Abstract
Engineering metabolism to efficiently produce chemicals from multi-step pathways requires optimizing multi-gene expression programs to achieve enzyme balance. CRISPR-Cas transcriptional control systems are emerging as important tools for programming multi-gene expression, but poor predictability of guide RNA folding can disrupt expression control. Here, we correlate efficacy of modified guide RNAs (scRNAs) for CRISPR activation (CRISPRa) in E. coli with a computational kinetic parameter describing scRNA folding rate into the active structure (rS = 0.8). This parameter also enables forward design of scRNAs, allowing us to design a system of three synthetic CRISPRa promoters that can orthogonally activate (>35-fold) expression of chosen outputs. Through combinatorial activation tuning, we profile a three-dimensional design space expressing two different biosynthetic pathways, demonstrating variable production of pteridine and human milk oligosaccharide products. This RNA design approach aids combinatorial optimization of metabolic pathways and may accelerate routine design of effective multi-gene regulation programs in bacterial hosts.
Subject terms: Metabolic engineering, Expression systems, Genomic engineering, Synthetic biology
Guide RNA folding affects functionality of CRISPR-Cas transcriptional control systems. Here, the authors report computational gRNA design together with creation of synthetic CRISPRa promoters for orthogonal expression control and demonstrate the application in pteridine and human milk oligosaccharide production in E. coli.
Introduction
Synthetic biology and metabolic engineering have great potential for enabling chemical bioproduction from sustainable feedstocks as part of a circular bioeconomy1–3. Efficient microbial conversion of simple substrates into valuable chemicals and materials often requires precise expression control across multiple genes to optimize enzyme levels and stoichiometry. Despite recent advances in gene expression technologies, it remains challenging to engineer and optimize multi-step metabolic pathways4–6. CRISPR-Cas transcriptional control systems have emerged as promising routes for programming the precise expression of multiple genes, which could accelerate the development of engineered organisms for a wide variety of applications7–10. We recently developed an approach for the construction of multi-gene CRISPR transcriptional control programs in bacteria, with activation (CRISPRa) or repression (CRISPRi) functions specified through the regulated expression of multiple guide RNAs (gRNAs)11,12. Recent demonstrations of dynamic multi-layer CRISPRa/i gene regulatory network designs in E. coli13,14 and CRISPR-based metabolic pathway engineering in the soil microbe Pseudomonas putida15–17 highlight the versatility of these systems for programmable multi-gene control. However, gaps in knowledge and technique continue to prevent the routine design of CRISPRa/i programs capable of quantitatively tuning activated expression from multiple bacterial genes at the same time9,18.
Quantitatively tunable multi-gene expression programs are particularly useful for microbial metabolic engineering applications19. It is important to identify gene expression programs that minimize enzyme imbalances in multi-gene heterologous pathways and tune endogenous networks to redirect metabolic flux towards the desired output4,6,20. Balanced enzyme expression helps minimize bottlenecks, prevent excess metabolic burden, and avoid accumulation of toxic intermediates. Identifying these programs is challenging, in part because we lack tools to systematically explore large, multi-dimensional spaces of gene expression programs. Addressing this challenge with CRISPRa/i systems requires reliable and tunable regulation of gene expression, in turn requiring predictive gRNA design tools for bacterial hosts. Significant progress has been made in gRNA design using folding energetics predictions, cell-based screens, and machine learning, although these methods have been applied primarily for gene editing applications in mammalian cells21. General design strategies for tunable CRISPRi with modified gRNAs have been reported for both mammalian and bacterial systems19,22. However, many bacterial CRISPRa systems use gRNAs with additional structured elements11,12,23, and it is unknown whether design rules for effective gRNA function are generalizable across applications and organisms.
Here, we identify structural properties that enable routine guide RNA design for tunable multi-gene bacterial CRISPRa programs. Our CRISPRa system uses modified single guide RNAs (sgRNAs) that are extended with hairpin sequences, termed scaffold RNAs (scRNAs), to recruit the transcriptional activator SoxS upstream of a promoter11,12. This recruitment results in activation of a weak minimal promoter to high expression levels. To identify design variables affecting CRISPRa, we investigate a set of thermodynamic and kinetic guide RNA folding parameters. We find that the largest impact comes from the size of the energy barrier separating the most stable scRNA structure from the active scRNA structure: this single kinetic parameter accurately predicts about 80% of the variation in CRISPR-activated expression. By comparison, we find that commonly used computational tools for gRNA design cannot consistently identify scRNAs for effective bacterial CRISPRa. We expect that our computational approach could be generalized to identify effective gRNAs for a broad range of CRISPR applications, because the parameters are intrinsic to the RNA sequence. Starting from highly effective and orthogonal scRNAs, we then generate predictable variations in gene activation by truncating scRNA spacer sequences. Using these design strategies, we engineer multi-guide programs that simultaneously direct tunable variations in CRISPRa from multiple promoters independently. We apply a combinatorial set of these CRISPRa programs to drive the design of engineered metabolic pathways producing valuable biopterins and oligosaccharide molecules in E. coli. Screening productive variants from these multi-gene programs is a simple method of engineering efficient microbial bioproduction, here indicating enzyme expression combinations producing up to 2.3-fold higher titer than that produced by maximal expression. This approach to biosynthetic profiling enables quantitative tuning of various pathways, and therefore is a versatile approach for a broad range of bioproduction applications. Furthermore, the capacity to reliably implement tunable, multiplexed gene expression will improve the ability to precisely implement perturbations computationally predicted24,25 to optimize production strains.
Results
scRNA target site sequences have variable effects on gene activation
To build multi-gene CRISPRa programs for metabolic engineering, we need promoters that can be selectively targeted for activation through the expression of a matched, or cognate, scRNA (Fig. 1). The rules for effective CRISPRa from bacterial promoters are known to be complex12. In particular, the 20 bp scRNA target site must be precisely positioned relative to the transcription start site for effective gene activation. We previously identified a highly effective promoter (J3) with an appropriately-positioned target site12. By altering only the target site sequence of the J3 promoter, we expected to generate orthogonal promoters that retain high levels of gene expression.
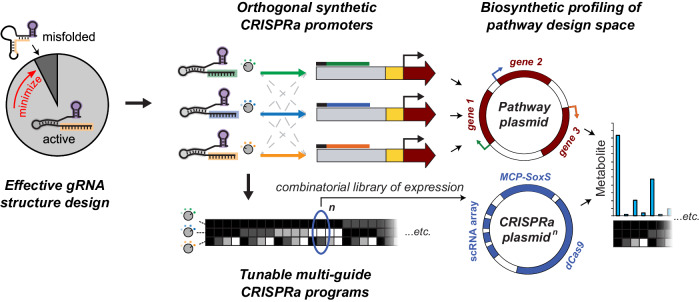
Fig. 1. Structure-based guide RNA design and synthetic promoters enable design space mapping with tunable CRISPRa.
Computational analysis of scRNA sequence identified a kinetic parameter describing the rate of conversion between the most stable structure and the active structure for CRISPRa, and scRNAs screened using this parameter predictably activated bacterial expression from a set of synthetic promoters. Tuning the activation of these promoters by truncating their scRNA spacer sequences—and again computationally verifying their efficacy—allows combinations of activation level at each promoter. The promoters can be paired with chosen output ORFs, including metabolic pathways. This method of controlling pathway gene expression allows for profiling of pathway design spaces for metabolic engineering using a combinatorial library of CRISPR-activated expression levels.
We modified the J3 target site sequence to generate 14 additional synthetic promoters with fully randomized target sites, each paired with its cognate scRNA (Fig. 2a). Targeting the CRISPRa complex in this way to each of the 15 promoters activated expression of a downstream fluorescent reporter gene (Fig. 2b) in E. coli MG1655 (Supplementary Table 1). All of the promoter variants showed measurable activation compared to the off-target scRNA control (Supplementary Fig. 1), but there was significant variability over a 3-fold range in expression levels (Fig. 2b). Consistent with previous findings12, these results suggest that the target site sequence identity can have unexpectedly large effects on gene activation.
Fig. 2. CRISPRa is sensitive to scRNA target sequence.
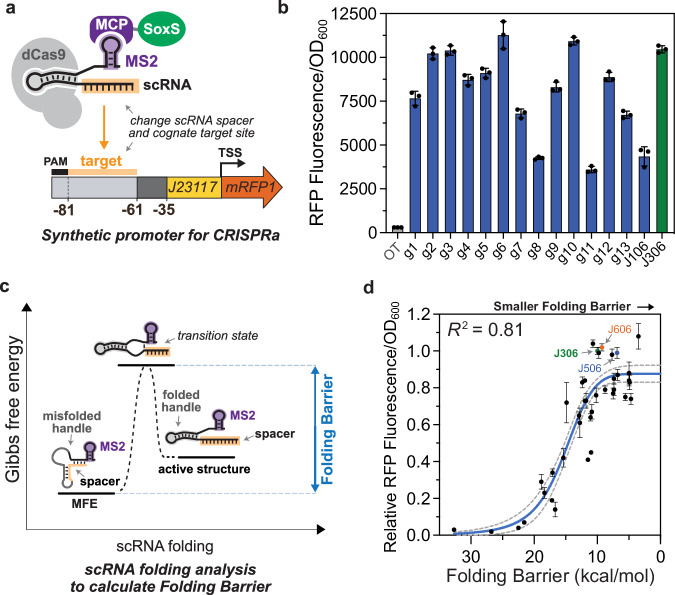
a Experimental system for testing the role of scRNA target site sequence on CRISPRa activity. Orthogonal 20 bp target sequences (Supplementary Data 1) were selected at random from the human genome. These sequences replaced the J306 target sequence in the previously described J3 promoter12, and the cognate scRNAs contained the complementary spacer sequences. b CRISPR-activated RFP expression from each promoter variant. In the presence of the cognate scRNA, sequence-dependent expression variation was measured across the set. Bars (blue for g1-J106, green for J306) represent the Fluorescence/OD600 of strains harboring each synthetic promoter and the cognate scRNA. The gray bar (OT) represents the baseline expression of the J3 promoter, obtained by expressing an off-target scRNA (J206). c Folding Barrier (FB) as a critical parameter determining CRISPR-activated expression. Additional kinetic and thermodynamic parameters are described in Supplementary Fig. 2 and Supplementary Method 1. Folding Barrier can be calculated as the height of the energy barrier separating the minimum free energy (MFE) secondary structure of a scRNA from the active structure for CRISPRa. d Folding Barrier predicts the CRISPR-activated expression of promoter-scRNA pairs based on sequence. In addition to the 15 promoters from panel b, 24 new synthetic promoter-scRNA pairs were designed with FBs ranging from 4.7 kcal/mol to 32.7 kcal/mol (Supplementary Data 1). The y-axis values represent Fluorescence/OD600 of strains harboring each promoter variant and expressing the cognate scRNA, relative to the Fluorescence/OD600 of the J3 promoter and the J306 scRNA (green). Blue and red dots respectively indicate the values of the strains expressing the J506 and J606 scRNAs targeting their cognate promoters (Fig. 3). The blue line represents a Hill function fit to the data, and the gray dotted lines represent the 95% confidence interval for the fit. R2 represents the coefficient of determination for the fit. Values in panels b and d represent the average ± standard deviation calculated from n = 3 biologically independent samples. Source data are provided as a Source Data file.
The kinetic folding barrier predicts scRNA activity for CRISPRa
Variable activation from the orthogonal synthetic promoters could occur if the corresponding 20 base scRNA spacer sequences have different effects on folding. Changes to the spacer sequence could lead to scRNA misfolding that disrupts binding to dCas9, recruitment of the SoxS activator, or binding to DNA. We reasoned that the kinetic and thermodynamic properties associated with the conversion of a misfolded scRNA into the correctly-folded structure could be important determinants of CRISPRa activity. Scaffold RNAs could be more effective in a kinetic sense if they readily transition to the correctly-folded state, or could be more effective in a thermodynamic sense if they are more likely to occupy the correctly-folded state.
