Abstract
The function of an uncharacterized protein is usually inferred either from its homology to, or its interactions with, characterized proteins. Here, we use both sequence similarity and protein interactions to identify relationships between remotely related protein sequences. We rely on the fact that homologous sequences share similar interactions, and, therefore, the set of interacting partners of the partners of a given protein is enriched by its homologs. The approach was benchmarked by assigning the fold and functional family to test sequences of known structure. Specifically, we relied on 1,434 proteins with known folds, as defined in the Structural Classification of Proteins (SCOP) database, and with known interacting partners, as defined in the Database of Interacting Proteins (DIP). For this subset, the specificity of fold assignment was increased from 54% for position-specific iterative blast to 75% for our approach, with a concomitant increase in sensitivity for a few percentage points. Similarly, the specificity of family assignment at the e-value threshold of 10-8 was increased from 70% to 87%. The proposed method would be a useful tool for large-scale automated discovery of remote relationships between protein sequences, given its unique reliance on sequence similarity and protein-protein interactions.
Keywords: remote homology, fold assignment, family assignment, protein function annotation, protein-protein interactions
Functional annotation of protein sequences by computation is essential in leveraging the impact of the genome-sequencing projects. To characterize the function of a protein sequence, it is often useful to identify its homologs and interacting proteins of known function. This task is facilitated by the classifications of protein domain families (1, 2), lists of protein-protein interactions (3, 4), and databases of protein structures (5-7). Protein domains are organized into folds (if sharing a similar structure), superfamilies (with evidence of homology in addition to structure similarity), and families (for homologs with similar function, sequence, and structure) (6). The vast majority of homologous sequences are expected to share the same fold.
The most sensitive algorithms for detecting homology between remotely related protein sequences rely on multiple sequence and protein structure information. The former group includes the sequence profile-based methods (8, 9) and hidden Markov models (10) that construct a multiple sequence alignment of the close homologs of the query, followed by scanning the corresponding profile against a database of sequences. The latter group includes sequence-structure threading methods that can sometimes reveal more distant relationships than purely sequence-based methods (11). Threading methods assign the fold by assessing the energy of coarse models corresponding to all of the possible ways of threading the sequence through each of the structures in a library of all known folds. Despite the increased coverage and accuracy of fold assignment when using multiple sequence and structure information, two major problems remain for sequences related at approximately <25% sequence identity (12), (i) finding remote homologs that are undetectable by sequence similarity alone and (ii) identifying the functional family even when the fold can be detected (13, 14). Of the known protein sequences, ≈60% have at least one domain with a reliable fold assignment, covering ≈35% of the amino acid residues in the known protein sequences (15, 16).
Even when two sequences share little or no sequence similarity, their structures and functions may be similar (17, 18). Therefore, similarity in function may indicate a similar structure. An indicator of related functions is similar protein-protein interaction patterns. One such special case are the “interlogs” (i.e., pairs of interacting proteins that interact identically in two species) (19). It has already been demonstrated that the information about the interacting partners can be used to predict the fold (7, 20-22) or function (14, 23-27) of a protein without considering its sequence. The usefulness of these approaches should grow with time, given the increasing amount of data about protein-protein interactions (20, 24, 28), collected in databases such as the Biomolecular Interaction Network Database (BIND; ref. 3), the Munich Information Center for Protein Sequences (MIPS; ref. 29), and the Database of Interacting Proteins (DIP) (4).
Here, our objective is to demonstrate that the combined use of protein interactions and sequence similarity improves detection of remote similarity. We implemented our method by using the position-specific iterative (PSI) blast program, but any other method for detection of remote sequence similarity could be used. We begin by describing the approach. Next, we benchmark the method by relying on a set of nonredundant domains from the Structural Classification of Proteins (SCOP) database that have known interacting partners defined in DIP. We conclude by discussing the implications of our results for protein structure modeling and functional annotation.
