Abstract
Tomato bushy stunt virus (TBSV) is a positive-strand RNA virus and is the prototype member of the genus Tombusvirus. The genomes of members of this genus are not polyadenylated, and prevailing evidence supports the absence of a 5′ cap structure. Previously, a 167-nucleotide-long segment (region 3.5) located near the 3′ terminus of the TBSV genome was implicated as a determinant of translational efficiency (S.K. Oster, B. Wu and K. A. White, J. Virol. 72:5845–5851, 1998). In the present report, we provide evidence that a 3′-proximal segment of the genome, which includes region 3.5, is involved in facilitating cap-independent translation. Our results indicate that (i) a 5′ cap structure can substitute functionally for the absence of region 3.5 in viral and chimeric reporter mRNAs in vivo; (ii) deletion of region 3.5 from viral and chimeric mRNAs has no appreciable effect on message stability; (iii) region 3.5 represents part of a larger 3′ proximal element, designated as the 3′ cap-independent translational enhancer (3′CITE), that is required for proficient cap-independent translation; (iv) the 3′CITE also facilitates cap-dependent translation; (v) none of the major viral proteins are required for 3′CITE activity; and (vi) no significant 3′CITE-dependent stimulation of translation was observed when mRNAs were tested in vitro in wheat germ extract under various assay conditions. This latter property distinguishes the 3′CITE from other characterized plant viral 3′-proximal cap-independent translational enhancers. Additionally, because the 3′CITE overlaps with cis-acting replication signals, it could potentially participate in regulating the initiation of genome replication.
Initiation of translation in eukaryotic cells involves various interactions between cis-acting signals in mRNAs and trans-acting factors (5, 24, 30). The 5′ cap and poly(A) tail represent general structures in mRNAs that contribute to translational regulation (11, 13, 25, 37). Similar to cellular messages, eukaryotic viral mRNAs contain elements which facilitate efficient and/or regulated synthesis of viral proteins in host cells (12, 18). Many plant positive-strand RNA viruses have adapted their genome structures to include a 5′ cap structure and/or a poly(A) tail; however, others do not contain either of these modifications (3, 12). In the absence of traditional regulatory elements, viruses are forced to adopt alternative strategies to ensure satisfactory levels of translation of their encoded products (3, 12).
Cap-independent translation has been described for a number of positive-strand RNA plant viruses (12), satellite tobacco necrosis virus (STNV) and barley yellow dwarf virus (BYDV) representing two of the most extensively studied examples (7, 38, 40). The STNV and BYDV genomes are able to efficiently express their encoded viral products even though they lack both a 5′ cap and a poly(A) tail (1, 7, 38–40). Efficient translation in vitro of the STNV genome requires a 120-nucleotide (nt)-long sequence, the translation enhancer domain, located in its 3′ untranslated region (UTR), whereas for the BYDV genome, a 109-nt-long sequence located at a 3′-proximal position, the 3′ translational enhancer (3′TE), is necessary (7, 38, 39). Both of these elements stimulate translation in vitro to a level similar to those for capped messages (22, 39). In vivo, similar stimulatory effects are observed (22, 39); however, for BYDV a 3′ sequence larger than that defined in vitro is required (39).
Tomato bushy stunt virus (TBSV) is the prototype member of the genus Tombusvirus. Its 4.8-kb-long positive-strand RNA genome encodes five open reading frames (ORFs) of known function, with a sixth small 3′-proximal ORF of unknown function (encoding pX) located near its 3′ terminus (Fig. 1A) (4, 16, 31). The genomes of members of this genus are not polyadenylated (29) and, based on the analysis of the tombusvirus carnation Italian ringspot virus (28, 29), are predicted to lack a 5′ cap structure. This latter concept is further supported by the finding that uncapped in vitro-generated transcripts of different tombusvirus genomes are as infectious as corresponding native virion-derived genomes (16, 29). In TBSV, the 5′-encoded p33 and p92 products are translated directly from the genome, and both are required for replication of viral RNAs (Fig. 1) (23, 33). The ORFs positioned more 3′ in the genome encode encapsidation- and movement-related proteins (32), and these products are translated from two subgenomic mRNAs that are synthesized in a regulated manner during infections (45).
FIG. 1.
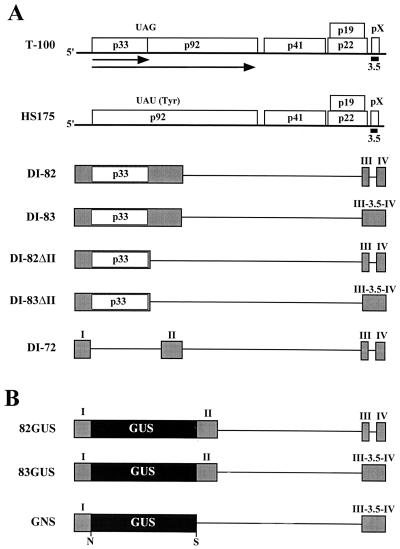
(A) Schematic representation of TBSV genome and various defective viral RNAs. The wild-type TBSV genome (T-100) is shown at the top as a thick horizontal line, with coding regions depicted as boxes which include the approximate molecular masses (in thousands) of the encoded proteins (16). The translation product p33 and its readthrough product p92 are presented as arrows below the genome, and the approximate position of region 3.5 is shown as a small black box near the 3′ end of the genome (labeled 3.5). HS175 is a mutant of the genome in which the p33 termination codon has been replaced by a tyrosine codon so as to allow for expression of p92 but not p33 (33). Below, various smaller defective viral RNAs are depicted; shaded boxes correspond to regions of the genome retained in these molecules, whereas thin horizontal lines correspond to genomic segments which are absent. DI-82 and DI-83 both encode p33 (open box) and are identical except that region 3.5 is absent in DI-82 (42). DI-82ΔII and DI-83ΔII are derivatives of DI-82 and DI-83, respectively, in which a segment, which includes region II, is deleted. DI-72 represents a prototypical DI RNA and is composed of four noncontiguous regions of the genome (i.e., regions I through IV) (42). (B) Schematic representation of GUS-virus hybrid mRNAs with the GUS coding region depicted as a black box. 82GUS and 83GUS are identical except for the absence of region 3.5 in the former. GNS is a derivative of 83GUS which lacks region II and contains introduced NcoI (N) and SacI (S) sites.
