Abstract
The three-dimensional structure of expressed VP2 capsids of Aleutian mink disease parvovirus strain G (ADVG-VP2) has been determined to 22 Å resolution by cryo-electron microscopy and image reconstruction techniques. A structure-based sequence alignment of the VP2 capsid protein of canine parvovirus (CPV) provided a means to construct an atomic model of the ADVG-VP2 capsid. The ADVG-VP2 reconstruction reveals a capsid structure with a mean external radius of 128 Å and several surface features similar to those found in human parvovirus B19 (B19), CPV, feline panleukopenia virus (FPV), and minute virus of mice (MVM). Dimple-like depressions occur at the icosahedral twofold axes, canyon-like regions encircle the fivefold axes, and spike-like protrusions decorate the threefold axes. These spikes are not present in B19, and they are more prominent in ADV compared to the other parvoviruses owing to the presence of loop insertions which create mounds near the threefold axes. Cylindrical channels along the fivefold axes of CPV, FPV, and MVM, which are surrounded by five symmetry-related β-ribbons, are closed in ADVG-VP2 and B19. Immunoreactive peptides made from segments of the ADVG-VP2 capsid protein map to residues in the mound structures. In vitro tissue tropism and in vivo pathogenic properties of ADV map to residues at the threefold axes and to the wall of the dimples.
Aleutian mink disease parvovirus (ADV) causes chronic disease and persistent infection in mink as manifested by marked hypergammaglobulinemia, plasmacytosis, and immune complex disease (18, 48, 49). Though a large fraction of the elevated gammaglobulins in infected mink is directed against the viral particle, the virus is not neutralized in vivo but circulates as infectious virus-antibody complexes. Disease caused by ADV differs significantly from that caused by other parvoviruses in which infection leads to a protective host response. For ADV, the presence of the capsid elicits a host response that actually contributes to the disease process (50, 51). Recent studies, in which the ADV capsid proteins were expressed in a series of nonoverlapping fragments, indicate that the unusual antibody response is targeted to specific epitopes (19).
The ADV capsid is a critical determinant of cellular host range and pathogenicity. For example, ADV-Utah 1 is highly pathogenic and does not grow in cell culture, whereas ADV-G replicates permissively in cell culture and is nonpathogenic in mink (6, 15, 20, 37). Analysis of full-length molecular clones that are chimeric between ADV-G and ADV-Utah has mapped the determinants which regulate these characteristics to capsid gene sequences (16, 17). Similar differences in tissue tropism and pathogenicity have also been mapped to the capsid genes for other parvoviruses, such as porcine parvovirus (PPV), minute virus of mice (MVM), and the closely homologous canine parvovirus (CPV) and feline panleukopenia virus (FPV) (3, 5, 11, 12, 14, 22, 35, 36, 40, 46, 47, 62).
The central role of ADV virions in the genesis of immune disorder and determination of host range and pathogenicity has focused attention on the ADV capsid. ADV contains two viral capsid proteins, VP1 (85 kDa) and VP2 (75 kDa), which are translated from the same mRNA (7) and have identical sequences with the exception of 42 additional amino acids at the amino terminus of the minor capsid protein, VP1. Native ADV virions contain 60 protein subunits, with VP1 and VP2 comprising, respectively, about 10 and 90% of the total. ADV capsid proteins expressed in recombinant vaccinia or baculovirus self-assemble into particles which appear to have the physicochemical and antigenic properties of native virions, though the ratio of VP1 to VP2 in the expressed capsids is reversed (26, 27, 68). Expressed VP2, the major capsid protein, can self-assemble into native-like capsids in the absence of VP1 (26), as has also been observed for other parvoviruses in mammalian cells (64).
The atomic structures of CPV, FPV, and MVM have been determined by X-ray crystallography techniques (3, 5, 41, 63, 69). Particles of these viruses contain a total of 60 copies of the capsid proteins, VP1, VP2, and VP3, in full (DNA-containing) virions (59). VP3 is formed in full virions (but not empty capsids) by cleavage of approximately 20 amino acids from the amino terminus of VP2 (28, 45, 60, 65). This cleavage process does not occur in ADV or human parvovirus B19 (B19). The capsid structures of CPV, FPV, and MVM consisted of 60 subunits of the region common to VP2. The unique VP1 amino termini of these capsids were not observed in their structures due to their low copy number (6 to 9%) in the capsid, which results in signal weakening during the structure determination averaging procedure utilizing noncrystallographic symmetry for refinement and phase extension (54). The core of VP2 is composed of an eight-stranded (strands B through I), antiparallel, β-barrel motif (55). Four large loop insertions between the β strands primarily constitute the capsid surface structure. In addition, a cylindrical channel lies at each of the 12 fivefold icosahedral axes. The walls of the channel are formed by insertions between the D and E strands of the β-barrel. It is postulated that one capsid protein at each fivefold axis has its N terminus externalized via the channel, and this enables maturation cleavage of VP2 to VP3 in virions, consistent with biochemical data (28, 45, 60, 65). The capsids of these parvoviruses have a small, spike-like protrusion (referred to hereafter as the spike) along each icosahedral threefold axis, a dimple-like depression (hereafter, the dimple) at each icosahedral twofold axis, and a canyon-like depression (hereafter, the canyon) around each fivefold axis. B19 differs slightly from these parvoviruses. Though it has the β-barrel motif and the variable surface loops, surface depressions rather than spikes occur at the threefold axes and there is density on the fivefold axes (4, 25). This correlates with the observation that VP2 cleavage to VP3 does not occur in B19 and hence supports the hypothesis that the fivefold channel is the site of externalization of N termini during the cleavage process that occurs in the other parvoviruses.
The available structural information indicates that differences in the capsid surface are what determine tissue tropism, pathogenicity, and antigenicity among different parvoviruses and among the strains of a particular virus. For example, the spike region is highly immunogenic in CPV and FPV (58, 67), and the dimple and canyon features are postulated to be possible sites for receptor interactions (5, 13, 24, 61). Residues that confer tissue tropism for CPV, FPV, and MVM are located at the top and shoulder of the spikes (5, 40) and in or near the dimples (3). The cellular receptor for B19, globoside, docks into the depression at the threefold axes of the B19 capsid (25), which is surrounded by regions where neutralizing antibodies are thought to bind, thus possibly explaining the mechanism for B19 neutralization.
Here, we present a cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction study of the structure at 22 Å resolution of baculovirus-expressed VP2 particles of ADV strain G (ADVG-VP2). We also report a modified amino acid sequence alignment of the ADV-G and CPV VP2 proteins, based in part on knowledge of the atomic structure of CPV. This alignment provided a rationale for generating a pseudoatomic model of the ADVG-VP2 capsid and fitting it into the cryo-EM density map. Analysis of this model shows that protrusions on the capsid surface, like that of other parvoviruses, serve as antigenic sites. The model also reveals possible sites near or at the twofold and threefold axes of the capsid that may contribute to tissue tropism and disease pathogenicity.
