Abstract
Background:
IgA antibodies against few Epstein-Barr virus (EBV) proteins are established serological markers for nasopharyngeal carcinoma (NPC). We recently validated a novel, comprehensive EBV marker panel and showed that IgA, but also IgG antibodies against multiple EBV proteins are highly sensitive and specific for EBV-positive NPC at diagnosis. However, data about these novel biomarkers as prospective markers for NPC are sparse.
Methods:
This study included 30 incident NPC cases and 60 matched controls from the Norwegian Janus Serum Bank. For 21 NPCs, molecular EBV and human papillomavirus (HPV) status were assessed by EBER-ISH and HPV DNA/RNA testing by PCR, respectively. IgA and IgG serum antibodies against 17 EBV antigens were analyzed in prediagnostic sera of cases (median lead time 14 years) and controls using multiplex serology. Sensitivities were calculated using receiver operating characteristic analysis pre-specified to yield 90% specificity in the control group. From 10 cases, serial samples were available.
Results:
Quantitative EBV antibody levels were significantly elevated among all cases (p < 0.05) for three IgA and six IgG antibodies. The highest sensitivities for defining 12 EBER-ISH-positive NPCs were observed for BGLF2 IgA (67%) and BGLF2 IgG (83%). Increased IgA and IgG antibody levels between the first and last draw before diagnosis were observed for EBER-ISH positive, but not for EBER-ISH negative NPCs. Among 21 molecularly analyzed NPCs, 4 EBER-ISH negative NPCs showed concomitant positivity to HPV type-specific DNA and RNA; 3 NPCs were HPV16 and 1 NPC was HPV18 positive.
Conclusion:
Both, EBV IgA and IgG antibody levels are significantly elevated many years before diagnosis of EBV-positive NPCs in Norway, an NPC low-incidence region. This study provides insights into one of the largest available prospective sample collections of NPCs in a non-endemic country.
Keywords: Nasopharyngeal carcinoma, Epstein-Barr virus, Multiplex serology, Prospective biomarker, Nested case/control study
1. Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor, which is distributed heterogeneously throughout the world. NPC is endemic in Southeast Asia, where incidence rates are as high as 20–30 per 100.000 person-years (PY) in men in highly affected regions [1]. Epstein-Barr virus (EBV) is a major risk factor for NPC and is uniformly associated with NPC development in endemic areas, although EBV infection alone is not sufficient to cause NPC [2].
In regions where NPC is not endemic, incidence rates are below 1 per 100,000 PY [3] and NPCs are not uniformly associated with EBV infection. Approximately two out of three NPCs are positive for EBV [4–8]. Whereas NPCs within NPC endemic areas show a characteristic incidence peak at the age of 50–59 years and decline afterwards, NPC incidence in NPC low-incidence areas is virtually absent below an age of 30, starts increasing from 40 years of age onwards and continuously rises with increasing age without any characteristic incidence peak [9].
EBV serum antibodies in NPC patients have been extensively studied since 1966 [10]. Immunofluorescence was used in early case control studies to examine IgA antibodies against the viral capsid antigen (VCA), early antigen (EA) and Epstein-Barr nuclear antigen 1 (EBNA1) [11–13]. Evidence for the presence of EBV IgA antibodies more than 5 years before NPC diagnosis was provided in a prospective study with around 10,000 Taiwanese men [14]. IgA antibodies against EBNA1 and VCAp18 have been shown to be the markers best differentiating between NPC cases and controls [15] and are currently investigated for screening purposes in a cluster randomized controlled trial in Southern China [16].
Serological markers other than IgA antibodies against these few antigens have never been examined until 2018, when an EBV proteome array was utilized for novel antigen discovery [17]. The newly discovered EBV antibody risk stratification signature consisting of 13 IgA and IgG markers against 11 antigens showed a significant improvement for predicting NPCs in comparison to EBNA1/VCAp18 IgA antibodies alone. We successfully validated the expanded EBV antigen panel for the use in multiplex serology, a bead-based high-throughput serological assay [18]. Not only IgA, but also IgG antibodies performed well in defining EBV-positive NPCs. Two IgG antibodies alone, LF2 and BGLF2, have been shown to be highly specific markers for EBV-positive NPCs, both in high and low incidence regions [8,18]. However, there is little knowledge about EBV serology as a predictive biomarker in low incidence regions since NPCs are very rare and therefore, NPC screening is not feasible.
As a unique characteristic of NPCs in low incidence regions, specific human papillomavirus (HPV) types have been shown to be present in those NPCs that are negative for EBV. Five studies from the UK, the US and Finland reported HPV attributable fractions in NPCs of 9–18% [4–8]. The characterization of HPV status, especially in EBV negative NPCs, helps to understand NPC etiology in non-endemic regions, where not all NPCs are positive to EBV infection.
The aim of our study was to characterize a broad panel of EBV antibodies in prediagnostic sera from NPC patients in a non-endemic region, and compare those to the molecular EBV and HPV tumor status. This is challenging, due to the rarity of NPC, and requires a large population-based biobanking infrastructure maintained over several decades. In the present study, we utilized serum of incident NPC cases up to 41 years prior to diagnosis matched to controls from a large prospective Norwegian study, the Janus Serum Bank [19]. EBV and HPV status were determined from tumor tissue and prediagnostic serum samples were examined for IgA and IgG antibodies against a comprehensive EBV antigen panel. The availability of serial samples allowed us to describe the kinetics of EBV antibody levels years prior to NPC diagnosis.
2. Methods
2.1. Study population
The Janus Serum Bank was established in 1973 with the purpose of collecting and storing population blood samples for future use in cancer research. Most individuals who contributed serum samples participated in regional health studies or were Red Cross blood donors. The detailed cohort profile is described elsewhere [19]. The cohort is annually linked to the Cancer Registry of Norway (CRN) database for the identification of new cancer cases. 30 incident NPC cases (International Classification of Diseases for Oncology (ICD-O-3) topography code C11) have been identified and were matched individually to two healthy controls by gender, date of birth and date of blood collection based on the HPV Cancer Cohort Consortium (HPVC3) protocol [20]. All participants from this study were white. The classification for cancer stage included 7 cases with local disease, 20 cases with regional spreading, one case with distant metastasis and two cases without assessment. Classification for TNM stage was not available. Data on smoking habits and alcohol consumption were collected from a questionnaire administered at baseline [19,21]. For 10 cases, blood from several time points was available. An overview of the study population is provided in Table 1.
Table 1.