Methods
A protein-interaction network can be represented by a graph with nodes as proteins and edges as protein interactions. In such a graph, a set of proteins connected to protein X (i.e., physically interacting with X) is named “partners of X.” Moreover, we define successive levels of partnership as follows: the set of partners of X is named “partners of X at level 1,” and the set of partners of the partners of X at level 1 forms the set of partners at level 2, etc. Given the commutative relation of the interactions (i.e., if B is found in the set of partners of A, then A is found in the set of partners of B), protein X should be in the set of partners at level 2 of itself. In fact, protein X should occur in all sets of partners at even levels. Therefore, given the fact that homologous proteins perform similar functions associated with similar interaction partners, the sets of partners of protein X at even levels contain more sequences homologous to protein X than a randomly selected set of sequences of the same size (see Results). Furthermore, partners of protein X at levels 1 and 3 may also include some of its homologs because some proteins interact with their homologs, or they evolved by means of a fusion of two genes of interacting ancestors (30). Here, we exploited these considerations in combination with sequence similarity to improve the assignment of a given protein sequence into the correct fold class and functional family.
We relied on the following three databases: the TrEMBL database of protein sequences (release, 23.6; April 2003) (31), the SCOP database of protein structure classification (version, 1.65; December 2003) (32), and the DIP of experimentally identified protein interactions (release, 20040113; January 2004) (33).
The DIP contains 16,903 protein sequences that are involved in 43,742 documented binary interactions. Fold, superfamily, and family domain codes of SCOP were assigned to a total of 4,324 proteins in DIP that could be matched by blast to a protein in SCOP, covering one-sixth of all proteins in DIP (i.e., DIP-SCOP group). More precisely, one or more domain codes were assigned to a protein sequence in DIP when the alignment between the two sequences had an e value of <10-8 over at least 75% of the residues in the SCOP domain. A total of 4,743 binary interactions had SCOP codes for both proteins, whereas 14,813 interactions had the SCOP code for only one partner. This initial set of proteins was reduced to 1,434 query proteins to remove redundancies so that no two proteins in the set share >25% sequence identity after aligning them with the blast program.
Next, we added extrapolated links to the protein-interaction network. Two proteins were linked by extrapolation if any members from their SCOP families interacted with each other. To enable benchmarking, the extrapolation was not performed for the query proteins in the benchmark. It was also not performed for “hub” proteins (34) that were defined here as proteins interacting with proteins in >10 different SCOP families. The hub proteins were excluded from extrapolation to minimize false positives. Thus, the list of reference links included both known interactions between pairs of domains as well as extrapolated links. Similarly, the term partner was expanded to include proteins connected by extrapolated links in addition to physical interactions.
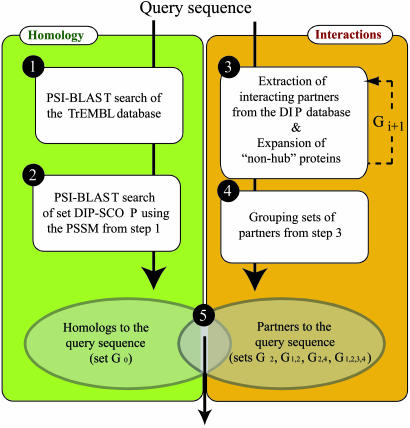
The assignment of a fold or a family to a query sequence involves five steps (Fig. 1). First, a profile [position-specific scoring matrix (PSSM)] is constructed of the query sequence by searching for its homologs in the TrEMBL database (31) by PSI-blast (35) for a maximum of five iterations. Second, query homologs are detected in the DIP-SCOP group by PSI-blast using the query profile from step 1 (set G0). Third, partners of the query at levels 1-4 are extracted by using the reference links. Fourth, the sets of partners obtained in step 3 are grouped into four main groups, formed by the set of partners at level 2 (G2), the union of the partners at levels 1 and 2 (G1,2), the union of the partners at levels 2 and 4 (G2,4), and the union of the four sets (G1,2,3,4). Fifth, the members of each of the groups in step 4 are ranked based on the e value calculated in step 2. Additional combinations of partner levels are either redundant or complex, and they are not reported in this study.
Fig. 1.
Flowchart for detection of remotely related proteins based on both sequence similarity and protein interactions. First, for a query protein, a PSSM is built by five iterations of scanning the TrEMBL database by PSI-blast. Second, the PSSM is used in another PSI-blast run to obtain the e values between the query and the proteins in DIP-SCOP. Third, the interaction partners of the query are extracted from DIP and may be expanded through SCOP family codes. This step is repeated, resulting in set Gi in iteration i. Fourth, partners at different levels are grouped as described in Methods. Fifth, proteins in the intersection are ranked by the PSI-blast e value to the query, obtained in the second step.
