Abstract
Insights into mechanisms driving either activation or inhibition of immune response are crucial in understanding the pathology of various diseases. The differentiation of viral from endogenous RNA in the cytoplasm by pattern-recognition receptors, such as retinoic acid-inducible gene I (RIG-I), is one of the essential paths for timely activation of an antiviral immune response through induction of type I interferons (IFN). In this mini-review, we describe the most recent developments centered around RIG-I's structure and mechanism of action. We summarize the paradigm-changing work over the past few years that helped us better understand RIG-I's monomeric and oligomerization states and their role in conveying immune response. We also discuss potential applications of the modulation of the RIG-I pathway in preventing autoimmune diseases or induction of immunity against viral infections. Overall, our review aims to summarize innovative research published in the past few years to help clarify questions that have long persisted around RIG-I.
Keywords: RIG-I, RNA, structural flexibility of RIG-I, antiviral immune response, RNA-binding proteins
Introduction
Retinoic acid-inducible gene I (RIG-I) like receptors (RLRs) are cytoplasmic viral RNA sensors, which induce an antiviral immune response by activating type I interferon (IFN) signaling pathways after binding to double-stranded RNA (dsRNA). 1 In total, there are three known RLRs: RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), and they are characterized by the presence of a DExD/H RNA helicase domain that exhibits ATPase and helicase activities. 2 In addition to the helicase core and a C-terminal domain (CTD), RIG-I, and MDA5 have two caspase activation and recruitment domains (CARDs) on the N-terminus. The CARDs are responsible for transmitting downstream signaling, which are absent only in the LGP2 receptor making it unable to recruit molecules to transmit signals, hence the LGP2's role in the regulation of the viral nucleic acid recognition by RIG-I and MDA5.1,3
Because RIG-I and MDA5 are essential for interferon production and host antiviral immune response, their malfunction can lead to higher susceptibility to RNA virus infections. It is also known that RLRs can be involved in the innate immune response against parasite and bacterium infections. 4 Notably, even though RIG-I and MDA5 show many structural similarities, they have different preferences for RNA ligands, and as a result, they detect distinct spectrums of viruses. 5 Since the discovery that RIG-I can induce type I interferon production, 6 researchers have been trying to better understand the molecular mechanisms of RLRs. Hence, multiple functional and structural studies on these receptors have been performed over the past 20 years. The recently published reviews of RLRs were centered around summarizing structural characteristics and their regulation and role in RNA sensing.7,8 Therefore, in this mini-review, we will mainly focus on the most recent developments in the mechanistic understanding of RIG-I function and, to a lesser extent, on the understanding of RIG-I structure or pathology and therapy.
Advances in understanding structural flexibility of RIG-I
Over the past 3 years, an increasing number of structures were deposited to the Protein Data Bank (PDB) for RIG-I proteins in complex with different ligands and cofactors, driven by the implementation of the cryo-EM technology.9,10 The most recent work on short dsRNA with a variety of 5′-end modifications and cofactors has greatly improved our understanding of the RIG-I/RNA machinery and provided novel insights into the structures of those complexes. 10 In this section we will briefly describe work from the last decade and center around structural information that became available in the past 3 to 4 years.
