Abstract
Recently, we demonstrated that inverted repeat sequences inserted into first-generation adenovirus (Ad) vector genomes mediate precise genomic rearrangements resulting in vector genomes devoid of all viral genes that are efficiently packaged into functional Ad capsids. As a specific application of this finding, we generated adenovirus–adeno-associated virus (AAV) hybrid vectors, first-generation Ad vectors containing AAV inverted terminal repeat sequences (ITRs) flanking a reporter gene cassette inserted into the E1 region. We hypothesized that the AAV ITRs present within the hybrid vector genome could mediate the formation of rearranged vector genomes (ΔAd.AAV) and stimulate transgene integration. We demonstrate here that ΔAd.AAV vectors are efficiently generated as by-products of first-generation adenovirus-AAV vector amplification. ΔAd.AAV genomes contain only the transgene flanked by AAV ITRs, Ad packaging signals, and Ad ITRs. ΔAd.AAV vectors can be produced at a high titer and purity. In vitro transduction properties of these deleted hybrid vectors were evaluated in direct comparison with first-generation Ad and recombinant AAV vectors (rAAVs). The ΔAd.AAV hybrid vector stably transduced cultured cells with efficiencies comparable to rAAV. Since cells transduced with ΔAd.AAV did not express cytotoxic viral proteins, hybrid viruses could be applied at very high multiplicities of infection to increase transduction rates. Southern analysis and pulsed-field gel electrophoresis suggested that ΔAd.AAV integrated randomly as head-to-tail tandems into the host cell genome. The presence of two intact AAV ITRs was crucial for the production of hybrid vectors and for transgene integration. ΔAd.AAV vectors, which are straightforward in their production, represent a promising tool for stable gene transfer in vitro and in vivo.
Vectors for the treatment of genetic deficiencies require the stable transduction of target cells without associated toxic or immunologic side effects. First-generation adenoviruses (Ads) have a number of properties that make them an attractive vehicle for gene transfer (12, 15). These include the ability to produce purified virus at high titers in concert with highly efficient gene transfer of up to 8-kb expression cassettes into a large variety of cell types in vivo, including nondividing cells. The major limitation has been short-term expression in vivo due to the development of immune responses to expressed viral proteins, resulting in toxicity and viral clearance as well as the episomal status of Ad DNA within transduced cells. Stable integration of Ad DNA into the host genome has been reported only for wild-type forms of specific subtypes and appears not to occur in a detectable manner with the E1- and E3-deleted Ad5 vectors widely used for gene transfer in vitro and in vivo (4).
Recombinant adeno-associated virus vectors (rAAV) have been developed by substituting the viral rep and cap genes with a therapeutic minigene (up to 5 kb) while retaining the inverted terminal repeat sequences (ITRs) (25). In cultured cells, rAAV integrates stably into host chromosomes with a relatively low frequency of 1 × 10−4 to 3 × 10−4 genomes per cell (34); however, the integration efficiency can be increased by stimulation of the host DNA repair machinery (1, 32). Based on a number of reports, it appears that the only requirement for rAAV integration and episomal concatemerization is the presence of AAV ITRs and as yet unknown cellular factors (2, 38, 41). In the single-stranded AAV genome, the palindromic 145-bp ITRs form a T-shaped secondary structure, which is probably the substrate for concatemerization and chromosomal integration. Analysis of rAAV integration junctions demonstrated that rAAV integrates randomly based on microhomology between ITRs and host sequences at the crossover point (34, 41). The functional importance of the AAV ITRs was underscored by in vitro transfection experiments with double-stranded circular plasmids. The presence of two ITRs in transfected plasmid DNA was sufficient to rescue the rAAV genome from the plasmid backbone and to mediate the integration of rAAV vector DNA (2, 38). Recently, Recchia et al. demonstrated that helper-dependent, “gutless” Ad vectors carrying a transgene flanked by AAV ITRs stably transduced hepatoma cells with the transgene integrated into the host genome (31).
These studies suggest that the presence of AAV ITRs in a double-stranded DNA, including the Ad vector genome, may mediate vector integration into the host DNA. This hypothesis assumes that the characteristic secondary structures are formed by AAV ITRs present in double-stranded DNA and that these structures are recognized by cellular enzymes, which mediate the excision of the AAV cassette and the integration into the host genome. In this context, structure-specific cellular recombination enzymes have been described to be associated with recombination processes in pro- and eukaryotes (22, 29).
Clearly, a vector that combines the advantages of Ad (high titer, high infectivity, and large capacity) with the integration capability of AAV would be advantageous for gene therapy approaches requiring stable gene transfer. In addition, this hybrid vector should be devoid of all Ad genes whose expression may cause immunological or toxic side effects. On the way to reach this goal, we utilized our earlier finding that inverted repeat sequences (IRs) inserted into Ad vector genomes mediated predictable genomic rearrangements resulting in vector genomes devoid of all viral genes (36). These genomes (named ΔAd.IR) contained only the transgene cassette flanked on both sides by precisely duplicated IRs, Ad packaging signals, and Ad ITRs. The formation of ΔAd.IR genomes appeared to involve recombination between homologous IRs and Ad replication. The generation of ΔAd.IR genomes was efficient (with ∼5 × 104 genomes produced per cell) and did not depend on the sequence within or adjacent to the IRs. These small vector genomes devoid of all viral genes were efficiently packaged into functional Ad particles. These particles could be separated from virions with full-length genomes based on their lighter buoyant density in CsCl gradients. ΔAd.IR vectors infected cultured cells with the same efficiency as first-generation vectors; however, transgene expression was only transient (∼7 days) due to the instability of the deleted genomes within transduced cells.
As a specific application of ΔAd.IR vectors, we designed E1-deleted Ad vectors containing two AAV ITRs flanking reporter gene cassettes as IRs. Our hypotheses behind the generation of these hybrid vectors were (i) that the AAV ITRs would mediate the formation of vector genomes devoid of all Ad genes that are packaged into Ad particles, (ii) that these particles would be produced at high titer and purity and would allow for efficient gene transfer into target cells, and (iii) that the presence of AAV ITRs in these genomic derivatives could mediate transgene integration into the host genome, resulting in stable gene expression.
MATERIALS AND METHODS
Production and characterization of viral vectors. Plasmids.
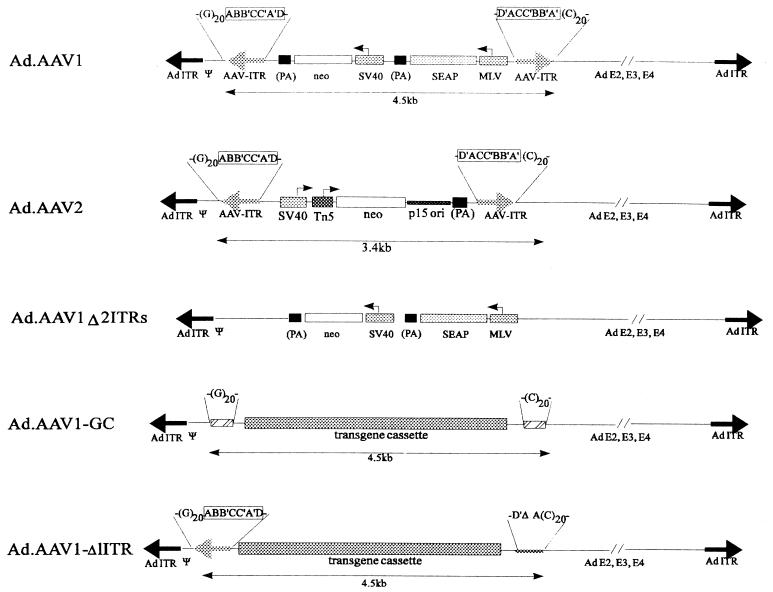
The AAV1 vector cassette containing AAV ITRs and secreted human placental alkaline phosphatase (SEAP) and neomycin phosphotransferase (neo) expression units was obtained by AseI/ScaI digestion of the plasmid pALSAPSN (1) (gift from David Russell, University of Washington). The 4.4-kb AAV vector fragment was cloned via NotI adapter linkers into pXJCL1 (Microbix, Toronto, Canada) (pAd.AAV1) (Fig. 1). Another shuttle vector (pAd.AAV1-Δ2ITRs) with the same transgene cassette, but lacking the AAV ITRs, was generated by inserting the 3.7-kb AflII/BsmI fragment of pALSAPSN into pXJCL1. For generating the Ad.AAV1-GC vector (Fig. 1), synthetic double-stranded oligonucleotides containing a (dG)20 or a (dC)20 stretch were linked to the transgene cassette lacking AAV ITRs and were cloned as an HindIII/XbaI insert into a modified pXJCL1. For pAd.AAV1-Δ1ITR (Fig. 1), a construct was used where a spontaneous deletion in the left AAV ITRs between the A and A′ regions had occurred. To create a second hybrid vector (Ad.AAV2) (Fig. 1), the AAVSNori cassette developed by E. A. Rutledge was used (34). AAV vector DNA was obtained from pASNori2 as a 3.4-kb BsaI/ScaI fragment and was inserted into the EcoRV site of pXCJL1. As is generally the case for AAV vector plasmids, the AAV ITRs are prone to rearrangements. To minimize deletions in these functionally critical regions, all constructs for generation of hybrid vectors were assembled in low-copy-number plasmids, which were grown in Escherichia coli XL1Blue (Stratagene, La Jolla, Calif.). Furthermore, after each cloning step or large-scale plasmid amplification, both AAV ITRs were carefully mapped by restriction analysis with enzymes that cut inside or adjacent to the ITRs (BssHII, AhdI, SmaI, BglI, BsmI, AflII, and ScaI).