Previous studies implicated a 3′-proximal segment in the TBSV genome (region 3.5; Fig. 1) as a determinant of translation (23). In the present study, we show that region 3.5 is a key component of a larger 3′-terminal portion of the genome which functions as a cap-independent translational enhancer. The relevance of this finding is discussed in relation to viral translation and replication.
MATERIALS AND METHODS
Oligonucleotides used in this study.
The oligonucleotides used were as follows (underlined and nonunderlined residues correspond to nonviral and viral sequences, respectively): P9, 5′GGCGGCCCGCATGCCCGGGCTGCATTTCTGCAATGTTCC (TBSV, minus sense, 4754 to 4776); P25, 5′GGCCCTCTAGACAGATTTACACTCATCTCCAC (TBSV, minus sense, 4430 to 4450); P49, 5′CCTAACATCCAGAACCCAACAAGAG (TBSV, minus sense, 4464 to 4492); PB21, 5′GAACTAGGTCGAGAAATCCTGGAGAATTTC (TBSV, minus sense, 1 to 30); PB32, 5′GTAACGACGCCAGTGAATTGGTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTCG (TBSV, plus sense, 1 to 21); PB33, 5′TAAGCTTACCATGGTCGCTTGTTTGTTGGAAG (TBSV, minus sense, 147 to 169 with one G insertion between 165 and 166 to generate NcoI site); PB34, 5′GGCGGCGAGCTCAGCGAGTAAGACAGACTCTTC (TBSV plus sense, 4398 to 4419); PB39, 5′GGGTGAGATCAACCGTGTCTGGGG (TBSV, minus sense, 4687 to 4707); PB40, 5′GGGCCAATTCATGCTGGCTGGTATTGC (TBSV, minus sense, 4622 to 4646); PB41, 5′GGGGAGACCCTTTCCATATCTCCATCC (TBSV, minus sense, 4536 to 4560); PB42, 5′GGGACCCAACAAGAGTAACCTGTATGC (TBSV, minus sense, 4455 to 4479); PB43, 5′GGCGGCGAGCTCTGTAAATCTGGCATAGCATACAGG (TBSV, plus sense, 4441 to 4464); PB44, 5′GGCGGCGAGCTCTCTGGATGTTAGGATGACGAG (TBSV, plus sense, 4480 to 4500); PB45, 5′GGCGGCGAGCTCGTGTGGTATCAGTCGGTCGAAG (TBSV, plus sense, 4561 to 4583); PB46, 5′GGCGGCGAGCTCATTCCTGTTTACGAAAGTTAGG (TBSV, plus sense, 4647 to 4668); PGUS3, 5′TCATTGTTTGCCTCCCTGCTGCGGTTTTTCACCG (GUS, minus sense, 1779 to 1812).
Viral constructs.
Plasmid construct T-100, containing cDNA corresponding to the full-length viral genome of TBSV, has been described previously (16). Defective interfering (DI) RNA clones DI-82, DI-83, and DI-72 are deletion derivatives of T-100 (42). HS175, PGUS1162, and PGUS1162BF have been described previously (33, 39).
DI-82ΔII was constructed by removing a 3′ viral fragment containing regions III and IV from clone RTD-22 (43) by digestion with SfiI (the termini of which were subsequently rendered blunt) and SphI. DI-83 was digested with StuI and SphI, and the small 3′ viral fragment released was replaced with that derived from RTD-22, thereby generating DI-82ΔII. DI-83ΔII was constructed in a similar manner except that the 3′ viral fragment used for replacement contained region 3.5 and was derived from clone RTD-23 (43). DI-82GUS was constructed by ligating the SalI/NcoI fragment from pAGUS1-TN2 (34) containing the β-glucuronidase (GUS) ORF with an XbaI-digested derivative of DI-72XP (26), DI-72XP(-AUG). The termini of both fragments were rendered blunt prior to ligation. DI-83GUS was generated by digesting DI-83 with BstXI and replacing the smaller fragment released with the corresponding GUS-containing fragment from BstXI-digested DI-82GUS. GNS was constructed by digesting 82GUS with BstXI and SphI and ligating the larger fragment with the following: (i) and BstXI/NcoI-digested PCR product generated with oligonucleotides P32 and P33, using 83GUS as the template; (ii) an SacI/SphI-digested PCR product generated with oligonucleotides PB34 and P9, using 83GUS as the template; and (iii) pAGUS1-TN2 digested with NcoI and SacI. Plasmids GNST1 to GNST15 were generated by the insertion of different PCR products (see below) into SacI- and SmaI-digested GNS plasmid. The template used for the PCRs described below was 83GUS. The construct name, the primer pairs used to generate the PCR product, and the restriction enzyme(s) used to digest the PCR product are as follows: GNST1, PB34-PB39, and SmaI; GNST4, PB34-PB40, and SmaI; GNST5, PB34-PB41, and SmaI; GNST7, PB34-PB42, and SmaI; GNST9, PB43-P9, and SacI/SmaI; GNST12, PB44-P9, and SacI/SmaI; GNST13, PB45-P9, and SacI/SmaI; and GNST15, PB46-P9, and SacI/SmaI.
In vitro transcription.
Viral transcripts were synthesized in vitro by transcription of SmaI-linearized template DNAs, using an Ampliscribe T7 RNA polymerase transcription kit (Epicentre Technologies) as described previously (23). Capped transcripts were prepared with the recommended 10:1 ratio of cap analog [m7G(5′)ppp(5′)G; New England Biolabs] to GTP.
