Abstract
Hairy leukoplakia (HL) is a proliferative lesion of the tongue that supports abundant Epstein-Barr virus (EBV) replication. Previous work showed high-level expression of the EBV BMRF2 gene in HL. To characterize the regulation of BMRF2 expression in HL, we mapped the 5′ ends of the BMRF1 and BMRF2 transcripts and showed that BMRF2 is expressed from a novel internal promoter within the BMRF1 coding region. Mechanisms of BMRF2 regulation were compared in oral epithelial cells and B lymphocytes, as were those of BMRF1 and BDLF3, early and late EBV transcripts, respectively, that are also known to be expressed in HL. Basal activity of the putative BMRF2 promoter was 10-fold higher in HSC-3 epithelial cells than in B lymphocytes. The BMRF2 and the BDLF3 promoters were responsive to induction by phorbol ester, but unlike the BMRF1 promoter, they were not responsive to BZLF1 transactivation. By mutational analysis, the major activity of the BMRF2 promoter mapped to a 50-bp region, which includes a TATA-like element and a GC box. The BMRF2 promoter may be regulated differentially from the BMRF1 promoter and more closely resembles that of BDLF3. This novel BMRF2 promoter likely belongs to a class of viral promoters that is more responsive to mechanisms known to induce epithelial cell differentiation, consistent with its high level of expression in HL.
Hairy leukoplakia (HL) is a white lesion typically found on the lateral border of the tongues of immunosuppressed individuals, primarily those with human immunodeficiency virus (HIV) infection (10). HL is associated with high levels of Epstein-Barr virus (EBV) replication in the tongue epithelial tissue. The histopathology of the lesion includes thickening of the epithelium, hyperparakeratosis, and ballooning of cells in the stratum spinosum or prickle cell layer. The lesion contains large numbers of EBV particles in the nuclei of the upper cell layers of the oral epithelium (30). In HL, the full repertoire of EBV gene expression and EBV’s role in the pathogenesis of HL have not been elucidated.
Among the viral transcripts highly expressed in HL are BZLF1, BMRF1, BMRF2, and BDLF3. BZLF1 (Zebra) is the major immediate-early transactivator of EBV early genes and acts as a switch from latency to replication (22, 37). BMRF1 is an early protein, which acts as an accessory protein for the viral polymerase (4, 5, 13, 16). BMRF1 is also a transactivator of the BHLF1 (oriLyt) promoter (39, 40). BMRF2 is an early protein of unknown function. The predicted BMRF2 protein contains multiple hydrophobic domains surrounding a central region of hydrophilic amino acids. Contained within the hydrophilic region is an arginine-glycine-aspartic acid (RGD) motif (27). BDLF3 is a late, virion-associated glycoprotein (17, 26).
Previous work suggested that BMRF1 and BMRF2 may be expressed from the same promoter as part of a bicistronic message (28). However, others have reported that BMRF1 and BMRF2, expressed in induced B lymphocytes, have different cap sites (34). In this report, we performed fine mapping of the start sites of BMRF1 and BMRF2 RNA from an HL biopsy specimen by 5′ rapid amplification of cDNA end (RACE) technology and have defined the natural start sites used by EBV during replication in the tongue. We show evidence that BMRF2 is expressed from a novel, highly active promoter within the BMRF1 open reading frame (ORF).
To determine whether BMRF2 is differentially regulated in different cell types, we compared the regulation of this novel BMRF2 promoter in oral epithelial cells and in B lymphocytes. To ascertain whether BMRF2 regulation was consistent with that of an early or late protein, we compared its regulation to that of BMRF1 and BDLF3 in oral epithelial cells and B lymphocytes. BMRF1 is an early EBV protein whose promoter has been previously described (12). BDLF3 is a virion-associated protein (17, 26); consistent with this, the BDLF3 transcript represents a late viral message and therefore may be regulated differently from early messages.
HL is found predominantly in the setting of HIV infection, which is associated with alterations in patterns of inflammatory cytokine expression. Expression of interleukin-2 (IL-2), IL-6, tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) has each been shown to be altered in HIV infection (14), but the role of these factors in modulating EBV promoter activity and in the pathogenesis of HL is not known. In addition, HIV type 1 (HIV-1) proteins such as Tat have been shown to transactivate gene expression of other viruses (35). We therefore examined the effect of these factors on regulation of the BMRF1, BMRF2, and BDLF3 promoters. Since EBV gene expression is upregulated in the more differentiated cell layers of HL, we also examined the effect of basic fibroblast growth factor (FGF), and transforming growth factor β (TGF-β) on promoter activity, since these factors modulate epithelial cell differentiation (24).
MATERIALS AND METHODS
Cell culture.
