Abstract
All polynucleotide polymerases have a similar structure and mechanism of catalysis, consistent with their evolution from one progenitor polymerase. Viral RNA-dependent RNA polymerases (RdRp) are expected to have properties comparable to those from this progenitor and therefore may offer insight into the commonalities of all classes of polymerases. We examined RNA synthesis by the brome mosaic virus RdRp on DNA, RNA, and hybrid templates and found that precise initiation of RNA synthesis can take place from all of these templates. Furthermore, initiation can take place from either internal or penultimate initiation sites. Using a template competition assay, we found that the BMV RdRp interacts with DNA only three- to fourfold less well than it interacts with RNA. Moreover, a DNA molecule with a ribonucleotide at position −11 relative to the initiation nucleotide was able to interact with RdRp at levels comparable to that observed with RNA. These results suggest that relatively few conditions were needed for an ancestral RdRp to replicate DNA genomes.
RNA molecules are thought to have performed catalytic as well as genomic functions in the precellular “RNA world” (7, 10, 14, 17). Support for this hypothesis includes (i) the discovery of the catalytic RNAs (12, 21); (ii) the requirement for RNA in many essential, and presumably ancient, cellular processes such as translation, splicing, and priming of DNA synthesis; (iii) the presence of ribonucleotides or derived components thereof in most biological coenzymes; and (iv) the biosynthesis of deoxyribonucleotides by the reduction of ribonucleotides rather than by a de novo pathway. To overcome the problem of efficiently and accurately copying genetic material, it has been postulated that one of the earliest proteins would have been an RNA replicase (22). Since DNA was eventually selected as the preferred carrier of genetic information, the preexisting RNA-dependent RNA polymerase (RdRp) probably evolved to fulfill the new function of replicating DNA genomes, in addition to generating mRNAs for protein synthesis. One probable early step during this transition would have been the ability of the ancestral RNA replicase to recognize its cognate promoter sequence in a deoxyribose form.
This proposed line of development implies a common ancestor for all polynucleotide polymerases. Fundamental similarities in the three-dimensional structure and the basic mechanism of nucleotidyl transfer in the four classes of polymerases, based on whether the template and synthesized product are DNA or RNA, support this view (16, 30). It has also been argued that an important vestige of the original RNA replicase is evolutionarily conserved in the modern-day eubacterial β′ subunit of DNA-dependent RNA polymerase (DdRp) and its homologues in archaeal and eukaryotic polymerases (4, 23, 25). Viral RdRps are the only extant class of polymerases that recapitulate the replication requirements of the RNA world. Presently, they have the arduous task of recognizing different promoters located both internally and on the termini (8), a situation that was also probably present in the RNA world before the advent of circularized genomes or telomerase functions. Therefore, viral RdRps offer a unique vantage point to gain a better understanding of the progenitor polymerase.
In our investigations of the mechanism of RNA-directed RNA synthesis, we study brome mosaic virus (BMV), the type member of the bromovirus group of plant viruses in the alphavirus-like superfamily of plus-strand RNA viruses (11). Three RNAs, designated RNA1, RNA2, and RNA3, and a subgenomic RNA4 that is initiated from minus-strand RNA3 comprise the BMV genome. The viral and cellular proteins that comprise the RdRp complex are responsible for directing viral RNA synthesis from the infecting RNA templates, a process which requires specific recognition of salient RNA features. In vitro, the BMV subgenomic promoter efficiently and accurately directs RNA synthesis by using highly enriched BMV RdRp preparations from infected barley (1, 24). We have studied BMV subgenomic RNA initiation from RNAs of minimal lengths, designated “proscripts” since they contain both the promoter (the 20 nucleotides [nt] 3′ of the subgenomic initiation site) and template for plus-strand RNA synthesis (1, 26).
In this study, we have tested the ability of the BMV RdRp to recognize and initiate RNA synthesis from a deoxyribose version of the subgenomic proscript. We observed accurate initiation of RNA synthesis from either an internal or penultimate initiation site on a DNA template. Studies of proscripts containing both ribose and deoxyribose nucleotides indicated that the ribose at position −11 but not the one at position −17 was important for RNA synthesis (27). Therefore, DNA proscripts containing various C2′ substitutions at position −11 were chemically synthesized and suggested the manner by which RdRp recognizes this specific hydroxyl group in the subgenomic promoter. Finally, a template competition analysis revealed that RdRp has only a moderately reduced affinity for the DNA version of the subgenomic promoter and that the insertion of only one ribose, at position −11, virtually restored binding to the level obtained with a wild-type (WT) RNA template.
