Abstract
Machine learning (ML) has transformed protein engineering by constructing models of the underlying sequence-function landscape to accelerate the discovery of new biomolecules. ML-guided protein design requires models, trained on local sequence-function information, to accurately predict distant fitness peaks. In this work, we evaluate neural networks’ capacity to extrapolate beyond their training data. We perform model-guided design using a panel of neural network architectures trained on protein G (GB1)-Immunoglobulin G (IgG) binding data and experimentally test thousands of GB1 designs to systematically evaluate the models’ extrapolation. We find each model architecture infers markedly different landscapes from the same data, which give rise to unique design preferences. We find simpler models excel in local extrapolation to design high fitness proteins, while more sophisticated convolutional models can venture deep into sequence space to design proteins that fold but are no longer functional. We also find that implementing a simple ensemble of convolutional neural networks enables robust design of high-performing variants in the local landscape. Our findings highlight how each architecture’s inductive biases prime them to learn different aspects of the protein fitness landscape and how a simple ensembling approach makes protein engineering more robust.
Subject terms: Protein engineering, Machine learning, Protein design, Molecular biophysics, Computational models
Machine learning accelerates protein engineering by predicting sequence-function relationships. Here, authors evaluate neural network architectures’ ability to extrapolate beyond training data, finding simpler models excel in local design while convolutional models explore deeper sequence spaces.
Introduction
Protein engineering can be envisioned as a search over the sequence-function landscape to discover new proteins with useful properties1. Machine learning (ML) accelerates the protein engineering process by integrating experimental data into predictive models in an automated fashion to direct the landscape search2. These models improve the quality of protein variants tested and reduce the total number of experiments needed to engineer a protein3. ML-assisted protein engineering approaches have expedited engineering of diverse proteins such as viral capsids, improved fluorescent proteins, and highly efficient biocatalysts4–7.
Broad classes of machine learning and deep learning techniques approach protein design from different perspectives8–11. We focus on supervised learning, which learns from experimental sequence-function examples to predict new sequence configurations with some intended biochemical/biophysical properties. There are many different classes of models that each make different assumptions about the underlying landscape, influencing the way sequence-function relationships are learned2. For example, linear models assume additive contributions from individual mutations and are unable to capture epistatic effects. More sophisticated convolutional neural networks use convolving kernels to extract meaningful patterns from the input data that capture long-range interactions and complex, non-linear functions12. Many studies have assessed the predictive performance of ML models on existing protein sequence-function datasets11–16, but there is little work that rigorously benchmarks performance in real-world protein design scenarios with experimental validation5,7,17. ML-guided protein design is inherently an extrapolation task that requires making predictions far beyond the training data, and evaluating models in this task is challenging due to the massive number of sequence configurations that must be searched and tested18.
In this paper, we evaluate different neural network architectures’ ability to extrapolate beyond training data for protein design. We develop a general protein design framework that uses an ML model to guide an in silico search over the sequence-function landscape. We use this approach with different model architectures developed in previous work from our lab to design thousands of protein G (GB1) variants that sample a massive sequence space far outside the model’s training regime. The different models prioritize distinct regions of the landscape and display unique preferences for the sequence positions mutated and types of amino acid substitutions. We then experimentally test the designs using a high-throughput yeast display assay that evaluates variant foldability and IgG binding. We find all models show the ability to extrapolate to 2.5-5x more mutations than the training data, but the design performance decreases sharply with further extrapolation. The simple fully connected neural networks showed the best performance for designing variants with improved binding relative to wild-type GB1. Intriguingly, we also found that the parameter-sharing convolutional models could design folded, but non-functional, proteins with sequence identity as low as 10% from wild type, suggesting these models are capturing more fundamental biophysical properties related to protein folding. Our high-throughput screen identified multiple designs with improved binding relative to wild-type GB1 and previously designed variants. Our work provides a rigorous assessment of model architectures’ capacities for protein design and will support ongoing advances in ML-driven protein engineering.
Results
Extrapolating learned protein fitness landscapes
Protein sequence space is nearly infinite and experimental methods can only characterize a very localized and minuscule fraction of this space1. Machine learning (ML) models trained on sparse sequence-function data infer the full fitness landscape and can make predictions for previously unobserved protein sequences. These models can guide a search through sequence space to discover protein designs with high predicted fitness (Fig. 1a). Many of these predictions extend far beyond the training data and thus are extrapolations on the fitness landscape. Although model performance is known to degrade as predictions are made further from the training regime, it remains unclear how far these models can extrapolate to design high-fitness sequences5,7.
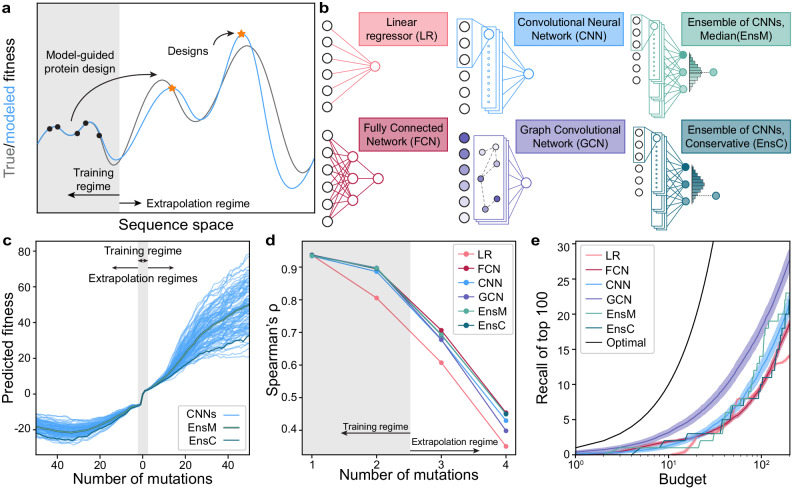
Fig. 1. Extrapolation of sequence-function models.
a Supervised sequence-function models are trained on experimental data and can make predictions across the fitness landscape. ML-guided protein design seeks to identify high-fitness sequences and often involves model extrapolation beyond the training regime. b We tested five model architectures that capture distinct aspects of the underlying sequence-function landscape. c A collection of 100 CNN models and their divergence when predicting deep into sequence space along a mutational trajectory. The ensemble predictor EnsM represents the median of the 100 CNNs, while EnsC is the 5th percentile. d We trained models on GB1 single and double mutants and predicted the fitness of 1-, 2-, 3-, 4-mutants. The Spearman’s rank correlation was determined between the model’s predicted fitness and experimental fitness. e Model recall of the top 100 protein variants within a design budget. Recall represents the number of the true top 100 4-mutants that are present in a model’s top N predictions, where N is the design budget. Optimal represents a theoretical model that always predicts the top N proteins. Shading represents 95% confidence intervals across 100 individually trained models, excluding EnsM and EnsC. The confidence interval is centered on the mean recall. Source data are provided as a Source Data file.
The B1 binding domain of streptococcal protein G (GB1) is a small 8-kDa domain that binds to the mammalian Immunoglobin G (IgG) fragment crystallizable (Fc) region with nM affinity and to the fragment antigen-binding (Fab) region with M affinity19,20. GB1 consists of 56 amino acids. We use GB1 as a model protein due to its extensively characterized IgG Fc-binding fitness landscape containing nearly all single and double mutations that was collected using a high-throughput mRNA display assay20. In previous work, we trained four different classes of supervised neural networks on GB1 double mutation data12. Briefly, these models were trained on sequence-function data from ~500k single and double mutants and the sequences were encoded using a fixed length of 56 amino acids and a physiochemical and categorical (one-hot) amino acid embedding. The networks included a linear model (LR) that considers each residue position independently, a fully connected network (FCN) that can capture nonlinear behavior and epistasis, sequence-based convolutional neural networks (CNN) that share parameters across the protein sequence, and a structure-based graph convolutional neural network (GCN) that considers residues’ context within the protein structure (Fig. 1b). We previously evaluated these models’ predictive performance on held out double mutant data and demonstrated the feasibility of ML-guided protein design. However, it remains unclear how these models perform when extrapolating deeper into sequence space.
