Abstract
To examine the early events in the life cycle of Moloney murine leukemia virus (MoMLV), we analyzed the intracellular complexes mediating reverse transcription. Partial purification of the reverse transcription complexes (RTCs) by equilibrium density fractionation and velocity sedimentation indicated that three distinct species of intracellular complexes are formed shortly after cell infection. Only one of these species is able to start and complete reverse transcription in the cell cytoplasm. This RTC is composed of at least the viral genome, capsid, integrase, and reverse transcriptase proteins. The RTC becomes permeable to micrococcal nuclease but not to antibodies. Shortly after initiation of reverse transcription, the viral strong stop DNA within the RTC is protected from nuclease digestion. The sedimentation velocity of the RTC decreases during reverse transcription. After entry into the nucleus, most capsid proteins are lost from the RTC and its sedimentation velocity decreases further.
All retroviruses synthesize a double-stranded DNA copy of their RNA genome that is subsequently integrated into the host cell chromosomal DNA. The process of reverse transcription of the RNA genome into DNA is carried out in the cytoplasm soon after viral penetration into the cell and is generally completed within 8 to 12 h (30). Little is known about the structure and the protein composition of the intracellular complex in which reverse transcription occurs, particularly during the early steps after virus internalization. In the murine leukemia virus (MLV), the intracellular viral structure that contains the fully reverse transcribed viral DNA (also called the preintegration complex [PIC]) retains components of the virion core (including CA protein), has a relatively large size, sedimenting at 160S, and is competent to integrate the DNA in vitro (5). The PIC of human immunodeficiency virus type 1 (HIV-1) appears to have a different organization since it contains no capsid proteins, but it contains at least reverse transcriptase (RT), integrase (IN), and a portion of matrix (MA) proteins (6, 9, 16, 23). In addition, two cellular proteins have been found to associate with the PIC. They increase the efficiency of integration of the viral genome in vitro (10) and/or prevent self-integration of the viral DNA, which would result in an nonproductive infection (17).
MLVs are widely used as vectors for gene therapy because of their relatively simple genome organization and their ability to infect a wide variety of cell types and integrate DNAs into the host cell genome (21). However, a major limitation of MLV-based vectors is their inability to infect nondividing cells (18, 22, 28), as opposed to vectors based on lentiviruses (24). The reasons for the inability of MLV-based vectors to infect nondividing cells are uncertain; the large size of their PIC or the lack of appropriate nuclear targeting signals may be responsible. A more detailed knowledge of the organization of the intracellular viral complex in which reverse transcription occurs would improve our understanding of the interactions between the incoming virus and the infected cells. It may also allow the design of new MLV-based vectors in which the reverse transcription complex (RTC) is targeted to the nucleus of nondividing cells. To characterize the dynamics of the early steps of virus life cycle, we have analyzed detergent-free cytoplasmic and nuclear extracts at various time points after acute infection. These studies revealed the existence of three distinct species of intracellular complexes. Two species are incompetent for reverse transcription in vivo, while a third species starts and completes reverse transcription in the cell cytoplasm.
MATERIALS AND METHODS
Cells and viruses.
The retroviral vector LNPOZ (kind gift of A. D. Miller, Fred Hutchinson Cancer Research Center, Seattle, Wash.) contains the neomycin phosphotransferase and lacZ genes separated by the poliovirus ribosome entry site (1). NIH 3T3 mouse fibroblasts and the producer cell clone AmpliGPE LNPOZ (11, 12) were grown in 175-cm2 flasks in Dulbecco’s modified Eagle’s medium supplemented with 2 mM glutamine and 10% fetal calf serum at 37°C in an atmosphere containing 5% CO2. Culture medium was replaced when cells reached confluence; the virus-containing supernatant was collected 16 to 18 h later and filtered through a 0.45-μm-pore-size filter. Viral titers were determined by infecting NIH 3T3 cells with serial dilutions of virus containing supernatant in the presence of 8 μg of polybrene per ml. Approximately 36 h after infection, cells were fixed in 0.5% glutaraldehyde in phosphate-buffered saline (PBS) containing 1 mM MgCl2 for 10 min at room temperature and stained 8 to 10 h with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 37°C in an humidified atmosphere (19).
Cell extracts.
Approximately 107 NIH 3T3 fibroblasts were infected in 175-cm2 flasks with 30 ml of freshly collected virus-containing supernatant (3 × 106 CFU/ml) in the presence of 8 μg of polybrene per ml. Infected cells were rapidly cooled to 4°C and incubated for 2 h to allow viral adhesion to the cell receptor but not viral internalization. Cells were then incubated at 37°C for 1, 2, 4, 7, and 16 h, washed once in PBS–0.5 mM EDTA, trypsinized, and washed once again with PBS. All subsequent manipulations were carried out at 4°C. The pellet containing the infected cells was resuspended in 5 volumes of hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 5 mM, dithiothreitol [DTT], 20 μg of aprotinin per ml, 2 μg of leupeptin per ml) and centrifuged for 5 min at 5,000 rpm in an Eppendorf model 5415C centrifuge. The pellet was resuspended in 3 volumes hypotonic buffer and incubated for 10 min. Cells were homogenized with 10 to 15 strokes in a Dounce homogenizer, and nuclei and unbroken cells were pelleted by centrifugation at 3,300 × g for 15 min (corresponding to 6,000 rpm in an Eppendorf Microfuge). The supernatant (called the cytoplasmic extract) was clarified by centrifugation at 7,500 × g for 20 min, and the pellet was discarded. The nuclear pellet from the previous centrifugation was resuspended in approximately 600 μl of isotonic buffer (10 mM Tris HCl [pH 7.4], 160 mM KCl, 5 mM MgCl2, 1 mM DTT, 20 μg of aprotinin per ml, 2 μg of leupeptin per ml) and homogenized in a ball-bearing homogenizer. The homogenate was then centrifuged at 7,500 × g for 20 min, and the supernatant (called the nuclear extract) was collected. Nuclear and cytoplasmic extracts were adjusted to 8% sucrose, snap frozen in liquid N2, and stored at −70°C.
