Abstract
Clinical success with poly (ADP-ribose) polymerase inhibitors (PARPi) is impeded by inevitable resistance and associated cytotoxicity. Depletion of Amplified in Liver Cancer 1 (ALC1), a chromatin-remodeling enzyme, can overcome these limitations by hypersensitizing BReast CAncer genes 1/2 (BRCA1/2) mutant cells to PARPi. Here, we demonstrate that PARPi hypersensitivity upon ALC1 loss is reliant on its role in promoting the repair of chromatin buried abasic sites. We show that ALC1 enhances the ability of the abasic site processing enzyme, Apurinic/Apyrimidinic endonuclease 1 (APE1) to cleave nucleosome-occluded abasic sites. However, unrepaired abasic sites in ALC1-deficient cells are readily accessed by APE1 at the nucleosome-free replication forks. APE1 cleavage leads to fork breakage and trapping of PARP1/2 upon PARPi treatment, resulting in hypersensitivity. Collectively, our studies reveal how cells overcome the chromatin barrier to repair abasic lesions and uncover cleavage of abasic sites as a mechanism to overcome limitations of PARPi.
Subject terms: Homologous recombination, Base excision repair, Stalled forks
Loss of Amplified in Liver Cancer 1 (ALC1) has been shown to confer PARP inhibitor hypersensitivity in BRCA-mutant cells. Here, the authors show that in ALC1-deficient BRCA-mutant cells, APE1 cleaves abasic sites at the forks resulting in DNA breaks and thereby enhances PARP inhibitor sensitivity.
Introduction
Synthetic lethality between the loss of BReast CAncer genes 1/2 (BRCA1/2)-mediated homologous recombination (HR) and Poly (ADP-ribose) polymerase inhibitors (PARPi) has been harnessed as FDA-approved therapeutics for various HR-deficient (HRD) cancers1,2. Multiple mechanisms have been linked to PARPi-induced cytotoxicity in HRD cells3. Cytotoxic chromosomal fusions via non-homologous end joining (NHEJ) of DNA double-strand breaks (DSBs) have been proposed to underlie PARPi sensitivity in BRCA mutant settings. This model is supported by findings demonstrating that concomitant loss of NHEJ and BRCA proteins restore PARPi resistance4,5. Another mechanism entails the nuclease-mediated degradation of replication forks in the absence of BRCA proteins. Support for this model emerges from the findings that loss of nucleases implicated in degrading forks confers PARPi resistance in BRCA mutant cells6,7. Recently, a gap model was proposed, whereby the frequency of replication gaps was shown to correlate with PARPi response8. Known mechanisms of replication gap formation include primase and DNA-directed polymerase (PrimPol) mediated repriming9 and failure to ligate Okazaki fragments. In support of this model, loss of proteins involved in Okazaki fragment maturation have been shown to confer PARPi sensitivity8,10, albeit whether PrimPol-mediated gap formation contributes to PARPi response remains unclear. Given the multitude of PARP1 and 2 functions in DNA repair and replication, it is evident that multiple mechanisms may underlie PARPi sensitivity depending on the cellular context11–13. Despite our knowledge of these varied mechanisms of PARPi sensitivity, a fundamental question that remains unanswered is what intrinsic cellular genomic lesion(s) triggers the generation of DSBs, fork degradation, or gap formation that leads to PARPi sensitivity. Identifying the endogenous lesion(s) and how their repair is altered upon PARPi treatment is of paramount importance to improve the efficacy of this therapy and to rationally predict the scenarios where PARPi would be most effective.
The emergence of resistance and side effects associated with PARPi has necessitated the development of approaches to improve the efficacy of this therapy14–16. Using clustered regulatory interspaced short palindromic repeats (CRISPR) genetic screens, we and others recently showed that loss of an ATP-dependent chromatin-remodeling enzyme, Amplified in Liver Cancer 1 (ALC1), hypersensitizes BRCA mutant cancer cells to PARPi17–20. Owing to this phenotype, ALC1 loss results in the expedited killing of BRCA mutant cancer cells upon treatment with PARPi and also helps overcome several clinically relevant resistance mechanisms of this targeted therapy. However, the molecular basis of PARPi hypersensitivity in ALC1-deficient BRCA mutant cancer cells remains unresolved.
ALC1-deficient cells are sensitive to various base-damaging agents21,22 and ultraviolet (UV) radiation23,24, highlighting its potential role in base- and global genome nucleotide-excision repair (BER and GG-NER). Whether ALC1 function in BER or GG-NER contributes to PARPi response in BRCA mutant cells remains unclear. One of the base excision repair intermediates called abasic sites has been implicated in enhanced PARPi sensitivity in BRCA mutant cells25. Abasic sites can also form spontaneously in single-stranded DNA (ss-DNA) upon hydrolysis of the N-glycosylic bond26,27. Abasic sites are predominantly repaired by Apurinic/Apyrimidinic Endonuclease 1 (APE1) nuclease. However, unlike ALC1, APE1 has not emerged as a high-confidence hit in most CRISPR screens for PARPi sensitivity28,29. This conundrum led us to speculate that the mere presence of abasic sites might not underlie the PARPi hypersensitivity in ALC1-deficient BRCA mutant cancer cells. We, therefore, set out to determine the mechanistic basis by which ALC1 loss augments PARPi response in BRCA mutant cells. Using cellular and in vitro reconstitution studies, we establish the role of ALC1 in promoting the repair of abasic sites buried in the chromatin. We show that in ALC1-deficient cells, abasic sites are cleaved by APE1 at the nucleosome-free replication forks resulting in DSBs, fork stalling, and hence PARPi hypersensitivity. Our studies, therefore, uncover that cleaved, rather than intact abasic sites, are a key cell-intrinsic lesion that can augment PARPi response in BRCA mutant cells. ALC1 targeting can, therefore, allow the exploitation of endogenous base damage in cancer cells and, hence, enhance the therapeutic efficacy of PARPi in the absence of external genotoxic damage.
Results
PrimPol loss abolishes replication gaps but does not rescue PARPi hypersensitivity in ALC1-deficient BRCA mutant cells
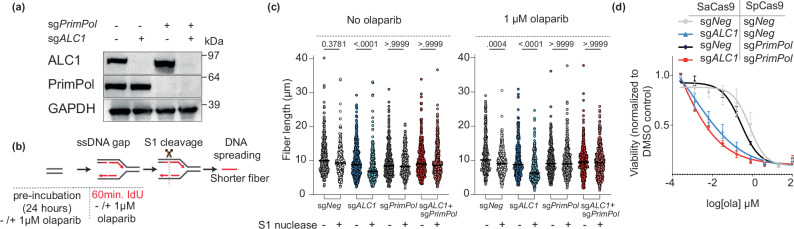
We and others recently reported that ALC1 loss results in an increased generation of replication gaps17,18. The frequency of gaps further increases upon treatment with olaparib, a PARPi. We, therefore, examined whether de novo repriming by PrimPol underlies replication gap formation in ALC1-deficient BRCA1 mutant SUM149PT cells. We generated PrimPol knockout (KO) SUM149PT cells (Fig. 1a) and assessed the impact on gap formation in ALC1-deficient settings using the S1 nuclease coupled DNA fiber assay (Fig. 1b)30. We observed that while the length of DNA fibers in ALC1-deficient cells were reduced upon treatment with S1 nuclease, indicating the presence of gaps, this phenotype was lost upon concomitant depletion of PrimPol (Fig. 1c). These data reveal that replication-coupled gaps observed in ALC1-deficient BRCA mutant cells are reliant on PrimPol. Moreover, we observe that olaparib-induced gaps in both ALC1-deficient and proficient BRCA1 mutant cells are also generated in a PrimPol-dependent manner (Fig. 1c). We established the on-target specificity of the PrimPol KO cells by complementing back with a Cas9 resistant V-5 tagged PrimPol complementary DNA (cDNA) construct (Supplementary Fig. 1a) and found a rescue in gap formation at levels equivalent to Wild Type (WT)-cells (Supplementary Fig. 1b, c).
Fig. 1. Role of PrimPol in replication gaps and olaparib sensitivity in ALC1-deficient BRCA1 mutant SUM149PT cells.
a Immunoblot showing depletion of ALC1 and PrimPol. Representative of three blots. b Schematic of S1-nuclease fiber assay. IdU: 5-iodo-2’-deoxyuridine. c Analysis of IdU tract length in the indicated cell lines in the absence (left) and presence (right) of olaparib. Median values are indicated. P values derived by Kruskal–Wallis test from >300 fibers/sample collected over three biologically independent experiments. d Sensitivities of the indicated cell lines to olaparib. Data are mean ± s.e.m from three biologically independent experiments. Source data are provided as a Source data file.
Because replication-coupled gaps can contribute to PARPi sensitivity31, we next assessed the impact of PrimPol loss on PARPi sensitivity in ALC1-deficient BRCA mutant cancer cells. We observed that PrimPol loss did not rescue the PARPi sensitivity (Fig. 1d), suggesting that PrimPol-mediated gap formation is not the key underlying mechanism for PARPi sensitivity in ALC1-deficient BRCA mutant cells.
Functions of ALC1 are restricted to the repair of damaged bases and nucleotide lesions
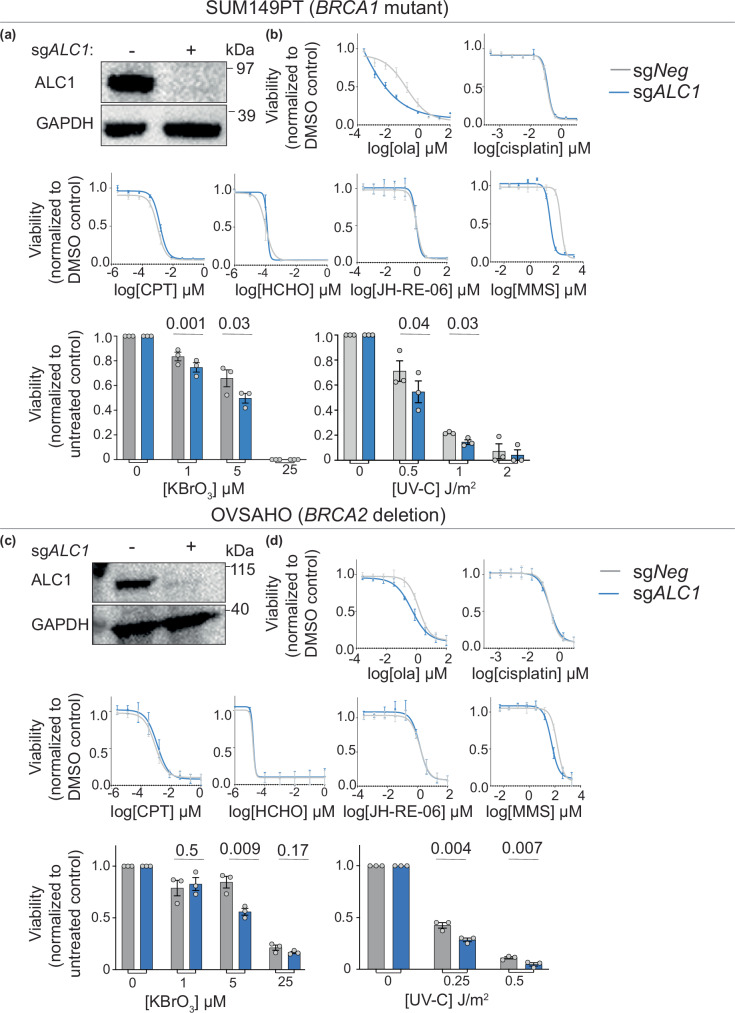
We next examined the nature of genomic lesions that rely on ALC1-mediated repair in BRCA mutant cancer cells. To do so, we profiled the sensitivity of ALC1-deficient BRCA1 mutant SUM149PT and BRCA2-deleted OVSAHO cells to various genotoxic agents (Fig. 2a–d). Consistent with previous studies, ALC1 loss enhanced olaparib response but did not increase sensitivity to the single- and double-strand break-inducing agents cisplatin and camptothecin17. Additionally, we observed that ALC1 loss does not enhance sensitivity to formaldehyde or JH-RE-06 (Rev1 inhibitor), highlighting the dispensability of this enzyme in transcription-coupled nucleotide-excision repair (TC-NER) and translesion synthesis, respectively. We and others have previously reported that ALC1 deficiency confers sensitivity to the base alkylating agent, methyl methane sulfonate (MMS)22. We could recapitulate this finding in both SUM149PT and OVSAHO cells. We further extended the role of ALC1 in base damage repair by demonstrating the sensitivity of ALC1-deficient cells to the oxidizing agent, potassium bromate, KBrO3. BRCA WT ALC1-deficient U-2 OS cells have been reported to be sensitive to UV radiation, suggesting a role of ALC1 in GG-NER23,24. We found a modest enhancement in the sensitivity of ALC1-deficient BRCA mutant cancer cells to UV-C. Together, our studies establish that ALC1 loss in BRCA mutant cells predominantly confers sensitivity to base-damaging agents and modest sensitization to UV radiation, suggesting a role in BER and GG-NER, respectively.
