Abstract
The gp41 envelope protein of human immunodeficiency virus type 1 (HIV-1) contains an α-helical core structure responsible for mediating membrane fusion during viral entry. Recent studies suggest that a conserved hydrophobic cavity in the coiled coil of this core plays a distinctive structural role in maintaining the fusogenic conformation of the gp41 molecule. Here we investigated the importance of this cavity in determining the structure and biological activity of the gp41 core by using the N34(L6)C28 model. The high-resolution crystal structures of N34(L6)C28 of two HIV-1 gp41 fusion-defective mutants reveal that each mutant sequence is accommodated in the six-helix bundle structure by forming the cavity with different sets of atoms. Remarkably, the mutant N34(L6)C28 cores are highly effective inhibitors of HIV-1 infection, with 5- to 16-fold greater activity than the wild-type molecule. The enhanced inhibitory activity by fusion-defective mutations correlates with local structural perturbations close to the cavity that destabilize the six-helix bundle. Taken together, these results indicate that the conserved hydrophobic coiled-coil cavity in the gp41 core is critical for HIV-1 entry and its inhibition and provides a potential antiviral drug target.
Infection by retroviruses and other enveloped viruses is initiated by fusion of the viral and cellular membranes, with the subsequent delivery of the viral genome into the host cell. Viral envelope glycoproteins play a critical role in this infectious process, because membrane fusion is an essential step for viral entry. They are also major targets in attempts to elicit the antiviral immune response in infected hosts. The envelope glycoprotein of human immunodeficiency virus type 1 (HIV-1) consists of two noncovalently associated subunits, gp120 and gp41, which are displayed on the lipid bilayer of the virion as well as on the plasma membranes of infected cells (42). The envelope glycoprotein is responsible for promoting membrane fusion between the virus and target cell during infection and between infected cells and uninfected cells during multinucleated giant cell (syncytium) formation (44). Binding of the surface gp120 subunit to the CD4 glycoprotein and a chemokine receptor on the T-cell surface triggers the membrane fusion activity of the transmembrane gp41 subunit (17, 37, 43, 44, 50). This receptor binding-mediated activation of membrane fusion is postulated to involve a conformational change in the envelope protein complex from a native (nonfusogenic) to a fusion-active (fusogenic) state. This conformational transition likely facilitates exposure of the hydrophobic, glycine-rich sequence referred to as the fusion peptide at the amino terminus of gp41, leading to insertion into the bilayer of the target membrane and initiating fusion (2, 6, 26).
The mechanism by which the gp41 molecule mediates membrane fusion has been the subject of intense investigation (8). As seen in most viral fusion proteins, the fusion peptide region of the gp41 ectodomain is followed by two regions with a 4-3 hydrophobic (heptad) repeat, a sequence feature characteristic of coiled coils (Fig. 1) (7, 16, 24). Protein dissection studies demonstrated that these two heptad-repeat regions from a soluble, α-helical core consisting of a trimer of antiparallel dimers (39, 41). Crystallographic analysis of this gp41 core confirmed that it is a six-helix bundle (10, 56, 57). The N (amino)-terminal helices form an interior trimeric coiled coil, while the C (carboxyl)-terminal helices pack in an antiparallel manner into three highly conserved hydrophobic grooves on the surface of the coiled coil. Given that this core structure is exceedingly stable to thermal denaturation and resembles the fusion-pH-induced conformation of influenza virus hemagglutinin, it was suggested that the six-helix bundle represents the conformation of fusion-active gp41 (10, 41, 56, 57). This view has been confirmed by recent antibody-binding studies which show that a monoclonal antibody specifically recognizing the gp41 core binds to the surface of HIV-1-infected cells only after interaction of the envelope protein complex with soluble CD4 (29).
FIG. 1.
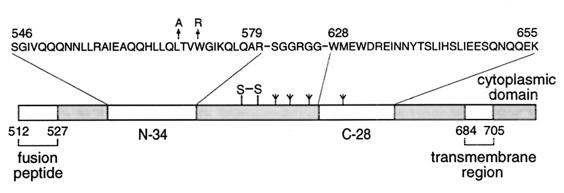
Schematic diagram of HIV-1 gp41. The important functional features of the gp41 ectodomain and the amino acid sequences of the N34 and C28 segments are shown. The N34(L6)C28 model consists of N34 and C28 plus a six-residue linker. Two point mutations, L568A and W571R, that abolish membrane fusion are indicated above the sequence. The disulfide bond and four potential N-glycosylation sites are depicted. The residues are numbered according to their position in gp160.
Synthetic peptides corresponding to the N- and C-terminal helical regions of the gp41 ectodomain have been shown to be potent inhibitors of HIV-1 infection and syncytium formation (30, 60, 62). It is striking that the C peptide T-20 has high efficacy in suppressing HIV-1 replication in clinical trials (35). Although the mechanism of action of these peptide inhibitors is not known, considerable evidence suggests that they act by binding to viral gp41, through a dominant-negative mechanism, to block formation of its fusion-active core (10, 11, 23, 34, 41, 45, 61). The structural and biophysical characteristics of the gp41 core and experiments on the inhibition of HIV-1 infectivity by gp41 peptides have led to the proposal that formation of the helical structure brings the viral and cellular membranes into close proximity and thus overcomes the energy barrier for membrane fusion (23, 27, 57).