Equilibrium density gradients.
Continuous linear sucrose gradients (5 ml) were poured with a two-chamber Hoefer SG gradient maker, using 20% sucrose solution in hypotonic buffer and 70% sucrose solution in D2O, and kept on ice. The pH of D2O was adjusted to 7.4 by dropwise addition of 10 mM NaOH. Gradients were overlaid with 0.5 ml of cytoplasmic or nuclear extracts and centrifuged at 35,000 rpm at 4°C for 20 h in a Beckman SW55 rotor. For analysis of the viral core, 1 ml of virus-containing supernatant was treated with the indicated detergent for 30 min at room temperature with gentle shaking. Treatment with ethyl ether (Baxter) was performed for 20 min at 4°C. Samples were loaded onto a 20 to 60% continuous sucrose gradients in 50 mM sodium phosphate buffer (pH 7.4) containing 2 mM DTT and centrifuged to equilibrium at 25,000 rpm for 20 h at 4°C in a Beckman SW41 rotor. Gradients were fractionated by puncturing the bottom of the tube and collecting 12 fractions. The density was calculated by weighing 100 μl of each fraction.
Sedimentation velocity gradients.
Continuous gradients were poured as described above, using 5 and 20% sucrose solutions in 50 mM sodium phosphate buffer (pH 7.4) containing 2 mM DTT, 20 μg of aprotinin per ml, and 2 μg of leupeptin per ml. Approximately 150 μl of the equilibrium density fraction containing the peak of the viral DNA was diluted with 1.2 ml of hypotonic buffer, loaded onto a Centricon 50 concentrator (Amicon), and centrifuged at 4,000 × g for 30 min at 4°C in a Sorvall centrifuge. The concentrate (50 μl) was resuspended in 300 μl of 50 mM sodium phosphate buffer, loaded on a 5 to 20% continuous sucrose gradient, and centrifuged at 23,000 rpm for 1 h at 4°C in a Beckman SW55 rotor. Fractions (0.4 ml each) were collected by puncturing the bottom of the tube, and the density was measured by weighing 100 μl of each fraction. The S value was calculated by the method of McEwen (20). Calibration of the system was performed by independently running 32S-labeled poliovirus, intact MoMLV, and naked viral DNA through identical sucrose gradients.
PCR.
PCR was performed in a final volume of 50 μl containing 1× PCR buffer, 100 μM each deoxynucleoside triphosphate (dNTP), 2.5 mM MgCl2, 5 U of Taq polymerase (Perkin-Elmer), and 30 pmol of each primer. Primer sequences were as follows: strong-stop forward primer, 5′-GCGCCAGTCTTCCGATAGAC-3′; strong-stop reverse complementary primer, 5′-AATGAAAGACCCCCGTCGTGG-3′; extended minus-strand forward primer, 5′-CACGACGCGCTGTATCGCTGG-3′; extended minus-strand reverse complementary primer, 5′-CATACAGAAATGGCGATCGTTC-3′; plus-strand forward primer, 5′-GTGATTGACTACCCACGACG-3′; and plus-strand reverse complementary primer, 5′-GACCTTGATCTTAACCTGGG-3′. Five microliters of the equilibrium density fractions or 1.5 μl of the sedimentation velocity fractions was used as the template for the PCR. Cycle parameter were as follows: 94°C for 3 min the first cycle; 94°C for 1 min, 55°C for 30 s, and 68°C for 1 min for 35 to 45 cycles; and one final extension cycle at 68°C for 10 min. PCR products were resolved on a 1% agarose–2% NuSieve gel and visualized by ethidium bromide staining. Semiquantitative PCR was performed in duplicate in the same conditions as described above, using threefold serial dilutions of the DNA template. The number of cycles was adjusted in each individual experiment to ensure linearity of amplification. Following amplification, the bands were resolved on a 1% agarose–2% NuSieve gel, visualized by ethidium bromide staining, and quantified by a Molecular Dynamics 300A densitometer.
Antibodies and Western blotting.
Goat polyclonal antibodies against MLV whole virus (81S000044) and against MLV CA protein (79S–804) were from the national Cancer Institute (Frederick, Md.). Rabbit polyclonal antibodies against MLV RT were as described previously (3). Chicken polyclonal antibodies against Moloney MLV (MoMLV) IN were raised against the multiple antigenic peptide HO - Ala - Lys7 - N - (C - L - T - W - R - V - Q - R - S - Q - N - P - L - K - I - R - L - T - R - E - A - P)8 (26), which corresponds to the last 21 amino acids of the MoMLV IN (34). Chicken immunization was performed by Gallina Biotechnology Inc. (Edmonton, Alberta, Canada). Briefly, 300 μg of peptide diluted in 100 μl of PBS was injected into two chickens in the presence of Freund’s adjuvant. Chickens were injected three times at 4-week intervals. Eggs were collected 4 weeks after the last injection, and antibodies were purified from the yolks by precipitation in 12% polyethylene glycol 8000 according to the method of Polson et al. (25). Purified antibodies were stored at −70°C in the presence of 0.01% sodium azide. For Western blot analyses, 300 μl of each fraction from the density equilibrium sucrose gradients was diluted in 1.2 ml of ice-cold 50 mM sodium phosphate buffer (pH 7.4) in the presence of 2 μg of bovine serum albumin (Sigma) and 10% (vol/vol) trichloroacetic acid. Fractions were incubated at −20°C for 16 h and centrifuged for 30 min at 4°C at maximum speed in an Eppendorf Microfuge. The pellets were washed once in a solution of ice-cold 80% acetone in distilled H2O and resuspended in 20 μl of sodium dodecyl sulfate (SDS) loading buffer (0.5 M Tris HCl [pH 6.8], 1% SDS, 10% glycerol, 0.1% bromophenol blue, 1 mM EDTA, 10 mM DTT, 20 μg of aprotinin per ml, 2 μg of leupeptin hemisulfate per ml, 10 μg of phenylmethylsulfonyl fluoride). The pH was adjusted to ∼7.0 by addition of 1 μl of 1.5 M Tris HCl (pH 8.8). Samples were boiled for 5 min and loaded on an SDS–12.5% polyacrylamide gel. After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, Calif.) and probed with the goat antiserum diluted 1:5,000 or with the chicken polyclonal antibody against MLV IN diluted 1:1,000. Horseradish peroxidase-conjugated secondary antibodies were used diluted 1:10,000 (anti-goat; Boehringer Mannheim) or 1:6,000 (anti-chicken; Promega) in 10% nonfat milk (Nestlé, Glendale, Calif.). Enhanced chemiluminescence (ECL-Plus; Amersham) was used to develop the blots as described by the manufacturer. Autoradiography films were exposed for different periods of time to ensure linearity of the signal and then analyzed by a Molecular Dynamics 300A densitometer.