Lymphoblastoid cell lines (LCLs) were maintained in RPMI 1640 medium supplemented with 10% newborn calf serum. Oral epithelial cells were maintained in 3:1 medium (three parts keratinocyte growth medium [Clonetics, San Diego, Calif.] to one part Dulbecco modified Eagle medium) supplemented with 10% newborn calf serum. Unless otherwise noted, all media were purchased from the University of California San Francisco (UCSF) Tissue Culture Facility, San Francisco, Calif. B95-8 is an LCL from marmoset lymphocytes transformed with a human EBV strain of infectious mononucleosis (1). P3HR-1 is an LCL strain of Burkitt lymphoma with spontaneous production of EBV particles (11), and BJAB LCL is a Burkitt lymphoma EBV-negative cell line (21). HSC-3 cells are a human oral squamous cell carcinoma line (23).
5′ RACE.
5′ RACE analysis (Gibco BRL-Life Technologies, Inc., Rockville, Md.) was performed with RNA from an HL biopsy specimen to map the 5′ ends of the BMRF1 and BMRF2 transcripts in the HL lesion. For comparison, RNAs from the EBV-positive P3HR-1 and the EBV-negative BJAB lymphocyte cell lines were studied as well. Briefly, RNA was isolated from an HL biopsy specimen, induced P3HR-1 cells, or BJAB cells by using the Qiagen RNA kit (Qiagen, Inc., Chatsworth, Calif.). The four gene-specific primers synthesized for use in 5′ RACE are shown below. 5′ RACE was performed with the protocols provided by the manufacturer (Gibco BRL). Briefly, cDNA was generated from total cell RNA with gene-specific primer 1 (GSP-1). The RNA template was removed by the addition of RNase H and RNase T1. The cDNA was purified, and a homopolymeric tail was added with TdT. The tailed cDNA was amplified with the GSP-2 primer and a 5′ RACE-abridged anchor primer. Finally, this product was amplified again with GSP-4 and 5′ RACE-abridged universal amplification primers. GSP-2 and GSP-3 are nested primers, which were then used to confirm that the product was gene specific. Primers were as follows: GSP-1 reverse, 5′-GATCAAGGCGCTGGCAAT-3′ GSP-2 reverse, 5′-GGCCCTCACCGCGTGCTT-3′ GSP-3 forward, 5′-GGGCCTGTGTCTTCTGTC-3′ GSP-4 reverse, 5′-GGGTACCAGCGTCCAGAGG-3′
Cloning of viral promoters.
Cosmids of the EBV M-ABA nasopharyngeal carcinoma (NPC) strain (29) and EBV from the HL lesion were used to generate viral promoter constructs. By use of the nucleotide sequence from the B95-8 strain, regions upstream of BMRF1 (bp 79296 to 79895), BMRF2 (bp 80640 to 81117), and BDLF3 (bp 131067 to 131246) were chosen to include minimal promoter sequences and were amplified by PCR. To facilitate cloning, GC tails and restriction endonuclease site SacI, XhoI, HindIII, or BglII were added to the 5′ ends of the primers. The oligonucleotides prepared for use as primers for PCR are shown below. The added restriction sites are underlined. BDLF3 forward, 5′-CCGCTCGAGGAGCTCGGTCCTAACTGGTCAGGGGC-3′ BDLF3 reverse, 5′-GGCCCAAGCTTCGCGTCTGGACGCAGAAGGC-3′ BMRF2 forward, 5′-CCCAAGCTTGGTGTTAAATGAGGGGGTTA-3′ BMRF2 reverse, 5′-CCGCTCGAGGAGCTCCTGGCGGCGGCGCTTAGCCT-3′ BMRF1 forward, 5′-CCGCTCGAGGAGCTCCTCCGTGACAAGGTGGCGGC-3′ BMRF1 reverse, 5′-GGCCCAAGCTTGATCACAAGCAGCAGCAGAA-3′
The PCR-generated BMRF1, BMRF2, and BDLF3 promoters were cloned into the reporter gene, pGL2 Basic, which contains a promoterless luciferase (luc) gene (Promega Corporation, Madison, Wis.), digested with XhoI and HindIII or SacI and BglII. The three promoter construct plasmids were named BMRF1-luc, BMRF2-luc (2p1), and BDLF3-luc, respectively. All restriction enzymes were purchased from Boehringer Mannheim (Indianapolis, Ind.).
Mutational analysis of the BMRF2 promoter.
One deletion was made in the BMRF2 promoter by removal of two EagI sites (2p3). Additional deletions in the BMRF2 promoter were made by PCR. The BMRF2-luc 2p4 plasmid was made with BMRF2-3 forward (bp 80820) and BMRF2 reverse primer. The BMRF2-luc 2p6, 2p7, and 2p8 plasmids were made with 2p1 and the Erase-a-Base system (Promega). Plasmid 2p6 begins at bp 80731, 2p7 begins at bp 80799, and 2p8 begins at bp 80749. Mutations were introduced into 2p9 and 2p10 by PCR. Changes in the native sequence are shown in lowercase letters. Plasmid 2p9 was made with T mutation and BMRF2 reverse primer. Plasmid 2p10 was made with GC-box mutation and BMRF2 reverse primer. The sequences are as follows: BMRF2-3 forward, 5′-CCGCTCGAGTCTGAGCCAGAAGATAAAAG-3′ T mutation, 5′-GATCTTAGTGTTtTTTTtTTTttCCACG-3′ GC-box mutation, 5′-TTATTTTATTTAAggAgGggaCCGAAGAGG-3′
DNA sequencing.
