Abstract
DA strain and other members of the TO subgroup of Theiler’s murine encephalomyelitis virus (TMEV) produce a chronic demyelinating disease in which the virus persists but has a restricted expression. We previously reported that TO subgroup strains, in addition to synthesizing the picornaviral polyprotein, use an alternative initiation codon just downstream from the polyprotein’s AUG to translate an 18-kDa protein called L* that is out of frame with the polyprotein (H. H. Chen et al., Nat. Med. 1:927–931, 1995; W. P. Kong and R. P. Roos, J. Virol. 65:3395–3399, 1991). L* is critically important for virus persistence and the induction of the demyelinating disease (Chen et al., 1995; G. D. Ghadge et al. J. Virol. 72:8605–8612, 1998). We have proposed that variations in the amount of translation initiation from the L* AUG versus the polyprotein AUG may occur in different cell types and therefore affect the degree of expression of viral capsid proteins. We now demonstrate that ribosomal translation initiation at the polyprotein’s initiation codon affects initiation at the L* AUG, suggesting that ribosomes land at the polyprotein’s initiation codon before scanning downstream and initiating at the L* AUG. We also find that the viral 5′ untranslated region affects utilization of the L* AUG. Surprisingly, mutant DA cDNAs were found to be infectious despite the presence of mutations of the polyprotein initiation codon or placement of a stop codon upstream of the L* AUG in the polyprotein’s reading frame. Sequencing studies showed that these viruses had a second site mutation, converting the reading frame of L* into the polyprotein’s reading frame; the results suggest that translation of the polyprotein during infection of these mutant viruses can be initiated at the L* AUG. These data are important in our understanding of translation initiation of TMEV and other RNAs that contain an internal ribosome entry site.
DA strain and other members of the TO subgroup of Theiler’s murine encephalomyelitis virus (TMEV) cause a biphasic disease in certain strains of mice (e.g., H2s haplotype), with an early acute subclinical neuronal infection (encephalomyelitis) followed by inflammatory white matter disease with chronic demyelination. There are large amounts of infectious virus present within the central nervous system (CNS) during the early disease. Viral titers decrease over the first few weeks but persist for the life of the mouse, with a restricted expression of viral antigens. In contrast to TO strains, GDVII strain and other members of the GDVII subgroup of TMEV are more virulent, inducing an acute fatal encephalomyelitis with no demyelination or evidence of virus persistence. In this study, we investigated translation of an alternatively initiated protein of TO subgroup strains that is critically important for the demyelinating disease and viral persistence.
Picornaviruses have a distinctive strategy for translation. There is a long 5′ untranslated region (5′UTR) that contains a large region that serves as an internal ribosome entry site (IRES) and has extensive secondary structure and a downstream conserved oligopyrimidine tract. Ribosomes enter the picornaviral genome at or very close to an AUG codon at the 3′ end of the IRES and then generally synthesize a single long polyprotein that is subsequently cleaved into viral structural and nonstructural proteins. Translation of the polyprotein is initiated at the AUG near the 3′ end of the IRES in the case of TMEV and other members of the Cardiovirus genus of Picornaviridae and of foot-and-mouth-disease (FMDV), a member of the Aphthovirus genus. In the case of members of the other picornavirus genera, ribosomes enter at the IRES but then scan or are transferred to the AUG used for translation initiation of the polyprotein, which is located downstream from the 3′ end of the IRES.
We previously identified a unique feature of translation of the TO subgroup strains of TMEV. In addition to synthesizing the picornaviral polyprotein, TO subgroup strains translate an 18 kD protein called L* which is out of frame with the polyprotein and is initiated from an AUG at nucleotide (nt) 1079 (in the case of DA strain), 13 nt downstream from the polyprotein’s initiation codon (3, 13). In contrast, GDVII subgroup strains have an ACG rather than an AUG corresponding to nt 1079 and therefore do not synthesize L*. We subsequently reported that L* is critical for the demyelinating activity and persistence of DA since a mutant virus which does not synthesize L* (DAL*-1 virus, which has an ACG rather than an AUG at the L* initiation codon) fails to cause white matter disease and to persist (3, 8). Mouse strains normally susceptible to the demyelinating disease have an antiviral cytotoxic T-lymphocyte (CTL) response following infection with DAL*-1 mutant virus but not wild-type DA. These results suggest that the synthesis of L* in wild-type DA interferes with the antiviral CTL, allowing for virus persistence; persistent virus leads to demyelination (2), perhaps by triggering a pathogenic cytokine and/or inflammatory cell response. DA L* also has antiapoptotic activity in certain cells, e.g., mouse macrophage cell lines (8), but it remains unclear whether the role of L* in mediating the demyelination or in inhibiting the CTL response is related to its antiapoptotic activity.
The above studies highlighted the importance of L* in interfering with virus clearance and fostering the persistent CNS infection. We wondered whether the presence of L* might also be important in regulating whether the CNS infection was a productive one or one with a restricted virus expression. We hypothesized that early after infection, initiation of viral translation in neuronal cells might primarily occur at the polyprotein AUG, with the subsequent synthesis of the polyprotein (which is cleaved into capsid and nonstructural proteins) leading to the production of a large amount of virus. In contrast, late after infection, ribosomes in microglia (which are the major reservoir for virus during the persistent infection [15]) might primarily initiate translation at the L* AUG, leading to a restricted infection. In other words, an increased utilization of the L* AUG for translation initiation would lead to a decreased use of the polyprotein’s AUG, with a resultant decrease in synthesis of the polyprotein (and of the capsid proteins that are cleaved from it). We hypothesized that certain cells might have RNA binding factors that could affect how much synthesis of the polyprotein versus L* occurs. Our recently published study demonstrated that there are in fact differences in L* synthesis among various cell types in culture (8). The present study investigates the utilization of the AUG of L* versus the polyprotein both in vitro in reticulocyte lysates and within cells. These studies have implications on our understanding of translation initiation in both TMEV as well as in other IRES-containing viruses.
