Abstract
Purpose:
The study of cell-free DNA (cfDNA) enables sequential analysis of tumor cell–specific genetic alterations in patients with neuroblastoma.
Experimental Design:
Eighteen patients with relapsing neuroblastoma having received lorlatinib, a third-generation ALK inhibitor, were identified (SACHA national registry and/or in the institution). cfDNA was analyzed at relapse for nine patients and sequentially for five patients (blood/bone marrow plasma) by performing whole-genome sequencing library construction followed by ALK-targeted ddPCR of the hotspot mutations [F1174L, R1275Q, and I1170N; variant allele fraction (VAF) detection limit 0.1%] and whole-exome sequencing (WES) to evaluate disease burden and clonal evolution, following comparison with tumor/germline WES.
Results:
Overall response rate to lorlatinib was 33% (CI, 13%–59%), with response observed in 6/10 cases without versus 0/8 cases with MYCN amplification (MNA). ALK VAFs correlated with the overall clinical disease status, with a VAF < 0.1% in clinical remission, versus higher VAFs (>30%) at progression. Importantly, sequential ALK ddPCR detected relapse earlier than clinical imaging. cfDNA WES revealed new SNVs, not seen in the primary tumor, in all instances of disease progression after lorlatinib treatment, indicating clonal evolution, including alterations in genes linked to tumor aggressivity (TP53) or novel targets (EGFR). Gene pathway analysis revealed an enrichment for genes targeting cell differentiation in emerging clones, and cell adhesion in persistent clones. Evidence of clonal hematopoiesis could be observed in follow-up samples.
Conclusions:
We demonstrate the clinical utility of combining ALK cfDNA ddPCR for disease monitoring and cfDNA WES for the study of clonal evolution and resistance mechanisms in patients with neuroblastoma receiving ALK-targeted therapy.
Translational Relevance.
In ALK-altered high-risk neuroblastoma, ALK inhibitor treatment is frequently proposed at relapse or progression, but secondary disease progression frequently occurs. Our real-world data highlight a higher response rate in cases without MYCN amplification. We now demonstrate the importance of sequential cfDNA (cell-free DNA) analysis by combining cfDNA digital droplet PCR and whole-exome sequencing (WES). We show that highly sensitive digital droplet PCR targeting ALK can detect disease progression earlier than imaging. Furthermore, cfDNA WES provides evidence of clonal evolution after ALK-targeted therapy, providing insights into mechanisms of resistance. Our work underlines the importance of systematic sequential cfDNA analyses for all patients with neuroblastoma receiving ALK inhibitor treatment, both for disease monitoring and identification of new/emerging genetic alterations.
Introduction
Neuroblastoma, the most common extracranial pediatric solid tumor, is highly heterogeneous in clinical presentation and biological features (1, 2).
In neuroblastoma, copy number alterations including MYCN amplification or large-scale segmental chromosome alterations have a prognostic impact, but gene-specific mutations are less frequent (3, 4). The most frequently altered gene is anaplastic lymphoma kinase ALK, with genomic amplification or constitutively activating mutations leading to active signaling of multiple downstream pathways (5–8). Single base missense mutations are found at three major hotspots of the kinase domain at positions R1275, F1174, and F1245, accounting for 85% of neuroblastoma ALK mutations (5, 7–9). ALK mutations are observed in approximately 10% of cases at diagnosis but in more than 25% at relapse, due to either expansion of subclonal ALK mutations or acquisition of new mutations upon relapse (10, 11).
ALK mutations exhibit differential sensitivity to ALK inhibitors. With a limited sensitivity to the first-generation ALK inhibitor Crizotinib, subsequent second- and third-generation ALK inhibitors were designed to overcome primary resistance (12–14). Recent studies indicate single-agent response rates of 20% to Ceritinib, >30% to lorlatinib, and >60% to lorlatinib in combination with chemotherapy (15–17). However, progression under targeted therapy occurs frequently, with the development of either on-target ALK resistance mutations in a subset of cases or off-target mutational events in particular in the RAS-MAPK pathway, highlighting the importance of sequential analyses (18–21).
Liquid biopsies and the study of circulating tumor DNA (ctDNA), a fraction of cell-free DNA (cfDNA), enable sequential analysis of tumor cell–specific genetic alterations in patients with neuroblastoma (22, 23).
ALK mutations can be detected using digital droplet PCR (ddPCR) at high sensitivity and specificity in the cfDNA of patients with neuroblastoma, highlighting their potential for disease surveillance (22–27). cfDNA can also be used for larger-scale copy-number variation (CNV) or SNV detection, and the identification of genetic alterations not identified in tissue biopsies indicates the potential of liquid biopsies to better capture genetic heterogeneity (25, 28–30).
Recently, serial cfDNA analyses from patients with neuroblastoma receiving lorlatinib have enabled the tracking of evolutionary dynamics. Using gene panel sequencing approaches, off-target resistance mutations were identified in 27% and secondary compound ALK mutations in 15% of cases (18).
We now analyze the response to ALK inhibitor treatment during neuroblastoma progression in real-world data. We demonstrate the importance of sequential cfDNA analyses by a combination of highly sensitive ddPCR and cfDNA whole-exome sequencing (WES). We show that highly sensitive ddPCR can detect disease progression earlier than imaging. Furthermore, cfDNA WES evidences clonal evolution after ALK-targeted therapy, providing insights into potential mechanisms of resistance.
Materials and Methods
Patients and samples
Patients with relapsed high-risk neuroblastoma were included in this study if they received lorlatinib treatment within a compassionate/off-label setting following the identification of an ALK genetic alteration. Lorlatinib could be provided by the pharmaceutical company Pfizer on a case-by-case basis, as lorlatinib is not approved for this indication. Clinical information of the 18 patients included in this study is summarized in Table 1. Clinical data of an adult patient have been partially published previously (17). All patients were initially treated according to national/international treatment protocols (31). Informed written consent from parents, guardians, or patients was obtained for the use of biological samples and the use of data, and the study was approved by the Institutional Review Board of Institut Curie (DATA210225, DATA220167). The study was conducted in accordance with the ethical guidelines of the Declaration of Helsinki.
Table 1.
Summary of patient characteristics.
| Patient characteristics | Overall (n = 18) |
|---|---|
| Gender, numbers (%) | |
| Female | 9 (50) |
| Male | 9 (50) |
| Age at start of lorlatinib therapy, median (range), y | 5.7 (2.1–39.1) |
| N-MYC status, numbers (%) | |
| Amplified | 8 (44) |
| Nonamplified | 10 (56) |
| Number of previous relapse/progression, median (range) | 2 (1–6) |
| Type of ALK alteration | |
| Mutation | |
| F1174L | 7 (39) |
| R1275Q | 3 (17) |
| F1245C | 1 (6) |
| F1245I | 1 (6) |
| I1170N | 1 (6) |
| R1278 + Y1278 | 1 (6) |
| F1174L + F1245I + R1275Q | 1 (6) |
| F1174S + R1275Q | 1 (6) |
| Amplification | 1 (6) |
| Rearrangement | 1 (6) |
| Administered lorlatinib/lorlatinib + chemotherapy cycles, numbers | 155 |
| Median (range) | 4 (1–8) |
SACHA-France (NCT04477681) is a prospective national observational registry collecting data on compassionate or off-label innovative anticancer therapies in patients aged ≤25 years (32). SACHA-France has been open in all French pediatric oncology centers since March 2020. It is recognized as a real-world data source by the French Health Technology Assessment Institution and is supported by the French National Agency for the Safety of Medicines and Health Products. Information includes demographics, patient history, dose reductions, treatment delays or discontinuation, objective disease response, time to progression, and grade ≥2 clinical and grade ≥3 laboratory adverse drug reactions (ADR; ref. 32).
Tumor response was assessed by imaging, including meta-iodobenzylguanidine (123I-MIBG) scintigraphy or positron emission tomography scan (18F-FDG, for MIBG non-avid tumors), using the modified International Neuroblastoma Response Criteria (33). Overall response corresponded to the best tumor response (complete response, CR, or partial response, PR) and confirmed by review of pseudonymized imaging reports. Survival rates were determined using the Kaplan–Meier method. Progression-free survival (PFS) measured the time between the first dose of lorlatinib (±chemotherapy) and progressive disease (PD) or death.
Tumor samples and genomic DNA extraction
Tumor tissue was obtained by percutaneous biopsy at diagnosis and/or at relapse in 13 cases, with >30% of tumor cells confirmed by pathologic examination. Genomic DNA (gDNA) was extracted according to standard procedures, with matched germline material available in eight cases (34).
