Abstract
Recent FDA modernization act 2.0 has led to increasing industrial R&D investment in advanced in vitro 3D models such as organoids, spheroids, organ-on-chips, 3D bioprinting, and in silico approaches. Liver-related advanced in vitro models remain the prime area of interest, as liver plays a central role in drug clearance of compounds. Growing evidence indicates the importance of recapitulating the overall liver microenvironment to enhance hepatocyte maturity and culture longevity using liver-on-chips (LoC) in vitro. Hence, pharmaceutical industries have started exploring LoC assays in the two of the most challenging areas: accurate in vitro-in vivo extrapolation (IVIVE) of hepatic drug clearance and drug-induced liver injury. We examine the joint efforts of commercial chip manufacturers and pharmaceutical companies to present an up-to-date overview of the adoption of LoC technology in the drug discovery. Further, several roadblocks are identified to the rapid adoption of LoC assays in the current drug development framework. Finally, we discuss some of the underexplored application areas of LoC models, where conventional 2D hepatic models are deemed unsuitable. These include clearance prediction of metabolically stable compounds, immune-mediated drug-induced liver injury (DILI) predictions, bioavailability prediction with gut-liver systems, hepatic clearance prediction of drugs given during pregnancy, and dose adjustment studies in disease conditions. We conclude the review by discussing the importance of PBPK modeling with LoC, digital twins, and AI/ML integration with LoC.
Keywords: Liver-on-chips, Liver organoids, Liver spheroids, Hepatic clearance, DILI, Microphysiological systems
Graphical abstract
Highlights
-
•
The last 4 years witnessed heightened interest in liver-on-chips.
-
•
Liver-chips recapitulate complexity through co-culture, dynamic perfusion and vascularization.
-
•
Challenges in clearance prediction of metabolically stable compounds can be addressed.
-
•
Compound pharmacokinetics prediction will be possible through integration of liver-chips with gut and kidney chips.
-
•
Emerging areas include PBPK modeling with LoC, AI/ML integration, and development of digital twins.
1. Introduction
The staggering cost of drug discovery, high drug attrition rates, ethical concerns related to animal usage in drug development, and poor translatability of animal models led to the development of more human-relevant advanced in vitro models in drug discovery [1]. These advanced in vitro models include a combination of the following: 3D environment, organ-relevant stem cells/primary cells/pluripotent stem cells, extracellular matrix, signaling factors, mechanical forces, and multi-cellular arrangement [2]. Many of these factors can be incorporated in miniature dynamic biofluidic systems termed organ-on-chips. Major applications of organ-on-chips include cell culture [3], disease modeling [4,5], drug discovery [5,6], personalized medicine [6,7] and tissue engineering & regeneration [8].
The last 10 years witnessed significant advances in this field with growing interest from government regulatory agencies, and pharmaceutical companies. Several initiatives such as FDA modernization act 2.0, NIH-DARPA Tissuechip program, European organ-on-chip society (EUROoCS), Japanese agency for medical research and development (AMED) MPS program, and joint workshop between FDA and pharmaceutical industries (Innovation and quality microphysiological systems (IQ MPS) Affiliate) catalyzed the development of several organ-on-chips companies [2].
Improving pre-clinical in vitro liver models has been the prime focus of the researchers due to four main reasons: (1) central role of liver in drug clearance and detoxification, (2) metabolic enzyme differences between human and pre-clinical animal species [9], (3) high drug attrition rate due to hepatotoxicity (∼21 %) [10], and (4) poor prediction rate of human hepatotoxicity from the animal studies [11,12].
Liver-on-chip (LoC) technology represents a promising advancement in preclinical in vitro science. LoC has five main benefits over conventional suspension culture of hepatocytes and monolayer culture of hepatocytes-based models: (1) relatively longer metabolic and transporter activities [13], (2) enhanced viability of hepatocytes over an extended period [14], (3) inclusion of dynamic physiological flow, (4) ability to incorporate non-parenchymal cells, liver-specific extracellular matrix components, and in vivo-like multicellular spatial arrangements, and (5) incorporation of vasculature. The rising interest in this technology is undeniable from the significant increase in the number of publications related to LoC in the last 10 years (Figure S1).
As mentioned before, the primary limitation of conventional monolayer and suspension cultures of hepatocytes is the rapid loss of metabolic activity over time [15]. Hence, sandwich cultures [16] and micropatterned models [17] have been developed. However, the 3D architecture of the liver tissue, cell-cell, and cell-ECM (extracellular matrix) interactions can be better mimicked by more advanced in vitro models, including liver spheroids [18], organoids [19], and 3D bioprinted models [20]. Combining such advanced in vitro models with LoC maintains three important conditions: (1) dynamic exchange of nutrients and waste products, reported to enhance the survival and functional activity of hepatocytes [21,22], (2) accumulation of cell-secreted molecules (for autocrine and paracrine signaling) due to low volume to cell ratio in microchannels [23], and (3) various gradients including oxygen gradient to maintain in vivo-like metabolic conditions [24]. One of the major benefits of culturing hepatocytes under dynamic flow is the possibility of connecting liver-chip with other organ-chips such as gut-chip and kidney-chip to predict the pharmacokinetic (PK) profile of the drugs given orally [25]. Current in vitro assays based on liver microsomes and 2D hepatocytes often underpredict the in vivo drug clearance [26]. Hence, one of the major focus areas of industrial research is the accurate prediction of the in vivo clearance using advanced LoC systems [27].
Despite the numerous benefits provided by liver-chip cultures, the industrial adoption of these assays in the regular drug discovery framework has been low [28,29]. Recently published IQ-MPS survey results highlighted that the majority of the companies used liver-on-chips for mechanistic studies and internal decision process [30,31]. The survey results also demonstrated the cautious approach of the industries in adopting such models. Furthermore, data derived from such studies were not included in regulatory filings. Some of the key challenges have been discussed in the previous papers, including the lack of standard guidelines from regulatory bodies for organ-on-chips that can aid in their industrial adoption [32,33]. To address standardization needs, a group of representatives from pharmaceutical companies (IQ-MPS) provided a set of criteria against which LoC devices need to be tested for usage in the safety assessment of compounds in pharmaceutical R&D (Table 1) [34].
Table 1.
Stage I LoC characterization guidelines by IQ-MPS [34].
| Functional marker | Specifications |
|---|---|
| Albumin secretion | 37–105 μg/day/million cells (should be stable across 14 days of culture duration, less than 50 % change over a 14 days period with less than 30 % C.V. of mean daily production rates) |
| Urea secretion | 56–159 μg/day/million cells (should be stable across 14 days of culture duration, less than 50 % change over a 14 days period with less than 30 % C.V. of mean daily production rates) |
| Gene expression of enzymes and transporters should be stable over time (levels should be comparable to cryopreserved hepatocyte in freshly prepared suspension) Baseline enzyme activity can also be measured and compared with fresh hepatocytes (part of stage II guidelines) |
Phase I enzymes (CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4), phase II enzymes (UGT1A1, GSTA1), uptake transporters (SLCO1B1, SLCO1B3, SLC22A1), and efflux transporters (ABCC2, ABCG2, ABCB1, ABCB11) |
We present this review on LoC aimed at evaluating the factors impeding their widespread adoption in drug discovery and highlighting the translational aspects of LoC by focusing on collaborative studies with the pharmaceutical industry. Our emphasis is on two crucial areas: accurate in vitro to in vivo extrapolation (IVIVE) of hepatic clearance and hepatic toxicity predictions.
For reliable prediction of clearance and toxicity, the hepatic model should maintain a steady level of functional markers such as albumin and urea production that correlates with in vivo values [34,35]. Moreover, the stable expression of phase I and phase II metabolic enzymes and drug-relevant uptake and efflux transporters should be maintained and match the levels reported in freshly thawed hepatocytes [34,35]. Compared to conventional 2D hepatic models, LoC-based hepatic models offer several key features such as dynamic flow culture, co-culture, and in vitro vascularization to enhance and preserve the aforementioned properties. We first describe the published reports on LoC to highlight the role of each key feature in improving the functionality of cultured hepatic tissues. Besides, we also compare the functional parameters of these models against the standard guidelines given by pharmaceutical industries (IQ-MPS) [34,35]. Once the functional markers are improved and stable, the hepatic clearance and drug-induced hepatic toxicities can be predicted confidently. However, LoC models should also be appropriately scaled-down with suitable design parameters for accurate IVIVE. Hence, we describe various mathematical scaling approaches to properly design miniature LoC platforms, that can reliably predict in vivo human hepatic clearance and drug toxicities. We specifically highlight industry-specific studies of LoC platforms pertaining to these two areas. We also discuss the emerging applications of LoC platforms by integrating advanced organoid cultures and other organ-chips for toxicity, clearance, and bioavailability prediction. Previous reviews provided comprehensive overview of advanced in vitro liver models [36,37]. This review specifically focuses on LoC-related industrial studies and discusses unique capabilities of LoC enabling their adoption in industries. Furthermore, evaluation of the LoC studies with standard guidelines (IQ-MPS) was lacking in the previous reports [[38], [39], [40], [41]]. Finally, we discuss few challenges for industrial adoption of LoC assays. We also provide several futuristic applications of LoC platforms, including the clearance prediction of drugs given during pregnancy, and other disease conditions that were not discussed in the previous reviews [[38], [39], [40], [41]]. We conclude the review by discussing the importance of PBPK modeling with LoC, digital twins, and AI/ML integration with LoC.
2. Important features offered by LoC platforms
2.1. Improved liver physiological mimicry: the role of fluid flow
Previous reports pointed out three main benefits of microfluidic dynamic culture: (1) enhanced drug delivery to cultured cells [42], (2) inter-organ communication through multi-organs-on-chips [43], and (3) convection-mediated transport of nutrients for longer cellular survival [44]. In this section, we have explained the role of fluidic flow on liver tissue maturation using LoC platform.
Increased blood flow after partial hepatectomy leads to hepatocyte proliferation and regeneration in mice, as shown by previous report [45]. The release of a paracrine signaling molecule, hepatocyte growth factor (HGF) by liver sinusoidal endothelial cells (LSECs) under elevated shear stress leads to a higher growth rate of hepatocytes (Fig. 1(a)) [46]. Hence, researchers have attempted to recapitulate the effect of shear stress on LSECs and downstream effect on hepatocytes in microfluidic devices. Chabbra et al. used spheroids of primary human hepatocytes co-cultured with human dermal fibroblasts. They detected higher production of HGF and hepatic proliferation under a mean shear stress of 3.95 dyne/cm2 applied to endothelial cells-lined vessel than static condition (Fig. 1(b)) [47].
Fig. 1.
Better liver physiological mimicry: the role of fluid flow (a) Increased shear stress after partial hepatectomy leads to enhanced HGF production and proliferation of hepatocytes. (b) Higher HGF production was recapitulated on a chip with endothelial cells lined vessel subjected to shear stress of 3.95 dyne/cm2 (n = 3 devices, Mean ± SEM, significance level was determined by *p < 0.05, ***p < 0.001, ****p < 0.0001, one-way ANOVA with Tukey’s multiple comparisons test). (c) The two-channel LoC device commercialized by Emulate Inc., operated with the dynamic flow on both top and bottom channels separated by a membrane at the same flow rate (30 μl/h) for toxicity prediction. (d) Higher functional activity of sandwich cultured hepatocytes in LoC: (i) Species-specific albumin production rates captured in LoC, which were higher than static plate culture, (ii) Higher CYP3A activity in LoC. CYP3A data was compared with Sidak’s multiple comparisons test (n = 3∼17 independent chips). Albumin data was compared with Sidak’s multiple comparisons test (n = 3∼14 independent chips, n = 3∼9 independent wells in plate). Significance level was determined by: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. All error bars were presented as Mean ± SEM. Panels (b) is adapted under terms of the CC-BY license from Ref. [47] Copyright 2022, PNAS. Panel (b) was rearranged and redrawn after removing channel dimensions. Panel (c) is adapted under terms of the CC-BY license from Ref. [13] Copyright 2022, Nature. In Panel (c), the device designs were redrawn indicating the flow rates. Panel (d) is reproduced with permission from Ref. [55] Copyright 2019, AAAS.
Fenestrated endothelium allows interstitial fluid flow and rapid transit of macromolecules to the hepatocytes. This allows nearly unimpeded access to the macromolecules traveling through the systemic circulation. Researchers have tried to decipher this effect of interstitial flow-mediated shear stress on primary hepatocytes. Li et al. used a single-channel microfluidic device seeded with mouse hepatocytes and found the highest hepatocyte proliferation rate at shear stress of 0.05 dyne/cm2 than lower shear stress values [48]. Further, they also found a higher albumin production rate under dynamic flow with shear stress of 0.05 dyne/cm2 than static culture. They found that β1 integrin-YAP (yes-associated protein) signaling was responsible for the shear stress-mediated increase in cell proliferation. The two-channel LoC device commercialized by Emulate Inc. was exposed to the dynamic flow on both top and bottom channels separated by a membrane at the same flow rate (30 μl/h) (Fig. 1(c)) [13]. The study demonstrated comparable metabolic activity of LoC to freshly thawed hepatocytes. Most importantly, CYP3A4 activity was maintained higher than freshly thawed hepatocytes for 7 days in the LoC platform. Due to five-fold higher apical channel height than basal channel, the shear stress experienced by liver sinusoidal endothelial cells (LSECs) cultured in the bottom channel was 25 times higher (∼11.25 × 10−3 dyne/cm2) than hepatocytes (∼0.445 × 10−3 dyne/cm2) cultured in the top channel. Hence, the design of their LoC platform correlates with liver physiology wherein LSECs experience higher shear stress due to direct contact with hepatic blood flow. However, the shear stress on LSECs was significantly lower than the reported in vivo values (0.1–0.5 dyne/cm2) [49]. Contrary to Emulate’s cellular arrangement, a previous study by Du et al. cultured hepatocytes and stellate cells in a bottom channel and LSECs and kupffer cells in a top channel separated by a membrane [21]. They found a synergistic effect of shear stress (applied only on the top channel, 0.1 dyne/cm2) and co-culture with non-parenchymal cells (NPCs). Albumin secretion, CYP activity, and HGF secretion were enhanced in the dynamic conditions than static conditions with NPC co-culture. The device used by Du et al. [21], differed from the Emulate’s platform [13] in terms of the device dimensions, cellular arrangement, and shear stress applied. Du et al. designed both chambers to have a height of 100 μm, whereas Emulate’s device features a top chamber height of 1 mm and a bottom chamber height of 200 μm. Additionally, in the Du et al. paper, hepatocytes were cultured in the bottom channel, while endothelial cells were cultured in the top channel. This setup contrasts with Emulate’s device, where hepatocytes were cultured in the top channel and endothelial cells in the bottom channel. Furthermore, the flow rate and shear stress applied in the two studies differed significantly. Du et al. applied a shear stress of 0.1 dyne/cm2 exclusively to the endothelial cells in the top channel. In contrast, Emulate applied a shear stress of 0.01125 dyne/cm2 to the endothelial cells in the bottom channel and a lower shear stress of 0.000445 dyne/cm2 to the hepatocytes in the top channel.
Induced pluripotent stem cells (iPSC) based hepatocyte (iHep) cultures are gaining popularity, as they don’t suffer from limited availability of patient-derived liver tissues. Recently, Zhang et al. demonstrated higher albumin and CYP450 enzymes activity of iPSC-derived hepatocytes under dynamic culture than static culture in well-plate [50]. In another study, Wang et al. demonstrated higher functional activity of iPSC-derived hepatic aggregates under dynamic flow than static culture [51]. Although 3D iHep-based culture models are promising, their albumin secretion rate remains significantly lower than in vivo values over 5 weeks under dynamic flow (<1 μg/day/million cells) [51,52].
Sandwich cultures are known to maintain hepatocyte polarity in culture. However, increasing evidence suggests that static well-plate-based sandwich culture models eventually dedifferentiate as shown by reduced albumin, urea, and metabolic activity within 2 weeks [53]. Dynamic fluidic flow improved the functional markers of sandwich model such as albumin production, urea production, and CYP enzymes expression under a shear stress of 0.6 dyne/cm2 than the static sandwich model in 14 days culture [54]. Similarly, Jang et al. captured species-specific (human, dog, rat) albumin production rates in dynamic LoC (Emulate) platform, that were significantly higher than static sandwich cultures [55]. The albumin production reached ∼60 μg/day/million cells on day 7 in their study for human sandwich cultured hepatocytes (Fig. 1(d-i)). CYP3A activity was also higher than static cultures and comparable to freshly isolated hepatocytes during 14 days in dynamic culture (Fig. 1(d-ii)).
High throughput is necessary for rapid industrial adoption of LoC assays. However, microfluidic technology is generally considered low-throughput. To address limitations related to throughput, Tan et al. introduced 96 well-plate-based dynamic LoC platform and found higher albumin, VEGF secretion, HGF secretion, and CYP3A4 activity than static sandwich cultures [56]. One of the reasons for the higher functional activity of sandwich cultures under dynamic flow can be higher proline-induced collagen secretion of hepatocytes, as suggested by Hegde et al. [57].
Rubiano et al. from center for drug evaluation and research (US FDA) used CN Bio’s dynamic microphysiological system (MPS) and demonstrated higher albumin and CYP3A4 activity than static 3D spheroids and sandwich culture models derived from primary human hepatocytes (PHH) [29]. Further, functional activity was observed for a longer duration (>2 weeks) in the MPS than spheroid culture (12 days) and sandwich culture (7 days). The albumin production in MPS peaked on day 15 (∼50 μg/million cells/day), which fell within the range provided by IQ-MPS guidelines. The CN Bio system has on-chip pump for recirculating the media through ECM-coated scaffolds containing self-assembled 3D hepatic microtissues.
Recently, the fluidic flow has also been shown to significantly enhance the culture longevity, metabolic activity, and albumin secretion of organotypic liver tissues (core-needle biopsy samples of the liver) than static well-plate-based culture [58]. Albumin secretion could be observed for 31 days in chip whereas well plate-based culture only supported culture up to 72 h. Similar results with enhanced functional markers were demonstrated by Rennert et al. under dynamic conditions for HepaRG cells [59].
