Abstract
The interaction of human immunodeficiency virus (HIV)-derived vectors with wild-type virus was analyzed in transduced cells. Vector transcripts upregulated by infection had no measurable effect on HIV type 1 (HIV-1) expression but competed efficiently for encapsidation, inhibiting the infectivity and spread of HIV-1 in culture and leading to mobilization and recombination of the vector. These effects were abrogated with a self-inactivating vector.
Gene transfer vectors based on human immunodeficiency virus type 1 (HIV-1) have shown significant potential for gene therapy (1, 12, 20, 21, 28). However, several safety concerns must be addressed before their clinical testing (8, 14, 19, 26, 30). Among them are the possible adverse interactions with wild-type virus in recipients who are or become infected by HIV-1. cis-acting sequences in the vector are responsive to the regulatory proteins of HIV-1 (19; Fig. 1) and could affect HIV-1 replication and lead to the mobilization and recombination of the vector in infected cells. To address experimentally these issues we infected lymphocytes previously transduced by HIV-derived vectors and monitored HIV-1 replication and vector expression. Control cells were transduced by a murine leukemia virus (MLV) vector or an HIV vector with a deletion in the long terminal repeat (LTR) (ΔU3) that prevents transcription and activation by HIV-1 infection (31).
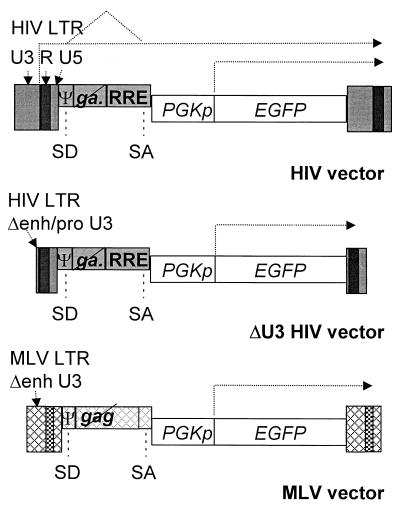
FIG. 1.
Schematic drawing of the types of vectors used in this work. The transduced proviral forms are shown. All vectors shown carry an internal expression cassette for EGFP driven by the PGK promoter (PGKp). The following viral cis-acting sequences are labeled and shown as dark boxes if derived from HIV-1 or as crosshatched boxes if derived from MLV: the LTRs with the constituent U3, R, and U5 regions, the leader sequence containing the major splice donor site (SD) and the packaging signal (Ψ), the portion of the gag gene included for optimal packaging efficiency—crossed out to indicate that the reading frame is closed by mutations—the env-derived splice acceptor sites (SA), and the RRE for the HIV vectors. The deletion of enhancer (Δenh) or enhancer and promoter sequences (Δenh/pro) from the U3 region of the LTR is indicated for the MLV and the self-inactivating ΔU3 HIV vectors, respectively. The types of vector transcripts discussed in the text are indicated by dotted arrows.
Replication of HIV-1 is inhibited in cells transduced by HIV-derived vectors.