MATERIALS AND METHODS
Synthesis of proscripts.
PCR was used to generate cDNA copies of the minus-strand BMV RNA3 encompassing the subgenomic promoter from the cDNA clone of RNA pB3TP8 (15). Pairs of primers, one of which contained a T7 promoter, allowed proscript RNAs to be generated by using T7 RNA polymerase (Ampliscribe; Epicentre) as described previously (1). RNAs were purified with Qiagen (Chatsworth, Calif.) columns by using the manufacturer’s protocol to remove nucleoside triphosphates and proteins remaining from the T7 transcription reaction. Oligonucleotides used as DNA templates were obtained from Operon Technologies, Inc. (Alameda, Calif.). Both RNA and DNA templates were visually inspected by denaturing polyacrylamide gel electrophoresis and quantified by measurement of UV absorbance.
Chemical synthesis of the proscripts containing base analogs was performed on a ABI 394 automated DNA synthesizer (Applied Biosystems, Inc., Foster City, Calif.) by using conventional phosphoramidite elongation cycles as described by Wincott et al. (34). Suitably protected 2′-fluoro-, 2′-o-methyl-, and 2′-amino-guanosine phosphoramidites were obtained from Glen Research (Sterling, Va.). After subsequent aqueous methylamine and triethylamine trihydrofluoride treatment to cleave the exocyclic amino and 2′-OH protecting groups, when present, the proscripts were purified and analyzed by anion-exchange high-pressure liquid chromatography (34). Mass spectral analysis of each chemically synthesized proscript was performed on a Voyager-DE MALDI-TOF spectrometer (Perseptive Biosystem, Framingham, Mass.). All RNAs were within 0.05% of the expected mass.
RdRp activity assay and product analysis.
BMV RdRp was prepared from infected barley as described previously (18, 31). Standard assay mixtures consisted of 25 nM template RNA or DNA (125 nM for templates with a penultimate initiation site) with 10 μl of RdRp in 40 μl containing 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM dithiothreitol, 0.5% (vol/vol) Triton X-100, 2 mM MnCl2, 200 μM ATP, 200 μM UTP, 500 μM GTP, and 250 nM [α-32P]CTP (Amersham). The reaction mixtures were incubated at 30°C for 90 min, and the reactions were stopped by phenol-chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen and 0.4 M ammonium acetate. The products were separated by electrophoresis on 20% denaturing polyacrylamide gels containing 8 M urea. The gels were wrapped in plastic and exposed to film at −80°C. Product bands were quantified with a PhosphorImager (Molecular Dynamics), and values were compared to the amount of product generated from the WT template (−20/13) to derive the relative percent activities of the various experimental templates. All values shown represent the means and standard deviations of at least three independent experiments.
Template competition assays were performed under the reaction conditions stated above, except that 25 nM proscript −20/15, directing the synthesis of a 15-nt product, was incubated with increasing concentrations of various competitors, all directing the synthesis of a 13-nt product. The product generated from the −20/15 proscript was quantitated as above, and the amount was plotted against the concentration of competitor to determine the concentration of competitor needed to reduce the 15-nt product by 50%; this was designated the IC50.
RESULTS
RdRp can synthesize RNA from a DNA template.
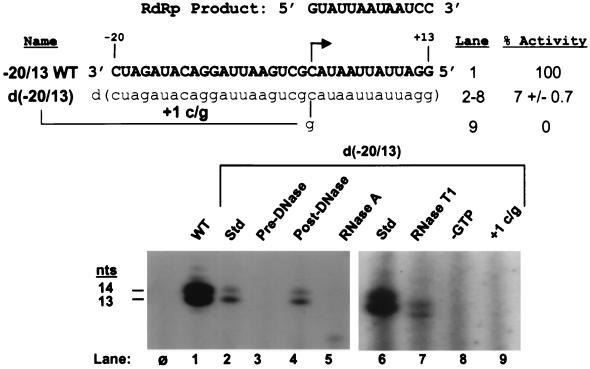
The sequence-specific recognition of the subgenomic RNA promoter by the BMV RdRp and the steps in viral RNA synthesis (2, 26) are analogous to those of DdRps. We therefore examined the ability of BMV RdRp to recognize and accurately initiate RNA synthesis from a DNA version of the subgenomic promoter. To evaluate the level of synthesis obtained with a DNA promoter, we first generated a WT control proscript composed entirely from ribonucleotides. This 33-nt proscript (designated −20/13 WT) contains the WT subgenomic promoter sequence directing the synthesis of a 13-nt product, whose first 11 nt is BMV sequence (complementary to viral plus-strand RNA3 from positions 1222 to 1252) and whose next nucleotides are two guanylates which allow the labeling of RdRp products with [α-32P]CTP. As judged from T7 DdRp-generated size markers, the predominant RdRp product was 14 nt due to the nontemplated addition of one residue, a phenomenon common to many polymerases (Fig. 1, lane 1).
