Abstract
We here explore the potential of the fungal genus Aureobasidium as a prototype for a microbial chassis for industrial biotechnology in the context of a developing circular bioeconomy. The study emphasizes the physiological advantages of Aureobasidium, including its polyextremotolerance, broad substrate spectrum, and diverse product range, making it a promising candidate for cost‐effective and sustainable industrial processes. In the second part, recent advances in genetic tool development, as well as approaches for up‐scaled fermentation, are described. This review adds to the growing body of scientific literature on this remarkable fungus and reveals its potential for future use in the biotechnological industry.
The extremotolerant fungus Aureobasidium chassis can utilize a broad spectrum of substrates to synthesize value‐added products including pullulan, polyol lipids, polymalate, and melanin. We furthermore describe available genetic engineering tools and up‐scaling approaches.
INTRODUCTION
In recent years, interest in developing a circular bioeconomy has increased significantly. The reasons are growing environmental awareness and a deeper understanding of the consequences of the overuse of natural resources and the accumulation of waste. The circular economy consists of a closed cycle of raw materials and energy over several phases (Franklin‐Johnson et al., 2016). This strategy involves converting side or waste streams into resources from which new products are created at the end of the cycle, thereby increasing resource utilization efficiency and reducing waste (Maina et al., 2017). However, replacing petroleum‐based products with bio‐based alternatives is still challenging and often not yet economically competitive. The main operation costs are generated by the substrate cost, the high consumption of energy and freshwater due to sterilization processes, bioreactor cooling, using batch processes instead of fully automated and continuous processes, and cost‐ and labour‐intensive downstream processing of the products (Chen & Jiang, 2018). It is therefore crucial to reduce production costs to create economically feasible and sustainable processes for a future bio‐based industry.
One approach to reducing production costs is the application of more robust production organisms that tolerate impurities and are less susceptible to contamination by other microbes (Chen & Jiang, 2018). Common industrial strains like Escherichia coli or Saccharomyces cerevisiae are grown under mild conditions with high nutrient supply, increasing the contamination risk since these environments also allow many other microorganisms to grow. Ideally, a production strain should be able to grow rapidly under harsh conditions to avoid contaminations and reduce the effort for sterilization (Chen & Wan, 2017). Moreover, it should grow on low‐cost and renewable substrates, tolerate impurities and different stresses during industrial processes, and molecular engineering tools for performance optimization should be available (Xu et al., 2020). Therefore, extremophile and extremotolerant microorganisms have received increasing attention in recent years since they can tolerate harsh environmental conditions like extreme pH values, high salt or sugar concentrations, or extreme temperatures. A prominent example is the NaCl‐tolerant Halomonas sp. used mainly in China for bioplastic production (Chen et al., 2022), besides other valuable products. These properties enable a significant reduction of production costs in industrial processes and perhaps also provide novel and robust bio‐molecules (Dumorné et al., 2017).
Yeasts and especially some fungi are already exploited under harsh conditions for their biotechnological potential, like Aspergillus niger at very low pH for citrate production (Książek, 2023). Here, we focus on the polyextremotolerant fungal genus Aureobasidium, which has great potential for application in an economically improved industrial biotechnology. It not only provides a large substrate and product spectrum but also tolerates extreme temperatures and pH values as well as high salt and sugar concentrations (Prasongsuk et al., 2018). Its broad substrate spectrum includes agricultural side streams (sugar beet/soy molasse, sweet whey, and oat hulls) (Viveka et al., 2021; Yegin et al., 2018), as well as various by‐products from the agricultural and food industry (corn steep liquor, olive oil mill wastewater, and whey protein) (Meneses et al., 2017; Wang et al., 2021). The metabolization of such cheap and sustainable carbon sources is a great advantage of the organism for an application in a sustainable and circular industrial biotechnology. The whole substrate spectrum of Aureobasidium spp. including enzymes involved was discussed in detail by Wang et al. (2022). The genus has a remarkable array of products, spanning from enzymes, to polysaccharides like pullulan and even biosurfactants. Furthermore, its use as active component in biocontrol agents and protective coatings in the construction industry has been proposed (Rensink et al., 2024).
This review aims to discuss the potential of the fungal genus Aureobasidium as microbial chassis for industrial biotechnology. Topics include physiological advantages, the variety of secondary metabolites of industrial interest, the status of tool development for efficient genome editing, and the perspective on scale‐up processes.
THE GENUS AUREOBASIDIUM
The first representative of the genus Aureobasidium was discovered by Viala and Boyer (1891) on grape leaves and called Aureobasidium vitis. The best‐known member, Aureobasidium pullulans, was first described in 1884, when it was still called Dematium pullulans (Bary, 1884). The genus is described as a group of extremotolerant yeast‐like fungi (Gostinčar et al., 2014). Taxonomically, the genus belongs to the order Dothideales inside the Ascomycetes and subdivision of Pezizomycotina (Thambugala et al., 2014). The phylogenetic relations of selected Aureobasidium species to some biotechnologically relevant representatives of the ascomycetes are shown in Figure 1. The first classification on the species level, based on morphological and physiological differences, suggested the subdivision into four subspecies (Zalar et al., 2008) that were later redefined as A. pullulans, A. melanogenum, A. namibiae, and A. subglaciale by genome analyses (Gostinčar et al., 2014). Since then, several strains were identified as new species (Arzanlou & Khodaei, 2012; Crous et al., 2011; Jiang et al., 2021, 2019; Lee et al., 2021; Onetto et al., 2020; Peterson et al., 2013; Wu et al., 2023).
FIGURE 1.
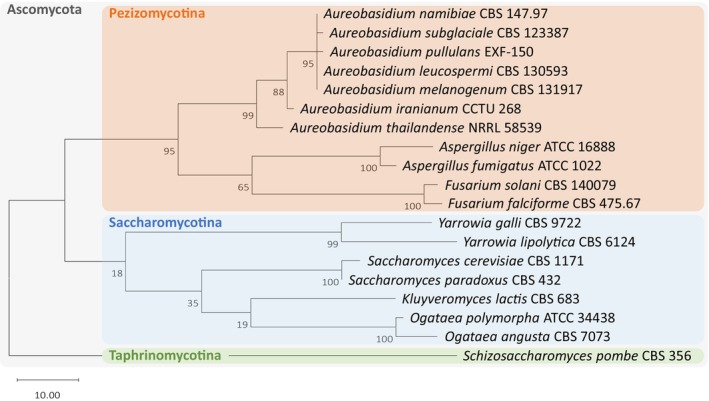
Phylogenetic tree of 19 Ascomycota species based on published internal transcribed spacer (ITS) regions. Sequence alignment was done with MUSCLE. The tree was constructed with MEGA11 and the numbers on the branches represent bootstrap values of maximum parsimony analyses from 1000 replicates. The partial‐deletion option was set to 90%. The scale bar indicates 10 base pair changes.
The members of the genus Aureobasidium are known to tolerate a variety of different stresses (Gostinčar et al., 2014) and, therefore, are considered polyextremotolerant. Strains have been isolated in diverse ecological niches, from cold or temperate regions across humid and tropical areas to warm and dry habitats, and show great adaptability to the different environments. Next to more common natural and anthropogenic habitats like plant surfaces, coastal seawater, wood or indoor areas, they also inhabit extreme environments like glacial ice, fuel tanks, salterns, or rock surfaces (Grube et al., 2011; Gunde‐Cimerman et al., 2000; Lotrakul et al., 2009; Rauch et al., 2006; Urzì et al., 1999; Zalar et al., 2008).
Morphologically, species of the genus show different colony structures and colours and form numerous different cell shapes. As shown in Figure 2, under beneficial conditions, many species show unicellular yeast‐like growth, which is beneficial for industrial processes since cells are more robust and less sensitive to sheer forces compared to hyphal cell forms (Klement & Büchs, 2013). Additionally, yeast‐like growth enables higher oxygen and nutrient transfer due to lower viscosity, resulting in higher growth‐rates and more efficient product formation (Gibbs et al., 2000). Depending on the nutrient availability and environmental conditions, Aureobasidium species also tend to convert to hyphal growth and formation of chlamydospores (Bermejo et al., 1981). A detailed description of different morphologies and a schematic life cycle of Aureobasidium spp. was addressed in more detail by Rensink et al. (2024).
FIGURE 2.
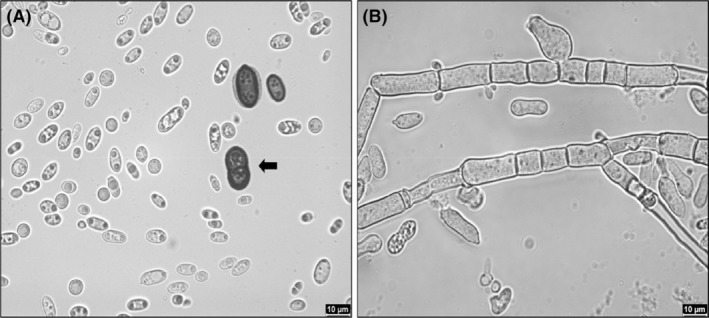
Microscopic pictures of the predominant yeast‐like growth form (A) and filamentous growth (B) of Aureobasidium species. A thick‐walled dark chlamydospore is indicated by the black arrow.
Aureobasidium pullulans is the most studied species from the genus, with currently 78 published genomes, and is best known for producing the exopolysaccharide pullulan, which is already exploited on the industrial scale (Leathers, 2003). Among the whole genus, it exhibits strong adaptive abilities since it can be found in polar, temperate, and tropical areas, rendering it robust against different stresses (Gunde‐Cimerman et al., 2000; Lotrakul et al., 2009; Peterson et al., 2013). Compared to other species of the genus, A. pullulans shows the highest salt tolerance, with a strain isolated from salterns in Slovenia tolerating salt concentrations up to 17% (Gunde‐Cimerman et al., 2000). Population genomics of 50 A. pullulans genomes revealed regular recombination within the species, yet despite the identification of a mating locus in all analysed genomes, it remains unclear whether recombination occurs sexually or asexually (Gostinčar et al., 2019; Gunde‐Cimerman et al., 2000).
In contrast to A. pullulans, the reproductive strategy of A. subglaciale is suggested to be strictly asexual without recombination and is also more specialized regarding the habitat (Zajc et al., 2022). To date, only a limited number of strains have been isolated with nine published genome sequences, predominantly from cold environments like glacial ice (Zalar et al., 2008). A. subglaciale strains are able to grow at temperatures ranging from 0 to 30°C which emphasizes their affinity for colder environments (Zajc et al., 2022). Besides low temperature, strains of the species also tolerate high salt concentrations, heavy‐metal stress, and radiation, making them extremely robust against environmental influences (Liu et al., 2017; Zajc et al., 2022).
The species A. melanogenum, similar to A. subglaciale, lacks recombination and reproduces strictly asexually. However, A. melanogenum strains occasionally form stable heterozygous diploid strains (Gostinčar et al., 2022). So far, the reason and mechanism for the hybridization remain unknown. Different from the other species, A. melanogenum strains can often grow at temperatures up to 37°C, though less well at low temperatures (Černoša et al., 2021; Zalar et al., 2008). As the name indicates, A. melanogenum is known to produce the black pigment melanin in greater amounts compared to other Aureobasidium species (Zalar et al., 2008).
Other species of the genus have not yet been intensively studied in terms of phenotypes and life cycle. Hence, there is still much research required to explore the differences in Aureobasidium biology.
PHENOTYPES BENEFICIAL FOR BIOTECHNOLOGY
Based on its physiological attributes, Aureobasidium represents a promising genus for the application as industrial chassis in an economically competitive bio‐based industry. We here aim to highlight advantageous properties of the genus for the application as a chassis in biotechnological processes.
One outstanding trait is the salt tolerance; as previously mentioned, some strains tolerate up to 17% salt (Gunde‐Cimerman et al., 2000). A high salt tolerance can be beneficial for the economy of bioprocesses since it facilitates the use of seawater, a resource many times more available than freshwater. This can significantly reduce costs, considering the high water consumption in industrial bioprocesses (Scapini et al., 2022). Yue et al. (2014), for example, have already developed a biotechnological process using seawater instead of fresh water for the production of bioplastics with the halophile Halomonas campanienses. Such processes could therefore potentially also be developed for Aureobasidium species. Next to the application in production processes, the wastewater treatment can often be a problem during industrial processes. Industrial wastewaters often contain elevated salt concentrations. Zeng et al. (2021) demonstrated the potential use of the strain Aureobasidium sp. MSP8 for the efficient removal of phosphorus from saline industrial wastewater. Thus, halotolerant microorganisms like Aureobasidum spp. can potentially also find application in the wastewater treatment.
Some Aureobasidium strains, especially A. melanogenum strains, are known to grow at higher temperatures up to 37°C (Černoša et al., 2021; Zalar et al., 2008). Growth at higher temperatures, hence fermentation at higher temperatures, like 37°C or even slightly higher, reduces cooling, contributing significantly to lowering the energy demand of industrial processes.
Aureobasidium spp. are also known to tolerate a wide pH range as well as oligotrophic conditions, making them highly robust against varying process conditions (Gostinčar et al., 2014; Zalar et al., 2008). This can reduce the need for extensive process monitoring and inhibit contamination. Thus, autosterility of the process can be achieved, eliminating the need for costly sterilization of production equipment (Wernick et al., 2016).
Next to potential economic process improvements and provision of robust chassis organisms, extremophile and extremotolerant microbes can also deliver novel and robust bio‐molecules like extremozymes. These enzymes are adapted to extreme conditions and open up new opportunities for applications, for example, in the detergent and food industries (Cavicchioli et al., 2011). Yegin (2017) characterized a highly robust xylanase from A. pullulans NRRL Y‐2311‐1 that has a wide pH stability, as well as a high salt and ethanol tolerance, which enables numerous possible applications, for example, for brewing and bioethanol production, processing of saline foods, or the feed industry.
SECONDARY METABOLITES PRODUCED BY AUREOBASIDIUM
Aureobasidium spp. are not only growing on a wide range of substrates, but they also produce a cornucopia of natural products that are potentially interesting for future industrial applications. The most prominent and extensively studied product of Aureobasidium is the polymer pullulan. Pullulan is a linear polysaccharide consisting of α‐1,6‐linked maltotriose subunits. It can form oxygen‐impermeable thin films that can be used, for example, for food preservation. Furthermore, the polymer shows adhesive properties, enabling pullulan to be utilized in the formulation of food pastes and binders (Cheng et al., 2011b). Pullulan is already commercially produced early in the biotechnological industry. Hayashibara Company Limited (Okayama, Japan) started the large‐scale production of pullulan in 1976. Since then, many other companies have also started to produce pullulan; however, Hayashibara Company Limited remains the market leader with a production of 1000 metric tonnes per year in 2009 (Chaen, 2009).
More detailed information about biosynthesis, regulation, and applications of pullulan can be found in detailed reviews by Cheng et al. (2011b), Leathers (2003), and Wei et al. (2021).
Other native products for future industrial applications are the biopolymers β‐glucan (Hirabayashi et al., 2016; Lotrakul et al., 2013) and polymalate (Chi, Liu, et al., 2016; Ma et al., 2013), the polyol lipid biosurfactants (a.k.a. liamocin) (Leathers et al., 2016; Tiso et al., 2024), siderophores (Wang et al., 2009), the pigment melanin (Zhou et al., 2023), organic acids like gluconic acid (Anastassiadis et al., 2005) and fumaric acid (Wang, Bai, et al., 2018), the antifungal antibiotic aureobasidin A (Slightom et al., 2009), fructooligosaccharides (Dominguez et al., 2012), sorbitol (Sasahara & Izumori, 2005), and erythritol (Guo et al., 2016). Additionally, A. pullulans can be used for the production of single‐cell protein (Baldwin et al., 2019). Details about the biosynthesis of the mentioned products and the interesting characteristics of some of these metabolites have been discussed in reviews (Chi et al., 2009; Prasongsuk et al., 2018; Wang et al., 2022). This review focuses on melanin, polyol lipids, and polymalate as prominent products of Aureobasidium. An overview of the respective biosynthesis pathways of these products is shown in Figure 3.
FIGURE 3.