Assays for reverse transcription.
To test for the ability of the rabbit polyclonal antibodies against MoMLV RT to inhibit RT activity, twofold serial dilutions of purified protein (1 ng to 0.3 ng) were incubated in the presence of antiserum or preimmune serum diluted 1:100 and 1:200 in PBS for 30 min at room temperature. Reverse transcriptase activity was then tested with the oligo(dT)-poly(rA) assay as previously described (14, 31). Briefly, 10 μl of samples was added to 40 μl of RT cocktail {60 mM Tris HCl (pH 8.0), 180 mM KCl, 6 mM MnCl2, 6 mM DTT, 0.05% Nonidet P-40, 6 μg of oligo(dT) per ml, 12 μg of poly(rA) per ml, 0.05 mM [α32]dTTP (800 Ci/mmol} for 1 h at 37°C. Samples were spotted onto DE-81 paper and washed three times with 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate). A PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) was used to quantitate the radioactivity incorporated. Endogenous reverse transcription reactions were carried out in 60 μl of buffer (50 mM Tris-HCl [pH 8.3], 50 mM NaCl, 6 mM MgCl2, 0.01% Nonidet P-40, 1 mM DTT, 2 mM each dNTP). Fifteen-microliter aliquots from the density equilibrium fractions were added to the buffer and incubated for 4 to 6 h at 37°C. The products of reverse transcription were detected by PCR using 5 μl of the endogenous reaction as the template. To test for the ability of the rabbit antisera against MoMLV RT to inhibit RT activity in the endogenous assay, samples were incubated as described above in the presence of either antiserum or preimmune serum diluted 1:100 and 1:200. After 4 to 6 h incubation at 37°C, serial dilutions of the samples were subjected to semiquantitative PCR to detect the product of reverse transcription.
Nuclease treatments.
All nuclease treatments were performed in 25 μl of isotonic buffer (1 mM DTT, 20 μg of aprotinin per ml, 2 μg of leupeptin hemisulfate per ml, 7 U of S7 micrococcal nuclease [Boehringer Mannheim], 2 mM CaCl2). Five-microliter aliquots from those equilibrium density fractions containing the peak of the retroviral DNA were added to the nuclease mixture, and samples were incubated on ice. The reaction was stopped by addition of 4 mM EGTA and kept on ice. Two aliquots of 5 μl each from the same reaction were analyzed by semiquantitative PCR with strong-stop and extended minus-strand primers. The nuclease sensitivity of the intact RTCs was compared with that of naked viral DNA. Control digests of naked viral DNA were carried out in the presence of 5-μl aliquots from equilibrium density fractions containing cytoplasmic extracts from uninfected NIH 3T3 fibroblasts and having the same density as the fractions containing the RTCs.
RESULTS
Disruption of virion cores by detergents.
Low yields of MoMLV cores can be released from intact virions by treatment with detergents, and the lipid-stripped core can then be separated from the virions by equilibrium density centrifugation on the basis of its higher density (4, 8). To test whether such protocols could be modified to permit efficient recovery of virion cores, virions were treated with a variety of detergents and then subjected to equilibrium density centrifugation through continuous sucrose gradients. The gradients were fractionated, and individual fractions were analyzed by both PCR (to detect the viral genome) and Western blotting (to detect viral Gag proteins). Untreated virions sedimented at a density of 1.16 g/ml (Table 1). Most detergents disrupted the virions, as indicated by the presence of >90% of the p30gag CA in the top fractions of the gradient. Treatment with low doses of some detergents resulted in the recovery of the immature Pr65gag at a density of 1.25 ± 0.017 g/ml, consistent with the density of the MLV core (4, 8). However, cores containing mature CA could never be recovered at densities higher than 1.16 g/ml. Thus, treatment of MoMLV with detergents at concentrations sufficient to remove the lipids could release intact immature cores composed of uncleaved Pr65gag but disrupted the mature core composed of p30gag CA. These results suggested that intracellular complexes might also be disrupted by detergents and should instead be isolated by hypotonic breakage of the cell membrane and Dounce homogenization.
TABLE 1.