FIG. 1.
BMV RdRp accurately initiates RNA synthesis from internal initiation sites on DNA proscripts. (Top) Proscript −20/13 WT is complementary to the viral plus-strand RNA3 from positions 1222 to 1252 and serves as the WT control. The initiation nucleotide is denoted by an arrow, and the sequence of the RdRp product is shown above. Schematics of the DNA constructs are displayed, and to the right are the lane numbers with the amount of RNA synthesis relative to that from the WT control. RNA sequences are denoted by bold capital letters, while DNA sequences are in lowercase letters. (Bottom) Autoradiograph of RdRp reaction products generated from 25 nM RNA proscript −20/13 WT (lane 1) or 25 nM all-deoxyribose proscript d(−20/13) (lanes 2 to 9). RNA synthesis and accurate initiation from proscript d(−20/13) were verified by the treatments indicated above the gel in lanes 3 to 5 and 7 to 9, respectively. Lane φ contains the products of a control reaction with no added template, while lanes Std contain products from the d(−20/13) proscript with no additional treatments. T7-generated size markers of the expected sequence of the RdRp products are denoted on the left. The autoradiograph containing lanes 6 to 9 is overexposed relative to that containing lanes 1 to 5.
The all-DNA proscript, designated d(−20/13), contained deoxyriboses in every position while retaining otherwise WT subgenomic promoter and template sequences. As would be required if an RNA replicase participated in the transition from an RNA to a DNA genome, the BMV RdRp was able to recognize the d(−20/13) proscript and initiate RNA synthesis (Fig. 1, lane 2). The level of RNA synthesis directed by this DNA construct was reduced to 7% relative to that obtained with the all-RNA control. While RdRp has a clear functional preference for RNA, it is nonetheless quite capable of utilizing a DNA version of the subgenomic promoter. Interestingly, the predominant product was now 13 rather than 14 nt, as it was with the RNA template (lanes 1 and 2). This change may reflect the need for 2′-OHs in the template to efficiently add the nontemplated nucleotide.
A number of enzymatic treatments were used to verify that RdRp was able to generate a RNA product from a DNA template. Treatment of the d(−20/13) proscript with DNase I abolished product synthesis (Fig. 1, lane 3). The product, however, was resistant to DNase I while being completely sensitive to RNase A (lanes 4 and 5). Product synthesis was also resistant to inhibitors of DdRps, such as actinomycin D and rifampin (data not shown). Accurate initiation was verified in a number of ways: comparisons of the product sizes to those from the −20/13 WT proscript (lanes 1 and 2), RNase T1 digestion (which cleaves after the initiating guanylate) resulting in labeled products 1 nt smaller than those without digestion (lanes 6 and 7), the absolute requirement for the GTP that is needed only for initiating accurate synthesis (lane 8), and the lack of synthesis from a proscript with a mutant initiation site (lane 9).
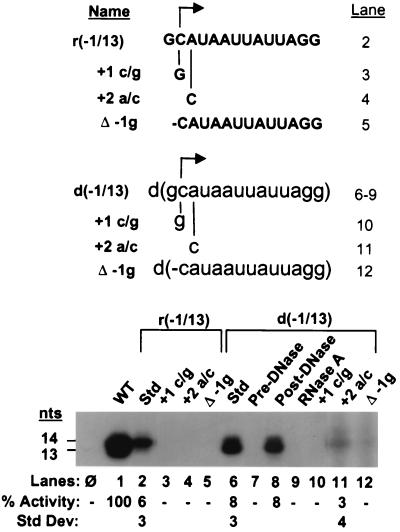
To copy the entire genomic template during viral replication, RdRp initiates RNA synthesis from the penultimate cytidylate of each of the three genomic RNAs. We next investigated whether the BMV RdRp could recognize the subgenomic initiation site when present at the penultimate position on a linear template in both RNA and DNA versions (Fig. 2). Constructs retaining nucleotides at positions −1 to +13 relative to the subgenomic initiation site were synthesized in both RNA and DNA versions, r(−1/13) and d(−1/13), respectively. The two versions of the −1/13 proscript were able to direct RNA synthesis by RdRp at approximately equal levels (6 and 8%, respectively [Fig. 2, lanes 2 and 6], of the amount of RNA synthesized from the −20/13 WT proscript [lane 1]). As observed for proscripts containing the subgenomic promoter, the predominant product was 14 nt for the RNA template, r(−1/13), and 13 nt for the DNA template, d(−1/13).
