Abstract
Infection with a replication-competent bovine leukemia virus structural gene vector (BLV SGV) is an innovative vaccination approach to prevent disease by complex retroviruses. Previously we developed BLV SGV that constitutively expresses BLV gag, pol, and env and related cis-acting sequences but lacks tax, rex, RIII, and GIV and most of the BLV long terminal repeat sequences, including the cis-acting Tax and Rex response elements. The novel SGV virus is replication competent and replicates a selectable vector to a titer similar to that of the parental BLV in cell culture. The overall goal of this study was to test the hypothesis that infection with BLV SGV is nonpathogenic in rabbits. BLV infection of rabbits by inoculation of cell-free BLV or cell-associated BLV typically causes an immunodeficiency-like syndrome and death by 1 year postinfection. We sought to evaluate whether in vivo transfection of BLV provirus recapitulates pathogenic BLV infection and to compare BLV and BLV SGV with respect to infection, immunogenicity, and clinical outcome. Three groups of rabbits were subjected to in vivo transfection with BLV, BLV SGV, or negative control DNA. The results of our 20-month study indicate that in vivo transfection of rabbits with BLV recapitulates the fatal BLV infection produced by cell-free or cell-associated BLV. The BLV-infected rabbits exhibited sudden onset of clinical decline and immunodeficiency-like symptoms that culminated in death. BLV and BLV SGV infected peripheral blood mononuclear cells and induced similar levels of seroconversion to BLV structural proteins. However, BLV SGV exhibited a reduced proviral load and did not trigger the immunodeficiency-like syndrome. These results are consistent with the hypothesis that BLV SGV is infectious and immunogenic and lacks BLV pathogenicity in rabbits, and they support the use of this modified proviral vector delivery system for vaccines against complex retroviruses like BLV.
The pathogenesis of complex retroviruses offers a distinct cancer paradigm that does not involve a cell-derived proto-oncogene. Instead, progression to neoplasia by bovine leukemia virus (BLV) and the related human T-cell leukemia viruses (HTLVs) is associated with long-term infection and indirect effects of virus-encoded oncoproteins on cell growth control (25, 34). BLV and HTLV encode the three classical retrovirus structural and enzymatic genes (gag, pol, and env), plus regulatory and accessory genes. The regulatory gene tax encodes a transcriptional trans activator that functions through the Tax responsive element (TRE) and transforms lymphocytes (1, 14, 17, 18, 32, 35, 38, 39). The second regulatory gene, rex, is a posttranscriptional trans activator that functions through the Rex responsive element (RxRE) that is positioned in long terminal repeat (LTR) RNA (14, 19, 21, 44). Although a specific role for Rex and RxRE in transformation is not documented, asymptomatic human immunodeficiency virus (HIV)-infected patients exhibit a correlation between viral latency and subthreshold availability of the functionally analogous trans activator, Rev (23). Furthermore, Rev-independent clones of HIV are attenuated for CD4 depletion in SCID-hu mice (37). While Tax and Rex are essential for infectivity in vivo and in vitro, BLV and HTLV accessory proteins (RIII, GIV/p12, p13, and p30) are dispensable in vitro and influence maintenance of virus load in vivo and, in the case of BLV, pathogenesis (2, 10, 12, 13, 15, 20, 26, 41, 42). BLV GIV acts as a cooperative immortalizing oncogene with Ras in rat embryo fibroblasts, and GIV mutant BLV proviruses exhibited reduced proviral load (by a factor ranging between 5 to over 125) and a lack of pathogenicity in sheep during a 40-month study (26).
A genetically simplified BLV derivative that constitutively expresses the BLV gag, pol, and env independently of tax, rex, RIII, and GIV and is replication competent has been proposed as a novel preventive vaccine against BLV disease (6, 7, 36). Previously we developed a hybrid spleen necrosis virus (SNV)-BLV structural gene vector (SGV) that is replication competent independently of tax, rex, RIII, and GIV (7). The requirement for Tax and TRE was relieved by substitution of the BLV promoter/enhancer sequences with analogous sequences in the LTRs of SNV (6). SNV is a genetically simple retrovirus that constitutively expresses the viral genes without trans-acting viral regulatory protein. To relieve the requirement for Rex, the vector lacks RxRE and the major BLV splice sites and instead contains an internal ribosome entry signal to facilitate translation of BLV env from polycistronic viral RNA (6, 7). In other studies, we have identified a unique element in the SNV 5′ LTR that facilitates HIV Rev/RRE-independent expression of HIV gag (9), and the possibility remains that the SNV 5′ LTR similarly facilitates the BLV Rex/RxRE-independent phenotype of BLV SGV. Experiments in a tissue culture system showed that BLV SGV is replication competent and replicates a selectable vector to a titer similar to that observed for BLV (7). Subsequent analysis in rats indicated that, again similar to BLV, BLV SGV infects peripheral blood mononuclear cells (PBMCs), induces a sustained BLV-specific antibody response, and lacks a disease endpoint in chronically infected rats (6-month study period) (7). The overall goal of this study was to test the hypothesis that BLV SGV lacks pathogenicity in rabbits, which exhibit a BLV disease endpoint.
Experimentally infected rabbits and sheep seroconvert to BLV shortly after inoculation, and antiviral antibody persists for life (4, 8, 28, 33, 43). BLV infection is characterized by cell-associated viremia, though virus is readily detectable upon culture of lymphocytes (32). In a study by Altaner et al., 21 of 23 newborn rabbits inoculated with cell-associated BLV became persistently infected and developed an immunodeficiency-like syndrome that culminated in death (4). Survival times ranged between 45 and 763 days, and the majority of rabbits died within 12 months. Inoculation of young rabbits with cell-associated BLV (four of four) or cell-free BLV (two of two) caused persistent infection and development of respiratory disease and severe weight loss within 18 months of inoculation (43). Experimental inoculation of sheep more closely follows the progression to lymphosarcoma that is observed in cattle, the natural host. However, sheep are more difficult to maintain in laboratory facilities, and time to disease onset can be as long as 7 years (16, 26, 28). Therefore, rabbits are more convenient than sheep for evaluation of the pathogenicity of BLV SGV. The limitation of the rabbit system is the differential outcome of BLV infection, which instead of neoplasia is an immunodeficiency-like syndrome.