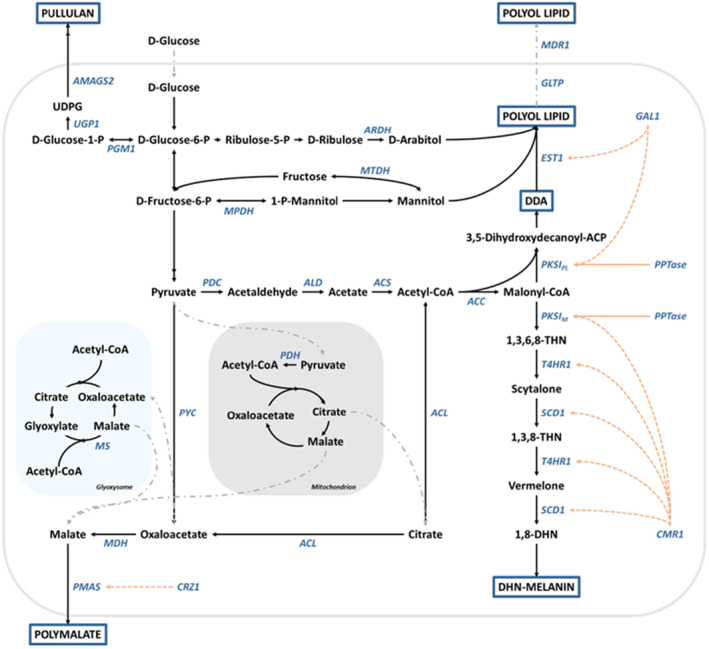
The biosynthetic pathways for pullulan, polyol lipids, polymalate, and DHN‐melanin in the genus Aureobasidium. Genes coding for the respective enzymes are shown in blue. 1,3,6,8‐THN, 1,3,6,8‐tetrahydroxynaphtalene; 1,3,8‐THN, 1,3,8‐trihydroxynaphtalene; 1,8‐DHN, 1,8‐dihydroxynaphtalene; ACC, acetyl‐CoA carboxylase; ACL, ATP‐citrate lyase; ACP, acyl carrier protein; ACS, acetyl‐CoA synthetase; ALD, acetaldehyde dehydrogenase; AMAGS2, a multidomain α‐glucan synthetase; ARDH, arabitol dehydrogenase; ATP, adenosine triphosphate; CMR1, transcription factor; CoA, coenzyme A; CRZ1, transcription factor; DDA, oligo‐dihydroxydecanoic acid; EST1, esterase 1; GAL1, transcription factor; GLTP, glycolipid transfer protein; MDH, malate dehydrogenase; MDR1, ATP‐binding cassette transporter; MPDH, mannitol‐1‐phosphate dehydrogenase; MS, malate synthase; MTDH, mannitol dehydrogenase; P, phosphate; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase; PGM1, phosphoglucomutase 1; PKSIM, polyketide synthase type I involved in melanin biosynthesis; PKSIPL, polyketide synthase type I involved in polyol lipid biosynthesis; PMAS, polymalate synthetase; PPTase, phosphopantetheinyl transferase; PYC, pyruvate carboxylase; SCD1, scytalone dehydratase 1; T4HR1, 1,3,6,8‐THN/1,3,8‐THN reductase; UDPG, uridine diphosphate glucose; UGP1, UDP‐glucose pyrophospholase.
MELANIN
Strains of the genus Aureobasidium are also called black yeasts due to the production of melanin, a dark brown to black polymeric pigment with high molecular weight and hydrophobic character. Different types of melanin can be found not only in the fungal kingdom but also in bacteria, plants, and animals, whereby the pigment classification is based on the monomer subunit structure (Suthar et al., 2023). The most prominent fungal melanin classes are 1,8‐dihydroxynaphthalene (DHN)‐melanin, also called allomelanin and L‐3,4‐dihydroxyphenylalanine (DOPA)‐melanin, also known as eumelanin (Tran‐Ly et al., 2020). For DHN‐melanin, a polyketide synthase (PKS) catalyses the reaction of five malonyl‐coenzyme A into 1,3,6,8‐tetrahydroxynaphthalene (THN). After a sequence of reactions, THN is transformed into DHN, followed by polymerization of DHN into DHN‐melanin (Figure 3). The biosynthesis of DOPA‐melanin involves the conversion of the precursor tyrosine into DOPA, which in turn is transformed into dopaquinone. These reactions are catalysed by tyrosinases and/or laccases. The last step, the polymerization of dopaquinone into DOPA‐melanin, occurs spontaneously (Eisenman & Casadevall, 2012).
Characteristic of all forms of melanin are their unique physicochemical properties, such as the absorption of a wide spectrum of ultraviolet and visible (UV–VIS) light, insolubility in organic solvents, and resistance to chemical degradation (Kumar et al., 2011; Suwannarach et al., 2019). These physicochemical properties provide melanized fungi with a robust resistance to a spectrum of stressors, including osmotic (sodium chloride) and oxidative stress (hydrogen peroxide), UV radiation, high temperatures, and antimicrobial agents (Campana et al., 2022; Jiang et al., 2017). A strong melanization of A. melanogenum XJ5‐1, for example, enables the fungus to survive in the harsh conditions of the Chinese Taklimakan Desert, characterized by limited water availability, extreme temperatures, high UV irradiation, and high osmotic stress (Jiang et al., 2020). The physicochemical properties of melanin offer benefits not solely for the fungal kingdom, but they also enable the pigment to be used for various applications that are of interest to humans, as detailed below.
The interest in natural pigments produced by microorganisms has increased in the last few years since they are considered less toxic than synthetic pigments and are more environmentally friendly (Narsing Rao et al., 2017). Melanin has multiple potential applications in various areas, such as the biomedical field. Here, one example is using the biomaterial as a nanocarrier for controlled drug release (Araújo et al., 2014; Caldas et al., 2020). Furthermore, the electronics industry has shown increased interest in melanin for bioelectronic applications. Melanin exhibits similar properties to amorphous semiconductors, making the pigment an organic semiconductor that is cheaper and easier to process compared to inorganic counterparts like silicon or germanium (Ligonzo et al., 2009; Suthar et al., 2023). More applications can be found in the dermo‐cosmetics, dyeing, textile, and packaging industries. In these sectors, the photoprotective, antibacterial, and antioxidant properties, along with its black colour, scavenging of reactive oxygen species, and biodegradable and eco‐friendly nature, play an important role (Roy & Rhim, 2022). Moreover, some melanized fungi have demonstrated potential in bioremediation. Melanin can effectively bind to a wide range of substances like heavy metals due to its chemical structure containing different functional groups. This characteristic can be used, for example, to remove heavy metal pollution in wastewaters (Mattoon et al., 2021).
The first study addressing Aureobasidium and melanin production was published in 1963 by Lingappa et al. (1963). They observed an effect of light and the cultivation medium on melanin formation in A. pullulans. More specifically, they found out that light can increase melanin production. However, this effect is sensitive to the respective cultivation medium. Since then, only a limited number of publications focusing on melanin production with Aureobasidium spp. have been published. Despite its interesting properties, melanin is often only a by‐product in pullulan production, the most prominent product of Aureobasidium.
In a detailed study investigating melanin biosynthesis of A. melanogenum XJ5‐1, the pivotal role of a non‐reducing fungal type I polyketide synthase (PKSI) was elucidated (Jiang et al., 2017). The PKSI responsible for DHN‐melanin production (PKSIM) contains six domains – a keto synthase, an acyl transferase, two acyl carrier proteins, a thioesterase, and one cyclase. The researchers could demonstrate that the PKSIM is activated by an Sfp‐type phosphopantetheinyl transferase (PPTase). Furthermore, it was found that the expression of both genes, PKSl M and PPtase, is suppressed by nitrogen and glucose. The study also highlighted the heightened sensitivity of the PKSI M knockout mutant, which is not producing melanin anymore, to harsh cultivation conditions like increased UV radiation (Jiang et al., 2017).
Additionally, the same group identified a central role of the cell wall integrity (CWI) signalling pathway in regulating DHN‐melanin biosynthesis in A. melanogenum. More particularly, the mitogen‐activated protein kinase (MAPK) Slt2 of the CWI signalling pathway was shown to regulate the activity of the transcription factor Swi4, which, in turn, positively controls the expression of the melanin‐specific transcriptional activator Cmr1. The CMR1 gene was found to cluster with the gene encoding PKSIM. Additionally, a knockout‐strain Δcmr1 showed no melanin production and a strongly reduced transcription level of PKSI M and other genes involved in DHN‐melanin synthesis (Jiang et al., 2020). In line with these findings, the overexpression of the transcription activator Cmr1 in A. pullulans Hit‐lcy3T led to an increased number of chlamydospores and a 1.4‐fold increase in melanin production, resulting in a titre of 13 g L−1 melanin after just 9 days of cultivation (Wang, Zhang, et al., 2023).
Other studies are more focused on increasing the melanin titre of Aureobasidium strains by testing different production media and fermentation strategies. Zhou et al. (2023) developed a simplified melanin production medium and an efficient fermentation strategy for A. melanogenum GXZ‐6 involving pH control, ammonium salt addition, and H2O2 stimulation, resulting in a melanin titre of 19 g L−1 and a productivity of 0.9 g L−1 d−1 in a 5 L stirred‐tank bioreactor. This titre is more than 2.8 times higher compared to the titre without any fermentation optimization (Zhou et al., 2023). Contrary to the previously mentioned results from other studies, the production of DOPA‐melanin instead of DHN‐melanin is speculated for A. melanogenum GXZ‐6 in this study. This assumption is based on the analytical approaches used to characterize the pigment. However, the stability, insolubility, and association of melanin with the fungal cell wall make a proper extraction and accurate characterization challenging. Moreover, due to different extraction methods and degrees of purity, the mentioned titres should only be compared with caution.
In another study, a multifaceted approach using response surface methodology and artificial neural networks to optimize melanin production with A. pullulans AKW was employed. The effect of three independent variables, sucrose, incubation time, and tyrosine, on melanin production was tested. Tyrosine is the starting molecule of DOPA‐melanin, but the addition of tyrosine showed no effect on pigment production, indicating the production of DHN‐melanin. In contrast, the concentration of sucrose and the incubation intervals showed an influence. By adjusting these variables with the help of artificial neural networks methodology, 10 g L−1 of melanin could be produced. This was 9% higher than the melanin amount achieved using response surface methodology. However, a high similarity between the prediction and experimental data was observed for both methods. This emphasizes the efficacy of these techniques in enhancing melanin production (Saber et al., 2023).
Furthermore, the use of more sustainable substrates, such as food waste, has been demonstrated for melanin production. Among different tested food wastes, carrot peel extract seems to be a suitable substrate for melanin production, reaching a titre of 3.7 g L−1 after 20 days of cultivation in a shake flask (Mujdeci, 2021). Once again, with the help of a response surface method, an increase in produced melanin of 9% was achieved. It was revealed that besides the fermentation time, the initial pH of the cultivation medium, the cultivation temperature, and the agitation rate impact melanin formation (Müjdeci, 2022).
Overall, the comprehensive insights from these studies provide a deeper understanding of melanin biosynthesis in Aureobasidium species. However, challenges persist in the purification and quantification of the polymer, and the detailed structure of the melanin polymer remains elusive, presenting opportunities for further research and exploration.
POLYOL LIPIDS
Polyol lipids (a.k.a. liamocins) (Tiso et al., 2024) are secondary metabolites produced and secreted by the genus Aureobasidium. They were first described in 1994 (Kurosawa et al., 1994), and their complete structure was published in 2013 (Price et al., 2013). Along with the polyol lipids, oligo‐dihydroxydecanoic acids (DDA) (a.k.a. exophilins) are synthesized (Tiso et al., 2024). The latter are polyesters composed of three or four 3,5‐dihydroxydecanoic acids. The first fatty acid can be O‐acetylated (Price et al., 2013). Polyol lipids are amphiphilic molecules composed of a single hydrophilic polyol head group linked to a DDA by an ester bond (Leathers et al., 2016; Price et al., 2017, 2013). The polyol head group is mainly mannitol or arabitol, but other polyols like glycerol or threitol were also published. Supplementation of polyols to the cultivation medium can vary congener distribution (Price et al., 2017). Due to their amphiphilic nature, these molecules have great potential as biosurfactants. Even at low concentrations, some polyol lipids lower the surface tension of water from 73 mN m−1 to around 30 mN m−1 (Kim et al., 2015; Manitchotpisit et al., 2011), which is comparable to the performance of rhamnolipids (Nitschke et al., 2005). Besides their possible application as biosurfactants, polyol lipids also have antibacterial activity against Streptococcus spp. (Bischoff et al., 2015) and show potential as anticancer agents (Manitchotpisit et al., 2011, 2014).
Regardless of their potential as biosurfactants, the biosynthesis of polyol lipids is not fully elucidated. However, some enzymes involved in the polyol lipid pathway in A. melanogenum are already known, and biosynthesis and applications have been reviewed in recent literature (Garay et al., 2018; Kang et al., 2022; Wan et al., 2022; Xue et al., 2018). As mentioned before, different polyols can form the head group of polyol lipids, with mannitol and arabitol being the most abundant (Price et al., 2017). As shown in Figure 3, the formation of mannitol from fructose in A. melanogenum is catalysed by a mannitol‐1‐phosphate‐dehydrogenase (Mpdh) and a mannitol dehydrogenase (Mtdh), encoded by MPDH and MTDH, respectively (Xue et al., 2020). The synthesis of arabitol from ribulose is catalysed by the arabitol dehydrogenase (Ardh), encoded by the gene ARDH (Xue et al., 2020). The key enzyme for polyol lipid biosynthesis was found to be an iterative type I polyketide synthase (PKSIPL). It is responsible for the formation of 3,5‐dihydroxydecanoic acids, which are the building blocks of DDAs (Price et al., 2013; Xue et al., 2020). This PKSIPL is composed of an acyl carrier protein (ACP), a ketosynthase (KS), an acyltransferase (AT), a ketoreductase (KR), a dehydratase (DH), and an enoylreductase (ER) domain (Xue et al., 2020). ACP, AT, and KS domains belong to the minimal set of domains of an iterative PKSI, and are responsible for polyketide elongation, while KR, DH, and ER domains are reducing domains (Sabatini et al., 2018). A thioesterase (TE) domain, responsible for releasing the polyketide by cleavage of the thioester bond, has not been found in the gene coding for PKSIPL. The deletion of the PKSIPL encoding gene PKSI PL in A. melanogenum 6‐1‐2 led to the prevention of polyol lipid and DDA formation, underlining its significance for polyol lipid biosynthesis (Xue et al., 2020). As for most PKS, the precursors used by the PKSIPL for synthesis of the 3,5‐dihydroxydecanoic acids are acetyl‐CoA and malonyl‐CoA, which are used as starter and extender units, respectively (Cheng et al., 2003; Nivina et al., 2019; Xue et al., 2020).
Polyketide synthases are activated by posttranslational modification by PPTases (Beld et al., 2014), and a PPTase associated with the PKSIPL involved in polyol lipid biosynthesis was also found in A. melanogenum (Xue et al., 2020). Another enzyme known to be involved is the esterase Est1 encoded by the gene EST1. This esterase is proposed to attach the polyol head group to the DDA backbone by formation of an ester bond. The genes encoding for the PKSIPL and the esterase are clustered in the genome of A. melanogenum and are regulated by the zinc finger protein Gal1. The gene GAL1 encoding for the transcriptional activator Gal1 is also clustered with PKSI PL and EST1 (Xue et al., 2020). Furthermore, the global transcriptional regulator Msn2 takes part in the regulation of polyol lipid production via cAMP‐PKA (cyclic adenosine monophosphate‐proteinkinase A) and HOG1 (high‐osmolarity glycerol 1) signalling pathways (Zhang, Gao, et al., 2021), similarly to the regulation of the synthesis of the exopolysaccharide pullulan in A. melanogenum (Yang et al., 2020). After synthesis, the polyol lipid congeners are transported by the intracellular glycolipid transfer protein (Gltp) and exported by an ABC transporter (Mdr1) (Xue et al., 2020).
Cultivations for polyol lipid production are usually carried out in shake flasks at 28–30°C and 150–200 rpm for 7 days (Wan et al., 2022). Different media are used for this. Leathers et al. (2015) compared various strains in four published media (Doshida et al., 1996; Manitchotpisit et al., 2011; Takafumi et al., 1994; Wang et al., 2014), which differ primarily in their initial pH values (5.5–7.0), carbon (glucose or sucrose), and nitrogen source. The highest titre of 8 g L−1 was achieved with sucrose, yeast extract, peptone, and an initial pH of 6.5 (Manitchotpisit et al., 2011) using A. pullulans NRRL 50384. By replacing sucrose with agriculture waste, polyol lipid production was tested using the same medium (Leathers et al., 2016). Furthermore, it was observed that the congener composition varied depending on the strain and medium (Leathers et al., 2015, 2016). Based on the medium developed by Manitchotpisit et al. (2011), Leathers et al. (2018) performed a medium optimization using design of experiments and achieved 22 g L−1 with A. pullulans NRRL 50384.
Based on the same medium, Haala et al. (2024) developed the first minimal medium for polyol lipid production. For this purpose, yeast extract and peptone were replaced by NH4NO3, trace elements, and vitamins, followed by a media optimization using a design of experiments approach. After optimization, the highest polyol lipid titre of 48 g L−1 was achieved using an Aureobasidium sp. wild‐type strain.
Using glucose, yeast extract, NH4NO3, K2HPO4, KCl, and MgSO4 Liu et al. (2014), Xue et al. (2020) achieved 26 g L−1 with A. melanogenum 6‐1‐2. With a similar medium (replacing yeast extract with corn steep liquor), the highest titre so far was achieved using a genetically optimized A. melanogenum 9‐1 V33 (Zhang et al., 2022). After the disruption of the CREA gene to relieve glucose repression, overexpression of the PK gene and PDC gene to enhance the supply of acetyl‐CoA, and overexpression of the VHb gene coding for Vitreoscilla haemoglobin to enhance the supply of ATP, the resulting strain A. melanogenum 91 V33 reached 55 g L−1 in a 10 L batch fermentation (Zhang et al., 2022). A. melanogenum 9‐1 wild type reached 31 g L−1 with the same setup. This highlights the potential for a combined approach of metabolic and bioprocess engineering to increase polyol lipid production.
