Abstract
Many adhesion proteins, evolutionarily related through gene duplication, exhibit distinct and precise interaction preferences and affinities crucial for cell patterning. Yet, the evolutionary paths by which these proteins acquire new specificities and prevent cross-interactions within their family members remain unknown. To bridge this gap, this study focuses on Drosophila Down syndrome cell adhesion molecule-1 (Dscam1) proteins, which are cell adhesion proteins that have undergone extensive gene duplication. Dscam1 evolved under strong selective pressure to achieve strict homophilic recognition, essential for neuronal self-avoidance and patterning. Through a combination of phylogenetic analyses, ancestral sequence reconstruction, and cell aggregation assays, we studied the evolutionary trajectory of Dscam1 exon 4 across various insect lineages. We demonstrated that recent Dscam1 duplications in the mosquito lineage bind with strict homophilic specificities without any cross-interactions. We found that ancestral and intermediate Dscam1 isoforms maintained their homophilic binding capabilities, with some intermediate isoforms also engaging in promiscuous interactions with other paralogs. Our results highlight the robust selective pressure for homophilic specificity integral to the Dscam1 function within the process of neuronal self-avoidance. Importantly, our study suggests that the path to achieving such selective specificity does not introduce disruptive mutations that prevent self-binding but includes evolutionary intermediates that demonstrate promiscuous heterophilic interactions. Overall, these results offer insights into evolutionary strategies that underlie adhesion protein interaction specificities.
Keywords: cell-cell recognition; adhesion proteins; gene duplication; protein–protein interactions; protein evolution; down syndrome cell adhesion molecule, DSCAM
Introduction
Adhesion proteins play a central role in cellular organization and communication within multicellular organisms (Dalva et al. 2007; Makrilia et al. 2009; Lele and Hindges 2023). These are often members of large protein families that have expanded through gene duplication and subsequent evolutionary divergence (i.e. paralog proteins). Due to their ancestral link, paralog proteins generally have similar sequences and nearly identical structures with a tendency for intrafamilial interactions (Ispolatov et al. 2005; Lukatsky et al. 2007; Pereira-Leal et al. 2007). Yet, adhesion proteins within the same family often display distinct binding specificities and affinities. These differences are central to their roles in cell patterning and organization. Some such roles include neural tube formation mediated by N- and E-cadherins (Taneyhill and Schiffmacher 2017), as well as the function of nectins within the inner ear cell patterning (Togashi et al. 2011), and clustered protocadherin and Dscam1 proteins in dendritic arborization (Zipursky and Sanes 2010; Honig and Shapiro 2020).
New protein–protein interaction specificities can evolve through mutations that impose negative constraints and prevent cross-interactions among family members (Zarrinpar et al. 2003; Reinke et al. 2013; Peleg et al. 2014; Cheng et al. 2019; Honig and Shapiro 2020; Sergeeva et al. 2020). There are two contrasting models that outline the evolutionary pathways proteins undergo from being identical duplicated copies to becoming paralogs with distinct binding specificities. The first model states that over extensive periods of time, proteins lose their functionality due to mutations that cause incompatible interacting interfaces until additional mutations lead to new interacting interfaces and binding specificities (Ohno 1970; Siddiq et al. 2017; McClune and Laub 2020). The second model suggests a continuous evolutionary shift, with intermediate proteins exhibiting promiscuous, nonspecific interactions (Sayou et al. 2014; Aakre et al. 2015). While the evolution of specific protein–protein interactions has been extensively studied within the context of enzyme–substrate specificities (Aharoni et al. 2005; Weinreich et al. 2006; Khersonsky and Tawfik 2010) and receptor–ligand systems (Chockalingam et al. 2005; Ortlund et al. 2007; Eick et al. 2012; Koehbach et al. 2013), it remains underexplored from the perspective of cell adhesion proteins.
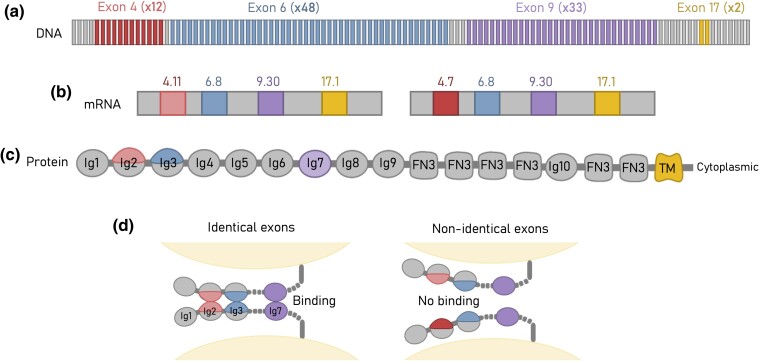
The Drosophila Dscam1 serves as an extraordinary example of a large paralogous protein family with highly precise cell surface adhesion interactions. The Dscam1 gene consists of 24 exons, 3 of which, exons 4, 6, and 9, have undergone extensive duplications (Fig. 1a). In alternative splicing, a single exon from each of the three clusters is stochastically selected, with the potential to encode an astounding array of 19,008 unique extracellular regions (Fig. 1b). Each extracellular region is characterized by distinct combinations of three alternative immunoglobulin (Ig) domains—Ig2, Ig3, and Ig7 encoded by exons 4, 6, and 9, respectively (Schmucker et al. 2000). The first four Ig domains (Ig1 to Ig4) form a horseshoe-like structure that positions the alternate Ig2 and Ig3 domains of two membrane-apposing Dscam1 proteins for homophilic interactions (Meijers et al. 2007; Sawaya et al. 2008). Dscam1 isoforms are also unique because of their strict homophilic binding specificity. Homodimerization occurs in a symmetric antiparallel fashion only when all three alternative Ig domains match up with each other (i.e. Ig2 binds to Ig2, Ig3 to Ig3, and Ig7 to Ig7; Fig. 1c; Wojtowicz et al. 2004, 2007; Sawaya et al. 2008). This is in contrast to many cell adhesion protein families that typically exhibit both homophilic and heterophilic binding between members of the same family (Katsamba et al. 2009; Vendome et al. 2014; Mosca 2015; Zinn and Özkan 2017; Brasch et al. 2018; Honig and Shapiro 2020; Sergeeva et al. 2020). The strict homophilic binding exhibited by Dscam1 enormous isoform repertoire is key for its ability to differentiate self from nonself cell–cell interactions. This ability is required for neural patterning within the developing Drosophila nervous system (Wang et al. 2002; Zhan et al. 2004; Zhu et al. 2006; Hughes et al. 2007; Matthews et al. 2007; Soba et al. 2007; Wojtowicz et al. 2007; Wu et al. 2012; Miura et al. 2013; Wilhelm et al. 2022).
Fig. 1.
Dscam1 gene has the capacity to transcribe tens of thousands of strictly homophilic isoforms. a) In insects, exons 4, 6, and 9 of the Dscam1 gene have undergone extensive tandem duplications, contributing to the vast diversity of isoforms produced by the gene. b) Through stochastic alternative splicing, a single exon from each cluster is retained in the mature transcript. In Drosophila, 38,016 unique isoforms can be expressed, with 19,008 unique extracellular domains. Each neuron expresses a different set of Dscam1 isoforms. c) Exons 4, 6, and 9 encode partial (exons 4 and 6) or an entire (exon 9) Ig domain. These domains determine the binding specificity of the Dscam1 protein. d) For a Dscam1 dimer to form between two cell membranes, all domains must fully match.
