The EZH2 inhibitor tulmimetostat achieves comprehensive target inhibition in ARID1A mutant solid tumor models and cancer patients that can be assessed with a pharmacodynamic gene signature in peripheral blood.
Abstract
Recurrent somatic mutations in the BRG1/BRM-associated factor (BAF) chromatin remodeling complex subunit ARID1A occur frequently in advanced urothelial, endometrial, and ovarian clear cell carcinomas, creating an alternative chromatin state that may be exploited therapeutically. The histone methyltransferase EZH2 has been previously identified as targetable vulnerability in the context of ARID1A mutations. In this study, we describe the discovery of tulmimetostat, an orally available, clinical stage EZH2 inhibitor, and it elucidates the aspects of its application potential in ARID1A mutant tumors. Tulmimetostat administration achieved efficacy in multiple ARID1A mutant bladder, ovarian, and endometrial tumor models and improved cisplatin response in chemotherapy-resistant models. Consistent with its comprehensive and durable level of target coverage, tulmimetostat demonstrated greater efficacy than other PRC2-targeted inhibitors at comparable or lower exposures in a bladder cancer xenograft mouse model. Tulmimetostat mediated extensive changes in gene expression, in addition to a profound reduction in global H3K27me3 levels in tumors. Phase I clinical pharmacokinetic and pharmacodynamic data indicated that tulmimetostat exhibits durable exposure and profound target engagement. Importantly, a tulmimetostat controlled gene expression signature identified in whole blood from a cohort of 32 patients with cancer correlated with tulmimetostat exposure, representing a pharmacodynamic marker for the assessment of target coverage for PRC2-targeted agents in the clinic. Collectively, these data suggest that tulmimetostat has the potential to achieve clinical benefit in solid tumors as a monotherapy but also in combination with chemotherapeutic agents, and may be beneficial in various indications with recurrent ARID1A mutations.
Significance: The EZH2 inhibitor tulmimetostat achieves comprehensive target inhibition in ARID1A mutant solid tumor models and cancer patients that can be assessed with a pharmacodynamic gene signature in peripheral blood.
Introduction
Cancer genomic alterations in certain chromatin modifier encoding genes result in dysregulation of chromatin regulatory pathways and changes in gene control that promote cancer progression. Recurrent mutations in histone methyltransferases and demethylases, chromatin remodelers, and histone acetyltransferases such as KMT2D, KMT2C, KDM6A, ARID1A, EP300 and CREBBP occur frequently in cancer (1, 2). When considered as a chromatin signaling pathway, alterations in chromatin modifier genes are as frequent as mutations in well-established oncogenic driver pathways such as the TP53-cell cycle and RTK-MAPK-PI3K pathways (3), which provides an attractive rationale to explore agents targeting chromatin modifiers as cancer therapeutics (4). Most of these “epigenetic gene alterations” result in loss of gene function (LOF) and may create a functional dependence on a related chromatin modifier. For instance, LOF mutations in the BAF (SWI/SNF) chromatin remodeling complex subunit ARID1A created a dependence on its paralog ARID1B (5). ARID1A is mutated in 25% of muscle-invasive bladder cancer cases, and a high frequency of ARID1A mutations has been reported in a number of indications, including ovarian clear cell carcinoma (OCCC; 46%–57%; refs. 6, 7), endometrial (30%–40%; refs. 8, 9), and gastric cancer (20%; ref. 10). In addition, pan-cancer studies suggest an overall ARID1A mutation frequency of approximately 7% in human cancer (11, 12). Although ARID1A mutations in certain contexts may predict responses to immunotherapy in metastatic urothelial carcinoma (13), no therapeutic approach has been developed to specifically exploit this potential cancer genomic vulnerability.
Enhancer of zeste homolog 2 (EZH2) is the catalytic component of the multisubunit polycomb repressive complex 2 (PRC2), which trimethylates histone H3 at lysine 27 (H3K27me3) to promote and maintain gene silencing (for review see ref. 14). Human EZH2 and its paralog EZH1 are the only enzymes known to catalyze H3K27 methylation. EZH2 is recurrently mutated in various cancer types, and depending on the type of mutation and context, it is considered both an oncogene and tumor suppressor (15). A synthetic lethal relationship has been described for PRC2 in the context of mutated BAF complex components such as SMARCB1 LOF in malignant rhabdoid tumor models (16) and SMARCA4 LOF alone (17) or with concurrent transcriptionally silenced SMARCA2 (18) in several cancer types. A functional link between ARID1A mutations and increased sensitivity to EZH2 inhibition has been reported in clear cell ovarian (19), gastric (20), and bladder cancer (21) cell models. In contrast, ARID1A deficiency did not appear associated with enhanced EZH2 inhibitor sensitivity or depletion of EZH2 in short-term bladder cancer cell growth assays (22).
Multiple PRC2 inhibitors targeting the EZH2 or EED subunits are undergoing clinical development (23, 24). The EZH2 inhibitor tazemetostat gained regulatory approval for the treatment of epithelioid sarcoma and follicular lymphoma (25, 26). Although first generation EZH2 inhibitors are generally well tolerated and do not show dose-limiting toxicities even at high doses these compounds induce their own metabolism, resulting in marked reduction in exposure upon repeat dose administration (27, 28). Thus, these compounds do not achieve complete target coverage over time (27–29). Although H3K27me3 is a proximal biomarker of EZH2 activity, it lacks sensitivity and connectivity to the downstream molecular gene regulatory consequences to be an effective clinical pharmacodynamic marker. In addition, there is currently no pharmacodynamic biomarker for PRC2 inhibitor activity with predictive potential.
Here, we report the discovery of tulmimetostat (CPI-0209), a potent, selective, orally available inhibitor of EZH2 that allows for comprehensive and durable target coverage, as well as superior performance in preclinical cancer models. We broadly investigated a bladder cancer cell panel and confirmed ARID1A LOF mutations as a context with preferential sensitivity to EZH2 inhibition. Restoration of ARID1A function in ARID1A mutant bladder cancer cells causes molecular and phenotypic consequences similar to those mediated by tulmimetostat treatment. Tulmimetostat outperforms cisplatin and combines well with it in bladder cancer models in vitro and in vivo. Importantly, to the best of our knowledge, we demonstrate for the first time EZH2 inhibitor efficacy in multiple ARID1A mutant endometrial tumor models in vivo, broadening therapeutic opportunities and clinical development paths for tulmimetostat. Tulmimetostat is currently being explored in a Phase I/II study for patients with advanced hematologic and solid tumors (ClinicalTrials.gov Identifier: NCT04104776). We report Phase I clinical pharmacokinetic (PK) and pharmacodynamic (PD) data, indicating that tulmimetostat exhibits durable exposure, profound target engagement, and a clear relationship between exposure and extent of target engagement in patients, support its potential for superior therapeutic performance.
Materials and Methods
Compound synthesis
The synthetic route of tulmimetostat is described in the Supplementary Materials and Methods. CPI-1205 was synthesized as described previously (30). GSK126, tazemetostat, MAK683, and valemetostat were purchased from Selleck Chemicals. PF-06821497 was purchased from Chemietek.
Biochemical assays
Reconstituted pentameric PRC2 composed of EZH2 (wildtype or mutant) or EZH1, plus EED, SUZ12, RbAp46, and RbAp48 was prepared in-house as described previously (31). Enzymatic assays with PRC2 containing wildtype EZH2 or EZH1, kinetic characterization of PRC2 containing EZH2 by TR-FRET assays, and differential scanning fluorimetry (DSF/TSA) assays for complexes containing either EZH2 or EZH1, were carried out as described previously (32).
Cell lines and culture
Cells were obtained from ATCC, DSMZ, European Collection of Cell Cultures (or through Sigma), or Japanese Collection of Research Biosources and were grown in media recommended by the vendor [Supplementary Table S1; includes research resource identifiers (RRID)] and maintained at 37°C in humidified incubators with 5% CO2. Cells were maintained in T75 flasks and subcultured by releasing from plates with TrypLE solution (Thermo Fisher Scientific/Invitrogen #12604021) every 2 to 4 days, depending on growth kinetics of the cell line, to maintain growth at subconfluent levels. Cells had undergone fewer than 10 passages from the purchased stock at the time of experiments. Furthermore, cells were tested regularly (at least monthly) for Mycoplasma infection using MycoAlert Mycoplasma Detection Kit (Lonza #LT07–318). Cell line authentication using STR genotyping was carried out through ATCC for all bladder cell lines, except JMSU1, RT4, J82, UMUC3, and KMBC2, and for all OCCC cell lines except TOV112D, A2780, OVISE, and ES2.
Cell viability assays
Cells were plated in the middle 60 wells of tulmimetostat or other EZH2 and EED inhibitor-containing 96-well plates at densities optimized to give <95% confluence after 6 days (bladder cancer panel) or 7 days (ovarian cancer panel) of culture. Compounds were plated in 2- to 3-fold dilution series in DMSO (9-point titrations of compounds) in at least duplicate per plate with either 1 or 10 µmol/L top concentrations, depending on the sensitivity of the cell line and compound potency. Cells were passed from the 96-well plates at days 4 and 7 for 11 total days of culture (KARPAS-422), days 6 and 12 for 18 total days of culture (bladder cancer panel) or days 7 and 14 for 21 total days of culture (ovarian cancer panel), replating each time with a split ratio that was determined for each cell line to restore the initial plating density. For cisplatin combination treatments, cell lines were pretreated for 7 days with tulmimetostat with two-fold, 8-point titrations from 0.25 to 1 µmol/L top concentrations in DMSO in six replicate rows, and then split and replated for cotreatment for 5 days with 3-fold, 4-point cisplatin titrations from 10 µmol/L top concentration vertically in 10 replicate columns in dimethylformamide (DMF). Single agent tulmimetostat and cisplatin titrations without combination were included on each plate for reference half-maximal growth inhibitory concentration (GI50) values. To assess cell viability in single agent and cisplatin combination assays, relative cell numbers were assessed by CellTiter-Glo 2.0 luminescent cell viability assay (Promega #G9243). 50 µL of CellTiter-Glo 2.0 reagent was added to each well containing 100 μL cell suspension and incubated for 30 minutes at room temperature, protected from light, with shaking at low speed. 100 µL of lysed cells were transferred to white-walled assay plates for readout using a Perkin Elmer EnVision Alpha Reader (model #2104). GraphPad Prism was used for curve fitting and GI50 determinations. Combination index values were generated using Genedata Screener (RRID:SCR_022506) program, with an effect level set at 70% of total effect.
Cell cycle analysis
Cells remaining postsplit from term viability assays at days 6 and 12 were transferred to v-bottom 96-well plates and pelleted, washed once with phosphate buffered saline (PBS), and fixed in ice-cold 70% ethanol for a minimum of 16 hours at 4°C. Fixed cells were pelleted, washed once with PBS, and stained with propidium iodide stain solution (propidium iodide (20 µg/mL, Sigma), RNAse A (25 µg/mL, Sigma), 0.1% Triton X-100 in PBS). Cell cycle data were obtained using a Guava Easycyte flow cytometer with the Express Pro module. Gated data were plotted in GraphPad Prism (RRID:SCR_002798).
H3K27me3 level assessment in cells and tissues
H3K27me3 and total H3 expression levels in cells and tumor tissues were analyzed by Meso Scale Discovery (MSD) ELISA assays, as previously described (32). For details, see Supplementary Materials and Methods.
In vitro washout assays
Cells were plated in T75 flasks for 4 days and treated with compounds at the following concentrations: tulmimetostat 25 nmol/L, CPI-1205 1 µmol/L, tazemetostat 1 µmol/L, valemetostat (DS-3201b) 25 nmol/L, PF-06821497 (PF-1497) 100 nmol/L, MAK683 (EEDi) 100 nmol/L or DMSO control. After 4 days, compound-containing culture media was removed, cells were washed twice with PBS and released from flasks with TrypLE solution. A portion of cells were removed and snap frozen for analysis of 4 days on-treatment by Western blot and qRT-PCR. Remaining cells were counted and plated in duplicate wells with continuing compound treatment (on-treatment) or no compound treatment (washout) in six-well plates for protein extraction and 24-well plates for RNA extraction, at a density that allowed for subconfluent growth for 1 to 4 additional days. Cells were harvested for both protein and RNA extraction from the on-treatment and washout wells for each compound at days 5, 6, 7, and 8 (Day 5 samples are 5 days on-treatment or 4 days on-treatment +1 day washout, etc.).