The first objective of this study was to evaluate whether in vivo transfection of rabbits with BLV proviral DNA causes disease comparable to cell-associated and cell-free BLV infection. Previous studies have shown that in vivo transfection is an effective approach to BLV infection of rat and sheep (7, 40, 41). The second objective was to compare infection and immunogenicity of BLV and BLV SGV provirus. The third objective was to compare the disease induction capacity of BLV and BLV SGV in order to test the hypothesis that BLV SGV, which lacks regulatory and accessory genes, lacks BLV pathogenicity. Our results indicate that in vivo transfection of rabbits with BLV proviral DNA produces pathogenic BLV infection. BLV SGV also produced chronic infection but failed to cause clinically measurable disease. Importantly, this unique retroviral vector system induced sustained immune response to structural gene products of the virus. Our data support the use of this modified proviral vector delivery system for vaccines against complex retroviruses like BLV.
MATERIALS AND METHODS
In vivo transfection and animal care.
Six-week-old outbred grey Chinchilla rabbits were inoculated at 10-day intervals with three 50-μg doses of DNA by intradermal injection at five sites between the mid-thoracic and inguinal regions of the dorsal side. Three animals received BLV provirus (pBL913) (Fig. 1A) (13), three animals received BLV SGV (pU5gag-pol-env) (Fig. 1B) (7), and two animals received mock DNA (pUC19). The rabbits were maintained in the approved animal care laboratory of the Cancer Research Institute, Slovak Academy of Sciences, Bratislava, Slovakia, and were housed in cages that provided unlimited access to food and water. None of the rabbits exhibited clinical signs of infection with two common bacterial pathogens, Pasturella multocida and Bordetella bronchiseptica; however, they were not tested for infection by these agents.
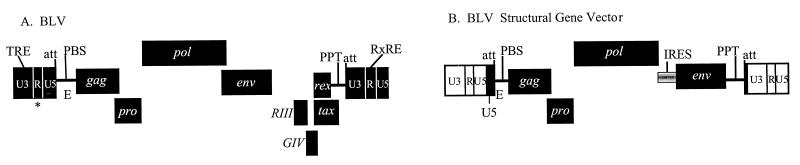
FIG. 1.
Genomic structures of BLV and BLV SGV with cis-acting replication sequences. (A) BLV genome. Labeled terminal black boxes, BLV LTRs with U3, R, and U5 regions separated by white lines; labeled rectangles, open reading frames; asterisk, major BLV splice donor. cis-acting replication sequences: TRE, Tax-responsive element; att, provirus integration sequence; E, viral RNA encapsidation signal. Reverse transcription sequences: PBS, primer binding site; PPT, polypurine tract. RxRE, Rex-responsive element. (B) BLV SGV (pU5gag-pol-env) (7). Labeled terminal white boxes, SNV LTRs with U3, R, and U5 regions separated by black lines; labeled black vertical lines, BLV U5 LTR region, which facilitates vector titer (6) (U5), and provirus integration signal (att). The internal ribosome entry sequence (IRES) promotes cap-independent translation of Env.
PCR analysis of PBMC DNA.
To prepare PBMCs samples for PCR, PBMCs were isolated by Ficoll-Paque (Pharmacia) gradient centrifugation and washed three times with phosphate-buffered saline. PBMCs (107) were lysed in 1 ml of PCR buffer (50 mM KCl, 10 mM Tris [pH 8.3], 2.5 mM MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20) containing 200 μg of proteinase K (Calbiochem-Behring, La Jolla, Calif.) per ml. The lysates were incubated at 55°C overnight and then at 90°C for 10 min. Aliquots of 5 to 20 μl of lysate (equivalent to 5 × 104 to 20 × 104 lysed PBMCs) were mixed in a 50-μl PCR mixture, incubated for 1 min at 94°C, 1 min at 62°C, and 1 min at 72°C for 35 cycles, and analyzed on agarose gels. Primary BLV pol primers KB2341 (GAA CGC CTC CAG GCC CTT CAA) and KB3175 (GTG GGA CAG GGC TTG TCG AAG) amplify an 854-bp sequence (designated the primary PCR product). Nested BLV pol primers KB560 (GGA GGT TTG TGC ATG ACC TAC) and KB561 (CAT TGG AGG TCT CCT AAG ACC) amplify a 591-bp PCR sequence. BLV env primers KB582 (CTG ACC TTA GGC CTA GCC) and KB567 (GTC GAC TCA AGG GCA GGG TCG) amplify a 636-bp sequence. Primers specific for reverse-transcribed BLV SGV are complementary to the BLV polypurine tract and the U5 region of the BLV LTR. These primers, KB504mod (CTG AGG GGG AGT CAT TTG TAT G) and KB572 (CGA GAA ACA GAA AGT AAG ACA GG), amplify a 650-bp sequence. The specificity of the PCR products was evaluated by Southern blot analysis with BLV- or SNV-specific DNA fragments that were [α32P]dCTP labeled by the random-primer method with Redi-Prime reagent (Amersham). Hybridization was performed in Rapid Hyb solution (Amersham) under stringent conditions, and signal was detected by autoradiography.
