Abstract
The human T-cell leukemia virus (HTLV) Rex protein is essential for efficient expression of the viral structural and enzymatic gene products. In this study, we assessed the role of the HTLV-2 rex gene in viral RNA expression and Gag protein production. Following transfection of human JM4 T cells with wild-type and rex mutant full-length proviral constructs, PCR was used for semiquantitative analysis of specific viral RNA transcripts. In the presence of Rex, the total amount of steady-state viral RNA was increased fourfold. Rex significantly up-regulated the level of incompletely spliced RNAs by increasing RNA stability and was associated with a twofold down-regulation of the completely spliced tax/rex RNA. PCR analysis of subcellular RNA fractions, isolated from transfected cells, indicated that the level of gag/pol and env cytoplasmic RNAs were increased 7- to 9-fold in the presence of Rex, whereas Gag protein production was increased 130-fold. These data indicate that HTLV-2 Rex increases the stability and promotes nucleus-to-cytoplasm transport of the incompletely spliced viral RNAs, ultimately resulting in increased structural protein production. Moreover, this model system provides a sensitive approach to further characterize HTLV gene expression from full-length proviral clones following transfection of human T cells.
Human T-cell leukemia virus types 1 and 2 (HTLV-1 and HTLV-2) are complex oncogenic retroviruses that transform primary human T cells in culture and are associated with leukemia and neurological disorders in humans (reviewed in reference 19). In addition to the essential gag, pol, and env structural and enzymatic genes expressed by all replication-competent retroviruses, the HTLVs contain at least two additional trans-regulatory genes that regulate expression of all viral genes. The HTLV provirus is expressed as three major RNA species which are derived from the full-length transcript by differential splicing. The completely spliced RNA codes for the regulatory gene products Tax and Rex (25, 30, 34, 36). Tax is an important modulator of both viral and cellular gene expression and is essential for HTLV-mediated transformation of human T lymphocytes in culture (18, 32). Tax localizes to the nucleus of infected cells (17, 38) and acts to increase the rate of transcription initiation by facilitating the binding of the CREB and ATF cellular proteins to the viral promoter (1, 2, 9, 15, 35, 39). The interaction of Tax with cellular proteins results in the activation of NFκB/Rel-, CREB/ATF-, and serum response factor-responsive genes (reviewed in references 12 and 16) and the dysregulation of cell cycle control (28, 31).
The Rex protein is required for the expression of the structural and enzymatic proteins that are translated from the unspliced gag/pol/genome and singly spliced env viral transcripts (23, 27, 29). Rex function is mediated by a cis-acting RNA Rex response element (RxRE) located in the R region of the viral long terminal repeat (5, 7, 40). Specific binding of Rex to the RxRE is correlated with function and is regulated by phosphorylation (6, 22). Previous studies have addressed the mechanism of HTLV-1 Rex (Rex-1) function using stably infected cells or by transfection of indicator plasmids or subgenomic constructs into cells in which amplification of the transfected DNA occurs (23, 24, 26). These approaches, necessitated by difficulties in detecting and quantitating low-abundance HTLV mRNA species, have demonstrated that Rex-1 increases the amounts of unspliced viral RNA by reducing the rates of splicing and degradation in the nucleus and stimulating the nucleocytoplasmic transport of incompletely spliced viral RNA. This study uses the transfection of infectious wild-type and mutant molecular proviral clones of HTLV-2 into JM4 human T cells, subcellular RNA fractionation, and semiquantitative PCR analysis of specific viral RNA species to evaluate the mechanism of HTLV-2 Rex (Rex-2) function. This approach allows us to study the regulation of HTLV gene expression in the context of proviral DNA transfected into T cells, the natural target for HTLV pathogenesis, and provides a basis for comparison of Rex-2, Rex-1, and the analogous human immunodeficiency virus type 1 (HIV-1) Rev function. Our results demonstrate that Rex increases the stability and promotes nucleus-to-cytoplasm transport of the gag/pol and env RNAs.
MATERIALS AND METHODS
Cells and plasmids.
B-cell line 729-6 (hereafter called 729), HTLV-2 chronically infected cell line 729pH6neo (37), and human leukemic T-cell line JM4 (33) were maintained in Iscove’s medium supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM glutamine.
The wild-type and rex mutant proviral plasmid clones of HTLV-2, pH6neo and pH6neoSph, have been described elsewhere (20) and are designated wtHTLV-2 and HTLV-2(rex−), respectively. The rex cDNA expression vector BCRex (20), tax expression vector BC20.2Sph (32), and the control and filler plasmids Sv2neo (21) and BC12 (11) were previously described.
Transfections.
Plasmid DNA was introduced into cells by electroporation as previously described (8). Briefly, cells were washed with phosphate-buffered saline and resuspended (2 × 107 cells/ml) in RPMI 1640 medium supplemented with 20% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM glutamine. A total of 107 cells were electroporated with 35 μg of total DNA (900-μF charge, 250-V potential) which included 5 μg of expression vector pCMVβGal. Cells were transferred to 3 ml of medium, incubated at 37°C, harvested and enumerated 48 to 72 h posttransfection, and subjected to a β-galactosidase (β-Gal) colorimetric assay to normalize for transfection efficiency. Briefly, 106 cells were lysed by sonication in 60 μl of 0.25 mM Tris (pH 7.8) and centrifuged 15 min at 4°C; 30 μl of extract was incubated for 1 to 5 h at room temperature in 1 mM MgCl2–50 mM β-mercaptoethanol–66 mM NaHPO4-Na2PO4–0.9 mg of o-nitrophenyl-β-d-galactopyranoside per ml. The reaction was stopped by addition of Na2CO3, and the absorbance was read at 410 nm. The remainder of the cells were used for Gag protein production analysis, total RNA isolation, or nuclear and cytoplasmic RNA isolation.
Gag protein analysis.