MATERIALS AND METHODS
Propagation and purification of ADVG-VP2 particles.
ADVG-VP2 particles were produced at 28°C in suspension cultures of Spodoptera frugiperda (Sf9) cells singly infected at a multiplicity of 1 PFU/cell with the recombinant baculovirus Autographa californica nuclear polyhedrosis virus (AcNPv), encoding the structural protein VP2 of ADV (26). The Sf9-infected cultures were harvested 72 h after inoculation by centrifugation at 15,300 × g for 15 min at 4°C, using a JA-14 rotor (Beckman). The cell pellets were suspended in 50 mM Tris buffer (pH 8.0) containing 0.15 M NaCl, 0.2% Triton X-100, and a cocktail of protease inhibitors (1 μg of pefabloc SC/ml, 1 μg of pepstatin/ml, 1 μg of aprotonin/ml, and 5 μg of leupeptin/ml). The suspension was subjected to four cycles of freeze-thawing with liquid nitrogen and three cycles of sonication for 15 s on ice followed by centrifugation at 15,300 × g for 30 min at 4°C with a JA-14 rotor. The supernatant was then clarified twice with an equal volume of chloroform and blended for 60 s at 4°C and sedimented by centrifugation at 15,300 × g for 30 min at 4°C, using a JA-14 rotor. ADVG-VP2 particles were then precipitated by adding solid polyethylene glycol 8000 (10% [wt/vol]) to the supernatant. After overnight incubation at 4°C, the precipitate was collected by centrifugation at 11,000 × g for 30 min at 4°C, using a JA-14 rotor. The pellets were resuspended in 50 mM Tris buffer (pH 8.0) containing 50 mM NaCl. The virus concentration was adjusted to 1 mg/ml, assuming an extinction coefficient of 1.0 mg/ml per cm at 280 nm.
Cryo-EM and 3D image reconstruction.
Small aliquots (3.5 μl) of the ADVG-VP2 sample were placed on holey carbon-coated, copper grids, blotted with filter paper, and plunged into liquid ethane to suspend the particles in a thin layer of vitreous ice as described previously (2, 33, 42, 43). The grids were inserted into a precooled, Gatan 626 cryotransfer holder (Gatan Inc., Warrendale, Pa.) that maintained a constant temperature of −186°C. Preliminary images of frozen-hydrated ADVG-VP2 samples were recorded with a CM120 electron microscope (Philips Electronic Instruments, Mahwah, N.J.) and a 3D reconstruction was computed. The initial orientation and origin parameters of the images for the reconstruction were determined by a model-based refinement approach (10) that used a 3D reconstruction of B19 (25) as the initial model. Subsequent capsid samples were examined with a Philips CM200 FEG transmission electron microscope, and images were recorded with a Gatan 794 Multiscan charge-coupled device (CCD) camera. The software used for automatically recording the images was the Low-Dose plug-in (version 1.0) in Gatan’s Digital Micrograph (version 2.5.9) program (43, 44). The size of the electron beam was adjusted to fall just within the limits of the 1K2 CCD detector (0.4 μm on the specimen) to allow images to be recorded without preirradiating adjacent regions (Fig. 1). A low-magnification (×4,560) survey image was captured on the CCD, and 10 separate areas for high-magnification imaging were identified and selected with a cursor on a computer graphics screen (Fig. 1). The 10 images were then automatically recorded at 200 kV, ×60,000 magnification (pixel size corresponds to 4.0 Å at the specimen), 1.4 μm underfocus, and with a dose of ∼14.5 e−/Å2 per image (43). Of the 106 individual particle images selected from the CCD images, 87 were used to compute a 3D reconstruction to 22 Å resolution. The resolution was estimated according to established procedures (9). The orientation and origin parameters for each particle image were determined as described above with the preliminary ADV reconstruction serving as the initial model (10). All calculated eigenvalues exceeded 10.0, which indicated that random and unique data were used for each reconstruction (31).
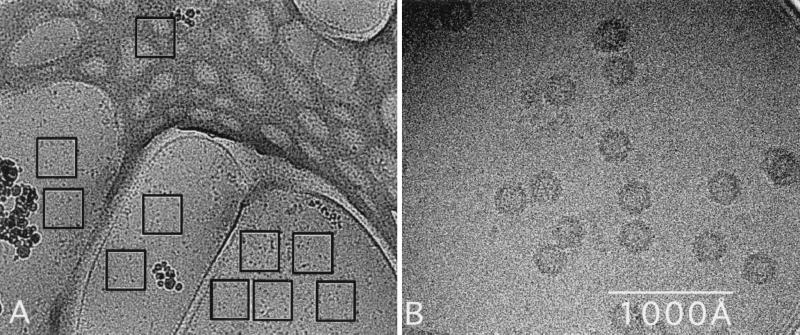
FIG. 1.
Micrograph of unstained ADVG-VP2 particles suspended in a layer of vitreous ice over holes in a carbon support film. (A) A low-magnification survey image of nine user-selected areas (black open squares) over holes in the carbon support film and one in the carbon film. (B) A portion of a 1K2 image recorded with spot-scan procedures on a slow-scan CCD (43).
A best fit between the ADVG-VP2 reconstruction (Fig. 2) and the atomic structure of the T=1 CPV particle (63) was achieved by comparing structure factors computed from the reconstructed density map (FcryoEM) at different map grid sizes with the structure factors computed from the CPV model (FCPV). Structure factors were calculated and compared using the CCP4 software package (29). The best fit was determined by adjusting the scale of the reconstruction to minimize the R factor, calculated as (Σ|FCPV −FcryoEM|/ΣFCPV) × 100.
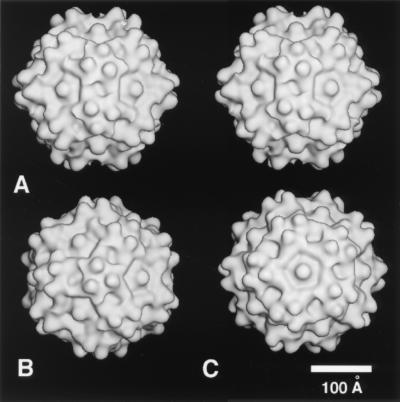
FIG. 2.
Shaded-surface representations of the ADVG-VP2 3D reconstruction viewed down a twofold icosahedral axis (in stereo) (A), down a threefold axis (B), and down a fivefold axis (C). The reconstruction was computed to 22 Å resolution from 87 different ADVG-VP2 particle images.
Alignment and modeling of ADVG-VP2 capsid protein.