To test these possibilities, we developed two coarse-grained parameters that describe the energetics of scRNA folding: Folding Barrier to capture kinetic properties and Folding Energy to capture thermodynamic properties (Fig. 2c and Supplementary Fig. 2). We defined the Folding Energy as the free energy difference between the most stable scRNA structure (Minimum Free Energy, or MFE) and the correctly-folded, CRISPR-active structure. The Folding Energy is large when the correctly-folded structure is less stable than the MFE, and approaches zero as the correctly-folded structure increases in stability. The Folding Barrier is the height of the activation energy barrier separating the MFE structure from the correctly-folded structure. When the MFE structure can easily overcome this barrier and rearrange into the correctly-folded structure, the Folding Barrier is low. The correctly-folded structure was defined as the conformation in which the spacer is unstructured and the Cas9-binding handle adopts the fold observed in the crystal structure of the Cas9-sgRNA-DNA complex26. Energetic parameters were calculated using custom algorithms that apply programs in the ViennaRNA folding package27,28 (see “Methods” section).
To probe the relationships between our calculated parameters and CRISPR-activated RFP expression, we experimentally tested a set of 39 scRNA-promoter pairs. This set includes the original J3 sequence, the 14 randomly selected targets described above, and 24 additional scRNAs designed to have Folding Barriers ranging from 5 to 35 kcal/mol (Supplementary Data 1 and Supplementary Method 1). High levels of CRISPR-activated expression correlated with smaller Folding Energies (rs = 0.7) and lower Folding Barriers (rs = 0.8) (Fig. 2d and Supplementary Fig. 3). Consistently, the MFE structures of the highest-activation scRNAs in our set closely resembled the active scRNA conformations, whereas the least effective scRNAs misfolded extensively (Supplementary Fig. 4 and Supplementary Table 2). Interestingly, we found that Folding Barrier alone may be sufficient for identifying highly effective scRNAs. The most effective scRNA in our 39-member set had the smallest Folding Barrier. In contrast, three of the worst-performing scRNAs, which generated 95% less gene activation than the J306 scRNA, had the largest Folding Barriers in the set. We also considered other thermodynamic and kinetic parameters for use in predicting scRNA folding, but found that Folding Barrier was the most effective predictor of CRISPRa function, with Folding Energy and Net Binding Energy providing limited additional predictive power for low-FB scRNAs. (Supplementary Figs. 3 and 5, and Supplementary Method 1).
Our data suggest that Folding Barrier analysis could be used to drive the design of scRNAs with a lower chance of weak activity. Out of the 24 rationally designed scRNAs, the 15 scRNAs with the lowest Folding Barrier all yielded effective CRISPRa (at least 50% of J306 output, or about 18-fold activation), and their CRISPR-activated expression levels showed less variability than those of the 15 randomly-designed scRNAs (Coefficient of variation = 12% vs. 31% for the random set) (Supplementary Fig. 5). We observed in our promoter set that high-performing scRNAs tended to have Folding Barriers ≤10 kcal/mol, and all defective scRNAs (<50% of J306 activation) were >10 kcal/mol. Therefore, a Folding Barrier threshold of <10 kcal/mol could provide a useful computational screening metric for rapid development of novel scRNAs (Supplementary Fig. 6 and Supplementary Table 3).
To further evaluate this kinetic parameter as a screening tool to design highly effective scRNAs, we compared Folding Barrier with pre-existing models currently in wide use for gRNA design. A common approach to analyze gRNAs involves calculating the free energy of binding a correctly-folded gRNA to its target DNA29,30 (termed Binding Energy in Supplementary Fig. 2a). In this approach, gRNAs with more negative Binding Energies have unstructured spacer sequences that should favor the DNA-bound state, and should therefore be more active. In our study, however, the scRNAs with the lowest Binding Energy included a significant fraction of defective scRNAs (33%), suggesting that Binding Energy is not sufficient to account for CRISPRa functionality (Supplementary Fig. 3 and Supplementary Fig. 5a). These failures might be explained by interactions between the spacer and the dCas9-binding handle, which are not accounted for in Binding Energy but are included in Folding Energy and Folding Barrier due to consideration of the entire scRNA sequence. The Folding Barrier metric correctly predicts these failures within the low-Binding-Energy set: defective scRNAs had relatively high Folding Barriers averaging 17.6 kcal/mol. Effective (≥50% of J306) scRNAs in this set had an average Folding Barrier of 9.3 kcal/mol, further supporting the use of a Folding Barrier threshold to screen functional scRNAs.
Several machine learning models have also been developed to predict gRNA activity21,31–36. These models were trained with supervised learning to extract gRNA design rules from large gene editing datasets and are widely used to aid the selection of gRNA target sites. Among the models we tested, none yielded predictions strongly correlated with observed CRISPR-activated expression from the scRNAs in our set. For example, the widely used Azimuth, Doench ‘1621, and Moreno-Mateos31 tools had correlation coefficients (rs) of 0.22, 0.02, and 0.09, respectively, and incorrectly selected several defective guides as the best (Supplementary Figs. 3 and 5). The top 15 scRNAs predicted by these tools contained both defective scRNAs (with consistently higher Folding Barriers, e.g. 21.6 kcal/mol average using Azimuth) and effective ones (7.3 kcal/mol average using Azimuth). Differences between gRNA-directed editing and scRNA-directed activation may account for the poor performance of these models in this application. A machine learning model trained on scRNAs used in bacteria could potentially be effective, but generating large enough bacterial CRISPRa datasets for such a model to account for the stringent target site requirements12 might be impractical. Given the predictive success and ease of calculation of the Folding Barrier, we proceeded with this kinetic parameter as a strategy to rapidly design highly effective scRNAs for bacterial CRISPRa.
Tunable CRISPRa expression from orthogonal synthetic promoters
By forward engineering scRNAs through computational folding design, our tools provide an avenue for developing synthetic promoters driving high levels of CRISPR-activated expression. To be useful for programming combinatorial variations in multi-gene expression, as in a metabolic engineering application, two additional capabilities are needed. First, the synthetic promoters must exhibit orthogonality with no cross-activation from other non-cognate scRNAs expressed in the cell. Second, a strategy is needed to tune expression levels from each of the promoters by independently modulating CRISPRa activity at each site. In this section, we show that promoter orthogonality is readily obtainable and that 5′ spacer sequence truncations enable quantitative and independent tuning of CRISPRa levels.
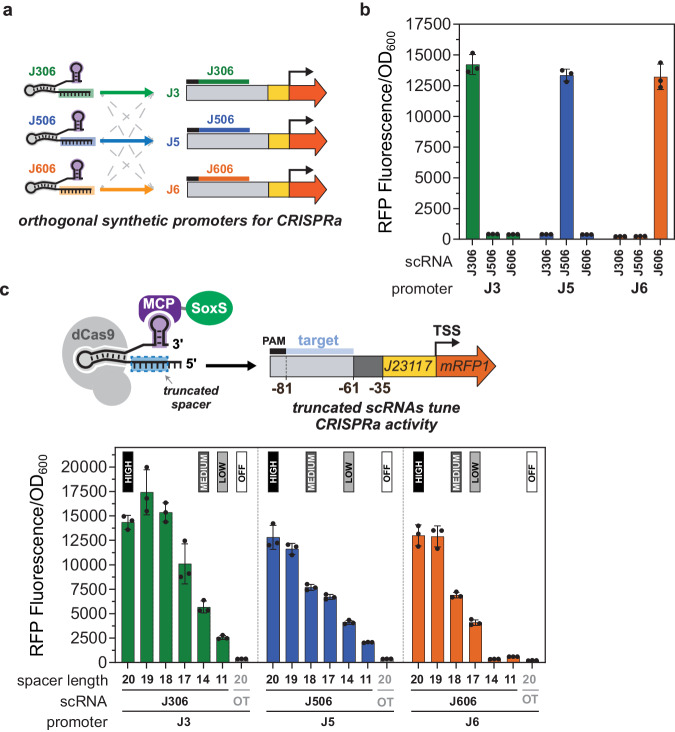
To construct three sequence-orthogonal synthetic promoters, we selected three high-performing scRNAs from the set identified through folding design. Because most randomly selected 20 base sequences will be orthogonal, we did not apply any explicit filters for orthogonality to select these sequences. The sequences included two new scRNAs, termed J506 and J606, and the previously described J306 scRNA with its cognate J3 promoter. All three scRNAs have low Folding Barriers (≤10 kcal/mol), consistent with the threshold criterion for effective scRNA selection. To construct cognate synthetic promoters for J506 and J606, termed J5 and J6, we inserted each target site at the optimal position 81 bases upstream of the transcription start site (Fig. 3a). To minimize repeating sequence elements between the promoters, we inserted distinct sequences in the intervening 26 bases between the target site and the minimal promoter (termed the UP-element), using sequences previously screened to permit high CRISPRa activity in this context12,37 (Supplementary Method 2). We also randomized about 120 bases upstream of the target site PAM in J5 and J6, without introducing additional dCas9 PAMs. From the new J5 and J6 promoters, we observe high levels of CRISPR-activated RFP expression, similar to the expression level from the J3 promoter (Fig. 3b). To confirm orthogonality of J3/J5/J6, we measured the response of each promoter paired with either non-cognate scRNA and observed no activation (Fig. 3b).
Fig. 3. CRISPR activation of orthogonal synthetic promoters can be tuned using truncated scRNAs.
a Orthogonal CRISPR activation was achieved for the J3, J5, and J6 synthetic promoters by the sequence orthogonality of their cognate scRNAs (J306, J506, J606, respectively). While J3 was previously described12, J5 and J6 were selected from our set of 38 synthetic promoters (Fig. 2d) because they generated similar CRISPR-activated expression levels as J3. b Synthetic promoters for CRISPRa can be selectively activated by expressing their cognate scRNAs. Bars represent the Fluorescence/OD600 of strains harboring the J3, J5, or J6 promoters and expressing the cognate or non-cognate scRNAs. c CRISPR-activated expression from the J3, J5, and J6 promoters can be tuned with truncated scRNAs by removing nucleotides from the 5′ end of the spacer. Bars represent the Fluorescence/OD600 of strains harboring J3, J5, or J6 and expressing the cognate scRNAs truncated to 19, 18, 17, 14, and 11 bases. Gray bars represent the baseline expression of the promoters, obtained from strains expressing an off-target scRNA (J206). Labels above bars indicate the spacer length chosen to encode high, medium, low and off expression levels in the combinatorial scRNA library (Fig. 4). Values in panels b and c represent the average ± standard deviation calculated from n = 3 biologically independent samples. The full sequences of the J3, J5, and J6 promoters are described in Supplementary Table 5 and Supplementary Data 4. Source data are provided as a Source Data file.
To generate independently tunable expression from our orthogonal CRISPRa promoters, we considered multiple strategies. Several approaches have been described, generally either by modulating gRNA expression level or by direct modification of gRNA sequence. For example, CRISPRi or CRISPRa activity can be tuned using different strengths of constitutive promoters to drive gRNA expression23,38. Alternatively, introducing mismatches in the gRNA spacer sequence can modulate CRISPRi gene repression39–42, and truncating the gRNA target sequence from the 5’ end has also been shown to reduce CRISPRi activity39. Here, we reasoned that truncation-based tuning would yield a more predictable response than spacer mismatches, and would allow us to keep the same constitutive promoter strength expressing each scRNA. This approach simplifies cloning and decreases the risk of dCas9-binding competition effects43,44.
