Summary
Although many host factors important for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection have been reported, the mechanisms by which the virus interacts with host cells remain elusive. Here, we identified tripartite motif containing (TRIM) 28, TRIM33, euchromatic histone lysine methyltransferase (EHMT) 1, and EHMT2 as proviral factors involved in SARS-CoV-2 infection by CRISPR-Cas9 screening. Our result suggested that TRIM28 may play a role in viral particle formation and that TRIM33, EHMT1, and EHMT2 may be involved in viral transcription and replication. UNC0642, a compound that specifically inhibits the methyltransferase activity of EHMT1/2, strikingly suppressed SARS-CoV-2 growth in cultured cells and reduced disease severity in a hamster infection model. This study suggests that EHMT1/2 may be a therapeutic target for SARS-CoV-2 infection.
Subject areas: Cell biology, Molecular biology, Virology
Graphical abstract
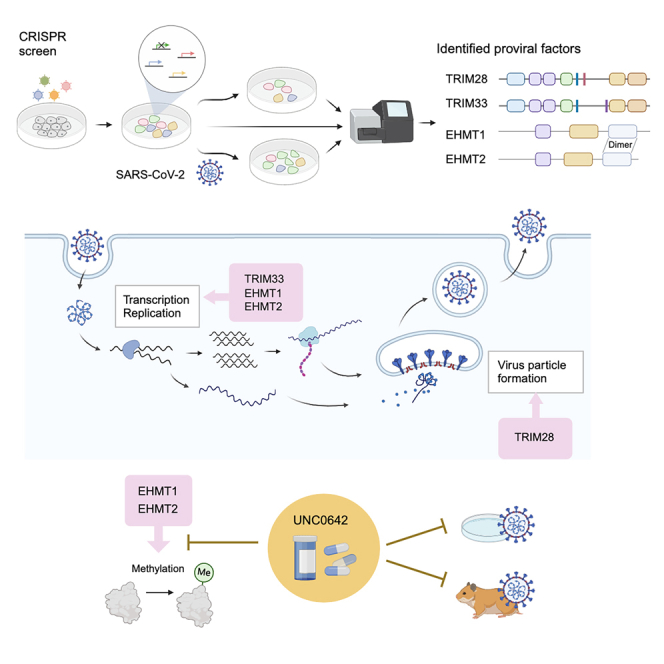
Highlights
-
•
TRIM28, TRIM33, EHMT1, and EHMT2 are proviral factors for SARS-CoV-2
-
•
TRIM28 is involved in viral assembly; TRIM33, EHMT1, and EHMT2 in transcription/replication
-
•
EHMT1/2 inhibitor significantly suppressed SARS-CoV-2 growth in vitro and in vivo
Cell biology; Molecular biology; Virology
Introduction
Therapeutics that target proviral factors have the potential to be effective against a broad spectrum of viruses. Genome-wide screens using CRISPR-Cas9 technology to identify host factors involved in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication have been performed in several studies.1,2,3,4,5,6,7,8,9,10,11,12,13 The only host factor identified in every screen is angiotensin-converting enzyme 2 (ACE2), and different genes are identified depending on conditions such as the cells used, the multiplicity of infection (MOI), and the time to sample collection.5,13
A549 cells, which are human lung epithelial type II cells, typically do not show clear cytopathic effects from SARS-CoV-2 infection. In previous studies, proviral factors were found by prolonging the sampling time1 or by using a mutant virus lacking the S1/S2 sites.2 We discovered that A549-hACE2 cells infected with purified SARS-CoV-2 caused clear cell death and applied this to our CRISPR-Cas9 screen.
In this study, we successfully identified four genes from the same family or complex as proviral factors of SARS-CoV-2. Tripartite motif containing (TRIM) 28 and TRIM33 are members of the same gene family that has common domains and regulates transcriptional pathways.14 These genes exhibited distinct functional roles in the life cycle of SARS-CoV-2. Euchromatic histone lysine methyltransferase (EHMT) 1/2, also known as G9a/GLP, forms heterodimers and catalyzes the methylation of histone or non-histone proteins.15,16 We found that depletion of their expression suppresses viral proliferation and that a specific inhibitor of EHMT1/2 suppressed viral growth in vitro and in vivo. This study contributes to the understanding of how SARS-CoV-2 interacts with host cells and to the development of methods to control the virus by targeting proviral factors.
Results
Identification of proviral factors for SARS-CoV-2 infection
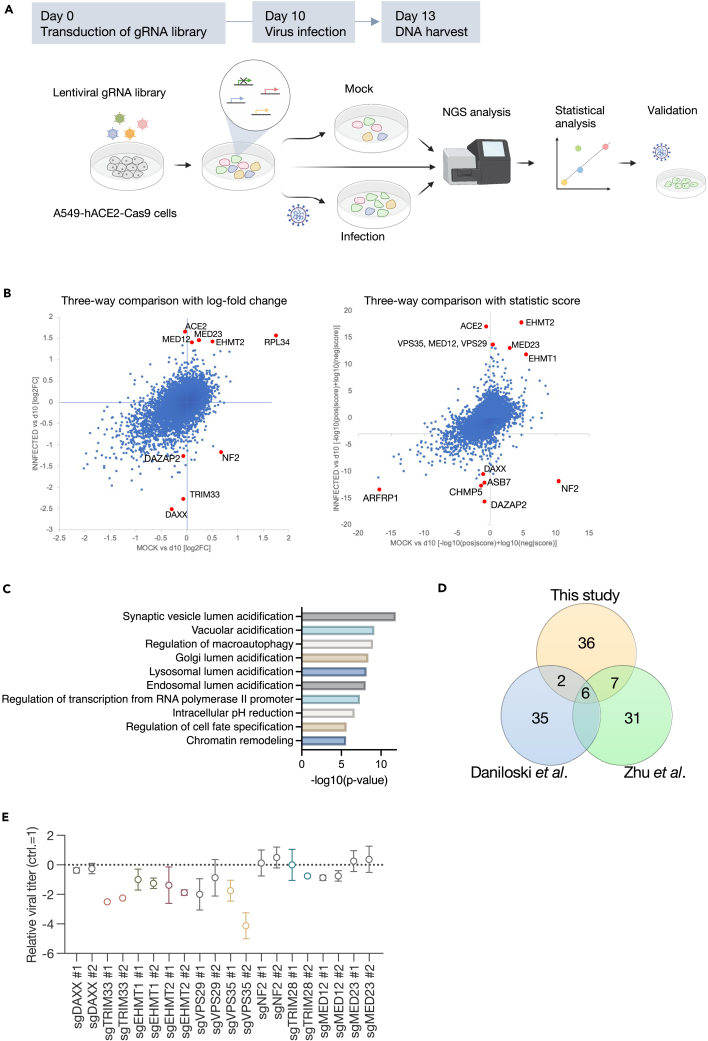
Following the confirmation that purified SARS-CoV-2 induces distinct cell death in A549-hACE2 cells (Figure S1), a genome-wide CRISPR-Cas9 screen was conducted using cell death as an indicator. We transduced a single-guide RNA (sgRNA) library targeting 18,365 human genes with 113,526 sgRNAs, including 1,004 nontargeting control guide RNAs (gRNAs),17,18 into A549-hACE2-Cas9 cells. On day 10 post-transduction, these cells were infected with purified SARS-CoV-2 UT-NCGM0219 at an MOI of 0.3. DNA was harvested from sgRNA-transduced cells at 10 days post-transduction and from the mock or infected cells at 3 days post-infection for next-generation sequencing analysis (Figure 1A). By comparing the number of sgRNA reads in the day 10 sample with mock or infected cells, we found that the control gRNA was linearly aligned, indicating that there was no bias due to the experimental technique (Figure S2A). MAGeCK analysis identified MED12, MED23, EHMT1, EHMT2, RPL34, and TRIM28 as target genes enriched in infected cells that showed resistance to infection-induced cell death by plotting the comparison of sgRNA read counts between the two groups based on log fold change or statistic score (Tables S1, S2, and S3; Figure 1B). Conversely, DAXX, NF2, DAZAP2, and TRIM33 were enriched in the mock cells (Tables S1, S2, and S3; Figure 1B). In the comparison of mock and infected cells, MED12 and TRIM28 were positively selected in addition to ACE2, whereas NF2 was under strong negative selection (Figure S2B). Gene set enrichment analysis revealed enrichment in six Gene Ontology (GO) categories related to acidification among the positively selected population (p < 0.05). Other categories were enriched for functions such as transcriptional regulation, autophagy, cell fate, and chromatin remodeling (Figure 1C). Six genes, ACE2, VPS35, COMMD4, CCDC22, RAB7A, and VPS29, were in the top 50 genes from the previous CRISPR-Cas9 screen for SARS-CoV-2 proviral factors in A549 cells1,2 and were also significant in this study (Figure 1D).
Figure 1.