The trimeric coiled-coil surface of the gp41 core is highly grooved and possesses a conserved hydrophobic cavity that is important for packing interactions between the N- and C-terminal helices (10). Mutations in the Leu 568 and Trp 571 residues, which are involved in the formation of this cavity, abolish membrane fusion activity (5). Here we have tested the importance of this cavity in determining the structure and function of the gp41 core by assessing the effects of Leu 568-to-Ala and Trp 571-to-Arg substitutions in the N34(L6)C28 model. The X-ray crystal structures of two single mutants at resolutions of 1.6 Å (L568A) and 2.1 Å (W571R) show that each mutant sequence is accommodated in the six-helix bundle structure by forming the cavity with different sets of atoms. Remarkably, L568A and W571R are effective inhibitors of HIV-1 infection, and they exhibit 16- and 5-fold greater activities than N34(L6)C28, respectively. Moreover, the double mutant (L568A/W571R) has the highest activity, 35-fold greater than that of the wild-type molecule. Our results provide evidence for the hypothesis that the conserved hydrophobic cavity in the coiled coil of the gp41 core plays a key role in HIV-1 entry and its inhibition.
MATERIALS AND METHODS
Protein production and purification.
Mutations were introduced into the N34(L6)C28 model by single-stranded mutagenesis (36) and verified by DNA sequencing. Standard recombinant DNA techniques were used (51). All recombinant peptides were expressed in Escherichia coli BL21(DE3)/pLysS by using the T7 expression system (54). Cells, freshly transformed with an appropriate plasmid, were grown to late log phase. Protein expression was induced by addition of 0.5 mM isopropylthio-β-d-galactoside (IPTG). After another 3 h of growth at 37°C, the bacteria were harvested by centrifugation, and the cells were lysed with glacial acetic acid as described previously (40). Peptides were purified from the soluble fraction to homogeneity by reverse-phase high-performance liquid chromatography, using a Vydac C18 preparative column and a linear gradient of acetonitrile containing 0.1% trifluoroacetic acid. Peptide identity was confirmed by mass spectrometry. Peptide concentrations were determined by absorbance at 280 nm in the presence of 6 M guanidinium hydrochloride (19).
CD spectroscopy.
Circular dichroism (CD) spectra were acquired at a peptide concentration of 10 μM in 50 mM sodium phosphate (pH 7.0)–150 mM NaCl (phosphate-buffered saline [PBS]) with an Aviv 62 DS spectrometer as described previously (40). The wavelength dependence of molar ellipticity, [θ], was monitored at 0°C as the average of five scans, using a 5-s integration time at 1.0-nm wavelength increments. Spectra were baseline corrected against the cuvette with buffer alone. The helix content was estimated from the CD signal by dividing the mean residue ellipticity at 222 nm by the value expected for 100% helix formation by helices of comparable size, i.e., −33,000° cm2 dmol−1 (13). Thermal stability was determined by monitoring the change in the CD signal at 222 nm as a function of temperature, and thermal melts were performed at intervals of 2°C with a 2-min equilibration at the desired temperature and an integration time of 30 s. Reversibility was checked by repeated scans. The thermal melts of L568A and W571R were reversible, while those of N34(L6)C28 and L568A/W571R were not reversible. The midpoint of the thermal unfolding transition (apparent melting temperature [Tm]) was determined from the maximum of the first derivative, with respect to the reciprocal of the temperature, of the [θ]222 values (4). The error in estimation of Tm is ±1°C.
Equilibrium ultracentrifugation.
Sedimentation equilibrium analysis was performed on a Beckman XL-A analytical ultracentrifuge as described previously (40). Protein solutions were dialyzed overnight against PBS; loaded at initial concentrations of 10, 30, and 100 μM; and analyzed at rotor speeds of 20 and 23 krpm at 20°C. Data sets were fitted simultaneously to a single-species model with the program NONLIN (31). The protein partial specific volume and solvent density were calculated with constants from Laue et al. (38). Molecular weights were all within 10% of those calculated for an ideal trimer, with no systematic deviation of the residuals.
Crystallization, data collection, and structure determination.
The N34(L6)C28 variants were crystallized by hanging-drop vapor diffusion at room temperature. To grow crystals, a 10-mg/ml stock of peptides was diluted 1:1 with a reservoir and allowed to equilibrate against the reservoir solution. Initial crystallization conditions were screened by using sparse matrix crystallization kits (Crystal Screen I and II; Hampton Research, Riverside, Calif.) and then optimized. Crystals of L568A in space group R3 (a = b = 49.68 Å, c = 60.20 Å) were grown from 0.1 M sodium acetate (pH 4.6)–0.2 M ammonium sulfate–20% (vol/vol) glycerol and transferred to a cryoprotectant solution containing 25% (vol/vol) glycerol in the corresponding mother liquor. Crystals of W571R in space group R3 (a = b = 53.39 Å; c = 59.56 Å) were obtained from 0.1 M sodium acetate (pH 4.6)–0.2 M ammonium sulfate–17% polyethylene glycol 4000–20% (vol/vol) glycerol. Cryoprotected crystals were frozen in propane before data collection. Data were collected at 95 K by using an R-axis IV image plate detector mounted on a Rigaku RU200 rotating anode X-ray generator at the X-ray Crystallography Facility at the Weill Medical College of Cornell University. All diffraction data were integrated and scaled with the HKL suite (48).
The structures of L568A and W571R were determined by molecular replacement with AMoRe (46). The 2.4-Å structure of the wild-type N34(L6)C28 trimer (PDB code ISTZ) was used in a combined rotation-translation search (with data for 15.0 to 3.5 Å) to yield solutions for L568A (correlation coefficient = 66.8%; R factor = 47.9%) and for W571R (correlation coefficient = 74.8%; R factor = 38.9%). The models were refined with simulated annealing and atomic displacement parameter refinements by using the program X-PLOR (1). The models were rebuilt to reflect the sequences of L568A and W571R by using conventional (2Fo − Fc)Φcalc and (Fo − Fc)Φcalc maps with the program O (33). Atomic coordinates for L568A (Protein Data Bank [PDB] file name 1QR9) and W571R (PDB file name 1QR8) have been deposited in the PDB.
Cell-cell fusion and infectivity assays.