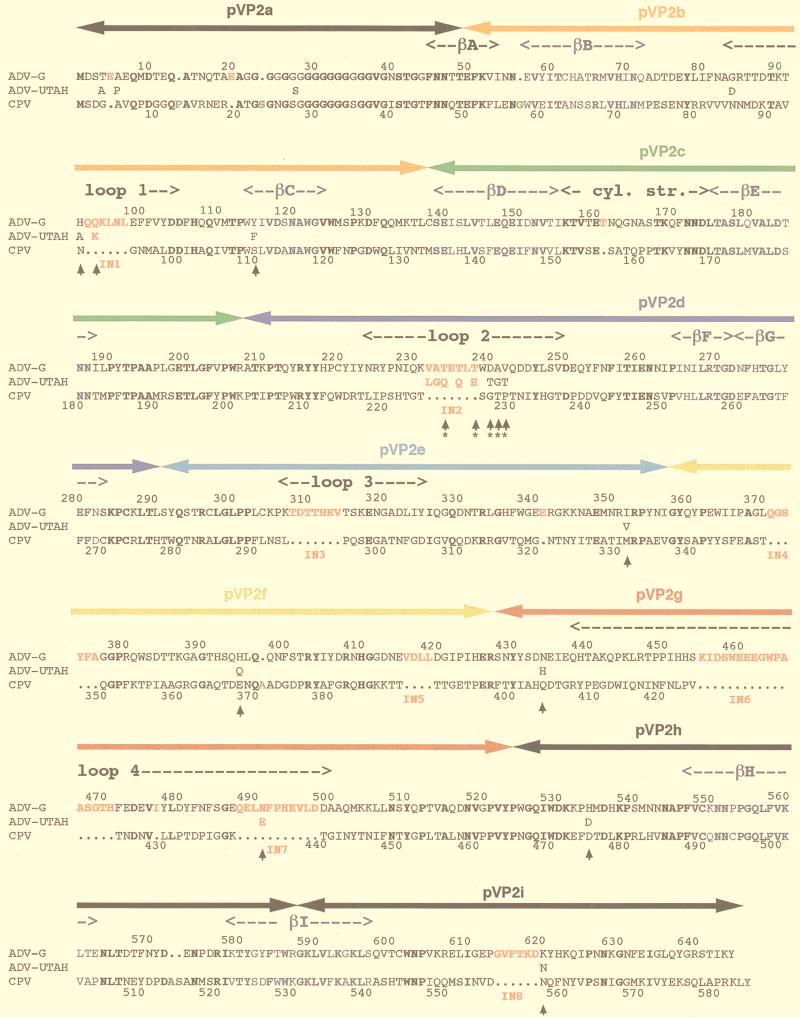
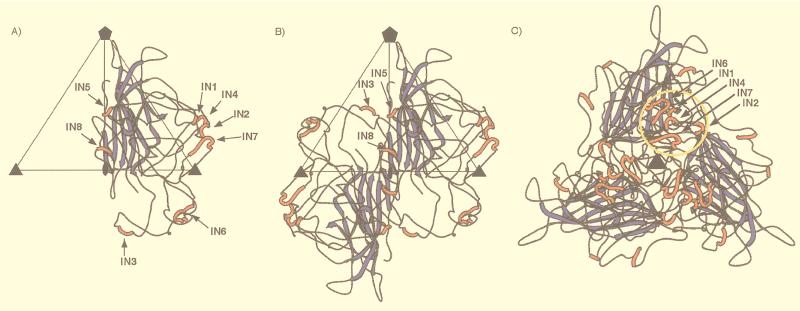
A modified version of the atomic structure of the CPV VP2 capsid protein was used as a template in the O program (38) to produce a model ADVG-VP2 structure. The pairwise alignment of the amino acid sequences of the ADV-G and CPV capsid proteins (24) was modified (32) based on two criteria, the molecular envelope of the cryo-EM reconstruction and the secondary structural elements of CPV (Fig. 3). The best agreement between these sequences occurred when eight additional peptides, IN1 to IN8, were inserted into the CPV structure. An insertion peptide was defined here as being greater than two residues long (Table 1, Fig. 3).
FIG. 3.
Sequence alignment of the VP2 capsid proteins of ADV-G, ADV-Utah 1, and CPV. Arrowheads demarcate residues that consistently differ between ADV-G and the pathogenic ADV strains. The ADV hypervariable region is indicated by asterisks (∗) under the arrowheads. Boldface lettering is used to highlight completely conserved residues. Colored lettering highlights residues that comprise the β-barrel structural motif (blue), insertions IN1-IN8 in the ADV capsid protein (red), and insertions in the CPV capsid protein relative to ADV (green). The sequence numberings are for ADV-G (top) and CPV (bottom). The peptides (pVP2a-i) used in a nonoverlapping peptide expression study (19) are indicated by bars with arrowheads at each end above the capsid sequence and are colored. Orange, pVP2b; green, pVP2c; dark blue, pVP2d; light blue, pVP2e; yellow, pVP2f; red, pVP2; and black, pVP2a, pVP2h, and pVP2i.
TABLE 1.
Peptide sequences in the ADVG-VP2 capsid protein that are not found in CPVa
| Insertion label | Starting position (aa) | Sequence | Length of insert (aa) |
|---|---|---|---|
| IN1 | 93 | QQKLNL | 6 |
| IN2 | 232 | VATETLT | 7 |
| IN3 | 308 | TDTTHKV | 7 |
| IN4 | 371 | QGSYEA | 6 |
| IN5 | 416 | VDLL | 4 |
| IN6 | 455 | KIDSWEEEGWPAASGTH | 17 |
| IN7 | 488 | QELNFPHEVLD | 11 |
| IN8 | 614 | GVFTKD | 6 |
aa, amino acids.
The secondary structures of the eight insertions in ADVG-VP2 were predicted with the aid of the PredictProtein server (56) and the BLAST network service (8). A model of the ADVG-VP2 capsid was built and fitted interactively into the ADVG-VP2 cryo-EM envelope by placing the modified CPV VP2 atomic model into the cryo-EM density map, using the O program (38). Seven insertions were modeled as a coil, but IN6 was modeled as an α-helical segment because the BLAST results indicated a strong correlation of the segment sequence with typical α-helical protein sequences (8). The O structural geometry library was used to constrain all bond geometries during modeling. CPV residues were mutated to the corresponding VP2 sequence for ADV-G in the pseudoatomic model.