Neural networks contain hundreds of thousands to millions of parameters, many of which are not constrained by the training data21. The values of these unconstrained parameters are greatly influenced by the random initialization during training, and we hypothesized these parameters may lead to model divergence when predicting away from the training data regime. We trained 100 CNNs with the same model architecture, hyperparameters, and training data but with different random initializations of model parameters. We then evaluated each model’s predictions along a mutational pathway across the fitness landscape (Fig. 1c). All models show close agreement in the training data regime within two mutations of wild-type GB1, but their predictions increasingly diverged the further they moved from the training regime. It is also notable that the fitness predictions in the extrapolated regime have values so extreme they are unlikely to be valid. To overcome model variation arising from random parameter initialization, we implemented neural network ensemble predictors EnsM and EnsC that input a sequence into all 100 CNNs and return the median or lower 5th percentile predictions for that sequence, respectively. We view EnsM as an average predictor, while EnsC is a conservative predictor because 95% of the models predict higher fitness values.
We evaluated the neural networks trained on single and double mutants on a separate GB1 dataset containing nearly all combinations of mutations at four positions known to have high epistatic interactions22. This combinatorial dataset used the same methods to evaluate fitness as the training dataset and reported a strong correlation (r = 0.97) for single and double mutants assessed in both experiments. In our assessment, single and double mutants are within the training regime, while prediction on the 3- and 4 mutants requires model extrapolation. We found all models displayed significantly decreased predictive performance when extrapolating away from the training data (Fig. 1d, Supplementary Fig. 1). Although model accuracy drops dramatically in the extrapolated regime, the Spearman’s correlation remains significantly above 0, suggesting potential for model-guided design at or beyond 4 mutations. All the nonlinear neural network models performed similarly, while the linear model displayed notably lower performance, presumably due to its inability to model epistatic interactions between mutations.
We additionally tasked the models with identifying the most fit variants from the set of 121,174 possible 4-mutants, a task similar to machine-learning-guided design. For a given testing budget N, each model ranks all 4-mutants and selects the top N. Recall is calculated as the proportion of the true top 100 that is represented in the model’s predicted top N sequences. For every budget tested, the GCN has the highest recall, indicating better extrapolation to identify high fitness variants (Fig. 1e). The FCN also has high recall with small design budgets but is quickly surpassed by the CNN.
ML-guided protein design for deep exploration of the fitness landscape
A 56-residue protein like GB1 has 2056 (>1070) possible sequence configurations, and we must search deep into sequence space to fully evaluate ML models’ performance for protein design. We developed a large-scale protein design pipeline that uses simulated annealing (SA) to optimize a model over sequence space to identify high fitness peaks. The approach executes hundreds of independent design runs to broadly search the landscape, clusters the final designs to remove redundant or similar solutions, and then selects the most fit sequence from each cluster to provide a diverse sampling of sequences predicted to have high fitness (Fig. 2a). The number of clusters can be adjusted to match any downstream gene synthesis or experimental budgets. We evaluated the convergence of our SA runs (Supplementary Fig. 2) and the performance of parallel tempering methods (Supplementary Fig. 3).
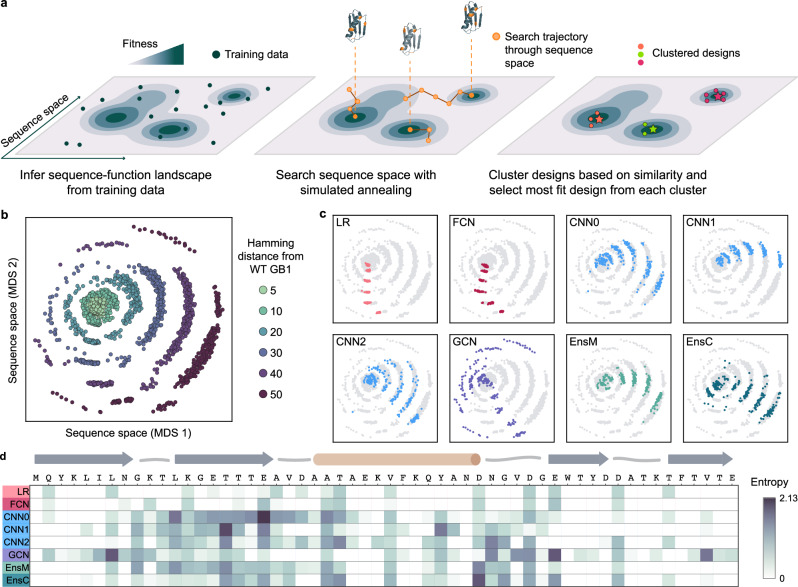
Fig. 2. ML-guided fitness landscape exploration.
a Supervised models infer the fitness landscape from sequence-function examples. We use simulated annealing (SA) to search through sequence space for designs with high predicted fitness. We perform hundreds of independent SA runs to broadly search sequence space, cluster designs to map distinct fitness peaks, and select the most fit sequence from each cluster (shown as a star). b We visualized all designs using multidimensional scaling (MDS) and found the designs occupy concentric rings emanating from wild-type GB1 with increasing number of mutations. c We colored the MDS visualization by model architecture and found individual models design sequences that occupy distinct regions of sequence space. d We calculated the sequence diversity across GB1 positions for the 10-mutant designs. We used Shannon entropy to quantify amino acid diversity at each position; low entropy indicates few amino acid options at a given site while high entropy indicates many amino acid options. The low entropy for the LR and FCN indicate that each model repeatedly proposes the same mutations at the same positions. The convolutional models propose sequences with more diversity spread across more positions, especially in regions of positive epistasis. Source data are provided as a Source Data file.
We applied our pipeline to design a diverse panel of GB1 variants testing different model architectures and spanning a range of extrapolation distances. We included eight models: LR, FCN, three CNNs with different initializations (CNN0, CNN1, CNN2), GCN, and the ensembles EnsM and EnsC. For each model, we designed variants at six extrapolation distances: 5, 10, 20, 30, 40, and 50 mutations from wild-type GB1. For each model-distance combination, we ran at least 500 design runs and clustered the designs into 41 clusters, to obtain 41 diverse sequences for each criterion. We visualized the design space using multi-dimensional scaling, which organizes sequences in a 2D space that attempts to preserve sequence interrelationships. We observe that sequences occupy concentric rings expanding outward from wild-type GB1, with each successive ring representing an increased mutation distance and the outermost ring corresponding to the 50-mutants (Fig. 2b).
We observed notable differences in the sequences designed by each model, suggesting each architecture prioritizes distinct regions of the landscape (Fig. 2c, Supplementary Fig. 4). The LR and FCN designs occupy similar regions of sequence space and tended to display less variation within the 41 designs, suggesting a smooth inferred landscape structure with a major prominent peak. The designs from the three CNNs were distinctly different than the LR and FCN designs and displayed a high degree of variation across each model, highlighting how random parameter initialization can greatly influence model extrapolation. The GCN designs were by far the most diverse, occupying all directions in sequence space and having the highest average Hamming distance, reminiscent of a landscape with many distinct fitness peaks. At lower mutational distances, EnsM and EnsC produced designs that were more similar to that of LR and FCN, but in higher mutational regimes, their designs were more similar to designs produced by individual CNNs (Supplementary Fig. 4). We also explored the influence of model initialization on the designs by randomly initializing five of each model (LR, FCN, CNN, GCN), designing sequences at all mutation distances, and comparing the similarity of the designs (Supplementary Fig. 5). We found that the sequences designed by the LR, FCN, and CNN architectures are more similar to other initializations of that architecture than other architectures, indicating the model architecture itself is a driving factor for the differences in the designs. The GCN models show high sequence variability across different model initializations.
We further explored the unique mutational variation of each architecture’s designs by analyzing the amino acid diversity at each site (Fig. 2d). Generally, the LR and FCN target only a few positions in the protein sequence and tend to propose the same mutations, as indicated by the low entropy value. We also looked at the BLOSUM scores of the designs and found most designs introduced non-conservative mutations with negative BLOSUM scores, with the exception of the EnsC designs with 5 mutations (Supplementary Fig. 6). Every mutation proposed by the LR and FCN are individually beneficial (Supplementary Fig. 7). In contrast, the CNNs and GCN target a much broader region of the GB1 sequence, including known sites of positive epistasis around residues 9–16 and 33–4020. This suggests the CNN and GCN models can exploit epistasis to design sequences composed of synergistic mutations.