Characteristics of NPC cases and controls in the Janus Serum Bank.
| Cases n (%) | Controls n (%) | p-value | |
|---|---|---|---|
| Incident cases and controls | 30 | 60 | |
| Analyzed blood samples | 48 | 60 | |
| Gender | 22 (73) | 44 (73) | 1 |
| Male | 8 (27) | 16 (27) | |
| Female | |||
| Smoking statusaNever | 5 (22) | 19 (34) | 0.56 |
| Former | 6 (26) | 13 (23) | |
| Current | 12 (52) | 24 (43) | |
| Alcohol use a | 2 (33) | 3 (13) | 0.24 |
| no current use | 4 (67) | 20 (87) | |
| current use | |||
| Case characteristics only | |||
| Cases with serial blood draws (N) | 10 | ||
| Median lead timeb in years for all cases (range) | 12 (−2, 41) | ||
| Median lead time in years for all incidentc cases (range) | 14 (3, 41) | ||
| Median age at diagnosis, (range) | 59 (29–83) | ||
| Median year of diagnosis (range) | 1998 (1983–2013) | ||
| Cases with valid tumor tissue results (N) | 21 | ||
| EBV positive by EBER-ISH | 12 (57) | ||
| HPV positive by HPV DNA and RNA | 4 (19) | ||
| EBV and HPV negative | 5 (24) | ||
| EBV and HPV positive | 0 (0) |
numbers may not add up to 100% due to missing data. Data on smoking was available for 23 cases and 56 controls. Data on alcohol use was available for 6 cases and 23 controls.
time between blood draw and diagnosis of 48 case sera, – 2 refers to two years post diagnosis.
incident cases include cases with a lead time of > 1 year.
The study was approved by the regional committee for medical and health research ethics, Oslo, Norway (no: 15944), and is based on a broad consent from participants in the Janus cohort.
2.2. Molecular tumor data
Tumor tissue of 22 NPCs in formalin-fixed paraffin-embedded (FFPE) blocks were retrieved from local pathologies by CRN. The blocks were sectioned with utmost care to avoid sample cross-contamination according to standard protocols [22]. Sections were used for performing EBV small RNA 1 (EBER-1) in situ hybridization (EBER-ISH) and HPV DNA/RNA testing by PCR. Each one additional section was cut before and after the section used for nucleic acid detection, to perform Hematoxylin and eosin (H&E) staining. HE sections were checked by a pathologist to confirm the sufficient presence of tumor tissue and the tumor origin.
EBV tumor status was assessed using EBER-ISH, the gold standard for detecting EBV RNA in tumor tissue [23]. Tumor sections were sent to an ISO-accredited medical laboratory (Severn Pathology) in Bristol (UK) for EBER-ISH staining and reading (Leica ISH EBER probe, automated BOND system) [8]. EBER-ISH status was obtained for 21 NPCs, one section did not have sufficient tumor tissue to assess EBV status.
HPV DNA and RNA testing was performed on 22 tumor samples as previously described [8]. Briefly, HPV DNA detection was performed using Multiplex Papillomavirus Genotyping [24,25]. DNA was analyzed for 51 HPV types including all known high-risk HPV types [24,25]. RNA detection of the HPV DNA positive samples was performed using RT-PCR and hybridization to detect HPV type-specific E6*I RNA [22]. Validity of the samples was assessed by cellular control genes. Valid tumor samples with combined HPV DNA and RNA positivity were considered as HPV positive. Three of 22 cases had invalid HPV DNA results, but were negative for HPV 16, 18, 31, 39, and 59 RNA, and thus categorized as HPV negative.
2.3. Serology
HPV16 E6 antibody level for all NPC cases and controls were previously generated in the framework of the HPVC3 consortium and were available for analysis [20].
Serological characterization of NPC cases and controls for EBV antibodies was performed using multiplex serology [26]. IgA and IgG serum antibodies against 17 antigens (Zebra, EBNA1trunc, EA-D, VCAp18, EBNA1pep, BXLF1, LF2, BZLF1, BORF1, BFRF1, BGLF2, BRLF1, BPLF1, VCAp40, BHRF1, BBRF1, and BaRF1) were analyzed as described before [18]. The novel EBV antigens VCAp40 (YP_401705.1–136418–137455), BHRF1 (YP_001129442.1–42204–42779), BBRF1 (YP_001129476.1–102746–104587) and BaRF1 (YP_0 01129453.1–66746–67654) derived from an array-based approach for identifying EBV antibodies specific for classical Hodgkin lymphoma and were expressed as full-length proteins for the use in multiplex serology [27]. As a specificity control, three antigens representing the major capsid protein VP1 of the three human polyomaviruses (HPyV) JC, BK and HPyV6 were included in the assay. These control antigens are not known to be associated with NPC development. All antigens were recombinantly expressed as glutathione-S-transferase (GST) fusion proteins in E. coli, as described previously [26].
To detect antibodies against these antigens, sera were preincubated at 1:50 dilution for IgA testing (final dilution 1:100) and at 1:500 dilution for IgG testing (final dilution 1:1000). The serum preincubation buffer was based on PBS with 2 g/L casein and additionally containing 2 g/L of E. coli protein overexpressing GST alone, 5 g/L polyvinyl alcohol and 8 g/L polyvinyl pyrrolidone [28].
Coefficients of variation for quality control samples tested in duplicate ranged from 1% to 7% (median 4%) for IgA testing and from 2% to 16% (median 6%) for IgG testing.
Goat anti-Human IgA-Biotin (1:1000, #109–065-011, Jackson ImmunoResearch) and goat anti-Human IgG-Biotin (1:1000, #109–065-098, Jackson ImmunoResearch) were used to detect the bound IgA and IgG antibodies, followed by staining with streptavidin-R-phycoerythrin (1:750, MOSS Inc.).
2.4. Statistics
All statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc., La Jolla, California), or SAS enterprise guide 7.1 (SAS Institute, Cary, North Carolina). A P-value of 0.05 was considered statistically significant.
The technical minimum cut-off above assay background for multiplex serology is a median fluorescence intensity value (MFI) of 30 and corresponds to the lower limit of quantitation. Thus, quantitative MFI values refer to those values with MFI values above 30. In our analysis, we focused on MFI values within the quantifiable range.
Serum samples with lead times > 1 year were referred to as incident NPCs and at least one sample fulfilling this definition was available for every case. Serum samples with a time of ≤ 1 year between blood draw and diagnosis were considered potential prevalent cases. In all analyses, except the analysis of serial samples, only the serum sample at the time closest to diagnosis of incident cases was considered for analysis.
Analyses stratified by lead times and cancer stage were not performed due to small case numbers and thus limited statistical power.
Differences in MFI values of all 30 cases and 60 matched controls were calculated by Mann-Whitney test (Table 2). Receiver operating characteristic (ROC) analysis on continuous MFI data was used to determine antibody-specific cut-offs to yield 90% seronegatives in the control group, i.e. 90% specificity. The resulting sensitivities were calculated for EBER-ISH positive cases (N = 12). Both, cut-offs and sensitivities are shown in Table 3.
Table 2.