FIG. 1.
Ad vectors with incorporated AAV cassettes. The genomic structures of first-generation vectors are shown. The length of these genomes is ∼35 kb. Arrows indicate promoters. Abbreviations: Ψ, Ad packaging signal; MLV, Moloney murine leukemia virus promoter; SV40, SV40 promoter; (PA), SV40 polyadenylation signal; Tn5, bacterial promoter; p15 ori, p15a bacterial plasmid origin with the direction of leading strand-strand DNA synthesis opposite to that of neo gene transcription. The palindromic AAV ITRs (ABB′CC′A′D and D′ACC′BB′A′) are framed. The following control vectors were generated: Ad.AAV1-Δ2ITRs (containing the transgene cassette without AAV ITRs), Ad.AAV1-GC (containing the transgene cassette flanked on both sides with an 20-mer dG and dC stretch), and Ad.AAV1-Δ1ITR (containing one intact AAV ITR upstream of the transgene cassette and an AAV ITR deleted for most of A′, the complete BB′/CC′ regions, and most of A located downstream of the transgene cassette).
Adenoviruses.
First-generation viruses with different transgene cassettes incorporated into their E1 regions were generated by recombination of pXCJL1-derived shuttle plasmids and pJM17 (Microbix) in 293 cells as previously described (20). For each virus, at least 20 plaques were picked, amplified, and analyzed by restriction digest. Viruses containing two AAV ITRs tended to have deletions within the ITRs or other Ad sequences and to recombine with Ad sequences present within the 293 cell genome. Only plaques from viruses with intact ITRs were amplified, CsCl banded, and titered as previously described (15, 20). All virus preparations tested negative for replication-competent adenovirus and bacterial endotoxin (21). Virus was stored at −80°C in a solution containing 10 mM Tris-Cl (pH 7.5), 1 mM MgCl2, and 10% glycerol.
To generate ΔAd.AAV, 293 cells were infected with Ad.AAV at a multiplicity of infection (MOI) of 25 and were harvested 40 h after infection. Cells were lysed in phosphate-buffered saline (PBS) by four cycles of freezing and thawing. Lysates were centrifuged to remove cell debris and were digested for 30 min at 37°C with 500 U of DNase I and 200 μg of RNase A per ml in the presence of 10 mM MgCl2. Five milliliters of lysate was layered on a CsCl step gradient (0.5 ml at 1.5 g/cm3, 2.5 ml at 1.35 g/cm3, and 4 ml at 1.25 g/cm3) and ultracentrifuged for 2 h at 35,000 rpm (rotor SW41). CsCl fractions were collected by puncturing the tube and were analyzed for viral DNA (20) or were subjected to ultracentrifugation at 35,000 rpm (rotor SW41) for 18 h in an equilibrium gradient with 1.32 g of CsCl per cm3. The band containing the deleted viruses (ΔAd.AAV) was clearly separated (0.5-cm distance) from other banded viral particles containing full-length Ad.AAV genomes. Fractions containing deleted virus were dialyzed against a solution containing 10 mM Tris-Cl (pH 7.5), 1 mM MgCl2, and 10% glycerol and were stored at −80°C. The genome titer of ΔAd.AAV preparations was determined based on quantitative Southern analysis of viral DNA purified from viral particles in comparison to different concentrations of a 4.4-kb AseI/ScaI fragment of pALSAPSN, according to a protocol previously described (20). Titers were routinely obtained in the range of 3 × 1012 to 8 × 1012 genomes per ml. Assuming one genome is packaged per capsid, the genome titer equals the particle titer. The level of contaminating Ad.AAV1 in ΔAd.AAV1 preparations was less than 0.1%, as determined by Southern analysis, which is consistent with results obtained by plaque assay of 293 cells (fewer than five plaques per 106 total genomes). Primers used for sequencing the left and right ITR-vector junctions in ΔAd.AAV1 genomes were specific to the SEAP-neo cassette (5′GGCGTTACTTAAGCTAGAGCTTATCG and 5′CTCTCTAGTTCTAGCCTCGATCTCAC). The recombinant AAV stock containing the SEAP-neo cassette (AV2/ALSAPSN [1]) used in these studies was obtained from Dusty Miller (Fred Hutchinson Cancer Research Center, Seattle, Wash.). The stock was free of replication-competent AAV (<50 particles/ml) and wild-type Ad (<100 particles/ml). The genome titer of the virus stock was obtained by quantitative Southern blotting as described by Russell et al. (33).
Cell culture.
SKHEP-1 cells (HTB-52, American Type Culture Collection, Rockville, Md.), an endothelial cell line derived from human liver (11), were grown in high-glucose Dulbecco’s modified Eagle medium with 10% fetal calf serum. SKHEP-1 cells were analyzed for integrated AAV provirus by Southern analysis of genomic DNA by using the AAV2 wild-type genome obtained from pAAV/Ad (35) (gift from David Russell) as a probe. No specific bands were detected in undigested genomic SKHEP-1 DNA or after digestion with HindIII. For viral infection, confluent cells were incubated with different viral doses for 2 h, followed by intensive washing. For G418 selection, 24 h after infection, SKHEP-1 cells were trypsinized and plated at different dilutions under G418 selection (900 μg of active compound per ml) (Boehringer-Mannheim, Mannheim, Germany). Culture medium containing G418 was changed every 3 days. The number of colonies with >104 cells was counted after 4 weeks of selection and then divided by the number of initially seeded cells. This ratio was used to express the integration frequency of viral vectors. Single colonies stably transduced with ΔAd.AAV1 were obtained by limiting dilutions of infected cells in 96-well plates. Colonies were grown to 106 cells in the presence of G418. Immunofluorescence analysis for Ad proteins expressed in SKHEP-1 cells was performed as previously described (20). Antibodies against SEAP were from DAKO Corp. (Carpinteria, Calif.).
Southern blotting.
Cultured cells were washed three times with PBS before harvesting. For analysis, 10-μg samples of genomic DNA were either left undigested or were digested with restriction endonucleases at 37°C overnight and then electrophoresed in a 0.8% agarose gel and transferred to a nylon membrane (Hybond N+; Amersham, Arlington Heights, Ill.). The blots were hybridized in a rapid hybridization buffer (Amersham) with [α-32P]dCTP-labeled DNA probes (>108 cpm/μg of DNA). The fragments used for labeling were the 1.7-kb BamHI SEAP fragment obtained from pALSAPSN, the 4.5-kb BsaI/SnaBI fragment of pAAV/Ad containing the AAV2 genome (ITRs, rep, and cap genes), and the 0.5-kb EcoRI/EcoRI fragment of pXCJL1 with the Ad ITR and packaging region.