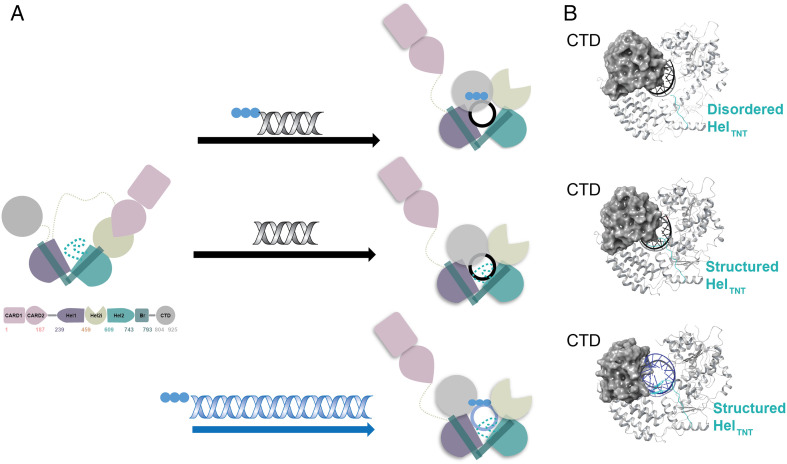
From early structural studies, we learned that RIG-I is composed of a DExD/H-box RNA helicase core, two CARDs on the N-terminus, and a C-terminal domain (CTD) on the C-terminus, Figure 1(a). 11 In the ligand-free form, which is incapable of signaling, RIG-I adopts an autorepressed conformation in which Hel2i-Hel2 form a rigid-body unit where Hel1 is not in direct contact with them, but an elbow-like helical structure emerges from Hel2 which bridges to Hel1 (bridging domain, Br). 11 In this conformation, the head-to-tail unit of the CARD1 and CARD2 domains while adapting a canonical CARD fold, is bound to the insertion domain of the helicase (Hel2i) and locked in an autorepressed conformation that makes the CARDs unavailable for signal transduction. 11 The domain arrangement in such autoinhibited states leaves the CTD unhindered from interactions with other domains, allowing it to sample RNAs in the cytoplasm, making the binding event of the CTD to RNA a first layer of RNA selectivity. 12
Figure 1.
Schematic representation of structural rearrangements of RIG-I upon binding distinct types of RNA. (a) Conformational changes and domain rearrangements that RIG-I can undergo when binding RNA. All RNA binding leads to the ejection of the N-terminal CARDs, independent of nucleotide presence. Top panel: Binding of short 5′-triphosphate dsRNA leads to rearrangement of the helicase core, an unstructured HELTNT loop (sequence N664-RGKTNQNTG-C672), and a CTD orientation that caps the 5′-end of RNA. Middle panel: Binding of short dsRNA ending with a 5′-hydroxy group leads to rearrangement of the helicase core, a structured HELTNT loop, and a CTD orientation that caps the 5′-end of RNA. Bottom panel: Binding of long dsRNA (here 112 bp) leads to rearrangement of the helicase core, a structured HELTNT loop, and the CTD rotated away from the central axis of RNA as in the case of MDA5. (b) Cartoon representation of differences seen for the CTD and HELTNT loop conformations when binding the respective types of RNA. The CTD is represented as a gray sphere, and short versus long RNA are color-coded black and blue, respectively. The apparent displacement of the CTD upon binding long RNA can be observed (bottom panel). The HELTNT loop is indicated in teal, and it is disordered when short 5′-triphosphate is bound to RIG-I (top panel); in the presence of short RNA ending with 5′-hydroxy group (middle panel) or long RNA (bottom panel), the loop becomes structured. CARDs: caspase activation and recruitment domains; CTD: C-terminal domain; dsRNA: double-stranded RNA; MDA5: melanoma differentiation-associated gene 5; RIG-I: retinoic acid-inducible gene I.
Upon viral RNA detection, the RIG-I undergoes pronounced conformational rearrangements to accommodate binding to various RNA substrates. The HEL1, HEL2i, and CTD reorganize to form an elaborate network of interactions dominated by polar contacts, resulting in a ring structure around dsRNA, Figure 1(a) and (b). 13 Unsurprisingly, the CTD, which contains a positively charged cavity at its inner face can bind to RNA and functions as an RNA sensor in solution, capping the RNA at the end of this ring-like helicase structure.13,14 In this RNA-bound conformation, the RIG-I is now competent to bind ATP, and in earlier works, it was proposed that either ATP binding or hydrolysis event can potentially promote CARD release and RIG-I activation.13,15 However, based on the most recent studies from multiple laboratories, it became clear that the CARD domains become exposed in solution because of RNA but not ATP binding. Thus, RNA binding is sufficient for CARD ejection, leading to productive interaction with Mitochondrial antiviral-signaling protein (MAVS) and subsequent initiation of the antiviral signaling cascade.16,17 More details on the function of ATP binding and hydrolysis can be found in “Recent developments in mechanistic understanding of RIG-I function” section.
