SUMMARY
The reduced effectiveness of COVID-19 vaccines due to the emergence of variants of concern (VOCs) necessitated the use of vaccine boosters to bolster protection against disease. However, it remains unclear how boosting expands protective breadth when primary vaccine platforms are distinct and how boosters containing VOC spike(s) broaden humoral responses. Here, we report that boosters composed of recombinant spike antigens of ancestral (prototype) and Beta VOCs elicit a robust, pan-VOC, and multi-functional humoral response in non-human primates largely independent of the primary vaccine series platform. Interestingly, Beta-spike-containing boosters stimulate immunoglobulin A (IgA) with a greater breadth of recognition in protein-primed recipients when administered with adjuvant system 03 (AS03). Our results highlight the utility of a component-based booster strategy for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for broad humoral recognition, independent of primary vaccine series. This is of high global health importance given the heterogeneity of primary vaccination platforms distributed.
Graphical Abstract
In brief
Cost-effective vaccine-boosting strategies to restore waned antibody responses and expand humoral breadth are of paramount global health interest as SARS-CoV-2 continues to evolve. Deng et al. demonstrate that subunit-based boosters can be used independent of primary series immunizations and that Beta-spike-containing formulations enhance IgA breadth, even to XBB lineages.
INTRODUCTION
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) has reduced protective immunity provided by previous vaccination or infection. Much of this is owed to mutations in the spike antigen by these variants, particularly within the receptor-binding domain (RBD). The spike RBD attaches to angiotensin-converting enzyme 2 (ACE2) and is the target for the majority of neutralizing antibodies.1–6 VOCs such as Beta (also known as B.1.351) or the more recently emerged Omicron (also known as B.1.529) and its associated sublineages contain mutations within the RBD that reduce or completely abrogate neutralizing antibody recognition directed against the original SARS-CoV-2 or previously circulating VOCs.7–10 However, protection against severe disease post-vaccination or recovery has not mimicked the precipitous fall in neutralizing antibody titers,11–15 pointing toward humoral responses outside of neutralizing antibodies and/or T cell responses as critical mediators of protection.
Fc-gamma (Fcγ) effector functions leveraged by non-neutralizing antibodies have been linked to attenuated disease states of COVID-19. Primary vaccines such as mRNA-, component-, and adenovirus-based platforms induce spike-reactive, Fcγ receptor (FcγR)-binding antibodies that mediate innate immune cell phagocytosis, natural killer cell degranulation, cytokine release, and complement deposition.16–22 Fcγ effector functions are expanded upon by boosters targeting ancestral spike antigens, with some notable decreases in elicited neutralization titers against VOCs such as Beta and Omicron. To that end, boosting strategies that incorporate a VOC’s spike antigen into the formulation have been proposed and are currently in use.12,23–27 However little information is available on how boosting with divergent VOC impacts the functional breadth of non-neutralizing responses established by distinct vaccination platforms. Moreover, it is unclear if the addition of adjuvant system 03 (AS03) impacts these non-neutralizing functions post-boost.28–30
In our study, we systematically compared the booster-induced antibody profiles in two groups of macaques; one group was primed with a two-dose mRNA vaccine and boosted with a prefusion protein vaccine, and the other group received a two-dose prefusion protein vaccine as the primary series and was subsequently boosted with prefusion protein vaccine. For each study group, we analyzed the antibody responses induced by vaccines boosted by different vaccine formulations, with and without AS03, and targeted to prototype, Beta, or a bivalent vaccine (prototype + Beta). Previously we reported a significant increase in cross-neutralization antibody titers across all VOCs tested in macaques boosted with prefusion protein booster vaccines and higher functional-to-binding antibody ratios.31 Here, we report that Beta-containing formulations, either by themselves or as part of a bivalent formulation, with AS03 significantly restore and expand antibody responses against all major SARS-CoV-2 VOCs, including Omicron. This recognition was linked to FcγR-binding antibodies, which were expanded by the boost, independent of the initial vaccine series platform. Moreover, Beta-spike-containing vaccinations induced a strong immunoglobulin A (IgA) response, critical to mucosal immunity against respiratory pathogens, particularly in macaques that received a protein-based primary vaccine series. Collectively, our results demonstrate that a Beta-spike-containing, component-based vaccine booster strategy can be employed to expand humoral breadth independent of the initial vaccine series type. We propose that humoral breadth can be expanded through the use of spike antigens containing mutations that exhibit antibody-recognition escape.
RESULTS
Subunit boosters improve antibody profiles in macaques primed with either mRNA or subunit vaccines
We compared the functional role of vaccine-induced antibodies against the prefusion spike of SARS-CoV-2 virus in macaques divided into two different cohorts (Figure 1A). To characterize the antibody response after a primary mRNA series with a subunit (protein) booster and after a full subunit vaccine series, we performed comprehensive antibody profiling across all macaques against the full-length ancestral antigen and VOCs including Alpha, Beta, Delta, and Omicron (Figure S1). Univariate comparison demonstrated that the booster elicited elevated levels of ancestral prefusion spike-specific IgG1-, FcγIIA-, and FcγIIIA-binding antibodies compared to the peak immunogenicity after a primary mRNA vaccine series and pre-boost time points. This was exclusively observed in AS03-containing vaccines (Figure 1B, top row; statistically significant differences from no-AS03 boosters are shown with an asterisk). In the cohort that received a protein primary series with AS03, prefusion spike-specific IgG1-, FcγIIA-, and FcγIIIA-binding antibodies showed a significantly higher response in the high-dose, bivalent-vaccine-boosted group that contained AS03. Other boosters showed different degrees of ancestral spike responsiveness, with some achieving significance relative to the no-AS03 booster (Figure 1B, bottom row). All groups showed a restoration of waned responses independent of the primary vaccine platform.
Figure 1.
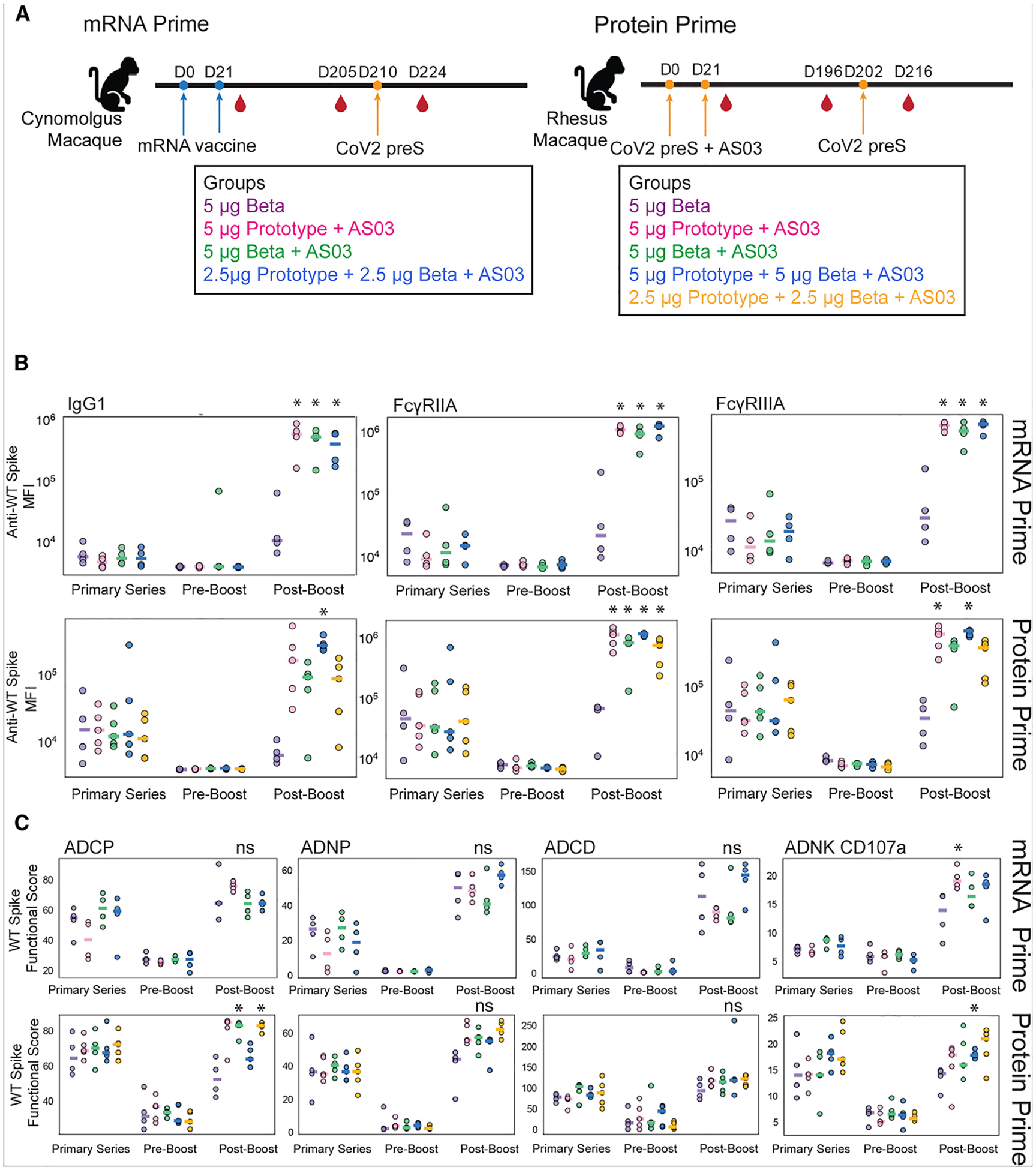
A subunit booster improves antibody profiles in both macaques primed with two-dose mRNA vaccines and macaques primed with two-dose subunit vaccines
(A) Schematic representation of the cohorts: in the mRNA-primed cohort, 16 macaques were evenly split into four groups, each group was primed with mRNA vaccines in different modalities on days 0 and 21 and boosted with a subunit booster on day 210, and blood samples were collected 2 weeks after the second primary series on day 35 and on days 205 and 224. In the protein-primed cohort, 24 macaques were split into five groups, each group was primed with subunit vaccines in different modalities on days 0 and 21 and boosted with a subunit booster on day 202, and blood samples were collected 2 weeks after the second primary series vaccination on day 35 and on days 196 and 216.