To analyze proviral load in PBMCs, proviral sequences were detected by PCR amplification of 10 μl of lysate (equivalent to 105 lysed PBMCs) with BLV pol-specific primers KB560 and KB561. The amplification of the β-actin gene sequence (product length, 594 bp) with primers Act+ (CCT TCT ACA ATG AGC) and Act− (GTA CTT CAC ACT GCA) was used as a control for semiquantitative analysis and performed with 2 μl of each lysate (equivalent to 2 × 104 lysed PBMCs) mixed in a 50-μl standard PCR mixture, incubated for 1 min at 94°C, 1 min at 42°C, and 1 min at 72°C for 30 cycles. One hundred nanograms of DNA isolated from either a BLV-producing cell clone derived from fetal lamb kidney [FLK(BLV) cells] (3) or BLV SGV-producing dog osteosarcoma cell line D17/5B (7) was used as a positive control.
RT-PCR analysis of RNA from activated PBMCs.
PBMCs were cultivated for 12 h in Dulbecco modified Eagle medium supplemented with lipopolysaccharide (LPS) from Salmonella minnesota (10 μg/ml; Sigma). Total RNA was prepared with the RNAgents total RNA isolation system (Promega) according to the manufacturer’s instruction with DNase treatment. One microgram of PBMC total RNA was subjected to reverse transcription for 1 h at 48°C and PCR amplification in the single-buffer Access reverse transcription-PCR (RT-PCR) system (Promega) with BLV pol primers KB2341 and KB3175. Nested PCR with 1/50 of the primary PCR product was performed with BLV pol primers KB560 and KB561. RNA samples without reverse transcriptase were used as negative control. Fifty nanogram of RNA isolated from BLV-producing rat cell line R(BLV) (5) was used as a positive control.
Western immunoblot analysis of rabbit sera.
As described previously (3, 7), disrupted BLV particles or immunoaffinity-purified BLV Env gp51 was applied to membrane strips and reacted with experimental rabbit sera, polyspecific anti-BLV rabbit serum, or preimmune rabbit sera prepared at various dilutions. Positive reactions were visualized by using Western blue stabilized substrate for alkaline phosphatase (Promega).
RESULTS
In vivo transfection of rabbits produces BLV and BLV SGV infection.
Chinchilla rabbits were inoculated with three 50-μg doses of BLV proviral DNA (pBL913) (Fig. 1A) (13), BLV SGV proviral DNA (pU5gag-pol-env) (Fig. 1B) (7), or negative control DNA (pUC19). BLV SGV is encoded by a hybrid retrovirus vector genome that is composed of SNV LTR sequences and BLV gag, pol, and env genes and related cis-acting replication sequences (Fig. 1B). The normal requirement for Tax- and Rex-regulated gene expression in BLV has been eliminated in BLV SGV and replaced with a constitutive pattern of gene expression (6). The vector genome maintains the BLV cis-acting sequences necessary for encapsidation, reverse transcription (primer binding site and polypurine tract) and integration, which interact with BLV structural and enzymatic proteins to replicate the novel viral genome.
PBMCs, the BLV target cell population (24, 29, 31, 33), were screened for replicated provirus sequences at 1, 4, and 10 months postinoculation, using PBMC lysates and PCR primers complementary to three regions: BLV pol, BLV env, and reverse-transcribed BLV SGV 3′ untranslated region. The specificity of the PCR products was evaluated by Southern blot hybridization. As summarized in Table 1, all BLV-inoculated rabbits or BLV SGV-inoculated rabbits, but not mock-inoculated rabbits (herein designated BLV rabbits, BLV SGV rabbits, and mock rabbits, respectively), exhibited BLV pol and env sequences. The pol sequences were consistently detected in both BLV and BLV SGV rabbits, but detection of env was not sustained. For BLV rabbits 44-8 and 44-10, env became undetectable at months 4 and 10 respectively (Table 1). Similarly, env became undetectable in BLV SGV rabbits at month 4 (rabbits 44-5 and 44-6) and month 10 (rabbit 44-4). This may be attributable to inefficiency of the env PCR or to instability of BLV and BLV SGV env sequences.
TABLE 1.
BLV sequences in rabbit PBMCs by PCR and Southern blot hybridization
| Treatment | Animal no. | Responsea
|
|||||
|---|---|---|---|---|---|---|---|
|
pol
|
env
|
||||||
| 1b | 4 | 10 | 1 | 4 | 10 | ||
| BLV SGV | 44-4 | ND | + | + | ND | + | − |
| 44-5 | + | + | + | + | − | − | |
| 44-6 | + | + | + | + | − | − | |
| BLV | 44-8 | + | + | NA | + | − | NA |
| 44-9 | + | + | NA | + | ND | NA | |
| 44-10 | + | + | NA | + | ND | − | |
| Mock | 46-7 | − | − | − | − | − | − |
| 46-8 | − | − | − | − | − | − | |
Presence (+) or absence (−) of positive signal after gene-specific PCR and Southern blot hybridization. ND, not determined because sample was not available for analysis. NA, sample not available because of death of the animal.
Month postinoculation.
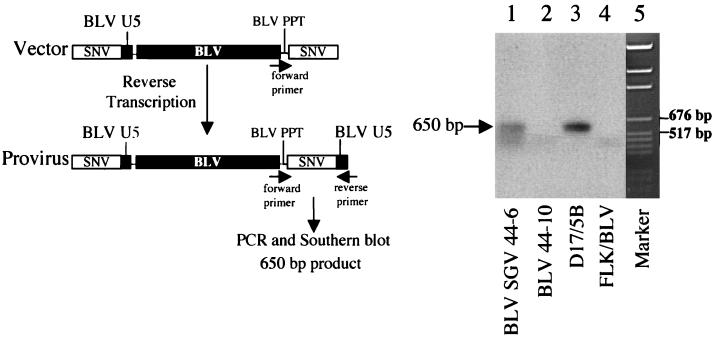
Experiments by Lew et al. (27) determined the half-life of in vivo-transfected plasmid DNA in blood to be less than 5 min, and at 1 month posttransfection, injected plasmid was not detectable in mice tissues except for muscle, where femtogram levels were detectable. Although the BLV SGV rabbits exhibited sustained pol provirus sequence at 1, 4, and 10 months postinoculation, we used PCR with primers specific for reverse-transcribed BLV SGV provirus to evaluate the possibility that the positive PCR signals are attributable to intradermally injected DNA that is residual in PBMC. As summarized in Fig. 2, the BLV U5 sequence is not present in the 3′ LTR in the transfected vector but would be present in the 3′ LTR of reverse-transcribed provirus. PCR and Southern blot analysis with 32P-labeled SNV LTR DNA indicated that the expected reverse-transcribed provirus is detectable in PBMC of BLV SGV rabbit 44-6 at 1 month postinoculation and in positive control tissue culture cells but not in negative control PBMC or negative control tissue culture cells (Fig. 2). These results confirm authentic BLV SGV infection, reverse transcription, and provirus formation.