At 96 h posttransfection, culture supernatants were analyzed by a specific enzyme-linked immunosorbent assay (ELISA) using a monoclonal antibody to either p24Gag (Coulter) or p19Gag (Cellular Products) for the presence of structural Gag antigen as described by the manufacturers. To assess cell-associated p24Gag levels in JM4 T cells 48 h posttransfection, cells were metabolically labeled with [35S]methionine-cysteine (Trans35S-label, 100 μCi/ml; ICN Biochemicals, Inc.) in methionine-cysteine-free RPMI 1640 medium supplemented with 10% dialyzed FCS. Cells were lysed in radioimmunoprecipitation assay buffer (0.05 M Tris-HCl [pH 8.0], 0.1% sodium dodecyl sulfate [SDS], 1.0% Triton X-100, 0.15 M NaCl, 2.0 mM phenylmethylsulfonyl fluoride), and lysates were clarified by centrifugation at 100,000 × g (1 h, 4°C). Various amounts of clarified extracts were immunoprecipitated with antisera specific for HTLV-2 p24Gag in the presence of protein A-Sepharose (Pharmacia). Immunoreactive proteins were fractionated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), visualized by autoradiography, and quantified by phosphorimage analysis.
Preparation and analysis of RNA.
Total cellular RNA was extracted from transfected 729, 729pH6neo, or JM4 T cells by the Tri Reagent procedure as described elsewhere (10). A three-step fractionation protocol (14) in conjunction with the Tri Reagent procedure was used to obtain one nuclear and two cytoplasmic RNA fractions. Briefly, cells were initially lysed by a low concentration of NP-40 (0.05%) to fractionate cytoplasmic fraction 1, which contains soluble cytoplasmic components and the bulk of the tRNA. The remaining pellet was treated with a higher concentration of NP-40 (0.65%) to release additional cytoplasmic RNA. This more stringent cytoplasmic fraction 2 contains less soluble cytoplasmic components, including much of the 18S and 28S RNAs and RNAs associated with membrane-bound polysomes. The remaining pellet contains the nuclear fraction. All RNA was treated three times with RNase-free DNase (Boehringer Mannheim), precipitated, and quantified by absorbance at 260 nm.
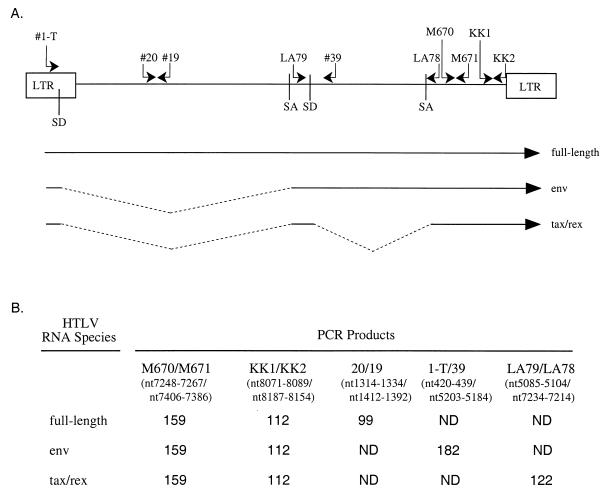
Approximately 200 ng of RNA (equivalent amounts of RNA based on transfection efficiency) was subjected to a coupled primer extension–25-cycle PCR using HTLV-2-specific oligonucleotide primer pairs. The 50-μl volume coupled primer extension-PCR mixture contained RNA, 0.25 mM deoxynucleoside triphosphates, 50 mM KCl, 10 mM Tris (pH 8.0), 1.5 mM MgCl2, 0.01% gelatin, 100 ng of 3′ (antisense) oligonucleotide, 50 ng of 5′ (sense) oligonucleotide end labeled with T4 DNA kinase to a specific activity of approximately 2 × 108 cpm/μg, and 2.5 U of Taq DNA polymerase (Promega) in the presence or absence of 5 U of murine leukemia virus reverse transcriptase (Amersham). The reaction was performed in a Perkin-Elmer model 9600 thermal cycler as follows: 65°C for 10 min, 50°C for 8 min, and 95°C for 5 min, followed by 25 cycles of 95°C for 1 min, 55°C for 2 min, and 72°C for 2 min. PCR-amplified products were separated on a 6% polyacrylamide gel and visualized by autoradiography or phosphorimage analysis (Fuji Imaging Systems). Sequences of the HTLV-2-specific oligonucleotides and proviral nucleotide locations, based on the pH6neo proviral clone (36), are as follows: 1-T, 5′ CTCGGCACCTCCTGAACTGC 3′ (nucleotides [nt] 420 to 439), 20, AGCCCCCAGTTCATGCAGACC 3′ (nt 1314 to 1334), 19, 5′ GAGGGAGGAGCATAGGTACTG 3′ (nt 1412 to 1392), LA79, 5′ CCGGTGGATCCCGTGGCGAT 3′ (nt 5085 to 5104), 39, 5′ AAAAGTAGGAAGAAAACATTA 3′ (nt 5203 to 5184), LA78, 5′ GTCCAAATCCTGGGAAATGGG 3′ (nt 7234 to 7214), M670, 5′ CGGATACCCAGTCTACGTGT 3′ (nt 7248 to 7267), M671, 5′ GAGCTGACAACGCGTCCATCA 3′ (nt 7406 to 7386), KK1, 5′ CCCTCCTATCTACTCTCTC 3′ (nt 8071 to 8089), and KK2, 5′ CGCCTCTTCTTTATTAAATAAAATAGAGACAGGG 3′ (nt 8187 to 8154). The primer pairs (see Fig. 2) were designed to amplify total viral RNA detected by M670-M671 (159 bp) or KK1-KK2 (112 bp), env-specific RNA (182 bp), tax/rex-specific RNA (122 bp), and genome or gag/pol-specific RNA (99 bp). Oligonucleotide primer pairs that detect β-actin unspliced pre-RNA (ActE5 [5′ TGGCACCCAGCACAATGAAG 3′] and ActI5 [5′ TGGATGTGACAGCTCCCCAC 3′]) and GAPDH spliced RNA (gapdh+ [5′ GGTGAAGGTCGGAGTCAACG 3′] and gapdh-[5′ GTTGAGGTCAATGAAGGGGTC 3′]) were used to control for nuclear/cytoplasmic separation and/or RNA loading.
FIG. 2.