FIG. 2.
BMV RdRp can initiate RNA synthesis from a penultimate initiation site on RNA and DNA templates. (Top) The sequences of the constructs tested are listed, and the lane numbers containing the reaction products in the autoradiograph below are shown on the right. RNA sequences are denoted by bold capital letters, while DNA sequences are in lowercase letters. Changes in nucleotide identity are shown below each sequence. The Δ−1g proscripts lack the 3′-terminal guanylate at position −1 relative to the initiation site. (Bottom) Autoradiograph of RdRp reaction products. Template concentrations of 125 nM were used in all reactions (lanes 2 to 12) except for WT −20/13 RNA, which was present at 25 nM (lane 1). The relative percent activity of each construct compared to that from the −20/13 WT proscript is presented below the autoradiograph (Std Dev, standard deviation). The reaction products in lanes 11 to 12 are from a different autoradiograph from those in lanes 1 to 10. Dashes denote lanes with no detectable level of RdRp product. Lane φ contains the products of a control reaction with no added template, while lanes Std contain products with no additional treatments.
The requirements for initiation from these templates were determined by introducing mutations at positions surrounding the initiation site. Mutation of the +1 cytidylate or removal of the −1 guanylate abolished RNA synthesis in both r(−1/13) and d(−1/13) templates (Fig. 2, lanes 3 and 5 and lanes 10 and 12, respectively). These results demonstrate that initiation must occur from a cytidylate at the penultimate position in these truncated templates, as it does for full-length genomic synthesis. However, the +2 A-to-C mutation abolished the ability to direct RNA synthesis in the r(−1/13) template (Fig. 2, lane 4) and resulted in a reduced but detectable level of RNA synthesis in the d(−1/13) template (lane 11). RNA synthesis from the d(−1/13) template was verified as above; treatment with DNase I degraded the DNA template and abolished RNA synthesis, while the product was resistant to DNase I but degraded by RNase A (lanes 7 to 9). These results demonstrate the feasibility of full-length replication from a DNA template in the absence of any upstream sequences and are consistent with an ancestral RdRp being able to function during the transition from RNA to DNA templates.
Riboses that facilitate RNA synthesis.
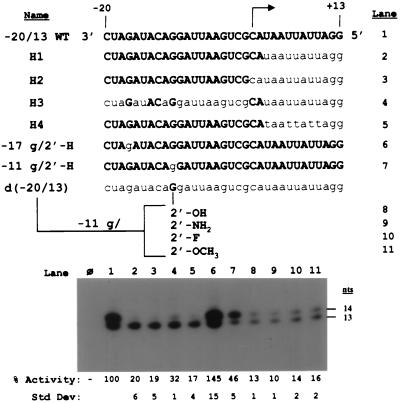
Since the level of synthesis directed by the all-DNA d(−20/13) proscript was decreased relative to that directed by the all-RNA −20/13 WT proscript, hybrids (containing both ribose and deoxyribose residues) were generated to determine the locations of residues that facilitate RNA synthesis by RdRp (Fig. 3). Hybrid H1, containing riboses only in the subgenomic promoter and the +1 and +2 positions, directed a two- to threefold increase in RNA synthesis (20%) relative to the d(−20/13) proscript (7%). However, synthesis was still four- to fivefold below that obtained from the −20/13 WT proscript (Fig. 3, lanes 1 and 2), indicating that riboses in the template may be important for efficient RNA synthesis. This preference for riboses in the template portion of the proscript does not include the initiation (+1) or +2 positions, as demonstrated by results obtained with hybrid H2. This construct extended the region of deoxyribose substitution to include the +1 and +2 positions of the template, and the relative activity of hybrid H2 was indistinguishable from that obtained from hybrid H1 (Fig. 3, lanes 2 and 3). Hybrid H4, in which deoxythymidines were used instead of deoxyuridines in the template from positions +3 to +13, also did not appreciably alter RNA synthesis relative to the H1 proscript (17 and 20%, respectively) (lanes 2 and 5).