Genome-wide CRISPR-Cas9 screening to identify host factors for SARS-CoV-2 infection
(A) Schematic diagram of the screening. ACE2- and Cas9-expressing A549 cells transduced with a lentiviral-packaged whole-genome sgRNA library were infected with purified SARS-CoV-2. DNA was extracted from surviving cells 3 days post-infection and analyzed for gRNA.
(B) Scatterplot of the results from screening. Comparison of gRNA changes between preinfection (day 10) versus mock and preinfection (day 10) versus infected.
(C) GO terms enrichment significance.
(D) Overlap of the top 50 ranked genes among 3 CRISPR screens conducted on A549 cells.
(E) Comparison of viral titers in target gene knockout cells. The knockout cells were infected with SARS-CoV-2. After 3 days, the supernatant was harvested, and the viral titer was evaluated by TCID50. The titer collected from control cells was used as a standard. Data are presented as the mean ± the standard deviation (SD) of three independent experiments.
It is noteworthy that genes that form families or complexes were identified as host factors associated with SARS-CoV-2 infection. To validate this result, individual knockout cells were generated by transducing A549-hACE2-Cas9 cells with sgRNA. These knockout cells were then infected with UT-NCGM02 at an MOI of 0.001, and the virus titer in the supernatant was measured 3 days later. The viral titer was found to be reduced in cells transduced with sgRNAs targeting TRIM28, TRIM33, EHMT1, and EHMT2, but not in cells with sgRNAs targeting MED12, MED23, NF2, and DAXX (Figure 1E). Although DAXX, NF2, and TRIM33 were enriched in mock cells in the screen, suggesting they might be negative regulators, the results were contrary to expectations. Inhibition of TRIM33 expression suppressed viral proliferation, while DAXX and NF2 had no effect on viral growth.
The identified proviral factors are not involved in the entry step of SARS-CoV-2
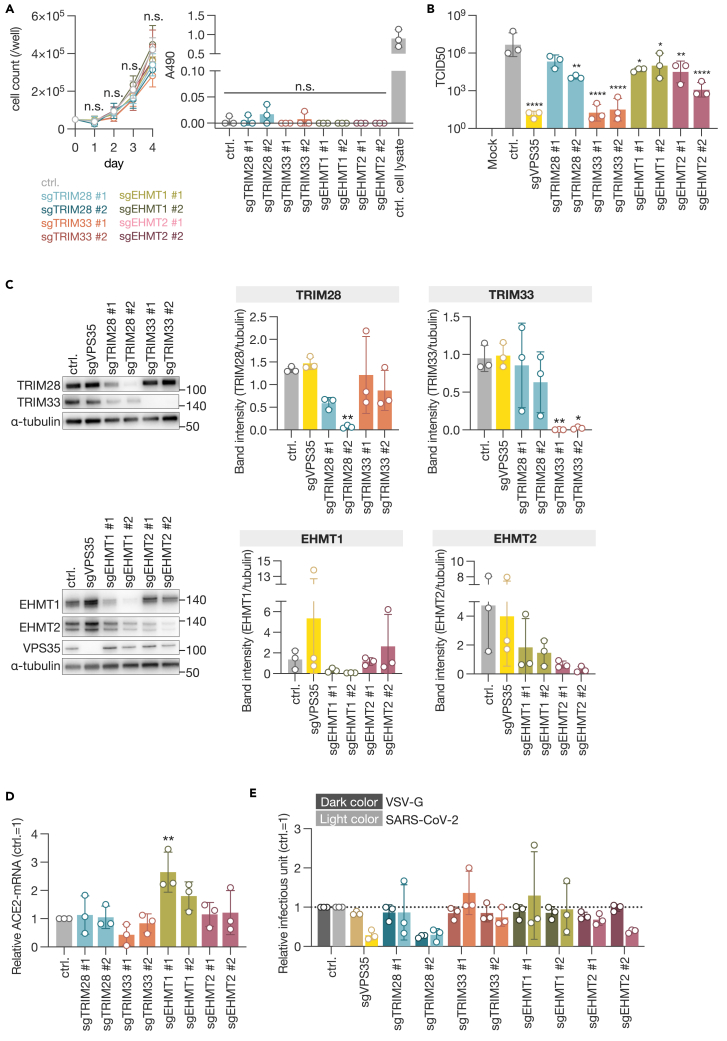
Consequently, we elected to concentrate our efforts on TRIM28, TRIM33, EHMT1, and EHMT2 as potential proviral factors. The effects of gene knockout on cell proliferation and cytotoxicity were evaluated, and no change was observed compared to the negative control (Figure 2A). When assessing the knockout efficiency and virus replication, it was observed that in cells with low knockout efficiency, the inhibitory effect on virus replication was also low (Figures 2B and 2C). Previous reports have indicated that the knockdown of TRIM28 with siRNA induces ACE2 expression, thereby facilitating the entry of SARS-CoV-2.20 To ascertain whether the suppression of these four genes affects ACE2 expression levels, we quantified ACE2 mRNA in Cas9-expressing A549 cells transduced with gRNA using quantitative reverse-transcription PCR (Figure S3). Although there was a trend toward increased ACE2 expression in cells targeted with gRNA against EHMT1, no significant changes were observed in cells targeted with gRNA against the other genes (Figure 2D). In a pseudotype assay using vesicular stomatitis virus (VSV)ΔG, a decreasing trend was observed in clone 2 of EHMT2 knockout cells. However, the efficiency of SARS-CoV-2 spike-mediated infection was not significantly reduced in cells targeted with gRNA against any of the genes (Figure 2E). Taken together, the identified genes TRIM28, TRIM33, EHMT1, and EHMT2 do not appear to be involved in the ACE2-mediated entry process of SARS-CoV-2.
Figure 2.
Entry steps of SARS-CoV-2 in proviral factor-knockout cells
(A) Cell growth kinetics and cytotoxicity assessment in knockout cells. Cell proliferation was evaluated daily over a period of 4 days by counting cell numbers. On the final day, an LDH assay was performed.
(B) Titration of viral production in selected gene knockout cells for validation.
(C) Western blot analysis of selected genes. Expression levels were assessed by band intensity and normalized to α-tubulin.
(D) Quantification of ACE2 mRNA in selected gene-knockout A549-Cas9 cells. Cells were harvested 18 days post-sgRNA transduction.
(E) VSV-based pseudotype assay. VSVΔG-GFP enveloped with VSV-G or SARS-CoV-2-S was used to infect knockout cells, and GFP-positive cells were counted. Data are presented as the mean ± SD of three independent experiments. Statistical analysis was performed using Dunnett’s multiple comparison test. n.s., not significant; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.
The reduced expression of the proviral factors does not result in the induction of an innate immune response
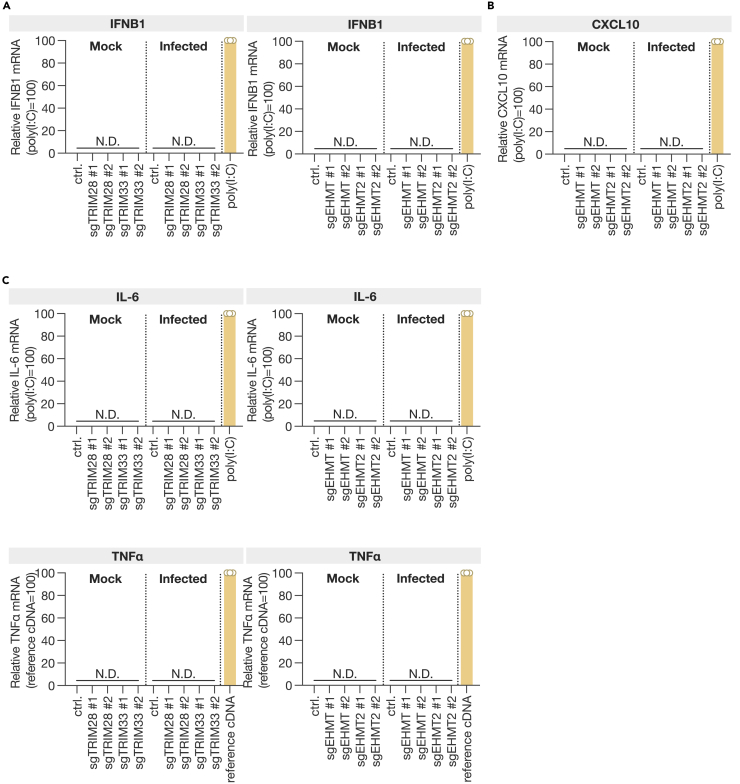
It has been reported that TRIM28 and TRIM33 negatively regulate the production of interferon β (IFNβ).21,22,23 EHMT1 and EHMT2 are crucial for suppressing IFNγ-induced chemokine transcription.24,25 In bovine cells, the inhibition of EHMT2 has been demonstrated to enhance IFNβ transcription.26 Therefore, we examined whether the expression of IFNB1 was induced in cells with targeted gRNAs against these four genes (sgTRIM28, sgTRIM33, sgEHMT1, and sgEHMT2 cells) (Figure 3A). Furthermore, the expression of CXCL10, an IFNγ-induced protein, was investigated in cells with knockout of EHMT1 or EHMT2. The results demonstrated that the suppression of any of these genes did not result in an increase in IFNB1 expression and that the knockout of EHMT1 and EHMT2 did not induce the expression of CXCL10 (Figure 3B). Furthermore, the expression of interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) was evaluated, and it was found that knocking out any of these genes did not induce their expression (Figure 3C). These findings indicate that the proviral factors identified in this study do not facilitate the replication of SARS-CoV-2 by suppressing the innate immune response.