Due to recent technical developments in cryo-EM, two interesting publications could address questions that were previously challenging to answer by X-ray crystallography alone. In the first body of work, a structure of fusion filaments of RIG-I could be solved, in which a trimeric segment of RIG-I in complex with RIPLET E3 ligase was bound to a 112 bp dsRNA, Figure 1(a) and (b). 9 This was the first time that a RIG-I/RNA complex structure was solved where the CTD moves away from the central axis of dsRNA, allowing RIG-I to adopt a ring confirmation around dsRNA, strongly resembling a conformation observed previously for MDA5. In the second publication, authors created an impressive workflow for solving cryo-EM structures of full-length RIG-I protein in the presence of agonist and antagonist dsRNA terminated with either 5′-triphosphate, 5′-diphosphate, 5′-monophosphate, or 5′-hydroxy groups. 10 By obtaining structures with mimics of both viral and host RNAs, the authors showed that RIG-I is capable of adopting two different conformations, depending on the type of RNA bound. 10 RIG-I's intricate RNA recognition is accomplished by adopting two mutually exclusive conformations depending on whether agonistic or antagonistic RNA is bound.10,18 In the conformation compatible to activate immune response, the CTD is tightly bound to 5′-triphosphate, 5′-diphosphate of RNA, preventing the Hel2 autoinhibitory loop from interacting with the CTD. 10 On the contrary, when host RNA binds to RIG-I, the Hel2 autoinhibitory loop can still interact with CTD residues that are binding to the same amino acids as the diphosphate group, weakening RNA's affinity and stabilizing RIG-I's conformation with active motor activity. 10
As seen from the above paragraphs, the accumulation of X-ray and cryo-EM structures in conjunction with data from solution-based technologies enable us to finally better understand the structural basis of RNA-driven conformational flexibility of RIG-I.
Recent developments in mechanistic understanding of RIG-I function
RIG-I can recognize viral RNA in cells and trigger innate immune responses while evading endogenous RNAs to prevent aberrant activation of downstream signaling. This multistep process of successful discrimination between triggering or preventing immune response strongly relies on RIG-I's ability to differentiate between molecular determinants of RNA. Multiple studies showed that the RIG-I signaling pathway can be activated by short RNA duplexes that are at least 10 nucleotides in length.19,20 We also learned that RIG-I strongly prefers blunt-ended dsRNAs terminated with a 5′-triphosphate, over nonblunt-ended dsRNAs representing the cellular pool of RNA. 12 However, RIG-I shows no preferences for the nucleotides present in the Watson-Crick base pair at the blunt end of RNAs terminated with 5′-triphosphate or 5′-diphosphate groups. 21 The 5′-triphosphate modification is typically present in pathogenic RNAs; however, to prevent triggering an innate immune response by endogenous mRNA, which can also be 5′-triphosphated, eukaryotes use N1-2′O-methylation and 5′-ppp-linked methylguanosine (m7G) cap to avoid immunorecognition.22,23 The 5′-end caps and N1-2′O-methylation are not the only mechanisms by which RIG-I activation can be prevented; the presence of any 5′-end or 3′-end overhangs on dsRNAs, the G-U wobble pairs, G-A pairs, as well as other mismatches also eliminate RNA's ability to bind RIG-I. 21 On the contrary, the most recent work by Schweibenz et al. showed that 5′-diphosphate dsRNAs capped with metabolites such as Nicotinamide Adenine Dinucleotide (NAD)+, Flavin Adenine Dinucleotide (FAD), and dephosphoCoA could induce interferon signaling at comparable levels to 5′-triphosphated dsRNA. 24