Isolation and inoculation of protoplasts.
Protoplasts were prepared from 6- to 8-day-old cucumber cotyledons (var. Straight 8) as described previously (42). Purified protoplasts were inoculated with viral transcripts by using the polyethylene glycol (PEG)-CaCl2 method described previously (42), and virus-GUS hybrid transcripts were introduced into protoplasts via electroporation (10). Approximately 5 μg of each transcript was used (except for DI-72, where 1 μg was used) in PEG-CaCl2-mediated inoculations. For electroporation, 106 protoplasts and 24 pmol of RNA transcript were suspended in 200 μl of electroporation solution (10), and samples were treated with one pulse at 150 V and 500-μF capacitance in 0.4-cm cuvette in a Gene Pulser (Bio-Rad). Incubation medium (1 ml) (42) was added to the electroporated protoplasts, which were then incubated at 22°C in growth chambers under fluorescent lighting.
Analysis of RNAs from protoplasts.
Total nucleic acid was harvested from protoplasts as described previously (42). Aliquots of the total nucleic acid preparation were separated in nondenaturing 1.4% agarose gels, and viral RNAs were detected by electrophoretic transfer to nylon (Hybond-N; Amersham) followed by Northern blot analysis using complementary 32P-end-labeled oligonucleotides. For RNA stability assays, RNAs were introduced into protoplasts by either PEG-CaCl2 treatment (DI-82ΔII, DI-83ΔII, or cDI-82ΔII [capped transcripts are designated by the prefix “c”] or electroporation (virus-GUS hybrid transcripts), and a fraction of the protoplasts were isolated at various times postinoculation. Total nucleic acids were prepared and analyzed by Northern blot analysis. For stability experiments, RNase A was added to the incubation medium at a final concentration of 30 μg/ml to degrade any RNA remaining outside the protoplasts following inoculation (26).
GUS assays.
GUS activity was quantified by a standard protocol (20). Briefly, 250 μl of GUS extraction buffer (20) was added to cucumber protoplasts isolated at various times postinoculation; the mixture was vortexed for 30 s and then centrifuged at 12,000 × g for 5 min. The supernatant containing soluble GUS protein was transferred to a new tube, and fluorogenic assays were performed (20). Values derived were standardized with respect to total protein content as determined by using a Bradford analysis kit (Bio-Rad); where indicated, the background values for mock-inoculated samples were subtracted.
In vitro translation and RNA analysis.
Translation of various viral and hybrid transcripts were carried out in nuclease-treated wheat germ extract under conditions suggested by the manufacturer (Promega). Reactions were carried out for 60 min in the presence of [35S]methionine (1000 Ci/mmol), using empirically determined subsaturating concentrations of transcripts. Products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were quantified by radioanalytical scanning of dried gels with an InstantImager (Packard Instrument Co.). For RNA stability assays, reactions were performed with nonradioisotopic methionine. Aliquots were removed from the reaction at specified time intervals, and the RNA was analyzed by Northern blotting.
RESULTS
A 5′ cap structure can substitute functionally for the absence of region 3.5 in viral mRNAs.
Our earlier studies of TBSV using a trans-expression system that allows for the independent synthesis of viral RNA replication proteins p33 and p92 suggested that the 3′ region of the genome may harbor a determinant for efficient translation (23). Using this system, we observed that amplification of a viral replicon corresponding to a DI RNA, DI-72 (Fig. 1), occurred in coinoculations that included an uncapped p92-encoding viral mRNA, HS175 (Fig. 1), and an uncapped p33-encoding viral mRNA, DI-83 (Fig. 1) (23). As both p92 and p33 are necessary for viral RNA replication, the amplification of DI-72 confirmed the expression of these proteins from HS175 and DI-83, respectively. In contrast, when DI-72 was coinoculated with uncapped HS175 and uncapped DI-82 (which is identical in structure to DI-83 except for a 3′-proximal deletion corresponding to region 3.5 [Fig. 1]), no DI-72 amplification was observed (23). However, when this coinoculation was repeated with capped DI-82 transcripts, the replicon was amplified (23). The ability to rescue the functional activity of DI-82 via capping implicated region 3.5 as a determinant of cap-independent translation.
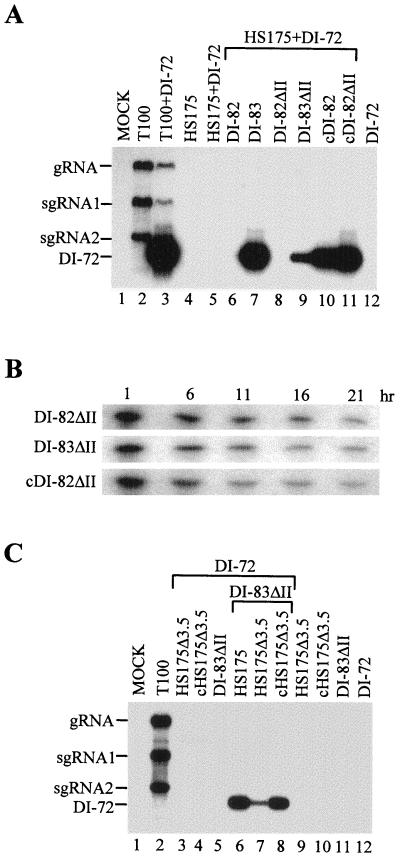
Our initial trans-expression assay used DI-82 and DI-83 for expression of p33. An undesirable feature of these mRNAs is that they are both replicable, a property which could influence the abundance of the messages and/or lead to alternatively structured progeny. To eliminate this variable, we prepared derivatives of these viral messages, DI-82ΔII and DI-83ΔII, which were rendered replication defective by the removal of an essential cis-acting replication element (i.e., region II [Fig. 1]). Coinoculation of protoplasts with genome transcripts and either DI-82ΔII or DI-83ΔII resulted in no detectable accumulation of progeny derived from the modified viral messages (data not shown); therefore, these viral RNAs act principally as mRNAs. Coinoculation of protoplasts with HS175, DI-72 and either DI-82ΔII or DI-83ΔII resulted in DI-72 amplification only in the coinoculation including DI-83ΔII (Fig. 2A, lanes 8 and 9). The level of amplification observed was lower than that observed for the corresponding HS175–DI-72–DI-83 coinoculation (Fig. 2A, lane 7), and this difference is likely related to the inability of DI-83ΔII to be amplified in vivo. When cDI-82ΔII, the capped version of the message, was tested in coinoculations, efficient amplification of DI-72 was observed (Fig. 2A, lane 11). Taken together, these results support a role for region 3.5 in an mRNA-related function such as mediating efficient translation or message stabilization. The analysis of the degradation kinetics of DI-82ΔII, DI-83ΔII, and cDI-82ΔII in vivo (Fig. 2B) suggests that region 3.5 does not confer enhanced message stability and instead facilitates cap-independent translation.