Figure 3.
SARS-CoV-2 and innate immune response in proviral factor-knockout cells
(A–C) Quantification of IFNβ (A), CXCL10 (B), IL-6, and TNF-α (C) mRNA in TRIM28, TRIM33, EHMT1, or EHMT2 knockout cells at 24 h post-infection. Poly(I:C) (high molecular weight) was transfected as a positive control in control cells, and cells were harvested 24 h post-infection. For TNF-α, human reference cDNA (30 ng equiv. total RNA) was used as a positive control for qPCR detection. Data are presented as the mean ± SD of three independent experiments. N.D., not detected.
TRIM28 and TRIM33 are involved in the replication of SARS-CoV-2 through different mechanisms
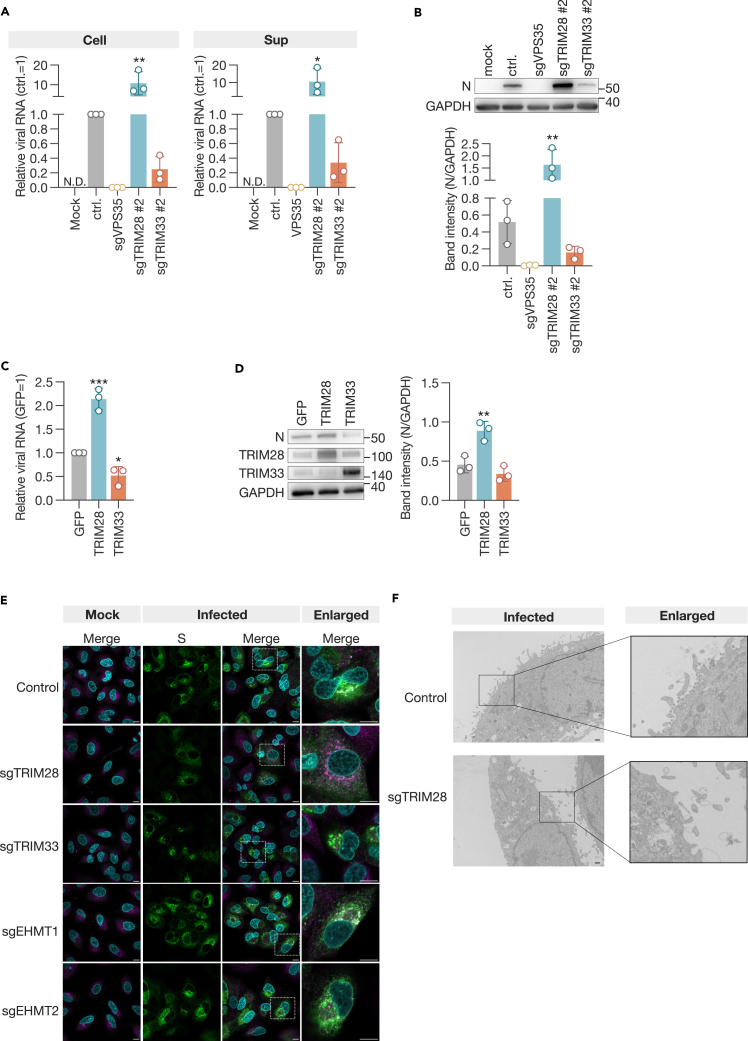
To assess the role of the identified proviral factors in the replication of SARS-CoV-2, we infected sgTRIM28 and sgTRIM33 cells with UT-NCGM02 and quantified the expression of viral RNA and protein in the cells and supernatants. In comparison to control cells, sgTRIM33 cells exhibited a reduction in the expression of viral RNA and protein in both cell lysates and supernatants. In contrast, the expression of viral RNA and protein was elevated in sgTRIM28-induced cells (Figures 4A and 4B). Subsequently, we introduced plasmids expressing TRIM28 or TRIM33 into A549-hACE2 cells, infected them with the virus at an MOI of 1.0 the following day, and quantified the viral RNA and protein within the cells 3 days later. In contrast to expectations, overexpression of TRIM28 resulted in increased levels of viral RNA and protein, while overexpression of TRIM33 led to decreased levels of both (Figures 4C and 4D). These results indicate that alterations in the expression levels of TRIM28 and TRIM33 influence fluctuations in viral transcription and/or replication.
Figure 4.
TRIM28 and TRIM33 as proviral factors for SARS-CoV-2 infection
(A) qPCR analysis of viral RNA.
(B) Western blot analysis of the viral nucleoprotein (N). The expression of the viral protein was evaluated by the band intensity of N, normalized to that of GAPDH. Cell lysates and culture supernatants were harvested 3 days post-infection.
(C) qPCR analysis of viral RNA in A549-hACE2 cells overexpressing TRIM28 or TRIM33.
(D) Western blot analysis of the viral N. The expression of the viral protein was evaluated by the band intensity of N, normalized to that of GAPDH. Cells were infected at an MOI of 1.0 the day after transfection with expression plasmids and harvested 3 days later.
(E) Subcellular localization of the S protein in infected cells. Cells were inoculated at an MOI of 2.0 and fixed the following day. Cells were stained with anti-S antibody and anti-ERGIC53. Bar, 10 μm.
(F) Electron microscopic analysis of virus particle formation in infected cells. Bar, 5.0 μm. Data are presented as the mean ± SD of three independent experiments. Statistical analysis was performed using the Dunnett test. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
The SARS-CoV-2 spike (S) protein was identified within the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), which is the site of viral assembly.27,28,29,30 Given the potential involvement of proviral factors in viral assembly, we conducted a confocal laser microscopy analysis to examine the localization of the S protein in knockout cells. In control cells, the S protein was observed to accumulate in the ERGIC, in accordance with previous reports (Figure 4E). However, in sgTRIM28 cells, the S protein was observed to be distributed throughout the cell (Figure 4E). Cells with other gene knockouts exhibited localization of the S protein that was similar to that observed in control cells. Furthermore, electron microscopy analysis revealed that no viral particle was detected in sgTRIM28 cells (Figure 4F). These findings indicate that TRIM28 may be involved in the formation of virus particles, while TRIM33 may be implicated in the transcription and/or replication process within the cell.
EHMT1/2 inhibitor suppresses the proliferation of SARS-CoV-2 in vitro
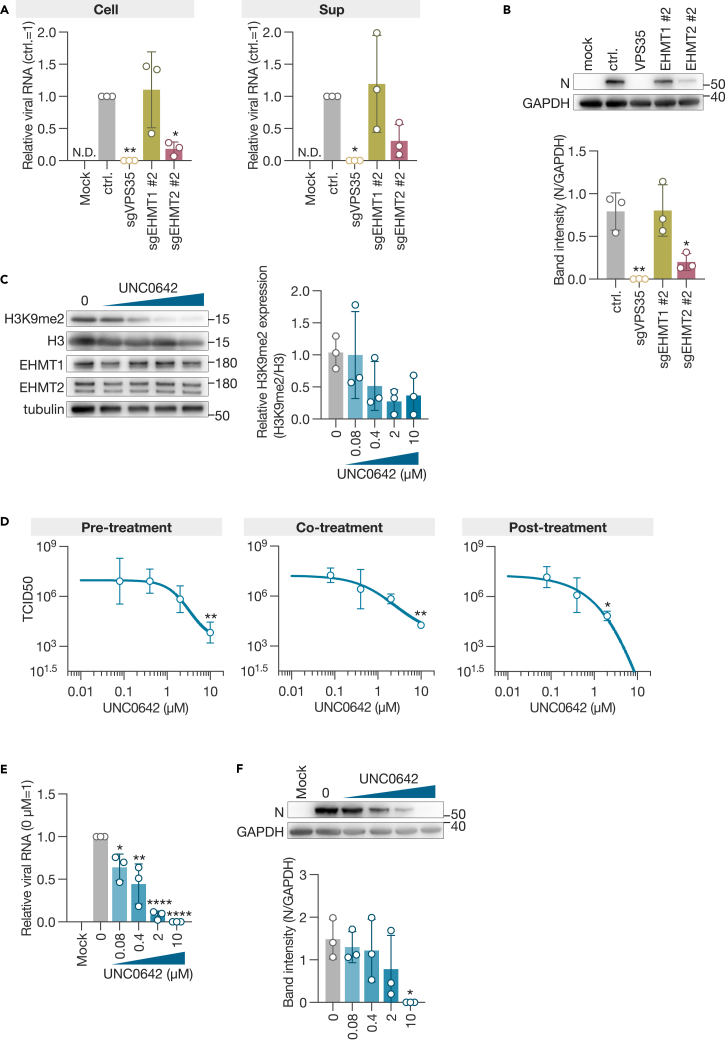
EHMT1 and EHMT2 (also referred to as GLP and G9a) form heterodimers in cells and methylate histone H3 as well as nonhistone proteins.15,16,31,32,33 We quantified viral RNA and protein in the cell lysate and supernatant of SARS-CoV-2-infected cells and found that both viral RNA and protein expression were downregulated in sgEHMT2 cells, while no differences were observed in sgEHMT1 cells compared to control cells (Figures 5A and 5B).