MATERIALS AND METHODS
Cells.
BHK-21 (baby hamster kidney) cells were obtained from the American Type Culture Collection and were grown in Dulbecco’s modified Eagle medium (DMEM; GIBCO BRL) supplemented with 1% l-glutamine, 1% gentamicin, and 5% fetal bovine serum (FBS). These cells were used for transfection, plaque assays, and the preparation of stock virus.
Constructs and viruses.
All constructs were generated from a parental infectious cDNA clone of DA strain (pDAFL3) or GDVII strain (pGDVIIFL2) which has been previously described (7, 22).
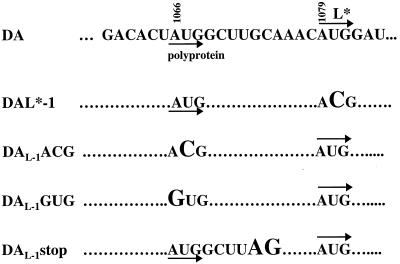
pDAL*-1 has a mutation in the AUG at nt 1079 (which serves as the initiation codon of L*) into ACG with no change in the amino acid sequence of the polyprotein (Fig. 1) (13).
FIG. 1.
Region of the wild-type and mutant DA genome which shows the sites for translation initiation of the polyprotein (nt 1066) and L* (nt 1079). Dots show nucleotides that are identical with those in wild-type DA. Mutations are shown in a larger size font. DAL*-1 has a change of the L* AUG to ACG. DAL-1AUG and DAL-1GUG have a change of the polyprotein AUG to ACG and GUG, respectively. DAL-1stop has a change of GC to AG at nts 1073 and 1074, generating a stop codon in the reading frame of the polyprotein.
We were interested in determining whether utilization of the polyprotein’s AUG for translation initiation affects the efficiency of translation initiation at the L* AUG. For this reason, we made a series of mutations upstream from the L* AUG that were predicted to interfere with synthesis of the polyprotein (Fig. 1). pDAL-1ACG contains an ACG rather than an AUG at the polyprotein initiation codon at nt 1066, preventing synthesis of the polyprotein (which includes L, the most amino located protein of the polyprotein). The construction of this mutant was previously described (13). pDAL-1GUG is similar to pDAL-1ACG but contains a GUG rather than an AUG at the polyprotein’s initiation codon at nt 1066. pDAL-1stop contains UAG rather than UGC at nt 1072, which is just 6 nt downstream from the polyprotein initiation codon and upstream of the L* AUG; the UAG is predicted to act as a stop codon in the reading frame of the polyprotein. The latter two constructs were generated through use of a U.S.E. mutagenesis kit (Pharmacia Biotech) according to directions of the manufacturer.
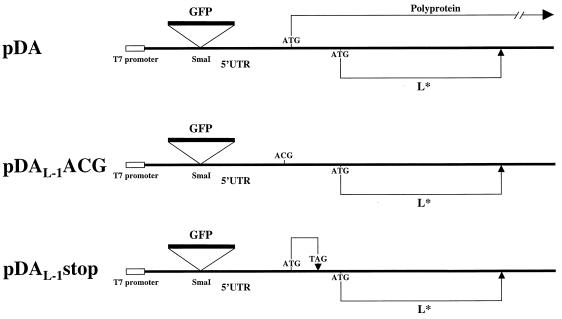
For some in vitro translation experiments (see below), bicistronic constructs of pDAFL3, pDAL-1ACG, and pDAL-1stop were prepared (Fig. 2). The bicistronic constructs contained an insertion of green fluorescent protein (GFP) cDNA (Clontech) into the SmaI restriction enzyme site at nt 14 in pDAFL3. Primers which included a SmaI restriction enzyme site at both ends were used to amplify the GFP cDNA (sense primer, 5′-CAGGTCGACTCTAGAGGATC-3′; antisense primer, 5′ ACTACCCGGGCTATTTGTATAGTTCATCCATGC-3′). The amplified DNA was then digested with SmaI and inserted into the SmaI site of pDAFL3, pDAL-1ACG, and pDAL-1stop.
FIG. 2.
Diagram of bicistronic constructs of wild-type and mutant DA plasmids. GFP cDNA was inserted in the SmaI restriction enzyme site at nt 14 in the 5′UTR. The T7 RNA polymerase promoter is just upstream of the DA genome (22). The location of the AUG codon for translation of the wild-type polyprotein or L* is shown. pDAL-1ACG has an ACG rather than AUG at the first start codon so that no polyprotein is translated. pDAL-1stop has a stop codon (UAG) in the polyprotein’s reading frame upstream from the L* AUG.