Western blotting, qRT-PCR analysis
Using standard methodologies; antibodies used for Western blotting are described in Supplementary Table S2 (includes available RRIDs). Reagents used for qRT-PCR are described in more detail in Supplementary Material and Methods.
Tumor xenograft studies
Cell line derived xenograft tumor experiments were performed at WuXi AppTec. Patient-derived xenograft (PDX) experiments were performed at Crown Bioscience. All the procedures related to animal handling, care, and the treatment in the xenograft studies were performed according to the guidelines approved by the Institutional Animal Care and Use Committee of WuXi AppTec or Crown Bioscience following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care. Female CB17 SCID mice were inoculated subcutaneously in the right flank with tumor cells in 0.2 mL PBS mixed with Matrigel (BD Biosciences); 5 × 106 cells per injection. Mice were randomized and drug treatment started 11 to 15 days post inoculation. Tumor size was measured three times weekly in two dimensions using a caliper, and the volume was expressed in mm3 using the formula: V = 0.5 a × b2 where a and b are the long and short diameters of the tumor, respectively. Tumor growth inhibition (TGI) was calculated for each group using the formula: TGI (%) = [1−(Ti−T0)/ (Vi−V0)] ×100; Ti is the average tumor volume of a treatment group on a certain day, T0 is the average tumor volume of the treatment group on the day the treatment was commenced, Vi is the average tumor volume of the vehicle control group on the same day with Ti, and V0 is the average tumor volume of the vehicle group on the day the treatment was commenced. Samples were collected at indicated interim timepoints on-treatment or at study endpoint for analysis at 1-hour postdosing for PK and PD studies, unless indicated otherwise (see Supplementary Materials and Methods for further details).
RNA-seq and analysis
Patient whole blood was collected using PAXgene blood RNA tubes. Processing and sequencing of RNA samples were performed by ALMAC. The RNA extractions were performed using PAXgene Blood RNA Kit following manufacturer’s instructions. Library preparation proceeded with rRNA and hemoglobin depletion using KAPA RNA HyperPrep Kit with RiboErase (HMR) Globin. Paired end 75 base pair sequencing were performed on an Illumina platform. Prior to sequencing, RNA integrity was assessed by 2100 Bioanalyser (Agilent), and concentration was assayed by Nanodrop. Processing and sequencing of mouse whole blood RNA samples were performed by GENEWIZ. The RNA extractions were performed using PAXgene Blood RNA Kit following manufacturer’s instructions. Library preparation with rRNA depletion and paired end 150 base pair sequencing on an Illumina HiSeq was performed by GENEWIZ. Prior to sequencing, RNA integrity was assessed by 2100 Bioanalyser (Agilent), and concentration was assayed by Qubit. RNA from cultured cell lines and xenograft models was isolated using with TRIzol (Thermo Fisher Scientific), following the manufacturer’s instructions. Library preparation with rRNA depletion and paired end 150 base pair sequencing on an Illumina HiSeq was performed by GENEWIZ. Prior to sequencing, RNA integrity was assessed by 2100 Bioanalyser (Agilent), and concentration was assayed by Qubit. Sequencing reads were mapped to the genome using STAR2, and the transcript and gene read count quantification were performed using RSEM with default settings. Gene set enrichment analysis (GSEA) was used to explore the overlap with annotated gene sets in the Molecular Signatures Database (33).
Chromatin immunoprecipitation sequencing
A total of 1 × 107 HT1376 cells treated with or without tulmimetostat (250 nmol/L) for 8 days were fixed by adding formaldehyde to final concentration of 1%. Fixation was done for 10 minutes at room temperature. Cross-linking was stopped by adding glycine to a final concentration of 125 mmol/L. The fixed cells were snap frozen and shipped by Active Motif for chromatin immunoprecipitation sequencing (ChIP-seq) processing. Briefly, ChIP reactions were performed with 30 μg of extracted chromatin with antibodies to EZH2 (Active Motif #39901, RRID:AB_2614956), H3K27ac (Active Motif #39133, RRID:AB_2561016) and H3K27me3 (Active Motif #39155, RRID:AB_2561020). Sequencing was performed with an Illumina NextSeq 500 platform to generate 75-nt sequence reads mapped to the genome using the BWA algorithm with default settings. For comparative analysis, Drosophila spike-in protocols and normalization were used. The number of test tags were adjusted by a factor that would result in the same number of usable spike-in Drosophila tags for each sample within a normalization group. Peaks were called using either the MACS or SICER algorithms. The MACS default cutoff was P-value 1e−7 for narrow peaks and 1e−1 for broad peaks, and SICER default cutoff was FDR 1e−10, with gap parameter of 600 bp. Peak filtering was performed by removing false ChIP-seq peaks, as defined in the ENCODE blacklist.
Clinical trial and PK and PD data
PK of tulmimetostat was evaluated based on plasma concentration data from the Phase I portion of the Phase I/II study of tulmimetostat in patients with advanced tumors (clinicaltrials.gov identifier: NCT04104776, data cut July 16, 2022). Phase I was a seven-cohort dose escalation study in patients receiving tulmimetostat once daily (QD) at doses between 50 and 375 mg (34). All patients gave written informed consent, and the trial was conducted in accordance with the International Council for Harmonization E6 Guideline for Good Clinical Practice, the Declaration of Helsinki, and applicable local regulations. This study was approved by an institutional review board. Tulmimetostat AUC was calculated using standard noncompartmental methods in Phoenix 8.3.1.5014.
Data availability
Transcriptomic and epigenomic data including patient RNA-seq data reported in this study are publicly available in Gene Expression Omnibus at GSE264383 and GSE176493. Mutations noted for ARID1A, KDM6A, and BAF complex members in cell lines were obtained from the Broad Institute’s Cancer Cell Line Encyclopedia (CCLE) portal at https://portals.broadinstitute.org/. Biochemical and in vitro cell data generated in this study are available upon request from the corresponding author. Raw data pertaining to animal studies were generated at WuxiApptec and Crown Biosciences. Derived data supporting the findings are available from the corresponding author upon request. Clinical trial data reported in this study will be made available by MorphoSys Inc. upon request.
Results
Discovery and characterization of second generation EZH2 inhibitor tulmimetostat
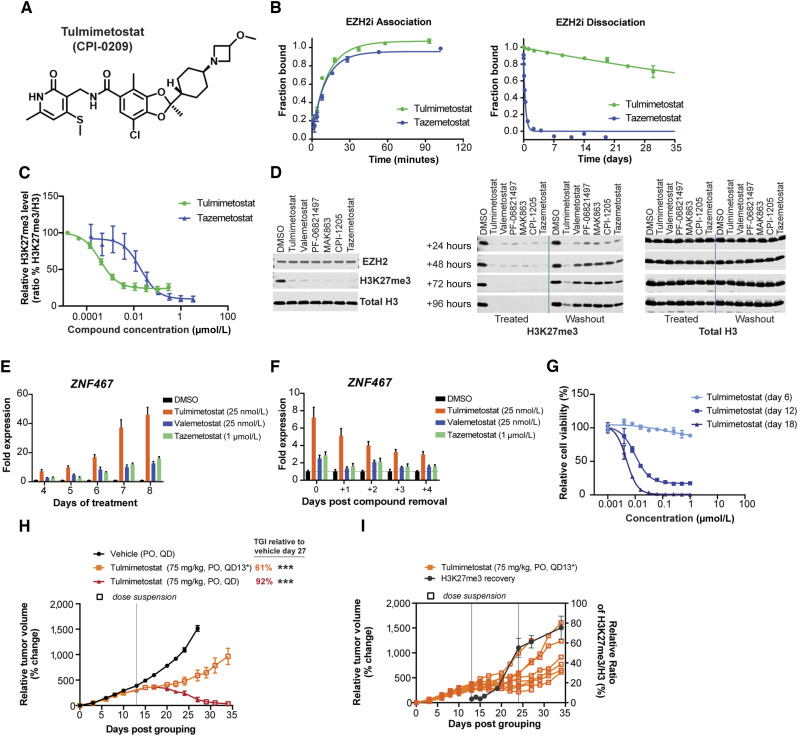
To extract the full potential of EZH2 inhibition as a therapeutic mechanism for hematology and oncology applications, we identified a thiomethyl substitution on the pyridone “warhead” that significantly increased potency and residence time (35). Further optimization of potency and physicochemical properties resulted in the discovery of tulmimetostat (Fig. 1A; Supplementary Fig. S1A). This compound exhibited a Ki of approximately 140 fmol/L by kinetic analyses and an extrapolated Ki of approximately 180 fmol/L by thermal shift assays (Supplementary Table S3). When profiled against a panel of over 30 protein and DNA methyltransferases at concentrations of up to 10 µmol/L, tulmimetostat only inhibited EZH2 and the closely related EZH1 enzyme (Supplementary Fig. S1B). Comparison of tulmimetostat with other EZH2 inhibitors in enzymatic and biophysical assays suggested that tulmimetostat is among the most potent EZH2 and EZH1 inhibitors identified to date. Tulmimetostat did not inhibit or activate any target in a safety pharmacology profiling panel (Cerep; Supplementary Fig. S1C). Although the on-rate of tulmimetostat was comparable to EZH2 inhibitor tazemetostat, the off-rates were very different (Fig. 1B; Supplementary Fig. S1D). Tulmimetostat had an exceptionally long residence time when compared in the same kinetic assays to other EZH2 inhibitors in the context of both allosterically activated and nonactivated PRC2 complex (Supplementary Fig. S1E) and retained a S-adenosyl-L-methionine competitive, reversible mechanism of inhibition (Supplementary Fig. S1F).
Figure 1.
Identification and characterization of tulmimetostat, a highly potent, long residence time EZH2 inhibitor. A, Chemical structure of tulmimetostat. B, Determination of tulmimetostat and tazemetostat-binding kinetics for the PRC2 protein complex. Representative data for association (left) and dissociation (right) rate determinations by TR-FRET are shown for both compounds and reported from two or more independent determinations ± SD. For association, tulmimetostat kon = 8.7 (±2) × 105 (mol/L)−1·second−1, tazemetostat kon = 5.6 (±1) × 105 (mol/L)−1·second−1. The residence times (t) are reported for koff [τ = (1/koff); for tulmimetostat, τ ∼ 96 (±10) days; for tazemetostat, τ ∼ 0.32 (±0.2) days]. C, HeLa cells were treated with increasing concentrations of tulmimetostat or tazemetostat for 72 hours, and global H3K27me3 levels were measured and normalized to total histone H3 levels to determine half-maximal inhibitory concentration (IC50) values calculated from the mean of duplicate experiments ± SEM. D, HT1376 cells were treated with tulmimetostat (25 nmol/L), valemetostat (25 nmol/L), PF-06821497 (100 nmol/L), MAK863 (EEDi; 100 nmol/L), CPI-1205 (1 µmol/L), and tazemetostat (1 µmol/L) for 4 days and then split for Western blot analysis (left) for replating and culturing in the presence of compounds (Treated) and for replating and culturing in the absence of compounds (Washout). Culturing was continued for indicated time points (24, 48, 72, and 96 hours), cells were harvested, and cell extracts subjected to Western blotting. Antibodies for Western blot analysis are indicated on the right and bottom of top and middle and right panels, respectively. E, HT1376 cells were cultured in the presence of tulmimetostat (25 nmol/L), valemetostat (25 nmol/L), and tazemetostat (1 µmol/L) for 4, 5, 6, 7, and 8 days, cells were harvested, total RNA extracted and analyzed by qPCR for the magnitude of ZNF467 transcript level change. Gene expression changes are represented as the fold change over control from the mean of internal quadruplicates ± SD. F, HT1376 cells were cultured as in E for 4 days, compounds were removed by media change, and cell culturing continued for various time periods (1, 2, 3, and 4 days). Cells were harvested, total RNA extracted and analyzed by qPCR for the magnitude of ZNF467 transcript level change. Gene expression changes are represented as the fold change over control from the mean of quadruplicates ± SD. G, Viability in response to tulmimetostat treatment at 6, 12, and 18 days in HT1376 cells. Data are represented as the mean of triplicate wells ± SD from one of three independent experiments. H,In vivo efficacy experiment in a HT1376 subcutaneous xenograft model was carried out to assess the impact of tulmimetostat on tumor growth. Tulmimetostat was dosed at 75 mg/kg orally (PO), QD for 13 or 27 days. Data are represented as the mean relative tumor volume per cohort and time point ± SEM, and n = 9 for vehicle and tulmimetostat QD13* arms, n = 6 for QD arm. TGI noted for day 27 and 34 relative to day 27 vehicle arm. P values calculated using two-way ANOVA up to day 27, ***, P < 0.0001. I, Tumor growth in individual animals of the cohort treated with tulmimetostat for 13 days from the experiment described in H is shown. Tumors (n = 3 for all time points except day 34, which n = 6) at various time points were analyzed for global H3K27me3 levels relative to total histone H3 levels. Vertical lines indicate tumor growth lag period post last tulmimetostat dose.