(B) The dot plots show the SARS-CoV-2 ancestral spike-specific IgG1 titer and the ability of the spike-specific antibodies to bind to the low-affinity Fcγ receptors (FcγRIIA and FcγRIIIA) across the macaques primed with mRNA vaccines and boosted with a subunit booster in mRNA prime (top) at different time points: peak (day 35), pre-boost (day 205), and post-boost (day 224), and the macaques primed with subunit vaccines and boosted with a subunit booster in protein prime (bottom) at different time points: peak (day 35), pre-boost (day 205), and post-boost (day 224). Each dot represents a different macaque. Each bar represents the median of each group.
(C) The dot plots show antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent complement deposition (ADCD), and antibody-dependent natural killer (NK) cell activity (ADNK), as measured by CD107a degranulation, against SARS-CoV-2 ancestral spike in macaques primed with mRNA vaccines and boosted with a subunit booster in mRNA prime (top) at different time points: peak (day 35), pre-boost (day 205), and post-boost (day 224), and the macaques primed with subunit vaccines and boosted with a subunit booster in protein prime (bottom) at different time points: peak (day 35), pre-boost (day 196), and post-boost (day 216). *p < 0.05 against the no-AS03 post-boost group, ns, not significant.
To determine whether the differences in FcγR binding translate to functional differences, we next compared several Fc-mediated effector activities of vaccine-induced antibodies. These included antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent cellular phagocytosis (ADCP), antibody-dependent complement deposition (ADCD), and antibody-dependent natural killer cell degranulation, as measured by CD107a expression, a degranulation marker (ADNK CD107a) (Figure 1C). We found that a subunit booster results in an upregulation of functional activity across both cohorts, but there was no clear trend for which formulation gave the broadest functional protection. For the mRNA-primed group, only boosting with the prototype resulted in a significant increase in ADNK activity compared with the no-AS03 boosted group (Figure 1C, top row). For the protein-primed group, a Beta and a bivalent booster yielded higher ADCP, and a high-dose bivalent gave higher ADNK compared with the no-AS03 reference group (Figure 1C, bottom row). Ultimately, restorative effects were observed across all functional assays after the booster (Figure 1C). In all cases, boosting with a component-based vaccine restored waned functionality and, in many cases, increased functionality compared to peak immunogenicity after the primary series.
Subunit boosters with AS03 enhance antibody profiles against the SARS-CoV-2 spike of VOCs and across some subdomains of the ancestral spike
To better understand the changes in the expansion of post-booster antibody responses against other VOCs, we analyzed IgG1 responses for each cohort for all SARS-CoV-2 spike VOCs, including Omicron. Boosting exponentially expanded IgG1 recognition of the full-length spikes of Alpha (Figures 2A and 2B, top left, B.1.1.7), Beta (Figures 2A and 2B, top right, B.1.351), Delta (Figures 2A and 2B, bottom left, B.1.617.2), and Omicron BA.1 (Figures 2A and 2B, bottom right, B.1.1.529) for both mRNA-primed and protein-primed cohorts. Interestingly, antibody responses after the subunit booster in the absence of AS03 displayed little expansion to spikes of all VOCs (purple line, shaded colored regions represent the standard error of the mean). However, in the presence of AS03, all protein boosters exhibited pan-VOC spike reactivity (Figures 2A and 2B; pre-boost values were set to 1 to calculate fold expansion for each booster for each VOC). For the protein-primed cohort, the bivalent booster demonstrated a dose-dependent effect on spike-specific IgG1 expansion, with the highest dose eliciting the highest titers for each VOC spike tested (Figure 2B, compare blue and yellow lines). Similar patterns were also observed in the fold expansion of IgG1 titers against the RBD of the VOC (Figure S2).
Figure 2.
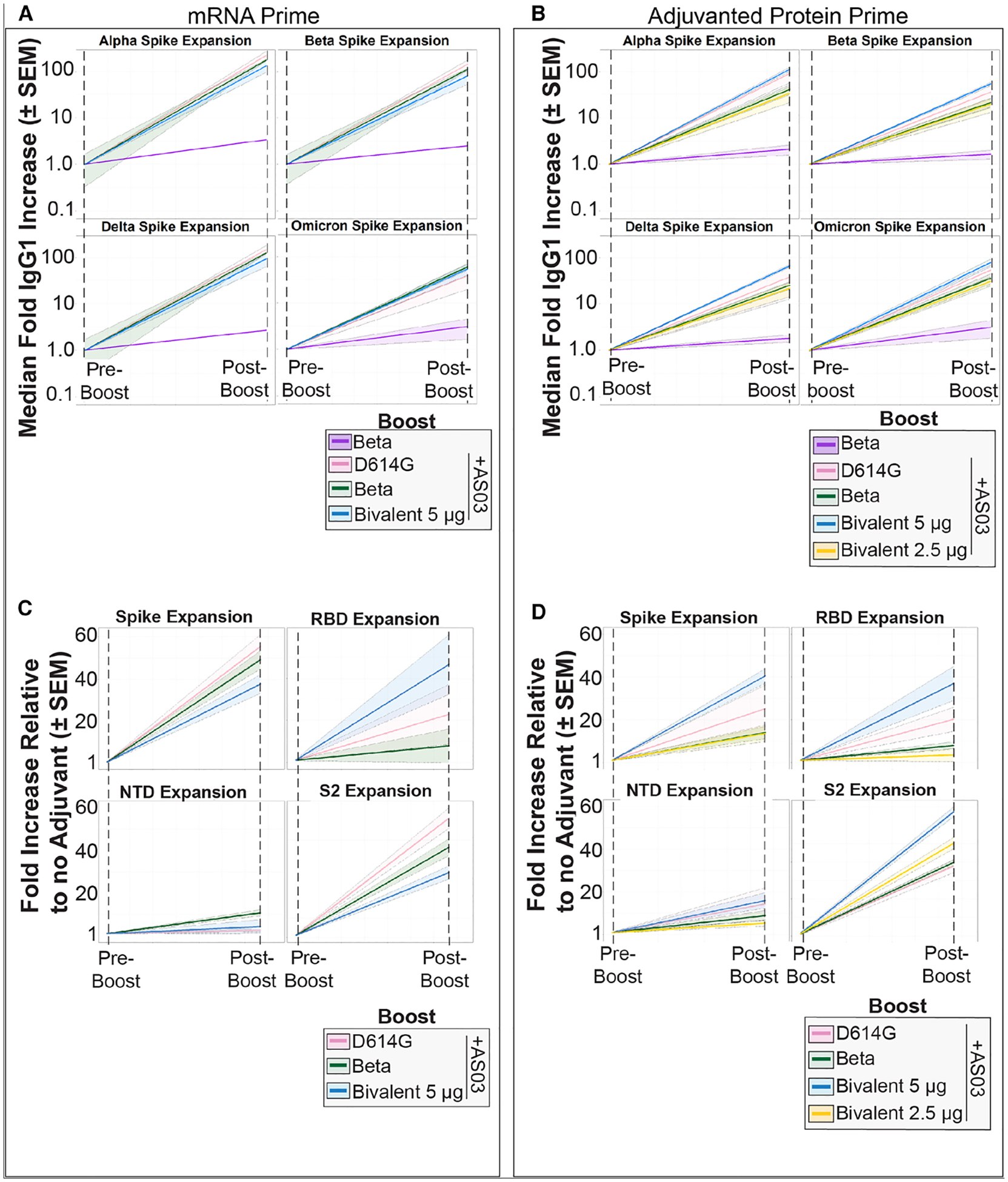
Protein boosters with AS03 expand IgG1 recognition of spikes in a VOC-independent manner
(A) Fold expansions of antibodies against the Alpha, Beta, Delta, and Omicron VOCs for the boosted mRNA-primed cohort. Pre-boost time points were standardized to 1 across boosting strategies (color legend shown in the bottom right), and post-boost increases were plotted relative to their pre-boost values. The median fold increases (solid, colored line) and the standard error of the mean (SEM; shaded region of the same color) are shown.
(B) Same as (A) but for the protein-primed cohort. Note that this cohort has two bivalent groups distinguished by the amount of protein in the dose (blue, 5 μg; gold, 2.5 μg).
(C) Subdomain expansion in the mRNA-primed cohort was plotted similarly to (A). Fold expansion against full-length ancestral spike, RBD, NTD, and S2 was quantified with each pre-boost point standardized to 1 across boosting strategies. All values are shown relative to the no-AS03 group.
(D) Same as (C) but for the protein-primed cohort.
