Abstract
To date, nearly one-quarter of colorectal cancer (CRC) patients develop liver metastases (CRCLM), and its aggressiveness can be correlated to defined histopathological growth patterns (HGP). From the three main HGPs within CRCLM, the replacement HGP emerges as particularly aggressive, characterized by heightened tumor cell motility and vessel co-option. Here, we investigated the correlation between the expression of calcium- and integrin-binding protein 1 (CIB1), a ubiquitously expressed gene involved in various cellular processes including migration and adhesion, and disease-free (DFS) and overall survival (OS) in primary CRC patients. Additionally, we explored the correlation between CIB1 expression and different HGPs of CRCLM. Proteomic analysis was used to evaluate CIB1 expression in a cohort of 697 primary CRC patients. Additionally, single-cell and spatial RNA-sequencing datasets, along with publicly available bulk sequencing data were used to evaluate CIB1 expression in CRCLM. In silico data were further validated by formalin-fixed paraffin-embedded immunohistochemical stainings. We observed that high CIB1 expression is independently associated with worse DFS and OS, regardless of Union Internationale Contre le Cancer stage, gender, or age. Furthermore, the aggressive replacement CRCLM HGP is significantly associated with high CIB1 expression. Our findings show a correlation between CIB1 levels and the clinical aggressiveness of CRC. Moreover, CIB1 may be a novel marker to stratify HGP CRCLM.
Keywords: MT: Regular Issue, CIB1, colorectal cancer, histopathological growth patterns of liver metastasis, single-cell RNA sequencing, spatial transcriptomics, patient-derived organoids
Graphical abstract
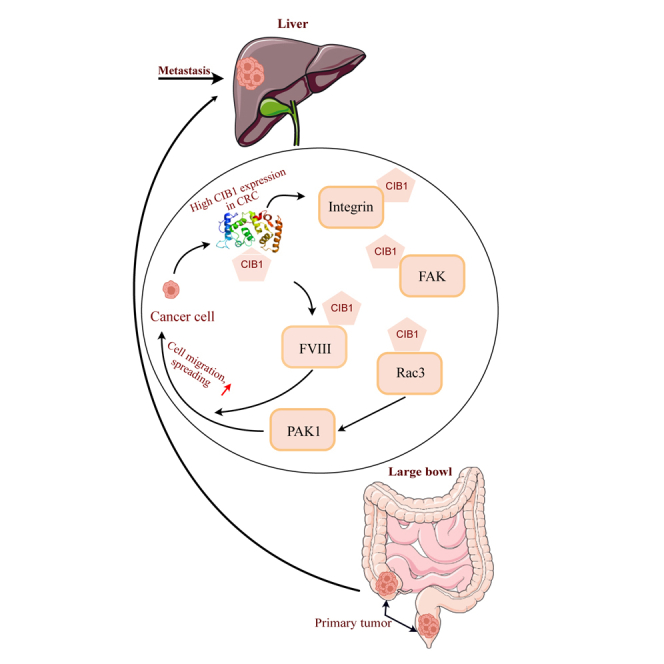
Conradi and colleagues have found that high CIB1 protein expression is a significant predictor of poor disease-free and overall survival in colorectal cancer liver metastases (CRCLM), particularly in aggressive replacement histopathological growth patterns. Elevated CIB1 expression correlates with enhanced tumor aggressiveness and may serve as a novel biomarker for stratifying CRCLM prognosis.
Introduction
According to global cancer statistics, in 2020 colorectal cancer (CRC) ranked third among the most prevalent tumor entities worldwide, with rising trends in its incidence and mortality in many countries in South America, Asia, and Eastern Europe.1 CRC patients with localized tumors exhibit a more favorable prognosis and higher survival rates compared with individuals with metastatic disease.2 To date, nearly one-quarter of CRC patients develop liver metastases (CRCLM), and their microscopic morphology can be used to define their histopathological growth patterns (HGP).3,4 The two most prevalent HGPs are (1) the desmoplastic (dHGP) and (2) the replacement (rHGP). The dHGP is characterized by a dense and fibrotic stroma surrounding the tumor cells and is associated with angiogenesis (new blood vessel formation). Desmoplastic CRCLMs are accompanied by a hepatic inflammatory response, characterized by the formation of a fibrous tissue reaction called desmoplasia.5 In the rHGP, the tumor cells infiltrate and replace the normal liver tissue, co-opting pre-existing blood vessels. The rHGP is well-known for its aggressiveness and is associated with resistance to anti-angiogenic therapy.6 Recently, it has been shown that any percentage of non-dHGP in CRCLMs is associated with reduced overall survival (OS) rates.7
The calcium- and integrin-binding protein 1 (CIB1) has been previously described as a predictor for the development of CRCLM, and previous studies have shown a strong association between elevated CIB1 gene expression and tumorigenesis across multiple tumor types.8,9,10,11 Molecularly, apart from binding soluble and transmembrane proteins,12 CIB1 expression has been linked to antiapoptotic effects via Sphingosein Kinase 1 and nuclear factor (NF)-κB signaling. Furthermore, cancer cell survival is enhanced in CIB1-expressing cells due to its interaction with the AKT and ERK pathways.13,14,15
In this study, we assessed CIB1 protein expression levels in a cohort of 697 patients with primary CRC and examined its correlation with disease-free survival (DFS) and OS, as well as other clinical parameters, including age, gender, Union Internationale Contre le Cancer (UICC) stage, and primary tumor location. Additionally, we performed in silico analyses to investigate whether CIB1 expression correlates with the observation of rHGP in CRCLM using single-cell, spatial, and bulk RNA-sequencing datasets. Furthermore, to validate these findings, we performed immunohistochemical (IHC) and immunofluorescence (IF) analyses of CIB1 on paraffin-embedded CRCLM tissue and patient-derived organoids (PDOs). Our investigation provides evidence that a higher expression of CIB1 is linked to a more aggressive behavior and with the rHGP CRCLM.
Results
High CIB1 protein expression levels inversely correlate with survival outcome in CRC patients
In this study, we investigated the relationship between CIB1 protein expression level and survival outcomes in CRC patients. The protein expression data utilized for this analysis was generated from untargeted proteomic analysis conducted on the tumor specimens obtained from the cohort of 697 patients with colorectal carcinomas. Hyper-reaction monitoring mass spectrometry technique was employed to detect and quantify proteins within the formalin-fixed paraffin-embedded (FFPE) tissue samples. Therefore, we established a cohort of 697 treatment-naïve patients with primary CRC tumors, accompanied by follow-up data. Additionally, we collected data on gender, age, UICC stage, and primary tumor location (Figures S1–S3). Subsequently, we categorized the cohort into high and low CIB1 protein expression, with a cutoff of 21,103.8, corresponding to a Youden index of 0.132. Patient baseline characteristics are shown in Table 1.