FIG. 3.
Ribose moieties which facilitate RNA synthesis by RdRp. (Top) The sequence of the RNA −20/13 WT proscript is shown, with the initiation site marked by an arrow. The sequences of chimeric proscripts, containing both ribose and deoxyribose residues, are listed below, with bold capital letters representing RNA and lowercase letters representing DNA. Proscripts containing substitutions of the C-2′ position at the −11 guanylate in an otherwise all-DNA proscript were also constructed. The lane number containing the RdRp product generated from each proscript in the autoradiograph below is shown to the right. (Bottom) Autoradiograph of the BMV reaction products from the hybrid proscripts. The amount of RNA synthesis from 25 nM proscript is shown in lanes 1 to 11. Product sizes are denoted on the side. Lane φ contains the products of a control reaction with no added template. Values listed represent the means and standard deviations (Std Dev) of at least five independent experiments.
Hybrid H3 was extensively substituted with deoxyriboses within the subgenomic core promoter. This hybrid contained riboses only at positions −17, −14, −13, and −11 in the core promoter and the +1 and +2 initiation nucleotides. Interestingly, hybrid H3 reproducibly directed an elevated level of RNA synthesis with respect to hybrid H1 (32 and 20%, respectively, compared to the WT control) (Fig. 3, lanes 2 and 4). The level of synthesis observed with hybrid H3 coupled with the result obtained with hybrid H2 indicated that ribose residues may be important only at positions −17, −14, −13, and −11 or a subset thereof in the subgenomic promoter. Previously (27) we had found that a deoxyguanosine at position −17 in an otherwise RNA proscript had no adverse effect on RNA synthesis; however, in stark contrast, a deoxyguanosine at position −11 reduced synthesis by over half relative to the −20/13 WT control (Fig. 3, lanes 6 and 7, respectively). These results argue strongly that the ribose at position −11 is important for RNA synthesis.
This disparity (−17 versus −11) prompted us to examine the role of the 2′-OH at position −11 in the initiation of RNA synthesis by the BMV RdRp. DNA proscripts with various C-2′ substitutions at position −11 (-OH, -NH2, -F, or -OCH3) were chemically synthesized to determine how this functional group is recognized by the BMV RdRp. The amounts of RNA synthesized from the proscripts containing these replacements were determined and compared to the amounts obtained with the RNA −20/13 WT proscript (Fig. 3, compare lanes 8 to 11 with lane 1). Surprisingly, all of the DNA proscripts containing the C-2′ substitutions had similar abilities to direct RNA synthesis relative to one another (ranging from 10 to 16% relative to that from −20/13 WT). In addition, proscript with each of these minor substitutions at position −11 were able to direct RNA synthesis approximately twofold better than the all-DNA proscript, d(−20/13) (data not shown).
RdRp-DNA interaction.
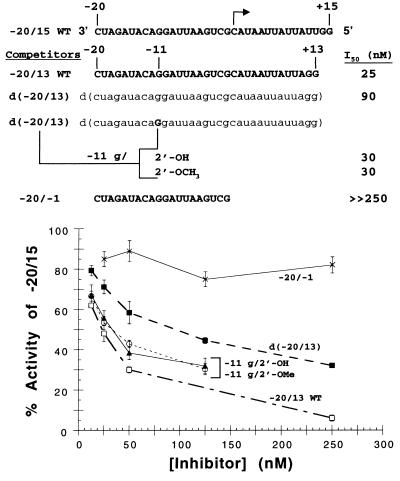
A template competition assay was used to evaluate whether the presence of deoxyriboses in the subgenomic promoter had an adverse effect on the ability to be directly recognized by RdRp, as would be expected from the functional results presented above. The amount of synthesis from a WT promoter directing the production of a 15-nt product (designated −20/15) was determined in the absence and presence of various competitor templates (Fig. 4). The concentration of the competitor required to reduce the activity from the −20/15 proscript by 50% was termed the IC50. Competitors that are able to easily interact with RdRp will reduce synthesis from −20/15 at lower concentrations and result in a lower IC50. In these experiments, the concentration of RdRp was limiting, ensuring competition between the various templates.
FIG. 4.