Figure 5.
EHMT1 and EHMT2 as proviral factors for SARS-CoV-2 infection
(A) qPCR analysis of viral RNA.
(B) Western blot analysis of the viral N. The expression of the viral protein was evaluated by the band intensity of N, normalized to that of GAPDH. Cell lysates and culture supernatants were harvested 3 days post-infection.
(C) Western blot analysis of the inhibitory effect of UNC0642 on histone methylation. A549-hACE2 cells were treated with UNC0642 and harvested the following day. DMSO was used as a negative control.
(D) Inhibitory effect of the EHMT1/2-specific inhibitor UNC0642 on SARS-CoV-2 infection. Viral titers were measured 3 days post-inoculation at an MOI of 0.001. Treatments included pre-treatment (1 h before inoculation), co-treatment during (1 h absorption), and post-treatment (after inoculation).
(E) qPCR analysis of viral RNA in cells treated post-infection with UNC0642.
(F) Western blot analysis of viral N protein in UNC0642-treated cells. The expression of viral protein was evaluated by the band intensity of N, normalized to that of GAPDH. Data are presented as the mean ± SD of three independent experiments. Statistical analysis was performed using Dunnett’s multiple comparison test. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗∗, p < 0.0001.
UNC0642 is a methyltransferase inhibitor with high selectivity for EHMT1/2 and low cytotoxicity (Figure S4A).34,35 Using A549-hACE2 cells, we evaluated the inhibitory effect of UNC0642 on H3K9 demethylation (H3K9me2) and observed significant inhibition at concentrations of 2 and 10 μM (Figure 5C). To evaluate whether the enzymatic activity of EHMT1/2 is involved in the proliferation of SARS-CoV-2, we treated the cells with the inhibitor 1 h before virus inoculation (pre-treatment), during the 1-h virus absorption (co-treatment), and after virus inoculation (post-treatment) and evaluated the virus titer in the supernatant. The results showed that remdesivir, which inhibits viral RNA-dependent RNA polymerase,36,37 suppressed virus proliferation only after inoculation (Figure S4B), whereas UNC0642 consistently and dose dependently suppressed SARS-CoV-2 proliferation under all conditions (Figure 5D). Specifically, when we treated with 10 μM UNC0642 after virus infection, virus proliferation was suppressed by approximately 106 (Figure 5D), a concentration consistent with the demonstrated inhibitory effect on H3K9me2. Post-treatment with UNC0642 also suppressed viral RNA and protein expression in cells in a dose-dependent manner (Figures 5E and 5F). UNC0642 has been demonstrated to target both EHMT1 and EHMT2.34 When sgEHMT1 or sgEHMT2 cells were treated with 10 μM UNC0642 following virus infection, a stronger tendency to suppress virus proliferation was observed (Figure S4C). These results demonstrate that EHMT1/2, through its enzymatic activity, is involved in the transcription and/or replication of SARS-CoV-2 in the cell.
EHMT1/2 inhibitor reduces SARS-CoV-2 growth and disease severity in infected animals
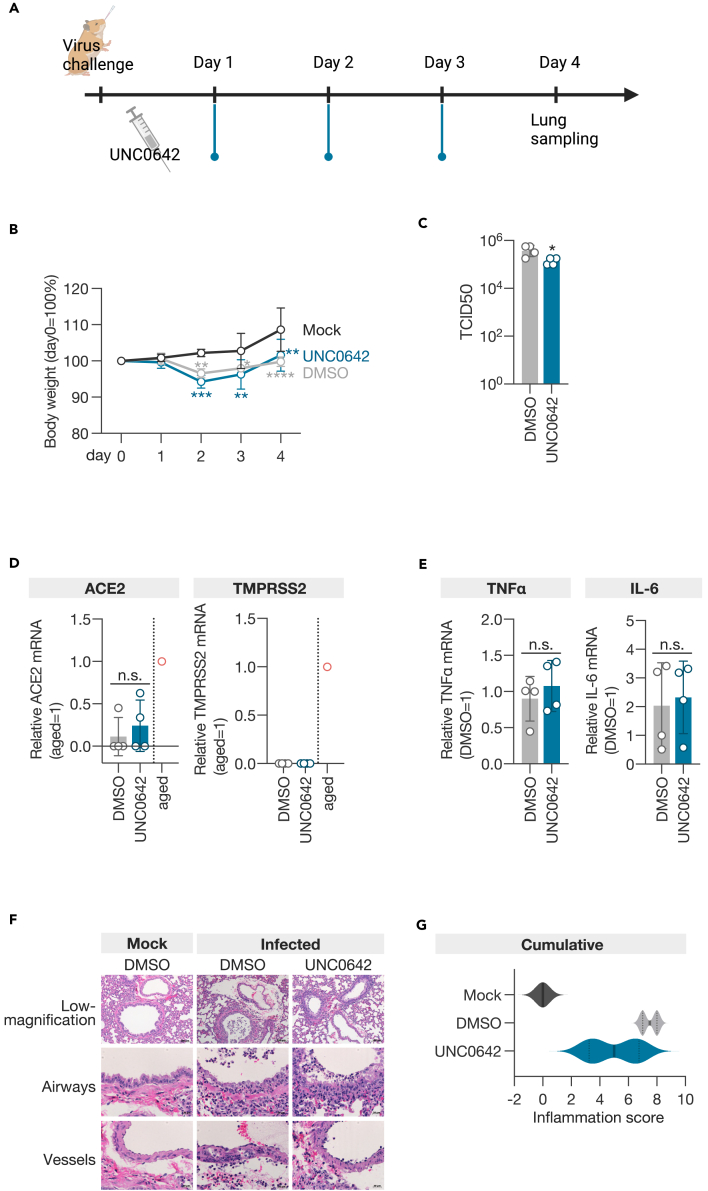
To assess whether UNC0642 also suppresses the growth of SARS-CoV-2 in vivo, we inoculated four 4-week-old male Syrian hamsters with the virus and administered the inhibitor intraperitoneally at a dose of 5 mg/kg once daily (Figure 6A). On day 4 post-inoculation, the lungs of the animals were examined for virus titration and pathological analysis. The UNC0642-treated group experienced weight loss due to viral infection, similar to the DMSO-treated group but showed greater recovery after 4 days of infection than the DMSO-treated group (Figure 6B). Viral growth in the lungs was suppressed in the UNC0642-treated group compared to the DMSO-treated group (Figure 6C). Similar to the observations in sgEHMT1 and sgEHMT2 cells, treatment with UNC0642 did not alter the expression of ACE2 and TMPRSS2, nor did it increase the expression of TNF-α and IL-6 (Figures 6D and 6E). These findings suggest that EHMT1/2 may not be involved in the cellular entry process of SARS-CoV-2 or the induction of innate immunity in vivo. Using a semiquantitative pathological scoring system38,39 to evaluate the infected animals, the cumulative score was significantly lower in the UNC0642-treated group than in the DMSO-treated group (Figures 6F and 6G). In particular, the scores for vessels and regeneration were lower in the UNC0642-treated group (Figure S5). These results demonstrate that the selective inhibitor of EHMT1/2, UNC0642, also suppresses the growth of SARS-CoV-2 and reduces the severity of infection in vivo.
Figure 6.
EHMT1/2 inhibitor reduces SARS-CoV-2 growth and disease severity in infected animals
(A) Timeline of virus challenge and inhibitor administration.
(B) Body weight change of hamsters (n = 4).
(C) Viral load in the lungs of hamsters 4 days post-infection (n = 4).
(D) qPCR analysis of ACE2 and TMPRSS2 mRNA in the lungs of hamsters 4 days post-infection (n = 4).
(E) qPCR analysis of TNF-α and IL-6 mRNA in the lungs of hamsters 4 days post-infection (n = 4).
(F) Representative histopathological images of the lungs of hamsters.
(G) Violin plot of the cumulative inflammation score. Data are presented as the mean ± SD of four individuals. Statistical analysis was performed using Dunnett’s multiple comparison test for body weight changes (B) and an unpaired t test for viral load and mRNA analyses (C–E). n.s., not significant; ∗, p < 0.05.