Table 1.
Baseline patient characteristics
| CIB1 low expression n (%) | CIB1 high expression n (%) | Total | |
|---|---|---|---|
| Patients | 342 | 355 | 697 |
| Sex | |||
| Female | 146 (43) | 139 (39) | 285 |
| Male | 196 (57) | 216 (61) | 412 |
| Age | |||
| 0-49 years | 36 (11) | 22 (6) | 58 |
| 50-64 years | 80 (23) | 101 (28) | 181 |
| 65+ years | 226 (66) | 232 (65) | 458 |
| Location of primary tumor | |||
| Left | 174 (51) | 182 (51) | 356 |
| Right | 168 (49) | 173 (49) | 341 |
| UICC | |||
| I | 63 (18) | 62 (17) | 125 |
| II | 111 (32) | 104 (29) | 215 |
| III | 103 (30) | 102 (29) | 205 |
| IV | 65 (19) | 87 (25) | 152 |
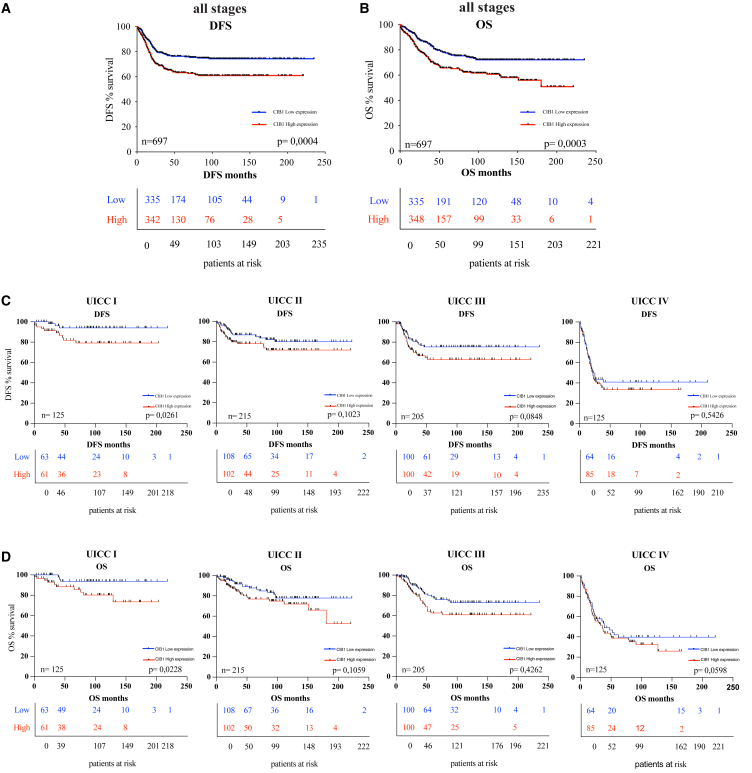
Following Kaplan-Meier survival analyses, we identified a significant inverse correlation between high CIB1 protein expression and DFS, as well OS in CRC patients. Specifically, patients with low CIB1 expression had higher DFS and OS rates compared with those with high CIB1 expression (Figures 1A and 1B).
Figure 1.
Correlation of CIB1 protein expression and survival outcomes in CRC patients
(A) Kaplan-Meier curve: DFS in a cohort of 697 CRC patients stratified for high and low protein expression of CIB1. (B) Kaplan-Meier curve: OS in a cohort of 697 CRC patients classified for high and low protein expression of CIB1. (C and D) Kaplan-Meier curves: DFS and OS in a cohort of 697 CRC patients stratified for high and low protein expression of CIB1 classified according to their pUICC stage.
Subsequently, we investigated whether this correlation persisted across patients with different stages of CRC by performing survival analyses of CIB1 expression with DFS and OS, according to their pathological UICC stages (pUICC) (Figures 1C and 1D). In line with this, patients in UICC I stage who exhibited low CIB1 expression demonstrated significantly higher DFS and OS rates compared with those with high CIB1 expression (Figures 1C and 1D). This trend persisted across pUICC stages II, III, and IV, where high CIB1 protein expression was associated with worse survival outcomes, although these correlations were not statistically significant (Figures 1C and 1D).
Overall, these findings indicate that high CIB1 protein expression correlates with worse survival outcomes, and that its expression is especially important in early tumor stages.
CIB1 protein expression in right-sided tumors has prognostic significance
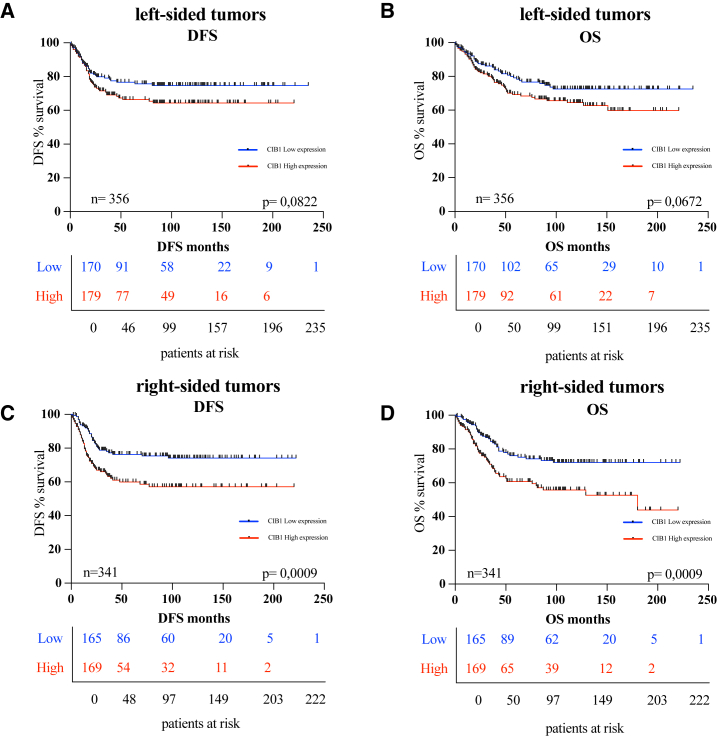
Given the distinct molecular and clinical characteristics associated with left-sided and right-sided colon cancers, exploring this correlation across both subsets contributes valuable insights into the role of CIB1 in CRC prognosis. Next, we investigated whether the correlation of high CIB1 expression is independent from the tumor location. Out of the total 697 CRC patients included in this study, 356 had tumors located in the colon descendens, sigmoid, and rectum, collectively referred as left-sided colon cancers, and 341 patients had tumors in the cecum, colon ascending, right flexure, and colon transverse, grouped as right-sided colon cancers (Table 1 and Figures S1E and S1F). Although in the left-sided colon cancers no significant statistical difference in survival rates was observed (Figures 2A and 2B), DFS and OS analysis revealed that in right-sided tumors, patients with low CIB1 expression showed higher survival rates (Figures 2C and 2D), matching with the current clinical knowledge that tumors prevenient from this site are more aggressive.16
Figure 2.