RESULTS
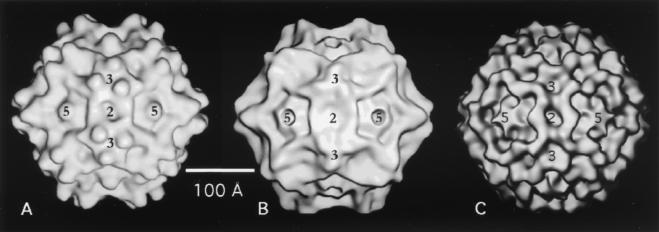
The 3D reconstruction of ADVG-VP2, computed to 22 Å resolution from images of vitrified particles, reveals a capsid structure with mean external radii of 115, 136, and 132 Å at the icosahedral twofold, threefold, and fivefold symmetry axes, respectively (Fig. 2 and 4A). Several surface features of ADVG-VP2 are similar to those found in the VP2 capsid structures of CPV (Fig. 4B), FPV, MVM, and B19 (Fig. 4C). Prominent features include depressions ∼15 Å deep and ∼30 Å wide (dimples) at the icosahedral twofold axes, canyons ∼15 Å deep and ∼15 Å wide around the fivefold axes, and protrusions (spikes) at the threefold axes (Fig. 4). The spikes are not present in B19 (Fig. 4C), and they are more prominent in ADVG-VP2 (Fig. 4A) than in CPV (Fig. 4B) or in FPV and MVM (not shown). The ADVG-VP2 spikes are more prominent because each one contains three mounds that lie 22 Å from the icosahedral threefold axes and extend to a maximum radius of 155 Å. Ridges separate the dimples and canyons and appear to make these regions the least accessible areas on the ADV capsid surface (Fig. 4) in much the same way as has been observed for CPV, FPV, MVM, and B19. Knobs of density at the fivefold axes of ADVG-VP2 and B19 (Fig. 4A and C) distinguish these viruses from CPV, FPV, and MVM, which have channels formed by five symmetry-related β-ribbons that provide access to the virus interior (Fig. 4B).
FIG. 4.
Shaded-surface representations of ADVG-VP2 (A), CPV (B), and B19 (C), all viewed down a twofold icosahedral axis of symmetry. Panels A and C show the 3D cryo-EM reconstruction of ADVG-VP2 and B19 VP2 capsids (25) to 22 and 26 Å resolution, respectively, and panel B shows the atomic structure of CPV (63), rendered at 21 Å resolution. Locations of some two- (2), three- (3), and fivefold (5) icosahedral axes are indicated.
A pseudoatomic model of the ADVG-VP2 capsid was built based on a modified alignment of the ADV-G and CPV VP2 capsid proteins. This new alignment used the cryo-EM envelope of the ADVG-VP2 particle and the secondary structural elements of CPV as a guide. The 38% sequence identity of the modified alignment is slightly better than the 35% identity for the previously reported alignment (24), and residues confined to the eight-stranded, antiparallel, β-barrel motif show 63% identity (Fig. 3). The new alignment incorporates eight insertions (labeled IN1 to IN8 in Fig. 3) in the ADV-G VP2 sequence relative to that of CPV. These insertions, which range from 4 to 17 amino acids long, all occur in the large loops between the strands of the β-barrel.
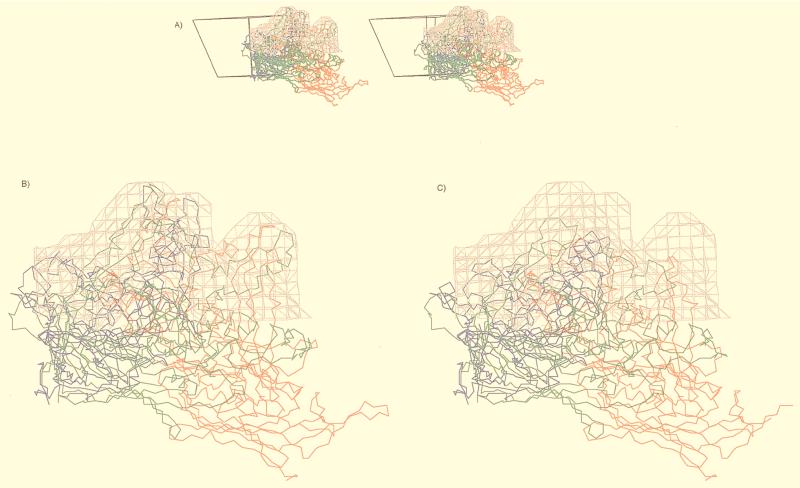
The ADVG-VP2 model places the β-barrel domain in the same position in the viral asymmetric unit as in the CPV capsid with the eight insertions clustered in two regions on the external surface of the capsid (Fig. 5). Insertions IN5 and IN8 from one VP2 subunit (Fig. 5A) and IN3 from a twofold symmetry-related subunit (Table 1, Fig. 3 and 5B) are clustered on the surface ridge that separates the canyon and dimple depressions (Fig. 5B). These three insertions create a wider ridge in ADVG-VP2 than in CPV and thereby result in a narrower dimple in ADVG-VP2, as is also observed in B19 (Fig. 4). Insertions IN1, IN2, IN4, and IN6 in one subunit (Fig. 5A), and IN7 from a threefold, symmetry-related subunit (Table 1, Fig. 3 and 5C) interact to form the mounds adjacent to the threefold axes (Fig. 4A and 5C). Each mound, consisting of a total of 47 amino acids contributed by the five insertion peptides, has no equivalent in the CPV structure (Fig. 4A and B and 6).
FIG. 5.
Ribbon drawings of the ADVG-VP2 pseudoatomic model. (A) VP2 monomer, viewed down an icosahedral twofold axis. (B) VP2 dimer, viewed as in panel A. (C) VP2 trimer, viewed down a threefold axis. The blue and red colors identify, respectively, the β-barrel motif and the insertions (IN1 to IN8) relative to the CPV capsid protein (Fig. 3). An icosahedral asymmetric unit (large open triangle) includes the region bounded by one fivefold (filled pentagon) and two threefold axes (filled triangles) separated by a twofold axes (filled oval) in panels A and B. The yellow circle in panel C highlights the clustering of insertion loops IN1, IN2, IN4, IN6, and IN7, which form the mounds near the threefold axes. The figure was generated with the program MOLSCRIPT (39).
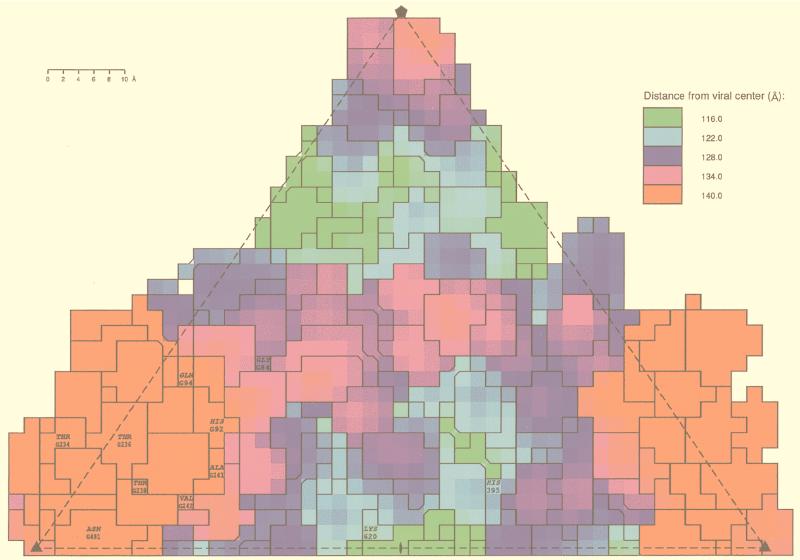
Comparison of the ADV-G VP2 sequence with the sequences of VP2 from ADV strains that are pathogenic in mink, such as ADV-Utah 1, ADV-TR, and ADV-Pullman, shows that strain-specific amino acids (Fig. 3) that occur throughout the linear sequence (17) cluster at or near the mounds and dimples (Fig. 7). More than half (9 of 14) of the strain-specific amino acid differences (Fig. 3) occur on the capsid surface (Fig. 7). The hypervariable antigenic region of ADV (residues 232 to 241) (Fig. 3) is mostly exposed at the mounds (Fig. 7), and residues implicated in tissue tropism and disease pathogenicity (16, 17) occur near the threefold axes, in the dimple, or on the wall of the dimple (Fig. 7).