Earlier investigations of Dscam1 evolution traced the extensive exon expansion to the last common ancestor (LCA) of the Pancrustacea (Lee et al. 2010; Armitage et al. 2012), which diverged into insects and crustaceans approximately 500 million years ago (Misof et al. 2014). These studies used a sample of representative genomes and identified a significant variability in the count of exon duplications among different species, indicating duplication events also occurred in subsequent evolutionary lineages (Lee et al. 2010; Armitage et al. 2012). However, these studies did not experimentally test whether newly duplicated Dscam1 isoforms, aside from Drosophila, bind strictly homophilically. The functionality of Dscam1 ancestral proteins also remains unknown, resulting in a knowledge gap in the evolution of adhesion specificity for this unique protein family.
Here, we extensively investigate the evolutionary expansion of Dscam1 exon 4 in insects. We predict the mutational pathways that connect ancestral to recent Dscam1 duplications. Our analysis reveals a clear pattern: as Dscam1 paralogous exons diverge from identical duplicates, some initially exhibit heterophilic interactions, which diminish with additional mutations, ultimately resulting in exclusive homophilic binding. We did not observe a “nonfunctional” ancestral isoform lacking homophilic binding. By demonstrating this evolutionary progression, we shed light on the key process of increasing the number of highly specific adhesion molecules, deepening our understanding of this critical aspect of molecular evolution.
Results
Phylogenetic Analysis of Dscam1 Exon 4 in Insects
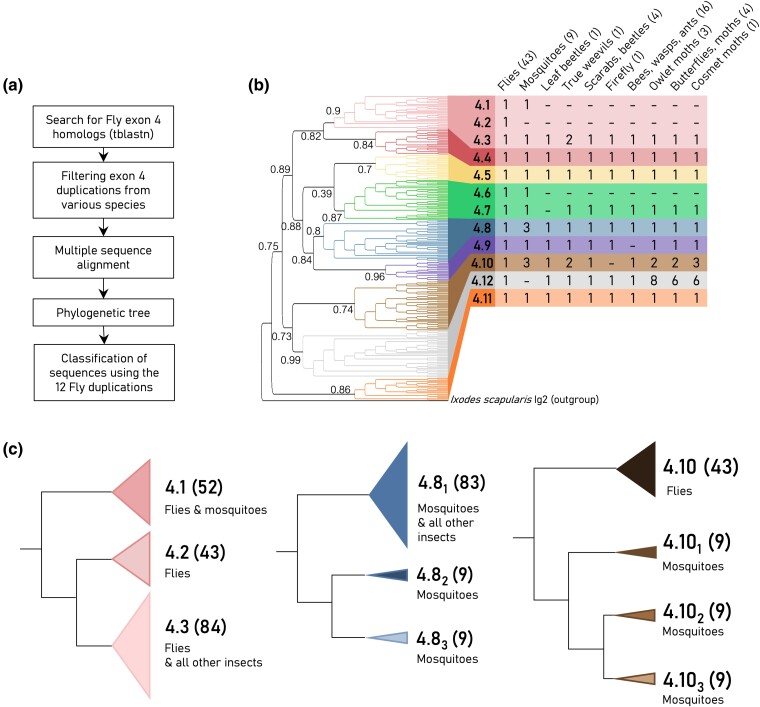
In this study, we investigated the evolutionary trajectory of the Dscam1 gene, focusing specifically on exon 4, which encodes a segment of the Ig2 domain that is involved in homophilic dimerization. A tblastn search was performed against the RefSeq genome database (Johnson et al. 2008; NCBI Resource Coordinators 2016; O’Leary et al. 2016) for sequences with homology to Drosophila melanogaster exon 4.7 (Fig. 2a). This comprehensive search covered 83 insect species, resulting in the identification of 962 homologous sequences. A nonredundant set of these sequences, including a Dscam2 sequence from Chelicerata Ixodes scapularis as an outgroup, was aligned and used to construct a phylogenetic tree (for additional details, see Materials and Methods). The resulting tree revealed nine distinct clusters of exons, each representing orthologous sequences from various species (Fig. 2b). These orthologous sequences maintain high levels of sequence conservation, particularly in the Ig2 dimer interface, with an average similarity of over 90% within each ortholog cluster. Notably, we found that all clusters contain representative sequences from most species, indicating paralogous relationships between clusters (Fig. 2b). For example, the beetle exon 4 sequences can be found in all nine clusters. While previous work suggests that the LCA of insects possessed nine exon 4 paralogs (Lee et al. 2010; Armitage et al. 2012), our current findings provide strong support for this notion, mainly due to the increased availability of genomic data.
Fig. 2.
Phylogenetic analysis of insect exon 4. a) Illustration of the workflow for phylogenetic analysis. b) The phylogenetic tree is constructed from 266 exon 4 sequences from 83 representative species (left). The tree topology preserves the organization of the major ortholog clusters, which are highlighted by different colors and are notated according to the Drosophila exon 4 nomenclature. The tree was generated using FastTree with FastTree local support values shown. The table summarizes the number of duplications (paralogs) per species group for each cluster (right). The number of species per group is denoted within brackets. c) Simplified phylogenetic subtrees of three recent duplications (4.1 to 4.3, 4.81-3, and 4.101-3, colors corresponding to the main tree) for both flies and mosquitoes. The number of sequences per cluster is denoted within the brackets. This number includes the redundant sequences that were not used in reconstructing the main tree.
Next, using the 12 exon 4 paralogs of D. melanogaster (4.1 to 4.12) as a reference, we focused on more recent exon 4 duplication events occurring in specific insect lineages. These analyses uncovered the absence of Drosophila exons 4.1, 4.2, and 4.6 in most insect species. These exons are unique to the Diptera lineage and encompass flies and mosquitoes, which diverged from other insects approximately 260 million years ago (Wiegmann et al. 2011). Sequence homology and branch support values strongly indicate recent duplications and divergence of exons 4.1 and 4.2 from exon 4.3 (Fig. 2c), as well as the divergence of exon 4.6 from exon 4.7 (Fig. 2b). We also discovered more recent lineage-specific duplications, including duplications of exons 4.8 and 4.10 found in mosquitoes (Figs. 2c and 3) and duplications of exons 4.10 and 4.12 in the Lepidoptera (i.e. moths and butterflies; Fig. 2b).
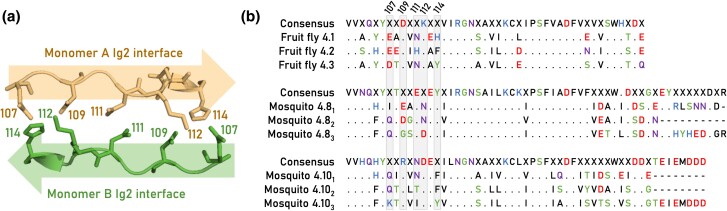
Fig. 3.
Three recent duplications for both mosquitos and flies. a) Structure of the Ig2 homodimer interface (PDB 3DMK) encoded by Drosophila exon 4.1. The interface spans amino acid positions 107 to 114 and aligns in an antiparallel fashion. b) Multiple sequence alignments of the fruit fly D. melanogaster exons 4.1 to 4.3 and the yellow fever mosquito A. aegypti exons 4.81 to 3 and 4.101 to 3. A period (i.e. “.”) in the multiple sequence alignments indicates invariant positions, and only residues that deviate from the consensus sequence are shown. The Ig2:Ig2 interface residues are highlighted in gray, and the sequence position is indicated at the top of the alignment.