Median fluorescence intensity (MFI) values of all incident cases and controls.
| Antigen |
IgA antibodies |
IgG antibodies |
||||
|---|---|---|---|---|---|---|
| Median MFI (cases) | Median MFI (controls) | p-valuea | Median MFI (cases) | Median MFI (controls) | p-valuea | |
| Zebra | 77 | 61 | 0.38 | 1168 | 711 | 0.08 |
| EBNA1trunc | 608 | 513 | 0.18 | 7475 | 7104 | 0.34 |
| EAD | 105 | 68 | 0.13 | 678 | 271 | 0.13 |
| VCAp18 | 1797 | 1957 | 0.70 | 6608 | 5888 | 0.21 |
| EBNA1pep | 233 | 169 | 0.04 | 4332 | 3152 | 0.07 |
| BXLF1 | 168 | 89 | < 0.01 | 454 | 414 | < 0.01 |
| LF2 | 26 | 21 | 0.16 | 21 | 10 | < 0.01b |
| BZLF1 | 5 | 3 | 0.17 | 22 | 2 | 0.01b |
| BORF1 | 40 | 24 | 0.02b | 19 | 14 | 0.04b |
| BFRF1 | 50 | 41 | 0.08 | 52 | 25 | 0.01b |
| BGLF2 | 114 | 38 | 0.001 | 939 | 315 | < 0.001 |
| BRLF1 | 136 | 116 | 0.49 | 103 | 53 | 0.03 |
| BPLF1 | 215 | 142 | 0.15 | 578 | 371 | 0.03 |
| VCAp40 | 465 | 398 | 0.16 | 868 | 568 | 0.03 |
| BHRF1 | 513 | 390 | 1.00 | 2501 | 2934 | 0.83 |
| BBRF1 | 47 | 47 | 0.60 | 23 | 19 | 0.02b |
| BaRF1 | 282 | 83 | 0.06 | 750 | 208 | 0.04 |
| JC VP1 | 61 | 58 | 0.30 | 376 | 476 | 0.87 |
| BK VP1 | 106 | 104 | 0.88 | 749 | 656 | 0.41 |
| HPyV6 VP1 | 197 | 197 | 0.67 | 2447 | 2352 | 0.74 |
Bold font = p-value significant (Mann-Whitney test).
despite statistical significance, the differences between cases and controls were not considered to be biologically relevant, as the median MFI values are not in the quantifiable range, e.g. above the lower limit of quantification of 30 MFI.
Table 3.
Antigen-specific cutoffs for all IgA and IgG antibodies.
| IgA | IgG | |||
|---|---|---|---|---|
|
|
|
|
||
| Cut-off | Sensitivitya (95% CI) | Cut-off | Sensitivitya (95% CI) | |
| Zebra | 590 | 16.7 (3.0–44.8%) | 3079 | 25.0 (8.9–53.2%) |
| EBNA1trunc | 1226 | 41.7 (19.3–68.1%) | 9007 | 16.7 (3.0–44.8%) |
| EA-D | 696 | 25.0 (8.9–53.2%) | 2257 | 16.7 (3.0–44.8%) |
| VCAp18 | 10561 | 16.7 (3.0–44.8%) | 9462 | 25.0 (8.9–53.2%) |
| EBNA1pep | 454 | 33.3 (13.8–60.9%) | 6793 | 25.0 (8.9–53.2%) |
| BXLF1 | 366 | 41.7 (19.3–68.1%) | 966 | 25.0 (8.9–53.2%) |
| LF2 | 59 | 33.3 (13.8–60.9%) | 81 | 50.0 (25.4–74.6%) |
| BZLF1 | 30b | 33.3 (13.8–60.9%) | 223 | 25.0 (8.9–53.2%) |
| BORF1 | 120 | 25.0 (8.9–53.2%) | 89 | 50.0 (25.4–74.6%) |
| BFRF1 | 90 | 16.7 (3.0–44.8%) | 173 | 33.3 (13.8–60.9%) |
| BGLF2 | 290 | 66.7 (39.1–86.2%) | 859 | 83.3 (55.2–97.0%) |
| BRLF1 | 660 | 16.7 (3.0–44.8%) | 257 | 25.0 (8.9–53.2%) |
| BPLF1 | 565 | 25.0 (8.9–53.2%) | 1246 | 25.0 (8.9–53.2%) |
| VCAp40 | 1856 | 41.7 (19.3–68.1%) | 1511 | 16.7 (3.0–44.8%) |
| BHRF1 | 2353 | 25.2 (8.9–53.2%) | 5772 | 16.7 (3.0–44.8%) |
| BBRF1 | 113 | 33.3 (13.8–60.9%) | 58 | 41.7 (19.3–68.1%) |
| BaRF1 | 1005 | 25.0 (8.9–53.2%) | 2566 | 16.7 (3.0–44.8%) |
CI: confidence interval.
sensitivity in Epstein-Barr virus small RNA 1 in situ hybridization (EBER-ISH) positive NPC cases at 90% specificity in the control group.
technical minimum cut-off (lower limit of quantitation).
Following the HPVC3 protocol [20] unadjusted (crude) odds ratios and 95% confidence intervals (CI) for the association of EBV antibodies with EBV positive NPC were calculated using unconditional logistic regression. These analyses were restricted to EBER-ISH positive cases and their matched controls. For those antibodies where none of the controls showed MFI values above the cut-off, one control was assigned as seropositive to allow the calculation. The imputation of a positive control was independent of the MFI value. Of note, conditional logistic regression was not possible due to power issues.
Differences in the median number of positive IgA and IgG antibodies in EBER-ISH positive cases and controls were assessed using Mann-Whitney test. The term “positive antibodies” refers to MFI values above the antibody-specific cut-off.
2.5. Serial sample analysis
For serial sample analysis, all available serum samples from each case were considered. This included samples taken prior to diagnosis, as well as for some cases at most one sample taken after diagnosis. As a descriptive measure for meaningful antibody level changes, we descriptively analyzed a 2-fold increase in quantitative MFI values in-between the first draw and the draw closest to diagnosis.
3. Results
3.1. Study and participant characteristics
Thirty incident NPC cases and 60 matched controls were included in this study (Table 1). Ten cases had serial blood draws (between two and six), resulting in a total number of 48 blood samples for the cases. The median time between blood draw and NPC diagnosis was 14 years (ranging from 41 years to 3 years prior to diagnosis) for incident cases, and 12 years (ranging from 41 prior to diagnosis to 2 years post diagnosis) for all cases including prevalent cases, which were included in the serial sample analysis. Cancer diagnosis years ranged from 1983 until 2013. The cases and matched controls did not significantly differ in alcohol consumption (p = 0.24) and smoking (p = 0.56). The median age at diagnosis was 59 years, ranging from 29 to 83 years. A detailed overview of the study participants is shown in Table 1.
3.2. EBV serology of all cases and controls
Prediagnostic serum samples from incident cases and controls were investigated for the presence of IgA and IgG antibodies against 17 EBV antigens (Zebra, EBNA1trunc, EA-D, VCAp18, EBNA1pep, BXLF1, LF2, BZLF1, BORF1, BFRF1, BGLF2, BRLF1, BPLF1, VCAp40, BHRF1, BBRF1, and BARF1) (Table 2).
When investigating antibodies in the serum closest to diagnosis of all incident cases in comparison to the control sera, MFI signals for four IgA antibodies and 11 IgG antibodies were significantly higher in the case than the control group (Table 2). Cases and/or controls showed MFI values below the lower limit of quantification for one IgA and five IgG antibodies, thus the quantitative comparison of MFI values between cases and controls among these antibodies was limited. However, three IgA and six IgG antibodies showed higher MFI values within the quantitative range for cases than for controls.