We tested family and fold assignment for different thresholds on the PSI-blast e value with proteins in sets G0, G2, G1,2, G2,4, and G1,2,3,4. The number of positive assignments is defined as the number of sequences that align with the query sequence with an e value smaller or equal to the threshold. Among these positives, we define the number of true positives as the number of sequences with the same SCOP code as the query sequence.
Results
Quantifying the Enrichment Afforded by Protein Interactions. Our method for detecting remote relationships by both sequence similarity and protein-protein interactions depends on the enrichment of the homologs among the set of partners of the partners of the query protein (i.e., the G2 set). Therefore, we quantified this enrichment as follows. First, we calculated the proportion of the correct fold assignments by dividing the sum of the correct fold assignments in each G2 set by the sum of the G2 set sizes (Table 1). Next, we compared this proportion with the corresponding proportion in the DIP-SCOP group. There was a significant enrichment of the proteins with the correct fold assignment in the G2 set relative to DIP-SCOP. The same assessment was also performed for family assignment instead of fold assignment, revealing an even larger enrichment than that for fold assignment. Reflecting the homodimers, the corresponding statistics for the G1 set also shows significant enrichment for homologs over a random selection from the DIP-SCOP set.
Table 1. Enrichment for the correct folds and families.
| Assignment | G1 | G2 | DIP-SCOP |
|---|---|---|---|
| Fold | 0.137 | 0.041 | 0.018 |
| Family | 0.107 | 0.018 | 0.003 |
Proportions of the correct fold and family assignments in the G1 and G2 sets are compared with the proportion of the correct folds in the DIP-SCOP set.
To quantify the statistical significance of enrichment in the G2 set, we calculated the P value with the Wilcoxon test (36). For each query, we compared the enrichment in G2 and in 1,000 random sets with the same number of proteins as in group G2, obtained from the DIP-SCOP group. The corresponding P value of 0.0064 quantifies the high statistical significance of enrichment in the G2 set.
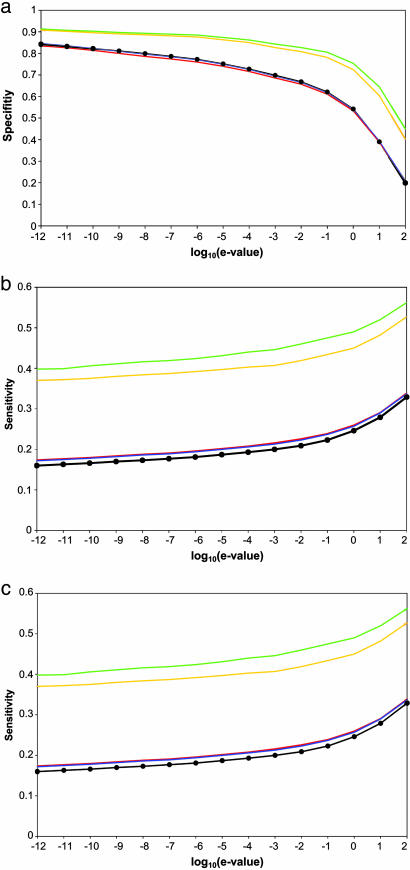
Improved Specificity of Fold and Family Assignment. The specificity is defined as the number of true positives over the total number of positives. For an e-value cutoff of <1, our approach achieved ≈75% specificity for group G1,2 (Fig. 2a). This relatively high specificity can be compared with the specificity of 54% for group G0, obtained by PSI-blast alone; simultaneously, sensitivity is also improved for several percentage points (below). The improvement in specificity justifies the use of less significant e-value cutoffs in the filtered groups of sequences (G2 and G1,2) than with PSI-blast (G0). The difference in performance between the traditional PSI-blast approach based on sequence matching alone and our approach, which also includes information about protein-protein interactions, increases as the e-value cutoff is raised.
Fig. 2.
Specificity, sensitivity, and applicability of fold assignment based on a combination of sequence similarity and protein interactions. The specificity (a), sensitivity (b), and applicability (c) are plotted as a function of the threshold on the PSI-blast e value for groups G2 (orange), G1,2 (green), G2,4 (blue), G1,2,3,4 (red), and G0 (black with filled circles).
The sets G2 and G1,2 were enriched for the correct family codes relative to the set G0, demonstrating an improvement relative to searching by PSI-blast alone (data not shown). The specificities obtained from groups G2 and G1,2 were ≈80% for the e-value cutoff of 10-3, whereas PSI-blast sequence search without consideration of interactions had a specificity of only ≈60%.