Recent Duplications in Mosquito Dscam1 Exhibit Strict Homophilic Binding
To date, the highly specific homophilic dimerization of the Dscam1 protein has been observed exclusively between D. melanogaster isoforms. This study aimed to investigate whether the binding specificities of recent exon 4 duplications, not present in Drosophila, would also maintain homophilic specificity. We focused on mosquito exons 4.8 and 4.10, along with their respective duplications, referred to here as 4.81, 4.82, 4.83, 4.101, 4.102, and 4.103. To assess whether the newly duplicated isoforms evolved new homophilic binding specificities, we used site-directed mutagenesis to swap the interface residues of Drosophila Dscam1 to match those found in the mosquito isoforms (Fig. 3). We implemented this strategy based on a previous study showing that isoform specificity could be altered by the substitution of residues in positions 107 to 114 on the Ig2 domain dimer interface (Wojtowicz et al. 2007).
We assessed the binding preferences of the mosquito interfaces via cell aggregation assays using HEK293F-suspended cells. Cell aggregation is a well-established method used to determine the binding specificity of adhesion proteins (Matthews et al. 2007; Schreiner and Weiner 2010; Boucard et al. 2014; Thu et al. 2014; Rubinstein et al. 2015; Bisogni et al. 2018; Zhou et al. 2020; Hou et al. 2022; Cheng et al. 2023; Wiseglass et al. 2024). Each protein is tagged with either red or green fluorescent markers and transfected into separate cell populations. The two cell populations are then mixed and allowed to aggregate based on the binding specificities of the adhesion proteins they express. If the two proteins are strictly homophilic, the cells will form separate red or green aggregates. In contrast, if the two proteins are heterophilic, the cells will form mixed red and green aggregates. To quantify the extent of aggregate mixing or separation, we employed a customized Python script (Wiseglass et al. 2024), calculating the ratio of proximate red and green cells (see Materials and Methods). The resulting ratio is displayed in the corner of each image, with a value exceeding 0.1 indicating visibly mixed aggregates (Fig. 4).
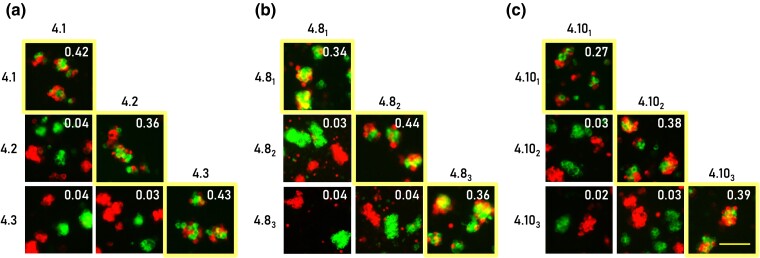
Fig. 4.
Recent duplications in mosquito Dscam1 engage in highly specific homophilic interactions. Pairwise combinations within each exon paralog cluster were assessed for their interaction specificity. HEK293F cells expressing identical isoforms formed mixed red and green aggregates (as marked by both a yellow boundary and an aggregation score exceeding 0.1), while cells expressing different isoforms formed separate red and green aggregates. a) Binding assay for the fruit fly D. melanogaster exons 4.1 to 4.3. b and c) Binding assays for the yellow fever mosquito A. aegypti exons 4.81 to 3 and 4.101 to 3, respectively. The aggregate mixing score is presented in the right corner of each image. Scale 100 µm.
Cell aggregation assay was performed in pairwise combinations of the yellow fever mosquito Aedes aegypti isoforms 4.81 to 4.83, 4.101 to 4.103, and the D. melanogaster isoforms 4.1 to 3. We observed that only cells expressing identical isoforms formed mixed aggregates, while all combinations of nonidentical isoform pairs resulted in separate aggregates (Fig. 4). These results demonstrate a strict homophilic binding preference for each isoform. Importantly, our findings indicate that the recent mosquito exons evolved to encode adhesion receptors with highly specific homophilic cell recognition. This suggests mosquito Dscam1 has a similar function to Drosophila Dscam1 in mediating the distinction between self and nonself in neurons.
Tracing Evolutionary Paths of Binary Specificities in Dscam1
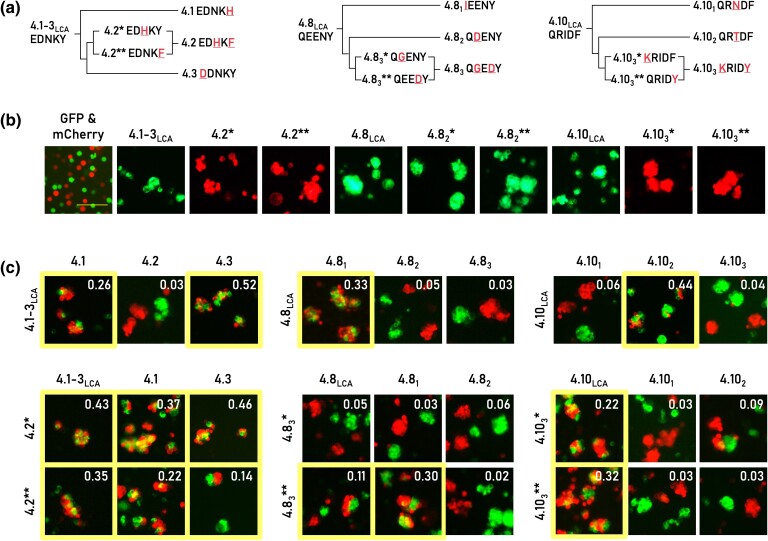
The evolution of Dscam1 provides a unique opportunity to explore the challenges associated with diversifying homophilic interfaces. Following exon duplication, alterations in the dimer interface can ultimately lead to the establishment of a new homophilic specificity. However, during this evolutionary process, intermediate changes may result in a nonspecific binding or potential loss of binding altogether. To gain deeper insights into the evolutionary trajectory of Dscam1 and to predict intermediate isoforms, we performed an ancestral sequence reconstruction (ASR). Utilizing two ASR programs, PaML (Yang 2007; Xu and Yang 2013) and GRASP (Ross et al. 2022), we predicted the LCA proteins prior to the recent mosquito duplication of exons 4.8 and 4.10 and the duplication in Drosophila exon 4.3, referred to here as 4.8LCA, 4.10LCA, and 4.1-3LCA. The reconstruction of the three ancestor proteins achieved high confidence of 0.85, 0.91, and 0.84 (for 4.1-3LCA, 4.8LCA, and 4.10LCA, respectively) with predicted ancestral interface residues similar, but not identical, to current sequences (Fig. 5a; supplementary file S4, Supplementary Material online).
Fig. 5.
Tracing evolutionary paths of binary specificities of Dscam1. a) Predicted mutational pathways from ancestor to contemporary extant isoforms. The Ig2:Ig2 interface residues corresponding to sequence positions 107, 109, 111, 112, and 114 are shown for each isoform. Exon duplications are indicated with diverging lines. Mutations are highlighted in red and is also underscored. b) Resurrected ancestral (“LCA”) and intermediate (“*”/”**”) proteins mediate cell aggregation, while the control cells expressing GFP and mCherry do not mediate aggregation. c) Binding preferences of resurrected and contemporary isoforms demonstrate heterophilic interactions in many cases (mixing score >0.1, highlighted by the yellow boundary). The aggregate mixing score is presented in the right corner of each image. Scale 100 µm.
