhibitors enhances the efficacy of immune checkpoint blockade therapy [178]. In addition, USP22, and USP9X stabilize PD-L1 through deubiquitination, promoting cancer development and migration [187, 193, 355]. Studies also show a positive correlation between PD-L1 and USP12 levels, with USP12 knockout desensitizing mouse lung tumor cells to anti-PD-1 therapy [192]. Additionally, USP14 knockout or inhibition enhances cell response to PD-1 inhibitors [191]. UPS also plays a vital role in cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) [185]. TRAF6 is involved in CD8 + T-cell-mediated antitumor immunity by promoting the ubiquitination and degradation of CTLA-4 [185]. The role of the UPS in immune checkpoints, particularly the PD-1/PD-L1 pathway, suggests a potential for novel therapeutic strategies aimed at enhancing antitumor immunity.
Unlocking phenotypic plasticity
During organ development, cells organize into tissues and undergo terminal differentiation, often leading to irreversible growth cessation. This process acts as a barrier to the sustained proliferation needed for tumor formation. Phenotypic plasticity can lead to changes such as dedifferentiation, blocked terminal differentiation, and transdifferentiation [356]. Subsequently, the impact of ubiquitination on surface plasticity was explored through an examination of its effects on dedifferentiation and transdifferentiation, along with a discussion of the processes of ubiquitination and deubiquitination.
The first step is dedifferentiation from the mature state to the progenitor cell state. Dedifferentiated liposarcoma (DDLPS) is an invasive adipose cell carcinoma. Characterized by a low tumor mutation burden and frequent chromosomal structural abnormalities, DDLPS often exhibits amplification of the MDM2 gene. MDM2, an E3 ligase, is responsible for targeting the degradation of p53. The overexpression of MDM2 in human cancers has been associated with a poor prognosis [202]. Targeting MDM2 is a promising therapeutic strategy, as demonstrated by the growing number of MDM2 inhibitors, such as RG7112, AMG-232, and APG-115, undergoing clinical trials [357]. Second, transdifferentiation refers to the transformation of tissue cells into cells of different lineages. The ubiquitin proteasome system is involved in the transdifferentiation process of specific cancers. The transformation of adventitial fibroblasts (AFs) into myofibroblasts is pivotal in the vascular restructuring observed in conditions such as atherosclerosis, restenosis, and aortic aneurysm. Notably, NADPH oxidase 4 (Nox4) undergoes ubiquitination through direct engagement with the ubiquitin-specific protease domain of CYLD. Elevated levels of CYLD and Nox4 in the adventitia due to hyperhomocysteinemia significantly enhance AF transformation, exacerbating CaPO4-induced abdominal aortic aneurysm progression in mice [203]. Furthermore, neuroendocrine prostate cancer represents a deadly subtype of prostatic cancer that is distinguished by the attenuation of AR signaling during neuroendocrine transdifferentiation. This alteration ultimately contributes to the development of drug resistance to AR-targeted therapies. Researchers have shown that the differentially expressed gene Rac GTPase-activating protein 1 (RACGAP1) is involved in the NE transdifferentiation of prostatic cancer. The underlying mechanism is that RACGAP1 promotes the neuroendocrine transdifferentiation of prostatic cancer by stabilizing the expression of EZH2 in the ubiquitin proteasome pathway [358].
Nonmutational epigenetic reprogramming
In recent years, as research on chromatin and histones has deepened, epigenetics has gradually emerged. Epigenetics encompasses diverse modifications in gene expression that occur without modifying the gene’s DNA sequence, resulting in hereditary changes in the gene’s function [359, 360]. Epigenetic phenomena include DNA methylation, histone modification, chromatin remodeling, and noncoding RNAs [361, 362]. Studies on epigenetic phenomena related to ubiquitination and deubiquitination include DNA methylation, histone modification, and chromatin remodeling (Fig. 3).
DNA methylation
DNA hypermethylation of the promoter at the cytosine-phosphate-guanine (CpG) sequence is a clearly defined epigenetic marker in all human tumor types and leads to the silencing of tumor suppressor genes (TSGs) and other genes associated with cancer, thus giving precancerous cells a selective advantage [363, 364]. Recent research on ubiquitination and deubiquitination in DNA methylation focuses on DNA methyltransferases (DNMTs). The E3 ligase RNF180 ubiquitinates DNMT1, significantly reducing PCDH10 methylation levels and increasing its expression in gastric cancer. Furthermore, the positive co-expression of RNF180 and PCDH10 is associated with a favorable clinical prognosis in gastric cancer patients, suggesting that PCDH10 and RNF180 could be potential biomarkers for gastric cancer diagnosis [206]. A similar study revealed the RNF180/DNMT3A/ADAMTS9 axis in gastric cancer. ADAMTS9 significantly inhibits cell viability and motility both in vitro and in vivo. RNF180 ubiquitinates DNMT3A, markedly reducing ADAMTS9 methylation levels and increasing its expression in gastric cancer [207]. Therefore, finding an inhibitor that targets RNF180 could potentially inhibit the progression of gastric cancer from multiple angles. Fructose-1,6-bisphosphatase 1 (FBP1) is an enzyme that catalyzes a key step in gluconeogenesis, converting fructose-1,6-bisphosphate to fructose-6-phosphate [365]. Nuclear FBP1 has been found to interact with DNMT1 and to recruit PARP1 to chromatin, enhancing the sensitivity of pancreatic cancer to the poly ADP-ribose polymerase (PARP) inhibitor Olaparib [211]. Significantly, USP7 can reverse this by deubiquitinating FBP1, thereby inhibiting this interaction. Consequently, USP7 inhibitors enhanced the anti-tumor effects of PARP inhibitors in an FBP1-dependent manner. Therefore, combining USP7 inhibitors with PARP inhibitors might yield a more potent anti-tumor response than using PARP inhibitors alone, potentially offering a more effective treatment approach for PC [211].
Histone ubiquitination
Histone octamer is composed of H2A, H2B, H3, and H4, which are connected with DNA to form nucleosomes [366]. Histone ubiquitination contributes to proper DSB repair and plays a significant role in the interaction with transcription and replication [367]. The E3 ligase RNF40 can interact with the E3 ligase complex DNA binding protein 1 (DDB1)-Cullin 4a (CUL4A), inhibiting the ubiquitination of H2A by DDB1-CUL4A [208]. Notably, RNA demethylase ALKB Homolog 5 (ALKBH5)-mediated m6A deficiency in osteosarcoma leads to increased expression of USP22 and RNF40, suppressing H2A ubiquitination and promoting gene expression related to replication and DNA repair, driving osteosarcoma progression [208]. Therapeutic strategies could focus on creating modulators to enhance or mimic ALKBH5 activity, targeting the overexpression of USP22 and RNF40, offering a new potential treatment for cancers with m6A abnormalities. Cyclin B1, encoded by the CCNB1 gene, is regulated by H2B ubiquitination at its promoter by the E2 enzyme radiation sensitive 6 (RAD6) in esophageal squamous cell carcinoma (ESCC), affecting ESCC cell proliferation [205, 368]. Additionally, a novel RAD6 selective small molecule inhibitor targeting the catalytic site of RAD6 (SMI#9) enhanced the sensitivity of cancer cells resistant to cisplatin or oxaliplatin in triple-negative breast cancer and colorectal cancer [369]. Using SMI#9 could provide a strategy to overcome drug resistance in chemotherapy, offering a promising avenue for enhancing the effectiveness of existing cancer treatments in various resistant tumors. Therefore, specific human proteins can affect histone ubiquitination by regulating E3 ligases or E2 ubiquitin binding enzymes, impacting the cell cycle and cancer development. Refer to section II.A for additional details on histone ubiquitination.
Chromatin remodeling
Nucleosomes, linker histones, and nonhistones undergo further assembly into highly organized chromatin structures, restricting access to DNA [370]. The main subfamilies of chromatin remodeling complexes are switch/sucrose nonfermentable (SWI/SNF), imitation SWI (ISWI), chromodomain-helicase DNA-binding protein (CHD), and inositol-requiring mutant 80 (INO80) [371, 372]. The SWI/SNF complex was the first remodeling complex discovered [373]. In prostate cancer, OTUD6A deubiquitinates the SWI/SNF ATPase subunit Brg1, promoting cancer progression [212]. AT-rich interactive domain protein 1A (ARID1A), a SWI/SNF complex component, acts as a tumor suppressor [374]. In gastric cancer and hepatocellular carcinoma, the E3 ubiquitin ligase complex SCF promotes degradation of ARID1A, which is triggered by ATM activation due to DNA damage in gastric cancer and by mTORC1 activation in hepatocellular carcinoma [186, 209]. Some progress has been made in the clinical application of targeted anticancer therapies focusing on SCF complexes, particularly through inhibitors of cullin neddylation and Skp2, such as MLN4924 and Erioflorin [375, 376]. During normal DNA synthesis, BAP1 stabilizes INO80 through deubiquitination. Additionally, BAP1 recruits INO80 to replication forks by interacting with H2Aub, thereby facilitating fork progression. This process underscores the molecular basis of BAP1’s tumor suppressor function [377]. Additionally, recent research indicates that the E3 ligase CHIP also can stabilize INO80 through nondegradable ubiquitination. Therefore, CHIP and BAP1 collaborate to regulate the ubiquitination of INO80, thereby facilitating DNA replication [210]. These findings are noteworthy for the investigation of chromatin remodeling in cancer.
Polymorphic microbiomes
Increasing evidence suggests that the polymorphic variability of the microbiome between individuals impacts cancer phenotypes, and distinct characteristic microbiota have been recognized in different tumors [378, 379]. Micromonas infection can enhance the proliferation and inflammatory response of colorectal cancer cells, and Parvimonas micra (P. micra) was found to affect protein expression in colorectal cancer intestinal epithelial cells. The upregulation of proteasome β4 (PSMB4) protein indicates the critical role of the ubiquitin proteasome pathway in colorectal cancer [213]. In addition, propionate, a microbial metabolite, was shown to target euchromatic histone lysine methyltransferase 2 (EHMT2) by coordinating proteasomal degradation through the upregulation of HECT domain E3 ubiquitin protein ligase 2 (HECTD2). Propionate treatment initially increases the expression of HECTD2, which then facilitates the proteasomal degradation of EHMT2 through posttranslational modification. EHMT2, through H3K9 dimethylation, forms heterochromatin structures and negatively regulates tumor necrosis factor alpha-induced protein 1 (TNFAIP1) [214]. Consequently, the degradation of EHMT2 reduces H3K9 dimethylation in the TNFAIP1 promoter region, leading to the upregulation of TNFAIP1 and apoptosis of colorectal cancer cells [214]. Furthermore, treatment of colorectal cancer cells with Clostridium butyricum (C. butyricum) decreased MYC-mediated resistance to 5-FU and enhanced the effectiveness of anti-PD-1 immunotherapy [380]. In summary, the microbiota and its metabolites can regulate tumor development and therapeutic efficacy by affecting the UPS. These experiments suggest potential therapeutic strategies and highlight the importance of microbiota mechanisms of action in cancer research.
Senescent cells
Senescence is an irreversible state of cell cycle arrest that occurs when cells respond to various stress factors, including DNA damage or activation of oncogenes [381]. While senescence can exert a tumor suppressive effect by preventing the proliferation of damaged or mutated cells, it also has potential impacts on the development and progression of cancer [7]. Senescence in cancer may rely on oncogene-induced senescence (OIS) and tumor suppressor gene loss-induced senescence (TIS). The P16INK4A-RB and p53-p21-RB pathways are crucial mechanisms for initiating and maintaining growth arrest [381, 382]. Melanoma is a malignant tumor that originates from melanocytes in the skin and is rich in senescent cells. In melanoma, overexpression of mutant BRAF promotes excessive proliferation of melanocytes, inducing the expression of P16INK4A, which subsequently inhibits the activity of CDK4 and CDK6, leading to hypophosphorylation of RB, cell cycle arrest, and thus inhibiting cancer development [383]. In a study, primary mouse and human cells lacking BRCA2 exhibited senescence characteristics, which reversed upon the loss of ARF [98]. This may be due to the ability of ARF to inhibit the activity of E3 ligase MDM2, thus enabling normal p53 function. Targeting the interaction between MDM2 or ARF-MDM2 using small molecule inhibitors may help restore p53 function in the impaired p53 signaling pathway, promoting senescence or apoptosis in cancer cells [384]. ALRN-6924 reactivates p53 function by inhibiting two proteins, MDM2 and MDMX, subsequently inhibiting tumor cell growth [384]. Compounds like ALRN-6924, which induce inflammatory responses and reduce immune evasion in the tumor microenvironment, could be effectively combined with immunotherapies, especially in melanomas rich in senescent cells [384]. Exploring the interplay between ubiquitination, deubiquitination, and cellular senescence opens promising avenues for developing targeted therapies that can selectively modulate cell fate in cancer.
Cancer treatment strategies
Proteinase inhibitors
The proteasome is a highly anticipated target in cancer therapy. Proteasome inhibitors (PIs), like bortezomib, carfilzomib, oprozomib, and ixazomib, have been successfully developed for clinical treatment and have shown good efficacy (Fig. 7) [184, 385, 386].
Fig. 7.
Schematic diagram of ubiquitin–proteasome system regulating proteins and its corresponding treatment strategies. Drugs targeting proteasome: bortezomib, carfilzomib, oprozomib and ixazomib. Drugs targeting El enzyme: TAK-243, pevonedistat, TAK-981 andTAS4464. Drugs targeting E2 enzyme: Leucetta A, manado sterols A, manado sterols B andCC0651. Drugs targeting E3 ligase: S-phase kinase-associated protein 2 (SKP2) inhibitors and homologous to the E6AP C-terminus (HECT)-type E3 ligase inhibitors, HOlL-1 interacting protein (HOIP) inhibitors, mouse double minute 2 (MDM2) inhibitors and IAPs inhibitors. Drugs targeting deubiquitinase (DUB): Broad-range DUB inhibitors, inhibitors targeting USP7, inhibitors targeting USP14, and inhibitors targeting UCH-L1
Bortezomib is used to treat patients with MM by inhibiting the chymotrypsin-like activity of the 26S proteasome [387, 388]. Although its use alone may cause a wide range of unintended side effects, its toxicity can be reduced when used in combination with pomalidomide and dexamethasone [389, 390]. In addition, the FDA has approved bortezomib for treating mantle cell lymphoma [391]. Currently, bortezomib is in clinical trials for treating other types of cancer, such as autoimmune hemolysis and COAD [392, 393].
Carfilzomib is a tetrapeptide epoxide that selectively binds to the 26S proteasome and inhibits protease activity. It was approved by the FDA in 2012 for the treatment of multiple myeloma [394, 395]. Carfilzomib is undergoing clinical trials for various cancers, including renal cell carcinoma, lymphoma, acute myeloid leukemia, lymphocytic leukemia, and small-cell lung cancer.
Oprozomib is a derivative of carfilzomib designed to have better oral bioavailability. Oprozomib has shown similar antitumor activity and efficacy to carfilzomib in the treatment of multiple myeloma. Therefore, it can be used for the treatment of multiple myeloma resistant to bortezomib, dexamethasone, or lenalidomide [396]. Oprozomib induces apoptosis by upregulating the proapoptotic proteins Bcl-2 interacting killer (BIK) and MCL-1. It can be used to treat solid tumors [397]. However, oprozomib has high gastrointestinal toxicity and unstable pharmacokinetics.
Ixazomib was used to inhibit the activity of the 20S proteasome [398, 399] and its antitumor effect is superior to bortezomib [399]. Ixazomib combined with lenalidomide/dexamethasone significantly improves the survival of patients with myeloma [400]. Ixazomib has completed phase I clinical trials for glioblastoma multiforme (GBM) and phase II clinical trials for malignant myeloid and lymphatic blood cancers. The patients who exhibited resistance to bortezomib demonstrated a favorable response to isazomil. In addition to these inhibitors, other proteasome inhibitors, such as marizomib and delanzomib, are also undergoing clinical trials. For example, some studies have tested the effects of malizomil on glioblastoma through its ability to cross the blood brain barrier [395].
E1 and E2 inhibitors
The E1 enzyme functions to activate ubiquitin and transfer it to the E2 enzyme [1]. Adenosine 3', 5' monophosphate (AMP) is tightly bound to the E1 enzyme in the catalytic cascade of ubiquitin activation, and some AMP-related drugs can act as E1 inhibitors. The major sodium adenosulfonate E1 inhibitors include perazone, TAK-243, ML-792, TAK-981, acetyl-DL-carnitine, ABPA3, and ABP1 (Fig. 7) [401]. Among the E1 inhibitors in clinical trials, the only one with published clinical trial data is pevonedistat. Combination therapy with risperidone, such as risperidone and azacitidine, has shown more promising efficacy in the treatment of AML patients, and carboplatin and paclitaxel have shown better clinical benefits in the treatment of advanced solid tumors. In patients with AML with TP53 mutations, the composite CR/PR rate was 80% with pevonedistat and azacitidine combination therapy [402, 403].
E2 enzymes primarily facilitate the binding of ubiquitin to substrates. Current efforts are focused on identifying inhibitors that disrupt the interaction between E1 enzymes and E2 enzymes or between E2 enzymes and E3 ligases. For instance, Leucetta A has been shown to inhibit the interaction between ubiquitin-conjugating enzyme 13 (UBC13) and ubiquitin-like protein one activating enzyme (UEV1A), thereby preventing complex formation [404]. Alternatively, manadosterols A and B, isolated from the sponge Lissodendoryx fibrosa, target the same molecular interaction as Ubc13-Uev1A [405]. In addition, the E2 enzyme Cdc34 inhibitor CC0651 blocks the ubiquitination and degradation of p27, thereby inhibiting tumor cell proliferation [406].
E3 inhibitors
The E3 ligase interacts with the ubiquitin-activating enzyme E1 and the ubiquitin-conjugating enzyme E2 to complete the ubiquitination process [407]. Drugs targeting E3 ligases play an essential role in cancer therapy by linking ubiquitin to specific protein amino acids. Clinical research information on these inhibitors has been obtained from https://clinicaltrials.gov/ and https://pubchem.ncbi.nlm.nih.gov/, as listed in Table 2.
Table 2.
Summary of pharmacological strategies direct targeting the ubiquitin proteasome system for cancer therapy in clinical trials (information was obtained from https://www.clinicaltrials.gov/)
| Identifier | Phase | Drug | Target | Cancer | Treatment | Status | References |
|---|---|---|---|---|---|---|---|
| NCT02045095 | Phase 1 | Uba1 | Uae | Solid tumors | Monotherapy | Terminated | NA |
| NCT04074330 | Phase 1/phase 2 | TAK-981 | Sae | Lymphoma | Combination with rituximab | Completed | [408] |
| NCT04065555 | Early phase 1 | TAK-981 | Sae | Head and neck cancer | Monotherapy and combination | Completed | [409] |
| NCT03770260 | Phase 1 | Pevonedistat | NEDD8 | Multiple myeloma | Combination with ixazomib | Completed | NA |
| NCT04712942 | Phase 2 | Pevonedistat | NEDD8 | Myeloid leukemia | Combination with azacitidine | Completed | NA |
| NCT03319537 | Phase 1/phase 2 | Pevonedistat | NEDD8 | Mesothelioma | Combination with standard chemotherapy, | Completed | NA |
| NCT03330106 | Phase 1 | Pevonedistat | NEDD8 | Solid neoplasm | Combination with standard chemotherapy | Completed | [410] |
| NCT03814005 | Phase 1 | Pevonedistat | NEDD8 | Myeloid leukemia | Combination with standard chemotherapy | Completed | NA |
| NCT03349281 | Phase 1 | Pevonedistat | NEDD8 | Lymphoblastic leukemia | Combination with vxld | Completed | NA |
| NCT02610777 | Phase 2 | Pevonedistat | NEDD8 | Myeloid leukemia | Combination with azacitidine | Completed | [411] |
| NCT02782468 | Phase 1 | Pevonedistat | NEDD8 | Myeloid leukemia | Combination with azacitidine | Completed | [412] |
| NCT03486314 | Phase 1 | Pevonedistat | NEDD8 | Advanced solid neoplasm | Monotherapy | Completed | [413] |
| NCT03459859 | Phase 1 | Pevonedistat | NEDD8 | Myeloid leukemia | Combination with low dose cytarabine | Completed | NA |
| Nct01862328 | Phase 1 | Pevonedistat | NEDD8 | Solid tumors | Combination | Completed | [402] |
| NCT01814826 | Phase 1 | Pevonedistat | NEDD8 | Myelogenous leukemia | Combination with azacitidine | Completed | [403] |
| NCT03057366 | Phase 1 | Pevonedistat | NEDD8 | Advanced solid tumors | Monotherapy | Completed | [414] |
| NCT02122770 | Phase 1 | Pevonedistat | NEDD8 | Advanced solid tumors | Monotherapy | Completed | [415] |
| NCT00911066 | Phase 1 | Pevonedistat | NEDD8 | Myeloid leukemia | Monotherapy | Completed | [416] |
| NCT00722488 | Phase 1 | Pevonedistat | NEDD8 | Multiple myeloma | Monotherapy | Completed | [417] |
| NCT00677170 | Phase 1 | Pevonedistat | NEDD8 | Nonhematologic malignancies | Monotherapy | Completed | [418] |
| NCT01011530 | Phase 1 | Pevonedistat | NEDD8 | Metastatic melanoma | Monotherapy | Completed | [419] |
| NCT02935907 | Phase 1 | APG115 | MDM2 | Solid tumor or lymphoma | Monotherapy | Completed | NA |
| NCT01877382 | Phase 1 | Milademetan | MDM2 | Advanced solid tumor | Monotherapy | Completed | [420] |
| NCT03671564 | Phase 1 | Milademetan | MDM2 | Myeloid leukemia | Monotherapy | Completed | [421] |
| NCT03614455 | Early phase 1 | Milademetan | MDM2 | Pharmacokinetics | Combination | Completed | [422] |
| NCT02890069 | Phase 1 | Siremadlin | MDM2 | Colorectal cancer | Combination | Completed | NA |
| NCT02143635 | Phase 1 | Siremadlin | MDM2 | Solid and hematological tumors | Monotherapy | Completed | [423] |
| NCT02343172 | Phase 1 | Siremadlin | MDM2 | Liposarcoma | Combination with lee011 | Completed | NA |
| NCT01723020 | Phase 1 | AMG 232 | MDM2 | Malignancy | Monotherapy | Completed | [241] |
| NCT02110355 | Phase 1 | AMG 232 | MDM2 | Malignancy | Combination with trametinib and dabrafenib | Completed | [424] |
| NCT02016729 | Phase 1 | AMG 232 | MDM2 | Malignancy | Combination with trametinib | Completed | [425] |
| NCT01677780 | Phase 1 | RG7112 | MDM2 | Myelogenous leukemia | Monotherapy | Completed | NA |
| NCT00559533 | Phase 1 | RG7112 | MDM2 | Neoplasms | Monotherapy | Completed | NA |
| NCT00623870 | Phase 1 | RG7112 | MDM2 | Hematologic neoplasms | Monotherapy | Completed | NA |
| NCT01143740 | Phase 1 | RG7112 | MDM2 | Sarcoma | Monotherapy | Completed | NA |
| NCT01164033 | Phase 1 | RG7112 | MDM2 | Neoplasms | Monotherapy | Completed | [426] |
| NCT01605526 | Phase 1 | RG7112 | MDM2 | Sarcoma | Combination with doxorubicin | Completed | NA |
| NCT01636479 | Phase 1 | SAR405838 | MDM2 | Neoplasm malignant | Monotherapy | Completed | [427] |
| NCT01985191 | Phase 1 | SAR405838 | MDM2 | Neoplasm malignant | Combination with pimasertib | Completed | [428] |
| Nct02670044 | Phase 1 | RG7388 | MDM2 | Acute myeloid leukemia | Combination with venetoclax | Completed | [429] |
| NCT03362723 | Phase 1 | RG7388 | MDM2 | Solid tumors | Monotherapy | Completed | NA |
| NCT02828930 | Phase 1 | RG7388 | MDM2 | Solid tumors | Monotherapy | Completed | [430] |
| NCT01773408 | Phase 1 | RG7388 | MDM2 | Acute myeloid leukemia | Combination with cytarabine | Completed | [431] |
| NCT01462175 | Phase 1 | RG7388 | MDM2 | Neoplasms | Monotherapy | Completed | [432] |
| NCT01901172 | Phase 1 | RG7388 | MDM2 | Neoplasms | Combination with posaconazole | Completed | NA |
| NCT01760525 | Phase 1 | CGM097 | MDM2 | Solid tumor | Monotherapy | Completed | [433] |
| NCT02890069 | Phase 1 | LCl161 | IAPs | Colorectal cancer | Combination | Completed | NA |
| NCT03111992 | Phase 1 | LCl161 | IAPs | Multiple myeloma | Combination with pdr001 | Completed | NA |
| NCT01240655 | Phase 1 | LCl161 | IAPs | Solid tumors | Combination with paclitaxel | Completed | NA |
| NCT01968915 | Phase 1 | LCl161 | IAPs | Neoplasms | Monotherapy | Completed | [434] |
| NCT01617668 | Phase 2 | LCl161 | IAPs | Breast cancer | Combination with paclitaxel | Completed | NA |
| NCT01098838 | Phase 1 | LCl161 | IAPs | Solid tumors | Monotherapy | Completed | [435] |
| NCT04122625 | Phase 1/phase 2 | AT-406 | IAPs | Solid tumor | Combination with nivolumab | Completed | NA |
| NCT03871959 | Phase 1 | AT-406 | IAPs | Adenocarcinoma of the pancreas | Combination with pembrolizumab | Completed | [436] |
| NCT02022098 | Phase 2 | AT-406 | IAPs | Squamous cell carcinoma | Combination with cisplatin and radiotherapy | Completed | [437] |
| NCT03270176 | Phase 1 | AT-406 | IAPs | Neoplasms | Combination with avelumab | Completed | NA |
| NCT01078649 | Phase 1 | AT-406 | IAPs | Solid tumors | Monotherapy | Completed | [438] |
| NCT01940172 | Phase 1 | Tl-32711 | IAPs | Ovarian cancer | Combination with conatumumab | Completed | [439] |
| NCT03386526 | Phase 1 | APG-1387 | IAPs | Solid Tumors | Monotherapy | Completed | [440] |
PROTACs
Small molecule drugs are designed to specifically target disease-related proteins using lock-and-key mechanisms. This approach relies on the presence of suitable pocket regions in the target protein structure as small molecule binding sites. PROTAC technology provides an essential platform for inducing the degradation of target proteins. The PROTAC molecule consists of two components, a ligand capable of specifically binding to the target protein and a ligand that recruits an E3 ligase to promote ubiquitination of the captured protein, leading to target protein degradation [441, 442]. ARV-110 and ARV-471 have progressed to phase II clinical trials. Much of the subsequent discussion has focused on these two pharmaceutical compounds.