FIG. 7.
The locations of amino acids that differ between ADV-G and pathogenic ADV strains (strain-specific amino acids), projected onto a roadmap (23) representation of the residues on the capsid surface of the ADVG-VP2 pseudoatomic model colored according to radial distance (in angstroms) from the viral center. The orientation is the same as in Fig. 5A and B. The asymmetric unit contains contributions from several symmetry-related VP2 subunits. A letter preceding the sequence number identifies a symmetry operation used to generate the residue from the unlettered, reference VP2 subunit. G denotes a clockwise fivefold operation with respect to the threefold axis at the bottom right of the diagram. The boundaries of the projected surface-exposed amino acids are labeled according to residue type and sequence number. Residues 84 and 236 differ only between ADV-G and ADV-Utah 1.
DISCUSSION
ADV pathogenesis dramatically differs from that of other parvoviruses, such as CPV, MVM, and B19. Hence, an important issue is the extent to which defined structural features in ADV and other parvoviruses correlate with biological differences. The ADV-G VP2 sequence is 38% identical to CPV at the amino acid level and, at 22 Å resolution, the capsids of these viruses share features that might be expected to coexist among members of the same virus family (Fig. 4A and B). Similarities include the canyons that encircle the fivefold axes, the dimples at each of the twofold axes, and spikes centered at each of the threefold axes. However, three main features distinguish the external surface structure of ADV and CPV: (i) the channels at the fivefold axes in CPV appeared to be closed in ADV-G like B19 (Fig. 4A and C); (ii) each ADV-G spike has three mounds (Fig. 4A); and (iii) the ridges that separate dimples and canyons are wider in ADV-G (like B19) than in CPV (Fig. 4).
Differences in postassembly processing of parvovirus VP2 subunits might explain the closed channels in ADV and B19 (Fig. 4A and C). A third capsid protein, VP3, is detected in the “rat-like” parvoviruses, such as CPV, FPV, and MVM (59). It is formed by postassembly cleavage of about 20 amino acids from the N terminus of VP2 in DNA-containing virions but not in empty particles (28, 45, 60, 65). This cleavage does not occur in ADV or B19. Density present in the fivefold channels of CPV (63, 70) and MVM (3) capsids have been modeled as the glycine-rich, N-terminal region of those VP2 subunits that become externalized and accessible for cleavage to VP3. The absence of channels and of cleavage in ADV and B19 is consistent with the notion that the N terminus of VP2 becomes exposed at the fivefold axes in the rat-like parvoviruses.
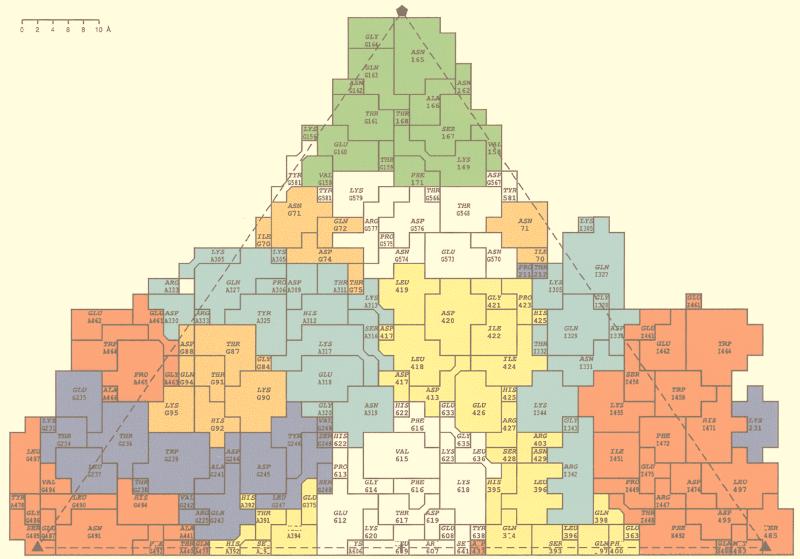
The spikes of ADVG-VP2 extend to a higher radius than the pinwheel-like spikes of CPV, owing to the extra protrusions (mounds) on ADV-G (Fig. 4A). The pseudoatomic model of the ADVG-VP2 structure predicted that each mound consisted of five short peptide insertions in the flexible loops that make up the capsid surface (including IN1, IN2, IN4, and IN6 from one VP2 polypeptide and IN7 from a threefold-related VP2 molecule [Table 1 and Fig. 3 and 5]). The spikes of CPV and FPV, which contain sequences from loops 1, 2, 3, and 4 (5, 63), have been shown to be dominant, neutralizing antigenic regions (58). A hallmark of ADV infection is the massive antibody response directed against the virus which enhances disease severity. The atomic model of ADVG-VP2 provides a useful means to relate antigenic features of the virion to the capsid structure. Recent work, in which defined segments of ADVG-VP2 (Fig. 3; pVP2a to pVP2i) were expressed as nonoverlapping, maltose-binding fusion proteins, indicates that specific regions of ADV-G are immunodominant (19). For example, linear polypeptides that align with CPV surface loops 3 and 4 (pVP2e, pVP2f, and pVP2g from ADV-G) are highly immunoreactive (19). Peptide pVP2d, which aligns with CPV loop 2 and contains the ADV hypervariable region, is also involved in the antigenic response and is thought to contain a strain-type specific epitope (16, 19). The current alignment of the ADV-G and CPV VP2 proteins based on sequence and structure show that the nonoverlapping peptides pVP2d, pVP2e, pVP2f, and pVP2g contain, respectively, insertions IN2, IN3, IN4 and IN5, and IN6 and IN7 (Fig. 3). IN2, IN4, IN6, and IN7 comprise most of the mound density (Fig. 5). A roadmap (23) that depicts the nonoverlapping peptide sequences (pVP2b-pVP2g) on the capsid surface of the ADVG-VP2 model (Fig. 8) confirms previous reports that the immunogenic linear peptides are located on or near the icosahedral threefold axes, or on the surface ridges that separate dimples and canyons (19). The most immunogenic peptide is pVP2g, which is the most highly exposed region of the capsid (Fig. 8). Hence, these observations suggest that the insertions in ADV that form the mounds are important determinants of capsid immunogenicity. It is noteworthy that parvoviruses CPV, FPV, and B19, which can all be neutralized by a host-antibody response, lack the ADV mounds (Fig. 4 and 6). The unusual pathogenicity of ADV might arise if the mounds act as immunodominant decoys that somehow prevent a protective response from being generated during infection.