Resurrected Ancestral Proteins Bind Homophilically
We resurrected these ancestral interfaces by mutagenesis of extant interface residues and examined their ability to self-bind using the cell aggregation assay. We found that all three ancestors effectively mediated cell aggregation, demonstrating their ability to function as homophilic adhesion receptors (Fig. 5b). Next, we compared the binding preferences of these ancestor sequences with their extant descendants. We observed that cells expressing the ancestral 4.8LCA formed mixed aggregates exclusively with cells expressing the extant 4.81 isoform, but not with 4.82 and 4.83. Similarly, 4.10LCA was observed to interact solely with one of its current descendants, 4.101, and not with the remaining two isoforms (Fig. 5c). These results implicate that postduplication, the ancestral 4.8 and 4.10 exons evolved into their respective current isoforms (4.81 and 4.101, respectively), while the other duplicates diverged and developed new binding specificities. Interestingly, cells expressing the 4.1-3LCA ancestor recognized cells expressing either 4.3 or 4.1 extant isoforms, but not 4.2 expressing cells (Fig. 5c, left). These results indicate that these extant proteins diverged via a subfunctionalization mechanism (McClune and Laub 2020), by which the ancestor protein binds to a wider range of partners (in this case, two distinct isoforms) compared to its descendants (which here bind strictly homophilically).
Next, we examined whether divergence of the Ig2 domain interface occurred through intermediate proteins that maintain homophilic cell adhesion or whether mutations in intermediate proteins disrupt self-binding. We observed that in all three studied examples, the ancestor sequence differs from two extant exons by a single residue and by two residues from the third extant exon. For example, the 4.1-3LCA interface is composed of the following five residues: “EDNKY.” A single substitution from tyrosine at position 114 to histidine is sufficient to reach the interface of exon 4.1 (EDNKH). Similarly, a single substitution of the 4.1-3LCA interface at position 107 from glutamate to aspartate would generate an exon 4.3 interface (DDNKY; Fig. 5a). Editing the 4.1-3LCA interface to the interface of exon 4.2 (EDHKF) requires at least two mutations, N111H and Y114F, with two possible intermediate interfaces, EDHKY and EDNKF, depending on the mutation order. Using similar logic, we identified interface intermediates from the 4.10LCA and 4.8LCA to 4.103 and 4.83, respectively (Fig. 5a).
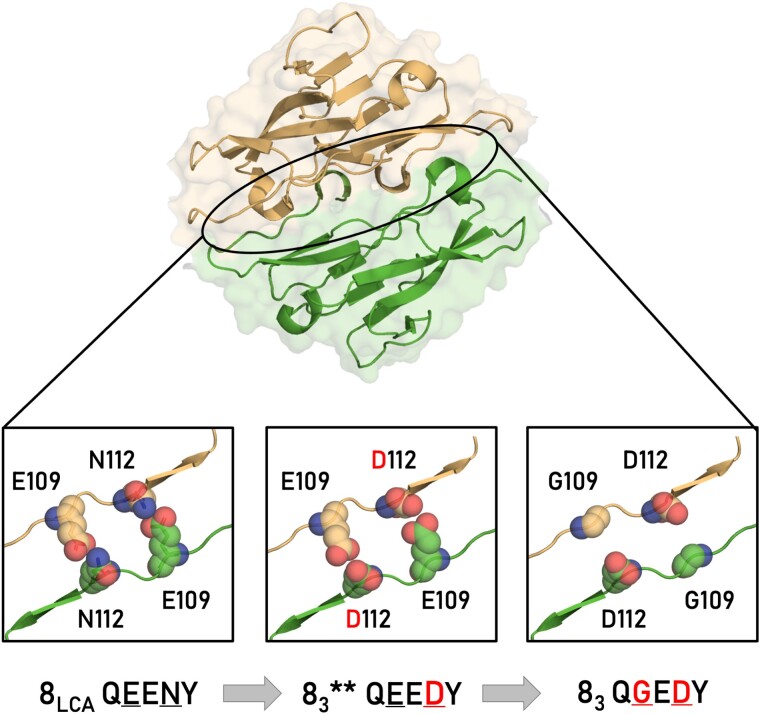
We then examined the self-aggregation abilities of cells expressing each intermediate isoform with the aim of testing the self-binding capabilities of intermediate states. Surprisingly, we observed that all intermediate proteins engaged in homophilic interactions despite having interfaces that appeared incompatible for such interactions (Fig. 5b). For example, within the homodimer interface of one 4.8 intermediate isoform, two negatively charged aspartate residues are positioned in proximity upon dimerization, where they could potentially lead to electrostatic repulsion (Fig. 6). These types of incompatibilities are thought to be central in preventing unwanted cross-talk between different Dscam1 isoforms (Fig. 4; Sawaya et al. 2008). To assess changes in the binding affinity (ΔΔGbind) of this intermediate, we used two computational methods, FoldX and SSIPe (Schymkowitz et al. 2005; Huang et al. 2020). Both methods predicted that the N112D mutation would significantly destabilize homophilic interactions, with ΔΔGbind exceeding 2 kcal/mol. These predictions indicate that negative constraints weaken the self-interaction of the intermediate isoforms. Yet, our cell aggregation results show these constraints do not entirely disrupt adhesive functionality.
Fig. 6.
Dscam1 exon 83 mutational path. The top image presents the structural model of the Ig2:Ig2 dimer complex with Ig2 encoded by exon 4.8 ancestor and highlighted by a black oval. At the bottom, three close-up views of the homophilic Ig2:Ig2 interface for 4.8 LCA (left), 4.8 intermediate isoform (middle), and 4.83 current isoform (right). The structural models' backbones are shown as cartoon, and mutated residues are shown with Van der Waals spheres and colored based on chain origin and atom type. Ig2:Ig2 interface residues are noted at the bottom.
Resurrected Ancestral Proteins Bind Heterophilically with Extant Isoforms
We then tested whether nonspecific binding could occur between ancestral, intermediate, and extant isoforms. We found that both intermediate proteins that may lead to exon 4.2 formed mixed aggregates with extant isoforms 4.1, 4.3, and their ancestor, demonstrating heterophilic binding specificities (Fig. 5c, left). Thus, both mutations leading to exon 4.2 are necessary to prevent cross-interactions with closely related paralogs, generating strict homophilic binding. One of the intermediate proteins leading to isoform 4.83 recognized both the ancestor and one paralog extant sequence (4.81). Finally, both intermediate proteins lead to 4.103 promiscuously bound to the ancestor, with one intermediate binding weakly to one of the extant paralogs (4.102; Fig. 5c, right). Overall, of the six intermediate proteins we tested, five exhibit promiscuous cross-interactions, demonstrating a gradual transition in specificity. Our experimental observations also explain the continued evolution of intermediate Dscam1 isoforms reconstructed here, as they generally exhibit nonspecific cross-interactions with other isoforms.