None of the three control antigens (human Polyomaviruses JC, BK, HPyV6) showed differences in MFI values between the case and control group (Table 2).
3.3. Molecular EBV and HPV tumor data
EBER-ISH and HPV DNA and RNA testing for 21 NPC cases revealed twelve (57%) EBER-ISH positive and nine (43%) EBER-ISH negative cases (Table 1). Four of the EBER-ISH negative cases showed concomitant positivity to HPV type-specific DNA and RNA: three cases for HPV16 and one for HPV18. There was no overlap in positivity to EBV and HPV. The overall number of EBV positive cases was 12 (57%), the number of HPV positive cases 4 (19%) and the number of both EBV and HPV negative cases was 5 (24%). The median time between blood draw and diagnosis for HPV positive cases was 28 years (range: 12–41 years). EBER-ISH positive cases showed a median lead time of 12.5 years (range: 6–27 years) between blood draw and diagnosis.
3.4. EBV serology stratified by EBER-status
Of all 30 NPC cases, 12 were EBER-ISH positive, 9 were EBER-ISH negative and 9 had an unknown EBV status. MFI values of the serum closest to diagnosis from all incident cases stratified by EBER-ISH status are shown in Fig. 1 for two selected antigens (BGLF2 and BXLF1) and in S1 Fig for all antigens. Based on the MFI values of all matched controls, an antibody-specific cut-off was applied with 90% specificity, allowing 10% (n = 6) positive controls. The cut-off values and the resulting sensitivities for EBER-ISH positive cases of the individual antibodies are shown in Table 3. Sensitivities for IgA antibodies ranged from 17% (Zebra, VCAp18, BFRF1, and BRLF1) to 67% for BGLF2. For IgG antibodies, the highest sensitivity was 83% (BGLF2) and the lowest 17% (EBNA1trunc, EA-D, VCAp40, BHRF1, and BARF1).
Fig. 1.
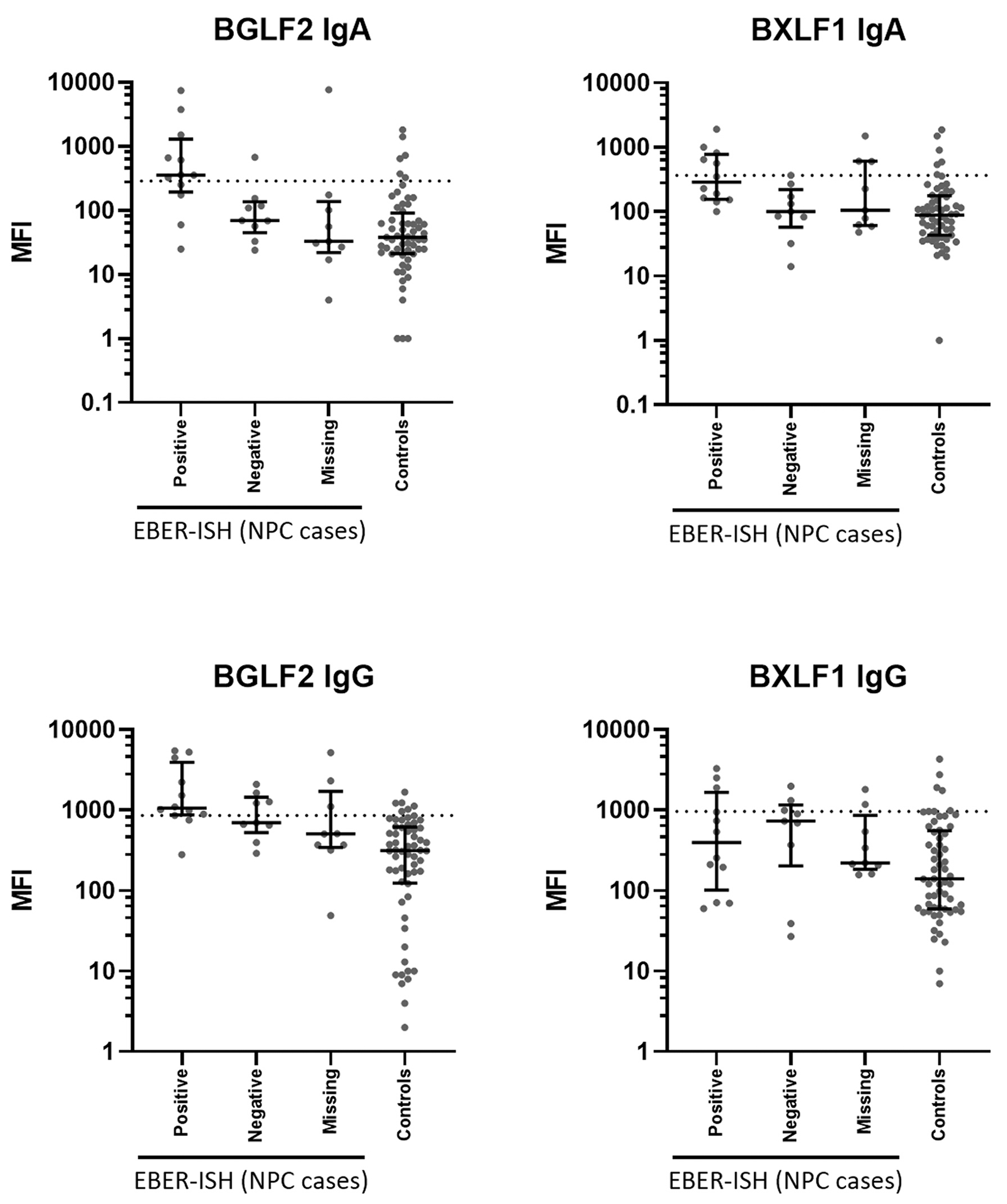
Median fluorescence intensity (MFI) values for BGLF2 and BXLF1 IgA and IgG antibodies. MFI values for all incident cases were split into Epstein-Barr virus small RNA 1 in situ hybridization (EBER-ISH) positive cases, EBER-ISH negative cases, cases with unknown EBER-ISH status and controls. Cut-offs are based on receiver operating characteristic (ROC) analysis to yield 90% seronegatives in the control group, i.e. 90% specificity.
Odds ratios (OR) of EBER-ISH positive cases versus matched controls showed a strong association of BGLF2 IgG antibodies and NPC (OR 115, 95% confidence interval (CI) 9.3 - ∞). A higher risk of NPC was likewise observed for IgG antibodies against LF2 (OR 23, 95% CI 2.3–229), BBRF1 (OR 16, 95% CI 1.6–165) and BORF1 (OR 7.0, 95% CI 1.3–37), as well as for IgA antibodies against BXLF1 (OR 16, 95% CI 1.6–165), BGLF2 (OR 14 95% CI 2.6–77), BBRF1 (OR 12, 95% CI 1.1–119) and EBNA1trunc (OR 7.9, 95% CI 1.2–50). Odds ratios for IgA and IgG antibodies against all antigens are shown in Fig. 2.