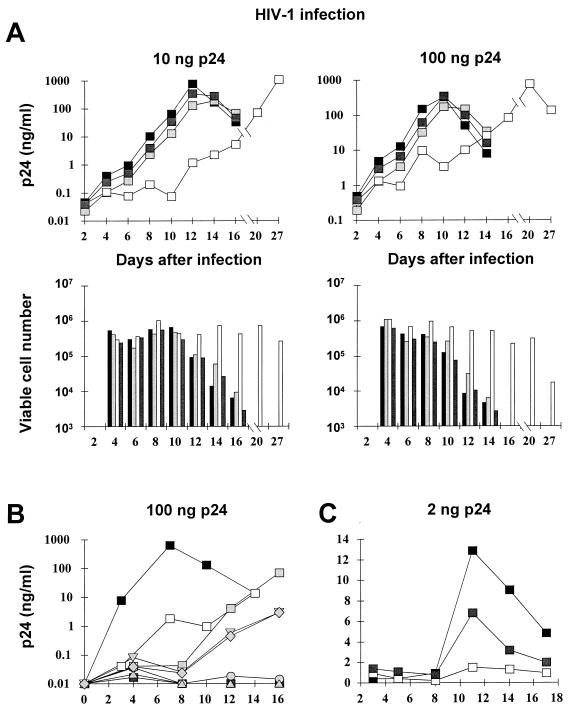
Human SupT1 lymphoid cells were transduced by vectors expressing the enhanced green fluorescent protein (EGFP) from an internal phosphoglycerokinase (PGK) promoter (Fig. 1). Vectors were produced by transient transfection of 293T cells with matched transfer and gag pol packaging constructs, derived either from HIV-1 or MLV, and a third plasmid expressing the vesicular stomatitis virus (VSV) envelope (21, 30). The ΔU3 HIV vector was produced by a transfer construct in which the 3′ LTR sequence from base pair −418 to −18 relative to the beginning of R was deleted. The transduction of this vector replicates the deletion in the upstream LTR (self-inactivation [31]). Vector titers were determined in HeLa cells and used at similar multiplicities of infection (ranging from 5 to 20 in different experiments). Pools of EGFP-positive transductants and control nontransduced cells were challenged with two different amounts of HIV-1 (Fig. 2A). Virus stock was produced by the transfection of 293T cells with the proviral plasmid R8, a lymphocytotropic hybrid of the HXB2D and NL43 isolates, which expresses all HIV reading frames (9). No significant difference in HIV replication was observed in nontransduced cells and in cells transduced by MLV or ΔU3 HIV vector. However, in HIV-transduced cultures, the peak of p24 antigen accumulation in the supernatant was delayed in an infection dose-dependent manner, and the cells remained viable much longer after infection.
FIG. 2.
HIV-1 replication is inhibited in lymphocytes transduced by HIV-derived vectors. The inhibitory activity is mediated by the vector LTR. (A) SupT1 cells transduced by HIV-derived (□), ΔU3 HIV-derived ( ), and MLV-derived (
), and MLV-derived ( ) vectors encoding EGFP or nontransduced cells (■) were infected with the indicated p24 equivalent of HIV-1 and scored for the accumulation of p24 antigen in the culture supernatant (upper panels) and cell viability (lower panels). (B) SupT1 cells were transfected (grey markers, with each marker representing an individual clone) or transduced (□) with HIV-derived vector encoding the neomycin resistance gene, selected in G418-containing medium, and infected with HIV-1 as above; control cells are also shown (■). HIV-1 replication was inhibited in all the cultures containing the vector, independent of the mode of delivery. (C) Primary CD4-positive lymphocytes were transduced with HIV-derived (□) and ΔU3 HIV-derived (
) vectors encoding EGFP or nontransduced cells (■) were infected with the indicated p24 equivalent of HIV-1 and scored for the accumulation of p24 antigen in the culture supernatant (upper panels) and cell viability (lower panels). (B) SupT1 cells were transfected (grey markers, with each marker representing an individual clone) or transduced (□) with HIV-derived vector encoding the neomycin resistance gene, selected in G418-containing medium, and infected with HIV-1 as above; control cells are also shown (■). HIV-1 replication was inhibited in all the cultures containing the vector, independent of the mode of delivery. (C) Primary CD4-positive lymphocytes were transduced with HIV-derived (□) and ΔU3 HIV-derived ( ) vectors encoding EGFP and selected for transgene expression or were not transduced and mock sorted (■) and infected as described above. The averages of duplicate determinations are shown for a representative experiment of three performed.
) vectors encoding EGFP and selected for transgene expression or were not transduced and mock sorted (■) and infected as described above. The averages of duplicate determinations are shown for a representative experiment of three performed.