Sensitivity of Fold and Family Assignment. Our method cannot correctly assign a fold to a protein sequence when a stringent threshold on the e-value filters out correct predictions or when there are no experimental data about relevant protein interactions. To estimate the sensitivity, we defined the undetected members of the same group that have the same domain fold as the query protein as false negatives. Sensitivity for groups G2 and G1,2 is consistently better for several percentage points than for G0 (Fig. 2b).
Applicability of Fold and Family Assignment. Our combined approach is not as general as the sequence comparison methods, which can be applied to all protein sequences. The reason is that the combined approach depends on the availability of protein-interaction data. Therefore, to gauge the practical utility of the combined approach, we estimated its applicability to fold assignment for proteins in the sets G2, G1,2, G2,4, and G1,2,3,4 (Fig. 2c).
Extrapolating Interactions to Increase the Coverage. To assess the effect of the extrapolation of protein interactions (Methods), we compared the number of true positives at the fold level obtained with and without extrapolation, respectively. When used without extrapolation, our method was able to find only 286 true positives with 81% specificity at the PSI-blast e-value cutoff of 1, compared with 2,885 true positives and 75% specificity when using extrapolation. Thus, extrapolation yields a 10-fold increase in coverage with a relatively small loss in specificity. However, even with extrapolation, only ≈50% of the proteins in the DIP-SCOP group have a partner at level 2.
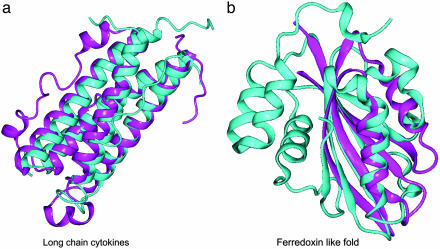
Example of Fold and Family Assignment. To illustrate the ability of our approach to detect relationships between members of the same family in the absence of significant sequence similarity, we describe an example of the Swiss-Prot sequences CNTF_HUMAN (ciliary neurotrophic factor) and ONCM_HUMAN (oncostatin M). CNTF_HUMAN is a survival factor for various neuronal cell types, and ONCM_HUMAN is a growth regulator that inhibits the proliferation of a number of tumor cell lines. The two proteins share the same fold (four-helical cytokines) and family (long-chain cytokines). However, sequence similarity is very low (PSI-blast e value, ≈0.1; sequence identity, 16%).
According to DIP, both proteins interact with a member of the cytokine receptor family, LIFR_HUMAN (leukemia inhibitory factor receptor), as revealed by immunoprecipitation experiments (DIP entries 10988E and 10064E for the interactions of CNTF_HUMAN and ONCM_HUMAN, respectively). Moreover, the PDB ID code 1I1R structure reveals a physical interaction between a member of the cytokine family (viral IL-6) and a member of the cytokine receptor family (human gp130). Thus, our method predicts with an e value of 0.1 in group G2 that CNTF_HUMAN has the same fold as ONCM_HUMAN (Fig. 3a).
Fig. 3.
Fold assignment by the combined approach. (a) Structural superposition of the human ciliary neurotrophic factor (chain 1 of PDB ID code 1CNT; cyan) and the human oncostatin M (chain A of PDB ID code 1EVS; magenta). Structures were superposed with CE (53), with a Cα rms deviation of 1.7 Å and 15% sequence identity. (b) Structural superposition of two members of the ferredoxin-like fold, the C-terminal domains of human elongation factor 1γ (chain A of PDB ID code 1PBU; cyan) and yeast elongation factor 1β (chain B of PDB ID code 1G7C; magenta). The structures were superposed with CE, obtaining a Cα rms deviation of 3.6 Å and 7.5% sequence identity.
Example of Fold Assignment. To illustrate the ability of our approach to detect remote relationships at the fold level, we describe here an example of the Swiss-Prot sequences EF1G_YEAST and EF1B_YEAST. The C-terminal domains of these sequences adopt a ferredoxin-like fold. Nevertheless, EF1G_YEAST is an elongation factor 1γ of the eEF1γ domain superfamily, whereas EF1B_YEAST is an elongation factor 1β of the eEF-1β-like superfamily. Both structures share a core formed by a sheet of three β-strands and an external helix, and could be superimposed with an rms deviation of 3.6 Å (Fig. 3b). The e value of the PSI-blast alignment between EF1G_YEAST and EF1B_YEAST is 0.036, obtained by querying the TrEMBL database with EF1G_YEAST for five iterations with default parameters.
