Abstract
A new variety of hornwort from northern Thailand, Phaeocerosperpusillusvar.scabrellus is described based on morphological characters and molecular phylogenetic analyses. In this study, phylogenetic analyses supported that the new variety is closely related to P.perpusillusvar.perpusillus. Morphologically, it is distinguished from the autonimic variety in nearly smooth spores under light microscope. A taxonomic description, illustrations, and light and scanning electron micrographs are provided. In addition, the new variety is assessed as Endangered (EN), demonstrating its rarity by being currently known from only three subpopulations.
Key words: Endangered, hornwort, low-copy nuclear markers, new variety, spore ornamentation
Introduction
Phaeoceros Prosk. (Notothyladaceae) is the third largest genus of hornwort with about 34 currently accepted species worldwide (Söderström et al. 2016) and widely distributed in both the Northern and Southern Hemispheres (Cargill and Fuhrer 2008). The genus is defined by a smooth solid thallus, single chloroplast per cell, presence of a pyrenoid, antheridial chambers with usually (1–)2–6(–8) antheridia, irregularly arranged jacket cells of the antheridia, and yellow spores (Duff et al. 2007; Cargill and Fuhrer 2008; Chantanaorrapint 2009; Villarreal et al. 2010). In Thailand three species have been reported: P.carolinianus (Michx.) Prosk., P.himalayensis (Kashyap) Prosk. ex Bapna & G.G.Vyas, and P.perpusillus Chantanaorr. (Lai et al. 2008; Chantanaorrapint 2009; Chantanaorrapint et al. 2015).
During the bryological surveys in Chiang Mai Province, northern Thailand, some interesting specimens of the hornwort genus Phaeoceros were collected. These specimens resemble P.perpusillus, an endemic species of northern Thailand, in having small gametophytes, short sporophytes (usually less than 1 cm long), yellow spores, and subquadrate pseudoelater cells. Following a detailed comparison with closely related taxa, we here describe and illustrate these specimens as a new variety of P.perpusillus. We also used for the first time three hornwort specific low-copy nuclear markers. In theory, low-copy nuclear genes tend to have higher mutation rates than organellar genes, resulting in more variable sites that can be used for phylogenetic reconstruction, especially at species-level (Sang 2002; Feliner and Rosselló 2007). However, despite the advantages of biparental inheritance, which provides a more comprehensive view of the genetic history and evolution, low-copy nuclear genes have not been widely used (Zhang et al. 2012). In this study, we combined selected low-copy nuclear sequences with chloroplast sequences to enhance the resolution of our phylogenetic analyses. By using both chloroplast and nuclear markers, our study aims to explore alternative genetic regions for species-level phylogenies, thus providing a greater understanding of hornwort evolution.
Materials and methods
Morphological study
This study is based on recent collections from Thailand. Voucher specimens of the new species are deposited in BKF, NICH, and PSU herbaria. Morphological and anatomical characters were studied using stereo- and compound microscopes. The distinctive characters of the species were photographed using an Olympus BX51 microscope equipped with a DP74 digital camera and illustrated with the aid of an Olympus drawing tube. Mature spores were coated with a thin layer of gold and examined under a FEI Quanta 400 scanning electron microscope operating at 20 kV. The preliminary conservation status was assessed following the International Union for Conservation of Nature (IUCN) Red List criteria (IUCN 2022) and using GeoCAT (Bachman et al. 2011) to calculate the area of occupancy (AOO) and extent of occurrence (EOO). In addition, distribution and ecological data were compiled; descriptions and illustrations are provided.
Taxon sampling
Twenty-seven samples of Phaeoceros spp. were included in our molecular dataset. Additionally, Notothylaslevieri Schiffn. ex Steph. and Paraphymatoceros sp. were employed as the outgroup. List of newly generated sequence used in the phylogeny with voucher information and GenBank accession numbers are provided in Table 1.
Table 1.
List of newly generated sequence used in the phylogeny with voucher information and GenBank accession numbers.