FIG. 2.
PCR and Southern blot analysis specific for BLV SGV provirus. (Left) Reverse transcription converts the 3′ SNV LTR in vector DNA to a 3′ hybrid SNV-BLV LTR in BLV SGV provirus. The hybrid LTR is amplified by PCR with primers specific for BLV polypurine tract (PPT), which functions in the reverse transcription step of replication (forward primer KB504mod), and the U5 region of the BLV LTR (reverse primer KB572). Southern blot hybridization analysis with a 32P-labeled SNV LTR probe is expected to detect a 650-bp product. (Right) Detection of 650-bp 3′ hybrid LTR PCR product by Southern blot analysis with a 32P-labeled SNV LTR probe. Lanes are labeled with the sources of DNA. Lanes 3 and 4 contain 100 ng of cells. Lane 5, pGem DNA size standard (Promega).
BLV SGV exhibits lower proviral load than BLV.
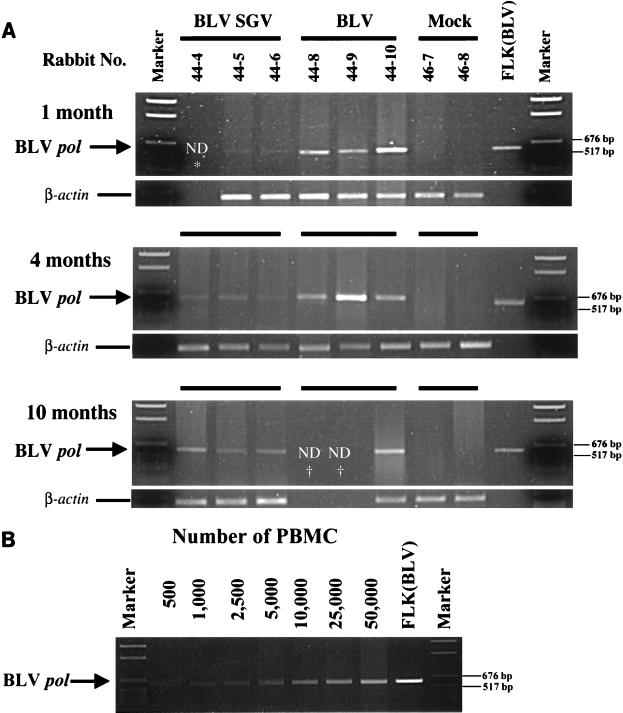
The significant differences in genomic structure between BLV and BLV SGV could have dramatic effects on proviral load in infected rabbits. One possible explanation is that replacement of BLV Tax/Rex-regulated gene expression with a constitutive pattern of gene expression drives the rabbit immune response to depletion of cells expressing BLV SGV. Another possibility is that the lack of accessory genes RIII and GIV in BLV SGV eliminates virus-host interactions important for viral load. In sheep, deletion of GIV in BLV provirus correlates with a reduction in proviral load by a factor ranging between 5 to over 125, and with lack of progression to neoplasia (12, 26, 42). In BLV SGV, the lack of RIII and GIV may function alone or in combination with the constitutive pattern of gene expression to modulate BLV SGV proviral load. To evaluate proviral load, PBMC lysates were subjected to semiquantitative PCR with BLV pol primers and β-actin primers to control for sample variation. As shown in Fig. 3A, similar control β-actin signals were detected among the PBMC samples. At each time point, the pol signal was strong for BLV, weak for BLV SGV, and negative for mock rabbits. These results indicate that BLV and BLV SGV proviruses are maintained through month 10, but that BLV SGV proviral load is consistently lower. Comparison to a PCR panel of a serially diluted BLV lysate indicates that the difference in signal intensity between BLV and BLV SGV samples represents a reduction in BLV SGV proviral load by an approximate factor of 10 to 100 (Fig. 3B).
FIG. 3.
Semiquantitative PCR analysis to evaluate differences in proviral load. (A) PCR to detect BLV pol in PBMC from treated rabbits. Rabbit PBMC were harvested at 1, 4, and 10 months postinoculation and subjected to PCR with BLV pol primers KB560 and KB561 (10 × 104 PBMC) or β-actin to control for sample variation (2 × 104 PBMC). Each panel is labeled with month of sample harvest. Lanes are labeled with the source of PBMC DNA by rabbit number and treatment, FLK(BLV) (positive control DNA [100 ng] from BLV-producing fetal lamb kidney cells), or marker (pGem DNA size standard [Promega]). In lower panels, the corresponding rabbit samples are designated by the matching parallel black lines. Positions of BLV pol amplicon (591 bp) and β-actin amplicon (594 bp) are designated. *, not determined (ND) because sample was not available; †, not determined because the animal died before harvest. (B) PCR standard curve. PBMC lysate harvested at 1 month postinoculation from BLV rabbit 44-10 was serially diluted in a range of 5 × 102 to 5 × 104 PBMC and subjected to PCR with BLV pol primers KB560 and KB561. Each lane is labeled with the number of cells used for PCR amplification, FLK(BLV) (positive control DNA [100 ng] from BLV-producing fetal lamb kidney cells), or marker (pGem DNA size standard [Promega]). The arrow on the left indicates the position of BLV pol amplicon (591 bp), and the lines on the right indicate positions of 676- and 571-bp DNA size markers.