Distribution of Gag proteins after equilibrium density fractionation of virions treated with different detergentsa
| Treatment | % p30gag in virions (at 1.16 g/ml) | % Pr65gag in cores (at 1.25 ± 0.017 g/ml) | % p30gag in top fractions |
|---|---|---|---|
| None | 100 | <1 | <1 |
| Digitonin | |||
| 0.025% | <1 | 1 | 99 |
| 0.0025% | 50 | <1 | 50 |
| Ether (2:1) | <1 | 1 | 99 |
| Nonidet P-40 | |||
| 1% | <1 | 0.5 | 99.5 |
| 0.1% | 50 | <1 | 50 |
| Triton X-100 (1%) | 1 | 3 | 96 |
| SDS | |||
| 0.05% | <1 | <1 | 100 |
| 0.01% | <1 | 0.2 | 99.8 |
No p30gag was detected in cores with a density of 1.25 ± 0.017 g/ml.
Extraction of the MoMLV RTC in hypotonic buffer.
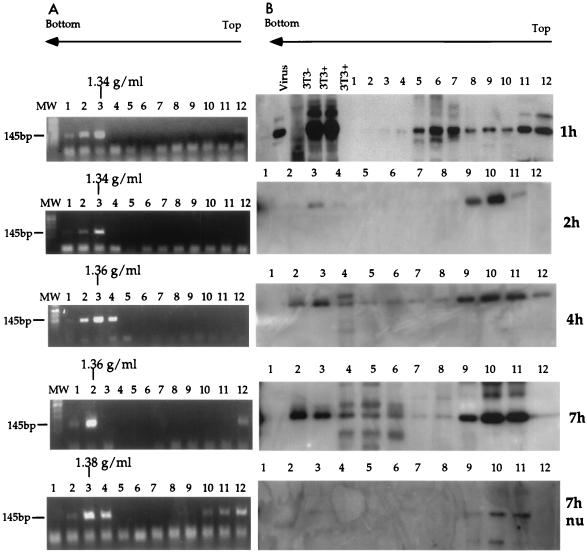
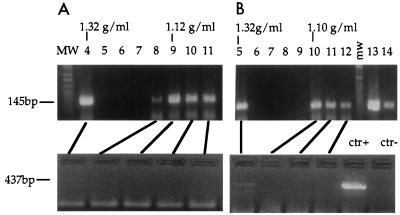
To prepare the RTC, NIH 3T3 fibroblasts were infected with an ecotropic MoMLV-based vector (11, 12) at a multiplicity of infection of 10. Infection was synchronized by preincubating the cells for 2 h at 4°C to allow for virus binding to the receptor. Cells were lysed by hypotonic breakage of the cell membrane and Dounce homogenization (see Materials and Methods). Control extractions performed in the presence of SDS indicated that approximately 60 to 70% of total viral DNA could be extracted by this method. Cytoplasmic extracts were prepared 1, 2, 4, 7, and 16 h postinfection, and nuclear extracts were prepared 7 and 16 h postinfection. The cell extracts were subjected to equilibrium density sedimentation, and the fractions were analyzed by PCR and Western blotting. The viral strong-stop DNA, which is the earliest product of reverse transcription, could be detected as a discrete peak at a density of 1.35 g/ml, consistent with the density of a nucleoprotein complex (Fig. 1). The strong-stop DNA was found at the same density in cytoplasmic and nuclear extracts, and this density did not change significantly with time (Table 2). Viral CA proteins were distributed over a wide density range in cytoplasmic extracts obtained 1 h after infection. At later time points, CA proteins were found distributed in two discrete peaks of 1.35 and 1.12 g/ml (Fig. 1). The 1.35-g/ml peak containing CA proteins consistently cofractionated with the strong-stop DNA, and the quantity of CA in this peak reached a maximum 7 h postinfection. Trace amounts of CA cofractionated with the strong-stop DNA in the nuclear extracts collected 7 h after infection (Fig. 1). Semiquantitative PCR analyses indicated that 7 h after infection, the quantity of strong-stop DNA in the peak fractions from the nuclear extracts was approximately 1/3 of that of cytoplasmic extracts, while the quantity of CA cofractionating with the strong-stop DNA in nuclear extracts was approximately 1/10 of that of cytoplasmic extracts (not shown). This finding suggested that most CA proteins were lost from the complex after entry into the cell nucleus. Neither the strong-stop DNA nor CA proteins were detected in equilibrium density gradients containing extracts from uninfected cells (not shown).
FIG. 1.
PCR and Western blot analyses of cytoplasmic and nuclear extracts after equilibrium density fractionation. NIH 3T3 fibroblasts were infected with a MoMLV-based vector; cell extracts were prepared 1, 2, 4, and 7 h postinfection, loaded on a 20 to 70% linear sucrose gradient, and centrifuged at 4°C for 20 h at 35,000 rpm in a Beckman SW55 rotor. After centrifugation, gradients were collected in 12 fractions and analyzed. Arrows indicate the direction of the gradient from the lowest (top) to the highest (bottom) density. (A) PCR analyses of the equilibrium density fractions using primers specific for the strong-stop DNA (expected band size is 145 bp). The density of the fraction containing the peak of the viral DNA is indicated for each time point. MW, DNA molecular weight standards. Lanes 1 to 12 correspond to fractions 1 to 12. The rapidly migrating bands are PCR artifacts. (B) The same fractions were precipitated in 10% trichloroacetic acid and analyzed by Western blotting using goat polyclonal antibodies against whole MLV. Purified virus (Virus) and infected NIH 3T3 cells (3T3+) were used as positive controls. Uninfected 3T3 cells (3T3−) were used as negative controls.
TABLE 2.