To assay which spike subdomains were expanding within each group, we analyzed recognition of the spike N-terminal domain (NTD), RBD, and S2 domains before and after the boost. Fold expansions were measured relative to the no-AS03 group. For both the mRNA and protein primary series, NTD-specific IgG1 showed little expansion post-boost (Figures 2C and 2D, bottom left). Interestingly, expansion toward the wild-type (WT) RBD was highly heterogeneous for both cohorts (Figures 2C and 2D, top right). However, in each group, the bivalent booster yielded the highest expansion (blue line); this increase was lost in lower doses in the protein-primed cohort (Figures 2B and 2D, gold line; the mRNA-primed cohort did not receive a 5 μg dose). In both cohorts, the most consistent expansion was toward S2, which is the more conserved subdomain of spike across betacoronaviruses (Figures 2C and 2D, bottom right). Expansion of S2-binding antibodies accounted for the majority of spike expansion of the domains tested across boosting types. Overall, correlates differed in each group after the boost, further highlighting that initial priming platforms can influence overall humoral architecture. Notably, a higher dose of the bivalent booster showed a tighter correlative humoral response than the lower dose, further supporting a dose-dependent phenotype (Figure S3).
Subunit booster vaccines increase humoral breadth against VOC independent of primary series
We then examined the breadth of the boosted antibody responses afforded by the protein boosters compared with those at the pre-boost time point. We plotted the responses as radar plots to visually demonstrate the effect of the boosters relative to the pre-boost time points. All data shown are the fold increase of the mean antibody or the functional feature relative to the corresponding groups’ pre-boost mean values. We found that protein boosters containing AS03 provided expanded humoral profiles in terms of antibody titers (IgA, IgG1, IgG3, IgM) and FcγR-binding titers (FcγRIIA and FcγRIIIA) against the ancestral, Beta, Delta, and Omicron spikes in the mRNA-primed cohort (Figure 3A). The Beta booster with AS03 showed the lowest breadth in fold expansions relative to pre-boost of antibody-dependent functional score (ADCP, ADCP, ADNK, ADNP) boosting among the AS03-containing boosters in the mRNA-primed cohort. Spike-specific, post-booster antibody responses in the mRNA-primed cohort (Figure S4A) also shared the same pattern, that protein boosters with AS03 tend to elicit higher antibody titers and FcγR binding, but this did not necessarily translate to functionality differences.
Figure 3.
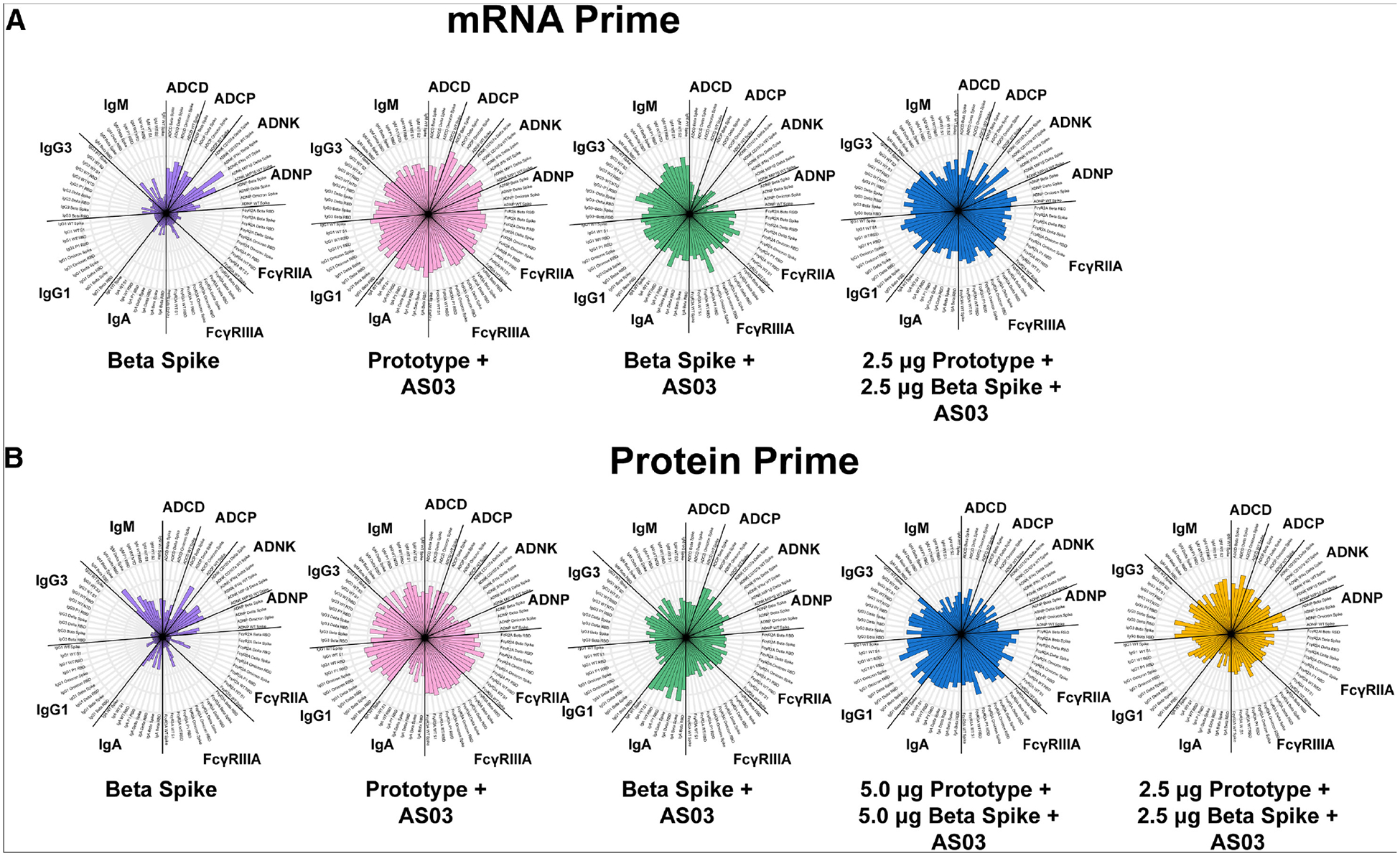
Boosters with AS03 induce a broader response between the post-booster and the pre-booster time points
(A) Median ranked percentiles of fold changes (post-boost relative to pre-boost) in antibody titers (IgA, IgG1, IgG3, IgM), FcγR-binding titers (FcγRIIA and FcγRIIIA), and antibody-dependent functional scores (ADCP, ADCP, ADNK, ADNP) against the ancestral, Beta, Delta, and Omicron antigens for the mRNA-primed cohort.
(B) Median ranked percentiles of fold changes (post-boost relative to pre-boost) in antibody titers (IgA, IgG1, IgG3, IgM), FcγR-binding titers (FcγRIIA and FcγRIIIA), and antibody-dependent functional scores (ADCP, ADCP, ADNK, ADNP) against the ancestral, Beta, Delta, and Omicron antigens for the protein-primed cohort.
Subunit boosters with AS03, monovalent or bivalent, exhibited a robust expansion of antibody binding compared with the no-AS03 booster (Figure 3B). Additionally, boosters with AS03 also induced a stronger boosting effect in terms of most antibody titers and FcγR-binding titers. Interestingly, the no-AS03 Beta booster induced higher post-booster IgM titers against Beta and Delta spikes than protein boosters with AS03 (Figure S4B). Despite having relatively high natural killer cell-produced interferon γ (IFN-γ) against Delta and ADCP against Omicron spikes, the no-AS03 protein booster did not show broader antibody responses compared with the protein booster with AS03 (Figure S4B). By focusing on the bivalent groups in the protein-primed cohort at the post-booster time point (Figure S4B), we found that the higher-dose group elicited broader responses in terms of antibody titers and FcγR-binding titers, while the lower-dose group induced broader and stronger functional activities.
Protein-based boosters expand the breadth of IgA binding
Notably, protein-based boosters with AS03 elicited consistently higher IgA responses (Figures S1 and S4) compared with the other vaccine formulations tested. At the VOC level, a consistent restoration of waned spike-reactive IgA was observed in the mRNA prime series (Figure 4A). For all VOCs, IgA responses post-boost either fully restored waned IgA titers or expanded breadth. This was particularly evident for WT and Delta spike IgA responses. Reactivity was not observed at any time for tetanus antigen, which serves as a specificity control. Interestingly, for the protein-primed cohort, IgA expansions were disproportionately observed in Beta-containing formulations. This was true for Beta + AS03 (Figure 4B, green line) as well as the high-dose bivalent vaccine recipients (Figure 4B, blue line). Boosts with prototype (Figure 4B, pink) and lower-dose bivalent (Figure 4B, gold) showed only a rescue of waned responses. Tetanus antigen again was used as an antigen specificity control and showed no responses at any time point for any group.
Figure 4.