Tumor molecular characterization was performed within the MICCHADO study (NCT03496402; n = 2), MAPPYACTS (NCT02613962, n = 1; ref. 34), within France Medicine Génomique PFMG2025 SeqOIA/Auragen program (https://pfmg2025.aviesan.fr/; n = 3), or by next-generation sequencing (NGS) panel sequencing. For five other cases, no primary tumor sequencing was performed, with ALK mutations identified on cfDNA by NGS panel sequencing at diagnosis (one case) or at relapse (four cases).
Reference genomic DNA was extracted from the neuroblastoma cell lines CLB-GA (RRID:CVCL_9529), SH-SY5Y (RRID:CVCL_0019) and SJNB1 (RRID:CVCL_8812), and the patient-derived xenograft pPDX-IC-75 (35) using the QIAamp DNA Mini Kit (Qiagen; refs. 35, 36).
Plasma samples and cell free DNA extraction
cfDNA analysis to identify ALK mutations could be performed in nine patients at relapse, and sequential samples for cfDNA analyses were available for five patients treated at Institut Curie (Supplementary Table S1). For the sequential cfDNA analysis, blood or bone marrow was collected in EDTA tubes and centrifuged (10 minutes, 1,000 × g, rtp) within 2 hours, followed by immediate separation of the plasma from the cellular pellet, flash freezing in liquid nitrogen and storage at −80°C. Plasma samples were collected at diagnosis, during treatment, follow-up, and relapse or progression, with a mean of nine time points per patient (range 5–13).
For each sample, between 250 µL and 1 mL of plasma was used for downstream analysis (Supplementary Table S1). cfDNA was extracted using the QIAamp MinElute ccfDNA mini kit (Qiagen) and quantified using the Qubit High Sensitivity dsDNA assay (ThermoFisher).
Digital droplet PCR
ddPCR primers and probes were designed for the ALK mutational hotspots F1174L, F1245C, R1275Q, and I1170N (Supplementary Table S2). ddPCR was performed on 10 ng of genomic or cfDNA, with 450 nmol/L of each primer, and 250 nmol/L of each probe. Samples were fractioned into approximately 20,000 droplets (QX100 Droplet Generator; Bio-Rad). Cycles for PCR DNA amplification were: 95°C (10 minutes), 40× (30 seconds - 94°C; 1 minute - 62.5°C for ALK R1275Q; 55°C for ALK F1174L/ALK I1170N), 10 minutes at 98°C (T100 Thermo Cycler; Bio-Rad). The fluorescence signal of each droplet was quantified (QX100 Droplet Reader; Bio-Rad), with wild-type DNA reporter probes containing Victoria (VIC) fluorophore, and mutated DNA-specific probes containing fluorescein amidite (FAM) fluorophore. All probes contained an MGB-EDQ TaqMan quencher (Eurogentec). Variant allele fractions (VAF) were analyzed using QuantaSoftAnalysisPro software, version 1.0.596.525 (Bio-Rad) including appropriate nontemplate, positive and negative controls.
VAF was calculated as follows:
Limit of detection of ddPCR
To determine the lower limit of detection by ddPCR, genomic DNA from neuroblastoma cell lines harboring known ALK mutations (CLB-GA: ALK R1275Q; SH-SY5Y: ALK F1174L), with ALK VAFs of 34% consistent with chromosome 2p copy number status were serially diluted (VAF range: 10%–0.1%) in ALK wild-type cell line SJNB1. Diluted samples were fragmented to 150 bp using Adaptive Focused Acoustics technology (Covaris), to mimic the length of cfDNA, and analyzed by ddPCR (Supplementary Fig. S1). A strong correlation between the calculated and measured number of copies per µL was observed, down to a mean of 0.03 copies per µL, corresponding to a VAF limit of detection of 0.19% and 0.12%, respectively.
WGS library construction and WES sequencing
Whole-genome sequencing (WGS) libraries of genomic DNA from primary tumors, paired germline DNA, neuroblastoma cell lines, or cfDNA extracted from plasma samples were constructed to enable either ddPCR on WGS libraries, or WES.
Indexed sequencing libraries were prepared using 10 to 200 ng of genomic DNA fragmented to 150 bp using Adaptive Focused Acoustics technology (Covaris), followed by SureSelect Low input dual index kit (Agilent) library construction, using eight cycles of PCR amplification.
For cfDNA extracted from plasma, no fragmentation step was required. The total extracted cfDNA was <10 ng in 17/40 plasma samples, requiring two to three additional PCR cycles (Supplementary Table S1).
Following library construction, whole exome capture was performed using the SureSelect Clinical Research Exome v2 kit (Agilent) as described previously (34).
The libraries were quantified using Qubit dsDNA assays (ThermoFisher), aiming for a concentration of 200 ng/µL of WGS library and for 2 ng/µL of library capture. For library quality control, 1 µL of WES capture libraries were analyzed using the Bioanalyzer high sensitivity DNA kit (Agilent), aiming for a mean library size of 300 bp. WES capture libraries were then sequenced according to previously published methods, aiming for a depth of coverage of 100× (Illumina technology, PE 100pb; refs. 29, 34).
Comparison of ALK VAFs determined by ddPCR on DNA pre-and post-library construction
To compare the ALK VAF determined by ddPCR on prelibrary (preamplification) and postlibrary construction genomic DNA, genomic DNA from CLB-GA (ALK R1275Q; VAF 34%) and pPDX-IC-75 (ALK F1174L, 58% VAF) were serially diluted in healthy donor genomic DNA (ALK wild type). Dilutions were fragmented to 150 bp using Adaptive Focused Acoustics technology (Covaris) to mimic cfDNA fragment size. Subsequently, 200 ng of DNA from each dilution was used for WGS library preparation as indicated previously. The ALK VAF was determined by ddPCR, using 10 ng of the prelibrary construction, or 10 ng of postlibrary construction DNA.
Sequential analysis of ALK VAF for patients with high-risk neuroblastoma using ddPCR and WES
To determine the cfDNA ALK VAF during treatment and follow-up of five high-risk patients with neuroblastoma, we performed ddPCR on WGS libraries of sequential cfDNA samples, at all available time points. Furthermore, WES was performed on gDNA extracted from the primary tumor, with a matched germline, and on cfDNA obtained at the time of relapse, and time points corresponding to lorlatinib treatment (Supplementary Fig. S2). In four cases, a cell fraction obtained at a later follow-up time point was sequenced in addition to the diagnostic germline sample for the study of clonal hematopoiesis.
Bioinformatics analysis
The WES sequencing raw reads were mapped to the reference human genome assembly GRCh37 using BWA v0.7.15 (default parameters; RRID:SCR_010910). The duplicate reads were marked, and the base scores were recalibrated.
Variants were called with Mutect2 (GATK 4.1.7.0; RRID:SCR_001876) applying the joint calling method on all the samples of the same patient [matched normal/germline, tumor, and plasma sample(s)].
Variants with ≤2 reads in germline and ≥2 reads in tumor/plasma (≥1 read per alternative strand), with a mapping or base quality scores ≥20, a median distance from the end of the read ≥3, ≤2 alternative alleles per position, ≤2 events in a single assembly, and ≤5 reference bases in short tandem repeat (STR) regions were kept.
The variants were annotated with SnpEff 5.0 (dbNSFP2.9.3, COSMIC (v92), gnomAD (r.2.1.1), clinvar_20200824; RRID:SCR_005191) (37). Only SNVs with a Moderate or High predicted impact were selected. Variants with a population prevalence than ≤1/2,000 were filtered out. A panel of normals was used for further filtering.
Copy number analysis were performed with snp-pileup_0.5.14 and FACETS v0.5.11 applying a threshold of ≥20 for mapping and base quality and minimum read counts per position to be outputted (38, 39).
Clonal evolution analysis was performed using Pyclone (RRID:SCR_016873) to infer cancer cell fraction (CCF) from allelic count data using total copy number and tumor content prediction from FACETS for each of the somatic SNV called by mutect2. Beta-binomial distribution was used as a model, and 10 random restarts were performed (40, 41).
The clonal ordering and clonal evolution visualization were obtained using the clonevol package from R. The cancer evolution model used was monoclonal, the subclonal test performed was bootstrap and the model was nonparametric. The number of boots was set to 1,000 and the funding cluster to 1 (the cluster with the highest CCF in the first time point). The minimum cluster CCF was set to 0.001. The function generateFishplotInputs was used to generate a Fish object (42). Finally, the clonal architecture of populations was generated by using the fishplot R package (40, 42).