Discussion
We used CRISPR-Cas9-mediated genome-wide screening to identify TRIM23, TRIM33, and EHMT1/2 as proviral factors of SARS-CoV-2. Using purified virus, we were able to perform a genetic screen using clear death of A549 cells as an indicator. This allowed us to shorten the time between inoculation and cell collection and led to the identification of previously unreported proviral factors. On the other hand, the set of proviral factors identified in this study included numerous genes involved in the acidification discovered in previous A549 screens,1,2 as well as five common factors associated with ACE2 (Figures 1C and 1D). This screening also extracted DAXX decreased in the infected cells (Figure 1B), which has been reported as a restriction factor.12 However, when we transduced sgRNA into A549-hACE2-Cas9 cells and assessed virus replication, we did not observe any promotion of virus replication (Figure 1E). This may be due to differences in knockout efficiency or cell types used (Figure 2C).
TRIM28 and TRIM33 are E3 ubiquitin ligases involved in the transcriptional regulation of various cellular responses, including innate immunity.21,22,23,40 However, in A549-hACE2-Cas9 cells in which TRIM28 or TRIM33 was knocked out, we did not observe an increase in the mRNA expression levels of IFNB1, IL-6, and TNF-α (Figures 3A and 3C). These results suggest that TRIM28 and TRIM33 do not promote SARS-CoV-2 proliferation by suppressing the antiviral response but are involved in the viral growth cycle. Previous reports indicated that TRIM28 knockdown upregulated ACE2,20 but our results showed no statistically significant change in ACE2 expression with TRIM28 knockout (Figure 2D). The discrepancies between these previous studies and our results may be due to whether the suppression of TRIM expression was transient or stable. Indeed, transient overexpression of TRIM28 and TRIM33 using plasmids yielded results contrary to expectations (Figures 4C and 4D), suggesting that cellular responses to changes in TRIM expression may differ significantly between transient and long-term effects. Rather than directly affecting viral proliferation, it is possible that transcriptionally regulated factors or post-transcriptional modifications mediated by these TRIMs may influence SARS-CoV-2 proliferation. Several SARS-CoV-2 proteins have been reported to be ubiquitinated,41,42,43,44,45 but further analysis is needed to determine whether TRIM28 and TRIM33 are involved in the ubiquitination of viral proteins.
EHMT1/2 catalyzes the methylation of histones and nonhistone proteins, and the substrates of the enzymes that contribute to SARS-CoV-2 replication may be either host or viral proteins or both. There are reports that EHMT2 is involved in numerous gene pathways that promote translation, many of which are also involved in SARS-CoV-2 replication.46,47 Moreover, ORF8 of SARS-CoV-2 mimics histones through the ARKS motif and disrupts epigenetic control in infected cells.48 Inhibitors that block viral replication, whether applied before or after infection (Figure 5D), suggest that EHMT1/2 is involved in SARS-CoV-2 replication in a complex manner.
The development of host-targeted antiviral drugs has been limited in the past due to concerns about potential side effects. However, there is an increasing need for diverse strategies to treat viral infections. In this study, UNC0642, which selectively inhibits the enzymatic activity of EHMT1/2, significantly suppressed SARS-CoV-2 replication in vitro and demonstrated the ability to limit, although not completely inhibit, viral replication and disease severity in vivo (Figure 6). The modest effects observed in vivo may be attributed to intraperitoneal administration of the drug and suboptimal pulmonary concentration due to its distribution throughout the body. Despite these challenges, we believe that our findings provide valuable insights for the future development of antiviral drugs. In particular, they highlight the potential for structurally improved EHMT1/2 inhibitors. This study advances the development of antiviral methods targeting proviral factors and suggests a promising direction for therapeutic intervention.
Limitations of the study
This study has several limitations. First, the CRISPR-Cas9 screen was limited to the A549-hACE2 cell line, and results may vary with other cell lines or tissues. Additionally, differences in knockout efficiency and cell types could affect the assessment of viral replication, which may influence data interpretation. Moreover, transient and long-term effects of TRIM28 and TRIM33 expression yielded different cellular responses, suggesting the need for further studies on this aspect. The EHMT1/2 inhibitor, UNC0642, demonstrated significant suppression of viral replication in vitro but had limited efficacy in vivo. This may be due to the challenges in achieving optimal pulmonary concentration through intraperitoneal administration. Furthermore, the mechanisms by which the identified proviral factors (TRIM28, TRIM33, EHMT1, and EHMT2) facilitate SARS-CoV-2 replication remain largely unclear. It is not yet known whether these factors directly modify viral proteins or indirectly promote viral replication through other cellular pathways. Finally, it is uncertain whether the proviral factors identified in this study have similar effects on other viruses, and further research is required to establish their broader applicability. Despite these limitations, our findings provide valuable insights for future antiviral drug development, highlighting the potential of targeting proviral factors.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-TRIM28 antibody | Santa Cruz Biotechnology | Catalog # sc-515790 |
| anti-TRIM33 antibody | Abcam | Catalog # ab47062; RRID:AB_956365 |
| anti-EHMT1 antibody | Cell Signaling Technology | Catalog # 35005S; RRID:AB_2799068 |
| anti-EHMT2 antibody | Abcam | Catalog # ab185050; RRID: AB_2792982 |
| anti-VPS35 antibody | Santa Cruz Biotechnology | Catalog # sc-374372; RRID: AB_10988942 |
| anti-SARS-CoV-2 N antibody | Abcam | Catalog # 33717; RRID: AB_2941972 |
| anti-tubulin antibody | Merck | Catalog # T5168 |
| anti-H3 antibody | Cell Signaling Technology | Catalog # 4499 |
| anti-H3K9me2 antibody | Cell Signaling Technology | Catalog # 4658; RRID:AB_10544405 |
| anti-SARS-CoV-2 S antibody | Abcam | Catalog # ab272504; RRID: AB_2847845 |
| anti-ERGIC53 antibody | Santa Cruz Biotechnology | Catalog # sc-365158; RRID: AB_10709004 |
| anti-HCoV-229E nucleoprotein antibody | Sino Biological, Beijing | Catalog # 40640-T62 |
| Peroxidase AffiniPure™ Donkey Anti-Mouse IgG (H + L) | Jackson Immuno Research | Catalog # 715-035-150; RRID: AB_2340770 |
| Peroxidase AffiniPure™ Donkey Anti-Rabbit IgG (H + L) | Jackson Immuno Research | Catalog # 711-035-152; RRID: AB_10015282 |
| Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 555 | Thermo Fisher Scientific | Catalog # A-11034; RRID: AB_2576217 |
| Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Thermo Fisher Scientific | Catalog # A-21422; RRID: AB_2535844 |
| Bacterial and virus strains | ||
| SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo (EPI_ISL_418809) | The University of Tokyo | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| UNC0642 | MedChemExpress | Catalog # HY-13980 |
| DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride) | Thermo Fisher Scientific | Catalog #D1306 |
| Dimethyl Sulfoxide, Nuclease and Protease tested | Nacalai Tesque | Catalog # 08904-14 |
| Dulbecco’s modified Eagle’s medium (DMEM), high glucose | Thermo Fisher Scientific | Catalog # 11965118 |
| Fetal calf serum (FCS) | Thermo Fisher Scientific | Catalog # 10270106 |
| Penicillin-Streptomycin Mixed Solution | Nacalai Tesque | Catalog # 09367-34 |
| Puromycin(solution) | INVIVOGEN | Catalog # ant-pr-1 |
| Critical commercial assays | ||
| Primer/Probe N2 | NIHON GENE RESEARCH LABORATORIES, INC | N/A |
| LDH cytotoxicity assay kit | Nacalai Tesque | Catalog # 18250-35 |
| Experimental models: Cell lines | ||
| Human: A549 | ATCC | Catalog # CCL-185 |
| African green monkey: VeroE6/TMPRSS2 | National Institutes of Biomedical Innovation, Health and Nutrition, Osaka, Japan | Catalog # JCRB1819 |
| 293T/17 | ATCC | Catalog # CRL-11268 |
| Experimental models: Organisms/strains | ||
| Syrian hamsters: Slc:Syrian | Japan SLC Inc | N/A |
| Oligonucleotides | ||
| sgRNA targeting sequence for target genes, see Table S4 | This paper | N/A |
| Primers used for RT-qPCR, see Table S5 | This paper | N/A |
| Recombinant DNA | ||
| pKLV2-EF1a-Cas9Bsd-W | Addgene | Catalog # 68343 |
| pKLV2-U6gRNA5(BbsI)-PGKpuro2ABFP-W | Addgene | Catalog # 67974 |
| The Human CRISPR Library v.3 | Addgene | Catalog # 67989 |
| pcDNA3 | Thermo Fisher Scientific | Catalog # A-150228 |
| Software and algorithms | ||
| MAGeCK v0.5.9.5 | Li et al.56 | https://anaconda.org/bioconda/mageck |
| ImageJ | Open source | N/A |
| Prism 10 software | GraphPad | N/A |
| Other | ||
| 96-well cell culture plates | Corning | Catalog # 3596 |
| 12-well cell culture plates | Corning | Catalog # 3513 |
| Fusion Solo instrument | Vilber-Lourmat | N/A |
| CFX96 | Bio-Rad | N/A |
| Ti-E inverted microscope with a C1 confocal laser scanning system | Nikon | N/A |
| Eclipse TE-2000-U inverted microscope | Nikon | N/A |
| HT-7700 instrument | Hitachi | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Akiko Makino (makino@infront.kyoto-u.ac.jp).