Cells infected by HIV-1 may become resistant to subsequent challenge with the virus due to interference with viral entry by downmodulation of the CD4 receptor from the cell surface (17) or possibly at later steps preceding viral gene expression (29). However, it is unlikely that any one of these mechanisms was responsible for the inhibition observed in transduced cells, as the vector does not transfer viral genes and was packaged by a construct with all HIV gene products known to induce CD4 downmodulation deleted (30). Moreover, direct immunostaining showed no difference in CD4 levels between parental and transduced cells (data not shown). To rule out the possibility that the process of infection by vector particles establishes the interference, we introduced an HIV vector expressing the neomycin resistance gene into SupT1 cells by electroporation of the corresponding plasmid and cloned the transfectants in a selective medium. HIV-1 replication was significantly inhibited both in the transfectant clones and in a pooled population of transduced cells obtained as described above (Fig. 2B). These results indicate that the inhibition of HIV-1 replication in transduced cells is independent of the mode of vector delivery and of transgene type and is mediated by functional HIV LTRs in the vector.
The inhibition of HIV-1 replication and improved cell survival were also observed in primary lymphocytes (Fig. 2C). CD4-positive lymphocytes were obtained from healthy blood donors by Ficoll gradient centrifugation and negative immunoselection of CD8-positive cells, stimulated with anti-CD3- and anti-CD28-coated Dynal beads for 1 day, transduced with EGFP vectors as described above, selected by fluorescence-activated cell sorting (FACS), and cultured in AIM-V medium (Gibco) with 100 U of interleukin 2 (Chiron) per ml for virus challenge. HIV-1 replication was inhibited only in cells transduced by the HIV vector with intact LTRs.
The initial steps of HIV-1 infection are not inhibited in transduced cells.
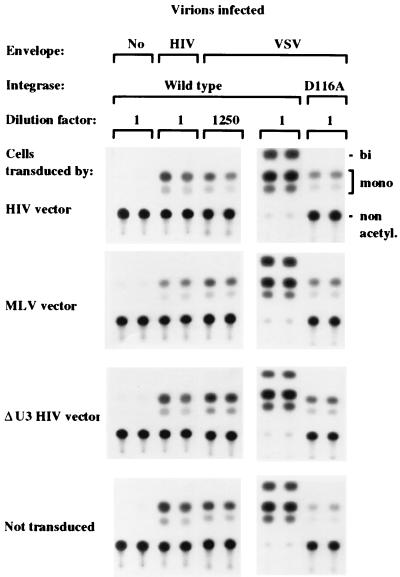
To clarify the mechanism of inhibition we analyzed the steps of HIV-1 replication in cells transduced by different types of vectors. We used virions transiently complemented by HIV-1 gp120 (pEnvHXB plasmid [25]) or VSV G glycoprotein (VSV.G) in a single-round infection assay (10). VSV.G was used to increase the efficiency of infection (3, 5). The viral genome had a deletion in the env gene and contained the bacterial chloramphenicol acetyltransferase (CAT) gene in place of the nef gene (HXBH10ΔenvCAT plasmid [25]). All infected cells had similar levels of CAT activity 3 days after infection (Fig. 3). CAT activity was not detected in cells exposed to virus produced without an envelope, and it was dramatically reduced in cells infected by a virus carrying a defective integrase (25). These data indicate that the inhibitory activity of the vector on HIV-1 replication is exerted at a postintegration step.
FIG. 3.
Lymphocytes transduced by HIV-derived vectors are permissive to all stages of HIV-1 infection, leading to the integration and expression of the early genes. SupT1 cells, transduced as indicated on the left, were infected by HIV-1 virions containing the CAT gene in place of nef and pseudotyped by the HIV-1 or VSV envelope. Duplicate cultures infected by the types of virions shown on top were extracted and assayed for CAT activity after the indicated dilution. All the cultures infected by the same viral pseudotype had similar levels of CAT activity, indicating that the integration and expression of the early genes were not affected by the vector. The VSV pseudotype was much more infectious than the HIV pseudotype, as shown by the higher dilution of extract yielding similar CAT activity. No activity was detected in cells exposed to virions produced without envelopes. CAT expression was dependent on the integration of the HIV genome, as shown by the comparison between cells infected by virus carrying wild-type or mutant defective integrase (D116A). The migration of the different acetylated forms of chloramphenicol is indicated on the right. A representative experiment of three performed is shown.