Fig. 2.
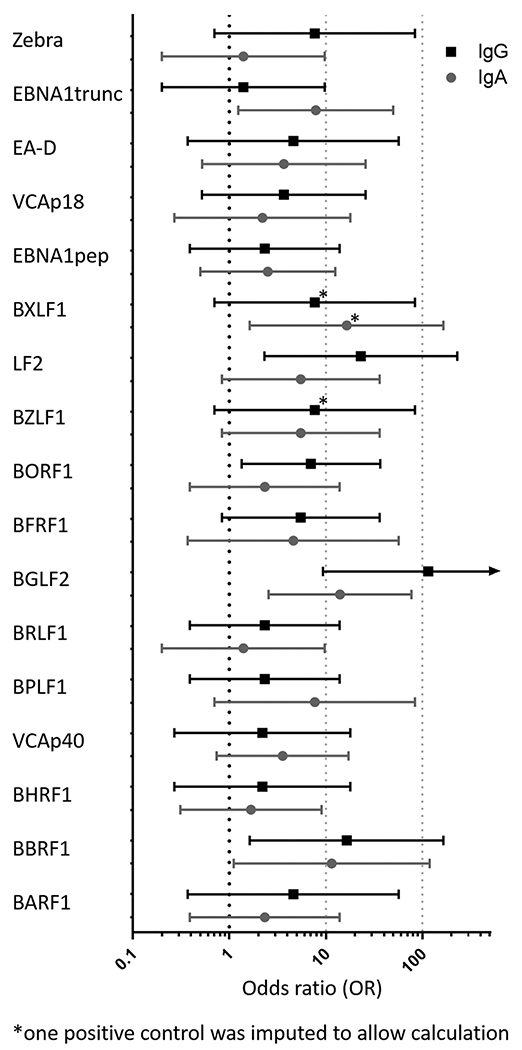
Odds ratios of Epstein-Barr virus small RNA 1 in situ hybridization (EBER-ISH) positive NPCs associated with EBV IgA and IgG antibodies. Crude odds ratios and 95% confidence intervals are shown.
To evaluate the role of EBV IgA and IgG antibodies in NPC development independently from particular antigens, we compared the overall number of positive IgA and IgG antibodies in all EBER-ISH positive cases, and the control group. The median number of positive IgA antibodies was 5 (range 1–12) in the incident cases and 1 (range 0–12) in the control group (p = 0.0002). For IgG antibodies, the median number of positive antibodies was 4 (range 0–15) in the incident cases and 1 (range 0–9) in the controls (p = 0.0003).
3.5. HPV16 E6 serology
None of the NPC cases had HPV16 E6 antibody level above 1000 MFI and was thus classified being HPV16 E6 seropositive.
3.6. Serial sample analysis
For 10 cases, several blood draws were available. The number of blood draws ranged from two to six, with one case having six blood draws, four cases having three draws and five cases having two draws. Among these 10 cases, three were EBER-ISH positive, three were EBER-ISH negative and four had an unknown EBER-ISH status. The course of antibody progression over time is shown in Fig. 3 for one EBER-ISH positive case with six blood draws before diagnosis. As a descriptive measure of antibody kinetics, we analyzed the data for a doubling of MFI values between the first and the last available sample. 13 IgA and 11 IgG antibodies had at least twice as high MFI values in the last sample (18 years prior to diagnosis) than in the first (31 years prior to diagnosis) sample for the EBER-ISH positive case shown in Fig. 3. The second EBER-ISH positive case had 1 IgA and 4 IgG antibodies with doubled MFI values between 23 and 13 years prior to diagnosis; the third EBER-ISH positive case had 11 IgA and 8 IgG antibodies with doubled MFI values between 8 years prior to and 2 years post diagnosis.
Fig. 3.
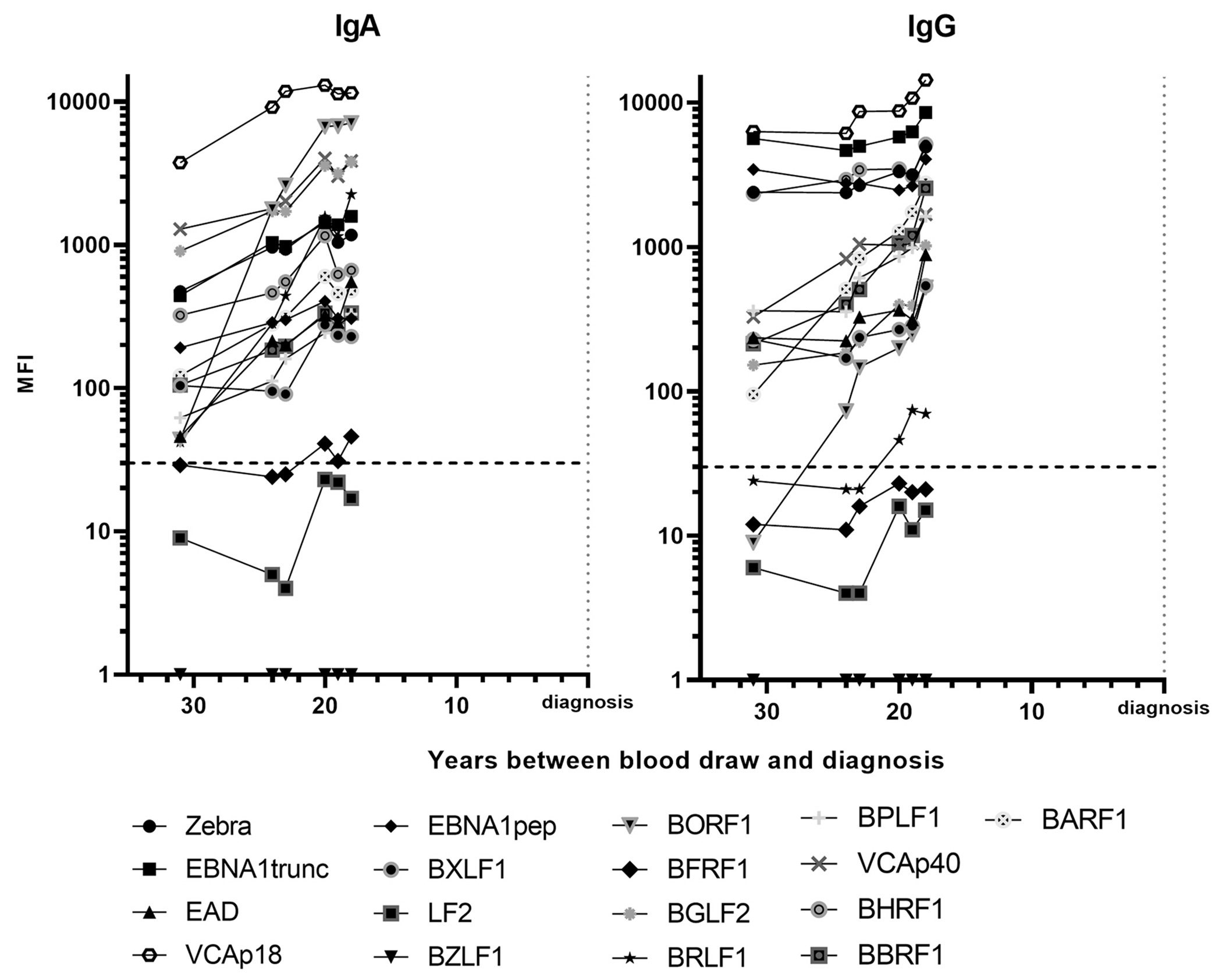
Serial serum samples analysis showing IgA and IgG antibody progression for one patient with Epstein-Barr virus small RNA 1 in situ hybridization (EBER-ISH) positive tumor sample. The progression shows prediagnostic IgA and IgG antibody levels in a single patient over time. Blood was drawn 31, 24, 23, 20, 19 and 18 years prior to diagnosis. The dashed line indicates the lower limit of quantification of 30 MFI.