To address uncertainties in the ancestral protein reconstruction process, we focused on three cases where the reconstruction's posterior probability for a particular interface residue was below 0.85. In these instances, we generated an alternative ancestor by incorporating the second most likely residue. These alternative ancestors were then tested for their binding preferences. All alternative ancestors displayed homophilic binding, as evidenced by their ability to mediate cell aggregation (supplementary fig. S2, Supplementary Material online). One of the alternative ancestors (the LCA of 4.82 and 4.83) has the same dimer interface as extant isoform 4.82 and binds only to 4.82, thus differing from the binding preferences of the primary ancestor, which has a 4.81-like interface (supplementary fig. S2, Supplementary Material online, left). Nevertheless, the transition of this alternative ancestor to extant isoform 4.83 exhibits the same promiscuous binding preferences as the primary ancestor (supplementary fig. S2, Supplementary Material online, middle). The two additional alternative ancestors exhibited the same promiscuous binding as the primary ancestors (supplementary fig. S2, Supplementary Material online). These findings show that the alternative ancestors produced similar binding preferences to the primary ancestors, thereby further supporting our conclusions.
Discussion
This study examined the evolution of Dscam1 isoform binding specificities, with a focus on the exon 4 cluster. This exon encodes the second Ig domain (Ig2), one of three domains involved in Dscam1 homophilic binding. We utilized a phylogenetic analysis, ASR, and cell aggregation experiments and revealed several key insights: we identified relatively recent Dscam1 duplications that evolve strict homophilic binding in mosquitos. We also observed that Dscam1 proteins have maintained their fundamental functionality throughout their evolutionary trajectory, as both ancestral and evolutionary intermediate proteins mediate homophilic cell recognition. Finally, we discovered that in contrast to extant Dscam1 proteins that interact only homophilically, ancestral and intermediate proteins exhibit promiscuous interactions and are able to engage in both homophilic and heterophilic binding.
Conservation of Dscam1 Exon 4 Cluster
With the goal of tracking evolutionary trajectories of the Ig2 dimer interface, our initial step involved a comprehensive search for current exon 4 sequences. Similar to past findings, we observed that exon 4 duplications are relatively conserved (Graveley et al. 2004; Lee et al. 2010; Armitage et al. 2012), comprising nine invariant exons (Lee et al. 2010). In addition to the nine conserved exons, we identified other exon 4 duplications across various insect lineages (Fig. 2). These duplications expand the Dscam1 isoform repertoire by diversifying the Ig2 dimer interface sequences, albeit to a lesser extent than the significant expansions observed in exons 6 and 9. In all these instances, a single ortholog can consistently be identified through the conserved Ig2 dimer interface, while the other duplicated exons undergo nonsynonymous substitutions, consistent with Ohno’s (1970) evolutionary model.
The conservation of most exon 4 variants could possibly be attributed to an unknown functional role these exon variants might have. Alternatively, it is possible that this conservation could be attributed to inherent constraints imposed by the small Ig2 dimer interface, comprising of only five residues.
Strict Homophilic Binding in Dscam1 Isoforms
To our knowledge, studies into the binding preferences of insect Dscam1 had previously been confined to D. melanogaster isoforms. However, both the current and previous studies identified duplication events outside of the Drosophila lineage for which there yet to have been experimental investigations of their binding preferences (Graveley et al. 2004; Lee et al. 2010; Armitage et al. 2012). Our findings reveal that even relatively recent duplications, occurring subsequent to the divergence of mosquitoes from flies, have evolved into isoforms exhibiting strict homophilic binding preferences (Fig. 4b and c). Overall, these findings underscore the remarkable ability of Dscam1 proteins to generate self-binding domains via exon duplication and sequence divergence. These results also highlight the evolutionary pressure to generate isoforms that can accurately differentiate self from nonself-interactions, which is central in neuronal patterning.
A previous study using enzyme-linked immunosorbent assay (ELISA) showed that a small subset of exons, including exons 4.1 and 4.3, engage in both homophilic and significantly lower-affinity promiscuous heterophilic interactions (Wojtowicz et al. 2007), in contrast to our findings. Interestingly, the authors themselves have observed such discrepancies in results between ELISA and cell aggregation assays. This difference in findings likely stems from the fundamental methodological differences between the assays. ELISA assays are more quantitative and sensitive to slight variations in protein binding affinities. On the other hand, cell aggregation assays offer an advantage by facilitating binding interactions within the context of native cellular membranes, potentially providing a more physiologically relevant measure of adhesion specificity.
Ancient Dscam1 Proteins Bind Promiscuously
Understanding the emergence of new binding specificities is a central question in biochemistry and evolution. Our study investigates the evolutionary trajectories of insect Dscam1 exon 4 by focusing on recent duplications in flies and mosquitoes to reconstruct ancestral isoforms. When reconstructing the evolutionary paths from ancestral proteins to current isoforms, we found that as little as one or two residue changes were sufficient to alter the ancestral binding specificity. We reconstructed intermediate isoforms by examining the shortest mutational paths and specifically testing the impact of the two mutations that altered binding specificity. While the reconstruction of intermediate isoforms did not rely on statistical data, our conclusion that these intermediate isoforms retain self-binding capability is likely to be robust to this uncertainty. This is because both identified mutations are likely to be responsible for insolating the isoform from cross heterophilic interactions with its closely related paralogs. By introducing one mutation at a time, we test the maximal impact of these alterations on interface stability without allowing for additional bridging mutations. Therefore, although the actual evolutionary trajectory may have been more complex, our analysis of the most extreme mutations that could impact binding specificity on this path strongly supports the validity of our conclusion.
We discovered that across various evolutionary trajectories, Dscam1 isoforms consistently maintained their self-binding capabilities. In addition to self-binding, several evolutionary intermediate isoforms demonstrated promiscuous heterophilic interactions. These results were particularly surprising considering the necessity for Dscam1 paralogs to achieve highly precise, strict homophilic interaction specificities for their role in neuronal self-avoidance. Previous studies have shown that Dscam1 isoforms evolve interfaces compatible with only self-interactions, while interfaces generated by heterophilic interactions contain noncomplementary electrostatic charges and shapes, thereby preventing heterophilic interactions (Sawaya et al. 2008).
Given these insights, one might expect intermediate isoforms to possess a noncomplementary interface that would hinder homophilic binding rather than the interface that allows for cross-interactions (Fig. 6, middle). While an isoform with abolished homophilic binding would not mediate neuronal self-recognition and patterning, the potential fitness penalty for a dysfunctional isoform may be mitigated by the presence of up to 50 randomly expressed isoforms in each neuron (Neves et al. 2004; Zhan et al. 2004). Our study challenges these expectations by demonstrating that, in fact, intermediate Dscam1 isoforms retain self-binding through their evolutionary development.
A possible explanation for this unexpected observation lies in the high degree of conservation of the exon 4 cluster, suggesting a specialized functional role, potentially imposing an additional evolutionary constraint. It would therefore be interesting to study the evolution of the exon 6 or exon 9 clusters, which have been shown to be nonconserved and therefore could potentially exhibit functional disruptive mutations.
In summary, our study into the evolutionary history of Dscam1 exon 4 across various insect lineages provides insights into the mechanisms by which cell adhesion proteins evolve distinct binding specificities crucial for the regulation of complex cellular processes. Mutations leading to promiscuous binding have previously been documented in other protein systems evolved under selection against cross-talk, including the toxin–antitoxin systems, hormone receptors, and enzymes (Voordeckers et al. 2012; Aakre et al. 2015; Devamani et al. 2016; Siddiq et al. 2017; Lite et al. 2020; Ghose et al. 2023). Our findings suggest parallel phenomenon in adhesion proteins, where promiscuous intermediates constitute a likely evolutionary step toward achieving highly precise binding specificity.