ARV-110 utilizes PROTAC technology and has potential antitumor activity. ARV-110 can bind to the AR ligand recognition domain of the E3 ligase [12]. ARV-110 successfully completely degrades AR in various cell lines (DC50 < 1 nM) [389], and oral administration of ARV-110 (10 mg/kg) successfully inhibits the growth of enzalutamide-insensitive tumors in hepatocellular carcinoma patient-derived xenograft (PDX) models [390].
ARV-471 is an oral heterobifunctional molecule that uses PROTAC technology to target estrogen receptor (ER) α and has potential antitumor activity [443]. A phase I study involving patients with ER + and HER2-BC found that ARV-471 significantly reduced ER expression by up to 90% in tumor tissue. The phase I data indicated that ARV-471 performs well at any dose, substantially degrades ER, and is well tolerated. ARV-471 degrades both wild-type and mutant ER proteins. ARV-471 is undergoing a phase II clinical trial evaluating its efficacy in patients with ER + /HER2 + locally advanced and metastatic breast cancer [444, 445]. Oral ARV-471 monotherapy showed promising antitumor activity in estrogen-dependent MCF7 xenografts and a significant reduction in ER protein levels. Enhanced anticancer effects were noted when combined with the CDK4/6 inhibitor, palbociclib [446]. ARV-471 also showed a good inhibitory effect on a hormone-independent PDX model of estrogen receptor 1 (ESR1) mutants [447]. These results demonstrate the feasibility of the PROTAC approach in patients.
Molecular glue
Although PROTAC technology offers significant potential for drug development, the designed molecules are typically large. An alternative effective strategy involves the use of molecular glue degraders, small molecules capable of facilitating novel interactions between target proteins and E3, ultimately leading to ubiquitination-mediated degradation of the target protein [448]. Unlike PROTACs, molecular glues have better chemical properties and smaller molecular volumes (Table 3).
Table 3.
Representative small molecules targeting protein degradation under clinical evaluation (information was obtained from https://www.clinicaltrials.gov/)
| Identifier | Phase | Drug | Target | Cancer | Treatment | Status | References |
|---|---|---|---|---|---|---|---|
| NCT02372240 | Phase 1/phase 2 | VLX1570 | UCHL5 and USP14 | Multiple myeloma | Combination with dexamethasone | Terminated | [449] |
| NCT01049841 | Phase 1 | Perifosine | UCHL3 | Pediatric solid tumors | Combination with temsirolimus | Completed | [450] |
| NCT00873457 | Phase 2 | Perifosine | UCHLl3 | Refractory tumors | Monotherapy | Completed | [451] |
| NCT00391560 | Phase 2 | Perifosine | UCHL3 | Leukemia | Monotherapy | Completed | NA |
| NCT00054145 | Phase 2 | Perifosine | UCHL3 | Breast cancer | Monotherapy | Completed | [452] |
| NCT01097018 | Phase 3 | Perifosine | UCHL3 | Colorectal cancer | Monotherapy | Completed | [453] |
| NCT05240898 | Phase 1 | KSQ-4279 | USP1 | Advanced solid tumors | Monotherapy and combination therapy | Active, not recruiting | NA |
| NCT04336982 | Phase 1/phase 2 | CC-90009 | CUL4-DDB1-CRBM-RBX1 E3 complex | Acute myeloid leukemia | Combination therapy | Active, not recruiting | [13] |
| NCT00676910 | Phase 1 | JNJ-26854165 | MDM2 | Neoplasms | Monotherapy | Completed | [454] |
| NCT03041688 | Phase 1 | AMG-232 | MDM2 | Acute myeloid leukemia | Combination with decitabine and venetoclax | Recruiting | NA |
| NCT03654716 | Phase 1 | ALRN-6924 | MDM2 | Leukemia | Monotherapy | Completed | [455] |
| NCT02579824 | Phase 1 | DS-3032b | MDM2 | Myeloma | Monotherapy | Terminated | NA |
| NCT02098967 | Phase 1 | RO6839921 | MDM2 | Acute myeloid leukemia | Monotherapy | Completed | [456] |
| NCT01462175 | Phase 1 | RO5503781 | MDM2 | Advanced malignancies except leukemia | Monotherapy | Completed | [457] |
| NCT03449381 | Phase 1 | BI907828 | MDM2 | Different types of advanced cancer | Monotherapy | Recruiting | NA |
| NCT05376800 | Phase 1 | BI907828 | MDM2 | Glioblastoma | Monotherapy | Recruiting | NA |
| NCT03964233 | Phase 1 | BI907828 | MDM2 | Different types of advanced cancer | Combination with ezabenlimab | Recruiting | NA |
| NCT05107674 | Phase 1 | NX-1607 | Cbl-b | Advanced malignancies | Monotherapy and combination with paclitaxel | Recruiting | NA |
| NCT04283097 | Phase 1 | KPG-818 | CRL4 | Hematological malignancies | Monotherapy | Recruiting | NA |
Progress has been made in the development of drugs containing the E1 and E2 enzymes. However, because E3 ligases can bind to target proteins more precisely and specifically, drugs that act on E3 ligases are expected to be developed [458]. CC-90009 can recruit SPT1 to the CRL4CRBN E3 complex and promote the ubiquitination of GSP for proteasomal degradation [13]. Serdemetan (JNJ-26854165) is an antagonist of the human double minute 2 (HDM2) E3 ligase, that blocks p53 degradation by inhibiting the ubiquitination of HDM2. In addition, serdemetan can inhibit cholesterol transport. Clinical research on human cell lymphoma and multiple leiomyomas is underway [454, 459]. Notably, several small molecule drugs targeting MDM2 have been identified, as shown in Table 3. In addition, phase I clinical trials are currently underway for KPG-818, a potential therapeutic agent targeting Cullin-RING ligase 4 (CRL4) to treat hematological malignancies (NCT04283097) [460].
Other inhibitors
SKP2 inhibitors
CpaA blocks SKP2 assembly in SCF complexes, leading to G1/S cell cycle arrest and SCFSKP2/ p27-dependent cell death while overcoming multidrug resistance [461]. In another experiment, thiazolidinedione derivatives C1, C2, C16, and C20 were shown to target the SKP2-Cks1/p27 binding interface by selectively inhibiting p27 ubiquitination [462]. SZL-P1-41 can effectively inhibit SKP2 and enhance the sensitivity of glioma cells to temozolomide (TMZ) [463]. Dt204 reduces myeloma growth by reducing the binding of SKP2 to cullin and commd1 [464].
HECT-type E3 ligase inhibitors
Research indicates that heclin, a small molecule inhibitor, can alter the conformation of the HECT domain, significantly suppressing the activity of HECT-type E3 ligases and demonstrating antitumor properties [465]. In addition, a high-throughput screen identified that clomipramine, an inhibitor of the HECT ubiquitin E3 ligase ITCH, acts as an autophagy modulator to inhibit the growth of breast cancer, prostate adenocarcinoma, and bladder urothelial carcinoma cells [466]. Alternatively, a molecular model of the WW domain containing E3 ubiquitin protein ligase 2 (WWP2) inhibitor complex, which combines saturation transfer differential nuclear magnetic resonance (STD NMR), DEEP-STD NMR methods, and docking calculations, has recently been proposed to provide a method for the development of novel inhibitors [467].
MDM2 inhibitors
MDM2 can ubiquitinate and degrade p53 and is an ideal target for cancer therapy [468, 469]. APG115, which has a high affinity for MDM2 and significantly promotes tumor regression, is currently undergoing clinical trials for cancer therapy [470]. In addition, the MDM2 inhibitor APG-115 showed a synergistic effect with PD-1 blockade to enhance antitumor immunity within the TME [471]. APG-115 has shown potent antitumor activity in preclinical models of acute myeloid leukemia [472]. Inhibitors of MDM2 include milademetan, milademetan tosylate, siremadlin, siremadlin succinate, AMG 232, RG7112, SAR405838, RG7388, CGM097, and Nutlin-3A, which are in clinical trials to investigate their therapeutic effects on cancer [357, 473].
IAP inhibitors
IAP inhibitors were created by mimicking Smac/Diablo, a natural antagonist of IAPs, to induce the proteasome-dependent degradation of cIAP1, cIAP2, and X-linked IAPs [474]. Various small molecule inhibitors targeting the IAP are clinically available, as shown in Table 2 [473]. LCL161 treatment can induce an acute inflammatory response and activate phagocytes. In addition, LCL161 treatment can stimulate myeloma cells to secrete soluble factors through MΦs and induce tumor cell phagocytosis, thereby enhancing innate and adaptive immune responses and effectively stimulating antitumor immunity [475]. In another study, APG-1387 exerted dual anti-tumor effects on ciAP2-overexpressing HBV-positive hepatocellular carcinoma cells by inducing apoptosis and enhancing antitumor immunity [173]. Currently, several inhibitors of IAPs remain under evaluation in clinical trials (NCT04568265 and NCT04643405), and IAP inhibitors are promising novel effective immunomodulators for cancer treatment.
DUB inhibitors
Ubiquitination is a dynamic process managed by DUBs, which facilitate the removal and alteration of UB or polyubiquitin chains from ubiquitinated proteins [476]. Many DUBs are involved in the cell cycle process, regulation of genomic instability, and various events in tumorigenesis [477]. As a result, numerous inhibitors of DUBs have been developed, including both broad-spectrum and targeted varieties, all of which are recognized as promising candidates for cancer therapy [478, 479].
G5 and F6 are broad-spectrum DUB inhibitors discovered through cell-based drug screening [480]. These chalcone DUB inhibitors are known for their ability to induce apoptosis in BCL-2-independent cells [480, 481]. Through active chemical proteomics, compound PR619 was suggested as a broad-spectrum DUB inhibitor [482]. NSC632839, another potent inhibitor with broad-spectrum activity against deubiquitinases, selectively targets USP2 and USP7, inducing apoptosis in cancer cells [480]. As a specific USP1 inhibitor, pimozide can block the maintenance and radiation resistance of glioma stem cells [483]. WP1130 inhibits several DUBs, including USP9X, USP5, USP14, ubiquitin carboxyl-terminal hydrolase isozyme L5 (UCHL5), and UCH37. It decreases the MCL-1 level and increases the p53 level, showing antitumor effects [484]. Betulinic acid, derived from various plants, has been recognized as a broad-spectrum DUB inhibitor, that triggers an aberrant transmembrane potential and apoptosis in cancer cells [485, 486]. Nevertheless, these inhibitors might amplify their impacts and nonspecific toxicity through diverse mechanisms, underscoring the clinical preference for specific DUB inhibitors [487].
USP7 is widely recognized as a target for drug development due to its key role in regulating p53 stability. USP7 antagonists, such as p5091 and p50429, have been developed to promote the ubiquitination and degradation of MDM2, leading to bortezomib resistance in MM cells [482, 488, 489]. Moreover, FT671 and FT827 target dynamic pockets near the USP7 catalytic center via self-inhibiting apolipoproteins [490], thereby disrupting the stability of the USP7 substrate, increasing the protein levels of p53 and its related genes, and ultimately inhibiting tumor growth [490, 491]. A series of small molecules, including HBX 19818, HBX28,258, P22077, and P50429, were shown by biochemical tests and protein mass spectrometry to have specific inhibitory effects on USP7 [489, 492]. USP14, which is related to WNT/β-catenin signal transduction [493, 494], is overexpressed in various cancer types and is positively associated with a poor prognosis [495, 496]. The inhibitor IU1 can effectively inhibit the activity of the USP14 enzyme by blocking its binding to the proteasome and enhancing its function [497].
Various new screening methods have been used to identify inhibitors and related compounds that target DUBs. For instance, high-throughput screening techniques were employed to identify selective inhibitors targeting UCH-L1, resulting in the identification of LDN-57444. This compound has been shown to trigger apoptosis in lung cell lines [498]. Furthermore, a cell-based screening methodology has been employed to identify compounds that can induce apoptosis across various tissue types, with b-AP15 serving as a prominent example [499]. B-AP15 triggers the accumulation of high molecular weight ubiquitin (Ub) complexes within cells and acts as an inhibitor of 19S regulatory particles. It selectively targets the ubiquitination activity of USP14 and other ubiquitin enzymes without impacting proteasome function [500]. In addition, b-AP15 inhibits the degradation of proteasome substrate proteins, leading to the accumulation of ubiquitin, which induces significant protein stress and mitochondrial damage [501, 502]. In various solid tumors and multiple myeloma, it may lead to tumor apoptosis through a c-MYC-NoXa-mediated pathway [500, 501, 503].
Conclusion
In the past few decades, significant progress has been achieved in the study of the UPS. In this review, we comprehensively reviewed the research progress on the UPS regarding tumor characteristics and treatment strategies. Regarding tumor characteristics, the specific mechanisms by which ubiquitination influences cancer progression remain unclear. Therefore, future investigations must delve into the specific mechanisms involved and endeavor to elucidate the efficacy of the UPS. In terms of treatment strategies, although we have collected some clinical drug information, we also noticed that some knowledge gaps persist, especially regarding E1- and E2-targeted drugs. These findings suggest that future drug research should focus more on these aspects.
Based on the role of the UPS in cancer, potential therapeutic targets have been identified, and the corresponding inhibitors have been further studied. Proteasome inhibitors, such as bortezomib, carfilzomib, oprozomib, and ixazomib, have been approved by the FDA. They have achieved good clinical results, but their widespread application is limited by the side effects caused by the abnormal accumulation of some upstream proteins. Therefore, targeted inhibitors of E1 enzymes, E2 enzymes, E3 ligases, deubiquitinases, and other targets, including MDM2 inhibitors, IAPs inhibitors, and SKP2 inhibitors, are being investigated [473]. PROTACs and molecular glues are also being developed for the treatment of cancer. At the same time, high-throughput screening also helps researchers screen suitable inhibitors [467]. Furthermore, during tumor proliferation, the UPS may influence various oncogenic signaling cascades, concurrently dysregulating multiple pathways and thereby complicating the development of targeted therapeutic strategies. Therefore, multitarget combination therapy is a direction for future development. Finally, further exploration of UPS function and clinical studies will provide important implications for the development of new cancer treatment strategies.
Acknowledgements
We would like to acknowledge the assistance of FigDraw in the creating cartoon illustrations.