POLYMALATE
Polymalate (PMA), a biodegradable and linear anionic homopolyester with many versatile pendant carboxyl groups, is composed of repetitive L‐malic acid subunits linked via ester bonds between the α‐hydroxyl group and β‐carboxyl group (Chi, Liu, et al., 2016; Ma et al., 2013). It has promising properties such as water‐solubility, biocompatibility, biodegradability, non‐immunogenicity, non‐toxicity, and chemical processability (Ding et al., 2013; Holler et al., 1992; Portilla‐Arias et al., 2008; Qi et al., 2021). PMA and its derivatives have raised considerable attention from researchers as a result of its potential application as a novel drug delivery platform to generate various derivatives like nanoconjugates, nanoparticles, and nanocarriers in the biomedical field (Arif et al., 2017; Huang et al., 2022, 2012; Zhang, Chen, et al., 2021). Particularly, Polycefins, a type of nanoconjugates based on natural PMA developed in the early 2000s, have been employed in preclinical studies and might be of great interest as anti‐cancer treatment (Lee et al., 2006; Ljubimova et al., 2008; Loyer & Cammas‐Marion, 2014). Additionally, PMA can be easily decomposed into L‐malic acid, which is an important dicarboxylic acid widely used as an acidulant in food and beverage industry and is regarded as a potential C4 chemical building block in biorefinery engineering (Cheng et al., 2017; Chi, Wang, et al., 2016; Dai et al., 2018; Kövilein et al., 2020). PMA has been reported to be chemically synthesized by debenzylating poly (β‐malic acid benzyl ester) obtained through the ring‐opening polymerization of benzyl malolactonate or direct polycondensation of L‐malic acid (Kajiyama et al., 2004, 2003; Vert, 1998). However, these traditional synthesis routes are energy‐costly, time‐consuming, and environmentally unfriendly, and their raw material is maleic anhydride derived from petroleum (Kajiyama et al., 2004). In recent years, it has been found that different strains of Aureobasidium spp. can secrete large quantities of β‐type linear PMA, and some of them are capable of producing over 30 g L−1 of PMA, while other known producers like Physarum polycephalum only produced small amounts (2.7 g L−1) of PMA (Lee & Holler, 1999; Qi et al., 2021; Rathberger et al., 1999).
The intracellular accumulation of L‐malic acid, the precursor for the polymerization of PMA, is proposed to be attributed to the TCA (tricarboxylic acid) cycle, the cytosolic reductive TCA (rTCA) pathway, and glyoxylate shunt in fungal cells (Iyyappan et al., 2019; Kövilein et al., 2020; West, 2017; Wu et al., 2022). The three anabolic pathways for intracellular malic acid synthesis are displayed in Figure 3. Notably, the rTCA pathway, which occurs in the cytoplasm, can provide the maximum theoretical yield of malic acid among these three pathways if pyruvate is produced via glycolysis. Specifically, the rTCA begins with the carboxylation of pyruvate to oxaloacetate under the catalysis of pyruvate carboxylase with CO2 fixation. Oxaloacetic acid is subsequently converted into malic acid catalysed by cytosolic malate dehydrogenase, resulting in a theoretical glucose‐to‐malic acid conversion rate of 2 mol/mol without consuming ATP (Yin et al., 2015; Zelle et al., 2008). The anabolic pathway of intracellular malic acid for PMA production was found to be strain‐specific. For instance, it was reported that malic acid from the TCA cycle was the main source for PMA biosynthesis in A. melanogenum ATCC 62921 using a genome‐wide deletion mutant analysis (Wang, Chi, Liu, et al., 2020). It was proposed that malic acid might derive from the glyoxylate cycle in A. pullulans GXZ‐6 by adding metabolic intermediates and inhibitors in batch fermentation (Zeng, Zhang, Chen, et al., 2019). Furthermore, metabolomics combined with in silico analysis of a genome‐scale metabolic model suggested that a considerable amount of carbon flux was from pyruvate into malic acid via the rTCA cycle, and the pyruvate carboxylase‐encoded PYC gene was regarded as a significant molecular engineering target for the high production of PMA (Feng et al., 2017, 2018).
It was not discovered until 2020 that the PMA synthetase encoded by the PMAS gene is a key enzyme responsible for polymerizing malic acid into PMA in A. melanogenum ATCC 62921 (Wang, Chi, Liu, et al., 2020). Recently, two PMAS gene homologues were found to also contribute to PMA biosynthesis in the whole genome‐duplicated strain A. melanogenum OUC (Qi et al., 2022). In addition, PMA synthetase is a non‐ribosomal peptide synthetase (NRPS) with six transmembrane regions in its N‐terminus which contains one adenylation (A) like domain, an adjacent thiolation (T) domain and a concentration (C) like domain. It was proposed that malic acid is first activated as a malyl‐O‐AMP by the A‐like domain and then tethered onto the 4′‐phosphopantetheine (4′‐PP) arm of the T domain along with AMP release, thereby leading to the formation of a malyl‐S‐enzyme. Finally, an ester bond is formed by the C‐like domain between two activated malic acids or the oligomer and the activated malic acid (Wang, Chi, Liu, et al., 2020). It should be stressed that the signal peptide sequence guiding the PMA synthetase to the membrane and thioester domain for catalysing the dissociation of the newly made PMA chain from the synthetase has not been found yet, according to bioinformatics prediction (Qi et al., 2021; Wang, Chi, Liu, et al., 2020).
It was reported that the supplement of CaCO3 is necessary for PMA biosynthesis by Aureobasidium spp. (Qi et al., 2021; Zhang et al., 2011). Recently, a comparative transcriptome sequencing analysis indicated that the TCA cycle and glyoxylate pathway were up‐regulated, and a gene encoding an NRPS‐like protein was highly upregulated with CaCO3 addition (Wang, Yin, et al., 2023). Moreover, it has been documented that the PMA synthetase gene in A. melanogenum ATCC 62921 was controlled by the transcriptional activator Crz1 in the Ca2+‐signalling pathway. It could be explained by the existence of the conserved sequences 5′‐CAGCCAC‐3′ and 5′‐GNGGCKCA‐3′ on the promoter of the PMAS gene, which are the transcription factor binding sites of the Crz1 (Chi et al., 2022; Wang, Chi, Liu, et al., 2020; Yoshimoto et al., 2002). CaCO3 can maintain the fermentation broth at a neutral pH value, thereby avoiding the hydrolysis of PMA at low pH and reducing the unwanted production of extracellular polysaccharides and the formation of chlamydospore (Holler et al., 1992; Li et al., 2009). Moreover, CO2 released from CaCO3 can be fixed via the reductive TCA cycle, which is one pathway for the biosynthesis of the intermediate malic acid (Zou et al., 2019). A high initial C/N ratio (nitrogen starvation) in the PMA production medium is another significant factor beneficial for PMA production by upregulating the key genes involved in the PMA biosynthesis pathway (Qi et al., 2021). The GATA‐family transcriptional factor Gat1, involved in nitrogen catabolite repression, and the concentration of nitrogen (glutamine or proline) might cross‐regulate the glucokinase in the glycolytic pathway and malate synthase in the glyoxylate shunt, thereby affecting PMA biosynthesis in A. pullulans CCTCC M2012223 (Song et al., 2020). However, it is unknown if the transcription regulation of Crz1 and Gat1 is strain or species‐specific.
Some isolates of Aureobasidium spp. sampled from different natural locations have shown their potential to synthesize high levels of PMA. For example, two strains of A. pullulans var. melanogenum GXZ‐6 and A. pullulans ZD‐3d isolated from fresh plant samples produced 63 g L−1and 62 g L−1 PMA in the respective optimized media (Zeng, Zhang, Chen, et al., 2019; Zhang et al., 2011). Aureobasidium sp. P6 and A. pullulans var. pullulans MCW isolated from mangrove systems produced 118 g L−1 and 153 g L−1 of Ca2+‐PMA, respectively (Ma et al., 2013; Wang et al., 2015). Recently, much has been reported on the sustainable production of PMA from low‐cost renewable feedstocks by native or engineered strains of Aureobasidium pullulans (Table 1). For example, A. pullulans YJ 6‐11 strain can produce 29 g L−1 of PMA in corncob hydrolysate (Zou et al., 2016). Sugarcane juice, without any extra pretreatment, can be utilized by A. pullulans ZX‐10 to synthesize PMA with a concentration of 116 g L−1 in a fed‐batch fermentation (Wei et al., 2017). Other renewable biomass and agricultural side streams such as raw sweet potato hydrolysate (Zan & Zou, 2013), barley straw hydrolysate (Yegin et al., 2019), Jerusalem artichoke (Cao et al., 2019; Xia et al., 2017), malt syrup (Zeng, Zhang, Li, et al., 2019), bagasse hydrolysates (Cao et al., 2020), and cassava hydrolysate (Liu et al., 2022) were used to produce PMA by Aureobasidium spp.
TABLE 1.
Polymalate production from renewable biomass and food processing wastes produced by varied Aureobasidium pullulans.
| Strain | Substrate | Fermentation mode | Titre (g L−1) | YP/S (g g−1) | Vol. prod. (g L−1 h−1) | References |
|---|---|---|---|---|---|---|
| A. pullulans GXL‐1 | Liquefied corn starch | Batch/repeated‐batch | 49 | 0.50 | 0.34 | Zeng et al. (2020) |
| A. pullulans YJ 6‐11 | Corncob hydrolysate | Batch | 29 | 0.40 | Zou et al. (2016) | |
| A. pullulans ZX‐10 | Sugarcane juice | Fed‐batch | 116 | 0.41 | 0.43 | Wei et al. (2017) |
| Repeated‐batch | 40 | 0.34 | 0.41 | |||
| Soy molasses | Fed‐batch | 62 | 0.69 | 0.29 | Cheng et al. (2017) | |
| Repeated‐batch | 20 | 0.39 | 0.22 | |||
| Soybean hull | Fed‐batch | 27 | 0.42 | 0.48 | ||
| A. pullulans CCTCC M2012223 | Raw sweet potato hydrolysate | Batch | 30 | 0.28 | 0.25 | Zan and Zou (2013) |
| Fed‐batch | 44 | 0.19 | 0.28 | |||
| Fibrous bed bioreactor | 58 | 0.20 | 0.37 | |||
| A. pullulans NRRL Y‐2311‐1 | Barley straw hydrolysate | Batch | 44 | 0.48 | 0.30 | Yegin et al. (2019) |
| A. pullulans HA‐4D | Jerusalem artichoke | Fed‐batch | 114 | 0.74 | Xia et al. (2017) | |
| A. pullulans HA‐4D & Kluyveromyces marxianus | Cheese whey | Fed‐batch | 97 | 0.56 | 0.51 | Xia et al. (2021) |
| A. pullulans ipe‐1 | Jerusalem artichoke | Repeated‐batch | 25 | 0.78 | Cao et al. (2019) | |
| Bagasse hydrolysates | Batch | 23 | Cao et al. (2020) | |||
| A. pullulans GXZ‐6 | Malt syrup | Repeated‐batch | 64 | 0.81 | 0.56 | Zeng, Zhang, Li, et al. (2019) |
| A. pullulans ZD‐3d | Cassava hydrolysate | Fed‐batch | 102 | 0.40 | 0.77 | Liu et al. (2022) |
| A. pullulans mutant FJ‐D2 | Waste xylose mother liquor | Batch | 57 | 0.77 | 0.58 | Feng et al. (2019) |
| A. pullulans AE‐59 | Waste xylose mother liquor | Batch | 49 | 0.33 | Li et al. (2023) | |
| A. pullulans NRRL 50383 | Pretreated corn fibre | Batch | 10 | Leathers and Manitchotpisit (2013) | ||
| Pretreated wheat straw | Batch | 24 |
Furthermore, other strategies have been employed to valorize agro‐industrial by‐products to PMA products. For example, Xia et al. (2021) designed a mixed culture of A. pullulans HA‐4D and permeabilized Kluyveromyces marxianus to achieve a high concentration of 97 g L−1 PMA with a in fed‐batch fermentation from cheese whey. Feng et al. (2019) constructed transfer‐DNA‐based mutant libraries and obtained A. pullulans FJ‐D2, yielding 57 g L−1 PMA from untreated waste xylose mother liquor (WXML) in batch fermentation. Li et al. (2023) employed adaptive evolution and overexpressed the exogenous CNB gene (calcineurin subunit B) from Beauveria bassiana to obtain an engineered strain AE‐59 of A. pullulans, which synthesized PMA at a concentration of 49 g L−1 at low pH values with Na2CO3 in a 5‐L fermenter using WXML.
Over the past few years, many endeavours have been attempted to realize high‐titre PMA production because large‐scale industrial production of PMA is the prerequisite for its commercial applications. First, a supplement of exogenous stimulatory agents such as Tween 80 (Tu et al., 2015; Yin et al., 2019), soybean oil (Xia et al., 2022), ethanol (Yang et al., 2018), and corn steep liquor (Wang et al., 2015; Wang, Shi, Zhang, et al., 2020) at certain concentrations have been added to overproduce PMA. Response surface methodology has been applied to optimize the fermentation medium formulation, resulting in a higher PMA production compared to the control condition (Qiao et al., 2015, 2012).
Some efforts in metabolic and genetic engineering have also been invested to improve the synthesis of PMA. For instance, the engineered strain Crz46 in which the PKS gene encoding melanin synthesis‐related PKS was removed and the PYC1 gene encoding pyruvate carboxylase, the VGB gene encoding haemoglobin synthesis, and the CRZ2 gene encoding the transcriptional activator were overexpressed, can produce 35 g L−1 Ca2+‐PMA while its parental strain A. melanogenum OUC produced 17 g L−1 Ca2+‐PMA (Qi et al., 2022). Overexpression of the MLS gene coding for malate synthase, a key enzyme in the glyoxylate shunt, enhanced the PMA concentration by 16% (Yang et al., 2018). Overexpression of the PYC gene in the native strain A. pullulans CCTCC M2012223 improved the PMA titre by 15% compared with the native strain (Feng et al., 2018).
It should be noted that not only the final concentration of PMA but also its MW is critical for PMA bioproduction because the weight average molar mass (MW) determines the application. For instance, PMA with a low MW of 5 kDa can be used as an inhibitor of proteases (Shimada et al., 1969). PMA with an MW of 50 kDa is generally utilized for creating drug carriers (Ljubimova et al., 2008). It has been documented that strains of Aureobasidium spp. are generally synthesizing PMA with an MW of 3–200 kDa (Zou et al., 2019). Recently, a super‐high MW PMA of 490 kDa produced by the engineered strain Crz46 of A. melanogenum OUC was reported. The polymer was tested as a fruit‐coating film (Qi et al., 2023).
Cao et al. (2016) revealed that supplementing 0.1 g L−1 CaCl2 to the fermentation broth could increase the MW by 26% when Na2CO3 was applied as the neutralizer, and the highest MW of up to 20 kDa was achieved in Ca2+ added repeated batch fermentation mode in which the fungal cells were maintained in their exponential growth phase. Moreover, Qi et al. (2022) constructed the engineered strain Crz46, which can synthesize PMA with an MW of 490 kDa, higher than the MW of 390 kDa from its wild‐type strain. Another factor that determines the MW of PMA is the PMA hydrolase (PMase). PMase, a glycoprotein, produced by P. polycephalum in diverse forms was reported very early (Chi, Liu, et al., 2016; Gasslmaier & Holler, 1997). Intriguingly, it has been found that A. melanogenum ATCC 62921 and A. melanogenum OUC contained homologous genes coding for PMase, which shared a similar protein domain with that in P. polycephalum (Qi et al., 2022). Additionally, quantitative real‐time PCR analysis suggested that the expression level of the PMase gene in the high PMA MW‐producing strain A. melanogenum OUC was much lower than that of the PMase gene in A. melanogenum ATCC 62921 grown under the same growth conditions. It may indicate that the lower expression level of the PMase gene led to a weaker activity of the PMase in A. melanogenum OUC, thereby resulting in a higher MW of PMA than those in A. melanogenum ATCC 62921 (Chi et al., 2022). Thus, it is necessary to understand how to regulate the polymerization degree of PMA at different levels, and the identification of relevant regulatory elements to control the MW of PMA precisely is being awaited.
MOLECULAR ENGINEERING OF AUREOBASIDIUM
Many genomes of various strains belonging to the genus Aureobasidium have been sequenced and analysed recently (Gostinčar et al., 2014, 2019; Rueda‐Mejia et al., 2021; Vignolle et al., 2021; Xiao et al., 2023). However, as for most non‐model organisms, the development of molecular tools for efficient genome editing of Aureobasidium spp. still needs further expansion. Strategies based on gene disruption by integration of a selection marker by homologous recombination have been used successfully in different studies (Chi et al., 2012; Guo et al., 2017; Slightom et al., 2009; Wang et al., 2017). While these systems hold the advantage of a straightforward design, selection markers remain in the genome of the modified strains, thus limiting further genome editing. A combination of these tools with the Cre/loxP system (alternatively Flp/FRT), where the specific loxP sequences are recognized and recombined by a Cre recombinase, is established for model organisms like Saccharomyces cerevisiae (Gueldener et al., 2002), but also for non‐conventional yeasts (Wagner & Alper, 2016; Zhang, Li, et al., 2019), or filamentous fungi (Li et al., 2017). Recombination of the loxP sites flanking the selection marker after genetic modification facilitates the generation of marker‐less strains, enabling repeated use of this system for combined modifications (Sternberg & Hamilton, 1981). Zhang, Lu, et al. (2019) developed such a Cre/loxP system based on homologous recombination for genome editing of Aureobasidium spp., and performed several gene deletions as well as integrations in various A. melanogenum strains. A similar Cre/loxP‐based system was used for several studies on elucidating the pullulan biosynthesis pathway in A. melanogenum P16 (Chen, Liu, Chen, et al., 2020; Chen, Liu, Wei, et al., 2020).