| Taxa | Collector | rbcL | L138 | L178 | L315 |
|---|---|---|---|---|---|
| Paraphymatoceros sp. Mexico | Morales 22 | OR943578 | PP481902 | PP471573 | PP471590 |
| Phaeoceroscarolinianus Thailand1 | Chantanaorrapint & Suwanmala 3955 | OR943588 | PP481909 | PP471580 | PP471598 |
| P.carolinianus Thailand2 | Chantanaorrapint & Suwanmala 3909 | OR943586 | PP481911 | PP471581 | PP471600 |
| P.carolinianus Thailand3 | Chantanaorrapint & Suwanmala 4057 | OR943585 | PP481913 | PP471583 | PP471602 |
| P.carolinianus India1 | Villarreal & Uniyal1314 | OR943596 | PP481901 | PP471572 | PP471589 |
| P.carolinianus India3 | Villarreal 1233 | OR943593 | PP481904 | PP471575 | PP471593 |
| P.carolinianus India2 | Duckett IE45 | OR943592 | PP481905 | PP471576 | PP471594 |
| P.carolinianus Czech Republic | Kopal s.n. | OR943591 | PP481906 | PP471577 | PP471595 |
| P.carolinianus Indonesia | Gradstein 12362 | OR943595 | - | - | PP471591 |
| P.carolinianus Vietnam | Suwanmala 849 | OR943582 | PP481916 | PP471585 | PP471604 |
| P.exiguus Thailand2 | Chantanaorrapint & Suwanmala 4129 | OR943580 | PP481918 | PP471587 | PP471606 |
| P.himalayensis India | Duckett IW15 | OR943594 | PP481903 | PP471574 | PP471592 |
| P.kashyapii Thailand | Chantanaorrapint & Suwanmala 3901 | OR943589 | PP481908 | PP471579 | PP471597 |
| P.mohrii USA | Doyle 11341 | OR943590 | PP481907 | PP471578 | PP471596 |
| P.perpusillusvar.perpusillus Thailand2 | Chantanaorrapint & Suwanmala 3883 | OR943587 | PP481910 | - | PP471599 |
| P.perpusillusvar.perpusillus Thailand3 | Chantanaorrapint & Suwanmala 4076 | OR943584 | PP481914 | PP471584 | PP471603 |
| P.perpusillusvar.scabrellus Thailand1 | Chantanaorrapint & Suwanmala 4077 | OR943583 | PP481915 | - | - |
| P.perpusillusvar.scabrellus Thailand2 | Chantanaorrapint & Suwanmala 4116 | OR943581 | PP481917 | PP471586 | PP471605 |
| Phaeoceros sp. Thailand | Chantanaorrapint & Suwanmala 4488 | OR943579 | PP481919 | PP471588 | PP471607 |
DNA extraction, amplification, and sequencing
Total genomic DNA from silica gel-dried sporophytes was extracted using E.Z.N.A. Plant DNA kit (Omega Bio-Tek, USA) following manufacturer’s protocols. An alignment of more than 400 loci from a probe developed and explained by Breinholt et al. (2021) was used to reconstruct the phylogeny of all hornworts (Peñaloza-Bojacá et al. submitted). From the alignment we selected three loci found in Phaeoceros species (L138, L178, and L315) and designed internal primers using Geneious 2021.1.1. Amplification was accomplished using four primers listed in Table 2, one for the rbcL gene as described in Duff et al. (2004) and three primers newly designed for Phaeoceros nuclear loci (L138, L178 and L315). The conditions for PCR were as follows: (1) for rbcL, L138 and L315: initial denaturation for 3 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 30 s at 55 °C, 1 min at 72 °C, and final extension for 10 min at 72 °C, (2) for L178: initial denaturation for 3 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 30 s at 58 °C, 1 min at 72 °C, and final extension for 10 min at 72 °C. The final products were incubated at 10 °C to complete the reaction. The PCR products were purified and sequenced by Plate-forme d’analyses génomiques (Quebec, Canada), except for O. Suwanmala 849, S. Chantanaorrapint & O. Suwanmala 4116, 4129, 4488 which were performed by the Macrogen sequencing service (Macrogen, Korea).
Table 2.
Primer sequence used for PCR amplification and sequencing.
| Region | Sequence 5’-3’ | Reference |
|---|---|---|
| L138 | ||
| Phaeoceros_L138_58F | TTG TCC TGA ATT CAC GTG GT | This study |
| Phaeoceros_L138_607R | GCT TTG CTA GGG TCT GGT AAG A | This study |
| L178 | ||
| Phaeoceros_L178_232F | CTC GGG GAT GAG CGG GAC | This study |
| Phaeoceros_L178_1088R | GCT TCA AGA GAT GGC TCC TT | This study |
| L315 | ||
| Phaeoceros_L315_676F | GGA TTT TGG GGA CTT GCA CA | This study |
| Phaeoceros_L315_1325R | CTT CTG CCC AAC AAC AGG AG | This study |
| rbcL | ||
| rbcL2_16F | GAG ACT AAA GCA GGT GTT GGA | Duff et al. (2004) |
| rbcL_976R | ACA CGA AAG TGA ATA CCA TG | Duff et al. (2004) |
Forward and reverse sequences were edited initially and assembled using Geneious 2021.1.1. We gathered published data from six samples generated by Breinholt et al. (2021), UFG_393201_P02_WH01, UFG_393201_P02_WA02, UFG_393201_P02_WB02, UFG_393201_P02_WH02, UFG_393201_P02_WG02, UFG_393201_P02_WD01, one sample of UFG_393202_P054_WD04 generated by Bechteler et al. (2023), and three samples, UFG_393202_P033_WD01, UFG_393202_P054_WE04, UFG_393202_P033_WC01, generated by Peñaloza-Bojacá et al. (submitted). Nineteen newly generated sequences (Table 1) and ten published sequences were aligned using the Geneious alignment algorithm with default settings. Uncertain alignment positions and columns displaying a large number of gaps were excluded from the phylogenetic assessments. Any incomplete sequence segments and nucleotide gaps were treated as missing data.