Large-scale experimental characterization of ML-designed GB1 variants
We used yeast surface display to characterize the expression and IgG binding of the designed GB1 variants. We sorted variants into display and IgG binding populations using fluorescence-activated cell sorting (FACS) (Fig. 3a, b, Supplementary Fig. 8) and sequenced these sorted populations to determine which variants fell into each bin. We used enrichment to devise display and IgG binding enrichment scores, and , respectively, for each variant. The display and binding enrichments show good reproducibility between experimental replicates and internal standards with varied nucleotide sequences but identical amino acid sequences. The original experiment used RNA display to measure binding affinity. We included 25 sequences (all double-mutants) from the training dataset with a uniform distribution across experimental fitness as a fitness calibration set and observed good correlation between our results and the original dataset (Supplementary Fig. 9). The display score captures a combination of protein expression, trafficking, folding, and stability, while the IgG binding score is more directly related to IgG binding affinity. Additionally, display is a prerequisite for binding because the GB1 protein must reach the cell surface to interact with IgG.
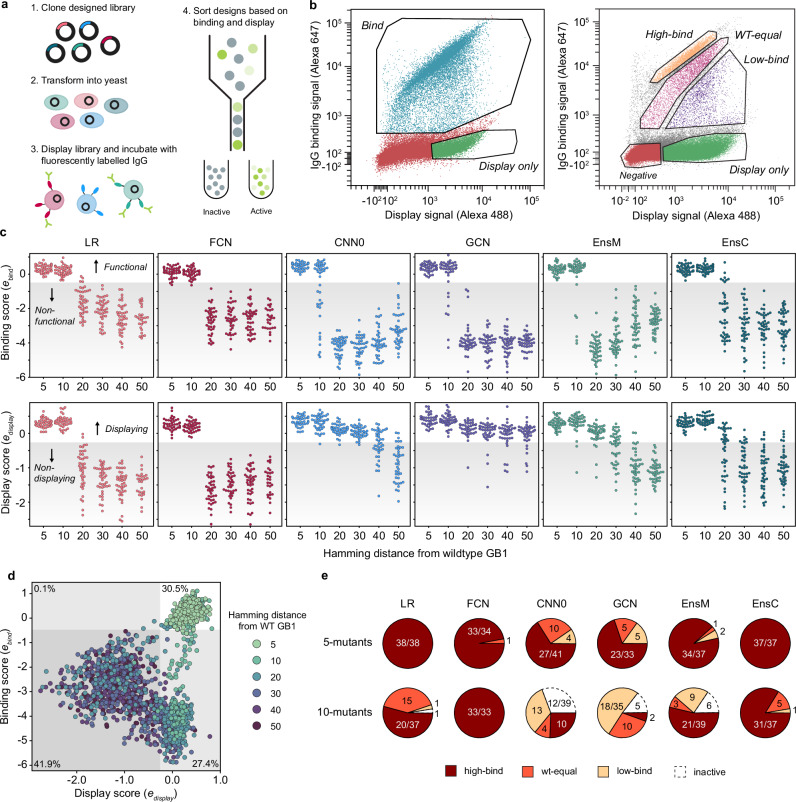
Fig. 3. Experimental characterization of ML-designed GB1s.
a An overview of our yeast surface display method to measure IgG-binding of GB1 designs. b FACS scatter plots and sorting gates for the library sorting experiments. In our first qualitative experiments, we sorted variants into bind (blue) and display only (green) populations. Events outside of these gates (red) were not sorted. In our second quantitative experiment, the library was sorted into display-only (green), low-bind (purple), wt-equal binding (pink), or high-bind (tan) categories. The negative population (red) and events falling outside of gates (gray) were not sorted. c The binding and display scores for each model as a function of the Hamming distance from wild-type GB1. Hamming distance reports the number of positions where the amino acids of two aligned sequence are different. In the plot, each point corresponds to a single design and the shaded region specifies the threshold between functional/nonfunctional and displaying/non-displaying. d A scatter plot of display and binding scores for each design. The gray-shaded regions specify the thresholds between functional/nonfunctional and displaying/non-displaying. The percentage of designs falling within each quadrant is specified in each quadrant’s corner. e The distribution of 5-mutant and 10-mutant designs categorized as high-bind, wt-equal, low-bind, or inactive from the quantitative experiment. Most designs beyond 10 mutations were inactive. Source data are provided as a Source Data file.
We found all models were successful at designing functional GB1 variants that displayed and bound IgG (Fig. 3c). The design success rate was high at five and ten mutations and significantly decreased with further extrapolation from the training data regime. The simple LR and FCN models outperformed the more sophisticated CNN and GCN models for designing functional sequences that bind to IgG. Interestingly, the ensembles, which were composed of CNNs, showed design performance comparable to the LR/FCN models. This suggests the CNN random initialization process may result in models with subpar performance, but this effect can be marginalized by averaging over many models. CNN ensembles combine the strong generalization performance of parameter-sharing models12 with the extrapolative design ability of simpler LR/FCN models.
The model design results for the display score were strikingly different than for IgG binding. While the LR and FCN design success decreased sharply with increasing extrapolation, the sequences designed by the CNN and especially the GCN displayed out to 50 mutations. The picture becomes more complete when we jointly visualize the binding and display scores over extrapolation distances (Fig. 3d). The sequences can be broadly categorized into the four quadrants binds/doesn’t bind and displays/doesn’t display. The non-binding LR and FCN designs tended to not display, while the non-binding CNN and GCN designs displayed, suggesting two distinct modes of functional inactivation (Supplementary Figs. 10 and 11).
We can contextualize these results if we consider that stable, folded proteins are more likely to display on the yeast surface, while unstable proteins are prone to proteolytic degradation23. In this case, the LR/FCN designs are inactive because they are unfolded, while the CNN/GCN designs are folded but may have a defective IgG-binding interface. The model architectures and their intrinsic inductive biases may explain the differences in each model’s design behavior. The LR/FCN models put an emphasis on specific sites and how those sites contribute to function. In contrast the CNN/GCN models have parameter-sharing architectures that learn general patterns across the protein sequence. The site-focused designs from the LR/FCN models will maintain an intact IgG-binding interface, at the potential cost of losing protein stability. While the CNN/GCN models learn more general rules related to protein folding, but in the process, put less emphasis on the IgG binding interface.
We observed that some of the yeast display results may be explained by the presence of KEX2 protease sites in the designed proteins. KEX2 is located in the yeast Golgi and cleaves secreted proteins with exposed Lys/Arg-Arg sites. Unfolded proteins with KEX2 sites will be cleaved and not display, while folded proteins with KEX2 sites will typically display24. We found designs with 20 or more mutations are less likely to display if they have a KEX2 site (Supplementary Fig. 12). We also found the LR and FCN models had a higher likelihood of designing sequences with KEX2 sites (Supplementary Fig. 13).
We performed an additional yeast display experiment to resolve more quantitative differences in IgG binding by sorting variants into four bins corresponding to greater than WT binding (high-bind), WT-like binding (WT-equal), lower than WT binding (low-bind), and no IgG binding (display only) (Fig. 3b). Our results reveal that all model architectures succeed in designing high-binding GB1 variants with five and ten mutations (Fig. 3e). When considering the individual models, the LR and FCN significantly out-perform the CNNs and GCN. Nearly 100% of LR and FCN 5-mutant designs are categorized as high-bind. At 10 mutations, all FCN designs and ~50% LR designs are high-binding. While the GCN and CNNs design functional 10-mutants, many are low-bind or inactive. In contrast to the individual CNNs, the ensembles are as successful as the LR and FCN in designing high-bind GB1 variants, suggesting that ensembles of models confer some benefit when extrapolating beyond the training regime.