The inhibitory activities of N34(L6)C28 and variants thereof were determined by using a dye transfer fusion assay as described previously (28). HIV-1IIIB-infected H9 cells were labeled with 2′,7′-bis-(2-carboxyethyl)-5 (and 6)-carboxyfluorescein-acetoxymethyl ester (BCECF-AM). Fluorescently labeled HIV-1IIIB-infected H9 cells (104) and MT-2 cells were cocultured at a 1:10 ratio at various peptide concentrations for 2 h at 37°C in a 96-well microplate to obtain a dose-response curve. The fused cells were scored for dye transfer under fluorescence microscopy. In the infectivity assay, as described previously (28), HIV-1IIIB was inoculated with 104 MT-2 cells at multiplicity of infection of 0.0045 in RPMI 1640 containing 10% fetal bovine serum in the presence of various peptide concentrations. After incubation at 37°C for 1 and 24 h, half of the culture medium was changed. HIV-1IIIB-mediated cytopathic effect (CPE) was assessed by viability assay 6 days after infection.
RESULTS
Characterization of the mutant N34(L6)C28 cores.
The recombinant N34(L6)C28 model of the gp41 ectodomain core, consisting of residues 546 to 579 and 628 to 655 joined by a linker of six hydrophilic residues (Fig. 1), forms a stable six-helix bundle (40, 56). Earlier genetic studies indicate that the Leu 568-to-Ala and Trp 571-to-Arg substitutions inhibit membrane fusion (5). X-ray crystallographic analyses of the gp41 core show that these two residues are involved in forming a large hydrophobic cavity on the surface of the trimeric coiled coil (10, 56, 57). To define the structural and functional importance of these residues and, hence, the cavity in promoting membrane fusion, we replaced Leu 568 with Ala and Trp 571 with Arg in N34(L6)C28, either separately or simultaneously, to yield two single mutants (L568A and W571R) and a double mutant (L568A/W571R).
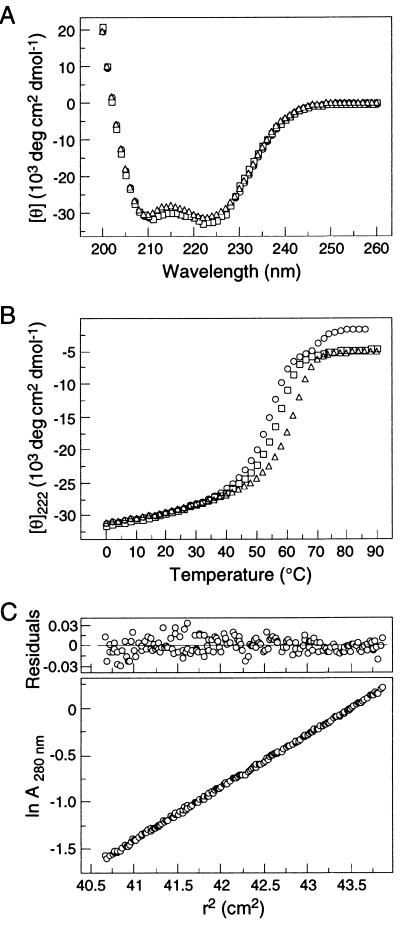
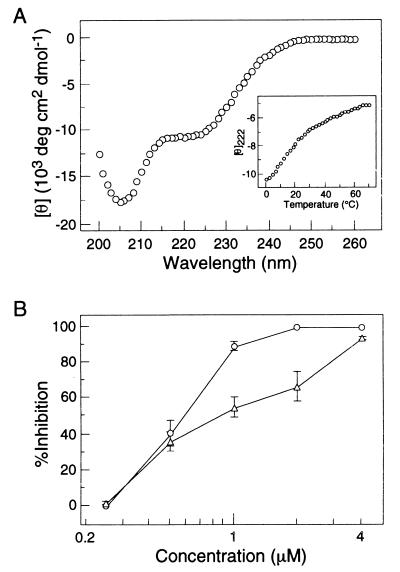
The CD spectra of L568A, W571R, and L568A/W571R exhibit the characteristic signature of an α-helical conformation, with minima at 222 and 208 nm (Fig. 2A). Each folded mutant core contains >90% α-helical structure at 0°C in PBS at a peptide concentration of 10 μM. Under these conditions, the apparent Tms of N34(L6)C28 (wild type), L568A, W571R, and L568A/W571R are 70, 56, 61, and 54°C, respectively (Fig. 2B; Table 1). Thus, the mutant molecules are less stable than the wild-type counterpart. Moreover, sedimentation equilibrium demonstrates that all the three mutant cores are trimeric in solution (Fig. 2C; Table 1). These results indicate that the Leu 568-to-Ala and Trp 571-to-Arg mutations do not alter structural features sufficiently to disrupt the six-helix bundle formation. The destabilization effect of these mutations observed here may be related to the fusion-defective phenotype (see below).
FIG. 2.
Folding of the N34(L6)C28 mutants as helical trimers. (A) CD spectra of L568A (squares), W571R (triangles), and L568A/W571R (circles) at 0°C in PBS (pH 7.0) at a peptide concentration of 10 μM. (B) Thermal melts monitored by CD at 222 nm for L568A (squares), W571R (triangles), and L568A/W571R (circles) in PBS (pH 7.0) at a peptide concentration of 10 μM. (C) Sedimentation equilibrium studies of the mutant N34(L6)C28 cores indicate that all species are trimeric. Representative analytical ultracentrifugation data (20 krpm) for L568A/W571R collected at 20°C in PBS (pH 7.0) at a peptide concentration of ∼30 μM are shown. The natural logarithm of the absorbance at 280 nm is plotted against the square of the radial position. Deviations from the calculated values are plotted as residuals in the upper panel.
TABLE 1.