Abbreviations
- ACC
Acetyl-CoA carboxylase
- ACLY
ATP- citrate lyase
- AF
Adventitial fibroblast
- AHNAK
Neuroblast differentiation-associated protein
- AIP4
Atrophin-interacting protein 4
- ALDH2
Aldehyde dehydrogenase 2
- ALKBH5
ALKB Homolog 5
- AML
Acute myeloid leukemia
- AMP
Adenosine 3', 5' monophosphate
- ANLN
Anillin
- APC
Anaphase-promoting complex
- AR
Androgen receptor
- ARF
ADP-ribosylation factor
- ARID1A
AT-rich interactive domain protein 1A
- ASK1
Apoptosis signal-regulating kinase 1
- ATM
Ataxia telangiectasia mutated
- ATR
Ataxia telangiectasia mutated Rad3-related kinase
- AXIN
Axis inhibitory protein
- BAP1
BRCA1 associated protein 1
- BECN1
Beclin 1
- BIK
BCL2 interacting killer
- BRCA1
Breast cancer susceptibility gene 1
- BRG1
Brahma-related gene 1
- CAF
Cancer-associated fibroblast
- CASTOR1
Cytosolic arginine sensor for mTORC1 subunit 1
- Cbl
Casitas B-lineage lymphoma
- CCF
Cytoplasmic chromatin fragments
- CCL2
CC-chemokine ligand 2
- CCNB1
Cyclin B1
- CDH1
E-cadherin
- CDK
Cyclin-dependent kinase
- CDKN2A
Cyclin-dependent kinase inhibitor 2A
- CHD1
Chromodomain helicase DNA binding protein 1
- CHIP
C-terminus of Hsp70-interacting protein
- CHK1
Cell cycle checkpoint kinase 1
- CK1a
Casein kinase 1a
- CLK2
CDC2-like kinase 2
- COAD
Colon adenocarcinoma
- CPG
Cytosine phosphate guanine
- CRL4CRBN
Cul4-DDB1-CRBN-RBX1
- CSN6
COP9 signalosome subunit 6
- CtIP
CtBP-interacting protein
- CTLA4
Cytotoxic T-lymphocyte-associated antigen 4
- CUL3
Cullin-3
- CYLD
Cylindromatosis
- DAXX
Death-associated protein
- DCAF1
DDB1- and CUL4-associated factor1
- DDLPS
Dedifferentiated liposarcoma
- DDR
DNA damage response
- DDX39B
DExD-box helicase 39B
- DLK1
Delta-like canonical notch ligand 1
- DNMT
DNA methyltransferase
- DSB
Double-strand break
- DUB
Deubiquitinase
- EGFR
Epidermal growth factor receptor
- EHMT2
Euchromatic histone lysine methyltransferase 2
- EMT
Epithelial-mesenchymal transition
- ENO2
Enolase 2
- ER
Estrogen receptor
- ESCC
Esophageal squamous cell carcinoma
- ESR1
Estrogen receptor 1
- EZH2
Enhancer of zeste homolog 2
- FASN
Fatty acid synthase
- FBP1
Fructose -1,6- bisphosphatase 1
- FBW7
F-box and WD repeat domain-containing 7
- FBXL18
F-box and leucine-rich repeat protein 18
- FBXO22
F-box protein 22
- FBXW2
F-box and WD repeat domain containing 2
- FZR1
Fizzy-related 1
- G-6-PD
Glucose 6-phosphate dehydrogenase
- GAC
Glutaminase C
- GAK
Cyclin G-associated kinase
- GBM
Glioblastoma multiforme
- GCN5
General control nonderepressible-5
- GDH
Glutamate dehydrogenase
- GPX4
Glutathione peroxidase 4
- GS
Glutamine synthetase
- GSDEM
Gasdermin E
- GSK3β
Glycogen synthase kinase 3β
- GSPT1
G1-to-S phase transition 1
- H2Aub
H2A ubiquitination
- H2BK120
Lysine 120 on histone H2B
- H2Bub1
Histone H2B monoubiquitination
- HCF-1
Host-cell factor 1
- HECT
Homologous to E6AP C-terminus
- HECTD2
HECT domain E3 ubiquitin protein ligase 2
- HERC1
HECT domain and RCC-1 like domain 1 gene
- HIF
Hypoxia-inducible factor
- HK2
Hexokinase 2
- HNSC
Head and neck squamous cell carcinoma
- HOIL-1L
Heme-oxidized IRP2 ubiquitin ligase 1
- HOIP
HOIL-1 interacting protein
- HR
Homologous recombination
- HUWE1
HECT, UBA, and WWE domain containing E3 ligase 1
- IAP
Inhibitor of apoptosis protein
- IBRDC2
IBR domain containing 2
- ICC
Intrahepatic cholangiocarcinoma
- IFN
Interferon
- IMiDs
Immunomodulatory drugs
- INO80
INOsitol-requiring mutant 80
- INSIG1
Insulin-induced gene 1
- IRS-1
Insulin receptor substrate-1
- ISG15
Interferon-stimulated gene 15
- ISWI
Imitation SWI
- ITGAV
Integrin αv
- KDM
αKG-dependent lysine demethylase
- KIRC
Kidney renal clear cell carcinoma
- KLF5
Kruppel-like factor 5
- KRT18
Keratin 18
- LDHA
Lactate dehydrogenase A
- LKB1
Liver kinase B1
- lncRNA
Long non‐coding RNA
- LPP
Lipoma preferred partner
- LRR
Leucine-rich repeat domain
- LUBAC
Linear ubiquitin chain assembly complex
- m6A
N6-methyladenosine
- MARCH
Membrane-associated RING-CH protein
- MDA-7
Melanoma differentiation-associated gene-7
- MDM2
Mouse double minute 2
- METTL5
Methyltransferase 5, N6-adenosine
- MIB2
Mind bomb homolog 2
- MKK4
Mitogen-activated protein kinase kinase 4
- MKRN1
Makorin ring finger protein 1
- MLK3
Mixed-lineage protein kinase 3
- MM
Multiple myeloma
- MMP
Mitochondrial membrane potential
- MOMP
Mitochondrial outer membrane permeabilization
- MRE11
Meiotic recombination 11
- MRN
Mre11-Rad50-Nbs1
- MTSS1
Metastasis suppressor protein 1
- MULAN
Mitochondrial ubiquitin ligase activator NF-κB
- NBS1
Nijmegen breakage syndrome 1
- NCOA4
Nuclear receptor coactivator 4
- NED
Neuroendocrine differentiation
- NEDD8
Neural precursor cells expressed developmentally down-regulated 8
- NEMO
NF-κB essential regulator
- NFAT2
Nuclear factor of activated T cell 2
- NHEJ
Non-homologous end joining
- NLR
NOD-like receptor
- NLRP3
NOD-like receptor protein 3
- NOD
Nucleotide-binding oligomerization domain
- NOX4
NADPH oxidase 4
- NPC
Nasopharyngeal carcinoma
- NRF-1
Nuclear respiratory factor 1
- NTP
Non-thermal plasma
- NuRD
Nucleosome remodeling and deacetylation
- OIS
Oncogene-induced senescence
- OTUB2
OTU domain-containing ubiquitin aldehyde-binding protein 2
- OTULIN
OTU deubiquitinase with linear linkage specificity
- OXPHOS
Oxidative phosphorylation
- P. micra
Parvimonas micra
- PABPC1
Poly(A)-binding protein, cytoplasmic 1
- PARP
Poly ADP-ribose polymerase
- PCDH10
Protocadherin 10
- PDAC
Pancreatic ductal adenocarcinoma
- PDGFRβ
Platelet-derived growth factor receptor ß
- PDK1
Phosphatidylinositol-dependent protein kinase 1
- PD-L1
Programmed cell death ligand 1
- PD-1
Programmed cell death 1
- PDX
Patient-derived xenograft
- PEBP
Phosphatidylethanolamine-binding protein
- PELI1
Pellino1
- PEP
Phosphoenolpyruvate
- PFK1
Phosphofructokinase 1
- PI
Proteasome inhibitor
- PINK1
PTEN-induced kinase 1
- PKB
Phosphokinase B
- PKM2
Pyruvate kinase M2
- POT1
Protection of telomeres 1
- PP2A
Protein phosphatase 2A
- PRAD
Prostate adenocarcinoma
- p-RB
Phosphorylated RB protein
- PR-DUB
Polycomb repressive deubiquitinase
- PROTAC
Proteolysis targeting chimera
- PRR
Pattern recognition receptor
- PSMB
Proteasome beta
- PSMD14
Proteasome non-ATPase regulatory subunit 14
- RACGAP1
Rac GTPase-activating protein 1
- RAP1
Ras-related protein 1
- RB
Retinoblastoma
- RBCK1
RANBP2-type and C3HC4-type zinc finger containing 1
- RCC
Renal cell carcinoma
- RFP2
Ret finger protein 2
- RIPK1
Receptor-interacting protein kinase 1
- RLIM
Ring finger LIM domain-interacting protein
- RNF2
Ring finger protein 2
- SAG
Sensitive to apoptosis gene
- SAGA
Spt-Ada-Gcn5 acetyltransferase
- SAMHD1
SAM and HD domain containing protein 1
- SETDB1
SET domain bifurcated 1
- SHARPIN
SHANK-associated RH domain-interacting protein
- SIAH2
Seven in absentia homolog 2
- SKP2
S-phase kinase-associated protein 2
- SLC7A11
Solute carrier family seven member 11
- SMAD7
Mothers against decapentaplegic homolog 7
- SPOP
Speckle-type POZ protein
- STD NMR
Saturation transfer differential nuclear magnetic resonance
- STUB1
STIP1 homology and U-box-containing protein 1
- SUMO
Small ubiquitin-related modifiers
- SUZ12
Suppressor of zeste 12
- SWI/SNF
Switch/sucrose non-fermentable
- SYVN1
Synovial apoptosis inhibitor 1
- TAB2
TAK1-binding protein 2
- TAK1
Transforming growth factor ß-activated kinase 1
- T-ALL
T-cell acute lymphoblastic leukemia
- TBK1
TANK-binding kinase 1
- TERT
Telomerase reverse transcriptase
- TIN2
TRF-interacting nuclear protein 2
- TIS
Tumor suppressor gene loss-induced senescence
- TLR
Toll-like receptors
- TME
Tumor microenvironment
- TMM
Telomere DNA maintenance mechanism
- TMUB1
Transmembrane ubiquitin-like domain 1
- TMZ
Temozolomide
- TNFAIP1
Tumor necrosis factor alpha-induced protein 1
- TNFR2
TNF receptor 2
- TPP1
Telomere protection protein 1
- TRAF4
TNF receptor-associated factor 4
- TRAP1
Tumor necrosis factor receptor-related protein 1
- TRF1
Telomere repeat binding factor 1
- TRIM7
Tripartite motif protein 7
- TSGs
Tumor suppressor genes
- U2AF65
U2 auxiliary factor 65
- Ub
Ubiquitin
- UBC13
Ubiquitin-conjugating enzyme 13
- UBE2B
Ubiquitin-binding enzyme E2B
- UBE2T
Ubiquitin-conjugating enzyme E2T
- UBE3C
Ub-protein Ligase E3C
- UBLs
Ubiquitin-like proteins
- UBR5
Ubiquitin ligase E3 component N-recognition protein 5
- UCHL5
Ubiquitin carboxyl-terminal hydrolase isozyme L5
- UEV1A
Ubiquitin-like protein 1 activating enzyme
- ULF1
Unexpected low fertilization
- UPS
Ubiquitin–proteasome system
- USP2
Ubiquitin-specific protease 2
- VDAC1
Voltage-dependent anion channel 1
- VEGF
Vascular endothelial growth factor
- VHL
Von-hippel lindau
- VSMC
Vascular smooth muscle cells
- WWP2
WW domain containing E3 ubiquitin protein ligase 2
- XIAP
X-linked apoptosis inhibitory protein
- YTHDF2
YTH N6-methyladenosine RNA binding protein F2
- ZMYM2
Zinc finger MYM-type protein 2
- αKG
α-Ketoglutarate
- β-TRCP
β-Transducin repeat-containing protein
- γH2AX
Gamma-H2A histone family member X
Glossary
E3 ubiquitin ligases
- E3 ubiquitin ligases
enzymes that facilitate the transfer of ubiquitin from a ubiquitin conjugating enzyme (E2) to a specific substrate protein, thereby marking it for degradation. E3 ubiquitin ligases are primarily classified into three groups based on their structural domains: RING finger ligases, HECT ligases, and RBR ligases.
- Parkin
an E3 ubiquitin ligase from the RBR family, targets proteins for degradation. In cancer, Parkin ubiquitinates key proteins like p53, Cyclin E, HIF-1α, and PARIS, influencing cancer progression and tumor suppression.
- F-box and WD repeat domain-containing 7 (FBXW7)
a member of the F-box protein family, regulates cancer cell growth by ubiquitinating oncoproteins like c-Myc, Cyclin E, and Notch.
- F-box protein 22 (FBXO22)
an E3 ubiquitin ligase from the F-box protein family. It can ubiquitinate p21, LSD1, and ERα, leading to their degradation and influencing cell cycle progression, gene expression, and hormone receptor signaling.
- Tripartite motif protein 7 (TRIM7)
an E3 ubiquitin ligase from the TRIM family, targets proteins for degradation by ubiquitin-proteasome system. The target proteins of TRIM7 include β-catenin and p53.
- Ring finger protein 2 (RNF2)
an E3 ubiquitin ligase belonging to the RING finger protein family, impacts cancer progression by targeting proteins such as H2A and BMI1 for ubiquitination.
- E3 ligase complex Cul4-DDB1-CRBN-RBX1 (CRL4CRBN)
an E3 ubiquitin ligase from the Cullin-RING family, regulates cancer cell growth by ubiquitinating target proteins like IKZF1 and IKZF3.
- Linear ubiquitin chain assembly complex (LUBAC)
an E3 ubiquitin ligase complex composed of HOIP, HOIL-1, and SHARPIN that regulates signaling pathways and cellular processes by adding linear ubiquitin chains to specific target proteins. In cancer, its substrates include NEMO, CYLD, TNFR1, RIPK1, and MLKL, which play crucial roles in the NF-κB signaling pathway.
- Synovial apoptosis inhibitor 1 (SYVN1)
is an E3 ubiquitin ligase that plays a key role in the endoplasmic reticulum-associated degradation (ERAD) pathway. It targets misfolded proteins for ubiquitination and subsequent degradation. In cancer, its substrates include p53, IRE1α, MCL1, and HIF-1α.
- HECT domain and RCC-1-like domain one gene (HERC1)
a ubiquitin ligase containing HECT and RCC1-like domains, is involved in various cellular processes such as protein degradation, cell cycle regulation, and DNA damage response, by regulating the stability and function of HSP70 and Caveolin-1. Abnormal HERC1 function may lead to uncontrolled growth and anti-apoptosis of cancer cells.
- Neural precursor cells expressed developmentally downregulated 8 (NEDD8)
a small ubiquitin-like protein that regulates protein modification processes by neddylating target proteins, impacting crucial cellular processes such as cell cycle regulation, DNA repair, protein degradation, and signal transduction. In cancer, NEDD8 modifies substrates like cullin, which are part of the E3 ubiquitin ligase complex SKP1-CUL1-F-box protein (SCF).
- Breast cancer susceptibility gene 1 (BRCA1)
a tumor suppressor gene that plays a pivotal role in DNA repair, cell cycle regulation, and apoptosis. Mutations in BRCA1 significantly increase the risk of breast, ovarian, and other cancers, making it a biomarker for hereditary cancer susceptibility and guiding tailored treatment approaches. BRCA1’s substrates include proteins involved in DNA repair pathways, such as RAD51, and cell cycle regulators like p21 and cyclin D1.
- F-box and leucine-rich repeat protein 18 (Fbxl18)
as a member of the F-box protein family, FBXL18 plays a significant role in cancer by participating in the Skp, Cullin, and F-box containing (SCF) E3 ubiquitin ligase complex. In cancer, FBXL18 has been implicated in targeting substrates like cyclin D1, a key regulator of cell cycle progression, and c-Myc, an oncogenic transcription factor, thus affecting tumor growth and progression.
- Speckle-type POZ protein (SPOP)
an adaptor protein for the Cullin3-RING E3 ubiquitin ligase complex, SPOP plays a crucial role in cancer. It regulates protein degradation by targeting specific substrates for ubiquitination. In cancer, SPOP has been implicated in the degradation of substrates such as AR and ERG, impacting oncogenic signaling pathways and contributing to tumor development and progression.
- Ubiquitin-conjugating enzyme 13 (UBC13)
an E2 ubiquitin conjugating enzyme that forms K63-linked polyubiquitin chains. In cancer, UBC13 is involved in the regulation of substrates such as TRAF6 and RIPK1, which play key roles in NF-κB signaling and cell survival.
- Ubiquitin-like protein 1 activating enzyme (UEV1A)
a co-factor for UBC13, plays a crucial role in cancer by facilitating K63-linked polyubiquitination of proteins such as BRCA1 and FANCD2 for DNA repair, IκB kinase for NF-κB signaling, and cyclin-dependent kinase inhibitors like p21 and p27 for cell cycle regulation.
- Trib1-COP1 complex
an instrumental player in controlling cell proliferation, differentiation, and metabolism, the Trib1-COP1 complex significantly impacts cancer development by ubiquitinating and regulating the stability of proteins such as c-Jun, p53, and AKT.
Deubiquitinase (DUB)s
- DUB
an enzyme that removes ubiquitin molecules from proteins in a process called deubiquitination, which counteracts the effects of ubiquitination by preventing protein degradation and regulating protein function and localization.
- OTU domain-containing ubiquitin aldehyde-binding protein 2 (OTUB2)
an OTU domain deubiquitinating enzyme, regulates NF-κB signaling by deubiquitinating key components such as IKKγ and RIP1. This promotes inflammation and potentially acts as a tumor suppressor in cancer.
- OTU deubiquitinase with linear linkage specificity (OTULIN)
a deubiquitinase regulates the NF-κB signaling pathway by specifically removing linear ubiquitin chains, potentially affecting the stability of IKKγ and NEMO, thus influencing inflammatory responses and immune regulation in cancer.
- COP9 signalosome subunit 5 (CSN5)
a part of the COP9 signalosome complex, promotes the degradation of key proteins such as p27, c-Myc, and HIF-1α in cancer, enhancing cell cycle progression, cell growth, and angiogenesis.
- Cezanne-1
an important deubiquitinating enzyme, modulates protein stability and activity in cancer. Its target proteins include IKKα/β, TRAF2, and SMAD7.
Key Concepts
- Ubiquitin-specific protease (USP)
is a large family of enzymes that plays a critical role in deubiquitination, which is the removal of ubiquitin molecules from protein substrates. USPs are involved in various pathologies due to their central role in regulating protein stability and function. In cancer, for example, alterations in the activity or expression of certain USPs can lead to the dysregulation of oncogenes or tumor suppressors, contributing to cancer progression.
- Mitochondrial outer membrane permeabilization (MOMP)
a process involving the penetration of the mitochondrial outer membrane, usually triggered by apoptotic signals.
- 26S proteasome
comprising a 20S core particle and two 19S regulatory particles, and functions as a protein degradation system within cells. It is crucial for maintaining cellular homeostasis by degrading unwanted proteins.
- Cancer-Associated Fibroblasts (CAFs)
a subtype of fibroblast cells in the tumor microenvironment, significantly impact tumor growth, invasion, and metastasis by interacting with tumor cells through the secretion of growth factors, extracellular matrix proteins, and inflammatory mediators.
- Tumor Microenvironment (TME)
a critical component of tumor biology. TME encompasses blood vessels, immune cells, fibroblasts, and extracellular matrix, playing vital roles in tumor growth, progression, angiogenesis, immune evasion, and metastasis.
- Epithelial-Mesenchymal Transition (EMT)
an essential process in cancer, enhances tumor invasiveness and metastasis by causing the loss of epithelial markers like E-cadherin and the gain of mesenchymal markers such as N-cadherin, vimentin, and fibronectin. This process is regulated by transcription factors like Snail, Slug, Twist, and ZEB1/2, and driven by signaling pathways including TGF-β, Wnt/β-catenin, Notch, and Hedgehog.
- Dedifferentiation
a process in which mature, specialized cells revert to a more primitive, unspecialized state, often regaining the ability to divide and differentiate into other cell types.
- Transdifferentiation
a direct conversion of one differentiated cell type into another without reverting to a stem cell state.
- Proteolysis targeting chimeras (PROTACs)
are bifunctional small molecules consisting of two specific ligands: one for an E3 ubiquitin ligase and another that binds to a target protein. These ligands are connected by a linker, creating a trimeric complex-target protein ligand-Linker-E3 ligand. The E3 ligase tags the target protein with ubiquitin, leading to its specific degradation through the ubiquitin-proteasome pathway.
- Molecular glue
is used in the field of drug discovery and biochemistry to refer to a small molecule that promotes the interaction between two proteins, typically leading to a therapeutic effect. These molecules act by “gluing” a target protein and a ubiquitin ligase together, facilitating the ubiquitination and subsequent degradation of the target protein by the proteasome system.
Authors’ contributions
F.L., J.C., K.L., H.L., and Y.Z. wrote the manuscript and prepared all figures, Y.Z., B.L., Y.F., and Z.L. collected the data, and X.J., Z.D., and K.L. revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant number: 82203594, F. Liu), the Postdoctoral Fellowship Program of CPSF (grant number: GZB20230669, F. Liu), the China Postdoctoral Science Foundation (grant number: 2023M743179, F. Liu), the China Postdoctoral Science Foundation, No.17 Special Funding (F, Liu), the Scientific Research Foundation for Returned Scholar, Zhengzhou University (grant number: 32213234, F. Liu), the Natural Science Foundation of Henan province (grant number: 242300421308, X. Chen), the Postdoctoral Fellowship Program of CPSF (grant number: GZC20230741, X. Jia), and the Central Plains Science and Technology Innovation Leading Talents (grant number: 224200510015, K. Liu).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xuechao Jia, Email: xcjia@hci-cn.org.
Zigang Dong, Email: dongzg@zzu.edu.cn.
Kangdong Liu, Email: kdliu@zzu.edu.cn.
References
- 1.Cappadocia L, Lima CD. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem Rev. 2018;118:889–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antao AM, Tyagi A, Kim KS, Ramakrishna S. Advances in deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers (Basel). 2020;12:1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun T, Liu Z, Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer. 2020;19:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon YT, Ciechanover A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci. 2017;42:873–86. [DOI] [PubMed] [Google Scholar]
- 5.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 8.Hou B, Chen T, Zhang H, Li J, Wang P, Shang G. The E3 ubiquitin ligases regulate PD-1/PD-L1 protein levels in tumor microenvironment to improve immunotherapy. Front Immunol. 2023;14:1123244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuang Z, Liu X, Zhang N, Dong J, Sun C, Yin M, Wang Y, Liu L, Xiao D, Zhou X, et al. USP2 promotes tumor immune evasion via deubiquitination and stabilization of PD-L1. Cell Death Differ. 2023;30:2249–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai T, Zhang X, Zhou X, Hu X, Huang X, Xing F, Tian H, Li Y. Long non-coding RNA VAL facilitates PKM2 enzymatic activity to promote glycolysis and malignancy of gastric cancer. Clin Transl Med. 2022;12:e1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu S, Zang W, Qiu Y, Liao L, Zheng X. Deubiquitinase OTUB2 exacerbates the progression of colorectal cancer by promoting PKM2 activity and glycolysis. Oncogene. 2022;41:46–56. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Qin ZL, Li N, Jia MQ, Liu QG, Bai YR, Song J, Yuan S, Zhang SY. Annual review of PROTAC degraders as anticancer agents in 2022. Eur J Med Chem. 2024;267:116166. [DOI] [PubMed] [Google Scholar]
- 13.Surka C, Jin L, Mbong N, Lu CC, Jang IS, Rychak E, Mendy D, Clayton T, Tindall E, Hsu C, et al. CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood. 2021;137:661–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrylak DP, Gao X, Vogelzang NJ, Garfield MH, Taylor I, Moore MD, Peck RA, Burris HA. First-in-human phase I study of ARV-110, an androgen receptor (AR) PROTAC degrader in patients (pts) with metastatic castrate-resistant prostate cancer (mCRPC) following enzalutamide (ENZ) and/or abiraterone (ABI). J Clin Oncol. 2020;38:3500.