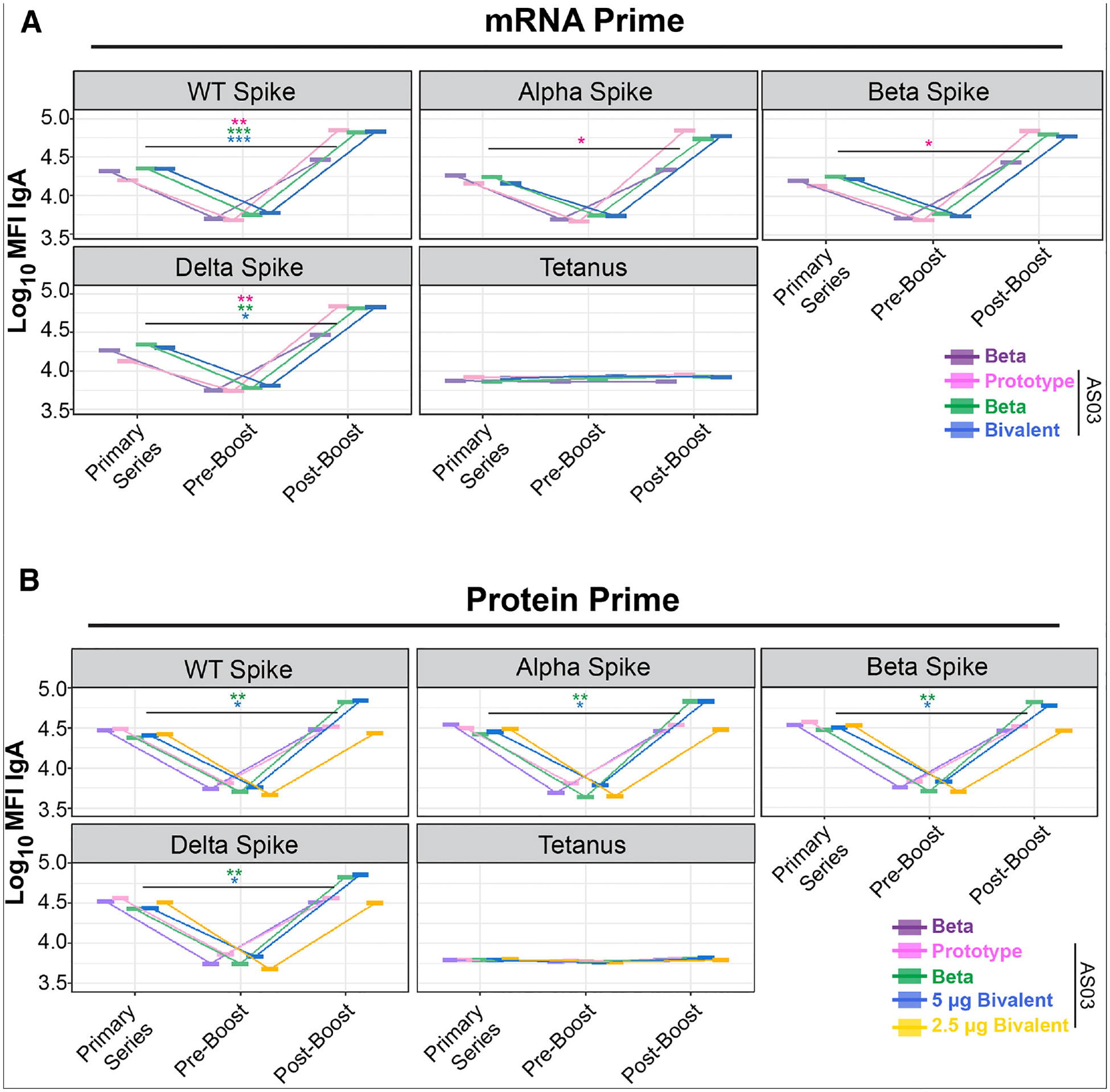
IgA toward SARS-CoV-2 spikes is expanded by Beta-containing component boosters in protein-primed NHPs
(A) Mean IgA binding to indicated SARS-CoV-2 VOCs in mRNA-primed NHPs. Shown are the values after the primary series, pre-boost, and post-boost against the indicated VOC spike or tetanus as a negative control. Shown on the y axis is the IgA mean fluorescent intensity (MFI) of binding to the antigen, and on the right is the color scheme.
(B) Same as (A) but for NHPs who received a protein primary vaccines series. Statistical significance (Mann-Whitney U test) of peak responses after primary series and post-boost was determined to show expansions of IgA binding breadth. *p < 0.05, **p < 0.01, and ***p < 0.001.
To assay for binding breadth of recently emerged variants, we tested IgG1 and IgA for binding against BA.2, BA.5, BM1.1, BQ1.1, CH1.1, XBB1.5, and XBB1.16. All of these variants are Omicron-lineage descendants that have caused surges in various regions, particularly in 2022–2023.32–36 Against all variants tested, IgG1 and IgA showed increased binding post-boost. This was especially noticeable for formulations containing AS03. Interestingly, IgA responses showed a higher degree of boosting than IgG1 for these variants and were again most strongly induced in formulations containing Beta spike, particularly in a bivalent form (Figure S5). This phenomenon was also primary series platform independent.
These findings further support a model in which boosting with protein-based formulations not only restores waned responses but can expand humoral breadth regardless of the primary platform immunization strategy. Notably, Beta-containing formulations demonstrate an ability to restore and expand IgA responses against VOCs in protein-primed non-human primates (NHPs). This is particularly important because IgA predominantly acts in mucosal regions.
DISCUSSION
Several studies have shown a waning of immunity against SARS-CoV-2 virus after a primary two-dose series, regardless of platform.37 The waning of effects can be linked to decreased concentrations in serum antibody levels, a lower initial response, underlying health conditions, or a combination of any of the aforementioned. Additionally, as SARS-CoV-2 has adapted to its new human host, mutations exhibiting selective sweeps have been observed, with many of these mutations showing various degrees of antibody-recognition escape. This is particularly notable for the Omicron VOC lineage, which contains numerous mutations that reduce or eliminate neutralizing antibody recognition in vaccinated or recovered patients. To that end, COVID-19 boosters have been deployed to restore antibody levels and expand recognition of spike antigens. It can be reasonably assumed that SARS-CoV-2 will continue to accumulate mutations that reduce humoral recognition, requiring periodic boosters in the future.7,38
Deployment of the COVID-19 vaccines has been affected by their production pipeline and storage conditions. For example, initial shipments of BNT162b2 and mRNA-1273 vaccines required ultracold storage, creating an obstacle to equitable distribution.39 Component-/protein-based vaccines benefit from their ease of scalability and production into good manufacturing practices and/or good clinical laboratory practices. Transport and distribution of protein-based vaccines is also more convenient since cold-chain storage is at 4°C–8°C. This can be of particular importance to resource-limited regions. Therefore, their utility as a booster platform that could restore and expand humoral function should be considered.
In this study, we systematically evaluated the humoral immune responses against SARS-CoV-2 VOCs elicited by a subunit booster in two study groups of macaques: one group was primed with a two-dose mRNA vaccine and the other with a two-dose subunit vaccine. In each study group, macaques were further separated into different groups receiving one-dose subunit boosters with different formulations. From the booster-induced antibody profiles against several VOCs of SARS-CoV-2, we observed that a subunit booster can induce high IgG1 titers and Fc-binding titers in all macaques at a comparable level or exceeding that of the peak immunogenicity after two-dose prime vaccines when used with AS03. Specifically, we observed that Beta-containing protein boosters, when administered with AS03, elicited robust recognition of all SARS-CoV-2 VOC spikes, restored waned functionality, and expanded antibody isotype recognition of divergent spikes. Our results are consistent with other reports of increased neutralization capacity of monovalent or bivalent Beta-spike-containing boosted NHPs, particularly against Omicron lineages.40,41 We have shown that these boosted humoral responses are not exclusive to neutralizing antibodies. Of particular interest was the highly expanded IgA responses we observed in Beta-spike-containing formulations.
Interestingly, the greatest fold expansion in IgG1 titer was against the S2 subdomain of the spike, which is a conserved region among all variants and a target for future SARS-CoV-2 vaccine designs.42 A consistent increase in IgA titers was observed in the subunit-boosted NHPs. Beta-variant-containing formulations gave cross-VOC increases in IgA recognition in the protein-primed cohort, consistent with the higher neutralization titers for these vaccines demonstrated in both NHP studies and several clinical trials.24,31,43 In humans, IgA has been shown to play a potent role in mucosal immunity through direct neutralization and opsonophagocytosis.44 This is notable given that the breadth of the cross-neutralization in the aforementioned NHP studies was also improved for the Beta-containing compared with prototype boosters. Collectively, this suggests that Beta-containing formulations may afford higher mucosal protection through the generation of broadly reactive, high-titer IgA responses that can leverage neutralizing and non-neutralizing functions at the site of infection. Importantly, these responses are not diminished in the presence of an ancestral-lineage spike.
In conclusion, our results suggest that variant-containing booster vaccines can bolster both neutralizing and non-neutralizing functions. Moreover, boosters containing the Beta spike, either by itself or in combination with other spikes, can restore waned functions and expand protective breadth. These results were largely independent of primary vaccination platforms, which is critical given the diversity of primary vaccination series platforms administered globally.
Limitations of the study
The present study was designed to deeply profile the humoral responses of NHPs boosted with various protein-based formulations. As the COVID-19 pandemic progressed, new variants continuously emerged, particularly Omicron-based lineages. We could not profile responses against all Omicron lineages due to the diversity within the lineage. Moreover, we could not perform live-virus neutralizations against the VOCs due to sample volume limitations.
The identification that IgA was disproportionately expanded in Beta-containing formulations was of high interest given its role in mucosal defenses. However, the IgA responses characterized in this study were from sera and not nasal washes or bronchiolar lavage fluid, per the allowed protocol design. This could also complicate our functional antibody assays, as IgA is known to have neutralizing and non-neutralizing roles, and these roles may be compartment specific.44–46 Further studies into compartment-based IgA responses, and how they correlate with serum-containing IgA, are thus imperative.
Another limitation of this study was our inability to correlate antibody profiles with T cell responses. Other groups have shown that a Beta spike subunit booster elicited a polyfunctional, TH-1 CD4+ response to Omicron peptides, in addition to stimulating a higher Omicron-specific neutralizing antibody response.24 To that end, a comprehensive systems-immunology-based approach to characterize immune responses to boosting strategies would be highly informative.