FIG. 2.
Northern blot analysis of progeny viral RNAs isolated from cucumber protoplasts inoculated with various combinations of viral RNA transcripts. (A) Analysis of trans-expression activities of DI-82ΔII and DI-83ΔII. The RNA transcripts used in the inoculations are indicated at the top; positions of the genomic (gRNA) and subgenomic (sgRNA1 and 2) mRNAs and of DI-72 are shown at the left. Approximately 3 × 105 protoplasts were inoculated with 5 μg of each transcript, except for DI-72, where 1 μg was used. Total nucleic acids were isolated after a 24-h incubation, separated in a nondenaturing 1.4% agarose gel, transferred to a nylon membrane, and hybridized with a 32P-end-labeled oligonucleotide P9 complementary to the 3′-terminal 23 nt of the TBSV genome. (B) Relative stabilities of DI-82ΔII, DI-83ΔII, and cDI-82ΔII in protoplasts. RNAs were isolated and analyzed by Northern blotting at the times indicated following inoculation of protoplasts with DI-82ΔII, DI-83ΔII, or cDI-82ΔII. Northern analysis was performed as described for panel A except that the blot was hybridized with 32P-labeled oligonucleotides P9, P25, PB21, and PB33, complementary to various regions of the TBSV genome (see Materials and Methods for coordinates). (C) Analysis of trans-expression activity in the context a full-length viral genome. Details are as for panel A.
The viral mRNAs analyzed thus far represent smaller derivatives of the viral genome. To address the role of this element in the context of a full-length genome, additional studies were performed. HS175 is a derivative of T-100 which contains a single base substitution that recodes the p33 termination codon as a tyrosine, thus allowing for the production of p92 only (Fig. 1) (33). HS175 has been shown previously to be replication defective both in cis and in trans (23, 33) and therefore acts primarily as an mRNA encoding p92. To determine if region 3.5 influences mRNA activity in a full-length genome context, HS175 and HS175Δ3.5 (a derivative in which region 3.5 is deleted) were tested individually in coinoculations with DI-83ΔII and DI-72 (Fig. 2C). The coinoculation containing HS175 allowed for efficient amplification of DI-72 as anticipated (Fig. 2C, lane 6). Interestingly, the coinoculation containing HS175Δ3.5 mediated low, but detectable, levels of amplification of DI-72 (Fig. 2C, lane 7). When the capped version of the message, cHS175Δ3.5, was tested in coinfections, DI-72 amplification was restored to a level similar to that for the coinfection containing HS175 (Fig. 2C, lane 8). These results are consistent with a primary role for region 3.5 in facilitating efficient cap-independent expression of viral proteins in the context of a full-length genome.
Cap-independent translation of a nonviral product is region 3.5 dependent.
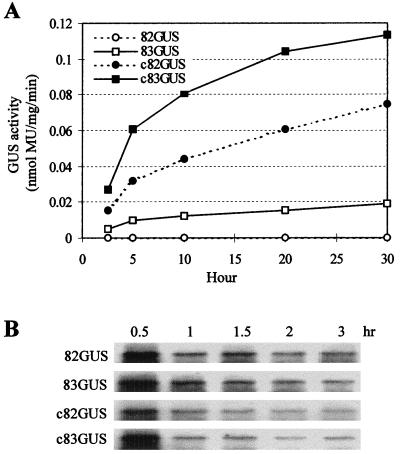
To determine if the 3′-proximal sequence could facilitate cap-independent translation of a nonviral product, virus-GUS hybrid mRNAs were constructed. The hybrid message 83GUS contained region 3.5, whereas 82GUS did not (Fig. 1B). Transcripts of these mRNAs were introduced into protoplasts via electroporation, and GUS accumulation was quantified. Consistent with the results obtained from the viral trans-expression system, no protein expression was detected from 82GUS whereas expression from 83GUS was readily detectable (Fig. 3A). Interestingly, the presence of region 3.5 also facilitated translation from capped messages (cf. c82GUS with c83GUS in Fig. 3A). For cap-dependent translation, region 3.5 further enhanced already high levels of activity, whereas for cap-independent translation, the sequence was essential for detectable product accumulation (Fig. 3A). Inspection of the graph in Fig. 3A reveals that the steady-state translation rates (which provide a measure of translational efficiency) (36) observed in the first 5 h are roughly proportional to product accumulation levels at later time points. This combined with the similar onset of detectable functional decay (i.e., 10-h time point) and continued accumulation of product at later time points (e.g., 10- and 20-h time points) suggest that the functional stabilities (i.e., length of time over which a message is translationally active) (36) of these messages do not differ substantially. Similarly, examination of the in vivo chemical stabilities of the mRNAs did not reveal any significant differences (Fig. 3B). For these analyses, shorter time points were used because the hybrid messages were significantly less stable than their viral counterparts (i.e., DI-82 and DI-83 [Fig. 2B]) and RNAs collected at later time points were difficult to detect (data not shown). Collectively, these results support further a role for region 3.5 in facilitating the rate of cap-independent translation and rule out an essential role for any of the major viral proteins.