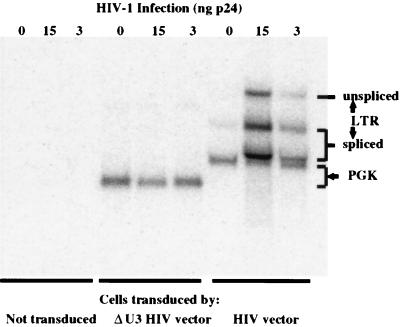
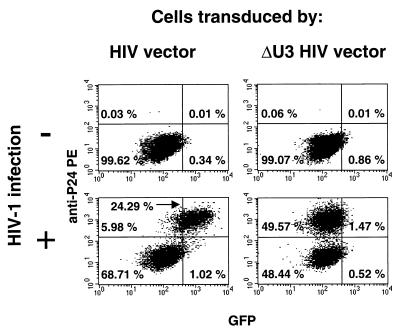
We then analyzed vector expression in the transduced SupT1 cells. As expected, Northern analysis showed that HIV infection stimulates vector transcription from the LTR and leads to the accumulation of both unspliced and spliced transcripts (Fig. 4). FACS analysis showed that some of these transcripts contribute to EGFP expression, as the superinfection of transduced cells results in the appearance of a distinct population of cells with increased expression of EGFP (Fig. 5). These cells represented the infected fraction as shown by secondary staining with phycoerythrin (PE)-conjugated anti-p24 antibodies (KC57-RD1; Coulter). In contrast, EGFP expression was unaffected by infection of cells transduced by the ΔU3 HIV vector.
FIG. 4.
Northern analysis of lymphocytes transduced by the EGFP vectors indicated on the bottom and infected by the p24 equivalent of HIV-1 virions indicated on the top, probed for expression of the EGFP gene. The expected positions of transcripts initiated from the LTR or the internal PGK promoter are indicated. HIV-1 infection increases transcription from the vector LTR. The transcripts accumulate partly unspliced and partly spliced into acceptor sites downstream from the RRE or in the PGK promoter sequence, thus contributing to transgene expression. Note the smaller size of the PGK-driven transcript and the lack of HIV-induced transcripts in cells transduced by the self-inactivating ΔU3 HIV vector. All lanes contained similar amounts of RNA by ethidium bromide staining (data not shown).
FIG. 5.
FACS analysis of SupT1 cells transduced by HIV-derived or ΔU3 HIV-derived vector expressing EGFP and infected by HIV-1 virions pseudotyped with (+) or without (−) the VSV envelope, after immunostaining for p24. All HIV-infected cells in the culture transduced by vector with intact LTRs overexpress EGFP, while transgene expression was unaffected in cells transduced by the ΔU3 vector. Note that all the cells shown were transduced by EGFP vectors and so are fluorescent; nontransduced cells, either infected or not with HIV-1, mapped to the left of the FACS diagram (data not shown).
Vector transcripts initiated from the LTR contain the transactivation response element (TAR) and the Rev response element (RRE), binding sites for the Tat and Rev proteins of HIV (17). Kim et al. (13) and Corbeau and Wong-Staal (6) reported a decreased expression of transfected HIV-1 proviral plasmid in cells transfected (13) or transduced with HIV vectors (6). Corbeau and Wong-Staal also showed a decreased expression of a Tat- or Rev-dependent reporter gene cotransfected with a Tat or Rev plasmid in HIV vector-transduced cells (6). These results indicated a decoy effect of the vector RNA as previously observed for TAR and RRE (4, 15, 16, 27). However, probing an actual infection, we did not find significant differences in expression of early (see CAT expression described above) or late genes of HIV-1 in cells transduced by HIV vectors. The expression of the late gene product p24 was similar in cells transduced by vector with wild-type or ΔU3 LTR by immunostaining (Fig. 5) and immunocapture in the supernatant (data not shown). Thus, the decoy activity of vector transcripts on the Tat and Rev proteins of the infecting virus was too small an effect to be noticeable in a single-cycle infection and consequently to explain the observed inhibition of HIV-1 replication.