Nucleotide sequence analysis of the BMRF1, BMRF2, and BDLF3 promoters was performed with Sequenase version 2.0 (Amersham, Arlington Heights, Ill.) or by the Biomolecular Resource Center (UCSF). Since the promoter clones were generated from cosmids of the M-ABA (NPC) strain and from an HL strain, we determined the nucleotide sequences of these clones and compared them to the published B95-8 strain sequence. The primers used to generate the minimal promoters for BMRF1, BMRF2, and BDLF3 were also used to determine the nucleotide sequence of the promoter clones. The base pair numbers correspond to the coordinates of the B95-8 strain.
Analysis of promoter luciferase activity.
The activity of the BMRF1, BMRF2, and BDLF3 promoters was measured in two cell lines, the epithelial HSC-3 human oral squamous carcinoma cell line (provided by Randall Kramer, UCSF) and the EBV-negative BJAB lymphocyte cell line (provided by Evelyne Lennette, Virolab Inc., Berkeley, Calif.). HSC-3 cells were transfected by a modified calcium phosphate method or by electroporation. The cells were transferred to Dulbecco modified Eagle medium prior to transfection and incubated with the calcium phosphate-DNA mixture for 16 to 18 h in a 5% CO2 incubator. Three micrograms of plasmid DNA was transfected into 5 × 105 cells. The medium was changed at 16 to 18 h posttransfection, and the cells were harvested at 24 h posttransfection in lysis buffer supplied in the Promega luciferase assay system. HSC-3 cells were also transfected by using the Gene Pulser II (Bio-Rad Laboratories, Richmond, Calif.) electroporation system. Ten micrograms of plasmid DNA was added to 106 cells in a 4-cm cuvette (Bio-Rad), incubated on ice for 10 min, and electroporated at 0.220 kV and 975 μF. Both techniques yielded similar results (data shown for cytomegalovirus [CMV] only).
BJAB cells were not successfully transfected by the calcium phosphate method but were electroporated by the Bio-Rad protocol as described above. Ten micrograms of DNA was added to 106 cells in a 4-cm cuvette (Bio-Rad), incubated on ice for 10 min, and electroporated at 0.250 kV and 975 μF. Cells were harvested at 24 h in lysis buffer supplied with the Promega luciferase assay.
To assess the ability of BZLF1 to transactivate the BMRF1, BMRF2, and BDLF3 promoters, HSC-3 cells or BJAB cells were cotransfected with a 2:1 ratio of effector (WZ-het), (provided by George Miller, Yale University, New Haven, Conn.) to target (promoter constructs) and harvested as described above. WZ-het plasmid contains a copy of the BZLF1 gene under positive regulation (22). In phorbol myristate acetate (PMA) (Sigma) induction experiments, the medium was removed from HSC-3 cells after 18 h and fresh medium containing a final concentration of 50 μM PMA was added. HSC-3 cells were analyzed 4 to 24 h after PMA addition.
The role of cytokines and growth factors as potential transactivators in HSC-3 cells was studied as well. In separate experiments, TNF-α (Boehringer Mannheim) was added to the transfected cells at a final concentration of 0.1 ng/ml, IL-2 (Boehringer Mannheim) was added at 1 ng/ml, IFN-γ (Boehringer Mannheim) was added at 1 U/ml, and IL-6 (Sigma) was added at 2 ng/ml. HSC-3 cells were analyzed 4 to 8 h after addition of these factors. Basic FGF (Boehringer Mannheim) was added to cells 72 h prior to transfection at a concentration of 20 ng/ml, and TGF-β (Gibco BRL) was added to cells 72 h prior to transfection at a concentration of 0.05 ng/ml. Lastly, HSC-3 cells were cultured in the presence of HIV-1 tat, by adding supernatants from the pANT retroviral cell line expressing tat (a gift from Mario Roederer, Stanford University, Stanford, Calif.) for 24 h prior to transfection. pANT7 cells are a retrovirus-producing line which produces a defective murine Moloney retrovirus that has a tat expression vector. Upon expression of pANT7 cells, the tat-expressing retrovirus produces sufficient tat to up-regulate an HIV-long terminal repeat reporter construct (36).
Promoter activity was measured by a luciferase assay according to the manufacturer’s instructions. The positive control for transfection and for the luciferase assay was the luciferase gene cloned under the control of the CMV major immediate-early promoter (CMV-luc). The negative control was the promoterless pGL Basic (Promega). Samples were analyzed on an ML2200 luminometer (Dynatech Laboratories, Chantilly, Va.), and promoter activity was expressed in relative light units (RLU).