Correlation of CIB1 protein expression and survival outcome in left- and right-sided CRC patients
(A and B) Kaplan-Meier curves: DFS and OS in a cohort of 697 CRC patients stratified for high and low protein expression of CIB1 in left-sided CRC. (C and D) Kaplan-Meier curves: DFS and OS in a cohort of 697 CRC patients stratified for high and low protein expression of CIB1 in right-sided CRC.
In summary, CIB1 expression levels are significantly associated with right-sided tumor patients’ survival outcomes, with CIB1 protein expression negatively correlating DFS and OS.
In silico CIB1 expression levels in replacement and desmoplastic HGP CRCLM
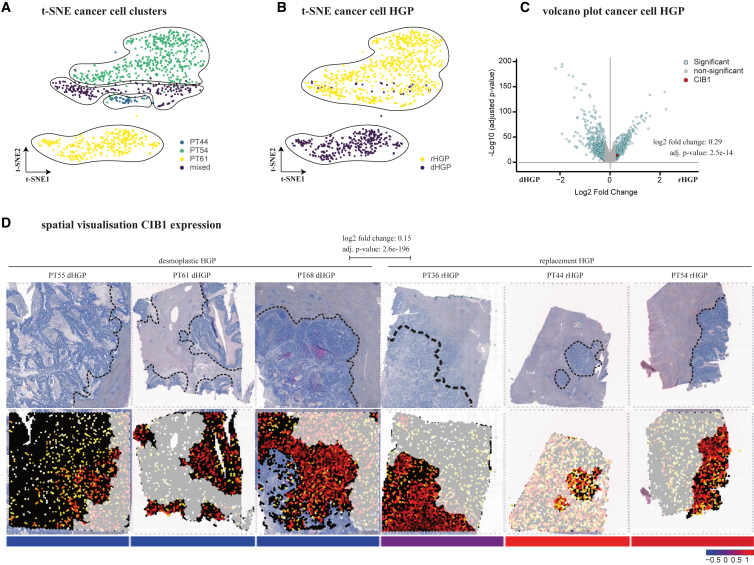
CRC patients with liver metastases have a poorer prognosis, reflected by a reduced 5-year survival rate of 14%.17 Moreover, only a subgroup of patients responds to anti-angiogenic therapy through anti-vascular endothelial growth factor (VEGF) antibody administration, as not all metastases use sprouting angiogenesis to secure their nutrient and oxygen supply.18 This specific mode of vascularization is followed by distinct HGPs, which have proven prognostic and predictive significance.3,4 Recently, we have shown that any amount of non-dHGP (e.g., rHGP) is associated with impaired survival rates in CRC.7 CIB1, a protein implicated in various cellular processes,9 including signal transduction and tumorigenesis, might play a role in the distinct microenvironments of the HGPs. Thus, we directed our attention to investigating the expression of CIB1 in CRCLM on the two most prevalent HGPs, the rHGP and the dHGP. Utilizing single-cell RNA-sequencing dataset (E-MTAB-12022), we identified an upregulation of CIB1 in cancer cells of the rHGP (Figures 3A–3C). To further validate our single-cell results, we performed a comparative analysis using our publicly available spatial RNA-sequencing data (E-MTAB-12043) and observed increased CIB1 gene expression in regions displaying rHGP over dHGP, thereby confirming the consistency of our results (Figure 3D). Additional evaluation of bulk CRCLM RNA sequencing (GSE151165) resulted in a similar trend in CIB1 expression (Figure S4A).
Figure 3.
High CIB1 gene expression in rHGP CRCLM
(A) t-SNE plot: unsupervised Louvain clustering of 2,541 cancer cells with cluster identification. (B) t-SNE plot: clustering of 2,541 cancer cells by HGPs. (C) Volcano plot: differential gene expression analysis of 2,541 single cancer cells of single-cell RNA sequencing (highlighting CIB1). (D) Spatial gene visualization: differential gene expression analysis of three dHGP vs. three rHGP cancer areas of spatial RNA sequencing (highlighting of CIB1).