We screened J3-, J5-, and J6-targeted scRNAs truncated 1–9 bases from the 5’ end to identify guides that encode discrete intermediate levels of CRISPR-activated gene expression. Across all three promoters, scRNA spacer truncation gradually reduced CRISPR-activated expression (Fig. 3c), and from those functions we selected high, medium, and low activation levels. The folding parameters predict similarly high efficacy for all truncations (Folding Barrier ≤10 kcal/mol), while the Net Binding Energy generally becomes less favorable with truncation (Supplementary Table 4). This effect is consistent with the smaller number of RNA bases available to pair with the DNA target, and loosely correlates with output activation (Supplementary Fig. 7). Specifically, the full-length J306 scRNA with a 20 base spacer generated 38-fold activation, and truncated scRNAs with 17, 14, or 11 base spacers tuned CRISPRa to 27-fold, 15-fold, and 7-fold activation, respectively. For the J506 and J606 scRNA truncations, the expected monotonically decreasing trends were observed, although the precise truncations to achieve similar activation levels were not the same (Fig. 3c). In particular, the J606 scRNA was more sensitive to truncation than J306 and J506. For instance, the 14-base J606 truncation activated gene expression by only 2-fold, while the 14-base J306 and J506 scRNAs activated their promoters by 15-fold and 11-fold, respectively. Consistent with previous work investigating DNA-level sequence context effects on CRISPRa37, sequences adjacent to the spacer targets in the J3/J5/J6 promoters might affect truncation response. Even if the energetic parameters here do not quantitatively explain the sensitivity of each promoter’s truncation response (Supplementary Fig. 7), they generally reflect the rank order of the tuned outputs (rs = 0.83 for J306, rs = 1 for J506, rs = 0.94 for J606).
Interestingly, the J306 scRNA with a 19 base spacer generated higher activation than the 20 base spacer (46-fold vs. 38-fold) even though the Net Binding Energy for the 20 base spacer (−32.3 kcal/mol) was similar to that of the 19 base spacer (−31.4). Taken together, the energetic parameters do not indicate impaired folding of the 20 base spacer or any other indication that the 19 base spacer should perform better for CRISPRa. It is possible that spacer truncations could affect transcription of the scRNA itself or could introduce scRNA folding characteristics not captured by our screening parameters. For practical applications, however, we can empirically choose the appropriate scRNA spacer length from within the truncation datasets to obtain tunable high, medium, or low activation from each of the three promoters.
Combinatorial CRISPRa library enables tuning of multi-gene expression programs
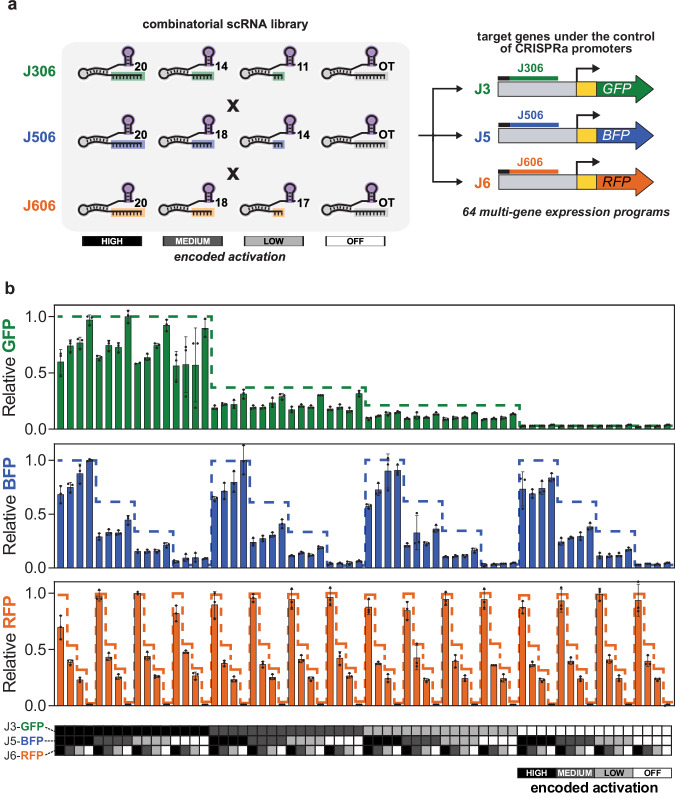
Encoding expression levels directly in multi-scRNA programs creates a straightforward way to implement combinatorial variations in the expression of multi-gene systems. Genes of interest can be cloned under the control of a set of synthetic CRISPRa promoters and tuned by simply changing the identity of the scRNAs transcribed in the cell. For example, driving the expression of three genes with the J3, J5, and J6 promoters and expressing a combination of a J306 scRNA with an 11 base spacer, J506 with a 20 base spacer and J606 with an 18 base spacer would result in low, high, and medium expression of the corresponding genes. By extending such a strategy to encompass all possible combinations of truncated J306, J506, and J606 scRNAs, we can rapidly explore large combinatorial spaces of gene expression under the control of CRISPRa promoters (Fig. 4a).
Fig. 4. Multi-gene expression can be rapidly tuned using combinatorial CRISPRa programs.
a Combinatorial library encoding all combinations of four CRISPR-activated expression levels across three genes. The library expresses three scRNAs (variants of J306, J506, and J606). Each scRNA is present in the library in three truncation variants to generate high, medium and low levels of expression of their target promoters (J3, J5, and J6, respectively). In addition to the three truncation variants, the library contains strains with an off-target scRNA in place of each of the J306, J506, and J606 scRNAs to encode a condition in which the target promoter remains unactivated. The lengths of the J306 scRNA variants are 20, 14, and 11 bases. The J506 scRNA variants are 20, 18, and 14 bases. The J606 scRNA variants are 20, 18, and 17 bases. b Use of the combinatorial scRNA library to specify the expression of multiple genes independently. Each member of the combinatorial scRNA library was delivered to a strain harboring a plasmid expressing J3-gfp, J5-bfp, and J6-rfp reporters, generating 64 strains expressing different combinations of the three fluorescent proteins. Points represent the flow cytometry median of GFP, BFP, and RFP from each strain, normalized to the average of the maximum strain across the experiment. The heatmap table below the plot indicates the encoded promoter expression for each strain, as described on the bottom right. Dashed lines represent the Relative Fluorescence/OD600 of strains harboring only one of the three fluorescent reporters and only the cognate scRNA (tested with RFP output; see Supplementary Data 2 for plasmids and Supplementary Fig. 9a for variation in single-channel expression), again normalized to the maximum value. Bars in panel b represent the average ± standard deviation calculated from n = 3 biologically independent samples, except strain #9, for which only n = 2 samples grew successfully. The sequence of each scRNA in the combinatorial library can be found in Supplementary Data 1. The sequence of the reporter plasmid expressing J3-gfp, J5-bfp, and J6-rfp is included in Supplementary Data 4. Source data are provided as a Source Data file.
We demonstrate the immediate utility of this design strategy by creating a set of genetic tools for combinatorial gene expression profiling. We constructed a library of multi-scRNA plasmids (program plasmids) (Supplementary Data 2 and Supplementary Fig. 8) that encode the expression levels from the set of synthetic CRISPRa promoters, which control a set of desired genes on an output plasmid. Three-gene combinatorial expression profiling is then enabled by simply combining an output plasmid with each member of the program library (Fig. 1), allowing the same scRNA library to be used for arbitrary outputs. We constructed a full library of scRNA plasmid variants to encode all possible combinations of high, medium, low (Fig. 3c) and basal expression of three target genes. Basal expression from each of the targeted promoters was minimal and resulted from an off-target scRNA. Together, the library is composed of 64 plasmids (43) that can be combined with any construct containing genes driven by the J3, J5, and J6 synthetic promoters, resulting in strains encoding 64 different combinations of multi-gene expression (Supplementary Table 5 and Supplementary Data 3).
As an initial validation of our strategy, we tested the combinatorial multi-scRNA library using fluorescent reporter expression. We delivered each of the 64 constructs from the library to an E. coli strain containing GFP, BFP, and RFP reporters under the control of the J3, J5, and J6 promoters, respectively. The resulting strains displayed every combination of high, medium, low, and basal expression for the three reporters. Across this set, the strains displayed variations in relative expression levels consistent with the multi-scRNA programs they contained (Fig. 4b and Supplementary Figs. 9 and 10). However, we also observed that tuning one gene could affect expression of the other genes. First, we found that total expression was reduced by 30–40% when high activation was simultaneously encoded for all three reporters, suggesting that high heterologous gene expression is limited by host expression capacity. Although these effects will vary with different target genes and ribosome binding site strengths, they indicate that maximal expression of multiple genes in a pathway can have unintended consequences that may result in suboptimal behavior. Second, we observed that high expression specifically of RFP had a deleterious effect on GFP and BFP levels (Supplementary Fig. 11). It is well-established that expression burden, metabolic burden, or toxicity can have effects on gene expression levels that are difficult to predict45,46. Our findings underscore the importance of systematically exploring the combinatorial design spaces of multi-gene expression programs to optimize engineered systems. Using this strategy, we applied our CRISPRa tools to build combinatorial expression programs to optimize flux through two engineered metabolic pathways.
Biosynthetic profiling of an engineered tetrahydrobiopterin pathway with combinatorial CRISPRa programs
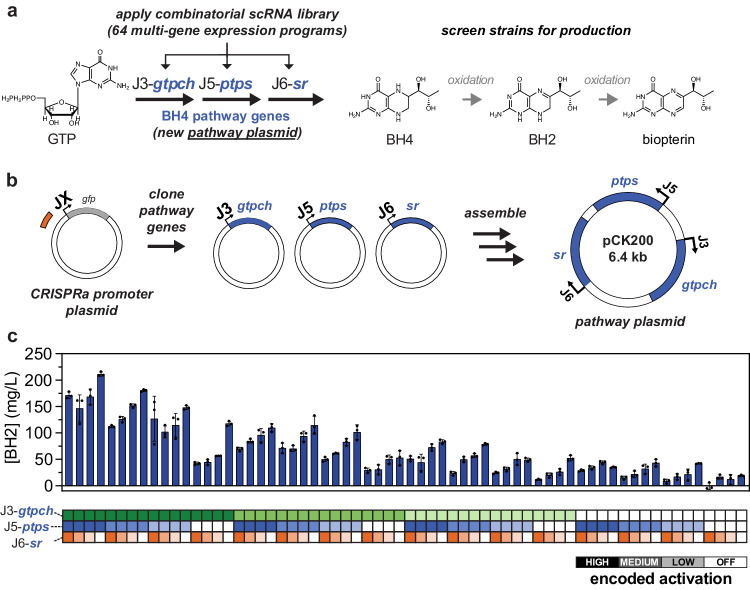
To determine if combinatorial optimization would be effective for metabolic engineering, we applied our CRISPRa promoters and library approach to regulate tetrahydrobiopterin (BH4) biosynthesis through a biopterins production pathway. BH4 is a central cofactor in aromatic amino acid metabolism and a treatment for life-threatening metabolic disorders, including a form of phenylketonuria47. It can be produced from a three-enzyme pathway48–50 using the E. coli gtpch and M. alpina ptps and sr genes, as described previously15. Production can be monitored with a fluorimetric assay48–50, providing a convenient model system for combinatorial screening. We placed codon-optimized gtpch, ptps, and sr genes in a BH4 pathway plasmid with enzyme expression controlled by the J3, J5, and J6 synthetic promoters, respectively (Fig. 5a, b). Co-transforming the BH4 pathway plasmid into E. coli with each member of our combinatorial multi-scRNA library resulted in 64 new strains, each encoding a different combination of high, medium, low, and basal expression of the BH4 pathway enzymes. We monitored biosynthetic flux through this pathway by measuring the fluorescence of the spontaneous BH4 oxidation products dihydrobiopterin (BH2) and biopterin15.
Fig. 5. Combinatorial CRISPRa programs can be applied to tune biosynthetic pathways.
a Tetrahydrobiopterin (BH4) production was tuned by delivering the combinatorial scRNA library to an E. coli strain harboring a BH4 pathway plasmid. BH4 is synthesized from GTP by expressing the gtpch gene from E. coli and the ptps and sr genes from M. alpina. BH4 then undergoes two oxidative decomposition steps yielding dihydrobiopterin (BH2) and biopterin. The BH4 pathway plasmid was constructed by placing the gtpch, ptps and sr genes under control of the J3, J5, and J6 promoters, respectively. b Tuning gene expression in new biosynthetic pathways only requires constructing a new pathway plasmid. The new plasmid is then cotransformed with the same scRNA library from Fig. 4. c Combinatorial tuning of BH4 pathway expression reveals that gtpch activity is limiting and that the sr gene is expressed in excess. Bars represent the average biopterins production of each strain in the combinatorial library harboring the BH4 pathway plasmid. Variations in biopterins production were measured by fluorescence (excitation: 340 nm, emission: 440 nm; see Supplementary Fig. 19). Baseline-subtracted normalized fluorescence values were converted into BH2 concentrations using the calibration curve in Supplementary Fig. 19a. The concentration values are given as BH2 concentration because >80% of the fluorescence signals generated from BH4 production strains have been previously shown to correspond to the BH2 oxidation state15. The x-axis heatmap is color coded to indicate the encoded promoter expression for each strain, as described on the bottom right. Values in panel c represent the average ± standard deviation calculated from n = 3 biologically independent samples. The sequence of the pathway plasmid containing J3-gtpch, J5-ptps and J6-sr is included in Supplementary Data 4. Source data are provided as a Source Data file.