FIG. 8.
The locations of nonoverlapping ADV-G peptides, pVP2b-pVP2g (Fig. 3), projected onto a roadmap (23) representation of the residues on the capsid surface of the ADVG-VP2 pseudoatomic model. The peptides are colored in the same scheme as in Fig. 3. The view is the same as in Fig. 5A and B and 7. Residue labeling is as explained in the legend for Fig. 7. A denotes a twofold operation, G denotes a clockwise fivefold operation, and I denotes a clockwise threefold operation with respect to the threefold axis at the bottom right of the diagram.
FIG. 6.
The pseudoatomic model of ADV-G VP2 fitted into the cryo-EM reconstruction (gray isodensity contour). (A) Stereo diagram of the mounds viewed parallel to an icosahedral twofold axis, showing the right-hand-side trimer with the reference VP2 in blue and the icosahedral threefold-related VP2s in red and green. Shown also is the viral asymmetric unit depicted as an open triangle. (B) Close-up view of panel A with inserted peptides IN1, IN2, IN4, IN6, and IN7 in ADV. (C) Close-up view of the ADV mound density with superimposed CPV atomic model, which has no equivalence structure to the inserted peptides in ADV. This figure was generated with the program MacInPlot (57).
Economic losses to the fur industry as a result of ADV infection of farmed mink are mainly a consequence of the unusual pathogenicity of ADV, in which infectivity is enhanced by capsid-antibody and capsid protein fragment-antibody complexes (1, 50, 51) formed during an infection. This property has thwarted numerous attempts at particle-based vaccine development. Current strategies for global development of vaccines against parvoviruses are focused on the use of virus-like particles or peptides produced in heterologous systems, since they form a cheap but safe alternative to the use of attenuated or inactivated live-virus vaccines. Our cryo-EM study of the baculovirus-expressed ADVG-VP2 capsids has led to a structural model that provides plausible correlations between the unusual antigenic properties of the virus and its capsid architecture. The model predicts that the mound features, comprised of inserted sequences relative to CPV and other parvoviruses that can be neutralized in vivo, harbor immunodominant sites on the ADV capsid surface. Thus, it may be possible to knock out dominant antigenic regions without affecting particle formation. Loss of such dominant antigenic regions could lead to recruitment of other minor antigenic epitopes in recombinant capsids. If such epitopes elicit a neutralization response, engineered viruses could function as effective vaccine vehicles in vivo.
The nonoverlapping ADV-G peptides pVP2b, pVP2c, pVP2h, and pVP2i (Fig. 3) had very little or no reaction against serum from infected mink (19), suggesting that these regions either contain conformational rather than linear epitopes or that they are not exposed at the capsid surface (19). The pseudoatomic model of ADVG-VP2 predicts that some of the residues from these nonreactive peptides are exposed at the capsid surface (Fig. 8). Thus, lack of reactivity of the peptides may indicate that the epitopes are not linear or that exposed residues are inaccessible to antibodies. Antibody inaccessibility may explain the lack of reactivity of peptides pVP2h and pVP2i, which have surface residues located in the canyons and dimples, respectively. The dimple has been postulated as being involved with receptor interactions, tissue tropism, and pathogenicity in parvoviruses (3, 5, 13, 61). Human parvovirus B19 (25) and other viruses, such as rhinoviruses (53), polioviruses (reviewed in reference 52), and influenza virus (66), have receptor recognition sites that are surface depressions surrounded by antigenic epitopes.
ADV residues which are implicated in tissue tropism in vitro and pathogenicity in vivo, cluster on or near the mounds and dimples (Fig. 7) and exhibit considerable sequence variability between ADV-G and the pathogenic strains of ADV (Fig. 3) (16, 17). Residues H92, Q94, and Y115, near the threefold axes, are located in the same region of the capsid as amino acid residues with analogous tissue tropism functions in CPV and FPV (5, 22, 47). Residues H395 and N434 on the dimple wall are topologically similar to residues of CPV and FPV, which determine differences in the interactions of these closely homologous viruses with erythrocyte receptors during hemagglutination (5, 13, 61). The residues in the dimple also have topological locations similar to those which confer tissue tropism differences between allotropic strains of MVM (3). Treatment of host cells with neuraminidase abolishes CPV and FPV hemagglutination (13, 61) and also MVM and ADV tissue infectivity (30, 34), suggesting that the dimple may be involved in recognition of host cell factors with terminal sialic acid moieties. However, sialic acid recognition appears not to be essential for CPV infection because, although residues in the wall of the CPV dimple are implicated in sialic acid recognition, CPV mutants which are unable to bind sialic acid in a hemagglutination assay are still competent to infect canine cells (13, 61). In contrast, for ADV and MVM, sialic acid recognition is important for infectivity (30, 34) and appears to be a prerequisite for successful host cell infection. In the latter viruses, the dimple, which is implicated in sialic acid recognition, is constricted compared to that in CPV (Fig. 4) (3) as a result of insertion IN8 in ADV (Table 1, Fig. 5) and a 5-amino-acid insertion in MVM VP2 relative to CPV VP2 (3). The proximity of these extra ADV and MVM residues to others in the dimple that determine tissue tropism suggests that the inserted sequences may be an adaptation which allows the viruses to utilize the binding of cellular factors with terminal sialic acids for productive host infection. It is of interest to note that in B19, the dimple is also constricted compared to that in CPV and is thus more like ADV (Fig. 4) as a result of an insertion analogous to the ADV IN8 in the B19 VP2 sequence (4, 24). However, while the infectious receptor for B19 consists of terminal N-acetylated sugars, sialic acid is not involved (21), and the receptor recognition site for B19 has been identified as a depression on the icosahedral threefold axis and not the twofold dimple (25). In view of these contrasts, more detailed analysis of the ADV capsid structure and additional mutational studies of ADV VP2 are needed to pinpoint the host receptor recognition site and to discern the mechanisms by which ADV recognizes host cells.
ACKNOWLEDGMENTS
This work was supported in part by University of Warwick Research and Teaching Initiative grant 0952 to R.M., National Institutes of Health grant GM33050 to T.S.B., and National Institutes of Health Program Project grant AI35212 to the Purdue University Structural Virology group.
We thank Colin R. Parrish for helpful discussions of the manuscript.