In the past years, CRISPR/Cas9‐based approaches have been developed for many organisms due to the exceptional flexibility and fast turnaround times to generate marker‐less strains (DiCarlo et al., 2013; Ran et al., 2013; Schuster et al., 2016; Stovicek et al., 2017). CRISPR/Cas9‐mediated methods for genome editing of Aureobasidium spp. have already been published, based on either the heterologous expression of cas9 or the direct use of Cas9 ribonucleoproteins (Kreuter et al., 2022; Zhang, Feng, et al., 2019).
Up to date, only few sites for the genomic integration of genes were tested in Aureobasidium spp. Ribosomal DNA is organized in tandem repeats and is thus a commonly used target for gene integration in many organisms (David & Siewers, 2015; Wang, Chi, Zou, et al., 2020; Wang, Deng, et al., 2018). It enables the expression of multiple genes without requiring extensive cloning and the identification of multiple integration sites. Expression of endogenous and heterologous genes by integration into the rDNA was shown to work in A. pullulans as well as in A. melanogenum (Guo et al., 2017; Li et al., 2019; Zhang, Lu, et al., 2019; Zhao et al., 2019). Furthermore, Feng et al. successfully used Agrobacterium tumefaciens‐mediated transformation (ATMT) for random integration of an endogenous gene in A. pullulans CCTCC M2012223 (Feng et al., 2018).
Altogether, several genetic tools for gene deletion or integration have been shown to work in Aureobasidium spp., promoting the implementation of this versatile fungus as biotechnologically relevant production organism. To potentially become a biotechnological chassis, a wild‐type strain should hold some remarkable qualities of interest, like the metabolic versatility and physiological robustness of many strains belonging to the genus Aureobasidium (de Lorenzo et al., 2021). The first steps from such a promising wild‐type organism towards a chassis organism include, for example, genome sequencing and genetic tool implementation before later steps like a streamlined genome or mutant collections can be approached (Calero & Nikel, 2019). Further expansion of the molecular toolbox, for example, regarding other integration sites or a broadened promoter/terminator set, is hence of great interest to unleash its full potential.
UP‐SCALING AUREOBASIDIUM PROCESSES
An essential part of developing a biotechnological process is the technology transfer and scale‐up from microtitre plates or shake flasks into stirred‐tank bioreactors. Optimal process control hinges primarily on factors such as product characteristics, strain behaviour, and cultivation conditions (Marques et al., 2010; Schmidt, 2005).
The potential of Aureobasidium spp. is already being exploited in the industrial production of the exopolysaccharide pullulan and is summarized by Singh et al. (2023). While producing pullulan as a main or by‐product and especially when scaling up into larger processes, the viscosity of the fermentation broth depends on the pullulan structure (Singh et al., 2008). The culture broth's viscosity strongly influences the oxygen transfer within the liquid and the energy input (Cheng et al., 2011b; Seviour et al., 2011). Oxygen is one of the most critical substrates in aerobic processes, as it is essential for growth, cell maintenance, and metabolite production (Büchs, 2001). Sufficient oxygen availability is, therefore, essential and can be a major challenge when scaling pullulan‐producing Aureobasidium spp. (Marques et al., 2010; Singh et al., 2008).
Besides oxygen supply, process conditions like pH value, temperature, and carbon‐to‐nitrogen (C/N) ratio strongly influence product formation. Literature shows that these parameters are strongly influenced by strain and product. In batch fermentations for the production of pullulan, a two‐phase process strategy is reported. Depending on the strain, either the pH value, the temperature, or both are adjusted, separating the growth and production phases (Wang et al., 2013; Wu et al., 2010; Xia et al., 2011). No general statement can be made as to whether higher or lower values are more advantageous. This depends on the respective production strain. A two‐phase batch fermentation has also been successfully used for polyol lipid production. Saur et al. (2019) induced polyol lipid production by a pH shift from 6.5 to 3.5. While very low pH values of 2.5 appear advantageous for producing pullulan and polyol lipids, a neutral pH value is preferred for PMA production to avoid hydrolysis. The pH value also has a considerable influence on melanin production. By controlling the pH at 2.4, Zhou et al. (2023) increased the melanin titre from 7 g L−1 to 10 g L−1.
The main reason for aiming for a two‐phase process is that pullulan, polyol lipids, and PMA are secondary metabolites produced in high titres. The two phases refer to an initial growth phase and a subsequent production phase. Nitrogen is essential for forming amino acids and, thus, for growth, while carbon is required for product formation in addition to biomass formation. A high C/N ratio ensures extended carbon availability and, thus, a long production phase, while biomass formation stops once the nitrogen has been depleted. Using statistical methods, an optimal C/N ratio of 25–28 was determined for pullulan production by Sugumaran et al. (2014) and Sugumaran and Ponnusami (2015).
A significantly higher ratio is preferred for polyol lipid production, resulting in high initial carbon concentrations (Leathers et al., 2018; Li et al., 2021; Tang et al., 2018). Leathers et al. (2018) optimized the production medium using statistical methods, resulting in a calculated C/N ratio of approximately 150. An exact calculation is difficult, as no defined minimal medium for polyol lipid production has been published (Garay et al., 2018). Complex nitrogen sources like yeast extract or peptone also impede precisely investigating the still relatively unknown metabolism and are economically disadvantageous when scaling up. Similar to polyol lipid production, high C/N ratios are also preferred for PMA production (Chi, Liu, et al., 2016). For example, Wang et al. (2015) produced 152 g L−1 Ca2+‐PMA in batch fermentations using 140 g L−1 glucose as carbon and 7.5 g L−1 corn steep liquor as nitrogen and further nutrient source.
The simplest bioreactor cultivations are batch fermentations. These do not involve adding media components and are primarily concerned with the technology transfer from shaken and small to stirred and bubble‐gassed larger systems. So far, for polyol lipid production, only batch fermentation strategies have been published. An overview can be seen in Table 2. With a titre of over 70 g L−1, a product‐to‐substrate yield YP/S of 0.54 g g−1, and a volumetric productivity of 0.45 g L−1 h−1, the best result to date was achieved in a 10 L batch fermentation with glucose, a genetically modified A. melanogenum 9‐1 V33, and a pH shift from 7 to 3 (Wang et al., 2024). This pH shift induces polyol lipid production and was first described by Saur et al. (2019). Furthermore, Brumano et al. (2017) conducted a central composite design to optimize gassing rate and initial carbon concentration. With an aeration rate of 1.1 vvm (volume air per volume medium per minute) and 80 g L−1 sucrose, the polyol lipid titre was increased to 1.5 g L−1 (Brumano et al., 2017). The comparatively low titres are probably due to the less potent strain A. pullulans LB 83.
TABLE 2.
Comparing polyol lipid production by titre, product‐to‐substrate yield YPS, and volumetric productivity of several bioreactor fermentations with different strains and substrates.
| Strain | Substrate | Fermentation mode | Titre (g L−1) | YP/S (g g−1) | Vol. prod. (g L−1 h−1) | References |
|---|---|---|---|---|---|---|
| A. pullulans NRRL 62042 | Glucose | 0.7 L batch | 41 | 0.20 | 0.17 | Haala et al. (2024) |
| A. pullulans NRRL 62031 | Sucrose | 0.75 L batch (pH shift) | 18 | – | 0.14 | Saur et al. (2019) |
| A. melanogenum M39 | Glucose | 10 L batch | 43 | 0.31 | 0.28 | Tang et al. (2018) |
| A. melanogenum 9‐1 | Inulin | 7 L batch | 38 | 0.24 | 0.32 | Xin et al. (2017) |
|
A. melanogenum 9‐1 transformant 88 |
Inulin | 7 L batch | 43 | 0.27 | 0.36 | |
| A. pullulans P5 | Glucose | 10 L batch | 32 | 0.28 | 0.19 a | Liu et al. (2014) |
| A. pullulans AH21 | WXML and WGML | 5 L batch | 28 | 0.25 | 0.17 | Li et al. (2021) |
| A. melanogenum 9‐1 V33 | Glucose | 10 L batch (pH shift) | 70 | 0.54 | 0.45 | Wang et al. (2024) |
Abbreviations: WGML, waste gluconate mother liquor; WXML, waste xylose mother liquor.
Calculated values.
In addition to batch fermentations, there are other options for process control in biotechnological processes. PMA production has already been implemented with other process strategies (Table 3). The implementation of a pulsed fed‐batch by adding glucose to keep substrate concentration above 50 g L−1 increased the productivity from 0.49 g L−1 h−1 to 0.61 g L−1 h−1 compared to the batch fermentation (Zou et al., 2013). Using a fibrous‐bed bioreactor, in which cells are immobilized in a fibrous matrix, increased productivity further to 0.74 g L−1 h−1 (Zou et al., 2013). Wei et al. (2017) also increased productivity using a pulsed fed‐batch with sugarcane molasses. First, they showed similar results regarding titre, YP/S, and volumetric productivity using sugarcane molasses instead of sucrose. Subsequently, repeated feeding of sugarcane molasses increased productivity from 0.28 g L−1 h−1 to 0.43 g L−1 h−1, and the titre doubled to 116 g L−1. Productivity was further optimized by repeated batch fermentation (Wei et al., 2017): At the end of each batch phase, cells were separated by centrifugation and resuspended in fresh medium. By shortening the fermentation time per batch from 90 h to 65 h in the third batch, while maintaining the same titre (40 g L−1) and YP/S (0.3 g g−1), productivity was increased from 0.43 g L−1 h−1 to 0.41–0.66 g L−1 h−1 compared to the fed‐batch fermentation (Wei et al., 2017). To avoid depolymerization of formed PMA and low concentrations of residual glucose, Cao et al. (2016) implemented a repeated batch. For this purpose, 3 of the 4 litres of culture broth were replaced with fresh medium, which increased the molecular weight to over 84%. On average, 57 g L−1 PMA could be produced with a productivity of 1.1 g L−1 h−1.
TABLE 3.
Comparing polymalate (PMA) production by titre, product‐to‐substrate yield YPS, and volumetric productivity of several bioreactor fermentations with different strains and substrates.
| Strain | Substrate | Fermentation mode | Titre (g L−1) | YPS (g g−1) | Vol. prod. (g L−1 h−1) | References |
|---|---|---|---|---|---|---|
| A. pullulans ZD‐3d | Glucose | 10 L batch | 57 | 0.48 a | 0.35 | Zhang et al. (2011) |
| A. pullulans P6 | Glucose | 10 L batch | 99 b | 0.85 b | 0.63 b | Ma et al. (2013) |
| A. pullulans ZX‐10 | Glucose | 5 L batch | 41 | 0.47 | 0.49 | Zou et al. (2013) |
| A. pullulans ZX‐10 | Glucose | 5 L pulsed fed‐batch | 76 | 0.49 | 0.61 | |
| A. pullulans ZX‐10 | Glucose | 5 L fed‐batch (fibrous bed) | 144 | 0.55 | 0.74 | |
| A. pullulans ZX‐10 | Sucrose | 5 L batch | 49 | 0.68 | 0.26 | Wei et al. (2017) |
| A. pullulans ZX‐10 | Sugarcane molasse | 5 L batch | 53 | 0.54 | 0.28 | |
| A. pullulans ZX‐10 | Sugarcane molasses | 5 L pulsed fed‐batch | 116 | 0.41 | 0.43 | |
| A. pullulans ZX‐10 | Sugarcane molasses | 5 L repeated batch | 40 | 0.34 | 0.41–0.66 | |
| A. pullulans ipe‐1 | Glucose | 7.5 L repeated batch | 44–57 | 0.23–0.27 | 0.91–1.3 | Cao et al. (2016) |
Calculated value.
Ca2+‐PMA.
The only publication about melanin production as the main product using a bioreactor was published by Zhou et al. (2023) (see Table 4), who initially used a one‐factor‐at‐a‐time method to determine a temperature of 30°C, a constant agitation rate of 200 min−1, and an aeration rate of 0.5 vvm as optimal process conditions. A titre of 6.6 g L−1, a YP/S of 0.11 g g−1, and a volumetric productivity of 0.33 g L−1 d−1 were achieved. By implementing a pH control at 2.4 using H2SO4 and subsequently (NH4)2SO4, the titre could be increased and, due to the higher biomass, also the productivity. Melanin is formed under environmental stress, artificially generated by adding H2O2 increasing the productivity to 0.93 g L−1 d−1.
TABLE 4.
Comparing melanin production by titre, yield YP/S, and volumetric productivity of several bioreactor fermentations with different strains and substrates.
| Strain | Substrate | Fermentation mode | Titre [g L−1] | YPS [g g−1] | Vol. prod. [g L−1 h−1] | Reference |
|---|---|---|---|---|---|---|
| A. melanogenum GXZ‐6 | Glucose | 5 L batch w/o pH control | 6.6 | 0.11 | 0.33 | Zhou et al. (2023) |
| A. melanogenum GXZ‐6 | Glucose | 5 L batch, pH control w/ H2SO4 | 9.8 | 0.20 | 0.44 | |
| A. melanogenum GXZ‐6 | Glucose | 5 L batch, pH control w/ (NH4)2SO4 | 14.0 | 0.32 | 0.72 | |
| A. melanogenum GXZ‐6 | Glucose | 5 L fed‐batch, pH control w/ (NH4)2SO4, suppl. H2O2 | 18.0 | 0.38 | 0.93 |
For pullulan production in bioreactors, the literature presents more suitable process strategies using Aureobasidium spp. Sugumaran and Ponnusami (2017) have already compiled the key performance parameters, which are only presented in excerpts. Most of these are batch fermentations in lab‐scale bioreactors of up to 5 L. In summary, Choudhury et al. (2012) and Sharma et al. (2013) achieved the best performances with pullulan titres of over 83 g L−1, YPS of 0.6–0.7 g g−1, and volumetric productivities of almost 18 g L−1 h−1 using batch fermentations. Also worth mentioning is a fed‐batch fermentation in the largest reported volume of 60 L medium in a 150 L bioreactor using Aureobasidium sp. (Moscovici et al., 1996). By increasing the stirrer speed from 280 min−1 to 340 min−1, the oxygen transfer was optimized, which led to a doubling of the specific productivity to 0.04 g gCDW −1 h−1. The first continuous fermentations using Aureobasidium sp. were also published for pullulan production. In 1993, Schuster et al. (1993) investigated the influence of different ammonium concentrations and dilution rates on continuous pullulan production using A. pullulans p56 and achieved the best result of 0.35 g L−1 h−1 at a dilution rate of 0.05 h−1. Cheng et al. (2011a) used a biofilm bioreactor with A. pullulans ATCC 201253 to investigate different nitrogen and carbon concentrations and dilution rates. The best production rate of 1.3 g L−1 h−1 was achieved with 15 g L−1 sucrose, 0.9 g L−1 ammonium sulphate, and 0.4 g L−1 yeast extract at a dilution rate of 0.16 h−1.
Low production costs are crucial for developing profitable and competitive processes. The most significant cost factors are substrate and purification costs (Roelants et al., 2019; Sharma & Oberoi, 2017). Accordingly, the aim is to form as much product as possible from the substrate used (high YP/S) and to realize purification with as little equipment and resources as possible. Many Aureobasidium spp. belong to the melanin‐producing yeasts, which causes product discoloration and thus impedes product purification (Leathers et al., 2015). Hilares et al. (2017) showed that by irradiating the culture broth with LED blue light with a specific wavelength, melanin production can be avoided using A. pullulans LB83, thus eliminating a purification step and reducing costs. Otherwise, melanin could be subsequently removed by the addition of 4% (w/w) H2O2 (Wu et al., 2009), activated carbon, or solvent/salt blends (Singh et al., 2008). On laboratory scale, polyol lipids have so far been separated by organic extraction. To separate polyol lipids from culture broth, they are first dissolved in methyl ethyl ketone, which is then evaporated using a rotary evaporator or surface gassing (Leathers et al., 2016; Manitchotpisit et al., 2011). Larger quantities of polyol lipids have not yet been purified, so practicability on larger scales cannot be assessed. However, large quantities of solvents have an economic and environmental impact based on their toxicity and require equipment for separation and recycling. A solvent‐free solution could be a three‐phase disk centrifuge separating cells, polyol lipids, and aqueous phase (Wan et al., 2022). For large‐scale production of PMA, a complete flowsheet including substrate pretreatment, fermentation, and downstreaming has been presented by Cheng et al. (2017): Purification of cell‐free culture broth involves first concentration by ultrafiltration, followed by alcoholic precipitation and hydrolysis of PMA to malic acid, which is then dried by spray drying. It should be noted that CaCO3 is widely used as a neutralizer in PMA production. Due to the low water solubility (0.013 g L−1 at 20°C), large quantities of solid waste are produced, which entails an additional downstreaming step (Zou et al., 2019). Therefore, it is necessary to consider utilizing water‐soluble neutralizers. In a 50 L batch bioreactor cultivation with the water‐soluble (230 g L−1 at 20°C) neutralizer, Na2CO3 in combination with biotin and A. pullulans CCTCC M2012223, 34 g L−1 PMA was obtained at a yield of 0.45 g g−1 and a volumetric productivity of 0.41 g L−1 h−1 (Zou et al., 2014). Even if the production is slightly lower compared to those with CaCO3 (see Table 3), there is the advantage of not having a solid in the reactor, especially when it comes to an industrial scale. A similar process is used to purify pullulan. After cell separation by centrifugation, pullulan was precipitated using solvents, separated by ultrafiltration, followed by freeze‐drying (Cheng et al., 2011b; Singh et al., 2008). Pullulan and PMA are both water‐soluble products that precipitate by adding solvents and are often produced simultaneously. However, due to the difference in molecular weight, both can be precipitated sequentially by fractional precipitation (Zou et al., 2019).