In sum, across all evaluated datasets, we observed that CIB1 expression was significantly elevated in rHGP CRCLM, compared with dHGP CRCLM, with the exception of bulk CRCLM RNA where only a trend could be found. These findings are in line with previous studies showing a correlation between elevated levels of CIB1 expression and increased tumorigenesis.8,10
Elevated CIB1 protein levels in rHGP CRCLM
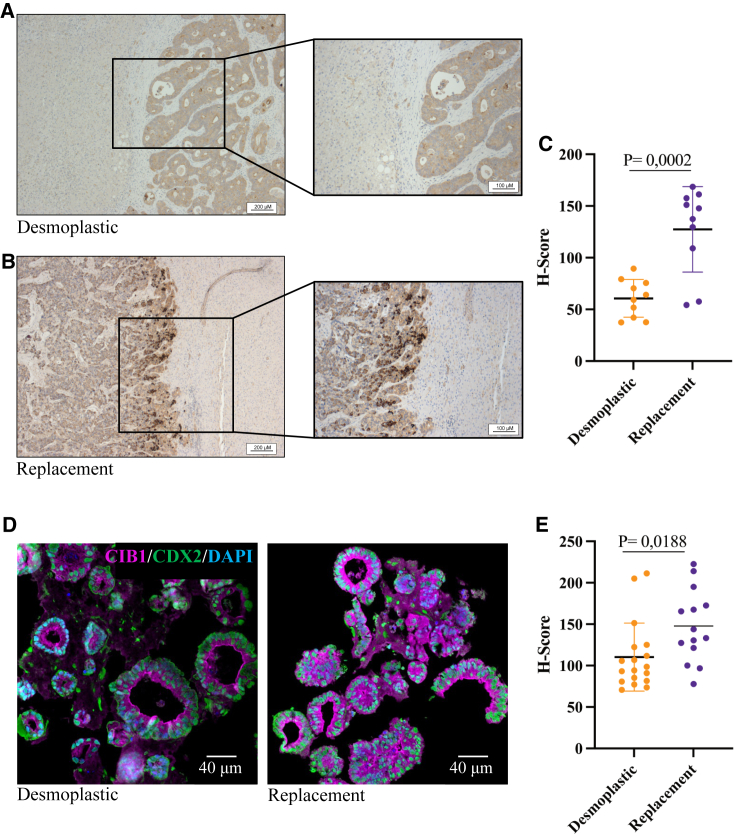
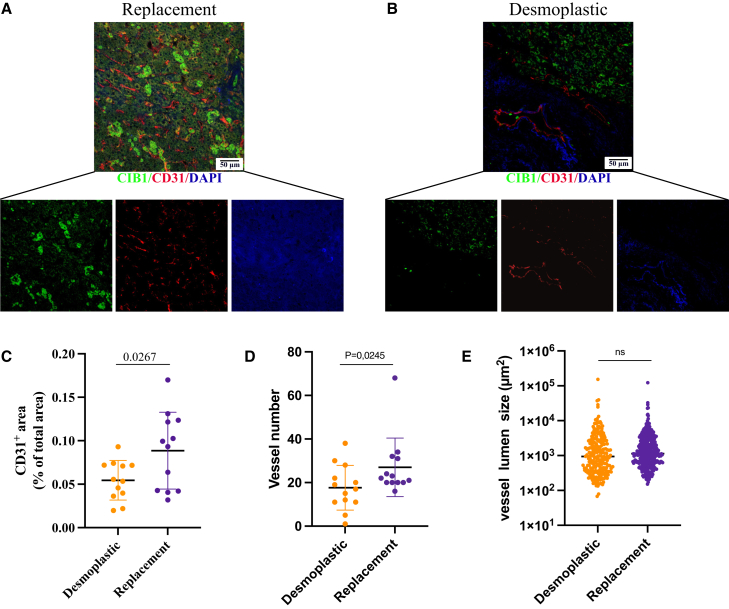
First, to corroborate the in silico findings of differential CIB1 expression levels between the dHGP and rHGP CRCLM groups, we performed immunohistochemical staining with FFPE samples from 20 different patients (10 rHGP and 10 dHGP). In agreement with our previous results, we observed a significant increase of CIB1 within the replacement group (H-score 127.40) (Figures 4A, 4B, and S5). Desmoplastic HGP samples exhibited an H-score of 60.66 (p < 0.001) (Figure 4C; Table 2). Second, to further validate these findings, we extended our investigation to CRCLM PDOs. Therefore, we established and expanded PDOs from 11 additional distinct patients (five rHGP CRCLM and six dHGP CRCLM) (Figure 4D). In line with the previous results, IF stainings confirmed increased CIB1 expression in the organoids extracted from the replacement group (Figures 4C and 4E). Last, to explore and characterize the vascular profile of our CRCLM samples, we have performed IF co-stainings of CD31 (a well-known endothelial marker) and CIB1 in extra 26 FFPE CRCLM samples. In line with previously reported data,18 our results show a higher CD31+ area (% of total area) in the replacement group, as well number of vessels, without differences in vessel lumen (Figures 5A–5E).
Figure 4.
High CIB1 protein expression in rHGP CRCLM
(A and B) IHC staining of CIB1 in either dHGP or rHGP CRCLM. (C) IHC quantification. Scatterplot: H-Score of CIB1 in dHGP and rHGP (n = 10 dHGP vs. n = 10 rHGP), unpaired t test. Data are presented as mean SD. (D) Representative IF analysis of CIB1 (magenta) and CDX2 (green). Nuclei were counterstained with DAPI. (E) Scatterplot of H-score of CIB1 in PDOs, unpaired t test. Data are presented as mean SD.
Table 2.
Immunohistochemical CIB1 expression distribution in CRC patients with different HGPs
| Desmoplastic | Replacement | |
|---|---|---|
| Number of values | 10 | 10 |
| Minimum | 37, 4 | 54, 25 |
| Maximum | 89, 4 | 168, 6 |
| Range | 52 | 114, 4 |
| Mean | 60, 66 | 127, 4 |
| Lower confidence limit (95%) | 47, 6 | 97, 82 |
| Upper confidence limit (95%) | 73, 72 | 157 |
Figure 5.
Evaluation of CRCLM vascular profile
(A and B) Immunofluorescence (IF) analysis of CIB1 (green) and CD31 (red) in CRCLM samples. Nuclei were counterstained with DAPI. (C–E) Quantification of CD31+ staining, scatterplot of vessel number and vessel lumen, in CRCLM (n = 13 dHGP vs. n = 13 rHGP), unpaired t test. Data are presented as mean SD, Student’s t test; ns = p > 0.05.
Overall, our consistent findings augmented the understanding of CIB1s role in the context of rHGP CRCLM across different platforms and experimental setups, suggesting the potential use of CIB1 as a promising biomarker for tumor aggressiveness and CRCLM HGP.
Discussion
Despite advancements in treatment regimens, CRC remains a significant global cause of cancer-related mortality.19,20 The clinical progression of advanced CRC varies significantly, making it challenging to stratify patients for treatment and surveillance. Some patients develop metastases, while others do not. Understanding the molecular markers that contribute to these differences is crucial for personalized treatment. Biomarkers such as genetic mutations or gene expression patterns can offer valuable insights into disease progression and prognosis.21 By analyzing the molecular landscape of CRC, researchers aim to identify key biomarkers to stratify patients into risk groups and optimize treatment strategies. Moreover, molecular markers also play vital roles in monitoring treatment response and detecting recurrences. Regular assessment of the tumor’s molecular profile allows for timely adjustments to treatment plans, ultimately improving patient outcomes.
Previous studies have demonstrated that CIB1, a small E-helix-loop-F helix (EF-hand) calcium-binding protein that is expressed ubiquitously within cells,22 is capable of binding soluble and transmembrane proteins that participate in numerous cellular processes, including calcium signaling, survival, adhesion, and apoptosis.12 Given these functional properties of CIB1, it is not surprising that it is also implicated in the development and progression of various types of cancer.13
Notably, it has been observed that CIB1 expression levels can affect the migration and invasive characteristics of tumor cells, which could potentially account for CRC patients expressing high CIB1 levels being more prone to develop liver metastases.8
Therefore, we have investigated CIB1 as possible molecular marker for CRC and CRCLM focusing on the more aggressive type of CRCLM, the rHGP. To evaluate its potential clinical relevance, we first performed a retrospective study on 697 CRC patients who received treatment between 2000 and 2020. Our study found that high CIB1 expression was linked to worse DFS and OS, particularly in patients with early-stage CRC and right-sided tumors. Therefore, CIB1 could be a promising early predictor for aggressive CRC tumors. We have further evaluated other important confounders, such as sex and age, confirming that the correlation remains consistent in every analysis.