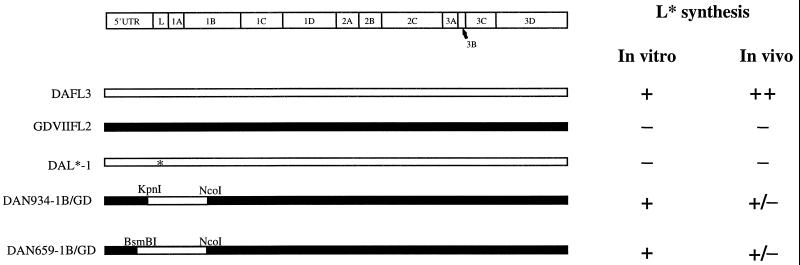
We also made use of chimeric cDNAs (pDAN934-1B/GD and pDAN659-1B/GD) which had been originally prepared for investigations of determinants of TMEV subgroup-specific disease. We specifically targeted chimeric cDNAs that contained the DA L* initiation codon and various parts of the DA IRES in a background of the GDVII genome in order to clarify whether the DA 5′UTR, rather than the GDVII 5′UTR, was important for L* synthesis. The chimeric cDNAs are named by listing the upstream and downstream boundaries of the substituted segment derived from the DA parental clone followed by a slash, and then GD (for GDVII). pDAN934-1B/GD (Fig. 3) was constructed by digestion of pDAFL3 cDNA with KpnI and NcoI followed by ligation of DA nt 934 to 1961 to the two large fragments of KpnI/NcoI-digested pGDVIIFL2. pDAN659-1B/GD (Fig. 3) was constructed by digestion of pDAFL3 cDNA with BsmBI and KpnI followed by ligation of DA nt 659 to 934 to the large fragment of BsmBI/KpnI-digested pDAN934-1B/GD. In the case of both of these chimeric cDNAs, the DA genomic segment contained the L* AUG. As controls, we also constructed similar chimeric cDNAs from pDAL*-1 (which contained a mutation of the L* AUG to ACG), generating pDAL*-1N934-1B/GD and pDAL*-1N659-1B/GD; these constructs do not synthesize L*.
FIG. 3.
Diagram of the parental, mutant, and chimeric TMEV genome and the TMEV coding region. The DA genome is shown as an open bar; the GDVII genome is shown as a black bar. The asterisk in DAL*-1 shows the site of the mutation of the L* AUG to ACG. The restriction enzyme sites bordering the chimeric segments are shown. The presence of L* synthesis in vitro in reticulocyte lysates or in vivo in infected BHK-21 cells is shown by a + or −.
In vitro translation.
Monocistronic or bicistronic cDNAs from wild-type or mutant DA were in vitro transcribed and translated in rabbit reticulocyte lysates as previously described (20). In brief, the DNA was digested with XbaI 3′ to the genome, in vitro transcribed in a T7 RNA polymerase reaction (Stratagene) for 45 min, and then incubated for 5 min with 10 U of RNase-free DNase (Stratagene) followed by a phenol-chloroform extraction and ethanol precipitation; 200 μg of in vitro-derived transcripts was used as a template for in vitro translation. The translation was performed for 3 h at 30°C in a 10-μl reaction (6.5 μl of rabbit reticulocyte lysate [Promega Biotech] with final concentrations of 10 mM potassium isothiocyanate, 0.04 mg of creatinine kinase [Boehringer Mannheim] per ml, and 0.5 mCi of [35S]methionine [Amersham] per ml). Translated lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5 to 20% gradient gels followed by autoradiography on XAR films (Kodak).
In the case of translation of bicistronic constructs, a ratio of L* synthesis to GFP synthesis was calculated as follows. Autoradiograms of electrophoresed lysates were scanned by Silverscanner II (Seiko Epson Corp.), and the density of the GFP and L* band was measured with NIH Image 1.6 in order to calculate the ratio of L* to GFP. The measurement was separately repeated on three separate gels for three separate experiments. Statistical analysis was carried out by means of Student’s paired t test.
Virus.
Wild-type and mutant viruses were generated by transfection of BHK-21 cells with in vitro-derived transcripts prepared from cDNAs as previously described (20). Plaque-purified stocks were prepared. Plaque assays for the quantitation of infectious virus were performed as previously described (21).
Sequencing.
Mutant plasmids and the viruses generated from these plasmids were partially sequenced to verify that the original mutation was present without any additional mutation. Sequencing was performed using an AmpliTaq FS dye terminator cycle sequencing kit (PE Applied Biosystems) and an ABI Prism 377 DNA sequencer.
Radiolabeling of infected cells.
For radiolabeling, 35-mm-diameter plates with BHK-21 cells were either mock infected or infected with a virus at a multiplicity of infection of 20 per cell. After 1 h of incubation at 37°C, culture medium containing 5% FBS and actinomycin D (2 μg/ml) was added, and the culture was incubated at 33°C for 9 h. The monolayers were then washed twice with Hanks balanced salt solution (GIBCO BRL) containing calcium, magnesium, and 2% FBS and incubated for 1 h with methionine-free DMEM containing 2% FBS; 50 μCi of [35S]methionine (ICN Biomedicals) was added, followed by incubation for 12 h at 33°C. Cells were harvested and then resuspended in Laemmli sample buffer. Radiolabeled proteins were separated by SDS-PAGE on 18% gels, and the gels were then dried and exposed to XAR film for autoradiography.
RESULTS
We engineered monocistronic (Fig. 1) and bicistronic (Fig. 2) constructs as well as chimeric cDNAs (Fig. 3) in order to identify TMEV genetic determinants that affect L* synthesis.
In vitro translation with initiation at the polyprotein start codon affects utilization of the L* AUG.
To test how translation of the polyprotein (Fig. 1) affects L* translation initiation, we disrupted the polyprotein’s translation initiation codon by (i) changing the AUG at nt 1066 to ACG or GUG (to generate pDAL-1ACG and pDAL-1GUG, respectively) and (ii) inserting a stop codon downstream of the polyprotein’s AUG but upstream of the L* AUG (to generate pDAL-1stop). We used pDA as a control. Sequencing of the cDNAs demonstrated that the original mutations were present and that there were no additional changes compared to wild type from approximately nt 450 to 1300.