BLV and BLV SGV RNA is expressed in activated PBMCs.
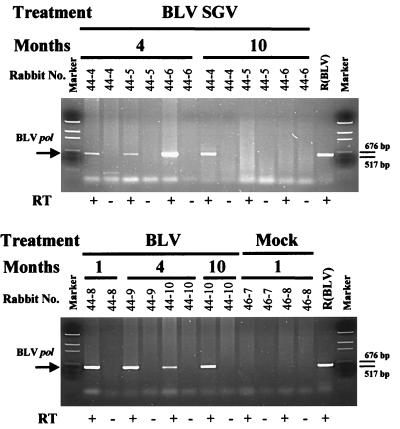
RT-PCR was used to confirm authentic BLV and BLV SGV RNA expression. Rabbit PBMCs were harvested at 1, 4, and 10 months postinoculation and stimulated with LPS (32), and total RNA was isolated. Aliquots of 1 μg were subjected to RT-PCR with BLV primary pol primers and Tfl polymerase, followed by a nested PCR amplification. Consistent with the provirus PCR results, the RT-PCR detected the expected 591-bp product in PBMC from BLV and BLV SGV rabbits but not mock rabbits (Fig. 4). The observation that these samples were negative in the absence of reverse transcription eliminated the possibility of DNA contamination. While each of the BLV SGV rabbits exhibited pol RNA at 4 months, only BLV SGV rabbit 44-4 exhibited pol RNA at 10 months even though each rabbit was positive for pol proviral sequence (Table 1; Fig. 3). This discrepancy may be attributable to PCR variation among the RNA samples or to differences in gene expression. In summary, the RT-PCR analysis confirms authentic BLV and BLV SGV gene expression in LPS-stimulated PBMC. These RNAs would be expected to produce viral proteins that induce BLV-specific antisera.
FIG. 4.
Detection of viral RNA in rabbit PBMC by RT-PCR. Rabbit PBMC were harvested at 1, 4, and 10 months postinoculation and subjected to overnight culture in medium containing LPS (10 μg/ml), followed by extraction of total cellular RNA. Aliquots of 1 μg were subjected to single-step RT-PCR with pol primers KB2341 and KB3175, followed by nested PCR of 1/50 of the primary reaction with pol primers KB560 and KB561. Each panel is labeled with rabbit treatment, month of harvest, and rabbit number: R(BLV) (positive control DNA [50 ng] from a BLV-producing rat cell line), or Marker (pGem DNA size standard [Promega]). The arrow on the left indicates the position of the BLV pol amplicon (591 bp), and the lines on the right indicate positions of 676- and 571-bp DNA size markers.
BLV SGV infection induces seroconversion to BLV structural proteins.
Our previous work in the rat system demonstrated that BLV and BLV SGV infections induce antisera against BLV Gag and Env (7). To evaluate seroconversion of the treated rabbits to the BLV antigens, we collected blood at 12 intervals and analyzed dilutions of the sera by Western immunoblot assay. As expected based on detection of provirus and viral RNA, each BLV or BLV SGV rabbit, but neither mock rabbit, seroconverted to BLV Gag and Env (Table 2). For BLV rabbits, the immune response typically persisted for life and anti-Gag levels increased in two (44-8 and 44-9) of three BLV rabbits in the sample preceding death. For BLV SGV rabbits, Gag seroconversion persisted to at least month 15, but Env-specific antisera diminished to an undetectable level by month 10. This trend to undetectable Env antibody level correlates with the loss of detectable env proviral sequences at month 10 in the BLV SGV rabbits (Table 1). Interestingly, the overall levels of seroconversion to Gag and Env were similar for BLV rabbits and BLV SGV rabbits even though significant differences were observed in provirus load (Fig. 3). This lack of correlation may be attributable to constitutive gene expression from the BLV SGV or to productive BLV SGV infection of a distinct subpopulation of activated PBMC.
TABLE 2.
Levels of BLV Gag and Env antisera in treated rabbits
| Treatment | Animal no. | Responsea
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BLV Gag antisera
|
BLV Env antisera
|
||||||||||||||||||
| 1b | 3 | 4 | 5 | 6 | 7 | 9 | 10 | 12 | 13 | 15 | 20 | 1 | 3 | 4 | 5 | 10 | 15 | ||
| BLV SGV | 44-4 | − | − | 4+ | 4+ | ND | ND | ND | ± | ± | ND | 4+ | NA | 5+ | 3+ | ND | − | − | − |
| 44-5 | 2+ | − | 2+ | 2+ | 2+ | 2+ | ND | 4+ | 2+ | ND | 4+ | − | 1+ | 3+ | 1+ | ± | − | − | |
| 44-6 | 2+ | 4+ | 4+ | 4+ | 4+ | ± | ND | 4+ | 4+ | 4+ | 4+ | ± | 1+ | 3+ | 1+ | 1+ | − | − | |
| BLV | 44-8 | 2+ | 4+ | 4+ | 6+ | NA | 3+ | 1+ | 1+ | 1+ | NA | ||||||||
| 44-9 | ± | 2+ | 2+ | ± | ND | ± | 7+ | NA | ND | 1+ | ND | − | NA | ||||||
| 44-10 | 4+ | 4+ | 2+ | 2+ | ND | 2+ | 4+ | 4+ | 4+ | ND | 4+ | 4+c | 3+ | 1+ | 1+ | 3+ | 3+ | 1+ | |
Western immunoblotting with alkaline phosphatase detection of BLV-specific sera. −, no visible signal at 50-fold dilution; 1+, detectable at 50-fold dilution; 2+, detectable at 100-fold dilution; 3+, detectable at 500-fold dilution; 4+, detectable at 1,000-fold dilution; 5+, detectable at 5,000-fold dilution; 6+, detectable at 10,000-fold dilution; 7+, detectable at 50,000-fold dilution; ±, weak but detectable at 50-fold dilution. Samples from mock-infected rabbit 46-7 and 46-8 were uniformly negative. ND, not determined because sample was not available for analysis. NA, sample not available because of the death of the animal.