Phylogenetic analysis
A maximum likelihood (ML) analysis was performed in RAxML HPC BlackBox v.8.2 (Stamatakis 2014) using GTR+I+GAMMA substitution model following default setting with 1000 bootstrap replications. The best model scheme of each partition was carried out in Partitionfinder 2 (Lanfear et al. 2016). Bayesian analysis was performed in MrBayes 3.2 (Ronquist et al. 2012) using Markov chain Monte Carlo (MCMC) searches with two runs and four chains of 3,500,000 generations. Trees were sampled every 1000th generation and the first 10% of sampled trees were discarded as a burn-in to ensure a convergence of the analyses. We used Tracer 1.5 (Rambaut et al. 2018) to evaluate the burn-in and convergence. Figtree was used to graph and edit trees (Rambaut 2017). Both maximum likelihood and Bayesian analyses were performed on CIPRES Science Gateway (Miller et al. 2010).
Results
A concatenated dataset of the coding region of one plastid and three nuclear markers (rbcL, L138, L178 and L315) contained 2856 characters (892, 549, 781, and 634 characters respectively). Tree topologies generated by Bayesian inference (BI) and maximum likelihood exhibited congruent patterns shown in Fig. 1, with posterior probabilities (PP) and maximum likelihood bootstrap values (MLBS) plotted on the branches. The monophyly of the genus Phaeoceros is well supported by posterior possibility (PP = 1) but weakly supported by maximum likelihood analysis (MLBS = 53). In the tree topology (Fig. 1), Phaeoceros was divided into two major lineages with strong support, clade A including twenty-three terminals, containing the new taxon and other papillate spore Phaeoceros (PP = 1, MLBS = 94), and clade B comprising four terminals of non-papillate spore Phaeoceros including P.himalayensis and P.kashyapii A.K. Asthana & S.C. Srivast. (PP = 1, MLBS = 100).
Figure 1.
Majority rule consensus tree of phylogenetic relationships of Phaeoceros derived from Bayesian analyses of the combined dataset of rbcL, L138, L178, and L315 genes. Bayesian posterior probability values (PP) and bootstrap percentages values (MLBS) are shown on branches respectively. Nucleotide substitution rates indicated below the tree. Clade A include papillate spore Phaeoceros and the new variety (highlighted), and Clade B include non-papillate spore Phaeoceros.
The inclusion of P.perpusillusvar.scabrellus and its autonimic variety in the data matrix resolves this species lineage as monophyletic with good support (PP = 1, MLBS = 90). The new variety is recovered as sister to the autonimic variety with less posterior probability and bootstrap support (PP = 0.56, MLBS = 80).
Taxonomic treatment
. Phaeoceros perpusillus Chantanaorr. var. scabrellus
Suwanmala & Chantanaorr. var. nov.
F7B035B5-CAA1-50E9-B59B-166914AA2CF5
Figure 2.
Phaeocerosperpusillusvar.scabrellusA plants in natural habitat B dorsal view of thallus showing marginal tubers (arrow) C ventral view of thallus showing ventral tubers (arrows) D cross-section of thallus showing the large dark lump of Nostoc colony (NC = Nostoc colony) E dorsal epidermal cells of thallus showing a single chloroplast with pyrenoid (arrows) per cell F spores and pseudoelaters. Photos by O. Suwanmala (A from S. Chantanaorrapint & O. Suwanmala 4116B–F from S. Chantanaorrapint & O. Suwanmala 4077).
Figure 3.