Fig. 2. Genotoxic sensitivity of ALC1-deficient cells.
a Immunoblot showing depletion of ALC1 in BRCA1 mutant SUM149PT cells. Representative of three blots. b Sensitivities of the indicated SUM149PT cell lines to various genotoxins. Data are presented as mean ± s.e.m from three biologically independent experiments. P values derived by unpaired two-tailed t-test. c Immunoblot showing depletion of ALC1 in BRCA2 deleted OVSAHO cells. Representative of three blots. d Sensitivities of the indicated OVSAHO cell lines to various genotoxins. Data are presented as mean ± s.e.m from three biologically independent experiments. P values derived by unpaired two-tailed t-test. Source data are provided as a Source data file.
Functions of ALC1 in PARPi response are not dependent on its role in resolving nucleotide adducts
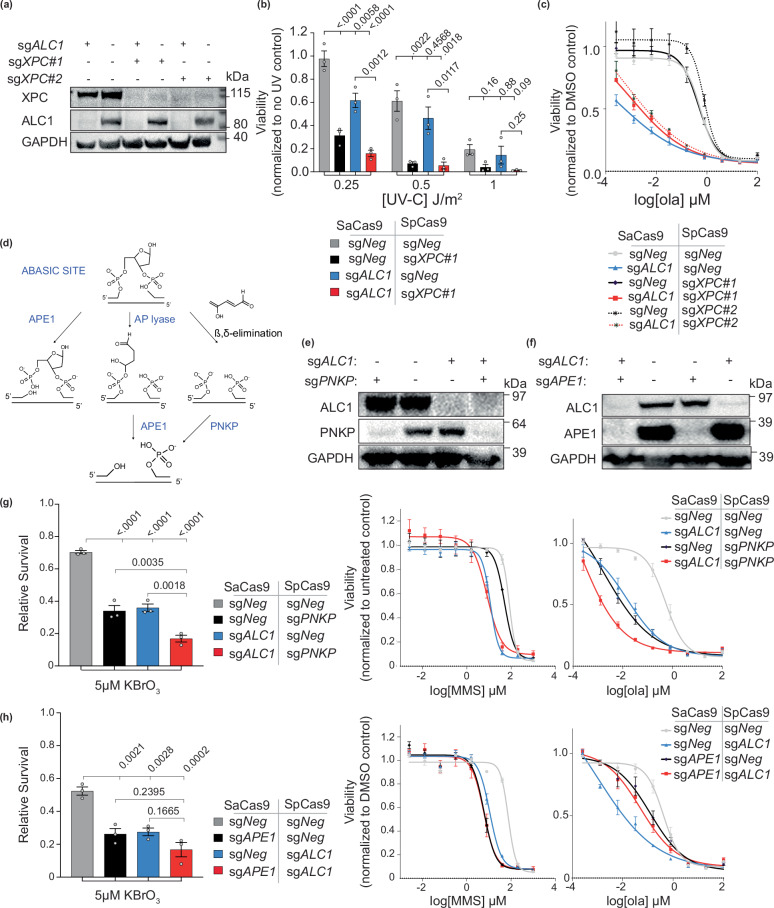
We next examined if ALC1 contributes to PARPi response via its role in repairing nucleotide adducts. We performed epistasis analysis between ALC1 and GG-NER factors for PARPi sensitivity. Recruitment of ALC1 to UV-damaged sites has been shown to be reliant on DNA damage-binding protein 2 (DDB2)23 and Xeroderma pigmentosum complementation group C (XPC)24. We, therefore, focused on these two proteins for the epistasis analysis. Consistent with the reported role of XPC in resolving a broad range of UV-induced lesions, XPC-deficient BRCA1 mutant cells were more sensitive to UV-C compared to the ALC1-deficient counterparts (Fig. 3a, b). ALC1 and DDB2 were epistatic in UV-C damage response (Supplementary Fig. 2a, b). However, XPC or DDB2 loss conferred minimal enhancement in PARPi sensitivity in either ALC1-proficient or -deficient SUM149PT cells (Fig. 3c and Supplementary Fig. 2c). These results were consistent across two different single guide RNAs (sgRNAs) targeting either XPC or DDB2, suggesting that the contribution of ALC1 to PARP inhibitor response is likely not reliant on its ability to process nucleotide adducts.
Fig. 3. Epistasis between ALC1 and GG-NER/BER factors in response to DNA damaging agents in BRCA1 mutant SUM149PT cells.
a Immunoblot showing depletion of ALC1 and XPC. Representative of three blots. b Sensitivities of the indicated cell lines to UV-C. Data presented as mean ± s.e.m from three biologically independent experiments. P values derived by ordinary one-way ANOVA. c Sensitivities of the indicated cell lines to olaparib. Data presented as mean ± s.e.m from three biologically independent experiments. Representative of three blots. d Schematic showing how the abasic site can be processed by either APE1 or PNKP. e Immunoblot showing depletion of ALC1 and PNKP. Representative of three blots. f Immunoblot showing depletion of ALC1 and APE1. Representative of three blots. g, h Sensitivities of the indicated cell lines to KBrO3 (left), MMS (middle), and olaparib (right). Data are presented as mean ± s.e.m from three biologically independent experiments. P value derived by ordinary one-way ANOVA. Source data are provided as a Source data file.
APE1 loss confers PARPi resistance in ALC1-deficient BRCA mutant cells
We next examined how the base damage repair function of ALC1 contributes to PARPi hypersensitivity in ALC1-deficient BRCA mutant cells. To do so, we examined epistasis between ALC1 and base damage repair factors for sensitivity to base-damaging agents and olaparib. Recent studies have highlighted abasic sites as the lesion that can potentially contribute to olaparib sensitivity25, albeit the underlying mechanism remains unclear. We therefore examined epistasis of ALC1 with APE1 and polynucleotide kinase-phosphatase (PNKP), two key enzymes implicated in the repair of abasic sites (Fig. 3d–f). PNKP loss enhanced the sensitivity of both WT and ALC1-deficient SUM149PT cells to potassium bromate but did not impact sensitivity to MMS, highlighting the specificity of this enzyme in repairing nitrogen bases damaged by oxidation. Depletion of PNKP sensitized both WT and ALC1-deficient SUM149PT cells to olaparib (Fig. 3g). Together, these findings uncover a non-epistatic relationship between PNKP and ALC1 in the repair of lesions generated by base damage and PARPi.
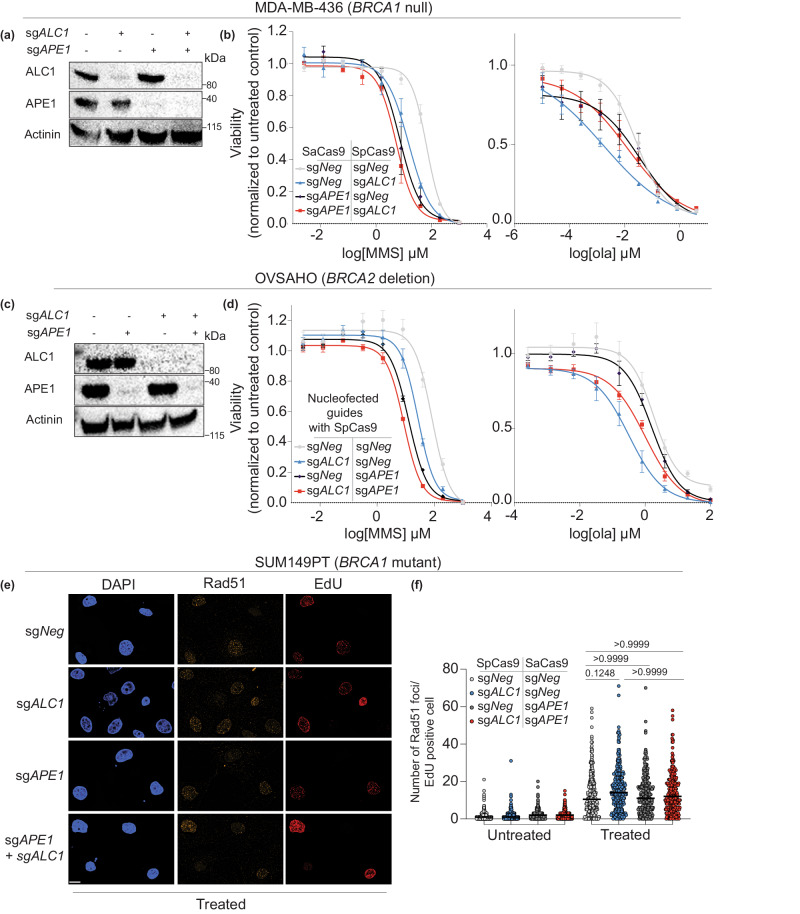
We next examined epistasis between APE1 and ALC1 for drug responses. Consistent with previous studies in BRCA-proficient cells18, we observed that ALC1 loss did not further enhance the sensitivity of APE1-deficient cells to either potassium bromate or MMS (Fig. 3h). Loss of APE1 conferred a modest enhancement in PARPi sensitivity compared to ALC1-deficient BRCA mutant cells. Unexpectedly, loss of APE1 rendered PARPi resistance in ALC1-deficient SUM149PT cells (Fig. 3h). Considering the pleiotropic phenotypes of different BRCA mutations, we validated our findings in cell lines of different tissue origin and BRCA mutation32. We chose UWB1.289 because, like SUM149PT, they express BRCA1-del11q mutant protein but are derived from high-grade serous ovarian cancer (HGSOC)32,33. We also tested our findings in MDA-MB-436 cells that have a BRCA1 5396 + 1 G > A mutation, which results in a complete loss of protein expression34. We observe that loss of APE1 confers olaparib resistance in ALC1-deficient settings across all three BRCA1 mutant cell lines (Fig. 4a, b and Supplementary Fig. 2d, e), highlighting the generalizability of our findings. We next tested our findings in BRCA2-deleted OVSAHO, a cell line with BRCA2 homozygous deletion35,36. Using nucleofection, we were able to successfully deplete both APE1 and ALC1 in OVSAHO cells. Consistent with our findings in BRCA1 mutant cells, we observe that while ALC1 is epistatic to APE1 for MMS response, loss of APE1 confers olaparib resistance in BRCA2 deleted OVSAHO cells (Fig. 4c, d). Together, our observations suggest that PARPi hypersensitivity in ALC1-deficient BRCA mutant cells is likely reliant on APE1.
Fig. 4. Epistasis analysis between ALC1 and APE1 for response to DNA damaging agents.
a Immunoblot showing depletion of ALC1 and APE1 in MDA-MB-436 cells. Representative of three blots. b Sensitivities of the indicated MDA-MB-436 cells to MMS (left) and olaparib (right). Data are presented as mean ± s.e.m from three biologically independent experiments. c Immunoblot showing depletion of ALC1 and APE1 in OVSAHO cells. Representative of three blots. d Sensitivities of the indicated OVSAHO cell lines to MMS (left) and olaparib (right). Data are presented as mean ± s.e.m from three biologically independent experiments. e Representative immunofluorescence image of Rad51 foci in indicated SUM149PT cells. Scale bar 10 microns. Representative from three biologically independent experiments. Source data are provided as a Source data file. f Quantification of the chromatin-bound for Rad51 foci/EdU-positive cells. Each dot represents a single cell. Median is indicated. P value determined by Kruskal–Wallis test derived from 300 cells sampled over three biologically independent experiments. Source data are provided as a Source data file.