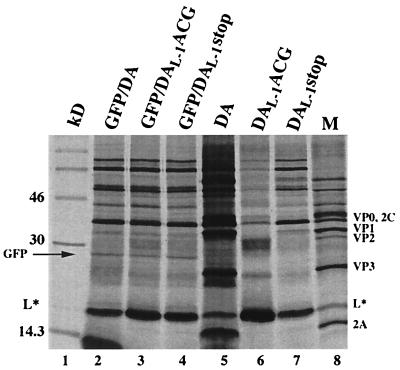
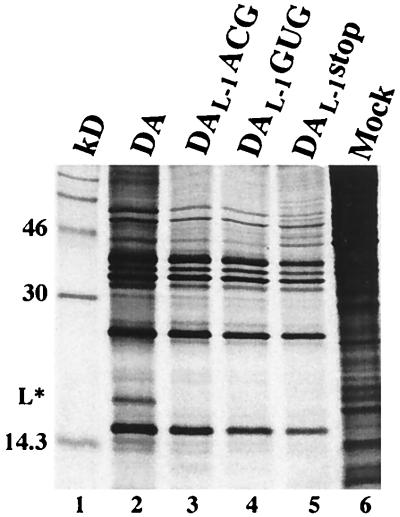
In vitro transcripts derived from pDAL-1ACG (Fig. 4, lane 6), pDAL-1GUG (data not shown), and pDAL-1stop (lane 7) synthesized L* in reticulocyte lysates, as expected, as well as a number of other protein products. Although some of these additional protein products seemed to align with authentic viral proteins (lane M), others clearly did not. These additional protein products were considered to result from aberrant translation initiation rather than initiation at the mutated polyprotein’s start codon (in the case of pDAL-1ACG or pDAL-1GUG) or read-through across the engineered L* stop codon at nt 1072 (in the case of pDAL-1stop) because the profile of synthesized proteins following in vitro translation did not change following the insertion of a stop codon at nt 1552 in the polyprotein’s reading frame (data not shown). Similar aberrant in vitro translation products have been described in the case of poliovirus (1, 5). We then examined differences in the translation of L* programmed by transcripts derived from mutant versus wild-type DA. We found that there was a significant increase in L* synthesis in pDAL-1ACG (lane 6) and pDAL-1GUG (data not shown) compared to the amount of L* synthesized in the case of wild-type pDA (lane 5). There was less of an increase in L* synthesis in the case of the pDAL-1stop mutant (lane 7), although the levels of this protein were still greater than that seen in the case of wild-type DA.
FIG. 4.
In vitro translation of transcripts derived from wild-type or mutant DA cDNAs in bicistronic (lanes 2 to 4) and monocistronic (lanes 5 to 7) constructs. The bicistronic constructs are shown in Fig. 3 and described in Materials and Methods. Samples were diluted in Laemmli sample buffer and electrophoresed. Lane 1, molecular weight markers; lanes 2 and 5, wild-type DA; lanes 3 and 6, DAL-1ACG; lanes 4 and 7, DAL-1stop; lane 8, [35S]methionine-labeled proteins from an extract of DA virus-infected BHK-21 cells harvested 12 h after infection. The electrophoretic mobility of GFP, shown with an arrow, corresponds to the mobility of a single protein species translated by a vector containing GFP alone (data not shown).
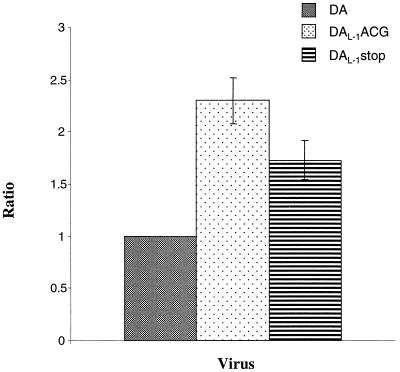
To provide a more quantitative assessment of the increase in L* synthesis following disruption of translation of the polyprotein, we prepared bicistronic constructs in which GFP cDNA was inserted in the 5′UTR of wild-type or mutant DA cDNA upstream of the IRES (Fig. 2). In this way, the amount of L* synthesis could be more precisely compared by determining the amount of L* synthesis relative to GFP synthesis. The translations of the bicistronic constructs did not have as abundant synthesis of viral proteins as in the case of the monocistronic constructs, perhaps as a result of the disturbance in the 5′UTR caused by the GFP cDNA insertion. Most important to the present study, however, the results demonstrated that there was a significant increase in the ratio of L* to GFP synthesis in the mutant that contained a disruption of the polyprotein AUG (pDAL-1ACG [Fig. 4, lane 3]) and a more moderate but definite increase in L*/GFP synthesis in pDAL-1stop (Fig. 4, lane 4) compared to wild-type DA (Fig. 4, lane 2). Analysis of these results (Fig. 5) showed that pDAL-1ACG had a mean L*/GFP ratio over twice that seen with wild-type DA (P ≤ 0.001) and that pDAL-1stop had a mean ratio over 1.7 times (P ≤ 0.005) that seen with pDAFL3. Similar results were obtained when the transcripts were capped (data not shown), although the translation of L* tended to be less efficient, and therefore more difficult to interpret.
FIG. 5.
In vitro translation and utilization of the L* initiation codon by in vitro-derived transcripts from wild-type and mutant DA bicistronic constructs (Fig. 2). Columns show mean values of the ratio of L* synthesis to GFP synthesis for translation of DA wild-type, DAL-1ACG, and DAL-1stop bicistronic cDNAs, using calculations that are detailed in Materials and Methods. The standard deviations are shown as error bars. The ratio of L* to GFP for DA was used as a standard and set as 1.
Mutant DA cDNAs are infectious despite the presence of a mutation of the translation initiation codon of the polyprotein or placement of a stop codon in the reading frame of the polyprotein.
We wondered how L* synthesis would be affected in the virus-infected cell following interference with synthesis of the polyprotein, either by mutating the polyprotein AUG (pDAL-1ACG and pDAL-1GUG) or prematurely terminating the polyprotein’s translation by insertion of a stop codon (pDAL-1stop). We suspected that transcripts of these mutant cDNAs might be infectious since DA strain can grow efficiently in BHK-21 cells in the absence of the L and L* proteins (12) and because there are two additional AUGs in the L coding region at nt 1243 and 1282 (with a good Kozak consensus sequence [14] and in frame with the polyprotein) that could be used for downstream translation initiation of the polyprotein.