Overall, culturing 3D spheroids, sandwich models, 3D microtissues, and organotypic models in LoC platforms has demonstrated marked improvement in hepatic functionality than conventional tissue plate cultures. Next, we discuss the role of co-culture in better physiological mimicry of liver in LoC platforms.
2.2. Improved liver physiological mimicry: the role of co-culture
The liver is majorly composed of hepatocytes (parenchymal cells), stellate cells, kupffer cells, hepatic biliary epithelial cells, and liver sinusoidal endothelial cells. All of them play a key role in liver regeneration after partial hepatectomy by secreting important growth factors, well-documented in previous literature [60,61]. Briefly, hepatocyte growth factor (HGF) produced by stellate cells and endothelial cells during regeneration is known to induce proliferation of hepatocytes. Moreover, kupffer cells start producing tumor necrosis factor α (TNFα) and IL-6. Hepatocytes produce pro-angiogenic factors (fibroblast growth factor 1 (FGF-1), and vascular endothelial growth factor (VEGF)) to induce vascularization. Hence, co-culture can be an effective way to induce liver maturation in vitro. Co-culture can be implemented by direct methods such as 3D spheroids, organoids, and micro-patterned co-culture or more complex strategies with 3D bioprinting and liver-on-chips.
HepatoPac (micro-patterned co-culture (MPCC)) is an excellent example of a direct co-culture system wherein the co-culture of fibroblasts with primary human hepatocytes enhances the culture longevity up to 6 weeks than hepatocytes monocultures [17]. Further, co-culture with fibroblasts is also known to improve the morphology and functionality of iHep [62]. However, HepatoPac offers limited spatial control over the cellular arrangement to mimic liver sinusoids. Further, the HepatoPac system usually lacks other non-parenchymal cells such as kupffer cells and liver sinusoidal endothelial cells that are known to play a significant role in drug-induced liver injury (DILI) mechanisms [36]. Bioprinting based approaches are popular to realize precise spatial arrangement of multiple cells to mimic hexagonal liver lobule structures. Using DLP (digital light processing), Ma et al. precisely deposited iPSC derived hepatocytes surrounded by non-parenchymal cells such as endothelial cells and adipose stem cells in a hexagonal shape [63]. They found the tri-culture model expressed higher albumin, urea and CYP3A4 activity compared to monoculture of iHeps.
3D liver spheroids can be co-cultured with non-parenchymal cells to enhance the functional activity, investigate drug-induced liver injury, and construct liver disease models [64]. InSphero’s hanging drop technology ensures matrix-free assembly of multiple cells including hepatocytes, biliary epithelial cells, kupffer cells, endothelial cells and stellate cells in 3 days with bile canaliculi formation [[65], [66], [67]]. Usually, co-cultured kupffer cells and stellate cells were observed throughout the spheroids in InSphero’s model [66]. However, biliary epithelial cells were located in periphery due to adhesion-dependent cell sorting [67]. Additionally, co-cultured spheroids with iHeps and endothelial cells demonstrated capillary like network formation and higher functional activity than monoculture of iHep spheroids [68].
Recently, Harrison et al. demonstrated that induced pluripotent stem cells derived liver organoids contained the non-parenchymal population including kupffer cells, cholangiocyte and stellate cells [69]. Moreover, liver organoids also included vascular luminal structures, confirming the presence of sinusoidal endothelial cells. Additionally, in contrast to spheroids, organoids demonstrated partial zonation shown by single cell RNA sequencing analysis in their study.
Similar to 3D spheroids and organoids models, microfluidic devices incorporating kupffer cells, stellate cells, and endothelial cells along with hepatocytes, have been reported to mimic the overall liver microenvironment. In contrast to 3D spheroids and organoids models, liver-on-chips can mimic spatial arrangement of cells in accordance with the liver sinusoidal structure. Ewart et al. co-cultured hepatocytes with all NPCs and captured idiosyncratic DILI due to trovafloxacin-mediated kupffer cells activation [13]. Further, the albumin production rate reached 25 μg/day/million cells on day 7 but it was slightly lower than the IQ MPS guidelines [34]. Besides, the device also failed to culture stellate cells in a space of Disse-like ECM environment. Despite the two drawbacks, the device successfully flagged hepatotoxic drugs with 87 % sensitivity. In another report, Du et al. demonstrated neutrophil recruitment through ICAM-1 expressing LSECs in a similar membrane based two compartment liver-chip using lipopolysaccharide (LPS) stimulation [21]. Such platforms can be potentially used to predict immune-mediated DILI of compounds.
Co-culture with NPCs enhances functional markers of hepatocytes. Recently, a study in collaboration with Sanofi and Merck demonstrated co-culture of iHeps with NPCs enhanced albumin secretion, urea secretion, and CYP3A4 activity than monocultured iHeps in high-throughput 384 well format Organoplate developed by Mimetas [70]. However, the albumin production rate was <20 μg/day/million cells, which is lower than in vivo values. In another study, liver spheroids co-cultured through fluidic connection with hepatic stellate cells on a microfluidic platform demonstrated higher albumin, urea secretion, and CYP450 activity than monoculture [71].
Co-culturing is an effective way for improving the functional maturity of hepatic tissues in LoC, as discussed in this section. Various design strategies have been implemented to allow spatial patterning and co-culture of cells in microfluidic platforms such as: (1) capillary pinning-based organ-on-chips (Fig. 2(a)), including micro-posts-based three channel devices (Fig. 2(a-i)) [72], and phaseguide-based three channel devices (Fig. 2(a-ii)) [73], (2) sacrificial material [74] or non-sacrificial material (needle) based strategy [47] (Fig. 2(b)). In 2nd strategy, a hollow space is generated inside a hydrogel material using either sacrificial material printed with bioprinting (Fig. 2(b–i)) or a needle-based strategy (Fig. 2(b-ii)). The tubular space can be endothelialized or epithelialized for dynamic perfusion culture of tissues, (3) Milli-compartments or micro-compartments based strategy (Fig. 2(c)), wherein standard transwells can be inserted and tissue epithelium and endothelium can be cultured on opposite sides of transwell’s membrane under flow (Fig. 2(c-i)). These chambers or compartments can also include 3D scaffolds. 3D scaffold-based strategy involves co-culturing various organ specific cells that are attached to 3D scaffold and each other under dynamic recirculating flow (Fig. 2(c-ii)). The devices in 3rd strategy can contain multiple organ compartments connected through recirculating flow, induced by on-chip micropump. Hence, they can be used for multi-organ cultures. Furthermore, they can also be used to culture multi-cellular 3D spheroids, 3D sandwich cultures, and 2D monolayers under recirculating flow. (4) membrane-based compartmentalization strategy ((Fig. 2(d)) [13]. In the 4th strategy the two channels can be controlled independently while maintaining the cross-talk between them through the micro-porous membrane sandwiched between the two channels. Table 2 lists several typical applications of each of these design strategies for liver organ modeling. Next, we discuss the strategies to incorporate vascularization in LoC models.
Fig. 2.
Various design strategies implemented to allow spatial patterning and co-culture of cells in microfluidic platforms: (a) Capillary pinning based: (i) Microposts based, (ii) Phaseguides based. (b) Sacrificial or non-sacrificial materials based strategy: (i) 3D bioprinting based strategy wherein hollow space is created inside hydrogel by removal of bioprinted sacrificial material, (ii) Needle based strategy wherein hollow space is created inside the hydrogel. In both (i) and (ii) the hydrogel can contain cells and hollow space can be endothelialized or epithelialized. (c) Milli-compartments or micro-compartments based strategy: (i) Transwell-based dynamic organ-on-chips, (ii) 3D scaffold-based dynamic organ-on-chips. (d) Membrane-based compartmentalization strategy. Figure (c–i) is adapted under terms of the CC-BY license from Ref. [75] Copyright 2022, Springer.
Table 2.
Typical applications reported for each design strategy.
| Design strategy | Reported applications of LoC |
|---|---|
| Capillary pinning-based devices | Primarily used for 3D culture of hepatic tissues inside hydrogels for toxicity prediction and generating vascularized liver tissues for disease modeling [76,77] |
| Sacrificial or non-sacrificial materials-based devices | To mimic the tubular structures of bile ducts and blood vessels. This approach is used to culture 3D hepatic tissues under dynamic perfusion, model cholestatic liver diseases, and study liver biology [47,78,79] |
| Milli-compartments or micro-compartments-based strategy | It can typically be used for conducting multi-organ interaction studies with liver. Several applications include PK profile prediction, transwell-based dynamic studies, and culture of various forms of 3D hepatic tissues for toxicity and clearance prediction [[80], [81], [82], [83]] |
| Membrane-based compartmentalization strategy | For sandwich culture of liver hepatocytes and co-culture with other liver specific cells for toxicity prediction [13] |
2.3. Improved liver physiological mimicry: vascularized models & bile ducts-on-chips
One of the major drawbacks of the current 3D models is the lack of vascularization, which starts during embryogenesis to ensure adequate nutrient supply during the later stages of organ development [84]. Given that the liver is a highly vascularized organ, the development of in vitro vascularized 3D models could be crucial for accurately mimicking the intricate processes of drug distribution and clearance in the liver.
One of the main benefits of organ-on-chip cultures is the ability to integrate vascular networks with organ parenchyma. Several methods to form vascularized organ-on-chips have been employed: (1) endothelialization of the hollow tube created by sacrificial inks (Fig. 2(b–i)) [74] or needles Fig. 2(b-ii)) [47], (2) facilitating confluent blood vessel formation covering the microfluidic channel by gravity-assisted adhesion to hydrogel and dynamic flow (Fig. 3(a-i)) [85], (3) endothelial lining at the interface of tissue epithelium separated by a membrane in middle (Fig. 3(a-ii)) [55], and (4) incorporation of endothelial cells with supporting matrix, and pro-angiogenic factors to allow natural sprouting (Fig. 3(a-iii)) [77].
Fig. 3.
(a) Strategies to vascularize organ-on-chips: (i) Facilitating confluent endothelial vessel formation covering the microfluidic channel by gravity-assisted adhesion to hydrogel and dynamic flow, (ii) Endothelial lining at the interface of tissue epithelium separated by a membrane in the middle, (iii) Incorporation of endothelial cells along with supporting matrix, and angiogenetic factors to allow natural sprouting. (b) Vascularized hepatic spheroids-on-chips: (i) OrganoPlate Graft (Mimetas) having an open-top chamber to introduce hepatic spheroid on pre-vascularized beds, (ii) The hepatic spheroid was stained with albumin (green) and microvessels were stained with CD31 (red) (Scale bar: 200 μm), (iii) The functionality and integration of vascular network were validated with perfusion of FITC-dextran through the left channel and retrieving solution from the right channel (shown by dotted ellipse). (c) primary sclerosing cholangitis-on-chip: (i, ii) Vascular and bile-duct on chip to study primary sclerosing cholangitis (VE-cadherin (red) shows endothelial cells, fibroblasts are shown in magenta, and cholangiocytes stained with Keratin-19 (green)) (Scale bar: 200 μm), (iii) Immune cells migration from the vascular channel (tracked in green), (iv) Quantification of enhanced migration of PBMCs from vascular channel under IL-17A stimulation (one way ANOVA or ANOVA on RANKs, Mean ± SD, n > 3 devices) ((E): endothelial cells, (C): cholangiocytes, (F): fibroblasts). Panel (b) is adapted under terms of the CC-BY license from Ref. [77] Copyright 2022, Springer. The device design in panel (b–i) was redrawn for better clarity and resolution. Panel (c) is reproduced under terms of the CC-BY license from Ref. [79] Copyright 2023, IOP science. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
On-chip vascularization has been reported to increase cell proliferation and viability of 3D culture models [86]. Using the sacrificial ink approach, on-chip vascularization has been able to support culture of 1 cm thick tissues for 6 weeks, which is unlikely with passive diffusion in conventional static culture [74].
Using a needle-based approach, a hollow space was endothelialized and perfused to culture PHH spheroids encapsulated in a matrix [47]. Liu et al. devised a strategy for hepatic aggregates culture with sacrificial gelatin methacrylate (GelMA) ink [87]. However, neither of the 3D spheroid studies could fabricate interconnected complex hepatic capillary networks reminiscent of in vivo structures. Lee et al. used sacrificial poly(N-isopropylacrylamide) (PNIPAM) fibers-based strategy to make such convolute structures with an average size of 17.78 ± 8.37 μm [88]. Furthermore, perfusion through the network induced higher albumin secretion, and CYP3A4 activity of hepatic spheroids during 10 days of culture than static group. Although the smaller size of microchannels was achieved, they remained unendothelialized.
Majority of the previous studies didn’t report the integration of the vascular network with 3D hepatic spheroids and tissues. However, recently Bonanini et al. used high-throughput open-top pre-vascularized beds-on-chips (OrganoPlate Graft, Mimetas) to integrate hepatic spheroids with vascular network (Fig. 3(b-i)) [77]. Initially, confluent endothelial vessels were established. Subsequently, natural vessel sprouting was induced by introducing a medium containing pro-angiogenic growth factors into the graft chamber. Finally, hepatic spheroid was introduced from the open-top chamber. The hepatic spheroid was stained with albumin (green) and microvessels were stained with CD31 (red) (Fig. 3(b-ii)). Further, the functionality and integration of the vascular network with spheroid were validated with perfusion of FITC-dextran from the left channel. Upon perfusion from the left channel, FITC-dextran was found in the right channel proving successful grafting with the spheroid (shown by a dotted ellipse) (Fig. 3(b-iii)).
The needle-based approach (Fig. 2 (b-ii)) has been used to form bile ducts-on-chips. Du et al. used this strategy to form a hollow tubular space lined by mouse cholangiocytes [78]. They successfully evaluated the barrier properties of the bile duct and glycocalyx-mediated protection of cholangiocytes from bile acid. In another study, the same group demonstrated recapitulation of primary sclerosing cholangitis by incorporating vascular and bile ducts using the same needle-based approach on a chip (Fig. 3(c-i, ii)) [79]. The endothelial cells were stained by VE-cadherin (red), cholangiocytes by keratin-19 (green), and fibroblasts were tracked in magenta. When cholangiocytes were stimulated with IL-17A, transmigration of PBMCs and CD 4 T cells could be observed (tracked in green) from the vascular channel (Fig. 3(c-iii, iv)).
However, current LoC platforms do not replicate the countercurrent bile flow alongside the existing sinusoidal flow. Bile juices are crucial for the digestion and absorption of fats. The absence of bile flow in current LoC platforms represents a significant unmet challenge that must be addressed for accurate prediction of drug clearance, PBPK (physiology-based pharmacokinetics) modeling from LoC, and predicting drug-induced cholestasis. Bile is a major route of clearance for several drugs/metabolites with higher molecular weight of more than 500 Da. Phase II reactions such as glucuronidation and conjugation with glutathione result in increased molecular weight of metabolites and lead to their biliary excretion. Lack of bile flow on LoC platforms severely limits accurate clearance prediction of such compounds. Furthermore, it also prevents study of enterohepatic cycling of drugs through liver-gut-on-chips. Therefore, future efforts should focus on integrating bile flow with hepatocytes to evaluate bile clearance of drugs without altering the media composition [89].
3. Prediction of hepatic clearance
Liver microsomes and suspension cultures of PHH are conventional in vitro models used in hepatic clearance prediction of compounds [90]. However, the major limitation related to these models is significant underprediction of in vivo CLint by more than 3-fold [91]. Further, they are not suitable for predicting clearance of metabolically stable compounds, since their metabolic activity is lost rapidly within a few hours. Additionally, traditional well-plate-based 2D PHH cultures also show sharp decline in metabolic activity within 24 h [15]. Bonn et al. demonstrated that conventional PHH culture could only predict clearance of two out of six low-clearance compounds within a 2-fold range [92].
A more advanced model of sandwich cultured hepatocytes eventually dedifferentiates, as shown by reduced albumin, urea, and metabolic activity within 2 weeks [53]. Additionally, sandwich model also failed to predict clearance of metabolically stable compounds within a 2-fold range [93,94]. Short incubation time with drugs (<2 days) and declining metabolic activity of hepatocytes in sandwich cultures are some of the possible reasons behind the failure of clearance prediction of metabolically stable compounds. These limitations promoted the development of complex in vitro models, including co-culture models, 3D spheroids and LoC models [95].
Liver spheroids allowed drug depletion monitoring for over 7 days and maintained significantly higher metabolic activity than sandwich cultured and 2D cultured hepatocytes [18,53]. Spheroid cultures predicted clearance of six out of seven low-clearance compounds within a 3-fold range and three compounds within a 2-fold deviation [18].
In a remarkable study, Chan et al. used the co-culture or MPCC model (HepatoPac) for ten low-clearance compounds and predicted hepatic clearance for all ten compounds within a 3-fold deviation and seven compounds within a 2-fold range [96]. Furthermore, they could measure compound depletion for 7 days like the spheroids model. HepatoPac has shown excellent functional activity with albumin production rate closer to in vivo condition in the liver [97]. Hence, a correction factor for albumin facilitated drug uptake mechanism further improved the overall clearance prediction ability of HepatoPac compared to direct and conventional correction methods [97]. Moreover, HepatoPac could also reasonably predict the clearance within 2–3 fold for the various compounds classified according to extended clearance classification system (ECCS) [98]. In addition to its robust phase I metabolic activity, HepatoPac also demonstrated strong phase II metabolic capabilities. A report by Docci et al. suggested HepatoPac could reasonably predict clearance of 69 % of the UGT substrates within 3 fold variation [99].