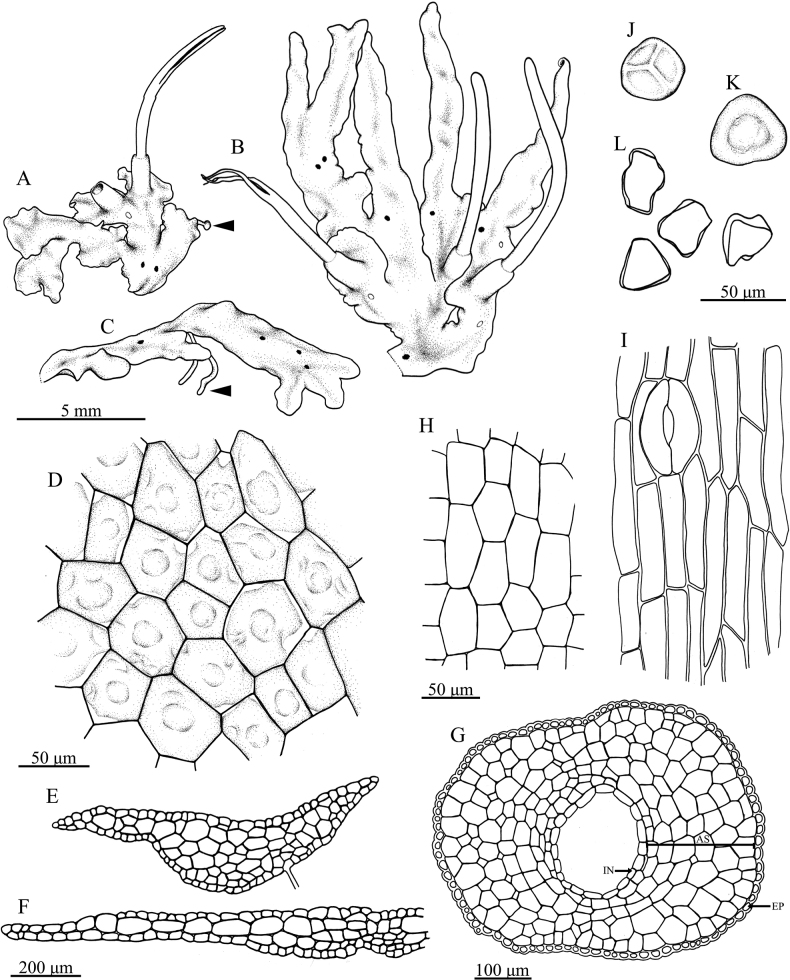
Phaeocerosperpusillusvar.scabrellusA gametophyte forming half-rosettes with sporophyte (arrow indicate tuber) B ensiform thalli and sporophytes C gametophyte showing ventral tuber (arrow) D dorsal epidermal cells of thallus E, F cross sections of thalli G cross section of sporangium (AS = assimilative tissue, EP = epidermal cell of capsule, IN = inner most sporangium wall) H inner most cells of sporangium wall I epidermal cells of capsule with stoma J proximal view of spore K distal view of spore L pseudoelaters. All from holotype and drawings by O. Suwanmala. (All drawing from S. Chantanaorrapint & O. Suwanmala 4116).
Figure 4.
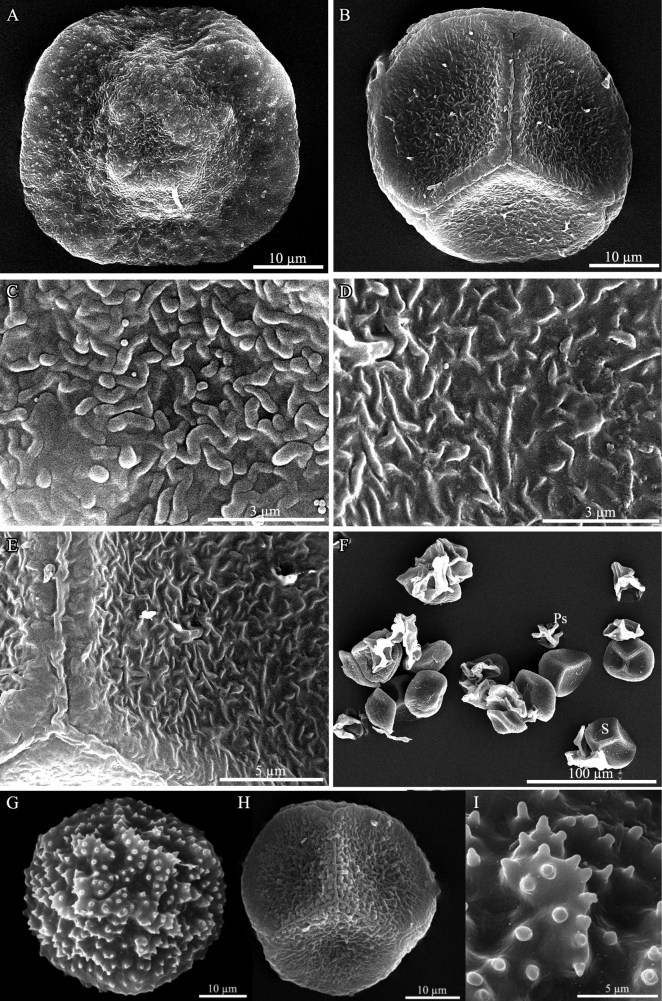
Scanning electron micrographs of spores A–FPhaeocerosperpusillusvar.scabrellusA distal view of spore showing a central hump-like projection B proximal view of spore with a distinct triradiate mark C close-up of distal surface showing packed vermiculae D close-up of proximal surface showing loosely arranged vermiculae E proximal surface showing trilete mark and loosely arranged vermiculae F spores and pseudoelaters G–IP.perpusillusvar.perpusillusG distal view of spore H proximal view of spore I close-up of distal surface showing pluripapillae. Photos by O. Suwanmala. (A–F images from S. Chantanaorrapint & C. Promma 3129G–I images from S. Chantanaorrapint & O. Suwanmala 3883).