Oligonucleotide primer pairs for RT-PCR using HTLV-2 RNAs. (A) Schematic representation of the HTLV genome showing locations and orientations of the oligonucleotides used for PCR. For sequences of the oligonucleotides and proviral nucleotide locations, see Materials and Methods. The major splice donor (SD) and splice acceptor (SA) sites and long terminal repeat (LTR) are shown. The three major species of HTLV RNA are depicted below the schematic diagram. (B) Sizes of the predicted amplified products generated by RT-PCR with pairs of oligonucleotide primers specific for HTLV-2 RNAs. ND, RNA species for which specific products were not detected because of their large theoretical size or absence of complementary binding site in the specific RNA.
To determine the decay rates of HTLV-2 gag/pol and tax/rex RNAs in the presence and absence of Rex, RNA polymerase II-dependent de novo transcription was inhibited by adding actinomycin D (5 μg/ml) to the culture supernatant of JM4 T cells 48 h post-transfection. Total RNA was isolated at different times after the addition of drug and subjected to reverse transcriptase PCR (RT-PCR) analysis. The estimates of transcript stability are based on phosphorimage analysis of the specific PCR-amplified product over the time course. RNA stability analysis was performed multiple times in independent experiments without significant differences.
RESULTS
HTLV-2 Rex is necessary for production of Gag antigen in human B and T cells.
The function of Rex in the expression of Gag proteins was determined by using the previously characterized proviral plasmid clones wtHTLV-2 and HTLV-2(rex−) along with Rex and Tax cDNA expression vectors. To compare Rex function in two human cell lines, plasmids were transfected into 729 B cells or JM4 CD4+ T cells. At 96 h posttransfection, culture supernatants were analyzed by p24Gag or p19Gag ELISA for the presence of structural Gag antigen. The results are summarized in Table 1. wtHTLV-2 gave rise to high levels of Gag antigen in both cell types tested. No viral Gag antigen was detected with vector controls or HTLV-2(rex−). Complementation experiments were performed to determine whether it was possible to rescue Gag protein production from HTLV-2(rex−). Gag proteins were produced when the HTLV-2(rex−) was coelectroporated with a cytomegalovirus promoter-driven rex expression vector. In contrast, coelectroporation of a tax expression vector did not rescue Gag protein production, indicating that the decreased Gag production is not influenced by the transactivator protein Tax. These results indicate that Rex is essential for efficient Gag production in the cell supernatant and that Rex functions in trans to allow Gag expression in human B and T cells.
TABLE 1.
Analysis of Rex function by HTLV Gag protein ELISAa
| Plasmid(s) | Gag (pg/ml)
|
|
|---|---|---|
| 729 B cells | JM4 T cells | |
| wtHTLV-2 | 400 | 550 |
| HTLV-2(rex−) | <15 | <30 |
| HTLV-2(rex−) + rex | 250 | 600 |
| HTLV-2(rex−) + tax | <15 | 45 |
| SV2neo | <15 | <30 |
| Mock | <15 | <30 |
Cells (107) were transfected by electroporation with 40 μg of wtHTLV-2, HTLV-2(rex−) or the vector control (SV2neo) plus 10 μg of the indicated cDNA expression vector expressing rex or tax or filler plasmid (mock). Culture supernatants were assayed at 96 h posttransfection for Gag protein production as suggested by the manufacturer. For 729 B cells, Gag protein was detected by ELISA (p24; Coulter), and values of <15 are below the sensitivity of the assay. For JM4 T cells, Gag protein was detected by ELISA (p19; Cellular Products), and values of <30 are below the sensitivity of the assay.
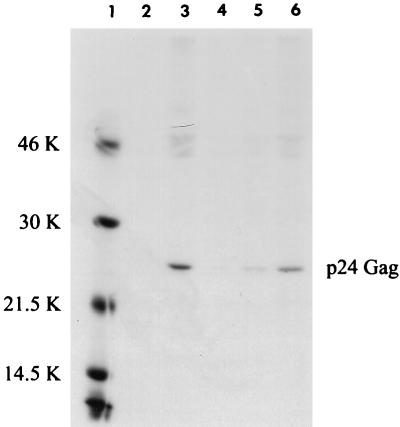
A threshold level of cell-associated Gag (necessary for budding) is likely required for significant levels of Gag to be detected in the culture supernatant. Therefore, we next assessed the amount of cell-associated p24 Gag in JM4 T cells 48 h after transfection of wtHTLV-2 or HTLV-2(rex−). We failed to detect cell-associated Gag protein with the p19Gag ELISA in the absence of Rex (data not shown). However, immunoprecipitation of metabolically labeled cell lysates with a p24Gag-specific antisera allowed detection of a low level of p24Gag in the absence of Rex, but only with 20-fold more lysate than for wtHTLV-2-transfected cells (Fig. 1; compare lanes 3 and 5). Phosphorimage analysis indicated that p24Gag protein production was increased on average 130-fold in the presence of Rex. These results indicate that a low level of Gag is produced in the absence of Rex but efficient Gag expression requires Rex.
FIG. 1.
Quantitation of intracellular radiolabeled p24 Gag in transfected T cells. JM4 human T cells (107) were cotransfected by electroporation with 5 μg of pCMVβGal and 25 μg of wtHTLV-2 proviral clone/5 μg of BC12 (control), 25 μg of HTLV-2(rex−) proviral clone/5 μg of BC12, or 25 μg of HTLV-2(rex−) proviral clone/5 μg of BCRex (rex cDNA expression vector); 48 h posttransfection, 106 cells were subjected to a β-Gal colorimetric assay to normalize for transfection efficiency. Cells were normalized for transfection efficiency and labeled for 3 h with [35S]methionine-cysteine, and cell lysates were made. Various amounts of cell lysate [100 μl of mock, wtHTLV, and HTLV(rex−) + rex (lanes 2, 3, and 6), 200 μl of HTLV(rex−) (lane 4), and 2 ml of HTLV(rex−) (lane 5)] were immunoprecipitated with human HTLV-2-specific antisera that detect primarily p24 Gag and Gag precursors in the presence of protein A-Sepharose. Immunoprecipitated proteins were resolved by SDS-PAGE, visualized by autoradiography, and quantified by phosphorimage analysis. Quantification indicates 130-fold increase in p24Gag in the presence of Rex. The sizes in kilodaltons (K) of markers (lane 1) are indicated on the left.