- 15.Liu F, Wu Q, Han W, Laster K, Hu Y, Ma F, Chen H, Tian X, Qiao Y, Liu H, et al. Targeting integrin αvβ3 with indomethacin inhibits patient-derived xenograft tumour growth and recurrence in oesophageal squamous cell carcinoma. Clin Transl Med. 2021;11:e548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Liu H, Wang P, Hu Y, Yang R, Liu F, Kim HG, Dong Z, Liu K. Honokiol inhibits melanoma growth by targeting keratin 18 in vitro and in vivo. Front Cell Dev Biol. 2020;8:603472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 2012;64:136–42. [DOI] [PubMed] [Google Scholar]
- 20.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. [DOI] [PubMed] [Google Scholar]
- 21.Yan Q, Chen BJ, Hu S, Qi SL, Li LY, Yang JF, Zhou H, Yang CC, Chen LJ, Du J. Emerging role of RNF2 in cancer: from bench to bedside. J Cell Physiol. 2021;236:5453–65. [DOI] [PubMed] [Google Scholar]
- 22.Yao L, Li J, Jiang B, Zhang Z, Li X, Ouyang X, Xiao Y, Liu G, Wang Z, Zhang G. RNF2 inhibits E-Cadherin transcription to promote hepatocellular carcinoma metastasis via inducing histone mono-ubiquitination. Cell Death Dis. 2023;14:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Zhu Z, Li W, Shen M, Cao C, Sun Q, Guo Z, Liu L, Wu D. UBE2T-regulated H2AX monoubiquitination induces hepatocellular carcinoma radioresistance by facilitating CHK1 activation. J Exp Clin Cancer Res. 2020;39:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Jia Z, Liang C, He Y, Cong M, Wu Q, Tian P, He D, Miao X, Sun B, et al. MTSS1 curtails lung adenocarcinoma immune evasion by promoting AIP4-mediated PD-L1 monoubiquitination and lysosomal degradation. Cell Discov. 2023;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng X, Zheng W, Sheng Y, Ma H. UBE2B promotes ovarian cancer growth via promoting RAD18 mediated ZMYM2 monoubiquitination and stabilization. Bioengineered. 2022;13:8000–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buneeva O, Medvedev A. Atypical ubiquitination and Parkinson’s disease. Int J Mol Sci. 2022;23:3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hrdinka M, Gyrd-Hansen M. The Met1-linked ubiquitin machinery: emerging themes of (De)regulation. Mol Cell. 2017;68:265–80. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Li M, Ponnusamy S, Chi Y, Xue J, Fahmy B, Fan M, Miranda-Carboni GA, Narayanan R, Wu J, Wu ZH. ABL1-dependent OTULIN phosphorylation promotes genotoxic Wnt/β-catenin activation to enhance drug resistance in breast cancers. Nat Commun. 2020;11:3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jo T, Nishikori M, Kogure Y, Arima H, Sasaki K, Sasaki Y, Nakagawa T, Iwai F, Momose S, Shiraishi A, et al. LUBAC accelerates B-cell lymphomagenesis by conferring resistance to genotoxic stress on B cells. Blood. 2020;136:684–97. [DOI] [PubMed] [Google Scholar]
- 30.Song K, Cai X, Dong Y, Wu H, Wei Y, Shankavaram UT, Cui K, Lee Y, Zhu B, Bhattacharjee S, et al. Epsins 1 and 2 promote NEMO linear ubiquitination via LUBAC to drive breast cancer development. J Clin Invest. 2021;131:e129374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Z, Wei W, Xiao W, Al-Saleem ED, Nejati R, Chen L, Yin J, Fabrizio J, Petrus MN, Waldmann TA, Yang Y. Essential role of the linear ubiquitin chain assembly complex and TAK1 kinase in A20 mutant Hodgkin lymphoma. Proc Natl Acad Sci U S A. 2020;117:28980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu Z, Fan J, Chen F, Yang H, Li X, Zhuang T, Guo C, Cao Q, Zhu J, Wang H, Huang Q. RBCK1 regulates the progression of ER-positive breast cancer through the HIF1α signaling. Cell Death Dis. 2022;13:1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu L, Qin C, Li T, Ma X, Qiu Y, Lin Y, Ma D, Qin Z, Sun C, Shen X, et al. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2020;27:1819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong L, Liu HS, Zhou C, Yang X, Huang L, Jie HQ, Zeng ZW, Zheng XB, Li WX, Liu ZZ, et al. A novel protein encoded by circINSIG1 reprograms cholesterol metabolism by promoting the ubiquitin-dependent degradation of INSIG1 in colorectal cancer. Mol Cancer. 2023;22:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang M, Wu HK, Pei Y, Zhang Y, Gao X, He Y, Chen G, Lv F, Jiang P, Li Y, et al. E3 ligase MG53 suppresses tumor growth by degrading cyclin D1. Signal Transduct Target Ther. 2023;8:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia P, Zhang H, Lu H, Xu K, Jiang X, Jiang Y, Gongye X, Chen Z, Liu J, Chen X, et al. METTL5 stabilizes c-Myc by facilitating USP5 translation to reprogram glucose metabolism and promote hepatocellular carcinoma progression. Cancer Commun (Lond). 2023;43:338–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtake F, Tsuchiya H. The emerging complexity of ubiquitin architecture. J Biochem. 2017;161:125–33. [DOI] [PubMed] [Google Scholar]
- 38.Zhong L, Liao D, Li J, Liu W, Wang J, Zeng C, Wang X, Cao Z, Zhang R, Li M, et al. Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes. Signal Transduct Target Ther. 2021;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng C, Zhong L, Liu W, Zhang Y, Yu X, Wang X, Zhang R, Kang T, Liao D. Targeting the lysosomal degradation of Rab22a-NeoF1 fusion protein for osteosarcoma lung metastasis. Adv Sci (Weinh). 2023;10:e2205483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Jia S, Wang G, Liang M, Guo T, Du H, Li S, Li X, Huangfu L, Guo J, et al. TRIM28 promotes the escape of gastric cancer cells from immune surveillance by increasing PD-L1 abundance. Signal Transduct Target Ther. 2023;8:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X, Li W, Liu H, Wang X, Coarfa C, Cheng C, Yu X, Zeng Z, Cao Y, Young KH, Li Y. PD-L1 translocation to the plasma membrane enables tumor immune evasion through MIB2 ubiquitination. J Clin Invest. 2023;133:e160456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao YF, Xie L, Tong BB, Chu MY, Shi WQ, Li X, He JZ, Wang SH, Wu ZY, Deng DX, et al. Targeting USP10 induces degradation of oncogenic ANLN in esophageal squamous cell carcinoma. Cell Death Differ. 2023;30:527–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Huang S, Xu P, Li Y. Progress in atypical ubiquitination via K6-linkages. Sheng Wu Gong Cheng Xue Bao. 2022;38:3215–27. [DOI] [PubMed] [Google Scholar]
- 44.Cundiff MD, Hurley CM, Wong JD, Boscia JAT, Bashyal A, Rosenberg J, Reichard EL, Nassif ND, Brodbelt JS, Kraut DA. Ubiquitin receptors are required for substrate-mediated activation of the proteasome’s unfolding ability. Sci Rep. 2019;9:14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Wang Y, Li Y, Yin W, Mo L, Qian X, Zhang Y, Wang G, Bu F, Zhang Z, et al. Ube2s stabilizes β-Catenin through K11-linked polyubiquitination to promote mesendoderm specification and colorectal cancer development. Cell Death Dis. 2018;9:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao K, Peng S, Lu J, Zhou T, Hong X, Chen S, Liu G, Li H, Huang J, Chen X, Lin T. UBE2S interacting with TRIM21 mediates the K11-linked ubiquitination of LPP to promote the lymphatic metastasis of bladder cancer. Cell Death Dis. 2023;14:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin X, Liu Q, Liu F, Tian X, Yan T, Han J, Jiang S. Emerging roles of non-proteolytic ubiquitination in tumorigenesis. Front Cell Dev Biol. 2022;10:944460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li T, Wang X, Ju E, da Silva SR, Chen L, Zhang X, Wei S, Gao SJ. RNF167 activates mTORC1 and promotes tumorigenesis by targeting CASTOR1 for ubiquitination and degradation. Nat Commun. 2021;12:1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu R, Hochstrasser M. Recent progress in ubiquitin and ubiquitin-like protein (Ubl) signaling. Cell Res. 2016;26:389–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida T, Kim JH, Carver K, Su Y, Weremowicz S, Mulvey L, Yamamoto S, Brennan C, Mei S, Long H, et al. CLK2 Is an oncogenic kinase and splicing regulator in breast cancer. Cancer Res. 2015;75:1516–26. [DOI] [PubMed] [Google Scholar]
- 51.Li TJ, Jin KZ, Zhou HY, Liao ZY, Zhang HR, Shi SM, Lin MX, Chai SJ, Fei QL, Ye LY, et al. Deubiquitinating PABPC1 by USP10 upregulates CLK2 translation to promote tumor progression in pancreatic ductal adenocarcinoma. Cancer Lett. 2023;576:216411. [DOI] [PubMed] [Google Scholar]
- 52.Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP. Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol Rev. 2012;92:273–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–48. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z, Stiegler AL, Boggon TJ, Kobayashi S, Halmos B. EGFR-mutated lung cancer: a paradigm of molecular oncology. Oncotarget. 2010;1:497–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson H, Del Rosario AM, Bryson BD, Schroeder MA, Sarkaria JN, White FM. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol Cell Proteomics. 2012;11:1724–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng M, Liu B, Song H, Yu R, Zou D, Chen Y, Ma Y, Lv F, Xu L, Zhang Z, et al. β-Elemene inhibits the metastasis of multidrug-resistant gastric cancer cells through miR-1323/Cbl-b/EGFR pathway. Phytomedicine. 2020;69:153184. [DOI] [PubMed] [Google Scholar]
- 58.Zhao H, Zheng C, Wang Y, Hou K, Yang X, Cheng Y, Che X, Xie S, Wang S, Zhang T, et al. miR-1323 promotes cell migration in lung adenocarcinoma by targeting Cbl-b and is an early prognostic biomarker. Front Oncol. 2020;10:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pareja F, Ferraro DA, Rubin C, Cohen-Dvashi H, Zhang F, Aulmann S, Ben-Chetrit N, Pines G, Navon R, Crosetto N, et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene. 2011;31:4599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Han B, Lu H, Zhao Y, Chen X, Meng Q, Cao M, Cai L, Hu J. USP22 promotes resistance to EGFR-TKIs by preventing ubiquitination-mediated EGFR degradation in EGFR-mutant lung adenocarcinoma. Cancer Lett. 2018;433:186–98. [DOI] [PubMed] [Google Scholar]
- 61.Niño CA, Wollscheid N, Giangreco G, Maspero E, Polo S. USP25 regulates EGFR fate by modulating EGF-induced ubiquitylation dynamics. Biomolecules. 2020;10:1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pareja F, Ferraro DA, Rubin C, Cohen-Dvashi H, Zhang F, Aulmann S, Ben-Chetrit N, Pines G, Navon R, Crosetto N, et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene. 2012;31:4599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhibo Q, Lianxin L. Ubiquitin-specific protease 22 is associated with poor prognosis in neuroblastoma. Adv Clin Exp Med. 2020;29:295–300. [DOI] [PubMed] [Google Scholar]
- 64.Peng J, Jiang K, Sun X, Wu L, Wang J, Xi X, Tan X, Liang T, Tan C, Zhang P. Identification of a class of potent USP25/28 inhibitors with broad-spectrum anti-cancer activity. Signal Transduct Target Ther. 2022;7:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hua H, Xie H, Zheng J, Lei L, Deng Z, Yu C, Adnan M. RNF7 facilitated the tumorigenesis of pancreatic cancer by activating PI3K/Akt signaling pathway. Oxid Med Cell Longev. 2023;2023:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turowec JP, Lau EWT, Wang X, Brown KR, Fellouse FA, Jawanda KK, Pan J, Moffat J, Sidhu SS. Functional genomic characterization of a synthetic anti-HER3 antibody reveals a role for ubiquitination by RNF41 in the anti-proliferative response. J Biol Chem. 2019;294:1396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Xu S, Huang Y, Liu S, Xu Z, Wei M, Liu J. MARCH6 promotes Papillary Thyroid Cancer development by destabilizing DHX9. Int J Biol Sci. 2021;17:3401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee Y, Lim B, Lee SW, Lee WR, Kim Y-I, Kim M, Ju H, Kim MY, Kang S-J, Song J-J, et al. ANKRD9 is associated with tumor suppression as a substrate receptor subunit of ubiquitin ligase. Biochim Biophys Acta Mol Basis Dis. 2018;1864:3145–53. [DOI] [PubMed] [Google Scholar]
- 69.Lee M-S, Jeong M-H, Lee H-W, Han H-J, Ko A, Hewitt SM, Kim J-H, Chun K-H, Chung J-Y, Lee C, et al. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun. 2015;6:7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruschmann J, Ho V, Antignano F, Kuroda E, Lam V, Ibaraki M, Snyder K, Kim C, Flavell RA, Kawakami T, et al. Tyrosine phosphorylation of SHIP promotes its proteasomal degradation. Exp Hematol. 2010;38(392–402):402.e391. [DOI] [PubMed] [Google Scholar]
- 71.Wang FF, Zhang XJ, Yan YR, Zhu XH, Yu J, Ding Y, Hu JL, Zhou WJ, Zeng ZC, Liao WT, et al. FBX8 is a metastasis suppressor downstream of miR-223 and targeting mTOR for degradation in colorectal carcinoma. Cancer Lett. 2017;388:85–95. [DOI] [PubMed] [Google Scholar]
- 72.Tan M, Xu J, Siddiqui J, Feng F, Sun Y. Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis. Mol Cancer. 2016;15:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SY, Kim HJ, Kang SU, Kim YE, Park JK, Shin YS, Kim YS, Lee K, Kim CH. Non-thermal plasma induces AKT degradation through turn-on the MUL1 E3 ligase in head and neck cancer. Oncotarget. 2015;6:33382–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joo HM, Kim JY, Jeong JB, Seong KM, Nam SY, Yang KH, Kim CS, Kim HS, Jeong M, An S, Jin YW. Ret finger protein 2 enhances ionizing radiation-induced apoptosis via degradation of AKT and MDM2. Eur J Cell Biol. 2011;90:420–31. [DOI] [PubMed] [Google Scholar]
- 75.Xiang T, Ohashi A, Huang Y, Pandita TK, Ludwig T, Powell SN, Yang Q. Negative regulation of AKT Activation by BRCA1. Cancer Res. 2008;68:10040–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su CH, Wang CY, Lan KH, Li CP, Chao Y, Lin HC, Lee SD, Lee WP. Akt phosphorylation at Thr308 and Ser473 is required for CHIP-mediated ubiquitination of the kinase. Cell Signal. 2011;23:1824–30. [DOI] [PubMed] [Google Scholar]
- 77.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li W, Peng C, Lee MH, Lim D, Zhu F, Fu Y, Yang G, Sheng Y, Xiao L, Dong X, et al. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Res. 2013;73:6938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, Yang Z, Ou J, Xia X, Zhi F, Cui J. The F-box protein FBXL18 promotes glioma progression by promoting K63-linked ubiquitination of Akt. FEBS Lett. 2017;591:145–54. [DOI] [PubMed] [Google Scholar]
- 80.Sarri N, Wang K, Tsioumpekou M, Castillejo-López C, Lennartsson J, Heldin C-H, Papadopoulos N. Deubiquitinating enzymes USP4 and USP17 finetune the trafficking of PDGFRβ and affect PDGF-BB-induced STAT3 signalling. Cell Mol Sci. 2022;79:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xia X, Huang C, Liao Y, Liu Y, He J, Shao Z, Hu T, Yu C, Jiang L, Liu J, Huang H. The deubiquitinating enzyme USP15 stabilizes ERα and promotes breast cancer progression. Cell Death Dis. 2021;12:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Han Y, Xiao W, Gao Y, Sui Z, Ren P, Meng F, Tang P, Yu Z. USP4 promotes the proliferation, migration, and invasion of esophageal squamous cell carcinoma by targeting TAK1. Cell Death Dis. 2023;14:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia X, Liao Y, Huang C, Liu Y, He J, Shao Z, Jiang L, Dou QP, Liu J, Huang H. Deubiquitination and stabilization of estrogen receptor α by ubiquitin-specific protease 7 promotes breast tumorigenesis. Cancer Lett. 2019;465:118–28. [DOI] [PubMed] [Google Scholar]
- 84.Sacco JJ, Yau TY, Darling S, Patel V, Liu H, Urbe S, Clague MJ, Coulson JM. The deubiquitylase Ataxin-3 restricts PTEN transcription in lung cancer cells. Oncogene. 2014;33:4265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, Wang M, Chen D, Sun Y, Hung MC, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15:1486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lu C, Ning Z, Wang A, Chen D, Liu X, Xia T, Tekcham DS, Wang W, Li T, Liu X, et al. USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma. Cancer Lett. 2018;436:139–48. [DOI] [PubMed] [Google Scholar]
- 87.Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H, Jiang X. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol Cell Biochem. 2018;441:1–7. [DOI] [PubMed] [Google Scholar]
- 88.Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, Xing G, Kong X, Wang L, Li Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17:1169–81. [DOI] [PubMed] [Google Scholar]
- 89.Li X, Stevens PD, Yang H, Gulhati P, Wang W, Evers BM, Gao T. The deubiquitination enzyme USP46 functions as a tumor suppressor by controlling PHLPP-dependent attenuation of Akt signaling in colon cancer. Oncogene. 2013;32:471–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhiqiang Z, Qinghui Y, Yongqiang Z, Jian Z, Xin Z, Haiying M, Yuepeng G. USP1 regulates AKT phosphorylation by modulating the stability of PHLPP1 in lung cancer cells. J Cancer Res Clin Oncol. 2012;138:1231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li W, Cui K, Prochownik EV, Li Y. The deubiquitinase USP21 stabilizes MEK2 to promote tumor growth. Cell Death Dis. 2018;9:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gangula NR, Maddika S. WD repeat protein WDR48 in complex with deubiquitinase USP12 suppresses Akt-dependent cell survival signaling by stabilizing PH domain leucine-rich repeat protein phosphatase 1 (PHLPP1). J Biol Chem. 2013;288:34545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McClurg UL, Summerscales EE, Harle VJ, Gaughan L, Robson CN. Deubiquitinating enzyme Usp12 regulates the interaction between the androgen receptor and the Akt pathway. Oncotarget. 2014;5:7081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hassin O, Oren M. Drugging p53 in cancer: one protein, many targets. Nat Rev Drug Discov. 2022;22:127–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jin JO, Lee GD, Nam SH, Lee TH, Kang DH, Yun JK, Lee PCW. Sequential ubiquitination of p53 by TRIM28, RLIM, and MDM2 in lung tumorigenesis. Cell Death Differ. 2020;28:1790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, Han L, Jiang G, Zhang L, Gao C, Zhao W. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7:13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiang S, Shi X, Chen P, Chen Y, Bing S, Jin X, Cao J, Wang J, Yang B, Shao X, et al. Targeting Cul3-scaffold E3 ligase complex via KLHL substrate adaptors for cancer therapy. Pharmacol Res. 2021;169:105616. [DOI] [PubMed] [Google Scholar]
- 98.Seo J, Seong D, Lee SR, Oh D-B, Song J. Post-translational regulation of ARF: perspective in cancer. Biomolecules. 2020;10:1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee Y-R, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19:547–62. [DOI] [PubMed] [Google Scholar]
- 100.Aghabozorgi AS, Bahreyni A, Soleimani A, Bahrami A, Khazaei M, Ferns GA, Avan A, Hassanian SM. Role of adenomatous polyposis coli (APC) gene mutations in the pathogenesis of colorectal cancer; current status and perspectives. Biochimie. 2019;157:64–71. [DOI] [PubMed] [Google Scholar]
- 101.Jin MH, Oh D-Y. ATM in DNA repair in cancer. Pharmacol Ther. 2019;203:107391. [DOI] [PubMed] [Google Scholar]
- 102.Stolarova L, Kleiblova P, Janatova M, Soukupova J, Zemankova P, Macurek L, Kleibl Z. CHEK2 germline variants in cancer predisposition: stalemate rather than checkmate. Cells. 2020;9:2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 Localization and Stability by deubiquitinating p53. Cell. 2010;140:384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harakandi C, Nininahazwe L, Xu H, Liu B, He C, Zheng YC, Zhang H. Recent advances on the intervention sites targeting USP7-MDM2-p53 in cancer therapy. Bioorg Chem. 2021;116:105273. [DOI] [PubMed] [Google Scholar]
- 105.Roberts JZ, Crawford N, Longley DB. The role of ubiquitination in apoptosis and necroptosis. Cell Death Differ. 2021;29:272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benard G, Neutzner A, Peng G, Wang C, Livak F, Youle RJ, Karbowski M. IBRDC2, an IBR-type E3 ubiquitin ligase, is a regulatory factor for Bax and apoptosis activation. EMBO J. 2010;29:1458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yan L, Cui Y, Feng J. Biology of Pellino1: a potential therapeutic target for inflammation in diseases and cancers. Front Immunol. 2023;14:1292022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Allen IC, TeKippe EM, Woodford RMT, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JPY. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li X, Yang K-B, Chen W, Mai J, Wu X-Q, Sun T, Wu R-Y, Jiao L, Li D-D, Ji J, et al. CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy. 2021;17:4323–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu Y, Zhang C, Huang M, Lin J, Fan X, Ni T. TRIM26 induces ferroptosis to inhibit hepatic stellate cell activation and mitigate liver fibrosis through mediating SLC7A11 ubiquitination. Front Cell Dev Biol. 2021;9:644901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roedig J, Kowald L, Juretschke T, Karlowitz R, AhangarianAbhari B, Roedig H, Fulda S, Beli P, van Wijk SJL. USP22 controls necroptosis by regulating receptor-interacting protein kinase 3 ubiquitination. EMBO Rep. 2020;22:e50163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He H, Yi L, Zhang B, Yan B, Xiao M, Ren J, Zi D, Zhu L, Zhong Z, Zhao X, et al. USP24-GSDMB complex promotes bladder cancer proliferation via activation of the STAT3 pathway. Int J Biol Sci. 2021;17:2417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ren Y, Feng M, Hao X, Liu X, Li J, Li P, Gao J, Qi Q, Du L, Wang C, et al. USP48 stabilizes gasdermin E to promote pyroptosis in cancer. Cancer Res. 2023;83:1074–93. [DOI] [PubMed] [Google Scholar]
- 114.Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: a non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer. 2019;26:R1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Her YR, Chung IK. Ubiquitin ligase RLIM modulates telomere length homeostasis through a proteolysis of TRF1. J Biol Chem. 2009;284:8557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J Biol Chem. 2006;281:759–68. [DOI] [PubMed] [Google Scholar]