PFGE.
Control SKHEP-1 cells or cells infected with ΔAd.AAV1 or Ad.AAV1 were trypsinized and washed once with PBS. SKHEP-1 cells (107) were resuspended in 1 ml of PBS and sealed in 1% SeaPlaque-low-melting agarose (FMC Bioproducts, Rockland, Maine) plugs. A fragment of these plugs containing approximately 106 cells was lysed in situ with the method described by Peterson et al. (30). For digestion, chromosomal DNA was incubated with 10 U of restriction endonuclease overnight. Analytical pulsed-field gel electrophoresis (PFGE) was performed at 200 V with 35/70-ms cycles for 16 h at 12°C in a 1% agarose gel. DNA transfer and hybridization were as described for Southern blotting.
RESULTS
Generation and characterization of adenovirus-AAV hybrid vectors devoid of all viral genes.
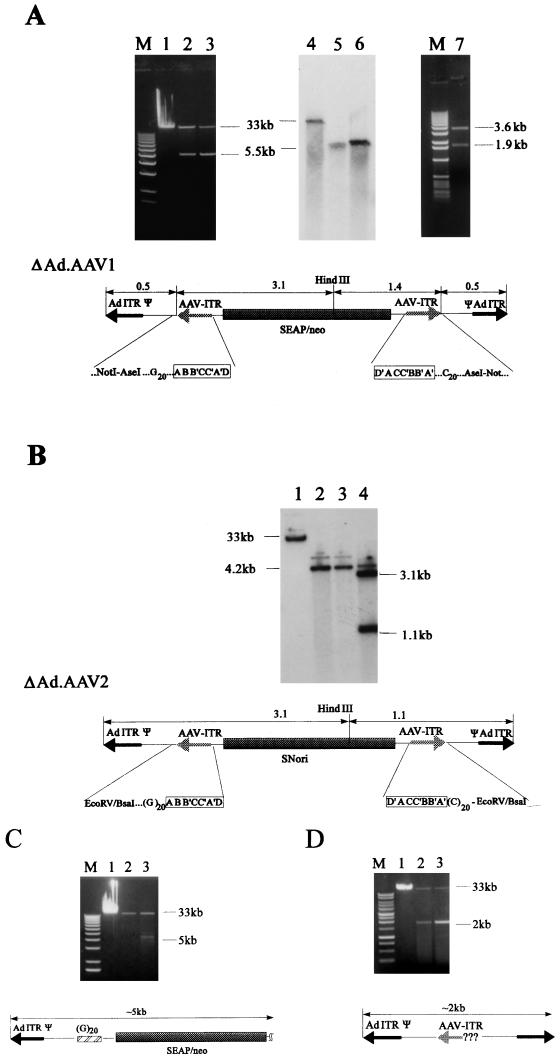
Previously, we demonstrated that IRs inserted into first-generation Ad vector genomes mediate precise genomic rearrangements resulting in vector genomes devoid of all viral genes that are efficiently packaged into functional Ad capsids (36). We hypothesized that these deleted genomes were formed by intermolecular homologous recombination between the IRs. This hypothesis was confirmed by coinfection with two viruses, each providing one inverse homology element (36). As a specific application of these replication derivatives, which can be used as efficient gene transfer vehicles, we designed an adenovirus-AAV hybrid vector (Ad.AAV1) (Fig. 1). This vector contained AAV ITRs as IRs that should allow for the formation of the specific genomes depleted of viral genes. Another rationale behind the generation of this hybrid vector was to utilize the potential of AAV ITRs to stimulate episomal concatemerization and chromosomal vector integration. Considering the transient character of gene expression from ΔAd.IR vectors (36), integration is important as a means of vector stabilization to provide persistent transgene expression. Therefore, we incorporated AAV vector DNA containing both AAV ITRs flanking a SEAP and neo expression cassette into the E1 region of recombinant E1- and E3-deficient Ads. To test whether the ∼200-bp fragments containing the AAV ITRs would mediate the formation of vectors devoid of all viral genes, viral material from infected 293 cells was banded in CsCl step gradients. In addition to the banded virus containing full-length (Ad.AAV1) genomes, a band with lower density that contained viral material appeared in CsCl gradients. Analysis of purified particles from this band revealed packaged viral DNA with a length of 5.5 kb (Fig. 2A), hereafter referred as to ΔAd.AAV1. ΔAd.AAV1 particles can be purified by additional CsCl equilibrium gradients to preparations with less than 0.1% contaminating first-generation Ad.AAV1, as analyzed by quantitative Southern blotting (Fig. 2A, lanes 4 to 6) and plaque assay on 293 cells (see Materials and Methods). After restriction and sequence analysis of viral DNA from purified particles, the structure of ΔAd.AAV1 was deduced as shown in Fig. 2A. The 5.5-kb ΔAd.AAV1 genome contains the left 540 nucleotides of Ad DNA with the left Ad ITR and packaging signal followed by the AAV vector cassette with intact left and right AAV ITRs linked to a second copy of the left-hand Ad genome (packaging signal and ITR) in reverse orientation. As determined by quantitative Southern analysis of viral DNA from purified particles, the titer for ΔAd.AAV1 routinely obtained was 5 × 1012 genomes per ml or ∼104 packaged genomes per 293 cell (data not shown). The scheme for generating high-titer ΔAd.AAV hybrid viruses was the same as for first-generation Ads. All functions for ΔAd.AAV1 replication and particle formation are provided from Ad.AAV1 genomes amplified in the same cell.
FIG. 2.
Production and characterization of Ad-AAV hybrid vectors. (A) Ad.AAV1 was amplified in 293 cells and banded in a CsCl step gradient. Undigested viral DNA purified from CsCl gradient fractions was analyzed in 0.8% agarose gels: lane 1, DNA isolated from 10 μl of banded Ad.AAV1 particles (full-length, 35-kb genome); lanes 2 and 3, DNA isolated from 10 μl of the banded ΔAd.AAV1 particles (5.5-kb genome) after one CsCl step gradient (from two different virus preparations). Viral material containing ΔAd.AAV1 genomes was purified by additional CsCl equilibrium gradients. Viral DNA (100 ng) from purified particles was analyzed by Southern blotting with a SEAP-specific probe. Lane 4, full-length Ad.AAV1 DNA; lanes 5 and 6, ΔAd.AAV1 particles purified by one (lane 5) or two (lane 6) rounds of ultracentrifugation in equilibrium gradients. Viral DNA (1 μg) from the 5.5-kb product of Ad.AAV1 isolated from purified particles was digested with HindIII and was analyzed in a 0.8% agarose gel (lane 7). The structure of the 5.5-kb genome (ΔAd.AAV1) shown at the bottom of panel A was deduced after restriction and sequence analysis. The position of the HindIII site is indicated. The AAV ITR-vector junctions were sequenced with primers specific to regions within the transgene cassette (see Materials and Methods). The palindromic composition of the AAV ITRs is framed. M, 1-kb DNA ladder (GIBCO-BRL). (B) Ad.AAV2 was amplified and CsCl banded in the same way as described for Ad.AAV1. Viral material was analyzed as for Ad.AAV1 (lanes 1 to 3). The amount of DNA per lane corresponds to 5 μl of viral material. The faint bands that run above Ad.AAV2 (lane 1) and ΔAd.AAV2 DNA (lanes 2 and 3) may represent a different conformational structure, which was seen often with linear plasmid DNA containing AAV ITRs. Lane 4; HindIII digest of ΔAd.AAV2 DNA. (C and D) Amplification products of Ad.AAV-GC and Ad.AAV1-Δ1ITR, respectively. Viral DNA in lanes 1 was isolated from 5-μl samples of banded viruses with full-length genomes. Viral DNA separated in lanes 2 and 3 was isolated from 1-ml gradient fractions with densities lighter than 1.32 g/cm3. No banded virus was visible in these fractions.