The relationship between both sequences could be extracted through the interaction of EF1G_YEAST with an elongation factor 1α (EF1A_YEAST) in the G protein family, obtained from tandem affinity purification experiments (DIP entry 17026E, between nodes 6813N and 2250N). In addition, DIP contains an interaction between EF1B_YEAST and TEM1_YEAST (a G protein) with the DIP entry code of 13895E (between nodes 6445N and 1691N), revealed by immunoprecipitation experiments. Therefore, EF1B_YEAST is a partner of EF1G_YEAST at level 2 (G2). Table 2 shows the set of proteins found in G0 of EF1G_YEAST with e values between 10-3 and 1; there is only a single analog sharing the ferredoxin-like fold with EF1G_YEAST and two false positives that do not appear in group G1,2. In this case, our method is both sensitive and specific, because the correct fold of EF1G_YEAST appears in group G1,2 without any false positives.
Table 2.
Partial results from a search for homologs of EF1G_YEAST (folds, 47615, 52832, and 54861; superfamilies, 47616, 52833, and 89942) by PSI-blast
| Swiss-Prot code | e Value | Shares fold | SCOP fold | SCOP superfamily | Appearance in G1,2 |
|---|---|---|---|---|---|
| SYEC_YEAST | 0.027 | No | 52373 | 52374 | No |
| EF1B_YEAST | 0.036 | Yes | 54861 | 54984 | Yes |
| SC14_YEAST | 0.83 | No | 46928, 52086 | 46938, 52087 | No |
Swiss-Prot codes of the sequences found in G0 and aligned with the sequence of EF1G_YEAST with e values between 10-3 and 0.1 (Methods). “e value” indicate the corresponding PSI-blast e values. “Shares fold” indicates whether or not the sequence shares a fold with EF1G_YEAST (ferredoxin-like fold). “SCOP fold” and “SCOP superfamily” indicate the SCOP fold and superfamily codes, respectively (multidomain proteins have multiple codes). “Appears in G1,2” indicates whether or not a sequence is found in the G1,2 set of EF1G_YEAST.
Discussion
We described, implemented, and benchmarked a method that uses information about both sequence similarity and protein-protein interactions to detect homology between remotely related protein sequences. The method was validated by a benchmark involving 1,434 query proteins of known structure (Figs. 1 and 2), and it was illustrated by two examples (Fig. 3). Although the method was benchmarked by using known protein structures, it is equally applicable to detection of remote relationships between protein sequences without known structures because it does not rely on protein structure information.
Generally, the function of uncharacterized proteins can be annotated in two fundamentally different ways (37). It can be done by establishing (i) a sequence and/or structure similarity to another characterized protein and (ii) a functional link to another characterized protein. The first group of methods includes sequence matching and threading (9, 11). The second group includes both experimental and computational methods, such as clustering by physical interactions (26), mRNA array expression profiles (38), analysis of gene fusion (30), phylogenetic profiles (39), and genomic association of genes (40). Our approach is unique in that it discovers homology by explicitly combining both sequence similarity and experimentally determined protein interactions. Therefore, it benefits from the databases of protein sequences, structures, and interactions. Another method infers fold and family membership from protein interactions (21) but not in combination with sequence similarity. Although a few other sequence similarity-based methods, such as 3D-PSSM (22), also use functional information, this information is mined from scientific texts and not from lists of protein interactions.
The benchmark clearly suggests that protein-interaction data increase the specificity and sensitivity of fold and family assignment (Fig. 2). Consequently, our method allows the assignment of fold and family to a higher percentage of known protein sequences without loss of accuracy. For example, the specificity of fold assignment at the PSI-blast e-value cutoff of 1 was increased from 54% for PSI-blast to 75% when combining sequence similarity and protein-protein interactions, with a concomitant increase of sensitivity for several percentage points. Similarly, the specificity of family assignment at the e-value threshold of 10-8 was increased from 70% to 87%, also with a slight increase in sensitivity. Moreover, at the e-value cutoff of 1, >90% of the correct fold assignments share the same family as the query, whereas only 65% of the correct fold assignments with PSI-blast correspond to proteins with the same family code. This result was expected, given that our approach benefits from the conservation of interaction patterns usually related to the protein function and, thus, family classification.