Type.
Thailand. Chiang Mai Province: Doi Suthep-Pui, Bhu Bing Palace, 1400 m, 18 October 2020, S. Chantanaorrapint & O. Suwanmala 4077 (holotype: PSU!; isotype: BKF!, NICH!).
Diagnostic.
Phaeocerosperpusillusvar.scabrellus is similar to the autonimic variety but differs in nearly smooth spores under light microscope (or vermiculate under SEM), whereas the autonimic variety have pluripapillae on the distal surface and vermiculate on the proximal.
Description.
Thallus yellowish-green to dark green in fresh material, dull green to blackish- brown in dry material, prostrate or moderately adhering to the substratum, solid, ecostate, orbicular to sub-orbicular, dichotomously branched into several lobes, with a smooth dorsal surface; lobes ensiform or sometimes fan-shaped, up to 0.8 mm long, 1–3 mm wide; margins wavy, nearly entire to shallowly crenulate; apex flat, rarely ascending, occasionally tapering into apical tubers; tubers sometimes present on ventral surface. Thallus in cross section plano-convex to concave-convex, 4–10 cells thick in the middle region, without mucilage cavities. Dorsal epidermal cells rectangular to heptagonal, 28–75 × 25–50 µm, thin-walled, smooth. Chloroplast one per cell, large, occupying almost entire cell, variable in shape; pyrenoid present. Nostoc colonies scattered through the ventral side of thallus, appearing as dark dots. Rhizoids hyaline or pale brown along ventral surface, inner wall smooth or tuberculate. Sexuality monoicous. Androecia scattered and slightly raised over the dorsal surface of thallus, 2–3 antheridia per chamber; antheridia subglobose to globose, exposed at maturity, irregularly arranged jacket cells, shortly stalked, stalk with quadriseriate cells. Archegonia embedded in thallus, connected to the upper surface, scattered near the lobe of thallus. Involucre solitary, conical-cylindrical, up to 2 mm long, 2–4 cells thick, mouth smooth to crenulate. Sporophytes capsule somewhat inclined, stout to narrowly cylindrical, 0.5–1(–1.2) cm long, yellow at apex, dehiscing from top toward base, bivalves rarely twisted when dry; epidermal cells of capsule elongate-rectangular, 68–200 × 12–30 µm, thick-walled, stomata present with two reniform guard cells, surrounded by 5–8 epidermal cells; assimilative layers 3–6 cells thick in cross section; the innermost capsule cells dark brown, subquadrate to rectangular; 27–67 × 22–53 µm; columella well-developed, red-brown, consisting of 16 cells (4 × 4 lines of cells) in cross section. Spores unicellular, yellow, rounded-triangular in polar view, equatorial diameter 32–50 µm in diameter, nearly smooth under light microscope (LM), proximal surface with a distinct trilete mark, bordered by vermiculate strip on each side of trilete mark, each facet covered with fine vermiculate pattern; distal surface with a slightly dome-like region at the center, more densely vermiculate than proximal surface, sometimes with minute granules. Pseudoelaters light brown or yellowish-brown at maturity, thin-walled, occasionally branched; pseudoelater cells subquadrate to short rectangular, 30–45 × 25–30 μm, without helicoidal band.
Etymology.
The epithet of the variety refers to scabrate ornamentation observed under light microscope.
Habitat and distribution.
Phaeocerosperpusillusvar.scabrellus is currently known only from northern Thailand. It grows on disturbed soil and sandstone in open site in grassland, pine-oak mixed montane deciduous forests at elevation of 1390–2100 m. It may grow associated with other bryophytes such as Anthocerossubtilis Steph., Notothylaslevieri, N.orbicularis (Schwein.) Sull. ex A.Gray, and P.carolinianus.
Conservation status.