Some of the limitations of MPCC assays include: (1) the requirement of stromal-only control, as stromal cells demonstrate drug dependent UDP-glucuronosyltransferase (UGT) activity [100], (2) underprediction of intermediate to high clearance compounds when plasma protein binding correction was applied [96], (3) low activity of non-CYP enzymes such as flavin-containing monooxygenases (FMO) and aldehyde oxidase (AO) [101], (4) a non-physiological ratio of hepatocytes to stellate cells of 4:1, deviating from the human liver composition where hepatocytes constitute approximately 80 % and stellate cells about 8 %, suggesting a more physiological ratio would be closer to 10:1 [36]. Despite some limitations, HepatoPac remains one of the most reliable systems for predicting hepatic clearance. Besides, in a recent study multi-well array culture of spheroids and HepatoPac demonstrated exceptional performance in predicting clearance of slowly metabolized compounds [90]. Therefore, conducting benchmarking studies against HepatoPac and 3D spheroids is essential for LoC models aiming to enhance clearance prediction accuracy. Furthermore, future efforts in LoC should focus on value addition to the existing state-of-the-art in clearance prediction. Incorporating biliary flow on the LoC could add significant value and enhance the predictive capability of pre-clinical in vitro models for the hepatic clearance of compounds.
In contrast to static models such as HepatoPac and spheroids, LoC models include physiological flow. The selection of the flow rate should be reflective of the in vivo conditions. Apart from media flow rate, hepatocyte cell density, and media volume should reflect the in vivo liver conditions properly in a scaled-down LoC system for reliable prediction of human in vivo clearance (IVIVE). The usual steps followed for IVIVE of hepatic clearance are [102]: (1) measurement of (intrinsic clearance) CLint, in vitro from in vitro assays (equation (1)), (2) calculation of scaled CLint, scaled by considering factors of hepatocellularity and liver weight (equation (2)), and (3) prediction of hepatic clearance (CLH, predicted) by considering appropriate clearance model (well-stirred (equation (3)) or parallel-tube model (equation (4))) to incorporate the effect of liver blood flow.
| (1) |
| (2) |
| (3) |
| (4) |
K = elimination rate constant, V = incubation medium volume, NH = Number of hepatocytes, HC = hepatocellularity, HLW = human liver weight, CLint = intrinsic clearance, Qh = liver blood flow rate, CLH = hepatic clearance, fub = fraction unbound in blood, fuinc = fraction unbound in incubation.
The scaling approaches are critical to extrapolate the findings of LoC to in vivo human liver (IVIVE). The goal of scaling approaches is to select the LoC design and operational parameters, such as media volume, hepatocyte cell number and flow rate so that the results produced by such systems translate well to in vivo systems. Several scaling approaches have been reported to select LoC designing parameters: (1) allometric scaling [103], (2) direct scaling [104], (3) parametric scaling [105], (4) similarity scaling [106], and (5) functional scaling [104]. As the CLint, in vitro is a function of hepatocyte cell number (can be controlled by the user and depends on LoC design) and media volume (can be controlled by the user and depends on LoC design), selection of these scaled-down parameters directly affects the clearance prediction accuracy of the LoC model. Moreover, the media flow rate in LoC also needs to be carefully scaled down.
A conventional allometric scaling approach based on body mass has been widely used to estimate human hepatic clearance from animal studies. Allometric scaling has also been proposed to design microfluidic systems. LoC systems can be viewed as micron level versions of humans, scaled down by a factor of 106. Using this microhuman (μHu) scaling factor, the average body weight of a μHu can be approximated as 70 mg (adult body weight/106). Using reported allometric powers and allometric coefficients for the liver, various parameters such as liver blood volume, liver blood flow rate, and number of hepatocytes can be calculated for 70 mg μHu [107]. The biggest benefit of using this approach to design LoC systems is the extensive availability of reference materials detailing allometric relationships for various parameters [107,108]. However, for multi-organ-chip systems, linear allometric scaling based on organ volume and organ surface area often produces channel dimensions difficult to achieve using existing fabrication methods [103]. Further, it produces higher brain mass and organ blood volume than μHu while designing body-on-chip systems [103,107]. Hence, the allometric scaling approach is more appropriate for designing single organ-on-chips than multi-organs-on-chips. However, designing single organ systems like LoC based on allometric scaling results in liver blood volume of less than 5 μl (with an allometric power of 0.86). This small volume is prone to rapid evaporation, posing challenges for regular sampling required for LC/MS analysis [107]. Hence, allometric scaling may not be practically suitable for designing LoC.
Another approach includes linear direct scaling, wherein LoC parameters are directly scaled down by 106 for μHu. Major advantage of direct scaling approach is that it is very simple to implement [108]. However, both allometric and direct approaches have been criticized in the literature, as they are based on organ-specific physical properties such as surface area, organ volume, flow rates, organ mass and not based on organ-specific functions (e.g. for liver: drug clearance) [104,109]. Furthermore, the biggest disadvantage is that they provide a single set of design parameters for single or multi-organ-on-chips independent of the application or context of use (CoU) [104].
To overcome the limitations related to traditional allometric and direct scaling approaches, functional scaling and multifunctional scaling approaches have emerged. Wikswo et al. defined the functional approach as an iterative design process based on key functional aspects of the organ [107]. The chip designed based on this approach recapitulates the biological function [104]. For functional scaling, a mechanistic modeling of the biological processes provides a foundation. Here, a goal is to match the observed in vitro clearance on the LoC platform to reported in vivo clearance values. Based on the mathematical governing equations, objective function minimization, and specified constraints, LoC design parameters can be iteratively obtained [104].
The objective function represents the desired biological functions that the system should reproduce and estimates the platform design parameters such as flow rate, media volume, and cell density that best satisfy the objective function. The objective function is defined by a weighted squared difference between a model outcome (prediction) and corresponding measurements (clinical data that need to be reproduced for training set of drugs). Maass et al. designed a liver-gut system using the objective function approach, which needs to reproduce clinical or in-vivo time-concentration profile of drugs by considering the reported in vitro liver metabolism and gut absorption values [104]. First, they defined the mathematical governing equations for biological processes of liver-gut system. Next, they specified known or desired model parameters. Next, they defined the objective function representing main biological functions of liver-gut system (metabolism for liver and absorption for gut). For optimization process they selected available experimental data (in vitro liver metabolism and gut absorption values). Finally, they selected the design parameters based on minimization of objective function and defined constraints. Finding the minima of the objective function will ensure that the model reproduces the in vivo time-concentration profiles of drugs and the suitable design parameters are established. In the end, the virtually designed liver-gut system was assessed by predicting concentration profiles of a test set or unknown set of drugs not included in training the model. The area under the normalized time-concentration curve (AUNC) was calculated from the predicted time-concentration profile of drugs. More specifically, AUNC was calculated from time of administration (t = 0) to the last measurement point of the drug specific time-concentration profile by integration [104]. The predicted AUNC for a test set of five drugs remained within a 2-fold deviation. Furthermore, it was found that, in contrast to the objective function minimization approach, both direct and allometric scaling approaches resulted in lower drug exposure and more rapid clearance compared to observations in vivo. Despite the promising results, several limitations of the study were: (1) a limited set of test drugs, (2) difficulty in applying functional approach for unknown new drug molecule and (3) usage of clearance data of suspended hepatocytes in the training set, which may not translate well for predicting clearance and time-concentration profiles of low clearance compounds.
Shuler lab introduced a set of design criteria based on functional scaling for organ-on-chips [105]. Contrary to the approach based on objective function minimization, Shuler defined a set of parametric equations for microfluidic chips that need to match the reported in vivo values. Apart from matching the drug concentration vs time curves, they also mentioned maintaining similar fu (unbound drug fraction), drug partitioning coefficient/blood to plasma ratio (Kp/B:P), drug residence time, and cardiac output rate for an organ cultured on microfluidic device. However, it is challenging to match the drug-specific physicochemical properties such as unbound drug fraction, blood: plasma ratio, and drug partitioning coefficient as observed in vivo.
Shuler has also used residence time-based scaling extensively in his work for designing multi-organ chips [108]. The volume/flow rate ratios (V/Q) for the organ-on-chips are matched with physiological values to ensure that the chemical cues are exposed to the organ for the same duration in vitro as they would be in the body. According to his theory, the in vitro intrinsic clearance should match in vivo clearance if the hepatocytes maintain similar metabolic activity in vitro as hepatocytes in the liver and liquid to cell ratio is maintained close to 1:2 [108].
Contrary to the functional approaches, Feng et al. demonstrated a similarity scaling approach based on π-theorem, which is used widely in the mechanical engineering domain [106]. The process involves defining a set of key parameters and non-dimensional ratios for the intended application of organ-on-chips. These ratios for the model (LoC) are matched to the prototype (a fully functionalized liver organ) to obtain design parameters. Although this approach can be useful for single-organ chip design, it may not satisfy all ratios for multi-organ system designs. Moreover, a few of the dimensionless ratios defined for LoC systems may not match in vivo values (e.g. fu), which leads to partial similarity.
A seminal paper by Herland et al. used mathematical governing equations to describe biological processes on multi-organ system consisting of liver, gut and kidney [110]. The in silico model, predictive of the experimental data on chip, could translate the in vitro findings and replicate the clinical PK profiles of drugs using scaled up parameters to represent full organ volumes, blood vessel length, and blood flow rates.
From the discussion, it can be concluded that allometric and direct scaling approaches are not appropriate for LoC designing, since they produce blood volume less than 5 μl. On the contrary, functional approach is more appropriate, since it recapitulates the organ function and produces the designing parameters based on the CoU [104,109]. Similarity scaling approach is a promising method but incorporates several dimensionless ratios that are often not practically achievable. Overall, functional scaling approach seems to be the most reliable method to design single and multi-organ-on-chips. Table 3 compares various design parameters of LoC reported by several scaling approaches discussed above. The liver tissue weight ranged from 1.52 to 3.87 mg, media flow rate values ranged from 12.8 to 1.45 μl/min, incubation medium volume ranged from 1.8 μl to 300 μl, and hepatocyte number ranged from 0.3 to 0.8 M. Furthermore, it should be noted that the liver blood volume, flow rate, and hepatocellularity can be effectively scaled down but drug-specific properties and extracellular matrix-specific properties (fu, porosity of matrix, permeability of matrix, effective diffusivity of matrix) are difficult to match the human in vivo values. Potential approaches in order to overcome some of these issues include liver slice and liver decellularized extracellular matrix-based (dECM) culture models. This is because liver slice and liver decellularized matrix help to preserve the native ECM components, which are crucial for mimicking the in vivo environment, leading to similar diffusion coefficient, drug binding factor to ECM, and porosity. However, further research will be required to validate the same by employing dECM and liver slice models. Moreover, for dECM all the physical properties will highly depend on the dECM concentration and crosslinking mechanisms.
Table 3.
LoC design parameters for various scaling approaches.
| In vivo values | Allometric scaling | Functional scaling | Direct scaling | |
|---|---|---|---|---|
| Liver tissue weight(g) | 1520 | 3.87 × 10−3 [107] | 1.52 × 10−3 [107] | 1.52 × 10−3 |
| blood flow rate (ml/min) | 1450 | 1.28 × 10−2 [107] | 1.45 × 10−3 [107] | 1.45 × 10−3 |
| Blood volume (ml) | 1800 | 3.9 × 10−3 [104] | 0.3 [104] | 1.8 × 10−3 |
| Number of hepatocytes | 3 × 1011 | 8 × 105 [104] | 5 × 105 [104] | 3 × 105 |
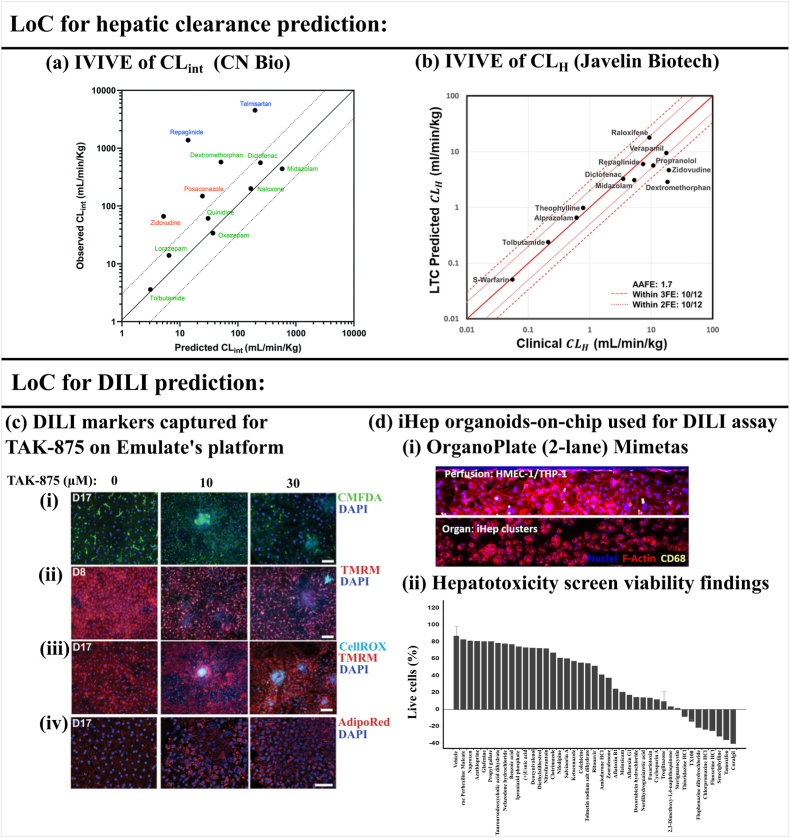
Baudoin et al. used a dynamic perfusion culture model with rat primary hepatocytes for hepatic clearance prediction in rats [111]. However, the model was based on 2D culture without matrix overlay, which only supported the observation of drug clearance up to 24 h and clearance prediction for medium to high clearance compounds. This was evident from poor hepatic clearance prediction for one low-clearance compound tested in their study (tolbutamide, within 5-fold). Hence, focus among the researchers has been on accurately predicting the clearance of metabolically stable compounds using advanced LoC models. Docci et al. from Roche used PHH-based 3D microtissue model (CN Bio) to predict intrinsic hepatic clearance of compounds [82]. The average albumin production rate was 39 ± 8 μg/day/million cells for 48 h. Further, they measured the drug depletion up to 4 days (for low clearance compounds) and found predicted CLint, predicted for 58 % of the 12 compounds within a 3-fold deviation (Fig. 4(a)). Moreover, predicted in vivo intrinsic clearance for metabolically stable compound tolbutamide was within the 1.5-fold range. Overall, 10 out of 12 compounds remained underpredicted from the chip. Some of the possible reasons can be: (1) declining metabolic activity of hepatocytes with time, (2) declining activity of transporters, (3) comparison of predicted CLint instead of CLH to in vivo values.
Fig. 4.
(a) Intrinsic clearance prediction (IVIVE of CLint) from CN Bio’s 3D microtissue-based model. The solid line shows 100 % prediction accuracy and the dotted line shows the ±3 fold range. Class 1a and 2 compounds are shown in green, class 3 compounds in blue, and class 4 compounds in red (according to ECCS: extended clearance classification system). (b) Hepatic clearance prediction from Javelin Biotech’s platform using parallel tube model. (c) Idiosyncratic DILI investigation of TAK-875 on LoC: (i) MRP2 transporter activity upon treatment with TAK-875 measured by efflux of MRP2 substrate: CMFDA, (ii) Mitochondrial membrane potential shown by TMRM on day 8, (iii) ROS formation shown by CellRox on day 17, (iv) Lipid accumulation shown by AdipoRed on day 17. (Scale bars = 100 μm) (d) High-throughput OrganoPlate developed by Mimetas cultured with iHep organoids: (i) Organ channel seeded with iHep and perfusion channel contains HMEC-1 and THP-1, (ii) Cell viability (%) results for 40 out of 159 compounds with evidence of hepatotoxicity are rank ordered going left to right from least to most toxic. The viability of the remaining compounds were at vehicle control levels. Error bars are shown for vehicle and troglitazone controls (n = 8). All cultures were exposed to test compounds for 72 h at 50 μM (n = 1). Panel (a) is adapted under terms of the CC-BY license from Ref. [82] Copyright 2022, RSC. The dotted line showing the ±3 fold range was added. Panel (b) is reproduced under terms of the CC-BY license from Ref. [81] Copyright 2023, Springer. Panel (c) is adapted with permission from Ref. [55] Copyright 2019, AAAS. Panel (d) is reproduced under terms of the CC-BY license from Ref. [76] Copyright 2021, Elsevier. We have rewritten the names of the compounds in Figure (d-ii) for the better clarity of the readers. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
On the contrary, Rajan et al. from Javelin Biotech, in collaboration with Pfizer used their sandwich culture model and compared predicted CLH from the LoC system to reported in vivo hepatic clearance values. Interestingly, they found hepatic clearance of 83 % of the 12 compounds fell within the 2-fold range (Fig. 4(b)) [81]. Furthermore, the platform could maintain sandwich cultured hepatocytes for 15 days with albumin production of 40 μg/day/million cells (day 15) and urea secretion rate of 100 μg/day/million cells (day 15). The reported values were comparable with the IQ-MPS guidelines. According to the same guidelines, they also found that genes related to CYPs, UGTs, and drug transporters (SLCs, ABCs) expressed for 15 days. The platform could predict the clearance of five metabolically stable compounds with an average absolute fold error (AAFE) of 1.1. However, three high-clearance compounds produced significantly higher AAFE (3.7). Overall, the number of reports on LoC-based hepatic clearance prediction assays has remained very low over the last 10 years. Therefore, chip makers and researchers should dedicate more effort to this area.
4. Prediction of hepatic drug safety
Historical data suggest that nonrodents failed to flag 37 % of the compounds, and rodent species missed detecting 57 % of the 150 compounds, that were found toxic to humans [112]. Drug-induced liver injury (DILI) continues to be one of the major causes of drug attrition in the 21st century [113]. This indicates an urgent requirement for human-relevant DILI prediction assays in drug discovery. A comprehensive overview of DILI predictions using pre-clinical in vitro models, including complex in vitro models, can be found in several excellent previously published reviews [14,113]. Here, we focus on the studies jointly published by commercial chip manufacturers and pharmaceutical companies.