In our study, vaccines were administered with and without AS03; however, study design did not permit us to have an AS03 group alone. How AS03 was potentiating effects is thus very difficult to characterize. Additionally, our study is limited by the number of macaques within each treatment arm. Therefore, small effects between antigens (for example, prototype vs. Beta spike) were not likely to be captured in this study. It should be noted that all vaccine boosters contained the same concentration of AS03. Therefore, the observed changes between prototype alone or a lower-dose bivalent compared with higher-dose Beta-spike-containing formulations cannot be explained solely by AS03. How AS03 or other adjuvants specifically leverage certain antibody functions is an emerging field. Numerous groups have shown that adjuvants can modify neutralization breadth and Fc-effector functions, particularly with subunit-based vaccines.47–52 Fully capturing how small changes within the antigen itself (e.g., prototype vs. Beta spike) interplay with a specific adjuvant would be paradigm shifting for next-generation vaccination platforms and strategies.
Lastly, strict comparisons between the two major arms in this study (mRNA vaccine primed and subunit protein primed) cannot be done due to the different species used. Other groups have shown that humoral profiles between rhesus macaques and cynomolgus macaques can have distinctions,53 although their responses to SARS-CoV-2 appear similar.54 We therefore only showed fold expansions post-boost within each of the major arms and did not conduct cross-species analyses.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Information related to this manuscript including datasets, protocols, and procuring of reagents and resources should be directed to the lead contact, Ryan McNamara (rpmcnamara@mgh.harvard.edu).
Materials availability
All materials and reagents used in this study are commercially available and can be found in the key resources table. No new unique reagents were generated in this study.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | DENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD66β (clone G10F5), Pacific Blue | BioLegend | AB_2563294 |
| Mouse anti-human CD3 (clone UCHT1), Alexa Fluor 700 | BD Biosciences | AB_396952 |
| Mouse anti-human CD107α (clone H4A3), PE-Cy5 | BD Biosciences | AB_396136 |
| Mouse anti-human IFNγ (clone B27), FITC | BD Biosciences | AB_398580 |
| Mouse anti-human MIP-1β (clone D21–1351), PE | BD Biosciences | AB_393549 |
| Mouse anti-human CD56 (clone B159), PE-Cy7 | BD Biosciences | AB_396853 |
| Mouse anti-human CD14 (clone MφP9), APC-Cy7 | BD Biosciences | AB_396889 |
| Mouse anti-human CD16 (clone 3G8), APC-Cy7 | BD Biosciences | AB_396864 |
| Anti-guinea pig complement C3 goat IgG fraction, FITC | MP Biomedicals | Cat# 0855385; RRID:AB_2334913 |
| Mouse anti-IgG1-PE | SouthernBiotech | Cat# 9054–09; RRID:AB_2796628 |
| Mouse anti-IgG3-PE | SouthernBiotech | Cat# 9210–09; RRID:AB_2796701 |
| Mouse anti-IgA-PE | SouthernBiotech | Cat# 9130–09; RRID:AB_2796656 |
| Anti-rhesus IgG1 | NHPRR | AB_2819310 |
| Anti-rhesus IgG3 | NHPRR | PR-1130 |
| Anti-rhesus IgA | NHPRR | AB_2819303 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 WT Spike | Sino Biological | 40589-V08H4 |
| SARS-CoV-2 WT S1 | Sino Biological | 40591-V08H |
| SARS-CoV-2 WT RBD | Sino Biological | 40592-V08H |
| SARS-CoV-2 WT S2 | Sino Biological | 40590-V08B |
| SARS-CoV-2 WT NTD | Sino Biological | 40591-V49H |
| SARS-CoV-2 D614G Spike | Sino Biological | 40589-V08B4 |
| SARS-CoV-2 Alpha Spike | Sino Biological | 40589-V08H12 |
| SARS-CoV-2 Alpha RBD | Sino Biological | 40592-V08H82 |
| SARS-CoV-2 Beta Spike | Sino Biological | 40589-V08B7 |
| SARS-CoV-2 Beta RBD | Sino Biological | 40592-V08H59 |
| SARS-CoV-2 Gamma Spike | Sino Biological | 40589-V08B10 |
| SARS-CoV-2 Gamma RBD | Sino Biological | 40592-V08H86 |
| SARS-CoV-2 Delta Spike | Sino Biological | 40589-V08B16 |
| SARS-CoV-2 Delta RBD | Sino Biological | 40592-V08H115 |
| SARS-CoV-2 Omicron BA.1 Spike | Sino Biological | 40589-V08H26 |
| SARS-CoV-2 Omicron BA.2 Spike | Sino Biological | 40589-V08H28 |
| SARS-CoV-2 Omicron BA.5 Spike | Sino Biological | 40592-V08H32 |
| SARS-CoV-2 Omicron BM.1.1 Spike | Sino Biological | 40589-V08H43 |
| SARS-CoV-2 Omicron BQ.1.1 Spike | Sino Biological | 40589-V08H41 |
| SARS-CoV-2 CH.1.1 Spike | Sino Biological | 40589-V08H46 |
| SARS-CoV-2 XBB.1.5 Spike | Sino Biological | 40589-V08H45 |
| SARS-CoV-2 XBB.1.16 Spike | Sino Biological | 40589-V08H49 |
| NHP FcγRIIA | Duke Human Vaccine Institute | Custom Order |
| NHP FcγRIIIA | Duke Human Vaccine Institute | Custom Order |
| Streptavidin-PE | Agilent Technologies | PB32-10 |
| Ebola Virus Glyoprotein | IBT Bioservices | 0501–015 |
| Influenza HA | Sino Biological | 11687-V08H |
| Brefeldin A | Sigma | B7651 |
| Guinea Pig Complement | Cedarlane | CL4051 |
| Protein Transport Inhibitor | BD Biosciences | 554724 |
| LC-LC-Sulfo-NHS Biotin | ThermoFisher | A35358 |
| Brefeldin A | Sigma Aldrich | B7651 |
| GolgiStop | BD Biosciences | 554724 |
| Streptavidin-R-Phycoerythrin | Prozyme | PJ31S |
| Critical commercial assays | ||
| RosetteSep NK cell enrichment kit | Stem Cell Technologies | 5025 |
| Experimental models: Cell lines | ||
| THP-1 monocytes | ATCC | CVCL_0006 |
| Deposited Data | ||
| RagonSystemSerology: CR20230925 | GitHub | N/A |
| Experimental models: Organisms/strains | ||
| Cynomolgus macaques (Mauritius) | New Iberia Research Center | N/A |
| Rhesus macaques | New Iberia Research Center | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad Software, Inc. | Ragon License |
| R Studio V. 4.0.4 | R Project for Statistical Computing | Open Source |
| Flow Jo | BD Bioscience | Ragon License |
| Python V 3.8.8 | MathWorks | Open Source |
| Matplotlib V 3.3.3 | Mathworks with Python | Open Source |
| Other | ||
| FluoSpheres® NeutrAvidin®-Labeled Microspheres, 1.0 μm, yellow-green fluorescent (505/515), 1% solids | Life Technologies | F-8776 |
| FluoSpheres® NeutrAvidin®-Labeled Microspheres, 1.0 μm, red fluorescent (580/605), 1% solids | Life Technologies | F8775 |
| MagPlex microspheres | Luminex corporation | MC12001-01 |
Data and code availability
Systems serology raw datasets have been deposited on GitHub on RagonSystemSerology: CR20230925.
All original code used for analysis in this manuscript can be found in the systemsseRology on GitHub (LoosC/systemsseRology). All other codes used are open source and available upon request to the lead contact. This manuscript does not report original code unique to this study.
Any additional information required for analysis of the data reported in this manuscript is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Animals and study design
Animal experiments were conducted in compliance with all pertinent US National Institutes of Health regulations according to approved animal protocols from the Institutional Animal Care and Use Committee (IACUC) at the research facilities. The NHP study was performed at the University of Louisiana at Lafayette New Iberia Research. Macaques between the ages of two and eight years-old were randomized based on sex, age, and weight (Figure 1A) and divided into two study groups, mRNA Prime and Protein Prime. In the mRNA Prime study group, 16 Cynomolgus macaques received two doses of mRNA vaccine primers on Day 0 and Day 21, respectively, and subsequently a subunit booster on Day 205. Blood samples were collected at peak immunogenicity after the first two doses, Day 210, as well as Day 224. The group of 16 animals was further divided into groups of four, with each group receiving one of the four different booster schemes. Group 1 received 5 μg of soluble prefusion-stabilized Beta spike timer (PANGO lineage B.1.351), group 2 received 5 μg prefusion-stabilized Prototype spike timer with AS03, group 3 received 5 μg Beta prefusion-stabilized spike timer with AS03, and group 4 received a bivalent 5 μg prefusion-stabilized spike timer (Prototype + Beta) with AS03. In the Protein Prime study group, 24 Rhesus macaques received two doses of Prototype prefusion-stabilized spike timer with AS03 vaccine as the primer on Day 0 and Day 21, respectively, and subsequently a subunit booster on Day 202. Blood samples were collected at peak immunogenicity after the first two doses, Day 196, as well as Day 216.