Beyond PNKP and APE1, abasic sites can also be processed by the AP lyase activity of the bifunctional glycosylases to form 3’ unsaturated aldehyde, which is processed by APE1 to 3’OH (Fig. 3d)37–39. Amongst the bifunctional glycosylases, 8-oxoguanine glycosylase (OGG1) has been reported to primarily function as a monofunctional glycosylase in vivo37,38. We, therefore, focused on the depletion of endonuclease III-like protein 1 (NTHL1), Nei endonuclease VIII-like 1 (NEIL1), and Nei endonuclease VIII-like 2 (NEIL2) glycosylases which function on oxidized bases. Given the substrate redundancy of these glycosylases, we used a Cas12a-based approach to simultaneously deplete all three enzymes and examined their epistasis with ALC1 for olaparib response40. We observed that NTHL1/NEIL1/NEIL2 deficiency did not have any impact on olaparib sensitivity in either ALC1 proficient or deficient settings. This observation suggests that substrates of NTHL1/NEIL1/NEIL2 may not be present in BRCA mutant cancer cells at a high-enough frequency to contribute to olaparib sensitivity (Supplementary Fig. 2f, g). Of note, we were not able to deplete APE1 in NTHL1/NEIL1/NEIL2-deficient cells and hence we cannot exclude the possibility that AP lyase activity of bifunctional glycosylases can contribute to PARPi sensitivity in APE1/ALC1-deficient BRCA mutant cells.
APE1 loss does not restore homologous recombination in ALC1-deficient cells
A potential mechanism that confers PARP inhibitor resistance in BRCA1 mutant cells entails the restoration of end-resection, enabling Rad51 loading and HR4. To examine whether APE1 loss restored HR in ALC1-deficient BRCA mutant cells, we quantified damaged-induced Rad51 foci formation. We chose SUM149PT cells for these assays because this cell line expresses hypomorphic BRCA1-del11q protein, which partially retains the ability to load Rad5141. Hence, Rad51 foci can be visualized in these BRCA mutant cells when treated with high levels of DNA damage. We observe that loss of ALC1 and APE1 alone or in combination did not impact the ability of these cells to form damage-induced-Rad51 foci (Fig. 4e, f). These data highlight that ALC1 or APE1 do not function in homology repair, and APE1 loss does not restore HR in ALC1-deficient cells. We further corroborated these findings by examining the sensitivity of ALC1-, APE1- and ALC1/APE1-deficient cells to camptothecin and platinum (Supplementary Fig. 3a, b). Both these drugs necessitate reliance on HR for repair and survival. Consistent with our conclusion from the Rad51 foci assay, we did not observe an impact on ALC1-, APE1-, and ALC1/APE1-deficiency on camptothecin or platinum sensitivity. In line with the dispensability of ALC1 or APE1 for HR, we did observe enhanced sensitivity of these ALC1-, APE1-, and ALC1/APE1-deficient cells BRCA WT DLD1 cells to olaparib, platinum, or camptothecin (Supplementary Fig. 3c, d). Together, our results rule out the possibility that APE1 confers olaparib resistance in ALC1-deficient BRCA mutant cells owing to restoration of HR.
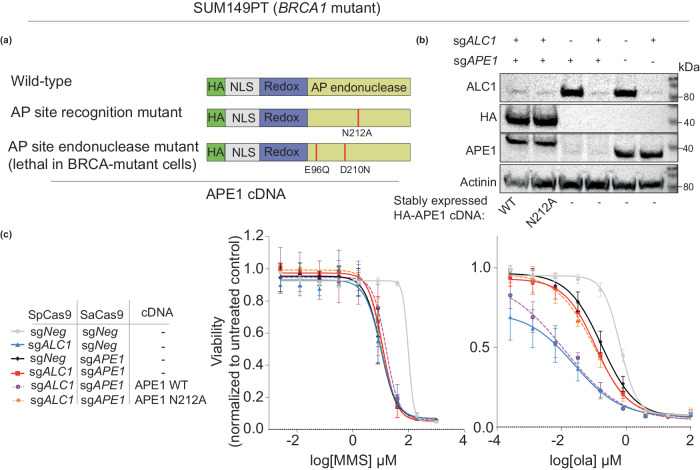
Recognition of abasic sites by APE1 is essential for PARPi hypersensitivity in ALC1-deficient BRCA mutant cancer cells
We next determined whether abasic site processing by APE1 is required for PARPi hypersensitivity in ALC1-deficient BRCA mutant cancer cells. We generated stable cell lines expressing sgRNA-resistant APE1 WT and E96Q/D210N APE1 endonuclease-dead42 constructs to perform the complementation studies (Fig. 5a). APE1 WT could be expressed in cells at a level comparable to the endogenous protein (Fig. 5b) and it restored PARPi hypersensitivity of sgAPE1sgALC1 deficient BRCA mutant cells (Fig. 5c). However, expression of either the E96Q or E96Q/D210N endonuclease-dead APE1 mutant in BRCA mutant cells compromised viability. We hypothesize that in the absence of endonucleolytic cleavage, the endonuclease-dead APE1 mutants are retained on the chromatin, resulting in DNA damage and lethality. Consistent with our hypothesis, we observe that unlike WT constructs, transient expression of APE1 endonuclease mutants resulted in increased γH2AX signal in BRCA mutant cells within 96 h of transduction (Supplementary Fig. 4a). This cytotoxicity induced by APE1 endonuclease-dead mutants precluded our ability to examine the impact of APE1 nuclease activity in long-term viability assays. As an alternative approach to assess how abasic site processing by APE1 impacts PARPi hypersensitivity in ALC1-deficient BRCA mutant cancer cells, we generated an N212A APE1 mutant. This mutation abrogates the ability of APE1 to recognize abasic sites in vitro43. Consistent with these reports, the N212A mutant failed to rescue MMS and olaparib sensitivity of APE1-deficient cells (Supplementary Fig. 4b). Complementation of sgALC1sgAPE1 cells with a N212A APE1 construct failed to restore PARPi hypersensitivity in ALC1-deficient BRCA mutant cancer cells (Fig. 5c). Together, these results suggest that recognition of abasic sites by APE1 is responsible for the enhanced PARPi sensitivity in ALC1-deficient BRCA mutant cancer cells.
Fig. 5. Processing of abasic sites by APE1 confers PARPi hypersensitivity in BRCA mutant cancer cells.
a Schematics for APE1 constructs. b Immunoblot showing expression levels of exogenous APE1 constructs and depletion of endogenous APE1 and ALC1. Representative of three blots. c Sensitivities of the indicated SUM149PT cell lines to MMS (left) and olaparib (right). Data are presented as mean ± s.e.m from three biologically independent experiments. Source data are provided as a Source data file.
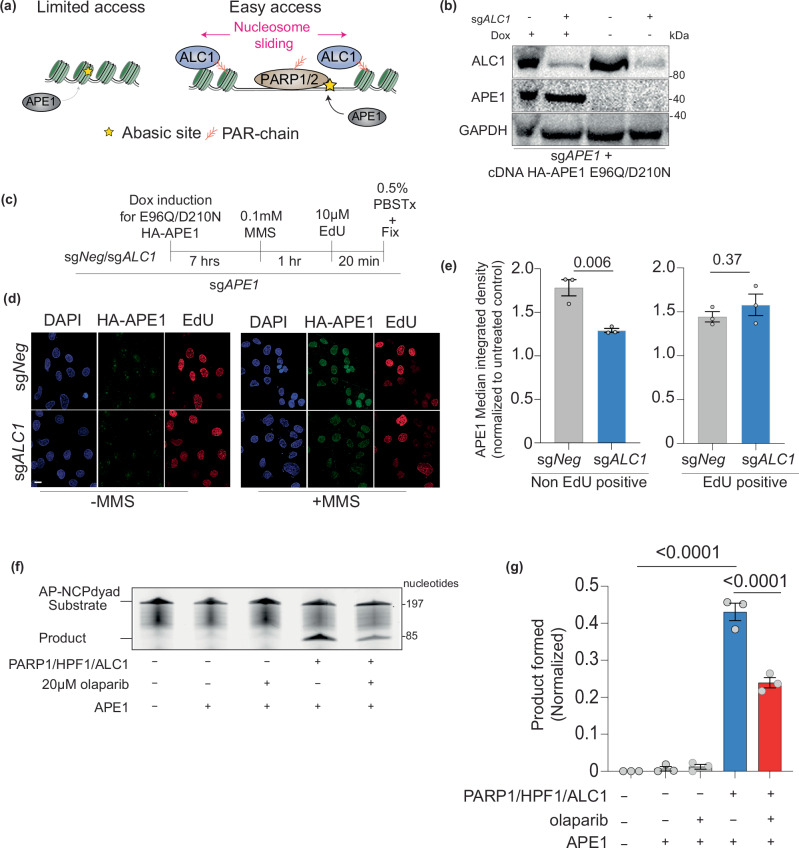
ALC1 promotes the association of APE1 with the chromatin
We and others have previously suggested that nucleosome sliding by ALC1 could be required for the recruitment of APE1 to damaged sites (Fig. 6a). However, examining APE1 localization at the chromatin is challenging owing to its rapid recruitment and dissociation while processing abasic sites44. We argue that endonuclease-deficient APE1 mutants may get trapped at the abasic sites and hence facilitate localization studies. Consistent with this idea, the E96Q/D210N APE1 endonuclease mutant was cytotoxic when overexpressed in BRCA mutant cancer cells. We, therefore, generated a cell line expressing doxycycline-inducible E96Q/D210N APE1 cDNA (Fig. 6b–d). Using this system, we were able to reliably detect a ~ 1.8-fold increased APE1 chromatin localization upon treatment with MMS (Fig. 6e, left panel). We next examined how ALC1 loss impacted the chromatin localization of APE1. We labeled cells with 5-ethynyl-2’-deoxyuridine (EdU) to distinguish between non-S and S-phase cells. Non-EdU-positive sgALC1 cells showed a ~1.4-fold decrease in APE1 signal at the chromatin. Of note, we did not observe a difference in APE1 localization between ALC1-proficient and deficient EdU-positive cells (Fig. 6e). These data suggest that ALC1 can facilitate APE1 chromatin localization, albeit this impact is not evident in S-phase cells. This difference could likely be due to eviction of nucleosomes and chromatin opening during replication, hence ALC1 would become dispensable for APE1 localization to damage genomic sites.
Fig. 6. ALC1 promotes the repair of abasic sites by APE1 at the chromatin.
a Schematic showing the hypothesis that ALC1 can enhance APE1 accessibility to nucleosome-buried abasic sites. b Immunoblot showing expression levels of Dox-inducible APE1 constructs and levels of ALC1 depletion. Representative of three blots. c Schematic for quantifying MMS-induced APE1 localization to chromatin using E96Q/D210N mutant. 0.5% PBS Tx: PBS + 0.5% Triton X-100. d Representative image of chromatin-bound HA-APE1 E96Q/D210N. Scale: 10 microns. e Quantification of MMS-induced chromatin bound HA-APE1 E96Q/D210N normalized to untreated control. Data are presented as mean ± s.e.m from three biologically independent experiments. P values derived by unpaired student’s t-test. f Representative gel for APE1 cleavage assay with AP-NCP dyad substrate and product bands detected using the 6-FAM label. Representative of three gels. g Quantification of the AP-NCP-6 product formation assays for the indicated reactions. The data shown are the mean ± s.e.m from the three independent experiments. P value derived by one-way ANOVA. Source data are provided as a Source data file.