Subsequently, we conducted single-cell and spatial RNA-sequencing analyses of rHGP and dHGP CRCLM, to investigate whether CIB1 expression may also be altered in patients with advanced disease, who are already known to differ in survival prognosis. Recently, our group demonstrated that rHGP exhibits higher malignancy (compared with dHGP) and that the presence of any non-dHGP is associated with decreased OS.7 Remarkably, our in silico transcriptome analyses demonstrated an elevated expression of CIB1 in rHGP, which was further confirmed in PDOs and FFPE from rHGP CRCLM samples. Furthermore, the results of our vascular profile analysis of CRCLM aligns with the results of Anthoula Lazaris et al.,18 who observed similar patterns when assessing micro-vessel density. Accordingly, we show vessel density to be higher in the replacement group (CIB1 high) compared with the desmoplastic group (CIB1 low), while vessel lumen size was not altered. These findings are corroborated with the work of Ibrahim et al., who as well show higher endothelial cell counts in replacement HGP by using endothelial cell marker proteins such as Tie-2 and Angiopoietin-1.23
Replacement HGP CRCLMs are known to rely on vessel co-option to achieve their supply of oxygen and nutrients3,18 and exhibit enhanced cancer cell motility and adhesion, which might be the underlying mechanisms driving this process.6,24 CIB1 is known for regulating platelet integrin activity, cell adhesion, and migration by binding to IIb3, FVIII, Rac3, and FAK.9 Integrins are a major adhesion receptor that controls cell migration, differentiation, and proliferation. Additionally, CIB1 interacts with the small GTPase Rac3 to promote IIb3-mediated cell adhesion and spreading.25 It also has been suggested that CIB1 activates FAK by direct binding, leading to Rac3 and PAK1-induced adhesion and spreading. FAK attachment to focal adhesions can influence cell migration and motility, and PAK1 is a crucial component of cell migration.26,27 Therefore, it is feasible that tumors with a higher CIB1 expression may have the capability to adhere to preformed vessels and undergo co-option, while tumors exhibiting relatively lower CIB1 may rely on sprouting angiogenesis through the HIF1a/VEGF pathway, as observed in dHGP.
Altogether, our study further elucidates the possible prognostic value of CIB1 in CRC and provides the first insight into the correlation between CIB1 expression and DFS as well as OS in CRC patients. Moreover, our results provide evidence for the possible role of CIB1 overexpression as a predictor for the aggressive rHGP. Nevertheless, the retrospective nature of the study imposes limitations. Although the correlation suggests a potential relationship between CIB1 expression, different HGP types, and survival outcomes, further research is needed to establish a causal link and explore the underlying mechanisms involved in these processes. Further prospective studies are required to validate our findings and to assess the potential utility of CIB1 expression in guiding personalized treatment strategies for CRC patients.
Materials and methods
Patient cohort
In this study, we conducted a survival analysis on patients with primary and metastatic CRC, who underwent colorectal surgery at the University Medical Center Göttingen between 2000 and 2020. Patients were stratified into two groups according to their CIB1 protein expression in the primary tumor, determined via proteomic analysis (see below). A total of 342 patients were assigned to the low CIB1 expression group and 355 patients were assigned to the high expression group, computing the cutoff using SPSS statistics software (details outlined below). For further analysis, patients were also stratified according to their gender, age, tumor location, and UICC classification. All patients included in the analysis were treatment-naïve.
Follow-up survey
Data collection took place from April 2020 to January 2021 and was performed by follow-up documentation at the University Medical Center Göttingen and via telephone consultation with the attending family physicians. Collected data were digitally recorded using SecuTrial (see below). Clinicopathological parameters were initially recorded at the time of the operation within the patient file, and later transferred to SecuTrial. A total of 697 patients underwent a systematic follow-up, investigating survival, the occurrence of distant metastases, and local recurrences based on the clinicopathological parameters. All procedures were done under the ethical approval of the University Medical Center Göttingen (UMG), votes Nr. 25/3/17, Nr. 24/4720, and Nr. 23/4/22.
Data collection in SecuTrial
Data collection, handling, and processing were facilitated by the web-based electronic data capture software SecuTrial (InterActive Systems),28 enabling standardized and secure collection of patient data using electronic case report forms (eCRFs).
Kaplan-Meier survival analysis
Kaplan-Meier method of survival analysis29 defined DFS and OS as the endpoints of the analysis. DFS describes the period after therapy without progression, recurrence, or metastasis. OS refers to the period from diagnosis to the patient’s death regardless of the diagnosis.30 Patients withdrawing from the study or those lost to follow-up were censored.
Proteomics
Proteomic analysis was conducted by Biognosys AG, using FFPE tissue samples obtained from 697 resected CRC patients. The specimens underwent processing and were subjected to hyper-reaction monitoring mass spectrometry for untargeted proteomic analysis as outlined in Piazza et al.31 This analysis resulted in the quantification of 7,611 proteins, 99,822 peptides, and 179,949 peptide variants.
Immunofluorescence
Paraffin-embedded CRCLM 2-μm slide tissue sections were initially deparaffinized using xylene and ethanol dilutions, followed by high-pH antigen retrieval through cooking/steaming. Subsequently, the slides were cooled and rinsed in distilled water. A blocking step of 30 min with blocking reagent (BR) was performed, followed by an overnight incubation at 4°C with CDX2 (mouse) primary antibody diluted 1:2 in BR. On the second day, the slides underwent a series of washes with Tris-buffered saline (TBS) and TBS-Tween (TBST), followed by a 44-min incubation with biotin-conjugated donkey anti-mouse secondary antibody (1:200 in TNB). This was succeeded by a 30-min incubation with streptavidin-horseradish peroxidase (HRP) (1:100 in TBST), a 10-min exposure to Cy5 amplification diluent (1:50), and a subsequent methanol-TBS wash. Then transitioned to antibody CIB1 (rabbit, 11823-1-AP, Proteintech) at a concentration of 1:50 in BR for 2 h at room temperature (RT), followed by the same series of washes with TBS and TBST. The rabbit antibody was detected using biotin-conjugated donkey anti-rabbit secondary antibody (1:200 in TNB), streptavidin-HRP (1:100 in TBST), and Cy5 amplification diluent (1:50). Hoechst staining (1:500 in PBS) for 30 min and a final TBS wash were conducted before covering the slides with DAPI ProLong Gold antifade reagent and a cover glass. The stained slides were then stored in the dark at +4°C. Additionally, CD31 (BD Biosciences, 1:500) and CIB1 (1:50) co-staining immunofluorescence were conducted following the same protocol as described above.