Gene set enrichment analysis was performed using (https://www.gsea-msigdb.org) ontology gene sets C5, selecting up to the top 25 significant gene sets with an FDR q-value of less than 0.05.
Statistical analysis
In this study, all the graph and correlation analyses were performed by using the GraphPad PRISM9 software (version 9; RRID:SCR_002798). Survival was calculated using the Kaplan–Meier method, from the time of lorlatinib treatment, and comparisons were performed using a logrank test. P-values of <0.05 were considered significant.
Data availability
The data generated in this study are available within the article and its supplementary data files. Raw data for this study were generated at the ICGex Sequencing Platform of Institut Curie, Paris, France. Raw data supporting the findings of this study are available from the corresponding author upon request.
Results
Response to lorlatinib
Eighteen patients with relapsed high-risk neuroblastoma received lorlatinib on a compassionate-use basis between 2018 and 2022 (Table 1). Among these, 15 patients were included in the SACHA-France study until October 2023 and were treated across 11 pediatric oncology centers in France. One patient had received lorlatinib before the SACHA-France study, and for one patient inclusion in SACHA was ongoing at data cutoff. One adult patient treated with off-label lorlatinib was also included (17).
Median age at lorlatinib therapy was 5.7 years (range 2.1–39.1 years), with only one patient older than 18 years. MYCN was amplified in 8/18 tumors. All cases except two presented with ALK mutations, mainly F1174L (7/16) followed by R1275Q (3/16), identified in the tumor for 11 patients (six targeted NGS panels, five WES) and in cfDNA in five patients (four at relapse, one at diagnosis). The ALK VAF was known in 10 tumors (range: 14%–89%). Two cases presented with ALK compound mutations (F1174S + R1275Q; F1174L + F1245I + R1275Q), both identified in cfDNA. One patient presented with an ALK rearrangement and another patient had an ALK amplification (but no ALK mutation).
All patients had previously received high-risk neuroblastoma treatment with a median time of 1.6 years since initial diagnosis (range: 0.3–6.3) and a median of two previous lines of therapy (range: 1–6). Only the adult patient had previously received a prior ALK inhibitor (crizotinib; ref. 17). Twelve patients received lorlatinib as a single agent, and six others lorlatinib combined with chemotherapy [topotecan/cyclophosphamide (5); topotecan/temozolomide (1)].
All patients had actively progressing disease at the time of lorlatinib therapy. The global objective response rate (ORR) was 33% (6/18; CI, 95%: 13%–59%), with similar ORR between patients receiving single-agent lorlatinib (4/12 patients) compared with patients receiving lorlatinib with chemotherapy (2/6 patients). Noteworthily, an objective response was achieved more frequently in patients with MYCN nonamplified (6/10 cases, independently of the treatment received [seven single agents, three combined with TC)] than with MYCN-amplified tumors (0/8 cases; Fig. 1). The 4-month PFS was 44%, the 6-month PFS was 33%, and the 12-month PFS was 22%.
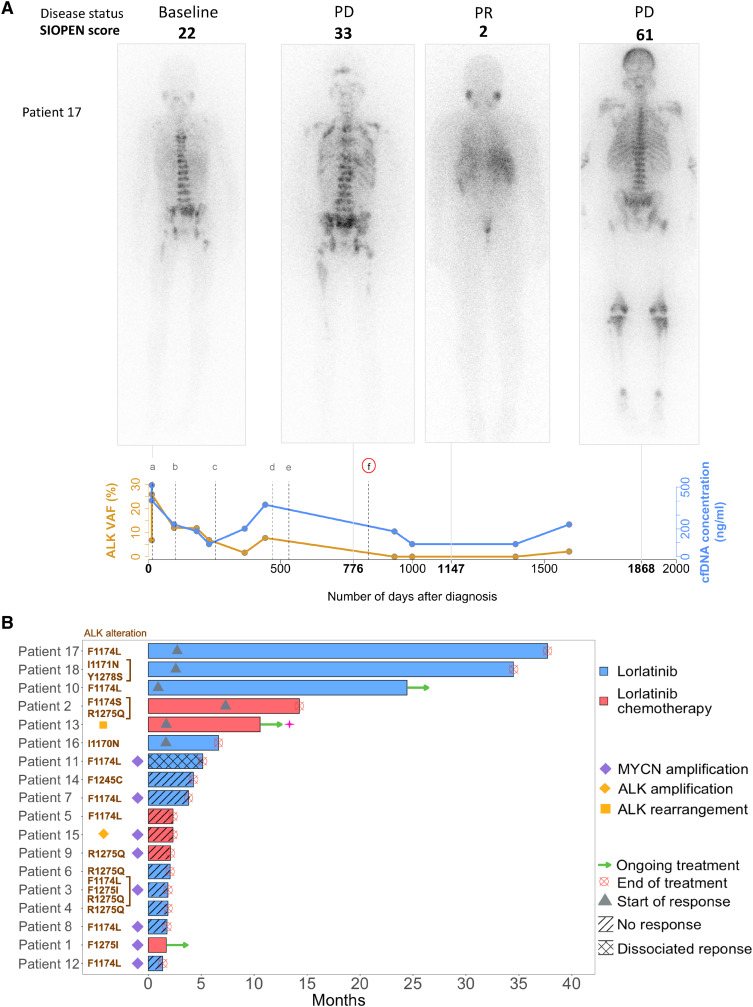
Figure 1.
Clinical efficacy of lorlatinib. A, Clinical evolution in a 10-year-old boy with neuroblastoma, INRGSS M, without MYCN amplification (patient 17). Successive 123I-MIBG scans indicate widespread osteomedullary disease at diagnosis. Initial therapy consisted of rapid COJEc (PD), CAV (PD), BEACON irinotecan-temozolomide (SD), pazopanib (PD), etoposide (PD), and cyclophosphamide (PD). Lorlatinib (single agent) resulted in an important reduction of the SIOPEN score (PR according to INRC). Following progression, trametinib was empirically associated with lorlatinib following (PD), based on a possibility of sensitivity to MEK inhibition in combination, while awaiting cfDNA analysis. Palliative irradiation to symptomatic metastatic sites followed by etoposide was given (PD). The patient passed away 67 months after diagnosis. The cfDNA concentrations (ng/mL, in blue) and ALK VAF (%, in orange) are indicated. PD, progressive disease; SD, stable disease; PR, partial response. B, Efficacy of lorlatinib in 18 patients with high-risk neuroblastoma treated with lorlatinib, with or without chemotherapy, achieving SD or PR. The bar indicates the duration of lorlatinib treatment. Triangles indicate the start of response to treatment. Discontinuation of treatment by lorlatinib occurred at relapse/progression.
Among the patients enrolled in SACHA, 15 patients received at least one dose of lorlatinib: nine (60%) as a single agent and six (40%) in combination with chemotherapy. Among patients treated with lorlatinib alone [median treatment duration: 62 days (range 53–733 days)], 3/9 (33%) experienced an ADR including grade 2 CTCAE constipation, diarrhea, and weight gain. No serious nor grade ≥3 ADR was reported in those patients.
In patients treated with lorlatinib combined with chemotherapy [median treatment duration: 70 days (range 41–428 days)], 5/6 (83%) experienced at least one ADR. The main observed toxicity was febrile neutropenia (three patients, 50%). Other ADRs included hypertension, oral mucositis, hypertriglyceridemia, and weight gain (one patient each; 16%). Febrile neutropenic events in three patients were considered serious, leading to hospitalization and delayed therapy. No patient required dose reduction or a definitive treatment discontinuation due to ADR. All mentioned ADRs were expected as listed in the SmPC (Supplementary Table S3).
ddPCR on genomic DNA sequencing libraries enables robust detection of ALK hotspot mutations
An approach combining highly sensitive ddPCR with WES on cfDNA was established. The ddPCR limit of detection of ALK mutations in a wild-type background was determined at 0.03 copies per µL, corresponding to a VAF of 0.19% and 0.12%, respectively (Supplementary Fig. S1).