Detection of HTLV-2 RNA species by quantitative PCR analysis.
To provide a sensitive and quantitative measure of RNA species, we developed a PCR approach to detect viral RNA transcripts in HTLV-2-transfected or -infected cells. Low-level viral gene expression and low transfection efficiency of full-length proviral constructs into human T cells necessitate the use of RT-PCR. We used specific oligonucleotide primer pairs to generate a profile of viral RNA expression. Oligonucleotide primers were designed to detect (i) all transcripts/total viral RNA, (ii) full-length gag/pol/genomic RNA, (iii) singly spliced env RNA, and (iv) doubly spliced tax/rex RNA. The locations of the oligonucleotide pairs in the HTLV-2 genome with respect to the major splice donor and acceptor sites and the predicted sizes of the HTLV-2-specific PCR products generated with each oligonucleotide pair are shown in Fig. 2. Oligonucleotide primer pairs M670-M671 and KK1-KK2 are designed to detect all HTLV-2 RNAs. Oligonucleotide primers 19 and 20 should generate a product specific for full-length gag/pol or genomic RNA. Oligonucleotide primers 1-T and 39 should direct the synthesis of a product corresponding to the singly spliced env RNA, and primer pair LA79-LA78 will amplify a product specific for the doubly spliced tax/rex RNA.
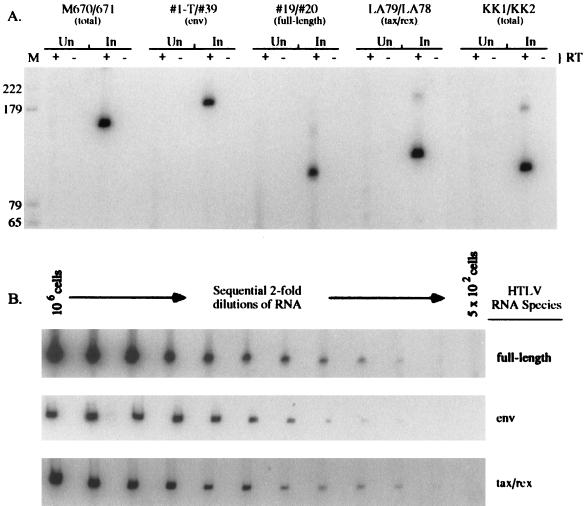
Total RNAs were isolated from uninfected and HTLV-2 chronically infected human lymphocytes. Equivalent amounts of uninfected and HTLV-2 chronically infected cell RNAs were subjected to a reverse transcriptase step coupled to 25 cycles of PCR amplification. Each reaction contained the appropriate primer pair with the oligonucleotide corresponding to the sense strand end labeled with 32P. This allowed direct detection of the amplified products following SDS-PAGE and autoradiography or phosphorimage analysis. A major product of the predicted size was RT-PCR amplified from HTLV-2-infected cell RNA by using each of the HTLV-specific oligonucleotide pairs (Fig. 3A). These products are not detected in uninfected total-cell RNA (Fig. 3A). Oligonucleotide pairs M670-M671, KK1-KK2, and 19-20 can specifically detect HTLV-2 plasmid or proviral DNA. However, reactions in which reverse transcriptase was omitted showed that no amplified product resulted from DNA contaminated RNA.
FIG. 3.
Detection of HTLV-2-specific RNAs by RT-PCR using RNA isolated from chronically infected and uninfected human lymphocytes. RNA was extracted from HTLV-2-infected (In) and uninfected (Un) 729 B cells. (A) RNA from 5 × 105 cells was subjected to coupled primer extension–25-cycle PCR as described in Materials and Methods in the presence (+) or absence (−) of 5 U of murine leukemia virus reverse transcriptase (Amersham). The primer pairs were designed to amplify total viral RNA detected by M670-M671 (159 bp) or KK1-KK2 (112 bp), env-specific RNA (182 bp), tax/rex-specific RNA (122 bp), and genome or gag/pol-specific RNA (99 bp). PCR-amplified products were separated on a 6% polyacrylamide gel and visualized by autoradiography or phosphorimage analysis (Fuji Imaging Systems). M, size markers (positions are indicated in base pairs). (B) Quantitation of RT-PCR data. HTLV-2-infected-cell RNA (106 to 500 cell equivalents) was subjected to RT-PCR and analyzed as described for panel A.
To determine whether HTLV-2-specific RNAs could be detected quantitatively, total cellular RNA was isolated from the HTLV-2 chronically infected cell line 729pH6neo. Sequential twofold dilutions of total RNA (from 106 to 500 cell equivalents) were subjected to 25-cycle RT-PCR analyses (Fig. 3B). The signals produced were quantitative across a wide range of RNA concentrations (2,000-fold) with all oligonucleotide pairs tested. In all cases, increasing signal intensities were detected from increasing amounts of RNA. However, at high concentrations of target RNA, the increase in signal was not linear and reached a plateau likely resulting from nucleotide or oligonucleotide concentration limitations. Therefore, this assay specifically detects HTLV-2 RNA from as few as 1,000 to 2,000 cells and will allow detection of low-level RNA species expressed following the introduction of proviral constructs into cells by transfection.
Effect of Rex on steady-state levels of gag/pol, env, and tax/rex RNAs.