Immunohistochemistry
IHC was performed as previously described.7 Briefly, samples of CRCLM were cut into 2-μm slides, deparaffinized in xylol, and subjected to a descending series of ethanol, followed by a standard hematoxylin and eosin (H&E) staining protocol. Subsequently, the slides were heated in a steamer with Tris-EDTA (pH 8.5) for 20 min. After blocking endogenous peroxidase activity and incubating with 5% bovine serum albumin diluted in PBS, the samples were washed with TBST. The primary CIB1 antibody (11823-1-AP, Proteintech) was diluted 1:50 and incubated for 60 min at RT. Following incubation and washing, secondary antibody anti-rabbit peroxidase polymer concentrate was used for 30 min at RT. After another two washes with TBST for 5 min each, the slides were treated with 3,3′-diaminobenzidine (DAB) chromogen (BS04-110, ImmunoLogic) at a dilution of 1:25 for 10 min at RT. After DAB staining, the slides were rinsed with distilled water for 2 min. To visualize the nuclei, the slides were briefly incubated with hematoxylin for 2 min, followed by a rinse with distilled water for 5 min. Subsequently, samples were dehydrated using an ascending ethanol series and xylol for 2 min. Finally, Vitro-Clud was used for mounting, and a cover glass was added.
IF image analysis using QuPath
The IF image analysis was conducted using QuPath version 0.4.3.32 To assess CIB1 expression in individual tumor cells, the “Positive cell detection” command was used. Tumor cell identification relied on CDX2, a colon cancer marker, with an 8-μm expansion radius. For scoring CIB1 expression, the algorithm categorized cells as CIB1-negative (intensity below 64), weakly positive (intensity between 65 and 128), moderately positive (intensity between 129 and 192), and strongly positive (intensity between 193 and 255). The analysis script generated outputs indicating cell counts and their distribution across high, medium, and low categories. Subsequently, the H-score was calculated.
IHC scoring and analysis
IHC scoring was conducted as previously described.8,33 IHC analysis of the tissue samples was performed in a blinded manner using standard light microscopy. For each slide, staining intensity was assessed on a scale of 0–3 (I0, I1-3), with I0 indicating no staining, I1 indicating a weak staining, I2 indicating moderate staining, and I3 indicating strong staining (Figure S5). The percentage of tumor staining at each intensity level was recorded in 5% increments, ranging from 0 to 100 (P0, P1-3). The final H-score (ranging from 0 to 300) was obtained by adding the products of the intensity score and the percentage of tumor staining for each intensity level (H-score = I1 × P1 + I2 × P2 + I3 × P3).
Single-cell RNA sequencing
For single-cell sequencing, resected tissue (malignant and normal hepatic tissue) from six patients with CRCLM (3 dHGP vs. 3rHGP) was collected. Sample preparation, library construction, and sequencing were performed as previously described7 using chromium single-cell gene expression technology (10X Genomics) and DNBSEQ technology (BGI). After data pre-processing with CellRanger software (10X Genomics) and quality filtering,7 a total of 22,419 cells were included in further analyses. For single-cell and spatial sequencing, subsequent analysis and visualization were conducted using the UniApp (Unicle Biomedical Data Science, Belgium).
Single-cell data were auto-scaled and principal-component analysis (PCA) was conducted to visualize the first 20 principal components in t-distributed stochastic neighbor embedding (t-SNE). Clusters identified by graph-based clustering (resolution = 1) were annotated via identification of uniquely upregulated genes and expression of canonical marker genes in each cluster.7 A total of 2,541 cancer cells were subsequently displayed by t-SNE calculating the first 20 principal components and subclustered at a resolution of 1.5. Cancer cells were annotated for patient and HGP of origin. Differentially expressed genes between rHGP cancer cells and dHGP cancer cells were visualized in a volcano plot and position of CIB1 was highlighted.
Spatial RNA sequencing
For spatial sequencing, FFPE tissue (5-μm thickness, roughly 6.5 × 6.5 mm size) from six patients with CRCLM (3 dHGP vs. 3 rHGP) was placed on a Visium spatial gene expression slide (10X Genomics, PN-2000233). All tissue slices showed tumor areas as well as adjacent normal hepatocyte areas. Each area of the gene expression slide is composed of 5,000 spots with a diameter of 55 μm each. Slides were stained and imaged and a sequencing library was prepared as previously published,7 using the Visium Spatial Gene Expression for FFPE Kit (10X Genomics, PN-1000338). Libraries were sequenced using DNBSEQ technology (BGI) and data were pre-processed with SpaceRanger software (10X Genomics).7 After quality filtering, 21,804 spots were included in further analyses. Data from all spots of each patient were auto-scaled, reduced in dimensionality by PCA, and displayed in t-SNE plots. After graph-based clustering, clusters were annotated according to their predominant cell types via identification of uniquely upregulated genes and expression of canonical marker genes. Biological annotation was confirmed by overlay on the H&E slide.7 Relative expression of CIB1 in each spot was determined and spatially visualized on the H&E overlay. Differential gene expression analysis between cancer areas of rHGP and dHGP was performed to identify the rank of CIB1. Expression of CIB1 in cancer areas of each patient was visualized as a heatmap based on cluster-averaged gene expression and auto-scaled for visualization.
Bulk RNA sequencing
The publicly available bulk sequencing dataset GSE151165,34 which stratifies CRCLMs for HGP (9 dHGP vs. 6 rHGP) was chosen as a result of NIH GEO DataSets search for the terms “Colorectal cancer liver metastases” and “HGP.” After TMM normalization,35 conducted using the edgeR package in Bioconductor (version 3.14),36 all samples from normal adjacent liver were excluded. CIB1 expression was evaluated by the number of normalized CIB1 reads per sample. Differential gene expression analysis between dHGP and rHGP cancer samples was performed to identify the rank of CIB1.