Various mechanisms of drug-induced liver injury (DILI) can be [113]: (1) mitochondrial dysfunction, (2) inhibition of biliary efflux (cholestasis), (3) lysosomal impairment, (4) production of reactive metabolites, (5) endoplasmic reticulum stress, and (6) immune-mediated DILI. The drug induced inhibition of mitochondrial fatty acid oxidation is one of the common causes of intracellular lipid accumulation and leads to hepatic steatosis [114]. Furthermore, inflammation combined with lipid accumulation leads to more serious disorder known as non-alcoholic steatohepatitis (NASH) [114]. Cholestasis in many cases is developed due to the drug induced inhibition of membrane transporters [113]. Bile salt export pump (BSEP) is the transporter involved in efflux of bile salts into the bile. Hence, drug inhibiting BSEP is one of the major mechanisms leading to cholestasis. Drug-induced inhibition of lysosomal enzymes involved in degradation of cellular membranes can lead to phospholipidosis.
Liver detoxification can lead to formation of chemically reactive metabolites [113]. These reactive metabolites covalently bind to cellular macromolecules such as proteins, enzymes, lipids and DNA. When these reactive metabolites are combined with protein molecules (hepten-like drug-protein adducts), they are recognized as foreign materials, leading to immune system activation and response. The accumulation of such reactive metabolites can initiate liver function disruption, involving oxidative stress (ROS accumulation), inflammation, mitochondrial function disruption, impaired bile acid efflux, and in severe cases, liver failure. Detailed DILI mechanisms have been discussed in the recent review paper with a focus on idiosyncratic liver injury [115].
As mentioned by Walker et al., extended exposure of drugs through repeated dosing to the in vitro hepatocyte model holds a key to accurate DILI prediction in humans [116]. Hence, research efforts are focused on developing 3D liver models capable of maintaining their functionality over an extended period, in contrast to conventional 2D models that have a limited culture lifespan. PHH-based 3D spheroid models have provided promising results in DILI prediction with repeated drug exposure and culture duration of 2–4 weeks [67,116,117]. As shown by Bell et al., the toxicity of fialuridine was not observed in PHH spheroids even at 100 × Cmax for the short-term exposure of 48 h [67]. However, the toxicity was evident after the long-term exposure of fialuridine to PHH spheroids for 7 days. Further, the IC50 approached close to the reported clinical Cmax values with 7–28 days of exposure of five hepatotoxic drugs in their study.
Moreover, 3D spheroids have been extensively characterized to detect drug-induced cholestasis and steatosis [67,118]. 3D spheroids could also capture species-specific hepatic toxicities as shown by Vorrink et al. [117]. They could demonstrate the acetaminophen toxicity observed in mouse and human hepatic spheroids but not in rat and monkey spheroids correlating with in vivo data. The spheroid model could identify 69 % of the 123 toxic drugs with 100 % specificity. Their spheroid model also proved to be better than sandwich-cultured hepatocytes and MPCC models in DILI prediction. Although 3D spheroids models have been greatly helpful, they usually take around a week time to form in low-attachment plates, making the process time consuming.
Apart from 3D spheroids, LoC based models have also been implemented for DILI predictions. Poor specificity of LoC, particularly due to oversimplified models that exclude non-parenchymal cells (NPCs), is a significant limitation. However, a recent collaborative study among Emulate Inc, Abbvie, and Janssen pharmaceuticals demonstrated excellent DILI detection sensitivity (87 %) and specificity (100 %) of LoC with drug dosing for a week [13]. This high level of accuracy can be attributed to the inclusion of all relevant NPCs alongside hepatocytes and the application of dynamic perfusion flow. The endpoints included albumin and alanine transaminase (ALT) assays. Foster et al. from AstraZeneca compared liver spheroids and LoC (Emulate Inc.) with equal endpoints and drug exposure. Their findings indicated comparable DILI sensitivity across both platforms [119].
Cox et al. from UCB Biopharma observed similar functional activity in static liver spheroid model and liver spheroids-on-chip (TissUse platform) for a week [80]. However, higher metabolite turnover of diclofenac was observed in liver spheroids-on-chip [80]. For longer culture duration (>2 weeks), liver-chip (CN Bio) outperformed static spheroid cultures shown by higher albumin production and CYP3A4 activity [29]. Cox et al. could detect hepatotoxicity of tolcapone demonstrated by reduced viability and CYP3A4 activity [80]. However, its structural analogue entacapone did not provoke a similar response. Although there are very few conclusive reports on a systematic comparison of liver spheroids and liver-chip cultures, both models seem equally promising.
A collaborative study among Emulate Inc, AstraZeneca, and Janssen pharmaceuticals captured species-specific toxicity of fialuridine (10 days treatment) and Janssen proprietary compound (JNJ-2) (2 weeks treatment) on the LoC platform [55]. Furthermore, the LoC predicted the most sensitive endpoint markers, providing insights into the DILI mechanism of compounds. In their 2 weeks study, idiosyncratic DILI investigation of TAK-875 suggested reactive metabolites formation (acyl glucuronide metabolite (TAK-875AG)) and MRP2 inhibition (Fig. 4(c-i)). Upon probing their downstream effect, compromised mitochondrial membrane (detected after 1 week) (Fig. 4(c-ii)), ROS formation (detected after 2 weeks) (Fig. 4(c)–iii)) and lipid droplet accumulation (detected after 2 weeks) (Fig. 4(c-iv)) were observed. This indicated that the drug and reactive metabolites first disturbed the cellular mitochondria, followed by ROS formation and lipid droplet accumulation. Further, TAK-875 induced inflammatory cytokine release and activation of NPCs. This shows the mechanistic investigations of idiosyncratic DILI are also possible with LoC models inclusive of parenchymal and non-parenchymal components. As elaborated by Godoy et al., NPCs exhibit secondary response after initial damage to hepatocytes amplifying the overall effect [36]. Hence, LoC models with NPCs can be an ideal choice for a detailed investigation of the DILI mechanisms of hepatotoxins [36]. Furthermore, co-culturing in LoC with stellate cells and kupffer cells is also required for making disease models of NASH [120].
5. Emerging applications of LoC in industries
5.1. Liver organoids and organoids-on-chips
Organoids-on-chips have become one of the most rapidly growing fields in the last few years [121]. By integrating the LoC technology with complex human liver physiology captured by liver organoid models, researchers can create one of the most powerful models in clearance and toxicity prediction. Mimetas and Hubrecht organoid technology (founded by Hans Clevers) had announced a collaboration to explore the potential of both technologies in organoids-on-chips. In this section, we highlight the published reports at the interface of liver organoids and microfluidics culture.
As iPSC organoids capture the organ regeneration process from an embryonic stage, they show organ-specific structure and morphology, functional features, organ-specific parenchymal and non-parenchymal population, vascularization, and various gradients. Since the first publication of the organoids in 2009, the field has evolved significantly providing great insights into developmental biology, disease models, personalized medicine, organ regeneration and tissue engineering, and recently drug discovery and development [122,123].
Primary mouse and human hepatocytes-derived liver organoids can be expanded up to 2.5–6 months in culture, which is significantly longer than other reported 3D culture models of the liver [124,125]. While liver organoids can be cultured for extended periods, a systematic comparison of functional markers, both passage-wise and time-wise, is missing in the published literature.
One of the widely used cell sources for liver organoids is iPSC-derived iHep. Using iHep, Mun et al. demonstrated dose-dependent trovafloxacin-mediated toxicity in a 6 days study [126]. Besides, its non-toxic analog levofloxacin didn’t show toxicity in organoids. However, the albumin secretion was significantly lower than in vivo values. We believe that the dynamic culture of organoids can improve their maturation and functional activity in LoC models [127,128]. This is especially relevant for iHep organoids, which are known to show fetal liver markers as stated by Mun et al. [126].
Recently, iHep organoids were cultured under the gravity-induced recirculating flow of 0–71 μl/h to screen 159 known toxic compounds in a high throughput 96-well plate-based LoC platform (Mimetas) [76]. Liver organoids were cultured in an organ channel and HMEC-1 endothelial cells and THP-1 monocytes were cultured in a perfusion channel (Fig. 4(d-i)). Expected shear stress based on the dimensions of perfusion channel (300 × 220 μm) and flow rate (0–71 μl/h) was in the range of 0–0.0725 dyne/cm2. However, the estimated shear stress experienced by HMEC-1 endothelial cells in the plate was significantly lower than the reported in vivo values (0.1–0.5 dyne/cm2) [49]. Liver organoids maintained albumin production of 20 μg/day/million cells after 5 days. Further, CYP3A4 activity was maintained for 15 days. Although albumin secretion was lesser than the in vivo values, it was significantly higher than previously reported matrix-free 3D iHep aggregates cultures on chip [51]. Further, the model failed to detect 31 high-risk DILI compounds, which can be attributed to the low maturity of iHep. The cell viability (%) results for 40 toxic compounds from the 159 compounds have been shown in (Fig. 4(d-ii)). One of the major limitations of the study was only one replicate per compound performed on the Mimetas plate.
Jin et al. cultured iHep and HUVEC (human umbilical cord vein endothelial cells) cells derived organoids in a decellularized extracellular matrix (dECM) using a recirculating microfluidic system [129]. They found higher albumin secretion, CYP3A4 activity, vessel sprouting, E-cadherin expression, and urea synthesis under flow than static organoids cultures. Further, dose-dependent acetaminophen toxicity was captured by ROS formation with higher sensitivity in dynamic culture than static culture.
By leveraging the extended expansion capacity of organoids and their matured phenotypes in LoC models, repeated drug dosing will be possible, enabling more accurate DILI and hepatic clearance prediction of the compounds. However, the field of organoids-on-chips is evolving and detailed studies on such topics will be of great interest to industries. Besides, LoC studies based on primary hepatocytes derived organoids can be explored, as iHep based LoC studies described here have suffered from low hepatocyte maturation.
5.2. Integration of LoC with gut-chip & kidney-chip in ADME
The integration of multiple organ components through fluidic flow in multi-organs-on-chips is a promising area of research, where microfluidic systems are anticipated to have a significant impact. Eventually, the ambitious vision of the scientific community is to make fully functional body-on-chip systems. While these systems are projected to be utilized for preclinical drug trials, achieving comparable throughput and cost-effectiveness to preclinical animal models could take several years [130]. Currently, the majority of the organ-on-chip companies are focusing on developing the multi-organ systems in ADME&T comprising of liver, gut, and kidney.
Herland et al. presented the liver-gut-kidney-on-a-chip with an arteriovenous mixing reservoir to replicate the PK profiles of oral (nicotine) and IV route drugs (cisplatin) [110]. The fluid was transferred among the organ-chips at specific time points using a liquid handling system. Unlike animal models, the concentration of nicotine could be measured in each organ-chip with time, which is one of the significant benefits of performing PK studies on multi-organs-on-chips. Based on the computational framework and scaled-up parameters, the system predicted hepatic intrinsic clearance of both drugs within a 2-fold range. In another study, Maschmeyer et al. demonstrated long-term maintenance of the liver-gut-kidney system for 28 days with stable expression of liver-specific CYP3A4 [83].
The gut is one of the important sites of extrahepatic metabolism with significant phase I and phase II metabolic activity [131]. As the intestine is known to have significant CYP3A4 activity, drugs like midazolam require in vitro liver-gut system for predicting bioavailability [132]. Bioavailability refers to the fractional unchanged drug present in the systemic circulation. Animal models have been poor predictors of bioavailability in humans [133]. Hence, there is a need to develop human-relevant predictive bioavailability models. Current in vitro models based on liver and gut are treated with compounds separately [134] or integrate liver and gut compartments in a single hybrid transwell platform [135] to predict bioavailability. However, the fluidic connection is necessary between the two compartments to recapitulate the dynamic human system.
Recently, Roche used a liver-gut system developed by CN Bio to study the metabolism of prodrug mycophenolate mofetil [43]. The system allowed to track the concentration of prodrug, active drug mycophenolic acid, and inactive metabolite mycophenolic acid glucuronide in gut and liver compartments. Using in silico model the human intrinsic gut and liver clearances were predicted. Nonetheless, additional system validation will be necessary with a wider range of drugs for human bioavailability prediction. Arakawa et al. used a liver-gut system to predict concentration profiles of phase I and phase II metabolites of triazolam in humans, which closely matched the reported human plasma profiles [136].
Prot et al. used primary hepatocytes and a Caco-2-based liver-gut fluidic system to predict the clearance and bioavailability of paracetamol [137]. They could predict the hepatic clearance within a 2-fold range. However, bioavailability was under-predicted. Bricks et al. overpredicted bioavailability using a gut-liver fluidic system for omeprazole and phenacetin [138]. This might be attributed to the overall usage of 2D culture models with lower hepatic metabolic activity. Furthermore, usage of Caco-2 cells in their bioavailability prediction model is questionable, as they poorly express phase I metabolic enzymes. The aforementioned studies prove the necessity for better gut-liver MPS for bioavailability prediction especially for compounds with low bioavailability. Apart from bioavailability prediction, the liver-gut system has also been applied to study various diseases such as inflammatory bowel disease [139], air pollution-related metabolic disturbances [140], nanoparticle-induced liver injury [141], and hepatic steatosis [142].
6. Challenges and future perspectives
In this review, we have elaborated on the unique features and applications of liver-on-chips (LoC) models, emphasizing their promise as highly human relevant technologies. The primary aim of this review was to evaluate the translational potential of current LoC models for clearance and toxicity prediction. To this end, we discovered that several collaborative studies have been conducted between pharmaceutical industries and LoC manufacturers, as listed in Table 4. Apart from liver-chips, several collaborative studies in kidney-chips, gut-chips, and bone marrow-chips are being conducted [28]. This demonstrates that the technology has quickly gained initial traction among industrial users. However, based on the current state of the art, it can be concluded that the field is still in its early stages. Several limitations need to be addressed before these models can be regularly used in the drug development process, as discussed below.
Table 4.
Collaborative studies between pharma industries and LoC makers.
| Pharmaceutical industries involved | Chip maker | Study objective | Outcomes of study | Reference |
|---|---|---|---|---|
| Sanofi and Merck | Mimetas | Evaluation of the reproducibility and robustness of the high-throughput LoC platform in PK and toxicity studies | Eight or more replicates required to address high variability of LoC | [70] |
| Roche | CN Bio | Intrinsic hepatic clearance prediction | Predicted CLint for 58 % of the 12 compounds within the 3-fold deviation | [82] |
| Roche | CN Bio | To study gut and liver clearances of the prodrug mycophenolate mofetil | Human liver and gut clearances were successfully predicted. Comparison with in vivo data was missing. | [143] |
| Pfizer | Javelin Biotech | Hepatic clearance prediction | Predicted hepatic clearance of 83 % of the 12 compounds within 2-fold | [81] |
| Abbvie and Janssen Pharmaceuticals | Emulate Inc | DILI prediction | Sensitivity = 87 %, Specificity = 100 % | [13] |
| AstraZeneca | Emulate Inc | To compare liver spheroids and liver chip culture for DILI prediction | Both models were equally promising with similar DILI sensitivity | [119] |
| UCB Biopharma | TissUse | To compare liver spheroids and liver chip culture and evaluate for drug safety and metabolism | Similar functional activity but higher metabolite turnover of diclofenac in liver chips | [80] |
| AstraZeneca, and Janssen Pharmaceuticals | Emulate Inc. | Evaluation of human and cross-species liver toxicity | Platform predicted species-specific toxicity of several compounds correctly | [55] |
One of the important factors is the lack of thorough validation of LoC platforms against the standard guidelines provided by IQ-MPS. As shown in Table 5, very few LoC platforms have been fully validated completely for basic stage I MPS markers. Furthermore, many LoC studies failed to report the albumin and urea secretion values normalized to the cell number, which is required to compare them with in vivo values given in the guidelines.
Table 5.
Comparison of LoC platforms against IQ-MPS phase I guidelines.
| LoC model | Maximum albumin synthesis value (μg/day/million cells) (study duration) | Maximum urea synthesis value (μg/day/million cells) (study duration) | Gene expression of enzymes and transporters (comparison with freshly thawed hepatocytes) | Reference |
|---|---|---|---|---|
| Matrix free iHep-on-chips | 0.1 (35 days) | 6 (35 days) | Gene expression reported only for CYP3A4, CYP2B6, CYP2C9 (comparison not reported) | [51] |
| PHH sandwich culture-on-chips (with LSEC) (Emulate) | 60 (14 days) | Not reported | Gene expression not reported, CYP activity compared (CYP3A and CYP2B higher, CYP1A lower than fresh hepatocytes) |
[55] |
| PHH sandwich culture-on-chips (with LSEC, stellate cells, and kupffer cells) (Emulate) | 60 (7 days) (donor 2) | 250 (7 days) (donor 2) | Gene expression reported (CYP3A4 significantly higher, 13 out of 17 genes with higher expression than fresh hepatocytes) | [13] |
| 3D microtissues-on-chips (CN Bio) | 50 (19 days) | Not reported | Gene expression reported for metabolic enzymes and CYP3A4 activity was also measured (comparison not reported) | [29] |
| iHep-on-chips (with endothelial cells and THP-1 cells) (Mimetas) | 15-20 (15–17 days) | 20-40 (15–17 days) | Gene expression not reported but measured metabolic activity (comparison not reported) | [70,76] |
| PHH sandwich culture-on-chips (Javelin Biotech) | 40 (15 days) | 160 (15 days) | Reported stable gene expression for 15 days and measured CYP450 activity (comparison not reported) | [81] |
For industrial users, assay throughput is a primary concern. As the creation of a microphysiological system is driven by the goal to mimic the complexity of the human system, achieving the throughput levels of the simpler, traditional well-plate-based static in vitro models poses a considerable challenge. Mimetas and AIM Biotech introduced well-plate based high-throughput 2-lane and 3-lane organ-on-plates models to address the same. However, user-friendliness and reproducibility are some of the bottlenecks that need to be carefully evaluated [70]. It is expected that the CRO (contract research organization) services offered by the majority of organ-chip makers will address the limitations related to complex operation procedures and user-friendliness. This approach would reduce the need for direct involvement of industrial users in managing the platforms. Furthermore, recent developments in automated liquid handler integration with LoC cultures may address limitations related to repeatability.