The superior target affinity and residence time of tulmimetostat resulted in potent suppression of global H3K27me3 levels in cell-based assays (tulmimetostat IC50 = 0.38 nmol/L vs. tazemetostat IC50 = 19.5 nmol/L; difference: >50-fold; Fig. 1C). A key cancer cell model for the assessment of EZH2 inhibitor performance is the lymphoma cell line KARPAS-422 harboring an activating EZH2 Y641N mutation. Tulmimetostat was effective in suppressing KARPAS-422 cell proliferation in a dose-dependent manner (tulmimetostat GI50 = 6 nmol/L vs. tazemetostat IC50 = 322 nmol/L; difference: >50-fold; Supplementary Fig. S1G). Consistent with the mechanism of action and our prior data suggesting that turnover of H3K27me3 upon EZH2 inhibition is slow (36), the growth inhibitory effect was time-dependent. In a KARPAS-422 mouse xenograft model, tulmimetostat was well tolerated and led to complete tumor regression, whereas tazemetostat at the same dose and schedule showed modest TGI (Supplementary Fig. S1H and S1I). No tumor regrowth was observed in tulmimetostat-treated animals after treatment cessation. Consistent with superior efficacy, tulmimetostat demonstrated a greater level of H3K27me3 level reduction in KARPAS-422 tumors compared with tazemetostat (Supplementary Fig. S1J). Exposures were similar on days 1 and 12 of treatment (Supplementary Table S4), suggesting that there was no induction of tulmimetostat metabolism over the course of treatment.
We chose the ARID1A mutant bladder cancer model HT1376 to study the impact of tulmimetostat residence time in driving the downstream molecular and phenotypic consequences of EZH2 inhibition in a solid tumor context. Consistent with other cell models, tulmimetostat potently reduced global H3K27me3 levels in a dose-dependent manner (Supplementary Fig. S1K). To explore how residence time impacts H3K27me3 levels, we performed washout experiments in HT1376 cells treated with EZH2 or EED inhibitors. Treatment for 4 days resulted in almost complete loss of global H3K27me3 levels (Fig. 1D, top). Next, cells were analyzed for time-dependent recovery of H3K27me3 after removal of compound; continuous compound treatment for up to 4 additional days was included as a control (Fig. 1D, bottom). Tulmimetostat-treated cells maintained ≥50% H3K27me3 loss for at least 4 days after compound removal, whereas cells treated with other EZH2 or EED inhibitors recovered H3K27me3 levels within 2 days. PRC2 functions in transcriptional repression and EZH2 inhibitors have been shown to induce target gene expression (36–38). Treatment with tulmimetostat resulted in time-dependent induction of target gene expression over 4 to 8 days and demonstrated greater overall level of gene induction compared with tazemetostat or valemetostat (Fig. 1E; Supplementary Fig. S1L). Consistent with the time-delayed recovery of the H3K27me3 mark after compound removal, tulmimetostat-induced genes returned to baseline expression status with slower kinetics (Fig. 1F; Supplementary Fig. S1L), indicating that the longer residence time of tulmimetostat also results in prolonged transcriptional effects. These tulmimetostat-induced changes in H3K27me3 levels and gene expression preceded potent growth phenotypes after extended treatment periods (Fig. 1G). In a HT1376 mouse xenograft model, administration of tulmimetostat for 27 days resulted in tumor regression with no tumor regrowth following the cessation of treatment (at least 7 days post last dose). Further, treatment for only 13 days resulted in tumor stasis for 12 days post last dose before tumor growth resumed (Fig. 1H). Global H3K27me3 levels were assessed on the last day of treatment (day 13) and at several timepoints after dosing was stopped to monitor H3K27me3 recovery. Significantly reduced H3K27me3 levels were maintained for up to 6 days post last dose (Fig. 1I; Supplementary Fig. S1M), indicating extended suppression of EZH2 activity in the absence of repeat dosing. Recovery of tumor H3K27me3 levels preceded tumor regrowth, which was seen in most animals after day 25 (Fig. 1I). Collectively, these data suggest that tulmimetostat has the potential to comprehensively and durably inhibit EZH2 activity, leading to significant and sustained TGI (Fig. 1H and I; Supplementary Fig. S1H).
Tulmimetostat phenotypic responses are enriched in the context of ARID1A LOF mutations
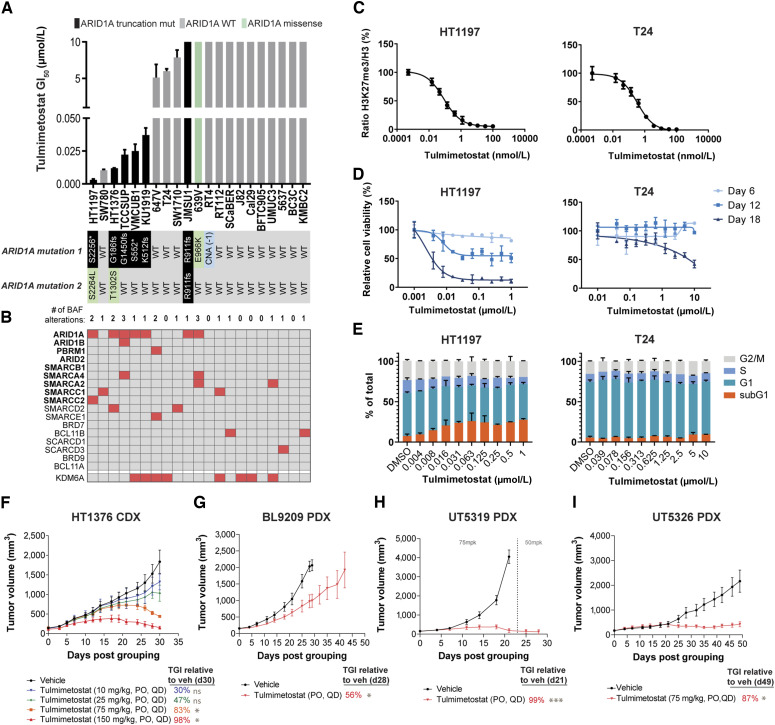
To expand on the results observed in HT1376 cells and further understand molecular contexts in bladder cancers that govern sensitivity to EZH2 inhibition, we evaluated a panel of 21 bladder cancer cell lines. Tulmimetostat inhibited the growth of a subset of the lines with GI50 of 3 to 37 nmol/L after 18 days of treatment (Fig. 2A; Supplementary Table S5). The growth inhibitory effects were significantly enriched (P = 3.7e−6, χ2 test) in bladder cancer lines carrying at least one ARID1A LOF allele, with 83% (5/6) sensitive cell lines harboring a truncation mutation (frameshift or nonsense) in ARID1A. By contrast, only 6% (1/15) of unresponsive lines with an 18-day GI50 >5 µmol/L, referred to as “resistant” from here on, had an ARID1A LOF allele. We observed no association between tulmimetostat sensitivity and baseline levels of EZH1, EZH2, H3K27me3, ARID1A, or ARID1B by Western blot (Supplementary Fig. S2A). The mutation status of a broader set of frequently mutated BAF complex components across cancer types (39) was also evaluated, and it was found that only ARID1A genomic alterations segregate with tulmimetostat response (Fig. 2B). This finding supported the notion that ARID1A LOF mutation may be an encouraging biomarker to enrich for response to EZH2 inhibition. Tulmimetostat is equally effective at reducing H3K27me3 levels in both resistant and sensitive cell lines (Supplementary Table S5), irrespective of phenotype or ARID1A mutation status (Fig. 2C; Supplementary Fig. S2B). Cell viability effects in tulmimetostat-sensitive bladder cancer cell lines were time-dependent. Although most bladder cancer cell lines show minimal viability effects after 6 days of treatment, prolonged treatment for 12 and 18 days substantially increased the sensitivity of ARID1A mutant cell lines (Figs. 1G and 2D). Phenotypically responsive cell lines such as HT1376 and HT1197 showed induction of cell death on Day 12, as evidenced by an increase in the subG1 population, whereas the cell cycle profiles of resistant cell lines such as T24 remain unchanged even after prolonged treatment (Fig. 2E; Supplementary Fig. S2C).
Figure 2.
Tulmimetostat-mediated phenotypic responses are enriched in the context of ARID1A LOF mutations. A, Eighteen-day viability assay GI50 values for tulmimetostat in a panel of bladder cancer cell lines. Black bar indicates cell line carrying at least one ARID1A stop-gain (denoted by an *) or frameshift (fs) allele (from here on out denoted as ARID1A LOF mutant), as detailed below the chart. Green bar indicates line carrying a single missense mutation, whereas gray bars indicate those lines with no mutations in the coding region of ARID1A. Data represented as an average of duplicate wells ± SD and are representative of duplicate independent experiments. B, Summary of the mutation status of the major components of the BAF complex as well as KDM6A in the bladder cancer panel. Those noted in bold are the most frequently mutated in cancer (40). Red box, presence of a mutation; gray, wildtype for a given gene. C, Normalized global H3K27me levels in HT1197 (left) and T24 (right) cell lines following 72 hours of treatment across a dose range of tulmimetostat. Data represented as average of triplicate wells ± SD and are representative of quadruplicate independent experiments. D, Cell viability dose response curves in HT1197 (left) and T24 (right) cell lines over 18 days of treatment. Data represented as an average of duplicate wells ± SD and are representative of duplicate independent experiments. E, Cell cycle stage distribution in HT1197 (left) and T24 (right) cell lines treated with tulmimetostat for 12 days. Data are represented as average of duplicate wells ± SD and are representative of duplicate independent experiments. F, TGI after treatment with 10, 25, 75, and 150 mg/kg tulmimetostat orally, QD or vehicle in HT1376 bladder cancer xenografts. Data represented as mean tumor volume ± SEM, with n = 5 mice per group for all groups except 150 mg/kg, which had n = 3. P values calculated using two-way ANOVA up to day 30, ns, nonsignificant, P > 0.05; *, P < 0.05. G, TGI in an ARID1A mutant PDX model of bladder cancer (BL9209) treated with tulmimetostat at 75 mg/kg orally, QD. Data represented as mean tumor volume ± SEM, with n = 3 mice per group. TGI calculated using tumor volumes at day 28, *, P < 0.05 using two-way ANOVA through day 28, when vehicle reached endpoint. H, TGI in an ARID1A mutant PDX model of endometrial cancer (UT5319) treated with tulmimetostat. Mice were initially treated with 75 mg/kg orally, QD; dose was reduced to 50 mg/kg orally, QD after day 23. Data represented as mean tumor volume ± SEM, with n = 3 mice per group. TGI calculated using tumor volumes at day 21, ***, P < 0.0001 using two-way ANOVA through day 21, when vehicle reached endpoint. I, TGI in an ARID1A mutant PDX model of endometrial cancer (UT5326) treated with tulmimetostat at 75 mg/kg orally, QD. Data represented as mean tumor volume ± SEM, with n = 5 mice per group. TGI calculated using tumor volumes at day 49, *, P < 0.05 using two-way ANOVA through day 49, when study reached endpoint.