The accuracy and coverage of our method are limited by false positives and negatives of sequence matching by PSI-blast (41, 42), as well as by false and missing interactions in DIP (43). To minimize sequence matching problems, additional methods, such as profile-profile searches (9), hidden Markov models (10), threading (44), and intermediate sequence search (41) can be used. With the interactions, false positives rate and coverage can be improved by probabilistic methods that rely on multiple sources of information about protein interactions (45) and by performing more experiments. Clearly, the coverage of the method will rise with the increase in the number of known protein-protein interactions that link query proteins to other proteins.
There are also intrinsic limitations of the method. For example, some of the proteins in the same SCOP family do not share the same interactions (46), resulting in false positives of our method. In addition, current interaction databases, including the DIP database, list protein-protein interactions and not domain-domain interactions. Therefore, the lack of distinction between a protein and a domain may also increase false positives when extrapolating links through the existence of common domains within proteins. This problem is reduced, but not eliminated, by not applying the extrapolation procedure to hub proteins.
The combined method is applicable only to protein sequences and their homologs for which protein-interaction data are available, in contrast to sequence comparison alone, which is applicable to all protein sequences. This limitation is quantified by the following two examples. First, ≈20-50% of the proteins in the benchmark have a partner in the G2 set (Fig. 2c). Second, for specificity of 75%, sequence comparison by PSI-blast makes 30,302 pairs with correct fold assignments, whereas our combined method finds 2,885 true positives of which 188 were not reported by PSI-blast. Two of these assignments are shown in Fig. 3. We suggest that even the comparatively small coverage of the combined method is already useful in practice, given the 2 million known protein sequences that need to be related to each other; very few methods for characterization of proteins, experimental or computational, are applicable to most protein sequences, and many proven methods are applicable only to a small fraction of all proteins. Moreover, the usefulness of our combined method is clearly increasing with the growth of the databases of known protein sequences and their interactions. We also expect that the idea of combining protein sequence comparison and protein interactions could enable additional improvements in the matching of remotely related protein sequences.
There are several fold assignment methods, such as profile-profile matching, hidden Markov models, and threading, that are more sensitive than PSI-blast. We did not assess the performance of our approach against these methods because we focused on the relative usefulness of protein interactions when added to the consideration of sequence similarity. However, we do suggest that our use of protein interactions will sometimes result in correct fold assignments when all other methods fail, especially when the most sensitive fold assignment methods are used instead of PSI-blast in our approach.
The proposed method is as applicable to establishing remote sequence-sequence matches as it is to fold assignment. However, we focused on fold assignment because of its importance in comparative protein structure modeling and structural genomics. The structural genomics initiative aims to experimentally determine carefully selected protein structures, such that most of the remaining sequences can be modeled with useful accuracy by comparative modeling (47). The number of experimentally determined structures for comparative modeling of most proteins based on at least 30% sequence identity to a known structure is estimated to be ≈16,000 (48). A reduction of this number, while keeping the accuracy of the corresponding models constant, would reduce both the cost and time required by structural genomics to fulfill its aim (49-52). This reduction can be partly achieved by using more sensitive fold detection methods, such as the method described here.
Our method could be made applicable to large-scale comparative protein structure modeling and, thus, increase the number of modeled proteins in MODBASE, our comprehensive database of comparative models for all known protein sequences that are detectably related to a known structure (15). The proposed method is expected to be a useful tool for large-scale automated discovery of remote protein similarities, given its unique reliance on sequence similarity and protein-protein interactions.
Acknowledgments
We thank D. Jaeggi, M. S. Madhusudhan, R. B. Russell, P. Aloy, C. von Mering, F. Davis, and H. Moss Sali for their comments. B.O. was supported by a “Salvador de Madariaga” fellowship from the Spanish Ministerio de Educación, Cultura, y Deporte (MECD), grants from Fundación Ramón Areces, Spanish Ministerio de Ciencia y Tecnología (MCyT) Grant BIO2002-03609, and the “Programa Gaspar de Portolà del Departament d'Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya (DURSI).” A.S. was supported by National Institutes of Health Grant R01 GM54762, the Sandler Family Foundation, Sun Academic Equipment Grant EDUD-7824-020257-US, an IBM Shared University Research grant, and an Intel computer hardware gift. J.E. and R.A. were supported by student fellowships from DURSI and MCyT, respectively. F.X.A. and E.Q. were supported by MECD Grants BIO2004-05829, BIO2004-20005E, and BFU2004-06327-CO2-01.