ALC1 promotes the ability of APE1 to cleave nucleosome-buried abasic sites
Previous studies have reported that APE1 can efficiently cleave abasic sites on nucleosome-free DNA and solvent-exposed abasic sites in the nucleosome. However, APE1 has a strong reduction in enzymatic activity on solvent-occluded abasic sites in the nucleosome42. Based on our data showing that ALC1 contributes to APE1 chromatin localization, we examined whether ALC1 can enhance the ability of APE1 to cleave solvent-occluded abasic sites. We assembled a nucleosome with DNA substrate containing a solvent-occluded abasic site and quantified APE1 endonuclease activity by monitoring the generation of a cleaved DNA product. Given that ATPase activity of ALC1 is enhanced in the presence of PARylated histone substrate, we included PARP1/histone PARylation factor 1 (HPF1) in the reaction mix with ALC1 (Fig. 6f, g)45,46. Consistent with previous studies42, APE1 alone displayed minimal endonuclease activity on the solvent-occluded nucleosomal abasic site. In the presence of ALC1/PARP1/HPF1, we observe a 36-fold increase in APE1 product formation, revealing that ALC1 can promote the enzymatic activity of APE1 on solvent-occluded nucleosomal abasic sites (Fig. 6f, g). Notably, the inclusion of PARPi reduced the ALC1-stimulated APE1 cleavage product roughly twofold, which aligns with previous findings that chromatin remodeling by ALC1 is enhanced in the presence of PARylated histones45,46.
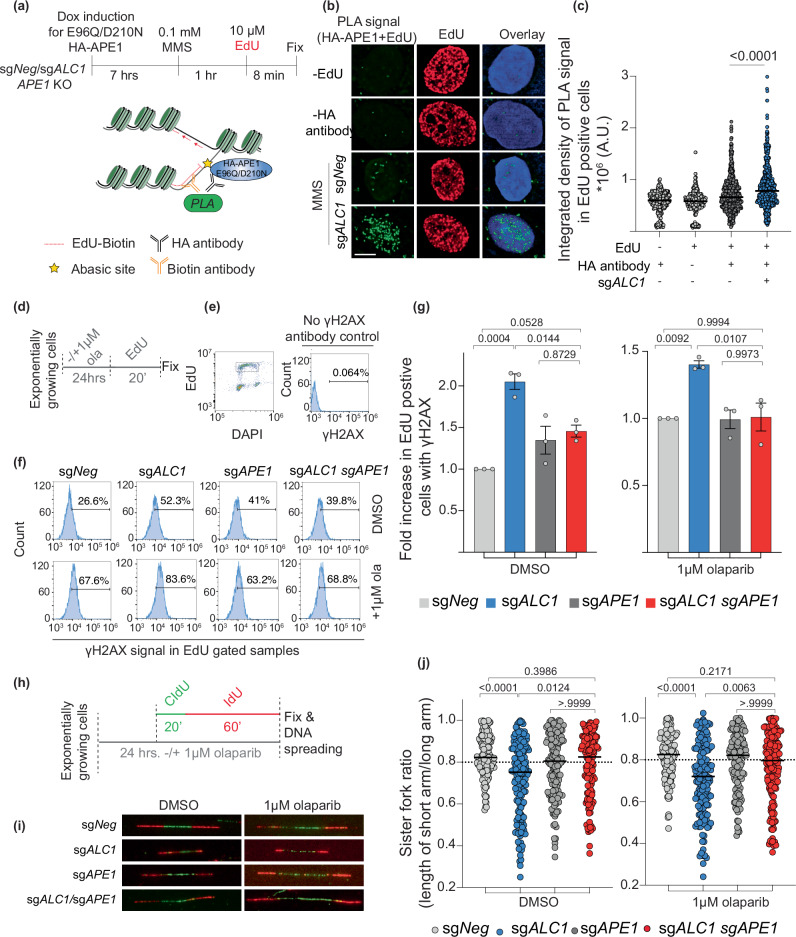
Loss of ALC1 increases localization of APE1 to active replication forks
Our cellular localization and in vitro reconstitution studies suggest that ALC1 loss would result in the accumulation of abasic sites in chromatin due to restricted access to APE1. However, we reasoned that APE1 could gain access to these abasic sites in front of the replication forks, where nucleosomes are evicted. Consistent with this hypothesis, recent studies have provided evidence for the ability of APE1 to act on single-stranded DNA and fork-like structures47. To examine whether ALC1 loss results in increased APE1 localization at the active forks, we performed in situ analysis of protein interactions at DNA replication forks (SIRF experiments)48 to assess proximity ligation assay (PLA) signal that emerged owing to proximity between biotin-conjugated EdU and hemagglutinin (HA)-tagged E96Q/D210N APE1 (Fig. 7a). We quantified the PLA signal specifically in EdU-positive cells to negate the background owing to endogenous biotin in cells. Consistent with our hypothesis, we observed increased colocalization between APE1 and EdU upon ALC1 depletion (Fig. 7b, c). Together, our studies reveal that ALC1-deficient cells display decreased APE1 localization to nucleosome-bound chromatin but increased APE1 localization at the replication forks. This observation may explain why ALC1 loss did not result in a discernible difference in APE1 chromatin localization in S-phase cells (Fig. 6e), which consists of both nucleosome-bound DNA and nucleosome-free replication forks.
Fig. 7. Localization of APE1 at forks in ALC1-deficient cells results in replication-coupled DSBs and fork stalling.
a Schematic of the PLA experiment to quantify APE1 localization to the replication fork. b Representative image of E96Q/D210N HA-APE1/EdU PLA signal. Scale is 5 microns. c Quantification of E96Q/D210N HA-APE1/EdU PLA signal. Median values are indicated. P values derived by Kruskal–Wallis test from >450 cells/condition collected over three biologically independent experiments. AU arbitrary units. d Schematic of the experiment to quantify replication-coupled DSBs. e Gating scheme to examine γH2Ax signal in EdU-positive cells. f Representative histograms showing γH2AX levels in indicated cell lines. g Normalized levels of EdU-positive cells with γH2AX in the absence (left) and presence of olaparib (right). Data are presented as mean ± s.e.m from three biologically independent experiments. P values derived by ordinary one-way ANOVA. h Schematic of the experiment to label nascent DNA for assessing fork symmetry. i Representative images of the nascent bi-directional fork in the indicated cell lines. j Quantification of sister fork ratio in the absence (left) and presence of olaparib (right). Median values are indicated. P values derived by Kruskal–Wallis test from >120 fibers/condition collected over three biologically independent experiments. Source data are provided as a Source data file.
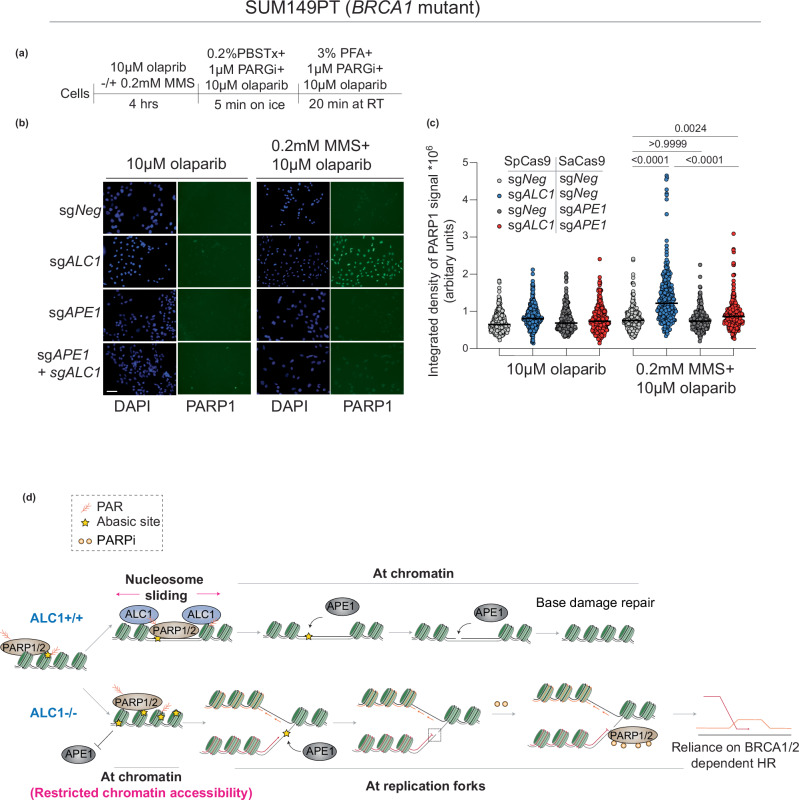
Loss of APE1 rescues replication-coupled DSBs, stalled forks, and PARP inhibitor trapping in ALC1-deficient BRCA mutant cancer cells
We and others have previously reported that ALC1 loss results in increased replication-coupled DSBs and fork stalling17,18. The frequency of breaks and stalling in ALC1-deficient cells is further enhanced upon PARPi treatment. Given our observation that ALC1-deficient cells display increased APE1 at the forks, we hypothesized that APE1-mediated fork cleavage would be the underlying source for replication-coupled DSBs and fork stalling previously reported in ALC1-deficient cells. To quantify DSBs, we used flow cytometry analysis to quantify γH2AX levels in EdU-positive cells upon ALC1 and APE1 loss in SUM149PT cells (Fig. 7d, e). While ALC1 loss resulted in increased DSBs, depletion of APE1 rescued the breaks both in the absence and presence of PARPi (Fig. 7f, g). We next used a single-molecule DNA fiber assay to assess sister fork ratios as a readout for fork stalling (Fig. 7h). ALC1 loss resulted in increased fork stalling, which was again rescued upon concomitant depletion of APE1. This trend remained consistent both in the absence and presence of PARPi (Fig. 7i, j). We have previously shown that PARPi hypersensitivity in ALC1-deficient BRCA mutant cells relies on trapping of PARP1 and 2 by PARP inhibitors17. To examine whether APE1-generated breaks were the lesions for trapping PARP1, we quantified chromatin-bound PARP1 using a previously reported immunofluorescence assay (Fig. 8a)49. APE1-deficient cells displayed no enhancement in PARP1 trapping, which is consistent with single-molecule in vitro assays demonstrating that olaparib does not support PARP1 retention on intact abasic sites50. Intriguingly, upon depletion of APE1 in ALC1-deficient cells we observe a significant reduction in PARPi-trapped PARP1 (Fig. 8b, c). We, therefore, surmise that APE1-generated breaks result in enhanced PARP1/2 trapping in ALC1-deficient BRCA mutant cells. Together, our results uncover that upon ALC1 loss, APE1 nuclease activity results in DSB formation and fork stalling. Increased DSBs and fork stalling result in the trapping of PARP1/2 enzymes by PARPi at the forks17, resulting in delayed fork repair and enhanced PARPi hypersensitivity in ALC1-deficient BRCA mutant cancer cells11.
Fig. 8. APE1 promotes PARPi mediated-PARP1 trapping and hypersensitivity in ALC1-deficient BRCA mutant cells.
a Schematic of the assay. b Representative images of the chromatin-bound PARP1 in indicated conditions. Scale bar 50 microns. c Quantification of the chromatin-bound signal for PARP1. Each dot represents a single cell. Median is indicated. P value determined by Kruskal–Wallis test derived from 300 cells sampled over three biologically independent experiments. d Model proposing the basis of PARPi hypersensitivity upon ALC1 loss in BRCA mutant cells. In WT cells, PAR-dependent nucleosome sliding by ALC1 promotes accessibility of APE1 to the abasic site buried in the chromatin. In contrast, in ALC1-deficient cells, abasic sites accumulate owing to restricted chromatin accessibility. At the replication forks, APE1 can gain access to abasic sites, resulting in replication-coupled DSBs and fork stalling. Trapping of PARP1/2 enzymes at the breaks by PARPi further delay fork re-start resulting in fork collapse and increased reliance on BRCA1/2 protein for repair and survival. Source data are provided as a Source data file.