To ensure that hybrid vectors with different transgene cassettes can be produced, we generated a second vector, Ad.AAV2 (Fig. 1). In this vector, the AAV cassette contained the neo gene under the control of both the simian virus 40 (SV40) early promoter and the transposon 5 promoter for expression in human and bacterial cells. This vector also contained a bacterial replication origin (34). Amplification of Ad.AAV2 in 293 cells yielded the production of Ad particles containing a small, 4.2-kb genome (ΔAd.AAV2) at an efficiency of ∼10% of the total amount of packaged genomes (Ad.AAV2 and ΔAd.AAV2) (Fig. 2B). Analysis of the viral genomes isolated from ΔAd.AAV2 revealed the same structure as seen for ΔAd.AAV1 genomes, with the AAV cassette flanked by Ad packaging signals and ITRs.
In Ad.AAV1, the AAV ITRs were flanked by oligo-dG or -dC tracts. To evaluate the role of these complementary regions in the formation of ΔAd.AAV1 genomes, the vector Ad.AAV-GC (Fig. 1) was constructed. Although during amplification of Ad.AAV-GC a smaller genome species was generated, the yield of this replication product was several orders of magnitude lower than that of ΔAd.AAV1 genomes (Fig. 2C, lanes 2 and 3). Moreover, the small Ad.AAV-GC derivative did not contain the duplicated Ad sequences. To investigate the role of AAV ITRs in ΔAd.AAV genome formation, we constructed a vector deleted for both ITRs (Ad.AAV1-Δ2ITRs) and a vector where the central part of the right AAV ITR (deleted for most of A′, the complete BB′ and CC′ regions, and most of A) was removed (Ad.AAV1-Δ1ITR) (see Fig. 1). No other viral genomes besides the full-length Ad DNA were observed after amplification of the first-generation vector lacking both ITRs and G/C tracts (Ad.AAV1-Δ2ITRs, not shown). For Ad.AAV1-Δ1ITR, a minor amplification product which was found in viral material from CsCl fractions of ∼1.30 g/cm3 was ∼2 kb and had the structure shown in Fig. 2D, lane 2. The formation of this 2-kb product was ∼1,000-fold less efficient than the generation of ΔAd.AAV1 genomes.
In the process of generating Ad.AAV hybrid vectors, we noticed that AAV ITRs are prone to rearrangements and deletions at the level of plasmid DNA and when they are incorporated into Ad vectors. These mutations included partial deletions within the AAV ITR(s) or the complete loss of the AAV ITR(s). This inherent instability required permanent testing for intactness of AAV ITRs to ensure the efficient formation of ΔAd.AAV genomes. The yield of ΔAd.AAV produced in 293 cells after infection with Ad.AAV was significantly reduced when Ad.AAV vector preparations were used that contained a high percentage of genomes with deleted AAV-ITRs.
Taken together, our data show that the presence of two intact AAV ITRs flanking a reporter gene cassette was required for the effective formation of ΔAd.AAV genomes. This process did not efficiently occur with partially deleted ITRs or oligo-dC and oligo-dG stretches flanking the expression cassette.
Transduction studies with deleted ΔAd.AAV hybrid vectors in SKHEp-1 cells.
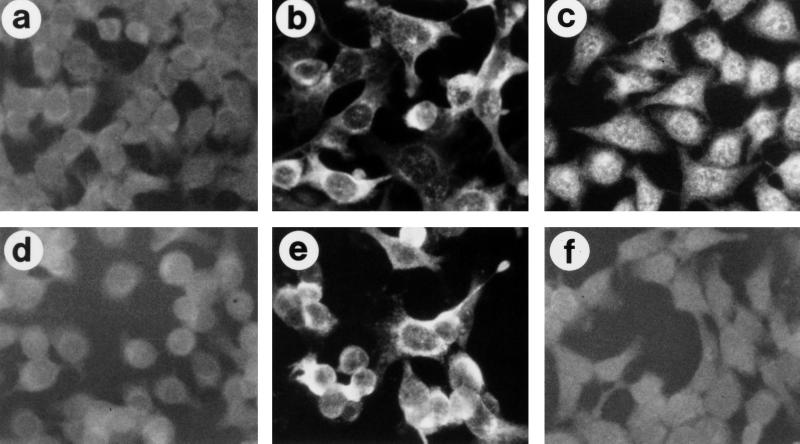
To evaluate the in vitro transduction efficiency of ΔAd.AAV1, SKHEP-1 cells were infected with hybrid viruses. SKHEP-1 cells (11) do not support replication of E1-deleted Ad vector DNA, unlike other transformed human cell lines, including HeLa and 293 cells (20, 27). This is important for long-term integration studies, which could be affected by episomal viral DNA replication of the small amount of contaminating (E1-deleted) Ad.AAV1 present in ΔAd.AAV1 preparations. In a first experiment, viral DNA was labeled with bromodeoxyuridine (BrdU) during amplification of virus to investigate cellular and nuclear vector uptake in situ. For these studies, confluent SKHEP-1 cells were infected with 2,000 genomes of ΔAd.AAV1 or Ad.AAV1 per cell. BrdU-tagged viral DNA was detected in 100% of nuclei at 3 h postinfection for both viruses (data not shown). The absence of expressed Ad gene products in ΔAd.AAV1-transduced cells at day 3 postinfection was demonstrated by immunofluorescence with antibodies to one of the major early proteins (E4-orf6). Expressed Ad proteins were detected only in cells infected with Ad.AAV1 (Fig. 3). The signals specific for the transgene product SEAP were similar in intensity between cells infected with Ad.AAV1 or ΔAd.AAV1.
FIG. 3.
Expression of viral E4 proteins and SEAP in transduced cells. SKHEP-1 cells were infected with first-generation vector Ad.AAV1 (b and c) and the deleted vector ΔAd.AAV1 (e and f) at MOIs of 2,000 genomes per cell. Two days after infection, the expression of viral E4 (open reading frame 3/4) protein (b and e) and SEAP (c and f) was analyzed by immunofluorescence with specific mouse monoclonal antibodies. All samples (a through f) were incubated with anti-mouse immunoglobulin-fluorescein isothiocyanate-conjugated antibodies. Panels a and d show nontransduced cells.
To test whether or not the AAV ITRs can mediate ΔAd.AAV1 integration into the host genome, SKHEP-1 cells were infected with different genome titers of ΔAd.AAV1, Ad.AAV1, and Ad.AAV1-Δ2ITRs, and the number of G418-resistant colonies that formed after 4 weeks of selection was determined (Table 1). For direct comparison, a rAAV1 virus containing exactly the same SEAP-neo cassette was included in these transduction studies. rAAV1 infection of SKHEP-1 cells with MOIs of 103 or 104 genomes/cell conferred G418 resistance in 2.7 and 90.8% of infected cells, respectively. The evaluation of higher MOIs was limited by the titer of the rAAV preparation (1010 genomes/ml). Compared with the rAAV1, the formation of G418-resistant cells after infection with ΔAd.AAV1 (MOIs 103 and 104) was half as efficient. However, since this virus had a titer 500 times higher than that of rAAV1, a 100% colony formation efficiency was obtained with ΔAd.AAV1 infections at MOIs greater than 105 genomes/cell. Although infection with the first-generation vector containing AAV vector DNA (Ad.AAV1) allowed for long-term G418 resistance as well, the transduction rate was clearly affected by dose-dependent toxicity. Cells infected with Ad.AAV1 at MOIs greater than 103 developed cytopathic effects by day 4 after infection and were not able to form colonies after 4 weeks of G418 selection. No G418-resistant colonies were present after infection with the first-generation vector lacking both AAV ITRs, which is consistent with the reported inability of Ad5-based recombinant Ad vectors to integrate at a detectable efficiency (4) or to persist episomally in dividing cells.
TABLE 1.