To further explore the potential dose dependence of tulmimetostat on TGI, HT1376 cell line–derived xenograft (CDX) studies were carried out administrating tulmimetostat doses from 10 to 150 mg/kg. Dose-dependent TGI was achieved and ranged from 30% with 10 mg/kg to 98% with 150 mg/kg by 30 days. All dose levels ≥75 mg/kg resulted in substantial reductions in tumor volume compared with vehicle (Fig. 2F; Supplementary Fig. S2D). As expected, global H3K27me3 levels in HT1376 tumors were reduced in response to tulmimetostat treatment in a dose-dependent manner, with at least an 80% reduction at 10 mg/kg and >90% in 25 mg/kg and higher doses at Day 10 and maintained at comparable levels at Day 31 (Supplementary Fig. S2E). A subsequent HT1376 CDX study with an extended duration of treatment showed reproducible levels of TGI at 30 days followed by tumor volume reduction at both 35 and 50 mg/kg QD dose levels at timepoints beyond 35 days (Supplementary Fig. S2F and S2G). Tulmimetostat also achieved significant anti-tumor activity as monotherapy in PDX models of ARID1A LOF bladder and endometrial cancers (Fig. 2G–I; Supplementary Fig. S2H–S2N). Consistent with a prior publication (19), tulmimetostat also demonstrated in vitro and in vivo efficacy in the ARID1A mutant OCCC TOV21G CDX model (Supplementary Fig. S2O–S2Q). Of note, investigation of ovarian cancer cell lines suggested that bi-allelic ARID1A mutations rather than presence of any ARID1A mutation may better indicate ovarian cancer cell sensitivity to tulmimetostat treatment (Supplementary Fig. S2Q). Together, these solid tumor CDX and PDX model data suggest that tulmimetostat is highly efficacious at well tolerated doses across tumor types with ARID1A mutations.
Tulmimetostat treatment increases the expression of PRC2 occupied and repressed genes in ARID1A mutant bladder cancer cells
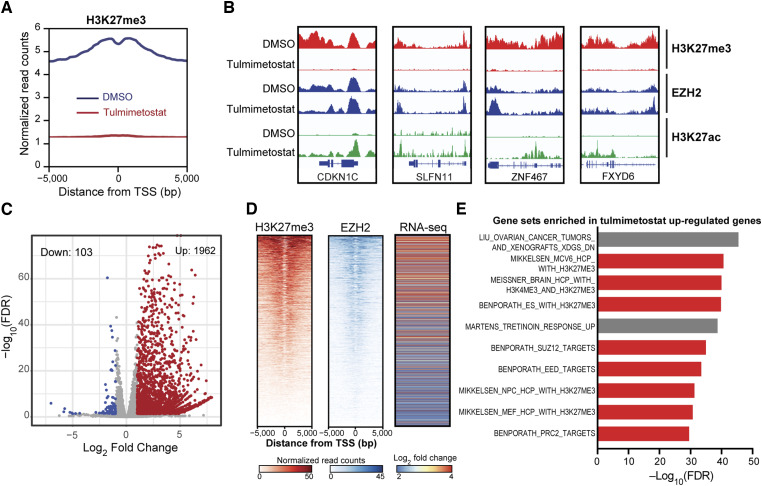
To study molecular changes underpinning the increased potential for EZH2 dependency in bladder cancer cell contexts we carried out ChIP-seq in ARID1A mutant HT1376 cells in the absence or presence of tulmimetostat. As expected, upon tulmimetostat treatment, H3K27me3 levels were substantially reduced at transcriptional start sites across the entire genome (Fig. 3A). At representative gene loci, H3K27me3 loss coincided with increased histone H3 lysine 27 acetylation (H3K27ac), whereas EZH2 occupancy remained largely unchanged or increased at certain loci (Fig. 3B). Because H3K27ac is a chromatin modification usually associated with active genes, we carried out RNA-sequencing to study whether these changes in chromatin states correlate with gene expression changes. The vast majority (95%) of genes changed by tulmimetostat treatment were significantly upregulated [Log2 fold change (L2FC) ≥ 1], consistent with the role of PRC2 in gene repression (Fig. 3C). The magnitude of gene induction correlated with the baseline level of H3K27me3 and EZH2 occupancy at these genes (Fig. 3D). GSEA that tulmimetostat-inducedgenes mostly fall into functional categories that are related to PRC2 (Fig. 3E). On the contrary, tulmimetostat downregulated genes lack H3K27me3 at baseline and likely represent indirect transcriptional effects.
Figure 3.
Tulmimetostat treatment increases the expression of PRC2 occupied and repressed genes in ARID1A mutant bladder cancer cells. A, Quantification of genome-wide H3K27me3 ChIP-seq enrichment on transcription start site (TSS) regions in HT1376 cells treated with 250 nmol/L tulmimetostat for 8 days. B, Integrative Genomics Viewer snapshots of H3K27me3, EZH2, and H3K27ac chromatin binding detected by ChIP-seq around four representative PRC2 target genes (CDKN1C, SLFN11, ZNF467, and FXYD6) following DMSO or tulmimetostat treatment. C, Volcano plot showing gene expression changes in HT1376 cells treated with DMSO or 250 nmol/L tulmimetostat for 4 days. Colored dots indicate absolute log2-fold change ≥ 1 and FDR ≤ 0.05. D, Heatmaps of H3K27me3 and EZH2 ChIP-seq data in HT1376 cells showing baseline (DMSO treated) occupancy of TSS regions for genes that are upregulated following tulmimetostat treatment in HT1376 cells. Right, RNA-seq heatmap of log2-fold changes of these same genes (in the same order) after treatment. E, GSEA of tulmimetostat upregulated genes showing the top 10 enriched gene signatures. Red bars, gene sets related to H3K27me3/PRC2.
To explore whether tulmimetostat mediated gene expression changes are similar to those induced by restoration of BAF complex function, we expressed wildtype ARID1A in the ARID1A mutant cell model HT1376 using a doxycycline-inducible system (Supplementary Fig. S3A). A significant reduction of global H3K27me3 levels in HT1376 cells treated with tulmimetostat was observed irrespective of ectopic ARID1A expression (Supplementary Fig. S3B). Restoration of ARID1A function in HT1376 cells resulted in loss of cell viability to a similar degree as treatment with tulmimetostat. Combination of ARID1A re-expression and tulmimetostat treatment did not result in combinatorial growth defects (Supplementary Fig. S3C), suggestive of EZH2 inhibition and ARID1A restoration having potentially overlapping mechanisms to promote cell death. This possibility is supported by prior observations in other cell contexts that PRC2 and BAF complexes frequently cotarget the same genes with opposing gene regulatory functions (19, 40). We next sought to compare changes in chromatin binding profiles and gene expression in HT1376 cells following re-expression of ARID1A and tulmimetostat treatment. On representative target genes, tulmimetostat treatment resulted in reduced H3K27me3 and increased H3K4me3 levels but did not promote BAF complex recruitment. Ectopic expression of ARID1A resulted in increased occupancy of BAF complex components ARID1A and SMARCA4 irrespective of tulmimetostat treatment (Supplementary Fig. S3D).
Principal component analysis (PCA) of global gene expression patterns among samples showed that tulmimetostat treatment and ARID1A re-expression (Supplementary Fig. S3E) are the major drivers of gene expression changes. ARID1A re-expression caused variable changes in gene expression, with ∼62% of significantly altered genes being downregulated and ∼38% upregulated by ARID1A, consistent with ARID1A’s role in both transcriptional repression and activation (Supplementary Fig. S3F; ref. 41). GSEA of genes upregulated by ARID1A re-expression revealed enrichment of direct EZH2 targets (defined in Supplementary Fig. S3G). Likewise, GSEA of genes upregulated after tulmimetostat treatment showed enrichment in ARID1A re-expression targets (Supplementary Fig. S3H). Comprehensive GSEA analysis with the Hallmark collection (42) indicated that more than half of enriched gene sets are commonly enriched by both tulmimetostat treatment and re-expression of ARID1A (Supplementary Fig. S3I), including pathways regulating cell differentiation, immune signaling, and inflammation.
Tulmimetostat treatment results in greater upregulation of PRC2 target gene expression in phenotypically sensitive ARID1A LOF cell lines
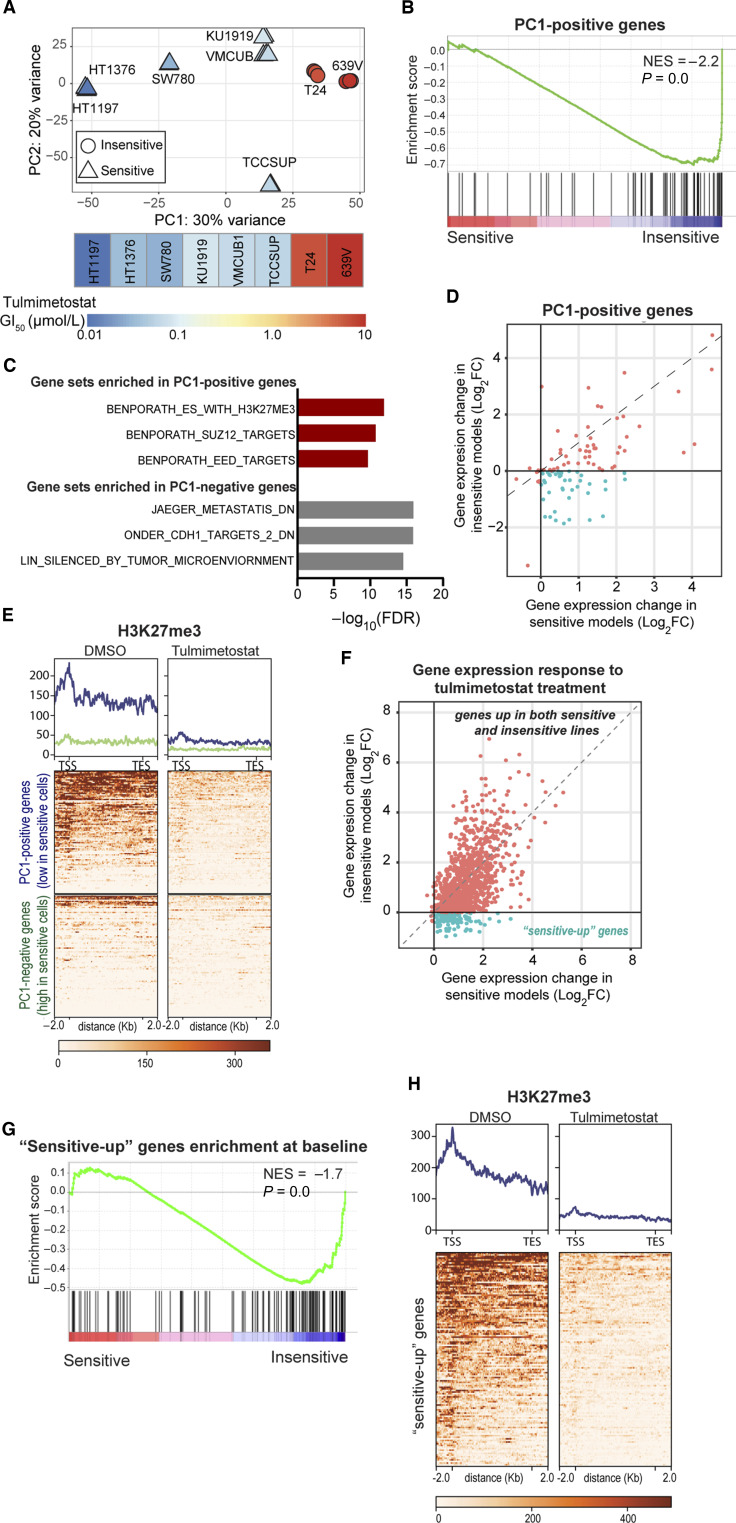
To more broadly understand gene expression changes in response to tulmimetostat, genome-wide expression profiling was performed on a panel of eight bladder cancer cell lines, composed of six sensitive and two insensitive models, with 5/6 sensitive lines harboring ARID1A mutations. Short-term (4 day) treatment resulted largely in gene upregulation and minimal gene downregulation in all eight bladder cancer cell lines, regardless of phenotypic sensitivity to tulmimetostat (Supplementary Fig. S4A and S4B). PCA of the baseline global gene expression patterns in these eight-cell lines revealed that the expression of genes within PC1 can distinguish cell lines with respect to their sensitivity to tulmimetostat (Fig. 4A), with the two insensitive lines clustering to the far right on PC1 and the two most sensitive lines clustering to the far left. The top 100 genes positively contributing to PC1 in this eight-cell line panel were referred to as “PC1-positive genes.” Next, we tested if this PC1-positive gene set would predict sensitivity across a larger panel of cell lines, expanding our analysis to all 21 bladder cancer cell lines tested for tulmimetostat response. Using CCLE-derived expression data for these lines (43), it was found that the PC1-positive genes were more highly expressed at baseline in insensitive cell lines (Fig. 4B). Conversely, the top 100 genes with negative contributions to PC1 (referred to as “PC1-negative genes”) were expressed at higher levels in sensitive cell lines (Supplementary Fig. S4C). This observation suggests that these gene sets may represent transcriptomic differences between tulmimetostat-sensitive and insensitive bladder cancer cell lines. Interestingly, the “PC1-positive genes” showed enrichment for PRC2 targets by GSEA (Fig. 4C), and tulmimetostat treatment resulted in greater upregulation of these “PC1-positive genes” in sensitive compared with insensitive cell lines (Fig. 4D; Supplementary Fig. S4D), suggesting that these targets may be more strongly repressed by EZH2 activity in sensitive cell lines. PC1-positive genes had high baseline H3K27me3 levels in tulmimetostat-sensitive HT1376 cells (Fig. 4E). H3K27me3 levels on these gene targets were substantially reduced, and expression of these genes increased with tulmimetostat treatment (Fig. 4E). PC1-negative genes behaved in the opposite way; they had lower baseline H3K27me3 levels on their promoters in HT1376, which was marginally reduced upon treatment (Fig. 4E), consistent with both higher baseline gene expression level (Supplementary Fig. S4C) and marginal induction by tulmimetostat (Supplementary Fig. S4E).