To sum up, great advancements have been achieved in process development for PMA and pullulan production, displayed in high productivities and feasible purification methods. As mentioned, the high potential of Aureobasidium spp. has been recognized, and pullulan is already being produced industrially.
CONCLUSIONS
Overall, it can be summarized that Aureobasidium spp. are versatile fungi that are highly interesting for industrial processes due to their anabolic synthesis capacities, their diverse and robust physiology, and their broad substrate spectrum. A range of enzymes to break down complex carbon sources facilitate the usage of low‐cost side and waste streams from several industries. Their extremophilic traits enable fermentation at extreme conditions, such as extreme pH values and high salt concentrations, reducing operating costs (Chen & Jiang, 2018). Furthermore, the ability to produce several industrially interesting compounds at the mid‐two‐digit titre range offers quick implementation of respective processes in the chemical industry. Of particular importance in this regard is the fact that large‐scale industrial processes already exist (for pullulan production). Many challenges of up‐scaling have thus already been addressed.
This review contributes to the growing collection of scientific publications on this remarkable fungus, highlighting some of the many features that make Aureobasidium a promising candidate for future exploitation in the biotechnological industry in the framework of the circular bioeconomy.
AUTHOR CONTRIBUTIONS
Difan Xiao: Writing – original draft; writing – review and editing. Marielle Driller: Writing – original draft; writing – review and editing. Marie Dielentheis‐Frenken: Writing – original draft; writing – review and editing. Frederick Haala: Writing – original draft; writing – review and editing. Philipp Kohl: Writing – original draft; writing – review and editing. Karla Stein: Writing – original draft; writing – review and editing. Lars M. Blank: Writing – review and editing; funding acquisition. Till Tiso: Writing – review and editing; funding acquisition; supervision; writing – original draft.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
DX was partially funded by the China Scholarship Council (no. 201906910036). MD and KS were funded by the German Bundesministerium für Bildung und Forschung (BMBF) within the project AureoFACTory (no. 031B1203). MDF and FH were funded by the Fachagentur Nachwachsender Rohstoffe e.V. of the German Bundesministerium für Ernährung und Landwirtschaft (BMEL) as part of the project ViRIDi Aurum (No. 2220NR262X). PK was funded by the Bioeconomy Science Center, which is financially supported by the Ministerium für Kultur und Wissenschaft (MKW) of the State of North Rhine‐Westphalia (NRW) within the framework of the NRW Strategieprojekt BioSC (No. 313/323‐400‐00213) in the project SurfIn. TT and LMB acknowledge funding from the European Union's Horizon 2020 research and innovation program under grant agreement no. 870294 for the project MIX‐UP. TT furthermore acknowledges funding by the Bundesagentur für Sprunginnovation (SPRIN‐D) in the framework of the Circular Biomanufacturing challenge in the Quantum Leap project. Open Access funding enabled and organized by Projekt DEAL.
Xiao, D. , Driller, M. , Dielentheis‐Frenken, M. , Haala, F. , Kohl, P. , Stein, K. et al. (2024) Advances in Aureobasidium research: Paving the path to industrial utilization. Microbial Biotechnology, 17, e14535. Available from: 10.1111/1751-7915.14535
Difan Xiao, Marielle Driller, Marie Dielentheis‐Frenken, Frederick Haala, Philipp Kohl, Karla Stein contributed equally to this work.
REFERENCES
- Anastassiadis, S. , Aivasidis, A. , Wandrey, C. & Rehm, H.J. (2005) Process optimization of continuous gluconic acid fermentation by isolated yeast‐like strains of Aureobasidium pullulans . Biotechnology and Bioengineering, 91, 494–501. [DOI] [PubMed] [Google Scholar]
- Araújo, M. , Viveiros, R. , Correia, T.R. , Correia, I.J. , Bonifácio, V.D. , Casimiro, T. et al. (2014) Natural melanin: A potential pH‐responsive drug release device. International Journal of Pharmaceutics, 469, 140–145. [DOI] [PubMed] [Google Scholar]
- Arif, M. , Raja, M.A. , Zeenat, S. , Chi, Z. & Liu, C. (2017) Preparation and characterization of polyelectrolyte complex nanoparticles based on poly (malic acid), chitosan. A pH‐dependent delivery system. Journal of Biomaterials Science, Polymer Edition, 28, 50–62. [DOI] [PubMed] [Google Scholar]
- Arzanlou, M. & Khodaei, S. (2012) Aureobasidium iranianum, a new species on bamboo from Iran. Mycosphere, 3, 404–408. [Google Scholar]
- Baldwin, E.L. , Karki, B. , Zahler, J.D. , Rinehart, M. & Gibbons, W.R. (2019) Submerged vs. solid‐state conversion of soybean meal into a high protein feed using Aureobasidium pullulans . Journal of the American Oil Chemists' Society, 96, 989–998. [Google Scholar]
- Bary, A. (1884) Vergleichende morphologie und biologie der pilze, mycetozoen und bacterien. Leipzig: Engelmann. [Google Scholar]
- Beld, J. , Sonnenschein, E.C. , Vickery, C.R. , Noel, J.P. & Burkart, M.D. (2014) The phosphopantetheinyl transferases: catalysis of a post‐translational modification crucial for life. Natural Product Reports, 31, 61–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo, J. , Dominguez, J. , Goni, F. & Uruburu, F. (1981) Influence of pH on the transition from yeast‐like cells to chlamydospores in Aureobasidium pullulans . Antonie Van Leeuwenhoek, 47, 385–392. [DOI] [PubMed] [Google Scholar]
- Bischoff, K.M. , Leathers, T.D. , Price, N.P. & Manitchotpisit, P. (2015) Liamocin oil from Aureobasidium pullulans has antibacterial activity with specificity for species of streptococcus. The Journal of Antibiotics, 68, 642–645. [DOI] [PubMed] [Google Scholar]
- Brumano, L.P. , Antunes, F.A.F. , Souto, S.G. , Dos Santos, J.C. , Venus, J. , Schneider, R. et al. (2017) Biosurfactant production by Aureobasidium pullulans in stirred tank bioreactor: new approach to understand the influence of important variables in the process. Bioresource Technology, 243, 264–272. [DOI] [PubMed] [Google Scholar]
- Büchs, J. (2001) Introduction to advantages and problems of shaken cultures. Biochemical Engineering Journal, 7, 91–98. [DOI] [PubMed] [Google Scholar]
- Caldas, M. , Santos, A.C. , Veiga, F. , Rebelo, R. , Reis, R.L. & Correlo, V.M. (2020) Melanin nanoparticles as a promising tool for biomedical applications–a review. Acta Biomaterialia, 105, 26–43. [DOI] [PubMed] [Google Scholar]
- Calero, P. & Nikel, P.I. (2019) Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non‐traditional microorganisms. Microbial Biotechnology, 12, 98–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campana, R. , Fanelli, F. & Sisti, M. (2022) Role of melanin in the black yeast fungi Aureobasidium pullulans and Zalaria obscura in promoting tolerance to environmental stresses and to antimicrobial compounds. Fungal Biology, 126, 817–825. [DOI] [PubMed] [Google Scholar]
- Cao, W. , Cao, W. , Shen, F. , Luo, J. , Yin, J. , Qiao, C. et al. (2020) Membrane‐assisted β‐poly (L‐malic acid) production from bagasse hydrolysates by Aureobasidium pullulans ipe‐1. Bioresource Technology, 295, 122260. [DOI] [PubMed] [Google Scholar]
- Cao, W. , Chen, X. , Luo, J. , Yin, J. , Qiao, C. & Wan, Y. (2016) High molecular weight β‐poly (l‐malic acid) produced by A. pullulans with Ca2+ added repeated batch culture. International Journal of Biological Macromolecules, 85, 192–199. [DOI] [PubMed] [Google Scholar]
- Cao, W. , Wang, Y. , Shen, F. , Luo, J. , Yin, J. , Qiao, C. et al. (2019) Efficient β‐poly (L‐malic acid) production from Jerusalem artichoke by Aureobasidium pullulans ipe‐1 immobilized in luffa sponge matrices. Bioresource Technology, 288, 121497. [DOI] [PubMed] [Google Scholar]
- Cavicchioli, R. , Charlton, T. , Ertan, H. , Omar, S.M. , Siddiqui, K. & Williams, T. (2011) Biotechnological uses of enzymes from psychrophiles. Microbial Biotechnology, 4, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Černoša, A. , Sun, X. , Gostinčar, C. , Fang, C. , Gunde‐Cimerman, N. & Song, Z. (2021) Virulence traits and population genomics of the black yeast Aureobasidium melanogenum . Journal of Fungi, 7, 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaen, H. (2009) Pullulan. In: Imeson, A. (Ed.) Food stabilisers, thickeners and gelling agents. Oxford, UK: Wiley‐Blackwell, pp. 266–274. [Google Scholar]
- Chen, G.‐Q. & Jiang, X.‐R. (2018) Next generation industrial biotechnology based on extremophilic bacteria. Current Opinion in Biotechnology, 50, 94–100. [DOI] [PubMed] [Google Scholar]
- Chen, G.‐Q. , Zhang, X. , Liu, X. , Huang, W. , Xie, Z. , Han, J. et al. (2022) H alomonas spp., as chassis for low‐cost production of chemicals. Applied Microbiology and Biotechnology, 106, 6977–6992. [DOI] [PubMed] [Google Scholar]
- Chen, T.‐J. , Liu, G.‐L. , Chen, L. , Yang, G. , Hu, Z. , Chi, Z.‐M. et al. (2020) Alternative primers are required for pullulan biosynthesis in Aureobasidium melanogenum P16. International Journal of Biological Macromolecules, 147, 10–17. [DOI] [PubMed] [Google Scholar]
- Chen, T.‐J. , Liu, G.‐L. , Wei, X. , Wang, K. , Hu, Z. , Chi, Z. et al. (2020) A multidomain α‐glucan synthetase 2 (AmAgs2) is the key enzyme for pullulan biosynthesis in Aureobasidium melanogenum P16. International Journal of Biological Macromolecules, 150, 1037–1045. [DOI] [PubMed] [Google Scholar]
- Chen, Z. & Wan, C. (2017) Non‐sterile fermentations for the economical biochemical conversion of renewable feedstocks. Biotechnology Letters, 39, 1765–1777. [DOI] [PubMed] [Google Scholar]
- Cheng, C. , Zhou, Y. , Lin, M. , Wei, P. & Yang, S.‐T. (2017) Polymalic acid fermentation by Aureobasidium pullulans for malic acid production from soybean hull and soy molasses: fermentation kinetics and economic analysis. Bioresource Technology, 223, 166–174. [DOI] [PubMed] [Google Scholar]
- Cheng, K.‐C. , Demirci, A. & Catchmark, J.M. (2011a) Evaluation of medium composition and fermentation parameters on pullulan production by Aureobasidium pullulans . Food Science and Technology International, 17, 99–109. [DOI] [PubMed] [Google Scholar]
- Cheng, K.‐C. , Demirci, A. & Catchmark, J.M. (2011b) Pullulan: biosynthesis, production, and applications. Applied Microbiology and Biotechnology, 92, 29–44. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.‐Q. , Tang, G.‐L. & Shen, B. (2003) Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proceedings of the National Academy of Sciences, 100, 3149–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Z. , Kong, C.‐C. , Wang, Z.‐Z. , Wang, Z. , Liu, G.‐L. , Hu, Z. et al. (2022) The signaling pathways involved in metabolic regulation and stress responses of the yeast‐like fungi Aureobasidium spp. Biotechnology Advances, 55, 107898. [DOI] [PubMed] [Google Scholar]
- Chi, Z. , Liu, G.‐L. , Liu, C.‐G. & Chi, Z.‐M. (2016) Poly (β‐l‐malic acid) (PMLA) from Aureobasidium spp. and its current proceedings. Applied Microbiology and Biotechnology, 100, 3841–3851. [DOI] [PubMed] [Google Scholar]
- Chi, Z. , Wang, F. , Chi, Z. , Yue, L. , Liu, G. & Zhang, T. (2009) Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Applied Microbiology and Biotechnology, 82, 793–804. [DOI] [PubMed] [Google Scholar]
- Chi, Z. , Wang, X.‐X. , Ma, Z.‐C. , Buzdar, M.A. & Chi, Z.‐M. (2012) The unique role of siderophore in marine‐derived Aureobasidium pullulans HN6. 2. Biometals, 25, 219–230. [DOI] [PubMed] [Google Scholar]
- Chi, Z. , Wang, Z.‐P. , Wang, G.‐Y. , Khan, I. & Chi, Z.‐M. (2016) Microbial biosynthesis and secretion of l‐malic acid and its applications. Critical Reviews in Biotechnology, 36, 99–107. [DOI] [PubMed] [Google Scholar]
- Choudhury, A.R. , Sharma, N. & Prasad, G. (2012) Deoiledjatropha seed cake is a useful nutrient for pullulan production. Microbial Cell Factories, 11, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous, P.W. , Summerell, B.A. , Swart, L. , Denman, S. , Taylor, J. , Bezuidenhout, C. et al. (2011) Fungal pathogens of Proteaceae. Persoonia‐Molecular Phylogeny and Evolution of Fungi, 27, 20–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Z. , Zhou, H. , Zhang, S. , Gu, H. , Yang, Q. , Zhang, W. et al. (2018) Current advance in biological production of malic acid using wild type and metabolic engineered strains. Bioresource Technology, 258, 345–353. [DOI] [PubMed] [Google Scholar]
- David, F. & Siewers, V. (2015) Advances in yeast genome engineering. FEMS Yeast Research, 15, 1–14. [DOI] [PubMed] [Google Scholar]
- de Lorenzo, V. , Krasnogor, N. & Schmidt, M. (2021) For the sake of the bioeconomy: define what a synthetic biology chassis is! New Biotechnology, 60, 44–51. [DOI] [PubMed] [Google Scholar]
- DiCarlo, J.E. , Norville, J.E. , Mali, P. , Rios, X. , Aach, J. & Church, G.M. (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR‐Cas systems. Nucleic Acids Research, 41, 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, H. , Helguera, G. , Rodríguez, J.A. , Markman, J. , Luria‐Pérez, R. , Gangalum, P. et al. (2013) Polymalic acid nanobioconjugate for simultaneous inhibition of tumor growth and immunostimulation in HER2/neu‐positive breast cancer. Journal of Controlled Release: Official Journal of the Controlled Release Society, 171, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, A. , Nobre, C. , Rodrigues, L.R. , Peres, A.M. , Torres, D. , Rocha, I. et al. (2012) New improved method for fructooligosaccharides production by Aureobasidium pullulans . Carbohydrate Polymers, 89, 1174–1179. [DOI] [PubMed] [Google Scholar]
- Doshida, J. , Hasegawa, H. , Onuki, H. & Shimidzu, N. (1996) Exophilin A, a new antibiotic from a marine microorganism Exophiala pisciphila. The Journal of Antibiotics, 49, 1105–1109. [DOI] [PubMed] [Google Scholar]
- Dumorné, K. , Córdova, D.C. , Astorga‐Eló, M. & Renganathan, P. (2017) Extremozymes: a potential source for industrial applications. Journal of Microbiology and Biotechnology, 27, 649–659. [DOI] [PubMed] [Google Scholar]
- Eisenman, H.C. & Casadevall, A. (2012) Synthesis and assembly of fungal melanin. Applied Microbiology and Biotechnology, 93, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J. , Li, T. , Zhang, X. , Chen, J. , Zhao, T. & Zou, X. (2019) Efficient production of polymalic acid from xylose mother liquor, an environmental waste from the xylitol industry, by a T‐DNA‐based mutant of Aureobasidium pullulans . Applied Microbiology and Biotechnology, 103, 6519–6527. [DOI] [PubMed] [Google Scholar]
- Feng, J. , Yang, J. , Li, X. , Guo, M. , Wang, B. , Yang, S.‐T. et al. (2017) Reconstruction of a genome‐scale metabolic model and in silico analysis of the polymalic acid producer Aureobasidium pullulans CCTCC M2012223. Gene, 607, 1–8. [DOI] [PubMed] [Google Scholar]
- Feng, J. , Yang, J. , Yang, W. , Chen, J. , Jiang, M. & Zou, X. (2018) Metabolome‐and genome‐scale model analyses for engineering of Aureobasidium pullulans to enhance polymalic acid and malic acid production from sugarcane molasses. Biotechnology for Biofuels, 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin‐Johnson, E. , Figge, F. & Canning, L. (2016) Resource duration as a managerial indicator for circular economy performance. Journal of Cleaner Production, 133, 589–598. [Google Scholar]