Structural diversity of ML-designed GB1s
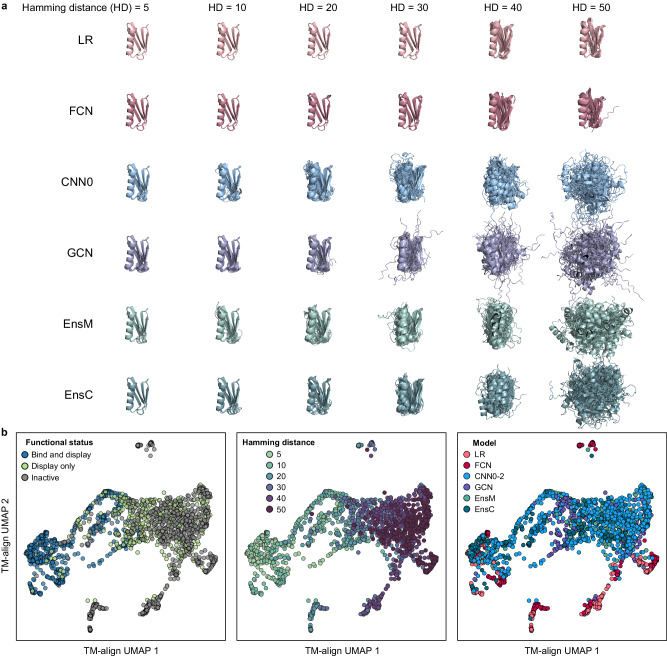
We predicted the structures of the designed GB1 using AlphaFold224,25 and found that designs further from wild-type GB1 showed increased variability in their predicted three-dimensional structures (Fig. 4a). There were notable differences between the models with the LR and FCN models designing sequences with much tighter conformational distributions across all Hamming distances that closely resemble the WT GB1 structure. To analyze structural differences in the library, we used TM-align26 to calculate the degree of alignment between each pairwise structure (TMscore), and used UMAP27 to visualize high-dimensional structure space as a 2D projection (Fig. 4b). The predicted structures tend to cluster by the designs’ functional status with designs that bind IgG tending to have structures similar to WT GB1.
Fig. 4. AlphaFold predictions of ML-designed GB1s.
a Predicted structures across models and Hamming distances. Each model-distance combination has 41 overlayed structures corresponding to each design. b UMAP visualization of predicted structures showing clustering and organization with functional status, Hamming distance, and design model. Source data are provided as a Source Data file.
ML-designed GB1s show improved display and IgG binding
We used the high-throughput screening data to identify interesting designs for more detailed functional and structural analysis (Supplementary Table 1). There were no 30/40/50-mutants that bound to IgG in our high-throughput screen (Fig. 3c), so we chose five 5/10-mutants that showed high IgG binding and five additional 20-mutants that showed moderate IgG binding. We also chose ten distant designs with 40–50 mutations that displayed but did not show IgG binding to assess their display relative to WT and confirm these designs lacked detectable IgG binding. We quantitatively evaluated display and IgG binding for each of these 20 designs in a clonal yeast display assay (Fig. 5a).
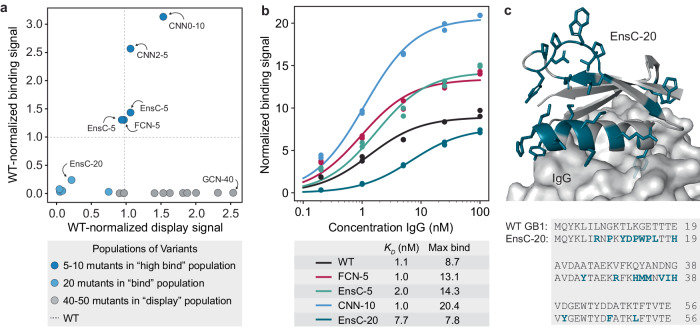
Fig. 5. Validation of high-throughput yeast display screen.
a Clonal yeast display assay to verify designs’ display and IgG binding properties. The display and binding signals were normalized to wild-type GB1. b IgG binding curves for wild-type GB1 and designs FCN-5 (a 5-mutant designed by FCN), EnsC-5 (a 5-mutant designed by EnsC), CNN-10 (a 10-mutant designed by a CNN) and EnsC-20. We analyzed cells using flow cytometry and the normalized binding signal is the ratio of IgG binding to display level. We estimated the and max binding signal parameters by fitting the data to the Hill equation. c AlphaFold2 predicted structure of the binding 20-mutant variant EnsC-20. The mutated residues are shown as sticks and highlighted in teal. IgG was included from the GB1 crystal structure (PDB: 1FCC). Source data are provided as a Source Data file.
All five 5/10-mutant high-binding designs showed IgG binding greater than wild type, while several of the 20-mutant binding designs showed IgG binding, but to a diminished level relative to WT. We performed IgG titrations to assess the binding curves for several designs (Fig. 5b). While the designs do not have significantly different KDs from wild type, the maximum binding signals are significantly larger for the designs, suggesting our designs have decreased koff rates in our yeast display assay. We hypothesize the initial GB1 training data mRNA display experiments20 may have captured koff effects rather than equilibrium KD measurements and thus resulted in trained models that design GB1 variants with improved binding kinetics but not necessarily thermodynamics. The most distant functional design was a 20-mutant designed by EnsC, termed EnsC-20, that showed significant IgG binding, although much weaker than wild type. Many of the mutations in EnsC-20 are at the IgG interface (Fig. 5c), while others, including three mutations to proline, were on the distal side of the protein, likely inducing structural changes that could affect binding.
All ten 40/50-mutant display designs displayed on the yeast surface equal to or greater than wild type GB1, including a 40-mutant designed by the GCN that displayed over 2× higher than wild type. This variant, called GCN-40, shares less than 30% sequence identity with wild-type GB1 and its structure is predicted to have some similar secondary structure elements, but with a completely new helical bundle fold (Supplementary Fig. 14).
Discussion
Protein engineering has broad applications across biocatalysis, biomanufacturing, agriculture, and human health, but engineering proteins is slow and requires expensive rounds of experimental characterization to search the fitness landscape. Machine learning accelerates the protein engineering process by inferring the underlying landscape structure and reducing the experimental burden required to explore protein sequence space. In this work, we assessed different supervised machine learning architectures’ ability to extrapolate beyond the training data for protein design. Adapting previously developed models trained on a large GB1 sequence-function dataset, we performed an ML-guided search to design sequences predicted to have high fitness. We found each model had markedly different perceptions of the underlying landscape that gave rise to unique preferences for the designed sequences. All models showed strong potential to extrapolate to 2.5-5x more mutations than represented in the training data, but the simpler fully connected architecture performed the best for designing functional and highly fit proteins.
Our work rigorously evaluates a common protein engineering setting wherein a supervised sequence-function model is trained on a local fitness landscape and tasked with extrapolating to new regions of sequence space5,6,11,28,29. These models become increasingly inaccurate further from the training data due to the challenge of generalizing local sequence rules to the global landscape, which is exacerbated by factors like landscape ruggedness, data sparsity, and magnitude of epistasis. Models that effectively capture epistasis and that can learn generalizable biophysical concepts will be more capable of extrapolating on the fitness landscape. We found inconsistences between in silico metrics calculated by holding out variants from a fixed dataset and experimental design outcomes. We considered how each model trained on single and double mutants ranks quadruple mutants (Fig. 1d) and found that the LR significantly underperformed relative to the more sophisticated models. This contrasts with the experimental design results (Fig. 3e), where the LR consistently designed high-bind variants with 5 and 10 mutations and was one of the top models. Similarly, our analysis found CNN/GCN models that perform well in predicting 4-mutants (Fig. 1d) tend to underperform when extrapolating to 5, 10, and 20 mutations (Fig. 3a, e). These findings highlight differences between local and global landscape structures and the need to design and experimentally test sequences to evaluate model performance. In silico metrics are widely used to guide modeling and design choices and define new state-of-the-art standards, but may not always translate to practical protein engineering settings.
A given model’s extrapolation ability will depend on the size and quality of the training data, the model’s inductive biases and its capacity to learn sequence-function relationships, and the ruggedness of the optimization surface, which can limit search methods’ ability to identify top solutions. Despite simulated annealing’s (SA) ability to bypass local optima, we observed notable differences in the spread of each model’s designs (Fig. 2c, Supplementary Fig. 2), suggesting varied degrees of ruggedness in the underlying optimization surface. The SA trajectories for the LR designs tended to converge toward the same sequences, as expected for a smooth landscape with a single global peak. In contrast, the GCN designs were broadly distributed across sequence space, indicating each SA trajectory had arrived at distinct local optima due to the difficulty in traversing a highly rugged optimization surface.