Discussion
Based on our findings, we propose the following model: at the chromatin, nucleosome remodeling by ALC1 makes solvent-occluded abasic sites accessible to APE1. Therefore, ALC1-deficient cells would accumulate abasic sites in the chromatin because of inefficient repair. These abasic sites would become accessible to APE1 at the forks where nucleosomes are evicted. Nucleolytic cleavage of abasic sites at the forks results in replication-coupled double-strand breaks and fork stalling. Given that PARPi can trap PARP1/2 enzymes at breaks11, this further delays the resumption of stalled forks51, resulting in PARP inhibitor hypersensitivity (Fig. 8d). This intriguing mechanism of action explains why APE1 loss has a minimal impact on PARPi sensitivity compared to ALC1 in BRCA mutant cancer cells. Our studies, therefore, reveal cleaved abasic sites as cell-intrinsic DNA lesions that can profoundly augment PARPi response. Notably, abasic sites can also be cleaved by bifunctional glycosylases and nucleases, such as the endonuclease/exonuclease/phosphatase family domain containing 1 (EEPD1)37,52. While EEPD1 has been shown to act on AP sites at forks52, it remains unclear if bifunctional glycosylases can access abasic sites at ss-DNA substrates. We were unable to deplete APE1 in NTHL1/NEIL1/NEIL2-deficient cells perhaps reflecting functional redundancy between the abasic site processing enzymes. It is possible that these bifunctional glycosylases may cleave abasic sites at forks in the absence of APE1 and contribute to PARPi response in ALC1-deficient cells. Together, our studies provide compelling evidence that cleaved, but not intact abasic sites, are the underlying cause of PARPi hypersensitivity in ALC1-deficient BRCA mutant cells.
During base damage repair, PARP1 has been primarily implicated in catalyzing PARylation at the nicks generated upon the cleavage of abasic sites and helps recruit downstream repair factors. Previous studies have shown that PARP1 can also catalyze PARylation at intact abasic sites, albeit with a lower efficiency53. However, the physiological relevance of PARP1 activity at abasic sites remains enigmatic. We propose that low PARylation levels at an abasic site are sufficient for the recruitment of ALC1 via its macrodomain. This is because the macrodomain of ALC1 has a nanomolar binding affinity (10 nM) for PAR46. This strong binding affinity of ALC1 to PAR makes it a unique chromatin remodeler in promoting the repair of abasic sites.
Forks stalled by abasic sites can be rescued by PrimPol-mediated repair, which also results in the generation of replication-coupled gaps9,54. Perhaps a fraction of abasic sites in ALC1-deficient cells are tolerated via PrimPol-mediated repriming55,56, accounting for the gaps in ALC1-deficient cells. However, abrogating PrimPol-dependent gap formation does not rescue PARPi response, suggesting a minimal role of replication gaps in rendering PARPi sensitivity in either BRCA1 mutant cells or ALC1-deficient BRCA1 mutant cells. Notably, recent studies in BRCA2 mutant cells align with our findings that the formation of PrimPol-dependent gaps may not be the key basis of PARPi sensitivity in BRCA mutant settings57. Our findings are more consistent with a model whereby increased DSBs generated upon APE1 cleavage augment PARPi response in ALC1-deficient BRCA mutant cells. This is also supported by data showing that depletion of NHEJ regulating proteins, 53BP1 and Rev1, which reduce cytotoxic joining of DSBs in BRCA1 mutant cells, renders PARP inhibitor resistance in ALC1-deficient cells17.
Recent studies uncovered a role for the replisome protein, 5-hydroxymethylcytosine binding, ES cell specific (HMCES), in shielding abasic sites from nuclease cleavage58, albeit APE1 loss did not rescue DSBs in HMCES-deficient cells59. While the interplay between HMCES and APE1 at forks needs further investigation, our data suggest a key role of APE1 in generating replication-coupled DSBs in ALC1-deficient BRCA mutant cells. One possibility is that the number of abasic sites in ALC1-deficient cells exceeds the level that can be protected by the limited HMCES protein in cells, allowing APE1 to access the forks. This possibility is consistent with recent reports demonstrating the presence of APE1 at the replication forks in HMCES-proficient cells52. In our unpublished work, we observe that overexpression of HMCES is cytotoxic in BRCA mutant cells, suggesting that cellular levels of HMCES might be tightly regulated.
A previous model suggested a role for ALC1 in removing PARP1 from chromatin and thereby allowing NHEJ and HR repair to occur efficiently19. However, we did not observe enhanced sensitivity of ALC1-deficient cells to multiple agents that necessitate HR or NHEJ for repair (Fig. 2), suggesting that ALC1 is not a key player in any of these repair pathways. Of note, our previous studies have ruled out a role of ALC1 in evicting PARP2 in PARPi hypersensitivity by demonstrating that PARP2 loss has no impact on either PARPi or MMS response in ALC1-deficient cells. These data suggest that PARPi and MMS sensitivity of ALC1-deficient cells is not reliant on its proposed role in displacing chromatin bound PARP1 or PARP219,60.
ALC1 is epistatic to APE1 in response to exogenous base-damaging agents, MMS and KBrO3. In contrast, loss of APE1 confers PARPi resistance in ALC1-deficient cells. This distinction likely reflects the level of abasic sites induced by exogenous versus endogenous damage. Excess abasic sites on DNA and RNA would necessitate APE1 for repair and survival. In contrast, endogenous levels of abasic sites can be tolerated and become cytotoxic only when processed into DSBs in HR-repair deficient BRCA mutant cells. The limited amount of abasic sites in cells may also explain why the impact of ALC1 loss shows a more profound response in BRCA-deficient than proficient cells17,18. This is in contrast to other sensitizers of PARPi, such as RNAseH228, XRCC1, or Fen1, which act on more predominant lesions such as genome-embedded ribonucleotides or unligated Okazaki fragments28,61. The selectivity of ALC1 in repairing abasic sites and not other lesions likely accounts for the dispensability of this protein in the survival and cellular fitness of a BRCA-proficient mouse model18 and suggests that it will be a safe therapeutic target.
Methods
Cell culture
SUM149PT cells were cultured in Ham’s F-12 (Thermo Scientific 11765070) medium supplemented with 5% fetal bovine serum (FBS) (Sigma F0926), 1X penicillin and streptomycin (P/S) (Genesee Scientific 25-512), 1 mg/mL hydrocortisone (Sigma H0888) and insulin (5 mg/ml, Sigma). UWB1.289 cells were cultured in 1:1 Mammary Epithelial Cell Basal Medium (Thermo Scientific M-171) supplemented with Mammary Epithelial Growth Supplement (Thermo Scientific S0155): RPMI 1640 with l-glutamine (Genesee Scientific 25-506) and supplemented with 10% fetal bovine serum and 1X P/S. OVSAHO cells were procured from Sigma (SCC294) and were cultured in RPMI 1640 with l-glutamine supplemented with 10%FBS and 1x P/S. MDA-MB-436 cells were cultured in DMEM supplemented with 10% FBS and 1x P/S. DLD1 cells were cultured in RPMI 1640 with l-glutamine supplemented with 10%BCS and 1x P/S.HEK293T cells were cultured in DMEM (Sigma D5796) supplemented with 10% BCS (Sigma SH3007203) and 1X P/S. Lentiviruses were produced using HEK293T. HEK293T cells in a 10 cm dish were transfected with Opti-MEM solution containing 22.5 µg PAX2, 15 µg VSVG, and a 31.5 µg plasmid of interest and 240 µl of 1 mg/mL polyethylenimine (PEI). Media was changed 7 h following transfection, then collected four times over the course of 3 days. Viruses were stored at −80 ˚C until use. Cells were transduced with lentivirus in the presence of 8 µg/ml polybrene and centrifuged at 600×g for 30 min to increase transduction efficiency. Media was changed 24 h after transduction and selection (Puromycin 2 µg/mL, Blasticidin 5 µg/mL, Neomycin 400 µg/mL, and Hygromycin 100 µg/mL) was applied 48 h after transduction.
Cloning
All cDNAs and vector backbones were amplified using Phusion Flash PCR Mix (Thermo F548L) and cloned using In-Fusion Snap Assembly Master Mix (Takara Bio 638497). All mutagenesis was performed by overlapping PCR and product assembly using NEBuilder HiFi DNA Assembly Master Mix (NEB E2621L). All sgRNAs were cloned by traditional ligation-based cloning in BsmBI digested vectors. Cas12a guide designing and cloning was performed as described before62. All cDNA constructs will be deposited in Addgene. The sequence for Primers and oligos is detailed in Supplementary Table 1. All cDNA generated in this study are listed in Supplementary Table 2.
Nucleofection
OVSAHO ALC1 and APE1 knock-out cells were generated using Lonza’s SF Cell Line 4D-Nucleofector kit (Lonza V4XC-2032). Both Alt-R CRISPR-Cas9 sgRNA guides and purified Alt-R™ S.p. Cas9 Nuclease V3 (IDT 1081059) were ordered through IDT. 5 µM Cas9, and 15 µM sgRNA were incubated together for 20 min at room temperature and was added to 1 million cells resuspended in 100 µl of nucleofection solution (82 ul nucleofector solution + 18 ul Supplement). The solution was transferred to 100 µL cuvette. Cells were electroporated with code FE-132 on a 4D-Nucleofector X Unit (Lonza AAF-1003X). The sequence for sgRNAs is detailed in Supplementary Table 1.
Cell viability assay
For all epistasis analysis, APE1 was depleted first, followed by ALC1. 1000 SUM149PT cells in a volume of 100 μl were plated into each well of 96-well clear bottom plates on Day 0. On Day 1, 100 μl of 2X drug dilution was added to cells in technical replicates. While other drugs were retained, MMS-treated cells were released into fresh media after 24 h. Seven days post drug addition, the media was replaced with phenol red-free DMEM (Thermo Scientific, 31053028) supplemented with 10% BCS, 1x penicillin-streptomycin, 2 mM l-glutamine freshly reconstituted with resazurin (Sigma 199303) to a final concentration of 10 ug/mL. Cells were then incubated for 3–4 h at 37 °C or until the media in solvent-treated wells developed a pink color. Fluorescence was measured at 560 nm excitation and 590 nm emission using the SoftMax Pro 6.4 software on the SpectraMax i3x microplate reader. For UWB1.289, OVSAHO, and MDA-MB-436 cell lines, cells were plated in solid white flat bottom plates and viability assessment was performed CellTiter Glo 2.0 Assay (Promega, PRG9242). When plotting the survival curves, the fluorescence/luminescence of the drug-treated population was normalized to solvent-exposed cells. The data were fitted to a curve in GraphPad Prism using the following equation: y = min+ (max-min)/1 + 10logEC50-x.
Clonogenic survival assay
For clonogenic experiments with SUM149PT and OVSAHO cells, 500 cells were plated in technical replicates in 6-well plates and analyzed after 10–14 days. For clonogenic experiments with potassium bromate, cells were plated on Day 0, potassium bromate was added on Day 1, and media was changed on Day 8. For clonogenic experiments with UV, cells were plated on Day 0, treated with UV on Day 2, and media was changed on Day 9. In each of these experiments, after 10–14 days, cells were washed with PBS and stained with 0.4% crystal violet in 20% methanol for 30 min at room temperature. Plates were then washed with deionized water and air-dried. Colonies were manually counted.
Immunoblotting
The cell pellet was lysed by suspending in 5x volume of RIPA buffer supplemented with 1 mM DTT, 1x cOmplete EDTA-free Protease inhibitor cocktail (Sigma 11873580001), 0.5 ul benzonase (EMD Millipore 70746-3) and 1 mM PMSF. After 20 min incubation on ice, the cell suspension was centrifuged at 4 ○C for 20 min, followed by collection of the supernatant. Protein concentration in the supernatant was quantified using Bradford assay. About 10–20 µg of the protein lysate was resolved Nupage 4–12% Tris protein Gels (Thermo Scientific NP0336) in 1x MOPS buffer followed by transfer to 0.2 micron Nitrocellulose membrane (Cytiva 10600004) for 2 h at 400 mA using wet transfer. Blots were then blocked using 5% Milk in PBS with 0.2% Tween for an hour. Blot were probed overnight with respective primary antibodies (details and dilution of antibodies is compiled in Supplementary Table 3) followed by washes with PBS with 0.2% Tween, probing with 1:3000 dilution of HRP-conjugated anti-mouse (Cytiva NA931) or anti-rabbit (Cytiva NA934) for an hour at room temperature followed by washes, development using Western lightning Plus ECL kit (Perkin Elmer NEL10500) and imaging on iBright CL1500 Imaging system (Thermo Scientific A44240).