Author contributions: M.A.M.-R., A.S., and B.O. designed research; J.E., R.A., and B.O. performed research; N.E. contributed new reagents/analytic tools; J.E., R.A., M.A.M.-R., A.S., and B.O. analyzed data; and J.E., R.A., M.A.M.-R., A.S., and B.O. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SCOP, Structural Classification of Proteins; PSI, position-specific iterative; DIP, Database of Interacting Proteins; PSSM, position-specific scoring matrix.
References
- 1.Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., et al. (2004) Nucleic Acids Res. 32, D138-D1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., Ponting, C. P. & Bork, P. (2004) Nucleic Acids Res. 32, D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader, G. D., Betel, D. & Hogue, C. W. (2003) Nucleic Acids Res. 31, 248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salwinski, L., Miller, C. S., Smith, A. J., Pettit, F. K., Bowie, J. U. & Eisenberg, D. (2004) Nucleic Acids Res. 32, D449-D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearl, F. M., Bennett, C. F., Bray, J. E., Harrison, A. P., Martin, N., Shepherd, A., Sillitoe, I., Thornton, J. & Orengo, C. A. (2003) Nucleic Acids Res. 31, 452-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreeva, A., Howorth, D., Brenner, S. E., Hubbard, T. J., Chothia, C. & Murzin, A. G. (2004) Nucleic Acids Res. 32, D226-D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietmann, S., Park, J., Notredame, C., Heger, A., Lappe, M. & Holm, L. (2001) Nucleic Acids Res. 29, 55-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mittelman, D., Sadreyev, R. & Grishin, N. (2003) Bioinformatics 19, 1531-1539. [DOI] [PubMed] [Google Scholar]
- 9.Marti-Renom, M. A., Madhusudhan, M. S. & Sali, A. (2004) Protein Sci. 13, 1071-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karchin, R., Cline, M., Mandel-Gutfreund, Y. & Karplus, K. (2003) Proteins 51, 504-514. [DOI] [PubMed] [Google Scholar]
- 11.Jones, D. T. (1997) Curr. Opin. Struct. Biol. 7, 377-387. [DOI] [PubMed] [Google Scholar]
- 12.Heger, A. & Holm, L. (2003) J. Mol. Biol. 328, 749-767. [DOI] [PubMed] [Google Scholar]
- 13.Devos, D. & Valencia, A. (2001) Trends Genet. 17, 429-431. [DOI] [PubMed] [Google Scholar]
- 14.Hegyi, H. & Gerstein, M. (2001) Genome Res. 11, 1632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pieper, U., Eswar, N., Braberg, H., Madhusudhan, M. S., Davis, F. P., Stuart, A. C., Mirkovic, N., Rossi, A., Marti-Renom, M. A., Fiser, A., et al. (2004) Nucleic Acids Res. 32, D217-D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherkasov, A. & Jones, S. J. (2004) BMC Bioinformatics 5, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian, W. & Skolnick, J. (2003) J. Mol. Biol. 333, 863-882. [DOI] [PubMed] [Google Scholar]
- 18.Devos, D. & Valencia, A. (2000) Proteins 41, 98-107. [PubMed] [Google Scholar]
- 19.Matthews, L. R., Vaglio, P., Reboul, J., Ge, H., Davis, B. P., Garrels, J., Vincent, S. & Vidal, M. (2001) Genome Res. 11, 2120-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180-183. [DOI] [PubMed] [Google Scholar]
- 21.Lappe, M., Park, J., Niggemann, O. & Holm, L. (2001) Bioinformatics 17, S149-S156, Suppl. 1. [DOI] [PubMed] [Google Scholar]
- 22.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2000) J. Mol. Biol. 299, 499-520. [DOI] [PubMed] [Google Scholar]
- 23.Letovsky, S. & Kasif, S. (2003) Bioinformatics 19, i197-i204, Suppl. 1. [DOI] [PubMed] [Google Scholar]
- 24.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141-147. [DOI] [PubMed] [Google Scholar]