We found the LR and FCN models strongly outperformed the convolutional models in designing functional 5- and 10-mutants that bound IgG tighter than wild-type GB1. These models have a simple architecture where each position in the amino acid sequence contributes to function and highlights the relative simplicity of the local landscape for a given property. Our LR model is similar to Addition models that predict variant fitness by summing individual mutational effects30 (Supplementary Fig. 15). Additive mutational effects have been known and leveraged in protein engineering for decades31,32, and the LR and FCN’s inductive biases are set up to capture these simple relationships. FCNs have the additional advantage that they can learn epistatic relationships between residues, which resulted in increased performance relative to LR. The primary distinguishing feature of the convolutional models is their parameter-sharing architecture that learns filters that are applied across the protein sequence/structure. Convolutional models have the capacity to learn more general relationships, but their inductive biases are not primed to capture the simpler additive and epistatic relationships that dominate local landscapes.
We interpret our results primarily through the lens of protein engineering where our goal is to design GB1 variants with improved binding to IgG. But we found the CNN, and especially the structure-based GCN, were highly effective at designing GB1s with up to 50 mutations that display on the yeast surface and presumably fold, but do not bind IgG. It’s worth noting that this phenomenon doesn’t necessarily indicate subpar model performance, and instead we postulate the parameter-sharing architectures may have focused more on learning general rules of protein folding and, in the process, ignored IgG binding activity. While the original sequence-function training data was based on IgG binding, this experimental mapping is dominated by folding effects because a majority of a protein’s residues are involved with maintaining structural stability, while relatively few are directly involved with binding. In other words, a mutation can inactivate a protein due to disrupted folding or disrupted binding, but the disrupted folding mechanism is statistically much more likely. It’s well known that most mutations destabilize proteins33 and this observation was noted in the original GB1 deep mutational scanning manuscript20. We observed that the CNN and GCN propose mutations in regions of positive epistasis. Considering that epistatic interactions are critical to stability, this feature likely contributes to the CNN and GCN’s capacity to design variants that fold despite lacking binding activity. The combination of data that’s dominated by folding effects and the convolutional models’ inductive bias to recognize patterns led to models that learned more general rules of protein folding.
We found ensembles of CNNs outperform individual CNNs in designing high-fitness variants. Ensembles highlight model variation arising from random parameter initialization and can identify regions of sequence space that are poorly understood by the model. Being able to identify where a model is not confident is critical for robust design. Model confidence can be estimated by inputting a single sequence into an ensemble of models and evaluating the agreement in prediction across the models34. If the models tend to agree, we can be more confident in the prediction, while if the ensembles’ predictions are highly variable, then the random parameter initialization may be influencing the models’ prediction. Evaluating this consensus between models is important for neural network models that may have millions or more internal parameters that are not fully determined by the data. Other work has used model uncertainty to guide designs toward regions of sequence space that are generally more confident21,35–37. For our EnsC predictor, we do not restrict design space based on uncertainty, but rather use the ensemble to make a conservative fitness prediction where 95% of the models agree the fitness is above a certain value. Designing sequences that maximize EnsC result in designs that most models agree have high fitness. We found the EnsC design strategy was the best overall with similar performance to LR and FCN for designing highly fit 5- and 10-mutants and some ability to design functional sequences with 20 mutations.
Our models were trained on GB1-IgG binding data with no explicit structure, folding, or stability information. For many proteins including GB1, folding is a prerequisite for function. The protein must fold into a stable structure before it can perform its function. For this reason, folding is implicitly captured by a functional assay. Every GB1 variant that binds IgG is also a stably folded protein. Function is often dictated by a handful of specific residues, while folding is a more global property of many residues interacting. Statistically, there are many more residues that contribute to folding versus function, so most high-throughput mutagenesis datasets have a major folding component. The inductive biases of LR and FCN models directly capture how specific residues contribute to function. In contrast, parameter-sharing models such as CNNs/GCNs learn patterns that map to the output function. We postulate the inductive biases of CNNs/GCNs to learn patterns, in addition to the fact that IgG binding has a major folding component dictated by underlying sequence patterns, causes the CNN/GCN models to pick up on folding signals. In other words, the measured IgG binding function has both protein-protein binding and folding components and the CNN/GCN models are more primed to learn the folding component. This naturally leads to the idea that these differing architectures could be used together to design functional proteins more robustly. A collection of model architectures, each with their own inductive biases, could be used together in an ensemble, similar to EnsC. Another design strategy could aim to jointly optimize multiple models simultaneously using multi-objective criteria38.
Our work reveals general guidelines for protein engineering. Despite the highly epistatic underlying landscape, the simpler LR and FCN models outperform the convolution-based models in local design tasks. This may be attributed to the training data, which comprehensively characterizes all single and double mutants. In this setting, models with a strong linear bias may be sufficient for engineering proteins near the training data and lends further support to the general observation that mutation effects are largely additive. We also found that CNN ensembles improve model design performance over the individual component CNNs, likely because it reduces the aleatoric uncertainty associated with random parameter initialization. Based on this finding and other work, we expect model ensembles will generally improve performance over individual models. This approach also provides the opportunity to ensemble different model architectures, which may improve design robustness; ensembles of different model classes have been successfully used for protein and metabolic engineering7,39. Our work also demonstrates that, regardless of architecture, learning a landscape from purely local data is not sufficient to extrapolate function to distant sequences and it’s best to stay close to the training data. Combining the above findings, we postulate the most reliable design method would consist of an ensemble of FCNs, designing sequence according to the EnsC predictor, and staying within 5-10 mutations from wild type. This optimal design strategy is likely dependent on the particular protein of interest, the function being assayed, and the quality and size of the training data.
Biophysical and molecular mechanics-based models have dominated the fields of rational protein engineering and design for decades. Proteins are challenging to model from a physical perspective because they are composed of thousands of atoms that are dynamically coupled over multiple length and time scales and the molecular basis of function is often poorly understood. Machine learning approaches overcome these limitations by directly learning the relationships between protein sequence and function from experimental data. Our work in this paper focuses on supervised neural network models to learn sequence-function relationships and design new proteins. Other recent work in the field uses pretrained protein representations that are learned from vast existing sequence and structure databases. Models such as UniRep and ESM2 are pretrained using self-supervised learning to identify general patterns across millions of natural protein sequences. These models transform input sequences into context-aware protein representations that are input into downstream supervised models to learn sequence-function relationships. An interesting future research direction would be exploring the ability of these pretrained models for extrapolative design on the protein fitness landscape7,39.
Deep learning and artificial intelligence are revolutionizing the fields of protein science and engineering. These tools can process, learn from, and make sense of large quantities of data to decode the complex inner workings of proteins with a scale and resolution beyond human comprehension. Continued advances will help realize the potential of protein design to address society’s most pressing problems and future global challenges.
Methods
Neural network model training
We used the Olson et al. GB1-IgG binding dataset20 and the Gelman et al. model architectures12 with the same parameter initializations, train-validation-test splits, and hyperparameters. We additionally trained 100 models of each architecture with different random initializations to assess the effects of parameter initialization and for ensemble learning. We constructed two CNN ensemble predictors: a median predictor (EnsM) that inputs a sequence into 100 CNN models and outputs the median fitness prediction, and a conservative predictor (EnsC) that inputs a sequence into 100 CNN models and outputs the 5th percentile fitness prediction.
For model training and library design, protein sequences were embedded as a concatenated vector containing a physicochemical and categorical (one-hot) representation of the amino acid sequence. Embeddings are a fixed length across all sequences and contain no gap characters.
Evaluation of model extrapolation on 3- and 4-mutant fitness landscapes
We evaluated the models’ ability to extrapolate to 3- and 4-mutants using the four-site combinatorial GB1 dataset from Wu et al. 22 (Dataset: 10.7554/eLife.16965.024) This dataset consists of experimental measurements for 93% of variants from a 4-site combinatorial saturation mutagenesis GB1 library screened for binding to IgG. The models were trained on single and double mutant data from Olson et al. 20 and were used to predict the fitness values of the 3- and 4-mutants. We also tested the ability of the models to identify the 4-mutants with the highest fitness values. The model recall was calculated by giving a model a design budget n, having the model rank all 121,174 characterized 4-mutants and select the top n, and then determining what percentage of the true top 100 variants were in this top n set. An optimal model would achieve 100% recall with a design budget n = 100.