- Garay, L.A. , Sitepu, I.R. , Cajka, T. , Xu, J. , Teh, H.E. , German, J.B. et al. (2018) Extracellular fungal polyol lipids: a new class of potential high value lipids. Biotechnology Advances, 36, 397–414. [DOI] [PubMed] [Google Scholar]
- Gasslmaier, B. & Holler, E. (1997) Specificity and direction of depolymerization of β‐poly (L‐malate) catalysed by polymalatase from Physarum polycephalum: fluorescence labeling at the Carboxy‐terminus of β‐poly (l‐malate). European Journal of Biochemistry, 250, 308–314. [DOI] [PubMed] [Google Scholar]
- Gibbs, P. , Seviour, R. & Schmid, F. (2000) Growth of filamentous fungi in submerged culture: problems and possible solutions. Critical Reviews in Biotechnology, 20, 17–48. [DOI] [PubMed] [Google Scholar]
- Gostinčar, C. , Ohm, R.A. , Kogej, T. , Sonjak, S. , Turk, M. , Zajc, J. et al. (2014) Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics, 15, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostinčar, C. , Sun, X. , Černoša, A. , Fang, C. , Gunde‐Cimerman, N. & Song, Z. (2022) Clonality, inbreeding, and hybridization in two extremotolerant black yeasts. GigaScience, 11, giac095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostinčar, C. , Turk, M. , Zajc, J. & Gunde‐Cimerman, N. (2019) Fifty Aureobasidium pullulans genomes reveal a recombining polyextremotolerant generalist. Environmental Microbiology, 21, 3638–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube, M. , Schmid, F. & Berg, G. (2011) Black fungi and associated bacterial communities in the phyllosphere of grapevine. Fungal Biology, 115, 978–986. [DOI] [PubMed] [Google Scholar]
- Gueldener, U. , Heinisch, J. , Koehler, G. , Voss, D. & Hegemann, J. (2002) A second set of loxP marker cassettes for Cre‐mediated multiple gene knockouts in budding yeast. Nucleic Acids Research, 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunde‐Cimerman, N. , Zalar, P. , de Hoog, S. & Plemenitaš, A. (2000) Hypersaline waters in salterns–natural ecological niches for halophilic black yeasts. FEMS Microbiology Ecology, 32, 235–240. [DOI] [PubMed] [Google Scholar]
- Guo, J. , Li, J. , Chen, Y. , Guo, X. & Xiao, D. (2016) Improving erythritol production of Aureobasidium pullulans from xylose by mutagenesis and medium optimization. Applied Biochemistry and Biotechnology, 180, 717–727. [DOI] [PubMed] [Google Scholar]
- Guo, J. , Wang, Y. , Li, B. , Huang, S. , Chen, Y. , Guo, X. et al. (2017) Development of a one‐step gene knock‐out and knock‐in method for metabolic engineering of Aureobasidium pullulans . Journal of Biotechnology, 251, 145–150. [DOI] [PubMed] [Google Scholar]
- Haala, F. , Dielentheis‐Frenken, M. , Brandt, F.M. , Karmainski, T. , Blank, L.M. & Tiso, T. (2024) DoE‐based medium optimization for improved biosurfactant production with Aureobasidium pullulans . Frontiers in Bioengineering and Biotechnology, 12, 1379707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilares, R.T. , Orsi, C.A. , Ahmed, M.A. , Marcelino, P.F. , Menegatti, C.R. , da Silva, S.S. et al. (2017) Low‐melanin containing pullulan production from sugarcane bagasse hydrolysate by Aureobasidium pullulans in fermentations assisted by light‐emitting diode. Bioresource Technology, 230, 76–81. [DOI] [PubMed] [Google Scholar]
- Hirabayashi, K. , Kondo, N. & Hayashi, S. (2016) Characterization and enzymatic hydrolysis of hydrothermally treated β‐1, 3–1, 6‐glucan from Aureobasidium pullulans . World Journal of Microbiology and Biotechnology, 32, 1–7. [DOI] [PubMed] [Google Scholar]
- Holler, E. , Angerer, B. , Achhammer, G. , Miller, S. & Windisch, C. (1992) Biological and biosynthetic properties of poly‐L‐malate. FEMS Microbiology Reviews, 9, 109–118. [Google Scholar]
- Huang, X. , Xu, L. , Qian, H. , Wang, X. & Tao, Z. (2022) Polymalic acid for translational nanomedicine. Journal of Nanobiotechnology, 20, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z.W. , Laurent, V. , Chetouani, G. , Ljubimova, J.Y. , Holler, E. , Benvegnu, T. et al. (2012) New functional degradable and bio‐compatible nanoparticles based on poly (malic acid) derivatives for site‐specific anti‐cancer drug delivery. International Journal of Pharmaceutics, 423, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyyappan, J. , Baskar, G. , Gnansounou, E. , Pandey, A. , Raaman, J.K. , Bharathiraja, B. et al. (2019) Recent advances in microbial production of malic acid from renewable by products. Reviews in Environmental Science and Bio/Technology, 18, 579–595. [Google Scholar]
- Jiang, H. , Chi, Z. , Liu, G.‐L. , Hu, Z. , Zhao, S.‐Z. & Chi, Z.‐M. (2020) Melanin biosynthesis in the desert‐derived Aureobasidium melanogenum XJ5‐1 is controlled mainly by the CWI signal pathway via a transcriptional activator Cmr1. Current Genetics, 66, 173–185. [DOI] [PubMed] [Google Scholar]
- Jiang, H. , Liu, G.‐L. , Chi, Z. , Wang, J.‐M. , Zhang, L.‐L. & Chi, Z.‐M. (2017) Both a PKS and a PPTase are involved in melanin biosynthesis and regulation of Aureobasidium melanogenum XJ5‐1 isolated from the Taklimakan desert. Gene, 602, 8–15. [DOI] [PubMed] [Google Scholar]
- Jiang, N. , Fan, X. & Tian, C. (2021) Identification and characterization of leaf‐inhabiting fungi from Castanea plantations in China. Journal of Fungi, 7, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. , Liang, Y.‐M. & Tian, C.‐M. (2019) Aureobasidium pini sp. nov. from pine needle in China. Phytotaxa, 402, 199–206. [Google Scholar]
- Kajiyama, T. , Kobayashi, H. , Taguchi, T. , Saito, H. , Kamatsu, Y. , Kataoka, K. et al. (2004) Synthesis of activated poly (α, β‐malic acid) using N‐hydroxysuccinimide and its gelation with collagen as biomaterials. Materials Science and Engineering: C, 24, 815–819. [Google Scholar]
- Kajiyama, T. , Taguchi, T. , Kobayashi, H. , Kataoka, K. & Tanaka, J. (2003) Synthesis of high molecular weight poly (α, β‐malic acid) for biomedical use by direct polycondensation. Polymer Degradation and Stability, 81, 525–530. [Google Scholar]
- Kang, X.‐X. , Jia, S.‐L. , Wei, X. , Zhang, M. , Liu, G.‐L. , Hu, Z. et al. (2022) Liamocins biosynthesis, its regulation in Aureobasidium spp., and their bioactivities. Critical Reviews in Biotechnology, 42, 93–105. [DOI] [PubMed] [Google Scholar]
- Kim, J.S. , Lee, I.K. & Yun, B.S. (2015) A novel biosurfactant produced by Aureobasidium pullulans L3‐GPY from a tiger lily wild flower, Lilium lancifolium thunb. PLoS One, 10, e0122917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement, T. & Büchs, J. (2013) Itaconic acid–a biotechnological process in change. Bioresource Technology, 135, 422–431. [DOI] [PubMed] [Google Scholar]
- Kövilein, A. , Kubisch, C. , Cai, L. & Ochsenreither, K. (2020) Malic acid production from renewables: a review. Journal of Chemical Technology & Biotechnology, 95, 513–526. [Google Scholar]
- Kreuter, J. , Stark, G. , Mach, R.L. , Mach‐Aigner, A.R. & Zimmermann, C. (2022) Fast and efficient CRISPR‐mediated genome editing in Aureobasidium using Cas9 ribonucleoproteins. Journal of Biotechnology, 350, 11–16. [DOI] [PubMed] [Google Scholar]
- Książek, E. (2023) Citric acid: properties, microbial production, and applications in industries. Molecules, 29, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, C.G. , Mongolla, P. , Pombala, S. , Kamle, A. & Joseph, J. (2011) Physicochemical characterization and antioxidant activity of melanin from a novel strain of Aspergillus bridgeri ICTF‐201. Letters in Applied Microbiology, 53, 350–358. [DOI] [PubMed] [Google Scholar]
- Kurosawa, T. , Sakai, K. , Nakahara, T. , Oshima, Y. & Tabuch, T. (1994) Extracellular accumulation of the polyol lipids, 3, 5‐dihydroxydecanoyl and 5‐hydroxy‐2‐decenoyl esters of arabitol and mannitol, by Aureobasidium sp. Bioscience, Biotechnology, and Biochemistry, 58, 2057–2060. [Google Scholar]
- Leathers, T.D. (2003) Biotechnological production and applications of pullulan. Applied Microbiology and Biotechnology, 62, 468–473. [DOI] [PubMed] [Google Scholar]
- Leathers, T.D. & Manitchotpisit, P. (2013) Production of poly (β‐L‐malic acid)(PMA) from agricultural biomass substrates by Aureobasidium pullulans . Biotechnology Letters, 35, 83–89. [DOI] [PubMed] [Google Scholar]
- Leathers, T.D. , Price, N.P. , Bischoff, K.M. , Manitchotpisit, P. & Skory, C.D. (2015) Production of novel types of antibacterial liamocins by diverse strains of Aureobasidium pullulans grown on different culture media. Biotechnology Letters, 37, 2075–2081. [DOI] [PubMed] [Google Scholar]
- Leathers, T.D. , Price, N.P. , Manitchotpisit, P. & Bischoff, K.M. (2016) Production of anti‐streptococcal liamocins from agricultural biomass by Aureobasidium pullulans . World Journal of Microbiology and Biotechnology, 32, 1–7. [DOI] [PubMed] [Google Scholar]
- Leathers, T.D. , Skory, C.D. , Price, N.P. & Nunnally, M.S. (2018) Medium optimization for production of anti‐streptococcal liamocins by Aureobasidium pullulans . Biocatalysis and Agricultural Biotechnology, 13, 53–57. [Google Scholar]
- Lee, B. & Holler, E. (1999) Effects of culture conditions on β‐poly (L‐malate) production by Physarum polycephalum . Applied Microbiology and Biotechnology, 51, 647–652. [Google Scholar]
- Lee, B.‐S. , Fujita, M. , Khazenzon, N.M. , Wawrowsky, K.A. , Wachsmann‐Hogiu, S. , Farkas, D.L. et al. (2006) Polycefin, a new prototype of a multifunctional nanoconjugate based on poly (β‐L‐malic acid) for drug delivery. Bioconjugate Chemistry, 17, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.‐H. , Cho, S.‐E. , Oh, J.Y. , Cho, E.‐J. & Kwon, S. (2021) A novel species of Aureobasidium (Dothioraceae) recovered from Acer pseudosieboldianum in Korea. Journal of Asia‐Pacific Biodiversity, 14, 657–661. [Google Scholar]
- Li, B. , Li, B. , Wang, P. , Feng, Y. , Xu, X. , Zhang, Y. et al. (2023) Bio‐refinery of xylose processing wastes for green polymalic acid production and l‐malic acid recovery by engineered Aureobasidium pullulans in a non‐waste‐disposal system. Chemical Engineering Journal, 454, 140533. [Google Scholar]
- Li, B.‐x. , Zhang, N. , Peng, Q. , Yin, T. , Guan, F.‐f. , Wang, G.‐l. et al. (2009) Production of pigment‐free pullulan by swollen cell in Aureobasidium pullulans NG which cell differentiation was affected by pH and nutrition. Applied Microbiology and Biotechnology, 84, 293–300. [DOI] [PubMed] [Google Scholar]
- Li, D. , Tang, Y. , Lin, J. & Cai, W. (2017) Methods for genetic transformation of filamentous fungi. Microbial Cell Factories, 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Yang, W. , Xu, X. , Zhang, Y. , Chen, J. & Zou, X. (2021) Coproduction of polymalic acid and liamocins from two waste by‐products from the xylitol and gluconate industries by Aureobasidium pullulans . Bioprocess and Biosystems Engineering, 44, 1965–1974. [DOI] [PubMed] [Google Scholar]
- Li, Y.‐F. , Jiang, H. , Hu, Z. , Liu, G.‐L. , Chi, Z.‐M. & Chi, Z. (2019) Overexpression of an inulinase gene in an oleaginous yeast, Aureobasidium melanogenum P10, for efficient lipid production from inulin. Microbial Physiology, 28, 190–200. [DOI] [PubMed] [Google Scholar]
- Ligonzo, T. , Ambrico, M. , Augelli, V. , Perna, G. , Schiavulli, L. , Tamma, M. et al. (2009) Electrical and optical properties of natural and synthetic melanin biopolymer. Journal of Non‐Crystalline Solids, 355, 1221–1226. [Google Scholar]
- Lingappa, Y. , Sussman, A.S. & Bernstein, I.A. (1963) Effect of light and media upon growth and melanin formation in Aureobasidium pullulans (de By.) Arn. (= pullularia pullulans). Mycopathologia et Mycologia Applicata, 20, 109–128. [DOI] [PubMed] [Google Scholar]
- Liu, T. , Zhu, L. , Zhang, Z. , Huang, H. , Zhang, Z. & Jiang, L. (2017) Protective role of trehalose during radiation and heavy metal stress in Aureobasidium subglaciale F134. Scientific Reports, 7, 17586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Si, Z. , Zhang, H. , Wei, P. & Xu, Z. (2022) Efficient poly (β‐L‐malic acid) production from cassava hydrolysate by cell recycle of Aureobasidium pullulans . Applied Microbiology and Biotechnology, 106, 2855–2868. [DOI] [PubMed] [Google Scholar]
- Liu, Y.‐Y. , Chi, Z. , Wang, Z.‐P. , Liu, G.‐L. & Chi, Z.‐M. (2014) Heavy oils, principally long‐chain n‐alkanes secreted by Aureobasidium pullulans var. melanogenum strain P5 isolated from mangrove system. Journal of Industrial Microbiology and Biotechnology, 41, 1329–1337. [DOI] [PubMed] [Google Scholar]
- Ljubimova, J.Y. , Fujita, M. , Khazenzon, N.M. , Lee, B.‐S. , Wachsmann‐Hogiu, S. , Farkas, D.L. et al. (2008) Nanoconjugate based on polymalic acid for tumor targeting. Chemico‐Biological Interactions, 171, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotrakul, P. , Deenarn, P. , Prasongsuk, S. & Punnapayak, H. (2009) Isolation of Aureobasidium pullulans from bathroom surfaces and their antifungal activity against some Aspergilli. African Journal of Microbiology Research, 3, 253–257. [Google Scholar]
- Lotrakul, P. , Unhapattaratitikul, P. , Seelanan, T. , Prasongsuk, S. & Punnapayak, H. (2013) An aubasidan‐like β‐glucan produced by Aureobasidium pullulans in Thailand. ScienceAsia, 39, 363–368. [Google Scholar]
- Loyer, P. & Cammas‐Marion, S. (2014) Natural and synthetic poly (malic acid)‐based derivates: a family of versatile biopolymers for the design of drug nanocarriers. Journal of Drug Targeting, 22, 556–575. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Wang, G.‐Y. , Liu, G.‐L. , Wang, Z.‐P. & Chi, Z.‐M. (2013) Overproduction of poly (β‐malic acid)(PMA) from glucose by a novel Aureobasidium sp. P6 strain isolated from mangrove system. Applied Microbiology and Biotechnology, 97, 8931–8939. [DOI] [PubMed] [Google Scholar]
- Maina, S. , Kachrimanidou, V. & Koutinas, A. (2017) A roadmap towards a circular and sustainable bioeconomy through waste valorization. Current Opinion in Green and Sustainable Chemistry, 8, 18–23. [Google Scholar]
- Manitchotpisit, P. , Price, N.P. , Leathers, T.D. & Punnapayak, H. (2011) Heavy oils produced by Aureobasidium pullulans . Biotechnology Letters, 33, 1151–1157. [DOI] [PubMed] [Google Scholar]
- Manitchotpisit, P. , Watanapoksin, R. , Price, N.P. , Bischoff, K.M. , Tayeh, M. , Teeraworawit, S. et al. (2014) Aureobasidium pullulans as a source of liamocins (heavy oils) with anticancer activity. World Journal of Microbiology and Biotechnology, 30, 2199–2204. [DOI] [PubMed] [Google Scholar]
- Marques, M.P. , Cabral, J.M. & Fernandes, P. (2010) Bioprocess scale‐up: quest for the parameters to be used as criterion to move from microreactors to lab‐scale. Journal of Chemical Technology & Biotechnology, 85, 1184–1198. [Google Scholar]
- Mattoon, E.R. , Cordero, R.J. & Casadevall, A. (2021) Fungal melanins and applications in healthcare, bioremediation and industry. Journal of Fungi, 7, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneses, D.P. , Gudiña, E.J. , Fernandes, F. , Gonçalves, L.R. , Rodrigues, L.R. & Rodrigues, S. (2017) The yeast‐like fungus Aureobasidium thailandense LB01 produces a new biosurfactant using olive oil mill wastewater as an inducer. Microbiological Research, 204, 40–47. [DOI] [PubMed] [Google Scholar]