We found that transfection of transcripts from all three of these mutant constructs, pDAL-1ACG, pDAL-1GUG, and pDAL-1stop, led to a typical TMEV-like cytopathic effect in BHK-21 cells. The cytopathic effect was slightly prolonged compared to that seen following transfection of transcripts derived from wild-type pDA, suggesting that these mutant viruses had had a reversion of the original mutation or a new second site mutation. Although sequencing demonstrated that all of the mutants maintained the nucleotide change that had been originally engineered, we found an additional insertion of one nucleotide or deletion of two nucleotides in the L coding region in each of the virus isolates. Interestingly, each of the additional mutations changed the reading frame of L* so that it converted to the reading frame of the polyprotein (Fig. 6). For example, plaques of virus derived from separate transfections of all three of these mutant constructs, pDAL-1ACG, pDAL-1GUG, and pDAL-1stop cDNAs, had an insertion of an A at nt 1238, enabling ribosomes that initiate at the L* AUG at nt 1079 to change their reading frame at nt 1239 to the same reading frame as the polyprotein. (Note that in the latter mutant and the ones that follow, the nucleotide inserted is given as the number of the first nucleotide in a repeated sequence; i.e., although the insertion of the A is listed at nt 1238, it may actually be an insertion at nt 1239 or 1240.) Similarly, the following sequence data from recovered viruses suggested that ribosomes initiated translation at the L* AUG and then changed their reading frame to that of the polyprotein: for DAL-1ACG virus, a deletion of nt 1159 and 1160; for DAL-1GUG virus, an insertion of an A at nt 1181 or a U at nt 1132 (in two separate plaque isolates) or a U at 1178 (in two separate plaque isolates) or a U at nt 1140 or a C at 1167. The larger number of mutations found in the case of DAL-1GUG virus is because more plaques were intentionally picked for this particular virus.
FIG. 6.
Sequences of DA wild-type and mutant viruses. Dots show nucleotides that are identical with those in wild-type DA. The mutations that were intentionally engineered in DAL-1ACG, DAL-1GUG and DAL-1stop are shown in italics in a large-size font. Inserted nucleotides are shown in a nonitalicized large-size font, and deleted nucleotides are shown with dashes. Nucleotide numbers of the polyprotein AUG (nt 1066), the L* AUG (nt 1079), and site for insertions (nt 1132, 1140, 1167, 1178, 1181, and 1238) and deletion (nt 1159 and 1160) are shown. Note that the actual nucleotide number in the case of an A insertion in an A-rich area or of a U insertion in a U-rich area or of a C insertion in a C-rich area may not be correct since it is unclear exactly where the insertion or deletion occurred.
To confirm that these mutant viruses synthesized the polyprotein by initiation at the L* AUG and then converting the L* reading frame to the polyprotein’s reading frame (and therefore no longer synthesized L*), we radiolabeled the infected cells with [35S]methionine. We found that representative mutant viruses synthesized viral structural and nonstructural proteins (Fig. 7, lanes 3 to 5) but as expected did not synthesize L*, as did wild-type DA (lane 2).
FIG. 7.
Autoradiogram of BHK-21 cell lysates following infection with wild-type and mutant DA viruses. Lysates were from radiolabeled cells following infection with wild-type DA (lane 2), DAL-1ACG (lane 3), DAL-1GUG (lane 4), and DAL-1stop (lane 5) virus or mock infection (lane 6). The DAL-1ACG mutant virus used for this experiment contained a deletion of nt 1159 and 1160, the DAL-1GUG mutant had an insertion of an A at nt 1181, and the DAL-1stop mutant had an insertion of an A at nt 1238 (Fig. 6). L* protein is synthesized in wild-type DA-infected BHK-21 cells, but not in mock-infected cells or cells infected with DAL-1ACG, DAL-1GUG, and DAL-1stop viruses.
The 5′UTR affects utilization of the L* AUG for translation initiation.
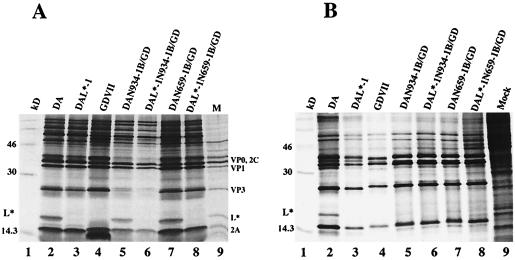
To determine the importance of the DA 5′UTR in selection of the L* AUG for ribosomal initiation both in vitro and in vivo, we investigated chimeric cDNAs that contained various amounts of the DA 5′UTR, L, and capsid coding region (including the L* coding region) substituted into the GDVII 5′UTR (Fig. 3). In vitro-derived transcripts of pDAN659-1B/GD (Fig. 8A, lane 7) and pDAN934-1B/GD (lane 5) synthesized L* in reticulocyte lysates to levels that were similar to those seen with wild-type DA (lane 2). As expected, L* was not synthesized in transcripts derived from pGDVII (lane 4), pDAL*-1 (lane 3) and from chimeric DAL*-1 cDNAs (pDAL*-1N659-1B/GD [lane 8] and pDAL*-1N934-1B/GD [lane 6]). Interestingly, the situation was very different following infection of BHK-21 cells with DAN659-1B/GD (Fig. 8B, lane 7) and DAN934-1B/GD viruses (lane 5), where there was little if any synthesis of DA L* compared to that seen following infection with wild-type DA virus (lane 2) or in vitro translation with the respective mutant cDNAs or wild-type cDNA (Fig. 8A, lanes 2, 5, and 7). As expected, control lanes of GDVII (Fig. 8B, lane 4), DAL*-1 (lane 3), DAL*-1N659-1B/GD (lane 6), and DAL*-1N934-1B/GD (lane 8) viruses failed to synthesize L*. These results suggest that the presence an upstream segment of the DA 5′UTR influences translation initiation of L* and that rabbit reticulocyte lysates, but not BHK-21 cells, have adequate amounts of a factor that is required for L* synthesis in the presence of these chimeric DA 5′UTR segments.