RESULTS
5′ RACE.
The 5′ ends of the BMRF1 and BMRF2 transcripts in HL were mapped by a 5′ RACE method. Two products were detected from the 5′ RACE corresponding to 5′ ends of the BMRF1 and BMRF2 transcripts. The BMRF1 product corresponds to the larger, 1,416-bp, band, and the BMRF2 product corresponds to the smaller, 453-bp, band (Fig. 1A). Nested primers GSP-2 and GSP-3 amplified a 189-bp gene-specific product. No bands were amplified with BJAB RNA (Fig. 1A). The 5′ end of the BMRF2 transcript mapped to bp 80862, within the BMRF1 ORF, 29 bp from a predicted TATA box (Fig. 1B). The 5′ end of the BMRF1 transcript mapped to bp 79899, 31 bp from a predicted TATA box (Fig. 1B). 5′ RACE mapping of the BMRF2 transcript from PMA-induced P3HR-1 B lymphocytes yielded two bands; the larger band mapped to bp 80896, 63 bp from the predicted TATA box, and the smaller band mapped to bp 81034, 201 bp from the predicted TATA box (data not shown).
FIG. 1.
(A) Agarose gel analysis of 5′ RACE products. Lane 1, molecular size markers (1-kb ladder); lane 2, HL 5′ RACE; lane 3, control for gene-specific product; lane 4, BJAB 5′ RACE. (B) Diagram of 5′ RACE analysis of HL RNA. The top lines diagram a region of the BamHI restriction fragment M of EBV. The predicted TATA boxes, cap sites, and initiating methionine (Met) are noted. The lower lines indicate the bands from the 5′ RACE analysis with arrows indicating the 5′ start site of BMRF1 (bp 79871) and BMRF2 (bp 80862).
Viral promoter characterization.
The nucleotide sequence of the putative BMRF2 promoter from an HL biopsy specimen-isolated EBV strain was determined, as were the sequences of the BMRF1, BMRF2, and BDLF3 promoter constructs amplified from the M-ABA strain. All were identical to the published nucleotide sequence of the B95-8 strain and the Raji strain (data not shown). The BMRF1, BMRF2, and BDLF3 promoters are shown schematically in Fig. 2.
FIG. 2.
Schematic diagram of the BMRF1, BMRF2, and BDLF3 promoters used to make luciferase reporter constructs. TATA boxes, DNA binding elements, GC-rich regions, predicted cap sites, the polyadenylation site, and in-frame stop codons are noted. Numbers correspond to the coordinates from the published sequence of the B95-8 EBV lymphocyte strain.
To determine whether the BMRF1, BMRF2, and BDLF3 transcripts were differentially regulated in epithelial cells and lymphocytes, we compared the basal activities of the putative promoters of these genes in the HSC-3 oral cancer cell line and the BJAB lymphocyte cell line. Measurement of luciferase expression under control of the CMV promoter was performed with each assay. In both the epithelial cells and the B lymphocytes, the BMRF2 promoter showed the highest basal activity of the promoters tested (Fig. 3A and B). In epithelial cells, the BMRF2 promoter was generally only 10-fold less active than the CMV promoter control. The basal activity of the BMRF2 promoter in epithelial cells was, on the average, 10-fold higher than the basal activity in B lymphocytes. The BMRF1 and BDLF3 promoters had low basal activity in both epithelial cells and lymphocytes (Fig. 3A and B). All methods of DNA introduction gave similar results in experiments with the CMV promoter (Fig. 3C).
FIG. 3.
Promoter activity of BMRF1, BMRF2, and BDLF3 in epithelial and lymphocyte cell lines. Cells were transfected with promoterless luciferase plasmid or plasmid constructs containing minimal promoter sequences from BMRF1 (bp 79296 to 79895), BMRF2 (bp 80640 to 81117), and BDLF3 (bp 131067 to 131246) as described in Materials and Methods. Basal activity is shown. (A and B) HSC-3 epithelial cells (A) or BJAB lymphocyte cell lines (B) were transfected with the BMRF1, BMRF2, or BDLF3 promoter construct or with pGL plasmid alone. (C) CMV-luciferase control plasmid was transfected by various methods and with various cell types. (D) Promoter constructs were cotransfected with the WZ plasmid expressing BZLF1 or induced with PMA. Promoter activity was reported as luciferase RLU or fold induction.
The activity of the three promoters was determined following cotransfection with the WZ-het (BZLF1) viral transactivator plasmid. In HSC-3 epithelial cells, cotransfection with WZ-het caused a 30-fold induction of BMRF1 (Fig. 3D). In B lymphocytes, WZ-het caused a 22-fold induction of BMRF1 (Fig. 3D). In contrast, WZ-het had little effect on the BMRF2 and BDLF3 promoters in either epithelial cells or B lymphocytes (Fig. 3D).