A pattern of rising EBV IgA and IgG antibody level was not observed for three EBER-ISH negative cases; only one of these showed 2 IgG antibodies with doubled MFI values within 12 years. The antibody progression of all EBER-ISH positive and EBER-ISH negative cases and remaining cases with unknown EBER-ISH status is shown in S2 Fig.
4. Discussion
In the present study, we characterized EBV IgA and IgG antibodies in prediagnostic sera of 30 individuals from Norway who developed NPC later on, and 60 matched controls.
Comparing EBV IgA and IgG antibodies in all prediagnostic case and control sera, we found that cases had significantly higher levels of EBV antibodies than controls, which was not observed for any of the Polyomavirus control antigens. In total, 3 IgA and 6 IgG antibodies were detected at significantly higher quantitative levels in cases compared to controls (Table 2), emphasizing the role of IgG antibodies for NPC early detection, as suggested previously [17,18].
Our results are based on NPCs from a large Norwegian population based cohort study, the JANUS serum bank. This is likely one of the largest sample collections of prediagnostic NPC sera in a region with low NPC incidence. Particular strengths of this study are the high tumor retrieval rate, which is necessary to determine the molecular viral tumor status, and the number of serial samples, which is probably unique to this study design [19] and allowed us to monitor antibody levels over time. As a further strength, we focused our investigation on the analysis of IgA and IgG antibodies against several EBV proteins. Prediagnostic EBV antibodies have only been studied for very few IgA antibodies so far, where elevated IgA antibodies against the viral capsid antigen and the EBV DNase were shown to be strong predictors for NPC development 5 years prior to diagnosis [14]. To the best of our knowledge, there is no published literature about other antigens and their IgA and IgG antibodies in a region with low NPC incidence.
Investigating EBV positivity in FFPE tumor blocks, 12 of 21 available tumors (57%) were solely positive to EBV, while 9 tumors (43%) were negative to the gold standard EBER-ISH. Odds ratios showed a positive association for the presence of all EBV antibodies with EBV positive NPC. The highest odds ratios were observed for BGLF2 IgG (OR 115, 95% CI 9.3 - ∞) and LF2 IgG (OR 23, 95% CI 2.3–229). These results match with the results from a British study of 98 newly diagnosed NPC cases, where LF2 and BGLF2 IgG had likewise the highest odds ratios among IgG antibodies at diagnosis [8]. We have also characterized LF2 and BGLF2 IgG antibodies in a case control study in Taiwan, an NPC endemic region, where a combination of both markers has been shown to define NPCs with 98.4% accuracy [18]. Within this study, we show for the first time that LF2 and BGLF2 IgG antibodies are likewise among the best EBV antibody markers in defining prospective NPC samples in a low-incidence region.
In EBER-ISH positive cases, LF2 and BGLF2 IgG antibodies reached sensitivities of 50% and 83%, respectively at a specificity of 90% in the control group. The low sensitivity of LF2 might be a consequence of the low antibody level of LF2, especially many years prior to diagnosis. In parallel, high titer LF2 IgG antibody level are virtual absent among the control group, as similarly observed in our previous studies [8,18] and shows that already low-level LF2 IgG antibodies are specific for NPC. We can only speculate that LF2 antibodies may appear later than other markers, which show already high MFI values in healthy EBV infected individuals. While BGLF2 is showing a high sensitivity for the cases and a high specificity in the control group, its specificity to EBER-ISH negative tumors is not as high (Fig. 1), as also observed for other antibodies (S1 Fig). The background of this phenomena is currently not sufficiently understood, however, it requires larger numbers of EBV negative NPCs to investigate the meaning. We observed previously that EBER-ISH negative NPC patients have similar EBV antibody level as the controls [8]. To evaluate the hypothesis that these two IgG markers might be good prognostic NPC markers rather than only being able to define EBV positive NPCs, the potential of LF2 and BGLF2 IgG antibodies needs to be investigated in large prospective studies.
Of note, we did not find significantly elevated IgA antibodies against VCAp18 within this study. EBNA1 and VCAp18 IgA antibodies are the preferred combination of EBV markers for NPC screening in Southern China [15]. However, to assess the use of IgA antibodies against EBNA1 and VCAp18 as markers for early NPC detection in Norway, sera drawn closer to diagnosis would need to be investigated.
Using sera at several time point prior or post diagnosis, we studied the antibody progression of some EBER-ISH positive and EBER-ISH negative cases. The antibody progression in Fig. 3 suggests that both IgA antibodies and IgG antibodies rise prior to NPC diagnosis. Presumably, the majority of all EBV antibodies, IgA and IgG, rise to high levels at some point in time prior to diagnosis, as indicated by one prevalent EBER-ISH positive case that showed positivity to 16 IgA and 16 IgG antibodies two years post diagnosis. We were not able to define the time point of seroconversion prior to NPC diagnosis in this study, as this would require a higher number of sera with serial blood draws closer to diagnosis. Unfortunately, no case with multiple serial samples drawn closer to diagnosis was available for analysis. Taken together, we observed a very early rise of both, EBV IgA and IgG antibodies many years prior to NPC diagnosis. However, as the disease etiology is not fully understood yet, the clinical relevance of this early increase in EBV antibody levels cannot be evaluated.
Next to studying EBV antibodies in EBV positive NPC cases, the FFPE tumor tissue of all available tumors was investigated for the presence of HPV RNA. Four tumors showed solely positivity to HPV (19%). Although the tumor tissue showed that EBV and HPV positivity are mutually exclusive, the analyzed sera derived from different time points prior to diagnosis, with EBV-positive cases having a median lead time of 12.5 years and HPV-positive cases having a median lead time of 28 years. Likewise, the long lead times of HPV-positive cases are likely the reason for the lack of detectable HPV16 E6 antibodies. The mutually exclusive presence of EBV and HPV and the fraction of HPV positive cases is in concordance with the growing number of publications describing the presence of HPV in NPCs. Studies from Europe and the US presented fractions between 9% and 18% of HPV positive NPCs [4–8], which were previously discussed to be likely extensions from the oropharynx [6]. Within the four HPV positive NPC cases, three were positive for HPV16 and one case positive for HPV18. Although these numbers are very small, they are in line with the HPV type distribution in HPV-positive NPCs from the British Head & Neck 5000 study, where among 18 HPV-positive NPCs, 13 were HPV16 positive (72%), four were HPV18 positive (22%) and one was HPV39 positive (6%) [8]. This distribution might be unique for NPCs, as HPV positive oropharyngeal carcinoma cases (OPCs) are mostly HPV16 positive, with few HPV33 positive cases but only very rarely other HPV types including HPV18 [29].