Materials and Methods
Dscam1 Exon 4 Tree Construction
The translated sequence encoded by exon 4 of D. melanogaster Dscam1 transcript variant BE (isoform 7.9.30, NCBI annotation NM001043041) was used as the query for identifying exon 4 duplications in other insect genomes. The tblastn algorithm was used with the default parameters against the RefSeq Genome Database in the NCBI portal (Johnson et al. 2008; NCBI Resource Coordinators 2016; O’Leary et al. 2016). The search set was limited to Coleoptera (taxid:7041), Diptera (taxid:7147), Hymenoptera (taxid:7399), and Lepidoptera (taxid:7088). Each order was searched separately due to the data set's favorable bias toward Drosophila species sequences, which led to other species sequences to be omitted from the search results. Notably, the query sequence did not limit the results to any specific Dscam1 isoform, as the blast results showed a consistently high sequence identity for exon 4 across all isoforms. Additionally, a second tblastn search was conducted using a different D. melanogaster Dscam1 exon 4 isoform (isoform 1.10.3, NCBI annotation NP_001036494.1) revealing identical results.
The search resulted in 962 sequences from 83 species (see supplementary file S1, Supplementary Material online). From the 83 species, 37 are Drosophilidae species with highly similar (>95% identity) exon 4 duplications to D. melanogaster. To avoid overrepresentation biases, we included only the 12 D. melanogaster exon 4 duplications in further analyses (see supplementary file S1, Supplementary Material online, for sequence list). Five hundred eighteen high-confidence exon 4 sequences retrieved in this search were clustered by cd-hit-v4.8.1-2019-0228 (Li and Godzik 2006; Fu et al. 2012) using 95% threshold, to remove identical sequences. The 266 representative sequences were aligned with an outgroup sequence, a translated exon 4 of I. scapularis (deer tick) Dscam2 (NCBI accession XP_042144217). Ixodes scapularis was previously used as an outgroup organism for Dscam1 insect alignments (Armitage et al. 2012). Alignment was performed by MAFFT v7.490 plugin in Geneious Prime (Katoh et al. 2002; Katoh and Standley 2013), using the progressive method FFT-NS-2 algorithm, legacy gap penalty, and default settings (see supplementary file S2, Supplementary Material online). Phylogenetic tree was constructed using FastTree 2.1.11 plugin in Geneious Prime (Price et al. 2009) with default parameters and the Whelan and Goldman (WAG) 2001 amino acid substitution model with 20 rate categories. The FastTree local support values were computed using the Shimodaira–Hasegawa test. The FastTree tree was rooted using Geneious Prime branch rooting feature (see supplementary file S3, Supplementary Material online). Sequence similarity of the five Ig2 dimer interface positions (107, 109, 111, 112, and 114) was calculated using the BLOSUM62 matrix.
Ancestry Sequence Reconstruction
Based on the rooted FastTree, ancestral sequences were predicted using two independent ASR programs: PaMLX 1.3.1 CodeML (Yang 2007; Xu and Yang 2013) and GRASP 2020.05.05 (Ross et al. 2022). With PaMLX, default parameters were used and two alternative models were applied: (i) the Poisson model, which assumes equal rates for any amino acid substitutions with ncatG = 5, and (ii) the WAG model with ncatG = 20. In the GRASP analyses, the Jones–Taylor–Thornton (JTT) and WAG evolutionary models were used. Both models corroborated the predictions made by PaMLX, with all predicting the same ancestors at the focus of this study. For detailed accuracy estimations using both methods, refer to supplementary file S4, Supplementary Material online.
Cloning
The plasmid pCMVi-Dscam1_7.27.25-AP (Addgene 72062) encoding Ig1-9 and the first FNIII domain of Dscam1 was purchased. To construct pCMVi-Dscam1_2.27.25-AP, bases 375 to 681 from clone IP15321 (DGRC Stock 1602902; https://dgrc.bio.indiana.edu//stock/1602902 from the Drosophila Genomics Resource Center, NIH Grant 2P40OD010949) were incorporated into pCMVi-Dscam1_7.27.25-AP using Gibson assembly (NEBuilder HiFi DNA Assembly Cloning Kit E5520S). Lentivirus (pLV) transfer plasmid was modified using Gibson to include full-length Dscam1 isoform 7.27.25 using the first ten domains from pCMVi-Dscam1_7.27.25-AP and the remaining ectodomain, transmembrane, and cytoplasmic domains from clone RE54695 (DGRC Stock 9407; https://dgrc.bio.indiana.edu//stock/9407 from the Drosophila Genomics Resource Center, NIH Grant 2P40OD010949).
Mutations were introduced into the Ig2 dimer interface sequence of Dscam1_7.27.25, creating extant and ancestor isoforms 4.8 and 4.10 using the pCMVi construct. Similarly, mutations on Dscam1_2.27.25 generated extant and ancestor isoforms 4.1 to 3. This was performed using QuikChange Lightning Site-Directed Mutagenesis Kit 210518. Mutagenesis primers were designed using the QuikChange primer design program.
To express the mutants as membrane-attached proteins, positions 1 to 3011 of the mutant pCMVi constructs were amplified and cloned into plv-Dscam1_7.27.25(Δ1-3011)-GFP and plv-Dscam1_7.27.25(Δ1-3011)-mCherry amplicons using Gibson assembly.
Cell Aggregation Assay
FreeStyle 293-F cells (Thermo Fisher R79007) were separately transfected using PEI MAX—linear polyethylenimine hydrochloride (MW 40,000, 49553-93-7) as follows: 1 million cells per milliliter were grown in FreeStyle 293 Expression Medium at six-well nontreated plates (SPL #32006), 2 mL per well at 37 °C, 8% CO2, 135 rpm. A total of 1.25 µg plasmid DNA was mixed in Opti-MEM I-reduced serum media (Thermo Fisher 11058021) to a final volume of 62.5 µL. A total of 3.12 µL PEI was added with Opti-MEM I media to a final volume of 62.5 µL. Both mixtures were incubated at room temperature for 15 min. The PEI mix was then added to the DNA mix, immediately vortexed and incubated at room temperature for an additional 15 min, then added to the cells. Three hours later, 500 µL FreeStyle 293 Expression Medium was added to the cells. One milliliter of each reaction was mixed with 1 mL of the complementary reaction for testing binding preferences of desired isoforms. Forty-eight hours post transfection, eight images were acquired per well using an Eclipse Ts2 inverted microscope with a 10× objective. Three replications for each isoform combination were performed.
Aggregate Mixing Quantification
Figures 4 and 5 illustrate the aggregation or separation of cells, quantified using a custom Python script available at https://github.com/Rubinstein-Lab/Mixing-Score. This script evaluates the ratio of red and green cells in close proximity. Each image is captured in a red and a green channel, and these images are segmented into black and white using Otsu thresholding. Initially, the script divides these segmented images into squares of a specified size (24.32 µm2, equivalent to 1.5-2 HEK293F cells). Subsequently, the script counts the number of squares with white pixels in both the red and green channel images. It scans each square for white pixels, updating counters accordingly. The mixing index score is then calculated as the ratio of squares with white pixels in both images to the total number of squares parsed. This score offers insight into the degree of overlap or proximity between red and green cells. Each image receives a score between 0 (no red cells in the proximity of green cells) and 1 (all red cells are proximate to green cells). Images with a score higher than 0.1 typically display mixed aggregation, and images with a lower than 0.1 score show visibly separate red or green aggregates.