A recent report published by TEX-VAL tissue chip testing consortium highlighted the superior role of quality of cell source in realizing reproducibility and repeatability of the results produced from LoC and microphysiological systems [144]. Primary human hepatocytes (PHH) are considered gold-standard in in vitro science. However, lot-to-lot variation, proliferation inability, and high cost are some of the limitations with PHH [145]. Induced pluripotent stem cells derived hepatocytes are gaining popularity, as they do not suffer from limited availability. However, as mentioned before in section 2.1, they often demonstrate incomplete maturation in vitro. This is evident from the previous studies that compared the performance of iHeps and PHH and demonstrated inferior albumin, urea and phase I and II metabolic enzymes activities in iHeps than PHH [101,146,147]. However, some of the potential benefits of iPSC include drug testing in subpopulations of patients with rare genetic polymorphisms, the genetic manipulability of iHeps, the ability to generate isogenic liver cells for testing, and the development of personalized body-on-chips or multi-organs-on-chips from patient-derived iPSCs for personalized medicine. Hence, until we develop robust protocols for improving maturity and differentiation of iHeps, PHH will continue to be the preferred choice of cell source for clearance and DILI predictions [144].
Although there is sufficient evidence that dynamic perfusion enhances the functional activity of hepatocytes, as discussed in Section 2.1, it is still unclear whether the benefits of flow outweigh the system’s low throughput and high cost, which are due to the external setup required to induce flow [144]. Additionally, the shear stresses applied to cellular systems are far from the physiologically relevant values in most of the cases [144]. Other static systems, such as 3D spheroids and HepatoPac, perform equally well or, in some cases, better than dynamic LoC systems for predicting hepatic clearance of drugs [82,90]. Currently, multi-organs-on-chip systems benefit most from the inclusion of dynamic flow, which facilitates inter-organ exchange of biomacromolecules, drugs, and metabolites. Additionally, in striving to create more physiologically relevant models, a common opinion advocates for including dynamic perfusion to accurately estimate human-relevant processes in single organ systems [144].
As discussed in section 2.2, co-culture with non-parenchymal cells demonstrated enhanced functional activity of hepatocytes. However, the co-culture method involves complex cell culture protocols with the use of multiple cell culture media. Additionally, sourcing non-parenchymal cells can be challenging in some cases. Hence, co-culture models can be more suitable for building liver related disease models such as NASH. Additionally, DILI investigation with immune system involvement can be realized with co-culture LoC models. However, hepatic clearance prediction assays, enzyme inhibition and induction assays, and metabolite identification assays can be conducted with mono-culture of hepatocytes as shown by some of the previous studies [81,82].
Integrating vasculature into LoC may enhance proliferation and viability of hepatic tissues; however, their adoption in industries is likely to remain low for DILI and clearance predictions due to high complexity involved in imaging and culturing the system with endothelial cells [86]. However, it can be a great model for studying liver biology and diseases. Bonanini et al. reported modeling of veno-occlusive disease using vascularized liver spheroids on chips [77]. Moreover, Chhabra et al. recapitulated the liver regeneration process by modeling interactions between liver and vascular cells as discussed before [47].
Overall, static models, including conventional microsomes, plated hepatocytes, and suspension cultures will continue to contribute in the early stages of drug discovery by screening a greater number of compounds for understanding hepatic clearance, enzyme inhibition and induction, and metabolite identification. In contrast, complex in vitro models including dynamic LoC platforms are expected to contribute in understanding complex problems in ADME such as clearance of metabolically stable compounds, chronic DILI prediction, mechanistic studies in toxicology, and immune-mediated toxicity [82].
Liver organoids-on-chips are one of the most promising technologies for clearance and toxicity prediction, as discussed in section 5.1. There are several benefits of culturing organoids on microfluidic platforms than static well-plates. On-chip culture of organoids ensures a consistent gel volume than traditional static well-plate cultures where manual pipetting results in varying sizes of gel droplets. As the human-derived recombinant proteins used in organoid culture medium are expensive, microfluidic culture can be more economical option than well-plate cultures.
However, several challenges related to organoid cultures such as unwanted drug binding to the matrix, drug clearance due to NPCs, and inadequate drug diffusion to the central part of organoids are required to be carefully assessed. Additionally, with the same cell seeding number, the number of organoids inside the matrix may vary, causing variability among repeats. Moreover, normalizing functional characterization results to the total number of cells is also challenging for organoids-on-chips, because it involves efficient harvesting of the cells from the gel. However, non-permanent bonding strategies can be beneficial for organoids-on-chips, as they permit access to tissues at the end of culture for immunohistochemistry, molecular studies and cell counting for data normalization [148].
Despite the growing number of collaborative studies between industries and commercial or academic chip makers, the prospect of completely replacing animal testing in drug discovery still appears to be a distant goal. Convincing the industry to use these assays in the preclinical stage will require demonstrating the superiority of MPS models over the current animal models. Therefore, in addition to human hepatocytes, hepatocytes from other species should be utilized to directly compare LoC data with historical or newly generated animal data [6]. Furthermore, deciding the context of use (CoU) is indispensable for the industrial adoption of MPS [2]. Hence, the majority of the commercial organ-on-chip companies are developing the platforms for specific fit for purpose/context of use.
For faster adoption of organ-on-chips in drug discovery, a clear value proposition is required by identifying the white spaces. Some of the distinct applications of MPS need to be demonstrated in the publications, where conventional in vitro models haven’t produced convincing results. As highlighted, immune-mediated DILI prediction seems to be a strong capability of LoC, which should be demonstrated in detail to persuade industry stakeholders. Furthermore, LoC models show great potential in predicting hepatic clearance of metabolically stable compounds, which can be a significant part of company’s portfolio operating in organ-on-chip sector. Additionally, the inclusion of biliary flow is an underdeveloped area where organ-on-chips developers can focus. Some of the other unique areas where LoC models will make a significant impact are liver drug metabolism in pregnancy, and disease conditions.
Fetal drug exposure is a matter of great concern leading to the exclusion of pregnant women from drug trials. Significant alterations in phase I and phase II enzymes have been well documented due to an increase in multiple reproductive hormones such as estrogen, progesterone, estradiol, cortisol, placental lactogen, and prolactin in pregnancy [149,150]. Most importantly, CYP3A4 activity may increase by 50–100 % during pregnancy. Additionally, a reduction in CYP1A2 and CYP2C19 activity can lead to excessive accumulation of relevant drugs in plasma, resulting in toxicity [151]. Further, increase in hepatic blood flow and decrease in plasma protein binding of drugs may result into altered drug clearance during pregnancy [151]. LoC models can be potentially employed to predict hepatic clearance of drugs in pregnancy. This can be achieved using a culture medium containing reproductive hormones and primary hepatocytes from female donors [150]. Alternative cell source can also be pregnant mice-derived hepatocytes, which are known to express altered CYP450 enzymes expression [152].
The gut microbiome is known to play an important role in drug metabolism [153]. Further, gut bacteria also alter hepatic CYP3A activity, thus affecting hepatic drug clearance [153]. Jalili-Firoozinezhad et al. demonstrated complex gut-on-chips by co-culturing intestinal epithelium and microbiome [154]. Recently, co-culture with gut microbiome enhanced albumin and urea secretion activity of HepG2 spheroids on a chip [155]. However, systematic studies on drug clearance are lacking using such systems. We believe liver-gut-microbiome-on-chips will provide further insights into the role of gut bacteria in drug clearance [156]. Moreover, the overgrowth of gut bacteria on gut-on-chips has been linked with inflammatory bowel disease (IBD) [157]. Therefore, gut-liver systems can be used to investigate the alterations in clearance and adjust drug dosing under IBD conditions.
Drug metabolism in patients with chronic kidney disease (CKD) needs to be properly investigated. Apart from reduced renal drug clearance, reports suggest alleviated expression of several hepatic CYP450 enzymes in CKD [158]. Hence, liver-kidney-on-chips may help in deciphering the complex cross-talk between the two in CKD. Moreover, such systems may help in calculating dose adjustment for over 75 commonly used drugs with substantially altered bioavailability due to impaired clearance in CKD [158]. Several kidney-on-chips models have been reported with the potential to recapitulate the pathophysiology of CKD [159,160]. Such models when integrated with healthy human LoC will advance our current understanding of clearance prediction in kidney ailments.
The potential to reduce R&D costs is the biggest incentive for industries to use MPS in drug discovery [161]. The report suggests that industries can increase their annual revenue by $3 billion with the integration of LoC in the present workflow [13]. Ewart et al. presented a detailed framework to combine LoC models with pre-clinical animal studies in current drug discovery DILI prediction pipeline. They presented a simple decision matrix where LoC data can help advancing the most promising candidates after conducting toxicity studies in two animals as per the FDA requirement [162]. More of such frameworks will be needed to assist the industries in determining where such predictive systems can fit into their existing drug discovery workflow.
Finding a common medium to nourish multiple organs connected through a shared media channel has posed challenges. Researchers have developed several strategies to address this issue, including mixing organ-specific media in specific ratios, incorporating organ-specific supplements into a common medium, and utilizing membrane-based compartmentalized cultures [163].
PBPK involves usage of underlying biological and physiological mechanisms represented by differential equations. It models each organ as a separate compartment and connects them through arterial and venous blood flow. PBPK models assist in the translation of preclinical data to meaningful clinical outcomes. With the advent of multi-organs-on-chips, prediction of PK profile and PK parameters are possible through PBPK modeling of the tissue-chips [164]. Furthermore, PBPK models based on LoC data will be able to accurately estimate hepatic clearance of the drugs for disease treatments across sub-populations not directly studied in clinical trials. These models can account for variations based on genetic polymorphisms, co-morbidities, and lifestyle factors.
Digital twins based on PBPK models are an emerging field and hold great promise for accurate human liver clearance prediction [110,165]. A recent report developed a digital twin based on the data from the published LoC systems [165]. They introduced a three-compartment based model to describe the drug concentration in media, interstitium, and intracellular space. These three compartments have been modeled based on mathematical governing equations and serve as the dynamic environment where the drug interact with hepatocytes, gets distributed and finally gets metabolized in the intracellular compartment. By adopting this approach, they could address the common issue of clearance underprediction associated with the traditional one-compartment modeling of LoC systems [82,166]. Apart from one-compartment modeling other possible reasons for LoC based clearance underprediction are lack of biliary clearance [82], medium evaporation during longer incubations [82], limited transporter and metabolic activities [26], inappropriate scaling method [167], and non-specific protein and chip material binding [26].
Finally, integration of machine learning with organ-on-chips is a promising area, which offers several potential benefits including automated analysis of large dataset generated from tissue-chips especially microscopic data, automatic identification of cells of interest from the multi-cell population, guidance in device design and material selection, real-time cell tracking, and automatic non-destructive analysis from brightfield images of chips [168,169]. In case of organoids-on-chips, usage of machine learning can enable automated organoids analysis spread across multiple planes in a hydrogel [170]. This automated analysis encompasses analysis of size, number, and morphology. Moreover, morphological changes in organoids, predictive of the drug responses can also be unveiled though AI/ML [171]. Finally, AI models trained from the rich human-relevant OMICS and imaging data produced from LoC systems will provide unparalleled insights during the early stages of drug discovery particularly for new drug modalities with limited available data. For instance, a large dataset of hepatic clearance and liver toxicity generated from LoC for the large drug molecules such as PROTACS, anti-body drug conjugates, peptides can be used to gain predictive insights in the pre-clinical stage of drug discovery for a new chemical entity.
7. Conclusion
The review has provided a comprehensive overview of the current status of LoC models in chronic DILI predictions, clearance prediction for metabolically stable compounds, species-specific DILI predictions, bioavailability prediction, and immune-medicated DILI assessments. We have emphasized the critical role of dynamic culture, co-culture systems, and vascularization in the enhanced functional maturation of hepatic tissues within LoC environments. We have found several industrial studies conducted on LoC platforms for DILI and clearance prediction, but their integration into the current drug discovery framework might take several years. Challenges associated with the industrial adoption of LoC, such as lack of characterization according to guidelines, drug binding to extracellular matrices and device materials, low throughput of LoC assays, problems with data reproducibility and repeatability, problems with accessing the hepatic tissues from LoC, and the user-friendliness of platforms, have been discussed in depth. Further, a need to conduct more collaborative studies for increasing confidence in LoC-based clearance prediction assays was identified. Finally, we have identified several important emerging areas in the LoC field, such as liver organoids-on-chips, bioavailability prediction using gut-liver systems, hepatic clearance prediction in pregnancy, hepatic clearance studies in disease conditions such as chronic kidney disease (CKD) and inflammatory bowel disease (IBD) conditions using multi-organs systems, PBPK modeling & digital twins, hepatic clearance studies for disease treatments in sub-populations based on genetic polymorphisms, co-morbidities, life style, and AI/ML integration with LoC.
CRediT authorship contribution statement
Viraj Mehta: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Conceptualization. Guruswamy Karnam: Writing – review & editing, Supervision, Writing – original draft. Vamsi Madgula: Supervision, Resources, Project administration, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge Biorender.com for providing icons for illustrations in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2024.101143.
Contributor Information
Viraj Mehta, Email: virajmehta.h@sailife.com.
Guruswamy Karnam, Email: guruswamy.k@sailife.com.