Model-guided protein design
We designed GB1 variants with eight different models at 5, 10, 20, 30, 40, and 50 mutations from wild-type GB1 (Supplementary Data 1). For each model-distance combination, we designed 41 diverse GB1 variants with high predicted fitness. For a given model-distance combination, we used simulated annealing (SA) to maximize the model’s predicted fitness constraining the number of mutations allowed in the designed protein. To ensure the number of mutations in a design were fixed, we exchanged random mutations. For example, when designing a 5-mutant, we might randomly choose two current mutations, mutate those back to the wild-type amino acids, and then choose two new random mutations to keep the total mutations fixed at five. At each step of simulated annealing, one or more mutations (sampled from a Poisson distribution) were randomly exchanged and predicted fitness of the given model was evaluated. Mutations that improved predicted fitness were automatically accepted while all other mutations were accepted with probability where T is decreased along a logarithmic temperature gradient ranging from 103 to 10−5 over 15,000 to 50,000 steps. We performed 500 independent simulated annealing runs for each model-distance combination, which typically resulted in more than 41 unique candidate sequences. The individual SA trajectories showed strong convergence to similar fitness values indicating a sufficient number of optimization steps (Supplementary Fig. 2). For some model-distance combinations, 50,000 SA steps converged on fewer than 41 candidate sequences, in which case, we decreased the number of SA steps until more than 41 unique candidate sequences were designed in 500 simulations. The number of SA steps for these specific categories were: (LR 5-mutants) and (FCN 5-mutants) = 10,000 steps and (LR-10 mutants), (FCN-10 mutants), (EnsM-5 mutants), and (EnsC-5 mutants) = 25,000 steps. All sequence designs were run in parallel using high-throughput computing40. To select a diverse and representative set of designs, we performed K-means clustering on the 500 independent designs using 41 clusters and selected the variant from each cluster with the highest predicted fitness for experimental characterization. This resulted in 41 unique GB1 designs for 8 models at 6 distances, for a total of 1968 designs.
BLOSUM analysis
We calculated the mean site-wise BLOSUM score for each design category using the BLOSUM62 substitution matrix. All BLOSUM scores were calculated with respect to WT. Since our objective was to illustrate whether mutations from WT were conservative or non-conservative, non-mutated residues were excluded from analyses. As such, reported mean BLOSUM scores only reflect the nature of mutations and not the effect of non-mutated residues. The mean site-wise BLOSUM score is the average BLOSUM score for mutated residues at a given site.
UMAP visualization of GB1 variant library structure space
We used Alphafold24,25 (AlphaFold Colab v2.3.2) to predict the structure for each GB1 variant. AlphaFold uses an input MSA to predict the query sequence structure. We used WT GB1 to build an MSA using a Jackhmmer search on the Uniref90, Small BFD, and Mgnify sequence databases as recommended by AlphaFold Colab. This MSA was used as the input MSA for all structure predictions. We generated six AlphaFold predictions for each sequence; the structure with the highest mean pLDDT (measure of structure confidence) was chosen as the representative model.
We generated a UMAP projection of the GB1 library structure space according to methods previously described41. Briefly, we calculated TMscores for all pairwise structures using TM-Align26 and used TMscore-1 as a distance metric for the UMAP projection. We performed a hyperparameter scan for n_neighbors and min_dist and selected a representative projection (n_neighbors = 15, min_dist = 0.1). General spatial patterns are conserved across hyperparameter settings.
High-throughput characterization of GB1 designs via yeast surface display
We codon optimized our GB1 designs for expression in Saccharomyces cerevisiae using the GenSmart Codon Optimization tool (GenScript) and excluded the BsaI, NheI, BamHI, MluI, and XhoI restriction enzyme sites. We also identified 25 sequences from the training data with a broad range of fitness values to correlate our fitness measurements with the original data from Olson et al. 20. Each of these variants was designed with two different synonymous codon sequences to provide internal controls to ensure reproducibility of our fitness measurements. The designed genes and control sequences were synthesized as an oligonucleotide pool by Twist Biosciences with flanking sequences to allow PCR amplification and downstream cloning.
The GB1 gene library was cloned into the yeast surface display vector pCTCON2 (provided by Dane Wittrup, MIT)42. To prepare the vector for library cloning, we (1) removed the BsaI restriction site from the AmpR gene using site directed mutagenesis (for: 5′-AGCGTGGGTCGCGCGGTATCA-3′; rev: 5′-CACCGGCTCCAGATTTATCAGC-3′) and (2) added in BsaI sites for cloning library sequences (for: 5′-GAAGGGTCTCTGATCCGAACAAAAGCTTATTTCTGAAG-3′; rev: 5′-GTATTGGTCTCTCTAGCCGACCCTCCGC-3′). We amplified the oligonucleotide pool using either Phusion Hot Start Flex 2X Master Mix (New England Biolabs, #M0536L) or KAPA HiFi HotStart ReadyMix (Roche, #KK2602) (for: 5′-ACTCAAGGTCTCGCTA-3′; rev: 5′-GTCAAGGTCTCGGAT-3′), cloned the gene library into the modified pCTCON2 vector using Golden Gate assembly (37 °C 5 min → 16 °C 5 min ×30 cycles → 60 °C 10 min), and transformed into 10G supreme electrocompetent Escherichia coli (Lucigen, #60080-2). The transformed library was cultured in Luria Broth (LB) (DOT Scientific Inc., #D5L24400-2000) media at 37 °C to an optical density of ~0.5, at which point plasmid DNA was harvested using the Qiaprep Spin miniprep kit (Qiagen, #27104). We then transformed the GB1 library into yeast display S. cerevisiae strain EBY100 made competent using the Zymo Research Frozen EZ Yeast Transformation II kit (Zymogen, #T2001). We grew the transformed library in Sabouraud Dextrose Casamino Acid media (SDCAA, pH 4.5: Components per liter - 20 g dextrose (Sigma Aldrich, CAS# 50-99-7, #47249), 6.7 g yeast nitrogen base (VWR Scientific, 97064-162), 5 g casamino acids (VWR, 97063-378), 10.4 g sodium citrate (Sigma Aldrich, CAS# 6132-04-3, C8532), 7.4 g citric acid monohydrate (Sigma Aldrich, CAS# 5949-29-1, C7129)) at 30 °C and 250 rpm for two days. We plated an aliquot of the transformant pool on synthetic dropout (SD)-Trp agar (Teknova, C3060) to quantify the number of library transformants.
We analyzed and sorted the GB1 library using florescence-activated cell sorting (FACS). We induced the library expression in Sabouraud Galactose Casamino Acid media (SGCAA, pH 7.4: Components per liter - 8.6 g NaH2PO*H2O (Sigma Aldrich, CAS# 10049-21-5, 71507), 5.4 g Na2HPO4 (Sigma Aldrich, CAS# 7558-79-4), 20 g galactose (Sigma Aldrich, CAS# 59-23-4, G0750-10G), 6.7 g yeast nitrogen base, 5 g Casamino Acids) overnight, harvested approximately 3 × 106 yeast cells by centrifugation, washed once in pH 7.4 Phosphate Buffered Saline (PBS) (Crystalgen, 221-133-40) containing 0.2% (w/v) bovine serum albumin (BSA) (Sigma Aldrich, 1071145400), and incubated overnight at 4 °C on a tube rotator at 18 rpm in 800 µL of PBS/0.2% BSA containing 15 nM human IgG1 (BioLegend, 403501) that had been conjugated with Alexa647 using NHS chemistry (Thermo Fisher Scientific, A37573) and 3 µg/mL anti-myc IgY (Aves Labs, ET-MY100) conjugated with Alexa488 using NHS chemistry (Thermo Fisher Scientific, A20000). Following the overnight incubation, we washed the yeast in PBS/0.2% BSA and resuspended in ice-cold PBS for FACS. We performed FACS using a FACS Aria II (Becton Dickinson) and analyzed yeast cells for display at 488 nm and IgG binding at 647 nm. Our qualitative experiments sorted cells into display-only and IgG bind populations, while our quantitative experiments sorted cells into display-only, low-bind, WT-like bind, and high-bind populations.