FIG. 3.
(A) GUS translation from hybrid viral mRNAs in protoplasts. Protoplasts were electroporated with 24 pmol of each capped or uncapped hybrid transcript, total soluble plant protein was isolated at the indicated times after electroporation, and GUS activity was quantified in nanomoles of methylumbelliferone (MU) produced per milligram of cellular protein per minute in a 2-h reaction. Results from a representative experiment are shown. (B) Relative stabilities of hybrid mRNAs in protoplasts. RNAs were introduced into protoplasts via electroporation and subsequently analyzed by Northern blot analysis as described in the legend to Fig. 2B).
Efficient cap-independent translation also requires sequences adjacent to region 3.5.
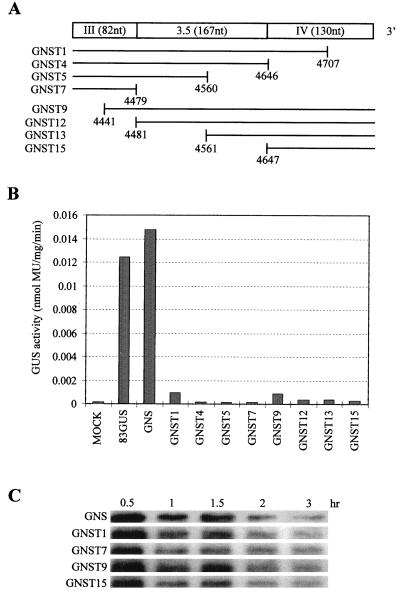
In the previous experiments, only mRNAs containing and lacking region 3.5 were compared. We therefore wanted to determine whether the cap-independent translation-enhancing activity associated with the 3′ end of the genome was localized entirely within region 3.5. To accomplish this, we generated a derivative of 83GUS into which useful restriction enzyme sites were introduced. The resulting construct GNS (Fig. 1B), which also lacked region II, produced GUS at levels comparable to those for 83GUS (Fig. 4B). GNS was used in the construction of a number of 3′-proximal deletion derivatives (Fig. 4A). When tested, all of the deletion mutants showed dramatically reduced levels of GUS production (Fig. 4B) despite similar chemical stabilities (Fig. 4C). These data indicate that in the context of this hybrid reporter mRNA, region 3.5 alone is not sufficient for cap-independent translational enhancement and that the critical sequences extend into the segments flanking this region.
FIG. 4.
(A) Schematic representation of the 3′-terminal virus derived portion of hybrid mRNA GNS (Fig. 1). The segments of this sequence which are deleted (blank) in mutant derivatives of GNS mRNA are indicated below. (B) GUS expression from mutant GNS mRNAs in protoplasts. Protoplasts were electroporated, and GUS expression was quantified 20 h postinoculation as described in the legend to Fig. 3. Results from a representative experiment are shown. (C) Relative stabilities of GNS and selected derivatives in protoplasts. RNAs were isolated and analyzed by Northern blotting as described in the legend to Fig. 2B except that the blot was hybridized with 32P-labeled oligonucleotides PGUS3, PB21, and PB33 complementary to the GUS coding region and regions the TBSV genome.
Region 3.5-mediated cap-independent translation is not observed in vitro.
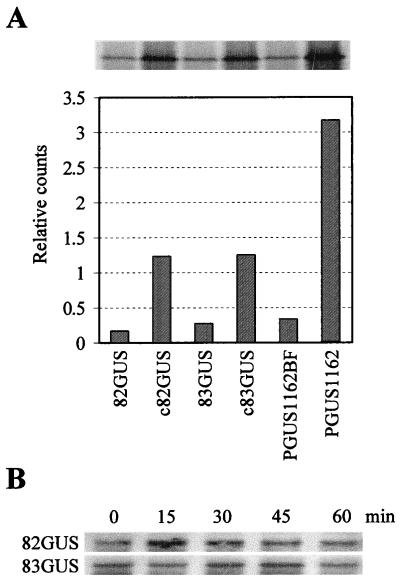
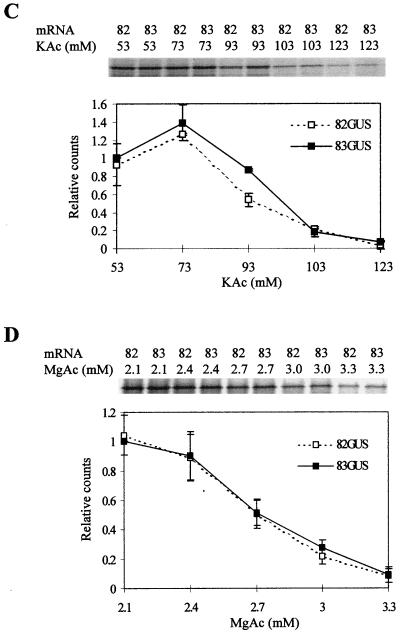
Various assays were performed within wheat germ extract to determine whether the 3′-proximal region of the TBSV genome could facilitate cap-independent translation in vitro. Both 82GUS and 83GUS were found to direct low levels of translation (Fig. 5A). The results from some experiments suggested that translation of 83GUS was slightly higher than that of 82GUS (Fig. 5A), but this difference was not observed consistently (data not shown). Analysis of the integrity of the mRNAs during the course of the assay indicated similar decay rates (Fig. 5B). The notable difference in cap-independent translation observed in vivo was therefore not reproducible in vitro under the conditions tested. Similarly, the consistent difference observed between capped transcripts c82GUS and c83GUS in vivo (Fig. 3A) was not observed in vitro (Fig. 5A). The inability to detect cap-independent differences in vitro differs from results obtained with BYDV and STNV, for each of which a greater than 20-fold increase in cap-independent translational enhancement was observed (22, 39). To address whether our assay conditions or extract preparations were capable of detecting such differences, we analyzed BYDV mRNAs encoding GUS. The BYDV message PGUS1162BF, containing a mutated nonfunctional 3′TE, showed low levels of translation, whereas PGUS1162, containing the wild-type 3′TE, showed an approximately 10-fold increase in translational efficiency (Fig. 5A). The somewhat lower increase observed in our experiments, as opposed to previously reported increases (39, 40), is likely the result of the assay conditions used (i.e., the conditions were not optimized for maximum activity of the BYDV 3′TE). These results do, however, demonstrate that the conditions tested are capable of clearly distinguishing differences in efficiencies of cap-independent translation. To address whether assay conditions which would allow for definitive detection of differences between the 82GUS and 83GUS messages could be achieved, different ionic strengths were tested. Although some differences were observed, altering the potassium (Fig. 5C) and magnesium (Fig. 5D) concentrations did not provide conditions capable of reproducing the substantial difference in translational efficiency observed in vivo.