Vamsi Madgula, Email: vamsi.m@sailife.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Marshall L.J., Bailey J., Cassotta M., Herrmann K., Pistollato F. Poor translatability of biomedical research using animals — a narrative review. Altern. Lab. Anim. 2023;51:102–135. doi: 10.1177/02611929231157756/ASSET/IMAGES/LARGE/10.1177_02611929231157756-FIG1.JPEG. [DOI] [PubMed] [Google Scholar]
- 2.Baran S.W., Brown P.C., Baudy A.R., Fitzpatrick S.C., Frantz C., Fullerton A., Gan J., Hardwick R.N., Hillgren K.M., Kopec A.K., Liras J.L., Mendrick D.L., Nagao R., Proctor W.R., Ramsden D., Ribeiro A.J.S., Stresser D., Sung K.E., Sura R., Tetsuka K., Tomlinson L., Van Vleet T., Wagoner M.P., Wang Q., Arslan S.Y., Yoder G., Ekert J.E. Perspectives on the evaluation and adoption of complex in vitro models in drug development: workshop with the FDA and the pharmaceutical industry (IQ MPS affiliate) ALTEX. 2022;39:297–314. doi: 10.14573/ALTEX.2112203. [DOI] [PubMed] [Google Scholar]
- 3.Mehta V., Vilikkathala Sudhakaran S., Rath S.N. Facile route for 3D printing of transparent PETg-based hybrid biomicrofluidic devices promoting cell adhesion. ACS Biomater. Sci. Eng. 2021;7:3947–3963. doi: 10.1021/ACSBIOMATERIALS.1C00633/SUPPL_FILE/AB1C00633_SI_003.AVI. [DOI] [PubMed] [Google Scholar]
- 4.Mehta V., Rath S.N. 3D printed microfluidic devices: a review focused on four fundamental manufacturing approaches and implications on the field of healthcare. Biodes. Manuf. 2021;4:311–343. doi: 10.1007/s42242-020-00112-5. [DOI] [Google Scholar]
- 5.Sankar S., Mehta V., Ravi S., Sharma C.S., Rath S.N. A novel design of microfluidic platform for metronomic combinatorial chemotherapy drug screening based on 3D tumor spheroid model. Biomed. Microdev. 2021. 2021;23(4 23):1–10. doi: 10.1007/S10544-021-00593-W. [DOI] [PubMed] [Google Scholar]
- 6.Ingber D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022. 2022;23(8 23):467–491. doi: 10.1038/s41576-022-00466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sukanya V.S., Mehta V., Jilla S., Narayan Rath S. Differential osteo-specific invasion of patient-derived cancer cells in a microfluidic co-culture model. Chem. Eng. J. 2024;489 doi: 10.1016/J.CEJ.2024.151202. [DOI] [Google Scholar]
- 8.Lin H., Lozito T.P., Alexander P.G., Gottardi R., Tuan R.S. Stem cell-based microphysiological osteochondral system to model tissue response to interleukin-1Β. Mol. Pharm. 2014;11:2203–2212. doi: 10.1021/MP500136B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martignoni M., Groothuis G.M.M., de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expet Opin. Drug Metabol. Toxicol. 2006;2:875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- 10.Nahle Z. A proof-of-concept study poised to remodel the drug development process: liver-Chip solutions for lead optimization and predictive toxicology. Front. Med. Technol. 2022;4 doi: 10.3389/FMEDT.2022.1053588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monticello T.M., Jones T.W., Dambach D.M., Potter D.M., Bolt M.W., Liu M., Keller D.A., Hart T.K., Kadambi V.J. Current nonclinical testing paradigm enables safe entry to First-In-Human clinical trials: the IQ consortium nonclinical to clinical translational database. Toxicol. Appl. Pharmacol. 2017;334:100–109. doi: 10.1016/J.TAAP.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W., Dorato M., Van Deun K., Smith P., Berger B., Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/RTPH.2000.1399. [DOI] [PubMed] [Google Scholar]
- 13.Ewart L., Apostolou A., Briggs S.A., V Carman C., Chaff J.T., Heng A.R., Jadalannagari S., Janardhanan J., Jang K.-J., Joshipura S.R., Kadam M.M., Kanellias M., Kujala V.J., Kulkarni G., Le C.Y., Lucchesi C., V Manatakis D., Maniar K.K., Quinn M.E., Ravan J.S., Rizos A.C., Sauld J.F.K., Sliz J.D., Tien-Street W., Ramos Trinidad D., Velez J., Wendell M., Irrechukwu O., Mahalingaiah P.K., Ingber D.E., Scannell J.W., Levner D., John H. Performance assessment and economic analysis of a human Liver-Chip for predictive toxicology. Commun. Med. 2022. 2022;2(1 2):1–16. doi: 10.1038/s43856-022-00209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jadalannagari S., Ewart L. Beyond the hype and toward application: liver complex in vitro models in preclinical drug safety. Expet Opin. Drug Metabol. Toxicol. 2024 doi: 10.1080/17425255.2024.2328794. [DOI] [PubMed] [Google Scholar]
- 15.Smith C.M., Nolan C.K., Edwards M.A., Hatfield J.B., Stewart T.W., Ferguson S.S., Lecluyse E.L., Sahi J. A comprehensive evaluation of metabolic activity and intrinsic clearance in suspensions and monolayer cultures of cryopreserved primary human hepatocytes. J. Pharmaceut. Sci. 2012;101:3989–4002. doi: 10.1002/jps.23262. [DOI] [PubMed] [Google Scholar]
- 16.Dunn J.C.Y., Tompkins R.G., Yarmush M.L. Hepatocytes in collagen sandwich: evidence for transcriptional and translational regulation. JCB (J. Cell Biol.) 1992;116:1043–1053. doi: 10.1083/JCB.116.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khetani S.R., Bhatia S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26 doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 18.Riede J., Wollmann B.M., Molden E., Ingelman-Sundberg M. Primary human hepatocyte spheroids as an in vitro tool for investigating drug compounds with low hepatic clearances. Drug Metabol. Dispos. 2021;49:501. doi: 10.1124/DMD.120.000340/-/DC1. [DOI] [PubMed] [Google Scholar]
- 19.Peng W.C., Logan C.Y., Fish M., Anbarchian T., Aguisanda F., Álvarez-Varela A., Wu P., Jin Y., Zhu J., Li B., Grompe M., Wang B., Nusse R. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell. 2018;175:1607–1619.e15. doi: 10.1016/J.CELL.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X., Qu X., Zhu W., Li Y.S., Yuan S., Zhang H., Liu J., Wang P., Lai C.S.E., Zanella F., Feng G.S., Sheikh F., Chien S., Chen S. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U.S.A. 2016;113:2206–2211. doi: 10.1073/PNAS.1524510113/SUPPL_FILE/PNAS.1524510113.SM01.AVI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Y., Li N., Yang H., Luo C., Gong Y., Tong C., Gao Y., Lü S., Long M. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab Chip. 2017;17:782–794. doi: 10.1039/C6LC01374K. [DOI] [PubMed] [Google Scholar]
- 22.Esch M.B., Prot J.M., Wang Y.I., Miller P., Llamas-Vidales J.R., Naughton B.A., Applegate D.R., Shuler M.L. Multi-cellular 3D human primary liver cell cultures elevate metabolic activity under fluidic flow. Lab Chip. 2015;15:2269. doi: 10.1039/C5LC00237K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haque A., Gheibi P., Gao Y., Foster E., Son K.J., You J., Stybayeva G., Patel D., Revzin A. Cell biology is different in small volumes: endogenous signals shape phenotype of primary hepatocytes cultured in microfluidic channels. Scient. Rep. 2016. 2016;6(1 6):1–15. doi: 10.1038/srep33980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonon F., Giobbe G.G., Zambon A., Luni C., Gagliano O., Floreani A., Grassi G., Elvassore N. In vitro metabolic zonation through oxygen gradient on a chip. Scient. Rep. 2019. 2019;9(1 9):1–10. doi: 10.1038/s41598-019-49412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herland A., Maoz B.M., Das D., Somayaji M.R., Prantil-Baun R., Novak R., Cronce M., Huffstater T., Jeanty S.S.F., Ingram M., Chalkiadaki A., Chou D.B., Marquez S., Delahanty A., Jalili-Firoozinezhad S., Milton Y., Sontheimer-Phelps A., Swenor B., Levy O., Parker K.K., Przekwas A., Ingber D.E. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat. Biomed. Eng. 2020;4:421–436. doi: 10.1038/s41551-019-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiba M., Ishii Y., Sugiyama Y. Prediction of hepatic clearance in human from in vitro data for successful drug development. AAPS J. 2009;11:262–276. doi: 10.1208/S12248-009-9103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cirit M., Stokes C.L. Maximizing the impact of microphysiological systems with in vitro–in vivo translation. Lab Chip. 2018;18:1831–1837. doi: 10.1039/C8LC00039E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marx U., Akabane T., Andersson T.B., Baker E., Beilmann M., Beken S., Brendler-Schwaab S., Cirit M., David R., Dehne E.M., Durieux I., Ewart L., Fitzpatrick S.C., Frey O., Fuchs F., Griffith L.G., Hamilton G.A., Hartung T., Hoeng J., Hogberg H., Hughes D.J., Ingber D.E., Iskandar A., Kanamori T., Kojima H., Kuehnl J., Leist M., Li B., Loskill P., Mendrick D.L., Neumann T., Pallocca G., Rusyn I., Smirnova L., Steger-Hartmann T., Tagle D.A., Tonevitsky A., Tsyb S., Trapecar M., Van de Water B., Van den Eijnden-van Raaij J., Vulto P., Watanabe K., Wolf A., Zhou X., Roth A. Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. ALTEX - Alternat. Anim. Exp. 2020;37:365–394. doi: 10.14573/ALTEX.2001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubiano A., Indapurkar A., Yokosawa R., Miedzik A., Rosenzweig B., Arefin A., Moulin C.M., Dame K., Hartman N., Volpe D.A., Matta M.K., Hughes D.J., Strauss D.G., Kostrzewski T., Ribeiro A.J.S. Characterizing the reproducibility in using a liver microphysiological system for assaying drug toxicity, metabolism, and accumulation. Clin. Transl. Sci. 2021;14:1049–1061. doi: 10.1111/CTS.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker T.K., Van Vleet T.R., Mahalingaiah P.K., Grandhi T.S.P., Evers R., Ekert J., Gosset J.R., Chacko S.A., Kopec A.K. The current status and use of microphysiological systems by the pharmaceutical industry: the international consortium for innovation and quality microphysiological systems affiliate survey and commentary. Drug Metab. Dispos. 2024;52:198–209. doi: 10.1124/DMD.123.001510. [DOI] [PubMed] [Google Scholar]
- 31.Stresser D.M., Kopec A.K., Hewitt P., Hardwick R.N., Van Vleet T.R., Mahalingaiah P.K.S., O’Connell D., Jenkins G.J., David R., Graham J., Lee D., Ekert J., Fullerton A., Villenave R., Bajaj P., Gosset J.R., Ralston S.L., Guha M., Amador-Arjona A., Khan K., Agarwal S., Hasselgren C., Wang X., Adams K., Kaushik G., Raczynski A., Homan K.A. Towards in vitro models for reducing or replacing the use of animals in drug testing. Nat. Biomed. Eng. 2023;2023:1–6. doi: 10.1038/s41551-023-01154-7. [DOI] [PubMed] [Google Scholar]
- 32.Piergiovanni M., Leite S.B., Corvi R., Whelan M. Standardisation needs for organ on chip devices. Lab Chip. 2021;21:2857–2868. doi: 10.1039/D1LC00241D. [DOI] [PubMed] [Google Scholar]
- 33.Low L.A., Tagle D.A. Organs-on-chips: progress, challenges, and future directions. Exp. Biol. Med. 2017;242:1573. doi: 10.1177/1535370217700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baudy A.R., Otieno M.A., Hewitt P., Gan J., Roth A., Keller D., Sura R., Van Vleet T.R., Proctor W.R. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip. 2020;20:215–225. doi: 10.1039/C9LC00768G. [DOI] [PubMed] [Google Scholar]
- 35.Fowler S., Chen W.L.K., Duignan D.B., Gupta A., Hariparsad N., Kenny J.R., Lai W.G., Liras J., Phillips J.A., Gan J. Microphysiological systems for ADME-related applications: current status and recommendations for system development and characterization. Lab Chip. 2020;20:446–467. doi: 10.1039/C9LC00857H. [DOI] [PubMed] [Google Scholar]
- 36.Godoy P., Hewitt N.J., Albrecht U., Andersen M.E., Ansari N., Bhattacharya S., Bode J.G., Bolleyn J., Borner C., Böttger J., Braeuning A., Budinsky R.A., Burkhardt B., Cameron N.R., Camussi G., Cho C.S., Choi Y.J., Craig Rowlands J., Dahmen U., Damm G., Dirsch O., Donato M.T., Dong J., Dooley S., Drasdo D., Eakins R., Ferreira K.S., Fonsato V., Fraczek J., Gebhardt R., Gibson A., Glanemann M., Goldring C.E.P., Gómez-Lechón M.J., Groothuis G.M.M., Gustavsson L., Guyot C., Hallifax D., Hammad S., Hayward A., Häussinger D., Hellerbrand C., Hewitt P., Hoehme S., Holzhütter H.G., Houston J.B., Hrach J., Ito K., Jaeschke H., Keitel V., Kelm J.M., Kevin Park B., Kordes C., Kullak-Ublick G.A., Lecluyse E.L., Lu P., Luebke-Wheeler J., Lutz A., Maltman D.J., Matz-Soja M., McMullen P., Merfort I., Messner S., Meyer C., Mwinyi J., Naisbitt D.J., Nussler A.K., Olinga P., Pampaloni F., Pi J., Pluta L., Przyborski S.A., Ramachandran A., Rogiers V., Rowe C., Schelcher C., Schmich K., Schwarz M., Singh B., Stelzer E.H.K., Stieger B., Stöber R., Sugiyama Y., Tetta C., Thasler W.E., Vanhaecke T., Vinken M., Weiss T.S., Widera A., Woods C.G., Xu J.J., Yarborough K.M., Hengstler J.G. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013;87:1315–1530. doi: 10.1007/S00204-013-1078-5/TABLES/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youhanna S., Kemas A.M., Preiss L., Zhou Y., Shen J.X., Caka S.D., Paqualini F.S., Goparaju S.K., Shafagh R.Z., Lind J.U., Sellgren C.M., Lauschke V.M. Organotypic and microphysiological human tissue models for drug discovery and development—current state-of-the-art and future perspectives. Pharmacol. Rev. 2022;74:141–206. doi: 10.1124/PHARMREV.120.000238. [DOI] [PubMed] [Google Scholar]
- 38.Yang S., Ooka M., Margolis R.J., Xia M. Liver three-dimensional cellular models for high-throughput chemical testing. Cell Rep. Method. 2023;3 doi: 10.1016/J.CRMETH.2023.100432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otumala A.E., Hellen D.J., Luna C.A., Delgado P., Dissanayaka A., Ugwumadu C., Oshinowo O., Islam M.M., Shen L., Karpen S.J., Myers D.R. Opportunities and considerations for studying liver disease with microphysiological systems on a chip. Lab Chip. 2023;23:2877–2898. doi: 10.1039/D2LC00940D. [DOI] [PubMed] [Google Scholar]
- 40.Qiu L., Kong B., Kong T., Wang H. Recent advances in liver-on-chips: design, fabrication, and applications. Smart Med. 2023;2 doi: 10.1002/SMMD.20220010. [DOI] [Google Scholar]
- 41.Yang Z., Liu X., Cribbin E.M., Kim A.M., Li J.J., Yong K.T. Liver-on-a-chip: considerations, advances, and beyond. Biomicrofluidics. 2022;16 doi: 10.1063/5.0106855/2835536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang J., Zhang J., Wu M., Zhang Y., Zhuang J., Zhang J., Zhang Y., Wu M. A dynamic 3D tumor spheroid chip enables more accurate nanomedicine uptake evaluation. Adv. Sci. 2019;6 doi: 10.1002/ADVS.201901462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milani N., Parrott N., Ortiz Franyuti D., Godoy P., Galetin A., Gertz M., Fowler S. Application of a gut–liver-on-a-chip device and mechanistic modelling to the quantitative in vitro pharmacokinetic study of mycophenolate mofetil. Lab Chip. 2022;22:2853–2868. doi: 10.1039/D2LC00276K. [DOI] [PubMed] [Google Scholar]
- 44.Jun Y., Lee J.S., Choi S., Yang J.H., Sander M., Chung S., Lee S.H. In vivo–mimicking microfluidic perfusion culture of pancreatic islet spheroids. Sci. Adv. 2019;5:4520–4547. doi: 10.1126/SCIADV.AAX4520/SUPPL_FILE/AAX4520_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niiya T., Murakami M., Aoki T., Murai N., Shimizu Y., Kusano M. Immediate increase of portal pressure, reflecting sinusoidal shear stress, induced liver regeneration after partial hepatectomy. J. Hepatobil. Pancreat Surg. 1999;6:275–280. doi: 10.1007/S005340050118/METRICS. [DOI] [PubMed] [Google Scholar]
- 46.Lorenz L., Axnick J., Buschmann T., Henning C., Urner S., Fang S., Nurmi H., Eichhorst N., Holtmeier R., Bódis K., Hwang J.H., Müssig K., Eberhard D., Stypmann J., Kuss O., Roden M., Alitalo K., Häussinger D., Lammert E. Mechanosensing by β1 integrin induces angiocrine signals for liver growth and survival. Nature. 2018:128–132. doi: 10.1038/s41586-018-0522-3. 2018 562:7725 562. [DOI] [PubMed] [Google Scholar]
- 47.Chhabra A., Song H.H.G., Grzelak K.A., Polacheck W.J., Fleming H.E., Chen C.S., Bhatia S.N. A vascularized model of the human liver mimics regenerative responses. Proc. Natl. Acad. Sci. U.S.A. 2022;119 doi: 10.1073/PNAS.2115867119/SUPPL_FILE/PNAS.2115867119.SM01.AVI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W., Wu Y., Hu W., Zhou J., Shu X., Zhang X., Zhang Z., Wu H., Du Y., Lü D., Lü S., Li N., Long M. Direct mechanical exposure initiates hepatocyte proliferation. JHEP Rep. 2023 doi: 10.1016/J.JHEPR.2023.100905. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W., Li P., Li N., Du Y., Lü S., Elad D., Long M. Matrix stiffness and shear stresses modulate hepatocyte functions in a fibrotic liver sinusoidal model. Am. J. Physiol. Gastrointest. Liver Physiol. 2021;320:G272–G282. doi: 10.1152/AJPGI.00379.2019/ASSET/IMAGES/LARGE/AJ-AGIP200058F005.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang C.J., Meyer S.R., O’Meara M.J., Huang S., Capeling M.M., Ferrer-Torres D., Childs C.J., Spence J.R., Fontana R.J., Sexton J.Z. A human liver organoid screening platform for DILI risk prediction. J. Hepatol. 2023;78:998–1006. doi: 10.1016/j.jhep.2023.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Wang H., Deng P., Chen W., Guo Y., Tao T., Qin J. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip. 2018;18:3606–3616. doi: 10.1039/C8LC00869H. [DOI] [PubMed] [Google Scholar]