Formation of G418-resistant colonies after infection with hybrid viruses in comparison with rAAVa
| MOI (genomes per cell) | % Formation of G418-resistant colonies after 4 weeks of selection (SEM)
|
|||
|---|---|---|---|---|
| rAAV1 | ΔAd.AAV1 | Ad.AAV1 | Ad.AAV1-Δ2ITRs | |
| 101 | <0.001 | <0.001 | <0.001 | <0.001 |
| 102 | 0.01 | 0.01 | 0.06 | <0.001 |
| 103 | 2.7 (1.6) | 1.3 (1) | 5.4 (3.0) | <0.001 |
| 104 | 90.8 (7.0) | 48.0 (8.9) | 12.9 (7.2) | <0.001 |
| 105 | N/A | 93.1 (5.4) | 3.8 (2.1) | <0.001 |
| 106 | N/A | 100 | <0.001 | <0.001 |
| 107 | N/A | 100 | <0.001 | <0.001 |
Confluent SKHEP-1 cells (12-well plates) were infected with different MOIs of rAAV1 (stock, 1010 genomes per ml), ΔAd.AAV1 (stock, 5 × 1012 genomes per ml), Ad.AAV1 (stock, 1013 genomes per ml), and Ad.AAV1Δ1ITR (stock, 9 × 1012 genomes per ml) in a volume of 100 μl. Twenty four hours after infection, cells were washed, trypsinized, and plated at different dilutions. G418 was added 24 h after plating, and selection was performed for 4 weeks. G418-resistant colonies contained, on average, >5 × 104 cells (at least 16 cell divisions). A significant number of small colonies visible at 2 weeks postinfection did not survive continued selection, probably due to episomal vector expression. Cells infected with first-generation Ads with MOIs greater than 104 developed cytopathic effects during the first week of selection. The rAAV titer was not high enough to perform infection studies with MOIs greater than 104. The colony formation is expressed as percentage of the number of colonies after selection to the number of cells initially seeded for selection. Results are based upon three independent experiments. N/A, nonapplicable.
Approximately 2 × 104 ΔAd.AAV1 genomes or 0.9 × 104 rAAV genomes were required to yield one stable transfectant. Since all stable colonies contained integrated ΔAd.AAV1 vector DNA (see below) or rAAV DNA, this number represents the minimal integration frequency for SKHEP-1 cells. The number of G418-resistant colonies does not necessarily reflect the total frequency of integration events, because not all integrated copies may express neo due to chromosomal positional effects or rearrangements within the vector cassette.
In conclusion, (i) ΔAd.AAV stably transduced an immortalized human cell line with a low frequency comparable to rAAV; however, transduction rates can be scaled up by using greater MOIs of ΔAd.AAV1, which is produced at higher titers than rAAV1. (ii) In contrast to infection with the first-generation vector, Ad.AAV1, infection with ΔAd.AAV1 was not associated with dose-dependent cytotoxicity. (iii) A comparison of the number of G418-resistant colonies after infection with ΔAd.AAV1, Ad.AAV1, or the vector lacking AAV ITRs, Ad.AAV-Δ2ITRs, supports the hypothesis that the presence of two intact AAV ITRs is crucial for hybrid vector integration.
Integration studies with ΔAd.AAV1.
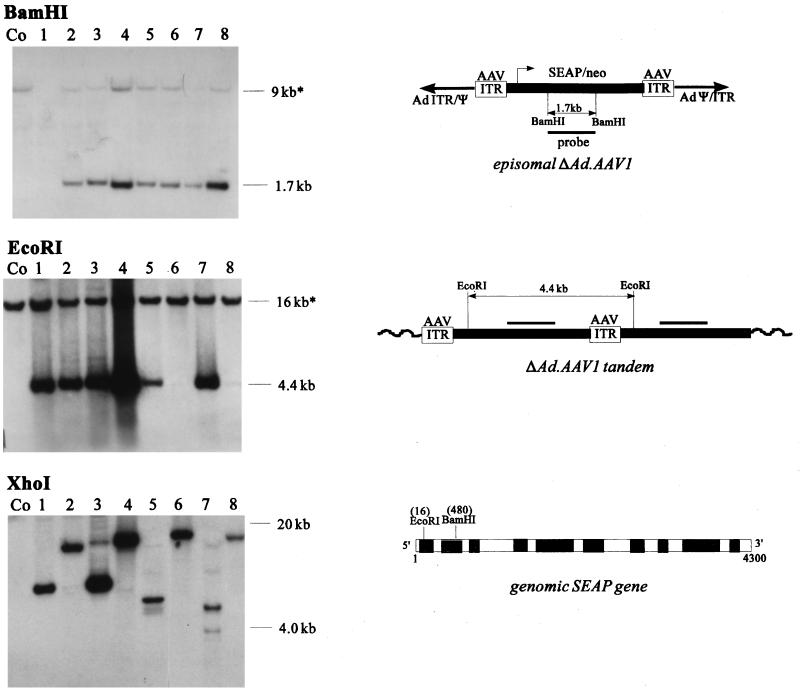
To investigate the structure of transduced ΔAd.AAV1 genomes in G418-resistant colonies, genomic DNA was isolated from single clones grown to 106 cells. Genomic DNA was digested with BamHI, which cuts in the AAV cassette twice, to confirm the presence and intactness of vector DNA, EcoRI, which cuts the cassette once near the 5′ end, to evaluate the presence of vector concatemers, and XhoI, which does not cut in ΔAd.AAV, to determine the integration pattern. Genomic Southern blots were hybridized with a SEAP-specific probe (Fig. 4). Nontransduced SKHEP-1 cells contain an endogenous SEAP gene, resulting in specific bands which can serve as internal controls to evaluate the copy number of transduced vector genomes.
FIG. 4.
Analysis of ΔAd.AAV1 genomes after transduction of SKHEP-1 cells by Southern blotting with a SEAP-specific probe. (A) SKHEP-1 cells were infected with 105 genomes per cell. Genomic DNA from single colonies was expanded to 106 cells and was analyzed by Southern blotting with a SEAP-specific probe. Genomic DNA (10 μg) was digested with BamHI, EcoRI, or XhoI and was separated in 0.8% agarose gels. Genomic DNA from diploid SKHEP-1 (11) contains two copies of the SEAP gene resulting in endogenous signals, labeled by asterisks. XhoI fragments were between 4 and 20 kb. The localizations of BamHI, EcoRI, and XhoI sites in the ΔAd.AAV1 genome (containing the SEAP cDNA) and in the endogenous genomic SEAP gene are indicated. The XhoI digestion product of the endogenous SEAP gene was larger than 23 kb and does not appear on the blot. The diagram illustrates the structure of episomal ΔAd.AAV1 vector DNA and integrated tandems forming junctions with chromosomal DNA. The complementary region for the SEAP-specific probe is indicated.
In all analyzed clones, digestion of genomic DNA with BamHI released an internal 1.7-kb fragment which was slightly more intense than the two copies of the endogenous SEAP gene in diploid SKHEP-1 cells (11). Digestion with EcoRI resulted in a 4.4-kb band in six of eight clones which would be released if the AAV cassette had formed a head-to-tail concatemer(s) (see graphic in Fig. 4). Digestion of the genomic DNA with XhoI produced bands of various sizes, ranging from 4 to 20 kb, which is consistent with random integration. The absence of integrated Ad sequences was confirmed by rehybridization of the same blot with a probe containing the Ad ITR and packaging region, which revealed no specific signals (data not shown). Since no Ad-specific signals were detected in DNA samples from clones transduced with ΔAd.AAV, we concluded that only AAV vector DNA had integrated without the flanking Ad sequences. Taken together, the data suggest that AAV vector DNA integrated randomly, mostly in the form of head-to-tail tandems. However, from this data, it remained unclear whether there were multiple integration sites consisting of single tandems or a single integration site consisting of concatemers with multiple vector copies.