We recovered the sorted yeast populations, as well as the initial unsorted library, and grew them in SDCAA media overnight. The following morning, we expanded the cultures into SDCAA media at an optical density of 0.1, grew them until reaching density of ~1.0, harvested and centrifuged the cultures, and extracted the plasmids using the Zymo Research Yeast Plasmid Miniprep II kit (Zymo Research, D2004). We transformed the extracted plasmid DNA into 10 G supreme electrocompetent E. coli (Lucigen, #60080-2), cultured overnight in LB + carbenicillin (GoldBio, #C1035) media shaking at 250 rpm at 30 °C, and harvested the plasmid DNA using the Qiaprep Spin miniprep kit (Qiagen, #27104). We cut out the GB1 gene insert using XhoI (NEB, R0146S) and PstI (NEB, R0140S) restriction enzymes, excised the corresponding band using agarose gel extraction, and purified using the Zymo Research gel DNA recovery kit (Zymo Research, #D4001). The UW-Madison Biotechnology Center DNA sequencing core prepared a sequencing library using the NEBNext Ultra II kit (NEB, E765S) and sequenced the samples using an Illumina NovaSeq6000 with 2 × 150 bp reads.
Illumina data processing and analysis
We aligned forward and reverse Illumina reads to wild-type GB1, using a predetermined offset, and merged the two reads by selecting the base with the higher quality score in overlapping regions. A design’s count was equal to the number of sequencing reads that exactly matched the designed nucleotide sequence and we filtered out any designs if they had fewer than 10 counts in the unsorted population.
The initial experiments consisted of two replicates each of unsorted (u), display only (d), and binding (b) populations. For each population, we divided by the total number of reads to obtain the proportion of each design. We refer to pu,i, pd,i, and pb,i as the proportion of design i in the unsorted, display only, and binding populations, respectively. We calculated a binding score as the enrichment of a design in the binding population relative to the display only population, where pb,wt and pd,wt are the proportion of wild-type GB1 in the binding and display only populations, respectively (Eq. 1). We also calculated a display score as the enrichment of a design in the full displaying population (b + d) relative to the unsorted population (Eq. 2). We observed a bimodal distribution for both binding and display scores that correspond to active and inactive populations. We used these distributions to classify that designs with display scores ≥ −0.26 display and designs with a binding score ≥ −0.5 bind IgG. Here, the 0.6 and 0.4 correspond the relative proportion of the binding and display only populations from the FACS experiment. The numerator estimates the full displaying population from the binding and display only populations.
For the quantitative screening of designs, we obtained unsorted (u), display only (d), low-binding (l), wild-type-like binding (w), and high-binding (h) populations. For each population we calculated the enrichment relative to the unsorted population where x corresponds to any sorted population l, w, h (Eq. 3). Each sequence was categorized into display only, low-binding, wild-type-like binding, and high-binding based on thresholds determined by calibration sequences with known fitness values (Supplementary Fig. 16). All data analysis can be found in Jupyter notebooks in the supplementary material.
| 1 |
| 2 |
| 3 |
Characterization of individual designs for display and binding
We identified 20 designs to test more thoroughly for binding and display in clonal yeast display assays (Supplementary Table 1). We chose five 5- and 10-mutants with high binding across all replicates, five 20 -mutants with some binding activity across all replicates, and ten 40 and 50-mutants with high levels of display across all replicates. We resynthesized their DNA sequences as individual gene fragments (Twist Biosciences) and cloned them into the same yeast display vector used for library screening. We transformed the plasmid DNA into EBY100 made competent using the Zymo Research Frozen EZ Yeast Transformation II (Zymo Research, T2001) kit and grew the transformants on SD -Trp agar plates (Teknova, C3060) for two days at 30 °C. After 2 days, we picked individual colonies into 4 mL SDCAA and grew them overnight at 30 °C and 250 rpm. We induced GB1 display in a 5 mL SGCAA culture started at an optical density as measured at 600 nm of 0.5 and shaken overnight at 250 rpm and 20 °C.
For IgG binding titrations, we harvested approximately 2 × 105 induced yeast cells for each titration data point, washed once in pH 7.4 PBS containing 0.2% (w/v) BSA, and incubated for three hours at 4 °C on a tube rotator at 18 rpm in between 100 µL and 1 mL of PBS/0.2% BSA containing various concentrations of Alexa647 human IgG1 and 3 µg/mL Alexa 488 anti-myc IgY. We varied the volumes of Alexa647 IgG-containing incubation solution to prevent ligand depletion from occurring at the lowest IgG concentrations. Following incubation, we washed the yeast once in PBS/0.2% BSA and resuspended in ice-cold PBS for flow cytometric analysis. We analyzed the samples using a Fortessa analyzer (Becton Dickinson).
For each design, we subtracted background display and binding activity from an unlabeled yeast negative control from raw MFU display and binding measurements. These activity measurements were normalized against WT GB1 by dividing by the WT activity with background subtracted out.
Statistics and reproducibility
No statistical method was used to predetermine the sample size.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank the University of Wisconsin Carbone Cancer Center Flow Cytometry Laboratory, supported by P30CA14520, 1S10OD018202-01, and 1S10OD018202-01, and the University of Wisconsin-Madison Next Generation Sequencing Core for assistance with experimental characterization of designs. This research was supported by the United States National Institutes of Health (R35GM119854 (P.R.), T32HG002760 (C.F.)), the UW-Madison Biochemistry Undergraduate Summer Research Award (S.F.), the UW-Madison Hilldale Undergraduate Research Award (S.F.), and the UW-Madison Biochemistry Mary Shine Peterson Undergraduate Award (S.F.).
Author contributions
S.F., C.F., and P.R. conceived of study design and analyzed data. S.F. and C.F. designed library sequences. P.H. performed all yeast display experiments. S.F., C.F., and P.R. analyzed the data and wrote the manuscript with feedback from P.H.
Peer review
Peer review information
Nature Communications thanks Mingyue Zheng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All relevant data supporting the key findings of this study are available within the article and its Supplementary Information files. The Illumina sequencing data generated in this study have been deposited in the NCBI BioProject database under accession code PRJNA111787743. The processed Illumina sequencing data are available in the Supplementary Information/Source Data file. The GB1 4-site combinatorial library data used in this study are available in the supplementary information of Wu et al. 22 [10.7554/eLife.16965.024]. Fitness values for single-mutant GB1 variants are from Gelman et al. 12. Source data are provided with this paper.
Code availability
All code for analyzing data and generating figures is available in the supplementary materials and on GitHub under the GPL3 license44. All code was archived on Zenodo (10.5281/zenodo.12518821)45.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chase R. Freschlin, Sarah A. Fahlberg.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50712-3.