Densities of fractions in which the peak of viral DNA is found
| Fraction | Time (h) after infection | Density (g/ml; mean ± SD) | n |
|---|---|---|---|
| Cytoplasmic | 1 | 1.356 ± 0.01 | 6 |
| 2 | 1.348 ± 0.03 | 7 | |
| 4 | 1.364 ± 0.01 | 7 | |
| 7 | 1.353 ± 0.02 | 3 | |
| 16 | 1.361 ± 0.01 | 2 | |
| Nuclear | 7 | 1.373 ± 0.01 | 2 |
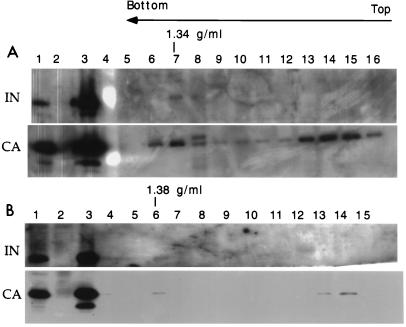
Equilibrium density fractions containing cytoplasmic and nuclear extracts were analyzed by Western blotting to detect the presence of IN. A peak of CA and IN was found to cofractionate at the same buoyant density of the strong-stop DNA (1.35 g/ml) (Fig. 2). The CA/IN ratio in the cytoplasmic extracts was higher than in the nuclear extracts, reinforcing the view that most CA proteins were lost from the RTC after entry into the cell nucleus (Fig. 2).
FIG. 2.
Western blot analyses of equilibrium density fractions containing cytoplasmic extracts collected 4 h postinfection (A) or nuclear extracts collected 7 h postinfection (B). Following SDS-polyacrylamide gel electrophoresis and protein transfer, the polyvinylidene difluoride membrane was probed with the goat polyclonal antibodies against MLV CA or the chicken polyclonal antibodies against MoMLV IN. Lane 1, purified virus; lane 2, uninfected NIH 3T3 cells; lane 3, infected NIH 3T3 cells before equilibrium density fractionation; lane 4, molecular weight standards; lanes 5 to 16, fractions 1 to 12 of the density gradient. The arrow indicates the direction of the gradient from the lowest (top) to the highest (bottom) density.
Ability of the RTCs to synthesize DNA.
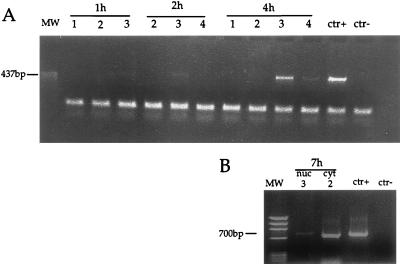
To evaluate whether the fractions containing the strong-stop DNA also contained elongated reverse transcription products, aliquots from the same fractions were subjected to PCR, using primers specific for the viral elongated minus-strand DNA and for the plus-strand DNA, which represent intermediate and late reverse transcription products, respectively. As shown in Fig. 3, the elongated minus strand could be detected 2 h after cell infection, and its quantity increased 4 h after infection. The plus strand could be detected only 7 h after infection, and it was 20-fold less abundant than the elongated minus strand, as determined by semiquantitative PCR. This finding suggested that reverse transcription was not completed in a large number of RTCs.
FIG. 3.
PCR analyses of equilibrium density fractions containing cell extracts collected 1, 2, 4, and 7 h postinfection. (A) Selected fractions from the same equilibrium density gradients shown in Fig. 1 were subjected to PCR using primers specific for the viral elongated minus-strand DNA (expected band size is 437 bp). Fraction numbers are the same as in Fig. 1. Naked viral DNA was used as a positive control (ctr+), and one reaction contained no viral DNA template (ctr−). (B) Fraction 3 of the density gradient containing the nuclear extracts (nuc 3) and fraction 2 of the density gradient containing the cytoplasmic extracts (cyt 2) were subjected to PCR using primers specific for the viral plus-strand DNA (expected band size is 700 bp). Fraction numbers are the same as in Fig. 1. Naked viral DNA was used as a positive control (ctr+), and one reaction contained no viral DNA template (ctr−). The rapidly migrating bands are PCR artifacts.
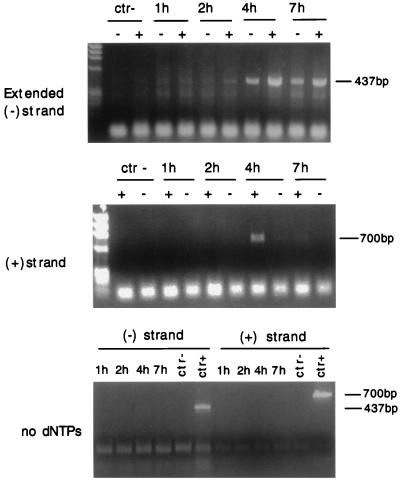
To test which of these complexes were competent to carry out reverse transcription in vitro, we performed an endogenous reverse transcription assay on 10-fold dilutions of the equilibrium density fractions containing the peak of the strong-stop DNA. The endogenous reverse transcription assay examines not only polymerase activity but also the integrity of the reverse transcription machinery and its ability to synthesize full-length viral DNA. Samples were incubated at 37°C in the presence or absence of 0.01% Nonidet P-40, and aliquots from the reactions were subjected to PCR using primers specific for the elongated minus strand and for the plus strand. No elongated reverse transcription product was detectable in the absence of exogenous dNTPs (Fig. 4). The amount of elongated minus strand increased progressively in the cytoplasmic extracts collected 1, 2, and 4 h after infection but decreased at 7 h after infection, perhaps because of partial intracellular degradation of the RTCs. Similarly, the maximum amount of the plus strand was detected in the fractions of the cell extracts collected 4 h after infection. The presence of Nonidet P-40 increased the efficiency of the endogenous reverse transcription assay; controls showed that it did not affect either the efficiency of the PCR or the activity of purified MoMLV RT (not shown). Thus, the complexes sedimenting at 1.35 g/ml are competent for DNA synthesis.
FIG. 4.