Figure 4.
Tulmimetostat treatment results in greater upregulation of PRC2 target gene expression in phenotypically sensitive ARID1A LOF cell lines. A, PCA of eight bladder cancer cell line samples treated with DMSO (baseline), colored based on their 18-day GI50 (as described in Fig. 2A), with the heatmap shown below. Top 100 genes contributing negatively to PC1 are referred to as “PC1-negative” and top 100 genes contributing positively to PC1 are referred to as “PC1-positive” here and throughout. B, GSEA showing enrichment of expression of the PC1-positive genes in A in resistant cell lines compared with sensitive cell lines in larger 21 cell line panel. C, Top gene sets significantly enriched (FDR < 0.01) from the C2 curated gene set list from MSigDB in the PC1-positive genes and PC1-negative genes. Red bars, PRC2-related gene sets. D, Scatter plot of gene expression changes of PC1-positive genes in response to tulmimetostat treatment in sensitive and resistant cell lines. Those genes that were induced (log2-fold change > 0) in sensitive cells but unchanged or down in resistant cells (log2-fold change ≤ 0) are highlighted in green. E, Heatmap of H3K27me3 ChIP-seq enrichment on gene body and flanking regions of the PC1-positive genes (blue; top) or the PC1-negative genes (green; bottom) in HT1376 cells treated with 250 nmol/L tulmimetostat for 4 days. Quantification of peaks within gene sets represented by histogram across the top. F, Scatter plot of gene expression changes in response to tulmimetostat treatment in sensitive and resistant cell lines. Those genes that were induced (log2-fold change > 0) in sensitive cells but unchanged or down in resistant cells (log2-fold change ≤ 0; 132 “sensitive-up” genes) are highlighted in green. G, GSEA showing enrichment of expression of “sensitive-up” gene set at baseline (DMSO treated) in resistant cell lines compared with sensitive cell lines. H, Heatmap of H3K27me3 ChIP-seq enrichment on gene body and flanking regions of “sensitive-up” gene set at baseline in HT1376 cells (DMSO treated). Quantification of peaks represented by histogram across the top.
To expand these findings, we investigated baseline expression and treatment-induced changes of EZH2/tulmimetostat target genes in bladder cancer cell models in an unbiased manner. The majority of EZH2 target genes (defined in Supplementary Fig. S3G) are induced by tulmimetostat treatment in both sensitive and resistant cell lines (upper right quadrant, Fig. 4F), but are expressed at lower baseline levels (Fig. 4G) and show greater degrees of induction in sensitive cell lines (Supplementary Fig. S4F). Notably, we identified 132 genes that were induced in sensitive cell lines and remained unchanged or were repressed in insensitive cell lines (hereinafter, “sensitive-up genes,” lower right, Fig. 4F; Supplementary Table S6). These genes, which are highly enriched for transcription factors (Supplementary Fig. S4G), had low baseline expression in sensitive cell lines (Fig. 4G), likely due to elevated H3K27me3 levels at their promoters (Fig. 4H). This differential degree of transcriptional activation in response to EZH2 inhibitor treatment in sensitive versus insensitive cell lines was previously observed in lymphoma cells (Supplementary Fig. S4H and S4I; ref. 37). Re-analysis of these data also showed that the magnitude of transcriptional activation is greater in sensitive cell lines compared with insensitive lymphoma cells (Supplementary Fig. S4J), consistent with our findings in bladder cancer cells.
Tulmimetostat improves cisplatin responsiveness in chemotherapy-resistant bladder cancer cells
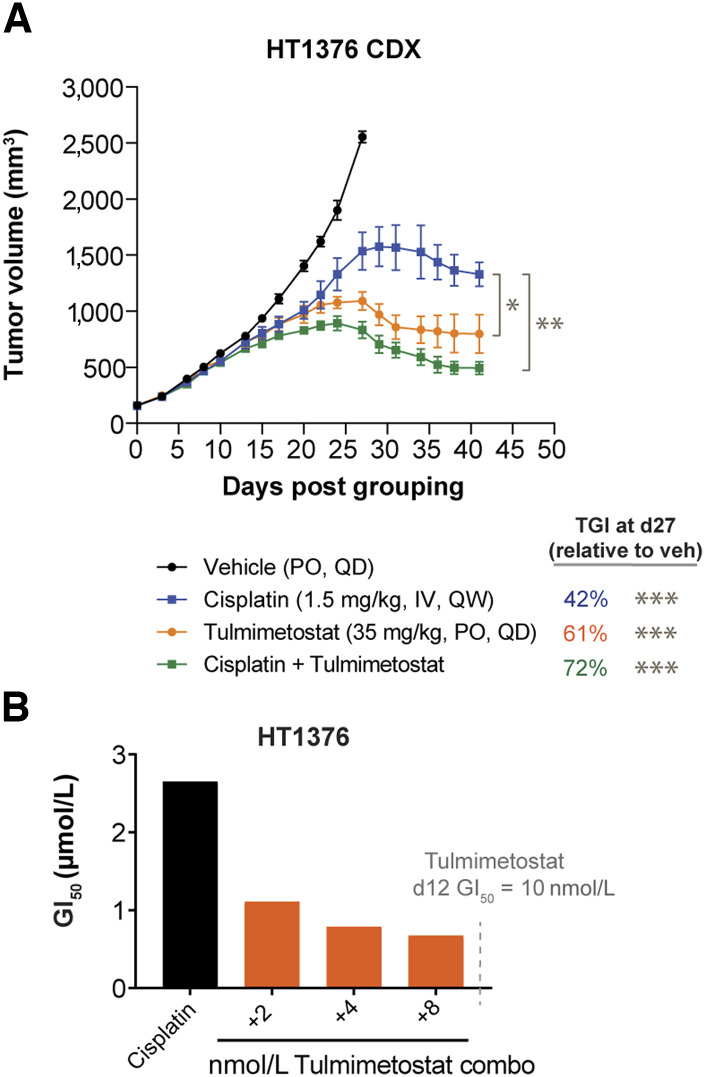
Because platinum-based chemotherapy is one of the main therapeutic options for metastatic bladder cancer, tulmimetostat combination treatment in cisplatin-sensitive and insensitive bladder cancer models was examined to determine whether tulmimetostat could augment intrinsic responsiveness to this chemotherapy. In the cisplatin-insensitive HT1376 xenograft model, administration of tulmimetostat resulted in a greater degree of TGI (63% vs. 42% TGI at day 27, P = 0.048) than cisplatin dosed intravenously at the maximum tolerated dose (1.5 mg/kg weekly for this model; Fig. 5A; Supplementary Fig. S5A). Additionally, when combined with cisplatin, tulmimetostat significantly deepened the response compared with cisplatin alone (P = 0.002). Evaluation of pretreatment with tulmimetostat alone followed by tulmimetostat and cisplatin co-treatment showed a moderately synergistic effect, as the GI50 decreased and the corresponding subG1 cell population increased (Fig. 5B; Supplementary Fig. S5B and S5C). Expanding the analysis to both cisplatin-sensitive (GI50 < 1.5 µmol/L) and insensitive (GI50 > 1.5 µmol/L) bladder cancer cell lines, we found that 5/6 cisplatin-insensitive lines and 2/7 cisplatin-sensitive models demonstrated synergistic effects of cisplatin and tulmimetostat cotreatment (Table 1; Supplementary Fig. S5D).
Figure 5.
Tulmimetostat improves cisplatin responsiveness in chemotherapy-resistant bladder cancer cells. A, TGI of tulmimetostat monotherapy, cisplatin monotherapy, or the combination in HT1376 bladder cancer xenografts. Data represented as mean ± SEM. n = 9 mice for vehicle; n = 6 mice each for tulmimetostat, cisplatin, and the combination arms. TGI values noted were calculated for all arms using day 27 tumor volumes, relative to vehicle. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 using two-way ANOVA up to day 27 (for TGIs) or through day 41 (for P values on the graph). B, Cisplatin GI50 shifts in HT1376 cells following pretreatment (7 days) and combination (5 days) with various nmol/L concentrations of tulmimetostat in 12-day assay. Dashed line shows the tulmimetostat GI50 at 12 days when assay was repeated (see Supplementary Fig. S5D), indicating that all shifts were occurring at sub-GI50 doses of tulmimetostat.
Table 1.
Summarization of the response of 13 bladder cancer cell lines to tulmimetostat, cisplatin, and their combination.
| Cell line | Tulmimetostat GI50 (µmol/L, d12) | Cisplatin GI50 (µmol/L, d5) | Combination index | Effect |
|---|---|---|---|---|
| TCCSUP | 0.07 | 0.22 | 0.55 | Synergism |
| JMSU1 | >1 | 0.73 | 1.02 | Nearly additive |
| T24 | >1 | 0.81 | 0.66 | Synergism |
| BFTC905 | >1 | 0.76 | 0.94 | Nearly additive |
| J82 | >1 | 0.45 | 1.12 | Slight antagonism |
| UMUC3 | >1 | 0.35 | 1.70 | Antagonism |
| SCaBER | >1 | 1.2 | 1.28 | Moderate antagonism |
| HT1197 | 0.05 | 3.88 | 0.58 | Synergism |
| HT1376 | 0.01 | 1.95 | 0.72 | Moderate synergism |
| KMBC2 | >1 | 3.64 | 0.66 | Synergism |
| RT4 | >1 | 3.05 | 0.91 | Nearly additive |
| Cal29 | >1 | 3.01 | 0.42 | Synergism |
| SW1710 | >1 | 2.15 | 0.64 | Synergism |
NOTE: Tulmimetostat (GI50 >1 μmol/L, 12‐day viability assay) and cisplatin (GI50 >1.5 μmol/L, 5‐day viability assay) insensitive lines are indicated by values in bold. Synergism based on Genedata Screener analysis of combination indexes with an effect threshold of 75%.