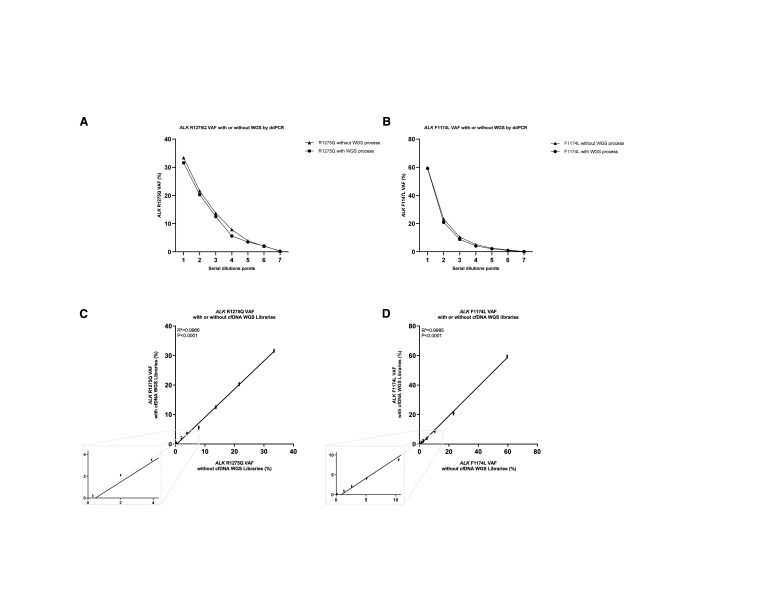
The feasibility of ddPCR to measure VAF of ALK hotspot mutations in WGS libraries was then evaluated. Serial dilutions of genomic DNA harboring different ALK hotspot mutations were fragmented and subjected to standardized WGS library construction. ALK VAFs were then determined by ddPCR either directly in the gDNA serial dilutions or following the WGS library construction. A strong correlation was observed between the VAFs of ALK hotspot mutations measured directly in gDNA and, following WGS library construction, both for the ALK R1275Q and ALK F1174L mutation (R2 = 0.9964 and R2 = 0.9986 with a P-value < 0.0001; Fig. 2).
Figure 2.
Droplet digital PCR maintains linearity for ALK F1174L and ALK R1275Q with or without WGS libraries process. A, Comparison analysis of measured ALK R1275Q VAFs’ (%) average (n = 4) in ddPCR between two conditions: with WGS libraries process or no WGS libraries process for each group of dilutions. B, Comparison analysis of measured ALK F1174L VAFs’ (%) average (n = 4) in ddPCR between two conditions, with WGS libraries process or no WGS libraries process for each group of dilutions. Comparison of the VAF means of the quadruplicates of each dilution demonstrated that no statistically significant difference was observed between the two conditions (Mann–Whitney test; P < 0.05). C, Correlation analysis of measured ALK R1275Q VAFs (%) without or with WGS libraries process obtained by calculating an average on four replicates (n = 4) represented respectively in the x-axis and the y-axis (as mean ± SD; n ≥ 3). Pearson’s correlation coefficient (r) and P-value are indicated. D, Correlation analysis of measured ALK F1174L VAFs (%) without or with WGS libraries process obtained by calculating an average on four replicates (n = 4) represented respectively in the x-axis and the y-axis (as mean ± SD; n ≥ 3). Pearson’s correlation coefficient (r) and P-value are indicated.
Detection of ALK mutation in serial cfDNA from patients with high-risk neuroblastoma correlates with clinical disease status
cfDNA for detection of ALK mutations could be analyzed for nine patients at relapse (patients 1, 2, 3, 4, 6, 7, 16, 17, and 18; Supplementary Figs. S2 and S3). For five patients, sequential cfDNA samples could be analyzed using the combination of ddPCR and WES (patients 6, 7, 16, 17, and 18).
Analysis of tumor tissue at diagnosis and/or at relapse revealed the ALK hotspot mutations F1174L (two cases), R1275Q, Y1278S, and I1170N, respectively (Table 1; Supplementary Table S1). ALK VAFs in tumor tissue assessed by WES was 15% to 35%, in accordance with chromosome 2 copy number status and tumor cellularity of the studied tumor samples.
The VAF of the ALK hotspot mutation previously identified in the tumor was then determined in serial plasma samples obtained from blood or bone marrow at different time points during the disease course of each patient, before and after lorlatinib treatment (Fig. 3). To enable additional cfDNA analyses, ALK ddPCR was performed on WGS libraries prepared from cfDNA of each plasma sample, rather than on cfDNA directly, as described above (Supplementary Figs. S2 and S3).
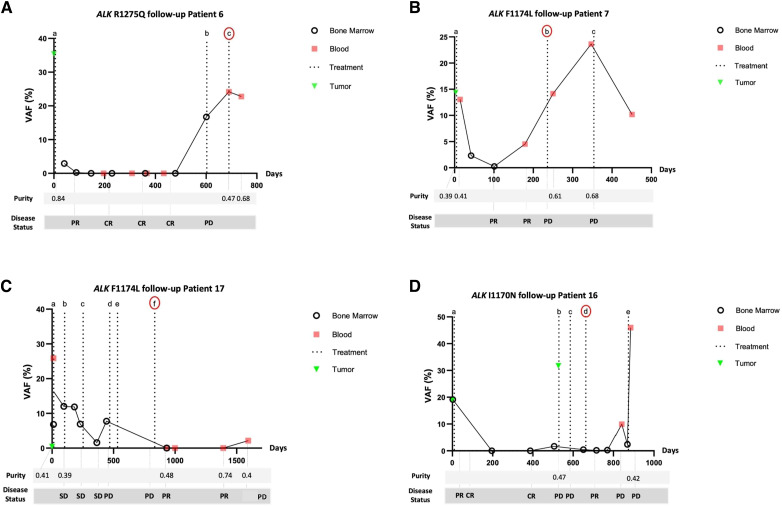
Figure 3.
ALK hotspot mutations in cfDNA extracted from plasma samples of patients with neuroblastoma and comparison with disease status measured by ddPCR and WES. A, Patient 6 (5 years, INRG M, no MNA): ALK R1275Q VAFs (%; y-axis) by ddPCR analysis on WGS libraries of cfDNA extracted from sequential blood or bone marrow plasma. The number of days after diagnosis is represented in the x-axis. Treatments are indicated by letter: (a) HR-NBL 1.7, (b) Temodal, and (c) lorlatinib. Treatment according to HR-NBL1.7 was administered (a) with a decrease of ALK R1275Q VAF between D6, to 0% at D 41, D88, and D479, correlating with clinical complete remission. Upon relapse (D603) an increase of ALK R1275Q VAF to 17% was observed, corresponding to PD. Treatment with Temodal (b) resulted in further PD, with a further increase of ALK R1275Q VAF to 24%. After the start of lorlatinib (c), a small decrease of ALK R1275Q VAF to 22% at D739 was observed. The patient passed away due to PD. B, Patient 7 (14 years, INRG M, MNA): ALK F1174L VAFs (%; y-axis) by ddPCR analysis on WGS libraries of cfDNA extracted from sequential blood or bone marrow plasma. The number of days after diagnosis is represented in the x-axis. Treatments are indicated by letter: (a) HR-NBL 1.7, (b) lorlatinib, and (c) TOTEM. Initial treatment HR-NBL 1.7 (a) resulted in a decrease of ALK F1174L VAF from 13% at D5 to almost 0% at D101, correlating with clinical very good PR. On D179, ALK F1174L VAF increased to 4.5%, whereas imaging (CT scan, MIBG) concluded persistent VGPR. Relapse was confirmed at D222 (by local imaging and MIBG), indicating that ddPCR detected relapse 43 earlier than imaging. Lorlatinib (b) treatment was commenced D236 with a further increase of ALK F1174L VAF to 24%, with new PD on D349. TOTEM (c) commenced on D354 resulted in ALK F1174L VAF 10%, indicating clinically stable disease. The patient passed away due to PD. C, Patient 17 (10 years, INRG M, no MNA): ALK F1174L VAFs (%; y-axis) by ddPCR analysis on WGS libraries of cfDNA extracted from sequential blood or bone marrow plasma. The number of days after diagnosis is represented in the x-axis. Treatments are indicated by letter: (a) HR-NBL 1.7, (b) Beacon, (c) Pazopanib, (d) RT + Etoposide, (e) cyclophosphamide, (f) lorlatinib. ALK F1174L VAFs at D12 was 26% (blood) versus 7% (bone marrow). Induction of HR-NBL1 resulted in stable disease, with an ALK F1174L VAF persisting at 12% at D96. Beacon (b) commenced on D102 and resulted in SD, with ALK F1174L VAF 7% on D229. Pazopanib (c) treatment began on D253 with a temporary decrease of ALK F1174L VAF to 1.5% at D364 followed by a renewed increase to 7.755% at (D) lorlatinib (f) was commenced on D834, with ALK F1174L VAF subsequently attaining nearly 0%. A reemergence of ALK F1174L to 2% concomitant with a single MIBG lesion, preceding multimetastatic relapse. The patient passed away due to PD. D, Patient 16 (3 years, INRG M with no identified primary, no MNA) ALK I1170N VAFs (%; y-axis) by ddPCR analysis on WGS libraries of cfDNA extracted from sequential blood or bone marrow plasma. The number of days after diagnosis is represented in the x-axis. Treatments are indicated by letter: (a) HR-NBL 1.7, (b) Beacon, (c) CAdO, (d) lorlatinib, and (e) etoposide. ALK I1170N VAFs, at 19% at diagnosis decreased to 0% at D196 and D387 following HR-NBL 1.7 (a) treatment. At D506, ALK I1170N VAF increased to 2% with confirmation of PD by imaging on D514. ALK I1170N VAF was 0.4% at the beginning of lorlatinib treatment on D665 with a decrease to 0.169% at D716 and 0.260 at D770 but a new increase to ALK I1170N 10% on D840 coinciding with new PD. Salvage treatment proved inefficient, with ALK I1170N VAF at 45%. The patient passed away due to PD.