Interestingly, even though the levels of uncapped 5′-monophosphate are much higher in cells than those of 5′-triphosphate, its binding to RIG-I does not trigger an immune response. This is because RIG-I has a razor-sharp specificity toward distinct types of RNAs, being able to discriminate between host RNAs that circulate in the cytoplasmic environment which differ only by a single phosphate group from pathogenic RNAs, as mentioned in the previous section.10,18 Briefly, the conformation forming high affinity interactions with viral RNA leads to a partially unfolded state of the Hel2 autoinhibitory loop and stimulates an antiviral immune response. In contrast, the alternative conformation induced by 5′-monophosphate and 5′-hydroxy dsRNA results in fewer contacts between RIG-I and RNA ligands which enables rapid release of RNA. These differences in conformations induced by viral and host RNA explain an accurate proofreading mechanism of RIG-I and why uncapped 5′-monophosphate dsRNAs do not induce interferon signaling despite being present at higher levels in cells than pathogenic 5′-triphosphate dsRNAs. 10
Recent work on long noncoding RNAs demonstrated that RIG-I can not only detect differences in 5′-end motifs to differentiate between agonistic and antagonistic RNA, but also recognize endogenous lncRNA (lnc-Lsm3b) that serves as a molecular decoy and competes with viral RNA. 25 Subsequent to this work, another example of lncRNA was published where lncATV was found to also act as a negative regulator of RIG-I, highlighting the ability of lncRNA to act as an antagonist of interferon production. 26
Recognizing diverse types of RNA with such high precision becomes critically important because the RNA binding event is a key driver of CARDs ejection and induction of downstream signaling pathways leading to production of interferons. Discrimination between distinct types of RNA with high precision is only one of the key events in this multistep control mechanism, another being RIG-I's ATPase activity. From the description above we have seen that the plethora of 5′-end RNA modifications enable the CTD to discriminate between RNA through differential dissociation kinetics. 12 However, it is the ATP hydrolysis that adds another layer of proofreading by driving faster dissociation of host versus pathogenic 5′-triphosphate RNAs. 27 Even though the role of ATP hydrolysis in RNA release was understood before, the question of whether RNA binding or ATP hydrolysis drives CARDs release was only recently answered. The FRET experiments, 16 followed by bioNMR studies, clearly showed that neither ADP nor ATP binding have any significant effect on RIG-I's ability to eject CARDs. 17
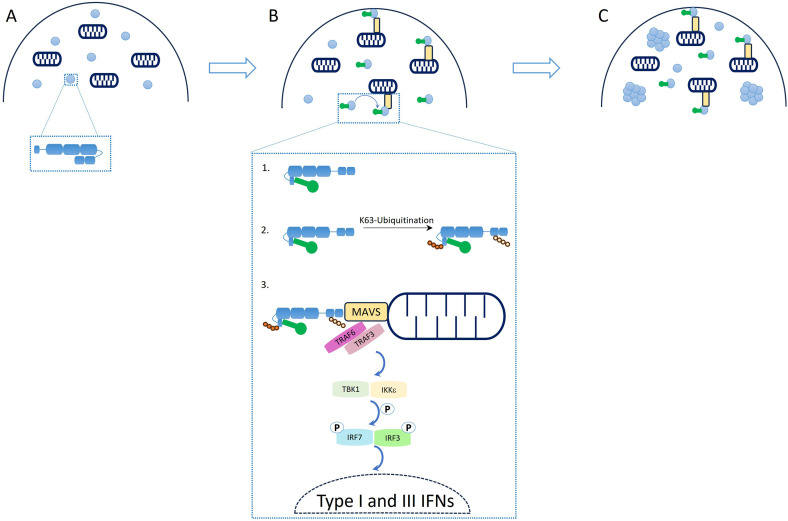
In addition to gaining a better understanding of the function of ATP hydrolysis, recent papers provided insights into RIG-I oligomerization mechanism and function. It became clear that 10 to 24 nucleotides long phosphorylated synthetic RNA duplexes can induce RIG-I signaling, contradicting the idea that RIG-I oligomerization on RNA is required for its function, thus revealing that additional mechanisms could lead to RIG-I multimerization, such as ubiquitination. 20 A follow-up study on the dependency of RIG-I signaling on aggregation at the mitochondrial membrane showed clearly that RIG-I‘s ability to create a transient signaling complex with MAVS is responsible for inducing interferon-related signaling, rather than RIG-I oligomerization as previously believed, Figure 2. 28 As the authors highlight, the timing of RIG-I response needs to be rapid to match viral infection and replication timelines, questioning the previous “mass aggregate” model for RIG-I signaling built on earlier in vitro data. 29 From this recent work it became clear that an immune response needs to be triggered immediately after viral infection, and that the entire process is dependent only on the resident pool of constitutively expressed RIG-I, rather than additional rounds of RIG-I expression that would slow down such a response. Therefore, it was proposed that large RNA-protein complex bodies may function as a location for postsignaling RIG-I to accumulate prior to degradation.