Promoter activity was also examined following induction with PMA. In contrast to the effect of WZ-het, PMA had no effect on BMRF1 in epithelial cells (Fig. 3D) but did induce expression of both the BMRF2 and the BDLF3 promoters by 10-fold in epithelial cells and lymphocytes (Fig. 3D).
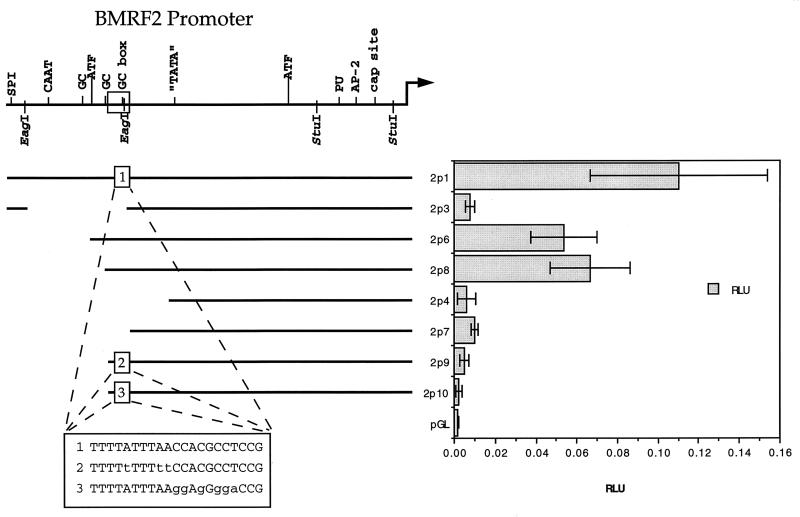
BMRF2 promoter analysis and deletions.
We investigated the region responsible for the high level of activity in the BMRF2 promoter in HSC-3 cells. Removal of a 50-bp region (80749 to 80799) shown as a box in Fig. 4 resulted in a 10-fold reduction in BMRF2 promoter activity. Contained within this region were two predicted transcription elements, a TATA-like region and a GC box. We separately mutated either the TATA region or the GC box, each of which is shown at the bottom of Fig. 4 compared to the native sequence. Both mutations resulted in loss of promoter activity (Fig. 4).
FIG. 4.
Deletion analysis of the BMRF2 promoter. The BMRF2 promoter is shown on top, and the promoter deletions are shown below. Luciferase activity is reported in RLU to the right of the promoter constructs. A boxed region encompasses the area responsible for the high level of promoter activity. The box below shows the changes in the TA-rich region and GC box.
Analysis of growth factors, cytokines, and tat.
The cytokines (IL-2, IL-6, TNF-α, and IFN-γ), growth factors (basic FGF and TGF-β), and HIV-1 tat that were tested had no effect on the expression of the BMRF1, BMRF2, and BDLF3 promoters when tested in oral epithelial cells (data not shown).
DISCUSSION
HL is a unique lesion characterized by abundant lytic viral replication. Unlike the restricted repertoire of EBV genes expressed during latency in B lymphocytes, the repertoire of EBV genes expressed in HL includes both lytic and latency genes, similar to primary infection. Therefore, transcription of viral messages in HL represents a natural state of lytic infection. Consistent with this, mapping the 5′ ends of both BMRF1 and BMRF2 transcripts from HL revealed start sites about 30 bp from the predicted TATA box.
There were no nucleotide sequence changes noted in the BMRF1, BMRF2, and BDLF3 promoter regions from both the M-ABA strain and an HL strain compared to B95-8 and Raji sequences. This suggests that there is a high level of conservation of the promoter regions, in contrast to the changes that were noted in the coding sequence of the BDLF3 gene in HL strains (27).
Little is known about the factors that regulate the lytic transcripts of EBV such as BMRF2 and BDLF3. We have shown that BMRF2 is expressed from a novel, highly active promoter within the BMRF1 ORF. However, this does not preclude the possibility that BMRF2 is expressed from the BMRF1 promoter as well. It is interesting to note that the DNA binding and polymerase processivity domains of BMRF1 are located within the first 900 bp (2, 15). Recently, Zhang et al. reported that the carboxy-terminal 26 amino acids (the last 78 bp) of BMRF1 are required for BHLF1 promoter activation and nuclear localization (38). Our data show that this 3′ region (the last 360 bp of BMRF1) includes the functional promoter for BMRF2. Internal promoter usage in EBV for the LMP1 transcript was previously described (3).
Others have suggested that the early transcript BMRF1 is regulated in a cell-specific manner (12). Our data show that the BMRF1 promoter has low basal activity and is transactivated by WZ-het (the BZLF1 gene under positive regulation) in oral epithelial cells, consistent with earlier reports for HeLa and HEp-2 cells (12). Holley-Guthrie et al. reported that both BZLF1 and BRLF1 were required for transactivation in lymphocytes (12). However, in our studies, WZ-het alone transactivated the BMRF1 promoter when transfected into BJAB cells. This may reflect differences in cell types used for the assays, since Holley-Guthrie et al. used Jurkat and Louckes cells.