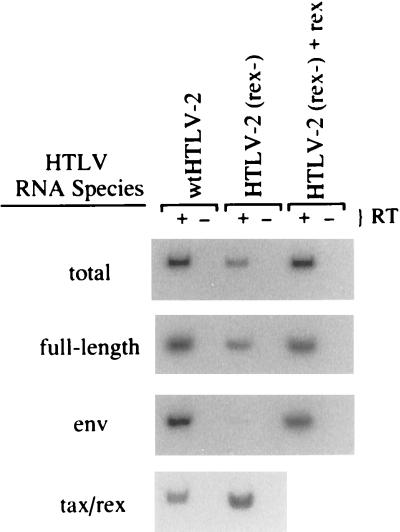
To examine the block to Gag production exhibited by HTLV-2(rex−) the expression of HTLV-specific RNAs in electroporated JM4 human T cells was analyzed by RT-PCR (Fig. 4). RNA, normalized for transfection efficiency, was subjected to RT-PCR using the panel of HTLV-2-specific oligonucleotide primer pairs. The steady-state RNA profile of the rex mutant clone was distinct from that of the wild-type clone. The rex mutant produced reduced levels of the full-length gag/pol and singly spliced env RNAs and a slight increase in the completely spliced tax/rex RNA relative to wild-type RNA (Fig. 4). The magnitude of the differences was determined by phosphorimage analyses of the PCR signals (Table 2). The level of unspliced and singly spliced transcripts (gag/pol and env, respectively) was 2.5- to 20-fold lower in the mutant, whereas the level of tax/rex RNA was reproducibly 2-fold higher than in the wild-type construct. Coelectroporation of a rex expression vector with the rex mutant proviral clone resulted in the restoration of wild-type RNA levels, indicating that Rex functions in trans (Fig. 4 and Table 2). In addition, the overall amount of HTLV-2 RNA was increased fourfold in the presence of Rex. Given that Rex functions posttranscriptionally, this result suggests that Rex has a positive effect on RNA stability.
FIG. 4.
Effect of Rex on HTLV-2 RNA accumulation. JM4 human T cells (107) were cotransfected by electroporation with 5 μg of pCMVβGal and 25 μg of wtHTLV-2 proviral clone/5 μg of BC12 (control), 25 μg of HTLV-2(rex−) proviral clone/5 μg of BC12, or 25 μg of HTLV-2(rex−) proviral clone/5 μg of BCRex (rex cDNA expression vector); 48 h posttransfection, 106 cells were subjected to a β-Gal colorimetric assay to normalize for transfection efficiency. Total cellular RNA was isolated from the remainder of the cells. Approximately 200 ng of RNA (equivalent amounts of RNA based on transfection efficiency) was subjected to a coupled primer extension–25-cycle PCR in the presence (+) or absence (−) of reverse transcriptase as described in Materials and Methods. The primer pairs were designated to amplify total viral mRNA, full-length gag/pol/genome RNA, env RNA, and tax/rex RNA. PCR products were separated on a 6% polyacrylamide gel and visualized by autoradiography. RNA was quantified by phosphorimage analysis and presented as experiment 1 in Table 2.
TABLE 2.
Quantitation of HTLV-2 RNA accumulation in transfected T cells
| HTLV RNA species | Relative unitsa
|
|||
|---|---|---|---|---|
| HTLV(rex−)
|
HTLV(rex−) + rex
|
|||
| Expt 1b | Expt 2c | Expt 1 | Expt 2 | |
| Total | 0.24 | 0.24 | 0.9 | 1.1 |
| Full length | 0.41 | 0.54 | 0.83 | 0.93 |
| env | 0.05 | 0.09 | 0.82 | 0.94 |
| tax/rex | 1.83 | 2.58 | NDd | ND |
Specific viral RNA levels were determined by phosphorimage analysis using a Fuji imaging system, and values in each row of RNA species are normalized to the value for wtHTLV (set at 1).
RNA levels in Fig. 3.
Data from an independent transfection, RT-PCR, and phosphorimage analysis.
ND, not determined since RT-PCR could not distinguish between tax/rex transcripts expressed from the transfected proviral construct and the rex cDNA expression vector.
Rex affects viral RNA stability.
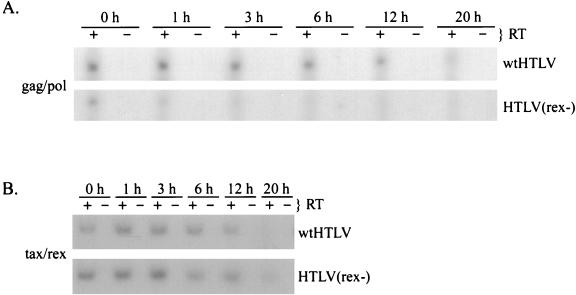
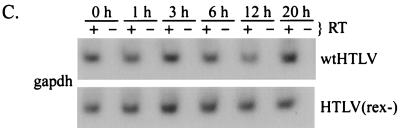
We next determined the stability of viral RNAs in the presence and absence of Rex expression by inhibiting RNA polymerase II-dependent de novo transcription by treatment with actinomycin D. JM4 T cells transfected with proviral clones were treated with actinomycin D beginning 48 h posttransfection. At different times after drug addition, total RNA was isolated and subjected to RT-PCR. RNA stability was determined by phosphorimage analysis of the RT-PCR-amplified product over time. The half-life of the completely spliced tax/rex RNA was approximately 10 h and was not affected by the presence of Rex (Fig. 5B). In contrast, the half-life of the unspliced gag/pol RNA was 10 h in the presence of Rex and approximately 1 h in the absence of Rex (Fig. 5A). These results clearly demonstrate that Rex stabilizes unspliced HTLV-2 transcripts in transfected T cells.
FIG. 5.
Rex increases the stability of unspliced gag/pol RNA in transfected JM4 T cells. JM4 human T cells (107) were transfected by electroporation with 25 μg of wtHTLV-2 or 25 μg of HTLV-2(rex−); 48 h posttransfection, cells were divided equally into a six-well tissue culture plate. Total cellular RNA was extracted at various times (0, 1, 3, 6, 12, and 20 h) following incubation with actinomycin D (5 μg/ml). Approximately 200 ng of RNA from each time point was subjected to a coupled primer extension–25-cycle PCR in the presence (+) or absence (−) of reverse transcriptase as described in Materials and Methods. The primer pairs were designated to amplify full-length gag/pol/genome RNA (A), tax/rex RNA (B), and control gapdh RNA (C). PCR products were separated on a 6% polyacrylamide gel and visualized by autoradiography. RNA was quantified by phosphorimage analysis.
Rex affects cellular distribution of HTLV RNAs.