The images shown in Figs. 4 and 5 are cropped to depict representative aggregates. However, the quantification of mixing was carried out across the entire image area for all acquired images. The proportion displayed represents the average score obtained from three replications, with eight images for each replication. For further statistical details, refer to supplementary fig. S1, Supplementary Material online.
Computational Evaluation of Binding Affinities
Structural model of Dscam1 8LCA Ig1 to Ig4 dimers was prepared using AlphaFold2 (Jumper et al. 2021; Mirdita et al. 2022). The structure was refined using at least five rounds of the “RepairPDB” utility in FoldX (Schymkowitz et al. 2005). Mutations were generated using the “BuildModel” utility and analyzed using “AnalyseComplex” utility. Additionally, we used the SSIPe server with default parameters for calculating the difference in binding energy upon mutation (Huang et al. 2020).
Supplementary Material
Acknowledgments
We thank Prof. Dinorah Friedmann-Morvinski from the Department of Biochemistry and Molecular Biology at Tel Aviv University for providing the lentivirus (pLV) transfer plasmid. We thank Dr. Karin Smorodinsky-Atias for her assistance during the experimental stages of this work.
Contributor Information
Gil Wiseglass, School of Neurobiology, Biochemistry and Biophysics, The George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel.
Rotem Rubinstein, School of Neurobiology, Biochemistry and Biophysics, The George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel; Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
G.W. and R.R. designed the research, analyzed the data, and wrote the paper. G.W. performed the research.
Funding
This work was supported by the Israel Science Foundation (1463/19 to R.R.).
Data Availability
The data underlying this article is available in the article and also in its online supplementary material.
References
- Aakre CD, Herrou J, Phung TN, Perchuk BS, Crosson S, Laub MT. Evolving new protein-protein interaction specificity through promiscuous intermediates. Cell. 2015:163(3):594–606. 10.1016/j.cell.2015.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, Gaidukov L, Khersonsky O, Gould SM, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005:37(1):73–76. 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- Armitage SAO, Freiburg RY, Kurtz J, Bravo IG. The evolution of Dscam genes across the arthropods. BMC Evol Biol. 2012:12(1):53. 10.1186/1471-2148-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisogni AJ, Ghazanfar S, Williams EO, Marsh HM, Yang JY, Lin DM. Tuning of delta-protocadherin adhesion through combinatorial diversity. Elife. 2018:7:e41050. 10.7554/eLife.41050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Maxeiner S, Südhof TC. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. J Biol Chem. 2014:289(1):387–402. 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch J, Katsamba PS, Harrison OJ, Ahlsén G, Troyanovsky RB, Indra I, Kaczynska A, Kaeser B, Troyanovsky S, Honig B, et al. Homophilic and heterophilic interactions of type II cadherins identify specificity groups underlying cell-adhesive behavior. Cell Rep. 2018:23(6):1840–1852. 10.1016/j.celrep.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Ashley J, Kurleto JD, Lobb-Rabe M, Park YJ, Carrillo RA, Özkan E. Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila. Elife. 2019:8:e41028. 10.7554/eLife.41028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Yu Y, Wang X, Zheng X, Liu T, Hu D, Jin Y, Lai Y, Fu T-M, Chen Q. Structural basis for the self-recognition of sDSCAM in Chelicerata. Nat Commun. 2023:14(1):2522. 10.1038/s41467-023-38205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chockalingam K, Chen Z, Katzenellenbogen JA, Zhao H. Directed evolution of specific receptor–ligand pairs for use in the creation of gene switches. Proc Natl Acad Sci USA. 2005:102(16):5691–5696. 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007:8(3):206–220. 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devamani T, Rauwerdink AM, Lunzer M, Jones BJ, Mooney JL, Tan MAO, Zhang Z-J, Xu J-H, Dean AM, Kazlauskas RJ. Catalytic promiscuity of ancestral esterases and hydroxynitrile lyases. J Am Chem Soc. 2016:138(3):1046–1056. 10.1021/jacs.5b12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick GN, Colucci JK, Harms MJ, Ortlund EA, Thornton JW. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet. 2012:8(11):e1003072. 10.1371/journal.pgen.1003072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012:28(23):3150–3152. 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneious Prime . Version 2020.2.4. https://www.geneious.com.
- Ghose DA, Przydzial KE, Mahoney EM, Keating AE, Laub MT. Marginal specificity in protein interactions constrains evolution of a paralogous family. Proc Natl Acad Sci USA. 2023:120(18):e2221163120. 10.1073/pnas.2221163120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Kaur A, Gunning D, Zipursky SL, Rowen L, Clemens JC. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA (New York, N.Y.). 2004:10(10):1499–1506. 10.1261/rna.7105504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B, Shapiro L. Adhesion protein structure, molecular affinities, and principles of cell-cell recognition. Cell. 2020:181(3):520–535. 10.1016/j.cell.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Li G, Xu B, Dong H, Zhang S, Fu Y, Shi J, Li L, Fu J, Shi F. Trans-splicing facilitated by RNA pairing greatly expands sDscam isoform diversity but not homophilic binding specificity. Sci Adv. 2022:8(27):eabn9458. 10.1126/sciadv.abn9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zheng W, Pearce R, Zhang Y. SSIPe: accurately estimating protein–protein binding affinity change upon mutations using evolutionary profiles in combination with an optimized physical energy function. Bioinformatics. 2020:36(8):2429–2437. 10.1093/bioinformatics/btz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Bäumer P, Kondo M, Uemura T, Schmucker D. Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron. 2007:54(3):417–427. 10.1016/j.neuron.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ispolatov I, Yuryev A, Mazo I, Maslov S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 2005:33(11):3629–3635. 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008:36(Web Server):W5–W9. 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021:596(7873):583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002:30(14):3059–3066. 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013:30(4):772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell T, Ben-Shaul A. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc Natl Acad Sci USA. 2009:106(28):11594–11599. 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010:79(1):471–505. 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Koehbach J, Stockner T, Bergmayr C, Muttenthaler M, Gruber CW. Insights into the molecular evolution of oxytocin receptor ligand binding. Biochem Soc Trans. 2013:41(1):197–204. 10.1042/BST20120256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Kim N, Roy M, Graveley BR. Massive expansions of Dscam splicing diversity via staggered homologous recombination during arthropod evolution. RNA. 2010:16(1):91–105. 10.1261/rna.1812710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele Z, Hindges R. Editorial: Cell adhesion molecules in neural development and disease. Frontiers Media SA. 2023:16:1112300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006:22(13):1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lite T-LV, Grant RA, Nocedal I, Littlehale ML, Guo MS, Laub MT. Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library. Elife. 2020:9:e60924. 10.7554/eLife.60924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukatsky D, Shakhnovich B, Mintseris J, Shakhnovich E. Structural similarity enhances interaction propensity of proteins. J Mol Biol. 2007:365(5):1596–1606. 10.1016/j.jmb.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrilia N, Kollias A, Manolopoulos L, Syrigos K. Cell adhesion molecules: role and clinical significance in cancer. Cancer Invest. 2009:27(10):1023–1037. 10.3109/07357900902769749. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007:129(3):593–604. 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- McClune CJ, Laub MT. Constraints on the expansion of paralogous protein families. Curr Biol. 2020:30(10):R460–R464. 10.1016/j.cub.2020.02.075. [DOI] [PubMed] [Google Scholar]