N34(L6)C28 variants from six-helix bundle structures
| Core model | −[θ]222 (deg cm2 dmol−1) | Tm (°C) | Molecular mass (kDa) |
|---|---|---|---|
| N34(L6)C28 | 31,300 | 70 | 24.4 |
| L568A | 32,500 | 56 | 24.1 |
| W571R | 31,000 | 61 | 24.6 |
| L568A/W571R | 31,600 | 54 | 23.8 |
| L568A/W571R/I573S | 10,600 | <20 |
Crystal structures of L568A and W571R.
To obtain a high-resolution view of the conserved hydrophobic cavity in the mutant cores, the X-ray crystal structures of L568A and W571R were determined. Crystals of the two mutants were grown by hanging-drop vapor diffusion (see Materials and Methods). The L568A and W571R structures were determined by molecular replacement, using a 2.4-Å structure of N34(L6)C28 (56). The electron density map of L568A allows the completion of the structure of 68 amino acid residues and the placement of 49 water molecules (Fig. 3A and B), while that of W571R reveals the positions of all of the amino acid residues except a few disordered side chains at the helix termini and in the linker region (Fig. 3C and D). The structure of L568A was refined against 15.0- to 1.6-Å data to a 1.6-Å resolution to yield crystallographic and free R factors of 20.2 and 26.1%, respectively (Table 2). The structure of W571R was refined to a 2.1-Å resolution, with crystallographic and free R factors of 21.2 and 26.2%, respectively, in the resolution range of 15.0 to 2.1 Å (Table 2). Details of the data collection and refinement statistics are presented in Table 2.
FIG. 3.
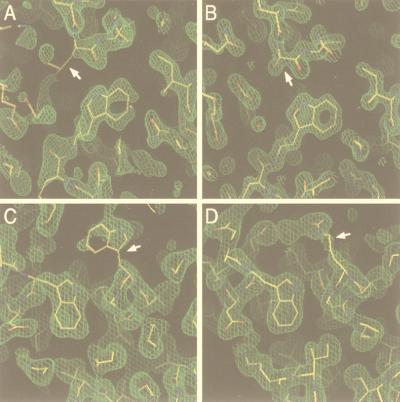
Electron density maps at the substitution sites. (A) Initial 2Fo − Fc map of L568A after density modification and phase improvement with DM (14). The initial molecular replacement solution model is superimposed. (B) Final 2Fo − Fc map of L568A with the refined model superimposed. (C) Initial 2Fo − Fc map of W571R as described for panel A. (D) Final 2Fo − Fc map of W571R as described for panel B. The side chains of the mutated residues are indicated by white arrows. Water molecules are indicated by small red balls. The maps of L568A and W571R are contoured at 1.8 and 1.2 ς, respectively. Figures were generated with the program O (33).
TABLE 2.
X-ray data collection and refinement statistics
| Parameter | L568A | W571R |
|---|---|---|
| Processing statistics | ||
| Resolution (Å) | 30.0–1.6 | 30.0–2.1 |
| Measured reflections | 29,806 | 7,391 |
| Unique reflections | 7,043 | 3,505 |
| Completeness (%) | 96.6 | 94.0 |
| Rmergea (%) | 3.6 | 6.0 |
| Refinement statistics | ||
| Resolution (Å) | 15.0–1.6 | 15.0–2.1 |
| Protein nonhydrogen atoms | 552 | 552 |
| Water molecules | 49 | 17 |
| Rfreeb (%) | 26.1 | 26.2 |
| Rcrystb (%) | 20.2 | 21.2 |
| Average B factor (Å2) | 20.6 | 48.9 |
| rms deviations from ideality | ||
| Bond lengths (Å) | 0.005 | 0.004 |
| Bond angles (°) | 1.245 | 0.894 |
| Torsion angles (°) | 19.093 | 23.389 |
Rmerge = Σ|I − <I<|/ΣI, where I is the intensity of an individual measurement and 〈I〉 is the mean recorded intensity over multiply recordings.
R = Σ|Fo − Fc|/ΣFo, where the crystallographic (cryst) and free R factors are calculated by using the working and free reflection sets, respectively.
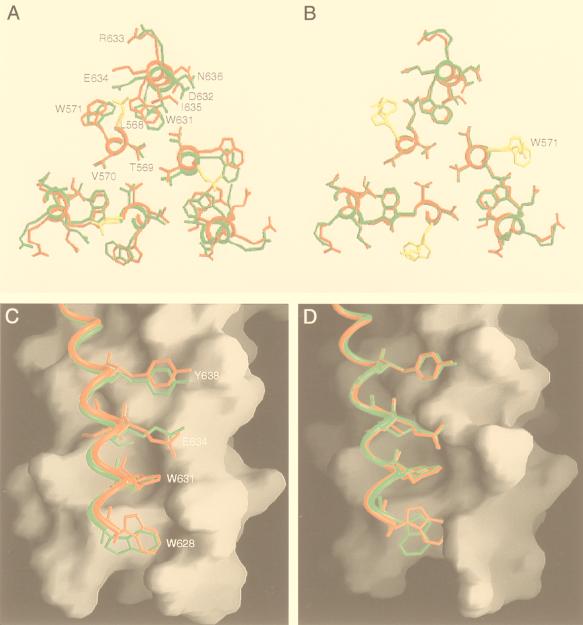
The overall structures of L568A and W571R are very similar to that of the wild-type core. In all cases, three hairpin-like molecules pack together on the crystallographic threefold-symmetry axis to form a six-helix bundle. The N-terminal helices within the bundle form a parallel, three-stranded coiled coil in the characteristic acute “knobs-into-holes” arrangement (Fig. 4) (15, 25). Three C-terminal helices pack in an antiparallel orientation into three hydrophobic grooves on the surface of the central coiled-coil trimer (Fig. 4). The Leu 568 and Trp 571 residues replaced in L568A and W571R are at each of the grooves, where they form the right wall of a deep cavity that accommodates the side chains of two tryptophans and a isoleucine from the C-terminal helices (Fig. 5). The root mean square (rms) deviations between all Cα atoms of the trimeric coiled coil in the wild-type and mutant molecules are 0.43 Å for L568A and 0.27 Å for W571R. The C-terminal helices in L568A and W571R can also be superimposed upon the wild-type counterpart with rms deviations of 0.78 and 0.52 Å, respectively. Thus, it appears that the fusion-defective Leu 568-to-Ala and Trp 571-to-Arg mutations each do not affect the overall protein fold of the gp41 core.