Figure 2.
Schematic representation of RIG-I's regulatory mechanisms controlling antiviral response. (a) Auto-repressed state of RIG-I with CARD domains buried in the helicase core. (b) IFNs activation cascade triggered by RNA binding: (b1) schematic representation of RNA binding to RIG-I and CARD release to solution; (b2) RIG-I's K63-linked ubiquitination mediated by several E3 ligases; (b3) downstream signaling cascade triggered by RIG-I induced MAVS oligomerization. (c) Formation of signaling-unrelated cytosolic aggregates of RIG-I. Figure is adapted from publication “A rapid RIG-I signaling relay mediates efficient antiviral response” 28 with permission from Elsevier. CARDs: caspase activation and recruitment domains; IFNs: interferons; RIG-I: retinoic acid-inducible gene I.
While binding of RIG-I to RNA is crucial for RIG-I activation, as mentioned in the previous paragraphs, post-translational modifications were shown to add an additional layer of control for RIG-I activation, with two modifications researched predominantly in the RIG-I context being phosphorylation and ubiquitination. Those post-translational modifications are proposed to trap RIG-I in conformations where CARDs are ejected to the solution or prevent their re-association, resulting in directing the signaling cascade forward. 10 The phosphorylation of CARDs is required for the addition of K63 linked polyubiquitin at residue K172 of RIG-I after CARDs release from its autoinhibited form, facilitating RIG-I interaction with MAVS and subsequent interferon production. 30 The opposite is observed for residues T770/S854/S855, whose phosphorylation leads to RIG-I inhibition. 31 In the case of K63-linked ubiquitination of RIG-I four E3 ligases were identified to perform this function: Tripartite motif-containing protein 25 (TRIM25), RING finger protein leading to RIG-I activation (RIPLET), RNA-binding E3 ubiquitin-protein ligase MEX3C (Mex3c), and Tripartite motif-containing protein 4 (TRIM4). 32 Interestingly, recent studies of TRIM25 described an involvement in the RIG-I pathway showing that even though this E3 ubiquitin ligase was proposed to play a key role in RIG-I activation by CARD's K172 ubiquitination, another E3 ubiquitin ligase RIPLET is sufficient to ubiquitinate and activate RIG-I.33,34 TRIM25 indeed seems to be involved in restricting viral infections but through mechanisms independent of the RIG-I pathway. 33 RIPLET however, is able to promote RIG-I-based activation of interferons not only through ubiquitination of dsRNA bound RIG-I complexes on CTD residues K849, K851 K888, K907, K909, and linker's region K788, 35 but also through induction of RIG-I's aggregate-like assemblies. 36 In addition MEX3C mediates K63-linked polyubiquitination of CARDs on K48/K99/K169 leading to a similar outcome as K172 modification.36,37 Moreover, TRIM4 was shown to mediate polyubiquitination of CARDs at K164 and K172 residues. 35
From this brief overview of the most recent developments to better understand RIG-I's signaling mechanism, an intricate interplay between rapid sensing of appropriate RNA, ATP hydrolysis, and posttranslational modifications became evident as processes that guide RIG-I activation. This multilayer regulatory mechanism ensures a timely and concise control of the antiviral response triggered by pathogenic RNA, while preventing unintentional hyperactivation of RIG-I's activity by cellular RNA.
RIG-I in pathology and therapy
As described in the previous sections, gaining a better understanding of RIG-I's mechanism driving the induction of interferons presents an opportunity for medical and pharmaceutical researchers to explore its function in developing novel therapeutic interventions. Multiple pathogen-sensing pathways can trigger the induction of interferons, which is important not only for antiviral immunity but also for the induction of antitumor immunity. 38 On the other hand, the dysregulation of RIG-I signaling can lead to a wide variety of autoimmune diseases.38,39 In this section, we will cover only emerging RIG-I related diseases and new strategies harnessing the RIG-I pathway in autoimmune diseases and in induction of the immunity against viral infections.