Role of ribose 2′-OHs in stable interactions with RdRp. (Top) The sequence of the −20/15 WT proscript, directing the synthesis of a 15-nt product from the initiating cytidylate (arrow), is shown. Below are the sequences of various competitors, all containing a WT subgenomic promoter sequence. RNA sequences are denoted by bold capital letters, while DNA sequences are in lowercase letters. The −20/−1 proscript contains the WT subgenomic promoter from positions −20 to −1 relative to the initiation site and served as a negative control. The IC50s are listed to the right. (Bottom) Determination of IC50s for RNA and DNA subgenomic promoters. The amount of 15-nt product generated from the −20/15 RNA proscript was measured and is plotted as a function of the concentration of each competitor (up to 10-fold molar excess). The identities of the competitors are shown to the right of the graph. Datum points represent the means of three independent experiments, and standard deviations are shown as error bars.
As a positive control, the −20/13 WT proscript (composed entirely of ribose residues) reduced the level of synthesis of the 15-nt product by half when present in the same molar ratio as the −20/15 proscript, generating an IC50 of 25 nM (Fig. 4). The ability of the d(−20/13) proscript to be recognized by RdRp was only mildly affected. The DNA proscript had an IC50 of 90 nM, a three- to fourfold reduction relative to the −20/13 WT proscript (Fig. 4). Next, proscripts containing either the -OH or -OCH3 group at the C2′ position of the −11 guanylate in an otherwise all-DNA proscript were used as competitors. The presence of either of these functional groups at this position generated results that were indistinguishable from one another, with IC50s of 30 nM (Fig. 4). The insertion of either of these functional groups at position −11 also virtually restored the ability to be recognized by RdRp to that observed with the −20/13 WT RNA proscript; i.e., the IC50s were 30 and 25 nM, respectively. An RNA containing the WT sequences from positions −20 to −1, previously shown to be unable to stably interact with the BMV RdRp since this construct lacks the subgenomic initiation site (27), was used as a negative control. As expected, the −20/−1 RNA proscript was not able to effectively inhibit the synthesis of the 15-nt product even when present at 10-fold molar excess with respect to −20/15 (IC50 > 250 nM).
DISCUSSION
We have demonstrated that RdRp has the ability to recognize and initiate accurate RNA synthesis from either an internal or penultimate initiation site on a DNA or hybrid template containing the same nucleotide sequences as the natural RNA counterparts, indicating that no single proscript ribose is absolutely required for RNA synthesis (Fig. 1 and 2). We also have shown that RdRp can tolerate the insertion of deoxythymidines in the template portion of a chimeric proscript (Fig. 3, lane 5). This demonstrated ability to use DNA templates was limited to oligonucleotides containing the appropriate promoter and/or initiation sequence, since unrelated oligonucleotides were unable to serve as templates for RNA synthesis (data not shown). Therefore, the sequence requirements for initiation of RNA synthesis are maintained by RdRp even in the presence of deoxyriboses. Most importantly, these studies revealed that RdRp interacts with the DNA proscript d(−20/13) only slightly less well than the RNA proscript −20/13, with IC50s of 90 and 25 nM, respectively (Fig. 4). These results were surprising, given that d(−20/13) was reduced in its ability to direct RNA synthesis by over 15-fold relative to that from the −20/13 WT proscript (Fig. 1), and indicate that the presence of deoxyriboses in the proscript can differentially affect the level of RNA synthesis and proscript recognition by RdRp.
Chimeric proscripts were used to discern the effects of the deoxyribose substitutions (Fig. 3 and 4). The presence of riboses in the subgenomic promoter increased the level of RNA synthesis by RdRp; however, ribonucleotides in the template portion of the proscript (positions +3 to +13) still seem to be needed to direct WT levels of RNA synthesis (Fig. 3, lane 2). More extensively substituted proscripts yielded similar levels of RNA synthesis. Deductive reasoning identified positions −17, −14, −13, and/or −11 as putatively containing one or more of the riboses important for RNA synthesis within the subgenomic promoter. We previously demonstrated that a deoxyribose substitution at position −17 did not have an adverse effect on the ability to direct RNA synthesis whereas the same substitution at position −11 reduced synthesis.