This variety is currently known from three subpopulations, which are in protected areas (Chiang Dao Wildlife Sanctuary and Doi Suthep-Pui National Park). One of the subpopulations is located in a camping area, which is a common visiting site for tourists and dominated by Ageratinaadenophora (Spreng.) R.M.King & H.Rob. (invasive species). Therefore, habitat quality is threatened by trampling and other destructive activities potentially caused by regular visits by tourists to the area, and invasive plant species. Together, these have the potential to cause a population reduction. The other subpopulation is also somewhat disturbed by human activities such as shifting cultivation. The extent of occurrence (EOO) of P.perpusillusvar.scabrellus is estimated to be 262.925 km2 and its area of occupancy (AOO) is estimated to be 12 km2, which falls within the limits for Endangered status under criterion B1 and B2 of IUCN Red List Categories and Criteria. Conservation efforts should focus on implementing strict regulations to reduce the impact of human activity and controlling invasive species, while also raising awareness among local communities about the importance of protecting the habitat.
Additional specimens examined.
Thailand. Chiang Mai Province: Chiang Dao Wildlife Sanctuary, 1700–2000 m, 1 November 2013, S. Chantanaorrapint & C. Promma 3125B, 3129, 3216 (PSU); Doi Suthep-Pui National Park, Doi Mon Long Viewpoint, 1390 m, 4 November 2015, S. Chantanaorrapint & W. Juengprayoon 143B (PSU); 15 November 2020, S. Chantanaorrapint & O. Suwanmala 4089, 4090 (PSU); Bhu Ping Palace, 1400 m, 8 September 2013, S. Rattanamanee 3 (PSU); 18 October 2020, S. Chantanaorrapint & O. Suwanmala 4077 (PSU); 5 October 2021, S. Chantanaorrapint & O. Suwanmala 4116 (PSU).
Discussions
Phaeocerosperpusillusvar.scabrellus is morphologically similar to the autonimic variety which is endemic to northern Thailand (Chantanaorrapint 2009). These two varieties share some common features, viz. small orbicular gametophytes (Fig. 2A, B), monoicous sexual condition, very short capsules (usually less than 1 cm long) (Figs 2B, 3A, B), yellow spores, and pseudoelater cells being subquadrate to short rectangular (Figs 2F, 3L). The new variety also resembles P.exiguus (Steph.) J. Haseg., a species found in Indonesia, New Caledonia and Taiwan (Hasegawa 1986, 1993; Siagian et al. 2021). They are monoicous, and have a small thallus, very short capsules, and small pseudoelaters. However, they can be distinguished by the spore ornamentation. Phaeocerosperpusillusvar.scabrellus is distinct from the autonimic variety and P.exiguus in nearly smooth spores under light microscope or vermiculate spores under SEM (Fig. 4A–F). In contrast, spores of P.perpusillusvar.perpusillus are pluripapillose on distal face and finely vermiculate on proximal face (Fig. 4G–I), while P.exiguus have button-like papillae on distal face and minutely papillae on proximal face.
In addition, the small plants of P.carolinianus, a common species, can be confused with P.perpusillus or P.exiguus in general appearance. The comparisons of morphological characters between these three monoicous species are summarized in Table 3.
Table 3.
The comparisons of characters between P.perpusillusvar.scabrellus, P.perpusillusvar.perpusillus, P.exiguus and P.carolinianus.
| Characters | P.perpusillusvar.scabrellus | P.perpusillusvar.perpusillus | P.exiguus | P.carolinianus |
|---|---|---|---|---|
| Thallus | 4–10 cells thick in the middle | 6–9 cells thick in the middle | 6–7 cells thick in the middle | 8–13 cells thick in the middle |
| Capsule placement | oblique | oblique | usually erect | Erect |
| Capsule length | usually less than 1 cm | less than 1 cm | up to 1.5 cm | usually more than 1.5 cm |
| Involucre | up to 2 mm high | 1–2 mm high | 1–2 mm high | 2–4 mm high |
| Spore diameter | 32–50 µm | 40–47 µm | 40–42 µm | 30–37 µm |
| Distal surface of spore | densely vermiculate, with minute granules | pluripapillose | dense clusters of button-like papillae | densely spinose |
| Proximal surface of spore | loosely vermiculate in each facet | finely vermiculate in each facet | minutely papillate throughout each facet | minutely papillate in central part of each facet |
| Pseudoelaters (length/width ratio) | 1–1.5 × | 1.5–2.5 × | 1.2–2 × | ≥4 × |
Although both varieties of P.perpusillus have been reported only from the northern part of Thailand, P.perpusillusvar.perpusillus seems to have a wider range of distribution and is more abundant than the new variety. The new variety has been found in only three subpopulations, overlapping with the autonimic variety, which is assessed as Endangered (EN) according to IUCN Red List.