For plasma sample at diagnosis, the ALK mutation identified in the tumor was detected in cfDNA in all cases (cfDNA ALK VAF 12%–20%). Importantly, the cfDNA ALK VAF decreased during the initial treatment phase (Fig. 3). Patients 6, 7, and 16 achieved complete remission during first-line treatment. For these three patients, the previously identified ALK mutation could not be detected in plasma samples obtained at complete remission. Conversely, for patient 17 primary refractory disease was observed, with only partial local and metastatic remission obtained following first-line treatment, and the ALK cfDNA VAF remained >1%. An increase of ALK cfDNA VAF was observed during the disease course in all patients and corresponded to disease relapse or progression. Importantly, in patient 7, at the time of increased ALK cfDNA VAF, the patient was considered in stable disease based on clinical and radiological evaluation (CT scan, MIBG scan; Supplementary Fig. S6). Disease progression was then confirmed on an MIBG scan 4 weeks later, indicating that the ALK cfDNA VAF might be more sensitive than imaging for detection of disease progression.
All five patients received lorlatinib treatment for disease progression. In 3/5 patients (patients 16,17,18; Fig. 3; Supplementary Fig. S4) the ALK cfDNA VAF decreased during lorlatinib treatment, consistent with a response to lorlatinib treatment confirmed by imaging. Patient 17 experienced very good partial remission under lorlatinib treatment, with ALK cfDNA VAF at 0 for >36 months. Furthermore, in patient 6 a transitory period of improvement of several weeks was observed (decrease in pain, clinical decrease of a metastatic lesion), but no blood or bone marrow sample could be obtained, and clinical disease progression occurred rapidly. However, in two other patients, no clinical benefit from lorlatinib was observed, and in these two patients, ALK VAF remained high in all subsequent samples. All five patients have died from disease progression, with the cfDNA ALK VAF >15% at the last time point.
cfDNA WES indicates an increase of SNVs at progression of neuroblastoma
cfDNA samples were analyzed by cfDNA WES, in addition to ALK ddPCR, at the time of diagnosis, at relapse, before or after lorlatinib treatment, and at the last time point available (Supplementary Fig. S2). cfDNA WES enables the estimation of ctDNA in cfDNA content. At diagnosis, in all cases, a high ctDNA content (mean 52.75%, 39%–84%) was observed (Supplementary Table S1). The ctDNA content also strongly correlated with overall disease status, with a lower ctDNA observed in samples obtained at partial or complete remission and an increase of ctDNA content at relapse. cfDNA WES confirmed the cfDNA ALK VAFs at the known hotspots, and no statistically significant difference between cfDNA ALK VAF measured by WES and ddPCR analysis cfDNA libraries was observed (Mann–Whitney text, P = 0.62; Supplementary Fig. S5).
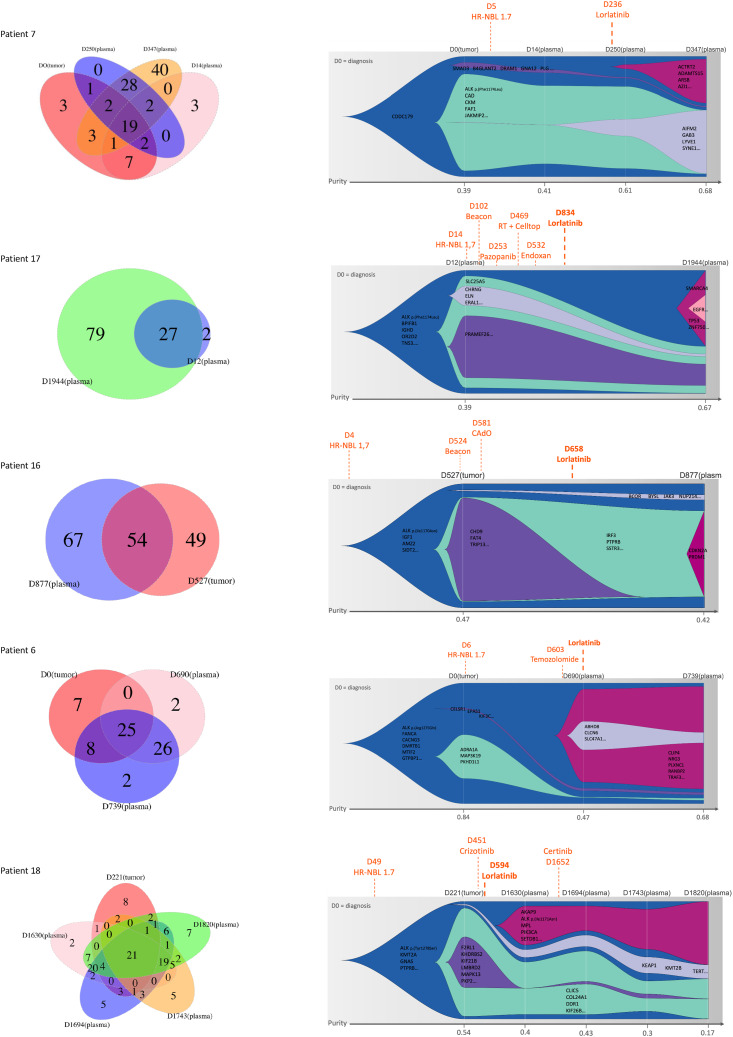
At diagnosis, the number of SNVs detected by cfDNA WES was compared with the number of SNVs detected in the primary tumor for patient 7 (Fig. 4). A total of 29 SNVs were common to the primary tumor and plasma cfDNA, whereas nine SNVs were seen only in the tumor and five only in the plasma cfDNA at diagnosis.
Figure 4.
Venn diagrams and clonal evolution based on cfDNA analysis via WES in sequential plasma samples of patients with neuroblastoma treated with lorlatinib. For each patient, the graph on the (left) indicates the number of somatic SNVs detected by cfDNA WES in each analyzed sample. CCFs were calculated to determine clonal composition and clonal evolution. The graph on the (right) indicates the clones according to the cancer cell fraction of all somatic SNVs identified in sequential plasma samples. The clone of origin is depicted in (blue), shrinking clones are indicated in (green, gray, and purple), and emerging clones are indicated in (red) and (pink).