The main limitation of this study was the low sample size of incident NPC cases, a result of the low incidence of NPCs in Europe. The small number of cases limited the ability to more thoroughly investigate lead time periods between blood draw and diagnosis, or stratify the cases by lead time. Due to the study design, very few cases were available with blood draws close to the diagnosis date. Further, we lacked detailed information of participant smoking status. Smoking is a known risk factors for NPC [30] and smoking and EBV infection have been described to be independent factors for NPC development [31].
In summary, we have characterized EBV IgA and IgG antibodies in prospective sera from NPC patients from an NPC low-incidence region and have shown that both, EBV IgA and IgG antibodies start to rise many years before NPC diagnosis. The two IgG markers, LF2 and BGLF2, which have shown to be good markers to define NPCs in the past, are likely to be good prospective markers for NPC. These markers should be further investigated in incident NPCs from both, NPC low-incidence regions and especially in large NPC studies from NPC endemic regions.
Supplementary Material
Acknowledgments
We would like to acknowledge Ditte Staldgaard for assistance regarding retrieval of formalin fixed paraffin embedded tissue blocks and Marianne Lauritzen for retrieval of blood samples.
This work was supported by HPVC3 grant funded by the US National Cancer Institute (Grant: 5U01CA195603), with additional support from the intramural program of the Division of Cancer Epidemiology and Genetics, US NCI.
Abbreviations:
- NPC
nasopharyngeal carcinoma
- EBV
Epstein-Barr virus
- EBER-ISH
EBV small RNA 1 (EBER-1) in situ hybridization
- PY
person-years
- VCA
viral capsid antigen
- EA
early antigen
- EBNA1
Epstein–Barr nuclear antigen 1
- HPV
human papillomavirus
- FFPE
formalin-fixed paraffin-embedded
- ROC
receiver operating characteristic
- MFI
median fluorescence intensity
- OR
odds ratio
Footnotes
CRediT authorship contribution statement
Julia Simon: Formal analysis, Visualization, Writing – original draft. Nicole Brenner: Formal analysis, Writing – review & editing. Sibylle Reich: Investigation, Writing – review & editing. Hilde Langseth: Resources, Investigation, Writing – review & editing. Bo T. Hansen: Resources, Investigation, Writing – review & editing. Giske Ursin: Formal analysis, Investigation, Writing – review & editing. Aida Ferreiro-Iglesias: Data curation, Writing – review & editing. Paul Brennan: Resources, Writing – review & editing. Aimée R. Kreimer: Resources, Writing – review & editing. Mattias Johansson: Resources, Writing – review & editing. Miranda Pring: Methodology, Writing – review & editing. Mari Nygard: Conceptualization, Resources, Writing – review & editing. Tim Waterboer: Conceptualization, Methodology, Resources, Supervision, Writing – review & editing.
Conflicts of interest: none.
Disclaimer
Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.canep.2022.102117.
References
- [1].Carioli G, Negri E, Kawakita D, Garavello W, La Vecchia C, Malvezzi M, Global trends in nasopharyngeal cancer mortality since 1970 and predictions for 2020: focus on low-risk areas, Int. J. Cancer 140 (10) (2017) 2256–2264. [DOI] [PubMed] [Google Scholar]
- [2].Wu L, Li C, Pan L, Nasopharyngeal carcinoma: a review of current updates, Exp. Ther. Med 15 (4) (2018) 3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F, Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012, Int. J. Cancer 136 (5) (2015) E359–E386. [DOI] [PubMed] [Google Scholar]
- [4].Robinson M, Suh YE, Paleri V, Devlin D, Ayaz B, Pertl L, Thavaraj S, Oncogenic human papillomavirus-associated nasopharyngeal carcinoma: an observational study of correlation with ethnicity, histological subtype and outcome in a UK population, Infect. Agents Cancer 8 (1) (2013) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dogan S, Hedberg ML, Ferris RL, Rath TJ, Assaad AM, Chiosea SI, Human papillomavirus and Epstein-Barr virus in nasopharyngeal carcinoma in a low-incidence population, Head & Neck 36 (4) (2014) 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Singhi AD, Califano J, Westra WH, High-risk human papillomavirus in nasopharyngeal carcinoma, Head & Neck 34 (2) (2012) 213–218. [DOI] [PubMed] [Google Scholar]
- [7].Ruuskanen M, Irjala H, Minn H, Vahlberg T, Randen-Brady R, Hagström J, Syrjänen S, Leivo I, Epstein-Barr virus and human papillomaviruses as favorable prognostic factors in nasopharyngeal carcinoma: a nationwide study in Finland, Head & Neck 41 (2) (2019) 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Simon J, Schroeder L, Ingarfield K, Diehl S, Werner J, Brenner N, Liu Z, Pawlita M, Pring M, Butt J, Ness A, Waterboer T, Epstein-Barr virus and human papillomavirus serum antibodies define the viral status of nasopharyngeal carcinoma in a low endemic country, Int. J. Cancer 147 (2) (2020) 461–471. [DOI] [PubMed] [Google Scholar]
- [9].Chang ET, Adami H-O, The enigmatic epidemiology of nasopharyngeal carcinoma, Cancer Epidemiol. Biomark. Prev 15 (10) (2006) 1765–1777. [DOI] [PubMed] [Google Scholar]
- [10].Old LJ, Boyse EA, Oettgen HF, Harven ED, Geering G, Williamson B, Clifford P, Precipitating antibody in human serum to an antigen present in cultured burkitt’s lymphoma cells, Proc. Nat. Acad. Sci. United States of America 56 (6) (1966), 1699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Henle G, Henle W, Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma, Int. J. Cancer 17 (1) (1976) 1–7 [DOI] [PubMed] [Google Scholar]
- [12].Lanier AP, Bornkamm GW, Henle W, Henle G, Bender TR, Talbot ML, Dohan PH, Association of Epstein-Barr virus with nasopharyngeal carcinoma in Alaskan native patients: serum antibodies and tissue EBNA and DNA, Int. J. Cancer 28 (3) (1981) 301–305 [DOI] [PubMed] [Google Scholar]