SPSS statistics
The survival data were formatted for SPSS (version 26.0), including variables such as time of observation, event occurrence (death), and other relevant factors. Receiver operating characteristic (ROC) curve analysis was performed using the SPSS software. In the "ROC Curve" dialogue box, the following settings were applied: "CIB1 value" was selected as the test outcome variable, and the "patient died or not" variable was selected as the status variable, with a value of “1” indicating death and a value of “0” indicating survival. The optimal cutoff value was determined using the Youden index, calculated in Excel by the following formula: sensitivity – (1 – specificity). The Youden index values were ranked to find the best cutoff value, which was found to be 21,103.8, corresponding to a Youden index of 0.132. The sensitivity ≈0.604, and the (1 – specificity) ≈ 0.472.
Statistical analysis
Cutoff values were calculated by IBM SPSS Statistics. Survival analysis curves were created using GraphPad Prism 9 (GraphPad Software), unless otherwise indicated in the figure legends; p < 0.05 was considered statistically significant. Data are presented as means and standard deviations.
Data and code availability
All spatial RNA-sequencing and single-cell RNA-sequencing data (including accession codes), are available in the ArrayExpress database, (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-12022 (scRNA-seq) and E-MTAB-12043 (spaRNAseq). Bulk mRNA-sequencing expression data (GSE151165) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
Acknowledgments
We gratefully acknowledge the financial support provided by the University Medical Center Göttingen’s intramural funding, the German Federal Ministry of Education and Research-funded consortia SATURN3 (BMBF: 0316173A; 01KD2206N) and CancerScout (BMBF: 13GW0451A), and by the Novo Nordisk Foundation, DK (NNF22OC0073440). S.F. was funded by the CSC, China Nr. 202108080171.
We acknowledge the approval of the study by the UMG Ethics Committee, Göttingen, and the collection of patient samples under the ethical approval of the UMG, Göttingen, Germany (Nr. 25/3/17, Nr. 24/4720, and Nr. 23/4/22). We extend our gratitude to Birgit Jünemann and Moritz Blume from the University Medical Center Göttingen, Germany, for their technical assistance.
Special thanks to Dr. Andreas Leha from the University Medical Center Göttingen, Germany, for his invaluable support in the statistical analyses. Additionally, we express our appreciation to Prof. Dr. Junichi Takagi from the Institute of Protein Research, Osaka University, Japan, for generously providing the WNT3A/Afamin cells.
Author contributions
Conceptualization: L.-C.C.; Supervision: L.-C.C. and T.D.O.; Methodology and Resources: L.-C.C., T.D.O., M.G., and J.M.K.; Manuscript Writing and Editing: J.R.F, S.F., T.D.O, L.-C.C.; L.D., and F.B.G.; Data Acquisition and Analysis: S.F., J.R.F., L.-A.E.R., T.D.O., L.-C.C., A.M.S., F.B.G., L.D., H.B., L.S.B., F.B., and X.X.; Figure Layout: J.R.F., S.F., T.D.O., and L.D. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omton.2024.200828.
Supplemental information
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Wagle N.S., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics, 2023. CA. Cancer J. Clin. 2023;73:233–254. doi: 10.3322/caac.21772. [DOI] [PubMed] [Google Scholar]
- 3.Vermeulen P.B., Colpaert C., Salgado R., Royers R., Hellemans H., Van Den Heuvel E., Goovaerts G., Dirix L.Y., Van Marck E. Liver metastases from colorectal adenocarcinomas grow in three patterns with different angiogenesis and desmoplasia. J. Pathol. 2001;195:336–342. doi: 10.1002/path.966. [DOI] [PubMed] [Google Scholar]
- 4.Latacz E., Hoppener D., Bohlok A., Leduc S., Tabaries S., Fernandez Moro C., Lugassy C., Nystrom H., Bozoky B., Floris G., et al. Histopathological growth patterns of liver metastasis: updated consensus guidelines for pattern scoring, perspectives and recent mechanistic insights. Br. J. Cancer. 2022;127:988–1013. doi: 10.1038/s41416-022-01859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández Moro C., Geyer N., Harrizi S., Hamidi Y., Söderqvist S., Kuznyecov D., Tidholm Qvist E., Salmonson Schaad M., Hermann L., Lindberg A., et al. An idiosyncratic zonated stroma encapsulates desmoplastic liver metastases and originates from injured liver. Nat. Commun. 2023;14:5024. doi: 10.1038/s41467-023-40688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frentzas S., Simoneau E., Bridgeman V.L., Vermeulen P.B., Foo S., Kostaras E., Nathan M., Wotherspoon A., Gao Z.-H., Shi Y., et al. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat. Med. 2016;22:1294–1302. doi: 10.1038/nm.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleischer J.R., Schmitt A.M., Haas G., Xu X., Zeisberg E.M., Bohnenberger H., Küffer S., Teuwen L.-A., Karras P.J., Beißbarth T., et al. Molecular differences of angiogenic versus vessel co-opting colorectal cancer liver metastases at single-cell resolution. Mol. Cancer. 2023;22:17. doi: 10.1186/s12943-023-01713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacob S., Bösch F., Schoenberg M.B., Pretzsch E., Lampert C., Haoyu R., Renz B.W., Michl M., Kumbrink J., Kirchner T., et al. Expression of CIB1 correlates with colorectal liver metastases but not with peritoneal carcinomatosis. BMC Cancer. 2021;21:1243. doi: 10.1186/s12885-021-08927-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahab M., Wadood A., Zheng G. Strategies for Targeting CIB1: A Challenging Drug Target. Curr. Pharm. Des. 2022;28:2343–2348. doi: 10.2174/1381612828666220728101812. [DOI] [PubMed] [Google Scholar]
- 10.Zhu W., Gliddon B.L., Jarman K.E., Moretti P.A.B., Tin T., Parise L.V., Woodcock J.M., Powell J.A., Ruszkiewicz A., Pitman M.R., Pitson S.M. CIB1 contributes to oncogenic signalling by Ras via modulating the subcellular localisation of sphingosine kinase 1. Oncogene. 2017;36:2619–2627. doi: 10.1038/onc.2016.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leisner T.M., Moran C., Holly S.P., Parise L.V. CIB1 prevents nuclear GAPDH accumulation and non-apoptotic tumor cell death via AKT and ERK signaling. Oncogene. 2013;32:4017–4027. doi: 10.1038/onc.2012.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Peng X., Zhang X., Xu H., Lu C., Liu L., Song J., Zhang Y. The Emerging Roles of CIB1 in Cancer. Cell. Physiol. Biochem. 2017;43:1413–1424. doi: 10.1159/000481873. [DOI] [PubMed] [Google Scholar]