Endogenous reverse transcription assay of equilibrium density fractions containing the peak of viral DNA from cytoplasmic extracts collected 1, 2, 4, and 7 h postinfection. Samples diluted 1:10 were incubated with exogenous dNTPs for 6 h at 37°C in the presence (+) or absence (−) of Nonidet P-40 and then subjected to PCR with primers specific for the elongated minus-strand [(−) strand] DNA (expected band size is 437 bp) or the plus-strand [(+) strand] DNA (expected band size is 700 bp). An aliquot of the same fractions was incubated in the absence of dNTPs (no dNTPs). Some samples contained the same amount of cytoplasmic extracts from uninfected cells (ctr−). The rapidly migrating bands are PCR artifacts.
Different species of RTC are found after viral entry into the cells.
Western blot analysis of the equilibrium density fractions containing the cytoplasmic extracts from infected cells showed small amounts of viral CA proteins distributed over a wide density range. Nevertheless, the viral DNA was found in a single discrete peak (Fig. 1). We reasoned that some of the fractions containing CA proteins might also have contained the viral RNA genome which could not be detected by standard PCR. Therefore, an endogenous reverse transcription assay was performed on fractions from the density gradients containing the cytoplasmic extracts collected 1 and 4 h after infection. As shown in Fig. 5, strong-stop DNA was now detectable in some of those fractions which scored negative in previous assays (compare Fig. 1 with Fig. 5). In particular, the elongated minus strand could be detected in the lower-density shoulder of the main peak seen previously (Fig. 1). These fractions have a buoyant density near 1.32 g/ml and thus are lighter than the preexisting DNA found at 1.34 to 1.36 g/ml. This finding suggested that different species of RTC existed which were unable to start reverse transcription after viral entry into the cells. Some of these RTCs had a relatively high density (approximately 1.32 g/ml) and were able to synthesize the elongated minus strand DNA upon incubation with exogenous dNTPs and Nonidet P-40.
FIG. 5.
Endogenous reverse transcription assay of equilibrium density gradient fractions containing cytoplasmic extracts collected 1 and 4 h postinfection. Aliquots of the density gradients fractions were incubated at 37°C for 4 h in the presence of dNTPs and Nonidet P-40 and then analyzed by PCR using primers specific for the strong-stop DNA (top; expected band size is 145 bp) or the elongated minus-strand DNA (bottom; expected band size is 437 bp). (A) MW, DNA molecular weight standards; lanes 4 to 11, fractions 4 to 11 of the gradient shown in Fig. 1. (B) Lanes 5 to 12, fractions 5 to 12 of the gradient shown in Fig. 1; mw, DNA molecular weight standards; lane 13, virus subjected to the endogenous reverse transcription assay; lane 14, virus incubated in the absence of dNTPs. Naked DNA was used as positive control for amplification of the elongated minus-strand DNA (ctr+). One reaction contained no viral DNA template (ctr−). The rapidly migrating bands are PCR artifacts.
The size of the RTC changes during reverse transcription.
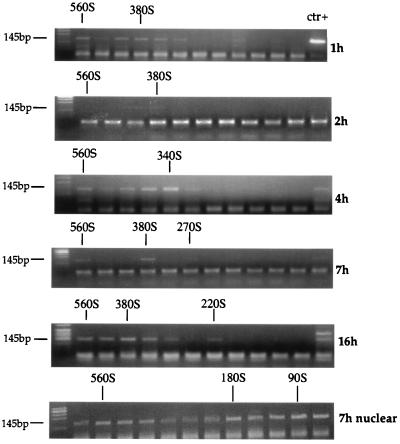
To determine whether there were changes in the size of the RTC with a buoyant density of 1.35 g/ml during reverse transcription, we subjected partially purified RTCs to velocity sedimentation through linear sucrose gradients. The equilibrium density fractions containing the peak of the viral DNA were dialyzed and concentrated by passage through a Centricon filter. The concentrate was then analyzed by sedimentation velocity, and the position of the viral DNA in the gradients was detected by PCR. In the cytoplasmic extracts, two discrete peaks with sedimentation velocities of 560S and 380S to 340S were consistently found 1, 2, and 4 h after infection. At later time points, a peak with a sedimentation velocity of 270S to 220S also appeared (Fig. 6). The 220S to 270S species were always less abundant than the 380S species. In the nuclear extracts, viral DNA was distributed in all fractions, although three discrete peaks with a sedimentation velocities of 560S, 180S, and 90S were consistently observed (Fig. 6). If the cytoplasmic extracts were prepared in the presence of 0.025% digitonin, a single RTC species was observed, and its sedimentation velocity was reduced to about 150S, in good agreement with previous reports (5). This suggested that RTC was partially disrupted by treatment with low concentrations of digitonin.
FIG. 6.
Analysis of RTCs by sedimentation velocity. Cytoplasmic extracts collected 1, 2, 4, 7, and 16 h postinfection and nuclear extracts collected 7 h postinfection were subjected to equilibrium density centrifugation. Fractions containing the peak of retroviral DNA were further purified and concentrated in a Centricon filter and centrifuged through a 5 to 20% linear sucrose gradient for 1 h at 23,000 rpm in a Beckman SW55 rotor. Twelve fractions were collected, and the viral DNA in each fraction was detected by PCR using primers specific for the strong-stop DNA (expected band size is 145 bp). The rapidly migrating bands are PCR artifacts.
The RTC is permeable to small but not to large macromolecules.