- [13].Lin TM, Yang CS, Chiou JF, Tu SM, Chen TY, Tu YC, Lin PJ, Kawamura A Jr., Hirayama T, Antibodies to Epstein-Barr virus capsid antigen and early antigen in nasopharyngeal carcinoma and comparison groups, Am. J. Epidemiol 106 (4) (1977) 336–339. [DOI] [PubMed] [Google Scholar]
- [14].Chien Y-C, Chen J-Y, Liu M-Y, Yang H-I, Hsu M-M, Chen C-J, Yang C-S, Serologic markers of Epstein–Barr virus infection and nasopharyngeal carcinoma in Taiwanese Men, New England J. Med 345 (26) (2001) 1877–1882. [DOI] [PubMed] [Google Scholar]
- [15].Liu Y, Huang Q, Liu W, Liu Q, Jia W, Chang E, Chen F, Liu Z, Guo X, Mo H, Chen J, Rao D, Ye W, Cao S, Hong M, Establishment of VCA and EBNA1 IgA-based combination by enzyme-linked immunosorbent assay as preferred screening method for nasopharyngeal carcinoma: a two-stage design with a preliminary performance study and a mass screening in southern China, Int. J. Cancer 131 (2) (2012), 406–16. [DOI] [PubMed] [Google Scholar]
- [16].Liu Z, Ji MF, Huang QH, Fang F, Liu Q, Jia WH, Guo X, Xie SH, Chen F, Liu Y, Mo HY, Liu WL, Yu YL, Cheng WM, Yang YY, Wu BH, Wei KR, Ling W, Lin X, Lin EH, Ye W, Hong MH, Zeng YX, Cao SM, Two Epstein-Barr virus-related serologic antibody tests in nasopharyngeal carcinoma screening: results from the initial phase of a cluster randomized controlled trial in Southern China, Am. J. Eepidemiol 177 (3) (2013), 242–50. [DOI] [PubMed] [Google Scholar]
- [17].Coghill AE, Pfeiffer RM, Proietti C, Hsu W-L, Chien Y-C, Lekieffre L, Krause L, Teng A, Pablo J, Yu KJ, Lou P-J, Wang C-P, Liu Z, Chen C-J, Middeldorp J, Mulvenna J, Bethony J, Hildesheim A, Doolan DL, Identification of a novel, EBV-based antibody risk stratification signature for early detection of nasopharyngeal Carcinoma in Taiwan, Clin. Cancer Res 24 (6) (2018) 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Simon J, Liu Z, Brenner N, Yu KJ, Hsu W-L, Wang C-P, Chien Y-C, Coghill AE, Chen C-J, Butt J, Proietti C, Doolan DL, Hildesheim A, Waterboer T, Validation of an Epstein-Barr virus antibody risk stratification signature for nasopharyngeal carcinoma by use of multiplex serology, J. Clin. Microbiol 58 (5) (2020) e00077–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Langseth H, Gislefoss RE, Martinsen JI, Dillner J, Ursin G, Cohort profile: the janus serum bank cohort in Norway, Int. J. Epidemiol 46 (2) (2017) 403–404g. [DOI] [PubMed] [Google Scholar]
- [20].Kreimer AR, Ferreiro-Iglesias A, Nygard M, Bender N, Schroeder L, Hildesheim A, Robbins HA, Pawlita M, Langseth H, Schlecht NF, Tinker LF, Agalliu I, Smoller SW, Ness-Jensen E, Hveem K, D’Souza G, Visvanathan K, May B, Ursin G, Weiderpass E, Giles GG, Milne RL, Cai Q, Blot WJ, Zheng W, Weinstein SJ, Albanes D, Brenner N, Hoffman-Bolton J, Kaaks R, Barricarte A, Tjønneland A, Sacerdote C, Trichopoulou A, Vermeulen RCH, Huang WY, Freedman ND, Brennan P, Waterboer T, Johansson M, Timing of HPV16-E6 antibody seroconversion before OPSCC: findings from the HPVC3 consortium, Ann. Oncol 30 (8) (2019) 1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hjerkind KV, Gislefoss RE, Tretli S, Nystad W, Bjørge T, Engeland A, Meyer HE, Holvik K, Ursin G, Langseth H, Cohort profile update: the janus serum bank cohort in Norway, Int. J. Epidemiol 46 (4) (2017) 1101–1102f. [DOI] [PubMed] [Google Scholar]
- [22].Halec G, Schmitt M, Dondog B, Sharkhuu E, Wentzensen N, Gheit T, Tommasino M, Kommoss F, Bosch FX, Franceschi S, Clifford G, Gissmann L, Pawlita M, Biological activity of probable/possible high-risk human papillomavirus types in cervical cancer, Int. J. Cancer 132 (1) (2013) 63–71. [DOI] [PubMed] [Google Scholar]
- [23].Ambinder RF, Mann RB, Epstein-Barr-encoded RNA in situ hybridization: diagnostic applications, Human Pathol. 25 (6) (1994) 602–605. [DOI] [PubMed] [Google Scholar]
- [24].Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T, Bead-based multiplex genotyping of human papillomaviruses, J. Clin. Microbiol 44 (2) (2006), 504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schmitt M, Dondog B, Waterboer T, Pawlita M, Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5+ and GP6+ primers, J. Clin. Microbiol 46 (3) (2008) 1050–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, Templin MF, Pawlita M, Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins, Clin. Chem 51 (10) (2005), 1845–53. [DOI] [PubMed] [Google Scholar]
- [27].Liu Z, Jarrett RF, Hjalgrim H, Proietti C, Chang ET, Smedby KE, Yu KJ, Lake A, Troy S, McAulay KA, Pfeiffer RM, Adami HO, Glimelius B, Melbye M, Hildesheim A, Doolan DL, Coghill AE, Evaluation of the antibody response to the EBV proteome in EBV-associated classical Hodgkin lymphoma, Int. J. Cancer 147 (3) (2020) 608–618. [DOI] [PubMed] [Google Scholar]
- [28].Waterboer T, Sehr P, Pawlita M, Suppression of non-specific binding in serological Luminex assays, J. Immunol. Methods 309 (1–2) (2006) 200–204. [DOI] [PubMed] [Google Scholar]
- [29].Carlander A-LF, Grønhøj Larsen C, Jensen DH, Garnæs E, Kiss K, Andersen L, Olsen CH, Franzmann M, Høgdall E, Kjær SK, Norrild B, Specht L, Andersen E, van Overeem Hansen T, Nielsen FC, von Buchwald C, Continuing rise in oropharyngeal cancer in a high HPV prevalence area: a Danish population-based study from 2011 to 2014, Eur. J. Cancer 70 (2017) 75–82. [DOI] [PubMed] [Google Scholar]
- [30].Ji X, Zhang W, Xie C, Wang B, Zhang G, Zhou F, Nasopharyngeal carcinoma risk by histologic type in central China: impact of smoking, alcohol and family history, Int. J. Cancer 129 (2011). [DOI] [PubMed] [Google Scholar]
- [31].Hsu WL, Chen JY, Chien YC, Liu MY, You SL, Hsu MM, Yang CS, Chen CJ, Independent effect of EBV and cigarette smoking on nasopharyngeal carcinoma: a 20-year follow-up study on 9,622 males without family history in Taiwan, Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive, Oncology 18 (4) (2009) 1218–1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