DNA fiber analysis
For fiber experiments in Fig. 1, exponentially growing cells were pulse-labeled with 250 µM 5-Iodo-2′-deoxyuridine (IdU) for 60 min. For fiber experiments in Supplementary Fig. 1 and Fig. 6, exponentially growing cells were first pulsed with 50 µM 5-chloro-2′-deoxyuridine (CldU) for 20 min, followed by 250 µM 5-Iodo-2′-deoxyuridine (IdU) for 40 min for fork asymmetry analysis or for 60 min for the S1 nuclease experiment. Cells were then collected by trypsinization, counted, and diluted in PBS to a concentration of 500,000 cells/mL. For the S1 nuclease experiment, diluted cells were treated with a CSK buffer (10 mM Pipes pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100) for 15 min on ice to pre-extract the nuclei. Samples were then divided into two: untreated or treated with 20 U/mL S1 nuclease in S1 nuclease buffer pH 4.6 (3 M acetic acid, 1 mL 1 M zinc acetate, 10 mL 50% glycerol, 1.25 mL 4 M sodium chloride, milli-Q water up to 100 mL). Cells were lysed on the slides by mixing with spreading buffer (20 mL 1X Tris-Cl pH 7.5, 2.5 mL 20% SDS, 10 mL 0.5 M EDTA, 67.5 mL milli-Q water) followed by tilting the slide at a 15–45 ° angle to spread the DNA. Once the slides were completely dry, DNA was fixed in 3:1 methanol: acetic acid overnight. The next day, slides were re-hydrated using two 5-min PBS washes. DNA was then denatured by incubating slides in 2.5 N hydrochloric acid for 1 h at room temperature. Slides were then washed three times with PBS and followed by incubation in the blocking buffer (5% BSA in 0.2% PBS-Tween). Primary antibodies for CldU and IdU made 1:200 each in blocking buffer were then added to the slides and incubated for 2 h at room temperature. After the 2-h incubation, slides were washed four times with 0.2% PBS-Tween. Slides were then treated with secondary antibodies for anti-mouse 594 and anti-rat 488 made 1:200 each in a blocking buffer for 1 h at room temperature. Slides were washed four times with 0.2% PBS-Tween. Coverslips were mounted to the slides using VECTASHIELD Vibrance Antifade Mounting Medium. Slides were stored at 4 °C until imaging. Slides were imaged using a Nikon Ti2 microscope. For fork asymmetry, fibers with red-green-red patterns were analyzed. Each red arm of the origin was measured, and then a ratio of the short to the long arm was calculated. For S1 nuclease analysis total length was calculated when pulsing was done only with IdU, and length of red attached to green was calculated when pulsing was done with CIdU followed by IdU. All analysis was blinded.
ɣH2AX-EdU FACS analysis
Exponentially growing cells were pulsed with 10 µM EdU for 20 min. Cells were then collected by trypsinization followed by PBS wash. Fixation was performed by dropwise addition of 90% cold methanol to gently vortexed cell pellets followed by overnight incubation at −20 °C. EdU was conjugated to Alexa-Fluor using manufacturer protocol. ɣH2AX was detected using 1:500 dilution of ɣH2AX antibody (05-636-I, Sigma) followed by 1:500 dilution of Alexa-Fluor conjugated anti-mouse antibody (Life technologies). Post staining for EdU and ɣH2AX, cells were incubated with 20 µg/ml 4’,6-diamidino-2-phenylindole (DAPI) made in 1xPBS with 1 mg/ml RNaseA for an hour before data acquisition. Data was acquired on a Beckman Cytoflex equipped with a 96-well plate loader. Images were analyzed using FlowJo.
Biotin-HA PLA
sgAPE1 cells were engineered with a dox-inducible HA-APE1 E96Q/D210N cDNA. Forty-eight hours prior to the experiment, 50,000 cells were plated on each Poly-lysine-coated coverslip in a 24-well plate. Expression of APE1 mutant protein was induced by the addition of 1 µg/ml doxycycline. Seven hours post induction, cells were treated with 100 µM MMS for 1 h, followed by pulsing with 10 µM EdU for 8 min. Cells were fixed with 3% PFA for 10 min followed by PBS washes. EdU was labeled both with Biotin-Azide and Alexa-Fluor Azide. Samples were incubated with 1:200 dilution of rabbit anti-biotin antibody (Cell Signaling, 5597 S) and 1:500 dilution of mouse anti-HA antibody (BioLegend, 901514) overnight. Negative controls included a no biotin-azide and no HA antibody-treated samples. Coverslips were mounted using Prolong Diamond Antifade mounting media with DAPI (Thermo Fisher Scientific, P36962). All images were acquired on a Nikon Ti-2 60x objective. Z-stacks were taken at every 0.2 microns. The stacks were subjected to deconvolution followed by extended depth focus analysis to obtain the 3D projections. The ROI for each nucleus was defined in the DAPI channel using the magic wand tool. The total integrated density of the PLA signal in each ROI was calculated using the control M function in ImageJ. All analysis was blinded.
HA-EdU Immunofluorescence staining
sgAPE1 SUM149PT cells were engineered with a dox-inducible HA-APE1 E96Q/D210N cDNA. Forty-eight hours prior to the experiment, 50,000 cells were plated on each poly-lysine-coated coverslip in a 24-well plate. Expression of APE1 mutant protein was induced by the addition of 1 µg/ml doxycycline. Seven hours post induction, cells were treated with 100 µM MMS for an hour followed by pulsing with 10 µM EdU for 8 min. Cells were pre-extracted on ice for 2 min with CSK buffer, followed by fixation with 3% PFA for 10 min. After PBS washes, cells were permeabilized with ice-cold 1:1 methanol-acetic acid for 5 min and blocked in 3% BSA made in PBS for 30 min at room temperature. EdU was labeled with Alexa-Fluor Azide using Click Reaction by following the manufacturer’s instructions (Fisher Scientific, 502108139). Cells were washed once with 3% BSA in PBS, thrice with 0.2% PBS-Tween, and once with PBS. Cells were fixed again with 3% PFA for 10 min, followed by PBS washes. Samples were incubated with 1:1000 dilution of mouse anti-HA antibody (BioLegend, 901514) for 1 h at room temperature, followed by washes with 0.2% PBS-Tween. Samples were incubated with 1:1000 dilution of Alexa-Fluor 488 conjugated anti-mouse secondary (Thermo, A32766) for 1 h at room temperature. Coverslips were mounted using Prolong Diamond Antifade mounting media with DAPI (Thermo, P36962). All images were acquired on a Nikon Ti-2 60x objective. For cells, z-stacks were taken at every 0.2 microns. The stacks were subjected to deconvolution followed by extended depth focus analysis to obtain the 3D projections. ROI for each nucleus was defined using a magic wand tool in the DAPI channel. The total integrated density of the HA signal for each ROI was calculated using the control M function in ImageJ. All analysis was blinded.
Preparation of recombinant human PARP1, HPF1, ALC1, and APE1
A pET24a vector containing H. sapiens PARP1 (full-length) was purchased from GenScript. The PARP1 protein was expressed in T7 Express lysY/Iq E. coli cells (New England Biolabs). The cells were grown at 37 °C to an OD600 = 1.0, induced with 0.2 mM IPTG overnight at 16 °C, and cells were harvested via centrifugation for 20 min at 3700×g. The harvested cells were lysed via sonication in a buffer containing 50 HEPES (pH 8.0), 500 mM NaCl, 0.5 mM DTT, 0.1% NP-40, 10 mM Benzamide (PARPi), 20 mM Imidazole, and a protease inhibitor cocktail (PMSF, leupeptin, pepstatin, antipain, aprotinin, benzamidine) at 4 oC. The cell lysate was cleared for 1 h at 24,242×g, and the supernatant loaded over a Histrap HP (Cytiva) equilibrated with a buffer containing 50 mM HEPES (pH 8.0), 500 mM NaCl, 0.5 mM DTT, and 20 mM Imidazole. The Histrap HP column was washed with a buffer 50 mM HEPES (pH 8.0), 1 M NaCl, 0.5 mM DTT, and 20 mM Imidazole, prior to elution of PARP1 with 50 mM HEPES (pH 8.0), 500 mM NaCl, 0.5 mM DTT, and 400 mM Imidazole. The PARP1 protein was subsequently loaded onto a Heparin HP column (Cytiva) equilibrated in a buffer containing 50 mM Tris (pH 7.0), 375 mM NaCl, 1 mM EDTA, and 1 mM DTT, and eluted from the column with a linear salt gradient to 1 M NaCl. The PARP1 protein was further purified using a HiPrep 26/60 Sephacryl S-200 HR (Cytiva) in a buffer containing 50 mM HEPES (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 0.5 mM DTT. The purified PARP1 protein was concentrated at 5 mg/mL and stored long-term at −80 °C.
A pET-6xHis-GFP-TEV vector (addgene #29663) containing the H. sapiens HPF1 (full-length) was purchased from GenScript. The GFP-HPF1 protein was expressed in One Shot BL21(DE3) plysS E. coli cells (Invitrogen). The cells were grown at 37 °C to an OD600 = 0.6, induced with 0.5 mM IPTG overnight at 18 °C, and cells were harvested via centrifugation for 20 min at 3700×g. The harvested cells were lysed via sonication in a buffer containing 50 mM HEPES (pH 7.4), 500 mM NaCl, 1 mM EDTA, 20 mM imidazole, and a protease inhibitor cocktail (AEBSF, leupeptin, benzamidine, pepstatin A) at 4 oC, and the cell lysate cleared for 1 h at 24,242×g. The supernatant containing GFP-HPF1 was purified via a Histrap HP (Cytiva) equilibrated with containing 50 mM HEPES (pH 7.4), 500 mM NaCl, and 20 mM imidazole, and GFP-HPF1 eluted off the column with a linear gradient to 500 mM Imidazole. HPF1 was then liberated from the GFP-tag using tobacco etch virus (TEV) protease for 2 h at 37 oC. The cleaved HPF1 was then purified by anion-exchange chromatography using a HiTrap Q HP column (Cytiva) equilibrated with 50 mM HEPES (pH 7.4), 50 mM NaCl, 1 mM EDTA, and 1 mM DTT, and eluted from the column with a linear salt gradient to 1 M NaCl. The HPF1 protein was then polished via gel filtration on a HiPrep 16/60 Sephacryl S-200 HR (Cytiva) in a buffer containing 50 mM HEPES (pH 7.4), 100 mM NaCl, and 1 mM TCEP. The purified HPF1 protein was concentrated to 10 mg/mL and stored long term at −80 °C.
A pET-6xHis-GFP-TEV vector (addgene #29663) containing H. sapien ALC1 (AA residues 16–879) was purchased from GenScript. The GFP-ALC1 protein was expressed in One Shot BL21(DE3) plysS E. coli cells (Invitrogen). The cells were grown at 37 °C to an OD600 = 0.6, induced with 0.5 mM IPTG overnight at 18 °C, and cells were harvested via centrifugation for 20 min at 3700×g. The harvested cells were lysed via sonication in a buffer containing 50 mM HEPES (pH 7.4), 500 mM NaCl, 1 mM EDTA, 10% glycerol, 20 mM imidazole, and a protease inhibitor cocktail (AEBSF, leupeptin, benzamidine, pepstatin A) at 4 oC, and the cell lysate cleared for 1 h at 24,242×g. The supernatant containing GFP-ALC1 was purified via a Histrap HP (Cytiva) equilibrated with containing 50 mM HEPES (pH 7.4), 500 mM NaCl, 10% glycerol, and 20 mM imidazole, and GFP-ALC1 eluted off the column with a linear gradient to 500 mM Imidazole. The GFP-ALC1 was then purified by cation-exchange chromatography using a RESOURCE S column (Cytiva) equilibrated with 50 mM HEPES (pH 7.4), 200 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10% glycerol, and eluted from the column with a linear salt gradient to 1 M NaCl. ALC1 was then liberated from the GFP-tag using TEV protease for 2 h at 37 oC, and the ALC1 protein was purified via gel filtration on a HiPrep 16/60 Sephacryl S-200 HR (Cytiva) in a buffer containing 50 mM HEPES (pH 7.4), 200 mM NaCl, 1 mM TCEP, and 10% glycerol. The purified ALC1 protein was concentrated to 5 mg/mL and stored long term at −80 °C.