Month postinoculation.
Rabbit 44-10 died 2 weeks after sample harvest.
In vivo transfection of BLV but not BLV SGV induces an immunodeficiency-like disease.
Rabbits infected with cell-associated BLV or cell-free BLV develop sudden onset of severe weight loss, bronchopneumonia, abscesses, leg paralysis, and spleen atrophy, and BLV sequences are detectable in spleen and other organs (4, 43). Similar to rabbits inoculated with cell-associated or cell-free BLV, our rabbits inoculated with BLV proviral DNA developed the sudden onset of clinical decline during a 2-week period that resulted in death. The rabbits experienced severe weight loss (20% or more), diarrhea, paralysis of the legs, and spleen atrophy and died at 5, 9, and 20 months postinoculation (rabbits 44-8, 44-9, 44-10, respectively). BLV-specific sequences were detectable by PCR in genomic DNA isolated from spleen, lung, kidney, and liver but not heart nor brain, and proviral load was consistently higher in spleen than in the other positive tissues (data not shown). These results indicate that in vivo transfection of BLV provirus recapitulates the pathogenic BLV infection observed in response to cell-associated or cell-free BLV inoculation (4, 43).
In contrast to the pathogenicity observed in the BLV rabbits, all of the BLV SGV rabbits and mock rabbits remained clinically healthy within the 20-month period and grew to an average weight of 4.65 kg. In summary, rabbits in vivo transfected with BLV but not BLV SGV succumbed to an immunodeficiency syndrome that culminated in death.
DISCUSSION
In vivo transfection is an effective approach to BLV infection of rats and sheep (7, 40, 41), and the first objective of this study was to validate in vivo transfection as an effective approach to BLV infection of rabbits. Our results establish that in vivo transfection recapitulates the clinical outcome that is produced by inoculation of rabbits with cell-associated and cell-free BLV. Three of three rabbits in vivo transfected with BLV succumbed to an immunodeficiency-like syndrome that culminated in death within the time frame observed for the cell-associated and cell-free infections (4, 43).
Our second objective, to compare infection and immunogenicity of BLV and BLV SGV, revealed that both viruses infect PBMC, form proviruses that express viral RNA in LPS-stimulated PBMC, and produce viral proteins that induce antisera to BLV Gag and Env. Interestingly, the levels of BLV-specific antisera were similar for BLV and BLV SGV rabbits even though proviral load was lower for BLV SGV by a factor ranging between 10 to 100. Possible explanations are that productive BLV SGV infection of a subpopulation of stimulated PBMC and/or the constitutive pattern of gene expression by BLV SGV drives depletion of the virus-producing cells. Interestingly, pol sequences persisted in BLV SGV rabbits at 10 months postinoculation, but env sequences became undetectable by 10 months. At month 10, we observed a correlation between the loss of env sequences and the loss of detectable Env-specific antisera to Env. Future experiments will address whether this is attributable to insensitivity of the env PCR assay compared to the pol assay or instability of BLV SGV env sequences.
Our third objective, to compare the disease induction capacity of BLV and BLV SGV, determined that the immunodeficiency-like syndrome caused by BLV is not caused by BLV SGV. While each of the BLV rabbits succumbed to fatal BLV disease, the BLV SGV rabbits and mock rabbits exhibited a healthy clinical condition during the course of the 20-month study. We note that BLV SGV rabbit 44-4 died suddenly at month 21 of an unknown cause. The rabbit lacked the clinical signs present in the BLV rabbits, was of average weight, and exhibited no spleen atrophy, and no BLV SGV proviral sequences were detectable in tissue as assessed by PCR, whereas proviral sequences were detected in tissue from BLV rabbits (data not shown). Because this rabbit exhibited sustained seroconversion to Gag at month 20, we suspect that the proviral load was below the limits of detection of the PCR assay. While it appears that death of the animal was a random event not related to BLV SGV infection, our sample size is not suitable for statistical prediction of a random death. Significantly, the other BLV SGV rabbits and the mock rabbits remain clinically healthy at over 34 months postinoculation. In conclusion, BLV SGV infection of rabbits did not cause the immunodeficiency-like syndrome observed in three of three rabbits infected with BLV.
Finally, it remains to be determined whether the BLV SGV rabbits do not progress to BLV disease because viral burden is insufficient or because the virus is inherently less cytopathic; a similar question remains with respect to SCID-hu mice infected with Rev-independent HIVs that exhibit low proviral load and lack cytopathicity (37). The second possibility of less cytopathicity is consistent with our observation of similar levels of BLV-specific antisera in BLV and BLV SGV rabbits.
In summary, our results indicate that the BLV SGV is infectious and immunogenic in rabbits but lacks the pathogenicity caused by BLV in rabbits. This study constitutes a necessary and important step in evaluation of our modified proviral delivery system as a preventative vaccine approach against BLV and other complex retroviruses including HTLV and HIV. Further experiments will compare the induction of BLV-specific cytotoxic T lymphocytes responses by BLV and BLV SGV because cell-mediated immunity is a central component of a protective immune response against BLV and other complex retroviruses (22, 30).
ACKNOWLEDGMENTS
We thank Patrick Green, Stacey Hull, Gary Kociba, Micheal Lairmore, and Lawrence Mathes for critical comments on the manuscript.
This work was supported in part by VEGA Grant Agency of the Slovak Academy of Sciences (C.A.), American Cancer Society, Ohio Division (K.B.-L.), Elsa Pardee Foundation (K.B.-L.), and National Institutes of Health (P30CA16058 and AI40851; K.B.-L.).