We observed the highest production—211 mg/L BH2—in strains with high expression of the first enzyme in the pathway, GTPCH, indicating that gtpch expression is a sensitive control point in this system (Fig. 5c). Reducing J3-gtpch activation from high to low decreased production by an average of 66%. Changes in expression of the second enzyme, PTPS, had relatively little impact on production across the whole set of combinatorial programs (J5-ptps high to low reduced production by an average of 29%), except for conditions in which its expression was basal (high to basal reduced production by an average of 59%). Interestingly, basal expression of the SR enzyme was not only sufficient for biopterins production, but increasing its expression led to reduction in product titers. For example, increasing J6-sr activation from off-target to high reduced production by an average of 51%. This reduction was widespread and consistent, with 14 out of 16 J6-high strains producing significantly less biopterins than their off-target counterparts. Previous kinetic characterization of SR renders this result unsurprising51, because even basal SR expression provides a vast excess of activity relative to the flux delivered by the upstream pathway. Additional SR beyond the basal level presumably only contributes additional expression burden without increasing overall pathway flux. Taken together, these results identify effective enzyme levels for BH4 biosynthesis through this pathway and highlight that maximal expression of all enzymes is not optimal.
Applying biosynthetic profiling for efficient production of a human milk oligosaccharide
We next applied our CRISPRa system to perform combinatorial expression analysis of a multi-gene pathway for producing the valuable oligosaccharide lacto-N-tetraose (LNT)52,53. Human milk oligosaccharides (HMOs) are major components of human milk54 with substantial effects on infant immune development55, microbiome establishment56,57, anti-inflammation58,59, and more60. Microbial production may provide routes to obtain scalable quantities of HMOs for research, nutrition, and therapeutic applications that are otherwise difficult to obtain using traditional chemical synthesis61,62. LNT is a highly abundant HMO, a valuable formula additive, and a core structure of several other structurally diverse HMOs61,63.
A three-gene pathway consisting of the LacY lactose permease and two heterologous enzymes, LgtA64 and WbgO65, can produce LNT in E. coli52,53 (Fig. 6a). Starting from a lactose feedstock supplied in the media, E. coli LacY imports the lactose into the cell, where LgtA, a β−1,3-N-acetylglucosaminyltransferase from Neisseria meningitidis, produces the intermediate metabolite lacto-N-triose II (LNT II) using the hexose sugar from endogenous UDP-N-acetylglucosamine. WbgO, a β−1,3-galactosyltransferase from E. coli O55:H7, then produces LNT using LNT II and endogenous UDP-galactose. Knocking out endogenous β-galactosidase activity (lacZ) is also necessary to prevent cleavage of the lactose feedstock into its constituent monosaccharides glucose and galactose, which would divert flux away from LNT biosynthesis and toward glycolysis52,61,66–69.
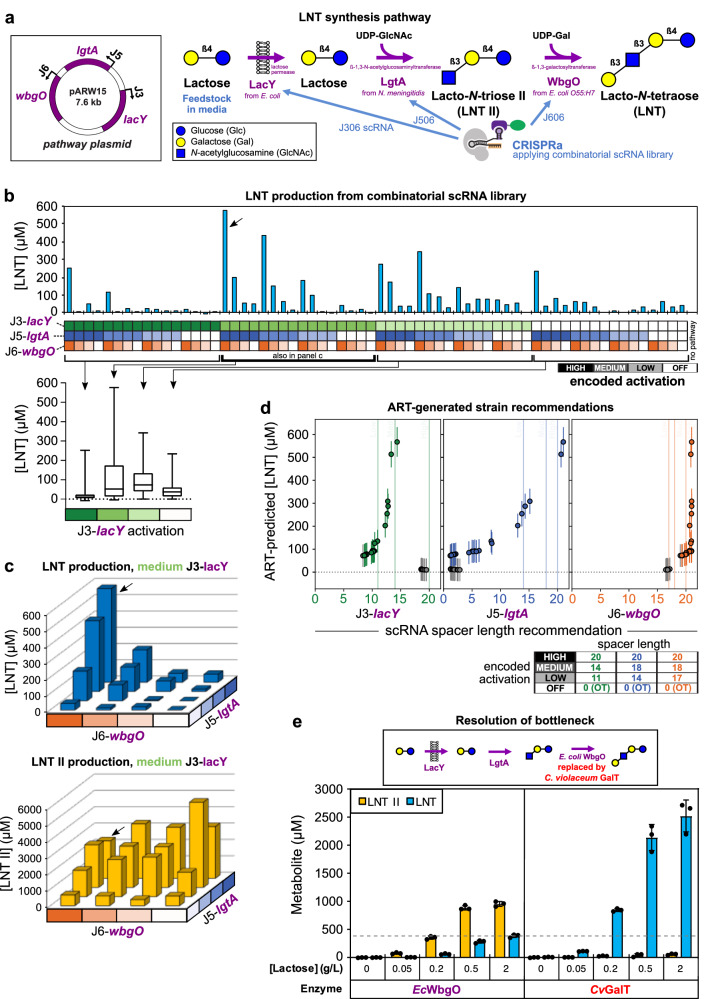
Fig. 6. Combinatorial CRISPRa library applied to an HMO biosynthesis pathway identifies high-producing strains and pathway bottlenecks.
a The LNT pathway consists of lacY, lgtA, and wbgO overexpression controlled by the J3, J5, and J6 promoters, respectively. The substrates UDP-GlcNAc and UDP-Gal come from endogenous metabolism. b HPLC analysis of supernatant from singlicate cultures indicates LNT production levels across the scRNA library. The highest producing strain (#17, black arrow) was used in the galactosyltransferase comparison in e. The x-axis heatmap is color coded to indicate the encoded promoter expression for each strain, as described on the bottom right. The no-pathway culture carries an empty vector. For comparison, LNT II levels are shown in Supplementary Fig. 12. c Dependence of LNT (top) and LNT II (bottom) production on lgtA and wbgO activation highlights sensitivity to wbgO activation and accumulation of LNT II. Only medium-lacY strains are shown here, due to their rich variance across the subset (box plot in b: center line, median; box limits, upper and lower quartiles; whiskers, range). The arrow again indicates strain #17. d Computational strain recommendations from the Automated Recommendation Tool (ART) and their predicted LNT titers. Strains are defined by their scRNA spacer lengths (measured in nucleotides), which determine degree of CRISPR activation (lower right). The 20 strains with highest predicted titer are highlighted in color on each subgraph, with the rest shown in gray. The same 32 strains are shown on each subgraph. Spacer lengths defined as high, medium, and low expression in experimental data are indicated as vertical lines. Points in d represent each recommended strain’s specific truncation for that scRNA, while error bars indicate the 95% credible interval of the predictive posterior distribution. See Supplementary Fig. 15 for how recommendations are combined within each strain. e A more active enzyme from C. violaceum73 (right) resolves accumulation of LNT II (left), at various initial feedstock concentrations. The horizontal line indicates LNT titer from WbgO and 2 g/L initial lactose; CvGalT achieves similar titer using only 0.05–0.2 g/L initial lactose. Bar values in e represent the average ± standard deviation calculated from n = 3 biologically independent samples. Source data are provided as a Source Data file.
To establish CRISPRa control of LNT production, we generated an output plasmid in which expression of the codon-optimized lacY, lgtA and wbgO genes are independently controlled by the J3, J5, and J6 synthetic promoters, respectively (Fig. 6a). We delivered this LNT pathway plasmid, together with our existing multi-scRNA library, to the lacZ knockout E. coli strain JM109. Using HPLC to quantify accumulation in the culture supernatant of LNT and intermediate metabolite LNT II, we found a wide range of extracellular titers across the library, from zero to nearly 600 μM LNT (nearly 425 mg/L) (Fig. 6b and Supplementary Figs. 12 and 13). A majority of the strains produced low or no LNT in supernatant, including some of the highest-expressing variants. For example, the strain with maximal expression (high-lacY, high-lgtA, high-wbgO) produced only 252 μM LNT (178 mg/L), while a strain with reduced lacY activation (medium-lacY, high-lgtA, high-wbgO) produced 576 μM LNT (408 mg/L). In general, we found that LNT production was compromised in the strains where lacY expression was highest, with only two out of 16 high-lacY strains producing >50 μM LNT (Fig. 6b, left). This finding is consistent with toxic proton transport resulting from LacY activity70,71, and exemplifies an underlying mechanism of non-monotonic genotype-phenotype relationship. When lacY is reduced to medium levels, there is a large spread in LNT production, with eight out of 16 strains producing >50 μM LNT (Fig. 6b). The J3-lacY local maximum highlights the importance of exploring a wide combinatorial space of enzyme expression, and the high variation of medium-lacY LNT production indicates the need for additional optimization of the other enzymes.
To understand the relative importance of LgtA and WbgO, we focused on the subset of medium lacY strains. In the medium-lacY sublibrary (Fig. 6c), LNT production appeared to be more sensitive to variation of J6-wbgO expression than to variation of J5-lgtA expression. High LNT production (>400 uM) required high wbgO expression, indicating a steep expression-production relationship. For lgtA, high production was possible at high or medium expression, indicating a more gradual expression-production relationship. Reducing wbgO expression from high to low decreased titer from 576 μM to 56 μM (90.3% reduction compared to the maximum), but reducing lgtA expression from high to low only decreased titer to 182 μM (68.4% reduction) (Fig. 6c). In most of these expression combinations, we also observed significant extracellular accumulation of the LNT II intermediate, the substrate for WbgO to convert into LNT. This accumulation was only avoided when lgtA was not activated (basal expression). When LNT II did accumulate, its titer did not depend strongly on low, medium, or high lgtA activation (Fig. 6c). High LNT II titers were much more widespread across the library than high LNT titers (35 strains with LNT II titer above 25% maximal, compared to 10 strains for LNT) (Supplementary Fig. 12). Taken together, these results suggest that limited β−1,3-galactosyltransferase activity of WbgO is a metabolic bottleneck in this pathway, confirming previous observations53. Our use of a combinatorial library to profile a multi-enzyme design space allowed us to easily characterize bottlenecks by probing for sensitive control points in the pathway.
A machine-learning analysis further validated the wbgO bottleneck. We used scRNA truncation levels from the library strains as inputs to the Automated Recommendation Tool (ART)72 to predict LNT production as a response variable, achieving high prediction accuracy (R2 = 0.71, Supplementary Fig. 14) after training with the experimental LNT production data from the library. ART then used the predictions and uncertainties to make recommendations of the most productive enzyme expression levels. The most highly recommended strains consistently prioritized maximal wbgO expression to achieve high LNT production. ART did not provide similarly stringent recommendations for lacY and lgtA (Fig. 6d and Supplementary Fig. 15), allowing substantial expression variation among LNT-productive strain recommendations. In agreement with the experimental library screen, these recommendations identify the wbgO bottleneck as a high priority for optimization, despite ART being unaware of LNT II accumulation. Furthermore, when allowed to recommend any spacer length up to 21 nucleotides, whether tested experimentally or not, ART frequently recommended wbgO levels above the highest experimentally tested level. Collectively, these data underscore the idea that WbgO (β−1,3-galactosyltransferase) activity should be increased beyond maximal CRISPR activation of wbgO in this context.