The placement of the new variety falls into the papillate spore Phaeoceros lineage (Fig. 1, clade A), despite the absence of spines or papillae on its spore surface which sets the new variety apart from other taxa. Within an assemblage of autonimic variety P.perpusillus and the new variety clade, the two taxa share a sister relationship with low support, and they show only one morphological difference in spore morphology. The vermiculate spore ornamentation observed in P.perpusillusvar.scabrellus seems to be an unusual form of the autonimic variety. However, based on careful investigation, it becomes evident that the absence of papillae on the spore surface is consistently observed throughout the entire capsule and reveals a uniform pattern in each population. Spore morphology serves as a key trait to differentiate hornwort species, allowing two distinct spore ornamentations to be considered as separate taxa.
This proposal for the new variety’s classification was made due to its gametophyte and sporophyte morphological similarity to the autonimic variety with the exception of the spore ornamentation, and was also supported by phylogenetic inference, and the shared distribution area.
Supplementary Material
Acknowledgments
We would like to thank the curators and staff of EGR, G, HSNU, LWG, LWU, NICH, PSU and TNS for making specimens, including types, available for study through loans or visits. The first author would like to express her sincere appreciation to the Science Achievement Scholarship of Thailand (SAST) and the Canada-ASEAN Scholarships and Educational Exchanges for Development (SEED) program for funding. JCV thanks the Canada Research Chair on Genomics and Metabolomics. The authors thank David Bell (E) and Gabriel Peñaloza‐Bojacá for sharing the raw sequences, and D. Christine Cargill and the anonymous reviewers for their constructive comments on the manuscript.
Citation
Suwanmala O, Villarreal A. JC, Li F-W, Chantanaorrapint S (2024) Phaeoceros perpusillus var. scabrellus (Notothyladaceae, Anthocerotophyta), a new taxon from northern Thailand. PhytoKeys 244: 271–283. https://doi.org/10.3897/phytokeys.244.124080
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This work was supported by the government budget of Prince of Songkla University (no. SCI6302219S), Science Achievement Scholarship of Thailand (SAST) and the Canada-ASEAN Scholarships and Educational Exchanges for Development (SEED) program .
Author contributions
All authors have contributed equally.
Author ORCIDs
Orawanya Suwanmala https://orcid.org/0000-0002-1113-7614
Juan Carlos Villarreal A. https://orcid.org/0000-0002-0770-1446
Fay-Wei Li https://orcid.org/0000-0002-0076-0152
Sahut Chantanaorrapint https://orcid.org/0000-0002-9739-0994
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Bachman S, Moat J, Hill AW, De La Torre J, Scott B. (2011) Supporting Red List threat assessments with GeoCAT: Geospatial conservation assessment tool. ZooKeys 150: 117–126. 10.3897/zookeys.150.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechteler J, Peñaloza‐Bojacá G, Bell D, Burleigh JG, McDaniel SF, Davis C, Sessa EB, Bippus A, Cargill DC, Chantanoarrapint S, Draper I, Lee GE, Endara L, Forrest LL, Garilleti R, Graham SW, Huttunen S, Lazo JJ, Lara F, Larraín J, Lewis LR, Long D, Quandt D, Renzaglia KS, Schäfer-Verwimp A, Sierra AM, von Konrat M, Zartman CE, Goffinet B, Villarreal JC. (2023) Comprehensive phylogenomic time tree of bryophytes reveals deep relationships and uncovers gene incongruences in the last 500 million years of diversification. American Journal of Botany 110(11): e16249. 10.1002/ajb2.16249 [DOI] [PubMed]
- Breinholt JW, Carey SB, Tiley GP, Davis EC, Endara L, McDaniel SF, Neves LG, Sessa EB, von Konrat M, Chantanaorrapint S, Fawcett S, Ickert-Bond S, Labiak PH, Larraín J, Lehnert M, Lewis LR, Nagalingum NS, Patel N, Rensing SA, Testo W, Vasco A, Villarreal JC, Williams EW, Burleiggh JG. (2021) A target enrichment probe set for resolving the flagellate land plant tree of life. Applications in Plant Sciences 9(1): e11406. 10.1002/aps3.11406 [DOI] [PMC free article] [PubMed]
- Cargill DC, Fuhrer BA. (2008) Taxonomic studies of the Australian Anthocerotophyta II: The genus Phaeoceros. Fieldiana. Botany 47(1): 239–253. 10.3158/0015-0746-47.1.239 [DOI] [Google Scholar]