- 13.Armacki M., Joodi G., Nimmagadda S.C., de Kimpe L., Pusapati G.V., Vandoninck S., Van Lint J., Illing A., Seufferlein T. A novel splice variant of calcium and integrin-binding protein 1 mediates protein kinase D2-stimulated tumour growth by regulating angiogenesis. Oncogene. 2014;33:1167–1180. doi: 10.1038/onc.2013.43. [DOI] [PubMed] [Google Scholar]
- 14.Jarman K.E., Moretti P.A.B., Zebol J.R., Pitson S.M. Translocation of Sphingosine Kinase 1 to the Plasma Membrane Is Mediated by Calcium- and Integrin-binding Protein 1. J. Biol. Chem. 2010;285:483–492. doi: 10.1074/jbc.M109.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtrich U., Wolf G., Yuan J., Bereiter-Hahn J., Karn T., Weiler M., Kauselmann G., Rehli M., Andreesen R., Kaufmann M., et al. Adhesion induced expression of the serine/threonine kinase Fnk in human macrophages. Oncogene. 2000;19:4832–4839. doi: 10.1038/sj.onc.1203845. [DOI] [PubMed] [Google Scholar]
- 16.Tejpar S., Stintzing S., Ciardiello F., Tabernero J., Van Cutsem E., Beier F., Esser R., Lenz H.-J., Heinemann V. Prognostic and Predictive Relevance of Primary Tumor Location in Patients With RAS Wild-Type Metastatic Colorectal Cancer: Retrospective Analyses of the CRYSTAL and FIRE-3 Trials. JAMA Oncol. 2017;3:194–201. doi: 10.1001/jamaoncol.2016.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA. Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 18.Lazaris A., Amri A., Petrillo S.K., Zoroquiain P., Ibrahim N., Salman A., Gao Z.H., Vermeulen P.B., Metrakos P. Vascularization of colorectal carcinoma liver metastasis: insight into stratification of patients for anti-angiogenic therapies. J. Pathol. Clin. Res. 2018;4:184–192. doi: 10.1002/cjp2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA. Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 20.Shinji S., Yamada T., Matsuda A., Sonoda H., Ohta R., Iwai T., Takeda K., Yonaga K., Masuda Y., Yoshida H. Recent Advances in the Treatment of Colorectal Cancer: A Review. J. Nippon Med. Sch. 2022;89:246–254. doi: 10.1272/jnms.JNMS.2022_89-310. [DOI] [PubMed] [Google Scholar]
- 21.Filip S., Vymetalkova V., Petera J., Vodickova L., Kubecek O., John S., Cecka F., Krupova M., Manethova M., Cervena K., Vodicka P. Distant Metastasis in Colorectal Cancer Patients-Do We Have New Predicting Clinicopathological and Molecular Biomarkers? A Comprehensive Review. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21155255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leisner T.M., Freeman T.C., Black J.L., Parise L.V. CIB1: a small protein with big ambitions. FASEB J. 2016;30:2640–2650. doi: 10.1096/fj.201500073R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibrahim N.S., Lazaris A., Rada M., Petrillo S.K., Huck L., Hussain S., Ouladan S., Gao Z.H., Gregorieff A., Essalmani R., et al. Angiopoietin1 Deficiency in Hepatocytes Affects the Growth of Colorectal Cancer Liver Metastases (CRCLM) Cancers. 2019;12 doi: 10.3390/cancers12010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada M., Kapelanski-Lamoureux A., Petrillo S., Tabariès S., Siegel P., Reynolds A.R., Lazaris A., Metrakos P. Runt related transcription factor-1 plays a central role in vessel co-option of colorectal cancer liver metastases. Commun. Biol. 2021;4:950. doi: 10.1038/s42003-021-02481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuboi S., Nonoyama S., Ochs H.D. Wiskott-Aldrich syndrome protein is involved in alphaIIb beta3-mediated cell adhesion. EMBO Rep. 2006;7:506–511. doi: 10.1038/sj.embor.7400665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haataja L., Kaartinen V., Groffen J., Heisterkamp N. The small GTPase Rac3 interacts with the integrin-binding protein CIB and promotes integrin alpha(IIb)beta(3)-mediated adhesion and spreading. J. Biol. Chem. 2002;277:8321–8328. doi: 10.1074/jbc.M105363200. [DOI] [PubMed] [Google Scholar]
- 27.Leisner T.M., Liu M., Jaffer Z.M., Chernoff J., Parise L.V. Essential role of CIB1 in regulating PAK1 activation and cell migration. J. Cell Biol. 2005;170:465–476. doi: 10.1083/jcb.200502090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright P., Haynes A., Markovic M. secuTrialR: Seamless interaction with clinical trial databases in R. J. Open Source Softw. 2020;5:2816. doi: 10.21105/joss.02816. [DOI] [Google Scholar]
- 29.Kaplan E.L., Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- 30.Ulm K. Primary endpoints in cancer trials. J. Thorac. Dis. 2011;3:82–83. doi: 10.3978/j.issn.2072-1439.2011.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piazza I., Beaton N., Bruderer R., Knobloch T., Barbisan C., Chandat L., Sudau A., Siepe I., Rinner O., de Souza N., et al. A machine learning-based chemoproteomic approach to identify drug targets and binding sites in complex proteomes. Nat. Commun. 2020;11:4200. doi: 10.1038/s41467-020-18071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camp R.L., Rimm E.B., Rimm D.L. Met expression is associated with poor outcome in patients with axillary lymph node negative breast carcinoma. Cancer. 1999;86:2259–2265. doi: 10.1002/(sici)1097-0142(19991201)86:11<2259::aid-cncr13>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Palmieri V., Lazaris A., Mayer T.Z., Petrillo S.K., Alamri H., Rada M., Jarrouj G., Park W.-Y., Gao Z.-H., McDonald P.P., Metrakos P. Neutrophils expressing lysyl oxidase-like 4 protein are present in colorectal cancer liver metastases resistant to anti-angiogenic therapy. J. Pathol. 2020;251:213–223. doi: 10.1002/path.5449. [DOI] [PubMed] [Google Scholar]
- 35.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All spatial RNA-sequencing and single-cell RNA-sequencing data (including accession codes), are available in the ArrayExpress database, (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-12022 (scRNA-seq) and E-MTAB-12043 (spaRNAseq). Bulk mRNA-sequencing expression data (GSE151165) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).