REFERENCES
- 1.Aasted B, Race R E, Bloom M E. Aleutian disease virus, a parvovirus, is proteolytically degraded during in vivo infection in mink. J Virol. 1984;51:7–13. doi: 10.1128/jvi.51.1.7-13.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrian M, Dubochet J, Lepault J, McDowall A W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 3.Agbandje-McKenna M, Llamas-Saiz A L, Wang F, Tattersall P, Rossmann M G. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 4.Agbandje M, Kajigaya S, McKenna R, Young N S, Rossmann M G. The structure of human parvovirus B19 at 8 Å resolution. Virology. 1994;203:106–115. doi: 10.1006/viro.1994.1460. [DOI] [PubMed] [Google Scholar]
- 5.Agbandje M, McKenna R, Rossmann M G, Strassheim M L, Parish C R. Structure determination of feline panleukopenia virus empty particles. Proteins. 1993;16:155–171. doi: 10.1002/prot.340160204. [DOI] [PubMed] [Google Scholar]
- 6.Alexandersen S. Pathogenesis of disease caused by Aleutian mink disease parvovirus. Acta Pathol Microbiol Immunol Scand. 1990;98:1–32. [PubMed] [Google Scholar]
- 7.Alexandersen S, Bloom M E, Perryman S. Detailed transcription map of Aleutian mink disease parvovirus. J Virol. 1988;62:3684–3694. doi: 10.1128/jvi.62.10.3684-3694.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Baker T S, Newcomb W W, Olson N H, Cowsert L M, Olson C, Brown J C. Structures of bovine and human papillomaviruses: analysis of cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker T S, Cheng R H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:20–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 11.Ball-Goodrich L J, Moir R D, Tattersall P. Parvoviral target cell specificity: acquisition of fibrotropism by a mutant of the lymphotropic strain of minute virus of mice involves multiple amino acid substitutions within the capsid. Virology. 1991;184:175–186. doi: 10.1016/0042-6822(91)90834-x. [DOI] [PubMed] [Google Scholar]
- 12.Ball-Goodrich L J, Tattersall P. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J Virol. 1992;66:3415–3423. doi: 10.1128/jvi.66.6.3415-3423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbis D P, Chang S-F, Parrish C R. Mutations adjacent to the dimple of the canine parvovirus capsid structure affect sialic acid binding. Virology. 1992;191:301–308. doi: 10.1016/0042-6822(92)90192-r. [DOI] [PubMed] [Google Scholar]
- 14.Bergeron J, Hébert B, Tijssen P. Genome organization of the Kresse strain of porcine parvovirus, identification of the allotropic determinant and comparison with those of NADL2 and field isolates. J Virol. 1995;70:2508–2515. doi: 10.1128/jvi.70.4.2508-2515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom M E, Alexandersen S, Garon C F, Mori S, Wei W, Perryman S, Wolfinbarger J B. Nucleotide sequence of the 5′-terminal palindrome of Aleutian mink disease parvovirus and construction of an infectious molecular clone. J Virol. 1990;64:3551–3565. doi: 10.1128/jvi.64.7.3551-3556.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom M E, Berry B D, Wei W, Perryman S, Wolfinbarger J B. Characterization of chimeric full-length molecular clones of Aleutian mink disease parvovirus (ADV): identification of a determinant governing replication of ADV in cell culture. J Virol. 1993;67:5976–5988. doi: 10.1128/jvi.67.10.5976-5988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloom M E, Fox J M, Berry B D, Oie K L, Wolfinbarger J B. Construction of pathogenic molecular clones of Aleutian mink disease parvovirus (ADV) that replicate both in vivo and in vitro. Virology. 1998;251:288–296. doi: 10.1006/viro.1998.9426. [DOI] [PubMed] [Google Scholar]
- 18.Bloom M E, Kanno H, Mori S, Wolfinbarger J B. Aleutian mink disease: puzzles and paradigms. Infect Agents Dis. 1994;3:279–301. [PubMed] [Google Scholar]
- 19.Bloom M E, Martin D A, Oie L L, Huhtanen M E, Costello F, Wolfinbarger J B, Hayes S F, Agbandje-McKenna M. Expression of Aleutian mink disease parvovirus capsid proteins in defined segments: localization of immunoreactive sites and neutralizing epitopes to specific regions. J Virol. 1997;71:705–714. doi: 10.1128/jvi.71.1.705-714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloom M E, Race R E, Wolfinbarger J B. Characterization of Aleutian disease virus as a parvovirus. J Virol. 1980;35:836–843. doi: 10.1128/jvi.35.3.836-843.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown K E, Anderson S M, Young N S. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–117. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 22.Chang S-F, Sgro J-Y, Parrish C R. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J Virol. 1992;66:6858–6867. doi: 10.1128/jvi.66.12.6858-6867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman M. Mapping the surface properties of macromolecules. Protein Sci. 1993;2:459–469. doi: 10.1002/pro.5560020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman M S, Rossmann M G. Structure, sequence, and function correlations among parvoviruses. Virology. 1993;194:491–508. doi: 10.1006/viro.1993.1288. [DOI] [PubMed] [Google Scholar]
- 25.Chipman P R, Agbandje-McKenna M, Kajigaya S, Brown K E, Young N S, Baker T S, Rossmann M G. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc Natl Acad Sci USA. 1996;93:7502–7506. doi: 10.1073/pnas.93.15.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christensen J, Storgaard T, Bloch B, Alexandersen S, Aasted B. Expression of Aleutian mink disease parvovirus proteins in a baculovirus vector system. J Virol. 1993;67:229–238. doi: 10.1128/jvi.67.1.229-238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemens D L, Wolfinbarger J B, Mori S, Berry B D, Hayes S F, Bloom M E. Expression of Aleutian mink disease parvovirus capsid proteins by recombinant vaccinia virus: self-assembly of capsid proteins into particles. J Virol. 1992;66:3077–3085. doi: 10.1128/jvi.66.5.3077-3085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinton G M, Hayashi M. The parvovirus MVM: a comparison of heavy and light particle infectivity and their density in vitro. Virology. 1976;79:57–63. doi: 10.1016/0042-6822(76)90127-6. [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project Number 4. The CCP4 suite: programs for crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 31.Crowther R A, DeRosier D J, Klug A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc R Soc Lond Ser A. 1970;317:319–340. doi: 10.1098/rspb.1972.0068. [DOI] [PubMed] [Google Scholar]