- 117.Wang C, Xiao H, Ma J, Zhu Y, Yu J, Sun L, Sun H, Liu Y, Jin C, Huang H. The F-box protein β-TrCP promotes ubiquitination of TRF1 and regulates the ALT-associated PML bodies formation in U2OS cells. Biochem Biophys Res Commun. 2013;434:728–34. [DOI] [PubMed] [Google Scholar]
- 118.Wang L, Chen R, Li G, Wang Z, Liu J, Liang Y, Liu J-P. FBW7 mediates senescence and pulmonary fibrosis through telomere uncapping. Cell Metab. 2020;32:860-877.e869. [DOI] [PubMed] [Google Scholar]
- 119.Fujita K, Horikawa I, Mondal AM, Jenkins LM, Appella E, Vojtesek B, Bourdon JC, Lane DP, Harris CC. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat Cell Biol. 2010;12:1205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rai R, Li JM, Zheng H, Lok GT, Deng Y, Huen MS, Chen J, Jin J, Chang S. The E3 ubiquitin ligase Rnf8 stabilizes Tpp1 to promote telomere end protection. Nat Struct Mol Biol. 2011;18:1400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.de Heer EC, Jalving M, Harris AL. HIFs, angiogenesis, and metabolism: elusive enemies in breast cancer. J Clin Investig. 2020;130:5074–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang F, Xu J, Li H, Tan M, Xiong X, Sun Y. FBXW2 suppresses migration and invasion of lung cancer cells via promoting β-catenin ubiquitylation and degradation. Nat Commun. 2019;10:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y, Liu P, Li H, Tan M, Xiong X, Sun Y. The β-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat Commun. 2017;8:14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou T, Chen T, Lai B, Zhang W, Luo X, Xia D, Fu W, Xu J. FBXW2 inhibits prostate cancer proliferation and metastasis via promoting EGFR ubiquitylation and degradation. Cell Mol Sci. 2022;79:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xia S, Ji L, Tao L, Pan Y, Lin Z, Wan Z, Pan H, Zhao J, Cai L, Xu J, Cai X. TAK1 is a novel target in hepatocellular carcinoma and contributes to sorafenib resistance. Cell Mol Gastroenterol Hepatol. 2021;12:1121–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Zhang X, Ye M, Jing P, Xiong J, Han Z, Kong J, Li M, Lai X, Chang N, et al. FBW7 loss promotes epithelial-to-mesenchymal transition in non-small cell lung cancer through the stabilization of Snail protein. Cancer Lett. 2018;419:75–83. [DOI] [PubMed] [Google Scholar]
- 127.Huang L-Y, Zhao J, Chen H, Wan L, Inuzuka H, Guo J, Fu X, Zhai Y, Lu Z, Wang X, et al. SCFFBW7-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat Commun. 2018;9:3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, Zhou X, Wu X. FBW7 suppresses ovarian cancer development by targeting the N6-methyladenosine binding protein YTHDF2. Mol Cancer. 2021;20:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Q, Li Y, Li J, Ma Y, Dai W, Mo S, Xu Y, Li X, Cai S. FBW7 suppresses metastasis of colorectal cancer by inhibiting HIF1α/CEACAM5 functional axis. Int J Biol Sci. 2018;14:726–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Davis Ryan J, Welcker M, Clurman Bruce E. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiong J, Wei B, Ye Q, Liu W. MiR-30a-5p/UBE3C axis regulates breast cancer cell proliferation and migration. Biochem Biophys Res Commun. 2019;516:1013–8. [DOI] [PubMed] [Google Scholar]
- 132.Williams BO, Wen JL, Wen XF, Li RB, Jin YC, Wang XL, Zhou L, Chen HX. UBE3C promotes growth and metastasis of renal cell carcinoma via activating Wnt/β-catenin pathway. PLoS One. 2015;10:e0115622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang J-H, Liu Y-F, Ke A-W, Gu F-M, Yu Y, Dai Z, Gao Q, Shi G-M, Liao B-Y, Xie Y-H, et al. Clinical significance of the ubiquitin ligase UBE3C in hepatocellular carcinoma revealed by exome sequencing. Hepatology. 2014;59:2216–27. [DOI] [PubMed] [Google Scholar]
- 134.Ge MK, Zhang N, Xia L, Zhang C, Dong SS, Li ZM, Ji Y, Zheng MH, Sun J, Chen GQ, Shen SM. FBXO22 degrades nuclear PTEN to promote tumorigenesis. Nat Commun. 2020;11:1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang L, Chen J, Ning D, Liu Q, Wang C, Zhang Z, Chu L, Yu C, Liang HF, Zhang B, Chen X. FBXO22 promotes the development of hepatocellular carcinoma by regulating the ubiquitination and degradation of p21. J Exp Clin Cancer Res. 2019;38:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Deng R, Guo Y, Li L, He J, Qiang Z, Zhang H, Chen R, Wang Y, Zhao X, Yu J. BAP1 suppresses prostate cancer progression by deubiquitinating and stabilizing PTEN. Mol Oncol. 2020;15:279–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen X-X, Yin Y, Cheng J-W, Huang A, Hu B, Zhang X, Sun Y-F, Wang J, Wang Y-P, Ji Y, et al. BAP1 acts as a tumor suppressor in intrahepatic cholangiocarcinoma by modulating the ERK1/2 and JNK/c-Jun pathways. Cell Death Dis. 2018;9:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Park SH, Fong KW, Kim J, Wang F, Lu X, Lee Y, Brea LT, Wadosky K, Guo C, Abdulkadir SA, et al. Posttranslational regulation of FOXA1 by Polycomb and BUB3/USP7 deubiquitin complex in prostate cancer. Sci Adv. 2021;7:eabe2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng N, Chu M, Lin M, He Y, Wang Z. USP7 stabilizes EZH2 and enhances cancer malignant progression. Am J Cancer Res. 2020;10:299–313. [PMC free article] [PubMed] [Google Scholar]
- 140.Duan D, Shang M, Han Y, Liu J, Liu J, Kong SH, Hou J, Huang B, Lu J, Zhang Y. EZH2–CCF–cGAS axis promotes breast cancer metastasis. Int J Mol Sci. 2022;23:1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.He L, Liu X, Yang J, Li W, Liu S, Liu X, Yang Z, Ren J, Wang Y, Shan L, et al. Imbalance of the reciprocally inhibitory loop between the ubiquitin-specific protease USP43 and EGFR/PI3K/AKT drives breast carcinogenesis. Cell Res. 2018;28:934–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu W, Zheng M, Zhang R, Jiang Q, Du G, Wu Y, Yang C, Li F, Li W, Wang L, et al. RNF126-mediated MRE11 ubiquitination activates the DNA damage response and confers resistance of triple-negative breast cancer to radiotherapy. Adv Sci (Weinh). 2023;10:e2203884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen Y, Zhao Y, Yang X, Ren X, Huang S, Gong S, Tan X, Li J, He S, Li Y, et al. USP44 regulates irradiation-induced DNA double-strand break repair and suppresses tumorigenesis in nasopharyngeal carcinoma. Nat Commun. 2022;13:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu X, Lu R, Yang Q, He J, Huang C, Cao Y, Zhou Z, Huang J, Li L, Chen R, et al. USP7 reduces the level of nuclear DICER, impairing DNA damage response and promoting cancer progression. Mol Oncol. 2024;18:170–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu J, Zhou T, Dong X, Guo Q, Zheng L, Wang X, Zhang N, Li D, Ren L, Yi F, et al. De-ubiquitination of SAMHD1 by USP7 promotes DNA damage repair to overcome oncogenic stress and affect chemotherapy sensitivity. Oncogene. 2023;42:1843–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Murphy M, Xiong Y, Pattabiraman G, Qiu F, Medvedev AE. Pellino-1 positively regulates Toll-like Receptor (TLR) 2 and TLR4 signaling and is suppressed upon induction of endotoxin tolerance. J Biol Chem. 2015;290:19218–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cao C, An R, Yu YY, Dai HY, Qu ZH, Gao MC, Wang JW. BICP0 negatively regulates TRAF6-mediated NF-kappa B and interferon activation by promoting K48-linked polyubiquitination of TRAF6. Front Microbiol. 2020;10:3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Okamoto M, Kouwaki T, Fukushima Y, Oshiumi H. Regulation of RIG-I activation by K63-linked polyubiquitination. Front Immunol. 1942;2018:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang LP, Zhang BQ, Wei MY, Xu Z, Kong WY, Deng K, Xu XX, Zhang L, Zhao XB, Yan L. TRIM22 inhibits endometrial cancer progression through the NOD2/NF-kB signaling pathway and confers a favorable prognosis. Int J Oncol. 2020;56:1225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Asano N, Imatani A, Watanabe T, Fushiya J, Kondo Y, Jin X, Ara N, Uno K, Iijima K, Koike T, et al. Cdx2 expression and intestinal metaplasia Induced by H. pylori infection of gastric cells is regulated by NOD1-mediated innate immune responses. Can Res. 2016;76:1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gopalan B, Shanker M, Scott A, Branch CD, Chada S, Ramesh R. MDA-7/IL-24, a novel tumor suppressor/cytokine is ubiquitinated and regulated by the ubiquitin-proteasome system, and inhibition of MDA-7/IL-24 degradation enhances the antitumor activity. Cancer Gene Ther. 2008;15:1–8. [DOI] [PubMed] [Google Scholar]
- 152.Damgaard RB, Jolin HE, Allison MED, Davies SE, Titheradge HL, McKenzie ANJ, Komander D. OTULIN protects the liver against cell death, inflammation, fibrosis, and cancer. Cell Death Differ. 2020;27:1457–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhao X, Zhou T, Wang Y, Bao M, Ni C, Ding L, Sun S, Dong H, Li J, Liang C. Trigred motif 36 regulates neuroendocrine differentiation of prostate cancer via HK2 ubiquitination and GPx4 deficiency. Cancer Sci. 2023;114:2445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang Z, Wang MM, Geng Y, Ye CY, Zang YS. Membrane-associated RING-CH protein (MARCH8) is a novel glycolysis repressor targeted by miR-32 in colorectal cancer. J Transl Med. 2022;20:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li M, Gao F, Zhao Q, Zuo H, Liu W, Li W. Tanshinone IIA inhibits oral squamous cell carcinoma via reducing Akt-c-Myc signaling-mediated aerobic glycolysis. Cell Death Dis. 2020;11:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhao G, Yuan H, Li Q, Zhang J, Guo Y, Feng T, Gu R, Ou D, Li S, Li K, Lin P. DDX39B drives colorectal cancer progression by promoting the stability and nuclear translocation of PKM2. Signal Transduct Target Ther. 2022;7:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wu H, Jiao Y, Guo X, Wu Z, Lv Q. METTL14/miR-29c-3p axis drives aerobic glycolysis to promote triple-negative breast cancer progression though TRIM9-mediated PKM2 ubiquitination. J Cell Mol Med. 2024;28:e18112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Han J, Zhao Z, Zhang N, Yang Y, Ma L, Feng L, Zhang X, Zuo J, Fan Z, Wang Y, et al. Transcriptional dysregulation of TRIM29 promotes colorectal cancer carcinogenesis via pyruvate kinase-mediated glucose metabolism. Aging (Albany NY). 2021;13:5034–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu H, Guo X, Jiao Y, Wu Z, Lv Q. TRIM35 ubiquitination regulates the expression of PKM2 tetramer and dimer and affects the malignant behaviour of breast cancer by regulating the Warburg effect. Int J Oncol. 2022;61:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen B, Hong Y, Gui R, Zheng H, Tian S, Zhai X, Xie X, Chen Q, Qian Q, Ren X, et al. N6-methyladenosine modification of circ_0003215 suppresses the pentose phosphate pathway and malignancy of colorectal cancer through the miR-663b/DLG4/G6PD axis. Cell Death Dis. 2022;13:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shao J, Shi T, Yu H, Ding Y, Li L, Wang X, Wang X. Cytosolic GDH1 degradation restricts protein synthesis to sustain tumor cell survival following amino acid deprivation. EMBO J. 2021;40:e107480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ito H, Nakamae I, Kato JY, Yoneda-Kato N. Stabilization of fatty acid synthesis enzyme acetyl-CoA carboxylase 1 suppresses acute myeloid leukemia development. J Clin Invest. 2021;131:e141529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wei W, Qin B, Wen W, Zhang B, Luo H, Wang Y, Xu H, Xie X, Liu S, Jiang X, et al. FBXW7β loss-of-function enhances FASN-mediated lipogenesis and promotes colorectal cancer growth. Signal Transduct Target Ther. 2023;8:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang T, Lu Z, Han T, Wang Y, Gan M, Wang JB. Deacetylation of glutaminase by HDAC4 contributes to lung cancer tumorigenesis. Int J Biol Sci. 2022;18:4452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zheng X, Shao J, Qian J, Liu S. circRPS19 affects HK2-mediated aerobic glycolysis and cell viability via the miR-125a-5p/USP7 pathway in gastric cancer. Int J Oncol. 2023;63:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Han X, Ren C, Lu C, Qiao P, Yang T, Yu Z. Deubiquitination of MYC by OTUB1 contributes to HK2 mediated glycolysis and breast tumorigenesis. Cell Death Differ. 2022;29:1864–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.He Y, Jiang S, Zhong Y, Wang X, Cui Y, Liang J, Sun Y, Zhu Z, Huang Z, Mao X. USP7 promotes non-small-cell lung cancer cell glycolysis and survival by stabilizing and activating c-Abl. Clin Transl Med. 2023;13:e1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Maddalena F, Condelli V, Matassa DS, Pacelli C, Scrima R, Lettini G, Li Bergolis V, Pietrafesa M, Crispo F, Piscazzi A, et al. TRAP1 enhances Warburg metabolism through modulation of PFK1 expression/activity and favors resistance to EGFR inhibitors in human colorectal carcinomas. Mol Oncol. 2020;14:3030–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sun T, Liu Z, Bi F, Yang Q. Deubiquitinase PSMD14 promotes ovarian cancer progression by decreasing enzymatic activity of PKM2. Mol Oncol. 2021;15:3639–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lv T, Zhang B, Jiang C, Zeng Q, Yang J, Zhou Y. USP35 promotes hepatocellular carcinoma progression by protecting PKM2 from ubiquitination-mediated degradation. Int J Oncol. 2023;63:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nguyen TV. USP15 antagonizes CRL4(CRBN)-mediated ubiquitylation of glutamine synthetase and neosubstrates. Proc Natl Acad Sci U S A. 2021;118:e2111391118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ning Z, Guo X, Liu X, Lu C, Wang A, Wang X, Wang W, Chen H, Qin W, Liu X, et al. USP22 regulates lipidome accumulation by stabilizing PPARγ in hepatocellular carcinoma. Nat Commun. 2022;13:2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pan W, Luo Q, Yan X, Yuan L, Yi H, Zhang L, Li B, Zhang Y, Sun J, Qiu M-Z, Yang D-J. A novel SMAC mimetic APG-1387 exhibits dual antitumor effect on HBV-positive hepatocellular carcinoma with high expression of cIAP2 by inducing apoptosis and enhancing innate anti-tumor immunity. Biochem Pharmacol. 2018;154:127–35. [DOI] [PubMed] [Google Scholar]
- 174.Zhang H, Xia Y, Wang F, Luo M, Yang K, Liang S, An S, Wu S, Yang C, Chen D, et al. Aldehyde dehydrogenase 2 mediates alcohol-induced colorectal cancer immune escape through stabilizing PD-L1 expression. Adv Sci (Weinh). 2021;8:2003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Meng F, Su Y, Xu B. Rho-associated protein kinase-dependent moesin phosphorylation is required for PD-L1 stabilization in breast cancer. Mol Oncol. 2020;14:2701–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu W, Ren D, Xiong W, Jin X, Zhu L. A novel FBW7/NFAT1 axis regulates cancer immunity in sunitinib-resistant renal cancer by inducing PD-L1 expression. J Exp Clin Cancer Res. 2022;41:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Meng X, Liu X, Guo X, Jiang S, Chen T, Hu Z, Liu H, Bai Y, Xue M, Hu R, et al. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature. 2018;564:130–5. [DOI] [PubMed] [Google Scholar]
- 178.De S, Holvey-Bates EG, Mahen K, Willard B, Stark GR. The ubiquitin E3 ligase FBXO22 degrades PD-L1 and sensitizes cancer cells to DNA damage. Proc Natl Acad Sci U S A. 2021;118:e2112674118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jing W, Wang G, Cui Z, Xiong G, Jiang X, Li Y, Li W, Han B, Chen S, Shi B. FGFR3 destabilizes PD-L1 via NEDD4 to control T-cell-mediated bladder cancer immune surveillance. Cancer Res. 2022;82:114–29. [DOI] [PubMed] [Google Scholar]
- 180.Karwacz K, Bricogne C, MacDonald D, Arce F, Bennett CL, Collins M, Escors D. PD-L1 co-stimulation contributes to ligand-induced T cell receptor down-modulation on CD8+ T cells. EMBO Mol Med. 2011;3:581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu Y, Xu Q, Deng F, Zheng Z, Luo J, Wang P, Zhou J, Lu X, Zhang L, Chen Z, et al. HERC2 promotes inflammation-driven cancer stemness and immune evasion in hepatocellular carcinoma by activating STAT3 pathway. J Exp Clin Cancer Res. 2023;42:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yang H, Xue M, Su P, Zhou Y, Li X, Li Z, Xia Y, Zhang C, Fu M, Zheng X, et al. RNF31 represses cell progression and immune evasion via YAP/PD-L1 suppression in triple negative breast Cancer. J Exp Clin Cancer Res. 2022;41:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pitarresi JR, Liu X, Avendano A, Thies KA, Sizemore GM, Hammer AM, Hildreth BE 3rd, Wang DJ, Steck SA, Donohue S, et al. Disruption of stromal hedgehog signaling initiates RNF5-mediated proteasomal degradation of PTEN and accelerates pancreatic tumor growth. Life Sci Alliance. 2018;1:e201800190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee JH, Jung SM, Yang KM, Bae E, Ahn SG, Park JS, Seo D, Kim M, Ha J, Lee J, et al. A20 promotes metastasis of aggressive basal-like breast cancers through multi-monoubiquitylation of Snail1. Nat Cell Biol. 2017;19:1260–73. [DOI] [PubMed] [Google Scholar]
- 185.Yu J, Cui J, Zhang X, Xu H, Chen Z, Li Y, Niu Y, Wang S, Ran S, Zou Y, et al. The OX40-TRAF6 axis promotes CTLA-4 degradation to augment antitumor CD8(+) T-cell immunity. Cell Mol Immunol. 2023;20:1445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang S, Zhou YF, Cao J, Burley SK, Wang HY, Zheng XFS. mTORC1 promotes ARID1A degradation and oncogenic chromatin remodeling in hepatocellular carcinoma. Cancer Res. 2021;81:5652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Huang X, Zhang Q, Lou Y, Wang JL, Zhao XY, Wang L, Zhang XZ, Li SS, Zhao YL, Chen Q, et al. USP22 deubiquitinates CD274 to suppress anticancer immunity. Cancer Immunol Res. 2019;7:1580–90. [DOI] [PubMed] [Google Scholar]
- 188.Wang Y, Sun Q, Mu N, Sun X, Wang Y, Fan S, Su L, Liu X. The deubiquitinase USP22 regulates PD-L1 degradation in human cancer cells. Cell Commun Signal. 2020;18:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yang H, Zhang X, Lao M, Sun K, He L, Xu J, Duan Y, Chen Y, Ying H, Li M, et al. Targeting ubiquitin-specific protease 8 sensitizes anti-programmed death-ligand 1 immunotherapy of pancreatic cancer. Cell Death Differ. 2023;30:560–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shi D, Wu X, Jian Y, Wang J, Huang C, Mo S, Li Y, Li F, Zhang C, Zhang D, et al. USP14 promotes tryptophan metabolism and immune suppression by stabilizing IDO1 in colorectal cancer. Nat Commun. 2022;13:5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang Z, Xu G, Wang B, Liu Y, Zhang L, Jing T, Tang M, Xu X, Jiao K, Xiang L, et al. USP12 downregulation orchestrates a protumourigenic microenvironment and enhances lung tumour resistance to PD-1 blockade. Nat Commun. 2021;12:4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jingjing W, Wenzheng G, Donghua W, Guangyu H, Aiping Z, Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Cancer Med. 2018;7:4004–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wu X, Liu M, Zhu H, Wang J, Dai W, Li J, Zhu D, Tang W, Xiao Y, Lin J, et al. Ubiquitin-specific protease 3 promotes cell migration and invasion by interacting with and deubiquitinating SUZ12 in gastric cancer. J Exp Clin Cancer Res. 2019;38:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang W, Wang J, Yan H, Zhang K, Liu Y. Upregulation of USP11 promotes epithelial-to-mesenchymal transition by deubiquitinating Snail in ovarian cancer. Oncol Rep. 2019;41:1739–48. [DOI] [PubMed] [Google Scholar]
- 196.Garcia DA, Baek C, Estrada MV, Tysl T, Bennett EJ, Yang J, Chang JT. USP11 enhances TGFβ-induced epithelial-mesenchymal plasticity and human breast cancer metastasis. Mol Cancer Res. 2018;16:1172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kit Leng Lui S, Iyengar PV, Jaynes P, Isa Z, Pang B, Tan TZ, Eichhorn PJA. USP26 regulates TGF-β signaling by deubiquitinating and stabilizing SMAD7. EMBO Rep. 2020;21:e49618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Eichhorn PJ, Rodón L, Gonzàlez-Juncà A, Dirac A, Gili M, Martínez-Sáez E, Aura C, Barba I, Peg V, Prat A, et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med. 2012;18:429–35. [DOI] [PubMed] [Google Scholar]
- 199.Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu CX, ten Dijke P. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat Cell Biol. 2012;14:717–26. [DOI] [PubMed] [Google Scholar]
- 200.Lambies G, Miceli M, Martínez-Guillamon C, Olivera-Salguero R, Peña R, Frías CP, Calderón I, Atanassov BS, Dent SYR, Arribas J, et al. TGFβ-Activated USP27X deubiquitinase regulates cell migration and chemoresistance via stabilization of snail1. Cancer Res. 2019;79:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wu Y, Wang Y, Lin Y, Liu Y, Wang Y, Jia J, Singh P, Chi YI, Wang C, Dong C, et al. Dub3 inhibition suppresses breast cancer invasion and metastasis by promoting Snail1 degradation. Nat Commun. 2017;8:14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Traweek RS, Cope BM, Roland CL, Keung EZ, Nassif EF, Erstad DJ. Targeting the MDM2-p53 pathway in dedifferentiated liposarcoma. Fronti Oncol. 2022;12:1006959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yu B, Liu Z, Fu Y, Wang Y, Zhang L, Cai Z, Yu F, Wang X, Zhou J, Kong W. CYLD deubiquitinates nicotinamide adenine dinucleotide phosphate oxidase 4 contributing to adventitial remodeling. Arterioscler Thromb Vasc Biol. 2017;37:1698–709. [DOI] [PubMed] [Google Scholar]