While other nucleotides may contribute to the level of RNA synthesis, template competition analysis identified nucleotide −11 as a key recognition position for the BMV RdRp (Fig. 4). While the competition assay does not directly demonstrate binding by RdRp to any given template, it is the most parsimonious interpretation. WT levels of interaction were virtually restored by inserting either the -OH or -OCH3 moiety at the C-2′ position of the −11 guanylate. RdRp could recognize the 2′-OH at this position as a hydrogen bond acceptor, as a hydrogen bond donor, or by the orientation of the sugar. The C-2′ substitutions -OH, -NH2, -F, and -OCH3 are speculated to be able to accept a hydrogen bond, while only the -OH and -NH2 moieties can also serve as a hydrogen bond donor (13, 33). Additionally, all but the 2′-NH2 substitution are expected to predominantly form the ribose C-3′ endo sugar conformation (13). The fact that all four of these substitutions directed similar levels of RNA synthesis (Fig. 3, lanes 8 to 11) and the -OH and -OCH3 substitutions resulted in identical IC50s (Fig. 4) eliminates the ability to donate a hydrogen for a hydrogen bond and the orientation of the sugar as factors mediating the recognition of the 2′-OH at this position. Since the 2′-H [present in the all-DNA d(−20/13) proscript] is the only substitution at position −11 which cannot accept a hydrogen bond, a possible explanation for the importance of the −11 ribose is that it provides a hydrogen bond acceptor site which is required for the proper positioning of RdRp or that the presence of the 2′-OH (and the other bulkier substitutions) simply prevents, by steric interference, some unknown deleterious structure from occurring.
Although the insertion of the single ribose at position −11 restored the ability to interact with RdRp to nearly WT levels, this substitution was still unable to direct RNA synthesis at a level comparable to that obtained with the WT RNA proscript. This apparent discrepancy led us to hypothesize that the additional riboses act to facilitate RNA synthesis in a mechanistic fashion rather than by contributing to the binding interaction with RdRp. A mutational study of the features within the BMV genomic promoter yielded results consistent with this claim (28). It is intriguing to speculate that the template riboses (+3 to +13) somehow stabilize or increase the likelihood of a conformational change in RdRp as it shifts from the initiation stage into an elongating complex. This preference for template riboses was not observed in the truncated proscripts initiating synthesis from the penultimate position. This is perhaps due to RdRp being able to adjust its conformation and/or activity in the absence of the core promoter. An induced fit mechanism facilitating the differentiation between RNA promoters by the BMV RdRp has recently been described (3).
We also tested whether the BMV RdRp can use the deoxynucleotide reaction (9, 29, 32). Therefore, a mutation in the catalytic domain and/or the substrate nucleotide binding pocket of the BMV RdRp would be expected to remove the nucleotide discriminator mechanism and allow the incorporation of deoxynucleoside triphosphates. The end result of this mutation would evolve the ancestral RdRp into a reverse transcriptase required to complete the transition from the RNA to DNA world.
The results obtained during the course of this study are consistent with the hypothesis that all modern polynucleotide polymerases could have derived from a progenitor polymerase present during the RNA world. Other single-subunit or multisubunit polymerases recognize and utilize an “unnatural” template and/or nucleotide substrate (5, 6, 29), including the recombinant RdRp from bovine viral diarrhea virus (35). Konarska and Sharp (19) demonstrated the ability of the T7 DdRp to use an RNA template for RNA synthesis. However, a nonspecific nucleic acid component was required to induce the initial synthesis of X RNA and the sequence of this RNA template did not resemble the natural T7 promoter. In fact, a predicted secondary structure of the X RNA bears a striking resemblance to the tRNAs found to initiate minus-strand viral RNA synthesis (20), further suggesting that components of RNA viruses may have played a role in the evolution of DdRps. Building upon these results, the fact that the BMV RdRp was able to synthesize RNA from a DNA complement of the subgenomic promoter under identical experimental conditions to those used for synthesis from the WT RNA proscript further supports the possibility that a primitive RdRp could use a DNA template without major changes in the active-site architecture of the polymerase, the environmental conditions, or the identity of the regulatory sequences controlling synthesis.
Since modern RdRps (or a conserved vestige thereof) best reflect the biochemical properties of the primitive RNA replicase, experiments with viral RdRp allow potential insights into the biological events surrounding the change from RNA to DNA genomes. The demonstration that the BMV RdRp interacts with the DNA version of the subgenomic promoter only slightly less well than with the RNA version strongly argues that no significant decrease in binding would have occurred during the transition from RNA to DNA templates. The removal of this potential penalty suggests a possible mode by which an ancestral RdRp could have survived this transition and evolved to transcribe and replicate DNA genomes required in the emerging DNA world.
ACKNOWLEDGMENTS
We thank our colleagues at RPI, V. Mokler and L. Maloney. We also thank the IU Cereal Killers for helpful discussions.
Funding was provided by USDA grant 9702126 and NSF grant MCB9507344. R.W.S. was supported by an NIH Genetics training grant to the IU Biology Department.