With the recent increase of work around SARS-CoV infection, we learned that the production of interferons is impaired in cells infected with SARS-, SARS-2-, and MERS-CoV viruses. 40 One of the interferon suppression mechanisms by ORF9 of SARS-CoV is triggering TRIM25 dependent ubiquitination of RIG-I and inhibiting its function. 41 In addition to attenuating RIG-I directly, the SARS-COVID ORF9b suppresses innate immunity by promoting the degradation of MAVS by the E3 ubiquitin ligase E3 ubiquitin-protein ligase Itchy homolog (ITCH),/Atrophin-1-interacting protein 4 (AIP4). 40 With the accumulation of data on SARS-CoV driving suppression of RIG-I and RLR-mediated signaling, it is possible that we will soon learn about the utilization of this signaling pathway in the design of effective therapies for COVID-19 treatment. 40
Although RIG-I is best known for the detection of viral RNA, in recent years, work on Listeria monocytogenes and Mycobacterium tuberculosis showed that also bacterial RNA and RNA-protein complexes released to the cytosol can lead to interferon β release through RIG-I signaling pathway further supporting RIG-I's important role in fighting pathogens.42,43
RIG-I agonists can be designed as potent antiviral agents to trigger antipathogenic immunity. Examples of nucleotides and RNA-based therapeutics were published recently. 44 Among dinucleotide-derived compounds, SB9200 can successfully induce interferon through RIG-I and Nucleotide-binding oligomerization domain-containing protein 2 (NOD2) pathways, and SB9200 treatment of adults with chronic hepatitis C, showed an association between the decline in viral RNA and the detection peak of the compound in plasma in a phase 1 clinical trial. 44 Interesting work aiming to induce immunity against viral infection with engineered RNA was published by Chiang et al. which demonstrated that one could design RNA agonists with enhanced antiviral and inflammatory properties against selected viruses, that is, influenza, dengue, or chikungunya, showing protective antiviral responses in vitro and in vivo. 45 This approach was further successfully implemented to design 5′-triphosphate dsRNA to inhibit duplication of Human enterovirus 71, Human poliovirus 1, Human coxsackievirus B5, and Influenza A virus in cells. Moreover, this study showed that prophylactic administration of designed RNA reduced mouse death in the lethal SARS-CoV-2 mouse model. 46
In addition to RIG-I's involvement in antipathogenic responses, diseases with autoinflammatory pathology can also be rooted in RIG-I's abnormalities and nucleic acid metabolism. RIG-I's single-nucleotide polymorphisms (SNPs) were shown to be associated with increased risks of developing autoimmune diseases. Especially, mutations in RIG-I's DDX58 gene were shown to be associated with Singleton–Merten syndrome (E373A mutation) and Congenital glaucoma (C268F). 39 In addition to SNPs, mRNA levels of RIG-I were also correlated to autoimmune diseases. In the case of multiple sclerosis (MS) patients, mRNA analysis indicated a significant decrease in RIG-I and MDA5 levels in the affected population versus control, and interferon β therapy decreased pathogenic inflammatory responses and upregulated RIG-I expression, especially in the female group. 47 Authors speculated that the downregulation of RIG-I and MDA-5 in MS patients can be responsible for MS patients’ defective immune responses against pathogens, suggesting a potential path for RIG-I therapy in that group.
Limitations of the study
This mini-review only covers the most recent developments in the mechanistic understanding of RIG-I function and, to a lesser extent, the understanding of RIG-I structures or pathology and therapy. The authors purposefully centered this work on recent discoveries released within the past 3 to 4 years, since in 2021 to 2022 multiple in-depth reviews were published that covered RIG-I's pathway regulation, mechanistic studies, post-translational modifications, and its therapeutic potential.4,8,30,32,39,40,48
Concluding remarks
Recent years strongly enhanced our understanding of the structural basis of processes leading to RIG-I's homeostasis. We now better comprehend the structural basis of RNA-driven conformational flexibility of RIG-I, the mechanistic functions of its domains, and the RNA recognition mechanism. However, we still need to learn how best to leverage this pathway for antiviral, antibacterial, or autoimmune diseases therapies. One advantage of therapies targeting host proteins, such as RIG-I, rather than pathogenic components is that they can provide a means to bypass frequently observed variability and genetic mutations in pathogens. But, targeting RIG-I comes with its own challenges because desired antiviral response requires careful stimulation of this pathway, while keeping RIG-I's ability to drive autoimmune diseases in check. Hence, an improved understanding of the requirements for immune cell programming by elucidating mechanisms downstream of the RIG-I pathway can potentially help design better therapies in the future.