Vector transcripts compete for encapsidation and reduce transfer of the viral genome.
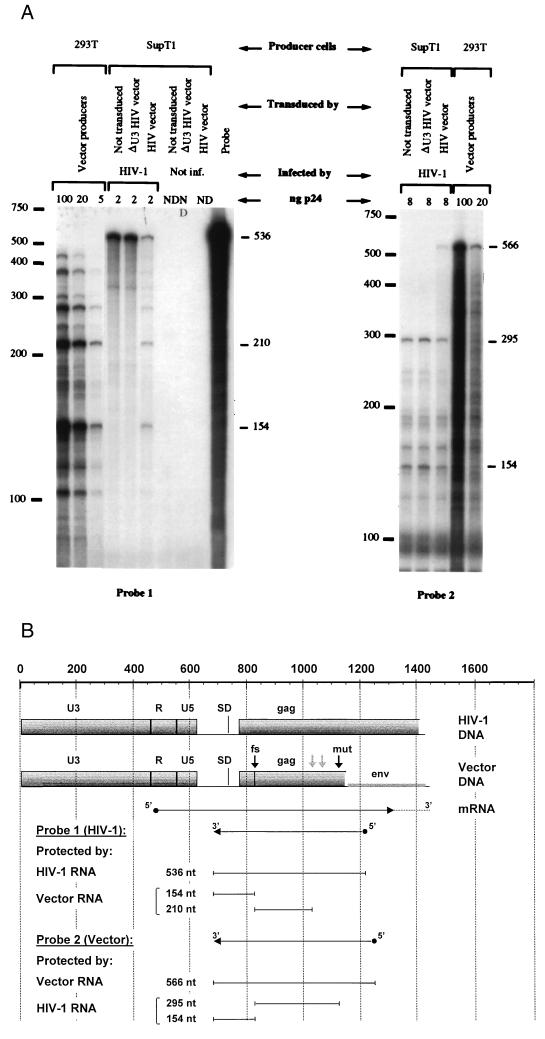
We then rescued HIV-1 particles from the infected transduced cells to measure the encapsidation and transfer of the viral and vector genomes. SupT1 cells transduced with EGFP vectors were infected by virions generated by 293T cells transfected with the plasmid pR8.7, an NL43-derived construct with all four accessory genes deleted, and a plasmid encoding VSV.G to make the first round of infection more efficient. The newly released virions were collected and assayed for RNA content and infectivity. RNA was purified from p24-matched viral pellets and analyzed by RNase protection (HybSpeed RPA kit; Ambion) (Fig. 6). The templates for the riboprobes were prepared by using the Lig’nScribe kit (Ambion). A 556-bp fragment (nucleotides 677 to 1233 of HIV-1 proviral DNA) was amplified from the R8 plasmid by PCR with primers 1s (5′-GCTCTCTCGACGCAGGACTCGG-3′) and 2as (5′-GTGATATGGCCTGATGTACCAT-3′), and a 566-bp fragment was amplified from pHR2 vector plasmid with primers 1s and 3as (5′-GTGCTACTCCTAATGGTTCAA-3′). The fragments were ligated to an adapter containing the T7 promoter and used as templates for a second PCR with primer 1s and a T7 primer provided with the kit. 32P-labeled antisense riboprobes 1 and 2 (Fig. 6B) were then generated with T7 RNA polymerase, purified, hybridized with virion RNA, digested with RNase A/T1 mix (0.5/20 U), and analyzed by electrophoresis and autoradiography. The protection patterns of vector and virus RNA are easily distinguished by the difference in length of the protected fragments as explained in the legend to Fig. 6. Only in virions released by cells transduced with HIV-derived vector with wild-type LTR the protection pattern of the vector RNA was superimposed on that of the viral genomic RNA (Fig. 6A). As the virion samples were adjusted to an equal content of p24 and had similar amounts of total RNA determined by phosphorimager analysis, it is evident that the vector RNA competes efficiently for encapsidation and reduces significantly the amount of HIV-1 RNA in the particles (approximately sixfold).