Comparison of the number of SNVs detected by WES in the primary tumor and the plasma cfDNA at the time of lorlatinib treatment was possible for patient 7 (plasma obtained 14 days after the beginning of lorlatinib treatment) and patient 6 (plasma obtained on the day of begin of lorlatinib treatment), with a mean of 27 SNVs (range 25–29) common to the primary tumor and plasma cfDNA, versus a mean of 12 SNVs (range 9–15) and 16 SNVS (range 5–28) seen only in the tumor and the cfDNA, respectively, indicating spatial genetic heterogeneity. At the latest analyzed plasma cfDNA time point, for the five patients (mean 549 days after the beginning of lorlatinib treatment; range 90–1,226 days), a mean of 39 new SNVs (range 2–79) not seen previously in the primary tumor or any previous plasma cfDNA was detected. Accordingly, comparison of the number of SNVs detected by cfDNA WES at diagnosis and the last cfDNA time point showed an increase in cfDNA WES SNVs in all cases (Supplementary Tables S1 and S4).
cfDNA WES indicates clonal evolution following lorlatinib treatment
To assess clonal evolution following lorlatinib treatment, the CCF was determined for all SNVs detected by tumor or cfDNA WES. SNVs were categorized into clonal (if CCF > 90%) or subclonal (if CCF < 90%), with a proposition of models of clonal evolution. In all five patients, sequential cfDNA WES revealed evidence of clonal evolution (Fig. 4). At diagnosis, the ALK mutation was attributed to the parental clone in all cases but one. In patient 7, the ALK mutation was identified in a subclone at diagnosis. In all cases, first-line treatment by chemotherapy led to a decrease of subclones seen at diagnosis. Importantly, following ALK-targeted treatment with lorlatinib, emergence of new SNVs, corresponding to an emergence of new subclones, was observed in all cases but one. Emerging subclones following lorlatinib treatment harbored SNVs in genes including SMARCA4, EGFR, and TP53 (patient 17; Fig. 4), an alteration in CDKN2A (patient 16), or PIK3CA (patient 18), representing a CCF of at least 0.3 in all cases. In patient 18 having received lorlatinib over a long period of time (>1,100 days), a new ALK mutation I1171N previously described as a resistance mutation to ALK-targeted treatment was also detected (Supplementary Fig. S4). Only in one case was no new subclone identified following lorlatinib treatment (patient 6), but the last follow-up plasma was obtained after a short period of 49 days following lorlatinib treatment. However, in this patient, nearly complete eradication of a subclone harboring among others an SNV of MAP3K19 was observed following chemotherapy, with the emergence of a new subclone following temozolomide treatment (Fig. 4).
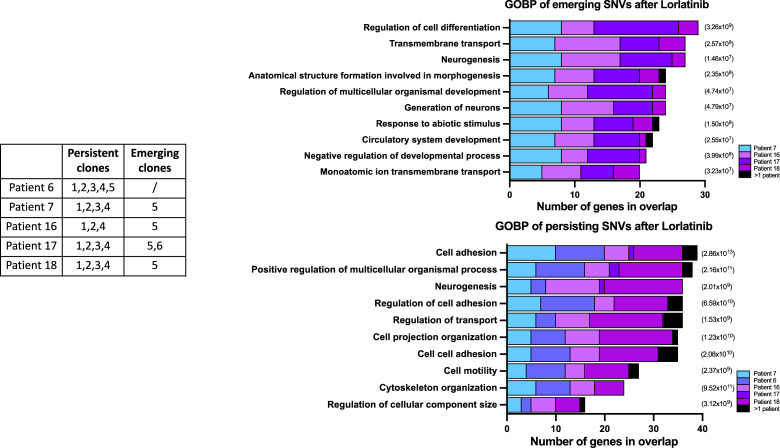
No SNVs targeting a single gene or gene family could be identified in the emerging or persistent subclones. The SNVs of emerging or persistent subclones were submitted to a gene ontology and biological pathway analysis to search for cellular pathways and cellular components impacted by these SNVs (Fig. 5). In the subclones emerging after lorlatinib treatment, an enrichment for SNVs targeting genes that play a role in the regulation of cell differentiation, neurogenesis, and transmembrane transport could be documented. In the persisting subclones, an enrichment for SNVs targeting genes of cell adhesion or multicellular organismal processes was identified.
Figure 5.
Top enriched gene sets after Gene Set Enrichment Analysis (GSEA) (http://www.gsea-msigdb.org/gsea/index.jsp, ontology gene sets C5—biological processes) for emerging (top) and persisting (bottom) SNVs after lorlatinib treatment. Depicted is the number of genes in the ontology overlap (x-axis), colored by patient contribution, and the P-value significance indicated (right).
Alterations possibly linked to clonal hematopoiesis were observed in cfDNA samples during follow-up, in genes such as DNMT3B or SETDB1 in patient 18, or BCOR or JAK3 in patient 16, with a VAF remaining below 2% in the cfDNA (43). Although no whole blood cell (WBC) pellet was available to provide proof of the hematologic origin of these alterations in patient 18, an enrichment of the VAF of BCOR and JAK3 in the WBC pellet obtained after treatment was observed for patient 16, strongly suggesting clonal hematopoiesis.
Discussion
Neuroblastoma is characterized by a mutational burden at diagnosis, with an enrichment of targetable alterations at relapse (10, 44). The most frequent targetable alteration is ALK, with genomic amplification or activating mutations seen in approximately 15% of patients at diagnosis at a clonal level, but mutations also occur subclonally and/or emerge at relapse (10, 11, 18, 45). Interesting ORRs to third-generation ALK inhibitors upon relapse have now been reported, and these treatment approaches are now proposed in frontline treatment strategies for patients with ALK-altered high-risk neuroblastoma (15, 16). However, secondary resistance to ALK-targeted treatment frequently occurs.
Our real-world data collected within the prospective SACHA-France study confirm the significant and long-lasting antitumor activity of lorlatinib in a subset of patients. In our study, 6/18 patients had an objective response, and in three patients, this antitumor response was prolonged (>12 months). Nevertheless, only one patient remains with controlled disease after 2 years of lorlatinib single-agent therapy.
Based on our small series, no differences in ORR were observed when lorlatinib was given as a single agent or in combination with chemotherapy. Although not randomized, the NANT2015-02 study showed higher ORR following lorlatinib combined with chemotherapy (16). We show that MYCN amplification correlates with poorer outcomes in this population, with no response to lorlatinib among eight patients with MYCN-amplified tumors. We confirm the favorable toxicity profile of lorlatinib. Nevertheless, more prospective information is needed, mainly when lorlatinib is combined with other chemotherapy backbones/immunotherapy or after high-dose chemotherapy with autologous stem cell transplantation due to the risk of potential overlapping toxicities.
cfDNA from blood or bone marrow has become an important surrogate for sequential analyses of tumor-specific genetic alterations. Previous studies have shown high overall cfDNA concentrations, and high ctDNA fractions in high-risk patients with neuroblastoma (18, 25, 28, 29). cfDNA might be used as a marker of treatment response based on the ALK VAF and also for wider analyses to elucidate genetic alterations upon resistance. Thus, it is crucial to combine highly sensitive detection of ALK alterations with an overview of all genetic alterations in cfDNA samples, which are frequently limited in quantity.
We applied a combination of ddPCR and WES to study cfDNA in five patients with ALK-altered high-risk neuroblastoma having received lorlatinib for relapse, confirming sensitivities of 0.1% for ddPCR and 1% for WES (24, 25). We validated the use of gDNA WGS libraries to perform ddPCR and capture WES analysis to enable a combined approach on frequently limited amounts of cfDNA. No statistically significant difference of the ALK VAF determined by ddPCR in gDNA versus postlibrary WGS construction versus WES capture of the WGS libraries could be seen. Nonetheless, due to the higher quantities of WGS libraries (200–300 ng/μL) than the WES captures (1–2 ng/μL), preference was given to ddPCR on the more abundant WGS libraries for further clinical routine development.
ddPCR requires mutation-specific set-up. Our designs targets four of the most frequent ALK mutations in neuroblastoma and will be useful for cfDNA analysis for patients with these mutations; however, our design does not target other rarer mutations or CNVs. Conversely, besides calling SNVs, WES might also enable the detection of CNVs, in particular in case of ctDNA content >0.3 (28).
As reported previously, a correlation between cfDNA ddPCR ALK VAFs and disease status could be documented. We provide further evidence that cfDNA analysis can be more sensitive than clinical imaging for the detection of disease progression (18, 25). Future prospective trials integrating cfDNA with highly sensitive analytic techniques are needed to determine the benefit to patients.
Sequential analysis by cfDNA WES in five patients treated with lorlatinib led to important novel insights. Clonal evolution occurred in all patients, with a significant increase of SNVs at relapse (10, 18, 44). Our findings further underline the importance of tracking overall mutational burden. Future prospective studies will determine whether this increase is a stochastical event or whether SNVs might emerge following certain chemotherapy treatments such as temozolomide.