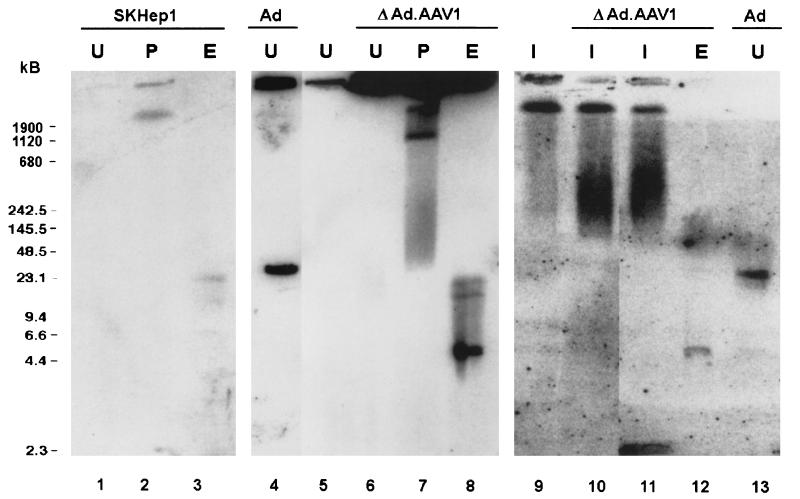
To confirm the integrated status of ΔAd.AAV1 DNA, after 4 weeks of selection high-molecular-weight chromosomal DNA from pools of transduced cells was separated by PFGE, followed by Southern analysis with a SEAP-specific probe (Fig. 5). Pulsed-field gels can separate DNA fragments with sizes ranging from 2 kb to several megabases and can, therefore, be used to demonstrate the stable association of vector DNA with host cell chromosomes. We analyzed undigested DNA and DNA digested with the intron-encoded endonucleases I-CeuI (9-bp recognition sequence) or PI-SceI (>11-bp recognition sequence) (Gibco BRL). As shown by ethidium bromide staining of PFGE gels, these enzymes produce fragments of human cellular DNA in the range of 300 to 800 kb (I-CeuI) and 1 to 2 Mb (PI-SceI), allowing the separation of high-molecular-weight episomal vector forms from chromosomal DNA and the detection of random integration events. In undigested DNA samples from cells stably transduced with ΔAd.AAV1, no large episomal forms of ΔAd.AAV1 DNA were detected, whereas a distinct 35-kb band was visible in DNA samples from SKHEP-1 cells isolated 3 days after infection with first-generation adenovirus, Ad.AAV1 (lanes 4 and 13). Digestion with EcoRI revealed a 4.4-kb fragment, specific for integrated tandem copies of the AAV cassette (lanes 8 and 12). Digestion with PI-SceI yielded a smear with a distinct >2-Mb endogenous SEAP fragment in SKHEP-1 cells (lane 2) and an additional distinct signal in the range of ∼1 to 2 Mb in G418-resistant colonies transduced with ΔAd.AAV1 (lane 7). We hypothesize that the ∼2-Mb signal represents an accumulation of random genomic DNA fragments carrying integrated ΔAd.AAV1 vector DNA. In a study by Miao et al., a similar signal was seen with PI-SceI-digested chromosomal DNA from liver transduced with rAAV vectors (24). The random integration status of these samples was later confirmed by sequencing the rAAV integration junctions (26). In support of random integration of ΔAd.AAV1, we demonstrated that I-CeuI digestion, which cuts chromosomal DNA more often than PI-SceI, resulted in a smear between 250 and 1,000 kb in ΔAd.AAV1-transduced SKHEP-1 cells (lanes 10 and 11), whereas only a specific high-molecular-weight band was observed in control SKHEP-1 cells (lane 9), representing the endogenous SEAP gene. In conclusion, these results confirm that ΔAd.AAV1 DNA integrated randomly into chromosomal DNA in vitro.
FIG. 5.
PFGE. Control SKHEP-1 cells (SKHEP1) (106) (lanes 1 to 3, 5, and 9), 106 SKHEP-1 cells from G418-resistant pools infected with ΔAd.AAV1 and selected for 4 weeks (ΔAd.AAV1) (lanes 6 to 8 and 10 to 12), or 106 SKHEP-1 cells collected 3 days after infection with 2,000 genomes of Ad.AAV1 (Ad) (lanes 4 and 13) were sealed in agarose plugs, lysed in situ, and subjected to PFGE with or without prior digestion with restriction endonucleases. U, undigested; P, digested with PI-SceI; I, digested with I-CeuI; E, digested with EcoRI. (The transfer of fragments of less than 10 kb from low-melting-agarose gels after PFGE is inefficient; therefore, the intensity of the high-molecular-weight band of the endogenous SEAP fragment cannot be compared with the signal from the 4.4-kb vector tandem-specific band in order to draw conclusions about the number of integrated vector copies.)
DISCUSSION
Previously, we demonstrated that IRs inserted into first-generation Ad vector genomes could be employed to mediate precise rearrangements within the Ad genome. The resulting rearranged vector genomes were devoid of all viral genes and were efficiently packaged into Ad capsids. These vectors infected cells with the same efficiency as first-generation vectors; however, the deleted genomes were unstable in transduced cells and allowed only for transient gene expression.
As a specific application of IR-mediated genomic rearrangements, we generated Ad.AAV hybrid vectors, representing first-generation Ad vectors containing two AAV ITRs flanking reporter gene cassettes inserted into the E1 region. We hypothesized that the AAV ITRs present in Ad.AAV could be used, on one hand, to mediate the formation of vector genomes devoid of all viral genes, and, on the other hand, as a means to stimulate transgene integration and thus provide stable gene expression.
This study demonstrates that deleted genomes (ΔAd.AAV) are efficiently formed during replication of Ad.AAV vectors in 293 cells. ΔAd.AAV genomes contain the transgene cassette flanked on both sides by an identical sequence comprising the AAV ITR, Ad packaging signal, and Ad ITR. We speculated that the mechanism(s) underlying this precise duplication involves homologous recombination and Ad replication (36).
We demonstrated here that ΔAd.AAV vector genomes were packaged into Ad capsids because DNase I-treated ΔAd.AAV particles efficiently transferred their genomes into cells, as shown by analysis of BrdU-tagged genomes and transgene expression. The 5.5-kb ΔAd.AAV genomes that contain two packaging signals were about fivefold less efficiently packaged than the corresponding full-length genomes. This is in agreement with a study by Parks and Graham (28), where plasmids carrying Ad genomes of different sizes were used in combination with helper virus to determine the lowest packaging size for Ad vectors. Vectors of less than 27 kb were recovered with about half the efficiency of larger vectors. Interestingly, a 15-kb genome was packaged at a higher efficiency than 20- to 25-kb vectors. However, the work of Parks and Graham demonstrated a clear disadvantage in the amplification of genomes of less than 25 kb during multiple virus passages. From this experiment, it was not clear whether the smaller vectors were less efficiently replicated or less efficiently packaged. This study is difficult to compare with our ΔAd.AAV vectors, which start out full length (Ad.AAV) and are deleted in the producer cells (perhaps after a critical event required for packaging has occurred) during one round of large-scale amplification. Packaging of a 9-kb mini-Ad vector generated by Cre-lox recombination (20) and encapsidated Ad minigenomes (8, 10, 17) has been previously reported.
With ΔAd.AAV1 as an example, this study shows that ΔAd.AAV vectors can be produced at a high titer and purity using standard techniques for first-generation Ad amplification and purification. All the functions required for ΔAd.AAV replication and particle formation are provided from Ad.AAV genomes amplified in the same cell. The purification of ΔAd.AAV particles is possible by ultracentrifugation in CsCl gradients based on their lighter buoyant density. The contamination of ΔAd.AAV1 with first-generation vector, Ad.AAV1, is less than 0.1%. This is within the range of helper virus contamination seen in preparations of gutless Ad vectors, which represent a new episomal vector type that allowed for stable transgene expression without hepatotoxicity and antiviral immune responses. The efficiency of vector production measured on a genome-per-cell basis is comparable or higher than labor-intensive techniques for rAAV production (3, 13, 16, 23, 35, 39). In our experiments, the transducing titers, expressed as G418-resistant colonies per milliliter, were 9 × 105 for rAAV and 2.5 × 108 for ΔAd.AAV1 with ratios of transducing to genome titers of 1:9,000 and 1:20,000, respectively.