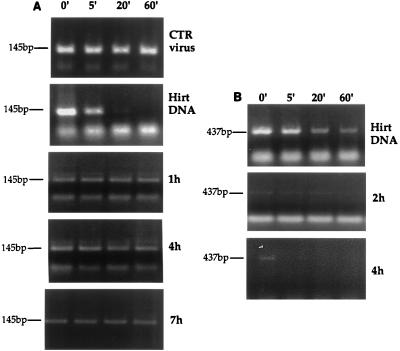
To evaluate the permeability of the RTC to macromolecules, we assayed the sensitivity to micrococcal nuclease of the viral DNA within the RTCs collected at various time points after cell infection. The equilibrium density fractions from cytoplasmic extracts containing the peak of the viral DNA were treated with micrococcal nuclease, and the reaction was stopped at the time point indicated in Fig. 7 by addition of EGTA. Nuclease-treated samples were then analyzed by PCR with primers specific for the strong-stop DNA and for the elongated minus-strand DNA, respectively. When naked viral DNA was treated with micrococcal nuclease, the strong-stop DNA appeared more sensitive to digestion than the minus-strand DNA. However, when the viral DNA within the RTC was tested for nuclease sensitivity, the strong-stop DNA was almost completely protected, while the minus-strand DNA was digested (Fig. 7). We also tested the ability of rabbit antiserum against MoMLV RT to inhibit reverse transcription of the viral genome within the RTC. This antiserum was able to inhibit the activity of purified RT about 10-fold, as detected by the oligo(dT)-poly(rA) assay (not shown). Equilibrium density fractions containing the peak of the viral DNA were subjected to endogenous reverse transcription assay in the presence of the antiserum or rabbit preimmune serum. Samples were then analyzed by semiquantitative PCR using primers specific for the plus-strand DNA. The antiserum was unable to inhibit reverse transcription of the viral genome in the RTC collected 1, 4, and 7 h after cell infection (not shown). The same antiserum was able to immunoprecipitate purified RT but was unable to immunoprecipitate RT in the RTC collected 7 h after cell infection (not shown). These data indicated that the RTC was permeable to small macromolecules like micrococcal nuclease but not to larger macromolecules like immunoglobulin G antibodies.
FIG. 7.
The viral strong-stop DNA in the RTC is protected from micrococcal nuclease digestion. Equilibrium density fractions containing the peak of the viral DNA from the cytoplasmic extracts collected 1, 4, and 7 h postinfection were incubated on ice in the presence of micrococcal nuclease and 2 mM CaCl2. The reactions were stopped by addition of 4 mM EGTA at the indicated time points and analyzed by PCR using primers specific for the strong-stop DNA (expected band size is 145 bp) or for the elongated minus-strand DNA (expected band size is 437 bp). Intact virions (CTR virus) incubated in the presence of micrococcal nuclease were used as a negative control. Naked viral DNA was incubated as described above in the presence of micrococcal nuclease and cytoplasmic extracts from uninfected cells (Hirt DNA). The fast-migrating bands are PCR artifacts. To ensure the linearity of amplification, the samples were subjected to two independent amplification rounds of 30 and 40 cycles, respectively.
DISCUSSION
In this study we have analyzed the dynamics of the MoMLV RTC from shortly after virus internalization to completion of reverse transcription and entrance into the cell nucleus. Analysis of the cytoplasmic extracts indicated that at least three different species of RTC are formed upon viral entry into the cells. All species appeared to contain at least the viral genome, CA proteins, and RT as detected by Western blotting, PCR, and endogenous reverse transcription assays. One RTC species had a buoyant density of approximately 1.12 g/ml, which is lower than the density of intact virions (13), and was unable to start reverse transcription, although it could synthesize strong-stop DNA if exogenous dNTPs and Nonidet P-40 were provided. A second RTC species fractionated at a density of 1.32 g/ml, contained RNA but no DNA, and was able to synthesize at least the minus-strand DNA in vitro in the presence of added dNTPs and Nonidet P-40. A third type of RTC fractionated at a density of 1.35 g/ml and contained both minus- and plus-strand DNAs, indicating that it was competent for reverse transcription. We suggest that the RTC with a buoyant density of 1.12 g/ml was associated with membranes and may or may not be on a productive pathway. The RTC with a buoyant density of 1.32 g/ml must be free of most lipids, but its lack of DNA suggests that it may not be functional. Nevertheless, this RTC could synthesize the minus-strand DNA in the endogenous reverse transcription assay. The RTC with a buoyant density of 1.35 g/ml contained the partially reverse transcribed viral genome, CA proteins, RT, and IN. This fraction presumably contained functional RTCs. This RTC species was also found in the nuclear extracts. Western blot analyses after equilibrium density fractionation showed that much of the CA protein was lost from the nuclear RTCs and that the CA/IN ratio of the cytoplasmic RTCs was higher than that of the nuclear RTCs. Most of the RTCs with a density of 1.35 g/ml did not contain the plus-strand DNA, even 16 h after infection. They were nevertheless able to synthesize this DNA in an endogenous reverse transcription assay, indicating that the structure of the RTCs was well preserved. These data are consistent with the possibility that this RTC is not very permeable to dNTPs in vivo.
The size of the RTC species with a density of 1.35 g/ml appeared to change with time. At early stages after infection, two species with sedimentation velocities of 560S and 380S were found. At later stages, a complex with a sedimentation velocity of 280S to 220S was also observed. In the nuclear extracts, three species with sedimentation velocities of 560S, 180S, and 90S were found consistently. These RTCs with different sedimentation velocities may represent discrete steps of uncoating which take place in the cell cytoplasm and nucleus. The results suggest the existence of an organized dissociation process of the RTC which occurs during reverse transcription. CA proteins are probably a major component which is lost during the process of uncoating of the RTC. CA proteins are almost completely lost from the PIC after entry into the nucleus, correlating with the lower sedimentation velocity observed in the RTCs isolated from nuclear extracts. The presence of CA in the RTCs may explain genetic and structural findings which support the involvement of CA protein in early steps of the viral life cycle. Indeed, mutants altered in CA often produce normal levels of virions but are unable to initiate reverse transcription in infected cells (15, 27, 29). Fv1 restriction in mouse cells, which blocks infection during the early stages, depends on specific sequences in the CA protein of MLV (7).