Acknowledgements
Due to the space constrains we were not able to discuss all most recent developments in the field, so we would like to apologize to all whose findings were not addressed in our manuscript.
Footnotes
Author Contributions: JS and DFW reviewed the literature, drafted the work, and reassessed all the paragraphs. No (AI)-assisted technologies were used in the production of the submitted work. Both authors contributed to the manuscript equally.
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Both the authors are employees or former employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA, and may hold stock or stock options in Merck & Co., Inc., Rahway, NJ, USA.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA.
ORCID iDs: Justyna Sikorska https://orcid.org/0000-0002-3836-6708
Daniel F Wyss https://orcid.org/0000-0002-5005-6104
References
- 1.Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther 2021; 6: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barral PM, Sarkar D, Su ZZ, et al. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol Ther 2009; 124: 219–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Matsumoto K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immun 2005; 175: 2851–2858. [DOI] [PubMed] [Google Scholar]
- 4.Song J, Li M, Li C, et al. Friend or foe: RIG- I like receptors and diseases. Autoimmun Rev 2022; 21: 103161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loo YM, Fornek J, Crochet N, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 2008; 82: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 2004; 5: 730–737. [DOI] [PubMed] [Google Scholar]
- 7.Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 2020; 20: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thoresen D, Wang W, Galls D, et al. The molecular mechanism of RIG-I activation and signaling. Immunol Rev 2021; 304: 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato K, Ahmad S, Zhu Z, et al. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol Cell 2021; 81: 599–613.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Pyle AM. The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands. Mol Cell 2022; 82: 4131–4144.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalinski E, Lunardi T, McCarthy A, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 2011; 147: 423–435. [DOI] [PubMed] [Google Scholar]
- 12.Ramanathan A, Devarkar SC, Jiang F, et al. The autoinhibitory CARD2-Hel2i interface of RIG-I governs RNA selection. Nucleic Acids Res 2015; 44: 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo D, Ding Steve C, Vela A, et al. Structural insights into RNA recognition by RIG-I. Cell 2011; 147: 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang F, Ramanathan A, Miller MT, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 2011; 479: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald ME, Rawling DC, Potapova O, et al. Selective RNA targeting and regulated signaling by RIG-I is controlled by coordination of RNA and ATP binding. Nucleic Acids Res 2016; 45: 1442–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickey TH, Song B, Pyle AM. RNA binding activates RIG-I by releasing an autorepressed signaling domain. Sci Adv 2019; 5: eaax3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sikorska J, Hou Y, Chiurazzi P, et al. Characterization of RNA driven structural changes in full length RIG-I leading to its agonism or antagonism. Nucleic Acids Res 2023; 51: 9356–9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren X, Linehan MM, Iwasaki A, et al. RIG-I selectively discriminates against 5′-monophosphate RNA. Cell Rep 2019; 26: 2019–2027.e4. [DOI] [PubMed] [Google Scholar]
- 19.Kohlway A, Luo D, Rawling DC, et al. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep 2013; 14: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linehan MM, Dickey TH, Molinari ES, et al. A minimal RNA ligand for potent RIG-I activation in living mice. Sci Adv 2018; 4: e1701854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren X, Linehan MM, Iwasaki A, et al. RIG-I recognition of RNA targets: the influence of terminal base pair sequence and overhangs on affinity and signaling. Cell Rep 2019; 29: 3807–3815.e3. [DOI] [PubMed] [Google Scholar]
- 22.Schuberth-Wagner C, Ludwig J, Bruder Ann K, et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity 2015; 43: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devarkar SC, Wang C, Miller MT, et al. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci 2016; 113: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweibenz BD, Solotchi M, Hanpude P, et al. RIG-I recognizes metabolite-capped RNAs as signaling ligands. Nucleic Acids Res 2023; 51: 8102–8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang M, Zhang S, Yang Z, et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 2018; 173: 906–919.e13. [DOI] [PubMed] [Google Scholar]