REFERENCES
- 1.Adkins S, Siegel R W, Sun J H, Kao C C. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA. 1997;3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins S, Stawicki S, Faurote G, Siegel R, Kao C C. Mechanistic analysis of RNA synthesis RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerase. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 3.Adkins S, Kao C C. RNA dictates the mode of promoter use by bromoviral RNA-dependent RNA polymerases. Virology. 1999;252:1–8. doi: 10.1006/viro.1998.9449. [DOI] [PubMed] [Google Scholar]
- 4.Allison L A, Moyle M, Shales M, Ingles C J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 5.Biebricher C, Luce R. Template-free generation of RNA species that replicate with bacteriophage-T7 RNA-polymerase. EMBO J. 1996;15:3458–3465. [PMC free article] [PubMed] [Google Scholar]
- 6.Biebricher C K, Orgel L E. An RNA that multiplies indefinitely with DNA-dependent RNA polymerase selection from a random co-polymer. Proc Natl Acad Sci USA. 1973;70:934–938. doi: 10.1073/pnas.70.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomberg C. On the appearance of function and organization in the origin of life. J Theor Biol. 1997;187:541–554. doi: 10.1006/jtbi.1996.0388. [DOI] [PubMed] [Google Scholar]
- 8.Buck K W. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618. [Google Scholar]
- 11.Goldbach R, LeGall O, Wellink J. Alpha-like viruses in plants. Semin Virol. 1991;2:19–25. [Google Scholar]
- 12.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1982;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 13.Guschlbauer W, Jankowski K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res. 1980;8:1421–1433. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James K, Ellington A D. The search for missing links between self-replicating nucleic acids and the RNA world. Origins Life Evol Biosphere. 1995;25:515–530. doi: 10.1007/BF01582021. [DOI] [PubMed] [Google Scholar]
- 15.Janda M, French R, Ahlquist P. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of the 5′ extensions on transcript infectivity. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 16.Joyce C-M, Steitz T A. Polymerase structures and function: variations on a theme. J Bacteriol. 1995;177:6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyce G F. RNA evolution and the origins of life. Nature. 1989;338:217–224. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- 18.Kao C C, Sun J-H. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J Virol. 1996;70:6826–6830. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konarska M M, Sharp P A. Replication of RNA by the DNA-dependent RNA-polymerase of phage-T7. Cell. 1989;57:423–431. doi: 10.1016/0092-8674(89)90917-3. [DOI] [PubMed] [Google Scholar]
- 20.Konarska M M, Sharp P A. Structure of RNAs replicated by the DNA-dependent T7 RNA polymerase. Cell. 1990;63:609–618. doi: 10.1016/0092-8674(90)90456-o. [DOI] [PubMed] [Google Scholar]
- 21.Kruger K, Grabowski P J, Zaug A J, Sands J, Gottschling D E, Cech T R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal intervening sequence of tetrahymena. Cell. 1982;31:147–158. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 22.Lazcano A, Guerrero R, Magulis L, Oro J. On the early evolution of RNA polymerase. J Mol Evol. 1988;27:283–290. doi: 10.1007/BF02101189. [DOI] [PubMed] [Google Scholar]
- 23.Lazcano A, Fastag J, Gariglio P, Ramirez C, Oro J. The evolutionary transition from RNA to DNA in early cells. J Mol Evol. 1988;27:365–376. [Google Scholar]
- 24.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA by internal initiation on (−)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 25.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic RNA promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel R W, Bellon L, Beigelman L, Kao C C. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1998;95:11613–11618. doi: 10.1073/pnas.95.20.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivakumaran K, Kao C C. Initiation of genomic plus-strand RNA synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J Virol. 1999;73:6415–6423. doi: 10.1128/jvi.73.8.6415-6423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sousa R, Padilla R. Mutant T7 RNA-polymerase as a DNA-polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steitz T A. A mechanism for all polymerases. Nature. 1998;391:231. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 31.Sun J-H, Adkins S, Faurote G, Kao C C. Initiation of minus-strand RNA synthesis catalyzed by BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 32.Tabor S, Richardson C C. A single residue in the DNA polymerase of the DNA polymerase I family is critical for distinguishing between deoxy and dideoxynucleotides. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uesugi S, Miki H, Ikehara M, Iwahashi H, Kyogoku Y. A linear relationship between electronegativity of 2′-substituents and conformation of adenine nucleosides. Tetrahedron Lett. 1979;42:4073–4076. [Google Scholar]
- 34.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and Ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]