FIG. 6.
RNase protection analysis of virions collected from transduced cells showing the efficient encapsidation of the vector and displacement of the viral genome. (A) Virions were collected from HIV-1-infected SupT1 cells previously transduced by HIV-derived vector with wild-type or U3-deleted LTR or not transduced, as indicated on top. Vector particles were from 293T transfectants. RNA was extracted from the indicated amounts of particles (by p24 content) and analyzed for RNase protection of two probes derived from the virus sequence (NL43 isolate [probe 1]) or the vector sequence (HXB2 isolate [probe 2]). Each probe encompasses the HIV-1 leader sequence and the first portion of the gag gene and gives a distinct protection pattern of vector versus virus RNA. (B) The vector sequence differs from the virus genome for the truncation, frameshift (fs), and additional mutations (mut) introduced in gag for vector construction and for the indicated 2-bp mismatches between the sequence of the two viral isolates (pale arrows). As these differences cause fragmentation of the probe, the expected size in base pairs of the protected fragments is shown in panel B, and the actual size of the major protected bands is indicated to the right of the gels in panel A. The protection pattern of the vector RNA is superimposed on that of the viral genome only in virions released by cells transduced by vector with the wild-type LTR. A decreased encapsidation of the viral genome is concurrently observed. The additional lanes in the gel to the left were loaded with mock-purified pellets from noninfected (Not inf.) transduced SupT1 as negative controls (ND, p24 not detectable), showing also the digestion of unprotected probe, and with the untreated probe. One of two similar experiments performed is shown. nt, nucleotide; SD, splice donor site.
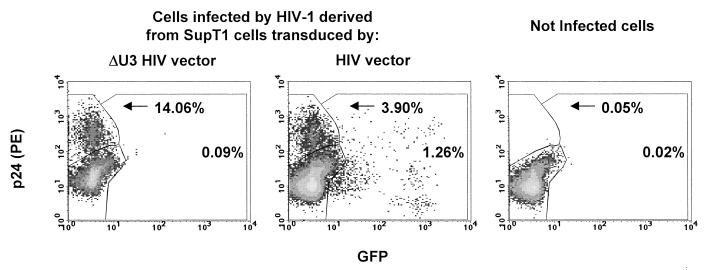
Rescued virions were then tested for gene transfer by adding an equal amount of p24 antigen to cultures of C8166, a T-lymphoblastoid cell line with high sensitivity to HIV infection. Four hours after infection unadsorbed virus was removed and the cells were further cultured in the presence of 5 μM ritonavir (Abbott Laboratories) and the CD4 [Cys(Bzl)]84-Fragment 81-92 (C2796; Sigma) to limit viral spread and syncytium formation. After 3 days, the infected cultures were scored for EGFP and Gag expression by FACS analysis (Fig. 7). The number of Gag-positive cells per nanogram of p24 antigen infected was reduced approximately fourfold for the virions released from cells transduced by HIV vector with wild-type LTR compared to the virions released from all other cells. Concurrently, a fraction of cells infected by this virus expressed EGFP, which indicated efficient vector mobilization. No EGFP-positive cells were found in the culture infected by virus derived from cells transduced with the ΔU3 HIV vector. Thus, only some of the virions produced by transduced cells propagate the virus, while others transfer the vector. The observed reduction in transfer of the viral genome should be amplified during each round of infection and significantly decrease the rate of HIV-1 spreading in the culture.
FIG. 7.
HIV-derived vectors are mobilized by HIV-1 infection of transduced cells. C8166 lymphocytes were used to score the virions for transfer of the HIV-1 gag gene, by immunostaining with anti-p24 antibodies, and of the vector, by EGFP fluorescence. Virions were collected from SupT1 cells transduced by the EGFP vectors indicated on top and adjusted to a p24 content of 8 ng for infection. Three days after infection C8166 cells were analyzed by FACS after immunostaining for p24. Virions derived from cells transduced by vector with the wild-type LTR transferred the viral genome less efficiently, as indicated by the percent of p24-positive cells, but mobilized the vector, as indicated by the appearance of EGFP-positive cells. No vector mobilization was detectable from cells transduced by the self-inactivating ΔU3 vector. Immunostained, uninfected C8166 cells are shown in the right panel. A representative experiment of three performed is shown.