FIG. 8.
In vitro and in vivo synthesis of L* in wild-type TMEV, chimeric TMEV cDNAs, and recombinant TMEV derived from them. (A) In vitro translation of transcripts derived from pDA, pDAL*-1, pGDVII, and various chimeric constructs. Samples were diluted in Laemmli sample buffer and electrophoresed. L* is synthesized in wild-type DA (lane 2), DAN934-1B/GD (lane 5), and DAN659-1B/GD (lane 7) but not DAL*-1 (lane 3), GDVII (lane 4), DAL*-1N934-1B/GD (lane 6), or DAL*-1N659-1B/GD. (B) Autoradiogram of [35S]methionine-labeled proteins from an extract of BHK-21 cells infected with various viruses. The presence of L* is demonstrated in DA (lane 2) but not in DAL*-1 (lane 3), GDVII (lane 4), DAN934-1B/GD (lane 5), DAL*-1N934-1B (lane 6), DAN659-1B/GD (lane 7), and DAL*-1N659-1B/GD (lane 8).
DISCUSSION
DA and other members of the TO subgroup of TMEV cause a chronic demyelinating disease with a persistent CNS infection and a restricted expression of the virus. In contrast, members of the GDVI subgroup neither demyelinate nor persist. TO strains but not GDVII strains have an initiation codon for L*, an alternatively initiated protein out of frame with the viral polyprotein, suggesting the importance of L* in the persistent infection and demyelinating disease. In the present study, we sought to further characterize the alternative translation initiation of L* and to identify determinants of the synthesis of L* versus the polyprotein.
We have previously shown that L* is synthesized in TO subgroups strains of TMEV from an AUG just downstream from the polyprotein’s initiation codon in an alternative reading frame from the polyprotein (3, 13). Our recent published studies demonstrated that L* inhibits the antiviral CTL response, presumably allowing virus to persist and the subsequent development of demyelination (16). We have proposed that L* (rather than the viral polyprotein) is preferentially synthesized in certain neural cells following infection by TO subgroup strains, leading to a restricted expression of the virus (since only small amounts of viral capsid proteins will be produced).
Some other picornaviruses have an alternative initiation codon downstream from the polyprotein. However, in contrast to TMEV, these alternatively synthesized proteins are in frame with the polyprotein; i.e., these picornaviruses synthesize a truncated in addition to a full-length polyprotein in vitro (hepatitis A virus and encephalomyocarditis virus [EMCV]) or in vivo (FMDV) (11, 23, 25). The synthesis of DA L* is unique among picornaviruses since L* is out of frame with the polyprotein. The translation strategy of DA and the synthesis of the alternatively initiated protein may be responsible for the relatively unique phenotype of TO subgroup strains: demyelination and virus persistence with a restricted expression of the virus.
We used a molecular genetics approach to assess how translation initiation of the polyprotein affects initiation at the L* AUG. In vitro translation of monocistronic as well as bicistronic constructs of pDAL-1ACG and pDAL-1GUG, demonstrated that mutation of the polyprotein AUG led to an increase in selection of the L* start codon over the polyprotein AUG. This result suggests that ribosomes normally travel from the IRES to the polyprotein AUG before they pass on to the L* AUG, rather than jumping directly from the IRES to nt 1079 in order to initiate translation of L*. The results are most consistent with leaky scanning, in which ribosomes that first land on the polyprotein’s mutated AUG do not initiate there but continue downstream to initiate at the L* AUG and carry out synthesis of L*.
We next examined this in more detail and questioned whether ribosomes are passed from the polyprotein’s AUG to the L* AUG or whether they scan through the intervening nt 1066 to 1079 sequence, a region which is predicted to have a stem loop configuration (24). To investigate this, we mutated a sequence in this stem loop so that a stop codon was generated (pDAL-1stop). We found that there was an increase in utilization of the L* AUG in translation of the mutant cDNA in reticulocyte lysates compared to that seen with wild-type DA. This result suggests that ribosomes initiate translation at the polyprotein AUG at nt 1066 and continue to scan to the engineered stop codon at nt 1072; following this, they presumably remain associated with the viral genomic RNA, finally reinitiating translation at nt 1079. A similar reinitiation event occurs in the case of the GCN4 gene of Saccharomyces cerevisiae, which follows two short open reading frames (9); the efficiency of reinitiation of ribosomes at the GCN4 AUG depends on A+U-rich sequences that are present near the termination codon of upstream open reading frame 1, possibly because stable base pairing of this region leads to ribosomal dissociation from the RNA. Although our results suggest that at least some ribosomes that initiate at nt 1066 have the capacity to scan the stem loop and then reinitiate at nt 1079, it remains unclear whether and how frequently this occurs in the wild-type DA genome (i.e., a DA genome which does not contain a stop codon between nt 1066 and 1079). In summary, our data from these mutant studies suggest that at least some translation initiation occurs as a result of leaky scanning, in which ribosomes that eventually initiate at the L* AUG first land at the polyprotein AUG and then scan through the stem loop from nt 1066 to 1078.