Materials availability statement
Newly generated materials are available from the corresponding author upon reasonable request.
Data and code availability
The datasets generated during this study are provided in the supplementary tables. This study does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cells
A549 (CCL-185), VeroE6/TMPRSS2 (JCRB1819), and 293T (CRL-11268) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal calf serum (FCS) (Thermo Fisher Scientific) and 1% penicillin‒streptomycin (Nacalai Tesque, Kyoto, Japan). The cells were incubated at 37°C with 5% CO2.
Plasmids and compound
Cas9 and gRNA plasmids were purchased from Addgene (Watertown, MA, USA) (Cas9 vector, 68343; gRNA vector, 67974). These CRISPR-related lentiviruses, including the human v3 library, were produced by cotransfection of 293T cells with the transfer vectors psPAX2 and pMD2.G using Lipofectamine LTX (Thermo Fisher Scientific, Waltham, MA, USA). hACE2 cDNA was inserted between the XhoI and XbaI sites of CSII-CMV-RfA. Lentiviral vectors were rescued through the transfection of these plasmids with pCAG-HIVgp, pCMV-VSVG, and pCMV-Rev into 293T cells using TranIT-293 (TaKaRa, Shiga, Japan) or the ViraPower Lentiviral Expression System (Thermo Fisher Scientific). TRIM28, TRIM33, and EGFP cDNA was inserted between the EcoRV and NotI sites of pcDNA3 (Thermo Fisher Scientific). UNC0642 (MCE, HY-13980) was dissolved in DMSO to 10 mM and stored at −80°C. The working solution was diluted in DMSO to 1 mM and stored at −30°C.
Method details
Virus preparation
SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo was propagated in VeroE6/TMPRSS2 cells. For CRISPR‒Cas9 screening, SARS-CoV-2 was purified through ultracentrifugation with 20% sucrose at 27,000 rpm for 2 h at 4°C. The pellets were suspended in NTE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and centrifuged at 3,000 rpm for 10 min at 4°C. The supernatants were stored at −80°C until used for screening. All experiments using SARS-CoV-2 were performed in a biosafety level 3 containment laboratory at Kyoto University, which is approved for use by the Ministry of Agriculture Forestry and Fisheries, Japan.
Generation of knockout cells using the CRISPR‒Cas9 system
A549-hACE2-Cas9 or A549-Cas9 cells were transduced with sgRNA targeting 10 or 4 genes using lentiviral vectors. The sequences of the sgRNAs used are shown in Table S4. The transduced cells were subjected to puromycin selection (1 or 2 μg/mL) (INVIVOGEN) at 72 h after transduction. The knockout efficacy of target genes was assessed by Western blotting 18 days after the introduction of gRNA.
Genome-scale CRISPR‒Cas9 screen
The Human CRISPR Library v.3, which targets 18,365 genes with 113,526 sgRNAs, including 1,004 nontargeting control sgRNAs (Addgene, 67989),18 was used in this study. A total of 3.3 × 107 A549-hACE2-Cas9 cells were transduced with the lentiviral-packaged whole-genome sgRNA library to achieve 30% transduction efficiency (100× library coverage). The cells were subjected to puromycin selection (1 μg/mL) (Thermo Fisher Scientific) at 72 h after transduction. At 10 days after transduction, 3.5× 107 cells were inoculated with purified SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo at a multiplicity of infection (MOI) of 0.3. At 3 days post-infection, cells were harvested and subjected to DNA extraction.
DNA extraction, gRNA PCR amplification, Illumina sequencing and gRNA counting
Genomic DNA was extracted from cell pellets using the Blood & Cell Culture DNA Maxi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. PCR amplification, sgRNA sequencing (19-bp single-end sequencing with custom primers) and gRNA counting were performed.17 The DNB-SEQ platform was used for sgRNA sequencing.
CRISPR‒Cas9 screen data analysis
To identify hit genes, statistical analysis was performed using MAGeCK v0.5.9.556 by comparing the infected population vs. day 10 population (preinfection), the mock-infected population vs. day 10 population, and the infected and the mock-infected populations. GO enrichment analysis was performed by using Database for Annotation, Visualization and Integrated Discovery.
Virus titration
The SARS-CoV-2 titer was determined by the median tissue culture infectious dose (TCID50) of VeroE6/TMPRSS2. The infectious unit of HCoV-229E was titrated as follows: HuH-7 cells seeded in 96-well plates were inoculated with a serial 10-fold dilution of the virus and incubated for one day. Antigen-positive cells were detected by indirect immunofluorescence assay (IFA) using an anti-HCoV-229E nucleoprotein antibody (Sino Biological, Beijing, China).
Western blotting
Cell lysates were prepared in SDS sample buffer, separated by SDS‒PAGE using e-PAGEL (ATTO Corporation, Tokyo, Japan), and transferred to polyvinylidene difluoride membranes from a Trans-Blot Turbo PVDF Transfer Pack (Bio-Rad, Richmond, USA). The membranes were blocked with 5% skim milk (Wako Pure Chemical Industries) in TBS-T (Tris-buffered saline, 0.1% Tween 20) and incubated with anti-TRIM28 antibody (1:3,000) (sc-515790, Santa Cruz Biotechnology, Dallas, TX, USA), anti-TRIM33 antibody (1:1,000) (ab47062, Abcam, Cambridge, UK), anti-EHMT1 antibody (1:500) (35005S, Cell Signaling Technology, Danvers, MA, USA), anti-EHMT2 antibody (1:1,000) (ab185050, Abcam), anti-VPS35 antibody (1:1,000) (sc-374372, Santa Cruz Biotechnology), anti-SARS-CoV-2 N antibody (1:1.000) (#33717, CST), anti-tubulin antibody (1:2,000) (T5168, Merck, Darmstadt, Germany), anti-H3 antibody (1:2,000) (#4499, CST), or anti-H3K9me2 (1:1,000) (#4658, CST) diluted with Can Get Signal Immunoreaction Enhancer Solution 1 (Toyobo, Osaka, Japan) at 4°C overnight. After washing with TBS-T, the membranes were incubated with a 50,000-fold-diluted HRP-labelled anti-mouse or rabbit IgG antibody (Merck) at room temperature for 2 h or more. After washing with TBS-T, Clarity Western ECL Substrate (Bio-Rad) was used for detection by chemiluminescence reaction. Bands were detected and photographed by a Fusion Solo instrument (Vilber-Lourmat, Marne-la-Vallée Cedex, France). Band intensities were analyzed using ImageJ.57
RT‒qPCR
The virus RNA in the cell or culture supernatant was extracted with nucleospin RNA plus (TaKaRa) and quantified by RT‒qPCR using Primer/Probe N2 (NIHON GENE RESEARCH LABORATORIES, INC. Miyagi, Japan) and One Step PrimeScript III RT‒qPCR Mix (TaKaRa) with CFX96 (Bio-Rad, Hercules, CA, U.S.A.) for the N gene. RNA was extracted from sgTRIM28, sgTRIM33, sgEHMT1, and sgEHMT2 cells using nucleospin RNA plus (TaKaRa), and RT‒qPCR was performed using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) and Luna Universal qPCR Master Mix (New England Biolabs, New England Biolabs, Ipswich, MA, USA) and CFX96 (Bio-Rad) to quantify the mRNA levels. As a positive control for the detection of TNFα, qPCR human reference cDNA, oligo(dT)-primed (TaKaRa) was used. The sequences of the primer sets are listed in Table S5. RNA was extracted from infected hamsters using the RNeasy kit (QIAGEN, Hilden, Germany). Viral RNA and mRNA levels were detected as described above. Before qPCR, cDNA was preamplified using SsoAdvanced PreAmp Supermix (Bio-Rad). Lungs from 9-month-old male Slc:Syrian hamsters were used as a positive control.
Indirect immunofluorescence assay
Cells were fixed with 4% paraformaldehyde (Nacalai Tesque) at room temperature and treated with PBS containing 0.5% Triton X-100 and 2% FCS for 15 min, followed by incubation with anti-SARS-CoV-2 S antibody (1:1,000) (ab272504, Abcam) and anti-ERGIC53 antibody (1:20) (sc-365158, Santa Cruz Biotechnology) for 1 h. After washing, the cells were reacted with a 1000-fold diluted Alexa Fluor mouse 555- or rabbit 488-conjugated secondary antibody (Thermo Fisher Scientific) and 300 nM DAPI (Thermo Fisher Scientific) for 1 h. After washing with PBS, Immunostained cells were observed using a Ti-E inverted microscope with a C1 confocal laser scanning system (Nikon, Tokyo, Japan).
Cell growth kinetics
A total of 5.0 x 104/well of sgTRIM28, sgTRIM33, sgEHMT1, and sgEHMT2-trasnduced A549-hACE-Cas9 cells were seeded in a 12 well plate. Starting the next day after cell seeding, the number of cells was counted daily for 3 days.