- 52.Schepers A., Li C., Chhabra A., Seney B.T., Bhatia S. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip. 2016;16:2644. doi: 10.1039/C6LC00598E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bell C.C., Dankers A.C.A., Lauschke V.M., Sison-Young R., Jenkins R., Rowe C., Goldring C.E., Park K., Regan S.L., Walker T., Schofield C., Baze A., Foster A.J., Williams D.P., van de Ven A.W.M., Jacobs F., van Houdt J., Lähteenmäki T., Snoeys J., Juhila S., Richert L., Ingelman-Sundberg M. Comparison of hepatic 2D sandwich cultures and 3D spheroids for long-term toxicity applications: a multicenter study. Toxicol. Sci. 2018;162:655–666. doi: 10.1093/TOXSCI/KFX289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dash A., Simmers M.B., Deering T.G., Berry D.J., Feaver R.E., Hastings N.E., Pruett T.L., LeCluyse E.L., Blackman B.R., Wamhoff B.R. Hemodynamic flow improves rat hepatocyte morphology, function, and metabolic activity in vitro. Am. J. Physiol. Cell Physiol. 2013;304:1053–1063. doi: 10.1152/AJPCELL.00331.2012/ASSET/IMAGES/LARGE/ZH00101372350007.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jang K.-J., Otieno M.A., Ronxhi J., Lim H.-K., Ewart L., Kodella K.R., Petropolis D.B., Kulkarni G., Rubins J.E., Conegliano D., Nawroth J., Simic D., Lam W., Singer M., Barale E., Singh B., Sonee M., Streeter A.J., Manthey C., Jones B., Srivastava A., Andersson L.C., Williams D., Park H., Barrile R., Sliz J., Herland A., Haney S., Karalis K., Ingber D.E., Hamilton G.A. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aax5516. eaax5516. [DOI] [PubMed] [Google Scholar]
- 56.Tan K., Keegan P., Rogers M., Lu M., Gosset J.R., Charest J., Bale S.S. A high-throughput microfluidic microphysiological system (PREDICT-96) to recapitulate hepatocyte function in dynamic, re-circulating flow conditions. Lab Chip. 2019;19:1556–1566. doi: 10.1039/C8LC01262H. [DOI] [PubMed] [Google Scholar]
- 57.Hegde M., Jindal R., Bhushan A., Bale S.S., McCarty W.J., Golberg I., Usta O.B., Yarmush M.L. Dynamic interplay of flow and collagen stabilizes primary hepatocytes culture in a microfluidic platform. Lab Chip. 2014;14:2033–2039. doi: 10.1039/C4LC00071D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Hoyos-Vega J.M., Hong H.J., Loutherback K., Stybayeva G., Revzin A. A microfluidic device for long-term maintenance of organotypic liver cultures. Adv. Mater Technol. 2023;8 doi: 10.1002/ADMT.202201121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rennert K., Steinborn S., Gröger M., Ungerböck B., Jank A.M., Ehgartner J., Nietzsche S., Dinger J., Kiehntopf M., Funke H., Peters F.T., Lupp A., Gärtner C., Mayr T., Bauer M., Huber O., Mosig A.S. A microfluidically perfused three dimensional human liver model. Biomaterials. 2015;71:119–131. doi: 10.1016/J.BIOMATERIALS.2015.08.043. [DOI] [PubMed] [Google Scholar]
- 60.Michalopoulos G.K. Liver regeneration. J. Cell. Physiol. 2007;213:286–300. doi: 10.1002/JCP.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Böhm F., Köhler U.A., Speicher T., Werner S. Regulation of liver regeneration by growth factors and cytokines. EMBO Mol. Med. 2010;2:294–305. doi: 10.1002/EMMM.201000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berger D.R., Ware B.R., Davidson M.D., Allsup S.R., Khetani S.R. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. doi: 10.1002/HEP.27621. [DOI] [PubMed] [Google Scholar]
- 63.Ma X., Qu X., Zhu W., Li Y.S., Yuan S., Zhang H., Liu J., Wang P., Lai C.S.E., Zanella F., Feng G.S., Sheikh F., Chien S., Chen S. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. U.S.A. 2016;113:2206–2211. doi: 10.1073/PNAS.1524510113/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma Y., Hu L., Tang J., Guo W., Feng Y., Liu Y., Tang F. Three-dimensional cell Co-culture liver models and their applications in pharmaceutical research. Int. J. Mol. Sci. 2023;24 doi: 10.3390/IJMS24076248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Messner S., Agarkova I., Moritz W., Kelm J.M. Multi-cell type human liver microtissues for hepatotoxicity testing. Arch. Toxicol. 2013;87:209–213. doi: 10.1007/S00204-012-0968-2/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prestigiacomo V., Weston A., Suter-Dick L. Rat multicellular 3D liver microtissues to explore TGF-β1 induced effects. J. Pharmacol. Toxicol. Methods. 2020;101 doi: 10.1016/J.VASCN.2019.106650. [DOI] [PubMed] [Google Scholar]
- 67.Bell C.C., Hendriks D.F.G., Moro S.M.L., Ellis E., Walsh J., Renblom A., Fredriksson Puigvert L., Dankers A.C.A., Jacobs F., Snoeys J., Sison-Young R.L., Jenkins R.E., Nordling Å., Mkrtchian S., Park B.K., Kitteringham N.R., Goldring C.E.P., Lauschke V.M., Ingelman-Sundberg M. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016;6 doi: 10.1038/SREP25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ardalani H., Sengupta S., Harms V., Vickerman V., Thomson J.A., Murphy W.L. 3-D culture and endothelial cells improve maturity of human pluripotent stem cell-derived hepatocytes. Acta Biomater. 2019;95:371–381. doi: 10.1016/J.ACTBIO.2019.07.047. [DOI] [PubMed] [Google Scholar]
- 69.Harrison S.P., Siller R., Tanaka Y., Chollet M.E., de la Morena-Barrio M.E., Xiang Y., Patterson B., Andersen E., Bravo-Pérez C., Kempf H., Åsrud K.S., Lunov O., Dejneka A., Mowinckel M.C., Stavik B., Sandset P.M., Melum E., Baumgarten S., Bonanini F., Kurek D., Mathapati S., Almaas R., Sharma K., Wilson S.R., Skottvoll F.S., Boger I.C., Bogen I.L., Nyman T.A., Wu J.J., Bezrouk A., Cizkova D., Corral J., Mokry J., Zweigerdt R., Park I.H., Sullivan G.J. Scalable production of tissue-like vascularized liver organoids from human PSCs. Exper. Mol. Med. 2023. 2023;55(9 55):2005–2024. doi: 10.1038/s12276-023-01074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kato Y., Lim A.Y., Sakolish C., Valdiviezo A., Moyer H.L., Hewitt P., Bajaj P., Han G., Rusyn I. Analysis of reproducibility and robustness of OrganoPlate® 2-lane 96, a liver microphysiological system for studies of pharmacokinetics and toxicological assessment of drugs. Toxicol. Vitro. 2022;85 doi: 10.1016/J.TIV.2022.105464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee S.A., No D.Y., Kang E., Ju J., Kim D.S., Lee S.H. Spheroid-based three-dimensional liver-on-a-chip to investigate hepatocyte–hepatic stellate cell interactions and flow effects. Lab Chip. 2013;13:3529–3537. doi: 10.1039/C3LC50197C. [DOI] [PubMed] [Google Scholar]
- 72.Shin Y., Han S., Jeon J.S., Yamamoto K., Zervantonakis I.K., Sudo R., Kamm R.D., Chung S. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 2012;7:1247–1259. doi: 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jang M., Neuzil P., Volk T., Manz A., Kleber A. On-chip three-dimensional cell culture in phaseguides improves hepatocyte functions in vitro. Biomicrofluidics. 2015;9 doi: 10.1063/1.4922863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kolesky D.B., Homan K.A., Skylar-Scott M.A., Lewis J.A. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. U.S.A. 2016;113:3179–3184. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valverde M.G., Faria J., Sendino Garví E., Janssen M.J., Masereeuw R., Mihăilă S.M. Organs-on-chip technology: a tool to tackle genetic kidney diseases. Pediatr. Nephrol. 2022;37:2985–2996. doi: 10.1007/S00467-022-05508-2/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bircsak K.M., DeBiasio R., Miedel M., Alsebahi A., Reddinger R., Saleh A., Shun T., Vernetti L.A., Gough A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®. Toxicology. 2021;450 doi: 10.1016/J.TOX.2020.152667. [DOI] [PubMed] [Google Scholar]
- 77.Bonanini F., Kurek D., Previdi S., Nicolas A., Hendriks D., de Ruiter S., Meyer M., Clapés Cabrer M., Dinkelberg R., García S.B., Kramer B., Olivier T., Hu H., López-Iglesias C., Schavemaker F., Walinga E., Dutta D., Queiroz K., Domansky K., Ronden B., Joore J., Lanz H.L., Peters P.J., Trietsch S.J., Clevers H., Vulto P. In vitro grafting of hepatic spheroids and organoids on a microfluidic vascular bed. Angiogenesis. 2022;25:455–470. doi: 10.1007/S10456-022-09842-9/FIGURES/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Du Y., Khandekar G., Llewellyn J., Polacheck W., Chen C.S., Wells R.G. A bile duct‐on‐a‐chip with organ‐level functions. Hepatology. 2020;71:1350. doi: 10.1002/HEP.30918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du Y., de Jong I.E.M., Gupta K., Waisbourd-Zinman O., Har-Zahav A., Soroka C.J., Boyer J.L., Llewellyn J., Liu C., Naji A., Polacheck W.J., Wells R.G. Human vascularized bile duct-on-a chip: a multi-cellular micro-physiological system for studying cholestatic liver disease. Biofabrication. 2024;16 doi: 10.1088/1758-5090/AD0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cox B., Barton P., Class R., Coxhead H., Delatour C., Gillent E., Henshall J., Isin E.M., King L., Valentin J.P. Setup of human liver-chips integrating 3D models, microwells and a standardized microfluidic platform as proof-of-concept study to support drug evaluation. Biomater. Biosyst. 2022;7 doi: 10.1016/J.BBIOSY.2022.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rajan S.A.P., Sherfey J., Ohri S., Nichols L., Smith J.T., Parekh P., Kadar E.P., Clark F., George B.T., Gregory L., Tess D., Gosset J.R., Liras J., Geishecker E., Obach R.S., Cirit M. A novel milli-fluidic liver tissue chip with continuous recirculation for predictive pharmacokinetics applications. AAPS J. 2023. 2023;25(6 25):1–14. doi: 10.1208/S12248-023-00870-X. [DOI] [PubMed] [Google Scholar]
- 82.Docci L., Milani N., Ramp T., Romeo A.A., Godoy P., Franyuti D.O., Krähenbühl S., Gertz M., Galetin A., Parrott N., Fowler S. Exploration and application of a liver-on-a-chip device in combination with modelling and simulation for quantitative drug metabolism studies. Lab Chip. 2022;22:1187–1205. doi: 10.1039/D1LC01161H. [DOI] [PubMed] [Google Scholar]
- 83.Maschmeyer I., Lorenz A.K., Schimek K., Hasenberg T., Ramme A.P., Hübner J., Lindner M., Drewell C., Bauer S., Thomas A., Sambo N.S., Sonntag F., Lauster R., Marx U. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/C5LC00392J. [DOI] [PubMed] [Google Scholar]
- 84.Park S.E., Georgescu A., Huh D. Organoids-on-a-chip. Science. 2019;364:960. doi: 10.1126/SCIENCE.AAW7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Haan L., Suijker J., van Roey R., Berges N., Petrova E., Queiroz K., Strijker W., Olivier T., Poeschke O., Garg S., van den Broek L.J. A microfluidic 3D endothelium-on-a-chip model to study transendothelial migration of T cells in health and disease. Int. J. Mol. Sci. 2021;22 doi: 10.3390/IJMS22158234/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nashimoto Y., Okada R., Hanada S., Arima Y., Nishiyama K., Miura T., Yokokawa R. Vascularized cancer on a chip: the effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials. 2020;229 doi: 10.1016/J.BIOMATERIALS.2019.119547. [DOI] [PubMed] [Google Scholar]
- 87.Liu X., Wang X., Zhang L., Sun L., Wang H., Zhao H., Zhang Z., Liu W., Huang Y., Ji S., Zhang J., Li K., Song B., Li C., Zhang H., Li S., Wang S., Zheng X., Gu Q. 3D liver tissue model with branched vascular networks by multimaterial bioprinting. Adv. Healthcare Mater. 2021;10 doi: 10.1002/ADHM.202101405. [DOI] [PubMed] [Google Scholar]
- 88.Lee J.B., Park J.S., Shin Y.M., Lee D.H., Yoon J.K., Kim D.H., Ko U.H., Kim Y.T., Bae S.H., Sung H.J. Implantable vascularized liver chip for cross-validation of disease treatment with animal model. Adv. Funct. Mater. 2019;29 doi: 10.1002/ADFM.201900075. [DOI] [Google Scholar]
- 89.Hughes D.J., Kostrzewski T., Sceats E.L. Opportunities and challenges in the wider adoption of liver and interconnected microphysiological systems. Exp. Biol. Med. 2017;242:1593. doi: 10.1177/1535370217708976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Preiss L.C., Georgi K., Lauschke V.M., Petersson C. Comparison of human long-term liver models for clearance prediction of slowly metabolized compounds. Drug Metabol. Dispos. 2024 doi: 10.1124/DMD.123.001638. DMD-AR-2023-001638. [DOI] [PubMed] [Google Scholar]
- 91.Hallifax D., Foster J.A., Houston J.B. Prediction of human metabolic clearance from in vitro systems: retrospective analysis and prospective view. Pharm. Res. (N. Y.) 2010;27:2150–2161. doi: 10.1007/S11095-010-0218-3/METRICS. [DOI] [PubMed] [Google Scholar]
- 92.Bonn B., Svanberg P., Janefeldt A., Hultman I., Grime K. Determination of human hepatocyte intrinsic clearance for slowly metabolized compounds: comparison of a primary hepatocyte/stromal cell Co-culture with plated primary hepatocytes and HepaRG. Drug Metabol. Dispos. 2016;44:527–533. doi: 10.1124/DMD.115.067769. [DOI] [PubMed] [Google Scholar]
- 93.Lau Y.Y., Sapidou E., Cui X., White R.E., Cheng K.C. Development of a novel in vitro model to predict hepatic clearance using fresh, cryopreserved, and sandwich-cultured hepatocytes. Drug Metab. Dispos. 2002;30:1446–1454. doi: 10.1124/DMD.30.12.1446. [DOI] [PubMed] [Google Scholar]
- 94.Treijtel N., Van Helvoort H., Barendregt A., Blaauboer B.J., Van Eijkeren J.C.H. The use of sandwich-cultured rat hepatocytes to determine the intrinsic clearance of compounds with different extraction ratios: 7-ethoxycoumarin and warfarin. Drug Metab. Dispos. 2005;33:1325–1332. doi: 10.1124/DMD.105.004390. [DOI] [PubMed] [Google Scholar]
- 95.Di L. Recent advances in measurement of metabolic clearance, metabolite profile and reaction phenotyping of low clearance compounds. Expet Opin. Drug Discov. 2023 doi: 10.1080/17460441.2023.2238606. [DOI] [PubMed] [Google Scholar]
- 96.Chan T.S., Yu H., Moore A., Khetani S.R., Tweedie D. Meeting the challenge of predicting hepatic clearance of compounds slowly metabolized by cytochrome P450 using a novel hepatocyte model. HepatoPac, Drug Metab. Dispos. 2013;41:2024–2032. doi: 10.1124/DMD.113.053397. [DOI] [PubMed] [Google Scholar]
- 97.Da-silva F., Boulenc X., Vermet H., Compigne P., Gerbal-Chaloin S., Daujat-Chavanieu M., Klieber S., Poulin P. Improving prediction of metabolic clearance using quantitative extrapolation of results obtained from human hepatic micropatterned cocultures model and by considering the impact of albumin binding. J. Pharmaceut. Sci. 2018;107:1957–1972. doi: 10.1016/j.xphs.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 98.Umehara K., Cantrill C., Wittwer M.B., Di Lenarda E., Klammers F., Ekiciler A., Parrott N., Fowler S., Ullah M. Application of the Extended Clearance Classification System (ECCS) in drug discovery and development: selection of appropriate in vitro tools and clearance predictions. Drug Metabol. Dispos. 2020;48:849–860. doi: 10.1124/DMD.120.000133/-/DC1. [DOI] [PubMed] [Google Scholar]
- 99.Docci L., Klammers F., Ekiciler A., Molitor B., Umehara K., Walter I., Krähenbühl S., Parrott N., Fowler S. In vitro to in vivo extrapolation of metabolic clearance for UGT substrates using short-term suspension and long-term Co-cultured human hepatocytes. AAPS J. 2020;22 doi: 10.1208/S12248-020-00482-9. [DOI] [PubMed] [Google Scholar]
- 100.Lai Y., Chu X., Di L., Gao W., Guo Y., Liu X., Lu C., Mao J., Shen H., Tang H., Xia C.Q., Zhang L., Ding X. Recent advances in the translation of drug metabolism and pharmacokinetics science for drug discovery and development. Acta Pharm. Sin. B. 2022;12:2751–2777. doi: 10.1016/J.APSB.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kratochwil N.A., Meille C., Fowler S., Klammers F., Ekiciler A., Molitor B., Simon S., Walter I., McGinnis C., Walther J., Leonard B., Triyatni M., Javanbakht H., Funk C., Schuler F., Lavé T., Parrott N.J. Metabolic profiling of human long-term liver models and hepatic clearance predictions from in vitro data using nonlinear mixed-effects modeling. AAPS J. 2017;19:534–550. doi: 10.1208/S12248-016-0019-7. [DOI] [PubMed] [Google Scholar]
- 102.Obach R.S., Baxter J.G., Liston T.E., Silber B.M., Jones B.C., Macintyre F., Rance D.J., Wastall P. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J. Pharmacol. Exp. Therapeut. 1997;283 [PubMed] [Google Scholar]
- 103.Moraes C., Labuz J.M., Leung B.M., Inoue M., Chun T.H., Takayama S. On being the right size: scaling effects in designing a human-on-a-chip. Integr. Biol. 2013;5:1149. doi: 10.1039/C3IB40040A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maass C., Stokes C.L., Griffith L.G., Cirit M. Multi-functional scaling methodology for translational pharmacokinetic and pharmacodynamic applications using integrated microphysiological systems (MPS) Integr. Biol. 2017;9:290. doi: 10.1039/C6IB00243A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abaci H.E., Shuler M.L. Human-on-a-chip design strategies and principles for physiologically based pharmocokinetics/pharmacodynamics modeling. Integr. Biol. 2015;7:383. doi: 10.1039/C4IB00292J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Feng J.J., Hedtrich S. A similarity scaling approach for organ-on-chip devices. Lab Chip. 2022;22:3663–3667. doi: 10.1039/D2LC00641C. [DOI] [PubMed] [Google Scholar]
- 107.Wikswo J.P., Curtis E.L., Eagleton Z.E., Evans B.C., Kole A., Hofmeister L.H., Matloff W.J. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip. 2013;13:3496. doi: 10.1039/C3LC50243K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sung J.H., Wang Y., Shuler M.L. Strategies for using mathematical modeling approaches to design and interpret multi-organ microphysiological systems (MPS) APL Bioeng. 2019;3 doi: 10.1063/1.5097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stokes C.L., Cirit M., Lauffenburger D.A. Physiome-on-a-Chip: the challenge of “scaling” in design, operation, and translation of microphysiological systems. CPT Pharmacometrics Syst. Pharmacol. 2015;4:559. doi: 10.1002/PSP4.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Herland A., Maoz B.M., Das D., Somayaji M.R., Prantil-Baun R., Novak R., Cronce M., Huffstater T., Jeanty S.S.F., Ingram M., Chalkiadaki A., Benson Chou D., Marquez S., Delahanty A., Jalili-Firoozinezhad S., Milton Y., Sontheimer-Phelps A., Swenor B., Levy O., Parker K.K., Przekwas A., Ingber D.E. Quantitative prediction of human drug pharmacokinetic responses using multiple vascularized organ chips coupled by fluid transfer. Nat. Biomed. Eng. 2020;4:421. doi: 10.1038/S41551-019-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baudoin R., Legendre A., Jacques S., Cotton J., Bois F., Leclerc E. Evaluation of a liver microfluidic biochip to predict in vivo clearances of seven drugs in rats. J. Pharmaceut. Sci. 2014;103:706–718. doi: 10.1002/JPS.23796. [DOI] [PubMed] [Google Scholar]