Tulmimetostat has greater potency than other PRC2 inhibitors in a bladder cancer xenograft model
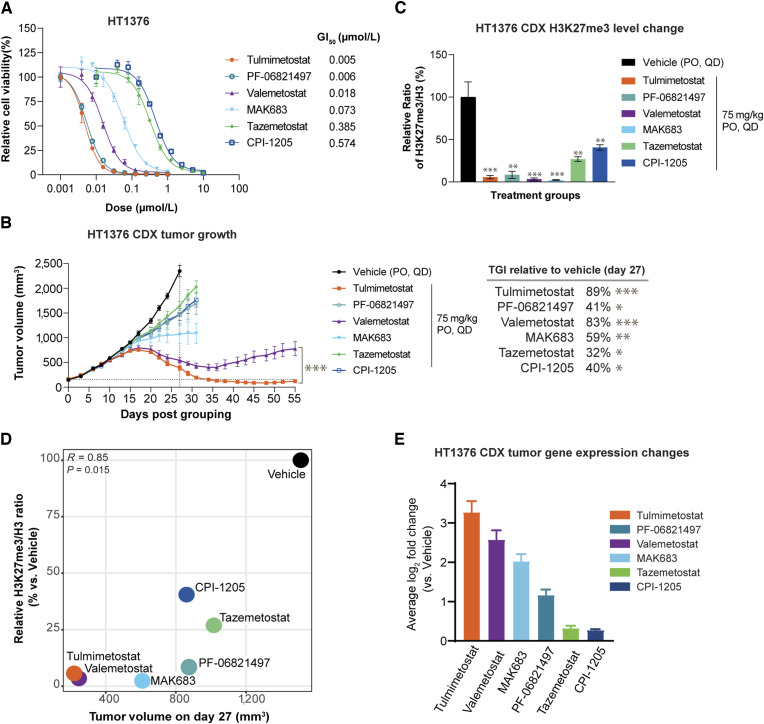
We next aimed to contextualize the responses observed both in vitro and in vivo with tulmimetostat monotherapy relative to other EZH2, dual EZH1/2, or EED inhibitors. In long-term cell viability assays, tulmimetostat had a comparable GI50 to PF-06821497 (∼5 nmol/L), whereas valemetostat and the EED inhibitor MAK683 were 3 to 10-fold less potent (GI50: 18 and 73 nmol/L, respectively). First generation EZH2 inhibitors such as tazemetostat (GI50 = 385 nmol/L) and CPI-1205 (GI50 = 574 nmol/L) were less potent in viability assays (Fig. 6A), which is consistent with their lower target affinities and shorter residence times. When administered at the same dose tulmimetostat, valemetostat, and MAK683 showed >50% TGI compared with vehicle in the HT1376 xenograft mouse model (Fig. 6B). At this dose, tulmimetostat exhibited significantly more robust and durable anti-tumor activity (P = 0.0001) than valemetostat, with tumor growth in the valemetostat arm rebounding after 35 days on-treatment, whereas tulmimetostat-treated tumors continued to regress. Tazemetostat, CPI-1205 and PF-06821497 resulted in <50% TGI (32%, 40%, and 41% TGI at day 27, respectively). All compounds were well tolerated at 75 mg/kg QD (Supplementary Fig. S6A), and plasma exposure of tulmimetostat was not greater than any of the other compounds (Supplementary Table S4). Compounds that resulted in >50% TGI have strong (>90%) reduction in H3K27me3 in tumors collected at Day 15, whereas those that had weaker TGI retained higher levels of H3K27me3 (Fig. 6C) demonstrating a link between tumor response and degree of H3K27me3 reduction (Fig. 6D). However, the magnitude of reduction in global H3K27me3 level changes >90% could not distinguish differences in TGI of the most efficacious compounds (tulmimetostat, valemetostat and MAK683). Gene expression profiling from tumors at Day 15 revealed that tulmimetostat induces significantly more EZH2/PRC2 target genes than any other compound and that overall changes in gene expression level are consistent with reduced H3K27me3 and tumor response (Fig. 6E; Supplementary Fig. S6B and S6C). Contrary to global changes in H3K27me3 levels, greater numbers of altered genes correlated with greater magnitude in TGI, suggesting that gene expression changes may be a preferred biomarker to relate target inhibition and efficacy.
Figure 6.
Tulmimetostat demonstrates superior level of tumor PRC2 target gene induction correlating with efficacy in the HT1376 bladder cancer CDX model. A, Cell viability dose response curve for various EZH2 and EED inhibitors in HT1376 bladder cancer cells on day 18. GI50s noted. Data are represented as mean ± SD. B, HT1376 xenograft efficacy study of various EZH2 and EED inhibitors. Data represented as mean ± SEM, with n = 6 mice per group for all arms except tulmimetostat, which had n = 12. TGI values noted were calculated for all arms using day 27 tumor volumes, relative to vehicle. P values calculated using two-way ANOVA, *, P < 0.05; **, P < 0.01; ***, P < 0.0001 using two-way ANOVA up to day 27 (for TGIs) or through day 55 (for P values on the graph). C, H3K27me3 levels from tumor samples collected at day 15 from study depicted in B. Data represented as mean ± SD, n = 3 tumors per group. P values calculated using unpaired Student t test, **, P < 0.01; ***, P < 0.0001. D, Scatter plot showing relationship between relative H3K27me3 levels and tumor size. The relative H3K27me3/H3 ratio from tumor samples collected at day 15 of each group was normalized to that of vehicle group. The tumor volume of each group was measured at day 27. P value was calculated by Pearson correlation coefficient. E, Bar plot of expression changes of EZH2 target genes (defined in Supplementary Fig. S3G) at day 15 relative to vehicle for the tumors from B. Data represented as mean log2-fold change ± 95% confidence interval.
Tulmimetostat demonstrates comprehensive target engagement in patients
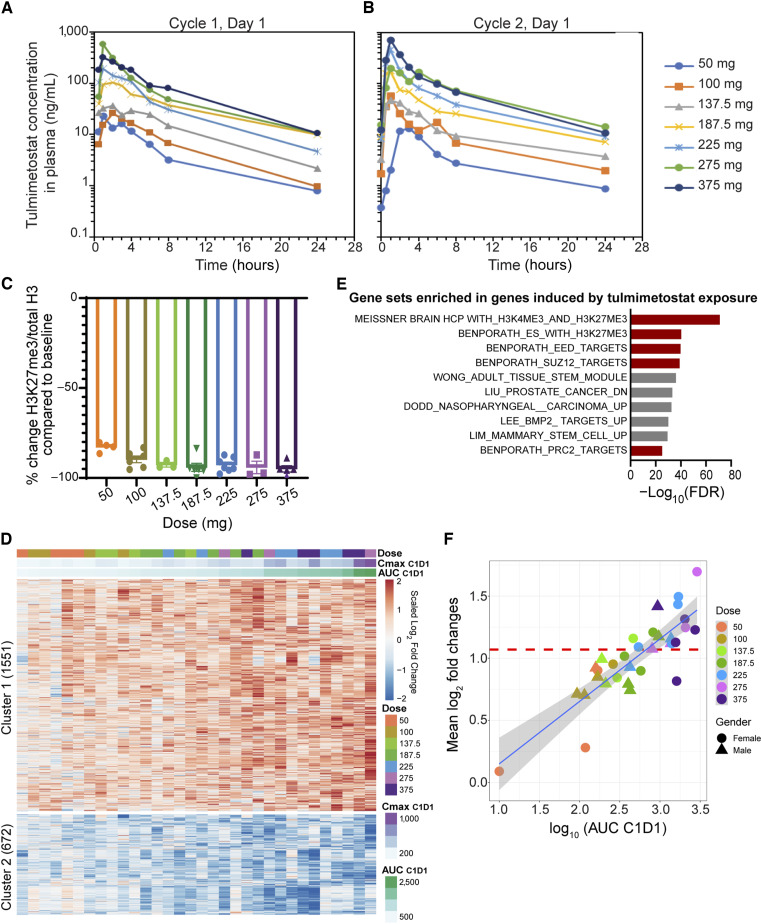
In the Phase I portion of the Phase I/II study of tulmimetostat in patients with advanced tumors (clinicaltrials.gov identifier: NCT04104776) tulmimetostat PK, PD, as well as the relationship between tulmimetostat plasma exposure and changes in blood cell pharmacodynamic markers in patients, were assessed. Tulmimetostat AUC and Cmax showed dose-dependent increases across the investigated dose range and plateaued at the highest dose levels (Fig. 7A and B). Administration of tulmimetostat resulted in slight accumulation at steady-state, consistent with the estimated half-life of approximately 8 hours. Importantly, as indicated by the similarity in exposure between Cycle 1, Day 1 (C1D1; Fig. 7A) and C2D1 (Fig. 7B), tulmimetostat PK showed no evidence of metabolic autoinduction after 28 days of administration, suggesting that exposure is durable and sustained following repeated dosing. Tulmimetostat treatment resulted in remarkable reduction of H3K27me3 levels in patient monocytes on Day 8 of the first treatment cycle in all dose cohorts (Fig. 7C). At dose levels >100 mg, a >90% mean reduction in H3K27me3 levels was observed, approaching the assay limit of detection for maximal target engagement. Given our preclinical data suggesting that removal of H3K27me3 from enhancers and promoters will increase PRC2 target gene expression, we conducted genome-wide gene expression profiling from patient whole blood, comparing pretreatment samples with samples from C1D22. We identified 1551 induced and 672 repressed genes following tulmimetostat treatment (Clusters 1 and 2, Fig. 7D). As expected, H3K27me3- or PRC2-related gene sets were over-represented in tulmimetostat-induced genes (Fig. 7E). Importantly, the magnitude of gene induction and repression showed exquisite correlation with tulmimetostat plasma exposure (r = 0.89; Supplementary Fig. S7A).
Figure 7.
In patients with cancer, tulmimetostat plasma exposure levels correlate with magnitude of PRC2 target gene expression changes in peripheral blood. A, Plasma concentration profiles of tulmimetostat in patients in CPI-0209-01 clinical trial at C1D1. Plasma concentration shown as the mean after oral administration of indicated dose of tulmimetostat. B, Plasma concentration profiles of tulmimetostat in patients in CPI-0209-01 clinical trial at C2D1. Plasma concentration shown as the mean after oral administration of indicated dose of tulmimetostat. C, Change of H3K27me3/total H3 ratio in monocytes at C1D8 compared with baseline (C1D1). Points whose values were <−100% were plotted as “−100%” to indicate maximal signal reduction. D, Heatmap of log2-fold changes of the gene expression associated with tulmimetostat exposure in whole blood from patients treated with tulmimetostat. Each column represents an individual patient ordered by increasing tulmimetostat plasma exposures at C1D1 (from left to right). E, GSEA of tulmimetostat-induced genes. Top gene sets significantly enriched from C2 curated gene set list (FDR ≤ 0.01) from MSigDB in the 1,551 genes induced by tulmimetostat treatment in patient whole blood samples. Red bars, PRC2-related gene sets. F, Scatter plot showing relationship between tulmimetostat-induced gene expression changes in the whole blood transcriptome of preclinical mouse models and patients enrolled in the CPI-0209-01 clinical trial. The average expression changes of 379 genes induced in clinical samples within each patient’s whole blood between C1D1 and C1D22 are plotted against the tulmimetostat exposure of that patient at C1D1. Dose levels of tulmimetostat are indicated in mg. The horizontal red dotted line indicates the average expression changes of these genes in whole blood of mice treated with 35 mg/kg tulmimetostat compared with vehicle.