A pet28a vector containing H. sapiens APE1 (full-length) was purchased from GenScript. The APE1 protein was expressed in One Shot BL21(DE3) plysS E. coli cells (Invitrogen). The cells were grown at 37 °C to an OD600 = 0.6, induced with 0.4 mM IPTG overnight at 20 °C, and cells were harvested via centrifugation for 20 min at 3700×g. The harvested cells were lysed via sonication in a buffer containing 50 mM HEPES (pH 7.4), 50 mM NaCl, and a protease inhibitor cocktail (AEBSF, leupeptin, benzamidine, pepstatin A) at 4 oC, and the cell lysate cleared for 1 h at 24,242×g. The supernatant containing APE1 was purified via a HiTrap Heparin HP (Cytiva) equilibrated with 50 mM HEPES (pH 7.4) and 50 mM NaCl, and APE1 eluted off the column with a linear salt gradient to 1 M NaCl. The APE1 was then purified by cation-exchange chromatography using a RESOURCE S column (Cytiva) equilibrated with 50 mM HEPES (pH 7.4) and 50 mM NaCl and eluted from the column with a linear salt gradient to 1 M NaCl. The APE1 protein was then polished via gel filtration on a HiPrep 16/60 Sephacryl S-200 HR (Cytiva) in a buffer containing 50 mM HEPES (pH 7.4) and 100 mM NaCl. The purified APE1 protein was concentrated to 25 mg/mL and stored long term at −80 °C.
Preparation of Cy5-labeled AP-DNA
The Cy5-labeled DNA substrate containing the 601 strong positioning sequence and a single AP-site was generated using a well-established ligation-based method63. In brief, three oligonucleotides (oligos) were resuspended to a final concentration of 1 mM in a buffer containing 10 mM Tris (pH 7.5) and 10 mM NaCl. The oligo sequences are listed in Supplementary Table 1. Equimolar amounts of the resuspended oligos were mixed and annealed by heating to 95 oC and cooling to 4 oC at a rate of 0.1 oC/sec. The annealed oligos were then diluted in a buffer containing 50 mM Tris (pH 7.5), 10 mM MgCl2, 10 mM DTT, and 1 mM ATP, and ligated with T4 DNA ligase (New England Biolabs). The ligation products were then separated by denaturing polyacrylamide gel electrophoresis (8 M urea, 10% 29:1 acrylamide: bis-acrylamide). The full-length Cy5-labeled AP-DNA substrate was extracted from the gel three times by soaking in a buffer containing 200 mM NaCl and 1 mM EDTA, and the purified substrate buffer was exchanged five times into 10 mM Tris (pH 7.5) and 1 mM EDTA. The Cy5-labeled AP-DNA substrate was then annealed by heating to 95 oC and cooling to 4 oC at a rate of 0.1 oC/sec. The annealed Cy5-labeled AP-DNA substrate was stored long-term at −20 oC.
Preparation of Cy5-labeled AP-NCP
The genes encoding H. sapiens histone H2A, histone H2B, histone H3.2 (C110A), and histone H4 were cloned into a pET3a vector. The vectors for histones H2A, H3, and H4 were transformed into T7 Express lysY competent cells (New England Biolabs), while histone H2B was transformed into Rosetta™ 2(DE3) pLysS competent cells (Novagen). For histone expression, the transformed cells were grown in M9 minimal media at 37 °C until an OD600 of 0.4 was reached, and histone expression induced with 0.2 mM (Histone H4) or 0.4 mM (Histone H2A, H2B, and H3) IPTG for 3–4 h at 37 °C. The histones were then purified under denaturing conditions using a well-established protocol63,64. In brief, the histones were extracted from inclusion bodies under denaturing conditions (6 M guanidinium chloride), and the extracted histones were purified using a combination of anion-exchange and cation-exchange chromatography. The purified histones were dialyzed into H2O five times, lyophilized, and stored long-term at –20 °C.
H2A/H2B dimers and H3/H4 tetramers were generated using a salt-dialysis approach63,64. In brief, individual histones were resuspended in a buffer containing 20 mM Tris (pH 7.5), 6 M guanidinium chloride, 10 mM DTT, and H2A-H2B or H3-H4 mixed at a 1:1 molar ratio. The H2A-H2B dimers and H3-H4 tetramers were refolded by dialyzing three times against a high salt buffer containing 2 M NaCl, 20 mM Tris (pH 7.5), and 1 mM EDTA. The refolded H2A/H2B dimers and H3/H4 tetramers were purified by gel filtration using a Sephacryl S-200 HR (GE Health Sciences) in a buffer with 2 M NaCl, 20 mM Tris (pH 7.5), and 1 mM EDTA. The purified H2A/H2B dimers and H3/H4 tetramers were mixed with 50% glycerol and stored long-term at –20 °C.
The Cy5-AP-NCP was generated using a well-established salt-dialysis method63,64. Initially, Cy5-AP-DNA, H2A/H2B dimer, and H3/H4 tetramer were mixed at a 1:2:1 molar ratio, respectively, in a buffer containing 20 mM Tris (pH 7.5), 2 M NaCl, and 1 mM EDTA. The Cy5-AP-NCP was then reconstituted using dialysis to incrementally decrease the salt to 1.5 M NaCl, 1.0 M NaCl, 0.66 M NaCl, 0.50 M NaCl, 0.25 M NaCl, and 0 M NaCl over 24 h. The reconstituted Cy5-AP-NCP was heat shocked at 55 °C and purified by ultracentrifugation through a sucrose gradient (10% − 40%). Nucleosome formation and purity was determined using native polyacrylamide gel electrophoresis (5% 59:1 acrylamide:bis-acrylamide). The Cy5-AP-NCP was concentrated to 1 µM and stored short-term at 4 °C.
APE1 cleavage assays
APE1 cleavage assays were performed by pre-incubating the AP-nucleosome substrate (25 nM) for 30 min at 37 oC with or without PARP1 (100 nM), HPF1 (100 nM), ALC1 (100 nM), Olaparib (20 µM, +/−) in a buffer containing 50 mM HEPES (pH 7.5), 100 mM KCl, 1 mM DTT, 5 mM MgCl2, 2 mM ATP, 50 uM NAD+, 0.1 mg ml−1 BSA, and 1% DMSO. The cleavage reactions were initiated through the addition of APE1 (1 nM) and the reactions quenched after 3 min with a loading dye containing 100 mM EDTA, 80% deionized formamide, 0.25 mg/ml bromophenol blue and 0.25 mg/ml xylene cyanol. The quenched reactions were incubated at 95 oC for 5 min, resolved on a denaturing polyacrylamide gel (15%, 29:1 acrylamide/bis-acrylamide), and the substrate and product bands visualized using an Amersham Typhoon RGB imager. The amount of substrate and product for each reaction was quantified using ImageJ. The APE1 cleavage assays were performed as three independent replicate experiments.
PARP1 trapping assay
The PARP1 trapping experiment was performed as described before49. Briefly, 3000 cells were plated on a poly-l-lysine-coated eight-well chamber slide. After 5 days, cells were either treated with 10 µM olaparib or 10 µM olaparib and 0.2 mM MMS for 4 h. Cells were pre-extracted on ice with 0 PBS supplemented with 0.2% Triton X-100, h 1 µM PARGi (PDD, Fisher, cat. no. 59-521-0) and 10 µM olaparib, for 5 min. Cells were then fixed with 3% PFA, supplemented with 1 µM PARGi and 10 µM olaparib, for 20 min at room temperature. Cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min on ice, then washed with PBS. Blocking was performed for 1 h at room temperature with 0.22 µ-filtered DMEM containing 10% FBS. Cells were incubated with 1:500 dilution of rabbit anti-PARP (Abcam, ab227244) for 1 h at room temperature, followed by washes with 0.02% PBS-Tween. Cells were next incubated with 1:500 dilution of Alexa-Fluor 488 conjugated anti-rabbit secondary (Thermo, A32766) for 1 h at room temperature. Finally, the chamber gasket was removed, and a coverslip was mounted using Prolong Diamond Antifade mounting media with DAPI (Thermo, P36962). All images were acquired on a Nikon Ti-2 using a 20x objective.
Rad51 foci assay
About 50,000 cells were plated on each Poly-lysine-coated coverslip in a 24-well plate. One day after plating, treatment was added to the cells. Twenty-four hours after treatment, cells were pre-extracted on ice for 3 min with ice-cold 0.5% Triton X-100 followed by fixation with 3% PFA for 15 min at room temperature. After PBS washes, cells were permeabilized with ice-cold 0.5% Triton X-100 for 5 min and blocked in 3% BSA made in PBS for 30 min at room temperature. EdU was labeled with Alexa-Fluor Azide using Click Reaction by following the manufacturer’s instructions (Fisher Scientific, 502108139). Cells were washed once with 3% BSA in PBS, thrice with 0.2% PBS-Tween, and once with PBS. Cells were fixed again with 3% PFA for 10 min, followed by PBS washes. Samples were incubated with 1:500 dilution of rabbit anti-Rad51 antibody (Cosmo Bio, BAM-70-001-EX) for 1 h at room temperature, followed by washes with 0.2% PBS-Tween. Samples were incubated with 1:1000 dilution of Alexa-Fluor 555 conjugated anti-rabbit secondary (Thermo, A32732) for 1 h at room temperature. Coverslips were mounted using Prolong Diamond Antifade mounting media with DAPI (Thermo, P36962). All images were acquired on a Nikon Ti-2 60x objective. For cells, z-stacks were taken at every 0.2 micron. The stacks were subjected to deconvolution followed by extended depth focus analysis to obtain the 3D projections. ROI for each nucleus was defined using a magic wand tool in the DAPI channel. The number of Rad51 foci for each ROI was quantified using the Find Maxima function in ImageJ. All analysis was blinded.
Software used
All figures were generated using Adobe Illustrator 2020. Chemdraw 23.1.1 was used for making chemical structures. Microscopy data was acquired using Nikon NIS-element Software. Absorbance and Luminescence data was acquired suing SoftMax Pro 6.4 and Flow data was acquired using CytExpert 2.4. Data was analysis and plotted using GraphPad PRISM 8, Microsoft Excel, Fiji 2.0 and FlowJo.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Dr. Roger A. Greenberg (University of Pennsylvania, PA) for sharing ALC1 sgRNA constructs and for insightful discussion on the project. We thank Vrutti Mehta (Washington University) for help with western blotting. This work was supported by the Inaugural Pedal the Cause Grant by the Alvin J. Siteman Cancer Center through The Foundation for Barnes-Jewish Hospital, the Rivkin Pilot Study Award, the Mary Kay Ash Cancer Research Grant, Early-career investigator Award from Ovarian Cancer Research Fund Alliance, Career Catalyst Grant from Susan G. Komen and R37-CA286908 to P.V. who is also supported by V-Scholar Grant, Early-career Investigator Award from Ovarian Cancer Academy, Department of Defense, ACS-IRG grants and Siteman Research Program Catalyst Award from Breast Cancer Research Program at Washington University. NR is supported by the NIH Cell and Molecular Biology training T32 grant to Washington University, St. Louis. L.N.A. is supported by the NIH Cancer Biology Pathway training T32 grant to Washington University, St. Louis. A.V. is supported by R01-CA237263 and R01-CA248526. B.D.F. and T.M.W. are supported by R35-GM128562 and R01-ES029203.