To increase β−1,3-galactosyltransferase activity, we replaced WbgO with the GalT enzyme from Chromobacterium violaceum (CvGalT), an enzyme with faster turnover73. We placed CvGalT under J6 control in the LNT pathway plasmid and paired it with the previously highest-producing scRNA library strain (medium-lacY, high-lgtA, high-CvGalT). Compared to the corresponding WbgO strain, the CvGalT strain produced a 5- to 10-fold increase in supernatant LNT titer, while LNT II accumulation decreased 5- to 20-fold, with the precise effect depending on the feedstock concentration (Fig. 6e). These paired effects reflect the higher ability of CvGalT to bind and convert LNT II before it is exported to accumulate in the supernatant74. The highest supernatant titer achieved from the CvGalT-containing system increased to 2.52 mM LNT (1.78 g/L), compared to 0.576 mM (0.407 g/L) from the WbgO-containing system. This improvement reflects a 4.4-fold increase in mol/mol yield on lactose from 0.099 to 0.432. Relieving the bottleneck identified by our biosynthetic profiling approach therefore resulted in significantly more LNT production by improving the efficiency of the β−1,3-galactosyltransferase reaction.
Biosynthetic profiling of the LNT pathway by combinatorial CRISPRa indicated both the effects of lacY overexpression and the relative sensitivity of production to wbgO expression, demonstrating the potential of this approach to rapidly optimize enzyme expression levels. Crucially, the library is readily portable to different pathways. Applying combinatorial CRISPRa to a different pathway only requires a new output plasmid with the pathway enzymes expressed by the existing synthetic promoters, followed by cotransformation with the existing library of scRNA program plasmids.
Discussion
Synthetic biology and metabolic engineering offer a route for sustainable bioproduction of chemicals from renewable feedstocks. Many of these products are metabolically complex, requiring precise control over multi-gene networks to effectively redirect metabolic flux. Combinatorial CRISPRa programs can provide precise control over multiple targets, but require predictable scRNA efficacy. Developing general bacterial gRNA design rules and avoiding the typical trial-and-error validation of gRNA functionality will be an important factor in advancing multi-gene regulation programs. By combining computational RNA folding and experimental analyses, we uncovered strong correlations (rs = 0.7–0.8) between CRISPR-activated expression and a set of thermodynamic and kinetic scRNA folding parameters75,76. Among the parameters examined, kinetic parameters associated with post-transcriptional RNA folding have the largest impacts on CRISPRa.
We found that a single kinetic parameter, Folding Barrier, can accurately predict bacterial CRISPRa across a broad range of expression levels, with a failure rate of zero for the set of 39 scRNA designs tested. We speculate that the predictive value of Folding Barrier may be higher than that of Folding Energy because binding to dCas9 may stabilize the active scRNA structure (Supplementary Figs. 2 and 3). The kinetic barrier to access the active structure determines the likelihood of dCas9 trapping the RNA in that structure, and is potentially more important than the intrinsic thermodynamic stability of the free RNA structure. dCas9 binding should also provide some resistance to RNA degradation77. The high predictability of scRNA design supplied by Folding Barrier should significantly facilitate the forward engineering of complex bacterial CRISPRa/i systems. Multi-guide applications that have remained inefficient or impractical with current gRNA failure rates, such as combinatorial expression screening78 or model- and data-driven strain engineering and optimization18, can therefore be accelerated. Recent metabolic engineering successes in related systems emphasize the value of predictive gRNA design22,79.
The Folding Barrier metric outperformed current state-of-the-art gRNA design tools in its ability to predict CRISPRa activity21,31. There are many possible explanations for the inability of existing models to apply to bacterial CRISPRa systems. It remains an open question whether guide RNA design rules derived from one function in one system, most commonly genome editing in eukaryotes, can be transferred to other functions and systems such as CRISPR gene regulation in prokaryotes. First, many of these models account for genome structure, which will vary greatly between eukaryotes and prokaryotes80,81. Second, in regression models trained on large gene editing datasets, it is difficult to decouple gRNA efficiency from feedback on gene expression as part of the overall gene regulatory network, and therefore the predictions of these models may not be readily transferable between organisms. Third, the models underlying these gRNA design tools were trained on unmodified gRNAs and do not capture potential folding effects of extended RNA elements included in scRNAs for bacterial CRISPRa. These models could likely be improved by incorporating biophysical parameters in their predictions. Finally, considerations of nucleic acid interactions in gRNA design models tend to focus on the thermodynamics of spacer-DNA interactions, and neglect other important aspects of gRNA folding30. For instance, a number of studies that model the thermodynamics of gRNA-Cas9-DNA complex formation employ parameters describing the impact of structure within the spacer sequence (e.g. ∆GU) and of spacer-target hybridization (e.g. ∆GH)30,82,83. Here, the conceptually similar parameter Binding Energy does not predict bacterial CRISPRa as well as Folding Energy and Net Binding Energy, which consider the spacer sequence in the context of the full scRNA sequence and structure (Supplementary Figs. 2-4). Developing models that combine solely sequence-based kinetic folding parameters with heuristics from large-scale functional screening should further improve our ability to design modified guide RNAs for bacterial CRISPRa.
Optimal multi-gene pathway expression could be influenced by many factors, possibly including total burden, enzyme imbalance, or toxic enzyme or metabolite effects. The difficulty in predicting these systems-level interactions means that finding global production optima often requires exploring large design spaces84. Toward this end, we successfully developed a scRNA library that can implement all combinations of four truncation-defined expression levels across three chosen genes, totaling 64 possible expression programs. For each of the pathways we examined, we found the optimal production to occur at non-maximal expression levels in at least one channel of expression (rfp, sr, and lacY in Figs. 4, 5, and 6, respectively). Production from these pathways therefore maps ruggedly to the underlying design space of enzyme expression, and systematically profiling these effects revealed high-producing strains and also pathway bottlenecks potentially sensitive to optimization. Pursuing bottleneck optimization in the LNT pathway with an improved enzyme variant pushed test-tube-scale titers into g/L magnitude (1.78 g/L). At the scale of test tubes typical of early-stage strain development, Sugita and Koketsu reported 2.96 g/L LNT74, a similar but higher titer than observed here. Notably, the previous study used 10 g/L lactose feedstock (0.143 mol/mol yield on lactose) compared to only 2 g/L in the present work (0.432 mol/mol), representing a 3-fold higher yield from the combinatorial CRISPRa system.
Well-tuned multi-gene expression programs identified through biosynthetic profiling provide starting points for later-stage optimization through genome engineering and process development25. A major challenge for the field is to effectively and efficiently optimize production from such starting points. Although beyond the scope of the current study, groups applying such efforts have often achieved 1–5 g/L LNT production titers in shake flasks and 5–50 g/L production in fed-batch bioreactors85. As an illustration, 8-fold increases in LNT titer (from 3.11 g/L to 25.4 g/L) and >2-fold increases in LNT yield on lactose (from 0.301 mol/mol to 0.773 mol/mol) were seen when scaling up a strain from 25 mL shake flask cultures to 1 L fed-batch bioreactor conditions, respectively86. We expect that similar increases in titer could be achieved by cultures of our optimized strain scaled up to similar fed-batch conditions.
Broadly speaking, biosynthetic profiling using trans-acting scRNAs can greatly reduce the time needed to tune multi-gene programs, compared to traditional cis-acting tools like promoter, RBS, or ribozyme libraries87,88. We expect that the combinatorial scRNA library described here will provide a straightforward approach to identifying production maxima and optimizing burdensome pathways or toxic intermediate accumulation, ahead of later-stage optimization. In the future, this approach could be extended to non-model hosts with metabolic and physiological capabilities suitable for next-generation bioproduction applications89–91.
Many bioproduction pathways and circuits of interest will require expression programs with more than three synthetic promoters or a combination of heterologous pathway control and genomic targeting. The scRNA design rules from this work can be applied alongside CRISPRa promoter design principles37 to generate a virtually unlimited supply of new, high dynamic range, CRISPR-activatable promoters. Beyond the three spacer targets that we focused on here (J306, J506, and J606), there are 16 additional scRNA spacer sequences with >75% of J306 activity (Fig. 2d and Supplementary Data 1) that are available for immediate use (Supplementary Fig. 16 and Supplementary Method 2). If desired, an arbitrary number of new scRNA spacer sequences can be designed using the Folding Barrier screening metric in the code accompanying this publication. Thus, additional nodes of heterologous control can be added as new scRNA-promoter pairs. In parallel, nodes of endogenous control can be added as scRNAs (CRISPRa) or gRNAs (CRISPRi) that target native genes.
Expanding beyond the three-node programs used here would allow activation of larger pathways, endogenously-targeted CRISPRa/i16,92 for flux optimization, or dynamic gene regulation through biosensors93,94. Combinatorial CRISPRa programs could also be extended to increase expression variation resolution or use alternative tuning methods19,22,95. There may be a practical limit on the size of functional scRNA/gRNA arrays, perhaps due to binding competition for a shared dCas9 pool43,44. Principles of gRNA design, including those reported in this work, and some autoregulatory circuit designs96 could be used to increase this limit and build larger multi-guide programs. Guide RNA engineering that minimizes the need for trial-and-error verification of CRISPR function should enable the construction of larger programs, which in turn should enable CRISPR control of larger metabolic pathways.
For large combinatorial libraries of genetic circuits, higher-throughput screening methods like biosensing technologies would be needed to screen through the added diversity18,97,98. For design spaces too large for current screening methods, data-driven and model-guided approaches like ART can be used to explore the full design space, informed by experimental efforts focused only on the most likely subsets of design parameters (Supplementary Fig. 17). An optimal subset size depends on the complexity of the pathway to be optimized, but the experimental CRISPRa profiling approach can ease the construction of these subsets.
Iterative cycles of model-guided optimization and data-driven model refinement present a promising path forward for rapid generation and optimization of biosynthetic pathways. The value of this approach is especially demonstrated when used together with combinatorial CRISPRa/i programs to access model predictions and build iteratively improved strains. Optimized metabolic engineering programs can help realize a circular bioeconomy that decreases our reliance on fossil feedstocks for production of industrial chemicals and materials. To help meet this challenge, synthetic biologists can use the tools presented in this work to rapidly optimize strains for bioproduction of valuable chemicals from renewable feedstocks.
Methods
Bacterial strains and plasmid construction
Bacterial strains used in this study are described in Supplementary Table 1. JM109 was a gift from Joachim Messing (Addgene plasmid #49761)99. Plasmids were cloned using standard molecular biology protocols and are described in Supplementary Data 2. Guide RNA target sequences are provided in Supplementary Data 1. Orthogonal target sequences replacing J306 were 20 bp sequences selected at random from the human genome. Plasmids expressing the CRISPRa components (dCas9, the activation domain MCP-SoxS, and one or more scRNAs) were constructed using a p15A vector. S. pyogenes dCas9 (Sp-dCas9) was expressed using the endogenous Sp.pCas9 promoter. The MCP-SoxS activation domain containing mutant SoxS (R93A and/or S101A; see Supplementary Data 2)12 was expressed using the BBa_J23107 promoter (http://parts.igem.org). The scRNAs were expressed using either the BBa_J23119 promoter or the BBa_J23105 (Supplementary Fig. 8), unless otherwise noted. scRNAs used the b2 design, in which the endogenous tracr terminator hairpin upstream of MS2 is removed11. Plasmids expressing target genes for CRISPRa were constructed using a low-copy pSC101** vector. mRFP1, sfGFP, mTagBFP2, or metabolic pathway genes were expressed from the weak BBa_J23117 minimal promoter preceded by synthetic DNA sequences containing the CRISPRa target sites. Pathway gene RBSs were selected from a previously reported list100 and predicted to have high strength101 in the new context. Transcriptional terminators used for scRNAs and output genes are listed in Supplementary Table 6.
Computational analysis of scRNA activity
Energetic parameters were generated using the RNAfold, RNAeval, RNAduplex, and Findpath programs from the ViennaRNA Package version 2.3.527. Sequences of full scRNAs were input to a custom script that returned the following parameters. Folding Barrier was calculated by using the folding trajectories identified by Findpath28 to predict the barrier height for the direct refolding pathway from the MFE structure to the active structure (Supplementary Fig. 2). The active structure is defined as the structure in which the Cas9-binding handle is correctly folded and the spacer is unstructured. Binding Energy was calculated by evaluating the RNA-RNA free energy of the spacer sequence binding to its reverse-complement sequence using RNAduplex. The Folding Energy, or free energy difference between the MFE structure and the active structure, was evaluated using RNAfold with constraint folding. Folding Energy was then added to the Binding Energy in order to estimate the net energetics of binding to a single-stranded target sequence. This sum yields the Net Binding Energy, or the free energy difference between the MFE and the bound state. All scRNA sequences were verified to have a prediction of correct folding of the MS2 aptamer at the 3’ end, to avoid confounding cases of target occupancy without bound MCP-SoxS.