Secondary compound mutations in ALK acquired at disease progression have recently been reported in neuroblastoma (18). We identified ALK resistance mutation (I1171N) in an adult neuroblastoma patient, >1,000 days after the beginning of lorlatinib treatment (17). This resistance mutation progressively increased to >0.5 CCF. ALK resistance mutations have been described in adult patients with NSCLC undergoing ALK-targeted treatment, in which ALK is activated by gene fusion. Long-read sequencing of nonfragmented DNA of tumor tissue would enable us to establish whether this resistance mutation occurs in cis or trans relative to the initially mutated allele (18).
Our results confirm spatial genetic heterogeneity and furthermore indicate that important clonal evolutionary dynamics exist in high-risk neuroblastoma (44–46). In all cases, subclones could be identified, which diminished with initial chemotherapy, suggesting subclones sensitive to chemotherapy. Furthermore, emerging subclones represent those resistant to previous treatment. In neuroblastoma, emergence of RAS-MAPK pathway mutations has been reported at disease progression or following treatment by lorlatinib (18, 44). Although no such alterations were observed in this study, other alterations in genes linked to disease progression or representing potential therapeutic targets such as EGFR or PIK3CA were identified. Clones emerging after lorlatinib treatment also harbored alterations in genes that might play a role in neuroblastoma oncogenesis such as SMARCA4, or which play a role in epithelial–mesenchymal transition and cell growth such as HAND2. The observation of clonal dynamics highlights the importance of identifying targetable molecular alterations to identify possibilities of combination therapies in case of resistance to third- or fourth-generation ALK inhibitors. Our findings also suggest that an overall accumulation of SNVs could be linked to higher aggressivity, even without specific resistance mutations. Further studies will enable the distinction of resistance-driving SNVs from drivers of aggressive relapse.
Our approach also for the first time provides evidence of the occurrence of clonal hematopoiesis (43). Further studies will enable us to determine the potential role or contribution of clonal hematopoiesis to tumor progression.
In conclusion, our real-world data confirm the antitumor activity and favorable safety profile of lorlatinib in patients with relapsed neuroblastoma. Sequential analysis of cfDNA combining ddPCR and WES highlights their potential for disease monitoring, with a potential to detect relapse earlier than clinical evaluation. The emergence of subclones after ALK-targeted treatment underlines the importance of developing new treatment strategies and treatment combinations in case of progression after ALK-targeted treatment.
Supplementary Material
SupplemetaryFigure12345
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table Legend
Acknowledgments
SACHA-France study is supported by the French National Agency for the Safety of Medicine and Health Products and the Société Française de lutte contre les Cancers et les leucémies de l’Enfant et l’adolescent (SFCE). SACHA-France is funded by Imagine for Margo, Association Hubert Gouin Enfance et Cancer, and Fondation des Entreprises du Medicament. The biological analyses were supported by the Annenberg Foundation, the Association Hubert Gouin Enfance et Cancer, the Fédération Enfants Cancers Santé, the Société Française de lutte contre les Cancers et les leucémies de l’Enfant et l’adolescent (SFCE), Imagine For Margo, Les Bagouz à Manon, Les amis de Claire, and the Fondation ARC pour la Recherche contre le Cancer (ARC). Funding was also obtained from SiRIC/INCa (Grant INCa-DGOS-4654), INCa grant PHRC IC2007-09, and INCa grant PRTK 2019-1-RT-02-ICR-1. High-throughput sequencing was performed by the ICGex NGS platform of the Institut Curie supported by the grants ANR-10-EQPX-03 (Equipex) and ANR-10-INBS-09-08 (France Génomique Consortium) from the Agence Nationale de la Recherche (“Investissements d’Avenir” program), by the Canceropole Ile-de-France and by the SiRIC-Curie program—SiRIC Grant “INCa-DGOS- 4654.” Some tumor sequencing data were provided from the MAPPYACTS (NCT02613962) or the MICCHADO studies (NCT03496402). Support was also provided by the Joint Transnational Call for Proposals 2017 (JTC 2017): “Translational research on rare cancers” (project entitled LIQUIDHOPE) and by the NBUK (registered Charity No 326385) and the Friends of Rosie (registered Charity No 1046278; project entitled “Blood-based Biomarker Testing to Guide the Diagnosis and Treatment of Neuroblastoma Patients”). This work has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska–Curie grant agreement, No. 956285 (VAGABOND). The authors thank Pfizer for providing lorlatinib for compassionate use for patients with ALK-altered progressive/relapsed neuroblastoma.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
C. Butterworth reports grants from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement, No. 956285—VAGABOND ITN, during the conduct of the study. P. Berlanga reports other support from EUSA Pharma and grants from Bayer outside the submitted work. G. Schleiermacher reports grants from Annenberg Foundation, Association Hubert Gouin Enfance et Cancer, Fédération Enfants Cancers Santé, Imagine For Margo, Fondation ARC pour la Recherche contre le Cancer, Les Bagouz à Manon, PHRC IC2007-09, PRTK 18-035, Joint Transnational Call for Proposals 2017 (JTC 2017): “Translational research on rare cancers,” and NBUK (registered Charity No. 326385) and the Friends of Rosie (registered Charity No. 1046278) during the conduct of the study, as well as grants from Roche, MSDavenir, BMS, and Pfizer outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
C. Bobin: Resources, formal analysis, investigation, visualization, methodology, writing—original draft, contributed equally. Y. Iddir: Formal analysis, validation, investigation, visualization, methodology. C. Butterworth: Data curation, software, formal analysis, investigation, visualization, methodology. J. Masliah-Planchon: Conceptualization, resources, data curation, validation, methodology, writing—review and editing. A. Saint-Charles: Conceptualization, resources, data curation, methodology, project administration, writing—review and editing. A. Bellini: Investigation, methodology, writing—review and editing. J. Bhalshankar: Software, writing—review and editing. G. Pierron: Resources, data curation, formal analysis, writing—review and editing. V. Combaret: Conceptualization, resources, data curation, formal analysis, writing–review and editing. V. Attignon: Formal analysis, writing—review and editing. N. André: Resources, writing—review and editing. N. Corradini: Resources, writing—review and editing. B. Dumont: Resources, writing—review and editing. L. Mansuy: Resources, writing—review and editing. C. Khanfar: Resources, writing—review and editing. S. Klein: Resources, writing—review and editing. C. Briandet: Resources, writing—review and editing. D. Plantaz: Resources, writing—review and editing. F. Millot: Resources, writing—review and editing. S. Thouvenin: Resources, writing—review and editing. I. Aerts: Resources, writing—review and editing. L.A. Ndounga-Diakou: Resources, formal analysis, writing—review and editing. S. Laghouati: Resources, formal analysis, writing—review and editing. S. Abbou: Resources, data curation, writing—review and editing. N. Jehanno: Resources, writing—review and editing. H. Tissot: Resources, formal analysis, writing—review and editing. S. Renault: Resources, data curation, formal analysis, validation, writing—review and editing. S. Baulande: Resources, data curation, project administration, writing—review and editing. V. Raynal: Resources, formal analysis. L. Bozec: Resources, writing—review and editing. I. Bieche: Resources, data curation, formal analysis, writing—review and editing. O. Delattre: Conceptualization, resources, data curation, writing—review and editing. P. Berlanga: Conceptualization, resources, data curation, formal analysis, supervision, writing–original draft, project administration, writing—review and editing. G. Schleiermacher: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing—original draft, project administration, writing–review and editing.