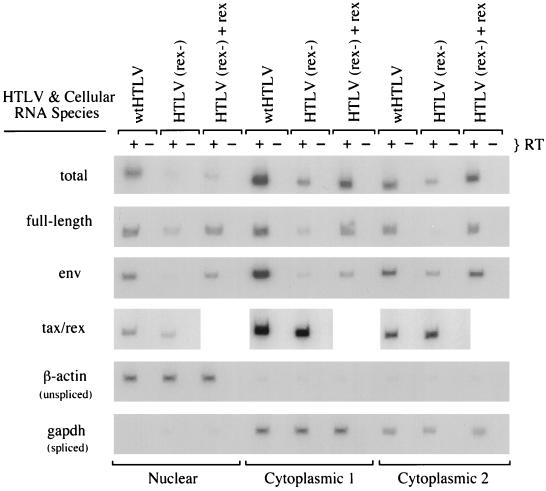
To determine whether Rex affects the subcellular distribution of HTLV-2 RNAs, electroporated JM4 T cells were fractionated into one nuclear and two cytoplasmic RNA fractions by a three-step fractionation protocol (14) as described in Materials and Methods. It is important to note that the majority of RNA in the cell is found in cytoplasmic fraction 2. RNAs prepared from these fractions were normalized for transfection efficiency and subjected to RT-PCR. To control for the fractionation of authentic nuclear and cytoplasmic RNA, PCR products were generated from unspliced β-actin pre-mRNA and spliced GAPDH RNA by complementary human oligonucleotide pairs. From the RT-PCR analysis (Fig. 6 and Table 3), it is clear that Rex increases the overall amount of HTLV-2 RNA in the three fractions three- to fivefold, consistent with a positive effect of Rex on RNA stability. In cells transfected with the rex proviral mutant, the full-length RNA (unspliced, gag/pol) was detectable at low levels in the nuclear fraction and cytoplasmic fraction 1 but not in cytoplasmic fraction 2. β-Actin and GAPDH RNAs were detected in the nuclear and cytoplasmic compartments as expected (Fig. 6 and Table 3). Expression of the full-length gag/pol RNA in the cytoplasmic fractions was restored by cotransfection of a rex cDNA expression vector along with the mutant rex proviral construct. env RNA was not detectable in the nucleus from the rex mutant provirus but was detected in both cytoplasmic fractions, indicating that once splicing occurs, transport to the cytoplasm is efficient. As was the case with full-length viral RNA, expression of env RNA was rescued in all fractions by cotransfection of a rex cDNA expression vector. The level of tax/rex RNA was only slightly varied in the cytoplasmic fractions in the absence of rex, with the major difference (1.4-fold increase) seen in cytoplasmic fraction 2. By phosphorimage analysis, very low levels of tax/rex RNA could be detected in the nuclear fraction; however, the majority of tax/rex RNA detected in cells transfected with both rex mutant and wild-type proviral constructs was found in the cytoplasmic fractions as expected.
FIG. 6.
Effect of Rex on nuclear and cytoplasmic RNA distribution. JM4 human T cells (107) were cotransfected with 5 μg of pCMVβGal and 25 μg of wtHTLV-2 proviral clone/5 μg of BC12 (control), 25 μg of HTLV-2(rex−) proviral clone/5 μg of BC12, or 25 μg of HTLV-2(rex−) proviral clone/5 μg of BCRex (rex cDNA expression vector); 48 h posttransfection, 106 cells were subjected to a β-Gal assay. One nuclear and two cytoplasmic RNA fractions were extracted from the remainder of the cells. Approximately 200 ng of RNA (equivalent amounts of RNA based on transfection efficiency) was subjected to a coupled primer extension–25-cycle PCR with oligonucleotide primer pairs to detect the indicated RNA species. Oligonucleotide primer pairs that detect β-actin unspliced pre-RNA and GAPDH spliced RNA were used to control for nuclear/cytoplasmic separation. PCR products were separated on a 6% polyacrylamide gel and visualized by autoradiography. RNA was quantified by phosphorimage analysis, and the data are presented in Table 3.
TABLE 3.
Quantitation of nuclear and cytoplasmic HTLV RNAs in transfected T Cells
| HTLV or cellular RNA species | Relative unitsa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nuclear
|
Cytoplasmic fraction 1
|
Cytoplasmic fraction 2
|
|||||||
| wt | (rex−) | (rex−) + rex | wt | (rex−) | (rex−) + rex | wt | (rex−) | (rex−) + rex | |
| Total | 1 | 0.2 | 0.3 | 2.0 | 0.5 | 1.3 | 0.86 | 0.3 | 0.94 |
| Full length | 1 | 0.34 | 0.83 | 1.55 | 0.27 | 0.96 | 0.93 | 0.09 | 0.94 |
| env | 1 | 0.08 | 0.64 | 3.9 | 0.12 | 0.46 | 1.3 | 0.44 | 1.4 |
| tax/rex | 1 | 0.6 | NDb | 3.9 | 3.2 | ND | 1.7 | 2.3 | ND |
| β-actin (unspliced) | 1 | 1.1 | 1.1 | 0.05 | 0.05 | 0.05 | 0.04 | 0.05 | 0.05 |
| GAPDH (spliced) | 0.08 | 0.09 | 0.08 | 1 | 1 | 0.9 | 0.66 | 0.56 | 0.67 |
RNA levels of subcellular fractions of JM4 T cells transfected with wtHTLV (wt) HTLV(rex−) [(rex−)], or HTLV(rex−) + BCRex [(rex−) + rex], presented in Fig. 4, were quantitated by phosphorimage analysis using Fuji imaging system. With the exception of GAPDH, values are normalized to the value for the wtHTLV nuclear fraction signal (set at 1). The GAPDH signals are normalized to the value for the wtHTLV cytoplasmic fraction 1 signal (set at 1).
ND, not determined since RT-PCR could not distinguish between tax/rex transcripts expressed from the transfected proviral construct and the rex cDNA expression vector.
DISCUSSION
In this study, we used transient transfection, in conjunction with a sensitive and semiquantitative RT-PCR assay, to examine Rex function in its natural context. This approach allowed the detection and quantitation of specific low-abundance viral RNAs expressed from HTLV-2 proviral clones transfected into human T cells, thus providing an opportunity to investigate the effects of Rex on viral RNA expression, stability, and cellular distribution prior to tissue culture and cellular selection processes. Our results demonstrate that Rex-2 increases the stability of the unspliced gag/pol RNA and promotes the nuclear-cytoplasmic transport of the incompletely spliced RNAs, ultimately resulting in efficient structural and enzymatic protein expression and virion production.