The reverse transcription-competent RTC was found to be permeable to small macromolecules like micrococcal nuclease but not to large ones like antibodies. This structural constraint may be advantageous for the completion of the viral life cycle, preventing dilution of RT and other factors in the cytoplasm but allowing dNTPs and other small cell proteins to enter the RTC. Treatment of the reverse transcription-competent RTC with micrococcal nuclease showed that the strong-stop DNA is almost completely protected from digestion whereas the minus-strand DNA is not. After completion of DNA synthesis, the DNA ends appear to form a large nucleoprotein complex called the intasome (32, 33). Our data showed that the strong-stop DNA is already protected 1 h after cell infection, indicating that the intasome may be at least partially organized early during the viral life cycle, well before reverse transcription is completed. It is not clear whether the two RTC species with buoyant densities of 1.12 and 1.32 g/ml represent functional intermediates or totally defective complexes. We suggest that they may be defective complexes arrested in intermediate steps of the uncoating process since they did not contain viral DNA even 4 h after infection. The system provided here may allow analysis of the RTCs from a variety of viral mutants defective in specific steps of the viral life cycle as well as cell mutants unable to sustain productive infection, thus helping to clarify many important issues related to the interactions between virus and cells.
ACKNOWLEDGMENTS
We are grateful to Marianna Orlova for advice and purified MoMLV RT, to Brian McDermott for preparing 32S-labeled poliovirus, and to Bernard Erlanger for advice on antibody peptide design. We are thankful to Eran Bacharach, Guanxia Gao, Jason Gonsky, and David Lim for advice and helpful discussions.
A.F. is a Wellcome Trust International Prize Research Fellow. S.P.G. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adam M A, Ramesh N, Miller A D, Osborne W R A. Internal initiation of translation in retroviral vectors carrying picornavirus 5′ nontranslated regions. J Virol. 1991;65:4985–4990. doi: 10.1128/jvi.65.9.4985-4990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alin K, Goff S P. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology. 1996;222:339–351. doi: 10.1006/viro.1996.0431. [DOI] [PubMed] [Google Scholar]
- 3.Blain S W, Goff S P. Nuclease activities of Moloney murine leukemia virus reverse transcriptase. J Biol Chem. 1993;268:23585–23592. [PubMed] [Google Scholar]
- 4.Bolognesi D P, Luftig R, Sharper J H. Localization of RNA tumor virus polypeptides. Virology. 1973;56:549–564. doi: 10.1016/0042-6822(73)90057-3. [DOI] [PubMed] [Google Scholar]
- 5.Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediated the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley W G, Stevenson M. Association of integrase, matrix and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DesGroseillers L, Jolicoeur P. Physical mapping of the Fv-1 tropism host range determinant of BALB/c murine leukemia virus. J Virol. 1983;48:685–696. doi: 10.1128/jvi.48.3.685-696.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durbin R K, Manning J S. The core of murine leukemia virus requires phosphate for structural stability. Virology. 1982;116:31–39. doi: 10.1016/0042-6822(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 9.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 11.Fassati A, Wells D J, Walsh F S, Dickson G. Efficiency of in vivo gene transfer using murine retroviral vectors is strain dependent in mice. Hum Gene Ther. 1995;6:1177–1183. doi: 10.1089/hum.1995.6.9-1177. [DOI] [PubMed] [Google Scholar]
- 12.Fassati A, Wells D J, Walsh F S, Dickson G. Transplantation of retroviral producer cells for in vivo gene transfer into mouse skeletal muscle. Hum Gene Ther. 1996;7:595–602. doi: 10.1089/hum.1996.7.5-595. [DOI] [PubMed] [Google Scholar]
- 13.Fink M A, Sibal L R, Wivel N A, Cowles C A, O’Conner T E. Some characteristics of an isolated group antigen common to most strains of murine leukemia virus. Virology. 1969;37:605–614. doi: 10.1016/0042-6822(69)90278-5. [DOI] [PubMed] [Google Scholar]
- 14.Goff S P, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu H-W, Schwartzberg P, Goff S P. Point mutations in the p30 domain of the gag gene of Moloney murine leukemia virus. Virology. 1985;142:211–214. doi: 10.1016/0042-6822(85)90435-0. [DOI] [PubMed] [Google Scholar]
- 16.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Hum Retroviruses. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 17.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis P, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim K, Chae C-B. A simple assay for DNA transfection by incubation of the cells in culture dishes with substrates for beta-galactosidase. BioTechniques. 1989;7:576–579. [PubMed] [Google Scholar]
- 20.McEwen C R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 21.Miller A D. Human gene therapy comes of age. Nature. 1992;357:455–460. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- 22.Miller D G, Adam M A, Miller M A. Gene transfer by retroviruses vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 25.Polson A, Coetzer T, Kruger J, von Maltzahn E, van der Merwe K J. Improvements in the isolation of IgY from the yolks of eggs laid by immunized hens. Immunol Investig. 1985;14:323–327. doi: 10.3109/08820138509022667. [DOI] [PubMed] [Google Scholar]
- 26.Posnett D N, McGrath H, Tam J P. A novel method for producing anti-peptide antibodies. J Biol Chem. 1988;263:1719–1725. [PubMed] [Google Scholar]
- 27.Reicin A, Ohagen A, Yin L, Hoglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps in viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roe T-Y, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartzberg P, Colicelli J, Gordon M L, Goff S P. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virus and reverse transcriptase. J Virol. 1984;49:918–924. doi: 10.1128/jvi.49.3.918-924.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 31.Telesnitsky A, Blain S, Goff S P. Assays for retroviral reverse transcriptase. Methods Enzymol. 1993;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 32.Wei S Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei S Q, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss R A, Teich N, Varmus H E, Coffin J M, editors. Molecular biology of tumor viruses: RNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1985. [Google Scholar]