- 112.Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W., Dorato M., Van Deun K., Smith P., Berger B., Heller A. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/RTPH.2000.1399. [DOI] [PubMed] [Google Scholar]
- 113.Weaver R.J., Blomme E.A., Chadwick A.E., Copple I.M., Gerets H.H.J., Goldring C.E., Guillouzo A., Hewitt P.G., Ingelman-Sundberg M., Jensen K.G., Juhila S., Klingmüller U., Labbe G., Liguori M.J., Lovatt C.A., Morgan P., Naisbitt D.J., Pieters R.H.H., Snoeys J., van de Water B., Williams D.P., Park B.K. Managing the challenge of drug-induced liver injury: a roadmap for the development and deployment of preclinical predictive models. Nat. Rev. Drug Discov. 2019. 2019;19(2 19):131–148. doi: 10.1038/s41573-019-0048-x. [DOI] [PubMed] [Google Scholar]
- 114.Cyprotex . Cyprotex; 2023. Mechanisms of Drug-Induced Toxicity. [Google Scholar]
- 115.Allison R., Guraka A., Shawa I.T., Tripathi G., Moritz W., Kermanizadeh A. Drug induced liver injury – a 2023 update. J. Toxicol. Environ. Health, Part A B. 2023;26:442–467. doi: 10.1080/10937404.2023.2261848. [DOI] [PubMed] [Google Scholar]
- 116.Walker P.A., Ryder S., Lavado A., Dilworth C., Riley R.J. The evolution of strategies to minimise the risk of human drug-induced liver injury (DILI) in drug discovery and development. Arch. Toxicol. 2020;94:2559–2585. doi: 10.1007/S00204-020-02763-W/FIGURES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vorrink S.U., Zhou Y., Ingelman-Sundberg M., Lauschke V.M. Prediction of drug-induced hepatotoxicity using long-term stable primary hepatic 3D spheroid cultures in chemically defined conditions. Toxicol. Sci. 2018;163:655. doi: 10.1093/TOXSCI/KFY058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hendriks D.F.G., Puigvert L.F., Messner S., Mortiz W., Ingelman-Sundberg M. Hepatic 3D spheroid models for the detection and study of compounds with cholestatic liability. Scient. Rep. 2016. 2016;6(1 6):1–12. doi: 10.1038/srep35434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Foster A.J., Chouhan B., Regan S.L., Rollison H., Amberntsson S., Andersson L.C., Srivastava A., Darnell M., Cairns J., Lazic S.E., Jang K.J., Petropolis D.B., Kodella K., Rubins J.E., Williams D., Hamilton G.A., Ewart L., Morgan P. Integrated in vitro models for hepatic safety and metabolism: evaluation of a human Liver-Chip and liver spheroid. Arch. Toxicol. 2019;93:1021–1037. doi: 10.1007/S00204-019-02427-4/FIGURES/6. [DOI] [PubMed] [Google Scholar]
- 120.Kostrzewski T., Snow S., Battle A.L., Peel S., Ahmad Z., Basak J., Surakala M., Bornot A., Lindgren J., Ryaboshapkina M., Clausen M., Lindén D., Maass C., Young L.M., Corrigan A., Ewart L., Hughes D. Modelling human liver fibrosis in the context of non-alcoholic steatohepatitis using a microphysiological system. Commun. Biol. 2021;4 doi: 10.1038/S42003-021-02616-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Z., He X., Qiao H., Chen P. Global Trends of Organoid and Organ-On-a-Chip in the Past Decade: A Bibliometric and Comparative Study. 2020;26:656–671. doi: 10.1089/TEN.TEA.2019.0251. [DOI] [PubMed] [Google Scholar]
- 122.Zhao Z., Chen X., Dowbaj A.M., Sljukic A., Bratlie K., Lin L., Fong E.L.S., Balachander G.M., Chen Z., Soragni A., Huch M., Zeng Y.A., Wang Q., Yu H. Organoids. Nature Rev. Method. Prim. 2022. 2022;2(1 2):1–21. doi: 10.1038/s43586-022-00174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sato T., Vries R.G., Snippert H.J., Van De Wetering M., Barker N., Stange D.E., Van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009. 2009;459(7244 459):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 124.Hu H., Gehart H., Artegiani B., Löpez-Iglesias C., Dekkers F., Basak O., van Es J., Chuva de Sousa Lopes S.M., Begthel H., Korving J., van den Born M., Zou C., Quirk C., Chiriboga L., Rice C.M., Ma S., Rios A., Peters P.J., de Jong Y.P., Clevers H. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606.e19. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 125.Peng W.C., Logan C.Y., Fish M., Anbarchian T., Aguisanda F., Álvarez-Varela A., Wu P., Jin Y., Zhu J., Li B., Grompe M., Wang B., Nusse R. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell. 2018;175:1607–1619.e15. doi: 10.1016/j.cell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mun S.J., Ryu J.S., Lee M.O., Son Y.S., Oh S.J., Cho H.S., Son M.Y., Kim D.S., Kim S.J., Yoo H.J., Lee H.J., Kim J., Jung C.R., Chung K.S., Son M.J. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019;71:970–985. doi: 10.1016/J.JHEP.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 127.Hofer M., Lutolf M.P. Engineering organoids. Nature Rev. Mater. 2021. 2021;6(5 6):402–420. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Homan K.A., Gupta N., Kroll K.T., Kolesky D.B., Skylar-Scott M., Miyoshi T., Mau D., Valerius M.T., Ferrante T., Bonventre J.V., Lewis J.A., Morizane R. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255. doi: 10.1038/S41592-019-0325-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jin Y., Kim J., Lee J.S., Min S., Kim S., Ahn D.H., Kim Y.G., Cho S.W. Vascularized liver organoids generated using induced hepatic tissue and dynamic liver-specific microenvironment as a drug testing platform. Adv. Funct. Mater. 2018;28 doi: 10.1002/ADFM.201801954. [DOI] [Google Scholar]
- 130.Dehne E.M., Marx U. Human body-on-a-chip systems, Organ-on-a-Chip. Eng. Microenviron. Safet. Eff. Test. 2020:429–439. doi: 10.1016/B978-0-12-817202-5.00013-9. [DOI] [Google Scholar]
- 131.Yau E., Petersson C., Dolgos H., Peters S.A. A comparative evaluation of models to predict human intestinal metabolism from nonclinical data. Biopharm Drug Dispos. 2017;38:163–186. doi: 10.1002/BDD.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thelen K., Dressman J.B. Cytochrome P450-mediated metabolism in the human gut wall. J. Pharm. Pharmacol. 2009;61:541–558. doi: 10.1211/JPP.61.05.0002. [DOI] [PubMed] [Google Scholar]
- 133.Musther H., Olivares-Morales A., Hatley O.J.D., Liu B., Rostami Hodjegan A. Animal versus human oral drug bioavailability: do they correlate? Eur. J. Pharmaceut. Sci. 2014;57:280–291. doi: 10.1016/J.EJPS.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zgair A., Dawood Y., Ibrahem S.M., Back H.M., Kagan L., Gershkovich P., Lee J.B. Predicting intestinal and hepatic first-pass metabolism of orally administered testosterone undecanoate. Appl. Sci. 2020. 2020;10:7283. doi: 10.3390/APP10207283. Page 7283 10. [DOI] [Google Scholar]
- 135.Li C., Liu T., Cui X., Uss A.S., Cheng K.C. Development of in vitro pharmacokinetic screens using Caco-2, human hepatocyte, and Caco-2/human hepatocyte hybrid systems for the prediction of oral bioavailability in humans. J. Biomol. Screen. 2007;12:1084–1091. doi: 10.1177/1087057107308892. [DOI] [PubMed] [Google Scholar]
- 136.Arakawa H., Sugiura S., Kawanishi T., Shin K., Toyoda H., Satoh T., Sakai Y., Kanamori T., Kato Y. Kinetic analysis of sequential metabolism of triazolam and its extrapolation to humans using an entero-hepatic two-organ microphysiological system. Lab Chip. 2020;20:537–547. doi: 10.1039/C9LC00884E. [DOI] [PubMed] [Google Scholar]
- 137.Prot J.M., Maciel L., Bricks T., Merlier F., Cotton J., Paullier P., Bois F.Y., Leclerc E. First pass intestinal and liver metabolism of paracetamol in a microfluidic platform coupled with a mathematical modeling as a means of evaluating ADME processes in humans. Biotechnol. Bioeng. 2014;111:2027–2040. doi: 10.1002/BIT.25232. [DOI] [PubMed] [Google Scholar]
- 138.Bricks T., Hamon J., Fleury M.J., Jellali R., Merlier F., Herpe Y.E., Seyer A., Regimbeau J.M., Bois F., Leclerc E. Investigation of omeprazole and phenacetin first-pass metabolism in humans using a microscale bioreactor and pharmacokinetic models. Biopharm Drug Dispos. 2015;36:275–293. doi: 10.1002/BDD.1940. [DOI] [PubMed] [Google Scholar]
- 139.Yang J., Imamura S., Hirai Y., Tsuchiya T., Tabata O., Kamei K.I. Gut-liver-axis microphysiological system for studying cellular fluidic shear stress and inter-tissue interaction. Biomicrofluidics. 2022;16 doi: 10.1063/5.0088232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Duan X., Zheng L., Zhang X., Wang B., Xiao M., Zhao W., Liu S., Sui G. A membrane-free liver-gut-on-chip platform for the assessment on dysregulated mechanisms of cholesterol and bile acid metabolism induced by PM2.5. ACS Sens. 2020;5:3483–3492. doi: 10.1021/ACSSENSORS.0C01524. [DOI] [PubMed] [Google Scholar]
- 141.Esch M.B., Mahler G.J., Stokol T., Shuler M.L. Body-on-a-Chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14:3081. doi: 10.1039/C4LC00371C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee S.Y., Sung J.H. Gut-liver on a chip toward an in vitro model of hepatic steatosis. Biotechnol. Bioeng. 2018;115:2817–2827. doi: 10.1002/BIT.26793. [DOI] [PubMed] [Google Scholar]
- 143.Milani N., Parrott N., Ortiz Franyuti D., Godoy P., Galetin A., Gertz M., Fowler S. Application of a gut–liver-on-a-chip device and mechanistic modelling to the quantitative in vitro pharmacokinetic study of mycophenolate mofetil. Lab Chip. 2022;22:2853–2868. doi: 10.1039/D2LC00276K. [DOI] [PubMed] [Google Scholar]
- 144.Rusyn I., Sakolish C., Kato Y., Stephan C., Vergara L., Hewitt P., Bhaskaran V., Davis M., Hardwick R.N., Ferguson S.S., Stanko J.P., Bajaj P., Adkins K., Sipes N.S., Hunter E.S., Baltazar M.T., Carmichael P.L., Sadh K., Becker R.A. Microphysiological systems evaluation: experience of TEX-VAL tissue chip testing consortium. Toxicol. Sci. 2022;188:143–152. doi: 10.1093/TOXSCI/KFAC061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Messelmani T., Morisseau L., Sakai Y., Legallais C., Le Goff A., Leclerc E., Jellali R. Liver organ-on-chip models for toxicity studies and risk assessment. Lab Chip. 2022;22:2423–2450. doi: 10.1039/D2LC00307D. [DOI] [PubMed] [Google Scholar]
- 146.Lim A.Y., Kato Y., Sakolish C., Valdiviezo A., Han G., Bajaj P., Stanko J., Ferguson S.S., Villenave R., Hewitt P., Hardwick R.N., Rusyn I. Reproducibility and robustness of a liver microphysiological system PhysioMimix LC12 under varying culture conditions and cell type combinations. Bioengineering. 2023;10:1195. doi: 10.3390/BIOENGINEERING10101195/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kvist A.J., Kanebratt K.P., Walentinsson A., Palmgren H., O’Hara M., Björkbom A., Andersson L.C., Ahlqvist M., Andersson T.B. Critical differences in drug metabolic properties of human hepatic cellular models, including primary human hepatocytes, stem cell derived hepatocytes, and hepatoma cell lines. Biochem. Pharmacol. 2018;155:124–140. doi: 10.1016/J.BCP.2018.06.026. [DOI] [PubMed] [Google Scholar]
- 148.Teixeira Carvalho D.J., Moroni L., Giselbrecht S. Clamping strategies for organ-on-a-chip devices. Nature Rev. Mater. 2023. 2023;8(3 8):147–164. doi: 10.1038/s41578-022-00523-z. [DOI] [Google Scholar]
- 149.Feghali M., Venkataramanan R., Caritis S. Pharmacokinetics of drugs in pregnancy. Semin. Perinatol. 2015;39:512. doi: 10.1053/J.SEMPERI.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jeong H., Stika C.S. Methods to study mechanisms underlying altered hepatic drug elimination during pregnancy. Semin. Perinatol. 2020;44 doi: 10.1016/J.SEMPERI.2020.151228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Eke A.C., Gebreyohannes R.D., Fernandes M.F.S., Pillai V.C. Physiologic changes during pregnancy and impact on small-molecule drugs, biologic (monoclonal antibody) disposition, and response. J. Clin. Pharmacol. 2023;63:S34–S50. doi: 10.1002/JCPH.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Koh K.H., Xie H., Yu A.M., Jeong H. Altered cytochrome P450 expression in mice during pregnancy. Drug Metabol. Dispos. 2011;39:165. doi: 10.1124/DMD.110.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsunoda S.M., Gonzales C., Jarmusch A.K., Momper J.D., Ma J.D. Contribution of the gut microbiome to drug disposition, pharmacokinetic and pharmacodynamic variability. Clin. Pharmacokinet. 2021. 2021;60(8 60):971–984. doi: 10.1007/S40262-021-01032-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jalili-Firoozinezhad S., Gazzaniga F.S., Calamari E.L., Camacho D.M., Fadel C.W., Bein A., Swenor B., Nestor B., Cronce M.J., Tovaglieri A., Levy O., Gregory K.E., Breault D.T., Cabral J.M.S., Kasper D.L., Novak R., Ingber D.E. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019;3:520. doi: 10.1038/S41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kang S.G., Choi Y.Y., Mo S.J., Kim T.H., Ha J.H., Hong D.K., Lee H., Park S.D., Shim J.J., Lee J.L., Chung B.G. Effect of gut microbiome-derived metabolites and extracellular vesicles on hepatocyte functions in a gut-liver axis chip. Nano Converg. 2023;10 doi: 10.1186/S40580-022-00350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lucchetti M., Kaminska M., Oluwasegun A.K., Mosig A.S., Wilmes P. Emulating the gut–liver axis: dissecting the microbiome’s effect on drug metabolism using multiorgan-on-chip models. Curr. Opin. Endocr. Metab. Res. 2021;18:94. doi: 10.1016/J.COEMR.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim H.J., Li H., Collins J.J., Ingber D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E7–E15. doi: 10.1073/PNAS.1522193112/SUPPL_FILE/PNAS.1522193112.SM03.AVI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yeung C.K., Shen D.D., Thummel K.E., Himmelfarb J. Effects of chronic kidney disease and uremia on hepatic drug metabolism and transport. Kidney Int. 2014;85:522–528. doi: 10.1038/KI.2013.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu D., Wu J., Liu H., Shi S., Wang L., Huang Y., Yu X., Lei Z., Ouyang T., Shen J., Wu G., Wang S. A biomimetic renal fibrosis progression model on-chip evaluates anti-fibrotic effects longitudinally in a dynamic fibrogenic niche. Lab Chip. 2023;23:4708–4725. doi: 10.1039/D3LC00393K. [DOI] [PubMed] [Google Scholar]
- 160.Ashammakhi N., Wesseling-Perry K., Hasan A., Elkhammas E., Zhang Y.S. Kidney-on-a-chip: untapped opportunities. Kidney Int. 2018;94:1073. doi: 10.1016/J.KINT.2018.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Franzen N., van Harten W.H., Retèl V.P., Loskill P., van den Eijnden-van Raaij J., Ijzerman M. Impact of organ-on-a-chip technology on pharmaceutical R&D costs. Drug Discov. Today. 2019;24:1720–1724. doi: 10.1016/J.DRUDIS.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 162.Levner D., Ewart L. Integrating Liver-Chip data into pharmaceutical decision-making processes. Expet Opin. Drug Discov. 2023 doi: 10.1080/17460441.2023.2255127. [DOI] [PubMed] [Google Scholar]
- 163.Picollet-D’hahan N., Zuchowska A., Lemeunier I., Le Gac S. Multiorgan-on-a-Chip: a systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 2021;39:788–810. doi: 10.1016/J.TIBTECH.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 164.Prantil-Baun R., Novak R., Das D., Somayaji M.R., Przekwas A., Ingber D.E. Physiologically based pharmacokinetic and pharmacodynamic analysis enabled by microfluidically linked organs-on-chips. Annu. Rev. Pharmacol. Toxicol. 2018;58:37–64. doi: 10.1146/ANNUREV-PHARMTOX-010716-104748. [DOI] [PubMed] [Google Scholar]
- 165.Aravindakshan M.R., Mandal C., Pothen A., Maass C. DigiLoCS: a leap forward in predictive organ-on-chip simulations. bioRxiv. 2024;2024(03.28) doi: 10.1101/2024.03.28.587123. [DOI] [Google Scholar]
- 166.Bowman C.M., Benet L.Z. In vitro-in vivo extrapolation and hepatic clearance-dependent underprediction. J. Pharmaceut. Sci. 2019;108:2500–2504. doi: 10.1016/J.XPHS.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schneider M.R., Oelgeschlaeger M., Burgdorf T., van Meer P., Theunissen P., Kienhuis A.S., Piersma A.H., Vandebriel R.J. Applicability of organ-on-chip systems in toxicology and pharmacology. Crit. Rev. Toxicol. 2021;51:540–554. doi: 10.1080/10408444.2021.1953439. [DOI] [PubMed] [Google Scholar]
- 168.Li J., Chen J., Bai H., Wang H., Hao S., Ding Y., Peng B., Zhang J., Li L., Huang W. An overview of organs-on-chips based on deep learning. Research 2022. 2022 doi: 10.34133/2022/9869518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bai L., Wu Y., Li G., Zhang W., Zhang H., Su J. AI-enabled organoids: construction, analysis, and application. Bioact. Mater. 2024;31:525–548. doi: 10.1016/J.BIOACTMAT.2023.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Haja A., Horcas-Nieto J.M., Bakker B.M., Schomaker L. Towards automatization of organoid analysis: a deep learning approach to localize and quantify organoid images. Comp. Method. Prog. Biomed. Update. 2023;3 doi: 10.1016/J.CMPBUP.2023.100101. [DOI] [Google Scholar]
- 171.Matthews J.M., Schuster B., Kashaf S.S., Liu P., Ben-Yishay R., Ishay-Ronen D., Izumchenko E., Shen L., Weber C.R., Bielski M., Kupfer S.S., Bilgic M., Rzhetsky A., Tay S. OrganoID: a versatile deep learning platform for tracking and analysis of single-organoid dynamics. PLoS Comput. Biol. 2022;18 doi: 10.1371/JOURNAL.PCBI.1010584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.