In contrast to BMRF1, both BMRF2 and BDLF3 were stimulated by PMA, but not WZ-het, and these likely represent a class of viral promoters that are more responsive to mechanisms known to induce epithelial cell differentiation. Although BMRF2 is an early promoter, it is not regulated in the same manner as the BMRF1 early promoter. The BMRF1 promoter contains two consensus AP1 binding sites, which BZLF1 binds during activation of the BMRF1 promoter (8). Neither the BMRF2 nor the BDLF3 promoter contains consensus AP1 sites.
Another EBV promoter, BHLF1, is activated by either BZLF1 or BMRF1; however, when both BZLF1 and BMRF1 are transfected, the level of BHLF1 promoter activity is greatly increased (40). We cannot exclude the possibility that the BMRF2 and BDLF3 promoters may be transactivated in vivo by a combination of WZ-het and another transactivator, which we have not tested in our system. Regulation of early and late promoters by multiple transactivators allows enhanced transcription as viral gene expression proceeds. Within the HL lesion, epithelial cell differentiation and changes in the level of certain transcription factors may also play a role in promoter regulation.
Overall, there was little difference in promoter activity of BMRF1 and BDLF3 between lymphocytes and epithelial cells. In contrast, the promoter activity of BMRF2 was 10-fold higher in epithelial cells than in lymphocytes, indicating that it may be modulated by specific cellular factors found in epithelial cells. Genes such as BMRF2 which are highly expressed in epithelial cells may have more notable effects in this cell type and thus may contribute to the pathology of the HL lesion. This is interesting because the putative BMRF2 protein contains an RGD motif. Although the function of the BMRF2-RGD motif is unknown, RGD motifs are found in proteins that function in cellular attachment. Both adenovirus and coxsackievirus contain proteins that use an RGD motif in penetration of cells via cellular integrins (6, 9, 19, 20, 32). Therefore, the BMRF2-RGD motif may function in a similar manner and contribute to viral spread within the HL lesion.
Analysis of the BMRF2 promoter revealed a 50-bp region responsible for a majority of the promoter activity. Contained within this region is a TA-rich region and a GC box. The GC box is a unique feature of the BMRF2 promoter and not found in the BDLF3 promoter, although the BDLF3 promoter does have GC-rich regions. Changes in either the GC box or the TA-rich region resulted in over a 10-fold loss of BMRF2 promoter activity. Although our 5′ RACE analysis suggested that this TA-rich region is distinct from the predicted TATA box, it may serve another modulatory role that we have not yet examined. GC boxes are found in a number of viral and cellular promoters and bind to the ubiquitous Sp family of transcription factors (18). Sp1 proteins have been shown to be important transactivators of regulated gene expression in keratinocytes (7) and thus may play a similar role in modulating the activity of the BMRF2 promoter in the oral epithelium.
Several EBV promoters, which are active in epithelial cells, contain Sp1 or Sp1-like binding sites. These include the BHLF1 (oriLyt) promoter, the BNLF1 (LMP1) promoter, and the promoter for LMP1 3.5-kb transcript (15, 25, 33, 39). The LMP1 3.5-kb transcript is the major transcript in HL (15) and is also expressed in the epithelial cells of NPC (33). The promoter for the LMP1 3.5-kb transcript is located in the terminal repeat region and contains four Sp1 sites. A second LMP1 transcript of 2.8 kb is expressed predominately in B-lymphocyte lines, and its promoter contains only one Sp1 site (33). This is further evidence that, in epithelial cells, promoters containing GC boxes may be more active. In addition to Sp1, other novel Sp1-like transcription factors, such as keratinocyte-specific factor, are active in squamous epithelial cells (25). Another transcription factor, GC-box binding protein (GBP-i), was inducible by phorbol ester (31), as was the activity of the BMRF2 promoter.
Factors that may be important specifically in the setting of HIV infection, such as IL-6 and HIV-1 tat, had no effect on the expression of the BMRF1, BMRF2, and BDLF3 promoters when tested in oral epithelial cells. It is likely that these factors do not play a direct role in the high level of expression of these genes in HL, but we cannot exclude a role in the activation of other promoters not examined in the study, such as that of BZLF1. Furthermore, the tongue provides a unique environment of differentiated epithelium, and while HSC-3 oral epithelial cells may provide some of the necessary transcription factors, others may be differentiation specific.
In summary, we and others have shown evidence that promoters of EBV are regulated in a cell-specific manner. Both latent and lytic promoters can be differentially regulated depending on the cell type infected by the virus. The transcription factor Sp1 may be playing a key role in EBV transcription in epithelial cells.
ACKNOWLEDGMENTS
We thank Jenny Berline for excellent technical assistance.
This work was supported by NIDR grant DEO7946 and by a grant from the UCSF AIDS Clinical Research Center (ACRC) funded by the University of California AIDS Research Program (UARP).