- Moscovici, M. , Ionescu, C. , Oniscu, C. , Fotea, O. , Protopopescu, P. & Hanganu, L.D. (1996) Improved exopolysaccharide production in fed‐batch fermentation of Aureobasidium pullulans, with increased impeller speed. Biotechnology Letters, 18, 787–790. [Google Scholar]
- Mujdeci, G.N. (2021) Natural melanin synthesized by Aureobasidium pullulans using food wastes and its characterization. Applied Food Biotechnology, 8, 307–318. [Google Scholar]
- Müjdeci, G.N. (2022) Experimental modeling and optimization of melanin production by Aureobasidium pullulans NBRC 100716 in carrot peel extract. Environmental Progress & Sustainable Energy, 41, e13919. [Google Scholar]
- Narsing Rao, M.P. , Xiao, M. & Li, W.‐J. (2017) Fungal and bacterial pigments: secondary metabolites with wide applications. Frontiers in Microbiology, 8, 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke, M. , Costa, S.G. & Contiero, J. (2005) Rhamnolipid surfactants: an update on the general aspects of these remarkable biomolecules. Biotechnology Progress, 21, 1593–1600. [DOI] [PubMed] [Google Scholar]
- Nivina, A. , Yuet, K.P. , Hsu, J. & Khosla, C. (2019) Evolution and diversity of assembly‐line polyketide synthases: focus review. Chemical Reviews, 119, 12524–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onetto, C.A. , Schmidt, S.A. , Roach, M.J. & Borneman, A.R. (2020) Comparative genome analysis proposes three new Aureobasidium species isolated from grape juice. FEMS Yeast Research, 20, foaa052. [DOI] [PubMed] [Google Scholar]
- Peterson, S.W. , Manitchotpisit, P. & Leathers, T.D. (2013) Aureobasidium thailandense sp. nov. isolated from leaves and wooden surfaces. International Journal of Systematic and Evolutionary Microbiology, 63, 790–795. [DOI] [PubMed] [Google Scholar]
- Portilla‐Arias, J.A. , García‐Alvarez, M. , de Ilarduya, A.M. , Holler, E. , Galbis, J.A. & Muñoz‐Guerra, S. (2008) Synthesis, degradability, and drug releasing properties of methyl esters of fungal poly (β, L‐malic acid). Macromolecular Bioscience, 8, 540–550. [DOI] [PubMed] [Google Scholar]
- Prasongsuk, S. , Lotrakul, P. , Ali, I. , Bankeeree, W. & Punnapayak, H. (2018) The current status of Aureobasidium pullulans in biotechnology. Folia Microbiologica, 63, 129–140. [DOI] [PubMed] [Google Scholar]
- Price, N.P. , Bischoff, K.M. , Leathers, T.D. , Cossé, A.A. & Manitchotpisit, P. (2017) Polyols, not sugars, determine the structural diversity of anti‐streptococcal liamocins produced by Aureobasidium pullulans strain NRRL 50380. The Journal of Antibiotics, 70, 136–141. [DOI] [PubMed] [Google Scholar]
- Price, N.P. , Manitchotpisit, P. , Vermillion, K.E. , Bowman, M.J. & Leathers, T.D. (2013) Structural characterization of novel extracellular liamocins (mannitol oils) produced by Aureobasidium pullulans strain NRRL 50380. Carbohydrate Research, 370, 24–32. [DOI] [PubMed] [Google Scholar]
- Qi, C.‐Y. , Chi, Z. , Liu, G.‐L. & Chi, Z.‐M. (2022) A high molecular weight polymalate is synthesized by the whole genome duplicated strain Aureobasidium melanogenum OUC. International Journal of Biological Macromolecules, 202, 608–619. [DOI] [PubMed] [Google Scholar]
- Qi, C.‐Y. , Chi, Z. , Liu, G.‐L. , Wang, P. & Chi, Z.‐M. (2023) A new high molecular weight polymalate coating film on grape. Industrial Crops and Products, 202, 116994. [Google Scholar]
- Qi, C.‐Y. , Jia, S.‐L. , Liu, G.‐L. , Chen, L. , Wei, X. , Hu, Z. et al. (2021) Polymalate (PMA) biosynthesis and its molecular regulation in Aureobasidium spp. International Journal of Biological Macromolecules, 174, 512–518. [DOI] [PubMed] [Google Scholar]
- Qiao, C. , Song, Y. , Zheng, Z. , Fan, X. & Yu, S. (2015) Synthesis of poly (β‐L‐malic acid) by the optimization of inorganic nitrogen complexing with growth factors using Aureobasidium pullulans CGMCC3337. In: Advances in applied biotechnology: proceedings of the 2nd international conference on applied biotechnology (ICAB 2014)‐volume I. Heidelberg: Springer, pp. 557–566. [Google Scholar]
- Qiao, C.S. , Zhong, K. , Hao, H.X. & Jia, Y.Y. (2012) Optimization of poly (β‐L‐malic acid) production using Aureobasidium pullulans by response surface methodology. Applied Mechanics and Materials, 108, 121–126. [Google Scholar]
- Ran, F.A. , Hsu, P.D. , Wright, J. , Agarwala, V. , Scott, D.A. & Zhang, F. (2013) Genome engineering using the CRISPR‐Cas9 system. Nature Protocols, 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathberger, K. , Reisner, H. , Willibald, B. , Molitoris, H.‐P. & Holler, E. (1999) Comparative synthesis and hydrolytic degradation of poly (L‐malate) by myxomycetes and fungi. Mycological Research, 103, 513–520. [Google Scholar]
- Rauch, M.E. , Graef, H.W. , Rozenzhak, S.M. , Jones, S.E. , Bleckmann, C.A. , Kruger, R.L. et al. (2006) Characterization of microbial contamination in United States air Force aviation fuel tanks. Journal of Industrial Microbiology and Biotechnology, 33, 29–36. [DOI] [PubMed] [Google Scholar]
- Rensink, S. , van Nieuwenhuijzen, E.J. , Sailer, M.F. , Struck, C. & Wösten, H.A. (2024) Use of Aureobasidium in a sustainable economy. Applied Microbiology and Biotechnology, 108, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants, S. , Solaiman, D.K. , Ashby, R.D. , Lodens, S. , Van Renterghem, L. & Soetaert, W. (2019) Production and applications of sophorolipids. Biobased Surfactants, 65–119. 10.1016/C2016-0-03179-0 [DOI] [Google Scholar]
- Roy, S. & Rhim, J.‐W. (2022) New insight into melanin for food packaging and biotechnology applications. Critical Reviews in Food Science and Nutrition, 62, 4629–4655. [DOI] [PubMed] [Google Scholar]
- Rueda‐Mejia, M.P. , Nägeli, L. , Lutz, S. , Hayes, R.D. , Varadarajan, A.R. , Grigoriev, I.V. et al. (2021) Genome, transcriptome and secretome analyses of the antagonistic, yeast‐like fungus Aureobasidium pullulans to identify potential biocontrol genes. Microbial Cell, 8, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, M. , Comba, S. , Altabe, S. , Recio‐Balsells, A.I. , Labadie, G.R. , Takano, E. et al. (2018) Biochemical characterization of the minimal domains of an iterative eukaryotic polyketide synthase. The FEBS Journal, 285, 4494–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saber, W.I. , Ghoniem, A.A. , Al‐Otibi, F.O. , El‐Hersh, M.S. , Eldadamony, N.M. , Menaa, F. et al. (2023) A comparative study using response surface methodology and artificial neural network towards optimized production of melanin by Aureobasidium pullulans AKW. Scientific Reports, 13, 13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahara, H. & Izumori, K. (2005) Production of L‐sorbitol from L‐fructose by Aureobasidium pullulans LP23 isolated from soy sauce mash. Journal of Bioscience and Bioengineering, 100, 223–226. [DOI] [PubMed] [Google Scholar]
- Saur, K.M. , Brumhard, O. , Scholz, K. , Hayen, H. & Tiso, T. (2019) A pH shift induces high‐titer liamocin production in Aureobasidium pullulans . Applied Microbiology and Biotechnology, 103, 4741–4752. [DOI] [PubMed] [Google Scholar]
- Scapini, T. , Dalastra, C. , Camargo, A.F. , Kubeneck, S. , Modkovski, T.A. , Júnior, S.L.A. et al. (2022) Seawater‐based biorefineries: a strategy to reduce the water footprint in the conversion of lignocellulosic biomass. Bioresource Technology, 344, 126325. [DOI] [PubMed] [Google Scholar]
- Schmidt, F. (2005) Optimization and scale up of industrial fermentation processes. Applied Microbiology and Biotechnology, 68, 425–435. [DOI] [PubMed] [Google Scholar]
- Schuster, M. , Schweizer, G. , Reissmann, S. & Kahmann, R. (2016) Genome editing in Ustilago maydis using the CRISPR–Cas system. Fungal Genetics and Biology, 89, 3–9. [DOI] [PubMed] [Google Scholar]
- Schuster, R. , Wenzig, E. & Mersmann, A. (1993) Production of the fungal exopolysaccharide pullulan by batch‐wise and continuous fermentation. Applied Microbiology and Biotechnology, 39, 155–158. [Google Scholar]
- Seviour, R.J. , McNeil, B. , Fazenda, M.L. & Harvey, L.M. (2011) Operating bioreactors for microbial exopolysaccharide production. Critical Reviews in Biotechnology, 31, 170–185. [DOI] [PubMed] [Google Scholar]
- Sharma, N. , Prasad, G. & Choudhury, A.R. (2013) Utilization of corn steep liquor for biosynthesis of pullulan, an important exopolysaccharide. Carbohydrate Polymers, 93, 95–101. [DOI] [PubMed] [Google Scholar]
- Sharma, R. & Oberoi, H.S. (2017) Biosurfactant‐aided bioprocessing: industrial applications and environmental impact. Recent Advances in Applied Microbiology, 55–88. 10.1007/978-981-10-5275-0_3 [DOI] [Google Scholar]
- Shimada, K. , Matsushima, K.I. , Fukumoto, J. & Yamamoto, T. (1969) Poly‐(L)‐malic acid; a new protease inhibitor from Penicillium cyclopium . Biochemical and Biophysical Research Communications, 35, 619–624. [DOI] [PubMed] [Google Scholar]
- Singh, R.S. , Kaur, N. , Singh, D. , Purewal, S.S. & Kennedy, J.F. (2023) Pullulan in pharmaceutical and cosmeceutical formulations: a review. International Journal of Biological Macromolecules, 231, 123353. [DOI] [PubMed] [Google Scholar]
- Singh, R.S. , Saini, G.K. & Kennedy, J.F. (2008) Pullulan: microbial sources, production and applications. Carbohydrate Polymers, 73, 515–531. [DOI] [PubMed] [Google Scholar]
- Slightom, J.L. , Metzger, B.P. , Luu, H.T. & Elhammer, A.P. (2009) Cloning and molecular characterization of the gene encoding the Aureobasidin A biosynthesis complex in Aureobasidium pullulans BP‐1938. Gene, 431, 67–79. [DOI] [PubMed] [Google Scholar]
- Song, X. , Wang, Y. , Wang, P. , Pu, G. & Zou, X. (2020) GATA‐type transcriptional factor Gat1 regulates nitrogen uptake and polymalic acid biosynthesis in polyextremotolerant fungus Aureobasidium pullulans . Environmental Microbiology, 22, 229–242. [DOI] [PubMed] [Google Scholar]
- Sternberg, N. & Hamilton, D. (1981) Bacteriophage P1 site‐specific recombination: I. Recombination between loxP sites. Journal of Molecular Biology, 150, 467–486. [DOI] [PubMed] [Google Scholar]
- Stovicek, V. , Holkenbrink, C. & Borodina, I. (2017) CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Research, 17, fox030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugumaran, K. & Ponnusami, V. (2015) Statistical modeling of pullulan production and its application in pullulan acetate nanoparticles synthesis. International Journal of Biological Macromolecules, 81, 867–876. [DOI] [PubMed] [Google Scholar]
- Sugumaran, K. & Ponnusami, V. (2017) Review on production, downstream processing and characterization of microbial pullulan. Carbohydrate Polymers, 173, 573–591. [DOI] [PubMed] [Google Scholar]
- Sugumaran, K. , Shobana, P. , Balaji, P.M. , Ponnusami, V. & Gowdhaman, D. (2014) Statistical optimization of pullulan production from Asian palm kernel and evaluation of its properties. International Journal of Biological Macromolecules, 66, 229–235. [DOI] [PubMed] [Google Scholar]
- Suthar, M. , Dufossé, L. & Singh, S.K. (2023) The enigmatic world of fungal melanin: a comprehensive review. Journal of Fungi, 9, 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwannarach, N. , Kumla, J. , Watanabe, B. , Matsui, K. & Lumyong, S. (2019) Characterization of melanin and optimal conditions for pigment production by an endophytic fungus, Spissiomyces endophytica SDBR‐CMU319. PLoS One, 14, e0222187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takafumi, K. , Kanichi, S. , Yoshiteru, O. & Takeshi, T. (1994) Extracellular accumulation of the polyol lipids, 3, 5‐dihydroxydecanoyl and 5‐hydroxy‐2‐decenoyl esters of arabitol and mannitol, by Aurebasidium sp. Bioscience, Biotechnology, and Biochemistry, 58, 2057–2060. [Google Scholar]
- Tang, R.‐R. , Chi, Z. , Jiang, H. , Liu, G.‐L. , Xue, S.‐J. , Hu, Z. et al. (2018) Overexpression of a pyruvate carboxylase gene enhances extracellular liamocin and intracellular lipid biosynthesis by Aureobasidium melanogenum M39. Process Biochemistry, 69, 64–74. [Google Scholar]
- Thambugala, K.M. , Ariyawansa, H.A. , Li, Y.‐M. , Boonmee, S. , Hongsanan, S. , Tian, Q. et al. (2014) Dothideales. Fungal Diversity, 68, 105–158. [Google Scholar]
- Tiso, T. , Welsing, G. , Lipphardt, A. , Sauer, D.F. , Chi, Z. , Blank, L.M. et al. (2024) Proposal for a systematic naming convention for liamocins. Journal of Surfactants and Detergents, 27, 317–461. [Google Scholar]
- Tran‐Ly, A.N. , Reyes, C. , Schwarze, F.W. & Ribera, J. (2020) Microbial production of melanin and its various applications. World Journal of Microbiology and Biotechnology, 36, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, G. , Wang, Y. , Ji, Y. & Zou, X. (2015) The effect of Tween 80 on the polymalic acid and pullulan production by Aureobasidium pullulans CCTCC M2012223. World Journal of Microbiology and Biotechnology, 31, 219–226. [DOI] [PubMed] [Google Scholar]
- Urzì, C. , De Leo, F. , Passo, C.L. & Criseo, G. (1999) Intra‐specific diversity of Aureobasidium pullulans strains isolated from rocks and other habitats assessed by physiological methods and by random amplified polymorphic DNA (RAPD). Journal of Microbiological Methods, 36, 95–105. [DOI] [PubMed] [Google Scholar]
- Vert, M. (1998) Chemical routes to poly (β‐malic acid) and potential applications of this water‐soluble bioresorbable poly (β‐hydroxy alkanoate). Polymer Degradation and Stability, 59, 169–175. [Google Scholar]
- Viala, P. & Boyer, G. (1891) Sur un Basidiomycète inferérieur, parasite des grains de raisins. Comptes Rendues Hebdomaires des Séances de l'Académie de Sciences, Paris, 112, 1148–1150. [Google Scholar]
- Vignolle, G.A. , Mach, R.L. , Mach‐Aigner, A.R. & Derntl, C. (2021) Genome sequence of the black yeast‐like strain Aureobasidium pullulans var. aubasidani CBS 100524. Microbiology Resource Announcements, 10, e01293‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveka, R. , Varjani, S. & Ekambaram, N. (2021) Valorization of cassava waste for pullulan production by Aureobasidium pullulans MTCC 1991. Energy & Environment, 32, 1086–1102. [Google Scholar]
- Wagner, J.M. & Alper, H.S. (2016) Synthetic biology and molecular genetics in non‐conventional yeasts: current tools and future advances. Fungal Genetics and Biology, 89, 126–136. [DOI] [PubMed] [Google Scholar]
- Wan, C. , Min, L. , Qin, F. , Liang, S. , Pan, Y. , Yi, T. et al. (2022) Production of liamocins by Aureobasidium spp. with potential applications. Biochemical Engineering Journal, 188, 108687. [Google Scholar]
- Wang, C.‐L. , Li, Y. , Xin, F.‐H. , Liu, Y.‐Y. & Chi, Z.‐M. (2014) Evaluation of single cell oil from Aureobasidium pullulans var. melanogenum P10 isolated from mangrove ecosystems for biodiesel production. Process Biochemistry, 49, 725–731. [Google Scholar]