METHOD DETAILS
Vaccines
Vaccine candidates were SARS-CoV-2 prefusion spike antigens based on the ancestral (Prototype) or Beta variant sequences with AS03 adjuvant (referred to as AS03 throughout the text and figures) and formulated as monovalent or bivalent vaccines as described previously.31,47
Messenger RNA incorporating coding sequences containing either the wild type (WT) sequence, stabilized pre-fusion mutant (2P),55 furin cleavage site mutant (GSAS),56,57 double mutant (2P, GSAS)), triple mutant (2P/GSAS/ALAYT), hexamutant (6P) and hexamutant with furin cleavage mutant (6P/GSAS) of the full-length SARS-CoV-2 spike glycoprotein were synthesized by in vitro transcription employing RNA polymerase with a plasmid DNA template encoding the desired gene using unmodified nucleotides. The resulting purified precursor mRNA was reacted further via enzymatic addition of a 5′ cap structure (Cap 1) and a 3′ poly(A) tail of approximately 200 nucleotides in length as determined by gel electrophoresis. The vaccine sequence is based on the Wuhan Hu-1 strain (GenBank accession MN908947). Preparation of mRNA/lipid nanoparticle (LNP) formulations was described previously.58 Briefly, an ethanolic solution of a mixture of lipids (ionizable lipid, phosphatidylethanolamine, cholesterol, and polyethylene glycol-lipid) at a fixed lipid and mRNA ratio were combined with an aqueous buffered solution of target mRNA at an acidic pH under controlled conditions to yield a suspensionn of uniform LNPs. Upon ultrafiltration and diafiltration into a suitable diluent system, the resulting nanoparticle suspensions were diluted to final concentration, filtered, and stored frozen at −80°C until use.
Ig Subclassing/Isotyping and FcγR binding
Antigen-specific antibody isotype titer and Fc-gamma receptor (FcγR)-binding was determined by a multiplex Luminex assay, as previously described.59 Carboxylated magplex microspheres (Luminex) were covalently linked to antigens using NHS-ester linkages by the addition of Sulfo-NHS and EDC (Thermo Fisher). Immune complexes were formed by adding the diluted serum to antigen-coupled microspheres, and plates were incubated overnight at 4°C, shaking at 700 rpm. The following day, plates were washed with 0.1% BSA and 0.02% Tween 20. After the wash, the immune complexes were then shaken and incubated with isotype-specific mouse anti-rhesus antibodies (NHPRR and Life Diagnostics) at room temperature for 2 h. For the detection of antibody isotype titer, PE-coupled goat anti-mouse detection antibodies (Invitrogen) were added to the plates. For the detection of FcγR-binding, Avi-tagged rhesus FcγRs (Duke Human Vaccine Institute) were biotinylated using BirA500 kit (Avidity) per manufacturer’s instructions and tagged with streptavidin-PE. The PE-tagged FcγR was added to the immune complex. Fluorescence was acquired using an iQue (Intellicyt), and the data represent the median fluorescence intensity (MFI). The Luminex assay was run in duplicate, and the data reported represents the average of the duplicates. Materials and resources can be found on the key resources table.
Antibody-dependent cellular phagocytosis (ADCP)
THP-1 cells (ATCC) were maintained in RPMI-1640 (Sigma Aldrich) supplemented with 10% fetal bovine serum (FBS), 5% penicillin/streptomycin (Corning, 50 μg/mL), 5% L-glutamine (Corning, 4 mM), 5% HEPES buffer (pH 7.2) (Corning, 50 mM) and 0.5% 2-Mercaptoethanol (Gibco, 275 μM) at 37°C, 5% CO2. ADCP was performed as previously described.60 Briefly, D614G, B.1.351 or Omicron spike (Sino Biological) were biotinylated and coupled to yellow-green NeutrAvidin FluoSpheres (Invitrogen). Immune complexes were formed by mixing antigen-coupled beads with serum diluted 1:100 in PBS. Immune complexes were incubated for 2 h at 37°C and washed in PBS. THP-1 cells were added to immune complexes at a concentration of 1.25×105 cells/mL and incubated overnight at 37°C, 5% CO2. The ability of antibodies to drive bead uptake by THP-1 cells was assessed by flow cytometry using a BD LSR II cytometer. PhagoScores were calculated as follows: (% bead+ cells x GeoMean of cells)/10000. Samples were run in duplicate, and the data represent the average of the duplicates.
Antibody-dependent neutrophil phagocytosis (ADNP)
ADNP was performed as previously.61 Peripheral whole blood was collected by the Ragon Institute from healthy volunteers. Volunteers provided signed consent, were over 18 years old, and were deidentified. The study was approved by the MGH Institutional Review Board. Red blood cells were lysed by ammonium chloride potassium (ACK) lysis. White blood cells were washed with PBS and maintained in RPMI-1640 (Sigma Aldrich) media supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich), 5% penicillin/streptomycin (Corning, 50 μg/mL), 5% L-glutamine (Corning, 4 mM), 5% HEPES buffer (pH 7.2) (Corning, 50 mM)) and 37°C, 5% CO2 for the duration of the assay. Yellow-green NeutrAvidin FluoSpheres were coupled to antigen as described for ADCP. Immune complexes were formed by mixing serum diluted 1:50 in PBS with coupled beads and incubating for 2 h at 37°C. Immune complexes were washed and white blood cells were added at a concentration of 2.5×105 cells/mL. Cells were incubated with immune complexes for 1 h at 37°C. PacBlue anti-CD66b (BioLegend, clone: UCH71) was used to stain for neutrophils. Phagocytosis was measured by flow cytometry using an iQue (Intellicyt). Phagocytosis by neutrophils (CD66b+) was calculated as described for ADCP. The experiment was performed with two donors and the reported value is the average of the two donors.
Antibody-dependent complement deposition (ADCD)
ADCD was performed as previously described.62 Red NeutrAvidin FluoSpheres were coupled to antigen as described for ADCP. Immune complexes were formed by mixing serum diluted 1:10 in PBS with coupled beads and incubating for 2 h at 37°C. Immune complexes were washed and lyophilized guinea pig complement (Cedarlane) diluted in gelatin veronal buffer with calcium and magnesium (Sigma Aldrich) was added to immune complexes and incubated for 20 min at 37°C. C3 deposition was measured by anti-guinea pig C3 FITC (MpBio). Fluorescence was acquired using a BD LSR II cytometer. C3 deposition is reported as the median fluorescence intensity of FITC. The experiment was performed in duplicate, and the reported value is the average of the two replicates.
Antibody-dependent natural killer cell (NK) activation (ADNKA)
ADNKA was performed as described previously.63 ELISA plates were coated with 2 μg/mL of antigen and incubated for 2 h at 37°C. Plates were washed with PBS and blocked overnight with 5% bovine serum albumin (BSA) at 4°C. Buffy coats were collected by Massachusetts General Hospital from healthy donors who were over 18 years old and provided signed consent. Samples were deidentified before use. NK cells were isolated from buffy coats using RosetteSep (STEMCELL Technologies) and then separated using a ficoll gradient. NK cells were rested overnight at 37°C, 5% CO2 in R10 (RPMI-1640 (Sigma Aldrich) media supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich), 5% penicillin/streptomycin (Corning, 50 μg/mL), 5% L-glutamine (Corning, 4 mM), 5% HEPES buffer (pH 7.2) (Corning, 50 mM)) supplemented with 2 ng/mL IL-15. The following day, plates were washed, and samples were diluted 1:25 in PBS were added to the plates. Plates were incubated for 2 h at 37°C, washed, and NK cells were added at a concentration of 2.5×105 cells/mL in R10 media supplemented with anti-CD107a–phycoerythrin (PE)–Cy5 (BD Biosciences, lot # 0149826, 1:1000 dilution), brefeldin A (10 mg/mL) (Sigma-Aldrich), and GolgiStop (BD Biosciences). Plates were incubated for 5 h at 37°C. Following the incubation, cells were stained for surface markers with anti-CD3 Pacific Blue (BD Biosciences, clone G10F5)), anti-CD16 allophycocyanin (APC)-Cy5 (BD Biosciences, clone 3G8), and anti-CD56 PE-Cy7 (BD Biosciences, clone B159) for 15 min at room temperature. Cells were fixed with PermA (Life Technologies) and permeabilized with PermB (Life Technologies) and stained with anti-MIP-1β PE (BD Biosciences) and anti-IFN-γ FITC, and for 15 min at room temperature. Fluorescence was analyzed by flow cytometry using a BD LSR II. NK cells were gated as CD56+/CD16+/CD3-and activity was determined as the percent of NK cells positive for CD107a or MIP-1b. The assay was performed with two donors and the data reported represents the average of the two donors.
QUANTIFICATION AND STATISTICAL ANALYSIS
All figures were plotted using Python version 3.8.8, with package matplotlib 3.3.3, or with R studio version 4.0.4. For statistical groupings, an initial Mann-Whitney U test, a non-parametric testing method, was performed using multiple corrections to assess statistical significance when necessary. For correlation plots, a Spearman’s Rank correlation was performed between paired samples. To show differences in antibody binding and functional activity in one heatmap, all data was z-scored and plotted using R studio as elsewhere described.20
Supplementary Material
Highlights.
Subunit-based boosters with AS03 restore waned humoral responses to VOC spikes
Expansion of antibody binding breadth is independent of the primary series platform
Beta-containing boosters stimulate high IgA to VOC spikes including XBB lineages
ACKNOWLEDGMENTS
We thank Nancy Zimmerman, Mark and Lisa Schwartz, an anonymous donor (financial support), Terry and Susan Ragone, and the SAMANA Kay MGH Research Scholars award for support. We acknowledge support from the Ragon Institute of MGH, MIT, and Harvard, the Massachusetts Consortium on Pathogen Readiness (MassCPR), the Bill and Melinda Gates Foundation, and the National Institutes of Health (3R37AI080289-11S1, R01AI146785, U19AI42790-01, 4U01CA260476 - 02, 1P01AI65072-01, and P01AI1650721 to R.P.M. and G.A. and 2U19AI135995-06 to R.P.M., D.A.L., and G.A.). This study was funded by Sanofi and by federal funds from the Biomedical Advanced Research and Development Authority, part of the office of the Administration for Strategic Preparedness and Response at the US Department of Health and Human Services in collaboration with the US Department of Defense Joint Program Executive Office for Chemical, Biological, Radiological, and Nuclear Defense under contract number W15QKN-16-9-1002. The views presented here are those of the authors and do not purport to represent those of the Department of the Army.