- 204.Zheng Z, Shang Y, Xu R, Yan X, Wang X, Cai J, Bai Z, Liu X, Yin J, Zhang J, Zhang Z. Ubiquitin specific peptidase 38 promotes the progression of gastric cancer through upregulation of fatty acid synthase. Am J Cancer Res. 2022;12:2686–96. [PMC free article] [PubMed] [Google Scholar]
- 205.Deng Y, Li Y, Wu T, Chen X, Li X, Cai K, Wu X. RAD6 positively affects tumorigenesis of esophageal squamous cell carcinoma by regulating histone ubiquitination of CCNB1. Biol Proced Online. 2022;24:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang N, Gao X, Yuan Q, Fu X, Wang P, Cai F, Liu H, Zhang J, Liang H, Nie Y, Deng J. E3 ubiquitin ligase RNF180 prevents excessive PCDH10 methylation to suppress the proliferation and metastasis of gastric cancer cells by promoting ubiquitination of DNMT1. Clin Epigenetics. 2023;15:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sun W, Ma G, Zhang L, Wang P, Zhang N, Wu Z, Dong Y, Cai F, Chen L, Liu H, et al. DNMT3A-mediated silence in ADAMTS9 expression is restored by RNF180 to inhibit viability and motility in gastric cancer cells. Cell Death Dis. 2021;12:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yadav P, Subbarayalu P, Medina D, Nirzhor S, Timilsina S, Rajamanickam S, Eedunuri VK, Gupta Y, Zheng S, Abdelfattah N, et al. M6A RNA methylation regulates histone ubiquitination to support cancer growth and progression. Cancer Res. 2022;82:1872–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jiang ZH, Peng T, Qian HL, Lu CD, Qiu F, Zhang SZ. DNA damage-induced activation of ATM promotes β-TRCP-mediated ARID1A ubiquitination and destruction in gastric cancer cells. Cancer Cell Int. 2019;19:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Seo HR, Jeong D, Lee S, Lee HS, Lee SA, Kang SW, Kwon J. CHIP and BAP1 act in concert to regulate INO80 ubiquitination and stability for DNA replication. Mol Cells. 2021;44:101–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Cheng X, Zhang B, Guo F, Wu H, Jin X. Deubiquitination of FBP1 by USP7 blocks FBP1-DNMT1 interaction and decreases the sensitivity of pancreatic cancer cells to PARP inhibitors. Mol Oncol. 2022;16:1591–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Fu X, Zhao J, Yu G, Zhang X, Sun J, Li L, Yin J, Niu Y, Ren S, Zhu Y, et al. OTUD6A promotes prostate tumorigenesis via deubiquitinating Brg1 and AR. Commun Biol. 2022;5:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hatta Muhammad Nur A, Mohamad Hanif Ezanee A, Chin SF, Low Teck Y, Neoh HM. Parvimonas micra infection enhances proliferation, wound healing, and inflammation of a colorectal cancer cell line. Biosci Rep. 2023;43:BSR20230609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ryu TY, Kim K, Han T-S, Lee M-O, Lee J, Choi J, Jung KB, Jeong E-J, An DM, Jung C-R, et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022;16:1205–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lee S, Rauch J, Kolch W. Targeting MAPK signaling in cancer: mechanisms of drug resistance and sensitivity. Int J Mol Sci. 2020;21:1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hong SW, Jin DH, Shin JS, Moon JH, Na YS, Jung KA, Kim SM, Kim JC, Kim KP, Hong YS, et al. Ring finger protein 149 is an E3 ubiquitin ligase active on wild-type v-Raf murine sarcoma viral oncogene homolog B1 (BRAF). J Biol Chem. 2012;287:24017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yeh CH, Bellon M, Wang F, Zhang H, Fu L, Nicot C. Loss of FBXW7-mediated degradation of BRAF elicits resistance to BET inhibitors in adult T cell leukemia cells. Mol Cancer. 2020;19:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chen Q, Hang Y, Zhang T, Tan L, Li S, Jin Y. USP10 promotes proliferation and migration and inhibits apoptosis of endometrial stromal cells in endometriosis through activating the Raf-1/MEK/ERK pathway. Am J Physiol Cell Physiol. 2018;315:C863-c872. [DOI] [PubMed] [Google Scholar]
- 219.Ito Y, Sasaki Y, Horimoto M, Wada S, Tanaka Y, Kasahara A, Ueki T, Hirano T, Yamamoto H, Fujimoto J, et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. 1998;27:951–8. [DOI] [PubMed] [Google Scholar]
- 220.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. [DOI] [PubMed] [Google Scholar]
- 221.Delire B, Stärkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur J Clin Invest. 2015;45:609–23. [DOI] [PubMed] [Google Scholar]
- 222.Kumari N, Jaynes PW, Saei A, Iyengar PV, Richard JLC, Eichhorn PJA. The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim Biophys Acta Rev Cancer. 2017;1868:456–83. [DOI] [PubMed] [Google Scholar]
- 223.Byun S, Lee SY, Lee J, Jeong CH, Farrand L, Lim S, Reddy K, Kim JY, Lee MH, Lee HJ, et al. USP8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin Cancer Res. 2013;19:3894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yan M, Zhao C, Wei N, Wu X, Cui J, Xing Y. High expression of Ubiquitin-Specific Protease 8 (USP8) is associated with poor prognosis in patients with cervical squamous cell carcinoma. Med Sci Monit. 2018;24:4934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tian Y, Liu K, Liu R, Qiu Z, Xu Y, Wei W, Xu X, Wang J, Ding H, Li Z, Bian J. Discovery of potent small-molecule USP8 inhibitors for the treatment of breast cancer through regulating ERα expression. J Med Chem. 2022;65:8914–32. [DOI] [PubMed] [Google Scholar]
- 226.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–49. [DOI] [PubMed] [Google Scholar]
- 227.Zhou Q, Cheng C, Wei Y, Yang J, Zhou W, Song Q, Ke M, Yan W, Zheng L, Zhang Y, Huang K. USP15 potentiates NF-κB activation by differentially stabilizing TAB2 and TAB3. FEBS J. 2020;287:3165–83. [DOI] [PubMed] [Google Scholar]
- 228.Maruyama T, Araki T, Kawarazaki Y, Naguro I, Heynen S, Aza-Blanc P, Ronai Z, Matsuzawa A, Ichijo H. Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Sci Signal. 2014;7:ra8. [DOI] [PubMed] [Google Scholar]
- 229.Tobiume K, Matsuzawa A, Takahashi T, Nishitoh H, Morita K, Takeda K, Minowa O, Miyazono K, Noda T, Ichijo H. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001;2:222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhao Y, Conze DB, Hanover JA, Ashwell JD. Tumor necrosis factor receptor 2 signaling induces selective c-IAP1-dependent ASK1 ubiquitination and terminates mitogen-activated protein kinase signaling. J Biol Chem. 2007;282:7777–82. [DOI] [PubMed] [Google Scholar]
- 231.Glaviano A, Foo ASC, Lam HY, Yap KCH, Jacot W, Jones RH, Eng H, Nair MG, Makvandi P, Geoerger B, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bae S, Kim SY, Jung JH, Yoon Y, Cha HJ, Lee H, Kim K, Kim J, An IS, Kim J, et al. Akt is negatively regulated by the MULAN E3 ligase. Cell Res. 2012;22:873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hu L, Cheng X, Binder Z, Han Z, Yin Y, O’Rourke DM, Wang S, Feng Y, Weng C, Wu A, Lin Z. Molecular and clinical characterization of UBE2S in glioma as a biomarker for poor prognosis and resistance to chemo-radiotherapy. Front Oncol. 2021;11:640910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhang M, Wang J, Guo Y, Yue H, Zhang L. Activation of PI3K/AKT/mTOR signaling axis by UBE2S inhibits autophagy leading to cisplatin resistance in ovarian cancer. J Ovarian Res. 2023;16:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jiang Q, Zheng N, Bu L, Zhang X, Zhang X, Wu Y, Su Y, Wang L, Zhang X, Ren S, et al. SPOP-mediated ubiquitination and degradation of PDK1 suppresses AKT kinase activity and oncogenic functions. Mol Cancer. 2021;20:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zou Z, Tao T, Li H, Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Alvarez-Garcia V, Tawil Y, Wise HM, Leslie NR. Mechanisms of PTEN loss in cancer: it’s all about diversity. Semin Cancer Biol. 2019;59:66–79. [DOI] [PubMed] [Google Scholar]
- 238.Li J, Yang D, Lin Y, Xu W, Zhao SM, Wang C. OTUD3 suppresses the mTORC1 signaling by deubiquitinating KPTN. Front Pharmacol. 2023;14:1337732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.de Mel S, Hue SSS, Jeyasekharan AD, Chng WJ, Ng SB. Molecular pathogenic pathways in extranodal NK/T cell lymphoma. J Hematol Oncol. 2019;12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Liu Y, Wang X, Wang G, Yang Y, Yuan Y, Ouyang L. The past, present and future of potential small-molecule drugs targeting p53-MDM2/MDMX for cancer therapy. Eur J Med Chem. 2019;176:92–104. [DOI] [PubMed] [Google Scholar]
- 241.Gluck WL, Gounder MM, Frank R, Eskens F, Blay JY, Cassier PA, Soria JC, Chawla S, de Weger V, Wagner AJ, et al. Phase 1 study of the MDM2 inhibitor AMG 232 in patients with advanced P53 wild-type solid tumors or multiple myeloma. Invest New Drugs. 2020;38:831–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lu Y, Su F, Cheng Z, Yang J, Dai H, Yang J, Zhang T, Bai Y. Nickel chloride promotes lung cancer invasion and metastasis by up-regulating the expression of E3 ubiquitin ligase TRIM31 through the IL-6/STAT3 signaling axis. Life Sci. 2023;332:122111. [DOI] [PubMed] [Google Scholar]
- 243.Yu C, Chen S, Guo Y, Sun C. Oncogenic TRIM31 confers gemcitabine resistance in pancreatic cancer via activating the NF-κB signaling pathway. Theranostics. 2018;8:3224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yang H, Liu Y, Yang J, Zhang Q, Wang H, Chen Y, Zhou K. Upregulation of USP25 promotes progression of human diffuse large B-cell lymphoma through blocking the ubiquitinated degradation of MDM2. Biochem Biophys Res Commun. 2023;676:21–9. [DOI] [PubMed] [Google Scholar]
- 245.Engeland K. Cell cycle regulation: p53–p21-RB signaling. Cell Death Differ. 2022;29:946–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kon S, Kobayashi N, Satake M. Altered trafficking of mutated growth factor receptors and their associated molecules. Cell Logist. 2014;4:e28461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Czerwińska P, Mazurek S, Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J Biomed Sci. 2017;24:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Huang Z, Li X, Tang B, Li H, Zhang J, Sun R, Ma J, Pan Y, Yan B, Zhou Y, et al. SETDB1 modulates degradation of phosphorylated RB and anticancer efficacy of CDK4/6 inhibitors. Cancer Res. 2023;83:875–89. [DOI] [PubMed] [Google Scholar]
- 249.Lagopati N, Belogiannis K, Angelopoulou A, Papaspyropoulos A, Gorgoulis V. Non-canonical functions of the ARF tumor suppressor in development and tumorigenesis. Biomolecules. 2021;11:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sui XY, Ma XY, Hou Y, Cao SW, Wang ZQ, Jia LJ, Fan L, Shao ZM, Zhang WJ. Elongin B promotes breast cancer progression by ubiquitinating tumor suppressor p14/ARF. Cell Biol Toxicol. 2024;40:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Fischer S, Trinh VT, Simon C, Weber LM, Forné I, Nist A, Bange G, Abendroth F, Stiewe T, Steinchen W, et al. Peptide-mediated inhibition of the transcriptional regulator Elongin BC induces apoptosis in cancer cells. Cell Chem Biol. 2023;30:766-779.e711. [DOI] [PubMed] [Google Scholar]
- 252.Zhang W, Li L, Cai L, Liang Y, Xu J, Liu Y, Zhou L, Ding C, Zhang Y, Zhao H, et al. Tumor-associated antigen Prame targets tumor suppressor p14/ARF for degradation as the receptor protein of CRL2(Prame) complex. Cell Death Differ. 2021;28:1926–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Bertheloot D, Latz E, Franklin BS. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell Mol Immunol. 2021;18:1106–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Liu J, Hong M, Li Y, Chen D, Wu Y, Hu Y. Programmed Cell Death Tunes Tumor Immunity. Front Immunol. 2022;13:847345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Xu X, Lai Y, Hua Z-C. Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep. 2019;39:BSR20180992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Tummers B, Green DR. Caspase-8: regulating life and death. Immunol Rev. 2017;277:76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lu S, Zhou J, Jian H, Wu L, Cheng Y, Fan Y, Fang J, Chen G, Zhang Z, Lv D, et al. Befotertinib (D-0316) versus icotinib as first-line therapy for patients with EGFR-mutated locally advanced or metastatic non-small-cell lung cancer: a multicentre, open-label, randomised phase 3 study. Lancet Respir Med. 2023;11:905–15. [DOI] [PubMed] [Google Scholar]
- 258.Moustafa-Kamal M, Gamache I, Lu Y, Li S, Teodoro JG. BimEL is phosphorylated at mitosis by Aurora A and targeted for degradation by βTrCP1. Cell Death Differ. 2013;20:1393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yan J, Wan P, Choksi S, Liu ZG. Necroptosis and tumor progression. Trends Cancer. 2022;8:21–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, Liu J, Yu X, Shi S. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. 2022;15:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Luo M, Wang X, Wu S, Yang C, Su Q, Huang L, Fu K, An S, Xie F, To KKW, et al. A20 promotes colorectal cancer immune evasion by upregulating STC1 expression to block “eat-me” signal. Signal Transduct Target Ther. 2023;8:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Hjelmeland AB, Wu Q, Wickman S, Eyler C, Heddleston J, Shi Q, Lathia JD, Macswords J, Lee J, McLendon RE, Rich JN. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol. 2010;8:e1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wei W, Lin Y, Song Z, Xiao W, Chen L, Yin J, Zhou Y, Barta SK, Petrus M, Waldmann TA, Yang Y. A20 and RBX1 regulate brentuximab vedotin sensitivity in hodgkin lymphoma models. Clin Cancer Res. 2020;26:4093–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang Y, Yu T, Wu X, Shi Y, Ma P, Shu Y. Pyroptosis: a new frontier in cancer. Biomed Pharmacother. 2020;121:109595. [DOI] [PubMed] [Google Scholar]
- 265.Wei X, Xie F, Zhou X, Wu Y, Yan H, Liu T, Huang J, Wang F, Zhou F, Zhang L. Role of pyroptosis in inflammation and cancer. Cell Mol Immunol. 2022;19:971–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Han S, Lear TB, Jerome JA, Rajbhandari S, Snavely CA, Gulick DL, Gibson KF, Zou C, Chen BB, Mallampalli RK. Lipopolysaccharide primes the NALP3 inflammasome by inhibiting its ubiquitination and degradation mediated by the SCFFBXL2 E3 ligase. J Biol Chem. 2015;290:18124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Tang J, Tu S, Lin G, Guo H, Yan C, Liu Q, Huang L, Tang N, Xiao Y, Pope RM, et al. Sequential ubiquitination of NLRP3 by RNF125 and Cbl-b limits inflammasome activation and endotoxemia. J Exp Med. 2020;217:e20182091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Jiang Q, Zhu Z, Mao X. Ubiquitination is a major modulator for the activation of inflammasomes and pyroptosis. Biochim Biophys Acta Gene Regul Mech. 2023;1866:194955. [DOI] [PubMed] [Google Scholar]
- 269.Tezcan G, Garanina EE, Alsaadi M, Gilazieva ZE, Martinova EV, Markelova MI, Arkhipova SS, Hamza S, McIntyre A, Rizvanov AA, Khaiboullina SF. Therapeutic potential of pharmacological targeting NLRP3 inflammasome complex in cancer. Front Immunol. 2020;11:607881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Arimoto KI, Miyauchi S, Troutman TD, Zhang Y, Liu M, Stoner SA, Davis AG, Fan JB, Huang YJ, Yan M, et al. Expansion of interferon inducible gene pool via USP18 inhibition promotes cancer cell pyroptosis. Nat Commun. 2023;14:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Huang T, Song X, Yang Y, Wan X, Alvarez AA, Sastry N, Feng H, Hu B, Cheng S-Y. Autophagy and hallmarks of cancer. Crit Rev Oncog. 2018;23:247–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Liu T, Wang L, Liang P, Wang X, Liu Y, Cai J, She Y, Wang D, Wang Z, Guo Z, et al. USP19 suppresses inflammation and promotes M2-like macrophage polarization by manipulating NLRP3 function via autophagy. Cell Mol Immunol. 2020;18:2431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Jao TM, Fang WH, Ciou SC, Yu SL, Hung YL, Weng WT, Lin TY, Tsai MH, Yang YC. PCDH10 exerts tumor-suppressor functions through modulation of EGFR/AKT axis in colorectal cancer. Cancer Lett. 2021;499:290–300. [DOI] [PubMed] [Google Scholar]
- 275.Meng Y, Sun H, Li Y, Zhao S, Su J, Zeng F, Deng G, Chen X. Targeting ferroptosis by ubiquitin system enzymes: a potential therapeutic strategy in cancer. Int J Biol Sci. 2022;18:5475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2020;31:107–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Tang R, Xu J, Zhang B, Liu J, Liang C, Hua J, Meng Q, Yu X, Shi S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2020;12:599–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Li D, Wang Y, Dong C, Chen T, Dong A, Ren J, Li W, Shu G, Yang J, Shen W, et al. CST1 inhibits ferroptosis and promotes gastric cancer metastasis by regulating GPX4 protein stability via OTUB1. Oncogene. 2023;42:83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85:6622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–10. [DOI] [PubMed] [Google Scholar]
- 282.Xu L, Li S, Stohr BA. The role of telomere biology in cancer. Annu Rev Pathol. 2013;8:49–78. [DOI] [PubMed] [Google Scholar]
- 283.Oh BK, Choi Y, Choi JS. Telomere shortening and expression of TRF1 and TRF2 in uterine leiomyoma. Mol Med Rep. 2021;24:606. [DOI] [PubMed] [Google Scholar]
- 284.Pal D, Sharma U, Singh SK, Kakkar N, Prasad R. Over-expression of telomere binding factors (TRF1 & TRF2) in renal cell carcinoma and their inhibition by using SiRNA induce apoptosis, reduce cell proliferation and migration invitro. PLoS One. 2015;10:e0115651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Bejarano L, Schuhmacher AJ, Méndez M, Megías D, Blanco-Aparicio C, Martínez S, Pastor J, Squatrito M, Blasco MA. Inhibition of TRF1 telomere protein impairs tumor initiation and progression in glioblastoma mouse models and patient-derived xenografts. Cancer Cell. 2017;32:590-607.e594. [DOI] [PubMed] [Google Scholar]
- 286.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35:352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Li J, Yuan S, Norgard RJ, Yan F, Yamazoe T, Blanco A, Stanger BZ. Tumor cell-intrinsic USP22 suppresses antitumor immunity in pancreatic cancer. Cancer Immunol Res. 2020;8:282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zemp I, Lingner J. The shelterin component TPP1 is a binding partner and substrate for the deubiquitinating enzyme USP7. J Biol Chem. 2014;289:28595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Fan YH, Cheng J, Vasudevan SA, Dou J, Zhang H, Patel RH, Ma IT, Rojas Y, Zhao Y, Yu Y, et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013;4:e867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Yamaguchi M, Miyazaki M, Kodrasov MP, Rotinsulu H, Losung F, Mangindaan RE, de Voogd NJ, Yokosawa H, Nicholson B, Tsukamoto S. Spongiacidin C, a pyrrole alkaloid from the marine sponge Stylissa massa, functions as a USP7 inhibitor. Bioorg Med Chem Lett. 2013;23:3884–6. [DOI] [PubMed] [Google Scholar]
- 291.Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci. 2019;77:1745–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176:1248–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Paredes F, Williams HC, San Martin A. Metabolic adaptation in hypoxia and cancer. Cancer Lett. 2021;502:133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Al Kawas H, Saaid I, Jank P, Westhoff CC, Denkert C, Pross T, Weiler KBS, Karsten MM. How VEGF-A and its splice variants affect breast cancer development – clinical implications. Cell Oncol. 2022;45:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Shaw P, Dwivedi SKD, Bhattacharya R, Mukherjee P, Rao G. VEGF signaling: role in angiogenesis and beyond. Biochim Biophys Acta Rev Cancer. 2024;1879:189079. [DOI] [PubMed] [Google Scholar]
- 296.Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, He X, Ma J, Xiang J, Jiang G, et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322–34. [DOI] [PubMed] [Google Scholar]
- 297.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–26. [DOI] [PubMed] [Google Scholar]
- 298.Sun R, Xie HY, Qian JX, Huang YN, Yang F, Zhang FL, Shao ZM, Li DQ. FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res. 2018;78:5274–86. [DOI] [PubMed] [Google Scholar]
- 299.Wang X, Wei XL, Cao Y, Xing P. Mcl-1 inhibition overcomes BET inhibitor resistance induced by low FBW7 expression in breast cancer. J Cell Mol Med. 2022;26:1672–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Gu J, Mao W, Ren W, Xu F, Zhu Q, Lu C, Lin Z, Zhang Z, Chu Y, Liu R, Ge D. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett. 2019;443:125–34. [DOI] [PubMed] [Google Scholar]
- 301.Aloni-Grinstein R, Shetzer Y, Kaufman T, Rotter V. p53: The barrier to cancer stem cell formation. FEBS Lett. 2014;588:2580–9. [DOI] [PubMed] [Google Scholar]
- 302.Zhu XN, He P, Zhang L, Yang S, Zhang HL, Zhu D, Liu MD, Yu Y. FBXO22 mediates polyubiquitination and inactivation of LKB1 to promote lung cancer cell growth. Cell Death Dis. 2019;10:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Zheng Y, Chen H, Zhao Y, Zhang X, Liu J, Pan Y, Bai J, Zhang H. Knockdown of FBXO22 inhibits melanoma cell migration, invasion and angiogenesis via the HIF-1α/VEGF pathway. Invest New Drugs. 2020;38:20–8. [DOI] [PubMed] [Google Scholar]
- 304.Li S, Shi L, Wang Y, Zhang L, Chu S, Li M, Bai J, Zhu W. FBXO22 inhibits proliferation and metastasis of cervical cancer cells by mediating ubiquitination-dependent degradation of GAK. Exp Cell Res. 2023;430:113719. [DOI] [PubMed] [Google Scholar]