- Wang, D. , Yu, X. & Gongyuan, W. (2013) Pullulan production and physiological characteristics of Aureobasidium pullulans under acid stress. Applied Microbiology and Biotechnology, 97, 8069–8077. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Bai, T. , Miao, Z. , Ning, W. & Liang, W. (2018) Simultaneous production of single cell oil and fumaric acid by a newly isolated yeast Aureobasidium pullulans var. aubasidani DH177. Bioprocess and Biosystems Engineering, 41, 1707–1716. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Shi, B. , Zhang, P. , Zhao, T. , Yin, H. & Qiao, C. (2020) Effects of corn steep liquor on β‐poly (l‐malic acid) production in Aureobasidium melanogenum . AMB Express, 10, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Yin, H. , Zhao, T. , Yang, D. , Jia, S. & Qiao, C. (2023) De novo transcriptome assembly of Aureobasidium melanogenum CGMCC18996 to analyze the β‐poly (L‐malic acid) biosynthesis pathway under the CaCO3 addition. Food Science and Human Wellness, 12, 1248–1256. [Google Scholar]
- Wang, K. , Chi, Z. , Liu, G.‐L. , Qi, C.‐Y. , Jiang, H. , Hu, Z. et al. (2020) A novel PMA synthetase is the key enzyme for polymalate biosynthesis and its gene is regulated by a calcium signaling pathway in Aureobasidium melanogenum ATCC62921. International Journal of Biological Macromolecules, 156, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Deng, A. , Zhang, Y. , Liu, S. , Liang, Y. , Bai, H. et al. (2018) Efficient CRISPR–Cas9 mediated multiplex genome editing in yeasts. Biotechnology for Biofuels, 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Chi, P. , Zou, Y. , Xu, Y. , Xu, S. , Bilal, M. et al. (2020) Metabolic engineering of Yarrowia lipolytica for thermoresistance and enhanced erythritol productivity. Biotechnology for Biofuels, 13, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Jia, S.‐L. , Liu, G.‐L. , Chi, Z. & Chi, Z.‐M. (2022) Aureobasidium spp. and their applications in biotechnology. Process Biochemistry, 116, 72–83. [Google Scholar]
- Wang, P. , Zhang, M. , Zhao, S.F. , Zhang, Z.R. , Liu, G.L. , Chi, Z. et al. (2024) Liamocins overproduction via the two‐pH stage fermentation and anti‐Aspergillus flavus activity of Massoia lactone. Biotechnology Journal, 19, 2300675. [DOI] [PubMed] [Google Scholar]
- Wang, Q.‐Q. , Lu, Y. , Ren, Z.‐Y. , Chi, Z. , Liu, G.‐L. & Chi, Z.‐M. (2017) CreA is directly involved in pullulan biosynthesis and regulation of Aureobasidium melanogenum P16. Current Genetics, 63, 471–485. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Chi, Z. , Chi, Z. , Li, J. & Wang, X. (2009) Siderophore production by the marine‐derived Aureobasidium pullulans and its antimicrobial activity. Bioresource Technology, 100, 2639–2641. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Zhang, K. , Lin, C. , Zhao, S. , Guan, J. , Zhou, W. et al. (2023) Influence of Cmr1 in the regulation of antioxidant function melanin biosynthesis in Aureobasidium pullulans . Food, 12, 2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.‐K. , Chi, Z. , Zhou, H.‐X. , Liu, G.‐L. & Chi, Z.‐M. (2015) Enhanced production of Ca2+‐polymalate (PMA) with high molecular mass by Aureobasidium pullulans var. pullulans MCW. Microbial Cell Factories, 14, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.‐P. , Zhang, X.‐Y. , Ma, Y. , Ye, J.‐R. , Jiang, J. , Wang, H.‐Y. et al. (2021) Whole conversion of agro‐industrial wastes rich in galactose‐based carbohydrates into lipid using oleaginous yeast Aureobasidium namibiae . Biotechnology for Biofuels, 14, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, P. , Cheng, C. , Lin, M. , Zhou, Y. & Yang, S.‐T. (2017) Production of poly (malic acid) from sugarcane juice in fermentation by Aureobasidium pullulans: kinetics and process economics. Bioresource Technology, 224, 581–589. [DOI] [PubMed] [Google Scholar]
- Wei, X. , Liu, G.‐L. , Jia, S.‐L. , Chi, Z. , Hu, Z. & Chi, Z.‐M. (2021) Pullulan biosynthesis and its regulation in Aureobasidium spp. Carbohydrate Polymers, 251, 117076. [DOI] [PubMed] [Google Scholar]
- Wernick, D.G. , Pontrelli, S.P. , Pollock, A.W. & Liao, J.C. (2016) Sustainable biorefining in wastewater by engineered extreme alkaliphile Bacillus marmarensis . Scientific Reports, 6, 20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, T.P. (2017) Microbial production of malic acid from biofuel‐related coproducts and biomass. Fermentation, 3, 14. [Google Scholar]
- Wu, F. , Feng, Z. , Wang, M. & Wang, Q. (2023) Proposal of four new Aureobasidium species for exopolysaccharide production. Journal of Fungi, 9, 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, N. , Zhang, J. , Chen, Y. , Xu, Q. , Song, P. , Li, Y. et al. (2022) Recent advances in microbial production of L‐malic acid. Applied Microbiology and Biotechnology, 106, 7973–7992. [DOI] [PubMed] [Google Scholar]
- Wu, S. , Chen, H. , Jin, Z. & Tong, Q. (2010) Effect of two‐stage temperature on pullulan production by Aureobasidium pullulans . World Journal of Microbiology and Biotechnology, 26, 737–741. [Google Scholar]
- Wu, S. , Jin, Z. , Kim, J.M. , Tong, Q. & Chen, H. (2009) Downstream processing of pullulan from fermentation broth. Carbohydrate Polymers, 77, 750–753. [Google Scholar]
- Xia, J. , He, J. , Xu, J. , Liu, X. , Qiu, Z. , Xu, N. et al. (2021) Direct conversion of cheese whey to polymalic acid by mixed culture of Aureobasidium pullulans and permeabilized Kluyveromyces marxianus . Bioresource Technology, 337, 125443. [DOI] [PubMed] [Google Scholar]
- Xia, J. , Liu, S. , Jiao, J. , Qiu, Z. , Liu, X. , He, A. et al. (2022) Evaluation of enhancing effect of soybean oil on polymalic acid production by Aureobasidium pullulans HA‐4D. Bioprocess and Biosystems Engineering, 45, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Xia, J. , Xu, J. , Liu, X. , Xu, J. , Wang, X. & Li, X. (2017) Economic co‐production of poly (malic acid) and pullulan from Jerusalem artichoke tuber by Aureobasidium pullulans HA‐4D. BMC Biotechnology, 17, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Z. , Wu, S. & Pan, S. (2011) Effect of two‐stage controlled pH and temperature on pullulan production by Auerobasidium pullulans . Carbohydrate Polymers, 86, 1814–1816. [Google Scholar]
- Xiao, D. , Blank, L.M. & Tiso, T. (2023) Draft whole‐genome sequence of the Black yeast Aureobasidium pullulans NRRL 62031. Microbiology Resource Announcements, 12, e0045822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, F.‐H. , Zhang, Y. , Xue, S.‐J. , Chi, Z. , Liu, G.‐L. , Hu, Z. et al. (2017) Heavy oils (mainly alkanes) over‐production from inulin by Aureobasidium melanogenum 9‐1 and its transformant 88 carrying an inulinase gene. Renewable Energy, 105, 561–568. [Google Scholar]
- Xu, X. , Liu, Y. , Du, G. , Ledesma‐Amaro, R. & Liu, L. (2020) Microbial chassis development for natural product biosynthesis. Trends in Biotechnology, 38, 779–796. [DOI] [PubMed] [Google Scholar]
- Xue, S.‐J. , Chi, Z. , Zhang, Y. , Li, Y.‐F. , Liu, G.‐L. , Jiang, H. et al. (2018) Fatty acids from oleaginous yeasts and yeast‐like fungi and their potential applications. Critical Reviews in Biotechnology, 38, 1049–1060. [DOI] [PubMed] [Google Scholar]
- Xue, S.‐J. , Liu, G.‐L. , Chi, Z. , Gao, Z.‐C. , Hu, Z. & Chi, Z.‐M. (2020) Genetic evidences for the core biosynthesis pathway, regulation, transport and secretion of liamocins in yeast‐like fungal cells. Biochemical Journal, 477, 887–903. [DOI] [PubMed] [Google Scholar]
- Yang, G. , Liu, G.‐L. , Wang, S.‐J. , Chi, Z.‐M. & Chi, Z. (2020) Pullulan biosynthesis in yeast‐like fungal cells is regulated by the transcriptional activator Msn2 and cAMP‐PKA signaling pathway. International Journal of Biological Macromolecules, 157, 591–603. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Yang, W. , Feng, J. , Chen, J. , Jiang, M. & Zou, X. (2018) Enhanced polymalic acid production from the glyoxylate shunt pathway under exogenous alcohol stress. Journal of Biotechnology, 275, 24–30. [DOI] [PubMed] [Google Scholar]
- Yegin, S. (2017) Xylanase production by Aureobasidium pullulans on globe artichoke stem: bioprocess optimization, enzyme characterization and application in saccharification of lignocellulosic biomass. Preparative Biochemistry and Biotechnology, 47, 441–449. [DOI] [PubMed] [Google Scholar]
- Yegin, S. , Altinel, B. & Tuluk, K. (2018) A novel extremophilic xylanase produced on wheat bran from Aureobasidium pullulans NRRL Y‐2311‐1: effects on dough rheology and bread quality. Food Hydrocolloids, 81, 389–397. [Google Scholar]
- Yegin, S. , Saha, B.C. , Kennedy, G.J. & Leathers, T.D. (2019) Valorization of egg shell as a detoxifying and buffering agent for efficient polymalic acid production by Aureobasidium pullulans NRRL Y‐2311‐1 from barley straw hydrolysate. Bioresource Technology, 278, 130–137. [DOI] [PubMed] [Google Scholar]
- Yin, H. , Gao, C. , Ye, K. , Zhao, T. , Sun, A. & Qiao, C. (2019) Evaluation of surfactant effect on β‐poly (L‐malic acid) production by Aureobasidium pullulans . Biotechnology & Biotechnological Equipment, 33, 954–966. [Google Scholar]
- Yin, X. , Li, J. , Shin, H.‐D. , Du, G. , Liu, L. & Chen, J. (2015) Metabolic engineering in the biotechnological production of organic acids in the tricarboxylic acid cycle of microorganisms: advances and prospects. Biotechnology Advances, 33, 830–841. [DOI] [PubMed] [Google Scholar]
- Yoshimoto, H. , Saltsman, K. , Gasch, A.P. , Li, H.X. , Ogawa, N. , Botstein, D. et al. (2002) Genome‐wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. Journal of Biological Chemistry, 277, 31079–31088. [DOI] [PubMed] [Google Scholar]
- Yue, H. , Ling, C. , Yang, T. , Chen, X. , Chen, Y. , Deng, H. et al. (2014) A seawater‐based open and continuous process for polyhydroxyalkanoates production by recombinant Halomonas campaniensis LS21 grown in mixed substrates. Biotechnology for Biofuels, 7, 1–12.24387051 [Google Scholar]
- Zajc, J. , Černoša, A. , Sun, X. , Fang, C. , Gunde‐Cimerman, N. , Song, Z. et al. (2022) From glaciers to refrigerators: the population genomics and biocontrol potential of the black yeast Aureobasidium subglaciale . Microbiology Spectrum, 10, e0145522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalar, P. , Gostinčar, C. , De Hoog, G. , Uršič, V. , Sudhadham, M. & Gunde‐Cimerman, N. (2008) Redefinition of Aureobasidium pullulans and its varieties. Studies in Mycology, 61, 21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan, Z. & Zou, X. (2013) Efficient production of polymalic acid from raw sweet potato hydrolysate with immobilized cells of Aureobasidium pullulans CCTCC M2012223 in aerobic fibrous bed bioreactor. Journal of Chemical Technology & Biotechnology, 88, 1822–1827. [Google Scholar]
- Zelle, R.M. , De Hulster, E. , Van Winden, W.A. , De Waard, P. , Dijkema, C. , Winkler, A.A. et al. (2008) Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Applied and Environmental Microbiology, 74, 2766–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, W. , Zhang, B. , Chen, G. , Li, M. & Liang, Z. (2019) Efficient production of polymalic acid by a novel isolated Aureobasidium pullulans using metabolic intermediates and inhibitors. Applied Biochemistry and Biotechnology, 187, 612–627. [DOI] [PubMed] [Google Scholar]
- Zeng, W. , Zhang, B. , Jiang, L. , Liu, Y. , Ding, S. , Chen, G. et al. (2020) Poly (malic acid) production from liquefied corn starch by simultaneous saccharification and fermentation with a novel isolated Aureobasidium pullulans GXL‐1 strain and its techno‐economic analysis. Bioresource Technology, 304, 122990. [DOI] [PubMed] [Google Scholar]
- Zeng, W. , Zhang, B. , Li, M. , Ding, S. , Chen, G. & Liang, Z. (2019) Development and benefit evaluation of fermentation strategies for poly (malic acid) production from malt syrup by Aureobasidium melanogenum GXZ‐6. Bioresource Technology, 274, 479–487. [DOI] [PubMed] [Google Scholar]
- Zeng, X. , Huang, J.J. & Hua, B. (2021) Efficient phosphorus removal by a novel halotolerant fungus Aureobasidium sp. MSP8 and the application potential in saline industrial wastewater treatment. Bioresource Technology, 334, 125237. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Cai, J. , Dong, J. , Zhang, D. , Huang, L. , Xu, Z. et al. (2011) High‐level production of poly (β‐L‐malic acid) with a new isolated Aureobasidium pullulans strain. Applied Microbiology and Biotechnology, 92, 295–303. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Chen, D. , Liang, G. , Xu, W. & Tao, Z. (2021) Biosynthetic polymalic acid as a delivery nanoplatform for translational cancer medicine. Trends in Biochemical Sciences, 46, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Gao, Z.‐C. , Chi, Z. , Liu, G.‐L. , Hu, Z. & Chi, Z.‐M. (2021) cAMP‐PKA and HOG1 signaling pathways regulate liamocin production by different ways via the transcriptional activator Msn2 in Aureobasidium melanogenum . Enzyme and Microbial Technology, 143, 109705. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Wang, Z. , Chi, Z. , Liu, G.‐L. & Chi, Z.‐M. (2022) Metabolic engineering of Aureobasidium melanogenum 9–1 for overproduction of liamocins by enhancing supply of acetyl‐CoA and ATP. Microbiological Research, 265, 127172. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Li, J. , Li, F. & Wang, S.a. (2019) Selectable marker recycling in the nonconventional yeast Xanthophyllomyces dendrorhous by transient expression of Cre on a genetically unstable vector. Applied Microbiology and Biotechnology, 103, 963–971. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Feng, J. , Wang, P. , Xia, J. , Li, X. & Zou, X. (2019) CRISPR/Cas9‐mediated efficient genome editing via protoplast‐based transformation in yeast‐like fungus Aureobasidium pullulans . Gene, 709, 8–16. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Lu, Y. , Chi, Z. , Liu, G.‐L. , Jiang, H. , Hu, Z. et al. (2019) Genome editing of different strains of Aureobasidium melanogenum using an efficient Cre/loxp site‐specific recombination system. Fungal Biology, 123, 723–731. [DOI] [PubMed] [Google Scholar]
- Zhao, S.‐F. , Jiang, H. , Chi, Z. , Liu, G.‐L. , Chi, Z.‐M. , Chen, T.‐J. et al. (2019) Genome sequencing of Aureobasidium pullulans P25 and overexpression of a glucose oxidase gene for hyper‐production of Ca2+−gluconic acid. Antonie Van Leeuwenhoek, 112, 669–678. [DOI] [PubMed] [Google Scholar]
- Zhou, R. , Ma, L. , Qin, X. , Zhu, H. , Chen, G. , Liang, Z. et al. (2023) Efficient production of melanin by Aureobasidium melanogenum using a simplified medium and pH‐controlled fermentation strategy with the cell morphology analysis. Applied Biochemistry and Biotechnology, 196, 1122–1141. [DOI] [PubMed] [Google Scholar]
- Zou, X. , Cheng, C. , Feng, J. , Song, X. , Lin, M. & Yang, S.‐T. (2019) Biosynthesis of polymalic acid in fermentation: advances and prospects for industrial application. Critical Reviews in Biotechnology, 39, 408–421. [DOI] [PubMed] [Google Scholar]
- Zou, X. , Tu, G. & Zan, Z. (2014) Cofactor and CO2 donor regulation involved in reductive routes for polymalic acid production by Aureobasidium pullulans CCTCC M2012223. Bioprocess and Biosystems Engineering, 37, 2131–2136. [DOI] [PubMed] [Google Scholar]
- Zou, X. , Yang, J. , Tian, X. , Guo, M. , Li, Z. & Li, Y. (2016) Production of polymalic acid and malic acid from xylose and corncob hydrolysate by a novel Aureobasidium pullulans YJ 6–11 strain. Process Biochemistry, 51, 16–23. [Google Scholar]
- Zou, X. , Zhou, Y. & Yang, S.T. (2013) Production of polymalic acid and malic acid by Aureobasidium pullulans fermentation and acid hydrolysis. Biotechnology and Bioengineering, 110, 2105–2113. [DOI] [PubMed] [Google Scholar]