FIG. 4.
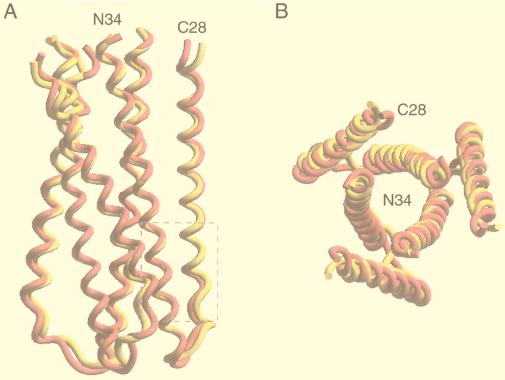
Overall views of the mutant L568A and W571R cores. (A) Side view. The amino termini of N34 and the carboxyl termini of C28 are at the top. Helices in L568A (yellow) and W571R (pink) were used for the superposition. The bottom of the central N34 coiled-coil surface contains three symmetry-related hydrophobic cavities (one is outlined by the box). (B) View from the top, looking down the threefold axis of the trimer. The same color coding as in panel A is used. Figures were generated with the program SETOR (21).
FIG. 5.
The conserved hydrophobic cavity in the L568A and W571R structures. (A) Cross-section of helix packing near the conserved cavity in L568A. The structures of the wild-type molecule (red) and L568A (green) are overlaid. The side chains of the mutated residues are in yellow. (B) Cross-section of helix packing in W571R. The same superposition as in panel A was used. (C) Interactions of the C28 helix with a deep cavity on the surface of the N34 coiled coil in L568A. The C28 helices of the wild-type molecule (red) and L568A (green), represented as ribbons, are shown against a surface representation of the N34 coiled coil in L568A. (D) Interactions of the C28 helix with a deep cavity on the surface of the N34 coiled coil in W571R. The same superposition as in panel C was used. Figures were generated with the programs SETOR (21) and GRASP (47).
The replacement of Leu 568 with Ala leads to local structural rearrangements in packing interactions between the N34 and C28 helices; the side chains of several key residues, including Tyr 638, Glu 634, Ile 635, Trp 631, and Trp 628, deviate substantially near the conserved cavity (Fig. 5A and C). In contrast, the Arg 571 residue in W571R is accommodated without significantly altering the six-helix bundle structure. A local distortion occurs at Trp 628 (Fig. 5B and D), which has high atomic B factors and may be poorly ordered due to chain terminus effects. In addition, the Arg 571 side chain is oriented toward the carboxyl group of the Glu 632 side chain from the abutting C28 helix, and the two side chains can interact via a water-mediated hydrogen bond (2.81 and 2.70 Å). This polar interaction appears to be responsible for the ordering of the Arg 573 side chain.
Effect of fusion-defective mutations on HIV-1 inhibition.
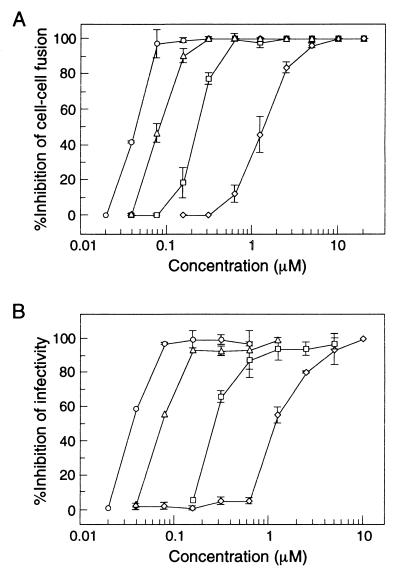
We have previously demonstrated that N34(L6)C28 can block syncytium formation at micromolar concentrations (40). To test if the Leu 568-to-Ala and Trp 571-to-Arg mutations affect this inhibitory activity, we examined the relative abilities of L568A, W571R, and L568A/W571R to inhibit HIV-1 infection by using both cell-cell fusion and infectivity assays. Figure 6 shows the inhibition of HIV-1IIIB-infected H9 cell-mediated syncytium formation and HIV-1 infectivity by different concentrations of the wild-type and mutant N34(L6)C28 molecules. The double mutant is strikingly the most effective inhibitor, with 50% inhibitory concentrations (IC50s) of 48 nM for syncytium formation and 44 nM for CPE, compared with IC50s of 1.51 and 1.49 μM, respectively, for N34(L6)C28 (Fig. 6; Table 3). L568A also potently inhibits cell-cell fusion and infection, with IC50s of 95 and 82 nM, respectively, while W571R does so with IC50s of 0.25 and 0.32 μM, respectively (Fig. 6; Table 3). Thus, the fusion-defective mutations cause the N34(L6)C28 core to have 5-, 16-, and 35-fold greater inhibitory activity for W571R, L658A, and L568/W571R, respectively.
FIG. 6.
Inhibition of HIV-1 infection by N34(L6)C28 variants. (A) Inhibition of HIV-1IIIB-infected H9 cell-induced cell-cell fusion by N34(L6)C28 (diamonds), L568A (triangles), W571R (squares), and L568A/W571R (circles). Error bars indicate standard deviations from quadruplicate experiments. (B) Inhibition of HIV-1IIIB-mediated CPE by N34(L6)C28 (diamonds), L568A (triangles), W571R (squares), and L568A/W571R (circles). Error bars indicate standard deviations from triplicate experiments.