Author contributions
P.V. conceptualized the study. N.R. and L.N.A. performed and analyzed most of the cell biology experiments with assistance from A.W., T.J., and K.S. under guidance from P.V. T.M.W. and B.D.F. designed the in vitro experiments. T.M.W. performed and analyzed the in vitro experiments in Fig. 6f, g. A.V. provided insightful suggestions. P.V. wrote the manuscript with contributions from N.R., T.M.W., L.N.A., A.V., and B.D.F.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information files and can be obtained from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50673-7.
References
- 1.Kim, D.-S., Camacho, C. V. & Kraus, W. L. Alternate therapeutic pathways for PARP inhibitors and potential mechanisms of resistance. Exp. Mol. Med.53, 42–51 (2021). 10.1038/s12276-021-00557-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slade, D. PARP and PARG inhibitors in cancer treatment. Genes Dev.34, 360–394 (2020). 10.1101/gad.334516.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Andrea, A. D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair71, 172–176 (2018). 10.1016/j.dnarep.2018.08.021 [DOI] [PubMed] [Google Scholar]
- 4.Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell141, 243–254 (2010). 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenberg, R. A. Assembling a protective shield. Nat. Cell Biol.20, 862–863 (2018). 10.1038/s41556-018-0152-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray Chaudhuri, A. et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature535, 382–387 (2016). 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liptay, M., Barbosa, J. S. & Rottenberg, S. Replication fork remodeling and therapy escape in DNA damage response-deficient cancers. Front. Oncol.10, 670 (2020). 10.3389/fonc.2020.00670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong, K. et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell81, 3227 (2021). 10.1016/j.molcel.2021.07.015 [DOI] [PubMed] [Google Scholar]
- 9.Mourón, S. et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol.20, 1383–1389 (2013). 10.1038/nsmb.2719 [DOI] [PubMed] [Google Scholar]
- 10.Vaitsiankova, A. et al. PARP inhibition impedes the maturation of nascent DNA strands during DNA replication. Nat. Struct. Mol. Biol.29, 329–338 (2022). 10.1038/s41594-022-00747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murai, J. et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res.72, 5588–5599 (2012). 10.1158/0008-5472.CAN-12-2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray Chaudhuri, A. & Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol.18, 610–621 (2017). 10.1038/nrm.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azarm, K. & Smith, S. Nuclear PARPs and genome integrity. Genes Dev.34, 285–301 (2020). 10.1101/gad.334730.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noordermeer, S. M. & van Attikum, H. PARP inhibitor resistance: a tug-of-war in BRCA-mutated cells. Trends Cell Biol.29, 820–834 (2019). 10.1016/j.tcb.2019.07.008 [DOI] [PubMed] [Google Scholar]
- 15.Gogola, E., Rottenberg, S. & Jonkers, J. Resistance to PARP inhibitors: lessons from preclinical models of BRCA-associated cancer. Annu. Rev. Cancer Biol.3, 235–254 (2019). 10.1146/annurev-cancerbio-030617-050232 [DOI] [Google Scholar]
- 16.Morice, P.-M. et al. Myelodysplastic syndrome and acute myeloid leukaemia in patients treated with PARP inhibitors: a safety meta-analysis of randomised controlled trials and a retrospective study of the WHO pharmacovigilance database. Lancet Haematol.8, e122–e134 (2021). 10.1016/S2352-3026(20)30360-4 [DOI] [PubMed] [Google Scholar]
- 17.Verma, P. et al. ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells. Nat. Cell Biol.23, 160–171 (2021). 10.1038/s41556-020-00624-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hewitt, G. et al. Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol. Cell81, 767–783.e11 (2021). 10.1016/j.molcel.2020.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juhász, S. et al. The chromatin remodeler ALC1 underlies resistance to PARP inhibitor treatment. Sci. Adv.6, eabb8626 (2020). [DOI] [PMC free article] [PubMed]
- 20.Verma, P. & Greenberg, R. A. Communication between chromatin and homologous recombination. Curr. Opin. Genet. Dev.71, 1–9 (2021). 10.1016/j.gde.2021.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuda, M. et al. ALC1/CHD1L, a chromatin-remodeling enzyme, is required for efficient base excision repair. PLoS ONE12, e0188320 (2017). 10.1371/journal.pone.0188320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahel, D. et al. Poly (ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science325, 1240–1243 (2009). 10.1126/science.1177321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pines, A. et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol.199, 235–249 (2012). 10.1083/jcb.201112132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blessing, C. et al. XPC-PARP complexes engage the chromatin remodeler ALC1 to catalyze global genome DNA damage repair. Nat. Commun.13, 4762 (2022). 10.1038/s41467-022-31820-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fugger, K. et al. Targeting the nucleotide salvage factor DNPH1 sensitizes BRCA-deficient cells to PARP inhibitors. Science372, 156–165 (2021). 10.1126/science.abb4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tubbs, A. & Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell168, 644–656 (2017). 10.1016/j.cell.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, P. S. & Cortez, D. New insights into abasic site repair and tolerance. DNA Repair90, 102866 (2020). 10.1016/j.dnarep.2020.102866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann, M. et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature559, 285–289 (2018). 10.1038/s41586-018-0291-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X. et al. ERCC6L2 promotes DNA orientation-specific recombination in mammalian cells. Cell Res.30, 732–744 (2020). 10.1038/s41422-020-0328-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinet, A., Carvajal-Maldonado, D., Lemacon, D. & Vindigni, A. DNA fiber analysis: mind the gap! Methods Enzymol.591, 55–82 (2017). [DOI] [PubMed]
- 31.Cong, K. & Cantor, S. B. Exploiting replication gaps for cancer therapy. Mol. Cell10.1016/j.molcel.2022.04.023 (2022). [DOI] [PMC free article] [PubMed]
- 32.Wang, Y. et al. The BRCA1-Δ11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res.76, 2778–2790 (2016). 10.1158/0008-5472.CAN-16-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DelloRusso, C. et al. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol. Cancer Res.5, 35–45 (2007). 10.1158/1541-7786.MCR-06-0234 [DOI] [PubMed] [Google Scholar]
- 34.Johnson, N. et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc. Natl Acad. Sci. USA110, 17041–17046 (2013). 10.1073/pnas.1305170110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura, N. et al. Specific mechanisms of chromosomal instability indicate therapeutic sensitivities in high-grade serous ovarian carcinoma. Cancer Res.80, 4946–4959 (2020). 10.1158/0008-5472.CAN-19-0852 [DOI] [PubMed] [Google Scholar]
- 36.Domcke, S., Sinha, R., Levine, D. A., Sander, C. & Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun.4, 2126 (2013). 10.1038/ncomms3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krokan, H. E. & Bjørås, M. Base excision repair. Cold Spring Harb. Perspect. Biol.5, a012583 (2013). 10.1101/cshperspect.a012583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill, J. W., Hazra, T. K., Izumi, T. & Mitra, S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res.29, 430–438 (2001). 10.1093/nar/29.2.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, R., Hao, W., Pan, L., Boldogh, I. & Ba, X. The roles of base excision repair enzyme OGG1 in gene expression. Cell. Mol. Life Sci.75, 3741–3750 (2018). 10.1007/s00018-018-2887-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs, A. L. & Schär, P. DNA glycosylases: in DNA repair and beyond. Chromosoma121, 1–20 (2012). 10.1007/s00412-011-0347-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nacson, J. et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep.24, 3513–3527.e7 (2018). 10.1016/j.celrep.2018.08.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weaver, T. M. et al. Structural basis for APE1 processing DNA damage in the nucleosome. Nat. Commun.13, 5390 (2022). 10.1038/s41467-022-33057-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothwell, D. G. & Hickson, I. D. Asparagine 212 is essential for abasic site recognition by the human DNA repair endonuclease HAP1. Nucleic Acids Res.24, 4217–4221 (1996). 10.1093/nar/24.21.4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitaker, A. M. & Freudenthal, B. D. APE1: a skilled nucleic acid surgeon. DNA Repair71, 93–100 (2018). 10.1016/j.dnarep.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann, L. C. et al. Mechanistic insights into autoinhibition of the oncogenic chromatin remodeler ALC1. Mol. Cell68, 847–859.e7 (2017). 10.1016/j.molcel.2017.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh, H. R. et al. A poly-ADP-ribose trigger releases the auto-inhibition of a chromatin remodeling oncogene. Mol. Cell68, 860–871.e7 (2017). 10.1016/j.molcel.2017.11.019 [DOI] [PubMed] [Google Scholar]
- 47.Hoitsma, N. M. et al. AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop. Nucleic Acids Res.48, 7345–7355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy, S. & Schlacher, K. SIRF: a single-cell assay for protein interaction with nascent DNA replication forks. Bio Protoc.9, e3377 (2019). 10.21769/BioProtoc.3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michelena, J. & Altmeyer, M. Cell cycle resolved measurements of poly (ADP-ribose) formation and DNA damage signaling by quantitative image-based cytometry. Methods Mol. Biol.1608, 57–68 (2017). 10.1007/978-1-4939-6993-7_5 [DOI] [PubMed] [Google Scholar]
- 50.Liu, L. et al. PARP1 changes from three-dimensional DNA damage searching to one-dimensional diffusion after auto-PARylation or in the presence of APE1. Nucleic Acids Res.45, 12834–12847 (2017). 10.1093/nar/gkx1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serrano-Benitez, A. et al. Unrepaired base excision repair intermediates in template DNA strands trigger replication fork collapse and PARP inhibitor sensitivity. EMBO J.42, e113190 (2023). 10.15252/embj.2022113190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaiswal, A. S. et al. EEPD1 promotes repair of oxidatively-stressed replication forks. NAR Cancer5, zcac044 (2023). 10.1093/narcan/zcac044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khodyreva, S. N. et al. Apurinic/apyrimidinic (AP) site recognition by the 5’-dRP/AP lyase in poly (ADP-ribose) polymerase-1 (PARP-1). Proc. Natl Acad. Sci. USA107, 22090–22095 (2010). 10.1073/pnas.1009182107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinet, A. et al. PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-deficient cells. Mol. Cell77, 461–474.e9 (2020). 10.1016/j.molcel.2019.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taglialatela, A. et al. REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. Mol. Cell81, 4008–4025.e7 (2021). 10.1016/j.molcel.2021.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawale, A. S. et al. APOBEC3A induces DNA gaps through PRIMPOL and confers gap-associated therapeutic vulnerability. Sci. Adv.10, eadk2771 (2024). 10.1126/sciadv.adk2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim, P. X., Zaman, M., Feng, W. & Jasin, M. BRCA2 promotes genomic integrity and therapy resistance primarily through its role in homology-directed repair. Mol. Cell84, 447–462.e10 (2024). 10.1016/j.molcel.2023.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohni, K. N. et al. HMCES maintains genome integrity by shielding abasic sites in single-strand DNA. Cell176, 144–153.e13 (2019). 10.1016/j.cell.2018.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehta, K. P. M., Lovejoy, C. A., Zhao, R., Heintzman, D. R. & Cortez, D. HMCES maintains replication fork progression and prevents double-strand breaks in response to APOBEC deamination and abasic site formation. Cell Rep.31, 107705 (2020). 10.1016/j.celrep.2020.107705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blessing, C. et al. The oncogenic helicase ALC1 regulates PARP inhibitor potency by trapping PARP2 at DNA breaks. Mol. Cell80, 862–875.e6 (2020). 10.1016/j.molcel.2020.10.009 [DOI] [PubMed] [Google Scholar]
- 61.Caldecott, K. W. Causes and consequences of DNA single-strand breaks. Trends Biochem. Sci. 10.1016/j.tibs.2023.11.001 (2023). [DOI] [PubMed]
- 62.Gier, R. A. et al. High-performance CRISPR-Cas12a genome editing for combinatorial genetic screening. Nat. Commun.11, 3455 (2020). 10.1038/s41467-020-17209-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryan, B. J., Weaver, T. M., Spencer, J. J. & Freudenthal, B. D. Generation of recombinant nucleosomes containing site-specific DNA damage. Methods Mol. Biol.2701, 55–76 (2023). 10.1007/978-1-0716-3373-1_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol.375, 23–44 (2004). 10.1016/S0076-6879(03)75002-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information files and can be obtained from the corresponding author upon request. Source data are provided with this paper.