References
- 1. Cohn SL, Pearson ADJ, London WB, Monclair T, Ambros PF, Brodeur GM, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG task force report. J Clin Oncol 2009;27:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Primers 2016;2:16078. [DOI] [PubMed] [Google Scholar]
- 3. Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7. [DOI] [PubMed] [Google Scholar]
- 4. Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013;45:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bellini A, Bernard V, Leroy Q, Rio Frio T, Pierron G, Combaret V, et al. Deep sequencing reveals occurrence of subclonal ALK mutations in neuroblastoma at diagnosis. Clin Cancer Res 2015;21:4913–21. [DOI] [PubMed] [Google Scholar]
- 6. Bellini A, Pötschger U, Bernard V, Lapouble E, Baulande S, Ambros PF, et al. Frequency and prognostic impact of ALK amplifications and mutations in the European neuroblastoma study group (SIOPEN) high-risk neuroblastoma trial (HR-NBL1). J Clin Oncol 2021;39:3377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 2008;455:967–70. [DOI] [PubMed] [Google Scholar]
- 8. Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008;455:930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. George RE, Sanda T, Hanna M, Fröhling S, Luther W, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 2008;455:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Padovan-Merhar OM, Raman P, Ostrovnaya I, Kalletla K, Rubnitz KR, Sanford EM, et al. Enrichment of targetable mutations in the relapsed neuroblastoma genome. PLoS Genet 2016;12:e1006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schleiermacher G, Javanmardi N, Bernard V, Leroy Q, Cappo J, Rio Frio T, et al. Emergence of new ALK mutations at relapse of neuroblastoma. J Clin Oncol 2014;32:2727–34. [DOI] [PubMed] [Google Scholar]
- 12. Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell 2014;26:682–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen L, Humphreys A, Turnbull L, Bellini A, Schleiermacher G, Salwen H, et al. Identification of different ALK mutations in a pair of neuroblastoma cell lines established at diagnosis and relapse. Oncotarget 2016;7:87301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guan J, Tucker ER, Wan H, Chand D, Danielson LS, Ruuth K, et al. The ALK inhibitor PF-06463922 is effective as a single agent in neuroblastoma driven by expression of ALK and MYCN. Dis Model Mech 2016;9:941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer M, Moreno L, Ziegler DS, Marshall LV, Zwaan CM, Irwin MS, et al. Ceritinib in paediatric patients with anaplastic lymphoma kinase-positive malignancies: an open-label, multicentre, phase 1, dose-escalation and dose-expansion study. Lancet Oncol 2021;22:1764–76. [DOI] [PubMed] [Google Scholar]
- 16. Goldsmith KC, Park JR, Kayser K, Malvar J, Chi YY, Groshen SG, et al. Lorlatinib with or without chemotherapy in ALK-driven refractory/relapsed neuroblastoma: phase 1 trial results. Nat Med 2023;29:1092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vasseur A, Cabel L, Geiss R, Schleiermacher G, Pierron G, Kamal M, et al. Efficacy of Lorlatinib in primary Crizotinib-resistant adult neuroblastoma harboring ALK Y1278S mutation. JCO Precis Oncol 2019;3:1–5. [DOI] [PubMed] [Google Scholar]
- 18. Berko ER, Witek GM, Matkar S, Petrova ZO, Wu MA, Smith CM, et al. Circulating tumor DNA reveals mechanisms of Lorlatinib resistance in patients with relapsed/refractory ALK-driven neuroblastoma. Nat Commun 2023;14:2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Durand S, Pierre-Eugene C, Mirabeau O, Louis-Brennetot C, Combaret V, Colmet-Daage L, et al. ALK mutation dynamics and clonal evolution in a neuroblastoma model exhibiting two ALK mutations. Oncotarget 2019;10:4937–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang HY, Ho CC, Shih JY. Multiple acquired resistance mutations of the ALK tyrosine kinase domain after sequential use of ALK inhibitors. J Thorac Oncol 2017;12:e49–51. [DOI] [PubMed] [Google Scholar]
- 21. Yoda S, Lin JJ, Lawrence MS, Burke BJ, Friboulet L, Langenbucher A, et al. Sequential ALK inhibitors can select for Lorlatinib-resistant compound ALK mutations in ALK-positive lung cancer. Cancer Discov 2018;8:714–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426–37. [DOI] [PubMed] [Google Scholar]
- 23. Van Paemel R, Vlug R, De Preter K, Van Roy N, Speleman F, Willems L, et al. The pitfalls and promise of liquid biopsies for diagnosing and treating solid tumors in children: a review. Eur J Pediatr 2020;179:191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Combaret V, Iacono I, Bellini A, Bréjon S, Bernard V, Marabelle A, et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med 2015;4:540–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lodrini M, Sprussel A, Astrahantseff K, Tiburtius D, Konschak R, Lode HN, et al. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget 2017;8:85234–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kahana-Edwin S, Cain LE, McCowage G, Darmanian A, Wright D, Mullins A, et al. Neuroblastoma molecular risk-stratification of DNA copy number and ALK genotyping via cell-free circulating tumor DNA profiling. Cancers (Basel) 2021;13:3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peitz C, Sprussel A, Linke RB, Astrahantseff K, Grimaldi M, Schmelz K, et al. Multiplexed quantification of four neuroblastoma DNA targets in a single droplet digital PCR reaction. J Mol Diagn 2020;22:1309–23. [DOI] [PubMed] [Google Scholar]
- 28. Chicard M, Boyault S, Colmet Daage L, Richer W, Gentien D, Pierron G, et al. Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin Cancer Res 2016;22:5564–73. [DOI] [PubMed] [Google Scholar]
- 29. Chicard M, Colmet-Daage L, Clement N, Danzon A, Bohec M, Bernard V, et al. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res 2018;24:939–49. [DOI] [PubMed] [Google Scholar]
- 30. Van Paemel R, Vandeputte C, Raman L, Van Thorre J, Willems L, Van Dorpe J, et al. The feasibility of using liquid biopsies as a complementary assay for copy number aberration profiling in routinely collected paediatric cancer patient samples. Eur J Cancer 2022;160:12–23. [DOI] [PubMed] [Google Scholar]
- 31. Ladenstein R, Potschger U, Pearson ADJ, Brock P, Luksch R, Castel V, et al. Busulfan and melphalan versus carboplatin, etoposide, and melphalan as high-dose chemotherapy for high-risk neuroblastoma (HR-NBL1/SIOPEN): an international, randomised, multi-arm, open-label, phase 3 trial. Lancet Oncol 2017;18:500–14. [DOI] [PubMed] [Google Scholar]
- 32. Berlanga P, Ndounga-Diakou LA, Aerts I, Corradini N, Ducassou S, Strullu M, et al. Measuring safety and outcomes for the use of compassionate and off-label therapies for children, adolescents, and young adults with cancer in the SACHA-France study. JAMA Netw Open 2023;6:e2321568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park JR, Bagatell R, Cohn SL, Pearson AD, Villablanca JG, Berthold F, et al. Revisions to the international neuroblastoma response criteria: a consensus statement from the national cancer institute clinical trials planning meeting. J Clin Oncol 2017;35:2580–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berlanga P, Pierron G, Lacroix L, Chicard M, Adam de Beaumais T, Marchais A, et al. The European MAPPYACTS trial: precision medicine program in pediatric and adolescent patients with recurrent malignancies. Cancer Discov 2022;12:1266–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thirant C, Peltier A, Durand S, Kramdi A, Louis-Brennetot C, Pierre-Eugène C, et al. Reversible transitions between noradrenergic and mesenchymal tumor identities define cell plasticity in neuroblastoma. Nat Commun 2023;14:2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tucker ER, Jimenez I, Chen L, Bellini A, Gorrini C, Calton E, et al. Combination therapies targeting ALK-aberrant neuroblastoma in preclinical models. Clin Cancer Res 2023;29:1317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet 2012;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arora A, Shen R, Seshan VE. FACETS: fraction and allele-specific copy number estimates from tumor sequencing. Methods Mol Biol 2022;2493:89–105. [DOI] [PubMed] [Google Scholar]
- 39. Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dang HX, White BS, Foltz SM, Miller CA, Luo J, Fields RC, et al. ClonEvol: clonal ordering and visualization in cancer sequencing. Ann Oncol 2017;28:3076–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gillis S, Roth A. PyClone-VI: scalable inference of clonal population structures using whole genome data. BMC Bioinformatics 2020;21:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller CA, McMichael J, Dang HX, Maher CA, Ding L, Ley TJ, et al. Visualizing tumor evolution with the fishplot package for R. BMC Genomics 2016;17:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Buttigieg MM, Rauh MJ. Clonal hematopoiesis: updates and implications at the solid tumor-immune interface. JCO Precis Oncol 2023;7:e2300132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eleveld TF, Oldridge DA, Bernard V, Koster J, Colmet Daage L, Diskin SJ, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet 2015;47:864–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosswog C, Fassunke J, Ernst A, Schömig-Markiefka B, Merkelbach-Bruse S, Bartenhagen C, et al. Genomic ALK alterations in primary and relapsed neuroblastoma. Br J Cancer 2023;128:1559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bosse KR, Giudice AM, Lane MV, McIntyre B, Schürch PM, Pascual-Pasto G, et al. Serial profiling of circulating tumor DNA identifies dynamic evolution of clinically actionable genomic alterations in high-risk neuroblastoma. Cancer Discov 2022;12:2800–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SupplemetaryFigure12345
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
Supplementary Table 4
Supplementary Table Legend
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Raw data for this study were generated at the ICGex Sequencing Platform of Institut Curie, Paris, France. Raw data supporting the findings of this study are available from the corresponding author upon request.