The deleted hybrid vector efficiently infected cells, resulting in transgene expression at a level comparable to first-generation vectors. The results of PFGE and Southern analysis after infection with ΔAd.AAV1 in vitro suggested random integration as head-to-tail tandems typically seen for rAAV. The integration frequency of ΔAd.AAV1 in vitro was comparable to that reported for rAAV (34). The mechanisms of ΔAd.AAV1 integration and concatemerization are still unclear. As shown in Table 1, hybrid vector integration was dependent on the presence of AAV ITRs. The tandem arrangement of integrated AAV cassettes observed requires hairpin resolution in the double-stranded genome by cellular factors with Rep-like activity (i.e., able to generate breakpoints at the Rep binding site) and a DNA replication step prior to integration similar to rAAV or AAV ITR-containing plasmids (2, 37, 38). In rAAV integration studies using cultured cell lines, both concatemeric and single-copy vector proviruses have been described (7, 34, 41). The parameters for vector concatemerization could include cell-type-specific functions and MOI. Furthermore, ΔAd.AAV1 tandem formation in vitro may be linked to genomic DNA replication in proliferating cell cultures during G418 selection because AAV ITRs can serve as origins for replication (40). Our results from Southern blotting do not allow for definitive conclusions about the number of vector copies per integration site.
Recently, we demonstrated that vectors containing small Ad genomes generated by Cre-lox recombination (20) or genomic rearrangements mediated by IRs (36) transduced cells efficiently. However, these small genomes were only short-lived and were degraded by day 7 after infection in vitro. We suggested that the expression of certain viral proteins, including the Ad precursor to the terminal protein, is required to stabilize Ad genomes in transduced cells (19). In contrast, infection with ΔAd.AAV1 allows for stable transduction, suggesting that transgene integration occurs shortly after infection or that AAV ITRs can stabilize the viral genome as an episome until transgene integration occurs. Theoretically, concatemeric episomal vector forms with covalently closed hairpins at either end may have extended nuclear stability. More detailed studies of integration kinetics as well as transduction studies in stationary versus proliferating cells are required to answer this question.
The new ΔAd.AAV hybrid vector has a number of advantages compared to recombinant Ads and rAAV. Compared to first-generation Ad vectors, ΔAd.AAV vectors are less cytotoxic, since no Ad genes are expressed within transduced cells. Furthermore, it appears that viral proteins present in the incoming ΔAd.AAV1 particles are not problematic in the dose range used in this study. The absence of viral gene expression within transduced cells in vivo may reduce the antiviral immune response, which is a prerequisite for vector persistence. We demonstrated that ΔAd.AAV1 stably transduced cells, most likely by random vector integration into chromosomal DNA. Integration of the transgene is important for gene transfer into proliferating cells where Ad, as an episomal vector, would be lost after several cell divisions.
Compared to rAAV vectors, the potential advantages of ΔAd.AAV vectors include the technically easy production of high-titer virus and the larger transgene size that can be accommodated by ΔAd.AAV vectors. Furthermore, we speculate that Ad structural proteins present in ΔAd.AAV particles together with the double-stranded nature of incoming ΔAd.AAV genomes may allow for the transduction of nondividing cells. Finally, the composite structure of the Ad capsid and the accumulated knowledge on virus-cell interaction may allow for retargeting Ad5-based ΔAd.AAV vectors to new cellular receptors by modification of their fiber knobs. Modification of vector tropism is more problematic with rAAV vectors, where the capsid is composed of only three proteins.
The concept of combining elements from different viruses is not new. Viral vector chimeras were generated, for example, between Ad and retrovirus (5), Ad and Epstein-Barr virus (18), Ad and SV40 (9), and Ad and AAV (6). Johnston et al. (14) constructed hybrid vectors based on a herpes simplex virus type 1 amplicon vector in which the transgene cassette was flanked by AAV ITRs to induce episomal amplification and chromosomal integration. The vector genome was packaged into herpes simplex virus type 1 virions, resulting in hybrid vectors with titers of up to 105 infectious units per ml, which allowed for extended transgene expression in human glioma cells. In a study by Fisher et al. (6), rAAV vector DNA was incorporated into first-generation Ads in order to improve rAAV production. These hybrid vectors were conjugated via poly-l-lysine with rep-expression plasmids to stimulate rAAV DNA rescue and replication. However, the authors did not analyze viral material, which banded at a lighter density in CsCl gradients, and failed to identify the ΔAd.AAV vector species. Furthermore, the infection and purification conditions in that study differed significantly from ours, which were optimized to increase the output of ΔAd.AAV1. In contrast to our study, these Ad-based hybrid vectors contained viral genes that express potentially cytotoxic and immunogenic viral proteins in infected cells. Shortly after completion of the presented report, Recchia et al. described the generation of helper-dependent Ads with incorporated AAV vector DNA or AAV rep genes (31). Their data confirm our observation that AAV ITRs present in Ad genomes mediate vector integration in the absence of Rep expression. The efficiency of stable transduction of hepatoma cells after infection with the gutless vectors carrying a hygromycin resistance transgene flanked by AAV ITRs was comparable to the transduction frequency obtained with ΔAd.AAV vectors described in this study. The authors demonstrated by in situ hybridization that the transgene integrated into the chromosomes of hepatoma cells. Coexpression of Rep78 increased targeted insertion of AAV ITR-flanked DNA into the AAV S1 site at chromosome 19; however, Rep78 had no effect on the overall integration frequency in hepatoma cells. Recchia et al. did not perform dose-dependent transduction studies in direct comparison to rAAV or first-generation Ads. Notably, the production of helper-dependent Ad vectors includes labor-intensive plasmid transfection and multiple rounds of vector passages to obtain vectors with minimal helper contamination. The titers of vectors used by Recchia et al. were more than 1,000-fold lower than the titers that can be obtained for ΔAd.AAV vectors after one round of infection of 293 cells followed by a standard CsCl gradient purification protocol. In this context, our ΔAd.AAV vector system devoid of all viral genes is technically more straightforward and allows for the production of vectors with higher titers.
Integrating Ad.AAV hybrid vectors devoid of all viral genes represent a promising tool for gene therapy approaches. Future developments of the ΔAd.AAV vector system are directed toward testing their properties for in vivo transduction and modifying the vector tropism.
ACKNOWLEDGMENTS
We thank Cheng-Yi He and Zong-Yi Li for technical assistance. We are grateful to David Russell for providing plasmid constructs and for helpful discussions. We thank Dusty Miller for providing the rAAV vector stock and Jeff Engler for providing the Ad E4 antibodies.
This work was supported by the Cystic Fibrosis Foundation and NIH grants R01 CA80192-01 (to A.L.) and R01 DK49022 (to M.A.K.). D.S.S. is a recipient of a predoctoral DAAD fellowship.