FIG. 5.
(A) Translation of hybrid viral RNAs in wheat germ extract in the presence of [35S]Met. The accumulation of GUS protein following separation of samples via SDS-PAGE is shown at the top. Below, GUS accumulation levels were quantified by radioanalytical scanning of the gel, and the corresponding relative values are presented graphically. pGUS1162BF and pGUS1162 contain inactive and active BYDV 3′TEs, respectively, and were included in the experiment as internal controls. (B) Stability of transcripts in wheat germ extract. Aliquots were removed from reactions at the times indicated, and RNAs were analyzed by Northern blotting. (C and D) Effect of ionic strength on translational activity. Transcripts GUS82 (labeled as 82) and GUS83 (labeled as 83) were translated in wheat germ extract under various concentrations of potassium (C) or magnesium (D). The products were analyzed and quantified as described for panel A. The values shown represent means (with standard errors) from two separate experiments.
DISCUSSION
Cap-independent translation of viral proteins.
Previous studies implicated region 3.5 as a determinant of translation (23), and the present study provides evidence in support of this concept. Our results indicate that cap-independent translation of nonamplifiable viral mRNAs can direct viral protein synthesis at biologically relevant levels and that the efficiency of this process is mediated by a 3′-proximal region of the genome. The 3′ element is also active in the context of the full-length genome, a finding which is consistent with evidence suggesting that tombusvirus genomes are uncapped (28, 29). Interestingly, deletion of region 3.5 in the context of the full-length genome (i.e., HS175Δ3.5) did not completely eliminate protein synthesis. This suggests that other regions of the genome may harbor additional less efficient cap-independent enhancer elements or that the deletion of region 3.5 is less detrimental to the 3′-proximal element when it is in a larger natural context. Alternatively, very weak translation of the message may be sufficient to allow for accumulation of p92 to levels capable of directing minimal viral RNA amplification. In wild-type TBSV infections, the translation of p92 via readthrough of the p33 termination codon results in comparatively low levels of accumulation of p92 (i.e., p33:p92 ≈ 20:1) (33). However, the absence of the p33 termination codon in HS175Δ3.5 would allow for more efficient production of p92 following translational initiation; thus, even inefficient initiation on this message could potentially allow for the accumulation of functionally relevant levels of p92.
The deletion analyses carried out with GUS-encoding viral messages suggest that the 3′-proximal sequence necessary for efficient cap-independent translation in vivo extends out of region 3.5 in both directions. We have designated this functional segment the 3′ cap-independent translational enhancer (3′CITE). The sequences flanking region 3.5 may constitute part of the active site of the element and/or may participate indirectly in some structural capacity. The 3′CITE-mediated expression of a foreign protein indicates that no viral proteins, for which functions have been assigned, are essential for cap-independent translation. However, this finding does not preclude their involvement in some regulatory capacity. The 3′CITE does contain within it a small ORF encoding an ∼8-kDa product, pX, of unknown function (4, 31). Although similar small ORFs are conserved in other tombusviruses (4), studies of TBSV and cymbidium ringspot tombusvirus in which the pX ORF was inactivated or disrupted suggest that the product plays no critical role in the viral reproductive cycle (6, 31).
General and specific mechanistic aspects of the 3′CITE activity remain to be investigated. However, based on the activity of most other translational enhancers, the element likely increases the rate of initiation (12). This function would, in turn, require some form of communication between the 3′CITE and the 5′ end of the message. For eukaryotic cellular mRNAs, 5′-3′ interactions are mediated by proteins which bind to the 5′ cap and poly(A) tail (14). In the case of the TBSV genome, different RNA structures, and possibly proteins, would have to be involved. The 3′-proximal enhancers of STNV and BYDV both require their homologous viral 5′ termini for optimal activity, supporting a requirement for 5′-3′ communication (22, 39). We are currently investigating the possible functional relationship between the 3′CITE and the TBSV 5′ UTR.
Interestingly, although translation of BYDV and STNV genomes rely on nontraditional elements, these activities are dependent on translation initiation factor 4F; therefore, the cap-independent mechanisms employed may share some features of cap-dependent translation (8, 39). Our finding that the 3′CITE also facilitates cap-dependent translation raises the possibility that the 3′CITE could possess poly(A) tail-like properties. In this respect, a number of nonpolyadenylated viral 3′ UTRs can substitute functionally for a poly(A) tail in cap-dependent translation (13).
The 3′CITE and viral RNA replication.
The importance of the 3′CITE in facilitating translation of viral proteins involved in replication is evident. Less obvious are possible alternative roles for the element in influencing the efficiency of viral RNA replication. Region 3.5 is clearly not essential for viral RNA replication in trans since numerous defective viral RNAs lacking this region are amplified efficiently when coinfected with helper (41). In fact, the region 3.5 deletion was first identified as a naturally occurring deletion in efficiently replicating prototypical DI RNAs (21). Previous studies on DI RNA species containing or lacking region 3.5 have revealed that the latter species is more competitive in coinfections (23, 42). This enhanced competitiveness could not be attributed to increased RNA stability; therefore, it was suggested that region 3.5 somehow reduces replication efficiency (42). Conversely, the present study indicates that region 3.5 increases translational efficiency. Taken together, these results suggest that a functional 3′CITE could be inhibitory to viral RNA replication.