- Chantanaorrapint S. (2009) Phaeocerosperpusillus (Notothyladaceae), a new species of hornwort from Thailand. Acta Botanica Hungarica 51(1–2): 29–33. 10.1556/ABot.51.2009.1-2.6 [DOI] [Google Scholar]
- Chantanaorrapint S, Penjor P, Villarreal JC. (2015) Taxonomic notes on Phaeoceroshimalayensis, with lectotypification of Anthoceroshimalayensis. Phytotaxa 231(2): 193–196. 10.11646/phytotaxa.231.2.9 [DOI] [Google Scholar]
- Duff RJ, Cargill DC, Villarreal JC, Renzaglia KS. (2004) Phylogenetic relationships of the hornworts based on rbcL sequence data: Novel relationships and new insights. Monographs in Systematic Botany from the Missouri Botanical Garden 98: 41–58. [Google Scholar]
- Duff RJ, Villarreal JC, Cargill DC, Renzaglia KS. (2007) Progress and challenges toward developing a phylogeny and classification of the hornworts. The Bryologist 110(2): 214–243. 10.1639/0007-2745(2007)110[214:PACTDA]2.0.CO;2 [DOI]
- Feliner GN, Rosselló JA. (2007) Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics and Evolution 44(2): 911–919. 10.1016/j.ympev.2007.01.013 [DOI] [PubMed] [Google Scholar]
- Hasegawa J. (1986) A collection of the Anthocerotae from Seram and Ambon. Acta Phytotaxonomica et Geobotanica 37: 9–15. [Google Scholar]
- Hasegawa J. (1993) Taxonomical studies on Asian Anthocerotae V. A short revision of Taiwanese Anthocerotae. Acta Phytotaxonomica et Geobotanica 44: 97–112. [Google Scholar]
- IUCN (2022) Guidelines for Using the IUCN Red List Categories and Criteria. Version 15. https://www.iucnredlist.org/resources/redlistguidelines [Accessed 10.10.2023]
- Lai MJ, Zhu RL, Chantanaorrapint S. (2008) Liverworts and hornworts of Thailand: An updated checklist and bryofloristic accounts. Annales Botanici Fennici 45(5): 321–341. 10.5735/085.045.0501 [DOI] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. (2016) PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34(3): 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), November 2010. Institute of Electrical and Electronics Engineers (IEEE), New Orleans, Louisiana, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Peñaloza-Bojacá GF, Maciel-Silva A, Goffinet B, Cargill C, Bell D, Sessa E, Li F, Burleigh G, Endara L, Allen NS, Schafran P, Chantanaorrapint S, McDaniel S, Davis C, Duckett J, Pressel S, Lemus CS, Renzaglia K, Villarreal JC. (submitted) Ancient reticulation and incomplete lineage sorting at the dawn of hornwort diversification and origin of the pyrenoid in the Carboniferous. Annals of Botany.
- Rambaut A. (2017) FigTree (version 1.4.4), a graphical viewer of phylogenetic trees. http://tree.bio.ed.ac.uk/software/figtree/ [Accessed 30.09.2023]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67(5): 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T. (2002) Utility of low-copy nuclear gene sequences in plant phylogenetics. Critical Reviews in Biochemistry and Molecular Biology 37(3): 121–147. 10.1080/10409230290771474 [DOI] [PubMed] [Google Scholar]
- Siagian AU, Ariyanti NS, Djuita NR. (2021) Diversity of hornwort in Mount Slamet (Central Java). Floribunda 6(7): 264–272. 10.32556/floribunda.v6i7.2021.357 [DOI] [Google Scholar]
- Söderström L, Hagborg A, Von Konrat M, Bartholomew-Began S, Bell D, Briscoe L, Brown E, Cargill DC, Costa DP, Crandall-Stotler BJ, Cooper ED, Dauphin G, Engel JJ, Feldberg K, Glenny D, Gradstein SR, He X, Ilkiu-Borges AL, Heinrichs J, Hentschel J, Katagiri T, Konstantinova NA, Larraín J, Long DG, Nebel M, Pócs T, Puche F, Reiner-Drehwald ME, Renner MAM, Sass-Gyarmati A, Schäfer-Verwimp A, Segarra Moragues JG, Stotler RE, Sukkharak P, Thiers BM, Uribe J, Vána J, Villarreal JC, Wigginton M, Zhang L, Zhu RL. (2016) World checklist of hornworts and liverworts. PhytoKeys 59: 1–828. 10.3897/phytokeys.59.6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal JC, Cargill DC, Hagborg A, Söderström L, Renzaglia KS. (2010) A synthesis of hornwort diversity: Patterns, causes and future work. Phytotaxa 9(1): 150–166. 10.11646/phytotaxa.9.1.8 [DOI] [Google Scholar]
- Zhang N, Zeng L, Shan H, Ma H. (2012) Highly conserved low‐copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. The New Phytologist 195(4): 923–937. 10.1111/j.1469-8137.2012.04212.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.