For the purpose of comparison to this work’s scRNA efficacy predictions, the Doench ‘16, Azimuth in vitro, and Moreno-Mateos tools for CRISPR guide design and evaluation were implemented using the CRISPOR webserver (http://crispor.tefor.net/)102. The 20 bp variable target sites for scRNA-directed CRISPRa flanked by 50 bp of upstream and 50 bp of downstream sequence (120 bp total) were used as target DNA inputs (Upstream flanking sequence, variable target site, PAM site, downstream flanking sequence: CCCTAGGACTGAGCTAGCTGTCAATCTATAATCGCAACTTCAAGACGACGNNNNNNNNNNNNNNNNNNNNAGGAGAAGTGAGGAGACGAGCGAACGCGTCGTACGAGCTTTATGCATCTT). Analysis was carried out with the default settings for “No Genome” and Protospacer Adjacent Motif (PAM) set to “20bp-NGG - SpCas9, SpCas9-HF1, eSpCas9 1.1”. Each 20 bp target was evaluated using the “predicted guide efficiency” outputs generated by the respective CRISPR guide design tools.
Construction of combinatorial scRNA library
To encode high, medium, and low activation of the J3, J5, and J6 promoters, we selected the 20, 14, and 11 nucleotide variants of J306; the 20, 18, and 14 nucleotide variants of J506; and the 20, 18, and 17 nucleotide variants of J606, respectively. For all three promoters, a fourth, unactivated condition was included via an off-target scRNA with a spacer sequence not complementary to any of the synthetic promoters. In the CRISPRa component plasmid library, a three-member array of scRNA expression, each with its own BBa_J23105 promoter and terminator, was constructed for every possible combination of the J306, J506, and J606 truncation variants. Including the off-target versions, this resulted in a 64-member combinatorial library of CRISPRa component plasmids, accounting for all combinations of high, medium, low, and baseline expression of all three synthetic promoters (Supplementary Data 3).
Plate reader experiments
Single colonies from LB-agar plates were inoculated in triplicate in 500 μL EZ-RDM (Teknova, M2105) with 2 g/L glucose supplemented with appropriate antibiotics and grown in 96-deep-well plates at 37 °C and shaking on a microplate orbital shaker (Heidolph Titramax 1000) overnight. For mRFP1 detection, 150 μL of the overnight culture were transferred into a flat, clear-bottomed black 96-well plate and the OD600 and fluorescence (excitation wavelength: 540 nm; emission wavelength: 600 nm) were measured in a Biotek Synergy HTX plate reader for Figs. 2 and 3, and Supplementary Figs. 1, 3, 5–8, and 9a. For sfGFP (ex 485 nm, em 528 nm), mTagBFP2 (ex 400 nm, 455 nm), and mRFP1 (ex 540 nm, em 600 nm) detection in Supplementary Fig. 9b, 150 µL of the overnight culture were transferred into a flat, clear-bottomed black 96-well plate and measured in a monochromator-equipped plate reader (Biotek Synergy H1). Kinetic growth data in Supplementary Fig. 10 were obtained from 200 µL cultures set up in a flat, clear-bottomed black 96-well plate, avoiding edge wells, and measured in the Biotek Synergy H1 plate reader at 37 °C with shaking for 18 h.
Flow cytometry
Single colonies from LB-agar plates were inoculated in triplicate in 500 μL EZ-RDM (Teknova, M2105) with 2 g/L glucose supplemented with appropriate antibiotics and grown in 96-deep-well plates at 37 °C and shaking on a microplate orbital shaker (Heidolph Titramax 1000). Overnight cultures were diluted in 1:100 in DPBS and analyzed on a MACSQuant VYB flow cytometer (Miltenyi Biotec) using the following strategy to gate for single cells11. A side scatter threshold trigger (SSC-H) was applied to enrich for single cells. A narrow gate along the diagonal line on the SSC-H vs SSC-A plot was selected to exclude the events where multiple cells were grouped together. Within the selected population, events that appeared on the edges of the FSC-A vs. SSC-A plot and the fluorescence histogram were excluded. We observed that this cytometer offered clearer separation and quantification of the three colors than a monochromator-equipped plate reader (Biotek Synergy H1) (Supplementary Fig. 18). For sfGFP detection, the excitation wavelength was 488 nm and emission wavelength was 525 nm (50 nm bandpass). For mTagBFP2 detection, the excitation wavelength was 405 nm and emission wavelength was 450 nm (50 nm bandpass). For mRFP1 detection, the excitation wavelength was 561 nm and emission wavelength was 615 nm (20 nm bandpass). Data were analyzed using FlowJo 10.0.7. Median values were normalized to the highest observed value within each channel and were baseline-subtracted using a strain lacking the genes encoding the fluorescent proteins.
Biopterin production experiments
Single colonies from LB-agar plates were inoculated in triplicate in 500 μL EZ-RDM (Teknova, M2105) with 2 g/L glucose supplemented with appropriate antibiotics and grown overnight in 96-deep-well plates at 37 °C with shaking. 100 μL of the overnight culture were transferred into a flat, clear-bottomed black 96-well plate and the OD600 and fluorescence (excitation wavelength: 340 nm; emission wavelength: 440 nm) were measured in a monochromator-equipped plate reader (Tecan Infinite M1000) to assess pteridine production15,103–105. Fluorescence values were normalized across different experimental days (Supplementary Fig. 19), then baseline-subtracted using a strain harboring an empty output plasmid. In a previous report15, the majority of BH4 produced from this pathway was found to be spontaneously oxidized into BH2 (>80%). Therefore, we attributed all of the fluorescence output to BH2 species and used spiked-in standards to calculate BH2 concentration. Standard curves were generated by spiking the commercially available BH2 standard (Cayman Chemical, 81882) into cultures of the strain harboring an empty output plasmid (Supplementary Fig. 19).
Lacto-N-tetraose production experiments
Single colonies from LB-agar plates were inoculated in singlicate in 2 mL EZ-RDM (Teknova, M2105) with 10 g/L glucose, 2 g/L lactose and supplemented with appropriate antibiotics. For the JM109 strain, agar plates used 100 μg/mL chloramphenicol and 100 μg/mL carbenicillin to avoid slightly chloramphenicol-resistant background growth, but liquid cultures used the more typical concentrations of 25 μg/mL chloramphenicol and 100 μg/mL carbenicillin. Cultures were grown in 14 mL polypropylene culture tubes at 37 °C with shaking for 48 h. 500 μL of supernatant from each culture were loaded onto 10 kDa microcentrifuge filters (Millipore, UFC501096) and spun for 20 min at 14,000 rcf. 1 μL of filtered supernatants were assayed with a Shimadzu HPLC using UV-vis detection at 210 nm. Lacto-N-tetraose (LNT) was separated using a Rezex ROA-Organic Acid H+ column (Phenomenex, 00H-0138-K0) and a 20 mM H2SO4 isocratic mobile phase. A standard curve was prepared by spiking known amounts of LNT or LNT II into supernatants derived from cultures of JM109 E. coli transformed with empty vectors. Product LNT was observed at 10.6 minutes, and intermediate LNT II, a triose, was observed at 11.4 minutes. LNT and LNT II peak areas were normalized by the area of an endogenous peak observed at 9.1 minutes. Normalized peak areas were baseline-subtracted using a control strain lacking the pathway genes. Cell pellets also contained significant LNT, as previously reported53 and verified in pellets lysed by boiling, but the difficulty of consistently quantifying lysis efficiency and the rich variation in supernatant titers led us to consider mainly supernatant data for comparative analysis.
ART predictions and recommendations
The Automated Recommendation Tool (ART)72 was trained on the 64 experimental LNT strains, with J3-lacY, J5-lgtA, and J6-wbgO CRISPRa variations as input variables and LNT production as the response variable. ART is an ensemble model that linearly combines a variety of machine learning models. Models are cross-validated individually on the data, and the weight for each model represents its performance (higher for better-performing models, lower for worse-performing ones). These weights are considered as random variables with probability distributions obtained through Monte Carlo sampling. This approach enables quantification of both the prediction mean and uncertainty for any given input data. Predictions are possible at any point in the possible design space, not limited to the discrete high, medium, low, and off-target activation levels comprising the experimental library. ART was trained, however, using the exact activation levels from the experimental library, expressed as spacer length in nucleotides (e.g. 20 for high, 14 for medium, and 11 for low in the J3 case). In all cases, off-target spacers were expressed as an input of 0. Cross-validation correlations were also computed using exact library activation levels.
For the strain recommendations, strains are defined by their recommended input levels, expressed in scRNA spacer length for that channel. ART was allowed to recommend any spacer length from 0 to 21 nucleotides (non-integers allowed), with the constraint that new designs had to be at least one nucleotide away (in at least one dimension) from other recommendations and from training data. The 32 recommended strains resulting in the highest predicted LNT concentration were obtained from ART. In this work, recommendations were fully exploitative (α = 0), meaning that they prioritized maximizing LNT as opposed to minimizing the uncertainty in LNT predictions.
Statistics
Statistical significance was calculated using two-tailed unpaired Welch’s t-tests. Quantitative correlations are expressed as Pearson correlations. Rank-order correlations are expressed as Spearman correlations. Hill function (Fig. 2d) was fitted as the following nonlinear function in GraphPad Prism 8.4.3.686, using least squares regression:
| 1 |
Dose-response function (Supplementary Fig. 8) was fitted as the following nonlinear function in GraphPad Prism, using least squares regression:
| 2 |
Simple linear and exponential fits (Supplementary Figs. 1, 7, 13, and 17a) were performed using default settings in GraphPad Prism or Microsoft Excel 15.17.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Semira Beraki for technical assistance and Venkata P. Chavali for technical assistance and helpful discussions. We thank members of the Zalatan and Carothers groups for advice. This work was supported by U.S. National Science Foundation Awards MCB 1817623 (awarded to J.G.Z. and J.M.C.), MCB 2032794 (J.M.C. and J.G.Z.), CBET 1844152 (J.M.C.), and U.S. Department of Energy Awards DE-EE0008927 and DE-SC0023091 (J.M.C., J.G.Z., and H.G.M.) and a grant from BASF, Inc. (J.M.C.).
Author contributions
J.F., D.S.Y., I.F., H.G.M., J.G.Z., and J.M.C. designed experiments. J.F., D.S.Y., I.F., R.C., C.K., A.W., T.G.P., and P.C.K. performed experiments and analyzed data. J.F., D.S.Y., I.F., R.C., C.K., P.C.K., J.G.Z., and J.M.C. wrote and edited the manuscript with input from all of the authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
A reporting summary for this article is available as a Supplementary Information file. Data supporting the findings of this work are available within the paper and its Supplementary Information files. Source data are provided with this paper.
Code availability
Custom Python code to analyze input RNA and generate the energetic parameters described in this work is available on GitHub [https://github.com/carothersresearch/gRNA_screen_docker]106 and can be run directly in that environment using a Codespace or locally using a Docker image. Jupyter notebooks to view and reproduce the ART results from this paper are available on GitHub [https://github.com/carothersresearch/art_lnt]107. These notebooks can be viewed on GitHub or run in an ART Docker container after acquiring a license. See https://github.com/JBEI/ART for software and licensing details.
Competing interests
The University of Washington has filed a patent (WO2022150311A1) covering the scRNA analysis, scRNA forward design, and combinatorial CRISPRa, and listing J.M.C., J.G.Z., D.S.Y., and J.F. as inventors. J.M.C., J.G.Z., D.S.Y., and J.F. have financial interests in Wayfinder Biosciences, Inc. The remaining authors declare no competing interests related to this work.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jason Fontana, David Sparkman-Yager, Ian Faulkner.