Because we successfully accessed target engagement in patient whole blood by gene expression profiling, next, we compared the magnitude of whole blood gene expression changes in patients with those in mice with HT1376 bladder cancer xenografts treated with tulmimetostat at an efficacious dose of 35 mg/kg QD (Supplementary Fig. S2F). We identified 365 genes induced by tulmimetostat treatment in patients who had a similar average fold change in mice (Supplementary Table S7). We found that patients treated with 225 mg or greater dose levels showed a magnitude of expression increase in these genes similar to or greater than that seen in mice treated with 35 mg/kg tulmimetostat (Fig. 7F). Our current clinical data suggest that tulmimetostat can induce PRC2 target gene expression in patient whole blood in a dose-dependent manner, which to our knowledge has not been demonstrated clinically for any other EZH2 inhibitor. Importantly, our data also suggest that we achieve PD levels in the clinic that exceed those resulting in single agent efficacy in preclinical models of solid tumors.
Discussion
EZH2 inhibition in advanced solid tumors potentially represents a new therapeutic opportunity to exploit a specific, recurrent genomic aberration—loss of function mutations in ARID1A. Here, we describe tulmimetostat, a second generation EZH2 inhibitor with a best-in-class potential and dual selectivity for EZH2 and EZH1. Improved potency, long residence time, no metabolic liability, and QD dosing schedule differentiate it from other inhibitors targeting the PRC2 complex. These properties lead to a robust and durable reduction of global H3K27me3 levels followed by gene expression changes in PRC2 target genes that are associated with significant and durable TGI in urothelial carcinoma, OCCC, endometrial cancer, and lymphoma tumor models that can persist extended periods of time after cessation of tulmimetostat treatment (Supplementary Fig. S1H).
Consistent with data previously reported for OCCC (19), gastric cancer (20), and bladder cancer (21), we demonstrated that EZH2 inhibition with tulmimetostat preferentially impacts viability of ARID1A LOF mutant bladder cancer cell lines, despite significant reduction in H3K27me3 levels in both ARID1A wildtype and mutant cell lines. Interestingly, our data are in contrast with a prior study that suggested ARID1A-deficient bladder cancer cells show no enhanced sensitivity toward EZH2 inhibition (22). In this previous study, cell viability was only assessed in short-term growth assays with a first generation EZH2 inhibitor, which would limit responses in these cell lines. We clearly demonstrate here, and consistent with the collective knowledge on EZH2 inhibitor mechanism of action that phenotypes with tulmimetostat treatment are time-dependent and require longer-term assays. Moreover, tulmimetostat not only potently inhibits EZH2 but also EZH1, which may provide an advantage over EZH2-selective inhibitors, consistent with reports where dual EZH1 and EZH2 inhibitors demonstrated superior performance (38, 44, 45). Although in other cancer contexts EZH2 inhibitor activity was associated with SMARCB1 (46) and SMARCA4 (18) mutations, in bladder cancer, functional dependency on the PRC2 complex in the context of BAF complex mutations appears to be unique to ARID1A (Fig. 2A).
Importantly, we observe significant TGI in vivo in both ARID1A mutant CDX and PDX models, with treatment arms showing tumor stasis, and in certain instances regression, in response to tulmimetostat treatment. These data support the concept that ARID1A LOF mutations have the propensity to increase sensitivity to treatment with tulmimetostat. Profound tulmimetostat single agent efficacy was also achieved in CDX and PDX models of OCCC and endometrial cancer, in keeping with the idea that ARID1A LOF mutations may predict EZH2 inhibitor sensitivity irrespective of cancer type. Future preclinical and clinical studies with tulmimetostat may shed additional light on the predictive potential of ARID1A LOF mutations for EZH2 inhibitor cancer therapy.
Combining tulmimetostat with cisplatin in bladder cancer models increases tumor cell death. Although tulmimetostat as a single agent can outperform cisplatin in an ARID1A mutant CDX model, it also combines with cisplatin to deepen the impact on tumor growth. Importantly, we find that both ARID1A wildtype and mutant bladder cancer cell models show enhanced cell viability defects when tulmimetostat and cisplatin treatments are combined. These combinatorial effects were preferentially observed in bladder cancer lines with limited baseline sensitivity to cisplatin alone. This may be due in part to the reported effect of EZH2 inhibitor treatment to sensitize cells to genotoxic stress (47). Interestingly, four of six cisplatin-insensitive cell lines tested are ARID1A wildtype, and three of those four showed synergistic responses to treatment with tulmimetostat and cisplatin. Although the ARID1A wildtype bladder cancer cell models are usually insensitive to tulmimetostat alone, our data suggest combining tulmimetostat with chemotherapeutic agents such as cisplatin may provide new avenues for the treatment of advanced, chemotherapy-resistant urothelial carcinoma.
Tulmimetostat is currently in clinical development investigating the safety, tolerability, and clinical activity in patients with multiple types of advanced solid tumors or hematologic malignancies. A recommended Phase II dose (RP2D) of tulmimetostat as monotherapy was chosen as 350 mg QD, as part of the primary objective of the Phase I dose escalation part of the study (34). Tulmimetostat was well tolerated by patients on this QD dosing schedule, exhibited dose-dependent increases in exposure until the highest doses, showed no evidence of induction of metabolism, and demonstrated substantial levels of target engagement as assessed with multiple pharmacodynamic markers in patient samples. The Phase II expansion portion of the study is evaluating tulmimetostat’s RP2D in a continuous, dose once a day in six tumor-based cohorts including three cohorts evaluated for various ARID1A-mutated solid tumors (48). We also report a novel, tulmimetostat-responsive gene signature from patient blood that can be used as a sensitive, quantitative pharmacodynamic biomarker for target engagement. With the enhanced target coverage and residence time afforded by tulmimetostat, to the best of our knowledge, we show for the first time a correlation between an EZH2 inhibitor’s plasma exposure with gene expression changes in patient blood. Our substantial preclinical and preliminary clinical data suggest that tulmimetostat has the potential to achieve clinical benefit in solid tumors as monotherapy but also in combination with chemotherapeutic agents and may be beneficial in various indications with recurrent ARID1A mutations. The Phase II expansion cohorts will help clarify whether magnitude and specificity of the gene expression change in peripheral blood as a novel pharmacodynamic biomarker also has predictive potential to guide further development of tulmimetostat.
Supplementary Material
Additional method information to describe technical aspects of the experiments described in this manuscript
Table with cell line information including source and RRID
Table with antibody source information including RRID if available
Table listing experimentally determined compound binding affinities for EZH2 and EZH1 containing PRC2 protein complexes
Summary of PK data for EZH2 inhibitors used in KARPAS-422 and HT1376 xeograft mmouse models
Summary of cell viability data for bladder cancer cells treated with tulmimetostat
List of gene names corresponding to 132 tulmimetostat upregulated genes in responder versus non responder cell lines
List of 365 genes (used in main Fig. 7F) that are induced in patient’s whole blood upon treatment with tulmimetostat
Synthetic pathway and characterization of tulmimetostat, a long residence time EZH2 inhibitor.
Tulmimetostat mediated cancer cell phenotypic responses are increased in ARID1A LOF contexts.
Tulmimetostat increases PRC2-repressed gene expression in ARID1A mutant bladder cancer cells. (
Tulmimetostat treatment results in greater PRC2 target gene expression induction in phenotypically sensitive ARID1A LOF cell lines
Tulmimetostat improves cisplatin responsiveness in chemotherapy-resistant bladder cancer cells.
Tulmimetostat demonstrates superior level of tumor PRC2 target gene inducon and correlation with efficacy in a bladder cancer xenograft mouse model.
In cancer patients tulmimetostat plasma exposure levels correlate with PRC2 target gene expression changes in peripheral blood.
Acknowledgments
We acknowledge Florence Poy and Christine Petersen for technical and administrative assistance. We would like to thank the patients and caregivers for their invaluable participation in the CPI-0209-01 study. We acknowledge the Broad Institute Cancer Cell Line Encyclopedia for making RNA and DNA sequencing data publicly available and declare that those who carried out the original analysis and collection of the data bear no responsibility for the analysis or interpretation of the data presented in this manuscript.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Disclosures
P.J. Keller reports a patent for 63/445,024 pending to Constellation Pharmaceuticals and was an employee of Constellation Pharmaceuticals, Inc., during time of work on publication. E.J. Adams reports a patent for WO2024015566(A1) pending; and E.J. Adams was an employee of Constellation Pharmaceuticals and MorphoSys US during time of work on publication. Third party funding of Constellation Pharmacueticals and MorphoSys AG can be found in their respective SEC filings. R. Meyer reports other support from Constellation Pharmaceuticals and MorphoSys during the conduct of the study. J.A. Mertz reports other support from Flare Therapeutics outside the submitted work. V. Gehling reports personal fees and other support from Odyssey Therapeutics and personal fees from Kura Oncology outside the submitted work; in addition, V. Gehling has a patent for WO/2019/204490 issued. J. Cui reports other support from Constellation Pharmaceuticals outside the submitted work and is an employee of Constellation Pharmaceuticals. J.I. Stuckey reports a patent for WO2019204490—Modulators of Methyl Modifying Enzymes, Compositions and Uses Thereof issued. A. Khanna reports a patent 20240116905 issued. Z. Yu reports a patent for EZH2 inhibition therapies for the treatment of AT-rich interactive domain-containing protein 1A (ARID1A) mutated cancers pending. N.J. Lakhani reports nonfinancial support and other support from MorphoSys/Constellation Pharma during the conduct of the study; other support from Arcus Biosciences, Artios, GSK, Celgene/BMS, Gilead, Ikena, InhibRx, Janssen, Jounce, KSQ/Roche, LOXO/Lilly, Macrogenics, Merck, Alkermes/Mural Oncology, Regeneron, Revolution Medicines, Repare Theraepeutics, Symphogen/Servier, Seagen/Pfizer, Shattuck Labs, Sapience, Tizona, SK Lifesciences, and Volastra; nonfinancial support and other support from Incyte. D. Rasco reports other support from MorphoSys during the conduct of the study and other support from 23 & Me, Arcus Biosciences, Adcentrx, Aulos, Bolt, BMS, Cullinan, Compugen USA, Inc., Flare, GSK, Eikon, JNJ, Kronos, AMAL, PureTech Health, Molecular Templates, and Takeda outside the submitted work. M. Gutierrez reports personal fees from Guardant Health, COTA Healthcare, Cellularity, Merck, BMS, Sanofi, and Incyte outside the submitted work; and research funding/institutional relationship with no financial interest: Acerta Pharma; Adlai Nortye; Arcus Biosciences; Array BioPharma; Bayer; Bellicum Pharmaceuticals; BMS; Boehringer Ingelheim; Celgene; Checkpoint Therapeutics; Compass Therapeutics; Constellation Pharmaceuticals; Cullinan Oncology; Cyteir; Daiichi Sankyo Company; Eisai; EMD Serono; EMD Serono; Erasca, Inc.; Fate Therapeutics; Georgetown University; GSK; GSB Pharma; Hackensack Meridian Health; Imugene; Incyte; Infinity Pharmaceuticals; ITeos Therapeutics; Janssen; Johnson & Johnson; KSQ Therapeutics; MedImmune; Memorial Sloan-Kettering Cancer Center; Merck; Millennium; Mirati Therapeutics; Moderna Therapeutics; NextCure; NextCure; Nimbus Therapeutics; Pfizer; Pharmacyclics; Rapa Therapeutics; Regeneron; Roche/Genentech; Sanofi; Seagen; Silenseed; Synlogic; Tesaro; Turning Point Therapeutics; Vedanta Biosciences; VelosBio; Verastem; and Vincerx Pharma. L. Duska reports grants from MorphoSys during the conduct of the study and personal fees from Aadi Bioscience and Regeneron outside the submitted work. J. Wang reports other support from Constellation Pharmaceutical outside the submitted work. K. Sun reports a patent for 63/455,024 pending. P. Trojer reports personal fees from Constellation Pharmaceuticals, Inc., and MorphoSys during the conduct of the study and personal fees from MorphoSys, Inc. and Fulcrum Therapeutics outside the submitted work; in addition, P. Trojer has a patent for WO2024015566(A1) pending. No disclosures were reported by the other authors.