The 3′CITE extends into adjacent regions III and IV, which harbor cis-acting replication elements (Fig. 1) (26, 27). This localized organization of replication and translation elements in the 3′ region of the TBSV genome could provide a mechanism for controlling the use of the genome as either a mRNA for translation or template for replication. Similarly, the 5′ UTR of the TBSV genome contains important cis-acting elements for replication (44); therefore, if it is determined that the 3′CITE does require its corresponding 5′ UTR for activity, the 5′ UTR could also represent a target for regulation. The 5′ UTR is a modulator of translation and replication in poliovirus (14). In the poliovirus genome, the 5′ UTR contains a cloverleaf structure that contains both replication and translation elements (2), and a viral protein interacts with this structure to mediate downregulation of translation and stimulation of negative-strand synthesis (14). The overlapping nature of translation and replication signals within the cloverleaf therefore provides a strategy to temporally regulate the transition of the poliovirus genome from an mRNA for translation to a template for replication (14). For poliovirus, the key viral protein involved in facilitating the switch is 3CD (14). In brome mosaic virus, the 1a viral protein may play a somewhat analogous role as it is able to both stabilize genomic RNA3 and downregulate its translation (19, 35). Correspondingly, the activity of 1a is dependent on an RNA element within RNA3 which also functions as a replication enhancer (9). In the case of TBSV, the role of viral proteins with respect to genome translation and stability have not yet been investigated.
Comparison of the 3′CITE with other plant viral 3′-proximal cap-independent translational enhancers.
Several features of the TBSV 3′CITE are distinct from the 3′-proximal cap-independent translational enhancers reported for other plant positive-strand RNA viruses. A primary difference is that significant 3′CITE-mediated enhancement is not discernible in vitro in wheat germ extracts. For both STNV and BYDV, a clear and striking enhancement is observed when these elements are analyzed in vitro in this system (7, 39). By including BYDV mRNAs containing and lacking an active 3′TE in our experiments, we were able to rule out general assay conditions as the reason for this discrepancy. One possible explanation for the contrasting results is that the wheat germ extracts lack a factor which interacts specifically with the 3′CITE. This possibility is not without precedence. In picornaviruses, two distinct classes of internal ribosome entry sites (IRESs) have been identified (18). Messages containing the cardiovirus/aphthovirus IRESs are efficiently translated in rabbit reticulocyte lysates, but the enterovirus/rhinovirus IRESs do not function efficiently in this in vitro system (18). However, supplementation of the rabbit reticulocyte lysate with extracts from HeLa cells (or L cells or Krebs II ascites cells) allows enterovirus/rhinovirus IRESs to function effectively (18).
As an alternative to a missing factor, the lack of 3′CITE activity in vitro may be related to assay conditions which do not allow for distinction between differences in affinity for a rate-limiting factor. Such differences may not be discernible unless the factor is present at an appropriate concentration (i.e., approaching the dissociation constants of the factor-mRNA complexes) (17). Saturating levels of a factor can be reduced through competition by adding exogenous mRNA, and this strategy was used successfully to reveal differences in translational efficiencies of alfalfa mosaic virus subgenomic mRNAs (15). Preliminary experiments using capped α-globin mRNA as a competitor have not revealed 3′CITE-dependent translation in vitro (43). Alternatively, the concentration of a rate-limiting factor may be far below the threshold required for productive association. Such situations can be remedied by increasing the concentration of the factor through supplementation (8).
Additional properties which distinguish the TBSV 3′CITE from the enhancers of BYDV and STNV are that (i) the relative position of the 3′CITE is more 3′-proximal, (ii) the 3′CITE overlaps replication elements, and (iii) no extensive sequence identity with known 3′-proximal enhancers is evident. Taken together, our data suggest that we have identified a novel viral 3′-proximal cap-independent translational enhancer which represents the first such element to be described for the genus Tombusvirus.
ACKNOWLEDGMENTS
We thank members of our laboratory for reviewing the manuscript. DI-82ΔII and DI-83ΔII were constructed by M. Ostrovsky and K. Ireland, respectively. We are also grateful to W. A. Miller for providing pGUS1162 and pGUS1162BF, H. B. Scholthof for supplying HS175, and J. Skuzeski for providing pAGUS1-TN2.