Interestingly, a reduced capacity of the virions to transfer the viral genome was not fully compensated by the transfer of the vector. This could be explained if the vector RNA cannot accomplish all preintegration steps as efficiently as the viral genome, for example, for lack of some cis-acting elements. Also, it is expected that some of the virions produced by the transduced cells are heterodimeric and that template switching during reverse transcription may generate recombinants between the vector and the virus (11). Some of these recombinants will score negative both for Gag and EGFP expression. Of note, in Fig. 7 a distinct population of bright EGFP-positive, p24-negative cells is observed in the culture infected by virions derived from HIV-transduced cells. In the experiment shown in Fig. 5, all EGFP-bright cells are p24 positive, as expected from the presence of the HIV-1 genome expressing the Tat protein. The unique population of Fig. 7 may therefore have received a recombinant genome coding for both EGFP and a functional Tat protein but unable to express Gag. Taken together, these results indicate that the encapsidation of vector RNA displaces the viral genome and leads to reduced particle infectivity, vector mobilization, and recombination, all of which further reduce the delivery of intact viral genomes to target cells.
Given the high degree of conservation among different HIV-1 strains within the sequences involved in encapsidation, it is expected that the interactions described in this study will apply to most primary isolates of the virus. Therefore, in view of anti-HIV therapy, it is attractive to think that following vector-mediated transduction, HIV-1 itself may deliver antiviral therapeutics to physiologically relevant targets in vivo through vector mobilization. In this case, however, adjustments in the design of the vector may be required if the therapeutics target functions also necessary for vector production. On the other hand, it is difficult to predict whether the inhibitory effect of HIV vectors described here will be similarly significant in vivo, where the transduced cells may be highly diluted.
An important aspect of this study is that no interaction of HIV-1 with a self-inactivating vector was detected even under permissive in vitro conditions. These data provide direct experimental evidence of the increased biosafety achieved by this type of vector (18, 31). It is thus expected that the use of a self-inactivating vector would greatly reduce the risks associated with recombination between vector and viral genomes and allow a more stringent control of transgene expression. Of note, the risks associated with recombination must be considered as a function of both the vector sequence and the transgene delivered. In the case of vectors derived from HIV-1 or HIV-2 (2, 7, 24), the vector sequences are derived from laboratory strains of the viruses and have a limited potential of increasing the diversity or pathogenetic potential of the existing viral clades. A different case may apply to vectors derived from lentiviruses of nonhuman origin (22, 23). In this case, the consequences of their possible recombination with HIV-1 and of the introduction of novel sequences in the viral pool infecting humans should be carefully considered.
Acknowledgments
Anatoly A. Bukovsky and Jin-Ping Song contributed equally to this work.
We are indebted to Didier Trono for suggestions, Tom Dull, Michael Scott, Minh Nguyen, and Michael Kelly for help with some of the experiments, and Heinrich Göttlinger for the HIV-CAT constructs.