Authors’ Contributions
P.J. Keller: Conceptualization, formal analysis, supervision, validation, investigation, visualization, writing–original draft. E.J. Adams: Formal analysis, validation, investigation, visualization, writing–original draft, writing–review and editing. R. Wu: Formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Côté: Conceptualization, supervision, investigation, project administration. S. Arora: Supervision, investigation, project administration. N. Cantone: Resources, data curation, investigation, visualization. R. Meyer: Resources, data curation, validation, investigation, visualization, methodology. J.A. Mertz: Conceptualization, supervision, project administration. V. Gehling: Conceptualization, investigation, project administration. J. Cui: Resources, data curation, formal analysis, investigation, methodology. J.I. Stuckey: Conceptualization, supervision, investigation, project administration. A. Khanna: Conceptualization, investigation, visualization, project administration. F. Zhao: Data curation, supervision, validation, investigation, visualization, writing–original draft, project administration. Z. Chen: Formal analysis, visualization. Z. Yu: Data curation, investigation. R.T. Cummings: Conceptualization, supervision, visualization, writing–original draft, project administration. M. Taimi: Resources, data curation, investigation. N.J. Lakhani: Conceptualization, resources, data curation, investigation. D. Rasco: Conceptualization, resources, data curation, investigation, visualization. M. Gutierrez: Resources, data curation, investigation. L. Duska: Resources, data curation, investigation. M. Devitt: Resources, data curation, investigation. R. Rippley: Data curation, supervision, visualization, writing–original draft, project administration. J. Levell: Conceptualization, supervision, writing–original draft, project administration. J. Truong: Conceptualization, supervision, writing–original draft, project administration, writing–review and editing. J. Wang: Conceptualization, resources, data curation, supervision, investigation, project administration. K. Sun: Supervision, project administration. P. Trojer: conceptualization, resources, data curation, supervision, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Gonzalez-Perez A, Jene-Sanz A, Lopez-Bigas N. The mutational landscape of chromatin regulatory factors across 4,623 tumor samples. Genome Biol 2013;14:r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017;171:540–56.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conery AR, Rocnik JL, Trojer P. Small molecule targeting of chromatin writers in cancer. Nat Chem Biol 2022;18:124–33. [DOI] [PubMed] [Google Scholar]
- 5. Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med 2014;20:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones S, Wang T-L, Shih IM, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010;330:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010;363:1532–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takeda T, Banno K, Okawa R, Yanokura M, Iijima M, Irie-Kunitomi H, et al. ARID1A gene mutation in ovarian and endometrial cancers (Review). Oncol Rep 2016;35:607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wiegand KC, Lee AF, Al-Agha OM, Chow C, Kalloger SE, Scott DW, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol 2011;224:328–33. [DOI] [PubMed] [Google Scholar]
- 10. Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573–82. [DOI] [PubMed] [Google Scholar]
- 11. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goswami S, Chen Y, Anandhan S, Szabo PM, Basu S, Blando JM, et al. ARID1A mutation plus CXCL13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in mUCC. Sci Transl Med 2020;12:eabc4220. [DOI] [PubMed] [Google Scholar]
- 14. Laugesen A, Hojfeldt JW, Helin K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol Cell 2019;74:8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res 2018;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010;18:316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y, Chen SY, Karnezis AN, Colborne S, Santos ND, Lang JD, et al. The histone methyltransferase EZH2 is a therapeutic target in small cell carcinoma of the ovary, hypercalcaemic type. J Pathol 2017;242:371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Januario T, Ye X, Bainer R, Alicke B, Smith T, Haley B, et al. PRC2-mediated repression of SMARCA2 predicts EZH2 inhibitor activity in SWI/SNF mutant tumors. Proc Natl Acad Sci U S A 2017;114:12249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 2015;21:231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamada L, Saito M, Thar Min AK, Saito K, Ashizawa M, Kase K, et al. Selective sensitivity of EZH2 inhibitors based on synthetic lethality in ARID1A-deficient gastric cancer. Gastric Cancer 2021;24:60–71. [DOI] [PubMed] [Google Scholar]
- 21. Rehman H, Chandrashekar DS, Balabhadrapatruni C, Nepal S, Balasubramanya SAH, Shelton AK, et al. ARID1A-deficient bladder cancer is dependent on PI3K signaling and sensitive to EZH2 and PI3K inhibitors. JCI Insight 2022;7:e155899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garczyk S, Schneider U, Lurje I, Becker K, Vogeli TA, Gaisa NT, et al. ARID1A-deficiency in urothelial bladder cancer: No predictive biomarker for EZH2-inhibitor treatment response? PLoS One 2018;13:e0202965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levell JR. Recent progress in the discovery and clinical application of small-molecule inhibitors EZH2 and EED. Med Chem Rev 2020;55:319–43. [Google Scholar]
- 24. Martin MC, Zeng G, Yu J, Schiltz GE. Small molecule approaches for targeting the polycomb repressive complex 2 (PRC2) in cancer. J Med Chem 2020;63:15344–70. [DOI] [PubMed] [Google Scholar]
- 25. Hoy SM. Tazemetostat: first approval. Drugs 2020;80:513–21. [DOI] [PubMed] [Google Scholar]
- 26. Morschhauser F, Tilly H, Chaidos A, McKay P, Phillips T, Assouline S, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol 2020;21:1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Italiano A, Soria JC, Toulmonde M, Michot JM, Lucchesi C, Varga A, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol 2018;19:649–59. [DOI] [PubMed] [Google Scholar]
- 28. Harb W, Abramson J, Lunning M, Goy A, Maddocks K, Lebedinsky C, et al. A phase 1 study of CPI-1205, a small molecule inhibitor of EZH2, preliminary safety in patients with B-cell lymphomas. Ann Oncol 2018;29:37. [Google Scholar]
- 29. Yap TA, Winter JN, Giulino-Roth L, Longley J, Lopez J, Michot JM, et al. Phase I study of the novel enhancer of Zeste homolog 2 (EZH2) inhibitor GSK2816126 in patients with advanced hematologic and solid tumors. Clin Cancer Res 2019;25:7331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaswani RG, Gehling VS, Dakin LA, Cook AS, Nasveschuk CG, Duplessis M, et al. Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1 -(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J Med Chem 2016;59:9928–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garapaty-Rao S, Nasveschuk C, Gagnon A, Chan EY, Sandy P, Busby J, et al. Identification of EZH2 and EZH1 small molecule inhibitors with selective impact on diffuse large B cell lymphoma cell growth. Chem Biol 2013;20:1329–39. [DOI] [PubMed] [Google Scholar]
- 32. Stuckey JI, Cantone NR, Cote A, Arora S, Vivat V, Ramakrishnan A, et al. Identification and characterization of second-generation EZH2 inhibitors with extended residence times and improved biological activity. J Biol Chem 2021;296:100349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lakhani NJ, Gutierrez M, Duska LR, Do KT, Sharma M, Gandhi L, et al. Phase 1/2 first-in-human (FIH) study of CPI-0209, a novel small molecule inhibitor of enhancer of zeste homolog 2 (EZH2) in patients with advanced tumors. Chicago (IL): American Society of Clinical Oncology; 2021. [Google Scholar]
- 35. Khanna A, Cote A, Arora S, Moine L, Gehling VS, Brenneman J, et al. Design, synthesis, and pharmacological evaluation of second generation EZH2 inhibitors with long residence time. ACS Med Chem Lett 2020;11:1205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bradley WD, Arora S, Busby J, Balasubramanian S, Gehling VS, Nasveschuk CG, et al. EZH2 inhibitor efficacy in non-Hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem Biol 2014;21:1463–75. [DOI] [PubMed] [Google Scholar]
- 37. McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012;492:108–12. [DOI] [PubMed] [Google Scholar]
- 38. Yamagishi M, Hori M, Fujikawa D, Ohsugi T, Honma D, Adachi N, et al. Targeting excessive EZH1 and EZH2 activities for abnormal histone methylation and transcription network in malignant lymphomas. Cell Rep 2019;29:2321–37.e7. [DOI] [PubMed] [Google Scholar]
- 39. Mittal P, Roberts CWM. The SWI/SNF complex in cancer—biology, biomarkers and therapy. Nat Rev Clin Oncol 2020;17:435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kadoch C, Copeland RA, Keilhack H. PRC2 and SWI/SNF chromatin remodeling complexes in Health and disease. Biochemistry 2016;55:1600–14. [DOI] [PubMed] [Google Scholar]
- 41. Kelso TWR, Porter DK, Amaral ML, Shokhirev MN, Benner C, Hargreaves DC. Chromatin accessibility underlies synthetic lethality of SWI/SNF subunits in ARID1A-mutant cancers. Elife 2017;6:e30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the cancer cell line Encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Honma D, Kanno O, Watanabe J, Kinoshita J, Hirasawa M, Nosaka E, et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci 2017;108:2069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujita S, Honma D, Adachi N, Araki K, Takamatsu E, Katsumoto T, et al. Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia 2018;32:855–64. [DOI] [PubMed] [Google Scholar]
- 46. Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A 2013;110:7922–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liao Y, Chen CH, Xiao T, de la Pena Avalos B, Dray EV, Cai C, et al. Inhibition of EZH2 transactivation function sensitizes solid tumors to genotoxic stress. Proc Natl Acad Sci U S A 2022;119:e2105898119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Drescher C, Walter HS, Gastinne T, Lakhani NJ, Ribrag V, Rasco DW, et al. EZH2/EZH1 inhibitor tulmimetostat (CPI-0209) in patients with advanced solid tumors or hematologic malignancies: preliminary phase II results. J Clin Oncol 2023;41:3094. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional method information to describe technical aspects of the experiments described in this manuscript
Table with cell line information including source and RRID
Table with antibody source information including RRID if available
Table listing experimentally determined compound binding affinities for EZH2 and EZH1 containing PRC2 protein complexes
Summary of PK data for EZH2 inhibitors used in KARPAS-422 and HT1376 xeograft mmouse models
Summary of cell viability data for bladder cancer cells treated with tulmimetostat
List of gene names corresponding to 132 tulmimetostat upregulated genes in responder versus non responder cell lines
List of 365 genes (used in main Fig. 7F) that are induced in patient’s whole blood upon treatment with tulmimetostat
Synthetic pathway and characterization of tulmimetostat, a long residence time EZH2 inhibitor.
Tulmimetostat mediated cancer cell phenotypic responses are increased in ARID1A LOF contexts.
Tulmimetostat increases PRC2-repressed gene expression in ARID1A mutant bladder cancer cells. (
Tulmimetostat treatment results in greater PRC2 target gene expression induction in phenotypically sensitive ARID1A LOF cell lines
Tulmimetostat improves cisplatin responsiveness in chemotherapy-resistant bladder cancer cells.
Tulmimetostat demonstrates superior level of tumor PRC2 target gene inducon and correlation with efficacy in a bladder cancer xenograft mouse model.
In cancer patients tulmimetostat plasma exposure levels correlate with PRC2 target gene expression changes in peripheral blood.
Data Availability Statement
Transcriptomic and epigenomic data including patient RNA-seq data reported in this study are publicly available in Gene Expression Omnibus at GSE264383 and GSE176493. Mutations noted for ARID1A, KDM6A, and BAF complex members in cell lines were obtained from the Broad Institute’s Cancer Cell Line Encyclopedia (CCLE) portal at https://portals.broadinstitute.org/. Biochemical and in vitro cell data generated in this study are available upon request from the corresponding author. Raw data pertaining to animal studies were generated at WuxiApptec and Crown Biosciences. Derived data supporting the findings are available from the corresponding author upon request. Clinical trial data reported in this study will be made available by MorphoSys Inc. upon request.