- 305.Nakagawa S, Miyashita M, Maeda I, Goda A, Tada H, Amari M, Kojima Y, Tsugawa K, Ohi Y, Sagara Y, et al. Potential role of Fbxo22 in resistance to endocrine therapy in breast cancer with invasive lobular carcinoma. Breast Cancer Res Treat. 2024;204:453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, Shao M, You D, Fan Z, Xia H, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.He Y, Ramesh A, Gusev Y, Bhuvaneshwar K, Giaccone G. Molecular predictors of response to pembrolizumab in thymic carcinoma. Cell Rep Med. 2021;2:100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Xue Y, Li M, Hu J, Song Y, Guo W, Miao C, Ge D, Hou Y, Wang X, Huang X, et al. Ca(v)2.2-NFAT2-USP43 axis promotes invadopodia formation and breast cancer metastasis through cortactin stabilization. Cell Death Dis. 2022;13:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Pei LP, Zhao F, Zhang Y. USP43 impairs cisplatin sensitivity in epithelial ovarian cancer through HDAC2-dependent regulation of Wnt/β-catenin signaling pathway. Apoptosis. 2024;29:210–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–17. [DOI] [PubMed] [Google Scholar]
- 311.Pikor L, Thu K, Vucic E, Lam W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev. 2013;32:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Osborne HC, Irving E, Forment JV, Schmidt CK. E2 enzymes in genome stability: pulling the strings behind the scenes. Trends Cell Biol. 2021;31:628–43. [DOI] [PubMed] [Google Scholar]
- 313.Koo SY, Park EJ, Noh HJ, Jo SM, Ko BK, Shin HJ, Lee CW. Ubiquitination links DNA damage and repair signaling to cancer metabolism. Int J Mol Sci. 2023;24:8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 2016;17:379–94. [DOI] [PubMed] [Google Scholar]
- 315.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. [DOI] [PubMed] [Google Scholar]
- 316.Ha GH, Ji JH, Chae S, Park J, Kim S, Lee JK, Kim Y, Min S, Park JM, Kang TH, et al. Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks. Nat Commun. 2019;10:1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Qiu S, Huang J. MRN complex is an essential effector of DNA damage repair. J Zhejiang Univ Sci B. 2021;22:31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Daddacha W, Koyen AE, Bastien AJ, Head PE, Dhere VR, Nabeta GN, Connolly EC, Werner E, Madden MZ, Daly MB, et al. SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep. 2017;20:1921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. [DOI] [PubMed] [Google Scholar]
- 320.Chang SC, Ding JL. Ubiquitination and SUMOylation in the chronic inflammatory tumor microenvironment. Biochim Biophys Acta Rev Cancer. 2018;1870:165–75. [DOI] [PubMed] [Google Scholar]
- 321.Kensche T, Tokunaga F, Ikeda F, Goto E, Iwai K, Dikic I. Analysis of nuclear factor-κB (NF-κB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J Biol Chem. 2012;287:23626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Cockram PE, Kist M, Prakash S, Chen SH, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28:591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Courtois G, Fauvarque M-O. The many roles of ubiquitin in NF-κB signaling. Biomedicines. 2018;6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Song K, Li S. The role of ubiquitination in NF-κB signaling during virus infection. Viruses. 2021;13:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Jang H-D, Hwang HZ, Kim H-S, Lee SY. C-Cbl negatively regulates TRAF6-mediated NF-κB activation by promoting K48-linked polyubiquitination of TRAF6. Cell Mol Biol Lett. 2019;24:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Ji J, Ding K, Luo T, Zhang X, Chen A, Zhang D, Li G, Thorsen F, Huang B, Li X, Wang J. TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα. Cell Death Differ. 2021;28:367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Chang SC, Ding JL. Ubiquitination by SAG regulates macrophage survival/death and immune response during infection. Cell Death Differ. 2014;21:1388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Chang SC, Choo WQ, Toh HC, Ding JL. SAG-UPS attenuates proapoptotic SARM and Noxa to confer survival advantage to early hepatocellular carcinoma. Cell Death Discov. 2015;1:15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Callao V, Montoya E. Toxohormone-like factor from microorganisms with impaired respiration. Science. 1961;134:2041–2. [DOI] [PubMed] [Google Scholar]
- 330.Ye M, Chen J, Lu F, Zhao M, Wu S, Hu C, Yu P, Kan J, Bai J, Tian Y, Tang Q. Down-regulated FTO and ALKBH5 co-operatively activates FOXO signaling through m6A methylation modification in HK2 mRNA mediated by IGF2BP2 to enhance glycolysis in colorectal cancer. Cell Biosci. 2023;13:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Liang C, Wang SQ, Qin C, Bao ML, Cheng G, Liu BJ, Shao PF, Lv Q, Song NH, Hua LX, et al. TRIM36, a novel androgen-responsive gene, enhances anti-androgen efficacy against prostate cancer by inhibiting MAPK/ERK signaling pathways. Cell Death Dis. 2018;9:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Wang Q, Chen Q, Zhu L, Chen M, Xu W, Panday S, Wang Z, Li A, Roe OD, Chen R, et al. JWA regulates TRAIL-induced apoptosis via MARCH8-mediated DR4 ubiquitination in cisplatin-resistant gastric cancer cells. Oncogenesis. 2017;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Zheng YL, Li L, Jia YX, Zhang BZ, Li JC, Zhu YH, Li MQ, He JZ, Zeng TT, Ban XJ, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics. 2019;9:796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Yu M, Xue S, Chen X, Wu K, Ju L, Tang J, Xiong A, Chen X, Ying X. Long non-coding RNA UCA1a promotes proliferation via PKM2 in cervical cancer. Reprod Sci. 2023;30:601–14. [DOI] [PubMed] [Google Scholar]
- 335.Röhrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16:732–49. [DOI] [PubMed] [Google Scholar]
- 336.Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. [DOI] [PubMed] [Google Scholar]
- 338.He Y, Jin YJ, Zhang YH, Meng HX, Zhao BS, Jiang Y, Zhu JW, Liang GY, Kong D, Jin XM. Ubiquitin-specific peptidase 22 overexpression may promote cancer progression and poor prognosis in human gastric carcinoma. Transl Res. 2015;165:407–16. [DOI] [PubMed] [Google Scholar]
- 339.Sun T, Zhang KQ, Li WD, Liu YZ, Pangeni RP, Li AM, Arvanitis L, Raz DJ. Transcription factor AP2 enhances malignancy of non-small cell lung cancer through upregulation of USP22 gene expression. Cell Commun Signal. 2022;20:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Liang JX, Ning Z, Gao W, Ling J, Wang AM, Luo HF, Liang Y, Yan Q, Wang ZY. Ubiquitin-specific protease 22-induced autophagy is correlated with poor prognosis of pancreatic cancer. Oncol Rep. 2014;32:2726–34. [DOI] [PubMed] [Google Scholar]
- 341.Liu B, Jiang S, Li M, Xiong X, Zhu M, Li D, Zhao L, Qian L, Zhai L, Li J, et al. Proteome-wide analysis of USP14 substrates revealed its role in hepatosteatosis via stabilization of FASN. Nat Commun. 2018;9:4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Yang JS, Yoon N, Kong M, Jung BH, Lee H, Park J. USP14 regulates cancer cell growth in a fatty acid synthase-independent manner. Int J Mol Sci. 2021;22:13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Mara P, Fragiadakis GS, Gkountromichos F, Alexandraki D. The pleiotropic effects of the glutamate dehydrogenase (GDH) pathway in Saccharomyces cerevisiae. Microb Cell Fact. 2018;17:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Marsico M, Santarsiero A, Pappalardo I, Convertini P, Chiummiento L, Sardone A, Di Noia MA, Infantino V, Todisco S. Mitochondria-mediated apoptosis of HCC cells triggered by knockdown of glutamate dehydrogenase 1: perspective for its inhibition through quercetin and permethylated anigopreissin A. Biomedicines. 2021;9:1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Häberle J, Görg B, Rutsch F, Schmidt E, Toutain A, Benoist JF, Gelot A, Suc AL, Höhne W, Schliess F, et al. Congenital glutamine deficiency with glutamine synthetase mutations. N Engl J Med. 2005;353:1926–33. [DOI] [PubMed] [Google Scholar]
- 346.Niederkorn M, Ishikawa C, M. Hueneman K, Bartram J, Stepanchick E, R. Bennett J, E. Culver-Cochran A, Bolanos LC, Uible E, Choi K, et al. The deubiquitinase USP15 modulates cellular redox and is a therapeutic target in acute myeloid leukemia. Leukemia. 2022;36:438–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wang Z, Liu F, Fan N, Zhou C, Li D, Macvicar T, Dong Q, Bruns CJ, Zhao Y. Targeting glutaminolysis: new perspectives to understand cancer development and novel strategies for potential target therapies. Front Oncol. 2020;10:589508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Certo M, Tsai CH, Pucino V, Ho PC, Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat Rev Immunol. 2021;21:151–61. [DOI] [PubMed] [Google Scholar]
- 349.Tognon CE, Rafn B, Cetinbas NM, Kamura T, Trigo G, Rotblat B, Okumura F, Matsumoto M, Chow C, Davare M, et al. Insulin-like growth factor 1 receptor stabilizes the ETV6-NTRK3 chimeric oncoprotein by blocking its KPC1/Rnf123-mediated proteasomal degradation. J Biol Chem. 2018;293:12502–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Xiang X, Niu YR, Wang ZH, Ye LL, Peng WB, Zhou Q. Cancer-associated fibroblasts: vital suppressors of the immune response in the tumor microenvironment. Cytokine Growth Factor Rev. 2022;67:35–48. [DOI] [PubMed] [Google Scholar]
- 351.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Liu H, Liu M, He B, Li Q. Inhibition of USP11 sensitizes gastric cancer to chemotherapy via suppressing RhoA and Ras-mediated signaling pathways. Clin Res Hepatol Gastroenterol. 2022;46:101779. [DOI] [PubMed] [Google Scholar]
- 353.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Zhang H, Jin XF, Huang HJ. Deregulation of SPOP in Cancer. Can Res. 2023;83:489–99. [DOI] [PubMed] [Google Scholar]
- 355.Xiong W, Gao X, Zhang T, Jiang B, Hu MM, Bu X, Gao Y, Zhang LZ, Xiao BL, He C, et al. USP8 inhibition reshapes an inflamed tumor microenvironment that potentiates the immunotherapy. Nat Commun. 2022;13:1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Yuan S, Norgard RJ, Stanger BZ. Cellular plasticity in cancer. Cancer Discov. 2019;9:837–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Konopleva M, Martinelli G, Daver N, Papayannidis C, Wei A, Higgins B, Ott M, Mascarenhas J, Andreeff M. MDM2 inhibition: an important step forward in cancer therapy. Leukemia. 2020;34:2858–74. [DOI] [PubMed] [Google Scholar]
- 358.Guo Y, Ruan H, Wang Y, Chen K, Li G, Peng S, Luo G, Zhu C, Lou N, Li X, et al. Overexpression of RACGAP1 by E2F1 promotes neuroendocrine differentiation of prostate cancer by stabilizing EZH2 expression. Aging Dis. 2023;14:1757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Yang J, Xu J, Wang W, Zhang B, Yu X, Shi S. Epigenetic regulation in the tumor microenvironment: molecular mechanisms and therapeutic targets. Signal Transduct Target Ther. 2023;8:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Rodenhiser D, Mann M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ. 2006;174:341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Barkess G. Chromatin remodeling and genome stability. Genome Biol. 2006;7:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Ducasse M, Brown MA. Epigenetic aberrations and cancer. Mol Cancer. 2006;5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. [DOI] [PubMed] [Google Scholar]
- 364.Grady WM, Yu M, Markowitz SD. Epigenetic alterations in the gastrointestinal tract: current and emerging use for biomarkers of cancer. Gastroenterology. 2021;160:690–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Jin X, Pan Y, Wang L, Ma T, Zhang L, Tang AH, Billadeau DD, Wu H, Huang H. Fructose-1,6-bisphosphatase inhibits ERK activation and bypasses gemcitabine resistance in pancreatic cancer by blocking IQGAP1-MAPK interaction. Cancer Res. 2017;77:4328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.McGhee JD, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–56. [DOI] [PubMed] [Google Scholar]
- 367.Sekiguchi M, Matsushita N. DNA damage response regulation by histone ubiquitination. Int J Mol Sci. 2022;23:8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12:658–65. [DOI] [PubMed] [Google Scholar]
- 369.Sanders MA, Haynes B, Nangia-Makker P, Polin LA, Shekhar MP. Pharmacological targeting of RAD6 enzyme-mediated translesion synthesis overcomes resistance to platinum-based drugs. J Biol Chem. 2017;292:10347–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Luo RX, Dean DC. Chromatin remodeling and transcriptional regulation. J Natl Cancer Inst. 1999;91:1288–94. [DOI] [PubMed] [Google Scholar]
- 371.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Mathur R. ARID1A loss in cancer: towards a mechanistic understanding. Pharmacol Ther. 2018;190:15–23. [DOI] [PubMed] [Google Scholar]
- 375.Hussain M, Lu YZ, Liu YQ, Su K, Zhang JC, Liu JS, Zhou GB. Skp 1: Implications in cancer and SCF-oriented anti-cancer drug discovery. Pharmacol Res. 2016;111:34–42. [DOI] [PubMed] [Google Scholar]
- 376.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13:889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun. 2014;5:5128. [DOI] [PubMed] [Google Scholar]
- 378.Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, Trinchieri G. Microbes and cancer. Annu Rev Immunol. 2017;35:199–228. [DOI] [PubMed] [Google Scholar]
- 379.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25:377–88. [DOI] [PubMed] [Google Scholar]
- 380.Xu H, Luo H, Zhang J, Li K, Lee MH. Therapeutic potential of Clostridium butyricum anticancer effects in colorectal cancer. Gut Microbes. 2023;15:2186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169:1000–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Childs BG, Gluscevic M, Baker DJ, Laberge R-M, Marquess D, Dananberg J, van Deursen JM. Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov. 2017;16:718–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Calcinotto A, Kohli J, Zagato E, Pellegrini L, Demaria M, Alimonti A. Cellular senescence: aging, cancer, and injury. Physiol Rev. 2019;99:1047–78. [DOI] [PubMed] [Google Scholar]
- 384.Kung CP, Weber JD. It’s getting complicated-A fresh look at p53-MDM2-ARF triangle in tumorigenesis and cancer therapy. Front Cell Dev Biol. 2022;10:818744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Kouroukis TC, Baldassarre FG, Haynes AE, Imrie K, Reece DE, Cheung MC. Bortezomib in multiple myeloma: systematic review and clinical considerations. Curr Oncol. 2014;21:e573-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS-341): a novel, first-in-class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control. 2003;10:361–9. [DOI] [PubMed] [Google Scholar]
- 387.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–93. [DOI] [PubMed] [Google Scholar]
- 388.Yamamoto S, Egashira N. Pathological mechanisms of bortezomib-induced peripheral neuropathy. Int J Mol Sci. 2021;22:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Neklesa T, Snyder LB, Willard RR, Vitale N, Pizzano J, Gordon DA, Bookbinder M, Macaluso J, Dong HQ, Ferraro C, et al. ARV-110: an oral androgen receptor PROTAC degrader for prostate cancer. J Clin Oncol. 2019;37:259.
- 390.Wang WY, Yang J, Liao YY, Cheng G, Chen J, Mo SW, Yuan L, Cheng XD, Qin JJ, Shao ZZ. Aspeterreurone A, a cytotoxic dihydrobenzofuran-phenyl acrylate hybrid from the deep-sea-derived fungus aspergillus terreus CC-S06-18. J Nat Prod. 2020;83:1998–2003. [DOI] [PubMed] [Google Scholar]
- 391.Cheah CY, Seymour JF, Wang ML. Mantle cell lymphoma. J Clin Oncol. 2016;34:1256–69. [DOI] [PubMed] [Google Scholar]
- 392.Härtel H, Theiß J, Abdelaziz MO, Raftery MJ, Pecher G, Bogner E. HCMV-mediated interference of bortezomib-induced apoptosis in colon carcinoma cell line caco-2. Viruses. 2021;13:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Pasquale R, Giannotta JA, Barcellini W, Fattizzo B. Bortezomib in autoimmune hemolytic anemia and beyond. Ther Adv Hematol. 2021;12:20406207211046428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Stewart AK, Rajkumar SV, Dimopoulos MA, Masszi T, Spicka I, Oriol A, Hájek R, Rosiñol L, Siegel DS, Mihaylov GG, et al. Carfilzomib, lenalidomide, and dexamethasone for relapsed multiple myeloma. N Engl J Med. 2015;372:142–52. [DOI] [PubMed] [Google Scholar]
- 395.Fricker LD. Proteasome inhibitor drugs. Annu Rev Pharmacol Toxicol. 2020;60:457–76. [DOI] [PubMed] [Google Scholar]
- 396.Chauhan D, Singh AV, Aujay M, Kirk CJ, Bandi M, Ciccarelli B, Raje N, Richardson P, Anderson KC. A novel orally active proteasome inhibitor ONX 0912 triggers in vitro and in vivo cytotoxicity in multiple myeloma. Blood. 2010;116:4906–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Zang Y, Thomas SM, Chan ET, Kirk CJ, Freilino ML, DeLancey HM, Grandis JR, Li C, Johnson DE. Carfilzomib and ONX 0912 inhibit cell survival and tumor growth of head and neck cancer and their activities are enhanced by suppression of Mcl-1 or autophagy. Clin Cancer Res. 2012;18:5639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Augello G, Modica M, Azzolina A, Puleio R, Cassata G, Emma MR, Di Sano C, Cusimano A, Montalto G, Cervello M. Preclinical evaluation of antitumor activity of the proteasome inhibitor MLN2238 (ixazomib) in hepatocellular carcinoma cells. Cell Death Dis. 2018;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Kupperman E, Lee EC, Cao Y, Bannerman B, Fitzgerald M, Berger A, Yu J, Yang Y, Hales P, Bruzzese F, et al. Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res. 2010;70:1970–80. [DOI] [PubMed] [Google Scholar]
- 400.Moreau P, Masszi T, Grzasko N, Bahlis NJ, Hansson M, Pour L, Sandhu I, Ganly P, Baker BW, Jackson SR, et al. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med. 2016;374:1621–34. [DOI] [PubMed] [Google Scholar]
- 401.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–6. [DOI] [PubMed] [Google Scholar]
- 402.Lockhart AC, Bauer TM, Aggarwal C, Lee CB, Harvey RD, Cohen RB, Sedarati F, Nip TK, Faessel H, Dash AB, et al. Phase Ib study of pevonedistat, a NEDD8-activating enzyme inhibitor, in combination with docetaxel, carboplatin and paclitaxel, or gemcitabine, in patients with advanced solid tumors. Invest New Drugs. 2019;37:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Swords RT, Coutre S, Maris MB, Zeidner JF, Foran JM, Cruz J, Erba HP, Berdeja JG, Tam W, Vardhanabhuti S, et al. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood. 2018;131:1415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Tsukamoto S, Takeuchi T, Rotinsulu H, Mangindaan RE, van Soest RW, Ukai K, Kobayashi H, Namikoshi M, Ohta T, Yokosawa H. Leucettamol A: a new inhibitor of Ubc13-Uev1A interaction isolated from a marine sponge, Leucetta aff. microrhaphis. Bioorg Med Chem Lett. 2008;18:6319–20. [DOI] [PubMed] [Google Scholar]
- 405.Ushiyama S, Umaoka H, Kato H, Suwa Y, Morioka H, Rotinsulu H, Losung F, Mangindaan RE, de Voogd NJ, Yokosawa H, Tsukamoto S. Manadosterols A and B, sulfonated sterol dimers inhibiting the Ubc13-Uev1A interaction, isolated from the marine sponge Lissodendryx fibrosa. J Nat Prod. 2012;75:1495–9. [DOI] [PubMed] [Google Scholar]
- 406.Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–87. [DOI] [PubMed] [Google Scholar]
- 407.Yang Q, Zhao J, Chen D, Wang Y. E3 ubiquitin ligases: styles, structures and functions. Mol Biomed. 2021;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Assouline S, Mehta A, Phillips T, Alinari L, Danilov AV, Doucet S, Park SI, Berg D, Gomez-Pinillos A, Martinez A, et al. TAK-981, a first-in-class SUMO-activating enzyme inhibitor, combined with rituximab in adult patients (Pts) with CD20-positive Relapsed/Refractory (R/R) Non-Hodgkin Lymphoma (NHL): phase 1 data. Blood. 2021;138:2488. [Google Scholar]
- 409.Gundle KR, Rajasekaran K, Houlton J, Deutsch GB, Ow TJ, Maki RG, Pang J, Nathan CO, Clayburgh D, Newman JG, et al. Early, precise, and safe clinical evaluation of the pharmacodynamic effects of novel agents in the intact human tumor microenvironment. Front Pharmacol. 2024;15:1367581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Zhou XF, Richardson DL, Dowlati A, Goel S, Sahebjam S, Strauss J, Chawla S, Wang D, Mould DR, Samnotra V, et al. Effect of pevonedistat, an investigational NEDD8-activating enzyme inhibitor, on the QTc interval in patients with advanced solid tumors. Clin Pharmacol Drug Dev. 2023;12:257–66. [DOI] [PubMed] [Google Scholar]
- 411.Sekeres MA, Watts J, Radinoff A, Sangerman MA, Cerrano M, Lopez PF, Zeidner JF, Campelo MD, Graux C, Liesveld J, et al. Randomized phase 2 trial of pevonedistat plus azacitidine versus azacitidine for higher-risk MDS/CMML or low-blast AML. Leukemia. 2021;35:2119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Handa H, Cheong JW, Onishi Y, Iida H, Kobayashi Y, Kim HJ, Chiou TJ, Izutsu K, Tsukurov O, Zhou X, et al. Pevonedistat in East Asian patients with acute myeloid leukemia or myelodysplastic syndromes: a phase 1/1b study to evaluate safety, pharmacokinetics and activity as a single agent and in combination with azacitidine. J Hematol Oncol. 2022;15:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Zhou XF, Vaishampayan U, Mahalingam D, Harvey RD, Chung KY, Sedarati F, Dong C, Faller DV, Venkatakrishnan K, Gupta N. Phase 1 study to evaluate the effects of rifampin on pharmacokinetics of pevonedistat, a NEDD8-activating enzyme inhibitor in patients with advanced solid tumors. Invest New Drugs. 2022;40:1042–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Zhou XF, Sedarati F, Faller DV, Zhao D, Faessel HM, Chowdhury S, Bolleddula J, Li YX, Venkatakrishnan K, Papai Z. Phase I study assessing the mass balance, pharmacokinetics, and excretion of 14C -pevonedistat, a NEDD8-activating enzyme inhibitor in patients with advanced solid tumors. Invest New Drugs. 2021;39:488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Faessel H, Nemunaitis J, Bauer TM, Lockhart AC, Faller DV, Sedarati F, Zhou X, Venkatakrishnan K, Harvey RD. Effect of CYP3A inhibitors on the pharmacokinetics of pevonedistat in patients with advanced solid tumours. Br J Clin Pharmacol. 2019;85:1464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Swords RT, Erba HP, DeAngelo DJ, Bixby DL, Altman JK, Maris M, Hua Z, Blakemore SJ, Faessel H, Sedarati F, et al. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br J Haematol. 2015;169:534–43. [DOI] [PubMed] [Google Scholar]