- 32.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubochet J, Adrian M, Chang J-J, Homo J-C, Lepault J, McDowall A W, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 34.Fox J M, Bloom M E. Identification of a cell surface protein from Crandell feline kidney cells that specifically binds Aleutian mink disease parvovirus. J Virol. 1999;73:3835–3842. doi: 10.1128/jvi.73.5.3835-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardiner E M, Tattersall P. Evidence that developmentally regulated control of gene expression by a parvoviral allotropic determinant is particle mediated. J Virol. 1988;62:1713–1722. doi: 10.1128/jvi.62.5.1713-1722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardiner E M, Tattersall P. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J Virol. 1988;62:2605–2613. doi: 10.1128/jvi.62.8.2605-2613.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahn E, Ramos C L, Kenyon A J. Expression of Aleutian mink disease antigen in cell culture. Infect Immun. 1977;15:204–211. doi: 10.1128/iai.15.1.204-211.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 39.Kraulis P. MOLSCRIPT: a program to produce both detail and schematic plots of proteins structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 40.Llamas-Saiz A L, Agbandje-McKenna M, Parker J S L, Wahid A T M, Parrish C R, Rossmann M G. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology. 1996;225:65–71. doi: 10.1006/viro.1996.0575. [DOI] [PubMed] [Google Scholar]
- 41.Llamas-Saiz A L, Agbandje-McKenna M, Wikoff R W, Bratton J, Tattersall P, Rossmann M G. Structure determination of minute virus of mice. Acta Crystallogr Sect D Biol Crystallogr. 1997;53:93–102. doi: 10.1107/S0907444996010566. [DOI] [PubMed] [Google Scholar]
- 42.Olson N H, Baker T S, Johnson J E, Hendry D A. The three-dimensional structure of frozen-hydrated Nudaurelia capensis β virus, a T=4 insect virus. J Struct Biol. 1990;105:111–122. doi: 10.1016/1047-8477(90)90105-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson N H, Chipman P R, Bloom M E, McKenna R, Agbandje-McKenna M, Booth T F, Baker T S. Automated CCD data collection and 3D reconstruction of Aleutian mink disease parvovirus. Microsc Microanal. 1997;3:1117–1118. [Google Scholar]
- 44.Pan M, Olson N H, Baker T S. Automated acquisition of cryo-electron microscope images with slow-scan CCD camera and microscope control. Microsc Microanal. 1997;3:1113–1114. [Google Scholar]
- 45.Paradiso P R. Infectious process of the parvovirus H-1: correlation of protein content, particle density, and viral infectivity. J Virol. 1981;39:800–807. doi: 10.1128/jvi.39.3.800-807.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker J S L, Parrish C R. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J Virol. 1997;71:9214–9222. doi: 10.1128/jvi.71.12.9214-9222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parrish C R, Aquadro C F, Carmichael L E. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink and raccoon parvoviruses. Virology. 1988;166:293–307. doi: 10.1016/0042-6822(88)90500-4. [DOI] [PubMed] [Google Scholar]
- 48.Porter D D. Aleutian disease: a persistent parvovirus infection of mink with a maximal but effective host immune response. Prog Med Virol. 1986;33:42–60. [PubMed] [Google Scholar]
- 49.Porter D D, Dixon F J, Porter H G. Metabolism and function of gammaglobulin in Aleutian disease of mink. J Exp Med. 1965;121:889–900. doi: 10.1084/jem.121.6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porter D D, Larsen A E. Aleutian disease of mink: infectious virus-antibody complexes in the serum. Proc Soc Exp Biol Med. 1967;126:680–682. [Google Scholar]
- 51.Porter D D, Larsen A E, Porter H G. The pathogenesis of Aleutian disease of mink. I. In vivo viral replication and the host antibody response to viral antigen. J Exp Med. 1969;130:575–593. doi: 10.1084/jem.130.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Racanielo V R. Interaction of poliovirus with its cell receptor. Semin Virol. 1992;3:473–481. [Google Scholar]
- 53.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 54.Rossmann M G, McKenna R, Tong L, Xia D, Dai J-B, Wu H, Choi H-K, Lynch R E. Molecular replacement real-space averaging. J Appl Crystallogr. 1992;25:166–180. [Google Scholar]
- 55.Rossmann M G, Johnson J E. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 56.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 57.Smith T J. MacInPlot: a program to display electron density and atomic models on the Macintosh personal computer. J Appl Crystallogr. 1990;23:141–142. [Google Scholar]
- 58.Strassheim M L, Gruenberg A, Veijalainen P, Sgro J-Y, Parrish C R. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the capsid. Virology. 1994;198:175–184. doi: 10.1006/viro.1994.1020. [DOI] [PubMed] [Google Scholar]
- 59.Tattersall P, Cawte P J, Shatkin A J, Ward D C. Three structural polypeptides coded for by minute virus of mice, a parvovirus. J Virol. 1976;20:273–289. doi: 10.1128/jvi.20.1.273-289.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tattersall P, Shatkin A J, Ward D C. Sequence homology between the structural polypeptides of minute virus of mice. J Mol Biol. 1977;111:375–394. doi: 10.1016/s0022-2836(77)80060-0. [DOI] [PubMed] [Google Scholar]
- 61.Tresnan D B, Southard L, Weichert W, Sgro J-Y, Parrish C R. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology. 1995;211:123–132. doi: 10.1006/viro.1995.1385. [DOI] [PubMed] [Google Scholar]
- 62.Truyen U, Agbandje M, Parrish C. Characterization of feline host range and a specific epitope of feline panleukopenia virus. Virology. 1994;200:494–503. doi: 10.1006/viro.1994.1212. [DOI] [PubMed] [Google Scholar]
- 63.Tsao J, Chapman M S, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith T J, Rossmann M G, Compans R W, Parrish C R. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 64.Tullis G E, Burger L R, Pintel D J. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but it is required for infectivity. J Virol. 1993;67:131–141. doi: 10.1128/jvi.67.1.131-141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weichert W S, Parker J S L, Wahid A T M, Chang S-F, Meier E, Parrish C R. Assay for structural variation in the parvovirus capsid and its role in infection. Virology. 1998;150:106–177. doi: 10.1006/viro.1998.9352. [DOI] [PubMed] [Google Scholar]
- 66.Weiss W, Brown J H, Cusack S, Paulson J C, Skekel J J, Wiley D C. Structure of influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 67.Wikoff W R, Wang G, Parrish C R, Cheng R H, Strassheim M L, Baker T S, Rossmann M G. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure. 1994;2:595–607. doi: 10.1016/s0969-2126(00)00062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu W-H, Bloom M E, Berry B D, McGinley M J, Platt K B. Expression of Aleutian mink disease parvovirus capsid proteins in a baculovirus expression system for potential diagnostic use. J Vet Diagn Investig. 1994;6:23–29. doi: 10.1177/104063879400600105. [DOI] [PubMed] [Google Scholar]
- 69.Wu H, Rossmann M G. The canine parvovirus empty capsid structure. J Mol Biol. 1993;233:231–244. doi: 10.1006/jmbi.1993.1502. [DOI] [PubMed] [Google Scholar]
- 70.Xie Q, Chapman M S. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J Mol Biol. 1996;264:497–520. doi: 10.1006/jmbi.1996.0657. [DOI] [PubMed] [Google Scholar]