We show that Rex increases the full-length unspliced viral RNA in the cytoplasm but has a less dramatic effect on the singly spliced env RNA. Thus, Rex functions to promote nuclear-cytoplasmic transport of incompletely spliced RNAs. Although not dramatic, this increase in the incompletely spliced RNAs is associated with a slight reduction in completely spliced tax/rex RNA. This is clearly apparent by comparison of the tax/rex RNA signals in total cellular RNA from T cells transfected with wild-type and rex mutant proviral clones (Fig. 4). Although less apparent, this reduction is also observed in cytoplasmic fraction 2 (Fig. 6), where the bulk of total cellular RNA fractionates. One possibility consistent with this result is that Rex inhibits RNA splicing. One report has provided evidence that HTLV-2 Rex inhibits pre-mRNA splicing in vitro (4). However, another likely possibility is that Rex increases RNA stability, resulting in a redirection of the incompletely spliced RNA pools. Indeed, analysis of RNA half-lives indicated that gag/pol/genome transcripts are unstable in the absence of Rex (approximately 1 h). In contrast, these RNAs had a half-life of approximately 10 h in the presence of Rex, whereas Rex had no effect on the half-life of the completely spliced RNAs (Fig. 5). A similar effect on the stability of viral RNA was exerted by Rex-1 in T-cell lines stably transformed by recombinant rhadinoviruses expressing Tax and/or Rex (23). Therefore, not only can we conclude that Rex-2 has a function similar to that of Rex-1, but more importantly the findings obtained with our approach help validate results of previous studies using reporter constructs and stably transfected or infected and transformed T-cell lines.
We show that Rex increases the incompletely spliced RNAs in the cytoplasm 7- to 9-fold (Fig. 6 and Table 3), while Gag protein production increases 130-fold (Fig. 2). It has been reported that HIV-1 Rev significantly (800-fold) increases the utilization or translation efficiency of the incompletely spliced RNA (3, 13). Our results may suggest that Rex has an effect on translation efficiency, but if so, it appears to be less than that reported for Rev and HIV-1. Further studies will be required to determine the effect of Rex on translation efficiency. We feel that this sensitive experimental approach should facilitate Rex structure-function analysis in the context of a full-length proviral clone, which will likely be required to more precisely determine the mechanism of Rex action.
ACKNOWLEDGMENTS
We thank Zhi-Yu Fang for technical assistance and Kathleen Boris-Lawrie and Michael Lairmore for critical comments.
This work was supported by grants from the National Institutes of Health (CA59581 and P30CA16058) and Leukemia Society of America (1030-94). P.L.G. is a scholar of the Leukemia Society of America.
REFERENCES
- 1.Adya N, Zhao L-J, Huang W, Boros I, Giam C-Z. Expansion of CREB’s DNA recognition specificity by Tax results from interaction with Ala-Ala-Arg at positions 282-284 near the conserved DNA-binding domain of CREB. Proc Natl Acad Sci USA. 1994;91:5642–5646. doi: 10.1073/pnas.91.12.5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson M G, Dynan W S. Quantitative studies of the effect of HTLV-1 Tax protein on CREB protein-DNA binding. Nucleic Acids Res. 1994;22:3194–3201. doi: 10.1093/nar/22.15.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo S J, Chen I S Y. Rev is necessary for the translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991;5:808–818. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 4.Bakker A L X, Ruland C T, Stephens D W, Black A C, Rosenblatt J D. Human T-cell leukemia virus type 2 Rex inhibits pre-mRNA splicing in vitro at an early stage of spliceosome formation. J Virol. 1996;70:5511–5518. doi: 10.1128/jvi.70.8.5511-5518.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballaun C, Farrington G R, Dobrovnik M, Rusche J, Hauber J, Bohnlein E. Functional analysis of human T-cell leukemia virus type I Rex-response element: direct RNA binding of Rex protein correlates with in vivo binding activity. J Virol. 1991;65:4408–4413. doi: 10.1128/jvi.65.8.4408-4413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black A C, Ruland C T, Yip M T, Luo J, Tran B, Kalsi A, Quan E, Chen I S Y, Rosenblatt J D. Human T-cell leukemia virus type II Rex binding and activity requires an intact splice donor site and a specific RNA secondary structure. J Virol. 1991;65:6545–6653. doi: 10.1128/jvi.65.12.6645-6653.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd H P, Huckaby G L, Ahmed Y F, Hanly S M, Greene W C. The type 1 human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements. Proc Natl Acad Sci USA. 1991;88:5704–5708. doi: 10.1073/pnas.88.13.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cann A J, Koyanagi Y, Chen I S Y. High efficiency transfection of primary human lymphocytes and studies of gene expression. Oncogene. 1988;3:123–128. [Google Scholar]
- 9.Cann A J, Rosenblatt J D, Wachsman W, Shah N P, Chen I S Y. Identification of the gene responsible for human T-cell leukemia virus transcriptional regulation. Nature. 1985;318:571–574. doi: 10.1038/318571a0. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P. A reagent for the single step simultaneous isolation of RNA, DNA, and proteins from cell and tissue samples. BioTechniques. 1993;15:532–537. [PubMed] [Google Scholar]
- 11.Cullen B. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 12.Cullen B R. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Agostino D, Felber B, Harrison J, Pavlakis G. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favaro J P, Arrigo S J. Characterization of Rev function using subgenomic and genomic constructs in T and COS cells. Virology. 1997;228:29–38. doi: 10.1006/viro.1996.8374. [DOI] [PubMed] [Google Scholar]
- 15.Felber B K, Paskalis H, Kleinman-Ewing C, Wong-Staal F, Pavlakis G N. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science. 1985;229:675–679. doi: 10.1126/science.2992082. [DOI] [PubMed] [Google Scholar]
- 16.Franklin A A, Nyborg J K. Mechanisms of Tax regulation of human T-cell leukemia virus type I gene expression. J Biomed Sci. 1995;2:17–29. doi: 10.1007/BF02257921. [DOI] [PubMed] [Google Scholar]