Contributor Information
Jesse G. Zalatan, Email: zalatan@uw.edu
James M. Carothers, Email: jcaroth@uw.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50528-1.
References
- 1.Hodgson, A., Alper, J. & Maxon, M. TheU. S. Bioeconomy: Charting a Course for a Resilient and Competitive Future. Schmidt Futures 10.55879/d2hrs7zwc (2022).
- 2.Intasian, P. et al. Enzymes, in vivo biocatalysis, and metabolic engineering for enabling a circular economy and sustainability. Chem. Rev.121, 10367–10451 (2021). 10.1021/acs.chemrev.1c00121 [DOI] [PubMed] [Google Scholar]
- 3.Lee, S. Y. et al. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal.2, 18–33 (2019). 10.1038/s41929-018-0212-4 [DOI] [Google Scholar]
- 4.Nielsen, J. & Keasling, J. D. Engineering cellular metabolism. Cell164, 1185–1197 (2016). 10.1016/j.cell.2016.02.004 [DOI] [PubMed] [Google Scholar]
- 5.Han, T., Nazarbekov, A., Zou, X. & Lee, S. Y. Recent advances in systems metabolic engineering. Curr. Opin. Biotechnol.84, 103004 (2023). 10.1016/j.copbio.2023.103004 [DOI] [PubMed] [Google Scholar]
- 6.Jung, S.-W., Yeom, J., Park, J. S. & Yoo, S. M. Recent advances in tuning the expression and regulation of genes for constructing microbial cell factories. Biotechnol. Adv.50, 107767 (2021). 10.1016/j.biotechadv.2021.107767 [DOI] [PubMed] [Google Scholar]
- 7.Xu, X. & Qi, L. S. A CRISPR–dCas toolbox for genetic engineering and synthetic biology. J. Mol. Biol.431, 34–47 (2019). 10.1016/j.jmb.2018.06.037 [DOI] [PubMed] [Google Scholar]
- 8.Shi, S., Qi, N. & Nielsen, J. Microbial production of chemicals driven by CRISPR-Cas systems. Curr. Opin. Biotechnol.73, 34–42 (2022). 10.1016/j.copbio.2021.07.002 [DOI] [PubMed] [Google Scholar]
- 9.Vigouroux, A. & Bikard, D. CRISPR tools to control gene expression in bacteria. Microbiol. Mol. Biol. Rev.84, e00077–19 (2020). 10.1128/MMBR.00077-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casas-Mollano, J. A., Zinselmeier, M. H., Sychla, A. & Smanski, M. J. Efficient gene activation in plants by the MoonTag programmable transcriptional activator. Nucleic Acids Res.51, 7083–7093 (2023). 10.1093/nar/gkad458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong, C., Fontana, J., Patel, A., Carothers, J. M. & Zalatan, J. G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun.9, 2489 (2018). [DOI] [PMC free article] [PubMed]
- 12.Fontana, J. et al. Effective CRISPRa-mediated control of gene expression in bacteria must overcome strict target site requirements. Nat. Commun.11, 1–11 (2020). 10.1038/s41467-020-15454-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tickman, B. I. et al. Multi-layer CRISPRa/i circuits for dynamic genetic programs in cell-free and bacterial systems. Cell Syst.13, 215–229.e8 (2022). 10.1016/j.cels.2021.10.008 [DOI] [PubMed] [Google Scholar]
- 14.Barbier, I. et al. Synthetic gene circuits combining CRISPR interference and CRISPR activation in E. coli: importance of equal guide RNA binding affinities to avoid context-dependent effects. ACS Synth. Biol.12, 3064–3071 (2023). 10.1021/acssynbio.3c00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiattisewee, C. et al. Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metab. Eng.66, 283–295 (2021). 10.1016/j.ymben.2021.04.002 [DOI] [PubMed] [Google Scholar]
- 16.Fenster, J. A. et al. Dynamic and single cell characterization of a CRISPR-interference toolset in Pseudomonas putida KT2440 for β-ketoadipate production from p-coumarate. Metab. Eng. Commun.15, e00204 (2022). 10.1016/j.mec.2022.e00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozaeva, E. et al. Model-guided dynamic control of essential metabolic nodes boosts acetyl-coenzyme A–dependent bioproduction in rewired Pseudomonas putida. Metab. Eng.67, 373–386 (2021). 10.1016/j.ymben.2021.07.014 [DOI] [PubMed] [Google Scholar]
- 18.Fontana, J., Sparkman-Yager, D., Zalatan, J. G. & Carothers, J. M. Challenges and opportunities with CRISPR activation in bacteria for data-driven metabolic engineering. Curr. Opin. Biotechnol.64, 190–198 (2020). 10.1016/j.copbio.2020.04.005 [DOI] [PubMed] [Google Scholar]
- 19.Byun, G., Yang, J. & Seo, S. W. CRISPRi-mediated tunable control of gene expression level with engineered single-guide RNA in Escherichia coli. Nucleic Acids Res.10.1093/nar/gkad234 (2023). [DOI] [PMC free article] [PubMed]
- 20.Tian, T., Kang, J. W., Kang, A. & Lee, T. S. Redirecting metabolic flux via combinatorial multiplex CRISPRi-mediated repression for isopentenol production in Escherichia coli. ACS Synth. Biol.8, 391–402 (2019). 10.1021/acssynbio.8b00429 [DOI] [PubMed] [Google Scholar]
- 21.Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol.34, 184–191 (2016). 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jost, M. et al. Titrating gene expression using libraries of systematically attenuated CRISPR guide RNAs. Nat. Biotechnol.38, 355–364 (2020). 10.1038/s41587-019-0387-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Y., Wan, X. & Wang, B. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria. Nat. Commun.10, 3693 (2019). 10.1038/s41467-019-11479-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volk, M. J. et al. Biosystems design by machine learning. ACS Synth. Biol.9, 1514–1533 (2020). 10.1021/acssynbio.0c00129 [DOI] [PubMed] [Google Scholar]
- 25.Lawson, C. E. et al. Machine learning for metabolic engineering: a review. Metab. Eng.63, 34–60 (2021). 10.1016/j.ymben.2020.10.005 [DOI] [PubMed] [Google Scholar]
- 26.Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell156, 935–949 (2014). 10.1016/j.cell.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol.6, 26 (2011). 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flamm, C., Fontana, W., Hofacker, I. L. & Schuster, P. RNA folding at elementary step resolution. RNA6, 325–338 (2000). 10.1017/S1355838200992161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corsi, G. I. et al. CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nat. Commun.13, 1–14 (2022). 10.1038/s41467-022-30515-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, Y. et al. Improved prediction of bacterial CRISPRi guide efficiency from depletion screens through mixed-effect machine learning and data integration. Genome Biol.25, 1–22 (2024). 10.1186/s13059-023-03153-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods12, 982–988 (2015). 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, H. et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res.25, 1147–1157 (2015). 10.1101/gr.191452.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong, N., Liu, W. & Wang, X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol.16, 218 (2015). 10.1186/s13059-015-0784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science343, 80–84 (2014). 10.1126/science.1246981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fusi, N., Smith, I., Doench, J. & Listgarten, J. In silico predictive modeling of CRISPR/Cas9 guide efficiency. bioRxiv10.1101/021568 (2015).
- 36.Chari, R., Mali, P., Moosburner, M. & Church, G. M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods12, 823–826 (2015). 10.1038/nmeth.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alba Burbano, D. et al. Engineering activatable promoters for scalable and multi-input CRISPRa/i circuits. Proc. Natl Acad. Sci. USA120, e2220358120 (2023). 10.1073/pnas.2220358120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontana, J., Dong, C., Ham, J. Y., Zalatan, J. G. & Carothers, J. M. Regulated expression of sgRNAs tunes CRISPRi in E. coli. Biotechnol. J.13, 1800069 (2018). 10.1002/biot.201800069 [DOI] [PubMed] [Google Scholar]
- 39.Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell152, 1173–1183 (2013). 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vigouroux, A., Oldewurtel, E., Cui, L., Bikard, D. & Van Teeffelen, S. Tuning dCas9’s ability to block transcription enables robust, noiseless knockdown of bacterial genes. Mol. Syst. Biol.14, e7899 (2018). 10.15252/msb.20177899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathis, A. D., Otto, R. M. & Reynolds, K. A. A simplified strategy for titrating gene expression reveals new relationships between genotype, environment, and bacterial growth. Nucleic Acids Res.49, e6–e6 (2021). 10.1093/nar/gkaa1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawkins, J. S. et al. Mismatch-CRISPRi reveals the co-varying expression-fitness relationships of essential genes in Escherichia coli and Bacillus subtilis. Cell Syst.11, 523–535.e9 (2020). 10.1016/j.cels.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, S. & Voigt, C. A. Engineered dCas9 with reduced toxicity in bacteria: implications for genetic circuit design. Nucleic Acids Res.10.1093/nar/gky884 (2018). [DOI] [PMC free article] [PubMed]
- 44.Clamons, S. & Murray, R. Modeling predicts that CRISPR-based activators, unlike CRISPR-based repressors, scale well with increasing gRNA competition and dCas9 bottlenecking. bioRxiv10.1101/719278 (2019).