REFERENCES
- 1.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Chen L W, Lin L S, Chang Y S, Liu S T. Functional analysis of EA-D of Epstein-Barr virus. Virology. 1995;211:593–597. doi: 10.1006/viro.1995.1443. [DOI] [PubMed] [Google Scholar]
- 3.Chen M L, Hsu N C, Liu S T, Chang Y S. Identification of an internal promoter of the latent membrane protein 1 gene of Epstein-Barr virus. DNA Cell Biol. 1995;14:205–211. doi: 10.1089/dna.1995.14.205. [DOI] [PubMed] [Google Scholar]
- 4.Chiou J F, Li J K, Cheng Y C. Demonstration of a stimulatory protein for virus-specified DNA polymerase in phorbol ester-treated Epstein-Barr virus-carrying cells. Proc Natl Acad Sci USA. 1985;82:5728–5731. doi: 10.1073/pnas.82.17.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho M S, Milman G, Hayward S D. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J Virol. 1985;56:860–866. doi: 10.1128/jvi.56.3.860-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuzange A, Chroboczek J, Jacrot B. The penton base of human adenovirus type 3 has the RGD motif. Gene. 1994;146:257–259. doi: 10.1016/0378-1119(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 7.Eckert R L, Crish J F, Banks E B, Welter J F. The epidermis: genes on—genes off. J Investig Dermatol. 1997;109:501–509. doi: 10.1111/1523-1747.ep12336477. [DOI] [PubMed] [Google Scholar]
- 8.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman M J, Wilson J M. Expression of αvβ5 integrin is necessary for efficient adenovirus-mediated gene transfer in the human airway. J Virol. 1995;69:5951–5958. doi: 10.1128/jvi.69.10.5951-5958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenspan J S, Greenspan D, Lennette E T, Abrams D I, Conant M A, Petersen V, Freese U K. Replication of Epstein-Barr virus within the epithelial cells of oral “hairy” leukoplakia, an AIDS-associated lesion. N Engl J Med. 1985;313:1564–1571. doi: 10.1056/NEJM198512193132502. [DOI] [PubMed] [Google Scholar]
- 11.Hinuma Y, Konn M, Yamaguchi J, Wudarski D J, Blakeslee J R, Jr, Grace J T., Jr Immunofluorescence and herpes-type virus particles in the P3HR-1 Burkitt lymphoma cell line. J Virol. 1967;1:1045–1051. doi: 10.1128/jvi.1.5.1045-1051.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holley-Guthrie E A, Quinlivan E B, Mar E C, Kenney S. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallin B, Sternas L, Saemundssen A K, Luka J, Jornvall H, Eriksson B, Tao P Z, Nilsson M T, Klein G. Purification of Epstein-Barr virus DNA polymerase from P3HR-1 cells. J Virol. 1985;54:561–568. doi: 10.1128/jvi.54.2.561-568.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kehrl J H, Rieckmann P, Kozlow E, Fauci A S. Lymphokine production by B cells from normal and HIV-infected individuals. Ann NY Acad Sci. 1992;651:220–227. doi: 10.1111/j.1749-6632.1992.tb24617.x. [DOI] [PubMed] [Google Scholar]
- 15.Kiehl A, Dorsky D I. Bipartite DNA-binding region of the Epstein-Barr virus BMRF1 product essential for DNA polymerase accessory function. J Virol. 1995;69:1669–1677. doi: 10.1128/jvi.69.3.1669-1677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiehl A, Dorsky D I. Cooperation of EBV DNA polymerase and EA-D(BMRF1) in vitro and colocalization in nuclei of infected cells. Virology. 1991;184:330–340. doi: 10.1016/0042-6822(91)90849-7. [DOI] [PubMed] [Google Scholar]
- 17.Kurilla M G, Heineman T, Davenport L C, Kieff E, Hutt-Fletcher L M. A novel Epstein-Barr virus glycoprotein gp150 expressed from the BDLF3 open reading frame. Virology. 1995;209:108–121. doi: 10.1006/viro.1995.1235. [DOI] [PubMed] [Google Scholar]
- 18.Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 19.Liebermann H, Dolling R, Schmidt D, Thalmann G. RGD-containing peptides of VP1 of foot-and-mouth disease virus (FMDV) prevent virus infection in vitro. Acta Virol. 1991;35:90–93. [PubMed] [Google Scholar]
- 20.Mathias P, Wickham T, Moore M, Nemerow G. Multiple adenovirus serotypes use αv integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menezes J, Leibold W, Klein G, Clements G. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine. 1975;22:276–284. [PubMed] [Google Scholar]
- 22.Miller G. The switch between latency and replication of Epstein-Barr virus. J Infect Dis. 1990;161:833–844. doi: 10.1093/infdis/161.5.833. [DOI] [PubMed] [Google Scholar]
- 23.Momose F, Araida T, Negishi A, Ichijo H, Shioda S, Sasaki S. Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J Oral Pathol Med. 1989;18:391–395. doi: 10.1111/j.1600-0714.1989.tb01570.x. [DOI] [PubMed] [Google Scholar]