- 17.Goh W C, Sodroski J, Rosen C, Essex M, Haseltine W A. Subcellular localization of the product of the long open reading frame of human T-cell leukemia virus type I. Science. 1985;227:1227–1228. doi: 10.1126/science.2983419. [DOI] [PubMed] [Google Scholar]
- 18.Grassmann R, Berchtolds S, Radant I, Alt M, Fleckenstein B, Sodroski J G, Haseltine W A, Ramstedt U. Role of the human T-cell leukemia virus type 1 X region proteins in immortalization of primary human lymphocytes in culture. J Virol. 1992;66:4570–4575. doi: 10.1128/jvi.66.7.4570-4575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green P L, Chen I S Y. Molecular features of the human T-cell leukemia virus: mechanisms of transformation and leukemogenicity. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 227–311. [Google Scholar]
- 20.Green P L, Ross T M, Chen I S Y, Pettiford S. Human T-cell leukemia virus type II nucleotide sequences between env and the last exon of tax/rex are not required for viral replication or cellular transformation. J Virol. 1995;69:387–394. doi: 10.1128/jvi.69.1.387-394.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green P L, Xie Y, Chen I S Y. The internal methionine codons of the human T-cell leukemia virus type-II rex gene are not required for p24Rex production or virus replication and transformation. J Virol. 1990;64:4914–4921. doi: 10.1128/jvi.64.10.4914-4921.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green P L, Yip M T, Xie Y, Chen I S Y. Phosphorylation regulates RNA binding by the human T-cell leukemia virus Rex protein. J Virol. 1992;66:4325–4330. doi: 10.1128/jvi.66.7.4325-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gröne M, Koch C, Grassmann R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology. 1996;218:316–325. doi: 10.1006/viro.1996.0200. [DOI] [PubMed] [Google Scholar]
- 24.Hanly S M, Rimsky L T, Malim M H, Kim J H, Hauber J, Duc Dodon M, Le S-Y, Maizel J V, Cullen B R, Greene W C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989;3:1534–1544. doi: 10.1101/gad.3.10.1534. [DOI] [PubMed] [Google Scholar]
- 25.Haseltine W A, Sodroski J, Patarca R, Briggs D, Perkins D, Wong-Staal F. Structure of 3′ terminal region of type II human T lymphotropic virus: evidence of new coding region. Science. 1984;225:419–421. doi: 10.1126/science.6330894. [DOI] [PubMed] [Google Scholar]
- 26.Inoue J, Itoh M, Akizawa T, Toyoshima H, Yoshida M. HTLV-1 Rex protein accumulates unspliced RNA in the nucleus as well as in cytoplasm. Oncogene. 1991;6:1753–1757. [PubMed] [Google Scholar]
- 27.Inoue J I, Yoshida M, Seiki M. Transcriptional (p40x) and post-transcriptional (p27xIII) regulators are required for the expression and replication of human T-cell leukemia virus type I genes. Proc Natl Acad Sci USA. 1987;84:3653–3657. doi: 10.1073/pnas.84.11.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin D Y, Spencer F, Jeang K T. Human T-cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:1–20. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 29.Kiyokawa T, Seiki M, Iwashita S, Imagawa K, Shimizu F, Yoshida M. p27xIII and p21xIII proteins encoded by the pX sequence of human T-cell leukemia virus type I. Proc Natl Acad Sci USA. 1985;82:8359–8363. doi: 10.1073/pnas.82.24.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee T H, Coligan J E, Sodroski J G, Haseltine W A, Salahuddin S Z, Wong-Staal F, Gallo R C, Essex M. Antigens encoded by the 3′-terminal region of human T-cell leukemia virus: evidence for a functional gene. Science. 1984;226:57–61. doi: 10.1126/science.6089350. [DOI] [PubMed] [Google Scholar]
- 31.Low K G, Dorner L F, Fernando D B, Grossman J, Jeang K T, Comb M J. Human T-cell leukemia virus type I Tax releases cell cycle arrest induced by p16INK4a. J Virol. 1997;71:1956–1962. doi: 10.1128/jvi.71.3.1956-1962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross T M, Pettiford S M, Green P L. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J Virol. 1996;70:5194–5202. doi: 10.1128/jvi.70.8.5194-5202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider U, Schwenk H, Bornkamm G. Characterization of EBV genome-negative “null” and T cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkins lymphoma. Int J Cancer. 1977;19:621–626. doi: 10.1002/ijc.2910190505. [DOI] [PubMed] [Google Scholar]
- 34.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seiki M, Inoue J, Takeda T, Yoshida M. Direct evidence that p40xI of human T-cell leukemia virus type I is a trans-acting transcriptional activator. EMBO J. 1986;5:561–565. doi: 10.1002/j.1460-2075.1986.tb04247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimotohno K, Takahashi Y, Shimizu N, Goiobori T, Chen I S Y, Golde D W, Miwa M, Sugimura T. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type I and type II long terminal repeats for trans-activation of transcription. Proc Natl Acad Sci USA. 1985;82:3101–3105. doi: 10.1073/pnas.82.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimotohno K, Wachsman W, Takahashi Y, Golde D W, Miwa M, Sugimura T, Chen I S Y. Nucleotide sequence of the 3′ region of an infectious human T-cell leukemia virus type II genome. Proc Natl Acad Sci USA. 1984;81:6657–6661. doi: 10.1073/pnas.81.21.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slamon D J, Boyle W J, Keith D E, Press M F, Golde D W, Souza L M. Subnuclear localization of the trans-acting protein of human T-cell leukemia virus type I. J Virol. 1988;62:680–686. doi: 10.1128/jvi.62.3.680-686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodroski G J, Rosen C A, Haseltine W A. Trans-acting transcriptional activation of the long terminal repeat of human T-lymphotropic viruses in infected cells. Science. 1984;225:381–385. doi: 10.1126/science.6330891. [DOI] [PubMed] [Google Scholar]
- 40.Yip M T, Dynan W S, Green P L, Black A C, Arrigo S J, Torbati A, Heaphy S, Ruland C, Rosenblatt J D, Chen I S Y. Human T-cell leukemia virus (HTLV) type II Rex protein binds specifically to RNA sequences of the HTLV long terminal repeat but poorly to the human immunodeficiency virus type 1 Rev-responsive element. J Virol. 1991;65:2261–2272. doi: 10.1128/jvi.65.5.2261-2272.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]