References
- 1.Romero, P. A. & Arnold, F. H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol.10, 866–876 (2009). 10.1038/nrm2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freschlin, C. R., Fahlberg, S. A. & Romero, P. A. Machine learning to navigate fitness landscapes for protein engineering. Curr. Opin. Biotechnol.75, 102713 (2022). 10.1016/j.copbio.2022.102713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wittmann, B. J., Johnston, K. E., Wu, Z. & Arnold, F. H. Advances in machine learning for directed evolution. Curr. Opin. Struct. Biol.69, 11–18 (2021). 10.1016/j.sbi.2021.01.008 [DOI] [PubMed] [Google Scholar]
- 4.Hawkins-Hooker, A. et al. Generating functional protein variants with variational autoencoders. PLoS Comput Biol.17, 1–23 (2021). 10.1371/journal.pcbi.1008736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, D. H. et al. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol.39, 691–696 (2021). 10.1038/s41587-020-00793-4 [DOI] [PubMed] [Google Scholar]
- 6.Greenhalgh, J. C., Fahlberg, S. A., Pfleger, B. F. & Romero, P. A. Machine learning-guided acyl-ACP reductase engineering for improved in vivo fatty alcohol production. Nat. Commun.12, 1–10 (2021). 10.1038/s41467-021-25831-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas, S., Khimulya, G., Alley, E. C., Esvelt, K. M. & Church, G. M. Low-N protein engineering with data-efficient deep learning. Nat. Methods18, 389–396 (2021). 10.1038/s41592-021-01100-y [DOI] [PubMed] [Google Scholar]
- 8.Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature620, 1089–1100 (2023). 10.1038/s41586-023-06415-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc Natl Acad Sci USA. 118, e2016239118 (2021). [DOI] [PMC free article] [PubMed]
- 10.Hie, B., Bryson, B. D. & Berger, B. Leveraging uncertainty in machine learning accelerates biological discovery and design. Cell Syst.11, 461–477.e9 (2020). 10.1016/j.cels.2020.09.007 [DOI] [PubMed] [Google Scholar]
- 11.Wittmann, B. J., Yue, Y. & Arnold, F. H. Informed training set design enables efficient machine learning-assisted directed protein evolution. Cell Syst.12, 1026–1045.e7 (2021). 10.1016/j.cels.2021.07.008 [DOI] [PubMed] [Google Scholar]
- 12.Gelman, S., Fahlberg, S. A., Heinzelman, P., Romero, P. A. & Gitter, A. Neural networks to learn protein sequence-function relationships from deep mutational scanning data. Proc. Natl. Acad. Sci. USA118, e2104878118 (2021). [DOI] [PMC free article] [PubMed]
- 13.Yang, K. K., Wu, Z., Bedbrook, C. N. & Arnold, F. H. Learned protein embeddings for machine learning. Bioinformatics34, 2642–2648 (2018). 10.1093/bioinformatics/bty178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alley, E. C., Khimulya, G., Biswas, S., AlQuraishi, M. & Church, G. M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods16, 1315–1322 (2019). 10.1038/s41592-019-0598-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu, C., Nisonoff, H., Fannjiang, C. & Listgarten, J. Learning protein fitness models from evolutionary and assay-labeled data. Nat. Biotechnol.40, 1114–1122 (2022). [DOI] [PubMed]
- 16.Xu, Y. et al. Deep dive into machine learning models for protein engineering. J. Chem. Inf. Model60, 2773–2790 (2020). 10.1021/acs.jcim.0c00073 [DOI] [PubMed] [Google Scholar]
- 17.Li, L. et al. Machine learning optimization of candidate antibody yields highly diverse sub-nanomolar affinity antibody libraries. Nat. Commun.14, 1–12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fannjiang, C. & Listgarten, J. Is novelty predictable? 1–30. Preprint at https://arxiv.org/abs/2306.00872 (2023). [DOI] [PMC free article] [PubMed]
- 19.Bailey, L. J. et al. Applications for an engineered Protein-G variant with a pH controllable affinity to antibody fragments. J. Immunol. Methods415, 24–30 (2014). 10.1016/j.jim.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson, C. A., Wu, N. C. & Sun, R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr. Biol.24, 2643 (2014). 10.1016/j.cub.2014.09.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brookes, D., Park, H. & Listgarten, J. Conditioning by adaptive sampling for robust design. In Proc. of the 36th International Conference on Machine Learning (eds Chaudhuri, K. & Salakhutdinov, R.) vol. 97, 773–782 (PMLR, 2019).
- 22.Wu, N. C., Dai, L., Olson, C. A., Lloyd-Smith, J. O. & Sun, R. Adaptation in protein fitness landscapes is facilitated by indirect paths. Elife5, e16965 (2016). [DOI] [PMC free article] [PubMed]
- 23.Li, Q. et al. Profiling protease specificity: combining yeast ER Sequestration Screening (YESS) with Next Generation Sequencing. ACS Chem. Biol.12, 510–518 (2017). 10.1021/acschembio.6b00547 [DOI] [PubMed] [Google Scholar]
- 24.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods19, 679–682 (2022). 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res.33, 2302–2309 (2005). 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
- 28.Bedbrook, C. N. et al. Machine learning-guided channelrhodopsin engineering enables minimally invasive optogenetics. Nat. Methods16, 1176–1184 (2019). 10.1038/s41592-019-0583-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rapp, J. T., Bremer, B. J. & Romero, P. A. Self-driving laboratories to autonomously navigate the protein fitness landscape. Nat. Chem. Eng.1, 97–107 (2024). [DOI] [PMC free article] [PubMed]
- 30.Chen, L. et al. Learning protein fitness landscapes with deep mutational scanning data from multiple sources. Cell Syst.14, 706–721.e5 (2023). 10.1016/j.cels.2023.07.003 [DOI] [PubMed] [Google Scholar]
- 31.Wells, J. A. Additivity of mutational effects in proteins. Biochemistry29, 8509–8517 (1990). 10.1021/bi00489a001 [DOI] [PubMed] [Google Scholar]
- 32.Skinner, M. M. & Terwilliger, T. C. Potential use of additivity of mutational effects in simplifying protein engineering. Proc. Natl Acad. Sci. USA93, 10753–10757 (1996). 10.1073/pnas.93.20.10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokuriki, N., Stricher, F., Schymkowitz, J., Serrano, L. & Tawfik, D. S. The stability effects of protein mutations appear to be universally distributed. J. Mol. Biol.369, 1318–1332 (2007). 10.1016/j.jmb.2007.03.069 [DOI] [PubMed] [Google Scholar]
- 34.Lakshminarayanan, B., Pritzel, A. & Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. In Advances in Neural Information Processing Systems (eds Guyon, I. et al.) vol. 30 (Curran Associates, Inc., 2017).
- 35.Gruver, N. et al. Effective surrogate models for protein design with Bayesian optimization. ICML Workshop on Computational Biology (2021).
- 36.Zeng, H. & Gifford, D. K. Quantification of uncertainty in peptide-MHC binding prediction improves high-affinity peptide selection for therapeutic design. Cell Syst.9, 159–166.e3 (2019). 10.1016/j.cels.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fannjiang, C. & Listgarten, J. Autofocused oracles for model-based design (2020).
- 38.Makowski, E. K. et al. Co-optimization of therapeutic antibody affinity and specificity using machine learning models that generalize to novel mutational space. Nat. Commun.13, 3788 (2022). [DOI] [PMC free article] [PubMed]
- 39.Radivojević, T., Costello, Z., Workman, K., & Garcia Martin, H. A machine learning automated recommendation tool for synthetic biology. Nat. Commun.11, 1–14 (2020). 10.1038/s41467-020-18008-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Center for High Throughput Computing. Center for High Throughput Computing. 10.21231/GNT1-HW21 (2006).
- 41.Basanta, B. et al. An enumerative algorithm for de novo design of proteins with diverse pocket structures. Proc. Natl Acad. Sci. USA117, 22135–22145 (2020). 10.1073/pnas.2005412117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao, G. et al. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc.1, 755–768 (2006). 10.1038/nprot.2006.94 [DOI] [PubMed] [Google Scholar]
- 43.Freschlin, C. R., Fahlberg, S. A., Heinzelman, P. & Romero, P. A. Neural network extrapolation to distant regions of the protein fitness landscape. NCBI BioProject, Accession: PRJNA1117877 (2024). [DOI] [PMC free article] [PubMed]
- 44.Freschlin, C. R., Fahlberg, S. A., Heinzelman, P. & Romero, P. A. Neural network extrapolation to distant regions of the protein fitness landscape. GitHub. https://github.com/RomeroLab/nn-extrapolation (2024). [DOI] [PMC free article] [PubMed]
- 45.Freschlin, C. R., Fahlberg, S. A., Heinzelman, P. & Romero, P. A. Neural network extrapolation to distant regions of the protein fitness landscape. Zenodo. 10.5281/zenodo.12518821 (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All relevant data supporting the key findings of this study are available within the article and its Supplementary Information files. The Illumina sequencing data generated in this study have been deposited in the NCBI BioProject database under accession code PRJNA111787743. The processed Illumina sequencing data are available in the Supplementary Information/Source Data file. The GB1 4-site combinatorial library data used in this study are available in the supplementary information of Wu et al. 22 [10.7554/eLife.16965.024]. Fitness values for single-mutant GB1 variants are from Gelman et al. 12. Source data are provided with this paper.
All code for analyzing data and generating figures is available in the supplementary materials and on GitHub under the GPL3 license44. All code was archived on Zenodo (10.5281/zenodo.12518821)45.