REFERENCES
- 1.Anderson W F. Human gene therapy. Nature. 1998;392:25–30. doi: 10.1038/32058. [DOI] [PubMed] [Google Scholar]
- 2.Arya S K, Zamani M, Kundra P. Human immunodeficiency virus type 2 lentivirus vectors for gene transfer: expression and potential for helper virus-free packaging. Hum Gene Ther. 1998;9:1371–1380. doi: 10.1089/hum.1998.9.9-1371. [DOI] [PubMed] [Google Scholar]
- 3.Bartz S R, Rogel M E, Emerman M. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevec D, Volc-Platzer B, Zimmermann K, Dobrovnik M, Hauber J, Veres G, Bohnlein E. Constitutive expression of chimeric neo-Rev response element transcripts suppresses HIV-1 replication in human CD4+ T lymphocytes. Hum Gene Ther. 1994;5:193–201. doi: 10.1089/hum.1994.5.2-193. [DOI] [PubMed] [Google Scholar]
- 5.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbeau P, Wong-Staal F. Anti-HIV effects of HIV vectors. Virology. 1998;243:268–274. doi: 10.1006/viro.1998.9089. [DOI] [PubMed] [Google Scholar]
- 7.Corbeau P, Kraus G, Wong-Staal F. Transduction of human macrophages using a stable HIV-1/HIV-2-derived gene delivery system. Gene Ther. 1998;5:99–104. doi: 10.1038/sj.gt.3300563. [DOI] [PubMed] [Google Scholar]
- 8.Dull T, Zufferey R, Kelly M, Mandel R J, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 10.Helseth E, Kowalski M, Gabuzda D, Olshevsky U, Haseltine W, Sodroski J. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J Virol. 1990;64:2416–2420. doi: 10.1128/jvi.64.5.2416-2420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu W S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 12.Kafri T, Blömer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 13.Kim J H, McLinden R J, Mosca J D, Vahey M T, Greene W C, Redfield R R. Inhibition of HIV replication by sense and antisense rev response elements in HIV-based retroviral vectors. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;12:343–351. doi: 10.1097/00042560-199608010-00003. [DOI] [PubMed] [Google Scholar]
- 14.Kim V N, Mitrophanous K, Kingsman S M, Kingsman A J. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S-W, Gallardo H F, Gilboa E, Smith C. Inhibition of human immunodeficiency virus type 1 in human T cells by a potent Rev response element decoy consisting of the 13-nucleotide minimal Rev-binding domain. J Virol. 1994;68:8254–8264. doi: 10.1128/jvi.68.12.8254-8264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lisziewicz J, Sun D, Smythe J, Lusso P, Lori F, Louie A, Markham P, Rossi J, Reitz M, Gallo R C. Inhibition of human immunodeficiency virus type 1 replication by regulated expression of a polymeric Tat activation response RNA decoy as a strategy for gene therapy in AIDS. Proc Natl Acad Sci USA. 1993;90:8000–8004. doi: 10.1073/pnas.90.17.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luciw P A. Human immunodeficiency viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1881–1975. [Google Scholar]
- 18.Miyoshi H, Blömer U, Takahashi M, Gage F H, Verma I M. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naldini L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr Opin Biotechnol. 1998;9:457–463. doi: 10.1016/s0958-1669(98)80029-3. [DOI] [PubMed] [Google Scholar]
- 20.Naldini L, Blömer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 22.Olsen J C. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 1998;5:1481–1487. doi: 10.1038/sj.gt.3300768. [DOI] [PubMed] [Google Scholar]
- 23.Poeschla E M, Wong-Staal F, Looney D J. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- 24.Poeschla E, Corbeau P, Wong-Staal F. Development of HIV vectors for anti-HIV gene therapy. Proc Natl Acad Sci USA. 1996;93:11395–11399. doi: 10.1073/pnas.93.21.11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reil H, Bukovsky A, Gelderblom H R, Gottlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiser J, Harmison G, Kluepfel-Stahl S, Brady R O, Karlsson S, Schubert M. Transduction of nondividing cells by pseudotyped defective high-titer HIV type 1 particles. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullenger B A, Gallardo H F, Ungers G E, Gilboa E. Analysis of trans-acting response decoy RNA-mediated inhibition of human immunodeficiency virus type 1 transactivation. J Virol. 1991;65:6811–6816. doi: 10.1128/jvi.65.12.6811-6816.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 29.Volsky D J, Simm M, Shahabuddin M, Li G, Chao W, Potash M J. Interference to human immunodeficiency virus type 1 infection in the absence of downmodulation of the principal virus receptor, CD4. J Virol. 1996;70:3823–3833. doi: 10.1128/jvi.70.6.3823-3833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 31.Zufferey R, Dull T, Mandel R J, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]