Our data also suggest that the IRES is important in ribosome binding and subsequent translation initiation both at the L* AUG and at the polyprotein AUG. Transcripts derived from pDAN659-1B/GD and pDAN934-1B/GD synthesized levels of L* in vitro that were similar to those seen with in vitro translation of wild-type pDAFL3, and also in vivo following wild-type DA virus infection of BHK-21 cells; however, levels of L* were extremely low in DAN659-1B/GD and DAN934-1B/GD virus-infected cells. These results suggest that interactions of host cell-specific factors with the DA IRES affect the efficiency of L* translation initiation. The host cell factors may be more available in reticulocyte lysates than in BHK-21 cells. We are presently investigating the efficiency of L* translation initiation in neural cells as well as studying the disease phenotype of viruses that have different efficiencies of L* synthesis.
It is clear that there are many additional determinants important in selection of the polyprotein AUG versus L* AUG. The spacing of the L* AUG from the IRES in the case of the BeAn strain, a TO subgroup strain similar to DA, affects the efficiency of TMEV translation initiation (18). Initiation of translation at the BeAn L* AUG increased (and translation of the polyprotein AUG decreased) following deletion of 4 to 11 nt in the region between the IRES and the polyprotein AUG. Similarly, the distance of the AUG from upstream IRES elements (including the oligopyrimidine tract) in the case of another cardiovirus, EMCV, affects selection of translation initiation at the 11th, rather than the 10th and 12th, AUG from the 5′ end of this virus (10). The distance between the initiation site and upstream IRES elements also appears important in the selection of noncardioviral picornaviruses (19, 25).
Our results with TMEV differ from those recently published by López de Quinto and Martinez-Salas (17) on translation initiation in FMDV. FMDV translation naturally occurs at two in-frame AUGs separated by 84-nt, with usage varying among different viral isolates; the two AUGs encode either a full-length (Lab) or truncated leader (Lb) in the L coding region of the polyprotein. Using antisense oligonucleotides to interfere with the first AUG, these investigators found no change in frequency of translation initiation at the second start codon, suggesting that recognition at the second AUG was not dependent on the accessibility of the first AUG to the translational machinery. They also found that initiation at the second AUG in bicistronic constructs was unchanged following insertion of a stop codon in frame with the first AUG and upstream from the second start codon. These results suggest that FMDV translation initiation at the second AUG occurs as a result of a separate alternative entry site. In contrast, our studies suggest that at least some translation initiation of DA at the second (L*) AUG is a result of leaky scanning from the first (polyprotein) start codon. A similar situation to DA may exist with EMCV, where mutating the surrounding nucleotides of the start codon so that there is a poor sequence context results in increased utilization of the next AUG (4).
The differences in determinants for alternative translation initiation in DA versus FMDV may be related to differences in the organization of the two AUGS in these two viruses. There is a much larger separation between the two AUGs in the case of FMDV (over 80 nt) than is the case with DA (13 nt). It may be that this difference is the reason that the selection of the second FMDV AUG can depend on control elements that are present in the large RNA sequence separating the two start codons (17). The small separation between the two DA start codons may not allow a control element to be included in the short intervening sequence, and for this reason a leaky scanning mechanism is necessary for translation of the second AUG. Another difference between the organization of the two AUGs in DA and FMDV is that L* is out of frame with the DA polyprotein whereas both FMDV AUGs are in frame. This difference, however, is probably not the reason for the use of different mechanisms by the two viruses underlying selection of the second AUG, since initiation at the second FMDV AUG was independent of whether the second AUG was out of frame with the first (17).
The disease caused by FMDV is very different from that induced by DA. It is not unlikely that differences in genome organization and in the alternative initiation are related to the different roles of the alternatively initiated proteins in the two virus-induced diseases.
To further examine the importance of initiation at the polyprotein AUG versus the L* AUG, we tried to generated viruses from mutant DA cDNAs which had disruptions of the initiation codon of the polyprotein or a stop codon in the polyprotein’s reading frame just upstream from the L* AUG. Interestingly, we found that all three of these mutant cDNAs were infectious. Sequencing of the genome of these viruses demonstrated that the original mutations were maintained; however, in all cases we found that there had been either a deletion of two nucleotides or an insertion of one nucleotide in the L coding region downstream from the L* AUG. Insertions (which we found in 10 of 11 plaques that were analyzed) were far more common than deletions, perhaps partly because the deletion involved two nucleotides while the insertion involved only one nucleotide. These second site mutations enabled translation initiation to occur at the L* AUG, with a subsequent conversion of the L* reading frame to the polyprotein’s reading frame. These results emphasize the capability of the L* AUG to function efficiently as the initiation codon; i.e., high titers of stock virus can be produced with initiation of the polyprotein occurring via the L* AUG. These observations also support our previous findings that DA can efficiently grow in BHK-21 cells despite mutation and deletion of L (12) and L* (3).
The insertion and deletion of nucleotides that we found in the L coding regions of these mutant DA viruses are similar to changes described during in vitro growth of FMDV following serial plaque transfer (6). A mutant FMDV with an extremely low plating efficiency accumulated a number of mutations, including nucleotide insertions and deletions, between the two functional AUGs that encode the two different-size leaders. In the case of some of these mutants, the open reading frame of the polyprotein (and capsid proteins) was disrupted downstream of the first AUG; therefore, translation of the polyprotein occurred only after a separate initiation at the second (Lb) AUG. The mutations that changed the FMDV reading frame occurred in the region of a homopolymeric poly(A) extension in the L coding region. Interestingly, the insertions we found occurred primarily in regions that were A rich or U rich (which is A rich in the case of the negative strand); there was a C insertion in only one of 10 separate plaque isolates that had insertions. At least one plaque from each of the three different mutant viruses that disrupted DA polyprotein synthesis had an insertion of an A in an A-rich area at nt 1238. In addition, two separate plaque isolates had insertions of a U at nt 1132 and a U at nt 1178. These results suggest that there are areas of hotspots for mutation, similar to the situation in FMDV. Errors in replication of homopolymeric regions, especially ones rich in purines, have been reported in a number of systems (reviewed by Escarmis et al. [6]) and may relate to slippage of the template during RNA replication.