- 25.Samanta, M. P. & Liang, S. (2003) Proc. Natl. Acad. Sci. USA 100, 12579-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser, A. G. & Marcotte, E. M. (2004) Nat. Genet. 36, 559-564. [DOI] [PubMed] [Google Scholar]
- 27.Vazquez, A., Flammini, A., Maritan, A. & Vespignani, A. (2003) Nat. Biotechnol. 21, 697-700. [DOI] [PubMed] [Google Scholar]
- 28.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- 29.Mewes, H. W., Amid, C., Arnold, R., Frishman, D., Guldener, U., Mannhaupt, G., Munsterkotter, M., Pagel, P., Strack, N., Stumpflen, V., et al. (2004) Nucleic Acids Res. 32, D41-D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcotte, E., Pellegrini, M., Ho-Leung, Rice, D. W., Yeates, T. O. & Eisenberg, D. (1999) Science 285, 751-753. [DOI] [PubMed] [Google Scholar]
- 31.Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., Martin, M. J., Michoud, K., O'Donovan, C., Phan, I., et al. (2003) Nucleic Acids Res. 31, 365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreeva, A., Howorth, D., Brenner, S. E., Hubbard, T. J. P., Chothia, C. & Murzin, A. G. (2004) Nucl. Acids. Res. 32, D226-D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salwinski, L., Miller, C., Smith, A., Pettit, F., Bowie, J. & Eisenberg, D. (2004) Nucleic Acids Res. 32, D449-D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong, H., Mason, S. P., Barabasi, A. L. & Oltvai, Z. N. (2001) Nature 411, 41-42. [DOI] [PubMed] [Google Scholar]
- 35.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilcoxon, F. (1945) Biometrics 1, 80-83. [Google Scholar]
- 37.Sali, A. (1999) Nature 402, 23-26. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, X., Kao, M. & Wong, W. (2002) Proc. Natl. Acad. Sci. USA 99, 12783-12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. (1999) Proc. Natl. Acad. Sci. USA 96, 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dandekar, T., Snel, B., Huynen, M. & Bork, P. (1998) Trends Biochem. Sci. 23, 324-328. [DOI] [PubMed] [Google Scholar]
- 41.John, B. & Sali, A. (2004) Protein Sci. 13, 54-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner, S. E., Chothia, C. & Hubbard, T. J. (1998) Proc. Natl. Acad. Sci. USA 95, 6073-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deane, C. M., Salwinski, L., Xenarios, I. & Eisenberg, D. (2002) Mol. Cell Proteomics 1, 349-356. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, C., Liu, S., Zhou, H. & Zhou, Y. (2004) Protein Sci. 13, 400-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards, A. M., Kus, B., Jansen, R., Greenbaum, D., Greenblatt, J. & Gerstein, M. (2002) Trends Genet. 18, 529-536. [DOI] [PubMed] [Google Scholar]
- 46.Aloy, P. & Russell, R. B. (2002) Trends Biochem. Sci. 27, 633-638. [DOI] [PubMed] [Google Scholar]
- 47.Baker, D. & Sali, A. (2001) Science 294, 93-96. [DOI] [PubMed] [Google Scholar]
- 48.Vitkup, D., Melamud, E., Moult, J. & Sander, C. (2001) Nat. Struct. Biol. 8, 559-566. [DOI] [PubMed] [Google Scholar]
- 49.Sali, A. (1998) Nat. Struct. Biol. 5, 1029-1032. [DOI] [PubMed] [Google Scholar]
- 50.Burley, S. K., Almo, S. C., Bonanno, J. B., Capel, M., Chance, M. R., Gaasterland, T., Lin, D., Sali, A., Studier, F. W. & Swaminathan, S. (1999) Nat. Genet. 23, 151-157. [DOI] [PubMed] [Google Scholar]
- 51.Burley, S. K. & Bonanno, J. B. (2003) Methods Biochem. Anal. 44, 591-612. [PubMed] [Google Scholar]
- 52.Terwilliger, T. C., Park, M. S., Waldo, G. S., Berendzen, J., Hung, L. W., Kim, C. Y., Smith, C. V., Sacchettini, J. C., Bellinzoni, M., Bossi, R., et al. (2003) Tuberculosis (Edinb) 83, 223-249. [DOI] [PubMed] [Google Scholar]
- 53.Shindyalov, I. N. & Bourne, P. E. (1998) Protein Eng. 11, 739-747. [DOI] [PubMed] [Google Scholar]