Cytotoxicity LDH assay
A total of 4.0 × 102 sgTRIM28, sgTRIM33, sgEHMT1, and sgEHMT2-transduced A549-hACE2-Cas9 cells per well were seeded into a 96-well plate. Three days after seeding, cytotoxicity was assessed by measuring the absorbance at 490 nm using an LDH cytotoxicity assay kit (Nacalai Tesque).
Entry assay using VSV vector pseudotyped with SARS-CoV-2-S
293T cells were transfected with pCMV-VSV-G (Addgene) expressing VSV-G or pEF4/HisA (Thermo) expressing codon-optimized SARS-CoV-2 spike with a 19 amino acid deletion at the C-terminus using TransIT-293. Twenty-four hours later, G-complemented VSVΔG-GFP58,59 was inoculated at an MOI of 0.5. The supernatant was collected 24 h after inoculation, and debris was removed by centrifugation and stored at −80°. The efficiency of cell entry mediated by each envelope protein was calculated by counting the number of GFP-expressing cells with an Eclipse TE-2000-U inverted microscope (Nikon) after inoculating the generated pseudotyped virus into target cells.
The overexpression of TRIM28 and TRIM33
A total of 2.0 × 105 A549-Cas9-hACE2 cells per well were seeded in a 12-well plate. The next day, 1 μg of GFP, TRIM28, or TRIM33 expression plasmids were introduced using Lipofectamine LTX. The following day, the cells were infected at an MOI of 1.0, and harvested three days later. Viral RNA and viral proteins were evaluated by RT-qPCR and Western blotting, respectively.
Electron microscopic analysis
A549-hACE2-Cas9 cells infected with SARS-CoV-2 were fixed with 2.5% glutaraldehyde (TAAB Laboratories Equipment Ltd) and postfixed with 1% osmium tetroxide (TAAB Laboratories Equipment Ltd) at 4°C. After en bloc staining with 1% uranyl acetate, the cells were dehydrated with a series of ethanol gradients, followed by propylene oxide, and embedded in Epon 812 resin (TAAB Laboratories Equipment Ltd). The ultrathin sections were stained with 2% uranyl acetate and lead citrate and observed using a Hitachi HT-7700 instrument at 80 kV.
Evaluation of the inhibitory effect of UNC0642 on H3K9me2
A total of 1.0 × 105 A549-hACE2 cells per well were seeded in a 24-well plate. The following day, UNC0642 was added at concentrations of 0.08, 0.4, 2, and 10 μM. After 24 h, cells were harvested and the expression levels of EHMT1/2, histone H3, and H3K9me2 were assessed by Western blotting.
Animal experiments
SARS-CoV-2 was intranasally inoculated into four 4-week-old male Syrian hamsters (Japan SLC Inc, Shizuoka, Japan) at a dose of 102 TCID50 per animal using a mixture of 3 anaesthetics. For mock infection, DMEM with 2% FCS was used. UNC0642 at a dose of 5 mg/kg or 0.1% DMSO was administered intraperitoneally once daily starting from one day after infection. Body weight was measured daily from the day of virus inoculation. Four days after infection, the animals were euthanized, and the lungs were removed for virus titer measurement and pathological analysis. The animal experiments were approved by the Animal Experiment Committee at Kyoto University (#A22-8).
Pathologic assessment
Lungs collected from animals 4 days postinfection were fixed in 10% neutral buffered formalin (Wako), embedded in paraffin (Thermo), sectioned, and stained with haematoxylin (Merck) and eosin (Wako). Pathological features in airways, interstitium, vessels, alveolar spaces, oedema, and alveolar epithelial regeneration were scored as previously reported,37,38 with a score of 0 (no lesions), 1 (mild), 2 (moderate), and 3 (severe).
Quantification and statistical analysis
Statistical significance values were calculated using GraphPad Prism 10 software. The statistical tests used for the data are indicated in the figure legends.
Acknowledgments
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI grant numbers JP19K22530 (K.T.), JP20H00662 (K.T.), JP21K19909 (K.T.), and JP18K05991 (A.M.); National BioResource Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI grant number 19H04834 (A.M.); Japan Agency for Medical Research and Development grant number JP20wm0325011h0001 (A.M. and K.Y.); Japan Science and Technology Agency (JST) Start-ups from Advanced Research and Technology (START) Project Promotion Type (Commercialization Support), grant number JPMJST2113 (K.T.); JSPS Core-to-Core Program JPJSCCA20190008 (T.N. and K.T.); 2021 Kaketsuken Research grant (K.T.); JST Core Research for Evolutional Science and Technology grant number JPMJCR20HA (T.N.); and the Joint Usage/Research Center Program on Institute for Life and Medical Sciences, Kyoto University, Japan. CSII-CMV-RfA, pCAG-HIVgp, pCMV-VSVG, and pCMV-Rev were provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan. The illustrations in this paper were created using BioRender.
Author contributions
A.M. designed and managed the overall project. M.S., Y.M., Yusuke.T., M.I., R.K., K.Y., K.T., and A.M. performed and analyzed the in vitro experiments. Yugo.T. and T.N. performed electron microscope analysis. M.S. and A.M. conducted animal experiments. N.A. and Y.K. performed the pathological assessment.
Declaration of interests
The authors declare no competing interests.
Published: July 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110475.
Supporting Citations
The following references appear in the Supplemental information: 49,50,51,52,53,54,55.
Supplemental information
References
- 1.Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Y., Feng F., Hu G., Wang Y., Yu Y., Zhu Y., Xu W., Cai X., Sun Z., Han W., et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12:961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehfeld F., Eitson J.L., Ohlson M.B., Chang T.C., Schoggins J.W., Mendell J.T. CRISPR screening reveals a dependency on ribosome recycling for efficient SARS-CoV-2 programmed ribosomal frameshifting and viral replication. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rebendenne A., Roy P., Bonaventure B., Chaves Valadão A.L., Desmarets L., Arnaud-Arnould M., Rouillé Y., Tauziet M., Giovannini D., Touhami J., et al. Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. Nat. Genet. 2022;54:1090–1102. doi: 10.1038/s41588-022-01110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann H.H., Sánchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280.e5. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Le Pen J., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biering S.B., Sarnik S.A., Wang E., Zengel J.R., Leist S.R., Schäfer A., Sathyan V., Hawkins P., Okuda K., Tau C., et al. Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection. Nat. Genet. 2022;54:1078–1089. doi: 10.1038/s41588-022-01131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baggen J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021;53:435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- 10.Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell. 2021;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mac Kain A., Maarifi G., Aicher S.M., Arhel N., Baidaliuk A., Munier S., Donati F., Vallet T., Tran Q.D., Hardy A., et al. Identification of DAXX as a restriction factor of SARS-CoV-2 through a CRISPR/Cas9 screen. Nat. Commun. 2022;13:2442. doi: 10.1038/s41467-022-30134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan K., Farias A.G., Lee H., Guvenc F., Mero P., Brown K.R., Ward H., Billmann M., Aulakh K., Astori A., et al. Survival-based CRISPR genetic screens across a panel of permissive cell lines identify common and cell-specific SARS-CoV-2 host factors. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2022.e12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baggen J., Vanstreels E., Jansen S., Daelemans D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021;6:1219–1232. doi: 10.1038/s41564-021-00958-0. [DOI] [PubMed] [Google Scholar]
- 14.Alexander S.P.H., Fabbro D., Kelly E., Mathie A., Peters J.A., Veale E.L., Armstrong J.F., Faccenda E., Harding S.D., Pawson A.J., et al. THE CONCISE GUIDE TO PHARMACOLOGY 2021/22: Enzymes. Br. J. Pharmacol. 2021;178:S313–S411. doi: 10.1111/bph.15542. [DOI] [PubMed] [Google Scholar]
- 15.Tachibana M., Matsumura Y., Fukuda M., Kimura H., Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda J., Tachibana M., Ikura T., Shinkai Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 2006;281:20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- 17.Tzelepis K., Koike-Yusa H., De Braekeleer E., Li Y., Metzakopian E., Dovey O.M., Mupo A., Grinkevich V., Li M., Mazan M., et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016;17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong S.H., Li Y., Koike-Yusa H., Yusa K. Optimised metrics for CRISPR-KO screens with second-generation gRNA libraries. Sci. Rep. 2017;7:7384–7410. doi: 10.1038/s41598-017-07827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Fan Y., Huang Y., Du T., Liu Z., Huang D., Wang Y., Wang N., Zhang P. TRIM28 regulates SARS-CoV-2 cell entry by targeting ACE2. Cell. Signal. 2021;85 doi: 10.1016/j.cellsig.2021.110064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferri F., Parcelier A., Petit V., Gallouet A.S., Lewandowski D., Dalloz M., Van Den Heuvel A., Kolovos P., Soler E., Squadrito M.L., et al. TRIM33 switches off Ifnb1 gene transcription during the late phase of macrophage activation. Nat. Commun. 2015;6:8900. doi: 10.1038/ncomms9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vahid E., Pinto R.M., Sandi C., Sandi M., Al-Mahdawi S., Hein te Riele M.A.P. TRIM28 is a SUMO E3 Ligase and Negative Regulator of Interferon Regulatory Factor 7. NIH public acess. 2012;23:1–7. doi: 10.4049/jimmunol.1101704.TRIM28. [DOI] [Google Scholar]