- Meijers R, Puettmann-Holgado R, Skiniotis G, Liu JH, Walz T, Wang JH, Schmucker D. Structural basis of Dscam isoform specificity. Nature. 2007:449(7161):487–491. 10.1038/nature06147. [DOI] [PubMed] [Google Scholar]
- Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022:19(6):679–682. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters Ralph S, Donath A, Mayer C, Frandsen Paul B, Ware J, Flouri T, Beutel Rolf G, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014:346(6210):763–767. 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- Miura SK, Martins A, Zhang KX, Graveley BR, Zipursky SL. Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell. 2013:155(5):1166–1177. 10.1016/j.cell.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca TJ. On the Teneurin track: a new synaptic organization molecule emerges. Front Cell Neurosci. 2015:9:204. 10.3389/fncel.2015.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators . Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016:44(D1):D7–D19. 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. 2004:36(3):240–246. 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016:44(D1):D733–D745. 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007:317(5844):1544–1548. 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg O, Choi J-M, Shakhnovich EI. Evolution of specificity in protein-protein interactions. Biophys J. 2014:107(7):1686–1696. 10.1016/j.bpj.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007:8(4):1–12. 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009:26(7):1641–1650. 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science. 2013:340(6133):730–734. 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CM, Foley G, Boden M, Gillam EM. Using the evolutionary history of proteins to engineer insertion-deletion mutants from robust, ancestral templates using graphical representation of ancestral sequence predictions (GRASP). Methods Mol Biol. 2022:2397:85–110. 10.1007/978-1-0716-1826-4_6. [DOI] [PubMed] [Google Scholar]
- Rubinstein R, Thu CA, Goodman KM, Wolcott HN, Bahna F, Mannepalli S, Ahlsen G, Chevee M, Halim A, Clausen H, et al. Molecular logic of neuronal self-recognition through protocadherin domain interactions. Cell. 2015:163(3):629–642. 10.1016/j.cell.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Wojtowicz WM, Andre I, Qian B, Wu W, Baker D, Eisenberg D, Zipursky SL. A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell. 2008:134(6):1007–1018. 10.1016/j.cell.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayou C, Monniaux M, Nanao MH, Moyroud E, Brockington SF, Thévenon E, Chahtane H, Warthmann N, Melkonian M, Zhang Y, et al. A promiscuous intermediate underlies the evolution of LEAFY DNA binding specificity. Science. 2014:343(6171):645–648. 10.1126/science.1248229. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000:101(6):671–684. 10.1016/S0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Weiner JA. Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc Natl Acad Sci U S A. 2010:107(33):14893–14898. 10.1073/pnas.1004526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005:33(Web Server):W382–W388. 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva AP, Katsamba PS, Cosmanescu F, Brewer JJ, Ahlsen G, Mannepalli S, Shapiro L, Honig B. DIP/Dpr interactions and the evolutionary design of specificity in protein families. Nat Commun. 2020:11(1):2125. 10.1038/s41467-020-15981-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq MA, Hochberg GK, Thornton JW. Evolution of protein specificity: insights from ancestral protein reconstruction. Curr Opin Struct Biol. 2017:47:113–122. 10.1016/j.sbi.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. 2007:54(3):403–416. 10.1016/j.neuron.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Schiffmacher AT. Should I stay or should I go? Cadherin function and regulation in the neural crest. Genesis. 2017:55(6):e23028. 10.1002/dvg.23028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu CA, Chen WV, Rubinstein R, Chevee M, Wolcott HN, Felsovalyi KO, Tapia JC, Shapiro L, Honig B, Maniatis T. Single-cell identity generated by combinatorial homophilic interactions between α, β, and γ protocadherins. Cell. 2014:158(5):1045–1059. 10.1016/j.cell.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Kominami K, Waseda M, Komura H, Miyoshi J, Takeichi M, Takai Y. Nectins establish a checkerboard-like cellular pattern in the auditory epithelium. Science. 2011:333(6046):1144–1147. 10.1126/science.1208467. [DOI] [PubMed] [Google Scholar]
- Vendome J, Felsovalyi K, Song H, Yang Z, Jin X, Brasch J, Harrison OJ, Ahlsen G, Bahna F, Kaczynska A. Structural and energetic determinants of adhesive binding specificity in type I cadherins. Proc Natl Acad Sci USA. 2014:111(40):E4175–E4184. 10.1073/pnas.1416737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, Maere S, Verstrepen KJ. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012:10(12):e1001446. 10.1371/journal.pbio.1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee C-HJ, Lee T. Drosophila dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002:33(4):559–571. 10.1016/S0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006:312(5770):111–114. 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim J-W, Lambkin C, Bertone MA, Cassel BK, Bayless KM, Heimberg AM, et al. Episodic radiations in the fly tree of life. Proc Natl Acad Sci USA. 2011:108(14):5690–5695. 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm N, Kumari S, Krick N, Rickert C, Duch C. Dscam1 has diverse neuron type specific functions in the developing Drosophila CNS. eNeuro. 2022:9(4):ENEURO.0255–22.2022. 10.1523/ENEURO.0255-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseglass G, Boni N, Smorodinsky-Atias K, Rubinstein R. Clustered protocadherin cis-interactions are required for combinatorial cell–cell recognition underlying neuronal self-avoidance. Proc Natl Acad Sci. 2024:121(29):e2319829121. 10.1073/pnas.2319829121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004:118(5):619–633. 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007:130(6):1134–1145. 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Ahlsen G, Baker D, Shapiro L, Zipursky SL. Complementary chimeric isoforms reveal Dscam1 binding specificity in vivo. Neuron. 2012:74(2):261–268. 10.1016/j.neuron.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Yang Z. PAMLX: a graphical user interface for PAML. Mol Biol Evol. 2013:30(12):2723–2724. 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007:24(8):1586–1591. 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A, Park S-H, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003:426(6967):676–680. 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- Zhan X-L, Clemens JC, Neves G, Hattori D, Flanagan JJ, Hummel T, Vasconcelos ML, Chess A, Zipursky SL. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron. 2004:43(5):673–686. 10.1016/j.neuron.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Zhou F, Cao G, Dai S, Li G, Li H, Ding Z, Hou S, Xu B, You W, Wiseglass G, et al. Chelicerata sDscam isoforms combine homophilic specificities to define unique cell recognition. Proc Natl Acad Sci U S A. 2020:117(40):24813–24824. 10.1073/pnas.1921983117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Hummel T, Clemens JC, Berdnik D, Zipursky SL, Luo L. Dendritic patterning by Dscam and synaptic partner matching in the Drosophila antennal lobe. Nat Neurosci. 2006:9(3):349–355. 10.1038/nn1652. [DOI] [PubMed] [Google Scholar]
- Zinn K, Özkan E. Neural immunoglobulin superfamily interaction networks. Curr Opin Neurobiol. 2017:45:99–105. 10.1016/j.conb.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010:143(3):343–353. 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article is available in the article and also in its online supplementary material.