- 24.Moses H L. TGF-beta regulation of epithelial cell proliferation. Mol Reprod Dev. 1992;32:179–184. doi: 10.1002/mrd.1080320215. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa H, Inomoto T, Rustgi A K. A CACCC box-like cis-regulatory element of the Epstein-Barr virus ED-L2 promoter interacts with a novel transcriptional factor in tissue-specific squamous epithelia. J Biol Chem. 1997;272:16688–16699. doi: 10.1074/jbc.272.26.16688. [DOI] [PubMed] [Google Scholar]
- 26.Nolan L A, Morgan A J. The Epstein-Barr virus open reading frame BDLF3 codes for a 100–150 kDa glycoprotein. J Gen Virol. 1995;76:1381–1392. doi: 10.1099/0022-1317-76-6-1381. [DOI] [PubMed] [Google Scholar]
- 27.Peñaranda M E, Lagenaur L A, Pierik L T, Berline J W, MacPhail L A, Greenspan D, Greenspan J S, Palefsky J M. Expression of Epstein-Barr virus BMRF-2 and BDLF-3 genes in hairy leukoplakia. J Gen Virol. 1997;78:3361–3370. doi: 10.1099/0022-1317-78-12-3361. [DOI] [PubMed] [Google Scholar]
- 28.Pfitzner A J, Strominger J L, Speck S H. Characterization of a cDNA clone corresponding to a transcript from the Epstein-Barr virus BamHI M fragment: evidence for overlapping mRNAs. J Virol. 1987;61:2943–2946. doi: 10.1128/jvi.61.9.2943-2946.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polack A, Hartl G, Zimber U, Freese U K, Laux G, Takaki K, Hohn B, Gissmann L, Bornkamm G W. A complete set of overlapping cosmid clones of M-ABA virus derived from nasopharyngeal carcinoma and its similarity to other Epstein-Barr virus isolates. Gene. 1984;27:279–288. doi: 10.1016/0378-1119(84)90072-6. [DOI] [PubMed] [Google Scholar]
- 30.Rabanus J P, Greenspan D, Petersen V, Leser U, Wolf H, Greenspan J S. Subcellular distribution and life cycle of Epstein-Barr virus in keratinocytes of oral hairy leukoplakia. Am J Pathol. 1991;139:185–197. [PMC free article] [PubMed] [Google Scholar]
- 31.Raj G V, Khalili K. Identification and characterization of a novel GGA/C-binding protein, GBP-i, that is rapidly inducible by cytokines. Mol Cell Biol. 1994;14:7770–7781. doi: 10.1128/mcb.14.12.7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roivainen M, Hyypia T, Piirainen L, Kalkkinen N, Stanway G, Hovi T. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J Virol. 1991;65:4735–4740. doi: 10.1128/jvi.65.9.4735-4740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadler R H, Raab-Traub N. The Epstein-Barr virus 3.5-kilobase latent membrane protein 1 mRNA initiates from a TATA-less promoter within the first terminal repeat. J Virol. 1995;69:4577–4581. doi: 10.1128/jvi.69.7.4577-4581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sample J, Lancz G, Nonoyama M. Mapping of genes in BamHI fragment M of Epstein-Barr virus DNA that may determine the fate of viral infection. J Virol. 1986;57:145–154. doi: 10.1128/jvi.57.1.145-154.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sieczkowski L, Chandran B, Wood C. The human immunodeficiency virus tat gene enhances replication of human herpesvirus-6. Virology. 1995;211:544–553. doi: 10.1006/viro.1995.1436. [DOI] [PubMed] [Google Scholar]
- 36.Staal F J, Roederer M, Herzenberg L A, Herzenberg L A. Intracellular thiols regulate activation of nuclear factor kappa B and transcription of human immunodeficiency virus. Proc Natl Acad Sci USA. 1990;87:9943–9947. doi: 10.1073/pnas.87.24.9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young L S, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F, Rowe D T, Greenspan D, Greenspan J S, Rickinson A B, Farrell P J. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol. 1991;65:2868–2874. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Holley-Guthrie E, Dorsky D, Kenney S. Identification of transactivator and nuclear localization domains in the Epstein-Barr virus DNA polymerase accessory protein, BMRF1. J Gen Virol. 1999;80:69–74. doi: 10.1099/0022-1317-80-1-69. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Q, Holley-Guthrie E, Ge J Q, Dorsky D, Kenney S. The Epstein-Barr virus (EBV) DNA polymerase accessory protein, BMRF1, activates the essential downstream component of the EBV oriLyt. Virology. 1997;230:22–34. doi: 10.1006/viro.1997.8470. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q, Hong Y, Dorsky D, Holley-Guthrie E, Zalani S, Elshiekh N A, Kiehl A, Le T, Kenney S. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J Virol. 1996;70:5131–5142. doi: 10.1128/jvi.70.8.5131-5142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]