- 417.Shah JJ, Jakubowiak AJ, O’Connor OA, Orlowski RZ, Harvey RD, Smith MR, Lebovic D, Diefenbach C, Kelly K, Hua Z, et al. Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin Cancer Res. 2016;22:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Sarantopoulos J, Shapiro GI, Cohen RB, Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD, Faessel HM, et al. Phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin Cancer Res. 2016;22:847–57. [DOI] [PubMed] [Google Scholar]
- 419.Bhatia S, Pavlick AC, Boasberg P, Thompson JA, Mulligan G, Pickard MD, Faessel H, Dezube BJ, Hamid O. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Invest New Drugs. 2016;34:439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Gounder MM, Bauer TM, Schwartz GK, Weise AM, LoRusso P, Kumar P, Tao B, Hong Y, Patel P, Lu Y, et al. A first-in-human Phase I study of Milademetan, an MDM2 inhibitor, in patients with advanced liposarcoma, solid tumors, or lymphomas. J Clin Oncol. 2023;41:1714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Sekiguchi N, Kasahara S, Miyamoto T, Kiguchi T, Ohno H, Takagi T, Tachibana M, Sumi H, Kakurai Y, Yamashita T, Usuki K. Phase I dose-escalation study of milademetan in patients with relapsed or refractory acute myeloid leukemia. Int J Hematol. 2023;117:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Hong Y, Ishizuka T, Watanabe A, Tachibana M, Lee M, Ishizuka H, LaCreta F, Abutarif M. Model-based assessments of CYP3A-mediated drug-drug interaction risk of milademetan. Clin Transl Sci. 2021;14:2220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Stein EM, DeAngelo DJ, Chromik J, Chatterjee M, Bauer S, Lin CC, Suarez C, de Vos F, Steeghs N, Cassier PA, et al. Results from a First-in-Human Phase I Study of Siremadlin (HDM201) in patients with advanced wild-type TP53 solid tumors and acute leukemia. Clin Cancer Res. 2022;28:870–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Moschos SJ, Sandhu S, Lewis KD, Sullivan RJ, Puzanov I, Johnson DB, Henary HA, Wong H, Upreti VV, Long GV, Flaherty KT. Targeting wild-type TP53 using AMG 232 in combination with MAPK inhibition in Metastatic Melanoma; a phase 1 study. Invest New Drugs. 2022;40:1051–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Erba HP, Becker PS, Shami PJ, Grunwald MR, Flesher DL, Zhu M, Rasmussen E, Henary HA, Anderson AA, Wang ES. Phase 1b study of the MDM2 inhibitor AMG 232 with or without trametinib in relapsed/refractory acute myeloid leukemia. Blood Adv. 2019;3:1939–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Patnaik A, Tolcher A, Beeram M, Nemunaitis J, Weiss GJ, Bhalla K, Agrawal M, Nichols G, Middleton S, Beryozkina A, et al. Clinical pharmacology characterization of RG7112, an MDM2 antagonist, in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2015;76:587–95. [DOI] [PubMed] [Google Scholar]
- 427.de Jonge M, de Weger VA, Dickson MA, Langenberg M, Le Cesne A, Wagner AJ, Hsu K, Zheng W, Macé S, Tuffal G, et al. A phase I study of SAR405838, a novel human double minute 2 (HDM2) antagonist, in patients with solid tumours. Eur J Cancer. 2017;76:144–51. [DOI] [PubMed] [Google Scholar]
- 428.de Weger VA, de Jonge M, Langenberg MHG, Schellens JHM, Lolkema M, Varga A, Demers B, Thomas K, Hsu K, Tuffal G, et al. A phase I study of the HDM2 antagonist SAR405838 combined with the MEK inhibitor pimasertib in patients with advanced solid tumours. Br J Cancer. 2019;120:286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Daver NG, Dail M, Garcia JS, Jonas BA, Yee KWL, Kelly KR, Vey N, Assouline S, Roboz GJ, Paolini S, et al. Venetoclax and idasanutlin in relapsed/refractory AML: a nonrandomized, open-label phase 1b trial. Blood. 2023;141:1265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Pápai Z, Chen LC, Da Costa D, Blotner S, Vazvaei F, Gleave M, Jones R, Zhi J. A single-center, open-label study investigating the excretion balance, pharmacokinetics, metabolism, and absolute bioavailability of a single oral dose of [(14)C]-labeled idasanutlin and an intravenous tracer dose of [(13)C]-labeled idasanutlin in a single cohort of patients with solid tumors. Cancer Chemother Pharmacol. 2019;84:93–103. [DOI] [PubMed] [Google Scholar]
- 431.Yee K, Papayannidis C, Vey N, Dickinson MJ, Kelly KR, Assouline S, Kasner M, Seiter K, Drummond MW, Yoon SS, et al. Murine double minute 2 inhibition alone or with cytarabine in acute myeloid leukemia: results from an idasanutlin phase 1/1b study⋆. Leuk Res. 2021;100:106489. [DOI] [PubMed] [Google Scholar]
- 432.Italiano A, Miller WH Jr, Blay JY, Gietema JA, Bang YJ, Mileshkin LR, Hirte HW, Higgins B, Blotner S, Nichols GL, et al. Phase I study of daily and weekly regimens of the orally administered MDM2 antagonist idasanutlin in patients with advanced tumors. Invest New Drugs. 2021;39:1587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Bauer S, Demetri GD, Halilovic E, Dummer R, Meille C, Tan DSW, Guerreiro N, Jullion A, Ferretti S, Jeay S, et al. Pharmacokinetic-pharmacodynamic guided optimisation of dose and schedule of CGM097, an HDM2 inhibitor, in preclinical and clinical studies. Br J Cancer. 2021;125:687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Morita S, Minami H, Mitsuma A, Toyoda M, Kiyota N, Ando Y. A phase I study of LCL161, a novel oral pan-inhibitor of apoptosis protein (IAP) antagonist, in Japanese patients with advanced solid tumors. Asia Pac J Clin Oncol. 2022;18:e427–34. [DOI] [PubMed] [Google Scholar]
- 435.Infante JR, Dees EC, Olszanski AJ, Dhuria SV, Sen S, Cameron S, Cohen RB. Phase I dose-escalation study of LCL161, an oral inhibitor of apoptosis proteins inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2014;32:3103–10. [DOI] [PubMed] [Google Scholar]
- 436.Crawford N, Stott KJ, Sessler T, McCann C, McDaid W, Lees A, Latimer C, Fox JP, Munck JM, Smyth T, et al. Clinical positioning of the IAP Antagonist Tolinapant (ASTX660) in colorectal cancer. Mol Cancer Ther. 2021;20:1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Tao Y, Sun XS, Pointreau Y, Le Tourneau C, Sire C, Kaminsky MC, Coutte A, Alfonsi M, Calderon B, Boisselier P, et al. Extended follow-up of a phase 2 trial of xevinapant plus chemoradiotherapy in high-risk locally advanced squamous cell carcinoma of the head and neck: a randomised clinical trial. Eur J Cancer. 2023;183:24–37. [DOI] [PubMed] [Google Scholar]
- 438.Hurwitz HI, Smith DC, Pitot HC, Brill JM, Chugh R, Rouits E, Rubin J, Strickler J, Vuagniaux G, Sorensen JM, Zanna C. Safety, pharmacokinetics, and pharmacodynamic properties of oral DEBIO1143 (AT-406) in patients with advanced cancer: results of a first-in-man study. Cancer Chemother Pharmacol. 2015;75:851–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Noonan AM, Bunch KP, Chen JQ, Herrmann MA, Lee JM, Kohn EC, O’Sullivan CC, Jordan E, Houston N, Takebe N, et al. Pharmacodynamic markers and clinical results from the phase 2 study of the SMAC mimetic birinapant in women with relapsed platinum-resistant or -refractory epithelial ovarian cancer. Cancer. 2016;122:588–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Rasco DW, Lakhani NJ, Tang YF, Wang HB, Ji J, Chen J, Liang ZY, Amaya A, Yang DJ, Zhai YF. Phase Ib study of a novel bivalent IAP antagonist APG-1387 in combination of pembrolizumab for patients with advanced solid tumors. J Clin Oncol. 2020;38:3508.
- 441.Schneekloth JS Jr, Fonseca FN, Koldobskiy M, Mandal A, Deshaies R, Sakamoto K, Crews CM. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc. 2004;126:3748–54. [DOI] [PubMed] [Google Scholar]
- 442.Nalawansha DA, Crews CM. PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem Biol. 2020;27:998–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Qin H, Zhang Y, Lou Y, Pan Z, Song F, Liu Y, Xu T, Zheng X, Hu X, Huang P. Overview of PROTACs targeting the estrogen receptor: achievements for biological and drug discovery. Curr Med Chem. 2022;29:3922–44. [DOI] [PubMed] [Google Scholar]
- 444.Hamilton EP, Schott AF, Nanda R, Lu HL, Keung CF, Gedrich R, Parameswaran J, Han HS, Hurvitz SA. ARV-471, an estrogen receptor (ER) PROTAC degrader, combined with palbociclib in advanced ER plus /human epidermal growth factor receptor 2-negative (HER2-) breast cancer: phase 1b cohort (part C) of a phase 1/2 study. J Clin Oncol. 2022;40:16. [Google Scholar]
- 445.Hamilton E, Vahdat L, Han HS, Ranciato J, Gedrich R, Keung CF, Chirnomas D, Hurvitz S. First-in-human safety and activity of ARV-471, a novel PROTAC (R) estrogen receptor degrader, in ER+/HER2-locally advanced or metastatic breast cancer. Cancer Res. 2022;82:PD13–8. [Google Scholar]
- 446.Flanagan J, Qian Y, Gough S, Andreoli M, Bookbinder M, Cadelina G, Bradley J, Rousseau E, Willard R, Pizzano J, et al. Abstract P5–04–18: ARV-471, an oral estrogen receptor PROTAC degrader for breast cancer. Cancer Res. 2019;79:P5-04-18-P05-04–18. [Google Scholar]
- 447.Flanagan JJ, Qian Y, Gough SM, Andreoli M, Bookbinder M, Cadelina G, Bradley J, Rousseau E, Willard R, Pizzano J, et al. ARV-471, an oral estrogen receptor PROTAC degrader for breast cancer. Cancer Res. 2019;79:P5-04-18-P05-04-18.
- 448.Schapira M, Calabrese MF, Bullock AN, Crews CM. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov. 2019;18:949–63. [DOI] [PubMed] [Google Scholar]
- 449.Rowinsky EK, Paner A, Berdeja JG, Paba-Prada C, Venugopal P, Porkka K, Gullbo J, Linder S, Loskog A, Richardson PG, Landgren O. Phase 1 study of the protein deubiquitinase inhibitor VLX1570 in patients with relapsed and/or refractory multiple myeloma. Invest New Drugs. 2020;38:1448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Becher OJ, Gilheeney SW, Khakoo Y, Lyden DC, Haque S, De Braganca KC, Kolesar JM, Huse JT, Modak S, Wexler LH, et al. A phase I study of perifosine with temsirolimus for recurrent pediatric solid tumors. Pediatr Blood Cancer. 2017;64:e26409. [DOI] [PubMed] [Google Scholar]
- 451.Friedman DR, Lanasa MC, Davis PH, Allgood SD, Matta KM, Brander DM, Chen Y, Davis ED, Volkheimer AD, Moore JO, et al. Perifosine treatment in chronic lymphocytic leukemia: results of a phase II clinical trial and in vitro studies. Leuk Lymphoma. 2014;55:1067–75. [DOI] [PubMed] [Google Scholar]
- 452.Leighl NB, Dent S, Clemons M, Vandenberg TA, Tozer R, Warr DG, Crump RM, Hedley D, Pond GR, Dancey JE, Moore MJ. A Phase 2 study of perifosine in advanced or metastatic breast cancer. Breast Cancer Res Treat. 2008;108:87–92. [DOI] [PubMed] [Google Scholar]
- 453.Bendell JC, Ervin TJ, Senzer NN, Richards DA, Firdaus I, Lockhart AC, Cohn AL, Saleh MN, Gardner LR, Sportelli P, Eng C. Results of the X-PECT study: a phase III randomized double-blind placebo-controlled study of perifosine plus capecitabine (P-CAP) versus placebo plus capecitabine (CAP) in patients (pts) with refractory metastatic colorectal cancer (mCRC). J Clin Oncol. 2012;30:LBA3501-LBA3501.
- 454.Chargari C, Leteur C, Angevin E, Bashir T, Schoentjes B, Arts J, Janicot M, Bourhis J, Deutsch E. Preclinical assessment of JNJ-26854165 (Serdemetan), a novel tryptamine compound with radiosensitizing activity in vitro and in tumor xenografts. Cancer Lett. 2011;312:209–18. [DOI] [PubMed] [Google Scholar]
- 455.Saleh MN, Patel MR, Bauer TM, Goel S, Falchook GS, Shapiro GI, Chung KY, Infante JR, Conry RM, Rabinowits G, et al. Phase 1 trial of ALRN-6924, a dual inhibitor of MDMX and MDM2, in patients with solid tumors and lymphomas bearing wild-type TP53. Clin Cancer Res. 2021;27:5236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Uy GL, Assouline S, Young AM, Blotner S, Higgins B, Chen LC, Yee K. Phase 1 study of the MDM2 antagonist RO6839921 in patients with acute myeloid leukemia. Invest New Drugs. 2020;38:1430–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Blotner S, Chen LC, Ferlini C, Zhi J. Phase 1 summary of plasma concentration-QTc analysis for idasanutlin, an MDM2 antagonist, in patients with advanced solid tumors and AML. Cancer Chemother Pharmacol. 2018;81:597–607. [DOI] [PubMed] [Google Scholar]
- 458.Dale B, Cheng M, Park KS, Kaniskan HU, Xiong Y, Jin J. Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer. 2021;21:638–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Jones RJ, Gu D, Bjorklund CC, Kuiatse I, Remaley AT, Bashir T, Vreys V, Orlowski RZ. The novel anticancer agent JNJ-26854165 induces cell death through inhibition of cholesterol transport and degradation of ABCA1. J Pharmacol Exp Ther. 2013;346:381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Ge CS, Liao BS, Zhang L. KPG-818, a novel cereblon modulator, inhibits hematological malignancies in preclinical models. Cancer Res. 2020;80:6367. [Google Scholar]
- 461.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, et al. Targeting the p27 E3 ligase SCFSkp2 results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Wu L, Grigoryan Arsen V, Li Y, Hao B, Pagano M, Cardozo Timothy J. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Chan C-H, Morrow John K, Li C-F, Gao Y, Jin G, Moten A, Stagg Loren J, Ladbury John E, Cai Z, Xu D, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Malek E, Abdel-Malek MAY, Jagannathan S, Vad N, Karns R, Jegga AG, Broyl A, van Duin M, Sonneveld P, Cottini F, et al. Pharmacogenomics and chemical library screens reveal a novel SCFSKP2 inhibitor that overcomes Bortezomib resistance in multiple myeloma. Leukemia. 2016;31:645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Mund T, Lewis MJ, Maslen S, Pelham HR. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci. 2014;111:16736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G, Bernassola F, Cavasotto CN, Knight RA, Ciechanover A, Melino G. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis. 2014;5:e1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Watt JE, Hughes GR, Walpole S, Monaco S, Stephenson GR, Bulman Page PC, Hemmings AM, Angulo J, Chantry A. Discovery of small molecule WWP2 ubiquitin ligase inhibitors. Chem Eur J. 2018;24:17677–80. [DOI] [PubMed] [Google Scholar]
- 468.Zafar A, Wang W, Liu G, Xian W, McKeon F, Zhou J, Zhang R. Targeting the p53-MDM2 pathway for neuroblastoma therapy: rays of hope. Cancer Lett. 2021;496:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Klein AM, de Queiroz RM, Venkatesh D, Prives C. The roles and regulation of MDM2 and MDMX: it is not just about p53. Genes Dev. 2021;35:575–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Aguilar A, Lu J, Liu L, Du D, Bernard D, McEachern D, Przybranowski S, Li X, Luo R, Wen B, et al. Discovery of 4-((3′R,4′S,5′R)-6″-Chloro-4′-(3-chloro-2-fluorophenyl)-1′-ethyl-2″-oxodispiro[cyclohexane-1,2′-pyrrolidine-3′,3″-indoline]-5′-carboxamido)bicyclo[2.2.2]octane-1-carboxylic Acid (AA-115/APG-115): a potent and orally active Murine Double Minute 2 (MDM2) inhibitor in clinical development. J Med Chem. 2017;60:2819–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Fang DD, Tang Q, Kong Y, Wang Q, Gu J, Fang X, Zou P, Rong T, Wang J, Yang D, Zhai Y. MDM2 inhibitor APG-115 synergizes with PD-1 blockade through enhancing antitumor immunity in the tumor microenvironment. J Immunother Cancer. 2019;7:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Fang DD, Tang Q, Kong Y, Rong T, Wang Q, Li N, Fang X, Gu J, Xiong D, Yin Y, et al. MDM2 inhibitor APG-115 exerts potent antitumor activity and synergizes with standard-of-care agents in preclinical acute myeloid leukemia models. Cell Death Discov. 2021;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Ye P, Chi X, Cha J-H, Luo S, Yang G, Yan X, Yang W-H. Potential of E3 ubiquitin ligases in cancer immunity: opportunities and challenges. Cells. 2021;10:3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Cossu F, Milani M, Mastrangelo E, Lecis D. Targeting the BIR domains of Inhibitor of Apoptosis (IAP) proteins in cancer treatment. Comput Struct Biotechnol J. 2019;17:142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Chesi M, Mirza NN, Garbitt VM, Sharik ME, Dueck AC, Asmann YW, Akhmetzyanova I, Kosiorek HE, Calcinotto A, Riggs DL, et al. IAP antagonists induce anti-tumor immunity in multiple myeloma. Nat Med. 2016;22:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. [DOI] [PubMed] [Google Scholar]
- 477.Fang Y, Fu D, Shen XZ. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim Biophys Acta. 2010;1806:1–6. [DOI] [PubMed] [Google Scholar]
- 478.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Liu J, Shaik S, Dai X, Wu Q, Zhou X, Wang Z, Wei W. Targeting the ubiquitin pathway for cancer treatment. Biochim Biophys Acta. 2015;1855:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Aleo E, Henderson CJ, Fontanini A, Solazzo B, Brancolini C. Identification of new compounds that trigger apoptosome-independent caspase activation and apoptosis. Cancer Res. 2006;66:9235–44. [DOI] [PubMed] [Google Scholar]
- 481.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, et al. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KG, Konietzny R, Fischer R, Kogan E, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–12. [DOI] [PubMed] [Google Scholar]
- 483.Lee JK, Chang N, Yoon Y, Yang H, Cho H, Kim E, Shin Y, Kang W, Oh YT, Mun GI, et al. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro Oncol. 2016;18:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Kapuria V, Peterson LF, Fang D, Bornmann WG, Talpaz M, Donato NJ. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–76. [DOI] [PubMed] [Google Scholar]
- 485.Fulda S, Scaffidi C, Susin SA, Krammer PH, Kroemer G, Peter ME, Debatin KM. Activation of mitochondria and release of mitochondrial apoptogenic factors by betulinic acid. J Biol Chem. 1998;273:33942–8. [DOI] [PubMed] [Google Scholar]
- 486.Reiner T, Parrondo R, de Las PA, Palenzuela D, Perez-Stable C. Betulinic acid selectively increases protein degradation and enhances prostate cancer-specific apoptosis: possible role for inhibition of deubiquitinase activity. PLoS One. 2013;8:e56234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. [DOI] [PubMed] [Google Scholar]
- 488.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, McDermott JL, Leach CA, Fulcinniti M, Kodrasov MP, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Pozhidaeva A, Valles G, Wang F, Wu J, Sterner DE, Nguyen P, Weinstock J, Kumar KGS, Kanyo J, Wright D, Bezsonova I. USP7-specific inhibitors target and modify the Enzyme’s active site via distinct chemical mechanisms. Cell Chem Biol. 2017;24:1501-1512.e1505. [DOI] [PubMed] [Google Scholar]
- 490.Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, Tonkin LM, Townsend EC, Buker SM, Lancia DR, et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Lamberto I, Liu X, Seo HS, Schauer NJ, Iacob RE, Hu W, Das D, Mikhailova T, Weisberg EL, Engen JR, et al. Structure-guided development of a potent and selective non-covalent active-site inhibitor of USP7. Cell Chem Biol. 2017;24:1490-1500.e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, Colland F. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19:467–77. [DOI] [PubMed] [Google Scholar]
- 493.Shinji S, Naito Z, Ishiwata S, Ishiwata T, Tanaka N, Furukawa K, Suzuki H, Seya T, Matsuda A, Katsuta M, Tajiri T. Ubiquitin-specific protease 14 expression in colorectal cancer is associated with liver and lymph node metastases. Oncol Rep. 2006;15:539–43. [PubMed] [Google Scholar]
- 494.Wu N, Liu C, Bai C, Han YP, Cho WC, Li Q. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of β-catenin. Int J Mol Sci. 2013;14:10749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Zhu Y, Zhang C, Gu C, Li Q, Wu N. Function of deubiquitinating enzyme USP14 as oncogene in different types of cancer. Cell Physiol Biochem. 2016;38:993–1002. [DOI] [PubMed] [Google Scholar]
- 496.Zhang B, Li M, Huang P, Guan XY, Zhu YH. Overexpression of ubiquitin specific peptidase 14 predicts unfavorable prognosis in esophageal squamous cell carcinoma. Thorac Cancer. 2017;8:344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Liu Y, Lashuel HA, Choi S, Xing X, Case A, Ni J, Yeh LA, Cuny GD, Stein RL, Lansbury PT Jr. Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line. Chem Biol. 2003;10:837–46. [DOI] [PubMed] [Google Scholar]
- 499.Berndtsson M, Beaujouin M, Rickardson L, Havelka AM, Larsson R, Westman J, Liaudet-Coopman E, Linder S. Induction of the lysosomal apoptosis pathway by inhibitors of the ubiquitin-proteasome system. Int J Cancer. 2009;124:1463–9. [DOI] [PubMed] [Google Scholar]
- 500.Tian Z, D’Arcy P, Wang X, Ray A, Tai YT, Hu Y, Carrasco RD, Richardson P, Linder S, Chauhan D, Anderson KC. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood. 2014;123:706–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.D’Arcy P, Brnjic S, Olofsson MH, Fryknäs M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R, Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636–40. [DOI] [PubMed] [Google Scholar]
- 502.Zhang X, Pellegrini P, Saei AA, Hillert EK, Mazurkiewicz M, Olofsson MH, Zubarev RA, D’Arcy P, Linder S. The deubiquitinase inhibitor b-AP15 induces strong proteotoxic stress and mitochondrial damage. Biochem Pharmacol. 2018;156:291–301. [DOI] [PubMed] [Google Scholar]
- 503.Sha B, Chen X, Wu H, Li M, Shi J, Wang L, Liu X, Chen P, Hu T, Li P. Deubiquitylatinase inhibitor b-AP15 induces c-Myc-Noxa-mediated apoptosis in esophageal squamous cell carcinoma. Apoptosis. 2019;24:826–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.