DECLARATION OF INTERESTS
G.A. is a founder and equity holder for Seromyx Systems, Inc., an employee and equity holder for Leyden Labs, and has received financial support from AbbVie, BioNtech, GSK, Gilead, Merck, Moderna, Novartis, Pfizer, and Sanofi. G.A.’s interests were reviewed and managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. T.T., V.L., and R.M.C. are Sanofi employees and may own Sanofi shares. R.P.M. receives financial support from AbbVie, Pfizer, GSK, the Bill and Melinda Gates Foundation, the Wellcome Trust, the Department of Defense, and the NIH.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113292.
REFERENCES
- 1.Noy-Porat T, Makdasi E, Alcalay R, Mechaly A, Levy Y, Bercovich-Kinori A, Zauberman A, Tamir H, Yahalom-Ronen Y, Israeli M, et al. (2020). A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun 11, 4303. 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, et al. (2020). Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963. 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, et al. (2020). Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310.e20. 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joyce MG, Sankhala RS, Chen WH, Choe M, Bai H, Hajduczki A, Yan L, Sterling SL, Peterson CE, Green EC, et al. (2020). A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Preprint at bioRxiv. 10.1101/2020.03.15.992883. [DOI] [Google Scholar]
- 5.Baum A, Ajithdoss D, Copin R, Zhou A, Lanza K, Negron N, Ni M, Wei Y, Mohammadi K, Musser B, et al. (2020). REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 370, 1110–1115. 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, et al. (2020). Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449. 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, De la Cruz E, Jorge A, et al. (2022). Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med 28, 481–485. 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, Huang W, Li Q, Wang P, An R, et al. (2022). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663. 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu L, Mok BWY, Chen LL, Chan JMC, Tsang OTY, Lam BHS, Chuang VWM, Chu AWH, Chan WM, Ip JD, et al. (2022). Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin. Infect. Dis 75, e822–e826. 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, et al. (2022). Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 11.Costa Clemens SA, Weckx L, Clemens R, Almeida Mendes AV, Ramos Souza A, Silveira MBV, da Guarda SNF, de Nobrega MM, de Moraes Pinto MI, Gonzalez IGS, et al. (2022). Heterologous versus homologous COVID-19 booster vaccination in previous recipients of two doses of CoronaVac COVID-19 vaccine in Brazil (RHH-001): a phase 4, non-inferiority, single blind, randomised study. Lancet 399, 521–529. 10.1016/S0140-6736(22)00094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pajon R, Doria-Rose NA, Shen X, Schmidt SD, O’Dell S, McDanal C, Feng W, Tong J, Eaton A, Maglinao M, et al. (2022). SARS-CoV-2 Omicron Variant Neutralization after mRNA-1273 Booster Vaccination. N. Engl. J. Med 386, 1088–1091. 10.1056/NEJMc2119912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauring AS, Tenforde MW, Chappell JD, Gaglani M, Ginde AA, McNeal T, Ghamande S, Douin DJ, Talbot HK, Casey JD, et al. (2022). Clinical severity of, and effectiveness of mRNA vaccines against, covid-19 from omicron, delta, and alpha SARS-CoV-2 variants in the United States: prospective observational study. BMJ 376, e069761. 10.1136/bmj-2021-069761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nyberg T, Ferguson NM, Nash SG, Webster HH, Flaxman S, Andrews N, Hinsley W, Bernal JL, Kall M, Bhatt S, et al. (2022). Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet 399, 1303–1312. 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altarawneh HN, Chemaitelly H, Ayoub HH, Tang P, Hasan MR, Yassine HM, Al-Khatib HA, Smatti MK, Coyle P, Al-Kanaani Z, et al. (2022). Effects of Previous Infection and Vaccination on Symptomatic Omicron Infections. N. Engl. J. Med 387, 21–34. 10.1056/NEJMoa2203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartsch YC, St Denis KJ, Kaplonek P, Kang J, Lam EC, Burns MD, Farkas EJ, Davis JP, Boribong BP, Edlow AG, et al. (2022). SARS-CoV-2 mRNA vaccination elicits robust antibody responses in children. Sci. Transl. Med 14, eabn9237. 10.1126/scitranslmed.abn9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartsch YC, Tong X, Kang J, Avendaño MJ, Serrano EF, García-Salum T, Pardo-Roa C, Riquelme A, Cai Y, Renzi I, et al. (2022). Omicron variant Spike-specific antibody binding and Fc activity are preserved in recipients of mRNA or inactivated COVID-19 vaccines. Sci. Transl. Med 14, eabn9243. 10.1126/scitranslmed.abn9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaudoin-Bussières G, Chen Y, Ullah I, Prévost J, Tolbert WD, Symmes K, Ding S, Benlarbi M, Gong SY, Tauzin A, et al. (2022). A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep. 38, 110368. 10.1016/j.celrep.2022.110368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowman KA, Stein D, Shin S, Ferbas KG, Tobin NH, Mann C, Fischinger S, Ollmann Saphire E, Lauffenburger D, Rimoin AW, et al. (2022). Hybrid Immunity Shifts the Fc-Effector Quality of SARS-CoV-2 mRNA Vaccine-Induced Immunity. mBio 13, e0164722. 10.1128/mbio.01647-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplonek P, Cizmeci D, Fischinger S, Collier AR, Suscovich T, Linde C, Broge T, Mann C, Amanat F, Dayal D, et al. (2022). mRNA-1273 and BNT162b2 COVID-19 vaccines elicit antibodies with differences in Fc-mediated effector functions. Sci. Transl. Med 14, eabm2311. 10.1126/scitranslmed.abm2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atyeo C, DeRiso EA, Davis C, Bordt EA, De Guzman RM, Shook LL, Yonker LM, Fasano A, Akinwunmi B, Lauffenburger DA, et al. (2021). COVID-19 mRNA vaccines drive differential antibody Fc-functional profiles in pregnant, lactating, and nonpregnant women. Sci. Transl. Med 13, eabi8631. 10.1126/scitranslmed.abi8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorman MJ, Patel N, Guebre-Xabier M, Zhu AL, Atyeo C, Pullen KM, Loos C, Goez-Gazi Y, Carrion R, Tian JH, et al. (2021). Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep. Med 2, 100405. 10.1016/j.xcrm.2021.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sridhar S, Chicz RM, Warren W, Tartaglia J, Savarino S, Gurunathan S, and Toussaint JF (2022). The potential of Beta variant containing COVID booster vaccines for chasing Omicron in 2022. Nat. Commun 13, 5794. 10.1038/s41467-022-33549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Launay O, Cachanado M, Luong Nguyen LB, Ninove L, Lachâtre M, Ben Ghezala I, Bardou M, Schmidt-Mutter C, Lacombe K, Laine F, et al. (2022). Immunogenicity and Safety of Beta-Adjuvanted Recombinant Booster Vaccine. N. Engl. J. Med 387, 374–376. 10.1056/NEJMc2206711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalkias S, Eder F, Essink B, Khetan S, Nestorova B, Feng J, Chen X, Chang Y, Zhou H, Montefiori D, et al. (2022). Safety, immunogenicity and antibody persistence of a bivalent Beta-containing booster vaccine against COVID-19: a phase 2/3 trial. Nat. Med 28, 2388–2397. 10.1038/s41591-022-02031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou J, Kurhade C, Patel S, Kitchin N, Tompkins K, Cutler M, Cooper D, Qi Y, Cai H, Alexander M, et al. Improved Neutralization of Omicron BA.4/5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent BA.4/5 Vaccine. Cold Spring Harbor Laboratory. [Google Scholar]
- 27.Francica JR, Flynn BJ, Foulds KE, Noe AT, Werner AP, Moore IN, Gagne M, Johnston TS, Tucker C, Davis RL, et al. (2021). Protective antibodies elicited by SARS-CoV-2 spike protein vaccination are boosted in the lung after challenge in nonhuman primates. Sci. Transl. Med 13, eabi4547. 10.1126/scitranslmed.abi4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munro APS, Janani L, Cornelius V, Aley PK, Babbage G, Baxter D, Bula M, Cathie K, Chatterjee K, Dodd K, et al. (2021). Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): a blinded, multicentre, randomised, controlled, phase 2 trial. Lancet 398, 2258–2276. 10.1016/S0140-6736(21)02717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guirakhoo F, Wang S, Wang CY, Kuo HK, Peng WJ, Liu H, Wang L, Johnson M, Hunt A, Hu MM, et al. (2022). High Neutralizing Antibody Levels Against Severe Acute Respiratory Syndrome Coronavirus 2 Omicron BA.1 and BA.2 After UB-612 Vaccine Booster. J. Infect. Dis 226, 1401–1406. 10.1093/infdis/jiac241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frederiksen LSF, Zhang Y, Foged C, and Thakur A (2020). The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. Front. Immunol 11, 1817. 10.3389/fimmu.2020.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavot V, Berry C, Kishko M, Anosova NG, Huang D, Tibbitts T, Raillard A, Gautheron S, Gutzeit C, Koutsoukos M, et al. (2022). Protein-based SARS-CoV-2 spike vaccine booster increases cross-neutralization against SARS-CoV-2 variants of concern in non-human primates. Nat. Commun 13, 1699. 10.1038/s41467-022-29219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, Subramoney K, Makatini Z, Moyo S, Amoako DG, et al. (2022). Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat. Med 28, 1785–1790. 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J, Hachmann NP, Collier ARY, Lasrado N, Mazurek CR, Patio RC, Powers O, Surve N, Theiler J, Korber B, and Barouch DH (2023). Substantial Neutralization Escape by SARS-CoV-2 Omicron Variants BQ.1.1 and XBB.1. N. Engl. J. Med 388, 662–664. 10.1056/NEJMc2214314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Planas D, Bruel T, Staropoli I, Guivel-Benhassine F, Porrot F, Maes P, Grzelak L, Prot M, Mougari S, Planchais C, et al. (2023). Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat. Commun 14, 824. 10.1038/s41467-023-36561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Iketani S, Li Z, Liu L, Guo Y, Huang Y, Bowen AD, Liu M, Wang M, Yu J, et al. (2023). Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286.e8. 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamasoba D, Uriu K, Plianchaisuk A, Kosugi Y, Pan L, Zahradnik J, Genotype to Phenotype Japan G2P-Japan Consortium; Ito J, and Sato K (2023). Virological characteristics of the SARS-CoV-2 omicron XBB.1.16 variant. Lancet Infect. Dis 23, 655–656. 10.1016/S1473-3099(23)00278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varese A, Mazzitelli B, Erra Díaz F, Kjolhede MV, Ojeda D, Vellicce A, Arto P, Cicero C, Pascowsky M, Figueras L, et al. (2022). Omicron Breakthrough Infection After Heterologous Prime-Boost Vaccination Induces a Vigorous Antibody Response. J. Infect. Dis 226, 1717–1720. 10.1093/infdis/jiac250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipsitch M, Krammer F, Regev-Yochay G, Lustig Y, and Balicer RD (2022). SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat. Rev. Immunol 22, 57–65. 10.1038/s41577-021-00662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan X, Shi J, Hu X, Wu Y, Zeng L, Yao Y, Shang W, Liu K, Gao G, Guo W, et al. (2021). RBD-homodimer, a COVID-19 subunit vaccine candidate, elicits immunogenicity and protection in rodents and nonhuman primates. Cell Discov. 7, 82. 10.1038/s41421-021-00320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavot V, Berry C, Kishko M, Anosova NG, Li L, Tibbitts T, Huang D, Raillard A, Gautheron S, Gutzeit C, et al. (2023). Beta variant COVID-19 protein booster vaccine elicits durable cross-neutralization against SARS-CoV-2 variants in non-human primates. Nat. Commun 14, 1309. 10.1038/s41467-023-36908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Branche A, Rouphael N, Diemert D, Falsey A, Losada C, Baden LR, Frey S, Whitaker J, Little S, Anderson E, et al. (2023). Bivalent and Monovalent SARS-CoV-2 Variant Vaccine Boosters Improve coverage of the known Antigenic Landscape: Results of the COVID-19 Variant Immunologic Landscape (COVAIL) Trial. Res. Sq 10.21203/rs.3.rs-2653179/v1. [DOI] [Google Scholar]
- 42.Ng KW, Faulkner N, Finsterbusch K, Wu M, Harvey R, Hussain S, Greco M, Liu Y, Kjaer S, Swanton C, et al. (2022). SARS-CoV-2 S2-targeted vaccination elicits broadly neutralizing antibodies. Sci. Transl. Med 14, eabn3715. 10.1126/scitranslmed.abn3715. [DOI] [PubMed] [Google Scholar]
- 43.de Bruyn G, Wang J, Purvis A, Ruiz MS, Adhikarla H, Alvi S, Bonaparte MI, Brune D, Bueso A, Canter RM, et al. (2023). Safety and immunogenicity of a variant-adapted SARS-CoV-2 recombinant protein vaccine with AS03 adjuvant as a booster in adults primed with authorized vaccines: a phase 3, parallel-group study. EClinicalMedicine 62, 102109. 10.1016/j.eclinm.2023.102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maurer MA, Meyer L, Bianchi M, Turner HL, Le NPL, Steck M, Wyrzucki A, Orlowski V, Ward AB, Crispin M, and Hangartner L (2018). Glycosylation of Human IgA Directly Inhibits Influenza A and Other Sialic-Acid-Binding Viruses. Cell Rep. 23, 90–99. 10.1016/j.celrep.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantis NJ, Rol N, and Corthésy B (2011). Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 4, 603–611. 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aleyd E, van Hout MWM, Ganzevles SH, Hoeben KA, Everts V, Bakema JE, and van Egmond M (2014). IgA enhances NETosis and release of neutrophil extracellular traps by polymorphonuclear cells via Fcα receptor I. J. Immunol 192, 2374–2383. 10.4049/jimmunol.1300261. [DOI] [PubMed] [Google Scholar]
- 47.Sridhar S, Joaquin A, Bonaparte MI, Bueso A, Chabanon AL, Chen A, Chicz RM, Diemert D, Essink BJ, Fu B, et al. (2022). Safety and immunogenicity of an AS03-adjuvanted SARS-CoV-2 recombinant protein vaccine (CoV2 preS dTM) in healthy adults: interim findings from a phase 2, randomised, dose-finding, multicentre study. Lancet Infect. Dis 22, 636–648. 10.1016/S1473-3099(21)00764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva M, Kato Y, Melo MB, Phung I, Freeman BL, Li Z, Roh K, Van Wijnbergen JW, Watkins H, Enemuo CA, et al. (2021). A particulate saponin/TLR agonist vaccine adjuvant alters lymph flow and modulates adaptive immunity. Sci. Immunol 6, eabf1152. 10.1126/sciimmunol.abf1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arunachalam PS, Walls AC, Golden N, Atyeo C, Fischinger S, Li C, Aye P, Navarro MJ, Lai L, Edara VV, et al. (2021). Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594, 253–258. 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- 50.Barouch DH, Tomaka FL, Wegmann F, Stieh DJ, Alter G, Robb ML, Michael NL, Peter L, Nkolola JP, Borducchi EN, et al. (2018). Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13–19). Lancet 392, 232–243. 10.1016/S0140-6736(18)31364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boudreau CM, Yu WH, Suscovich TJ, Talbot HK, Edwards KM, and Alter G (2020). Selective induction of antibody effector functional responses using MF59-adjuvanted vaccination. J. Clin. Invest 130, 662–672. 10.1172/JCI129520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loos C, Coccia M, Didierlaurent AM, Essaghir A, Fallon JK, Lauffenburger D, Luedemann C, Michell A, van der Most R, Zhu AL, et al. (2023). Systems serology-based comparison of antibody effector functions induced by adjuvanted vaccines to guide vaccine design. NPJ Vaccines 8, 34. 10.1038/s41541-023-00613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sibley L, Daykin-Pont O, Sarfas C, Pascoe J, White AD, and Sharpe S (2021). Differences in host immune populations between rhesus macaques and cynomolgus macaque subspecies in relation to susceptibility to Mycobacterium tuberculosis infection. Sci. Rep 11, 8810. 10.1038/s41598-021-87872-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salguero FJ, White AD, Slack GS, Fotheringham SA, Bewley KR, Gooch KE, Longet S, Humphries HE, Watson RJ, Hunter L, et al. (2021). Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat. Commun 12, 1260. 10.1038/s41467-021-21389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schie L, VIB-CMB COVID-19 Response Team; and Pöhlmann S, et al. (2020). Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 181, 1004–1015.e15. 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing Y, Li X, Gao X, and Dong Q (2020). Natural Polymorphisms Are Present in the Furin Cleavage Site of the SARS-CoV-2 Spike Glycoprotein. Front. Genet 11, 783. 10.3389/fgene.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK, and Rodriguez-Morales AJ (2020). SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med 28, 174–184. [PubMed] [Google Scholar]
- 58.DeRosa F, Smith L, Shen Y, Huang Y, Pan J, Xie H, Yahalom B, and Heartlein MW (2019). Improved Efficacy in a Fabry Disease Model Using a Systemic mRNA Liver Depot System as Compared to Enzyme Replacement Therapy. Mol. Ther 27, 878–889. 10.1016/j.ymthe.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown EP, Dowell KG, Boesch AW, Normandin E, Mahan AE, Chu T, Barouch DH, Bailey-Kellogg C, Alter G, and Ackerman ME (2017). Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods 443, 33–44. 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ackerman ME, Barouch DH, and Alter G (2017). Systems serology for evaluation of HIV vaccine trials. Immunol. Rev 275, 262–270. 10.1111/imr.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karsten CB, Mehta N, Shin SA, Diefenbach TJ, Slein MD, Karpinski W, Irvine EB, Broge T, Suscovich TJ, and Alter G (2019). A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J. Immunol. Methods 471, 46–56. 10.1016/j.jim.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischinger S, Fallon JK, Michell AR, Broge T, Suscovich TJ, Streeck H, and Alter G (2019). A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods 473, 112630. 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, Jost S, Berger CT, Sciaranghella G, Liu Q, et al. (2011). A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366, 8–19. 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Systems serology raw datasets have been deposited on GitHub on RagonSystemSerology: CR20230925.
All original code used for analysis in this manuscript can be found in the systemsseRology on GitHub (LoosC/systemsseRology). All other codes used are open source and available upon request to the lead contact. This manuscript does not report original code unique to this study.
Any additional information required for analysis of the data reported in this manuscript is available from the lead contact upon request.