TABLE 3.
Inhibition of HIV-1 infection by N34(L6)C28 variants
| Peptide | Cell-cell fusion
|
Infectivity
|
||
|---|---|---|---|---|
| IC50 (nM) | IC90 (nM) | IC50 (nM) | IC90 (nM) | |
| N34(L6)C28 | 1,508 ± 160 | 2,841 ± 201 | 1,487 ± 87 | 2,920 ± 286 |
| L568A | 95 ± 8 | 139 ± 7 | 82 ± 7 | 134 ± 4 |
| W571R | 248 ± 11 | 405 ± 45 | 319 ± 10 | 574 ± 70 |
| L568A/W571R | 48 ± 7 | 69 ± 31 | 44 ± 10 | 67 ± 16 |
| L568A/W571R/I573S | 765 ± 14 | 1,220 ± 18 | 1,153 ± 77 | 2,500 ± 261 |
| I573S | 685 ± 9 | 766 ± 11 | 730 ± 20 | 965 ± 80 |
| N34 | 9,737 ± 498 | 15,563 ± 1,175 | 11,384 ± 338 | 17,133 ± 670 |
| C28 | 947 ± 67 | 1,663 ± 168 | 2,192 ± 19 | 4,865 ± 363 |
In contrast to the Leu 568-to-Ala and Trp 571-to-Arg mutations, the Ser substitution for Ile 573, an a heptad position, essentially disrupts the six-helix complex formation (40). Consequently, N34(L6)C28 bearing this mutation inhibits cell-cell fusion and infection with IC50 values of 0.69 and 0.73 μM, respectively (Table 3). This activity appears to reflect that of the unfolded C28 segment in the I573S mutant, since the isolated N34 and C28 peptides have IC50s of 9.74 and 0.95 μM for syncytium formation and 11.38 and 2.19 for CPE, respectively (Table 3). To examine whether the Ile 573-to-Ser mutation can eliminate the antiviral activity of L568A/W571R, we replaced Ile 573 with Ser in the double mutant to produce a triple mutant (L568A/W571R/I573S). CD experiments indicate that the triple mutant is predominantly unfolded at 0°C in PBS at a peptide concentration of 10 μM (Fig. 7A; Table 1). Interestingly, this molecule is significantly less active in blocking cell-cell fusion and infectivity, with IC50s of 0.77 and 1.15 μM, respectively (Fig. 7B; Table 3). Taken together, these results indicate that the inhibition of HIV-1 infection by the N34(L6)C28 core is both conformation and sequence specific and that the L568 and W571 residues are important determinants of the inhibitory activity.
FIG. 7.
An Ile 573-to-Ser mutation disrupts the six-helix bundle formation of L568A/W571R and abolishes its potent antiviral activity. (A) CD spectrum of L568A/W571R/I573S at 0°C in PBS (pH 7.0) at a peptide concentration of 10 μM. The inset shows a thermal melt monitored by CD at 222 nm for L568A/W571R/I573S in PBS (pH 7.0) at a peptide concentration of 10 μM. (B) Inhibition of HIV-1IIIB-infected H9 cell-induced cell-cell fusion (circles) and HIV-1IIIB-mediated CPE (triangles) by L568A/W571R/I573S. Error bars indicate standard deviations from quadruplicate and triplicate experiments for cell-cell fusion and infectivity assays, respectively.
DISCUSSION
Core structure and mechanism.
Recent descriptions of the gp41 ectodomain core structure provide detailed information about the fusogenic conformation of the HIV-1 envelope protein (3, 10, 56, 57). The structure reveals a three-stranded coiled coil adjacent to the N-terminal fusion peptide, surrounded by an outer layer of antiparallel α-helices. This structural organization is shared by the influenza virus and Moloney murine leukemia virus transmembrane envelope proteins (2, 22), suggesting conservation of a common core structure. The overall fusion mechanism of these viral envelope proteins is thought to involve a conformational change and subsequent antiparallel association of α-helices that lead to membrane apposition and fusion (23, 27, 57). A similar mechanism is also proposed for the SNARE proteins in the fusion of vesicles with their target membranes (52, 55).
Although the functional role of the gp41 core in promoting membrane fusion remains largely undefined, additional information has come from the functional analysis of engineered HIV-1 envelope protein variants. Mutagenesis studies demonstrate that residues that stabilize this core structure are critical for membrane fusion activity (5, 12, 18, 58). Moreover, the folding and stability of the N34(L6)C28 model in vitro correlate well with severity of the in vivo phenotypes observed in cells expressing envelope proteins bearing mutations at Ile 573 (18, 40). Thus, the gp41 core structure appears to play a direct role in membrane fusion. Interestingly, point mutations in the Leu 568 and Trp 571 residues lining the right wall of the conserved hydrophobic cavity in the coiled coil of gp41 result in mutant envelope proteins that are completely defective in mediating membrane fusion (5). Our results indicate that the N34(L6)C28 core structure is destabilized by the fusion-defective mutations. Remarkably, our results also indicate clearly that these substitutions do not prevent the six-helix bundle formation. The crystal structures of the wild-type and mutant molecules can be superimposed, with rms deviations of 0.67 Å for L568A and 0.45 Å for W571R. From the structures we can see that each mutant sequence is accommodated in the six-helix bundle, resulting in hydrophobic cavities that are surrounded by different sets of atoms. Taken together, these data suggest that a protein-folding defect in the gp41 core is unlikely to be a major cause of the fusion-defective phenotype of the mutant viruses. We propose that the conserved hydrophobic cavity within the fusogenic gp41 core plays a specific and key role during the membrane fusion step of HIV-1 infection.
Inhibition of HIV-1 infectivity.