- 26.Fan J, Cheng M, Chi X, et al. A human long non-coding RNA LncATV promotes virus replication through restricting RIG-I–mediated innate immunity. Front Immunol 2019; 10: 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devarkar SC, Schweibenz B, Wang C, et al. RIG-I uses an ATPase-powered translocation-throttling mechanism for kinetic proofreading of RNAs and oligomerization. Mol Cell 2018; 72: 355–368.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoresen DT, Galls D, Götte B, et al. A rapid RIG-I signaling relay mediates efficient antiviral response. Mol Cell 2023; 83: 90–104.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peisley A, Wu B, Yao H, et al. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol Cell 2013; 51: 573–583. [DOI] [PubMed] [Google Scholar]
- 30.Deng Y, Wang Y, Li L, et al. Post-translational modifications of proteins in cytosolic nucleic acid sensing signaling pathways. Front Immunol 2022; 13: 898724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Z, Ren H, Liu Y, et al. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J Virol 2011; 85: 1036–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S, Cheng A, Wang M, et al. Viruses utilize ubiquitination systems to escape TLR/RLR-mediated innate immunity. Front Immunol 2022; 25: 1065211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choudhury NR, Trus I, Heikel G, et al. TRIM25 inhibits influenza A virus infection, destabilizes viral mRNA, but is redundant for activating the RIG-I pathway. Nucleic Acids Res 2022; 50: 7097–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayman TJ, Hsu AC, Kolesnik TB, et al. RIPLET, and not TRIM25, is required for endogenous RIG-I-dependent antiviral responses. ICB 2019; 97: 840–852. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto M, Kouwaki T, Fukushima Y, et al. Regulation of RIG-I activation by K63-linked polyubiquitination. Front Immunol 2018; 05: 1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cadena C, Ahmad S, Xavier A, et al. Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 2019; 177: 1187–1200.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuniyoshi K, Takeuchi O, Pandey S, et al. Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I–mediated antiviral innate immunity. Proc Natl Acad Sci 2014; 111: 5646–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solstad A, Hogaboam O, Forero A, et al. RIG-I-like receptor regulation of immune cell function and therapeutic implications. J Immun 2022; 209: 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batool M, Kim MS, Choi S. Structural insights into the distinctive RNA recognition and therapeutic potentials of RIG-I-like receptors. Med Res Rev 2022; 42: 399–425. [DOI] [PubMed] [Google Scholar]
- 40.Onomoto K, Onoguchi K, Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol Immunol 2021; 18: 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y, Li W, Gao T, et al. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol 2017; 91: e02143–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagliuso A, Tham TN, Allemand E, et al. An RNA-binding protein secreted by a bacterial pathogen modulates RIG-I signaling. Cell Host Microbe 2019; 26: 823–835.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Y, Schorey JS. Mycobacterium tuberculosis-induced IFN-β production requires cytosolic DNA and RNA sensing pathways. J Exp Med 2018; 215: 2919–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yong HY, Luo D. RIG-I-like receptors as novel targets for pan-antivirals and vaccine adjuvants against emerging and re-emerging viral infections. Front Immunol 2018; 9: 1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang C, Beljanski V, Yin K, et al. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J Virol 2015; 89: 8011–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Z, Wang Q, Bian L, et al. A short 5′ triphosphate RNA nCoV-L induces a broad-spectrum antiviral response by activating RIG-I. Viruses 2022; 14: 2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asadikaram G, Meimand HAE, Noroozi S, et al. The effect of IFN-β 1a on expression of MDA5 and RIG-1 in multiple sclerosis patients. Biotechnol Appl Biochem 2021; 68: 267–271. [DOI] [PubMed] [Google Scholar]
- 48.Chowdhury A, Witte S, Aich A. Role of mitochondrial nucleic acid sensing pathways in health and patho-physiology. Front Cell Dev Biol 2022; 10: 796066. [DOI] [PMC free article] [PubMed] [Google Scholar]