- 45.Jayanthi, S., Nilgiriwala, K. S. & Del Vecchio, D. Retroactivity controls the temporal dynamics of gene transcription. ACS Synth. Biol.2, 431–441 (2013). 10.1021/sb300098w [DOI] [PubMed] [Google Scholar]
- 46.Qian, Y., Huang, H.-H., Jiménez, J. I. & Del Vecchio, D. Resource competition shapes the response of genetic circuits. ACS Synth. Biol.6, 1263–1272 (2017). 10.1021/acssynbio.6b00361 [DOI] [PubMed] [Google Scholar]
- 47.Lee, P. et al. Safety and efficacy of 22 weeks of treatment with sapropterin dihydrochloride in patients with phenylketonuria. Am. J. Med. Genet. Part A146A, 2851–2859 (2008). 10.1002/ajmg.a.32562 [DOI] [PubMed] [Google Scholar]
- 48.Carmona‐Martínez, V. et al. Therapeutic potential of pteridine derivatives: a comprehensive review. Med. Res. Rev.39, 461–516 (2019). 10.1002/med.21529 [DOI] [PubMed] [Google Scholar]
- 49.Ehrenworth, A. M., Sarria, S. & Peralta-Yahya, P. Pterin-dependent mono-oxidation for the microbial synthesis of a modified monoterpene indole alkaloid. ACS Synth. Biol.4, 1295–1307 (2015). 10.1021/acssynbio.5b00025 [DOI] [PubMed] [Google Scholar]
- 50.Trenchard, I. J., Siddiqui, M. S., Thodey, K. & Smolke, C. D. De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab. Eng.31, 74–83 (2015). 10.1016/j.ymben.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, H. et al. Biochemical characterization of the tetrahydrobiopterin synthesis pathway in the oleaginous fungus Mortierella alpina. Microbiology157, 3059–3070 (2011). 10.1099/mic.0.051847-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Priem, B., Gilbert, M., Wakarchuk, W. W., Heyraud, A. & Samain, E. A new fermentation process allows large-scale production of human milk oligosaccharides by metabolically engineered bacteria. Glycobiology12, 235–240 (2002). 10.1093/glycob/12.4.235 [DOI] [PubMed] [Google Scholar]
- 53.Baumgärtner, F., Conrad, J., Sprenger, G. A. & Albermann, C. Synthesis of the human milk oligosaccharide lacto-N-tetraose in metabolically engineered, plasmid-free E. coli. Chembiochem15, 1896–1900 (2014). 10.1002/cbic.201402070 [DOI] [PubMed] [Google Scholar]
- 54.Bode, L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology22, 1147–1162 (2012). 10.1093/glycob/cws074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill, D. R. & Newburg, D. S. Clinical applications of bioactive milk components. Nutr. Rev.73, 463–476 (2015). 10.1093/nutrit/nuv009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newburg, D. S. & Morelli, L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr. Res.77, 115–120 (2015). 10.1038/pr.2014.178 [DOI] [PubMed] [Google Scholar]
- 57.Asakuma, S. et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem.286, 34583–34592 (2011). 10.1074/jbc.M111.248138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kulinich, A. & Liu, L. Human milk oligosaccharides: the role in the fine-tuning of innate immune responses. Carbohydr. Res.432, 62–70 (2016). 10.1016/j.carres.2016.07.009 [DOI] [PubMed] [Google Scholar]
- 59.Peterson, R., Cheah, W. Y., Grinyer, J. & Packer, N. Glycoconjugates in human milk: protecting infants from disease. Glycobiology23, 1425–1438 (2013). 10.1093/glycob/cwt072 [DOI] [PubMed] [Google Scholar]
- 60.Moore, R. E., Townsend, S. D. & Gaddy, J. A. The diverse antimicrobial activities of human milk oligosaccharides against group B Streptococcus. ChemBioChem. 23, e202100423 (2022). 10.1002/cbic.202100423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sprenger, G. A., Baumgärtner, F. & Albermann, C. Production of human milk oligosaccharides by enzymatic and whole-cell microbial biotransformations. J. Biotechnol.258, 79–91 (2017). 10.1016/j.jbiotec.2017.07.030 [DOI] [PubMed] [Google Scholar]
- 62.Xu, L. L. & Townsend, S. D. Synthesis as an expanding resource in human milk science. J. Am. Chem. Soc.143, 11277–11290 (2021). 10.1021/jacs.1c05599 [DOI] [PubMed] [Google Scholar]
- 63.Urashima, T. et al. The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv. Nutr.3, 473S–482S (2012). 10.3945/an.111.001412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blixt, O., van Die, I., Norberg, T. & van den Eijnden, D. H. High-level expression of the Neisseria meningitidis lgtA gene in Escherichia coli and characterization of the encoded N-acetylglucosaminyltransferase as a useful catalyst in the synthesis of GlcNAcβ1→ 3Gal and GalNAcβ1→ 3Gal linkages. Glycobiology9, 1061–1071 (1999). 10.1093/glycob/9.10.1061 [DOI] [PubMed] [Google Scholar]
- 65.Liu, X. et al. Characterization and synthetic application of a novel β1,3-galactosyltransferase from Escherichia coli O55:H7. Bioorg. Med. Chem.17, 4910–4915 (2009). 10.1016/j.bmc.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 66.Baumgärtner, F., Seitz, L., Sprenger, G. A. & Albermann, C. Construction of Escherichia coli strains with chromosomally integrated expression cassettes for the synthesis of 2′-fucosyllactose. Microb. Cell Fact.12, 40 (2013). 10.1186/1475-2859-12-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, W. et al. Metabolic engineering of Escherichia coli for the production of Lacto-N-neotetraose (LNnT). Syst. Microbiol. Biomanuf.1, 291–301 (2021). 10.1007/s43393-021-00023-1 [DOI] [Google Scholar]
- 68.Lee, W.-H. et al. Whole cell biosynthesis of a functional oligosaccharide, 2′-fucosyllactose, using engineered Escherichia coli. Microb. Cell Fact.11, 48 (2012). 10.1186/1475-2859-11-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dumon, C. et al. In vivo fucosylation of lacto-N-neotetraose and lacto-N-neohexaose by heterologous expression of Helicobacter pylori a-1,3 fucosyltransferase in engineered Escherichia coli. Glycoconj. J.18, 465–474 (2001). [DOI] [PubMed]
- 70.Dykhuizen, D. & Hartl, D. Transport by the lactose permease of Escherichia coli as the basis of lactose killing. J. Bacteriol.135, 876–882 (1978). 10.1128/jb.135.3.876-882.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eames, M. & Kortemme, T. Cost-benefit tradeoffs in engineered lac operons. Science336, 911–915 (2012). 10.1126/science.1219083 [DOI] [PubMed] [Google Scholar]
- 72.Radivojević, T., Costello, Z., Workman, K. & Garcia Martin, H. A machine learning automated recommendation tool for synthetic biology. Nat. Commun.11, 4879 (2020). 10.1038/s41467-020-18008-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McArthur, J. B., Yu, H. & Chen, X. A bacterial β1–3-Galactosyltransferase enables multigram-scale synthesis of human milk lacto- N -tetraose (LNT) and its fucosides. ACS Catal.9, 10721–10726 (2019). 10.1021/acscatal.9b03990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sugita, T. & Koketsu, K. Transporter engineering enables the efficient production of lacto- N -triose II and lacto- N -tetraose in Escherichia coli. J. Agric. Food Chem.70, 5106–5114 (2022). 10.1021/acs.jafc.2c01369 [DOI] [PubMed] [Google Scholar]
- 75.Thimmaiah, T., Voje, W. E. & Carothers, J. M. Computational Methods in Synthetic Biology (ed. Marchisio, M. A.) p. 45–61 (Springer, 2015).
- 76.Burke, C. R., Sparkman-Yager, D. & Carothers, J. M. Multi-state design of kinetically-controlled RNA aptamer ribosensors. Preprint at 10.1101/213538 (2017).
- 77.Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol.33, 985–989 (2015). 10.1038/nbt.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Todor, H., Silvis, M. R., Osadnik, H. & Gross, C. A. Bacterial CRISPR screens for gene function. Curr. Opin. Microbiol.59, 102–109 (2021). 10.1016/j.mib.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang, L. et al. Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids. Nat. Commun.12, 4976 (2021). 10.1038/s41467-021-25243-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jensen, K. T. et al. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett.591, 1892–1901 (2017). 10.1002/1873-3468.12707 [DOI] [PubMed] [Google Scholar]
- 81.Weiss, T. et al. Epigenetic features drastically impact CRISPR–Cas9 efficacy in plants. Plant Physiol.190, 1153–1164 (2022). 10.1093/plphys/kiac285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alkan, F., Wenzel, A., Anthon, C., Havgaard, J. H. & Gorodkin, J. CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol.19, 177 (2018). 10.1186/s13059-018-1534-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiang, X. et al. Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun.12, 1–9 (2021). 10.1038/s41467-021-23576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hsu, S.-Y., Lee, J., Sychla, A. & Smanski, M. J. Rational search of genetic design space for a heterologous terpene metabolic pathway in Streptomyces. Metab. Eng.77, 1–11 (2023). 10.1016/j.ymben.2023.02.011 [DOI] [PubMed] [Google Scholar]
- 85.Liao, Y. et al. Metabolic engineering of Escherichia coli for high-level production of lacto-N-neotetraose and lacto-N-tetraose. J. Agric. Food Chem.71, 11555–11566 (2023). 10.1021/acs.jafc.3c02997 [DOI] [PubMed] [Google Scholar]
- 86.Zhu, Y. et al. Metabolic engineering of Escherichia coli for efficient biosynthesis of lacto-N-tetraose using a novel β−1,3-galactosyltransferase from pseudogulbenkiania ferrooxidans. J. Agric. Food Chem.69, 11342–11349 (2021). 10.1021/acs.jafc.1c04059 [DOI] [PubMed] [Google Scholar]
- 87.Copeland, M. F., Politz, M. C. & Pfleger, B. F. Application of TALEs, CRISPR/Cas and sRNAs as trans-acting regulators in prokaryotes. Curr. Opin. Biotechnol.29, 46–54 (2014). 10.1016/j.copbio.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teng, Y., Jiang, T. & Yan, Y. The expanded CRISPR toolbox for constructing microbial cell factories. Trends Biotechnol.10.1016/j.tibtech.2023.06.012 (2023). [DOI] [PMC free article] [PubMed]
- 89.Call, S. N. & Andrews, L. B. CRISPR-based approaches for gene regulation in non-model bacteria. Front. Genome Ed.4, 892304 (2022). 10.3389/fgeed.2022.892304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ameruoso, A., Villegas Kcam, M. C., Cohen, K. P. & Chappell, J. Activating natural product synthesis using CRISPR interference and activation systems in Streptomyces. Nucleic Acids Res.50, 7751–7760 (2022). 10.1093/nar/gkac556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ho, H., Fang, J. R., Cheung, J. & Wang, H. H. Programmable CRISPR‐Cas transcriptional activation in bacteria. Mol. Syst. Biol.16, e9427 (2020). 10.15252/msb.20199427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Han, Y., Li, W., Filko, A., Li, J. & Zhang, F. Genome-wide promoter responses to CRISPR perturbations of regulators reveal regulatory networks in Escherichia coli. Nat. Commun.14, 1–13 (2023). 10.1038/s41467-022-34464-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hartline, C. J., Schmitz, A. C., Han, Y. & Zhang, F. Dynamic control in metabolic engineering: theories, tools, and applications. Metab. Eng.63, 126–140 (2021). 10.1016/j.ymben.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ni, C., Dinh, C. V. & Prather, K. L. J. Dynamic control of metabolism. Annu. Rev. Chem. Biomol. Eng.12, 519–541 (2021). 10.1146/annurev-chembioeng-091720-125738 [DOI] [PubMed] [Google Scholar]
- 95.Wu, Y. et al. CRISPR–dCas12a-mediated genetic circuit cascades for multiplexed pathway optimization. Nat. Chem. Biol.19, 367–377 (2023). 10.1038/s41589-022-01230-0 [DOI] [PubMed] [Google Scholar]
- 96.Huang, H.-H. et al. dCas9 regulator to neutralize competition in CRISPRi circuits. Nat. Commun.12, 1692 (2021). 10.1038/s41467-021-21772-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Volk, M. J. et al. Metabolic engineering: methodologies and applications. Chem. Rev.123, 5521–5570 (2023). 10.1021/acs.chemrev.2c00403 [DOI] [PubMed] [Google Scholar]
- 98.Kaczmarek, J. A. & Prather, K. L. J. Effective use of biosensors for high-throughput library screening for metabolite production. J. Ind. Microbiol. Biotechnol.48, kuab049 (2021). 10.1093/jimb/kuab049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yanisch-Perron, C., Vieira, J. & Messing, J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene33, 103–119 (1985). 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- 100.Kosuri, S. et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl Acad. Sci. USA110, 14024–14029 (2013). 10.1073/pnas.1301301110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tian, T. & Salis, H. M. A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons. Nucleic Acids Res.43, 7137–7151 (2015). 10.1093/nar/gkv635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res.46, W242–W245 (2018). 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Espinosamansilla, A., Delapena, A., Canadacanada, F. & Dellanos, A. LC determination of biopterin reduced forms by UV-photogeneration of biopterin and fluorimetric detection. Talanta77, 844–851 (2008). 10.1016/j.talanta.2008.07.046 [DOI] [Google Scholar]
- 104.Cañada-Cañada, F., Espinosa-Mansilla, A., Muñoz De La Peña, A. & Mancha De Llanos, A. Determination of marker pteridins and biopterin reduced forms, tetrahydrobiopterin and dihydrobiopterin, in human urine, using a post-column photoinduced fluorescence liquid chromatographic derivatization method. Anal. Chim. Acta648, 113–122 (2009). 10.1016/j.aca.2009.06.045 [DOI] [PubMed] [Google Scholar]
- 105.Sugianto, W. et al. Gene expression dynamics in input-responsive engineered living materials programmed for bioproduction. Mater. Today Bio20, 100677 (2023). 10.1016/j.mtbio.2023.100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fontana, J. et al. Guide RNA structure design enables combinatorial CRISPRa programs for biosynthetic profiling (this paper). Github10.5281/zenodo.12558212 (2024).
- 107.Fontana, J. et al. Guide RNA structure design enables combinatorial CRISPRa programs for biosynthetic profiling (this paper). Github10.5281/zenodo.12559439 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
A reporting summary for this article is available as a Supplementary Information file. Data supporting the findings of this work are available within the paper and its Supplementary Information files. Source data are provided with this paper.
Custom Python code to analyze input RNA and generate the energetic parameters described in this work is available on GitHub [https://github.com/carothersresearch/gRNA_screen_docker]106 and can be run directly in that environment using a Codespace or locally using a Docker image. Jupyter notebooks to view and reproduce the ART results from this paper are available on GitHub [https://github.com/carothersresearch/art_lnt]107. These notebooks can be viewed on GitHub or run in an ART Docker container after acquiring a license. See https://github.com/JBEI/ART for software and licensing details.