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Allen E, Wang S, Miller W A. Barley yellow dwarf virus RNA requires a cap-independent translation sequence because it lacks a 5′ cap. Virology. 1999;253:139–144. doi: 10.1006/viro.1998.9507. [DOI] [PubMed] [Google Scholar]
- 2.Andino R, Boddeker N, Silvera D, Gamarnik A V. Intracellular determinants of picornavirus replication. Trends Microbiol. 1999;7:76–82. doi: 10.1016/s0966-842x(98)01446-2. [DOI] [PubMed] [Google Scholar]
- 3.Bailey-Serres J. Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci. 1999;4:142–148. doi: 10.1016/s1360-1385(99)01386-2. [DOI] [PubMed] [Google Scholar]
- 4.Boyko V P, Karasev A V. Tombusvirus genome may encode the sixth small protein near its 3′ terminus. Virus Genes. 1992;6:143–148. doi: 10.1007/BF01703063. [DOI] [PubMed] [Google Scholar]
- 5.Browning K S. The plant translational apparatus. Plant Mol Biol. 1996;32:107–144. doi: 10.1007/BF00039380. [DOI] [PubMed] [Google Scholar]
- 6.Dalmay T, Rubino L, Burgyan J, Kollar A, Russo M. Functional analysis of cymbidium ringspot virus genome. Virology. 1993;194:697–704. doi: 10.1006/viro.1993.1310. [DOI] [PubMed] [Google Scholar]
- 7.Danthinne X, Seurinck J, Meulewaeter F, Van Montagu M, Cornelissen M. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol Cell Biol. 1993;13:3340–3349. doi: 10.1128/mcb.13.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher L, Corbin S D, Browning K S, Ravel J M. The absence of a m7G cap on beta-globin mRNA and alfalfa mosaic virus RNA 4 increases the amounts of initiation factor 4F required for translation. J Biol Chem. 1990;265:19582–19587. [PubMed] [Google Scholar]
- 9.French R, Ahlquist P. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J Virol. 1987;61:1457–1465. doi: 10.1128/jvi.61.5.1457-1465.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fromm M, Callis J, Taylor L P, Walbot V. Electroporation of DNA and RNA into plant protoplasts. Methods Enzymol. 1987;153:351–366. [Google Scholar]
- 11.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 12.Gallie D R. Translational control of cellular and viral mRNAs. Plant Mol Biol. 1996;32:145–158. doi: 10.1007/BF00039381. [DOI] [PubMed] [Google Scholar]
- 13.Gallie D R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 14.Gamarnik A V, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hann L E, Webb A C, Cai J M, Gehrke L. Identification of a competitive translation determinant in the 3′ untranslated region of alfalfa mosaic virus coat protein mRNA. Mol Cell Biol. 1997;17:2005–2013. doi: 10.1128/mcb.17.4.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hearne P Q, Knorr D A, Hillman B I, Morris T J. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology. 1990;177:141–151. doi: 10.1016/0042-6822(90)90468-7. [DOI] [PubMed] [Google Scholar]
- 17.Herson D, Schmidt A, Seal S, Marcus A, van Vloten-Doting L. Competitive mRNA translation in an in vitro system from wheat germ. J Biol Chem. 1979;254:8245–8249. [PubMed] [Google Scholar]
- 18.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 19.Janda M, Ahlquist P. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc Natl Acad Sci USA. 1998;95:2227–2232. doi: 10.1073/pnas.95.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferson R A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 21.Knorr D A, Morris T J. De novo generation of defective interfering RNAs of tomato bushy stunt virus by high multiplicity passage. Virology. 1991;181:193–202. doi: 10.1016/0042-6822(91)90484-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meulewaeter F, Danthinne X, Van Montagu M, Cornelissen M. 5′- and 3′-sequences of satellite tobacco necrosis virus RNA promoting translation in tobacco. Plant J. 1998;14:169–176. doi: 10.1046/j.1365-313x.1998.00104.x. [DOI] [PubMed] [Google Scholar]
- 23.Oster S K, Wu B, White K A. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J Virol. 1998;72:5845–5851. doi: 10.1128/jvi.72.7.5845-5851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 25.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 26.Ray D, White K A. Enhancer-like properties of an RNA element that modulates Tombusvirus RNA accumulation. Virology. 1999;256:162–171. doi: 10.1006/viro.1999.9630. [DOI] [PubMed] [Google Scholar]
- 27.Ray, D., and K. A. White. Unpublished data.
- 28.Rubino L, Burgyan J, Russo M. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch Virol. 1995;140:2027–2039. doi: 10.1007/BF01322690. [DOI] [PubMed] [Google Scholar]
- 29.Russo M, Burgyan J, Martelli G P. Molecular biology of tombusviridae. Adv Virus Res. 1994;44:381–428. doi: 10.1016/s0065-3527(08)60334-6. [DOI] [PubMed] [Google Scholar]
- 30.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 31.Scholthof H B, Jackson A O. The enigma of pX: a host-dependent cis-acting element with variable effects on tombusvirus RNA accumulation. Virology. 1997;237:56–65. doi: 10.1006/viro.1997.8754. [DOI] [PubMed] [Google Scholar]
- 32.Scholthof H B, Scholthof K B, Kikkert M, Jackson A O. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 1995;213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- 33.Scholthof K-B G, Scholthof H B, Jackson A O. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology. 1995;208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 34.Skuzeski J M, Nichols L M, Gesteland R F. Analysis of leaky viral translation termination codons in vivo by transient expression of improved beta-glucuronidase vectors. Plant Mol Biol. 1990;15:65–79. doi: 10.1007/BF00017725. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan M L, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanguay R L, Gallie D R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol Cell Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarun S Z, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 38.Timmer R T, Benkowski L A, Schodin D, Lax S R, Metz A M, Ravel J M, Browning K S. The 5′ and 3′ untranslated regions of satellite tobacco necrosis virus RNA affect translational efficiency and dependence on a 5′ cap structure. J Biol Chem. 1993;268:9504–9510. [PubMed] [Google Scholar]
- 39.Wang S, Browning K S, Miller W A. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 1997;16:4107–4116. doi: 10.1093/emboj/16.13.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Miller W A. A sequence located 4.5 to 5 kilobases from the 5′ end of the barley yellow dwarf virus (PAV) genome strongly stimulates translation of uncapped mRNA. J Biol Chem. 1995;270:13446–13452. doi: 10.1074/jbc.270.22.13446. [DOI] [PubMed] [Google Scholar]
- 41.White K A. Formation and evolution of tombusvirus defective interfering RNAs. Semin Virol. 1996;7:409–416. [Google Scholar]
- 42.White K A, Morris T J. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J Virol. 1994;68:14–24. doi: 10.1128/jvi.68.1.14-24.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, B., and K. A. White. Unpublished data.
- 44.Wu B, White K A. Formation and amplification of a novel tombusvirus defective RNA which lacks the 5′ nontranslated region of the viral genome. J Virol. 1998;72:9897–9905. doi: 10.1128/jvi.72.12.9897-9905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang G, Slowinski V, White K A. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA. 1999;5:550–561. doi: 10.1017/s1355838299982080. [DOI] [PMC free article] [PubMed] [Google Scholar]