Synthetic N- and C-terminal peptides of gp41 inhibit HIV-1 infection and syncytium formation (30, 60, 62). Several considerations provide good evidence that these peptides act by binding to virus gp41 and preventing the fusogenic gp41 core formation (9, 11, 23, 34, 41, 45, 49, 61). First, inhibition of syncytium formation by the C43 peptide is markedly reduced when stoichiometric amounts of the N51 peptide are also present (41). Moreover, when a gp41 ectodomain maltose-binding chimeric protein contains a proline mutation in the trimeric coiled-coil region of gp41, the resulting mutant is a potent inhibitor of HIV-1 infection (11). Second, the inhibitory activity of a C-terminal peptide depends on its ability to interact with the N-terminal coiled coil (9, 61). Third, the structural features of the gp41 core are fully consistent with the simple dominant-negative model of inhibition (10, 41, 56, 57). The C-terminal peptides block membrane fusion by binding to the coiled coil of gp41, while the N-terminal peptides act as inhibitors by preventing the trimeric coiled-coil formation and/or associating with the endogenous C-terminal region of gp41. Finally, kinetic studies suggest that these peptides do not act on the native conformation of the HIV-1 envelope (23) but can act during its transition to the fusogenic state (23, 32, 45).
The use of the single-chain N34(L6)C28 model for the study of the gp41 core structure leads to the interesting result that N34(L6)C28 also inhibits HIV-1 infectivity. We have defined the following characteristics of this inhibitory activity. First, the inhibition is not attributable to the isolated N34 and C28 peptides, because the N34(L6)C28 trimer is still highly stable to thermal denaturation at its IC50, with an apparent Tm of approximately 63°C (40). Second, there is amino acid sequence-specific inhibition by N34(L6)C28. The inhibitory activity is dramatically increased by the fusion-defective Leu 568-to-Ala and Trp 571-to-Arg mutations; W571R, L568A, and L568A/W571R exhibit 5-, 16-, and 35-fold greater activity than the wild-type molecule, respectively. Third, the inhibition is also conformation specific; the presence of the Ile 573-to-Ser mutation in the double mutant essentially disrupts the six-helix bundle formation, while reducing its inhibitory activity 16-fold. Fourth, the enhanced inhibitory activity by these mutations correlates with local structural perturbations near the hydrophobic cavity which destabilize the N34(L6)C28 trimer. Further studies of the inhibition mechanism of the gp41 core should provide insights into the HIV-1 entry process and could open new perspectives in the search for effective antiviral therapies.
Implications for membrane fusion.
The hemagglutinin protein of influenza virus irreversibly switches from the native structure to the fusogenic conformation when exposed to the acidic environment of the cellular endosome (2, 6, 26, 63). This structural dimorphism is the basis for conformational changes that are crucial for activation of membrane fusion. The HIV-1 envelope protein is also thought to exist in two different conformations (for recent reviews, see references 8 and 52). It is generally accepted that the native conformation exists on the surface of free virions, while upon binding of gp120 to CD4 and particular coreceptors (e.g., CCR5 or CXCR4), the HIV-1 envelope protein undergoes a complex of structural changes to the fusogenic state. The current model for gp41-mediated membrane fusion suggests that formation of the six-helix bundle leads to colocalization of the viral and cellular membranes for fusion (23, 27, 57). While relatively little is known about how membrane apposition leads to complete fusion, there is evidence for the higher-order assembly of envelope protein trimers and the formation of fusion pores, as proposed to be required for influenza virus fusion (20, 53, 59).
Since the gp41 ectodomain core structure, with a Tm in excess of 90°C, is too stable to be disrupted by exogenous peptide binding, only during the gp41 conformational change to the fusogenic state does one anticipate that the targets for the peptides are available (11, 23, 34, 40, 41). This consideration has led to the proposal that gp41 can exist as a transiently populated intermediate after initiating the receptor-activated conformational change but prior to formation of the six-helix bundle (8, 23, 45). According to this view, synthetic peptides derived from the gp41 ectodomain inhibit membrane fusion in a dominant-negative manner by associating with their endogenous partners of viral gp41 at this intermediate stage.
Earlier genetic studies indicate that mutations in the Leu 568 and Trp 571 residues abolish membrane fusion activity, although the mutant HIV-1 envelope proteins appear to have no other defects, including cell surface expression, gp160 precursor processing, and CD4 binding (5). Our results indicate that these fusion-defective mutations destabilize the gp41 core structure although they still confer the six-helix bundle fold. Since the Leu 568 and Trp 571 residues form the right wall of a conserved coiled-coil cavity that provides a binding pocket for three C-terminal helices (9), our data suggest that the fusion-defective mutations introduce structural perturbations in the cavity that weaken helical packing interactions in the six-helix complex and thus inhibit its formation.
These fusion-defective mutations also exert striking effects on the inhibitory activity of N34(L6)C28; the L568A and W571R mutants exhibit 5- to 16-fold-greater activity than the wild-type molecule. Several lines of evidence suggest that this enhanced inhibitory activity results from the synergistic inhibition of the N34 and C28 peptides in the mutant molecules. First, while the L568A and W571R trimers are stable, with Tm values of 56 and 61°C, respectively, in PBS (pH 7.0) at a peptide concentration of 10 μM, L568A and W571R are predominantly unfolded at their IC50s (0.1 μM for L568A and 0.3 μM for W571R) under physiological conditions. The monomeric forms of the L568A and W571R molecules readily interact bivalently with virus gp41. Second, the Ile 573-to-Ser mutation that disrupts the N34 coiled-coil formation (40) can reduce the potency of the double mutant (L568A/W571R) in inhibiting membrane fusion close to that of the isolated C28 peptide. The nature of the multivalency in the N34(L6)C28 variants is likely to be responsible for their enhanced inhibitory activity. Finally, this synergy is fully consistent with the hypothesis that there is a populated intermediate of gp41 during transition to the fusogenic structure (8, 23, 45). Only in the intermediate state are the N- and C-terminal heptad-repeat regions of virus gp41 not associated, allowing the N34 and C28 peptides to bind to these regions with a high effective concentration.