Our findings and those of Escarmis et al. (6) suggest that insertion (and deletion) mutations of coding regions are not uncommon during picornavirus replication. The mutations are probably not usually identified because they are negatively selected against, either because they are nonviable or, if situated in the L coding region, lead to a change in the initiating codon and a less vigorously growing virus. The presence of an alternative initiation site in DA provides the opportunity for the virus to survive in the face of these frameshift mutations, as well as to synthesize an additional protein important in disease pathogenesis. One wonders whether there may be other alternatively initiated out-of-frame picornaviral proteins whose synthesis depends on factors and a milieu that may be unique to particular cell types.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institutes of Health and the National Multiple Sclerosis Society.
We thank E. Martinez-Salas for sharing unpublished data and E. Domingo for helpful comments.
REFERENCES
- 1.Celma M L, Ehrenfeld E. Translation of poliovirus RNA in vitro: detection of two different initiation sites. J Mol Biol. 1975;98:761–780. doi: 10.1016/s0022-2836(75)80009-x. [DOI] [PubMed] [Google Scholar]
- 2.Chamorro M, Aubert C, Brahic M. Demyelinating lesions due to Theiler’s virus are associated with ongoing central nervous system infection. J Virol. 1986;57:992–997. doi: 10.1128/jvi.57.3.992-997.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H H, Kong W P, Zhang L, Ward P L, Roos R P. A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nat Med. 1995;1:927–931. doi: 10.1038/nm0995-927. [DOI] [PubMed] [Google Scholar]
- 4.Davies M V, Kaufman R J. The sequence context of the initiation codon in the encephalomyocarditis virus leader modulates efficiency of internal translation initiation. J Virol. 1992;66:1924–1932. doi: 10.1128/jvi.66.4.1924-1932.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorner A J, Semler B L, Jackson R J, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escarmis C, Davila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller’s ratchet in an RNA virus. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 7.Fu J L, Stein S, Rosenstein L, Bodwell T, Routbort M, Semler B L, Roos R P. Neurovirulence determinants of genetically engineered Theiler viruses. Proc Natl Acad Sci USA. 1990;87:4125–4129. doi: 10.1073/pnas.87.11.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghadge G D, Ma L, Sato S, Kim J, Roos R P. A protein critical for a Theiler’s virus-induced immune system-mediated demyelinating disease has a cell type-specific antiapoptotic effect and a key role in virus persistence. J Virol. 1998;72:8605–8612. doi: 10.1128/jvi.72.11.8605-8612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant C M, Hinnebusch A G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol Cell Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaminski A, Belsham G J, Jackson R J. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaminski A, Howell M T, Jackson R J. Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J. 1990;9:3753–3759. doi: 10.1002/j.1460-2075.1990.tb07588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong W P, Ghadge G D, Roos R P. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc Natl Acad Sci USA. 1994;91:1796–1800. doi: 10.1073/pnas.91.5.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong W P, Roos R P. Alternative translation initiation site in the DA strain of Theiler’s murine encephalomyelitis virus. J Virol. 1991;65:3395–3399. doi: 10.1128/jvi.65.6.3395-3399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–41. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy M, Aubert C, Brahic M. Theiler’s virus replication in brain macrophages cultured in vitro. J Virol. 1992;66:3188–3193. doi: 10.1128/jvi.66.5.3188-3193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin X, Roos R P, Pease L R, Wettstein P, Rodriguez M. A Theiler’s virus alternatively initiated protein inhibits the generation of H-2K-restricted virus-specific cytotoxicity. J Immunol. 1999;162:17–24. [PubMed] [Google Scholar]
- 17.López de Quinto S, Martinez-Salas E. Involvement of the aphthovirus region located between the two functional AUGs in start codon selection. Virology. 1999;15(255):324–336. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- 18.Pilipenko E V, Gmyl A P, Maslova S V, Belov G A, Sinyakov A N, Huang M, Brown T D, Agol V I. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J Mol Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- 19.Pilipenko E V, Gmyl A P, Maslova S V, Svitkin Y V, Sinyakov A N, Agol V I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 20.Roos R P, Kong W P, Semler B L. Polyprotein processing of Theiler’s murine encephalomyelitis virus. J Virol. 1989;63:5344–5353. doi: 10.1128/jvi.63.12.5344-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roos R P, Richards O C, Ehrenfeld E. Analysis of Theiler’s virus isolates from persistently infected mouse nervous tissue. J Gen Virol. 1983;64:701–706. doi: 10.1099/0022-1317-64-3-701. [DOI] [PubMed] [Google Scholar]
- 22.Roos R P, Stein S, Ohara Y, Fu J L, Semler B L. Infectious cDNA clones of the DA strain of Theiler’s murine encephalomyelitis virus. J Virol. 1989;63:5492–5496. doi: 10.1128/jvi.63.12.5492-5496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sangar D V, Newton S E, Rowlands D J, Clarke B E. All foot and mouth disease virus serotypes initiate protein synthesis at two separate AUGs. Nucleic Acids Res. 1987;15:3305–5315. doi: 10.1093/nar/15.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein S B, Zhang L, Roos R P. Influence of Theiler’s murine encephalomyelitis virus 5′ untranslated region on translation and neurovirulence. J Virol. 1992;66:4508–4517. doi: 10.1128/jvi.66.7.4508-4517.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tesar M, Harmon S A, Summers D F, Ehrenfeld E. Hepatitis A virus polyprotein synthesis initiates from two alternative AUG codons. Virology. 1992;186:609–618. doi: 10.1016/0042-6822(92)90027-m. [DOI] [PubMed] [Google Scholar]