REFERENCES
- 1.Akagi T, Ono J, Nyunoya H, Shimotohno K. Characterization of peripheral blood T lymphocytes transduced with human T-cell leukemia virus type I Tax mutants with different trans-activating phenotypes. J Virol. 1997;14:2071–2078. doi: 10.1038/sj.onc.1201045. [DOI] [PubMed] [Google Scholar]
- 2.Alexandersen S, Carpenter S, Christensen J, Storgaard T, Viuff B, Wannemuehler Y, Belousov J, Roth J A. Identification of alternatively spliced mRNAs encoding potential new regulatory proteins in cattle infected with bovine leukemia virus. J Virol. 1993;67:39–52. doi: 10.1128/jvi.67.1.39-52.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altaner C, Merza M, Altanerova V, Morein B. Envelope glycoprotein gp51 of bovine leukemia virus is differently glycosylated in cells of various species and organ origin. Vet Immunol Immunopathol. 1993;36:163–177. doi: 10.1016/0165-2427(93)90105-d. [DOI] [PubMed] [Google Scholar]
- 4.Altanerova V, Ban J, Altaner C. Induction of immune deficiency syndrome in rabbits by bovine leukemia virus. AIDS. 1989;3:775–780. doi: 10.1097/00002030-198911000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Altanerova V, Portetelle D, Kettman R, Altaner C. Infection of rats with bovine leukemia virus: establishment of a virus-producing rat cell line. J Gen Virol. 1989;70:1929–1932. doi: 10.1099/0022-1317-70-7-1929. [DOI] [PubMed] [Google Scholar]
- 6.Boris-Lawrie K, Temin H M. Genetically simpler bovine leukemia virus derivatives can replicate independently of Tax and Rex. J Virol. 1995;69:1920–1924. doi: 10.1128/jvi.69.3.1920-1924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boris-Lawrie K, Altanerova V, Altaner C, Kucerova L, Temin H M. In vivo study of genetically simplified bovine leukemia virus derivatives that lack tax and rex. J Virol. 1997;71:1514–1520. doi: 10.1128/jvi.71.2.1514-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burny A, Cleuter Y, Kettman R, Mammerickx M, Marbaix G, Portetelle D, Van den Broeke A, Willems L, Thomas R. Bovine leukemia virus: facts and hypotheses derived from the study of an infectious cancer. Cancer Surv. 1987;6:139–159. [PubMed] [Google Scholar]
- 9.Butsch M, Hull S, Wang Y, Roberts T M, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J Virol. 1999;73:4847–4855. doi: 10.1128/jvi.73.6.4847-4855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cockerell G L, Rovnak J, Green P L, Chen I S Y. A deletion in the proximal untranslated pX region of human T-cell leukemia virus type II decreases viral replication but not infectivity in vivo. Blood. 1996;87:1030–1035. [PubMed] [Google Scholar]
- 11.Collins N D, Newbound G C, Albrecht B, Beard J L, Ratner L, Lairmore M D. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood. 1998;91:4701–4707. [PubMed] [Google Scholar]
- 12.Dequiedt F, Hanon E, Kerkhofs P, Pastoret P P, Portetelle D, Burny A, Kettman R, Willems L. Both wild-type and strongly attenuated bovine leukemia viruses protect peripheral blood mononuclear cells from apoptosis. J Virol. 1997;71:630–639. doi: 10.1128/jvi.71.1.630-639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987;61:2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derse D. trans-acting regulation of bovine leukemia virus mRNA processing. J Virol. 1988;62:1115–1119. doi: 10.1128/jvi.62.4.1115-1119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derse D, Mikovits J, Ruscetti F. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology. 1997;237:123–128. doi: 10.1006/viro.1997.8781. [DOI] [PubMed] [Google Scholar]
- 16.Djilali S, Parodi A-L. The BLV-induced leukemia-lymphosarcoma complex in sheep. Vet Immunol Immunopathol. 1989;22:233–244. doi: 10.1016/0165-2427(89)90010-x. [DOI] [PubMed] [Google Scholar]
- 17.Franchini G. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type I infection. Blood. 1995;86:3619–3639. [PubMed] [Google Scholar]
- 18.Grassman R, Berchtolds S, Radant I, Alt M, Fleckenstein B, Sodroski J G, Haseltine W A, Ramstedt U. Role of the human T-cell leukemia virus type 1 X region proteins in immortalization of primary human lymphocytes in culture. J Virol. 1992;66:4570–4575. doi: 10.1128/jvi.66.7.4570-4575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green P L, Yip M T, Xie Y, Chen I S Y. Phosphorylation regulates RNA binding by the human T-cell leukemia virus Rex protein. J Virol. 1992;66:4325–4330. doi: 10.1128/jvi.66.7.4325-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman W J, Kimata J T, Wong F H, Zutter M, Ley T J, Ratner L. Development of leukemia in mice transgenic for the tax gene of human T-cell leukemia virus type 1. Proc Natl Acad Sci USA. 1995;92:1057–1061. doi: 10.1073/pnas.92.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanley S M, Rimsky L T, Malim M H, Kim J H, Hauber J, Duc Dondon M, Le S-Y, Maizel J V, Cullen B R, Greene W C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989;3:1534–1544. doi: 10.1101/gad.3.10.1534. [DOI] [PubMed] [Google Scholar]
- 22.Hislop A D, Good M F, Mateo L, Gardner J, Gatei M H, Daniel R C W, Meyers B V, Lavin M, Schurbier A. Vaccine-induced cytotoxic T lymphocytes protect against retroviral challenge. Nat Med. 1998;4:1193–1196. doi: 10.1038/2690. [DOI] [PubMed] [Google Scholar]
- 23.Hope T, Pomerantz R J. The human immunodeficiency virus type 1 Rev protein: a pivotal protein in the viral life cycle. Curr Top Microbiol Immunol. 1995;193:91–105. doi: 10.1007/978-3-642-78929-8_5. [DOI] [PubMed] [Google Scholar]
- 24.Jensen W A, Rovnak J, Cockerell G L. In vivo transcription of bovine leukemia virus tax/rex region in normal and neoplastic lymphocytes of cattle and sheep. J Virol. 1991;65:2484–2490. doi: 10.1128/jvi.65.5.2484-2490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kettmann, R., A. Burny, I. Callebaut, L. Droogmans, M. Mammerickx, L. Willems, and D. Portetelle. Bovine leukemia virus, p 39–81. In J. A. Levy (ed.), The Retroviridae, vol. 3. Plenum Press, New York, N.Y.