ACKNOWLEDGMENTS
We thank Jun Dong for suggestions on structural refinement and Neville Kallenbach for critical reading of the manuscript.
This research was funded by NIH grants (AI-42693 to S.J. and AI-42382 to M.L.) and by the New York City Council Speaker’s Fund for Biomedical Research (to M.L.).
REFERENCES
- 1.Brünger A T. XPLOR version 3.1: a system for X-ray crystallography and NMR. New Harven, Conn: Yale University Press; 1992. [Google Scholar]
- 2.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 3.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. Three-dimensional solution of the 44 kDa ectodomain of SIV gp41. EMBO J. 1988;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantor C, Schimmel P. Biophysical chemistry, part III. New York, N.Y: W. H. Freeman and Company; 1980. pp. 1131–1132. [Google Scholar]
- 5.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 7.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 8.Chan D C, Kim P S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 9.Chan D C, Chutkowski C T, Kim P S. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen C H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S S, Lee C N, Lee W R, McIntosh K, Lee T H. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J Virol. 1993;67:3615–3619. doi: 10.1128/jvi.67.6.3615-3619.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y-H, Yang J T, Chau K H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 14.Cowtan K D. Joint CCP 4 and ESF-EACBM. Newsl Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 15.Crick F H C. The packing of α-helices: simple coiled coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 16.Delwart E J, Mosialos G, Gilmore T. Retroviral envelope glycoproteins contain a leucine zipper like repeat. AIDS Res Hum Retroviruses. 1990;6:703–706. doi: 10.1089/aid.1990.6.703. [DOI] [PubMed] [Google Scholar]
- 17.Dimitrov D S. How do viruses enter cells? The HIV coreceptors teach us a lesson of complexity. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 18.Dubay J W, Roberts S J, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 20.Ellens H, Bentz J, Mason D, Zhang F, White J M. Fusion of influenza hemagglutinin-expressing fibroblasts with glycophorin-bearing liposomes: role of hemagglutinin surface density. Biochemistry. 1990;29:9697–9707. doi: 10.1021/bi00493a027. [DOI] [PubMed] [Google Scholar]
- 21.Evans S V. SETOR: hardware lighted three-dimensional solid model representation of macromolecules. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 22.Fass D, Harrison S C, Kim P S. Structure of Moloney murine virus envelope domain at 1.7Å resolution. Nat Struct Biol. 1997;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 23.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 24.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 25.Harbury P B, Kim P S, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 27.Hughson F M. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 28.Jiang S, Lin K, Neurath A R. Enhancement of human immunodeficiency virus type 1 (HIV-1) infection by antisera to peptides from the envelope glycoprotein gp120/gp41. J Exp Med. 1991;174:1557–1563. doi: 10.1084/jem.174.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang S, Lin K, Lu M. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the HIV-1 envelope glycoprotein. J Virol. 1998;72:10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang S, Lin K, Strick N, Neurath A R. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 31.Johnson M L, Correia J J, Yphantis D A, Halvorson H R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones P L, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 33.Jones T A, Zou J W, Cowan S W, Kjelgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;D47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Judice J K, Tom J Y K, Huang W, Wrin T, Vennari J, Petropoulos C J, McDowell R S. Inhibition of HIV type 1 infectivity by constrained α-helical peptides: implications for the viral fusion mechanism. Proc Natl Acad Sci USA. 1997;94:13426–13430. doi: 10.1073/pnas.94.25.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge Y, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in human by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 37.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laue T M, Shah B D, Ridgeway T M, Pelletier S L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S E, Rowe A J, Horton J C, editors. Analytical ultracentrifugation in biochemistry and polymer science. Cambridge, United Kingdom: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 39.Lu M, Kim P S. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J Biomol Struct Dyn. 1997;15:465–471. doi: 10.1080/07391102.1997.10508958. [DOI] [PubMed] [Google Scholar]
- 40.Lu M, Ji H, Shen S. Subdomain folding and biological activity of the core structure from human immunodeficiency virus type 1 gp41: implications for viral membrane fusion. J Virol. 1999;73:4433–4438. doi: 10.1128/jvi.73.5.4433-4438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu M, Blacklow S C, Kim P S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 42.Luciw P A. Human immunodeficiency viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melinick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1881–1952. [Google Scholar]
- 43.Moore J P, Jameson B A, Dragic T. Co-receptor for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 44.Moore J P, Jameson B A, Weiss R A, Sattentau Q J. The HIV-cell fusion reaction. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press; 1993. pp. 233–289. [Google Scholar]
- 45.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1988;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 47.Nichols A, Sharp K, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Rimsky L T, Shugars D C, Matthews T J. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J Virol. 1998;72:986–993. doi: 10.1128/jvi.72.2.986-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J A. Conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 52.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 53.Spruce A E, Iwata A, White J M, Almers W. Patch clamp studies of single cell-fusion events mediated by a viral fusion protein. Nature. 1989;342:555–558. doi: 10.1038/342555a0. [DOI] [PubMed] [Google Scholar]
- 54.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 55.Sutton R B, Fasshauer D, Brunger T A. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1988;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 56.Tan K, Liu J, Wang J, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 58.Weng Y, Weiss D C. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1998;72:9676–9682. doi: 10.1128/jvi.72.12.9676-9682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 60.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive α-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wild C T, Greenwell T, Shugars D, Rimsky-Clarke L, Matthews T J. The inhibitory activity of an HIV-1 type peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res Hum Retroviruses. 1995;11:323–325. doi: 10.1089/aid.1995.11.323. [DOI] [PubMed] [Google Scholar]
- 62.Wild C T, Oas T, McDanal C B, Bolognesi D, Matthews T J. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]