- 23.Krischuns T., Günl F., Henschel L., Binder M., Willemsen J., Schloer S., Rescher U., Gerlt V., Zimmer G., Nordhoff C., et al. Phosphorylation of TRIM28 Enhances the Expression of IFN-β and Proinflammatory Cytokines During HPAIV Infection of Human Lung Epithelial Cells. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seier J.A., Reinhardt J., Saraf K., Ng S.S., Layer J.P., Corvino D., Althoff K., Giordano F.A., Schramm A., Fischer M., Hölzel M. Druggable epigenetic suppression of interferon-induced chemokine expression linked to MYCN amplification in neuroblastoma. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2020-001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehnertz B., Northrop J.P., Antignano F., Burrows K., Hadidi S., Mullaly S.C., Rossi F.M.V., Zaph C. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J. Exp. Med. 2010;207:915–922. doi: 10.1084/jem.20100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh N., Ramĩrez-Carvajal L., De Los Santos T., Golding M.C., Long C.R. Inhibition of EHMT2 Induces a Robust Antiviral Response Against Foot-and-Mouth Disease and Vesicular Stomatitis Virus Infections in Bovine Cells. J. Interferon Cytokine Res. 2016;36:37–47. doi: 10.1089/jir.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boson B., Legros V., Zhou B., Siret E., Mathieu C., Cosset F.L., Lavillette D., Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cattin-Ortolá J., Welch L.G., Maslen S.L., Papa G., James L.C., Munro S. Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation. Nat. Commun. 2021;12:1–11. doi: 10.1038/s41467-021-25589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherer K.M., Mascheroni L., Carnell G.W., Wunderlich L.C.S., Makarchuk S., Brockhoff M., Mela I., Fernandez-Villegas A., Barysevich M., Stewart H., et al. SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abl4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinson D.A., Stephens K.E., O’Meally R.N., Bhat S., Dancy B.C.R., Cole R.N., Yegnasubramanian S., Taverna S.D. De novo methylation of histone H3K23 by the methyltransferases EHMT1/GLP and EHMT2/G9a. Epigenet. Chromatin. 2022;15:36. doi: 10.1186/s13072-022-00468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chopra A., Cho W.C., Willmore W.G., Biggar K.K. Hypoxia-Inducible Lysine Methyltransferases: G9a and GLP Hypoxic Regulation, Non-histone Substrate Modification, and Pathological Relevance. Front. Genet. 2020;11:579636. doi: 10.3389/fgene.2020.579636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinkai Y., Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coates D.R., Chin J.M., Chung S.T.L. Discovery of an in vivo Chemical Probe of the Lysine Methyltransferases G9a and GLP. Bone. 2011;23:1–7. doi: 10.1021/jm401480r.Discovery. [DOI] [Google Scholar]
- 35.Cao Y.P., Sun J.Y., Li M.Q., Dong Y., Zhang Y.H., Yan J., Huang R.M., Yan X. Inhibition of G9a by a small molecule inhibitor, UNC0642, induces apoptosis of human bladder cancer cells. Acta Pharmacol. Sin. 2019;40:1076–1084. doi: 10.1038/s41401-018-0205-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tostanoski L.H., Yu J., Mercado N.B., Mcmahan K., Martinot A.J., Piedra-mora C., Anioke T., Giffin V.M., Hope D.L., Wan H., et al. Immunity elicited by natural infection or Ad26.COV2.S vaccination protects hamsters against SARS-CoV-2 variants of concern. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abj3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piedra-Mora C., Robinson S.R., Tostanoski L.H., Dayao D.A.E., Chandrashekar A., Bauer K., Wrijil L., Ducat S., Hayes T., Yu J., et al. Reduced SARS-CoV-2 disease outcomes in Syrian hamsters receiving immune sera: Quantitative image analysis in pathologic assessments. Vet. Pathol. 2022;59:648–660. doi: 10.1177/03009858221095794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng L., Mitoma H., Tricot C., Bao M., Liu Y., Zhang Z., Liu Y.-J. The E3 Ubiquitin Ligase Tripartite Motif 33 Is Essential for Cytosolic RNA–Induced NLRP3 Inflammasome Activation. J. Immunol. 2014;193:3676–3682. doi: 10.4049/jimmunol.1401448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valerdi K.M., Hage A., van Tol S., Rajsbaum R., Giraldo M.I. The role of the host ubiquitin system in promoting replication of emergent viruses. Viruses. 2021;13:369. doi: 10.3390/v13030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., Zheng H., Zhu J., Dong Q., Wang J., Fan H., Chen Y., Zhang X., Han X., Li Q., et al. Ubiquitin-Modified Proteome of SARS-CoV-2-Infected Host Cells Reveals Insights into Virus-Host Interaction and Pathogenesis. J. Proteome Res. 2021;20:2224–2239. doi: 10.1021/acs.jproteome.0c00758. [DOI] [PubMed] [Google Scholar]
- 43.Cao Z., Xia H., Rajsbaum R., Xia X., Wang H., Shi P.Y. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell. Mol. Immunol. 2021;18:746–748. doi: 10.1038/s41423-020-00603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan Z., Hu B., Xiao H., Tan X., Li Y., Tang K., Zhang Y., Cai K., Ding B. The E3 Ubiquitin Ligase RNF5 Facilitates SARS-CoV-2 Membrane Protein-Mediated Virion Release. mBio. 2021;13 doi: 10.1128/mbio.03168-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu G., Wu Y., Xiao T., Qi F., Fan L., Zhang S., Zhou J., He Y., Gao X., Zeng H., et al. Multiomics approach reveals the ubiquitination-specific processes hijacked by SARS-CoV-2. Signal Transduct. Target. Ther. 2022;7 doi: 10.1038/s41392-022-01156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., May B., De Freitas J.K., Glicksberg B.S., Mason C.E., et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L., Muneer A., Xie L., Zhang F., Wu B., Mei L., Lenarcic E.M., Feng E.H., Song J., Xiong Y., et al. Novel gene-specific translation mechanism of dysregulated, chronic inflammation reveals promising, multifaceted COVID-19 therapeutics. bioRxiv. 2020 doi: 10.1101/2020.11.14.382416. Preprint at. [DOI] [Google Scholar]
- 48.Kee J., Thudium S., Renner D.M., Glastad K., Palozola K., Zhang Z., Li Y., Lan Y., Cesare J., Poleshko A., et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature. 2022;610:381–388. doi: 10.1038/s41586-022-05282-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., Sasaki M., Takaoka A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 50.Tokunaga T., Yamamoto Y., Sakai M., Tomonaga K., Honda T. Antiviral activity of favipiravir (T-705) against mammalian and avian bornaviruses. Antiviral Res. 2017;143:237–245. doi: 10.1016/j.antiviral.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 51.Jaworska J., Gravel A., Fink K., Grandvaux N., Flamand L. Inhibition of Transcription of the Beta Interferon Gene by the Human Herpesvirus 6 Immediate-Early 1 Protein. J. Virol. 2007;81:5737–5748. doi: 10.1128/jvi.02443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Z., Ge Y., Ding L., Zhang Z., Qu Y., Jin C., Wang X.N., Wang Z. Synthesis and evaluation of alkoxy-substituted enamides against influenza A virus in vitro and in vivo. Bioorg. Chem. 2023;139 doi: 10.1016/j.bioorg.2023.106712. [DOI] [PubMed] [Google Scholar]
- 53.Suresh V., Parida D., Minz A.P., Sethi M., Sahoo B.S., Senapati S. Tissue Distribution of ACE2 Protein in Syrian Golden Hamster (Mesocricetus auratus) and Its Possible Implications in SARS-CoV-2 Related Studies. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.579330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao W., Lau H.E., Xie H., Poon V.K.M., Chan C.C.S., Chu H., Yuan S., Yuen T.T.T., Chik K.K.H., Tsang J.O.L., et al. SARS-CoV-2 infection induces inflammatory bone loss in golden Syrian hamsters. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-30195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang S.J., Wei T.C., Hsu C.H., Ho S.N., Lai C.Y., Huang S.F., Chen Y.Y., Liu S.J., Yu G.Y., Dou H.Y. Characterization of virus replication, pathogenesis, and cytokine responses in syrian hamsters inoculated with sars-cov-2. J. Inflamm. Res. 2021;14:3781–3795. doi: 10.2147/JIR.S323026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.National Institutes of Health (NIH). Turning Discovery Into Health https://www.nih.gov/.
- 58.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G. Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein. J. Virol. 2020;94 doi: 10.1128/jvi.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are provided in the supplementary tables. This study does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.