REFERENCES
- 1.Alexander I E, Russell D W, Spence A M, Miller A D. Effects of gamma irradiation on the transduction of nondividing cells in brain and muscle of rats by adeno-associated virus vectors. Hum Gene Ther. 1996;7:841–850. doi: 10.1089/hum.1996.7.7-841. [DOI] [PubMed] [Google Scholar]
- 2.Balague C, Kalla M, Zhang W-W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conway J E, Zolotuchin S, Muzyczka N, Hayward G S, Byrne B J. Recombinant adeno-associated virus type 2 replication and packaging is entirely supported by a herpes simplex virus type 1 amplicon expressing rep and cap. J Virol. 1997;71:8780–8789. doi: 10.1128/jvi.71.11.8780-8789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doerfler W. Adenoviral DNA integration and changes in DNA methylation patterns: a different view of insertional mutagenesis. Prog Nucleic Acid Res Mol Biol. 1993;46:1–37. doi: 10.1016/s0079-6603(08)61016-8. [DOI] [PubMed] [Google Scholar]
- 5.Feng M, Jackson W R, Goldman C K, Rancourt C, Wang M, Dusing S K, Siegal G, Curiel D T. Stable in vivo gene transduction via a novel adenoviral/retroviral chimeric vector. Nat Biotechnol. 1997;15:866–870. doi: 10.1038/nbt0997-866. [DOI] [PubMed] [Google Scholar]
- 6.Fisher K J, Kelley W M, Burda J F, Wilson J M. A novel adenovirus-adeno-associated virus hybrid vector that displays efficient rescue and delivery of the AAV genome. Hum Gene Ther. 1996;7:2079–2087. doi: 10.1089/hum.1996.7.17-2079. [DOI] [PubMed] [Google Scholar]
- 7.Fisher K J, Joos K, Alston J, Yang Y, Hacecker S E, High K, Pathak R, Raper S E, Wilson J M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 8.Fisher K J, Choi H, Burda J, Chen S, Wilson J M. Recombinant adenovirus deleted of all genes for gene therapy of cystic fibrosis. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 9.Gluzman Y, Van-Doren K. Palindromic adenovirus type 5-SV40 hybrids. J Virol. 1983;45:91–103. doi: 10.1128/jvi.45.1.91-103.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haecker S E, Stedman H H, Balice-Gordon R J, Smith D B J, Greelish J P, Mitchell M A, Wells A, Sweeney H L, Wilson J M. In vivo expression of full-length human dystrophin from adenoviral vectors deleted for all viral genes. Hum Gene Ther. 1996;7:1907–1914. doi: 10.1089/hum.1996.7.15-1907. [DOI] [PubMed] [Google Scholar]
- 11.Heffelfinger S C, Hawkins H H, Barrish J, Taylor L, Darlington G J. SKHEp1: a human cell line of endothelial origin. In Vitro Cell Dev Biol. 1992;28A:136–142. doi: 10.1007/BF02631017. [DOI] [PubMed] [Google Scholar]
- 12.Hitt M M, Addison C L, Graham F L. Human adenoviral vectors for gene transfer into mammalian cells. Adv Pharmacol. 1997;40:137–205. doi: 10.1016/s1054-3589(08)60140-4. [DOI] [PubMed] [Google Scholar]
- 13.Inoue N, Russell D W. Packaging cells based on inducible gene amplification for the production of AAV vectors. J Virol. 1998;72:7024–7031. doi: 10.1128/jvi.72.9.7024-7031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston K M, Jakoby D, Pechan P A, Fraefel C, Borghesani P, Schuback D, Dunn R J, Smith F I, Breakfield X O. HSV/AAV hybrid amplicon vectors extend transgene expression in human glioma cells. Hum Gene Ther. 1997;8:359–370. doi: 10.1089/hum.1997.8.3-359. [DOI] [PubMed] [Google Scholar]
- 15.Kay M A, Graham F, Leland F, Woo S L. Therapeutic serum concentrations of human alpha 1-antitrypsin after adenoviral-mediated gene transfer into mouse hepatocytes. Hepatology. 1995;21:815–819. [PubMed] [Google Scholar]
- 16.Kotin R M. Prospects for the use of adeno-associated virus vectors for human gene therapy. Hum Gene Ther. 1994;5:793–801. doi: 10.1089/hum.1994.5.7-793. [DOI] [PubMed] [Google Scholar]
- 17.Kumar-Singh R, Farber D B. Encapsidated adenovirus mini-chromosome-mediated delivery of genes to the retina: application to the rescue of photoreceptor degeneration. Hum Mol Genet. 1998;7:1893–1900. doi: 10.1093/hmg/7.12.1893. [DOI] [PubMed] [Google Scholar]
- 18.Leblois H, Orsini C, Yeh P, Perricaudet M. Adenovirus-mediated delivery of an EBV-based replicon via Cre/loxP recombination: a novel vector system for efficient and long-term gene expression, abstr. 705. 1998. p. 177a. . Presented at the 1st meeting of the American Society of Gene Therapy. American Society of Gene Therapy, Thorofare, N.J. [Google Scholar]
- 19.Lieber A, He C-Y, Kay M A. Adenoviral preterminal protein stabilizes mini-adenoviral genomes in vitro and in vivo. Nat Biotechnol. 1997;15:1383–1387. doi: 10.1038/nbt1297-1383. [DOI] [PubMed] [Google Scholar]
- 20.Lieber A, He C-Y, Kirillova I, Kay M A. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J Virol. 1996;70:8944–8960. doi: 10.1128/jvi.70.12.8944-8960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieber A, He C-Y, Meuse L, Schowalter D, Kirillova I, Winther B, Kay M A. The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors. J Virol. 1997;71:8798–8807. doi: 10.1128/jvi.71.11.8798-8807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyu Y L, Lin C-T, Liu L F. Inversion/dimerization of plasmids mediated by inverted repeats. J Mol Biol. 1999;285:1485–1501. doi: 10.1006/jmbi.1998.2419. [DOI] [PubMed] [Google Scholar]
- 23.Malik P, McQuiston S A, Yu X-J, Pepper K A, Krall W J, Podsakoff G M, Kurtzman G J, Kohn D B. Recombinant adeno-associated virus mediates a high level of gene transfer but less efficient integration in the K562 human hematopoietic cell line. J Virol. 1997;71:1776–1783. doi: 10.1128/jvi.71.3.1776-1783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao C H, Snyder R O, Schowalter D B, Patjin G A, Donahue B, Winther B, Kay M A. The kinetics of rAAV integration in the liver. Nat Genet. 1998;19:13–15. doi: 10.1038/ng0598-13. [DOI] [PubMed] [Google Scholar]
- 25.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 26.Nakai H, Iwaki Y, Kay M A, Couto L B. Isolation of recombinant AAV vector-cellular DNA junctions from mouse liver. J Virol. 1999;73:5438–5447. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson J, Kay M A. Persistence of recombinant adenovirus in vivo is not dependent on vector replication. J Virol. 1997;71:8902–8907. doi: 10.1128/jvi.71.11.8902-8907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks R J, Graham F L. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson C E, Zorbas H, Price G B, Zannis-Hadjopoulos M. Inverted repeats, stem loops, and cruciforms: significance for initiation of DNA replication. J Cell Biochem. 1996;63:1–22. doi: 10.1002/(SICI)1097-4644(199610)63:1%3C1::AID-JCB1%3E3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 30.Peterson K R, Clegg C H, Navas P A, Norton E J, Kimbrough T G, Stamatoyannopoulos G. Effect of deletion of 5′HS3 or 5′HS2 of the human B-globin locus control region on the developmental regulation of globin gene expression in B-globin locus yeast artificial chromosome transgenic mice. Proc Natl Acad Sci USA. 1996;93:6605–6609. doi: 10.1073/pnas.93.13.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recchia A, Parks R J, Lamartina S, Toniatti C, Pieroni L, Palombo F, Ciliberto G, Graham F L, Cortese R, La Monica N, Colloca S. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc Natl Acad Sci USA. 1999;96:2615–2620. doi: 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell D, Alexander I E, Miller A D. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc Natl Acad Sci USA. 1995;92:5719–5723. doi: 10.1073/pnas.92.12.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell D W, Miller A D, Alexander I E. AAV vectors preferentially transduce cells in S phase. Proc Natl Acad Sci USA. 1994;91:8915–8919. doi: 10.1073/pnas.91.19.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutledge E A, Russell D W. Adeno-associated virus vector integration junctions. J Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samulski R J, Chang L S, Shenk T. Helper free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3928. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinwaerder D, Carlson C A, Lieber A. Generation of adenovirus vectors devoid of all viral genes by recombination between inverted repeats. J Virol. 1999;73:9303–9313. doi: 10.1128/jvi.73.11.9303-9313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao X, Li J, Samulski R J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao X, Xiao W, Li J, Samulski R J. A novel 165-bp terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao X, Li J, Samulski R J. Production of hight-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yalkinoglu A O, Zentgraf H, Hubscher U. Origin of adeno-associated virus DNA replication is a target for carcinogen-inducible DNA amplification. J Virol. 1991;65:3175–3184. doi: 10.1128/jvi.65.6.3175-3184.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]