- 26.Kerkhofs P, Heremans H, Burny A, Kettmann R, Willems L. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J Virol. 1998;72:2554–2559. doi: 10.1128/jvi.72.3.2554-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew D, Parker S E, Latimer T, Abai A M, Kuwahara-Rundell A, Doh S G, Yang Z, Laface D, Gromkowski S H, Nabel G J, Manthorpe M, Norman J. Cancer gene therapy using plasmid DNA: pharmacokinetic study of DNA following injection in mice. Hum Gene Ther. 1995;6:553–564. doi: 10.1089/hum.1995.6.5-553. [DOI] [PubMed] [Google Scholar]
- 28.Mammerickx M, Palm R, Portetelle D, Burny A. Experimental transmission of enzootic bovine leukosis to sheep: latency period of the tumoral disease. Leukemia. 1988;2:103–107. [PubMed] [Google Scholar]
- 29.Mirsky M L, Olmstead C A, Da Y, Lewin H A. The prevalence of proviral bovine leukemia virus in peripheral blood mononuclear cells at two subclinical stages of infection. J Virol. 1996;70:2178–2183. doi: 10.1128/jvi.70.4.2178-2183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantification of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 31.Paul P S, Pomeroy K A, Johnson D W, Muscoplat C C, Handwerger B S, Soper F F, Sorenson D K. Evidence for the replication of bovine leukemia virus in B lymphocytes. Am J Vet Res. 1977;38:873–876. [PubMed] [Google Scholar]
- 32.Powers M A, Radke K. Activation of bovine leukemia virus transcription in lymphocytes from infected sheep: rapid transition through early to late gene expression. J Virol. 1992;66:4769–4777. doi: 10.1128/jvi.66.8.4769-4777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radke K, Grossman D, Kidd L C. Humoral immune response of experimentally infected sheep defines two early periods of bovine leukemia virus replication. Microb Pathog. 1990;9:159–171. doi: 10.1016/0882-4010(90)90019-m. [DOI] [PubMed] [Google Scholar]
- 34.Ressler S, Connor L M, Marriott S J. Cellular transformation by human T-cell leukemia virus type I. FEMS Microbiol Letters. 1996;140:99–109. doi: 10.1111/j.1574-6968.1996.tb08322.x. [DOI] [PubMed] [Google Scholar]
- 35.Ross T M, Pettiford S M, Green P L. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J Virol. 1996;70:5194–5202. doi: 10.1128/jvi.70.8.5194-5202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Temin H M. A proposal for a new approach to a preventive vaccine against human immunodeficiency virus type I. Proc Natl Acad Sci USA. 1993;9:4419–4420. doi: 10.1073/pnas.90.10.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentin A, Aldrovandi G, Zolotukhin A S, Cole S W, Zack J A, Pavlakis G N, Felber B K. Reduced viral load and lack of CD4 depletion in SCID-hu mice infected with Rev-independent clones of human immunodeficiency virus. J Virol. 1997;71:9817–9822. doi: 10.1128/jvi.71.12.9817-9822.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willems L, Gegonne A, Chen G, Kettmann R, Ghysdael J. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 1987;6:3385–3389. doi: 10.1002/j.1460-2075.1987.tb02661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willems L, Heremans H, Chen G, Portetelle D, Billiau A, Burny A, Kettmann R. Cooperation between bovine leukemia virus transactivator protein and Ha-ras oncogene in cellular transformation. EMBO J. 1990;9:1577–1581. doi: 10.1002/j.1460-2075.1990.tb08277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willems L, Portetelle D, Kerkhofs P, Chen G, Burny A, Mammerickx M, Kettmann R. In vivo transfection of bovine leukemia virus mutants into sheep. Virology. 1992;189:775–777. doi: 10.1016/0042-6822(92)90604-n. [DOI] [PubMed] [Google Scholar]
- 41.Willems L, Kettman R, Dequiedt R, Portetelle D, Voneche V, Cornil I, Kerkhofs P, Burny A, Mammerickx M. In vivo infection of sheep by bovine leukemia virus mutants. J Virol. 1993;67:4078–4085. doi: 10.1128/jvi.67.7.4078-4085.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willems L, Kerkhofs P, Dequiedt F, Portetelle D, Mammerickx M, Burny A, Kettmann R. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc Natl Acad Sci USA. 1994;91:11532–11536. doi: 10.1073/pnas.91.24.11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyatt C, Wingett D, White J, Buck C, Knowles D, Reeves R, Magnuson N. Persistent infection of rabbits with bovine leukemia virus associated with development of immune dysfunction. J Virol. 1989;63:4498–4506. doi: 10.1128/jvi.63.11.4498-4506.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yip M T, Dynan W S, Green P L, Black A C, Arrigo S J, Torbati A, Heaphy S, Ruland C, Rosenblatt J D, Chen I S Y. Human T-cell leukemia virus (HTLV) type II Rex protein binds specifically to RNA sequences of the HTLV long terminal repeat but poorly to the human immunodeficiency virus type 1 Rev-responsive element. J Virol. 1991;65:2261–2272. doi: 10.1128/jvi.65.5.2261-2272.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]